Exosomal microRNAs in breast cancer: towards theranostic applications
- 1Division of Medical Research, Faculty of Medicine and Health Sciences, SRM Institute of Science and Technology, Kattankulathur, Tamil Nadu, India
- 2Dietrich School of Arts and Sciences, University of Pittsburgh, Pittsburgh, PA, United States
- 3Department of Biochemistry and Molecular Biology, Mayo Clinic, Rochester, MN, United States
- 4Department of Obstetrics and Gynecology, Mayo Clinic, Rochester, MN, United States
Breast cancer is one of the top two reproductive cancers responsible for high rates of morbidity and mortality among women globally. Despite the advancements in the treatment of breast cancer, its early diagnosis remains a challenge. Recent evidence indicates that despite the adroit use of numerous strategies to facilitate rapid and precision-oriented screening of breast cancer at the community level through the use of mammograms, Fine-needle aspiration cytology (FNAC) and biomarker tracking, no strategy has been unequivocally accepted as a gold standard for facilitating rapid screening for disease. This necessitates the need to identify novel strategies for the detection and triage of breast cancer lesions at higher rates of specificity, and sensitivity, whilst taking into account the epidemiologic and social-demographic features of the patients. Recent shreds of evidence indicate that exosomes could be a robust source of biomaterial for the rapid screening of breast cancer due to their high stability and their presence in body fluids. Increasing evidence indicates that the Exosomal microRNAs- play a significant role in modifying the tumour microenvironment of breast cancers, thereby potentially aiding in the proliferation, invasion and metastasis of breast cancer. In this review, we summarize the role of ExomiRs in the tumour microenvironment in breast cancer. These ExomiRs can also be used as candidate biomarkers for facilitating rapid screening and triaging of breast cancer patients for clinical intervention.
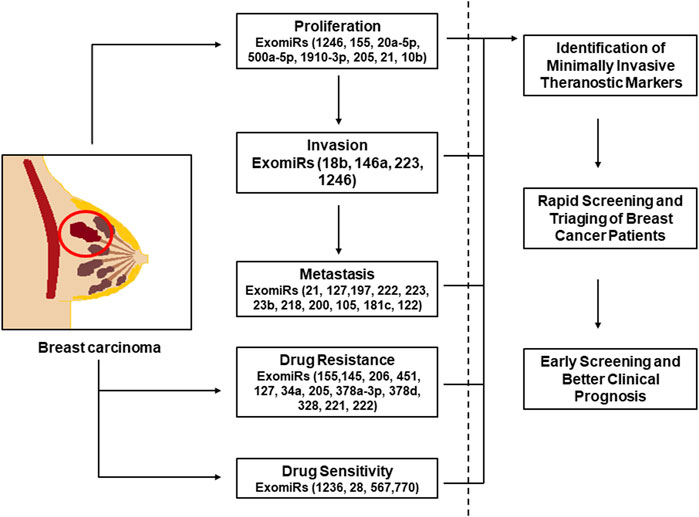
GRAPHICAL ABSTRACT | The graphical abstract shows the ExomiRs involved in the process of proliferation, metastasis and invasion of breast cancer along with the ExomiRs involved in the drug resistance and sensitivity. This implies the diagnostic as well as therapeutic potential of these ExomiRs with respect to breast cancer, indicating that these can be used a theranostic biomarkers for early diagnosis and treatment of breast cancer.
1 Introduction
Breast cancer is one of the most common cancers among women, with cases estimated to be 2.2 million in 2020 globally (Breast, 2020). Despite significant advances in the diagnosis and treatment of breast cancer, it remains one of the leading causes of morbidity among women. According to research, the 5-year survival rates for women with localized breast cancer, regional stage, and metastasis are 99 percent, 84 percent, and 23 percent, respectively (Desantis et al., 2012; Lowry et al., 2015). This suggests that early detection of the disease and treatment can lower breast cancer death rates. With a median overall survival of between 0.5–2.2 years, advanced-stage breast cancer is still incurable (Yeo et al., 2014). Histologically, breast cancer can be divided into invasive (ductal carcinoma in situ) and pre-invasive types (ductal carcinoma). Clinically significant classifications of the intrinsic, molecular subtypes of invasive breast carcinomas include luminal B-like HER2-positive, luminal B-like HER2-negative, and luminal A breast cancer, as well as triple-negative breast cancer (TNBC), HER2-enriched (nonluminal), and luminal B-like HER2-positive (Cheang et al., 2015; Nagini, 2017). Early detection of breast cancer, improving primary tumour treatment efficacy to prevent residual disease or metastatic seeding, and/or encouraging cell dormancy in metastatic niches are all the current strategies and roadblocks for reducing metastatic disease (Sempere et al., 2017).
Apart from women, breast cancer is also diagnosed among males, although it is a rare disease among them with an incidence rate of approximately 0.5%–1% (WHO, 2024). For male breast cancer, the median age of diagnosis is 68 years old, whilst for women it is 62 (Khan and Tirona, 2021). In terms of clinical presentation, histologic type distribution, and hormone-receptor expression, male breast cancer is distinct from female breast cancer (Gucalp et al., 2019; Fox et al., 2022). Male breast cancers usually appear clinically later and are primarily estrogen receptor (ERα) positive (up to 95%). Human epidermal growth factor receptor 2 (HER2, also known as ERBB2) expression is rare, and triple negativity is especially unusual in men (Chatterji et al., 2023). When compared to females with the same classification of the disease, males with breast cancer have a worse survival rate due to several factors that impact their prognosis (Ottini et al., 2010). There is a lack of studies exploring the mechanisms and risk factors involved in male breast cancer when compared to female breast cancer due to its low incidence and prevalence rates across the world. Researchers are working on identifying novel biomarkers for male breast cancer and have met some success, however, these biomarkers are not validated individually suggesting biomarkers for male breast cancer still remain under-investigated.
Extracellular vesicles (EVs) include a subtype called exosomes, which are coated in 30–100 nm lipid bilayers that contains a variety of bioactive substances such as lipids, proteins, nucleic acids, and small non-coding RNAs (Green et al., 2015; Wang M. et al., 2019). Almost all eukaryotic cells, including tumour cells and healthy stromal cells, have the ability to produce exosomes. Exosomes are released at higher concentrations by cancer cells compared to healthy proliferative cells (Sempere et al., 2017; Wang M. et al., 2019). Exosomes have crucial functions in intracellular and intercellular communication, including the elimination of toxic or damaged cell components and the stimulation of immune cells (Wang M. et al., 2019). Cancer-derived exosomes have been identified as key participants in the interaction between cancer cells and possible new microenvironments coined “pre-metastatic niches” because of their crucial involvement in cell-cell communication. Exosomes have recently piqued the interest of academics, primarily due to their potential as non-invasively available biomarkers, as they circulate in body fluids such as blood, urine, faeces, and saliva (Bahrami et al., 2018).
