- 1State Key Laboratory of Food Science and Technology, Jiangnan University, Wuxi, China
- 2School of Food Science and Technology, Jiangnan University, Wuxi, China
- 3State Key Laboratory of Biocatalysis and Enzyme Engineering, School of Life Sciences, Hubei University, Wuhan, China
- 4Key Laboratory of Carbohydrate Chemistry and Biotechnology, Ministry of Education, Jiangnan University, Wuxi, China
- 5China Biotech Fermentation Industry Association, Beijing, China
- 6Anhui Heiwa Food-Jiangnan University Joint R & D Center, Anhui Heiwa Food Technology Co., Ltd., Bozhou, China
Aflatoxins are naturally occurring high-toxic secondary metabolites, which cause worldwide environmental contaminations and wastes of food and feed resources and severely threaten human health. Thus, the highly efficient methods and technologies for detoxification of aflatoxins are urgently needed in a long term. In this work, we report the construction of recombinant Kluyveromyces lactis strains GG799(pKLAC1-Phsmnp), GG799(pKLAC1-Plomnp), GG799(pKLAC1-Phcmnp), and then the food-grade expression of the three manganese peroxidases in these strains, followed by the degradation of aflatoxin B1 (AFB1) using the fermentation supernatants. The expression of the manganese peroxidases was achieved in a food-grade manner since Kluyveromyces lactis is food-safe and suitable for application in food or feed industries. The inducible expression process of the optimal recombinant strain GG799(pKLAC1-Phcmnp) and the aflatoxin B1 degradation process were both optimized in detail. After optimization, the degradation ratio reached 75.71%, which was an increase of 49.86% compared to the unoptimized results. The degradation product was analyzed and determined to be AFB1-8,9-dihydrodiol. The recombinant strain GG799(pKLAC1-Phcmnp) supernatants degraded more than 90% of AFB1 in the peanut samples after twice treatments. The structural computational analysis for further mutagenesis of the enzyme PhcMnp was also conducted in this work. The food-grade recombinant yeast strain and the enzyme PhcMnp have potential to be applied in food or feed industries.
Introduction
Aflatoxins are naturally occurring highly toxic secondary metabolites, mainly produced by several species of fungus genus Aspergillus such as A. flavus, A. parasiticus, and A. nomius (Ismail et al., 2018; von Hertwig et al., 2020). Together with other mycotoxins, aflatoxins had caused worldwide contaminations and wastes of food supplies and severely threatened human health (Pitt and Miller, 2017; Eskola et al., 2020). There are more than 20 types of structurally similar but different molecules of aflatoxins, in which aflatoxin B1 is the most prominent and dangerous type (Abrar et al., 2013; Loi et al., 2016). In 2012, AFB1 was classified as the Group 1 carcinogen (carcinogenic to humans) by the International Agency for Research on Cancer (IARC) (Haque et al., 2020). Further studies and conclusions by IARC and World Cancer Research Fund (WCRF) strongly supported the linkage of AFB1 to liver cancer and other types of cancer risks (Marques et al., 2019; Claeys et al., 2020). In China, the risks of exposure to aflatoxin B1 in foodstuffs or feeds still needed long-term supervision and control (Zhang et al., 2020). Thus, the highly efficient methods and techniques for detoxification of aflatoxin are urgently needed in a long term, to continuously minimize the loss of foodstuffs worldwide and reduce the harmful effects to human beings.
Currently, there are mainly three types of mycotoxin detoxification methods and techniques: physical, chemical, and biological. The physical methods are mainly solvent extraction and absorption, high-temperature degradation, and radiation processing (Di Gregorio et al., 2014; Ismail et al., 2018), and the chemical methods include structural degradation of toxins by some organic reagents such as aldehydes, oxidizing agents (Ismail et al., 2018), and ozone (Wang et al., 2016a,b). The biological methods usually utilize the alive microorganisms (Styriak et al., 2001; Wang et al., 2018), the expressed enzymes (Cao et al., 2011; Wang et al., 2019), and the microbial metabolites (Ghazvini et al., 2016) for structural degradation of toxins. In comparison with the biological methods, the physical and chemical methods have some defects such as the loss of nutrition and the residue of absorbent and chemical compounds, so that the safety and negative effects of using these methods should be continuously investigated (Peng et al., 2018). The biological methods degrade the mycotoxins with mild parameters and in more environment-friendly ways (Loi et al., 2017). However, the direct use of microorganisms still has limitations. For example, many foodstuffs are not suitable for inoculation and proliferation of microorganisms (Assaf et al., 2019), or else the nutrition contents and the texture characteristics might be changed (Peng et al., 2018). Therefore, utilization of extracellular enzymes and culture supernatants might be the best strategies for biological detoxification of mycotoxins (Cao et al., 2011; Feltrin et al., 2017; Zhao et al., 2020).
In this work, we report a new and safe microbial approach for degradation of aflatoxin B1 (AFB1) by the culture supernatants of three food-grade recombinant yeasts, which contained inducibly expressed manganese peroxidases. Manganese peroxidase was originally used in industrial and agricultural applications such as enzymatic bleaching of pulp, treatment of agricultural waste, and treatment of dye wastewater (Ha et al., 2001); in this work, we achieved food-grade expression of the enzymes in yeast and applied the enzyme for the degradation of AFB1 in foodstuff peanuts. The coding genes for manganese peroxidases PhsMnp, PloMnp, and PhcMnp, which originated from Phanerochaete sordida, Pleurotus ostreatus, and Phanerochaete chrysosporium, were respectively cloned to expression vector pKLAC1 and then expressed in the food-grade expression host Kluyveromyces lactis GG799, which is widely regarded as a food-safe yeast strain and is a suitable organism for the production of food enzymes (van den Dungen et al., 2021). The recombinant strain GG799(pKLAC1-Phcmnp) with the highest AFB1 degradation efficiency was further studied, and the AFB1 degradation parameters were optimized. Besides, the structure of the manganese peroxidase PhcMnp was homology-modeling analyzed for further mutagenesis of this enzyme.
Materials and Methods
Chemicals, Strains, and Culture Techniques
AFB1 was purchased from Pribolab (Qingdao, China), which was made into a 1.0-mg/ml stock solution with methanol or acetonitrile solvent and was preserved and refrigerated in the darkness at −20°C. DNA marker, restriction enzymes (BglII, SalI, SacII, etc.), T4 DNA ligase, and plasmid extraction kits were purchased from Thermo Fisher Scientific (Shanghai, China). Sorbitol and hemin were purchased from Aladdin (Shanghai, China). Fungus genomic DNA extraction kit and growth media components were purchased from Sangon Biotech (Shanghai, China). Methanol, acetonitrile, and formic acid were purchased from Tedia (Fairfield, OH, United States). All other reagents and chemicals were of analytical reagent grade. The genome DNA of K. lactis recombinants was extracted and purified by the fungus genomic DNA extraction kit. All the DNA sequencing works were done in Sangon Biotech (Shanghai, China).
For cultivation of K. lactis GG799 and the recombinant strains GG799(pKLAC1-Phsmnp), GG799(pKLAC1-Plomnp), and GG799(pKLAC1-Phcmnp), the strains were grown in YEPD liquid (1.0% yeast extract, 2.0% peptone, 2.0% glucose, and pH 6.3) at 30°C and 200 rpm for 18 h. For the inducible expression of manganese peroxidases in the recombinant strains, the strains were cultivated in YEPG medium (1.0% yeast extract, 2.0% peptone, 2.0% galactose, 1.0 mmol/l MnSO4, 1.0 mmol/l hemin, pH 6.5) under aerobic conditions at 30°C and 200 rpm for 96 h.
Construction of Recombinant Plasmids and Expression Strains
Three manganese peroxidases known to be capable of degrading AFB1 in the original strains were selected. Two manganese peroxidase genes from P. sordida (Wang et al., 2011) and P. ostreatus (Yehia, 2014) were synthesized and named Phsmnp and Plomnp, respectively, and the manganese peroxidase gene from P. chrysosporium (Gowda et al., 2007) was constructed and named Phcmnp by adding a glutathione-S-transferase (GST) tag to its 5 ′-end, for the GST tag can be co-expressed with proteins to increase the solubility of easily aggregated proteins (Tarazi et al., 2021). The three synthesized genes were inserted in the pKLAC1 vector to the BglII and SalI sites and then linked by T4 DNA ligase to obtain the constructs (pKLAC1-Phsmnp, pKLAC1-Plomnp, and pKLAC1-Phcmnp). These plasmids were transformed into the host strain E. coli DH5α for screening of positive clones by PCR verification (detailed PCR program shown in Supplementary Table 1), restriction digestion of the plasmids, and DNA sequencing. The verified recombinant plasmids were amplified in the E. coli hosts and then transformed into the yeast host K. lactis GG799, for the construction of recombinant yeast strains. Detailed construction procedures for recombinant plasmids and yeast strains are shown in Figure 1. The result recombinant yeast strains were named GG799(pKLAC1-Phsmnp), GG799(pKLAC1-Plomnp), and GG799(pKLAC1-Phcmnp), respectively.
