- Laboratoire de Chimie Bactérienne, Institut de Microbiologie de la Méditerranée, CNRS-Aix Marseille Univ (UMR7283), Marseille, France
Guanosine penta- or tetraphosphate (known as (p)ppGpp) serves as second messenger to respond to nutrient downshift and other environmental stresses, a phenomenon called stringent response. Accumulation of (p)ppGpp promotes the coordinated inhibition of macromolecule synthesis, as well as the activation of stress response pathways to cope and adapt to harmful conditions. In Escherichia coli, the (p)ppGpp level is tightly regulated by two enzymes, the (p)ppGpp synthetase RelA and the bifunctional synthetase/hydrolase SpoT. We recently identified the small protein YtfK as a key regulator of SpoT-mediated activation of stringent response in E. coli. Here, we further characterized the regulation of ytfK. We observed that ytfK is subjected to catabolite repression and is positively regulated by the cyclic AMP (cAMP)-cAMP receptor protein (CRP) complex. Importantly, YtfK contributes to SpoT-dependent accumulation of (p)ppGpp and cell survival in response to glucose starvation. Therefore, regulation of ytfK by the cAMP-CRP appears important to adjust (p)ppGpp level and coordinate cellular metabolism in response to glucose availability.
Introduction
Bacteria have evolved efficient stress response mechanisms to quickly adjust cell growth and metabolism according to challenging environments. One of such bacterial responses is the near-universal stringent response. The hyperphosphorylated derivatives of GDP and GTP, guanosine tetra- and pentaphosphate (collectively named (p)ppGpp), are the central signaling molecules of the stringent response (Cashel and Gallant, 1969; Potrykus and Cashel, 2008). These alarmones allow rapid and robust stress adaptation by affecting gene expression and metabolism (Cashel, 1969; Potrykus and Cashel, 2008; Hauryliuk et al., 2015; Liu et al., 2015; Steinchen and Bange, 2016). Since, (p)ppGpp has also emerged as an important regulator of bacterial virulence, survival during host invasion (reviewed in Hauryliuk et al., 2015; Irving et al., 2021) and antibiotic resistance and tolerance (Nguyen et al., 2011; Amato et al., 2013; Helaine et al., 2014; Amato and Brynildsen, 2015).
The RelA–SpoT Homologue (RSH) family of bifunctional proteins is key players in synthesizing and degrading (p)ppGpp (Atkinson et al., 2011). Therefore, the tight balance between both reciprocal activities constitutes a crucial point of regulation for fine tuning (p)ppGpp homeostasis. The long RSH proteins share a similar domain architecture and can be divided into two regions of similar size. The N-terminal half of the protein harbors the catalytic synthetase and the hydrolase domains. The C-terminal half of the protein contains four regulatory (TGS, helical, CC and ACT) domains with essential role in sensing and transducing stress signal to the catalytic domains (Hogg et al., 2004; Hauryliuk et al., 2015; Pausch et al., 2020; Tamman et al., 2020). In most gamma and beta-proteobacteria, to which Escherichia coli belongs, the stringent response is driven by two paralogous RSH enzymes named RelA and SpoT. While SpoT has both functional synthetase and hydrolase domains, RelA is a monofunctional synthetase with a degenerated inactive hydrolase domain, making SpoT the primary source of (p)ppGpp hydrolysis (Xiao et al., 1991).
The (p)ppGpp synthetase activity of RelA is triggered in response to amino acid starvation via a ribosomal mechanism. Under this condition, deacylated tRNAs accumulate and activation occurs when RelA binds with an uncharged tRNA at an empty A-site of a stalled ribosome (Cashel and Gallant, 1969; Haseltine and Block, 1973; Arenz et al., 2016; Winther et al., 2018).
SpoT functions as a central protein which integrates various stress signals (Hernandez and Bremer, 1991; Xiao et al., 1991), other than amino acid starvation, such as fatty acid (Seyfzadeh et al., 1993), carbon (Xiao et al., 1991), iron (Vinella et al., 2005) and phosphate (Spira et al., 1995) starvations. Importantly, the hydrolysis activity of SpoT is crucial for balancing the basal activity of RelA. Indeed, disruption of the spoT gene in the presence of relA leads to a lethal accumulation of (p)ppGpp (Xiao et al., 1991; Baba et al., 2006). Therefore, a fine regulation of reciprocal SpoT activities is essential to correctly adjust intracellular (p)ppGpp level in response to bacterial surrounding.
Interaction of SpoT with other protein partners directly controls the balance between reciprocal activities. Indeed, it has been reported that the acyl carrier protein (ACP) binds the TGS domain of SpoT to promote (p)ppGpp accumulation during fatty acid starvation (Battesti and Bouveret, 2006). Interaction of SpoT with the CgtA/ObgE GTPase is proposed to modulate hydrolase activity during exponential growth (Wout et al., 2004; Jiang et al., 2007). In addition, SpoT hydrolase activity is promoted by binding to the anti-σ70 factor Rsd upon carbon downshift (Lee et al., 2018). Finally and more recently, we reported that the small protein YtfK can directly interact with the catalytic domains of SpoT to activate the stringent response under fatty acid or phosphate starvations by tilting the catalytic balance toward synthesis rather than hydrolysis (Germain et al., 2019). Moreover, the SpoT-YtfK ratio controls the switch of SpoT activities (Germain et al., 2019). Therefore, regulation of the level of YtfK protein is crucial for adjusting (p)ppGpp level in response to external stresses.
In this study, we dissected the ytfK promoter region and searched for new candidate genes involved in regulation of ytfK expression. Overexpression of one of these genes (cpdA), encoding a cAMP phosphodiesterase, strongly decreases ytfK expression. Importantly, our results show that the cAMP-CRP complex directly binds the ytfK promoter region to positively regulate its transcription in response to glucose availability. Moreover, we show that YtfK contributes to SpoT-dependent accumulation of (p)ppGpp and cell survival during glucose deprivation. Therefore, regulation of ytfK by the cAMP-CRP complex seems to play an important role in sensing and transducing signal to SpoT to coordinate cellular metabolism in response to glucose availability.
Materials and Methods
Bacterial Strains, Media, and Growth Conditions
Bacterial strains used in this study are listed in Supplementary Table S1. E. coli DH5α strain was the general cloning host. All E. coli strains were derived from MG1655 strain and grown at 37°C in LB (Lysogeny Broth) liquid medium from Oxoid (LP0021B and LP0042B; Clark and Maaløe, 1967) or NA (Nutrient Agar) solid medium from Oxoid (CM0003B). M9 minimal liquid medium was composed of M9 salt (60mM Na2HPO4, 22mM KH2PO4, 8mM NaCl, and 20mM NH4Cl), 1mM MgSO4, 100μM CaCl2, 1μg/ml thiamine, and 0.025% or 0.2% glucose. MOPS minimal liquid medium was prepared as previously described (Neidhardt et al., 1974) and was free of nucleobases and amino acids. When necessary, media were supplemented with 80μg/ml X-gal (5-bromo-4-chloro-3-indolyl-β-D-galactopyranoside) and antibiotics used at the following concentrations: 50μg/ml ampicillin, 50μg/ml chloramphenicol, and 25μg/ml kanamycin. P1 transductions were performed as previously described (Thomason et al., 2007).
Expression of lambda recombinase from pKD46 (Datsenko and Wanner, 2000) was induced by adding 0.2% of arabinose and by growing cells at 30°C during 1.5h, pKD46 plasmid was then eliminated by streaking colonies on NA solid medium and by incubating plates overnight at 37°C. Kanamycin resistance cassette was flipped out as previously described (Cherepanov and Wackernagel, 1995).
DNA Manipulations
Plasmids used in this work are listed in Supplementary Table S1 and were extracted using Monarch plasmid miniprep kit (Biolabs). PCRs were carried out from colonies with Phusion DNA polymerase (Thermo Scientific) to amplify DNA fragments used for cloning or strain constructions and Gotaq flexi DNA polymerase (Promega) for diagnostic PCR. PCR products were purified using the Monarch DNA gel extraction kit (Biolabs).
Plasmid Constructions
The plasmid derivatives used in this study (Supplementary Table S1) were constructed by amplifying genes by PCR from template chromosomal DNA using primers listed in Supplementary Table S2 and by digesting DNA with restriction enzymes indicated in Supplementary Table S2.
Construction of Reporter Strains
Transcriptional and translational fusions reporter strains were constructed by two-step λ red-mediated recombination, adapted from Blank et al. (2011). A chloramphenicol resistance cassette was amplified by PCR together with an I-SceI recognition site using pWRG100 plasmid as template with primers (357/358) for translational and (353/354 or 577/354) for transcriptional fusions (Supplementary Table S2) containing a 50bp homologous sequence with upstream and downstream of the ytfK locus. MG1655 strain harboring pKD46 plasmid was electroporated with the resulting fragment and the insertion of the I-SceI:cat fragment into the target locus was verified by PCR and then P1 transduced into the TB28 strain. The resulting strain was then transformed with pWRG99 harboring the I-SceI endonuclease under the control of an anhydrotetracycline inducible promoter. The chloramphenicol cassette was then removed by counter selection using lambda red recombination to insert a PCR product complementary to the flanking regions of the I-SceI:cat cassette on the chromosome. The PCR product of the different transcriptional fusions was generated with primers (355/356) and pGH254:PytfK P1+P2:lacZ or pGH254:PytfK P2:lacZ as templates or primers (708/356) and pGH254:PytfK P1:lacZ as template (Supplementary Tables S1, S2). The translational fusion PCR product was obtained by PCR amplification with primers (360/361) and pGH254 as template (Supplementary Tables S1, S2). Selection of successful recombinants was mediated by spreading cells on NA plates containing ampicillin, X-gal, and 1μg/ml anhydrotetracycline. The proper integration of transcriptional or translational fusions was confirmed by diagnostic PCR and then sequenced.
Construction of the Δcrp Mutant
Deletion of the crp gene was achieved by replacement of the crp locus with a kanamycin resistance cassette using λ red-mediated recombination as previously described (Datsenko and Wanner, 2000). The kanamycin resistance cassette was amplified from pKD4 plasmid template with primers (362/363; Supplementary Tables S1, S2) containing a 50bp homologous extension with upstream and downstream of the coding sequence of crp. The resulting PCR product was then used to transform, by electroporation, MG1655 strain harboring pKD46 plasmid. Deletion of crp was confirmed by diagnostic PCR.
Genetic Screening for the Identification of Genes Involved in Regulation of ytfK Expression
A collection of plasmids containing 6His-tagged genes (minus GFP) from the ASKA library (Kitagawa et al., 2005) was used to transform the TB28 translational fusion (ytfK TL P1+P2) reporter strain by electroporation. Cells were diluted and spread on NA plates supplemented with chloramphenicol, 50 or 200μM IPTG and 80μg/ml X-gal. Petri plates were incubated overnight at 37°C and colonies were screened for their dysregulated expression of ytfK (Supplementary Figure S1), then streaked and their plasmids were sequenced.