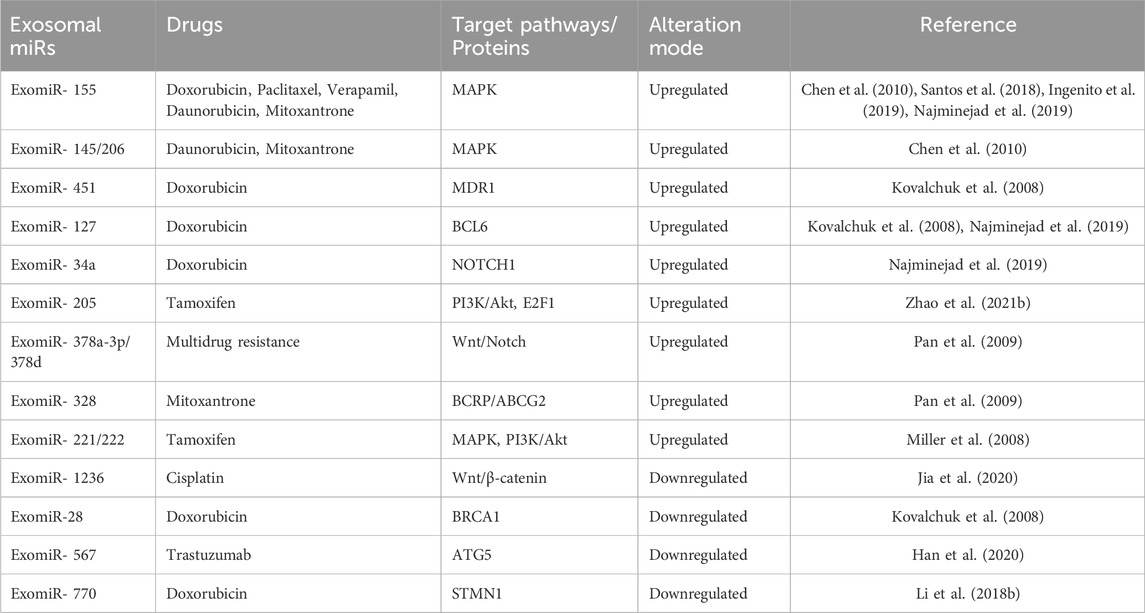
MicroRNAs (miRNAs) are endogenous noncoding RNAs that are short (21–23 nt) and have been demonstrated to regulate gene expression in a variety of physiological activities (O'Brien et al., 2018; Youngman and Claycomb, 2014). Numerous studies in recent years have demonstrated that different microRNAs have a role in direct or indirect interactions between breast cancer cells and tumour microenvironment (TME) components (Zhang et al., 2010; Challagundla et al., 2014). Although some of these studies hypothesized that miRNA had cell-extrinsic effects by regulating secreted factors (Pencheva and Tavazoie, 2013), the identification of miRNAs as a plentiful cargo in exosomes, called exosomal microRNAs (ExomiRs) (Graveel et al., 2015) raised the possibility of direct, miRNA-mediated regulation in recipient cells both close to and far from the donor cell expressing the miRNA(s). miRNAs have long been known to play a role in the diagnosis, onset, progression, prognosis, and response to treatment of breast cancer (Lu et al., 2005; Calin and Croce, 2006). Additionally, miRNAs are circulating in bodily fluids, and their expression in peripheral blood may serve as a biological marker for the prognosis and differential diagnosis of breast cancers (Kurozumi et al., 2017; Bahrami et al., 2018). Furthermore, several cell line studies show the effectiveness of ExomiRs in promoting both drug resistance as well as drug sensitivity. The ExomiRs that are generally upregulated in breast cancer cell lines are seen to be associated with an increased drug resistance to several drugs like doxorubicin, mitoxantrone, and tamoxifen, whereas the ExomiRs that are downregulated are seen to promote sensitivity to drugs like doxorubicin, cisplatin, and trastuzumab.
This review focuses on the need for prospecting ExomiRs as the potential theranostic markers to meet the unmet clinical need to both diagnoses as well as therapeutic treatment of breast cancer in women.
2 Diagnostic potential of exosomal microRNAs in breast cancer
Over recent years, it has been established that cancer patients have more ExomiRs than their healthy counterparts (Sempere et al., 2017; Wang M. et al., 2019). This shows that as opposed to exosomes formed from unaffected normal cells, ExomiRs in cancer patients more frequently reflect exosomes from cancer cells and/or other cell types in the tumour microenvironment (TME). Hence, detection/identification of these ExomiRs could help in better diagnosis or clinical prognosis of disease (Figure 1) (Table 1).
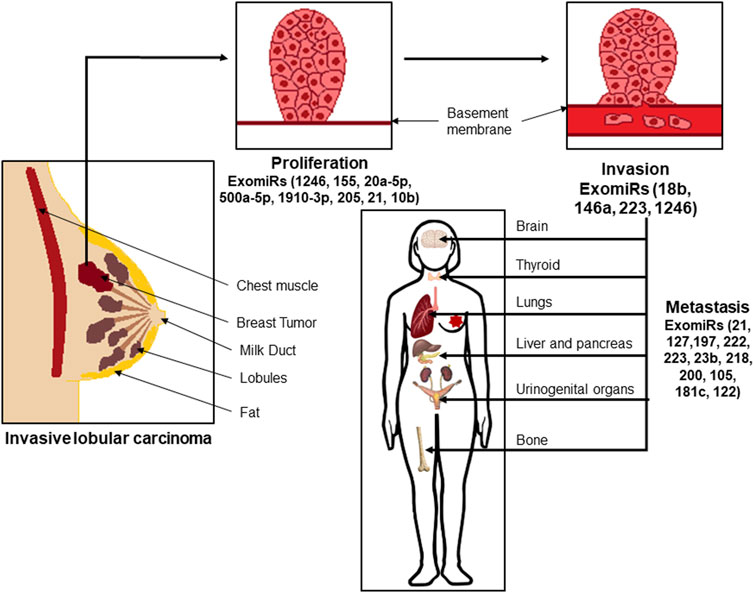
FIGURE 1. Exosomal microRNAs (ExoMiRs) play a critical role in the proliferation, invasion as well as metastasis of breast cancer. Such invasive lobular carcinomas are accompanied by the expression of different ExomiRs modulating the process of proliferation of the cells, followed by breaking the basement epithelial membrane leading to invasion and migration of the cells into the bloodstream, and metastasis to other organs like the brain, thyroid, lungs, etc.
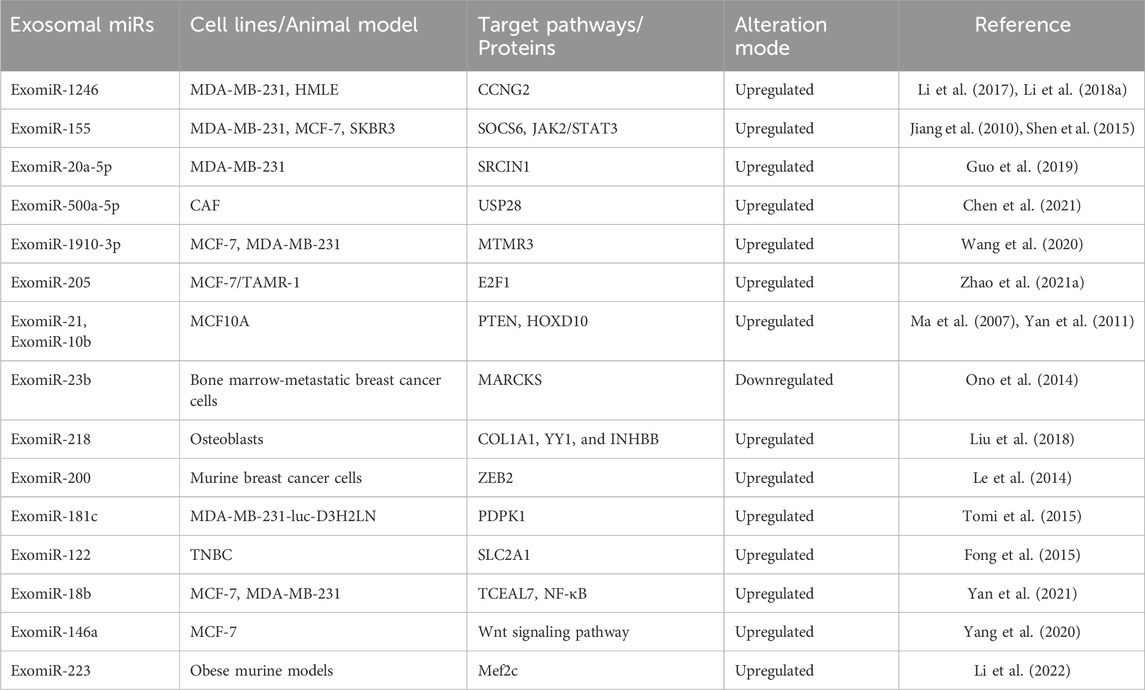
TABLE 1. Relationship of ExomiRs modulated by specific pathways in proliferation, invasion, and metastasis of breast cancer.
