A risk signature based on endoplasmic reticulum stress-associated genes predicts prognosis and immunity in pancreatic cancer
- 1The Second Clinical Medical School, Lanzhou University, Lanzhou, China
- 2Department of General Surgery, Lanzhou University Second Hospital, Lanzhou, China
- 3The Second Affiliated Hospital of Kunming Medical University, Kunming, China
- 4Wuhan Blood Center, Wuhan, China
- 5Department of General Surgery, The First Hospital of Lanzhou University, Lanzhou, China
- 6Department of Gastrointestinal Surgery, The Third People’s Hospital of Hubei Province, Wuhan, China
Introduction: The involvement of endoplasmic reticulum (ER) stress in cancer biology is increasingly recognized, yet its role in pancreatic cancer (PC) remains unclear. This study aims to elucidate the impact of ER stress on prognosis and biological characteristics in PC patients.
Methods: A bioinformatic analysis was conducted using RNA-seq data and clinicopathological information from PC patients in the TCGA and ICGC databases. The ER stress-associated gene sets were extracted from MSigDB. ER stress-associated genes closely linked with overall survival (OS) of PC patients were identified via log-rank test and univariate Cox analysis, and further narrowed by LASSO method. A risk signature associated with ER stress was formulated using multivariate Cox regression and assessed through Kaplan-Meier curves, receiver operating characteristic (ROC) analyses, and Harrell’s concordance index. External validation was performed with the ICGC cohort. The single-sample gene-set enrichment analysis (ssGSEA) algorithm appraised the immune cell infiltration landscape.
Results: Worse OS in PC patients with high-risk signature score was observed. Multivariate analysis underscored our ER stress-associated signature as a valuable and independent predictor of prognosis. Importantly, these results based on TCGA were further validated in ICGC dataset. In addition, our risk signature was closely associated with homeostasis, protein secretion, and immune regulation in PC patients. In particular, PC microenvironment in the high-risk cluster exhibited a more immunosuppressive status. At last, we established a nomogram model by incorporating the risk signature and clinicopathological parameters, which behaves better in predicting prognosis of PC patients.
Discussion: This comprehensive molecular analysis presents a new predictive model for the prognosis of PC patients, highlighting ER stress as a potential therapeutic target. Besides, the findings indicate that ER stress can have effect modulating PC immune responses.
1 Introduction
Pancreatic cancer (PC) is a relatively frequent malignant disease in digestive system, which mostly occurs as pancreatic adenocarcinoma. According to data from the GLOBOCAN, there exists an estimated 459,000 newly diagnosed cases all over the world with an upward trend (Globocan, 2020). Adding to the woes, the mortality rate of PC is almost equal to the morbidity rate, putting it the seventh cause in terms of cancer-associated mortality (Cai et al., 2021). PC has been recognized as a silent disease that hardly presents any clinical manifestations at the early stage. Most PC patients are at an advanced or metastatic stage and lost the opportunity of surgical treatment, when firstly diagnosed (Mizrahi et al., 2020). The advent of intraoperative revascularization and chemotherapy have led to some success in PC treatment (Conroy et al., 2018; Neoptolemos et al., 2018; Sohal et al., 2020). However, the sobering reality is that the overall 5-year survival rate of PC patients is still below 10% (Zeng et al., 2018; Siegel et al., 2020). Worse still, the clinical outcome of PC patients differs largely from a few months to several years. Traditional clinical indicators such as the AJCC stage have poor performance in predicting the survival time for PC, and it is hard for doctors to establish effective therapeutic strategies for PC patients. Thus, it is imperative and urgent to develop novel tools of predicting prognosis so as to provide effective and individualized therapies for PC patients.
Endoplasmic reticulum (ER) is the largest organelle characterized by a high dynamic sac or tube membrane structure that emerges from the nucleus (Schwarz and Blower, 2016). ER plays a vital role in proteostasis due to the exquisite activities of ER-assisted folding (Wiseman and Balch, 2005), ER-associated degradation (Allison, 2021), and COPⅡ export pathway (Zanetti et al., 2011). Abnormal accumulation of misfolded or unfolded protein and alteration of Ca2+ concentration will provoke ER stress in eukaryotic cells, subsequently triggering unfolded protein reaction (UPR) that is a homeostatic mechanism mainly mediated by three transmembrane proteins deemed as sensors of ER stress: protein kinase RNA-like ER kinase (PERK), inositol-required enzyme 1 (IRE1α) and activating transcription factor 6 (ATF6) (Hetz et al., 2020). A variety of perturbations in the tumour microenvironment (TME), such as hypoxia, deprivation, acidosis, and accumulation of reactive oxygen species (ROS), which is aggravated by oncogenic events in cancer cells, have been confirmed to drive persistent ER stress. In turn, signal transduction through ER sensors can not only induce UPR but also modulate UPR-independent transcriptional and metabolic pathways, resultantly contributing to tumour initiation, progression, immune evasion, and chemoradiotherapy resistance (Chen and Cubillos-Ruiz, 2021). Prior research has elucidated that ER stress exhibits a significant correlation with the survival outcomes in patients with osteosarcoma (Shi et al., 2022), colon cancer (Liu et al., 2018), renal carcinoma (Zhang et al., 2022), glioma (Zhang et al., 2021). In particular, recent studies show that ER stress can promote stemness and chemoresistance of PC by interfering fatty acid metabolism of tumour cells (Tadros et al., 2017; Dauer et al., 2019). Additionally, hypoxia-induced ER stress has been demonstrated to facilitate PC metastasis by inducing release of vascular endothelial growth factor A (Kong et al., 2015).
A body of work has indicated that ER stress can have effect modulating tumoural immune responses. The study by Pommier et al. unveils that ER stress can induce PC cell dormancy and downregulate the expression of major histocompatibility complex (MHC) class I on PC cells to escape from immune killing, consequently leading to tumour distant metastasis (Pommier et al., 2018). In a subsequent investigation on melanoma, it was discovered that the MHC class I polypeptide-related sequence A/B (MICA/B) - the cognate ligands of major NK activating receptor NK group 2 member D (NKG2D), is negatively regulated by ER stress-associated IRE1/XBP1 pathway in human melanoma cell lines (Obiedat et al., 2019). Beyond modulating the expression of immune-related molecules in cancer cells, the tumour cell-intrinsic ER stress can also reprogram the function of immune cells within the surrounding milieu, which facilitates immune evasion. The findings of Song and colleagues discerned that cell-free ascites from ovarian cancer patients instigate ER stress and triggers IRE1α/XBP1 signaling in activated T cells from healthy women; tumour-bearing mice model further showed that the IRE1α/XBP1 signaling in T cells impairs its mitochondrial respiration by regulating glutamine transporter, ultimately attenuating their anti-neoplastic efficacy (Song et al., 2018). This suggested that ER-stressed cancer cells can release additional factors to remodel the tumour milieu. Another compelling evidence (Yuan et al., 2022) showed that ER-stressed human head and neck squamous cell carcinoma cell line HN4 evades immune surveillance by releasing exosome PD-L1 and upregulating the expression of PD-L1 in macrophages to skew macrophages towards a pro-tumoural phenotype. Cubillos-Ruiz et al. revealed that persistent activation of XBP1 in cancer-associated dendritic cells hindered its capacity to instigate anti-tumour T cells, subsequently catalyzing the progression of ovarian malignancies. (Cubillos-Ruiz et al., 2015). Of note, severe ER stress tip the tumour cells towards releasing damage-associated molecular patterns (DAMPs), triggering immunogenic cell death (ICD) (Rufo et al., 2017). A most recent study revealed that B16 melanoma cells release externalized calreticulin (ExoCRT) during severe ER stress, which is subsequently recognized by the NKp46 receptor on NK cell membrane. And the binding of NKp46–CRT curtails B16 melanoma growth and lung metastasis through amplifying NK killing (Sen Santara et al., 2023). Such revelations underscore that ER stress wields multifaceted impacts on tumours, contingent on its intensity and duration in a context-dependent manner. To holistically understand the intricacies of ER stress in PC, we endeavored to devise a prognostic prediction model for PC patients, underpinned by ER stress-related genes, through an exhaustive exploration of large-scale genomic databases.
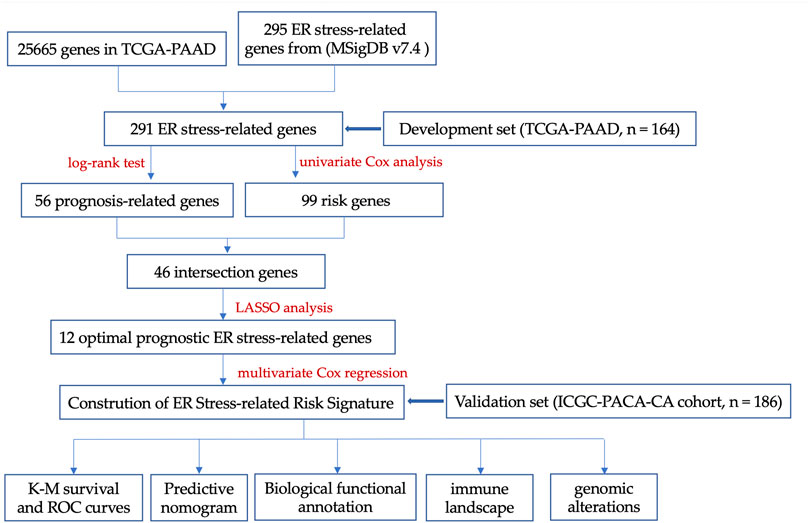
In current work, we first identified 46 ER stress-associated genes closely linked with OS of 164 PC patients from TCGA by log-rank test and univariate Cox regression, and further narrowed 12 optimal prognostic genes (SERP1, BCL2L1, RNF139, TSPYL2, ERO1A, BFAR, MAP3K5, ARFGAP1, ATP6V0D1, NCCRP1, PLA2G4B, USP 19) to build a risk signature via LASSO method. Next, these 164 PC patients were classified as two groups based on the risk signature score, and better OS in the individuals with low-risk score was observed by the Kaplan-Meier (K-M) survival analysis method. Meanwhile, the receiver operating characteristic (ROC) curves and multivariate hazard regression demonstrated that our ER stress-associated risk signature was a robust and independent prognostic biomarker. Furthermore, we established a nomogram model by integrating our risk signature and several clinicopathological parameters, which shows promise in predicting prognosis of PC patients. In addition, the risk signature based on ER stress-associated genes was tightly correlated with homeostasis, protein secretion, and immune regulation in PC. Particularly, PC microenvironment in high-risk group exhibited a more immunosuppressive status. In summary, this study suggested that ER stress is of much prognostic significance and may serve as promising therapeutic targets in PC.
2 Materials and methods
2.1 Datasets and samples
The RNA sequencing metrics of PC samples and normal pancreatic tissues and key clinicopathological characteristics were acquired from TCGA, which is considered as a training cohort. Similarly, we procured the transcriptome profile and clinical data from PACA-CA of the ICGC data portal as an external test cohort. All patients enrolled in this study met the following criteria: i availability of matched gene expression data; ⅱ complete clinical information; ⅲ survival time greater than 30 days. We opted to exclude individuals who, post neoadjuvant therapy, succumbed to extraneous factors such as non-cancerous ailments, other malignancies, or surgical complications. Donors’ transcriptome matrix from Genotype-Tissue Expression Project (GTEx) was used for comparing the expression landscape between PC and normal samples. As for the transcriptome profile, probe IDs were mapped to ENSEMBL IDs and gene symbols using the Annoprobe package (Xiang and Zeng, 2019) in R, and genes were filtered based on their expression in more than half of the samples. The counts were just used in the differentiated analysis and were log2-transformed in other cases. The genes involved in ER stress were extracted from MSigDB v7.4 in the two GO pathways (Huang et al., 2021). A list containing Pan-cancer Immune Metagenes of 28 immune cells was obtained from a widely acknowledged research (Charoentong et al., 2017) to reveal the PC immune microenvironment landscape.
