- 1Department of Biological Sciences, University of Cincinnati, Cincinnati, OH, United States
- 2Division of Hydrologic Sciences, Desert Research Institute, Las Vegas, Las Vegas, NV, United States
- 3Department of Molecular and Cell Biology, University of California, Berkeley, Berkeley, CA, United States
Terrestrial serpentinizing systems allow us insight into the realm of alkaliphilic microbial communities driven by geology in a way that is frequently more accessible than their deep subsurface or marine counterparts. However, these systems are also marked by geochemical and microbial community variation due to the interactions of serpentinized fluids with host geology and the surface environment. To separate the transient from the endemic microbes in a hyperalkaline environment, we assessed the Ney Springs terrestrial serpentinizing system microbial community and geochemistry at six time points over the span of a year. Using 16S rRNA gene surveys we observed 93 amplicon sequence variants (ASVs) that were found at every sampling event. This is compared to ~17,000 transient ASVs that were detected only once across the six sampling events. Of the resident community members, 16 of these ASVs were regularly greater than 1% of the community during every sampling period. Additionally, many of these core taxa experienced statistically significant changes in relative abundance with time. Variation in the abundance of some core populations correlated with geochemical variation. For example, members of the Tindallia group, showed a positive correlation with variation in levels of ammonia at the spring. Investigating the metagenome assembled genomes of these microbes revealed evidence of the potential for ammonia generation via Stickland reactions within Tindallia. This observation offers new insight into the origin of high ammonia concentrations (>70 mg/L) seen at this site. Similarly, the abundance of putative sulfur-oxidizing microbes like Thiomicrospira, Halomonas, and a Rhodobacteraceae species could be linked to changes observed in sulfur-oxidation intermediates like tetrathionate and thiosulfate. While these data supports the influence of core microbial community members on a hyperalkaline spring’s geochemistry, there is also evidence that subsurface processes affect geochemistry and may impact community dynamics as well. Though the physiology and ecology of these astrobiologically relevant ecosystems are still being uncovered, this work helps identify a stable microbial community that impacts spring geochemistry in ways not previously observed in serpentinizing ecosystems.
Introduction
Serpentinization is a globally relevant subsurface process caused by the hydration of iron and magnesium rich minerals within the Earth’s crust which subsequently releases hydrogen gas (Mccollom and Seewald, 2013). The hydrogen produced, in addition to other reduced compounds generated, can serve as the energetic basis for microbial food webs. However, the high pH fluids overall have a profound effect on habitability. The degree of serpentinized fluid input can greatly alter microbial community composition, with pH in particular cited as a significant driver in systems that experience a range of pH values (Rempfert et al., 2017; Twing et al., 2017; Fones et al., 2021). The interaction of high pH serpentinized fluids with local geology and other water sources can also result in variation at the microbial community level, even within the same system (Morrill et al., 2013; Rempfert et al., 2017; Ortiz et al., 2018). Lastly, time scale can also have an effect on community composition, as sites undergoing active serpentinization are more impacted by high pH fluids than inactive ones (Schrenk et al., 2004; Szponar et al., 2013). Despite these known broad effects on microbial community composition, we still have limited insight into the specific geochemical drivers that explain the differences seen in the microbial communities across these systems.
Remarkable variation is seen across continental serpentinizing systems, even when comparing ones that are located within the same geologic formation (Woycheese et al., 2015; Trutschel et al., 2022). For example, Ney Springs and The Cedars are both a part of the Franciscan Subduction complex, but feature very different levels of salinity and are dominated by different microorganisms (Suzuki et al., 2017; Cook et al., 2021; Trutschel et al., 2022). Ney Springs is a terrestrial system notable for its extremely high pH (12.3–12.7) and abundance of ammonia, methane, and sulfide compared to other serpentinizing systems (Cook et al., 2021; Trutschel et al., 2022). Despite its continental location it also has marine-like levels of sodium, potassium, and boron which are likely the result of serpentinized fluids mixing with connate seawater and/or the Franciscan subduction complex marine deposit (Feth et al., 1961; Barnes et al., 1972). Ney Springs also contains incredibly high amounts of silica (>4,000 mg/L) which is likely due to the hyperalkaline fluids dissolving nearby silica-rich volcanic rocks (Feth et al., 1961; García-Ruiz et al., 2017). Ney Springs is dominated by members belonging to Tindallia and Izimaplasma, which are not typically abundant or even observed within other characterized serpentinizing systems (Trutschel et al., 2022). In comparison, The Cedars is known for its low conductivity fluids and a shallow groundwater microbial community that is dominated by the alkaliphilic and hydrogenotrophic Serpentinomonas (Morrill et al., 2013; Suzuki et al., 2013, 2017). Conductivity values at The Cedars are much lower compared to Ney Springs (0.8–3.0 mS/cm vs. 32–39 mS/cm, respectively), and The Cedars is limited for terminal electron acceptors such as sulfate and nitrate (Suzuki et al., 2013, 2017). The geochemical differences observed in these environments are likely explained by local variation in geology and hydrology, which in turn shape the microbial community composition and the challenges these microorganisms face.
Though surface exposed terrestrial systems are generally more easily accessed compared to their marine or purely deep subsurface counterparts, they are also subject to greater exogenous inputs and/or may be more impacted by seasonality (e.g., through precipitation, temperature, or photoperiod). Thus, they likely contain a mixture of microorganisms sustained solely by deep subsurface fluid chemistry, and microorganisms that utilize nutrient inputs and/or oxygen resulting from surface exposure. Long term geochemical and microbial community monitoring has been used to study temporal changes and the surface influence on the microbial community composition of mines and soda lake environments (Boros et al., 2017; Osburn et al., 2019). This approach allows one to determine what the endemic microbial community members of these interface environments are, how they utilize both subsurface and surface resources, and how they are impacted by temporal or seasonal changes in the environment.
In this work we assess the microbial community and aqueous geochemistry at Ney Springs over several points in a year (May 2021 – June 2022). This work identifies geochemical parameters at Ney Springs that change seasonally and those that vary temporally and are not associated with seasonality. We also present data identifying a core microbial community with an average seasonal relative abundance greater than 1%. Using metagenomics, we then investigated the potential metabolic features of this core community and how microbial metabolism may link to geochemical variation observed in this environment.
Materials and methods
Sample collection and analysis of aqueous geochemistry
Samples and field work were conducted at Ney Springs roughly every two months starting in May of 2021 through June of 2022 for a total of six trips. All Ney Springs fluids were collected from a 1 m × 1 m concrete cistern which captures the spring discharge (Supplementary Figure 1). A Mettler-Toledo multimeter (Columbus, OH, United States) was used to measure temperature, pH, conductivity, total dissolved solids (TDS), resistivity, and oxidation–reduction potential (ORP). Geochemical analyses conducted on site for dissolved oxygen (DO), S2−, Fe2+, tetrathionate (S4O62−), and thiosulfate (S2O32−) were done with a HACH (Loveland, CO, United States) portable spectrometer as described previously (Trutschel et al., 2022).
Fluid samples for ion chromatograph (IC) analysis of anions (F−, Cl−, NO2−, Br−, NO3−, PO43−, and SO42−) and cations (Li+, Na+, NH4+, K+, Mg2+, and Ca2+) were collected using autoclaved MasterFlex® PharMed® BPT tubing (Cole-Palmer, Vernon Hills, IL, United States) with a Geopump™ peristaltic pump (GeoTech, Denver, CO, United States) to pump up water from the bottom of the cistern (Supplementary Figure 1). Fluids were passed through a polypropylene in-line filter housing (Millipore; Bedford, MA, United States) containing 0.1 μm polycarbonate membrane filters (47 mm diameter, Millipore, Tullagreen, Carrigtwohill Co. Cork, IRL) and kept on ice or refrigerated (4°C) until analysis on a Dionex Aquion Ion Chromatograph (Thermo Fisher Scientific, Waltham, MA, United States). All samples were run at a 1:10 dilution with MilliQ water, or at a 1:5 dilution after the sample had been mixed with Amberlite® MB20 H/OH resin beads (Sigma-Aldrich, United States, with a ratio of 80 mg of beads per 2 mL of sample) for chloride removal. This allowed for better detection of less abundant constituents such as NO3− and NO2−. Additional samples were collected for external analysis through ACZ Laboratories (acz.com) (Steamboat Springs, CO, United States) for metals (silicon, iron, sodium, etc.) and ion species (nitrate, nitrite, phosphate, sulfate, and sulfide) using sample bottles and protocols provided by the company. Briefly, filtered, and unfiltered fluids were added to 250 mL HDPE bottles that were empty or contained 2 mL of 50% HNO3. Bottles were kept on ice (~4°C) then shipped within 24 h and analyzed at ACZ. Inductively Coupled Plasma Spectroscopy (ICP) according to EPA Method 200.7 was used for metal analysis. EPA methods M353.2, M350.1 and M365.1 were used for nitrate/nitrite, ammonia, and phosphorous/phosphate, respectively. Methods D516-02/-07/-11 and SM4500s2-D were used for sulfate and sulfide (total sulfides or S). Samples for stable isotope analysis of hydrogen and oxygen in water were collected in glass exetainer vials filled without headspace or bubbles and capped to prevent evaporation and exchange of samples with atmospheric water vapor. Samples were analyzed at the Center for Stable Isotope Biogeochemistry at the University of California, Berkeley using Isotope Ratio Mass Spectrometry (IRMS). Stable isotope results were reported in parts per thousand (‰), using standard delta notation (δ2H and δ18O) and are relative to VSMOW (Vienna Standard Mean Ocean Water).
Sample collection, DNA extraction, 16S rRNA gene and metagenomic sequencing
Microbial biomass was collected using the aforementioned peristaltic pump, 0.1 μm polycarbonate membrane filters, and filter setup. Filter housings were kept on ice in the dark while pumping water during summer collection periods. Water was pumped through the filters until they clogged, which occurred over a range of 2–12 L, then filters were promptly harvested and preserved on dry ice and then later at −20°C. At minimum three filters were obtained from the Ney Springs cistern at each collection period. DNA was extracted from preserved filters alongside an unused filter from the same pack to serve as a blank control using a Qiagen DNAeasy Powersoil kit. DNA was then quantified using a Qubit fluorometer (ThermoFisher Scientific, United States). Samples were then sent to Novogene (en.novogene.com) (Beijing, CHN) for 16S rRNA NovaSeq PE250 amplicon sequencing targeting the V4 region (515F-806R) using the Earth Microbiome project primers and protocol (Thompson et al., 2017) or for paired end 150 shotgun metagenomic sequencing.
16S rRNA analysis
Raw sequence data was trimmed, chimera checked, and quality filtered in DADA2 (V. 1.22) (Callahan et al., 2016) for R (V. 4.1.2). Taxonomic classification was performed using a compatible Naive Bayesian classifier trained using the SILVA_nr99_V138 training set implemented for DADA2 (McLaren, 2020). Phyloseq (V. 1.38.0) was used to generate taxonomic bar charts for 16S rRNA gene data (Mcmurdie and Holmes, 2013). Contamination sequences were determined by using the blank filter controls and were removed from the dataset using the prevalence based method in Decontam, which compares the presence/absence of taxa found in contaminated control samples to that in actual samples (Davis et al., 2018). For determination of core community Amplicon Sequence Variants (ASVs), 23 samples were first pooled into six categories based on time of sampling and then assessed for ASV detection. ASVs that were found within all six sampling events were deemed resident community members, while ASVs found during only one of the six sampling events were deemed transient. Microbial community composition and seasonal taxa overlap were visualized by generating an upset plot in UpSetR (V. 1.4.0) (Conway et al., 2017). Comparisons of mean relative abundance of core community ASVs was done using a Kruskal-Wallis test from the Vegan R package (V. 2.6-2) (Oksanen et al., 2022) followed by a Dunn test adjusted with Benjamini-Hochberg correction. The MicroViz R package (Barnett et al., 2021) was used to performed a redundancy analysis (RDA) on community samples with ASVs only detected once over the six sampling periods removed from the sample pool. Changes in the relative abundance of core community ASVs were compared with changes in various geochemical species overtime by calculating the correlation coefficient in Excel. 16S rRNA gene amplicon sequences are available under the NCBI BioProject accession number PRJNA739719.
