- 1Department of Molecular Genetics, University of Groningen, Groningen, Netherlands
- 2Institute of Chemistry, Technical University of Berlin, Berlin, Germany
- 3Department of Chemistry, University of Manitoba, Winnipeg, MB, Canada
Coupling functional moieties to lantibiotics offers exciting opportunities to produce novel derivatives with desirable properties enabling new functions and applications. Here, five different synthetic hydrophobic polyproline peptides were conjugated to either nisin AB (the first two rings of nisin) or nisin ABC (the first three rings of nisin) by using click chemistry. The antimicrobial activity of nisin ABC + O6K3 against Enterococcus faecium decreased 8-fold compared to full-length nisin, but its activity was 16-fold better than nisin ABC, suggesting that modifying nisin ABC is a promising strategy to generate semi-synthetic nisin hybrids. In addition, the resulting nisin hybrids are not prone to degradation at the C-terminus, which has been observed for nisin as it can be degraded by nisinase or other proteolytic enzymes. This methodology allows for getting more insight into the possibility of creating semi-synthetic nisin hybrids that maintain antimicrobial activity, in particular when synthetic and non-proteinaceous moieties are used. The success of this approach in creating viable nisin hybrids encourages further exploring the use of different modules, e.g., glycans, lipids, active peptide moieties, and other antimicrobial moieties.
Introduction
Nisin is the first discovered and the best studied lantibiotic and it is produced by Lactococcus lactis (Rogers, 1928). In addition to its natural presence in fermented foods, nisin has been applied as a food preservative for many decades, because of its excellent activity against food spoilage (Hansen and Sandine, 1994; Gharsallaoui et al., 2016). Beyond its role in food safety and preservation, nisin has potential therapeutic applications. It is for instance effective against many Gram-positive antibiotic-resistant organisms, such as methicillin-resistant Staphylococcus aureus (MRSA) and vancomycin-resistant Enterococcus (VRE) (Shin et al., 2016). The exceptional activity of nisin is derived from a unique structure, containing one lanthionine and four methyllanthionine rings, which has a dual mode of action. The first two rings (AB) form a lipid II recognition site. By binding to the peptidoglycan precursor lipid II, nisin inhibits cell wall biosynthesis. The last two rings (DE), which are connected to rings ABC through a hinge region, constitute a membrane insertion domain. After rings AB dock to lipid II (Brötz et al., 1998), rings DE and the tail can insert into the bacterial membrane to create pores, where nisin forms pores (Lubelski et al., 2008). Nisin’s dual mode of action increases its antimicrobial activity, and decreases the chance of resistance development in target organisms. These features make nisin an attractive candidate for development into an antibiotic alternative. Unfortunately, nisin is readily degraded in the gut, which precludes oral delivery. Also, the administering of nisin by injection, especially at its full-length, is limited by the instability of its dehydro-residues. If possible, these problems should be addressed to broaden the scope of nisin application in the therapeutic setting. A promising strategy to achieve this has been provided by the chemical coupling of specific (protease resistant) moieties to nisin, and semi-synthetic fragments of nisin. This approach allows for the synthesis of novel derivatives with useful properties like increased stability, alleviating some of nisin’s characteristics that are problematic in its potential role as an antibiotic alternative.
The coupling of peptides to compatible moieties can be achieved through the widely used click chemistry method (Ahmad Fuaad et al., 2013). “Copper (I)-catalyzed azide-alkyne cycloaddition (CuAAC)” is the most common reaction representing the click chemistry. It is a region selective copper (I) catalytic cycloaddition reaction between an azide and an alkyne leading to the formation of a triazole. The molecules connected to the respective reagents are effectively “clicked” together (Kolb et al., 2001). Due to its reliability, specificity, biocompatibility, easiness to perform, and mild reaction conditions, click chemistry is being used increasingly in diverse areas, such as bioconjugation, drug design and polymer science (Thirumurugan et al., 2013; McKay and Finn, 2014; Jiang et al., 2019). The success of click chemistry in the field of peptide modification can be attributed to the resulting triazole ring which resembles an amide bond, and which increases the stability of the resulting molecule. This is achieved at least in part by increasing the molecule’s resistance to proteases, as the triazole aligns with the biological targets through hydrogen bonding and dipole interactions (Ahmad Fuaad et al., 2013). Peptide coupling through click chemistry has been the subject of several studies toward the development of target-specific bacterial probes and expanding application possibilities of this method (Arnusch, 2008; Yoganathan et al., 2011; Oldach et al., 2012; Slootweg et al., 2013a, 2014; Koopmans et al., 2015; Bolt et al., 2018). A prominent example of applying click chemistry to enhance lantibiotics, is that of coupling nisin AB to lipid moieties (Koopmans et al., 2015). The resulting hybrid molecules exhibited increased stability, as well as potent antimicrobial activity against drug-susceptible and -resistant strains of Gram-positive bacteria. In other studies, the lipid II-binding motif (rings AB) of nisin has been conjugated with various functional molecules (Arnusch, 2008; Koopmans et al., 2015; Bolt et al., 2018).
Here, a range of experiments was designed for the synthesis of nisin hybrids by coupling specific synthetic polyproline peptides, some of which containing cationic residues, to either nisin AB or nisin ABC. These polyproline peptides (Figure 1) were designed based on a polyproline structural skeleton using a proline analog [(2S,3aS,7aS)-octahydroindole-2-carboxylic acid, Oic] to display a linear and hydrophobic structure affine to a lipid membrane (Kubyshkin and Budisa, 2018; Kubyshkin et al., 2019). The selected properties of these nisin hybrids should aid in the membrane translocation of their C-terminal region and, as the molecular weight of the clicked polyproline moieties ranges between 0.5 and 1.5 kDa, the resulting molecules remain well under nisin in size. Hypothetically, the lipid-II targeting nisin AB would guide the conjugate to the bacterial membrane, where the hydrophobic tail would flip into the membrane core, tightly anchoring the conjugate. The newly synthesized nisin hybrids were compared to nisin with regard to their antimicrobial activity, and susceptibility to proteolytic degradation.
