- 1Institute of Bioengineering, Research Center of Biotechnology of the Russian Academy of Sciences, Moscow, Russia
- 2Department of Biochemistry and Cell Physiology, Voronezh State University, Voronezh, Russia
- 3New England Biolabs, Ipswich, MA, United States
- 4Department of Physical and Colloid Chemistry, Gubkin Russian State University of Oil and Gas, Moscow, Russia
- 5Laboratory of Biochemistry and Molecular Biology, Tomsk State University, Tomsk, Russia
Two strains of filamentous, colorless sulfur bacteria were isolated from bacterial fouling in the outflow of hydrogen sulfide-containing waters from a coal mine (Thiothrix sp. Ku-5) and on the seashore of the White Sea (Thiothrix sp. AS). Metagenome-assembled genome (MAG) A52 was obtained from a sulfidic spring in the Volgograd region, Russia. Phylogenetic analysis based on the 16S rRNA gene sequences showed that all genomes represented the genus Thiothrix. Based on their average nucleotide identity and digital DNA-DNA hybridization data these new isolates and the MAG represent three species within the genus Thiothrix with the proposed names Thiothrix subterranea sp. nov. Ku-5T, Thiothrix litoralis sp. nov. AST, and “Candidatus Thiothrix anitrata” sp. nov. A52. The complete genome sequences of Thiothrix fructosivorans QT and Thiothrix unzii A1T were determined. Complete genomes of seven Thiothrix isolates, as well as two MAGs, were used for pangenome analysis. The Thiothrix core genome consisted of 1,355 genes, including ones for the glycolysis, the tricarboxylic acid cycle, the aerobic respiratory chain, and the Calvin cycle of carbon fixation. Genes for dissimilatory oxidation of reduced sulfur compounds, namely the branched SOX system (SoxAXBYZ), direct (soeABC) and indirect (aprAB, sat) pathways of sulfite oxidation, sulfur oxidation complex Dsr (dsrABEFHCEMKLJONR), sulfide oxidation systems SQR (sqrA, sqrF), and FCSD (fccAB) were found in the core genome. Genomes differ in the set of genes for dissimilatory reduction of nitrogen compounds, nitrogen fixation, and the presence of various types of RuBisCO.
Introduction
At the end of the 19th century, Sergei Winogradsky described filamentous, colorless sulfur bacteria, often present as foulings in running waters rich in hydrogen sulfide, and gave this group of microorganisms the generic name Thiothrix. They can accumulate elemental sulfur in the form of intracellular inclusion bodies and are characterized by the ability to form rosettes and the presence of a pronounced mucous sheath in the filaments. They were formerly known as members of the Thiotrichaceae family, and are now classified into three different families Thiolineaceae, Thiofilaceae, and Thiotrichaceae (Boden and Scott, 2018). However, only a few cultivated representatives are known in this group of microorganisms and the taxonomy of filamentous, colorless sulfur bacteria, and in particular, of the genus Thiothrix is poorly developed (Boden and Scott, 2018).
Members of the genus Thiothrix are capable of both organoheterotrophic and lithoautotrophic growth in the presence of reduced sulfur compounds as well as of mixotrophic growth under appropriate conditions. Due to the flexible sulfur, nitrogen and carbon metabolism and the ability for aerobic and anaerobic growth, these bacteria can occupy various ecological niches. As a rule, they dominate microbial populations in sulfide-rich waters, forming powerful bacterial fouling (Larkin and Shinabarger, 1983; Chernousova et al., 2010; Rossmassler et al., 2016).
The known range of habitats of the genus Thiothrix is now actively expanding. Metagenomic analysis of groundwater and cave lakes has shown the presence of representatives of the genus Thiothrix (Macalady et al., 2008; Rossmassler et al., 2016; Panova et al., 2020). Also members of this genus were found in hydrothermal marine ecosystems, wastewater treatment bioreactors (Mardanov et al., 2020), and as ectosymbionts of invertebrates (Konkol et al., 2010; Bauermeister et al., 2012; Li et al., 2018). However, no pure cultures were obtained from these biotopes.
There are no reliable phylogenetic markers for the delineation of species within the genus Thiothrix since the 16S rRNA gene sequences are highly similar. For example, for Thiothrix lacustris BLT, Thiothrix fructosivorans QT, and Thiothrix caldifontis G1T these sequences are 98.6–98.9% identical. Therefore, the use of genome-based data like average nucleotide identity (ANI), average amino acid identity (AAI), and DNA-DNA hybridization distance (dDDH) is important for distinguishing between species and would be beneficial for building taxonomy of the genus Thiothrix.
After the last taxonomic revision (Boden and Scott, 2018) the genus Thiothrix includes five species: Thiothrix nivea (Larkin and Shinabarger, 1983), T. fructosivorans, Thiothrix unzii (Howarth et al., 1999), T. caldifontis, and T. lacustris (Chernousova et al., 2010). In addition, two Thiothrix MAGs were obtained from laboratory-scale enhanced biological phosphorus removal bioreactors (Mardanov et al., 2020). These MAGs represent two new species referred to as “Candidatus Thiothrix moscowensis” RT and “Candidatus Thiothrix singaporensis” SSD2 (Mardanov et al., 2020). Therefore, obtaining new isolates and MAGs, especially from atypical habitats, could expand our knowledge of the diversity of Thiothrix and would allow to identify new species of this genus.
In this work we describe three new species of the genus Thiothrix, Thiothrix subterranea sp. nov. Ku-5T, Thiothrix litoralis sp. nov. AST, and a new MAG representing a new candidate species “Candidatus Thiothrix anitrata” sp. nov. A52. We also determined complete genomic sequences for T. unzii A1T (ATCC 49747T) and T. fructosivorans QT (ATCC 49748T), that were isolated earlier (Howarth et al., 1999), but have not been sequenced until now. In addition, we performed comparative genome analysis of nine Thiothrix species to obtain an in-depth comparison of the metabolic potential of representatives of this genus.
Materials and Methods
Culture Media
Bacteria were cultivated in a medium containing (gL–1): (NH4)2SO4 – 0.5, NaNO3 – 0.3, CaCl2 – 0.03, KH2PO4 – 0.01, K2HPO4 – 0.022, Na2HPO4.7H2O – 0.035, MgSO4.7H2O – 0.05. Prior to inoculation, the following components were added to one liter of the medium as sterile solutions: trace elements and vitamins (Pfennig and Lippert, 1966; Kuznetsov and Dubinina, 1989), 1 ml; NaHCO3, 0.5 g; Na2S2O3.5H2O, 1.0 g; sodium lactate, 0.25 g and sodium acetate, 0.25 g. The pH of the medium was adjusted to 7.5. Cultures were incubated at 27°C.
Isolation of Pure Cultures and Growth Conditions
Physiological tests were carried out using a medium of the above composition using alternative carbon sources. Three passages of culture were carried out with each of the substrates to test bacterial growth. To study the ability to fix molecular nitrogen, bacteria were cultivated in a medium of the above-described composition, excluding nitrogen sources. The ability of the strains to fix nitrogen was confirmed by the acetylene reduction assay (Stewart et al., 1968).
Isolation of pure cultures of representatives of the genus Thiothrix was performed from the selected bacterial fouling, where bundles of filamentous sulfur bacteria were collected, washed with a large amount of sterile water, and transferred to a Potter manual homogenizer to prepare suspensions of colorless sulfur bacteria threads, in order to obtain Thiothrix gonidia. A number of tenfold dilutions were prepared by the method of limiting dilution from the starting material. To obtain individual colonies, serial dilutions were plated on a solid nutrient medium of the composition described above, which additionally contained (gL–1): agar Difco – 7.0; Na2S.9H2O – 0.3.
Fatty acid analysis was performed as described elsewhere (Slobodkina et al., 2020). Briefly, about 10 mg of freeze-dried biomass collected at the late exponential growth phase were treated with anhydrous HCl/MeOH, extracted with hexane, and analyzed with a Trace GC Ultra DSQ II GC-MS system (Thermo Fisher Scientific, HP-5MS column, EI70 eV).
Genome Sequencing of T. unzii A1T and Thiothrix sp. AS
Genomic DNA was isolated from Thiothrix sp. AS and T. unzii A1T (strain was obtained from ATCC collection ATCC 49747T) using a DNeasy PowerSoil DNA isolation kit (Mo Bio Laboratories, Carlsbad, CA, United States). Genomic DNA was sequenced using Illumina and Oxford Nanopore platforms. For Illumina sequencing, the shotgun genome libraries were prepared using the NEBNext Ultra II DNA library prep kit (New England Biolabs, United States). The libraries were sequenced on an Illumina MiSeq (Illumina, San Diego, CA, United States) in a paired reads mode (2 × 300 nt). Low quality sequences were trimmed using Sickle v.1.33 (q = 30)1. In addition, genomic DNA was sequenced on a MinION device (Oxford Nanopore, United Kingdom) using the ligation sequencing kit 1D and FLO-MIN110 cells. Sequencing statistics are shown in Supplementary Table 1. For each genome, nanopore reads were assembled into a single circular contig using Flye v. 2.8.2 (Kolmogorov et al., 2019). The consensus sequence of the assembled contig was corrected with two iterations of Pilon v.1.22 (Walker et al., 2014) using Illumina MiSeq reads.
