- 1Laboratory of Microbiology, Department of Biochemistry and Biotechnology, Faculty of Sciences, Ghent University, Ghent, Belgium
- 2BCCM/LMG Bacteria Collection, Department of Biochemistry and Biotechnology, Faculty of Sciences, Ghent University, Ghent, Belgium
- 3Division of Infectious Diseases, Department of Pediatrics, The University of British Columbia, Vancouver, BC, Canada
- 4Department of Pediatrics, University of Michigan Medical School, Ann Arbor, MI, United States
- 5Laboratory of Pharmaceutical Microbiology, Department of Pharmaceutical Analysis, Faculty of Pharmaceutical Sciences, Ghent University, Ghent, Belgium
The objective of the present study was to provide an updated classification for Burkholderia cepacia complex (Bcc) taxon K isolates. A representative set of 39 taxon K isolates were analyzed through multilocus sequence typing (MLST) and phylogenomic analyses. MLST analysis revealed the presence of at least six clusters of sequence types (STs) within taxon K, two of which contain the type strains of Burkholderia contaminans (ST-102) and Burkholderia lata (ST-101), and four corresponding to the previously defined taxa Other Bcc groups C, G, H and M. This clustering was largely supported by a phylogenomic tree which revealed three main clades. Isolates of B. contaminans and of Other Bcc groups C, G, and H represented a first clade which generally shared average nucleotide identity (ANI) and average digital DNA-DNA hybridization (dDDH) values at or above the 95–96% ANI and 70% dDDH thresholds for species delineation. A second clade consisted of Other Bcc group M bacteria and of four B. lata isolates and was supported by average ANI and dDDH values of 97.2 and 76.1% within this clade and average ANI and dDDH values of 94.5 and 57.2% toward the remaining B. lata isolates (including the type strain), which represented a third clade. We therefore concluded that isolates known as Other Bcc groups C, G, and H should be classified as B. contaminans, and propose a novel species, Burkholderia aenigmatica sp. nov., to accommodate Other Bcc M and B. lata ST-98, ST-103, and ST-119 isolates. Optimized MALDI-TOF MS databases for the identification of clinical Burkholderia isolates may provide correct species-level identification for some of these bacteria but would identify most of them as B. cepacia complex. MLST facilitates species-level identification of many taxon K strains but some may require comparative genomics for accurate species-level assignment. Finally, the inclusion of Other Bcc groups C, G, and H into B. contaminans affects the phenotype of this species minimally and the proposal to classify Other Bcc group M and B. lata ST-98, ST-103, and ST-119 strains as a novel Burkholderia species is supported by a distinctive phenotype, i.e., growth at 42°C and lysine decarboxylase activity.
Introduction
The Burkholderia cepacia complex (Bcc) is a group of closely related bacteria that occur naturally in a wide variety of ecological niches and that possess a remarkably versatile metabolism (Coenye et al., 2001; Parke and Gurian-Sherman, 2001; Depoorter et al., 2016; Kunakom and Eustáquio, 2019). Despite their agricultural potential, these organisms are perhaps best known as opportunistic pathogens, causing persistent infections in persons with cystic fibrosis (CF). The taxonomy of these bacteria is complex and continuously evolving. At present, Bcc consists of at least 22 validly named species (Peeters et al., 2013; De Smet et al., 2015; Bach et al., 2017; Martina et al., 2018). One additional species, i.e., “Burkholderia paludis” was effectively described but its name is awaiting validation (Ong et al., 2016).
One common group of Bcc bacteria, previously known as taxon K or group K, has presented a challenge in terms of classification (Vermis et al., 2002; Payne et al., 2005; Vanlaere et al., 2009). Taxon K now consists of two validly named species, Burkholderia contaminans and Burkholderia lata, and has been isolated from numerous human and environmental sources, including CF sputum, blood, cerebrospinal fluid, river water, sediment, soil, and various plants (Baldwin et al., 2005; Mahenthiralingam et al., 2006; Cesarini et al., 2009; da Silva et al., 2012). B. contaminans can be considered an emerging CF pathogen; it is the most common Bcc species in CF patients in Argentina (Cipolla et al., 2018) and it is increasingly isolated in CF centers in Spain (Medina-Pascual et al., 2015), the United Kingdom (Kenna et al., 2017), Portugal (Coutinho et al., 2015), and Ireland (Power et al., 2016). In addition, these organisms are frequently responsible for outbreaks of healthcare-associated infections due to contamination of pharmaceutical products such as nasal spray (CDC, 2004), dialysis water (Souza et al., 2004), moist washcloths (Martin et al., 2011), mouthwash (Zurita et al., 2014), and liquid docusate laxative (Glowicz et al., 2018).
A previous study that addressed the taxonomic position and structure of taxon K by means of recA gene sequence analysis and multilocus sequence typing (MLST) entailed the description of B. contaminans and B. lata (Vanlaere et al., 2009). However, the data presented indicated that the classification of taxon K bacteria was not completely resolved and would benefit from future study of a larger set of isolates. Today, many additional taxon K isolates have been collected through CF surveillance programs, environmental studies and outbreak investigations. In 2014, a study mining the Bcc PubMLST database using the 3% threshold value of average concatenated allele sequence divergence for species delineation (Vanlaere et al., 2009) revealed the presence of 16 putative novel Bcc species (Vandamme and Peeters, 2014), four of which (referred to as Other Bcc groups C, G, H, and M) grouped with B. contaminans and B. lata in a single taxon K clade that was supported by high bootstrap values (Vandamme and Peeters, 2014). The aim of the present study was to re-evaluate the taxonomy of taxon K bacteria. We performed a phylogenomic analysis to investigate established and potentially novel Bcc species within taxon K and we propose a revised classification of these common Bcc bacteria with considerable clinical relevance.
Materials and Methods
Bacterial Strains and Growth Conditions
Table 1 lists the sources of the 39 studied taxon K isolates. Isolates were grown aerobically on Tryptone Soya Agar (TSA, Oxoid CM0131) and incubated at 28°C. Cultures were preserved in MicroBankTM vials at −80°C.
Genome Sequencing, Assembly and Annotation
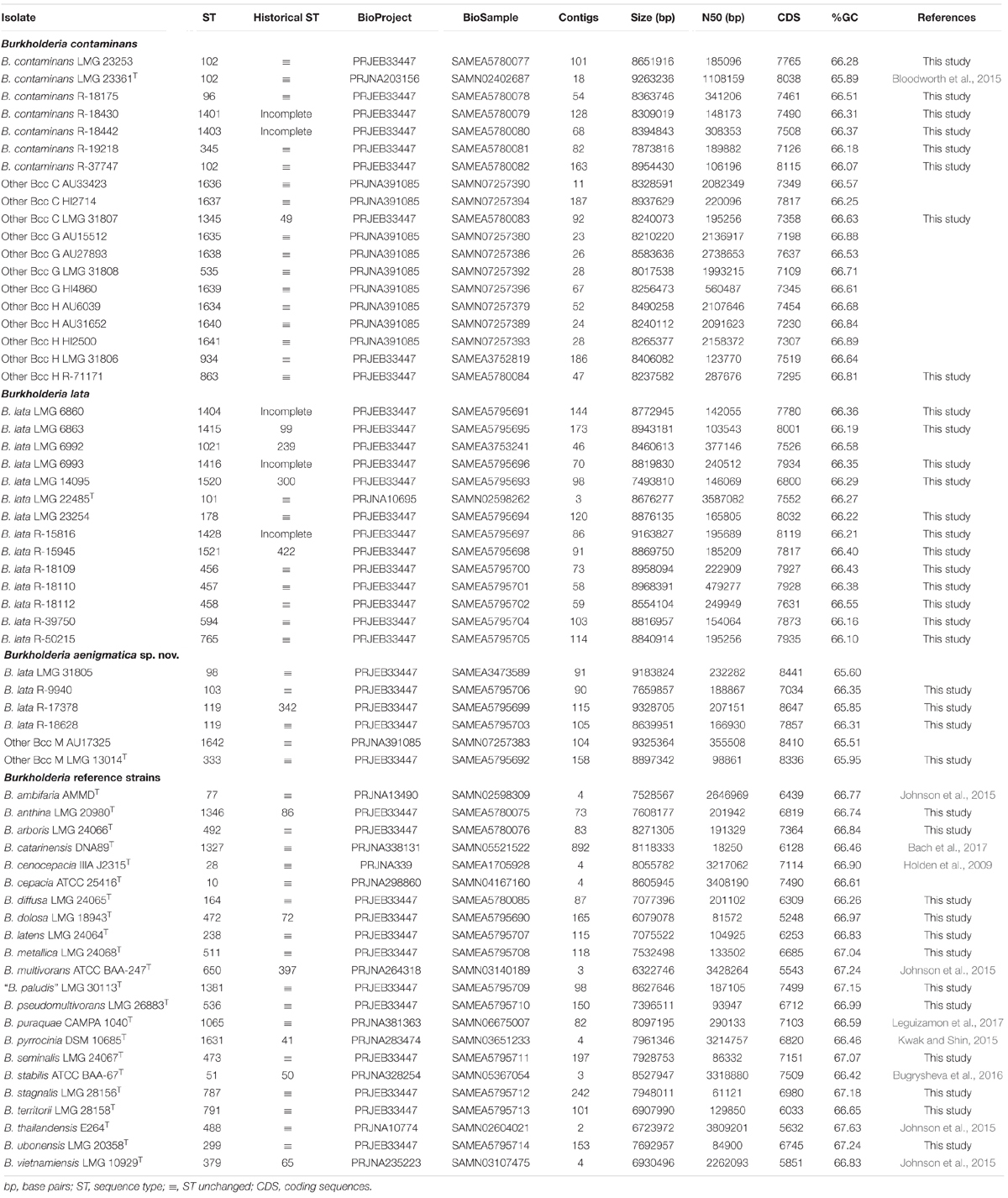
Genomic DNA of 36 isolates (24 taxon K isolates and 12 Bcc type strains) was prepared from a 48-h culture at 28°C on TSA. Cells were harvested using a 10 μl inoculation loop and suspended in 300 μl of 4 M guanidine isothiocyanate solution (Fischer Scientific). DNA extraction was performed using the Maxwell© 16 automated nucleic acid purification system using the Maxwell© 16 Tissue DNA purification kit (Promega) according to the manufacturer’s instructions. Paired-end 150 bp libraries were sequenced on an Illumina HiSeq4000 sequencer (Oxford Genomics Centre – Wellcome Centre for Human Genetics, Oxford, United Kingdom). Quality control, assembly and annotation were performed as described previously (Peeters et al., 2019). Annotation was performed using Prokka v1.12 (Seemann, 2014) with a genus-specific database based on reference genomes from the Burkholderia Genome Database v8.1 (Winsor et al., 2008).
