- 1Department of Social Medicine, Universidade Federal do Espírito Santo, Vitória, Brazil
- 2Postgraduate Programme in Infectious Diseases, Universidade Federal do Espírito Santo, Vitória, Brazil
- 3National Infection Service, Public Health England, London, United Kingdom
Mycobacterium lepromatosis was identified as a new species and second causal agent of Hansen’s disease (HD, or leprosy) in 2008, 150years after the disease was first attributed to Mycobacterium leprae. M. lepromatosis has been implicated in a small number of HD cases, and clinical aspects of HD caused by M. lepromatosis are poorly characterized. HD is a recognized zoonosis through transmission of M. leprae from armadillos, but the role of M. lepromatosis as a zoonotic agent of HD is unknown. M. lepromatosis was initially associated with diffuse lepromatous leprosy, but subsequent case reports and surveys have linked it to other forms of HD. HD caused by M. lepromatosis has been reported from three endemic countries: Brazil, Myanmar, and Philippines, and three non-endemic countries: Mexico, Malaysia, and United States. Contact with armadillos in Mexico was mentioned in 2/21 M. lepromatosis HD case reports since 2008. M. lepromatosis in animals has been investigated only in non-endemic countries, in squirrels and chipmunks in Europe, white-throated woodrats in Mexico, and armadillos in the United States. To date, there have only been a small number of positive findings in Eurasian red squirrels in Britain and Ireland. A single study of environmental samples found no M. lepromatosis in soil from a Scottish red squirrel habitat. Future studies must focus on endemic countries to determine the true proportion of HD cases caused by M. lepromatosis, and whether viable M. lepromatosis occurs in non-human sources.
Introduction
Hansen’s disease (HD) or leprosy has been attributed to Mycobacterium leprae (M. leprae) since the late nineteenth century, after the discoveries of Gerhard Armauer Hansen. In 2008, bacilli from two HD cases were identified as a new species, Mycobacterium lepromatosis (M. lepromatosis; Han et al., 2008). These first two cases manifested as a specific multibacillary form of HD, diffuse lepromatous leprosy (DLL), co-occurring with Lucio’s phenomenon (LP), a severe HD reaction. Subsequent case reports indicated that M. lepromatosis was not related specifically to DLL, but also with other multibacillary forms of HD. Dual infection with M. lepromatosis and M. leprae has also been reported in case reports from South America (Aldama Olmedo et al., 2020) and South East Asia (Han et al., 2012a; Widiatma and Sukanto, 2019) and in surveys of specimens from Mexico and Brazil (Han et al., 2012b, 2014; Kai et al., 2016; Torres-Guerrero et al., 2018; Sharma et al., 2020). Medical and scientific literature refers to both species as the causal agents of HD, but cases attributed to M. lepromatosis infection are infrequently reported, and the clinical aspects of HD caused by M. lepromatosis remain poorly characterized.
In addition to this apparent commonality of M. lepromatosis and M. leprae in causing a single disease in humans, both mycobacterium species have been detected in mammals presenting with and without clinical signs of disease. For M. leprae, experimental and natural infection in armadillos is well-described (Oliveira et al., 2019). Indeed, armadillo species have served as a model for HD in humans since the 1970s (Storrs, 1971; Sharma et al., 2013). A meta-analysis of M. leprae infection in wild armadillos in Brazil found a pooled prevalence from 8 PCR-based studies conducted in seven states equivalent to one in ten animals being infected, with three studies reporting no infection and one study in a hyperendemic area of a northeastern state reporting infection in 20/20 armadillos (Deps et al., 2020). Apart from armadillos, natural M. leprae infection in wild (non-captive) animals has been reported only in red squirrels from the British Isles and Ireland (Ploemacher et al., 2020), and more recently, in two chimpanzees in West Africa (Hockings et al., 2020).
From a One Health perspective, HD in the United States is recognized as a zoonosis (Centers for Disease Control and Prevention, 2020), with genomic evidence linking M. leprae from armadillos in south eastern states with sporadic cases of HD (Truman et al., 2011; Sharma et al., 2015). In Brazil, where HD is endemic and hunting and consumption of armadillos is widespread, zoonotic transmission is generally regarded to be of little or no concern given the presumed predominance of human-to-human transmission, despite evidence that exposure to armadillos confers additional HD risk even in highly endemic areas (Deps et al., 2021). M. leprae has also been detected in environmental samples (Ploemacher et al., 2020), including in soil and water in areas endemic for HD in India (Mohanty et al., 2016; Singh et al., 2020), and from water sources in endemic areas in Brazil (Holanda et al., 2017).
In this article, we review the emergence of M. lepromatosis as a cause of HD in humans and evidence to date for the existence of M. lepromatosis in animal hosts and environmental reservoirs, and we discuss, in the context of evidence for M. leprae as a zoonotic pathogen, what steps need to be taken to determine the prevalence of M. lepromatosis infection in human and animals and whether M. lepromatosis might be a zoonotic source of HD.
