- 1Institute of Urology, The Third Affiliated Hospital of Shenzhen University, Shenzhen, China
- 2Shenzhen Institute of Synthetic Biology, Shenzhen Institutes of Advanced Technology, Chinese Academy of Sciences, Shenzhen, China
- 3Teaching Center of Shenzhen Luohu Hospital, Shantou University Medical College, Shantou, China
- 4Medical Laboratory of Shenzhen Luohu People’s Hospital, Shenzhen, China
- 5School of Medicine, Anhui University of Science and Technology, Huainan, China
- 6Kobilka Institute of Innovative Drug Discovery, School of Medicine, The Chinese University of Hong Kong, Shenzhen, Guangdong, China
- 7South China Hospital, Health Science Center, Shenzhen University, Shenzhen, China
Introduction: Klebsiella pneumonia (K. pneumonia) is a Gram-negative bacterium that opportunistically causes nosocomial infections in the lung, bloodstream, and urinary tract. Extended-spectrum β-Lactamases (ESBLs)-expressed K. pneumonia strains are widely reported to cause antibiotic resistance and therapy failure. Therefore, early identification of K. pneumonia, especially ESBL-positive strains, is essential in preventing severe infections. However, clinical detection of K. pneumonia requires a time-consuming process in agar disk diffusion. Nucleic acid detection, like qPCR, is precise but requires expensive equipment. Recent research reveals that collateral cleavage activity of CRISPR-LbCas12a has been applied in nucleic acid detection, and the unique testing model can accommodate various testing models.
Methods: This study established a system that combined PCR with CRISPR-LbCas12a targeting the K. pneumoniae system. Additionally, this study summarized the antibiotic-resistant information of the past five years’ K. pneumoniae clinic cases in Luohu Hospital and found that the ESBL-positive strains were growing. This study then designs a crRNA that targets SHV to detect ESBL-resistant K. pneumoniae. This work is to detect K. pneumoniae and ESBL-positive strains’ nucleic acid using CRISPR-Cas12 technology. We compared PCR-LbCas12 workflow with PCR and qPCR techniques.
Results and Discussion: This system showed excellent detection specificity and sensitivity in both bench work and clinical samples. Due to its advantages, its application can meet different detection requirements in health centers where qPCR is not accessible. The antibiotic-resistant information is valuable for further research.
Introduction
Klebsiella pneumonia (K. pneumonia) is a family of Gram-negative bacteria that causes nosocomial infections in the bloodstream, wound, and urinary tract (Magill et al., 2014). Hypervirulent K. pneumoniae strains, such as K1, K2, and K5, have emerged worldwide and caused severe infections, including liver abscess and pneumonia, with a mortality rate as high as 20–30% (Podschun and Ullmann, 1998).
β-Lactamases (ESBLs) can degrade β-Lactam antibiotics into non-effective compounds, thus resulting in drug-resistant strains (Bush and Bradford, 2016). However, due to the excessive use of β-Lactam antibiotics, the prevalence of ESBL-producing K. pneumonia has primarily increased. To date, ESBL-producing K. pneumonia contributes to nearly 45% of K. pneumoniae nosocomial infections (Miftode et al., 2021) and 43% in the intensive care unit (Paterson et al., 2004; Calbo et al., 2011). More strikingly, ESBL-positive strains result in significantly higher mortality (Miftode et al., 2021); a recent study reported that ESBL-producing K. pneumonia is associated with over 55% mortality (Starzyk-Luszcz et al., 2017). Despite various genes encoding ESBLs, most ESBLs were derived from one or two amino acid substitutions of SHV-1 and TEM-1 (Ramdani-Bouguessa et al., 2011; Ben Achour et al., 2014). SHV-type ESBLs almost evolved from SHV-1; for example, SHV-2 harbors G238S (Zhong et al., 2021). Over 100 SHV variants have been found,1 and most of which are associated with ESBL-positive strains. Until 2016, the SHV-type ESBLs only accounted for 10% ESBLs. However, the majority are found in K. pneumoniae (Castanheira et al., 2016). Thus, the detection of SHV is valuable in identifying ESBLs in K. pneumoniae.
Klebsiella pneumonia colonies derived from clinical samples under 24–48 h of 37°C incubation culture after disk diffusion present features that could be diagnosed by well-trained personnel (Wagner et al., 2016). Usually, 1 week is required to determine the specific antibiotic resistance of bacterial strain (Giske et al., 2022). Despite the low cost and the simplicity of operation, disk diffusion is time-consuming and sometimes produces false-negative results especially for atypical colonies (Johnson et al., 2006; Hutchison et al., 2018). In contrast, nucleic acid detection methods that detect K. pneumoniae-specific DNA fragments are more advantageous. For example, quantitative real-time PCR or qPCR is the most widely used one (Hyun et al., 2019). The K. pneumonia and specific drug-resistant strains can be determined by qPCR (Yan et al., 2021). For ESBL testing, SHV-1, TEM, and CTX-M gene DNA fragments are used (Souverein et al., 2017). However, qPCR requires expensive equipment, which restricts its application in the healthcare center.
The CRISPR-Cas systems have been discovered to cleave target DNA or RNA under the guidance of crRNA in a base-pairing manner (Yan et al., 2019). Recent research shows that Cas13 and Cas12 exhibit collateral cleavage activity that could degrade probes if crRNA perfectly base-pairs targeted RNAs or DNAs (Chen et al., 2018; Gootenberg et al., 2018). Combined with DNA amplification and signal detection methods, Cas12 and Cas13 have been applied to detect nucleic acid (Li et al., 2019). The detection workflow only requires 37°C incubation and does not rely on complicated equipment. To combat COVID-19, lots of nucleic acid detection workflow based on Cas13 and Cas12 have been developed (Kostyusheva et al., 2021; Nouri et al., 2021). Both Cas12 and Cas13 require no expensive equipment and are compatible with various amplification or detection methods.
