- 1Department of Microbiology, National Taiwan University College of Medicine, Taipei, Taiwan
- 2Division of Infectious Diseases, Department of Internal Medicine, Fu Jen Catholic University Hospital, New Taipei City, Taiwan
- 3Division of Infectious Diseases, Department of Medicine, Taipei Veterans General Hospital, Taipei, Taiwan
- 4Department of Internal Medicine, National Taiwan University Hospital, Taipei, Taiwan
Infections caused by carbapenem-resistant Klebsiella pneumoniae (CRKP) are of significant clinical concern worldwide. Fosfomycin is one of the limited treatment options for CRKP. However, resistance to fosfomycin in CRKP has been observed. In this study, we aimed to investigate the fosfomycin resistance mechanism of CRKP. Fosfomycin-resistant Klebsiella pneumoniae isolates were collected from four medical centers in Taiwan from 2010 to 2018. The genes that contributed to fosfomycin resistance were amplified and sequenced. Carbohydrate utilization assays and mutagenesis studies were performed to determine the mechanisms underlying fosfomycin resistance. Forty fosfomycin-resistant CRKP strains were collected and used for further analysis. Fourteen strains exhibited low-level resistance (MIC = 256–512 mg/dl), while 26 strains showed high-level resistance (MIC ≥ 1,024 mg/dl). Chromosomal fosAKP I91V was detected in 39/40 fosfomycin-resistant CRKP strains. We observed that amino acid substitution of chromosomal fosAKP I91V increased the MIC of fosfomycin by approximately eight folds, and this was the only mechanism elucidated for low-level fosfomycin resistance. Among the 26 high-level resistance strains, fosAKP I91V combined with transporter deficiencies (18/26, 69.2%) was the most common resistant mechanism, and one strain showed transporter deficiency only. Plasmid-borne fosA3 accounted for 27.0% (7/26) of high-level resistance. Various G3P and G6P transporter gene mutations, including three novel single amino acid mutations (glpT E299D, glpT D274V, and uhpC A393V) were detected in 19 strains. No murA mutation was found in this study. Our study highlights the need for new therapeutic agents for CRKP infections in Taiwan.
Introduction
The emergence of multidrug-resistant bacteria has raised great clinical concern worldwide, and Klebsiella pneumoniae is one of the most important pathogens that cause several types of healthcare-associated infections (Podschun and Ullmann, 1998; Juan et al., 2019; Nordmann and Poirel, 2019). Carbapenems are considered as the last resort of treatment of multidrug-resistant K. pneumoniae, but an increasing prevalence of carbapenem-resistant K. pneumoniae (CRKP) has been recorded worldwide (Chiu et al., 2013; Logan and Weinstein, 2017).
Fosfomycin is one of the limited treatment options for CRKP (Morrill et al., 2015). Fosfomycin is a phosphonic acid antibiotic discovered in a fermentation broth of Streptomyces fradiae in Spain (Gadebusch et al., 1992). It inhibits N-acetylglucosamine enolpyruvyl transferase (murA), which is an essential enzyme in the early stage of cell wall synthesis. In previous studies, fosfomycin was found to be active against most CRKP, with Clinical and Laboratory Standards Institute (CLSI) breakpoints, (Endimiani et al., 2010; Falagas et al., 2010) and several clinical studies have reported its promising efficacy and low toxicity (Michalopoulos et al., 2010; Pontikis et al., 2014). However, fosfomycin-resistant CRKP has been observed, and is a problem in clinical settings (Yang et al., 2019).
Currently, several mechanisms of resistance have been reported. First, mutation in murA reduces the affinity of fosfomycin (Kumar et al., 2009). Second, fosfomycin enters the cell through two different uptake pathways: the glycerol-3-phosphate (G3P) and the glucose-6-phosphate (G6P) transporter systems. Deficiency of these transporters decreases fosfomycin uptake, leading to resistance (Takahata et al., 2010). Third, three major types of fosfomycin-modifying enzymes, namely fosA, fosB, and fosX, were described to contribute to resistance. Glutathione S-transferase (GST) homologs are most commonly found in gram-negative bacteria, including K. pneumoniae, and they reduce fosfomycin susceptibility (Ito et al., 2017). Several subtypes of plasmid-mediated fosA homologs were reported to confer high-level resistance (Yang et al., 2019).
In this study, we aimed to evaluate the mechanisms of fosfomycin resistance in CRKP strains in Taiwan. We analyzed the capsular type distribution of fosfomycin-resistant CRKP and attempted to clarify the resistance mechanism. Chromosomal mutagenesis, plasmid transformation, and sole carbohydrate growth assays were performed to confirm that the phenotypic changes were caused by genetic changes.
Materials and Methods
Clinical Strains
Clinical fosfomycin-resistant CRKP strains which caused infections were retrospectively collected from the National Taiwan University Hospital, Taipei Veterans General Hospital, National Cheng Kung University Hospital, and Linkou Chang Gung Memorial Hospital from January 2010 to August 2018. If multiple strains were isolated from a same patient, only one strain will be enrolled. CRKP was defined as strains with a minimum inhibitory concentration (MIC) of ≥4 mg/L for imipenem or meropenem, based on the CLSI guidelines.
Antimicrobial Susceptibility Testing
Following the CLSI guidelines, the MICs of fosfomycin (Sigma-Aldrich, St. Louis, MO, United States) were determined by the agar dilution method using Mueller-Hinton agar plates (BD, France) supplemented with 25 mg/L G6P (Sigma-Aldrich). Escherichia coli ATCC 25922 was used as quality control for antimicrobial susceptibility testing. High-level fosfomycin-resistant K. pneumoniae strains were defined as those with MIC ≥ 1,024 mg/dL.
Sequence Analysis of Capsular Types, Fosfomycin-Related Genes, Carbapenemase Genes, and Multilocus Sequence Typing (MLST)
The capsular types of the CRKP strains were determined using wzc genotyping and whole sequencing, as previously reported (Pan et al., 2015). Enzymatic mediated fosfomycin resistance genes (fosA3, fosB, fosC, fosX, fomA, and fomB), and fosfomycin-related genes (murA, glpT, glpR, uhpT, uhpA, uhpB, uhpC, ptsI, and cyaA) were amplified and sequenced; the primers used are listed in Supplementary Table 1. Two fosfomycin-susceptible strains, NTUH-K2044 (Fang et al., 2004) (GenBank: AP006725.1) and MGH 78578 (GenBank: CP000647.1) were used as reference strains to identify amino acid substitutions, excluding polymorphisms, observed in the fosfomycin-susceptible strains. Most common carbapenemase genes (blaKPC, blaNDM, blaIMP, blaVIM, blaGES, blaOXA–23–like, blaOXA–48–like) were amplified for the presence of carbapenemase. If the carbapenemase genes were not detected by PCR method, The Simplified Carbapenem Inactivation Method (Jing et al., 2018) was performed to identify carbapenemase production. Sequence types (STs) were determined by sequence alignment using the Pasteur Institute database. Next generation sequencing was performed for strains in which the genes could not be amplified with the various primer pairs. The genomes were sequenced using the Pacific Biosciences RS II platform (Menlo Park, CA, United States). Assembly of the data was performed using the hierarchical genome assembly process (HGAP) compiled specifically for quality trimming, de novo assembly, and polishing of PacBio data.
