- 1Department of Thoracic Surgery, Yunnan Cancer Hospital, Kunming, China
- 2Department of Thoracic Surgery, First Affiliated Hospital of Kunming Medical University, Kunming, China
- 3Department of Thoracic Surgery, First People's Hospital of Yunnan Province, Kunming, China
- 4Department of Medical Oncology, First People's Hospital of Yunnan Province, Kunming, China
Xuanwei County in Southwest China shows the highest incidence and mortality rate of lung cancer in China. Although studies have reported distinct clinical characteristics of patients from Xuanwei, the molecular features of these patients with non-small cell lung cancer (NSCLC) remain unclear. Here, we comprehensively characterised such cases using next-generation sequencing (NGS). Formalin-fixed, paraffin-embedded tumour samples from 146 patients from Xuanwei with NSCLC were collected for an NGS-based target panel assay; their features were compared with those of reference Chinese and The Cancer Genome Atlas (TCGA) cohorts. Uncommon EGFR mutations, defined as mutations other than L858R, exon 19del, exon 20ins, and T790M, were the predominant type of EGFR mutations in the Xuanwei cohort. Patients harbouring uncommon EGFR mutations were more likely to have a family history of cancer (p = 0.048). A higher frequency of KRAS mutations and lower frequency of rearrangement alterations were observed in the Xuanwei cohort (p < 0.001). Patients from Xuanwei showed a significantly higher tumour mutation burden than the reference Chinese and TCGA cohorts (p < 0.001). Our data indicates that patients from Xuanwei with NSCLC harbouring G719X/S768I co-mutations may benefit from treatment with EGFR-tyrosine kinase inhibitors. Our comprehensive molecular profiling revealed unique genomic features of patients from Xuanwei with NSCLC, highlighting the potential for improvement in targeted therapy and immunotherapy.
Introduction
Lung cancer is the leading cause of cancer-related deaths in China (1). Many patients (57%) are diagnosed with metastatic disease, leading to a 5-year relative survival rate of 5% (2). Approximately 85% of lung cancers are non-small cell lung cancers (NSCLCs), of which lung adenocarcinoma and lung squamous cell carcinoma are the most common subtypes (3). Compared with other regions in China, Xuanwei County in Yunnan Province has the highest mortality rate of lung cancer. The age-standardised mortality rates of lung cancer patients from Xuanwei were six and three times higher than those of patients from rural areas of China, among females and males, respectively (4). Hospitals in Yunnan Province organise free CT examinations to ensure early detection of lung cancer, and many patients are diagnosed at stage I. Xuanwei is rich in smoky (bituminous) coal, which may be associated with the high mortality rate of lung cancer in this area. Retrospective studies have shown that a lifelong use of smoky coal is associated with a 36- and 99-fold increase in mortality in men and women, respectively, compared with smokeless coal use (5, 6). Notably, lung cancer in Xuanwei has some remarkable characteristics, such as higher incidence in non-smoking females, diagnosis at a younger age, rapid tumour progression, multiple lung lesions, poor overall prognosis, and family aggregation (7). Overall, lung cancer patients in Xuanwei may present a distinct subgroup globally, leading researchers to consider whether epidemiological and clinicopathological peculiarities can be interpreted based on genomic features. Recent studies suggested that the NSCLC cohort in Xuanwei harboured a significantly higher co-mutation rate in EGFR exons 18 and 20 (8). NSCLCs are often found to have a high tumour mutation burden (TMB), which has been associated with apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like (APOBEC) signatures (9). However, the detailed characteristics of these EGFR mutations, comprehensive molecular profiling, and TMB characteristics of patients with NSCLC in Xuanwei are unclear.
We performed comprehensive genomic testing in an NSCLC cohort from Xuanwei. The genomic features of this cohort were compared with those of a reference Chinese NSCLC cohort (1,802 patients, excluding patients from Xuanwei) and data from The Cancer Genome Atlas (TCGA) mainly comprising a Western population from Europe and the US (10).
Methods
Patient Enrolment
In total, 1948 Chinese patients diagnosed with NSCLC at the Yunnan Cancer Hospital, First People's Hospital of Yunnan Province, or First Affiliated Hospital of Kunming Medical University were recruited. Formalin-fixed paraffin-embedded tumour samples were collected between December 2017 and January 2019. Matched blood samples were collected as reference controls. Of these patients, 146 were from Xuanwei and defined as the Xuanwei cohort. The remaining 1,802 Chinese patients with NSCLC were defined as the reference Chinese cohort. This study was approved by the Institution Review Board of the First Hospital of Kunming Medical University and conducted according to the Declaration of Helsinki. Informed consent was obtained from all enrolled patients.
Next-Generation Sequencing (NGS)
All tumour tissues and matched blood samples underwent targeted NGS-based genomic testing (OrigiMed, Shanghai, China) in a College of American Pathologists-accredited and Clinical Laboratory Improvement Amendments-certified laboratory (11). Approximately 50 ng of cancer tissue DNA was extracted from 40 mm formalin-fixed paraffin-embedded tumour samples and blood samples using the DNA Extraction Kit (Qiagen, Hilden, Germany), according to the manufacturer's instructions. All coding exons and selected introns of targeted genes were captured for hybridisation capture panel and then sequenced on an Illumina NextSeq-500 Platform (Illumina Incorporated, San Diego, CA). For formalin-fixed paraffin-embedded samples, sequencing depth was 900 × mean coverage (minimum 700 ×); for matched blood samples, sequencing depth was 300 ×. Genomic alterations, including single nucleotide variants, short and long insertions/deletions, copy number variations, and gene rearrangements, were subjected to advanced analysis. TMB score was calculated from a 450-gene panel data (Supplementary Table 1) for each sample by counting the number of somatic mutations, including coding single nucleotide variants and insertions/deletions, per megabase (Mb) of the sequence examined. Known somatic mutations in the Catalogue of Somatic Mutations in Cancer (COSMIC; https://cancer.sanger.ac.uk/cosmic/signatures) and known germline polymorphisms in the U.S. National Centre for Biotechnology Information's Single Nucleotide Polymorphism Database were not counted (12). Particularly, 35 and 111 tumour samples were subjected to 37 and 450 cancer-related gene panel testing (Supplementary Tables 1, 2), respectively. TMB analysis was available for 111 cases. A high TMB (TMB-H) was defined as ≥10 muts/Mb, and a low TMB (TMB-L) was defined as <10 muts/Mb. Mutational signature analysis was conducted using the deconstructSigs package v1.8.0. All the detected somatic mutations, including synonymous mutations in the cohort, were imported for signature analysis. Finally, the weights of 30 known cancer mutation signatures in COSMIC were generated (13, 14). Uncommon EGFR mutations were defined as mutations other than L858R, exon 19del, exon 20ins, and T790M (15).