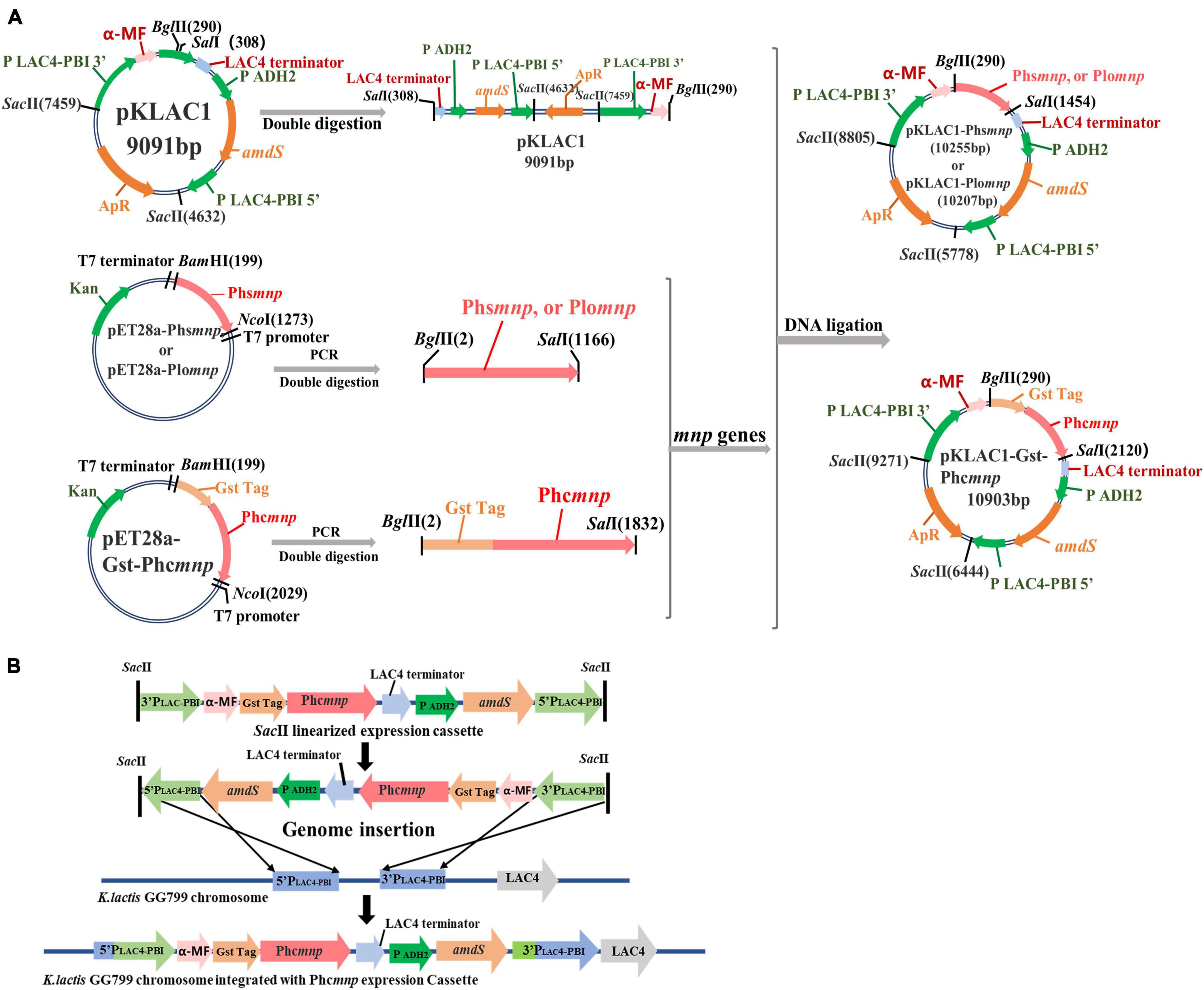
Figure 1. Flowchart for constructions of recombinant plasmids and strains. (A) Flowchart for constructions of recombinant plasmids. (B) The integration mechanism for construction of recombinants using K. lactis GG799(pKLAC1-Phcmnp) as an example.
Inducible Expression Methods of the Enzymes
The recombinant strains K. lactis GG799(pKLAC1-Phsmnp), GG799(pKLAC1-Plomnp), and GG799(pKLAC1-Phcmnp) were cultivated in YEPD liquid. When OD600 reached 1.0, 1% of the cultures were transferred into the YEPG liquid for the inducible expression of the manganese peroxidases, according to the methods described above. The supernatants of these cultures were collected by centrifugation and concentrated by 10-kDa ultrafiltration for subsequent experiments.
Degradation System of AFB1 by the Enzymes
The AFB1 degradation process was conducted with the supernatants obtained from the recombinant strains GG799(pKLAC1-Phsmnp), GG799(pKLAC1-Plomnp), and GG799(pKLAC1-Phcmnp). The total volume of the reaction system was 1.0 ml, containing 50.0 mmol/l malonic acid buffer (pH 4.5), 2.0 μg/ml AFB1, 1.0 mmol/l MnSO4, 2.0 mmol/l glucose, 1.0 U/ml glucose oxidase, and 1.0 mg/ml culture supernatant protein, while the protein was replaced with 50.0 mmol/l malonic acid buffer (pH 4.5) in blank control. The solution containing all the samples needed was incubated at 40°C for 40 h, followed by enzyme inactivation in a water bath at 80°C for 10 min. The samples were filtered through the 0.22-μm membranes for removal of the impurities and then subjected to UPLC-TQD analysis.
Quantitative Assay of AFB1
The UPLC-TQD assay was used for qualitative and quantitative analyses of the concentration of AFB1 in samples. The specific operating parameters were as follows, UPLC chromatographic conditions: column: C18; flow rate: 0.30 ml/min; column temperature: 40°C; mobile phases: H2O (buffer A) and acetonitrile (buffer B). MS (Mass) spectrometry conditions: ion source: electrospray ion source; MS spectrometry scanning mode: multiple reaction monitoring mode (MRM); cone hole voltage: 3.0 kV; heating gas temperature: 500°C; ion source temperature: 150°C; desolvation gas: 800 l/h.
The AFB1 standard samples were prepared in acetonitrile with the concentration gradients (100.0, 200.0, 500.0, 1,000.0, 2,000.0, and 5,000.0 ng/ml) to establish the standard curve for AFB1 concentration calculation.
Optimization of Inducible Expression Conditions for GG799(pKLAC1-Phcmnp)
The recombinant strain K. lactis GG799(pKLAC1-Phcmnp) was subjected to different induction conditions (temperature: 15–35°C, time: 24–120 h, rotation speed: 0–300 rpm, hemin concentration: 0.1–5.0 mmol/l, initial pH of induction medium: 3.0–9.0, MnSO4 concentration: 0.1–5.0 mmol/l, galactose concentration: 5.0–80.0 g/l). The enzyme was expressed and secreted to the supernatants as reported (Gnanamani et al., 2006). The expression levels of enzyme PhcMnp were optimized with these parameters univariately and characterized by the degradation ratio of AFB1.
On the basis of the single-factor test, seven factors (temperature, time, rotation speed, hemin concentration, initial pH of induction medium, MnSO4 concentration, and galactose concentration) with different levels were designed for the orthogonal test. The test factors and levels are shown in Supplementary Table 2.
Optimization of Degradation Parameters for AFB1 Degradation by Supernatants of GG799(pKLAC1-Phcmnp)
Under the optimal induced expression conditions, the degradation reaction conditions for AFB1 by the supernatants of recombinant strain GG799(pKLAC1-Phcmnp) were further optimized by setting the concentration gradient, according to the methods reported previously (Wang et al., 2011). For the optimization of single factors by testing the degradation ratio under different reaction conditions, reaction time (4–48 h), temperature (20–60°C), pH (3.0–5.5), protein concentration (0.4–8.0 g/l), MnSO4 concentration (0.2–10.0 mmol/l), glucose concentration (1.0–3.5 mmol/l), and glucose oxidase concentration (0.2–5.0 U/ml) were investigated.
Seven factors (time, temperature, pH, protein concentration, MnSO4 concentration, glucose concentration, and glucose oxidase concentration) and levels of the orthogonal test were also designed (Supplementary Table 3).
Degradation Methods of AFB1 in Peanuts by the Enzymes
According to the methods reported previously (Yang et al., 2020), the peanut samples were shelled, the impurities and dust of which were removed. The samples were ground into powders by homogeneity, with the particle size less than 2.0 mm. The powder samples (5.0 g) were added with 10.0 ml water, stirred thoroughly, followed by sterilization in an autoclave.
