- 1Antibiotics Research and Re-evaluation Key Laboratory of Sichuan Province, School of Pharmacy, Chengdu University, Chengdu, Sichuan, China
- 2Affiliated Hospital of Chengdu University, Chengdu University, Chengdu, Sichuan, China
Colonization of Pseudomonas aeruginosa in the lung environments frequently leads to the enrichment of strains displaying enhanced antibiotic resistance and reduced production of quorum-sensing (QS) controlled products. However, the relationship between the emergence of QS deficient variants and antibiotic resistance remains less understood. In this study, 67 P. aeruginosa strains were isolated from the lungs of 14 patients with chronic obstructive pulmonary disease, followed by determining their genetic relationship, QS-related phenotypes and resistance to commonly used antibiotics. The integrity of P. aeruginosa QS system was checked by DNA sequencing. The relationship between the QS system and antibiotic resistance was then assessed by correlation analyses. The function of the LasR protein and bacterial virulence were evaluated through homology modeling and nematode-infection assay. The influence of antibiotic on the development of extracellular protease production ability of P. aeruginosa was tested by an evolutionary experiment. The results showed that P. aeruginosa clinical strains displayed abundant diversity in phenotype and genotype. The production of extracellular proteases was significantly negatively correlated with antibiotic resistance. The strains with enhanced antibiotic resistance also showed a notable overlap with the mutation of lasR gene, which is the core regulatory gene of P. aeruginosa QS system. Molecular docking and Caenorhabditis elegans infection assays further suggested that P. aeruginosa with impaired LasR protein could also have varying pathogenicity. Moreover, in vitro evolution experiments demonstrated that antibiotic-mediated selective pressure, particularly from Levofloxacin contributed to the emergence of extracellular protease-negative strains. Therefore, this study provides evidence for the connection of P. aeruginosa QS system and antibiotic resistance, and holds significance for developing targeted strategies to address antibiotic resistance and improving the management of antibiotic-resistant infections in chronic respiratory diseases.
Introduction
Multidrug resistance (MDR) has been increased all over the world that is considered a public health threat (Magiorakos et al., 2012; Algammal et al., 2023a). Several recent investigations reported the emergence of multidrug-resistant bacterial pathogens from different origins that increase the necessity for the proper use of antibiotics (Algammal et al., 2022, 2023b; Elbehiry et al., 2022; Shafiq et al., 2022). The common and essential use of antibiotics in managing chronic bacterial infections, often involving concurrent administration of multiple antibiotics, inevitably leads to the emergence and proliferation of MDR bacterial strains.
Pseudomonas aeruginosa is a predominant pathogen in hospital-associated MDR events (King, 2019; Mac Aogáin et al., 2021). It can cause various infections, serving as a major contributor to sepsis and neutropenia in immunocompromised patients, and standing as a primary cause of hospital-acquired pneumonia and respiratory failure (Gallego et al., 2014; Rodrigo-Troyano et al., 2018). Pseudomonas aeruginosa is one of the most frequently isolated bacterial pathogens from airway samples (Garcia-Vidal et al., 2009). It poses a significant challenge as it secretes various virulence factors, such as elastase and lipase, which can destroy lung tissue and aggravate inflammatory reactions. Additionally, P. aeruginosa can form biofilms, which can resist elimination by the host immune system and inhibit the effects of antibiotics (Filloux, 2011; Hilker et al., 2015). Therefore, regular antimicrobial susceptibility testing is essential for selecting appropriate antibiotics and detecting emerging multidrug-resistant strains of P. aeruginosa. Simultaneously, a thorough investigation into the characteristics and pathogenicity of clinical P. aeruginosa is crucial for identifying and evaluating infection control strategies (Høiby et al., 2015; Miravitlles and Anzueto, 2017; Behzadi et al., 2021; Langendonk et al., 2021).
The quorum-sensing (QS) system of P. aeruginosa is known to be involved in cell-to-cell communication mediated by signaling molecules that respond to changes in cell density (Schuster and Greenberg, 2006; Jimenez et al., 2012). The essential communication facilitated by QS enables coordinated behavior and regulates key virulence factors in P. aeruginosa, enhancing pathogenicity and environmental adaptability (Wilder et al., 2011; Chevalier et al., 2017). For instance, pyocyanin, a phenazine, aids in redox reactions for interspecies competition; proteases facilitate tissue invasion and immune evasion; rhamnolipids, acting as surfactants, contribute to adhesion, biofilm formation, and migration (Hancock and Speert, 2000; Liang et al., 2014). Additionally, QS-controlled motility (swimming, swarming, and twitching) crucially influences biofilm formation and increases antibiotic resistance (Oshri et al., 2018; Pena et al., 2019). Currently, three QS systems, including las, rhl, and pqs, have been confirmed in P. aeruginosa. LasR, RhlR, and PqsR are three transcriptional regulatory proteins at the core of the interlinked QS system, interacting with homologous chemical molecules to induce the expression of downstream virulence factors (Kostylev et al., 2019).
Generally, the virulence factors of P. aeruginosa are not exclusively regulated by a single QS pathway. However, clinical experience suggests that P. aeruginosa in chronic respiratory infections exhibits phenotypic heterogeneity in cell motility and the production of characteristic virulence factors, indicating variations in QS system integrity (Heurlier et al., 2006; Feliziani et al., 2014; Jiricny et al., 2014; Vanderwoude et al., 2020). Previous research has indicated that this heterogeneity is influenced by environmental stress and the adaptability of P. aeruginosa, manifested in dissolved oxygen levels inside and outside the biofilm and low nutrient concentrations (Smith et al., 2006; Jorth et al., 2015; Zhao et al., 2016; Lozano et al., 2018; Figueiredo et al., 2021; Grekov et al., 2021). Theoretically, environmental pressures on the QS system, including antibiotics, may partially influence the rise in bacterial resistance. LasR mutants of P. aeruginosa isolated from cystic fibrosis patients exhibit a distinct QS regulatory hierarchy compared to the wild-type (Feltner et al., 2016). However, it is unclear if environmentally adaptive QS variants may be associated with clinical multidrug-resistant events.
Therefore, this study aims to explore the association between QS-related phenotypes and antibiotic resistance of P. aeruginosa clinical strains, with the expectation of providing valuable insights into the identification of clinical infection types and the application of antibiotic management. Firstly, we isolated 67 P. aeruginosa strains from the respiratory samples of 14 patients with chronic obstructive pulmonary disease (COPD). Subsequently, 26 representative strains were phenotypically identified through assays involving extracellular proteases, pyocyanin, biofilm, and cell motilities. Further analyses, including antimicrobial susceptibility testing, gene sequencing, and correlation assessments, revealed a connection between lasR mutations and phenotypic variations as well as antibiotic resistance. The functionality of the LasR protein and overall bacterial virulence were assessed through homology modeling and nematode infection assay. Finally, an evolutionary experiment simulated the impact of selective pressure from a single antibiotic on the QS system and the production of extracellular proteases.
Materials and methods
Ethics statement
Bronchoalveolar lavage fluids and sputum samples were obtained from 14 COPD patients hospitalized in the Department of Respiratory and Critical Care Medicine at the affiliated hospital of Chengdu University (Supplementary Table S1). Written informed consent was obtained from the patients or their immediate family members, and the study was approved by the Ethics Committee of the Affiliated Hospital of Chengdu University (Approval Code: PJ2020-021-03).
Identification of P. aeruginosa from clinical samples
The respiratory samples were stored on ice, liquefied by adding an equal volume of phosphate buffer solution (PBS), and cultured overnight at 37°C on lysogeny broth (LB; Hope Bio-Technology Co., Ltd, Qingdao, China) plates. Colonies with discernible variations in size, color, and surface smoothness were chosen and were subjected to 16S rRNA sequence-based species identification.
Phenotypic identification
Phenotypic characterization of P. aeruginosa COPD isolates was conducted using the methods described elsewhere (Zhao et al., 2020). Chemical reagents for the following experiments were purchased from Chengdu Kelong Chemical Co., Ltd. (Chengdu, China) unless specified otherwise. All the experiments were independently repeated three times and compared to the values of the reference strain wild-type (WT) PAO1, which was preserved in our laboratory (Zhao et al., 2020). In brief, the production of pyocyanin was determined by measuring the optical density at 520 nm (OD520) of the liquid after HCl-chloroform extraction. Biofilm production was assessed by staining with 0.1% ammonium oxalate-crystal violet dye, followed by elution with 95% ethanol. The eluate's optical density at 595 nm (OD595) was measured. For the cell motility assay, the bacterial solution is inoculated in the center of an LB plate with 0.5% agar and in the bottom plastic layer of an LB plate with 1% agar, followed by measuring the colony movement diameter. For the production of extracellular proteases, inoculate bacteria on a 0.5% M9-skim milk (Sigma-Aldrich, USA) plate and measure the protease hydrolysis zone's diameter. For the adenosine utilization assay, inoculate bacterial solution on the surface of M9 solid medium with 0.1% adenosine (Sigma-Aldrich, USA) and assess colony growth relative to WT PAO1. Growth equivalent to WT PAO1 is denoted as +, better than WT PAO1 as ++, worse than WT PAO1 as –, and no growth as null. For the mucoid transformation level assay, mucus transformation was evaluated by observing colony growth on LB plates after 24 h of incubation at 37°C. The results were categorized as follows: null for round colonies without mucus between adjacent colonies, – for round colonies with adjacent mucus, and + for deformed, liquid colonies with viscous adjacent colonies.
