Cardiolipin occupancy profiles of YidC paralogs reveal the significance of respective TM2 helix residues in determining paralog-specific phenotypes
- 1Department of Oral Biology, University of Florida, Gainesville, FL, United States
- 2Department of Chemistry and Biochemistry, Texas Tech University, Lubbock, TX, United States
YidC belongs to an evolutionarily conserved family of insertases, YidC/Oxa1/Alb3, in bacteria, mitochondria, and chloroplasts, respectively. Unlike Gram-negative bacteria, Gram-positives including Streptococcus mutans harbor two paralogs of YidC. The mechanism for paralog-specific phenotypes of bacterial YidC1 versus YidC2 has been partially attributed to the differences in their cytoplasmic domains. However, we previously identified a W138R gain-of-function mutation in the YidC1 transmembrane helix 2. YidC1W138R mostly phenocopied YidC2, yet the mechanism remained unknown. Primary sequence comparison of streptococcal YidCs led us to identify and mutate the YidC1W138 analog, YidC2S152 to W/A, which resulted in a loss of YidC2- and acquisition of YidC1-like phenotype. The predicted lipid-facing side chains of YidC1W138/YidC2S152 led us to propose a role for membrane phospholipids in specific-residue dependent phenotypes of S. mutans YidC paralogs. Cardiolipin (CL), a prevalent phospholipid in the S. mutans cytoplasmic membrane during acid stress, is encoded by a single gene, cls. We show a concerted mechanism for cardiolipin and YidC2 under acid stress based on similarly increased promoter activities and similar elimination phenotypes. Using coarse grain molecular dynamics simulations with the Martini2.2 Forcefield, YidC1 and YidC2 wild-type and mutant interactions with CL were assessed in silico. We observed substantially increased CL interaction in dimeric versus monomeric proteins, and variable CL occupancy in YidC1 and YidC2 mutant constructs that mimicked characteristics of the other wild-type paralog. Hence, paralog-specific amino acid- CL interactions contribute to YidC1 and YidC2-associated phenotypes that can be exchanged by point mutation at positions 138 or 152, respectively.
Introduction
Approximately one-third of all proteins are translocated to various biological membranes to support essential functions such as molecular transport, signaling, and energy transduction. In bacteria, protein translocation to the cytoplasmic membrane occurs through a heterotrimeric proteinaceous channel, SecYEG (Van den Berg et al., 2004). An accessory membrane protein, YidC, functions with SecYEG in the insertion of most of the membrane proteins (Beck et al., 2001; Urbanus et al., 2001; Sachelaru et al., 2017), except for a few cases in which smaller polypeptides with only one or two transmembrane (TM) helices are inserted by YidC in a SecYEG-independent manner (reviewed in (Steinberg et al., 2018)). YidC belongs to the YidC/Oxa1/Alb3 family of proteins that are ubiquitously present in all three domains of life and localized within bacterial, mitochondrial, and chloroplast membranes, respectively (reviewed in (Lewis and Brady, 2015)). Gram-negative bacteria contain a single YidC, whose deletion is lethal, while most Gram-positive bacteria contain two YidC paralogs. These were originally denoted YidC1 and YidC2 in the cariogenic dental pathogen Streptococcus mutans (Hasona et al., 2005). The presence of at least one YidC paralog is necessary to support the growth and survival of Gram-positive bacteria; however, different phenotypic consequences are observed upon the elimination of each one suggesting discrete functional activities (Murakami et al., 2002; Tjalsma et al., 2003; Hasona et al., 2005; Dong et al., 2008; Palmer et al., 2012; Palmer et al., 2019; Mishra and Brady, 2021). Paralog-specific protein-protein interactions and individual physiological roles have been identified for YidC homologs of eukaryotic organelles as well (Woolhead et al., 2001; Jia et al., 2003; Fiumera et al., 2007; Benz et al., 2009; Falk et al., 2010). Hence, sequence-structure features that impart distinct functional partitioning of bacterial YidC paralogs are of considerable interest.
Phylogenetic analysis of various yidC1 and yidC2 sequences suggests their origin stems from an independent gene duplication event that occurred early during evolution and likely led to the divergent amino acid sequences observed across the spectrum of Gram-positive YidC paralogs (Funes et al., 2009). Despite variation in amino acid sequences, the predicted 3-dimensional (3D) structures of representative Gram-positive YidC1 and YidC2 proteins closely resemble the resolved crystal structures of Bacillus halodurans YidC2 and the core domain of Escherichia coli YidC (Kumazaki et al., 2014a; Kumazaki et al., 2014b). For all bacterial YidCs, the core domain consists of five TM helices interspersed with two distinct cytoplasmic loops and ending with a C-terminal cytoplasmic tail (Figure 1, top). Notably, YidC2 of Gram-positive bacteria, mitochondrial Oxa1, and chloroplast Alb3 all contain a longer C-terminal tail compared to YidC1, Oxa2, and Alb4, respectively. Previous work from our laboratory showed the functional significance of the longer and more positively charged C-terminal tail of S. mutans YidC2 in that partial rescue of the signature acid and salt intolerance characteristic of a ΔyidC2 mutant was demonstrated following the introduction of chimeric YidC1 in which the YidC2 tail had been appended (Palmer et al., 2012). Furthermore, swapping YidC1 and YidC2 C1 and C2 cytoplasmic loops, singly or in combination with their tails, also revealed contributions to paralog-specific phenotypes (Mishra and Brady, 2021). This is not surprising based on multiple sequence alignment (MSA) of S. mutans YidC1 and YidC2 amino acid sequences that reveal that cytoplasmic domains are less well-conserved compared to their respective TM helices (Supplementary Figure S1). Acid sensitivity of the ΔyidC2 strain was also reversed by a spontaneous YidC1W138R gain of function mutation within the highly conserved TM2 helix (Mishra et al., 2019). This mutation within yidC1 was reproducibly observed following serial passage of the ΔyidC2 strain. The underlying mechanism for the intriguing acquisition of YidC2-like behavior by YidC1 following a single point mutation within TM2 led us to explore this question further.
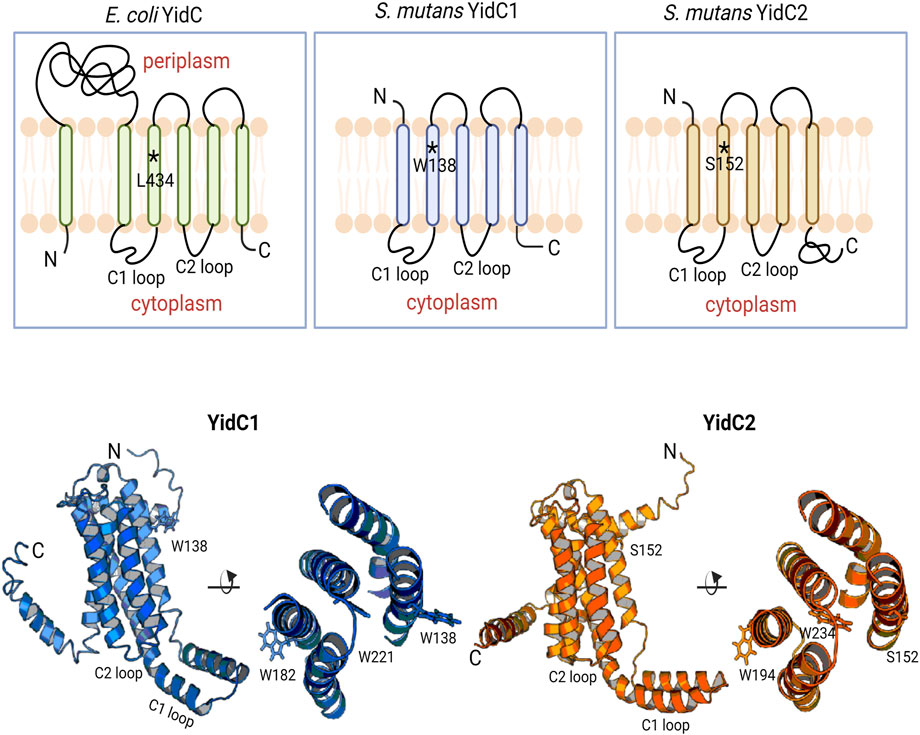
FIGURE 1. Schematic representation of bacterial YidCs’ topology and structure. Schematic representation of Escherichia coli YidC membrane topology compared to S. mutans YidC1 and YidC2. Side (left) and top down (right) views of predicted tertiary structures of S. mutans YidC1 (blue) and YidC2 (orange). Locations of relevant amino acid residues, and C1 and C2 cytoplasmic loops, are indicated.
MSA of various Gram-positive YidC1 sequences represented as sequence logo plots further reveals that while TM2 is generally well-conserved, residue W138 itself is not conserved among different species’ YidC1s (Supplementary Figure S2) (Crooks et al., 2004). In contrast, MSA of various Gram-positive YidC2 sequences showed the analogous position within their TM2s to be more highly conserved and usually occupied by serine or methionine (Supplementary Figure S2). This represents S152 in S. mutans YidC2, and the analogous position is occupied by serine in other streptococcal and enterococcal YidC2 as well. The analogous residue in E. coli YidC is L434. While YidCL434S is able to support the growth of an E. coli YidC depletion strain, impaired insertase activity was observed in vitro (Jiang et al., 2003). This result further highlights the significance of this specific position on bacterial YidC function. Studies of the abundance of amino acids within TM helices suggest that Trp, Ser, Arg, and Leu are all favored in terms of maintaining helix integrity, although positional conservation can vary due to respective location relative to the bilayer mid-plane (Mbaye et al., 2019).
I-TASSER predicted 3D structures of each S. mutans insertase illustrate the potential positional relevance of YidC1 W138 and YidC2 S152 (Figure 1, bottom). Both residues appear to be positioned away from the membrane mid-plane with their side chains facing outward into the lipid bilayer. We therefore hypothesized a contributory role of membrane lipids in YidC1W138R - and YidC2S152 - associated growth phenotypes. The lipid composition of the S. mutans cytoplasmic membrane was previously characterized by extensive lipidomic analysis carried out in our laboratory (Morales-Aparicio et al., 2020). Cardiolipin (CL) was identified as the predominant phospholipid (60%) in stationary phase cells followed by other anionic phospholipids including phosphatidylinositol (PI), phosphatidylserine (PS), and phosphatidylglycerol (PG). Increased bacterial CL content following acid- or osmotic-stress (Romantsov et al., 2009; MacGilvray et al., 2012; Woodall et al., 2021), as well as increased sensitivity of S. mutans lacking cls, the sole cardiolipin synthase encoding gene (MacGilvray et al., 2012), have been reported. Although, the mechanisms of CL- and YidC2-dependent protection of S. mutans from acid stress are not well understood, the findings that CL and YidC2 are both needed to support S. mutans growth at low pH suggests that they may work in concert.
Herein, we demonstrate an increase in cls and yidC2, but not yidC1, promoter activity upon exposure to acid and envelope stress suggesting the coordinate roles of CL and YidC2 in protection from environmental stressors. We also modeled YidC1 and YidC2 interactions with anionic phospholipids, specifically CL, PI, and PS, using a Martini2.2 Forcefield based coarse grain molecular dynamics (CGMD) approach to understand their potential contributions to paralog-specific phenotypic attributes of S. mutans. CGMD has proven to be an excellent tool in characterizing protein dynamics (Mustafa et al., 2015; Poma et al., 2017; Damre et al., 2019), oligomerization (Johnston et al., 2012; Prasanna et al., 2014; Pluhackova et al., 2016; Song et al., 2021), and lipid interactions (Ans et al., 2020; Duncan et al., 2020; Yekefallah et al., 2022). Our CGMD analyses demonstrated a preference for dimer formation by both S. mutans YidC paralogs, and their substitution variants, and the occupancy map of the YidC1W138R dimer showed a significant gain in CL residency within multiple TM helix residues similar to that observed for the YidC2 dimer. Furthermore, mutation of YidC2 S152 to W not only phenocopied elimination of yidC2, but an overall decrease in CL occupancy was observed for the YidC2S152W dimer. These results reveal the positional significance of the respective W138 and S152 TM2 residues in conferring YidC1 versus YidC2 paralog-specific phenotypic properties by virtue of characteristic interactions with a prevalent membrane phospholipid such as CL.
