- Sanbo Brain Hospital, Capital Medical University, Beijing, China
Background: The TCF7L2 gene is known as transcription factor 7-like 2 which has been identified as a novel transcription factor epithelial-mesenchymal transition (EMT) in tumor cells at 10q25.3. TCF7L2 may affect cancer progression and plays a central role in cancer proliferation, migration, and invasion. However, its clinical and prognostic value have not been researched in glioma. The purpose of our study was to research TCF7L2 expression and evaluate the clinical value of prognosis.
Method: We collected glioma specimens including low-grade glioma (n = 46) and glioblastoma (n = 51) from September 2015 to September 2017. Expression of TCF7L2 in 97 specimens was detected by quantitative real-time PCR (qRT-PCR). The chi-square test was applied to analyze the relationship between TCF7L2 expression and clinicopathological characteristics. The overall survival (OS) was estimated by log-rank tests among strata, and the survival curves were drawn by Kaplan-Meier. Univariate and multivariate analysis were utilized to analyze the relationship between prognosis and clinicopathological characteristics including TCF7L2 expression.
Results: Compared with the low-grade glioma group, the expression of TCF7L2 was significantly increased in the glioblastoma group (p = 0.001). TCF7L2 overexpression was associated with higher WHO grade (p = 0.001), isocitrate dehydrogenase (IDH) wild-type (p = 0.001), and lack of O(6)-methylguanine-DNA methyltransferase (MGMT) methylation (p = 0.001). Moreover, Kaplan-Meier analysis proved that overexpressed TCF7L2 was associated with poor OS (p = 0.010). The multivariate analysis suggested that TCF7L2 expression was an independent prognostic factor (p = 0.020).
Conclusions: Our research proved that TCF7L2 was overexpressed in glioblastoma, and related with tumor long-term prognosis, which, therefore, could be an independent prognostic factor for glioma patients.
Background
Glioblastoma is a common malignant brain tumor owing to its stronger invasive ability and resistance to treatment. The morbidity rate of glioblastoma is approximately 0.59–0.69/100000 people worldwide, affecting those aged 20 years or older, especially those aged 40–70 years (1). The morbidity is higher in men (3.97/100,000) than in women (2, 3) (2.53/100,000).
According to NCCN Guidelines, the standard therapies include surgery, radiotherapy with concomitant temozolomide (TMZ), adjuvant TMZ chemotherapy, and TTF therapies, but the 5-year overall survival (OS) is only 9.8% (3). Despite comprehensive therapy, the median survival time is ~12–15 months for GBM patients after diagnosis (4, 5).
An increasing number of molecular markers have been discovered, which improves our understanding of the mechanism of glioma and development. To date, the final histological diagnosis, final integrated diagnosis, pathological classification, and prognosis assessment are more accurate, which can help develop personalized therapy for glioma. Therefore, it is important to find novel biomarkers which can predict the prognosis, and explore its potential as a therapeutic target for glioma patients.
The Wnt/β-catenin signaling pathway is important in the majority of tumor progression. There is over 90% of aberrant activation of Wnt signaling in colorectal cancer (6). TCF7L2 is a member of the Wnt/β-catenin signaling pathway, which plays an important role in metabolism, cell differentiation/ proliferation, and cell death (7). Several meta-analyses have evaluated cancer risk induced by TCF7L2 gene variants (8–10).
A study (11) provided summary evidence that TCF7L2 genes are associated with the risk of breast, colorectal, and lung cancer and glioma. The loss of TCF7L2 enhances tumor cell growth, whereas a gain inhibits tumor cell growth (12). TCF7L2 plays a significant role in the pathogenesis of human cancers. A recent study (13) demonstrated that LEF/TCF-specific transcriptional regulation of Wnt target genes is associated with cancer progression and survival in human colorectal tumor samples.
However, the study of TCF7L2 mutation mainly focused on breast, colorectal, and lung cancer. Therefore, the aim of our study was to analyze the clinical value of TCF7L2 in glioma. We collected samples including 51 GBM tissues and 46 low-grade glioma tissues WHO (I-II). We focused on the TCF7L2 effect and the expression level in glioma tissue. Subsequently, we analyzed the relation between TCF7L2 expression level and clinicopathological characteristics. In our study, we performed a preliminary analysis between the expression level of TCF7L2 and overall survival risk of glioma among Chinese people.