6His-CRP Tagged Protein Purification
The pEG25:6His-crp plasmid expressing crp with an N-terminal 6His tag, under the control of the T5 lac promoter inducible by IPTG was used to transform BL21 (DE3) cells. Several transformants were grown at 37°C overnight in LB medium containing 100μg/ml of ampicillin. Culture was then diluted 50-fold in 2l of the same medium and incubated at 37°C with shaking until OD600nm 0.6. The expression of the 6His-CRP protein was induced by adding IPTG at a final concentration of 0.5mM for 2h. Bacteria were harvested (9,000 × g, 20min at 4°C) and the pellet was stored at – 80°C. Cells were resuspended and incubated in lysis buffer (50mM Tris-HCl pH 8, 300mM NaCl, 1mM EDTA, 10mM imidazole, 0.5mg/ml lysozyme, 1mM phenylmethylsulfonyl fluoride (PMSF), 20μg/ml DNase and 15mM MgCl2) for 1.5h at 4°C with gentle shacking and were disrupted using three cycles of French press lysis steps. The cleared lysate was recovered by centrifugation (6,080 × g, 25min) and 6His-CRP protein was purified by ion metal affinity chromatography using a 5ml Nickel (HiTrapHP) column on an AKTA pure 25 (GE healthcare) and desalted using Hiprep 26/10 Desalting column, as previously described (Germain et al., 2019; Supplementary Figure S2A). The last step in 6His-CRP purification was achieved by size-exclusion chromatography (SEC) using a HiLoad 26/600 Superdex 200pg. column pre-equilibrated with 50mM Tris-HCl pH 8, 500mM NaCl, 500mM KCl, 2mM β-mercaptoethanol, and 2% glycerol. The SEC chromatogram is visualized in Supplementary Figure S2B. The purity of the 6His-CRP was verified by SDS-gel electrophoresis (Supplementary Figure S2C). The 6His-CRP protein was stored at −80°C in storage buffer (40mM Tris-HCl pH 8, 238mM NaCl, 22% glycerol, and 1.6mM β-mercaptoethanol).
Electrophoretic Mobility Shift Assay
5' or 3'-Cy5-labeled DNA fragments of the ytfK promoter region were obtained by PCR amplification with appropriate oligonucleotides (Supplementary Table S2). DNA fragments (5nM) were incubated for 15min at 37°C with 6His-CRP (12.5, 25, 65, 130, 195, 300, or 500μM) or not, in 14μl of the binding buffer [10mM Tris-HCl pH 7.5, 50mM NaCl, 5mM MgSO4, 1mM DTT (dithiothreitol), 1mg/ml BSA (bovine serum albumin), 200μM cAMP, 10% glycerol and 12μg/ml poly (dI-dC)]. Ten microliters of the sample were then loaded into a 5% polyacrylamide (37.5/1 [wt/wt] acrylamide-bisacrylamide) gel containing 0.25X TBE (tris-borate EDTA) pH 7.5, 10% glycerol and 200μM cAMP. After migration (10 volts/cm, 1h, 4°C) in migration buffer (TBE 0.25X, 20μM cAMP), the bands were visualized using phosphoImaging (GE Healthcare). The DNA fragment of ytfK promoter region deleted for the putative CRP-binding site was obtained by PCR extension of overlapping DNA fragments using appropriate oligonucleotides (Supplementary Table S2).
β-Galactosidase Activity Assay
Cells expressing transcriptional fusion (ytfK P1+P2) were grown at 37°C in MOPS minimal medium containing 0.025% of glucose and 0.4mM KH2PO4. At the indicated time point, OD600nm was measured and 200μl of the culture was taken at indicated times and incubated with 800μl of Z-Buffer (0.06M Na2HPO4 7H2O, 0.04M NaH2PO4 H2O, 0.01M KCl, 0.001M MgSO4 7H2O and 0.05M β-mercaptoethanol). Twenty microliters of chloroform was added and cells were vortexed 3×10s followed by incubation at room temperature for 2min. Fifty microliters of the sample was incubated with 150μl of Z-Buffer pre-heated to 28°C. ONPG was added at a final concentration of 0.67mg/ml and the β-galactosidase activity was measured according to the Miller method (Miller, 1992) by following the OD420nm with TECAN microplate reader.
In vivo (p)ppGpp Assessment
Bacteria were grown at 37°C overnight in MOPS medium containing 2mM KH2PO4 and 0.2% glucose. Cells were then diluted 100-fold in 500μl of fresh MOPS medium containing 0.4mM phosphate (KH2PO4) and 0.025% glucose. Cells were continuously and uniformly labeled with 5μl of 32P (0.37MBq/μl, PerkinElmer) and grown at 37°C with shaking (440rpm). Fifty microliters of samples were taken at the indicated times and 20μl of 21M ice-cold formic acid was added to stop the reaction. Samples were kept on ice for 20min and then stored at −20°C. Cell extracts were recovered by centrifugation (14,000 × g for 60min at 4°C). Five microliters of each sample were spotted into PEI Cellulose TLC (thin layer chromatography) plates (purchased from MercK-Millipore) and resolved with 1.5M KH2PO4 pH 3.4. TLC plates were then revealed by PhosphoImaging (GE Healthcare) and analyzed using ImageQuant software (GE Healthcare). The amount of ppGpp was normalized by the amount of pppGpp, ppGpp and GTP for the wild-type strain and the ΔytfK mutant and by the amount of ppGpp and GTP for the ΔrelA and ΔrelAΔytfK mutants.
Cell Survival to Prolonged Carbon Starvation
Overnight cultures of M9 minimum medium containing 2% of glucose were diluted 100-fold in fresh M9 medium containing 0.025% of glucose. Bacteria were cultivated during 120h with shaking at 37°C. Evaporated water was measured and compensated by regularly adding the missing volume of water throughout the culture. Aliquots were taken at indicated hours, cells were serially diluted and plated on NA medium. Plates were then incubated at 37°C overnight and the number of CFU/ml was determined.
Results
ytfK Is Transcribed From Two-Independent Promoters and the cAMP Level Plays a Key Regulatory Role in ytfK Expression
ytfK is transcribed as a monocistronic unit (Figure 1A). One promoter, referenced in this study as P1 promoter, has been shown to be recognized by the sigma factor σS (Lacour and Landini, 2004), which is proposed to be involved in the induction of ytfK expression at the onset of stationary phase (Salgado et al., 2013). In addition, two pho boxes recognized by PhoB are located upstream the P1 transcriptional start site and are required for induction of ytfK expression in response to phosphate starvation (Baek and Lee, 2006; Yoshida et al., 2011; Figure 1A). Moreover, a second putative P2 promoter is predicted (Lacour and Landini, 2004) upstream the P1 promoter but has not yet been experimentally validated.
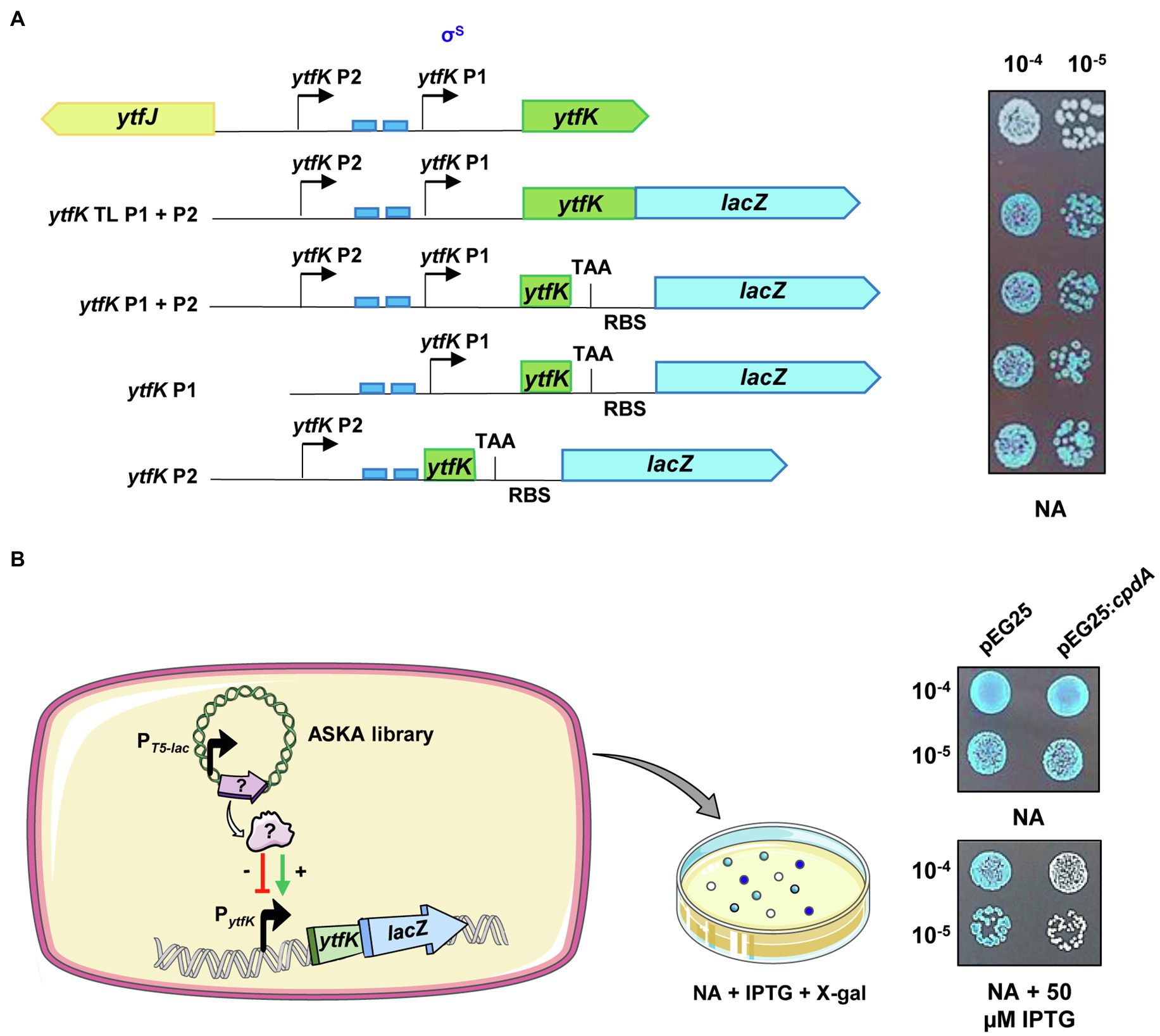
Figure 1. ytfK is transcribed from two distinct promoters. (A) ytfK promoter region. The divergent ytfK and ytfJ genes are represented by green and yellow arrows, respectively. The previously identified (P1) and the putative (P2) transcriptional start sites are indicated by black arrows. The two pho boxes recognized by the response regulator PhoB are visualized by blue rectangles. The following schematic representations describe translational and transcriptional lacZ fusions used in this study and on the right the corresponding expression level as judged by colorimetric visualization of lacZ activity on X-gal plate. Briefly, stationary-phase cultures of TB28 cells harboring translational (ytfK TL P1+P2) or transcriptional (ytfK P1+P2, ytfK P1, or ytfK P2) fusions were serially diluted and 5µl of 10−4 and 10−5 dilutions were spotted on NA rich solid medium containing X-gal. Results are representative of three independent experiments. (B) Schematic representation of the genetic screen based on overexpression of E. coli genes from a pooled plasmid of ASKA library (Kitagawa et al., 2005) in TB28 cells harboring translational fusion (ytfK TL P1+P2) and screening of candidates having dysregulated level of YtfK as judged by colorimetric visualization of lacZ level on X-gal NA plates. On the right, stationary-phase cultures of cells expressing translational fusion (ytfK TL P1+P2) and harboring pEG25 or pEG25:cpdA plasmids were serially diluted and spotted on NA rich solid medium supplemented or not with 50μM IPTG and X-gal. Results are representative of three independent experiments.