Despite these advantages, the CRISPR-Cas12 detection method targeting K. pneumonia has not been reported. We established a sensitive nucleic acid detection method based on CRISPR-Cas12 and PCR to detect K. pneumonia. It produces results in less than 2 h and requires less expensive equipment (Figure 1A). Furthermore, we tried to identify ESBLs by targeting SHV DNA fragments. Compared to the traditional detection methods, this trial may help community healthcare centers to accomplish nucleic acid detection.
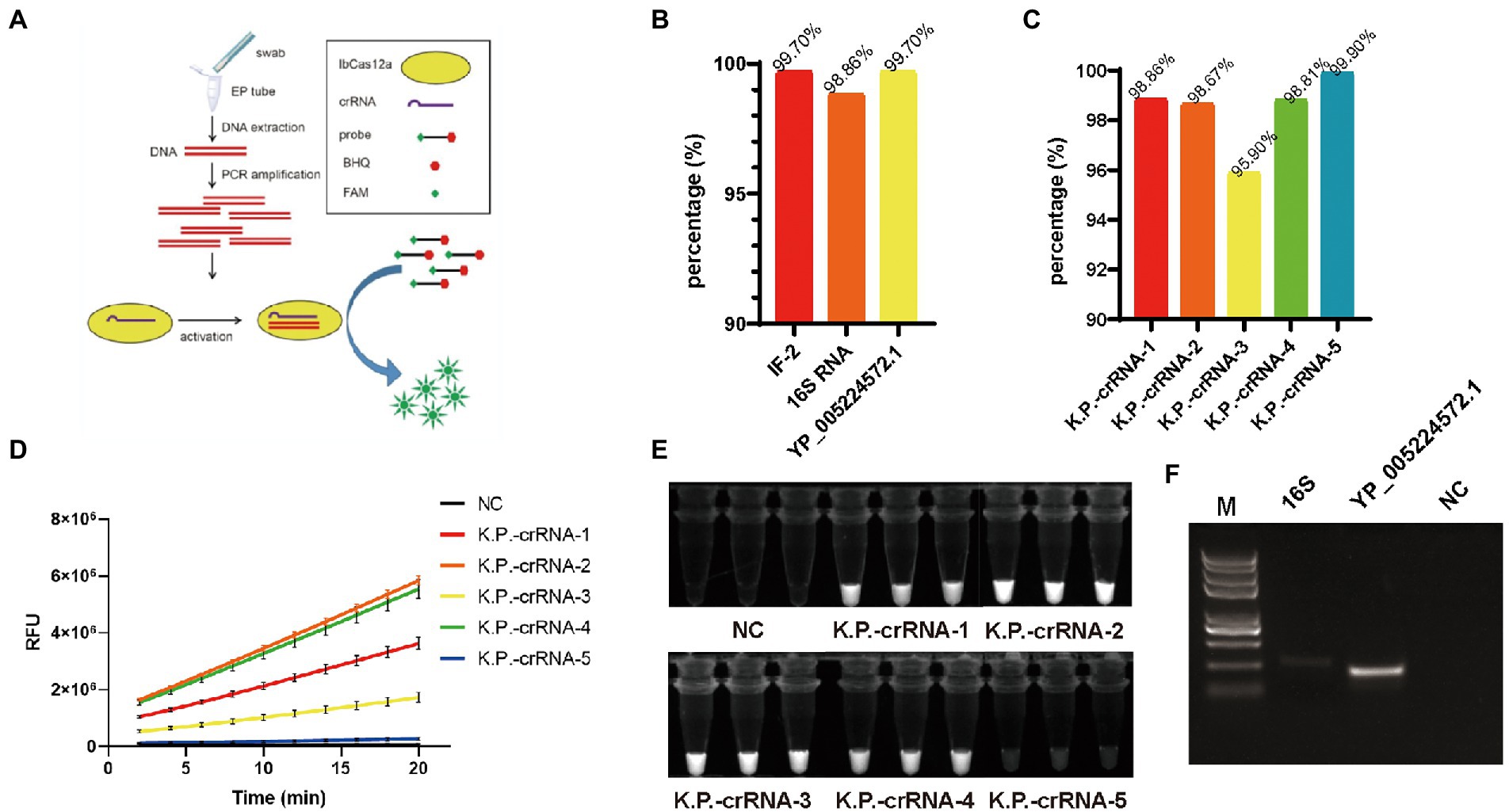
Figure 1. (A) Graphical illustration of PCR-LbCas12a detecting Klebsiella pneumonia nucleic acid. (B) The proportion of IF-216SRNA and YP_005224572.1 in the 2024 K. pneumonia genomes. (C) The percentage of each K.P.-crRNA hitting the 2024 K. pneumonia genomes. (D) FAM signal of five K.P.-crRNA activity in LbCas12a reaction. (E) PCR-LbCas12a reaction products of (D) were photographed under UV activation. (F) The PCR amplification efficiency of primers targeting16S and YP_005224572.1.
Results
Establishment of PCR-LbCas12a detection targeting Klebsiella pneumonia
To find a suitable target for nucleic acid detection, we used blast to screen the most relevant gene as a target for nucleic acid detection. We downloaded 14,288 genomic sequences from NCBI and removed the sequences under 5.2 M, for this is a complete genomic DNA size for K. pneumonia. As a result, 2,024 sequences survived for further analysis. We found that IF-2, 16S RNA, and YP_005224572.1 compose 99.70% (2018/2024), 98.86% (2001/2024), and 99.70% (2018/2024) of all the sequences (Figure 1B). Therefore, they are suitable targets for nucleic acid detection. Considering the TTTV PAM sequences required for LbCas12a targeting, we designed five 36-nt long crRNAs which share 19 nt standards nucleotides and have 17 nt unique sequences to base-pair different K. pneumoniae DNA targets. K.P.-crRNA-1,2 target 16S RNA, K.P.-crRNA-3,4 target YP_005224572.1, and K.P.-crRNA-5 targets IF-2. We also screened the K. pneumonia genomes and found that these five crRNA targets separately 98.86% (2001/2024), 98.67% (1997/2024), 95.90% (1941/2024), 98.81% (2000/2024), and 99.90% (2022/2024) of the genomes (Figure 1C). To quickly test the detecting efficiency of the crRNAs, chemically synthetic single-stranded DNA targets were added to the LbCas12a system. Compared to the negative control, all crRNAs generated a significantly high level of FAM signals (Figure 1D). Further, the free FAM group generated by digesting probes can generate 488 nm fluorescent under 360 nM UV light exposure (Wang et al., 2022). We photographed the tube after the Cas12a reaction (Figure 1E). The K.P.-crRNA-2 and K.P.-crRNA-4 generate potent and stable FAM signals.