Results
Expression of yidC1W138R phenocopies expression of yidC2
Previous characterization of S. mutans ΔyidC1 and ΔyidC2 deletion mutants revealed severe impairment of growth of the ΔyidC2 strain upon exposure to acid, osmotic, and oxidative stress (Hasona et al., 2005; Hasona et al., 2007; Funes et al., 2009). However, the ΔyidC2 mutant reproducibly resolved acid-sensitivity by acquiring a point mutation, W138R, in the TM2 domain of YidC1 (Mishra et al., 2019). Unlike deletion of yidC2, deletion of yidC1 does not manifest in sensitivity to environmental stress except under conditions of zinc-excess. Since, yidC1W138R expression conferred tolerance to acid-stress in a ΔyidC2 background, we hypothesized that this strain might show altered sensitivity to Zn-excess. We compared growth of S. mutans wild-type (WT), ΔyidC1, and ΔyidC2 strains, as well as strains expressing yidC1W138R or yidC1W138A in a double ΔyidC1/yidC2 background, under non-stress, acid stress, and Zn-excess conditions (Figure 2). As expected, deletion of yidC2 resulted in impaired growth and pronounced acid sensitivity. Growth of the ΔyidC2 strain under non-stress conditions was restored by expression of yidC1W138R, and growth at pH 5 was restored by expression of either yidC1W138R or yidC1W138A. Deletion of yidC1 impacted S. mutans growth in THYE containing 2.5 mM ZnCl2 far more than deletion of yidC2, and zinc tolerance was restored by expression of yidC1W138R but not by yidC1W138A. These results highlight the importance of S. mutans YidC1 residue 138 under several biologically relevant environmental conditions and demonstrate that expression of yidC1W138R phenocopies expression of yidC2 when either is present as the sole functional yidC.
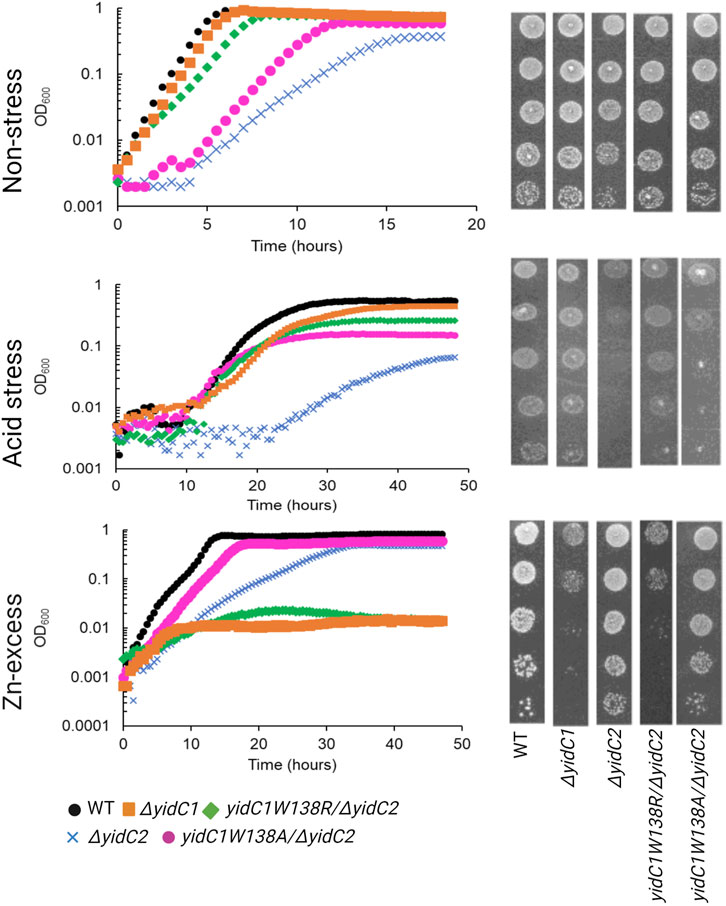
FIGURE 2. Comparison of growth of S. mutans wild-type and mutant strains under non-stress, acid-stress, and zinc-excess conditions. Growth of each indicated strain was monitored under varying environmental conditions in liquid culture (left) and by efficiency of plating on agar (right).
CL and YidC2 function in concert to protect S. mutans from acid and envelope stress
Often, genes that contribute to the same biological pathways will manifest similar phenotypes upon deletion. To test the degree to which a S. mutans Δcls mutant shares known ΔyidC2- or ΔyidC1-associated properties, WT, Δcls, ΔyidC1, and ΔyidC2 strains were compared for their ability to tolerate low pH, high salt, and Zn-excess conditions (Figure 3A). In addition to impaired growth at pH 5, the Δcls mutant demonstrated pronounced sensitivity to 3% NaCl, another ΔyidC2-associated phenotype. The Δcls strain also acquired moderate sensitivity to 2.5 mM ZnCl2, but resembled ΔyidC2 more than ΔyidC1 in this regard (Figure 3A).
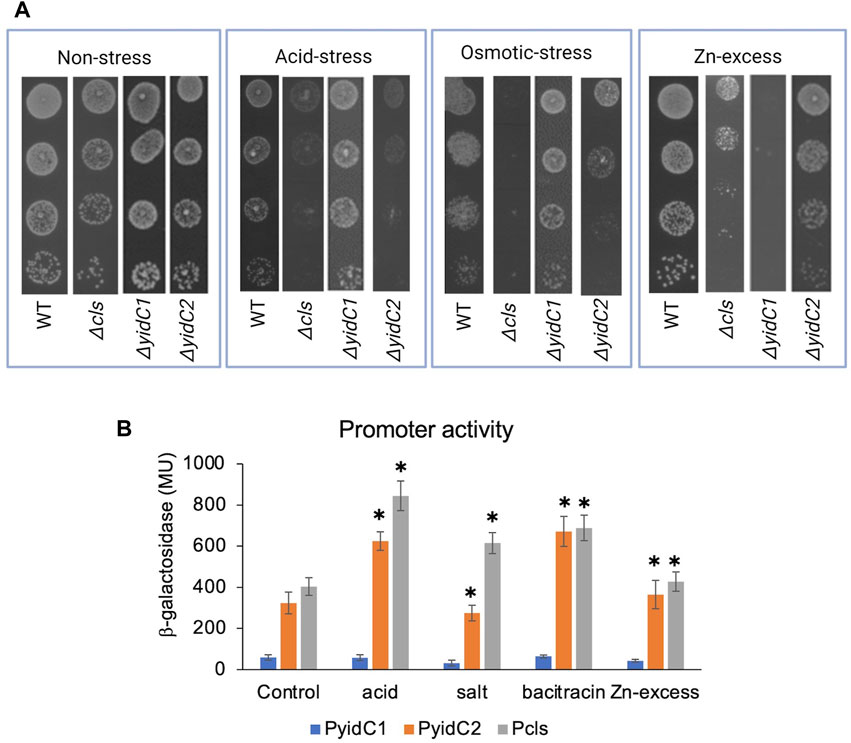
FIGURE 3. Effect of environmental stressors on growth and promoter activities of yidC1, yidC2, and cls. (A) Growth of each indicated strain was monitored under varying environmental by efficiency of plating on agar. (B) Promoter activities in mid-exponential phase cultures were measured following 45 min exposure to various environmental stressors. β-galactosidase activity was expressed in Miller Units (MU). The results are representative of three independent experiments performed in duplicate. *Significant difference (p-value <0.005), compared to untreated in each case (determined from Student’s t-test).
To further evaluate whether YidC2 and CL act in concert to protect against environmental stress, we hypothesized that yidC2 and cls would be transcriptionally upregulated by identical stressors. To test this question, promoter-reporter gene fusions of yidC1, yidC2, and cls were constructed. The genetic context of yidC1 is well conserved among Gram-positive bacteria with yidC1 in an operon with the gene encoding the protein subunit of ribonuclease P (rnpA). In S. mutans, 17 bp of rnpA, including the stop codon, overlaps with yidC1. The start codon of the downstream gene, jag, is located 12 bp from the stop codon of yidC1. Bacterial promoter prediction tools (BPROM) identified two distinct −10 and −35 sites upstream of yidC1 (Solovyev and Salamov, 2011). One set of −10 and −35 boxes is located 256 and 276 bp upstream of yidC1 and within the rnpA gene, while a second set is located 48 and 67 bp upstream of rnpA. No transcription termination sites were identified between rnpA, yidC1, and jag suggesting these genes comprise an operon. Based on these predictions, the 470 bp promoter region upstream of the yidC1 start site was used to construct a yidC1 promoter-lacZ fusion construct. Unlike yidC1, the genomic context of yidC2 is not conserved. In S. mutans, yidC2 is present 91 bp downstream of an acylphosphatase encoding gene, acp, and ∼400 bp upstream of the translation elongation factor, greA, which is oriented opposite to yidC2. The genetic context of yidC2 suggests it is likely transcribed from its own promoter. A−10 (TAAAAT) and a −35 (ATTGAGT) box were predicted 40 and 64 bp upstream of the yidC2 start site, respectively, and a 283 bp segment of the yidC2 promoter was used to make its lacZ promoter-reporter construct. Previous work from the Quivey lab identified the cls transcriptional start site 5’ to the upstream gene, wapA (MacGilvray et al., 2012). A 300 bp segment upstream of wapA was used to make the cls promoter-lacZ reporter construct. All three promoter-reporter gene fusions were integrated in a single copy at the phnA-mtlA2 locus within the S. mutans UA159 chromosome.
To determine the impact of varying environmental stressors on yidC1, yidC2, and cls expression, β-galactosidase activities were measured in response to acid stress (pH 5), osmotic stress (3% NaCl), Zn-excess (2.5 mM ZnCl2), and cell envelop stress (250 μg mL-1 bacitracin) conditions and compared with exponential growth-phase expression for each respective promoter (Figure 3B). Acid stress and bacitracin-induced envelope stress led to ∼2-fold induction of yidC2 and cls, while yidC1 promoter activity remained unchanged under all environmental stress conditions tested. Osmotic stress and Zn-excess had no measurable effect on the activity of any of the promoters. These results are consistent with the hypothesis that YidC2 and CL likely function together to enable acid and envelope stress tolerance. PI and PS biosynthetic pathways have not been characterized in S. mutans or other streptococci. Therefore, mutant strains are not available to explore potential phenotypic overlap between ΔyidC1 and ΔyidC2 strains and mutants lacking anionic phospholipids other than CL.
YidC1 and YidC2 likely function as dimers in a CL-rich membrane
Due to challenges in adequate expression and purification of recombinant YidC1 and YidC2 proteins in sufficient quantity for lipid-protein interaction studies, we used coarse grain molecular dynamics (CGMD) simulations to study the impact of membrane lipids on S. mutans YidC paralogs in our current study. CL, PI, and PS together comprised ∼90% of the total phospholipid content of cytoplasmic membrane samples prepared from S. mutans cells harvested from stationary phase cultures (Morales-Aparicio et al., 2020). CL, PI, and PS are anionic phospholipids with negatively charged polar head groups (Supplementary Figure S3). We modeled CL, PI, and PS at a 3:1:1 molar ratio mimicking the S. mutans lipid bilayer. To start, monomeric YidC1 or YidC2 simulations containing the CL-rich tri-lipid mixture were analyzed to identify potential interactions between monomeric forms of YidC1 and YidC2 with the membrane lipids. A potential monomeric organization of YidC1 and YidC2 was evaluated first based on crystallographic data of various YidCs from B. halodurans (Kumazaki et al., 2014a; Kumazaki et al., 2014c), E. coli (Kumazaki et al., 2014b), and Thermotoga maritima (Xin et al., 2018; Nass et al., 2022). Here, and for simulations described below, single systems were built using the CHARMM-GUI (Jo et al., 2008; Lee et al., 2016) Martini maker (Qi et al., 2015; Hsu et al., 2017) and were energy minimized and equilibrated in the Groningen Machine for Chemical Simulation (GROMACS) (Abraham et al., 2015). After equilibration, systems were cloned into 5 replicates (Knapp et al., 2018) to ensure equivalent starting energy profiles, and production was carried out for a total of 10 μs per simulation. The average RMSD of Cα-atoms provides insight into system stability for a given protein throughout the simulation. All systems that we assessed were observed to quickly reach equilibrium within ∼2 Å Cα RMSD compared to the final snapshot of respective equilibration steps (Supplementary Figure S4). PyLipID was then used to calculate lipid residency times by residue (Song et al., 2022). Lastly, we calculated the RMSF values for each amino acid residue during the course of the simulation of YidC1 and YidC2 in the 3CL:PI:PS mixture to understand environment-dependent protein dynamics (Supplementary Figure S5). RMSF values represent the extent of fluctuation for each residue around their average position. Interestingly, none of the YidC1 or YidC2 residues were significantly occupied (>3 μs) by CL in the simulated CL-rich bilayer (Supplementary Figure S6). Average RMSF values of residues comprising the cytoplasmic loops as well as the TM helices were less than 2 Å in both YidC1 and YidC2 monomers suggesting relatively inflexible structures. Collectively, our results support the notion that monomeric YidC1 and YidC2 do not represent the functional forms of the insertases in vivo. This observation is predicated upon the expectation that insertases exist in vivo as oligomers or in heteromeric complexes. Since CL interaction is likely related to function, and significant CL interaction is observed only for dimer simulations (discussed below), we conclude that insertase monomers are likely not functionally relevant units in a CL-rich environment.
