- 1State Key Laboratory of Biocontrol, Guangdong Provincial Key Laboratory of Plant Resources and Southern Marine Science and Engineering Guangdong Laboratory (Zhuhai), School of Life Sciences, Sun Yat-sen University, Guangzhou, China
- 2Department of Pathogenic Biology, School of Basic Medical Sciences, Binzhou Medical University, Yantai, China
- 3Henan Key Laboratory of Industrial Microbial Resources and Fermentation Technology, College of Biological and Chemical Engineering, Nanyang Institute of Technology, Nanyang, China
- 4Bioproducts Research Chair, Zoology Department, College of Science, King Saud University, Riyadh, Saudi Arabia
- 5Botany and Microbiology Department, Faculty of Science, Beni-Suef University, Beni-Suef, Egypt
- 6State Key Laboratory of Desert and Oasis Ecology, Xinjiang Institute of Ecology and Geography, Chinese Academy of Sciences, Urumqi, China
In the present study, physicochemical and microbial diversity analyses of seven Indian hot springs were performed. The temperature at the sample sites ranged from 32 to 67°C, and pH remained neutral to slightly alkaline. pH and temperature influenced microbial diversity. Culture-independent microbial diversity analysis suggested bacteria as the dominant group (99.3%) when compared with the archaeal group (0.7%). Alpha diversity analysis showed that microbial richness decreased with the increase of temperature, and beta diversity analysis showed clustering based on location. A total of 131 strains (divided into 12 genera and four phyla) were isolated from the hot spring samples. Incubation temperatures of 37 and 45°C and T5 medium were more suitable for bacterial isolation. Some of the isolated strains shared low 16S rRNA gene sequence similarity, suggesting that they may be novel bacterial candidates. Some strains produced thermostable enzymes. Dominant microbial communities were found to be different depending on the culture-dependent and culture-independent methods. Such differences could be attributed to the fact that most microbes in the studied samples were not cultivable under laboratory conditions. Culture-dependent and culture-independent microbial diversities suggest that these springs not only harbor novel microbial candidates but also produce thermostable enzymes, and hence, appropriate methods should be developed to isolate the uncultivated microbial taxa.
Introduction
Hot springs are the places where warm or hot groundwater comes out from the Earth (Narsing Rao et al., 2018). Hot springs were once considered a sterile area (Chaudhuri et al., 2017), but Thomas Brock’s groundbreaking work in discovering Thermus aquaticus from a thermal environment (Brock and Freeze, 1969; Brock, 1997) has fully altered our perception of the microbial diversity of hot springs. Researchers around the world have begun to study similar ecosystems using a culture-dependent approach to understand microbial diversity (Baker et al., 2001; Pathak and Rathod, 2014; Liu et al., 2016). For decades, microbial diversity analysis was carried out by the traditional culture-dependent method; however, this method has several disadvantages. Most of the microorganisms in this method remain hidden or difficult to grow (Kumar et al., 2004; Najar et al., 2018; Narsing Rao et al., 2018).
Next-generation sequencing allows for culture-free microbial diversity detection (Sabat et al., 2017), based on molecular phylogeny of the small-subunit ribosomal RNA gene (16S rRNA gene) (Myer et al., 2016). In the past few years, this method played an important role in understanding the microbial diversity of various ecological niches (Ghelani et al., 2015; De Mandal et al., 2017; Anguita-Maeso et al., 2020).
India harbors approximately 400 geothermal springs (Pednekar et al., 2011). People bathe in these hot springs to cure diseases (Pednekar et al., 2011; Narsing Rao et al., 2018). Culture-dependent microbial diversity analysis of Indian hot springs suggested that they harbor novel candidates and produce thermostable secondary metabolites. A novel genus, Emticicia, was reported from an Indian (Assam) warm water spring (Saha and Chakrabarti, 2006b). A moderately thermophilic, thiosulfate-oxidizing novel species Thiomonas bhubaneswarensis was reported from an Atri hot spring (Bhubaneswar, India) (Panda et al., 2009). The high arsenate-tolerant novel species Pannonibacter indica was reported from an Athamallik (Orissa, India) hot spring sediment (Bandyopadhyay et al., 2013). Antimicrobial ability of bacterial strains isolated from a Maharashtra (India) hot spring was also reported (Pednekar et al., 2011). A novel species, Thermus parvatiensis, isolated from the hot water spring of Manikaran, India, was reported to produce a thermostable enzyme (protease activity at 70°C) (Dwivedi et al., 2015).
Similarly, culture-independent microbial diversity analysis of Indian hot spring samples was also carried out suggesting that they have diverse microbial diversity and that many microbial communities are still unclassified (Ghelani et al., 2015; Chaudhuri et al., 2017). Although both culture-dependent and culture-independent microbial analyses have been carried out separately to understand the microbial diversity of Indian hot springs (Kumar et al., 2004; Ghelani et al., 2015; Chaudhuri et al., 2017), only a few studies performed both analyses together (Najar et al., 2018). In the present study, we performed both culture-dependent and culture-independent microbial analyses of seven Indian hot springs. We further analyzed the various physicochemical parameters that govern microbial diversity.
Materials and Methods
Description of the Sample Sites and Physicochemical Analysis
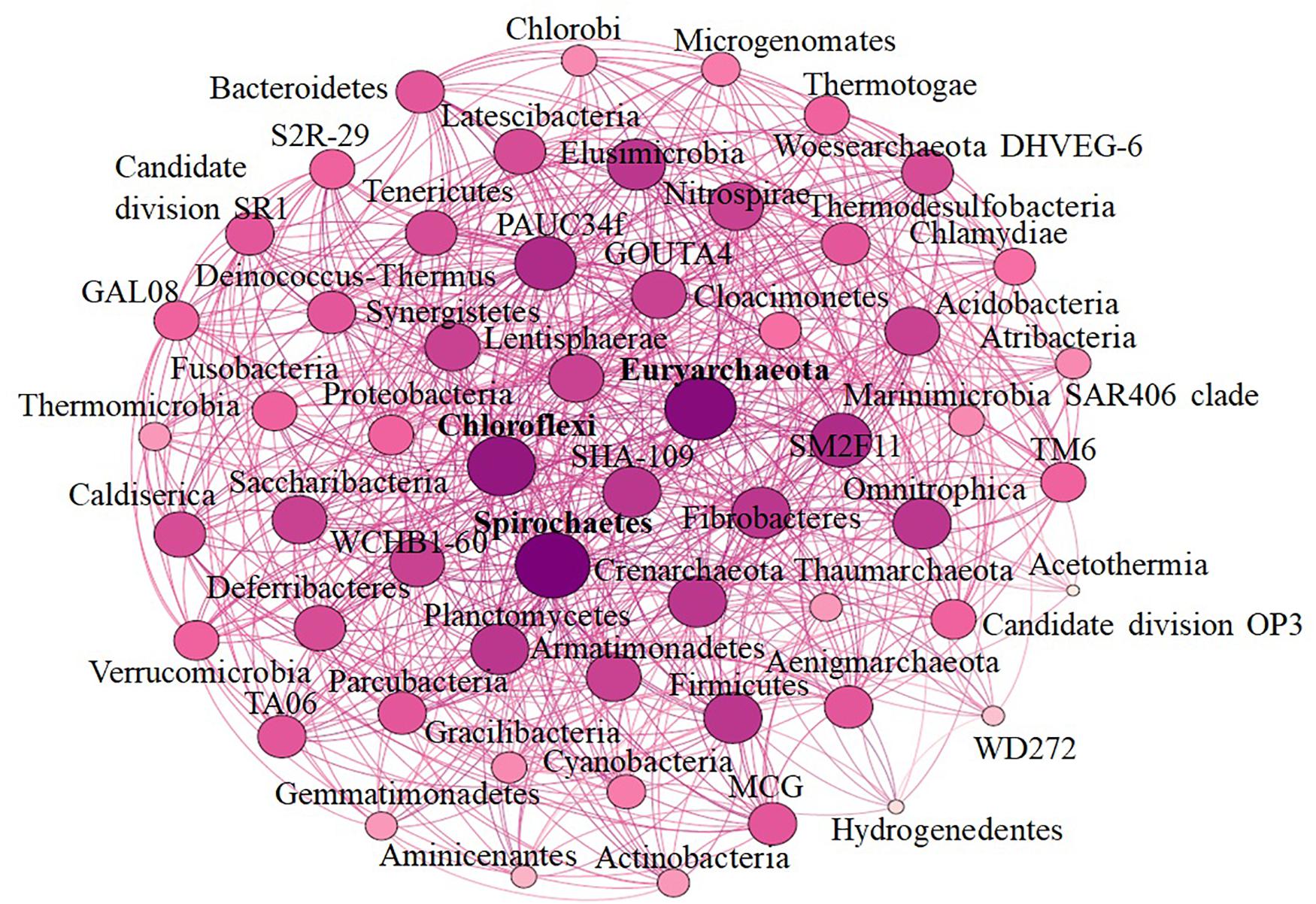
Seven hot spring samples from three Indian provinces, namely, Karnataka, Maharashtra, and Telangana, were included in the present study (Figure 1). Four hot spring samples were collected from Maharashtra Province located at Vajreshwari (VAJ) (19°29′13.2″N, 73°01′40.8″E), Akaloli (AKA) (19°29′24.7″N, 73°02′20.8″E), Ganeshpuri (GAN) (19°30′05.0″N, 73°00′47.5″E), and Sativali (SATI) (19°37′51.0″N, 72°54′32.4″E). The hot spring located in Karnataka Province is called Bendru Theertha (BT) located in the village Irde in Puttur Taluka (12°45′53.3″N, 75°11′03.1″E). The hot spring of Tuwa (TUW) (22°47′58″N, 73°27′37″E) is situated in Panchmahal District, Gujarat. The hot spring at Bhadrachalam (BHA) (17°40′07.7″N, 80°53′37.0″E), Telangana Province in Gundala is located on the bed of the Godavari River.