Mycobacterium lepromatosis as a Cause of Disease in Humans
Discovery and First Cases
The new mycobacterium species M. lepromatosis was identified and named by Han et al. (2008) in 2008 following the death of a patient of Mexican origin who had been diagnosed with DLL and LP (Han et al., 2008). Sequencing by PCR of the ~1500bp 16S ribosomal RNA (rRNA) gene in acid-fast bacilli (AFB) from frozen liver autopsy specimens showed that the AFB strain (designated FJ924) had a closest match with M. leprae (1475/1506bp, 97.9%) and a second closest match with M. haemophilum (1465/1505, 97.3%). Strain FJ924 had a 19bp AT-rich inserted sequence in its 16S rRNA gene which was not found in any other bacterial species, just as M. leprae has a unique short (16bp) AT-rich sequence inserted in its 16S rRNA gene. The researchers obtained archived biopsy specimens from a second patient of Mexican origin who had died 5years previously, also with DLL and LP. Gene sequences from this earlier case, including of the 16S rRNA gene, matched 100% with strain FJ924.
On the basis of these results, Han et al. (2012a) proposed a new species, M. lepromatosis, as a causal agent of DLL, while speculating that it might also cause lepromatous (LL) and borderline lepromatous (BL) forms of HD (Han et al., 2008). At the time of their first study, the researchers obtained archived specimens from two fatal cases of DLL in Singapore (both patients died in 1999). PCR using primers for the unique 16S rRNA inserted sequences, LPMF2 for M. lepromatosis and LERF2 for M. leprae, was positive for both species in both patients, indicating dual infection (Han et al., 2012a).
Subsequent Case Reports and Specimen Surveys
Case reports since 2008 have identified M. lepromatosis infection in a further 15 patients (Vera-Cabrera et al., 2011; Jessamine et al., 2012; Han and Jessurun, 2013; Han and Quintanilla, 2015; Sotiriou et al., 2016; Velarde-Felix et al., 2016; Cleary et al., 2017; Virk et al., 2017; Htet et al., 2018; Flores-Suarez et al., 2019; Trave et al., 2020; Watson et al., 2020), plus two patients with dual infections of M. lepromatosis and M. leprae (Widiatma and Sukanto, 2019; Aldama Olmedo et al., 2020). Of the 21 M. lepromatosis cases, 11 (52.4%) patients were from Mexico (seven were United States residents), 2 each from Singapore, Myanmar, and United States, and one each from Indonesia, Paraguay, Cuba, and Canada. Five of the 21 case investigations used DNA from archived biopsy specimens, the earliest of which was a patient from Mexico who was diagnosed with DLL in 1963 and treated at Carville, United States (Han and Jessurun, 2013). Where reported, clinical presentation was DLL in 10 cases including six with LP, four were LL, including two with erythema nodosum leprosum (ENL), and two were BL.
In addition to case reports, 10 retrospective specimen surveys have examined 1,260 archived biopsy specimens, detecting M. lepromatosis, M. leprae or dual infection in, respectively, 106 (15.8%), 798 (84.6%), and 28 (3.0%) of 943 PCR-positive specimens (Han et al., 2012b, 2014; Singh et al., 2015; Yuan et al., 2015; Zhang et al., 2015; Kai et al., 2016; Torres-Guerrero et al., 2018; Bezalel et al., 2019; Masood et al., 2019; Sharma et al., 2020). M. lepromatosis was detected in 42.2% (116/275) of PCR-positive specimens from Mexico, in 12.7% (10/79) from Brazil, 4.8% (5/105) from south East Asia (Malaysia, Myanmar, and Philippines), and 1.5% (3/195) from the United States (Table 1). All of 157 specimens from China (Yuan et al., 2015; Zhang et al., 2015), 48 from Mali, and 77 from Venezuela were positive only for M. leprae (Singh et al., 2015).
