Can Deep Learning Hit a Moving Target? A Scoping Review of Its Role to Study Neurological Disorders in Children
- Department of Engineering, College of Engineering and Natural Sciences, University of Tennessee at Martin, Martin, TN, United States
Neurological disorders dramatically impact patients of any age population, their families, and societies. Pediatrics are among vulnerable age populations who differently experience the devastating consequences of neurological conditions, such as attention-deficit hyperactivity disorders (ADHD), autism spectrum disorders (ASD), cerebral palsy, concussion, and epilepsy. System-level understanding of these neurological disorders, particularly from the brain networks' dynamic perspective, has led to the significant trend of recent scientific investigations. While a dramatic maturation in the network science application domain is evident, leading to a better understanding of neurological disorders, such rapid utilization for studying pediatric neurological disorders falls behind that of the adult population. Aside from the specific technological needs and constraints in studying neurological disorders in children, the concept of development introduces uncertainty and further complexity topping the existing neurologically driven processes caused by disorders. To unravel these complexities, indebted to the availability of high-dimensional data and computing capabilities, approaches based on machine learning have rapidly emerged a new trend to understand pathways better, accurately diagnose, and better manage the disorders. Deep learning has recently gained an ever-increasing role in the era of health and medical investigations. Thanks to its relatively more minor dependency on feature exploration and engineering, deep learning may overcome the challenges mentioned earlier in studying neurological disorders in children. The current scoping review aims to explore challenges concerning pediatric brain development studies under the constraints of neurological disorders and offer an insight into the potential role of deep learning methodology on such a task with varying and uncertain nature. Along with pinpointing recent advancements, possible research directions are highlighted where deep learning approaches can assist in computationally targeting neurological disorder-related processes and translating them into windows of opportunities for interventions in diagnosis, treatment, and management of neurological disorders in children.
1. Background and History
The brain, as the body commander in chief, evolves by passing through multiple developmental and maturation stages, from the neurogenesis (neuron production stage) and migration (neuron translocation to the neocortex stage) to the differentiation (neurons integration in specialized neural networks by forming axonal connections) (Stiles and Jernigan, 2010). Before transition into the postnatal phase, a massive systematic synaptic exuberance and pruning occurs. After birth, axonal myelination (the process of sheath formation) comes into play that significantly improves axonal integrity and conductance. In support of emerging brain connectivity, synaptic exuberance and pruning continue to occur in parallel with myelination. Tackling neurological disorders by studying the brain structure and function has long been in the neuroscientific research spotlight.
In 1906, an exciting milestone was set for the history of modern neuroscience when the Noble Prize of physiology and medicine was shared between two people, let us indeed reemphasize, between two opponent theories, proposed by Camillo Golgi (1843–1926) and Santiago Ramon y Cajal (1852–1934) (Swanson et al., 2007). Their opposition is rooted in their opinions of how neurons, as the critical units of nervous systems, intercommunicate. Golgi's reticular theory formulated the nervous system as a continuous organization, wherein Cajal's neuron doctrine expressed it as a contiguous organization. While scholarly activities have been on the rise more in favor of the contiguous organization, a growing belief in networks' role has persistently emerged in studying the brain under normal and pathology. The emergence of brain connectivity networks is an essential aspect of brain plasticity, also known as neuroplasticity, a brain adaptation process from experience and learning. During development, brain plasticity rises as it is exposed to environmental events (Rosenzweig and Bennett, 1996). Neuroimaging studies have confirmed the dynamic evolution of these cortical networks through use-induced plasticity to learn and improve functions (Hua and Smith, 2004).
Our current understanding of the connectivity role is the direct consequence of advancements in two main inter-related domains, hardware and computational algorithms. The breakthrough of network-level studies of brain activities was expedited by a series of technology inventions that began by Electroencephalography (EEG) (Britton et al., 2016) and continued with Magnetoencephalography (MEG) (Cohen, 1968), X-ray, Computed Tomography (CT), and Positron Emission Tomography (PET) in the 70's (Raichle, 2009). In the late 70s, Magnetic Resonance Imaging (MRI) was introduced, further boosting brain-related discoveries. The expedition offered by the MRI technology itself was obscured until the advent of different MRI sequences. The MRI sequence refers to a particular harmony set between radio-frequency pulses and magnetic gradients, leading to capturing a specific perspective from the tissue appearance. In 1990 (Ogawa et al., 1990), oxygen consumption and supply to cerebral regions was, for the first time, considered as an endogenous contrast agent for recording brain functional activities. The method, called Blood-Oxygen-Level-Dependant (BOLD) functional MRI, has since been a dominant MRI method of choice in resting-state functional MRI (rs-fMRI) task-based functional MRI, both based on the regional association of brain activity with oxygen supply and consumption. Shifting the focus from functional imaging, The knowledge of brain microstructure diffusion properties has driven diffusion-weighted imaging (Basser et al., 1994) to streamline structural imaging. In parallel to hardware-related technological advancements, computational and visualization power has immensely matured following the development of computing backbones such as C/C++ and python programming languages and the ease of virtual memorization and parallel processing. The development of tools like AFNI (Cox, 1996), FreeSurfer (Fischl, 2012), FSL (Jenkinson et al., 2012), and SPM (Penny et al., 2011) has complemented these inter-related efforts. The rising investment in portraying the brain and its role in diseases and disorders is reflected amid initiatives such as the Brain Initiative or the International Brain Initiative.
The field has been immensely grown since Golgi and Ramon y Cajal's neuroanatomical work that emphasized the role of the functional interplay among regional brain structures. Nevertheless, the growth has happened at a lower rate within discoveries related to the knowledge we have about children's brain networks. Figure 1 captures a synopsis of scholarly activities within the “brain network” domain over the years highlighted with milestones of technology advancements. Contrasted by the age of 19 years old, the graphs show the annual number of full-text articles found in the PubMed search engine with [(brain) AND (network)] term, normalized by the total number of publications found by [(brain)] keyword. From the early 1970s, there has been a steady increase in brain research's scholarly activities, particularly under the umbrella of the brain network. However, the increase in trends shows a slower rate for studies in 19 years old and younger age population.
