- 1Institute of Pathogen Biology, Taishan Medical College, Tai’an, China
- 2Department of Orthopedics, The Second Xiangya Hospital, Central South University, Changsha, China
- 3Department of Sports Medicine Research Center, Central South University, Changsha, China
- 4Department of Ji Nan Children’s Hospital, Jinan, China
Toxoplasma gondii is an intracellular parasite and causes a global epidemic parasitic disease. T. gondii-infection could inhibit the growth of tumor. In this study, the transcriptomes of samples were detected by deep sequencing analysis. The transcriptome data was compared with reference genome to perform sequence alignment and the further analysis. The analyses of differential expression and the differentially expressed genes were performed in the present study. Genes involved in P53 signaling pathway, COLORECTAL cancer pathway, NON-SMALL CELL LUNG cancer signaling pathway, and BREAST cancer signaling pathway were up-regulated or down-regulated among the samples. The KEGG analysis indicated that the cancer pathways changed after infection of T. gondii. Furthermore, tumor-related mRNAs from different samples had a large difference, which suggested that the difference might provide important information in resisting cancer. The protein results indicated that tumor-related protein changes occurred after infection of T. gondii. In conclusion, the infection changed the cancer pathways, which could possibly inhibit the growth of tumor.
Introduction
Toxoplasma gondii is a kind of obligate intracellular parasite protozoan that is distributed worldwide. Most of T. gondii infections are latent infections and do not show clinical symptoms. However, once the body’s immune function is impaired, T. gondii can cause opportunistic diseases (Martins-Duarte et al., 2010). The occurrence of tumors activates the reproduction of T. gondii in hosts. Tumor patients are more likely to suffer toxoplasmosis than non-tumor people (Jiang et al., 2015; Al-Mendalawi, 2017; Imam et al., 2017; Vittecoq and Thomas, 2017). Conversely, T. gondii infection can also promote tumorigenesis? The answer is no. Recent studies have indicated that T. gondii infection can’t cause tumors, but can inhibit the occurrence of tumors (Baird et al., 2013; Fox et al., 2013; Sanders et al., 2015, 2016; Pyo et al., 2016; Fox et al., 2017). Uracil-auxotrophic T. gondii confers long-term effective immunity to diffuse pancreatic cancer. T. gondii enhances the response of host anti-tumor CD8+ T cells and prolongs the survival time of tumor-inoculated mice (Sanders et al., 2015, 2016). Furthermore, uracil-auxotrophic T. gondii can preferentially invade tumor-associated antigen-presenting cells and can stimulate the anti-ovarian tumor response of anti-tumor CD8+ T cells (Baird et al., 2013).
Profilin-like protein (TgPLP) of T. gondii is a host toll-like receptor (TLR) activator that activates TLR11 and dendritic cells (DCs) (Yarovinsky et al., 2005; Hatai et al., 2016). Innate recognition plays an important role in immune response of host (Plattner et al., 2008; Mizel and Bates, 2010; Koblansky et al., 2013). The research indicates that TgPLP can enhance the immune response of autologous whole-tumor-cell vaccine (AWV). Similarly, T. gondii virulence-associated molecule of dense granule protein (ToxoGRA15II) can induce macrophage polarization to M1, which has a limiting effect on tumor growth (Li et al., 2017). In addition, rhoptry protein 5, 17, 18, 35, and 38 (ROP5, ROP17, ROP18, ROP35, and ROP38), dense granule protein 2, 12, and 24 (GRA2, GRA12, and GRA24) can induce the anti-tumor immune response (Fox et al., 2016). Although these molecules play a role in stimulating immunity in anti-tumor, the mechanism remains unclear. Is there any tumor-related factor participate in the response? So it is necessary to analyze the products of tumor-related factors before and after infection with T. gondii in mice.
Materials and Methods
Mice and Parasites
Eight-week-old female BALB/c mice were obtained from Shandong University Laboratory Animal Center. They were bred in groups of twelve per cage under specific-pathogen-free conditions and were free to diet and tap water. They were bred in 12 h of continuous lighting every day at 25°C. T. gondii (low virulent PRU strain) was maintained from our laboratory using passage of cysts in Kunming mice. The treated mice were challenged intragastrically with 20 cysts of T. gondii PRU strain. The mice without cysts-challenge were used as control group. All of the animal experiments were approved by the Ethics Committee on Animal Experiments of the Medical School of Shandong University. BALB/c mice were challenged with cysts and the spleens were collected for mRNA extraction after a month.
Total mRNA Extraction
The total mRNA of cell extracted used TRIZOL for isolation of total RNA according to the previous protocol. Smartspecplus (BioRad) was used to measure the absorption value 260/280 nm (A260/A280) to qualify and quantify the collected RNA. Lastly, the integrity of the extracted RNA was further detected using 1.5% agarose gel electrophoresis. Subsequently, the RNA was transcribed to first strand cDNA by the First Strand cDNA Synthesis Kit (TAKARA) for gene expression analysis.
RNA-Seq Library Construction and Sequencing
A total of 3 μg RNA per sample was used as input material for the RNA sample preparations. Sequencing libraries were generated using NEBNext®UltraTM RNA Library Prep Kit for Illumina® (NEB, United States), which were added to attribute sequences to each sample. Briefly, mRNA was purified from total RNA using poly-T oligo-attached magnetic beads. Fragmentation was carried out using divalent cations under elevated temperature in NEBNext First Strand Synthesis Reaction Buffer (5×. First strand cDNA was synthesized using random hexamer primers and M-MuLV Reverse Transcriptase (RNase H-). Second strand cDNA synthesis was subsequently performed using DNA Polymerase I and RNase H. Remaining overhangs were converted into blunt ends via exonuclease/polymerase activities. After adenylating the 3’ends of DNA fragments, NEBNext Adaptor with hairpin loop structure was ligated to prepare for hybridization. In order to preferentially select cDNA fragments ∼150-200 bp in length, the library fragments were purified with AMPure XP system (Beckman Coulter, Beverly, United States). Subssequently, 3 μl USER Enzyme (NEB, United States) was used with size-selected, adaptor-ligated cDNA at 37°C for 15 min followed by 5 min at 95°C before PCR. Then, PCR was performed with Phusion High-Fidelity DNA polymerase, Universal PCR primers and Index (X) Primer. Finally, PCR products were purified (AMPure XP system) and library quality was assessed with the Agilent Bioanalyzer 2100 system.
According to Sequencing by Synthesis (SBS) technology, the Illumina HiSeq2500 high-throughput sequencing platform was used to sequence the cDNA library and it obtained significant amounts of high-quality reads. The reads and the sequenced-bases were usually considered to be Raw Data and most of them got a Q30 score for base quality.
Annotation and Analysis of the Differentially Expressed Genes
On the basis of method described by Audic and Claverie, we defined the cut-off values to collect the differentially expressed genes. In order to quantify expression, the coverage from every transcript was analyzed with applying BEDtools v. 2.9.0 (Quinlan and Hall, 2010). The coverage of all libraries was then integrated into a single file. The file was predicted using R v. 3.0.1 with the bioconductor package ‘DEGseq’ v. 1.2 (Wang et al., 2010). A proven Benjamini-Hochberg correction method was used to correct the significant p-value in the analysis of differential expression, and then FDR was used as the key indicator of screening of the differentially expressed genes.
