- 1Departamento de Genética, Facultad de Biología, Universidad de Sevilla, Sevilla, Spain
- 2Julius Kühn-Institut – Bundesforschungsinstitut für Kulturpflanzen, Federal Research Centre for Cultivated Plants, Institute for Epidemiology and Pathogen Diagnostics, Brunswick, Germany
SrfJ is an effector of the Salmonella pathogenicity island 2-encoded type III secretion system. Salmonella enterica serovar Typhimurium expresses srfJ under two disparate sets of conditions: media with low Mg2+ and low pH, imitating intravacuolar conditions, and media with myo-inositol (MI), a carbohydrate that can be used by Salmonella as sole carbon source. We investigated the molecular basis for this dual regulation. Here, we provide evidence for the existence of two distinct promoters that control the expression of srfJ. A proximal promoter, PsrfJ, responds to intravacuolar signals and is positively regulated by SsrB and PhoP and negatively regulated by RcsB. A second distant promoter, PiolE, is negatively regulated by the MI island repressor IolR. We also explored the in vivo activity of these promoters in different hosts. Interestingly, our results indicate that the proximal promoter is specifically active inside mammalian cells whereas the distant one is expressed upon Salmonella colonization of plants. Importantly, we also found that inappropriate expression of srfJ leads to reduced proliferation inside macrophages whereas lack of srfJ expression increases survival and decreases activation of defense responses in plants. These observations suggest that SrfJ is a relevant factor in the interplay between Salmonella and hosts of different kingdoms.
Introduction
Salmonella enterica serovar Typhimurium (S. Typhimurium) is a facultative intracellular bacterial pathogen that can survive and proliferate in diverse hosts (Wiedemann et al., 2014) as well as non-host environments (Winfield and Groisman, 2003). This non-typhoidal Salmonella serovar can infect a broad range of animal species. In humans, it causes gastrointestinal infections with occasional secondary bacteremia (Chen et al., 2013). In mice, in contrast, it produces systemic infections that are similar to typhoid fever induced by S. enterica serovar Typhi in humans (Garai et al., 2012). It also causes acute enteritis and exudative diarrhea in calves, which are considered as a relevant model for non-typhoidal salmonellosis in humans (Costa et al., 2012). In addition, chickens and pigs can be asymptomatic carriers of these bacteria (Foley et al., 2007). As a consequence, these animals are important sources of S. Typhimurium in the food chain. S. Typhimurium and other serovars can also enter the agricultural production chain. This can happen on different levels, e.g., via animal feces used for soil amendments or as post-harvest contamination. Salmonella is able to adhere to plant surface, colonize plant organs, and suppress the plant immune system (Schikora et al., 2012; Neumann et al., 2014). Therefore, plants are considered as alternative hosts for these pathogens, and fresh fruits and vegetables are recognized as an important source of food-borne disease (Wiedemann et al., 2014; Holden et al., 2015).
Many virulence factors are involved during the interactions of Salmonella with its host. Prominent among them are two type III secretion systems (T3SS), heteromultimeric nanomachines specialized in the delivery of effector proteins into host cells (Deng et al., 2017). More than 30 effectors are translocated by Salmonella into the host cell through both T3SSs (Ramos-Morales, 2012).
Effectors secreted by the Salmonella Pathogenicity Island 1 (SPI1)-encoded system (T3SS1) (Galán and Curtiss, 1989) promote invasion of host cells by a trigger mechanism that involves remodeling the actin cytoskeleton to form membrane ruffles that internalize the bacteria in a vacuole known as Salmonella-Containing Vacuole (SCV). This system is also involved in modulation of epithelial tight junctions (Boyle et al., 2006; Liao et al., 2008; Zhang et al., 2015; Lin et al., 2016), induction of polymorphonuclear leukocytes transepithelial migration (Zhang et al., 2006; Wall et al., 2007), control of the initial stages of the SCV biogenesis (Bakowski et al., 2008; Steele-Mortimer, 2008), inhibition of host cell exocytosis (Perrett and Zhou, 2013), and induction of rapid pyroptosis in macrophages (Fink and Cookson, 2007). The SPI2-encoded system (T3SS2) is expressed several hours after internalization and is important for SCV biogenesis, intracellular survival and proliferation, generation of Salmonella-induced tubules, and systemic infection (Hensel, 2000; Knodler and Steele-Mortimer, 2003; Liss and Hensel, 2015).
SrfJ is a poorly characterized T3SS2 effector (Cordero-Alba et al., 2012). Interestingly, virulence of a S. Typhimurium strain with mutation in srfJ was mildly attenuated in mice (Ruiz-Albert et al., 2002). The SrfJ protein shares 30% amino acid sequence identity with human glucosylceramidase over 447 residues (Kim et al., 2009). Initially, it was proposed as a putative effector because the gene srfJ is positively regulated by SsrB (Worley et al., 2000), the main positive regulator of T3SS2 (Fass and Groisman, 2009). Surprisingly, we found that, in addition to the regulation by SsrB, srfJ is also negatively regulated by IolR (Cordero-Alba et al., 2012), the repressor of the myo-inositol (MI) utilization genes in S. Typhimurium (Kröger and Fuchs, 2009). Salmonella and other bacteria can use MI as the sole carbon source, and this substrate is ubiquitous in soil and plants.
Here, we analyzed the molecular basis and studied the ecological relevance of the dual regulation of srfJ. We found that two distinct promoters control the expression of srfJ. The proximal (PsrfJ) responds to intravacuolar signals in animal cells, whereas the second and distant PiolE is active during plant colonization.
Materials and Methods
Bacterial Strains, Bacteriophages, and Strain Construction
Escherichia coli and S. Typhimurium strains used in this study are described in Table 1. Salmonella strains derived from the mouse-virulent strain ATCC 14028. Transductional crosses using phage P22HT 105/1 int201 (Schmieger, 1972) were used for strain construction (Maloy, 1990). To obtain phage-free isolates, transductants were purified by streaking on green plates. Green plates were prepared as described previously (Chan et al., 1972), except that methyl blue (Sigma) substituted aniline blue. Phage sensitivity was tested by cross-streaking with the clear-plaque mutant P22 H5 (Chan et al., 1972).
Bacterial Culture
The standard culture medium for S. enterica and E. coli was Luria-Bertani (LB) broth. Solid LB contained 1.5% agar (final concentration). Antibiotics were used at the following concentrations: kanamycin (Km), 50 μg/ml; chloramphenicol (Cm), 20 μg/ml; ampicillin (Ap), 100 μg/ml; and tetracycline (Tc), 20 μg/ml. For some experiments, 55.5 mM MI was added for 4 h. For SPI1-inducing conditions, Salmonella strains were grown overnight at 37°C in LB 0.3 M NaCl medium in static conditions. For SPI2-inducing conditions, cells from cultures in LB were washed and diluted 1:125 with minimal medium at pH 5.8 (LPM) containing 80 mM 2-(N-morpholino)-ethanesulfonic acid (pH 5.8), 5 mM KCl, 7.5 mM (NH4)2SO4, 0.5 mM K2SO4, 0.1% Casamino acids, 38 mM glycerol, 337.5 μM K2HPO4–KH2PO4 (pH 7.4), and 8 μM MgCl2, and then incubated overnight at 37°C with shaking. Two different media with plants extracts were made: Lettuce Medium (LM) contained 25% lettuce plant extract sterilized with 0.22 μm filter and 20% M9-Minimal Salts (Sigma) (Fornefeld et al., 2017); Tomato Medium (TM) contained 25% tomato plant extract sterilized with 0.22 μm filter and 20% M9-Minimal Salts. Salmonella cells from saturated cultures in LB were washed with MgCl2 10 mM and diluted in each plant media at OD600 0.1, and then incubated at 37°C with shaking. All the experiments involving S. Typhimurium were carried out using the standard biosecurity procedures that include containment level 2 practices, and safety equipment and facilities.