In order to dissect the promoter region of ytfK and to analyze the genetic regulation of ytfK expression, we first generated (Figure 1A; see Materials and Methods) chromosomal translational (ytfK TL P1+P2) and several truncated transcriptional lacZ fusions (ytfK P1, ytfK P2, or ytfK P1+P2) to follow promoter activity on X-gal plates. As shown in Figure 1A, ytfK expression is driven from two-independent promoters when cells were spotted on nutrient rich agar, thus confirming the existence of a distal P2 promoter.
To gain further insight on how ytfK is regulated, we used cells harboring the translational fusion (ytfK TL P1+P2) and searched for activators and inhibitors of ytfK expression by overexpressing E. coli genes from the ASKA library pool (Kitagawa et al., 2005) and by using screening assay on X-gal plates for selection of clones with dysregulated lacZ activity (Figure 1B). The ASKA library encompasses almost all E. coli genes cloned into the high-copy-number vector pCA24N, under the control of a PT5-lac promoter inducible by IPTG (Kitagawa et al., 2005). Out of the approximately 40,000 clones screened, 29 candidates were selected and their plasmids were sequenced (Table 1; Supplementary Figure S1).
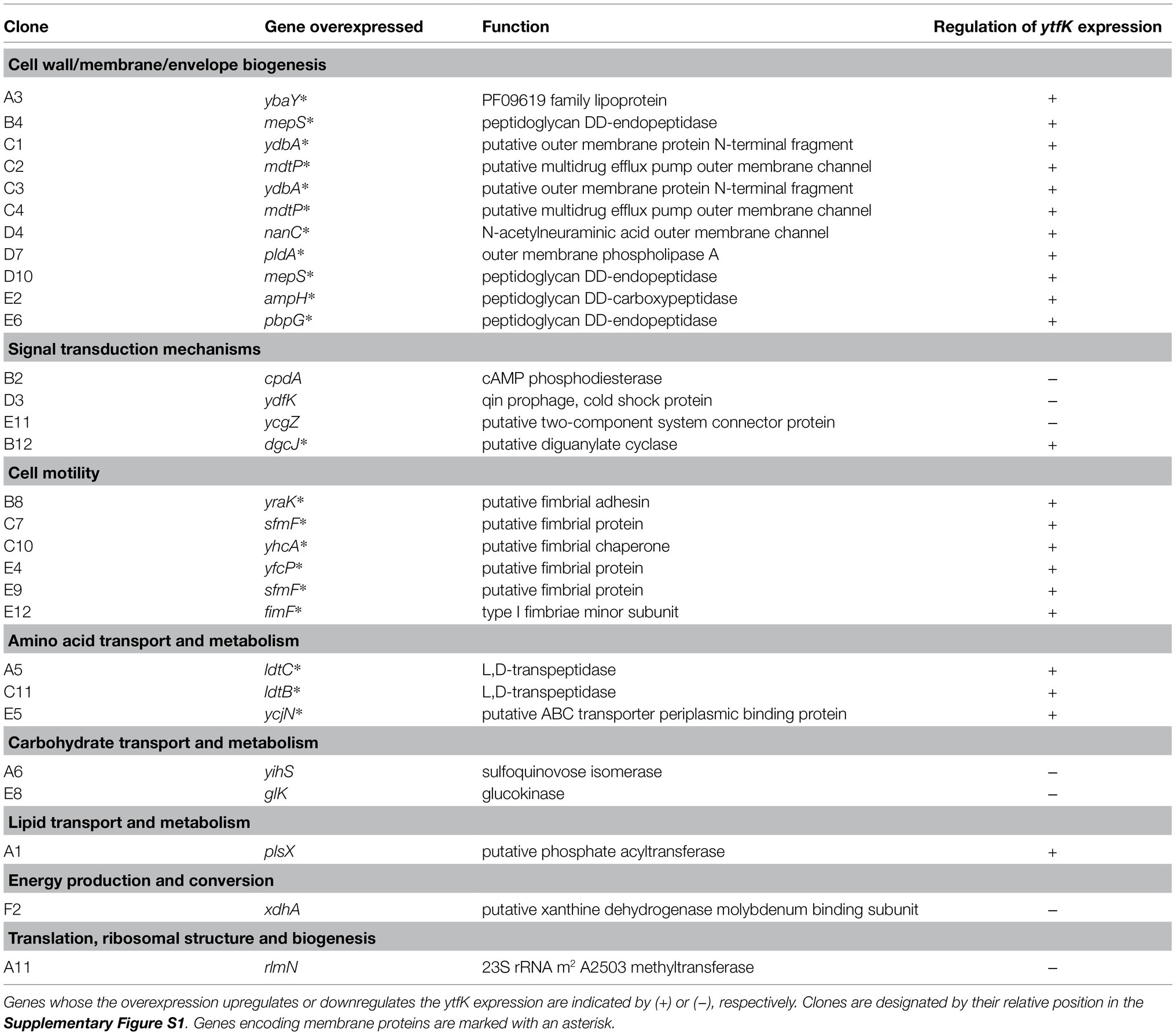
Table 1. Genes identified by overexpressing each E. coli gene from the ASKA library (Kitagawa et al., 2005) and by screening for the impaired regulation of translational fusion (ytfK TL P1+P2).
We confirmed, after re-cloning into a more suitable physiological plasmid harboring a tight IPTG-inducible PT5-lac promoter (pEG25), that the ectopic overexpression of cpdA, encoding a cAMP phosphodiesterase, strongly decreases ytfK expression level (Figure 1B). Therefore, this result suggests that the intracellular level of cAMP plays an important role in adjusting ytfK expression level.
ytfK Expression Level Is Induced by the cAMP-CRP Complex at the Transcriptional Level
In E. coli, cAMP is degraded by CpdA (Imamura et al., 1996) and is synthesized by the adenylate cyclase CyaA (Notley-McRobb et al., 1997). To understand how cAMP plays an essential role in controlling the YtfK level, we first followed lacZ activity of wild-type (TB28 strain) and ΔcyaA cells harboring translational or transcriptional lacZ fusions. We first observed that deletion of the adenylate cyclase cyaA dramatically reduces ytfK expression and this regulation occurs at the transcriptional level (Figure 2A). Moreover, adding 1mM of cAMP in the medium fully restored ytfK expression (Figure 2A). These results confirm that cAMP is necessary to induce ytfK transcription under standard rich conditions.
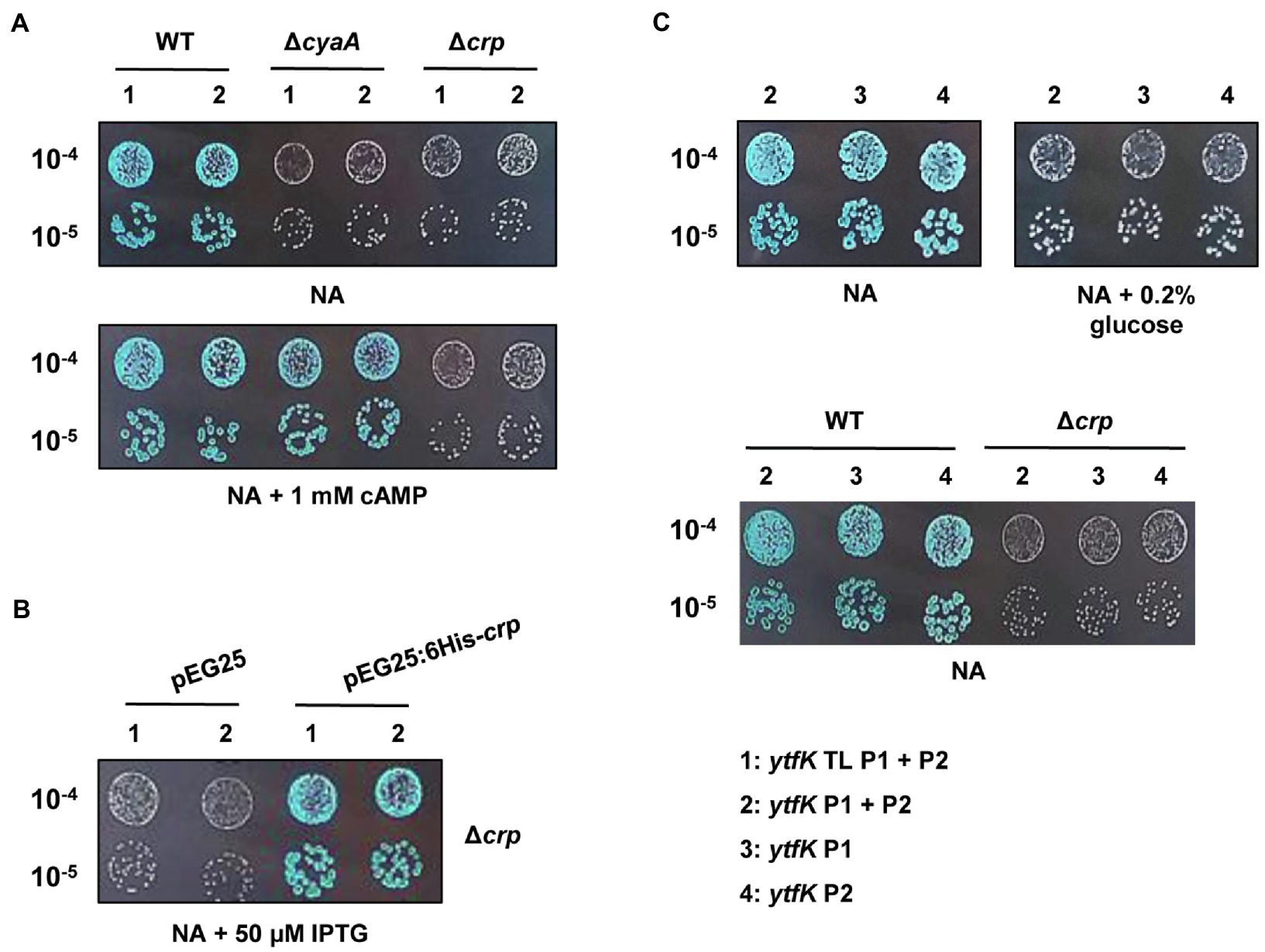
Figure 2. ytfK expression is controlled by cAMP level and requires the presence of the master regulator CRP. (A) Stationary-phase cultures of WT (TB28), ΔcyaA or Δcrp cells expressing translational (1: ytfK TL P1+P2) or transcriptional (2: ytfK P1+ P2) lacZ fusions were serially diluted and 5µl of 10−4 and 10−5 dilutions were spotted on X-gal NA plates supplemented or not with 1mM cAMP. (B) Stationary-phase cultures of TB28 Δcrp cells harboring pEG25 or pEG25:6His-crp plasmids and expressing translational (1: ytfK TL P1+P2) or transcriptional (2: ytfK P1+ P2) fusions were serially diluted and spotted on X-gal NA plates supplemented with 50μM IPTG. (C) Stationary-phase cultures of WT (TB28) or Δcrp cells expressing transcriptional (2: ytfK P1+ P2; 3: ytfK+P1 or 4: ytfK+P2) lacZ fusions were serially diluted and spotted on X-gal NA plates supplemented or not with 0.2% of glucose. Results are representative of three independent experiments.