We next tested whether the assembly of YidC1 or YidC2 as dimers within the simulated bilayer would change the potential lipid-interacting interfaces of the proteins. A dimeric YidC structure has been proposed in the literature based on bioinformatic analysis of an ancestral YidC-like protein as the progenitor of SecY (Lewis and Hegde, 2021), as well as experimental data on E. coli YidC and other Oxa1 family proteins, EMC3 and GET1 (Oliver and Paetzel, 2008; Ravaud et al., 2008; McDowell et al., 2020; O'Donnell et al., 2020; Pleiner et al., 2020). Recently a preprint supporting a functional dimeric pore-like structure of E. coli YidC, based on fluorescence correlation spectroscopy, Blue-Native PAGE, reconstitution of purified protein into vesicles, and single molecule microscopy, was published (Knyazev et al., 2023).
To model how dimeric YidC1 or YidC2 interact with membrane phospholipids, simulation systems containing the CL-rich tri-lipid mixtures were built around constructed homodimers of WT YidC1 and WT YidC2. Notably, the TM4 helices of S. mutans both YidC1 and YidC2 align with TM5 of E. coli YidC, which was proposed by Lewis and Hegde to be present at the dimer interface (Lewis and Hegde, 2021). Thus, dimer structures were built by cloning and placing a 180-degree rotated monomer respective to the original monomer with TM4-TM4 distance measured to be 7–8 Å using PyMol. Dimeric simulations were energy-minimized and equilibrated identically to monomeric simulations, and production was carried out for 10 μs prior to trajectory analysis. As with monomeric simulations, dimeric YidC1 and YidC2 displayed average RMSD values less than ∼2 Å for Cα-atoms when fit to the final equilibration frame of respective equilibration runs (Supplementary Figure S4). High CL densities were observed around multiple residues for both YidC1 and YidC2 dimeric structures (Figures 4A–C). Although the number of CL-occupying residues was comparable for YidC1 and YidC2, 38 and 32, respectively, CL occupied distinctly different locations within each dimer. Residues within TM1, TM4 and TM5 helices were strongly occupied by CL in dimeric YidC1 in comparison to multiple residues within TM2, TM3, and TM4 helices in dimeric YidC2. Notably, CL significantly occupied the TM4-TM4 dimer interface in both YidC1 and YidC2. In our simulations, cytoplasmic loop 1 residues at the junctions of TM1 and TM2 were significantly occupied by CL in the YidC1 dimer, but those sites were unoccupied in the YidC2 dimer. A lack of CL occupancy at the lipid-water interface in the TM1 and TM2 helices of YidC2 suggests a more flexible C1 loop in YidC2 compared to YidC1, which is supported by RMSF analysis (Supplementary Figure S5). In addition, RMSF analyses suggest structural plasticity in dimeric structures for both YidC1 and YidC2 when compared to their monomeric forms (Figures 4D, E). The coincident observations of increased dynamics and enhanced CL interaction as dimers is intriguing.
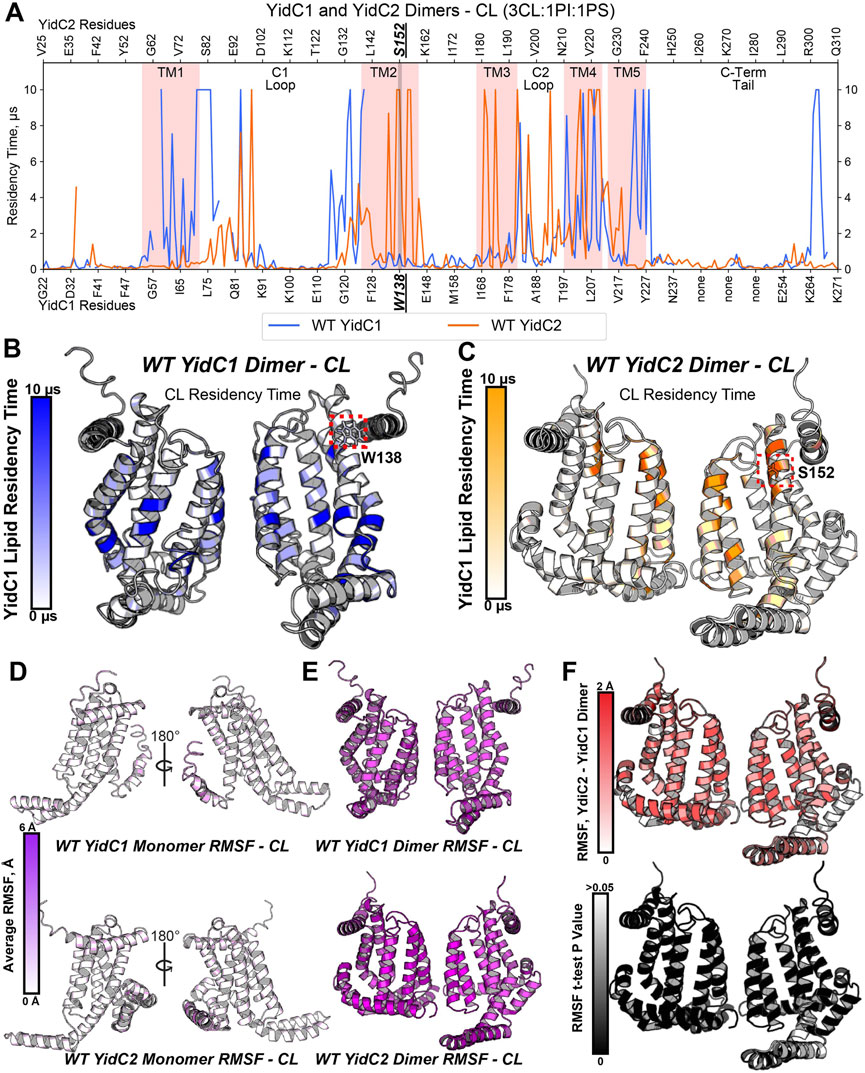
FIGURE 4. Phospholipid residency and molecular dynamics (RMSF) derived by coarse grain molecular dynamics (CGMD) analysis of YidC1 or YidC2 in simulated cardiolipin (CL)-rich bilayer. (A) Phospholipid residency plot of YidC1 and YidC2 dimer in CL-rich bilayer. Residency times of individual numbered residues of YidC1 (denoted at the bottom of each graph) or YidC2 (denoted at the top of each graph) are shown. Results indicate occupancy of CL when simulations were performed in a CL-rich (3CL:1PI:1PS). Pink shading indicates residues comprising transmembrane domains 1 through 5. Locations of the C1 and C2 cytoplasmic loops and the C-terminal cytoplasmic tail segments are also indicated. (B) CL residency plotted as heat map on YidC1 dimer 3D model structure, (C) CL residency plotted as heat map on YidC2 dimer 3D model structure. The intensity of color deepens with an increase in residency times. Dynamics of YidC1 and YidC2 (RMSF values) plotted as heat maps on modeled structure: (D) monomers, and (E) dimers. (F) p-values per residue of the RMSF differences between YidC2 and YidC1 dimers. T-test analyses were performed comparing RMSF values per residue from each simulation replicate of dimeric YidC2 to values derived from YidC1 simulations.
When assembled as dimeric forms in the CL-rich environment, differences in RMSF values between YidC1 and YidC2 were quite substantial. This information is illustrated by a YidC2 minus YidC1 heat map (Figure 4F). The YidC2 dimer appears more dynamic by approximately 1 Å compared to YidC1 for each aligned residue pair. Although a 1 Å RMSF difference in magnitude at a given individual position may not seem high, statistically significant differences (p<0.05) were observed for nearly every homologous residue. Collectively, these results reveal a level of structural flexibility for the YidC2 dimer in a CL-rich environment that is not as strongly shared by the YidC1 dimer. In addition, both YidC1 and YidC2 dimers acquired substantially increased CL occupancies compared to their respective simulated monomers, and the patterns of CL occupancy varied between the paralogs. A dimer-promoting role of CL was previously reported for SecYEG (Corey et al., 2018); therefore, it is not surprising that YidC1 and YidC2 both formed stable dimers in the simulated CL-rich environment. None of the YidC1 dimer residues showed significant PI or PS occupancy in the CL-rich tri-lipid mixture (Supplementary Figure S7). In contrast, the YidC2 dimer showed significant residency times with all three anionic phospholipids in the simulated bilayer (Supplementary Figure S7).
YidC1W138R and YidC2 dimers are similar in their interactions with CL
To understand how YidC1W138R might impact lipid interaction and protein dynamics, we also simulated this mutated protein in a CL-rich environment for 10 μs per replicate. Structure files were prepared by introducing the point mutation to the WT, membrane-aligned, YidC1 monomer or dimer model prior to system generation in CHARMM-GUI. When evaluated in either monomeric or dimeric forms in the simulated CL-rich bilayer, YidC1W138R became stable in <1 µs (Supplementary Figure S4). RMSF analysis and residue-wise comparison with the WT YidC1 dimer did not identify significant differences suggesting that the W138R point mutation did confer an overall dynamic structure like that observed for the YidC2 dimer. Both YidC1 and YidC1W138R dimers were less dynamic than the YidC2 dimer in the CL-rich environment (Supplementary Figure S5). Nevertheless, the W138R point mutation within TM2 of YidC1 resulted in a loss of CL interactions with multiple residues within TM1, and TM1-C1 loop and TM2-C1 loop junctions (Figure 5A). Lipid occupancy maps further showed gain of CL interactions with residues within TM2 and TM3, a feature characteristic of the YidC2 dimer (Figures 5A, B). Because it is negatively charged, CL would be expected to interact better with positively charged Arg, compared to hydrophobic Trp. This was evidenced by the increased CL residency time at R138 of the mutant YidC1 dimer compared to W138 of the wild-type YidC1 dimer. Notably, the lipid occupancy map of the YidC1W138R dimer acquired multiple similarities to that of the YidC2 dimer except at the C-terminal tail, which more closely resembled the YidC1 dimer (Figure 5A). Neither PI nor PS showed any significant occupancy with the YidC1W138R dimer (Supplementary Figure S8). Collectively, evaluation of lipid interactions with the varying YidC monomers and dimers reveals the significance of CL in the YidC1W138R acquisition of YidC2-like features, namely a gain in ability to support acid tolerance and a loss in tolerance to Zn-excess. Similar to monomeric YidC1 and YidC2, the YidC1W138R monomer likewise did not demonstrate significant lipid occupancies (Supplementary Figure S6), further supporting a dimeric structure as the functionally relevant form of YidC insertase paralogs.