Enterobacterial repetitive intergenic consensus-polymerase chain reaction
Enterobacterial repetitive intergenic consensus-polymerase chain reaction (ERIC-PCR) is a reliable method for analyzing genomic structure and genetic relationships among various bacteria, even non-enterobacteria (Stehling et al., 2010). Genomic DNA from clinical strains of P. aeruginosa and WT PAO1 was extracted using a Bacterial DNA Isolation Kit (Foregene Biotechnology, Co. Ltd., China), followed by PCR amplification using the single primer 5′-AAGTAAGTGACTGGGGTGAGCG-3′. Images were captured and processed using Image J v1.53a software. To construct a dendrogram, a binary matrix was created by designating the presence and absence of bands as 1 and 0, respectively, at the same positions. The matrix was then subjected to clustering analysis using the Cluster Vis platform at https://biit.cs.ut.e/ClustVis/.
Antimicrobial susceptibility testing
The minimum inhibitory concentrations (MICs) of commonly prescribed clinical antibiotics, including aminoglycosides (Amikacin, Gentamicin, and Tobramycin), antipseudomonal carbapenems (Imipenem), antipseudomonal cephalosporins (Cefotaxime, Cefepime), antipseudomonal fluoroquinolones (Levofloxacin, Ciprofloxacin), monobactams (Aztreonam), and polymyxins (Polymyxin B), were determined against P. aeruginosa isolates using the broth microdilution method according to the guidelines of the Clinical and Laboratory Standards Institute (CLSI, 2022). Ciprofloxacin and Levofloxacin were purchased from Shanghai Titan Technology Co., Ltd., and other antibiotics were purchased from Shanghai YuanYe Biotechnology Co., Ltd. The multiple antibiotic resistance (MAR) index was calculated following the method established by Krumperman (1983). Subsequently, the MAR index was categorized into high and low based on its median value. MAR values below the median were classified as low, while values equal to or exceeding the median were designated as high (Puspita et al., 2021).
PCR amplification
The complete sequences of the core regulatory genes of the P. aeruginosa QS system were initially amplified using PCR (lasR: 5′-TCAAACGCTGCGGTCTATT-3′ and 5′-CATCTCGCCCAGCAGTTT-3′, lasI: 5′-GCAGGGTTCTCGCCA TTCT-3′ and 5′-GCACCACCCACAGCATC-3, rhlR: 5′-GCTG GCATAACAGATAGGGT-3′ and 5′-CTCTCAGTCGGAGGACA TAC-3′, rhlI: 5′-ATCCGATGCTGATGTCCAA-3′ and 5′-TCTTCCGTGCGGTAGCTG-3′, pqsR: 5′-TTTCTTAGAACCGTT CCTGG-3′ and 5′-TGCTGGAGAACGCTCTACTC-3′), followed by DNA sequencing and sequence alignment to the genes of WT PAO1 (NCBI accession number: AE004091.2).
Homology modeling and molecular docking
We employed a multi-step approach that integrates homology modeling and molecular docking to investigate the binding interactions between LasR mutant variants and self-inducing ligands. Initially, the lasR gene sequences of seven distinct lasR mutants of P. aeruginosa COPD isolates in the present study, encompassing both point mutations and deletions, were modeled. To generate accurate 3D structures, we utilized the Swiss-Model (https://swissmodel.expasy.org/) and AlphaFold (https://alphafold.com/) platforms. Structural completion, energy minimization, and site-directed mutagenesis were conducted using SPDBV v4.10 software. Subsequently, we evaluated the structural disparities among the LasR mutant models to pinpoint unique features associated with each variant. Following model generation, we conducted molecular docking experiments with Dockey v0.8.2 (Du et al., 2023). Twenty docking runs were performed for each LasR mutant with self-inducing ligands to evaluate differences in binding free energies and interaction sites. PyMOL facilitated the visualization and analysis of results, offering insights into structural and functional distinctions among LasR variants.
Caenorhabditis elegans infection models
Caenorhabditis elegans serves as a valuable model organism for investigating the interplay between bacterial virulence systems and host defense mechanisms (Balasubramanian et al., 2016). For the fast-killing assay, we used the Peptone-Glucose-Sorbitol medium, while Nematode Growth Medium was employed for slow-killing assay. Nematodes were cultured at 37°C for 24 h. We introduced ten nematodes (for acute infection) or fifteen nematodes (for chronic infection), synchronized to the L4 stage, to plates with various bacterial strains and maintained them in a 25°C incubator. We monitored nematode survival at 4-h intervals for acute infection and 8-h intervals for chronic infection. Each experimental group included control groups fed with Escherichia coli OP50, a non-pathogenic strain commonly used to support nematode growth and reproduction.
In vitro evolution experiment under antibiotic stress
The evolution of WT PAO1 and its isogenic lasR mutant, PAO1-ΔlasR, in which the lasR gene was replaced by gentamicin cassette as described elsewhere (Zhao et al., 2019), under antibiotic pressure, was determined by using the methods established by Hernando-Amado et al. (2019) with slight modifications. Initially, the MICs of Polymyxin B, Cefepime, and Levofloxacin for WT PAO1 and PAO1-ΔlasR were determined (Supplementary Table S2). Sub-MIC concentrations were established by appropriately reducing antibiotic concentrations in LB broth, where growth rates were inhibited but viability remained unaffected. The initial antibiotic concentrations were as follows: Polymyxin B at 1.5 μg/ml, Cefepime at 1.5 μg/ml, and Levofloxacin at 0.25 μg/ml for WT PAO1. For PAO1-ΔlasR, the initial antibiotic concentrations were Polymyxin B at 0.25 μg/ml, Cefepime at 1 μg/ml, and Levofloxacin at 0.25 μg/ml. In the evolutionary experiment, WT PAO1 and PAO1-ΔlasR were cultured in 2 ml deep 96-well plates containing 1 ml of LB broth and Polymyxin B, Cefepime, or Levofloxacin at sub-MICs at 37°C with shaking (180 rpm), respectively. Subsequently, 100 μl of the culture liquid was transferred to fresh media with higher antibiotic concentrations every 3 days. The antibiotic concentrations were progressively increased until reaching final levels of Polymyxin B at 32 μg/ml, Cefepime at 128 μg/ml, and Levofloxacin at 32 μg/ml (Supplementary Table S3). The resulting cultures were diluted and plated on LB agar plates, from which 10 individual colonies were randomly selected per plate, yielding a total of 100 colonies per experimental group. Extracellular protease production was assessed and the lasR gene of the WT PAO1 group's cultured products was sequenced to investigate the genetic changes.
Statistical analysis
Statistical analysis was primarily conducted using GraphPad Prism v9.0. Quantitative experiments related to phenotypes were subjected to t-tests, and correlation analysis between drug resistance and phenotype was performed by calculating Spearman's rank correlation coefficient. Survival curves of nematodes were analyzed using the Log-rank (Mantel-Cox) test. The heatmaps for antibiotic resistance and bicomponent correlation analysis were generated using the online software package ChiPlot (https://www.chiplot.online/, Ji and Tang, 2022; Li et al., 2023). Molecular docking analysis was performed using Dockey and PyMOL software. Custom Python scripts were utilized for some composition ratio statistical analyses.
Results
COPD-derived P. aeruginosa strains display diversity in phenotype and genotype
We quantitatively or qualitatively measured QS-related phenotypes in clinically isolated P. aeruginosa strains, primarily to trace genetic and phenotypic differences among infection-causing strains, reflecting their diverse characteristics. The results of phenotypic identification revealed that about 80% (56/67) of P. aeruginosa clinical strains lost the capacity to produce extracellular proteases (Supplementary Figure S1). Moreover, 22% (15/67) of strains surpassed PAO1 in pyocyanin production, while 42% (28/67) produced less. In terms of biofilm production, 27% (18/67) exhibited a significant increase over PAO1, and 18% (12/67) showed a notable decrease. Regarding motility, 13% (9/67) had enhanced swarming compared to PAO1, while 40% (27/67) displayed reduced motility. In twitching motility, 9% (6/67) exceeded PAO1, and 43% (29/27) fell short.
Subsequently, 26 strains exhibiting distinct phenotypic variations (including colony shape, color, size, and surface characteristics) were chosen for further investigation. The levels of pyocyanin and biofilm production in P. aeruginosa from the same sample exhibited significant fluctuations. Genomic DNA-based fingerprinting was utilized to investigate the genetic relationships among the strains. Cluster analysis of the gel electrophoresis bands generated via ERIC-PCR identified 10 different types of P. aeruginosa, with four of them detected in various patient samples (Figure 1). The combination of phenotype identification and genotypic typing allows the selected clinical strains to be classified into two groups. The first group comprised 8 strains that exhibited genotypic similarity to WT PAO1. These strains, except C2 and J2, demonstrated positive extracellular protease production. In contrast, the second group displayed significant genotypic dissimilarity from WT PAO1 and exhibited negative extracellular protease production (except for strains B3, F8, E2, M1, and G4).
