- 1State Key Laboratory of Tibetan Plateau Earth System, Resources and Environment (TPESRE), Institute of Tibetan Plateau Research, Chinese Academy of Sciences, Beijing, China
- 2College of Resources and Environment, University of Chinese Academy of Sciences, Beijing, China
- 3Center for the Pan-Third Pole Environment, Lanzhou University, Lanzhou, China
- 4Institute of International Rivers and Eco-Security, Yunnan University, Kunming, China
Cryoconites harbor diverse microbial communities and are the metabolic hotspot in the glacial ecosystem. Glacial ecosystems are subjected to frequent climate disturbances such as precipitation (snowing), but little is known about whether microbial communities in cryoconite can maintain stability under such disturbance. Here, we investigated the bacterial community in supraglacial cryoconite before and after a snowfall event on the Laohugou Glacier (Tibetan Plateau), based on Illumina MiSeq sequencing of the 16S rRNA gene. Our results showed that the diversity of the microbial community significantly decreased, and the structure of the microbial community changed significantly after the disturbance of snowfall. This was partly due to the relative abundance increased of cold-tolerant bacterial taxa, which turned from rare into abundant sub-communities. After snowfall disturbance, the contribution of the deterministic process increased from 38 to 67%, which is likely due to the enhancement of environmental filtering caused by nitrogen limitation. These findings enhanced our understanding of the distribution patterns and assembly mechanisms of cryoconite bacterial communities on mountain glaciers.
Introduction
Cryoconite is dark sediment deposited on the glacial surfaces, comprising both mineral and biological materials. Supraglacial cryoconite serves as a habitat for diverse microbial communities, including viruses, bacteria, fungi, archaea, algae, and invertebrates, which are responsible for the glacier biogeochemical cycling (Anesio et al., 2007, 2010; Edwards et al., 2011, 2013; Zarsky et al., 2013). Supraglacial cryoconite microbes are directly exposed to environmental pressures or perturbations, such as frequent flushing by supraglacial meltwater and precipitation (Zawierucha et al., 2019b). Thus, understanding the response of microbial communities to climate disturbance is essential for the study of ecosystem functions (Steudel et al., 2012).
Microbial communities are affected by various physicochemical factors (Stibal et al., 2012; Cook et al., 2016; Anesio et al., 2017). Temperature and nutrient availability have been proposed to be the main factors controlling the microbial communities in polar and mountain glacier cryoconite (Zarsky et al., 2013; Ambrosini et al., 2016; Liu et al., 2017; Sommers et al., 2017; Lutz et al., 2018). Differences in physicochemical conditions can also indirectly influence microbial community structure in cryoconite by altering biotic interactions (Friedman and Gore, 2017; Khan et al., 2018; Bergk Pinto et al., 2019). For example, on Spitsbergen Island in Svalbard, the addition of organic carbon shifted microbial interactions from collaboration to competition (Bergk Pinto et al., 2019). Intensive collaboration can enhance complex organic carbon degradation and mineralization, which are particularly important for oligotrophic environments such as glaciers (Bergk Pinto et al., 2019; Krug et al., 2020). These interactions change can favor or discriminate certain microbial groups, thereby alter the microbial community structure (i.e., biofiltering).
Recently, a growing body of research has emphasized the ecological importance of rare taxa (Lynch and Neufeld, 2015; Jousset et al., 2017), and distinct succession patterns and functional characteristics have been found between the abundant and rare taxa (Jia et al., 2018; Jiao and Lu, 2020; Rocca et al., 2020). The abundant taxa are usually perceived to be involved in the active category in biogeochemical cycles, especially carbohydrate metabolism, and take up the core ecological niches (Jiao et al., 2017; Kurm et al., 2019; Liang et al., 2020). In addition, due to their high abundance, the abundant taxa might present greater tolerance to environmental stresses (Jiang et al., 2019; Jiao and Lu, 2020). The rare taxa serve as a reservoir of genetic and functional diversity for the entire community and contribute to the maintenance of microbial diversity (Jousset et al., 2017; Rocca et al., 2020). That is, when facing environmental disturbance, rare taxa could respond promptly to maintain community stability (Zhang et al., 2020). For example, some rare taxa may turn into dominant taxa in the community to compensate for the ecosystem function loss due to the disturbance (Jiang et al., 2019; Du et al., 2020). The interaction and transformation of abundant and rare bacterial are critical for maintaining functional redundancy and community stability (Liang et al., 2020). Therefore, additional studies are needed to investigate the relationship between abundant and rare bacterial groups in cryoconite communities after disturbance.
Previous studies have demonstrated that two different assembly mechanisms, stochastic and deterministic processes, affect abundant and rare bacterial sub-communities in different patterns. Deterministic processes include niche-based factors such as abiotic and biotic factors (Elshahed et al., 2008; Pedros-Alio, 2012; Sauret et al., 2014). Stochastic processes include probabilistic dispersal and random changes in the relative abundance of species (ecological drift) (Liu et al., 2015; Zawierucha et al., 2019a,b). Little is known about stochastic or deterministic processes structuring the rare and abundant microbial communities in the cryoconite ecosystem. A recent study found that stochastic weather events (rainstorms) changed the tardigrades community structure on an Arctic glacier (Zawierucha et al., 2019b). The authors also reported that the tardigrades in the Arctic cryoconite granules were passively dispersed by streams and melting water, while pH and electrical did not shape invertebrate communities (Zawierucha et al., 2019b). However, whether the community assembly of cryoconite microbial communities is affected by climate disturbance remains largely unknown.
Here we studied the changes and assembly mechanisms of microbial communities in supraglacial cryoconite before and after a snowfall event in the Laohugou Glacier (LHG), Tibetan Plateau. The Tibetan Plateau contains the largest area of glacial ice outside the polar regions and has an average altitude > 4,000 m above sea level (a.s.l) (Yao et al., 2012). Compared to other areas with alpine glaciers, the glaciers in the Tibetan Plateau are exposed to lower temperatures and lower allochthonous inputs of organic matter, due to the higher altitude (Liu et al., 2017). The aims of the present study are: (i) to explore the cryoconite microbial community structure and assembly process following snowfall disturbance; (ii) to investigate the response of the abundant and rare bacterial sub-communities after disturbance.
Materials and Methods
Sites and Sampling
The LHG (39°26.4′N, 96°32.5′E), with a length of 10.1 km and an area of 21.9 km2, is the largest glacier in the Qilian Mountains, in the northeastern Tibetan Plateau (Sun et al., 2012). It is a typical valley glacier, with elevations ranging from 4,200 m a.s.l at its terminus to 5,481 m a.s.l at its top. The region has a typical continental climate affected by desertification in central Asia, with an average temperature above 0°C in summer and long periods below freezing in winter. Precipitation is mainly concentrated in May-September, primarily affected by westerly winds (Wei et al., 2017).
In August 2016, we randomly selected four superficially deposited cryoconite mounds, which piled up due to glacial melting within the ablation zone for collecting samples (Supplementary Figure 1). Four sampling sites were 100–400 m apart, covering a large area in order to represent the glacier’s features. To ensure that collecting was made at the same locations, we determined the coordinates of each site using GPS and mounted bamboo sticks in the ice. Samples were collected from each of the observation sites on the 1st, 4th, 8th, 26th, and 31st of August, which are referred as day 1, 4, 8, 26, and 31 hereafter). The large gap between the third and fourth sampling days was due to heavy snowfall and flooding that occurred after 10th August. Before sampling, we scraped off the surface layer (top 1 cm) of cryoconite. Approximately 200 g of cryoconite debris were collected using a pre-cleaned stainless-steel spoon with polyethylene gloves worn all the time. The sample was placed into sterile 250 ml Nalgene ® plastic bottles that were first cleaned with tap water and then rinsed three times with Milli-Q water, followed by sterilization for 15 min at 120°C in steam autoclaving and drying at 55°C. Samples were transported in a dark container within 4 h to the Qilian Shan Station of Glaciology and Ecologic Environment, Chinese Academy of Sciences. All samples were kept frozen at −20°C until laboratory analysis.
Geochemical Measurements
The total organic carbon (TOC) contents of the samples were determined using a TOC-L (Shimadzu Corp., Kyoto, Japan) on lyophilized samples. Total nitrogen (TN) and total carbon (TC) were measured using an elemental analyzer (Vario MAX, Elementar, Germany). The elemental concentrations are reported as the percentage of dry weight (100°C for 24 h) of cryoconite material.
DNA Extraction and Illumina Sequencing
Cryoconite debris (0.5 g) was soaked in 1.5 mL of lysis buffer (20 mg mL–1 Proteinase K, 0.1 M EDTA, and 10% SDS) and incubated at 55°C for 2 h before DNA extraction. DNA extraction was performed using a Fast DNA ®SPIN Kit for Soil (MP Biomedicals, Santa Ana, CA, United States), according to the manufacturer’s instructions. The extracted DNA was eluted to 100 μL TE buffer. The raw DNA was purified using a 1% (w/v) agarose gel. DNA bands were removed from the gel and extracted using an Agarose Gel DNA purification kit (TaKaRa, Japan). The concentration of DNA extracts was measured by the NanoDrop 1000 spectrophotometer (Thermo-Scientific, Wilmington, DE, United States). The extracted DNA was stored at -80°C until amplification.
The V4 region of the bacterial 16S rRNA genes was amplified in triplicate by primer pair 515F (5′-GTGCCAGC MGCCGCGGTAA-3′) and 806R (5′- GGACTACHVGGGTW TCTAAT-3′) and sequenced on an Illumina MiSeq system (Illumina, Inc., San Diego, CA, United States) following the procedure by Caporaso et al. (2012). Briefly, PCR was performed under the condition of 94°C for 5 min, 30 cycles of 94°C for 30 s, 52°C for 30 s, 72°C for 30 s; followed by a final cycle of 10 min at 72°C. Each PCR reaction contained 25 μL 2x Premix Taq DNA polymerase (Takara Biotechnology, Dalian Co., Ltd., China), 3 μL DNA template (20 ng μL–1), 1 μL each primer (10 μM), and 20 μL nuclease-free water. PCR products of the triplicate reaction samples were pooled and quantified by PicoGreen staining. The sample libraries were generated with purified PCR products. The MiSeq 500 cycles kit was used for 2 × 250 bp paired-end sequencing.