Response Evaluation
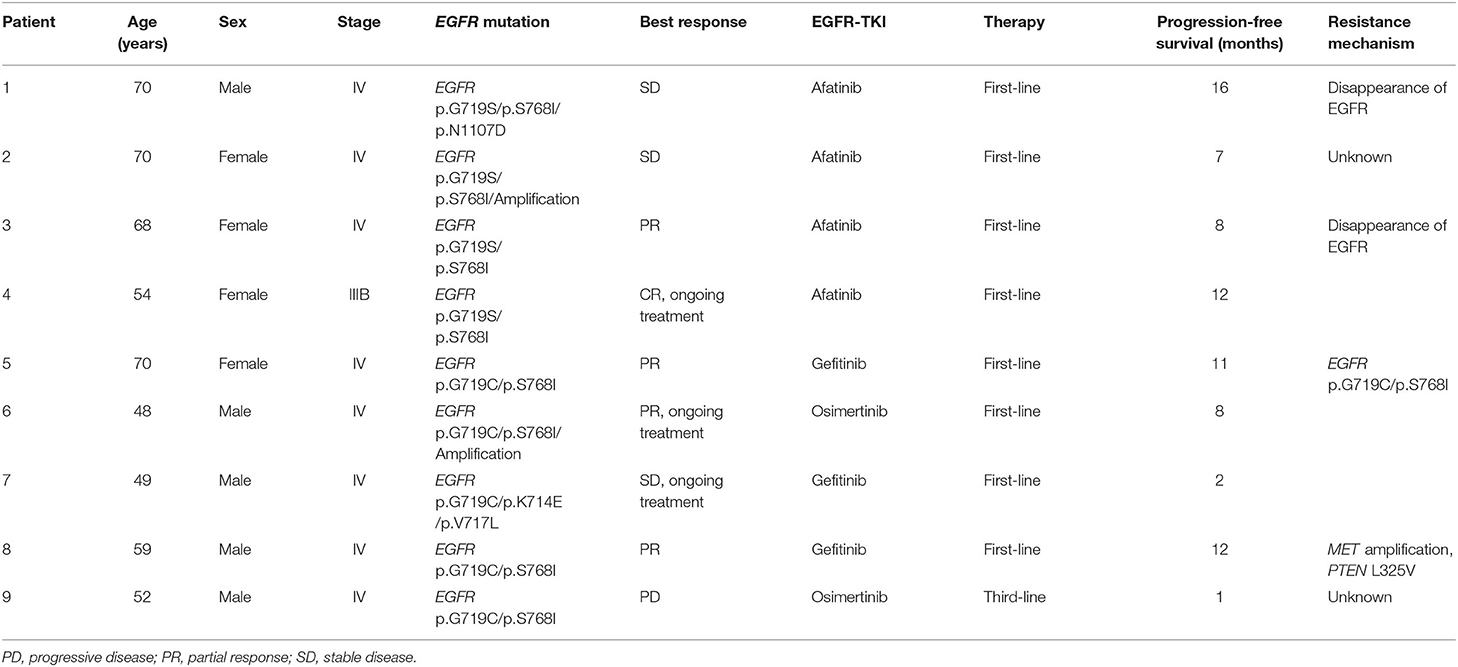
All nine patients received oral EGFR-tyrosine kinase inhibitor (TKI) treatment. Radiological follow-up was performed first, after 1 month, and then, once every 2 months, via computed tomography of the thorax and upper abdomen. Response was assessed according to the Response Criteria in Solid Tumours (RECIST) 1.1 (16). Progression-free survival was defined as the interval from the date of initiation of EGFR-TKI therapy to the date of disease progression or death from any cause, whichever occurred first.
Statistical Analyses
Statistical analyses were performed using the R Statistical Software package (R Foundation for Statistical Computing, Vienna, Austria). To analyse differences in continuous variables and TMB, the Wilcoxon test was performed when comparing each two groups, and the Kruskal-Wallis test was performed when comparing all three groups. The Chi-square or Fisher's exact tests was used for association of categorical variables. The threshold for statistical significance was set at p < 0.05. The significance associated with each symbol is as follows: ***p < 0.001; **p < 0.01; and *p < 0.05.
Results
Patients
Clinicopathological data of Xuanwei and reference Chinese patients with NSCLC are summarised in Table 1. The median age of the Xuanwei cohort was lower than that of the reference Chinese cohort (55 vs. 66 years, p < 0.001). Patients from Xuanwei showed less incidence of squamous cell carcinoma than reference Chinese patients (7.5 vs. 12.5%, p = 0.008). According to the pathology and medical history following the American Journal of Critical Care Cancer Staging Manual, patients were classified based on their main clinical stages (I–IV). The Xuanwei cohort contained more stage I–II patients than the reference Chinese cohort (68.5 vs. 45.7%, p = 0.0018). Moreover, Xuanwei cases had a greater cancer-related family history than reference Chinese cases (41.8 vs. 26.6%, p < 0.001), with most showing a family history of lung cancer (96.7%).
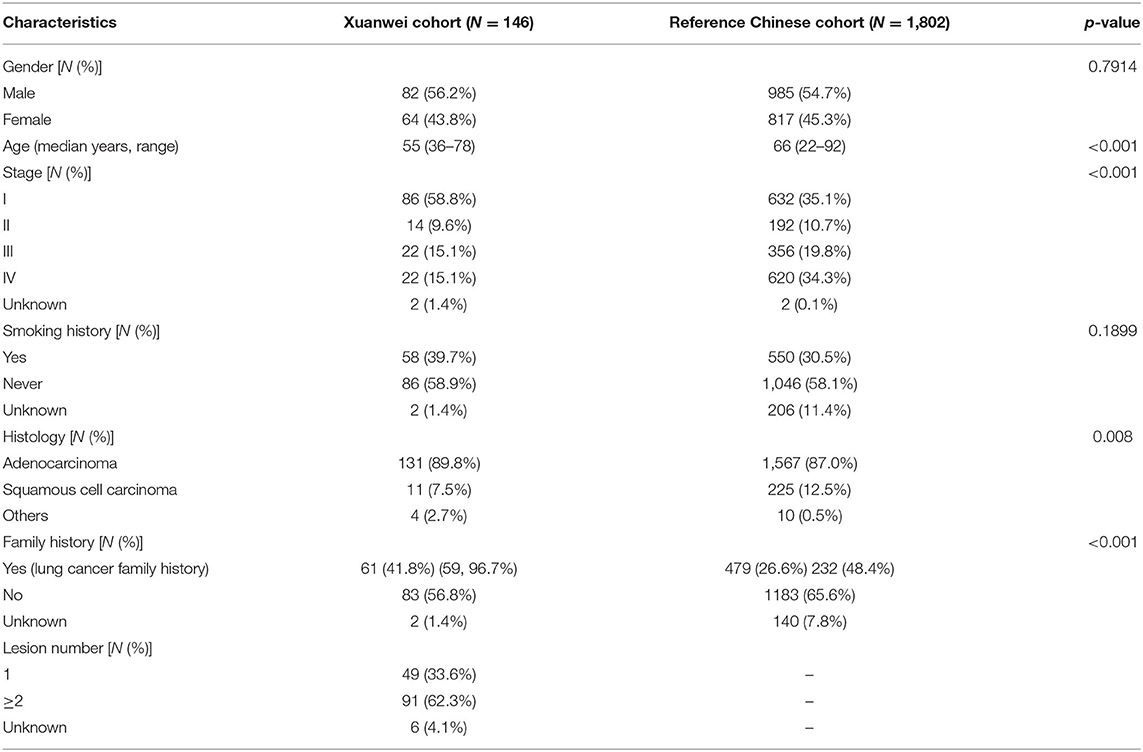
Table 1. Clinicopathological baseline characteristics of patients from Xuanwei and reference Chinese patients with NSCLC.