Plasmid Transformation
Nucleobond PC100 Plasmid Midiprep (Takara Bio, Mountain View, CA, United States) was used to purify the plasmid of fosA3-positive strains and controls. Purified plasmids were transformed into recipient E. coli DH10B cells through electroporation under the conditions of 2,000 V/200 Ω/25 μF. After 1 h of recovery, the E. coli cells were cultured on plates with 16 mg/dL fosfomycin. Insertion of fosA3 in the E. coli cells that grew on the plates was confirmed using PCR amplification.
Sole Carbohydrates Growth Assay
Utilization of carbohydrates, which indicated the activity of glpT and uhpT transporters, was performed as previously described (Tseng et al., 2015). Briefly, 0.2% (w/v) G6P or G3P was supplied in M9 minimal medium agar as the sole carbon source. Briefly, M9 minimal medium agar was supplemented with 0.2% (w/v) G6P or G3P as the sole carbon source. Overnight bacterial suspensions were washed with an equivalent volume of saline and McFarland No. 4. The suspension was diluted 10-fold with physiological saline, thoroughly mixed, plated (2 μL) on M9 minimal medium agar supplemented with different sole carbohydrates, and then incubated at 37°C for 48 h. A negative phenotype was defined as a lack of colonies on the plate.
Site-Directed Mutagenesis
Site-directed mutations were generated in the fosfomycin-susceptible strain NTUH-K2044 (MIC = 32) using the pKO3-km plasmid (Link et al., 1997). The DNA fragments, including point mutation sites and flanking regions (glpT, uhpA, uhpC, and fosAKP), were amplified using PCR (primers are listed in Supplementary Table 1) and then cloned into the pKO3-km plasmid. The resulting plasmids were used to generate point mutants as in a previous study (Lin et al., 2012), and the mutants were confirmed by sequencing.
Analysis of Chromosomal fosA
We named the chromosomal fosA in the genomes of K. pneumoniae as fosAKP, as previously described (Ito et al., 2017). Because no previous reported mechanism was detected in 14 low level resistant strains, sequences and mRNA expression level of fosAKP were analyzed in this study. We designed primer pairs to distinguish fosAKP from the plasmid fosA3 (Supplementary Table 1). There are 13 nucleotides differences between primer pairs of chromosomal fosA and fosA3. Furthermore, we also check surrounding elements of fosA3 (Primer pairs: IS and fosA3) and chromosomal fosA (Primer pairs: FosA frk) to confirm the fosA3 is located on plasmid. Then, fosAKP was amplified, sequenced, and compared with the reference strains.
Results
In total, 108 CRKP strains were collected in our study. Antibiotic susceptibility test revealed that 54 strains (50.0%) were susceptible to fosfomycin, 14 strains (13.0%) exhibited intermediate resistance, and 40 strains (37.0%) were resistant to fosfomycin. Among the 40 fosfomycin-resistant CRKP strains, 26 exhibited high levels of fosfomycin resistance (MIC > 1,024 mg/dl), while 14 showed low levels of fosfomycin resistance (MIC between 256 and 512 mg/dl) (Table 1). As presented in Table 1, capsular typing results showed that K47 was the most dominant capsular type (52.5%, 21/40), followed by K64 (27.5%, 11/40) and K62 (7.5%, 3/25). Capsular types K5, K15, K24, K54, and KN2 were each identified in one strain (2.5%, 1/40). KPC was detected in 21 of 40 fosfomycin-resistant CRKP strains, and the remaining 19 strains fosfomycin-resistant CRKP strains had no carbapenemase detected by both genotype and phenotype assay.
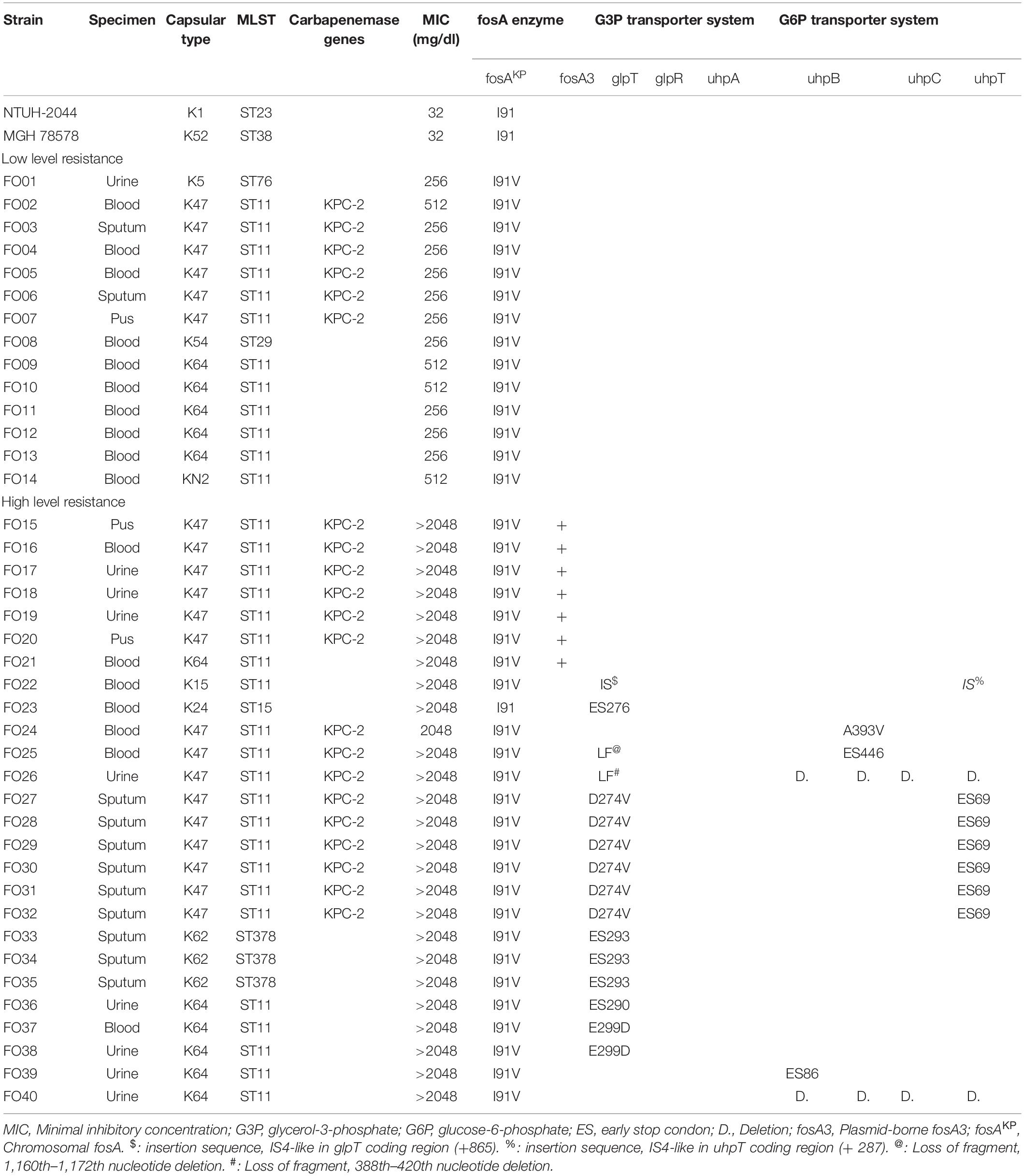
Table 1. Characteristics and detected mechanisms of fosfomycin-resistant CRKPs and reference strains.
