- 1Guangdong Provincial Key Laboratory of Marine Biology, Shantou University, Shantou, China
- 2Institute of Marine Sciences, Shantou University, Shantou, China
- 3STU-UMT Joint Shellfish Research Laboratory, Shantou University, Shantou, China
Dietary carbohydrates are anaerobically fermented by gut microbiota to short-chain fatty acids (SCFAs), conferring gut health benefits. Of all tested prebiotics, galactooligosaccharides (GOS) and resistant starch (RS) stimulated the SCFA production in mud crab (Scylla paramamosain), a crustacean model, to a greater extent than the other carbohydrates tested. Using in vitro anaerobic fermentation cultures, this study further explored the prebiotic potential of GOS and RS in mud crab by assessing their impacts on gut microbiota changes and SCFA production. Both GOS and RS significantly promoted SCFA production. Bacterial diversity in the GOS group was lower than in the RS or control group. GOS promoted the growth of Bacteroidetes, while RS promoted Tenericutes. A strong positive correlation was found between SCFA production and bacterial abundance; most bacteria per se correlated with each other. The findings demonstrated the prebiotic potential of GOS and RS in mud crab.
Introduction
The intestinal tract of aquatic animals is harbored by a large number of symbiotic microbes (Wang et al., 2018). Gut microbiota are important for host health and nutritional metabolism through multiple mechanisms (Li et al., 2018; Ringø et al., 2018). It has been found that, in aquatic animals, the composition of gut microbiota is influenced by several factors, including age, food type, feeding regimen of fish, as well as environmental and ecological factors (Ringø et al., 2006; Kashinskaya et al., 2014). Of these factors, the feeding regimen is one of the most important. In fact, consuming specific diets may affect the composition and activity of indigenous gut microbiota (Ringø et al., 2010). Prebiotics and non-digestible foods are some crucial components of diets that indirectly contribute to the health-promoting effects of the hosts (Gibson, 2004). In principle, prebiotics are known to promote the growth of beneficial bacteria, which stimulates metabolite production by gut microbes (Reichardt et al., 2018). In aquatic animals, short-chain fatty acids (SCFAs, including acetic, propionic, and butyric acids) produced by gut bacteria through anaerobic fermentation of dietary carbohydrates in the gut have been demonstrated to confer gut health benefits (Tran et al., 2020). The rate and amount of SCFA production depend on the species and amounts of gut microbiota, substrate sources, and gut transit time (Wong et al., 2006). Previously, some prebiotics such as α-starch, gelatinized starch, inulin, maize, barley, wheat, rye, lactosucrose, arabinoxylooligosaccharides, fructooligosaccharides, and pregelatinized tapioca starch were shown to affect the formation of SCFAs in the gut of aquatic animals in vivo (Tran et al., 2020).
Butyrate has been of great interest in human biology because of its roles in nourishing gut mucosa, preventing gut cancer by promoting cell differentiation, cell-cycle arrest, and apoptosis of transformed colonocytes, as well as inhibiting the enzyme histone deacetylase and decreasing the transformation of primary to secondary bile acids as a result of colonic acidification (Wong et al., 2006). Dietary supplementation of carbohydrates is a way to modulate microbial fermentation and promote butyrate production in the gut, which consequently improves health status (Sato et al., 2017). Recent studies in aquatic animals have demonstrated the roles of butyrate in improving growth performance, feed efficiency, immune responses, disease resistance, and survival rate, as well as in modulating gut microbiota (Gao et al., 2011; Robles et al., 2013; Liu B. et al., 2014; Estensoro et al., 2016; Rimoldi et al., 2016; Silva et al., 2016a, b; Terova et al., 2016; Ebrahimi et al., 2017; Piazzon et al., 2017; Ramírez et al., 2017; Tian et al., 2017; Ali et al., 2018; Rimoldi et al., 2018). For these reasons, the selection of a prebiotic capable of promoting the growth of butyrate-producing bacteria is necessary to enhance survival rate and improve aquaculture production. Supplementation with prebiotics might be a promising method that could confer positive health benefits in aquatic animals, including cultured crustaceans (Daniels et al., 2013; Chen et al., 2017; Duan et al., 2017; Elshopakey et al., 2018; Safari and Paolucci, 2018). In vitro anaerobic fermentation studies provide a primary approach for initial evaluation of the positive benefits of compounds through their effects on metabolic activities of gut microbiota (Pham et al., 2017). Many research efforts aimed at assessing the impacts of potential prebiotics on the dynamics of human gut microbiota have applied in vitro fermentation approaches (Scott et al., 2014; Pham et al., 2017; Sato et al., 2017; Baxter et al., 2019). These studies have demonstrated that prebiotics (i.e., L-sorbose, xylitol, and resistant starch) can selectively stimulate the growth of butyrate producers and increase the production of butyric acid in batch cultures. Accordingly, several substrates, such as arabinoxylan, whole wheat, soybean oligosaccharides, isomaltooligosaccharides, raffinose, gentiobiose, lactosucrose, arabinoxylanoligosaccharides, oligofructose, xylose, and fructose, have been shown to enhance the levels of SCFAs in batch cultures with gut microbes of common carp (Cyprinus carpio) (Kihara and Sakata, 2002), Nile tilapia (Oreochromis niloticus), and European seabass (Dicentrarchus labrax) (Leenhouwers et al., 2008), Siberian sturgeon (Acipenser baerii), and African catfish (Clarias gariepinus) (Geraylou et al., 2014). In this study, mud crab (Scylla paramamosain) was used as a model to investigate the effects of two kinds of carbohydrate (galactooligosaccharides [GOS] and resistant starch [RS]) on gut microbiota and SCFA production using in vitro anaerobic fermentation cultures. The results of this study provide an effective approach in the selection of prebiotic potential via stimulation of beneficial gut microbiota populations and the induction of metabolites.
Materials and Methods
Substrates and in vitro Fermentation
Two kinds of carbohydrates, including GOS and RS (manufactured by Yuanye Shengwu Co. Ltd., Shanghai, China) were investigated in this study. Both GOS and RS were demonstrated to stimulate the formation of much more butyric acid than the other kinds of carbohydrates tested (xylooligosaccharides, mannan-oligosaccharides, fructooligosaccharides, inulin, D-mannitol, D-sorbitol, L-sorbose, and xylitol) (Supplementary Text S1). The carbohydrate stock solutions were prepared in water, boiled for 1 min, and maintained anaerobically using O2-free N2. The saturated peptone-yeast extract (PY) (1 L) contained 5.0 g of peptone, 5.0 g of trypticase peptone, 10.0 g of yeast extract, 0.5 g of L-cysteine HCl⋅H2O, 4.0 g of Na2CO3, 10 mL of 0.05% hemin solution, 1.0 mL of 0.1% resazurin solution, 0.4 g of K2HPO4, 0.04 g of KH2PO4, 0.08 g of Na2HCO3, 0.04 g of NaCl, 8 mg of CaCl2, 1.9 mg of MgSO4⋅7H2O (pH 6.8), and 1 mg of vitamin K1 (Sato et al., 2017).
Gut contents were obtained from 18 healthy mud crabs (average weight 63.6 ± 8.8 g) without clinical disease purchased from a culture farm in Shantou (Guangdong, China). All mud crabs were randomly divided into three groups (of replications), chilled on ice, and then dissected. The intestinal tract was aseptically removed, and the contents from the posterior gut were gently squeezed out, placed into sterile Eppendorf tubes, and immediately transferred to an anaerobic (10% H2, 5% CO2, 85% N2) workstation (Whitley Workstation DG250, Don Whitley Scientific Ltd., Bingley, West Yorkshire, United Kingdom) upon delivery to carry out the experiment. Gut content samples were homogenized in a 10-fold dilution of anaerobic 0.1 M sodium phosphate buffer (pH 6.8) in a sterile Eppendorf tube. The mixtures (1 mL) were transferred to Hungate tubes sealed with butyl rubber stoppers and screw caps containing 4 mL of O2-free CO2-PY broth supplemented with 0.05 g of GOS or RS. Immediately after dilutions of the sample into tubes containing PY broth supplemented with carbohydrate, the mixture (2 mL) was collected from each test tube using a sterile syringe for time point 0 h. All steps for the cultures were conducted in an anaerobic chamber. Incubations for each carbohydrate and control (without carbohydrate supplementation) were performed in triplicate on a shaker (Shanghai Bluepard Instruments Co., Ltd.) at 140 × g for 24 h at 30°C. At 24 h after incubation, the samples being collected in the same manner. All samples collected were stored at −80°C until further analysis.
Short-Chain Fatty Acid Analysis
Fermentation samples were thawed at room temperature and centrifuged at 10,000 × g for 10 min at 4°C. After centrifugation, the culture supernatants (1.0 mL) of the sample was removed and used for assessing SCFA production via gas chromatography after a 24-h growth as described previously (Li et al., 2019). The pH of the supernatant was adjusted to 2–3 by addition of H2SO4 and mixed thoroughly (2 min). The supernatant was collected after centrifugation at 13,000 × g for 10 min (at 4°C). The supernatant was collected and filtered (using a 0.2-μm nylon syringe filter). The filtrate was used for analyzing SCFA production using Agilent 6890N Network Gas Chromatograph (Agilent Technologies, Inc., Santa Clara, CA, United States) with a flame ionization detector equipped with a polar HP-INNOWAX capillary column (30 mm × 0.25 mm, 0.25 μm). A 1-μL injection volume of the sample was automatically injected into an inlet (at 230°C, 10:1 split ratio). Nitrogen was used as the carrier gas (flow rate: 1.0 mL/min). The detector temperature was 250°C, and the injector temperature was 230°C. The flow rates of nitrogen, hydrogen, and air as auxiliary gases were 25, 40, and 450 mL/min, respectively. The initial oven temperature was 50°C, which was maintained for 6 min and then raised to 230°C at 15°C/min and held at that temperature for 5 min.
Quantification of SCFAs in the fermentation products was performed using the calibration standard curves method. Three calibration standards were prepared at eight levels of concentration ranging from 0.699 to 69.9 mM for acetic acid and 0.218 to 21.8 mM for propionic and butyric acids. SCFA standards with three replicates were also treated as with the fermentation samples. From the chromatograms, the peak area of each calibration standard was plotted against the concentration of the SCFAs. Calibration curves were built using linear regression implemented in SPSS 16.0, after which the slope a, y-intercept b, and square correlation coefficients (r2) for the tested acids were calculated. Concentrations of acetic, propionic, and butyric acids were determined using the standard curve of each SCFA and expressed as mean mM.