2.1 ExomiRs in breast cancer proliferation
Cancer cell growth and tumorigenesis are both affected by increased proliferation and decreased apoptosis. In comparison to microvesicles originating from healthy mammary epithelial cells, a study found that microvesicles from MDA-MB-231 breast cancer cells could provide a transformed-like phenotype to normal fibroblasts and epithelial cells, which is characterized by anchorage-independent proliferation and improved survival (Antonyak et al., 2011). Another study by Melo et al. showed that miRNA biogenesis in exosomes aids in the progression of the tumour (Melo et al., 2014). In a MDA-MB-231 cell line study, ExomiR-1246 was found to induce a tumour-promoting phenotype, including enhanced cell proliferation (Li X. J. et al., 2018). Furthermore, miR-1246 binds to the 3′UTR of Cyclin-G2(CCNG2) thereby directly targeting CCNG2 expression and leading to the downregulation in the expression of the tumour suppressor gene in non-malignant HMLE cells (Li X. J. et al., 2018). Ectopic ExomiR-155 overexpression boosted tumour proliferation in the breast cancer cell lines MDA-MB-231 and MCF-7 (Jiang et al., 2010). Another study involving ExomiR-155 showed that it inhibits SOCS6, activating the JAK2/STAT3 signaling pathway leading to the proliferation and differentiation of SKBR3 cells and MCF-7, which ultimately results in breast cancer cells becoming resistant to tamoxifen (Shen et al., 2015).
Exosomal miRNAs were shown to be much more abundant in metastatic breast cancer cells (MDA-MB-231 and 4T1) than in non-metastatic breast cancer cells or normal breast cells (MCF10A and NMuMG) (He et al., 2018). By inhibiting SRCIN1, a Src protein kinase involved in the control of cell migration, ExomiR-20a-5p could encourage proliferation and migration in Triple-negative breast cancer (TNBC) cells and accelerate osteoclastogenesis in mouse bone marrow-derived macrophages (Guo et al., 2019). A study by Chen et. al., demonstrated that cancer-associated fibroblasts (CAF)-derived exosomes substantially aided tumour proliferation in breast cancer cell lines (Chen et al., 2021). By binding to ubiquitin-specific peptidase 28 (USP28), ExomiR-500a-5p was transported from CAFs to the cancer cells, where it promoted proliferation and metastasis (Chen et al., 2021).
Wang et. al., showed that mammary epithelial cells and breast cancer cells exhibited proliferation and migration when ExomiR-1910-3p, which regulates myotubularin-related protein 3 (MTMR3), was overexpressed in vitro and in vivo, whereas miR-1910-3p silencing dramatically reduced these functions in both types of cells (Wang et al., 2020). ExomiR-205 can target E2F1 in vivo and in vitro to enhance tamoxifen resistance and proliferation in breast cancer (Zhao et al., 2021a). In MCF10A cells (normal breast cell line) treated with cancer exosomes, upregulation of miR-21 and miR-10b was seen (Ma et al., 2007; Yan et al., 2011). These two miRNAs have been linked to the progression of breast cancer. On the other hand, tumour suppressors PTEN and HOXD10, the known targets of miR-21 and miR-10b respectively, were found to be downregulated (Ma et al., 2007; Yan et al., 2011). It is important to understand that various ExomiRs involved in breast cancer population might vary among individuals of different origin as well as different breast cancer subtypes. An extensive research on these molecules is needed to better understand the diagnostic potential of the ExomiRs involved in the process of proliferation of tumour. Targeting these ExomiRs might provide an alternative approach for breast cancer intervention.
2.2 ExomiRs in breast cancer invasion and metastasis
The multistep process of metastasis includes the spread of tumour cells from the main tumour site, the development of a tumour microenvironment, and the colonization of distant organs by tumour cells (Turajlic and Swanton, 2016). Most deaths from breast cancer are the result of metastasis of the tumour, which highlights the significance of identifying novel biomarkers for metastasis for early diagnosis and effective treatment. In this section, we will focus on the ExomiRs involved in the metastasis of breast cancer.
Breast cancer has different subtypes, each having different characteristics with respect to mortality rate, aggressiveness, metastasis, and standard of intervention. Bone is the most preferred location for hormone receptor-positive (HR+) malignancies (Wu et al., 2017), where up to 70% of metastases originate (Chen et al., 2018). Although the methods by which primary breast tumours spread to the bone are still poorly understood, bone metastases have been found to reappear after 10years of remission, showing that metastatic breast cancer cells may remain dormant in the bone for long periods of time (Lim et al., 2011; Buonomo et al., 2017). According to a study by Yang et al., ExomiR-21 functions as exosomal cargo that is transported to prepare the bone metastatic location for breast cancer metastasis (Yang et al., 2020). Experiments revealed that by boosting osteoclast activity and bone lysis both in vitro and in vivo, exosomal miR-21 released by cancer cells serves as a crucial mediator for creating a favorable pre-metastatic environment. By targeting CXCL12, a stromal-cell-derived chemokine ligand, several ExomiRs (ExomiR-127, ExomiR-197, ExomiR-222, and ExomiR-223) contributed to a dormant phenotype in breast cancer bone metastases (Lim et al., 2011). Similarly, by inhibiting MARCKS, ExomiR-23b may promote the dormancy of bone marrow-metastatic breast cancer cells (Ono et al., 2014). ExomiR-218 can also influence the bone pre-metastatic niche by controlling the osteoblasts’ deposition of collagen through its targets COL1A1, YY1, and INHBB (Liu et al., 2018).
Most frequently connected with HR+ and Her2+ breast tumours, the liver is the second most frequent metastatic location after the brain, accounting for 40 to 50 percent of all cases (Zhao et al., 2018). Among all the subtypes of breast cancer, TNBC has the highest propensity to metastasize to the lungs first and then to other distant organs of the body (Van Mechelen et al., 2020). According to an in vivo study, ExomiR-200 contributed to the colonization of circulating murine breast cancer cells and promoted metastasis by transforming non-metastatic cells to metastatic by targeting ZEB2 (Le et al., 2014). An ExomiR-200 variation known as ExomiR-200c has also been linked to accelerated tumour development and subsequent lung metastasis over time (Zhang et al., 2017).
A higher metastatic potential and predictable brain metastasis are characteristics of aggressive breast carcinoma such as TNBC and HER-2 enriched (Wu et al., 2017). Patients with metastatic breast cancer are 10%–30% more likely to develop brain metastases, with breast cancer brain metastasis (BCBM) accounting for 15% of all cases (Ekici et al., 2016; Suh et al., 2020). A 2013 study suggested that nonmetastatic breast cancer cells could take up exosomes from brain metastasis, opening the door for exosomal functions in brain metastasis (Camacho et al., 2013). A study by Zhou et. al., demonstrated that ExomiR-105 plays a significant role in the host’s vascular endothelial barrier destruction during the early stages of premetastatic niche development by specifically targeting cellular tight junctions (Zhou et al., 2014). Migration of breast cancer cells over the blood-brain barrier (BBB) is a critical step in the disease spreading to the brain. A study assessed how ExomiR-181c helps breast cancer cells pass through the BBB. By targeting PDPK1 in brain endothelial cells and disturbing the actin filament localization, ExomiR-181c translocation from breast cancer BC cells facilitates the breakdown of the BBB in vitro (Tomi et al., 2015). Exosomal miR-122 has been demonstrated to improve the Warburg Effect in premetastatic niches, particularly the brain and lungs, in TNBC cells. This study showed that increased levels of ExomiR-122 facilitate metastasis in breast cancer patients by increasing the availability of nutrients in the premetastatic niche (Fong et al., 2015).
