- 1Department of Ecosustainable Marine Biotechnology, Stazione Zoologica Anton Dohrn, Napoli, Italy
- 2Department of Earth, Environmental and Life Sciences, University of Genoa, Genoa, Italy
- 3Bio-Fab Research Srl, Rome, Italy
- 4Department of Ecosustainable Marine Biotechnology, Stazione Zoologica Anton Dohrn, Calabria Marine Centre, Amendolara, Italy
- 5Dipartimento Di Scienze Biologiche, Chimiche, Farmaceutiche Ed Ambientali, Università Di Messina, Messina, Italy
- 6Department of Biology, University of Naples Federico II, Complesso Universitario di Monte Sant'Angelo, Naples, Italy
- 7Department of Ecosustainable Marine Biotechnology, Stazione Zoologica Anton Dohrn, Ischia Marine Centre, Naples, Italy
The oceans cover over 70% of our planet, hosting a biodiversity of tremendous wealth. Sponges are one of the major ecosystem engineers on the seafloor, providing a habitat for a wide variety of species to be considered a good source of bioactive compounds. In this study, a metataxonomic approach was employed to describe the bacterial communities of the sponges collected from Faro Lake (Sicily) and Porto Paone (Gulf of Naples). Morphological analysis and amplification of the conserved molecular markers, including 18S and 28S (RNA ribosomal genes), CO1 (mitochondrial cytochrome oxidase subunit 1), and ITS (internal transcribed spacer), allowed the identification of four sponges. Metataxonomic analysis of sponges revealed a large number of amplicon sequence variants (ASVs) belonging to the phyla Proteobacteria, Cloroflexi, Dadabacteria, and Poribacteria. In particular, Myxilla (Myxilla) rosacea and Clathria (Clathria) toxivaria displayed several classes such as Alphaproteobacteria, Dehalococcoidia, Gammaproteobacteria, Cyanobacteria, and Bacteroidia. On the other hand, the sponges Ircinia oros and Cacospongia mollior hosted bacteria belonging to the classes Dadabacteriia, Anaerolineae, Acidimicrobiia, Nitrospiria, and Poribacteria. Moreover, for the first time, the presence of Rhizobiaceae bacteria was revealed in the sponge M. (Myxilla) rosacea, which was mainly associated with soil and plants and involved in biological nitrogen fixation.
1 Introduction
The Mediterranean Sea is considered very significant in terms of biodiversity, as it hosts 7% of the world's marine biodiversity (with only 0.82% of the oceans' surface), including a large number of endemic species, which are also subject to anthropogenic stress destined to increase in the future (Coll et al., 2010). Among marine organisms, marine sponges emerged on the earth more than 600 million years ago (the Cambrian Age) and represented one of the most important components of benthic fauna. They are extensively distributed from polar to tropical waters and from intertidal regions to waters thousands of meters deep (Fusetani, 1996), representing dominant taxon in marine communities in terms of species biomass, richness, and spatial coverage (Corriero et al., 2000). A miscellaneous of archaea, heterotrophic bacteria, cyanobacteria, green algae, red algae, cryptophytes, dinoflagellates, and diatoms have been found together with sponges (Larkum et al., 1987; Santavy et al., 1990; Duglas, 1994). Their microbial community is very diverse, with species composition showing temporal and geographical differences (Webster and Hill, 2001). A sponge can be associated with diverse organisms, such as Theonella swinhoei, which hosted unicellular bacteria and cyanobacteria at the same time (Bewley et al., 1996). Additionally, a sponge belonging to the Aplysina genus included heterogeneous bacteria Micrococcus sp., Bacillus sp., Arthrobacter sp., Pseudoalteromonas sp., Vibrio sp., and others (Hentschel et al., 2001). The surfaces or internal sides of marine sponges are richer in nutrients than seawater and sediments; therefore, sponges offer both a feeding source and a secure habitat for their microorganisms (Bultel-Poncé et al., 1999). On the other hand, symbiotic microorganisms contribute to the nutritional process through intracellular digestion and/or translocation of metabolites, such as nitrogen fixation, nitrification, and photosynthesis (Wilkinson and Fay, 1979; Wilkinson and Garrone, 1980). Microorganisms also stabilize the skeleton of sponges (Wilkinson et al., 1981) and participate in the host chemical defense system against hunters and biofouling (Bakus et al., 1986; Paul, 1992; Proksch, 1994; Unson et al., 1994). Sponges and associated microorganisms represent very useful organisms as sources of bioactive compounds for marine biotechnology (Cooper and Yao, 2010; Karuppiah and Li, 2015; Thompson et al., 2017; Cerrano et al., 2022). In fact, one of the first peptides with antibacterial activity was isolated by the gram-negative bacterium Vibrio sp. associated with the sponge Hyatella sp. (Oclarit et al., 1994). Starting from this, several studies showed pharmacological applications of natural products from marine sponges and their associated biota (Newman and Cragg, 2020; Zhang et al., 2020; Amelia et al., 2022).
In the present study, we deeply explored the bacterial communities associated with four sponges collected in the Mediterranean Sea. These species were collected in Italy in two different areas: Faro Lake (Sicily) and Porto Paone (Gulf of Naples). Species were characterized by skeletal morphological observation and amplification of several conserved molecular markers, represented by RNA ribosomal genes (18S and 28S rRNA), mitochondrial cytochrome oxidase subunit 1 (CO1), and internal transcribed spacer (ITS). To analyze the biodiversity of symbiotic communities between the two sampling sites, we performed metataxonomic analysis of sponge samples using the Illumina MiSeq platform. More than 1,000 bacterial isolates from four samples were phylogenetically identified to understand if they are host-specific and/or location-specific. Subsequently, the ASV analysis was also applied to reveal the level of variability in the host-specific microbial communities, which were then discussed to assess the biotechnological potential of the sponges under analysis.
2 Materials and methods
2.1 Sponge sampling
Two sponge samples (indicated as S36 and S39) were collected in the Gulf of Naples by scuba divers from the Stazione Zoologica Anton Dohrn. The sampling site was Porto Paone (40°47′N, 14°9′E), and the specimens were collected at depths between 15 and 17 m. The other two sponges (indicated as S23 and S25) were collected at the Faro Lake in Messina, Sicily (38°16′N, 15°38′E) at depths between 2 and 3 m (Ruocco et al., 2021, see Table 1). This sampling site is located in the Natural Reserve of “Capo Peloro.” The Faro Lake has the shape of a funnel, and due toits maximum depth of 29 m, which more or less coincides with the center of the lake, it is considered the deepest coastal lake in Italy. The Faro Lake is a salty lake connected by two channels to the Strait of Messina and the Tyrrhenian Sea (northern and northeastern side). The abiotic lake parameters of the lake include a salinity range of 26–36 PSU, pH levels ranging from 7.0 to 8.6, and temperature ranging from 10 to 30°C (Saccà et al., 2008; Marra et al., 2016).
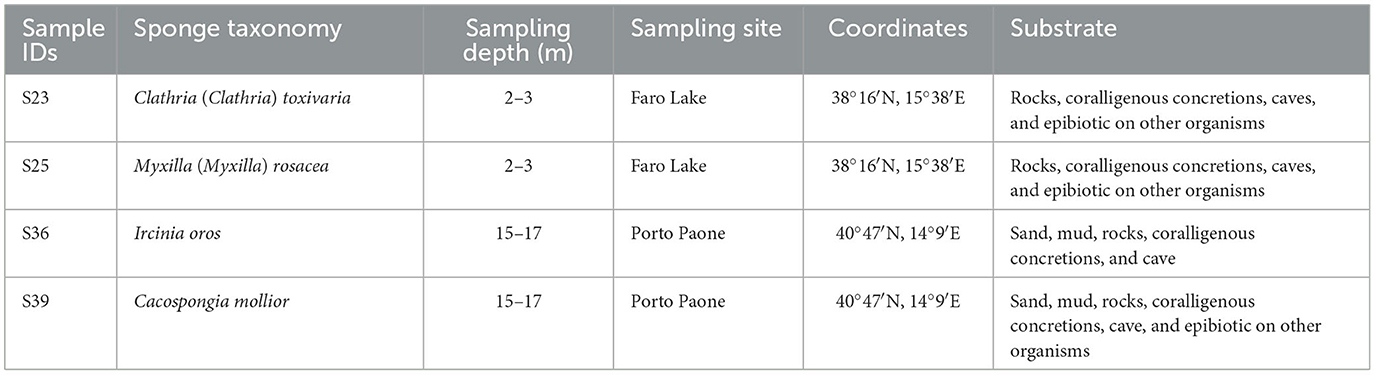
Table 1. Sample IDs, taxonomy, sampling depth (m), sites, geographical coordinates, and type of substrate of sponge specimens.
2.2 Morphological identification of the sponges
The taxonomic analysis was conducted by examining the skeletal architecture and spicule complement of each sponge specimen according to the protocols already published (Núñez-Pons et al., 2022). The taxonomic identification was made at species level as described in the World Porifera Database (WPD; Van Soest et al., 2023).
2.3 Molecular identification of sponges
The DNA extraction was performed starting from 10 mg of sponge tissue and employing QIAamp® DNA Micro kit (QIAGEN, Germany), according to the manufacturer's protocol. The DNA (ng/μL) was quantified using NanoDrop spectrophotometer. The C1000 Touch Thermal Cycler (Biorad) was used for PCR reaction processes in a 30 μL final volume of master mix with ~50–100 ng of genomic DNA. The reaction mixture was prepared using 6 μL of 5X Buffer GL (GeneSpin Srl, Milan, Italy), 3 μL of dNTPs (2 mM each), 2 μL each of forward and reverse primers (25 pmol/μL), and 0.5 μL of Xtra Taq Polymerase (5 U/μL, GeneSpin Srl, Milan, Italy) as follows:
i For 18S and 28S, the amplification conditions were 95°C for 2 min followed by 35 cycles of 95°C for 1 min, 60°C (A/B) and 55°C (18S-AF/18S-BR, NL4F/NL4R) for 1 min, and 72°C for 2 min, with a final extension step at 72°C for 10 min (Chombard et al., 1998; Manuel et al., 2003);
ii ITS primers (RA2/ITS2.2), with initial denaturation at 95°C for 2 min. This was followed by 35 cycles of 95°C for 1 min, 67°C for 1 min, and 72°C for 2 min, with a final extension step at 72°C for 10 min (Worheide et al., 2002; Schmitt et al., 2005);
iii CO1 primers (dgLCO1490/dgHCO2198), with initial denaturation at 94°C for 3 min, 35 cycles of denaturation at 94°C for 30 s, 45°C for 30 s, and a final step of elongation of 72°C for 1 min (Meyer et al., 2005).
The gel of agarose (1.5% electrophoresis in 40 mM Tris–acetate, 1 mM EDTA, pH 8.0, TAE buffer) was used to separate PCR products; the amplified fragments were identified using a DNA ladder of 100 bp (GeneSpin Srl, Milan, Italy). The QIAquick Gel Extraction Kit (Qiagen) was used to extract the fragments from the gel following the instructions of the manufacturer. The NanoDrop spectrophotometer evaluated the DNA quantity (ng/μL). Applied Biosystems (Life Technologies) 3730 Analyzer (48 capillaries) sequenced both strands of the obtained amplicons. All the sequences from PCR products were then aligned to GeneBank using BLAST (Basic Local Alignment Search Tool) to reach for high similarity sequences. Then, Multialign was used to confirm the alignment with high similarity sequences (http://multalin.toulouse.inra.fr/multalin/).
2.4 Metagenomic DNA extraction, illumina miseq sequencing, and diversity analysis
The DNeasy® PowerSoil® Pro Kit (QIAGEN) was used for the extraction of DNA following the protocol described by the manufacturer. For this extraction, a piece of the tissue weighing about 250 mg was used. Using the NanoDrop spectrophotometer it was possible to evaluate DNA quantity (ng/μL) and quality (A260/280, A260/230). To check for DNA integrity, the samples were loaded on 0.8% agarose gel electrophoresis immersed in TAE buffer. For metataxonomic analysis performed by Bio-Fab Research (Roma, Italy), a final concentration of 30 ng/μL of DNA was used. Illumina adapter overhang nucleotide sequences were added to the gene-specific primer pairs targeting the V3-V4 region (S-DBact-0341-b-S-17/S-D-Bact-0785-a-A-2), according to Klindworth et al. (2013). 16S PCR amplification was performed in a final volume of 25 μL, which was set up using the following quantities: 5 μL of microbial genomic DNA (10 ng/μL in 10 mM Tris pH 8.5), 1x PCRBIO HiFi Buffer (PCR BIOSYSTEMS, USA) composed by 1 mM of dNTPs and 3 mM of MgCl2, 0.5 units of PCRBIO HiFi Polymerase (PCR BIOSYSTEMS, USA), and 0.2 μM of each primer. Cycling conditions followed an initial denaturation at 95°C for 3 min, 25 cycles of 95°C for 30 s, 55°C for 30 s, 72°C for 30 s and a final extension step at 72°C for 5 min, hold at 4°C. After the amplification of the 16S, a PCR clean-up was done to purify the V3-V4 amplicon from free primers and primer dimers. After that, a limited-cycle amplification step using Nextera XT Index Kit was carried out to add multiplexing indices and illumine sequencing adapters. In addition, a further clean-up step was performed, followed by a normalization of the libraries and their pooling by denoizing processes. The sequencing on Illumina MiSeq Platform with 2×300 bp paired-end reads took place only after these steps were completed. Taxonomy assignments were done using a “home-made” Naive Bayesian Classifier trained on V3-V4 16S sequences of the SILVA 138.1 release database (Quast et al., 2013). The sample frequencies per feature and per sample are indicated in Supplementary Figures 1, 2. The metataxonomic analysis from raw DNA sequencing data was conducted on the Quantitative Insights Into Microbial Ecology (QIIME 2, Version: 2022.2) platform (Bolyen et al., 2019) by demultiplexing, quality filtering, chimera removal, taxonomic assignment, and both alpha and beta diversity analyses. With the use of R version 4.1.1 (Li, 2021) and Cairo graphics library (Urbanek and Horner, 2020), the taxonomy barplot was generated. To evaluate the species diversity, three different indices were took into account: Chao 1 index (Chao, 1984) is a qualitative species-based method (species richness in the sample that is the number of ASV) and Shannon (1948), Shannon and Weaver (1949), and Simpson (1949) indexes estimated the quantitative species-based measures, which indicated the community diversity as species richness and evenness. The estimation was made at three taxa levels (Level 5 = Family, Level 6 = Genus, Level 7 = Species). Statistically significant differences for alpha and beta diversities were explored using the Kruskal–Wallis test and pairwise PERMANOVA analysis, respectively. The distance matrix between each pair of sample (independently from environmental variables) was calculated using Bray–Curtis and “un-, weighted” UniFrac metrics.
The data were deposited in the SRA database (submission ID: SUB14019148; BioProject ID: PRJNA1049642).