DNA Extraction, PCR Amplification, and Sequencing
Genomic DNA was extracted using the TIANamp Stool DNA Kit (Tiangen Biotech Co., Ltd., Beijing, China) according to the manufacturer’s instructions. The quality and quantity of extracted DNA were assessed using 2% agarose gel electrophoresis and a NanoDrop 2000 spectrophotometer. The DNAs were stored at −20°C until use. The V4 region of the 16S rRNA gene was amplified using barcoded universal primers: 515F (5′-GTGCCAGCMGCCGCGGTAA-3′) and 806R (5′-GGACTACHVGGGTWTCTAAT-3′) (Caporaso et al., 2011) with the barcode. Polymerase chain reaction (PCR) reactions were conducted as described previously (Li et al., 2019). Briefly, each PCR reaction was carried out in a 30-μL volume containing 10 ng of DNA, 15 μL of Phusion® High-Fidelity PCR Master Mix with GC buffer (New England Biolabs, Ipswich, MA, United States), 0.2 μM forward and reverse primers, and 2 μL of distilled water. The PCR conditions were as follows: 1 min of initial denaturation at 98°C; 30 cycles of 98°C for 10 s, 50°C for 30 s, and 72°C for 30 s; and a final elongation step at 72°C for 5 min. The PCR products were separated using agarose gel electrophoresis and purified by application of a DNA Gel Extraction Kit (Axygen, Hangzhou, China). The concentration of the purified DNA was determined on a NanoDrop 2000 spectrophotometer. Sequencing libraries were synthesized using Ion Plus Fragment Library Kit 48 rxns (Thermo Fisher Scientific, Waltham, MA, United States) and assessed on a Qubit® 2.0 Fluorometer (Thermo Fisher Scientific). The libraries were sequenced on an IonS5TMXL platform (Thermo Fisher Scientific) and 400-bp single-end reads were generated.
Bioinformatics and Data Analysis
All sequence reads were sorted based on their unique barcodes. The sequence quality was filtered using Cutadapt 1.9.1 (Martin, 2011). Chimera sequences were removed using the UCHIME algorithm (Edgar et al., 2011). Sequences were clustered into operational taxonomic units (OTUs) using 97% identity as the cut-off value by Uparse software (Version 7.0.1001) (Edgar, 2013). The representative sequences from each OTU were taxonomically classified using the SILVA reference database with a confidence threshold of 80%. The alpha diversity indices, richness estimators (Chao1 and Abundance-based Coverage Estimator [ACE]), and diversity index (Simpson, Shannon index) were all calculated using QIIME 1.7.0 (Caporaso et al., 2010). For the beta diversity metrics, principal coordinates analysis (PCoA) was applied using the UniFrac web tool (Lozupone et al., 2006). Bacterial community profiling data were analyzed statistically with one-way analysis of variance (ANOVA) implemented in SPSS 16.0. Non-parametric permutational multivariate analysis of variance (PERMANOVA) was performed via PAST 2.16 (Hammer et al., 2001) to evaluate the significance of differences in bacterial community structure among groups. The association between the SCFA production and microbiota community was analyzed using SPSS 16.0. Differences obtained by these tests were considered statistically significant at P < 0.05. Heat maps showing the correlation between the relative abundance of microbial taxa and SCFA production were generated via Heml 1.01. Linear discriminant analysis effect size (LEfSe) analysis to determine microbial genera with significantly different abundances between groups was performed using Galaxy/Hutlab2.
Results
Potential Prebiotics and SCFA Analysis
To investigate the effects of GOS and RS on the metabolic activities of gut microbiota in mud crab, an in vitro anaerobic fermentation study was applied. The concentrations of SCFAs in cultures supplemented with either GOS or RS estimated at 0 and 24 h are shown in Figure 1. The results revealed that, compared with control, both GOS and RS significantly increased the concentrations of acetic acid (2.4- and 1.9-fold, respectively), propionic acid (6.3- and 2.3-fold, respectively), and butyric acid (6.4- and 4.3-fold, respectively) (P < 0.05). In addition, the concentrations of both acetic acid and propionic acid were significantly higher in the GOS compared with that in RS (P < 0.05), while that of butyric acid was not significantly different between the two groups (P = 0.29).
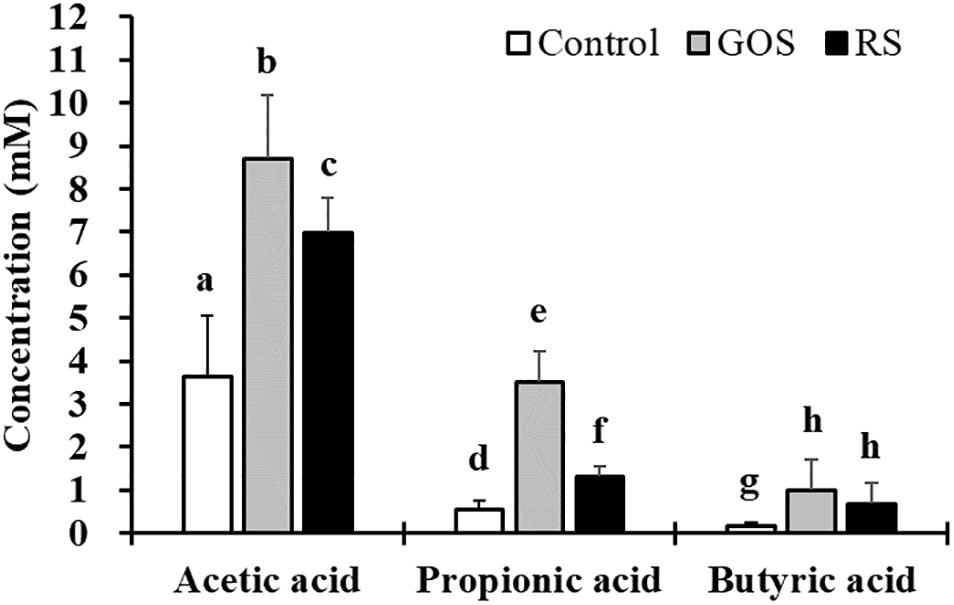
Figure 1. Production of short-chain fatty acids (SCFAs) in the in vitro fermentation of galactooligosaccharides (GOS) and resistant starch (RS) with gut contents of mud cab. Different letters indicate significant differences between groups within each kind of SCFAs (P < 0.05).
Microbial Sequencing and Community Structure
To investigate the influence of GOS or RS on gut microbial community structure, high-throughput sequence analysis of bacterial hypervariable V4 region of the 16S rRNA gene was performed. Raw sequencing reads have been deposited in GenBank under BioProject code PRJNA565751, BioSample numbers SAMN12767047, SAMN12767049, and SAMN12767050. After quality filtering and chimera removal, the 27 samples (9 for each group) yielded a total of 2,027,455 high-quality reads, with an average number of 75,090.93 reads per sample. The rarefaction curves reached the saturation plateau (Supplementary Figure S1A), indicating that saturation in sequencing was obtained. The rank abundance curves revealed a similarity in diversity in all samples (Supplementary Figure S1B). The Good’s coverage for each sample ranged from 99.9 to 100%, indicating that most of the bacteria present in the samples were identified (Supplementary Figure S1C). The reads were assigned to 741 OTUs at a 97% sequence identity threshold. Each sample contained 277–436 OTUs. Across all the samples, 97.98 and 63.83% of the total sequences were assigned into 18 phyla (control: 16; GOS: 15; RS: 16) (Supplementary Table S1) and 144 genera (control: 101; GOS: 90; RS: 108), respectively (Supplementary Table S2). At the phylum level, the samples predominantly consisted of five phyla: Bacteroidetes, Firmicutes, Proteobacteria, Tenericutes, and Fusobacteria, which together accounted for 99.53% of the OTUs (Figure 2A). Specifically, the genera Bacteroides, Vibrio, Candidatus Bacilloplasma, Proteocatella, Carboxylicivirga, Cetobacterium, Epulopiscium, and Shewanella were the most abundant gut microbiota (Figure 2B). All these genera together accounted for 81.92% of the total microbiota.