Methods
Patients and Tissue Samples
Patients who underwent initial surgery in the Sanbo Brain Hospital of Capital Medical University from September 2015 to September 2017 were retrospectively selected for this research. All samples were diagnosed by pathologists and stored in liquid nitrogen.
As for patients with low-grade glioma, we identified high-risk patients according to the literature (14), including at least three factors (age >40 years old, astrocytomas, tumor maximum diameter ≧6 cm, tumors across the midline, and preoperative neurological impairment). Those patients were treated with radiochemotherapy, while the patients at low risk were followed up. Patients with glioblastoma received radiotherapy with concomitant temozolomide (TMZ) and adjuvant TMZ chemotherapy.
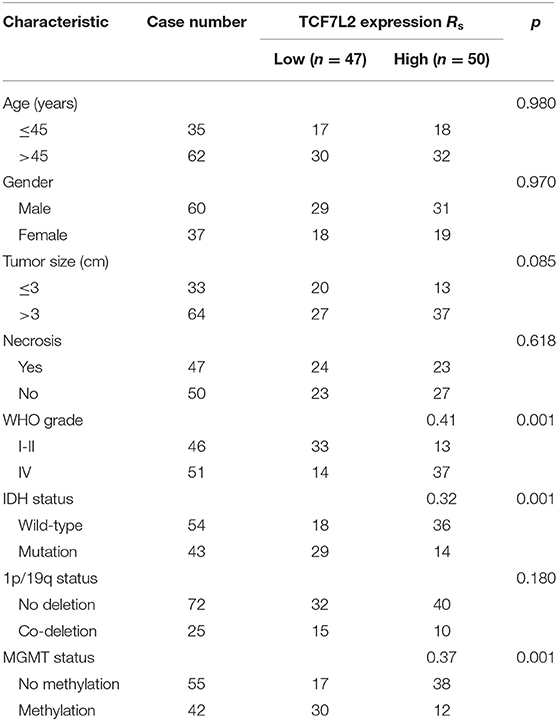
The patients' characteristics are described in Table 1. Tissue sample usage was approved by Ethics Committee of the Sanbo Brain Hospital of Capital Medical University and written informed consent was obtained from all study participants.
RNA Extraction and qRT-PCR Analyses
The total RNA was extracted from the frozen sample by TRIzol reagent (Invitrogen, Carlsbad, CA, USA). RNA was reverse-transcribed to complementary DNA (cDNA) using a PrimeScript RT reagent kit (Thermo, Beijing, China) according to the manufacturer's protocol. Its synthesis was conducted at 37°C for 15 min, then 85°C for 5 s according to the experimental protocols. Real-time PCR reactions were carried by Applied Biosystems 7500. Real-time PCR was carried in triplicate. We selected glyceraldehyde3-phosphate dehydrogenase (GAPDH) as a suitable endogenous reference gene. The relative TCF7L2 expression was computed and normalized using the 2-ΔCT method relative to GAPDH. The primers for TCF7L2: 5′-TGCTCTGCGGTTGCTATGTTGAC-3′, 3′-GCT GCGAGTCCTCACCAATGTC-5′ and for GAPDH: 5′-CAGACCACAG TCCATGCCATCAC-3′, 3′-GACGCCTGCTTCACCACCTTC-5′.
Data Analysis
The comparison of the TCF7L2 expression level between GBM and low-grade glioma was performed by the two-sample Student's t-test. The chi-square test was used to examine the associations between TCF7L2 expression and the clinicopathological characteristics.
In addition, survival curves were drawn by the Kaplan-Meier method and analyzed with log-rank test. Cox proportional-hazards regression analysis was applied to estimate univariate and multivariate hazard ratios for OS. A value of P < 0.05 was considered as statistically significant. SPSS software 20.0 was applied in the present study.
Results
TCF7L2 Was Upregulated in Glioblastoma Tissues
We measured TCF7L2 expression levels in 97 patients with glioma by qRT-PCR. The data revealed that TCF7L2 had a higher expression in GBM tissues than the low-grade group (p = 0.001) (Figure 1).