Next, we test the primers to amplify DNA fragments for crRNA-2 and crRNA-4. The amplification efficiency of YP_005224572.1 primers is far more efficient than that of 16S (Figure 1F). To test the specificity one step further, we blast the K.P.-crRNA-2,4, 16S, YP_005224572.1 in other Klebsiella strains (Supplementary Table 1). The results showed that the K.P.-crRNA-2 targets all of them, while the K.P.-crRNA-4 perfectly targets only Klebsiella oxytoca. Klebsiella michiganensis, Klebsiella variicola, and Klebsiella Africana have mutations on the targets, which might not be detected. As a result, K.P.-crRNA-4 and the primers targeting the YP_005224572.1 gene are used to establish this system to detect the K. pneumoniae strain (Figure 1A).
PCR-LbCas12a is sensitive and specific in Klebsiella pneumonia nucleic acid detection
To explore the minimum amount of DNA sample required for nucleic acid detection, serial diluted standard DNA samples were to test PCR, qPCR, and PCR-LbCas12a techniques. Basic PCR exhibited positive signals when target DNA was as few as 10 copies (Figures 2A–C). In contrast, the LbCas12a system and qPCR can display a signal in 40 min when the copy number is as few as one single copy (Figure 2A). Next, to confirm if the detection system’s specificity targets only K. pneumoniae, we applied PCR-LbCas12a detection in 10 commonly seen pathogens in laboratory department, including Escherichia coli (E. coli), Staphylococcus aureus (S. aureus), Shigella dysenteriae, Salmonella enterica, Pseudomonas aeruginosa, Proteus mirabilis, Stenotrophomonas maltophilia, Acinetobacter baumannii, Corynebacterium striatum, and Candida albicans (fungi).
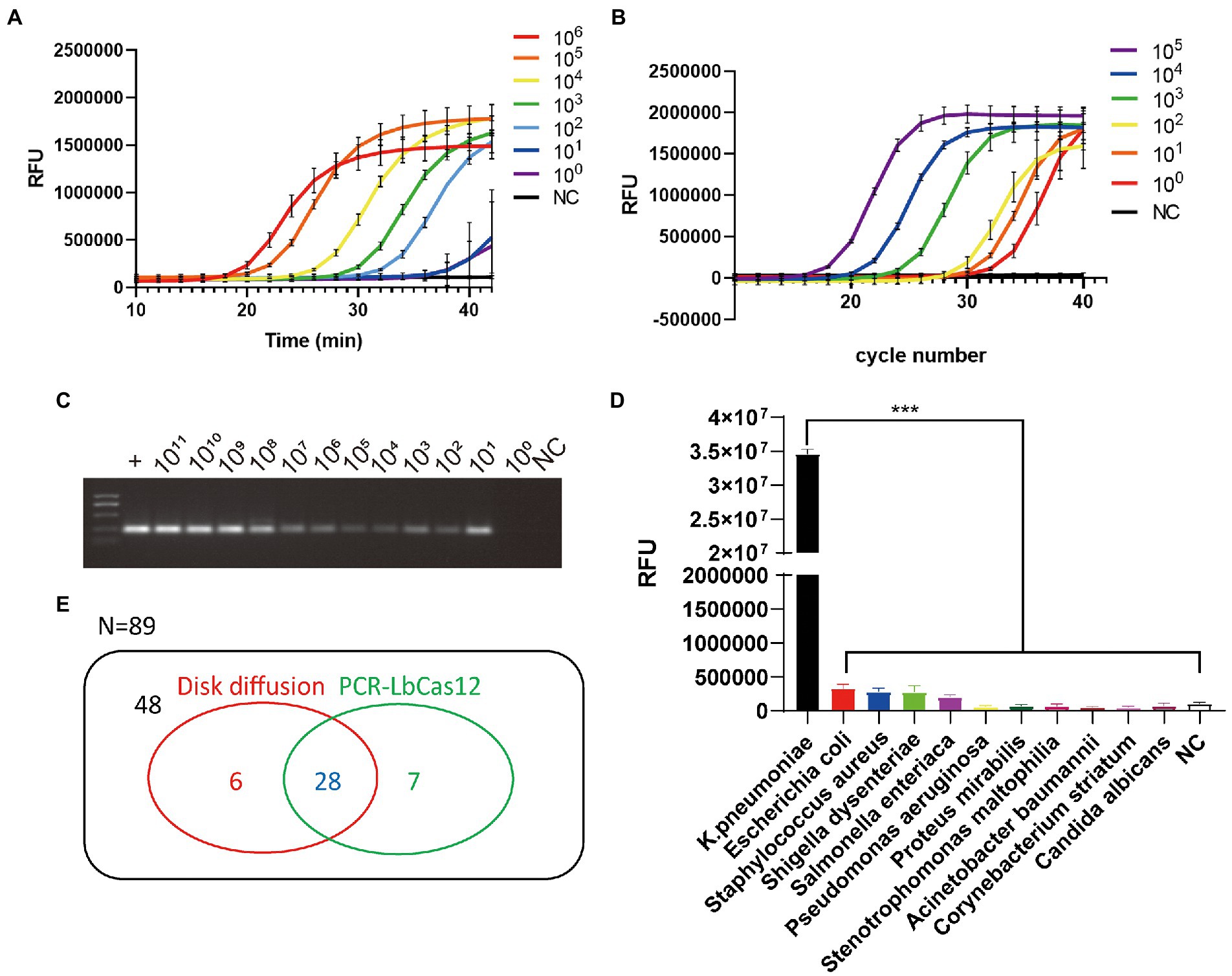
Figure 2. (A) The RFU signal generated by the PCR-LbCas12a system detecting the serially diluted standard YP_005224572.1 DNA. (B) The qPCR results of serial diluted standard DNA samples. (C) 1% agarose gel electrophoresis of the PCR product from serial diluted standard DNA samples. (D) Different pathogens was detected by PCR-Cas12a system. Only Klebsiella pneumoniae was successfully detected. Data are mean ± s. d. of n = 3 biological independent experiments. (E) Venn diagram of results of testing 89 sputum samples in disk diffusion assay and PCR-LbCas12a workflow.
