- 1Laboratory Sciences and Services Division, International Centre for Diarrhoeal Disease Research Bangladesh (icddr,b), Dhaka, Bangladesh
- 2Department of Biotechnology and Genetic Engineering, Jahangirnagar University, Dhaka, Bangladesh
- 3Department of Infection Biology, London School of Hygiene and Tropical Medicine, London, United Kingdom
- 4Department of Infectious Disease Epidemiology, London School of Hygiene and Tropical Medicine, London, United Kingdom
Atypical Escherichia coli forms exhibit unusual characteristics compared to typical strains. The H2S-producing variants of some atypical E. coli strains cause a wide range of illnesses in humans and animals. However, there are sparse reports on such strains worldwide. We performed whole-genome sequencing (WGS) and detailed characterization of four H2S-producing E. coli variants from poultry and human clinical sources in Dhaka, Bangladesh. All four isolates were confirmed as E. coli using biochemical tests and genomic analysis, and were multidrug-resistant (MDR). WGS analysis including an additional Chinese strain, revealed diverse STs among the five H2S-producing E. coli genomes, with clonal complex ST10 being detected in 2 out of 5 genomes. The predominant phylogroup detected was group A (n = 4/5). The blaTEM1B (n = 5/5) was the most predominant extended-spectrum beta-lactamase (ESBL) gene, followed by different alleles of blaCTX-M (blaCTX-M -55,-65,-123; n = 3/5). Multiple plasmid replicons were detected, with IncX being the most common. One E. coli strain was classified as enteropathogenic E. coli. The genomes of all five isolates harbored five primary and four secondary function genes related to H2S production. These findings suggest the potential of these isolates to cause disease and spread antibiotic resistance. Therefore, such atypical E. coli forms should be included in differential diagnosis to understand the pathogenicity, antimicrobial resistance and evolution of H2S-producing E. coli.
Introduction
Escherichia coli is one of the most genetically diverse and versatile organisms, varying from commensal/avirulent to highly specialized pathogenic strains. E. coli can thrive in several niches, including hosts and in the environment (Kaper et al., 2004; Braz et al., 2020). The variant strains of E. coli may act as facultative or obligate pathogens (Köhler and Dobrindt, 2011). The facultative strains of pathogenic E. coli survive in the intestinal tract and often cause opportunistic infections when reaching suitable extraintestinal sites (Nataro and Kaper, 1998; Kaper et al., 2004). In contrast, enteric obligate pathogens can cause infections in different conditions that range from moderate to severe diarrhea, and can sometimes cause lethal gastrointestinal infections (Nataro and Kaper, 1998).
Pathogenic variants of E. coli are responsible for infections in a variety of animals, more commonly in humans and poultry (Bélanger et al., 2011; Hussain et al., 2017). Pathogenic E. coli has been reported in livestock, including poultry, cattle, and swine (Bélanger et al., 2011). Animal reservoirs of pathogenic E. coli are responsible for diseases in animals, but can spread the infections to humans, including antimicrobial resistant (AMR) strains (Bélanger et al., 2011). Traditionally, biochemical tests have been used for differentiating and identifying members of Enterobacteriaceae, including hydrogen sulfide (H2S) gas (Zabransky et al., 1969). H2S can be synthesized by bacteria such as Campylobacter, Salmonella, Citrobacter, and Erwardsiella and Proteus species on TSI or KIA media (Blachier et al., 2019). This distinct characteristic feature of H2S production by certain bacteria within Enterobcateriacea is used as a bacterial identification test in diagnostic microbiology. E. coli generally does not produce H2S, which differentiates it from the other members of Enterobacteriaceae (Percival et al., 2004). However, a few studies worldwide have reported the presence of atypical H2S-producing E. coli forms in humans and animals (Darland and Davis, 1974; Maker and Washington, 1974; Magalhães and Vance, 1978). They have been isolated from poultry, swine and clinical human urine specimens.
The enzyme 3-mercaptopyruvate sulfurtransferase (3MST) is reported to be mainly responsible for the synthesis of endogenous H2S in Enterbacteriaceae (Mironov et al., 2017). Some studies have also demonstrated the transmissibility of H2S-producing traits between strains via plasmids (Jones and Silver, 1978; Magalhães and Vance, 1978). Although the physiological function of endogenously produced hydrogen sulphide is not clearly defined, recent studies have pointed out its role in protecting bacteria against antibiotics and host defence systems (Mironov et al., 2017; Rahman et al., 2020). A plausible explanation for this is that the antibiotics induce oxidative stress in bacteria by increasing the levels of reactive oxygen species; in response to this, the bacteria produces H2S which in turn stimulates enzymes such as superoxide dismutase (SOD) and catalase that alleviates the effect of reactive oxygen species, and thereby reduces the efficacy of antibiotics contributing to AMR (Eswarappaápradeep, 2017). Also, studies have demonstrated the role of bacterial H2S production in defence against host immunity by making them resistant to leukocytes- mediated killing via unknown mechanisms (Toliver-Kinsky et al., 2019; Rahman et al., 2020).
The accurate identification of H₂S-producing variants of E. coli in diagnostic laboratories is an important step for initiating effective infection management. There is a need to raise awareness of this unusual type of E. coli form that occurs frequently but differs in its inability to produce H2S compared to the typical E. coli forms. Therefore, this study aimed to perform bacteriological, biochemical and genomic characterization of H2S-producing variants of E. coli from healthy poultry and human clinical sources in Dhaka, Bangladesh. We present the first report on the genomic characterization of H2S-producing variants of E. coli from Bangladesh and that from South Asia.
Materials and methods
Ethics statement
The study protocols were approved by the Research Review committee and Ethics Review Committee of icddr,b, Dhaka, Bangladesh (PR-23045).
Bacterial strains
A surveillance study was conducted between 2019 and 2021 to investigate the genomic-based epidemiology of AMR Enterobacteriaceae in healthy poultry and human clinical samples in Dhaka, Bangladesh (Mazumder et al., 2020a, 2021, 2022). During that study, we detected four lactose fermenting E. coli colonies but with an atypical biochemical feature of H2S production. These were confirmed to be E. coli by the methods described hereafter. Three (BD7, BD8, BD9) of these H2S-producing E. coli originated from raw poultry meat and one isolate (BD_CL10) was cultured from a urine sample of a suspected urinary tract infection patient in Dhaka, Bangladesh. Thus, from a collection of 96 poultry E. coli isolates and 204 human clinical E. coli study isolates, we could obtain three and one H2S-producing E. coli isolates, respectively. These four H2S-producing E. coli isolates then formed the basis of this study, and underwent various tests and whole genome sequencing (WGS). One H2S positive E. coli genome from China (Biswas et al., 2020) was used for the in-silico analysis together with the four studied H2S positive E. coli genomes sequenced in this study.
Biochemical characterization and antimicrobial susceptibility
The complete bacteriological and biochemical characteristics of H2S-producing variants of E. coli strains are summarized (Table 1). The biochemical identification included the following tests; kligler iron agar (KIA) test, motility, indole and urease (MIU) test, citrate and acetate utilization test, catalase test, oxidase test, vogas-proskauer test, gelatin liquefaction and ONPG tests. In addition, colonies were plated on Muller-Hinton agar containing 0.68% of sodium thiosulfate plus 0.08% of ferric ammonium sulfate as previously described (Park et al., 2015). The isolates that mimic E. coli in all aspects except H2S-production in Kligler iron agar (KIA) and Muller-Hinton agar (with sodium thiosulfate and ferric ammonium sulfate) were carried forward in this study. These preliminary identified 4 H2S-positive E. coli isolates were subjected to additional tests, including fermentation of sugars and decarboxylation reaction of amino acids (Mazumder et al., 2022). Further, the possibility of Salmonella spp. was ruled out by slide agglutination test using O, O1 polyvalent and VI Salmonella antisera (Denka Seiken Co. Ltd. Tokyo, Japan). The API 20E kit (bioMérieux) was used to generate the analytical profile index (Table 1). Haemolysis was evaluated using 5% sheep blood agar plates. Disk diffusion method was employed to determine the antimicrobial susceptibility. The Clinical and Laboratory Standards Institute (CLSI) guidelines (Weinstein, 2019) were followed. Twenty commercially available antibiotic disks (Oxoid, US) covering 11 antimicrobial classes were tested (see Table 2). The intermediate susceptibility was described as non-susceptible. Isolates were termed multi-drug resistant (MDR) if refractory to at least one antibiotic from three or more antimicrobial classes (Magiorakos et al., 2012).
