- 1Key Laboratory of Animal Disease and Human Health of Sichuan, College of Veterinary Medicine, Sichuan Agricultural University, Chengdu, China
- 2New Ruipeng Pet Healthcare Group, Chengdu, China
- 3State Key Laboratory of Agricultural Microbiology, National Engineering Research Center of Microbial Pesticides, College of Life Science and Technology, Huazhong Agricultural University, Wuhan, China
Introduction: Proteus mirabilis is a multi-host pathogen that causes diseases of varying severity in a wide range of mammals, including humans. Proteus mirabilis is resistant to multiple antibiotics and has acquired the ability to produce expanded spectrum of β-lactamases, leading to serious public health problems. However, the available information on P. mirabilis isolated from feces of dogs, is still poorly understood, as is the correlation between its virulence-associated genes (VAGs) and antibiotic resistance genes (ARGs).
Method: In this study, we isolated 75 strains of P. mirabilis from 241 samples, and investigated the swarming motility, biofilm formation, antimicrobial resistance (AMR), distribution of VAGs and ARGs, as well as the presence of class 1, 2, and 3 integrons in these isolates.
Results: Our findings suggest a high prevalence of intensive swarming motility and strong biofilm formation ability among P. mirabilis isolates. Isolates were primarily resistant to cefazolin (70.67%) and imipenem (70.67%). These isolates were found to carry ureC, FliL, ireA, zapA, ptA, hpmA, hpmB, pmfA, rsbA, mrpA, and ucaA with varying prevalence levels of 100.00, 100.00, 100.00, 98.67, 98.67, 90.67, 90.67, 90.67, 90.67, 89.33, and 70.67%, respectively. Additionally, the isolates were found to carry aac(6′)-Ib, qnrD, floR, blaCTX-M, blaCTX-M-2, blaOXA-1, blaTEM, tetA, tetB and tetM with varying prevalence levels of 38.67, 32.00, 25.33, 17.33, 16.00, 10.67, 5.33, 2.67, 1.33, and 1.33%, respectively. Among 40 MDR strains, 14 (35.00%) were found to carry class 1 integrons, 12 (30.00%) strains carried class 2 integrons, while no class 3 integrons was detected. There was a significant positive correlation between the class 1 integrons and three ARGs: blaTEM, blaCTX-M, and blaCTX-M-2. This study revealed that P. mirabilis strains isolated from domestic dogs exhibited a higher prevalence of MDR, and carried fewer VAGs but more ARGs compared to those isolated from stay dogs. Furthermore, a negative correlation was observed between VAGs and ARGs.
Discussion: Given the increasing antimicrobial resistance of P. mirabilis, veterinarians should adopt a prudent approach towards antibiotics administration in dogs to mitigate the emergence and dissemination of MDR strains that pose a potential threat to public health.
1. Introduction
Proteus mirabilis, a member of the Enterobacteriaceae family, as an opportunistic pathogen can cause skin infection, respiratory tract infection, urinary tract infection (UTI), and gastrointestinal tract infection, and it has been suggested that it may be related to Crohn’s disease (Zhang J. W. et al., 2021). As a zoonotic bacteria, P. mirabilis can infect a variety of animals, such as chicken (Wong et al., 2013), ducks (Algammal et al., 2021), turtles (Pathirana et al., 2018), cattle (Sun et al., 2020), companion animals (Marques et al., 2019).
The virulence factors of P. mirabilis include flagella, pili, urease, hemolysin, metalloproteinase, which can help P. mirabilis to colonize, destroy tissues, and escape immunity (Coker et al., 2000). Proteus mirabilis is a model organism for urease+ bacteria (Norsworthy and Pearson, 2017). The main functional subunit of urease, the alpha subunit, is encoded by ureC, which breaks down urea into one carbonic acid and two ammonia molecules (Armbruster et al., 2018). Ammonia produced by urease can increase the pH of urine above 7.2 and precipitates calcium and magnesium compounds into crystals of magnesium ammonium phosphate (struvite) and calcium phosphate (apatite) (Broomfield et al., 2009). Struvite and apatite crystals are deposited in biofilms to form crystalline biofilms (Armbruster et al., 2018). The presence of crystalline biofilms makes antibiotic treatment more difficult (Maszewska et al., 2021). Proteus mirabilis has peritrichous flagella, which can carry out the typical swarming motility, called the “bull-eye” (Aygül et al., 2019). The swarming motility of P. mirabilis is a process in which vegetative cells periodically differentiate into swarming cells on solid surfaces.
The swarming motility is important because it is coupled to the expression of virulence-associated genes (VAGs) and the ability to invade cells (Rather, 2005). It has been shown that there is a specific correlation between virulence factors and antimicrobial resistance (AMR) in P. mirabilis (Filipiak et al., 2020). The AMR of clinical P. mirabilis isolates is gradually increasing. Studies have shown that in addition to intrinsic resistance to tetracycline and polymyxin, P. mirabilis has acquired resistance to the β-lactams (Shelenkov et al., 2020). Proteus mirabilis is usually susceptible to fluoroquinolones (Girlich et al., 2020). Multidrug-resistant (MDR) was defined as acquired non-susceptibility to at least one agent in three or more antimicrobial categories (Magiorakos et al., 2012). MDR isolates of P. mirabilis have been identified in both human and veterinary medicine, emphasizing on the need for continuous surveillance (Decôme et al., 2020). MDR may be mediated by mutations in a resistance gene, or resistance genes may be acquired through horizontal transfer. These resistant genes are widely present on plasmids and integrons, leading to the problems of rapid transmission and treatment failure (Leverstein-van Hall et al., 2003). Recent studies have also shown that the prevalence of β-lactams resistance genes in P. mirabilis is gradually increasing (Wong et al., 2013; Algammal et al., 2021). The antibiotic resistance gene (ARG) is one of the many mechanisms by which bacteria can develop AMR (Hughes and Andersson, 2017). However, the presence of a specific ARG does not necessarily lead to the corresponding AMR, as other genetic and environmental factors can influence the expression and function of the gene (Hughes and Andersson, 2017). Previous studies in Escherichia coli have revealed both positive and negative associations between VAGs and ARGs, with predominance of positive correlations (Zhao et al., 2021; Zhang S. et al., 2021). But little is known about the AMR, VAGs, and ARGs of P. mirabilis isolated from feces of dogs and the correlations among them.
Previous studies have shown that UTI in companion animals and humans may be caused by closely related P. mirabilis strains and that such strains from both origins share common ARGs and VAGs (Marques et al., 2019), which suggest that companion animals may serve as potential reservoirs of antibiotic-resistant bacteria in humans. Proteus mirabilis also was considered to be the host of storing ARGs and VAGs (Hu et al., 2020). With 50.85 million dogs living in Chinese cities as of 2018, dogs are the most popular companion animals in China (Yin et al., 2020). There have been some studies of P. mirabilis isolated from the urinary tract of dogs (Gaastra et al., 1996; Harada et al., 2014). Therefore, there is great significance in monitoring AMR, ARGs and VAGs carried by P. mirabilis isolates from dog feces for public health security. Additionally, prior studies overlooked P. mirabilis isolates carried by stray dogs. Therefore, this study aimed to determine the prevalence of P. mirabilis in the feces of domestic dogs and stray dogs. Furthermore, the swarming motility, biofilm formation, AMR, VAGs, and ARGs of P. mirabilis isolates were detected and their correlations were analyzed.
2. Materials and methods
2.1. Sample collection
From April 2021 to April 2022, we collected a total of 241 dog fecal swabs, comprising of 147 fecal swabs from domestic dogs obtained from 8 pet hospitals situated in Chengdu city, Sichuan province in southwestern China, as well as 94 fecal swabs from stray dogs sourced from the Center of Protect Beastie located in Sichuan Province. All stray dog fecal swabs were collected by laboratory staff and domestic dog fecal swabs were collected by veterinarians. To minimize the risk of contamination between samples, disinfection of hands and changing of disposable gloves were mandatory for staff involved in sample collection. The fecal swabs of the dogs were collected with sterile cotton swabs into sterile Eppendorf (EP) tubes and placed in a foam box with an ice pack, then sent to the laboratory by express as soon as possible for bacteriological examination.
2.2. Proteus mirabilis screening
The obtained samples were enriched in 1 mL of Luria-Bertani (LB) broth at 37°C, 120 r/min for 24 h. The bacteria suspension was streaked onto LB solid medium using an inoculation loop, and observed for colony morphology on LB agar medium after 24 h of static cultivation at 37°C. Suspected P. mirabilis colonies were selected and streaked on Salmonella-Shigella (SS) agar medium for further purification. The colony color on SS agar medium was observed, and then the single colony with a black center was selected for Gram staining and observed under microscope.