Metagenome Sequencing and Assembly of Thiothrix sp. A52 Genome
The total DNA was extracted from 200 μg of a microbial fouling sample using a DNeasy PowerSoil DNA isolation kit. Metagenomic DNA was sequenced using Illumina and Oxford Nanopore platforms as described above. Nanopore reads were de novo assembled using Flye v.2.7 (Kolmogorov et al., 2019). The obtained contigs were binned into metagenome-assembled genomes (MAGs) using MetaBAT v. 2.12.1 (Kang et al., 2015). The taxonomic assignment of the obtained MAGs was performed using the Genome Taxonomy Database Toolkit (GTDB-Tk) v.1.1.1 (Chaumeil et al., 2020) and Genome Taxonomy Database (GTDB; Parks et al., 2018). Completeness of the MAGs and their redundancy (contamination) were estimated using the CheckM v.1.05 tool (Parks et al., 2015).
One MAG, designated A52, and assigned to Thiothrix, was manually assembled into a single circular contig using the Flye assembly graph visualized in Bandage v. 0.8.1 tool (Wick et al., 2015). The assembly sequence was polished using Illumina reads with two iterations of Pilon v. 1.22 (Walker et al., 2014).
Single Molecule Real Time Sequencing of Thiothrix sp. Ku-5 and T. fructosivorans QT
Genomic DNAs from Thiothrix sp. Ku-5 and T. fructosivorans QT (strain was obtained from ATCC collection ATCC 49748T) were purified using a Monarch Genome Purification kit (T3010S, NEB, MA, United States) and sequenced using the Pacific Biosciences (PacBio) RSII sequencing platform. SMRTbell libraries were prepared using a modified PacBio protocol adapted for NEB reagents. Genomic DNA samples were sheared to an average size of ∼ 10–20 kb using the G-tube protocol (Covaris; Woburn, MA, United States), treated with FFPE, end repaired, and ligated with hairpin adapters. Incompletely formed SMRTbell templates were removed by digestion with a combination of exonuclease III and exonuclease VII (New England Biolabs, Ipswich, MA, United States). The qualification and quantification of the SMRTbell libraries were made on a Qubit fluorimeter (Invitrogen, Eugene, OR, United States) and a 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, United States). Single molecule real time sequencing (SMRT) sequencing was performed using a PacBio RSII (Pacific Biosciences; Menlo Park, CA, United States) based on standard protocols for 10--20 kb SMRTbell library inserts. Sequencing reads were collected and processed using the SMRT Analysis pipeline from Pacific Biosciences2 (Chin et al., 2013).
In addition, genomic DNA of T. fructosivorans QT was sequenced on a GridION device (Oxford Nanopore, United Kingdom) using the ligation sequencing kit 1D and R9.4 flow cell for 12 h run. Sequencing statistics are shown in Supplementary Table 1. Nanopore reads allowed us to overcome two large, approximately 50 kb repeats and the chromosome was assembled into a single circular contig using Basecaller 4.2.3 and MinKNOW 20.10.6 software.
Annotation and Analysis of the Genomes
Gene search and annotation were performed using the National Center for Biotechnology Information (NCBI) Prokaryotic Genome Annotation Pipeline (Tatusova et al., 2016; Haft et al., 2018). ANI was calculated using an online resource3 based on the OrthoANIu algorithm, using USEARCH (Lee et al., 2016). AAI between the genomes was determined using the aai.rb script from the enveomics collection (Rodriguez-R and Konstantinidis, 2016). dDDH calculation was performed using the GGDC4 online platform.
The core genome of analyzed strains was determined by clustering all genes selected by the BLASTclust v.2.2.26 program from the BLAST package (Korf and Gish, 2000). Clustering was carried out by the single link method. Genes whose predicted protein products had more than 70% amino acid sequence identity across more than 80% of the length were combined into one cluster. Clusters containing at least one protein from each genome formed the core genome.
Phylogenetic Analysis
The Genome Taxonomy Database toolkit (GTDB-Tk; Chaumeil et al., 2020), release 04-RS89 was used to identify the 120 single-copy phylogenetically informative conservative marker genes used in the GTDB classification system (Parks et al., 2018) in the genomes of T. lacustris BLT, Thiothrix sp. AS, T. fructosivorans QT, Thiothrix sp. Ku-5, T. caldifontis G1T, Thiothrix sp. A52, T. unzii A1T, “Ca. Thiothrix moscowensis RT,” “Ca. Thiothrix singaporensis SSD2,” Thiothrix singaporensis SSD2,” Thiothrix nivea JP2T, Thiolinea disciformis B3-1T, Thiothrix eikelboomii AP3T, Thiofilum flexile EJ2M-BT, and Leucothrix mucor DSM 2157T. These genes were used to construct a multiple alignment of concatenated amino acid sequences in GTDB-Tk. The multiple alignment was used to construct a maximum-likelihood phylogenetic tree in PhyML v. 3.3 (Guindon et al., 2010) with default parameters (LG amino-acid substitution model was used, equilibrium amino-acid frequencies are defined by the substitution model, gamma distribution with estimated shape parameter was used to model 4 substitution rate categories, no invariant sites). The levels of support for internal branches were assessed using the Bayesian test in PhyML.
Nucleotide Sequence Accession Numbers
The annotated genome sequences of Thiothrix sp. AS, T. unzii A1T, and Thiothrix sp. A52 were submitted to the NCBI GenBank database and are accessible via BioProject PRJNA719636. The annotated genome sequences of Thiothrix sp. Ku-5 and T. fructosivorans QT were submitted to the NCBI GenBank database and are accessible via the BioProject PRJNA631701.
Results
Biotopes Harboring New Species of Thiothrix
The samples for metagenomic analysis, as well as for the isolation of new Thiothrix isolates were taken from microbial fouling in the place of the outflow of sulfidic water from a drained well from the closed and flooded coal mine “Severnaya” in Kemerovo region of Russia (55°19.59′N, 84°4.59′E), a sulfidic spring in the Volgograd region of Russia (49°8.921′N, 44°6.236′E) and on the shore of the White Sea, Russia at the border of the freshwater stream flowing into the sea (66°33.17′N, 33°5.58′E) (Figures 1A,C,D). In the latter case, bacterial fouling was taken at low tide in the channel of a fresh stream; hydrogen sulfide came from the bottom sediments.
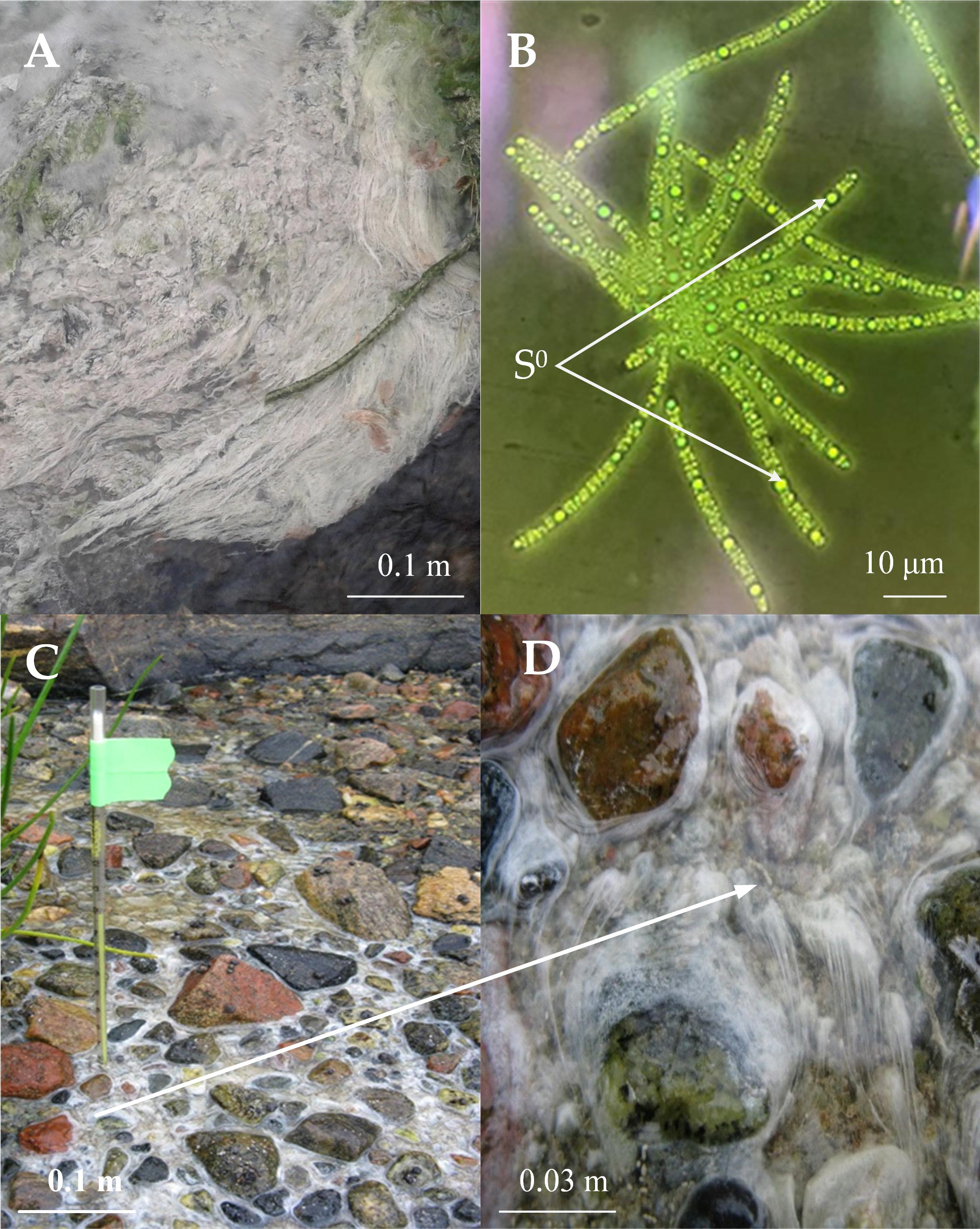
Figure 1. The biotope properties where Thiothrix samples of Thiothrix sp. Ku-5 (A) and AS (C) and (D) were collected and morphology of Thiothrix sp. Ku-5 cells: rosette formation; inclusion of elemental sulfur in cells (B). Cell morphology was observed by using an Olympus CX41 microscope equipped with a phase-contrast device. The white arrow indicates the scaled area in (C) with Thiothrix fouling.