Cancer cell invasion is the process where the tumour cells overcome the barriers of the extracellular matrix and spread into surrounding tissues (Krakhmal et al., 2015) followed by the formation of metastatic foci and metastasis (van Zijl et al., 2011). It is generally of two types, collective cell migration and individual cell migration, depending on the number of cancer cells participating in the invasive front (Krakhmal et al., 2015). This mesenchymal mechanism of invasion is a result of epithelial-mesenchymal transition (EMT) as the multicellular groups gain mesenchymal phenotypes and start to divide into single tumour cells by virtue of active dedifferentiation of a malignant epithelial tumour (Friedl et al., 2004). ExomiRs play a significant role in cell invasion and migration ultimately leading to metastasis (Li et al., 2022). ExomiR-18b is seen to induce tumour invasion and metastasis in mouse models as the overexpression of ExomiR-18b suppresses the expression of transcription elongation factor A-like 7 (TCEAL7) (Yan et al., 2021). TCEAL7 plays a key role in the reversing of Epithelial to Mesenchymal Transition (EMT) by suppression of the NF-κB pathway (Rattan et al., 2010). ExomiR-146a overexpression is also seen to regulate tumour invasion and migration by enhancing the Wnt signaling pathway in MCF-7 breast cancer cell lines (Yang et al., 2020). Overexpression of ExomiR-1246 is also seen to influence tumour cell proliferation and invasion in breast cancer cell lines as it suppresses the expression level of the target gene CCNG2 in MDA-MB-231 cells, an epithelial human breast cancer cell line (Li et al., 2017). The overexpression of ExomiR-223 also enhances migration and invasion of breast cancer by targeting the Mef2c gene thereby affecting the production of β-catenin in obese murine models (Li et al., 2022).
3 Therapeutic potential of exosomal microRNAs in breast cancer
miRNAs play critical roles in the progression of tumours; however, therapeutic miRNAs are making a run for FDA approval (Fire et al., 1998; Chakraborty et al., 2017). Several new biotech companies such as Miragen, MiRNA Therapeutics, and so on are trying to develop miRNA therapeutics however, drug delivery and resistance remain the biggest roadblocks in the field. MiRNAs can be packaged in liposomes, nanoparticles, and micelles similarly to conventional systemic drugs, but they have a limited ability to cross the blood-brain barrier (Krakhmal et al., 2015). However, no miRNA drug has yet to enter a phase III clinical trial because many phase II trials were stopped early due to serious adverse events, despite the fact that miRNAs can offer promising clinical applications (Austin, 2013; Chakraborty et al., 2017; Hanna et al., 2019).
Most primary breast cancers initially respond well to miRNA treatment; however, show resistance at a later stage (Pondé et al., 2019). In order to overcome this resistance, ExomiRs and exosomes transporting other molecular cargo have been prospected as second-line treatment for breast cancer due to their significance in tumour drug resistance (Del Re et al., 2019) (Figure 2).
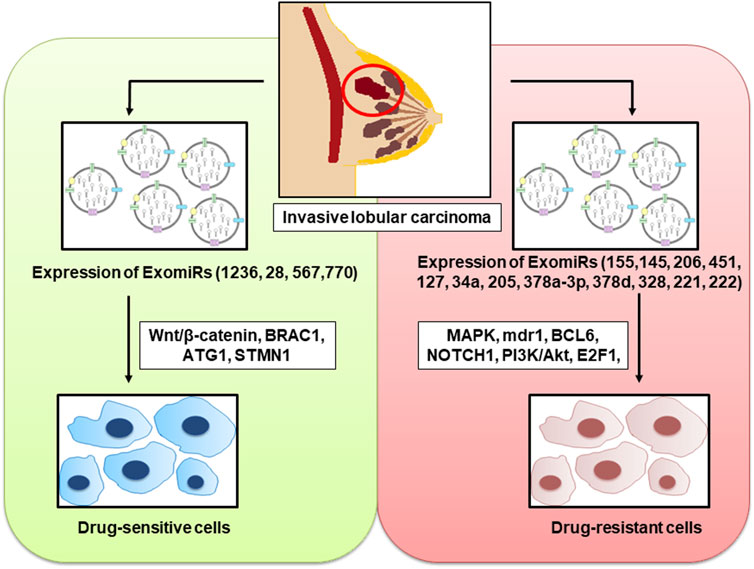
FIGURE 2. ExomiRs play a pivotal role in the incidence of both drug resistance and drug sensitivity in breast cancer leading to the alteration of several cellular metabolic pathways and proteins thereby influencing the progression, invasion, and metastasis of the carcinoma.
3.1 ExomiRs promoting drug resistance
An increase in the concentration of ExomiRs by several tumour cell types not only promotes tumour progression, angiogenesis, and metastasis but also plays a significant role in chemoresistance in all aggressive cancers, including breast cancer. This makes therapeutic treatments for breast cancer much more difficult (Cao et al., 2021). ExomiRs are highly stable molecules that are easily detectable in biological fluids like blood (Gallo et al., 2012), urine (Solé et al., 2019), and saliva (Gallo et al., 2012). Hence, they can act as potential cancer biomarkers useful both diagnostically and prognostically. These ExomiRs are seen to alter the expression of various proteins as they have strong binging affinity to the genes that are responsible for the expression of the protein thereby downregulating the protein expression in the body of the breast cancer patient, enhancing drug-resistance (Santos and Almeida, 2020). In breast cancer, miR-155 induces exosome-mediated chemoresistance to doxorubicin and paclitaxel, while simultaneously acting as an oncogenic signal reprogramming cancer metabolism (Santos et al., 2018; Ingenito et al., 2019). This resistance against cancer therapy is partially attributed to cancer progenitors as these cells arise from epithelial cells that undergo EMT (Santos et al., 2018). It was also found that the increase in chemoresistance was induced by the transfer of ExomiR-155 from resistant cancer cells to sensitive cells (Santos et al., 2018). Moreover, a study by Santos et al. shows the development of chemoresistance in MCF7 cell lines upon transfecting them with exosomal miRNA-155 mimics (Santos et al., 2018). ExomiR-155 is seen to produce doxorubicin resistance by inhibiting the MAPK signaling pathways in MCF7 breast cancer cell lines (Chen et al., 2010; Najminejad et al., 2019). The inhibition of the MAPK signaling pathway by ExomiR-155 also induces resistance to chemotherapeutic drug verapamil in MCF7 cell lines (Chen et al., 2010). A study by Chen et al. also showed that ExomiR-145 and ExomiR-206 interfere with the MAK signaling pathway in the MCF7 breast cancer cell line, leading to cross-resistance to daunorubicin and mitoxantrone (Chen et al., 2010). Overexpression of ExomiR-451 is also seen to induce doxorubicin resistance in MCF7 cell lines by inhibiting the expression of multidrug resistance R1 (mdr1) which in turn leads to pronounced downregulation of the Argonaute 2 protein, a member of the Argonaute protein family having an important role in RNA silencing (Kovalchuk et al., 2008). In the study by Kovalchuk et al., the overexpression of ExomiR-127 and ExomiR-34a was also seen to induce doxorubicin resistance (Najminejad et al., 2019). Western blot analysis shows that ExomiR-127 leads to an increase in the antiapoptotic protein BCL6 while ExomiR-34a increases the expression of the antiapoptotic protein NOTCH1 thereby inducing cell invasion and metastasis (Kovalchuk et al., 2008).