Relation Between TCF7L2 Expression and Clinicopathological Factors of GBM Patients
We found high TCF7L2 expression in GBM. The median expression level of TCF7L2 was used as a dividing point, and we divided 97 patients into two groups (high and low expression). Table 1 summarizes the relationship between TCF7L2 expression and clinicopathological parameters in glioma. The results showed that TCF7L2 high expression was significantly related to higher WHO grade (p = 0.001), isocitrate dehydrogenase (IDH) wild-type (p = 0.001), and lack of O(6)-methylguanine-DNA methyltransferase (MGMT) methylation (p = 0.001).
Upregulation of TCF7L2 Confers Poor Prognosis in Patients
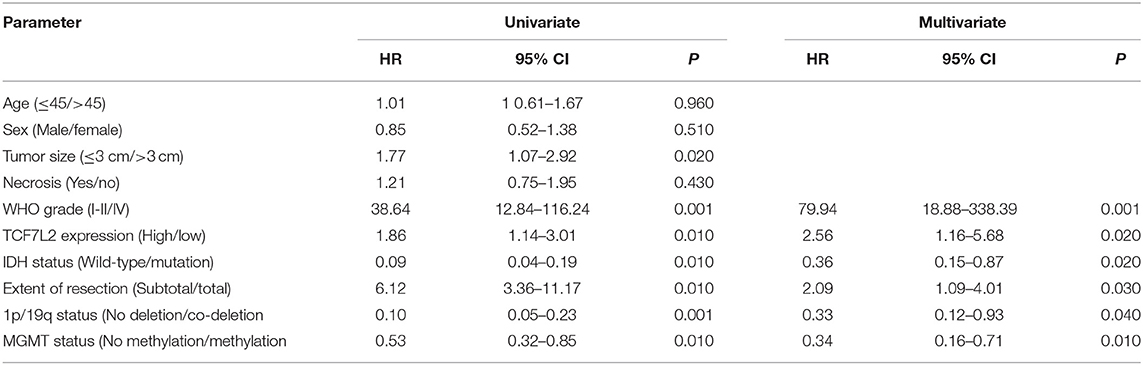
Univariate and multivariate analyses were utilized to evaluate the association between OS and various clinicopathological features including TCF7L2 expression level (Table 1).
The Kaplan-Meier method indicated that the 5-year OS of patients was significantly shorter in patients with high TCF7L2 expression than in those with low TCF7L2 expression (p = 0.010), but there were no significant differences in 2-year OS (p = 0.070). The OS was significantly longer with small tumor size (≦3 cm) compared to those with large tumor size (>3 cm) (p = 0.020). The OS was significantly shorter in patients with IDH wild-type than in those with IDH mutation (p = 0.010). The OS was significantly shorter in patients receiving subtotal resection than those receiving total resection (p = 0.010), but was significantly longer in patients in the low-grade group compared to those with glioblastoma (p = 0.001). The OS was significantly shorter in patients with no 1p/19q deletion than in those with 1p/19q codeletion (p = 0.001). The OS was significantly shorter in patients with no MGMT methylation than in those with MGMT methylation (p = 0.010). There were no significant differences between necrosis and non-necrosis patients (p = 0.420) (Figures 2, 3).