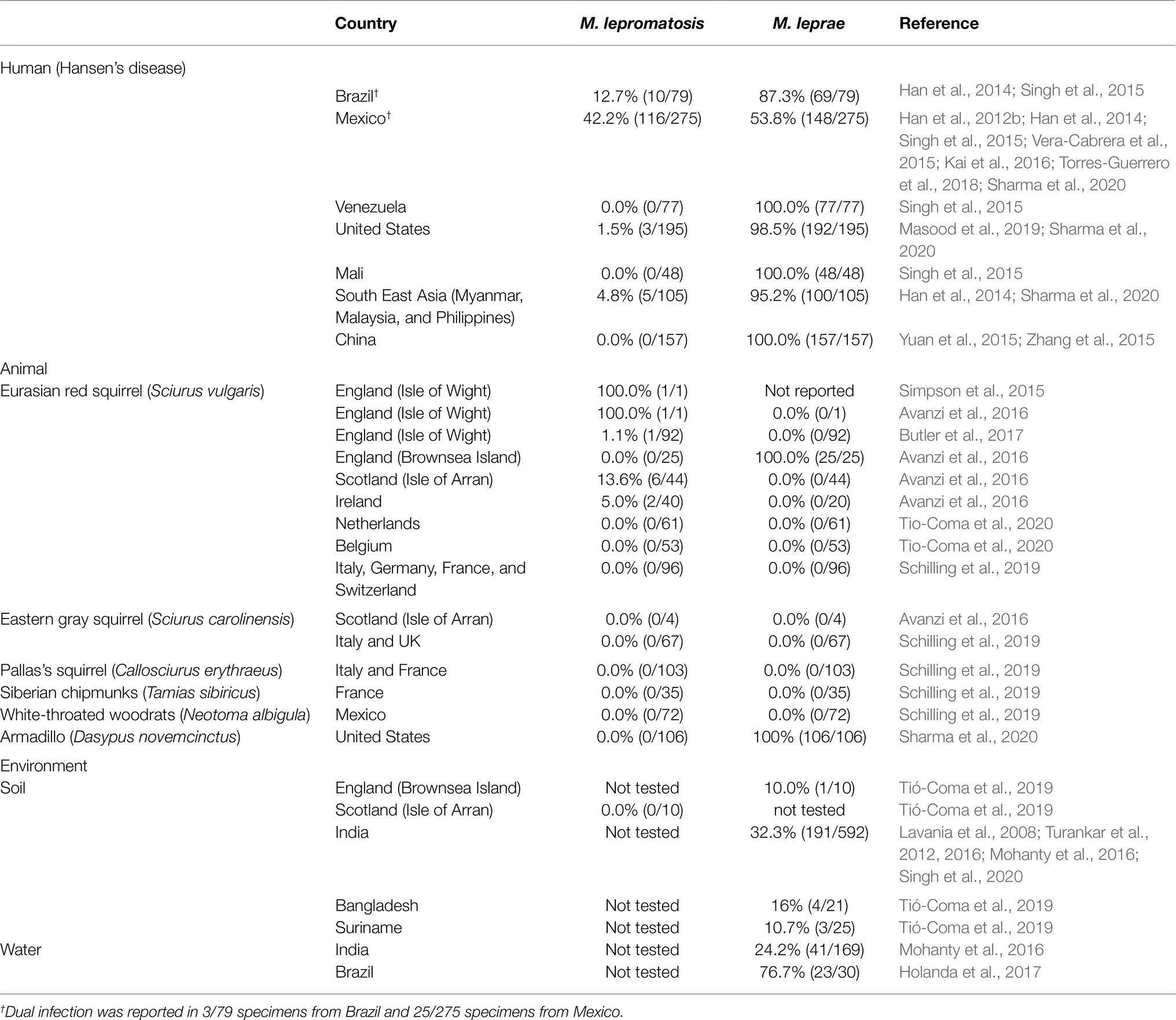
Table 1. Survey findings of Mycobacterium leprae and Mycobacterium lepromatosis in Hansen’s disease and animal and environmental reservoirs.
Case report and specimen survey studies have demonstrated that M. lepromatosis can be found in patients with different forms of HD, including DLL, LL, and BL, and with leprosy reactions, including ENL and LP. Biases in case reporting and archived specimen selection, particularly from regions, such as Mexico where DLL appears to be more common, compounded by possible case misclassification (misdiagnosis), mean that currently available data cannot be used to test the hypothesis that M. lepromatosis is disproportionately associated with DLL and/or LP.
A potentially important methodological limitation of some of these case studies and specimen surveys that warrants an in-depth systematic review with detailed quality assessment is that M. lepromatosis was detected using a range of M. lepromatosis specific primers without confirmation by sequencing.
Genomics and Phylogenetics
Phylogenetic analyses and genome sequencing of the original FJ924 strain (Han et al., 2009, 2015) [NCBI NZ_LAWX01000000] and of two strains of M. lepromatosis from HD cases in Mexico Mx1-22A (GenBank JRPY00000000.1) (Singh et al., 2015; and NHDP-385 (NCBI SAMN12872980) (Sharma et al., 2020), has determined that the M. leprae and M. lepromatosis genomes have near-perfect synteny and their protein-coding genes share 93% nucleotide sequence identity. Phylogenetic analysis indicates divergence from a most recent common ancestor approximately 10–14 millionyears ago (Han et al., 2009; Singh et al., 2015). M. lepromatosis is closest to M. leprae SNP type 3K strains, which represent the most ancestral lineage of M. leprae (Singh et al., 2015). Pseudogenes were ~80% identical between M. leprae and M. lepromatosis, and 84 genomic regions (>500 nucleotides) of M. lepromatosis, representing ~5% of the genome (~166kb) comprising mainly pseudogenes, have no counterparts in M. leprae (Singh et al., 2015). M. lepromatosis and M. leprae share the same repeat families (RLEP, REPLEP, LEPREP, and LEPRPT) at the same genomic locations, but their sequences have diverged substantially ranging from 75 to 90% sequence identity and proportional to copy number (Singh et al., 2015).
The few functional gene differences that have been identified to date do not provide evidence to support differences in the pathogenic properties of the two species, such as a putative greater propensity of M. lepromatosis to invade the cutaneous vascular endothelium (Singh et al., 2015). M. leprae genes encoding laminin-2 binding protein ML1683c and the six enzymes (ML0128, ML2348 ML0126, ML0127, ML23246c, and ML2347) required to produce the terminal trisaccharide moiety of phenolic glycolipid 1 (PGL-1) are highly conserved in M. lepromatosis (Singh et al., 2015), suggesting likely anti-PGL-1 seropositivity in M. lepromatosis HD cases and nerve involvement and damage through invasion of Schwann cells (Ng et al., 2000). One human and one squirrel case of M. lepromatosis infection were reported to be anti-PGL-1 seropositive (Avanzi et al., 2016). The folP1, rpoB, and gyrA drug resistance determining regions have high homology, but with sufficient mutations to require that drug sensitivity techniques developed for M. leprae are validated against M. lepromatosis (Kai et al., 2016; Araujo et al., 2017).