The AFB1 standards were added to the peanut samples at different concentrations (50.0, 500.0, and 2,000.0 μg/kg), and the samples were stirred thoroughly. For AFB1 degradation tests, the samples were mixed with 1.2 mmol/l MnSO4, 2.5 mmol/l glucose, 1.5 U/ml glucose oxidase, and the supernatants (containing proteins concentration: 3.0 g/l) for enzymatic degradation reactions. The final volume of the reaction system was 35.0 ml. The reaction was carried out at 40°C, pH of 4.5, for 40 h. All the samples were analyzed by HPLC-MS spectrum (UPLC-TQD) assay to detect the residual concentration of AFB1 in the samples.
Results
Construction of Recombinant
Strains Kluyveromyces lactis GG799(pKLAC1-Phsmnp), GG799(pKLAC1-Plomnp), and GG799(pKLAC1-Phcmnp).
Three recombinant plasmids pKLAC1-Phsmnp, pKLAC1-Plomnp, and pKLAC1-Phcmnp were constructed in this work according to the methods described above. The flowchart for the construction is shown in Figure 1A. The vector pKLAC1 and genes Phsmnp, Plomnp, and Phcmnp were 9,091, 1,164, 1,116, and 1,824 bp (with a GST tag fused on the 5 ′-terminal), respectively. Plasmid verification results using agarose gel electrophoresis are shown in Supplementary Figure 1, and the plasmids with correct digestion results were subjected to DNA sequencing verification.
The recombinant plasmids were transferred K. lactis GG799 host, and the transformants were selected and verified by PCR amplification, as shown in Supplementary Figure 2, which indicated successful construction of the recombinant strains of K. lactis GG799(pKLAC1-Phsmnp), GG799(pKLAC1-Plomnp), and GG799(pKLAC1-Phcmnp).
Inducible Expression of Enzymes and Degradation of AFB1 by the Culture Supernatants of the Recombinant Strains
To express inducibly for manganese peroxidases, the recombinant strains GG799(pKLAC1-Phsmnp), GG799(pKLAC1-Plomnp), and GG799(pKLAC1-Phcmnp) were firstly cultivated in YEPD liquid for accumulation of the biomass and then induced in YEPG liquid.
The activities of manganese peroxidases were characterized by AFB1 degradation ratios. The supernatants of the recombinant strains were used for AFB1 degradation, and the remaining contents of AFB1 were analyzed by HPLC-TQD. A comparison of the liquid chromatogram of AFB1 degradation results by the three recombinants’ supernatants is shown in Figure 2. It can be seen that the peak area of degradation residue by PhcMnp was the smallest in the three samples, which indicated that the enzyme PhcMnp has the best enzyme activity. The degradation ratios of AFB1 by the three recombinants’ supernatants are shown in Table 1. As shown in this table, the supernatant of recombinant strain GG799(pKLAC1-Phcmnp) had the highest degradation ratio for AFB1 (50.52 ± 3.69%), which was significantly higher than those from the recombinant strains GG799(pKLAC1-Phsmnp) (35.55 ± 3.30%) and GG799(pKLAC1-Plomnp) (40.02 ± 1.77%) (p < 0.05).
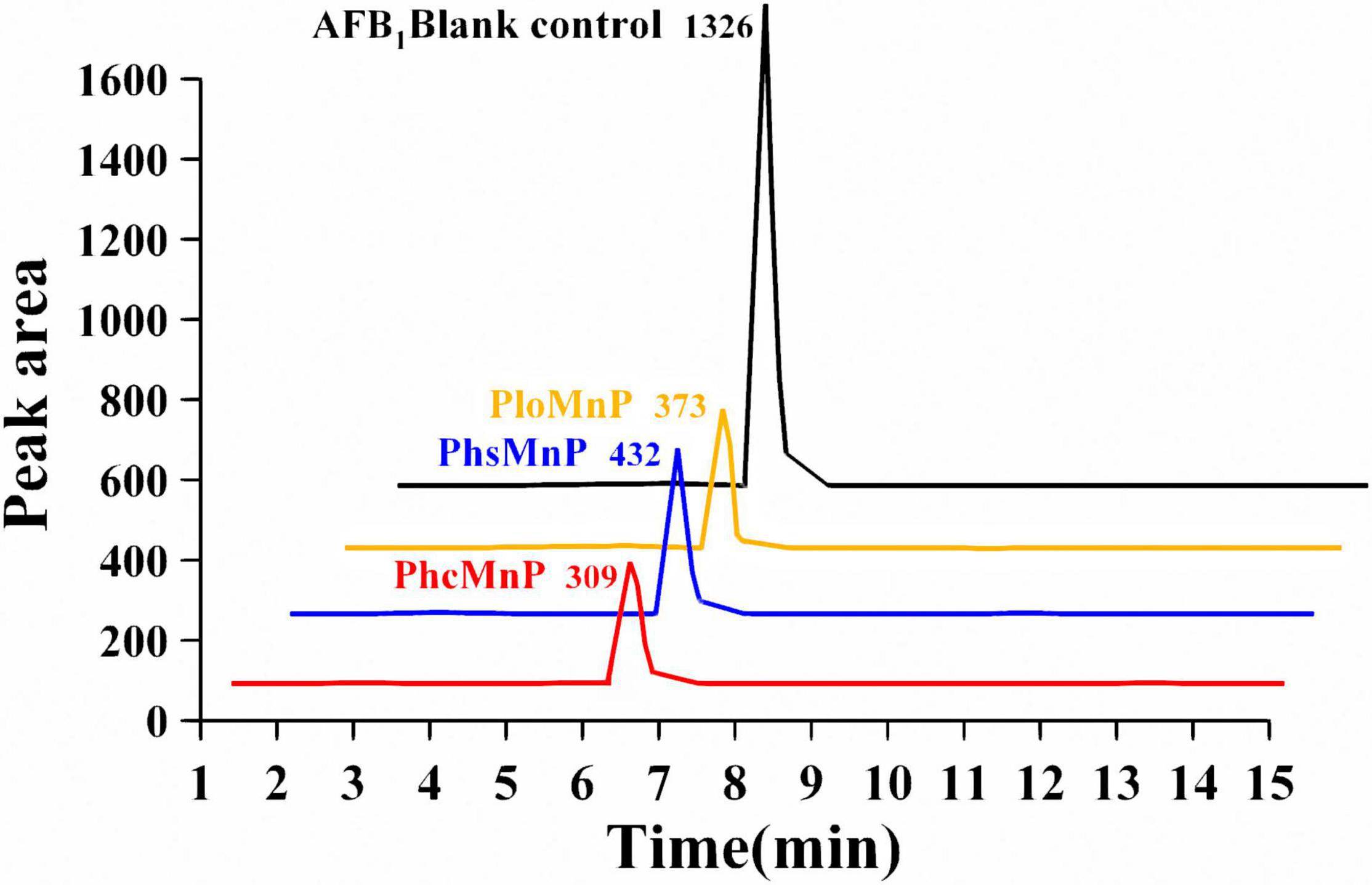
Figure 2. Comparison of liquid chromatogram of AFB1 degradation reaction by GG799(pKLAC1-Plomnp), GG799(pKLAC1-Phsmnp), and GG799(pKLAC1-Phcmnp). The addition amount of three enzymes in the system was 1.0 mg/ml. The black peak was AFB1 blank control; the orange peak was the residual of AFB1 degraded by enzyme PloMnp; the blue peak was the residual of AFB1 degraded by PhsMnp; and the red peak was the residual of AFB1 degraded by PhcMnp. All the peaks in this 3D visualization figure were appeared at the retention time of 6.3 min.
Optimization of Inducible Expression for GG799(pKLAC1-Phcmnp) and the Degradation Efficiency of AFB1
The influence of factors such as induction temperature, time, and rotation speed on degradation ratios of AFB1 was designed to analyze the expression level for the enzyme PhcMnp. As shown in Supplementary Figures 3, 4, the degradation ratios observed by the recombinants’ supernatants on AFB1 under the conditions of different induction factors had similar trends. With the increase in induction temperature, time, rotation speed, medium pH (Supplementary Figures 3A–D), and the increased concentrations of hemin, MnSO4, and galactose (Supplementary Figures 4A–C), the degradation ratios of AFB1 showed a trend of increasing at first and then decreasing.
In Supplementary Figure 4A, a high concentration of hemin inhibited the enzyme activity and degradation, causing low degradation efficiency of AFB1, which was similar to the previous results (Jiang et al., 2008).
The results in Supplementary Figure 4B indicated that with the increase in MnSO4 concentration in the medium, the degradation effect on AFB1 increased at first and then decreased, and the highest degradation ratio of AFB1 reached 58.32 ± 2.07% at the MnSO4 concentration of 1.0 mmol/l. Meanwhile, the expression level of the enzyme GG799(pKLAC1-Phcmnp) was also influenced by the concentration of the inducer galactose (Xia et al., 2021). In Supplementary Figure 4C, the highest degradation ratio of AFB1 reached 53.48 ± 1.63% at the MnSO4 concentration of 60 g/l.
From the above results, the optimal conditions for the inducible expression of the AFB1 degradation enzyme were as follows: YEPG medium containing 60.0 g/l galactose, 1.0 mmol/l MnSO4, 1.0 mmol/l hemin, and an initial medium pH of 6.0, with induction at 30°C and 200 rpm for 96 h.