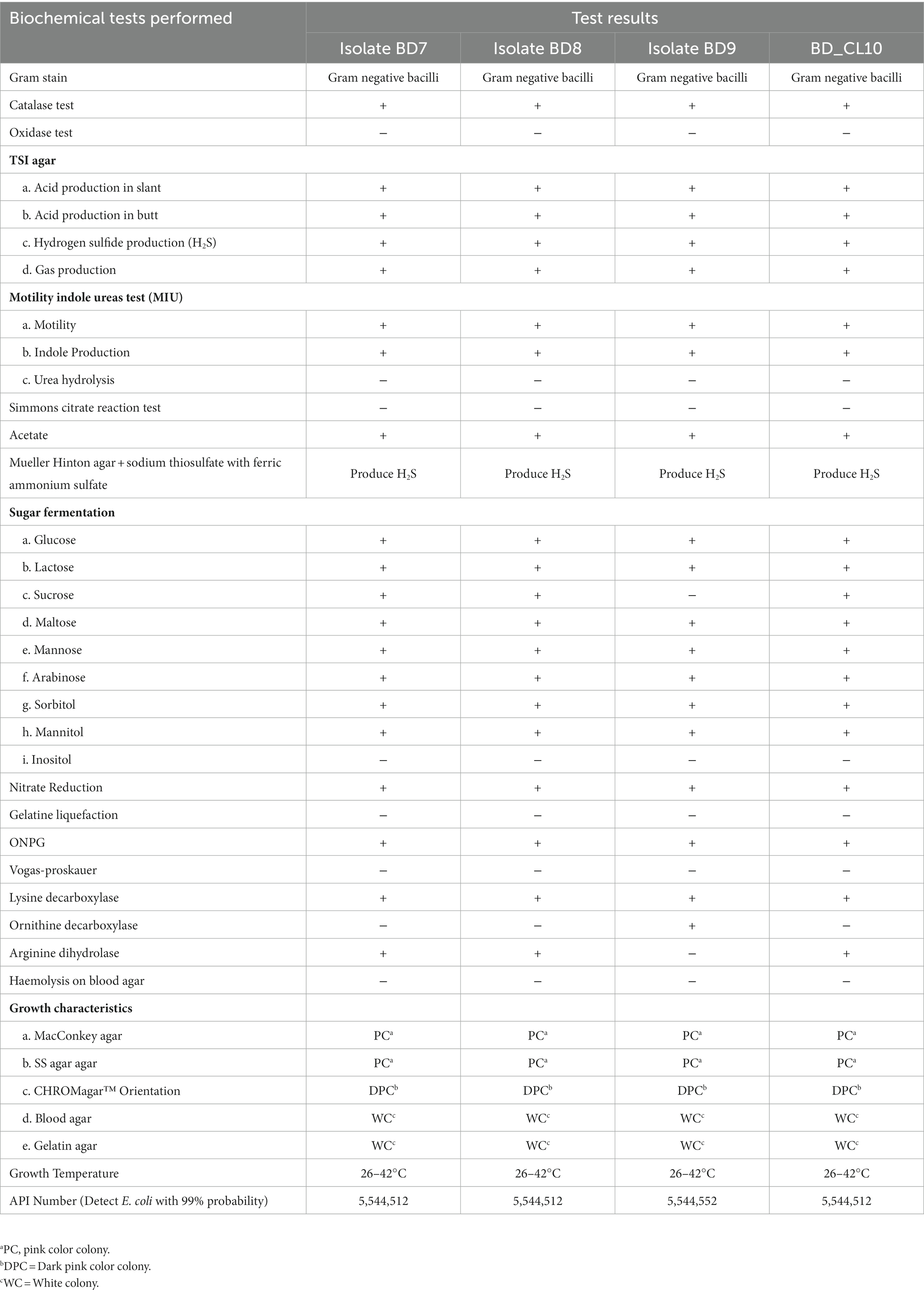
Table 1. Biochemical and growth characteristics of H2S-producing Escherichia coli from Dhaka, Bangladesh.
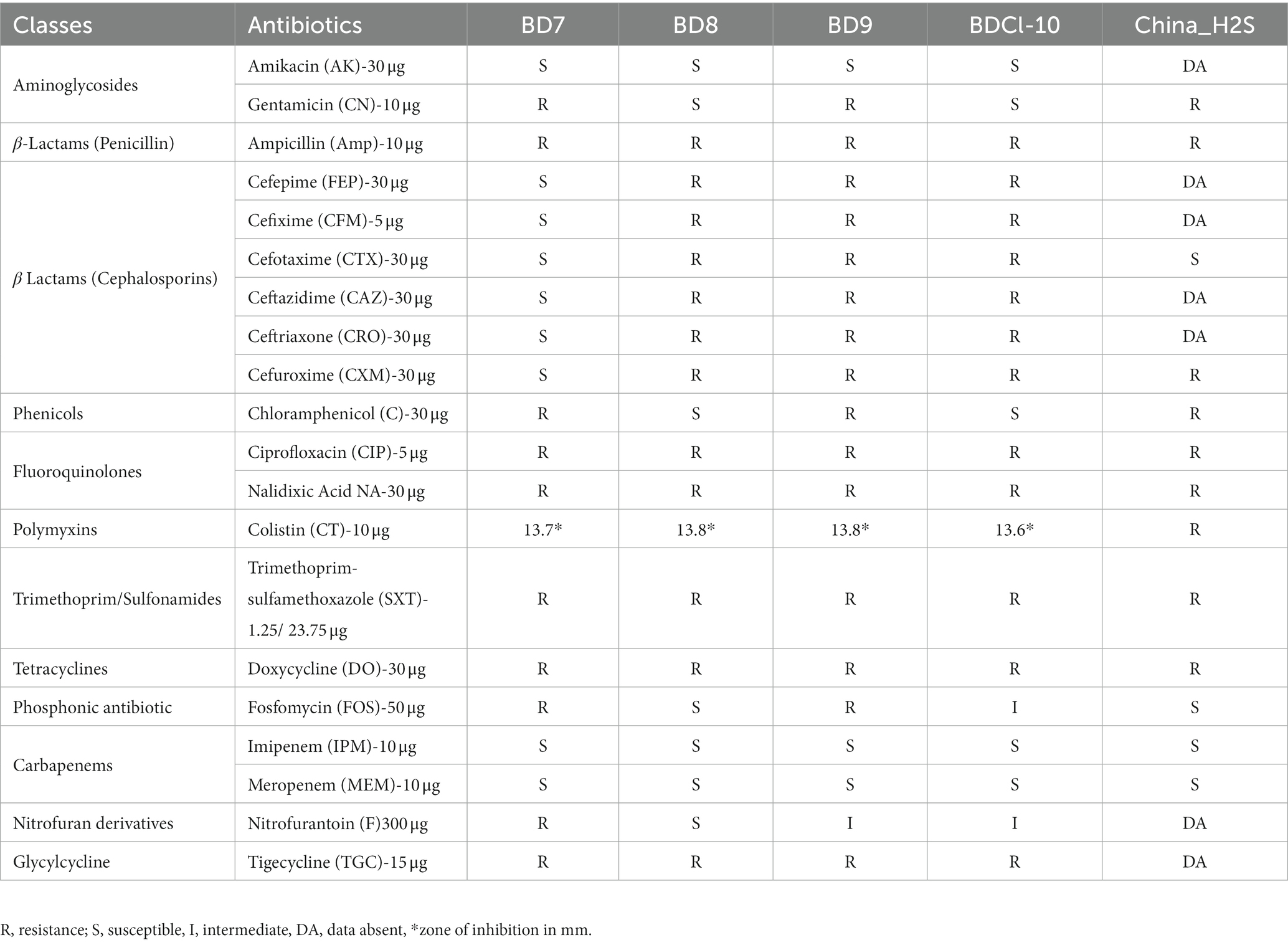
Table 2. Antimicrobial susceptibility profiles of H2S-producing Escherichia coli isolates from Dhaka, Bangladesh.
Whole-genome sequencing
Total bacterial DNA was extracted using the Maxwell RSC Instrument and Culture Cell DNA extraction Kit (Promega) for gram-negative bacteria with an additional RNaseA treatment (Baddam et al., 2020; Mazumder et al., 2020b,c). The DNA QC was assessed by Nanodrop spectrophotometer (Thermo Fisher Scientific, US), Quantus Fluorometer (Promega, US) and by 1% agarose gel electrophoresis. The paired-end bacterial WGS libraries were constructed from 220 to 250 ng of genomic DNA using the Illumina DNA Prep kit as per the manufacturer’s instructions (Mazumder et al., 2021). The pooled libraries thus obtained were sequenced at the icddr,b Genome Centre on Illumina NextSeq 500 system to obtain 100- to 150-fold coverage for each genome using a NextSeq v2.5 Mid Output reagent kit (2 × 150 bp) (Mazumder et al., 2022; Monir et al., 2023).
Sequence assembly and annotation
WGS data quality was examined using FastQC (Andrews, 2010). Trimmomatic software (v0.36) (Faircloth, 2013) was used to extract adapters and poor-quality bases (<Q30) from the unprocessed sequencing reads using the following parameters described elsewhere (Mazumder et al., 2020c, 2021, 2022). Deconseq software (v4.3) was used to eliminate contaminated sequences (Schmieder and Edwards, 2011). The processed reads were used to create de novo assemblies of each genome using SPAdes software (v3.11.1) (Bankevich et al., 2012). QUAST (v5.0) (Gurevich et al., 2013) was used to evaluate the assembly metrics of scaffold fasta files. The genomes were annotated using Prokka (v1.12) (Seemann, 2014) using E. coli MG1655 as the reference genome (GenBank accession number NC 000913.3). The genomic features of H2S-producing E. coli strains are summarized (Table 3).