A study by Zhao, et al. suggests that ExomiR-205 induces tamoxifen resistance, thereby promoting proliferation, migration, and invasion and the suppression of apoptosis which were associated with activating the caspase pathway and phosphorylating Akt. The study also shows the E2F Transcription Factor 1 (E2F1) to be a direct target of ExomiR-205, in turn affecting the cellular transcription machinery in human breast cancer cells (Zhao et al., 2021b). In vivo studies suggest that intratumoral injection of ExomiR-205 to mice is correlated not only with a decrease in chemosensitivity but also with larger tumour size (Zhao et al., 2021b) as a result of horizontal transfer of these miRs (Chen et al., 2014). In breast cancer, chemotherapy activates the EZH2/STAT3 axis leading to the production of ExomiR-378a-3p and ExomiR-378d, resulting in chemotherapy-elicited, exosome-induced drug resistance (Yang et al., 2021). This is seen to occur by activating the Wnt and Notch stem cell signaling pathways via the targeting of Dickkopf-3 (a heat shock transcription factor 1 protein) and NUMB (an endocytic adaptor protein) expression in rodents (Pan et al., 2009). The overexpression of ExomiR-328 is seen to negatively regulate the expression of breast cancer resistance protein (BCRP/ABCG2) in human cancer cells leading to mitoxantrone resistance (Pan et al., 2009). It was also seen in a study by Miller et al. that the upregulation of ExomiR-221 and ExomiR-222 in MCF7 breast cancer cell line confers tamoxifen resistance by targeting p27Kip1, which induces caspase 7 and poly (ADP-ribose) polymerase cleavage, leading to its degradation, thereby activating MAPK and Akt signaling pathways favoring cell proliferation and survival (Miller et al., 2008) (Table 2).
3.2 ExomiRs inhibiting drug resistance
Emerging evidence also suggests the possible role of certain ExomiRs in reducing chemoresistance in breast cancer cells. These ExomiRs are downregulated in breast cancer and both in vitro and in vivo evidence suggest their role in increasing the sensitivity of cells to the chemotherapeutic drugs, thereby not only decreasing the tumour size but also affecting the invasion, proliferation, and metastasis influencing the organotropism of breast cancer (Wong et al., 2020). ExomiR-1236 is seen to reduce cisplatin resistance by downregulating SLC9A1 and inactivating the Wnt/β-catenin pathway (Jia et al., 2020). Cisplatin is one of the most common chemotherapy regimens for several cancers including breast cancer, especially in the case of triple-negative breast cancers (Wang Y. et al., 2019). ExomiR-28 is seen to be downregulated in MCH7 breast cancer cell lines, leading to doxorubicin resistance (Kovalchuk et al., 2008). It was seen that upregulating ExomiR-28 leads to a loss of BRCA1 expression, which is associated with an increase in doxorubicin resistance in cell lines (Kovalchuk et al., 2008). Overexpression of ExomiR-567 in HER2-enriched cells was seen to directly target ATG5, thereby reversing the autophagy-dependent resistance to the targeted monoclonal antibody trastuzumab (Han et al., 2020). In vivo studies also suggest that overexpression of ExomiR-770 directly targets STMN1, a stathmin family phosphoprotein involved in intracellular signaling, thereby increasing doxorubicin sensitivity via inducing apoptosis and decreasing the tumour volume and metastasis (Li Y. et al., 2018) and (Table 2).
Taken together, ExomiRs not only act as an inducer of chemoresistance but also can aid as potential targets for endocrine therapies of breast cancer.
4 Conclusion
The strong regulatory effect of exosomal miRNA in the tumour microenvironment of breast cancer is evident and real. We are still in the early phase of research on exosomes in breast cancer, often encountering challenges despite the fact that this field has made considerable progress and certain sophisticated understandings have been attained. The ExomiRs mentioned in this review provides diagnostic as well as therapeutic potential against breast cancer. However, the combinatorial potential of these ExomiRs (theranostic potential) still remain an under-explored area in human beings. The studies that have identified ExomiRs with respect to breast cancer so far are majorly in cell lines/animal models, the very few ones in humans do not explore if they are niche-specific or are common for all populations. According to the statistical data by World Cancer Research Fund in 2020, the highest prevalence of breast cancer was recorded in European countries such as Belgium, Netherlands, Luxembourg, France, to name a few (WCRF, 2020), due to genetic reasons, lifestyle choices, as well as environmental factors. The world at large is known to have wide genetic diversity accompanied by divergent socio-cultural norms and lifestyle choices which are known to play a statutory role towards the creation of a highly heterogenous landscape of mutations associated with the onset, progression as well as clinical prognosis of breast cancer. The heterogeneity in the mutation landscape associated with breast cancer are expected to correspond to niche-specific variations in the ExomiRs whilst being prospected as theranostic markers for breast cancer. This niche-specific identification of ExomiRs will help in provisioning niche-specific theranostic regimens which cater to the needs of a specific community. Prospecting of ExomiRs as theranostic targets might be community-specific due to the extant variation in the genetic base of the populace. Hence, it might be imperative for the clinician researchers at large to take into account the perturbances at micro level which might influence the socio-cultural norms, diet and lifestyle choices of the populace in a niche-specific manner necessitating the need for big data with larger sample size having wide genetic base before prospecting the ExomiRs as theranostic targets.
Author contributions
RG: Conceptualization, Methodology, Writing–original draft, Writing–review and editing. TM: Conceptualization, Methodology, Writing–original draft, Writing–review and editing. RR: Writing–original draft. CP: Supervision, Writing–review and editing. EE: Supervision, Writing–review and editing. RJ: Conceptualization, Methodology, Supervision, Writing–review and editing.
Funding
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
Conflict of interest
Authors CP and EE were employed by Mayo Clinic.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmolb.2024.1330144/full#supplementary-material
References
Antonyak, M. A., Li, B., Boroughs, L. K., Johnson, J. L., Druso, J. E., Bryant, K. L., et al. (2011). Cancer cell-derived microvesicles induce transformation by transferring tissue transglutaminase and fibronectin to recipient cells. Proc. Natl. Acad. Sci. 108 (12), 4852–4857. doi:10.1073/pnas.1017667108
Austin, T. B. M. (2013). First microRNA mimic enters clinic. Nat. Biotechnol. 31 (7), 577. doi:10.1038/nbt0713-577
Bahrami, A., Aledavood, A., Anvari, K., Hassanian, S. M., Maftouh, M., Yaghobzade, A., et al. (2018). The prognostic and therapeutic application of microRNAs in breast cancer: tissue and circulating microRNAs. J. Cell. physiology 233 (2), 774–786. doi:10.1002/jcp.25813
Breast (2020). Cancer incidence and mortality statistics worldwide and by region. Available at: https://gco.iarc.fr/today/data/factsheets/cancers/20-Breast-fact-sheet.pdf.