Metagenome analysis
Metagenome sequencing data was pooled from the July 2021, January 2022, and March 2022 sampling trips as we were able to obtain sufficient biomass for metagenomics during these trips. Initial taxonomic classification of metagenome reads was performed with Kaiju (v1.7.4) (Menzel et al., 2016). Metagenomic reads were obtained and co-assembled by IDBA-ID (v1.1.3) (Peng et al., 2012), MEGAHIT (v1.2.9) (Li et al., 2015) and metaSPAdes (Nurk et al., 2017) (v 3.15.3) within the Kbase web platform (Arkin et al., 2018). The three assemblies were then binned using CONCOCT (v1.1) (Alneberg et al., 2014), MaxBin2 (V2.2.4) (Wu et al., 2016) and MetaBAT2 (v1.7) (Kang et al., 2019) for a total of nine different permutations. DAS tool (v1.1.2) (Sieber et al., 2018) was then used to merge any overlapping or redundant bins generated from CONCOCT, MaxBin2, and MetaBAT2 into one set of bins for each of the three assembly methods. These three assemblies were then classified using GTDB-tk (v1.7.0) (Parks et al., 2018) and extracted as bins. The bins were named for their number, taxonomic classification, and assembly method and were then merged as one large assembly set which was assessed in CheckM (v1.0.18) (Parks et al., 2015). A multiple sequence alignment (MSA) is generated in CheckM with HMMER1, which uses 43 single copy phylogenetic marker genes to assess bin completeness (Parks et al., 2015). The MSA obtained from the CheckM output was then used to generate a tree using FastTree2 (v2.1.9) (Price et al., 2010). Using the phylogenetic tree along with CheckM stats, bins were manually selected based on phylogenic classification, completeness, and contamination. In most instances the phylogenetic tree nodes were grouped in sets of three, representative of each assembly method (i.e., IDBA-ID, MEGAHIT, and metaSPAdes), which all contained the same marker lineage designation, number of genomes, number of markers, and number of marker sets. The representative bin was then chosen based on highest completeness and lowest contamination. These finalized metagenome assembled genomes (MAGs) were combined with previously obtained MAGs associated with dominant Ney Springs taxa (Trutschel et al., 2022) to investigate the core microbial community members. All assemblies were annotated or re-annotated using the KEGG GhostKoala (v2.2.) online interface (Kanehisa et al., 2016). Metabolic pathway completeness was assessed using the KeggDecoder package (Graham et al., 2018) and via manual search of KO terms for genes of interest not included in the KeggDecoder package. Metagenome data is available under the NCBI BioProject accession number PRJNA739719.
Results and discussion
93 ASVs constitute Ney Springs’ cistern resident community
Analysis of the pooled monthly seasonal samples revealed many transient ASVs found during only one of the sampling events, with approximately 17,000 out of the almost 20,000 ASVs detected falling into this category. These transient microorganisms are suspected to mostly come from input of debris from the surrounding environment (e.g., plants, insects, dust) into the cistern. The transient community is higher in diversity but much lower in abundance compared to the resident community, which was comprised of only 93 ASVs observed every sampling period. These 93 ASVs are referred to as the resident microbial members due to their persistent detection in the spring (Figure 1A). Notably, the resident community members were all bacteria, with Archaea only detected in low amounts in both the 16S RNA gene survey and metagenomic data for the cistern (Supplementary Datas 1, 2). This aligns with previous findings from Ney Springs which showed very little Archaeal presence (Trutschel et al., 2022). Of these resident community ASVs, the Tindallia and Izimaplasma genera consistently dominated the microbial community; seven Tindallia ASVs comprised 36–55% of the community and two Izimaplasma ASVs ranged from 3 to 36%. 16 of the resident community ASVs had an average annual abundance of greater than 1%. These 16 were deemed the core community ASVs and collectively comprised 63–87% of the community alone, while the 93 resident ASVs comprised 74–93% of the microbial community (Figure 1B).
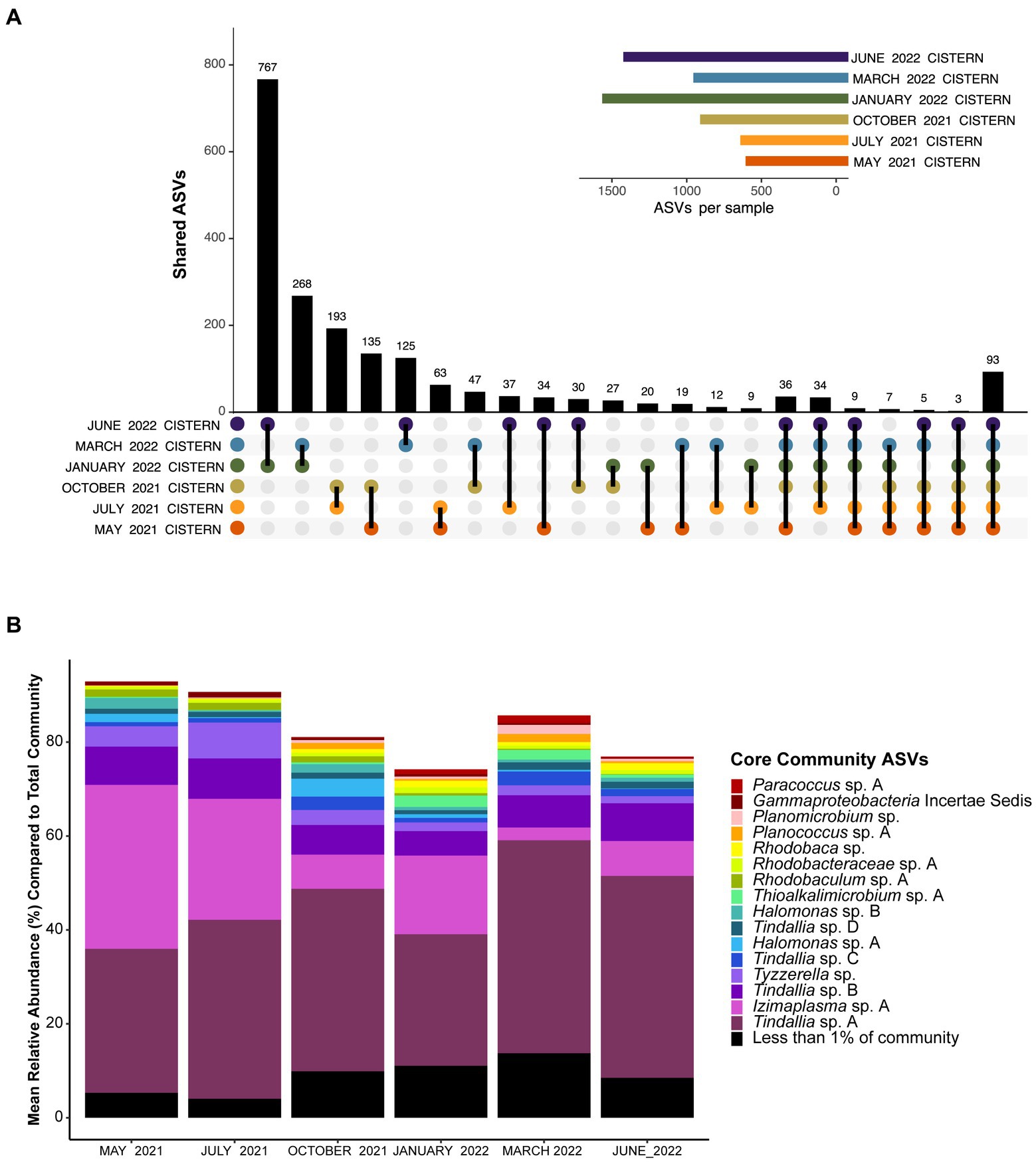
Figure 1. (A) UpSet plot showing how many unique ASVs overlap between pooled monthly samples. Plot shows overlap between all six months, at least five of the six months, and then ASVs which are only found in two of the six months sampled. The 93 ASVs found in all six months sampled represent the resident community members. The sample size for each month was n ≥ 3. (B) Barplot showing the mean relative abundance of the 93 resident community ASVs compared to the total community. Only the top 16 with a mean relative abundance of >1% are shown, which represent the core community members. The remaining 77 resident community members are grouped together. A complete list of the resident community members can be found in Supplementary Data 1.
The core community taxa found are from genera predominantly associated with alkaline environments, with many representatives previously detected in soda lakes. For example, the predominance of Tindallia and Izimaplasma species is distinct compared with other serpentinizing systems, though these taxa have been detected within soda lakes (Kevbrin et al., 1998; Vavourakis et al., 2018). Other predominant core community taxa are those belonging to the Halomonas and Rhodobacteraceae groups. Isolates from these groups have been cultured from multiple alkaline soda lake environments and have been shown to be heterotrophic sulfur oxidizers (Sorokin et al., 2000; Sorokin, 2003; Bryantseva et al., 2015; Kopejtka et al., 2017). Approximately 15% of the resident community ASVs belong to the Rhodobacteraceae and include the intermingled and poorly phylogenetically resolved Paracoccus, Rhodobaca, Rhodobaculum, Roseibaca, and Roseinatronobacter genera. The closest relative of the Gammaproteobacteria incertae sedis ASV is Wenzhouxiangella, another genus originally isolated from an alkaline soda lake (Sorokin et al., 2020). Thioalkalimicrobium (aka Thiomicrospira) is the only core community member also observed in high abundance in other serpentinizing system microbial communities-the Lost City and Prony Bay hydrothermal fields (Brazelton et al., 2012; Postec et al., 2015), though species have also been isolated from soda lakes as well (Sorokin et al., 2002). Ney Springs is located <640 km from Mono lake, a soda lake which shares many similar microbial members to those found in Ney Springs, such as Halomonas, Thiomicrospira, and Roseinatronobacter (Humayoun et al., 2003; Trutschel et al., 2022). The remaining core community taxa include Planomicrobium species, which are not known to be associated with alkaline environments, but the closely related Planococcus have been isolated from alkaline soils (Wang et al., 2015). There is also Tyzzerella, which is commonly found in the human gut microbiome, though the closest matches with our 16S rRNA sequence are from uncultured members detected in termite guts—which, are known for highly alkaline conditions that aid in digestion of plant material (Schmitt-Wagner et al., 2003). Overall, the core community taxa identified show precedence for being alkaliphiles, though this is the first time many have been detected in abundance within a serpentinizing system.
Seasonal evaporation occurs in the Ney Springs cistern
Our previous work used water isotopes to demonstrate that fluids from the Ney Springs primary cistern are distinct from other water sources in the Mt. Shasta/Dunsmuir, CA area as they diverge greatly from the meteoric water line (Trutschel et al., 2022). Our seasonal analysis has now identified fluctuations within the water isotope signatures, specifically within the δ18O (‰ VSMOW) isotopes of H2O (Figure 2). This change in oxygen isotope enrichment is likely due to evaporation as the highest δ18O (‰ VSMOW) values are observed in July 2021 and June 2022, corresponding to the highest site temperatures, and the lowest δ18O (‰ VSMOW) concentrations coinciding with the lowest temperature in January 2022 (Figures 2, 3A,B). The temperature extremes for the cistern were observed in January 2022 at 6°C (external daytime temperature −0.5 to 11.7°C) and in July 2021 at 13.9°C (external daytime temperature 13.3 to 32.8°C). A strong positive correlation is seen between δ18O (‰ VSMOW) values and cistern temperature (correlation coefficient of 0.96) as well as δ18O (‰ VSMOW) values when plotted alongside average monthly temperature for the region (correlation coefficient of 0.89) (Figure 3). When comparing average monthly precipitation to changes in water isotopes, we do not see a strong correlation. Very little precipitation is observed in the region, and a decrease in neither δ2H (‰ VSMOW) nor δ18O (‰ VSMOW) was observed in Ney water isotopes during October when precipitation was greatest (Figure 3). The cistern itself has a recharge rate of 3.88 L/h and its ability to refill quickly does not appear to be influenced by meteoric input. While evaporation appears to be the main driver of seasonal changes in water isotopic signatures, evaporation and precipitation do not appear to influence concentration in redox stable geochemical species such as silicon and sodium which may be more indicative of water rock-interactions (Figures 3E,F). Sodium levels at Ney Springs are elevated compared to typical marine geochemistry, making it a likely byproduct of subsurface water-rock interactions (Feth et al., 1961). At this point Ney Springs hydrogeology and specifically how this particular spring is isolated from meteoric water remains unknown. This in addition to the variation in geochemistry that may relate to active vs. mineralized serpentinized fluids is in question, but at present there is no evidence for fluid mixing in the Ney Springs cistern.