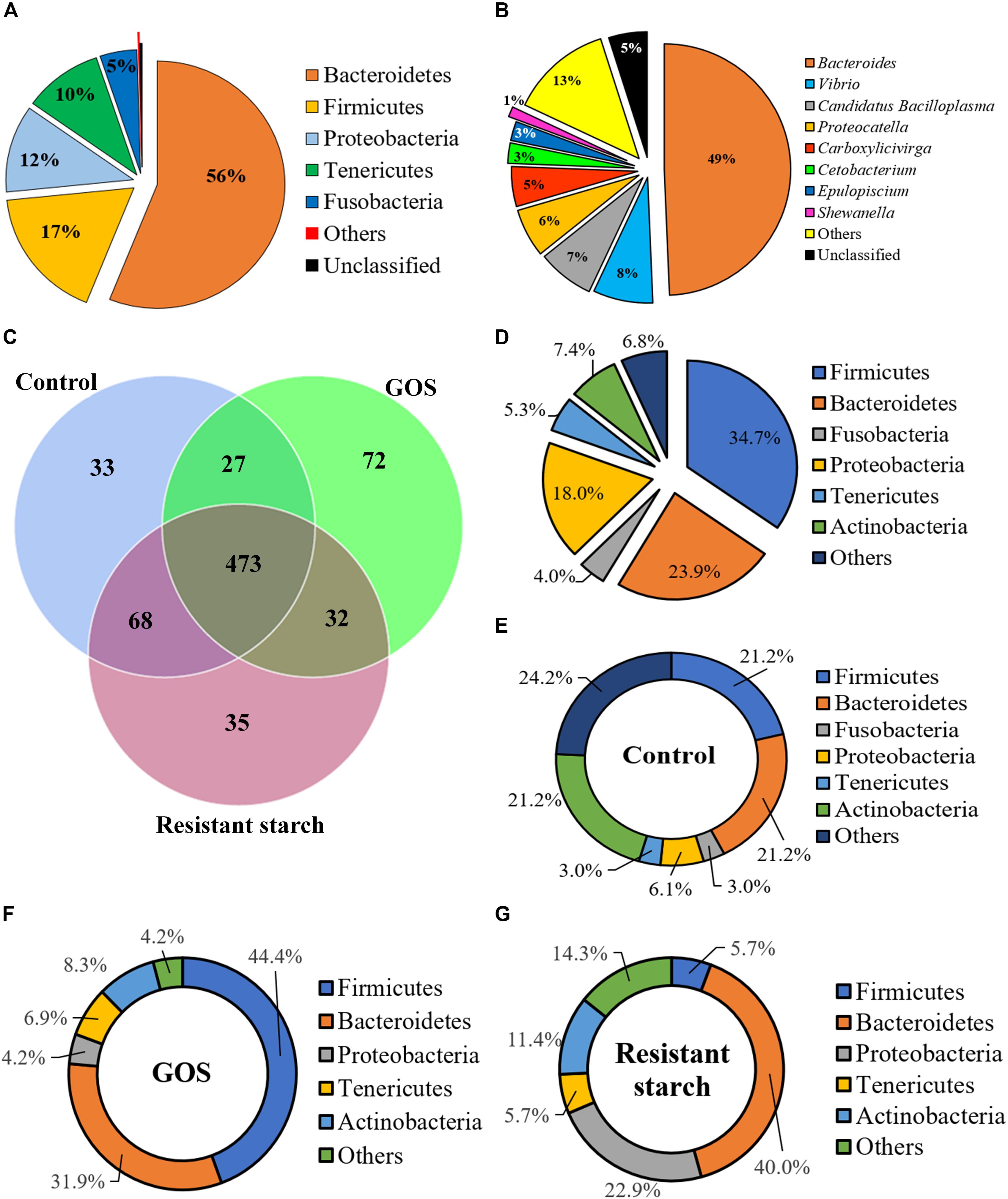
Figure 2. The composition of gut microbiota of galactooligosaccharides and resistant starch (using the combined data set) and the unique and shared OTUs in the cultures after a 24-h in vitro fermentation. (A) The five high-abundance phyla combined accounted for 99.53% of all OTUs, whereas low-abundance phyla accounted merely for 0.19% (marked as “others”) and 0.28% (“unclassified”). (B) The five genera with abundance ≥ 1% accounted for 81.92% of the total microbiota; the sequences that could not be classified into any known genus were marked as “unclassified” (accounting for 5.0%). (C) Venn diagram shows the number of shared and unique OTUs among the samples in different groups (the phylum level). (D–G) Pie charts show the characteristics of shared OTUs (D) and unique OTUs in the samples in controls (E), GOS (F), and resistant starch (G) groups (at the phylum level).
A Venn diagram revealed the numbers of unique or common OTUs (Figures 2C–G). Figure 2C showed that the numbers of unique OTUs in the control, GOS, and RS groups were 33, 72, and 35, respectively, and the numbers of shared OTUs among groups were 473 OTUs. The GOS and control samples shared 27 OTUs, while the RS and controls shared 68 OTUs and the GOS and RS groups shared 32 OTUs. At the phylum level, the most abundant unique and shared OTUs were determined as Firmicutes, Bacteroidetes, Proteobacteria, Tenericutes, Actinobacteria, and Fusobacteria (Figures 2D–G).
Microbial Diversity
The microbial complexity in the three groups was estimated using alpha diversity indices, including richness estimators (Chao1 and ACE) and diversity indexes (Simpson and Shannon index) (Figures 3A–D). Statistical testing revealed that the alpha diversity indices were significantly different between the GOS and controls or RS (P < 0.05), but not between RS and controls (P > 0.05). The beta diversity analysis, as estimated by Turkey and Wilcoxon tests, was significantly different between control and GOS (P = 0.00 and 0.00, respectively) or GOS and RS (P = 0.00 and 0.00, respectively), but not between control and RS (P > 0.05). The PERMANOVA analysis revealed a significant difference in overall bacterial community structure using the weighted UniFrac distance matrix between GOS and control (F = 53.25, P = 0.0001), GOS and RS (F = 55.7, P = 0.0002), and RS and control (F = 1.128, P = 0.331). A principal coordinate analysis (PCoA) (based on weighted UniFrac distance matrixes) was used to estimate the similarity in the microbial community composition of the samples (Figure 3E). A scatterplot based on PCoA scores revealed a difference in the bacterial community composition in the GOS in comparison to the RS and control and more similarity in that composition between the RS and control groups. This observation was also found in the weighted pair-group method with arithmetic means (WPGMA) clustering analysis (Figure 3F). The samples from the GOS group tend to cluster together, whereas the RS and control seemed to be more variable and grouped together.
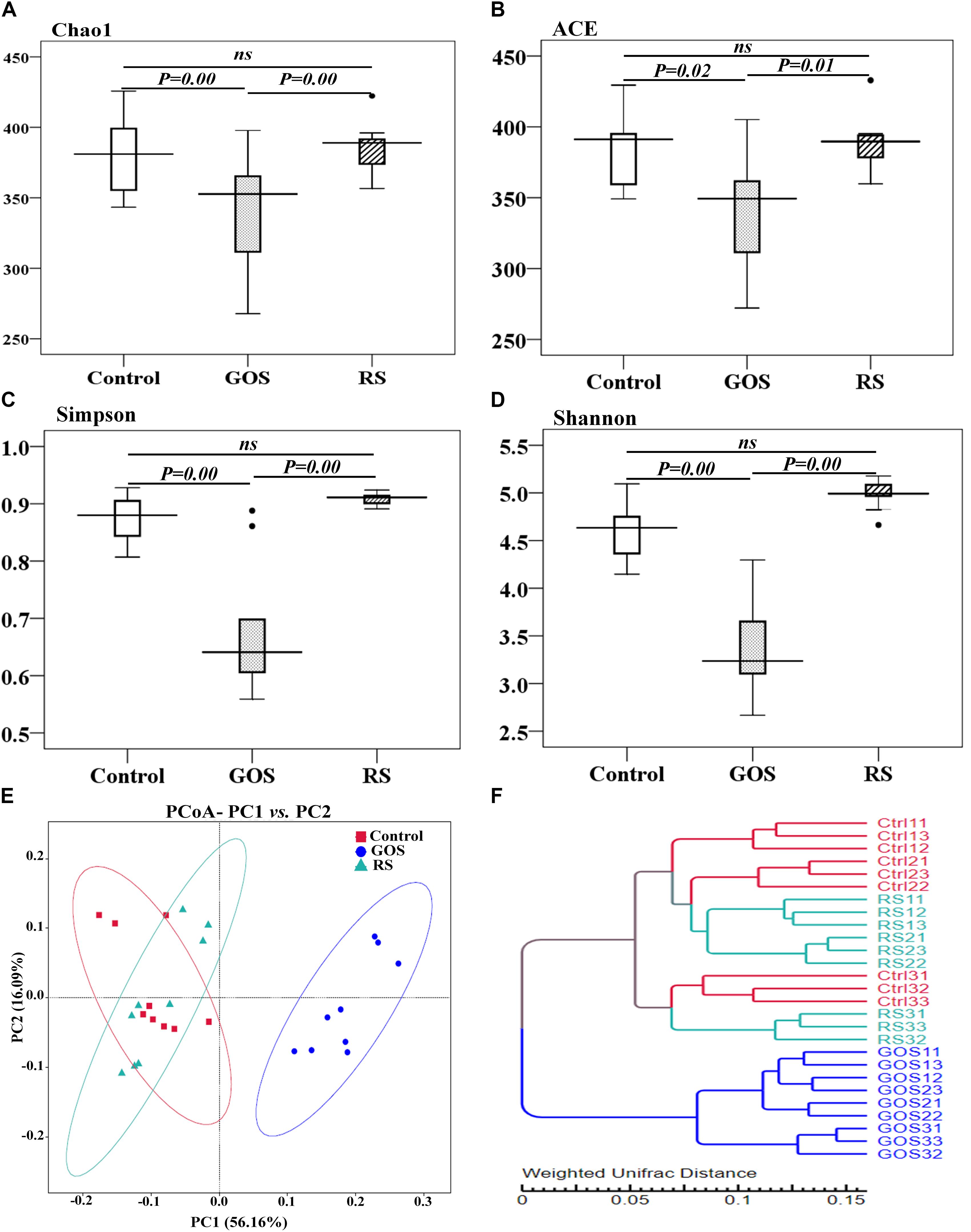
Figure 3. Alpha diversity estimators and bacterial community composition of gut microbiota populating the cultures after a 24-h in vitro fermentation of GOS or RS. (A–D) Alpha diversity estimators: Chao1 (A), ACE (B), Simpson (C), and Shannon (D). Differences are determined by ANOVA Turkey test (with P < 0.05). A non-significant difference is indicated with “ns.” (E) Principal coordinate analysis (PCoA) (PC1: 56.16% and PC2: 16.09% of the explained variance). (F) Cluster analysis using a weighted pair-group method with arithmetic mean. Each dot shows a single sample (Ctrl, GOS, and RS indicate the samples from the control, galactooligosaccharide, and resistant starch groups, respectively).
Microbial Composition
Differences in the taxonomical profiles [(Supplementary Table S1) and genus (Supplementary Table S2) levels] were analyzed. At the phylum level, Bacteroidetes, Firmicutes, Proteobacteria, Tenericutes, and Fusobacteria were predominant (together accounting for 99.4, 99.8, and 99.4% of the microbiota in the control, GOS, and RS, respectively). After 24 h of incubation with GOS or RS, the relative abundance of the dominant phyla changed (Supplementary Table S1 and Figure 4A). Compared to the control, GOS significantly increased the levels of Bacteroidetes and significantly decreased those of Firmicutes, Proteobacteria, Tenericutes, and Fusobacteria (P < 0.05). Also, the relative abundance of Bacteroidetes was significantly increased in the GOS compared to the RS, while that of Firmicutes, Proteobacteria, and Tenericutes was significantly decreased (P < 0.05). In the RS group, compared with control, the most notable changes were a significant increase in the levels of Tenericutes and a significant decrease in that of Fusobacteria (P < 0.05). Nevertheless, there was no significant difference in the remaining phyla (P > 0.05).