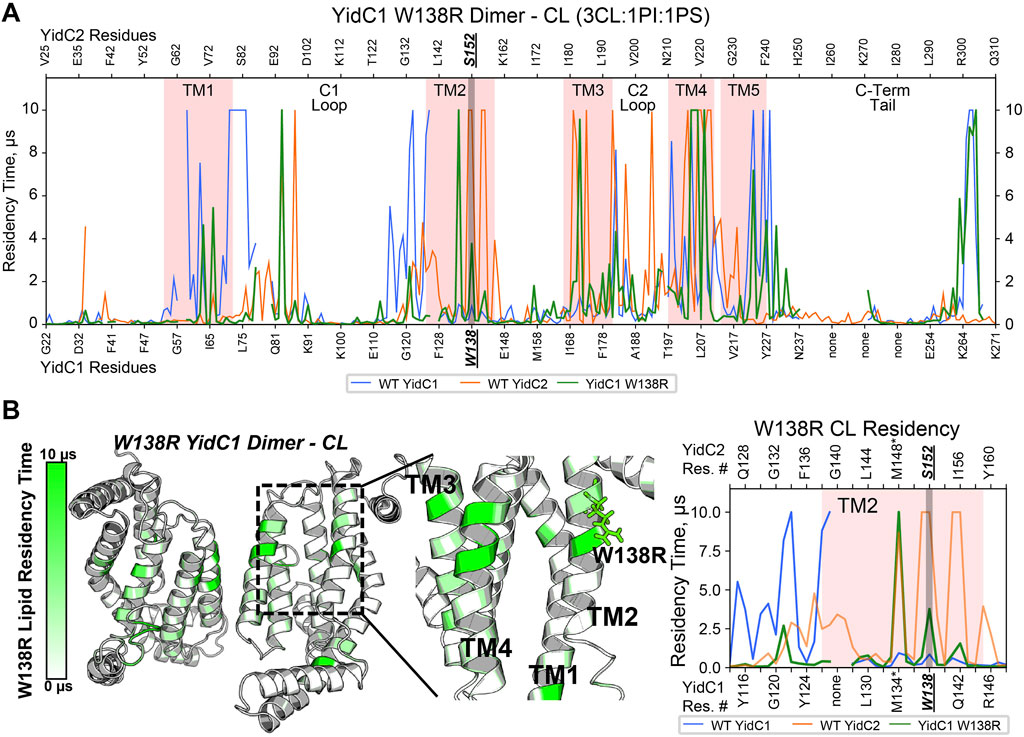
FIGURE 5. Comparison of CL occupancy of YidC1W138R dimer versus YidC1 and YidC2 dimers derived from CGMD in a simulated CL-rich bilayer. (A) Residency plot. Residency times of individual numbered residues of YidC1W138R (denoted at the bottom of each graph) or YidC2 (denoted at the top of each graph) are shown. Pink shading indicates residues comprising transmembrane domains 1 through 5. Locations of the C1 and C2 cytoplasmic loops and the C-terminal cytoplasmic tail segments are also indicated. (B) Heat map of CL residency of YidC1W138R dimer. Sites close to W138R mutation were zoomed in the heat map and residency plot.
Impact of YidC2 S152 point mutations on S. mutans phenotypic properties and modeled CGMD interactions with specific lipids
The acquisition of YidC2-associated behaviors by the YidC1W138R mutant prompted us to also engineer substitution mutations at position 152 of YidC2. These included S152W, S152A, and S152R mutations, with each gene variant produced as the sole yidC paralog in a ΔyidC1/yidC2 background. First, we assayed each strain for growth under non-stress conditions (Figure 6). As expected, the ΔyidC2 strain demonstrated an obvious growth defect compared to the ΔyidC1 strain. None of the amino acid substitutions at YidC2 position 152 conferred a severe growth defect of S. mutans in liquid culture, although growth of the YidC2S152A variant was somewhat more impacted than the YidC2S152R and YidC2S152W strains. Similar results were observed in the efficiency of plating assays. Importantly, when S. mutans growth was evaluated at pH 5, the YidC2S152A and YidC2S152W strains were both acid sensitive, as was the ΔyidC2 deletion mutant (Figure 6). This indicates that neither YidC1, nor YidC2S152A or YidC2S152W variants, can efficiently insert those substrates necessary for acid tolerance in S. mutans. In stark contrast, the YidC2S152R strain tolerated acidic pH even better than the ΔyidC1 strain that produces unmodified YidC2. That is, YidC2-mediated acid tolerance was improved by the YidC2S152R substitution. Again, results were recapitulated in efficiency of plating assays. Collectively, these results demonstrate the critical importance of residues 138 and 152 to YidC1 and YidC2s’ respective abilities to support acid tolerance, or not. Taken together, our results reveal that a Trp at this position is associated with acid sensitivity, while a Ser or Arg at this position enables the insertase to support acid tolerance as is observed for unmodified YidC2, YidC2S152R, and YidC1W138R.
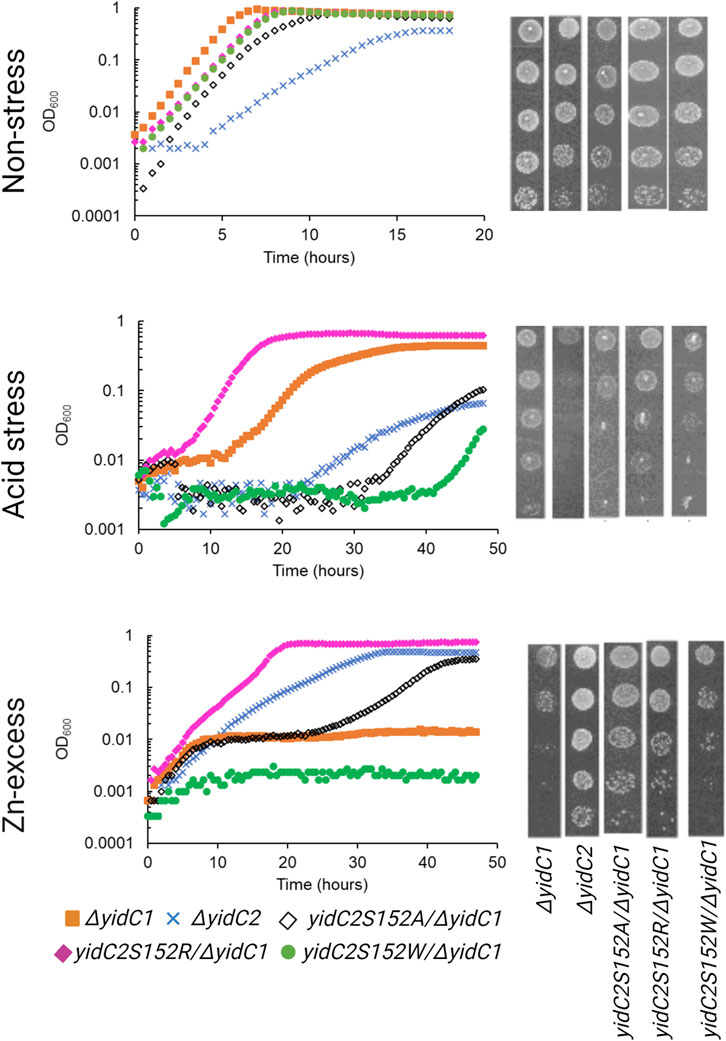
FIGURE 6. Comparison of growth of S. mutans YidC2 mutant strains with YidC1 and YidC2 harboring strains under non-stress, acid-stress, and zinc-excess conditions. Growth of each indicated strain was monitored under varying environmental conditions in liquid culture (left) and by the efficiency of plating on agar (right).
Lastly, S. mutans was grown in the presence of excess Zn (Figure 6). As expected, growth of the ΔyidC1 mutant was severely impaired in liquid culture and on agar plates. The YidC2S152W strain also acquired the severe ΔyidC1-associated zinc sensitivity phenotype. In other words, the YidC2S152W variant became YidC1-like. In addition, the YidC2S152A strain also became intolerant to excess zinc, though more so in liquid culture than on agar plates. Again, the result of the YidC2S152R mutation was opposite to that of the other amino acid substitutions tested. In this case, zinc tolerance was notably improved in liquid culture. When tested on agar plates, the YidC2S152W mutation was as severe as eliminating yidC1, but less impact was observed for YidC2S152R followed by YidC2S152A in efficiency of plating assays. Lack of complete concordance of results in liquid culture compared to those on agar plates could stem from differences in insertion efficiency or functionality of relevant substrates under conditions of varying aeration, or as yet unknown changes in lipid composition in the presence of excess zinc.
Because the yidC2S152W-expressing strain phenotypically resembled the strain expressing yidC1 only, we wanted to understand the impact of the S152W mutation on phospholipid interactions at the level of individual residues in comparison to dimeric unmodified YidC1 and YidC2. As with dimeric YidC1W138R simulations, YidC2S152W/R structure files were prepared by introducing the point mutation to the WT, membrane-aligned, YidC2 dimer model prior to system generation and production MD in the simulated CL/PI/PS tri-lipid bilayer. RMSD values equilibrated to <2.5 Å compared to the final snapshot of equilibration, indicating system stability (Supplementary Figure S4). After PyLipID analysis, the CL occupancy profile of YidC2S152W dimers clearly showed decreased intensity at all positions, including S152, compared to YidC1 or YidC2 dimers (Figures 7A, B; Supplementary Figure S9). The most apparent CL occupancy observed for the YidC2S152W dimer was at residues within the cytoplasmic C1 loop close to the TM1 interface. This would be expected to confer rigidity to the C1 loop region similar to that observed for the YidC1 dimer. PI occupancy for the YidC2S152W dimer followed a pattern generally similar to that of CL occupancy; however, PI occupancy was gained for residues within the C-terminal tail of the YidC2S152W mutant dimer (Supplementary Figure S9). Similar to results observed for CL occupancy, the YidC2S152W mutation also resulted in an overall loss of PS occupancy at numerous residues within the YidC2S152W dimer. This distinguished it from the unmodified YidC2 dimer with regard to phospholipid residency (Supplementary Figure S7). Comparison of RMSF values of YidC2S152W residues with those of the YidC2 dimer showed a ∼0.5–1 Å decrease in the RMSF values for approximately 2/3 of YidC2S152W residues (Figure 7C, left). Although these decreases in RMSF values in YidC2S152W were small compared to those in unmodified YidC2, they were statistically significant as determined by t-test indicating the S152W mutation resulted in a global decrease in YidC2 protein dynamics (Figure 7C, left, Supplementary Figure S5).
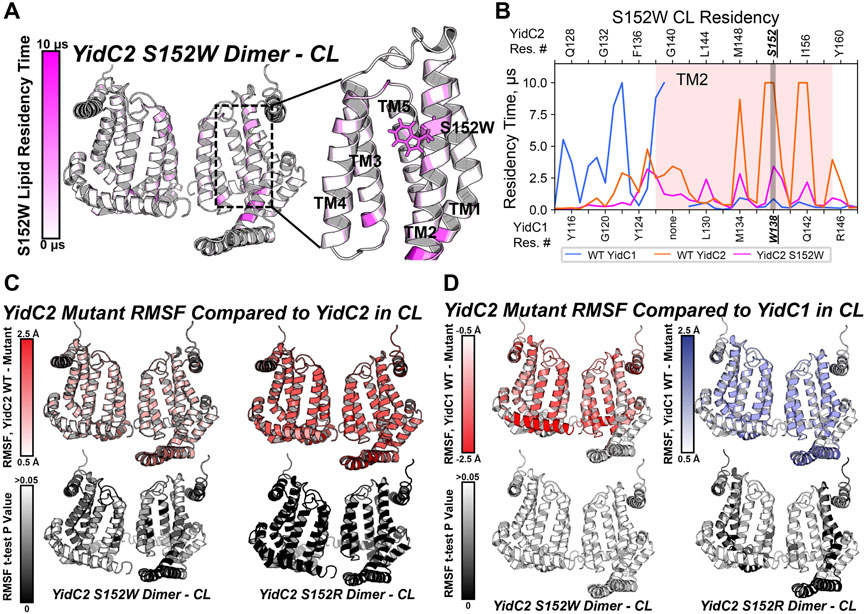
FIGURE 7. Comparison of mutant YidC2 CL interactions and dynamics, versus those of YidC1 and YidC2 dimers derived by CGMD in a simulated CL-rich bilayer. (A) Heat map showing CL occupancy of the YidC2S152W mutant zoomed in on the S152W region. (B) CL residency of TM2 helix residues presented as a plot illustrating that S152W severely reduced CL interaction at position 152 in YidC2. Dynamics of YidC2 mutants determined from RMSF values compared to WT YidC2 (C) and YidC1 (D) dimers, presented as heat maps (top). p-values of the RMSF differences per residue, compared to dimeric WT YidC2 (left) and YidC1 (right), are plotted as heat maps onto respective model structures. T-test analysis was performed comparing RMSF values per residue from each simulation replicate of respective datasets.
Phenotypic characterization of the panel of YidC2 point mutants showed that substitution of Arg for Ser at position 152 favored growth under all conditions tested. Similar to every YidC variant tested, YidC2S152R quickly equilibrated within the CL-rich membrane, and the CL occupancy profile of the YidC2S152R dimer was similar to that observed for the WT YidC2 dimer (Supplementary Figures S7, S9). However, S152R showed a lower CL residency time compared to S152. Residue-based RMSF analysis suggests an overall decrease in the dynamics of YidC2S152R in a CL-rich environment compared to the results for unmodified YidC2 (Figure 7C, right). Interestingly, YidC2S152W RMSF values compared to those of WT dimeric YidC1 are not statistically significant, which is taken as further supportive evidence that YidC2S152W phenocopies WT YidC1 (Figure 7D, left). In contrast, RMSF values of the YidC2S152R dimer suggest increased dynamics compared to unmodified dimeric YidC1 (Figure 7D, right). That is to say, the YidC2S152W dynamic profile was more similar to that of YidC1 than to that of WT YidC2, and the YidC2S152R dynamic profile was statistically significantly different from either WT YidC1 or WT YidC2. The latter observation was somewhat surprising since in other cases we were able to relate dynamic behavior observed in CGMD experiments to improved YidC2 functionality in vivo. An important caveat, however, is that our simulations were carried out in the absence of a confirmed substrate. Respective substrates of bacterial translocation systems, of which there are potentially hundreds, are largely unknown. Even in the most widely studied organism E. coli, only a handful of membrane-localized substrates are verified and utilized in model experimental systems.
