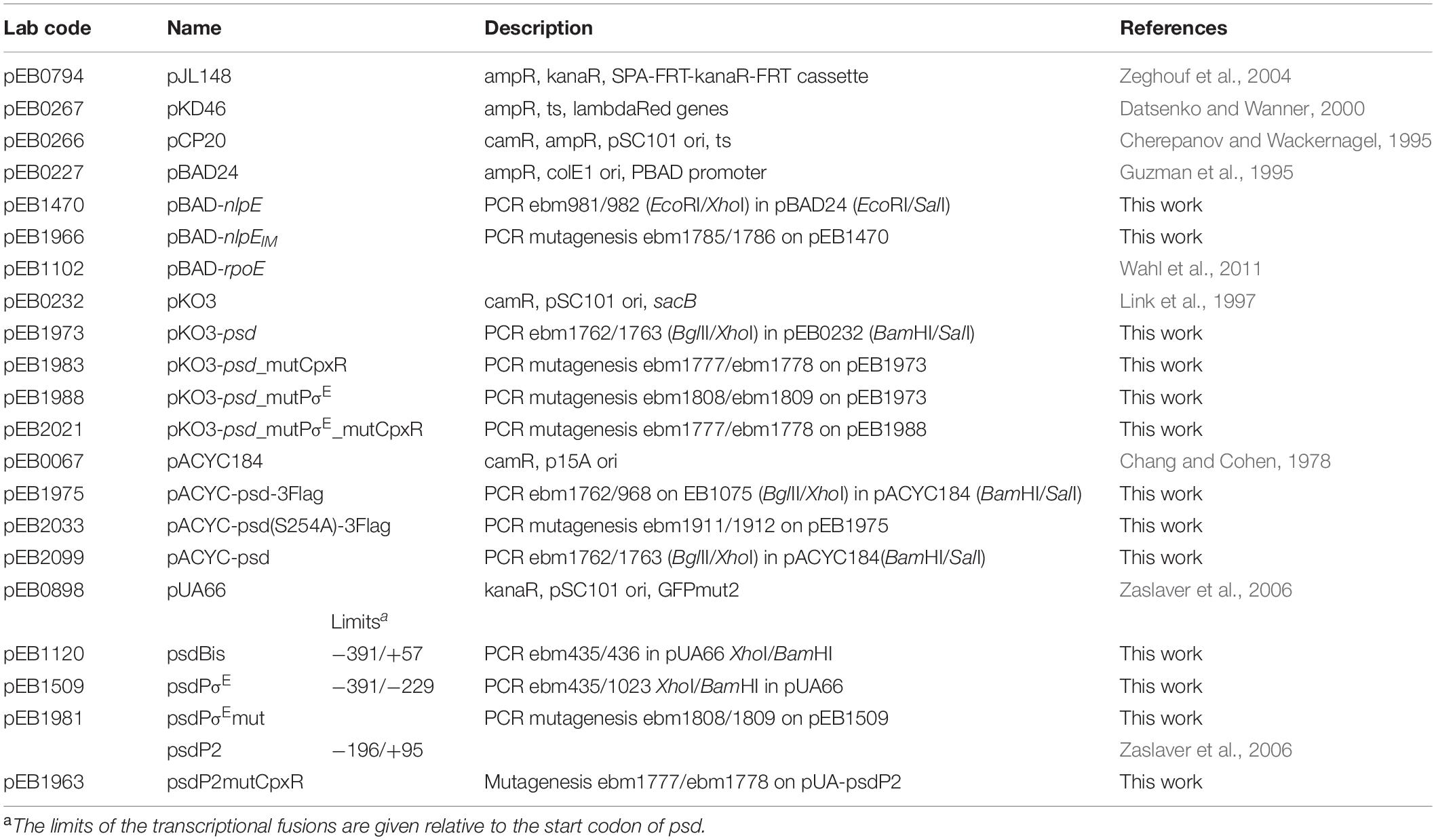
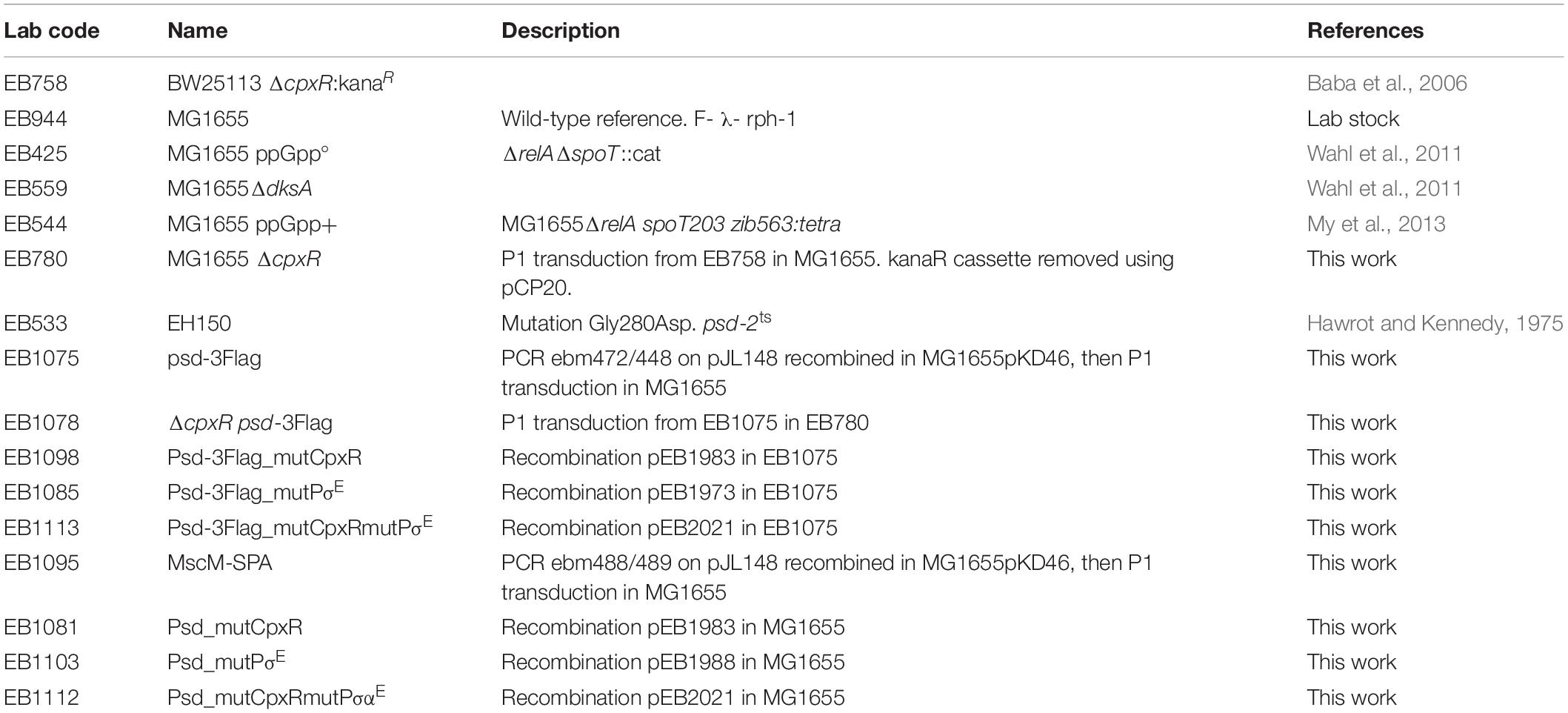
Dual Regulation of Phosphatidylserine Decarboxylase Expression by Envelope Stress Responses
- 1LISM, Institut de Microbiologie de la Méditerranée, UMR 7255, CNRS and Aix-Marseille Université, Marseille, France
- 2SAMe Unit, UMR 2001, Microbiology Department, Pasteur Institute, Paris, France
Bacteria adapt to versatile environments by modulating gene expression through a set of stress response regulators, alternative Sigma factors, or two-component systems. Among the central processes that must be finely tuned is membrane homeostasis, including synthesis of phospholipids (PL). However, few genetic regulations of this process have been reported. We have previously shown that the gene coding the first step of PL synthesis is regulated by σE and ppGpp, and that the BasRS (PmrAB) two component system controls the expression of the DgkA PL recycling enzyme. The gene coding for phosphatidylserine decarboxylase, the last step in phosphatidylethanolamine synthesis is another gene in the PL synthesis pathway susceptible of stress response regulation. Indeed, psd appears in transcriptome studies of the σE envelope stress Sigma factor and of the CpxAR two component system. Interestingly, this gene is presumably in operon with mscM coding for a miniconductance mechanosensitive channel. In this study, we dissected the promoter region of the psd-mscM operon and studied its regulation by σE and CpxR. By artificial activation of σE and CpxRA stress response pathways, using GFP transcriptional fusion and western-blot analysis of Psd and MscM enzyme production, we showed that the operon is under the control of two distinct promoters. One is activated by σE, the second is activated by CpxRA and also responsible for basal expression of the operon. The fact that the phosphatidylethanolamine synthesis pathway is controlled by envelope stress responses at both its first and last steps might be important for adaptation of the membrane to envelope perturbations.