Figure 2. Water isotope plot showing Ney Springs cistern samples collected seasonally compared to surface water proximal springs, Ney Springs Creek, snow melt, and the global meteoric water line. Samples are differentiated by color and shape.
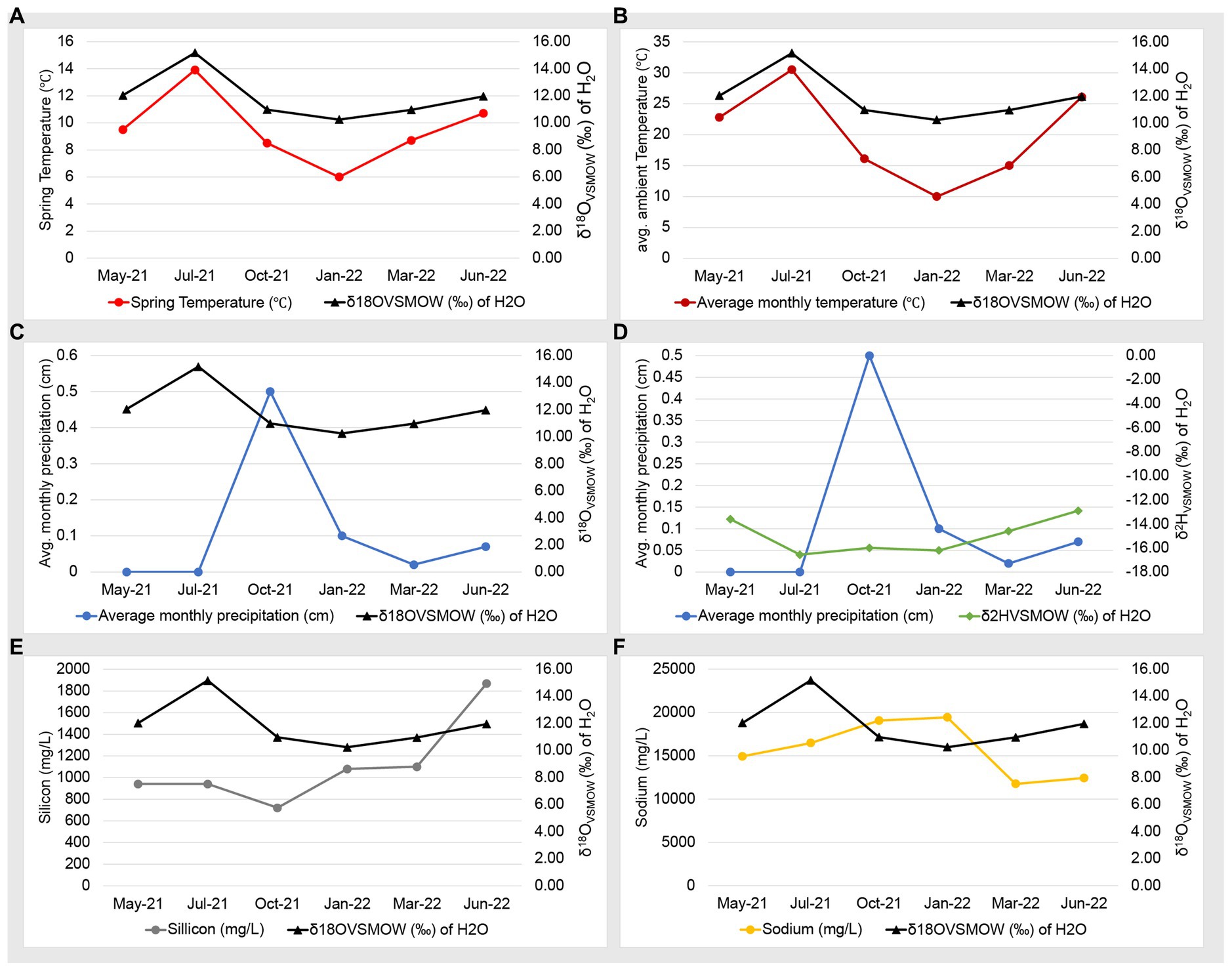
Figure 3. Geochemistry and water isotope time series plots. Plots consists of two Y axes, with data points listed in chronological order on the X axis. Panels are as follows: (A) Ney Springs cistern fluid temperature compared to δ18O (‰ VSMOW) of H2O. (B) Average ambient monthly temperature for greater Ney Springs area compared to δ18O (‰ VSMOW) of H2O. (C) Average monthly precipitation for greater Ney Springs area compared to δ18O (‰ VSMOW) of H2O. (D) Average monthly precipitation for greater Ney Springs area compared to δ2H (‰ VSMOW) of H2O. (E) Silicon concentration in Ney Springs cistern (mg/L) compared to δ2H (‰ VSMOW) of H2O. (F) Sodium concentration in Ney Springs cistern (mg/L) compared to δ2H (‰ VSMOW) of H2O.
Temporal variation is also observed in several redox active geochemical constituents, such as sulfur and nitrogen species. These species are more liable to be altered by microbial processes, and their variation may suggest that microbial community dynamics are driving changes that may or may not be related to other environmental parameters that change seasonally (i.e., temperature). In this system, sulfate is predicted to come from the connate nature of the deeper ground waters being influenced by the marine Franciscan subduction complex. It has previously been speculated that the sulfide present in the spring is potentially a product of microbial sulfate reduction, as it is not volcanic in nature (Feth et al., 1961). However, sulfur oxidation, which was previously shown to be a viable metabolism in this system, could also impact sulfate/sulfide concentrations (Trutschel et al., 2022). Interestingly, the balance of sulfur species changes over the course of our year sampling period. The abundance of sulfide vs. oxidized products supports the influence of microbial activity (Figure 4). While this change may be occurring at the surface level, deeper subsurface microbial activity and/or water-rock interactions could be influencing the sulfur species composition as well.
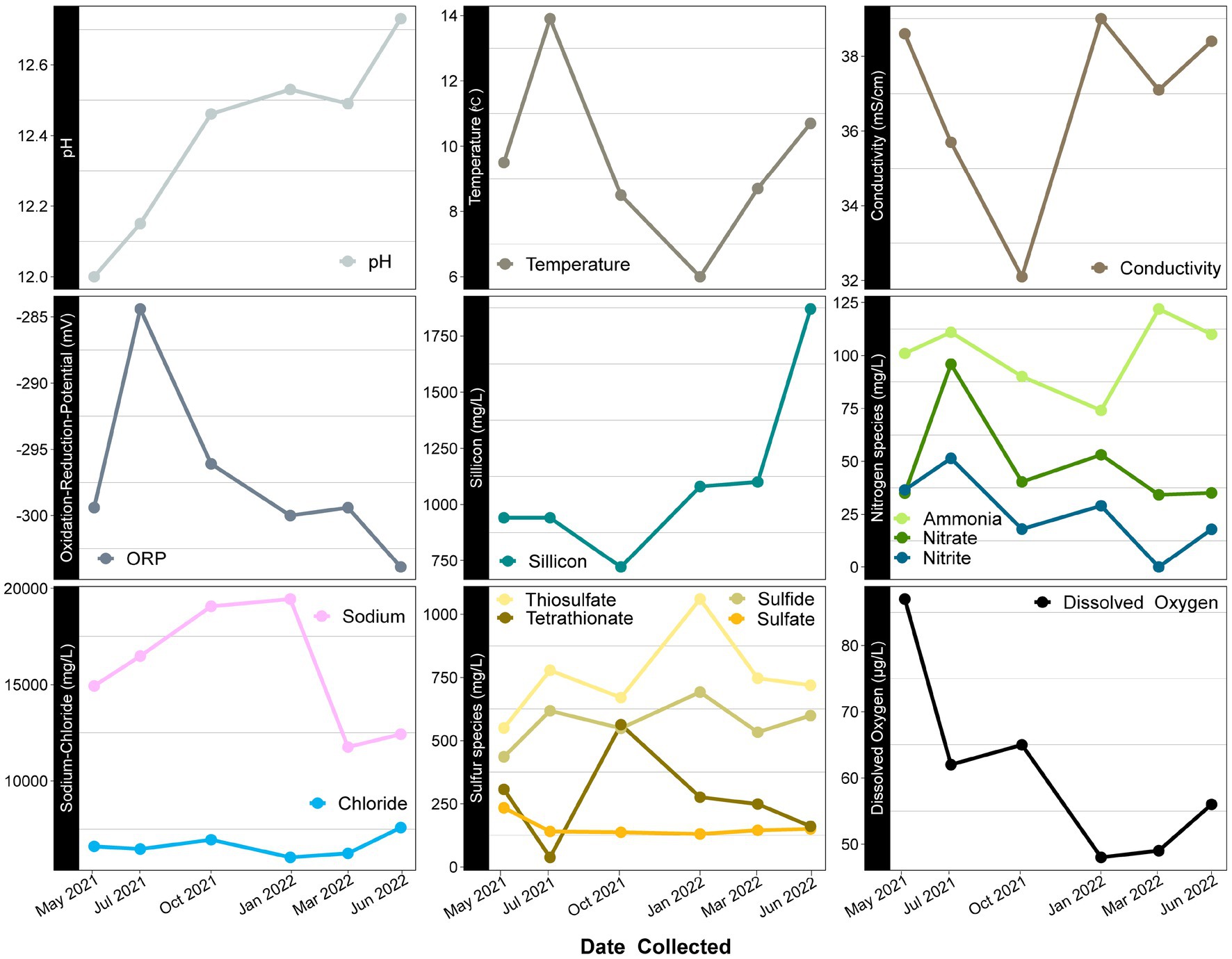
Figure 4. Time series scatter plots of geochemical constituents measured over a year at Ney Springs. X axis represents time sampled while Y axis specifies units for each constituent. Similar species likely to have relationships are grouped together.
The high ammonia concentration (74–122 mg/L) in this system has been anomalous, especially compared to other characterized serpentinizing systems (Trutschel et al., 2022). It has been hypothesized that the high ammonia in Ney Springs may originate from decaying organic matter, though it is currently unclear if ancient or modern material could be the source (Waring, 1915; Feth et al., 1961). Ammonia concentrations vary over the sampling period, as do other detected nitrogen species. Nitrate (34–95 mg/L) and nitrite (0.01–51 mg/L) are also much higher than what is seen in other serpentinizing systems (Cardace et al., 2015; Crespo-Medina et al., 2017; Cook et al., 2021). The high concentration of nitrogen species within Ney Springs could come from interactions with the Franciscan Subduction Complex, but as the values are much higher than other serpentinizing systems within the same host geology (Morrill et al., 2013), this suggests the presence of additional nitrogen sources as well. Temporal variation in input from ancient marine sediment rich in organic matter could be contributing to nitrogen concentrations, as could subsurface microbial dissimilatory nitrate reduction to ammonia. However, it is also worth noting that within our system, we see particularly elevated amounts of nitrogen species during May and July of 2021, which may be due to seasonal changes in proximal environmental factors such as vegetation.
Changes in geochemistry and abundance of core microbial community members help explain seasonal variation
While the overall microbial community composition of Ney Springs changes seasonally, all of the samples collected across the six sampling events have a similar degree of variance. Permanova/adonis results on Bray-Curtis distances calculated for the monthly samples revealed there is a significant difference in the centroids of monthly samples (pr (>F) = 0.001), and they maintain a similar homogeneity of dispersion between them and are not significantly different in dispersal pattern (betadisper, pr (>F) = 0.224). Interestingly, community structure does not appear to be solely a function of season. For example, not all summer months cluster similarly. While May 2021 and July 2021 samples cluster, the June 2022 community samples cluster near March 2022 (Figure 5). Structure is also not simply a product of linear divergence over time due to the placement of the January 2022 and October 2021 samples in between these clusters. However, a longer sampling period would be needed to determine if the community follows any sort of cyclical or oscillating pattern.