The studied biotopes were characterized by a low redox potential (from −112 to −120 mV), the oxygen concentration varied from 2 to 5 mg L–1, and the concentration of hydrogen sulfide was from 1 to 6 mg L–1. The water temperatures at all sampling sites was between 9 and 16°C (Table 1).
Thiothrix Genomes Assembly and Analysis
The Thiothrix sp. Ku-5 was isolated from bacterial fouling in the drainage water flowing out of the coal mine in Kemerovo, Russia. The genome sequence of the Thiothrix sp. Ku-5 assembled into four closed-circular contigs; one circular 4,036,668 bp long chromosome (CP053482) and three circular plasmids, – pTSUKu5_27_unt8 (CP053484), pTSUKu5_12_unt9 (CP053483), and pTSUKu5_2_unt34 (CP053485) with lengths of 26,592 bp, 12,118 bp, and 2,180 bp, respectively. Annotation of the genome of Thiothrix sp. Ku-5 revealed four copies of the 16S rRNA gene, 46 tRNA genes, and 3,965 potential protein-coding genes.
The Thiothrix sp. AS was isolated from bacterial fouling in the shore zone of the White Sea. The genome of the Thiothrix sp. AS was assembled into a single closed circular contig (CP072801) with a total length of 4,177,302 bp. Genome annotation revealed three copies of the 16S rRNA gene, 44 tRNA genes, and 4,045 potential protein-coding genes.
The genome of the T. unzii A1T was assembled into eight contigs; one circular 3,648,856 bp long chromosome (CP072793.1) and seven circular plasmids: pTunz1 (CP072796.1, 34,476 bp), pTunz2 (CP072795.1, 22,458 bp), pTunz3 (CP072792.1, 21,012 bp), pTunz4 (CP072799.1, 18,228 bp), pTunz5 (CP072797.1, 14,837 bp), pTunz6 (CP072798.1, 14,317 bp), and pTunz7 (CP072794.1, 14,102 bp). The genome annotation revealed two copies of the 16S rRNA gene, 44 tRNA genes, and 3,729 potential protein-coding genes.
The original polished assembly of T. fructosivorans QT generated three linear contigs (1,938,437; 1,467,464 and 312,998 bp) (JAFMPM000000000.1) for the main chromosome with 51.37% GC and four closed circular plasmids pTfr153 (CP072750, 153,578 bp), pTfr21 (CP072751, 21,013 bp), pTfr12 (CP072749, 12,657 bp), pTf2 (CP072752, 2,341 bp). Therefore, T. fructosivorans QT was additionally sequenced on a GridION device (Oxford Nanopore, United Kingdom). Nanopore reads allowed us to overcome two large, approximately 50 kb repeats and the chromosome was finally assembled into a single circular contig (CP072748, 4,137,062 bp). Annotation of the genome of T. fructosivorans QT revealed two copies of the 16S rRNA gene, 44 tRNA genes, and 4,071 potential protein-coding genes.
Metagenome-assembled genome of Thiothrix sp. A52 was assembled from the environmental DNA isolated from bacterial fouling from a sulfidic spring in the Volgograd region. The metagenome sequencing resulted in 11 MAGs with completeness higher than 70%; three of these MAGs were assigned to the genus Thiothrix. One MAG was sequenced at 201-fold coverage and represented about half of the entire metagenome. Using long reads obtained by nanopore sequencing, the complete closed circular sequence of the chromosome with a length of 3,546,315 bp (CP072800) was obtained. The genome annotation revealed two copies of the 16S rRNA gene, 30 tRNA genes, and 3,613 potential protein-coding genes.
The main characteristics of the obtained genomes are shown in Table 2.
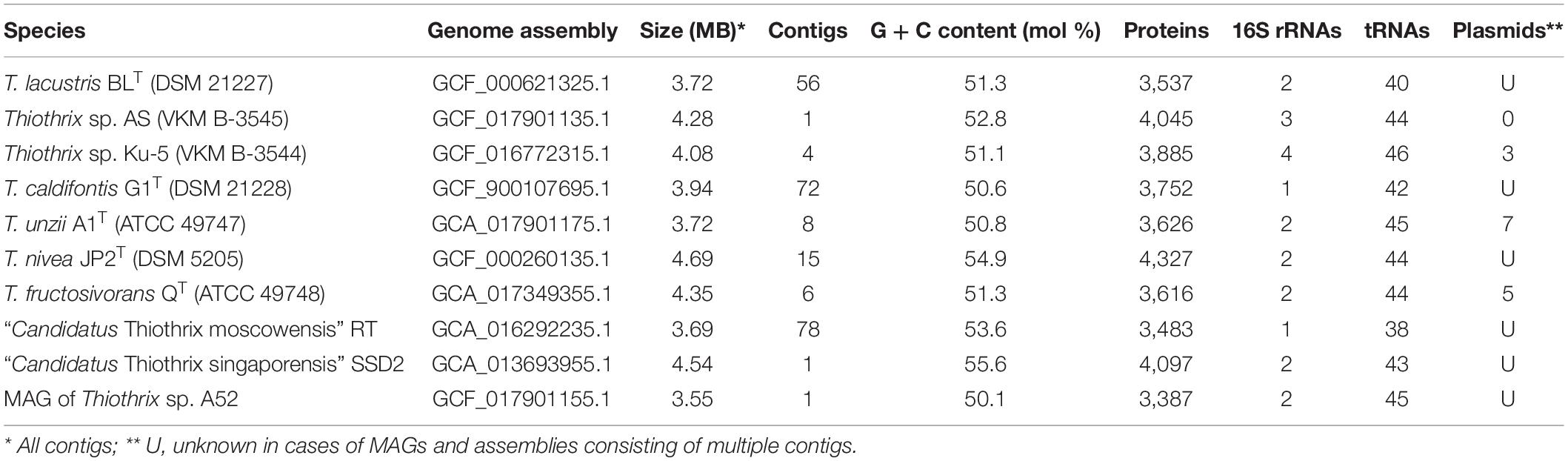
Table 2. The general properties of assembled Thiothrix genomes that were used for pangenome analysis.
Phylogenetic Analysis of the Genus Thiothrix
The sequence identities of the 16S rRNA genes among the members of the genus Thiothrix are in the range from 93.3 to 98.9% (Figure 2). For example, the 16S rRNA genes of Thiothrix sp. AS and T. lacustris BLT are completely identical (Figure 2). Strain AS previously was used as an example of the ability of representatives of the genus Thiothrix to grow anaerobically on nitrates (Trubitsyn et al., 2014). However, the ANI and dDDH values clearly show that Thiothrix sp. AS and T. lacustris BLT are different species. According to ANI and dDDH data Thiothrix sp. AS is sufficiently distant from other Thiothrix species (<92.3% and <56.2%, respectively) to be classified as a new species (Figure 2 and Supplementary Figure 1). The ANI values between MAG Thiothrix sp. A52, Thiothrix sp. Ku-5, and other Thiothrix genomes are less than 90.1%, which is below the species boundary cutoff of 95% (Konstantinidis and Tiedje, 2005; Figure 2). These data are also consistent with the results of dDDH (<46.9%) (Figure 2).