GO database is a standard structured biological annotation system, aimed at establishing a system of standard vocabulary and knowledge of genes and their products. The GO annotation system contains three main branches: Biological Process, Molecular Function, and Cellular Component. GO database was used to predict the functions of the differentially expressed genes in the study. Enrichment analysis of the differences between samples was carried out using Top GO. The cluster of orthologs groups of proteins (COG) database is constructed on the basis of phylogeny of bacteria, algae and eukaryotes to classify the orthologs of the gene product. To annotate the pathway of the differentially expressed genes, it is necessary to further analyze the function of the genes. The kyoto encyclopedia of genes and genomes (KEGG) database is a major metabolic pathways database. In the present study, COG and KEGG were used to analyze the differentially expressed genes.
Real-Time PCR Analysis
Total RNA was extracted from independent biological replicates using RNA extraction kit on the basis of the manufacturer’s protocol. 10 μg of total RNA was subjected to DNaseI treatment with 1 U DNaseI (NEB, United States). The reaction was carried out at 37°C for 10 min followed by heat inactivation at 65°C for10 min. 2.5 μg of DNase-treated RNA was used for cDNA synthesis with reverse transcriptase (BioRad, United States) according to the manufacturer’s protocol. The expression of Gapdh gene was detected to be stable in the transcriptome database, and it was used as the control in qRT-PCR. Primers were designed for selected transcripts from the transcriptome database and real time PCR was performed by SYBR green I master mix (Roche, GmBH) on CFX-ConnectTM Real time system (BioRad, United States). Relative expression of the transcripts was calculated by the ΔΔCt method.
Western Blot Analysis
Western blot analysis of spleen cells was performed as below: The cells were treated using RIPA Lyses Buffer (50mM Tris pH 7.4, 150mM NaCl, 1% Triton X-100, 1% Sodium deoxycholate, 0.1% SDS) containing 1mM protease inhibitor PMSF (phenylmethanesulfonyl fluoride) and centrifuged at 13,000 ×g for 10 min. Then supernatant was extracted and resuspended in 50 μl of SDS-PAGE sample buffer, and boiled for 5 min and 20 μl was loaded onto corresponding polyacrylamide gel. Proteins were transferred onto PVDF (polyvinylidene fluoride) membrane via electrophoresis, carried at 80 V for 3 h, using Bio-Rad transfer system (Bio-Rad, Hercules, CA, United States). The membrane was saturated for 2 h with sealing fluid at room temperature and probed with corresponding antibody (Goat) diluted 1:10,000 in saturation buffer. The membrane was incubated for 2 h with a HRP (horseradish peroxides)-labeledrabbit anti-goat IgG antibody (Sigma, United States) diluted 1:20,000 in saturation buffer, and signals were detected with super sensitive signal ECL (Enhanced Chemiluminescence) system.
Statistical Analysis
Data are presented as mean ± SD. We performed statistical analyses with ANOVA or the unpaired two tailed Student’s t-test. All statistical analyses were performed using SPSS 17.0 software, and values of P < 0.05 were considered statistically significant.
Results
mRNA Transcriptome Library Construction, Sequencing, and Sequence Analysis From Infected Mice
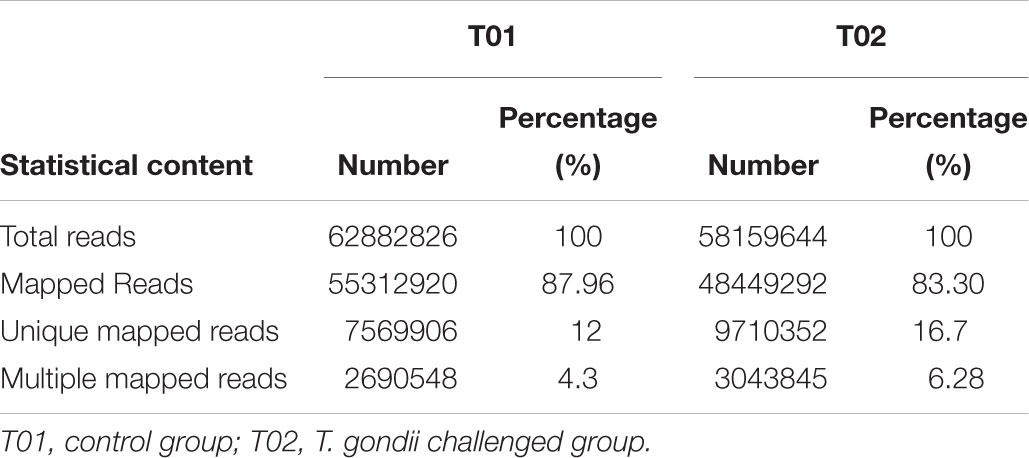
The sequencing results show that a total of 62,882,826 raw reads for the control group and 58,159,644 raw reads for treated group (Table 1), a total of 55,312,920 mapped reads for the control group and 48,449,292 mapped reads for treated group (Table 1) were detected. In control group, the numbers of unique mapped reads and multiple mapped reads were 7,569,906 and 2,690,548 separately, which make up 12 and 4.3% of the raw reads. A total of 9,710,352 unique mapped reads and 3,043,845 multiple mapped reads were selected from treated group in this study.
Differential Expression of Genes
In order to deeply explore the changes of tumor-related factors, RNA-seq was performed. In the result, 432 differences expression genes were identified. Compared with the control group, 307 up regulated genes and 125 down-regulated genes were found (Figure 1). The heatmap of DEGS is shown in Appendix Figure A1. The GO defines classes used to describe gene function, and relationships between these genes. Figure 2 presented the GO analysis of DEGs in the result. The GO terms of DEGs was mainly enriched in the biology regulation, biological process, cellular component, and molecular function. The KEGG PATHWAY is a collection of manually drawn pathway maps representing our knowledge on the molecular interaction and reaction networks. In this study, the KEGG PATHWAY analysis of DEGs is shown in the Figure 3. The pathway terms enriched by DEGs were mainly in the cancer pathways, RAS signaling pathway, RAP1 signaling pathway, calcium signaling pathway, MAPK signaling pathway and Oxytocin pathway, etc. The levels of expression of Gadd45 and Fas changed significantly after T. gondii-challenge. The levels of expression of APC, Smad2, Smad4, Bax, hMLH1, hMSH2, hMSH3, p53, hMSH6 changed significantly after T. gondii-challenge in colorectal cancer pathway. Similarly, the levels of expression of some genes changed significantly after T. gondii-challenge in non-small cell lung cancer and breast cancer pathways (Figure 3). The results indicated that T. gondii-infection may influence cancer signaling pathways.