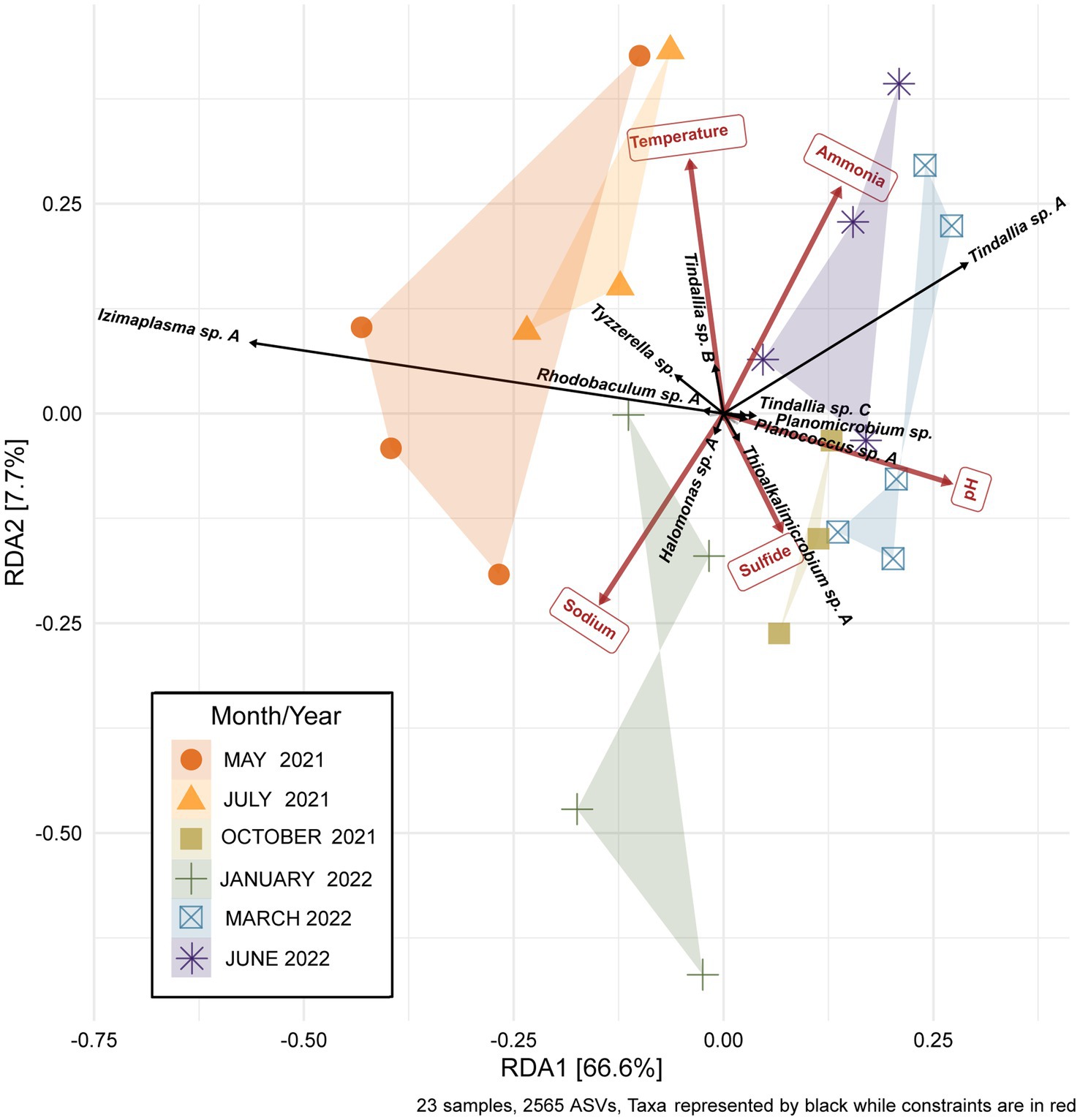
Figure 5. Redundancy analysis (RDA) plot of the Ney Springs cistern microbial community samples. 23 microbial community samples were collected during six different sampling events. Samples are devoid of transient ASVs, i.e., ASVs that were only encountered during one sampling event, in order to best represent the resident microbial community. Count data is transformed to be in terms of relative abundance per sample. Constrained elements were chosen based on their ability to explain variation within the microbial community and lack of overlap with one another.
The strongest correlations between taxa with particular sampling periods were seen in core community ASVs that experienced a significant increase in relative abundance within that sampling period (Figures 5, 6). Out of the 16 core community members, 12 underwent significant changes to their mean relative abundance seasonally (Kruskal-Wallis test, p value <0.05) (Figure 6). The greatest change in average relative abundance was observed in Izimaplasma sp. A, between May 2021 and March 2022 at 35% vs. 2.7% of the total microbial community, respectively, (Dunn test, p.adj. value = 0.0001). Previously, Izimaplasma had been observed as the most abundant microbial community member during the first sampling of Ney Springs in late May of 2019, reinforcing its observed strong association with the early summer month (Trutschel et al., 2022). Two other ASVs followed the inverse of this pattern, with their highest abundance and strongest correlation associated with March 2022 and their lowest abundance observed in May 2021. This included Planococcus sp. A (March 1.7% vs. May 0.05%) and the Planomicrobium sp. (March 1.91% vs. May 0.05%) (Dunn test, p.adj. values <0.01). Other ASVs experiencing a period of upsurge where their average relative abundance was significantly higher (Dunn test, p.adj. value<0.04) compared to two or more of the other sampling times included the Tyzzerella sp. during July 2021 with a maximum observed relative abundance of 7.6%, Halomonas sp. A in October 2021 at 3.84%, Thioalkalimicrobium sp. A in January 2022 at 2.41%, and Tindallia sp. C in March at 2.93%.
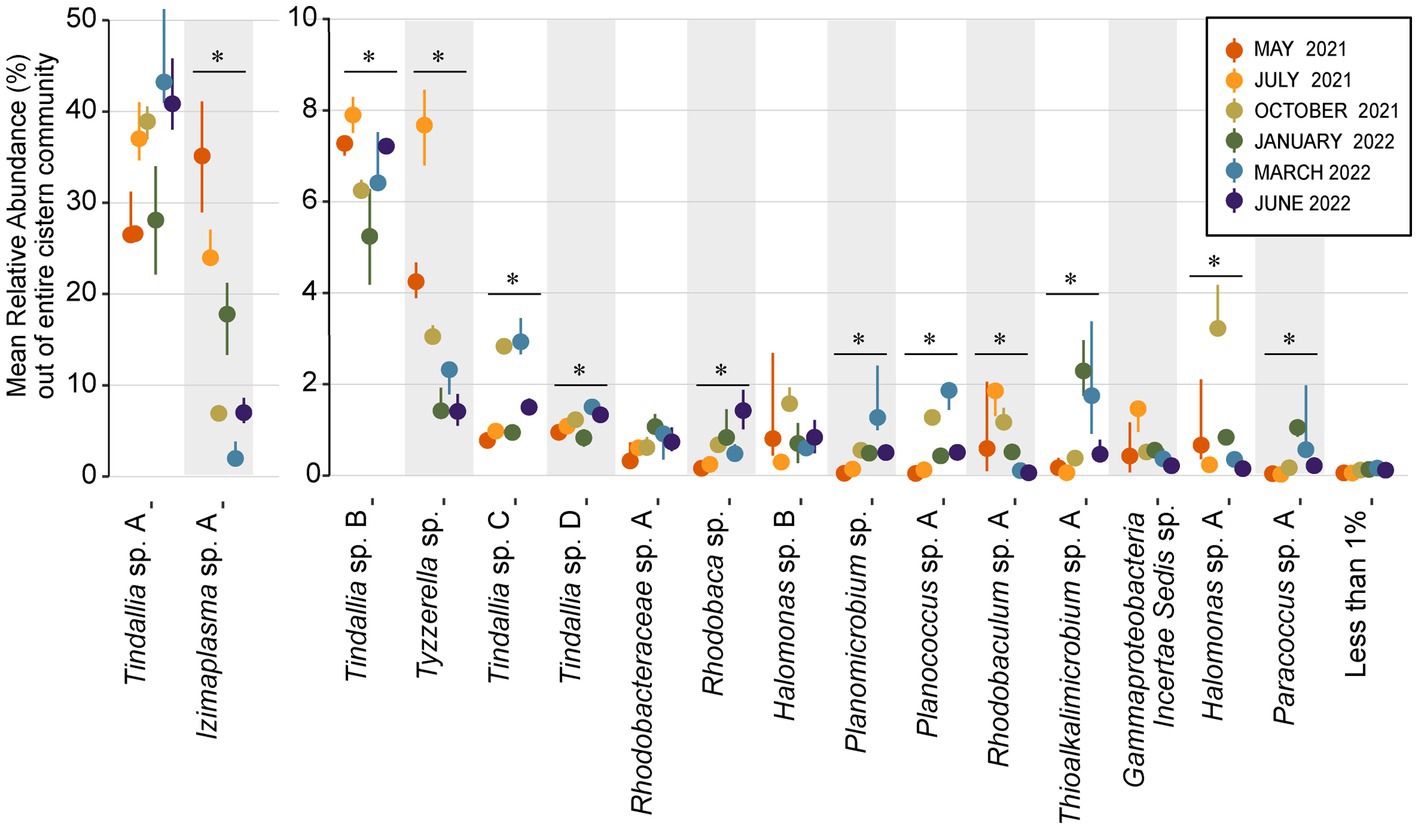
Figure 6. Dot plot showing change in mean relative abundance of sixteen core community ASVs that regularly comprise 1% or more of the total community. The remaining 77 resident community members are grouped together as the “Less than 1%” category. ASVs are organized by decreasing overall mean relative abundance, with groups split between two Y axes in order to better visualize changes in less abundant ASVs. The dots plotted represent the mean relative abundance of n ≥ 3 samples each month, while lines emitting from the dots represent the 95% confidence interval. ASVs that had a significant change in relative abundance between months (Kruskal-Wallis test, p value <0.05) are denoted with a bar and “*” above them.
In addition to determining sampling periods’ associations with specific ASVs, a redundancy analysis (RDA) was performed to determine how much seasonal variation within the microbial community may be explained by changes in geochemistry. Constrained elements were chosen based on their seasonal variation, potential for interaction with microbial metabolism, and on their unrelatedness to one another. These parameters included pH, temperature, sodium, ammonia, and sulfide. The five independent constrained variables explained 74.3% of the variation seen between the microbial community samples (Figure 5). This revealed several potential relationships between core community taxa and constrained elements associated with metabolism, such as Tindallia spp. with ammonia as well as Thioalkalimicrobium sp. A, and Halomonas sp. A with sulfide. Meanwhile, changes in pH, sodium, and temperature may cause a shift in favorable growth conditions for multiple core community members in a way that broadly alters structure. This could also explain why certain sampling periods are associated more closely with these parameters (e.g., sodium with January 2022).
Core community associated MAGs show adaptations to salinity and alkalinity
To understand the drivers of the observed correlations between species abundance, seasonality, and geochemistry, metagenome assembled genomes (MAGs) of the core microbial community were analyzed. In addition to seven previously obtained MAGs (Trutschel et al., 2022), we report nine additional MAGs used to investigate the metabolic potential of the core community members (Table 1). Three MAGs were below 95% complete (Planococcus bin 006, Lachnospirales bin 026, and Roseinatronobacter bin 022) and though all were present in the core microbial community, they have been omitted from further analysis due to the inability to confidently assess metabolism. Tindallia bin 004 was included despite its higher potential for contamination (9.27%) because it contained a 16S rRNA gene sequence that directly matched the most abundant ASV (Tindallia sp. A) and because all genes of interest matched Tindallia bin 001, which only contained 3% contamination. When assessing mechanisms for dealing with the stress of this environment, focus was placed on the organisms’ genetic potential for tolerating salinity and alkalinity. While temperature is potentially a driving feature of seasonal variation of the spring community, genome level adaptations to temperature were not investigated as the cistern temperature remained in the low mesophilic to psychrophilic range all year (6–13.9°C), and as such, we would not expect a strong genome level signature for temperature.