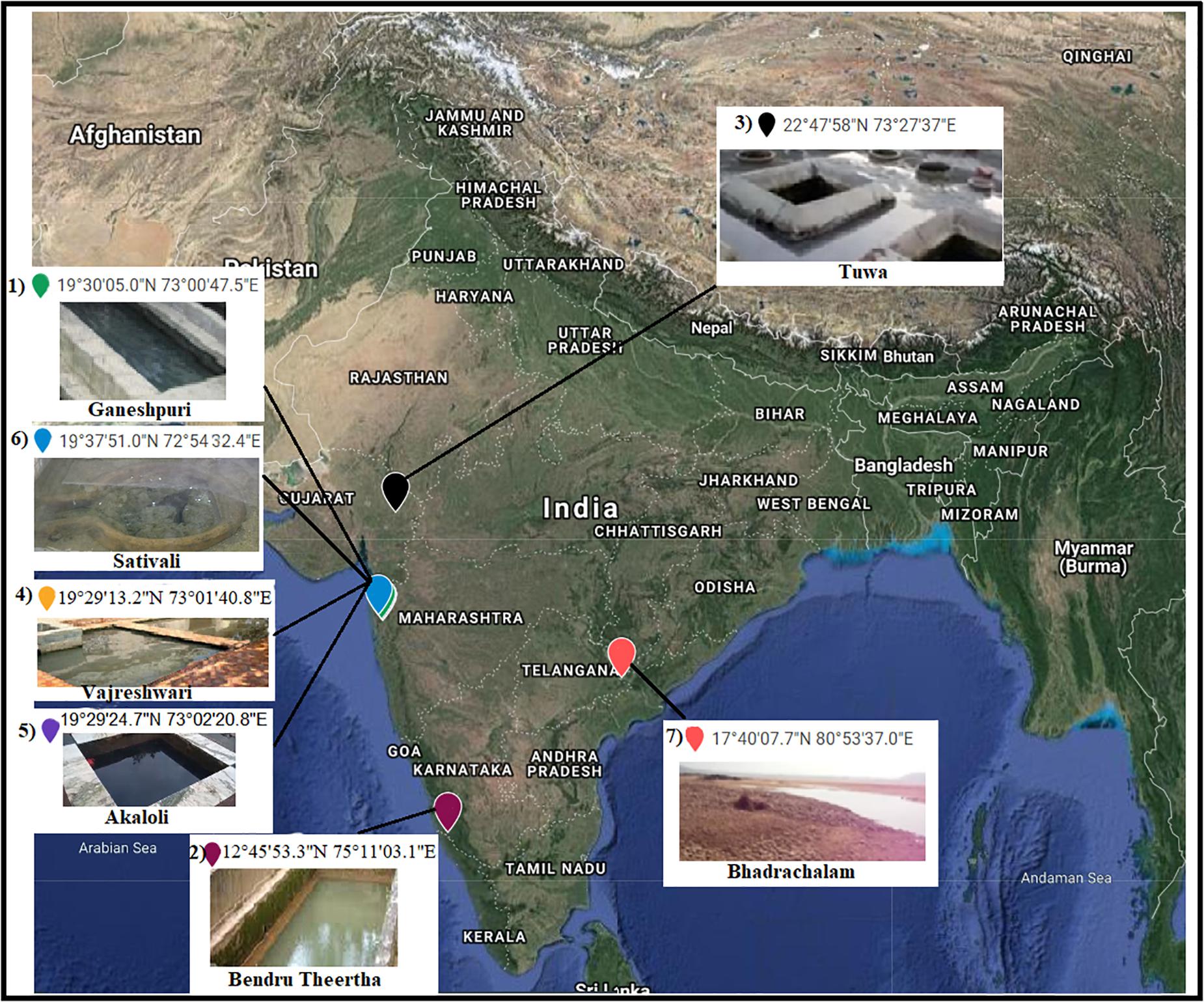
Figure 1. Sample sites. 1, Ganeshpuri; 2, Bendru Theertha; 3, Tuwa; 4, Vajreshwari; 5, Akaloli; 6 Sativali; 7, Bhadrachalam.
Water temperature and pH were measured using a portable digital thermometer and pH meter. Samples for culture-dependent analysis were collected in sterilized tubes kept in the dark, while samples for culture-independent analysis were kept in dry ice until brought to the laboratory.
Culture-Independent Microbial Diversity and Statistical Analysis
Community genomic DNA from the sediment samples were extracted using the PowerSoil DNA isolation kit (MoBio) as per the manufacturer’s instructions. PCR was carried out by the Phusion® High-Fidelity PCR Master Mix with GC Buffer (New England Biolabs) using specific primers (16S V4 region primers; 515F and 806R) (Caporaso et al., 2011) with a barcode (12 nt). The libraries were constructed using the TruSeq® DNA PCR-free sample preparation kit and sequenced using the HiSeq 2500 platform (Illumina). According to the barcode sequence, data samples were separated. The paired-end reads were assembled into single reads using USEARCH 11 (Edgar, 2010). The raw reads were filtered (Bokulich et al., 2013) referring to the quality control process of QIIME 2 (Bolyen et al., 2019). Chimeras were removed using UCHIME (Edgar et al., 2011). Zero radius operational taxonomic unit (ZOTU) clustering was performed using the UPARSE pipeline (Edgar, 2013), and the taxonomic assignment was performed against the SILVA database (release 132) (Quast et al., 2013). The multisequence alignment was performed using the MUSCLE (version 3.8.31) (Edgar, 2004) software to obtain a systematic relationship of all ZOTU representative sequences. The QIIME 2 software was used to calculate the observed species (Bolyen et al., 2019), and the R software (version 2.15.3)1 was used to draw the species accumulation curve. The UniFrac distance was calculated using the QIIME 2 software (Bolyen et al., 2019) and the UPGMA sample clustering tree was constructed. The principal coordinate analysis (PCoA) map was drawn by the R software (see text footnote 1) using the WGCNA, stats, and ggplot2 software packages (Zhang and Horvath, 2005; Wickham, 2016). The correlation between ZOTUs was made using the sparse correlations for the compositional (SparCC) data algorithm implemented in the Python module (Friedman and Alm, 2012) with bootstraps and p-values of 1,000 and 0.01, respectively. The corresponding network was plotted using the R package igraph (Csardi and Nepusz, 2006).