DNA Amplification with the PCR
Amplification reactions were carried out in a T100 Thermal Cycler (BioRad). For plasmid constructs, the final volume of reactions was 50 μl, and the final concentration of MgCl2 was 1.5 mM. Reagents were used at the following concentrations: deoxynucleoside triphosphates (dNTPs), 300 μM; primers, 0.3 μM; and Taq polymerase (KAPA HiFi DNA Polymerase; Kapa Biosystems), 1 U per reaction. The PCR program included the following steps: (i) initial denaturation for 5 min at 95°C; (ii) 25 cycles of denaturation (98°C, 20 s), annealing (57°C, 15 s), and extension (72°C, 30 s); and (iii) final incubation at 72°C for 5 min to complete the extension. For colony PCR, the final volume of reactions was 20 μl. Reagents were used at the following concentrations: 1× MyTaq Red Reaction Buffer; primers, 0.3 μM; and Taq polymerase (MyTaq Red DNA Polymerase; Bioline), 1 U per reaction. The program included the following steps: (i) initial denaturation for 3 min at 95°C; (ii) 25 cycles of denaturation (95°C, 15 s), annealing (57°C, 15 s), and extension (72°C, 30 s–2 min); and (iii) final incubation at 72°C for 5 min to complete the extension. Primers are listed in Table 2. PCR constructs were sequenced with an automated DNA sequencer (Stab Vida, Oeiras, Portugal) to confirm that the sequence was correct.
Plasmids
Plasmids used in this study are listed in Table 1. Plasmid pSB377 (Winson et al., 1998) was used to make transcriptional fusions of putative promoter regions with the luxCDABE operon from Photorhabdus luminescens. This operon encodes a bacterial luciferase whose product, the light, can be measured without disturbing the cell or adding any substrate. To make these constructions, DNA from strain 14028 was used as a template for PCR amplification with the primers listed in Table 2. The amplified fragments were digested with EcoRI and ligated with EcoRI digested and dephosphorylated pSB377. The ligation mixture was transformed into E. coli DH5α and transformants were selected in LB agar supplemented with Ap. Transformants with plasmids containing the correct transcriptional lux fusions were isolated and verified by PCR and sequencing (Stab Vida, Oeiras, Portugal).
Generation of a PiolE Mutant
Disruption and replacement of PiolE with a Cm resistance gene were performed as described previously (Datsenko and Wanner, 2000). Briefly, the Cm resistance gene from plasmid pKD3 was PCR amplified with primers PiolEH1P1fw and PiolEH2P2rev (Table 2). The PCR product was used to transform the wild-type strain carrying the Red recombinase expression plasmid pKD46.
Mammalian Cell Culture
RAW264.7 cells (murine macrophages; ECACC No. 91062702) were cultured in Dulbecco’s modified Eagle medium (DMEM) supplemented with 10% fetal calf serum and 2 mM L-glutamine. The 60 μg/ml penicillin and 100 μg/ml streptomycin were included in the culture media (except for bacterial infection experiments). Cells were maintained in a 5% CO2 humidified atmosphere at 37°C.
Luminescence Measurements and Infection of Cultured Cells
Salmonella strains were grown in triplicate in the media described above and samples of 150 μl of each culture were used to measure luminescence and OD600. Luminescence was read in white, clear bottom 96-well plates (Corning) using a Synergy HT microplate reader (BioTek) or a Sunrise reader (Tecan). To measure luminescence of intracellular bacteria, RAW264.7 cells were plated into white, clear bottom, 96-well plates at 3 × 104 cells per well, and were infected 24 h later with non-invasive bacteria. For that purpose, bacteria were grown in LB medium for 24 h at 37°C with shaking and were added at a multiplicity of infection (MOI) of 500. Bacteria were centrifuged onto the cell monolayer at 200 g for 5 min and then incubated at 37°C with 5% CO2. The cell culture was washed twice with phosphate-buffered saline (PBS) 30 min post-infection (p.i.), overlaid with DMEM containing 100 μg/ml gentamicin, and incubated for 1 h and 30 min. The culture was then washed twice with PBS, covered with DMEM with gentamicin (16 μg/ml), and incubated for 24 h. Luminescence was measured 2, 4, 8, and 24 h p.i. and the numbers of CFU per well were calculated after incubation with 1% Triton X-100 in PBS for 10 min at 37°C to release bacteria, plating appropriate dilutions in LB with Ap, and counting colonies after 24 h of incubation at 37°C. For proliferation assays, infections were carried out using a mix of two strains, as indicated in the Section “Result.” Competitive index for proliferation was calculated as previously described (Segura et al., 2004) after plating appropriate dilutions and enumerating colonies of both strains. Bacteria were recovered 1.25 h p.i. (input) and 24 h p.i. (output).
RNA Extraction and Reverse Transcription
Bacterial strains were grown overnight in LPM or LB. Thereafter, 4 ml of each strain were pelleted and resuspended in 100 μl of water containing 3 mg/ml lysozyme. RNA from these lysates was isolated with 1 ml of TRIzol reagent (Invitrogen) by using the protocol supplied by the manufacturer. An additional step of phenolization or the kit Direct-zolTM RNA MiniPrep Plus (Zymo Research) was carried out to obtain pure samples. RNA (∼1 μg) was reverse transcribed into cDNA with the Quantitect Reverse Transcriptase (Qiagen) before carrying out PCR with appropriate primers.
5′-RACE
Fifteen micrograms of RNA was used to determine the cDNA 5′-end (Bensing et al., 1996; Saito et al., 2009). RNAs were prepared either with or without RNA 5′-Pyrophosphohydrolase (RppH) (New England BioLabs) to distinguish primary transcript 5′-ends from internal 5′-processing sites. DNA primers Jrev and Erev were used for cDNA synthesis with SuperScript III Reverse Transcriptase (Invitrogen) after fusing the GeneRacer RNA Oligo to the isolated RNA. Additional primers for subsequent PCR amplification of cDNAs were GeneRacer 5′-nested primer, homologous to the adaptor GeneRacer RNA oligo, Erev2 and Jrev2. PCR products that were detected both with and without tobacco acid pyrophosphatase treatment were purified by using a PCR clean-up system kit (Promega) and cloned by using the pGEM-T Easy kit (Promega), and three clones of each candidate were sequenced.
Western Blotting and Antibodies
Protein lysates were prepared in SDS–PAGE sample buffer. Proteins were separated by SDS–PAGE in 12% polyacrylamide gels and electrophoretically transferred to nitrocellulose filters for Western blot analysis using anti-Flag M2 monoclonal antibodies (1:5000; Sigma) or anti-GroEL polyclonal antibodies (1:30,000; Sigma). Goat anti-mouse IgG IRDye 800CW and goat anti-rabbit IgG IRDye 680RD (LICOR) were used as secondary antibodies. Bands were detected using and Odyssey Fc infrared imaging system (LICOR).