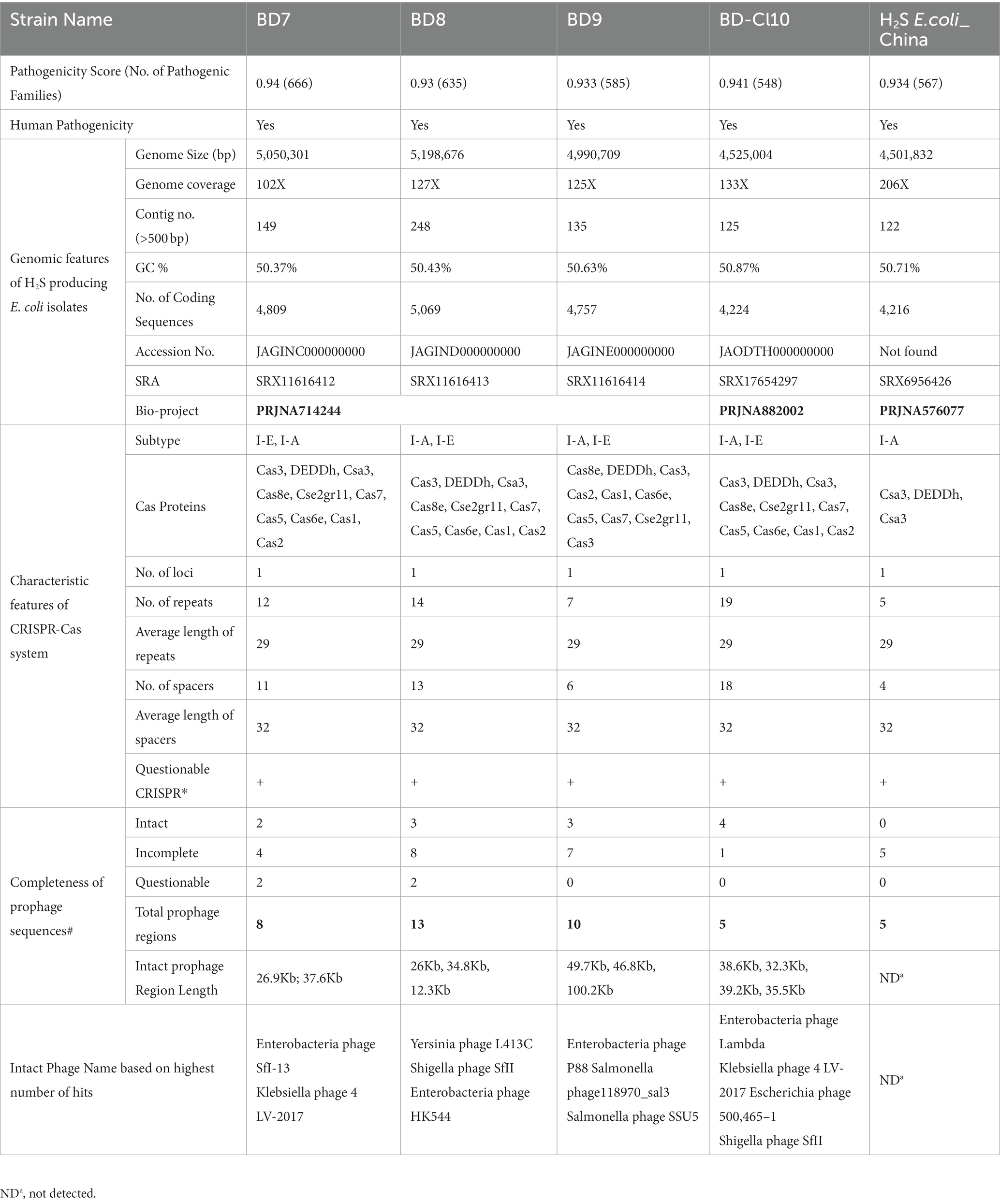
Table 3. Genomic features, the status of CRISPR-CAS system and prophage sequences in H2S-producing Escherichia coli isolates.
In silico sequence analysis
The reads of H2S-producing E. coli were uploaded to the KmerFinder v3.2 (Hasman et al., 2014; Larsen et al., 2014) for species confirmation. The phylogenetic groups were ascertained using Clermon Typing tool (Beghain et al., 2018). The sequence types (STs), clonal complex and pathovars were predicted employing the Achtman7 seven-locus scheme at EnteroBase v1.1.31 web tool. The O and H serotypes were determined employing SerotypeFinder v2.0 (Joensen et al., 2015). FimH and FumC types were determined by CH typer 1.0 (Roer et al., 2018). AMR determinants, virulence factors, and plasmid types were screened using the ABRicate tool v1.0.1 (Seemann, 2018), ResFinder (Zankari et al., 2012), Virulence Factor Database (VFDB) (Chen et al., 2005), and PlasmidFinder (Carattoli et al., 2014) databases, respectively. We used a cut-off of 80% query coverage and 98% identity for screening genes in the genomes analysed. Mobile Element Finder (v1.0.3) was utilized to identify mobile genetic elements linked with acquired antimicrobial resistance genes. Mutations encoding fluoroquinolone resistance were detected by PointFinder (Zankari et al., 2017). IntegronFinder (v2.0) was used to identify integrons (Néron et al., 2022). The chromosomal or plasmid origin of ESBLs genes were analysed by BLASTn analysis of contigs against NCBI database. Prophage sequences in E. coli genomes were determined using the Phage Search Tool Enhanced Release (PHASTER). Prophage regions were classified as intact, questionable, and incomplete based on prophage sequence scores of ≥90, 70–90, and ≤ 70, respectively. (Arndt et al., 2016). CRISPR-Cas system of H2S-producing E. coli strains were characterized using CRISPRone tool2 (Zhang et al., 2012). Cysteine-degradation genes in E. coli were identified based on genes described previously (Braccia et al., 2021). A threshold of 100% coverage and 98% identity were used. The pathogenic pontential of strain was predicted using the web-server PathogenFinder (Cosentino et al., 2013). Default parameters were used for the in-silico analysis unless otherwise stated.
Single nucleotide polymorphism-based core genome phylogeny
We used Snippy (v4.4.0) software (Seemann, 2015) with default parameters to obtain the reference-guided multi-fasta consensus alignment of 5 H2S-producing E. coli genomes using E. coli MG1655 as the reference. Gubbins software (v3.2 5) (Croucher et al., 2015) was used to filter true point mutations from those arising from recombination. The phylogenetic tree was determined using RaxML (v8.2.12), utilizing the Generalized Time Reversible substitution model and a GAMMA distribution to account for rate heterogeneity (Stamatakis, 2014). Finally, the phylogenetic tree was displayed using IToL (Letunic and Bork, 2016).
Accession numbers
The four genomes that were sequenced for this study can be identified by their GenBank accession numbers: JAGINC000000000 (BD7), JAGIND000000000 (BD8), JAGINE000000000 (BD9) and JAODTH000000000 (BD_CL10) (Table 3).
Results
Bacterial characteristics
Four H2S-positive E. coli variants were identified that formed a black precipitate after overnight incubation in an aerobic environment in the Kligler iron agar (KIA) and Mueller Hinton agar medium enriched with both sodium thiosulfate and ferric ammonium sulfate (Figure 1). Attempts to agglutinate the strains with polyvalent Salmonella antisera yielded negative results. When streaked on CHROMagar Orientation media, E. coli produced small, pink-red colonies that were characteristic of the species. Routine biochemical tests identified the strains as E. coli, except for their ability to reduce thiosulfate to H2S (Table 1). All four isolates were gram-negative rods, motile, oxidase-negative, catalase-positive and indole positive. All isolates tested showed a lack of urease and Voges-Proskauer reaction, and they did not grow on the Simmons Citrate agar medium. Nonetheless, all isolates exhibited a positive result for O-nitrophenyl-beta-D-galactopyranoside (ONPG) and carried out fermentation of glucose and lactose sugars, leading to gas production (Figure 1). The API results revealed two distinct profiles, 5,544,512 (n = 3), and 5,544,552 (n = 1) and confirmed isolates as E. coli with 99% certainty (Table 1). The optimum growth temperate ranged between 26° to 42°C and they produced gamma-haemolysis on sheep blood agar.
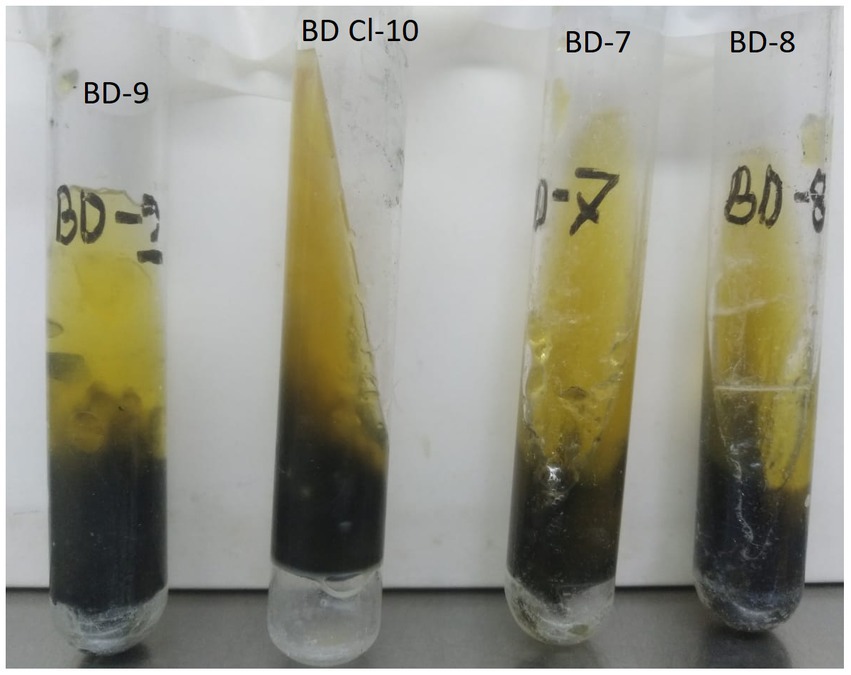
Figure 1. Hydrogen Sulfide (H2S)-producing variants of Escherichia coli strains: BD7, BD8, BD9, and BD-CL10 showing black precipitate of H2S on Kligler Iron Agar (KIA) tubes.
Molecular and phylogenomic analysis of H2S-positive E. coli genomes
This analysis included the four in-house strains and a genome of H2S-producing E. coli reported from China (China_H2S). WGS-based species identification confirmed all the isolates as E. coli. Across the five H2S-producing E. coli strains, the average genome size was 4,853,304 bp (range 4,501,832 to 5,198,676) with an average GC content of 50.6% (range: 50.4 to 50.9%). The genome assemblies had an average coverage of 138-fold, with a range of 102 to 206-fold (Table 3). They had five distinct STs, which comprised ST10, ST48, ST12434, ST189, and ST12066. We detected four clonal complexes that included CC10 (two strains from human sources) followed by CC155, CC165 and CC206, representing one strain each (Figure 2). We identified four isolates (80%) belonging to commensal phylogroup A and one isolate (20%) to B1 phylogroup. All H2S-producing E. coli isolates exhibited distinct serotypes and CH types. A phylogenetic tree was constructed for five H2S-producing E. coli genomes using the MG1655 genome as a reference, by aligning the core genome single nucleotide polymorphisms (SNPs). The studied H2S-producing E. coli strains were found to be relatively diverse. However, the three poultry H2S-positive strains from Bangladesh clustered together, with human strains adjacent to this cluster. The molecular characteristics did not correlate with the source of origin or the phylogenetic clustering of the H2S-producing E. coli isolates (Figure 2).