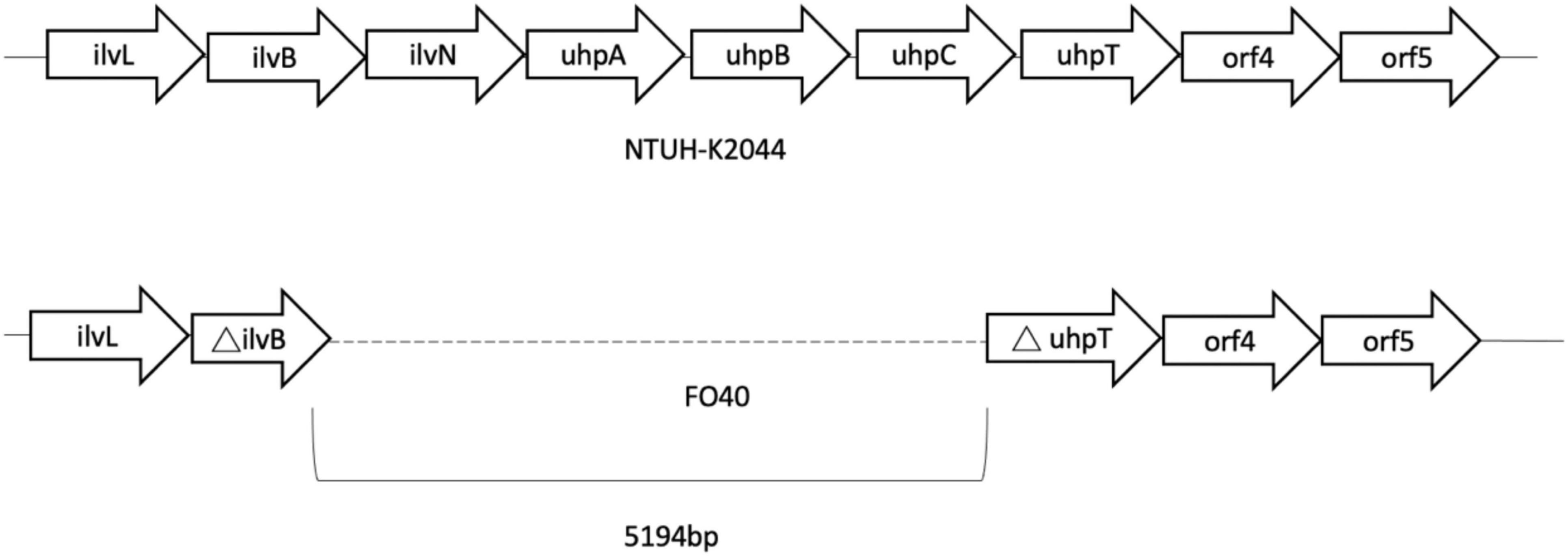
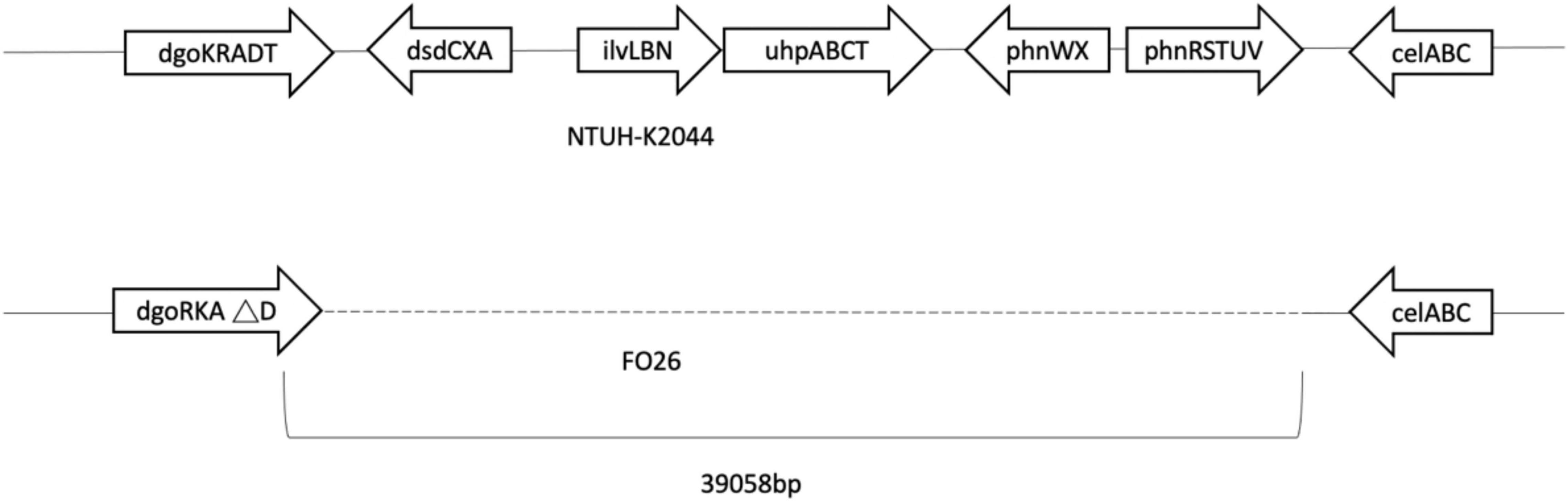
Genes of fosfomycin-targeting enzymes and transporters were examined using PCR amplification and sequencing (Table 1). We did not find any sequence changes or amino acid variations in murA in all 40 fosfomycin-resistant strains, while transporter gene mutations were observed in 19 strains. As growth impairment using a sole carbohydrate source among the 19 strains indicated the loss of function of transporter genes, we performed a growth assay using G3P and G6P to check the function of these transporters. All 19 strains had growth impairment on either G3P or G6P minimal medium agar, which was compatible with gene mutations (Table 2). In particular, eight strains were not able to grow on G3P minimal medium agar only, three isolates could not grow on G6P minimal medium agar only, and growth impairment was observed on both G3P and G6P minimal medium agar in eight strains. In the 15 strains that showed growth impairment on G3P minimal medium agar, an early stop codon was found in glpT of 5 strains, and amino acid substitutions in glpT (299D, 274V) were detected in 6 strains. Loss of small nucleotide fragment in glpT in 2 strains and an insertion sequence added in the glpT coding region in one strain were also detected. Mutations in glpR were not detected in this study. Among 11 isolates with growth impairment on G6P minimal medium agar, an early stop codon was detected in uhpA and uhpT in one strain and six strains, respectively. Amino acid substitution was detected in uhpB (393V) in one strain. We were not able to amplify uhpA, uhpB, uhpC, and uhpT in two strains (FO26, FO40) using the primer pairs. Therefore, we designed primer pairs for the outer area of the uhpABCT operon to amplify and sequence the fragments. Results showed that FO40 had lost a 5,154 bp-long fragment, including most of the uhpABCT operon and ilvNBL operon (Figure 1). Because we were unable to find suitable primer pairs, next generation sequencing revealed that FO26 (GenBank accession: JAHZSM000000000) had lost a large fragment (39,058 bp), including the entire uhpABCT operon (Figure 2).