Plant Cultivation
Tomato (Solanum lycopersicum) seeds were surface sterilized in 2% natrium hypochlorite solution (10 ml) for 10 min. The seeds were then washed vigorously six times with sterile distilled water. Seeds were germinated during 1 week in Petri dishes with sterile 0.5× Murashige and Skoog (MS) medium (Sigma). Seedlings were grown in sterile conditions in 0.25× MS medium (Sigma) in a cabinet with a light intensity of 150 μmol × m2/s (16 h photoperiod) for a further 2 weeks at 22°C.
Bacterial Colonization of Tomato Plants
Salmonella enterica serovar Typhimurium strain 14028 carrying derivatives of plasmid pSB377 (empty, PiolE, PsrfJ, PiolE–PsrfJ) was used to spray 3-week-old tomato plants grown in sterile conditions. Bacteria were grown 1 day before the infection on LM or TM plates. Three tomato plants were spray-inoculated with bacteria suspended in 10 mM MgCl2 at OD600 0.1. Tomato plants were imaged 2 days p.i. with an X-ray film exposed for 48 h.
Bacterial Survival in Plants
To prepare the bacterial inoculum, bacteria were grown on solid LM and then suspended in 10 mM MgCl2 and diluted to an OD600 = 0.01. Leaves of tomato and lettuce (Lactuca sativa) were syringe infiltrated with bacterial solutions, the inoculated leaf areas were sampled 3 h (day 0), 7 days, and 14 days after the inoculation. Serial dilutions were plated on XLD agar to determine the CFU numbers. The experiments were repeated three times with six plants per experiment.
Analysis of Defense Gene Expression Using Quantitative Real-Time PCR (qRT-PCR)
Total RNA from plant leaves was extracted with TRIZOL reagent (Ambion) and treated with DNase I (Quanta BioSciences) following the suppliers’ protocols. Poly A-tailed RNA (1 μg) was converted to cDNA using the qScript cDNA Synthesis Kit (Quanta BioSciences) and oligo-dT primers. qRT-PCR reactions were performed in triplicates with the Maxtra SYBR Green Master Mix (Fermentas) and run on a BioRad iCycler according to the manufacturer’s instructions. The primers used for the qRT-PCR are presented in Table 2. Relative gene expression was normalized to the expression of actin transcript. Expression levels were compared to the control (10 mM MgCl2). Data were processed with the iQ software (BioRad).
Statistical Analysis
Student’s t-test was used to analyze every competitive index against the null hypothesis that the mean is not significantly different from 1. This test was also used to compare mean survival of mutants and wild-type Salmonella strains in plants, as well as expression levels of defense response genes after colonization with different Salmonella strains. P-values of 0.05 or less were considered significant.
Results
Identification of Promoter Regions Driving the Expression of srfJ
Previous data showed expression of srfJ under two disparate conditions: culture medium imitating the intravacuolar environment (LPM) and culture medium supplemented with MI (Cordero-Alba et al., 2012). To understand this dual expression at the molecular level, we explored the genomic region around the srfJ gene. As shown in Figure 1A, this gene resides inside the MI utilization island (Kröger and Fuchs, 2009), with iolE and iolG1 upstream and iolI1 downstream of srfJ. Promoter activities for regions upstream of these genes (putatively called PiolE, PiolG1, PsrfJ, and PiolI1) were tested using plasmid pSB377 (Winson et al., 1998) that carries a promoterless version of the luxCDABE operon of P. luminescens that encodes the luciferase LuxAB subunits and a fatty acid reductase complex involved in synthesis of the fatty aldehyde substrate for the luminescence reaction (Meighen, 1991). This reporter system allows continuous monitoring of light production without disrupting the bacteria or the infected host. Plasmids were introduced in wild-type S. Typhimurium strain 14028 and the luminescence was measured after growth in three different culture conditions: LPM at pH 5.8 with high aeration for SPI2-inducing conditions, LB with 0.3 M NaCl without aeration for SPI1-inducing conditions, and the later medium supplemented with MI to induce expression of the iol genes. Only DNA fragments upstream of iolE and srfJ coding regions showed promoter activity (Figure 1B). Interestingly, PiolE was specifically active in the presence of MI whereas PsrfJ was only active upon SPI2-inducing conditions. These results suggest that expression of srfJ is driven by two promoters: a distal promoter, PiolE, and a proximal promoter, PsrfJ, depending on the environmental conditions. Additional support for these conclusions was obtained studying the production of a chromosomically tagged version of the protein SrfJ by immunoblot. As shown in Figure 1C, SrfJ-3xFLAG was detected in extracts from bacteria grown in minimal LPM medium and in rich LB medium supplemented with MI. In a ΔPiolE background, however, the protein was detected only in LPM, confirming that the distal promoter, PiolE, is specifically necessary for MI-dependent induction of srfJ.
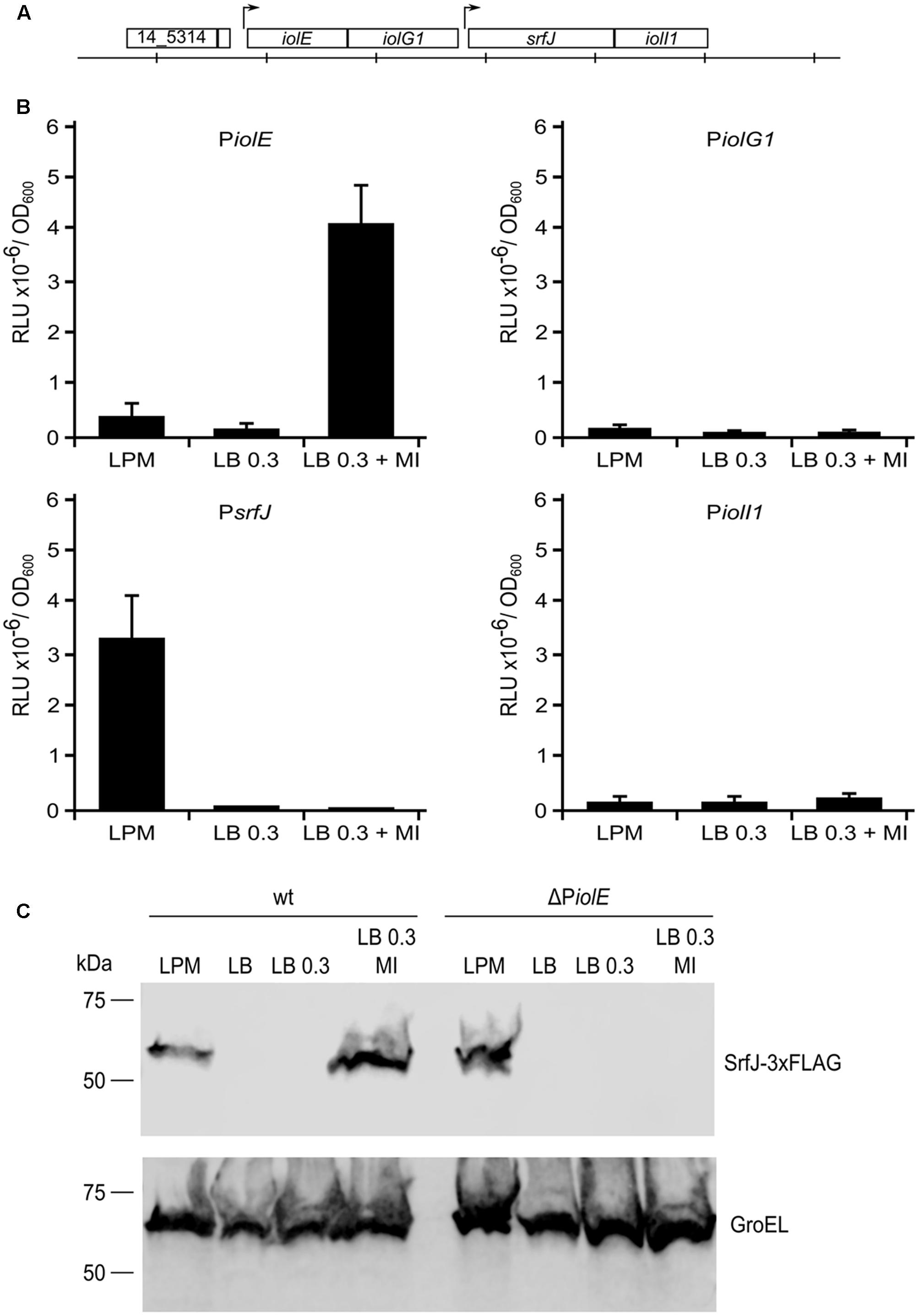
FIGURE 1. Activity of the putative promoter regions in response to LPM and myo-inositol (MI). (A) Representation of the coding regions of srfJ and surrounding genes in S. Typhimurium strain 14028. The distance between vertical lines represents 1 kb. (B) Fragments of DNA upstream the coding regions of genes iolE, iolG1, srfJ, and iolI1 were cloned into plasmid pSB377 to generate luxCDABE transcriptional fusions. These plasmids were introduced into S. Typhimurium strain 14028 and luminescence was measured in cultures grown until stationary phase in LPM, LB 0.3 M NaCl, and LB 0.3 M NaCl with MI. RLU, relative light units. (C) Extracts of a derivative of S. Typhimurium 14028 expressing 3xFLAG-tagged SrfJ were resolved by 12% SDS–PAGE. Immunoblotting was performed with monoclonal anti-FLAG antibodies. Anti-GroEL antibodies were used as loading control. Media tested were LPM, LB, LB with 0.3 M NaCl (LB 0.3), and LB 0.3 M NaCl with MI (LB 0.3 MI). Molecular mass markers are indicated on the left.
Differential Regulation of PiolE and PsrfJ
In order to study the regulation of both promoters, the corresponding plasmids were transferred into different genetic backgrounds. We tested the effect of null mutations in genes encoding relevant regulators: IolR, SsrB, PhoP, and RcsB. IolR is the negative regulator of the MI utilization island (Kröger and Fuchs, 2009). SsrB is encoded in SPI2 and is the main positive regulator of the island (Cirillo et al., 1998). PhoP positively regulates SPI2 through SsrB (Bijlsma and Groisman, 2005). RcsB is the response regulator of the Rcs phosphorelay system (Stout and Gottesman, 1990; Chen et al., 2001). In Salmonella, it positively or negatively regulates genes in SPI1 and SPI2 depending on the level of activation (Wang et al., 2007; Wang and Harshey, 2009). We also used the allele rcsC55 that causes constitutive activation of the Rcs system (García-Calderón et al., 2005). Analysis of the expression patterns in the different genetic backgrounds (Figure 2) indicates that IolR negatively regulates PiolE, whereas PsrfJ is positively regulated by PhoP and SsrB. In addition, PsrfJ is also negatively regulated by the Rcs global regulatory system, since the activating mutation rcsC55 abrogates expression of the lux reporter from this promoter. Interestingly, a transcriptional lux fusion with 2357 bp upstream of srfJ containing both promoters and the intervening genes (PiolE–PsrfJ) is regulated by IolR, PhoP, SsrB, and Rcs, recapitulating the regulation patterns observed with the isolated promoters (Figures 2B,D).
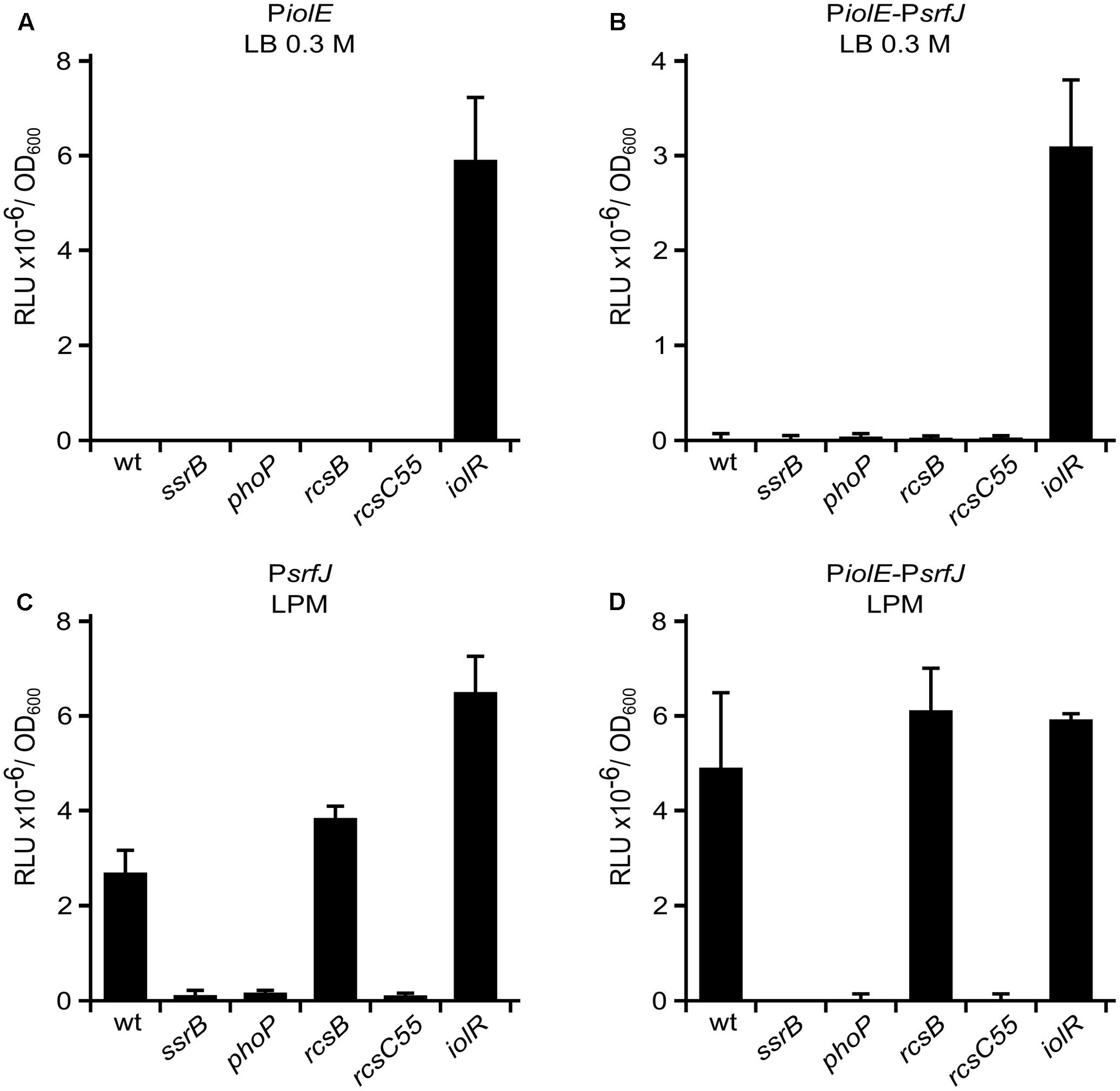
FIGURE 2. Regulation of PiolE and PsrfJ promoters. Fragments of DNA containing the promoters of iolE (PiolE) (A) or srfJ (PsrfJ) (C) or the region containing from the promoter of iolE to the promoter of srfJ, including genes iolE and iolG1 (PiolE–PsrfJ) (B,D) were cloned into plasmid pSB377 to generate luxCDABE transcriptional fusions. These plasmids were introduced into S. enterica serovar Typhimurium strain 14028 or derivatives with null mutations in iolR, ssrB, phoP, rcsB, or a point mutation in rcsC (rcsC55) that confers constitutive activation to the Rcs system. Luminescence was measured in cultures grown until stationary phase in LB 0.3 M NaCl (A,B) and LPM (C,D). RLU, relative light units.