Buonomo, O. C., Caredda, E., Portarena, I., Vanni, G., Orlandi, A., Bagni, C., et al. (2017). New insights into the metastatic behavior after breast cancer surgery, according to well-established clinicopathological variables and molecular subtypes. PLoS One 12 (9), e0184680. doi:10.1371/journal.pone.0184680
Calin, G. A., and Croce, C. M. (2006). MicroRNA signatures in human cancers. Nat. Rev. cancer 6 (11), 857–866. doi:10.1038/nrc1997
Camacho, L., Guerrero, P., and Marchetti, D. (2013). MicroRNA and protein profiling of brain metastasis competent cell-derived exosomes. PloS one 8 (9), e73790. doi:10.1371/journal.pone.0073790
Cao, J., Zhang, M., Wang, B., Zhang, L., Zhou, F., and Fang, M. (2021). Chemoresistance and metastasis in breast cancer molecular mechanisms and novel clinical strategies. Front. Oncol. 11, 658552. doi:10.3389/fonc.2021.658552
Chakraborty, C., Sharma, A. R., Sharma, G., Doss, C. G. P., and Lee, S. S. (2017). Therapeutic miRNA and siRNA: moving from bench to clinic as next generation medicine. Mol. Therapy-Nucleic Acids 8, 132–143. doi:10.1016/j.omtn.2017.06.005
Challagundla, K. B., Fanini, F., Vannini, I., Wise, P., Murtadha, M., Malinconico, L., et al. (2014). microRNAs in the tumor microenvironment: solving the riddle for a better diagnostics. Expert Rev. Mol. diagnostics 14 (5), 565–574. doi:10.1586/14737159.2014.922879
Chatterji, S., Krzoska, E., Thoroughgood, C. W., Saganty, J., Liu, P., Elsberger, B., et al. (2023). Defining genomic, transcriptomic, proteomic, epigenetic, and phenotypic biomarkers with prognostic capability in male breast cancer: a systematic review. Lancet Oncol. 24 (2), e74–e85. doi:10.1016/S1470-2045(22)00633-7
Cheang, M. C., Martin, M., Nielsen, T. O., Prat, A., Voduc, D., Rodriguez-Lescure, A., et al. (2015). Defining breast cancer intrinsic subtypes by quantitative receptor expression. Oncol. 20 (5), 474–482. doi:10.1634/theoncologist.2014-0372
Chen, B., Sang, Y., Song, X., Zhang, D., Wang, L., Zhao, W., et al. (2021). Exosomal miR-500a-5p derived from cancer-associated fibroblasts promotes breast cancer cell proliferation and metastasis through targeting USP28. Theranostics 11 (8), 3932–3947. doi:10.7150/thno.53412
Chen, G. Q., Zhao, Z. W., Zhou, H. Y., Liu, Y. J., and Yang, H. J. (2010). Systematic analysis of microRNA involved in resistance of the MCF-7 human breast cancer cell to doxorubicin. Med. Oncol. N. Lond. Engl. 27 (2), 406–415. doi:10.1007/s12032-009-9225-9
Chen, W., Hoffmann, A. D., Liu, H., and Liu, X. (2018). Organotropism: new insights into molecular mechanisms of breast cancer metastasis. NPJ Precis. Oncol. 2 (1), 4. doi:10.1038/s41698-018-0047-0
Chen, W. X., Liu, X. M., Lv, M. M., Chen, L., Zhao, J. H., Zhong, S. L., et al. (2014). Exosomes from drug-resistant breast cancer cells transmit chemoresistance by a horizontal transfer of microRNAs. PloS one 9 (4), e95240. doi:10.1371/journal.pone.0095240
Del Re, M., Bertolini, I., Crucitta, S., Fontanelli, L., Rofi, E., De Angelis, C., et al. (2019). Overexpression of TK1 and CDK9 in plasma-derived exosomes is associated with clinical resistance to CDK4/6 inhibitors in metastatic breast cancer patients. Breast Cancer Res. Treat. 178, 57–62. doi:10.1007/s10549-019-05365-y
Desantis, C., Siegel, R., and Jemal, A. (2012). Cancer treatment and survivorship facts and figures 2012-2013. Atlanta, GA: American Cancer Society.
Ekici, K., Temelli, O., Dikilitas, M., Dursun, I. H., Kaplan, N. B., and Kekilli, E. (2016). Survival and prognostic factors in patients with brain metastasis: single center experience. Age 230 (85), 73.
Fire, A., Xu, S., Montgomery, M. K., Kostas, S. A., Driver, S. E., and Mello, C. C. (1998). Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. nature 391 (6669), 806–811. doi:10.1038/35888
Fong, M. Y., Zhou, W., Liu, L., Alontaga, A. Y., Chandra, M., Ashby, J., et al. (2015). Breast-cancer-secreted miR-122 reprograms glucose metabolism in premetastatic niche to promote metastasis. Nat. Cell Biol. 17 (2), 183–194. doi:10.1038/ncb3094
Fox, S., Speirs, V., and Shaaban, A. M. (2022). Male breast cancer: an update. Virchows Arch. 480 (1), 85–93. doi:10.1007/s00428-021-03190-7
Friedl, P., Hegerfeldt, Y., and Tusch, M. (2004). Collective cell migration in morphogenesis and cancer. Int. J. Dev. Biol. 48 (5-6), 441–449. doi:10.1387/ijdb.041821pf
Gallo, A., Tandon, M., Alevizos, I., and Illei, G. G. (2012). The majority of microRNAs detectable in serum and saliva is concentrated in exosomes. PloS one 7 (3), e30679. doi:10.1371/journal.pone.0030679
Graveel, C. R., Calderone, H. M., Westerhuis, J. J., Winn, M. E., and Sempere, L. F. (2015). Critical analysis of the potential for microRNA biomarkers in breast cancer management. Breast cancer targets Ther. 7, 59–79. doi:10.2147/BCTT.S43799
Green, T. M., Alpaugh, M. L., Barsky, S. H., Rappa, G., and Lorico, A. (2015). Breast cancer-derived extracellular vesicles: characterization and contribution to the metastatic phenotype. BioMed Res. Int. 2015, 634865. doi:10.1155/2015/634865
Gucalp, A., Traina, T. A., Eisner, J. R., Parker, J. S., Selitsky, S. R., Park, B. H., et al. (2019). Male breast cancer: a disease distinct from female breast cancer. Breast cancer Res. Treat. 173, 37–48. doi:10.1007/s10549-018-4921-9
Guo, L., Zhu, Y., Li, L., Zhou, S., Yin, G., Yu, G., et al. (2019). Breast cancer cell-derived exosomal miR-20a-5p promotes the proliferation and differentiation of osteoclasts by targeting SRCIN1. Cancer Med. 8 (12), 5687–5701. doi:10.1002/cam4.2454
Han, M., Hu, J., Lu, P., Cao, H., Yu, C., Li, X., et al. (2020). Exosome-transmitted miR-567 reverses trastuzumab resistance by inhibiting ATG5 in breast cancer. Cell death Dis. 11 (1), 43. doi:10.1038/s41419-020-2250-5
Hanna, J., Hossain, G. S., and Kocerha, J. (2019). The potential for microRNA therapeutics and clinical research. Front. Genet. 10, 478. doi:10.3389/fgene.2019.00478
He, Y., Deng, F., Yang, S., Wang, D., Chen, X., Zhong, S., et al. (2018). Exosomal microRNA: a novel biomarker for breast cancer. Biomarkers Med. 12 (2), 177–188. doi:10.2217/bmm-2017-0305
Ingenito, F., Roscigno, G., Affinito, A., Nuzzo, S., Scognamiglio, I., Quintavalle, C., et al. (2019). The role of exo-miRNAs in cancer: a focus on therapeutic and diagnostic applications. Int. J. Mol. Sci. 20 (19), 4687. doi:10.3390/ijms20194687
Jia, Z., Zhu, H., Sun, H., Hua, Y., Zhang, G., Jiang, J., et al. (2020). Adipose mesenchymal stem cell-derived exosomal microRNA-1236 reduces resistance of breast cancer cells to cisplatin by suppressing SLC9A1 and the wnt/β-catenin signaling. Cancer Manag. Res. 12, 8733–8744. doi:10.2147/CMAR.S270200