3 Results
3.1 Morphological and molecular characterization
All the sponges were characterized by morphological and molecular approaches. Concerning the morphological analysis, all the analyzed sponges belonged to the class Demospongiae (see Table 1) and usually live in the Mediterranean Sea. Concerning C. (Clathria) toxivaria, collected in the Faro Lake (Sicily), molecular tool for characterization of this species identified by morphological analysis was not available in GenBank. The second sponge collected in the Faro Lake was identified by morphological analysis as M. (Myxilla) rosacea. In the case of the two sponge samples collected in “Porto Paone” (Gulf of Naples), CO1 was the best molecular marker with 98.5% identity, characterizing the sponge S36 as Ircinia oros with 98.5% identity. For Sample S39, this species was identified by ITS as Cacospongia mollior, reporting a percentage of sequence similarity of 99%.
3.2 Metataxonomic profile
As indicated by the ASVs analysis, most microbial species had a confidence percentage ≥75%. The complete taxonomy classification of bacterial communities of sponge samples is presented in Supplementary Tables 1–4. In Porto Paone, two sponges (Gulf of Naples) were found: (i) M. (M.) rosacea showed the largest number of features (519), especially Alphaproteobacteria, Dehalococcoidia, Gammaproteobacteria, and Cyanobacteria (Figure 1) and (ii) 250 ASVs in C. (C.) toxivaria, with a greater abundance of Alphaproteobacteria, Gammaproteobacteria, and Bacteroidia (Figure 1). In sponges I. oros and C. mollior, both collected in the Faro Lake (Sicily), 156 and 204 ASVs were detected, respectively. The most abundant bacterial classes in I. oros were Dadabacteriia, Anaerolineae, and Acidimicrobiia, while the bacterial community of C. mollior was dominated by Nitrospiria, Anaerolineae, Dadabacteriia, and Poribacteria (Figure 1). In particular, the taxonomic composition revealed an abundance of Proteobacteria present in C. toxivaria (36%) and M. (M.) rosacea (35%; Figure 1). In contrast, a low percentage (2%) of Proteobacteria was detected in two sponges collected from the Faro Lake (Sicily). In addition, C. toxivaria revealed 1% of Bacteroidia class. The sponge M. (M.) rosacea revealed low percentages of Cyanobacteria and Dehalococcoidia (1–0.9%, respectively; Figure 1). Interestingly, this sponge was the only species revealing a certain abundance of Rhizobiaceae, belonging to the phylum Proteobacteria (75.6 %; Figure 1). Concerning the sponges I. oros and C. mollior, low percentages of Dadabacteria (1%) and Poribacteria (0.9%) were present, respectively (Figure 1). As reported above, the sponges C. (C.) toxivaria and M. (M.) rosacea revealed a comparable composition in the distribution of microbial species. In fact, a great plethora of Proteobacteria was observed in these species (Figure 1). Similarly, the sponges I. oros and C. mollior showed the same distribution of bacteria and archaea.
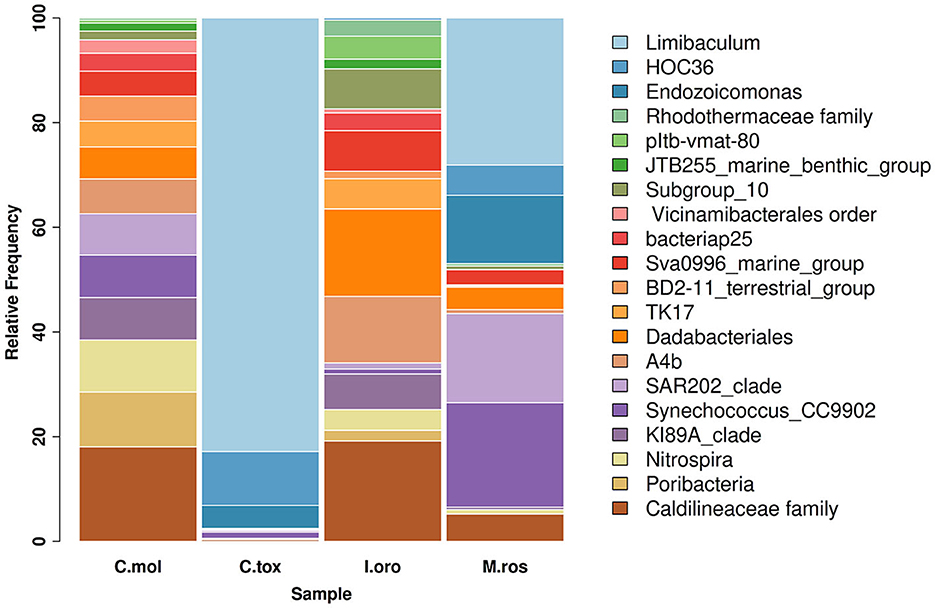
Figure 1. Taxonomy barplot comparing the most abundant bacterial phyla at the genus level in sponges C. mollior, C. (C.) toxivaria, I. oros, and M. (M.) rosacea. Data were normalized in each sample by median sequencing depth, filtered to low abundance and to sum rows >500.
3.3 Principal coordinates analyses (PCoA) of bacterial communities
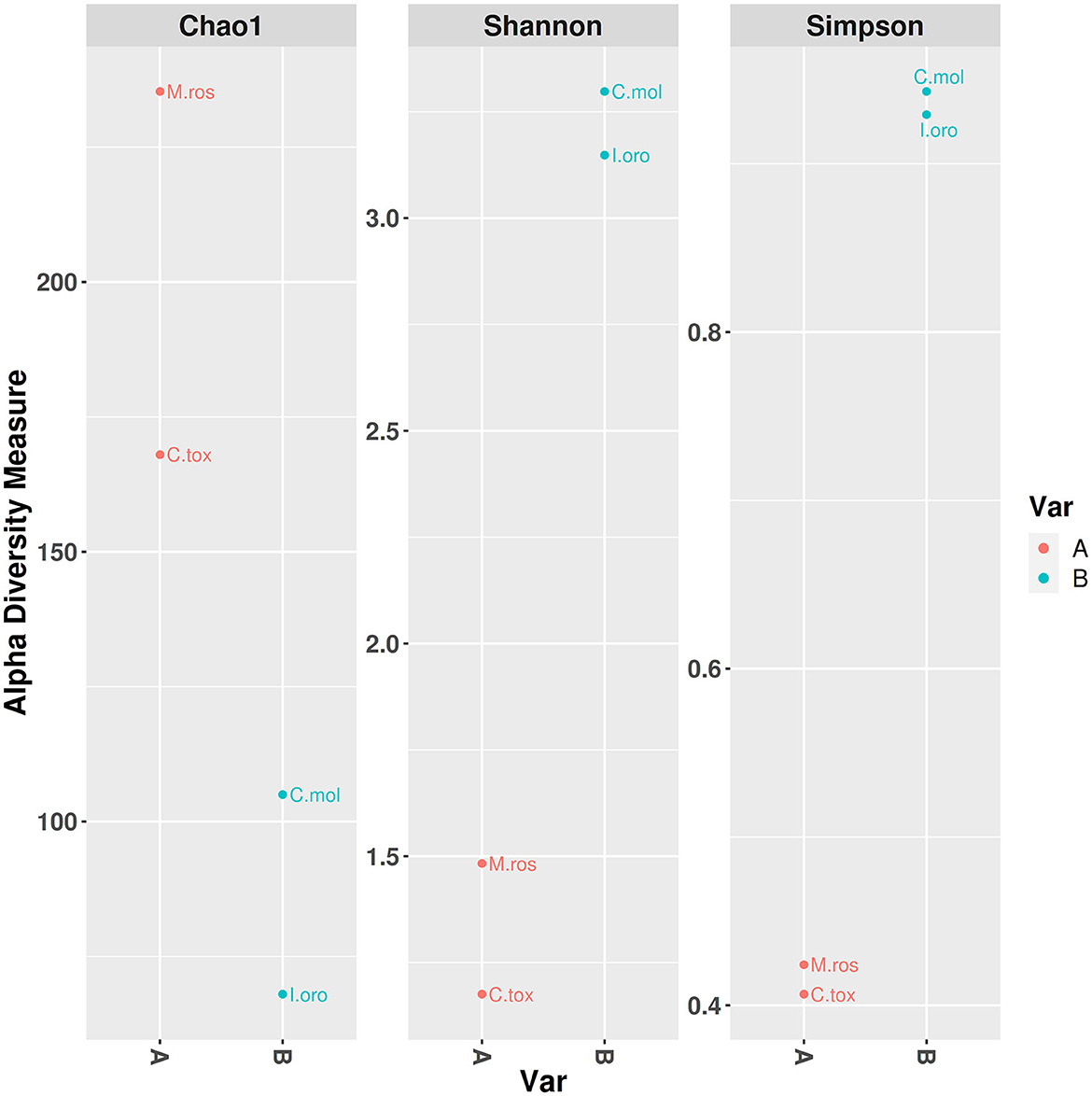
PCoA based on the relative abundance of genera revealed a significant separation in bacterial community composition between Group A (sponges collected in the Faro Lake) and Group B (sponges collected in Porto Paone), using both principal component scores of PC1 and PC2 (68.7 and 29.9%, respectively; Figure 2 and Table 2). Alpha diversity was used to analyze the samples, and three diversity indices (Chao1, Shannon, and Simpson) were used to determine whether each sample was sequenced at sufficient depth or not. As shown in Figure 3, the Chao index of Group B, including sponges collected in Porto Paone, was lower than that of the sponges collected in the Faro Lake (Group A). The results showed that the species richness of Group B was significantly lower than that of Group A. Moreover, Shannon and Simpson indexes of the Group B were higher than that of the Group A, demonstrating that the two groups had the same trend.