Culture-Dependent Microbial Diversity Analysis
According to our pre-laboratory experience of cultivating high-temperature microorganisms (Liu et al., 2016; Khan et al.,2017a,b; Habib et al., 2018), T5, R2A, ISP5, CC, TSA, and TH media were used for the isolation. Their composition details are mentioned in Supplementary Table S1.
For culture-dependent microbial diversity analysis, the samples were serially diluted. About 100 μl of the serially diluted samples were spread on six different media and incubated at temperatures of 37, 45, 55, and 70°C. The isolates obtained were picked and re-streaked on the same isolation media until pure colonies were obtained. The isolated strains were identified using 16S rRNA gene sequencing. Genomic DNA extraction and PCR amplification of the 16S rRNA gene sequence was performed as described by Li et al. (2007). The obtained 16S rRNA gene sequence was compared with available sequences of cultured species from the EzTaxon database (Yoon et al., 2017).
Screening for Enzyme Production
The isolated organisms were screened for the presence of industrially important enzymes like amylase, protease, cellulase, and xylanase. Amylase activity was determined by inoculating bacterial strains on starch agar (Difco). The hydrolysis of starch was detected by flooding the plates with an iodine solution. The clear zone around the colony was positive for amylase activity. Protease activity was determined on skim milk agar (Sigma-Aldrich). The zone of hydrolysis around the colony was regarded as positive for protease activity.ı Cellulase activity was determined by incorporating 1% carboxymethylcellulose in tryptic soy agar (Difco). Congo red was used as an indicator for determining carboxymethylcellulose hydrolysis (Sazci et al., 1986).
Data Availability and Accession Numbers
Raw reads were submitted to the NCBI Sequence Read Archive (SRA) under accession numbers SRR10420990–SRR10420996.
Results
Physicochemical Analysis of the Sample Sites
The temperature at the sample sites ranged from 32 to 67°C. The highest temperature was recorded at SATI (67°C), while the lowest was at VAJ (32°C) (Table 1). The pH at all the sample sites remained neutral to slightly alkaline (Table 1).
Culture-Independent Microbial Diversity Analysis
A total of 504,226 high-quality clean reads from 536,302 raw reads from seven samples were obtained and grouped into 2,965 ZOTUs. About 99.3% of ZOTUs were assigned as bacteria and 0.7% of ZOTUs were assigned as archaea. Figure 2 shows the relative abundance of the top 10 bacterial ZOTUs at different sample sites. Overall, in all the sample sites, phylum Proteobacteria was dominant. The other dominant phyla were Firmicutes, Actinobacteria, Chloroflexi, Deinococcus–Thermus, Bacteroidetes, Cyanobacteria, Acidobacteria, Armatimonadetes, and Spirochaetes, but their proportion in each sample varied (Figure 2A). The BHA hot spring sample had a high relative abundance of Proteobacteria and Firmicutes when compared with other samples. The GAN and SATI hot spring samples had a high relative abundance of Deinococcus–Thermus, while the VAJ and SATI samples had a high relative abundance of Cyanobacteria (Figure 2A). At the genus level, Escherichia and Shigella dominated the BHA sample site. Meiothermus dominated at the GAN and SATI sample sites. Acinetobacter dominated at the TUW and BT sample sites, while Bacillus dominated at the AKA and VAJ sample sites (Figure 2B).
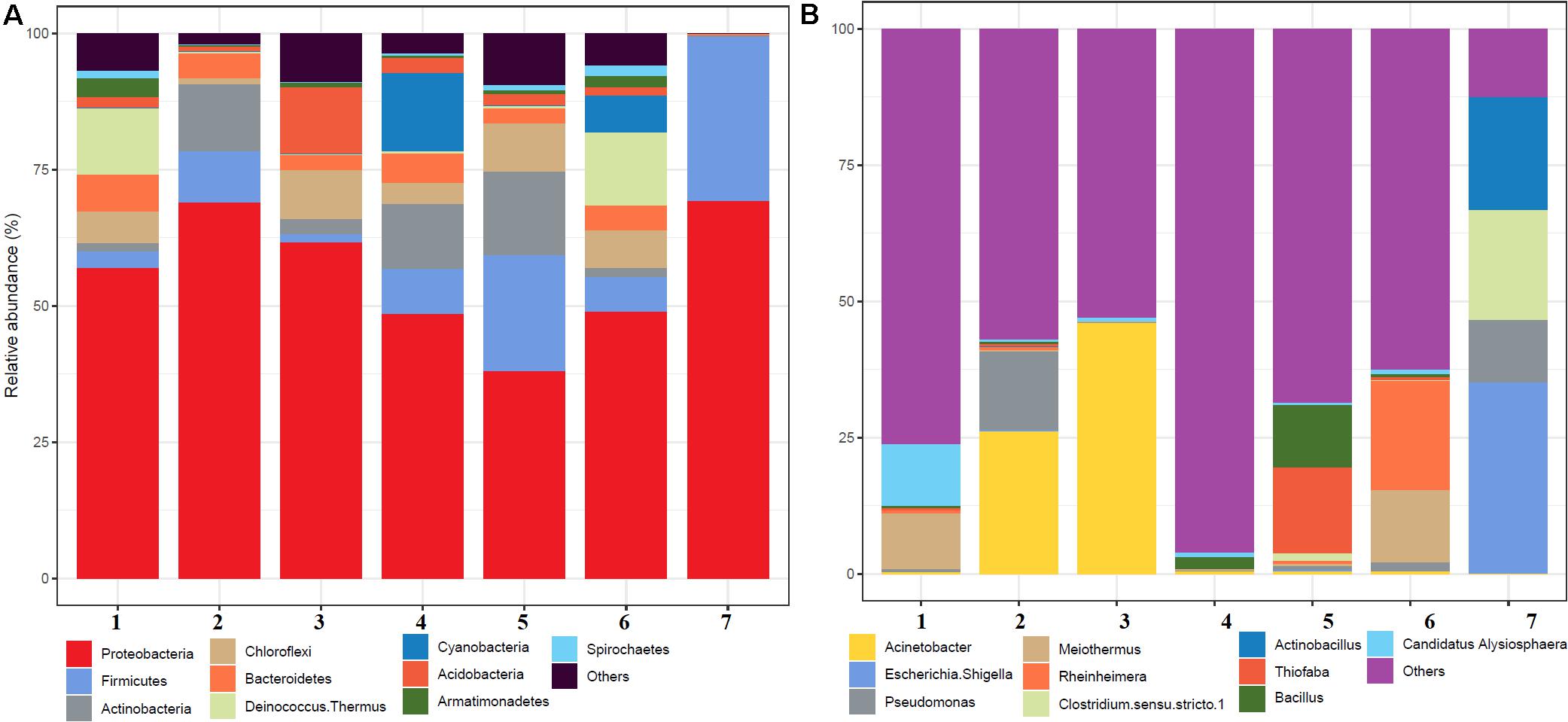
Figure 2. Relative abundance of the top 10 microbial communities based on the culture-independent method: (A) phylum level and (B) genus level. 1, Ganeshpuri; 2, Bendru Theertha; 3, Tuwa; 4, Vajreshwari; 5, Akaloli; 6 Sativali; 7, Bhadrachalam.