Notable genetic differences between the species included the presence in M. lepromatosis of the coproporphyrinogen III oxidase (hemN) gene, which is present in M. tuberculosis but absent from M. leprae, and relatively large variation in ESX-1 secreted protein genes associated with mycobacterial virulence (Ates et al., 2016). One of these, espA with 78% protein identity (Singh et al., 2015), codes for part of the LID-1 fusion protein developed as a serological test of HD (Duthie et al., 2020), raising the possibility that this test might be less sensitive for M. lepromatosis infection. Given concerns that the 98% identity of 16S rDNA sequences in the two species could yield unreliable PCR results when using primers for this region, the recent development and validation of a unique repetitive element PCR assay for M. lepromatosis (RLPM, equivalent to RLEP M. leprae) provide a reliable diagnostic method with which to investigate further this new species as a causative agent of HD (Sharma et al., 2020). RLPM is an approximately 200bp region of which there are 5–6 copies yielding an approximate limit of detection (LOD) of 3.0 M. lepromatosis bacilli per reaction (compared with LOD ≈0.8 for RLEP ~130bp sequence of which there are 29–36 copies in M. leprae; Sharma et al., 2020). The RLPM assay was demonstrated to be positive for M. lepromatosis but negative against 17 other mycobacterial species, including M. leprae and 10 other mycobacteria associated with human diseases (Sharma et al., 2020).
The RLPM and RLEP assays have been validated according to the Clinical Laboratory Improvement Amendments guidelines (Sharma et al., 2020), and their use should address the limitations described above regarding detection of M. lepromatosis in earlier studies.
Mycobacterium lepromatosis as a Zoonotic Agent
Animal Hosts
In animals, M. lepromatosis was first detected in Eurasian red squirrels (Sciurus vulgaris) from England, Ireland, and Scotland (Avanzi et al., 2016) (NCBI SRR3672737-SRR3672758, SRR3674396-SRR3674450, SRR3674451-SRR3674453, SRR3673933). These findings have not been repeated in Eurasian red, Eastern gray (Sciurus carolinensis), or Pallas’s (Callosciurus erythraeus) squirrels or in Siberian chipmunks (Tamias sibiricus) from other parts of Europe (Schilling et al., 2019; Tio-Coma et al., 2020). The prevalence of M. lepromatosis in the Scottish squirrel population 12.5% (6/48 squirrels) was lower than the 100% prevalence (25/25) of M. leprae in Brownsea Island squirrels (Tió-Coma et al., 2019; Table 1).
Phylogenetic analyses determined that the strain of M. leprae in squirrels in the British Isles was closest to strains that circulated in Medieval England (and which belong to the sequence type 3I branch of M. leprae found in wild armadillos in the United States), whereas the M. lepromatosis strain had diverged around 27,000years ago from a common ancestor of the strain recently identified in HD cases in Mexico (Avanzi et al., 2016).
In the United States, lymph node and spleen specimens from 106 wild 9-banded armadillos (Dasypus novemcinctus) found to be seropositive for M. leprae PGL-1 or LID-1 antigens in a previous study (Sharma et al., 2015) were all PCR positive for M. leprae and negative for M. lepromatosis (Sharma et al., 2020). Schilling et al. (2019) tested 72 Mexican white-throated woodrats (Neotoma albigula) obtained from a meat market in Monterrey, Mexico, for the presence of M. leprae and M. lepromatosis, and all PCR results were negative (Schilling et al., 2019). No studies to date have investigated wild animals in HD endemic countries (Table 1).
In summary, while evidence for armadillos as natural hosts of M. leprae is irrefutable, and natural M. leprae infection has also been detected in small numbers of red squirrels, the latter (from a single site in England) remain the only known natural hosts of M. lepromatosis.
Environmental Reservoirs
In the British Isles, M. leprae-specific DNA was found in 1/10 soil samples from Brownsea Island, an area where M. leprae infection in red squirrels had previously been identified (Avanzi et al., 2016), but M. lepromatosis DNA could not be detected in soil samples from the Isle of Arran, where squirrels had been infected by M. lepromatosis (Avanzi et al., 2016; Tió-Coma et al., 2019). All other studies of water and soil have used methods only for detecting M. leprae (Table 1), providing conclusive evidence for persistence of M. leprae in the environment that is currently lacking for M. lepromatosis.
Zoonotic Transmission
Among the case reports of HD caused by M. lepromatosis, zoonotic sources were suggested in two patients from Guerrero state, Mexico, who were diagnosed several years after emigrating to the United States, both with multibacillary forms of HD (one DLL and one BL). Both patients reported direct contact with armadillos (hunting, handling, and eating) when they lived in Mexico (Sotiriou et al., 2016; Cleary et al., 2017).
Outside the United States, history of contact with armadillos is not often asked of persons presenting with HD. However, in a survey of biopsy specimens from 38 HD patients currently under treatment in Nuevo Léon, Mexico, of 5 patients positive for M. lepromatosis, one DLL case reported eating armadillo and field rat (rattus rattus) meat and one BL case reported field rat meat consumption (Vera-Cabrera et al., 2015).