To further explore the optimal combination of the induction expression conditions of GG799(pKLAC1-Phcmnp), factors like induction temperature, induction time, rotation speed, hemin concentration, initial pH of the medium, MnSO4 concentration, and galactose concentration were designed in an orthogonal experiment, and the optimization results were characterized by the degradation ratio of AFB1. The results are shown in Supplementary Table 4. The optimum induction parameter combination was A2D2B2F2C2E3G1. Under this condition, the highest degradation ratio of AFB1 by the recombinant’s supernatant was 67.40 ± 0.74%, which was increased by 15.6% compared to the optimal result of the single-factor optimization test.
Optimization of Reaction Parameters for AFB1 Degradation and Determination of the Results
In this study, the reaction conditions for AFB1 degradation by supernatant of GG799(pKLAC1-Phcmnp) were optimized with the following factors: reaction time, reaction temperature, pH, protein concentration, MnSO4 concentration, glucose addition, and glucose oxidase addition, and the results are shown in Figures 3, 4. As shown in Figure 3, the trends of the degradation ratio of AFB1 under different reaction parameters were approximately similar. With the increase in induction time, temperature, pH, and protein concentration (Figures 3A–D), the degradation ratio of AFB1 by PhcMnp showed a trend of increasing at first and then decreasing.
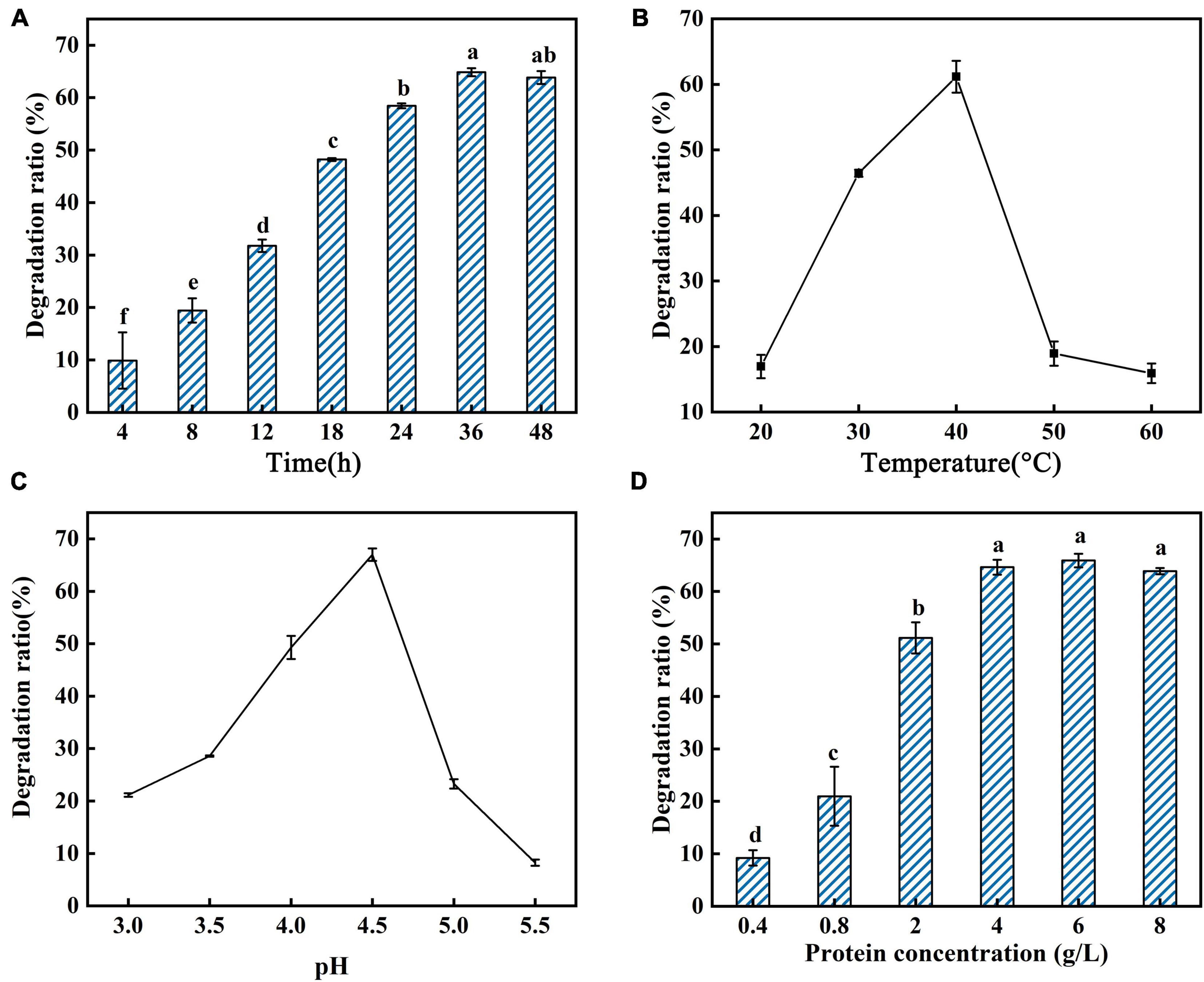
Figure 3. Degradation ratio of AFB1 by PhcMnp under different reaction parameters. (A) The effects of time on the degradation ratio of AFB1 by PhcMnp. (B) The effects of temperature on the degradation ratio of AFB1 by PhcMnp. (C) The effects of pH on the degradation ratio of AFB1 by PhcMnp. (D) The effects of protein concentrations on the degradation ratio of AFB1 by PhcMnp.
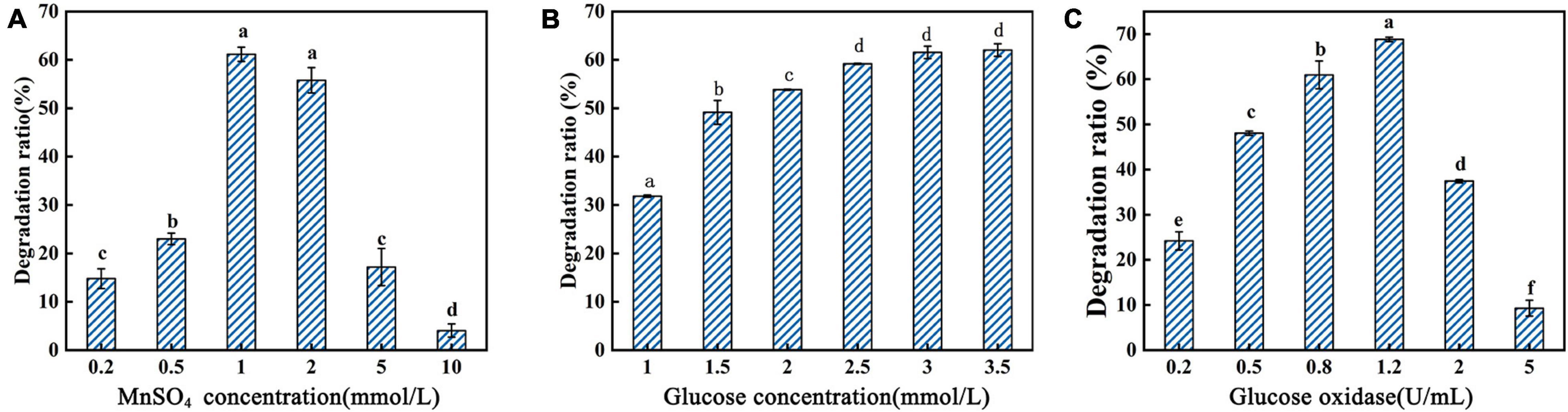
Figure 4. Degradation ratio of AFB1 by PhcMnp under different concentrations of reaction additives. (A) The effects of MnSO4 concentrations on the degradation ratio of AFB1. (B) The effects of glucose concentrations on the degradation ratio of AFB1. (C) The effects of glucose oxidase concentrations on the degradation ratio of AFB1.
As shown in Figure 4A, a high concentration of MnSO4 in the medium inhibited the degradation ratio of AFB1. The highest degradation ratio of AFB1 by the recombinant’s supernatant reached 61.16 ± 1.09% at the MnSO4 concentration of 1.0 mmol/l; however, when the MnSO4 concentration was 10.0 mmol/l, the degradation ratios of AFB1 were less than 10%. The degradation of AFB1 by the enzyme requires the participation of H2O2, but due to the instability of H2O2 and the reversible inactivation of manganese peroxidase under the condition of a high concentration of H2O2 (Wariishi et al., 1988), we chose to add glucose and glucose oxidase to the reaction system to release H2O2 slowly and steadily in this study, as shown in Figures 4B,C. In Figure 4B, the degradation ratio of AFB1 increased with the addition of glucose and then decreased when the glucose addition was greater than 2.5 mmol/l, probably because the H2O2 produced by the catalysis of 1.5 U/ml glucose oxidase at the beginning of the addition was not sufficient for the degradation of 2.0 μg/ml AFB1. Conversely, when the glucose addition reached 2.5 mmol/l, the amount of H2O2 produced by the catalysis of 1.5 U/ml glucose oxidase was saturated.