Jiang, S., Zhang, H. W., Lu, M. H., He, X. H., Li, Y., Gu, H., et al. (2010). MicroRNA-155 functions as an OncomiR in breast cancer by targeting the suppressor of cytokine signaling 1 gene. Cancer Res. 70 (8), 3119–3127. doi:10.1158/0008-5472.CAN-09-4250
Khan, N. A. J., and Tirona, M. (2021). An updated review of epidemiology, risk factors, and management of male breast cancer. Med. Oncol. 38, 39–48. doi:10.1007/s12032-021-01486-x
Kovalchuk, O., Filkowski, J., Meservy, J., Ilnytskyy, Y., Tryndyak, V. P., Chekhun, V. F., et al. (2008). Involvement of microRNA-451 in resistance of the MCF-7 breast cancer cells to chemotherapeutic drug doxorubicin. Mol. cancer Ther. 7 (7), 2152–2159. doi:10.1158/1535-7163.MCT-08-0021
Krakhmal, N. V., Zavyalova, M. V., Denisov, E. V., Vtorushin, S. V., and Perelmuter, V. M. (2015). Cancer invasion: patterns and mechanisms. Acta Naturae 7 (2), 17–28. doi:10.32607/20758251-2015-7-2-17-28
Kurozumi, S., Yamaguchi, Y., Kurosumi, M., Ohira, M., Matsumoto, H., and Horiguchi, J. (2017). Recent trends in microRNA research into breast cancer with particular focus on the associations between microRNAs and intrinsic subtypes. J. Hum. Genet. 62 (1), 15–24. doi:10.1038/jhg.2016.89
Le, M. T., Hamar, P., Guo, C., Basar, E., Perdigão-Henriques, R., Balaj, L., et al. (2014). miR-200–containing extracellular vesicles promote breast cancer cell metastasis. J. Clin. investigation 124 (12), 5109–5128. doi:10.1172/JCI75695
Li, C., Zhou, T., Chen, J., Li, R., Chen, H., Luo, S., et al. (2022). The role of Exosomal miRNAs in cancer. J. Transl. Med. 20 (1), 6–15. doi:10.1186/s12967-021-03215-4
Li, X. J., Ren, Z. J., Tang, J. H., and Yu, Q. (2017). Exosomal MicroRNA MiR-1246 promotes cell proliferation, invasion and drug resistance by targeting CCNG2 in breast cancer. Cell. physiology Biochem. 44 (5), 1741–1748. doi:10.1159/000485780
Li, X. J., Ren, Z. J., Tang, J. H., and Yu, Q. (2018a). Exosomal MicroRNA MiR-1246 promotes cell proliferation, invasion and drug resistance by targeting CCNG2 in breast cancer. Cell. Physiology Biochem. 44 (5), 1741–1748. doi:10.1159/000485780
Li, Y., Liang, Y., Sang, Y., Song, X., Zhang, H., Liu, Y., et al. (2018b). MiR-770 suppresses the chemo-resistance and metastasis of triple negative breast cancer via direct targeting of STMN1. Cell death Dis. 9 (1), 14. doi:10.1038/s41419-017-0030-7
Lim, P. K., Bliss, S. A., Patel, S. A., Taborga, M., Dave, M. A., Gregory, L. A., et al. (2011). Gap junction–mediated import of microRNA from bone marrow stromal cells can elicit cell cycle quiescence in breast cancer cells. Cancer Res. 71 (5), 1550–1560. doi:10.1158/0008-5472.CAN-10-2372
Liu, X., Cao, M., Palomares, M., Wu, X., Li, A., Yan, W., et al. (2018). Metastatic breast cancer cells overexpress and secrete miR-218 to regulate type I collagen deposition by osteoblasts. Breast Cancer Res. 20 (1), 127–212. doi:10.1186/s13058-018-1059-y
Lowry, M. C., Gallagher, W. M., and O'Driscoll, L. (2015). The role of exosomes in breast cancer. Clin. Chem. 61 (12), 1457–1465. doi:10.1373/clinchem.2015.240028
Lu, J., Getz, G., Miska, E. A., Alvarez-Saavedra, E., Lamb, J., Peck, D., et al. (2005). MicroRNA expression profiles classify human cancers. nature 435 (7043), 834–838. doi:10.1038/nature03702
Ma, L., Teruya-Feldstein, J., and Weinberg, R. A. (2007). Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature 449 (7163), 682–688. doi:10.1038/nature06174
Melo, S. A., Sugimoto, H., O’Connell, J. T., Kato, N., Villanueva, A., Vidal, A., et al. (2014). Cancer exosomes perform cell-independent microRNA biogenesis and promote tumorigenesis. Cancer Cell 26 (5), 707–721. doi:10.1016/j.ccell.2014.09.005
Miller, T. E., Ghoshal, K., Ramaswamy, B., Roy, S., Datta, J., Shapiro, C. L., et al. (2008). MicroRNA-221/222 confers tamoxifen resistance in breast cancer by targeting p27Kip1. J. Biol. Chem. 283 (44), 29897–29903. doi:10.1074/jbc.M804612200
Nagini, S. (2017). Breast cancer: current molecular therapeutic targets and new players. Anti-Cancer Agents Med. Chem. Former.17 (2), 152–163. doi:10.2174/1871520616666160502122724
Najminejad, H., Kalantar, S. M., Abdollahpour-Alitappeh, M., Karimi, M. H., Seifalian, A. M., Gholipourmalekabadi, M., et al. (2019). Emerging roles of exosomal miRNAs in breast cancer drug resistance. IUBMB life 71 (11), 1672–1684. doi:10.1002/iub.2116
O'Brien, J., Hayder, H., Zayed, Y., and Peng, C. (2018). Overview of microRNA biogenesis, mechanisms of actions, and circulation. Front. Endocrinol. 9, 402. doi:10.3389/fendo.2018.00402
Ono, M., Kosaka, N., Tominaga, N., Yoshioka, Y., Takeshita, F., Takahashi, R. U., et al. (2014). Exosomes from bone marrow mesenchymal stem cells contain a microRNA that promotes dormancy in metastatic breast cancer cells. Sci. Signal. 7 (332), ra63. doi:10.1126/scisignal.2005231
Ottini, L., Palli, D., Rizzo, S., Federico, M., Bazan, V., and Russo, A. (2010). Male breast cancer. Crit. Rev. oncology/hematology 73 (2), 141–155. doi:10.1016/j.critrevonc.2009.04.003
Pan, Y. Z., Morris, M. E., and Yu, A. M. (2009). MicroRNA-328 negatively regulates the expression of breast cancer resistance protein (BCRP/ABCG2) in human cancer cells. Mol. Pharmacol. 75 (6), 1374–1379. doi:10.1124/mol.108.054163
Pencheva, N., and Tavazoie, S. F. (2013). Control of metastatic progression by microRNA regulatory networks. Nat. Cell Biol. 15 (6), 546–554. doi:10.1038/ncb2769
Pondé, N., Aftimos, P., and Piccart, M. (2019). Antibody-drug conjugates in breast cancer: a comprehensive review. Curr. Treat. Options Oncol. 20, 37–22. doi:10.1007/s11864-019-0633-6
Rattan, R., Narita, K., Chien, J., Maguire, J. L., Shridhar, R., Giri, S., et al. (2010). TCEAL7, a putative tumor suppressor gene, negatively regulates NF-kappaB pathway. Oncogene 29 (9), 1362–1373. doi:10.1038/onc.2009.431
Santos, J. C., Lima, N. D. S., Sarian, L. O., Matheu, A., Ribeiro, M. L., and Derchain, S. F. M. (2018). Exosome-mediated breast cancer chemoresistance via miR-155 transfer. Sci. Rep. 8 (1), 829. doi:10.1038/s41598-018-19339-5
Santos, P., and Almeida, F. (2020). Role of exosomal miRNAs and the tumor microenvironment in drug resistance. Cells 9 (6), 1450. doi:10.3390/cells9061450
Sempere, L. F., Keto, J., and Fabbri, M. (2017). Exosomal microRNAs in breast cancer towards diagnostic and therapeutic applications. Cancers 9 (7), 71. doi:10.3390/cancers9070071
Shen, R., Wang, Y., Wang, C. X., Yin, M., Liu, H. L., Chen, J. P., et al. (2015). MiRNA-155 mediates TAM resistance by modulating SOCS6-STAT3 signalling pathway in breast cancer. Am. J. Transl. Res. 7 (10), 2115–2126.
