- 1Department of Laboratory Medicine, State Key Laboratory of Complex Severe and Rare Diseases, Peking Union Medical College Hospital, Chinese Academy of Medical Science and Peking Union Medical College, Beijing, China
- 2Graduate School, Chinese Academy of Medical Science and Peking Union Medical College, Beijing, China
- 3Beijing Key Laboratory for Mechanisms Research and Precision Diagnosis of Invasive Fungal Diseases, Beijing, China
- 4Medical Research Center, Peking Union Medical College Hospital, Chinese Academy of Medical Science and Peking Union Medical College, Beijing, China
- 5Jinling Hospital Institute of Clinical Laboratory Science, School of Medicine, Nanjing University, Nanjing, Jiangsu, China
- 6Department of Dermatology, Hua Shan Hospital, Fudan University, Shanghai, China
- 7Department of Laboratory Medicine, Fujian Provincial Hospital, Fuzhou, China
- 8Department of Laboratory Medicine, West China Hospital, Sichuan University, Chengdu, China
- 9Department of Laboratory Medicine, Ruian People's Hospital, Wenzhou, China
- 10Department of Laboratory Medicine, The First Medicine Center, Chinese PLA General Hospital, Beijing, China
- 11Department of Laboratory Medicine, Zhejiang Provincial People's Hospital, Hangzhou, China
- 12Department of Laboratory Medicine, The People’s Hospital of Liaoning Province, Shenyang, China
- 13Department of Laboratory Medicine, No.908 Hospital of Joint Logistics Support Force, Nanchang, China
- 14Department of Laboratory Medicine and Internal Medicine, National Taiwan University Hospital, National Taiwan University College of Medicine, Taipei, Taiwan
- 15Department of Laboratory Medicine and Internal Medicine, China Medical University Hospital, China Medical University, Taichung, Taiwan
- 16Department of Infectious Diseases and Clinical Microbiology, Beijing Institute of Respiratory Medicine and Beijing Chao-Yang Hospital, Capital Medical University, Beijing, China
- 17Department of Laboratory Medicine, Peking University Third Hospital, Beijing, China
Candida duobushaemulonii, type II Candida haemulonii complex, is closely related to Candida auris and capable of causing invasive and non-invasive infections in humans. Eleven strains of C. duobushaemulonii were collected from China Hospital Invasive Fungal Surveillance Net (CHIF-NET) and identified using matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF), VITEK 2 Yeast Identification Card (YST), and internal transcribed spacer (ITS) sequencing. Whole genome sequencing of C. duobushaemulonii was done to determine their genotypes. Furthermore, C. duobushaemulonii strains were tested by Sensititre YeastOne™ and Clinical and Laboratory Institute (CLSI) broth microdilution panel for antifungal susceptibility. Three C. duobushaemulonii could not be identified by VITEK 2. All 11 isolates had high minimum inhibitory concentrations (MICs) to amphotericin B more than 2 μg/ml. One isolate showed a high MIC value of ≥64 μg/ml to 5-flucytosine. All isolates were wild type (WT) for triazoles and echinocandins. FUR1 variation may result in C. duobushaemulonii with high MIC to 5-flucytosine. Candida duobushaemulonii mainly infects patients with weakened immunity, and the amphotericin B resistance of these isolates might represent a challenge to clinical treatment.
Introduction
Candida duobushaemulonii belongs to the Candida haemulonii species complex, along with Candida haemulonii and Candida haemulonii var. vulnera. Yeasts belonging to this complex are closely related to the notorious Candida auris, which has attracted global attention with multi-drug resistant and widely disseminating (Du et al., 2020). Candida duobushaemulonii was initially classified as type II of Candida haemulonii complex. It was clearly identified as C. duobushaemulonii in 2012 (Cendejas-Bueno et al., 2012). The conventional panels used in routine microbiology laboratories often misidentify these species, making it hard to identify accurately (Fang et al., 2016; Ambaraghassi et al., 2019; Frias-De-Leon et al., 2019). Therefore, their actual incidence and global prevalence may be underestimated.
A retrospective study found that C. duobushaemulonii was first isolated in foot ulcers in 1996, where it was recovered from the toenail of a patient from Bizkaia, Spain (Jurado-Martin et al., 2020). The first isolate in China was collected under the China Hospital Invasive Fungal Surveillance Net (CHIF-NET) project in 2010 (Hou et al., 2016a). However, as an emerging species, it has been reported that fluconazole, amphotericin B, and echinocandins non-wild-type (non-WT) C. duobushaemulonii have been identified (Cendejas-Bueno et al., 2012), and the mechanism of C. duobushaemulonii with high MIC for antifungal drugs is still unclear.
Although currently reported cases of C. duobushaemulonii in China are few, hospital outbreaks of C. duobushaemulonii have been reported (Gade et al., 2020). Therefore, we conducted antifungal drug susceptibility testing and whole-genome sequencing of C. duobushaemulonii in China for 8 years. The aims were to confirm whether C. duobushaemulonii had broken out in China, and to discover the underlying mechanism of its resistance to antifungal drugs.
Materials and methods
Ethics statement
This study was approved by the Human Research Ethics Committee of Peking Union Medical College Hospital (No. S-263). Written informed consent was obtained from all the patients who participated in this study, aimed at culturing and studying the isolates obtained from them for scientific research.