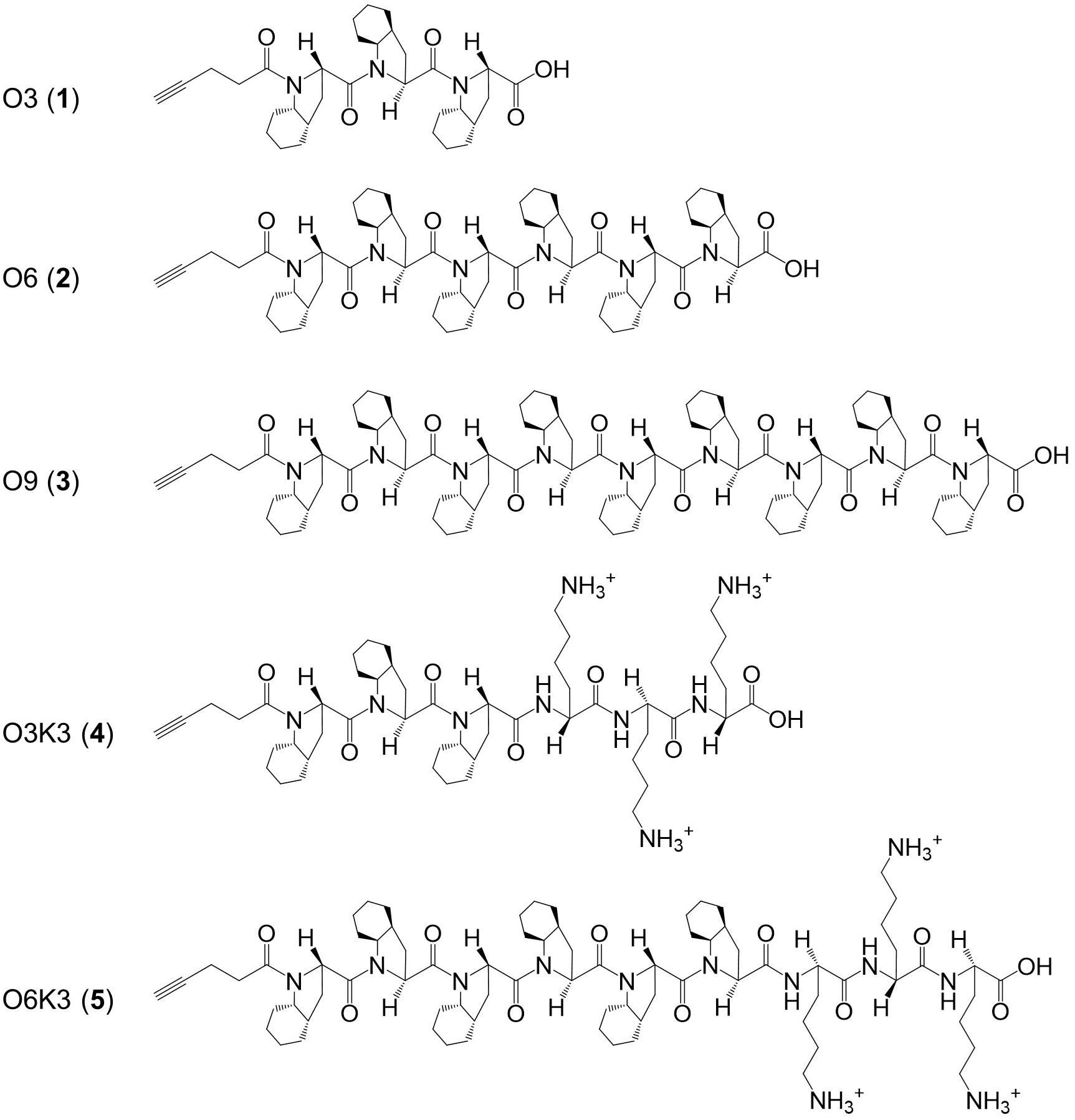
Figure 1. Structure of the hydrophobic polyproline peptides O3, O6, O9, O3K3, and O6K3 for putative membrane anchoring. The peptides were constructed with a hydrophobic proline analog (2S,3aS,7aS)-octahydroindole-2-carboxylic acid (Oic, designated as O) and lysine residues (Lys, K).
Materials and Methods
Bacterial Strains and Plasmids
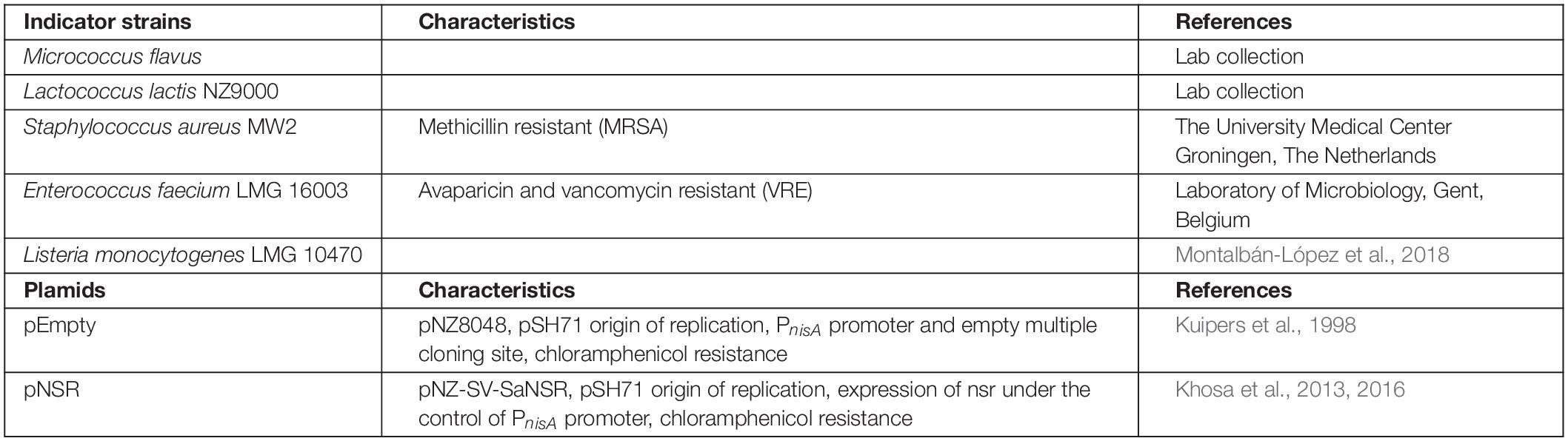
Indicator strains and plasmids used in this work are given in Table 1.