2.2 Identification candidate genes with prognostic potential
For determining the ER stress-related genes that own prognostic significance, the univariate Cox regression and log-rank tests were carried out (Terry and Patricia, 2001; Therneau, 2021) based on the TCGA-PAAD dataset. A p-value threshold of 0.05 was instituted as the criterion for statistical significance. Then the candidate genes associated with PC patients’ survival were then selected based on the p-values of the above two test results and included in the following analyses.
2.3 Establishment and confirmation of the ER stress-associated risk signature
To assess the link of ER stress with OS of PC patients, we tried to build a risk signature according to the ER stress-associated genes. LASSO penalty algorithm avoiding the overfitting concern (Friedman et al., 2010) was employed to further filter prognostic genes associated with ER stress. In the LASSO regression, the optimal tuning parameter was identified by harnessing the minimal lamda value and the random seed is set to 15091102. Subsequently, multivariate Cox analysis was conducted to yield the ER stress-associated risk signature as previously reported (Song et al., 2022). Then, we scored each PC patient based on the risk signature and calculated the optimal cutoff point of the risk scores. PC patients were grouped low- and high-risk clusters. The OS time of the two clusters was compared by K-M analysis. In addition, the ROC curve was adopted for assessing the prognostic accuracy of our risk signature.
2.4 Development and assessment of a predictive nomogram
To better predict the prognosis of PC patients, we established a nomogram by integrating the ER stress-associated signature and several key clinicopathological parameters which have been reported elsewhere as risk factors in PC (Mizrahi et al., 2020; Cai et al., 2021).
2.5 Functional enrichment analyses
Differentially expressed genes (DEGs) were initially delineated utilizing the DESeq2 algorithm as (Love et al., 2014), with a criterion of |LogFC| ≥1 coupled with an adjusted p-value of < 0.05 to signify statistical relevance. Following this, these identified DEGs underwent comprehensive KEGG enrichment and GO analytic evaluations, applying an adjusted p-value benchmark of < 0.05 to ascertain statistical noteworthiness. Furthermore, the paramount enriched KEGG pathways and GO terms were delineated using the GOplot visualization tool.
2.6 Evaluation of immune cell infiltration landscape
Utilizing the ssGSEA algorithm, we quantified the abundance of TME immune cells based on gene sets delineated by Charoentong et al. (2017). In detail, 782 genes were regarded as the markers for 28 kinds of immune cells to detect the abundance of each kind of immune cell in PC samples. Additionally, the stromal and immune scores in the two different clusters were calculated via the estimate function (Kosuke et al., 2016), estimating tumour purity. Moreover, using the complot (Wei and Simko, 2021), we analyzed the correlation between the abundance of these 28 kinds of immune cells and risk scores, with correlation coefficients derived via the Pearson technique.
2.7 Relationship between the risk signature and genomic alterations
In consideration of the vital role of tumour somatic mutations in response to the TME and therapy-induced associated stresses that occur during evolution, we also investigate the association of risk signature with the mutation landscape. Somatic mutation data of PC patients in mutation annotation format, which has been processed under mutect software, was acquired at Genomic Data Commons (GDC) Data Portal. A genomic analysis of PC patients was conducted using the maftools. Tumour mutational burden was further estimated using tmb function to analyze the TMB in different groups.
2.8 Prediction of chemotherapy sensitivity for PC patients
To further elucidate the nexus between the ER stress-associated risk signature and therapeutic efficacy, we harnessed the Drug Sensitivity in Cancer (GDSC) database, a comprehensive resource boasting transcriptomic profiles of 805 cell lines and accompanying IC50 values for a staggering 198 bioactive compounds. Using oncoPredict R package to align PC transcriptome data in TCGA, we calculated the predicted IC50 value for each PC patient for several chemotherapeutic drugs commonly used in PC treatment. Subsequent comparative analysis between high and low-risk cohorts was facilitated using the Wilcoxon test.
2.9 Statistical analysis
The continuous data in two clusters were compared by conducting Wilcoxon test. As mentioned above, combined application of log-rank test and univariate Cox analysis were to screen genes linked with the PC patients’ survival. The LASSO penalty algorithm further narrowed prognosis genes associated with ER stress. Independent prognostic factors were further assessed by multivariate analysis, supplemented with Pearson’s correlation to elucidate the linear relationship between risk scores and immune cell abundance.
3 Results
3.1 Identification of ER stress-associated genes
The key design and procedures of this research were depicted in Figure 1. We finally included 164 PC patients and acquired 295 unique ER stress-associated genes (Supplementary Table S1). Univariate Cox analysis identified 99 risk genes affecting survival (Supplementary Table S2), while log-rank test determined 56 prognostic genes (Supplementary Table S3) within these 295 genes. Furthermore, these 46 intersection genes were considered to have significant relevance to the OS of PC patients and were introduced to LASSO analysis to screen ultimate genes with the best predictive performance. Finally, 12 optimal prognostic ER stress-associated genes with non-zero regression coefficients were determined to construct the risk signature (Figures 2A, B), namely stress-associated endoplasmic reticulum protein 1 (SERP1), B-cell lymphoma 2 like 1 (BCL2L1), ring finger protein 139 (RNF139), testis-specific protein Y-like 2 (TSPYL2), endoplasmic reticulum oxidoreductase 1-α (ERO1A), bifunctional apoptosis regulator (BFAR), Mitogen-Activated Protein 3 Kinase 5 (MAP3K5), ADP-ribosylation factor GTPase activating protein 1 (ARFGAP1), vacuolar proton ATPase subunit d 1 (ATP6V0D1), non-specific cytotoxic cell receptor protein 1 (NCCRP1), phospholipase A2, group IVB (PLA2G4B), ubiquitin specific peptidase 19 (USP 19). The K-M survival plot of every gene for PC patients was shown as Supplementary Figure S1, where the group is based on the best cut-off point for each gene. The mRNA profiles of these 12 genes were also compared between PC tissues and normal pancreatic tissues, and we found that eight of them (BCL2L1, ATP6V0D1, ERO1A, RNF139, BFAR, USP19, ARFGAP1, MAP3K5) were significantly upregulated, three (PLA2G4B, TSPYL2, NCCRP1) were downregulated, and there was no difference in SERP1 expression (Figure 2C).
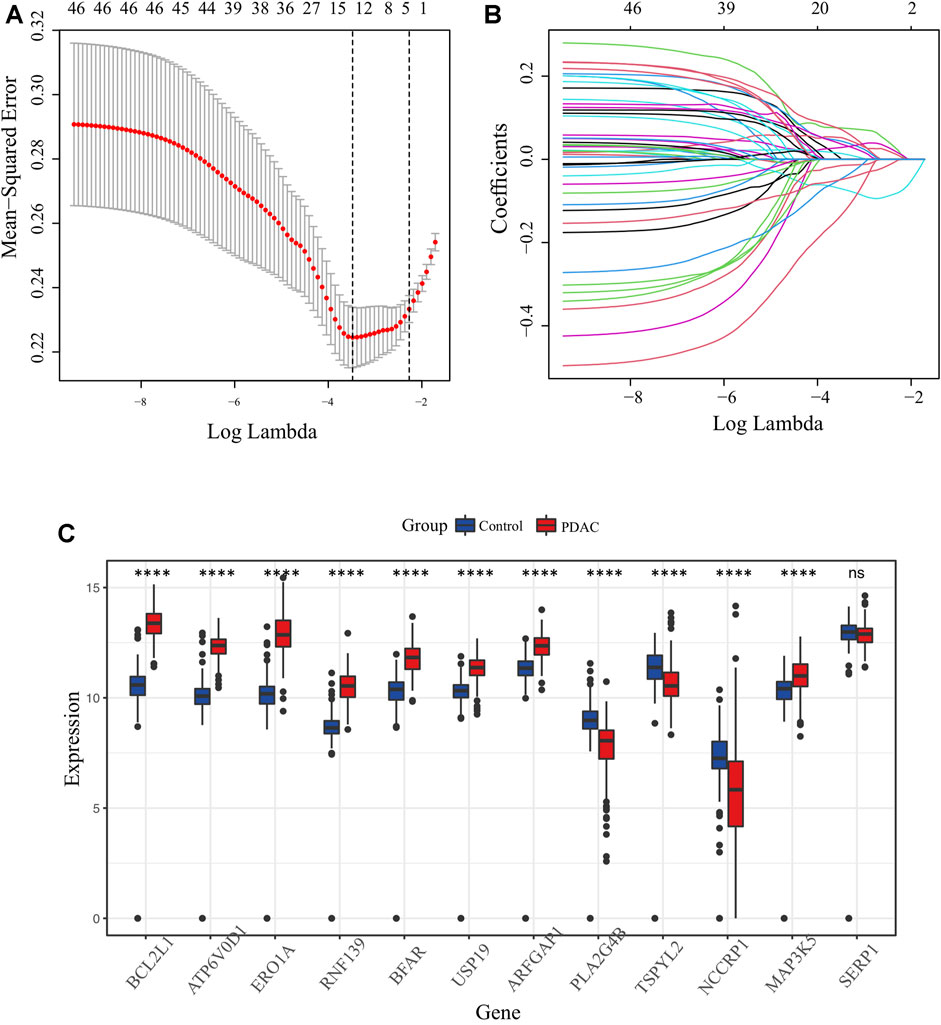
FIGURE 2. Identification of candidate ER stress-associated genes with prognostic potential. (A) Selection of lambda parameters for the LASSO model in TCGA-PAAD dataset. (B) Lambda-dependent coefficients for the 46 intersecting ER stress-associated genes. (C) Comparative expression profiling of the 12 prime ER stress-associated genes between pancreatic cancer and normal pancreatic tissues.
3.2 ER stress-associated risk signature was established, and the low-risk score PC patients owned longer OS time
The ER stress-associated risk signature was established using the above 12 markers, and the risk score equation was: riskscore = 0.813468268 × [(0.496637586 × SERP1) + (0.589598808 × BCL2L1) + (0.406009527 × RNF139) + (−0.521549002 × TSPYL2) + (−0.027972152 × ERO1A) + (0.323475966 × BFAR) + (0.476688839 × MAP3K5) + (0.025209694 × ARFGAP1) + (-0.92171365 × ATP6V0D1) + (0.152614999 × NCCRP1) + (0.005952034 × LA2G4B) + (-1.026914644 × USP19)]. We scored each PC patient with the above signature, and classified 164 PC patients into low-risk group (n = 87) and high-risk group (n = 77) according to the optimal cutoff point (1.322269) of the risk scores (Figure 3A). As Figure 3B showed, the profiles of gene expression in low- and high-risk clusters represented discrepant distribution patterns according to the principal components analysis (PCA), which implied that our ER stress-associated signature was correlated with a whole genome-wide difference. The K-M survival analysis is depicted in Figure 3C. It’s obvious that low-risk score PC patients owned longer OS time (p < 0.0001) than those patients with high-risk scores. The area under the ROC curve was 0.79, confirming the prognostic accuracy of our risk signature (Figure 3D). We did the same in the ICGC-PACA-CA cohort to validate the prognostic significance of our ER stress-associated risk signature. Survival analysis also confirmed a statistically significant survival advantage for low-risk score PC patients despite a slight decrease in the AUC value (Figures 3E, F). Moreover, a linked graph of a smooth curve, scatterplot, and heatmap intuitively indicated that as the risk score increases, the survival status of patients with PC from TCGA is more likely to be death and shorter survival time (Figure 3G). Furthermore, the differential expression landscape of these 12 risk factors between the two subgroups was also observed (Supplementary Figure S2).