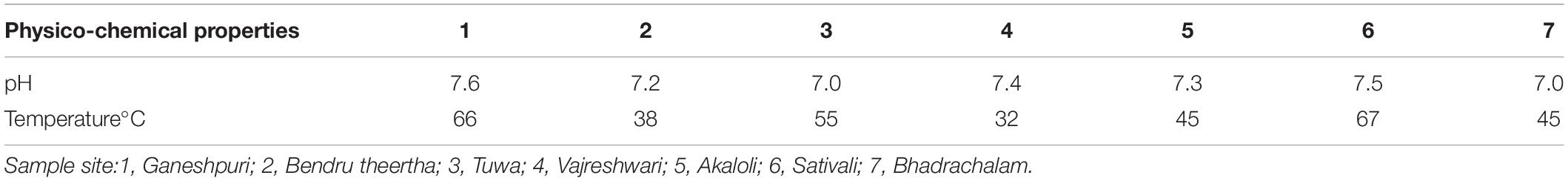
Alpha diversity analysis showed that sample sites with low temperatures had a high taxonomic richness and the richness decreased with the increase of temperature (Figure 3A), with one exception (BHA sample site). Further, the petal diagram (Figure 3B) showed that the sample sites with low temperature had more unique ZOTUs when compared with high-temperature sample sites. Beta diversity analysis (Figures 4A,B) showed that clustering based on location (GAN, VAJ, and SATI) and their relative abundance at the phylum level were almost the same. Network analysis (Figure 5) showed that phyla Chloroflexi, Spirochaetes, and Euryarchaeota had a high correlation with other members.
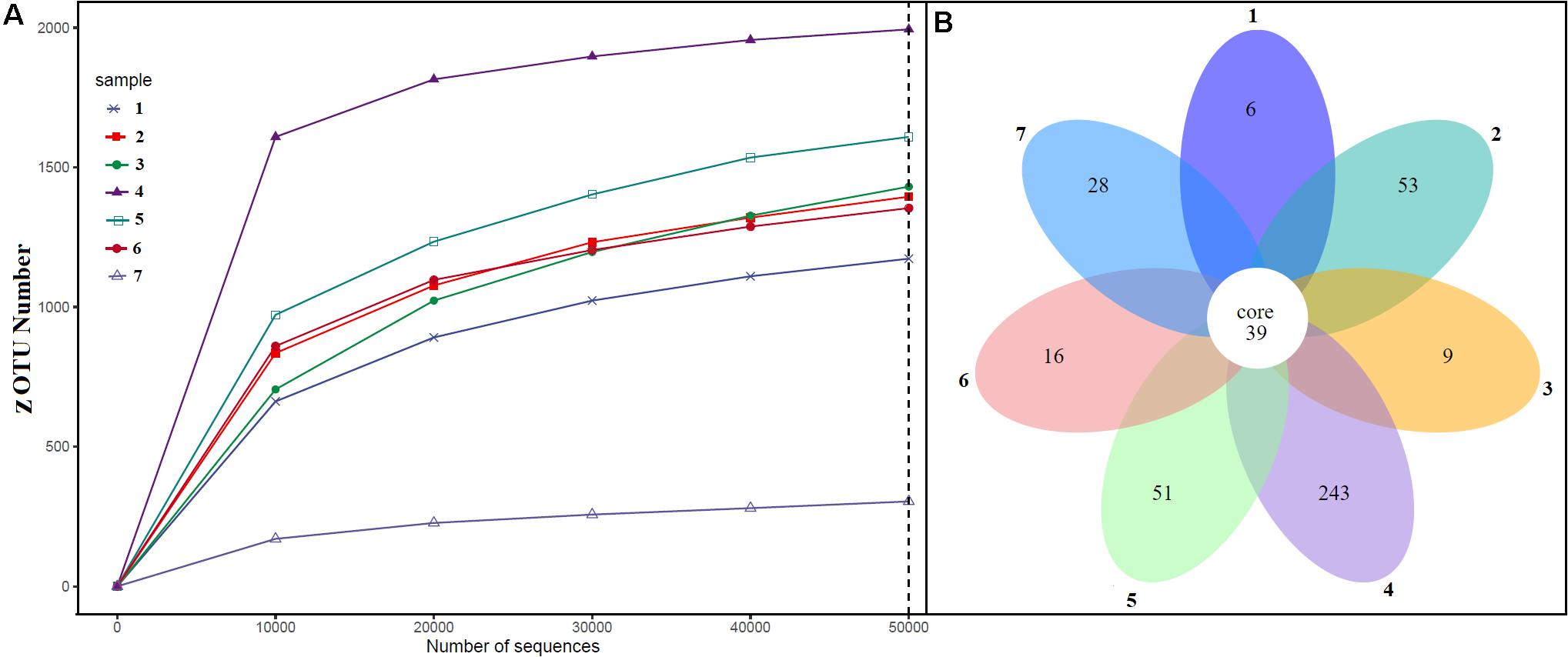
Figure 3. Alpha diversity. (A) ZOTUs per sequence number; (B) petal graph showing unique and common ZOTUs present in different sample sites. 1, Ganeshpuri; 2, Bendru Theertha; 3, Tuwa; 4, Vajreshwari; 5, Akaloli; 6 Sativali; 7, Bhadrachalam.
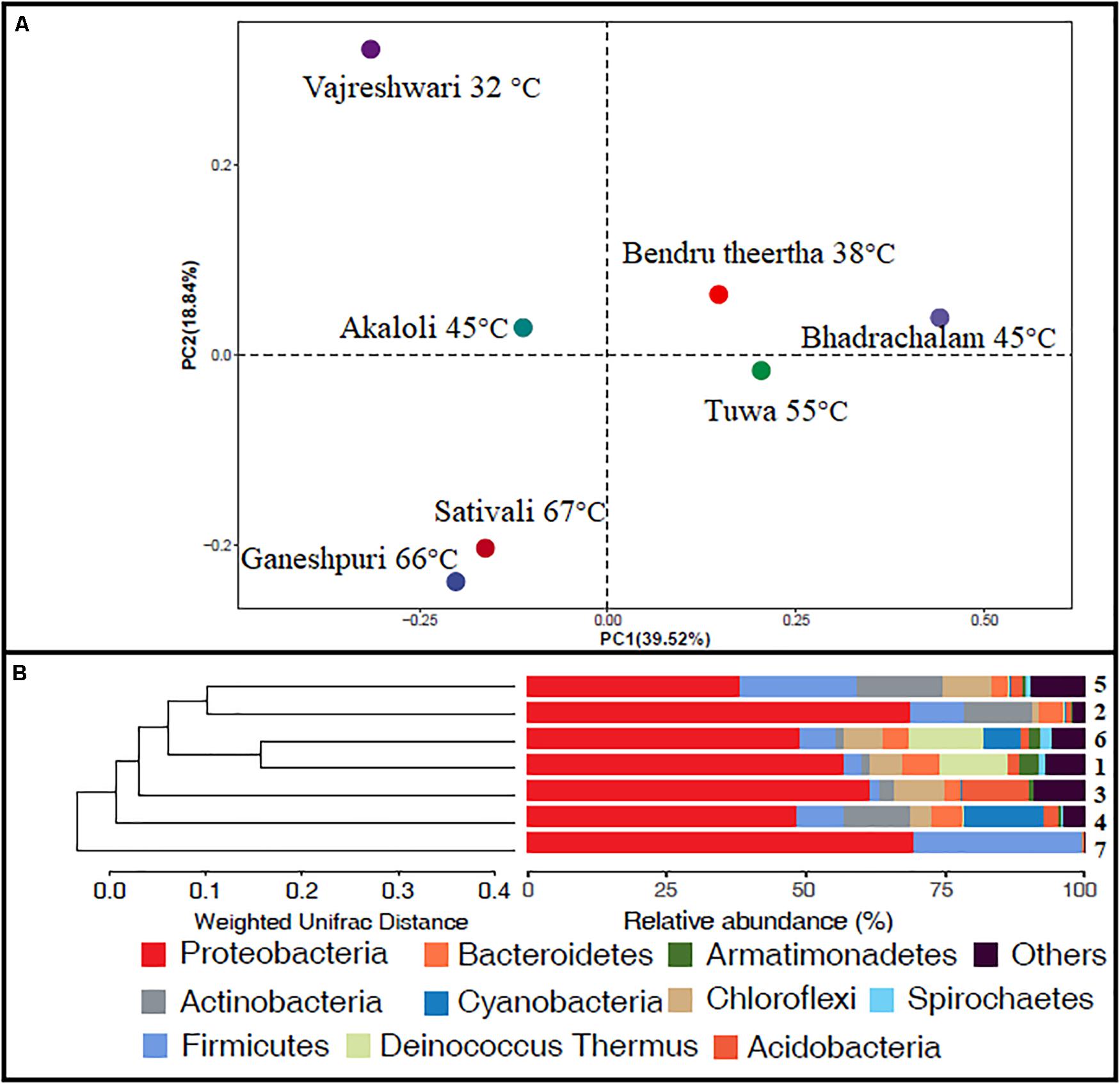
Figure 4. Beta diversity. (A) Weighted UniFrac distances calculated using amplicon reads and plotted on a PCoA plot; (B) hierarchical clustering based on weighted UniFrac distance (UPGMA) algorithm (1, Ganeshpuri; 2, Bendru Theertha; 3, Tuwa; 4, Vajreshwari; 5, Akaloli; 6 Sativali; 7, Bhadrachalam).