From the above results, the optimal reaction parameters for AFB1 degradation by GG799(pKLAC1-Phcmnp) supernatants were as follows: degradation system containing 6.0 g/l protein, 1.0 mmol/l MnSO4, 3.5 mmol/l glucose, and 1.2 U/ml glucose oxidase, pH of 4.5, with induction at 40°C for 36 h.
To further explore the optimal combination of the induction reaction parameters of GG799(pKLAC1-Phcmnp), factors were designed in an orthogonal experiment, and the optimization results were characterized by the degradation ratio of AFB1. The orthogonal experimental results are shown in Supplementary Table 5. The optimum induction proposal was C2B2D3F2A3E1G3. The highest degradation ratio of AFB1 by the recombinant’s supernatant was 75.71 ± 1.21% under this optimum induction proposal, which was increased by 9.1% compared to the optimal result of the single-factor optimization test.
Analysis of AFB1 Degradation Products by GG799(pKLAC1-Phcmnp) Supernatants
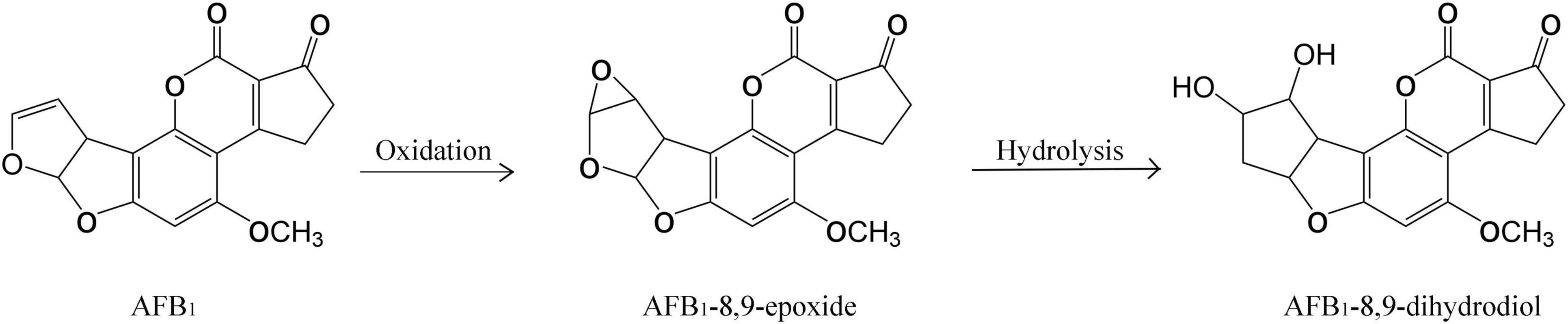
The AFB1 degradation products treated with fermentation broth of GG799(pKLAC1-Phcmnp) were identified by the mass spectrum, and the results are shown in Figure 5. The chromatogram of AFB1-degraded results by GG799(pKLAC1-Phcmnp) is shown in Figure 5A, in which the AFB1 retention time was found at 6.33 min. In addition, a large number of chromatogram peaks were found near the retention time of 11.15 min, which was considered to be the retention time of AFB1-8,9-dihydrodiol. This compound was regarded as the major degradation product of AFB1 treated with GG799(pKLAC1-Phcmnp) supernatants. To confirm this, the AFB1-8,9-dihydrodiol standards were subjected to UPLC-TQD analysis, and the retention time of this compound was found near 11.15 min (Figure 5B), whose result was consistent with that shown in Figure 5A. Theoretically, the molecular weights of AFB1 and AFB1-8,9-dihydrodiol are 312.27 and 346.288, respectively. The mass spectra of the samples of AFB1 treated with the supernatant of recombinant GG799 (pKLAC1-Phcmnp) in MRM scan mode are shown in Figure 5C, the [M + H]+ ions with a mass-to-charge ratio (m/z) of 313 were AFB1, and those slightly larger than 346 were presumed to be AFB1-8,9-dihydrodiol, which was consistent with the conclusion of Wang et al. (2011), who found that AFB1-8,9-dihydrodiol produced by the degradation of AFB1 was much less toxic than AFB1. The schematic for the degradation of AFB1 by manganese peroxidases is shown in Figure 6.
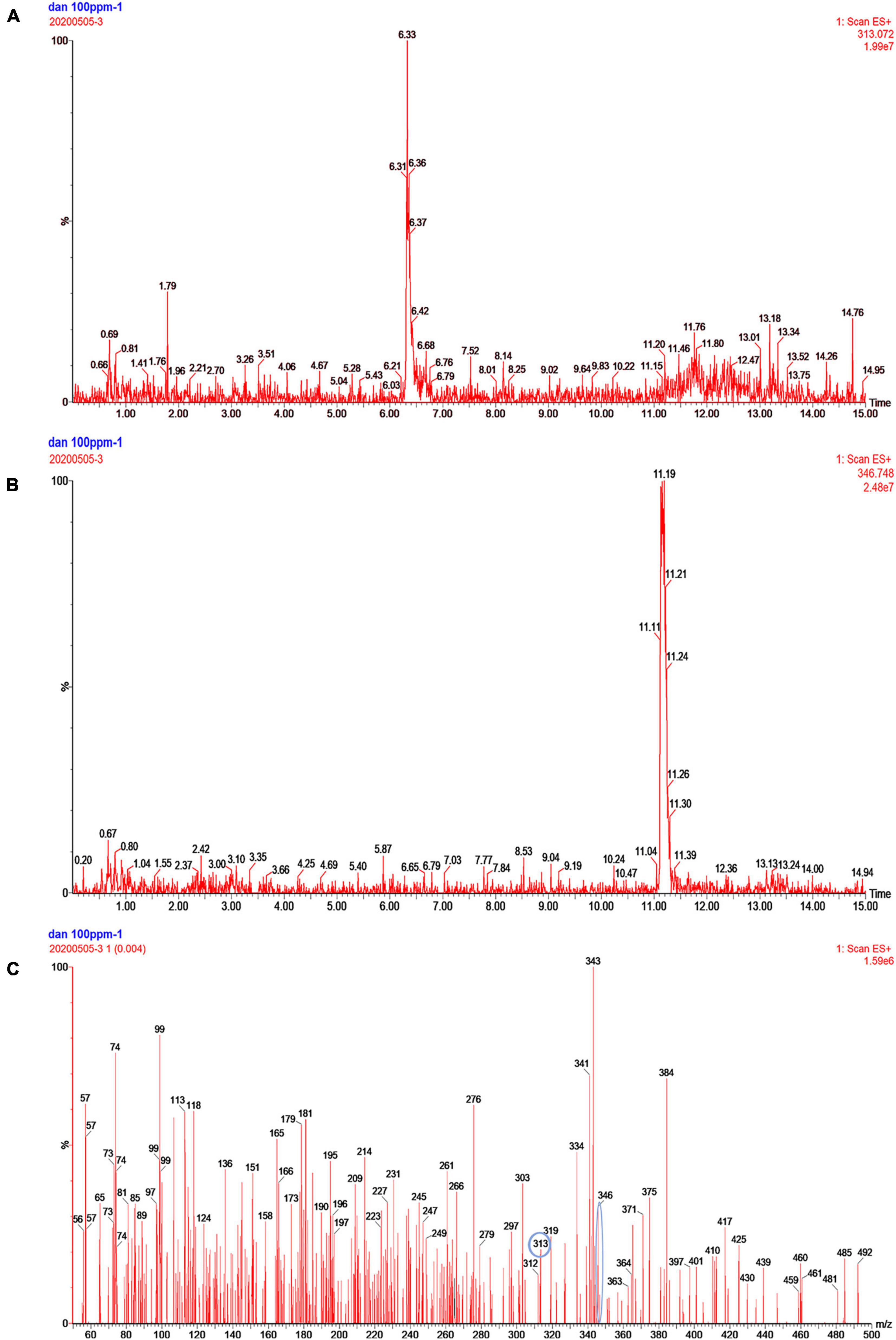
Figure 5. Analysis of the degradation products of AFB1 by GG799(pKLAC1-Phcmnp) fermentation broth. (A) Chromatogram of AFB1 metabolites degraded by fermentation broth of GG799(pKLAC1-Phcmnp). The chromatogram peak at retention time of 6.3 min was AFB1. (B) Chromatogram of the AFB1-8,9-dihydrodiol standard. The chromatogram peak near the retention time of 11.19 min was AFB1-8,9-dihydrodiol standard. (C) Mass spectrum of AFB1 degradation product by fermentation supernatants of GG799(pKLAC1-Phcmnp). [M + H]+ ions with a mass-to-charge ratio (m/z) of 313 (circled) were AFB1, and those of slightly larger than 346 (circled) were presumed to be AFB1-8,9-dihydrodiol.