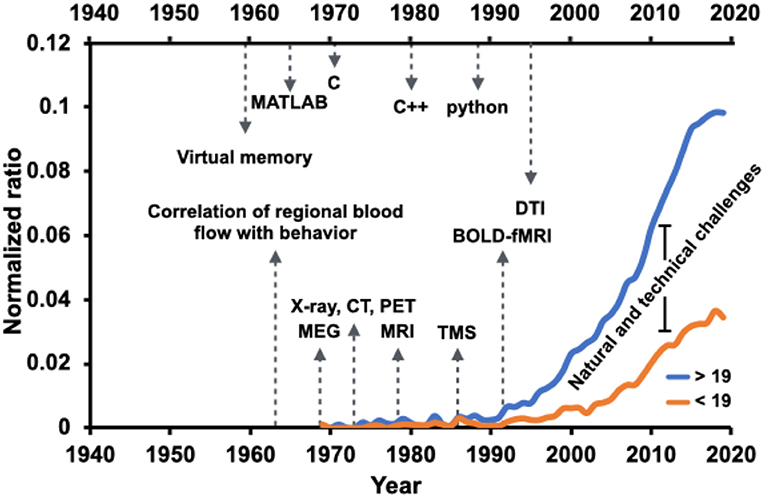
Figure 1. Brain network-related scholarly activities in the pediatric population (younger than 19 years old of age) have paced at a slower rate compared to the adult population (more aged than 19 years old of age), a possible reason being the natural and technical challenges. Normalized ratio of the PubMed-indexed full text articles including [(brain) AND (network)] term, normalized by the total number of publications found by [(brain)] keyword, overlaid with advancement milestones in the field of technology and computation. The annual ratio is visualized for two age groups, younger and older than 19 years old.
2. Challenge and Prospect
Natural and technical challenges (Figure 2) could be deemed for the slower trend of scholarly activities related to pediatric brain studies. The natural, or to better frame it, the inherent difficulty in studying the developmental brain is the development factor's attachment. The development of a complex biological entity, such as the human brain, entails significant synergies among phased development stages, from the genetic blueprint to the formation of brain structure on the foundation of millions of brain cells, including neuronal and glial cells (Gibb and Kovalchuk, 2018). It becomes even further intricate when adding the element of functional network formation to the developmental stages. Brain circuitry emergence is the result of brain cell interactions over time. It is a set of highly dynamic processes, including iterative formation and elimination of synapses and stabilization of relevant synaptic connections to mature functional brain activity in conjunction with brain structure (Kuczmarski, 2002; Hua and Smith, 2004). Aside from the genetic blueprint, it is recognized to depend upon other factors, including nutrition, environmental exposures, and experiences such as interactions with people (Robinson et al., 2017).
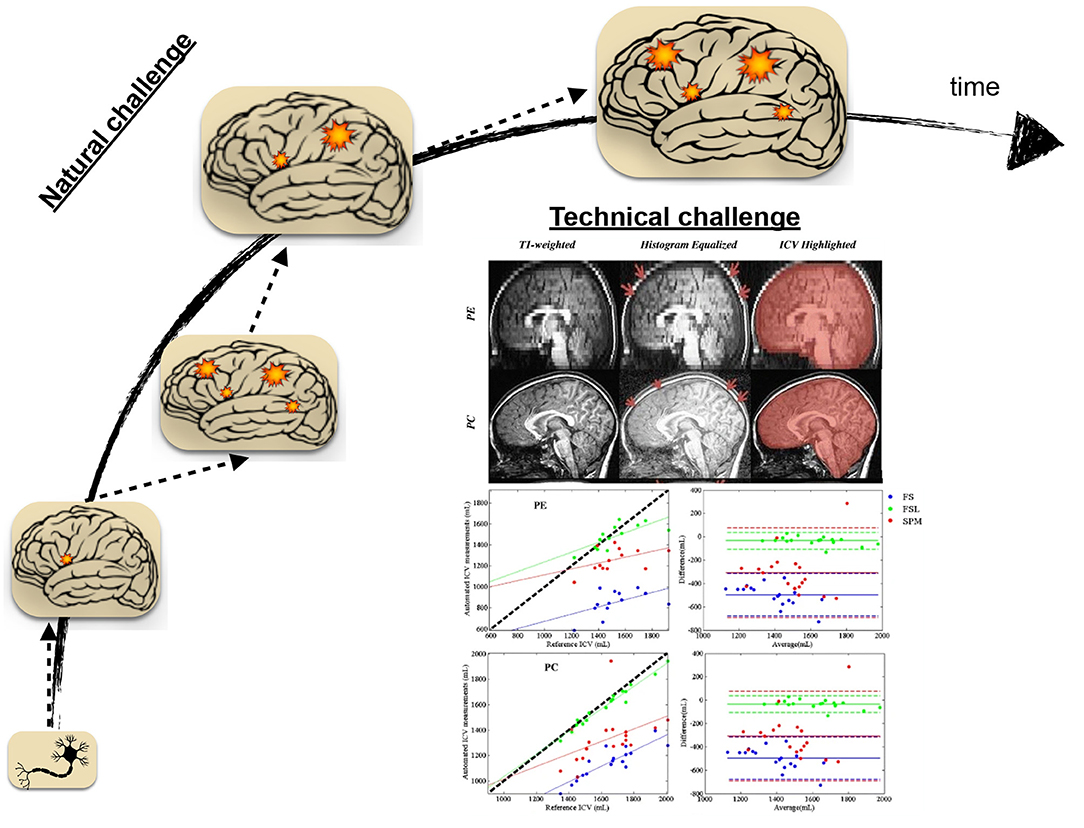
Figure 2. Natural and technical challenges in the face of studying neurological disorders in the pediatric population. Over time, brain development involves many ongoing processes from the cellular to the network level. In the presence of disease or disorder, The ensemble of normal brain development processes is occluded by dynamic processes led by disease or condition, making the clinical studies naturally challenging in pediatric age groups. Technical challenges, including procedural difficulties and data processing methodological limitations, are also surrounding the brain's studies in the pediatric population. The presented technical problem here is reproduced with permission from Figures 2, 4 of Sargolzaei et al. (2015) for the task of intracranial volume estimation (ICV) using automated tools in two pediatric age group of human subjects (PE, Pediatric Epilepsy; PC, Pediatric Control).