The CRP is the best-known cAMP target in E. coli (Postma et al., 1993). Once activated by cAMP, the cAMP-CRP complex has a central role in the integration of external signals, such as carbon starvation by regulating, in absence of glucose, the expression of several hundred genes involved in the uptake and catabolism of other carbon sources (Zheng et al., 2004; Shimada et al., 2011). We naturally investigated the involvement of CRP in regulation of ytfK. Interestingly and similarly to what was observed in a ΔcyaA strain, we found that transcriptional expression of ytfK is also strongly impaired in the Δcrp mutant (Figure 2A). Importantly, this phenotype is fully trans-complemented by pEG25:6His-crp (Figure 2B). However, addition of 1mM of cAMP does not restore expression of ytfK in the Δcrp mutant (Figure 2A). Therefore, these results show that the cAMP-CRP complex positively regulates transcription of ytfK.
Glucose is transported into the cell and phosphorylated to glucose-6-phosphate, by the phosphotransferase system (PTS), which is composed of several proteins (i.e., EI, HPr, and EIIAGlc; Postma et al., 1993; Bettenbrock et al., 2007). The phosphorylation state of the PTS is lower when glucose is available in the medium, whereas once glucose is consumed, phosphorylated PTS proteins accumulate. The phosphorylated EIIAGlc protein interacts with CyaA and stimulates its activity, thus increasing the intracellular cAMP concentration (Notley-McRobb et al., 1997). In agreement with these data, we observed that the ytfK expression is also highly reduced, when cells are grown in a NA rich medium supplemented with 0.2% of glucose (Figure 2C). In addition, we found that cells expressing transcriptional (ytfK P1) or (ytfK P2) fusions are both submitted to carbon catabolite repression (Figure 2C), suggesting that in the absence of glucose, the cAMP-CRP complex promotes ytfK transcription from both promoters.
Taken together, our results support that the cAMP-CRP complex is required to regulate ytfK transcription in response to glucose availability.
The cAMP-CRP Complex Directly Binds the ytfK P2 Promoter to Regulate Its Expression
To address whether the cAMP-CRP complex directly regulates ytfK expression, we first produced and purified the CRP protein by two consecutive chromatography steps (affinity and size exclusion; Supplementary Figure S2) and electrophoretic mobility shift assay (EMSA) were performed with 5' or 3'-Cy5-labeled DNA fragments selected from the ytfK promoter region (Figure 3A).
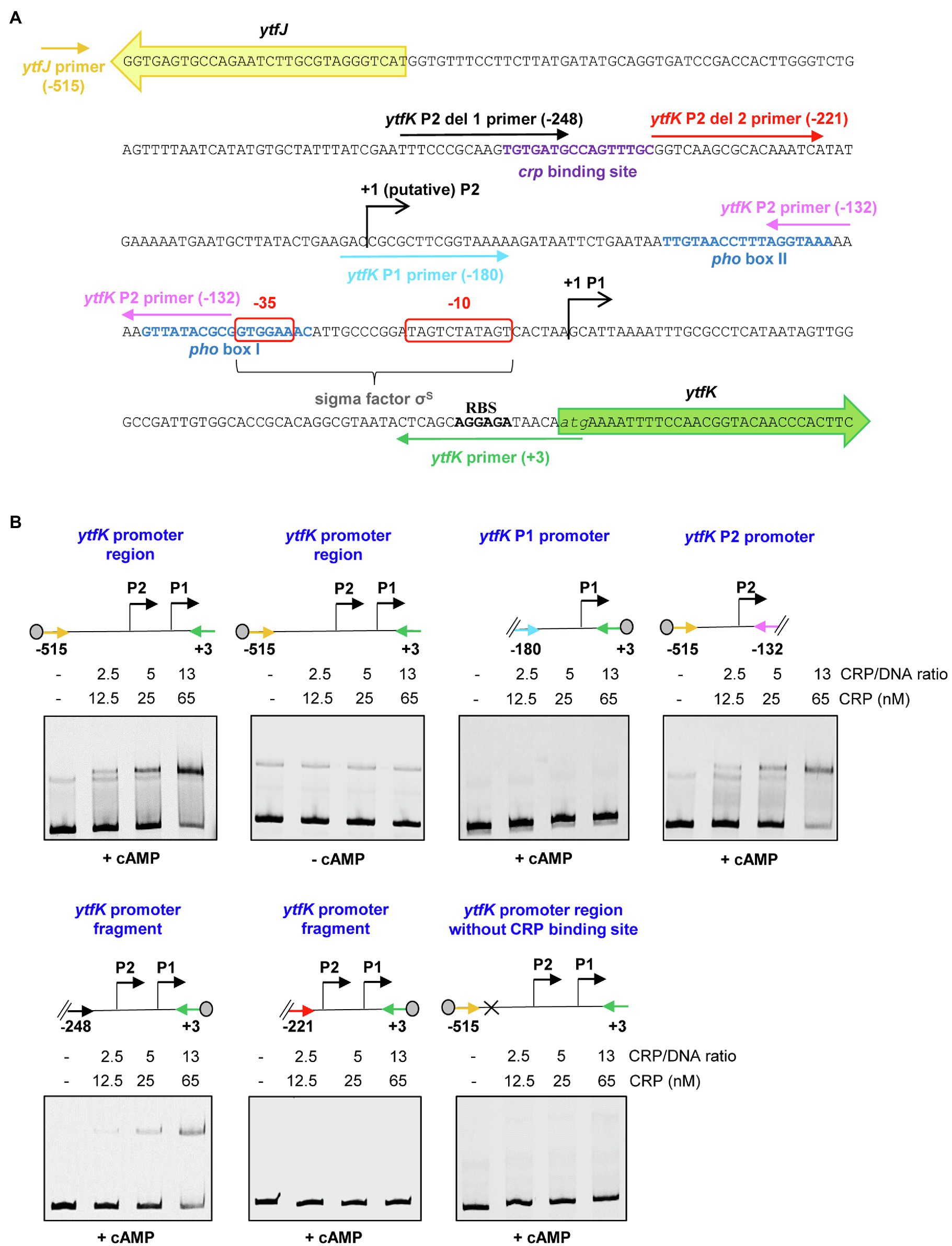
Figure 3. The cAMP-CRP complex forms a tight complex with the ytfK P2 promoter. (A) Intergenic sequence encompassing the ytfK promoter region. The coding sequences of ytfK and ytfJ are shown by green and yellow arrows, respectively. The −35 and−10 sequences of the P1 promoter are similar to the consensus sequences recognized by the sigma factor σS (Lacour and Landini, 2004; Salgado et al., 2013) and are boxed as red. The pho boxes (I and II) and the putative CRP-binding site are written in blue and purple, respectively. The transcriptional start sites of the P1 and putative P2 promoters are indicated by black arrows. The putative ribosome-binding site (RBS) located upstream of the ytfK coding sequence and its start codon is indicated in bold and in italic letters, respectively. Primers used to generate DNA fragments for electrophoretic mobility shift assay are indicated by colored arrows and the relative position to the start codon of ytfK is indicated. (B) EMSA using the CRP protein (0, 12.5, 25 or 65nM) and the indicated DNA fragments (with relative position to the ATG start codon of ytfK). Position of the Cy5 dye is represented by a gray circle. Samples were incubated for 15min at 37°C, separated by native 5% PAGE and bands were visualized using phosphoImaging (Typhoon, GE Healthcare). Results are representative of three independent experiments.
Using DNA covering the entire promoter sequence, a band shift is observed even in the presence of a large amount of poly (dI-dC) competitor (Figures 3A,B). Moreover, this interaction occurs only in the presence of cAMP (Figures 3A,B), as is commonly observed for E. coli CRP homologs (Green et al., 2014). Importantly, no band shift is detected when the intragenic sequence of hofB is used as internal negative control as previously shown (Cameron and Redfield, 2006; Supplementary Figure S3). In addition, the CRP binding to the ytfK promoter sequence appears upon addition of a 2.5-fold molar excess CRP/DNA and the estimated dissociation constant (KD) is around 25nM range suggesting that CRP has a strong affinity for the ytfK promoter region (Figures 3A,B). Moreover, no additional shifted band of lower mobility was observed even when the CRP/DNA mixing ratio was raised up to 100 (Supplementary Figure S4) suggesting existence of a single-binding site. Finally, and consistent with this observation, we show that CRP specifically interacts with the P2 promoter region and that no binding was observed with the P1 promoter region (Figures 3A,B).
To further determine the accurate localization of the CRP-binding site, we made several truncated deletions in the ytfK P2 promoter region (Figure 3A). Using this approach, we observed that cAMP-CRP complex binds a region located between 248 and 221 base pairs upstream the ytfK start codon (Figures 3A,B). We searched in this short region of interest for DNA sequence similar to the known consensus sequence (5'-TGTGAT-N6-TCACA-3') recognized by CRP (Shimada et al., 2011) and we found as a potential CRP-binding site, the 5'-TGTGATGCCAGTTTGC-3' sequence located at 229bp upstream the start codon (Figure 3A). To validate this prediction, the putative CRP-binding site has been deleted and we observed that CRP no longer binds to the ytfK promoter region (Figures 3A,B). These results suggest that CRP induces ytfK transcription by its binding to the 5'-TGTGATGCCAGTTTGC-3' sequence located in the P2 promoter and that induction of transcription from P1 promoter is likely to be indirect (Figures 3A,B).
ytfK Contributes to SpoT-Mediated (p)ppGpp Accumulation During Glucose Starvation
Under carbon starvation, SpoT promotes the (p)ppGpp accumulation (Xiao et al., 1991; Gentry and Cashel, 1996; Murray and Bremer, 1996), but little is known about how this environmental change is sensed by bacteria and linked to SpoT-dependent (p)ppGpp accumulation. As mentioned before, YtfK protein level plays an important role in adjusting intracellular (p)ppGpp level in E. coli (Germain et al., 2019). Given that cAMP-CRP complex plays an important role in carbon sources catabolism and positively regulates the ytfK expression, we naturally investigated the role of YtfK in SpoT-dependent (p)ppGpp accumulation under glucose starvation. For that purpose, we first followed expression of ytfK during glucose exhaustion, a condition known to trigger SpoT-dependent (p)ppGpp accumulation (Xiao et al., 1991; Gentry and Cashel, 1996). As shown in Figure 4A and consistent with the observed positive regulatory role of cAMP-CRP complex, ytfK expression gradually increased 30min after growth arrest due to glucose exhaustion. We then compared (p)ppGpp accumulation in the wild-type strain and the ΔytfK mutant in response to glucose exhaustion. However, and as shown in Supplementary Figure S5, both strains seem to have similar kinetics of (p)ppGpp accumulation in response to glucose exhaustion. Importantly, in the absence of amino acids, both RelA and SpoT contribute to (p)ppGpp accumulation during carbon source or diauxic growth transition (Gentry and Cashel, 1996; Fernández-Coll and Cashel, 2018). Therefore, we further decided to address the role of YtfK in response to glucose starvation in cells devoid of RelA. We observed that compared to the wild-type strain, deletion of relA causes earlier growth arrest and a 30min delayed kinetic of (p)ppGpp accumulation (Figures 4B,C and Supplementary Figures S5, S6). Interestingly, we observed that compared to the ΔrelA mutant, the ΔrelAΔytfK mutant exhibits a strong decrease in ppGpp accumulation in response to glucose exhaustion (Figures 4B,C). Hence, YtfK contributes to the full SpoT-dependent (p)ppGpp accumulation in response to carbon starvation.