Publicly Available Genomes
The publicly available whole-genome sequences of 12 taxon K isolates and 10 Burkholderia type strains were downloaded from the Burkholderia Genome Database and the NCBI database (Table 2). Publicly available raw Illumina reads from three taxon K isolates were downloaded from the European Nucleotide Archive (Table 2). These reads were assembled and annotated as described above.
recA Gene Sequence Analysis
Partial recA gene sequences (663 bp) of the 61 Burkholderia isolates in the present study were extracted from the assembled whole-genome sequences using BLAST searches. The sequences were aligned based on their amino acid sequences using Muscle (Edgar, 2004) in MEGA7 (Kumar et al., 2016). A phylogenetic tree was constructed using the maximum likelihood method in RAxML version 8.2.12 (Stamatakis, 2014). Rapid bootstrapping and ML search were performed using the general time reversible model with CAT approximation (GTRCAT). Visualization and annotation of the phylogenetic tree was performed using iTOL (Letunic and Bork, 2019).
MLST Analysis
Multilocus sequence typing allele sequences were extracted from assembled genomes through BLAST searches against the Bcc PubMLST website (Jolley and Maiden, 2010). Nucleotide sequences of each allele, allelic profiles and sequence types for all isolates in the present study are available on the Bcc PubMLST website1. For phylogenetic analysis, concatenated allele sequences were exported from the Bcc PubMLST database. Isolate metadata were processed using Rstudio and R version 3.4.4. A phylogenetic tree was constructed as described above.
Phylogenomic Analyses
Pairwise digital DNA-DNA hybridization (dDDH) values and their confidence intervals were determined using the Genome-to-Genome Distance Calculator 2.1 (GGDC) (Meier-Kolthoff et al., 2013). Average nucleotide identity (ANI) values were calculated through OrthoANI using USEARCH (Lee et al., 2016; Yoon et al., 2017). Protein sequences were extracted from annotated genomes using the SeqIO utility of Biopython (Cock et al., 2009). BcgTree was used to extract the amino acid sequences of 107 essential bacterial single-copy core genes and to construct a phylogenomic tree through partitioned maximum-likelihood analysis (Ankenbrand and Keller, 2016). Visualization and annotation of the phylogenetic tree was performed using iTOL (Letunic and Bork, 2019).
Matrix-Assisted Laser Desorption/Ionization Time-of-Flight Mass Spectrometry (MALDI-TOF MS) Analysis
All 39 taxon K isolates were grown on TSA (Oxoid CM0131) for 48 h at 28°C and were subcultivated twice prior to harvesting. Cell extracts were prepared as described previously (Wieme et al., 2014). For all isolates examined, both cell extracts (1 μl) and homogeneous cell smears of isolated colonies were transferred to the spot sites of a 96-well stainless steel target plate (Bruker Daltonics). Both cell smears and cell extracts were air-dried at room temperature. Each spot was overlaid with 1 μl of matrix solution (10 mg/ml α-cyano-4-hydroxycinnamic acid in acetonitrile:trifluoroacetic acid:MilliQ [50:2.5:47.5] water-solvent) and left to air-dry at room temperature. Cell smears were spotted in triplicate and cell extracts were spotted in duplicate to verify reproducibility. Prior to analysis, the mass spectrometer was externally calibrated using the Bacterial Test Standard (Bruker Daltonics) used for calibration. The target plate was measured twice automatically on a Bruker MicroflexTM LT/SH Smart platform (Bruker Daltonics). The spectra were obtained in linear, positive ion mode using FlexControl 3.4 software according to the manufacturer’s recommended settings (Bruker Daltonics).
The spectra of the cell smears and cell extracts were compared to those in the MALDI BioTyper in vitro diagnostics (MBT IVD) (DB-7712 MSP) library using the MBT Compass Explorer 4.1 software (Bruker Daltonics) according to the manufacturer’s default settings. The spectra were also compared with the combined spectra of the MBT IVD library and the LM-UGent in-house library, which consists of 298 main spectra (MSPs) representing 271 Burkholderia reference strains for which the identification has been verified through recA gene sequence analysis or MLST analysis, and which includes 17 taxon K strains (Supplementary Table 1). This in-house library was constructed by preparing a cell extract for each strain as described above, spotting each extract eight times on the target plate and measuring each spot four times to generate 32 spectra. FlexAnalysis 3.4 software (Bruker Daltonics) was used to assess the quality of the spectra generated. MBT Compass Explorer 4.1 software was used to generate MSPs based on a minimum of 24 spectra according to manufacturer’s instructions. Species-level identification was performed according to the Optimized Acceptance Criterium (OAC) (De Bel et al., 2011), meaning that the identification of the best matching spectrum was accepted in case of a log score ≥2.0 and when a 0.200 log difference was observed between the best matching spectrum and the highest-scoring spectrum of a different species.
Biochemical Characterization and Cellular Fatty Acid Analysis
After a 24 h incubation period at 28°C on TSA (BD), a loopful of well-grown cells was harvested and fatty acid methyl esters were prepared, separated and identified using the Microbial Identification System (Microbial ID) as described previously (Vandamme et al., 1992). Biochemical characterization was performed as described previously (Henry et al., 2001).
Results
Whole-Genome Sequencing
To further characterize the taxonomic structure of B. contaminans, B. lata and related organisms, 61 isolates were selected for genome sequence analysis, including 39 taxon K isolates (24 newly sequenced, 12 publicly available whole-genome sequences and 3 publicly available sequencing read sets) and 22 Burkholderia reference strains (12 newly sequenced and 10 publicly available whole-genome sequences). The type strain of Burkholderia thailandensis was included as an outgroup. The whole-genome sequence of 36 isolates for which no genome sequence was publicly available was determined. The assembly of the Illumina HiSeq 150 bp paired-end reads resulted in assemblies with 46–242 contigs with N50 values ranging from 61 to 479 Kbp and a total genome size of 6.1–9.3 Mbp (Table 2). Both the raw Illumina reads and the annotated assemblies of these 36 genomes were submitted to the European Nucleotide Archive and are publicly available through the GenBank/EMBL/DDBJ accession numbers listed in Table 2. The genome sequences of the remaining isolates were publicly available (Table 2).
MLST and recA Gene Sequence Analysis
Although 16S rRNA gene sequence analysis is commonly used to identify the nearest neighbors of potentially novel taxa (Chun et al., 2018), the taxonomic resolution of this gene has proven insufficient to discriminate Burkholderia bacteria, in particular members of the Bcc (Vandamme and Dawyndt, 2011). Isolates were therefore assigned to the Other Bcc groups C, G, H, and M by means of MLST analysis as described previously (Vandamme and Peeters, 2014). Phylogenetic analysis of concatenated allele sequences of 1120 Bcc strains with unique STs present in the Bcc PubMLST database showed that taxon K strains represented a monophyletic clade (Figure 1). Within this clade, at least six clusters were observed. The type strains of B. contaminans LMG 23361T and B. lata LMG 22485T each resided in a stable cluster with bootstrap values of 96%. The Other Bcc group C, G, H, and M strain clusters were mostly supported by high bootstrap values (i.e., 100, 60, 82, and 99, respectively). A phylogenetic tree based on recA gene sequences is presented in Supplementary Figure 1.

Figure 1. Phylogenetic tree based on the concatenated sequences (2760 bp) of seven housekeeping gene fragments of established Bcc species, taxon K strains and related Other Bcc isolates. Sequences [atpD (443 bp), gltB (400 bp), gyrB (454 bp), recA (393 bp), lepA (397 bp), phaC (385 bp), and trpB (301 bp)] corresponding to 1120 sequence types (STs) were downloaded from the Bcc PubMLST database (https://pubmlst.org/bcc/) (Jolley and Maiden, 2010) and aligned based on their amino acid sequences. Phylogeny was inferred using the Maximum Likelihood method and GTRCAT substitution model in RAxML. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1,000 replicates) are shown next to the branches if greater than 50%. The scale bar indicates the number of nucleotide substitutions per site. STs indicated in bold font were used for phylogenetic analyses and 39 taxon K STs indicated in bold were used for MALDI-TOF MS analyses. Gray circles indicate collapsed branches, the number of STs in such a collapsed branch is given after the species name between parentheses. STs that were reclassified into the novel species Burkholderia aenigmatica sp. nov. are marked in blue.