As a result, after 40 min PCR-Cas12 reactions, only K. pneumonia generated a significantly higher FAM signal (Figure 2D). The PCR-LbCas12a detection system is sensitive and specific.
PCR-LbCas12a detects clinic samples
To apply this PCR-LbCas12a detection system to clinic use, we collected 89 sputum samples tested using disk diffusion. Total DNA was extracted from the samples. Then, PCR was processed and followed by LbCas12a incubation at 37°C for 20 min. Results were collected by photographing samples under UV light exposure (Supplementary Figure S1). Most of the positive and the negative samples were identical. Six samples were only positive in disk diffusion assay, and seven samples were only positive in PCR-LbCas12a workflow (Figure 2E). To verify the controversial samples, we PCR amplified them and processed Sanger sequencing. The Sanger sequence results showed that they are nearly 100% identical to the K. pneumonia YP_005224572.1 gene fragment. Meanwhile, seven samples that are positive failed to be detected in this system. This failure might result from the corruption of the sputum samples, which were not well preserved after disk diffusion for diagnosis.
Detection of SHV In ESBL-producing Klebsiella pneumonia
To start the task, we looked up the drug-resistant information of K. pneumonia in the medical laboratory department’s record since 2018. The ESBL-positive strains’ percentage is increasing from 22.9% (2018) to 40.3% (2020) and then remains at a relatively high level (Figure 3A). This information suggests that the identification of ESBL-positive strains is valuable. SHV-associated ESBLs are the most commonly reported groups, so we designed four crRNAs that target SHV to detect ESBLs. We also screened the 184 reported SHV genes by this four crRNAs, and found that SHV-crRNA1-4 target 93.47% (172/184), 93.47% (172/184), 93.47% (172/184), and 98.91% (182/184) of 184 SHV genes (Figure 3B).
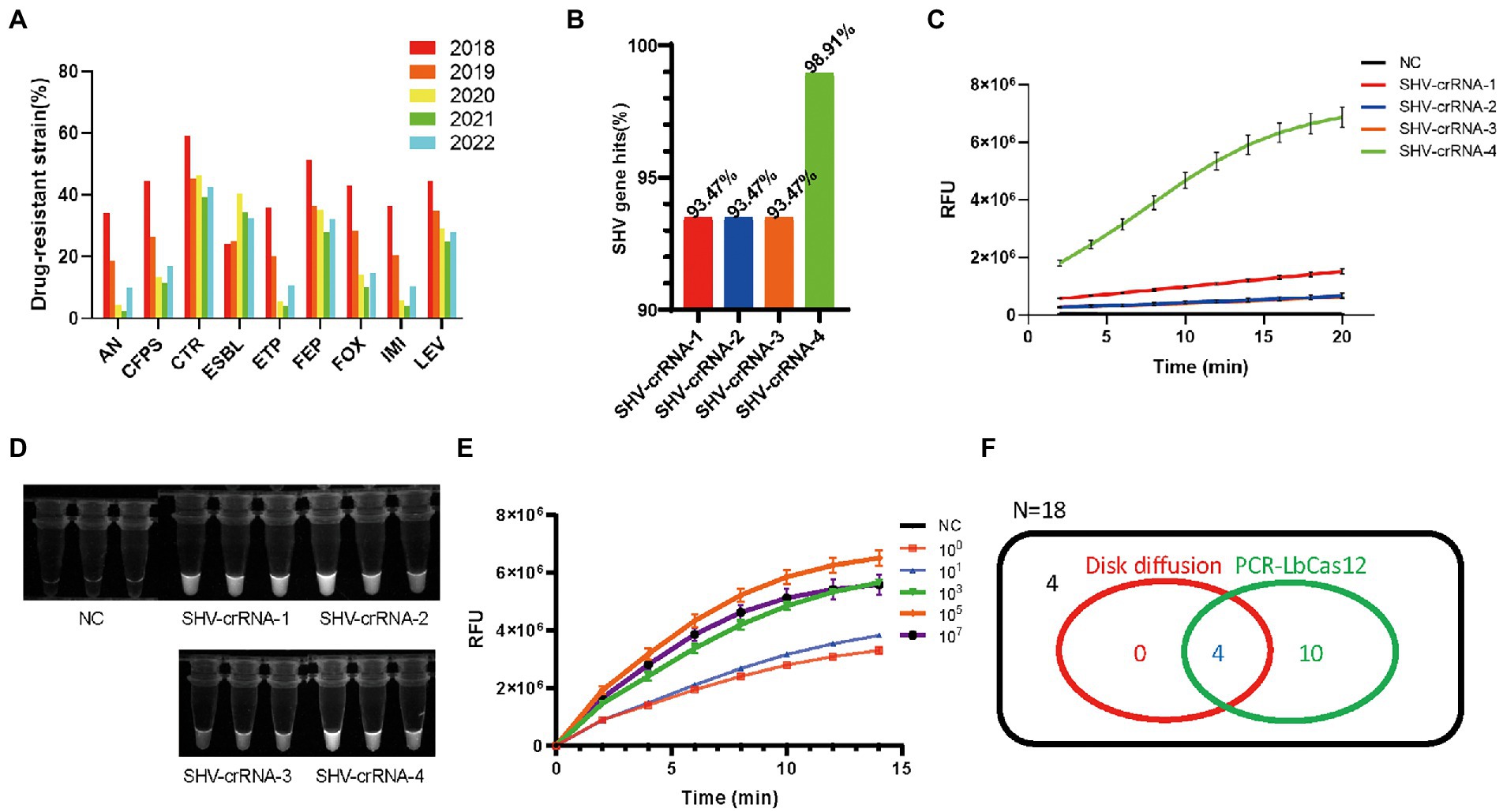
Figure 3. (A) The summary of drug-resistant Klebsiella pneumonia information in the Medical Laboratory Department of Shenzhen Luohu People’s Hospital from January 2018 to June 2022. (B) The percentage of SHV genes hit by four SHV-crRNAs. FAM signal, (C) and photograph under UV, (D) of four SHV-crRNAs activity in PCR-LbCas12a reaction. (E) The RFU signal generated by the PCR-LbCas12a system detecting the serially diluted standard SHV DNA. (F) Venn Diagram of results of testing ESBL of 18 K. pneumonia samples in PCR-LbCas12a and disk diffusion susceptibility test.