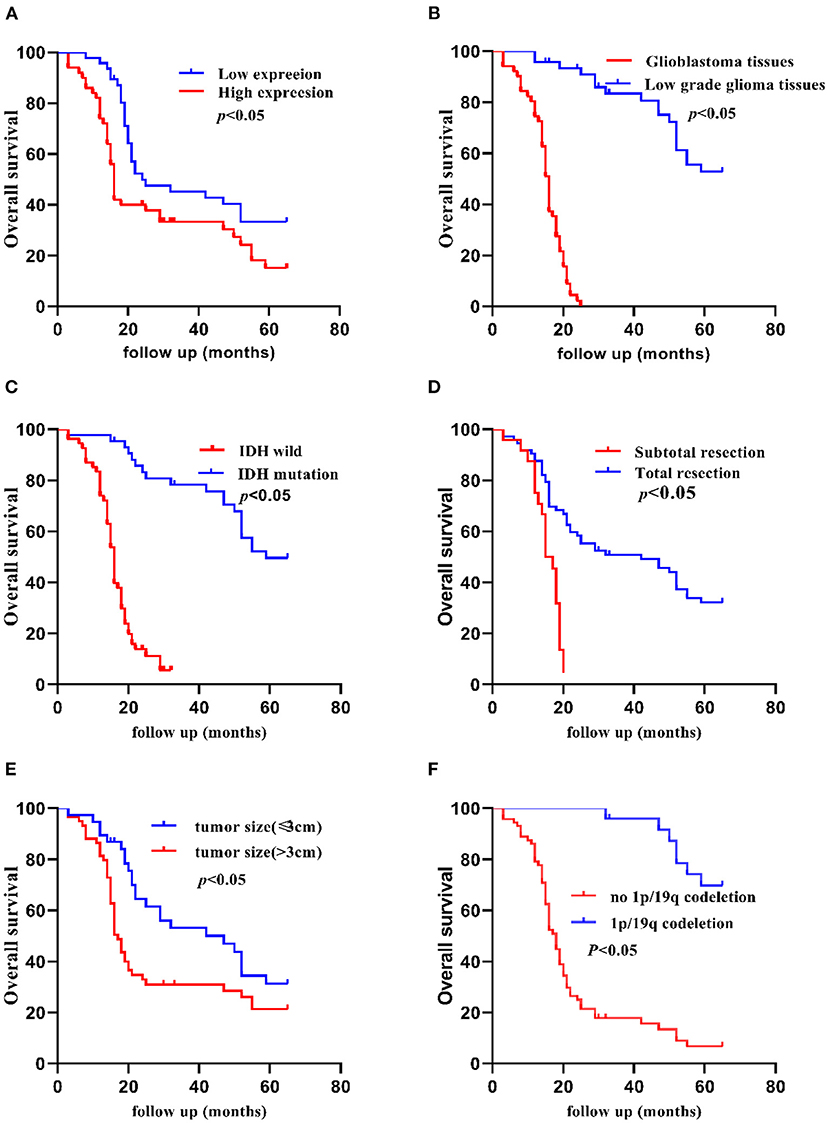
Figure 2. Kaplan-Meier 5-year OS curves of patients with glioma according to clinicopathological features. (A) Patients with higher TCF7L2 expression showed shorter OS compared with those with a lower expression (p = 0.010), (B) Glioblastoma patients showed worse OS compared with low-grade patients (p = 0.001). (C) Patients with IDH wild-type showed worse OS compared with those with an IDH mutation (p = 0.010). (D) Patients who had a subtotal resection showed worse OS compared with those who had a total resection (p = 0.010). (E) Patients with a larger tumor size (>3 cm) showed worse OS compared with those with a small tumor size (≤3 cm) (p = 0.020). (F) Patients with no 1p/19q codeletion showed worse OS compared with those with 1p/19q codeletion (p = 0.001).
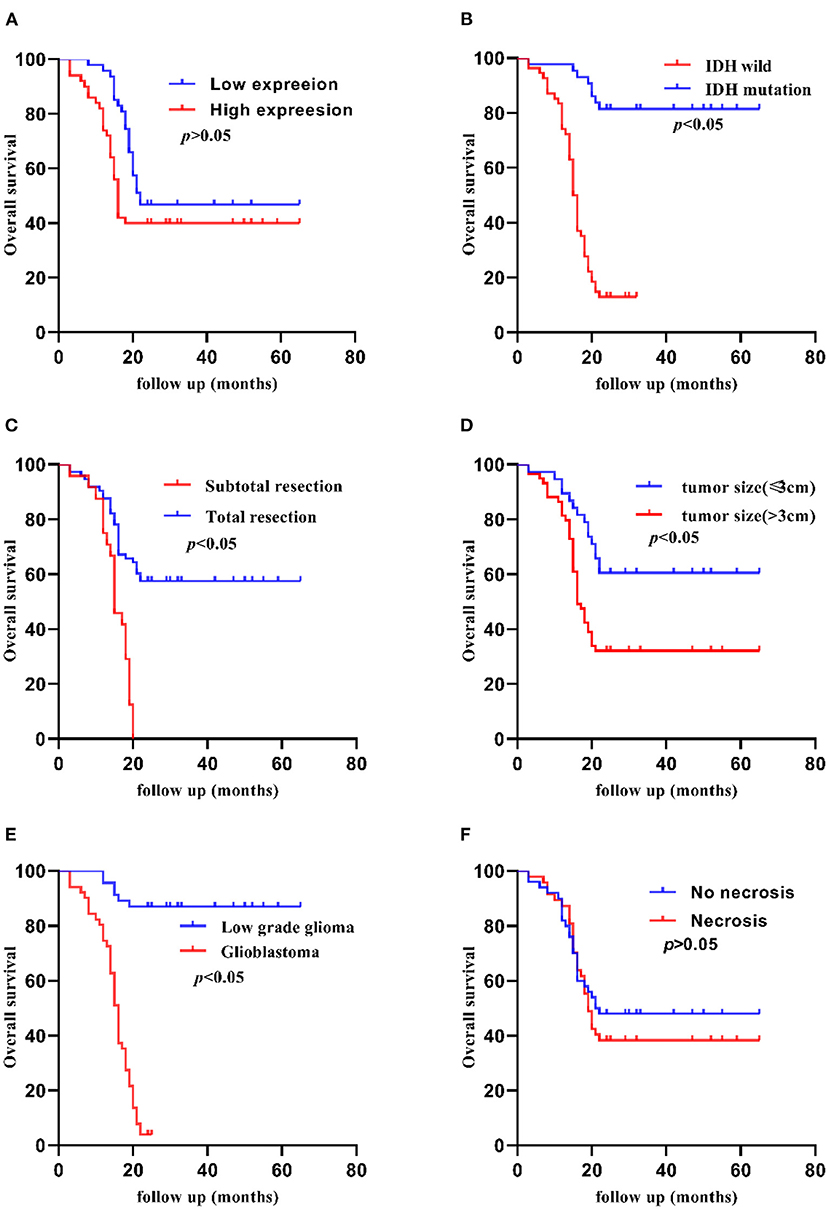
Figure 3. Kaplan-Meier 2-year OS curves of patients with glioma according to clinicopathological features. (A) Patients with higher TCF7L2 expression showed no significant differences compared with those with a lower expression (p = 0.008). (B) Patients with IDH wild-type showed worse OS compared with those with an IDH mutation (p = 0.001). (C) Patients who had a subtotal resection showed worse OS compared with those who had a total resection (p = 0.001). (D) Patients with a larger tumor size (>3 cm) showed worse OS compared with those with a small tumor size (≤3 cm) (p = 0.003). (E) Glioblastoma patients showed worse OS compared with low-grade patients (p = 0.001). (F) Patients with necrosis showed no significant differences compared with those with no necrosis (p = 0.486).