Introduction
Phospholipids (PL) are major structural and functional components of biomembranes and play a dynamic role in many regulatory processes. The biochemistry of the PL biosynthesis pathway is well described in bacteria (Parsons and Rock, 2013). It begins by two successive acylations of the positions 1 and 2 of a glycerol 3-phosphate backbone to give phosphatidic acid. Then, the third position of the future polar head group is activated with a CDP nucleotide. Finally, two separate pathways give rise in Escherichia coli to the zwitterionic phosphatidylethanolamine (PE), or to the anionic phospholipids phosphatidylglycerol (PG), and cardiolipin (CL). Bacterial species display specific membrane compositions, and the less frequent phosphatidylcholine, phosphatidylinositol, and a variety of other membrane lipids can be found in addition to the common PE, PG, and CL lipids (Sohlenkamp and Geiger, 2016).
Though the composition of the membrane is highly stable in E. coli, it can adapt to the growth conditions, through modification of fatty acid chains, but also of the ratio of the different lipids (Sohlenkamp and Geiger, 2016). For example, the proportion of unsaturated fatty acids varies with temperature to modify the fluidity of the membrane. Another example of adaptation is the increase of CL following increase of osmotic pressure. Also, in stationary phase, there is a relative increase in anionic phospholipids, together with the replacement of fatty acid unsaturations by cyclopropane groups. This conversion is performed by the Cfa enzyme, whose expression is activated by σS alternative Sigma factor (Wang and Cronan, 1994; Eichel et al., 1999). In addition, global transcriptome studies had suggested that expression of PL synthesis genes might be globally affected during the stringent response (Durfee et al., 2008), and that specific genes of PL synthesis might be regulated by envelope stress responses (De Wulf et al., 2002; Rhodius et al., 2006).
In bacteria, several regulating pathways are present to monitor and respond to envelope perturbations. In E. coli, the alternative Sigma factor σE responds to the accumulation of unfolded outer membrane proteins or altered forms of LPS in the periplasm through a proteolytic signaling cascade (Mitchell and Silhavy, 2019). In addition, two-component systems such as CpxRA, RcsBA, or BaeRS respond to various envelope perturbations such as defects in protein secretion, alterations in LPS or peptidoglycan, or exposure to toxic compounds, by a phosphorylation signaling cascade that ends up in the activation of a transcriptional regulator (Mitchell and Silhavy, 2019). We have previously shown that plsB, coding for the first acylation step in PL synthesis, is activated by the envelope stress response σE factor, and that dgkA, coding for a diacylglycerol kinase involved in PL recycling, is controlled by the two-component system BasRS (Wahl et al., 2011).
The psd gene coding for phosphatidylserine decarboxylase, the last step in phosphatidylethanolamine synthesis, appears in the regulon of σE like plsB, but also in the regulon of the CpxAR two-component system (De Wulf et al., 2002; Rezuchova et al., 2003; Rhodius et al., 2006). The psd gene is followed by mscM (previously called yjeP), and they probably form an operon. MscM protein is a miniconductance mechanosensitive channel of the MscS family, involved in the protection of the physical integrity of the cell during transitions from high to low osmolarity (Booth, 2014; Edwards et al., 2012).
In this study, we dissected the organization of the promoter region of the psd-mscM operon, and we studied the dual regulation of these genes by σE and the response regulator CpxR. We showed that the operon is under the control of two distinct promoters. One is activated by σE, the other by CpxRA. This second promoter is responsible for basal expression of the operon. Therefore, the PL synthesis pathway (and more specifically the PE pathway) is controlled by envelope stress responses at both its first and last steps, which might be important for adaptation of the membrane to envelope perturbations.
Materials and Methods
Plasmid and Strain Constructions
pUA66 and pUA-psdP2 plasmids were obtained from the library of E. coli promoters fused to GFP coding sequence (Zaslaver et al., 2006). The other psd transcriptional fusions were constructed using primers indicated in Supplementary Table 1, and cloned in XhoI/BamHI sites of pUA66. Expression plasmids for rpoE, nlpE, and nlpEIM were constructed using primers indicated in Supplementary Table 1, and cloned in EcoRI/SalI sites of pBAD24 vector (Guzman et al., 1995). A region of 1600 base pairs encompassing psd promoter was cloned in pKO3 vector (Link et al., 1997). Mutations were introduced in the pKO3-psd vector, in the pBAD-nlpE and in the transcriptional fusions by PCR mutagenesis on plasmid, using the oligonucleotides indicated (Table 1 and Supplementary Table 1).
In order to introduce the sequence coding for the Flag or SPA tags downstream of the coding sequences of interest on the chromosome, we amplified the corresponding cassette from the template plasmid pJL148 (Zeghouf et al., 2004). MG1655_Psd-Flag and MG1655_MscM-SPA strains were then constructed by PCR recombination at the locus followed by P1 transduction as described previously (Datsenko and Wanner, 2000) (Table 2). When required, the gene conferring resistance to Kanamycin was removed using the pCP20 plasmid (Cherepanov and Wackernagel, 1995), allowing the transformation by transcriptional fusion plasmids. Point mutations were introduced on the chromosome using the pKO3 vector (Link et al., 1997).
Transcriptional Fusions with GFP
We used several clones from the E. coli transcriptional fusions library (Zaslaver et al., 2006) and we constructed the required additional transcriptional fusions (see above for plasmid construction and Table 1). MG1655 wild-type E. coli strain or isogenic mutants were transformed with plasmids carrying the gfp transcriptional fusions and maintained with kanamycin. For co-transformation, compatible plasmids (pBAD24 and derivatives) were used, and ampicillin added. Selection plates were incubated at 37°C for 16 h. A total of 600 μl of LB medium supplemented with required antibiotics, and with 0.01% arabinose when necessary for pBAD-driven expression, were incubated (four to six replicate each assay) and grown for 16 h at 30°C in 96-well polypropylene plates of 2.2 ml wells under aeration and agitation. Fluorescent intensity measurement was performed in a TECAN infinite M200 plate reader. A total of 150 μl of each well was transferred into black Greiner 96-well plate for reading optical density at 600 nm and fluorescence (excitation: 485 nm; emission: 530 nm). The expression levels were calculated by dividing the intensity of fluorescence by the optical density at 600 nm, after subtracting the values of blank sample. These results are given in arbitrary units because the intensity of fluorescence is acquired with an automatic optimal gain and hence varies from one experiment to the other.
SDS-PAGE and Western Blotting
Total cell extracts were prepared by resuspending cell pellets in Laemli buffer 1X at a concentration of 0.3 uOD600 nm in 10 μl, and then heating for 10 min at 95°C. After separation of 8 μl of total cell extracts on SDS-PAGE, electrotransfer onto nitrocellulose membranes was performed using Trans-Blot turbo transfer system from Bio-Rad. After blocking in PBS 1X + milk 5%, SPA-tagged or 3Flag-tagged proteins were detected with monoclonal anti-Flag M2 antibody purchased from Sigma. IscS protein was used as an internal control and revealed with polyclonal anti-IscS antibodies (Gully et al., 2003). Fluorescent secondary antibodies were respectively IRDye 800 anti-mouse and IRDye 680 anti-rabbit purchased from Li-Cor. Scanning and quantification were performed on a Li-Cor Odyssey-Fc imaging system, reading at 700 nm (for IscS detection) or 800 nm (for Flag detection).