Characterization of Transcriptional Units Containing srfJ
Results presented above suggest that two different promoters can initiate the expression of srfJ. This would result in RNAs of different lengths. To test this hypothesis, RT-PCR was performed using primers designed to amplify different fragments in the srfJ region (Figure 3A). RNA was obtained from two sources: (i) wild-type S. Typhimurium incubated in LPM, where PsrfJ is expected to be active, and (ii) iolR mutant strain incubated in LB, where the absence of IolR repressor should lead to constitutive expression from PiolE. Positive and negative controls were carried out using genomic DNA and non-retrotranscribed RNA, respectively. As seen in Figure 3B, RT-PCR carried out on RNA from wild-type bacteria incubated in LPM yielded only an internal fragment of srfJ. In contrast, fragments partially expanding iolE–iolG1 and iolG1–srfJ were obtained when RT-PCR was carried out using RNA from the iolR mutant, indicating that these genes are transcriptionally linked when PiolE is derepressed. 5′-RACE was used for the determination of both transcriptional start sites. They were located 99 and 33 bp upstream of the coding regions of iolE and srfJ, respectively (Figure 3C). These results confirm that srfJ belongs to two different transcriptional units: a short transcriptional unit with PsrfJ as promoter and an operon including genes iolE, iolG1, and srfJ with PiolE as promoter.
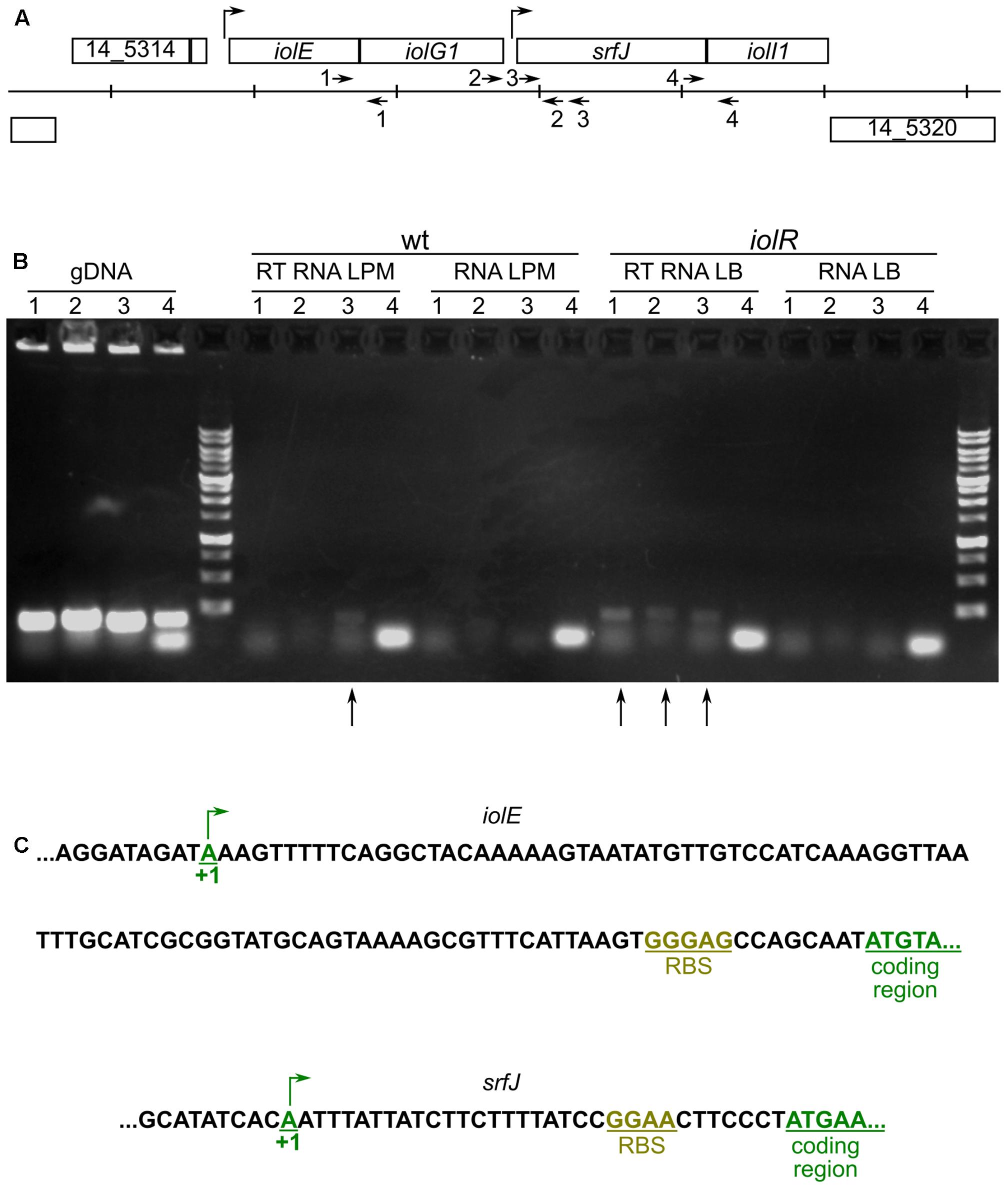
FIGURE 3. Transcriptional organization of the srfJ region. (A) Organization of the chromosomal region containing the srfJ gene in S. Typhimurium strain 14028. Vertical lines are separated by 1 kb. The arrows indicate the positions and orientations of the primers that were used for RT-PCR. (B) Agarose gel of the products obtained with the following primers: 1, Efw and G1rev; 2, G1fw and Jrev2; 3, Jfw and Jrev; and 4, Jfw2 and Jrev. RT-PCR was carried out on RNA isolated from cultures on LPM of the wild-type strain (wt, RT RNA LPM) and cultures in LB of the iolR mutant strain (iolR, RT RNA LB). PCR were also carried out on genomic DNA (gDNA) as positive control and non-retrotranscribed RNA as negative control (RNA LPM and RNA LB). Vertical arrows indicated lanes with amplified products after retrotranscription. The molecular weight marker is the 1 kb DNA ladder (NIPPON Genetics). (C) 5′-RACE was carried out on RNA isolated from cultures in LB of the iolR mutant to map the transcriptional start site of iolE and from cultures in LPM of the wild-type strain to map the transcriptional start site of srfJ. The sequences surrounding the transcriptional start sites (+1) and the start of the coding regions are shown. RBS, ribosomal binding site.