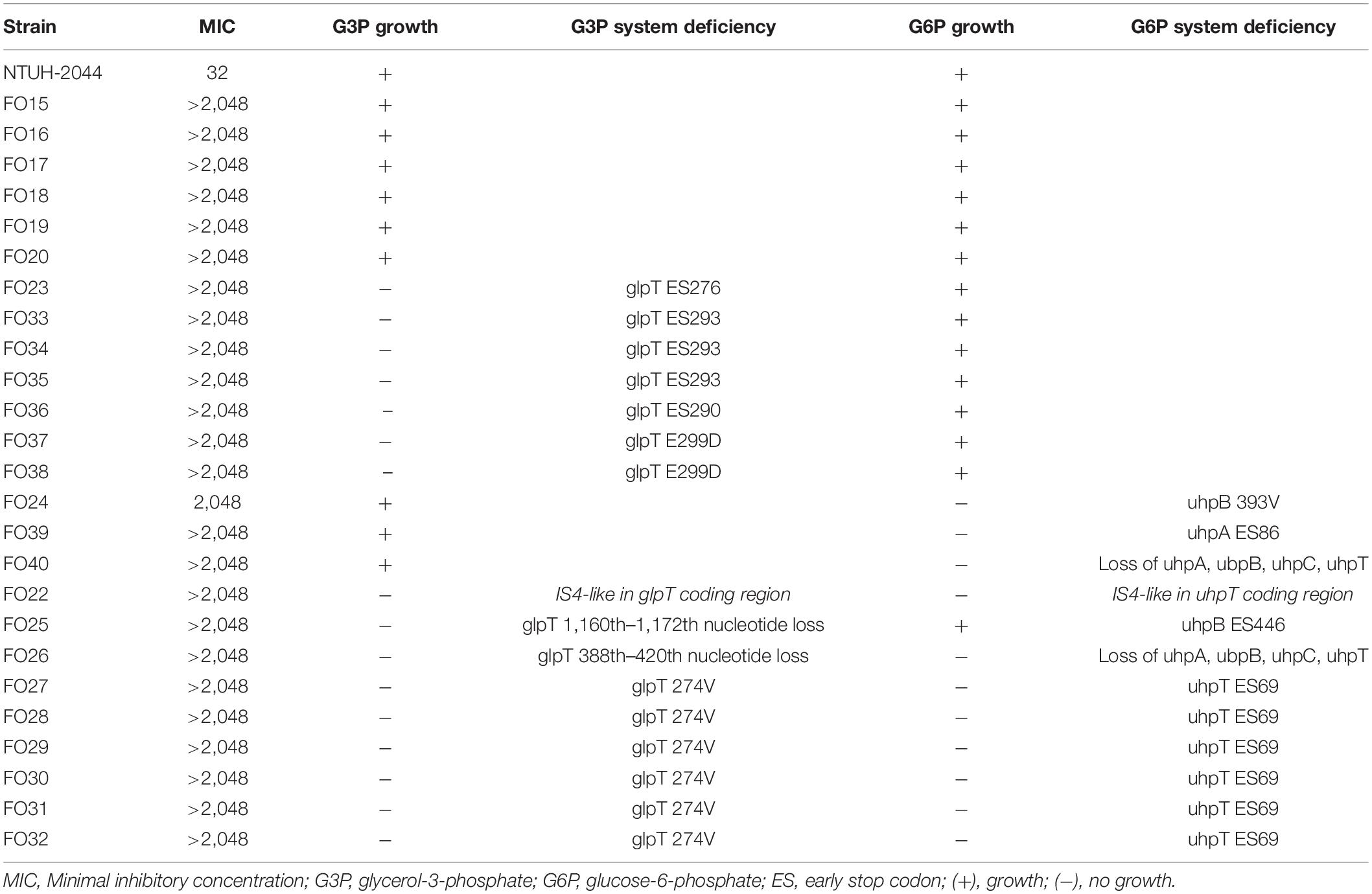
Table 2. Deficiency in G3P and G6P transport systems and consequent carbohydrate utilization among the fosfomycin high-level resistant CRKP strains.
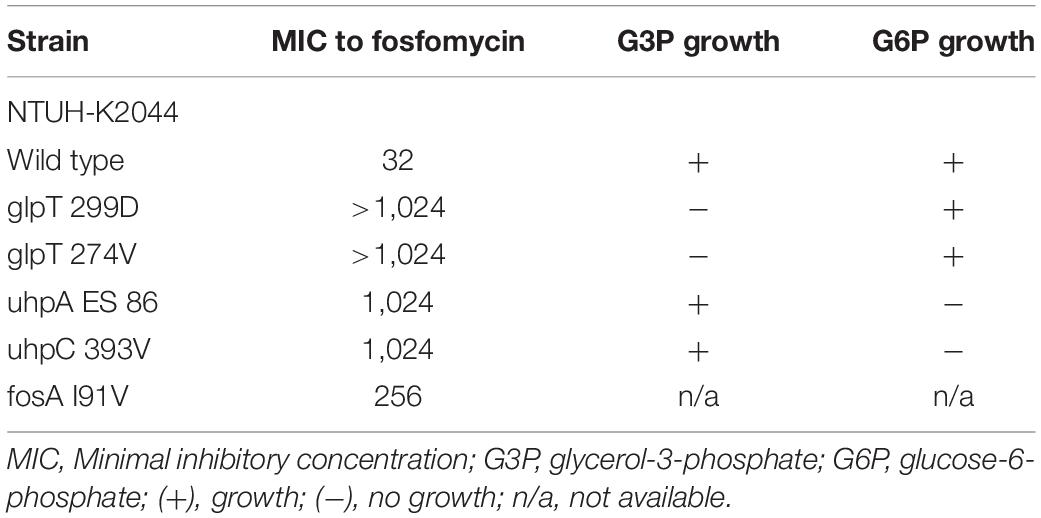
To confirm whether the transporter-related point mutations contribute to fosfomycin resistance, we performed site-directed mutagenesis. After the mutations were generated in NTUH-2044, the MICs of fosfomycin increased by 32-fold (1,024 mg/dL) (Table 3). Moreover, the NTUH-2004 mutants did not grow on either G3P or G6P minimal medium agar plates. These phenotypic changes were comparable with the gene mutations generated.