Driver Genes in the Xuanwei NSCLC Cohort
The nine driver genes of NSCLC found in patients from Xuanwei are summarised in Figure 1A. Comparison of the mutational profile of driver genes in the three cohorts showed significant differences (Figure 1B). The KRAS mutation frequency in the Xuanwei cohort was significantly higher than that in reference Chinese groups (26.3 vs. 11.2%, p < 0.001). The frequency of rearrangement alterations identified in the Xuanwei cohort was lower than that in the reference Chinese cohort (2.7 vs. 11.9%, p < 0.001). Rearrangement in ROS1 and NTRK1/2/3 were not found in the Xuanwei cohort (Figures 1B,C). The most common KRAS mutation in the Xuanwei cohort was KRAS G12C (53.8%), followed by KRAS G12V (23.1%) (Figure 1D).
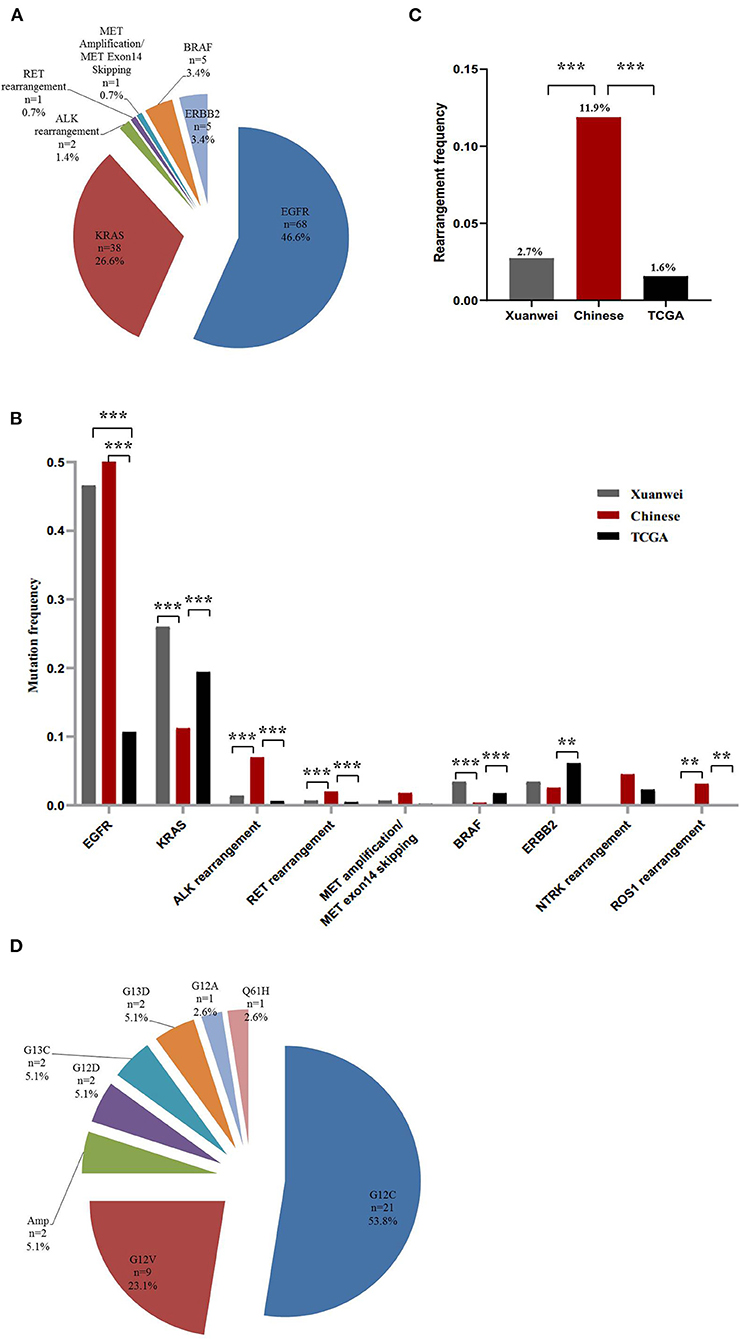
Figure 1. Driver gene mutation profile in the Xuanwei NSCLC cohort. (A) Mutation frequency of nine genes in Xuanwei cohorts. (B) Composition of the alteration type in the nine genes among the three groups, namely the Xuanwei cohort (left column), reference Chinese cohort (middle column), and TCGA cohort (right column). (C) Composition of the rearrangement alterations among the three groups. (D) Distribution of KRAS mutation subtypes in patients from Xuanwei. ***p < 0.001, **p < 0.01, and *p < 0.05.
EGFR Mutation Profile of the Xuanwei Cohort
A higher mutation frequency of EGFR was observed in the Xuanwei NSCLC cohort than in TCGA cases (46.6% vs. 10.7%, p < 0.001), although this frequency was comparable to that in reference Chinese cases (46.6 vs. 50.3%, p = 0.44) (Figures 1A,B). Notably, comparison of EGFR mutation subtypes demonstrated that patients from Xuanwei, compared to reference Chinese and Western patients, harboured a striking mutation pattern of higher EGFR G719X (47.6% vs. 5.0 vs. 3.1%, p < 0.001) and S768I (24.6% vs. 2% vs. 0.6%, p < 0.001) mutations, whereas classical EGFR-sensitive mutations, such as L858R (26.1% vs. 42.8% vs. 18.9%, p < 0.001) and exon 19del (10.1% vs. 40.6% vs. 25.8%, p < 0.001), showed significantly lower frequencies (Figures 2A,B; Supplementary Figures 1A,B).