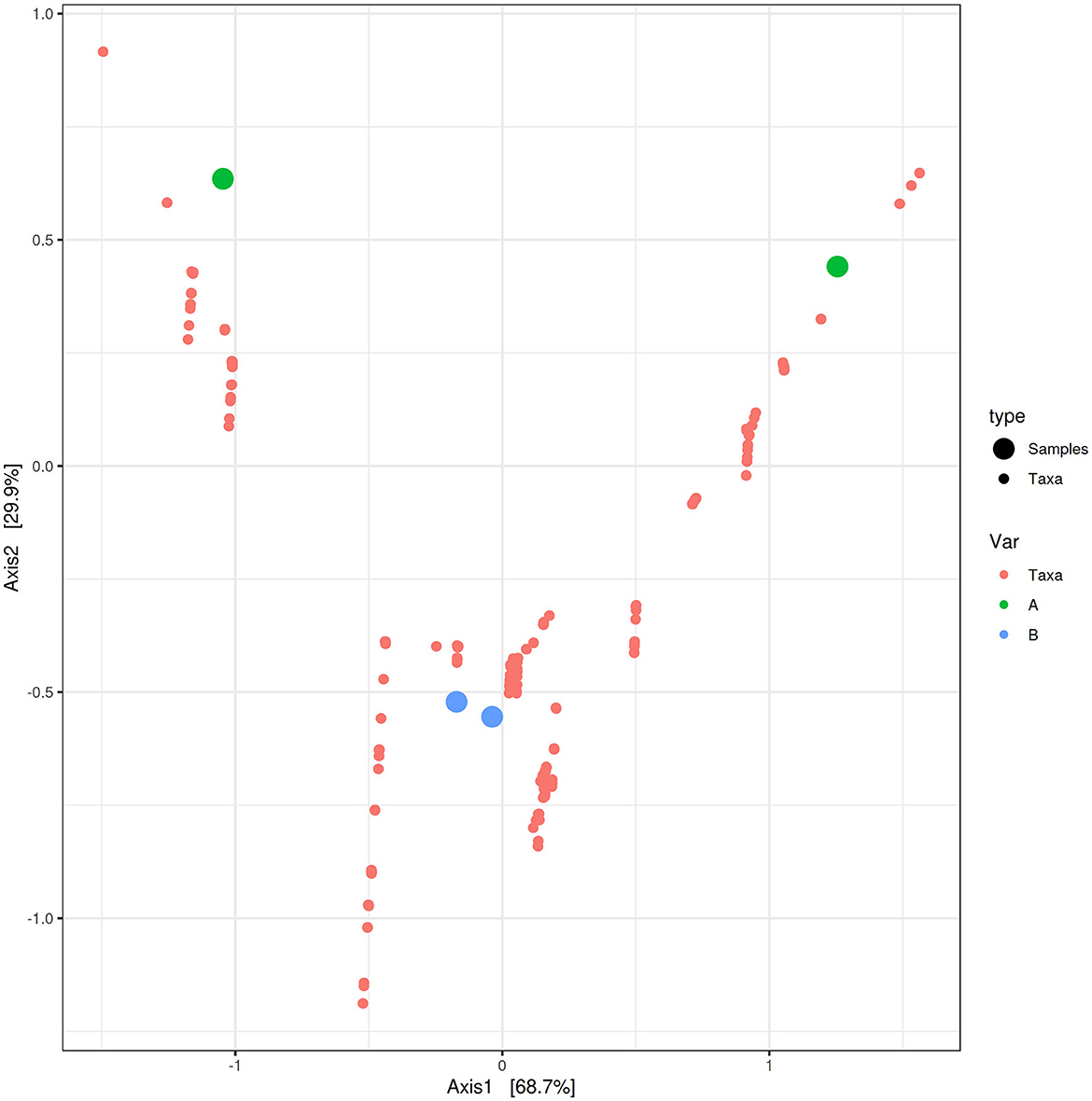
Figure 2. Principal coordinates analysis (PCoA) biplot between Group A and Group B for identifying features that contribute the most in terms of separating the samples in a PCoA plot.