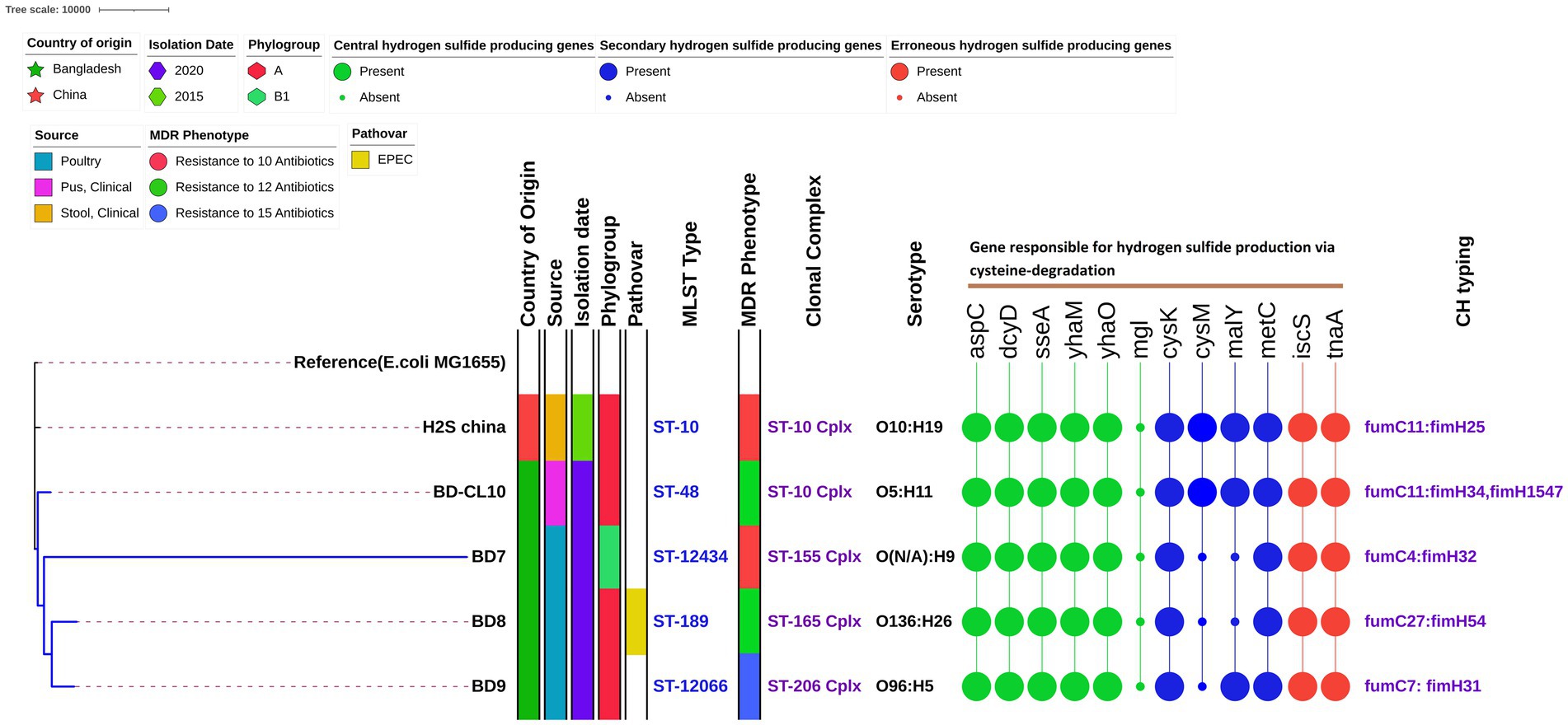
Figure 2. Phylogenetic relationships among sequenced Hydrogen Sulfide (H2S)-Producing Escherichia coli genomes. The maximum likelihood phylogenetic tree is based on the alignment of detected core genomes with the MG1655 genome strain as a reference genome. The country of origin, isolation date, sample source, phylogroups, pathovar, MDR status, STs, clonal complex, serogroups, genes responsible for hydrogen sulfide production via cysteine-degradation and CH types are shown next to the tree.
H2S-producing genes
All five H2S-producing E. coli genomes harbored five primary hydrogen sulfide-producing genes; cysteine aminotransferase (aspC), cysteine desulfhydrase (dcyD), 3-mercapto pyruvate sulfurtransferase (sseA), yhaOM operon (yhaM, yhaO). Whereas the methionine gamma-lyase (mgl) gene was completely absent. Two of the four secondary function genes, cysteine synthase A (cysK) and cystathionine beta-lyase (metC) were found in all four strains. However, the other genes such as cysteine synthase B (cysM), and cystathionine beta-lyase-like repressor of maltose regulon (malY) are sparingly present in poultry isolates. The erroneous H2S-producing genes, including cysteine desulfurase (iscS) and tryptophanase (tnaA) were observed in all isolates. As expected, all three class of cysteine-degradation genes were found on chromosomes (Figure 2; Table 4).
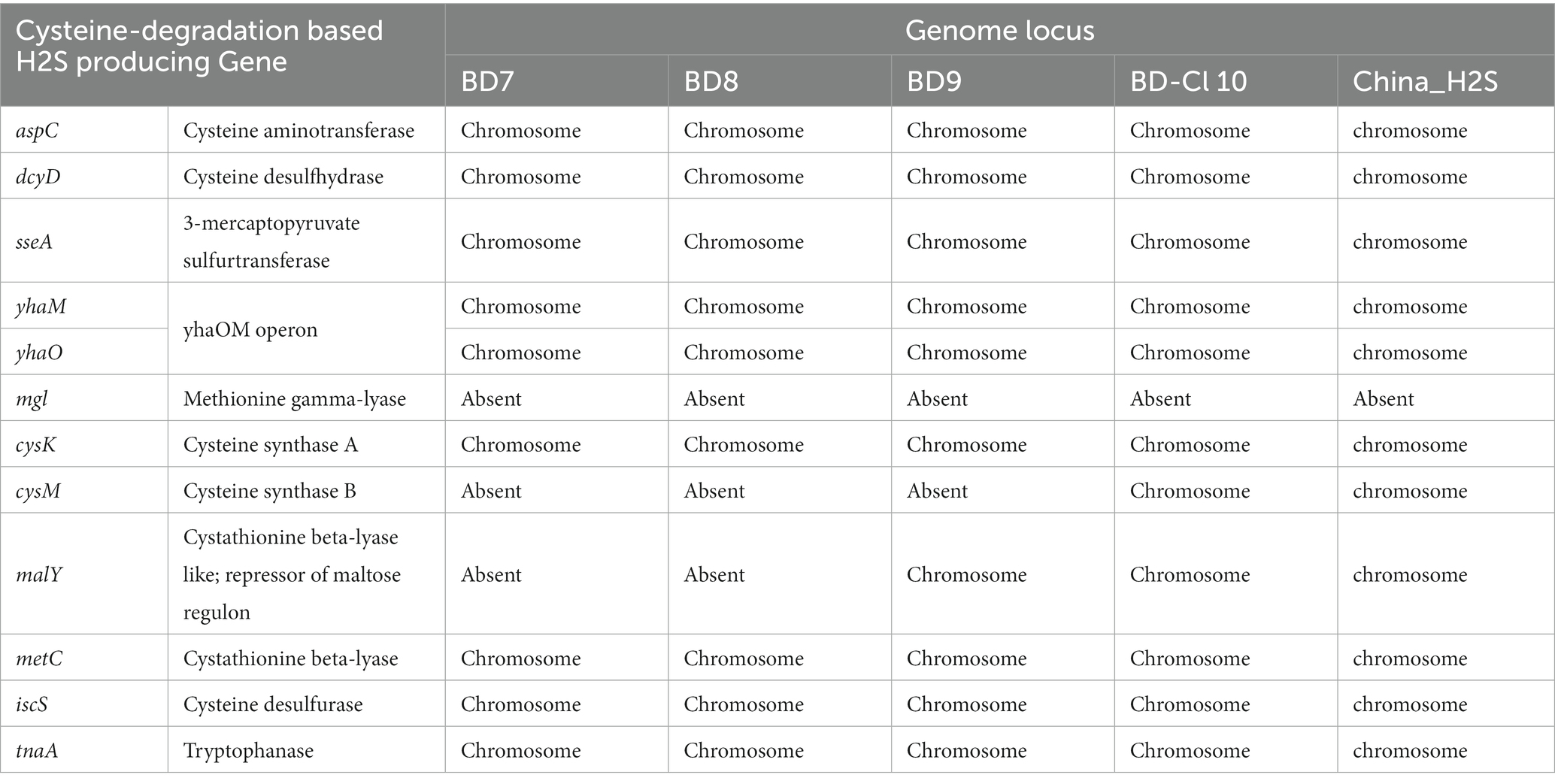
Table 4. Cysteine-degradation genes and their location in the H2S-producing Escherichia coli genomes.
Plasmid replicon types
PlasmidFinder identified 20 unique plasmid replicon groups (Table 5). All five isolates harbored multiple plasmid replicons. The plasmid replicons identified include IncFII (pHN7A8), IncFII (pSE11), IncFIA (HI1), IncFIB (K), IncFIB (pLF82-PhagePlasmid), IncFIB (pB171), IncHI2, IncHI2A, IncI (Gamma), IncN, IncQ1, IncR, IncX2, IncX1, IncY, ColE10, ColRNAI, Col (MG828), Col (pHAD28) and p0111 (Table 5). The majority of isolates (4/5; 80%) harbored the IncX, followed by (3/5; 60%) IncF (FII, FIB, FIA), IncN and Col. The H2S-producing E. coli strains that were positive for the blaCTX-M gene were significantly linked to IncF-type replicons (specifically FIA, FIB, and FII) and CoI plasmids.