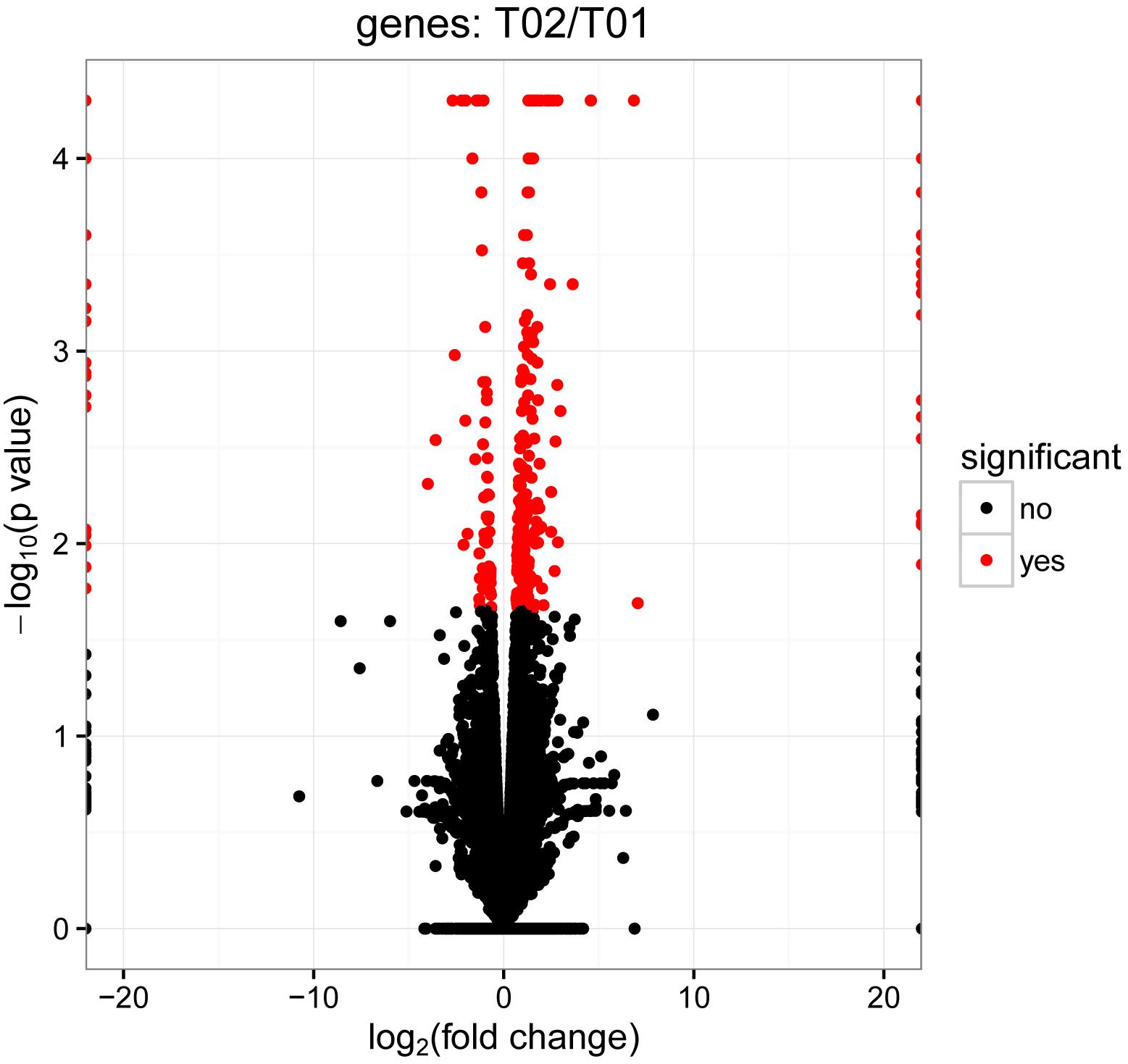
Figure 1. Volcano plots of differentially expressed genes between control group and infected group. In the volcano plots, each dot is a gene, the “red” genes are those that were significantly regulated in infected group compared with control group at a q-value < 0.05. However, the “black” genes shown there were no significantly difference between control group and infected group. In total, 432 unigenes were identified as differentially expressed, including 307 genes that were up-regulated and 125 genes that were down regulated.
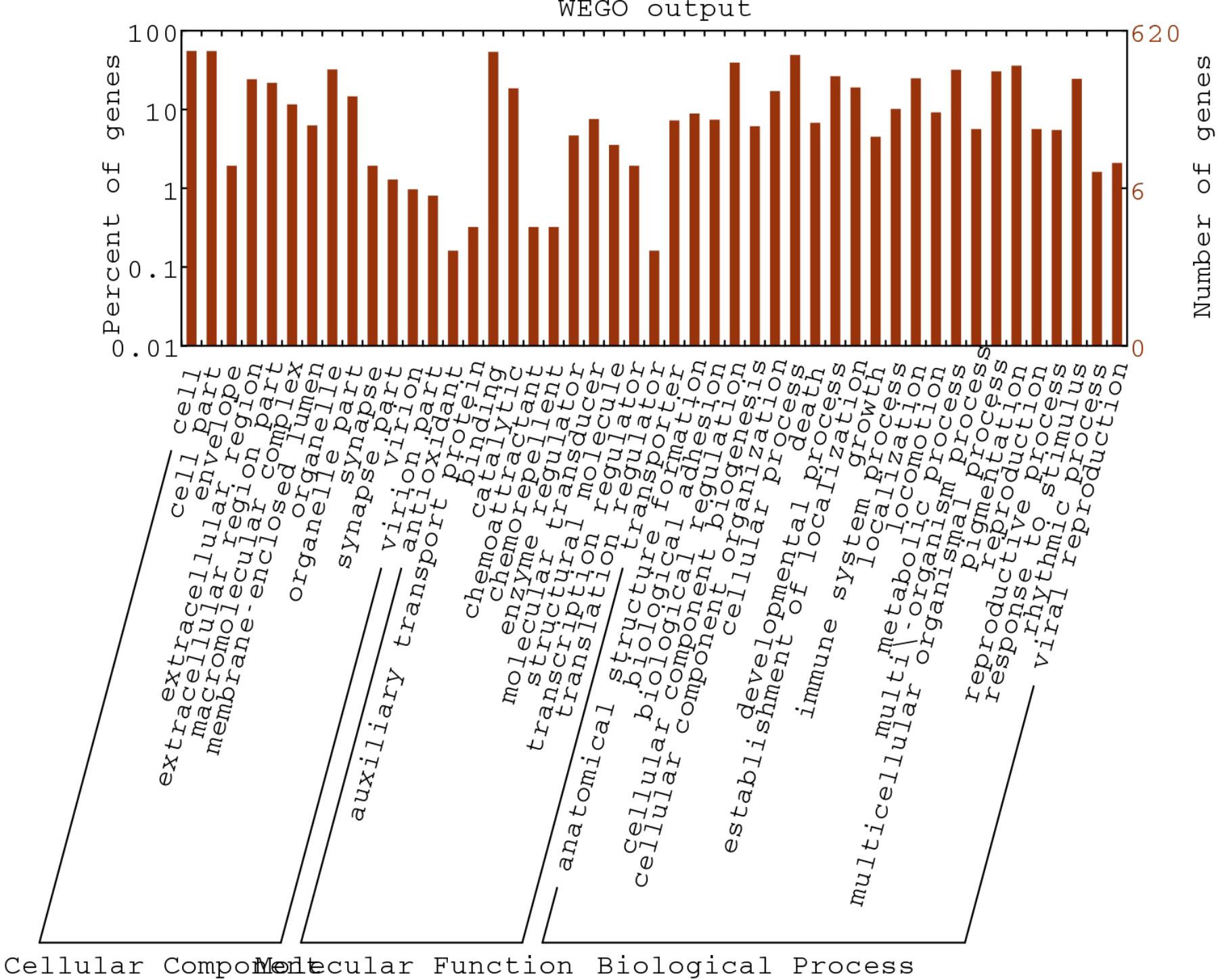
Figure 2. Gene Ontology classification of assembled unigenes. The DEGs genes were classified into 3 functional categories: molecular function, biological process, and cellular component. The y-axis indicates the number of genes in a category. The x-axis indicates the specific category of genes in that main category.