As for multivariate Cox regression analysis including TCF7L2 expression, WHO grade, extent of resection, 1p/19q status, IDH status, and MGMT status, we found a 5-year OS advantage of the low expression vs high expression of TCF7L2 (HR 2.56, 95% CI:1.16–5.68, p = 0.020), low-grade glioma vs. glioblastoma (HR 79.94, 95% CI:18.88–338.39, p = 0.001), total resection vs subtotal resection (HR 2.09, 95% CI:1.09–4.01, p = 0.030), IDH mutation vs IDH wild-type (HR 0.36, 95% CI:0.15–0.87, p = 0.020), 1p/19q codeletion vs no 1p/19q deletion (HR 0.33, 95% CI: 0.12–0.93, p = 0.040), and MGMT methylation vs no MGMT methylation (HR 0.34, 95% CI:0.16–0.71, p = 0.010) (Table 2).
Discussion
TCF7L2 has been reported to have an effect on the suppression of cell proliferation, migration, and invasion in the literature (15). The transcription factor 7-like 2 (TCF7L2) gene may affect cancer development and prognosis because the TCF7L2 gene plays an important role in the Wnt/β-catenin signaling pathway (16, 17).
To date, various biological markers have been reported in glioma (18, 19). TCF7L2 represents a central factor in metabolism, cell proliferation, and cell apoptosis (20, 21). Bo Yu et al. (15) found that TCF7L2 overexpression increased cell viability, migration, and invasion in cells with CRNDE inhibition. The TCF7L2 expression and prognostic value in glioma have rarely been reported.