Expression of srfJ Inside Macrophages
Salmonella enterica serovar Typhimurium is known to infect macrophages and express the T3SS2 several hours p.i. (Drecktrah et al., 2006). Since SrfJ is an effector of this secretion system, it is expected to be produced inside macrophages. To ascertain the relevance in this context of the two promoters that drive the expression of srfJ, Salmonella strains carrying PiolE::lux or PsrfJ::lux transcriptional fusions were used to infect RAW264.7 macrophages. The luminescence resulting from the activity of the lux operon driven by PsrfJ increased over time during the infection (Figure 4A). In these conditions the PiolE promoter was not active. Expression of srfJ in internalized bacteria was also studied by immunoblot using a strain of Salmonella that expresses a chromosomally 3xFLAG-tagged version of SrfJ. Intracellular expression was detected both in a wild-type background and in a strain lacking the distal promoter PiolE (Figure 4B). These results suggest that srfJ expression is induced in response to intravacuolar signals and that the induction depends specifically on the proximal promoter.
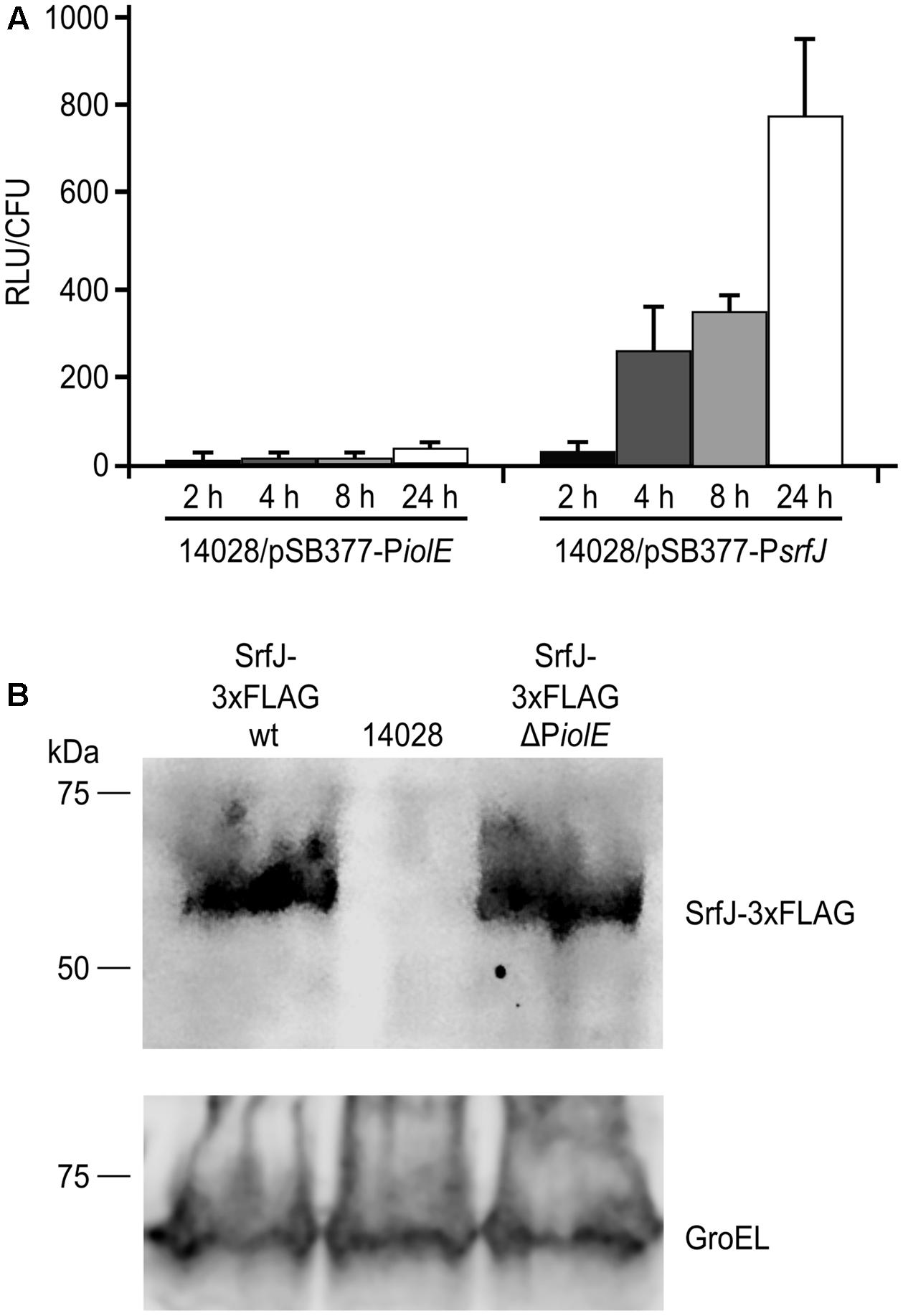
FIGURE 4. Activity of PiolE and PsrfJ during macrophage infection. (A) The wild-type strain of S. Typhimurium carrying a plasmid expressing PiolE::luxCDABE or PsrfJ::luxCDABE transcriptional fusions was grown for 24 h in LB at 37°C with aeration (non-invasive conditions). These bacteria were used to infect RAW264.7 murine macrophage-like cells and luminescence produced by intracellular bacteria was measured 2, 4, 8, and 24 h p.i. RLU, relative light units. (B) The wild-type strain of S. Typhimurium (14028) and derivatives expressing a 3xFLAG-tagged form of SrfJ in a wild-type (wt) background or in a ΔPiolE background were grown under non-invasive conditions and used to infect RAW264.7 cells. Expression of srfJ was measured 8 h p.i. by immunoblot using anti-FLAG antibodies. Anti-GroEL antibodies were used as loading control. Molecular mass markers, in kDa, are indicated on the left.
Contribution of SrfJ to Proliferation of Salmonella in Macrophages
Since the srfJ mutant is attenuated in mice (Ruiz-Albert et al., 2002), we decided to explore the possibility that this mutant could also have a defect in survival and proliferation inside macrophages. This was assessed calculating the competitive index in RAW264.7 macrophages of the srfJ mutant against the trg::MudJ strain, which is wild-type for intracellular proliferation (Segura et al., 2004). No significant defect was detected for this mutant (P > 0.05; Figure 5). We also tested the effect of a null mutation in iolR and we found a very significant defect in intracellular proliferation (Figure 5). Since the iolR mutation leads to derepression of srfJ transcription (Figure 2), we then measured the intracellular proliferation of the double null mutant iolR srfJ. Interestingly, the srfJ mutation suppressed the effect of the iolR mutation on macrophages (Figure 5), suggesting that the proper regulation of the expression of srfJ is essential for survival and/or proliferation of S. Typhimurium inside murine macrophages.