As previously mentioned, M. leprae in red squirrels in the British Isles is similar to that found in medieval human remains from England and Denmark (Avanzi et al., 2016). One hypothesis is that squirrels served as a zoonotic source for HD in medieval times (Inskip et al., 2017). There was substantial trade in squirrel fur, including between Scandinavia and the British Isles and further afield, with fur being highly prized for use in clothing (Veale, 2003), while squirrel meat was also valued. Only one paleopathological study to date has applied ancient DNA methods for M. leprae (RLEP primer) and M. lepromatosis (135bp hemN gene fragment primer). This was a study of the remains of four people from Ireland dated to the 10th–14th century and one person from the 15th–17th century, all of whom had osteoarchaeological signs of HD (Taylor et al., 2018). PCR was positive for M. leprae in three of the earlier remains and negative for M. lepromatosis in all five remains. Hansen’s disease declined in central Europe from the thirteenth century onward and was almost eliminated by the sixteenth century (Rawcliffe, 2006), leaving a zoonotic transmission hypothesis which is plausible but probably untestable either for M. leprae or M. lepromatosis.
Conclusion
The recent discovery of M. lepromatosis and the attribution to this new species of cases of HD raised some initial doubts (Gillis et al., 2011; Scollard, 2016). While there does now appear to be consensus that M. lepromatosis is a second causal agent of HD (Sharma et al., 2020), a multitude of questions remain. Most of these can be answered using genomic methods to differentiate M. leprae and M. lepromatosis infection (Avanzi et al., 2020; Sharma et al., 2020). The most important immediate question in terms of public health, human disease burden, and HD prevention and control is the extent of M. lepromatosis infection in HD endemic countries and its contribution to HD incidence. In terms of individual persons affected by HD, although treatment outcomes for HD caused by M. lepromatosis appear similar to outcomes for the same forms of HD when caused (presumably) by M. leprae, the small number of M. lepromatosis HD cases described to date leaves a substantial gap in clinical knowledge. On these and the specific question of M. lepromatosis as a possible zoonotic source of HD, we propose the following steps:
1. A systematic review and data synthesis to determine clinical and other characteristics of all reported cases of HD attributed to M. lepromatosis.
2. PCR-based surveillance studies in current and new HD patients (regardless of the form of the disease), particularly in Mexico and Brazil but ideally in all endemic countries.
3. PCR-based surveys in Latin American countries to detect M. lepromatosis in newly captured animals or archived specimens from earlier studies, focusing on species, such as armadillos which are known to harbor M. leprae and which come into contact with humans. Experience with studies investigating M. leprae in wild armadillos in Brazil suggests that substantial variation in prevalence of infection might be expected (Deps et al., 2020); therefore, sufficiently large samples of animals from different geographic locations will be needed.
4. Whole genome sequencing of specimens from animal surveys and surveillance studies in Latin American countries to elucidate strains and determine the relatedness of M. lepromatosis (and M. leprae) in animals and humans (Avanzi et al., 2020).
Although improving our understanding of M. lepromatosis as a cause of clinical disease in humans should be our highest priority, the role of zoonotic transmission in HD caused by M. leprae proves the need to adopt a One Health perspective. This is particularly so in non-endemic countries, such as Mexico, where a large proportion of the small total number of new cases each year (<100 in 2020) could be attributable to zoonotic transmission. Conversely, knowing what proportion of the very large number of cases (~30,000 cases per year) in an endemic country, such as Brazil, might be caused by M. lepromatosis rather than M. leprae, as has been assumed to date, is of fundamental importance to public health.
Author Contributions
PD conceived this article and the research question. SC reviewed the literature and wrote the first draft. PD and SC synthesized and interpreted data from the review, contributed to revisions, and approved the final version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Aldama Olmedo, O. M., Escobar, M., Martínez, M. J., Aldama, M., Montoya Bueno, C., Celias, L. F., et al. (2020). Necrotizing erythema nodosumin lepromatous leprosy associated with mixed infection by mycobacterium lepromatosis and mycobacterium leprae case report. Rev. Del Nacional 12, 107–115. doi: 10.18004/rdn2020.dic.02.107.115
Araujo, S., Goulart, L. R., Truman, R. W., Goulart, I. M. B., Vissa, V., Li, W., et al. (2017). qPCR-high resolution melt analysis for drug susceptibility testing of mycobacterium leprae directly from clinical specimens of leprosy patients. PLoS Negl. Trop. Dis. 11:e0005506. doi: 10.1371/journal.pntd.0005506
Ates, L. S., Houben, E. N. G., and Bitter, W. (2016). Type VII secretion: A highly versatile secretion system. Microbiol. Spectr. 4. doi: 10.1128/microbiolspec.VMBF-0011-2015
Avanzi, C., Del-Pozo, J., Benjak, A., Stevenson, K., Simpson, V. R., Busso, P., et al. (2016). Red squirrels in the British Isles are infected with leprosy bacilli. Science 354, 744–747. doi: 10.1126/science.aah3783
Avanzi, C., Singh, P., Truman, R. W., and Suffys, P. N. (2020). Molecular epidemiology of leprosy: An update. Infect. Genet. Evol. 86:104581. doi: 10.1016/j.meegid.2020.104581
Bezalel, S. A., Onajin, O., Gonzalez-Santiago, T. M., Patel, R., Pritt, B. S., Virk, A., et al. (2019). Leprosy in a Midwestern dermatology clinic: report of 9 patients. Mayo Clin. Proc. 94, 417–423. doi: 10.1016/j.mayocp.2018.11.022
Butler, H. M., Stevenson, K., Mcluckie, J., and Simpson, V. (2017). Further evidence of leprosy in isle of Wight red squirrels. Vet. Rec. 180, 407–407. doi: 10.1136/vr.j1920
Centers for Disease Control and Prevention (2020). Hansen's Disease (Leprosy) (Online). Atlanta (United States): CDC. Available at: https://www.cdc.gov/leprosy/index.html (Accessed July 18, 2021).