Phylogenomic Analyses
The MLST dendrogram was used to select 39 taxon K isolates for genome sequence analyses (Figure 1, selected STs indicated in bold character type). The Maximum Likelihood phylogenetic tree resulting from the concatenated amino acid alignment of 107 single copy core genes was well-resolved and showed high bootstrap support on most branches (Figure 2). The B. contaminans and B. lata type strains were well-separated and grouped into distinct major clades. A first major clade included the B. contaminans type strain LMG 23361T (ST-102) and consisted of four clusters (B. contaminans and Other Bcc groups C, G, and H), each supported by bootstrap values of 100%. A second major clade in the phylogenomic tree (Figure 2) included the B. lata type strain LMG 22485T (ST-101) and consisted of four clusters of two to five isolates, supported by bootstrap values of 100% each, and two isolates with separate positions (i.e., R-39750 [ST-594] and LMG 23254 [ST-178]). B. lata isolates LMG 31805 (ST-98) and R-9940 (ST-103), which were present in a stable cluster (supported by a bootstrap value of 96%) with the B. lata type strain LMG 22485T (ST-101) in the MLST tree (Figure 1), formed a distinct third clade together with the two ST-119 isolates R-17378 and R-18628 and the two Other Bcc M isolates in the phylogenomic tree. This third clade was supported by a bootstrap value of 100% (Figure 2).
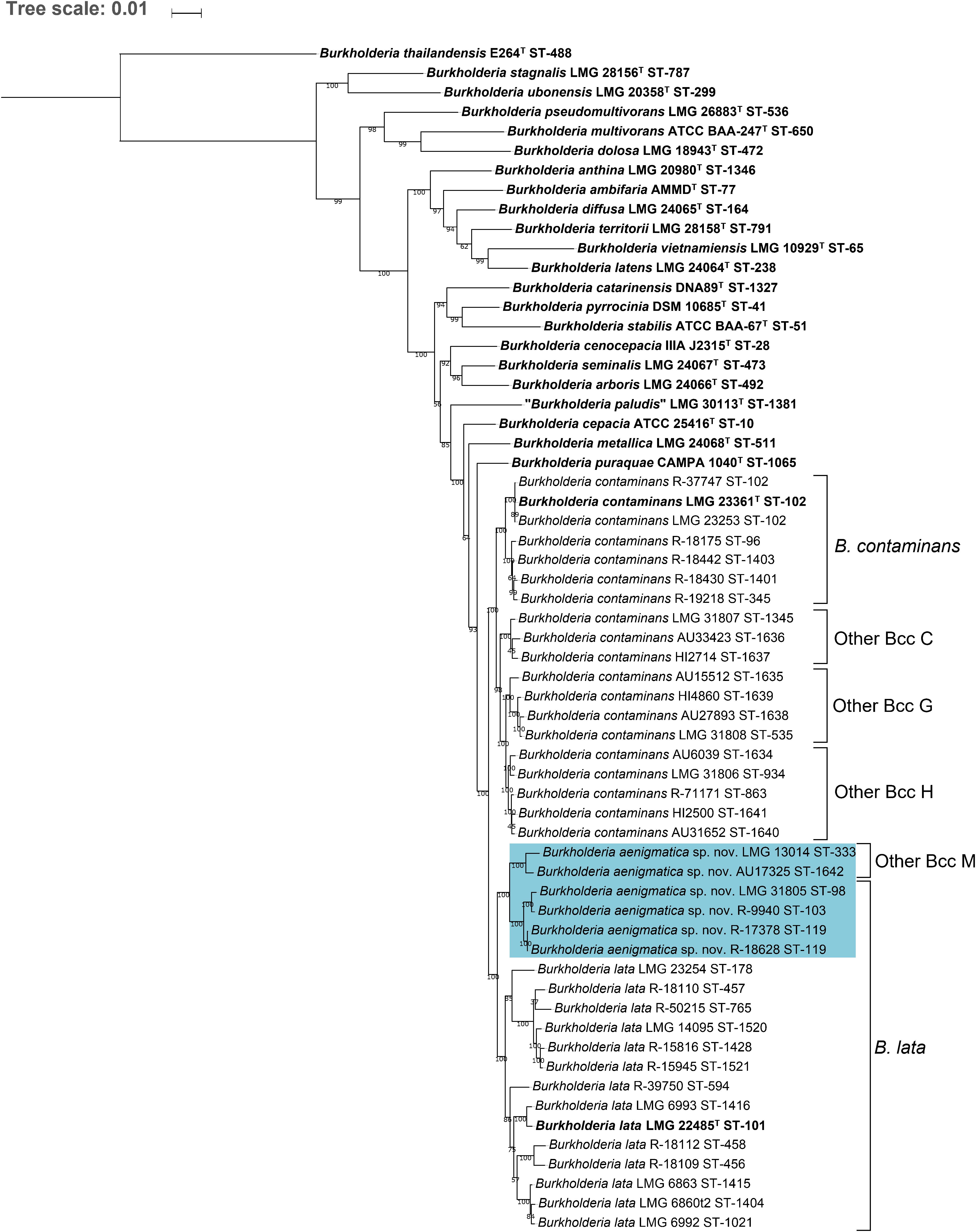
Figure 2. Whole-genome phylogeny of established Bcc species, taxon K strains and related Other Bcc group isolates. bcgTree was used to extract the amino acid sequences of 107 single-copy core genes from the genomes of 61 Burkholderia strains and to reconstruct their phylogeny through partitioned maximum-likelihood analysis (Ankenbrand and Keller, 2016). The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1,000 replicates) are shown next to the branches if greater than 50%. The scale bar indicates the number of amino acid substitutions per site. Species names on the branches correspond to the taxonomic proposals in the present study, previous nomenclature is indicated next to braces on the right. Bold font indicates type strains of established Bcc species. Strains that were reclassified into the novel species Burkholderia aenigmatica sp. nov. are marked in blue.
The pairwise ANI and dDDH values of the 39 taxon K isolates and 22 Burkholderia reference strain genomes are presented in Supplementary Tables 2, 3 and average within-cluster and between-cluster ANI and dDDH values are presented in Table 3. Within the first major clade containing B. contaminans and Other Bcc groups C, G, and H, the average within-cluster ANI and dDDH values ranged from 97.7% ANI and 79.7% dDDH in Other Bcc G to 98.6% ANI and 87.4% dDDH in Other Bcc C (Table 3). The average between-cluster ANI and dDDH values ranged from 95.4% ANI and 62.4% dDDH between B. contaminans and Other Bcc C to 96.7% ANI and 71.2% dDDH between Other Bcc G and Other Bcc H (Table 3).
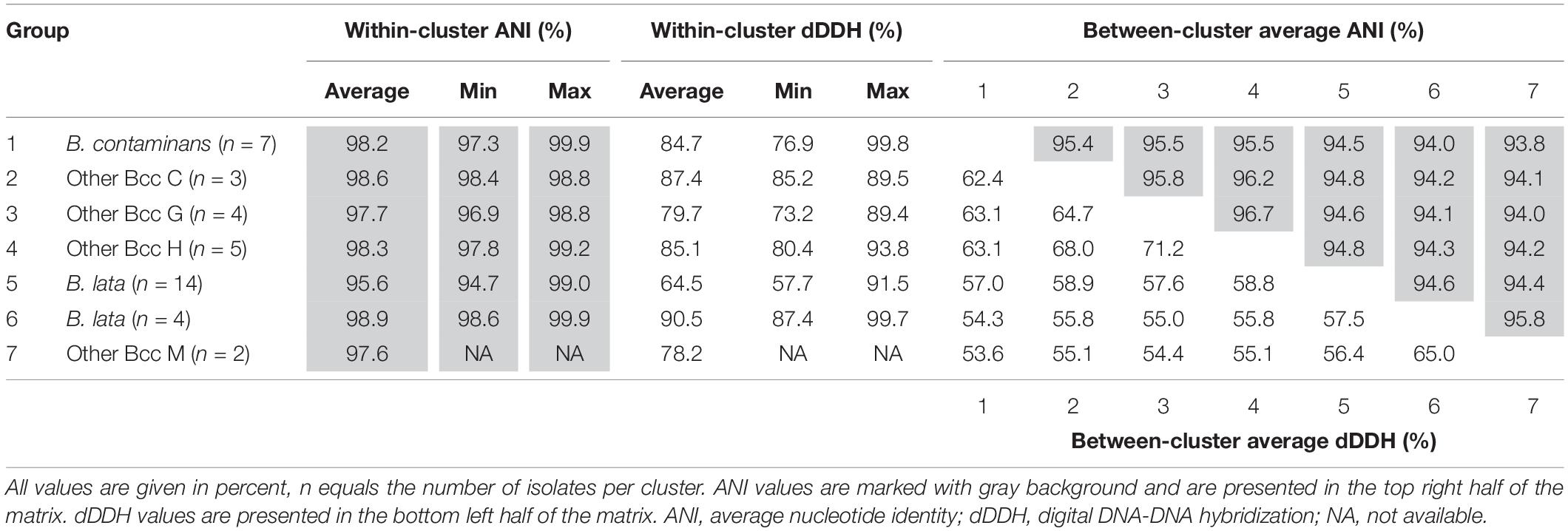
Table 3. Average within- and between-cluster ANI and dDDH values of seven clusters of taxon K isolates.