2.3. Identification of isolates
Firstly, biochemical tests such as gelatin liquefaction test, fermentation test of maltose, glucose, sucrose, lactose and mannose, VP test, nitrate dynamic test, MR Test, indole test, iron triosaccharide test, hydrogen sulfide test and citrate utilization were carried out on all isolates using biochemical tubes and interpreted according to Berger’s manual. Then, the 16S rRNA gene was amplified by polymerase chain reaction (PCR) and combined with biochemical result to identify P. mirabilis isolates (Gao et al., 2018). The PCR products were then sent to Beijing Tsingke Biotechnology Co., LTD for sequencing. BLAST (Basic Local Alignment Search Tool) was used to detect sequence homology between the sequencing results and GenBank accessions.
2.4. Hemolysin and urease production
The hemolysis properties of P. mirabilis isolates were determined by observing clear zones around bacterial colonies on blood agar supplemented with 5% defibrinated sheep blood after 24 or 48 h of static cultivation in 37°C. The bacterial suspension of P. mirabilis isolates was streaked on SS agar medium and incubated at 37°C for 24 h. A single colony was selected and streaked into the urease biochemical tubes and incubated at 37°C for 8–10 h to observe the color change. A change in color from orange to red observed in the biochemical tubes indicates the production of ureses by isolates.
2.5. Swarming motility testing
The swarming motility rates of P. mirabilis isolates were assessed by measuring the coverage scale of LB solid medium after incubating under the same conditions. LB solid medium with an agar concentration of 1.5% was prepared. After the medium was solidified, the plates were dried in an oven at 42°C for 60 min before use. The optical density of the bacterial suspension propagated to the logarithmic growth stage was measured at 600 nm and diluted to an optical density (OD) 600 of 0.4. 5 μL of the diluted bacterial suspension was inoculated into the center of LB solid plates. The plates were inverted and incubated at 37°C after the bacterial suspension was absorbed into the agar matrix (~5 min). Swarming motility of P. mirabilis isolates was observed and the coverage scale was measured after 9 h. The ability of swarming motility is divided into three categories: category 1, weak swarming (coverage ≤5%); category 2 medium swarming (5% < coverage ≤25%); category 3, dense swarming (coverage >25%; Filipiak et al., 2020). The assay was repeated three times for each strain.
2.6. Estimation of the biofilm formation
Biofilm formation was determined by using the crystal violet staining method in 96-well cell plates. After the OD600 of the bacterial solution culture to the logarithmic growth stage was detected and adjusted to 0.1, 200 μL diluted bacterial suspension was added to a 96-well cell culture plate and incubated for 24 h at 37°C. The experiment was repeated with 3 wells for each strain. Wells containing only LB broth medium were used as negative controls. After 24 h, the biofilm formed in 96-well cell culture plates was stained by crystal violet staining according to the literature (Sun et al., 2020). After removing the medium and washing the cells with phosphate-buffered saline (PBS), the bacteria were stained with 200 μL crystal violet for 5 min. The crystal violet dye was solubilized by the addition of 200 μL of organic solvent (anhydrous ethanol: acetone = 70:30, v/v), and was quantified by measuring absorbance at 590 nm with a microplate reader. When OD value exceeds 1, dilute with 33% glacial acetic acid, and multiply the obtained value by dilution ratio.
The mean OD of the negative control plus three times its standard deviation (SD) was defined as the cut-off value (ODc). Based on the ODc, the biofilm-forming ability of isolates can be divided into the following four types: OD590 ≤ ODc is a non-biofilm-forming strain (−), ODc < OD590 ≤ 2ODc is a weak biofilm-forming strain (+), and 2ODc < OD590 ≤ 4ODc is medium biofilm-forming strains (++), OD590 > 4ODc is strong biofilm-forming strains (+++) (Khoramian et al., 2015).
2.7. Antimicrobial susceptibility testing
Kirby-Bauer disc diffusion method was used to evaluate the AR of 75 P. mirabilis isolates to 15 antibiotics in 7 categories. These antibiotics include ampicillin (AMP) (10 μg), amoxicillin/clavulanic acid (AUG) (20/10 μg), ampicillin/sulbactam (SAM) (10/10 μg), cefazolin (CZO) (30 μg), cefepime (FEP) (30 μg), cefotaxime (CTX) (30 μg), imipenem (IPM) (10 μg), meropenem (MEM) (10 μg), aztreonam (ATM) (30 μg), gentamicin (GEN) (10 μg), tetracycline (TET) (30 μg), ciprofloxacin (CIP) (5 μg), sulfamethoxazole/trimethoprim (SXT) (25 μg), fosfomycin (FOS) (200 μg) and chloramphenicol (C) (30 μg). In brief, bacterium suspension of 0.5 McFarland was uniformly spread onto Mueller-Hinton (MH) agar plates and incubated at 37°C for 18 h. Escherichia coli ATCC 25922 was used as the control microorganism. The inhibitory zone around each disc was measured, and the results were interpreted according to the guidelines provided by the manufacturer and the Clinical and Laboratory Standards Institute (CLSI; Clinical and Laboratory Standards Institute, 2020). Isolates were considered non-susceptible if they were intermediate or resistant to a certain antibiotic (Decôme et al., 2020). MDR was defined as acquired non-susceptibility to at least one agent in three or more antimicrobial categories (Magiorakos et al., 2012). Non-MDR was defined as resistance to none or up to two antimicrobial categories (Qi et al., 2016).
2.8. Detection of VAGs, ARGs, and integrons
The distribution of VAGs and ARGs in 75 P. mirabilis isolates was investigated through screening of 11 VAGs, 18 ARGs, and class 1, 2, and 3 integrons using conventional PCR. The details of VAGs, ARGs, and integrons (gene names, primer sequences, product lengths and annealing temperatures) are shown in Supplementary Tables 1, 2. The reaction system containing 12.5 μL of 2 × Taq Master Mix, 1 μL each of upstream/downstream primers, 2 μL of bacterial-DNA, and then sterile ddH2O was added up to 25 μL. The cycling conditions of VAGs were as follows: initial degeneration at 94°C for 2 min; followed by 30 cycles of denaturation at 94°C for 2 min, anneal for 1 min and extension at 72°C for 1 min; 72°C, 5 min. The cycling conditions of ARGs and integrons were as follows: initial degeneration at 93°C for 5 min; followed by 30 cycles of denaturation at 93°C for 30s, anneal for 30s and extension at 72°C for 1 min; 72°C 5 min. At the same time, negative controls without DNA were set. PCR products with positive bands after electrophoresis screening were sent to Beijing Tsingke Biotechnology Co., Ltd. for sequencing. BLAST (Basic Local Alignment Search Tool) was used to detect sequence homology between the sequencing results and GenBank accessions.
2.9. Statistical analyses
The obtained data were analyzed using the Chi-square test (SPSS software, version 9.4; Significance-level; p < 0.05). Comparisons between groups were conducted by Fisher’s Exact test, with an alpha value of 0.05. The statistical analysis took into account standard deviations, and they are based on data obtained from repeated experiments.
3. Results
3.1. Prevalence of Proteus mirabilis in the examined samples
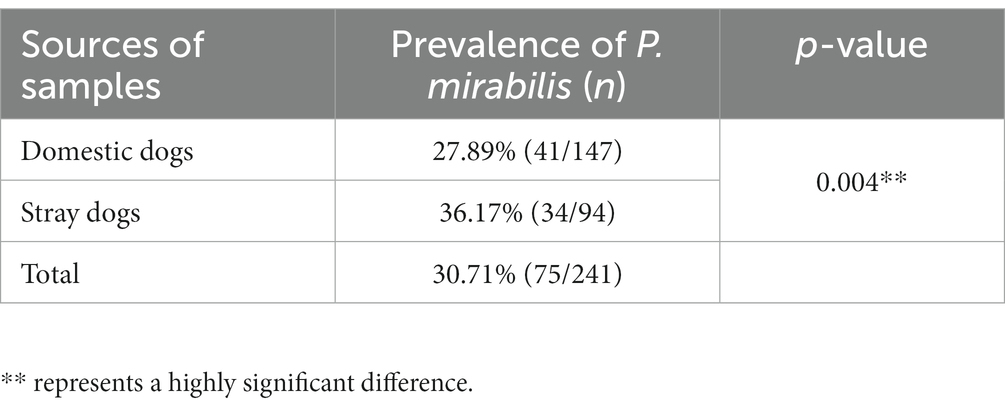
A total of 75 non-duplicate strains of P. mirabilis were isolated from 241 samples of dog feces, and the isolation rate was 31.12% (75/241). Among them, 41 strains of P. mirabilis isolates were from domestic dog feces collected from pet hospitals, and 34 strains of P. mirabilis isolates were from stray dog feces collected from the Center of Protect Beastie (Table 1). The detection rate of P. mirabilis in the feces of stray dogs was significantly higher than that of domestic dogs (p < 0.01).