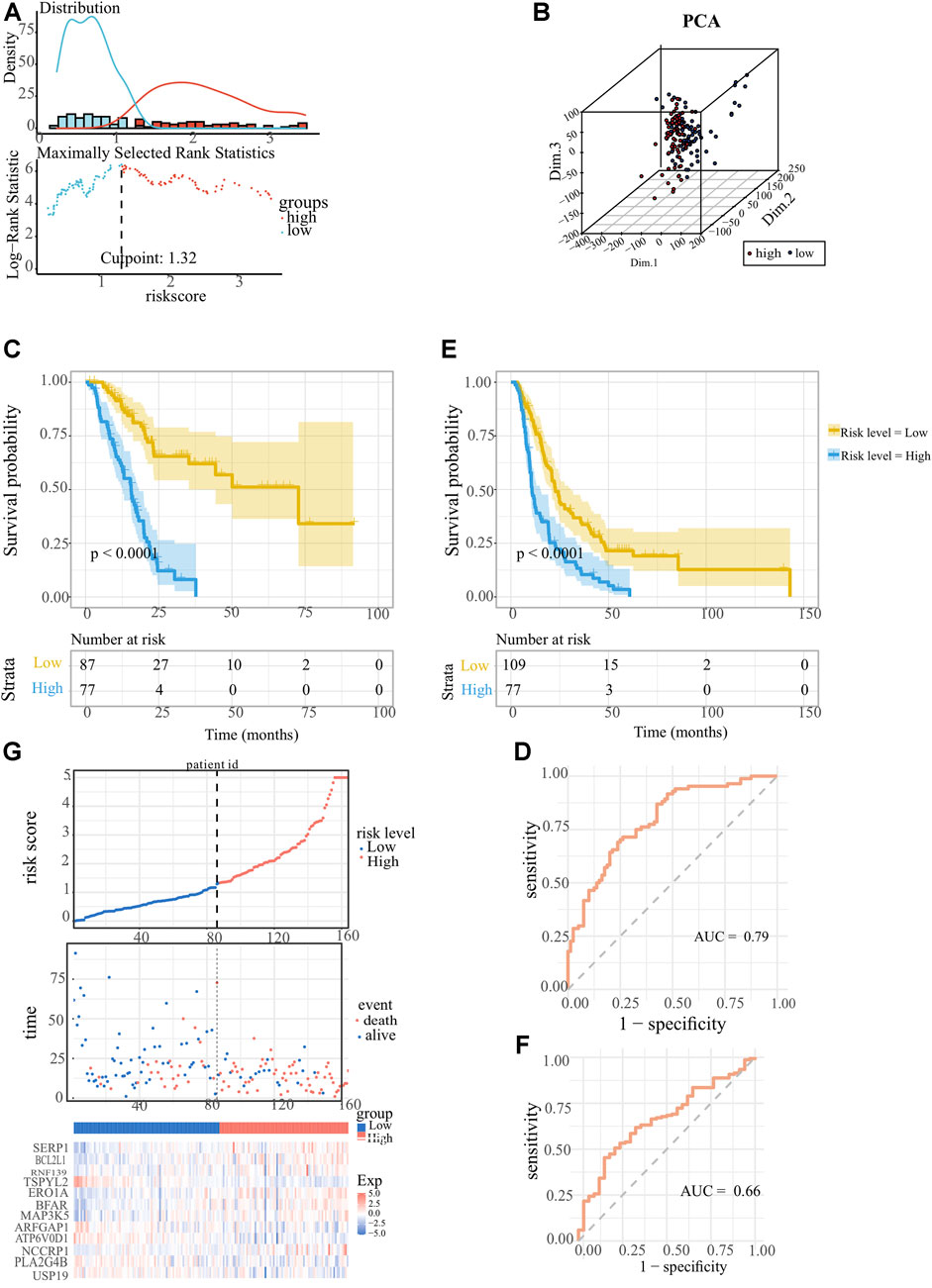
FIGURE 3. Establishment and confirmation of the ER stress-associated risk signature for pancreatic cancer. (A) Optimal cutoff ascertained via survival R package. (B) Genomic-wide expression-based Principal Component Analysis (PCA). (C, E) Kaplan-Meier survival curves showed indicating adverse overall survival (OS) in PC patients with lower risk indices, as observed in TCGA-PAAD and ICGC-PACA-CA datasets. (D, F) Receiver operating characteristic (ROC) curves Receiver Operating Characteristic (ROC) analyses confirming the robustness of the predictive model for pancreatic carcinoma patient survival within the TCGA-PAAD and ICGC-PACA-CA datasets. (G) Comprehensive display encompassing risk score distribution, survival status scatter plots, and a heatmap representation of the 12 optimal ER stress-correlated genes.
3.3 Establishment of nomogram model integrating ER stress-related risk signature and clinical parameters for predicting prognosis of PC patients
To better predict the survival probability of PC patients, we further built a nomogram based on our ER stress-associated risk signature and several readily available clinical parameters that recognized elsewhere as PC risk factors. Considering the number of patients with complete clinical information in the TCGA-PAAD cohort, our analysis did not include the M stage. Consequently, 91 patients provided complete information contributing to the predicting model. The forest plot showed that risk level and histological grade were independent prognostic indicators (Figure 4A). In particular, the risk level has the most apparent statistical significance (HR = 3.613, p < 0.001). Finally, we generated a nomogram model to predict the survival probability for each PC patient with a robust performance (Concordance Index = 0.72), as shown in Figure 4B. Total points can be calculated according to these parameters, and the 1-year, 3-year, and 5-year survival probability can be further obtained based on this total point.
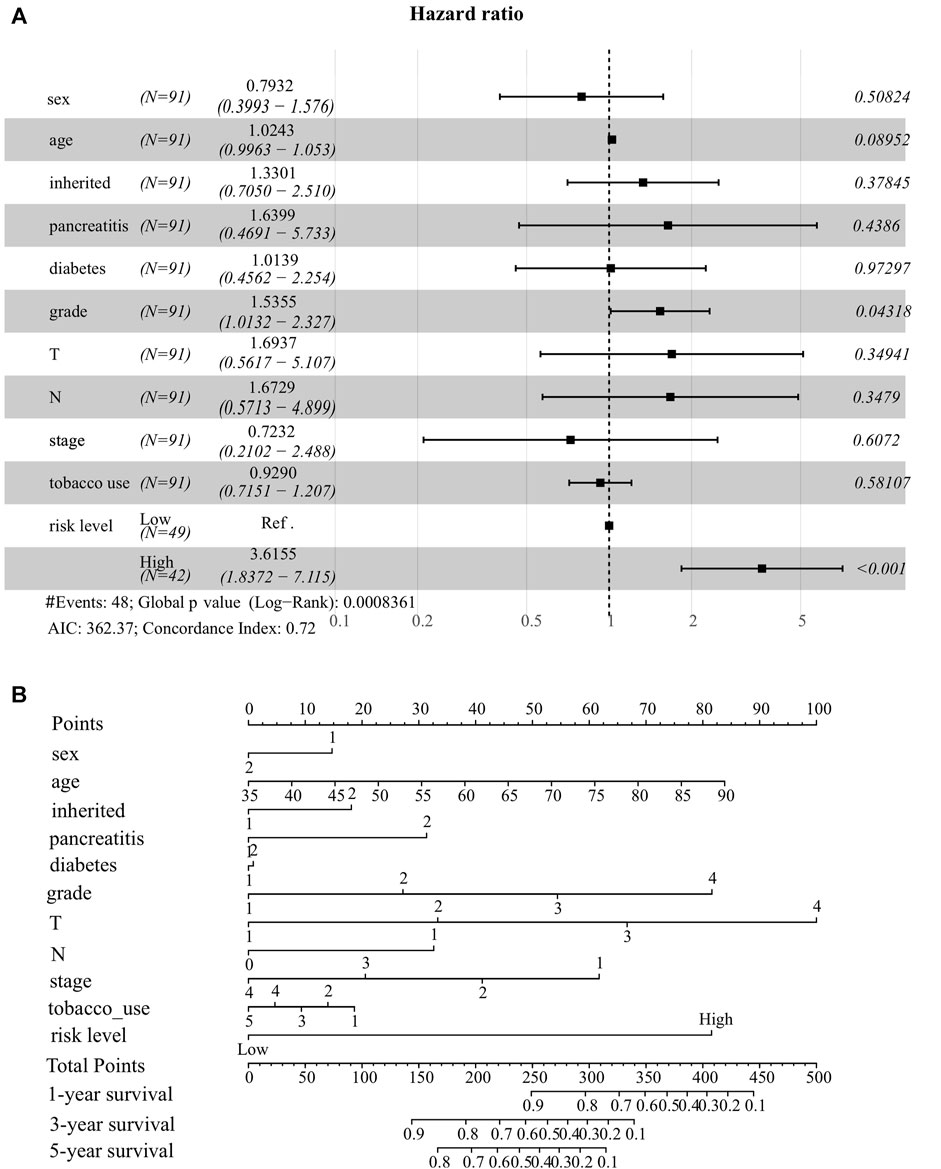
FIGURE 4. Establishment of nomogram model integrating ER stress-associated risk signature with clinicopathological parameters for prognostic predictions for pancreatic cancer. (A) Forest plot showing risk level as an independent prognostic biomarker for pancreatic cancer by multivariate analysis. (B) A prognostic nomogram amalgamating the ER stress-associated risk signature and multiple clinical determinants, predicting 1-, 2-, and 3-year overall survival likelihoods in pancreatic cancer patients.
3.4 GO and KEGG analyses of the risk signature reveal alteration of the homeostasis and immune signaling pathways
To investigate the underlying molecular mechanisms through which ER stress influences the prognosis of pancreatic cancer, we conducted a detailed functional enrichment analysis. Firstly, we identified DEGs between low- and high-risk clusters. A total of 1929 genes were differently expressed, containing 324 high-expressed and 1605 low-expressed genes. The enrichment of biological processes suggested that the ER stress-associated risk signature altered “membrane potential”, “chemical synaptic transmission”, “ trans-synaptic signaling”, “protein secretion” and “B-cell receptor signaling pathway” (Figure 5A). Interestingly, the genes enriched in biological processes terms of the “B cell receptor signaling pathway” were almost downregulated, indicating that high-risk patients may be “colder” regarding the immune microenvironment. The frontmost cellular components were “synaptic membrane,” “presynapse,” “neuronal cell body,” “transport complex,” and “ion channel complex” (Figure 5B). Molecular functions, including “passive transmembrane transporter activity,” “gated channel activity,” and “hormone activity,” were enriched (Figure 5C). More details of GO enrichment analyses can be found in Supplementary Table S4. The top 10 down and upregulated KEGG pathways are shown in bar graphs (Figure 5D). As we can see, the upregulated KEGG pathways mainly involve the exocrine function of the pancreas. Meanwhile, enriched downregulated pathways include “dopaminergic synapse,” “calcium ion signaling,” and “insulin secretion.” Thus, the KEGG and GO analysis results confirmed that the risk signature correlated with homeostasis and emphasized the role of ER status in during PC progression.