Fungal isolates
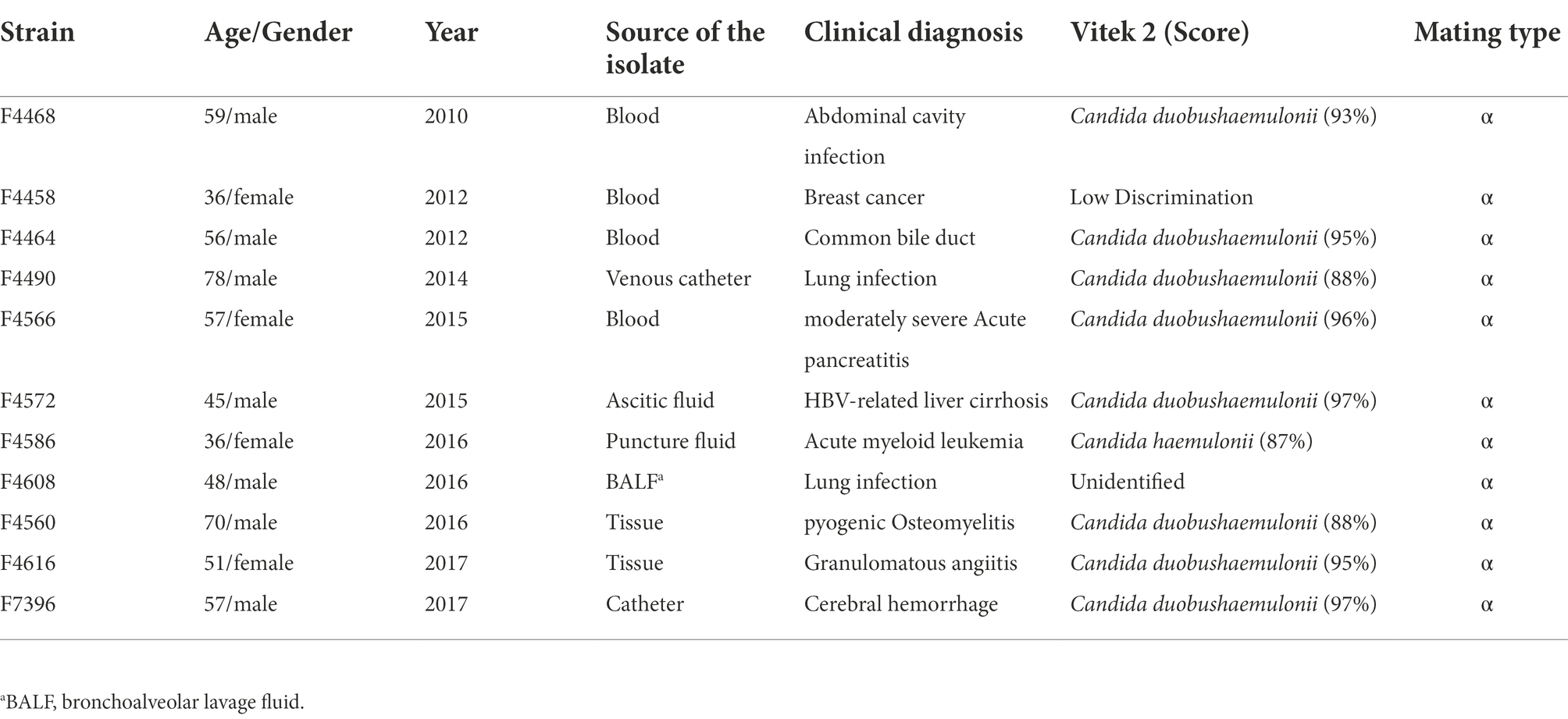
During the period from 2010 to 2017 (Table 1), 11 C. duobushaemulonii isolates were collected from nine different hospitals in eight provinces under the CHIF-NET. These isolates were mainly of invasive fungal infection specimens. Strains isolated before 2015 were identified and their susceptibility tested in our previous article (Hou et al., 2016a).
Species identification
All C. duobushaemulonii were identified at the species level using Autof-MS 1000 (Autobio, Zhengzhou, China) and Vitek MS (bioMérieux, Marcy l’Étoile, France), and confirmed by sequencing the rDNA internal transcribed spacer region (ABI 3730XL, Thermo Fisher Scientific, Cleveland, OH, United States). PCR and sequencing of the amplicons were performed using the former primers (Zhang et al., 2014; Hou et al., 2016b). All 11 isolates were also re-identified using the Vitek 2 YST Card by VITEK 2 (9.02 version, bioMérieux, Marcy l’Etoile, France) following the manufacturer’s instructions.
DNA extraction and whole-genome sequencing
The whole genomic DNA of C. duobushaemulonii was extracted by the sodium dodecyl sulfate (SDS) method (Lim et al., 2016). The DNA library was constructed using NEBNext® Ultra™, following the manufacturer’s instructions. Agilent 2100 Bioanalyzer was used for quality confirmation. Whole genome of C. duobushaemulonii was sequenced using Illumina NovaSeq 6000 at Beijing Novogene Bioinformatics Technology Co., Ltd. Illumina reads from this study were deposited at National Center for Biotechnology Information (NCBI) under BioProject PRJNA883504. In addition, we downloaded the genome data of C. duobushaemulonii from the NCBI SRA database as described by Gade et al. (2020).