Culture-Dependent Microbial Diversity Analysis
A total of 131 strains (Supplementary Table S2) were isolated from the hot spring samples which were divided into 12 genera and four phyla. At the phylum level, Firmicutes were dominant followed by Actinobacteria, Proteobacteria, and Bacteroidetes. At the genus level, Bacillus were dominant followed by Brevibacillus, Paenibacillus, Oceanibaculum, Micromonospora, Microbacterium, Terrimonas, Aneurinibacillus, Actinocorallia, Sphingomonas, Micrococcus, and Rhodoligotrophos. The highest number of strains was isolated using T5 media, followed by R2A, TH, ISP5, and CC media (Supplementary Table S2). Strains SYSU-M-14, SYSU-M-111, SYSU-M1-37, and SYSU-M1-3 shared < 97% 16S rRNA gene sequence similarity with Rhodoligotrophos appendicifer, Bacillus alkalitolerans, Paenibacillus albidus, and Terrimonas lutea, respectively. It was noticed that the incubation temperature of 37 and 45°C was better for isolation. It was further noticed that no strain was isolated above 55°C. Details about the number of strains, their 16S rRNA gene sequence similarity, sample location, and media used for isolation are mentioned in Supplementary Table S2.
The isolated strains were evaluated for their ability to produce enzymes. About 63, 35, 13, and 8% of strains were positive for protease, amylase, cellulase, and xylanase, respectively (Supplementary Table S2).
Comparison of Culture-Dependent and Culture-Independent Microbial Diversity Analysis
Figure 6 represents the culture-dependent and culture-independent microbial diversity analyses of seven hot spring samples. In the culture-dependent analysis, Firmicutes were dominant followed by Proteobacteria, whereas in the culture-independent analysis, Proteobacteria were dominant followed by Firmicutes. In both culture-dependent and culture-independent analyses, the microbial richness decreased with the increase of temperature. Culture-independent analysis suggests that seven hot spring samples harbored several bacterial phyla, while only four bacterial phyla were recovered using culture-dependent analysis.
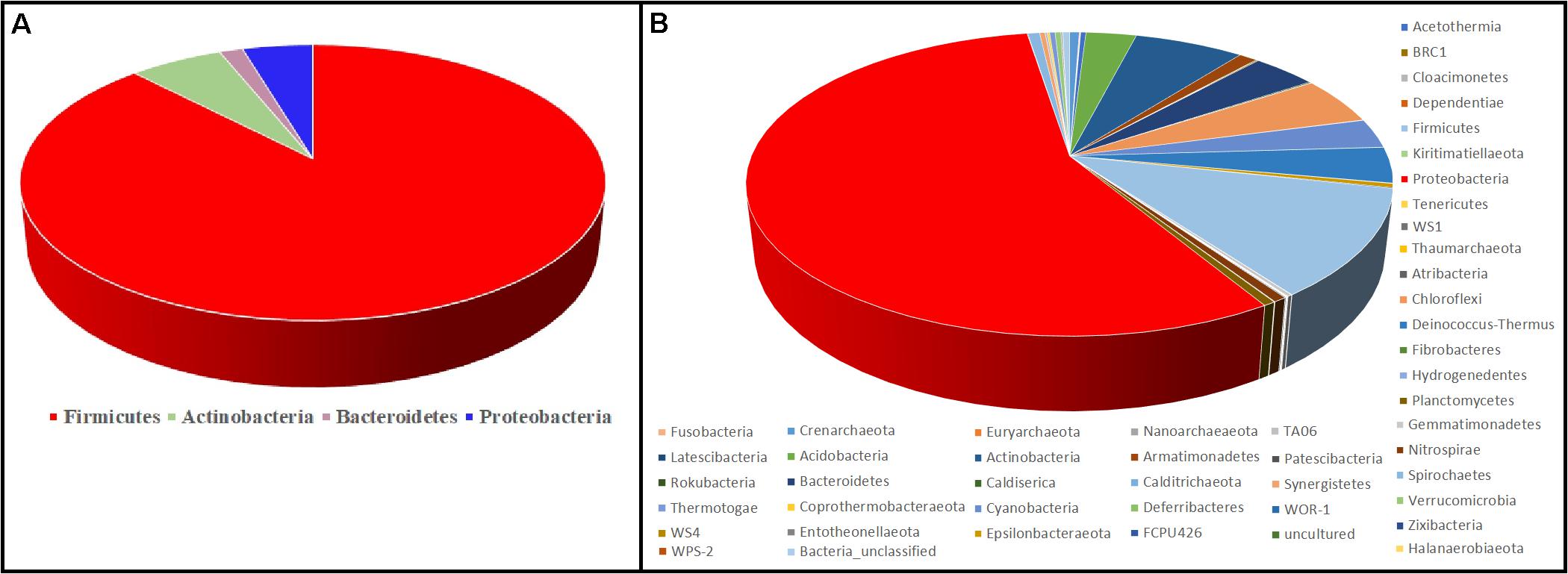
Figure 6. Comparison of microbial diversity at the phylum level. (A) Culture-dependent method; (B) culture-independent method.
Discussion
Hot springs were once perceived to be a sterile environment, but the pioneering work in discovering Taq DNA polymerase from Thermus aquaticus not only improved the economy of the industry but also brought a revolution in the field of molecular biology (Chien et al., 1976). The hot spring atmosphere was postulated to be close to the early chemical environment on Earth, and hot springs have thus become a model ecosystem for research on the origin and evolution of life (Li et al., 2015).
In the present study, physicochemical and microbial diversity analyses of seven Indian hot springs were carried out. The temperature of the seven hot springs ranged between 32 and 67°C. The highest temperature was recorded at SATI (67°C) and the lowest at VAJ (32°C). The pH of the hot spring sample ranged from neutral to alkaline which was similar to other Indian hot springs (Craig et al., 2013; Badhai et al., 2015).
Proteobacteria and Crenarchaeota (Figure 2A) were the dominant bacterial and archaeal phyla found in the present study and also in other Indian hot springs (Badhai et al., 2015; Sahoo et al., 2015; Saxena et al., 2017). Photosynthetic bacteria (Cyanobacteria) abundance was observed at VAJ and SATI (Figure 2A). The presence of phototrophs in the hot spring suggests the nutritive interaction among the microorganisms (Narsing Rao et al., 2018).