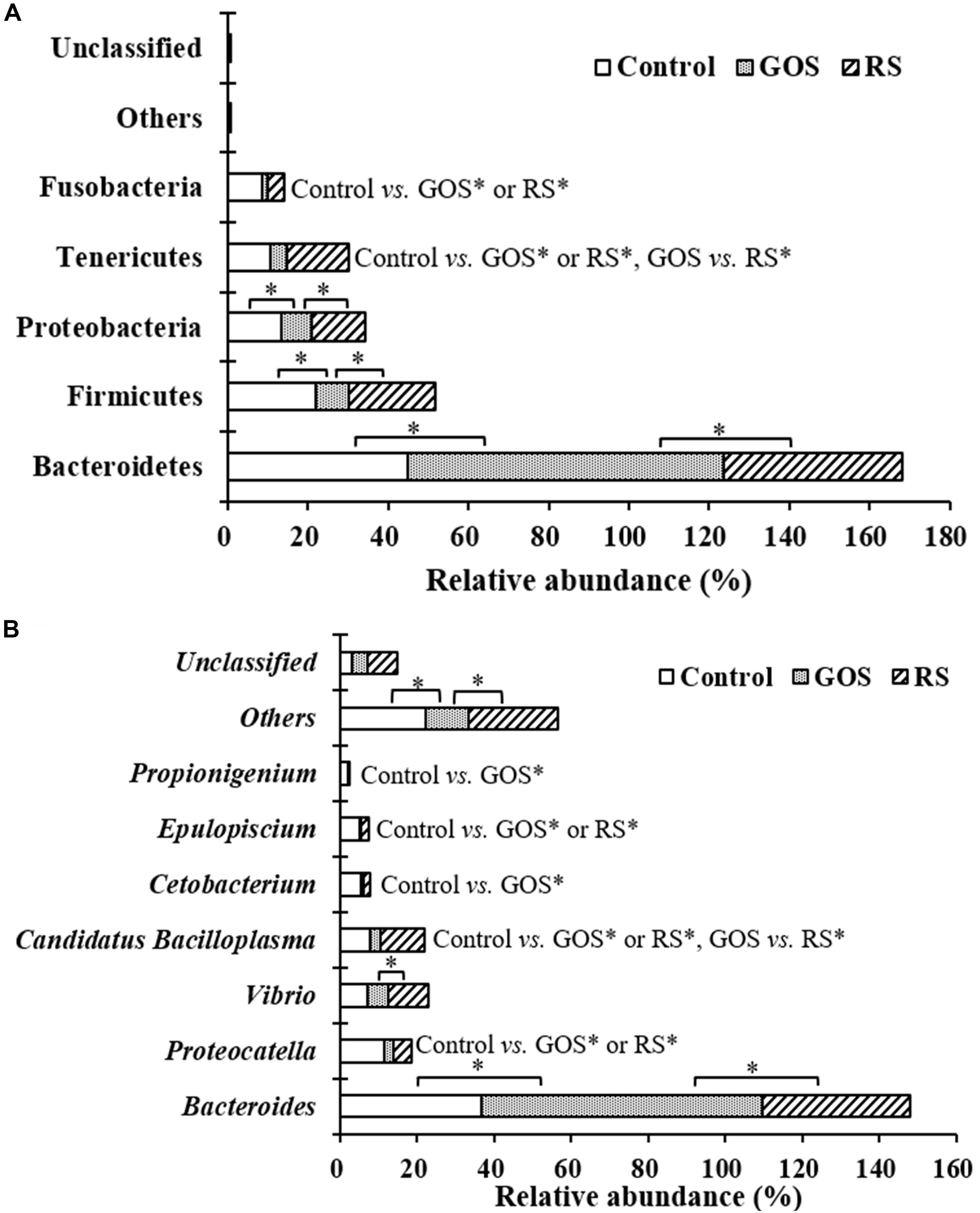
Figure 4. Composition of gut microbiota at the phylum (A) and genus (B) levels in the cultures after a 24-h in vitro fermentation of galactooligosaccharides (GOS) and resistant starch (RS), as well as in the controls. Differences determined by ANOVA Turkey test (with P < 0.05) are indicated with an asterisk.
At the genus level, the most abundant genera were classified as Bacteroides, Carboxylicivirga, Proteocatella, Vibrio, Shewanella, Candidatus Bacilloplasma, Cetobacterium, Epulopiscium, and Propionigenium (together accounting for 84.40, 89.32, and 74.60% of the microbiota in the control, GOS, and RS groups, respectively) (Figure 4B). After 24 h of incubation, the relative abundance of 25 of 144 classified genera changed across samples among the three groups (P < 0.05) (Supplementary Table S2). GOS dramatically increased the levels of Bacteroides and significantly decreased those of Proteocatella, Candidatus Bacilloplasma, Cetobacterium, Epulopiscium, and Propionigenium compared with the controls (Figure 4B). Bacteroides became more abundant, while Vibrio and Candidatus Bacilloplasma became less abundant in the GOS group compared to the RS (Figure 4B). In the RS group, compared to the controls, a significant increase in Candidatus Bacilloplasma abundance and a significant decrease in Proteocatella and Epulopiscium abundance were observed (Figure 4B).
Linear discriminant analysis (LDA) effect size (LEfSe) analysis (with LDA scores > 4.0) was performed to determine the specific taxa responsible for the differences among the three populations (Figure 5). A total of 50 bacterial taxa (24 in control; 6 in GOS; 20 in RS) showed differences among three populations (Figure 5A). Firmicutes and Fusobacteria were abundant in the control, while Bacteroidetes was abundant in the GOS, and Tenericutes and Proteobacteria in the RS group. Specifically, Bacteroides and Clostridium butyricum were abundant in the GOS, whereas Candidatus Bacilloplasma, Vibrio, Vagococcus, Dysgonomonas, Bacteroides xylanolyticus, Vagococcus fluvialis, and Dysgonomonas mossii were abundant in the RS. The cladogram analysis also revealed differences in bacterial taxa among the experimental groups (Figure 5B).
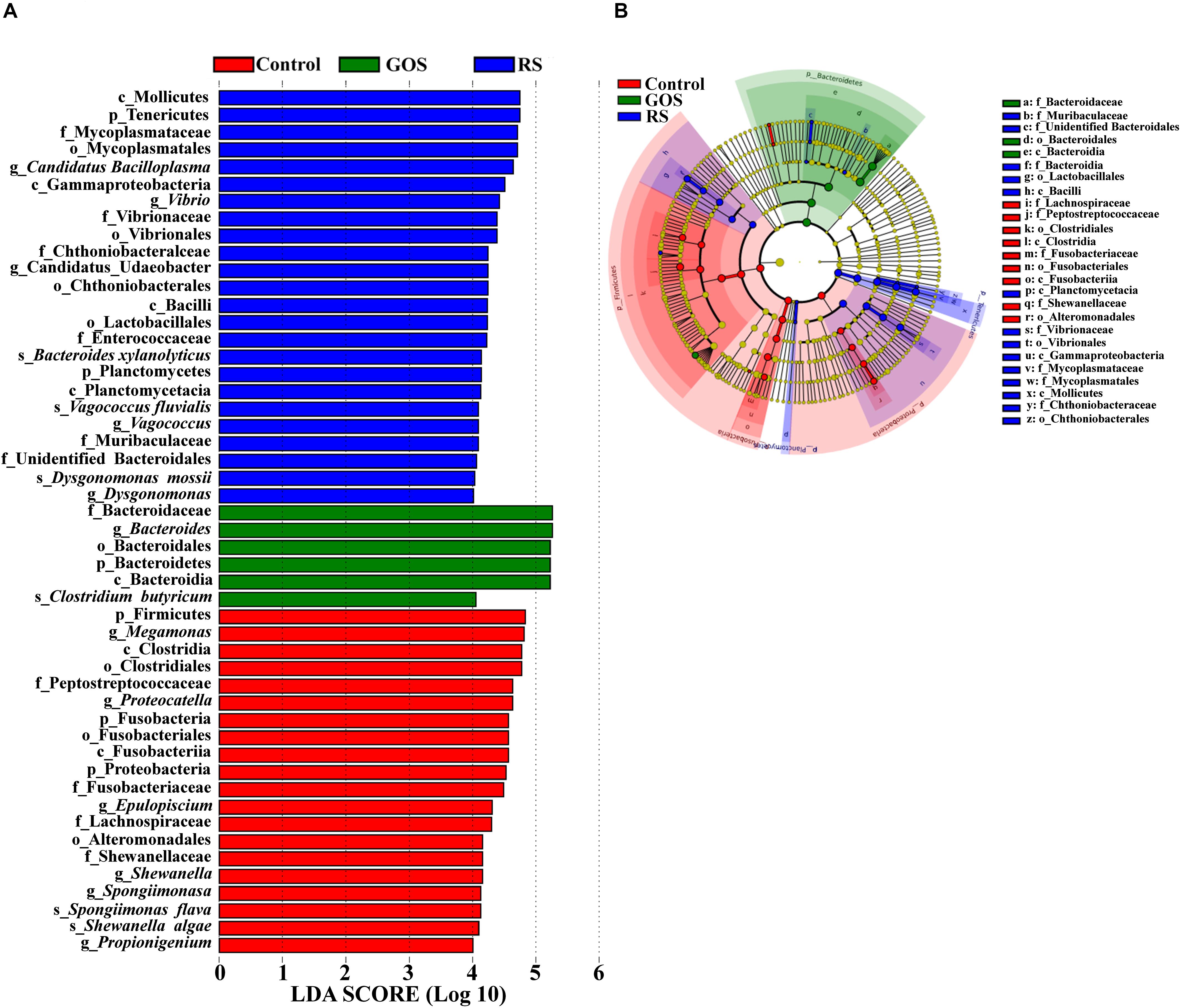
Figure 5. Characterization of gut microbiota in the cultures after a 24-h in vitro fermentation of GOS and RS, as well as in the controls by LDA and LEfSe analysis. (A) Histogram of the LDA scores (log10) calculated for features differentially abundant in control, GOS, and RS samples (with LDA scores > 4.0). (B) Bacterial taxa differentially represented among groups identified by LEfSe.