Cleary, L. C., Suraj, S., Haburchak, D., and Turrentine, J. E. (2017). The armadillo factor: Lepromatous leprosy. Am. J. Med. 130, 1163–1166. doi: 10.1016/j.amjmed.2017.06.001
Deps, P., Antunes, J., and Collin, S. M. (2021). Zoonotic risk of Hansen's disease from community contact with wild armadillos: A systematic review and meta-analysis. Zoo. Public Health 68, 153–164. doi: 10.1111/zph.12783
Deps, P., Antunes, J. M., Santos, A. R., and Collin, S. M. (2020). Prevalence of mycobacterium leprae in armadillos in Brazil: A systematic review and meta-analysis. PLoS Negl. Trop. Dis. 14:e0008127. doi: 10.1371/journal.pntd.0008127
Duthie, M. S., Pena, M. T., Khandhar, A. P., Picone, A., Mac, M. Z., Truman, R. W., et al. (2020). Development of LepReact, a defined skin test for paucibacillary leprosy and low-level M. leprae infection. Appl. Microbiol. Biotechnol. 104, 3971–3979. doi: 10.1007/s00253-020-10505-2
Flores-Suarez, L. F., Fernandez-Sanchez, M., Ahumada, V., and Charli-Joseph, Y. (2019). After All, still a magnificent impersonator. Rheumatology 60, e245–e246. doi: 10.1093/rheumatology/keab077
Gillis, T. P., Scollard, D. M., and Lockwood, D. N. J. (2011). What is the evidence that the putative mycobacterium lepromatosis species causes diffuse lepromatous leprosy? Lepr. Rev. 82, 205–209. doi: 10.47276/lr.82.3.205
Han, X. Y., Aung, F. M., Choon, S. E., and Werner, B. (2014). Analysis of the leprosy agents mycobacterium leprae and mycobacterium lepromatosis in four countries. Am. J. Clin. Pathol. 142, 524–532. doi: 10.1309/AJCP1GLCBE5CDZRM
Han, X. Y., and Jessurun, J. (2013). Severe leprosy reactions due to mycobacterium lepromatosis. Am J Med Sci 345, 65–69. doi: 10.1097/MAJ.0b013e31826af5fb
Han, X. Y., Mistry, N. A., Thompson, E. J., Tang, H. L., Khanna, K., and Zhang, L. (2015). Draft genome sequence of new leprosy agent mycobacterium lepromatosis. Genome Announc. 3:e00513-15. doi: 10.1128/genomeA.00513-15
Han, X. Y., and Quintanilla, M. (2015). Diffuse Lepromatous leprosy due to mycobacterium lepromatosis in Quintana Roo, Mexico. J. Clin. Microbiol. 53, 3695–3698. doi: 10.1128/JCM.01951-15
Han, X. Y., Seo, Y. H., Sizer, K. C., Schoberle, T., May, G. S., Spencer, J. S., et al. (2008). A new mycobacterium species causing diffuse lepromatous leprosy. Am. J. Clin. Pathol. 130, 856–864. doi: 10.1309/AJCPP72FJZZRRVMM
Han, X. Y., Sizer, K. C., and Tan, H. H. (2012a). Identification of the leprosy agent mycobacterium lepromatosis in Singapore. J. Drugs Dermatol. 11, 168–172.
Han, X. Y., Sizer, K. C., Thompson, E. J., Kabanja, J., Li, J., Hu, P., et al. (2009). Comparative sequence analysis of mycobacterium leprae and the new leprosy-causing mycobacterium lepromatosis. J. Bacteriol. 191, 6067–6074. doi: 10.1128/JB.00762-09
Han, X. Y., Sizer, K. C., Velarde-Felix, J. S., Frias-Castro, L. O., and Vargas-Ocampo, F. (2012b). The leprosy agents mycobacterium lepromatosis and mycobacterium leprae in Mexico. Int. J. Dermatol. 51, 952–959. doi: 10.1111/j.1365-4632.2011.05414.x
Hockings, K. J., Mubemba, B., Avanzi, C., Pleh, K., Düx, A., Bersacola, E., et al. (2020). Leprosy in wild chimpanzees. BioRxiv 80, 29–36. doi: 10.1101/2020.11.10.374371
Holanda, M. V., Marques, L. E. C., Macedo, M. L. B., Pontes, M. A. A., Sabadia, J. A. B., Kerr, L., et al. (2017). Presence of mycobacterium leprae genotype 4 in environmental waters in Northeast Brazil. Rev. Soc. Bras. Med. Trop. 50, 216–222. doi: 10.1590/0037-8682-0424-2016
Htet, L., Kai, M., and Miyamoto, Y. (2018). New etiology of leprosy in Myanmar: another two patients. Lepr. Rev. 89, 316–318. doi: 10.47276/lr.89.3.316
Inskip, S., Taylor, G. M., Anderson, S., and Stewart, G. (2017). Leprosy in pre-Norman Suffolk, UK: biomolecular and geochemical analysis of the woman from Hoxne. J. Med. Microbiol. 66, 1640–1649. doi: 10.1099/jmm.0.000606
Jessamine, P. G., Desjardins, M., Gillis, T., Scollard, D., Jamieson, F., Broukhanski, G., et al. (2012). Leprosy-like illness in a patient with mycobacterium lepromatosis from Ontario, Canada. J. Drugs Dermatol. 11, 229–233.