The amendment of disease-driven processes to the ensemble of normal brain development processes introduces further uncertainty and complexity, particularly for studies concerned with accurate delineation of normal development operations from those operations driven by pathology. The natural challenge is to an extent that cautious needs to be practiced before generalizing findings in neuroimaging studies of children and adolescents (Santosh, 2000). While neuroimaging, especially non-invasive ones, has become increasingly popular in the era of brain scientific investigation (Morita et al., 2016), its use is confined by issues such as non-compliance (Schlund et al., 2011; Raschle et al., 2012; Thieba et al., 2018). Non-compliance refers to the interruption or failure of the experiment, often due to subject movement or failure to abide by instruction due to anxiety. The issue arises even further for the clinical neuroimaging studies in children and adolescents to the extent that it necessitates proper considerations and study protocols (Greene et al., 2016).
The unique challenges of clinical studies currently extend beyond non-compliance to procedural difficulties, technical obstacles, and data processing methodological limitations. In this regard, The development and utilization of age-appropriate equipment can further streamline clinical studies in children and adolescents. Along with procedural and technical considerations, specific adjustments, age-dependent protocol developments, and analytical fine-tuning are essential for correct implications of neuroimaging studies in the pediatric population (Sargolzaei et al., 2015).
Figure 2 highlights an example of the technical considerations that must be practiced when the neuroimaging study population is pediatrics. It features the task of automatic intracranial volume (ICV) estimation in brain research. ICV estimation is a critically required task in neuroimaging studies. While the task may be fulfilled by manual inspection and landmarks identification, it is a tedious and laborious process. Automatic estimation involves the utilization of neuroimaging software packages and the implementation of a proper ICV estimation routine. As it is apparent from the shown figure, different software packages led into different ICV estimation when contrasted to the reference estimation of such quantity under different condition (control sample vs. pediatric patients with epilepsy)(Sargolzaei et al., 2015). The study emphasizes methodological challenges for pediatric neuroimaging studies, pointing out the necessity of guided decision-making in selecting developed tools under different circumstances.
To overcome the above-summarized challenges, indebted to the availability of high-dimensional data and increased computing capacities, approaches based on graph theory and machine learning have rapidly emerged a new trend to understand pathways better, accurately diagnose, and adequately manage the disorders. Deep learning has recently gained an ever-increasing role in the era of health and medical investigations under the umbrella of machine learning-based solutions to studies of neurological disorders. For the field of neurological disorders in children, our prospect is at the conjunction of applied deep learning engaged with graph theory on a personalized scale.
3. Deep Learning, Something New or the New Face of the Old
Granting machines the gift of intelligence is an ever-increasing appetite for humankind, and the learning skill is standing at the forefront of this intellectual gift. Through decades of artificial intelligence field evolution, machines learned through human-generated rules and accurately engineered features. This approach, conveniently referred to as the classical approach, has phenomenally succeeded in multiple application domains, yet extracting an optimized set of features and rules is not always cumbersome. Further tasks, such as recognition, which is intuitive for humans, remained a challenge for machines to learn via classical approaches that rely upon the existence of high-level abstract features (Goodfellow et al., 2016).
The deep learning approach's neurobiological spirit roots back in experimental studies (Hubel and Wiesel, 1959; Métin and Frost, 1989; Roe et al., 1992) discovering the mutually critical roles of intracortical circuit specifications and inputs to such circuits in the learning process of brain neural networks. Learning about the experience-dependant plasticity of the brain (Simons and Land, 1987; Kirkwood et al., 1995; Crist et al., 2001; Trachtenberg et al., 2002) inspires the methodological possibility of letting the machine also acquire knowledge by experience. Thorough experience-dependent knowledge acquisition involves exposure to a vast amount of such experiences, accompanied by corresponding outcomes, and the capacity to automatically form an intra-circuitry that maps these experiences to their corresponding outcome. Translating these requirements to artificial neural networks' jargon entails a specialized architecture, ultra-large datasets, and sophisticated linear and non-linear training algorithms, formulating the deep learning approach to the machine learning task. The specialized network architecture allows sequential processing of the supplied raw data units through multiple layers without the need for human-based feature design and engineering.
The deep learning approach (Figure 3) lets the network itself find an optimized allocation of credits to basic units (neurons) of these layers in sketching the desired outcome (LeCun et al., 2015; Schmidhuber, 2015). The network depth offers a way to break down a complex raw input into simplified units of information, mainly through convolution and sampling processes, and collectively map the input to the provided network output. Self-exploration mapping is the backbone of architectures such as deep convolutional neural networks (CNN) and recurrent neural networks (RNN), with the latter one empowering the learning of sequential data such as text and speech. Alongside other application sectors, healthcare has reported breakthroughs achieved with deep learning adoption in neuroimaging, genetics, oncology, radiation therapy, and drug discovery, to name a few (Wang et al., 2018, 2020; Boldrini et al., 2019; David et al., 2019; Serag et al., 2019; Tang et al., 2019; Zhu et al., 2019; Chen et al., 2020; Zhang et al., 2020).
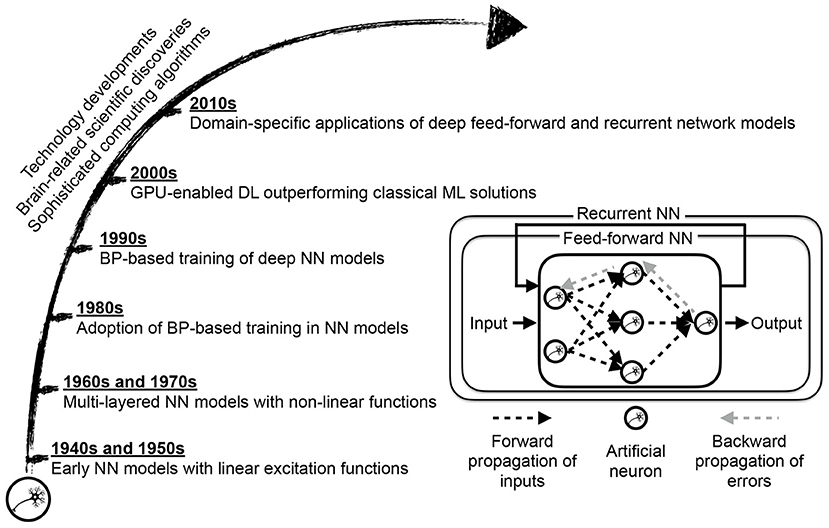
Figure 3. Deep learning (DL) approach maturation motivated by scientific discoveries of brain structure and function and enabled by computing technology and algorithms' immense progress. The core of a neural network (NN) model is an artificial neuron, resembling a biological neuron in a limited way, evaluating the collective input signals followed by conditional excitement. While early citations of NN models, as a linear regression variation, dates back to the 1800s, NN-based learning was not widespread until the development of sophisticated training algorithms based on back-propagation of error signals, and gradient descent optimization (Schmidhuber, 2015). The transition from classical NN-based learning, where the network examines extracted features from data to deep NN-based learning, where the network scans raw data representation, requires equipping NN with self-exploring capacity through multiple layers of abstraction. The graphical processing unit (GPU) based computing partially enabled such a self-exploratory multi-layered representation learning to the extent that deep learning becomes the choice method in numerous domain-specific applications (LeCun et al., 2015).