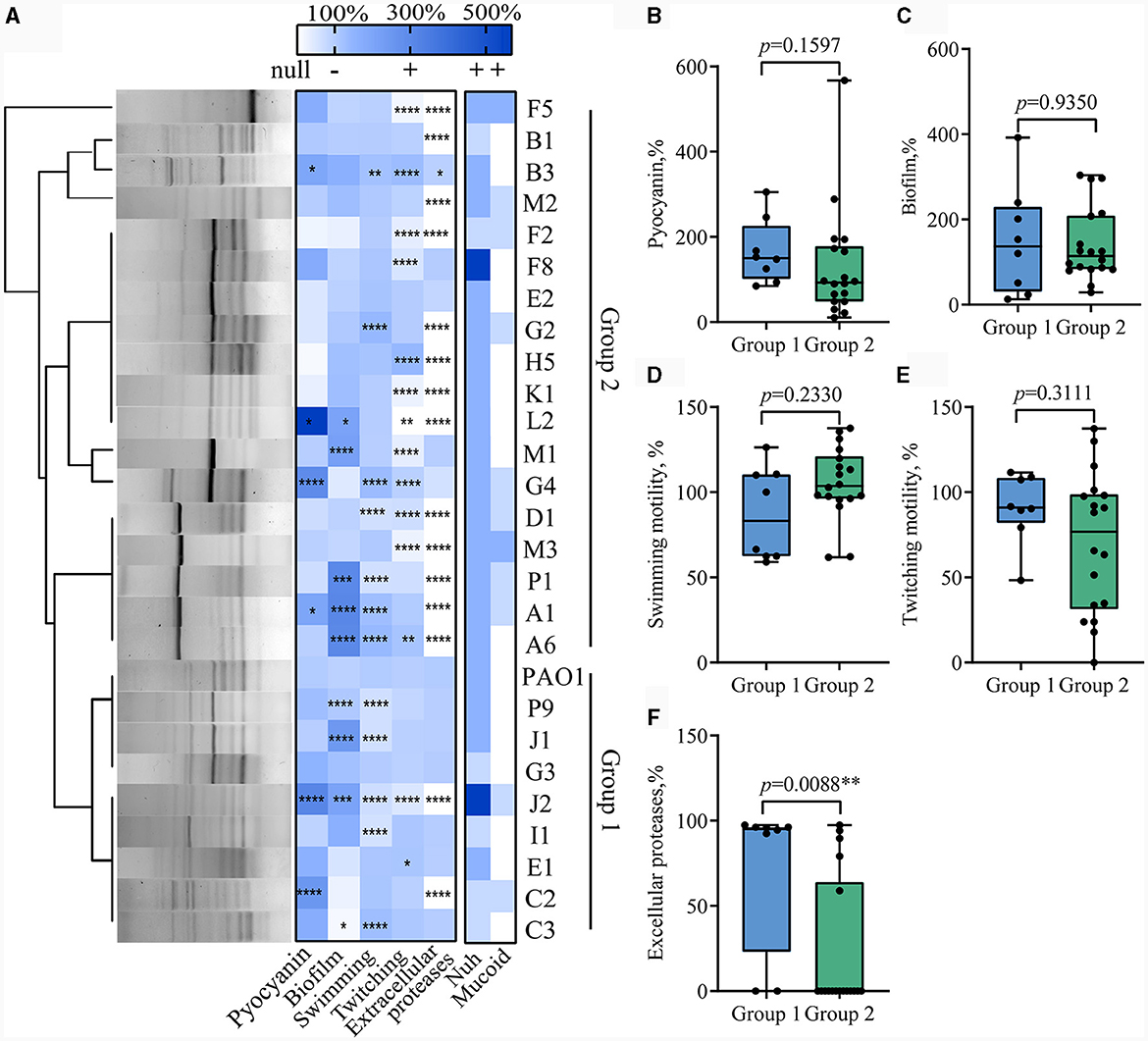
Figure 1. Diversity analysis of QS-related virulence phenotypes in clinical Pseudomonas aeruginosa isolates. (A) Genomic fingerprint typing and phenotypic thermography of the clinical P. aeruginosa isolates. The left side of the figure is the gel map of Eric PCR products after clustering, and the right side is the heat map of phenotypes related to the regulation of the QS system. t-test, Data are mean ± SD, n = 3 per group. (B–F) Box plots comparing extracellular proteases, pyocyanin, biofilm production, swimming motility, and twitching motility between Group 1 (n = 8) and Group 2 (n = 18). *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
Notably, even the strains with close genetic relationships isolated from the same patient displayed significant phenotypic differences and thus indicated substantial phenotypic heterogeneity among clinical strains. For instance, J1 and J2, despite having over 85% similarity in banding patterns, displayed distinct extracellular protease production capabilities, with J2 completely losing its extracellular protease production ability, while J1 retained the capacity to produce extracellular proteases similar to WT PAO1. Similar instances were observed with C2 and C3, B1 and B3, and F2 and F8 also.
Pseudomonas aeruginosa COPD isolates have a complex antibiotic resistance spectrum
To assess the antibiotic resistance of these clinical P. aeruginosa strains, in conjunction with the types of antibiotics administered to patients during hospitalization (Supplementary Table S1), this study selected 10 commonly used antibiotics and determined the MICs of the strains against these antibiotics. Antibiotic susceptibility tests indicated that the majority of P. aeruginosa clinical isolates in this study exhibited lower susceptibility to the tested antibiotics compared to the WT PAO1 (Supplementary Table S4). Among the 26 isolated P. aeruginosa strains, over half exhibited intermediate or resistant profiles to commonly used antibiotics for respiratory tract infection, such as Cefotaxime (54%, n = 14), Imipenem (54%, n = 14), and Aztreonam (50%, n = 13). Approximately 4%−15% of strains exhibit resistance to Tobramycin, Amikacin, Levofloxacin, and Polymyxin B.
Clustering of the antibiotic resistance profiles revealed that the P. aeruginosa clinical strains could primarily be categorized into three branches. The upper-left cluster (6/26) exhibited resistance to β-lactam antibiotics but sensitivity to quinolones and aminoglycoside antibiotics. The middle cluster (10/26) showed overall sensitivity to antibiotics, closely resembling the WT PAO1. The lower-right cluster (10/26) exhibited significantly higher antibiotic resistance, presenting a multi-antibiotic resistant profile to cephalosporins, carbapenems, and monobactam antibiotics (Figure 2). After calculating the MAR index, 26 strains were categorized into low MAR and high MAR groups (Figure 2). It's important to note that the classification of low MAR level and high MAR level was determined based on the median MAR index, where MAR values below 0.3 were considered low MAR, and values equal to or above 0.3 were considered high MAR. Heterogeneous resistance profiles were detected in the subgroups of P. aeruginosa within patients B, F, G, P, and J, where both low MAR and high MAR isolates coexisted. For instance, isolate B3 from patient B, with a MAR index of 0.2, was classified as low MAR, whereas B1, with a MAR index of 0.6, was classified as high MAR, displaying higher resistance profiles (Figure 2). Similar to the phenotypes of clinical P. aeruginosa strains, these findings highlight the considerable complexity in antibiotic susceptibilities among P. aeruginosa isolates from COPD airways.
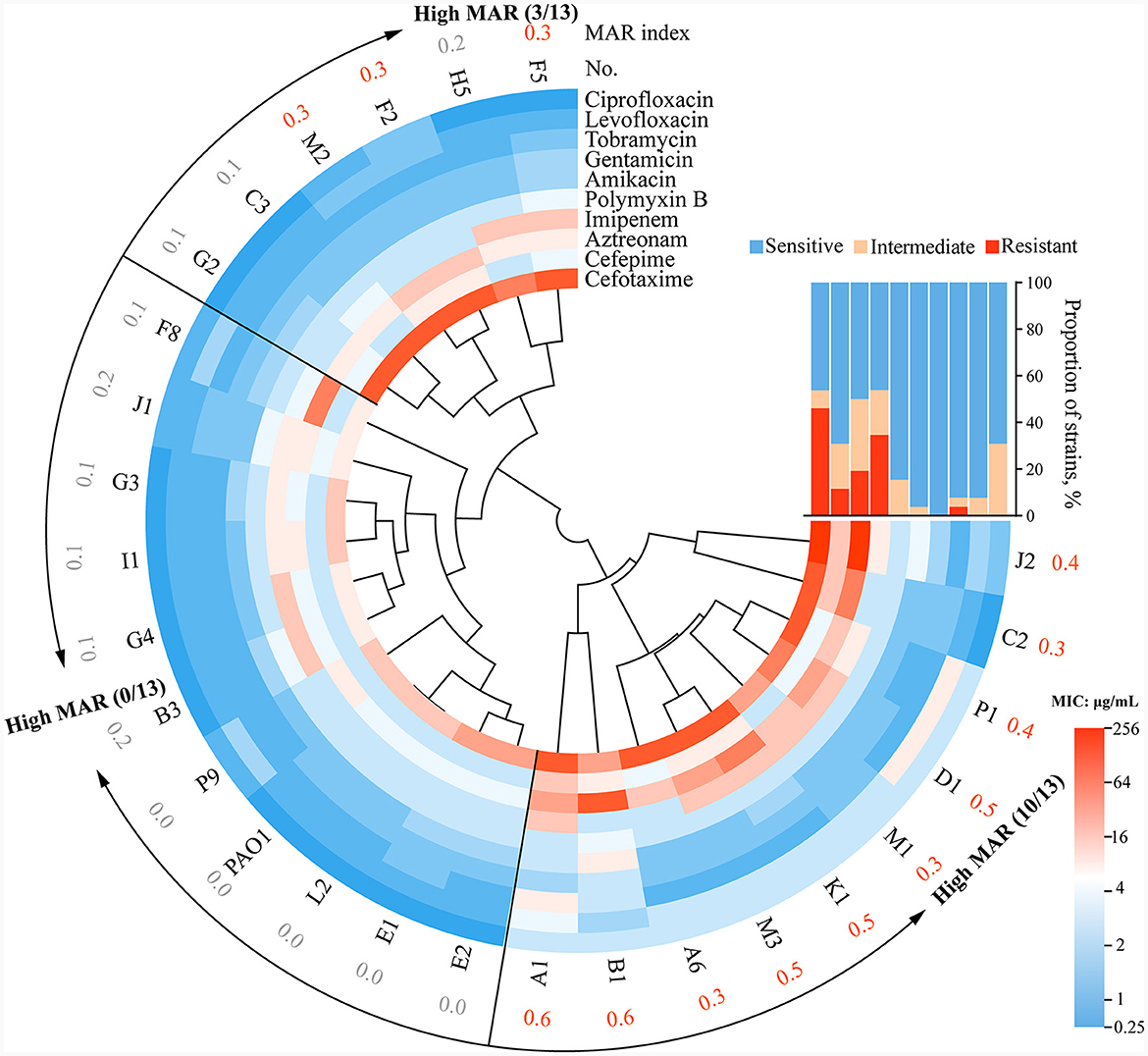
Figure 2. The circle diagram of drug resistance typing of clinical strains and the sensitivity rates to 10 antibiotics. Twenty-seven strains, including WT PAO1, were split into three groups based on the clustering results. MAR index = (total number of antibiotics tested)/(number of antibiotics resistant). Specifically, strains with a MAR index <0.3 were classified as low MAR, while those with a MAR index equal to or >0.3 were classified as high MAR, with the classification based on the median MAR index. The strains assigned the orange code based on the corresponding MAR index are defined as high MAR Group in this study. The interpretation scope is referenced from Supplementary Table S4: R, resistant, I intermediate, S, sensitive.