Discussion
Although initially evolved by gene duplication events, paralogous proteins achieve distinct physiological roles due to differences in amino acid sequences, cellular localization, and genetic regulation. In this study, we identified distinct phospholipid-protein interaction profiles as another feature that delineates paralogous proteins’ functions in a case in which they are membrane localized. Paralogs of membrane proteins, though less common than their cytoplasmic counterparts, include many relevant examples in bacteria such as SecY/SecY2, MpfA/MpfA2, and various DedA paralogs. These paralogs offer interesting examples for further investigation in this regard.
In this study, we showed that S. mutans YidC paralogs demonstrate distinctly different CL residency profiles despite remarkable similarity in their 3D structures. Phospholipid-protein interactions are known to modulate spatial localization, oligomerization, and protein-protein interactions within multiprotein complexes (reviewed in (Corradi et al., 2018)). Hence, we postulate that differences in S. mutans YidC1 and YidC2 lipid occupancy can lead to distinct protein-protein interactions and oligomeric status thereby contributing to their distinct phenotypic properties. We identified a specific residue within the TM2 domain of YidC1 and YidC2, W138/S152, which significantly influences CL residency across all residues in YidC1 and YidC2 dimers in a Cl-rich bilayer. To the best of our knowledge, these results represent the first report of the positional significance of a single TM domain residue that is significantly responsible for paralogous membrane protein phenotypes, and that when appropriately altered mimics the reciprocal paralog by modulating phospholipid residency profiles.
To date the functional oligomeric status of the most widely studied bacterial YidC, that from E. coli, is still debated. Early reports predicted a dimeric structure (Lotz et al., 2008; Kohler et al., 2009); however, crystal structure proposed a monomeric organization for both E. coli and Bacillus halodurance YidC (Kumazaki et al., 2014a; Kumazaki et al., 2014b). In an interesting insight on the evolution of SecY, YidC was predicted as a dimer (Lewis and Hegde, 2021). Lastly, a recent manuscript preprint reported dimeric assembly of an E. coli YidC ion-conducting pore (Knyazev et al., 2023). Because their functional oligomeric status in the S. mutans cytoplasmic membrane is unknown, we modeled YidC1 and YidC2, and their substitution derivatives, both as monomers and as dimers. Although monomeric and dimeric forms of both paralogs were stable within the CL-rich tri-lipid environment modeled in our studies, stronger affinity with anionic phospholipids was observed for dimers compared to monomers in all cases. Approximately 10%–15% of residues were occupied by CL in both YidC1 and YidC2 dimers, in contrast to nearly none in their monomeric counterparts. The potential significance of CL-membrane protein interactions in the stabilization of dimeric structures has been reported for the SecYEG translocon, the Na+/H+ antiporter NhaA, and the multidrug efflux pump EmrE (reviewed in (Corradi et al., 2018)). It is highly likely, therefore, that S. mutans YidC1 and YidC2 also function as dimers within a CL-rich membrane.
Several studies using human glycophorin A (GpA) as a model protein have shown that a GxxxG motif within integral membrane protein TMs often leads to dimerization (Brosig and Langosch, 1998). An important feature of GxxxG, or similar AxxxG or SxxxG motifs, is the formation of interhelical carbon-hydrogen bonds that occur across helix-helix interfaces between Cα-H donors and backbone carbonyl (O=C) acceptors (Senes et al., 2000). The motif’s xxx residues can be occupied by 9 amino acids: Gly, Ala, Ser, Thr, Val, Leu, Iso, and Pro. These amino acids are preferred because each contains small side chains thereby avoiding steric hindrance and allowing protein backbone residues to be close enough for a helix-helix interaction to occur. A GxxxG motif (GAPAG) occurs within the TM4 helix of YidC2, and appears to be located at the dimer interface within the simulated CL-rich membrane.
The preference of S. mutans YidC2 in particular for dimer formation was also evident based on our observation of the dynamic behavior of YidC2 within the simulated CL-rich bilayer. Crystal structures of B. halodurans YidC2 and E. coli YidC have suggested that their C1 loops represent dynamic segments that are likely to be involved in substrate identification (Kumazaki et al., 2014a; Kumazaki et al., 2014b; Kumazaki et al., 2014c). When the E. coli YidC crystal structure (PDB ID: 3WVF) (Kumazaki et al., 2014b) was modeled in a simulated POPE:POPG (3:1) bilayer, compaction of the protein structure and bilayer thinning was observed in the vicinity of the YidC molecule (Kumazaki et al., 2014b; Chen et al., 2017). The structural compaction of E. coli YidC was proposed to stem from an interaction between its C1 loop residues and the hydrophilic head groups of PE and PG, thereby bringing the C1 loop closer to the membrane (Chen et al., 2017). Thus, the presence of lipids appeared to render the E. coli YidC C1 loop less flexible compared to that observed within crystal structures that were determined in the absence of lipids. In another instance, a cryo-EM structure of the E. coli YidC-ribosome complex within DPPG/DPPC liposomes suggested large conformational changes within YidC’s membrane-embedded core, especially within the TM2 and TM3 helices, which were connected by the C1 loop and extramembrane domain N-terminal to the TM2 helix (Kedrov et al., 2016). The structural flexibility of membrane-localized protein transport machinery components becomes more apparent in the presence of their cognate substrates. In fact, MD simulation of E. coli YidC-mediated insertion of the Pf3 coat protein predicted a conformational change within the insertase’s structure during substrate insertion consistent with a requirement for flexibility (Polasa et al., 2022). CGMD analysis of the S. mutans YidC1W138R dimer in a simulated CL-rich bilayer resembled that of the YidC2 dimer with regard to a lack of CL-TM1 and CL-C1 loop contacts, as well as gain of CL-contacts in TM3 and TM4 helices. This configuration may enable conformational flexibility of the cytoplasmic domains of the YidC1W138R dimer in a CL-rich environment, although this was not readily apparent in RMSF calculations. As stated, our simulations were performed in the absence of substrates. We propose to confirm the dynamic behavior of YidC1W138R and YidC2 dimers in future studies once YidC2-specific substrates are identified.
Trp, Ser, Ala, and Arg are all favored residues within TM helices (Liu et al., 2002). Therefore, YidC1’s W138R or YidC2’s152W/R mutations would not be expected to result in large scale changes in protein structure. However, a Trp residue’s location at the water-lipid interface, such as YidC1 W138 or modified YidC2 W152, could impart both aromatic-lipid acyl chain hydrophobic interactions and indole N-lipid headgroup hydrogen bonding. Serine is a non-charged amino acid with a polar side chain, which is favored in interfacial regions, forming hydrogen bonds with other polar residues within TM helices or with polar head groups of membrane phospholipids (Li et al., 2012). The large side chain of Arg is positively charged, and its orientation within a TM helix can allow formation of salt-bridges with negatively charged lipid head groups or with negatively charged amino acids. Thus, Trp, Ser, and Arg have unique lipid-interaction profiles that are dependent on their locations within a given protein present in a lipid bilayer and would be expected to modulate the structure-function of membrane proteins like S. mutans YidC1 and YidC2. In CGMD simulations of dimeric YidC variants, YidC1 W138 demonstrated little CL residency (0.9 µs), which increased to 3.8 μs upon mutation to R138. Conversely, YidC2 S152 CL residency (10 μs) was reduced by mutation to R (6.1 μs), and almost abolished by mutation to W (0.7 μs). As summarized in Figures 8A, B, YidC2 or YidC1 dimeric forms demonstrate substantial CL residency when the residue at position 152 or 138, respectively, is Ser or to a lesser extent Arg, but not when the residue is Trp. Orientation of these residues relative to the dimer interface may also contribute to CL residency time (Supplementary Figures S7–S9). It is likely that YidC1 W138 and YidC2 S152W residues are embedded and contact at the dimer interface, rather than extending into the bilayer, thereby mechanically shielding these residues from lipid residency. YidC2 152R/S, and to a lesser extent YidC1 138R/W, appear partially exposed, while YidC2 S152 is generally unencumbered giving rise to CL interaction throughout the entire simulation trajectory.
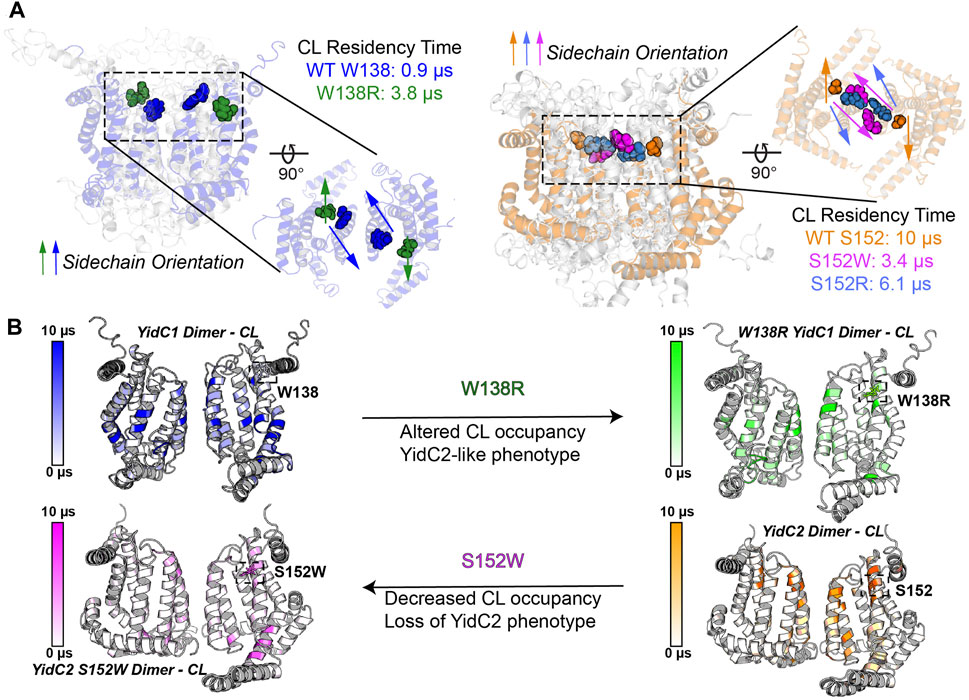
FIGURE 8. Schematic representation of the impact of YidC1 and YidC2 residues 138 and 152 on cardiolipin occupancy. (A) Sidechain orientation of indicated residues in the final CGMD trajectory frames is loosely predictive of CL residency time. Side and top-down views of YidC1 (left) and YidC2 (right) are illustrated. (B) Heat maps of CL occupancy derived by CGMD of YidC1 and YidC2 dimers compared to the corresponding W138R and S152W variants are shown. YidC1 W138R acquires YidC2-like properties, while YidC2 S152W loses CL occupancy and YidC2-associated functional activities.
Notably, Ser, Leu, and Arg are each encoded by six different genetic codons, the maximum observed for any amino acid. Trp is encoded by a single non-redundant genetic codon, TGG. Hence, any mutation to Trp, or from Trp to Ser/Arg/Leu, would be expected to have a substantial impact on the structure-function of a protein. Trp residues are often found in amphipathic helices thought to “slide” along the membrane during functional cycles of membrane proteins. Interestingly, multiple Trp residues (4–6) are common among Gram-positive YidC1 sequences, whereas YidC2 sequences contain only 2-3 Trp residues. S. mutans YidC1 contains five Trp residues (W34, W138, W157, W182, and W221), while YidC2 contains three (W37, W194, and W234). Of these, W34/37, W182/194, and W221/234 are conserved between the YidC1 and YidC2 paralogs, respectively. All Gram-positive YidCs contain W221 or W234 in TM5, which fall within the hydrophilic groove of YidC1 and YidC2, respectively. The analogous Tyr residue in E. coli YidC has been shown to be important for insertase activity (Chen et al., 2022). In contrast, W138 and W157 are unique to S. mutans YidC1 and are present in TM2 and the extramembrane domain that connects TM2 and TM3, respectively. The analogous positions within YidC2 are occupied by Ser and Gly, respectively, both of which contain small side chains and therefore, unlike aromatic residues, would not be expected to contribute to membrane anchoring. The presence of more Trp residues within YidC1 compared to YidC2 suggests that YidC1 is more firmly anchored within the membrane thereby conferring a more rigid structure. Substitution of Trp at position 138 of YidC1, and acquisition of Trp at position 152 of YidC2, would be expected to cause the former to gain flexibility and the latter to lose flexibility within a CL-rich membrane, consistent with their modified functional attributes. In a database analysis of membrane proteins related to human genetic diseases, a random mutation at Trp or Cys was found to have highest probability in translating to a disease state (Vitkup et al., 2003). The role of these residues in the structural stability of the proteins has been proposed to be the reason for the diseased state as a result of mutation. Phospholipid residency maps of WT and mutant proteins in a native-like simulated membrane are expected to provide important insights in this regard too.