Reverse Transcriptase – PCR
RNAs were purified using the NEB Monarch Total RNA MiniPrep Kit and further digested with Dnase I followed by clean up with the Qiagen Rneasy Mini kit. Reverse transcription was performed using Invitrogen SuperScript III First-Stand Synthesis System with random hexamers. Finally, PCR was performed using ebm446 and ebm2079 primers.
Results
In a global experimental study of all the transcriptional starting sites of E. coli (Thomason et al., 2015), two putative promoters were identified upstream the psd ORF (Figure 1). The first one, that we will call psdPσE, had been already reported as a promoter activated by the alternative sigma factor σE (Rezuchova et al., 2003; Rhodius et al., 2006). The second one, that we will call psdP2, is located 197 nucleotides downstream psdPσE. It has already been reported that psd expression was activated by CpxR, and a putative CpxR box was proposed (De Wulf et al., 2002). This CpxR box is located 41 nucleotides upstream the predicted transcription start site of the psdP2 promoter, which is a distance in perfect agreement with CpxR acting as an activator of the psdP2 promoter (Busby, 2019).

Figure 1. Promoter region of psd-mscM operon. (A) Positions of the predicted CpxR box, and of the two transcription start sites are indicated by a green box and gray arrows. Limits of the regions cloned in transcriptional fusion with gfp, in the pUA66 plasmid, are shown by green horizontal lines. End positions are given relative to the start codon of psd gene. (B) DNA sequences of the two promoters. Mutations introduced in the promoter regions or in the CpxRbox are indicated at the top of red stars.
Dissection of the Promoter Region
In order to dissect the promoter region of psd and to study the genetic regulation of psd expression, we first used transcriptional fusions with GFP, expressed from the low copy vector pUA66, which enable to follow promoter activity by measuring fluorescence directly in living cells (Zaslaver et al., 2006). A transcriptional fusion containing the upstream region of psd ORF was already available from a library of E. coli promoters (Zaslaver et al., 2006), but it only contained the putative psdP2 promoter. Therefore, we constructed additional transcriptional fusions, containing both promoters (called psdBis) or only the psdPσE promoter (Figure 1A).
We first studied the control of psd expression by σE. We artificially induced the σE response, by overproducing the σE factor from an inducible pBAD-rpoE plasmid as previously described (Wahl et al., 2011). We co-transformed an E. coli wild-type strain with the different GFP transcriptional fusions and with the pBAD-rpoE plasmid. Upon σE overproduction, only the transcriptional fusions containing the psdPσE promoter (psdBis and psdPσE, but not psdP2) showed a strong induction (Figure 2). We then mutated two nucleotide positions in the predicted −10 box of the psdPσE promoter. These mutations completely abolished the induction of the psdPσE transcriptional fusion by pBAD-rpoE (Figure 2).
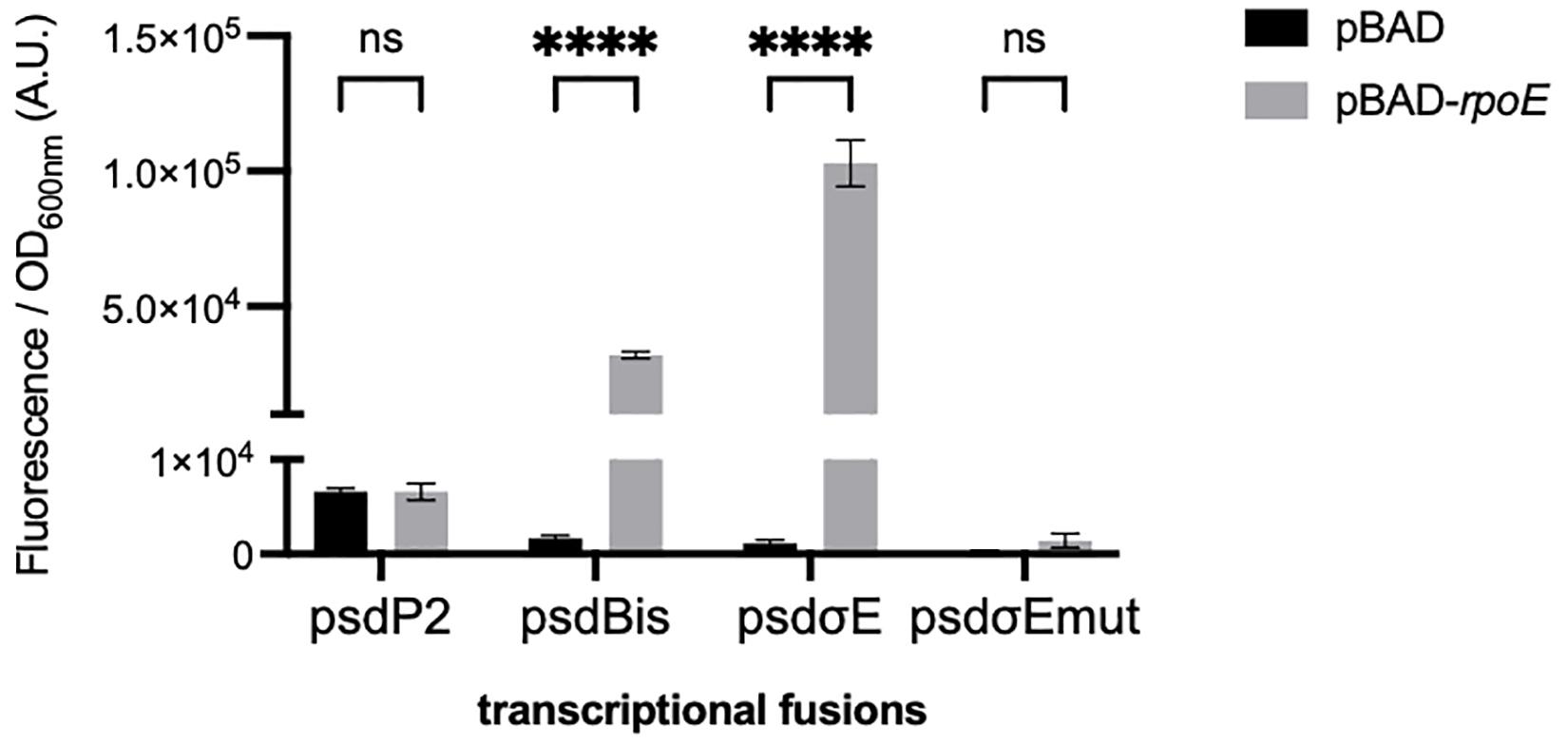
Figure 2. Effect of σE overproduction on the psdPσE promoter. MG1655 strain was transformed with plasmids carrying the transcriptional fusions pUA66, pUA-psdP2, pUA-psdBis, pUA-psdPσE, or pUA-psdPσEmut together with plasmids pBAD24 or pBAD-rpoE. Cultures were grown overnight at 30°C in LB supplemented with ampicillin, kanamycin, and 0.01% arabinose. The values show the mean ratio of GFP fluorescence over optical density at 600 nm, after subtraction of the pUA66 negative control, in arbitrary units (A.U.). The values are the mean of six replicas and the error bars show the SEM (standard error of the mean). ns, non-significant, ****p < 0.0001 in a two-way ANOVA statistical analysis.
We then tested if CpxR was a transcriptional activator of the psdP2 promoter. We first compared the activity of the psdP2 transcriptional fusion in wild-type and ΔcpxR strains. This activity was reduced in the ΔcpxR strain (Figure 3A), suggesting a potential role of CpxR in the regulation of psd expression, even in balanced growth conditions. In reverse, in order to induce the CpxR response, we overproduced the NlpE lipoprotein or its NlpEIM variant that triggers an even stronger response (Delhaye et al., 2016). We co-transformed an E. coli wild-type strain by the different transcriptional fusions and by the pBAD-nlpE or -nlpEIM plasmids. The psdP2 transcriptional fusion showed an activation when NlpE was overproduced, and the effect was reinforced with NlpEIM (Figure 3B). Furthermore, the activation was abolished when the experiment was repeated in a ΔcpxR strain (Figure 3C), which showed that this was indeed the result of an activation of the CpxR pathway. Finally, to prove that the activation was due to direct binding of CpxR response regulator to the psdP2 promoter, we introduced point mutations that destroy the CpxR binding motif in the psdP2 transcriptional fusion (Figure 1B). With this mutated transcriptional fusion, the activation of psdP2 promoter was abolished (Figure 3B).