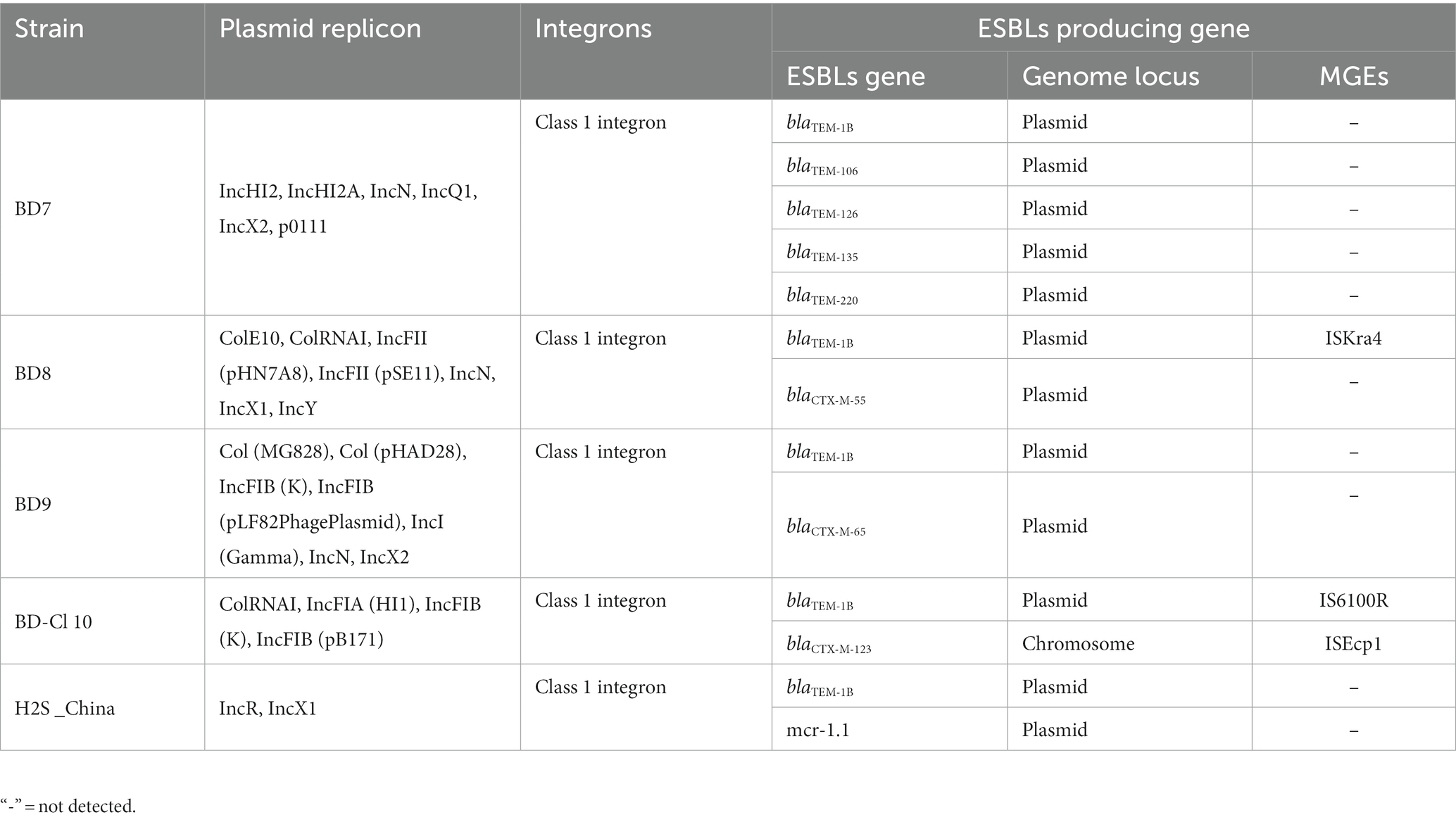
Table 5. Plasmid replicon, integrons, ESBL genes and genetic context of ESBL genes in the H2S-producing Escherichia coli isolates.
Prophage analysis
We detected intact prophages in the Bangladeshi H2S-producing E. coli strains, but not in the Chinese isolate. The poultry strains harbored two to three intact prophage sequences, while the human clinical strains carried four intact prophage sequences. Incomplete prophage sequences ranged from four to seven in poultry strains and one in human strains, while questionable prophage sequences ranged from zero to two in poultry strains and were absent in human strains identified in Bangladesh. However, five incomplete prophage sequences were detected in H2S-producing E. coli from China. The most common phages in the H2S-producing E. coli strains were Klebsiella phage 4 LV-2017 (2/5) and Shigella phage SfII (2/5), with both phages present together in a single H2S-producing human clinical E. coli strain (Table 3).
CRISPR-CAS system
The CRISPR-CAS system subtype I-A and I-E were found to be the most prevalent in the five H2S-producing E. coli genomes. All H2S-producing E. coli strains obtained from both poultry and human sources had only one CRISPR locus. The number, nucleotide sequence, and average length of repeats and spacers were similar in all H2S-producing E. coli strains, but they varied in the quantity of repeats and spacer units. The human clinical strain BDCl_10 was comparable to poultry strains, except that it had a higher number of repeats and spacers than the poultry strains (as shown in Table 3).
Virulome
The virulome analysis of H2S-producing E. coli isolates revealed the predominance of virulence factors (VFs) (Figure 3). The H2S-producing E. coli isolates showed a 93% mean probability of being human pathogens using the PathogenFinder web-server. The isolate BD8 harbored the highest number of VFs (57), followed by the isolates BD9 (48), BD7 (40), BD-Cl10 (32) and China_H2S (30). All isolates (5/5) harbored the type I fimbriae genes fim (A, C–D, E–H, l). All of the isolates showed the presence of invasin of brain endothelial cells locus B (ibeB) and invasin of brain endothelial cells locus C (ibeC) genes, which belong to the invasin virulence factor category (Figure 3). The E. coli laminin-binding fimbriae genes (ELF) elfA, elfC, elfD, elfG were also present (5/5) in all E. coli isolates. However, hemorrhagic E. coli pilus (HCP) genes associated with the production of type IV pili were highly prevalent of which hcpA gene was most predominant (100%, 5/5) followed by hcpC (80%, 4/5). Non-LEE encoded T3SS (Type III Secretion System) related genes specifically espL1, espL4, espR1, espX1, espX4, espX5 were observed in almost all H2S-producing E. coli isolates. The autotransporter genes such as aatA, cah, ehaB were also prevalent (80%, 4/5) (Figure 3). The hlyE/clyA, a pore-forming toxin was observed in 60% (3/5) of isolates. One H2S-producing E. coli strain BD8 harbored the Intimin related eae gene and was classified as Enteropathogenic E. coli (EPEC). Overall, the poultry E. coli isolates (87) harbored higher number of VFs than human isolates (44).

Figure 3. Heat map depicting the distribution of 89 virulence genes among 5 Hydrogen Sulfide (H2S)-producing Escherichia coli genomes. Dark violet blocks represent the presence, and light green blocks represent virulence gene absence.
Antimicrobial resistance phenotypes and genotypes
All four (100%) H2S-producing E. coli isolates from Bangladesh were resistant to ampicillin, nalidixic acid, ciprofloxacin, sulfamethoxazole-trimethoprim, doxycycline, and tigecycline. While they were all sensitive to amikacin, imipenem, meropenem, and colistin. However, three isolates (75%) were resistant to ceftazidime, cefotaxime, cefepime, cefuroxime, and ceftriaxone. Whereas 50% of isolates were resistant to chloramphenicol, fosfomycin, and gentamicin. All four H2S-producing E. coli isolates were classified as MDR.
We identified 43 distinct AMR gene alleles belonging to various classes (Figure 4). A minimum of seven AMR genes per genome were detected with some variation across strains (poultry E. coli 13–25; human E. coli 7–12). All (5/5) the H2S-producing E. coli genomes harbored beta-lactamase genes. All isolates were positive for the blaTEM1B gene (100%). The blaCTX-M gene alleles (blaCTX-M-55, blaCTX-M-65, and blaCTX-M-123) were detected in 3 out of 5 H2S-producing E. coli genomes (Figure 2). The blaTEM1B and blaCTX-M variants coexisted in three isolates (60%, 3/5). Among the 14 aminoglycoside resistance genes identified, aadA1 was predominant (60%, 3/5) followed by aadA2, aph (3′)-Ia, aph (3″)-Ib, and aph (6)-Id genes detected in 2 genomes (2/5). In addition, aadA5, aac (3)-IId, aac (3)-IV, and aac (6′)-Ib3 genes were found in one genome (20%, 1/5). All E. coli genomes harbored a tet (A) gene encoding tetracycline resistance. One isolate (BD7) harbored both tet (A) and tet (M) genes. The predominant sulfamethoxazole resistance gene was sul3 (4/5) followed by sul2 (1/5). Among the 4 different trimethoprim resistance genes identified, dfrA12 was predominant (40%, 2/5). Macrolide-associated resistance gene mph (A) was commonly detected (4/5). Phenicol resistance gene floR was predominantly (60%,3/5) found, followed by cmlA1 (40%, 2/5) gene. The efflux, small multidrug resistance transporter gene, qacL, was also detected in a poultry isolate (BD7). None of the isolates harbored carbapenemase genes and did not show phenotypic resistance to carbapenem antibiotics. Overall, the average number of AMR genes per genome was highest in poultry E. coli compared to human E. coli isolates (Table 2). The probable genome locus of the blaTEM1B and blaCTX-M-group genes were plasmids for two strains (Table 3). In BD-Cl10 isolate, the blaCTX-M gene was found on a chromosome with insertion element ISEcp1. The blaTEM1B gene in the BD8 strain and BD-Cl10 isolate was linked with insertion elements ISKra4 and IS6100R, respectively (Table 5). We identified amino acid substitutions in gyrA at codon positions S83L (4/5) and D87N (1/5), and in parC at S80I (2/5), S80R (1/5) and A56T (1/5). There was a significant correlation between the gyrA S83L mutation and resistance to ciprofloxacin. The ESBLs genes blaTEM1B, blaCTX-M-group, and gyrA S83L were associated with H2S-producing E. coli strains. Additionally, all the isolates harbored PMQR genes, including Qnrs1 (2/5), Qnrs4 (1/5) and Qnrs13 (1/5). The Qnrs13 gene in the BD8 strain and Qnrs4 in the BD-Cl10 strain consisted of the insertion element ISKra4 (Table 3). The β-lactamase genes, PMQRs and QRDRs were all strongly associated with the MDR phenotype.