QS phenotypes are correlated with antibiotic resistance
We then set out to investigate whether there were characteristic phenotypes indicative of drug resistance levels in clinical P. aeruginosa. Upon combining the quantified heatmap of phenotypes with ERIC-PCR typing (Figures 1A, 2), It was evident that most of the strains exhibiting a high MAR profile had lost their ability to produce extracellular proteases. Conversely, strains with a low MAR profile, except for L2, H5, and G2, were proficient in secreting extracellular proteases. Spearman rank correlation coefficient analysis was performed to assess the relationship between various quantitative virulence-related phenotypes and antibiotic resistance (Figure 3A). The results showed a significant negative correlation between extracellular protease production and antibiotic resistance, with major antibiotics including Gentamicin, Cefotaxime, Levofloxacin, and Aztreonam. Biofilm production showed a positive correlation with Ciprofloxacin resistances. Twitching motility exhibited a weak negative correlation with Aztreonam resistance (Figure 3A). Among several virulence phenotypes, only the production of extracellular proteases and twitching motility showed a significant difference between the high MAR and low MAR groups (Figures 3B–F).
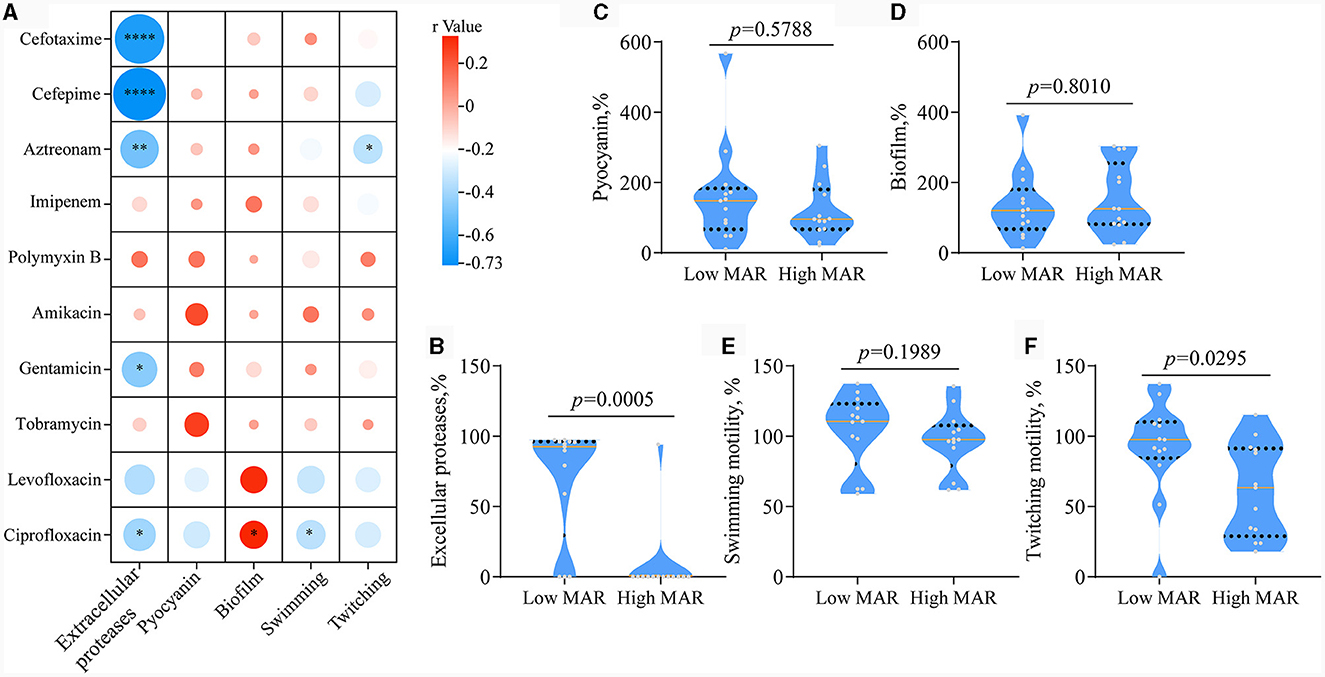
Figure 3. Analysis of the correlation between antibiotic resistance and QS-related virulence phenotypes. (A) Spearman's Rank Correlation Coefficient between minimum inhibitory concentration and quantitative phenotype. The code classifies correlation coefficients into four distinct categories based on their absolute values: WEAK (0.1–0.3), moderate (0.3–0.6), strong (0.6–0.8), and very strong (0.8–1.0). Phenotypes with weak or stronger correlations to drug resistance are highlighted with red boxes. *p < 0.05, **p < 0.01, and ***p < 0.0001. (B–F) Box plots comparing Extracellular Proteases, Pyocyanin, Biofilm Production, Swimming Motility, and Twitching Motility between the low MAR group (n = 13) and the high MAR group (n = 13). Unpaired two-tailed t-test.
The integrity of the lasR gene might be correlated with antibiotic resistance
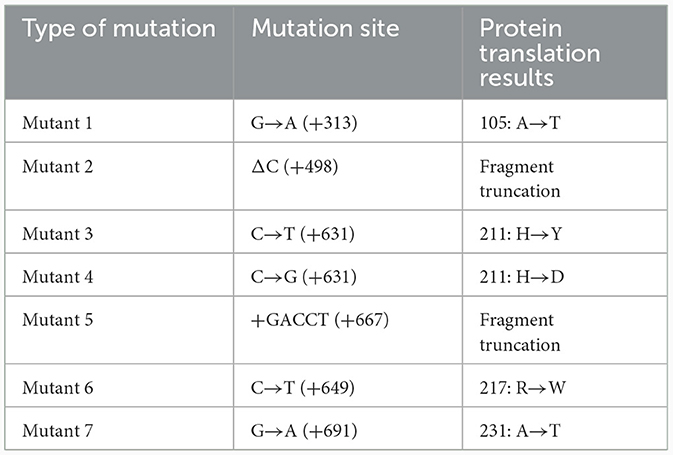
We then checked the integrity of QS regulatory genes in P. aeruginosa clinical strains to explore their potential connection to antibiotic resistance development. Among the 26 clinical strains of P. aeruginosa, we identified six synonymous base substitution mutations in the pqsR gene, 12 mutation sites, including five nonsynonymous mutations, in the rhlR gene, and 11 mutations, of which seven were nonsynonymous, in the lasR gene (Figure 4A and Table 1). Additionally, 5 synonymous mutations and no nonsynonymous mutations were detected in the lasI gene, and 13 mutations were detected in the rhlI gene, two of which were nonsynonymous mutations leading to amino acid substitutions, including 122AGG>AAG (F8), 184AGC>GGC (B3, C2, C3, G4, H5, J2, M1, K1, L2; Supplementary Table S5). In summary, nonsynonymous mutations primarily occurred in the lasR, rhlR, and rhlI genes, potentially influencing the hierarchy and regulatory capacity of the QS system, and no nonsynonymous mutations were detected in the pqsR and lasI genes. Moreover, 10 out of 14 P. aeruginosa COPD strains with intact lasR gene were found to be extracellular protease-positive (10 out of 11) and low MAR (10 out of 13), as well as the reference strain PAO1. On the other hand, 10 out of 12 P. aeruginosa COPD strains with mutated lasR gene were found to be extracellular protease-negative (10 out of 15) and high MAR (10 out of 13; Figure 4B). Such kind of gene integrity and antibiotic resistance nesting was not detected in rhlR and pqsR genes. These results suggested that the correlation between the QS regulatory genes and antibiotic resistance might be established by lasR gene integrity.