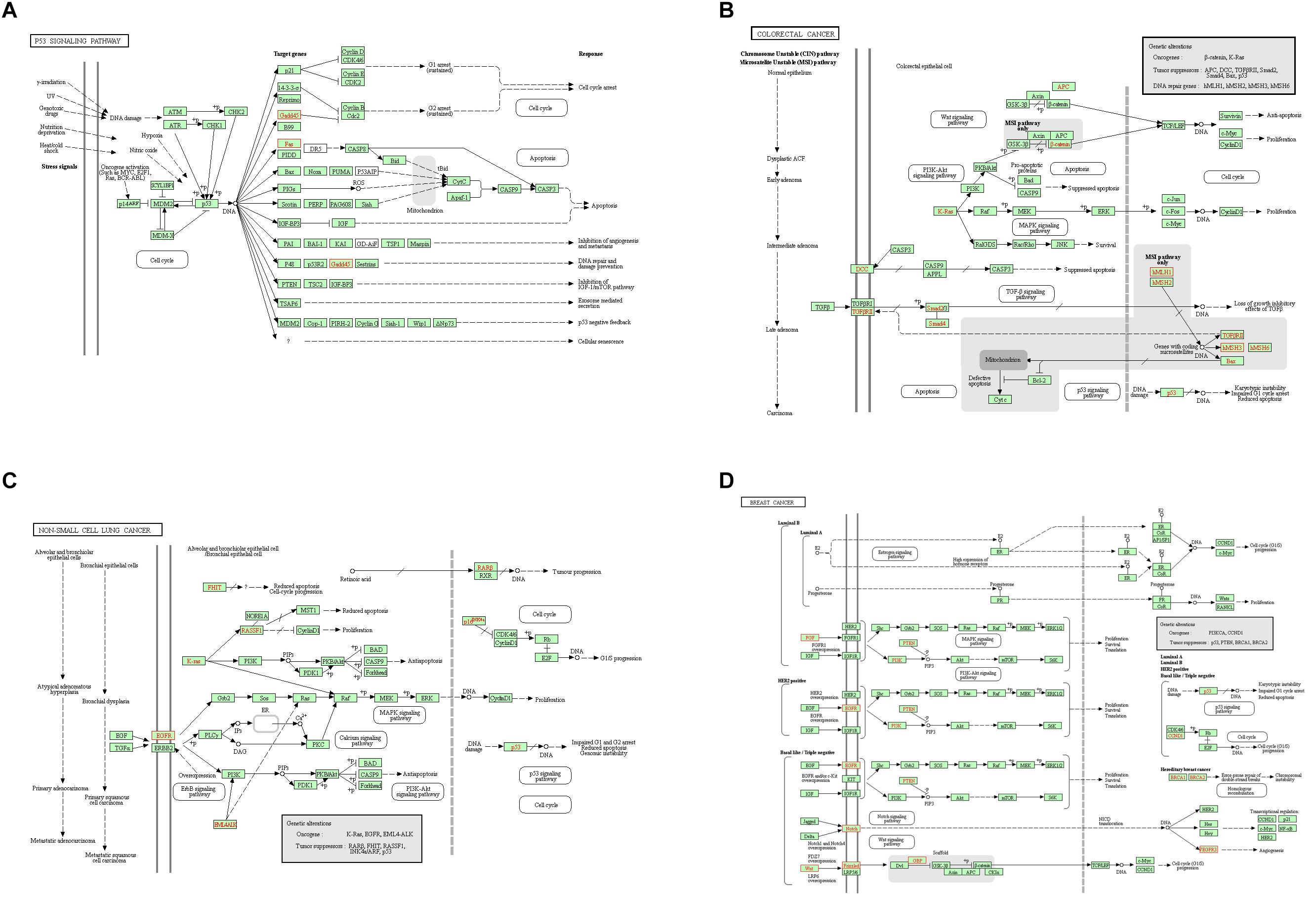
Figure 3. Unigenes predicted to be involved in the p53 signaling pathway (A), colorectal cancer pathway (B), non-small cell lung cancer pathway (C), and breast cancer pathway (D). Red indicates significantly different expression in infected group compared with control group.
Further Confirmation Using qRT-PCR
In order to verify the RNA-seq results, qRT-PCR was performed in this study. The candidate genes were selected from the cancer metabolic signaling pathway. Among these genes, CaM, RTK, PLA genes were involved in the RAS signaling pathway; Gadd45 and Fas genes belong to the P53 signaling pathway; APC, DCC, Smad2, Smad4, Bax, hMLH1, hMSH2, hMSH3 and hMSH6 genes were involved in the COLORECTAL cancer pathway; K-Ras, EGFR, FHIT, RASSF1 and RARβ genes belong to NON-SMALL CELL LUNG cancer signaling pathway; PTEN, BRCA1, BRCA2, PI3K, and CCND1were involved in the BREAST cancer signaling pathway. Figure 4 presented the RT-PCR results. Compared with control group, genes PLA, Gadd45, DCC, Smad2, Smad4,hMSH3, RASSF1 and BRCA2 were up-regulated, while the gene RTK, Fas, hMLH1, hMSH2, EGFR, and CCND1 were down-regulated in the treated samples. The results indicated that expressions of cancer-related genes obtained by qRT-PCR were nearly consistent with those obtained with RNA-seq.