Degradation Efficiency of AFB1 in Peanuts
To investigate the degradation ratio of GG799(pKLAC1-Phcmnp) supernatant in the actual samples, degradation experiments were conducted using peanut samples added with AFB1. The degradation results are shown in Table 2. The recombinant strain GG799(pKLAC1-Phcmnp) supernatant degraded more than 90% of AFB1 in the peanut samples after twice treatments, with no significant differences (p > 0.05). When the AFB1 concentration was 50.0 μg/kg, the standard deviation was larger due to the low AFB1 content in the contaminated peanut samples. Besides, the larger the AFB1 concentration used, the smaller the standard deviation was found. These consistent data indicated that the GG799(pKLAC1-Phcmnp) culture supernatants were suitable for practical usage.
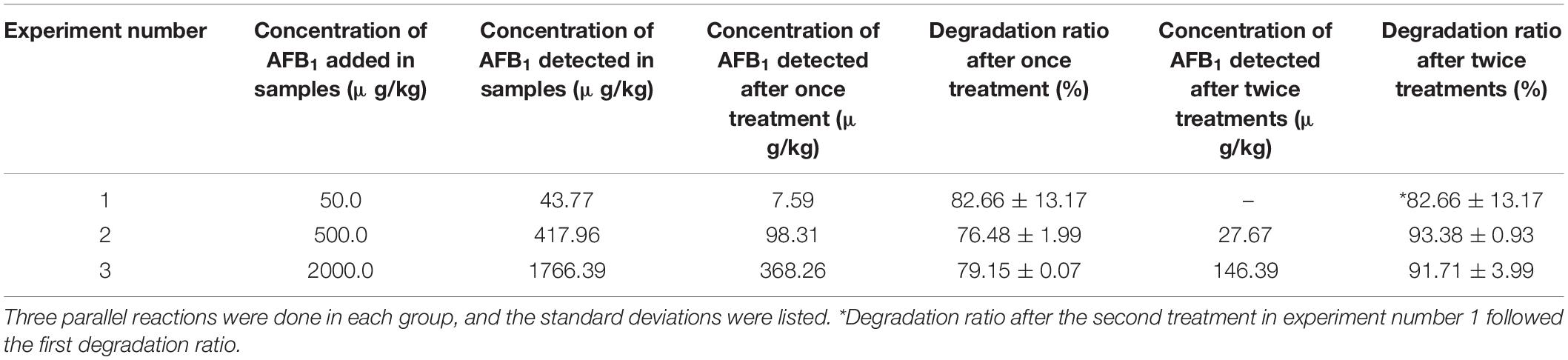
Table 2. — Degradation results of AFB1 in peanut samples by GG799(pKLAC1-Phcmnp) culture supernatants.
Mutagenesis Site Prediction of the Manganese Peroxidase PhcMnp
Single variants of PhcMnp were predicted based on the consensus approach, folding free energy calculations, and some structural considerations. On the one hand, amino acid evolutionary conservation distribution (Figure 7A) was obtained as follows: the amino acid sequence of PhcMnp was used to identify the homologous sequences (>40% sequence identity) from the NCBI database. After removing the redundant sequences through the CD-HIT program (Huang et al., 2010) with a 90% identity threshold, the 365 resulting sequences were aligned using MAFFT software (Katoh and Standley, 2013). The amino acid distributions at each position corresponding to that of PhcMnp were analyzed by WebLogo.1 On the other hand, we constructed a structural model of PhcMnp (Figure 7B) based on the crystal structures of its homolog enzyme MnP-CdII (PDB ID: 1YYG, 1.6 Å, 84% identity to PhcMnp). The software YASARA was used to perform the homology modeling. Alanine-scanning mutagenesis (Chauhan, 2017) was carried out on the homology model of PhcMnp using the Calculate Mutation Energy (Stability) protocol of Discovery Studio 2020 Client. Mutation energy prediction showed that six variants (70H, 110S, 174K, 178R, 192S, and 243T) had a much lower energy than others. These sites were then selected for full mutation scanning. Considering the conservation and energy, it was reasoned that the seven single variants (H70D, S110A, K174S, R178E, S192A, T243Q, and T243P) may have potentially enhanced activity. Besides, based on the criteria of non-mutation of highly conservative amino acids, low mutation energy, and being close to heme (<5 Å), it was reasoned that the 13 single variants (E59A, I65L, N105G, N105A, N105L, L138V, F179M, V199I, R201A, K204V, K204E, V205I, and L300M) may also have potentially enhanced activity.
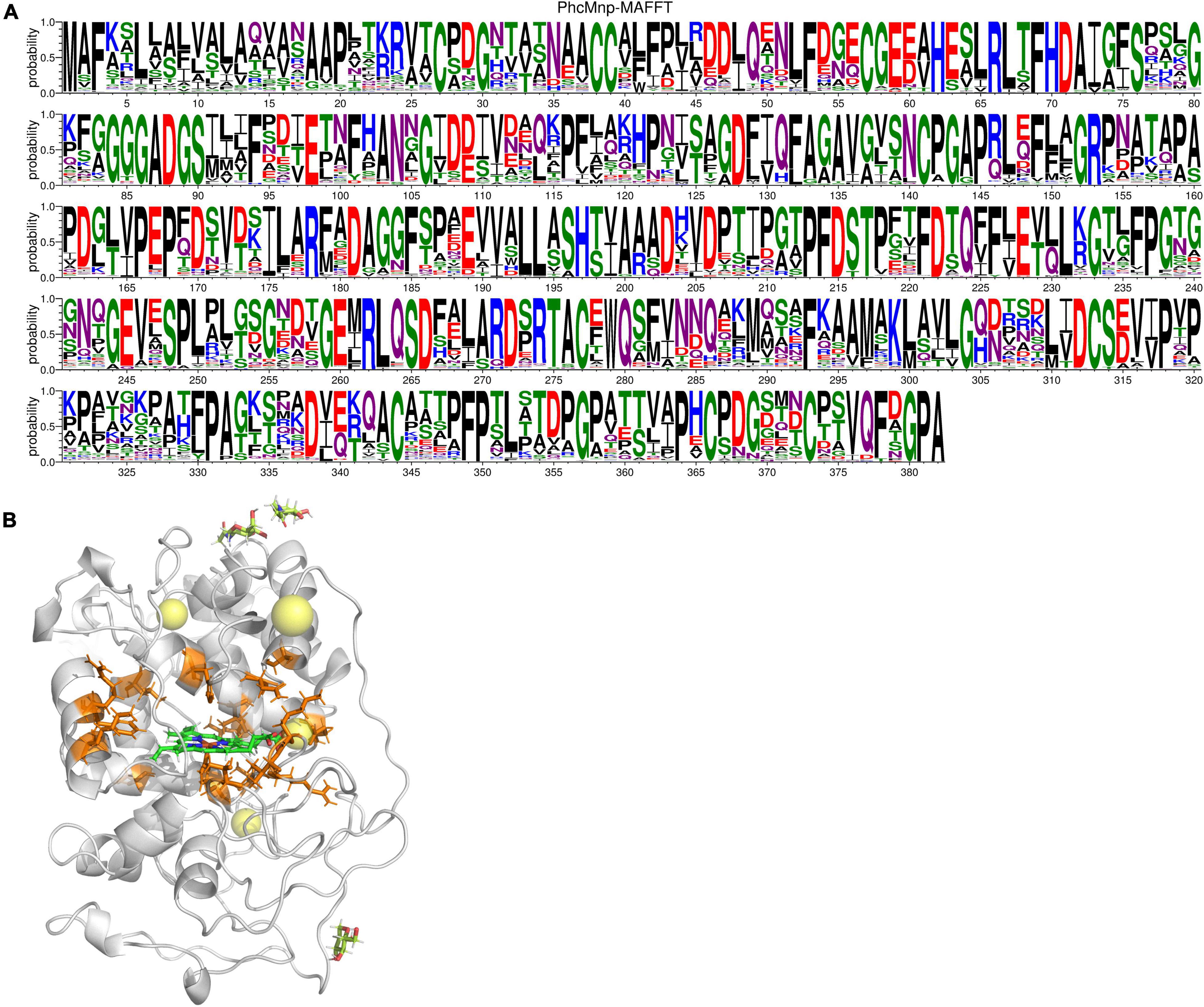
Figure 7. Amino acid sequence and structure analysis of PhcMnp. (A) Amino acid evolutionary conservation distribution of PhcMnp. The size of symbols within the stack indicates the relative frequency of each amino acid at that position. (B) Homology model of PhcMnp. The model structure of PhcMnp is constructed with the crystal structure of MnP-CdII (PDB ID: 1YYG) as a template. The predicted single mutant sites are shown in orange. The heme ring is shown in green.
Discussion
Manganese peroxidase is of increasing interest due to its potential for industrial applications, which was originally used in the treatment of agricultural waste and the lignin degradation (Ha et al., 2001). However, we used manganese peroxidase to achieve food-grade yeast expression in K. lactis GG799 for AFB1 degradation in this work.