In our study, we detected significantly higher TCF7L2 expression in GBM tissues than in the low-grade group. Moreover, TCF7L2 overexpression was significantly related with higher WHO grade, IDH wild-type, and no MGMT methylation, and the coefficients were 0.41, 0.32, and 0.37, respectively.
Our study reported shorter OS in patients with higher TCF7L2 expression, however, there was no significant difference in 2-year OS in the two groups. We found that the 2-year OS was relatively high in the patients who underwent standard therapies including radiotherapy with concomitant temozolomide (TMZ) and adjuvant TMZ after surgery in the high expression group. There was statistical significance in 5-year survival rate. On the other hand, TCF7L2 might be more advantageous in judging long-term prognosis.
Multivariate analysis revealed that TCF7L2, WHO grade, IDH status, extent of resection, 1p/19q status, and MGMT methylation status were independent prognostic factors for OS. The hazard ratio in the high TCF7L2 expression group was 2.56 times more than the low expression group (95% CI: 1.16–5.68, p = 0.020). TCF7L2 expression was independently related with OS, indicating that higher TCF7L2 level was a marker of poor prognosis for patients.
IDH mutation was found in both low-grade glioma and glioblastoma in our study, suggesting the IDH gene played an important role in the pathogenesis of tumors in glioma. The mutation rate was 15.6%, which was practically consistent with that reported in the literature (22). The multivariate analysis revealed that the patients of IDH mutation were closely associated with better prognosis, as compared with those of IDH wild-type.
Previous studies have reported that IDH mutations have been identified as one of the most important diagnostic and prognostic factors of gliomas (23). Due to intra-group heterogeneity, we need additional prognostic factors to subdivide the prognosis results in gliomas. There was a correlation between IDH status and TCF7L2 expression level, therefore, we would combine IDH status and TCF7L2 expression to further refine the stratified study and better judge the prognosis in a future study.
Previous studies have reported that patients with 1p/19q co-deletion have better prognosis and response to treatment (24, 25). 1p/19q co-deletion is a typical molecular genetic feature of oligodendroglioma, which provides an important reference for pathological diagnosis. We found that patients with 1p/19q co-deletion had a better prognosis than those no 1p/19q co-deletion. The results were consistent with literature reports.
Surgical resection plays a critical role in glioma therapy, which can reduce tumor load and provide an opportunity for postoperative adjuvant therapy. We found that patients receiving gross total resection had a better prognosis than those receiving subtotal resection. The results were consistent with other literature reports (26).
Glioblastoma patients showed worse OS compared with low-grade patients. In our study, we did not include WHO III patients, therefore, the hazard ratio was larger in the multivariate analysis. In addition, the sample size of data was relatively small, which might have introduced a bias. Currently, MGMT methylation is a widely accepted biomarker in glioblastoma, which can predict the effect of chemotherapeutic drugs (27). Patients with MGMT methylation showed a better prognosis than those with no methylation.
The sample size of data was relatively small. Some patients were reluctant to attend follow-up appointments, or the follow-up was interrupted in our study. We will increase the sample size for detailed study in the future.
TCF7L2 could be a potential prognostic factor and therapeutic target for patients with glioma. The underlying molecular mechanisms of TCF7L2 involvement in the Wnt/β-catenin signaling pathway needs to be investigated in future studies.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics Statement
The studies involving human participants were reviewed and approved by the Capital Medical University Hospital Ethics Committees. The patients/participants provided their written informed consent to participate in this study.
Author Contributions
LC, SH, and NL analyzed and interpreted the glioma patient data. SJ performed the qRT-PCR of the glioma tissues. CY and SJ were major contributors in writing the manuscript. All authors read and approved the final manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Reviewer YW declared a shared affiliation, with no collaboration, with the authors SJ, LC, SH, NL, MH, YY, and CY.
Acknowledgments
We appreciated W-Y. Deng for providing constructive suggestions to the manuscript.
Abbreviations
TCF7L2, Transcription factor 7-like 2; qRT-PCR, Real-time quantitative PCR; OS, Overall survival; TMZ, Temozolomide; EMT, Epithelial-mesenchymal transition; TTF, Tumor treating fields; GBM, Glioblastoma; cDNA, Complementary DNA; HR, Hazard ratios; CI, Confidence interval; MGMT, O(6)-methylguanine-DNA methyltransferase; IDH, Isocitrate dehydrogenase.