The GAN and SATI sample site temperatures were relatively high (66–67°C) when compared with those of the other sample sites with high abundance of Deinococcus–Thermus. Similarly, Jain et al. (2014) noticed a high abundance of Deinococcus–Thermus at the Tantloi hot spring, India, with a similar temperature. The sample sites’ temperature, ranging from 38 to 45°C (BT, AKA, and BHA), showed a high abundance of Firmicutes and their abundance decreased over this temperature range. At the genus level, Bacillus dominated at AKA and VAJ, while Acinetobacter dominated at the TUW and BT sample sites (Figure 2B). The abundance of the genus Bacillus has been reported in other Indian hot springs (Mangrola A. et al., 2015; Mangrola A. V. et al., 2015). Similarly, Saxena et al. (2017) noticed high Acinetobacter abundance at the Tattapani (India) hot spring. The genus Meiothermus, known for its thermophilic nature (Chen et al., 2002), dominated at sample sites (GAN and SATI) with a high temperature (66–67°C). The results suggest that temperature plays an important role in structuring hot spring microbial diversity.
In the BHA sample site, Escherichia and Shigella were dominant which was unusual when compared with the other sample sites in the present study. This might be due to environmental contamination as this sample site was located on the bed of a river.
Alpha diversity analysis showed that sample sites with low temperatures not only had high taxonomic richness but also had more unique ZOTUs (Figures 3A,B). In the beta diversity analysis, samples were clustered based on location (Figures 4A,B). Our results correlate with the study of Saxena et al. (2017), where the authors noticed high taxonomic richness at low temperatures and region-specific clustering.
Network analysis offers new insight into the structure of complex microbial communities, and such information is particularly valuable in environments, where the basic ecology and life history strategies of many microbial taxa remain unknown (Janssen, 2006). Further, it also helps to reveal the niche spaces shared by community members, direct symbioses between community members, potential biotic interactions, habitat affinities, or shared physiologies that could guide more focused studies or experimental settings (Proulx et al., 2005; Janssen, 2006; Barberán et al., 2012). In the present study, we noticed that phyla Chloroflexi, Spirochaetes, and Euryarchaeota had a high correlation with other members. Similarly, Zamkovaya et al. (2021) reported a high correlation between Chloroflexi and other members in the hot spring samples. Although the relative abundance of Proteobacteria was high (Figure 2A) when compared with Chloroflexi, Spirochaetes, and Euryarchaeota, however, the correlation of Proteobacteria with other members was less when compared with Chloroflexi, Spirochaetes, and Euryarchaeota (Figure 5). This suggests that the microbe’s interactions were specific irrespective of their abundance. However, further studies are needed to enlighten their interactions.
In the present study, Firmicutes at the phylum level and Bacillus at the genus level were dominant in the culture-dependent microbial diversity analysis. Similarly, Pathak and Rathod (2014) performed culture-dependent bacterial diversity analysis for the terrestrial thermal spring of Unkeshwar, India, and noticed Firmicutes at the phylum level and Bacillus at the genus level as the dominant bacterial groups.
Isolation medium and incubation temperature play an important role in determining microbial diversity through culture-dependent analysis. Pathak and Rathod (2014) during the bacterial diversity analysis of the Unkeshwar thermal spring noticed luxuriant bacterial growth on nutrient agar when compared with the other isolation medium. Sen and Maiti (2014) noticed that the optimum growth temperature for strains isolated from Odisha (India) hot springs ranged between 37 and 50°C. In the present study, the temperature range of 37 and 45°C and T5 media were better for isolation. Some of the sample sites’ temperature was above 60°C (Table 1), and culture-independent analysis showed the presence of microbial taxa (Figures 2A,B). However, we did not notice any microbial growth above 55°C using the culture-dependent method, suggesting that there are still many microbial taxa that remain uncultivated and there is still plenty of room for research.
Strains SYSU-M-14 and SYSU-M1-3 isolated from the TUW sample site and SYSU-M-111 and SYSU-M1-37 isolated from the BT sample site shared <97% 16S rRNA gene sequence similarity, suggesting that they could be novel candidates. Similarly, several novel bacterial candidates have been reported from Indian hot springs. Ruckmani et al. (2011) reported a novel genus Calidifontibacter from a hot spring sample of Irde (Mangalore, Karnataka State, India). Similarly, novel species Aeromonas sharmana and Flavobacterium indicum have been reported in an Indian (Assam) warm spring (Saha and Chakrabarti,2006a,c). The result suggests that Indian hot springs harbor many novel bacterial candidates. We will further characterize strains SYSU-M-14, SYSU-M1-3, SYSU-M-111, and SYSU-M1-37 to determine their taxonomic positions in our future work. The culture-dependent analysis showed that about 13 and 8% of the isolated strains were positive for cellulase and xylanase, respectively (Supplementary Table S2). Many bacterial strains isolated from Indian hot springs were reported to produce various enzymes like protease, lipase, esterase, amidase, caseinase, urease, amylase, oxidase, and gelatinase (Tripathy et al., 2016). The results suggest that Indian hot springs could be a valuable source for the enzymes.
Although the culture-dependent method is a classical approach for determining microbial diversity, this method has several disadvantages as most of the microorganisms remain hidden or are difficult to grow due to the lack of essential nutrients and optimal environmental conditions such as temperature, pH, and essential mixtures of gases (Narsing Rao et al., 2018). Culture-independent analysis suggests that seven hot spring samples harbored several bacterial phyla, while only four bacterial phyla were recovered using culture-dependent analysis (Figure 6); hence, appropriate methods should be developed to isolate these microbial taxa.
Conclusion
Hot springs are an important source for novel strains and bioactive molecules. In the present study, physicochemical and microbial diversity analyses of seven Indian hot springs were performed. The result suggests that physicochemical parameters play an important role in shaping the microbial community of hot springs, and hence, these parameters should be considered during isolation. Culture-dependent and culture-independent microbial diversity analyses suggest that microbial richness decreases with an increase in temperature. Culture-dependent analysis suggests that these hot springs have industrially important thermostable enzymes producing bacterial strains. Culture-independent microbial diversity analysis suggests that the hot springs of the present study harbored diverse and novel microbial populations, but only a few phyla were recovered using culture-dependent analysis, and hence, appropriate methods should be developed to isolate the uncultivated microbial taxa.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Author Contributions
W-JL, MX, WH, and S-XG designed the research and project outline. B-BL and M-ML performed the DNA extraction and physicochemical analysis. MN, Z-HL, and Z-YD performed the culture-dependent and culture-independent analyses. MN drafted the manuscript. All authors read and approved the final manuscript.