3.2. Hemolysin and urease production
On defibrinated sheep blood agar plates, all P. mirabilis isolates produced color changes, but none exhibited typical β-hemolysis. After 8–10 h of incubation, the urease biochemical tubes inoculated with P. mirabilis isolates changed from orange yellow to rose red, indicating that all 75 strains of P. mirabilis isolates could produce urease to decompose urea.
3.3. Swarming motility testing
In this experiment, all P. mirabilis isolates showed the ability of “fog creep” migration. The isolate with the strongest ability of swarming motility was PM55 with a velocity of 5.43 mm/h, while the isolate with the weakest ability of swarming motility was PM13, whose speed was only 0.125 mm/h. Additionally, 7 (6.67%) isolates of P. mirabilis exhibited weak swarming motility; 32 (42%) isolates exhibited moderate swarming motility, and 38 (50.67%) isolates exhibited strong swarming motility. There was no significant difference in swarming motility between P. mirabilis isolated from domestic and stray dog feces (p > 0.05).
3.4. Estimation of the biofilm formation
The test of biofilm formation showed that all P. mirabilis isolates could form biofilm. Only 7 strains (9.33%) of P. mirabilis isolates were medium biofilm producers (0.7 < OD590 ≤ 1.4), and 68 strains (90.67%) are intensive biofilm producers (OD590 > 1.4; Supplementary Figure 3). There was no significant difference in biofilm formation between P. mirabilis isolated from domestic and stray dog feces (p > 0.05).
3.5. Antimicrobial susceptibility
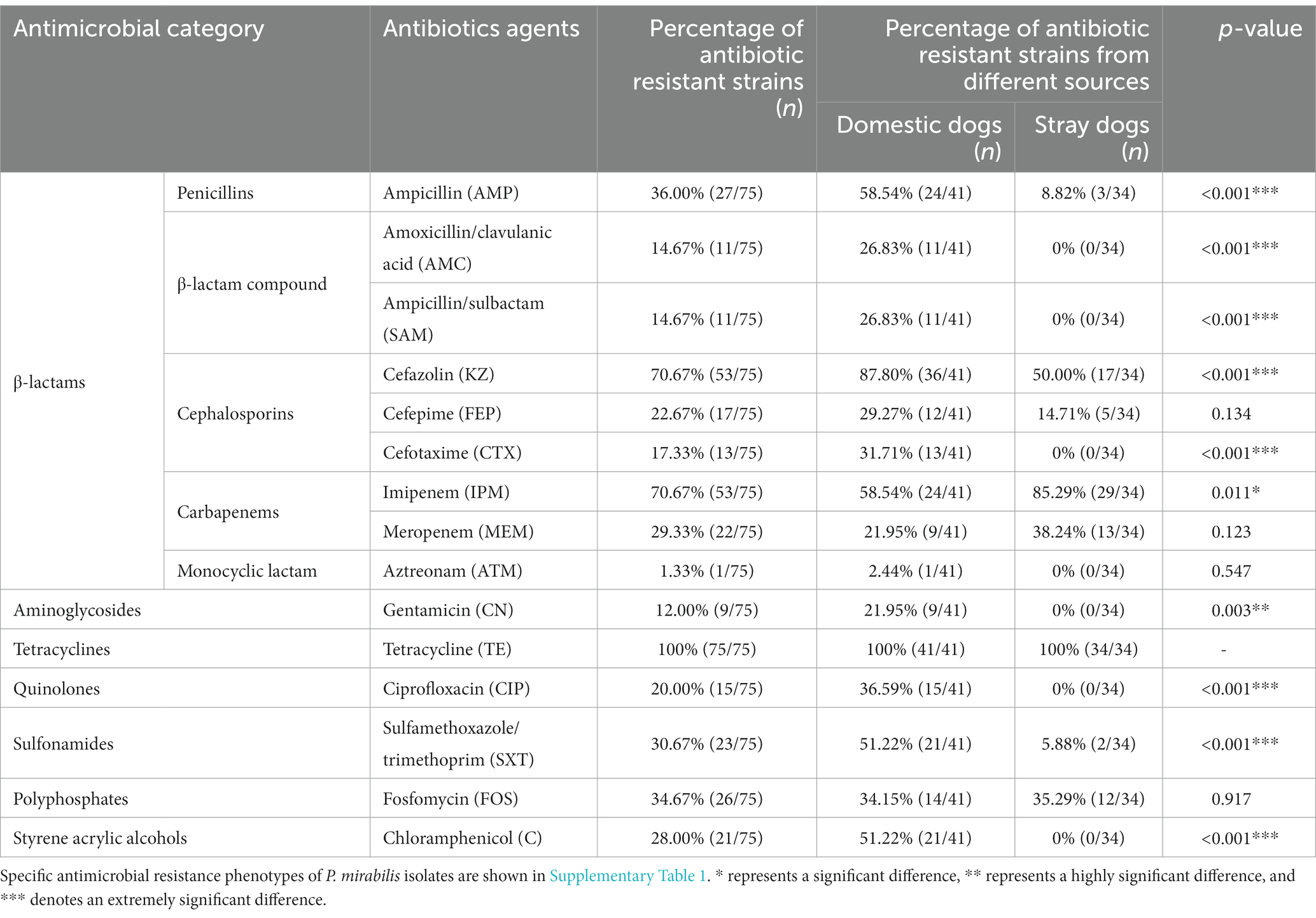
The AMR of the P. mirabilis isolates was assessed and is presented in Table 2. The results indicate that the resistance rate of these P. mirabilis isolates to tetracycline (TE) was the highest (75, 100%), followed by cefazolin (KZ) (53, 70.67%) and imipenem (IPM) (53, 70.67%), ampicillin (AMP) (27, 36.00%), fosfomycin (FOS) (26, 34.67%), sulfamethoxazole/trimethoprim (SXT) (23, 30.67%), meropenem (MEM) (22, 29.33%), chloramphenicol (C) (21, 28.00%), cefepime (FEP) (17, 22.67%), ciprofloxacin (CIP) (15, 20.00%), cefotaxime (CTX) (13, 17.33%), amoxicillin/clavulanic acid (AMC) (11, 14.67%), ampicillin/sulbactam (SAM) (11, 14.67%), gentamicin (CN) (9, 12%), aztreonam (ATM) (1, 1.33%). Moreover, the AMR of P. mirabilis isolates from the two sources was significantly different to different antibiotics (p < 0.05; Supplementary Figure 4).
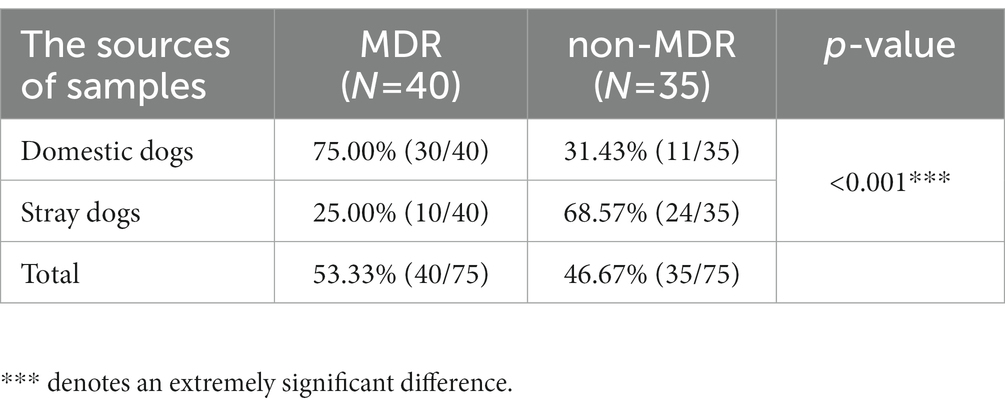
Out of 75 P. mirabilis isolates, 40 strains (53.33%) with MDR were identified, comprising of 30 strains sourced from domestic dogs and 10 from stray dogs (Table 3). Proteus mirabilis strains isolated from domestic dogs exhibited a significantly higher prevalence of MDR than those isolated from stray dogs (p < 0.001).
3.6. Detection of VAGs, ARGs, and integrons
This study identified 11 VAGs in 75 P. mirabilis isolates. Figure 1 shows that ureC, FliL and ireA were the universally present VAGs with a detection rate of 100%, followed by zapA (98.67%), ptA (98.67%), hpmA (90.67%), hpmB (90.67%), pmfA (90.67%), rsbA (90.67%), mrpA (89.33%) and ucaA (70.67%). The prevalence rates of VAGs (hpmA, hpmB, pmfA, mrpA, and rsbA) of P. mirabilis strains isolated from stray dogs were significantly higher than those isolated from domestic dogs (p < 0.05). Furthermore, the prevalence rates of these VAGs in non-MDR strains were significantly higher than those in MDR strains (p < 0.05).