Associations Between Gut Microbiota and SCFAs
Spearman’s correlation analysis was used to determine the relationships between differentially abundant taxa (at the genus levels) and SCFA production (Figures 6A,B). In the GOS group, butyric acid was positively correlated with the genera Vagococcus (Spearman’s ρ [rs] = 0.79, P = 0.01) and Lactococcus (rs = 0.84, P = 0.00) (Figure 6A). In the RS group (Figure 6B), acetic acid had a significant positive correlation with Vagococcus (rs = 0.70, P = 0.04) and unidentified Lachnospiraceae (rs = 0.72, P = 0.03). Propionic acid was found to be strongly positively correlated with Candidatus Bacilloplasma (rs = 0.75, P = 0.02) and Parabacteroides (rs = 0.71, P = 0.03). Furthermore, a strong positive correlation was observed between butyric acid and Vagococcus (rs = 0.83, P = 0.01), Macellibacteroide (rs = 0.88, P = 0.00), Robinsoniella (rs = 0.74, P = 0.02), Anaerosporobacter (rs = 0.88, P = 0.00), Roseimarinus (rs = 0.68, P = 0.05), Succinispira (rs = 0.85, P = 0.00), Haloimpatiens (rs = 0.71, P = 0.03), Butyricicoccus (rs = 0.84, P = 0.00), Mangrovibacterium (rs = 0.78, P = 0.01), Paenibacillus (rs = 0.76, P = 0.02), Microlunatus (rs = 0.73, P = 0.03), unidentified Clostridiales (rs = 0.93, P = 0.00), and unidentified Lachnospiraceae (rs = 0.73, P = 0.03).
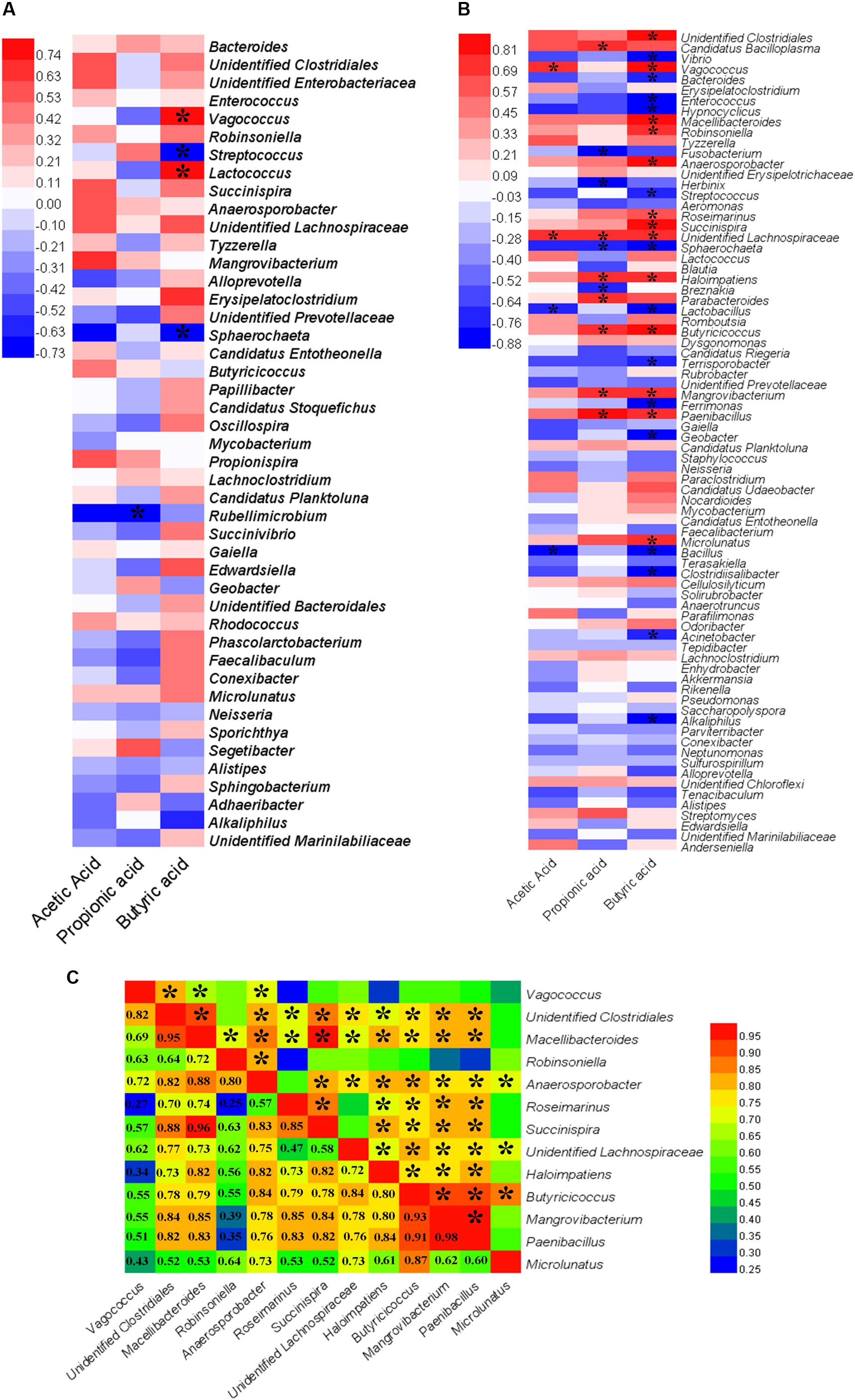
Figure 6. A heat map of Spearman’s correlation coefficients. (A,B) Correlation between the abundance of key microbial taxa (at the genus levels) and the production of short-chain fatty acids (SCFAs, including acetic, propionic, and butyric acids) in the cultures after a 24-h in vitro fermentation of galactooligosaccharides (A) and resistant starch (B). Red and blue colors indicate positive and negative correlation coefficients, respectively. Significant correlations (P < 0.05) are indicated with an asterisk. (C) Correlation among the bacterial genera that had a significantly positive correlation with the production of butyric acid in the cultures after a 24-h in vitro fermentation of resistant starch. Numbers indicate Spearman’s correlation coefficients. Significant correlations (P < 0.05) are indicated with an asterisk.
Correlations Among Bacteria Related to Butyric Acid Production
The correlations among bacterial taxa that had a significant positive correlation with the production of butyric acid were estimated. The results revealed that Vagococcus had a significant positive correlation with Lactococcus (rs = 0.93, P = 0.00) in the GOS. In the RS group, most genera were significantly positively correlated with others (P < 0.05) (Figure 6C). For example, Butyricicoccus were positively correlated with other taxa (rs > 0.55, P < 0.05), except Vagococcus (rs = 0.55, P = 0.12). The current findings indicated that the supplemented carbohydrates were fermented not only by a bacterium (or bacterial genus), but also by a population of bacteria to produce butyric acid.
Discussion
Dietary carbohydrate intake could influence the composition of gut microbiota and its metabolic products in the host intestinal tract (Scott et al., 2014). In aquatic animals, it has been shown that dietary carbohydrates can selectively promote the growth of beneficial microbes (including SCFA producers) inducing gut fermentation and SCFA formation (Tran et al., 2020). In this study, mud crab was used as a model organism to explore the effects of potential prebiotics (GOS and RS) selected from 10 kinds of carbohydrates on microbial composition and production of SCFAs under anaerobic fermentation with gut content samples in batch cultures. The production of SCFAs was measured using gas chromatography analysis while the composition of microbiota was determined using 16S rRNA sequencing.
SCFAs are the predominant end-products of bacterial fermentation of indigestible dietary carbohydrates, which confer beneficial health outcomes to the host (Piazzon et al., 2017; Tran et al., 2020). SCFAs are readily and quickly absorbed by host gut colonocytes, limiting the use of animal models; hence, in vitro fermentation cultures are useful for studying SCFA formation during fermentation (Inness et al., 2011). In this study, the concentrations of acetic, propionic, and butyric acids were significantly increased in batch cultures supplemented with either GOS or RS, indicating that both substrates could be fermented by microbes, as evidenced by SCFA production. This is similar to the results of a previous study in which cereal grains were used to stimulate the production of volatile fatty acids in Nile tilapia (O. niloticus) and European seabass (D. labrax) (Leenhouwers et al., 2008). Also, the supplementation of oligosaccharides increased the levels of total SCFA (sum of acetic, propionic, and n-butyric acids) in batch cultures with the gut contents of common carp (C. carpio) (Kihara and Sakata, 2002). Butyric acid shows protective effects, such as improving epithelial barrier function, gut permeability, and gut immune functions, on animals (Liu W. et al., 2014; Rimoldi et al., 2016; Safari et al., 2016; Piazzon et al., 2017; Tian et al., 2017). The term “prebiotics” has been defined as “ingredients of the non-digestible diet that is beneficial to the host for stimulating selectively the growth and/or the activity of one or more intestinal bacteria” (Gibson, 2004). Thus, a substrate that selectively stimulates the growth of butyrate-producing bacteria and is fermented to butyric acid meets the prebiotic definition criterion. It was observed that no significant difference in the concentration of butyric acid was found between the GOS and RS groups (P > 0.05), but a significant increase was observed between either GOS or RS and control (P < 0.05). The findings here indicate the prebiotic potential of GOS and RS in mud crab. Consistent with this view, it has been previously suggested that GOS and starch are potential prebiotics that affect growth performance, gut microbiota, and immune response in fish species, in vivo (Kihara and Sakata, 1997; Amirkolaie et al., 2006; Hoseinifar et al., 2013, 2017; Miandare et al., 2016; Romano et al., 2018). The effects of prebiotic-driven bacteria present in inocula on the relative proportions and rates of SCFAs produced in batch cultures have previously been reported (Sato et al., 2017; Reichardt et al., 2018; Baxter et al., 2019). Thus, this evidence supports the prebiotic potential of GOS and RS in modulating gut microbiota, which may indirectly improve the immune responses in mud crab. However, this needs to be confirmed in further studies.