Many of the MAGs encoded genes associated with salinity and alkalinity tolerance such as Na+/H+ antiporters Mrp and/or Nha (Figure 7). Mrp antiporters are often essential for maintaining an electrochemical gradient in alkaline and marine conditions by pumping sodium ions out while pumping protons in (Ito et al., 2017). Homologs of the Na+/H+ antiporter NhaD found in the Halomonas MAG do not exhibit activity below pH 8 and have thus far only been found in alkaliphiles (Nozaki et al., 1998). NhaC homologs, which are detected in the Tindallia and Wenzhouxiangella MAGs, have been shown to be necessary for growth in alkaliphilic conditions for several Bacillus sp. (Ito et al., 1997; Krulwich et al., 1997). Putative sodium pumping NADH-coQ reductase (Nqr) was also observed in many of the MAGs, which can help maintain the electrochemical gradient under alkaline conditions in conjunction with the H+/Na+ antiporters by pumping sodium out (Vorburger et al., 2016). As described previously, most of the MAGs appear to encode for H+ binding rather than Na+ binding ATPases based on amino acid sequence despite the low concentrations of H+ at pH 12 (Mulkidjanian et al., 2008; Trutschel et al., 2022). The exceptions to this are the Tindalliaceae and Izemoplasmataceae MAGs which are predicted to contain Na+ binding F-type ATPases and are notably the most abundant taxa in the system (Figure 7). Tindallia sp. A and D exhibit a slight negative correlation with sodium respectively, but no other core community ASVs have a suggested strong relationship with sodium (Figure 8). Notably, the relative abundance of Izimaplasma sp. A is negatively correlated with pH, while only the Rhodobaca sp. A ASV was strongly positively correlated with an increase in pH (Figures 8, 9A,B). Previously an isolate from the Roseinatronobacter-Rhodobaca cluster of the Rhodobacteraceae family was isolated from Ney Springs and was found capable of growth in pH 12.4 media (Trutschel et al., 2022), suggesting that some members of this clade may be better at tolerating high pH conditions.

Figure 7. Heatmap of core community associated MAGs with selected marker proteins relating to nitrogen, sulfur, methane, hydrogen, formate, and acetate metabolism or are associated with alkalinity and salinity tolerance. Only MAGs that had greater than 95% completeness are shown within the Heatmap. The black boxes for ATP synthase protein I have all necessary subunits for an F-type ATPase (AtpFBCHGDAE) and instead specify whether an organism is likely to encode for a Na+ or H+ binding ATP synthase based on amino acid sequence. For organisms that do contain the gene homolog, the black boxes for the Na+/H+ antiporter specify if the organism is likely to contain NhaA, C or D, with Nha C and D originally characterized in and often associated with alkaliphiles.
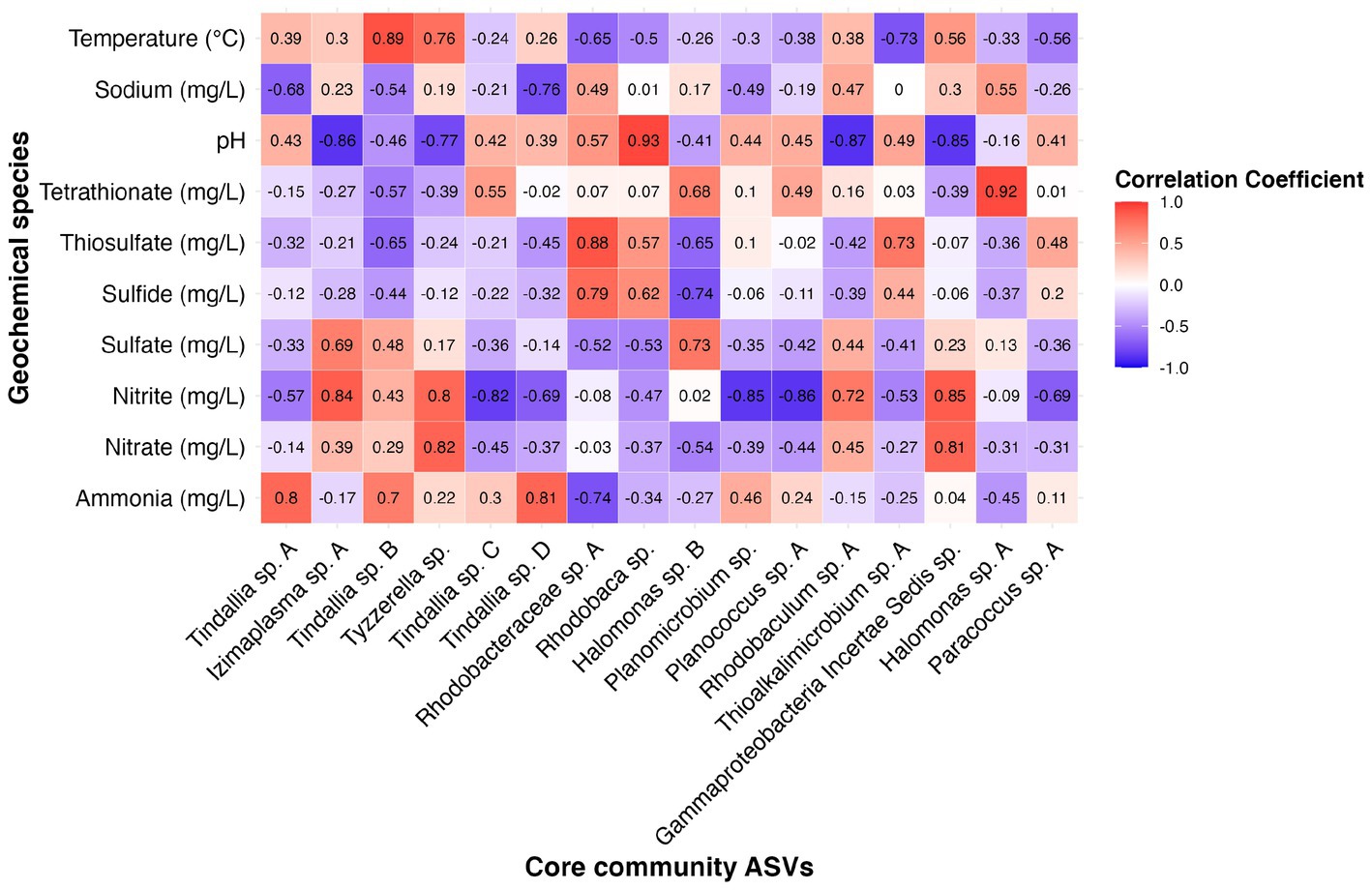
Figure 8. Heatmap showing correlation coefficient values between the relative abundance of 16 core community ASVs with geochemical constituents of interest as determined by results of RDA analysis.
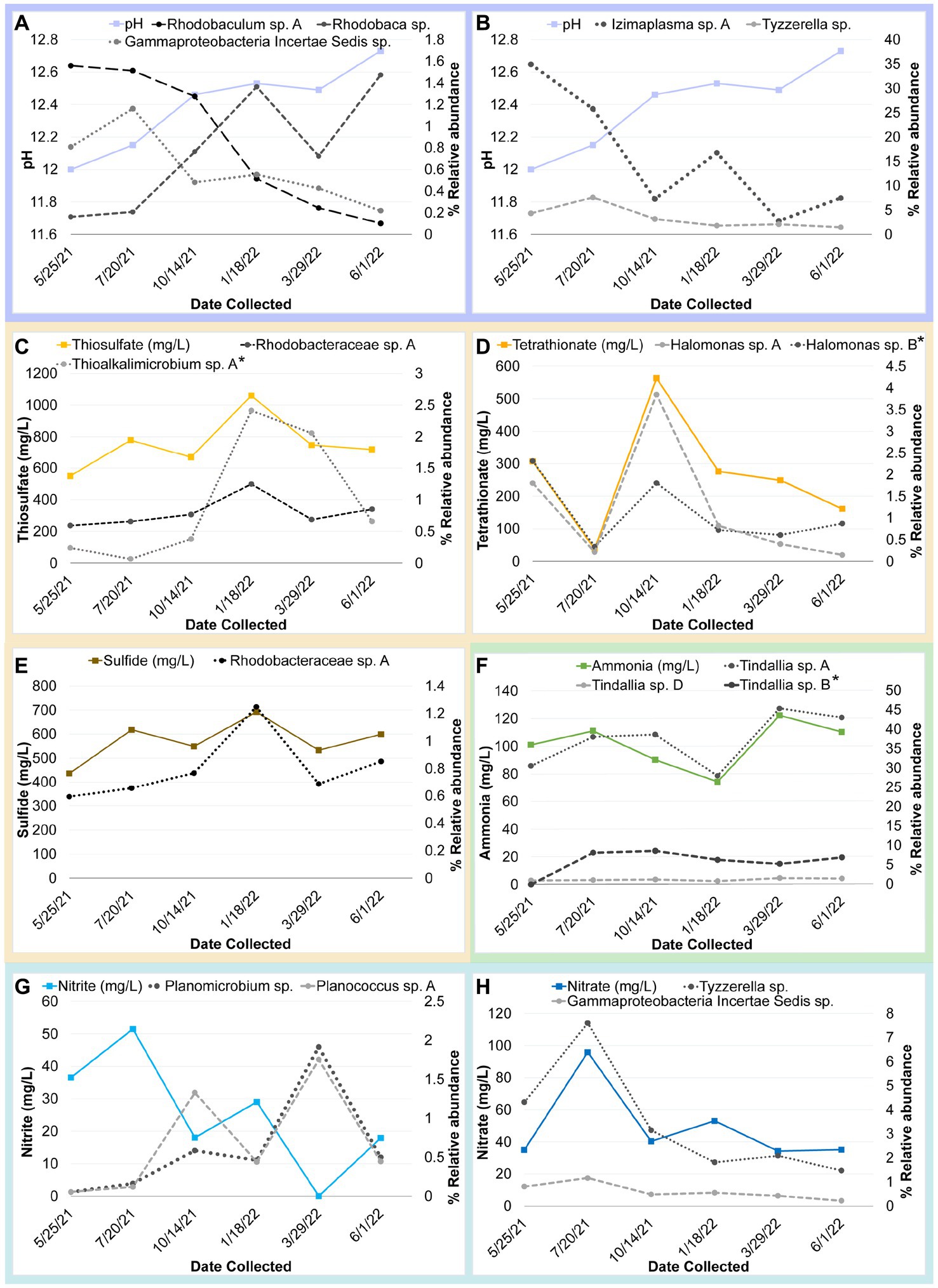
Figure 9. Timeseries plot of changes in core community ASV relative abundance that may be related seasonal changes in a geochemical constituent as identified by a correlation coefficient value at or above 0.80 unless otherwise noted (*). Each plot shows the relative abundance of one or more core community ASVs plotted alongside a different variable. Panels are as follows with correlation coefficient values for each ASV indicated in parentheses: (A) Change in pH compared to relative abundance of Gammaproteobacteria incertae Sedis sp. (−0.85), Rhodobaculum sp. (−0.87), and Rhodobaca sp. (0.93). (B) Change in pH compared to relative abundance of Izimaplasma sp. A (−0.86) and Tyzzerella sp. (−0.77) (C) Change in thiosulfate compared to relative abundance of Thioalkalimicrobium/Thiomicrospira sp. A (0.73*) and Rhodobacteraceae sp. A (0.88). (D) Change in tetrathionate compared to relative abundance of Halomonas sp. A and B (0.92 and 0.68*). (E) Change in sulfide compared to relative abundance of Rhodobacteraceae sp. A (0.79). (F) Change in ammonia compared to relative abundance of Tindallia sp. A, B and D (0.8, 0.7*, 0.81, respectively). (G) Change in nitrite compared to relative abundance of Planococcus sp. A (−0.86) and Planomicrobium sp. (−0.85). (H) Change in nitrate compared to relative abundance of Tyzzerella sp. (0.82) and Gammaproteobacteria incertae Sedis sp. (0.81).