Kai, M., Fafutis-Morris, M., Miyamoto, Y., Mukai, T., Mayorga-Rodriguez, J., Rodriguez-Castellanos, M. A., et al. (2016). Mutations in the drug resistance-determining region of mycobacterium lepromatosis isolated from leprosy patients in Mexico. J. Dermatol. 43, 1345–1349. doi: 10.1111/1346-8138.13483
Lavania, M., Katoch, K., Katoch, V. M., Gupta, A. K., Chauhan, D. S., Sharma, R., et al. (2008). Detection of viable Mycobacterium leprae in soil samples: insights into possible sources of transmission of leprosy. Infect. Genet. Evol. 8, 627–631. doi: 10.1016/j.meegid.2008.05.007
Masood, M., Altieri, L., Dasco, M., Sharma, R., and Ochoa, M. T. (2019). Mycobacterium lepromatosis: A novel cause of erythema nodosum leprosum. J. Am. Acad. Dermatol. 81:Ab243.
Mohanty, P. S., Naaz, F., Katara, D., Misba, L., Kumar, D., Dwivedi, D. K., et al. (2016). Viability of mycobacterium leprae in the environment and its role in leprosy dissemination. Indian J. Dermatol. Venereol. Leprol. 82, 23–27. doi: 10.4103/0378-6323.168935
Ng, V., Zanazzi, G., Timpl, R., Talts, J. F., Salzer, J. L., Brennan, P. J., et al. (2000). Role of the cell wall phenolic glycolipid-1 in the peripheral nerve predilection of mycobacterium leprae. Cell 103, 511–524. doi: 10.1016/S0092-8674(00)00142-2
Oliveira, I., Deps, P. D., and Antunes, J. (2019). Armadillos and leprosy: from infection to biological model. Rev. Inst. Med. Trop. Sao Paulo 61:e44. doi: 10.1590/s1678-9946201961044
Ploemacher, T., Faber, W. R., Menke, H., Rutten, V., and Pieters, T. (2020). Reservoirs and transmission routes of leprosy; A systematic review. PLoS Negl. Trop. Dis. 14:e0008276. doi: 10.1371/journal.pntd.0008276
Schilling, A. K., Avanzi, C., Ulrich, R. G., Busso, P., Pisanu, B., Ferrari, N., et al. (2019). British red squirrels remain the only known wild rodent host for leprosy bacilli. Front. Vet. Sci. 6:8. doi: 10.3389/fvets.2019.00008
Scollard, D. M. (2016). Infection with mycobacterium lepromatosis. Am. J. Trop. Med. Hyg. 95, 500–501. doi: 10.4269/ajtmh.16-0473
Sharma, R., Lahiri, R., Scollard, D. M., Pena, M., Williams, D. L., Adams, L. B., et al. (2013). The armadillo: a model for the neuropathy of leprosy and potentially other neurodegenerative diseases. Dis. Model. Mech. 6, 19–24. doi: 10.1242/dmm.010215
Sharma, R., Singh, P., Loughry, W. J., Lockhart, J. M., Inman, W. B., Duthie, M. S., et al. (2015). Zoonotic leprosy in the Southeastern United States. Emerg. Infect. Dis. 21, 2127–2134. doi: 10.3201/eid2112.150501
Sharma, R., Singh, P., Mccoy, R. C., Lenz, S. M., Donovan, K., Ochoa, M. T., et al. (2020). Isolation of mycobacterium lepromatosis and development of molecular diagnostic assays to distinguish mycobacterium leprae and M. lepromatosis. Clin. Infect. Dis. 71, e262–e269. doi: 10.1093/cid/ciz1121
Simpson, V., Hargreaves, J., Butler, H., Blackett, T., Stevenson, K., and Mcluckie, J. (2015). Leprosy in red squirrels on the Isle of Wight and Brownsea Island. Vet. Rec. 177, 206–207. doi: 10.1136/vr.h4491
Singh, P., Benjak, A., Schuenemann, V. J., Herbig, A., Avanzi, C., Busso, P., et al. (2015). Insight into the evolution and origin of leprosy bacilli from the genome sequence of mycobacterium lepromatosis. Proc. Natl. Acad. Sci. U. S. A. 112, 4459–4464. doi: 10.1073/pnas.1421504112
Singh, V., Turankar, R. P., and Goel, A. (2020). Real-time PCR-based quantitation of viable mycobacterium leprae strain from clinical samples and environmental sources and its genotype in multi-case leprosy families of India. Eur. J. Clin. Microbiol. Infect. Dis. 39, 2045–2055. doi: 10.1007/s10096-020-03958-w
Sotiriou, M. C., Stryjewska, B. M., and Hill, C. (2016). Two cases of leprosy in siblings caused by mycobacterium lepromatosis and review of the literature. Am. J. Trop. Med. Hyg. 95, 522–527. doi: 10.4269/ajtmh.16-0076
Storrs, E. E. (1971). The nine-banded armadillo: a model for leprosy and other biomedical research. Int. J. Lepr. Other Mycobact. Dis. 39, 703–714.