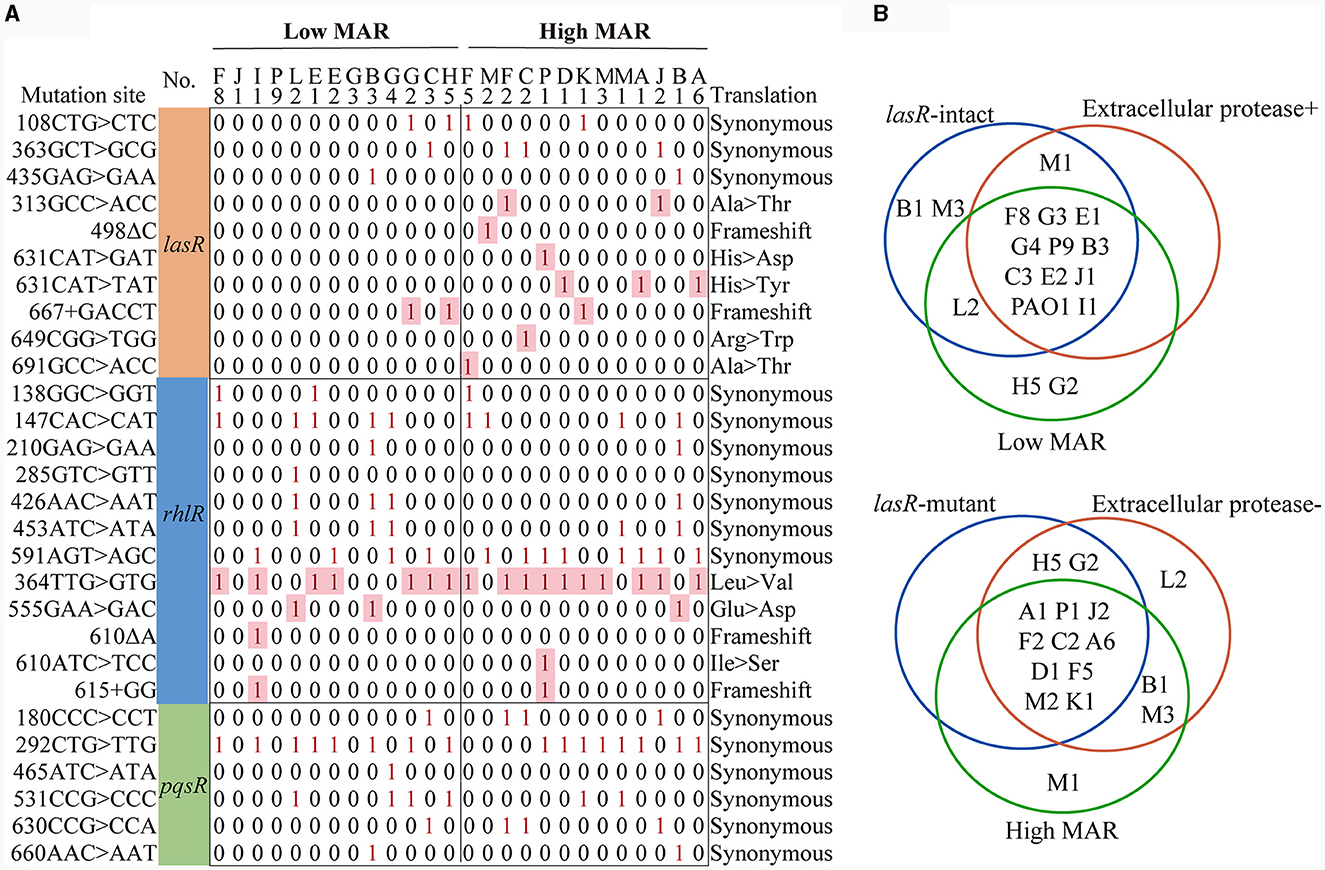
Figure 4. Multidrug resistance is consistently linked to lasR mutations and reduced extracellular protease production. (A) Matrix of lasR, rhlR, pqsR mutation sites. The sequencing results were compared with PAO1 in The Pseudomonas Genome Database. The matrix number 0 represents that no mutation was detected at this site, and 1 represents that mutation was detected at this site. (B) The Venn diagrams illustrate the classification of strains based on lasR integrity, MAR index, and extracellular protease characteristics.
Molecular docking-based structural and functional analyses of LasR mutants
We then evaluated the impact of mutation types on the function of the LasR protein by using homology modeling to predict the structural alterations in different LasR mutants. Subsequently, molecular docking was conducted to predict the potential effects of these mutations on the interactions of LasR with autoinducing small molecules. The crystal structure of the LasR protein complexed with N-(3-oxo-dodecanoyl)-L-homoserine lactone (3-oxo-C12-HSL) was resolved, revealing its dual binding domains: a hydrophobic pocket for self-induced small molecules and an effector region binding to the deep groove of DNA (Bottomley et al., 2007) (Figure 5). Nonsynonymous mutations occurred in both distinct regions of lasR gene (Figure 4 and Table 1). The results of homology modeling indicated that LasR Mutant 2 and Mutant 6 displayed the most significant structural deviations from WT LasR due to structural deletions and frameshift mutations, as evidenced by the Local Distance Difference Test (LDDT) analysis. In contrast, the remaining mutants mainly represented point mutation models, demonstrating structural similarities to WT LasR (Figure 5A).
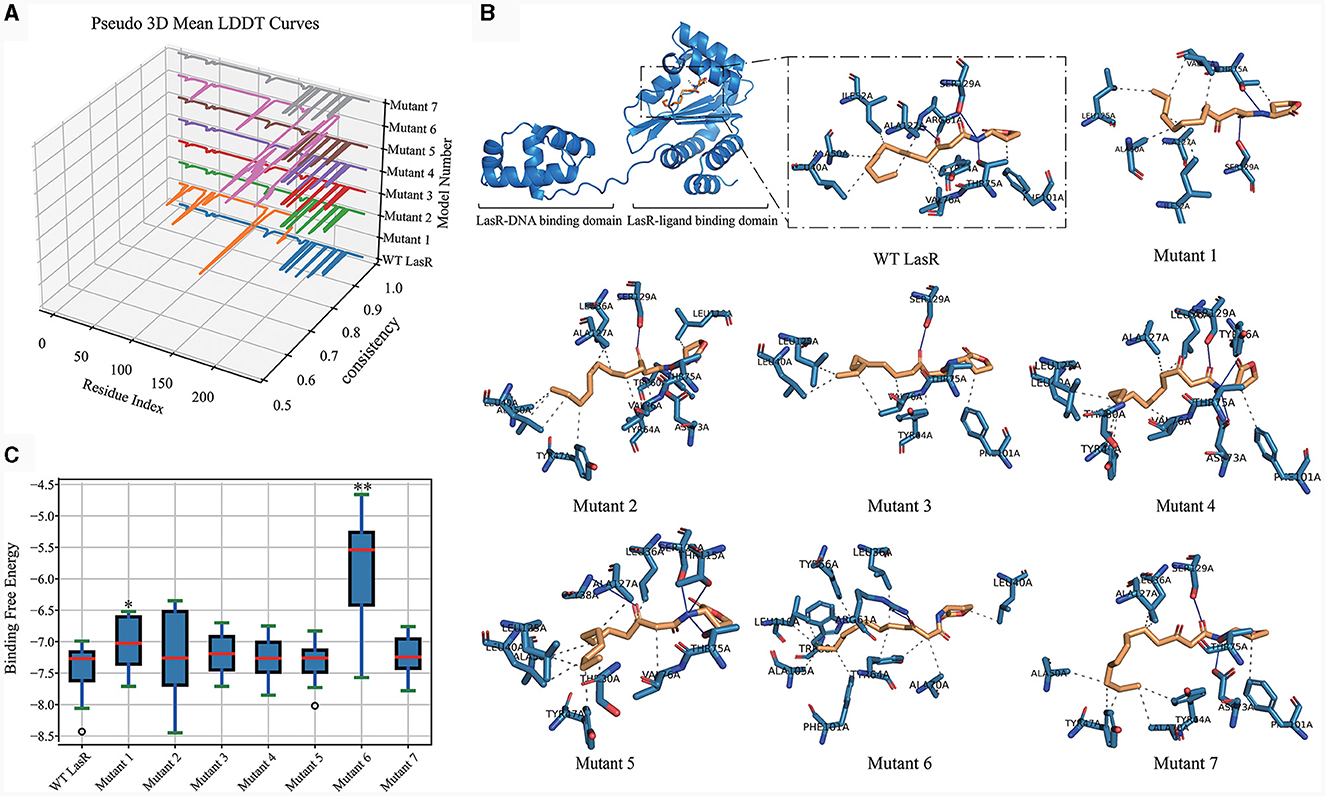
Figure 5. LasR Mutant Homology Modeling and Docking Analysis. (A) Comparative Pseudo-3D LDDT curves illustrating the quality of models generated for WT LasR and seven LasR mutants (1–7). They are performed using the Swiss-Model platform (https://swissmodel.expasy.org/) with Python, Matplotlib, and Pandas. The numbering corresponds to the mutation positions listed in Table 1. (B) Optimal binding of OHN (3-oxo-C12-HSL) ligand, compared for all mutants, accomplished using Dockey v0.8.2. Solid lines represent hydrogen bonds, while dashed lines represent hydrophobic or other interactions. (C) Box plot illustrating the distribution of binding free energies from 20 docking runs for WT and LasR mutants. t-test, *p <0.05, **p < 0.01.
Molecular docking analysis further revealed variations in the number of hydrogen bonds and other interaction forces between autoinducing ligands and different mutant models. For instance, Mutant 1 exhibited fewer hydrogen bonds compared to WT LasR (Figure 5B), possibly due to its mutation site at position 105, strategically located within the hydrophobic pocket center, where mutations exert a more pronounced impact on the docking interactions (Table 1). Mutant 6, characterized by a fragment deletion, demonstrated a more substantial effect on the binding of small molecules and ligands, while Mutant 2 displayed an impact that, though notable, lacked statistical significance. The remaining point mutants at the DNA binding sites exhibited binding free energies similar to WT LasR or slightly higher on average but lacked statistical significance (Figure 5C).
The virulence of lasR variants is multifaceted compared to WT PAO1
To validate the virulence differences between P. aeruginosa strains carrying lasR mutation and those without, this study employed C. elegans as an infection model and tested the fast-killing and slow-killing capabilities of six P. aeruginosa clinical strains. Among the tested strains, namely J1, I1, and P9, possessed intact lasR genes and exhibited a virulence factor expression profile similar to that of the reference strain WT PAO1. Strains J2 (mutant 1), D1 (mutant 4), and P1 (mutant 3) harbored point mutations in their lasR genes and failed to produce extracellular proteases (Figures 2, 4). Compared to WT PAO1, J2 (mutant 1) displayed significantly increased lethality in the fast-killing assay, resulting in complete nematode mortality 2 days post-inoculation. In contrast, P1 (mutant 3) exhibited lower virulence compared to WT PAO1, while D1 (mutant 4), I1, P9, and J1 displayed virulence levels similar to that of WT PAO1, consistent with previous virulence phenotype results (Figure 6A). In the context of the slow-killing model, D1 (mutant 4) and P1 (mutant 3) exhibited the weakest capabilities of killing nematodes, while the remaining strains, including J2 (mutant 1), showed a nematode-killing ability similar to WT PAO1 (Figure 6B). These results suggested that mutations in the lasR gene would influence the pathogenicity of P. aeruginosa, while the enhanced virulence of J2 (mutant 1) in the fast-killing assay might be attributed to the accumulation of unknown mutations in other genes during the evolution of this lasR mutant in COPD airway, as reported in our prior work (Zhao et al., 2023). The pathogenic mechanism of P. aeruginosa J2 will be explored in our further study.