4. Deep Learning and Neurological Disorders in Children
To review the scope of deep learning application in diagnosis, treatment and management of neurological disorders in children, we surveyed existing peer-reviewed literature. The search was performed using PubMed (https://pubmed.ncbi.nlm.nih.gov), and IEEE Xplore (https://ieeexplore.ieee.org/Xplore/home.jsp). The search queries were set to [(deep learning) AND ((children) OR (child) OR (pediatric) OR (paediatric))] for PubMed search, and to [((“All Metadata”:Deep learning) AND ((“All Metadata”:children) OR (“All Metadata”:child) OR (“All Metadata”:pediatric)))] for IEEE Xplore search. Setting the inclusion criteria to peer-reviewed journal articles reporting the deep learning approach utilization in studying, diagnosing, or managing neurological disorders in the pediatric (up to 19 years of age) population led to a total of 22 records. Included articles were examined to retrieve the following information, study neurological condition (Condition), study population age range (Age range), study goal (Goal), modality of the used data (Modality), used deep learning model (Model), and backbone implementation environment (Environment). Table 1 summarizes included studies.
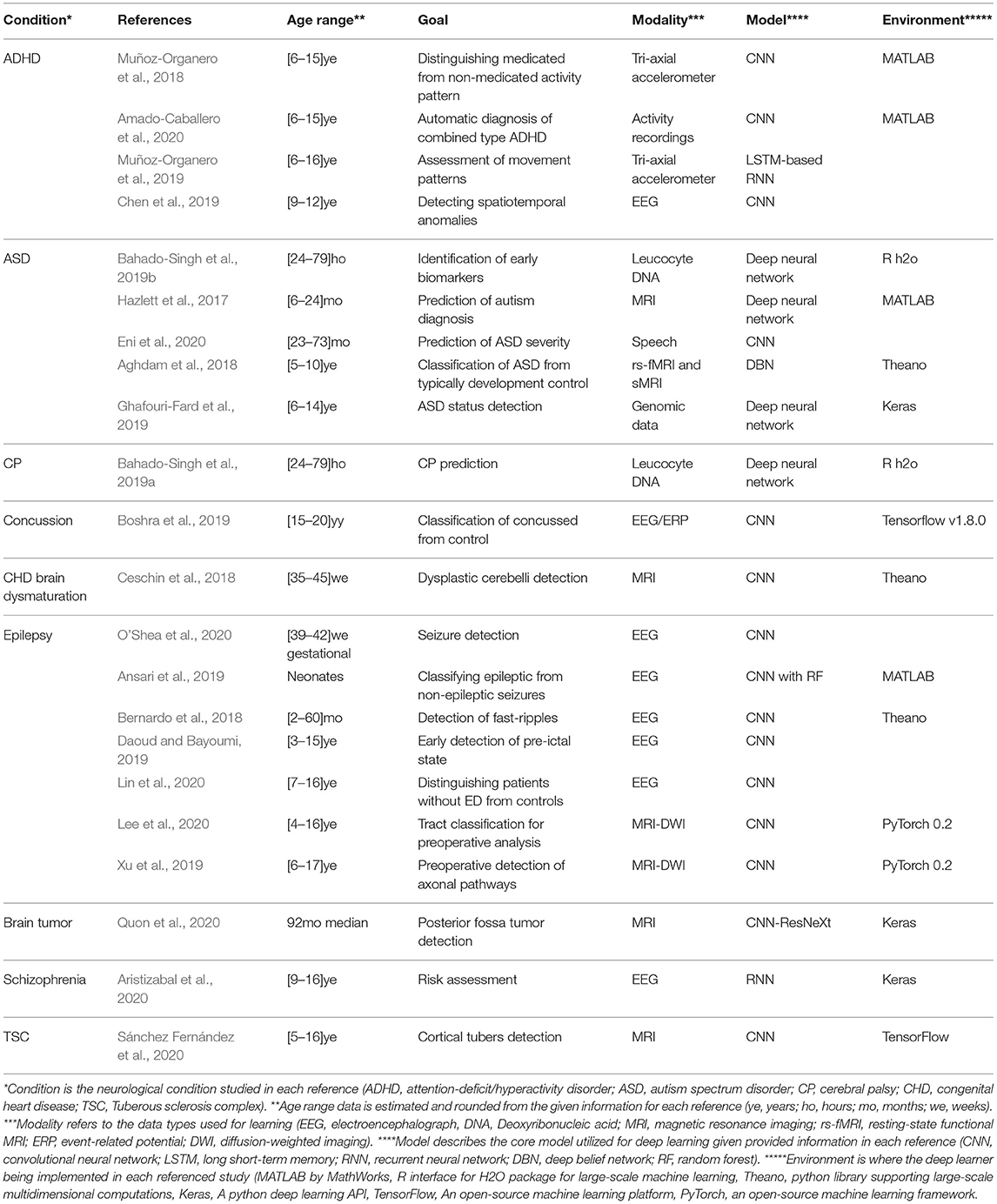
Table 1. A summary of found peer-reviewed journal articles utilizing deep learning to study, manage, diagnose, and prognosis of neurological conditions in pediatric (18 years or younger) populations.