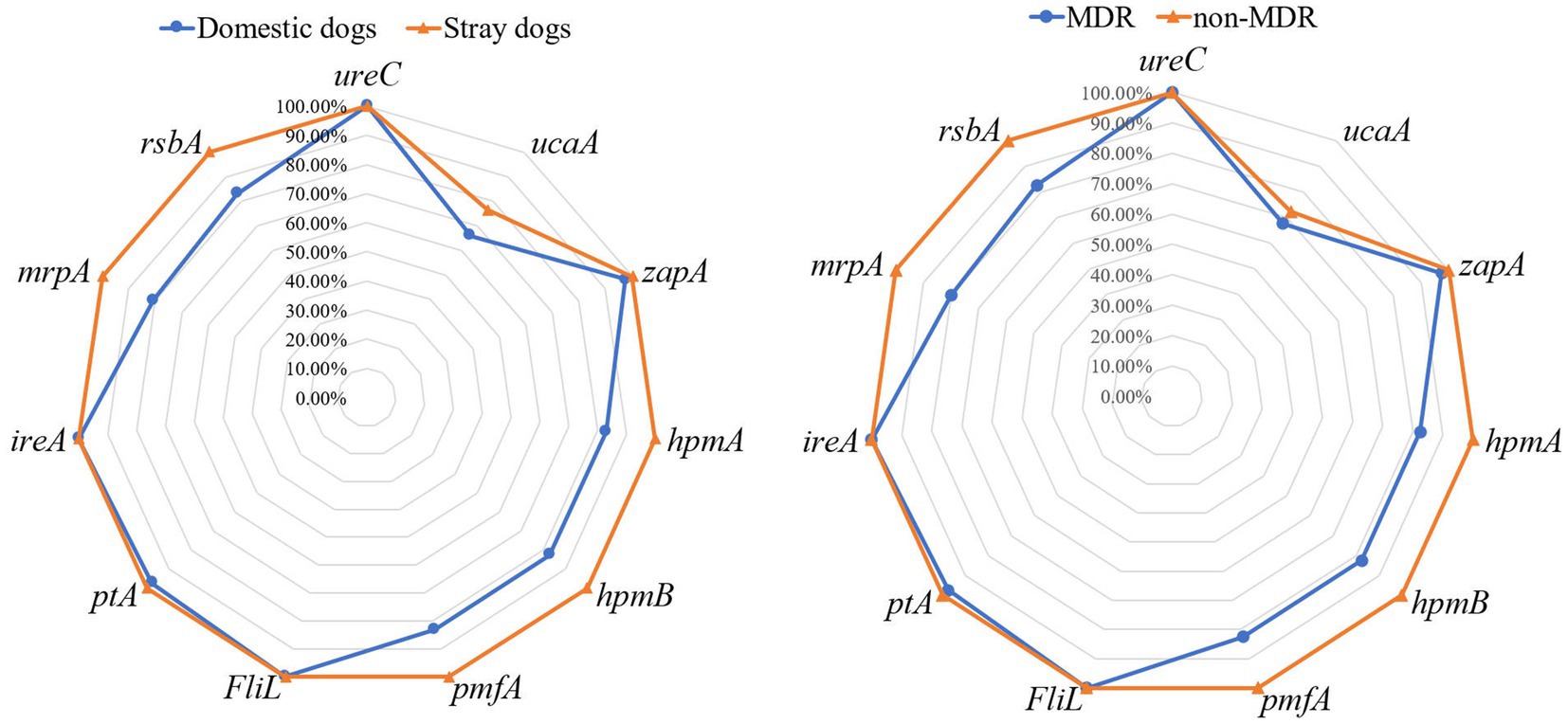
Figure 1. The prevalence of virulence-associated genes in Proteus mirabilis isolates. The radar map on the left shows the virulence-associated genes carried by P. mirabilis strains from domestic and stray dogs. The radar map on the right shows the virulence-associated genes carried by MDR and non-MDR strains.
In total, 10 out of 18 ARGs were identified in 75 P. mirabilis isolates. Among these, aac-(6′)-Ib was the universally present ARG with a prevalence rate of 38.67%, followed by qnrD (32.00%), floR (25.33%,), blaCTX-M (17.33%), blaCTX-M-2 (16.00%), blaOXA-1 (10.67%), blaTEM (5.33%), tetA (2.67%), tetB (1.33%) and tetM (1.33%; Figure 2). The detection rates of ARGs (blaOXA-1, blaCTX-M, blaCTX-M-2, floR and aac-(6′)-Ib) in P. mirabilis isolated from domestic dogs were significantly higher when compared to those isolated from stray dogs (p < 0.01). Furthermore, the detection rates of these ARGs in MDR strains were significantly higher when compared to non-MDR strains (p < 0.01). Out of 40 MDR strains, 14 (35.00%) were found to carry class 1 integrons, 12 (30.00%) strains carried class 2 integrons, while no class 3 integrons was detected. The abundance of the class 2 integrons was found to be significantly higher in P. mirabilis isolates from domestic dogs compared to that from stray dogs (p < 0.05), while no significant difference in class 1 integrons was observed between stray and domestic dogs.
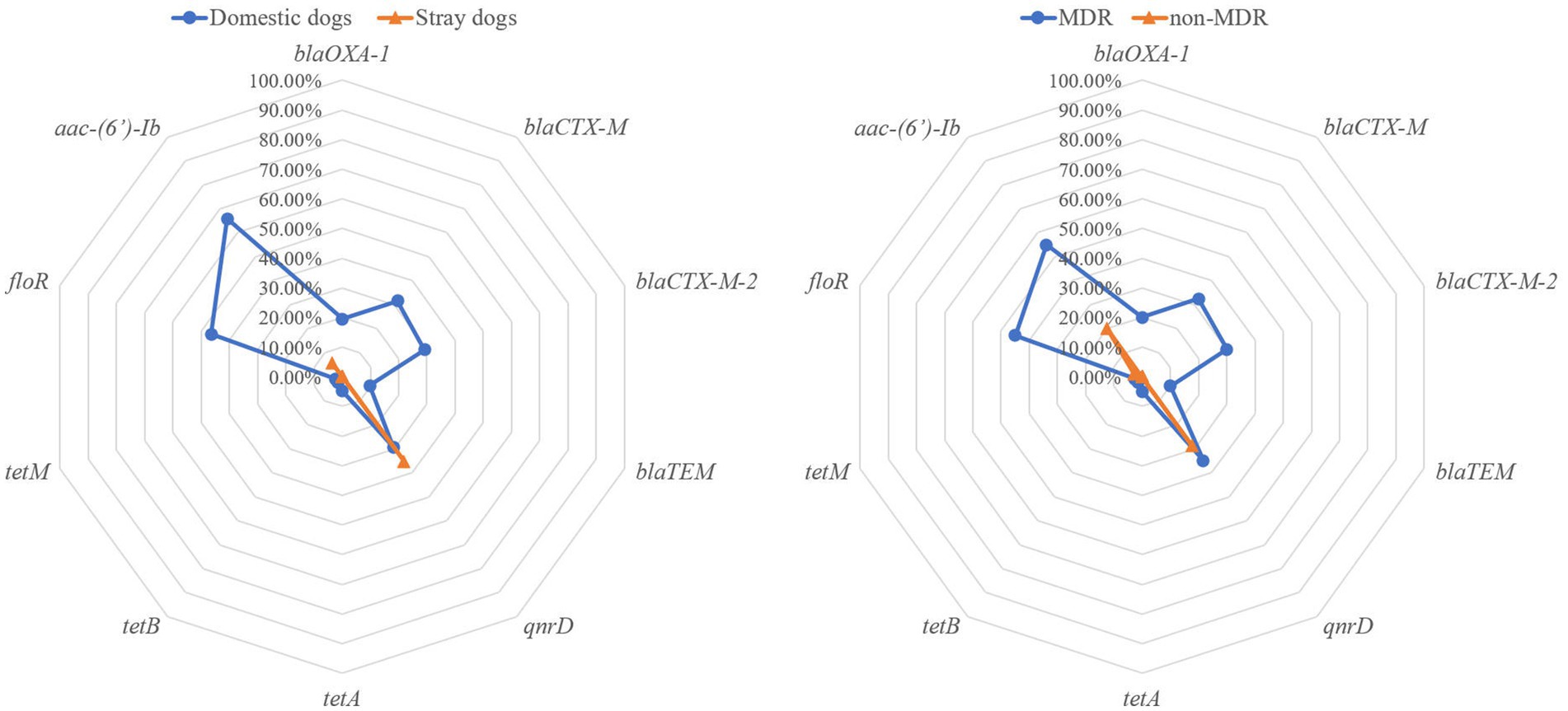
Figure 2. The prevalence of antibiotic resistance genes of the Proteus mirabilis isolates. The radar map on the left shows the antibiotic resistance genes carried by P. mirabilis strains from domestic and stray dogs. The radar map on the right shows the antibiotic resistance genes carried by MDR and non-MDR strains.