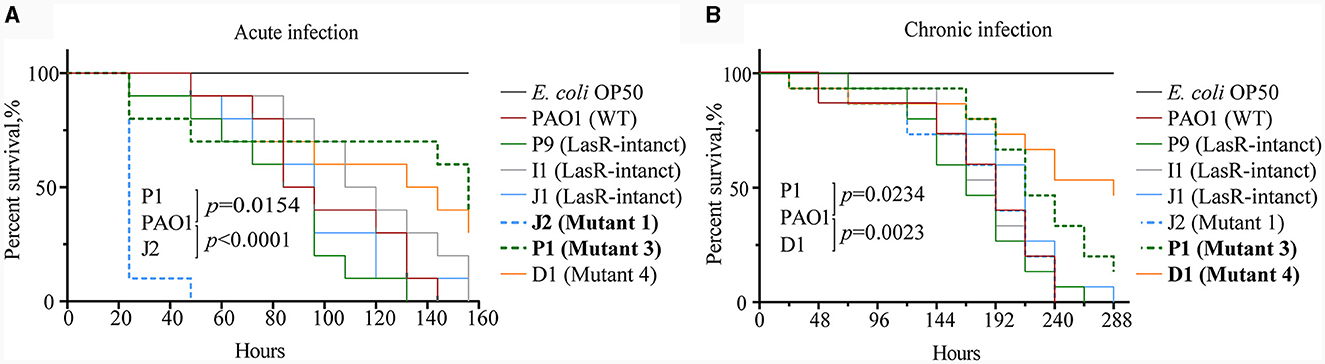
Figure 6. Differential virulence of Pseudomonas aeruginosa clinical strains with and without lasR mutations. (A) Survival curve of Caenorhabditis elegans with acute infection of P. aeruginosa. Acute infection model with three replicates per group, each containing 10 nematodes. Mantel-Cox test. (B) Survival curve of C. elegans with chronic infection of P. aeruginosa. Chronic infection model with three replicates per group, each containing 15 nematodes. Code labels with significant differences are bolded. Mantel-Cox test.
Levofloxacin and polymyxin B promote the emergence of extracellular protease-deficient strains
To assess the adaptive influence of P. aeruginosa under antibiotic stress, we employed WT PAO1 and PAO1-ΔlasR as the initial bacterial strains and separately treated them with Levofloxacin, Cefepime, and Polymyxin B. PAO1-ΔlasR showed a reduction in producing extracellular proteases compared to WT PAO1 (Figure 7A). WT PAO1 reached a growth plateau at ~12 h, whereas PAO1-ΔlasR required around 20 h to reach a similar plateau at the same initial inoculum. Under the initial antibiotic concentrations, the addition of Levofloxacin, Cefepime, or Polymyxin B did not affect extracellular protease production in WT PAO1 and PAO1-ΔlasR.
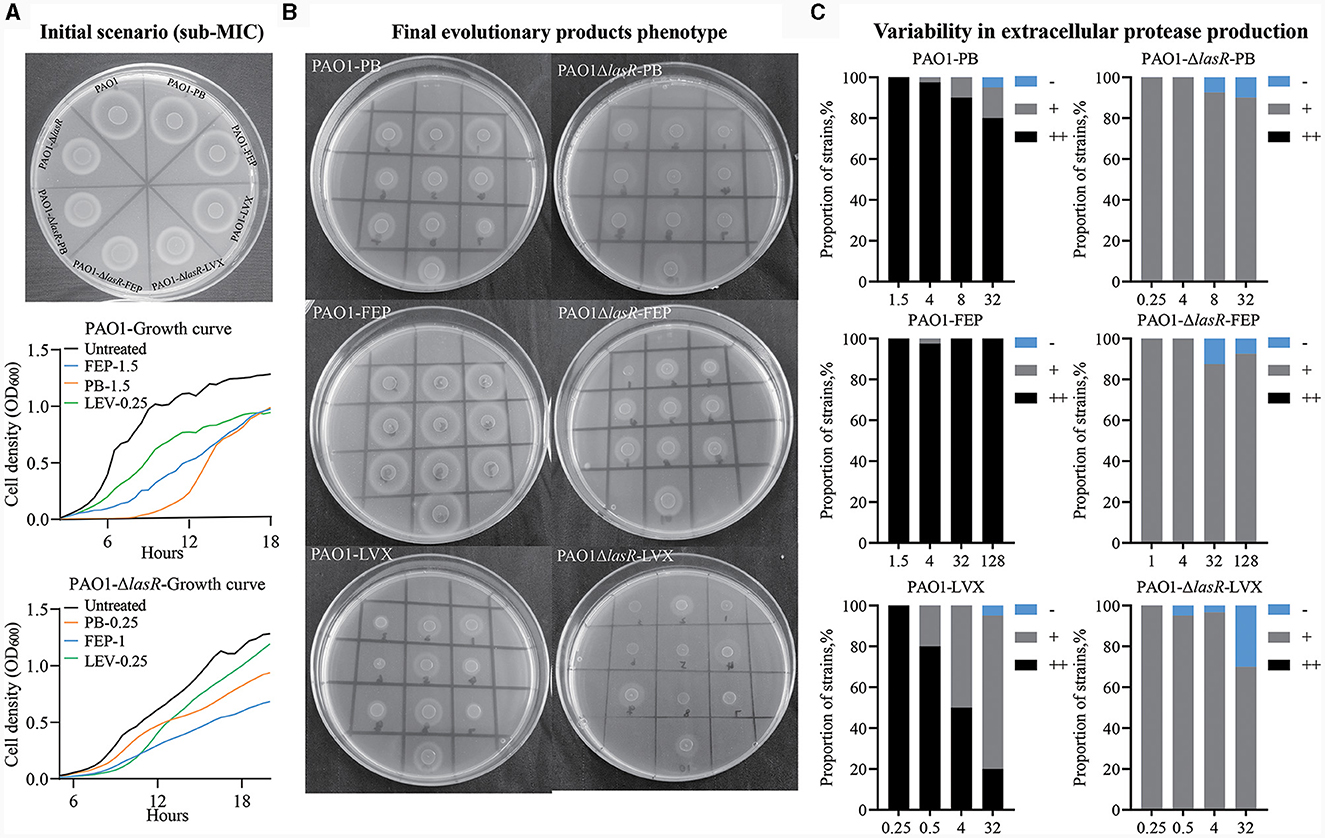
Figure 7. Long-term evolution of WT PAO1 and PAO1-ΔlasR under antibiotic pressure. (A) Extracellular protease production and growth curves were evaluated upon the initial application of three antibiotics at varying concentrations to PAO1 and PAO1-ΔlasR. (B) The production of extracellular protease in the final evolutionary products of each group. (C) Bar charts presenting the composition and statistical analysis of extracellular protease ring formation in 100 isolates. Comparison was made with the protein ring diameter of PAO1, denoted as “++” for consistency, “+” for smaller, and “–” for no protein ring production compared to PAO1. The unit of antibiotic concentration was μg/ml PB refers to Polymyxin B, FEP refers to Cefepime, and LVX refers to Levofloxacin.
Following 1 month of evolutionary culture without antibiotic treatment, the ability of WT PAO1 and PAO1-ΔlasR to produce extracellular proteases remained unchanged. The WT PAO1 population treated with Cefepime did not exhibit a notable reduction in extracellular protease expression, while the PAO1-ΔlasR group exhibited a low proportion of strains that did not produce extracellular protease. Heterogeneity was observed in both WT PAO1 and PAO1-ΔlasR populations after polymyxin B treatment, particularly in WT PAO1. The most pronounced heterogeneity was observed after exposure to Levofloxacin in both cases (Figures 7B, C). However, when we examined the lasR gene in the final evolved products of the WT PAO1 population, we detected significant A-to-G mutations at position 632 in the Levofloxacin treatment group, resulting in a histidine-to-arginine mutation at position 211 of the LasR protein. Coincidentally, clinical isolates P1, D1, A1, and A6 were found to exhibit mutations at this specific site (Figure 4). These results demonstrated that different classes of antibiotics might impose different selective pressures on the development of P. aeruginosa QS system. Levofloxacin and Polymyxin B, especially the former, could promote the emergence of extracellular protease-deficient strains.
Discussion
MDR P. aeruginosa remains a clinical challenge, necessitating vigilant strain monitoring and precise antibiotic management for effective resistance control. Pseudomonas aeruginosa exhibits intrinsic resistance to multiple antibiotics, while concurrently utilizing QS systems to mediate biofilm formation (García-Contreras et al., 2015). This coupled with mechanisms such as mutations in target sites, alterations in membrane permeability, and antibiotic efflux pumps, enhances its tolerance to antibiotics, ultimately leading to the emergence of MDR P. aeruginosa (Hancock and Speert, 2000; Langendonk et al., 2021; Elbehiry et al., 2022). This study primarily discusses the development of antibiotic resistance and its connection to QS in P. aeruginosa, particularly focusing on commonly used respiratory anti-infective antibiotics.
The results of antibiotic resistance typing and phenotypic identification in this study reflect the diversity and complexity of clinical P. aeruginosa strains sourced from the respiratory tract of COPD patients (Figure 1). Previous studies have shown that P. aeruginosa undergoes differentiation into low-virulence subpopulations, to produce specific virulence factors like extracellular proteases and pyocyanin during prolonged colonization (Marvig et al., 2015; Zhao et al., 2020, 2023). However, the significant differences in the production levels of virulence factors in high MAR and low MAR P. aeruginosa strains, highlight the prevalence of heterogeneous resistance resulting from multiclonal subpopulation colonization and monoclonal adaptation in COPD respiratory infections. Strains with negative extracellular protease typically exhibit multidrug resistance characteristics. Additionally, these strains often harbor mutations in the lasR gene. Conversely, strains with positive extracellular protease usually demonstrate lower antibiotic resistance and possess intact lasR genes (Figures 2–4). This also suggests a parallel trend between lasR mutations and the rise in antibiotic resistance (Oshri et al., 2018; Azimi et al., 2020).