Burkholderia lata isolates belonging to the second major clade containing the B. lata type strain (Figure 2) appeared to yield a continuum of ANI and dDDH values in which several clusters of closely related strains could be distinguished. Strains (i) LMG 22485T and LMG 6993, (ii) LMG 6860, LMG 6863 and LMG 6992, (iii) R-18109 and R-18112, and (iv) LMG 14095, R-18110, 15816, R-15945 and R-50215 revealed high pairwise ANI and dDDH each (i.e., values of at least 97.4 and 77.0%, respectively) (Supplementary Tables 2, 3). Values between strains of each of these four clusters and the two strains with unique positions (i.e., R-39750 and LMG 23254) ranged between 94.7 and 96.3% ANI, with an average of 95.2%; and between 57.9 and 68.5% dDDH, with an average of 60.9% (Supplementary Tables 2, 3).
Within the third major clade, the average within-cluster ANI and dDDH values ranged from 97.6% ANI and 78.2% dDDH in Other Bcc M to 98.9% ANI and 90.5% dDDH in the cluster containing B. lata isolates LMG 31805, R-9940, R-17378, and R-18628 (Table 3). Average between-cluster ANI and dDDH values of 95.8% ANI and 65.0% dDDH were observed between these two clusters (Table 3).
MALDI-TOF MS Analysis
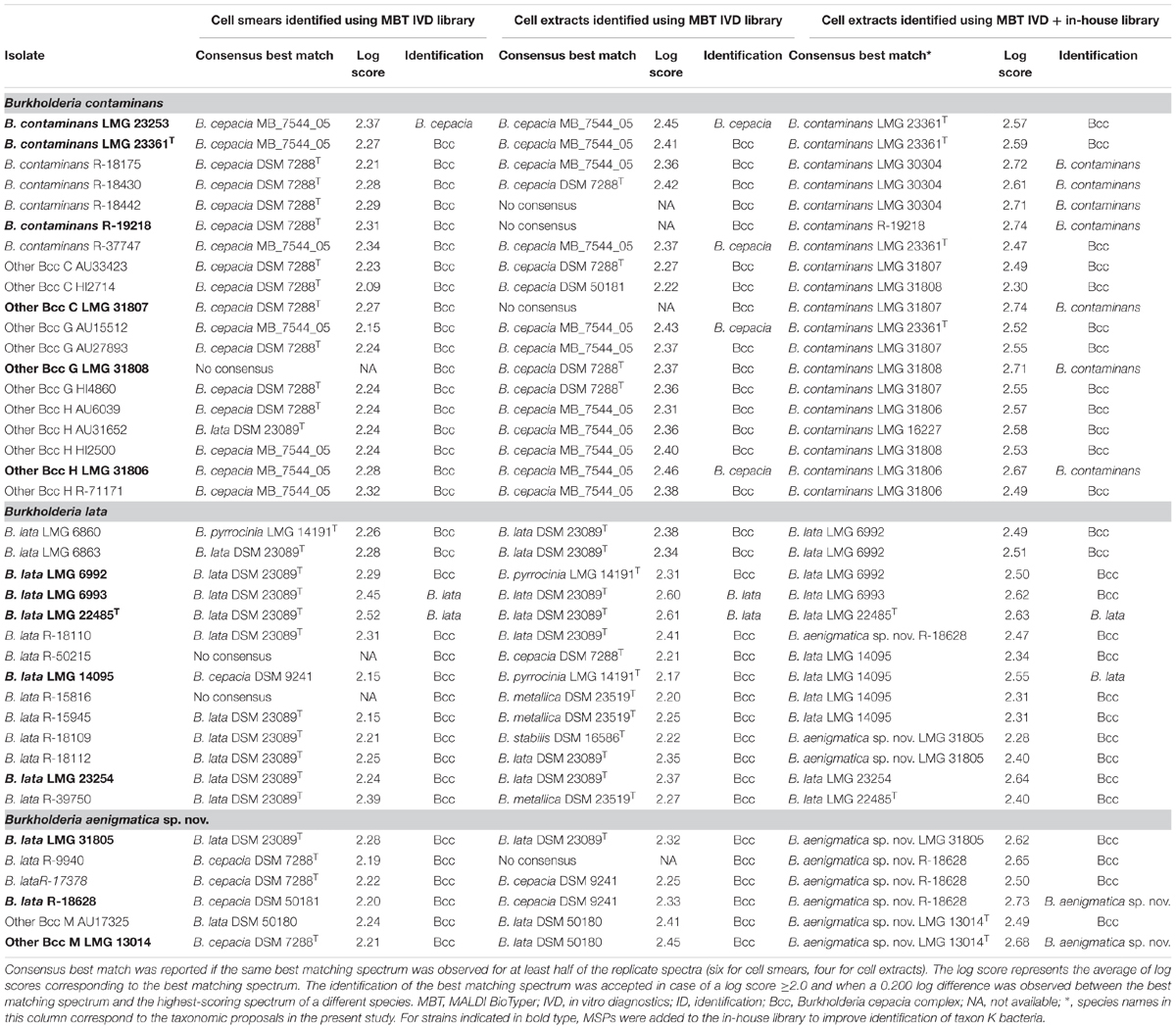
All 39 taxon K isolates included in the present study were analyzed and identified using MALDI-TOF MS in two ways. Analogous to the workflow in a routine clinical microbiology lab (Van Veen et al., 2010), isolates were analyzed by using the direct smear method and were identified by using the MBT IVD library. The resulting log scores of the best matching spectra are presented in Table 4 and ranged from 2.21–2.37 (B. contaminans), 2.09–2.27 (Other Bcc C), 2.15–2.24 (Other Bcc G), 2.24–2.32 (Other Bcc H), 2.15–2.52 (B. lata), and 2.21–2.24 (Other Bcc M). According to the optimized acceptance criteria for identification of Bcc bacteria (De Bel et al., 2011), only three taxon K strains could be identified to species level. B. lata strains LMG 22485T and LMG 6993 were correctly identified, whereas B. contaminans LMG 23253 was misidentified as B. cepacia. The remaining isolates could not reliably be identified to species level and were identified as B. cepacia complex.
Isolates were also analyzed using a cell extract method and were identified by using the MBT IVD library. The resulting log scores from the best matching spectra are presented in Table 4 and ranged from 2.36–2.45 (B. contaminans), 2.22–2.27 (Other Bcc C), 2.36–2.43 (Other Bcc G), 2.31–2.46 (Other Bcc H), 2.17–2.61 (B. lata), and 2.41–2.45 (Other Bcc M). Six taxon K strains were considered identified to species level, only two of which were correctly identified as B. lata (LMG 22485T and LMG 6993). Misidentification as B. cepacia was observed for two B. contaminans isolates (LMG 23253 and R-37747), one Other Bcc G strain (AU15512) and one Other Bcc H strain (LMG 31806). The remaining isolates could not reliably be identified to species level and were identified as B. cepacia complex.
To improve species-level identification, isolates were also analyzed using a cell extract method and were identified by using the spectra present in both the MBT IVD library and our in-house library of Burkholderia reference strains. The in-house library (Supplementary Table 1) included nine strains assigned below to B. contaminans, i.e., six B. contaminans strains (LMG 16227, LMG 23253, LMG 23361T, LMG 30304, R-18428 and R-19218), and one strain of each of the Other Bcc groups C (LMG 31807), G (LMG 31808) and H (LMG 31806); five strains belonging to the revised species B. lata (LMG 14095, LMG 22485T, LMG 23254, LMG 6992 and LMG 6993), and finally, three strains belonging to the proposed new taxon K species (LMG 13014T, LMG 31805 and R-18628) (see below). The resulting log scores from the best matching spectra are presented in Table 4 and ranged from 2.47–2.74 (B. contaminans), 2.30–2.74 (Other Bcc C), 2.52–2.71 (Other Bcc G), 2.49–2.67 (Other Bcc H), 2.28–2.73 (B. lata), and 2.49–2.68 (Other Bcc M). Eleven taxon K strains were correctly identified to species level, including four B. contaminans strains, one strain of each of the Other Bcc C, G, and H groups, two B. lata strains and two strains belonging to the proposed new species (Table 4). The remaining 28 isolates could not reliably be identified to the species level using the optimized acceptance criteria and were identified as B. cepacia complex. Nevertheless, the best matching spectra mostly represented isolates of the corresponding species.
Biochemical Characterization and Cellular Fatty Acid Analysis
Biochemical characteristics and cellular fatty acid profiles were determined for seven representative strains: one strain from each of the Other Bcc groups C, G, H, and M and three additional B. lata isolates (LMG 23254, LMG 14095, and R-39750) The results are listed in Supplementary Table 4. Other Bcc M strain LMG 13014 could be distinguished from B. lata strains through growth at 42°C and lysine decarboxylase activity. The main fatty acid components are C16:0, C17:0 cyclo and C18:1 ω7c, in correspondence with the values reported previously for taxon K strains and Bcc bacteria in general (Vanlaere et al., 2009).
Discussion
The Bcc PubMLST database serves as a framework to study the epidemiology and biodiversity of Bcc bacteria. To resolve the taxonomy of taxon K bacteria, a phylogenetic tree based on concatenated allele sequences of 1120 Bcc strains with unique STs was constructed. The resulting tree revealed the same four clusters of Other Bcc isolates, i.e., Other Bcc groups C, G, H, and M (names were adopted from Vandamme and Peeters, 2014) grouping with B. contaminans and B. lata, as reported earlier (Figure 1; Vandamme and Peeters, 2014). However, low bootstrap support for several nodes (Figure 1) and changing tree topologies upon analysis of different sets of STs from the PubMLST database (data not shown) prompted us to perform several whole-genome sequence-based comparisons. To this end, a total of 39 taxon K isolates (i.e., isolates belonging to B. contaminans, B. lata or Other Bcc groups C, G, H, and M) were selected based on the clustering result of the MLST tree (Figure 1) and their availability in our strain collections.