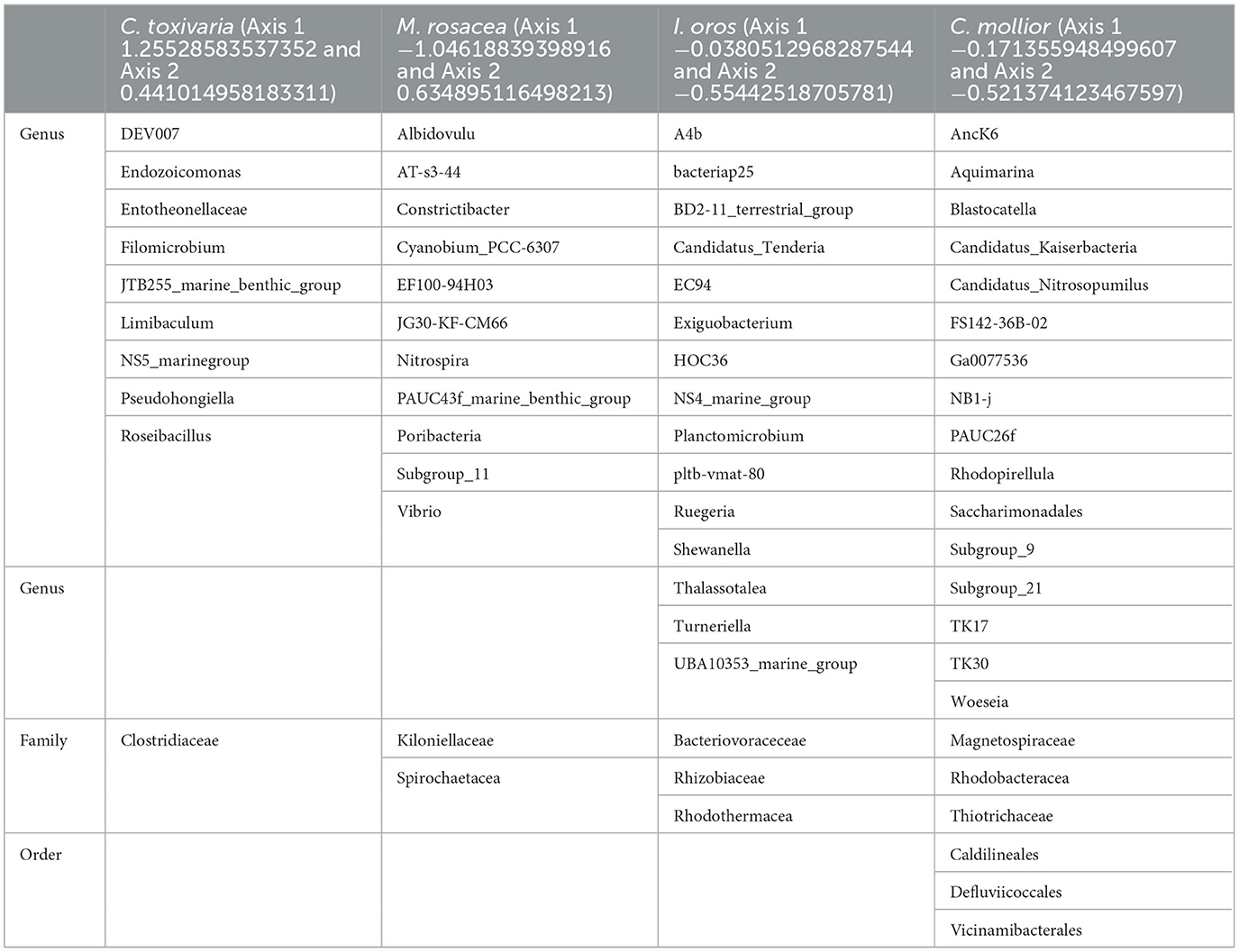
Table 2. Bacteria clustered with C. toxivaria, M. rosacea, I. oros, and C. mollior identified at genus, family, and order levels.
4 Discussion
This study builds upon previous investigations where we analyzed the microbial diversity by metataxonomic analysis of eight sponge samples from four sites: Oceanapia cf. perforate (Sarà 1960), Sarcotragus spinosulus (Schmidt 1862) and Erylus discophorus (Schmidt 1862) from Faro Lake in Sicily; Tethya aurantium (Pallas 1766) and Axinella damicornis (Esper 1794) from “Porto Paone” (Gulf of Naples); Geodia cydonium (Jamenson 1811) from “Secca delle fumose” (Gulf of Naples); Agelas oroides (Schmidt 1864) and Acanthella acuta (Schmidt 1862) from “Punta San Pancrazio” (Gulf of Naples). In fact, here we explored the sponge biodiversity, in terms of microorganisms, in the Mediterranean Sea, analyzing other species from the Strait of Messina and the Gulf of Naples.
We first identified four sponges, combining the identification by morphological features with a molecular approach, based on the sequences of 28S and 18S rDNA, ITS, and CO1. According to the literature, there is no single marker for all sponge species, and each primer pair displayed its own constraints and strengths (Yang et al., 2017; Ruocco et al., 2021). The difficulty in identifying them is also related to absent or incomplete sequences annotated in the database, thus limiting the phylogenetically based taxonomic methods used for species identification. For this reason, a molecular approach with multi-loci should be employed to ensure reliable sponge identification.
This represents the first record of metataxonomic analysis for the sponges, such as C. (C.) toxivaria, M. (M.) rosacea, Ircinia oros, and Cacospongia mollior.
An important finding concerning the species analyzed in this study concerned the fact that the sponges M. (M.) rosacea and C. (C.) toxivaria collected in the Faro Lake were already recorded in this lake in 2013 suggesting, their presence as persistent (Marra et al., 2016). The sponges collected in the Gulf of Naples are typical species in the Mediterranean Sea. In addition, we investigated the bacterial diversity of the Mediterranean sponges by performing the metataxonomic analysis. Our results revealed that the analyzed sponges hosted various bacterial communities according the sampling site. Interestingly, as reported in Ruocco et al. (2021), sponges collected in the Faro Lake had a more diverse phyla composition than sponges collected in the Gulf of Naples (Figure 4). In addition, PCA analysis revealed interesting results for the sponges collected in Faro Lake. In fact, M. (M.) rosacea is the only sponge with a significant presence of Rhizobiaceae bacteria. The community structure seems to be closely related to the order to which they belong, as demonstrated in the Krona plot and heat map. In fact, I. oros and C. mollior, belonging to Dictyoceratida order, were recorded as high microbial abundance (HMA) species, while M. (M.) rosacea and C. (C.) toxivaria, belonging to the Poecilosclerida order, were low microbial abundance (LMA) species. HMA sponges harbor a more diverse symbiotic community than LMA, which were found to be extremely stable at seasonal and interannual scales.
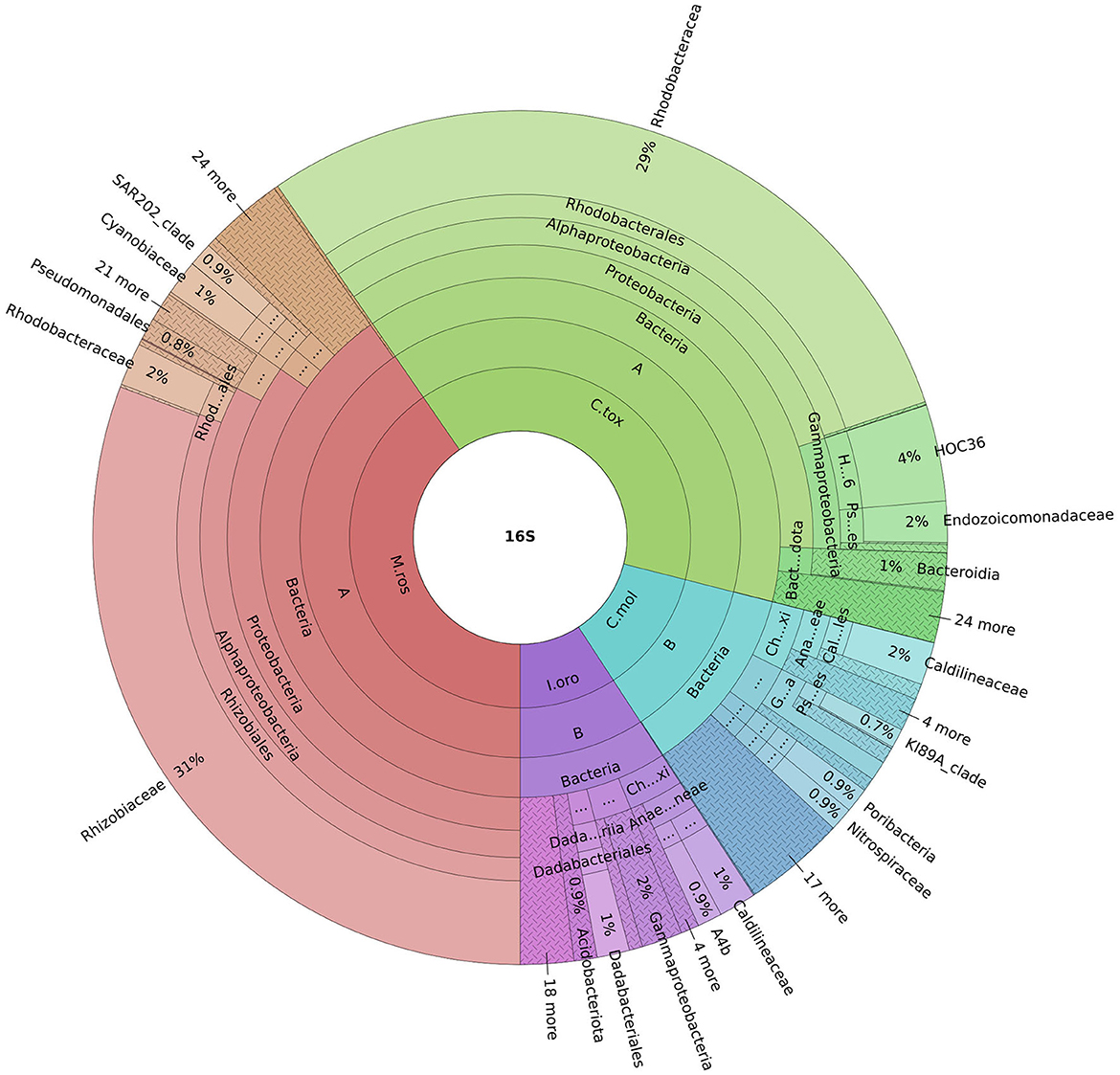
Figure 4. Krona plot represents the most abundant phyla for all sponges. Sample code: M.ros, M. (M.) rosacea; I.oro, I. oros; C.mol., C. mollior; C.tox., C. (C.) toxivaria.