Acid- and salt-sensitivity of the ΔyidC1 yidC2S152W -expressing strain is similar to that of the ΔyidC2 mutant that still expresses yidC1. Thus, a single amino acid residue at analogous positions within the TM2 helix of YidC1 and YidC2, respectively, confers paralog-specific phenotypic attributes. While the behavior of YidC2S152W and YidC2S152R mutants during growth in excess zinc is not explained by single amino acid substitution, this may stem from as yet unknown changes in cytoplasmic membrane lipid composition during growth in such an environment. The C1 loop of E. coli YidC was shown to contact SecY within the holo-translocon (Petriman et al., 2018), in addition to its contact with substrate (Kumazaki et al., 2014a). Our CGMD simulations did not include SecYEG or putative substrates; therefore, phenotypic characteristics of certain YidC2 point mutations are not yet fully understood and remain a future objective. Our previous studies have suggested that YidC1 generally functions as the housekeeping paralog in concert with SecYEG, while YidC2 functions in concert with the SRP pathway under conditions of acid, osmotic, and oxidative stress (Hasona et al., 2005; Lara Vasquez et al., 2021).
Taken together, the results of our CGMD simulations of interactions of S. mutans YidC1, YidC2, and variants thereof, with specific lipids within a CL-rich bilayer, coupled with phenotypic characterization of S. mutans harboring single YidC1 or YidC2 variants, has substantially advanced our understanding of how these two paralogs accomplish needed insertase activities under varying environmental conditions, and consequently varying membrane lipid compositions. Finally, this study has advanced our understanding of the coordinated roles played by CL and YidC2 in imparting tolerance to acid and osmotic stress, both defining characteristics of this highly resilient oral pathogen.
Materials and methods
Bacterial strains, plasmids, and growth conditions
Bacterial strains and plasmids used in this study are listed in Supplementary Table S1. All of the S. mutans mutants used in this study were derived from S. mutans strain UA159 (Ajdic et al., 2002), and were routinely grown in Todd-Hewitt broth (BBL, Becton Dickinson) supplemented with 0.3% yeast extract (THYE) at 37 °C in 5% CO2/95% air atmosphere (v/v). Spectinomycin (1 mg ml-1), kanamycin (1 mg ml-1), or erythromycin (10 μg ml-1) were added, where appropriate, for the growth and selection of S. mutans strains. E. coli strain C2987 (NEB) was used for genetic manipulations and was routinely grown in Luria–Bertani (LB) medium (10 g l-1 tryptone, 5 g l-1 yeast extract and 5 g l−1 NaCl) at 37°C with vigorous shaking or on LB agar (1.5%) with appropriate selection. Ampicillin (100 μg ml−1), or erythromycin (250 μg ml−1) were used for the growth and selection of E. coli transformants.
Site-directed mutagenesis of yidC1 and yidC2. A standard PCR-based mutagenesis technique was used for site-directed mutagenesis (Zheng et al., 2004). pSM15 and pSM20 (Mishra and Brady, 2021) were used as templates for the mutagenesis of yidC1 and yidC2 genes, respectively. Primers used for mutagenesis are listed in Supplementary Table S2.
Construction of S. mutans strains expressing single yidC1 or yidC2 variants
S. mutans strains were engineered to harbor the yidC1 or yidC2 variant of interest expressed under the regulatory control of the respective native promoter. Next, the residual yidC1 or yidC2 gene was deleted by allelic replacement with an antibiotic resistance cassette. Replacement cassettes consisting of the wild-type or mutant yidC1 or yidC2 gene were fused to an antibiotic resistance marker gene followed by the 3′-flanking regions of yidC1 or yidC2, respectively. To make the cassettes, PCR amplification products of the three regions were assembled by the NEB HiFi DNA assembly kit following the manufacturer’s instructions. The assembled products served as templates for PCR to generate gene the complete replacement cassettes using end primers. These were then used for transformation of S. mutans strain UA159. Once transformants were confirmed to harbor the desired yidC1 or yidC2 variant replacements, they were transformed with an allelic replacement construct that comprised the PCR-amplified 5′- and 3′-flanking regions of the residual yidC joined to an antibiotic resistance encoding gene. All mutations were confirmed by PCR and sequencing of the relevant genomic loci. Production of YidC1 or YidC2 and their derivatives was confirmed by Western blot using anti-C-terminal tail-specific rabbit antibodies (Mishra and Brady, 2021). Primers used for construction of deletion mutants are listed in Supplementary Table S2.
Construction of promoter-reporter gene fusions
Promoter-reporter gene fusions were constructed for yidC1, yidC2, and cls. For the yidC1 promoter, 470 bps of DNA located directly upstream of the yidC1 start codon, which included the promoter and partial sequence of rnpA was amplified using primers listed in Supplementary Table S2. For the yidC2 and cls promoter-reporter gene fusions, 289 and 300 bp segments upstream of yidC2 and wapA start codons were amplified, respectively. PCR products corresponding to promoters were subcloned between BamHI/SacI digested pMZ plasmid (Chen et al., 2002; Liu et al., 2009), which contains a promoterless lacZ gene derived from Streptococcus salivarius for integration within S. mutans genomic DNA by a double crossover event at mtlA-phnA locus. Promoter-lacZ fusion containing plasmids were transformed in S. mutans UA159 competent cells and the integrity of the gene fusions was verified by PCR amplification of the relevant locus followed by sequencing.
β-galactosidase assay
Exponential phase cultures (OD600 = 0.2) of promoter-reporter fusion strains grown in THYE were pelleted and resuspended in THYE pH 7.0 (control), THYE pH 5 (acid stress), THYE +3% NaCl (osmotic stress), THYE +2.5 mM ZnCl2 (Zn-excess), THYE +250 μg mL-1 bacitracin (cell envelope stress) for 45 min. Pellets were harvested and washed with Z-buffer ((60 mM Na2HPO4, 40 mM NaH2PO4, 10 mM KCl, 1 mM MgSO4, freshly-added 50 mM β-mercaptoethanol), and β-galactosidase activity was determined and expressed as described previously (Seaton et al., 2015).
Growth and stress tolerance assay in broth and on agar plates
Overnight cultures grown in THYE broth with appropriate selection agents were diluted 1:50 in THYE and grown to mid-exponential phase (OD600 = 0.2–0.4). Following 1:100 dilution of mid-log phase cultures, bacterial growth was measured using a Bioscreen C instrument as described (Mishra and Brady, 2021). Wells of the honeycomb plates used in the Bioscreen C instrument were covered with ∼50 μl of heavy mineral oil to achieve anaerobic growth under non-stress and acid stress conditions, except under conditions of Zn-excess in which mineral oil was not used because zinc toxicity is only observed under aerobic conditions. For efficiency of plating assays, mid-exponential phase cultures were adjusted to OD600 = 0.2. Serial ten-fold dilutions were made in THYE, and 4 μL aliquots of each dilution were spotted on THYE agar (pH 7.0, pH 5.0, 3% NaCl, or 2.5 mM ZnCl2). Plates were incubated at 37 °C in air with 5% CO2 for 2 days before documentation.
Coarse grain molecular dynamics (CGMD) simulations
Three-dimensional models of the monomeric YidC1 and YidC2 were built using the I-TASSER homology modeling webserver using B. halodurans YidC2 (PDB ID: 3WO6) and E. coli YidC (PDB ID: 3WVF) as templates (Kumazaki et al., 2014a; Kumazaki et al., 2014b; Yang et al., 2015; Yang and Zhang, 2015) (Figure 1B). Alpha Fold predicted models were similar to those derived by I-TASSER except that YidC1 and YidC2 C-terminal tail structures were not predicted with high confidence by Alpha-Fold. Head-to-head YidC1 or YidC2 dimer files were built by cloning and placing the 180-degree rotated monomer from the original monomer. TM3-4 of each monomer was designated as the interface with a minimum backbone-backbone distance of 7–8 Å at the closest point, measured via Pymol. Monomer and dimer structure files were aligned in a mock membrane using the PPM 2.0 webserver (Lomize et al., 2012). Point mutations were introduced into each WT insertase membrane aligned model structure using the PyMol mutagenesis wizard in order to build mutant systems. Groningen Machine for Chemical Simulation (GROMACS) (Abraham et al., 2015) system files of box size 200 nm, 310.15 K, and with 50 mM NaCl, and 1 bar pressure were built using the CHARMM-GUI (Jo et al., 2008; Lee et al., 2016) Martini maker (Qi et al., 2015; Hsu et al., 2017). Single systems of each protein, monomer or dimer, were built into lipid mixtures containing either 60 mol% CL or PE, with 20 mol% PI and 20 mol% PS. Lipid acyl chains were 1-palmitoyl-2-oleoyl (PO) in all cases. A schematic diagram of phospholipid molecules employed in this study is shown in Supplementary Figure S1. Each system was energy minimized and equilibrated according to default CHARMM-GUI mdp settings with the Martini2.2 forcefield (Monticelli et al., 2008; Periole et al., 2009; d et al., 2013), using the Berendensen Barostat (Berendsen et al., 1984) v-rescale thermostat, and the reaction-field method of electrostatics (Hünenburger and Gunsteren, 1998). After equilibration, each system was replicated for production to ensure equivalent starting energy profiles for a total of 5 replicates per system (Knapp et al., 2018). Production was carried out for 10 µs with identical parameters with the exception of the Parrinello-Rahman Barostat (Parrinello and Rahman, 1981) in place of the Berendensen Barostat. Production mdp files were altered only to influence simulation time length. Computational time was provided by the Texas Tech High Performance Computing Center (HPCC). Protein Cα root-mean-square deviation (RMSD) analysis was performed with the gmx rms tool and calculated as average Cα RMSD as a function of simulation time compared to the final structural snapshot from equilibration trajectories per simulation. RMSD is presented as mean ± s.d. from the 5 replicate systems. Average root-mean-square fluctuations (RMSF) per residue were calculated using the gmx rmsf tool. RMSF data is presented as mean ± s.d. of each residue throughout simulation across 5 replicates. Two-tailed t-tests from RMSF data were calculated between per replicate residue RMSF values across datasets. Lipid residency times were calculated using PyLipID (Song et al., 2022) with an interaction start distance of 0.55 nm and an end distance of 1.0 nm. Data are presented as the mean of 5 replicates.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author contributions