Funding
This work was financially supported by the Key Realm R&D Program of Guangdong Province (Grant No. 2018B020206001) and the National Natural Science Foundation of China (Grant Nos. 32061143043, 91951205, and 31800001). The Scientific Investigation Voyage supported by Southern Marine Science and Engineering Guangdong Laboratory (Zhuhai). The authors are grateful to the Deanship of Scientific Research, King Saud University for funding through Vice Deanship of Scientific Research Chairs.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.627200/full#supplementary-material
Footnotes
References
Anguita-Maeso, M., Olivares-García, C., Haro, C., Imperial, J., Navas-Cortés, J. A., and Landa, B. B. (2020). Culture-dependent and culture-independent characterization of the olive xylem microbiota: effect of sap extraction methods. Front. Plant Sci. 10:1708. doi: 10.3389/fpls.2019.01708
Badhai, J., Ghosh, T. S., and Das, S. K. (2015). Taxonomic and functional characteristics of microbial communities and their correlation with physicochemical properties of four geothermal springs in Odisha, India. Front. Microbiol. 6:1166. doi: 10.3389/fmicb.2015.01166
Baker, G. C., Gaffar, S., Cowan, D. A., and Suharto, A. R. (2001). Bacterial community analysis of Indonesian hot springs. FEMS. Microbiol. Lett. 200, 103–109. doi: 10.1111/j.1574-6968.2001.tb10700.x
Bandyopadhyay, S., Schumann, P., and Das, S. K. (2013). Pannonibacter indica sp. nov., a highly arsenate-tolerant bacterium isolated from a hot spring in India. Arch. Microbiol. 195, 1–8. doi: 10.1007/s00203-012-0840-z
Barberán, A., Bates, S. T., Casamayor, E. O., and Fierer, N. (2012). Using network analysis to explore co-occurrence patterns in soil microbial communities. ISME J. 6, 343–351. doi: 10.1038/ismej.2011.119
Bokulich, N. A., Subramanian, S., Faith, J. J., Gevers, D., Gordon, J. I., Knight, R., et al. (2013). Quality-filtering vastly juice diversity factors from illumina amplicon sequencing. Nat. Methods 10, 57–59. doi: 10.1038/nmeth.2276
Bolyen, E., Rideout, J. R., Dillon, M. R., Bokulich, N. A., Abnet, C. C., Al-Ghalith, G. A., et al. (2019). Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 37, 852–857. doi: 10.1038/s41587-019-0209-9
Brock, T. D. (1997). The value of basic research: discovery of Thermus aquaticus and other extreme thermophiles. Genetics 146, 1207–1210. doi: 10.1093/genetics/146.4.1207
Brock, T. D., and Freeze, H. (1969). Thermus aquaticus gen. n. and sp. n., a nonsporulating extreme thermophile. J. Bacteriol. 98, 289–297. doi: 10.1128/JB.98.1.289-297.1969
Caporaso, J. G., Lauber, C. L., Walters, W. A., Berg-Lyons, D., Lozupone, C. A., Turnbaugh, P. J., et al. (2011). Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl. Acad. Sci. U.S.A. 108, 4516–4522. doi: 10.1073/pnas.1000080107
Chaudhuri, B., Chowdhury, T., and Chattopadhyay, B. (2017). Comparative analysis of microbial diversity in two hot springs of Bakreshwar, West Bengal, India. Genom. Data 12, 122–129. doi: 10.1016/j.gdata.2017.04.001
Chen, M. Y., Lin, G. H., Lin, Y. T., and Tsay, S. S. (2002). Meiothermus taiwanensis sp. nov., a novel filamentous, thermophilic species isolated in Taiwan. Int. J. Syst. Evol. Microbiol. 52, 1647–1654. doi: 10.1099/00207713-52-5-1647
Chien, A., Edgar, D. B., and Trela, J. M. (1976). Deoxyribonucleic acid polymerase from the extreme thermophile Thermus aquaticus. J. Bacteriol. 127, 1550–1557. doi: 10.1128/jb.127.3.1550-1557.1976
Craig, J., Absar, A., Bhat, G., Cadel, G., Hafiz, M., Hakhoo, N., et al. (2013). Hot springs and the geothermal energy potential of Jammu & Kashmir State, N.W. Himalaya, India. Earth Sci. Rev. 126, 156–177. doi: 10.1016/j.earscirev.2013.05.004
Csardi, G., and Nepusz, T. (2006). The igraph software package for complex network research. Int. J. Complex Syst. 1695, 1–9.
De Mandal, S., Chatterjee, R., and Kumar, N. S. (2017). Dominant bacterial phyla in caves and their predicted functional roles in C and N cycle. BMC Microbiol. 17:90. doi: 10.1186/s12866-017-1002-x
Dwivedi, V., Kumari, K., Gupta, S. K., Kumari, R., Tripathi, C., Lata, P., et al. (2015). Thermus parvatiensis RLT sp. nov., isolated from a hot water spring, located a top the Himalayan ranges at Manikaran, India. Indian. J. Microbiol. 55, 357–365. doi: 10.1007/s12088-015-0538-4
Edgar, R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797. doi: 10.1093/nar/gkh340
Edgar, R. C. (2010). Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26, 2460–2461. doi: 10.1093/bioinformatics/btq461
Edgar, R. C. (2013). UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 10, 996–998. doi: 10.1038/nmeth.2604
Edgar, R. C., Haas, B. J., Clemente, J. C., Quince, C., and Knight, R. (2011). UCHIME Improves Speed and Sensitivity of Chimera Detection. Bioinformatics 27, 2194–2200. doi: 10.1093/bioinformatics/btr381
Friedman, J., and Alm, E. J. (2012). Inferring correlation networks from genomic survey data. PLoS Comput. Biol. 8:e1002687. doi: 10.1371/journal.pcbi.1002687
Ghelani, A., Patel, R., Mangrola, A., and Dudhagara, P. (2015). Cultivation-independent comprehensive survey of bacterial diversity in Tulsi Shyam hot springs, India. Genom. Data 4, 54–56. doi: 10.1016/j.gdata.2015.03.003
Habib, N., Khan, I. U., Salam, N., Xiao, M., Ahmed, I., Zhi, X. Y., et al. (2018). Tepidimonas sediminis sp. nov. and Tepidimonas alkaliphilus sp. nov., two novel moderately thermophilic species isolated from a hot spring. Antonie Van Leeuwenhoek 111, 1023–1031. doi: 10.1007/s10482-017-1002-8
Jain, P., Reza, H. M., and Pa, S. (2014). Molecular phylogenetic analysis of bacterial community and characterization of Cr(VI) reducers from the sediments of Tantloi hot spring, India. Aquat. Biosyst. 10:7. doi: 10.1186/2046-9063-10-7
Janssen, P. H. (2006). Identifying the dominant soil bacterial taxa in libraries of 16S rRNA and 16S rRNA genes. Appl. Environ. Microbiol. 72, 1719–1728. doi: 10.1128/AEM.72.3.1719-1728.2006
Khan, I. U., Habib, N., Hussain, F., Xian, W. D., Amin, A., Zhou, E. M., et al. (2017a). Thermus caldifontis sp. nov., a thermophilic bacterium isolated from a hot spring. Int. J. Syst. Evol. Microbiol. 67, 2868–2872. doi: 10.1099/ijsem.0.002037
Khan, I. U., Hussain, F., Tian, Y., Habib, N., Xian, W. D., Jiang, Z., et al. (2017b). Tibeticola sediminis gen. nov., sp. nov., a thermophilic bacterium isolated from a hot spring. Int. J. Syst. Evol. Microbiol. 67, 1133–1139. doi: 10.1099/ijsem.0.001777
Kumar, B., Trivedi, P., Mishra, A. K., Pandey, A., and Palni, L. M. (2004). Microbial diversity of soil from two hot springs in Uttaranchal Himalaya. Microbiol. Res. 159, 141–146. doi: 10.1016/j.micres.2004.01.004
Li, H., Yang, Q., Li, J., Gao, H., Li, P., and Zhou, H. (2015). The impact of temperature on microbial diversity and AOA activity in the Tengchong Geothermal Field, China. Sci. Rep. 5:17056. doi: 10.1038/srep17056