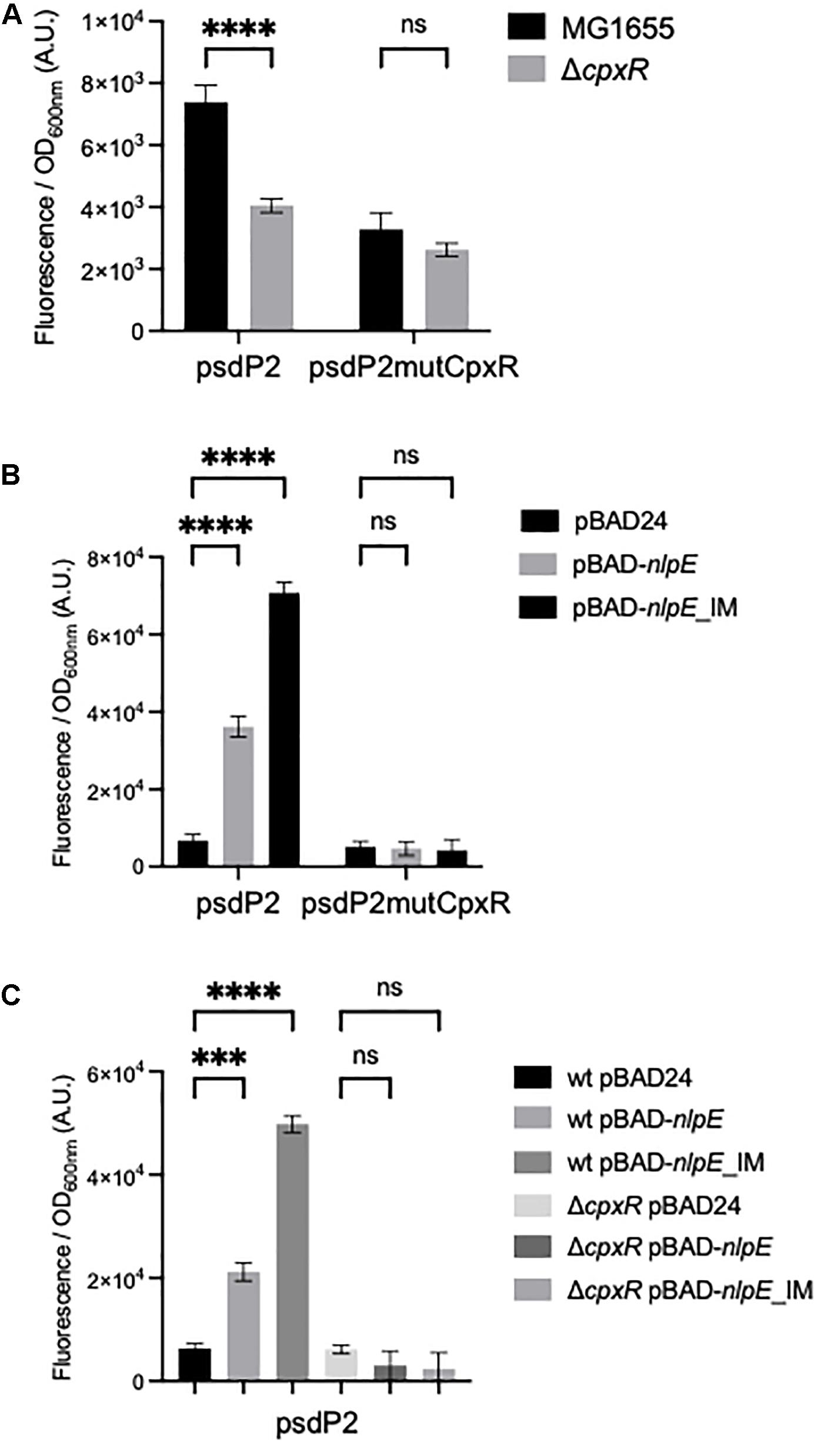
Figure 3. Control of the psdP2 promoter by CpxR. (A) MG1655 or ΔcpxR strains were transformed with the indicated transcriptional fusions. Cultures were grown overnight at 30°C in LB supplemented with kanamycin. (B,C) MG1655 or ΔcpxR strains were transformed with the indicated transcriptional fusions together with pBAD24, pBAD-nlpE, or pBAD-nlpE_IM plasmids. Cultures were grown overnight at 30°C in LB supplemented with ampicillin, kanamycin and 0.01% arabinose. The values show the mean ratio of GFP fluorescence over optical density at 600 nm, after subtraction of the pUA66 negative control, in arbitrary units (A.U.). The values are the mean of 12 (A) or 4 (B,C) replicas and the error bars show the SEM (standard error of the mean). ns, non-significant, ***p < 0.001, ****p < 0.0001 in a two-way ANOVA statistical analysis.
In conclusion, psd expression (and potentially mscM expression) is under the control of two distinct promoters. One is activated by σE, the other by CpxRA. Given that this promoter organization is very similar to the one we described previously for plsB (Wahl et al., 2011), we hypothesized that this second promoter might be regulated by ppGpp as shown for plsB and suggested by global study of the stringent response (Durfee et al., 2008). We tested this by measuring the activity of the transcriptional fusions in strains with modified ppGpp levels as previously described (Wahl et al., 2011). The ppGpp° (ΔrelAΔspoT) strain is devoid of ppGpp, the DksA cofactor is necessary for ppGpp action on RNAP hence a ΔdksA mutant mimics the absence of ppGpp, and finally the spoT203 mutant is impaired in ppGpp degradation, leading to ppGpp accumulation in the cell. The psdP2 promoter did not show a strong dependence on ppGpp levels (Supplementary Figure 1). The psdP2 expression did slightly increase in a ΔdksA mutant, but not in the ppGpp° mutant, and it did not show a corresponding repression in the ppGpp+ mutant. Interestingly, the activity of the psdPσE transcriptional fusion increased in the ppGpp accumulating mutant (Supplementary Figure 1), which is in agreement with the known positive effect of ppGpp on σE (Costanzo et al., 2008).
Impact of σE or CpxR Regulation on Psd and MscM Protein Amounts
Because psd expression is controlled by two promoters, we then wanted to study the relative involvement of these two promoters in the control of the amounts of Psd in the cell. For that, we constructed a strain where Psd is fused at its C-terminus with the 3Flag tag, in order to detect the protein by western blot with a monoclonal antibody. Psd is produced as a proenzyme, which after endoproteolysis gives rise to a mature two-domains enzyme (Li and Dowhan, 1990; Choi et al., 2015). Because the tag is fused to the C-terminus of the protein, we detected as expected the small C-terminal Psdα-3Flag subdomain, indicating that the cleavage occurred normally (it migrated at a higher position than the expected 12 kDa, but this is not exceptional, and might be influenced by the pyruvoyl N-terminal modification). Furthermore, the tagged strain did not display any obvious growth defect, suggesting that the Psd-3Flag fusion was functional. However, in order to prove the functionality of the Psd-3Flag tagged protein, we also cloned this construction or the wild-type psd gene under its own promoter in a plasmid, and tested the complementation of a psd-ts mutant (Hawrot and Kennedy, 1975). Expression of psd-3Flag or wild-type psd expressed from a plasmid similarly restored growth of the psdts mutant at 42°C, showing that the recombinant Psd-3Flag protein was functional (Figure 4). We then tested the effect of overproducing σE or inducing the CpxR response through NlpE expression. The levels of Psd-3Flag protein were clearly increased when σE was overproduced (Figure 5A). Similarly, levels of Psd-3Flag increased when NlpE or NlpEIM proteins were overproduced (Figure 5B). The increases observed when the CpxRA pathway was artificially induced were not as strong as the ones observed when σE was overproduced. However, they were clearly abolished when the experiment was repeated in a ΔcpxR genetic context (Figure 5B). It has to be noted that when Psd-3Flag amounts were strongly increased, a band at approximately 45 kDa was also detected (Figure 5A). We hypothesized that this band might correspond to the accumulation of an unmatured full length Psd-3Flag proenzyme. In order to test this, we introduced the point mutation S254A, which is known to prevent the maturation of Psd (Li and Dowhan, 1990), in the plasmid expressing psd-3Flag. As expected, this mutant form was detected at 45 kDa (Supplementary Figure 2). Interestingly, when we compared the production of Psd-3Flag with or without induction of rpoE, the band at 45 kDa accumulating when rpoE is induced migrated at the exact same size than the unmatured Psd(254A)-3Flag (Supplementary Figure 2), suggesting that endoproteolytic cleavage is indeed impaired when Psd-3Flag is overproduced.