Preparation of Nisin AB-Azide
Nisin AB was purified in accordance with protocols reported previously (Slootweg et al., 2013b). Briefly, nisin (180 mg) was dissolved in 150 mL Tris buffer (5 mmol Tris acetate, 5 mmol CaCl2, 25 mmol sodium acetate, pH 7.0) and the solution was cooled on ice for 15 min. Then trypsin (15 mg) was added and warmed up to room temperature for 15 min. The reaction was performed at 30°C for 16 h and an extra 15 mg trypsin was added. After 24 h incubation, another 15 mg trypsin was added and the reaction was performed for another 24 h. The reaction mixture was acidified with hydrochloric acid (1 M) to pH 4.0 followed by adding 3 mL acetonitrile and concentrated in vacuo. The pure nisin AB was purified from the mixture by RP-HPLC with the water-acetonitrile gradient mobile phase containing trifluoroacetic acid (0.1%) and lyophilized to obtain a white powder (20 mg). Nisin AB (10 mg, 8.6 μmol) was dissolved in N,N-dimethylformamide (DMF) (100 μl) and azidopropylamine (44 μl, 43.2 mg, 432 μmol), N,N-diisopropylethylamine (DIPEA, 6 μl, 34.8 μmol), and (benzotriazol-1-yloxy)-tris-(dimethylamino) phosphonium hexafluorophosphate (BOP, 7.6 mg, 17.2 μmol) or (benzotriazol-1-yl-oxy)-tris-(pyrrolidino)phosphonium hexafluorophosphate (PyBOP, 9 mg, 17.2 μmol) were added. The reaction was vortexed for 20 min and subsequently quenched with 5 mL buffer (water : acetonitrile, 95:5 vol+ 0.1% trifluoroacetic acid). The reaction mixture was purified by RP-HPLC with the water-acetonitrile gradient mobile phase containing trifluoroacetic acid (0.1%) and pure nisin AB-azide was lyophilized to obtain a white powder (8 mg).
Preparation of Nisin ABC-Azide
α-chymotrypsin was used to digest nisin to generate nisin ABC. Nisin (180 mg) was dissolved in 150 mL Tris buffer (25 mmol Tris acetate, pH7.5) and the solution was cooled on ice for 15 min. Then α-chymotrypsin (15 mg) was added and warmed up to room temperature for 15 min. The enzymatic digestion was performed same as described for nisin AB. Nisin ABC was purified from the mixture by RP-HPLC with the water-acetonitrile gradient mobile phase containing trifluoroacetic acid (0.1%) and then lyophilized to obtain a white powder (20 mg). Nisin ABC (10 mg, 6.5 μmol) was dissolved in DMF (50 μl). The azide-coupling reaction was performed same as described for nisin AB.
Preparation of the Hydrophobic Polyproline Peptides
The polyproline-containing peptides were prepared using a manual Fmoc-based solid-phase peptide synthesis scheme as described (Kubyshkin and Budisa, 2018). The sequences were grown on 2-clorotrityl resins pre-loaded with either Fmoc-Oic-OH or Fmoc-Lys (Boc)-OH (Fmoc = fluorenylmethyloxycarbonyl, Boc = tert-butyloxycarbonyl). The resin loading was estimated at 0.7–0.8 mmol/g. The synthesis was performed in DMF using 2.5 equiv. of the Fmoc-amino acid pre-activated with the mixture of 2.5 equiv. 2-(1H-benzotriazole-1-yl)-1,1,3,3-tetramethylaminium tetrafluoroborate (TBTU) and 2.5 equiv.1-hydroxybenzotriazole (HOBt) mixture under addition of 5 equiv. DIPEA. The N-terminal pentynyl moiety was installed under coupling with pentynoic acid under same activation conditions. The Fmoc group was removed by treatment with 22 vol% piperidine in DMF. The final peptides were cleaved off the resin by treatment with hexafluoroisopropanol : dichloromethane (1:3, vol: vol) mixture. The peptides were additionally purified on short silica gel columns using dichloromethane-methanol gradient elution. Pentynyl-(Oic)9-OH (O9) peptide was additionally purified by precipitation from methanol. The Boc-group was removed from the lysine side-chains by treatment with 4 M hydrogen chloride in dioxane. The identity and purity of the final peptides were confirmed by mass-spectra (ESI-Orbitrap) and 1H-NMR spectra (CD3OD, 700 MHz). The peptides were obtained in 10–50 mg quantities.
Click Chemistry
A stock solution of CuSO4 (10 mg, 100 mM), sodium ascorbate (200 mg, 1 M) 2-(4-((bis((1-(tert-butyl)-1H-1,2,3-triazol-4-yl)methyl)amino)methyl)-1H-1,2,3-triazol-1-yl)-acetic acid (BTTAA, 25 mg, 50 mM), and tris((1-hydroxy-propyl-1H-1,2,3-triazol-4-yl)methyl)amine (THPTA, 25 mg, 250 mM) in deionized water and a stock solution of O3 (1 mg, 36 mM), O6 (1.8 mg, 36 mM), O9 (2.6 mg, 36 mM), O3K3 (1.9 mg, 36 mM) and O6K3 (2.7 mg, 36 mM) in DMF (50 μl) were prepared, aliquoted and stored at −20°C for further use. Firstly, nisin AB-azide (25 μg, 0.02 μmol) or nisin ABC-azide (40 μg, 0.02 μmol) were dissolved in 100 mM phosphate buffer (pH7.0, final reaction volume: 200 μl). Then, the appropriate O3, O6, O9, O3K3, or O6K3 (5 μl, 0.18 μmol) and CuSO4 (4 μl, 0.4 μmol) : THPTA (8 μl, 2 μmol) or BTTAA (40 μl, 2 μmol)-premix were added followed by the addition of sodium ascorbate (20 μl, 20 μmol). The reaction was performed at 37°C for 1 h and purified directly by RP-HPLC with the water-acetonitrile gradient mobile phase containing trifluoroacetic acid (0.1%). The pure product-containing fractions were lyophilized to obtain nisin hybrids 6–15 as white fluffy powders (Figure 2). The reaction was further scaled up in ratio to obtain more products.