3.7. Correlations analysis
In this study, the correlation coefficient was utilized to evaluate the relationships between these factors. Figure 3 illustrates the intra-group and inter-group correlations of AMR, VAGs, and ARGs for 75 P. mirabilis isolates (excluding non-statistically significant data). There were 51 pairs of the 15 antibiotics that exhibited positive correlations, with the most significant positive correlation found between CIP and C (r = 0.80, p < 0.001). Negative correlations were shown between IPM and AMP (r = −0.25, p < 0.05), and between IPM and CTX (r = −0.32, p < 0.01). However, no significant association was observed between MEM and any of the other antibiotics. Analysis of the correlation between AMR and VAGs revealed that only AMP had a negative correlation with VAGs (hpmA, hpmB, pmfA, mrpA and rsbA), and the most significant correlation was found between AMP and mrpA (r = −0.46, p < 0.001). Meanwhile, there were 67 pairs of antimicrobial and ARGs positively correlated, and 3 pairs were negatively correlated. There was the most significant correlation between CTX of β-lactam and blaCTX-M, and between ATM and tetM (r = 1.00, p < 0.001).
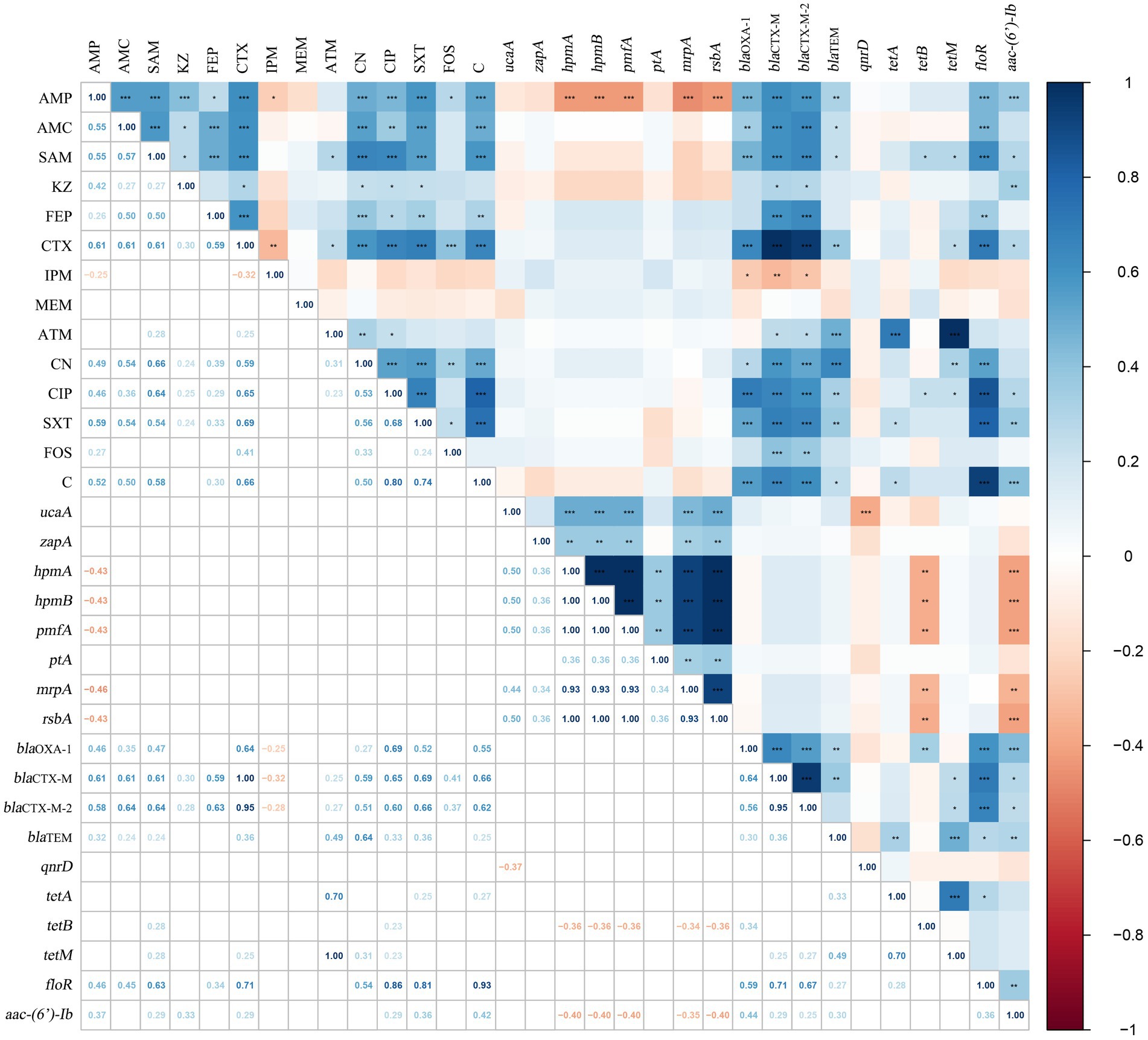
Figure 3. The correlations among AMR, VAGs and ARGs of Proteus mirabilis isolates. The numbers in the heat map represent the correlation coefficient (r). Positive numbers (blue) show a positive correlation and negative numbers (red) show a negative correlation.
Furthermore, 25 pairs of the 8 VAGs showed positive correlation with hmpA, hpmB, pmfA and rsbA exhibiting the strongest correlations (r = 1.00, p < 0.001). Similarly, 21 pairs of the 10 ARGs showed positive correlations, with the strongest correlation observed between blaCTX-M and blaCTX-M-2 (r = 0.95, p < 0.001). Additionally, the correlation analysis between VAGs and ARGs revealed that 11 pairs of VAGs and ARGs exhibited negative correlations. Among them, aac-(6′)-Ib had the strongest negative correlations with hpmA, hpmB, pmfA, and rsbA (r = −0.40, p < 0.01). Specific results of the correlation analysis can be found in Figure 3. In the present study, we examined the correlation between ARGs and the integrons of 40 MDR strains (Figure 4). Our results revealed a significant positive correlation between class 1 integrons and the β-lactam-resistant genes blaTEM (r = 0.31, p < 0.05), blaCTX-M (r = 0.35, p < 0.05), and blaCTX-M-2 (r = 0.35, p < 0.05). It is noteworthy that a significant negative correlation was observed between qnrD and the class 2 integrons.
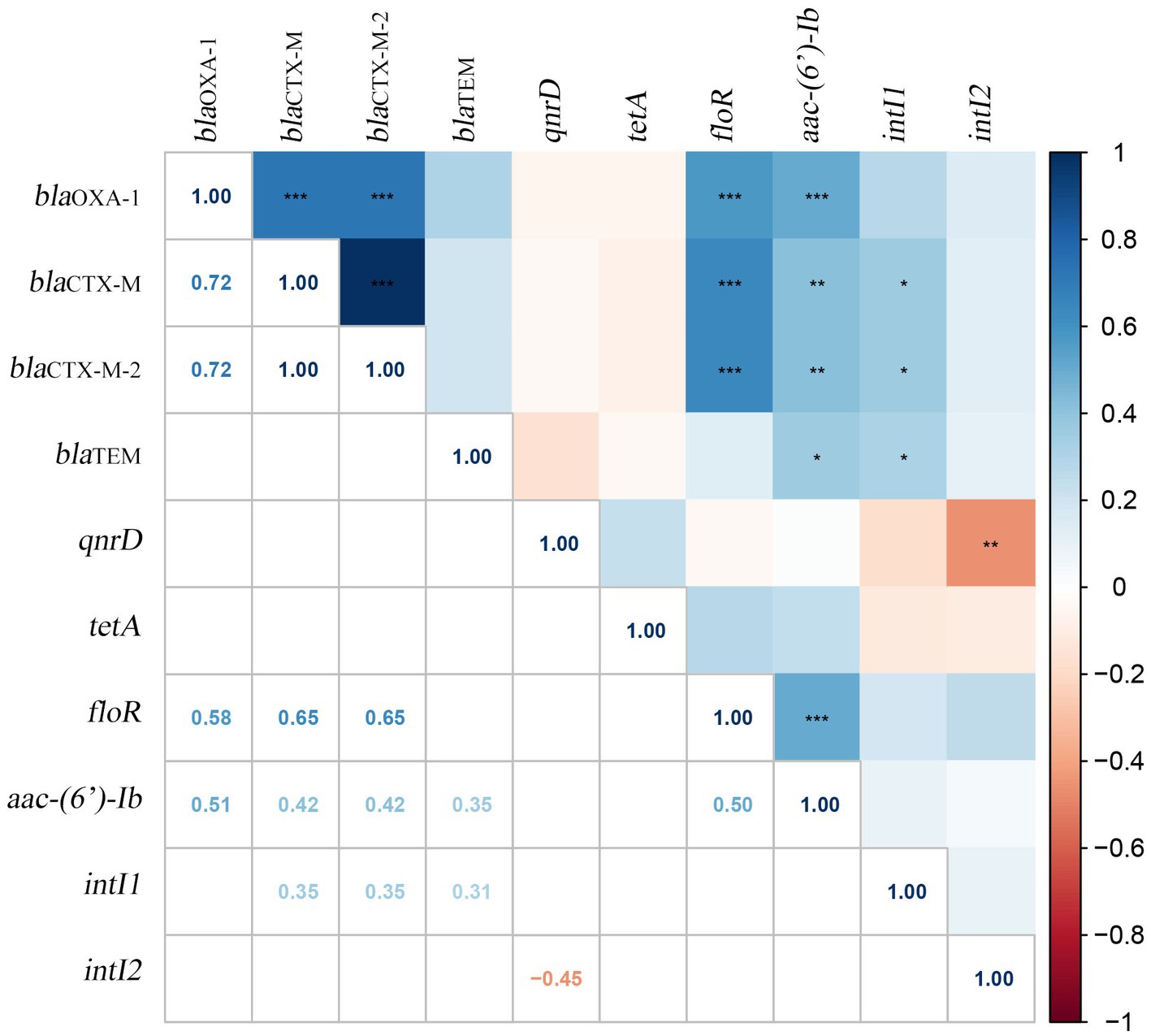
Figure 4. Correlation between ARGs and integrons in 40 MDR Proteus mirabilis strains. The numbers in the heat map represent the correlation coefficient (r). Positive numbers (blue) show a positive correlation and negative numbers (red) show a negative correlation.