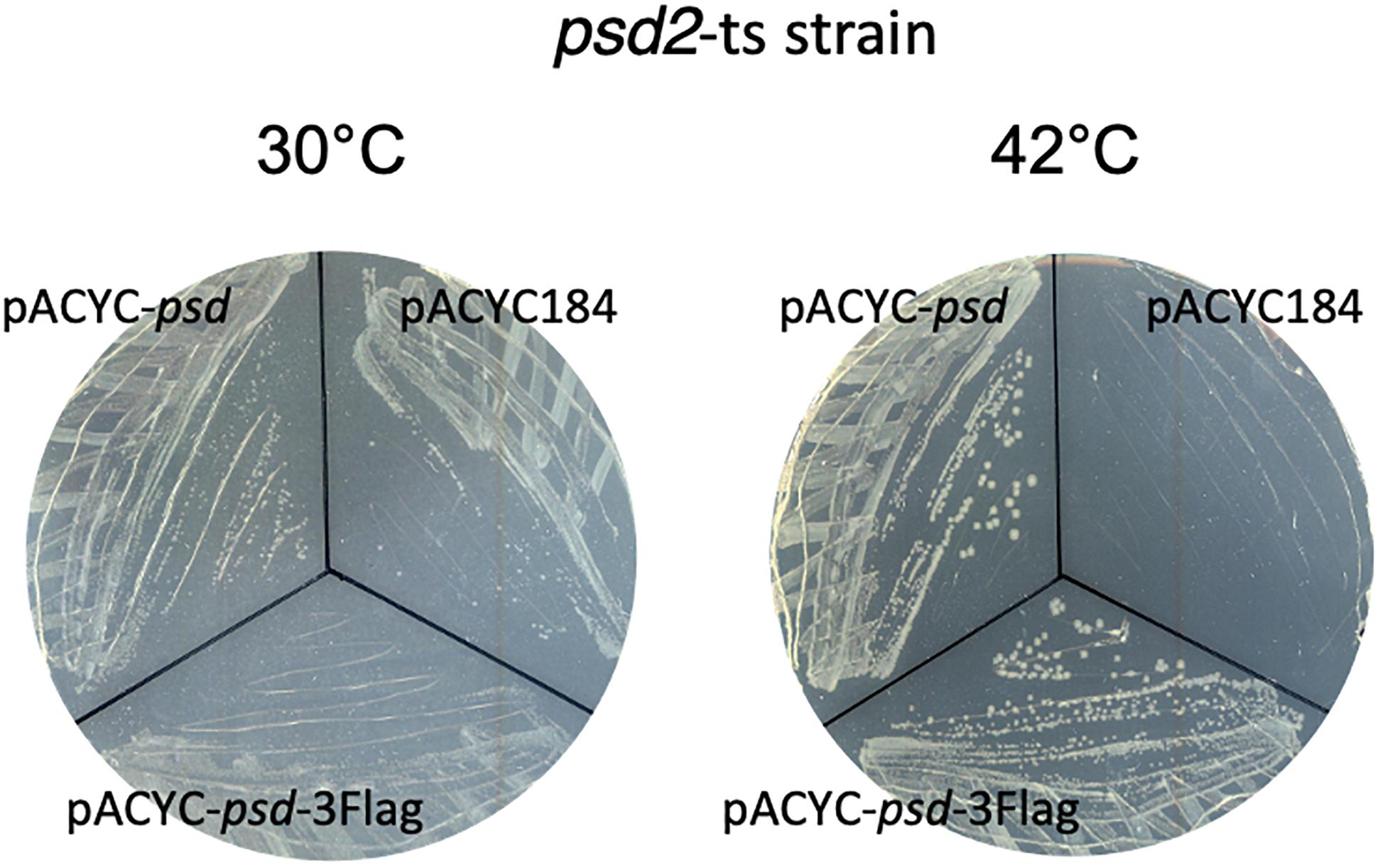
Figure 4. Strain EH150 psd2-ts (Hawrot and Kennedy, 1975) was transformed by pACYC184, pACYC-psd, or pACYC-psd-3Flag plasmids and the selection was performed at 30°C. Then, colonies were spread out on LB chloramphenicol plates at 30 or 42°C for 24 h.
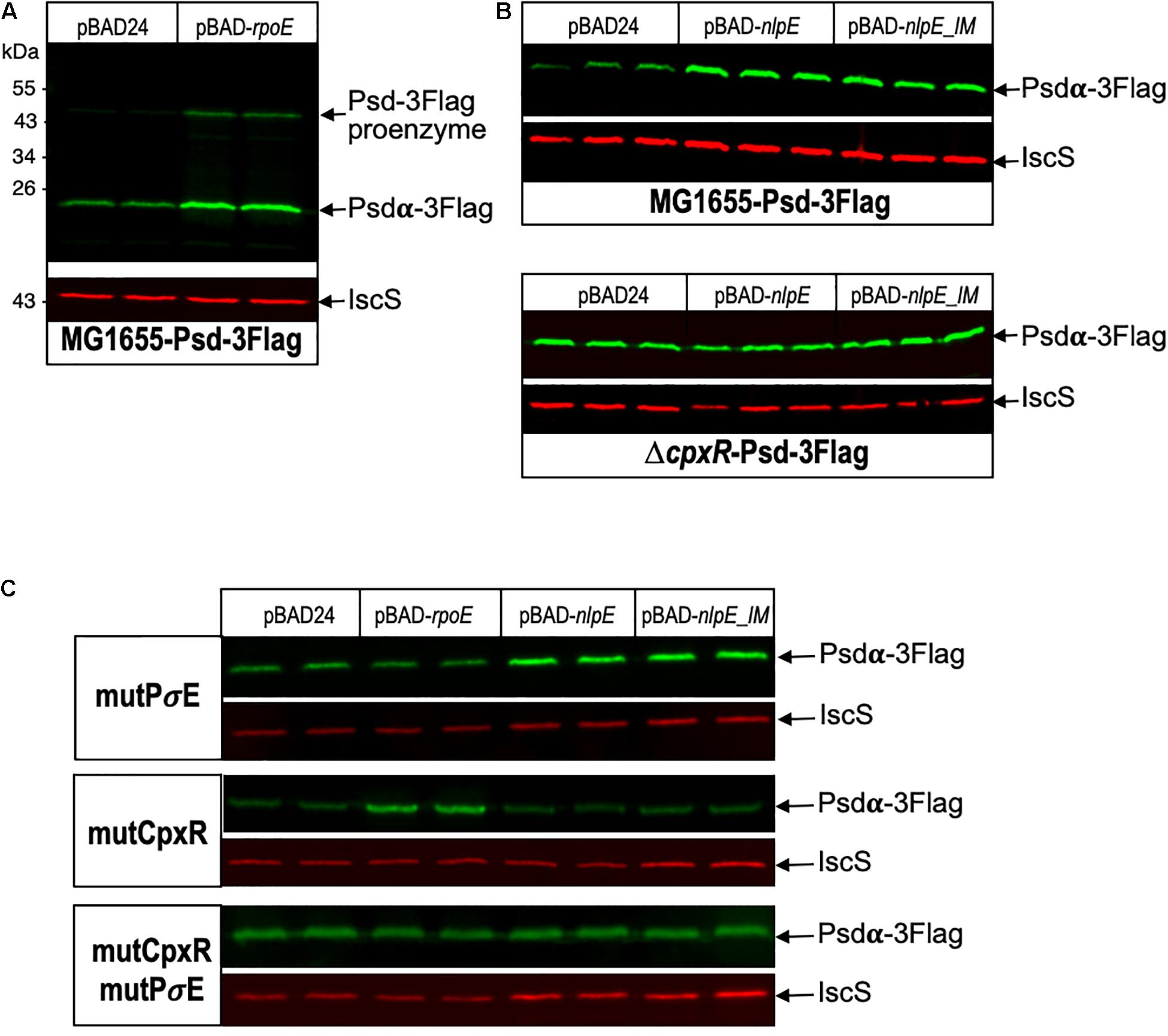
Figure 5. Psd protein levels are controlled by σE and CpxR. (A) E. coli Psd-3Flag strain was transformed by pBAD24 or pBAD-rpoE plasmids. Two replicas of each transformation were grown in LB supplemented with ampicillin until OD600 nm = 0.8, then cultures were induced with 0.2% arabinose for 2 h. (B) Psd-3Flag and ΔcpxR-Psd-3Flag strains were transformed by pBAD24, pBAD-nlpE, or pBAD-nlpE_IM plasmids. Three replicas of each transformation were grown in LB supplemented with ampicillin until OD600 nm = 0.8, then cultures were induced with 0.2% arabinose for 2 h. (C) Psd-3Flag_mutPσE, Psd-3Flag_mutCpxR, and Psd-3Flag_mutCpxRmutPσE strains containing mutations in the promoter region were transformed by pBAD24, pBAD-rpoE, pBAD-nlpE, or pBAD-nlpE_IM plasmids. Two replicas of each transformation were grown in LB supplemented with ampicillin until OD600 nm = 0.8, then cultures were induced with 0.2% arabinose for 2 h. Proteins were separated by SDS-PAGE 12% and detected by western blot using anti-Flag monoclonal antibody to detect Psd-3Flag or anti-IscS polyclonal antibody as an internal loading control. Quantification of the bands are shown in Supplementary Figure 4.