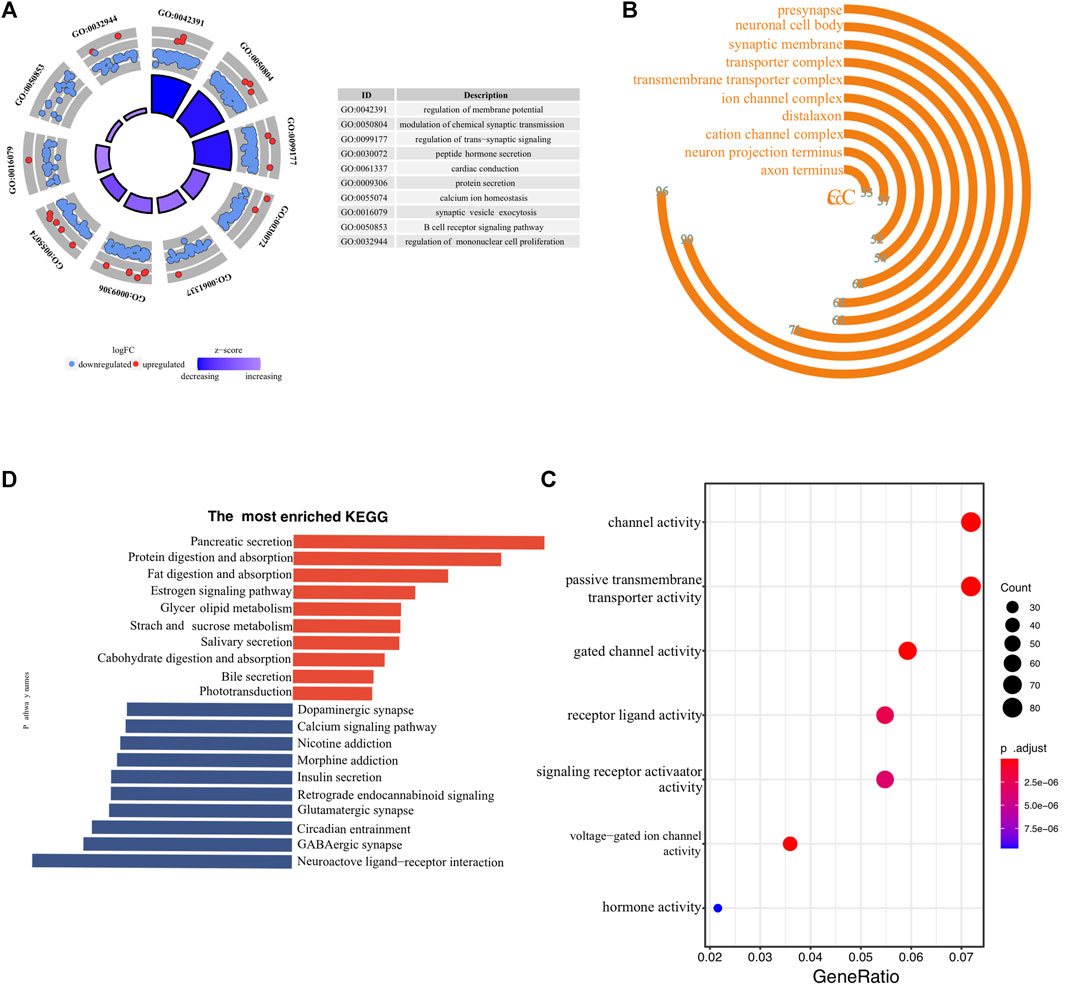
FIGURE 5. Biological functional annotation of differentially expressed genes between high- and low-risk samples. The front-most enriched biological processes (A), cellular components (B), and Molecular functions (C). (D) The most prominently upregulated and downregulated KEGG pathways in the TCGA-PAAD cohort.
3.5 Relationship between ER stress-associated risk signature and immune microenvironment
The landscape of the immune microenvironment of the two risk clusters were analyzed from the two aspects of immune cell abundance and immune scores. Four kinds of cells, including CD56 bright NK cell, Th2 cell, and Th17 cell, were infiltrated in the high-risk cluster; whereas other 14 types of cells were enriched in l enriched in the low-risk cluster, such as activated CD8+ T cells, NK cells, eosinophils, monocytes, activated DCs, as well as macrophages and MDSCs (Figure 6A). In addition, we calculated each PC patient’s stromal and immune scores at different risk levels. Unsurprisingly, the high-risk group got lower scores (Figure 6B). To further explore whether the ER stress-related signature affects PC immune microenvironment, a correlation heatmap was constructed to reflect the correlations between these differentially infiltrated immune cells and risk scores (Figure 6C). All these results demonstrated that the high-risk cluster has a more suppressive immune microenvironment that interacts with the risk signature. Considering the close link between our risk signature and the immune microenvironment and the therapeutic potential of immunotherapy in selected tumours, we also conducted a correlation analysis to identify PC patients who could benefit from immunotherapeutic modalities. Several immunotherapy targets, such as PDCD1, CTLA4, LMTK3, and LAG3, were also expressed in PC tissues. Interestingly, PDCD1 expression level had an inverse correlation with the risk score in PC patients with histopathological grade 2 or AJCC stage Ⅱ (Figures 6D, E).
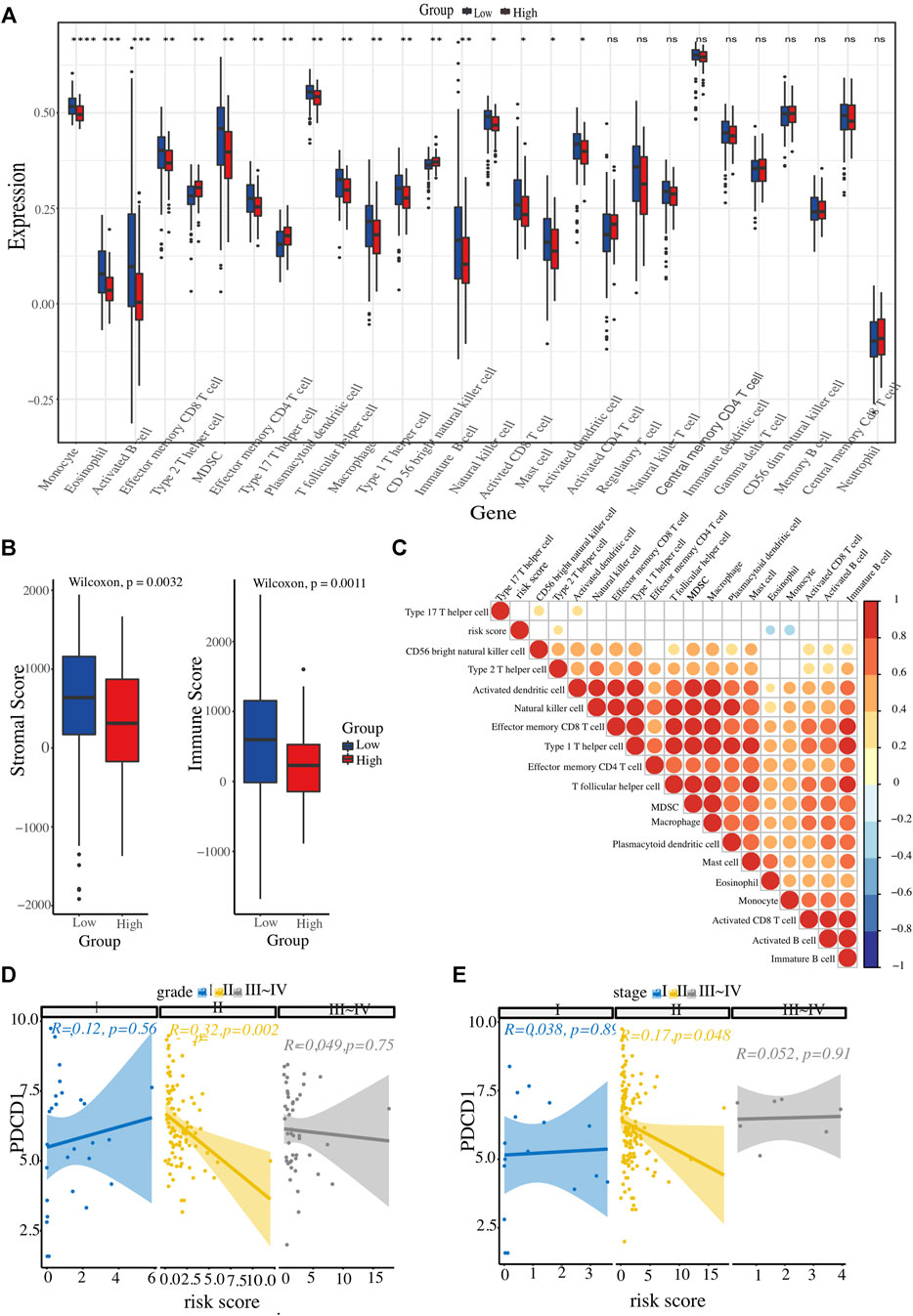
FIGURE 6. Evaluation of immune cell infiltration landscape. (A) Boxplot illustrating differential infiltrations of 28 immune cell subtypes between high- and low-risk groups. (B) Comparative evaluation of immune scores and stromal integrity between the two clusters. (C) Heatmap representation of risk score associations with 18 immune cell types displaying variable infiltration degrees. (D, E) The relationship between ER stress-associated risk score and immune checkpoint (PDCD1), stratified by histopathological grade and AJCC TNM stage, respectively.
3.6 Association of ER stress-associated risk with genomic alterations
Since somatic mutations are closely related to tumour initiation and progression, an integrated analysis was performed to establish an association of the status of genomic alterations with our ER stress-associated signature in PC. A total of 9,957 genes were mutated among 178 PC samples obtained from the GDC website, of which KRAS, TP53, SMAD4, and CDKN2A-the four main drive genes, were the most frequently mutated. The summary of the mutation landscape shows that missense mutation, nonsense mutation, frameshift deletion or insertions, and splice site mutation are the leading fourth variant types (Figure 7A). A waterfall plot depicted the top 10 mutated genes under ER stress signature-based clusters and other clinicopathological groups, and we found that 77% of PC patients experienced KRAS mutation (Figure 7B). As for the 12 ER stress-associated genes for constructing the risk model, only BFAR, MAP3K5, and ARFGAP1 have missense mutations or frameshift insertions, and USP19 has multiple mutations (multi-hits). The mutation ratio is less than 1%. In addition, the TMB was also calculated to reveal a possible mechanism for the different immune microenvironments in the two risk groups. However, our result showed no difference in TMB between the two clusters (Figure 7C).
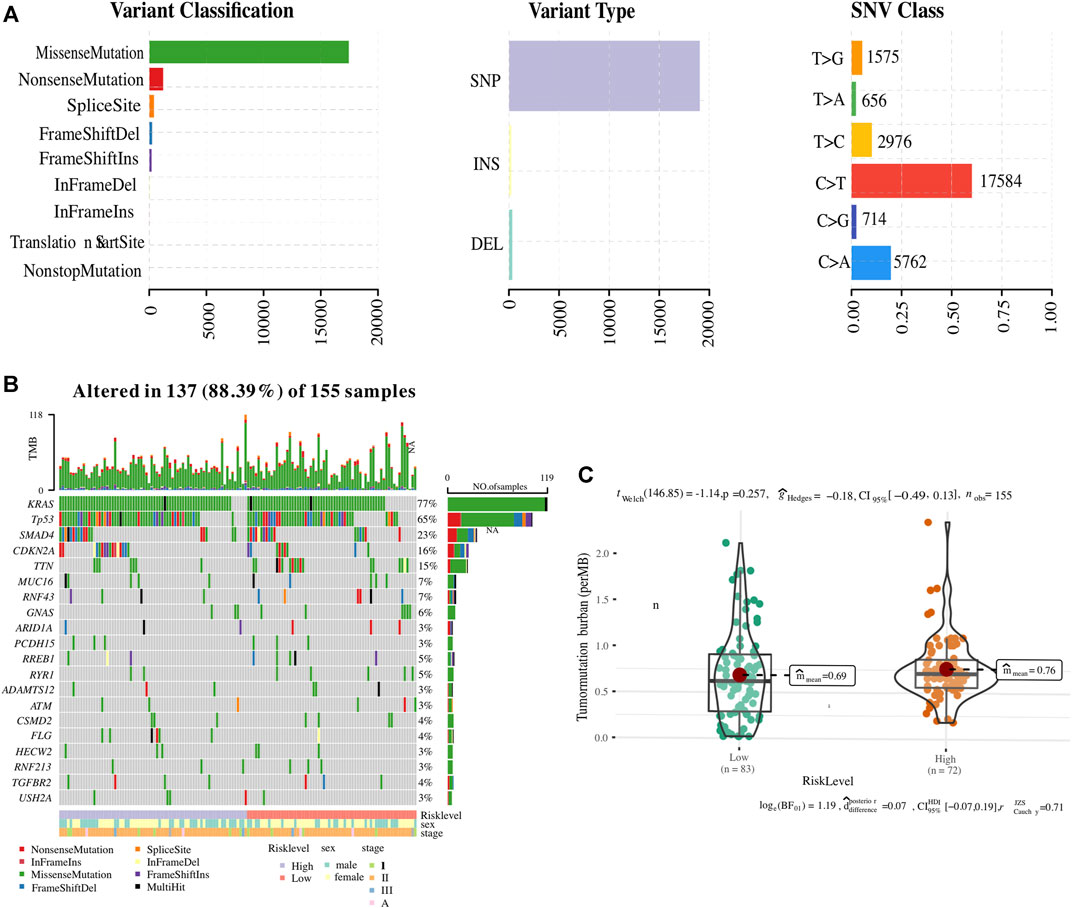
FIGURE 7. Association of ER stress-related risk signature with genomic alterations. (A) The summary of the mutation landscape of pancreatic cancer. (B) Waterfall representation highlighting the top 10 mutated genes. (C) Comparative tumour mutational burden (TMB) across the two ER stress-associated risk stratifications.