Perceived from Table 1, The deep learning architectures, in particular convolutional neural networks, have reported promises in a wide variety of applications, including automatic diagnosis, biomarker identification, early detection, within-condition type classification, and risk assessment for a variety of neurological conditions in children. The survey reveals the applicability potential of deep learning solutions beyond neuroimaging modalities, such as EEG and MRI, extending its use to other information sources such as speech and activity-based modalities within the pediatric clinical population. The latter inference from the survey is of utmost importance due to the challenges discussed before regarding methodological constraints of studies in pediatrics. While python-based libraries are the dominant library of choice reported in the current studies sample, MATLAB and R language libraries for large-scale machine learning implementations are also cited.
While deep learning solutions are relatively free of conventional feature engineering requirements, they are entangled with their choice of architecture and learning parameters, enforcing the need for careful implementation and selection of such internal architecture and its relevant parameters. The consequences of varying architecture are reported to have an impact on the deep learner performance for the use of convolutional neural network (Xu et al., 2019) and deep belief network (Aghdam et al., 2018). Therefore, the inclusion of benchmark routines, mainly reporting different architectures for optimum architecture selection, can enhance study findings.
Our survey search has also identified a rising interest in using deep learners to overcome inherent challenges, low tissue contrast, and dynamic appearance variation for brain imaging tasks in infants (Guo et al., 2014; Zhang et al., 2015; Kawahara et al., 2017; Mostapha and Styner, 2019; Sun et al., 2019) and fetus (Girault et al., 2019; Dou et al., 2020). A common goal for most surveyed articles is to conduct more extensive validation studies that involve multi-site investigations to make deep-learning-based computational tools more suitable as the non-invasive and inexpensive real-time clinical tool of choice.
5. Viewpoints and Conclusion
The current scoping review concentrated the scope on the rising use of deep learning to study, diagnose, and prognosis for children's neurological disorders. The hallmark of neurological disorders is the presence of brain dysfunction. Presentation of behavioral and psycho-behavioral symptoms is a consequence of such dysfunction. Neuroimaging studies have confirmed the dynamic evolution of these functional cortical networks through use-induced plasticity. In a similar context, neurological disorders have been shown to leave the pathophysiological signature on brain networks. Network neuroscience, in this regard, has significantly matured and been extensively utilized in studying neurological disorders; however, the pace of brain network understanding and its role in driving neurological conditions presentation in children falls behind that of the older age population.
We discussed inherent challenges in the face of studying neurological disorders in the pediatric population, contrasting it to the task of hitting a moving target. Brain plasticity is a complex and heterogeneous phenomenon, reflected as a multi-faceted maturation. The pace of brain development process variation occurs at a relatively higher rate in the early stages. Overlaying such a dynamic with uncertainties imposed by neurological disease and disorders turns the task of studying pediatric neurological studies into a difficult one. Inspired by the way our brain sees and learns the world, computational advancement gave birth to a new horizon, called deep learning, promising revolutionary insights into the field of medicine and biology.
Our scoping review of the current state of deep learning utilization in children's neurological disorders has identified a rising interest from scientific investigators for deep learning in various classification tasks related to children's neurological disorders. Data volume, along with, to a lesser extent, data quality, remains among major barriers to incorporate deep learning for studying neurological disorders (Miotto et al., 2018; Valliani et al., 2019). The emergence of deep learning will further continue in the era of pediatric clinical studies because of its lesser reliance upon the existence of engineered features.
Aiming at the development of the generalized deep learner to the level of clinical efficacy requires taking a steeper, and yet clear, road. It involves conducting more extensive multi-sites validation studies and performing benchmark evaluations of deep learning architectures to intensify their utilization further. Studying less utilized architectures such as recurrent neural networks and auto-encoders to learn the joint temporal dynamics of the disease and development in one shot will form a valuable complement to the existing efforts. Legal, privacy, and ethical challenges, with respect to the use of deep learners in healthcare (Miotto et al., 2018; Valliani et al., 2019), remain at place with greater emphasis for the vulnerable populations such as pediatrics. Efforts such as distributed deep learning schemes (Zeng et al., 2018; Remedios et al., 2019) can partially help resolving such dilemma. While deep learning is reported to outperform traditional machine learning algorithms, joined solutions need to be evaluated in cases where deep learners may supplement the established solutions.
Like hitting a moving target, studying neurological disorders in the developing brain is a cumbersome task. However, as tracking a moving target before hitting it can increase success, performing multi time-points longitudinal studies in the pediatric population can improve our chances of understanding and defeating the disease. Therefore, longitudinal modeling and analysis of children's neurological disorders using deep learning architectures can unravel windows of opportunities to hit the moving target.
Author Contributions
SS contributed to all stages of study, contributed to manuscript revision, read, and approved the submitted version.