SM: Conceptualization, Data curation, Formal Analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Supervision, Validation, Visualization, Writing–original draft, Writing–review and editing. EA: Writing–review and editing, Data curation, Formal Analysis, Investigation, Methodology, Software, Validation, Visualization. BW: Conceptualization, Formal Analysis, Funding acquisition, Investigation, Methodology, Resources, Software, Supervision, Validation, Visualization, Writing–review and editing. LB: Conceptualization, Formal Analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Supervision, Validation, Visualization, Writing–review and editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by NIH grant DE008007 to LB, GM124979 to BW, and DE031332 to SM.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmolb.2023.1264454/full#supplementary-material
References
Abraham, M. J. M., Schulz, R., Pall, S., Smith, J. C., Hess, B., Lindahl, E., et al. (2015). Gromacs: high performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 1-2, 19–25. doi:10.1016/j.softx.2015.06.001
Ajdic, D., McShan, W. M., McLaughlin, R. E., Savić, G., Chang, J., Carson, M. B., et al. (2002). Genome sequence of Streptococcus mutans UA159, a cariogenic dental pathogen. Proc. Natl. Acad. Sci. U. S. A. 99 (22), 14434–14439. doi:10.1073/pnas.172501299
Ansell, T. B., Song, W., and Sansom, M. S. P. (2020). The glycosphingolipid GM3 modulates conformational dynamics of the glucagon receptor. Biophys. J. 119 (2), 300–313. doi:10.1016/j.bpj.2020.06.009
Beck, K., Eisner, G., Trescher, D., Dalbey, R. E., Brunner, J., and Müller, M. (2001). YidC, an assembly site for polytopic Escherichia coli membrane proteins located in immediate proximity to the SecYE translocon and lipids. EMBO Rep. 2 (8), 709–714. doi:10.1093/embo-reports/kve154
Benz, M., Bals, T., Gügel, I. L., Piotrowski, M., Kuhn, A., Schünemann, D., et al. (2009). Alb4 of Arabidopsis promotes assembly and stabilization of a non chlorophyll-binding photosynthetic complex, the CF1CF0-ATP synthase. Mol. Plant 2 (6), 1410–1424. doi:10.1093/mp/ssp095
Berendsen, H. J. C., Postma, J. P. M., van Gunsteren, W. F., DiNola, A., and Haak, J. R. (1984). Molecular dynamics with coupling to an external bath. J. Chem. Phys. 81, 3684–3690. doi:10.1063/1.448118
Brosig, B., and Langosch, D. (1998). The dimerization motif of the glycophorin A transmembrane segment in membranes: importance of glycine residues. Protein Sci. 7 (4), 1052–1056. doi:10.1002/pro.5560070423
Chen, Y., Capponi, S., Zhu, L., Gellenbeck, P., Freites, J. A., White, S. H., et al. (2017). YidC insertase of Escherichia coli: water accessibility and membrane shaping. Structure 25 (9), 1403–1414. doi:10.1016/j.str.2017.07.008
Chen, Y. M., Betzenhauser, M. J., and Burne, R. A. (2002). cis-Acting elements that regulate the low-pH-inducible urease operon of Streptococcus salivarius. Microbiol. Read. 148 (11), 3599–3608. doi:10.1099/00221287-148-11-3599
Chen, Y., Sotomayor, M., Capponi, S., Hariharan, B., Sahu, I. D., Haase, M., et al. (2022). A hydrophilic microenvironment in the substrate-translocating groove of the YidC membrane insertase is essential for enzyme function. J. Biol. Chem. 298 (3), 101690. doi:10.1016/j.jbc.2022.101690
Corey, R. A., Pyle, E., Allen, W. J., Watkins, D. W., Casiraghi, M., Miroux, B., et al. (2018). Specific cardiolipin-SecY interactions are required for proton-motive force stimulation of protein secretion. Proc. Natl. Acad. Sci. U. S. A. 115 (31), 7967–7972. doi:10.1073/pnas.1721536115
Corradi, V., Mendez-Villuendas, E., Ingólfsson, H. I., Gu, R. X., Siuda, I., Melo, M. N., et al. (2018). Lipid-protein interactions are unique fingerprints for membrane proteins. ACS Cent. Sci. 4 (6), 709–717. doi:10.1021/acscentsci.8b00143
Crooks, G. E., Hon, G., Chandonia, J. M., and Brenner, S. E. (2004). WebLogo: A sequence logo generator. Genome Res. 14 (6), 1188–1190. doi:10.1101/gr.849004
de Jong, D. H., Singh, G., Bennett, W. F. D., Arnarez, C., Wassenaar, T. A., Schäfer, L. V., et al. (2013). Improved parameters for the Martini coarse-grained protein force field. J. Chem. Theory Comput. 9 (1), 687–697. doi:10.1021/ct300646g
Damre, M., Marchetto, A., and Giorgetti, A. (2019). Mermaid: dedicated web server to prepare and run coarse-grained membrane protein dynamics. Nucleic Acids Res. 47 (W1), W456–W461. doi:10.1093/nar/gkz416
Dong, Y., Palmer, S. R., Hasona, A., Nagamori, S., Kaback, H. R., Dalbey, R. E., et al. (2008). Functional overlap but lack of complete cross-complementation of Streptococcus mutans and Escherichia coli YidC orthologs. J. Bacteriol. 190 (7), 2458–2469. doi:10.1128/JB.01366-07
Duncan, A. L., Corey, R. A., and Sansom, M. S. P. (2020). Defining how multiple lipid species interact with inward rectifier potassium (Kir2) channels. Proc. Natl. Acad. Sci. U. S. A. 117 (14), 7803–7813. doi:10.1073/pnas.1918387117
Falk, S., Ravaud, S., Koch, J., and Sinning, I. (2010). The C terminus of the Alb3 membrane insertase recruits cpSRP43 to the thylakoid membrane. J. Biol. Chem. 285 (8), 5954–5962. doi:10.1074/jbc.M109.084996
Fiumera, H. L., Broadley, S. A., and Fox, T. D. (2007). Translocation of mitochondrially synthesized Cox2 domains from the matrix to the intermembrane space. Mol. Cell. Biol. 27 (13), 4664–4673. doi:10.1128/MCB.01955-06
Funes, S., Hasona, A., Bauerschmitt, H., Grubbauer, C., Kauff, F., Collins, R., et al. (2009). Independent gene duplications of the YidC/Oxa/Alb3 family enabled a specialized cotranslational function. Proc. Natl. Acad. Sci. U. S. A. 106 (16), 6656–6661. doi:10.1073/pnas.0809951106
Hasona, A., Crowley, P. J., Levesque, C. M., Mair, R. W., Cvitkovitch, D. G., Bleiweis, A. S., et al. (2005). Streptococcal viability and diminished stress tolerance in mutants lacking the signal recognition particle pathway or YidC2. Proc. Natl. Acad. Sci. U. S. A. 102 (48), 17466–17471. doi:10.1073/pnas.0508778102
Hasona, A., Zuobi-Hasona, K., Crowley, P. J., Abranches, J., Ruelf, M. A., Bleiweis, A. S., et al. (2007). Membrane composition changes and physiological adaptation by Streptococcus mutans signal recognition particle pathway mutants. J. Bacteriol. 189 (4), 1219–1230. doi:10.1128/JB.01146-06
Hsu, P. C., Bruininks, B. M. H., Jefferies, D., Cesar Telles de Souza, P., Lee, J., Patel, D. S., et al. (2017). CHARMM-GUI Martini Maker for modeling and simulation of complex bacterial membranes with lipopolysaccharides. J. Comput. Chem. 38 (27), 2354–2363. doi:10.1002/jcc.24895
Hünenburger, P., and Gunsteren, W. (1998). Alternative schemes for the inclusion of a reaction-field correction into molecular dynamics simulations: influence on the simulated energetic, structural, and dielectric properties of liquid water. J. Chem. Phys. 108, 6117–6134. doi:10.1063/1.476022
Jia, L., Dienhart, M., Schramp, M., McCauley, M., Hell, K., and Stuart, R. A. (2003). Yeast Oxa1 interacts with mitochondrial ribosomes: the importance of the C-terminal region of Oxa1. EMBO J. 22 (24), 6438–6447. doi:10.1093/emboj/cdg624
Jiang, F., Chen, M., de Gier, J. W., Kuhn, A., and Dalbey, R. E. (2003). Defining the regions of Escherichia coli YidC that contribute to activity. J. Biol. Chem. 278 (49), 48965–48972. doi:10.1074/jbc.M307362200
Jo, S., Kim, T., Iyer, V. G., and Im, W. (2008). CHARMM-GUI: A web-based graphical user interface for CHARMM. J. Comput. Chem. 29 (11), 1859–1865. doi:10.1002/jcc.20945
Johnston, J. M., Wang, H., Provasi, D., and Filizola, M. (2012). Assessing the relative stability of dimer interfaces in g protein-coupled receptors. PLoS Comput. Biol. 8 (8), e1002649. doi:10.1371/journal.pcbi.1002649
Kedrov, A., Wickles, S., Crevenna, A. H., van der Sluis, E. O., Buschauer, R., Berninghausen, O., et al. (2016). Structural dynamics of the YidC:ribosome complex during membrane protein biogenesis. Cell. Rep. 17 (11), 2943–2954. doi:10.1016/j.celrep.2016.11.059
Knapp, B., Ospina, L., and Deane, C. M. (2018). Avoiding false positive conclusions in molecular simulation: the importance of replicas. J. Chem. Theory Comput. 14 (12), 6127–6138. doi:10.1021/acs.jctc.8b00391
Knyazev, D. G., Winter, L., and Vogt, A. (2023). YidC from Escherichia coli forms an ion-conducting pore upon activation by ribosomes. arXiv.
Kohler, R., Boehringer, D., Greber, B., Bingel-Erlenmeyer, R., Collinson, I., Schaffitzel, C., et al. (2009). YidC and Oxa1 form dimeric insertion pores on the translating ribosome. Mol. Cell. 34 (3), 344–353. doi:10.1016/j.molcel.2009.04.019
Kumazaki, K., Chiba, S., Takemoto, M., Furukawa, A., Nishiyama, K. I., Sugano, Y., et al. (2014a). Structural basis of Sec-independent membrane protein insertion by YidC. Nature 509 (7501), 516–520. doi:10.1038/nature13167
Kumazaki, K., Kishimoto, T., Furukawa, A., Mori, H., Tanaka, Y., Dohmae, N., et al. (2014b). Crystal structure of Escherichia coli YidC, a membrane protein chaperone and insertase. Sci. Rep. 4, 7299. doi:10.1038/srep07299
Kumazaki, K., Tsukazaki, T., Nishizawa, T., Tanaka, Y., Kato, H. E., Nakada-Nakura, Y., et al. (2014c). Crystallization and preliminary X-ray diffraction analysis of YidC, a membrane-protein chaperone and insertase from Bacillus halodurans. Acta Crystallogr. F. Struct. Biol. Commun. 70 (8), 1056–1060. doi:10.1107/S2053230X14012540
Lara Vasquez, P., Mishra, S., Kuppuswamy, S. K., Crowley, P. J., and Brady, L. J. (2021). Protein interactomes of Streptococcus mutans YidC1 and YidC2 membrane protein insertases suggest SRP pathway-independent- and -dependent functions, respectively. mSphere 6 (2), e01308-20. doi:10.1128/mSphere.01308-20
Lee, J., Cheng, X., Swails, J. M., Yeom, M. S., Eastman, P. K., Lemkul, J. A., et al. (2016). CHARMM-GUI input generator for NAMD, GROMACS, AMBER, OpenMM, and CHARMM/OpenMM simulations using the CHARMM36 additive force field. J. Chem. Theory Comput. 12 (1), 405–413. doi:10.1021/acs.jctc.5b00935
Lewis, A. J. O., and Hegde, R. S. (2021). A unified evolutionary origin for the ubiquitous protein transporters SecY and YidC. BMC Biol. 19 (1), 266. doi:10.1186/s12915-021-01171-5
Lewis, N. E., and Brady, L. J. (2015). Breaking the bacterial protein targeting and translocation model: oral organisms as a case in point. Mol. Oral Microbiol. 30 (3), 186–197. doi:10.1111/omi.12088
Li, E., Wimley, W. C., and Hristova, K. (2012). Transmembrane helix dimerization: beyond the search for sequence motifs. Biochim. Biophys. Acta 1818 (2), 183–193. doi:10.1016/j.bbamem.2011.08.031
Liu, Y., Engelman, D. M., and Gerstein, M. (2002). Genomic analysis of membrane protein families: abundance and conserved motifs. Genome Biol. 3 (10), research0054. doi:10.1186/gb-2002-3-10-research0054
Liu, Y., Zeng, L., and Burne, R. A. (2009). AguR is required for induction of the Streptococcus mutans agmatine deiminase system by low pH and agmatine. Appl. Environ. Microbiol. 75 (9), 2629–2637. doi:10.1128/AEM.02145-08
Lomize, M. A., Pogozheva, I. D., Joo, H., Mosberg, H. I., and Lomize, A. L. (2012). OPM database and PPM web server: resources for positioning of proteins in membranes. Nucleic Acids Res. 40, D370–D376. doi:10.1093/nar/gkr703
Lotz, M., Haase, W., Kühlbrandt, W., and Collinson, I. (2008). Projection structure of yidC: A conserved mediator of membrane protein assembly. J. Mol. Biol. 375 (4), 901–907. doi:10.1016/j.jmb.2007.10.089