The expression level for the enzyme PhcMnp is mainly influenced by factors such as induction temperature, time, and rotation speed; meanwhile in this work, the inducible expression for the enzyme PhcMnp was also regulated by the concentration of hemin and Mn2+. Hemin is a non-covalently bound prosthetic group in MnP, which is crucial to activating soluble MnPs (Wang et al., 2019). Also, manganese peroxidase consists of a ferric heme group and Mn2+ as its active center, and previous reports (Whitwam and Tien, 1996; Gettemy et al., 1998) showed that the manganese peroxidase genes are regulated by Mn2+ at the transcriptional level, and a certain amount of Mn2+ may promote the synthesis of manganese peroxidase.
As for the degradation of AFB1, the catalytic reaction of manganese peroxidase (MnP) requires the participation of H2O2 and Mn2+. The Fe3+-containing MnP is oxidized by H2O2 to produce complex I (Fe4+–oxygen–porphyrin radical complex), and then complex I is reduced by Mn2+ to produce complex II (Fe4+–oxygen–porphyrin complex) by a single electron. Finally, complex II is reconverted to MnP and Mn3+ by using Mn2+ as an electron donor (Hofrichter, 2002). The product Mn3+ can form chelates with some organic acids (e.g., oxalic acid, malonic acid, and malic acid) to oxidize complex organics to achieve the degradation of AFB1 (Wang et al., 2019). Therefore, the addition of Mn2+ in the reaction system is necessary, but some reports (Xia et al., 2021) showed that the presence of excessive Mn2+ may also inhibit the enzymatic activity of manganese peroxidase.
The degradation product of AFB1 was analyzed and determined to be AFB1-8,9-dihydrodiol in this work. The main pathway of degradation of AFB1 was through the free radicals generated by the interaction of oxidized Mn3+ with the dicarboxylic acid malonate (Wang et al., 2019), which increased the redox potential for oxidization of AFB1. In the degradation process, the intermediate AFB1-8,9-epoxide was produced before AFB1-8, 9-dihydrodiol, and it would be rapidly hydrolyzed to AFB1-8,9-dihydrodiol. The degradation product AFB1-8,9-dihydrodiol was greatly less toxic than AFB1, since it has no binding sites for DNA, lipids, and other macromolecules, thus achieving the purpose of detoxification of AFB1 (Wang et al., 2019).
As for the degradation ratio of AFB1, the recombinant laccase expressed in Aspergillus niger (118.0 U/l) achieved 55% of the degradation ratio for AFB1 (Alberts et al., 2009), and manganese peroxidase produced by Phanerochaete sordida YK-624 achieved 86% of the degradation ratio for AFB1 (Wang et al., 2011). Doyle and Marth (1979) found that lactoperoxidase in Aspergillus parasiticus could degrade AFB1 with up to 30.4% degradation ratio. However, the degradation ratio of AFB1 in the peanuts by the manganese peroxidase in this work reached more than 90%, which was much higher than those results by the enzymes produced in wild-type strains.
In conclusion, the manganese peroxidases used in this study were all derived from food-safe microorganisms (Phanerochaete sordida, Pleurotus ostreatus, and Phanerochaete chrysosporium) and the recombinant enzyme PhcMnp performed well in the degradation of AFB1. The food-grade expression system including the host K. lactis GG799 and the vector pKLAC1 was used in this work, in which the yeast K. lactis has been widely used in the food, feed, and pharmaceutical industries without environmental risks (Spohner et al., 2016). Therefore, this study provides a basis for subsequent large-scale fermentation of food-grade recombinant enzymes for degradation of mycotoxins in foodstuffs or feedstuffs. Furthermore, results showed that the enzyme PhcMnp has potential for practical application in the food industry. Besides, mutagenesis site prediction of the manganese peroxidase PhcMnp provided the theoretical basis for the following experiments in improvement or property modification of the enzyme.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
YX conceived the research strategies, designed the experiments, gave important guidance to the experiments, and assisted in completing the final manuscript. RH designed the experiments, conducted the experiments, and prepared the drafts of the manuscript. YS conducted the experiments and analyzed the data. HZ conducted the protein structure analysis. MG participated in the mycotoxin degradation experiments in practical examples. XH, XC, and QC provided partial materials, investigation data, and some experimental technique supports. ZW provided guidance and partial financial supports for the experiments. All authors read and approved the final manuscript.
Funding
This work was supported by the Jiangsu Agriculture Science and Technology Innovation Fund (JASTIF) with the no. CX(19)3109 and the National Natural Science Foundation of China (31871881).
Conflict of Interest
XC is employed by Anhui Heiwa Food Technology Co., Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank the coauthors for their participation and cooperation during the complete study.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.821230/full#supplementary-material
Footnotes
References
Abrar, M., Anjum, F. M., Butt, M. S., Pasha, I., Randhawa, M. A., Saeed, F., et al. (2013). Aflatoxins: biosynthesis, occurrence, toxicity, and remedies. Crit. Rev. Food Sci. 53, 862–874. doi: 10.1080/10408398.2011.563154
Alberts, J. F., Gelderblom, W., Botha, A., and Zyl, W. (2009). Degradation of aflatoxin B1 by fungal laccase enzymes. Int. J. Food Microbiol. 135, 47–52. doi: 10.1016/j.ijfoodmicro.2009.07.022
Assaf, J. C., Nahle, S., Chokr, A., Louka, N., Atoui, A., and El Khoury, A. (2019). Assorted methods for decontamination of Aflatoxin M1 in milk using microbial adsorbents. Toxins (Basel) 11:304. doi: 10.3390/toxins11060304
Cao, H., Liu, D., Mo, X., Xie, C., and Yao, D. (2011). A fungal enzyme with the ability of aflatoxin B1 conversion: purification and ESI-MS/MS identification. Microbiol. Res. 166, 475–483. doi: 10.1016/j.micres.2010.09.002
Chauhan, N. M. (2017). “Aflatoxin: a risky menace for African’s food commodities,” in Analysis, Detection and Health Risks, ed. L. Abdulrauf (London: IntechOpen Press), 91–113.
Claeys, L., Romano, C., De Ruyck, K., Wilson, H., Fervers, B., Korenjak, M., et al. (2020). Mycotoxin exposure and human cancer risk: a systematic review of epidemiological studies. Compr. Rev. Food Sci. Food Saf. 19, 1449–1464. doi: 10.1111/1541-4337.12567
Di Gregorio, M. C., de Neeff, D. V., Jager, A. V., Corassin, C. H., de Pinho Carão, ÁC., Albuquerque, R., et al. (2014). Mineral adsorbents for prevention of mycotoxins in animal feeds. Toxin Rev. 33, 125–135. doi: 10.3109/15569543.2014.905604
Doyle, M. P., and Marth, E. H. (1979). Peroxidase activity in mycelia of Aspergillus parasiticus that degrade aflatoxin. Eur. J. Appl. Microbiol. 7, 211–217. doi: 10.1007/BF00505027
Eskola, M., Kos, G., Elliott, C. T., Hajslova, J., Mayar, S., and Krska, R. (2020). Worldwide contamination of food-crops with mycotoxins: validity of the widely cited ‘FAO estimate’ of 25%. Crit. Rev. Food Sci. 60, 2773–2789. doi: 10.1080/10408398.2019.1658570
Feltrin, A. C. P., Garcia, S. O., Caldas, S. S., Primel, E. G., Badiale-Furlong, E., and Garda-Buffon, J. (2017). Characterization and application of the enzyme peroxidase to the degradation of the mycotoxin DON. J. Environ. Sci. Health B 52, 777–783. doi: 10.1080/03601234.2017.1356672
Gettemy, J. M., Ma, B., Alic, R., and Gold, M. H. (1998). Reverse transcription PCR analysis of the regulation of the manganese peroxidase gene family. Appl. Environ. Microb. 64, 569–574. doi: 10.1089/oli.1.1998.8.67