A phylogenomic tree was constructed based on the amino acid sequences of 107 conserved single-copy genes, extracted from the assembled genomes of 61 Bcc strains including 39 taxon K isolates. This tree generally supported the six clusters observed in the MLST tree but also revealed some discrepancies. A first cluster contained the type strain, several B. contaminans reference strains (Vanlaere et al., 2009) and recent isolates that were identified as B. contaminans, and therefore corresponded with the recA-I/MLST-I taxon reported by Vanlaere et al. (2009). The three remaining clusters corresponded with the other Bcc groups C, G, and H (Figure 2; Vandamme and Peeters, 2014). To investigate the degree of relatedness of these four clusters, pairwise OrthoANI and dDDH values were calculated from the assembled genomes (Table 3 and Supplementary Tables 2, 3). The average within-cluster ANI and dDDH values of these four clusters clearly exceeded the 95–96% ANI and 70% dDDH thresholds for species delineation (Wayne et al., 1987; Goris et al., 2007; Lee et al., 2016), whereas, the average between-cluster ANI and dDDH values were at or above these thresholds. Moreover, both ANI and dDDH values within the B. contaminans clade were clearly higher than those observed between the B. contaminans clade and other validly named Bcc species, or even between any two other validly named Bcc species.
A second major clade in the MLST tree (Figure 1) included the B. lata type strain LMG 22485T (ST-101). Notably, the taxonomy of B. lata appeared unresolved in an earlier study (Vanlaere et al., 2009) with B. lata represented by two recA lineages (recA-II and recA-III) that were supported by low bootstrap values only. The same strains grouped in a single heterogeneous MLST lineage (MLST-II) and wet-lab DDH experiments revealed several values near the 70% DDH threshold for species delineation (Vanlaere et al., 2009). In the present study, the MLST tree revealed one major cluster of B. lata STs (Figure 1) and four additional STs that grouped with a low bootstrap support (51%) with the main B. lata cluster. Other Bcc M STs were well-separated from B. lata and all other taxon K strains and grouped with B. lata in the analysis shown in Figure 1, although this clustering was supported by a low bootstrap value only.
This same clustering of B. lata and Other Bcc M strains was not supported by the phylogenomic tree (Figure 2) where B. lata LMG 31805 (ST-98) and R-9940 (ST-103) no longer grouped with B. lata LMG 22485T (ST-101) but clustered with B. lata R-17378 (ST-119) and R-18628 (ST-119), and with Other Bcc M STs and represented a third major clade. The latter clustering was supported by an average ANI value of 97.2% and an average dDDH value of 76.1% among the six strains examined, highlighting their close genomic relatedness (Supplementary Tables 2, 3). Values toward other B. lata strains ranged between 94.1 and 95.1% ANI and between 55.2 and 60.4% dDDH (Supplementary Tables 2, 3). Values toward other Bcc species ranged between 87.0 and 94.9 % ANI, and between 32.9 and 59.1% dDDH. The highest ANI and dDDH values were observed toward Burkholderia puraquae CAMPA1040T, which also appeared as a nearest neighbor of taxon K bacteria in the phylogenomic tree (Figure 2).
The remaining B. lata strains examined appeared to yield a continuum of ANI and dDDH values in which several clusters of closely related strains could be distinguished. Values toward other Bcc species ranged between 87.3 and 94.3% ANI, and between 33.1 and 56.1% dDDH. Again, the comparisons with B. puraquae CAMPA1040T yielded the highest ANI and dDDH values (Supplementary Tables 2, 3).
Data of the present study therefore demonstrated that the taxon K bacteria examined represented three major genomic clades, and thus confirmed and extended earlier observations (Vanlaere et al., 2009). A first clade comprised B. contaminans and Other Bcc group C, G, and H strains; a second clade comprised the B. lata type strain and most other B. lata strains examined; and a third clade comprised the Other Bcc group M strains and some B. lata strains. While each of these major clades again comprised several clusters of closely related strains, especially the clade comprising the B. lata type strain appeared as a genomic continuum. Several genomic parameters have been proposed to replace the earlier wet-lab DDH for species delineation. ANI cutoffs of 94–95% (Konstantinidis and Tiedje, 2005) and 95–96% (Goris et al., 2007; Lee et al., 2016) and a dDDH cutoff of 70% (Wayne et al., 1987; Meier-Kolthoff et al., 2013) have been proposed and commonly allow straightforward interpretation of taxonomic data. Taxonomic proposals based on borderline values, such as many values obtained in the present study, are inevitably subjective. Small-scale comparisons of well-selected strains undoubtedly would allow to find some unique strains with a genomic divergence clearly beyond the threshold values and with a distinctive phenotype and would facilitate a straightforward description of several novel Burkholderia species (like for instance in reference Wallner et al., 2019). Yet, the complexity of the natural diversity as presented in this comprehensive dataset of taxon K bacteria argues against such practices.
Burkholderia contaminans and Other Bcc groups C, G, and H revealed ANI values that were near and above the species delineation threshold, while their dDDH values were near and below the species threshold (Supplementary Tables 2, 3). We previously introduced a 3% threshold value of average concatenated allele sequence divergence for species delineation within the B. cepacia complex (Vanlaere et al., 2009; Peeters et al., 2013). The inclusion of Other Bcc groups C, G, and H into B. contaminans would not affect this commonly used threshold as the average within-group divergence would become 2.2% [while 1.4% was reported earlier (Vanlaere et al., 2009)] and the average divergence between the nearest neighbor species of B. contaminans (including Other Bcc groups C, G, and H) would become 3.6% [while 3.5% was reported earlier (Vanlaere et al., 2009)].
The analysis of a considerable number of B. lata isolates in the present study further substantiated the complexity and the continued unresolved nature of the taxonomy of this species (Vanlaere et al., 2009). The discrepancies reported earlier between single and multilocus trees (Vanlaere et al., 2009) were extended into the discrepant topologies of the MLST and phylogenomic trees (Figures 1, 2). The ANI and dDDH analyses indicated that Other Bcc group M strains and B. lata ST-98, ST-103 and ST-119 strains should be classified as a novel Bcc species within taxon K. Although this was not apparent from the MLST tree (Figure 1), this was supported by the average concatenated allele divergence values: the average within-group divergence for this novel species was 2.8%; the average within-group divergence for B. lata was 2.4% [while 2.7% was reported earlier (Vanlaere et al., 2009)]. The average divergences between the three taxon K species were 3.1% between the novel species and B. lata, 3.3% between the novel species and B. contaminans (including Other Bcc groups C, G, and H) and 3.7% between B. lata and B. contaminans [including Other Bcc groups C, G, and H; while 3.5% was reported earlier (Vanlaere et al., 2009)].
The recA gene was introduced as a superior alternative to the 16S rRNA gene for discrimination of Bcc bacteria (Mahenthiralingam et al., 2000) and is still commonly used as a first-line identification method for putative Bcc bacteria. While recA gene sequence analysis remains a very useful tool for the identification of most Bcc species, the present study confirms that its use is problematic for the recognition of some Bcc species (Supplementary Figure 1).
In medical microbiology, MALDI-TOF MS commonly replaced traditional biochemical characterization as a first-line diagnostic instrument. We therefore analyzed the 39 taxon K isolates using the MALDI Biotyper system. By using the cell smear method and comparison of the resulting spectra with the MBT IVD database, we adhered to a routine workflow in a diagnostic laboratory. Of 39 isolates examined, only three were identified at the species level (all other isolates were identified as B. cepacia complex): B. lata isolates LMG 22485T and LMG 6993 were correctly identified, whereas B. contaminans LMG 23253 was misidentified as B. cepacia (Table 4). The latter was not surprising since the MBT IVD database does not contain B. contaminans reference spectra. In addition, the entry for B. lata strain DSM 50180 (=LMG 6993) in the MBT IVD database is mislabeled as B. cepacia, which may contribute to misidentification of field isolates. In an effort to improve species-level identification, all 39 taxon K isolates were also analyzed using a cell extract method. Overall this resulted in somewhat higher log scores but it did not affect species-level identification positively. Indeed, while the same identification results were obtained for the B. lata strains LMG 22485T and LMG 6993 (correctly identified), and B. contaminans LMG 23253 (misidentified), an additional three strains (B. contaminans R-37747, Other Bcc G AU15512 and Other Bcc H LMG 31806) were also misidentified as B. cepacia (Table 4). Finally, spectra derived from cell extracts were also identified by comparison with the spectra present in both the MBT IVD library and an in-house library of Burkholderia reference strains which included additional reference spectra of 17 taxon K bacteria (Supplementary Table 1) which were assigned to B. contaminans, B. lata and the novel taxon K Bcc species, as described below. This allowed correct species-level identification of seven B. contaminans strains, two B. lata strains and two strains belonging to the novel species; all remaining taxon K isolates were identified as B. cepacia complex. However, all but three strains examined had a strain corresponding to the correct species as their consensus best match (Table 4) confirming that the use of optimized MALDI-TOF MS databases can improve species-level identification of clinical Bcc bacteria (Degand et al., 2008; Fernández-Olmos et al., 2012; Alby et al., 2013).