3.7 Correlation between ER stress-associated risk signature and therapeutic response
To better connect our ER stress-related risk signature to clinical practice, we employed oncoPredict tool to calculate the IC50 value of several commonly used chemotherapeutic agents for each PC patients. The results showed that the predicted IC50 for agents such as “5-Fluorouracil”, “Oxaliplatin”, “Irinotecan”, “Paclitaxel”, and “Cisplatin” skews higher for the high-risk demographic., indicating that low-risk score patients may be more sensitive to chemotherapy (Figures 8A–F).
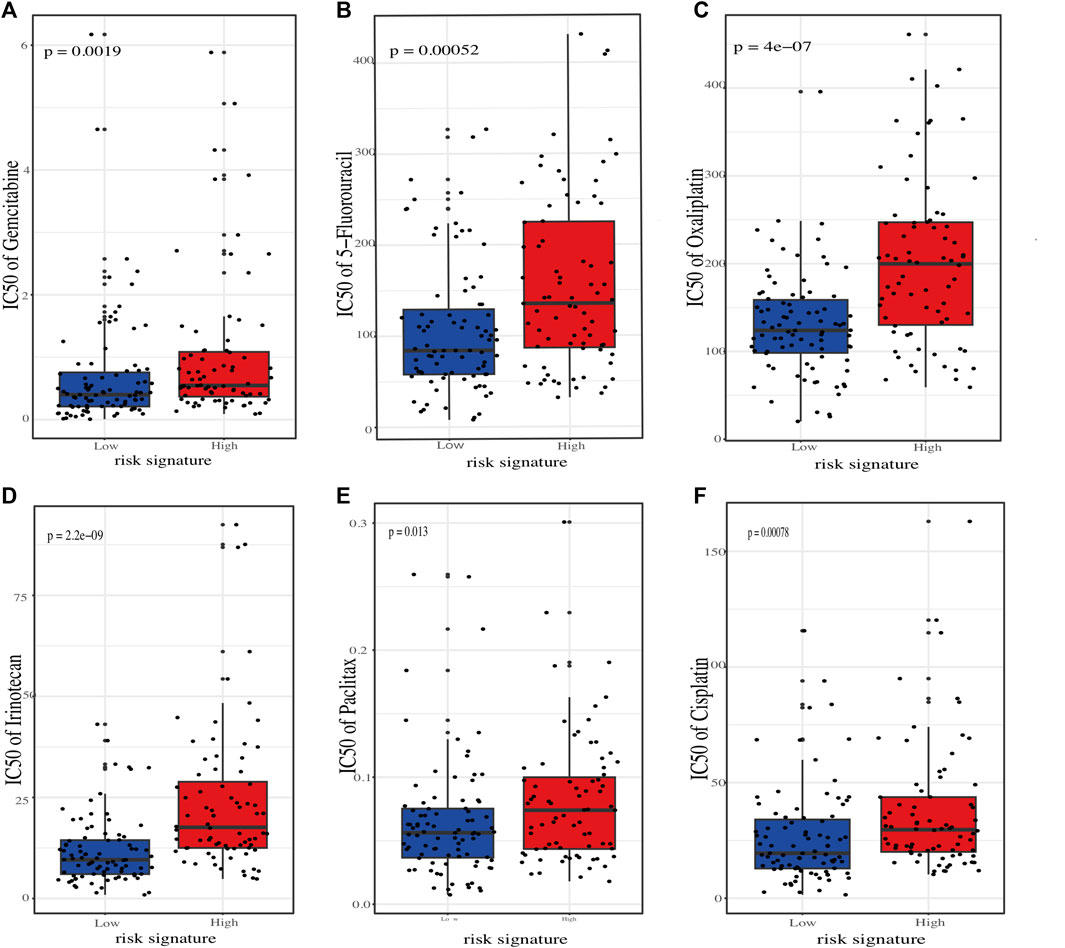
FIGURE 8. Estimation of chemotherapeutic sensitivity in stratified risk groups. Comparision of the predicted half-maximal inhibitory concentration (IC50) values for six chemotherapeutic agents: gemcitabine (A), 5-fluorouracil (B), oxaliplatin (C), irinotecan (D), paclitaxel (E), and cisplatin (F).
4 Discussion
The traditional clinical indicators are poor in predicting the survival time of PC, so it is imperative to develop new tools to predict survival and furnish effective individualized treatment for PC patients. Increasing evidence shows that ER stress plays a complex role in PC progression. Hence, we here performed a bioinformatic analysis based on TCGA database to study role of ER stress in prognosis and biological characteristics in PC patients. In this study, we initially established a twelve-gene risk signature in terms of ER stress according to the gene expression counts and corresponding survival time of 164 PC patients from TCGA. The twelve ER stress-related genes enrolled in the risk model could be divided into protective factors (TSPYL2, ARFGAP1, ATP6V0D1, PLA2G4B, USP19) and pernicious factors (SERP1, BCL2L1, RNF139, ERO1A, BFAR, MAP3K5, NCCRP1). The predictive potency of the ER stress-associated risk signature was further validated by another independent PC dataset from ICGC. Survival curve and forest plot demonstrated that our ER stress-associated signature was a valuable and independent prognostic biomarker, according to the ROC curves (AUC was 0.79) and the hazard ratio (HR was 3.613, p < 0.001, CI was 0.72). Besides, we constructed a credible nomogram integrated the risk signature and several clinicopathological parameters predicted the OS of individual PC patients. Function annotations elucidate the biological role of ER stress in PC, and the results indicated that the ER stress-related signature has an effect in shaping TME immune cell infiltration.
SERP1 has been uncovered to be overexpressed under stress, which helps to stabilize membrane proteins and functions as a translocon on the ER membrane (Yamaguchi et al., 1999). Evidence shows that SERP1 may act as an oncogene and could be developed as a valuable prognostic biomarker. For instance, a recent research uncovered that SERP1 can facilitate osteosarcoma progression through modulating circ_0085539/miR-526b-5p signalling axis (Liu et al., 2020). Additionally, SERP1 has been reported to maintain the viability of PC cells promote the survival of PC cells by down-regulating apoptosis-related protein SRPRB and activating the NF-κB signalling pathway (Ma et al., 2017). Consistently, our study demonstrated that SERP1 was upregulated in PC and its high expression predicted dismal patient survival prognosis. BCL2L1, also called Bcl-extra (Bcl-x), belongs to the B-cell lymphoma 2 apoptosis family. BCL2L1 translates two antagonistic variants including Bcl-XS and Bcl-XL due to alternative splicing. The short isoform Bcl-XS exhibits pro-apoptosis biological function, whereas the long isoform Bcl-XL contributes to survival of cells. Numerous studies have confirmed that the disequilibrium of BCL2L1 splice is related to the progression of multiple malignancies (Dou et al., 2021). In addition to promoting tumour cell survival, Bcl-XL can also boost metastasis and EMT of pancreatic neuroendocrine tumour cells via epigenetically enhancing TGFβ signalling (Choi et al., 2016). Furthermore, recent evidence shows in melanoma that Bcl-XL can positively regulate the expressions of IL-8 and CCL5, which not only recruit macrophages into tumour sites, but also polarize them into M2 phenotype (Lucianò et al., 2023). In PC, Bcl-XL is overexpressed, and correlated with worse prognosis of patients (Friess et al., 1998). Particularly, Bcl-XL has been identified as a crucial mediator of acquiring radioresistance and gemcitabine resistance (Lee et al., 1999; Thummuri et al., 2022). Similarly, we herein found that BCL2L1 was significantly increased in PC tissues and its high expression predicted unfavourable prognosis of patients. In general, our data and the literature evidence suggest that Bcl-XL may become a valuable prognostic biomarker and serve as a therapeutic target for PC patients.
RNF139, also known as TRC8, encodes a multi-membrane spanning protein resides in ER, and it has been revealed to act as a tumour suppressor. A study reported that RNF139 was significantly downregulated in tongue cancer tissue and experimentally that silencing RNF139 enhanced the viability and aggressiveness of tongue cancer cells (Wang et al., 2017). Intriguingly, our data shows that RNF139 is dramatically upregulated in PC tissues, and low-expressed RNF139 PC patients have shorter survival time, indicating a potential role of RNF139 in propagating PC progression. Hence, the roles of RNF139 in cancer may depend upon the tumour types, and cell-based and animal experiments are warranted to determine the specific roles of RNF139 in PC. ERO1A is an oxidoreductase located in ER lumen. Recently, accumulating data uncovers that ERO1A functions as an oncogene in many types of cancers. According to Yan and colleague, the expression of ERO1A is increased in cholangiocarcinoma and its overexpression predicts poor prognosis of patients (Yan et al., 2019). Zilli et al. (2021) revealed that upregulation of ERO1A significantly promotes primary breast tumour growth and lung metastatic colonization by enhancing HIF1α-VEGFA mediated angiogenesis. Zhang et al. corroborated that overexpression of ERO1A can contribute to tumour progression by accelerating hydrogen peroxide-associated epithelial-mesenchymal transition, and it may be used as a predictor for dismal prognosis in cervical cancer (Zhang et al., 2019). In PC, ERO1A has been demonstrated to boost tumour growth via augmenting aerobic glycolysis of tumour cells (Zhang et al., 2020). In agreement with these evidence, we sighted that mRNA of ERO1A is highly in PC and high ERO1A expression is accompanying worse prognosis. Altogether, ERO1A may be a useful prognostic biomarker and promising therapeutic target for PC patients.
BFAR has been recognized as an inhibitor of apoptosis with an alary helix DNA-binding structure and is tightly linked with tumour progression. Cheng et al. reported that BFAR is upregulated in glioma tissues and glioma patients with lower BFAR expression have worse long-term survival prognosis (Cheng et al., 2020). In gastric cancer, BFAR has been proven to rebound to the tumour metastasis via activating the PI3K/AKT/mTOR signalling axis (Chen et al., 2023). Similar to the previous studies, we observed that BFAR is highly expressed in PC and its increasing expression predicts worse outcomes of PC patients. Thus, BFAR may also act as an oncogene in PC. MAP3K5 has been known as an upstream protein of mitogen-activated protein kinase cascade signalling pathway and plays a vital tumour-suppressive role in many types of cancers. For instance, Cheng et al. showed that overexpression of MAP3K5 promotes PC cell apoptosis via activating the p38/MAPK axis (Cheng et al., 2014). Consistent with this, our data showed that MAP3K5 is upregulated in PC tissues and its high expression predicts poor prognosis of PC patients. NCCRP1 is a type III transmembrane protein containing an antigen recognition motif, also named as FBXO50. Evidence shows that NCCRP1 plays an essential role in tumour development, and whether it acts as a tumour suppressor gene or oncogene depend upon tumour types. A most recent study suggested that NCCRP1 was highly expressed in triple-negative breast cancers (TNBCs) and its overexpression could markedly enhance cell proliferation (Zhou et al., 2022). In gastric cancer, higher NCCRP1 expression is associated with shorter recurrence-free and overall survival time, as it promotes proliferation, movement, and incursion of gastric cancers (Miwa et al., 2017b). Inversely, another study (Miwa et al., 2017a) found that esophageal squamous cell carcinoma patients with lower NCCRP1 expression tended to have an increased risk of disease recurrence and dismal survival prognosis versus those with higher NCCRP1 expression, suggesting a putative tumour-suppressive role of NCCRP1. Notably, when it comes to PC, the roles of NCCRP1 seems rather complex. In our study, we found that NCCRP1 is downregulated in PC tissues compared to normal pancreatic tissues, but the high NCCRP1 expression patients have little survival time. Our paradoxical results may be due to the fact that NCCRP1 in PC tissues is not expressed mainly in tumour cells, but in stroma cells such as immune cells and fibroblasts. More research is thus required to further decipher the biological roles of NCCRP1 in PC and its clinical prognostic value.