4. Discussion
Proteus mirabilis has been extensively researched as a urinary tract pathogen in humans over the past decades (Durgadevi et al., 2020; Kot et al., 2021; Wilks et al., 2021). However, there is limited research focusing on P. mirabilis isolated from dog feces. This study aims to investigate the prevalence and biological characteristics of P. mirabilis in the feces of domestic and stray dogs in Chengdu, southwestern China, to determine the potential threat it poses to public health.
Out of 241 samples of dog feces, a total of 75 non-duplicate P. mirabilis strains were isolated, with a prevalence of 31.12%. This prevalence is similar to that found in a study of dogs with diarrhea in Northeast China (Sun et al., 2020), where 76 out of 232 samples had P. mirabilis infection, accounting for 32.76%. In the same study, Sun et al. also investigated the presence of P. mirabilis in the feces of other animals (mink, cattle, and fowl) with diarrhea. Notably, dogs had the highest infection rate among the animals tested. In our findings, the prevalence of P. mirabilis in the feces of stray dogs was significantly higher than that of domestic dogs (p < 0.01). This may be attributed to the fact that P. mirabilis is commonly present in soil and sewage, and stray dogs are more susceptible to exposure to these contaminated sources, resulting in the infection of P. mirabilis. This agrees with previous studies conducted on non-human species, such as Namibian cheetahs and chimpanzees, suggesting that gut microbiota can be influenced by the living environment (Vilson et al., 2018; Reese et al., 2021).
The 75 P. mirabilis isolates from this study exhibited the abilities of swarming motility and biofilm formation. There was no significant difference in swarming motility and biofilm formation between MDR and non-MDR isolates. This finding contrasts with a study conducted by Filipiak et al., which reported that multi-drug sensitive (MDS) strains had weaker biofilm formation but stronger swarming motility compared to MDR strains (Filipiak et al., 2020). Currently, AMR and MDR of bacteria derived from animals are the focus of attention. In this study, a high resistant rate was observed for most P. mirabilis isolates to KZ (70.67%) and IPM (70.67%). This resistance rate was higher than that reported in other studies, such as P. mirabilis isolated from dogs in Japan (Harada et al., 2014) and P. mirabilis isolated from dogs with bacterial urine (Decôme et al., 2020). Although current research on the AMR of P. mirabilis isolated from dogs reports varying results, a consistent trend across these studies is the gradual increase in AMR of P. mirabilis over time (Decôme et al., 2020). Furthermore, differences in AMR were observed among P. mirabilis strains from domestic and stray dogs. Except for natural resistance to tetracycline, the highest resistance to KZ (87.80%) was found in the P. mirabilis isolated from domestic dogs, followed by AMP (58.54%), IPM (58.54%), SXT (51.22%) and C (51.22%). There was no significant difference in AMR to FEP, MEM, ATM, F, and FOS between P. mirabilis isolated from domestic and stray dogs (p > 0.05). For the resistance to IPM, the AMR of P. mirabilis isolated from stray dogs was higher than that of domestic dogs. For the rest of the antibiotics, the opposite is true. The emergence of MDR bacteria is reflected as a risk factor for public health. A total of 30 (75.00%, 30/40) MDR strains were isolated from domestic dogs, which was significantly higher than 10 (25.00%, 10/40) MDR strains isolated from stray dogs. This phenomenon may be related the widespread use (and frequent misuse) of antibiotics in clinical, and stray dogs rarely receive antibiotic treatment from veterinarians (Liu C. et al., 2021; Liu Y. et al., 2021).
Aside from antimicrobial resistance, virulence factors are also receiving considerable attention. The current investigation identified a total of 11 VAGs, with detection rates exceeding 90% for all except mrpA (89.33%) and ucaA (70.67%). Notably, the ureC gene was detected in 100% of isolates, which was consistent with the result that all isolates were capable of producing urease. The detection rates of VAGs in this study were found to be higher compared to a prior study conducted in Northeast China, which fecal swabs collected from diarrhea animals (dogs, mink, cattle and poultry) demonstrated ureC as the most prevalent VAG, observed in 90.91% of samples (Sun et al., 2020). Similar results were obtained in a study conducted on P. mirabilis isolated from ducks, where 100% detection rates of ureC, 94.3% for rsbA, and 91.4% for zapA were recorded, consistent with our findings (Algammal et al., 2021). Interestingly, a high detection rate of ucaA (76.47%) was observed in 34 P. mirabilis isolates from stray dogs was, with all other VAGs demonstrating 100% detection rates. Comparatively, P. mirabilis strains isolated from stray dogs exhibited a greater number of VAGs when compared to those isolated from domestic dogs. Previous research has shown that the microbiota of captive animals harbors more VAGs in comparison to that of wild animals (Guo et al., 2019; Liu C. et al., 2021). Due to increased interactions between captive animals and humans, horizontal gene transfer from other bacteria in the environment (such as air and water) leads to virulence factor accumulation (Liu C. et al., 2021). Indeed, the environmental factors surrounding stray dogs, such as exposure to rotten food, sewage and other pollutants, along with inadequate living conditions, are likely to be contributing factors to the higher incidence of VAGs detected in P. mirabilis isolated from these animals. In contrast, wild animals and domestic dogs may have a broader range of dietary options and cleaner lifestyles, which might result in the reduced chance of acquisition of VAGs.
In addition, the present study also involved the examination of ARGs carried by P. mirabilis isolates. Out of the 18 screened ARGs, 10 were detected in the 75 isolates, with aac-(6′)-Ib (38.67%), qnrD (32.00%), floR (25.33%), blaCTX-M (17.33%), and blaCTX-M-2 (16.00%) being the top five ARGs detected. A Japanese study demonstated that clinical isolates of P. mirabilis did not exhibit positivity for qnrA, qnrB, qnrS, and aac (6′)-Ib-cr; however, 1.9% (2/105) of isolates were found to be positive for qnrD (Harada et al., 2014). Our study revealed higher rates of ARGs detection compared to a study from Brazil which detected only 4 ARGs (blaTEM, blaSHV, blaCTX-M-1, and blaOXA-1) in P. mirabilis isolated from dogs (Sfaciotte et al., 2021). However, the detection rates of ARGs in our study were lower than a previous study that detected 7 out of 14 ARGs in clinical isolates of P. mirabilis obtained from humanin northern Taiwan (Lin et al., 2019). The integrons system possesses the capability to capture and express exogenous ARGs. Bacteria have the perceptive capacity to acquire and aggregate ARGs from their surrounding environment through the integrase mediated by intI within the system, allowing for the formation of extensive drug-resistant gene arrays. Consequently, integrons assume a crucial role in the evolution of MDR bacteria and horizontal diffusion of ARGs determinants. Class 1 integrons and Class 2 integrons were detected in the MDR strain in this study, which was higher than the detection rate in a previous study on cooked meat products in China, but lower than the detection rate in Chinese human isolates (Yu et al., 2017; Lu et al., 2022). Significantly, the P. mirabilis isolates from domestic dogs were found to carry a significantly more ARGs compared to those isolated from stray dogs (p < 0.05). This observation is consistent with the result that P. mirabilis isolated from domestic dogs exhibited a more severe antimicrobial resistance phenotype than that isolated from stray dogs. In 2019, a study suggested that UTIs in companion animals (dogs and cats) and humans may could be caused by closely related strains of P. mirabilis, sharing similar ARGs in both sources (Marques et al., 2019), indicating a potential public health risk. To mitigate this risk, veterinarians must use antibiotics judiciously and avoid overuse. Additionally, researchers can explore alternative therapies such as phages and lactobacillus to combat P. mirabilis infections (Melo Luís et al., 2016; Shaaban et al., 2020).