Using the PCR-LbCas12a workflow, we tested the four crRNAs efficiencies. Although all the crRNAs are effective (Figures 3C, D), the SHV-crRNA-4 is the most efficient. The PCR-LbCas12a can test as few as one copy of the SHV-1 DNA fragment (Figure 3E).
Next, we detected the clinic samples. We collected 18 K. pneumoniae clinic samples that had been tested by disk diffusion susceptibility test. We tried it in our PCR-LbCas12a workflow and compared it with the clinic diagnosis (Figure 3F). All the ESBL (+) samples were positive in our workflow. Additionally, 10 more ESBL (−) were tested positive in PCR-LbCas12a detection. Sanger sequencing results suggest that three of them contain the SHV-1 DNA fragment; the rest seven samples failed to be detected by PCR.
Discussion
The excessive use of antibiotics increases drug-resistant bacteria. A recent discovery reveals that 73.1% of K. pneumonia are resistant to at least one antibiotic (Petrosillo et al., 2019; Sharahi et al., 2021). Multi-drug resistant and extensively drug-resistant K. pneumonia strains are increasing (Peng et al., 2020; Yan et al., 2021; Yang et al., 2021). Effectively controlling K. pneumonia requires the direct knowledge of drug-resistant information (Ludden et al., 2020).
The most used detection method is the disk diffusion antibiotic susceptibility test (Prastiyanto et al., 2020). However, the sensitivity and accuracy are only about 56% and 65% (Koyuncu and Haggblom, 2009). Furthermore, two rounds of tests are required to identify specific antibiotic information, which costs more time. In comparison, nucleic acid detection is more advantageous in terms of stability and accuracy. qPCR specifically and accurately identifies K. pneumonia from bacteria like E. coli and S. aureus (Kim et al., 2021) and its antibiotic-resistant gene (Castanheira et al., 2021). However, qPCR needs expensive equipment and skilled workers (Corman et al., 2020).
Recently, CRISPR-Cas12-mediated trans-collateral activity was widely applied to nucleic acid detection (Li et al., 2019; Ma et al., 2021; Selvam et al., 2021). The CRISPR-Cas12 detection system is accurate and specific and can also combine various readout and amplification technologies (Ali et al., 2020; Chen et al., 2020; Ramachandran et al., 2020).
In this study, we first established the PCR-LbCas12a system in this study to detect K. pneumoniae nucleic acid and SHV genes. We used PCR to harvest enough target DNA as a substrate for the LbCas12a reaction. The PCR-LbCas12a detection system can detect as low as only one copy of K. pneumonia. The results were highly consistent with the disk diffusion test. The clinic information on drug-resistant K. pneumonia showed that the ESBL (+) strains have been increasing over the past 5 years. We also tried the workflow to detect ESBL-resistant strains by detecting SHV fragments. The detection of SHV is successful. However, most of the positive strains are not ESBL (+) in disk diffusion tests. We speculate that this contradiction may result from the following reasons:
1. The disk diffusion might be too strict about detecting the K. pneumonia strains that are less resistant.
2. The K. pneumonia samples may not be well preserved to grow on the agar disk.
3. Many of the SHV variants do not encode ESBL enzymes.
Nevertheless, this workflow exhibits the potential to detect specific DNA fragments. Accurately detecting specific antibiotic-resistant strains needs more adjustment and knowledge of the mechanism. Compared to disk diffusion, PCR-LbCas12a detection, which takes no more than 2 h, is highly advantageous in time-consuming.
Materials and methods
Bioinformatics analysis and scripts
The source is downloaded from NCBI-genome. The R and Python scripts are prepared by Guoyu Peng. The detailed information is on https://github.com/GuoYu-Peng/GANAB_BLCA.
Nucleic acid preparation
crRNAs were designed to target 16sRNA, YP_005224572.1, and IF-2 gene according to the protocol (Chen et al., 2018). RNA nucleotides were chemically synthesized without 5′-phosphorylation (Transheep, China). crRNA consists of 19 nt common sequences and 17 nt for recognizing target (Li et al., 2018). DNA and RNA sequences used in this manuscript are in the Supplementary material.
DNA extraction and quantification
Clinical samples were swabs of sputa. According to the manufacturer’s protocol, swabs were dipped in cell lysate and processed genomic DNA extraction using the DNA extraction Kit (Tianlong science & technology, China). Extracted DNA samples were quantified by NanoDrop (Thermo Fisher Scientific, US) and preserved at −80°C before use.