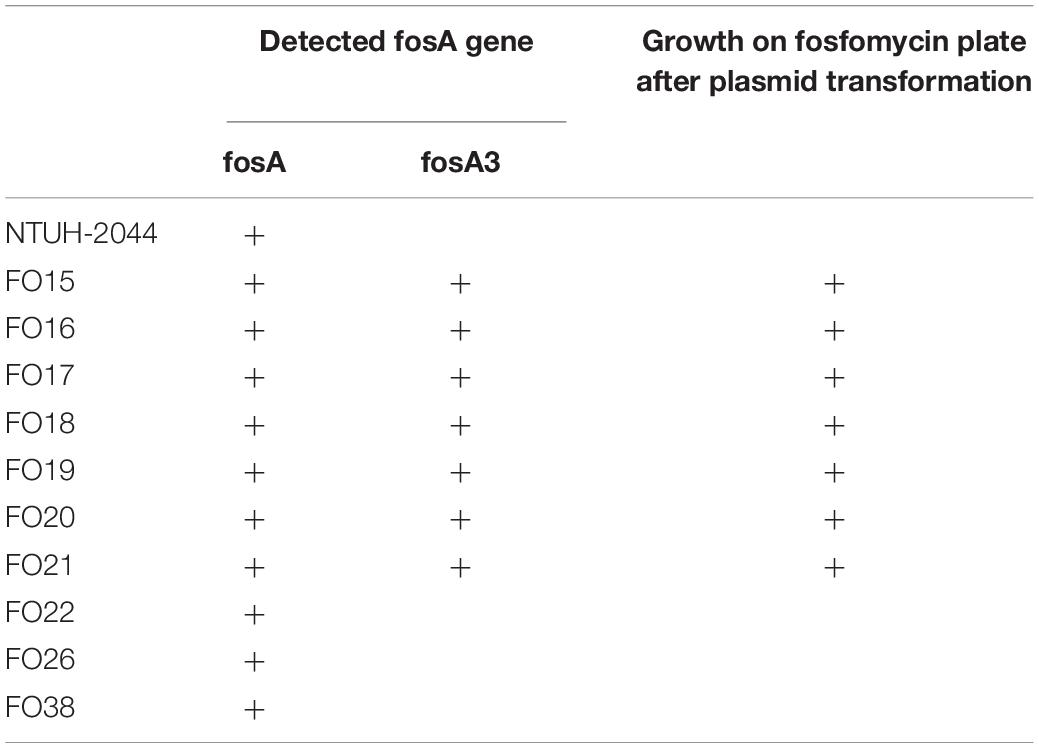
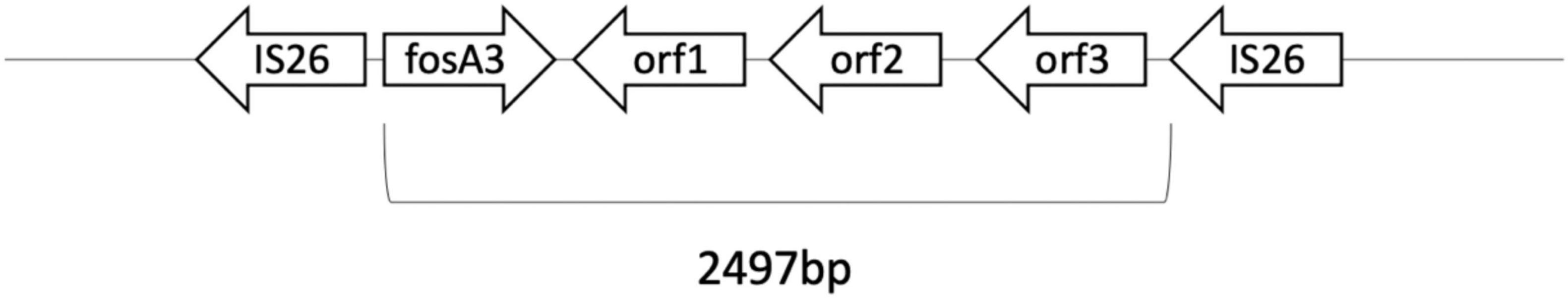
Plasmid-mediated fosA3 was detected in seven isolates (7/40, 24%) (Table 4). The flanking regions of the seven isolates were observed to be identical (Figure 3). Fragments with 2497 bp in total, which contained fosA3, orf1, orf2, and a truncated orf3, was located between two IS26 elements. Next generation sequencing was performed for FO15 (Genbank accession: CP073002) to read the plasmids. To further confirm that the fosA3 genes were located on the plasmid, we performed plasmid transformation study (Table 3). All fosA3 detected in the seven strains was successfully transformed into E. coli DH10B, which became resistant to fosfomycin. Three other randomly selected fosfomycin-resistant strains and NTUH-2044 were used to perform the same plasmid transformation procedures. The recipient E. coli DH10B did not grow on the fosfomycin plate after plasmid transformation.
Notably, mutations in murA or transporter gene or plasmid-mediated fosA3 were not detected in low-level fosfomycin-resistant strains (Table 1). Therefore, we examined the chromosomal fosAKP. The flanking regions of chromosomal fosAKP was shown on Figure 4. As fosAKP was detected in both fosfomycin-susceptible and fosfomycin-resistant strains, we also analyzed the gene sequence and mRNA expression levels of fosAKP in fosfomycin low-level resistant CRKP strains. Chromosomal fosAKP was found in all fosfomycin-resistant strains (40/40, 100%), and no significant difference in fosAKP mRNA expression between fosfomycin-resistant and fosfomycin-resistant strains was observed (Supplementary Figure 1). Compared with the reference strains (NTUH-2044, MGH 78578), mutation I91V was found in 39/40 (97.5%) fosfomycin-resistant CRKP. All 14 low-level resistant strains had I91V mutation in fosAKP. A site-directed mutation study was performed to confirm whether this mutation contributed to fosfomycin resistance. The MIC of fosfomycin in the NTUH-2044 fosAKP I91V mutant was 8-fold higher (from 32 to 256 mg/dl) than that in the wild type (Table 3).