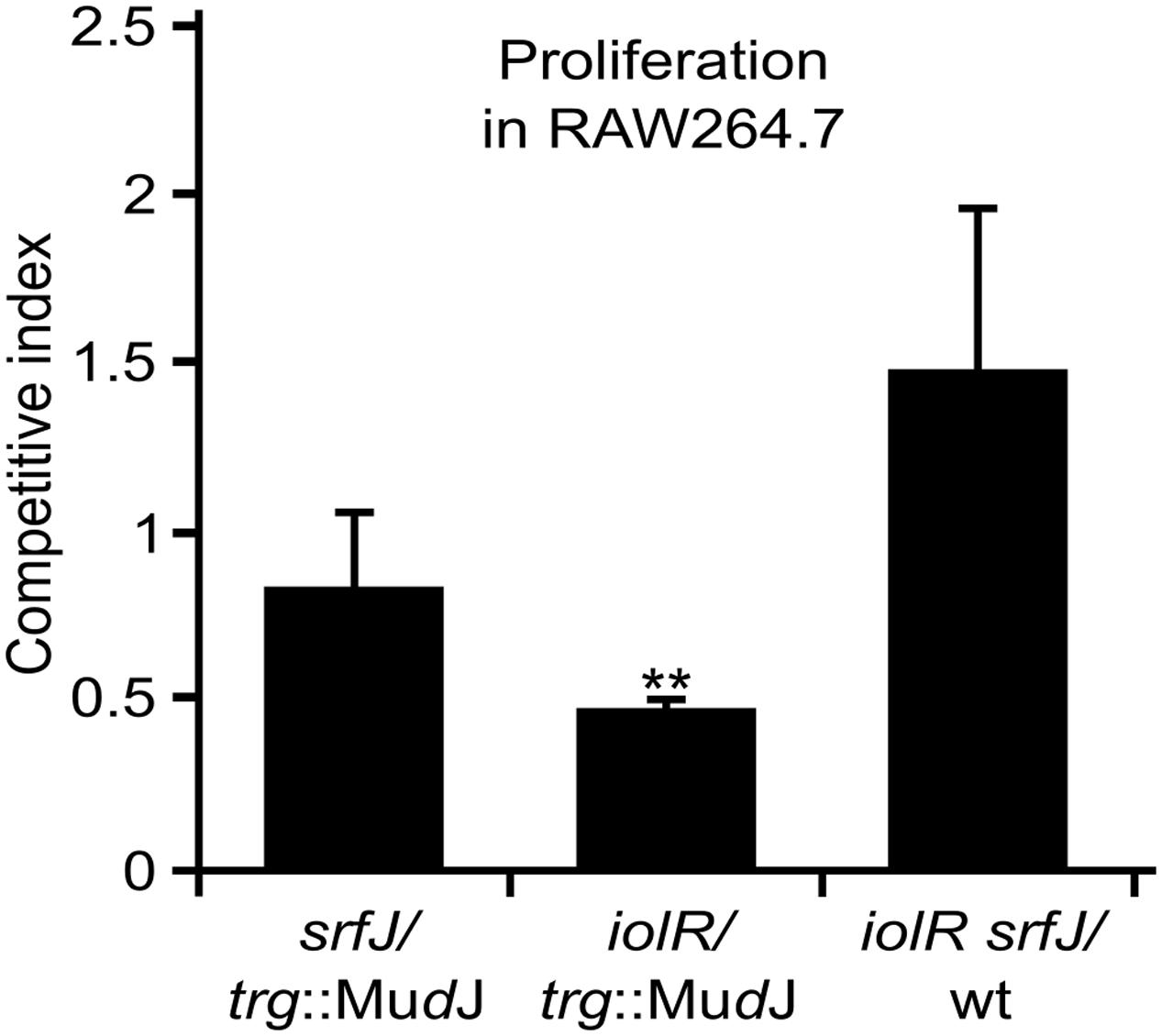
FIGURE 5. Effect of the expression of srfJ in intracellular proliferation. Analysis of intracellular proliferation of srfJ, iolR, and srfJ iolR mutants in mixed infections with a trg::MudJ mutant or the 14028 strain (wt) used as control strains. The competitive indices are the mean from three infections. Error bars represent the standard deviations. Asterisks denote that the indices are significantly different from 1 for a t-test: ∗P-value < 0.05, ∗∗P-value < 0.01.
Expression of srfJ in the Presence of Plant Extracts
The results obtained in macrophages confirmed our hypothesis and proved that SrfJ, as a T3SS2 effector, depends on PsrfJ, the promoter that is induced in media imitating intravacuolar conditions. In contrast, it is more difficult to understand the physiological role of the expression of srfJ from the distal promoter, PiolE. In order to investigate the significance of the double regulation we analyzed different environments known to host Salmonella, one of them are plants. Salmonella is able to thrive and proliferate in plants, including crop plants designated for direct consumption, e.g., lettuce or tomatoes. Thus, we reasoned that since most plants produce MI, PiolE could be relevant in allowing transcription of srfJ in these alternative hosts. We therefore explored the expression of srfJ in response to plant signals. Salmonella with lux transcriptional fusions were grown in LB or in media supplemented with lettuce (LM) or tomato (TM) extracts. We detected high level of luminescence 24 h after the inoculation of LM or TM media with Salmonella carrying the long fusion PiolE-srfJ::lux or the PiolE::lux fusion. Luminescence was not detected after inoculation with a strain carrying the PsrfJ::lux fusion (Figures 6A–C). Expression of srfJ was also studied at the protein level taking advantage of the chromosomal SrfJ-3xFLAG fusion. As shown in Figures 6D,E, SrfJ-3xFLAG was detected by immunoblot in extracts from bacteria grown in LM or TM for 24 h. However, the protein was not produced if PiolE was deleted. These results show that PiolE can drive expression of srfJ in response to plant extracts.