A previous study showed that the occurrence and the positive correlations of VAGs and ARGs can be used as a reference for the regulatory use of antibiotics to stop the direct or indirect transmission of these resistance and virulent microbes to the natural environment (Zhang S. et al., 2021). So, we further analyzed the correlation among AMR, VAGs, and ARGs of P. mirabilis isolates. In this study, there was a positive correlation between the resistance phenotypes of P. mirabilis isolates to various antibiotics. There was also a significant positive correlation between the ARGs they carried. In addition, there was a significant positive correlation between AMR and ARGs, which were consistent with the statement of Algammal et al. (2021). It is worth noting that there were only negative correlations between AMR and VAGs, which were also reflected in the correlations between ARGs and VAGs. A previous study showed that MDR strains of P. mirabilis isolated from ducks did not differ significantly from the VAGs carried by non-MDR strains (Algammal et al., 2021). Therefore, our results are different from previous studies. Moreover, class 1 integrons are positively correlated with β-lactam resistance genes (blaCTX-M, blaCTX-M-2, blaTEM), which means that these genes may be on class 1 integrons.
5. Conclusion
This study has shown that P. mirabilis strains isolated from domestic dogs carrying fewer VAGs but more ARGs compares to those isolated from stay dogs. In addition, we observed a negative correlation between VAGs and ARGs, which is different from previous studies and requires further study at a later stage. Increased antimicrobial resistance has been detected in MDR P. mirabilis isolates over the past 10 years, and therefore the need to control the use of antimicrobial agents in animals to minimize the emergence and eventual spread of resistant pathogens is necessary to protect human and animal health.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding authors.
Author contributions
GP, LL, MD, SA, and XQ: conceptualization, methodology, and software. LL, ZD, SC, SA, and QL: data curation and writing-original draft preparation. ZZ, HL, XM, ZR, and XQ: visualization and investigation. YH, ZR, HF, and GS: supervision. LL, MD, SA, and ZZ: software and validation. LL, ZD, SC, MD, XQ, and GP: writing-reviewing and editing. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by “Research on Prevention and Control Technology and Regulation Products of Important Nutritional Metabolic Diseases of Pets,” a key Special project of “13th Five-Year Plan,” Ministry of Science and Technology (2016YFD0501009).
Acknowledgments
The authors are grateful to the providing at the Center of Protect Beastie in Sichuan Province and the veterinaries at the pet hospitals for their help with collecting the samples.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1141418/full#supplementary-material
References
Algammal, A. M., Hashem, H. R., Alfifi, K. J., Hetta, H. F., Sheraba, N. S., Ramadan, H., et al. (2021). atpD gene sequencing, multidrug resistance traits, virulence-determinants, and antimicrobial resistance genes of emerging XDR and MDR-Proteus mirabilis. Sci. Rep. 11:9476. doi: 10.1038/s41598-021-88861-w
Armbruster, C. E., Mobley, H. L. T., and Pearson, M. M. (2018). Pathogenesis of Proteus mirabilis infection. EcoSal Plus 8:ecosalplus. ESP-0009-2017. doi: 10.1128/ecosalplus.ESP-0009-2017
Aygül, A., Öztürk, İ., Çilli, F. F., and Ermertcan, Ş. (2019). Quercetin inhibits swarming motility and activates biofilm production of Proteus mirabilis possibly by interacting with central regulators, metabolic status or active pump proteins. Phytomedicine 57, 65–71. doi: 10.1016/j.phymed.2018.12.014
Broomfield, R. J., Morgan, S. D., Khan, A., and Stickler, D. J. (2009). Crystalline bacterial biofilm formation on urinary catheters by urease-producing urinary tract pathogens: a simple method of control. J. Med. Microbiol. 58, 1367–1375. doi: 10.1099/jmm.0.012419-0
Clinical and Laboratory Standards Institute (2020). Performance standards for antimicrobial disk and dilution susceptibility tests for Bacteria isolated from animals, 5th ed, PA: Clinical and Laboratory Standards Institute (VET01SEd5E).
Coker, C., Poore, C. A., Li, X., and Mobley, H. L. T. (2000). Pathogenesis of Proteus mirabilis urinary tract infection. Microbes Infect. 2, 1497–1505. doi: 10.1016/S1286-4579(00)01304-6
Decôme, M., Cuq, B., Fairbrother, J.-H., Gatel, L., and Conversy, B. (2020). Clinical significance of Proteus mirabilis bacteriuria in dogs, risk factors and antimicrobial susceptibility. Can. J. Vet. Res. 84, 252–258.
Durgadevi, R., Kaleeshwari, R., Swetha, T. K., Alexpandi, R., Pandian, S. K., and Ravi, A. V. (2020). Attenuation of Proteus mirabilis colonization and swarming motility on indwelling urinary catheter by antibiofilm impregnation: an in vitro study. Colloids Surf. B: Biointerfaces 194:111207. doi: 10.1016/j.colsurfb.2020.111207
Filipiak, A., Chrapek, M., Literacka, E., Wawszczak, M., Głuszek, S., Majchrzak, M., et al. (2020). Pathogenic factors correlate with antimicrobial resistance among clinical Proteus mirabilis strains. Front. Microbiol. 11:579389. doi: 10.3389/fmicb.2020.579389
Gaastra, W., Van Oosterom, R. A. A., Pieters, E. W. J., Bergmans, H. E. N., Van Dijk, L., Agnes, A., et al. (1996). Isolation and characterisation of dog uropathogenic Proteus mirabilis strains. Vet. Microbiol. 48, 57–71. doi: 10.1016/0378-1135(95)00133-6
Gao, H., Gao, Y., Yang, C., Dong, D., Yang, J., Peng, G., et al. (2018). Influence of outer membrane vesicles of Proteus mirabilis isolated from boar semen on sperm function. Vet. Microbiol. 224, 34–42. doi: 10.1016/j.vetmic.2018.08.017
Girlich, D., Bonnin, R. A., Dortet, L., and Naas, T. (2020). Genetics of acquired antibiotic resistance genes in Proteus spp. Front. Microbiol. 11:256. doi: 10.3389/fmicb.2020.00256
Guo, W., Mishra, S., Wang, C., Zhang, H., Ning, R., Kong, F., et al. (2019). Comparative study of gut microbiota in wild and captive Giant pandas (Ailuropoda melanoleuca). Genes (Basel) 10:E827. doi: 10.3390/genes10100827
Harada, K., Niina, A., Shimizu, T., Mukai, Y., Kuwajima, K., Miyamoto, T., et al. (2014). Phenotypic and molecular characterization of antimicrobial resistance in Proteus mirabilis isolates from dogs. J. Med. Microbiol. 63, 1561–1567. doi: 10.1099/jmm.0.081539-0
Hu, R., Wang, X., Muhamamd, I., Wang, Y., Dong, W., Zhang, H., et al. (2020). Biological characteristics and genetic analysis of a highly pathogenic Proteus Mirabilis strain isolated from dogs in China. Front. Vet. Sci. 7:589. doi: 10.3389/fvets.2020.00589
Hughes, D., and Andersson, D. I. (2017). Environmental and genetic modulation of the phenotypic expression of antibiotic resistance. FEMS Microbiol. Rev. 41, 374–391. doi: 10.1093/femsre/fux0004
Khoramian, B., Jabalameli, F., Niasari-Naslaji, A., Taherikalani, M., and Emaneini, M. (2015). Comparison of virulence factors and biofilm formation among Staphylococcus aureus strains isolated from human and bovine infections. Microb. Pathog. 88, 73–77. doi: 10.1016/j.micpath.2015.08.007
Kot, B., Grużewska, A., Szweda, P., Wicha, J., and Parulska, U. (2021). Antibiotic resistance of Uropathogens isolated from patients hospitalized in district Hospital in Central Poland in 2020. Antibiotics 10:447. doi: 10.3390/antibiotics10040447