Conflict of Interest
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Aghdam, M. A., Sharifi, A., and Pedram, M. M. (2018). Combination of rs-fMRI and sMRI data to discriminate autism spectrum disorders in young children using deep belief network. J. Digit. Imaging 31, 895–903. doi: 10.1007/s10278-018-0093-8
Amado-Caballero, P., Casaseca-de-la Higuera, P., Alberola-Lopez, S., Andres-de Llano, J. M., Villalobos, J. A. L., Garmendia-Leiza, J. R., et al. (2020). Objective ADHD diagnosis using convolutional neural networks over daily-life activity records. IEEE J. Biomed. Health Inform. 24, 2690–2700. doi: 10.1109/JBHI.2020.2964072
Ansari, A. H., Cherian, P. J., Caicedo, A., Naulaers, G., De Vos, M., and Van Huffel, S. (2019). Neonatal seizure detection using deep convolutional neural networks. Int. J. Neural Syst. 29:1850011. doi: 10.1142/S0129065718500119
Aristizabal, D. A., Fernando, T., Denman, S., Robinson, J. E., Sridharan, S., Johnston, P. J., et al. (2020). Identification of children at risk of schizophrenia via deep learning and EEG responses. IEEE J. Biomed. Health Inform. 25, 69–76. doi: 10.1109/JBHI.2020.2984238
Bahado-Singh, R. O., Vishweswaraiah, S., Aydas, B., Mishra, N. K., Guda, C., and Radhakrishna, U. (2019a). Deep learning/artificial intelligence and blood-based dna epigenomic prediction of cerebral palsy. Int. J. Mol. Sci. 20:2075. doi: 10.3390/ijms20092075
Bahado-Singh, R. O., Vishweswaraiah, S., Aydas, B., Mishra, N. K., Yilmaz, A., Guda, C., et al. (2019b). Artificial intelligence analysis of newborn leucocyte epigenomic markers for the prediction of autism. Brain Res. 1724:146457. doi: 10.1016/j.brainres.2019.146457
Basser, P. J., Mattiello, J., and LeBihan, D. (1994). MR diffusion tensor spectroscopy and imaging. Biophys. J. 66, 259–267. doi: 10.1016/S0006-3495(94)80775-1
Bernardo, D., Nariai, H., Hussain, S. A., Sankar, R., Salamon, N., Krueger, D. A., et al. (2018). Visual and semi-automatic non-invasive detection of interictal fast ripples: a potential biomarker of epilepsy in children with tuberous sclerosis complex. Clin. Neurophysiol. 129, 1458–1466. doi: 10.1016/j.clinph.2018.03.010
Boldrini, L., Bibault, J.-E., Masciocchi, C., Shen, Y., and Bittner, M.-I. (2019). Deep learning: a review for the radiation oncologist. Front. Oncol. 9:977. doi: 10.3389/fonc.2019.00977
Boshra, R., Ruiter, K. I., DeMatteo, C., Reilly, J. P., and Connolly, J. F. (2019). Neurophysiological correlates of concussion: deep learning for clinical assessment. Sci. Rep. 9, 1–10. doi: 10.1038/s41598-019-53751-9
Britton, J. W., Frey, L. C., Hopp, J. L., Korb, P., Koubeissi, M. Z., Lievens, W. E., et al. (2016). Electroencephalography (EEG): An Introductory Text and Atlas of Normal and Abnormal Findings in Adults, Children, and Infants. Chicago, IL: American Epilepsy Society.
Ceschin, R., Zahner, A., Reynolds, W., Gaesser, J., Zuccoli, G., Lo, C. W., et al. (2018). A computational framework for the detection of subcortical brain dysmaturation in neonatal MRI using 3d convolutional neural networks. Neuroimage 178, 183–197. doi: 10.1016/j.neuroimage.2018.05.049
Chen, C., Qin, C., Qiu, H., Tarroni, G., Duan, J., Bai, W., et al. (2020). Deep learning for cardiac image segmentation: a review. Front. Cardiovasc. Med. 7:25. doi: 10.3389/fcvm.2020.00025
Chen, H., Song, Y., and Li, X. (2019). Use of deep learning to detect personalized spatial-frequency abnormalities in EEGs of children with ADHD. J. Neural Eng. 16:066046. doi: 10.1088/1741-2552/ab3a0a
Cohen, D. (1968). Magnetoencephalography: evidence of magnetic fields produced by alpha-rhythm currents. Science 161, 784–786. doi: 10.1126/science.161.3843.784
Cox, R. W. (1996). AFNI: software for analysis and visualization of functional magnetic resonance neuroimages. Comput. Biomed. Res. 29, 162–173. doi: 10.1006/cbmr.1996.0014
Crist, R. E., Li, W., and Gilbert, C. D. (2001). Learning to see: experience and attention in primary visual cortex. Nat. Neurosci. 4, 519–525. doi: 10.1038/87470
Daoud, H., and Bayoumi, M. A. (2019). Efficient epileptic seizure prediction based on deep learning. IEEE Trans. Biomed. Circ. Syst. 13, 804–813. doi: 10.1109/TBCAS.2019.2929053
David, L., Arús-Pous, J., Karlsson, J., Engkvist, O., Bjerrum, E. J., Kogej, T., et al. (2019). Applications of deep-learning in exploiting large-scale and heterogeneous compound data in industrial pharmaceutical research. Front. Pharmacol. 10:1303. doi: 10.3389/fphar.2019.01303
Dou, H., Karimi, D., Rollins, C. K., Ortinau, C. M., Vasung, L., Velasco-Annis, C., et al. (2020). A deep attentive convolutional neural network for automatic cortical plate segmentation in fetal MRI. arXiv preprint arXiv:2004.12847. doi: 10.1109/TMI.2020.3046579
Eni, M., Dinstein, I., Ilan, M., Menashe, I., Meiri, G., and Zigel, Y. (2020). Estimating autism severity in young children from speech signals using a deep neural network. IEEE Access 8, 139489–139500. doi: 10.1109/ACCESS.2020.3012532
Ghafouri-Fard, S., Taheri, M., Omrani, M. D., Daaee, A., Mohammad-Rahimi, H., and Kazazi, H. (2019). Application of single-nucleotide polymorphisms in the diagnosis of autism spectrum disorders: a preliminary study with artificial neural networks. J. Mol. Neurosci. 68, 515–521. doi: 10.1007/s12031-019-01311-1
Gibb, R., and Kovalchuk, A. (2018). “Chapter 1: brain development,” in The Neurobiology of Brain and Behavioral Development, eds R. Gibb, and B. Kolb (Academic Press), 3–27. doi: 10.1016/B978-0-12-804036-2.00001-7
Girault, J. B., Munsell, B. C., Puechmaille, D., Goldman, B. D., Prieto, J. C., Styner, M., et al. (2019). White matter connectomes at birth accurately predict cognitive abilities at age 2. Neuroimage 192, 145–155. doi: 10.1016/j.neuroimage.2019.02.060
Goodfellow, I., Bengio, Y., and Courville, A. (2016). Deep Learning. MIT Press. Available online at: http://www.deeplearningbook.org
Greene, D. J., Black, K. J., and Schlaggar, B. L. (2016). Considerations for MRI study design and implementation in pediatric and clinical populations. Dev. Cogn. Neurosci. 18, 101–112. doi: 10.1016/j.dcn.2015.12.005
Guo, Y., Wu, G., Commander, L. A., Szary, S., Jewells, V., Lin, W., et al. (2014). “Segmenting hippocampus from infant brains by sparse patch matching with deep-learned features,” in International Conference on Medical Image Computing and Computer-Assisted Intervention (Boston, MA: Springer), 308-315. doi: 10.1007/978-3-319-10470-6_39
Hazlett, H. C., Gu, H., Munsell, B. C., Kim, S. H., Styner, M., Wolff, J. J., et al. (2017). Early brain development in infants at high risk for autism spectrum disorder. Nature 542, 348–351. doi: 10.1038/nature21369
Hua, J. Y., and Smith, S. J. (2004). Neural activity and the dynamics of central nervous system development. Nat. Neurosci. 7, 327–332. doi: 10.1038/nn1218
Hubel, D. H., and Wiesel, T. N. (1959). Receptive fields of single neurones in the cat's striate cortex. J. Physiol. 148:574. doi: 10.1113/jphysiol.1959.sp006308
Jenkinson, M., Beckmann, C. F., Behrens, T. E., Woolrich, M. W., and Smith, S. M. (2012). FSL. Neuroimage 62, 782–790. doi: 10.1016/j.neuroimage.2011.09.015
Kawahara, J., Brown, C. J., Miller, S. P., Booth, B. G., Chau, V., Grunau, R. E., et al. (2017). Brainnetcnn: convolutional neural networks for brain networks; towards predicting neurodevelopment. Neuroimage 146, 1038–1049. doi: 10.1016/j.neuroimage.2016.09.046
Kirkwood, A., Lee, H.-K., and Bear, M. F. (1995). Co-regulation of long-term potentiation and experience-dependent synaptic plasticity in visual cortex by age and experience. Nature 375, 328–331. doi: 10.1038/375328a0
Kuczmarski, R. J. (2002). 2000 CDC Growth Charts for the United States: Methods and Development. Number 246. Department of Health and Human Services, Centers for Disease Control.