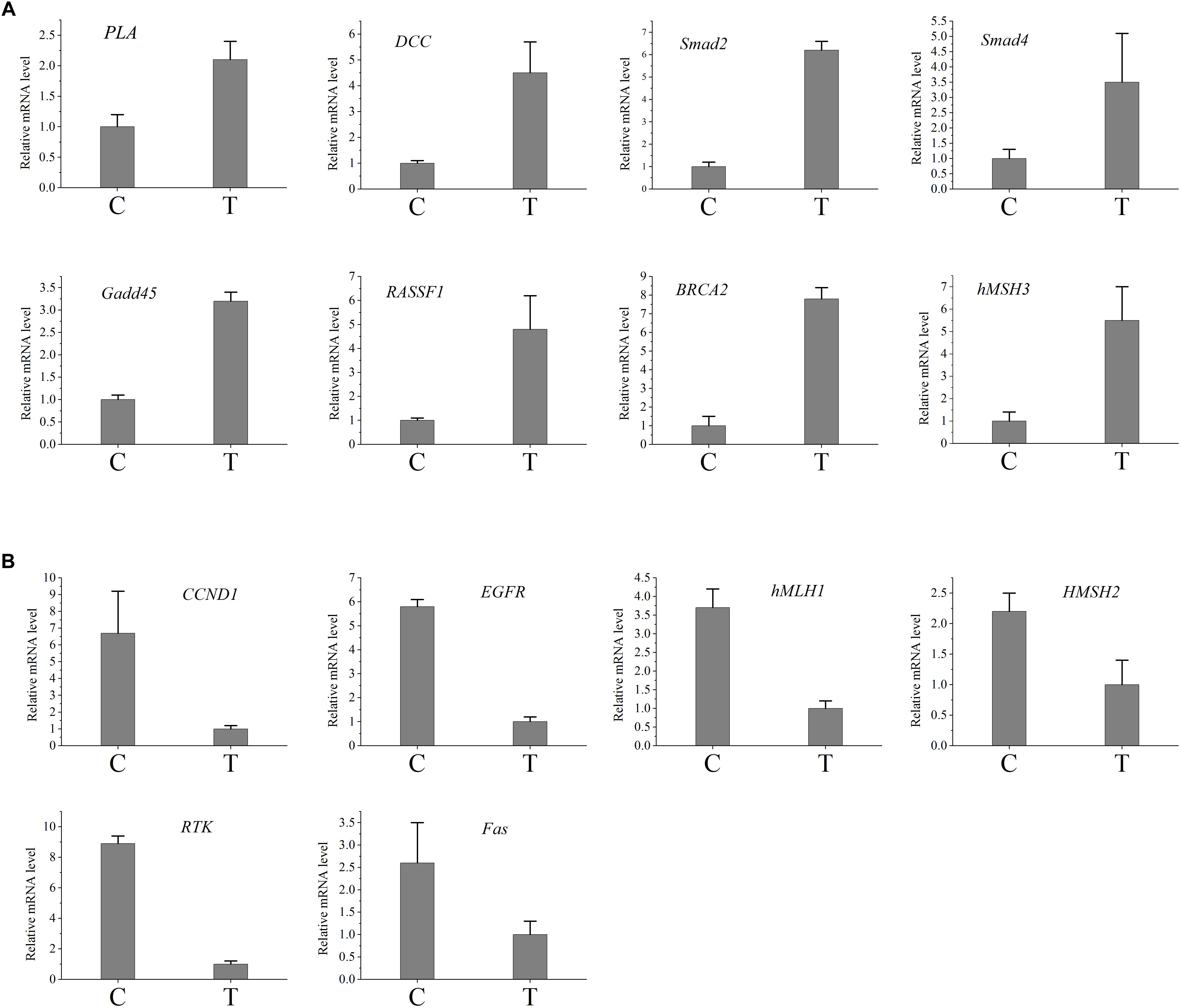
Figure 4. Validation of the difference in expression of the genes using qRT-PCR. (A) The qRT-PCR results of the up-regulated genes in infected group. (B) The qRT-PCR results of the down-regulated genes in infected group. C, control group; T, treated group.
Changes of Tumor-Related Factors
Western blotting analysis of PLA, Gadd45, DCC, Smad2, Smad4, hMSH3, RASSF1, BRCA2,RTK, Fas, hMLH1, hMSH2, EGF,R and CCND1 were performed. As shown in Figure 5, compared to control group, the levels of DCC, Smad2, BRCA2, and RASSF1 proteins from treated group were higher. Moreover, the levels of PLA, Gadd45, Smad4, and hMSH3 proteins of treated group were similar to those of control group. Compared to control group, the levels of RTK, hMLH1, EGFR, and CCND1 proteins from treated group were lower. Furthermore, the levels of Fas and hMSH2 proteins of treated group were similar to those of control.
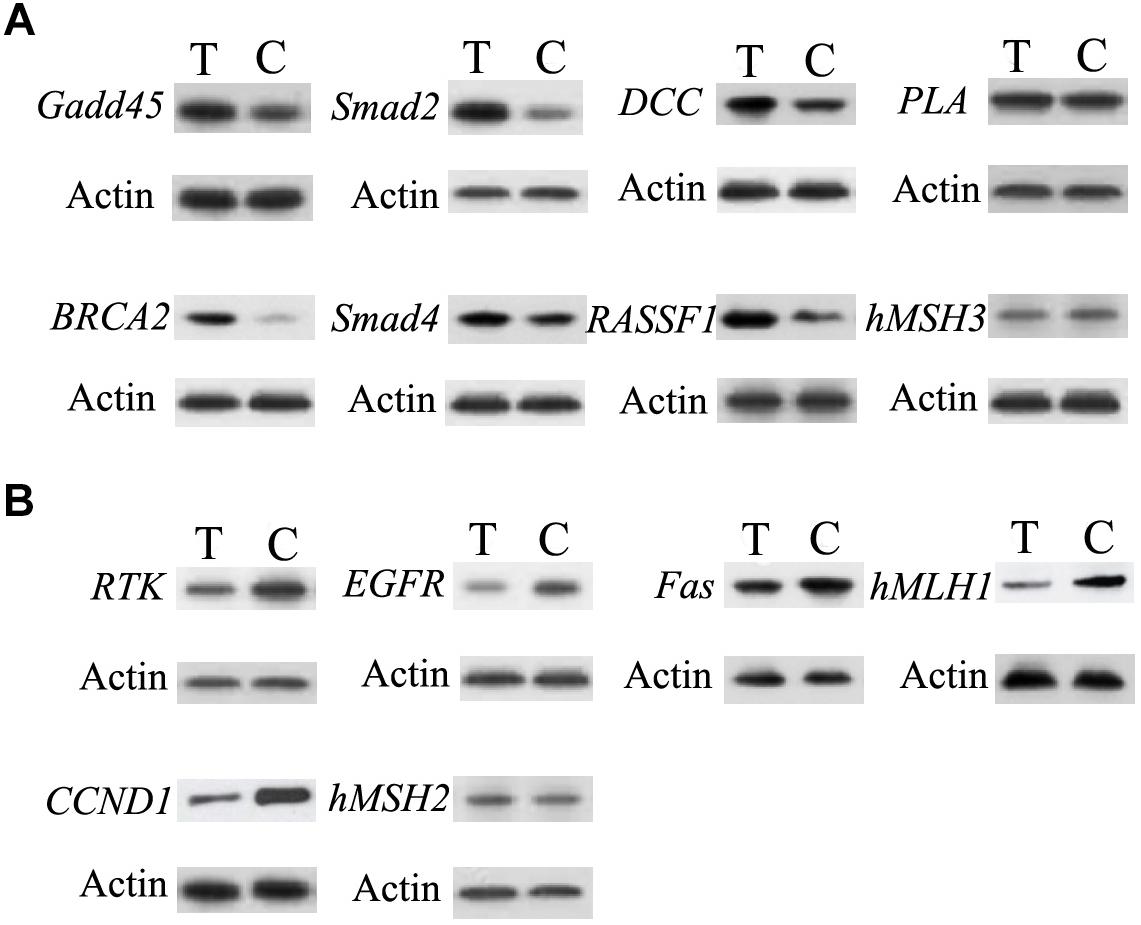
Figure 5. Validation of the difference in expression of the genes by western blot. (A) The western blot results of the up-regulated genes in infected group. (B) The western blot results of the down-regulated genes in infected group. C, control group; T, treated group.