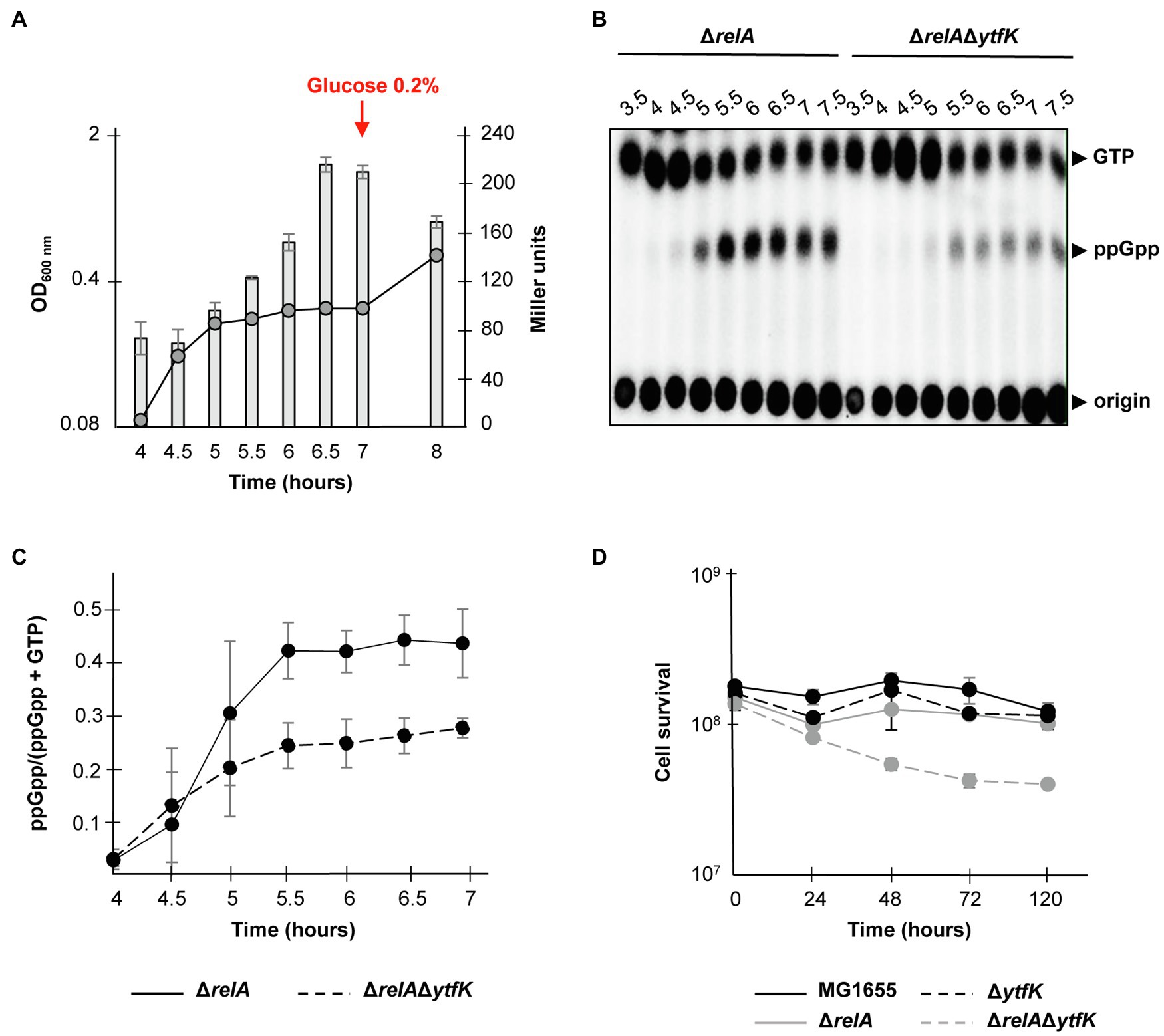
Figure 4. ytfK is required for SpoT-dependent (p)ppGpp accumulation during glucose exhaustion. (A) Growth curve and β-galactosidase activity assay of WT cells in glucose limited minimal medium. WT (TB28) cells harboring the transcriptional fusion (ytfK P1+ P2) were grown in MOPS minimum medium containing 0.025% glucose. The OD600nm values of the cultures (curve) were taken and β-galactosidase activity (bar chart) was measured at indicated times. After 7h of growth, glucose was added at a final concentration of 0.2%. Error bars indicate the standard deviations of averages of three independent experiments. (B) In vivo (p)ppGpp accumulation in response to glucose exhaustion. The ΔrelA and ΔrelAΔytfK mutants were labeled with 32P and grown at 37°C in low-phosphate MOPS medium containing 0.025% of glucose (see Materials and Methods). Samples were taken at indicated times prior to nucleotides extraction and are separated by TLC. Representative autoradiograph of the TLC plates is shown and quantification is provided in (C). Error bars indicate the standard deviations of averages of four independent experiments. (D) YtfK is required for long-term survival during carbon starvation in absence of relA. Cells of MG1655 (WT; black line) and isogenic deletion strains ΔrelA (gray line), ΔytfK (black dotted line) and the ΔrelAΔytfK double mutant (gray dotted line) were grown in M9 minimum medium containing limited concentration of glucose (0.025%). Cell survival (log scale) was determined at indicated time. Error bars indicate the standard deviations of averages of three independent experiments.
SpoT-dependent (p)ppGpp accumulation is also known to be essential for survival during prolonged glucose starvation (Nystöm, 1994). Therefore, we addressed the role of YtfK in cell survival during carbon starvation. While survival of the wild-type, ΔrelA and ΔytfK strains are not significantly affected after 5days of carbon starvation (Figure 4D), the ΔrelAΔytfK double mutant rapidly loses its viability after 24h and retains less than 30% of viability after 5days of prolonged carbon starvation (Figure 4D). Taken together, our results show that regulation of ytfK is a fine-tuned regulated process that allows cells to rapidly adapt and survive during long-term carbon starvation by triggering SpoT-dependent (p)ppGpp accumulation.
ytfK Coordinates Growth Resumption During Glucose-Lactose Diauxic Shift
When E. coli is cultivated in a medium containing various carbon sources, the glucose is preferentially consumed until its exhaustion. (p)ppGpp accumulation rapidly occurs resulting in a transitional growth arrest allowing the establishment of regulatory networks that coordinate the resumption of growth on another carbon source (Harshman and Yamazaki, 1971; Traxler et al., 2006). This phenomenon causes biphasic growth, well known as diauxie (Jacob and Monod, 1961). Importantly a tight control of (p)ppGpp level governs the length of the diauxic lag (Fernández-Coll and Cashel, 2018). We therefore addressed the role of YtfK in diauxic shift. For this purpose, we cultivated the wild-type strain, the simple mutants ΔrelA, ΔytfK and the double-mutant ΔrelAΔytfK in MOPS minimal medium containing a limiting concentration of glucose (0.025%) and an excess of lactose (0.4%). The diauxic lag times were calculated and normalized by generation times on glucose, as previously described (Fernández-Coll and Cashel (2018). We showed that the wild-type strain and the ΔytfK mutant display a similar diauxic lag times of 53min (Figure 5A) and a diauxic lag time/generation time on glucose close to 0.7 (Figure 5B). Moreover and as previously reported (Fernández-Coll and Cashel, 2018), we observed that compared to the wild-type strain, deletion of relA, significantly increases the diauxic lag time (64min; Figure 5A) and the ratio diauxic lag time/generation time on glucose (0.9; Figure 5B). Interestingly, the ΔrelAΔytfK double mutant displays an important extended diauxic lag time (92min; Figure 5A) and the ratio diauxic lag time/generation time on glucose reaches 1.2 (Figure 5B), showing that YtfK also plays an important role in SpoT-dependent (p)ppGpp accumulation during diauxie.