On the genome of E. coli, psd ORF is followed by mscM ORF, suggesting an organization of these two genes as an operon. To study the involvement of σE and CpxR on mscM expression, we constructed a strain where MscM is fused at its C-terminus with the SPA tag. MscM-SPA was faintly detected on Western blot at the expected size of 130 kDa, and MscM protein levels were increased when rpoE or nlpE were artificially expressed, similarly as for Psd-3Flag (Figure 6A). We then performed an RT-PCR experiment, which confirmed that psd and mscM are expressed as an operon, controlled by the same two promoters (Figure 6B).
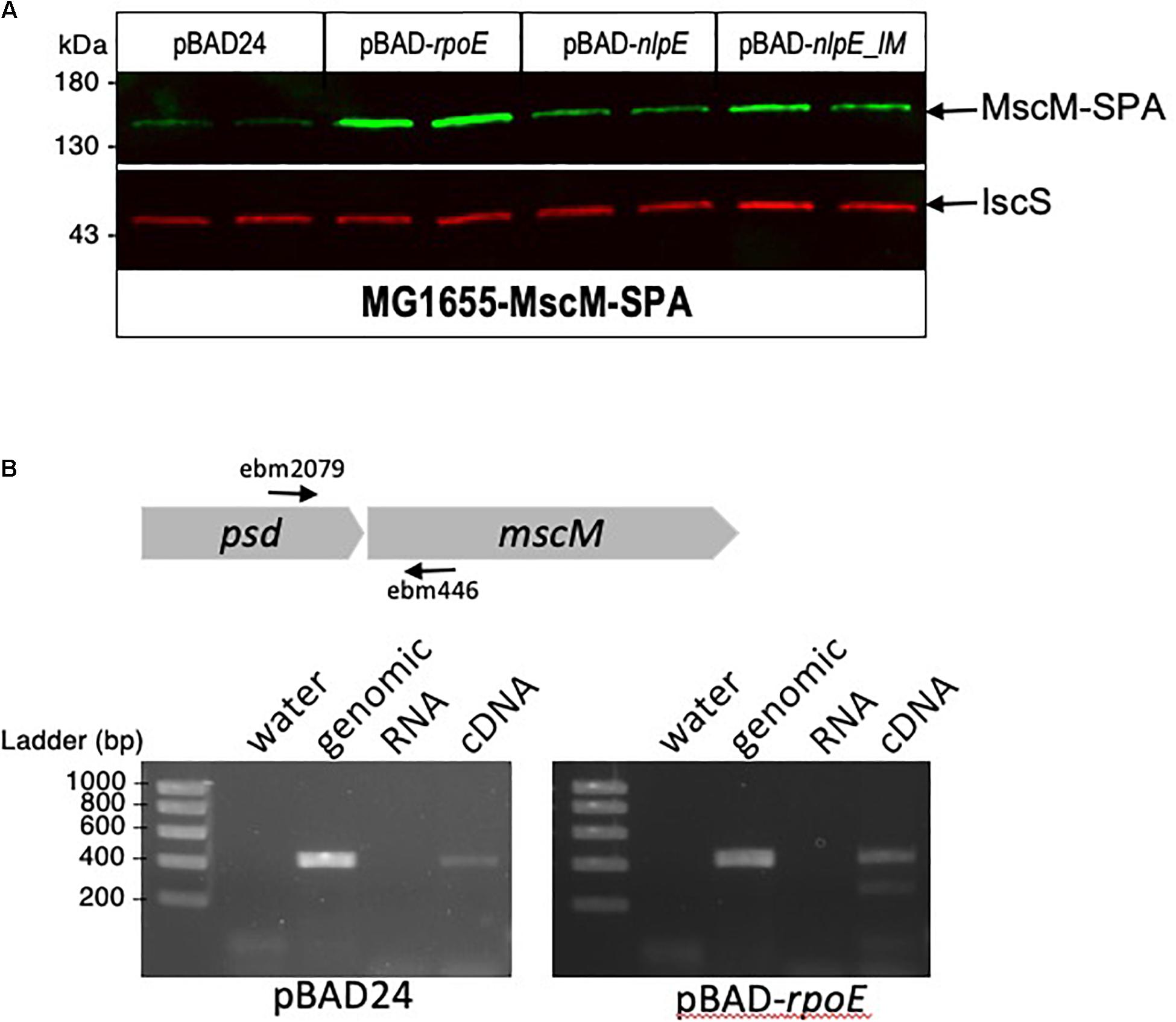
Figure 6. mscM is in operon with psd, under control of the σE and CpxR dependent promoters. (A) E. coli MscM-SPA strain was transformed by pBAD24, pBAD-rpoE, pBAD-nlpE, or pBAD-nlpE_IM plasmids. Two replicas of each transformation were grown in LB supplemented with ampicillin until OD600 nm = 0.8, then cultures were induced with 0.2% arabinose for 2 h. Proteins were separated by SDS-PAGE 10% and detected by western blot using anti-Flag monoclonal antibody for the detection of MscM-SPA or anti-IscS polyclonal antibodies as an internal loading control. (B) RT-PCR was performed on MG1655 strain transformed by pBAD or pBAD-rpoE induced in the above conditions. The black arrows indicate the positions of the primers used for the amplification. PCR controls on water, genomic DNA, and equivalent amounts of RNA as in the RT-PCR reaction were performed.
Characterization of Strains Devoid of psd-mscM Control by σE and CpxR
The above experiments using transcriptional fusions or following the amount of Psd and MscM proteins demonstrated the control of psd and mscM expression by the two envelope stress response pathways mediated by σE or CpxR. The next question was naturally to understand the physiological role of these regulations. To address this question, we chose to introduce point mutations in the promoters of psd, on the chromosome, in order to prevent specifically the activation of psd-mscM expression by σE, CpxR or both stress responses at the same time. We have shown above that the mutations chosen in the σE promoter and CpxR box completely abolished the induction of the transcriptional fusions by pBAD-rpoE or pBAD-nlpE, respectively (Figures 2, 3). Note that both mutations are localized inside the ORF of the upstream rsgA gene. The mutation in the PsdPσE promoter does not modify the amino acid sequence of RsgA, while the mutation in the CpxR box does modify residue Lys340 of RsgA in Glutamine. We introduced the mutations in wild-type MG1655 strain and in the Psd-3Flag strain using homologous recombination with the pKO3 plasmid (Link et al., 1997). The three types of mutants were readily obtained in the two strains (Table 2). The mutant strains did not display any obvious growth phenotype in LB (Supplementary Figure 3) showing that σE and CpxR regulations are not essential in balanced growth conditions. In the Psd-3Flag background strains, we observed that mutations in the PσE promoter and CpxR box had no effect on the basal amounts of Psd-3Flag protein (Figure 5C and data not shown). This suggests that the decreased expression observed with the psdP2muCpxR transcriptional fusion (Figure 3A) does not impact protein level, and shows that these regulations are not important to control basal Psd and MscM protein levels. However, when σE was induced, the amounts of Psd-3Flag protein did not increase anymore in the strains containing the PσE mutation (Figure 5C, first and third panel), while Psd-3Flag still increased in the CpxRbox mutant (Figure 5C, second panel). When the CpxRA pathway was induced, with pBAD-nlpE or pBAD-nlpE_IM plasmids, Psd-3Flag protein amounts did not increase anymore in the strains containing the mutation in the CpxR binding site (Figure 5C, second and third panel), while Psd-3Flag still increased in the PσE mutant (Figure 5C, first panel). These results confirmed what we observed with the transcriptional fusions (Figure 3A), and showed that the σE and CpxR control of psd promoters impact the amount of proteins produced in vivo upon stress induction.