Apart from the production of SCFAs from fermentation, prebiotics are expected to have the capacity to selectively promote the growth and/or activity of gut bacteria involved in health and well-being (Pham et al., 2017). Changes in the community composition due to the ingestion of dietary fibers occur as a result of differences in their physicochemical and metabolic properties (Umu et al., 2015). As shown in the Venn diagram, all samples in the control, GOS, and RS groups shared a large core of microbiota. This is consistent with previous findings in which gut microbiota of fish species on different trophic levels shared a large core microbiome (Liu et al., 2016), which might be important in the stability of the gut microbiota. In spite of this, each group still had a unique microbiome, suggesting that the supplemented carbohydrates mainly contributed to the growth of gut microbiota. Furthermore, the results of this study clearly revealed that GOS and RS induced shifts in the composition of gut microbiota, which correlated with the production of butyric acid in the batch cultures. These results are in agreement with previous studies, in which anaerobic fermentation with human fecal samples supplied with prebiotics (i.e., L-sorbose, xylitol, type 2 RS) was carried out in vitro (Scott et al., 2014; Pham et al., 2017; Sato et al., 2017; Reichardt et al., 2018; Baxter et al., 2019). Our data revealed that alpha diversity was lower in the GOS group than that in RS and the controls, which may be due to the selection of particular microbial communities within the prebiotic-supplemented samples. Consistent with this view, studies in young healthy humans treated with GOS for 14 days revealed worsened alpha diversity compared with those on day 0 (Liu et al., 2017). This supports the view that a more diverse community is coupled with a healthier and more stable ecosystem (Xiao et al., 2016). The decreased bacterial diversity, as shown in this study, may be explained by the carbohydrate-driven lowering of pH, which affects the survival capacity of acid-sensitive species and stimulates the overgrowth of acid-tolerant species, thereby suppressing bacterial diversity in batch cultures (Gross et al., 2010). Low pH (5.5) increased the growth of butyrate-producing bacteria, resulting in the elevation of butyric acid production (Reichardt et al., 2018). The results further support the selective stimulation of GOS on the growth of SCFA-producing bacteria. Notably, supplementation with RS led to an unchanged bacterial diversity in the batch cultures compared to the controls, indicating that the microbes were relatively constant, resulting in the stability of the living ecosystem on RS supplementation (Xiao et al., 2016). In contrast to our findings, alpha diversity within the microbiome was lower in pigs fed an RS diet compared to controls (Umu et al., 2015). The differences in results might be due to differences in experimental designs (in vitro vs. in vivo), study subjects, and microbial composition communities. Furthermore, the analyses of PERMANOVA, PCoA, and WPGMA clustering showed significant differences in beta diversity between the GOS group and RS or control (P < 0.05), suggesting that the diversity of gut microbiota in the batch cultures was affected by the supplemented carbohydrates.
Although the presence of bacterial communities (at the phylum level, with a dominance of Bacteroidetes, Firmicutes, Proteobacteria, Tenericutes, and Fusobacteria) was almost similar among the groups, the abundance of some phyla in the GOS and RS groups differed significantly from the controls. The composition of predominant bacterial phyla in this study was similar to those previously described for mud crab (S. paramamosain) (Li et al., 2012). This indicates that these bacterial members may be a core microbiome that harbors the gut of mud crab, conserving the stability of gut microbiota even when stimulated with supplemented carbohydrates under in vitro fermentation cultures. The increased level of the genera Bacteroides (GOS group) and Candidatus Bacilloplasma (RS group) suggests that the members of these genera may be important in degrading GOS and RS, respectively, in the batch cultures.
The benefits of butyric acid in the gut health of aquatic animals have been discussed previously (Tran et al., 2020). Remarkably, the relative proportions and rates of SCFAs produced are influenced by the bacteria present in the batch cultures (Inness et al., 2011). Herein, we tested whether the relative abundance of gut microbiota was related to the production of SCFAs through fermentation of the supplemented carbohydrates. In the GOS group, no bacterial genus was found to be associated with the production of acetic and propionic acids, suggesting that GOS herein is not an ideal substrate fermented by acetate- and propionate-producing bacteria. However, RS could be fermented by members of Vagococcus and unidentified Lachnospiraceae to acetic acid, as well as Candidatus Bacilloplasma and Parabacteroides to propionic acid. Interestingly, more bacterial genera were associated with the production of butyric acid in either the GOS or RS group. This finding supports the foregoing conclusions that GOS and RS are able to selectively promote the growth of butyrate producers, resulting in an increase of butyric acid production. Generally, the increased relative abundance of a bacterial taxon is inadequate to explain the association with butyrate production within samples (Baxter et al., 2019). Thus, the correlations among the bacterial genera that positively associated with the increased level of butyric acid within each group (GOS or RS) were investigated. Our results showed a significant positive correlation among the bacterial genera. This is similar to the results in a previous study that the correlation between Ruminococcus bromii and Eubacterium rectale was coupled with higher butyrate concentration in humans consuming unmodified starches (Baxter et al., 2019). The findings herein suggest that the carbohydrate-driven bacteria can interact syntrophically with others and change the metabolic pathways, which stimulated the production of butyric acid.
Conclusion
This in vitro fermentation study provides insight into the supplementation of GOS and RS, which is capable of altering gut microbiota and the production of SCFAs. Generally, GOS induced a decrease in the diversity of gut microbial communities compared with RS and control, while RS did not change the diversity of these compared with controls. An overview of the microbiota related to the pathways for SCFA production in response to the supplementation of GOS or RS is shown in Figure 7. The production of butyric acid was found to be related to the increased relative abundance of many bacterial genera in both GOS and RS groups. Most of these genera were positively correlated with others, suggesting that the carbohydrate-driven bacteria could interact syntrophically with others and change the metabolic pathways to produce butyric acid. As the current study was conducted using only an in vitro fermentation approach, the results need to be confirmed by an in vivo mud crab fermentation study.
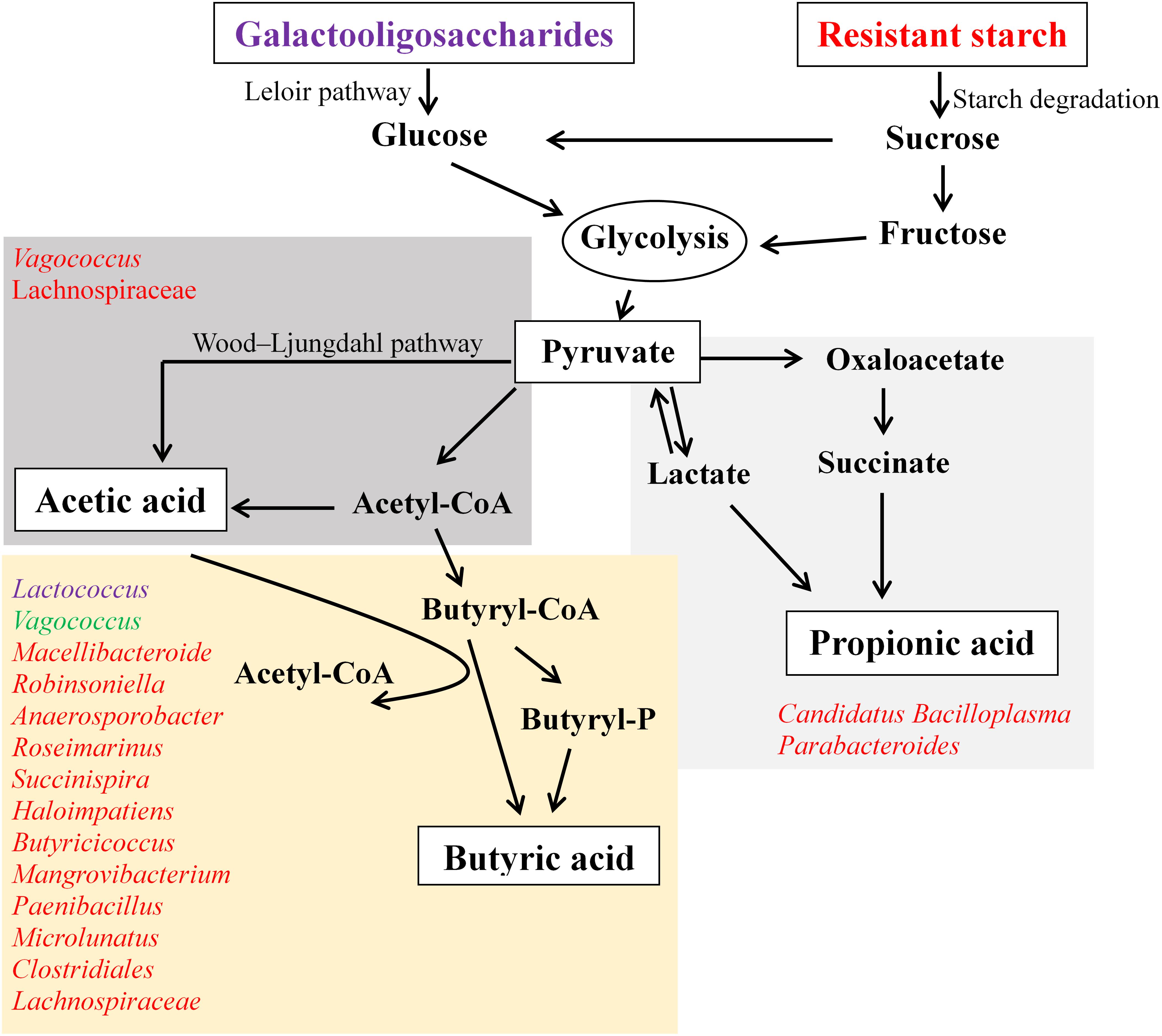
Figure 7. An overview of the production of acetic, propionic, and butyric acids in the in vitro fermentation of galactooligosaccharides (GOS) and resistant starch (RS) with gut contents of mud cab. Bacterial genera involved in the production of SCFAs were determined based on the Spearman’s correlation coefficients between the abundance of microbial taxa and the production of each kind of SCFA. Bacteria illustrated in purple indicate the correlations found in the GOS group, red indicates the correlations in the RS group, and green indicates the correlations in both GOS and RS groups.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics Statement
The animal study was reviewed and approved by all animal handling procedures in this study were reviewed and approved by the ethics committee of “The Regulations for The Administration of Affairs Concerning Experimental Animals.”
Author Contributions
NT and SL designed the study, analyzed the data, and wrote the manuscript. YT, ZL, and MZ attended the sample processing, DNA isolation, and sequencing of samples. XW and HM reviewed the manuscript and provided the guidance. All the authors read and approved the final manuscript.