USP19 is a kind of deubiquitinating protease. Similar to NCCRP1, USP19 is aberrantly expressed in multiple cancers and may act as a tumour suppressor gene or oncogene, which relies upon the tumour type (Rossi and Rossi, 2022). For example, USP19 was found to be upregulated in TNBC tissues and promote the invasion and metastasis of TNBC (Rossi et al., 2021). Besides, a recent study reported that upregulation of USP19 contributed to tumourigenicity of colorectal cancer cells by stabilizing Survivin protein (Chandrasekaran et al., 2022). On contrast, diminished USP19 expression in ovarian cancer correlates with an adverse prognosis (Kang et al., 2021). Adding to this complexity, (Hu et al., 2020), reported in clear cell renal cell carcinoma that overexpression of USP19 markedly curtails tumour cell proliferation and migration via ERK pathway inactivation. Aligning with these findings, our research discerned that elevated USP19 expression portends a favorable survival outcome in PC, and it is of clinical significance to investigate its specific biological functions and the relevant mechanisms in the progression of PC. TSPYL2 is the nucleosome component of chromatin remodelling (Cardano et al., 2023). Recent work by Pan et al. (2022) recently confirmed that TSPYL2 is downregulated in PC and overexpression of TSPYL2 can effectively overcome gemcitabine resistance in PC cells. Likewise, in this study we also found that TSPYL2 is significantly downregulated in PC and its low expression predicts shorter survival time of PC patients. Within the cohort of ER stress-associated risk signature genes, the roles of PLA2G4B, ARFGAP1, and ATP6V0D1 in human malignancies remain largely unexplored. Our research determined that PLA2G4B exhibits decreased expression in PC tissues, correlating lower expression levels with a more guarded prognosis. In contrast, both ARFGAP1 and ATP6V0D1 manifested heightened expression in PC tissues, resonating with an improved prognostic outlook for PC patients. Future endeavors should be directed towards elucidating the precise biological and clinical implications of PLA2G4B, ARFGAP1, and ATP6V0D1 in the context of PC.
The tumour immune microenvironment is intertwined with PC growth, metastasis, and therapeutic resistance (Bear et al., 2020; Ho et al., 2020; Sherman and Beatty, 2023). Increasing evidence reveals that ER stress can confer immunomodulatory capacity on cancer cells (Cubillos-Ruiz et al., 2017; Di Conza et al., 2023), prompting us to evaluate the correlation of our ER stress-associated signature with the PC immune microenvironment. The functional enrichment analysis showed that ER stress status in PC may have a close relationship to a multiple of immune signalling pathways including T cell proliferation, B cell proliferation and activation, and interleukin-4 production. A body of findings in oncological research indicate that ER stress can have effects on the trafficking of immune cell to the tumour milieu. For instance, Song et al. showed XBP1 is upregulated in T cells isolated from ovarian cancer patient samples in contrast with healthy donor-derived T cells, and the upregulation of XBP1 is correlated with decreased intratumoural T cell infiltration (Song et al., 2018). Work by Harnoss and colleagues demonstrated that in TNBC xenograft, knockout of IRE1α can reduce MDSC infiltration by downregulating the transcription of several genes involved in recruitment, such as PTGS2, CXCL8, and CXCR4 (Harnoss et al., 2020). Histological analyses revealed that in human hepatocellular carcinoma (HCC) specimens, the expression levels of ER stress markers such as BiP, ATF6, PERK, and IRE1α were closely associated with an elevated infiltration of CD68+PDL1+ macrophages. The elevating PD-L1 expression on macrophages is primarily achieved by exosomes containing miR-23a-3p released from HCC cells subjected to ER stress mediated PTEN/PI3K/AKT pathway, which in turn impacts T cell functions and survival (Liu et al., 2019). In our study, we observed that low-risk cluster has higher accumulation of T cells and NK cells, comparing with high-risk cluster. It has been demonstrated that low infiltration and dysfunction of NK cells in PC tissues are tightly associated with tumour recurrence and survival of patients (Hoshikawa et al., 2018; Xie et al., 2019). As such, our ER stress risk signature could serve as a valuable tool for evaluating NK cell immune status and predicting the efficacy of NK cell-based immunotherapy in PC. Since the cytotoxic and cytokine-producing effector molecules functions of NK cells are governed by a delicate balance between activating and inhibitory receptors, we sought correlations between NK cell ligands and our risk signature, albeit with inconclusive outcomes. Comparative analyses revealed that our high-risk and low-risk cohorts exhibited no discernible disparities in the mRNA expression levels of the MICA/B, cognate ligands of major NK activating receptor NKG2D (Cadoux et al., 2021), and the inhibitory receptors ligands HLA-E (Liu et al., 2023) and CEACAM1 (Park et al., 2020). Interestingly, relative to healthy pancreatic tissue, all the aforementioned molecules exhibited augmented expression in PC. This observation may be attributed to the impairment of IRE1α arm in PC, as it was reported that the MICA/B expression was suppressed by the IRE1α/XBP1 arm of the ER stress response in melanoma cells (Obiedat et al., 2019). Eosinophils, primitive cells of the innate immune system primarily studied in allergic diseases, have recently garnered attention for their role in promoting antitumour immune responses (Grisaru-Tal et al., 2022; O'Flaherty et al., 2017; Blomberg et al., 2023) demonstrated that eosinophils can enhance NK cell chemotaxis and activation via secretion of CCL5, CXCL10 and IL-12. Moreover, colorectal cancer-infiltrated eosinophils have been shown to promote IFN-γ and TNF-α production by CD8+ T cells via the GM-CSF-IRF5 signalling axis, thereby augmenting antitumour immune responses (Arnold et al., 2020). Intriguingly, our findings revealed a predominant concentration of tumour-infiltrating eosinophils in the low-risk cluster compared to its high-risk counterpart, potentially elucidating the abundant occurrence of NK and T cells within the low-risk group. It's hypothesized that ER stress might compromise the antitumour efficacy of NK and T cells by obstructing eosinophil recruitment and activation. Both macrophages and MDSCs, pivotal myeloid suppressive entities, can debilitate NK and T cell functionality, facilitating tumour immune evasion (Sui et al., 2022; Cassetta and Pollard, 2023). Our observations indicate an inverse relationship between MDSCs and ER stress scores, presenting an apparent paradox. Given the pronounced presence of NK and T cells within low-risk strata, this data intimates potential regulatory feedback exerted by MDSCs that modulates NK- and T-cell-mediated antitumour immune responses. Thus, ER stress’s influence on the TME seems intricate, making targeted interventions demanding. Therefore, the effect of ER stress on the TME may be multifaceted and targeting ER stress remains challenging.
In summary, we established a 12-gene ER stress-associated risk signature, holding promise as a prognostic tool for PC patients. Importantly, this prognostic risk signature is positively correlated with an immunosuppressive state in PC, indicating its potential of guiding clinical decision making in the immunotherapy of PC. The analyses of functional enrichment and quantification of the immune infiltration landscape yielded essential findings for initiating future research on the mechanisms underlying ER stress-related genes and tumour immunity in PC. Nonetheless, it's crucial to highlight that our investigation relied solely on transcriptomic data from surgical biopsies, so further clinical trials and experimental validations are required to confirm our findings.
Data availability statement
Publicly available datasets were analyzed in this study. This data can be found here: The Cancer Genome Atlas (https://xenabrowser.net/), International Cancer Genome Consortium (https://icgc.org), and Molecular Signature Database v7.4 (https://www.gsea-msigdb.org/). The full original source data used in this study has been downloaded from the above public datasets and uploaded to jianguoyun along with codes. Detailed information can be found here: “https://www.jianguoyun.com/p/DYpnVlIQ4qX6CxjQ3ZoFIAA.”
Ethics statement
Ethical approval was not required for the studies involving humans because the data were extracted from the public database. The studies were conducted in accordance with the local legislation and institutional requirements. The human samples used in this study were acquired from The Cancer Genome Atlas Program (TCGA). Written informed consent to participate in this study was not required from the participants or the participants’ legal guardians/next of kin in accordance with the national legislation and the institutional requirements.