Solé, C., Moliné, T., Vidal, M., Ordi-Ros, J., and Cortés-Hernández, J. (2019). An exosomal urinary miRNA signature for early diagnosis of renal fibrosis in lupus nephritis. Cells 8 (8), 773. doi:10.3390/cells8080773
Suh, J. H., Kotecha, R., Chao, S. T., Ahluwalia, M. S., Sahgal, A., and Chang, E. L. (2020). Current approaches to the management of brain metastases. Nat. Rev. Clin. Oncol. 17 (5), 279–299. doi:10.1038/s41571-019-0320-3
Tominaga, N., Kosaka, N., Ono, M., Katsuda, T., Yoshioka, Y., Tamura, K., et al. (2015). Brain metastatic cancer cells release microRNA-181c-containing extracellular vesicles capable of destructing blood–brain barrier. Nat. Commun. 6 (1), 6716. doi:10.1038/ncomms7716
Turajlic, S., and Swanton, C. (2016). Metastasis as an evolutionary process. Science 352 (6282), 169–175. doi:10.1126/science.aaf2784
Van Mechelen, M., Van Herck, A., Punie, K., Nevelsteen, I., Smeets, A., Neven, P., et al. (2020). Behavior of metastatic breast cancer according to subtype. Breast Cancer Res. Treat. 181, 115–125. doi:10.1007/s10549-020-05597-3
van Zijl, F., Krupitza, G., and Mikulits, W. (2011). Initial steps of metastasis: cell invasion and endothelial transmigration. Mutat. Research/Reviews Mutat. Res. 728 (1-2), 23–34. doi:10.1016/j.mrrev.2011.05.002
Wang, B., Mao, J. H., Wang, B. Y., Wang, L. X., Wen, H. Y., Xu, L. J., et al. (2020). Exosomal miR-1910-3p promotes proliferation, metastasis, and autophagy of breast cancer cells by targeting MTMR3 and activating the NF-κB signaling pathway. Cancer Lett. 489, 87–99. doi:10.1016/j.canlet.2020.05.038
Wang, M., Yu, F., Ding, H., Wang, Y., Li, P., and Wang, K. (2019a). Emerging function and clinical values of exosomal microRNAs in cancer. Mol. therapy-Nucleic acids 16, 791–804. doi:10.1016/j.omtn.2019.04.027
Wang, Y., Li, S., Zhu, L., Zou, J., Jiang, X., Chen, M., et al. (2019b). Letrozole improves the sensitivity of breast cancer cells overexpressing aromatase to cisplatin via down-regulation of FEN1. Clin. Transl. Oncol. 21 (8), 1026–1033. doi:10.1007/s12094-018-02019-1
WCRF (2020). Breast cancer statistics. Available at: https://www.wcrf.org/cancer-trends/breast-cancer-statistics/.
WHO (2024). Breast cancer. Available at: https://www.who.int/news-room/fact-sheets/detail/breast-cancer?gclid =CjwKCAiA4smsBhAEEiwAO6DEjRsKfkEfjT5mXfUXzgym2hYBSnNNmsIonfh_zUtx9FVmnrPyuRTNsRoCW4EQAvD_BwE.
Wong, G. L., Abu Jalboush, S., and Lo, H. W. (2020). Exosomal MicroRNAs and organotropism in breast cancer metastasis. Cancers 12 (7), 1827. doi:10.3390/cancers12071827
Wu, Q., Li, J., Zhu, S., Wu, J., Chen, C., Liu, Q., et al. (2017). Breast cancer subtypes predict the preferential site of distant metastases: a SEER based study. Oncotarget 8 (17), 27990–27996. doi:10.18632/oncotarget.15856
Yan, L. X., Wu, Q. N., Zhang, Y., Li, Y. Y., Liao, D. Z., Hou, J. H., et al. (2011). Knockdown of miR-21 in human breast cancer cell lines inhibits proliferation, in vitro migration and in vivotumor growth. Breast cancer Res. 13 (1), R2–R14. doi:10.1186/bcr2803
Yan, Z., Sheng, Z., Zheng, Y., Feng, R., Xiao, Q., Shi, L., et al. (2021). Cancer-associated fibroblast-derived exosomal miR-18b promotes breast cancer invasion and metastasis by regulating TCEAL7. Cell Death Dis. 12 (12), 1120. doi:10.1038/s41419-021-04409-w
Yang, Q., Zhao, S., Shi, Z., Cao, L., Liu, J., Pan, T., et al. (2021). Chemotherapy-elicited exosomal miR-378a-3p and miR-378d promote breast cancer stemness and chemoresistance via the activation of EZH2/STAT3 signaling. J. Exp. Clin. cancer Res. CR 40 (1), 120. doi:10.1186/s13046-021-01901-1
Yang, S. S., Ma, S., Dou, H., Liu, F., Zhang, S. Y., Jiang, C., et al. (2020). Breast cancer-derived exosomes regulate cell invasion and metastasis in breast cancer via miR-146a to activate cancer associated fibroblasts in tumor microenvironment. Exp. Cell Res. 391 (2), 111983. doi:10.1016/j.yexcr.2020.111983
Yeo, B., Turner, N. C., and Jones, A. (2014). An update on the medical management of breast cancer. Bmj 348, g3608. doi:10.1136/bmj.g3608
Youngman, E. M., and Claycomb, J. M. (2014). From early lessons to new frontiers: the worm as a treasure trove of small RNA biology. Front. Genet. 5, 416. doi:10.3389/fgene.2014.00416
Zhang, G., Zhang, W., Li, B., Stringer-Reasor, E., Chu, C., Sun, L., et al. (2017). MicroRNA-200c and microRNA-141 are regulated by a FOXP3-KAT2B axis and associated with tumor metastasis in breast cancer. Breast Cancer Res. 19 (1), 73–13. doi:10.1186/s13058-017-0858-x
Zhang, H., Li, Y., and Lai, M. (2010). The microRNA network and tumor metastasis. Oncogene 29 (7), 937–948. doi:10.1038/onc.2009.406
Zhao, H. Y., Gong, Y., Ye, F. G., Ling, H., and Hu, X. (2018). Incidence and prognostic factors of patients with synchronous liver metastases upon initial diagnosis of breast cancer: a population-based study. Cancer Manag. Res. 10, 5937–5950. doi:10.2147/CMAR.S178395
Zhao, Y., Jin, L. J., and Zhang, X. Y. (2021a). Exosomal miRNA-205 promotes breast cancer chemoresistance and tumorigenesis through E2F1. Aging (Albany NY) 13 (14), 18498–18514. doi:10.18632/aging.203298
Zhao, Y., Jin, L. J., and Zhang, X. Y. (2021b). Exosomal miRNA-205 promotes breast cancer chemoresistance and tumorigenesis through E2F1. Aging 13 (14), 18498–18514. doi:10.18632/aging.203298
Keywords: breast cancer, exosomal miRNAs, diagnostic markers, drug resistance, drug sensitivity
Citation: Gulati R, Mitra T, Rajiv R, Rajan EJE, Pierret C, Enninga EAL and Janardhanan R (2024) Exosomal microRNAs in breast cancer: towards theranostic applications. Front. Mol. Biosci. 11:1330144. doi: 10.3389/fmolb.2024.1330144
Received: 30 October 2023; Accepted: 29 January 2024;
Published: 22 February 2024.
Edited by:
Dwijendra K. Gupta, Department of Biochemistry, Allahabad University, IndiaReviewed by:
Jianqiang Xu, Dalian University of Technology, ChinaBeatrice Aramini, University of Bologna, Italy
Copyright © 2024 Gulati, Mitra, Rajiv, Rajan, Pierret, Enninga and Janardhanan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rajiv Janardhanan, rajivj@srmist.edu.in
 Richa Gulati
Richa Gulati Tridip Mitra
Tridip Mitra Rohan Rajiv
Rohan Rajiv Emilda Judith Ezhil Rajan1
Emilda Judith Ezhil Rajan1  Chris Pierret
Chris Pierret Elizabeth Ann L. Enninga
Elizabeth Ann L. Enninga Rajiv Janardhanan
Rajiv Janardhanan