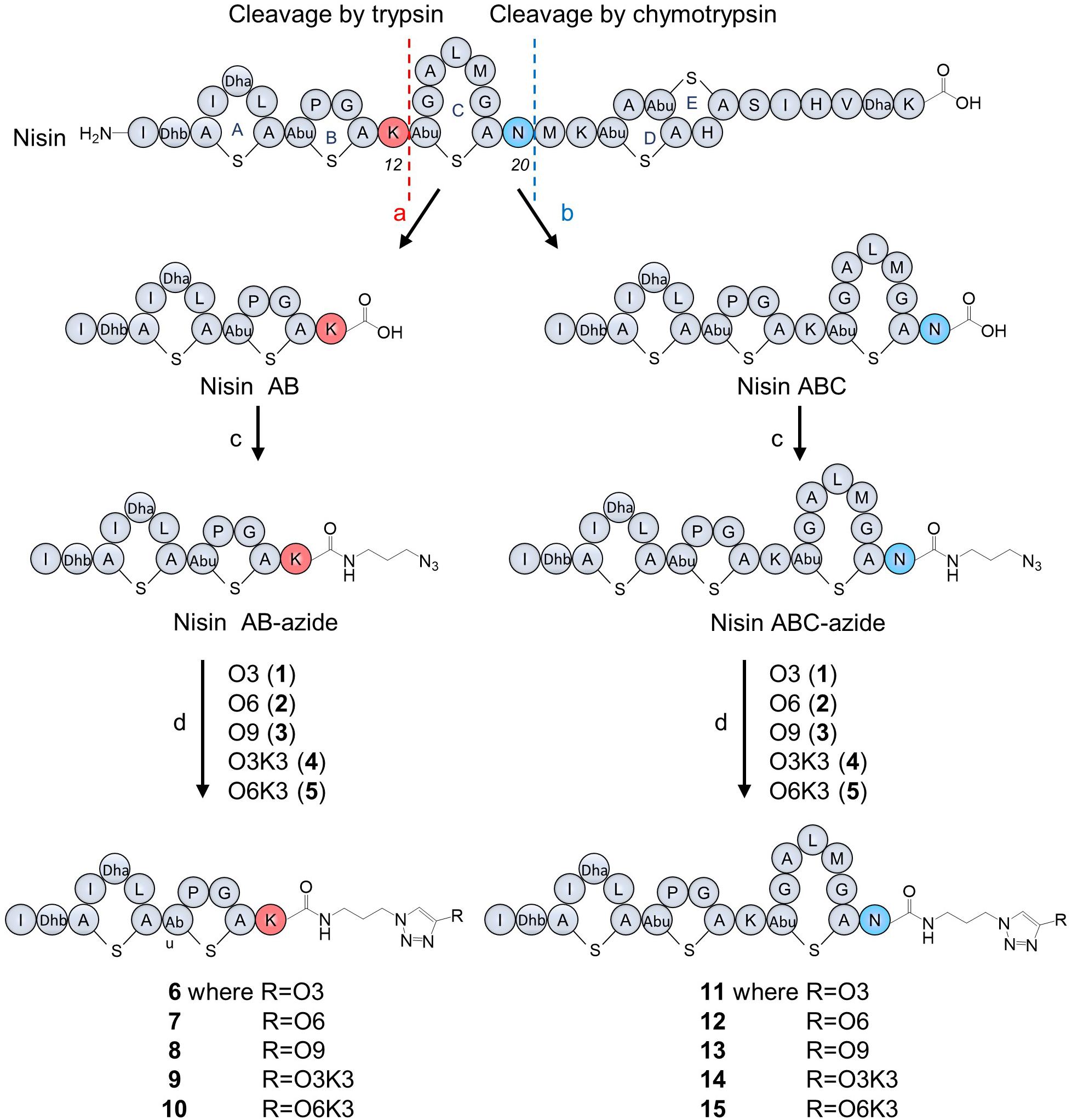
Figure 2. Nisin digestion and semi-synthesis of nisin AB and nisin ABC conjugates. (a) Trypsin, Tris buffer, pH 7.0, 30°C, 48 h; (b) chymotrypsin, Tris buffer, pH 7.5, 30°C, 48 h; (c) azidopropylamine, PyBOP, DIPEA, DMF, RT, 20 min; (d) CuSO4, BTTAA, sodium ascorbate in phosphate buffer, 37°C, 1 h.
Agar Well Diffusion Assay
Agar well diffusion assay against Micrococcus flavus was performed as described previously (van Heel et al., 2013). 0.15 nmol of each sample was added to each well. The agar plate was incubated at 30°C overnight, after which the zone of inhibition was measured.
Determination of the Minimal Inhibitory Concentration (MIC)
All samples were resuspended in 0.05% aqueous acetic acid solution and the peptide amount was quantified by HPLC according to Schmitt et al. (Schmitt et al., 2019). The indicator strains MW2-MRSA, Enterococcus faecium, Listeria monocytogenes, and Lactococcus lactis were first streaked on GM17 plate and cultured overnight. The peptide samples were diluted with 0.05% acetic acid to a concentration of 40–320 μM (depending on the estimated activity of the peptide and the strain tested). The MIC value test was performed according to Wiegand et al. (2008).
Results
Production of Nisin AB-Azide and Nisin ABC-Azide
Nisin was digested using trypsin and chymotrypsin, respectively, to generate nisin AB and nisin ABC fragments (Figure 2). The desired truncated nisin molecules were purified from the digestion mixture with yields in the milligram range, in accordance with protocols reported previously (Slootweg et al., 2013b). After purification, an azide linker was added to the C-terminus of the acquired nisin fragments. Since the truncated variants with the azide linker were needed in relatively large quantities for the generation of the semi-synthetic analogs, the previously reported peptide coupling procedure was optimized for this study. Initially, addition of the azide linker was done by coupling azidopropylamine to nisin AB in the presence of BOP as the coupling reagent. HPLC analysis of the reactions performed under these conditions showed substantial amounts of substrate remained unreacted, resulting in a reaction efficiency of only 7.4% (Supplementary Figure 1). Prolonging the reaction time to 1 h did not increase the conversion. However, by substituting the coupling reagent BOP for PyBOP, the reaction efficiency could be increased to 89% (Supplementary Figure 1). For reactions of this nature, PyBOP was shown to be a better coupling reagent than to BOP. Using the optimized conditions from the above experiment, azidopropylamine was coupled to nisin AB and nisin ABC in a reaction containing PyBOP/DIPEA to give nisin AB-azide and nisin ABC-azide in 89 and 87% yield, respectively, which was purified by HPLC and characterized by MALDI-TOF. The resulting nisin AB-azide and nisin ABC-azide contain convenient handles for ligation to alkynes via CuAAC.