LeCun, Y., Bengio, Y., and Hinton, G. (2015). Deep learning. Nature 521, 436–444. doi: 10.1038/nature14539
Lee, M.-H., O'Hara, N., Sonoda, M., Kuroda, N., Juhasz, C., Asano, E., et al. (2020). Novel deep learning network analysis of electrical stimulation mapping-driven diffusion MRI tractography to improve preoperative evaluation of pediatric epilepsy. IEEE Trans. Biomed. Eng. 67, 3151–3162. doi: 10.1109/TBME.2020.2977531
Lin, L.-C., Ouyang, C.-S., Wu, R.-C., Yang, R.-C., and Chiang, C.-T. (2020). Alternative diagnosis of epilepsy in children without epileptiform discharges using deep convolutional neural networks. Int. J. Neural Syst. 30:1850060. doi: 10.1142/S0129065718500600
Métin, C., and Frost, D. O. (1989). Visual responses of neurons in somatosensory cortex of hamsters with experimentally induced retinal projections to somatosensory thalamus. Proc. Natl. Acad. Sci. U.S.A. 86, 357–361. doi: 10.1073/pnas.86.1.357
Miotto, R., Wang, F., Wang, S., Jiang, X., and Dudley, J. T. (2018). Deep learning for healthcare: review, opportunities and challenges. Brief. Bioinform. 19, 1236–1246. doi: 10.1093/bib/bbx044
Morita, T., Asada, M., and Naito, E. (2016). Contribution of neuroimaging studies to understanding development of human cognitive brain functions. Front. Hum. Neurosci. 10:464. doi: 10.3389/fnhum.2016.00464
Mostapha, M., and Styner, M. (2019). Role of deep learning in infant brain MRI analysis. Magnet. Reson. Imaging 64, 171–189. doi: 10.1016/j.MRI.2019.06.009
Muñoz-Organero, M., Powell, L., Heller, B., Harpin, V., and Parker, J. (2018). Automatic extraction and detection of characteristic movement patterns in children with ADHD based on a convolutional neural network (CNN) and acceleration images. Sensors 18:3924. doi: 10.3390/s18113924
Muñoz-Organero, M., Powell, L., Heller, B., Harpin, V., and Parker, J. (2019). Using recurrent neural networks to compare movement patterns in ADHD and normally developing children based on acceleration signals from the wrist and ankle. Sensors 19:2935. doi: 10.3390/s19132935
Ogawa, S., Lee, T.-M., Nayak, A. S., and Glynn, P. (1990). Oxygenation-sensitive contrast in magnetic resonance image of rodent brain at high magnetic fields. Magnet. Reson. Med. 14, 68–78. doi: 10.1002/mrm.1910140108
O'Shea, A., Lightbody, G., Boylan, G., and Temko, A. (2020). Neonatal seizure detection from raw multi-channel EEG using a fully convolutional architecture. Neural Netw. 123, 12–25. doi: 10.1016/j.neunet.2019.11.023
Penny, W. D., Friston, K. J., Ashburner, J. T., Kiebel, S. J., and Nichols, T. E. (2011). Statistical Parametric Mapping: The Analysis of Functional Brain Images. Elsevier.