PCR and qPCR
PCR system was carried out in a 20 μl reaction system in the 0.2 ml EP tube. Each reaction contains 10 μl of PrimeSTAR (TAKARA, Japan) PCR premix, 1 μl of forward primer (10 nM) and 1 μl of reverse primer (10 nM), 10 ng of sample DNA, and ddH2O to supplement the volume to 20 μl. The PCR reactions were processed for 35 cycles on an Eppendorf thermocycler with denaturation at 94°C for 15 s, annealing at 58°C for 15 s, and extension at 72°C for 20 s. DNA electrophoresis was processed in 1% agarose gel in TAE buffer.
qPCR reactions were processed using Hieff UNICON Universal Blue qPCR SYBR Green Master Mix (Yeasen, China) on QuantStudio Dx (ABI, US). Program started with a 95°C for 2 min followed by 40 cycles of denaturation at 95°C for 10 s, annealing at 60°C for 10 s, and extension at 72°C for 15 s qPCR. Each reaction was repeated in three biologically independent experiments.
PCR-LbCas12a detection
The LbCas12a detection was carried out in a 20 μl system. The system contains 2 μl Buffer 3 (NEB, US), 50 nM LbCas12a protein, 60 nM crRNA, and 30 nM labeled probe, and 100 ng purified PCR product. Samples were mixed and then incubated at 37°C, and signals were obtained from QuantStudio Dx (ABI, US) every minute for 20–60 min. Each reaction was repeated in three biologically independent experiments.
For clinic detection, samples were incubated at 37°C for 20 min, and then photographed under the UV light exposure. Two independent experiments were processed for each sample.
Sample information
Sample information is available in Supplementary Table 2.
Study approval
The Luohu Ethics Committee approved the project [LLBGS (2021) 020].
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
Ethics statement
The Luohu Ethics Committee approved the project [LLBGS (2021) 020]. Written informed consent to participate in this study was provided by the participants’ legal guardian/next of kin.
Author contributions
ShangW conceived this idea. ShangW, ShanW, and YT processed the experiments. GP analyzed the data. TH, XQ, and JW collected the clinic samples. XW extracted the DNA. YL, SZ and SoW offered the funding and the platform. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by the National Natural Science Foundation of China (81802741) to YL, Shenzhen Science and Technology Innovation Commission (RCJC20200714114557005) to SoW, The China Postdoctoral Science Foundation (2020 M682949) to ShangW, and The Guangdong Basic and Applied Basic Research Foundation (2020A1515110908) to SZ.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1128261/full#supplementary-material
Abbreviations
PCR, Polymerase chain reaction; K. pneumonia, Klebsiella pneumonia; UTI, Urinary tract infection; BSI, Bloodstream infections; ESBL, Extended-spectrum β-lactamases.
Footnotes
References
Ali, Z., Aman, R., Mahas, A., Rao, G. S., Tehseen, M., Marsic, T., et al. (2020). iSCAN: an RT-LAMP-coupled CRISPR-Cas12 module for rapid, sensitive detection of SARS-CoV-2. Virus Res. 288:198129. doi: 10.1016/j.virusres.2020.198129
Ben Achour, N., Belhadj, O., Galleni, M., Ben Moussa, M., and Mercuri, P. S. (2014). Study of a natural mutant SHV-type beta-lactamase, SHV-104, from Klebsiella pneumoniae. Int. J. Microbiol. 2014:548656. doi: 10.1155/2014/548656
Bush, K., and Bradford, P. A. (2016). Beta-lactams and beta-lactamase inhibitors: an overview. Cold Spring Harb. Perspect. Med. 6:a025247. doi: 10.1101/cshperspect.a025247
Calbo, E., Freixas, N., Xercavins, M., Riera, M., Nicolas, C., Monistrol, O., et al. (2011). Foodborne nosocomial outbreak of SHV1 and CTX-M-15-producing Klebsiella pneumoniae: epidemiology and control. Clin. Infect. Dis. 52, 743–749. doi: 10.1093/cid/ciq238
Castanheira, M., Johnson, M. G., Yu, B., Huntington, J. A., Carmelitano, P., Bruno, C., et al. (2021). Molecular characterization of baseline Enterobacterales and Pseudomonas aeruginosa isolates from a phase 3 nosocomial pneumonia (ASPECT-NP) clinical trial. Antimicrob. Agents Chemother. 65:e02461-20. doi: 10.1128/AAC.02461-20
Castanheira, M., Mendes, R. E., Jones, R. N., and Sader, H. S. (2016). Changes in the frequencies of beta-lactamase genes among Enterobacteriaceae isolates in U.S. hospitals, 2012 to 2014: activity of Ceftazidime-Avibactam tested against beta-lactamase-producing isolates. Antimicrob. Agents Chemother. 60, 4770–4777. doi: 10.1128/AAC.00540-16
Chen, J. S., Ma, E., Harrington, L. B., Da Costa, M., Tian, X., Palefsky, J. M., et al. (2018). CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity. Science 360, 436–439. doi: 10.1126/science.aar6245
Chen, Y., Shi, Y., Chen, Y., Yang, Z., Wu, H., Zhou, Z., et al. (2020). Contamination-free visual detection of SARS-CoV-2 with CRISPR/Cas12a: a promising method in the point-of-care detection. Biosens. Bioelectron. 169:112642. doi: 10.1016/j.bios.2020.112642