This taxonomic analysis was useful for us, being strongly interested in the biotechnological potential of these sponges. In fact, as reported in the introduction, sponges in general are among the marine organisms richest in bioactive compounds of biotechnological interest. There is a strong debate in the literature whether the bioactive compounds isolated from sponges are produced by the sponges themselves, by the organisms associated with them, or by the interaction between sponges and microorganisms. The fact is that the microorganisms associated with sponges play a fundamental role in their production. Many studies reported that sponge-associated bacteria are good candidates for isolating natural compounds with their application in pharmacological, nutraceutical, and cosmeceutical fields. In this context, our study represents an important step in this field, representing one of the first assessments of the biotechnological potential of mentioned sponge-associated bacteria. In all species of sponges are present Alphaproteobacteria and Gammaproteobacteria, which showed antimicrobial activity making them suitable tools for pharmacological purposes (Thiel and Imhoff, 2003; Thakur et al., 2005; Taylor et al., 2007; Werner, 2010; Haber and Ilan, 2014; Brinkmann et al., 2017; Bibi et al., 2020). The phylum Cloreflexi (classes Anaerolineae and Dehaloccoidia) is typical of both collection sites, being in all sponge samples. However, no data had been reported so far for marine biotechnology applications for class Anaerolineae. Despite playing an important role in biochemical cycles in various environments, they are manifestly difficult to cultivate due to slow growth (Nakahara et al., 2019; De Castro-Fernández et al., 2023). On the contrary, the anaerobic Dehalococcoides showed an interesting capability to transform various chlorinated organics, which are normally released through industrial and agricultural activities (Bedard et al., 2007; Taş et al., 2010). In the case of the sponge M. (M.) rosacea, Rhizobiaceae was the most abundant family (order Alphaproteobacteria) among other bacteria isolated. These bacteria are generally associated with soil and plant hosts and are involved in the process of biological nitrogen fixation (Carrareto-Alves et al., 2015). Therefore, the family Rhizobiaceae, described in this study, was found associated with the sponge M. (M.) rosacea for the first time. Potentially, these bacteria can be used for bioremediation of heavy metals and biodegradation of toxic compounds due to their metabolic capabilities and ecological roles (Aoki et al., 2022). However, their role in marine environments remain unclear because of the limited availability of cultured marine isolates and their sequenced genomes (Kimes et al., 2015). In summary, our data drew attention to the biodiversity of species in the Mediterranean Sea and the 16S rRNA sequence dataset, allowing the detection of several resident microbiomes featured. The sponges and their associated microorganisms revealed good source to identify new compounds for biotechnological applications, and further analyses will be needed to investigate the potential role of the bacteria associated to these sponges. In the meantime, we analyzed the biotechnological potential of these sponges by bioassay-guided fractionation on several human cancer lines, and first results showed specific antimitotic activity against some cancers. We also tried identifying the chemical structure of the compounds responsible for this activity. We also performed metagenomic and transcriptomic analyses on these sponges, which could help us identify the gene/enzyme responsible for the production of the bioactive compounds. These –omic techniques represent an environmental-friendly approach mainly for organisms, such as the sponges, which cannot be cultivated and are difficult to keep in the laboratory to increase biomass.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding authors.
Ethics statement
The manuscript presents research on animals that do not require ethical approval for their study.
Author contributions
RE: Data curation, Investigation, Methodology, Writing – original draft, Writing – review & editing. SF: Data curation, Methodology, Writing – original draft. MS: Data curation, Formal analysis, Methodology, Writing – review & editing. SR: Data curation, Formal analysis, Methodology, Writing – review & editing. MB: Data curation, Formal analysis, Investigation, Methodology, Writing – review & editing. NR: Data curation, Methodology, Writing – review & editing. GZ: Data curation, Methodology, Writing – review & editing. MGi: Data curation, Methodology, Supervision, Writing – review & editing. MP: Data curation, Methodology, Supervision, Writing – review & editing. MGu: Data curation, Methodology, Writing – review & editing. VZ: Conceptualization, Resources, Writing – original draft, Writing – review & editing. MC: Conceptualization, Funding acquisition, Investigation, Methodology, Resources, Validation, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. SF received support through a Ph.D. fellowship co-funded by the Stazione Zoologica Anton Dohrn (Naples, Italy) and the University of Genoa.
Acknowledgments
We thank the scuba diving team of the Stazione Zoologica for their assistance in collecting sponges in the Gulf of Naples. This work was performed in the framework of the PRIN 2022 project “Mediterranean marine organisms as sources of new anti-inflammatory and pro-resolving compounds”.