Genome variation, phylogenetic, and population genetic analyses
Paired-end sequences with greater than 100X coverage were used for Bioinformatics analysis. Candida duobushaemulonii B09383 (GenBank accession number PKFP00000000.1) was used as the reference genome for analysis (Chow et al., 2018). We used BWA 0.5.9 and SAMtools and bcftools 0.1.19 to analyze single nucleotide polymorphism (SNP) and insertion-deletion (indel) (Li and Durbin, 2009a; Li et al., 2009b). SNP and Indel function annotation analysis were used snpeff 4.3 (Cingolani et al., 2012). Phylogenetic tree was constructed using RAxML 8.2.12 based on 1,000 bootstrap replicates by maximum likelihood method to investigate the C. duobushaemulonii genetic relationships (Stamatakis, 2014). The genome-wide nucleotide diversity (Pi) and the average Tajima’s D estimate were calculated by DNASP 6 (Rozas et al., 2017).
Chromosome structure analysis and mating type analysis
We used YMAP to perform Copy Number Variation (CNV) analysis of C. duobushaemulonii (Abbey et al., 2014). We checked the BAM (Binary Alignment Map) file of C. duobushaemulonii genome by SAMtools to determine the coverage depth of the region where the MTLα gene is located, and determine the mating type of C. duobushaemulonii.
Broth microdilution antifungal susceptibility testing
Candida duobushaemulonii strains were tested by Sensititre YeastOne (Thermo Scientific, Cleveland, OH, United States). In addition, the standard antifungal susceptibility testing was performed according to CLSI M27-A3. Essential agreement (EA) is defined as the percent of all Sensititre™ YeastOne™ MIC results within one 2-fold dilution of the CLSI MIC result. Candida krusei ATCC 6258 and Candida parapsilosis ATCC 22019 were selected for quality control. The epidemiological cutoff values (ECV) and clinical breakpoints of antifungals against C. duobushaemulonii in vitro have been established by the CLSI (CLSI, 2020). Among them, fluconazole MICs of greater than 32 μg/ml is considered as non-WT for C. duobushaemulonii and C. auris. Flucytosine MIC values (≥32 μg/ml) were interpreted according to the CLSI document M27-S3 (CLSI, 2008). In addition, MIC of ≥2 μg/ml was used for interpreting “resistance” of amphotericin B (Pfaller et al., 2012).
Identification of variations associated with high MICs for Candida duobushaemulonii
We analyzed the mutations in ERG11, FUR1, and other genes of interest in the pathways related to sterol metabolism and 5-flucytosine metabolism (Supplementary Table S1; Arendrup and Patterson, 2017; Berkow and Lockhart, 2017).
Review of Candida duobushaemulonii infections reported in PubMed
This literature review considered the available data regarding the susceptibility of the C. duobushaemulonii species to antifungals. The literature search was performed on June 26, 2022, using the following three databases: PubMed,1 Web of Science,2 and Embase.3 The terms “Candida duobushaemulonii” were entered in the category of “Title/Abstract” in the PubMed Advanced Search Builder, and “TS = (Candida duobushaemulonii)” was entered in the Web of Science databases. The search in Embase was conducted in the advanced search area, including the terms “‘Candida duobushaemulonii’: ab,ti..”
Results
Isolates information
Of all 11 cases, seven were male and four were female, with an average age of 54 years. Among the specimens, blood specimens accounted for four patients, tissue culture specimens for two patients, bronchoalveolar lavage fluid (BALF) culture specimens, ascitic fluid, catheter, venous catheter, and puncture fluid specimens for one patient, respectively (Table 1). The patients belonged to the following departments: medicine department (45.5%; 5/11), surgery department (45.5%; 5/11), and emergency intensive care unit (9.1%; 1/11).
Species identification of Candida duobushaemulonii using MALDI-TOF, ITS sequencing and Vitek 2
All 11 clinical isolates were identified as C. duobushaemulonii by the Autof MS 1000 and Vitek MS. The ITS sequences of the study isolates exhibited over 99.5% identity to the corresponding ITS sequences of the reference C. duobushaemulonii CBS7798T isolates. For Vitek 2 system, eight C. duobushaemulonii could be identified accurately, one could not be identified, one identified with low discrimination and the remaining one was misidentified as C. haemulonii (score = 87%; Table 1).
Phylogenetic relationships and genetic diversity among Candida duobushaemulonii
Candida duobushaemulonii isolated in China shows no clustering distribution, and its evaluation did not exhibit clustered outbreaks (Figure 1). Based on the number of SNPs that differ between the Chinese strains and the international strains with a very little difference, it can be seen that the evolution rate of C. duobushaemulonii is very slow. The average pairwise distance between C. duobushaemulonii isolates was 700 SNPs (range: 78–1,271). The average number of nucleotides is close to the previously reported average (Gade et al., 2020). All strains in the phylogenetic tree can be divided into two clades (Figure 1). The first clade includes strains isolated from China, the United States, Guatemala, Venezuela, and Panama. The second clade includes strains isolated from China, the United States, Guatemala, Colombia, and Panama. The strains around the world presented a scattered distribution. Genome-wide diversity estimates show reduced polymorphism in C. duobushaemulonii (Pi = 0.24013), but the average Tajima’s D estimate was −0.89987 expected population expansion.