Funding
This research work was supported by grants from the National Natural Science Foundation of China (41876152, 31850410487, and 31802341), China Postdoctoral Science Foundation (2018M643133), Guangdong provincial project of Science and Technology (2017B020204003), and by support from the Department of Education of Guangdong Province (2017KCXTD014).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.01352/full#supplementary-material
FIGURE S1 | Alpha diversity in rarefaction curves plot (A) and in rank abundance curves.
TABLE S1 | Bacterial composition of gut microbiota in the batch cultures after 24 h of fermentation of galactooligosaccharides (GOS) and resistant starch (RS) (at the phylum level).
TABLE S2 | Bacterial composition of gut microbiota in the batch cultures after 24 h of fermentation of galactooligosaccharides (GOS) and resistant starch (RS) (at the genus level).
TEXT S1 | Prescreening of potential prebiotics and SCFA analysis.
Footnotes
References
Ali, T. E.-S., El-Sayed, A. M., Eissa, M. A.-R., and Hanafi, H. M. (2018). Effects of dietary biogen and sodium butyrate on hematological parameters, immune response, and histological characteristics of Nile tilapia (Oreochromis niloticus) fingerlings. Aquacult. Int. 26, 139–150. doi: 10.1007/s10499-017-0205-3
Amirkolaie, A. K., Verreth, J. A. J., and Schrama, J. W. (2006). Effect of gelatinization degree and inclusion level of dietary starch on the characteristics of digesta and faeces in Nile tilapia (Oreochromis niloticus (L.). Aquaculture 260, 194–205. doi: 10.1016/j.aquaculture.2006.06.039
Baxter, N. T., Schmidt, A. W., Venkataraman, A., Kim, K. S., Waldron, C., and Schmidt, T. M. (2019). Dynamics of human gut microbiota and short-chain fatty acids in response to dietary interventions with three fermentable fibers. mBio 10:e02566-18. doi: 10.1128/mBio.02566-18
Caporaso, J. G., Kuczynski, J., Stombaugh, J., Bittinger, K., Bushman, F. D., Costello, E. K., et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336. doi: 10.1038/nmeth.f.303
Caporaso, J. G., Lauber, C. L., Walters, W. A., Berg-Lyons, D., Lozupone, C. A., Turnbaugh, P. J., et al. (2011). Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. PNAS 108, 4516–4522. doi: 10.1073/pnas.1000080107
Chen, W. W., Romano, N., Ebrahimi, M., and Natrah, I. (2017). The effects of dietary fructooligosaccharide on growth, intestinal short chain fatty acids level and hepatopancreatic condition of the giant freshwater prawn (Macrobrachium rosenbergii) post-larvae. Aquaculture 469, 95–101. doi: 10.1016/j.aquaculture.2016.11.034
Daniels, C. L., Merrifield, D. L., Ringø, E., and Davies, S. J. (2013). Probiotic, prebiotic and synbiotic applications for the improvement of larval European lobster (Homarus gammarus) culture. Aquaculture 416, 396–406. doi: 10.1016/j.aquaculture.2013.08.001
Duan, Y., Zhang, Y., Dong, H., Wang, Y., and Zhang, J. (2017). Effects of dietary poly-β-hydroxybutyrate (PHB) on microbiota composition and the mTOR signaling pathway in the intestines of Litopenaeus vannamei. J. Microbiol. 55, 946–954. doi: 10.1007/s12275-017-7273-y
Ebrahimi, M., Daeman, N. H., Chong, C. M., Karami, A., Kumar, V., Hoseinifar, S. H., et al. (2017). Comparing the effects of different dietary organic acids on the growth, intestinal short-chain fatty acids, and liver histopathology of red hybrid tilapia (Oreochromis sp.) and potential use of these as preservatives. Fish Physiol. Biochem. 43, 1195–1207. doi: 10.1007/s10695-017-0365-0
Edgar, R. C. (2013). UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 10, 996–998. doi: 10.1038/nmeth.2604
Edgar, R. C., Haas, B. J., Clemente, J. C., Quince, C., and Knight, R. (2011). UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27, 2194–2200. doi: 10.1093/bioinformatics/btr381
Elshopakey, G. E., Risha, E. F., Abdalla, O. A., Okamura, Y., Hanh, V. D., Ibuki, M., et al. (2018). Enhancement of immune response and resistance against Vibrio parahaemolyticus in kuruma shrimp (Marsupenaeus japonicus) by dietary supplementation of β-1, 4-mannobiose. Fish Shellfish Immunol. 74, 26–34. doi: 10.1016/j.fsi.2017.12.036
Estensoro, I., Ballester-Lozano, G., Benedito-Palos, L., Grammes, F., Martos-Sitcha, J. A., Mydland, L.-T., et al. (2016). Dietary butyrate helps to restore the intestinal status of a marine teleost (Sparus aurata) fed extreme diets low in fish meal and fish oil. PLoS One 11:e0166564. doi: 10.1371/journal.pone.0166564
Gao, Y., Storebakken, T., Shearer, K. D., Penn, M., and Øverland, M. (2011). Supplementation of fishmeal and plant protein-based diets for rainbow trout with a mixture of sodium formate and butyrate. Aquaculture 311, 233–240. doi: 10.1016/j.aquaculture.2010.11.048
Geraylou, Z., Rurangwa, E., van der Wiele, T., Courtin, C. M., Delcour, J. A., Buyse, J., et al. (2014). Fermentation of arabinoxylan-oligosaccharides, oligofructose and their monomeric sugars by hindgut bacteria from Siberian sturgeon and African catfish in batch culture in vitro. J. Aquac. Res. Dev. 5:1000230. doi: 10.4172/2155-9546.1000230
Gibson, G. R. (2004). Fibre and effects on probiotics (the prebiotic concept). Clin. Nutr. Suppl. 1, 25–31. doi: 10.1016/j.clnu.2004.09.005
Gross, E. L., Leys, E. J., Gasparovich, S. R., Firestone, N. D., Schwartzbaum, J. A., Janies, D. A., et al. (2010). Bacterial 16S sequence analysis of severe caries in young permanent teeth. J. Clin. Microbiol. 48, 4121–4128. doi: 10.1128/JCM.01232-10
Hammer, Ø, Harper, D. A. T., and Ryan, P. D. (2001). PAST: paleontological statistics software package for education and data analysis. Palaeontol. Electron. 4, 1–9.
Hoseinifar, S. H., Ahmadi, A., Raeisi, M., Hoseini, S. M., Khalili, M., and Behnampour, N. (2017). Comparative study on immunomodulatory and growth enhancing effects of three prebiotics (galactooligosaccharide, fructooligosaccharide and inulin) in common carp (Cyprinus carpio). Aquac. Res. 48, 3298–3307. doi: 10.1111/are.13156
Hoseinifar, S. H., Khalili, M., Rostami, H. K., and Esteban, M. Á (2013). Dietary galactooligosaccharide affects intestinal microbiota, stress resistance, and performance of Caspian roach (Rutilus rutilus) fry. Fish Shellfish Immunol. 35, 1416–1420. doi: 10.1016/j.fsi.2013.08.007
Inness, V. L., Khoo, C., Gross, K. L., Hoyles, L., Gibson, G. R., and McCartney, A. L. (2011). Use of static batch culture systems to investigate the fermentation effects of selected oligosaccharides and fibres by the canine faecal microbiota. Int. J. Probiotics Prebiotics 6, 57–64.