Author contributions
HC: Conceptualization, Data curation, Formal Analysis, Methodology, Visualization, Writing–original draft. NX: Conceptualization, Investigation, Methodology, Writing–original draft. JX: Conceptualization, Investigation, Methodology, Writing–original draft. CZ: Conceptualization, Investigation, Methodology, Writing–original draft. XL: Conceptualization, Data curation, Investigation, Validation, Writing–review and editing. HX: Conceptualization, Data curation, Formal Analysis, Investigation, Methodology, Writing–original draft. WZ: Formal Analysis, Investigation, Writing–original draft. JL: Conceptualization, Methodology, Writing–review and editing. DL: Conceptualization, Methodology, Writing–review and editing. WZ: Conceptualization, Data curation, Funding acquisition, Methodology, Project administration, Supervision, Writing–review and editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the National Natural Science Foundation of China (grant number 82260555); Medical Innovation and Development Project of Lanzhou University (grant number lzuyxcx-2022-177) and Major Science and Technology Projects of Gansu Province (grant number 22ZD6FA021-4).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmolb.2023.1298077/full#supplementary-material
References
Allison, S. (2021). ER-associated degradation of nephrin. Nat. Rev. Nephrol. 17 (5), 297. doi:10.1038/s41581-021-00421-7
Arnold, I. C., Artola-Boran, M., Gurtner, A., Bertram, K., Bauer, M., Frangez, Z., et al. (2020). The GM-CSF-IRF5 signaling axis in eosinophils promotes antitumor immunity through activation of type 1 T cell responses. J. Exp. Med. 217 (12), e20190706. doi:10.1084/jem.20190706
Bear, A. S., Vonderheide, R. H., and O'Hara, M. H. (2020). Challenges and opportunities for pancreatic cancer immunotherapy. Cancer Cell 38 (6), 788–802. doi:10.1016/j.ccell.2020.08.004
Blomberg, O. S., Spagnuolo, L., Garner, H., Voorwerk, L., Isaeva, O. I., van Dyk, E., et al. (2023). IL-5-producing CD4(+) T cells and eosinophils cooperate to enhance response to immune checkpoint blockade in breast cancer. Cancer Cell 41 (1), 106–123.e10. doi:10.1016/j.ccell.2022.11.014
Cadoux, M., Caruso, S., Pham, S., Gougelet, A., Pophillat, C., Riou, R., et al. (2021). Expression of NKG2D ligands is downregulated by β-catenin signalling and associates with HCC aggressiveness. J. Hepatol. 74 (6), 1386–1397. doi:10.1016/j.jhep.2021.01.017
Cai, J., Chen, H., Lu, M., Zhang, Y., Lu, B., You, L., et al. (2021). Advances in the epidemiology of pancreatic cancer: trends, risk factors, screening, and prognosis. Cancer Lett. 520, 1–11. doi:10.1016/j.canlet.2021.06.027
Cardano, M., Magni, M., Alfieri, R., Chan, S. Y., Sabbioneda, S., Buscemi, G., et al. (2023). Sex specific regulation of TSPY-Like 2 in the DNA damage response of cancer cells. Cell Death Dis. 14 (3), 197. doi:10.1038/s41419-023-05722-2
Cassetta, L., and Pollard, J. W. (2023). A timeline of tumour-associated macrophage biology. Nat. Rev. Cancer 23 (4), 238–257. doi:10.1038/s41568-022-00547-1
Chandrasekaran, A. P., Tyagi, A., Poondla, N., Sarodaya, N., Karapurkar, J. K., Kaushal, K., et al. (2022). Dual role of deubiquitinating enzyme USP19 regulates mitotic progression and tumorigenesis by stabilizing survivin. Mol. Ther. 30 (11), 3414–3429. doi:10.1016/j.ymthe.2022.07.019
Charoentong, P., Finotello, F., Angelova, M., Mayer, C., Efremova, M., Rieder, D., et al. (2017). Pan-cancer immunogenomic analyses reveal genotype-immunophenotype relationships and predictors of response to checkpoint blockade. Cell Rep. 18 (1), 248–262. doi:10.1016/j.celrep.2016.12.019
Chen, X., and Cubillos-Ruiz, J. R. (2021). Endoplasmic reticulum stress signals in the tumour and its microenvironment. Nat. Rev. Cancer 21 (2), 71–88. doi:10.1038/s41568-020-00312-2
Chen, G., Zhang, H., Sun, H., Ding, X., Liu, G., Yang, F., et al. (2023). Bufalin targeting BFAR inhibits the occurrence and metastasis of gastric cancer through PI3K/AKT/mTOR signal pathway. Apoptosis. doi:10.1007/s10495-023-01855-z
Cheng, X., Holenya, P., Can, S., Alborzinia, H., Rubbiani, R., Ott, I., et al. (2014). A TrxR inhibiting gold(I) NHC complex induces apoptosis through ASK1-p38-MAPK signaling in pancreatic cancer cells. Mol. Cancer 13, 221. doi:10.1186/1476-4598-13-221
Cheng, Z., Shu, H., Cui, Y., Zhang, Q., Zhao, B., Pan, D., et al. (2020). MiR-424-5p inhibits proliferation, invasion and promotes apoptosis and predicts good prognosis in glioma by directly targeting BFAR. Pathol. Oncols. Res. 26 (4), 2327–2335. doi:10.1007/s12253-020-00831-1
Conroy, T., Hammel, P., Hebbar, M., Ben Abdelghani, M., Wei, A. C., Raoul, J. L., et al. (2018). FOLFIRINOX or gemcitabine as adjuvant therapy for pancreatic cancer. N. Engl. J. Med. 379 (25), 2395–2406. doi:10.1056/NEJMoa1809775
Cubillos-Ruiz, J. R., Bettigole, S. E., and Glimcher, L. H. (2017). Tumorigenic and immunosuppressive effects of endoplasmic reticulum stress in cancer. Cell 168 (4), 692–706. doi:10.1016/j.cell.2016.12.004
Cubillos-Ruiz, J. R., Silberman, P. C., Rutkowski, M. R., Chopra, S., Perales-Puchalt, A., Song, M., et al. (2015). ER stress sensor XBP1 controls anti-tumor immunity by disrupting dendritic cell homeostasis. Cell 161 (7), 1527–1538. doi:10.1016/j.cell.2015.05.025
Dauer, P., Sharma, N. S., Gupta, V. K., Durden, B., Hadad, R., Banerjee, S., et al. (2019). ER stress sensor, glucose regulatory protein 78 (GRP78) regulates redox status in pancreatic cancer thereby maintaining "stemness". Cell Death Dis. 10 (2), 132. doi:10.1038/s41419-019-1408-5
Di Conza, G., Ho, P. C., Cubillos-Ruiz, J. R., and Huang, S. C. (2023). Control of immune cell function by the unfolded protein response. Nat. Rev. Immunol. 23, 546–562. doi:10.1038/s41577-023-00838-0
Dou, Z., Zhao, D., Chen, X., Xu, C., Jin, X., Zhang, X., et al. (2021). Aberrant Bcl-x splicing in cancer: from molecular mechanism to therapeutic modulation. J. Exp. Clin. Cancer Res. 40 (1), 194. doi:10.1186/s13046-021-02001-w
Friedman, J., Hastie, T., and Tibshirani, R. (2010). Regularization paths for generalized linear models via coordinate descent. J. Stat. Softw. 33 (1), 1–22. doi:10.18637/jss.v033.i01
Friess, H., Lu, Z., Andrén-Sandberg, A., Berberat, P., Zimmermann, A., Adler, G., et al. (1998). Moderate activation of the apoptosis inhibitor bcl-xL worsens the prognosis in pancreatic cancer. Ann. Surg. 228 (6), 780–787. doi:10.1097/00000658-199812000-00009
Globocan (2020). Cancer today. Available at: https://gco.iarc.fr/today/home.
Grisaru-Tal, S., Rothenberg, M. E., and Munitz, A. (2022). Eosinophil-lymphocyte interactions in the tumor microenvironment and cancer immunotherapy. Nat. Immunol. 23 (9), 1309–1316. doi:10.1038/s41590-022-01291-2
Harnoss, J. M., Le Thomas, A., Reichelt, M., Guttman, O., Wu, T. D., Marsters, S. A., et al. (2020). IRE1α disruption in triple-negative breast cancer cooperates with antiangiogenic therapy by reversing ER stress adaptation and remodeling the tumor microenvironment. Cancer Res. 80 (11), 2368–2379. doi:10.1158/0008-5472.Can-19-3108
Hetz, C., Zhang, K., and Kaufman, R. J. (2020). Mechanisms, regulation and functions of the unfolded protein response. Nat. Rev. Mol. Cell Biol. 21 (8), 421–438. doi:10.1038/s41580-020-0250-z
Ho, W. J., Jaffee, E. M., and Zheng, L. (2020). The tumour microenvironment in pancreatic cancer - clinical challenges and opportunities. Nat. Rev. Clin. Oncol. 17 (9), 527–540. doi:10.1038/s41571-020-0363-5
Hoshikawa, M., Aoki, T., Matsushita, H., Karasaki, T., Hosoi, A., Odaira, K., et al. (2018). NK cell and IFN signatures are positive prognostic biomarkers for resectable pancreatic cancer. Biochem. Biophys. Res. Commun. 495 (2), 2058–2065. doi:10.1016/j.bbrc.2017.12.083
Hu, W., Su, Y., Fei, X., Wang, X., Zhang, G., Su, C., et al. (2020). Ubiquitin specific peptidase 19 is a prognostic biomarker and affect the proliferation and migration of clear cell renal cell carcinoma. Oncol. Rep. 43 (6), 1964–1974. doi:10.3892/or.2020.7565
Huang, R., Li, G., Wang, K., Wang, Z., Zeng, F., Hu, H., et al. (2021). Comprehensive analysis of the clinical and biological significances of endoplasmic reticulum stress in diffuse gliomas. Front. Cell Dev. Biol. 9, 619396. doi:10.3389/fcell.2021.619396
Kang, H., Choi, M. C., Kim, S., Jeong, J. Y., Kwon, A. Y., Kim, T. H., et al. (2021). USP19 and RPL23 as candidate prognostic markers for advanced-stage high-grade serous ovarian carcinoma. Cancers (Basel) 13 (16), 3976. doi:10.3390/cancers13163976
Kong, B., Cheng, T., Wu, W., Regel, I., Raulefs, S., Friess, H., et al. (2015). Hypoxia-induced endoplasmic reticulum stress characterizes a necrotic phenotype of pancreatic cancer. Oncotarget 6 (31), 32154–32160. doi:10.18632/oncotarget.5168
Kosuke, Y., Hoon, K., and Roel, G. V. (2016). Stimate: estimate of stromal and immune cells in malignant tumor tissues from expression data. R package version 1.0.13/r21. Available at: https://rdrr.io/rforge/estimate/#:∼:text=from%20Expression%20Data-,estimate%3A%20Estimate%20of%20Stromal%20and%20Immune%20Cells%20in%20Malignant%20Tumor,tissues%20using%20gene%20expression%20data.
Lee, J. U., Hosotani, R., Wada, M., Doi, R., Kosiba, T., Fujimoto, K., et al. (1999). Role of Bcl-2 family proteins (Bax, Bcl-2 and Bcl-X) on cellular susceptibility to radiation in pancreatic cancer cells. Eur. J. Cancer 35 (9), 1374–1380. doi:10.1016/s0959-8049(99)00134-3
Liu, C. Y., Hsu, C. C., Huang, T. T., Lee, C. H., Chen, J. L., Yang, S. H., et al. (2018). ER stress-related ATF6 upregulates CIP2A and contributes to poor prognosis of colon cancer. Mol. Oncol. 12 (10), 1706–1717. doi:10.1002/1878-0261.12365
Liu, J., Fan, L., Yu, H., Zhang, J., He, Y., Feng, D., et al. (2019). Endoplasmic reticulum stress causes liver cancer cells to release exosomal miR-23a-3p and up-regulate programmed death ligand 1 expression in macrophages. Hepatology 70 (1), 241–258. doi:10.1002/hep.30607
Liu, W., Wang, D., Wang, X., Liu, P., and Yan, M. (2020). hsa_circ_0085539 promotes osteosarcoma progression by regulating miR-526b-5p and SERP1. Mol. Ther. Oncolytics 19, 163–177. doi:10.1016/j.omto.2020.09.009
Liu, X., Song, J., Zhang, H., Liu, X., Zuo, F., Zhao, Y., et al. (2023). Immune checkpoint HLA-E:CD94-NKG2A mediates evasion of circulating tumor cells from NK cell surveillance. Cancer Cell 41 (2), 272–287.e9. doi:10.1016/j.ccell.2023.01.001
Love, M. I., Huber, W., and Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome. Biol. 15 (12), 550. doi:10.1186/s13059-014-0550-8
Lucianò, A. M., Di Martile, M., Pérez-Oliva, A. B., Di Caprio, M., Foddai, M. L., Buglioni, S., et al. (2023). Exploring association of melanoma-specific Bcl-xL with tumor immune microenvironment. J. Exp. Clin. Cancer Res. 42 (1), 178. doi:10.1186/s13046-023-02735-9