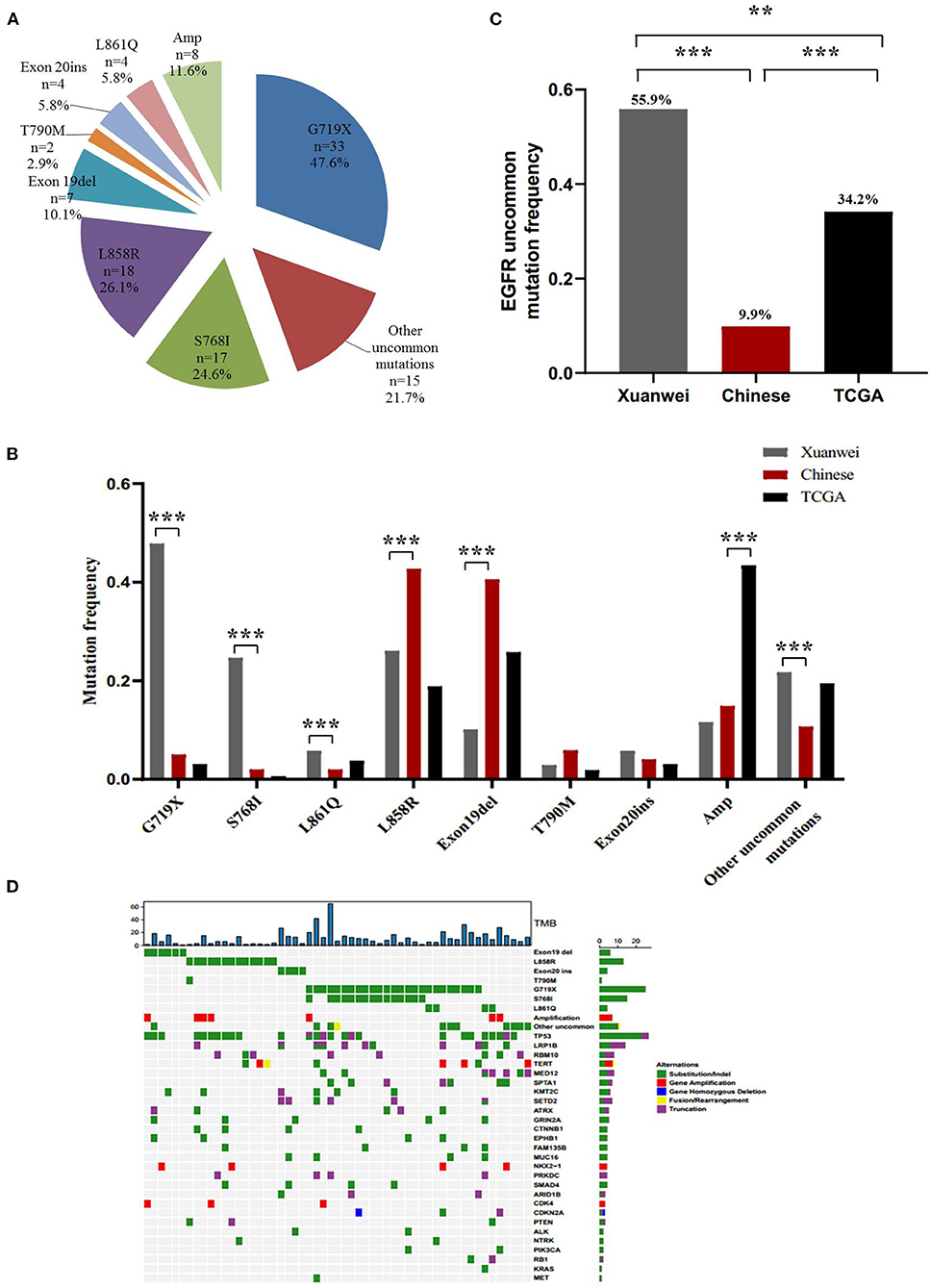
Figure 2. EGFR mutation spectrum in the Xuanwei, reference Chinese, and TCGA NSCLC cohorts. (A) EGFR mutation subtypes of patients from Xuanwei. (B) Comparison of EGFR mutation profile among the three groups. (C) Ratio of uncommon EGFR mutations in the three groups. (D) Mutation profiles of patients from Xuanwei with EGFR genomic alterations. Mutant frequencies in the cohort are shown on the right. TMB for each patient is shown at the top. ***p < 0.001, **p < 0.01, and *p < 0.05.
Uncommon EGFR mutations were the predominant EGFR mutation type in the Xuanwei cohort compared to the reference Chinese cohort (55.9 vs. 9.9%, p < 0.001) (Figure 2C). The most commonly co-mutated genes are shown in Figure 2D. Tumours harboured a higher ratio of EGFR G719X and S768I co-mutations, which were mutually exclusive of L858R and exon 19del (Figure 2D). The clinicopathological characteristics of the Xuanwei NSCLC cohort with either uncommon or common EGFR mutations are summarised in Table 2. Patients with uncommon EGFR mutations were more likely to have a family history of cancer than those with common EGFR mutations (p = 0.048).
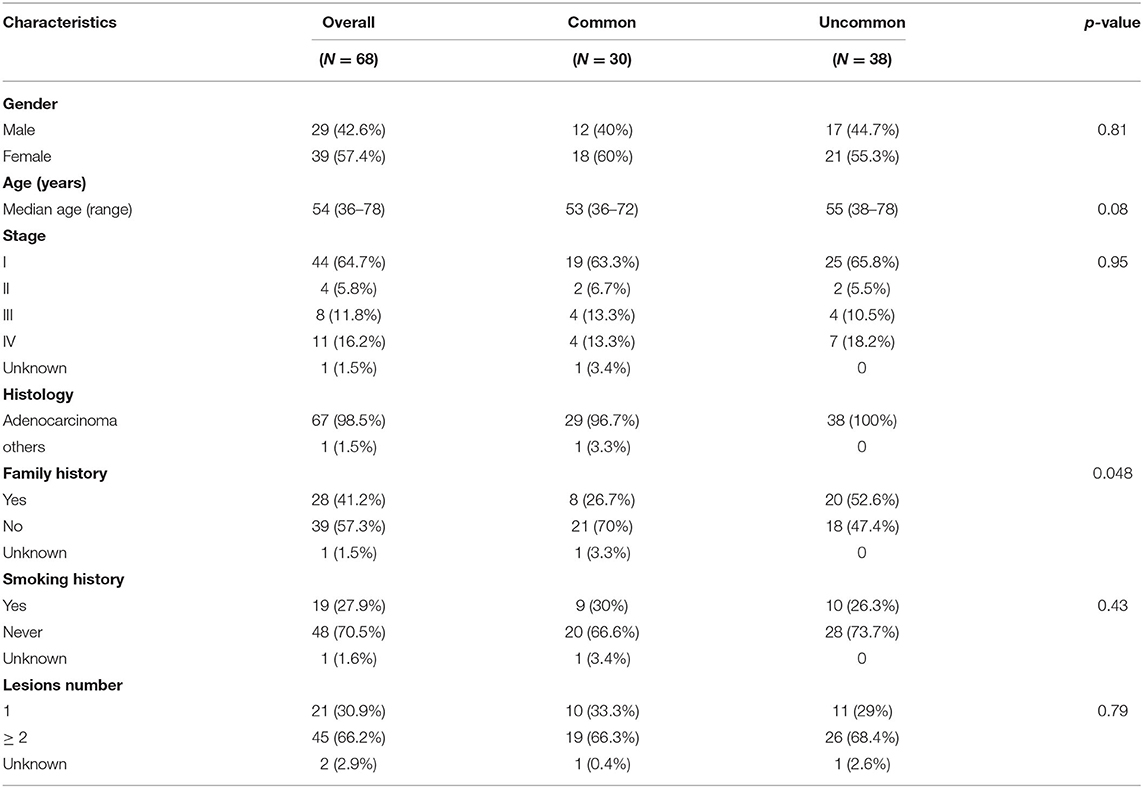
Table 2. Comparison of characteristics of patients from Xuanwei with NSCLC harbouring common and uncommon EGFR mutations.