Figure 2. The heat map of 16S rRNA and ANI pairwise values for assembled Thiothrix genomes. T. lacustris BLT, (GCF_000621325.1); Thiothrix sp. AS (GCF_017901135.1); T. fructosivorans QT (GCA_017349355.1); Thiothrix sp. Ku-5 (GCF_016772315.1); T. caldifontis G1T (GCF_900107695.1); T. unzii A1T (GCA_017901175.1); MAG of Thiothrix sp. A52 (GCF_017901155.1); “Ca. Thiothrix moscowensis” RT (GCA_016292235.1); T. nivea JP2T (GCF_000260135.1); “Ca. Thiothrix singaporensis” SSD2 (GCA_013693955.1).
The phylogenetic position of the new Thiothrix bacteria was also analyzed by building a phylogenetic tree based on concatenated sequences of 120 conservative marker genes. All new genomes appeared to form a distinct lineage within the genus Thiothrix, along with previously described species (Figure 3).
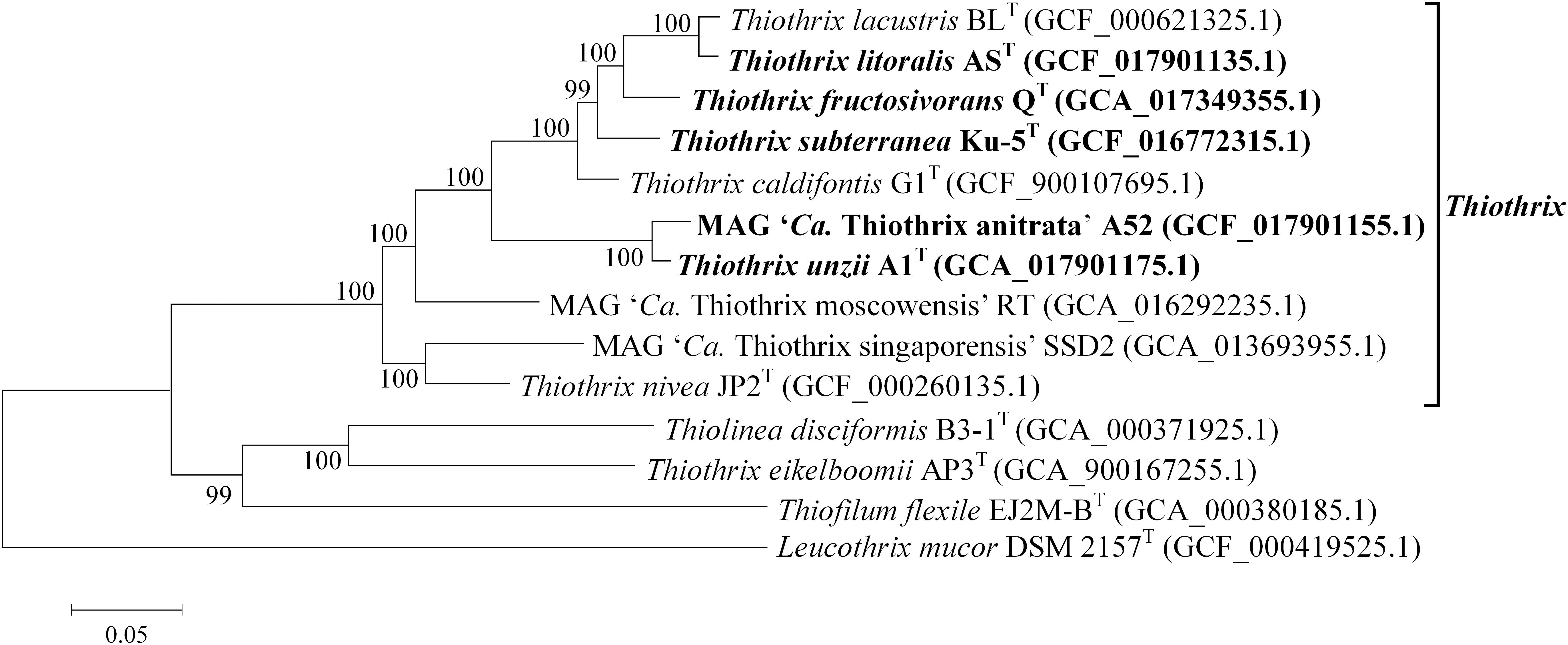
Figure 3. Phylogenetic tree of Thiothrix species. Position of Thiothrix genomes in the maximum-likelihood are determined by concatenated protein phylogeny. GenBank assembly accession numbers are shown after the genome names. The levels of support for internal branches assessed using the Bayesian test in PhyML are indicated at the nodes. The genome of Leucothrix mucor DSM 2157T was used to root the tree. Note that Thiothrix eikelboomii actually belongs to the genus Thiolinea (Boden and Scott, 2018) but was not yet formally reclassified. The species whose genomes are described in this work are highlighted in bold.
Overall, genome-based phylogenetic reconstructions and pairwise ANI and dDDH values clearly confirmed that Thiothrix sp. AS, Thiothrix sp. Ku-5, and MAG of Thiothrix sp. A52 can be classified as new species within the genus Thiothrix, for which we propose names Thiothrix litoralis sp. nov. AST, Thiothrix subterranea sp. nov. Ku-5T, and “Candidatus Thiothrix anitrata” sp. nov. A52.
Phenotypic and Physiological Properties of the New Species of Thiothrix
Thiothrix sp. Ku-5 and Thiothrix sp. AS are Gram negative rods forming immobile filaments (trichomes) surrounded by a mucous sheath. They form mobile gonidia, which give white colonies, which are rosettes with a diameter of 1–3 mm (Figure 1B).
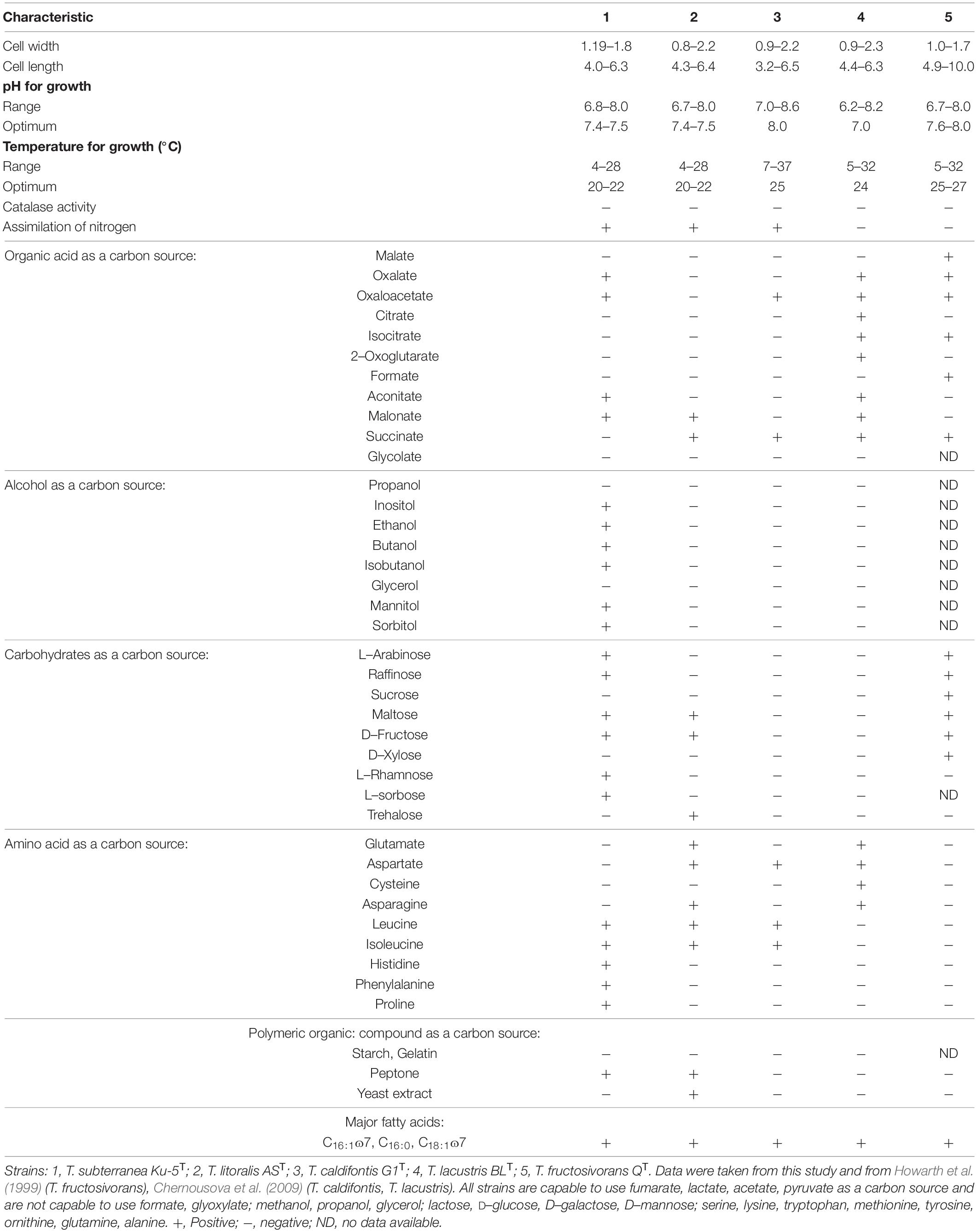
The bacteria can grow in a temperature range of 4–28°C with an optimum growth at 20–22°C. pH range 6.8–8.0, with an optimal at pH 7.4–7.5. The growth range with NaCl for Thiothrix sp. Ku-5 is up to 1%, however, Thiothrix sp. AS is able to survive up to 3% NaCl in the medium. Both are capable of lithotrophic growth in the presence of reduced sulfur compounds (H2S, Na2S2O3). When growing with thiosulfate or hydrogen sulfide, elemental sulfur accumulates in the cells (Figure 1B). They are also capable of lithoautotrophic, lithoheterotrophic, and organoheterotrophic growth. Comparative characteristics of the phenotypic and cultural properties of Thiothrix sp. Ku-5 and Thiothrix sp. AS with other phylogenetic closely related species of the genus Thiothrix are presented in Table 3. It should be noted that the phenotypic and cultural properties of all pure cultures of representatives of the genus Thiothrix are largely consistent. The composition of major fatty acids determined in this work is consistent with the data obtained earlier for other species (Chernousova et al., 2009; Supplementary Table 2 and Supplementary Figure 2).