Production of Nisin AB and Nisin ABC Conjugates
Five hydrophobic polyproline peptides (1–5) were coupled to nisin AB-azide and nisin ABC-azide (Figure 2). The first click reaction was attempted with O3 (1) and nisin AB-azide in the presence of THPTA as copper (I)-stabilizing ligand. Analysis by HPLC showed that a good amount of product was formed under these reaction conditions. Increasing the reaction temperature to 50°C and extending the reaction time to 2 h led to degradation rather than increased conversion. The conversion could however be increased by using BTTAA as substitute for THPTA as copper (I)-stabilizing ligand improved the conversion. The best results were obtained using 9 equiv. O3, 20 equiv. CuSO4, 100 equiv. BTTAA and 1,000 equiv. sodium ascorbate, reacted at 37°C for 1 h. Under these optimized conditions, the click reaction of nisin AB-azide and nisin ABC-azide with the five hydrophobic polyproline peptides (1–5) were carried out successfully to give semi-synthetic nisin hybrids 6–15 in 42–54% yields. The resulting semi-synthetic nisin hybrids 6–15 were further characterized by MALDI-TOF.
Antimicrobial Activity of Nisin AB and Nisin ABC Conjugates
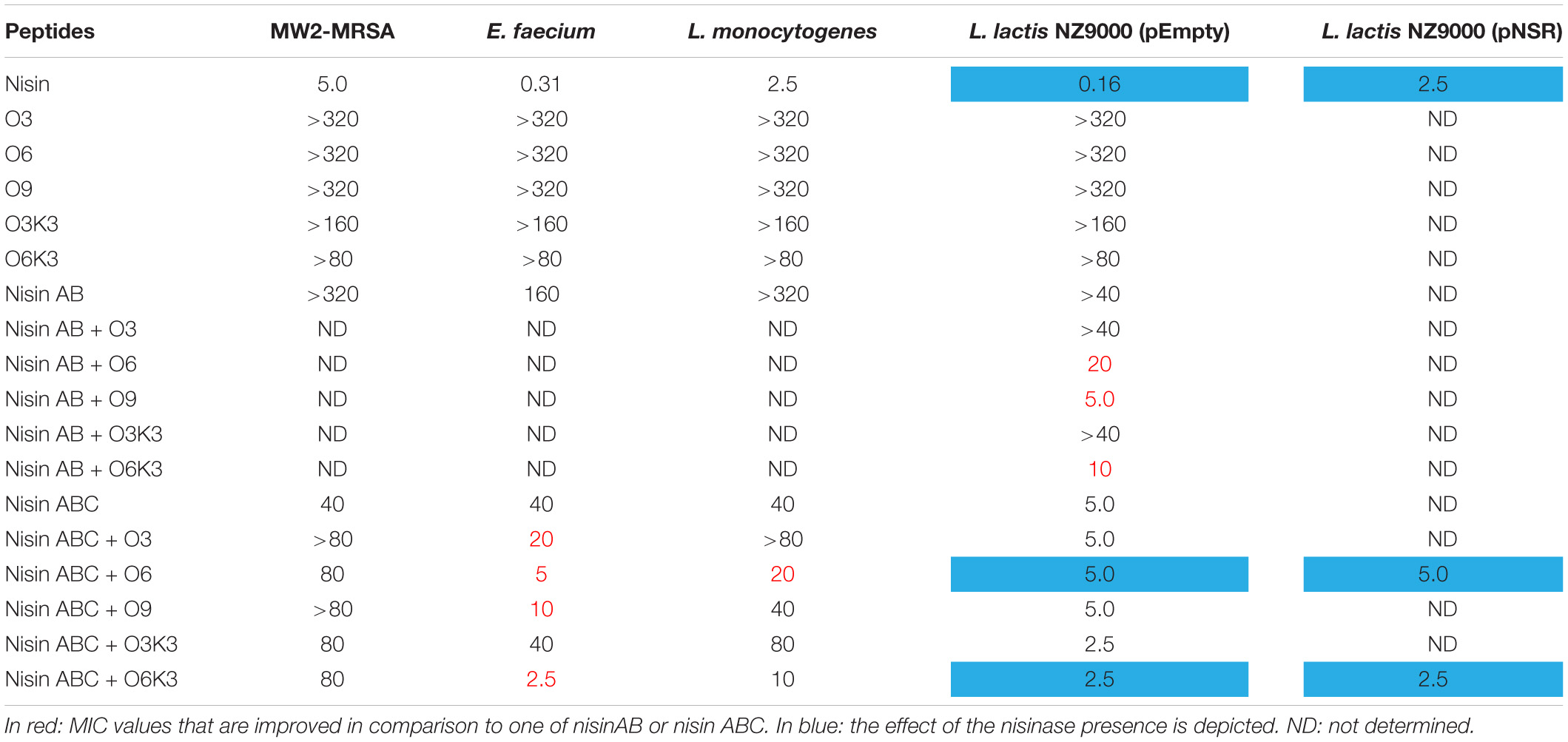
To investigate the biological activity of the nisin hybrids, an agar well diffusion assay and a growth inhibition assay were performed. M. flavus was used as the indicator strain for the agar well diffusion assay, and 0.15 nmol of each sample was added to each well (Figure 3). The results showed that nisin AB and five hydrophobic polyproline moieties (1–5) are not active alone and nisin has the highest activity. Of the nisin AB conjugates, nisin AB + O6K3 is the only active one. Notably, with the exception of nisin ABC + O9, all four nisin ABC conjugates showed activity. Most notably the activity of nisin ABC + O3K3 is considerably higher than that of nisin ABC. Antimicrobial activity of all compounds was tested by growth inhibition assays against two clinically relevant Gram-positive pathogens, i.e., methicillin resistant S. aureus and vancomycin resistant E. faecium, as well as L. monocytogenes, and L. lactis. Their minimal inhibitory concentration (MIC) was determined using an established broth microdilution assay (Table 2), using nisin as a positive control. Nisin AB was devoid of activity at the highest concentration tested except against E. faecium. Since of the nisin AB conjugates only nisin AB + O6K3 showed activity in the agar well diffusion assay, they were only tested against L. lactis. In this growth inhibition assay, nisin AB + O9 showed the best activity among the five nisin AB conjugates. Nisin ABC conjugates displayed a retained or even increased activity against E. faecium and L. lactis compared to nisin ABC alone, whereas activity against MW2-MRSA diminished. The antimicrobial activity of nisin ABC + O6K3 against E. faecium, L. monocytogenes, and L. lactis decreased only 8-, 4-, and 12-fold compared to full nisin, respectively. Strikingly, its antimicrobial activity against E. faecium, L. monocytogenes, and L. lactis increased 16-, 4-, and 2-fold compared to nisin ABC, respectively, and increased all twice compared to nisin ABC + O6, respectively. Compared to nisin ABC, nisin ABC + O6K3 displayed improved activity against E. faecium, L. monocytogenes, and L. lactis but decreased activity against MW2-MRSA while nisin ABC + O9 showed enhanced activity against E. faecium although activity against other strains was retained or even reduced. An additional test was performed to assess if the resistance to proteolytic degradation of the semi-synthetic nisin hybrids had improved compared to the parent compound. Nisin, nisin ABC + O6K3, and nisin ABC + O6 were exposed to the nisin resistance protein (NSR), a peptidase that cleaves the linear C-terminus of nisin. For this experiment, an activity test was performed against the NSR producing strain L. lactis NZ9000 (pNSR). In this test, NSR conferred its producing strain over 16-fold resistance toward nisin in MIC, caused by the proteolytic cleavage at the C-terminal tail of nisin. The hybrids Nisin ABC + O6K3 and nisin ABC + O6 bypassed this nisin resistance mechanism, having identical MICs against NZ9000 regardless of it producing NSR.