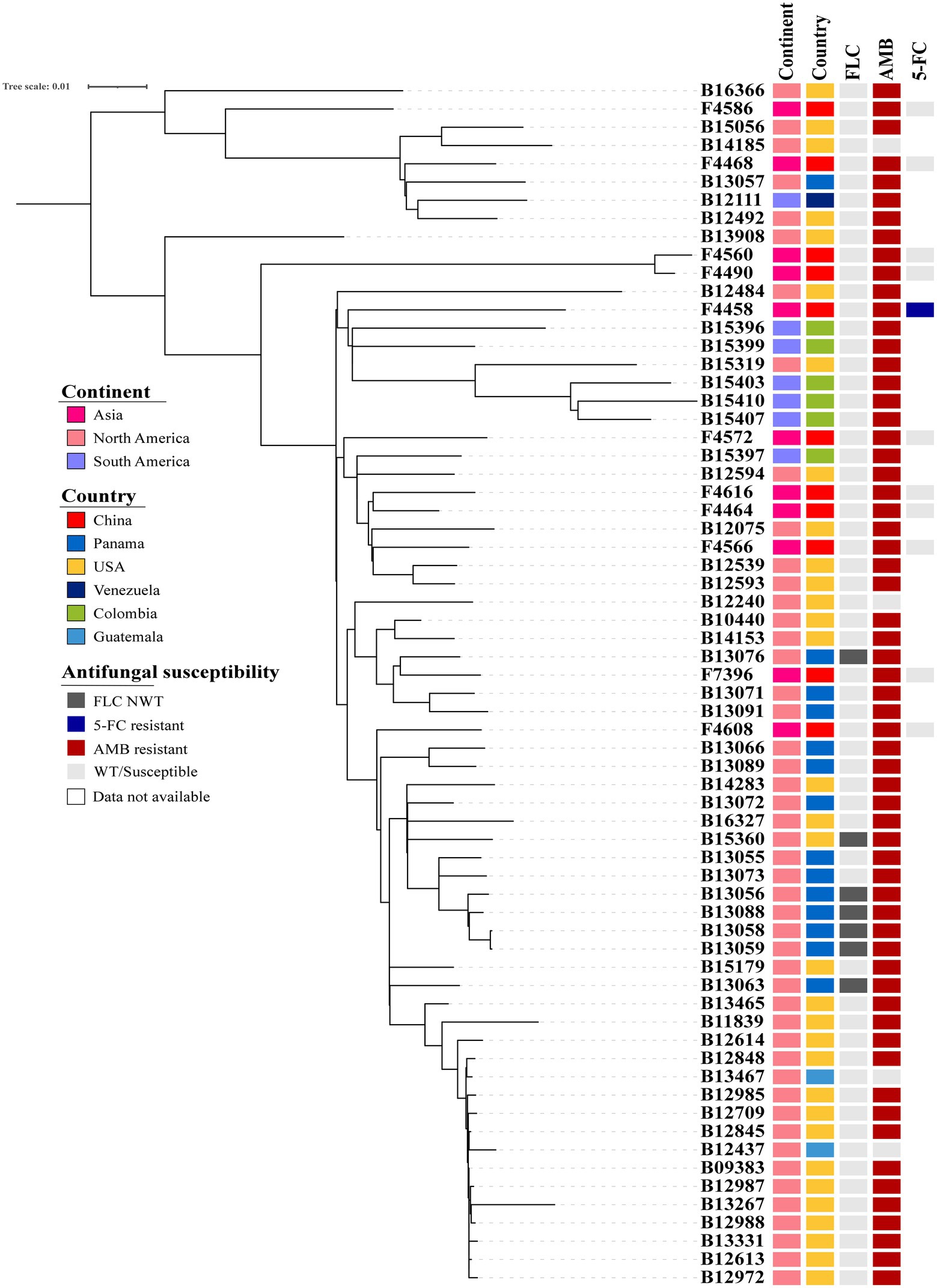
Figure 1. Phylogenetic tree of Candida duobushaemulonii. The phylogenetic tree was constructed with RAxML using the maximum likelihood method based on 8,117 SNPs. Phylogenetic tree detailing susceptibility to fluconazole (FLC), amphotericin B (AMB), and 5-flucytosine (5-FC).
Chromosome variation and mating type
Analysis of large fragments of C. duobushaemulonii chromosomes showed neither copy number variation nor aneuploidy in genome (Supplementary Figure S1). All C. duobushaemulonii isolates were mating type alpha.
Antifungal susceptibility
The quality control strains (Candida krusei ATCC 6258 and Candida parapsilosis ATCC 22019) showed MICs within the expected ranges. Aggregated MIC distributions of nine antifungal agents of C. duobushaemulonii isolates by YeastOne™ are shown in Table 2. All strains were WT to fluconazole, voriconazole, itraconazole, posaconazole, caspofungin, anidulafungin, and micafungin. MIC of 5-flucytosine for one strain was >64 μg/ml, while that of the remaining strains were all less than 0.12 μg/ml. All the 11 isolates tested showed high amphotericin B MICs (MIC ≥4 μg/ml).