Processing of Illumina Sequencing Data
The “fastq_filter” command in USEARCH was used to trim and filter the raw sequence reads (Edgar, 2016). The trimmed and filtered reads were used to call amplicon sequence variants (ASVs) using the “-cluster_ASVs” and “-unoise3” commands in USEARCH, respectively (Edgar, 2016). Singleton ASVs with fewer than nine reads were removed using the default “-minsize” values. The “-usearch_global” command with a minimum sequence identity of 99% was used to map ASVs sequences. Chimeric ASVs were removed using the “uchime_ref” command in USEARCH. Representative sequences were taxonomically classified using the “sintax” command along with the RDP training set (version 16) with confidence scores above the 0.5 cutoff (Chiellini et al., 2019). Normalization across samples was performed using a variance stabilizing transformation with the DESeq2 package in R (McMurdie and Holmes, 2014). After the taxonomy assignment, ASVs associated with chloroplasts, archaea, and unclassified sequences were excluded from subsequent analyses.
Definition of Abundant and Rare Taxa
Rare taxa were defined as the ASVs with mean relative abundance < 0.1% in all samples in one group, whereas abundant ASVs were defined as the ASVs with a relative abundance > 1% in one or more samples in one group (Ji et al., 2020; Zhang et al., 2020; Dong et al., 2021). Taxa that did not fall into either the abundant or rare categories were defined as the oscillating taxa.
Statistical Analysis
Wilcoxon rank-sum test and Kruskal-Wallis test were used to determine whether there were any statistically significant differences among groups of environmental variables. The alpha-diversity indices (Shannon-Wiener and Pielou’s Evenness) were calculated using the “vegan” R package (Oksanen et al., 2010). Wilcoxon rank-sum test and Kruskal-Wallis test were used to determine whether there were any statistically significant differences among groups of alpha-diversity. The significance of dissimilarity of community composition among groups was tested using permutational multivariate analysis of variance (PERMANOVA) based on Bray-Curtis distance metrics with the “adonis” function in the R package “vegan” (Oksanen et al., 2010). Test results with P < 0.05 were considered as statistically significant. The correlations between bacterial diversity and environmental variables were calculated using Spearman’s rank correlation, as implemented in the “ppcor” R package. Mantel test based on Spearman’s rank correlations was performed to show the correlation between variations of microbial communities and environmental variables using the “vegan” package of R (Oksanen et al., 2010).
Co-occurrence networks were explored using Molecular Ecological Network Analysis Pipeline (MENA2)1 based on the Random Matrix Theory (RMT) and Spearman correlation (Deng et al., 2012). The resulting correlation matrix was analyzed with the RMT-based network approach to determine the correlation threshold for network construction, and the same threshold was used for all networks, so the topological properties of all networks are comparable. Then global network properties were calculated including individual nodes’ centrality, degree, betweenness, and clustering coefficient. Integrating all the results obtained from MENA, co-occurrence network was visualized by Gephi.
The normalized stochasticity ratio (NST) based on the Bray–Curtis dissimilarity was calculated using the “NST” package in R to estimate the determinacy and stochasticity of the bacterial assembly processes with high accuracy and precision (Ning et al., 2019). The NST index used 50% as the boundary point between more deterministic (< 50%) and more stochastic (> 50%) assembly processes.
Results
Environmental Characteristics of the Cryoconite Samples
The TOC concentration ranged from 1.04 to 3.39%. The TC concentration ranged from 2.10 to 3.85%. The concentration of TOC and TC of samples after snowfalls were similar between the samples before and after snowfalls (Wilcoxon rank-sum test, P = 0.13, and P = 0.73, respectively). The TN concentrations ranged from 0.16 to 0.24%, and that before the snowfall was significantly higher than that after snowfall (Wilcoxon rank-sum test, P = 0.003). The C/N ratio ranged from 9.47 to 22.62, and the C/N ratio of samples after the snowfall was significantly higher than those before the snowfall (Wilcoxon rank-sum test, P = 0.02; Figure 1). All environmental characteristics showed no significant difference between sampling sites (Kruskal-Wallis test, all P > 0.05; Supplementary Figure 2).
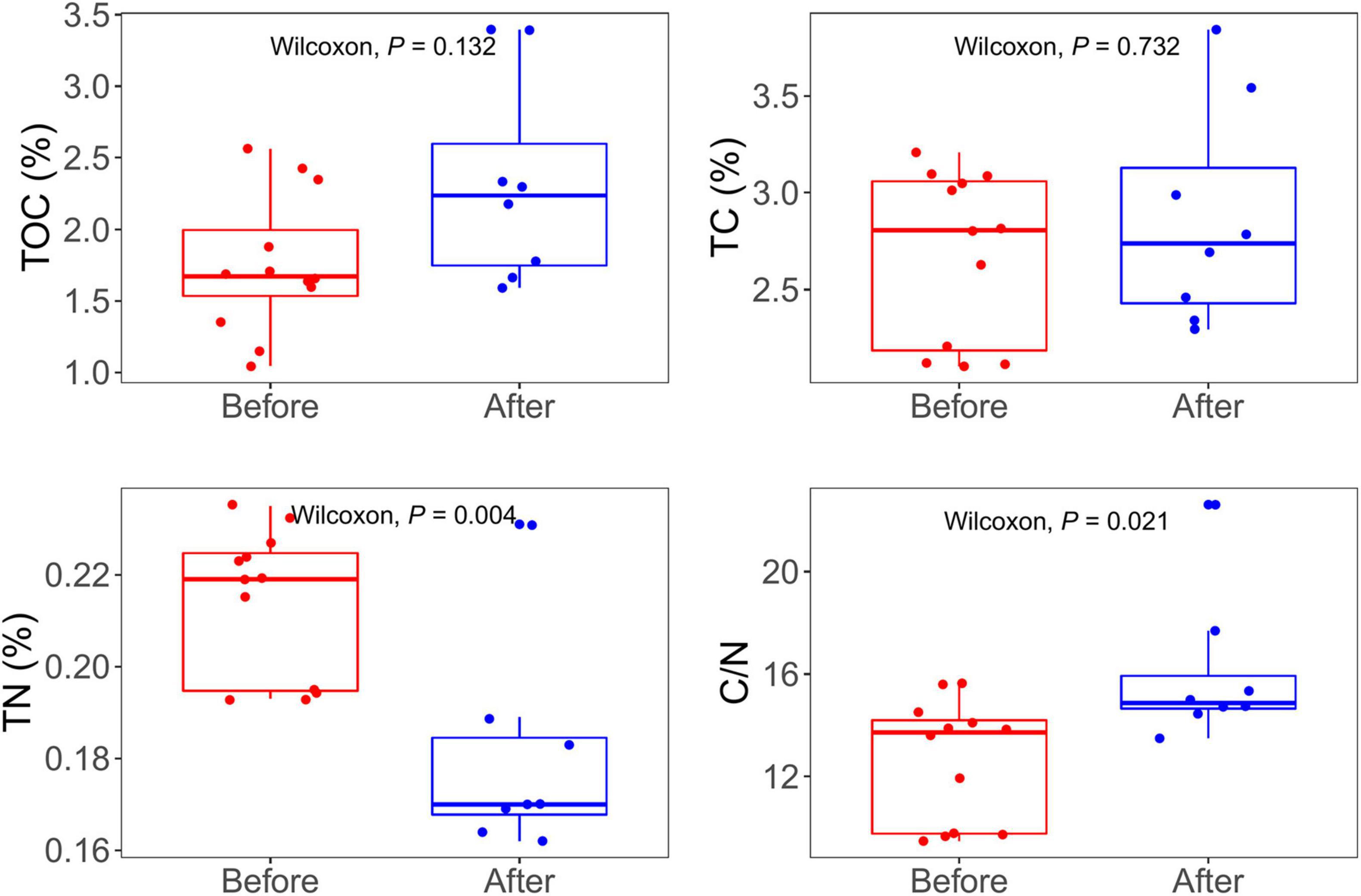
Figure 1. Total dissolve carbon (TOC), total carbon (TC), total nitrogen (TN), and rate of carbon nitrogen (C/N) concentrations in samples before and after snowfall disturb. Each dot represents an individual sample. Significantly higher concentrations of TN were observed in the samples before snowfall, while a significantly higher ratio of C/N was in the samples after snowfall (Wilcoxon rank-sum test, P < 0.05).
Taxonomic Compositions of Bacterial Community
After quality filtering and the removal of low-quality sequences, a total of 867,658 reads were obtained, and were clustered into 872 ASVs (99% sequence similarity). The most abundant bacterial phyla were Bacteroidetes, Cyanobacteria, Betaproteobacteria, Gammaproteobacteria, Actinobacteria, Alphaproteobacteria, Chloroflexi, and Firmicutes, which account for 86.2 and 95.0% of sequence abundance before and after snowfalls, respectively (Figure 2). The composition of the abundant sub-community was similar to the whole community. However, the rare and oscillating sub-communities differed from the abundant sub-communities. The rare and oscillating taxa contained more taxonomic groups than the abundant taxa. Bacteroidetes, Alphaproteobacteria, Verrucomicobia, Deltaproteobacteria, and Acidobacteria were more prevalent in rare and oscillating sub-communities, while Cyanobacteria and Betaproteobacteria were more prevalent in abundant sub-community (Figure 2).