Kashinskaya, E., Suhanova, E., Solov’ev, M., Izvekova, G., and Glupov, V. (2014). Diversity of microbial communities of the intestinal mucosa and intestinal contents of fish from Lake Chany (Western Siberia). Inland Water Biol. 7, 172–177. doi: 10.1134/S1995082914020047
Kihara, M., and Sakata, T. (1997). Fermentation of dietary carbohydrates to short-chain fatty acids by gut microbes and its influence on intestinal morphology of a detritivorous teleost tilapia (Oreochromis niloticus). Comp. Biochem. Physiol. A Physiol. 118, 1201–1207. doi: 10.1016/S0300-9629(97)00052-2
Kihara, M., and Sakata, T. (2002). Production of short-chain fatty acids and gas from various oligosaccharides by gut microbes of carp (Cyprinus carpio L.) in micro-scale batch culture. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 132, 333–340. doi: 10.1016/S1095-6433(02)00029-6
Leenhouwers, J. I., Pellikaan, W. F., Huizing, H. F. A., Coolen, R. O. M., Verreth, J. A. J., and Schrama, J. W. (2008). Fermentability of carbohydrates in an in vitro batch culture method using inocula from Nile tilapia (Oreochromis niloticus) and European sea bass (Dicentrarchus labrax). Aquac. Nutr. 14, 523–532. doi: 10.1111/j.1365-2095.2007.00558.x
Li, S., Sun, L., Wu, H., Hu, Z., Liu, W., Li, Y., et al. (2012). The intestinal microbial diversity in mud crab (Scylla paramamosain) as determined by PCR-DGGE and clone library analysis. J. Appl. Microbiol. 113, 1341–1351. doi: 10.1111/jam.12008
Li, X., Ringø, E., Hoseinifar, S. H., Lauzon, H. L., Birkbeck, H., and Yang, D. (2018). The adherence and colonization of microorganisms in fish gastrointestinal tract. Rev. Aquacult. 11, 603–618. doi: 10.1111/raq.12248
Li, Z., Tran, N. T., Ji, P., Sun, Z., Wen, X., and Li, S. (2019). Effects of prebiotic mixtures on growth performance, intestinal microbiota and immune response in juvenile chu’s croaker. Nibea coibor. Fish Shellfish Immunol. 89, 564–573. doi: 10.1016/j.fsi.2019.04.025
Liu, B., Qian, J., Wang, Q., Wang, F., Ma, Z., and Qiao, Y. (2014). Butyrate protects rat liver against total hepatic ischemia reperfusion injury with bowel congestion. PLoS One 9:e106184. doi: 10.1371/journal.pone.0106184
Liu, W., Yang, Y., Zhang, J., Gatlin, D. M., Ringø, E., and Zhou, Z. (2014). Effects of dietary microencapsulated sodium butyrate on growth, intestinal mucosal morphology, immune response and adhesive bacteria in juvenile common carp (Cyprinus carpio) pre-fed with or without oxidised oil. Br. J. Nutr. 112, 15–29. doi: 10.1017/S0007114514000610
Liu, F., Li, P., Chen, M., Luo, Y., Prabhakar, M., Zheng, H., et al. (2017). Fructooligosaccharide (FOS) and galactooligosaccharide (GOS) increase Bifidobacterium but reduce butyrate producing bacteria with adverse glycemic metabolism in healthy young population. Sci. Rep. 7:11789. doi: 10.1038/s41598-017-10722-2
Liu, H., Guo, X., Gooneratne, R., Lai, R., Zeng, C., Zhan, F., et al. (2016). The gut microbiome and degradation enzyme activity of wild freshwater fishes influenced by their trophic levels. Sci. Rep. 6:24340. doi: 10.1038/srep24340
Lozupone, C., Hamady, M., and Knight, R. (2006). UniFrac-an online tool for comparing microbial community diversity in a phylogenetic context. BMC Bioinformatics 7:371. doi: 10.1186/1471-2105-7-371
Martin, M. (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 17, 10–12. doi: 10.14806/ej.17.1.200
Miandare, H. K., Farvardin, S., Shabani, A., Hoseinifar, S. H., and Ramezanpour, S. S. (2016). The effects of galactooligosaccharide on systemic and mucosal immune response, growth performance and appetite related gene transcript in goldfish (Carassius auratus gibelio). Fish Shellfish Immunol. 55, 479–483. doi: 10.1016/j.fsi.2016.06.020
Pham, T., Savary, B. J., Teoh, K., Chen, M.-H., McClung, A., and Lee, S.-O. (2017). In vitro fermentation patterns of rice bran components by human gut microbiota. Nutrients 9:1237. doi: 10.3390/nu9111237
Piazzon, M. C., Calduch-Giner, J. A., Fouz, B., Estensoro, I., Simó-Mirabet, P., Puyalto, M., et al. (2017). Under control: how a dietary additive can restore the gut microbiome and proteomic profile, and improve disease resilience in a marine teleostean fish fed vegetable diets. Microbiome 5:164. doi: 10.1186/s40168-017-0390-3
Ramírez, N. C. B., Rodrigues, M. S., Guimarães, A. M., Guertler, C., Rosa, J. R., Seiffert, W. Q., et al. (2017). Effect of dietary supplementation with butyrate and probiotic on the survival of Pacific white shrimp after challenge with Vibrio alginolyticus. R. Bras. Zootec. 46, 471–477. doi: 10.1590/s1806-92902017000600001
Reichardt, N., Vollmer, M., Holtrop, G., Farquharson, F. M., Wefers, D., Bunzel, M., et al. (2018). Specific substrate-driven changes in human faecal microbiota composition contrast with functional redundancy in short-chain fatty acid production. ISME J. 12, 610–622. doi: 10.1038/ismej.2017.196
Rimoldi, S., Finzi, G., Ceccotti, C., Girardello, R., Grimaldi, A., Ascione, C., et al. (2016). Butyrate and taurine exert a mitigating effect on the inflamed distal intestine of European sea bass fed with a high percentage of soybean meal. Fish Aquat. Sci. 19:40. doi: 10.1186/s41240-016-0041-9
Rimoldi, S., Gliozheni, E., Ascione, C., Gini, E., and Terova, G. (2018). Effect of a specific composition of short-and medium-chain fatty acid 1-Monoglycerides on growth performances and gut microbiota of gilthead sea bream (Sparus aurata). PeerJ 6:e5355. doi: 10.7717/peerj.5355
Ringø, E., Hoseinifar, S. H., Ghosh, K., Doan, H. V., Beck, B. R., and Song, S. K. (2018). Lactic acid bacteria in finfish- An update. Front. Microbiol. 9:1818. doi: 10.3389/fmicb.2018.01818
Ringø, E., Olsen, R., Gifstad, T., Dalmo, R., Amlund, H., Hemre, G. I., et al. (2010). Prebiotics in aquaculture: a review. Aquac. Nutr. 16, 117–136. doi: 10.1111/j.1365-2095.2009.00731.x
Ringø, E., Sperstad, S., Myklebust, R., Refstie, S., and Krogdahl, Å (2006). Characterisation of the microbiota associated with intestine of Atlantic cod (Gadus morhua L.): the effect of fish meal, standard soybean meal and a bioprocessed soybean meal. Aquaculture 261, 829–841. doi: 10.1016/j.aquaculture.2006.06.030
Robles, R., Lozano, A., Sevilla, A., Márquez, L., Nuez-Ortin, W., and Moyano, F. (2013). Effect of partially protected butyrate used as feed additive on growth and intestinal metabolism in sea bream (Sparus aurata). Fish Physiol. Biochem. 39, 1567–1580. doi: 10.1007/s10695-013-9809-3
Romano, N., Kanmani, N., Ebrahimi, M., Chong, C. M., Teh, J. C., Hoseinifar, S. H., et al. (2018). Combination of dietary pre-gelatinized starch and isomaltooligosaccharides improved pellet characteristics, subsequent feeding efficiencies and physiological status in African catfish, Clarias gariepinus, juveniles. Aquaculture 484, 293–302. doi: 10.1016/j.aquaculture.2017.09.022
Safari, O., and Paolucci, M. (2018). Effect of in vitro selected synbiotics (galactooligosaccharide and mannanoligosaccharide with or without Enterococcus faecalis) on growth performance, immune responses and intestinal microbiota of juvenile narrow clawed crayfish, Astacus leptodactylus leptodactylus Eschscholtz, 1823. Aquac. Nutr. 24, 247–259. doi: 10.1111/anu.12553
Safari, R., Hoseinifar, S. H., and Kavandi, M. (2016). Modulation of antioxidant defense and immune response in zebra fish (Danio rerio) using dietary sodium propionate. Fish Physiol. Biochem. 42, 1733–1739. doi: 10.1007/s10695-016-0253-z
Sato, T., Kusuhara, S., Yokoi, W., Ito, M., and Miyazaki, K. (2017). Prebiotic potential of L-sorbose and xylitol in promoting the growth and metabolic activity of specific butyrate-producing bacteria in human fecal culture. FEMS Microbiol. Ecol. 93:fiw227. doi: 10.1093/femsec/fiw227
Scott, K. P., Martin, J. C., Duncan, S. H., and Flint, H. J. (2014). Prebiotic stimulation of human colonic butyrate-producing bacteria and bifidobacteria, in vitro. FEMS Microbiol. Ecol. 87, 30–40. doi: 10.1111/1574-6941.12186
Silva, B. C., Nolasco-Soria, H., Magallón-Barajas, F., Civera-Cerecedo, R., Casillas-Hernández, R., and Seiffert, W. (2016a). Improved digestion and initial performance of whiteleg shrimp using organic salt supplements. Aquac. Nutr. 22, 997–1005. doi: 10.1111/anu.12315
Silva, B. C., Vieira, F. D. N., Mouriño, J. L. P., Bolivar, N., and Seiffert, W. Q. (2016b). Butyrate and propionate improve the growth performance of Litopenaeus vannamei. Aquac. Res. 47, 612–623. doi: 10.1111/are.12520
Terova, G., Díaz, N., Rimoldi, S., Ceccotti, C., Gliozheni, E., and Piferrer, F. (2016). Effects of sodium butyrate treatment on histone modifications and the expression of genes related to epigenetic regulatory mechanisms and immune response in European sea bass (Dicentrarchus labrax) fed a plant-based diet. PLoS One 11:e0160332. doi: 10.1371/journal.pone.0160332
Tian, L., Zhou, X.-Q., Jiang, W.-D., Liu, Y., Wu, P., Jiang, J., et al. (2017). Sodium butyrate improved intestinal immune function associated with NF-κB and p38MAPK signalling pathways in young grass carp (Ctenopharyngodon idella). Fish Shellfish Immunol. 66, 548–563. doi: 10.1016/j.fsi.2017.05.049
Tran, N. T., Li, Z., Wang, S., Zheng, H., Aweya, J. J., Wen, X., et al. (2020). Progress and perspectives of short-chain fatty acids in aquaculture. Rev. Aquacult. 12, 283–298. doi: 10.1111/raq.12317
Umu, ÖC., Frank, J. A., Fangel, J. U., Oostindjer, M., Da Silva, C. S., Bolhuis, E. J., et al. (2015). Resistant starch diet induces change in the swine microbiome and a predominance of beneficial bacterial populations. Microbiome 3:16. doi: 10.1186/s40168-015-0078-5
Wang, A. R., Ran, C., Ringø, E., and Zhou, Z. G. (2018). Progress in fish gastrointestinal microbiota research. Rev. Aquacult. 10, 626–640. doi: 10.1111/raq.12191
Wong, J. M. W., de Souza, R., Kendall, C. W. C., Emam, A., and Jenkins, D. J. A. (2006). Colonic health: fermentation and short chain fatty acids. J. Clin. Gastroenterol. 40, 235–243. doi: 10.1097/00004836-200603000-00015
Keywords: butyric acid, gut microbiota, prebiotic, Scylla paramamosain, short-chain fatty acids
Citation: Tran NT, Tang Y, Li Z, Zhang M, Wen X, Ma H and Li S (2020) Galactooligosaccharides and Resistant Starch Altered Microbiota and Short-Chain Fatty Acids in an in vitro Fermentation Study Using Gut Contents of Mud Crab (Scylla paramamosain). Front. Microbiol. 11:1352. doi: 10.3389/fmicb.2020.01352
Received: 01 April 2020; Accepted: 26 May 2020;
Published: 30 June 2020.
Edited by:
Luisa I. Falcon, National Autonomous University of Mexico, MexicoReviewed by:
Adele Costabile, University of Roehampton London, United KingdomGabriela Gaxiola, Universidad Nacional Autónoma de México, Mexico
Copyright © 2020 Tran, Tang, Li, Zhang, Wen, Ma and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shengkang Li, lisk@stu.edu.cn
 Ngoc Tuan Tran
Ngoc Tuan Tran Yong Tang1,2,3
Yong Tang1,2,3 Ming Zhang
Ming Zhang Hongyu Ma
Hongyu Ma Shengkang Li
Shengkang Li