Conclusion
Burkholderia bacteria, and Bcc bacteria in particular, attracted a lot of interest because of their biocontrol and other biotechnological properties, but even more so as life-threatening pathogens in persons with CF (Mahenthiralingam et al., 2008; LiPuma, 2010). Among Bcc bacteria, taxon K became well-known as a notorious contaminant of pharmaceutical products (Torbeck et al., 2011; Cunningham-Oakes et al., 2020; Tavares et al., 2020). The present study was initiated to clarify the unresolved taxonomy of taxon K bacteria but revealed many borderline values which prevented a straightforward interpretation of data. While data from the present study can be used to argue that each of the clusters of taxon K strains with ANI and dDDH values well above the commonly applied cutoff levels corresponds with a distinct species or subspecies, such a classification would not bear a practical purpose and would render communication exceedingly complex. The classification of Other Bcc group C, G, and H bacteria as B. contaminans, and the proposal of a separate species status for Other Bcc group M bacteria and some strains now classified as B. lata acknowledges the genomic divergence between these three main clades but does not yield a simple identification scheme. The use of optimized MALDI-TOF MS databases for the identification of clinical Burkholderia isolates (Table 4) may provide accurate species-level identification for some of these bacteria; others will be identified as Bcc only. The public deposit of a reference strain of each of these Other Bcc groups into the BCCM/LMG Bacteria Collection2 (Table 1) will allow improvement of MALDI-TOF MS databases. Although MLST facilitates species-level identification of many taxon K strains, comparative genomics may be required to identify some recalcitrant strains. Finally, although phenotypic tests are rarely used today for species-level identification of Bcc bacteria (Vandamme and Dawyndt, 2011), the inclusion of Other Bcc groups C, G, and H into B. contaminans affects the phenotype of this species minimally (Supplementary Table 4) and the proposal of a novel species status for Other Bcc group M and B. lata ST-98, ST-103, and ST-119 strains is supported by their distinctive phenotype [i.e., growth at 42°C and lysine decarboxylase activity distinguish the novel species from B. lata (Supplementary Table 4)].
Description of Burkholderia aenigmatica sp. nov.
Burkholderia aenigmatica sp. nov. (ae.nig.ma’ti.ca. L. fem. adj. aenigmatica, enigmatic, referring to its unusual and complex taxonomy).
Burkholderia aenigmatica cells are Gram-negative, non-sporulating, straight rods. All strains grow on B. cepacia selective agar and MacConkey agar. Oxidase, ornithine decarboxylase and gelatinase activity is present. No lysine decarboxylase, β-galactosidase, arginine dihydrolase, urease, esculin hydrolase or nitrate reductase activity. Growth is observed at 30 and 42°C. Assimilation of glucose, L-arabinose, D-mannose, D-mannitol, N-acetyl-glucosamine, D-maltose, D-gluconate, caprate, adipate, L-malate, citrate, and phenylacetate. Acidification of glucose and lactose; not of maltose, xylose, sucrose and adonitol. The G+C content is between 65.5 and 66.3 mol%. The following fatty acids are present in all strains: C14:0, C16:0, cyclo C17:0, C16:0 2-OH, C16:0 3-OH, C16:1 2-OH, C18:1 2-OH, C18:1 ω7c, cyclo C19:0 ω8c, summed feature 2 (comprising C14:0 3-OH, iso C16:1 I, an unidentified fatty acid with equivalent chain-length of 10.928 or C12:0 ALDE or any combinations of these fatty acids), summed feature 3 (comprising C16:1 ω7c or iso C15 2-OH or both) and summed feature 7 (comprising C18:1 ω7c, C18:1 ω9t, C18:1 ω12t or any combination of these fatty acids). B. aenigmatica strains have been isolated from the environment and the respiratory tract of CF patients.
The type strain is LMG 13014T. Phenotypic characteristics of the type strain are the same as described above for the species. Its G+C content is 66.0 mol% and its 16S rRNA, recA and whole-genome sequences are publicly available through the accession numbers LR760817, LR761314, and CABVQC000000000, respectively. Strain LMG 13014T was isolated as a contaminant of hand cream in Belgium in 1991 (Vandamme et al., 1997).
Emended Description of Burkholderia contaminans Vanlaere et al., 2009
The description of B. contaminans is the one given by Vanlaere et al. (2009) with the following modifications: β-galactosidase activity and acidification of lactose and D-xylose are strain-dependent, rather than consistently present. B. contaminans includes the taxa known as Other Bcc groups C, G, and H. The G+C content of the type strain is 65.9 % and its whole-genome sequence is publicly available through the accession number GCA_000987075.1.
Emended Description of Burkholderia lata Vanlaere et al., 2009
The description of B. lata is the one given by Vanlaere et al. (2009) with the following modifications: acidification of maltose, lactose and D-xylose are strain-dependent, rather than consistently present. The G+C content of the type strain is 66.3% and its whole-genome sequence is publicly available through the accession number GCA_000012945.1.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/bioproject/PRJEB33447.
Author Contributions
ED and PV conceived the study and wrote the manuscript. ED, PV, and TC proofread the manuscript. ED and EDC performed all experiments except the biochemical characterization. MC and JZ performed the biochemical characterization. ED and CP carried out the genomic data analyses. ED and AW analyzed the MALDI-TOF MS data. JL isolated the strains. All authors read and approved the final manuscript.
Funding
ED is supported by fellowships from the Fund for Scientific Research Flanders (FWO, 1119517N, 1119519N). The BCCM/LMG Bacteria Collection is supported by the Federal Public Planning Service – Science Policy, Belgium. JZ and the Canadian Burkholderia cepacia complex research and referral repository were supported by a grant from Cystic Fibrosis Canada. JL receives support from the Cystic Fibrosis Foundation (United States).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This publication made use of the PubMLST website (https://pubmlst.org/) developed by Keith Jolley (Jolley et al., 2018) and sited at the University of Oxford. The development of that website was funded by the Wellcome Trust. We thank the Oxford Genomics Centre at the Wellcome Centre for Human Genetics (funded by Wellcome Trust grant reference 203141/Z/16/Z) for the generation and initial processing of the sequencing data. The computational resources (Stevin Supercomputer Infrastructure) and services used in this work were provided by the VSC (Flemish Supercomputer Center), funded by Ghent University, FWO and the Flemish Government – department EWI. We are grateful to all depositors of isolates who contributed to this study.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.01594/full#supplementary-material
Abbreviations
Bcc, Burkholderia cepacia complex; MLST, multilocus sequence typing; ST, sequence type; ANI, average nucleotide identity; dDDH, digital DNA-DNA hybridization; MALDI-TOF MS, matrix-assisted laser desorption/ionization time-of-flight mass spectrometry; CF, cystic fibrosis; GGDC, Genome-to-Genome Distance Calculator; MBT IVD, MALDI BioTyper in vitro diagnostics; MSP, main spectrum.
Footnotes
References
Alby, K., Gilligan, P. H., and Miller, M. B. (2013). Comparison of matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) mass spectrometry platforms for the identification of gram-negative rods from patients with cystic fibrosis. J. Clin. Microbiol. 51, 3822–3854. doi: 10.1128/JCM.01618-13
Ankenbrand, M. J., and Keller, A. (2016). BcgTree: automatized phylogenetic tree building from bacterial core genomes. edited by frédéric chain. Genome 59, 783–791. doi: 10.1139/gen-2015-0175
Bach, E., Sant’Anna, F. H., Magrich, dos Passos, J. F., Balsanelli, E., Baura, V. A., et al. (2017). Detection of Misidentifications of Species from the Burkholderia Cepacia complex and description of a new member, the soil bacterium Burkholderia Catarinensis Sp. Nov. Pathog. Dis. 75, 1–8. doi: 10.1093/femspd/ftx076
Baldwin, A., Mahenthiralingam, E., Kathleen, M., Honeybourne, D., Maiden, M. C. J., John, R., et al. (2005). Multilocus sequence typing scheme that provides both species and strain differentiation for the burkholderia cepacia complex. J. Clin. Microbiol. 43, 4665–4673. doi: 10.1128/JCM.43.9.4665
Bloodworth, R. A. M., Selin, C., López, De Volder, M. A., Drevinek, P., Galanternik, L., et al. (2015). Draft genome sequences of burkholderia contaminans, a burkholderia cepacia complex species that is increasingly recovered from cystic fibrosis patients. Genome Announc. 3, 3–4. doi: 10.1128/genomeA.00766-15
Bugrysheva, J. V., Cherney, B., Sue, D., Conley, A. B., Rowe, L. A., Knipe, K. M., et al. (2016). Complete genome sequences for three chromosomes of the Burkholderia Stabilis type strain (ATCC BAA-67). Genome Announc. 4, 4–5. doi: 10.1128/genomeA.01294-16
CDC (2004). Notice to Readers: Manufacturer’s Recall of Nasal Spray Contaminated with Burkholderia Cepacia Complex. Morbidity and Mortality Weekly Report. 2004. Available online at: https://www.cdc.gov/mmwr/preview/mmwrhtml/mm5311a8.htm (accessed December 13, 2019).