Quon, J., Bala, W., Chen, L., Wright, J., Kim, L., Han, M., et al. (2020). Deep learning for pediatric posterior fossa tumor detection and classification: a multi-institutional study. Am. J. Neuroradiol. 41, 1718–1725. doi: 10.3174/ajnr.A6704
Raichle, M. E. (2009). A brief history of human brain mapping. Trends Neurosci. 32, 118–126. doi: 10.1016/j.tins.2008.11.001
Raschle, N., Zuk, J., Ortiz-Mantilla, S., Sliva, D. D., Franceschi, A., Grant, P. E., et al. (2012). Pediatric neuroimaging in early childhood and infancy: challenges and practical guidelines. Ann. N. Y. Acad. Sci. 1252:43. doi: 10.1111/j.1749-6632.2012.06457.x
Remedios, S., Roy, S., Blaber, J., Bermudez, C., Nath, V., Patel, M. B., et al. (2019). “Distributed deep learning for robust multi-site segmentation of CT imaging after traumatic brain injury,” in Medical Imaging 2019: Image Processing (International Society for Optics and Photonics), 109490A. doi: 10.1117/12.2511997
Robinson, L. R., Bitsko, R. H., Thompson, R. A., Dworkin, P. H., McCabe, M. A., Peacock, G., et al. (2017). CDC grand rounds: addressing health disparities in early childhood. Morbidity Mortality Weekly Report 66:769. doi: 10.15585/mmwr.mm6629a1
Roe, A. W., Pallas, S. L., Kwon, Y. H., and Sur, M. (1992). Visual projections routed to the auditory pathway in ferrets: receptive fields of visual neurons in primary auditory cortex. J. Neurosci. 12, 3651–3664. doi: 10.1523/JNEUROSCI.12-09-03651.1992
Rosenzweig, M. R., and Bennett, E. L. (1996). Psychobiology of plasticity: effects of training and experience on brain and behavior. Behav. Brain Res. 78, 57–65. doi: 10.1016/0166-4328(95)00216-2
Sánchez Fernández, I., Yang, E., Calvachi, P., Amengual-Gual, M., Wu, J. Y., Krueger, D., et al. (2020). Deep learning in rare disease. Detection of tubers in tuberous sclerosis complex. PLoS ONE 15:e0232376. doi: 10.1371/journal.pone.0232376
Santosh, P. J. (2000). Neuroimaging in child and adolescent psychiatric disorders. Arch. Dis. Childhood 82, 412–419. doi: 10.1136/adc.82.5.412
Sargolzaei, S., Sargolzaei, A., Cabrerizo, M., Chen, G., Goryawala, M., Pinzon-Ardila, A., et al. (2015). Estimating intracranial volume in brain research: an evaluation of methods. Neuroinformatics 13, 427–441. doi: 10.1007/s12021-015-9266-5
Schlund, M. W., Cataldo, M. F., Siegle, G. J., Ladouceur, C. D., Silk, J. S., Forbes, E. E., et al. (2011). Pediatric functional magnetic resonance neuroimaging: tactics for encouraging task compliance. Behav. Brain Funct. 7, 1–10. doi: 10.1186/1744-9081-7-10
Schmidhuber, J. (2015). Deep learning in neural networks: an overview. Neural Netw. 61, 85–117. doi: 10.1016/j.neunet.2014.09.003
Serag, A., Ion-Margineanu, A., Qureshi, H., McMillan, R., Saint Martin, M.-J., Diamond, J., et al. (2019). Translational AI and deep learning in diagnostic pathology. Front. Med. 6:185. doi: 10.3389/fmed.2019.00185
Simons, D. J., and Land, P. W. (1987). Early experience of tactile stimulation influences organization of somatic sensory cortex. Nature 326, 694–697. doi: 10.1038/326694a0
Stiles, J., and Jernigan, T. L. (2010). The basics of brain development. Neuropsychol. Rev. 20, 327–348. doi: 10.1007/s11065-010-9148-4
Sun, L., Zhang, D., Lian, C., Wang, L., Wu, Z., Shao, W., et al. (2019). Topological correction of infant white matter surfaces using anatomically constrained convolutional neural network. Neuroimage 198, 114–124. doi: 10.1016/j.neuroimage.2019.05.037
Swanson, L. W., Grant, G., Hökfelt, T., Jones, E. G., and Morrison, J. H. (2007). A century of neuroscience discovery: reflecting on the nobel prize awarded to golgi and cajal in 1906. Brain Res. Rev. 55, 191–192. doi: 10.1016/j.brainresrev.2007.07.001
Tang, B., Pan, Z., Yin, K., and Khateeb, A. (2019). Recent advances of deep learning in bioinformatics and computational biology. Front. Genet. 10:214. doi: 10.3389/fgene.2019.00214
Thieba, C., Frayne, A., Walton, M., Mah, A., Benischek, A., Dewey, D., et al. (2018). Factors associated with successful MRI scanning in unsedated young children. Front. Pediatr. 6:146. doi: 10.3389/fped.2018.00146
Trachtenberg, J. T., Chen, B. E., Knott, G. W., Feng, G., Sanes, J. R., Welker, E., et al. (2002). Long-term in vivo imaging of experience-dependent synaptic plasticity in adult cortex. Nature 420, 788–794. doi: 10.1038/nature01273
Valliani, A. A.-A., Ranti, D., and Oermann, E. K. (2019). Deep learning and neurology: a systematic review. Neurol. Therapy 8, 351–365. doi: 10.1007/s40120-019-00153-8
Wang, C., Xu, P., Zhang, L., Huang, J., Zhu, K., and Luo, C. (2018). Current strategies and applications for precision drug design. Front. Pharmacol. 9:787. doi: 10.3389/fphar.2018.00787
Wang, M., Zhang, Q., Lam, S., Cai, J., and Yang, R. (2020). A review on application of deep learning algorithms in external beam radiotherapy automated treatment planning. Front. Oncol. 10:580919. doi: 10.3389/fonc.2020.580919
Xu, H., Dong, M., Lee, M.-H., O'Hara, N., Asano, E., and Jeong, J.-W. (2019). Objective detection of eloquent axonal pathways to minimize postoperative deficits in pediatric epilepsy surgery using diffusion tractography and convolutional neural networks. IEEE Trans. Med. Imaging 38, 1910–1922. doi: 10.1109/TMI.2019.2902073
Zeng, L.-L., Wang, H., Hu, P., Yang, B., Pu, W., Shen, H., et al. (2018). Multi-site diagnostic classification of schizophrenia using discriminant deep learning with functional connectivity MRI. EBiomedicine 30, 74–85. doi: 10.1016/j.ebiom.2018.03.017
Zhang, L., Wang, M., Liu, M., and Zhang, D. (2020). A survey on deep learning for neuroimaging-based brain disorder analysis. Front. Neurosci. 14:779. doi: 10.3389/fnins.2020.00779
Zhang, W., Li, R., Deng, H., Wang, L., Lin, W., Ji, S., et al. (2015). Deep convolutional neural networks for multi-modality isointense infant brain image segmentation. Neuroimage 108, 214–224. doi: 10.1016/j.neuroimage.2014.12.061
Keywords: ADHD, ASD, cerebral palsy, concussion, deep learning, epilepsy, network neuroscience, neurological disorders
Citation: Sargolzaei S (2021) Can Deep Learning Hit a Moving Target? A Scoping Review of Its Role to Study Neurological Disorders in Children. Front. Comput. Neurosci. 15:670489. doi: 10.3389/fncom.2021.670489
Received: 21 February 2021; Accepted: 09 April 2021;
Published: 05 May 2021.
Edited by:
Xiaoli Li, Beijing Normal University, ChinaCopyright © 2021 Sargolzaei. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Saman Sargolzaei, saman@utm.edu
 Saman Sargolzaei
Saman Sargolzaei