Conflict of interest
MS and SR were employed by Bio-Fab Research Srl.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1295459/full#supplementary-material
References
Amelia, T. S. M., Suaberon, F. A. C., Vad, J., Fahmni, A. D. M., Saludes, J. P., and Bhubalan, K. (2022). Recent advances of marine sponge-associated microorganisms as a source of commercially viable natural products. Mar. Biotechnol. 24, 492–512. doi: 10.1007/s10126-022-10130-2
Aoki, M., Nakahara, N., Kusube, M., and Syutsubo, K. (2022). Metagenome-assembled genome sequence of marine Rhizobiaceae sp. strain MnEN-MB40S, obtained from manganese-oxidizing enrichment culture. Am. Soc. Microbiol. 11, 16–18. doi: 10.1128/mra.00645-22
Bakus, G. J., Targett, N. M., and Schulte, B. (1986). Chemical ecology of marine organisms: an overview. J. Chem. Ecol. 12, 951–987. doi: 10.1007/BF01638991
Bedard, D. L., Ritalahti, K. M., and Löffler, F. E. (2007). The Dehalococcoides population in sediment-free mixed cultures metabolically dechlorinates the commercial polychlorinated biphenyl mixture aroclor 1260. Appl. Environ. Microbiol. 73, 2513–2521. doi: 10.1128/AEM.02909-06
Bewley, C. A., Holland, N. D., and Faulkner, D. J. (1996). Two classes of metabolites from Theonella swinhoei are localized in distinct populations of bacterial symbionts. Experientia 52, 716–722. doi: 10.1007/BF01925581
Bibi, F., Alvi, S. A., Al-Sofyani, A., Naseere, M. I., Yasir, M., and Azhar, E. I. (2020). Pyrosequencing reveals sponge specific bacterial communities in marine sponges of Red Sea, Saudi Arabia. Saudi J. Biol. Sci. 27, 67–73. doi: 10.1016/j.sjbs.2019.05.002
Bolyen, E., Rideout, J. R., Dillon, M. R., Bokulich, N. A., Abnet, C. C., Al-Ghalith, G. A., et al. (2019). Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 37, 852–857. doi: 10.1038/s41587-019-0209-9
Brinkmann, C. M., Marker, A., and Kurtböke, D. I. (2017). An overview on marine sponge-symbiotic bacteria as unexhausted sources for natural product discovery. Diversity 9, 1–31. doi: 10.3390/d9040040
Bultel-Poncé, V., Berge, J. P., Debitus, C., Nicolas, J. L., and Guvot, M. (1999). Metabolites from the sponge-associated bacterium Pseudomonas species. Mar. Biotechnol. 1, 384–390. doi: 10.1007/PL00011792
Carrareto-Alves, L. M., Marcondes de Souza, J. A., de Mello Varani, A., and de Macedo Lemos, E. G. (2015). The Family Rhizobiaceae. (Berlin; Heidelberg: Springer-Verlag), 419–437.
Cerrano, C., Giovine, M., and Steindler, L. (2022). Petrosia ficiformis (Poiret, 1789): an excellent model for holobiont and biotechnological studies. Curr. Opin. Biotechnol. 74, 61–65. doi: 10.1016/j.copbio.2021.10.022
Chao, A. (1984). Nonparametric estimation of the number of classes in a population. Scand. J. Stat. 1984, 265–270.
Chombard, C., Boury-Esnault, N., and Tillier, S. (1998). Reassessment of homology of morphological characters in tetractinellid sponges based on molecular data. Syst. Biol. 47, 351–366. doi: 10.1080/106351598260761
Coll, M., Piroddi, C., Steenbeek, J., Kaschner, K., Ben Rais Lasram, F., Aguzzi, J., et al. (2010). The biodiversity of the Mediterranean Sea: estimates, patterns, and threats. PLoS ONE 5, e11842. doi: 10.1371/journal.pone.0011842
Cooper, E. L., and Yao, D. (2010). Diving for drugs: tunicate anticancer compunds. Drug Discov. Today 17, 636–648. doi: 10.1016/j.drudis.2012.02.006
Corriero, G., Liaci, L. S., Ruggiero, D., and Pansini, M. (2000). The sponge community of a semi-submerged Mediterranean cave. Mar. Ecol. 21, 85–96. doi: 10.1046/j.1439-0485.2000.00655.x
De Castro-Fernández, P., Ballesté, E., Angulo-Preckler, C., Biggs, J., Avila, C., and Garcìa-Aljaro, C. (2023). How does heat stress affect sponge microbiomes? Structure and resilience of microbial communities of marine sponges from different habitats. Front. Mar. Sci. 9, 1–23. doi: 10.3389/fmars.2022.1072696
Haber, M., and Ilan, M. (2014). Diversity and antibacterial activity of bacteria cultured from Mediterranean Axinella spp. sponges. J. Appl. Microbiol. 116, 519–532. doi: 10.1111/jam.12401
Hentschel, U., Schmid, M., Wagner, M., Fieseler, L., Gernert, C., and Hacker, J. (2001). Isolation and phylogenetic analysis of bacteria with antimicrobial activities from the Mediterranean sponges Aplysina aerophoba and Aplysina cavernicola. FEMS Microbiol. Ecol. 35, 305–312. doi: 10.1111/j.1574-6941.2001.tb00816.x
Karuppiah, V., and Li, Z. (2015). “Marine sponge metagenomics,” in Handbook of Marine Biotechnology (Berlin; Heidelberg: Springer Handbooks), 457–473.
Kimes, N. E., Lopez-Perez, M., Flores-Felix, J. D., Ramìrez-Bahena, M. H., Igual, J. M., Peix, A., et al. (2015). Pseudorhizobium pelagicum gen. nov., sp. nov. isolated from a pelagic Mediterranean zone. Syst. Appl. Microbiol. 38, 293–299. doi: 10.1016/j.syapm.2015.05.003
Klindworth, A., Pruesse, E., Schweer, T., Peplies, J., Quast, C., Horn, M., et al. (2013). Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucl. Acids Res. 41, 1–11. doi: 10.1093/nar/gks808
Larkum, A. W. D., Cox, G. C., Hiller, R. G., Parry, D. L., and Dibbayawan, T. P. (1987). Filamentous cyanophytes containing phycourobilin and in symbiosis with sponges and an ascidian of coral reefs. Mar. Biol. 95, 1–13. doi: 10.1007/BF00447479
Li, J. (2021). The exploration of the approach to data preparation for Chinese text analysis based on R language. Open Access Library J. 8, 1–8. doi: 10.4236/oalib.1107821
Manuel, M., Borchiellini, C., Alivon, E., Le Parco, Y., Vacelet, J., and Boury-Esnault, N. (2003). Phylogeny and evolution of calcareous sponges: monophyly of calcinea and calcaronea, high level of morphological homoplasy, and the primitive nature of axial symmetry. Syst. Biol. 52, 311–333. doi: 10.1080/10635150390196966
Marra, M. V., Bertolino, M., Pansini, M., Giacobbe, S., Manconi, R., and And Pronzato, R. (2016). Long-term turnover of the sponge fauna in Faro Lake (North-East Sicily, Mediterranean Sea). Ital. J. Zool. 83, 579–588. doi: 10.1080/11250003.2016.1251981
Meyer, C. P., Geller, J. B., and Paulay, G. (2005). Fine scale endemism on coral reefs: archipelagic differentiation in turbinid gastropods. Evolution 59, 113–125. doi: 10.1111/j.0014-3820.2005.tb00899.x
Nakahara, N., Nobu, M. K., Takaki, Y., Miyazaki, M., Tasumi, E., Sakai, S., et al. (2019). Aggregatilinea lenta gen. nov., sp. nov., a slow-growing, facultatively anaerobic bacterium isolated from subseafloor sediment, and proposal of the new order aggregatilineales ord. nov. within the class anaerolineae of the phylum Chloroflexi. Int. J. Syst. Evol. Microbiol. 69, 1185–1194. doi: 10.1099/ijsem.0.003291
Newman, D. J., and Cragg, G. M. (2020). Natural products as sources of new drugs over the nearly four decades from 01/1981 to 09/2019. J. Nat. Prod. 83, 770–803. doi: 10.1021/acs.jnatprod.9b01285
Núñez-Pons, L., Mazzella, V., Rispo, F., Efremova, J., and Calcinai, B. (2022). DNA barcoding procedures of taxonomical and phylogenetic studies in marine animals: porifera as a case study. Mar. Genomics Methods Protoc. 10, 195–223. doi: 10.1007/978-1-0716-2313-8_10
Oclarit, J. M., Okada, H., Ohta, S., Kaminura, K., Yamaoka, Y., Iizuka, T., et al. (1994). Anti-bacillus substance in the marine sponge, Hyatella species, produced by an associated Vibrio species bacterium. Microbios 78, 7–16.