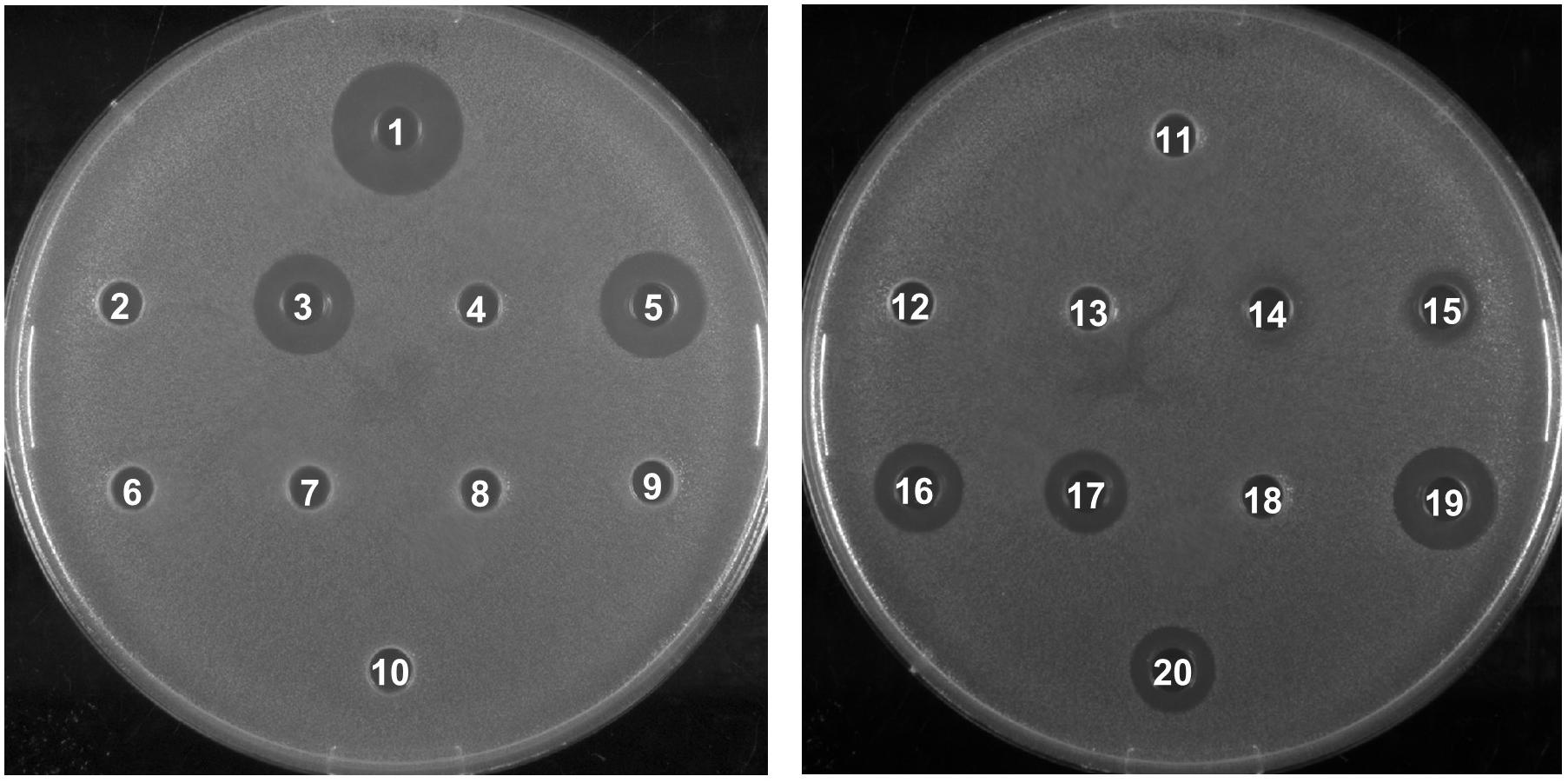
Figure 3. Antimicrobial activity of semi-synthetic nisin hybrids against M. flavus by agar well diffusion assay. 1: Nisin; 2: Nisin AB; 3: Nisin ABC; 4: Nisin AB-azide; 5: Nisin ABC-azide; 6: O3; 7: O6; 8: O9; 9: O3K3; 10: O6K3; 11: Nisin AB + O3; 12: Nisin AB + O6; 13: Nisin AB + O9; 14: Nisin AB + O3K3; 15: Nisin AB + O6K3; 16: Nisin ABC + O3; 17: Nisin ABC + O6; 18: Nisin ABC + O9; 19: Nisin ABC + O3K3; 20: Nisin ABC + O6K3.