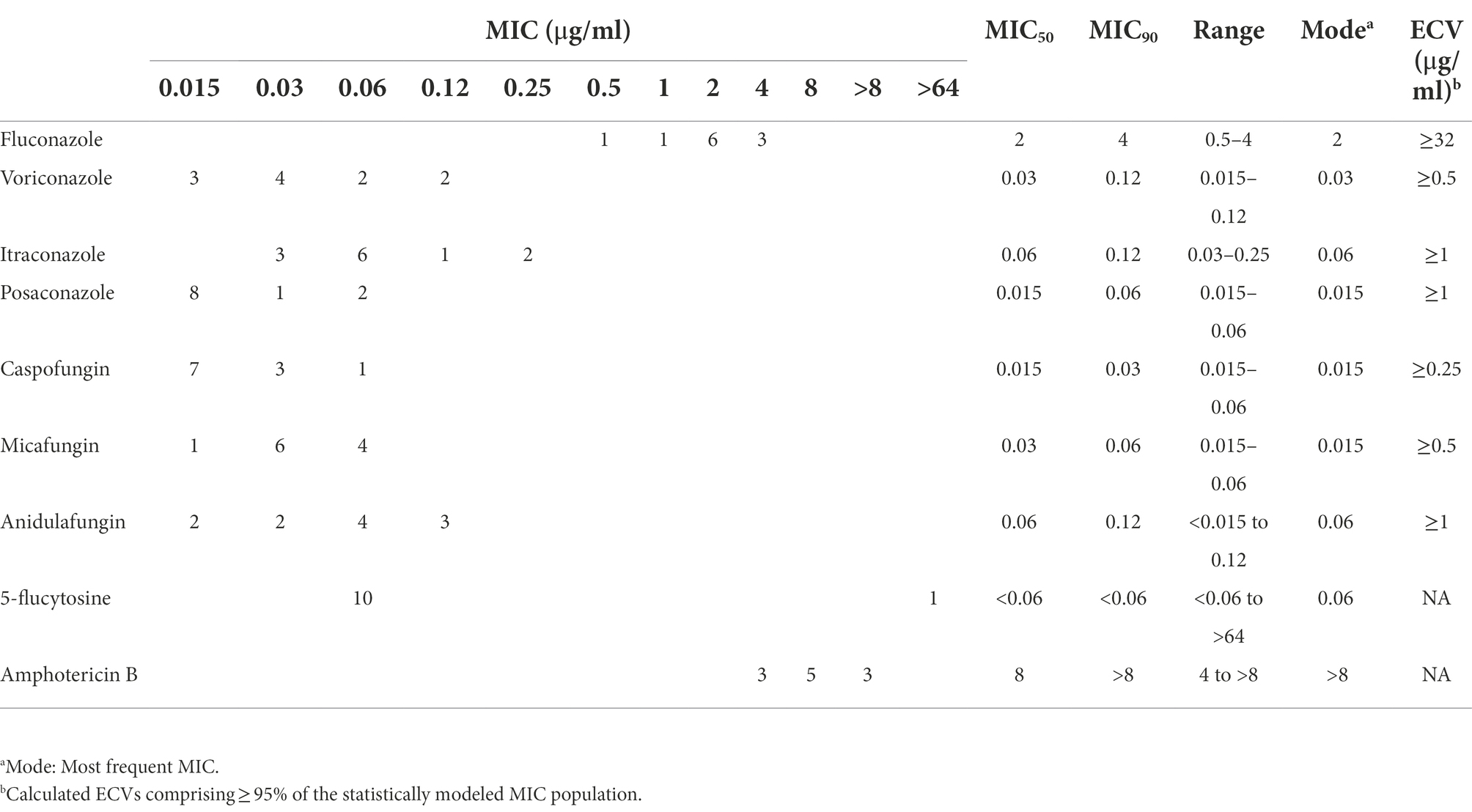
Table 2. Epidemiological cutoff values (ECV) of nine antifungal agents based on aggregated minimum inhibitory concentration distributions for C. duobushaemulonii.
Agreement between the CLSI method and sensititre YeastOne™
The EA values of the MICs between the CLSI method and YeastOne™ for most of the antifungal drugs tested were > 90%. 100% EA values were obtained for amphotericin B and 5-flucytosine. EA values for anidulafungin and micafungin were 36.4 (4/11) and 27.3% (3/11), respectively (Supplementary Table S2).
Potential variation linked to 5-flucytosine and amphotericin B resistance
Compared with the reference genome of B09383, which is sensitive to azoles and echinocandins, we found that the amphotericin B-resistant C. duobushaemulonii isolated from China shows unique variation. We found that F4490 and F4560 have a novel mutation (V907A) in the HMG1 gene and F4468 and F4586 has a previously reported mutation of S54N. In the ERG20 gene, we found two novel mutations, K347N in F4468 and M101T in F4608. In the UPC2 gene, both F4490 and F4608 possess A100T mutation. Interestingly, we discovered a novel mutation in the initiation codon (ATG-- > ATA) of FUR1 gene in a strain (F4458) with high MIC for 5-flucytosine (Table 2). In addition, it is interesting that we found that seven strains carried A626Y, T637I or P1042A substitutions in FKS1p and V30M, A485V and/or H352R in FKS2p. However, all strains were WT to echinocandins (Supplementary Table S1).
Literature review
Relatively limited data of 15 articles on antifungal susceptibility information for C. duobushaemulonii were reviewed. Some strains exhibited high MICs to fluconazole alone or to all azoles, and carried varation in Erg11p (Gade et al., 2020). In addition, there are three research reported emergence of echinocandin-resistant C. duobushaemulonii. To date there has been only one report on emergence of flucytosine-resistant C. duobushaemulonii strains (Supplementary Table S2).
Discussion
Candida duobushaemulonii, belongs to type II Candida haeumlonii complex, is relative of C. auris and Candida pseudohaemulonii. Literature reveals that C. duobushaemulonii was wrongly identified as C. haemulonii, Candida intermedia, and Debaryomyces hansenii (Desnos-Ollivier et al., 2008; Fang et al., 2016; Jurado-Martin et al., 2020). Previous studies have also shown that the identification ability of MALDI-TOF needs to be improved (Hou et al., 2016a). In the present study, although ITS sequencing, Autof MS 1000, and Vitek MS system have achieved good identification results, but three C. duobushaemulonii strains could not be identified by the Vitek 2 Compact system, which database includes C. auris, C. duobushaemulonii, and C. haemulonii var. vulnera. Considering, MALDI-TOF and ITS sequencing techniques are not all available in routine microbiology laboratories and C. duobushaemulonii actual incidence might be underestimated.