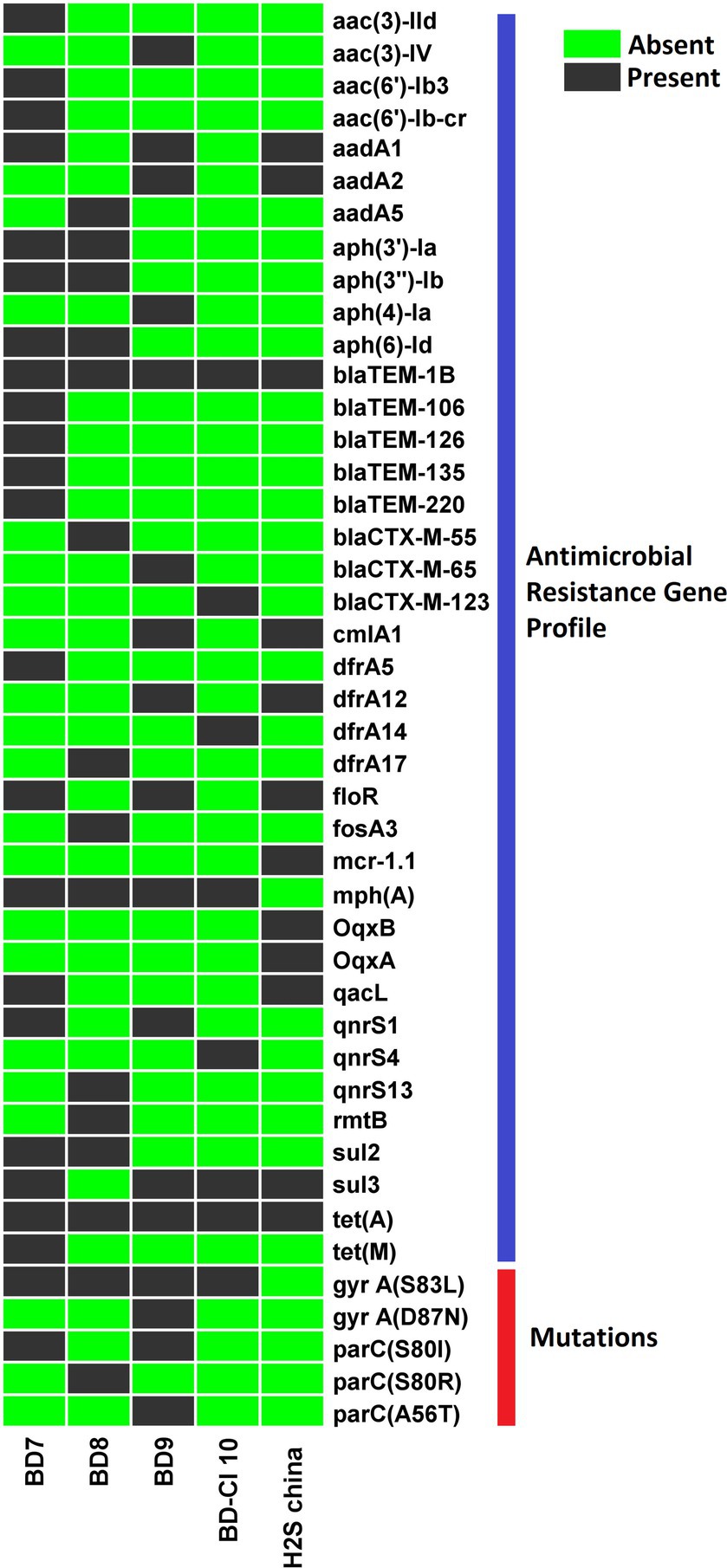
Figure 4. Heat map demonstrating the antimicrobial resistance (AMR) genes profile of 5 Hydrogen Sulfide (H2S)-producing Escherichia coli genomes. Dark black blocks represent the presence of AMR genes, and light green blocks represent the absence of particular genes.
Discussion
Production of hydrogen sulphide (H2S) is seen in many members of Enterobacteriaceae. However, it is well established that E. coli strains are H2S non-producers. H2S non-production is one of the key characteristics used to identify E. coli in laboratory tests. Nonetheless, a fraction of H2S-producing E. coli variants has previously been identified in animal and human infections (Maker and Washington, 1974; Sogaard, 1975; Magalhães and Vance, 1978). By studying H2S-producing E. coli, researchers can better understand the biology and behavior of such variants and develop improved diagnostic tests. Further, a comprehensive characterization of H2S-producing E. coli including analysis of genomic features was needed. To address this, we conducted a thorough investigation of four H2S-producing E. coli variants by utilizing whole-genome sequencing (WGS) in combination with comprehensive microbiological and biochemical testing.
The four bacterial isolates were recovered from poultry and human clinical samples in Dhaka, Bangladesh, as part of a larger surveillance study. These isolates biochemically mimic typical E. coli for all reactions except for one reaction, the H2S production. The prevalence of H2S-producing variants in our study can be estimated at 3% (3/96) in poultry and 0.5% (1/204) in clinical E. coli isolates. However, this may not reflect the true prevalence figures, as in this study, the primary specimens were not screened targeting H2S-producing E. coli. But only the archived E. coli isolates were tested. However, our estimates of prevalence are similar to those previously reported (Maker and Washington, 1974; Sogaard, 1975; Weber et al., 1981). Our and other reports reveal that H2S-positive strains of E. coli are not uncommon among poultry and human clinical samples (Braunstein and Mladineo, 1974; Maker and Washington, 1974; Sogaard, 1975; Traub and Kleber, 1975; Magalhães and Vance, 1978; Weber et al., 1981; Barbour et al., 1985). Many such variants are probably misidentified in laboratories, such as Citrobacter, Arizona and Salmonella (Darland and Davis, 1974). This misidentification stems from the production of black precipitate on KIA or TSI medium. It is also possible that acid production sometimes masks H2S production due to lactose fermentation (Magalhães and Vance, 1978). Muller-Hinton agar supplemented with sodium thiosulfate and ferric ammonium sulfate media is considered superior to KIA agar media for identifying H2S production. The utility of the same has been demonstrated in this study. However, the CHROMagar Orientation media could not differentiate between typical E. coli and H2S-producing E. coli variants. Primary screening with this media can effectively screen typical E. coli and H2S-producing E. coli variants in a single step.
The studied H2S-producing E. coli strains mainly belonged to the commensal phylogenetic groups A (80%,4/5) and B1 (20%, 1/5). Several reports confirm that phylogroups A and B1 were the most prevalent among E. coli isolates, particularly in the gut microbiome (Li et al., 2010; Stoppe et al., 2017). The H2S-producing E. coli strains of human origin, isolated from Bangladesh and China, belonged to the worldwide predominant clonal complex CC10. CC10 group of strains belong to emerging clone of extra-intestinal pathogenic E. coli (ExPEC) (Manges et al., 2019). They are isolated from a wide range of niches including clinical settings, food animals and environment (Manges et al., 2019). They are also known to be associated with wide range of AMR and virulence genotypes (Massella et al., 2021). This group of E. coli needs close monitoring to safeguard public health (Hussain et al., 2023). We identified 8–13 prophage regions in H2S-producing E. coli, of which 2–4 were found intact. Klebsiella phage 4 LV-2017 and Shigella phage SfII were the predominant bacteriophages detected. The existence of a higher number of phage elements (8 to 13) in poultry strains compared to the human clinical strain (5) may indicate more horizontal gene transfer (HGT) events that brought in more toxin genes in poultry strains than in the human clinical strain. The CRISPR-Cas system confers immunity against viruses and plasmids (Horvath and Barrangou, 2010). Investigation of the CRISPR-Cas system in H2S-producing E. coli strains indicated that it was conserved in both poultry and human clinical H2S-producing E. coli isolates.
Previous work has identified cysteine-degradation genes in H2S-producing bacteria and classified them into primary, secondary and erroneous categories based on their functions (Braccia et al., 2021). Most primary producer genes (aspC, dcyD, sseA, yhaOM operon) were present in all H2S-producing E. coli strains. In the case of secondary producer genes, we observed inconsistent results. But all erroneous genes were present in the study isolates. We found all genes related to H2S production on chromosomes, which is in line with the previous report (Braccia et al., 2021).
The patterns of antibiotic resistance were similar for human and poultry isolates. High resistance rates were observed for ampicillin, ciprofloxacin, nalidixic acid, trimethoprim and sulfamethoxazole, doxycycline and cephalosporin. Our findings show partial agreement with the previous report on H2S-producing E. coli (Braunstein and Mladineo, 1974; Maker and Washington, 1974; Sogaard, 1975; Traub and Kleber, 1975; Magalhães and Vance, 1978; Weber et al., 1981; Barbour et al., 1985; Park et al., 2015). The H2S-producing E. coli isolates contained multiple plasmids. The major replicon types were IncX (4/5; 80%) and IncF (3/5; 60%). As per earlier reports, these plasmid replicons were associated with fluoroquinolone resistance and blaCTX-M-group in humans and livestock E. coli (Phan et al., 2015; Sun et al., 2017). As healthy animals and humans were found to harbor H2S-producing E. coli (Sogaard, 1975; Biswas et al., 2020), the presence of these plasmids may contribute as careers of antibiotic resistance in microbiomes. The results of our study suggests that aminoglycosides and carbapenem antibiotics are effective candidates against these strains. However, this cannot be generalized due to several limitations of our study and it is always better to initiate evidence-based treatment of diseases arising from infectious agents.
All isolates were predicted as human pathogens as per their pathogenicity score determined by in silico analysis. The studied H2S-producing E. coli isolates harbored at least 30 virulence factors. Among them, poultry isolates had more virulence genes (40–57 VFs) than human samples. The H2S-producing E. coli isolates harbored a wide range of virulence factors encoding E. coli laminin-binding fimbriae (ELF) (elfA,C,D,G), Hemorrhagic E. coli pilus (HCP) (hcpA-B), Type I fimbriae (fimD, fimF, fimG, fimH) and Non-LEE encoded TTSS effectors (espL1, espL4, espR1, espX4). The intimin (eae) gene, a marker for enteropathogenic E. coli, was observed in one H2S-producing E. coli isolate (BD8) belonging to ST189. This indicates that E. coli pathotypes also exhibit H2S production features or vice versa. Therefore, virulence genes play an important role in the pathogenicity of H2S-producing E. coli strains. Also, the convergence of wide range of AMR and virulence genotypes is a cause of great concern (Massella et al., 2021). These observations warrant studying the role of H2S-producing E. coli isolates in different infections for developing effective treatments and preventive measures.