Discussion
The goal of this work was to decipher the genetic control of psd-mscM operon and its regulation by the two envelope stress responses mediated by the alternative Sigma factor σE and by the two-component system CpxRA. We found that psd promoter region comprises two promoters, confirming the +1 transcription start sites identified previously (Thomason et al., 2015): a distal σE promoter and a proximal P2 promoter, activated by CpxRA, which is likely to be also responsible for the basal expression of psd. This organization is very similar to what we observed previously for the promoter of plsB, which comprises a distal σE promoter and a proximal P2 promoter, responsible for the basal expression of plsB (Wahl et al., 2011). However, plsB expression was not controlled by CpxR (data not shown). We also systematically tested if other genes of PL synthesis were regulated by σE or CpxR but it was not the case (data not shown). Therefore, it appears that envelope stress responses control the first step of PL synthesis (plsB) and the last step of PE synthesis (psd). This suggests that in response to envelope stress, an increase in the pathway for PE might be important, in order to help in envelope biogenesis processes such as LPS or outer membrane proteins trafficking. An enhanced PE biosynthesis might be obtained by “pushing” from the top with PlsB and “pulling” from the bottom of the pathway with Psd. However, enzymes of PL synthesis are believed to be present in excess and it is not clear how increasing enzyme amounts might have an effect on the flux in the PL synthetic pathway. This regulation might be a long-term adaptation rather than an immediate response to the encountered stress. An alternative hypothesis is that the enzymes might be damaged during envelope stress and need to be replaced.
We previously showed that the basal promoter of the plsB gene was repressed by ppGpp (Wahl et al., 2011). Furthermore, it was proposed that the genes for PE pathway, including psd, might be downregulated during the stringent response, while the genes for PG/CL were upregulated (Durfee et al., 2008), which would be consistent with an increase in PG and CL in the membrane during growth arrest. However, we did not detect a strong regulation of psdP2 promoter by ppGpp (Supplementary Figure 1).
It is striking to find psd in operon with a gene encoding a mechanosensitive channel. There is no obvious direct functional link between these two genes, apart from a function in membrane homeostasis. Mechanosensitive channels open in response to transition from high to low osmolarity in order to protect cells from bursting (Booth, 2014). There are seven mechanosensitive channels in E. coli, MscL and six channels of the MscS family, including MscM. This redundancy might be explained by a need to respond to different rates or intensities in osmolarity variations (Booth, 2014). As for psd, a genetic regulation might appear irrelevant for a rapid response to physical stress. However, the number and types of channels are important factors to determine the protective ability and cell fate (Bialecka-Fornal et al., 2015), and it has been shown that over-expression of MscM confers protection to hypo-osmotic shock in a strain otherwise devoid of all the other mechanosensitive channels (Edwards et al., 2012). Therefore, increased expression of mscM by σE and CpxR in response to envelope stress might be a bet hedging strategy, as it might confer adaptation for future additional envelope injuries. Similarly, mscL and mscS genes are regulated by the alternative Sigma factor σS (Stokes et al., 2003). Also, in Pseudomonas aeruginosa, the cmpX gene coding for a putative mechanosensitive channel is part of the SigX envelope stress response regulon (Gicquel et al., 2013). Furthermore, these mechanosensitive channels possess diverse periplasmic and cytoplasmic domains that might play additional functions in stress responses, which for example was suggested recently for YbdG in E. coli (Amemiya et al., 2019).
Unfortunately, we were not able yet to find growth conditions or stress conditions where the σE or CpxRA regulations on plsB and psd-mscM had a role for cell physiology. We tried to subject the mutant strains to stresses known to induce the σE or CpxRA pathways, i.e., osmotic stress, alkaline stress, envelope porin misfolding or mislocalization, or heat shock, but we did not detect any effects in these experiments. As mentioned above, these regulations are certainly involved in long-term adaptation rather than being an immediate response to the encountered stress. This might be why we did not detect any obvious phenotypic defects in the regulatory mutant strains when testing for immediate resistance to stress. Combining stresses, long-term, or competition experiments might be needed to be able to unravel a phenotypic advantage of the wild-type over the regulatory mutant strains. Interestingly, and in a reversed way, several studies have shown that PL synthesis mutants, which affect membrane composition, activate envelope stress responses, including σE and CpxRA signaling pathways (Mileykovskaya and Dowhan, 1997; Itou et al., 2012; Rowlett et al., 2017; Nepper et al., 2019). Therefore, the σE or CpxRA regulations of plsB and psd-mscM might also be part of a feedback loop involved in maintaining an optimum PL synthesis balance for membrane homeostasis.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
Author Contributions
YH, EB, and JV contributed to conception and design of the study. YH, JB, and AW performed experiments. EB wrote the manuscript. All authors contributed to manuscript revision, read, and approved the submitted version.
Funding
This work was funded by the Centre National de la Recherche Scientifique (CNRS), the Pasteur Institute, and grant ANR-19-CE13-0009-02. YH was supported by a doctoral fellowship from the French Ministry of Higher Education and Research.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Celia Torrielli for the construction of E. coli strains.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmolb.2021.665977/full#supplementary-material
References
Amemiya, S., Toyoda, H., Kimura, M., Saito, H., Kobayashi, H., Ihara, K., et al. (2019). The mechanosensitive channel YbdG from Escherichia coli has a role in adaptation to osmotic up-shock. J. Biol. Chem. 294, 12281–12292. doi: 10.1074/jbc.ra118.007340
Baba, T., Ara, T., Hasegawa, M., Takai, Y., Okumura, Y., Baba, M., et al. (2006). Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2:2006.0008.
Bialecka-Fornal, M., Lee, H. J., and Phillips, R. (2015). The rate of osmotic downshock determines the survival probability of bacterial mechanosensitive channel mutants. J. Bacteriol. 197, 231–237. doi: 10.1128/jb.02175-14
Booth, I. R. (2014). Bacterial mechanosensitive channels: progress towards an understanding of their roles in cell physiology. Curr. Opin. Microbiol. 18, 16–22. doi: 10.1016/j.mib.2014.01.005
Busby, S. J. W. (2019). Transcription activation in bacteria: ancient and modern. Microbiology 165, 386–395. doi: 10.1099/mic.0.000783
Chang, A. C., and Cohen, S. N. (1978). Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J. Bacteriol. 134, 1141–1156. doi: 10.1128/jb.134.3.1141-1156.1978
Cherepanov, P. P., and Wackernagel, W. (1995). Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene 158, 9–14. doi: 10.1016/0378-1119(95)00193-a
Choi, J. Y., Duraisingh, M. T., Marti, M., Ben Mamoun, C., and Voelker, D. R. (2015). From Protease to Decarboxylase: the molecular metamorphosis of phosphatidylserine decarboxylase. J. Biol. Chem. 290, 10972–10980.
Costanzo, A., Nicoloff, H., Barchinger, S. E., Banta, A. B., Gourse, R. L., and Ades, S. E. (2008). ppGpp and DksA likely regulate the activity of the extracytoplasmic stress factor sigmaE in Escherichia coli by both direct and indirect mechanisms. Mol. Microbiol. 67, 619–632. doi: 10.1111/j.1365-2958.2007.06072.x
Datsenko, K. A., and Wanner, B. L. (2000). One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U.S.A. 97, 6640–6645. doi: 10.1073/pnas.120163297
De Wulf, P., McGuire, A. M., Liu, X., and Lin, E. C. (2002). Genome-wide profiling of promoter recognition by the two-component response regulator CpxR-P in Escherichia coli. J. Biol. Chem. 277, 26652–26661. doi: 10.1074/jbc.m203487200
Delhaye, A., Collet, J. F., and Laloux, G. (2016). Fine-tuning of the Cpx envelope stress response is required for cell wall homeostasis in Escherichia coli. mBio 7:e00047-16.