In the in vitro evolution experiments, the present study demonstrated that different antibiotics might impose diverse influences on the development of P. aeruginosa QS system. This can be supported by the remarkably increased frequencies of extracellular protease-deficient strains in Polymyxin B- or Levofloxacin-treated WT PAO1 and the lasR mutant identified from Levofloxacin-treated WT PAO1 (Figure 7). Additionally, antibiotic is the sole factor that brings the main selective pressure during the evolution of P. aeruginosa in LB medium containing an antibiotic. However, the PAO1-ΔlasR strain used in this study showed no increase in antibiotic resistance, and its resistance to Polymyxin B was even decreased compared to the parental WT PAO1 (Supplementary Table S2). Therefore, it is hard to simply conclude that antibiotics exert selective pressure on lasR gene or that lasR mutations facilitate the development of resistance. It is also important to note that the production of extracellular proteases by P. aeruginosa is not exclusively governed by the lasR gene. Hence, while antibiotic exposure may select for extracellular protease-deficient phenotype, the direct causation by lasR remains uncertain. The indirect contribution of antibiotic-driven selective pressure on the utilization of sharable extracellular products to the emergence of lasR-deficient strains (Oshri et al., 2018), causing diversity in the QS system integrity within the population, still necessitates further validation.
In conclusion, this study dissects the negative correlation between antibiotic resistance and QS system in clinical P. aeruginosa, establishing a connection mediated by extracellular protease-deficient phenotypes. While adaptation theory generally suggests that P. aeruginosa in chronic infections may evolve toward decreasing QS regulation and virulence (Jiricny et al., 2014), our results indicate that the QS mutants, especially the strains deficient in producing extracellular proteases, exhibit increasing levels of resistance and present distinct risks in comparison to QS-intact strains. These findings also provide an important reference for further research and development of novel antimicrobial agents, and for the selection and application of clinical antibiotics to treat pseudomonal infections.
Data availability statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics statement
The studies involving humans were approved by Affiliated Hospital of Chengdu University. The studies were conducted in accordance with the local legislation and institutional requirements. The participants provided their written informed consent to participate in this study.
Author contributions
XY: Investigation, Methodology, Writing – original draft. QZ: Investigation, Project administration, Resources, Writing – review & editing. SG: Investigation, Writing – review & editing. YW: Investigation, Writing – review & editing. XM: Investigation, Writing – review & editing. HZ: Investigation, Resources, Writing – review & editing. KZ: Conceptualization, Data curation, Formal analysis, Funding acquisition, Methodology, Project administration, Supervision, Validation, Writing – review & editing, Writing – original draft.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the National Natural Science Foundation of China (32270121 and 31970131) and the Sichuan Province Science and Technology Support Program (2021JDJQ0042).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1327675/full#supplementary-material
References
Algammal, A., Hetta, H. F., Mabrok, M., and Behzadi, P. (2023a). Editorial: Emerging multidrug-resistant bacterial pathogens “superbugs”: a rising public health threat. Front. Microbiol. 14:1135614. doi: 10.3389/fmicb.2023.1135614
Algammal, A. M., Abo Hashem, M. E., Alfifi, K. J., Al-Otaibi, A. S., Alatawy, M., ElTarabili, R.M., et al. (2022). Sequence analysis, antibiogram profile, virulence and antibiotic resistance genes of XDR and MDR Gallibacterium anatis isolated from layer chickens in Egypt. Infect. Drug Resist. 15, 4321–4334. doi: 10.2147/IDR.S377797
Algammal, A. M., Eidaroos, N. H., Alfifi, K. J., Alatawy, M., Al-Harbi, A. I., Alanazi, Y. F., et al. (2023b). oprL gene sequencing, resistance patterns, virulence genes, quorum sensing and antibiotic resistance genes of XDR Pseudomonas aeruginosa isolated from broiler chickens. Infect. Drug Resist. 16, 853–867. doi: 10.2147/IDR.S401473
Azimi, S., Roberts, A. E., Peng, S., Weitz, J. S., and Diggle, S. (2020). Allelic polymorphism shapes community function in evolving Pseudomonas aeruginosa populations. ISME J. 14, 1929–1942. doi: 10.1038/s41396-020-0652-0
Balasubramanian, V., Sellegounder, D., Suman, K., and Krishnaswamy, B. (2016). Proteome analysis reveals translational inhibition of Caenorhabditis elegans enhances susceptibility to Pseudomonas aeruginosa PAO1 pathogenesis. J. Proteomics 145, 141–152. doi: 10.1016/j.jprot.2016.04.025
Behzadi, P., Baráth, Z., and Gajdács, M. (2021). It's not easy being green: a narrative review on the microbiology, virulence and therapeutic prospects of multidrug-resistant Pseudomonas aeruginosa. Antibiotics 10:42. doi: 10.3390/antibiotics10010042
Bottomley, M. J., Muraglia, E., Bazzo, R., and Carfi, A. (2007). Molecular insights into quorum sensing in the human pathogen Pseudomonas aeruginosa from the structure of the virulence regulator LasR bound to its autoinducer. J. Biol. Chem. 282, 13592–13600. doi: 10.1074/jbc.M700556200
Chevalier, S., Bouffartigues, E., Bodilis, J., Maillot, O., Lesouhaitier, O., Feuilloley, M. G. J., et al. (2017). Structure, function, and regulation of Pseudomonas aeruginosa porins. FEMS Microbiol. Rev. 41, 698–722. doi: 10.1093/femsre/fux020
CLSI (2022). Performance Standards for Antimicrobial Susceptibility Testing; 32nd Informational Supplement. Wayne, PA: Clinical and Laboratory Standards Institute. CLSI Document, M100–S25.
Du, L., Geng, C., Zeng, Q., Huang, T., Tang, J., Chu, Y., et al. (2023). Dockey: a modern integrated tool for large-scale molecular docking and virtual screening. Brief. Bioinform. 24:bbad047. doi: 10.1093/bib/bbad047
Elbehiry, A., Marzouk, E., Aldubaib, M., Moussa, I., Abalkhail, A., Ibrahem, M., et al. (2022). Pseudomonas species prevalence, protein analysis, and antibiotic resistance: an evolving public health challenge. AMB Express 12:53. doi: 10.1186/s13568-022-01390-1
Feliziani, S., Marvig, R. L., Luján, A. M., Moyano, A. J., Di Rienzo, J. A., Krogh Johansen, H., et al. (2014). Coexistence and within-host evolution of diversified lineages of hypermutable Pseudomonas aeruginosa in long-term cystic fibrosis infections. PLoS Genet. 10:e1004651. doi: 10.1371/journal.pgen.1004651
Feltner, J. B., Wolter, D. J., Pope, C. E., Groleau, M. C., Smalley, N. E., Greenberg, E. P., et al. (2016). LasR variant cystic fibrosis isolates reveal an adaptable quorum-sensing hierarchy in Pseudomonas aeruginosa. MBio 7:e01513-16. doi: 10.1128/mBio.01513-16
Figueiredo, A. R. T., Wagner, A., and Kümmerli, R. (2021). Ecology drives the evolution of diverse social strategies in Pseudomonas aeruginosa. Mol. Ecol. 30, 5214–5228. doi: 10.1111/mec.16119
Filloux, A. (2011). Protein secretion systems in Pseudomonas aeruginosa: an essay on diversity, evolution, and function. Front. Microbiol. 2:155. doi: 10.3389/fmicb.2011.00155
Gallego, M., Pomares, X., Espasa, M., Castañer, E., Solé, M., Suárez, D., et al. (2014). Pseudomonas aeruginosa isolates in severe chronic obstructive pulmonary disease: characterization and risk factors. BMC Pulm. Med. 14:103. doi: 10.1186/1471-2466-14-103
García-Contreras, R., Nuñez-López, L., Jasso-Chávez, R., Kwan, B. W., Belmont, J. A., Rangel-Vega, A., et al. (2015). Quorum sensing enhancement of the stress response promotes resistance to quorum quenching and prevents social cheating. ISME J. 9, 115–125. doi: 10.1038/ismej.2014.98
Garcia-Vidal, C., Almagro, P., Romaní, V., Rodríguez-Carballeira, M., Cuchi, E., Canales, L., et al. (2009). Pseudomonas aeruginosa in patients hospitalized for COPD exacerbation: a prospective study. Eur. Respir. J. 34, 1072–1078. doi: 10.1183/09031936.00003309
Grekov, I., Thöming, J. G., Kordes, A., and Häussler, S. (2021). Evolution of Pseudomonas aeruginosa toward higher fitness under standard laboratory conditions. ISME J. 15, 1165–1177. doi: 10.1038/s41396-020-00841-6
Hancock, R. E., and Speert, D. P. (2000). Antibiotic resistance in Pseudomonas aeruginosa: mechanisms and impact on treatment. Drug Resist. Update Univ. S C. Dep. Music 3, 247–255. doi: 10.1054/drup.2000.0152