In addition, we observed that several strains shared the same resistance mechanism. To distinguish these strains, we analyzed the capsular types using the available nucleotide sequences and performed pulsed-field gel electrophoresis (PFGE) analysis. Among the 14 low-level fosfomycin-resistant strains with the fosA I91V mutation, we detected five different capsular types (K5, K47, K54, K64, and KN2). However, we could not distinguish six strains with capsular type K47 using the available methods. All of the available nucleotide sequences among these six K47 CRKP strains were identical, and the results of MLST showed that all six strains belonged to ST11. Six strains shared the same gene deficiency of glpT D274V and uhpT ES 69 (FO27, 28, 29, 30, 31, 32). FO31 had one nucleotide change in uhpB, and FO29 had two fragments different from other strains of PFGE analysis (Supplementary Figure 2). The MLST results also showed that all six strains belonged to ST11.
Discussion
Treatment of CRKP remains a challenge, with fosfomycin as one of the limited options available. A recent report showed that the fosfomycin resistance rate (approximately 32–48.5%) is high among CRKP (Kaase et al., 2014; Yang et al., 2019; Liu et al., 2020). Several studies have reported that the fosA family is the main mechanism of CRKP resistance, especially in East Asian countries. Among several variants of the fosA family, fosA3 was the most commonly reported subtype of plasmid-mediated fosA based on reports from Taiwan (Tseng et al., 2017), Korea (Lee et al., 2012), and China (Wang et al., 2020). Huang et al. (2017) and Tseng et al. (2017) reported that fosA5 was also prevalent among CRKP. However, according to a previous literature (Ma et al., 2015), fosA5 showed high similarity to the chromosomal fosAKP of K. pneumoniae, which can be found in most of K. pneumoniae (Ito et al., 2017). In the present study, we designed suitable primers to distinguish all K. pneumoniae strains, including fosfomycin-susceptible strains such as NUTH-K2044. Therefore, the detection of fosA5 by amplification may not contribute to plasmid-mediated high level fosfomycin resistance. The results of analysis of surrounding elements of fosA3 and plasmid transformation support that only fosA3, and not fosAKP, was located on the plasmid.
Chromosomal fosAKP is present in most K. pneumoniae, and fosAKP I91V has also been reported previously by Ito et al. (2017). However, the relationship between the mutation and MIC of fosfomycin was not studied. A previous study suggested that chromosomally encoded fosA was associated with a higher MIC distribution in CRKP (Elliott et al., 2019). However, most K. pneumoniae harboring fosA still exhibited MIC below CLSI breakpoints (64 mg/dL), and the difference between resistant and susceptible strains was not clear. To the best of our knowledge, the genetic or functional mechanism underlying this phenomenon is not yet reported. Our study is the first to document that chromosomal fosAKP I91V is related to low-level fosfomycin resistance. Mutagenesis of fosA increased the MIC by eight-fold (from 32 to 256 mg/dL) in NTUH-K2044. Furthermore, genetic studies revealed that 39/40 of the fosfomycin-resistant CRKP contained chromosomal fosAKP I91V, and this is the only mechanism observed in low-level fosfomycin-resistant strains. We detected approximately 25% of the fosfomycin-susceptible strains carrying chromosomal fosAKP I91V, suggesting that chromosomal fosAKP I91V might not be sufficient to convert strains with very low intrinsic fosfomycin-resistant strains into resistant strains. The chromosomal fosAKP did not raised concern in previous studies (Yang et al., 2019; Liu et al., 2020) of fosfomycin resistant mechanism. However, the chromosomally encoded fosA may also increase risk of development of drug resistance during treatment, which should be taken into consideration on choosing therapeutic option.
Aside from the chromosomal fosA mutation, transporter gene deficiencies were the most common mechanism identified among the high-level resistant strains in our study. Although transporter gene deficiency-related resistance has been well studied in E. coli, there are only limited reports on K. pneumoniae. Lu et al. (2016) reported that glpT and uhpT were found in 97% of fosfomycin-resistant extended spectrum β-lactamase (ESBL)-producing K. pneumoniae. Various amino acid substitutions were found in the study. However, most of the strains in their study exhibited intermediate resistance, and genetic studies were not performed to confirm this relationship. In the study conducted by Liu et al. (2020), only 3 of 48 strains harbored deletion or mutation of glpT. No gene deficiency or functional changes were found in the G6P transporter system. Furthermore, an antimicrobial susceptibility testing above 512 mg/dL was not performed. In our study, deficient transporters were associated with various kinds of gene mutations, including early stop codons, insertion in the coding region, and single amino acid mutations in transporters and their regulatory genes. The entire uhpABCT operon was also deleted in two of the strains. Deficiencies in the transporter genes in these strains contributed to high fosfomycin resistance (MIC > 1,024 mg/dL). As these transporters are not essential for K. pneumoniae, it is not surprising that deficient transporters with various gene mutations were frequently observed under fosfomycin selective pressure. Notably, we found that three single amino acid mutations (glpT E299D, glpT D274V, and uhpC A393V) were related to fosfomycin resistance, and mutagenesis of deficiency transporter genes to NTUH-K2044 increased the MIC to 1,024 mg/dL. Although a previous biochemical study revealed that D274 and E299 are conserved sites in E. coli glpT essential to its function (Law et al., 2008), these mutations, as well as mutations in uhpC, have not yet been reported in K. pneumoniae.
We did not observe any murA mutations in this study. This was different from previous findings (Lu et al., 2016), wherein variant murA modifications accounted for 70% (21/30) of fosfomycin-resistant ESBL-producing K. pneumoniae. Liu et al. (2020) detected murA mutations in only one strain among 48 fosfomycin-resistant strains. As murA is an essential gene for cell wall synthesis, the possibility of losing murA function is low. Previous studies have observed that most of the strains with the murA mutation carried transporter gene mutations as well, and the MICs were not very high (Lu et al., 2016; Liu et al., 2020). However, mutagenesis was not performed, and the mechanism underlying it was not elucidated. Therefore, the murA mutation might be polymorphisms, and other mechanisms may be responsible for the resistance.
In our previous study (Pan et al., 2015), we demonstrated that capsular type-specific bacteriophages and capsule depolymerases could be novel therapies for eradicating CRKP. Due to limited options for treating CRKP infections, investigating the capsular type is essential. In the present study, K47 and K64 were the predominant capsular types of fosfomycin-resistant CRKP, but K47 was not frequently seen in our previous study. This finding suggests that novel capsular type-specific therapies, such as bacteriophage therapy or vaccine development, should be elucidated further.