Metagenomic information shows potential for ammonia production by most abundant core community member
The source of ammonia within Ney Springs is unknown, but may be linked to current or past microbial activity. The potential for generation of ammonia through denitrification (DNRA) is observed in the Rhodobacteraceae and Halomonas MAGs, which each encode nitrate and nitrite reductases (NarGH/NapAB and NirBD) (Figure 7), however none of the Rhodobacteraceae or Halomonas ASVs exhibit a strong correlation coefficient with nitrite, nitrate, or ammonia (Figure 8). Only Planococcus sp. A and the Planomicrobium sp. were observed to have a relationship with nitrite (Figure 9G), while the Tyzzerella and Gammaproteobacteria Incertae Sedis spp. were the only ASVs positively correlated with nitrate (Figure 9H). Conversely, Tindallia sp. A, B and D are all positively correlated with ammonia (Figures 8, 9F). The Tindallia MAGs encode the enzymes necessary for Stickland reactions from glycine and ornithine (GrdABE and Ord), which have been shown to produce ammonia (Sangavai and Chellapandi, 2017). Tindallia magadii, the type-strain of the genus, has been observed producing upwards of 30 mM of ammonium over a 60 h period when grown in culture with 2 g/L arginine and ornithine as the initial substrate (Kevbrin et al., 1998). Stickland reaction in members of the Peptostreptococcaceae are cited as the most abundant ammonia producing organisms within the rumen, with several strains capable of producing up to 0.4 mM per mg of protein per minute (Paster et al., 1993; Sangavai and Chellapandi, 2017). Given the high concentrations of ammonia generally produced by these groups, it is predicted that these organisms have adaptations for ammonia tolerance, though there is little insight into what these genetic adaptations may be.
Lack of hydrogen and methane metabolism amongst core community members
Ammonia is hypothesized to be one of key driving factors of community composition within this environment, and the likely reason we do not observe methanogens or methane oxidizers typically associated with serpentinizing systems within Ney Springs (Trutschel et al., 2022). The abundance of free molecular ammonia (NH3 as opposed to NH4+) potentially places strong selective pressure on microbial inhabitants due to its increased membrane passivity (Kayhanian, 1999). Both ammonia and methane associated metabolisms are known to be inhibited by high ammonia concentrations (Lehtovirta-Morley, 2018; Yan et al., 2020). No evidence of potential ammonia oxidation (AmoA or Hzo), nor methanogenesis or methane oxidation (McrA, MmoA or PmoA) was observed within the core community MAGs (Figure 7), which concurs with previous results showing a lack of evidence for these metabolisms (Cook et al., 2021; Trutschel et al., 2022). Similarly, there were very few potential hydrogenases detected within the core community MAGs (Figure 7). Partially complete NAD (HoxFUY) and NADP-reducing (HndBCD or HndCD) hydrogenases were found in five of the MAGs. A partially complete NiFe hydrogenase (HyaBC) was found within Rhodobacteraceae bin 004, but it was missing the small subunit (HyaA). This could suggest a loss of gene function in these organisms. Hydrogen has been measured at exceptionally low concentrations at Ney Springs when compared to other serpentinizing systems. Bubbles that arise from the bottom of the cistern have consistently contained around 0.02 atm hydrogen by volume, while dissolved hydrogen was measured at <0.01 mg/L (Mariner et al., 2003; Trutschel et al., 2022). Although thermodynamically favorable in this system, hydrogen oxidation is likely limited due to the low concentration of hydrogen available within the cistern (Trutschel et al., 2022). Acetate and formate represent other potential energy sources that may be formed via serpentinization. Many of the MAGs did encode for putative formate dehydrogenases (FdoGHI) (Figure 7), with many of the Rhodobacteraceae MAGs containing multiple copies. The Izemoplasmataceae/Tenericutes MAGs also contained formate C-acetyltransferase (PflAD). Other than the Tindalliaceae and Izemoplasmataceae/Tenericutes MAGs, the core community members all contained Acetyl-CoA synthetase (ACS). Rhodobacteraceae and Halomonas spp. have been isolated from the system previously and have been observed using acetate as a carbon/energy source (Trutschel et al., 2022).
Temporal fluctuation in sulfur species concentrations associated with putative sulfur-oxidizing core community members
Another peculiar aspect of Ney Springs is the abundance of sulfide, which is not commonly found in terrestrial serpentinizing systems. Sulfide is found in marine serpentinizing systems such as the Lost City [2–32 mg/L (Schrenk et al., 2004)], but it is often orders of magnitude higher at Ney Springs (430–700 mg/L). Despite the abundance of sulfide and theoretical energy available for sulfate-reducing metabolic reactions, we have once again found little genetic evidence of microbial sulfide production via dissimilatory sulfate reduction or anaerobic methane oxidation using sulfate as a terminal electron acceptor (Trutschel et al., 2022). We did not detect methyl-coenzyme M reductase (McrA) or dissimilatory sulfate reductase (DsrAB) within the core microbial community associated MAGs, though a putative DsrAB was previously found in a MAG associated with resident community member Desulfurivibrio (Figure 7; Trutschel et al., 2022). Two Rhodobacteraceae MAGs putatively contain sulfate adenylyltransferase (Sat), which is likely to be involved in sulfur assimilation but has also been implicated in dissimilatory sulfur oxidation in this organism (Yu et al., 2007; Parey et al., 2013). Evidence of sulfur oxidation is much more prevalent in the core community members, as all the core community MAGs except those belonging to the Tindalliaceae and Izemoplasmataceae have the potential to engage in some form of sulfur species oxidation. The Thiomicrospira/Thioalkalimicrobium MAG contains Sqr (sulfide:quinone oxidoreductase) along with SoxXYZABCD (sulfur oxidation operon) and is predicted to oxidize sulfur species completely to sulfate. Thioalkalimicrobium sp. A relative abundance has a slight positive correlation with thiosulfate (Figure 8) and is most abundant when sulfide and thiosulfate are highest in January 2022. MAGs classified as Rhodobacteraceae all contain Sqr, have varying degrees of completeness of the Sox sulfur oxidation pathway, and all contain a complete or almost complete SoeABC (quinone sulfite dehydrogenase). Despite this putative evidence, only Rhodobacteraceae sp. A exhibits a positive correlation with sulfide and thiosulfate concentrations within the cistern (Figure 8), as its abundance is highest when thiosulfate and sulfide are also at their highest and conversely low when these concentrations are also low (Figures 9C,E). The Halomonas MAG only contains SoxZ (thiosulfate oxidation carrier protein), but does contain thiosulfate dehydrogenase (TsdA), an alternate thiosulfate oxidizing protein (Denkmann et al., 2012). This pathway produces tetrathionate as an end-product, which is not observed in organisms only utilizing the Sox system (Kelly et al., 1997; Grabarczyk and Berks, 2017). The changes in relative abundance of the Halomonas sp. A and B ASVs tracks well with changes in tetrathionate concentration within the cistern over time (Figure 9D) and Halomonas sp. A has a very high positive correlation coefficient with tetrathionate (Figure 8). A Halomonas isolate from Ney Springs has previously been shown to oxidize thiosulfate to tetrathionate in vitro as well, confirming this as a likely product produced by these organisms in the environment (Trutschel et al., 2022).
Implications
Since the discovery of active serpentinization in the Coast Range ophiolite many serpentinizing systems have been identified by Barnes in Northern California, including what is now the Coast Range Ophiolite Microbial Observatory, The Cedars, and Ney Springs (Barnes et al., 1967, 1972; Barnes and O’Neil, 1969). The investigation of Ney Springs has allowed us greater insight into the ecology of terrestrial serpentinizing systems and the role host geology and microbial metabolism have on shaping geochemistry. Serpentinizing springs like Ney are commonly studied as windows into subsurface microbial communities and food webs that subsist on the reduced compounds generated by the serpentinzation reaction. Notably, these systems maintain their high pH and much of their geochemistry despite surface exposure, which results in a specialized microbial community. This can be seen within Ney Springs, with the resident community members making up the overwhelming majority of this interface microbial community. Using ASVs as the final denominator may produce an artifact of a seemingly large introduced community, but this more conservative method is preferred since it allowed us to focus on a limited number of well-established core taxa adapted to the polyextreme conditions of Ney Springs. By further identifying the putative adaptations and metabolic capabilities of these core community members, we could then assess the potential influence these organisms have on their environment and how that may explain temporal variation observed in the geochemistry.
The putative role of these core community members at Ney Springs is of interest, as they are likely driving temporal geochemical changes in the spring through their metabolisms. Within the core community, a few members had strong associations with changing geochemical parameters, and the metabolic potential we observed in their corresponding MAGs supports the capacity to use or produce these geochemical species. This was seen clearly with the Tindallia taxa and their correlation with ammonia concentrations. While additional experiments will be necessary to confirm that the Tindallia species detected are capable of excess ammonia generation, these findings represent the first plausible explanation with evidence for the profuse ammonia found within this environment. Though ammonia is a stressor, and is not common in many naturally occurring alkaline environments, it has been shown to inhibit microbial activities in bioreactors that experience ammonia buildup over time (Kayhanian, 1999; Leejeerajumnean et al., 2000). Similarly, while we have not yet observed this in other serpentinizing systems on Earth, the co-occurrence of high ammonia concentrations and serpentinization end products (e.g., hydrogen) have been detected on icy moons such as Enceladus (Vance et al., 2007; Waite et al., 2009). Understanding how ammonia impacts microbial metabolism and viability is an astrobiologically relevant question that could be further investigated at Ney Springs.
While the source of the sulfide at Ney Springs remains unclear, this work points to a metabolically diverse group of sulfur-oxidizing microbes that may use sulfide, thiosulfate, or elemental sulfur found within the spring. The complex role of sulfur intermediates within hyperalkaline environments is understudied, though many species, such as polysulfides and thiosulfate, have increased stability at high pH and are much more abundant and biologically available under these conditions (Van den Bosh et al., 2008; Findlay, 2016). Though best observed in the case of Halomonas and tetrathionate, other core community species may be producing and consuming these less studied sulfur intermediates. Thiomicrospira and members of the Rhodobacteraceae were more abundant when sulfide and thiosulfate were at their highest, supporting a potential link between the energy available for sulfur oxidation and these populations. Organisms like Thiomicrospira are obligate chemolithoautotrophs and the majority of Rhodobacteraceae from this environment are likely chemolithoheterotrophs. A Rhodobacteraceae isolate from this cistern, as well as closely related members of this family isolated from soda lakes, have been previously described as chemolithoheterotrophs and their ability to supplement energy through sulfur oxidation could explain their increased abundance during times of higher reduced sulfur species availability (Sorokin, 2003; Trutschel et al., 2022). As such, Rhodobacteraceae populations may be more linked to carbon pools rather than sulfur species, though at present we have only low-resolution measurements for DOC/TOC from Ney Springs and cannot identify which carbon species are present and potentially bioavailable.
Future work within Ney Springs will focus on the role of carbon speciation and how it shapes the microbial community, as many of the core community species identified did not appear to have a strong association with the geochemical constraints chosen, such as seen with Izimaplasma. These organisms have been twice observed having a period of significantly increased abundance within late May, but the driving factor for this bloom has yet to be identified. Potentially increased organic availability via exogenous carbon input from detritus could explain this, but further investigation is required. Other potential impacts on organism abundance are their relationships with one another. Organisms with similar metabolisms, like Izimaplasma and Planocococcacae species, which are both putative simple sugar fermenters, may face competition with one another. Similarly, a decrease in exogenous organic carbon input utilized by many of the abundant heterotrophic and/or fermentative taxa may then allow for the increased abundance of autotrophic organisms like Thiomicrospira. Additional work with enrichments and in situ activity assays may help identify which organisms are most active within this environment and are in direct competition with one another for resources. The role subsurface processes play in introducing or supporting different microbial taxa observed in this system remains to be explored. Certain geochemical parameters vary temporally with no seasonal pattern, and could be a function of differences in host rock interactions. In addition, subsurface microbial processes that are feasible but not observed in the surface community (e.g., sulfate reduction or anaerobic methane oxidation) could also be impacting the spring, though we currently lack evidence for these activities.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
Author contributions
LT, BK, JS, GC, and AR performed field sampling and data collection for Ney Springs. LT and BK performed geochemical analyses and interpretations. LT performed metagenomic analyses primarily with assistance from GC. LT performed statistical analyses and visualization. LT and AR are the primary authors of the manuscript with editing by BK, JS, and GC. AR and BK performed funding acquisition. All authors contributed to the article and approved the submitted version.