Cesarini, S., Bevivino, A., Tabacchioni, S., Chiarini, L., and Dalmastri, C. (2009). RecA gene sequence and multilocus sequence typing for species-level resolution of burkholderia cepacia complex isolates. Lett. Appl. Microbiol. 49, 580–588. doi: 10.1111/j.1472-765X.2009.02709.x
Chun, J., Oren, A., Ventosa, A., Christensen, H., Arahal, D. R., Costa, M. S., et al. (2018). Proposed minimal standards for the use of genome data for the taxonomy of prokaryotes. Int. J. Syst. Evol. Microbiol. 68, 461–466. doi: 10.1099/ijsem.0.002516
Cipolla, L., Rocca, F., Martinez, C., Aguerre, L., Barrios, R., and Prieto, M. (2018). Prevalence of Burkholderia Cepacia complex species in cystic fibrosis patients in argentina during the period 2011–2015. Enfermedades Infec. Microbiol. Clín. 36, 431–434. doi: 10.1016/j.eimc.2017.09.002
Cock, P. J. A., Antao, T., Chang, J. T., Chapman, B. A., Cox, C. J., Dalke, A., et al. (2009). Biopython: freely available python tools for computational molecular biology and bioinformatics. Bioinformatics 25, 1422–1423. doi: 10.1093/bioinformatics/btp163
Coenye, T., Mahenthiralingam, E., Henry, D., LiPuma, J. J., Laevens, S., Gillis, M., et al. (2001). Burkholderia ambifaria Sp. Nov., a novel member of the burkholderia cepacia complex including biocontrol and cystic fibrosis-related isolates. Int. J. Syst. Evol. Microbiol. 51, 1481–1490. doi: 10.1099/00207713-51-4-1481
Coutinho, C. P., Barreto, C., Pereira, L., Lito, L., Melo Cristino, J., and Sá-Correia, I. (2015). Incidence of Burkholderia contaminans at a cystic fibrosis centre with an unusually high representation of burkholderia cepacia during 15 years of epidemiological surveillance. J. Med. Microbiol. 64, 927–935. doi: 10.1099/jmm.0.000094
Cunningham-Oakes, E., Weiser, R., Pointon, T., and Mahenthiralingam, E. (2020). Understanding the challenges of non-food industrial product contamination. FEMS Microbiol. Lett. 366, 1–9. doi: 10.1093/femsle/fnaa010
da Silva, K., Souza Cassetari, A., de, Silva Lima, A., Brandt, E., and De, et al. (2012). Diazotrophic Burkholderia species isolated from the amazon region exhibit phenotypical, functional and genetic diversity. Syst. Appl. Microbiol. 35, 253–262. doi: 10.1016/j.syapm.2012.04.001
De Bel, A., Wybo, I., Vandoorslaer, K., Rosseel, P., Lauwers, S., and Pierard, D. (2011). Acceptance criteria for identification results of gram-negative rods by mass spectrometry. J. Med. Microbiol. 60, 684–686. doi: 10.1099/jmm.0.023184-0
De Smet, B., Mayo, M., Peeters, C., Zlosnik, J. E. A., Spilker, T., Hird, T. J., et al. (2015). Burkholderia stagnalis Sp. Nov. and Burkholderia territorii Sp. Nov., two novel burkholderia cepacia complex species from environmental and human sources. Int. J. Syst. Evol. Microbiol. 65, 2265–2271. doi: 10.1099/ijs.0.000251
Degand, N., Carbonnelle, E., Dauphin, B., Beretti, J. L., Bourgeois, M., and Le, et al. (2008). Matrix-assisted laser desorption ionization-time of flight mass spectrometry for identification of nonfermenting gram-negative bacilli isolated from cystic fibrosis patients. J. Clin. Microbiol. 46, 3361–3367. doi: 10.1128/JCM.00569-08
Depoorter, E., Bull, M. J., Peeters, C., Coenye, T., Vandamme, P., and Mahenthiralingam, E. (2016). Burkholderia: an update on taxonomy and biotechnological potential as antibiotic producers. Appl. Microbiol. Biotechnol. 100, 5215–5229. doi: 10.1007/s00253-016-7520-x
Edgar, R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797. doi: 10.1093/nar/gkh340
Fernández-Olmos, A., García-Castillo, M., Morosini, M.-I. I., Lamas, A., Máiz, L., and Cantón, R. (2012). MALDI-TOF MS improves routine identification of non-fermenting gram negative isolates from cystic fibrosis patients. J. Cyst. Fibr. 11, 59–62. doi: 10.1016/j.jcf.2011.09.001
Glowicz, J., Crist, M., Gould, C., Moulton-Meissner, H., Noble-Wang, J., Man, T. J. B., et al. (2018). A multistate investigation of health care-associated burkholderia cepacia complex infections related to liquid docusate sodium contamination, January-October 2016. Am. J. Infect. Control 46, 1–7. doi: 10.1016/j.ajic.2017.11.018
Goris, J., Konstantinidis, K. T., Klappenbach, J. A., Coenye, T., Vandamme, P., and Tiedje, J. M. (2007). DNA-DNA hybridization values and their relationship to whole-genome sequence similarities. Int. J. Syst. Evol. Microbiol. 57, 81–91. doi: 10.1099/ijs.0.64483-0
Henry, D., Mahenthiralingam, E., Vandamme, P., Coenye, T., and Speert, D. P. (2001). Phenotypic methods for determining genomovar status of the Burkholderia Cepacia complex. J. Clin. Microbiol. 39, 1073–1078. doi: 10.1128/JCM.39.3.1073-1078.2001
Holden, M. T. G., Seth-Smith, H. M. B., Crossman, L. C., Sebaihia, M., Bentley, S. D., Cerdeño-Tárraga, A. M., et al. (2009). The genome of Burkholderia Cenocepacia J2315, an epidemic pathogen of cystic fibrosis patients. J. Bacteriol. 91, 261–277. doi: 10.1128/JB.01230-08
Johnson, S. L., Bishop-Lilly, K. A., Ladner, J. T., Daligault, H. E., Davenport, K. W., Jaissle, J., et al. (2015). Complete genome sequences for 59 Burkholderia isolates, both pathogenic and near neighbor. Genome Announc. 3, 18–20. doi: 10.1128/genomeA.00159-15
Jolley, K. A., Bray, J. E., and Maiden, M. C. J. (2018). Open-access bacterial population genomics: BIGSdb software, the PubMLST.Org website and their applications. Wellcome Open Res. 3:124. doi: 10.12688/wellcomeopenres.14826.1
Jolley, K. A., and Maiden, M. C. (2010). BIGSdb: scalable analysis of bacterial genome variation at the population level. BMC Bioinform. 11:595. doi: 10.1186/1471-2105-11-595
Kenna, D. T. D., Lilley, D., Coward, A., Martin, K., Perry, C., Pike, R., et al. (2017). Prevalence of Burkholderia species, including members of burkholderia cepacia complex, among uk cystic and non-cystic fibrosis patients. J. Med. Microbiol. 66, 490–501. doi: 10.1099/jmm.0.000458
Konstantinidis, K. T., and Tiedje, J. M. (2005). Genomic insights that advance the species definition for prokaryotes. Proc. Natl. Acad. Sci. U.S.A. 102, 2567–2572. doi: 10.1073/pnas.0409727102
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Kunakom, S., and Eustáquio, A. S. (2019). Burkholderia as a source of natural products. J. Nat. Prod. 82, 2018–2037. doi: 10.1021/acs.jnatprod.8b01068
Kwak, Y., and Shin, J.-H. (2015). Complete genome sequence of burkholderia pyrrocinia 2327T, the first industrial bacterium which produced antifungal antibiotic pyrrolnitrin. J. Biotechnol. 211, 3–4. doi: 10.1016/j.jbiotec.2015.06.420
Lee, I., Kim, Y. O., Park, S.-C., and Chun, J. (2016). OrthoANI: an improved algorithm and software for calculating average nucleotide identity. Int. J. Syst. Evol. Microbiol. 66, 1100–1103. doi: 10.1099/ijsem.0.000760
Leguizamon, M., Draghi, W. O., Montanaro, P., Schneider, A., Prieto, C. I., Martina, P., et al. (2017). Draft Genome Sequence of Burkholderia Puraquae type strain CAMPA 1040, isolated from hospital settings in córdoba, argentina. Genome Announc. 5, 15–16. doi: 10.1128/genomea.01302-17
Letunic, I., and Bork, P. (2019). Interactive tree of life (ITOL) v4: recent updates and new developments. Nucleic Acids Res. 47, W256–W259. doi: 10.1093/nar/gkz239
LiPuma, J. J. (2010). The changing microbial epidemiology in cystic fibrosis. Clin. Microbiol. Rev. 23, 299–323. doi: 10.1128/CMR.00068-09
Mahenthiralingam, E., Baldwin, A., and Dowson, C. G. (2008). Burkholderia Cepacia complex bacteria: opportunistic pathogens with important natural biology. J. Appl. Microbiol. 104, 1539–1551. doi: 10.1111/j.1365-2672.2007.03706.x
Mahenthiralingam, E., Baldwin, A., Drevinek, P., Vanlaere, E., Vandamme, P., LiPuma, J. J., et al. (2006). Multilocus sequence typing breathes life into a microbial metagenome. PLoS ONE 1:e17. doi: 10.1371/journal.pone.0000017
Mahenthiralingam, E., Bischof, J., Byrne, S. K., Radomski, C., Davies, J. E., Av-gay, Y., et al. (2000). DNA-based diagnostic approaches for identification of burkholderia cepacia complex, burkholderia vietnamiensis, burkholderia vietnamiensis, burkholderia multivorans, burkholderia stabilis and burkholderia cepacia genomovars I and III. J. Clin. Microbiol. 38, 3165–3173. doi: 10.1128/jcm.38.9.3165-3173.2000