Ma, Q., Wu, X., Wu, J., Liang, Z., and Liu, T. (2017). SERP1 is a novel marker of poor prognosis in pancreatic ductal adenocarcinoma patients via anti-apoptosis and regulating SRPRB/NF-κB axis. Int. J. Oncol. 51 (4), 1104–1114. doi:10.3892/ijo.2017.4111
Miwa, T., Kanda, M., Koike, M., Iwata, N., Tanaka, H., Umeda, S., et al. (2017a). Identification of NCCRP1 as an epigenetically regulated tumor suppressor and biomarker for malignant phenotypes of squamous cell carcinoma of the esophagus. Oncol. Lett. 14 (4), 4822–4828. doi:10.3892/ol.2017.6753
Miwa, T., Kanda, M., Tanaka, H., Tanaka, C., Kobayashi, D., Umeda, S., et al. (2017b). FBXO50 enhances the malignant behavior of gastric cancer cells. Ann. Surg. Oncol. 24 (12), 3771–3779. doi:10.1245/s10434-017-5882-7
Mizrahi, J. D., Surana, R., Valle, J. W., and Shroff, R. T. (2020). Pancreatic cancer. Lancet 395 (10242), 2008–2020. doi:10.1016/s0140-6736(20)30974-0
Neoptolemos, J. P., Kleeff, J., Michl, P., Costello, E., Greenhalf, W., and Palmer, D. H. (2018). Therapeutic developments in pancreatic cancer: current and future perspectives. Nat. Rev. Gastroenterol. Hepatol. 15 (6), 333–348. doi:10.1038/s41575-018-0005-x
Obiedat, A., Seidel, E., Mahameed, M., Berhani, O., Tsukerman, P., Voutetakis, K., et al. (2019). Transcription of the NKG2D ligand MICA is suppressed by the IRE1/XBP1 pathway of the unfolded protein response through the regulation of E2F1. Faseb J. 33 (3), 3481–3495. doi:10.1096/fj.201801350RR
O'Flaherty, S. M., Sutummaporn, K., Häggtoft, W. L., Worrall, A. P., Rizzo, M., Braniste, V., et al. (2017). TLR-stimulated eosinophils mediate recruitment and activation of NK cells in vivo. Scand. J. Immunol. 85 (6), 417–424. doi:10.1111/sji.12554
Pan, H., Yang, L., Bai, H., Luo, J., and Deng, Y. (2022). Ginsenoside Rg3 increases gemcitabine sensitivity of pancreatic adenocarcinoma via reducing ZFP91 mediated TSPYL2 destabilization. J. Ginseng Res. 46 (5), 636–645. doi:10.1016/j.jgr.2021.08.004
Park, D. J., Sung, P. S., Kim, J. H., Lee, G. W., Jang, J. W., Jung, E. S., et al. (2020). EpCAM-high liver cancer stem cells resist natural killer cell-mediated cytotoxicity by upregulating CEACAM1. J. Immunother. Cancer 8 (1), e000301. doi:10.1136/jitc-2019-000301
Pommier, A., Anaparthy, N., Memos, N., Kelley, Z. L., Gouronnec, A., Yan, R., et al. (2018). Unresolved endoplasmic reticulum stress engenders immune-resistant, latent pancreatic cancer metastases. Science 360 (6394), eaao4908. doi:10.1126/science.aao4908
Rossi, F. A., Enriqué Steinberg, J. H., Calvo Roitberg, E. H., Joshi, M. U., Pandey, A., Abba, M. C., et al. (2021). USP19 modulates cancer cell migration and invasion and acts as a novel prognostic marker in patients with early breast cancer. Oncogenesis 10 (3), 28. doi:10.1038/s41389-021-00318-x
Rossi, F. A., and Rossi, M. (2022). Emerging role of ubiquitin-specific protease 19 in oncogenesis and cancer development. Front. Cell Dev. Biol. 10, 889166. doi:10.3389/fcell.2022.889166
Rufo, N., Garg, A. D., and Agostinis, P. (2017). The unfolded protein response in immunogenic cell death and cancer immunotherapy. Trends Cancer 3 (9), 643–658. doi:10.1016/j.trecan.2017.07.002
Schwarz, D. S., and Blower, M. D. (2016). The endoplasmic reticulum: structure, function and response to cellular signaling. Cell Mol. Life Sci. 73 (1), 79–94. doi:10.1007/s00018-015-2052-6
Sen Santara, S., Lee, D. J., Crespo, Â., Hu, J. J., Walker, C., Ma, X., et al. (2023). The NK cell receptor NKp46 recognizes ecto-calreticulin on ER-stressed cells. Nature 616 (7956), 348–356. doi:10.1038/s41586-023-05912-0
Sherman, M. H., and Beatty, G. L. (2023). Tumor microenvironment in pancreatic cancer pathogenesis and therapeutic resistance. Annu. Rev. Pathol. 18, 123–148. doi:10.1146/annurev-pathmechdis-031621-024600
Shi, C., Zhao, F., Zhang, T., Xu, D., Hao, Z., Cui, F., et al. (2022). A novel prognostic signature in osteosarcoma characterised from the perspective of unfolded protein response. Clin. Transl. Med. 12 (3), e750. doi:10.1002/ctm2.750
Siegel, R. L., Miller, K. D., and Jemal, A. (2020). Cancer statistics, 2020. CA Cancer J. Clin. 70 (1), 7–30. doi:10.3322/caac.21590
Sohal, D. P. S., Kennedy, E. B., Cinar, P., Conroy, T., Copur, M. S., Crane, C. H., et al. (2020). Metastatic pancreatic cancer: ASCO guideline update. J. Clin. Oncol. 38, 3217–3230. doi:10.1200/jco.20.01364
Song, M., Sandoval, T. A., Chae, C. S., Chopra, S., Tan, C., Rutkowski, M. R., et al. (2018). IRE1α-XBP1 controls T cell function in ovarian cancer by regulating mitochondrial activity. Nature 562 (7727), 423–428. doi:10.1038/s41586-018-0597-x
Song, P., Li, W., Guo, L., Ying, J., Gao, S., and He, J. (2022). Identification and validation of a novel signature based on NK cell marker genes to predict prognosis and immunotherapy response in lung adenocarcinoma by integrated analysis of single-cell and bulk RNA-sequencing. Front. Immunol. 13, 850745. doi:10.3389/fimmu.2022.850745
Sui, H., Dongye, S., Liu, X., Xu, X., Wang, L., Jin, C. Q., et al. (2022). Immunotherapy of targeting MDSCs in tumor microenvironment. Front. Immunol. 13, 990463. doi:10.3389/fimmu.2022.990463
Tadros, S., Shukla, S. K., King, R. J., Gunda, V., Vernucci, E., Abrego, J., et al. (2017). De novo lipid synthesis facilitates gemcitabine resistance through endoplasmic reticulum stress in pancreatic cancer. Cancer Res. 77 (20), 5503–5517. doi:10.1158/0008-5472.Can-16-3062
Terry, T., and Patricia, G. (2001). Modeling Survival Data: extending the Cox Model. Terry M. Therneau and Patricia M. Grambsch, Springer-Verlag, New York, 2000. No. of pages: xiii + 350. Price: $69.95. ISBN 0-387-98784-3. Statistics Med. 20 (13), 2053–2054. doi:10.1002/sim.956
Therneau, T. (2021). A package for survival analysis in R. R package version 3.2-13. Available at: https://cran.r-project.org/web/packages/survival/vignettes/survival.pdf.
Thummuri, D., Khan, S., Underwood, P. W., Zhang, P., Wiegand, J., Zhang, X., et al. (2022). Overcoming Gemcitabine Resistance in Pancreatic Cancer Using the BCL-X(L)-Specific Degrader DT2216. Mol. Cancer Ther. 21 (1), 184–192. doi:10.1158/1535-7163.Mct-21-0474
Wang, L., Yin, W., and Shi, C. (2017). E3 ubiquitin ligase, RNF139, inhibits the progression of tongue cancer. BMC Cancer 17 (1), 452. doi:10.1186/s12885-017-3438-7
Wei, T., and Simko, V. (2012). “R package ‘corrplot’: Visualization of a Correlation Matrix”. R package version 0.90 ed.
Wiseman, R. L., and Balch, W. E. (2005). A new pharmacology--drugging stressed folding pathways. Trends Mol. Med. 11 (8), 347–350. doi:10.1016/j.molmed.2005.06.011
Xiang, Y., and Zeng, J. (2019). AnnoProbe: annotate the gene symbols for probes in expression array. R package version 0.1.0. Available at: https://rdrr.io/cran/AnnoProbe/.
Xie, X., Ma, L., Zhou, Y., Shen, W., Xu, D., Dou, J., et al. (2019). Polysaccharide enhanced NK cell cytotoxicity against pancreatic cancer via TLR4/MAPKs/NF-κB pathway in vitro/vivo. Carbohydr. Polym. 225, 115223. doi:10.1016/j.carbpol.2019.115223
Yamaguchi, A., Hori, O., Stern, D. M., Hartmann, E., Ogawa, S., and Tohyama, M. (1999). Stress-associated endoplasmic reticulum protein 1 (SERP1)/Ribosome-associated membrane protein 4 (RAMP4) stabilizes membrane proteins during stress and facilitates subsequent glycosylation. J. Cell Biol. 147 (6), 1195–1204. doi:10.1083/jcb.147.6.1195
Yan, W., Wang, X., Liu, T., Chen, L., Han, L., Xu, J., et al. (2019). Expression of endoplasmic reticulum oxidoreductase 1-α in cholangiocarcinoma tissues and its effects on the proliferation and migration of cholangiocarcinoma cells. Cancer Manag. Res. 11, 6727–6739. doi:10.2147/cmar.S188746
Yuan, Y., Jiao, P., Wang, Z., Chen, M., Du, H., Xu, L., et al. (2022). Endoplasmic reticulum stress promotes the release of exosomal PD-L1 from head and neck cancer cells and facilitates M2 macrophage polarization. Cell Commun. Signal 20 (1), 12. doi:10.1186/s12964-021-00810-2
Zanetti, G., Pahuja, K. B., Studer, S., Shim, S., and Schekman, R. (2011). COPII and the regulation of protein sorting in mammals. Nat. Cell Biol. 14 (1), 20–28. doi:10.1038/ncb2390
Zeng, H., Chen, W., Zheng, R., Zhang, S., Ji, J. S., Zou, X., et al. (2018). Changing cancer survival in China during 2003-15: a pooled analysis of 17 population-based cancer registries. Lancet Glob. Health 6 (5), e555–e567. doi:10.1016/s2214-109x(18)30127-x
Zhang, J., Yang, J., Lin, C., Liu, W., Huo, Y., Yang, M., et al. (2020). Endoplasmic Reticulum stress-dependent expression of ERO1L promotes aerobic glycolysis in Pancreatic Cancer. Theranostics 10 (18), 8400–8414. doi:10.7150/thno.45124
Zhang, Q., Guan, G., Cheng, P., Cheng, W., Yang, L., and Wu, A. (2021). Characterization of an endoplasmic reticulum stress-related signature to evaluate immune features and predict prognosis in glioma. J. Cell Mol. Med. 25 (8), 3870–3884. doi:10.1111/jcmm.16321
Zhang, Y., Li, T., Zhang, L., Shangguan, F., Shi, G., Wu, X., et al. (2019). Targeting the functional interplay between endoplasmic reticulum oxidoreductin-1α and protein disulfide isomerase suppresses the progression of cervical cancer. EBioMedicine 41, 408–419. doi:10.1016/j.ebiom.2019.02.041
Zhang, Y., Song, Y., Dai, J., Wang, Z., Zeng, Y., Chen, F., et al. (2022). Endoplasmic Reticulum Stress-Related Signature Predicts Prognosis and Drug Response in Clear Cell Renal Cell Carcinoma. Front. Pharmacol. 13, 909123. doi:10.3389/fphar.2022.909123
Zhou, J., Qian, W., Huang, C., Mai, C., Lai, Y., Lin, Z., et al. (2022). Combined targeting of KRT23 and NCCRP1 as a potential novel therapeutic approach for the treatment of triple-negative breast cancer. Gland. Surg. 11 (10), 1673–1682. doi:10.21037/gs-22-486
Keywords: ER stress, immune microenvironment, prognosis, bioinformatics, pancreatic cancer
Citation: Chen H, Xu N, Xu J, Zhang C, Li X, Xu H, Zhu W, Li J, Liang D and Zhou W (2023) A risk signature based on endoplasmic reticulum stress-associated genes predicts prognosis and immunity in pancreatic cancer. Front. Mol. Biosci. 10:1298077. doi: 10.3389/fmolb.2023.1298077
Received: 21 September 2023; Accepted: 15 November 2023;
Published: 29 November 2023.
Edited by:
Li Chen, Huazhong University of Science and Technology, ChinaReviewed by:
Boxuan Zhou, Third Affiliated Hospital of Sun Yat-sen University, ChinaYancheng Song, Qingdao University, China
Copyright © 2023 Chen, Xu, Xu, Zhang, Li, Xu, Zhu, Li, Liang and Zhou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jinze Li, 1576328807@qq.com; Daoming Liang, daomingliangkm@sina.com; Wence Zhou, zhouwc129@163.com
†These authors share first authorship
 Haofei Chen
Haofei Chen Ning Xu3†
Ning Xu3†  Xin Li
Xin Li Weixiong Zhu
Weixiong Zhu Jinze Li
Jinze Li Wence Zhou
Wence Zhou