According to polyphase analysis, the new isolates of Thiothrix sp. Ku-5 and Thiothrix sp. AS were assigned as new species of the genus Thiothrix, T. subterranea sp. nov. Ku-5T, and T. litoralis sp. nov. AST, respectively.
Description of Thiothrix subterranea sp. nov.
Thiothrix subterranea (sub.ter.ra′ne.a. L. fem. adj. subterranea underground, below the earth surface, from where the strain was isolated). The cell size is 1.19–1.8 × 4.0–6.3 μm. Growth is observed in the temperature range 4–28°C with an optimum of growth at 20–22°C, pH 6.8–8.0 with an optimum at pH 7.4–7.5. It can tolerate NaCl up to 1%. It is catalase negative, oxidase positive, Gram negative and can sustain aerobic, microaerobic and anaerobic growth. Anaerobic growth is possible in the presence of nitrates. It is capable of all types of lithoautotrophic, lithoheterotrophic and organoheterotrophic growth. This strain uses alcohols (ethanol, butanol, isobutanol, mannitol, sorbitol, inositol), organic acids (acetate, lactate, pyruvate, aconitate, oxaloacetate, fumarate, malonate), amino acids (leucine, isoleucine, histidine, phenylalanine, proline), carbohydrates (arabinose, raffinose, maltose, fructose, rhamnose, sorbose) as carbon and energy sources, while during lithoheterotrophic growth, they are used mainly as a source of carbon. During lithotrophic growth, reduced sulfur compounds like H2S, Na2S2O3 can be used as an electron donor. It also capable of fixing molecular nitrogen. Major fatty acids are C16:1ɷ7, C16:0, C18:1ɷ7. The G + C content of the genome of T. subterranea Ku-5T strain is 51.1%.
The type strain Thiothrix subterranea Ku-5T (=VKM B-3544, =UQM 41459, =DSM 113265) was obtained from microbial fouling in the place of the outflow of drainage water from the coal mine “Severnaya,” Kemerovo, Kuzbass, Russia.
Description of Thiothrix litoralis sp. nov.
Thiothrix litoralis (li.to.ra′lis. L. fem. adj. litoralis of or belonging to the seashore). The cell size is 0.8–2.2 × 4.3–6.4 μm. Growth is observed in the temperature range 4–28°C with an optimum at 20–22°C, pH 6.7–8.0 with an optimal at pH 7.4–7.5. It can tolerate NaCl up to 3%, catalase negative, oxidase positive, Gram negative and can sustain aerobic, microaerobic and anaerobic growth. Anaerobic growth is possible in the presence of nitrates. The strain is also capable of lithoautotrophic, lithoheterotrophic and organoheterotrophic growth. It uses organic acids (acetate, lactate, pyruvate, fumarate, malonate, succinate), amino acids (glutamate, aspartate, asparagine, leucine, isoleucine), and carbohydrates (maltose, fructose, trehalose) as carbon and energy sources, while during lithoheterotrophic growth, they are used mainly as a source of carbon. During lithotrophic growth, reduced sulfur compounds like H2S and Na2S2O3 can be used as an electron donor. It is capable of fixing molecular nitrogen. Major fatty acids are C16:1ɷ7, C16:0, C18:1ɷ7. The G + C content in the genome of T. litoralis AST is 52.8%.
The type strain, Thiothrix litoralis AST (=VKM B-3545, =UQM 41458, =DSM 113264), was isolated from bacterial growths in the seashore zone of the White Sea, Murmansk region, Russia.
“Candidatus Thiothrix anitrata” sp. nov. [a.ni.tra′ta. Gr.pref. a, not; N.L. masc. n. nitras (genitive: nitratis), nitrate; N.L. fem.adj. anitrata, not reducing nitrate], MAG was obtained from the metagenome of bacterial fouling of a hydrogen sulfide brook in the Volgograd region, Russia.
The Pangenome of the Genus Thiothrix
In this work, representatives of the genus Thiothrix were studied using a pangenomic approach. We analyzed all publicly available genomes of taxonomically described species of the genus Thiothrix. In addition, the newly obtained genome sequences of T. subterranea Ku-5T, T. litoralis AST, “Ca. Thiothrix anitrata” A52, T. fructosivorans QT and T. unzii A1T were included in the analysis (Table 2). All genomes were of high quality except for MAG of “Ca. Thiothrix singaporensis” SSD2 which contained multiple frameshifts and therefore was excluded from the analysis.
The pangenome analysis of nine species indicated that the genus Thiothrix comprises 11,549 gene clusters. The core genome of the genus Thiothrix contains 1,355 genes and the auxiliary genome comprises 3,894 gene clusters present in 2–8 genomes. The number of unique genes varies in different species from 397 to 1,360 (Figure 4). These genes were predicted to encode mostly hypothetical proteins with unknown functions, transporters, transcriptional regulators, methyltransferases, transposases, and the toxin-antitoxin systems.
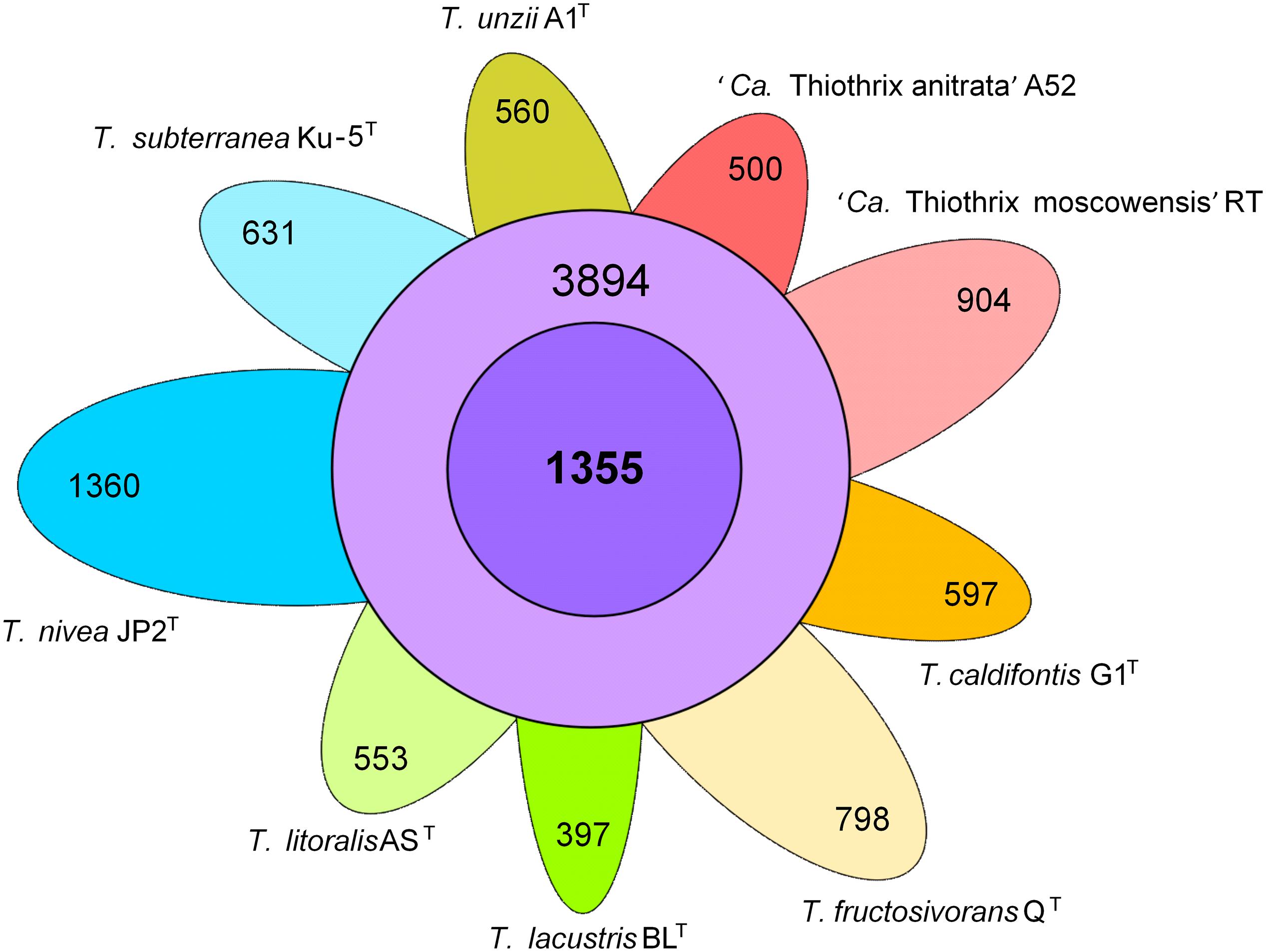
Figure 4. Pangenome of the genus Thiothrix. Venn diagram demonstrating the distribution of common (purple), additional (violet), and unique (different colors) genes among analyzed Thiothrix species.