Paul, V. J. (1992). Chemical defenses of benthic marine invertebrates: ecological roles of marine natural products. Ithaca, NY: Comstock Publishing Associates.
Proksch, P. (1994). Defensive roles for secondary metabolites from marine sponges and sponge-feeding nudibranchs. Toxicon 32, 639–655. doi: 10.1016/0041-0101(94)90334-4
Quast, C., Pruesse, E., Yilmaz, P., Gerken, J., Schweer, T., Yarza, P., et al. (2013). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucl. Acids Res. 41, 590–596. doi: 10.1093/nar/gks1219
Ruocco, N., Esposito, R., Zagami, G., Bertolino, M., De Matteo, S., Sonnessa, M., et al. (2021). Microbial diversity in Mediterranean sponges as revealed by metataxonomic analysis. Sci. Rep. 11, 1–12. doi: 10.1038/s41598-021-00713-9
Saccà, A., Guglielmo, L., and Bruni, V. (2008). Vertical and temporal microbial community patterns in a meromictic coastal lake influenced by the Straits of Messina upwelling system. Hydrobiologia 600, 89–104. doi: 10.1007/s10750-007-9179-x
Santavy, D. L., Willenz, P., and Colwell, R. R. (1990). Phenotypic study of bacteria associated with the Caribbean sclerosponge, Ceratoporella nicholsoni. Appl. Environ. Microbiol. 56, 1750–1762. doi: 10.1128/aem.56.6.1750-1762.1990
Schmitt, S., Hentschel, U., Zea, S., Dandekar, T., and Wolf, M. (2005). ITS-2 and 18S rRNA gene phylogeny of aplysinidae (verongida, demospongiae). J. Mol. Evol. 60, 327–336. doi: 10.1007/s00239-004-0162-0
Shannon, C. E. (1948). The Bell system technical journal. J. Franklin. Inst. 196, 379–423. doi: 10.1002/j.1538-7305.1948.tb01338.x
Shannon, C. E., and Weaver, W. (1949). A Mathematical Model of Communication. Champaign, IL: University of Illinois Press.
Taş, N., Van Eekert, M. H. A., De Vos, W. M., and Smidt, H. (2010). The little bacteria that can diversity, genomics and ecophysiology of “Dehalococcoides” spp. in contaminated environments. Microb. Biotechnol. 3, 389–402. doi: 10.1111/j.1751-7915.2009.00147.x
Taylor, M. W., Radax, R., Steger, D., and Wagner, M. (2007). Sponge-associated microorganisms: evolution, ecology, and biotechnological potential. Microbiol. Mol. Biol. Rev. 71, 295–347. doi: 10.1128/MMBR.00040-06
Thakur, A. N., Thakur, N. L., Indap, M. M., Pandit, R. A., Datar, V., and Müller, W. E. (2005). Antiangiogenic, antimicrobial, and cytotoxic potential of sponge-associated bacteria. Mar. Biotechnol. 7, 245–252. doi: 10.1007/s10126-004-4085-y
Thiel, V., and Imhoff, J. F. (2003). Phylogenetic identification of bacteria with antimicrobial activities isolated from Mediterranean sponges. Biomol. Eng. 20, 421–423. doi: 10.1016/S1389-0344(03)00069-8
Thompson, C. C., Kruger, R. H., and Thompson, F. L. (2017). Unlocking marine biotechnology in the developing world. Trends Biotechnol. 35, 1119–1121. doi: 10.1016/j.tibtech.2017.08.005
Unson, M. D., Holland, N. D., and Faulkner, D. J. (1994). A brominated secondary metabolite synthesized by the cyanobacterial symbiont of a marine sponge and accumulation of the crystalline metabolite in the sponge tissue. Mar. Biol. 119, 1–11. doi: 10.1007/BF00350100
Urbanek, S., and Horner, J. (2020). Cairo: R Graphics Device Using Cairo Graphics Library for Creating High-Quality Bitmap (PNG, JPEG, TIFF), Vector (PDF, SVG, PostScript) and Display (X11 and Win32) Output. Available online at: https://cran.r-project.org/ (accessed January 31, 2022).
Van Soest, R. W. M., Boury-Esnault, N., Hooper, J. N. A., Rützler, K. D., De Voogd, N. J., Alvarez, B., et al (2023). World Porifera Database. Available online at: https://www.marinespecies.org/porifera (accessed June 21, 2023).
Webster, N. S., and Hill, R. T. (2001). The culturable microbial community of the great barrier reef sponge Rhopaloeides odorabile is dominated by an α-Proteobacterium. Mar. Biol. 138, 843–851. doi: 10.1007/s002270000503
Werner, T. (2010). Next generation sequencing in functional genomics. Brief Bioinform. 2, 499–511. doi: 10.1093/bib/bbq018
Wilkinson, C., and Garrone, R. (1980). Nutrition of Marine Sponges. Involvement of Symbiotic Bacteria in the Uptake of Dissolved Carbon. Oxford: Pergamon Press Ltd.
Wilkinson, C. R., and Fay, P. (1979). Nitrogen fixation in coral reef sponges with symbiotic cyanobacteria. Nature 279, 527–529. doi: 10.1038/279527a0
Wilkinson, C. R., Nowak, M., Austin, B., and Colwell, R. R. (1981). Specificity of bacterial symbionts in Mediterranean and Great Barrier Reef sponges. Microb. Ecol. 7, 13–21. doi: 10.1007/BF02010474
Worheide, G., Degnan, B., Hooper, J., and Reitner, J. (2002). Phylogeography and taxonomy of the indo-Pacific reef cave dwelling coralline demosponge Astrosclera willeyana: new data from nuclear internal transcribed spacer sequences. Proc. 9th Int. Coral. Reef. Symp. 1, 339–346.
Yang, Q., Franco, C. M. M., Sorokin, S. J., and Zhang, W. (2017). Development of a multilocus-based approach for sponge (phylum Porifera) identification: refinement and limitations. Sci. Rep. 7, 1–14. doi: 10.1038/srep41422
Keywords: bacteria, metataxonomic analysis, molecular identification, morphological identification, sponges
Citation: Esposito R, Federico S, Sonnessa M, Reddel S, Bertolino M, Ruocco N, Zagami G, Giovine M, Pozzolini M, Guida M, Zupo V and Costantini M (2024) Characterizing the bacterial communities associated with Mediterranean sponges: a metataxonomic analysis. Front. Microbiol. 14:1295459. doi: 10.3389/fmicb.2023.1295459
Received: 02 October 2023; Accepted: 20 December 2023;
Published: 11 January 2024.
Edited by:
Zhiyong Li, Shanghai Jiao Tong University, ChinaReviewed by:
Cara Fiore, Appalachian State University, United StatesKumar Saurav, Academy of Sciences of the Czech Republic (ASCR), Czechia
Copyright © 2024 Esposito, Federico, Sonnessa, Reddel, Bertolino, Ruocco, Zagami, Giovine, Pozzolini, Guida, Zupo and Costantini. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Maria Costantini, maria.costantini@szn.it
 Roberta Esposito
Roberta Esposito Serena Federico
Serena Federico Michele Sonnessa
Michele Sonnessa Sofia Reddel3
Sofia Reddel3 Marco Bertolino
Marco Bertolino Nadia Ruocco
Nadia Ruocco Marco Giovine
Marco Giovine Marina Pozzolini
Marina Pozzolini Valerio Zupo
Valerio Zupo Maria Costantini
Maria Costantini