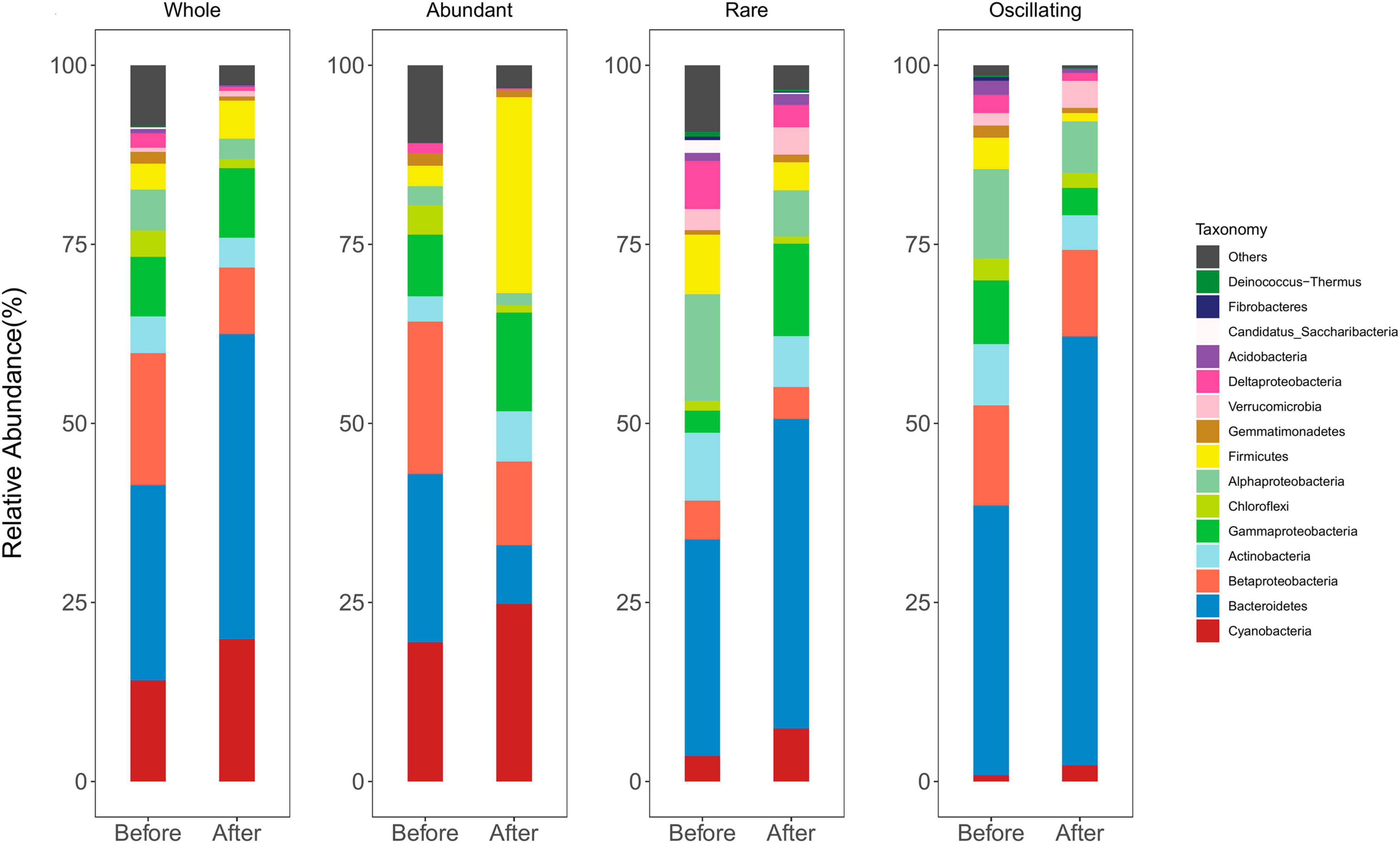
Figure 2. Taxonomic composition of bacteria before and after snowfalls in whole, abundant, and rare communities. The communities were dominated by Cyanobacteria, Bacteroidetes, Betaproteobacteria, Actinobacteria, Gammaproteobacteria, Chloroflexi, Alphaproteobacteria, and Firmicutes.
In the whole community, the relative abundance of Betaproteobacteria, Chloroflexi, Alphaproteobacteria, Gemmatimonadetes, and Deltaproteobacteria were significantly higher before snowfall than after snowfalls (Wilcoxon rank-sum test, all P < 0.05; Supplementary Figure 3). In the abundant sub-community, the relative abundance of Bacteroidetes, Chloroflexi, Alphaproteobacteria, and Deltaproteobacteria were significantly higher before snowfall than after snowfalls (Wilcoxon rank-sum test, all P < 0.05), while Actinobacteria were significantly higher after snowfall than before snowfalls (Wilcoxon rank-sum test, P < 0.01). In the rare sub-community, the relative abundance of Alphaproteobacteria and Deltaproteobacteria were significantly higher before snowfall than after snowfalls (Wilcoxon rank-sum test, all P < 0.05). In the oscillating sub-community, the relative abundance of Gammaproteobacteria, Firmicutes, Gemmatimonadetes, and Deltaproteobacteria were significantly higher before snowfall than after snowfalls (Wilcoxon rank-sum test, all P < 0.05; Supplementary Figure 3).
The abundant taxa (34-48 ASVs) accounted for 62.1–94.5% of all sequences. The rare taxa (167-495 ASVs) only accounted for 2.6–13.6% of all sequences. The oscillating taxa (40-128 ASVs) accounted for 2.2–29.4% of all sequences (Supplementary Table 1). After snowfall disturbance, the proportion of abundant bacterial taxa did not change significantly, while the percentage of rare taxa significantly increased, and the percentage of oscillating taxa significantly decreased (Supplementary Figure 4). Approximately 59% of abundant bacterial taxa before snowfall were still abundant after the snowfall, and 8% became rare bacterial taxa, and 35% became oscillating bacterial taxa. After the snowfall disturbance, a large proportion (93%) of rare bacterial taxa still being the rare bacterial taxa, while 1% became abundant bacterial taxa, and 1% became oscillating bacterial taxa, and an additional 5% disappeared. Only 29% of oscillating bacterial taxa in samples before snowfall were still oscillating in bacterial taxa after the snowfall, 2% became abundant taxa, and 69% became rare bacterial taxa (Supplementary Table 2). The abundant bacterial taxa that came from the rare and oscillating bacterial taxa after snowfall accounted for 23 and 19% of the community. They are primarily taxonomically classified as Flavobacterium (Bacteroidetes), Massilia (Betaproteobacteria), Polaromonas (Betaproteobacteria), Psychrobacter (Gammaproteobacteria), Cellulomonas (Actinobacteria), and Carnobacterium (Firmicutes) (Supplementary Table 3).
Diversity of Bacterial Community
The Shannon-Wiener diversity of the whole, abundant, rare, and oscillating community were 1.74–4.90, 1.47–3.67, 3.79–5.38, and 2.72–4.56, respectively. The Pielou’s Evenness of the whole, abundant, rare, and oscillating community were 0.02–0.22, 0.07–0.60, 0.46–0.68, and 0.33–0.85, respectively. The Shannon diversity indices of the rare sub-community were significantly higher than those of the abundant and oscillating sub-communities (Wilcoxon rank-sum test, all P < 0.001; Supplementary Figure 5). After the snowfall, the Shannon-Wiener and Pielou’s Evenness indices were significantly reduced in all subcommunities (Wilcoxon rank-sum test, all P < 0.05; Figure 3). The Shannon and Evenness indices positively correlated with the TN concentration (Spearman’s correlation: r = 0.45, P < 0.05; r = 0.45, P < 0.05; Table 1).
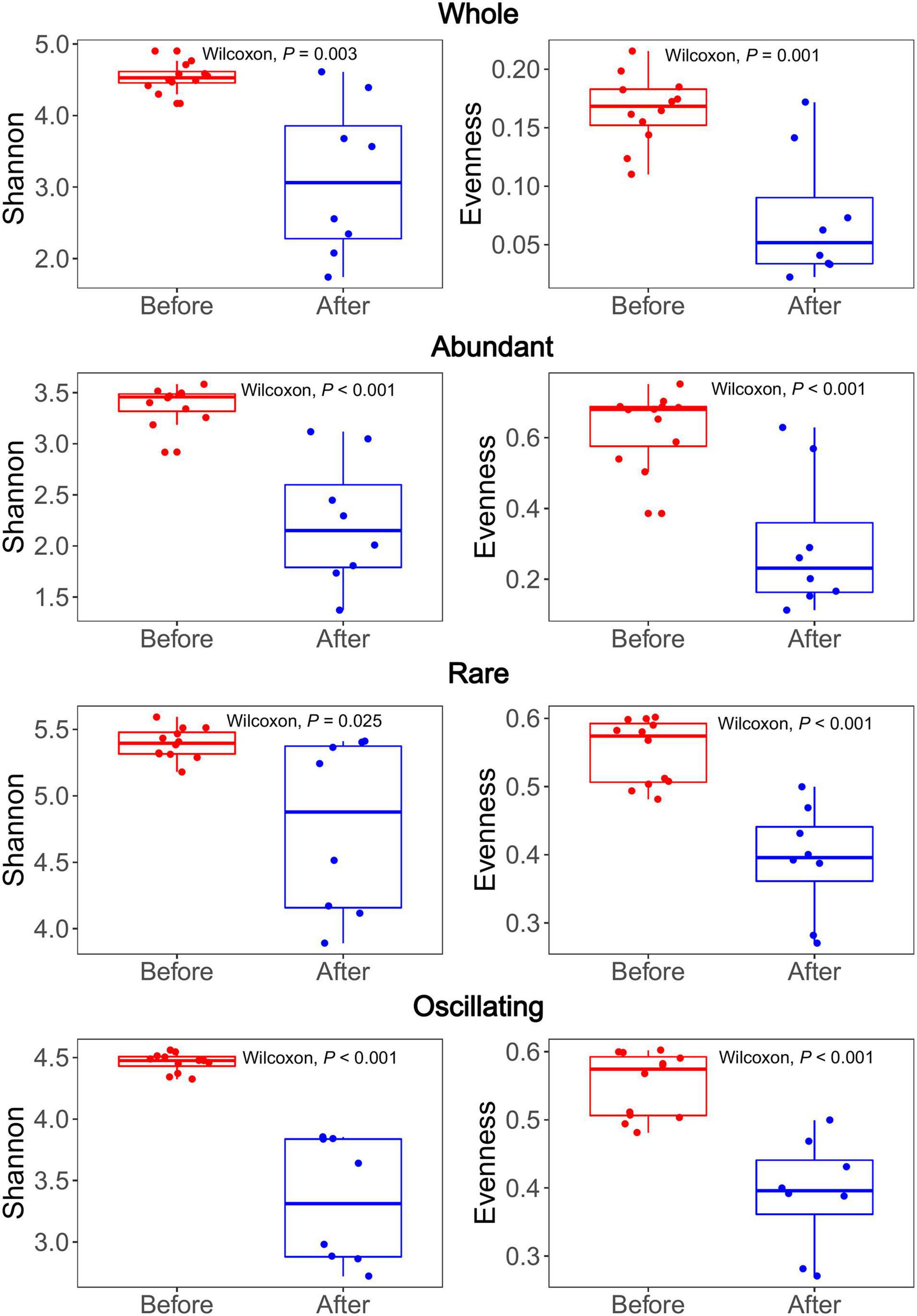
Figure 3. Bacterial alpha diversity comparison between the communities before and after snowfall. Each dot represents an individual sample. For both Shannon and Evenness indices, significant higher diversity was observed in communities before snowfall in the whole, abundant, rare and oscillating communities (Wilcoxon rank-sum test, P < 0.05).