One case of hospital transmission of C. duobushaemulonii has been reported (Gade et al., 2020). Therefore, we conducted a genetic relationship analysis of C. duobushaemulonii isolated from China. We found that there was no obvious hospital infection transmission in China. However, considering the low isolation rate of C. duobushaemulonii in China, a large data sample is needed for analysis. In the overall genome evolution, the average SNP of C. duobushaemulonii is similar to that described by Gade et al. (2020).
In the drug susceptibility test, Gade et al reported that only 12.7% (7/55) strain as non-WT to fluconazole, and the MICs of these strains ranged from 64 to 256 μg/ml, with six isolates from Panama and one isolate from Texas, United States (Gade et al., 2020). De Almeida et al found that four C. duobushaemulonii isolated in Brazil has high MICs to azole and amphotericin B (de Almeida et al., 2016). Ramos et al has been reported echinocandins-resistance strains isolated in Brazil (Ramos et al., 2022). Regretfully, previous studies lacked 5-flucytosine antifungal drug sensitivity and only 18.2% (2/11) C. duobushaemulonii tested were 5-flucytosine-resistance (Cendejas-Bueno et al., 2012; de Almeida et al., 2016; Ramos et al., 2022). In the literature review, we can see that despite the low isolation rate of C. duobushaemulonii, strains resistant to azoles, echinocandins, amphotericin B, or 5-flucytosine have been emerging. However, our research found that all C. duobushaemulonii are WT to all azoles. Although there were seven isolates have missense mutations in the FKS1 and FKS2 genes, all strains were WT to echinocandins. In addition, our study might be the first to report the high MIC of 5-flucytosine for a strain isolated from China (MIC >64 μg/ml). All strains were with high MIC range to amphotericin B (4 to >8 μg/ml), which is consist with previous reports of C. duobushaemulonii high MIC to amphotericin B (Ramos et al., 2022). Although 5-flucytosine and amphotericin B lack the interpretation breakpoint, C. duobushaemulonii is notable for the high MICs of 5-flucytosine and amphotericin B.
Compared with C. auris, C. duobushaemulonii fails to attract the attention of the whole world. C. duobushaemulonii is resistant to amphotericin B and 5-flucytosine, but the resistance mechanism is not well understood. In our study, there is no missense mutation in the common drug resistance gene ERG11, and aneuploidy and multiple copies were also not found in C. duobushaemulonii, which may be different from that in C. auris and C. haemulonii, with a quite different resistance mechanism as reported previously (Gade et al., 2020). Candida duobushaemulonii not only shows high MIC of 5-flucytosine, but also shows high MIC of amphotericin B. For 5-flucytosine, there is a missense mutation G3A (M1I) in the FUR1 gene in the drug-resistant strains. This mutation is a completely new site and has not been reported. In addition, the resistance mechanism of amphotericin B is also worthy of attention. The mechanism of C. duobushaemulonii with high MIC to amphotericin B remains to be elusive. Although we found mutations involving sterol synthesis pathway genes in five strains, there were still six strains without mutations. In literature review, only 11.1% (11/99) C. duobushaemulonii had a lower amphotericin B MIC (<4 μg/ml). In addition, Carolus et al. founded the cell membrane sterols profile of C. duobushaemulonii was similar to amphotericin B-resistant species with mutations in ERG2, ERG3, ERG6, and ERG11 (Silva et al., 2020). Thus, C. duobushaemulonii maybe possess high MIC to amphotericin B. Our study is similar to the previous studies in terms of shortcomings. Due to the lack of clinical treatment information, the correlation between the non-WT C. duobushaemulonii and the clinical treatment and prognosis needs further study.
In conclusion, the emergence of C. duobushaemulonii, a rare amphotericin B, and 5-flucytosine resistant fungus, is a potential threat. The phenotype of C. duobushaemulonii resistant to 5-flucytosine might be due to the variations in FUR1. Although the invasive infection of C. duobushaemulonii is very rare, it still needs our attention due to its drug resistance. Due to the lack of clear clinical treatment data, it is necessary to study in vitro the relationship between drug resistance and clinical treatment effect in the future.
Data availability statement
The whole genome sequence raw reads presented in the study are deposited in the NCBI, BioProject PRJNA883504.
Ethics statement
Written informed consent was not obtained from the individual(s) for the publication of any potentially identifiable images or data included in this article.
Author contributions
X-FC, HZ, and X-MJ conceived and designed the experiment. JC, LL, X-LH, NL, Y-LX, FX, L-YY, Q-FH, X-LW, L-PN provided isolates. X-FC, HZ, X-MJ, XF, XH, SY-Y, J-JH, W-HY, X-LX, Y-XL, GZ, J-JZ, S-MD, WK, TW, and JL performed the experiments. X-FC, HZ, and X-MJ analyzed the data and wrote the manuscript. MX, Y-CX, XH and P-RH revised the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Natural Science Foundation of China (82002178), National High Level Hospital Clinical Research Funding (2022-PUMCH-B-074), CAMS Innovation Fund for Medical Sciences (2021-I2M-1-038 and 2021-I2M-1-044), and Beijing Key Clinical Specialty for Laboratory Medicine-Excellent Project (No. ZK201000).