The genus Thiothrix is characterized by the presence of a number of genes for dissimilatory sulfur metabolism. Thiothrix species are classical lithotrophs, capable of using reduced sulfur compounds as an energy source. The hydrogen sulfide oxidation systems, SQR (sqrA and sqrF) and FCSD (fccAB), are present in the core genome and were found in all species. The branched SOX system, represented by the soxAXYZB genes and involved in the oxidation of thiosulfate to sulfur and sulfate, was also found in all members of the genus. The genes of the reverse Dsr (rDsr) dsrABEFHÑEMKLJONR complex involved in the oxidation of sulfur to sulfite were also present in all species, as well as genes for direct (soeABC) and indirect (aprAB, sat) pathways of oxidation of sulfite to sulfate. Thus, the presence of genes of several systems of dissimilatory sulfur metabolism in all known species of the genus Thiothrix indicates their exceptional role in the life of this group of sulfur bacteria.
The core genome contains the hyaA and hyaB genes encoding the large and small catalytic subunits of membrane-bound [NiFe] -hydrogenase of type I, and the hyaC gene, which encodes the cytochrome b subunit, which attaches hydrogenase to the cytoplasmic membrane. In addition, the core genome contains hypABCDEF genes, which are involved in the maturation of [NiFe] – hydrogenases.
The core genome includes all genes for the glycolysis, the Krebs cycle and the glyoxylate cycle. All members of the genus Thiothrix lack the classical NAD-dependent malate dehydrogenase (mdh), but instead contain genes encoding FAD-dependent malate: quinone oxidoreductase (mqo). The glk gene for glucokinase was found in all members of the genus, however, it was not included in the core genome due to the lower sequence similarity, which suggests horizontal transfer or an increase rate of evolution of this gene.
All Thiothrix species have a complete set of genes for the Calvin-Benson-Bassham cycle of carbon fixation. Genes for the type IAq RuBisCO were in the core genome, while type IAc was found in all genomes with the exception of T. lacustris BLT and T. fructosivorans QT. All the species except “Ca. Thiothrix anitrata” A52 also have type II RuBisCO (Figure 5).
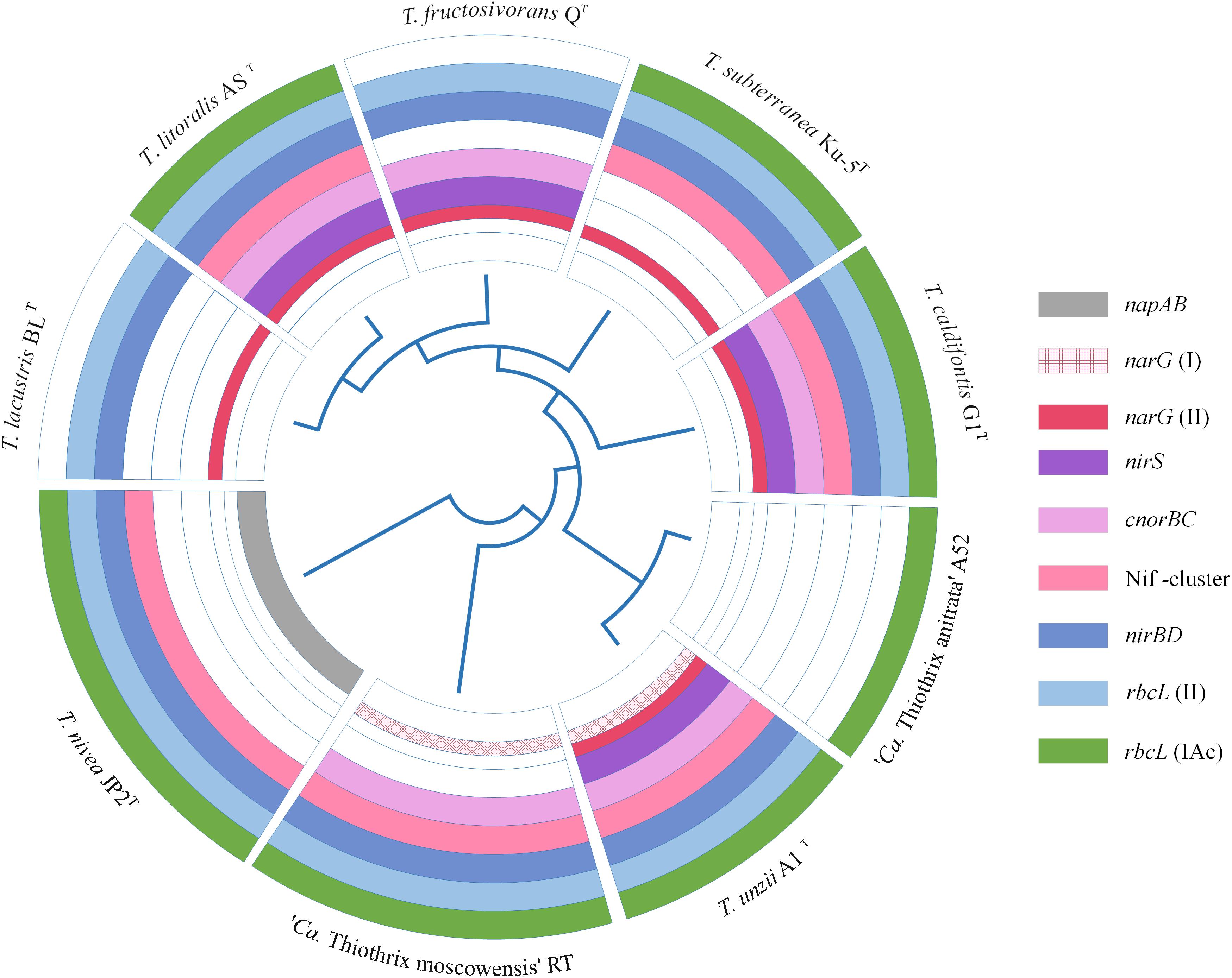
Figure 5. The presence of genes for nitrogen metabolism and various types of RuBisCO in the genomes of Thiothrix sp. Dissimilatory nitrate reduction and nitrogen assimilation genes: napAB, periplasmic nitrate reductase; narG, membrane-bound nitrate reductase; nirS, nitrite reductase; cnorB, NO-reductase; nirBD, ammonia forming assimilatory nitrite reductase; nifDKH, nitrogenase genes; rbcL (II) and rbcL (IAc), genes encoding the large subunit of RuBisCO.
The electron transport chain (ETC) was represented in all species by complex I NuoABCDEFGHIJKLMN, complex II SdhABCD, complex III (CytB, Cyc1, Ryp1), and cytochrome c oxidase of complex IV (CcoNOPGQS). The presence of all ETC genes in all genomes additionally confirms the respiratory type of metabolism in the genus Thiothrix (Chernousova et al., 2009).
All members of the genus Thiothrix except for “Ca. Thiothrix anitrata” A52, are capable of anaerobic respiration with nitrogen compounds. However, the genes for dissimilatory nitrogen metabolism were not found in the core genome, and their set differs markedly among Thiothrix species (Figure 5). The nirS nitrite reductase gene is presented in T. litoralis AST, T. unzii A1T, T. fructosivorans QT, T. caldifontis G1T, but was probably lost in T. lacustris BLT, T. nivea JP2T, “Ca. Thiothrix moscowensis” RT, T. subterranea Ku-5T, and “Ca. Thiothrix anitrata” A52. Assimilatory ammonium-forming nitrite reductase nirBD is absent only in “Ca. Thiothrix anitrata” A52. The cnorBC nitric oxide (NO) reductase genes were found in T. litoralis AST, T. unzii A1T, T. fructosivorans QT, T. caldifontis G1T, and “Ca. Thiothrix moscowensis” RT. All members of the genus Thiothrix lack the nosZ gene encoding the dissimilatory N2O reductase. In the cases of nirS, nirBD and cnorBC, the corresponding genes found in different Thiothrix species were highly homologous; therefore, their absence in individual species probably indicates a loss in course of evolution rather than an independent acquisition as a result of horizontal gene transfer.