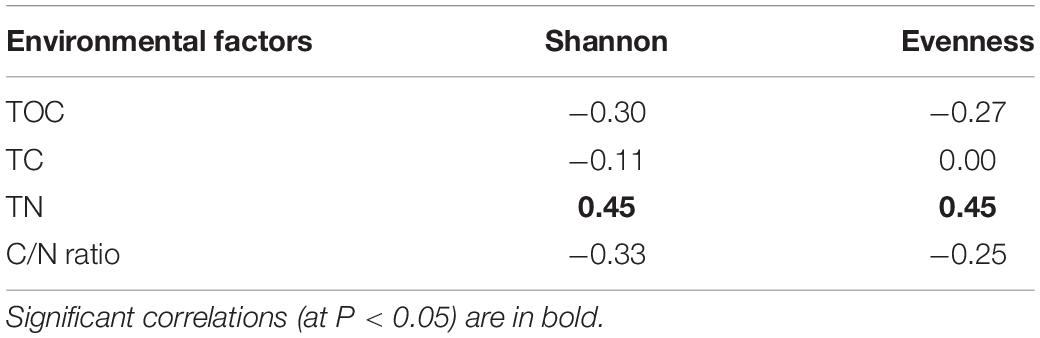
Table 1. Correlation between cryoconite community Shannon and Evenness indices and environmental factors.
The PCoA plot showed clear segregation between the samples before and after snowfall (Figure 4). The result was also confirmed by the PERMANOVA analysis (PERMANOVA: F = 7.57, P < 0.001). Mantel tests revealed significant correlations of the community composition with the TN concentration (Mantel test: r = 0.45, P = 0.002; Table 2).
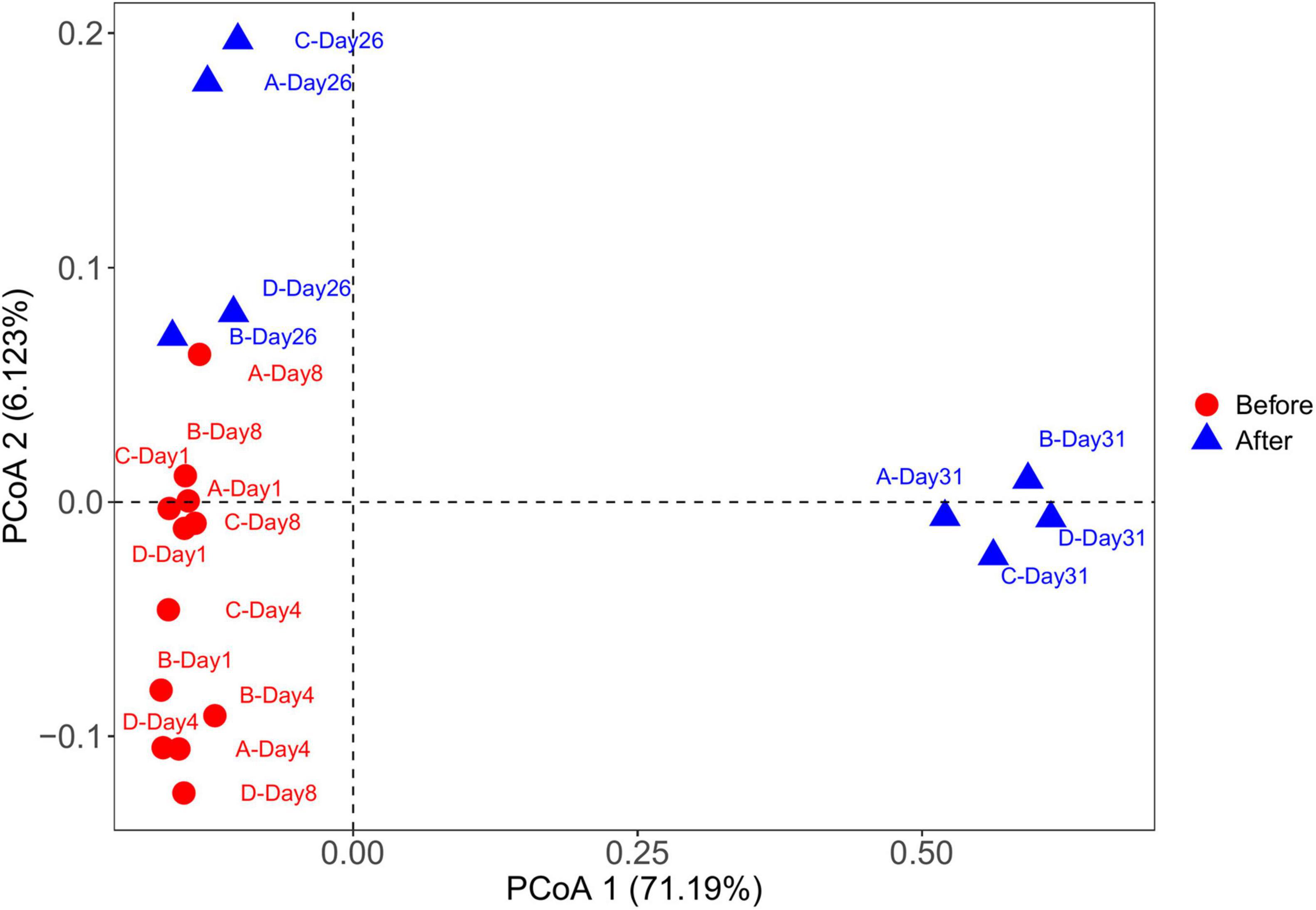
Figure 4. Principal coordinate analysis (PCoA) of microbial communities from cryoconite samples based on Bray-Curtis distance. The PCoA plot showed clear segregation between the before and after snowfall samples.
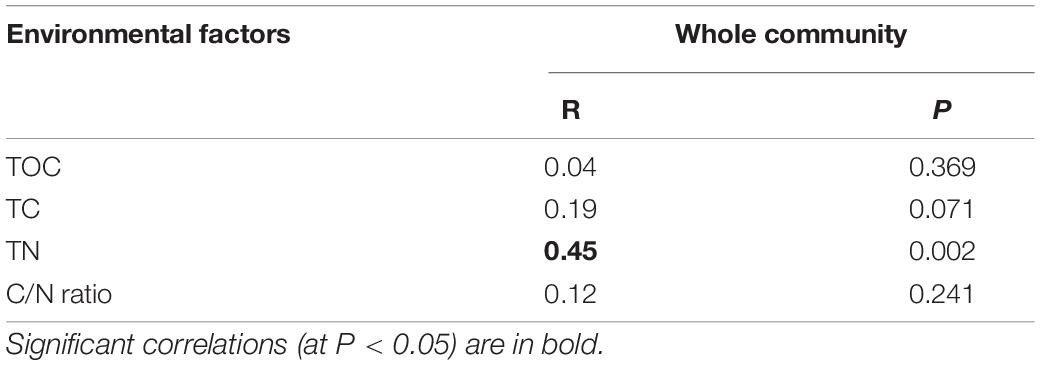
Table 2. Correlation relationships between the community compositional dissimilarity and the measured environmental factors by Mantel test.
Co-occurrence Patterns of Bacterial Community
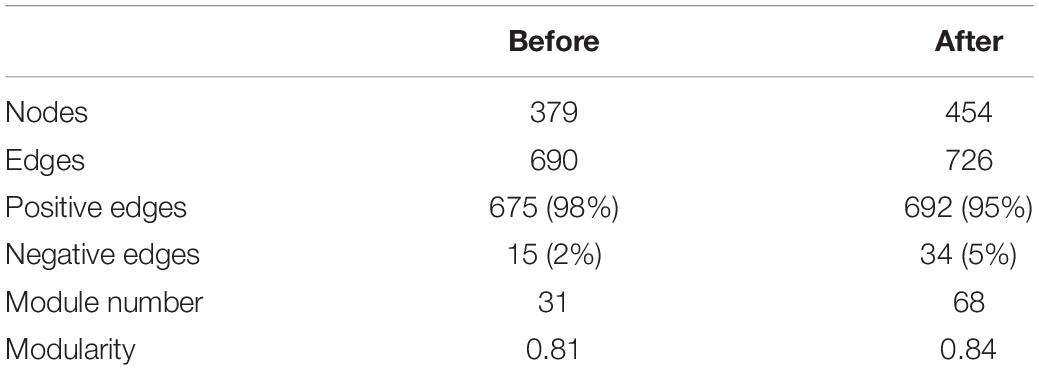
The co-occurrence network of bacterial communities was performed at the ASV level based on Spearman’s correlation relationships to examine the interactions between microorganisms. The co-occurrence network of bacterial communities was more complex after snowfall (Figure 5). Before the snowfall, the network contained 379 nodes which were connected by 690 edges, while after the snowfall the network contained 454 nodes and 726 edges. In addition, the networks before and after snowfall had 675 and 692 positive edges, respectively, but only 15 and 34 negative edges, respectively. The network after snowfall showed higher values of module number (68) and modularity (0.84) than the network before snowfall (31 and 0.82, respectively), indicating increased degree of modularity (Table 3). The nodes in the network before snowfall were composed of abundant ASVs (12.0%), rare ASVs (57.6%), and oscillating ASVs (30.4%). The nodes in the post snowfall network were composed of a lower proportion of abundant and oscillating ASVs (7.0 and 11.7%), but a higher proportion of rare ASVs (81.3%).