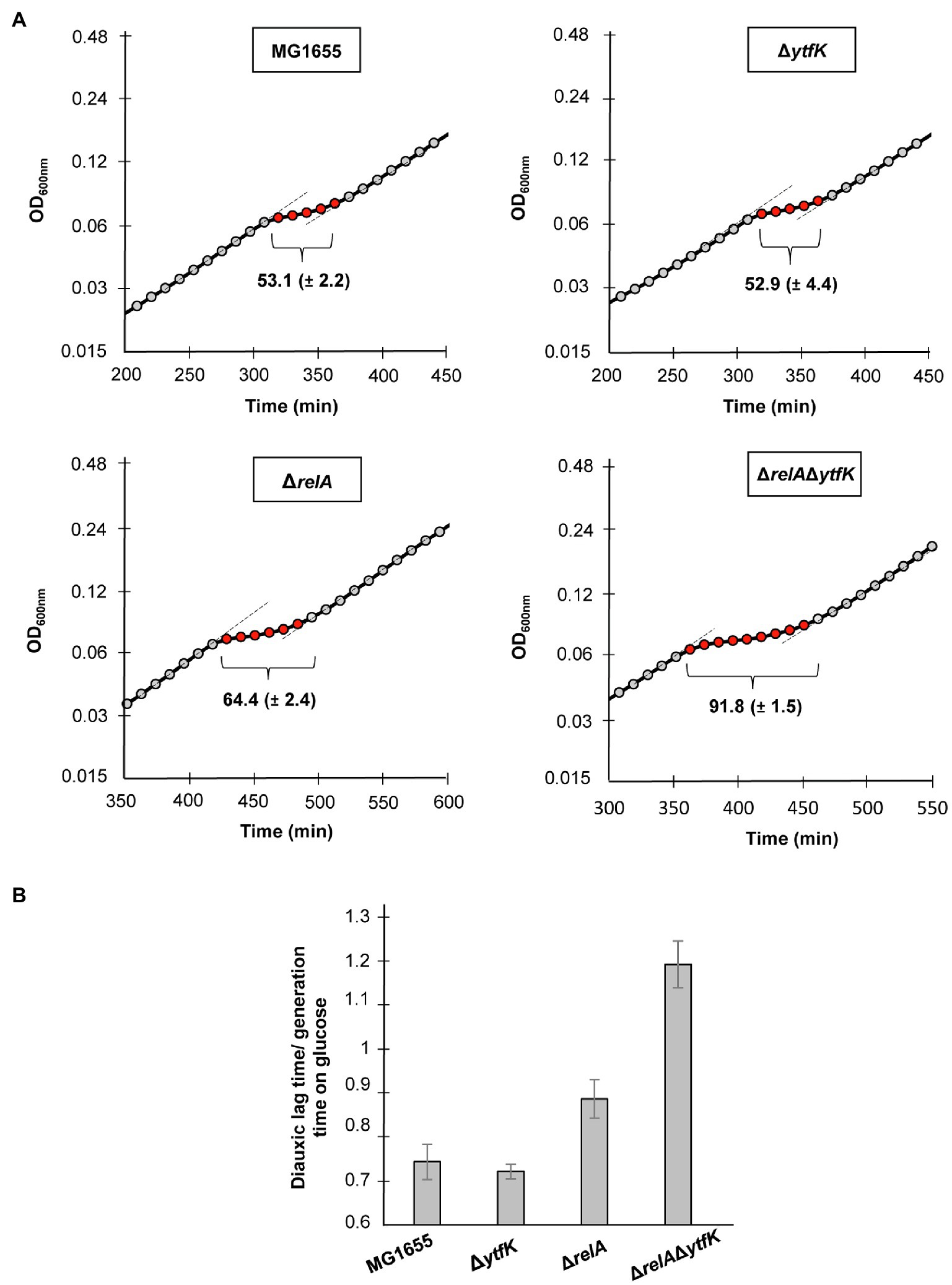
Figure 5. YtfK contributes to diauxic shift adaptation. (A) Cells of MG1655 (WT) and isogenic deletion strains ΔrelA, ΔytfK and the ΔrelAΔytfK were grown in MOPS minimum medium containing 0.025% of glucose and 0.4% of lactose. Growth was monitored every 10min using Tecan microplate reader. Representative growth is provided. The length of diauxic lag times (visualized by brackets) is calculated from three independent experiments and was normalized to the generation time during growth on glucose (B). Error bars indicate the standard deviations of averages of three independent experiments.
Taken together, these results showed that YtfK plays a key role in sensing carbon starvation though the cAMP-CRP complex and transducing the signal to SpoT to orchestrate (p)ppGpp accumulation and thus coordinate cellular metabolism.
Discussion
In E. coli, two homologous enzymes work in concert to control (p)ppGpp level: the (p)ppGpp synthetase RelA and the bifunctional synthetase/hydrolase SpoT. While RelA possesses only (p)ppGpp synthetic activity responding primarily to amino acid starvation (or other stresses that would ultimately cause amino acid starvation), SpoT has both hydrolytic and synthetic activities and functions as a central protein responding to an extreme variety of stress (Xiao et al., 1991; Seyfzadeh et al., 1993; Spira et al., 1995; Vinella et al., 2005). Tight regulation of the synthetic and hydrolytic intracellular activities is crucial for rapidly adjusting (p)ppGpp level and several molecular mechanisms have been reported (Wout et al., 2004; Battesti and Bouveret, 2006; Jiang et al., 2007; Lee et al., 2018; Germain et al., 2019). We previously observed that ectopic production of YtfK is sufficient to trigger SpoT-dependent (p)ppGpp accumulation in absence of external stress and that the YtfK/SpoT ratio controls the intracellular amount of (p)ppGpp (Germain et al., 2019). Therefore, the aim of this work was to further decipher the genetic control involved in the regulation of ytfK expression and its impact on the cell physiology. We first confirmed experimentally that the ytfK promoter region comprises two promoters (Figure 1A), as previously expected (Lacour and Landini, 2004; Salgado et al., 2013). We then systematically searched for trans-regulatory element of ytfK expression using screening assay. This approach leads us to the observation that cAMP level is critical for ytfK expression (Figures 1B, 2A and Supplementary Figure S1). In addition, we showed that ytfK is submitted to carbon catabolite repression and that in the absence of glucose, the cAMP-CRP complex promotes ytfK transcription from both promoters (Figures 2A,C). Further analysis highlights that CRP directly binds the 5'-TGTGATGCCAGTTTGC-3' sequence located in the P2 promoter. Moreover CRP probably regulates ytfK expression via the P1 promoter by an indirect unknown mechanism (Figures 3A,B).
Interestingly, earlier studies have observed an interplay between the (p)ppGpp and the cAMP-CRP regulon in response to carbon starvation, where (p)ppGpp is at the apex of the signaling pathway and maximizes induction of CRP-activated genes (Traxler et al., 2006). Importantly, while regulatory mechanisms orchestrating carbon catabolite repression in E. coli have been intensively characterized, signaling pathway regulating SpoT-dependent (p)ppGpp accumulation in response to carbon availability remains poorly understood. Here, we partially elucidated this mechanism by showing that the YtfK contributes to SpoT-dependent (p)ppGpp accumulation (Figures 4B,C) and cell viability (Figure 4D) in response to glucose starvation. We also observed that a residual (p)ppGpp accumulation persists in the ΔrelAΔytfK double mutant during carbon starvation. Upon fatty acid starvation, the acyl carrier protein (ACP) interacts with SpoT to promote the accumulation of (p)ppGpp (Battesti and Bouveret, 2006; Figure 6). Importantly, ACP and fatty acid metabolism could also be a relay for responding to carbon source starvation and be responsible for the residual SpoT activity observed in the ΔrelAΔytfK cells. Indeed, carbon exhaustion would lead to fatty acid starvation through shrinkage of the acetyl-CoA pool produced during glycolysis. It is interesting to note that YtfK and ACP interact with two different regions of SpoT. YtfK binds the catalytic domains in the N-terminal region (Germain et al., 2019), while ACP binds the TGS domain in C-terminal regulatory region (Battesti and Bouveret, 2006; Figure 6). It is therefore tempting to speculate that binding of the both proteins may work in concert to maximize (p)ppGpp level in response to glucose starvation. It seems to be a striking opposite parallel between the mechanisms of SpoT regulation upon fatty acid starvation via ACP and the SpoT regulation by the anti-σ70 Rsd during carbon downshift. Indeed, Rsd binding to the TGS domain of SpoT promotes hydrolase activity upon carbon downshift and plays a physiological role in controlling cell growth recovery during diauxic shift (Figure 6). Our observation that YtfK contributes to the SpoT-dependent (p)ppGpp accumulation during diauxie and seems also to be important for growth resumption during glucose-lactose diauxic shift (Figure 5) is consistent with the notion that a tightly coordinated balance between synthesis versus hydrolysis activity rather than the absolute (p)ppGpp level is important for faster adaption during diauxic shift.
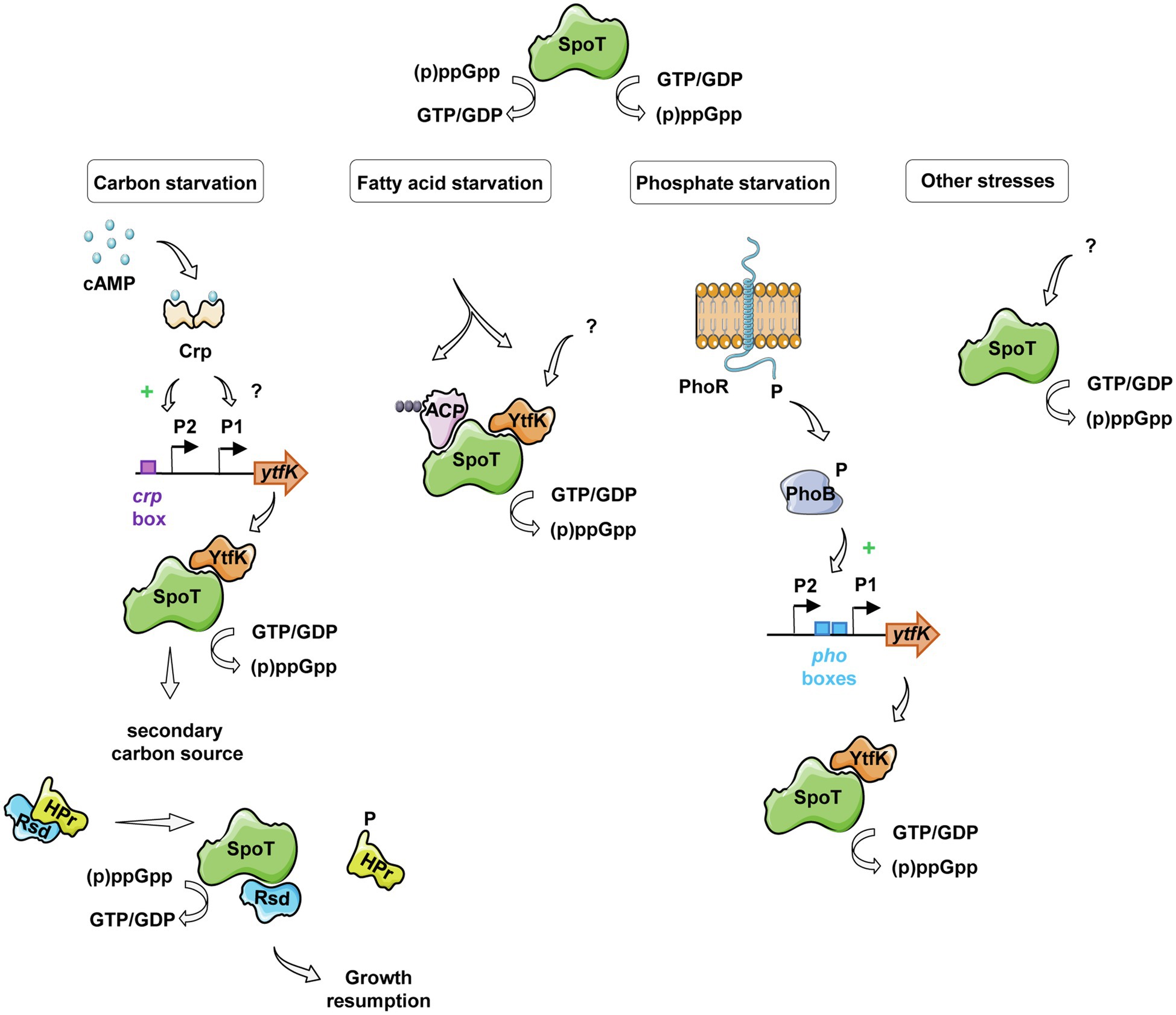
Figure 6. Schematic view of SpoT integration signals and its regulation in response to various starvations in E. coli. Under glucose starvation, an increase of cAMP concentration activates the CRP protein, which induces ytfK transcription by direct binding to the ytfK P2 promoter and by an unknown and probably indirect mechanism for the ytfK P1 promoter. YtfK thus interacts with the catalytic domains of SpoT (Germain et al., 2019), promoting the accumulation of (p)ppGpp. The availability of a secondary carbon source releases the anti-σ70 Rsd protein from HPr, which can interact with the C-terminal regulatory domain of SpoT, rebalancing its (p)ppGpp synthesis activity toward hydrolysis allowing growth resumption (Lee et al., 2018). In response to fatty acid starvation, YtfK also promotes SpoT-dependent accumulation of (p)ppGpp (Germain et al., 2019) and on the other hand, the nature of the fatty acid intermediates bound to ACP differs and induces conformational changes in the protein leading to SpoT-dependent (p)ppGpp accumulation (Battesti and Bouveret, 2006). During phosphate starvation, the PhoR-PhoB two-component system mediates signal transduction that results to induce ytfK transcription which could orchestrate SpoT-dependent (p)ppGpp accumulation (Spira et al., 1995; Spira and Yagil, 1998; Baek and Lee, 2006; Yoshida et al., 2011; Iwadate and Kato, 2017).