Li, W. J., Xu, P., Schumann, P., Zhang, Y. Q., Pukall, R., Xu, L. H., et al. (2007). Georgenia ruanii sp. nov., a novel actinobacterium isolated from forest soil in Yunnan (China), and emended description of the genus Georgenia. Int. J. Syst. Evol. Microbiol. 57, 1424–1428. doi: 10.1099/ijs.0.64749-0
Liu, L., Salam, N., Jiao, J. Y., Jiang, H. C., Zhou, E. M., Yin, Y. R., et al. (2016). Diversity of culturable thermophilic actinobacteria in hot springs in Tengchong, China and studies of their biosynthetic gene profiles. Microb. Ecol. 72, 150–162. doi: 10.1007/s00248-016-0756-2
Mangrola, A., Dudhagara, P., Koringa, P., Joshi, C. G., Parmar, M., and Patel, R. (2015). Deciphering the microbiota of Tuwa hot spring, India using shotgun metagenomic sequencing approach. Genom. Data 4, 153–155. doi: 10.1016/j.gdata.2015.04.014
Mangrola, A. V., Dudhagara, P., Koringa, P., Joshi, C. G., and Patel, R. K. (2015). Shotgun metagenomic sequencing based microbial diversity assessment of Lasundra hot spring, India. Genom. Data 4, 73–75. doi: 10.1016/j.gdata.2015.03.005
Myer, P. R., Kim, M., Freetly, H. C., and Smith, T. P. L. (2016). Evaluation of 16S rRNA amplicon sequencing using two next-generation sequencing technologies for phylogenetic analysis of the rumen bacterial community in steers. J. Microbiol. Methods 127, 132–140. doi: 10.1016/j.mimet.2016.06.004
Najar, I. N., Sherpa, M. T., Das, S., Das, S., and Thakur, N. (2018). Microbial ecology of two hot springs of Sikkim: predominate population and geochemistry. Sci. Total Environ. 63, 730–745. doi: 10.1016/j.scitotenv.2018.05.037
Narsing Rao, M. P., Liu, L., Jiao, J. Y., Xiao, M., and Li, W. J. (2018). “Hot springs of india: occurrence and microbial diversity,” in Extremophiles in Eurasian Ecosystems: Ecology, Diversity, and Applications. Microorganisms for Sustainability, Vol. 8, eds D. Egamberdieva, N. K. Birkeland, H. Panosyan, and W. J. Li (Singapore: Springer), 29–55. doi: 10.1007/978-981-13-0329-6_2
Panda, S. K., Jyoti, V., Bhadra, B., Nayak, K. C., Shivaji, S., Rainey, F. A., et al. (2009). Thiomonas bhubaneswarensis sp. nov., an obligately mixotrophic, moderately thermophilic, thiosulfate-oxidizing bacterium. Int. J. Syst. Evol. Microbiol. 59, 2171–2175. doi: 10.1099/ijs.0.007120-0
Pathak, A. P., and Rathod, M. G. (2014). Cultivable bacterial diversity of terrestrial thermal spring of Unkeshwar, India. J. Biochem. Technol. 5, 814–818.
Pednekar, P., Jain, R., and Mahajan, G. (2011). Anti-infective potential of hotspring bacteria. J. Glob. Infect. Dis. 3, 241–245. doi: 10.4103/0974-777X.83529
Proulx, S. R., Promislow, D. E., and Phillips, P. C. (2005). Network thinking in ecology and evolution. Trends Ecol. Evol. 20, 345–353. doi: 10.1016/j.tree.2005.04.004
Quast, C., Pruesse, E., Yilmaz, P., Gerken, J., Schweer, T., Yarza, P., et al. (2013). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41, D590–D596. doi: 10.1093/nar/gks1219
Ruckmani, A., Kaur, I., Schumann, P., Klenk, H. P., and Mayilraj, S. (2011). Calidifontibacter indicus gen. nov., sp. nov., a member of the family Dermacoccaceae isolated from a hot spring, and emended description of the family Dermacoccaceae. Int. J. Syst. Evol Microbiol. 61, 2419–2424. doi: 10.1099/ijs.0.025593-0
Sabat, A. J., van Zanten, E., Akkerboom, V., Wisselink, G., van Slochteren, K., de Boer, R. F., et al. (2017). Targeted next-generation sequencing of the 16S-23S rRNA region for culture-independent bacterial identification - increased discrimination of closely related species. Sci. Rep. 7:3434. doi: 10.1038/s41598-017-03458-6
Saha, P., and Chakrabarti, T. (2006a). Aeromonas sharmana sp. nov., isolated from a warm spring. Int. J. Syst. Evol. Microbiol. 56, 1905–1909. doi: 10.1099/ijs.0.63972-0
Saha, P., and Chakrabarti, T. (2006b). Emticicia oligotrophica gen. nov., sp. nov., a new member of the family ‘Flexibacteraceae’, phylum Bacteroidetes. Int. J. Syst. Evol. Microbiol. 56, 991–995. doi: 10.1099/ijs.0.64086-0
Saha, P., and Chakrabarti, T. (2006c). Flavobacterium indicum sp. nov., isolated from warm spring water in Assam. India. Int. J. Syst. Evol. Microbiol. 56, 2617–2621. doi: 10.1099/ijs.0.64309-0
Sahoo, R. K., Subudhi, E., and Kumar, M. (2015). Investigation of bacterial diversity of hot springs of Odisha, India. Genom. Data 6, 188–190. doi: 10.1016/j.gdata.2015.09.018
Saxena, R., Dhakan, D. B., Mittal, P., Waiker, P., Chowdhury, A., Ghatak, A., et al. (2017). Metagenomic analysis of hot springs in Central India reveals hydrocarbon degrading thermophiles and pathways essential for survival in extreme environments. Front. Microbiol. 7:2123. doi: 10.3389/fmicb.2016.02123
Sazci, A., Radford, A., and Erenle, K. (1986). Detection of cellulolytic fungi by using Congo red as an indicator: a comparative study with the dinitrosalicyclic acid reagent method. J. Appl. Bacteriol. 61, 559–562. doi: 10.1111/j.1365-2672.1986.tb01729.x
Sen, R., and Maiti, N. K. (2014). Genomic and functional diversity of bacteria isolated from hot springs in Odisha. India. Genomicrobiol. J. 31, 541–550. doi: 10.1080/01490451.2013.850560
Tripathy, S., Padhi, S. K., Sen, R., Maji, U., Samanta, M., Mohanty, S., et al. (2016). Draft genome sequence of Brevibacillus borstelensis cifa_chp40, a thermophilic strain having biotechnological importance. J. Genom. 4, 4–6. doi: 10.7150/jgen.14036
Yoon, S. H., Ha, S. M., Kwon, S., Lim, J., Kim, Y., Seo, H., et al. (2017). Introducing EzBio-Cloud: a taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int. J. Syst. Evol. Microbiol. 67, 1613–1617. doi: 10.1099/ijsem.0.001755
Zamkovaya, T., Foster, J. S., de Crécy-Lagard, V., and Conesa, A. (2021). A network approach to elucidate and prioritize microbial dark matter in microbial communities. ISME J. 15, 228–244. doi: 10.1038/s41396-020-00777-x
Keywords: Indian hot springs, culture-dependent microbial diversity analysis, culture-independent microbial diversity analysis, alpha diversity, beta diversity
Citation: Narsing Rao MP, Dong Z-Y, Luo Z-H, Li M-M, Liu B-B, Guo S-X, Hozzein WN, Xiao M and Li W-J (2021) Physicochemical and Microbial Diversity Analyses of Indian Hot Springs. Front. Microbiol. 12:627200. doi: 10.3389/fmicb.2021.627200
Received: 08 November 2020; Accepted: 08 February 2021;
Published: 03 March 2021.
Edited by:
André Antunes, Macau University of Science and Technology, ChinaReviewed by:
Brandon Briggs, University of Alaska Anchorage, United StatesShawn E. McGlynn, Tokyo Institute of Technology, Japan
Copyright © 2021 Narsing Rao, Dong, Luo, Li, Liu, Guo, Hozzein, Xiao and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bing-Bing Liu, liubingbing200666@163.com; 454873856@qq.com; Wen-Jun Li, liwenjun3@mail.sysu.edu.cn
 Manik Prabhu Narsing Rao1
Manik Prabhu Narsing Rao1 Zhen-Hao Luo
Zhen-Hao Luo Meng-Meng Li
Meng-Meng Li