Discussion
In this research, an efficient and direct method for the preparation of nisin hybrids was developed. Nisin AB and nisin ABC fragments were obtained by enzymatic digestion of nisin and these fragments were subsequently C-terminally functionalized with azidopropylamine. Five hydrophobic polyproline peptides (without and with cationic residues) were synthesized and coupled to nisin AB-azide and nisin ABC-azide by using click chemistry. Ten newly synthesized nisin hybrids were obtained and their antimicrobial activities were tested. The agar diffusion assay showed that the activity of nisin ABC conjugates are much better than nisin AB conjugates. These results are in line with previous studies that showed that variants lacking ring C, or where ring C is not closed, lack antimicrobial activity (Chan et al., 1996). It is noteworthy that while nisin AB is inactive having a lysine at the C-terminus, it gains higher activity through conjugation with O6K3 than with the more hydrophobic O6. The growth inhibition experiments showed that the activity of nisin ABC + O6K3 are better than nisin ABC + O6 and nisin ABC + O9, again indicating that addition of lysines (positive charge) at the C-terminal region can improve the activity. The antimicrobial activity of nisin ABC + O6K3 against E. faecium was 8-fold less active than full-length nisin. However, the activity was 16-fold better than nisin ABC, suggesting that modifying nisin ABC is a promising strategy to generate semi-synthetic nisin hybrids. It is notable that the inhibition activities of the semi-synthetic hybrids did not fully correlate when comparing solid media tests and broth MIC tests. However, this effect has been described for many nisin mutants, e.g., nisin A and nisin Z (de Vos et al., 1993). These compounds have an identical MIC, but a single amino acid difference leads to different halo sizes on diffusion assays, which is likely caused by altered diffusion properties. The five polyproline moieties that were tested in this study have varying hydrophobicities. Therefore, the lack of correlation between both essay results are likely caused by their distinct diffusion characteristics. In addition to their increased activity, these variants are not prone to degradation at the C-terminus by NSR, as was observed for nisin. Although the full proteolytic resistance of the conjugates was not tested, polyproline chains commonly have resistance against proteolysis in general, and convey this property to the nisin hybrids, providing a proof of concept. Notably, the method described in this study can be applied to conjugate other compatible (synthetic and non-proteinaceous) moieties that can provide the desired resistance to specific proteases. Nisin AB-azide and nisin ABC-azide can be readily generated with yields in the milligram range according to our optimized protocol. Future studies may focus on coupling peptides, especially anti-Gram-negative peptides, with nisin ABC-azide. Overall, this study highlights how lantibiotic fragments can be used as lead structures to create novel variants with altered properties (e.g., stability, activity, and specificity) via chemical coupling.
Data Availability Statement
All datasets generated for this study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author Contributions
JD and OK planned, conceived, and analyzed the experiments. VK designed the polyproline moieties. JD, JV, and VK performed the experiments. JD, VK, NB, and OK drafted the manuscript and contributed to the data interpretation. All authors read, critically revised, and approved the final manuscript.
Funding
JD was supported by the Chinese Scholarship Council (CSC). JV was supported by the Netherlands Organization for Scientific Research (NWO, ALWOP. 214). VK was supported by the DFG-funded research group 1805.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Manuel Montalban-Lopez (Department of Microbiology, Faculty of Sciences, University of Granada, Spain) for helpful discussions and thank Sander H. J. Smits for the pNSR construct (Institute of Biochemistry, Heinrich Heine University).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.575334/full#supplementary-material
References
Ahmad Fuaad, A. A. H., Azmi, F., Skwarczynski, M., and Toth, I. (2013). Peptide conjugation via CuAAC ‘click’ chemistry. Molecules 18, 13148–13174. doi: 10.3390/molecules181113148
Arnusch, C. J. (2008). The vancomycin-nisin (1-12) hybrid restores activity against vancomycin resistant enterococci. Biochemistry 47, 12661–12663. doi: 10.1021/bi801597b
Bolt, H. L., Kleijn, L. H. J., Martin, N. I., and Cobb, S. L. (2018). Synthesis of antibacterial nisin (-) peptoid hybrids using click methodology. Molecules 23:1566. doi: 10.3390/molecules23071566
Brötz, H., Josten, M., Wiedemann, I., Schneider, U., Götz, F., Bierbaum, G., et al. (1998). Role of lipid-bound peptidoglycan precursors in the formation of pores by nisin, epidermin and other lantibiotics. Mol. Microbiol. 30, 317–327. doi: 10.1046/j.1365-2958.1998.01065.x
Chan, W. C., Leyland, M., Clark, J., Dodd, H. M., Lian, L.-Y., Gasson, M. J., et al. (1996). Structure-activity relationships in the peptide antibiotic nisin antibacterial activity of fragments of nisin. FEBS Lett. 390, 129–132. doi: 10.1016/0014-5793(96)00638-2
de Vos, W. M., Mulders, J. W., Siezen, R. J., Hugenholtz, J., and Kuipers, O. P. (1993). Properties of nisin Z and distribution of its gene, nisZ, in Lactococcus lactis. Appl. Environ. Microbiol. 59, 213–218. doi: 10.1128/AEM.59.1.213-218
Gharsallaoui, A., Oulahal, N., Joly, C., and Degraeve, P. (2016). Nisin as a food preservative: part 1: physicochemical properties, antimicrobial activity, and main uses. Crit. Rev. Food Sci. Nutr. 56, 1262–1274. doi: 10.1080/10408398.2013.763765