Discussion
Hosts always allow development of latent infections of T. gondii, which is necessary for the parasite to spread to new hosts (Denkers et al., 2012). T. gondii could secrete special proteins to extensively manipulate the host cells (Hunter and Sibley, 2012). Interestingly, some secreted proteins could improve the immunity against tumors in hosts (Baird et al., 2013; Fox et al., 2013, 2017; Sanders et al., 2015, 2016; Pyo et al., 2016). Previous results indicated the secreted proteins were able to resist tumors through improving the immunity of hosts. Importantly, the present result suggested T. gondii-infection changed the expressions of tumor-related factors, which might improve the anti-tumor ability of hosts.
The differential expression from the control and infected group had been detected using the comparative analysis of their transcriptomes. In infected sample, compared with control material, more genes were up-regulated, which suggested that infection could change the host’s genes expression. Moreover, compared to our study, more different expression genes were found in Zhou’s study (Zhou et al., 2016). Obviously, Zhou’s study had been obtained from infected pK-15 cells, while our experimental samples were from mice, which might be the reason causing different expression genes. He et al. (2016) revealed the mRNA change of mouse splenocyte organelle components after T. gondii infection, which only analyzed a little mRNAs from mouse spleen. In this study, we had systematically analyzed the mRNAs of mouse spleen, especially tumor-related factors.
As a classic cancer suppressor, p53 could encode a special protein which inhibits oncogenesis by inducing senescence, apoptosis, growth arrest, and angiogenesis inhibition. Mutations in the p53 gene (TP53) are common associated with an increased susceptibility to form cancer, and inactivation of p53-regulated pathways has been described in over 50% of all human cancers (Bullock and Fersht, 2001). P53 mutation is a driver of lung cancer and inactivation is a common situation in lung cancer (Jamal-Hanjani et al., 2017). Regain activation of p53 is favorable for lung cancer treatment (Wang and Sun, 2010). P53 could accumulate breast cancer cells, which plays an important role in breast cancer growth (Ferraz da Costa et al., 2018). So p53 signaling pathway plays an important role in human cancers. In the present study, p53 signaling pathway had been changed after T. gondii infection. After T. gondii challenge, the expression of Gadd45 protein was up-regulated, while the expression of Fas protein was down-regulated, which affected p53 signaling pathway. Moreover, Fas protein plays an important role in suppressing colon cancer immune evasion (Xiao et al., 2018). Human surfactant protein D induces apoptosis in pancreatic cancer cell lines via Fas-mediated pathway (Kaur et al., 2018). Gadd45 protein could suppress the development of leukemia (Wiest, 2018), while up-regulation of GADD45 induced cell cycle arrest and apoptosis in colorectal cancer cells (Huang et al., 2017). Therefore, Fas down-regulation and Gadd45 up-regulation in the treated mice may change the resistance for cancer.
In our study, the expressions of DCC, Smad2, Smad4, hMLH1, hMSH2, and hMSH3 proteins had been changed, which regulated the colorectal cancer pathway. As one of genetic marker for diagnosis of colorectal cancer, DCC could inhibit the growth of colorectal cancer (Hao et al., 2017). The expression of DCC protein was up-regulated after T. gondii infection, which suggested that T. gondii infection might improve the resistance of host against colorectal cancer through increasing the expression of DCC protein. Similarly, Smad2 and Smad4 proteins had ability to inhibit the growth of colorectal cancer (Cai et al., 2018; Hirsch et al., 2018). Our result showed that the expression of Smad2 (not Smad4) protein was up-regulated after T. gondii infection, which indicated the enhanced ability against colorectal cancer. In addition, hMLH1, hMSH2, and hMSH3 proteins play important roles in colorectal cancer pathway. Their methylation affect survival in patients with colorectal cancer (Kim et al., 2018). The expression of hMLH1 protein was down-regulated, while the expressions of hMSH2 and hMSH3 proteins had no change after infection, which further suggested the relevance between T. gondii infection and resistance to colorectal cancer.
The expressions of proteins from NON-SMALL CELL LUNG cancer signaling pathway had been analyzed in this study. RASSF1, one of NON-SMALL CELL LUNG tumor suppressors, play an important role in resisting the growth of NON-SMALL CELL LUNG cancer (Dubois et al., 2016; Wang H. et al., 2018). The expression of RASSF1 protein was up-regulated after challenge in the present study, which indicated that T. gondii infection might improve the resistance for NON-SMALL CELL LUNG cancer. Conversely, EGFR could improve the growth of NON-SMALL CELL LUNG tumor (Saboundji et al., 2018; Wang Y. et al., 2018). Compared to control group, the expression of EGFR protein of challenged group was down-regulated, which suggested that T. gondii infection might inhibit the growth of NON-SMALL CELL LUNG tumor by reducing the expression of EGFR protein.
PTEN, BRCA1, BRCA2, PI3K, and CCND1 genes were related to the BREAST cancer signaling pathway. PTEN, BRCA1, and BRCA2 proteins play an important role in resisting the growth of BREAST cancer (Barnes-Kedar et al., 2018; El Botty et al., 2018; Sirisena et al., 2018), while PI3K and CCND1 proteins could improve the growth of BREAST cancer (Ortiz et al., 2017; Sirisena et al., 2018). In this study, the expression of BRCA2 was up-regulated, while CCND1 protein was down-regulated, which indicated that the infection might inhibit the growth of BREAST cancer.
BRCA2, Smad4, EGFR genes play important role in pancreatic cancer pathway. As shown in Appendix Figure A1, the expressions of tumor suppressors (p16, p53, BRCA2, and Smad4) had been changed after T. gondii infection. Importantly, the expressions of BRCA2 and Smad4 genes were up-regulated after challenge, which suggested the infection might also inhibit the developing of pancreatic cancer of host. Sanders et al. revealed that attenuated T. gondii could treat disseminated pancreatic cancer through generating long-lasting immunity (Sanders et al., 2016). Given the results of the present study, T. gondii could not only generate immunity but also change the tumor-related factors of host. Furthermore, lots of tumor-related factors of glioma pathway, prostate cancer pathway, basal cell carcinoma pathway, melanoma pathway, bladder cancer pathway, and endometrial cancer pathway had been changed after challenge (Appendix Figure A2), suggesting the infection of T. gondii might widely regulate the expressions of tumor-related proteins in host.
Infection of T. gondii could cause many changes of physiological phenomenon in host. Previous researches verified the enhanced immunity against tumor in infected hosts. In the present study, lots of tumor-related proteins were detected and many of them had been changed after challenge. Importantly, the changes were possibly beneficial to the suppression of tumor.
Author Contributions
LW conceived and designed the study, and critically revised the manuscript. GL carried out the experiments and drafted the manuscript. JZ, YZ, QL, and YG contributed to the revision of the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by the National Foundation of Natural Science of China (Grant No. 81602455), the Science Foundation of Shandong Province (Grant No. ZR2013HM033), and the National College Students’ Innovation and Entrepreneurship Training Program (Grant No. 201710422116). This work was also supported by the Central South University Sports Medicine Scholarship.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.00181/full#supplementary-material
FIGURE A1 | The heat map of the differently expressed genes. T01, control group; T02, T. gondii-infected group.