In conclusion, this study investigated H2S-producing E. coli variants recovered from poultry and human clinical samples in Dhaka, Bangladesh. The isolates were confirmed as E. coli by routine biochemical tests and WGS-based species identification. The H2S-producing isolates exhibited relatively diverse molecular characteristics with no correlation between the source of origin or the phylogenetic clustering of the isolates. The study also found high rates of AMR and extensive virulence gene repertoire in these isolates. The findings of this study highlight that the genomic features, antibiotic resistance and virulence potential of H2S-producing E. coli resemble the typical E. coli forms. Therefore, we suggest the need for continued surveillance and genomic characterization of atypical E. coli forms like H2S-producing E. coli to better understand the characteristics of such variants and improve diagnostics and treatment outcomes.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/genbank/, PRJNA882002, PRJNA714244.
Author contributions
RM designed the study and conducted all microbiological tests, and whole genome sequencing. RM and AH carried out the bioinformatics analyses interpretation of results, prepared tables and figures, and drafted the manuscript. MR and RM performed the sample collections and initial sample processing. AH contributed to the discussions, manuscript writing, editing, and proofreading. AA, JP, SC, TC, and DM contributed to the discussions and reviewed the manuscript. DM managed the funds and supervised the study. All authors contributed to the article and approved the submitted version.
Funding
The work was funded through a Royal Society International Collaboration Award (ref. ICA\R1\191309).
Acknowledgments
Icddr,b is grateful to the governments of Bangladesh, Canada, Sweden, and the United Kingdom for providing core/unrestricted support for its operations and research.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Footnotes
References
Andrews, S. (2010). FastQC: a quality control tool for high throughput sequence data. Babraham Bioinformatics, Babraham Institute, Cambridge, United Kingdom.
Arndt, D., Grant, J. R., Marcu, A., Sajed, T., Pon, A., Liang, Y., et al. (2016). PHASTER: a better, faster version of the PHAST phage search tool. Nucleic Acids Res. 44, W16–W21. doi: 10.1093/nar/gkw387
Baddam, R., Sarker, N., Ahmed, D., Mazumder, R., Abdullah, A., Morshed, R., et al. (2020). Genome dynamics of Vibrio cholerae isolates linked to seasonal outbreaks of cholera in Dhaka, Bangladesh. MBio 11, e03339–e03319. doi: 10.1128/mBio.03339-19
Bankevich, A., Nurk, S., Antipov, D., Gurevich, A. A., Dvorkin, M., Kulikov, A. S., et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477. doi: 10.1089/cmb.2012.0021
Barbour, E. K., Nabbut, N. H., and Al-Nakhli, H. M. (1985). Production of H2 S by Escherichia coli isolated from poultry: an unusual character useful for epidemiology of Colisepticemia. Avian Dis. 29, 341–346. doi: 10.2307/1590494
Beghain, J., Bridier-Nahmias, A., Le Nagard, H., Denamur, E., and Clermont, O. (2018). ClermonTyping: an easy-to-use and accurate in silico method for Escherichia genus strain phylotyping. Microb. Genom. 4:e000192. doi: 10.1099/mgen.0.000192
Bélanger, L., Garenaux, A., Harel, J., Boulianne, M., Nadeau, E., and Dozois, C. M. (2011). Escherichia coli from animal reservoirs as a potential source of human extraintestinal pathogenic E. coli. FEMS Immunol Med Microbiol. 62, 1–10. doi: 10.1111/j.1574-695X.2011.00797.x
Biswas, S., Elbediwi, M., Gu, G., and Yue, M. (2020). Genomic characterization of new variant of hydrogen sulfide (H2S)-producing Escherichia coli with multidrug resistance properties carrying the mcr-1 gene in China. Antibiotics 9:80. doi: 10.3390/antibiotics9020080
Blachier, F., Beaumont, M., and Kim, E. (2019). Cysteine-derived hydrogen sulfide and gut health: a matter of endogenous or bacterial origin. Curr. Opin. Clin. Nutr. Metab. Care 22, 68–75. doi: 10.1097/MCO.0000000000000526
Braccia, D. J., Jiang, X., Pop, M., and Hall, A. B. (2021). The capacity to produce hydrogen sulfide (H2S) via cysteine degradation is ubiquitous in the human gut microbiome. Front. Microbiol. 12:705583. doi: 10.3389/fmicb.2021.705583
Braunstein, H., and Mladineo, M. A. (1974). Escherichia coli strains producing hydrogen sulfide in Iron-agar media. Am. J. Clin. Pathol. 62, 420–424. doi: 10.1093/ajcp/62.3.420
Braz, V. S., Melchior, K., and Moreira, C. G. (2020). Escherichia coli as a multifaceted pathogenic and versatile bacterium. Front. Cell. Infect. Microbiol. 10:548492. doi: 10.3389/fcimb.2020.548492
Carattoli, A., Zankari, E., García-Fernández, A., Voldby Larsen, M., Lund, O., Villa, L., et al. (2014). In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob. Agents Chemother. 58, 3895–3903. doi: 10.1128/AAC.02412-14
Chen, L., Yang, J., Yu, J., Yao, Z., Sun, L., Shen, Y., et al. (2005). VFDB: a reference database for bacterial virulence factors. Nucleic Acids Res. 33, D325–D328. doi: 10.1093/nar/gki008
Cosentino, S., Voldby Larsen, M., Møller Aarestrup, F., and Lund, O. (2013). PathogenFinder-distinguishing friend from foe using bacterial whole genome sequence data. PLoS One 8:e77302. doi: 10.1371/journal.pone.0077302
Croucher, N. J., Page, A. J., Connor, T. R., Delaney, A. J., Keane, J. A., Bentley, S. D., et al. (2015). Rapid phylogenetic analysis of large samples of recombinant bacterial whole genome sequences using Gubbins. Nucleic Acids Res. 43:e15. doi: 10.1093/nar/gku1196
Darland, G., and Davis, B. R. (1974). Biochemical and serological characterization of hydrogen sulfide-positive variants of Escherichia coli. Appl. Microbiol. 27, 54–58. doi: 10.1128/am.27.1.54-58.1974
Eswarappaápradeep, B. (2017). On demand redox buffering by H 2 S contributes to antibiotic resistance revealed by a bacteria-specific H 2 S donor. Chem. Sci. 8, 4967–4972. doi: 10.1039/C7SC00873B
Faircloth, B. (2013). Illumiprocessor: a trimmomatic wrapper for parallel adapter and quality trimming.
Gurevich, A., Saveliev, V., Vyahhi, N., and Tesler, G. (2013). QUAST: quality assessment tool for genome assemblies. Bioinformatics 29, 1072–1075. doi: 10.1093/bioinformatics/btt086
Hasman, H., Saputra, D., Sicheritz-Ponten, T., Lund, O., Svendsen, C. A., Frimodt-Møller, N., et al. (2014). Rapid whole-genome sequencing for detection and characterization of microorganisms directly from clinical samples. J. Clin. Microbiol. 52, 139–146. doi: 10.1128/JCM.02452-13
Horvath, P., and Barrangou, R. (2010). CRISPR/CAS, the immune system of bacteria and archaea. Science 327, 167–170. doi: 10.1126/science.1179555
Hussain, A., Mazumder, R., Asadulghani, M., Clark, T. G., and Mondal, D. (2023). "combination of virulence and antibiotic resistance: a successful bacterial strategy to survive under hostile environments," in bacterial survival in the hostile environment. Elsevier, 101–117. doi: 10.1016/B978-0-323-91806-0.00004-7
Hussain, A., Shaik, S., Ranjan, A., Nandanwar, N., Tiwari, S. K., Majid, M., et al. (2017). Risk of transmission of antimicrobial resistant Escherichia coli from commercial broiler and free-range retail chicken in India. Front. Microbiol. 8:2120. doi: 10.3389/fmicb.2017.02120
Joensen, K. G., Tetzschner, A. M., Iguchi, A., Aarestrup, F. M., and Scheutz, F. (2015). Rapid and easy in silico serotyping of Escherichia coli isolates by use of whole-genome sequencing data. J. Clin. Microbiol. 53, 2410–2426. doi: 10.1128/JCM.00008-15
Jones, R. T., and Silver, R. P. (1978). Genetic and molecular characterization of an Escherichia coli plasmid coding for hydrogen sulfide production and drug resistance. Antimicrob. Agents Chemother. 14, 765–770. doi: 10.1128/AAC.14.5.765
Kaper, J., Nataro, J., and Mobley, H. (2004). Nature reviews. Microbiology. Nat. Rev. Microbiol. 2, 123–140. doi: 10.1038/nrmicro818
Köhler, C.-D., and Dobrindt, U. (2011). What defines extraintestinal pathogenic Escherichia coli? Int. J. Med. Microbiol. 301, 642–647. doi: 10.1016/j.ijmm.2011.09.006
Larsen, M. V., Cosentino, S., Lukjancenko, O., Saputra, D., Rasmussen, S., Hasman, H., et al. (2014). Benchmarking of methods for genomic taxonomy. J. Clin. Microbiol. 52, 1529–1539. doi: 10.1128/JCM.02981-13
Letunic, I., and Bork, P. (2016). Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 44, W242–W245. doi: 10.1093/nar/gkw290
Li, B., Sun, J.-Y., Han, L.-Z., Huang, X.-H., Fu, Q., and Ni, Y.-X. (2010). Phylogenetic groups and pathogenicity island markers in fecal Escherichia coli isolates from asymptomatic humans in China. Appl. Environ. Microbiol. 76, 6698–6700. doi: 10.1128/AEM.00707-10