Leverstein-van Hall, M. A., Blok, H. E. M., Donders, A. R. T., Paaum, A., Fluit, A. C., and Verhoef, J. (2003). Multidrug resistance among Enterobacteriaceae is strongly associated with the presence of integrons and is independent of species or isolate origin. J. Infect. Dis. 187, 251–259. doi: 10.1086/345880
Lin, M., Liou, M., Kuo, C., Lin, Y., Chen, J., and Kuo, H. (2019). Antimicrobial Susceptibility and Molecular Epidemiology of Proteus mirabilis Isolates from Three Hospitals in Northern Taiwan. Microbial. Drug Resistance 25, 1338–1346. doi: 10.1089/mdr.2019.0066
Liu, C., Hu, J., Wu, Y., Irwin, D., Chen, W., Zhang, Z., et al. (2021). Comparative study of gut microbiota from captive and confiscated-rescued wild pangolins. J. Genet. Genomics 48, 825–835. doi: 10.1016/j.jgg.2021.07.009
Liu, Y., Liu, B., Liu, C., Hu, Y., Liu, C., Li, X., et al. (2021). Differences in the gut microbiomes of dogs and wolves: roles of antibiotics and starch. BMC Vet. Res. 17:112. doi: 10.1186/s12917-021-02815-y
Lu, W., Qiu, Q., Chen, K., Zhao, R., Li, Q., and Wu, Q. (2022). Distribution and molecular characterization of functional class 2 Integrons in clinical Proteus mirabilis isolates. Infect. Drug Resist. 15, 465–474. doi: 10.2147/IDR.S347119
Magiorakos, A.-P., Srinivasan, A., Carey, R. B., Carmeli, Y., Falagas, M. E., Giske, C. G., et al. (2012). Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 18, 268–281. doi: 10.1111/j.1469-0691.2011.03570.x
Marques, C., Belas, A., Aboim, C., Trigueiro, G., Cavaco-Silva, P., Gama, L. T., et al. (2019). Clonal relatedness of Proteus mirabilis strains causing urinary tract infections in companion animals and humans. Vet. Microbiol. 228, 77–82. doi: 10.1016/j.vetmic.2018.10.015
Maszewska, A., Moryl, M., Wu, J., Liu, B., Feng, L., and Rozalski, A. (2021). Amikacin and bacteriophage treatment modulates outer membrane proteins composition in Proteus mirabilis biofilm. Sci. Rep. 11, 1522–1512. doi: 10.1038/s41598-020-80907-9
Melo Luís, D. R., Veiga, P., Cerca, N., Kropinski, A. M., Almeida, C., Azeredo, J., et al. (2016). Development of a phage cocktail to control Proteus mirabilis catheter-associated urinary tract infections. Front. Microbiol. 7:1024. doi: 10.3389/fmicb.2016.01024
Norsworthy, A. N., and Pearson, M. M. (2017). From catheter to kidney stone: the Uropathogenic lifestyle of Proteus mirabilis. Trends Microbiol. 25, 304–315. doi: 10.1016/j.tim.2016.11.015
Pathirana, H. N. K. S., Silva, S. H. M. P., Hossain, S., and Heo, G. J. (2018). Comparison of virulence genes in Proteus species isolated from human and pet turtle. Iran J. Vet. Res. 19, 48–52.
Qi, L., Li, H., Zhang, C., Liang, B., Li, J., Wang, L., et al. (2016). Relationship between antibiotic resistance, biofilm formation, and biofilm-specific resistance in Acinetobacter baumannii. Front. Microbiol. 7:483. doi: 10.3389/fmicb.2016.00483
Rather, P. N. (2005). Swarmer cell differentiation in Proteus mirabilis. Environ. Microbiol. 7, 1065–1073. doi: 10.1111/j.1462-2920.2005.00806.x
Reese, A. T., Chadaideh, K. S., Diggins, C. E., Schell, L. D., Beckel, M., Callahan, P., et al. (2021). Effects of domestication on the gut microbiota parallel those of human industrialization. elife 10:e60197. doi: 10.7554/eLife.60197
Sfaciotte, R. A. P., Parussolo, L., Melo, F. D., Wildemann, P., Bordignon, G., Israel, N. D., et al. (2021). Identification and characterization of multidrug-resistant extended-Spectrum Beta-lactamase-producing Bacteria from healthy and diseased dogs and cats admitted to a veterinary Hospital in Brazil. Microbiol. Drug Resist 27, 855–864. doi: 10.1089/mdr.2020.0043
Shaaban, M., EI-Rahman, O. A. A., AI-Qaidi, B., and Ashour, H. M. (2020). Antimicrobial and Antibiofilm activities of probiotic lactobacilli on antibiotic-resistant Proteus mirabilis. Microorganisms 8:960. doi: 10.3390/microorganisms8060960
Shelenkov, A., Petrova, L., Fomina, V., Zamyatin, M., Mikhaylova, Y., and Akimkin, V. (2020). Multidrug-resistant Proteus mirabilis strain with Cointegrate plasmid. Microorganisms 8:1775. doi: 10.3390/microorganisms8111775
Sun, Y., Wen, S., Zhao, L., Xia, Q., Pan, Y., Liu, H., et al. (2020). Association among biofilm formation, virulence gene expression, and antibiotic resistance in Proteus mirabilis isolates from diarrhetic animals in Northeast China. BMC Vet. Res. 16:176. doi: 10.1186/s12917-020-02372-w
Vilson, Å., Ramadan, Z., Li, Q., Hedhammar, Å., Reynolds, A., Spears, J., et al. (2018). Disentangling factors that shape the gut microbiota in German shepherd dogs. PLoS One 13:e0193507. doi: 10.1371/journal.pone.0193507
Wilks, S. A., Koerfer, V. V., Prieto, J. A., Fader, M., and Keevil, C. W. (2021). Biofilm development on urinary catheters promotes the appearance of viable but Nonculturable Bacteria. mBio 12:e03584-20. doi: 10.1128/mBio.03584-20
Wong, M. H. Y., Wan, H. Y., and Chen, S. (2013). Characterization of multidrug-resistant Proteus mirabilis isolated from chicken carcasses. Foodborne Pathog. Dis. 10, 177–181. doi: 10.1089/fpd.2012.1303
Yin, D., Gao, Q., Zhu, H., and Li, J. (2020). Public perception of urban companion animals during the COVID-19 outbreak in China. Health Place 65:102399. doi: 10.1016/j.healthplace.2020.102399
Yu, T., Jiang, X., Liang, Y., Zhu, Y., Tian, J., Wang, X., et al. (2017). Characterization and horizontal transfer of antimicrobial resistance genes and Integrons in Bacteria isolated from cooked meat products in China. J. Food Prot. 80, 2048–2055. doi: 10.4315/0362-028X.JFP-17-119
Zhang, S., Chen, S., Rehman, M. U., Yang, H., Yang, Z., Wang, M., et al. (2021). Distribution and association of antimicrobial resistance and virulence traits in Escherichia coli isolates from healthy waterfowls in Hainan, China. Ecotoxicol. Environ. Saf. 220:112317. doi: 10.1016/j.ecoenv.2021.112317
Zhang, J. W., Hoedt, E. C., Liu, Q., Berendsen, E., Teh, J. J., Hamilton, A., et al. (2021). Elucidation of Proteus mirabilis as a key bacterium in Crohn’s disease inflammation. Gastroenterology 160, 317–330.e11. doi: 10.1053/j.gastro.2020.09.036
Zhao, X., Lv, Y., Adam, F. E. A., Xie, Q., Wang, B., Bai, X., et al. (2021). Comparison of antimicrobial resistance, virulence genes, Phylogroups, and biofilm formation of Escherichia coli isolated from intensive farming and free-range sheep. Front. Microbiol. 12:699927. doi: 10.3389/fmicb.2021.699927
Keywords: Proteus mirabilis , virulence-related factors, antimicrobial resistance, antibiotic resistance genes, virulence-associated genes, domestic and stray dogs
Citation: Liu L, Dong Z, Ai S, Chen S, Dong M, Li Q, Zhou Z, Liu H, Zhong Z, Ma X, Hu Y, Ren Z, Fu H, Shu G, Qiu X and Peng G (2023) Virulence-related factors and antimicrobial resistance in Proteus mirabilis isolated from domestic and stray dogs. Front. Microbiol. 14:1141418. doi: 10.3389/fmicb.2023.1141418
Edited by:
Weiqi He, Soochow University, ChinaReviewed by:
Piklu Roy Chowdhury, University of Technology Sydney, AustraliaAdriana Belas, Universidade Lusófona, Portugal
Copyright © 2023 Liu, Dong, Ai, Chen, Dong, Li, Zhou, Liu, Zhong, Ma, Hu, Ren, Fu, Shu, Qiu and Peng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Guangneng Peng, pgn.sicau@163.com; Xianmeng Qiu, 13008142361@126.com
†These authors have contributed equally to this work and share first authorship
 Lijuan Liu1†
Lijuan Liu1† Ziyao Zhou
Ziyao Zhou Haifeng Liu
Haifeng Liu Xiaoping Ma
Xiaoping Ma Zhihua Ren
Zhihua Ren Gang Shu
Gang Shu Guangneng Peng
Guangneng Peng