Corman, V. M., Landt, O., Kaiser, M., Molenkamp, R., Meijer, A., Chu, D. K., et al. (2020). Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 25:2000045. doi: 10.2807/1560-7917.ES.2020.25.3.2000045
Giske, C. G., Turnidge, J., Canton, R., Kahlmeter, G., and Committee, E. S. (2022). Update from the European Committee on antimicrobial susceptibility testing (EUCAST). J. Clin. Microbiol. 60:e0027621. doi: 10.1128/jcm.00276-21
Gootenberg, J. S., Abudayyeh, O. O., Kellner, M. J., Joung, J., Collins, J. J., and Zhang, F. (2018). Multiplexed and portable nucleic acid detection platform with Cas13, Cas12a, and Csm6. Science 360, 439–444. doi: 10.1126/science.aaq0179
Hutchison, J. R., Piepel, G. F., Amidan, B. G., Hess, B. M., Sydor, M. A., and Deatherage Kaiser, B. L. (2018). Comparison of false-negative rates and limits of detection following macrofoam-swab sampling of bacillus anthracis surrogates via rapid viability PCR and plate culture. J. Appl. Microbiol. 124, 1092–1106. doi: 10.1111/jam.13706
Hyun, M., Lee, J. Y., Ryu, S. Y., Ryoo, N., and Kim, H. A. (2019). Antibiotic resistance and clinical presentation of health care-associated Hypervirulent Klebsiella pneumoniae infection in Korea. Microb. Drug Resist. 25, 1204–1209. doi: 10.1089/mdr.2018.0423
Johnson, G., Millar, M. R., Matthews, S., Skyrme, M., Marsh, P., Barringer, E., et al. (2006). Evaluation of BacLite rapid MRSA, a rapid culture based screening test for the detection of ciprofloxacin and methicillin resistant S. aureus (MRSA) from screening swabs. BMC Microbiol. 6:83. doi: 10.1186/1471-2180-6-83
Kim, S. H., Lee, J. S., Lee, J. H., Kim, Y. J., Choi, J. G., Lee, S. K., et al. (2021). Development and application of a multiplex real-time polymerase chain reaction assay for the simultaneous detection of bacterial Aetiologic agents associated with equine venereal diseases. J. Equine. Vet. Sci. 105:103721. doi: 10.1016/j.jevs.2021.103721
Kostyusheva, A., Brezgin, S., Babin, Y., Vasilyeva, I., Glebe, D., Kostyushev, D., et al. (2021). CRISPR-Cas systems for diagnosing infectious diseases. Methods 203, 431–446. doi: 10.1016/j.ymeth.2021.04.007
Koyuncu, S., and Haggblom, P. (2009). A comparative study of cultural methods for the detection of salmonella in feed and feed ingredients. BMC Vet. Res. 5:6. doi: 10.1186/1746-6148-5-6
Li, S. Y., Cheng, Q. X., Liu, J. K., Nie, X. Q., Zhao, G. P., and Wang, J. (2018). CRISPR-Cas12a has both cis- and trans-cleavage activities on single-stranded DNA. Cell Res. 28, 491–493. doi: 10.1038/s41422-018-0022-x
Li, L., Li, S., Wu, N., Wu, J., Wang, G., Zhao, G., et al. (2019). HOLMESv2: a CRISPR-Cas12b-assisted platform for nucleic acid detection and DNA methylation quantitation. ACS Synth. Biol. 8, 2228–2237. doi: 10.1021/acssynbio.9b00209
Ludden, C., Moradigaravand, D., Jamrozy, D., Gouliouris, T., Blane, B., Naydenova, P., et al. (2020). A one health study of the genetic relatedness of Klebsiella pneumoniae and their mobile elements in the east of England. Clin. Infect. Dis. 70, 219–226. doi: 10.1093/cid/ciz174
Ma, L., Peng, L., Yin, L., Liu, G., and Man, S. (2021). CRISPR-Cas12a-powered dual-mode biosensor for ultrasensitive and cross-validating detection of pathogenic bacteria. ACS Sens. 6, 2920–2927. doi: 10.1021/acssensors.1c00686
Magill, S. S., Edwards, J. R., Bamberg, W., Beldavs, Z. G., Dumyati, G., Kainer, M. A., et al. (2014). Multistate point-prevalence survey of health care-associated infections. N. Engl. J. Med. 370, 1198–1208. doi: 10.1056/NEJMoa1306801
Miftode, I. L., Nastase, E. V., Miftode, R. S., Miftode, E. G., Iancu, L. S., Luncă, C., et al. (2021). Insights into multidrug-resistant K. pneumoniae urinary tract infections: from susceptibility to mortality. Exp. Ther. Med. 22:1086. doi: 10.3892/etm.2021.10520
Nouri, R., Tang, Z., Dong, M., Liu, T., Kshirsagar, A., and Guan, W. (2021). CRISPR-based detection of SARS-CoV-2: a review from sample to result. Biosens. Bioelectron. 178:113012. doi: 10.1016/j.bios.2021.113012
Paterson, D. L., Ko, W. C., Von Gottberg, A., Mohapatra, S., Casellas, J. M., Goossens, H., et al. (2004). International prospective study of Klebsiella pneumoniae bacteremia: implications of extended-spectrum beta-lactamase production in nosocomial infections. Ann. Intern. Med. 140, 26–32. doi: 10.7326/0003-4819-140-1-200401060-00008
Peng, Q., Fang, M., Liu, X., Zhang, C., Liu, Y., and Yuan, Y. (2020). Isolation and characterization of a novel phage for controlling multidrug-resistant Klebsiella pneumoniae. Microorganisms 8:542. doi: 10.3390/microorganisms8040542
Petrosillo, N., Taglietti, F., and Granata, G. (2019). Treatment options for Colistin resistant Klebsiella pneumoniae: present and future. J. Clin. Med. 8:934. doi: 10.3390/jcm8070934