Durfee, T., Hansen, A. M., Zhi, H., Blattner, F. R., and Jin, D. J. (2008). Transcription profiling of the stringent response in Escherichia coli. J. Bacteriol. 190, 1084–1096. doi: 10.1128/jb.01092-07
Edwards, M. D., Black, S., Rasmussen, T., Rasmussen, A., Stokes, N. R., Stephen, T. L., et al. (2012). Characterization of three novel mechanosensitive channel activities in Escherichia coli. Channels 6, 272–281. doi: 10.4161/chan.20998
Eichel, J., Chang, Y. Y., Riesenberg, D., and Cronan, J. E. (1999). Effect of ppGpp on Escherichia coli cyclopropane fatty acid synthesis is mediated through the RpoS sigma factor (sigmaS). J. Bacteriol. 181, 572–576. doi: 10.1128/jb.181.2.572-576.1999
Gicquel, G., Bouffartigues, E., Bains, M., Oxaran, V., Rosay, T., Lesouhaitier, O., et al. (2013). The extra-cytoplasmic function sigma factor sigX modulates biofilm and virulence-related properties in Pseudomonas aeruginosa. PLoS One 8:e80407. doi: 10.1371/journal.pone.0080407
Gully, D., Moinier, D., Loiseau, L., and Bouveret, E. (2003). New partners of acyl carrier protein detected in Escherichia coli by tandem affinity purification. FEBS Lett. 548, 90–96. doi: 10.1016/s0014-5793(03)00746-4
Guzman, L. M., Belin, D., Carson, M. J., and Beckwith, J. (1995). Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177, 4121–4130.
Hawrot, E., and Kennedy, E. P. (1975). Biogenesis of membrane lipids: mutants of Escherichia coli with temperature-sensitive phosphatidylserine decarboxylase. Proc. Natl. Acad. Sci. U.S.A. 72, 1112–1116. doi: 10.1073/pnas.72.3.1112
Itou, A., Matsumoto, K., and Hara, H. (2012). Activation of the Cpx phosphorelay signal transduction system in acidic phospholipid-deficient pgsA mutant cells of Escherichia coli. Biochem. Biophys. Res. Commun. 421, 296–300. doi: 10.1016/j.bbrc.2012.04.003
Li, Q. X., and Dowhan, W. (1990). Studies on the mechanism of formation of the pyruvate prosthetic group of phosphatidylserine decarboxylase from Escherichia coli. J. Biol. Chem. 265, 4111–4115. doi: 10.1016/s0021-9258(19)39709-1
Link, A. J., Phillips, D., and Church, G. M. (1997). Methods for generating precise deletions and insertions in the genome of wild-type Escherichia coli: application to open reading frame characterization. J. Bacteriol. 179, 6228–6237. doi: 10.1128/jb.179.20.6228-6237.1997
Mileykovskaya, E., and Dowhan, W. (1997). The Cpx two-component signal transduction pathway is activated in Escherichia coli mutant strains lacking phosphatidylethanolamine. J. Bacteriol. 179, 1029–1034. doi: 10.1128/jb.179.4.1029-1034.1997
Mitchell, A. M., and Silhavy, T. J. (2019). Envelope stress responses: balancing damage repair and toxicity. Nat. Rev. Microbiol. 17, 417–428. doi: 10.1038/s41579-019-0199-0
My, L., Rekoske, B., Lemke, J. J., Viala, J. P., Gourse, R. L., and Bouveret, E. (2013). Transcription of the Escherichia coli fatty acid synthesis operon fabHDG is directly activated by FadR and inhibited by ppGpp. J. Bacteriol. 195, 3784–3795. doi: 10.1128/jb.00384-13
Nepper, J. F., Lin, Y. C., and Weibel, D. B. (2019). Rcs phosphorelay activation in cardiolipin-deficient Escherichia coli reduces biofilm formation. J. Bacteriol. 201:e00804-18.
Parsons, J. B., and Rock, C. O. (2013). Bacterial lipids: metabolism and membrane homeostasis. Prog. Lipid Res. 52, 249–276. doi: 10.1016/j.plipres.2013.02.002
Rezuchova, B., Miticka, H., Homerova, D., Roberts, M., and Kormanec, J. (2003). New members of the Escherichia coli sigmaE regulon identified by a two-plasmid system. FEMS Microbiol. Lett. 225, 1–7. doi: 10.1016/s0378-1097(03)00480-4
Rhodius, V. A., Suh, W. C., Nonaka, G., West, J., and Gross, C. A. (2006). Conserved and variable functions of the sigmaE stress response in related genomes. PLoS Biol. 4:e2. doi: 10.1371/journal.pbio.0040002
Rowlett, V. W., Mallampalli, V. K. P. S., Karlstaedt, A., Dowhan, W., Taegtmeyer, H., Margolin, W., et al. (2017). Impact of membrane phospholipid alterations in Escherichia coli on cellular function and bacterial stress adaptation. J. Bacteriol. 199:e00849-16.
Sohlenkamp, C., and Geiger, O. (2016). Bacterial membrane lipids: diversity in structures and pathways. FEMS Microbiol. Rev. 40, 133–159. doi: 10.1093/femsre/fuv008
Stokes, N. R., Murray, H. D., Subramaniam, C., Gourse, R. L., Louis, P., Bartlett, W., et al. (2003). A role for mechanosensitive channels in survival of stationary phase: regulation of channel expression by RpoS. Proc. Natl. Acad. Sci. U.S.A. 100, 15959–15964. doi: 10.1073/pnas.2536607100
Thomason, M. K., Bischler, T., Eisenbart, S. K., Förstner, K. U., Zhang, A., Herbig, A., et al. (2015). Global transcriptional start site mapping using differential RNA sequencing reveals novel antisense RNAs in Escherichia coli. J. Bacteriol. 197, 18–28. doi: 10.1128/jb.02096-14
Wahl, A., My, L., Dumoulin, R., Sturgis, J. N., and Bouveret, E. (2011). Antagonistic regulation of dgkA and plsB genes of phospholipid synthesis by multiple stress responses in Escherichia coli. Mol. Microbiol. 80, 1260–1275. doi: 10.1111/j.1365-2958.2011.07641.x
Wang, A. Y., and Cronan, J. E. (1994). The growth phase-dependent synthesis of cyclopropane fatty acids in Escherichia coli is the result of an RpoS(KatF)-dependent promoter plus enzyme instability. Mol. Microbiol. 11, 1009–1017. doi: 10.1111/j.1365-2958.1994.tb00379.x
Zaslaver, A., Bren, A., Ronen, M., Itzkovitz, S., Kikoin, I., Shavit, S., et al. (2006). A comprehensive library of fluorescent transcriptional reporters for Escherichia coli. Nat. Methods 3, 623–628. doi: 10.1038/nmeth895
Keywords: psd, mscM, SigmaE, CpxRA, envelope stress response, E. coli, phospholipid synthesis
Citation: Hassoun Y, Bartoli J, Wahl A, Viala JP and Bouveret E (2021) Dual Regulation of Phosphatidylserine Decarboxylase Expression by Envelope Stress Responses. Front. Mol. Biosci. 8:665977. doi: 10.3389/fmolb.2021.665977
Received: 09 February 2021; Accepted: 09 April 2021;
Published: 07 May 2021.
Edited by:
Heidi Vitrac, University of Texas Health Science Center at Houston, United StatesReviewed by:
Stanley Spinola, Indiana University–Purdue University Indianapolis, United StatesTracy Raivio, University of Alberta, Canada
Copyright © 2021 Hassoun, Bartoli, Wahl, Viala and Bouveret. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Julie Pamela Viala, jviala@imm.cnrs.fr; Emmanuelle Bouveret, emmanuelle.bouveret@pasteur.fr
 Yasmine Hassoun
Yasmine Hassoun Julia Bartoli
Julia Bartoli Astrid Wahl1
Astrid Wahl1  Emmanuelle Bouveret
Emmanuelle Bouveret