The distribution pattern is more complex for dissimilatory nitrate reductase genes. In the Thiothrix genomes two types of genes for NarGHI reductase were found, the identity of their deduced amino acid sequences is less than 60%. Genomes of T. lacustris BLT, T. litoralis AST, T. fructosivorans QT, T. subterranea Ku-5T, and T. caldifontis G1T contain only one type of narG, the genome of “Ca. Thiothrix moscowensis” RT harbored only another type, and the genome of T. unzii A1 T contains both variants of the narG gene. The narG genes are missing in the genomes of “Ca. Thiothrix anitrata” A52 and T. nivea JP2T; in the later species the periplasmic nitrate reductase napAB is present instead of the narGHI (Figure 5). Such distribution pattern indicates the events of the acquisition of additional copies of the nar genes as a result of horizontal transfer with the subsequent loss of one of the two copies, as well as the replacement of nar operon with Nap type of nitrate reductase.
The nifASUBNXX2YB2ENQVWMHDKZTO gene cluster for molecular nitrogen fixation is present in all species of the genus, with the exception of T. fructosivorans QT, “Ca. Thiothrix anitrata” A52, and T. lacustris BLT. The amination process for all members of the Thiothrix genus is represented by glutamine synthetase (glnB), glutamate synthase (gltBD), and aspartate aminotransferase (aspB).
The genes pstDCABS and phoURB, involved in inorganic phosphorus transport into the cell, were found in the core genome. The core genome of the genus Thiothrix contains gene for polyphosphate kinases PPK1 that can catalyzes the synthesis of the poly-P chain. Also The core genome contains gene for poly-AMP phosphotransferase (PAP), which belongs to class II of polyphosphate kinases PPK2 and that can catalyze the synthesis of the ADF (Kimura and Kamatani, 2021). Most likely, in Thiothrix, PAP is responsible for ATP regeneration in the cell (Kameda et al., 2001). In addition, the epp gene for exopolyphosphatase, which enables the cleavage of polyphosphates, was found in all genomes. The presence of polyphosphate kinase PPK1 and exopolyphosphatase suggests that Thiothrix species could accumulate and degrade polyphosphates which are probably used for temporal ATP storage.
Discussion
The genus Thiothrix is relatively poorly studied due to the small number of described species, the complexity of cultivation, as well as the limited number of genomic sequences and high-quality MAGs. In this work and previously (Mardanov et al., 2020) we have obtained new pure cultures of new species of the genus Thiothrix, including T. litoralis sp. nov. AST, T. subterranea sp. nov. Ku-5T, and two MAGs, assigned to candidate species “Ca. Thiothrix moscowensis” RT, and “Ca. Thiothrix anitrata” A52. For the first time full genomic sequences were obtained for T. unzii A1T and T. fructosivorans QT, which were isolated back in 1999 (Howarth et al., 1999). Available genomes enabled us to perform comparative analysis of nine species of this genus and reveal their metabolic features.
Comparative genome analysis revealed that the key features common to all species of the genus Thiothrix are their ability for lithotrophic growth in the presence of reduced sulfur compounds, H2S, and thiosulfate, the latter being oxidized through the branched Sox pathway (SoxAXYZB); autotrophic carbon fixation (Calvin-Benson-Bassham cycle), and the presence of FAD-dependent malate: quinone oxidoreductase (EC 1.1.5.4) instead of the classical NAD-dependent malate dehydrogenase (EC 1.1.1.37). The ability for lithoautotrophic and lithoheterotrophic growth in the presence of reduced sulfur compounds, as well as for organoheterotrophic growth, has been previously proven experimentally (Larkin and Shinabarger, 1983; Chernousova et al., 2009).
All members of the genus Thiothrix have all the necessary genes encoding enzymes involved in H2 oxidation during lithotrophic growth. However, lithotrophic growth in the presence of H2 was assumed only on the basis of the genome analysis and requires experimental verification.
The main differences between the Thiothrix genomes included genes involved in dissimilatory reduction of nitrogen compounds and nitrogen fixation, as well as genetic determinants of various types of RuBisCO (Figure 5). Probably, oxidized forms of nitrogen that could be used in anaerobic respiration are not available in all ecosystems in which representatives of Thiothrix were found, which explain the patchy distribution of the corresponding genes in the genomes. Likewise, the presence of different forms of RuBisCOs in Thiothrix could be explained by the flexibility of the CO2-fixing lifestyles associated with environments where the levels of CO2 and O2 can fluctuate, and different forms of RuBisCO could be used under different environmental conditions (Badger and Bek, 2008).
Data Availability Statement
The datasets created for this study can be found at GenBank (https://www.ncbi.nlm.nih.gov/genbank) via the access numbers: GCF_017901135.1, GCF_016772315.1, GCA_017901175.1, GCA_017349355.1, and GCF_017901155.1.
Author Contributions
NR and MG: conceptualization, supervision, and funding acquisition. DS, TR, AR, NM, AF, and AN: investigation. OK: resources. NR and AB: data curation. AB, AF, LS, and RR: formal analysis. TR, DS, and NM: validation. NR, DS, TR, AF, and MG: writing – original draft preparation. NR, TR, AF, RR, and MG: writing – review and editing. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the Russian Science Foundation (project 20-14-00137 to MG) and the Ministry of Education and Science of the Russian Federation (AR and NR). Chemotaxonomic studies were performed by AN in Gubkin Russian State University of Oil and Gas (State Assignment project 0768-2020-0007).
Conflict of Interest
AF, LS, and RR are paid employees of New England Biolabs (https://www.neb.com/). There are no patents, products in development, or marketed products associated with this research to declare. This does not alter our adherence to Frontiers in Microbiology policies on sharing data and materials.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.760289/full#supplementary-material
Footnotes
- ^ https://github.com/najoshi/sickle
- ^ http://www.pacbiodevnet.com/SMRT-Analysis/Software/SMRT-Pipe
- ^ https://www.ezbiocloud.net/tools/ani
- ^ https://ggdc.dsmz.de/ggdc.php#
References
Badger, M. R., and Bek, E. J. (2008). Multiple Rubisco forms in proteobacteria: their functional significance in relation to CO2 acquisition by the CBB cycle. J. Exp. Bot. 59, 1525–1541. doi: 10.1093/jxb/erm297
Bauermeister, J., Ramette, A., and Dattagupta, S. (2012). Repeatedly evolved host-specific ectosymbioses between sulfur-oxidizing bacteria and amphipods living in a cave ecosystem. PLoS One 7:e50254. doi: 10.1371/journal.pone.0050254
Boden, R., and Scott, K. M. (2018). Evaluation of the genus Thiothrix Winogradsky 1888 (Approved Lists 1980) emend. Aruga et al. 2002: reclassification of Thiothrix disciformis to Thiolinea disciformis gen. nov., comb. nov., and of Thiothrix flexilis to Thiofilum flexile gen. nov., comb nov., with emended description of Thiothrix. Int. J. Syst. Evol. Microbiol. 68, 2226–2239. doi: 10.1099/ijsem.0.002816
Chaumeil, P. A., Mussig, A. J., Hugenholtz, P., and Parks, D. H. (2020). GTDB-Tk: a toolkit to classify genomes with the genome taxonomy database. Bioinformatics 36, 1925–1927. doi: 10.1093/bioinformatics/btz848
Chernousova, E., Gridneva, E., Grabovich, M., Dubinina, G., Akimov, V., Rossetti, S., et al. (2009). Thiothrix caldifontis sp. nov. and Thiothrix lacustris sp. nov., novel gammaproteobacteria isolated from sulfide springs. Int. J. Syst. Evol. Microbiol. 59, 3128–3135. doi: 10.1099/ijs.0.009456-0
Chernousova, E. Y., Akimov, V. N., Gridneva, E. V., Dubinina, G. A., and Grabovich, M. Y. (2010). Biodiversity and monitoring of colorless filamentous bacterial sulfide aquatic systems of north caucasus region. Microbiology 79, 682–687.