In addition to the regulation presented in this study, ytfK transcription is subjected to complex regulatory network. Indeed, ytfK also contains, upstream of its coding sequence, two pho boxes similar to the consensus sequence recognized by the response regulator PhoB and transcriptome analyses showed that ytfK is induced 15-fold by PhoB during phosphate starvation (Baek and Lee, 2006; Yoshida et al., 2011). Moreover, the PhoR-PhoB two-component system has been shown to play key role in triggering SpoT-dependent (p)ppGpp accumulation during phosphate starvation (Spira et al., 1995; Spira and Yagil, 1998: Figure 6). Importantly, deletion of ytfK also affects cell viability and (p)ppGpp accumulation under phosphate starvation (Iwadate and Kato, 2017; Germain et al., 2019). Interestingly, ytfK is also induced and involved in H2O2 tolerance (Iwadate and Kato, 2017). In addition, we previously described that ytfK is also induced during fatty acid starvation causing accumulation of (p)ppGpp and cell survival (Figure 6; Germain et al., 2019). However, the exact nature of this regulation remains to be determine. Finally, ytfK expression is also under the control of the ferric uptake regulator Fur (Zhang et al., 2005), involved in response to iron limitation (Stojiljkovic et al., 1994; Hantke, 2001, 2002; McHugh et al., 2003), another signal known to trigger SpoT-dependent (p)ppGpp accumulation in vivo (Vinella et al., 2005). Importantly, the amount of SpoT protein has been estimated around hundreds of molecules per cell (Pedersen and Kjeldgaard, 1977) and to our knowledge, no transcriptional regulation of spoT has been directly linked to the environmental control of SpoT activities. Therefore, the regulation of ytfK appears as an emerging key node in the SpoT-mediated activation of stringent response in E. coli (Figure 6).
Finally, aside from identification of the role of cAMP in the regulation of ytfK, our genetic screening assay enables the identification of many proteins, probably acting indirectly, involved in a variety of key functions, such as cell wall biogenesis, cell motility, amino acid metabolism, carbohydrates and lipids transport, energy maintenance and signal transduction (Table 1).
Therefore, any perturbation in these metabolic pathways may influence YtfK protein level and therefore could impact SpoT activities and modulate intracellular (p)ppGpp level. For instance, it is well known that the overexpression of membrane proteins disrupts membrane integrity by limiting the capacity of proteins translocation into the membrane and by increasing the aggregation of cytoplasmic proteins due to the titration of chaperones, which leads to broad perturbations of the proteome and is responsible for inefficient ATP synthesis (Wagner et al., 2007). Overexpression of membrane proteins highly induces ytfK expression (Table 1 and Supplementary Figure S1), suggesting that YtfK protein may play a role in response to cell envelope disruption which could impact SpoT activities and modulate the intracellular level (p)ppGpp.
Therefore, beyond a better understanding of the genetic regulation, our systematic analysis of the trans-regulatory elements can indirectly allow us to better understand the metabolic pathways behind the regulation of SpoT activities and can lead to the identification of new stressful conditions in which SpoT and its partners intervene (Figure 6).
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Author Contributions
EM, LM, and EG designed the study and discussed the results. LM performed the experiments. EM and LM wrote the manuscript. EM acquired the funding. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the European Research Council starting grant (ERC StG) under the European Union’s Horizon 2020 and innovation program grant agreement no. 714934 “Stringency” to EM.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank Mieli Morgane for her technical help in cloning transcriptional fusions in pGH254. We thank Servier Medical Art by Servier (smart.servier.com) for helping us to illustrate our Figures 1B, 6.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.775164/full#supplementary-material
References
Amato, S. M., and Brynildsen, M. P. (2015). Persister heterogeneity arising from a single metabolic stress. Curr. Biol. 25, 2090–2098. doi: 10.1016/j.cub.2015.06.034
Amato, S. M., Orman, M. A., and Brynildsen, M. P. (2013). Metabolic control of Persister formation in Escherichia coli. Mol. Cell 50, 475–487. doi: 10.1016/j.molcel.2013.04.002
Arenz, S., Abdelshahid, M., Sohmen, D., Payoe, R., Starosta, A. L., Berninghausen, O., et al. (2016). The stringent factor RelA adopts an open conformation on the ribosome to stimulate ppGpp synthesis. Nucleic Acids Res. 44, 6471–6481. doi: 10.1093/nar/gkw470
Atkinson, G. C., Tenson, T., and Hauryliuk, V. (2011). The RelA/SpoT homolog (RSH) superfamily: distribution and functional evolution of ppGpp Synthetases and hydrolases across the tree of life. PLoS One 6:e23479. doi: 10.1371/journal.pone.0023479
Baba, T., Ara, T., Hasegawa, M., Takai, Y., Okumura, Y., Baba, M., et al. (2006). Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2:ms4100050. doi: 10.1038/msb4100050
Baek, J. H., and Lee, S. Y. (2006). Novel gene members in the pho regulon of Escherichia coli. FEMS Microbiol. Lett. 264, 104–109. doi: 10.1111/j.1574-6968.2006.00440.x
Battesti, A., and Bouveret, E. (2006). Acyl carrier protein/SpoT interaction, the switch linking SpoT-dependent stress response to fatty acid metabolism. Mol. Microbiol. 62, 1048–1063. doi: 10.1111/j.1365-2958.2006.05442.x
Bettenbrock, K., Sauter, T., Jahreis, K., Kremling, A., Lengeler, J. W., and Gilles, E.-D. (2007). Correlation between growth rates, EIIACrr phosphorylation, and intracellular cyclic AMP levels in Escherichia coli K-12. J. Bacteriol. 189, 6891–6900. doi: 10.1128/JB.00819-07
Blank, K., Hensel, M., and Gerlach, R. G. (2011). Rapid and highly efficient method for scarless mutagenesis within the salmonella enterica chromosome. PLoS One 6:e15763. doi: 10.1371/journal.pone.0015763
Cameron, A. D. S., and Redfield, R. J. (2006). Non-canonical CRP sites control competence regulons in Escherichia coli and many other gamma-proteobacteria. Nucleic Acids Res. 34, 6001–6014. doi: 10.1093/nar/gkl734
Cashel, M. (1969). The control of ribonucleic acid synthesis in Escherichia coli. IV. Relevance of unusual phosphorylated compounds from amino acid-starved stringent strains. J. Biol. Chem. 244, 3133–3141.
Cashel, M., and Gallant, J. (1969). Two compounds implicated in the function of the RC gene of Escherichia coli. Nature 221, 838–841. doi: 10.1038/221838a0
Cherepanov, P. P., and Wackernagel, W. (1995). Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene 158, 9–14. doi: 10.1016/0378-1119(95)00193-A
Clark, D. J., and Maaløe, O. (1967). DNA replication and the division cycle in Escherichia coli. J. Mol. Biol. 23, 99–112. doi: 10.1016/S0022-2836(67)80070-6
Datsenko, K. A., and Wanner, B. L. (2000). One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97, 6640–6645. doi: 10.1073/pnas.120163297
Fernández-Coll, L., and Cashel, M. (2018). Contributions of SpoT hydrolase, SpoT Synthetase, and RelA Synthetase to carbon source diauxic growth transitions in Escherichia coli. Front. Microbiol. 9:1802. doi: 10.3389/fmicb.2018.01802
Gentry, D. R., and Cashel, M. (1996). Mutational analysis of the Escherichia coli spoT gene identifies distinct but overlapping regions involved in ppGpp synthesis and degradation. Mol. Microbiol. 19, 1373–1384. doi: 10.1111/j.1365-2958.1996.tb02480.x
Germain, E., Guiraud, P., Byrne, D., Douzi, B., Djendli, M., and Maisonneuve, E. (2019). YtfK activates the stringent response by triggering the alarmone synthetase SpoT in Escherichia coli. Nat. Commun. 10:5763. doi: 10.1038/s41467-019-13764-4
Green, J., Stapleton, M. R., Smith, L. J., Artymiuk, P. J., Kahramanoglou, C., Hunt, D. M., et al. (2014). Cyclic-AMP and bacterial cyclic-AMP receptor proteins revisited: adaptation for different ecological niches. Curr. Opin. Microbiol. 18, 1–7. doi: 10.1016/j.mib.2014.01.003
Hantke, K. (2001). Iron and metal regulation in bacteria. Curr. Opin. Microbiol. 4, 172–177. doi: 10.1016/S1369-5274(00)00184-3
Hantke, K. (2002). Members of the fur protein family regulate iron and zinc transport in E. coli and characteristics of the fur-regulated fhuF protein. J. Mol. Microbiol. Biotechnol. 4, 217–222.
Harshman, R. B., and Yamazaki, H. (1971). Formation of ppGpp in a relaxed and stringent strain of Escherichia coli during diauxie lag. Biochemistry 10, 3980–3982. doi: 10.1021/bi00797a027
Haseltine, W. A., and Block, R. (1973). Synthesis of guanosine tetra- and pentaphosphate requires the presence of a codon-specific, uncharged transfer ribonucleic acid in the acceptor site of ribosomes. Proc. Natl. Acad. Sci. U. S. A. 70, 1564–1568. doi: 10.1073/pnas.70.5.1564
Hauryliuk, V., Atkinson, G. C., Murakami, K. S., Tenson, T., and Gerdes, K. (2015). Recent functional insights into the role of (p)ppGpp in bacterial physiology. Nat. Rev. Microbiol. 13, 298–309. doi: 10.1038/nrmicro3448
Helaine, S., Cheverton, A. M., Watson, K. G., Faure, L. M., Matthews, S. A., and Holden, D. W. (2014). Internalization of salmonella by macrophages induces formation of nonreplicating Persisters. Science 343, 204–208. doi: 10.1126/science.1244705
Hernandez, V. J., and Bremer, H. (1991). Escherichia coli ppGpp synthetase II activity requires spoT. J. Biol. Chem. 266, 5991–5999.