FIGURE A2 | Unigenes predicted to be involved in endometrial cancer pathway (A), glioma pathway (B), prostate cancer pathway (C), basal cell carcinoma pathway (D), melanoma pathway (E), and bladder cancer pathway (F). Red indicates significantly different expression in infected group compared with control group.
References
Al-Mendalawi, M. D. (2017). Serologic evidence of Toxoplasma gondii infection among cancer patients: a prospective study from Qassim region, Saudi Arabia. Saudi Med. J. 38, 319–321. doi: 10.15537/smj.2017.6.19868
Baird, J. R., Fox, B. A., Sanders, K. L., Lizotte, P. H., Cubillos-Ruiz, J. R., Scarlett, U. K., et al. (2013). Avirulent toxoplasma gondii generates therapeutic antitumor immunity by reversing immunosuppression in the ovarian cancer microenvironment. Cancer Res. 73, 3842–3851. doi: 10.1158/0008-5472.CAN-12-1974
Barnes-Kedar, I., Bernstein-Molho, R., Ginzach, N., Hartmajer, S., Shapira, T., Magal, N., et al. (2018). The yield of full BRCA1/2 genotyping in Israeli high-risk breast/ovarian cancer patients who do not carry the predominant mutations. Breast Cancer Res. Treat. 172, 151–157. doi: 10.1007/s10549-018-4887-7
Bullock, A. N., and Fersht, A. R. (2001). Rescuing the function of mutant p53. Nat. Rev. Cancer 1, 68–76. doi: 10.1038/35094077
Cai, J., Xia, L., Li, J., Ni, S., Song, H., and Wu, X. (2018). Tumor-associated macrophages derived TGF-β–induced epithelial to mesenchymal transition in colorectal cancer cells through Smad2,3-4/Snail signaling pathway. Cancer Res. Treat. 51, 252–266. doi: 10.4143/crt.2017.613
Denkers, E. Y., Bzik, D. J., Fox, B. A., and Butcher, B. A. (2012). An inside job: hacking into Janus kinase/signaltransducer and activator of transcription signaling cascades by the intracellular protozoan Toxoplasmagondii. Infect. Immun. 80, 476–482. doi: 10.1128/IAI.05974-11
Dubois, F., Keller, M., Calvayrac, O., Soncin, F., Hoa, L., Hergovich, A., et al. (2016). RASSF1A suppresses the invasion and metastatic potential of human non-small cell lung cancer cells by inhibiting YAP activation through the GEF-H1/RhoB pathway. Cancer Res. 76, 1627–1640. doi: 10.1158/0008-5472.CAN-15-1008
El Botty, R., Coussy, F., Hatem, R., Assayag, F., Chateau-Joubert, S., Servely, J. L., et al. (2018). Inhibition of mTOR downregulates expression of DNA repair proteins and is highly efficient against BRCA2-mutated breast cancer in combination to PARP inhibition. Oncotarget 9, 29587–29600. doi: 10.18632/oncotarget.25640
Ferraz da Costa, D. C., Campos, N. P. C., Santos, R. A., Zanphorlin, L., Ramos, C., Rangel, L. P., et al. (2018). Resveratrol prevents p53 aggregation in vitro and in breast cancer cells. Oncotarget 9, 29112–29122. doi: 10.18632/oncotarget.25631
Fox, B. A., Butler, K. L., Guevara, R. B., and Bzik, D. J. (2017). Cancer therapy in a microbial bottle: uncorking the novel biology of the protozoan Toxoplasma gondii. PLoS Pathog. 13:e1006523. doi: 10.1371/journal.ppat.1006523
Fox, B. A., Sanders, K. L., Chen, S., and Bzik, D. J. (2013). Targeting tumors with nonreplicating Toxoplasma gondii uracil auxotroph vaccines. Trends Parasitol. 29, 431–437. doi: 10.1016/j.pt.2013.07.001
Fox, B. A., Sanders, K. L., Rommereim, L. M., Guevara, R. B., and Bzik, D. J. (2016). Secretion of rhoptry and dense granule effector proteins by nonreplicating Toxoplasma gondii uracil auxotrophs controls the development of antitumor immunity. PLoS Genet. 12:e1006189. doi: 10.1371/journal.pgen.1006189
Hao, Y., Zhang, J., Shan, G., Zhang, N., Jin, W., and Nan, K. (2017). Establishment of optimal regulatory network of colorectal cancer based on p42.3 protein. Saudi J. Biol. Sci. 24, 1781–1786. doi: 10.1016/j.sjbs.2017.11.012
Hatai, H., Lepelley, A., Zeng, W., Hayden, M. S., and Ghosh, S. (2016). Toll-Like Receptor 11 (TLR11) Interacts with Flagellin and Profilin through disparate mechanisms. PLoS One 11:e0148987. doi: 10.1371/journal.pone.0148987
He, J. J., Ma, J., Li, F. C., Song, H. Q., Xu, M. J., and Zhu, X. Q. (2016). Transcriptional changes of mouse splenocyte organelle components following acute infection with Toxoplasma gondii. Exp. Parasitol. 167, 7–16. doi: 10.1016/j.exppara.2016.04.019
Hirsch, D., Wangsa, D., Zhu, Y. J., Hu, Y., Edelman, D. C., Meltzer, P. S., et al. (2018). Dynamics of genome alterations in Crohn’s disease associated colorectal carcinogenesis. Clin. Cancer Res. 24, 4997–5011. doi: 10.1158/1078-0432.CCR-18-0630
Huang, W. S., Kuo, Y. H., Kuo, H. C., Hsieh, M. C., Huang, C. Y., Lee, K. C., et al. (2017). CIL-102-induced cell cycle arrest and apoptosis in colorectal cancer cells via upregulation of p21 and GADD45. PLoS One 12:e0168989. doi: 10.1371/journal.pone.0168989
Hunter, C. A., and Sibley, L. D. (2012). Modulation of innate immunity by Toxoplasma gondii virulence effectors. Nat. Rev. Microbiol. 10, 766–778. doi: 10.1038/nrmicro2858
Imam, A., Al-Anzi, F. G., Al-Ghasham, M. A., Al-Suraikh, M. A., Al-Yahya, A. O., and Rasheed, Z. (2017). Serologic evidence of Toxoplasma gondii infection among cancer patients. A prospective study from Qassim region, Saudi Arabia. Saudi Med. J. 38, 319–321. doi: 10.15537/smj.2017.3.18535
Jamal-Hanjani, M., Wilson, G. A., McGranahan, N., Birkbak, N. J., Watkins, T. B. K., Veeriah, S., et al. (2017). Tracking the evolution of non-small-cell lung cancer. N. Engl. J. Med. 376, 2109–2121. doi: 10.1056/NEJMoa1616288
Jiang, C., Li, Z., Chen, P., and Chen, L. (2015). The seroprevalence of Toxoplasma gondii in Chinese population with cancer: a systematic review and meta-analysis. Medicine 94:e2274. doi: 10.1097/MD.0000000000002274