Chin, C. S., Alexander, D. H., Marks, P., Klammer, A. A., Drake, J., Heiner, C., et al. (2013). Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat. Methods 10, 563–569. doi: 10.1038/nmeth.2474
Guindon, S., Dufayard, J. F., Lefort, V., Anisimova, M., Hordijk, W., and Gascuel, O. (2010). New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 59, 307–321. doi: 10.1093/sysbio/syq010
Haft, D. H., DiCuccio, M., Badretdin, A., Brover, V., Chetvernin, V., O’Neill, K., et al. (2018). RefSeq: an update on prokaryotic genome annotation and curation. Nucleic Acids Res. 46, D851–D860. doi: 10.1093/nar/gkw569
Howarth, R., Unz, R. F., Seviour, E. M., Seviour, R. J., Blackall, L. L., Pickup, R. W., et al. (1999). Phylogenetic relationships of filamentous sulfur bacteria (Thiothrix spp. and Eikelboom type 021N bacteria) isolated from wastewater-treatment plants and description of Thiothrix eikelboomii sp. nov., Thiothrix unzii sp. nov., Thiothrix fructosivorans sp. nov. and Thiothrix defluvii sp. nov. Int. J. Syst. Bacteriol. 49, 1817–1827. doi: 10.1099/00207713-49-4-1817
Kameda, A., Shiba, T., Kawazoe, Y., Satoh, Y., Ihara, Y., Munekata, M., et al. (2001). A novel ATP regeneration system using polyphosphate-AMP phosphotransferase and polyphosphate kinase. J. Biosci. Bioeng. 91, 557–563. doi: 10.1263/jbb.91.557
Kang, D. D., Froula, J., Egan, R., and Wang, Z. (2015). MetaBAT, an efficient tool for accurately reconstructing single genomes from complex microbial communities. PeerJ 3:e1165. doi: 10.7717/peerj.1165
Kimura, Y., and Kamatani, S. (2021). Catalytic activity profile of polyP:AMP phosphotransferase from Myxococcus xanthus. J. Biosci. Bioeng. 131, 147–152. doi: 10.1016/j.jbiosc.2020.09.016
Kolmogorov, M., Yuan, J., Lin, Y., and Pevzner, P. A. (2019). Assembly of long, error-prone reads using repeat graphs. Nat. Biotechnol. 37, 540–546. doi: 10.1038/s41587-019-0072-8
Konkol, N. R., Bruckner, J. C., Aguilar, C., Lovalvo, D., and Maki, J. S. (2010). Dominance of epiphytic filamentous Thiothrix spp. on an aquatic macrophyte in a hydrothermal vent flume in Sedge Bay, Yellowstone Lake, Wyoming. Microb. Ecol. 60, 528–538. doi: 10.1007/s00248-010-9656-z
Konstantinidis, K. T., and Tiedje, J. M. (2005). Towards a genome-based taxonomy for prokaryotes. J. Bacteriol. 87, 6258–6264. doi: 10.1128/JB.187.18.6258-6264.2005
Korf, I., and Gish, W. (2000). MPBLAST: improved BLAST performance with multiplexed queries. Bioinformatics 16, 1052–1053. doi: 10.1093/bioinformatics/16.11.1052
Kuznetsov, S. I., and Dubinina, G. A. (1989). Methods of investigation of aqueous microorganisms. Moscow: Nauka. 285
Larkin, J. M., and Shinabarger, D. L. (1983). Characterization of Thiothrix nivea. Int. J. Syst. Bacteriol. 33, 841–846.
Lee, I., Ouk, K. Y., Park, S. C., and Chun, J. (2016). OrthoANI: an improved algorithm and software for calculating average nucleotide identity. Int. J. Syst. Evol. Microbiol. 66, 1100–1103. doi: 10.1099/ijsem.0.000760
Li, Y., Tang, K., Zhang, L., Zhao, Z., Xie, X., Chen, C. A., et al. (2018). Coupled carbon, sulfur, and nitrogen cycles mediated by microorganisms in the water column of a shallow-water hydrothermal ecosystem. Front. Microbiol. 9:2718. doi: 10.3389/fmicb.2018.02718
Macalady, J. L., Dattagupta, S., Schaperdoth, I., Jones, D. S., Druschel, G. K., and Eastman, D. (2008). Niche differentiation among sulfur-oxidizing bacterial populations in cave waters. ISME J. 2, 590–601.
Mardanov, A. V., Gruzdev, E. V., Smolyakov, D. D., Rudenko, T. S., Beletsky, A. V., Gureeva, M. V., et al. (2020). Genomic and metabolic insights into two novel Thiothrix species from enhanced biological phosphorus removal systems. Microorganisms 8:2030. doi: 10.3390/microorganisms8122030
Panova, I. A., Rusanov, I. I., Kadnikov, V. V., Latygolets, E. A., Avakyan, M. R., Ivanov, M. V., et al. (2020). Sulfate Reduction in underground horizons of a flooded coal mine in Kuzbass. Microbiology 89, 542–550. doi: 10.1134/S0026261720050185
Parks, D. H., Chuvochina, M., Waite, D. W., Rinke, C., Skarshewski, A., Chaumeil, P. A., et al. (2018). A standardized bacterial taxonomy based on genome phylogeny substantially revises the tree of life. Nat. Biotechnol. 36, 996–1004. doi: 10.1038/nbt.4229
Parks, D. H., Imelfort, M., Skennerton, C. T., Hugenholtz, P., and Tyson, G. W. (2015). CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 25, 1043–1055. doi: 10.1101/gr.186072.114
Pfennig, N., and Lippert, K. D. (1966). Über das vitamin B12-Bedürfnis phototropher Schwefelbakterien. Arch. Microbiol. 55, 425–432.
Rodriguez-R, L. M., and Konstantinidis, K. T. (2016). The enveomics collection: a toolbox for specialized analyses of microbial genomes and metagenomes. PeerJ Prepr. 4:e1900v1. doi: 10.7287/PEERJ.PREPRINTS.1900V1
Rossmassler, K., Hanson, T. E., and Campbell, B. J. (2016). Diverse sulfur metabolisms from two subterranean sulfidic spring systems. FEMS Microbiol. Lett. 363:fnw162. doi: 10.1093/femsle/fnw162
Slobodkina, G. B., Merkel, A. Y., Novikov, A. A., Bonch-Osmolovskaya, E. A., and Slobodkin, A. I. (2020). Pelomicrobium methylotrophicum gen. nov., sp. nov. a moderately thermophilic, facultatively anaerobic, lithoautotrophic and methylotrophic bacterium isolated from a terrestrial mud volcano. Extremophiles 24, 177–185. doi: 10.1007/s00792-019-01145-0
Stewart, W. D., Fitzgerald, G. P., and Burris, R. H. (1968). Acetylene reduction by nitrogen-fixing blue-green algae. Arch. Mikrobiol. 62, 336–348. doi: 10.1007/BF00425639
Tatusova, T., DiCuccio, M., Badretdin, A., Chetvernin, V., Nawrocki, E. P., Zaslavsky, L., et al. (2016). NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res. 44, 6614–6624.
Trubitsyn, I. V., Belousova, E. V., Tutukina, M. N., Merkel, A. Y., Dubinina, G. A., and Grabovich, M. Y. (2014). Expansion of ability of denitrification within the filamentous colorless sulfur bacteria of the genus Thiothrix. FEMS Microbiol. Lett. 358, 72–80. doi: 10.1111/1574-6968.12548
Walker, B. J., Abeel, T., Shea, T., Priest, M., Abouelliel, A., Sakthikumar, S., et al. (2014). Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One 9:e112963. doi: 10.1371/journal.pone.0112963
Keywords: Thiothrix, genome, phylogeny, comparative genomics, metagenome-assembled genome
Citation: Ravin NV, Rudenko TS, Smolyakov DD, Beletsky AV, Rakitin AL, Markov ND, Fomenkov A, Sun L, Roberts RJ, Novikov AA, Karnachuk OV and Grabovich MY (2021) Comparative Genome Analysis of the Genus Thiothrix Involving Three Novel Species, Thiothrix subterranea sp. nov. Ku-5, Thiothrix litoralis sp. nov. AS and “Candidatus Thiothrix anitrata” sp. nov. A52, Revealed the Conservation of the Pathways of Dissimilatory Sulfur Metabolism and Variations in the Genetic Inventory for Nitrogen Metabolism and Autotrophic Carbon Fixation. Front. Microbiol. 12:760289. doi: 10.3389/fmicb.2021.760289
Received: 17 August 2021; Accepted: 28 September 2021;
Published: 22 October 2021.
Edited by:
Kira Makarova, National Center for Biotechnology Information (NLM), United StatesReviewed by:
Aharon Oren, Hebrew University of Jerusalem, IsraelKathleen Scott, University of South Florida, United States
Copyright © 2021 Ravin, Rudenko, Smolyakov, Beletsky, Rakitin, Markov, Fomenkov, Sun, Roberts, Novikov, Karnachuk and Grabovich. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Margarita Y. Grabovich, margarita_grabov@mail.ru
 Nikolai V. Ravin
Nikolai V. Ravin Tatyana S. Rudenko
Tatyana S. Rudenko Dmitry D. Smolyakov
Dmitry D. Smolyakov Alexey V. Beletsky
Alexey V. Beletsky Andrey L. Rakitin
Andrey L. Rakitin Nikita D. Markov
Nikita D. Markov Alexey Fomenkov
Alexey Fomenkov Luo Sun
Luo Sun Richard J. Roberts
Richard J. Roberts Andrey A. Novikov
Andrey A. Novikov Olga V. Karnachuk
Olga V. Karnachuk Margarita Y. Grabovich
Margarita Y. Grabovich