Acknowledgments
The authors thank all the hospitals involved in the CHIF-NET study and Vladimir Gritsenko for his technical help in YMAP.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer XL declared a shared affiliation with the authors X-FC, HZ, X-MJ, S-YY, J-JH, X-LX, W-HY, Y-XL, GZ, J-JZ, S-MD, WK, TW, JL, MX, and Y-CX to the handling editor at the time of review.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.1001845/full#supplementary-material
SUPPLEMENTARY FIGURE S1 | No obvious copy number variation in Candida duobushaemulonii.
Footnotes
References
Abbey, D. A., Funt, J., Lurie-Weinberger, M. N., Thompson, D. A., Regev, A., Myers, C. L., et al. (2014). YMAP: a pipeline for visualization of copy number variation and loss of heterozygosity in eukaryotic pathogens. Genome Med. 6:100. doi: 10.1186/s13073-014-0100-8
Ambaraghassi, G., Dufresne, P. J., Dufresne, S. F., Vallieres, E., Munoz, J. F., Cuomo, C. A., et al. (2019). Identification of Candida auris by use of the updated Vitek 2 yeast identification system, version 8.01: a multilaboratory evaluation study. J. Clin. Microbiol. 57:e00884-19. doi: 10.1128/JCM.00884-19
Arendrup, M. C., and Patterson, T. F. (2017). Multidrug-resistant Candida: epidemiology, molecular mechanisms, and treatment. J. Infect. Dis. 216, S445–S451. doi: 10.1093/infdis/jix131
Berkow, E. L., and Lockhart, S. R. (2017). Fluconazole resistance in Candida species: a current perspective. Infect. Drug Resist. 10, 237–245. doi: 10.2147/IDR.S118892
Cendejas-Bueno, E., Kolecka, A., Alastruey-Izquierdo, A., Theelen, B., Groenewald, M., Kostrzewa, M., et al. (2012). Reclassification of the Candida haemulonii complex as Candida haemulonii (C. haemulonii group I), C. duobushaemulonii sp. nov. (C. haemulonii group II), and C. haemulonii var. vulnera var. nov.: three multiresistant human pathogenic yeasts. J. Clin. Microbiol. 50, 3641–3651. doi: 10.1128/JCM.02248-12
Chow, N. A., Gade, L., Batra, D., Rowe, L. A., Juieng, P., Loparev, V. N., et al. (2018). Genome sequence of the amphotericin B-resistant Candida duobushaemulonii strain B09383. Genome Announc. 6:e00204-18. doi: 10.1128/genomeA.00204-18
Cingolani, P., Platts, A., Wang le, L., Coon, M., Nguyen, T., Wang, L., et al. (2012). A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Flying 6, 80–92. doi: 10.4161/fly.19695
CLSI (2008). Reference method for broth dilution anti fungal susceptibility testing of yeasts, third informational supplement. CLSI document M27-S3. 3rd Edn. Wayne, PA: Clinical and Laboratory Standards Institute.
CLSI (2020). Epidemiological Cutoff Values for Antifungal Susceptibility Testing. CLSI document M59. 3rd Edn. Wayne, PA: Clinical and Laboratory Standards Institute.
de Almeida, J. N., Assy, J. G. P. L., Levin, A. S., del Negro, G. M. B., Giudice, M. C., Tringoni, M. P., et al. (2016). Candida haemulonii complex species, Brazil, January 2010-march 2015. Emerg. Infect. Dis. 22, 561–563. doi: 10.3201/eid2203.151610
Desnos-Ollivier, M., Ragon, M., Robert, V., Raoux, D., Gantier, J. C., and Dromer, F. (2008). Debaryomyces hansenii (Candida famata), a rare human fungal pathogen often misidentified as Pichia guilliermondii (Candida guilliermondii). J. Clin. Microbiol. 46, 3237–3242. doi: 10.1128/JCM.01451-08
Du, H., Bing, J., Hu, T., Ennis, C. L., Nobile, C. J., and Huang, G. (2020). Candida auris: epidemiology, biology, antifungal resistance, and virulence. PLoS Pathog. 16:e1008921. doi: 10.1371/journal.ppat.1008921
Fang, S. Y., Wei, K. C., Chen, W. C., Lee, S. J., Yang, K. C., Wu, C. S., et al. (2016). Primary deep cutaneous candidiasis caused by Candida duobushaemulonii in a 68-year-old man: the first case report and literature review. Mycoses 59, 818–821. doi: 10.1111/myc.12540
Frias-De-Leon, M. G., Martinez-Herrera, E., Acosta-Altamirano, G., Arenas, R., and Rodriguez-Cerdeira, C. (2019). Superficial candidosis by Candida duobushaemulonii: an emerging microorganism. Infect. Genet. Evol. 75:103960. doi: 10.1016/j.meegid.2019.103960
Gade, L., Munoz, J. F., Sheth, M., Wagner, D., Berkow, E. L., Forsberg, K., et al. (2020). Understanding the emergence of multidrug-resistant candida: using whole-genome sequencing to describe the population structure of Candida haemulonii species complex. Front. Genet. 11:554. doi: 10.3389/fgene.2020.00554
Hou, X., Xiao, M., Chen, S. C., Wang, H., Cheng, J. W., Chen, X. X., et al. (2016a). Identification and antifungal susceptibility profiles of Candida haemulonii species complex clinical isolates from a multicenter study in China. J. Clin. Microbiol. 54, 2676–2680. doi: 10.1128/JCM.01492-16