MacGilvray, M. E., Lapek, J. D., Friedman, A. E., and Quivey, R. G. (2012). Cardiolipin biosynthesis in Streptococcus mutans is regulated in response to external pH. Microbiol. Read. 158 (8), 2133–2143. doi:10.1099/mic.0.057273-0
Mbaye, M. N., Hou, Q., Basu, S., Teheux, F., Pucci, F., and Rooman, M. (2019). A comprehensive computational study of amino acid interactions in membrane proteins. Sci. Rep. 9 (1), 12043. doi:10.1038/s41598-019-48541-2
McDowell, M. A., Heimes, M., Fiorentino, F., Mehmood, S., Farkas, Á., Coy-Vergara, J., et al. (2020). Structural basis of tail-anchored membrane protein biogenesis by the GET insertase complex. Mol. Cell. 80 (1), 72–86. doi:10.1016/j.molcel.2020.08.012
Mishra, S., and Brady, L. J. (2021). The cytoplasmic domains of Streptococcus mutans membrane protein insertases YidC1 and YidC2 confer unique structural and functional attributes to each paralog. Front. Microbiol. 12, 760873. doi:10.3389/fmicb.2021.760873
Mishra, S., Crowley, P. J., Wright, K. R., Palmer, S. R., Walker, A. R., Datta, S., et al. (2019). Membrane proteomic analysis reveals overlapping and independent functions of Streptococcus mutans Ffh, YidC1, and YidC2. Mol. Oral Microbiol. 34 (4), 131–152. doi:10.1111/omi.12261
Monticelli, L., Kandasamy, S. K., Periole, X., Larson, R. G., Tieleman, D. P., and Marrink, S. J. (2008). The MARTINI coarse-grained force field: extension to proteins. J. Chem. Theory Comput. 4 (5), 819–834. doi:10.1021/ct700324x
Morales-Aparicio, J. C., Lara Vasquez, P., Mishra, S., Barrán-Berdón, A. L., Kamat, M., Basso, K. B., et al. (2020). The impacts of sortase A and the 4'-phosphopantetheinyl transferase homolog sfp on Streptococcus mutans extracellular membrane vesicle biogenesis. Front. Microbiol. 11, 570219. doi:10.3389/fmicb.2020.570219
Murakami, T., Haga, K., Takeuchi, M., and Sato, T. (2002). Analysis of the Bacillus subtilis spoIIIJ gene and its Paralogue gene, yqjG. J. Bacteriol. 184 (7), 1998–2004. doi:10.1128/jb.184.7.1998-2004.2002
Mustafa, G., Nandekar, P. P., Yu, X., and Wade, R. C. (2015). On the application of the MARTINI coarse-grained model to immersion of a protein in a phospholipid bilayer. J. Chem. Phys. 143 (24), 243139. doi:10.1063/1.4936909
Nass, K. J., Ilie, I. M., Saller, M. J., Driessen, A. J. M., Caflisch, A., Kammerer, R. A., et al. (2022). The role of the N-terminal amphipathic helix in bacterial YidC: insights from functional studies, the crystal structure and molecular dynamics simulations. Biochim. Biophys. Acta Biomembr. 1864 (3), 183825. doi:10.1016/j.bbamem.2021.183825
O'Donnell, J. P., Phillips, B. P., Yagita, Y., Juszkiewicz, S., Wagner, A., Malinverni, D., et al. (2020). The architecture of EMC reveals a path for membrane protein insertion. Elife 9, e57887. doi:10.7554/eLife.57887
Oliver, D. C., and Paetzel, M. (2008). Crystal structure of the major periplasmic domain of the bacterial membrane protein assembly facilitator YidC. J. Biol. Chem. 283 (8), 5208–5216. doi:10.1074/jbc.M708936200
Palmer, S. R., Crowley, P. J., Oli, M. W., Ruelf, M. A., Michalek, S. M., and Brady, L. J. (2012). YidC1 and YidC2 are functionally distinct proteins involved in protein secretion, biofilm formation and cariogenicity of Streptococcus mutans. Microbiol. Read. 158 (7), 1702–1712. doi:10.1099/mic.0.059139-0
Palmer, S. R., Ren, Z., Hwang, G., Liu, Y., Combs, A., Söderström, B., et al. (2019). Streptococcus mutans yidC1 and yidC2 impact cell envelope biogenesis, the biofilm matrix, and biofilm biophysical properties. J. Bacteriol. 201 (1), e00396-18. doi:10.1128/JB.00396-18
Parrinello, M., and Rahman, A. (1981). Polymorphic transitions in single crystals: A new molecular dynamics method. J. Appl. Phys. 52, 7182–7190. doi:10.1063/1.328693
Periole, X., Cavalli, M., Marrink, S. J., and Ceruso, M. A. (2009). Combining an elastic network with a coarse-grained molecular force field: structure, dynamics, and intermolecular recognition. J. Chem. Theory Comput. 5 (9), 2531–2543. doi:10.1021/ct9002114
Petriman, N. A., Jauß, B., Hufnagel, A., Franz, L., Sachelaru, I., Drepper, F., et al. (2018). The interaction network of the YidC insertase with the SecYEG translocon, SRP and the SRP receptor FtsY. Sci. Rep. 8 (1), 578. doi:10.1038/s41598-017-19019-w
Pleiner, T., Tomaleri, G. P., Januszyk, K., Inglis, A. J., Hazu, M., and Voorhees, R. M. (2020). Structural basis for membrane insertion by the human ER membrane protein complex. Science 369 (6502), 433–436. doi:10.1126/science.abb5008
Pluhackova, K., Gahbauer, S., Kranz, F., Wassenaar, T. A., and Böckmann, R. A. (2016). Dynamic cholesterol-conditioned dimerization of the G protein coupled chemokine receptor type 4. PLoS Comput. Biol. 12 (11), e1005169. doi:10.1371/journal.pcbi.1005169
Polasa, A., Hettige, J., Immadisetty, K., and Moradi, M. (2022). An investigation of the YidC-mediated membrane insertion of Pf3 coat protein using molecular dynamics simulations. Front. Mol. Biosci. 9, 954262. doi:10.3389/fmolb.2022.954262
Poma, A. B., Cieplak, M., and Theodorakis, P. E. (2017). Combining the MARTINI and structure-based coarse-grained approaches for the molecular dynamics studies of conformational transitions in proteins. J. Chem. Theory Comput. 13 (3), 1366–1374. doi:10.1021/acs.jctc.6b00986
Prasanna, X., Chattopadhyay, A., and Sengupta, D. (2014). Cholesterol modulates the dimer interface of the β₂-adrenergic receptor via cholesterol occupancy sites. Biophys. J. 106 (6), 1290–1300. doi:10.1016/j.bpj.2014.02.002
Qi, Y., Ingólfsson, H. I., Cheng, X., Lee, J., Marrink, S. J., and Im, W. (2015). CHARMM-GUI Martini maker for coarse-grained simulations with the Martini force field. J. Chem. Theory Comput. 11 (9), 4486–4494. doi:10.1021/acs.jctc.5b00513
Ravaud, S., Stjepanovic, G., Wild, K., and Sinning, I. (2008). The crystal structure of the periplasmic domain of the Escherichia coli membrane protein insertase YidC contains a substrate binding cleft. J. Biol. Chem. 283 (14), 9350–9358. doi:10.1074/jbc.M710493200
Romantsov, T., Guan, Z., and Wood, J. M. (2009). Cardiolipin and the osmotic stress responses of bacteria. Biochim. Biophys. Acta 1788 (10), 2092–2100. doi:10.1016/j.bbamem.2009.06.010
Sachelaru, I., Winter, L., Knyazev, D. G., Zimmermann, M., Vogt, A., Kuttner, R., et al. (2017). YidC and SecYEG form a heterotetrameric protein translocation channel. Sci. Rep. 7 (1), 101. doi:10.1038/s41598-017-00109-8
Solovyev, A., and Salamov, V. S. (2011). “Automatic annotation of microbial genomes and metagenomic sequences,” in Metagenomics and its applications in agriculture, biomedicine and environmental studies. Editor R. W. Li (Hauppauge, NY: Nova Science Publishers).
Seaton, K., Ahn, S. J., and Burne, R. A. (2015). Regulation of competence and gene expression in Streptococcus mutans by the RcrR transcriptional regulator. Mol. Oral Microbiol. 30 (2), 147–159. doi:10.1111/omi.12079
Senes, A., Gerstein, M., and Engelman, D. M. (2000). Statistical analysis of amino acid patterns in transmembrane helices: the GxxxG motif occurs frequently and in association with beta-branched residues at neighboring positions. J. Mol. Biol. 296 (3), 921–936. doi:10.1006/jmbi.1999.3488
Song, W., Corey, R. A., Ansell, T. B., Cassidy, C. K., Horrell, M. R., Duncan, A. L., et al. (2022). PyLipID: A Python package for analysis of protein-lipid interactions from molecular dynamics simulations. J. Chem. Theory Comput. 18 (2), 1188–1201. doi:10.1021/acs.jctc.1c00708
Song, W., Duncan, A. L., and Sansom, M. S. P. (2021). Modulation of adenosine A2a receptor oligomerization by receptor activation and PIP2 interactions. Structure 29 (11), 1312–1325.e3. doi:10.1016/j.str.2021.06.015
Steinberg, R., Knüpffer, L., Origi, A., Asti, R., and Koch, H. G. (2018). Co-translational protein targeting in bacteria. FEMS Microbiol. Lett. 365 (11), 95. doi:10.1093/femsle/fny095
Tjalsma, H., Bron, S., and van Dijl, J. M. (2003). Complementary impact of paralogous Oxa1-like proteins of Bacillus subtilis on post-translocational stages in protein secretion. J. Biol. Chem. 278 (18), 15622–15632. doi:10.1074/jbc.M301205200
Urbanus, M. L., Scotti, P. A., Froderberg, L., Saaf, A., de Gier, J. W., Brunner, J., et al. (2001). Sec-dependent membrane protein insertion: sequential interaction of nascent FtsQ with SecY and YidC. EMBO Rep. 2 (6), 524–529. doi:10.1093/embo-reports/kve108
Van den Berg, B., Clemons, W. M., Collinson, I., Modis, Y., Hartmann, E., Harrison, S. C., et al. (2004). X-ray structure of a protein-conducting channel. Nature 427 (6969), 36–44. doi:10.1038/nature02218
Vitkup, D., Sander, C., and Church, G. M. (2003). The amino-acid mutational spectrum of human genetic disease. Genome Biol. 4 (11), R72. doi:10.1186/gb-2003-4-11-r72
Woodall, B. M., Harp, J. R., Brewer, W. T., Tague, E. D., Campagna, S. R., and Fozo, E. M. (2021). Enterococcus faecalis readily adapts membrane phospholipid composition to environmental and genetic perturbation. Front. Microbiol. 12, 616045. doi:10.3389/fmicb.2021.616045
Woolhead, C. A., Thompson, S. J., Moore, M., Tissier, C., Mant, A., Rodger, A., et al. (2001). Distinct Albino3-dependent and -independent pathways for thylakoid membrane protein insertion. J. Biol. Chem. 276 (44), 40841–40846. doi:10.1074/jbc.M106523200
Xin, Y., Zhao, Y., Zheng, J., Zhou, H., Zhang, X. C., Tian, C., et al. (2018). Structure of YidC from Thermotoga maritima and its implications for YidC-mediated membrane protein insertion. FASEB J. 32 (5), 2411–2421. doi:10.1096/fj.201700893RR
Yang, J., Yan, R., Roy, A., Xu, D., Poisson, J., and Zhang, Y. (2015). The I-tasser suite: protein structure and function prediction. Nat. Methods 12 (1), 7–8. doi:10.1038/nmeth.3213
Yang, J., and Zhang, Y. (2015). Protein structure and function prediction using I-tasser. Curr. Protoc. Bioinforma. 52, 5 8 1–5. doi:10.1002/0471250953.bi0508s52
Yekefallah, M., Rasberry, C. A., van Aalst, E. J., Browning, H. P., Amani, R., Versteeg, D. B., et al. (2022). Mutational insight into allosteric regulation of kir channel activity. ACS Omega 7 (48), 43621–43634. doi:10.1021/acsomega.2c04456
Keywords: Streptococcus mutans, YidC, cardiolipin, molecular dynamics, paralog
Citation: Mishra S, van Aalst EJ, Wylie BJ and Brady LJ (2023) Cardiolipin occupancy profiles of YidC paralogs reveal the significance of respective TM2 helix residues in determining paralog-specific phenotypes. Front. Mol. Biosci. 10:1264454. doi: 10.3389/fmolb.2023.1264454
Received: 20 July 2023; Accepted: 22 September 2023;
Published: 06 October 2023.
Edited by:
Fabio Polticelli, Roma Tre University, ItalyReviewed by:
Mrinal Shekhar, Broad Institute, United StatesGiray Enkavi, University of Helsinki, Finland
Copyright © 2023 Mishra, van Aalst, Wylie and Brady. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: L. Jeannine Brady, jbrady@dental.ufl.edu
†These authors have contributed equally to this work
 Surabhi Mishra
Surabhi Mishra Evan J. van Aalst
Evan J. van Aalst Benjamin J. Wylie
Benjamin J. Wylie L. Jeannine Brady
L. Jeannine Brady