References
1. Darlix A, Zouaoui S, Rigau V, Bessaoud F, Figarella-Branger D, Mathieu-Daudé H, et al. Erratum to: Epidemiology for primary brain tumors: a nationwide population-based study. J Neurooncol. (2017) 131:547. doi: 10.1007/s11060-016-2340-5
2. Ostrom QT, Gittleman H, Fulop J, Liu M, Blanda R, Kromer C, et al. CBTRUS statistical report: primary brain and central nervous system tumors diagnosed in the United States in 2008-2012. Neuro Oncol. (2015) 17(Suppl 4):iv1–62. doi: 10.1093/neuonc/nov189
3. Koshy M, Villano JL, Dolecek TA, Howard A, Mahmood U, Chmura SJ, et al. Improved survival time trends for glioblastoma using the SEER 17 population-based registries. J Neurooncol. (2012) 107:207–12. doi: 10.1007/s11060-011-0738-7
4. Stupp R, Hegi ME, Mason WP, van den Bent MJ, Taphoorn MJ, Janzer RC, et al. Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol. (2009) 10:459–66. doi: 10.1016/S1470-2045(09)70025-7
5. Stupp R, Mason WP, van den Bent MJ, Weller M, Fisher B, Taphoorn MJ, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. (2005) 352:987–96. doi: 10.1056/NEJMoa043330
6. Cancer Genome Atlas Network. Comprehensive molecular characterization of human colon and rectal cancer. Nature. (2012) 487:330–7. doi: 10.1038/nature11252
7. Hrckulak D, Kolar M, Strnad H, Korinek V. TCF/LEF transcription factors: an update from the internet resources. Cancers. (2016) 8:70. doi: 10.3390/cancers8070070
8. Chen J, Yuan T, Liu M, Chen P. Association between TCF7L2 gene polymorphism and cancer risk: a meta-analysis. PLoS ONE. (2013) 8:e71730. doi: 10.1371/journal.pone.0071730
9. Lu XP, Hu GN, Du JQ, Li HQ. TCF7L2 gene polymorphisms and susceptibility to breast cancer: a meta-analysis. Genet Mol Res. (2015). 14:2860–7. doi: 10.4238/2015.March.31.16
10. Wang F, Jiang L, Li J, Yu X, Li M, Wu G, et al. Association between TCF7L2 polymorphisms and breast cancer susceptibility: a meta-analysis. Int J Clin Exp Med. (2015) 8:9355–61.
11. Zhang M, Tang M, Fang Y, Cui H, Chen S, Li J, et al. Cumulative evidence for relationships between multiple variants in the VTI1A and TCF7L2 genes and cancer incidence. Int J Cancer. (2018) 142:498–513. doi: 10.1002/ijc.31074
12. Angus-Hill ML, Elbert KM, Hidalgo J, Capecchi MR. T-cell factor 4 functions as a tumor suppressor whose disruption modulates colon cell proliferation and tumorigenesis. Proc Natl Acad Sci USA. (2011) 108:4914–9. doi: 10.1073/pnas.1102300108
13. Mayer CD, Giclais SM, Alsehly F, Hoppler S. Diverse LEF/TCF expression in human colorectal cancer correlates with altered Wnt-regulated transcriptome in a meta-analysis of patient biopsies. Genes. (2020) 11:538. doi: 10.3390/genes11050538
14. Pignatti F, van den Bent M, Curran D, Debruyne C, Sylvester R, Therasse P, et al. Prognostic factors for survival in adult patients with cerebral low-grade glioma. J Clin Oncol. (2002) 20:2076–84. doi: 10.1200/JCO.2002.08.121
15. Yu B, Ye X, Du Q, Zhu B, Zhai Q, Li XX. The long non-coding RNA CRNDE promotes colorectal carcinoma progression by competitively binding miR-217 with TCF7L2 and Enhancing the Wnt/β-catenin signaling pathway. Cell Physiol Biochem. (2017) 41:2489–502. doi: 10.1159/000475941
16. Spranger S, Bao R, Gajewski TF. Melanoma-intrinsic β-catenin signalling prevents anti-tumour immunity. Nature. (2015) 523:231–5. doi: 10.1038/nature14404