Taylor, G. M., Murphy, E. M., Mendum, T. A., Pike, A. W. G., Linscott, B., Wu, H., et al. (2018). Leprosy at the edge of Europe-biomolecular, isotopic and osteoarchaeological findings from medieval Ireland. PLoS One 13:e0209495. doi: 10.1371/journal.pone.0209495
Tio-Coma, M., Sprong, H., Kik, M., Van Dissel, J. T., Han, X. Y., Pieters, T., et al. (2020). Lack of evidence for the presence of leprosy bacilli in red squirrels from North-West Europe. Transbound. Emerg. Dis. 67, 1032–1034. doi: 10.1111/tbed.13423
Tió-Coma, M., Wijnands, T., Pierneef, L., Schilling, A. K., Alam, K., Roy, J. C., et al. (2019). Detection of mycobacterium leprae DNA in soil: multiple needles in the haystack. Sci. Rep. 9:3165. doi: 10.1038/s41598-019-39746-6
Torres-Guerrero, E., Sanchez-Moreno, E. C., Atoche-Dieguez, C. E., Carrillo-Casas, E. M., Arenas, R., Xicohtencatl-Cortes, J., et al. (2018). Identification of mycobacterium leprae and mycobacterium lepromatosis in formalin-fixed and paraffin-embedded skin samples from Mexico. Ann. Dermatol. 30, 562–565. doi: 10.5021/ad.2018.30.5.562
Trave, I., Barabino, G., Cavalchini, A., and Parodi, A. (2020). Long-term ulcerations caused by mycobacterium lepromatosis. Int. J. Mycobacteriol. 9, 223–225. doi: 10.4103/ijmy.ijmy_40_20
Truman, R. W., Singh, P., Sharma, R., Busso, P., Rougemont, J., Paniz-Mondolfi, A., et al. (2011). Probable zoonotic leprosy in the southern United States. N. Engl. J. Med. 364, 1626–1633. doi: 10.1056/NEJMoa1010536
Turankar, R. P., Lavania, M., Singh, M., Sengupta, U., Siva Sai, K., and Jadhav, R. S. (2016). Presence of viable Mycobacterium leprae in environmental specimens around houses of leprosy patients. Indian J. Med. Microbiol. 34, 315–321. doi: 10.4103/0255-0857.188322
Turankar, M., Singh, M., Siva Sai, K. S., and Jadhav, R. S. (2012). Dynamics of Mycobacterium leprae transmission in environmental context: deciphering the role of environment as a potential reservoir. Infect. Genet. Evol. 12, 121–126. doi: 10.1016/j.meegid.2011.10.023
Velarde-Felix, J. S., Alvarado-Villa, G., and Vera-Cabrera, L. (2016). "Lucio's phenomenon" associated with mycobacterium lepromatosis. Am. J. Trop. Med. Hyg. 94, 483–484. doi: 10.4269/ajtmh.15-0439
Vera-Cabrera, L., Escalante-Fuentes, W. G., Gomez-Flores, M., Ocampo-Candiani, J., Busso, P., Singh, P., et al. (2011). Case of diffuse lepromatous leprosy associated with "mycobacterium lepromatosis". J. Clin. Microbiol. 49, 4366–4368. doi: 10.1128/JCM.05634-11
Vera-Cabrera, L., Escalante-Fuentes, W., Ocampo-Garza, S. S., Ocampo-Candiani, J., Molina-Torres, C. A., Avanzi, C., et al. (2015). Mycobacterium lepromatosis infections in Nuevo Leon, Mexico. J. Clin. Microbiol. 53, 1945–1946. doi: 10.1128/JCM.03667-14
Virk, A., Pritt, B., Patel, R., Uhl, J. R., Bezalel, S. A., Gibson, L. E., et al. (2017). Mycobacterium lepromatosis Lepromatous leprosy in US citizen who Traveled to disease-endemic areas. Emerg. Infect. Dis. 23, 1864–1866. doi: 10.3201/eid2311.171104
Watson, W., Vassantachart, J. M., and Luke, J. (2020). Clinicopathological challenge: acute blistering and dermal papules in a patient with scleroderma. Int. J. Dermatol. 59, e99–e101. doi: 10.1111/ijd.14571
Widiatma, R. R., and Sukanto, H. (2019). Diffuse lepromatous leprosy caused by dual infection of mycobacterium leprae and mycobacterium lepromatosis: A case report. Dermatol. Rep. 11, 180–182. doi: 10.4081/dr.2019.8094
Yuan, Y., Wen, Y., You, Y., Xing, Y., Li, H., Weng, X., et al. (2015). Characterization of mycobacterium leprae genotypes in China--identification of a new polymorphism C251T in the 16S rRNA gene. PLoS One 10:e0133268. doi: 10.1371/journal.pone.0144773
Keywords: Mycobacterium lepromatosis, Mycobacterium leprae, Hansen’s disease, zoonosis, leprosy
Citation: Deps P and Collin SM (2021) Mycobacterium lepromatosis as a Second Agent of Hansen’s Disease. Front. Microbiol. 12:698588. doi: 10.3389/fmicb.2021.698588
Edited by:
Moises Batista Da Silva, Federal University of Pará, BrazilReviewed by:
Linda B. Adams, The National Hansen's Disease Programs, United StatesVerena Schuenemann, University of Zurich, Switzerland
Pushpendra Singh, National Institute for Research in Tribal Health (ICMR), India
Copyright © 2021 Deps and Collin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Patrícia Deps, patricia.deps@ufes.br
 Patrícia Deps
Patrícia Deps Simon M. Collin
Simon M. Collin