In our study, detailed gene examination was performed, and serial functional assay and mutagenesis study confirmed that the detected gene mutations contribute to the high-level fosfomycin resistance. Three single amino acid mutations (glpT E299D, glpT D274V, and uhpC A393V) were observed for the first time to contribute to K. pneumoniae fosfomycin resistance. A series of strains shared same gene deficiency. Although the clinical strains were collected from different patients, it is possible that these strains belonged to a clone. We also performed capsular typing, nucleotide sequences analysis, MLST, and PFGE analysis. Most of the strains could be differentiated by these methods. However, some of them, such as six strains of K47 with fosAI91V (FO02-07), and four strains of K47 with glpT D274V (FOS27, 28, 30, 32), could not be excluded as a same clone.
In summary, we observed that chromosomal fosAKP I91V was present in 39/40 fosfomycin-resistant CRKP and contributed to low level fosfomycin resistance. Chromosomal fosAKP mutation is the only mechanism underlying the low-level resistance in CRKP strains, and most high-level resistance strains harbored this mutation. Transporter deficiency was the second most common resistance mechanism among high-level fosfomycin-resistant CRKP in Taiwan. Various G3P and G6P transporter gene mutations, including three novel single amino acid mutations (glpT E299D, glpT D274V, and uhpC A393V), were identified. Plasmid-mediated fosA3 only accounted for 27% of the high-level resistant mechanisms. No murA mutations were found. These findings may provide new therapeutic approaches against CRKP infection in Taiwan.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/genbank/, JAHZSM000000000; https://www.ncbi.nlm.nih.gov/genbank/, CP073002.
Author Contributions
Y-PW and Y-HChe: study concept and design. Y-PW, Y-HChe, I-CH, P-HC, and Y-HCha: carrying out the experiments. Y-PW: analysis and interpretation of data. Y-PW and Y-TL: drafting of the manuscript. Y-HChe, I-CH, P-HC, and Y-HCha: statistical analysis. Y-TL, H-CY, and J-TW: administrative, technical, and material support. H-CY and J-TW: study supervision. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by the Ministry of Science and Technology, the Excellent Translational Medicine Research Projects of National Taiwan University College of Medicine and National Taiwan University Hospital, Good Liver Foundation, and “Center of Precision Medicine” from the Featured Areas Research Center Program within the framework of the Higher Education Sprout Project by the Ministry of Education (MOE) in Taiwan.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.816806/full#supplementary-material
References
Chiu, S. K., Wu, T. L., Chuang, Y. C., Lin, J. C., Fung, C. P., Lu, P. L., et al. (2013). National surveillance study on carbapenem non-susceptible Klebsiella pneumoniae in Taiwan: the emergence and rapid dissemination of KPC-2 carbapenemase. PLoS One 8:e69428. doi: 10.1371/journal.pone.0069428
Elliott, Z. S., Barry, K. E., Cox, H. L., Stoesser, N., Carroll, J., Vegesana, K., et al. (2019). The role of fosA in challenges with Fosfomycin susceptibility testing of multispecies Klebsiella pneumoniae Carbapenemase-producing clinical isolates. J. Clin. Microbiol0 57:e00634-19. doi: 10.1128/jcm.00634-19
Endimiani, A., Patel, G., Hujer, K. M., Swaminathan, M., Perez, F., Rice, L. B., et al. (2010). In vitro activity of fosfomycin against blaKPC-containing Klebsiella pneumoniae isolates, including those nonsusceptible to tigecycline and/or colistin. Antimicrob. Agents Chemother. 54, 526–529. doi: 10.1128/aac.01235-09
Falagas, M. E., Maraki, S., Karageorgopoulos, D. E., Kastoris, A. C., Mavromanolakis, E., and Samonis, G. (2010). Antimicrobial susceptibility of multidrug-resistant (MDR) and extensively drug-resistant (XDR) Enterobacteriaceae isolates to fosfomycin. Int. J. Antimicrob. Agents 35, 240–243. doi: 10.1016/j.ijantimicag.2009.10.019
Fang, C. T., Chuang, Y. P., Shun, C. T., Chang, S. C., and Wang, J. T. (2004). A novel virulence gene in Klebsiella pneumoniae strains causing primary liver abscess and septic metastatic complications. J. Exp. Med. 199, 697–705. doi: 10.1084/jem.20030857
Gadebusch, H. H., Stapley, E. O., and Zimmerman, S. B. (1992). The discovery of cell wall active antibacterial antibiotics. Crit. Rev. Biotechnol. 12, 225–243. doi: 10.3109/07388559209069193
Huang, L., Hu, Y. Y., and Zhang, R. (2017). Prevalence of fosfomycin resistance and plasmid-mediated fosfomycin-modifying enzymes among carbapenem-resistant Enterobacteriaceae in Zhejiang, China. J. Med. Microbiol. 66, 1332–1334. doi: 10.1099/jmm.0.000578
Ito, R., Mustapha, M. M., Tomich, A. D., Callaghan, J. D., Mcelheny, C. L., Mettus, R. T., et al. (2017). Widespread Fosfomycin resistance in gram-negative bacteria attributable to the chromosomal fosA Gene. mBio 8:e00749-17. doi: 10.1128/mBio.00749-17
Jing, X., Zhou, H., Min, X., Zhang, X., Yang, Q., Du, S., et al. (2018). The Simplified carbapenem inactivation method (sCIM) for simple and accurate detection of carbapenemase-producing gram-negative Bacilli. Front. Microbiol. 9:2391. doi: 10.3389/fmicb.2018.02391
Juan, C. H., Chuang, C., Chen, C. H., Li, L., and Lin, Y. T. (2019). Clinical characteristics, antimicrobial resistance and capsular types of community-acquired, healthcare-associated, and nosocomial Klebsiella pneumoniae bacteremia. Antimicrob. Resist. Infect. Control. 8:1. doi: 10.1186/s13756-018-0426-x
Kaase, M., Szabados, F., Anders, A., and Gatermann, S. G. (2014). Fosfomycin susceptibility in carbapenem-resistant Enterobacteriaceae from Germany. J. Clin. Microbiol. 52, 1893–1897. doi: 10.1128/jcm.03484-13
Kumar, S., Parvathi, A., Hernandez, R. L., Cadle, K. M., and Varela, M. F. (2009). Identification of a novel UDP-N-acetylglucosamine enolpyruvyl transferase (MurA) from Vibrio fischeri that confers high fosfomycin resistance in Escherichia coli. Arch. Microbiol. 191, 425–429. doi: 10.1007/s00203-009-0468-9