17. Zhao C, Deng Y, Liu L, Yu K, Zhang L, Wang H, et al. Dual regulatory switch through interactions of Tcf7l2/Tcf4 with stage-specific partners propels oligodendroglial maturation. Nat Commun. (2016) 7:10883. doi: 10.1038/ncomms10883
18. Qu DW, Xu HS, Han XJ, Wang YL, Ouyang CJ. Expression of cyclinD1 and Ki-67 proteins in gliomas and its clinical significance. Eur Rev Med Pharmacol Sci. (2014) 18:516–9. doi: 10.1016/j.jep.2013.12.015
19. Li W, Xie P, Ruan WH. Overexpression of lncRNA UCA1 promotes osteosarcoma progression and correlates with poor prognosis. J Bone Oncol. (2016) 5:80–5. doi: 10.1016/j.jbo.2016.05.003
20. Chen C, Cao F, Bai L, Liu Y, Xie J, Wang W, et al. IKKβ Enforces a LIN28B/TCF7L2 positive feedback loop that promotes cancer cell stemness and metastasis. Cancer Res. (2015) 75:1725–35. doi: 10.1158/0008-5472.CAN-14-2111
21. Weng Q, Tan B, Wang J, Wang J, Zhou H, Shi J, et al. Corrigendum to “5-Fluorouracil causes severe CNS demyelination by disruption of TCF7L2/HDAC1/HDAC2 complex in adolescent mice” [Toxicology 325 (2014) 144-150]. Toxicology. (2020) 430:152342. doi: 10.1016/j.tox.2019.152342
22. Ichimura K, Pearson DM, Kocialkowski S, Bäcklund LM, Chan R, Jones DT, et al. IDH1 mutations are present in the majority of common adult gliomas but rare in primary glioblastomas. Neuro Oncol. (2009) 11:341–7. doi: 10.1215/15228517-2009-025
23. Mirchia K, Richardson TE. Beyond IDH-mutation: emerging molecular diagnostic and prognostic features in adult diffuse gliomas. Cancers. (2020) 12:1817. doi: 10.3390/cancers12071817
24. Buckley PG, Alcock L, Heffernan J, Woods J, Brett F, Stallings RL, et al. Loss of chromosome 1p/19q in oligodendroglial tumors: refinement of chromosomal critical regions and evaluation of internexin immunostaining as a surrogate marker. J Neuropathol Exp Neurol. (2011) 70:177–82. doi: 10.1097/NEN.0b013e31820c765b
25. Durand KS, Guillaudeau A, Weinbreck N, DeArmas R, Robert S, Chaunavel A, et al. 1p19q LOH patterns and expression of p53 and Olig2 in gliomas: relation with histological types and prognosis. Mod Pathol. (2010) 23:619–28. doi: 10.1038/modpathol.2009.185
26. Lee JH, Jung TY, Jung S, Kim IY, Jang WY, Moon KS, et al. Performance status during and after radiotherapy plus concomitant and adjuvant temozolomide in elderly patients with glioblastoma multiforme. J Clin Neurosci. (2013) 20:503–8. doi: 10.1016/j.jocn.2012.03.044
Keywords: glioma, TCF7L2, overall survival, prognosis, clinicopathological characteristics
Citation: Jing S, Chen L, Han S, Liu N, Han M, Yang Y and Yan C (2021) Expression of TCF7L2 in Glioma and Its Relationship With Clinicopathological Characteristics and Patient Overall Survival. Front. Neurol. 12:627431. doi: 10.3389/fneur.2021.627431
Received: 09 November 2020; Accepted: 13 May 2021;
Published: 08 July 2021.
Edited by:
Yaohua Liu, Shanghai First People's Hospital, ChinaReviewed by:
Alissa A. Thomas, University of Vermont, United StatesYinyan Wang, Capital Medical University, China
Copyright © 2021 Jing, Chen, Han, Liu, Han, Yang and Yan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Changxiang Yan, yancx65828@sina.com
 Shiyuan Jing
Shiyuan Jing Changxiang Yan
Changxiang Yan