Podschun, R., and Ullmann, U. (1998). Klebsiella spp. as nosocomial pathogens: epidemiology, taxonomy, typing methods, and pathogenicity factors. Clin. Microbiol. Rev. 11, 589–603. doi: 10.1128/CMR.11.4.589
Prastiyanto, M. E., Tama, P. D., Ananda, N., Wilson, W., and Mukaromah, A. H. (2020). Antibacterial potential of Jatropha sp. latex against multidrug-resistant bacteria. Int. J. Microbiol. 2020:8509650. doi: 10.1155/2020/8509650
Ramachandran, A., Huyke, D. A., Sharma, E., Sahoo, M. K., Huang, C., Banaei, N., et al. (2020). Electric field-driven microfluidics for rapid CRISPR-based diagnostics and its application to detection of SARS-CoV-2. Proc. Natl. Acad. Sci. U. S. A. 117, 29518–29525. doi: 10.1073/pnas.2010254117
Ramdani-Bouguessa, N., Manageiro, V., Jones-Dias, D., Ferreira, E., Tazir, M., and Canica, M. (2011). Role of SHV beta-lactamase variants in resistance of clinical Klebsiella pneumoniae strains to beta-lactams in an Algerian hospital. J. Med. Microbiol. 60, 983–987. doi: 10.1099/jmm.0.030577-0
Selvam, K., Najib, M. A., Khalid, M. F., Mohamad, S., Palaz, F., Ozsoz, M., et al. (2021). RT-LAMP CRISPR-Cas12/13-based SARS-CoV-2 detection methods. Diagnostics 11. doi: 10.3390/diagnostics11091646
Sharahi, J. Y., Hashemi, A., Ardebili, A., and Davoudabadi, S. (2021). Molecular characteristics of antibiotic-resistant Escherichia coli and Klebsiella pneumoniae strains isolated from hospitalized patients in Tehran. Iran. Ann. Clin. Microbiol. Antimicrob. 20:32. doi: 10.1186/s12941-021-00437-8
Souverein, D., Euser, S. M., van der Reijden, W. A., Herpers, B. L., Kluytmans, J., Rossen, J. W. A., et al. (2017). Clinical sensitivity and specificity of the check-points check-direct ESBL screen for BD MAX, a real-time PCR for direct ESBL detection from rectal swabs. J. Antimicrob. Chemother. 72, 2512–2518. doi: 10.1093/jac/dkx189
Starzyk-Luszcz, K., Zielonka, T. M., Jakubik, J., and Zycinska, K. (2017). Mortality due to nosocomial infection with Klebsiella pneumoniae ESBL<sup/>. Adv. Exp. Med. Biol. 1022, 19–26. doi: 10.1007/5584_2017_38
Wagner, S. J., Benjamin, R. J., Hapip, C. A., Kaelber, N. S., Turgeon, A. M., Skripchenko, A., et al. (2016). Investigation of bacterial inactivation in apheresis platelets with 24 or 30 hours between inoculation and inactivation. Vox Sang. 111, 226–234. doi: 10.1111/vox.12410
Wang, Z., Wang, Y., Lin, L., Wu, T., Zhao, Z., Ying, B., et al. (2022). A finger-driven disposable micro-platform based on isothermal amplification for the application of multiplexed and point-of-care diagnosis of tuberculosis. Biosens. Bioelectron. 195:113663. doi: 10.1016/j.bios.2021.113663
Yan, W. X., Hunnewell, P., Alfonse, L. E., Carte, J. M., Keston-Smith, E., Sothiselvam, S., et al. (2019). Functionally diverse type V CRISPR-Cas systems. Science 363, 88–91. doi: 10.1126/science.aav7271
Yan, X., Su, X., Ren, Z., Fan, X., Li, Y., Yue, C., et al. (2021). High prevalence of antimicrobial resistance and Integron gene cassettes in multi-drug-resistant Klebsiella pneumoniae isolates from captive Giant pandas (Ailuropoda melanoleuca). Front. Microbiol. 12:801292. doi: 10.3389/fmicb.2021.801292
Yan, W., Zhang, Q., Zhu, Y., Jing, N., Yuan, Y., Zhang, Y., et al. (2021). Molecular mechanism of Polymyxin resistance in multidrug-resistant Klebsiella pneumoniae and Escherichia coli isolates from Henan Province, China: a multicenter study. Infect. Drug Resist. 14, 2657–2666. doi: 10.2147/IDR.S314490
Yang, Y., Peng, Y., Jiang, J., Gong, Z., Zhu, H., Wang, K., et al. (2021). Isolation and characterization of multidrug-resistant Klebsiella pneumoniae from raw cow milk in Jiangsu and Shandong provinces, China. Transbound. Emerg. Dis. 68, 1033–1039. doi: 10.1111/tbed.13787
Keywords: CRISPR-Cas, nucleic acid detection, Klebsiella pneumonia (K. pneumonia), Extended-spectrum β-lactamases (ESBL), SHV
Citation: Wang S, Wang S, Tang Y, Peng G, Hao T, Wu X, Wei J, Qiu X, Zhou D, Zhu S, Li Y and Wu S (2023) Detection of Klebsiella pneumonia DNA and ESBL positive strains by PCR-based CRISPR-LbCas12a system. Front. Microbiol. 14:1128261. doi: 10.3389/fmicb.2023.1128261
Edited by:
Zhi Ruan, Zhejiang University, ChinaReviewed by:
Yi Li, University of New South Wales, AustraliaSiamak Heidarzadeh, Zanjan University of Medical Sciences, Iran
Copyright © 2023 Wang, Wang, Tang, Peng, Hao, Wu, Wei, Qiu, Zhou, Zhu, Li and Wu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yuqing Li,  liyuqing@email.szu.edu.cn; Song Wu,
liyuqing@email.szu.edu.cn; Song Wu,  wusong@mail.szu.edu.cn
wusong@mail.szu.edu.cn
†These authors share first authorship
 Shang Wang
Shang Wang Shan Wang1†
Shan Wang1† Ying Tang
Ying Tang Guoyu Peng
Guoyu Peng Xinying Qiu
Xinying Qiu Shimao Zhu
Shimao Zhu Yuqing Li
Yuqing Li Song Wu
Song Wu