Hogg, T., Mechold, U., Malke, H., Cashel, M., and Hilgenfeld, R. (2004). Conformational antagonism between opposing active sites in a Bifunctional RelA/SpoT homolog modulates (p)ppGpp metabolism during the stringent response. Cell 117, 57–68. doi: 10.1016/S0092-8674(04)00260-0
Imamura, R., Yamanaka, K., Ogura, T., Hiraga, S., Fujita, N., Ishihama, A., et al. (1996). Identification of the cpdA gene encoding cyclic 3',5'-adenosine monophosphate phosphodiesterase in Escherichia coli. J. Biol. Chem. 271, 25423–25429. doi: 10.1074/jbc.271.41.25423
Irving, S. E., Choudhury, N. R., and Corrigan, R. M. (2021). The stringent response and physiological roles of (pp)pGpp in bacteria. Nat. Rev. Microbiol. 19, 256–271. doi: 10.1038/s41579-020-00470-y
Iwadate, Y., and Kato, J.-I. (2017). Involvement of the ytfK gene from the PhoB regulon in stationary-phase H2O2 stress tolerance in Escherichia coli. Microbiology 163, 1912–1923. doi: 10.1099/mic.0.000534
Jacob, F., and Monod, J. (1961). Genetic regulatory mechanisms in the synthesis of proteins. J. Mol. Biol. 3, 318–356. doi: 10.1016/S0022-2836(61)80072-7
Jiang, M., Sullivan, S. M., Wout, P. K., and Maddock, J. R. (2007). G-protein control of the ribosome-associated stress response protein SpoT. J. Bacteriol. 189, 6140–6147. doi: 10.1128/JB.00315-07
Kitagawa, M., Ara, T., Arifuzzaman, M., Ioka-Nakamichi, T., Inamoto, E., Toyonaga, H., et al. (2005). Complete set of ORF clones of Escherichia coli ASKA library (a complete set of E. coli K-12 ORF archive): unique resources for biological research. DNA Res. 12, 291–299. doi: 10.1093/dnares/dsi012
Lacour, S., and Landini, P. (2004). SigmaS-dependent gene expression at the onset of stationary phase in Escherichia coli: function of sigmaS-dependent genes and identification of their promoter sequences. J. Bacteriol. 186, 7186–7195. doi: 10.1128/JB.186.21.7186-7195.2004
Lee, J.-W., Park, Y.-H., and Seok, Y.-J. (2018). Rsd balances (p)ppGpp level by stimulating the hydrolase activity of SpoT during carbon source downshift in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 115, E6845–E6854. doi: 10.1073/pnas.1722514115
Liu, K., Bittner, A. N., and Wang, J. D. (2015). Diversity in (p)ppGpp metabolism and effectors. Curr. Opin. Microbiol. 24, 72–79. doi: 10.1016/j.mib.2015.01.012
McHugh, J. P., Rodríguez-Quiñones, F., Abdul-Tehrani, H., Svistunenko, D. A., Poole, R. K., Cooper, C. E., et al. (2003). Global iron-dependent gene regulation in Escherichia coli. J. Biol. Chem. 278, 29478–29486. doi: 10.1074/jbc.M303381200
Miller, J. H. (1992). A short course in bacterial genetics: a laboratory manual and handbook for Escherichia coli and related bacteria/Jeffrey H. Miller.e. Cold Spring Harb. Lab. Press.
Murray, K. D., and Bremer, H. (1996). Control of spoT-dependent ppGpp synthesis and degradation in Escherichia coli. J. Mol. Biol. 259, 41–57. doi: 10.1006/jmbi.1996.0300
Neidhardt, F. C., Bloch, P. L., and Smith, D. F. (1974). Culture medium for enterobacteria. J. Bacteriol. 119, 736–747. doi: 10.1128/jb.119.3.736-747.1974
Nguyen, D., Joshi-Datar, A., Lepine, F., Bauerle, E., Olakanmi, O., Beer, K., et al. (2011). Active starvation responses mediate antibiotic tolerance in biofilms and nutrient-limited bacteria. Science 334, 982–986. doi: 10.1126/science.1211037
Notley-McRobb, L., Death, A., and Ferenci, T. (1997). The relationship between external glucose concentration and cAMP levels inside Escherichia coli: implications for models of phosphotransferase-mediated regulation of adenylate cyclase. Microbiology 143, 1909–1918. doi: 10.1099/00221287-143-6-1909
Nystöm, T. (1994). Role of guanosine tetraphosphate in gene expression and the survival of glucose or seryl-tRNA starved cells of Escherichia coli K12. Mol. Gen. Genet. 245, 355–362. doi: 10.1007/BF00290116
Pausch, P., Abdelshahid, M., Steinchen, W., Schäfer, H., Gratani, F. L., Freibert, S.-A., et al. (2020). Structural basis for regulation of the opposing (p)ppGpp Synthetase and hydrolase within the stringent response orchestrator Rel. Cell Rep. 32:108157. doi: 10.1016/j.celrep.2020.108157
Pedersen, F. S., and Kjeldgaard, N. O. (1977). Analysis of the relA gene product of Escherichia coli. Eur. J. Biochem. 76, 91–97. doi: 10.1111/j.1432-1033.1977.tb11573.x
Postma, P. W., Lengeler, J. W., and Jacobson, G. R. (1993). Phosphoenolpyruvate:carbohydrate phosphotransferase systems of bacteria. Microbiol. Rev. 57, 543–594.
Potrykus, K., and Cashel, M. (2008). (p)ppGpp: still magical? Annu. Rev. Microbiol. 62, 35–51. doi: 10.1146/annurev.micro.62.081307.162903
Salgado, H., Peralta-Gil, M., Gama-Castro, S., Santos-Zavaleta, A., Muñiz-Rascado, L., García-Sotelo, J. S., et al. (2013). RegulonDB v8.0: omics data sets, evolutionary conservation, regulatory phrases, cross-validated gold standards and more. Nucleic Acids Res. 41, D203–D213. doi: 10.1093/nar/gks1201
Seyfzadeh, M., Keener, J., and Nomura, M. (1993). spoT-dependent accumulation of guanosine tetraphosphate in response to fatty acid starvation in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 90, 11004–11008. doi: 10.1073/pnas.90.23.11004
Shimada, T., Fujita, N., Yamamoto, K., and Ishihama, A. (2011). Novel roles of cAMP receptor protein (CRP) in regulation of transport and metabolism of carbon sources. PLoS One 6:e20081. doi: 10.1371/journal.pone.0020081
Spira, B., Silberstein, N., and Yagil, E. (1995). Guanosine 3',5'-bispyrophosphate (ppGpp) synthesis in cells of Escherichia coli starved for pi. J. Bacteriol. 177, 4053–4058. doi: 10.1128/jb.177.14.4053-4058.1995
Spira, B., and Yagil, E. (1998). The relation between ppGpp and the PHO regulon in Escherichia coli. Mol. Gen. Genet. 257, 469–677. doi: 10.1007/s004380050671
Steinchen, W., and Bange, G. (2016). The magic dance of the alarmones (p)ppGpp. Mol. Microbiol. 101, 531–544. doi: 10.1111/mmi.13412
Stojiljkovic, I., Bäumler, A. J., and Hantke, K. (1994). Fur regulon in gram-negative bacteria. J. Mol. Biol. 236, 531–545. doi: 10.1006/jmbi.1994.1163
Tamman, H., Van Nerom, K., Takada, H., Vandenberk, N., Scholl, D., Polikanov, Y., et al. (2020). A nucleotide-switch mechanism mediates opposing catalytic activities of Rel enzymes. Nat. Chem. Biol. 16, 834–840. doi: 10.1038/s41589-020-0520-2
Thomason, L. C., Costantino, N., and Court, D. L. (2007). E. coli genome manipulation by P1 transduction. Curr. Protoc. Mol. Biol. 79, 1.17.1–1.17.8. doi: 10.1002/0471142727.mb0117s79
Traxler, M. F., Chang, D.-E., and Conway, T. (2006). Guanosine 3',5'-bispyrophosphate coordinates global gene expression during glucose-lactose diauxie in Escherichia coli. Proc. Natl. Acad. Sci. 103, 2374–2379. doi: 10.1073/pnas.0510995103
Vinella, D., Albrecht, C., Cashel, M., and D’Ari, R. (2005). Iron limitation induces SpoT-dependent accumulation of ppGpp in Escherichia coli. Mol. Microbiol. 56, 958–970. doi: 10.1111/j.1365-2958.2005.04601.x
Wagner, S., Baars, L., Ytterberg, A. J., Klussmeier, A., Wagner, C. S., Nord, O., et al. (2007). Consequences of membrane protein overexpression in Escherichia coli. Mol. Cell. Proteomics 6, 1527–1550. doi: 10.1074/mcp.M600431-MCP200
Winther, K. S., Roghanian, M., and Gerdes, K. (2018). Activation of the stringent response by loading of RelA-tRNA complexes at the ribosomal A-site. Mol. Cell 70, 95.e4–105.e4. doi: 10.1016/j.molcel.2018.02.033
Wout, P., Pu, K., Sullivan, S. M., Reese, V., Zhou, S., Lin, B., et al. (2004). The Escherichia coli GTPase CgtA E Cofractionates with the 50S ribosomal subunit and interacts with SpoT, a ppGpp Synthetase/hydrolase. J. Bacteriol. 186, 5249–5257. doi: 10.1128/JB.186.16.5249-5257.2004
Xiao, H., Kalman, M., Ikehara, K., Zemel, S., Glaser, G., and Cashel, M. (1991). Residual guanosine 3',5'-bispyrophosphate synthetic activity of relA null mutants can be eliminated by spoT null mutations. J. Biol. Chem. 266, 5980–5990.
Yoshida, Y., Sugiyama, S., Oyamada, T., Yokoyama, K., Kim, S.-K., and Makino, K. (2011). Identification of PhoB binding sites of the yibD and ytfK promoter regions in Escherichia coli. J. Microbiol. 49, 285–289. doi: 10.1007/s12275-011-0360-6
Zhang, Z., Gosset, G., Barabote, R., Gonzalez, C. S., Cuevas, W. A., and Saier, M. H. (2005). Functional interactions between the carbon and iron utilization regulators, Crp and fur, in Escherichia coli. J. Bacteriol. 187, 980–990. doi: 10.1128/JB.187.3.980-990.2005
Keywords: (p)ppGpp, stringent response, ytfK, cAMP, CRP, glucose starvation, E. coli
Citation: Meyer L, Germain E and Maisonneuve E (2021) Regulation of ytfK by cAMP-CRP Contributes to SpoT-Dependent Accumulation of (p)ppGpp in Response to Carbon Starvation YtfK Responds to Glucose Exhaustion. Front. Microbiol. 12:775164. doi: 10.3389/fmicb.2021.775164
Edited by:
Gert Bange, University of Marburg, GermanyReviewed by:
Shinji Masuda, Tokyo Institute of Technology, JapanDitlev Egeskov Brodersen, Aarhus University, Denmark
Copyright © 2021 Meyer, Germain and Maisonneuve. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Etienne Maisonneuve, emaisonneuve@imm.cnrs.fr
 Laura Meyer
Laura Meyer Elsa Germain
Elsa Germain Etienne Maisonneuve
Etienne Maisonneuve