Law, C. J., Almqvist, J., Bernstein, A., Goetz, R. M., Huang, Y., Soudant, C., et al. (2008). Salt-bridge dynamics control substrate-induced conformational change in the membrane transporter GlpT. J. Mol. Biol. 378, 828–839. doi: 10.1016/j.jmb.2008.03.029
Lee, S. Y., Park, Y. J., Yu, J. K., Jung, S., Kim, Y., Jeong, S. H., et al. (2012). Prevalence of acquired fosfomycin resistance among extended-spectrum β-lactamase-producing Escherichia coli and Klebsiella pneumoniae clinical isolates in Korea and IS26-composite transposon surrounding fosA3. J. Antimicrob. Chemother. 67, 2843–2847. doi: 10.1093/jac/dks319
Lin, T. L., Yang, F. L., Yang, A. S., Peng, H. P., Li, T. L., Tsai, M. D., et al. (2012). Amino acid substitutions of MagA in Klebsiella pneumoniae affect the biosynthesis of the capsular polysaccharide. PLoS One 7:e46783. doi: 10.1371/journal.pone.0046783
Link, A. J., Phillips, D., and Church, G. M. (1997). Methods for generating precise deletions and insertions in the genome of wild-type Escherichia coli: application to open reading frame characterization. J. Bacteriol. 179, 6228–6237. doi: 10.1128/jb.179.20.6228-6237.1997
Liu, P., Chen, S., Wu, Z. Y., Qi, M., Li, X. Y., and Liu, C. X. (2020). Mechanisms of fosfomycin resistance in clinical isolates of carbapenem-resistant Klebsiella pneumoniae. J. Glob. Antimicrob. Resist. 22, 238–243. doi: 10.1016/j.jgar.2019.12.019
Logan, L. K., and Weinstein, R. A. (2017). The epidemiology of carbapenem-resistant Enterobacteriaceae: the impact and evolution of a global menace. J. Infect. Dis. 215, S28–S36. doi: 10.1093/infdis/jiw282
Lu, P. L., Hsieh, Y. J., Lin, J. E., Huang, J. W., Yang, T. Y., Lin, L., et al. (2016). Characterisation of fosfomycin resistance mechanisms and molecular epidemiology in extended-spectrum β-lactamase-producing Klebsiella pneumoniae isolates. Int. J. Antimicrob. Agents 48, 564–568. doi: 10.1016/j.ijantimicag.2016.08.013
Ma, Y., Xu, X., Guo, Q., Wang, P., Wang, W., and Wang, M. (2015). Characterization of fosA5, a new plasmid-mediated fosfomycin resistance gene in Escherichia coli. Lett. Appl. Microbiol. 60, 259–264. doi: 10.1111/lam.12366
Michalopoulos, A., Virtzili, S., Rafailidis, P., Chalevelakis, G., Damala, M., and Falagas, M. E. (2010). Intravenous fosfomycin for the treatment of nosocomial infections caused by carbapenem-resistant Klebsiella pneumoniae in critically ill patients: a prospective evaluation. Clin. Microbiol. Infect. 16, 184–186. doi: 10.1111/j.1469-0691.2009.02921.x
Morrill, H. J., Pogue, J. M., Kaye, K. S., and Laplante, K. L. (2015). Treatment options for carbapenem-resistant Enterobacteriaceae infections. Open Forum Infect. Dis. 2:ofv050. doi: 10.1093/ofid/ofv050
Nordmann, P., and Poirel, L. (2019). Epidemiology and diagnostics of carbapenem resistance in gram-negative bacteria. Clin. Infect. Dis. 69, S521–S528. doi: 10.1093/cid/ciz824
Pan, Y. J., Lin, T. L., Lin, Y. T., Su, P. A., Chen, C. T., Hsieh, P. F., et al. (2015). Identification of capsular types in carbapenem-resistant Klebsiella pneumoniae strains by wzc sequencing and implications for capsule depolymerase treatment. Antimicrob. Agents Chemother. 59, 1038–1047. doi: 10.1128/aac.03560-14
Podschun, R., and Ullmann, U. (1998). Klebsiella spp. as nosocomial pathogens: epidemiology, taxonomy, typing methods, and pathogenicity factors. Clin. Microbiol. Rev. 11, 589–603.
Pontikis, K., Karaiskos, I., Bastani, S., Dimopoulos, G., Kalogirou, M., Katsiari, M., et al. (2014). Outcomes of critically ill intensive care unit patients treated with fosfomycin for infections due to pandrug-resistant and extensively drug-resistant carbapenemase-producing Gram-negative bacteria. Int. J. Antimicrob. Agents 43, 52–59. doi: 10.1016/j.ijantimicag.2013.09.010
Takahata, S., Ida, T., Hiraishi, T., Sakakibara, S., Maebashi, K., Terada, S., et al. (2010). Molecular mechanisms of fosfomycin resistance in clinical isolates of Escherichia coli. Int. J. Antimicrob. Agents 35, 333–337. doi: 10.1016/j.ijantimicag.2009.11.011
Tseng, S. P., Wang, S. F., Kuo, C. Y., Huang, J. W., Hung, W. C., Ke, G. M., et al. (2015). Characterization of Fosfomycin resistant extended-spectrum beta-lactamase-producing Escherichia coli isolates from human and pig in Taiwan. PLoS One 10:e0135864. doi: 10.1371/journal.pone.0135864
Tseng, S. P., Wang, S. F., Ma, L., Wang, T. Y., Yang, T. Y., Siu, L. K., et al. (2017). The plasmid-mediated fosfomycin resistance determinants and synergy of fosfomycin and meropenem in carbapenem-resistant Klebsiella pneumoniae isolates in Taiwan. J. Microbiol. Immunol. Infect. 50, 653–661. doi: 10.1016/j.jmii.2017.03.003
Wang, B., Pan, F., Wang, C., Zhao, W., Sun, Y., Zhang, T., et al. (2020). Molecular epidemiology of carbapenem-resistant Klebsiella pneumoniae in a paediatric hospital in China. Int. J. Infect. Dis. 93, 311–319. doi: 10.1016/j.ijid.2020.02.009
Keywords: Klebsiella pneumoniae, carbapenem resistance enterobacteriaceae, fosfomycin resistance mechanism, fosfomycin resistance gene, glpT and uhpT transporters
Citation: Wang Y-P, Chen Y-H, Hung I-C, Chu P-H, Chang Y-H, Lin Y-T, Yang H-C and Wang J-T (2022) Transporter Genes and fosA Associated With Fosfomycin Resistance in Carbapenem-Resistant Klebsiella pneumoniae. Front. Microbiol. 13:816806. doi: 10.3389/fmicb.2022.816806
Received: 17 November 2021; Accepted: 07 January 2022;
Published: 31 January 2022.
Edited by:
Miklos Fuzi, Semmelweis University, HungaryReviewed by:
Xiaofei Jiang, Fudan University, ChinaQi Wang, Peking University People’s Hospital, China
Copyright © 2022 Wang, Chen, Hung, Chu, Chang, Lin, Yang and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hung-Chih Yang, hcyang88@ntu.edu.tw; Jin-Town Wang, wangjt@ntu.edu.tw
 Yu-Ping Wang
Yu-Ping Wang Yen-Hao Chen1
Yen-Hao Chen1 Yi-Tsung Lin
Yi-Tsung Lin Hung-Chih Yang
Hung-Chih Yang Jin-Town Wang
Jin-Town Wang