Martin, M., Christiansen, B., Caspari, G., Hogardt, M., Thomsen, A. J., and von, et al. (2011). Hospital-wide outbreak of burkholderia contaminans caused by prefabricated moist washcloths. J. Hosp. Infect. 77, 267–270. doi: 10.1016/j.jhin.2010.10.004
Martina, P., Leguizamon, M., Prieto, C. I., Sousa, S. A., Montanaro, P., Draghi, W. O., et al. (2018). Burkholderia Puraquae Sp. Nov., a novel species of the burkholderia cepacia complex isolated from hospital settings and agricultural soils. Int. J. Syst. Evol. Microbiol. 68, 14–20. doi: 10.1099/ijsem.0.002293
Medina-Pascual, M. J., Valdezate, S., Carrasco, G., Villalón, P., Garrido, N., and Saéz-Nieto, J. A. (2015). Increase in isolation of Burkholderia contaminans from spanish patients with cystic fibrosis. Clin. Microbiol. Infect. 21, 150–156. doi: 10.1016/j.cmi.2014.07.014
Meier-Kolthoff, J. P., Auch, A. F., Klenk, H.-P., and Göker, M. (2013). Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinform. 14:60. doi: 10.1186/1471-2105-14-60
Ong, K. S., Aw, Y. K., Lee, L. H., Yule, C. M., Cheow, Y. L., and Lee, S. M. (2016). Burkholderia Paludis Sp. Nov., an antibiotic-siderophore producing novel burkholderia cepacia complex species, isolated from malaysian tropical peat swamp soil. Front. Microbiol. 7:1–14. doi: 10.3389/fmicb.2016.02046
Parke, J. L., and Gurian-Sherman, D. (2001). Diversity of the Burkholderia Cepacia complex and implications for risk assessment of biological control strains. Annu. Rev. Phytopathol. 39, 225–258. doi: 10.1146/annurev.phyto.39.1.225
Payne, G. W., Vandamme, P., Morgan, S. H., Lipuma, J. J., and Coenye, T. (2005). Development of a RecA gene-based identification approach for the entire burkholderia genus. Appl. Environ. Microbiol. 71, 3917–3927. doi: 10.1128/AEM.71.7.3917
Peeters, C., Canck, E., De, Cnockaert, M., Brandt, E., and De, et al. (2019). Comparative genomics of pandoraea, a genus enriched in xenobiotic biodegradation and metabolism. Front. Microbiol. 10:1–21. doi: 10.3389/fmicb.2019.02556
Peeters, C., Zlosnik, J. E. A., Spilker, T., Hird, T. J., LiPuma, J. J., and Vandamme, P. (2013). Burkholderia pseudomultivorans Sp. Nov., a novel burkholderia cepacia complex species from human respiratory samples and the rhizosphere. Syst. Appl. Microbiol. 36, 483–489. doi: 10.1016/j.syapm.2013.06.003
Power, R. F., Linnane, B., Martin, R., Power, N., Harnett, P., Casserly, B., et al. (2016). The first reported case of burkholderia contaminans in patients with cystic fibrosis in ireland: from the sargasso sea to irish children. BMC Pulmon. Med. 16:57. doi: 10.1186/s12890-016-0219-z
Seemann, T. (2014). Prokka: rapid prokaryotic genome annotation. Bioinformatics 30, 2068–2069. doi: 10.1093/bioinformatics/btu153
Souza, A. V., Moreira, C. S., Pasternak, J., Lurdes Hirata, M., de, Alves Saltini, D., et al. (2004). Characterizing uncommon Burkholderia cepacia complex isolates from an outbreak in a haemodialysis unit. J. Med. Microbiol. 53, 999–1005. doi: 10.1099/jmm.0.45702-0
Stamatakis, A. (2014). RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313. doi: 10.1093/bioinformatics/btu033
Tavares, M., Kozak, M., Balola, A., and Sá-Correia, I. (2020). Burkholderia cepacia complex bacteria: a feared contamination risk in water-based pharmaceutical products. Clin. Microbiol. Rev. 33, 1–25. doi: 10.1128/CMR.00139-19
Torbeck, L., Raccasi, D., Guilfoyle, D. E., Friedman, R. L., and Hussong, D. (2011). Burkholderia cepacia: this decision is overdue. PDA J. Pharm. Sci. Technol. 65, 535–543. doi: 10.5731/pdajpst.2011.00793
Van Veen, S. Q., Claas, E. C. J., and Kuijper, E. J. (2010). High-throughput identification of bacteria and yeast by matrix-assisted laser desorption ionization-time of flight mass spectrometry in conventional medical microbiology laboratories. J. Clin. Microbiol. 48, 900–907. doi: 10.1128/JCM.02071-09
Vandamme, P., and Dawyndt, P. (2011). Classification and identification of the Burkholderia Cepacia complex: past, present and future. Syst. Appl. Microbiol. 34, 87–95. doi: 10.1016/j.syapm.2010.10.002
Vandamme, P., Holmes, B., Vancanneyt, M., Coenye, T., Hoste, B., Coopman, R., et al. (1997). Occurrence of multiple genomovars of Burkholderia Cepacia in cystic fibrosis patients and proposal of Burkholderia Multivorans Sp. Nov. Int. J. Syst. Bacteriol. 47, 1188–1200. doi: 10.1099/00207713-47-4-1188
Vandamme, P., and Peeters, C. (2014). Time to revisit polyphasic taxonomy. Antonie Van Leeuwenhoek 106, 57–65. doi: 10.1007/s10482-014-0148-x
Vandamme, P., Vancanneyt, M., Pot, B., Mels, L., Hoste, B., Dewettinck, D., et al. (1992). Polyphasic taxonomic study of the emended genus Arcobacter with Arcobacter Butzleri Comb. Nov. and Arcobacter Skirrowii Sp. Nov., an aerotolerant bacterium isolated from veterinary specimens. Int. J. Syst. Bacteriol. 42, 344–356. doi: 10.1099/00207713-42-3-344
Vanlaere, E., Baldwin, A., Gevers, D., Henry, D., Brandt, E., and De, et al. (2009). Taxon K, a complex within the Burkholderia cepacia complex, comprises at least two novel species, Burkholderia Contaminans Sp. Nov. and Burkholderia lata Sp. Nov. Int. J. Syst. Evol. Microbiol. 59(Pt 1), 102–111. doi: 10.1099/ijs.0.001123-0
Vermis, K., Coenye, T., Mahenthiralingam, E., Nelis, H. J., and Vandamme, P. (2002). Evaluation of species-specific RecA-based PCR tests for genomovar level identification within the Burkholderia cepacia complex. J. Med. Microbiol. 51, 937–940. doi: 10.1099/0022-1317-51-11-937
Wallner, A., King, E., Ngonkeu, E. L. M., Moulin, L., and Béna, G. (2019). Genomic analyses of Burkholderia cenocepacia reveal multiple species with differential host-adaptation to plants and humans. BMC Genomics 20:803. doi: 10.1186/s12864-019-6186-z
Wayne, L. G., Brenner, D. J., Colwell, R. R., Grimont, P. A. D., Kandler, O., Krichevsky, M. I., et al. (1987). International committee on systematic bacteriology: announcement of the report of the ad hoc committee on reconciliation of approaches to bacterial systematics. Syst. Appl. Microbiol. 10, 463–464. doi: 10.1016/S0723-2020(88)80020-1
Wieme, A. D., Spitaels, F., Aerts, M., Bruyne, K., De, Landschoot, A., et al. (2014). Effects of growth medium on matrix-assisted laser desorption–ionization time of flight mass spectra: a case study of acetic acid bacteria. Appl. Environ. Microbiol. 80, 1528–1538. doi: 10.1128/AEM.03708-13
Winsor, G. L., Khaira, B., Rossum, T., Van, Lo, R., Whiteside, M. D., et al. (2008). The Burkholderia genome database: facilitating flexible queries and comparative analyses. Bioinformatics 24, 2803–2804. doi: 10.1093/bioinformatics/btn524
Yoon, S.-H., Ha, S., Lim, J., Kwon, S., and Chun, J. (2017). A large-scale evaluation of algorithms to calculate average nucleotide identity. Antonie Van Leeuwenhoek 110, 1281–1286. doi: 10.1007/s10482-017-0844-4
Zurita, J., Mejia, L., Zapata, S., Trueba, G., Vargas, A. C., Aguirre, S., et al. (2014). Healthcare-associated respiratory tract infection and colonization in an intensive care unit caused by Burkholderia Cepacia Isolated in mouthwash. Int. J. Infect. Dis. 29, e96–e99. doi: 10.1016/j.ijid.2014.07.016
Keywords: Burkholderia, novel species, genomic taxonomy, opportunistic pathogens, multilocus sequence typing
Citation: Depoorter E, De Canck E, Peeters C, Wieme AD, Cnockaert M, Zlosnik JEA, LiPuma JJ, Coenye T and Vandamme P (2020) Burkholderia cepacia Complex Taxon K: Where to Split? Front. Microbiol. 11:1594. doi: 10.3389/fmicb.2020.01594
Received: 07 May 2020; Accepted: 18 June 2020;
Published: 14 July 2020.
Edited by:
Narjol González-Escalona, United States Food and Drug Administration, United StatesReviewed by:
Edward Cunningham-Oakes, Cardiff University, United KingdomCarina Marie Hall, Northern Arizona University, United States
Copyright © 2020 Depoorter, De Canck, Peeters, Wieme, Cnockaert, Zlosnik, LiPuma, Coenye and Vandamme. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Peter Vandamme, peter.vandamme@ugent.be
 Eliza Depoorter
Eliza Depoorter Evelien De Canck1
Evelien De Canck1 Charlotte Peeters
Charlotte Peeters Anneleen D. Wieme
Anneleen D. Wieme James E. A. Zlosnik
James E. A. Zlosnik John J. LiPuma
John J. LiPuma Tom Coenye
Tom Coenye Peter Vandamme
Peter Vandamme