Comprehensive Profiling and Mutational Signatures in the Xuanwei Cohort
The most commonly mutated genes in the Xuanwei NSCLC cohort were TP53 (51%), EGFR (49%), KRAS (28%), LRP1B (26%), and SPTA1 (23%) (Figure 3A). The mutation statuses of NSCLC-related pathways were analysed. Genes involved in these signalling pathways that were included in the 450-gene panel are listed in Supplementary Table 3. Gene mutations in the Wnt/MAPK/ERBB signalling pathways were the most common in patients from Xuanwei with NSCLC (Figure 3B). The Xuanwei cohort showed a higher median TMB than the reference Chinese cohort and TCGA cases (13.1, 4.6, and 6.9 muts/Mb, respectively; p < 0.001) (Figure 3C). TMB-H was detected in 58.6% of the Xuanwei cohort and 23.5% of the reference Chinese cohort (p < 0.001) (Figure 3D). TMB and mutational signatures reflect the process of mutation accumulation in cancer. To gain further insights into the mutational process of patients from Xuanwei, we characterised the mutation signatures via analysis of TMB-H and TMB-L tumours using the somatic mutation data. The profile of somatic mutations is shown in Figure 3E. We identified 3,596 single nucleotide variants and 58 insertions/deletions from 111 paired sequences of NSCLC in Xuanwei cohorts. The six subtypes of base substitutions (C > A, C > G, C > T, T > A, T > C, and T > G) were unevenly represented in single nucleotide variants. C > A was the most common substitution (1,685, 49.8%), followed by C > T (752, 22.2%) (Figure 3E).
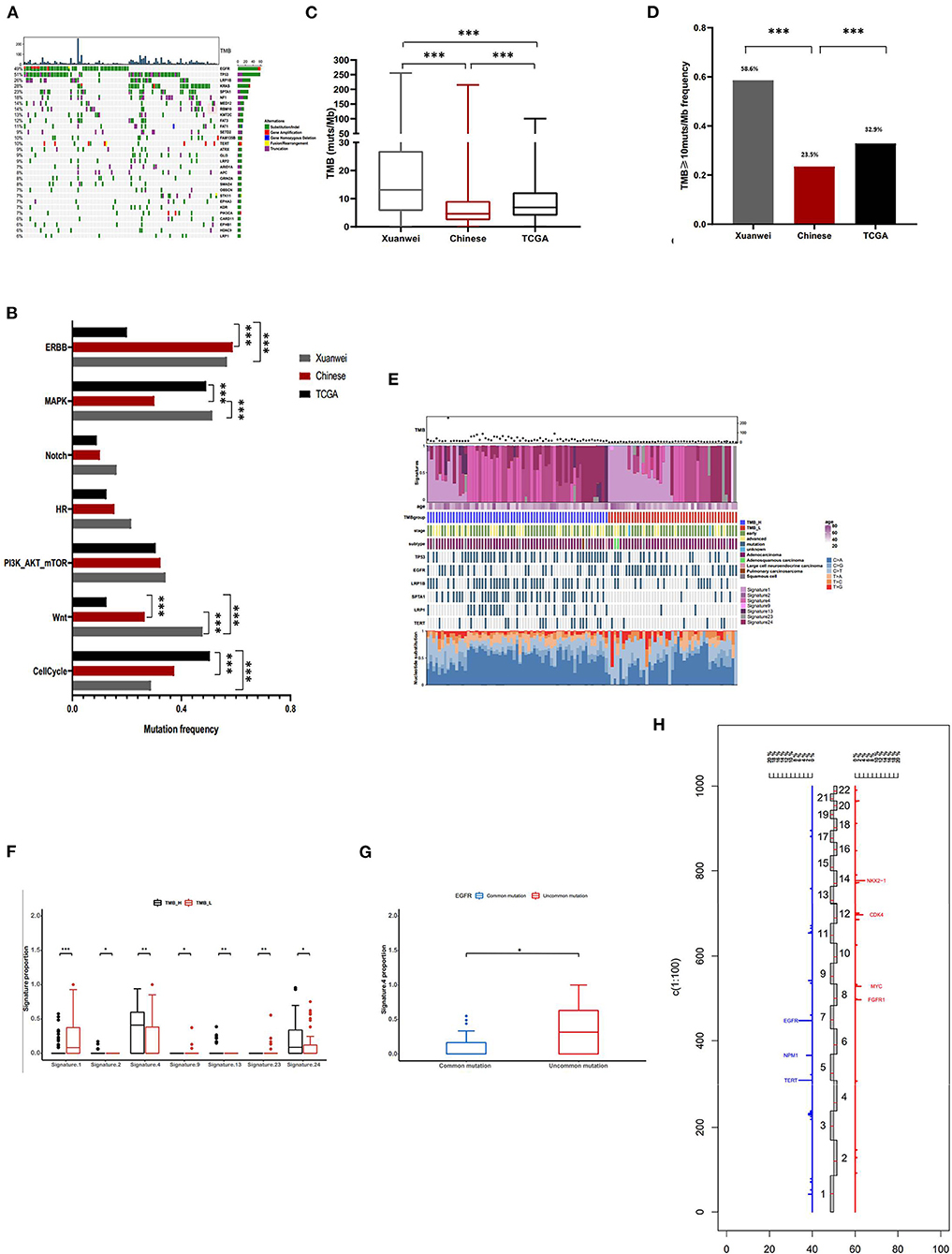
Figure 3. Comprehensive profiling of and somatic mutation signatures in patients from Xuanwei with NSCLC. (A) Comprehensive profiling of 111 patients from the Xuanwei cohort. (B) Comparison of the detection rate of gene mutations in pathways in the three cohorts. (C) Comparison of the proportion of the TMB value and (D) patients with TMB-H. (E) Somatic mutation signatures of the Xuanwei cohort. (F,G) Correlation between TMB value/EGFR mutation type and mutation signature. (H) Frequency of copy number variations of all chromosomal changes. Numbers 1–22 in the middle represent the human chromosome number. ***p < 0.001, **p < 0.01, and *p < 0.05.