Funding
Funding and salary support for LT, BK, and AR have been provided by the NSF-EAR LowTemp Geochemistry Geobiology award 2025687 and NASA-Roses Exobiology Program grant number (80NSSC21K0482). LT received a Lewis and Clark Field work in Astrobiology fellowship and the University of Cincinnati Dr. Stacy Pfaller memorial scholarship.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1182497/full#supplementary-material
Footnotes
References
Alneberg, J., Bjarnason, B. S., De Bruijn, I., Schirmer, M., Quick, J., Ijaz, U. Z., et al. (2014). Binning metagenomic contigs by coverage and composition. Nat. Methods 11, 1144–1146. doi: 10.1038/nmeth.3103
Arkin, A. P., Cottingham, R. W., Henry, C. S., Harris, N. L., Stevens, R. L., Maslov, S., et al. (2018). KBase: the United States department of energy systems biology knowledgebase. Nat. Biotechnol. 36, 566–569. doi: 10.1038/nbt.4163
Barnes, I., LaMarche, V. C. Jr., and Himmelberg, G. (1967). Geochemical evidence of present-day serpentinization. Science 156, 830–832. doi: 10.1126/science.156.3776.830
Barnes, I., and O’Neil, J. R. (1969). The relationship between fluids in some fresh alpine-type ultramafks and possible modern serpentinization, Western United States. Geol. Soc. Am. Bull. 80, 1947–1960. doi: 10.1130/0016-7606(1969)80[1947:TRBFIS]2.0.CO;2
Barnes, I., Rapp, J. B., O’Neil, J. R., Sheppard, R. A., and Gude, A. J. III (1972). Metamorphic assemblages and the direction of flow of metamorphic fluids in four instances of serpentinization. Contrib. Mineral. Petrol. 35, 263–276. doi: 10.1007/BF00371220
Barnett, D., Arts, I., and Penders, J. (2021). microViz: an R package for microbiome data visualization and statistics. J. Open Source Softw. 6:3201. doi: 10.21105/joss.03201
Boros, E., Balogh, K., Vörös, L., and Horváth, Z. (2017). Multiple extreme environmental conditions of intermittent soda pans in the Carpathian Basin (Central Europe). Limnologica 62, 38–46. doi: 10.1016/j.limno.2016.10.003
Brazelton, W. J., Nelson, B., Schrenk, M. O., Christner, B. C., and State, L. (2012). Metagenomic evidence for H 2 oxidation and H 2 production by serpentinite-hosted subsurface microbial communities. Front. Microbiol. 2:268. doi: 10.3389/fmicb.2011.00268
Bryantseva, I. A., Gaisin, V. A., and Gorlenko, V. M. (2015). Rhodobaculum claviforme gen. nov., sp. nov., a new alkaliphilic nonsulfur purple bacterium. Microbiol 84, 247–255. doi: 10.1134/S0026261715020022
Callahan, B. J., McMurdie, P. J., Rosen, M. J., Han, A. W., Johnson, A. J. A., and Holmes, S. P. (2016). DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–583. doi: 10.1038/nmeth.3869
Cardace, D., Meyer-dombard, D. A. R., Woycheese, K. M., Arcilla, C. A., Brazelton, W., and Carolina, E. (2015). Feasible metabolisms in high pH springs of the Philippines. Front. Microbiol. 6, 1–16. doi: 10.3389/fmicb.2015.00010
Conway, J. R., Lex, A., and Gehlenborg, N. (2017). UpSetR: An R package for the visualization of intersecting sets and their properties. Bioinformatics 33, 2938–2940. doi: 10.1093/bioinformatics/btx364
Cook, M. C., Blank, J. G., Suzuki, S., Nealson, K. H., and Morrill, P. L. (2021). Assessing geochemical bioenergetics and microbial metabolisms at three terrestrial sites of serpentinization: The Tablelands (NL, CAN), The Cedars (CA, USA), and Aqua de Ney (CA, USA). J. Geophys. Res. Biogeosciences 126, 1–16. doi: 10.1029/2019jg005542
Crespo-Medina, M., Twing, K. I., Sánchez-Murillo, R., Brazelton, W. J., McCollom, T. M., and Schrenk, M. O. (2017). Methane dynamics in a tropical serpentinizing environment: the Santa Elena Ophiolite, Costa Rica. Front. Microbiol. 8, 1–14. doi: 10.3389/fmicb.2017.00916
Davis, N. M., Proctor, D. M., Holmes, S. P., Relman, D. A., and Callahan, B. J. (2018). Simple statistical identification and removal of contaminant sequences in marker-gene and metagenomics data. Microbiome 6, 226–214. doi: 10.1186/s40168-018-0605-2
Denkmann, K., Grein, F., Zigann, R., Siemen, A., Bergmann, J., van Helmont, S., et al. (2012). Thiosulfate dehydrogenase: a widespread unusual acidophilic c-type cytochrome. Environ. Microbiol. 14, 2673–2688. doi: 10.1111/j.1462-2920.2012.02820.x
Feth, J. H., Rogers, S. M., and Roberson, C. E. (1961). Aqua de Ney, California, a spring of unique chemical character. Geochim. Cosmochim. Acta 26, 519–521. doi: 10.1016/0016-7037(62)90101-1
Findlay, A. J. (2016). Microbial impact on polysulfide dynamics in the environment. FEMS Microbiol. Lett. 363:fnw103. doi: 10.1093/femsle/fnw103
Fones, E. M., Colman, D. R., Kraus, E. A., Stepanauskas, R., Templeton, A. S., Spear, J. R., et al. (2021). Diversification of methanogens into hyperalkaline serpentinizing environments through adaptations to minimize oxidant limitation. ISME J. 15, 1121–1135. doi: 10.1038/s41396-020-00838-1
García-Ruiz, J. M., Nakouzi, E., Kotopoulou, E., Tamborrino, L., and Steinbock, O. (2017). Biomimetic mineral self-organization from silica-rich spring waters. Sci. Adv. 3, e1602285–e1602287. doi: 10.1126/sciadv.1602285
Grabarczyk, D. B., and Berks, B. C. (2017). Intermediates in the sox sulfur oxidation pathway are bound to a sulfane conjugate of the carrier protein SoxYZ. PLoS One 12:e0173395-15. doi: 10.1371/journal.pone.0173395
Graham, E. D., Heidelberg, J. F., and Tully, B. J. (2018). Potential for primary productivity in a globally-distributed bacterial phototroph. ISME J. 12, 1861–1866. doi: 10.1038/s41396-018-0091-3
Humayoun, S. B., Bano, N., and Hollibaugh, J. T. (2003). Depth distribution of microbial diversity in mono lake, a meromictic soda lake in California. Appl. Environ. Microbiol. 69, 1030–1042. doi: 10.1128/AEM.69.2.1030-1042.2003
Ito, M., Guffanti, A. A., Zemsky, J., Ivey, D. M., and Krulwich, T. A. (1997). Role of the nhaC-encoded Na+/H+ antiporter of alkaliphilic Bacillus firmus OF4. J. Bacteriol. 179, 3851–3857. doi: 10.1128/jb.179.12.3851-3857.1997
Ito, M., Morino, M., and Krulwich, T. A. (2017). Mrp antiporters have important roles in diverse bacteria and archaea. Front. Microbiol. 8:2325. doi: 10.3389/fmicb.2017.02325
Kanehisa, M., Sato, Y., and Morishima, K. (2016). BlastKOALA and GhostKOALA: KEGG tools for functional characterization of genome and metagenome sequences. J. Mol. Biol. 428, 726–731. doi: 10.1016/j.jmb.2015.11.006
Kang, D. D., Li, F., Kirton, E., Thomas, A., Egan, R., An, H., et al. (2019). MetaBAT 2: an adaptive binning algorithm for robust and efficient genome reconstruction from metagenome assemblies. PeerJ 7:e7359. doi: 10.7717/peerj.7359
Kayhanian, M. (1999). Ammonia inhibition in high-solids biogasification: an overview and practical solutions. Environ. Technol. 20, 355–365. doi: 10.1080/09593332008616828
Kelly, D. P., Shergill, J. K., Lu, W.-P., and Wood, A. P. (1997). Oxidative metabolism of inorganic sulfur compounds by bacteria. Antonie Van Leeuwenhoek 71, 95–107. doi: 10.1023/A:1000135707181
Kevbrin, V. V., Zhilina, T. N., Rainey, F. A., and Zavarzin, G. A. (1998). Tindallia magadii gen. nov., sp. nov.: An alkaliphilic anaerobic ammonifier from Soda Lake deposits. Curr. Microbiol. 37, 94–100. doi: 10.1007/s002849900345
Kopejtka, K., Tomasch, J., Zeng, Y., Tichý, M., Sorokin, D. Y., and Koblížek, M. (2017). Genomic analysis of the evolution of phototrophy among haloalkaliphilic rhodobacterales. Genome Biol. Evol. 9, 1950–1962. doi: 10.1093/gbe/evx141
Krulwich, T. A., Ito, M., Gimour, R., and Guffanti, A. A. (1997). Mechanisms of cytoplasmic pH regulation in alkaliphilic strains of Bacillus. Extremophiles 1, 163–170. doi: 10.1007/s007920050030
Leejeerajumnean, A., Ames, J. M., and Owens, J. D. (2000). Effect of ammonia on the growth of Bacillus species and some other bacteria. Lett. Appl. Microbiol. 30, 385–389. doi: 10.1046/j.1472-765x.2000.00734.x
Lehtovirta-Morley, L. E. (2018). Ammonia oxidation: ecology, physiology, biochemistry and why they must all come together. FEMS Microbiol. Lett. 365:fny058. doi: 10.1093/femsle/fny058
Li, D., Liu, C. M., Luo, R., Sadakane, K., and Lam, T. W. (2015). MEGAHIT: an ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics 31, 1674–1676. doi: 10.1093/bioinformatics/btv033
Mariner, R. H., Evans, W. C., Presser, T. S., and White, L. D. (2003). Excess nitrogen in selected thermal and mineral springs of the Cascade Range in northern California, Oregon, and Washington: sedimentary or volcanic in origin? J. Volcanol. Geotherm. Res. 121, 99–114. doi: 10.1016/S0377-0273(02)00414-6
Mccollom, T. M., and Seewald, J. S. (2013). Serpentinites, hydrogen, and life. Elements 9, 129–134. doi: 10.2113/gselements.9.2.129
McLaren, M. R. (2020). Silva SSU taxonomic training data formatted for DADA2 (Silva version 138). Zenodo. doi: 10.5281/ZENODO.3986799
Mcmurdie, P. J., and Holmes, S. (2013). phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One 8:e61217. doi: 10.1371/journal.pone.0061217
Menzel, P., Ng, K. L., and Krogh, A. (2016). Fast and sensitive taxonomic classification for metagenomics with Kaiju. Nat. Commun. 7:11257. doi: 10.1038/ncomms11257
Morrill, P. L., Kuenen, J. G., Johnson, O. J., Suzuki, S., Rietze, A., Sessions, A. L., et al. (2013). Geochemistry and geobiology of a present-day serpentinization site in California: the cedars. Geochim. Cosmochim. Acta 109, 222–240. doi: 10.1016/j.gca.2013.01.043
Mulkidjanian, A. Y., Galperin, M. Y., Makarova, K. S., Wolf, Y. I., and Koonin, E. V. (2008). Evolutionary primacy of sodium bioenergetics. Biol. Direct 3, 1–19. doi: 10.1186/1745-6150-3-13
Nozaki, K., Kuroda, T., Mizushima, T., and Tsuchiya, T. (1998). A new Na+/H+ antiporter, NhaD, of Vibrio parahaemolyticus. Biochim. Biophys. Acta Biomembr. 1369, 213–220. doi: 10.1016/S0005-2736(97)00223-X
Nurk, S., Meleshko, D., Korobeynikov, A., and Pevzner, P. A. (2017). MetaSPAdes: a new versatile metagenomic assembler. Genome Res. 27, 824–834. doi: 10.1101/gr.213959.116
Oksanen, J., Blanchet, F. G., Friendly, M., Kindt, R., Legendre, P., McGlinn, D., et al. (2022). Vegan: Community Ecology Package, R Package Version 2.6-2. Available online: https://CRAN.R-project.org/package=vegan
Ortiz, E., Tominaga, M., Cardace, D., Schrenk, M. O., Hoehler, T. M., Kubo, M. D., et al. (2018). Geophysical characterization of serpentinite hosted hydrogeology at the McLaughlin natural reserve, coast range Ophiolite. Geochem. Geophys. Geosyst. 19, 114–131. doi: 10.1002/2017GC007001
Osburn, M. R., Kruger, B., Masterson, A. L., Casar, C. P., and Amend, J. P. (2019). Establishment of the deep mine microbial observatory (DeMMO), South Dakota, USA, a geochemically stable portal into the deep subsurface. Front. Earth Sci. 7:196. doi: 10.3389/feart.2019.00196
Parey, K., Demmer, U., Warkentin, E., Wynen, A., Ermler, U., and Dahl, C. (2013). Structural, biochemical and genetic characterization of dissimilatory ATP sulfurylase from Allochromatium vinosum. PLoS One 8:e74707. doi: 10.1371/journal.pone.0074707
Parks, D. H., Chuvochina, M., Waite, D. W., Rinke, C., Skarshewski, A., Chaumeil, P. A., et al. (2018). A standardized bacterial taxonomy based on genome phylogeny substantially revises the tree of life. Nat. Biotechnol. 36, 996–1004. doi: 10.1038/nbt.4229
Parks, D. H., Imelfort, M., Skennerton, C. T., Hugenholtz, P., and Tyson, G. W. (2015). CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 25, 1043–1055. doi: 10.1101/gr.186072.114
Paster, B. J., Russell, J. B., Yang, C. M. J., Chow, J. M., Woese, C. R., and Tanner, R. (1993). Phylogeny of the ammonia-producing ruminal bacteria Peptostreptococcus anaerobius, Clostridium sticklandii, and Clostridium aminophilum sp. nov. Int. J. Syst. Bacteriol. 43, 107–110. doi: 10.1099/00207713-43-1-107
Peng, Y., Leung, H. C. M., Yiu, S. M., and Chin, F. Y. L. (2012). IDBA-UD: a de novo assembler for single-cell and metagenomic sequencing data with highly uneven depth. Bioinformatics 28, 1420–1428.