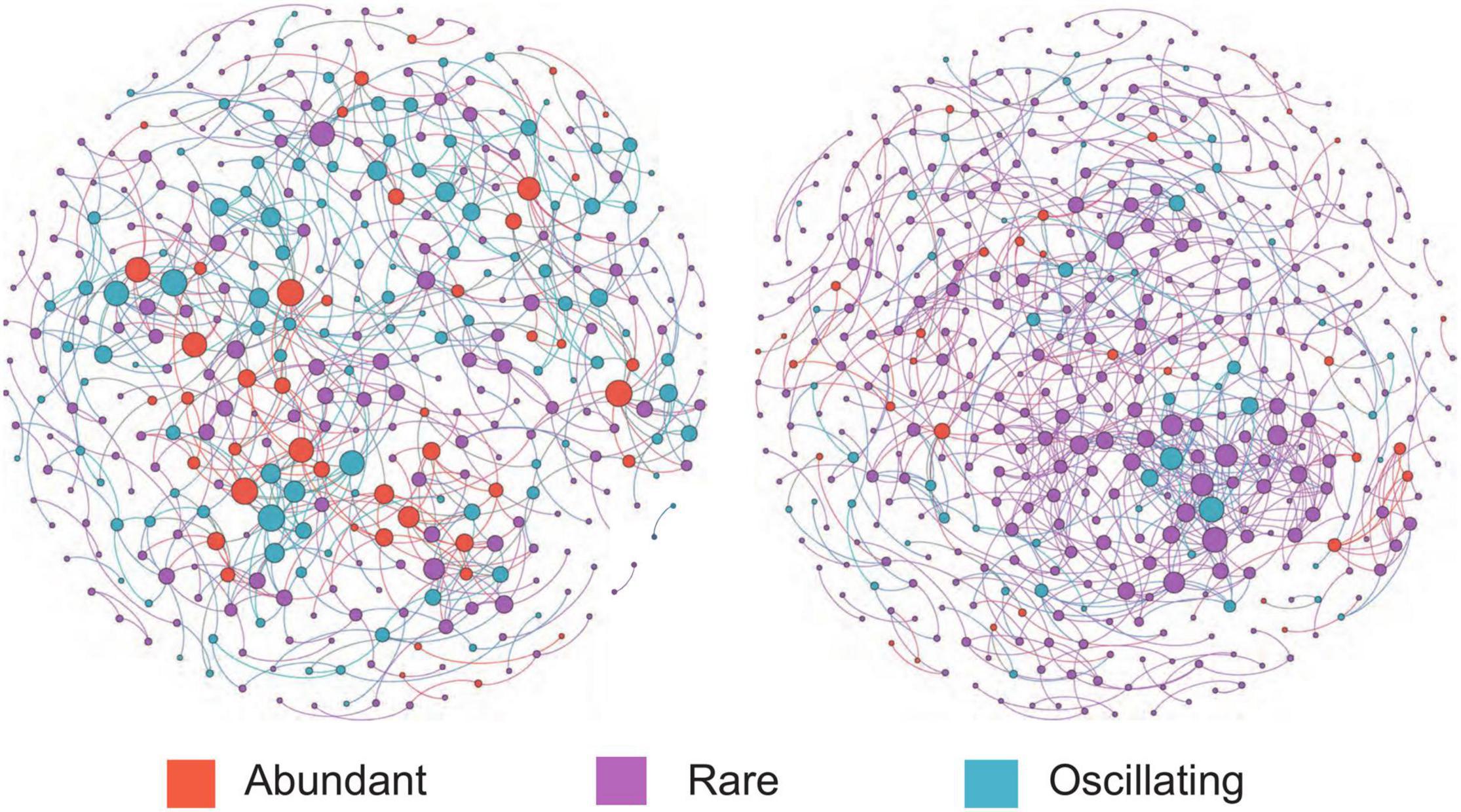
Figure 5. Bacterial Co-occurrence networks for the communities before and after snowfall. Each node represents an amplicon sequence variant (ASV). Nodes are colored by abundant, rare, and oscillating taxa, and the node sizes were proportional to their node degrees. The community network after snowfall is more complex with a higher connectedness than before snowfall.
In the community before snowfall, the node degree and betweenness of the abundant bacterial were significantly higher than that of rare bacterial and oscillating taxa (Figure 6), indicating that more information may be passed through abundant bacterial taxa. While in the community post snowfall, no significant differences were detected between abundant, rare and, oscillating taxa.
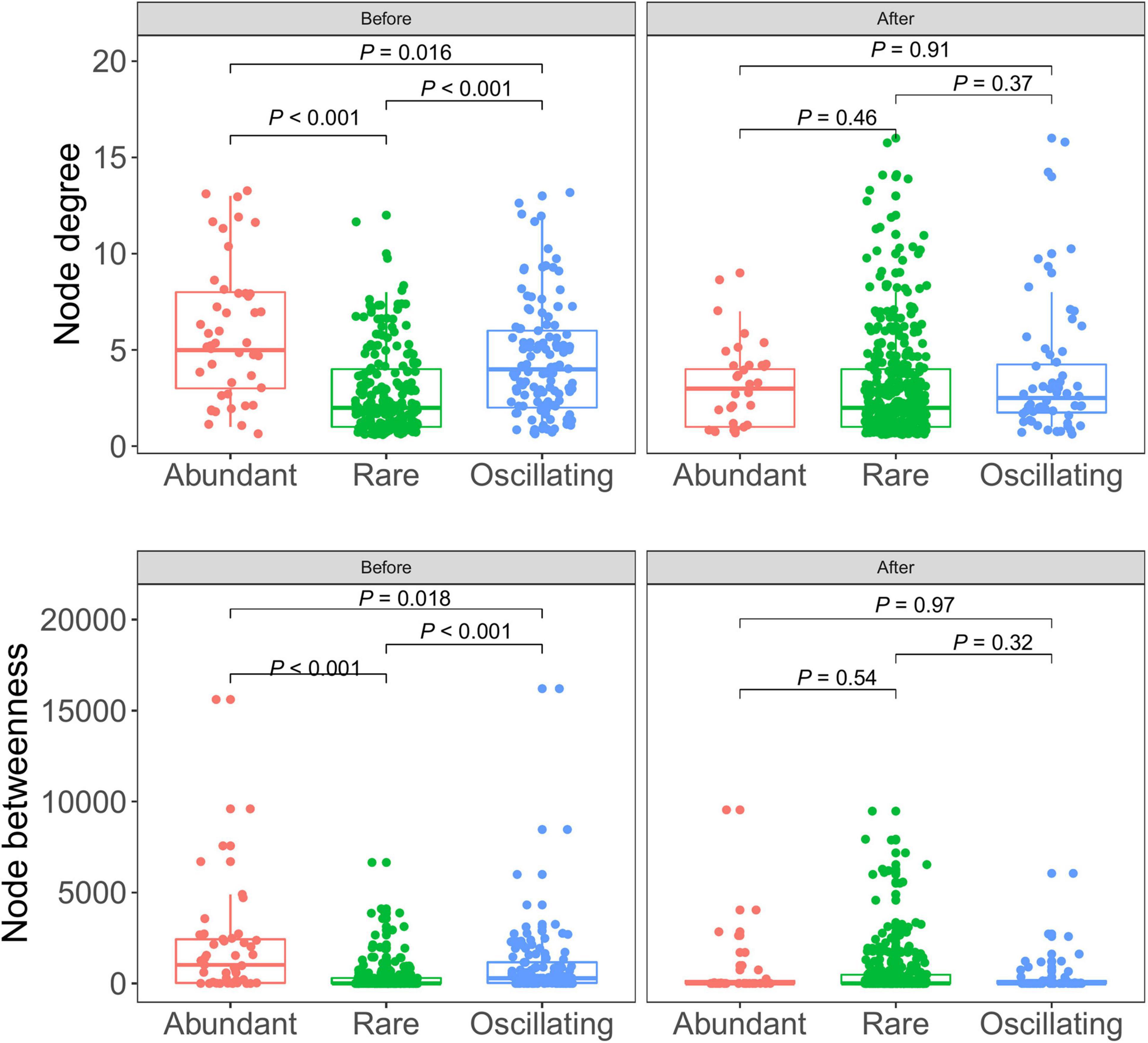
Figure 6. Node-level topological features between abundant, rare, and oscillating taxa in cryoconite communities before and after snowfall disturb. Each dot represents an individual sample. In the community before snowfall, the node degree and betweenness of the abundant bacterial were significantly higher than that of rare bacterial and oscillating taxa (Wilcoxon rank-sum test, P < 0.05).
Community Assembly Processes
NST was used to examine the relative contributions of stochasticity and determinism in shaping bacterial communities. The average NST values were 62 and 33% in the communities before and after snowfall (Figure 7). After the snowfall, the stochasticity processes significantly decreased for the whole community (Wilcoxon test, P < 0.001), suggesting the deterministic processes increased after snowfall disturbance.