We analysed the Spearman correlation coefficient between TMB and mutational signatures. Four predominant signatures (APOBEC, Smoking, Signature 13, and Signature 24) were observed in tumours with TMB-H, whereas Signature 1, Signature 9, and Signature 23 showed a correlation with TMB-L, suggesting the extensive accumulation of mutations due to smoking and exposure to aflatoxin. Although Signature 24 has been found in cancer samples from patients with known exposure to aflatoxin (supported by COSMIC data) (Figure 3F), further exploration of this association is necessary to confirm this hypothesis. Uncommon EGFR mutations showed high frequency about smoking-associated signature (Figure 3G).
Copy number variation analysis showed that the most frequently amplified genes were EGFR (6.3%), TERT (6.3%), NKX2-1 (4.5%), CDK4 (3.6%), FGFR1 (2.7%), MYC (2.7%), NMP1 (2.7%), and SDHA (2.7%) (Figure 3H). The association between gene mutations and TMB was analysed (Supplementary Figure 2A); among the common actionable mutations, EGFR mutation was significantly inversely correlated with TMB (6.9 vs. 17 muts/Mb, p < 0.001). Tumours with uncommon EGFR mutations showed a significantly higher TMB than those with common EGFR mutations (11.2 vs. 3.1 muts/Mb, p < 0.01) (Supplementary Figures 2A,B). KRAS mutation cases showed a higher median TMB than KRAS wild-type mutation cases (21.1 vs. 9.6 muts/Mb, p < 0.01). Tumours with KRAS G12C showed a lower TMB than those with non-KRAS G12C mutations (12 vs. 20.5 muts/Mb, p < 0.05) (Supplementary Figures 2A,B). TP53, LRP1B, SPTA1, NF1, and KMT2C mutations frequently occurred in patients with NSCLC and were significantly positively correlated with TMB (p < 0.001) (Supplementary Figure 2C).
Antitumor Activity of EGFR-TKI in Patients From Xuanwei With Uncommon EGFR Mutations
Nine patients from Xuanwei with advanced lung adenocarcinoma bearing uncommon EGFR mutations, with a median age of 59 years, started EGFR-TKI treatment. Detailed mutation characteristics and the outcome of EGFR-TKI treatment are shown in Table 3. Nine patients included in the efficacy assessment had the sensitive uncommon G719X (9/9, 100%) and S768I (8/9, 89%) mutations. Before EGFR-TKI treatment, five patients showed metastasis in the lungs, two were diagnosed with brain metastasis, and one was diagnosed with bone metastasis. Eight patients received EGFR-TKI as first-line therapy, and one patient received osimertinib as third-line therapy. As per RECIST 1.1 guidelines, five patients achieved partial response or complete response, three showed stable disease, and the one that received osimertinib as third-line therapy showed progressive disease. Treatment-related adverse events included rash (2/9), pruritus (2/9), headache (2/9), stomatitis (2/9), and constipation (1/9). No grade 3 or more adverse events were identified. At data cut-off (February 1, 2020), three patients presented sustained disease control and remained on EGFR-TKI treatment, while the other six showed progressive disease. Four patients provided serial plasma to characterise the acquired resistance mechanisms: EGFR mutation loss was observed in two patients at the time of EGFR-TKI treatment. MET amplification emerged post-EGFR-TKI treatment in one patient; no resistance mechanisms were detected in the remaining patients via NGS. As blood analysis is not 100% sensitive for the detection of acquired mutations, other genetic alterations may be associated with resistance.
Discussion
High mortality and incidence rates of lung cancer have been documented in Xuanwei County. Our study revealed that the Xuanwei cohort had a greater cancer-related family history compared with the reference Chinese cohort, and most patients from Xuanwei showed a family history of lung cancer (96.7%). However, we did not detect any germline mutations in patients with family history. Patients with uncommon EGFR mutations were more likely to have a family history of cancer than those with common EGFR mutations (p = 0.05). A multicentre case-control study suggested that environmental tobacco smoke exposure could influence the EGFR mutation profile of lung cancer in never smokers and that exposure during adulthood might reduce the probability of EGFR mutation (17). Uncommon EGFR mutations and TMB-H were the predominant genomic features of patients with NSCLC from Xuanwei. Mutational signatures analysis demonstrated that uncommon EGFR mutations and TMB-H were correlated with smoking signature and we speculate that the environment (smoky coal) in Xuanwei may influence molecular features of patients from Xuanwei with NSCLC, which requires further analysis.
Patients with NSCLC harbouring EGFR mutations may benefit from EGFR-TKI therapy (18–20). The “uncommon” EGFR mutations account for 10–18% of all EGFR mutations, and NGS testing can broaden the spectrum of aberrations within the “uncommon group” in patients with NSCLC (21). Patients with EGFR mutations, including L858R, ex19del, and T790M, show a good response rate to first- or third-generation EGFR-TKIs, whereas those with uncommon EGFR mutations generally exhibit less benefit from targeted therapy (22). In our study, the NGS-based analysis of patients from Xuanwei with NSCLC revealed their comprehensive and unique profile of genomic alterations; uncommon EGFR mutations, mainly including G719X, S768I, and L861Q, were the predominant EGFR mutation types in the Xuanwei cohort, forming a distinctive subgroup of NSCLC globally. For patients with such non-classical EGFR mutations, previous studies illustrated that progression-free survival was significantly longer after afatinib treatment than that after first-generation EGFR-TKI (gefitinib/erlotinib) treatment (11.3 vs. 3.6 months, p = 0.03) (23). Osimertinib shows favourable activity with manageable toxicity in patients with metastatic or recurrent NSCLC harbouring uncommon EGFR mutations, achieving a median progression-free survival of 8.2 months (95% CI, 5.9 to 10.5 months) (24). According to our study, patients from Xuanwei with NSCLC harbouring G719X/S768I co-mutations may benefit from first-, second-, and third-generation EGFR-TKI treatment. The efficacy of these EGFR-TKIs in advanced patients from Xuanwei with uncommon EGFR mutations requires further analysis. Adjuvant therapy with gefitinib led to significantly longer disease-free survival in patients with completely resected stage II–IIIA NSCLC with EGFR mutations (exon 19del and exon 21 L858R) than platinum-based chemotherapy (25). Whether patients from Xuanwei with uncommon EGFR mutations could benefit from adjuvant EGFR-TKI treatment is also worth further investigation.