Hernando-Amado, S., Sanz-García, F., and Martínez, J. L. (2019). Antibiotic resistance evolution is contingent on the quorum-sensing response in Pseudomonas aeruginosa. Mol. Biol. Evol. 36, 2238–2251. doi: 10.1093/molbev/msz144
Heurlier, K., Dénervaud, V., and Haas, D. (2006). Impact of quorum sensing on fitness of Pseudomonas aeruginosa. Int. J. Med. Microbiol. 296, 93–102. doi: 10.1016/j.ijmm.2006.01.043
Hilker, R., Munder, A., Klockgether, J., Losada, P. M., Chouvarine, P., Cramer, N., et al. (2015). Interclonal gradient of virulence in the Pseudomonas aeruginosa pangenome from disease and environment. Environ. Microbiol. 17, 29–46. doi: 10.1111/1462-2920.12606
Høiby, N., Bjarnsholt, T., Moser, C., Bassi, G. L., Coenye, T., Donelli, G., et al. (2015). ESCMID guideline for the diagnosis and treatment of biofilm infections 2014. Clin. Microbiol. Infect. 21, S1–S25. doi: 10.1016/j.cmi.2014.10.024
Ji, X., and Tang, J. (2022). Effects of salt stress on the morphology, growth and physiological parameters of Juglansmicrocarpa L. seedlings. Plants 11:2381. doi: 10.3390/plants11182381
Jimenez, P. N., Koch, G., Thompson, J. A., Xavier, K. B., Cool, R. H., Quax, W. J., et al. (2012). The multiple signaling systems regulating virulence in Pseudomonas aeruginosa. Microbiol. Mol. Biol. Rev. 76, 46–65. doi: 10.1128/MMBR.05007-11
Jiricny, N., Molin, S., Foster, K., Diggle, S. P., Scanlan, P. D., Ghoul, M., et al. (2014). Loss of social behaviors in populations of Pseudomonas aeruginosa infecting lungs of patients with cystic fibrosis. PLoS ONE 9:e83124. doi: 10.1371/journal.pone.0083124
Jorth, P., Staudinger, B. J., Wu, X., Hisert, K. B., Hayden, H., Garudathri, J., et al. (2015). Regional isolation drives bacterial diversification within cystic fibrosis lungs. Cell Host Microbe. 18, 307–319. doi: 10.1016/j.chom.2015.07.006
King, P. T. (2019). Pseudomonas aeruginosa cross-infection: is this important in bronchiectasis and COPD? Respirology 24, 926–927. doi: 10.1111/resp.13578
Kostylev, M., Kim, D. Y., Smalley, N. E., Salukhe, I., Greenberg, E. P., Dandekar, A. A., et al. (2019). Evolution of the Pseudomonas aeruginosa quorum-sensing hierarchy. PNAS 116, 7027–7032. doi: 10.1073/pnas.1819796116
Krumperman, P. H. (1983). Multiple antibiotic resistance indexing of Escherichia coli to identify high-risk sources of fecal contamination of foods. Appl. Environ. Microbiol. 46, 165–170. doi: 10.1128/aem.46.1.165-170.1983
Langendonk, R. F., Neill, D. R., and Fothergill, J. L. (2021). The building blocks of antimicrobial resistance in pseudomonas aeruginosa: implications for current resistance-breaking therapies. Front. Cell. Infect. Microbiol. 11:665759. doi: 10.3389/fcimb.2021.665759
Li, X., Li, J., Zhao, Q., Qiao, L., Wang, L., Yu, C., et al. (2023). Physiological, biochemical, and genomic elucidation of the Ensifer adhaerens M8 strain with simultaneous arsenic oxidation and chromium reduction. J. Hazard. Mater. 441:129862. doi: 10.1016/j.jhazmat.2022.129862
Liang, H., Deng, X., Li, X., Ye, Y., and Wu, M. (2014). Molecular mechanisms of master regulator VqsM mediating quorum-sensing and antibiotic resistance in Pseudomonas aeruginosa. Nucleic Acids Res. 42, 10307–10320. doi: 10.1093/nar/gku586
Lozano, C., Azcona-Gutiérrez, J. M., Van Bambeke, F., and Sáenz, Y. (2018). Great phenotypic and genetic variation among successive chronic Pseudomonas aeruginosa from a cystic fibrosis patient. PLoS ONE 13:e0204167. doi: 10.1371/journal.pone.0204167
Mac Aogáin, M., Narayana, J. K., Tiew, P. Y., Ali, N. A. B. M., Yong, V. F. L., Jaggi, T. K., et al. (2021). Integrative microbiomics in bronchiectasis exacerbations. Nat. Med. 27, 688–699. doi: 10.1038/s41591-021-01289-7
Magiorakos, A. P., Srinivasan, A., Carey, R. B., Carmeli, Y., Falagas, M. E., Giske, C. G., et al. (2012). Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 18, 268–281. doi: 10.1111/j.1469-0691.2011.03570.x
Marvig, R. L., Sommer, L. M., Molin, S., and Johansen, H. K. (2015). Convergent evolution and adaptation of Pseudomonas aeruginosa within patients with cystic fibrosis. Nat. Genet. 47, 57–64. doi: 10.1038/ng.3148
Miravitlles, M., and Anzueto, A. (2017). Chronic respiratory infection in patients with chronic obstructive pulmonary disease: what is the role of antibiotics? Int. J. Mol. Sci. 18:1344. doi: 10.3390/ijms18071344
Oshri, R. D., Zrihen, K. S., Shner, I., Omer Bendori, S., and Eldar, A. (2018). Selection for increased quorum-sensing cooperation in Pseudomonas aeruginosa through the shut-down of a drug resistance pump. ISME J. 12, 2458–2469. doi: 10.1038/s41396-018-0205-y
Pena, R. T., Blasco, L., Ambroa, A., González-Pedrajo, B., Fernández-García, L., López, M., et al. (2019). Relationship between quorum sensing and secretion systems. Front. Microbiol. 10:1100. doi: 10.3389/fmicb.2019.01100
Puspita, M., Wasito, E. B., and Alimsardjono, L. (2021). Association of blood isolate's multi antibiotic resistance-index on laboratory-confirmed bloodstream infection: a cross-sectional study. Ann. Med. Surg. 72:103086. doi: 10.1016/j.amsu.2021.103086
Rodrigo-Troyano, A., Melo, V., Marcos, P. J., Laserna, E., Peiro, M., Suarez-Cuartin, G., et al. (2018). Pseudomonas aeruginosa in chronic obstructive pulmonary disease patients with frequent hospitalized exacerbations: a prospective multicentre study. Respiration 96, 417–424. doi: 10.1159/000490190
Schuster, M., and Greenberg, E. P. (2006). A network of networks: quorum-sensing gene regulation in Pseudomonas aeruginosa. Int. J. Med. Microbiol. 296, 73–81. doi: 10.1016/j.ijmm.2006.01.036
Shafiq, M., Zeng, M., Permana, B., Bilal, H., Huang, J., Yao, F., et al. (2022). Coexistence of blaNDM-5 and tet(X4) in international high-risk Escherichia coli clone ST648 of human origin in China. Front. Microbiol. 13:1031688. doi: 10.3389/fmicb.2022.1031688
Smith, E. E., Buckley, D. G., Wu, Z., Saenphimmachak, C., Hoffman, L. R., D'Argenio, D. A., et al. (2006). Genetic adaptation by Pseudomonas aeruginosa to the airways of cystic fibrosis patients. PNAS 103, 8487–8492. doi: 10.1073/pnas.0602138103
Stehling, E. G., Leite, D. S., and Silveira, W. D. (2010). Molecular typing and biological characteristics of Pseudomonas aeruginosa isolated from cystic fibrosis patients in Brazil. Braz. J. Infect. Dis. 14, 462–467. doi: 10.1016/S1413-8670(10)70094-5
Vanderwoude, J., Fleming, D., Azimi, S., Trivedi, U., Rumbaugh, K. P., Diggle, S. P., et al. (2020). The evolution of virulence in Pseudomonas aeruginosa during chronic wound infection. Proc. Biol. Sci. 287:2272. doi: 10.1101/2020.05.29.124545
Wilder, C. N., Diggle, S. P., and Schuster, M. (2011). Cooperation and cheating in Pseudomonas aeruginosa: the roles of the las, rhl and pqs quorum-sensing systems. ISME J. 5, 1332–1343. doi: 10.1038/ismej.2011.13
Zhao, K., Huang, T., Lin, J., Yan, C., Du, L., Song, T., et al. (2020). Genetic and functional diversity of Pseudomonas aeruginosa in patients with chronic obstructive pulmonary disease. Front. Microbiol. 11:598478. doi: 10.3389/fmicb.2020.598478
Zhao, K., Li, W., Li, J., Ma, T., Wang, K., Yuan, Y., et al. (2019). TesG is a type I secretion effector of Pseudomonas aeruginosa that suppresses the host immune response during chronic infection. Nat. Microbiol. 4, 459–469. doi: 10.1038/s41564-018-0322-4
Zhao, K., Yang, X., Zeng, Q., Zhang, Y., Li, H., Yan, C., et al. (2023). Evolution of lasR mutants in polymorphic Pseudomonas aeruginosa populations facilitates chronic infection of the lung. Nat. Commun. 14:5976. doi: 10.1038/s41467-023-41704-w
Keywords: Pseudomonas aeruginosa, COPD, quorum-sensing, antibiotic resistance, mutation, evolution
Citation: Yang X, Zeng Q, Gou S, Wu Y, Ma X, Zou H and Zhao K (2024) Phenotypic heterogeneity unveils a negative correlation between antibiotic resistance and quorum sensing in Pseudomonas aeruginosa clinical isolates. Front. Microbiol. 15:1327675. doi: 10.3389/fmicb.2024.1327675
Received: 25 October 2023; Accepted: 29 January 2024;
Published: 12 February 2024.
Edited by:
Chun-Hsing Liao, National Yang Ming Chiao Tung University, TaiwanReviewed by:
Samara Paula Mattiello, University of Tennessee Southern, United StatesAbdelazeem Algammal, Suez Canal University, Egypt
Rolf Kümmerli, University of Zurich, Switzerland
Copyright © 2024 Yang, Zeng, Gou, Wu, Ma, Zou and Zhao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kelei Zhao, zhaokelei@cdu.edu.cn
 Xiting Yang
Xiting Yang Qianglin Zeng2
Qianglin Zeng2 Kelei Zhao
Kelei Zhao