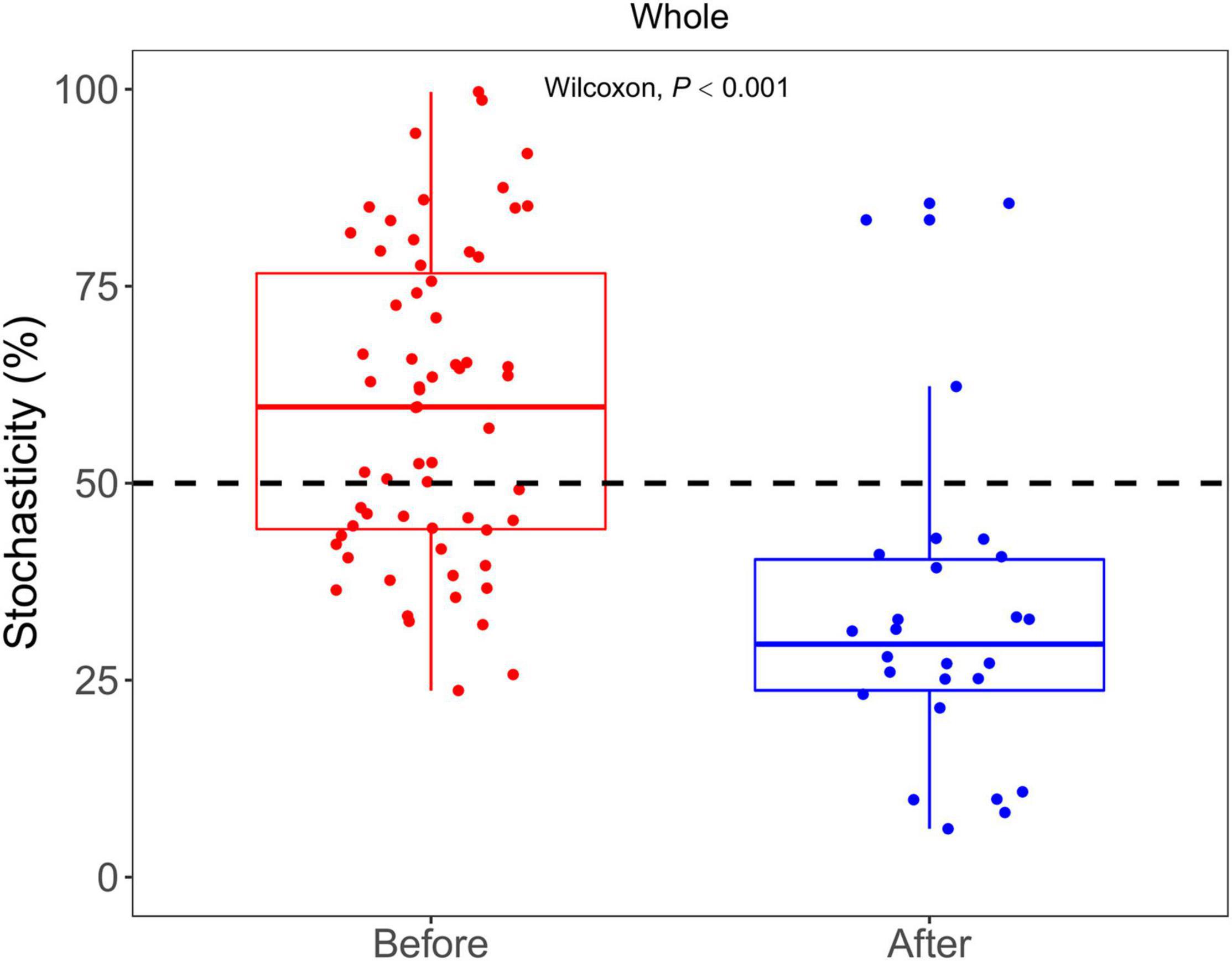
Figure 7. Bar plots show the comparison of NST between the bacterial communities before and after snowfall. The contribution of stochasticity was higher than 50% in the community before snowfall, indicating that the stochasticity process dominates the community assembly processes. The contribution of stochasticity was lower than 50% in the community after snowfall, indicating that the determinism process dominates the community assembly processes. The contribution of stochasticity in cryoconite communities was significantly decreased after snowfall disturbance.
Discussion
Nitrogen Drives the Bacterial Community Diversity and Community Structure
Bacterial diversity reduced significantly after snowfall disturbance, indicating that the cryoconite microbial community may be subjected to greater environmental filtering after snowfall. This is mostly likely due to nitrogen limitation, as nitrogen was the only measured environmental factors whose concentration significantly reduced post snowfall. This was consistent with the positive correlative relationships between nitrogen and bacterial diversity (Table 1). Nitrogen is an essential nutrient for microbial growth and plays important roles in controlling microbial diversity and ecosystem productivity (Vitousek et al., 2002; Xia et al., 2008; Sun et al., 2014). In the present study, the nitrogen concentration significantly reduced after the snowfall (Figure 1), which could be due to the snowmelt, which lead nitrogen to be flushed out. Generally, the rate at which chemical compounds are released at snowmelt depends on the way a chemical is incorporated in the snow grain (Pomeroy et al., 2005). Important microbial nutrients, such as NO3–, are not incorporated into the snow grain structure and are released easily (Tranter et al., 1986). The positive correlations between nitrogen availability and alpha diversity indices suggest that nitrogen limitation is an important determinant of bacterial diversity. Similar conclusions have been reported in three glaciers on Tibetan Plateau that TN concentrations were significantly correlated with the alpha diversity (Liu et al., 2017).
Consistent with the alpha-diversity indices, the bacterial community changed after snowfall disturbance (Figure 4), again indicating greater environmental filtering effect. The PCoA results showed that the samples on the 26th day were more similar to those before snowfall, while the samples on the 31st day were significantly different. This may be due to a delayed response to disturbance of the microbial community in cryoconite samples. This was also consistent with the increased deterministic processes in bacterial community structure after snowfall (Figure 7). Increased determinism is frequently attributed to the enhanced environmental filtering (Stegen et al., 2012). Our results demonstrated that TN significantly affected the bacterial community (Table 2). This is consistent with the previous findings that nitrogen availability strongly regulates microbial community structure in cryoconite and ice samples from Arctic and Tibetan Plateau glaciers (Christner et al., 2003; Mueller and Pollard, 2004; Edwards et al., 2011; Cameron et al., 2012; Liu et al., 2017). For example, a prior study indicated that nitrite and nitrate concentrations had important effects on the microbial community local scale variations in cryoconite on the Canada Glacier (Antarctic) (Mueller and Pollard, 2004).
Nitrogen Limitation Increases Network Complexity
Biotic interactions can explain a substantial proportion of the community structure variations (Hacquard et al., 2015; Dang and Lovell, 2016). A previous study of cryoconite from the High Arctic Foxfonna ice cap found that the assembly of the bacterial community of cryoconite was driven principally by biotic filtering (Gokul et al., 2016). Our results indicated that the community network was more complex after the snowfall disturbance, as evidenced by the higher nodes and edge numbers (Table 3). This is likely due to the enhanced environmental filtering, which has been observed in other systems subjected to nitrogen limitation (Wang et al., 2018). Positive correlations were substantial components in the networks before and after the snowfall (Table 3). Positive relationships could be used to infer commensal, mutualistic, obligate syntrophic, and parasitic relationships between bacterial species (Ji et al., 2019). The increased negative correlations after snowfall disturbance may be due to nutrient competition caused by nitrogen limitation (Zhao et al., 2016). It was proposed that the relationship between microorganisms occupying a similar ecological niche could change from commensalism (positive correlations) to competition (negative correlations) when nutrients become limited (Ji et al., 2019). Thus, the nitrogen limitation may influence bacterial diversity and community structure directly and indirectly.
Rare Bacteria Play an Important Role in the Microbial Community
The rare sub-community possessed a higher diversity than the abundant sub-community (Supplementary Figure 5). Thus, the rare taxa are the reservoir of genetic seeds to maintain community diversity and stability (Lennon and Jones, 2011; Rocca et al., 2020), providing biological buffering capacity to withstand environmental changes (Yachi and Loreau, 1999). Our results support prior studies that the rare bacterial taxa have an opportunity to be activated to maintain the stability of the bacterial community under nutrients limitations (Jousset et al., 2017). The snowfall disturbance leads to a decrease in nitrogen content and there is a decrease in temperature at the same time. After the snowfall disturbance, some rare species were adapted to the cold and oligotrophic environment and became abundant species, contributing 22% to the overall community. Four ASVs belonged to Flavobacterium (ASV_4, OTU_26, OTU_145, and OTU_146), one ASV belonged to Psychrobacter (ASV_9) belong to the rare species in samples before snowfall became abundant after snowfall disturbance (Supplementary Table 3). The genus Flavobacterium is known as exopolysaccharide (EPS) producing psychrophiles bacterium. EPS can substantially improve the tolerance of Flavobacterium from freeze–thaw cycles to survive in cold regions. When exposed to a cold environment suddenly, a number of physiological changes occur in the cells, and a set of small molecules are synthesized. The genes encoding these cold-shock-inducible proteins, such as cold shock protein (cspA), have been identified in Flavobacterium and Psychrobacter strains (Liu et al., 2019). They have been frequently reported in oligotrophic Antarctic freshwater systems (Michaud et al., 2012), Antarctic cryoconite holes from Canada Glacier (Christner et al., 2003), and Svalbard soils (Hansen et al., 2007), thus suggesting their bloom in oligotrophic and cold conditions after the snowfall.
Conclusion
This study explored the response of supraglacial cryoconite bacterial communities to a snowfall disturbance at the Laohugou Glacier (Tibetan Plateau). Our results showed that nitrogen concentration plays an important role in shaping the bacterial community diversity and structure in Tibetan glacial cryoconite. After the disturbance of a snowfall, the contribution of deterministic processes significantly increased, and the bacterial community assembly process changes were mainly caused by environmental filtration and bio-filtrating. Our results also revealed that some cold-tolerant rare taxa became abundant in the community after snowfall disturbance. Our results highlight the importance of rare taxa in maintaining glacial biodiversity and ecosystem stability. In addition, these results reveal that, from an ecological perspective, assembly processes play an important role in shaping the cryoconite bacterial community in Tibetan Plateau.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/bioproject/PRJNA509012.