Acquired resistance to EGFR-TKIs is a common event, and several mechanisms, including T790M, MET amplification, and PTEN down-regulation, have been reported for the common EGFR mutations exon 19del and L858R (26). However, mechanisms underlying EGFR-TKI resistance have not been investigated for uncommon EGFR mutations. Our study revealed that MET amplification and loss of EGFR mutation loss were related to EGFR-TKI resistance in patients harbouring EGFR G719X/S768I co-mutations. Nevertheless, further studies are required to confirm these mechanisms.
KRAS is a G-protein with intrinsic GTPase activity. KRAS mutations are associated with reduced responsiveness to EGFR-TKI therapy and poor survival (27). The KRAS mutation frequency in the Xuanwei cohort was significantly higher than that in the reference Chinese cohort. Furthermore, KRAS G12C accounted for 53.8% of KRAS-mutant patients in the Xuanwei cohort. AMG 510 is a novel, first-in-class, small molecule that specifically and irreversibly inhibits KRAS G12C by permanently locking it in an inactive GDP-bound state, and it demonstrated promising antitumor activity in patients with advanced NSCLC harbouring the KRAS G12C mutation (28). Patients from Xuanwei, who harbour the KRAS G12C mutation, may benefit from this inhibitor.
The TMB is an evolving biomarker for identifying eligible patients for checkpoint inhibitors in NSCLC (29, 30). Patients from Xuanwei with NSCLC showed a much higher median TMB than those in the reference Chinese and TCGA cohorts. Thus, patients from Xuanwei with NSCLC may benefit from immunotherapy. Furthermore, the TMB of patients with uncommon EGFR mutations was significantly higher than that of patients with common EGFR mutations in the Xuanwei cohort. Whether patients with uncommon EGFR mutations can benefit from immunotherapy requires further investigation.
This study comprehensively elucidated the molecular features of patients from Xuanwei with NSCLC via NGS. These patients appear to have a higher uncommon EGFR mutation ratio and a higher TMB value than reference Chinese patients and TCGA cohorts, and patients with uncommon EGFR mutations seem to show a good response to EGFR-TKI therapy. Studies with larger cohorts are needed to validate these observations, and further clinical research is warranted to provide insights into how comprehensive genomic profiling can guide treatment decisions for patients from Xuanwei with NSCLC.
Data Availability Statement
The datasets presented in this study can be found in online repositories. This data can be found here: https://db.cngb.org/cnsa/review/show/CNP0001608_20210319_9919b624/.
Ethics Statement
The studies involving human participants were reviewed and approved by First Hospital of Kunming Medical University. The patients/participants provided their written informed consent to participate in this study.
Author Contributions
YS, TS, and GG contributed to conception and design of the study. HL, QG, JL, and XY provided study or patients material. GL and YL collected and/or assembled data. GL, YL, and YS performed the statistical analysis and interpreted data. YS, TS, GG, and YL wrote the manuscript. All authors contributed to manuscript revision, read, and approved the submitted version.
Funding
This study was funded by the National Natural Science Foundation of China (81760554), Applied Basic Research in Yunnan Province (2017FE468-055), Kunming Medical Association Special Project for Applied Basic Research of Yunnan (2017FE467-142), and Yunnan Province Health and Family Planning Commission Medical Reserve Talents Plan (H-2017013).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank the patients for providing their samples for this study and OrigiMed for conducting genomic profiling.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2021.621422/full#supplementary-material
Abbreviations
APOBEC, apolipoprotein B mRNA editing enzyme catalytic polypeptide-like; COSMIC, Catalogue of Somatic Mutations in Cancer; Mb, megabase; NGS, next-generation sequencing; NSCLC, non-small cell lung cancer; RECIST, Response Criteria in Solid Tumours; TCGA, The Cancer Genome Atlas; TKI, tyrosine kinase inhibitor; TMB, tumour mutation burden; TMB-H, high TMB; TMB-L, low TMB; CT, computed tomography.
References
1. Cao M, Chen W. Epidemiology of lung cancer in China. Thorac Cancer. (2019) 10:3–7. doi: 10.1111/1759-7714.12916
2. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. (2020) 70:7–30. doi: 10.3322/caac.21590
3. Molina JR, Yang P, Cassivi SD, Schild SE, Adjei AA. Non-small cell lung cancer: epidemiology, risk factors, treatment, and survivorship. Mayo Clin Proc. (2008) 83:584–94. doi: 10.4065/83.5.584
4. Chen G, Sun X, Ren H, Wan X, Huang H, Ma X, et al. The mortality patterns of lung cancer between 1990 and 2013 in Xuanwei, China. Lung Cancer. (2015) 90:155–60. doi: 10.1016/j.lungcan.2015.08.006
5. Xiao Y, Shao Y, Yu X, Zhou G. The epidemic status and risk factors of lung cancer in Xuanwei City, Yunnan Province, China. Front Med. (2012) 6:388–94. doi: 10.1007/s11684-012-0233-3
6. Marx A, Chan JKC, Coindre J-M, Detterbeck F, Girard N, Harris NL, et al. The 2015 World Health Organization classification of tumors of the thymus: continuity and changes. J Thorac Oncol. (2015) 10:1383–95. doi: 10.1097/JTO.0000000000000654
7. Li R, Liu Y, Wang T, Tang J, Xie L, Yao Z, et al. The characteristics of lung cancer in Xuanwei County: a review of differentially expressed genes and noncoding RNAs on cell proliferation and migration. Biomed Pharmacother. (2019) 119:109312. doi: 10.1016/j.biopha.2019.109312
8. Chen Y, Ye L, Stanford RR, Zhang D, Zhang Z, Wei W. Distinct epithelial growth factor receptor mutation profile in non-small-cell lung cancer patients from the Xuanwei area of China. Mol Clin Oncol. (2016) 4:749–755. doi: 10.3892/mco.2016.805
9. Chen H, Chong W, Teng C, Yao Y, Wang X, Li X. The immune response-related mutational signatures and driver genes in non-small-cell lung cancer. Cancer Sci. (2019) 110:2348–56. doi: 10.1111/cas.14113