Ghazvini, R. D., Kouhsari, E., Zibafar, E., Hashemi, S. J., Amini, A., and Niknejad, F. (2016). Antifungal activity and aflatoxin degradation of Bifidobacterium Bifidum and Lactobacillus Fermentum against toxigenic Aspergillus Parasiticus. Open Microbiol. J. 10, 197–201. doi: 10.2174/1874285801610010197
Gnanamani, A., Jayaprakashvel, M., Arulmani, M., Sadulla, S. J. E., and Technology, M. (2006). Effect of inducers and culturing processes on laccase synthesis in Phanerochaete chrysosporium NCIM 1197 and the constitutive expression of laccase isozymes. Enzyme Microb. Technol. 38, 1017–1021. doi: 10.1016/j.enzmictec.2006.01.004
Gowda, N. K. S., Suganthi, R. U., Malathi, V., and Raghavendra, A. (2007). Efficacy of heat treatment and sun drying of aflatoxin-contaminated feed for reducing the harmful biological effects in sheep. Anim. Feed Sci. Tech. 133, 167–175. doi: 10.1016/j.anifeedsci.2006.08.009
Ha, H. C., Honda, Y., Watanabe, T., and Kuwahara, M. (2001). Production of manganese peroxidase by pellet culture of the lignin-degrading basidiomycete, Pleurotus ostreatus. Appl. Microbiol. Biotechnol. 55, 704–711. doi: 10.1007/s002530100653
Haque, M. A., Wang, Y., Shen, Z., Li, X., Saleemi, M. K., and He, C. (2020). Mycotoxin contamination and control strategy in human, domestic animal and poultry: a review. Microb. Pathog. 142:104095. doi: 10.1016/j.micpath.2020.104095
Hofrichter, M. (2002). Review lignin conversion by manganese peroxidase (MnP). Enzyme Microb. Technol. 30, 454–466. doi: 10.1016/S0141-0229(01)00528-2
Huang, Y., Niu, B. F., Gao, Y., Fu, L. M., and Li, W. Z. (2010). CD-HIT Suite: a web server for clustering and comparing biological sequences. Bioinformatics 26, 680–682. doi: 10.1093/bioinformatics/btq003
Ismail, A., Goncalves, B. L., de Neeff, D. V., Ponzilacqua, B., Coppa, C., Hintzsche, H., et al. (2018). Aflatoxin in foodstuffs: occurrence and recent advances in decontamination. Food Res. Int. 113, 74–85. doi: 10.1016/j.foodres.2018.06.067
Jiang, F., Kongsaeree, P., Charron, R., Lajoie, C., Xu, H., Scott, G., et al. (2008). Production and separation of manganese peroxidase from heme amended yeast cultures. Biotechnol. Bioeng. 99, 540–549. doi: 10.1002/bit.21590
Katoh, K., and Standley, D. M. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. doi: 10.1093/molbev/mst010
Loi, M., Fanelli, F., Liuzzi, V. C., Logrieco, A. F., and Mule, G. (2017). Mycotoxin biotransformation by native and commercial enzymes: present and future perspectives. Toxins (Basel) 9:111. doi: 10.3390/toxins9040111
Loi, M., Fanelli, F., Zucca, P., Liuzzi, V. C., Quintieri, L., Cimmarusti, M. T., et al. (2016). Aflatoxin B1 and M1 degradation by Lac2 from Pleurotus pulmonarius and redox mediators. Toxins (Basel) 8:245. doi: 10.3390/toxins8090245
Marques, M. M., de Gonzalez, A. B., Beland, F. A., Browne, P., Demers, P. A., Lachenmeier, D. W., et al. (2019). Advisory group recommendations on priorities for the IARC monographs. Lancet Oncol. 20, 763–764. doi: 10.1016/s1470-2045(19)30246-3
Peng, Z., Chen, L., Zhu, Y., Huang, Y., Hu, X., Wu, Q., et al. (2018). Current major degradation methods for aflatoxins: a review. Trends Food Sci. Technol. 80, 155–166. doi: 10.1016/j.tifs.2018.08.009
Pitt, J. I., and Miller, J. D. (2017). A concise history of mycotoxin research. J. Agr. Food Chem. 65, 7021–7033. doi: 10.1021/acs.jafc.6b04494
Spohner, S. C., Schaum, V., Quitmann, H., and Czermak, P. (2016). Kluyveromyces lactis: an emerging tool in biotechnology. J. Biotechnol. 222, 104–116. doi: 10.1016/j.jbiotec.2016.02.023
Styriak, I., Conkovd, E., Kmec, V., Bbhm, J., and Razzazi, E. (2001). The use of yeast for microbial degradation of some selected mycotoxins. Mycotoxin. Res. 17, 24–27. doi: 10.1007/BF03036705
Tarazi, S., Ahmadi, S., Ostvar, N., Ghafouri, H., Sarikhan, S., Mahmoodi, Z., et al. (2021). Enhanced soluble expression of glutathione S-transferase Mu from Rutilus kutum by co-expression with Hsp70 and introducing a novel inhibitor for its activity. Process Biochem. 111, 261–266. doi: 10.1016/j.procbio.2021.10.003
van den Dungen, M. W., Boer, R., Wilms, L. C., Efimova, Y., and Abbas, H. E. (2021). The safety of a Kluyveromyces lactis strain lineage for enzyme production. Regul. Toxicol. Pharm. 126:105027. doi: 10.1016/j.yrtph.2021.105027
von Hertwig, A. M., Iamanaka, B. T., Amorim Neto, D. P., Rezende, J. B., Martins, L. M., Taniwaki, M. H., et al. (2020). Interaction of Aspergillus flavus and A. parasiticus with Salmonella spp. isolated from peanuts. Int. J. Food Microbiol. 328:108666. doi: 10.1016/j.ijfoodmicro.2020.108666
Wang, J. Q., Ogata, M., Hirai, H., and Kawagishi, H. (2011). Detoxification of aflatoxin B1 by manganese peroxidase from the white-rot fungus Phanerochaete sordida YK-624. FEMS Microbiol. Lett. 314, 164–169. doi: 10.1111/j.1574-6968.2010.02158.x
Wang, L., Luo, Y., Luo, X., Wang, R., Li, Y., Li, Y., et al. (2016a). Effect of deoxynivalenol detoxification by ozone treatment in wheat grains. Food Control 66, 137–144. doi: 10.1016/j.foodcont.2016.01.038
Wang, L., Shao, H., Luo, X., Wang, R., Li, Y., Li, Y., et al. (2016b). Effect of ozone treatment on deoxynivalenol and wheat quality. PLoS One 11:e0147613. doi: 10.1371/journal.pone.0147613
Wang, X., Qin, X., Hao, Z., Luo, H., Yao, B., and Su, X. (2019). Degradation of four major mycotoxins by eight manganese peroxidases in presence of a dicarboxylic acid. Toxins (Basel) 11:566. doi: 10.3390/toxins11100566
Wang, Y., Zhao, C., Zhang, D., Zhao, M., Zheng, D., Peng, M., et al. (2018). Simultaneous degradation of aflatoxin B1 and zearalenone by a microbial consortium. Toxicon 146, 69–76. doi: 10.1016/j.toxicon.2018.04.007
Wariishi, H., Akileswaran, L., and Gold, M. H. (1988). Manganese peroxidase from the basidiomycete Phanerochaete chrysosporium: spectral characterization of the oxidized states and the catalytic cycle. Biochemistry 27, 5365–5370. doi: 10.1021/bi00414a061
Whitwam, R., and Tien, M. (1996). Heterologous expression and reconstitution of fungal Mn peroxidase. Arch. Biochem. Biophys. 333, 439–446. doi: 10.1006/abbi.1996.0413
Xia, Y., Wu, Z., He, R., Gao, Y., Qiu, Y., Cheng, Q., et al. (2021). Simultaneous degradation of two mycotoxins enabled by a fusion enzyme in food-grade recombinant Kluyveromyces lactis. Bioresour. Bioprocess. 8:62. doi: 10.1186/s40643-021-00395-1
Yang, B., Zhang, C., Zhang, X., Wang, G., Li, L., Geng, H., et al. (2020). Survey of aflatoxin B1 and heavy metal contamination in peanut and peanut soil in China during 2017–2018. Food Control 118:107372. doi: 10.1016/j.foodcont.2020.107372
Yehia, R. S. (2014). Aflatoxin detoxification by manganese peroxidase purified from Pleurotus ostreatus. Braz. J. Microbiol. 45, 127–133. doi: 10.1590/s1517-83822014005000026
Zhang, W., Liu, Y., Liang, B., Zhang, Y., Zhong, X., Luo, X., et al. (2020). Probabilistic risk assessment of dietary exposure to aflatoxin B1 in Guangzhou, China. Sci. Rep. 10:7973. doi: 10.1038/s41598-020-64295-8
Keywords: mycotoxins, aflatoxin B1, degradation, Kluyveromyces lactis, food-grade
Citation: Xia Y, He R, Sun Y, Zhou H, Gao M, Hu X, Cui X, Cheng Q and Wang Z (2022) Food-Grade Expression of Manganese Peroxidases in Recombinant Kluyveromyces lactis and Degradation of Aflatoxin B1 Using Fermentation Supernatants. Front. Microbiol. 12:821230. doi: 10.3389/fmicb.2021.821230
Received: 24 November 2021; Accepted: 14 December 2021;
Published: 14 February 2022.
Edited by:
Lei Chen, Tianjin University, ChinaReviewed by:
Xiao-Jun Ji, Nanjing Tech University, ChinaJin Wang, Shenzhen Second People’s Hospital, China
Copyright © 2022 Xia, He, Sun, Zhou, Gao, Hu, Cui, Cheng and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yu Xia, yuxia@jiangnan.edu.cn
 Yu Xia
Yu Xia Rui He2
Rui He2 Ying Sun
Ying Sun Minjie Gao
Minjie Gao Zhouping Wang
Zhouping Wang