Author Contributions
YQL designed the study and revised the manuscript. YC performed the lab experiments and wrote the draft. YL sampled the cryoconite from the Laohugou glacier. YC, KL, and MJ analyzed the data. All authors approved the final version.
Funding
This research was financially supported by the National Research and Development Program of China (Grant No. 2019YFC1509100), the National Natural Science Foundation of China (Grant No. 91851207), the Second Tibetan Plateau Scientific Expedition and Research (STEP) Program (Grant No. 2019QZKK0503), and the National Natural Science Foundation of China (Grant No. 42171138).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We greatly thank Ate Hendrik-Jan Jaarsma from Aarhus University for revising our manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.784273/full#supplementary-material
Footnotes
References
Ambrosini, R., Musitelli, F., Navarra, F., Tagliaferri, I., Gandolfi, I., Bestetti, G., et al. (2016). Diversity and assembling processes of bacterial communities in cryoconite holes of a Karakoram glacier. Microb. Ecol. 73, 827–837. doi: 10.1007/s00248-016-0914-6
Anesio, A. M., Lutz, S., Chrismas, N. A. M., and Benning, L. G. (2017). The microbiome of glaciers and ice sheets. NPJ Biofilms Microb. 3:10. doi: 10.1038/s41522-017-0019-0
Anesio, A. M., Mindl, B., Laybourn-Parry, J., Hodson, A. J., and Sattler, B. (2007). Viral dynamics in cryoconite holes on a high Arctic glacier (Svalbard). J. Geophys. Res. Biogeosci. 112:G04S31. doi: 10.1029/2006JG000350
Anesio, A. M., Sattler, B., Foreman, C., Telling, J., Hodson, A., Tranter, M., et al. (2010). Carbon fluxes through bacterial communities on glacier surfaces. Ann. Glaciol. 51, 32–40. doi: 10.3189/172756411795932092
Bergk Pinto, B., Maccario, L., Dommergue, A., Vogel, T. M., and Larose, C. (2019). Do organic substrates drive microbial community interactions in Arctic snow? Front. Microbiol. 10:2492. doi: 10.3389/fmicb.2019.02492
Cameron, K. A., Hodson, A. J., and Osborn, A. M. (2012). Structure and diversity of bacterial, eukaryotic and archaeal communities in glacial cryoconite holes from the Arctic and the Antarctic. FEMS Microbiol. Ecol. 82, 254–267. doi: 10.1111/j.1574-6941.2011.01277.x
Caporaso, J. G., Lauber, C. L., Walters, W. A. Berg-Lyons, D., Huntley, J., Fierer, N., et al. (2012). Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 6, 1621–1624. doi: 10.1038/ismej.2012.8
Chiellini, C., Chioccioli, S., Vassallo, A., Mocali, S., Miceli, E., Fagorzi, C., et al. (2019). Exploring the bacterial communities of infernaccio waterfalls: a phenotypic and molecular characterization of acinetobacter and Pseudomonas strains living in a red epilithic biofilm. Diversity 11:175.
Christner, B. C., Kvitko, B. H. II, and Reeve, J. N. (2003). Molecular identification of bacteria and Eukarya inhabiting an Antarctic cryoconite hole. Extremophiles 7, 177–183. doi: 10.1007/s00792-002-0309-0
Cook, J., Edwards, A., Takeuchi, N., and Irvine-Fynn, T. (2016). Cryoconite: the dark biological secret of the cryosphere. Prog. Phys. Geogr. 40, 66–111. doi: 10.1177/0309133315616574
Dang, H., and Lovell, C. R. (2016). Microbial surface colonization and biofilm development in marine environments. Microbiol. Mol. Biol. Rev. 80:91. doi: 10.1128/MMBR.00037-15
Deng, Y., Jiang, Y.-H., Yang, Y., He, Z., Luo, F., and Zhou, J. (2012). Molecular ecological network analyses. BMC Bioinformatics 13:113. doi: 10.1186/1471-2105-13-113
Dong, Y., Wu, S., Deng, Y., Wang, S., Fan, H., Li, X., et al. (2021). Distinct functions and assembly mechanisms of soil abundant and rare bacterial taxa under increasing pyrene stresses. Front. Microbiol. 12:689762. doi: 10.3389/fmicb.2021.689762
Du, S., Dini-Andreote, F., Zhang, N., Liang, C., Yao, Z., Zhang, H., et al. (2020). Divergent co-occurrence patterns and assembly processes structure the abundant and rare bacterial communities in a Salt Marsh ecosystem. Appl. Environ. Microbiol. 86:e00322-20. doi: 10.1128/AEM.00322-20
Edgar, R. C. (2016). UNOISE2: improved error-correction for Illumina 16S and ITS amplicon sequencing. bioRxiv [Preprint]. doi: 10.1101/081257
Edwards, A., Anesio, A. M., Rassner, S. M., Sattler, B., Hubbard, B., Perkins, W. T., et al. (2011). Possible interactions between bacterial diversity, microbial activity and supraglacial hydrology of cryoconite holes in Svalbard. ISME J. 5:150.
Edwards, A., Pachebat, J. A., Swain, M., Hegarty, M., Hodson, A. J., Irvine-Fynn, T. D., et al. (2013). A metagenomic snapshot of taxonomic and functional diversity in an alpine glacier cryoconite ecosystem. Environ. Res. Lett. 8:035003. doi: 10.1088/1748-9326/8/3/035003
Elshahed, M. S., Youssef, N. H., Spain, A. M., Sheik, C., Najar, F. Z., Sukharnikov, L. O., et al. (2008). Novelty and uniqueness patterns of rare members of the soil biosphere. Appl. Environ. Microbiol. 74, 5422–5428. doi: 10.1128/aem.00410-08
Friedman, J., and Gore, J. (2017). Ecological systems biology: the dynamics of interacting populations. Curr. Opin. Syst. Biol. 1, 114–121. doi: 10.1016/j.coisb.2016.12.001
Gokul, J. K., Hodson, A. J., Saetnan, E. R., Irvine-Fynn, T. D., Westall, P. J., Detheridge, A. P., et al. (2016). Taxon interactions control the distributions of cryoconite bacteria colonizing a high Arctic ice cap. Mol. Ecol. 25, 3752–3767. doi: 10.1111/mec.13715
Hacquard, S., Garrido-Oter, R., González, A., Spaepen, S., Ackermann, G., Lebeis, S., et al. (2015). Microbiota and host nutrition across plant and animal kingdoms. Cell Host Microbe 17, 603–616.
Hansen, A. A., Herbert, R. A., Mikkelsen, K., Jensen, L. L., Kristoffersen, T., Tiedje, J. M., et al. (2007). Viability, diversity and composition of the bacterial community in a high Arctic permafrost soil from Spitsbergen. Northern Norway. 9, 2870–2884. doi: 10.1111/j.1462-2920.2007.01403.x
Ji, M., Kong, W., Stegen, J., Yue, L., Wang, F., Dong, X., et al. (2020). Distinct assembly mechanisms underlie similar biogeographical patterns of rare and abundant bacteria in Tibetan Plateau grassland soils. Environ. Microbiol. 22, 2261–2272. doi: 10.1111/1462-2920.14993
Ji, M., Kong, W., Yue, L., Wang, J., Deng, Y., and Zhu, L. (2019). Salinity reduces bacterial diversity, but increases network complexity in Tibetan Plateau lakes. FEMS Microbiol. Ecol. 95:fiz190. doi: 10.1093/femsec/fiz190
Jia, X., Dini-Andreote, F., and Falcão Salles, J. (2018). Community assembly processes of the microbial rare biosphere. Trends Microbiol. 26, 738–747. doi: 10.1016/j.tim.2018.02.011
Jiang, Y., Song, H., Lei, Y., Korpelainen, H., and Li, C. (2019). Distinct co-occurrence patterns and driving forces of rare and abundant bacterial subcommunities following a glacial retreat in the eastern Tibetan Plateau. Biol. Fertil. Soils 55, 351–364.
Jiao, S., and Lu, Y. (2020). Soil pH and temperature regulate assembly processes of abundant and rare bacterial communities in agricultural ecosystems. Environ. Microbiol. 22, 1052–1065. doi: 10.1111/1462-2920.14815
Jiao, S., Luo, Y., Lu, M., Xiao, X., Lin, Y., Chen, W., et al. (2017). Distinct succession patterns of abundant and rare bacteria in temporal microcosms with pollutants. Environ. Pollut. 225, 497–505. doi: 10.1016/j.envpol.2017.03.015
Jousset, A., Bienhold, C., Chatzinotas, A., Gallien, L., Gobet, A., Kurm, V., et al. (2017). Where less may be more: how the rare biosphere pulls ecosystems strings. ISME J. 11, 853–862. doi: 10.1038/ismej.2016.174
Khan, N., Maezato, Y., McClure, R. S., Brislawn, C. J., Mobberley, J. M., Isern, N., et al. (2018). Phenotypic responses to interspecies competition and commensalism in a naturally-derived microbial co-culture. Sci. Rep. 8:297. doi: 10.1038/s41598-017-18630-1
Krug, L., Erlacher, A., Markut, K., Berg, G., and Cernava, T. (2020). The microbiome of alpine snow algae shows a specific inter-kingdom connectivity and algae-bacteria interactions with supportive capacities. ISME J. 14, 2197–2210. doi: 10.1038/s41396-020-0677-4
Kurm, V., Geisen, S., and Gera Hol, W. H. (2019). A low proportion of rare bacterial taxa responds to abiotic changes compared with dominant taxa. Environ. Microbiol. 21, 750–758. doi: 10.1111/1462-2920.14492
Lennon, J. T., and Jones, S. E. (2011). Microbial seed banks: the ecological and evolutionary implications of dormancy. Nat. Rev. Microbiol. 9:119.