10. Campbell JD, Alexandrov A, Kim J, Wala J, Berger AH, Pedamallu CS, et al. Distinct patterns of somatic genome alterations in lung adenocarcinomas and squamous cell carcinomas. Nat Genet. (2016) 48:607–16. doi: 10.1038/ng.3564
11. Cao J, Chen L, Li H, Chen H, Yao J, Mu S, et al. An accurate and comprehensive clinical sequencing assay for cancer targeted and immunotherapies. Oncologist. (2019) 24:e1294–302. doi: 10.1634/theoncologist.2019-0236
12. Tang B, Yan X, Sheng X, Si L, Cui C, Kong Y, et al. Safety and clinical activity with an anti-PD-1 antibody JS001 in advanced melanoma or urologic cancer patients. J Hematol Oncol. (2019) 12:7. doi: 10.1186/s13045-018-0693-2
13. Guo J, Huang J, Zhou Y, Zhou Y, Yu L, Li H, et al. Germline and somatic variations influence the somatic mutational signatures of esophageal squamous cell carcinomas in a Chinese population. BMC Genomics. (2018) 19:538. doi: 10.1186/s12864-018-4906-4
14. Rosenthal R, McGranahan N, Herrero J, Taylor BS, Swanton C. DeconstructSigs: delineating mutational processes in single tumors distinguishes DNA repair deficiencies and patterns of carcinoma evolution. Genome Biol. (2016) 17:31. doi: 10.1186/s13059-016-0893-4
15. Wen S, Dai L, Wang L, Wang W, Wu D, Wang K, et al. Genomic signature of driver genes identified by target next-generation sequencing in Chinese non-small cell lung cancer. Oncologist. (2019) 24:e1070–81. doi: 10.1634/theoncologist.2018-0572
16. Eisenhauer EA, Therasse P, Bogaerts J, Schwartz LH, Sargent D, Ford R, et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1). Eur J Cancer. (2009) 45:228–47. doi: 10.1016/j.ejca.2008.10.026
17. Torres-Duran M, Ruano-Ravina A, Kelsey KT, Parente-Lamelas I, Leiro-Fernández V, Abdulkader I, et al. Environmental tobacco smoke exposure and EGFR and ALK alterations in never smokers' lung cancer. Results from the LCRINS study. Cancer Lett. (2017) 411:130–5. doi: 10.1016/j.canlet.2017.09.042
18. Mitsudomi T, Morita S, Yatabe Y, Negoro S, Okamoto I, et al. Gefitinib versus cisplatin plus docetaxel in patients with non-small-cell lung cancer harbouring mutations of the epidermal growth factor receptor (WJTOG3405): an open label, randomised phase 3 trial. Lancet Oncol. (2010) 11:121–8. doi: 10.1016/S1470-2045(09)70364-X
19. Yang JC-H, Wu Y-L, Schuler M, Sebastian M, Popat S, Yamamoto N, et al. Afatinib versus cisplatin-based chemotherapy for EGFR mutation-positive lung adenocarcinoma (LUX-Lung 3 and LUX-Lung 6): analysis of overall survival data from two randomised, phase 3 trials. Lancet Oncol. (2015) 16:141–51. doi: 10.1016/S1470-2045(14)71173-8
20. Rosell R, Carcereny E, Gervais R, Vergnenegre A, Massuti B, Felip E, et al. Erlotinib versus standard chemotherapy as first-line treatment for European patients with advanced EGFR mutation-positive non-small-cell lung cancer (EURTAC): a multicentre, open-label, randomised phase 3 trial. Lancet Oncol. (2012) 13:239–46. doi: 10.1016/S1470-2045(11)70393-X
21. O'Kane GM, Bradbury PA, Feld R, Leighl NB, Liu G, Pisters K-M, et al. Uncommon EGFR mutations in advanced non-small cell lung cancer. Lung Cancer. (2017) 109:137–44. doi: 10.1016/j.lungcan.2017.04.016
22. Xu J, Jin B, Chu T, Dong X, Yang H, Zhang Y, et al. EGFR tyrosine kinase inhibitor (TKI) in patients with advanced non-small cell lung cancer (NSCLC) harboring uncommon EGFR mutations: a real-world study in China. Lung Cancer. (2016) 96:87–92. doi: 10.1016/j.lungcan.2016.01.018
23. Shen Y-C, Tseng G-C, Tu C-Y, Chen W-C, Liao W-C, Chen W-C, et al. Comparing the effects of afatinib with gefitinib or Erlotinib in patients with advanced-stage lung adenocarcinoma harboring non-classical epidermal growth factor receptor mutations. Lung Cancer. (2017) 110:56–62. doi: 10.1016/j.lungcan.2017.06.007
24. Cho JH, Lim SH, An HJ, Kim KH, Park KU, Kang EJ, et al. Osimertinib for patients with non-small-cell lung cancer harboring uncommon EGFR mutations: a multicenter, open-label, phase ii trial (KCSG-LU15-09). J Clin Oncol. (2020) 38:488–95. doi: 10.1200/JCO.19.00931
25. Zhong W-Z, Wang Q, Mao W-M, Xu S-T, Wu L, Shen, et al. Gefitinib versus vinorelbine plus cisplatin as adjuvant treatment for stage II-IIIA (N1-N2) EGFR-mutant NSCLC (ADJUVANT/CTONG1104): a randomised, open-label, phase 3 study. Lancet Oncol. (2018) 19:139–48. doi: 10.1016/S1470-2045(17)30729-5
26. Wu SG, Shih JY. Management of acquired resistance to EGFR TKI-targeted therapy in advanced non-small cell lung cancer. Mol Cancer. (2018) 17:38. doi: 10.1186/s12943-018-0777-1
27. Eberhard DA, Johnson BE, Amler LC, Goddard AD, Heldens SL, Herbst RS, et al. Mutations in the epidermal growth factor receptor and in KRAS are predictive and prognostic indicators in patients with non-small-cell lung cancer treated with chemotherapy alone and in combination with erlotinib. J Clin Oncol. (2005) 23:5900–9. doi: 10.1200/JCO.2005.02.857
28. Canon J, Rex K, Saiki AY, Mohr C, Cooke K, Bagal D, et al. The clinical KRAS(G12C) inhibitor AMG 510 drives anti-tumour immunity. Nature. (2019) 575:217–23. doi: 10.1038/s41586-019-1694-1
29. Hellmann MD, Ciuleanu T-E, Pluzanski A, Lee JS, Otterson GA, Audigier-Valette C, et al. Nivolumab plus Ipilimumab in Lung Cancer with a High Tumor Mutational Burden. N Engl J Med. (2018) 378:2093–104. doi: 10.1056/NEJMoa1801946
Keywords: NSCLC, tumour mutation burden, uncommon EGFR mutations, Xuanwei county, NGS
Citation: Guo G, Li G, Liu Y, Li H, Guo Q, Liu J, Yang X, Shou T and Shi Y (2021) Next-Generation Sequencing Reveals High Uncommon EGFR Mutations and Tumour Mutation Burden in a Subgroup of Lung Cancer Patients. Front. Oncol. 11:621422. doi: 10.3389/fonc.2021.621422
Received: 26 October 2020; Accepted: 15 February 2021;
Published: 06 April 2021.
Edited by:
Qiyuan Li, Xiamen University, ChinaReviewed by:
Haiying Cheng, Albert Einstein College of Medicine, United StatesJie Meng, Central South University, China
Copyright © 2021 Guo, Li, Liu, Li, Guo, Liu, Yang, Shou and Shi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tao Shou, yn_shoutao@hotmail.com; Yunfei Shi, km-syf@163.com
†These authors have contributed equally to this work and share first authorship
 Gang Guo1†
Gang Guo1† Yunfei Shi
Yunfei Shi