Magalhães, M., and Vance, M. (1978). Hydrogen sulphide-positive strains of Escherichia coli from swine. J. Med. Microbiol. 11, 211–214. doi: 10.1099/00222615-11-2-211
Magiorakos, A.-P., Srinivasan, A., Carey, R. B., Carmeli, Y., Falagas, M., Giske, C., et al. (2012). Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 18, 268–281. doi: 10.1111/j.1469-0691.2011.03570.x
Maker, M. D., and Washington, J. A. (1974). Hydrogen sulfide-producing variants of Escherichia coli. Appl. Microbiol. 28, 303–305. doi: 10.1128/am.28.2.303-305.1974
Manges, A. R., Geum, H. M., Guo, A., Edens, T. J., Fibke, C. D., and Pitout, J. D. (2019). Global extraintestinal pathogenic Escherichia coli (ExPEC) lineages. Clin. Microbiol. Rev. 32, e00135–e00118. doi: 10.1128/CMR.00135-18
Massella, E., Giacometti, F., Bonilauri, P., Reid, C. J., Djordjevic, S. P., Merialdi, G., et al. (2021). Antimicrobial resistance profile and ExPEC virulence potential in commensal Escherichia coli of multiple sources. Antibiotics 10:351. doi: 10.3390/antibiotics10040351
Mazumder, R., Abdullah, A., Ahmed, D., and Hussain, A. (2020a). High prevalence of Bla CTX-M-15 gene among extended-spectrum β-lactamase-producing Escherichia coli isolates causing extraintestinal infections in Bangladesh. Antibiotics 9:796. doi: 10.3390/antibiotics9110796
Mazumder, R., Abdullah, A., Hussain, A., Ahmed, D., and Mondal, D. (2020b). Draft genome sequence of Chromobacterium violaceum RDN09, isolated from a patient with a wound infection in Bangladesh. Microbiol. Resour. Announc. 9, e00957–e00920. doi: 10.1128/MRA.00957-20
Mazumder, R., Hussain, A., Abdullah, A., Islam, M. N., Sadique, M. T., Muniruzzaman, S., et al. (2021). International high-risk clones among extended-Spectrum β-lactamase–producing Escherichia coli in Dhaka, Bangladesh. Front. Microbiol. 12:736464. doi: 10.3389/fmicb.2021.736464
Mazumder, R., Hussain, A., Phelan, J. E., Campino, S., Haider, S., Mahmud, A., et al. (2022). Non-lactose fermenting Escherichia coli: following in the footsteps of lactose fermenting E. coli high-risk clones. Front. Microbiol. 13:1027494. doi: 10.3389/fmicb.2022.1027494
Mazumder, R., Sadique, T., Sen, D., Mozumder, P., Rahman, T., Chowdhury, A., et al. (2020c). Agricultural injury–associated Chromobacterium violaceum infection in a Bangladeshi farmer. Am. J. Trop. Med. Hyg. 103, 1039–1042. doi: 10.4269/ajtmh.20-0312
Mironov, A., Seregina, T., Nagornykh, M., Luhachack, L. G., Korolkova, N., Lopes, L. E., et al. (2017). Mechanism of H2S-mediated protection against oxidative stress in Escherichia coli. Proc. Natl. Acad. Sci. 114, 6022–6027. doi: 10.1073/pnas.1703576114
Monir, M. M., Islam, M. T., Mazumder, R., Mondal, D., Nahar, K. S., Sultana, M., et al. (2023). Genomic attributes of Vibrio cholerae O1 responsible for 2022 massive cholera outbreak in Bangladesh. Nat. Commun. 14:1154. doi: 10.1038/s41467-023-36687-7
Nataro, J. P., and Kaper, J. B. (1998). Escherichia coli diarrheagenic. Clin. Microbiol. Rev. 11, 142–201. doi: 10.1128/CMR.11.1.142
Néron, B., Littner, E., Haudiquet, M., Perrin, A., Cury, J., and Rocha, E. P. (2022). IntegronFinder 2.0: identification and analysis of integrons across bacteria, with a focus on antibiotic resistance in Klebsiella. Microorganisms 10:700. doi: 10.3390/microorganisms10040700
Park, H.-E., Shin, M.-K., Park, H.-T., Shin, S. W., Jung, M., Im, Y. B., et al. (2015). Virulence factors, antimicrobial resistance patterns, and genetic characteristics of hydrogen sulfide-producing Escherichia coli isolated from swine. Korean J. Vet. Res. 55, 191–197. doi: 10.14405/kjvr.2015.55.3.191
Percival, S.L., Chalmers, R., Hunter, P.R., Sellwood, J., and Wyn-Jones, P. (2004). Microbiology of waterborne diseases. Elsevier academic press San Diego, CA, USA:.
Phan, M. D., Forde, B. M., Peters, K. M., Sarkar, S., Hancock, S., Stanton-Cook, M., et al. (2015). Molecular characterization of a multidrug resistance IncF plasmid from the globally disseminated Escherichia coli ST131 clone. PLoS One 10:e0122369. doi: 10.1371/journal.pone.0122369
Rahman, M., Glasgow, J. N., Nadeem, S., Reddy, V. P., Sevalkar, R. R., Lancaster, J. R. Jr., et al. (2020). The role of host-generated H2S in microbial pathogenesis: new perspectives on tuberculosis. Frontiers in cellular and infection. Microbiology 10:586923. doi: 10.3389/fcimb.2020.586923
Roer, L., Johannesen, T. B., Hansen, F., Stegger, M., Tchesnokova, V., Sokurenko, E., et al. (2018). CHTyper, a web tool for subtyping of extraintestinal pathogenic Escherichia coli based on the fumC and fimH alleles. J. Clin. Microbiol. 56, e00063–e00018. doi: 10.1128/JCM.00063-18
Schmieder, R., and Edwards, R. (2011). Fast identification and removal of sequence contamination from genomic and metagenomic datasets. PLoS One 6:e17288. doi: 10.1371/journal.pone.0017288
Seemann, T. (2014). Prokka: rapid prokaryotic genome annotation. Bioinformatics 30, 2068–2069. doi: 10.1093/bioinformatics/btu153
Seemann, T. (2018). ABRicate: mass screening of contigs for antimicrobial and virulence genes. Department of Microbiology and Immunology, The University of Melbourne, Melbourne, Australia.
Sogaard, H. (1975). Hydrogen sulphide producing variants of Escherichia coli, widespread occurrence in animals and humans within a confined environment. Acta Vet. Scand. 16, 31–38. doi: 10.1186/BF03546693
Stamatakis, A. (2014). RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313. doi: 10.1093/bioinformatics/btu033
Stoppe, N. D. C., Silva, J. S., Carlos, C., Sato, M. I., Saraiva, A. M., Ottoboni, L. M., et al. (2017). Worldwide phylogenetic group patterns of Escherichia coli from commensal human and wastewater treatment plant isolates. Front. Microbiol. 8:2512. doi: 10.3389/fmicb.2017.02512
Sun, J., Fang, L.-X., Wu, Z., Deng, H., Yang, R.-S., Li, X.-P., et al. (2017). Genetic analysis of the IncX4 plasmids: implications for a unique pattern in the mcr-1 acquisition. Sci. Rep. 7:424. doi: 10.1038/s41598-017-00095-x
Toliver-Kinsky, T., Cui, W., Törö, G., Lee, S.-J., Shatalin, K., Nudler, E., et al. (2019). H2S, a bacterial defense mechanism against the host immune response. Infect. Immun. 87, e00272–e00218. doi: 10.1128/IAI.00272-18
Traub, W. H., and Kleber, I. (1975). Characterization of two H2S producing, multiple drug-resistant isolates of Escherichia colifrom clinical urine specimens. Pathobiology 43, 10–16. doi: 10.1159/000162789
Weber, A., Lembke, C., and Bertholdt, A. (1981). Isolation of H2S-producing escherichia coli from urine samples (author's transl). Zentralbl. Bakteriol. Mikrobiol. Hyg. A. Med. Mikrobiol. Infekt. Parasitol. 251, 185–189.
Weinstein, M. P. (2019). Performance standards for antimicrobial susceptibility testing, Wayne, PA: Clinical and Laboratory Standards Institute.
Zabransky, R. J., Hall, J. W., Day, F. E., and Needham, G. M. (1969). Klebsiella, Enterobacter, and Serratia: biochemical differentiation and susceptibility to ampicillin and three cephalosporin derivatives. Appl. Microbiol. 18, 198–203. doi: 10.1128/am.18.2.198-203.1969
Zankari, E., Allesøe, R., Joensen, K. G., Cavaco, L. M., Lund, O., and Aarestrup, F. M. (2017). PointFinder: a novel web tool for WGS-based detection of antimicrobial resistance associated with chromosomal point mutations in bacterial pathogens. J. Antimicrob. Chemother. 72, 2764–2768. doi: 10.1093/jac/dkx217
Zankari, E., Hasman, H., Cosentino, S., Vestergaard, M., Rasmussen, S., Lund, O., et al. (2012). Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 67, 2640–2644. doi: 10.1093/jac/dks261
Keywords: atypical H2S-producing E. coli variants, genomics, epidemiology, functional characterization, differential diagnosis, E. coli identification algorithm
Citation: Mazumder R, Hussain A, Rahman MM, Phelan JE, Campino S, Abdullah A, Clark TG and Mondal D (2023) Genomic and functional portrait of multidrug-resistant, hydrogen sulfide (H2S)-producing variants of Escherichia coli. Front. Microbiol. 14:1206757. doi: 10.3389/fmicb.2023.1206757
Edited by:
Qixia Luo, Zhejiang University, ChinaReviewed by:
Silpak Biswas, Calcutta School of Tropical Medicine, IndiaEswarappa Pradeep Bulagonda, Sri Sathya Sai Institute of Higher Learning, India
Copyright © 2023 Mazumder, Hussain, Rahman, Phelan, Campino, Abdullah, Clark and Mondal. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dinesh Mondal, din63d@icddrb.org
 Razib Mazumder
Razib Mazumder Arif Hussain
Arif Hussain Mohammad Mustafizur Rahman2
Mohammad Mustafizur Rahman2 Jody E. Phelan
Jody E. Phelan Susana Campino
Susana Campino Taane G. Clark
Taane G. Clark Dinesh Mondal
Dinesh Mondal