Liang, Y., Xiao, X., Nuccio, E. E., Yuan, M., Zhang, N., Xue, K., et al. (2020). Differentiation strategies of soil rare and abundant microbial taxa in response to changing climatic regimes. Environ. Microbiol. 22, 1327–1340. doi: 10.1111/1462-2920.14945
Liu, L., Yang, J., Yu, Z., and Wilkinson, D. M. (2015). The biogeography of abundant and rare bacterioplankton in the lakes and reservoirs of China. ISME J. 9:2068. doi: 10.1038/ismej.2015.29
Liu, Q., Liu, H.-C., Zhou, Y.-G., and Xin, Y.-H. (2019). Microevolution and adaptive strategy of psychrophilic species Flavobacterium bomense sp. nov. isolated from glaciers. Front. Microbiol. 10:1069. doi: 10.3389/fmicb.2019.01069
Liu, Y., Vick-Majors, T. J., Priscu, J. C., Yao, T., Kang, S., Liu, K., et al. (2017). Biogeography of cryoconite bacterial communities on glaciers of the Tibetan Plateau. FEMS Microbiol. Ecol. 93, 1–9.
Lutz, S., McCutcheon, J., McQuaid, J. B., and Benning, L. G. (2018). The diversity of ice algal communities on the Greenland Ice sheet as revealed by oligotyping. Microb. Genomics 4:e000159.
Lynch, M. D. J., and Neufeld, J. D. (2015). Ecology and exploration of the rare biosphere. Nat. Rev. Microbiol. 13, 217–229. doi: 10.1038/nrmicro3400
McMurdie, P. J., and Holmes, S. (2014). Waste not, want not: why rarefying microbiome data is inadmissible. PLoS Comput. Biol. 10:e1003531. doi: 10.1371/journal.pcbi.1003531
Michaud, L., Caruso, C., Mangano, S., Interdonato, F., Bruni, V., and Lo Giudice, A. (2012). Predominance of Flavobacterium, Pseudomonas, and Polaromonas within the prokaryotic community of freshwater shallow lakes in the northern Victoria Land, East Antarctica. FEMS Microbiol. Ecol. 82, 391–404. doi: 10.1111/j.1574-6941.2012.01394.x
Mueller, D. R., and Pollard, W. H. (2004). Gradient analysis of cryoconite ecosystems from two polar glaciers. Polar Biol. 27, 66–74. doi: 10.1007/s00300-003-0580-2
Ning, D., Deng, Y., Tiedje, J. M., and Zhou, J. (2019). A general framework for quantitatively assessing ecological stochasticity. Proc. Natl. Acad. Sci. 116, 16892–16898. doi: 10.1073/pnas.1904623116
Oksanen, J., Blanchet, F. G., Kindt, R., Legendre, P. O’hara, R., Simpson, G. L., et al. (2010). vegan: Community Ecology Package. R Package Version 1.17-2. Available online at: http://cran.r-project.org/web/packages/vegan/vegan.pdf (accessed June 20, 2011).
Pedros-Alio, C. (2012). The rare bacterial biosphere. Ann. Rev. Mar. Sci. 4, 449–466. doi: 10.1146/annurev-marine-120710-100948
Pomeroy, J. W., Jones, H. G., Tranter, M., and Lilbæ, G. (2005). “Hydrochemical processes in snow-covered basins,” in Encyclopaedia of the Hydrological Sciences, Vol. 4, ed. M. Anderson (London: John Wiley & Sons Ltd), 2525–2538.
Rocca, J. D., Simonin, M., Bernhardt, E. S., Washburne, A. D., and Wright, J. P. (2020). Rare microbial taxa emerge when communities collide: freshwater and marine microbiome responses to experimental mixing. Ecology 101:e02956. doi: 10.1002/ecy.2956
Sauret, C., Séverin, T., Vétion, G., Guigue, C., Goutx, M., Pujo-Pay, M., et al. (2014). ‘Rare biosphere’bacteria as key phenanthrene degraders in coastal seawaters. Environ. Pollut. 194, 246–253.
Sommers, P., Darcy, J. L., Gendron, E. M. S., Stanish, L. F., Bagshaw, E. A., Porazinska, D. L., et al. (2017). Diversity patterns of microbial eukaryotes mirror those of bacteria in Antarctic cryoconite holes. FEMS Microbiol. Ecol. 94:fix167. doi: 10.1093/femsec/fix167
Stegen, J. C., Lin, X., Konopka, A. E., and Fredrickson, J. K. (2012). Stochastic and deterministic assembly processes in subsurface microbial communities. ISME J. 6:1653.
Steudel, B., Hector, A., Friedl, T., Löfke, C., Lorenz, M., Wesche, M., et al. (2012). Biodiversity effects on ecosystem functioning change along environmental stress gradients. Ecol. Lett. 15, 1397–1405. doi: 10.1111/j.1461-0248.2012.01863.x
Stibal, M., Telling, J., Cook, J., Mak, K. M., Hodson, A., and Anesio, A. M. (2012). Environmental controls on microbial abundance and activity on the greenland ice sheet: a multivariate analysis approach. Microb. Ecol. 63, 74–84. doi: 10.1007/s00248-011-9935-3
Sun, W., Qin, X., Ren, J., Yang, X., Zhang, T., Liu, Y., et al. (2012). The surface energy budget in the accumulation zone of the Laohugou Glacier No. 12 in the western Qilian Mountains, China, in summer 2009. Arctic Antarctic Alpine Res. 44, 296–305.
Sun, Y., Shen, Y.-X., Liang, P., Zhou, J., Yang, Y., and Huang, X. (2014). Linkages between microbial functional potential and wastewater constituents in large-scale membrane bioreactors for municipal wastewater treatment. Water Res. 56, 162–171. doi: 10.1016/j.watres.2014.03.003
Tranter, M., Brimblecombe, P., Davies, T. D., Vincent, C. E., Abrahams, P. W., and Blackwood, I. (1986). The composition of snowfall, snowpack and meltwater in the Scottish highlands-evidence for preferential elution. Atmos. Environ. 20, 517–525. doi: 10.1016/0004-6981(86)90092-2
Vitousek, P. M., Hättenschwiler, S., Olander, L., and Allison, S. (2002). Nitrogen and nature. AMBIO 31, 97–102.
Wang, S., Wang, X., Han, X., and Deng, Y. (2018). Higher precipitation strengthens the microbial interactions in semi-arid grassland soils. Glob. Ecol. Biogeogr. 27, 570–580. doi: 10.1111/geb.12718
Wei, T., Dong, Z., Kang, S., Qin, X., and Guo, Z. (2017). Geochemical evidence for sources of surface dust deposited on the Laohugou glacier, Qilian Mountains. Appl. Geochem. 79, 1–8. doi: 10.1016/j.apgeochem.2017.01.024
Xia, S., Li, J., and Wang, R. (2008). Nitrogen removal performance and microbial community structure dynamics response to carbon nitrogen ratio in a compact suspended carrier biofilm reactor. Ecol. Eng. 32, 256–262.
Yachi, S., and Loreau, M. (1999). Biodiversity and ecosystem productivity in a fluctuating environment: the insurance hypothesis. Proc. Natl. Acad. Sci. U.S.A. 96, 1463–1468. doi: 10.1073/pnas.96.4.1463
Yao, T., Thompson, L., Yang, W., Yu, W., Gao, Y., Guo, X., et al. (2012). Different glacier status with atmospheric circulations in Tibetan Plateau and surroundings. Nat. Clim. Change 2, 663–667. doi: 10.1038/nclimate1580
Zarsky, J. D., Stibal, M., Hodson, A., Sattler, B., Schostag, M., Hansen, L. H., et al. (2013). Large cryoconite aggregates on a Svalbard glacier support a diverse microbial community including ammonia-oxidizing archaea. Environ. Res. Lett. 8:035044. doi: 10.1088/1748-9326/8/3/035044
Zawierucha, K., Buda, J., Azzoni, R. S., Niśkiewicz, M., Franzetti, A., and Ambrosini, R. (2019a). Water bears dominated cryoconite hole ecosystems: densities, habitat preferences and physiological adaptations of Tardigrada on an alpine glacier. Aquat. Ecol. 53, 543–556. doi: 10.1007/s10452-019-09707-2
Zawierucha, K., Buda, J., and Nawrot, A. (2019b). Extreme weather event results in the removal of invertebrates from cryoconite holes on an Arctic valley glacier (Longyearbreen, Svalbard). Ecol. Res. 34, 370–379. doi: 10.1111/1440-1703.1276
Zhang, H., Hou, F., Xie, W., Wang, K., Zhou, X., Zhang, D., et al. (2020). Interaction and assembly processes of abundant and rare microbial communities during a diatom bloom process. Environ. Microbiol. 22, 1707–1719. doi: 10.1111/1462-2920.14820
Keywords: cryoconite, snowfall, rare bacteria, deterministic processes, stochastic processes
Citation: Chen Y, Liu Y, Liu K, Ji M and Li Y (2022) Snowstorm Enhanced the Deterministic Processes of the Microbial Community in Cryoconite at Laohugou Glacier, Tibetan Plateau. Front. Microbiol. 12:784273. doi: 10.3389/fmicb.2021.784273
Received: 27 September 2021; Accepted: 27 December 2021;
Published: 27 January 2022.
Edited by:
Jianjun Wang, Nanjing Institute of Geography and Limnology, Chinese Academy of Sciences (CAS), ChinaReviewed by:
Ze Ren, Beijing Normal University, ChinaJuntao Wang, Western Sydney University, Australia
Copyright © 2022 Chen, Liu, Liu, Ji and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yongqin Liu, yqliu@itpcas.ac.cn
 Yuying Chen
Yuying Chen Yongqin Liu
Yongqin Liu Keshao Liu
Keshao Liu Mukan Ji
Mukan Ji Yang Li4
Yang Li4