Postec, A., Quéméneur, M., Bes, M., Mei, N., and Benaïssa, F. (2015). Microbial diversity in a submarine carbonate edifice from the serpentinizing hydrothermal system of the Prony Bay (New Caledonia) over a 6-year period. Front. Microbiol. 6:857. doi: 10.3389/fmicb.2015.00857
Price, M. N., Dehal, P. S., and Arkin, A. P. (2010). FastTree 2 – approximately maximum-likelihood trees for large alignments. PLoS One 5:e9490. doi: 10.1371/journal.pone.0009490
Rempfert, K. R., Miller, H. M., Bompard, N., Nothaft, D., Matter, J. M., Kelemen, P., et al. (2017). Geological and geochemical controls on subsurface microbial life in the Samail Ophiolite, Oman. Front. Microbiol 8:56. doi: 10.3389/fmicb.2017.00056
Sangavai, C., and Chellapandi, P. (2017). Amino acid catabolism-directed biofuel production in Clostridium sticklandii: an insight into model-driven systems engineering. Biotechnol. Rep. 16, 32–43. doi: 10.1016/j.btre.2017.11.002
Schmitt-Wagner, D., Friedrich, M. W., Wagner, B., and Brune, A. (2003). Phylogenetic diversity, abundance, and axial distribution of bacteria in the intestinal tract of two soil-feeding termites (Cubitermes spp.). Appl. Environ. Microbiol. 69, 6007–6017. doi: 10.1128/AEM.69.10.6007-6017.2003
Schrenk, M. O., Kelley, D. S., Bolton, S. A., and Baross, J. A. (2004). Low archaeal diversity linked to subseafloor geochemical processes at the lost city hydrothermal field, mid-atlantic ridge. Environ. Microbiol. 6, 1086–1095. doi: 10.1111/j.1462-2920.2004.00650.x
Sieber, C. M. K., Probst, A. J., Sharrar, A., Thomas, B. C., Hess, M., Tringe, S. G., et al. (2018). Recovery of genomes from metagenomes via a dereplication, aggregation and scoring strategy. Nat. Microbiol. 3, 836–843. doi: 10.1038/s41564-018-0171-1
Sorokin, D. Y. (2003). Oxidation of inorganic sulfur compounds by obligately organotrophic bacteria. Microbiology 72, 641–653. doi: 10.1023/B:MICI.0000008363.24128.e5
Sorokin, D. Y., Gorlenko, V. M., Tourova, T. P., Tsapin, A. I., Nealson, K. H., and Kuenen, G. J. (2002). Thioalkalimicrobium cyclium sp. nov. and Thioalkalivibrio jannaschii sp. nov., novel species of haloalkaliphilic, obligately chemolithoautotrophic sulfur-oxidizing bacteria from hypersaline alkaline Mono Lake (California). Int. J. Syst. Evol. Microbiol. 52, 913–920. doi: 10.1099/ijs.0.02034-0
Sorokin, D. Y., Mosier, D., Zorz, J. K., Dong, X., and Strous, M. (2020). Wenzhouxiangella strain AB-CW3, a proteolytic bacterium from hypersaline Soda Lakes that preys on cells of gram-positive Bacteria. Front. Microbiol. 11:597686. doi: 10.3389/fmicb.2020.597686
Sorokin, D. Y., Tourova, T. P., Kuznetsov, B. B., Bryantseva, I. A., and Gorlenko, V. M. (2000). Roseinatronobacter thiooxidans gen. nov., sp. nov., a new alkaliphilic aerobic bacteriochlorophyll a-containing bacterium isolated from a soda lake. Microbiology 69, 75–82. doi: 10.1007/BF02757261
Suzuki, S., Ishii, S., Hoshino, T., Rietze, A., Tenney, A., Morrill, P. L., et al. (2017). Unusual metabolic diversity of hyperalkaliphilic microbial communities associated with subterranean serpentinization at the cedars. Nat. Publ. Gr. 11, 2584–2598. doi: 10.1038/ismej.2017.111
Suzuki, S., Ishii, S., Wu, A., Cheung, A., Tenney, A., Wanger, G., et al. (2013). Microbial diversity in the cedars, an ultrabasic, ultrareducing, and low salinity serpentinizing ecosystem. Proc. Natl. Acad. Sci. 110, 15336–15341. doi: 10.1073/pnas.1302426110
Szponar, N., Brazelton, W. J., Schrenk, M. O., Bower, D. M., Steele, A., and Morrill, P. L. (2013). Geochemistry of a continental site of serpentinization, the Tablelands Ophiolite, Gros Morne National Park: a Mars analogue. Icarus 224, 286–296. doi: 10.1016/j.icarus.2012.07.004
Thompson, L. R., Sanders, J. G., Mcdonald, D., Amir, A., Ladau, J., Locey, K. J., et al. (2017). A communal catalogue reveals earth’ s multiscale microbial diversity. Nature 551, 457–463. doi: 10.1038/nature24621
Trutschel, L. R., Chadwick, G. L., Kruger, B., Blank, J. G., Brazelton, W. J., Dart, E. R., et al. (2022). Investigation of microbial metabolisms in an extremely high pH marine-like terrestrial serpentinizing system: Ney Springs. Sci. Total Environ. 836:155492. doi: 10.1016/j.scitotenv.2022.155492
Twing, K. I., Brazelton, W. J., Kubo, M. D. Y., Hyer, A. J., Cardace, D., Hoehler, T. M., et al. (2017). Serpentinization-influenced groundwater harbors extremely low diversity microbial communities adapted to high pH. Front. Microbiol. 8:308. doi: 10.3389/fmicb.2017.00308
Van den Bosh, P. L. F., Sorokin, D. Y., Buisman, C. J. N., and Janssen, A. J. H. (2008). The effect of pH on thiosulfate formation in a biotechnological process for the removal of hydrogen sulfide from gas streams. Environ. Sci. Technol. 42, 2637–2642. doi: 10.1021/es7024438
Vance, S., Harnmeijer, J., Kimura, J., Hussmann, H., Demartin, B., and Brown, J. M. (2007). Hydrothermal systems in small ocean planets. Astrobiology 7, 987–1005. doi: 10.1089/ast.2007.0075
Vavourakis, C. D., Andrei, A. S., Mehrshad, M., Ghai, R., Sorokin, D. Y., and Muyzer, G. (2018). A metagenomics roadmap to the uncultured genome diversity in hypersaline soda lake sediments 06 biological sciences 0605 microbiology 06 biological sciences 0604 genetics. Microbiome 6, 1–18. doi: 10.1186/s40168-018-0548-7
Vorburger, T., Nedielkov, R., Brosig, A., Bok, E., Schunke, E., Steffen, W., et al. (2016). Role of the Na+−translocating NADH:quinone oxidoreductase in voltage generation and Na+ extrusion in Vibrio cholerae. Biochim. Biophys. Acta Bioenerg. 1857, 473–482. doi: 10.1016/j.bbabio.2015.12.010
Waite, J. H., Lewis, W. S., Magee, B. A., Lunine, J. I., McKinnon, W. B., Glein, C. R., et al. (2009). Liquid water on Enceladus from observations of ammonia and 40Ar in the plume. Nature 460, 487–490. doi: 10.1038/nature08153
Wang, K., Zhang, L., Li, J., Pan, Y., Meng, L., Xu, T., et al. (2015). Planococcus dechangensis sp. nov., a moderately halophilic bacterium isolated from saline and alkaline soils in Dechang Township, Zhaodong City, China. Antonie van Leeuwenhoek 107, 1075–1083. doi: 10.1007/s10482-015-0399-1
Woycheese, K., Arcilla, C. A., Magnuson, T., and State, I. (2015). Out of the dark: transitional subsurface-to-surface microbial diversity in a terrestrial serpentinizing seep. Front. Microbiol. 6, 1–12. doi: 10.3389/fmicb.2015.00044
Wu, Y. W., Simmons, B. A., and Singer, S. W. (2016). MaxBin 2.0: an automated binning algorithm to recover genomes from multiple metagenomic datasets. Bioinformatics 32, 605–607. doi: 10.1093/bioinformatics/btv638
Yan, M., Treu, L., Zhu, X., Tian, H., Basile, A., Fotidis, I. A., et al. (2020). Insights into ammonia adaptation and methanogenic precursor oxidation by genome-centric analysis. Environ. Sci. Technol. 54, 12568–12582. doi: 10.1021/acs.est.0c01945
Keywords: serpentinization, Ney Springs, Illumina 16S rRNA, metagenome, sulfur oxidation intermediates
Citation: Trutschel LR, Kruger BR, Sackett JD, Chadwick GL and Rowe AR (2023) Determining resident microbial community members and their correlations with geochemistry in a serpentinizing spring. Front. Microbiol. 14:1182497. doi: 10.3389/fmicb.2023.1182497
Edited by:
Marianne Quéméneur, UMR7294 Institut Méditerranéen d'océanographie (MIO), FranceReviewed by:
Daniel Colman, Montana State University, United StatesOkamoto Akihiro, National Institute for Materials Science, Japan
Copyright © 2023 Trutschel, Kruger, Sackett, Chadwick and Rowe. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Annette R. Rowe, annette.rowe@uc.edu
 Leah R. Trutschel
Leah R. Trutschel Brittany R. Kruger
Brittany R. Kruger Joshua D. Sackett
Joshua D. Sackett Grayson L. Chadwick
Grayson L. Chadwick Annette R. Rowe
Annette R. Rowe