Kaur, A., Riaz, M. S., Murugaiah, V., Varghese, P. M., Singh, S. K., and Kishore, U. (2018). A recombinant fragment of human surfactant protein D induces apoptosis in pancreatic cancer cell lines via fas-mediated pathway. Front. Immunol. 9:1126. doi: 10.3389/fimmu.2018.01126
Kim, S. H., Park, K. H., Shin, S. J., Lee, K. Y., Kim, T. I., Kim, N. K., et al. (2018). CpG island methylator phenotype and methylation of Wnt pathway genes together predict survival in patients with colorectal cancer. Yonsei Med. J. 59, 588–594. doi: 10.3349/ymj.2018.59.5.588
Koblansky, A. A., Jankovic, D., Oh, H., Hieny, S., Sungnak, W., Mathur, R., et al. (2013). Recognition of profilin by Toll-like receptor 12 is critical for host resistance to Toxoplasma gondii. Immunity 38, 119–130. doi: 10.1016/j.immuni.2012.09.016
Li, Y., Poppoe, F., Chen, J., Yu, L., Deng, F., Luo, Q., et al. (2017). Macrophages polarized by expression of toxoGRA15II inhibit growth of hepatic carcinoma. Front. Immunol 8:137. doi: 10.3389/fimmu.2017.00137
Martins-Duarte, E. S., Lemgruber, L., de, Souza W, and Vommaro, R. C. (2010). Toxoplasma gondii: fluconazole and itraconazole activity against toxoplasmosis in a murine model. Exp. Parasitol. 124, 466–469. doi: 10.1016/j.exppara.2009.12.011
Mizel, S. B., and Bates, J. T. (2010). Flagellin as an adjuvant: cellular mechanisms and potential. J. Immunol. 185, 5677–5682. doi: 10.4049/jimmunol.1002156
Ortiz, A. B., Garcia, D., Vicente, Y., Palka, M., Bellas, C., and Martin, P. (2017). Prognostic significance of cyclin D1 protein expression and gene amplification in invasive breast carcinoma. PLoS One 12:e0188068. doi: 10.1371/journal.pone.0188068
Plattner, F., Yarovinsky, F., Romero, S., Didry, D., Carlier, M. F., Sher, A., et al. (2008). Toxoplasma profilin is essential for host cell invasion and TLR11-dependent induction of an interleukin-12 response. Cell Host Microbe 3, 77–87. doi: 10.1016/j.chom.2008.01.001
Pyo, K.-H., Lee, Y.-W., Lim, S. M., and Shin, E.-H. (2016). Immune adjuvant effect of a Toxoplasma gondii profilin-like protein in autologous whole-tumor-cell vaccination in mice. Oncotarget 7, 74107–74119. doi: 10.18632/oncotarget.12316
Quinlan, A. R., and Hall, I. M. (2010). BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842. doi: 10.1093/bioinformatics/btq033
Saboundji, K., Auliac, J. B., Pérol, M., François, G., Janicot, H., Marcq, M., et al. (2018). Efficacy of osimertinib in EGFR-mutated non-small cell lung cancer with leptomeningeal metastases pretreated with EGFR-tyrosine kinase inhibitors. Target. Oncol. 13, 501–507. doi: 10.1007/s11523-018-0581-2
Sanders, K. L., Fox, B. A., and Bzik, D. J. (2015). Attenuated Toxoplasma gondii stimulates immunity to pancreatic cancer by manipulation of myeloid cell populations. Cancer Immunol. Res. 3, 891–901. doi: 10.1158/2326-6066.CIR-14-0235
Sanders, K. L., Fox, B. A., and Bzik, D. J. (2016). Attenuated Toxoplasma gondii therapy of disseminated pancreatic cancer generates long-lasting immunity to pancreatic cancer. Oncoimmunology 5:e1104447. doi: 10.1080/2162402X.2015.1104447
Sirisena, N. D., Adeyemo, A., Kuruppu, A. I., Samaranayake, N., and Dissanayake, V. H. W. (2018). Genetic variants associated with clinicopathological profiles in sporadic breast cancer in Sri Lankan Women. J. Breast Cancer 21, 165–172. doi: 10.4048/jbc.2018.21.2.165
Vittecoq, M., and Thomas, F. (2017). Toxoplasmose et cancer : connaissances actuelles et perspectives de recherché. Pathol. Exot. 110, 76–79. doi: 10.1007/s13149-016-0518-x
Wang, H., Cao, D., and Wu, F. (2018). Long noncoding RNA UPAT promoted cell proliferation via increasing UHRF1 expression in non-small cell lung cancer. Oncol. Lett. 16, 1491–1498. doi: 10.3892/ol.2018.8829
Wang, L. K., Feng, Z. X., Wang, X., Wang, X. W., and Zhang, X. G. (2010). DEGseq: an R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics 26, 136–138. doi: 10.1093/bioinformatics/btp612
Wang, Y., Lai, H., Fan, X., Luo, L., Duan, F., Jiang, Z., et al. (2018). Gossypol inhibits non-small cell lung cancer cells proliferation by targeting EGFRL858R/T790M. Front. Pharmacol. 9:728. doi: 10.3389/fphar.2018.00728
Wang, Z., and Sun, Y. (2010). Targeting p53 for novel anticancer therapy. Transl. Oncol. 3, 1–12. doi: 10.1593/tlo.09250
Wiest, D. L. (2018). Gadd45 stress sensors in suppression of leukemia. Oncotarget 9, 34191–34192. doi: 10.18632/oncotarget.26154
Xiao, W., Ibrahim, M. L., Redd, P. S., Klement, J. D., Lu, C., Yang, D., et al. (2018). Loss of fas expression and function is coupled with colon cancer resistance to immune checkpoint inhibitor immunotherapy. Mol. Cancer Res. [Epub ahead of print] doi: 10.1158/1541-7786.MCR-18-0455
Yarovinsky, F., Zhang, D., Andersen, J. F., Bannenberg, G. L., Serhan, C. N., Hayden, M. S., et al. (2005). TLR11 activation of dendritic cells by a protozoan profilin-like protein. Science 308, 1626–1629. doi: 10.1126/science.1109893
Keywords: Toxoplasma gondii, RNA-Seq, tumor-related factors, changes, suppression
Citation: Lu G, Zhou J, Zhao Yh, Li Ql, Gao Yy and Wang L (2019) Transcriptome Sequencing Investigated the Tumor-Related Factors Changes After T. gondii Infection. Front. Microbiol. 10:181. doi: 10.3389/fmicb.2019.00181
Received: 11 August 2018; Accepted: 23 January 2019;
Published: 07 February 2019.
Edited by:
Rustam Aminov, University of Aberdeen, United KingdomReviewed by:
Juan Diego Maya, Universidad de Chile, ChileGeorge Grant, University of Aberdeen, United Kingdom
Copyright © 2019 Lu, Zhou, Zhao, Li, Gao and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lin Wang, wanglinfenghuaxue@163.com; linwang1988@outlook.com
 Gang Lu1
Gang Lu1 Jian Zhou
Jian Zhou Lin Wang
Lin Wang