Hansen, J. N., and Sandine, W. E. (1994). Nisin as a model food preservative. Crit. Rev. Food Sci. Nutr. 34, 69–93. doi: 10.1080/10408399409527650
Jiang, X., Hao, X., Jing, L., Wu, G., Kang, D., Liu, X., et al. (2019). Recent applications of click chemistry in drug discovery. Expert Opin. Drug Discov. 14, 779–789. doi: 10.1080/17460441.2019.1614910
Khosa, S., AlKhatib, Z., and Smits, S. H. (2013). NSR from Streptococcus agalactiae confers resistance against nisin and is encoded by a conserved nsr operon. Biol. Chem. 394, 1543–1549. doi: 10.1515/hsz-2013-0167
Khosa, S., Frieg, B., Mulnaes, D., Kleinschrodt, D., Hoeppner, A., Gohlke, H., et al. (2016). Structural basis of lantibiotic recognition by the nisin resistance protein from Streptococcus agalactiae. Sci. Rep. 6:18679. doi: 10.1038/srep18679
Kolb, H. C., Finn, M. G., and Sharpless, K. B. (2001). Click chemistry: diverse chemical function from a few good reactions. Angew. Chem. Int. Ed. 40, 2004–2021. doi: 10.1002/1521-3773(20010601)40:11<2004::AID-ANIE2004>3.0.CO;2-5
Koopmans, T., Wood, T. M., Hart, P. T., Kleijn, L. H. J., Hendrickx, A. P. A., Willems, R. J. L., et al. (2015). Semisynthetic lipopeptides derived from nisin display antibacterial activity and lipid II binding on par with that of the parent compound. J. Am. Chem. Soc. 137, 9382–9389. doi: 10.1021/jacs.5b04501
Kubyshkin, V., and Budisa, N. (2018). Exploring hydrophobicity limits of polyproline helix with oligomeric octahydroindole-2-carboxylic acid. J. Pept. Sci. 24:e3076. doi: 10.1002/psc.3076
Kubyshkin, V., Grage, S. L., Ulrich, A. S., and Budisa, N. (2019). Bilayer thickness determines the alignment of model polyproline helices in lipid membranes. Phys. Chem. Chem. Phys. 21:22396. doi: 10.1039/c9cp02996f
Kuipers, O. P., de Ruyter, P. G. G. A., Kleerebezem, M., and de Vos, W. M. (1998). Quorum sensing-controlled gene expression in lactic acid bacteria. J. Biotechnol. 64, 15–21. doi: 10.1016/S0168-1656(98)00100-X
Lubelski, J., Rink, R., Khusainov, R., Moll, G. N., and Kuipers, O. P. (2008). Biosynthesis, immunity, regulation, mode of action and engineering of the model lantibiotic nisin. Cell. Mol. Life Sci. 65, 455–476. doi: 10.1007/s00018-007-7171-2
McKay, C. S., and Finn, M. G. (2014). Click chemistry in complex mixtures: bioorthogonal bioconjugation. Chem. Biol. 21, 1075–1101. doi: 10.1016/j.chembiol.2014.09.002
Montalbán-López, M., Deng, J., van Heel, A. J., and Kuipers, O. P. (2018). Specificity and application of the lantibiotic protease NisP. Front. Microbiol. 9:160. doi: 10.3389/fmicb.2018.00160
Oldach, F., Al Toma, R., Kuthning, A., Caetano, T., Mendo, S., Budisa, N., et al. (2012). Congeneric lantibiotics from ribosomal in vivo peptide synthesis with noncanonical amino acids. Angew. Chem. Int. Ed. 51, 415–418. doi: 10.1002/anie.201106154
Rogers, L. A. (1928). The inhibiting effect of Streptococcus Lactios on Lactobacillus Bulgaricus. J. Bacteriol. 16, 321–325.
Schmitt, S., Montalbán-López, M., Peterhoff, D., Deng, J., Wagner, R., Held, M., et al. (2019). Analysis of modular bioengineered antimicrobial lanthipeptides at nanoliter scale. Nat. Chem. Biol. 15, 437–443. doi: 10.1038/s41589-019-0250-5
Shin, J. M., Gwak, J. W., Kamarajan, P., Fenno, J. C., Rickard, A. H., and Kapila, Y. L. (2016). Biomedical applications of nisin. J.Appl. Microbiol. 120, 1449–1465. doi: 10.1111/jam.13033
Slootweg, J. C., Peters, N., van Ufford, H. L. C. Q., Breukink, E., Liskamp, R. M. J., and Rijkers, D. T. S. (2014). Semi-synthesis of biologically active nisin hybrids composed of the native lanthionine ABC-fragment and a cross-stapled synthetic DE-fragment. Bioorg. Med. Chem. 22, 5345–5353. doi: 10.1016/j.bmc.2014.07.046
Slootweg, J. C., van der Wal, S., van Ufford, H. C. Q., Breukink, E., Liskamp, R. M. J., and Rijkers, D. T. S. (2013a). Synthesis, antimicrobial activity, and membrane permeabilizing properties of C-terminally modified nisin conjugates accessed by CuAAC. Bioconj. Chem. 24, 2058–2066. doi: 10.1021/bc400401k
Slootweg, J. C., Liskamp, R. M. J., and Rijkers, D. T. S. (2013b). Scalable purification of the lantibiotic nisin and isolation of chemical/enzymatic cleavage fragments suitable for semi-synthesis. J. Pept. Sci. 19, 692–699. doi: 10.1002/psc.2551
Thirumurugan, P., Matosiuk, D., and Jozwiak, K. (2013). Click chemistry for drug development and diverse chemical-biology applications. Chem. Rev. 113, 4905–4979. doi: 10.1021/cr200409f
van Heel, A. J., Mu, D., Montalbán-López, M., Hendriks, D., and Kuipers, O. P. (2013). Designing and producing modified, new-to-nature peptides with antimicrobial activity by use of a combination of various lantibiotic modification enzymes. ACS Synth. Biol. 2, 397–404. doi: 10.1021/sb3001084
Wiegand, I., Hilpert, K., and Hancock, R. E. W. (2008). Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat. Protoc. 3, 163–175. doi: 10.1038/nprot.2007.521
Keywords: click chemistry, RiPPs, lantibiotics, nisin, polyproline peptides
Citation: Deng J, Viel JH, Kubyshkin V, Budisa N and Kuipers OP (2020) Conjugation of Synthetic Polyproline Moietes to Lipid II Binding Fragments of Nisin Yields Active and Stable Antimicrobials. Front. Microbiol. 11:575334. doi: 10.3389/fmicb.2020.575334
Received: 23 June 2020; Accepted: 30 October 2020;
Published: 20 November 2020.
Edited by:
Paul David Cotter, Teagasc Food Research Centre, IrelandReviewed by:
Ratchaneewan Aunpad, Thammasat University, ThailandHilario C. Mantovani, Universidade Federal de Viçosa, Brazil
Copyright © 2020 Deng, Viel, Kubyshkin, Budisa and Kuipers. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Oscar P. Kuipers, o.p.kuipers@rug.nl
 Jingjing Deng1
Jingjing Deng1 Vladimir Kubyshkin
Vladimir Kubyshkin Nediljko Budisa
Nediljko Budisa Oscar P. Kuipers
Oscar P. Kuipers