Hou, X., Xiao, M., Chen, S. C., Wang, H., Zhang, L., Fan, X., et al. (2016b). Sequencer-based capillary gel electrophoresis (SCGE) targeting the rDNA internal transcribed spacer (ITS) regions for accurate identification of clinically important yeast species. PLoS One 11:e0154385. doi: 10.1371/journal.pone.0154385
Jurado-Martin, I., Marcos-Arias, C., Tamayo, E., Guridi, A., de Groot, P. W. J., Quindos, G., et al. (2020). Candida duobushaemulonii: an old but unreported pathogen. J. Fungi 6, 374–384. doi: 10.3390/jof6040374
Li, H., and Durbin, R. (2009a). Fast and accurate short read alignment with burrows-wheeler transform. Bioinformatics 25, 1754–1760. doi: 10.1093/bioinformatics/btp324
Li, H., Handsaker, B., Wysoker, A., Fennell, T., Ruan, J., Homer, N., et al. (2009b). The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079. doi: 10.1093/bioinformatics/btp352
Lim, H. J., Lee, E. H., Yoon, Y., Chua, B., and Son, A. (2016). Portable lysis apparatus for rapid single-step DNA extraction of Bacillus subtilis. J. Appl. Microbiol. 120, 379–387. doi: 10.1111/jam.13011
Pfaller, M. A., Espinel-Ingroff, A., Canton, E., Castanheira, M., Cuenca-Estrella, M., Diekema, D. J., et al. (2012). Wild-type MIC distributions and epidemiological cutoff values for amphotericin B, flucytosine, and itraconazole and Candida spp. as determined by CLSI broth microdilution. J. Clin. Microbiol. 50, 2040–2046. doi: 10.1128/JCM.00248-12
Ramos, L. S., Figueiredo-Carvalho, M. H. G., Silva, L. N., Siqueira, N. L. M., Lima, J. C., Oliveira, S. S., et al. (2022). The threat called Candida haemulonii species complex in Rio de Janeiro state, Brazil: focus on antifungal resistance and virulence attributes. J. Fungi 8, 574–593. doi: 10.3390/jof8060574
Rozas, J., Ferrer-Mata, A., Sanchez-DelBarrio, J. C., Guirao-Rico, S., Librado, P., Ramos-Onsins, S. E., et al. (2017). DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol. Biol. Evol. 34, 3299–3302. doi: 10.1093/molbev/msx248
Silva, L. N., Oliveira, S. S. C., Magalhães, L. B., Neto, V. V. A., Torres-Santos, E. C., Carvalho, M. D. C., et al. (2020). Unmasking the mmphotericin B resistance mechanisms in candida haemulonii species complex. ACS Infect Dis. 6, 1273–1282. doi: 10.1021/acsinfecdis.0c00117
Stamatakis, A. (2014). RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313. doi: 10.1093/bioinformatics/btu033
Zhang, L., Xiao, M., Wang, H., Gao, R., Fan, X., Brown, M., et al. (2014). Yeast identification algorithm based on use of the Vitek MS system selectively supplemented with ribosomal DNA sequencing: proposal of a reference assay for invasive fungal surveillance programs in China. J. Clin. Microbiol. 52, 572–577. doi: 10.1128/JCM.02543-13
Keywords: Candida duobushaemulonii, antifungal susceptibility, FUR1, whole genome sequence, drug resistance mechanisms
Citation: Chen X-F, Zhang H, Jia X-M, Cao J, Li L, Hu X-L, Li N, Xiao Y-L, Xia F, Ye L-Y, Hu Q-F, Wu X-L, Ning L-P, Hsueh P-R, Fan X, Yu S-Y, Huang J-J, Xie X-L, Yang W-H, Li Y-X, Zhang G, Zhang J-J, Duan S-M, Kang W, Wang T, Li J, Xiao M, Hou X and Xu Y-C (2022) Antifungal susceptibility profiles and drug resistance mechanisms of clinical Candida duobushaemulonii isolates from China. Front. Microbiol. 13:1001845. doi: 10.3389/fmicb.2022.1001845
Edited by:
Weihua Pan, Shanghai Changzheng Hospital, ChinaReviewed by:
Xiaofang Li, Chinese Academy of Medical Sciences and Peking Union Medical College, ChinaYinggai Song, First Hospital, Peking University, China
Shallu Kathuria, National Centre for Disease Control (NCDC), India
Mohammad Asadullah Asadzadeh, Kuwait University, Kuwait
Lalitha Gade, Centers for Disease Control and Prevention (CDC), United States
Copyright © 2022 Chen, Zhang, Jia, Cao, Li, Hu, Li, Xiao, Xia, Ye, Hu, Wu, Ning, Hsueh, Fan, Yu, Huang, Xie, Yang, Li, Zhang, Zhang, Duan, Kang, Wang, Li, Xiao, Hou and Xu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ying-Chun Xu, xycpumch@139.com; Xin Hou, houxinyffs@163.com
†These authors have contributed equally to this work and share first authorship
 Xin-Fei Chen
Xin-Fei Chen Han Zhang
Han Zhang Xin-Miao Jia
Xin-Miao Jia Jin Cao5
Jin Cao5 Yu-Ling Xiao
Yu-Ling Xiao Qing-Feng Hu
Qing-Feng Hu Po-Ren Hsueh
Po-Ren Hsueh Xin Fan
Xin Fan Shu-Ying Yu
Shu-Ying Yu Jing-Jing Huang
Jing-Jing Huang Wen-Hang Yang
Wen-Hang Yang Ge Zhang
Ge Zhang Jing-Jia Zhang
Jing-Jia Zhang Si-Meng Duan
Si-Meng Duan Meng Xiao
Meng Xiao Xin Hou
Xin Hou Ying-Chun Xu
Ying-Chun Xu