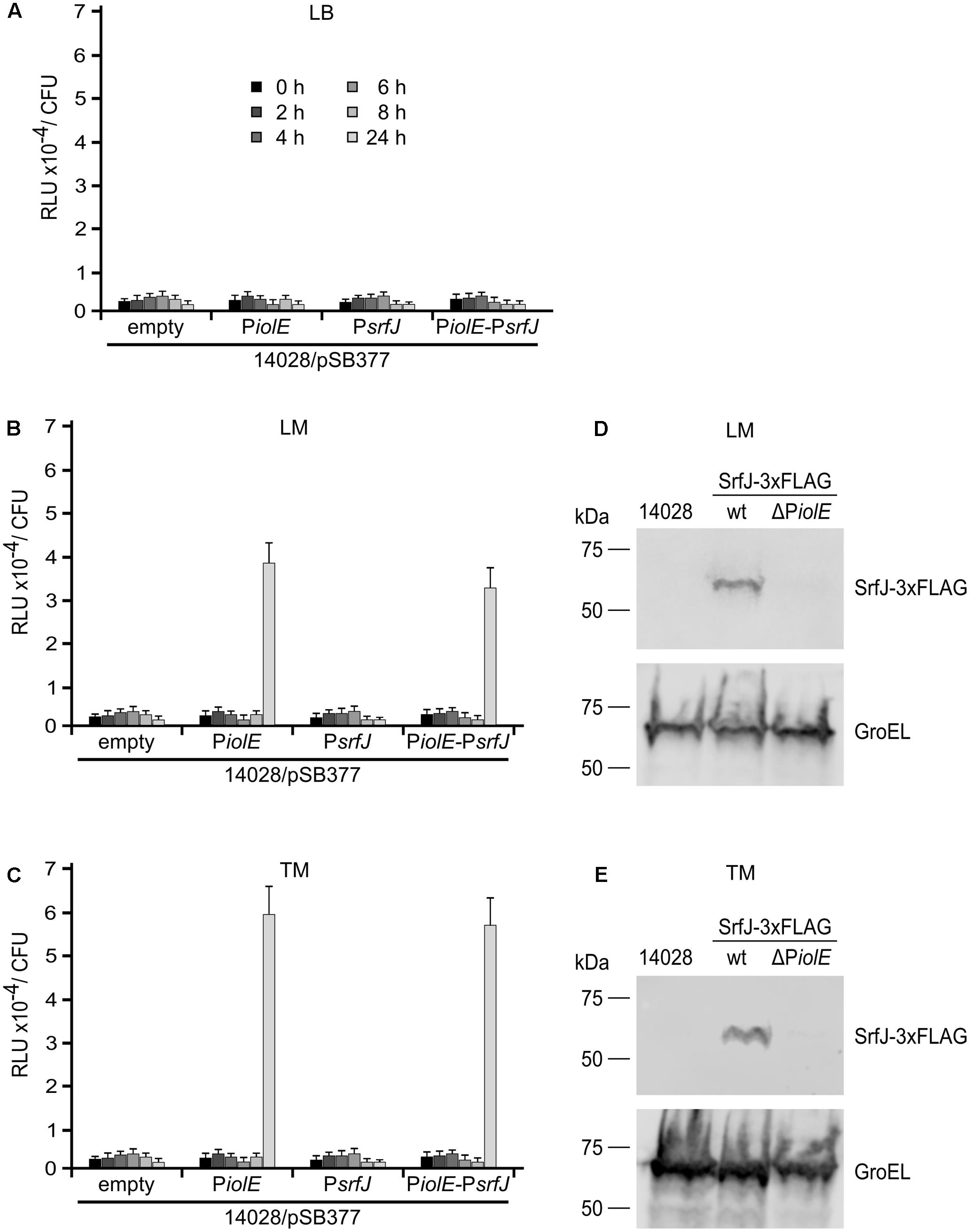
FIGURE 6. Activity of PiolE and PsrfJ in media with plant extracts. Fragments of DNA containing the promoters of iolE (PiolE) or srfJ (PsrfJ) or the region containing from the promoter of iolE to the promoter of srfJ, including genes iolE and iolG1 (PiolE–PsrfJ) were cloned into plasmid pSB377 to generate luxCDABE transcriptional fusions. These plasmids as well as the empty plasmid were introduced into S. Typhimurium strain 14028. Luminescence was measured at different time points (0, 2, 4, 6, 8, 10, and 24 h) in cultures grown in (A) LB, (B) Lettuce Medium (LM), and (C) Tomato Medium (TM). Bacteria were grown overnight in LB and diluted to OD600 0.1 in the different test media before 0 h time point. RLU, relative light units. The wt strain of S. Typhimurium (14028) and derivatives expressing a 3xFLAG-tagged form of SrfJ in a wt background or in a ΔPiolE background were grown in LM (D) or TM (E). Expression of srfJ was measured 8 h p.i. by immunoblot using anti-FLAG antibodies. Anti-GroEL antibodies were used as loading control. Molecular mass markers, in kDa, are indicated on the left.
Expression of srfJ in Plants
The results presented above suggest that srfJ could be expressed during Salmonella colonization of plants. To evaluate this hypothesis, 3-week-old tomato plants were spray-irrigated with suspensions of wild-type S. Typhimurium carrying derivatives of plasmid pSB377 to generate transcriptional luxCDABE fusions with PiolE, PsrfJ, or PiolE–srfJ. Tomato leaves were imaged 2 days post-inoculation using an X-ray film. Luminescence was detected in plants colonized with bacteria carrying PiolE::lux and PiolE-srfJ::lux fusions but not with PsrfJ::lux or the empty vector (Figure 7). Promoters of genes encoding effectors SlrP and SteA were also tested in this system but their expression was not detected (data not shown). These results reveal that Salmonella expresses srfJ together with the MI utilization island, during colonization of a plant host.
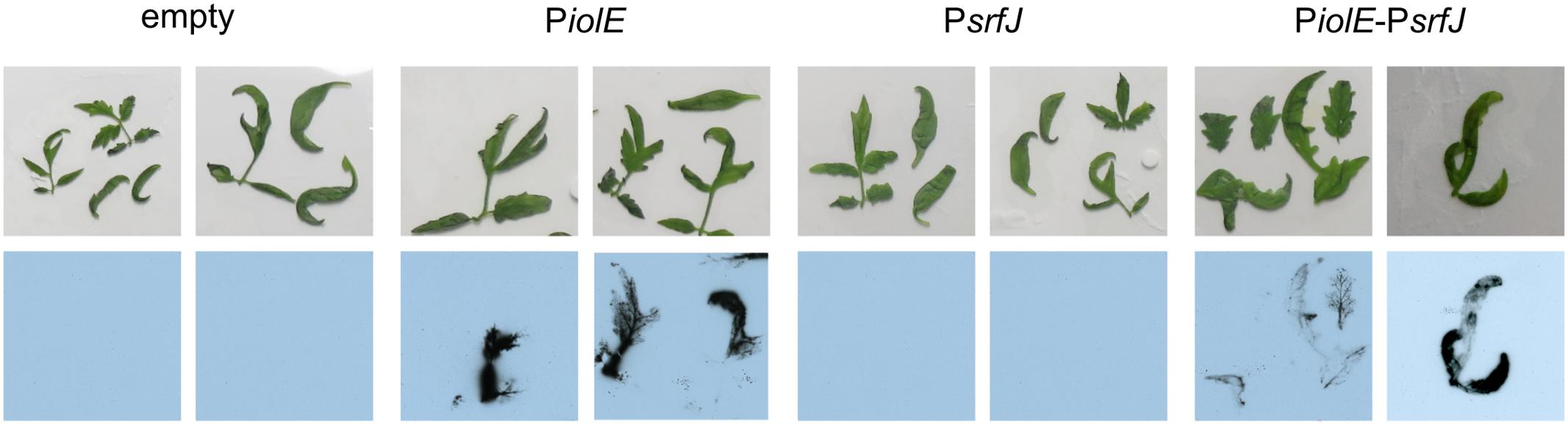
FIGURE 7. Expression in plants. S. Typhimurium strain 14028 carrying derivatives of plasmid pSB377 (empty, PiolE, PsrfJ, PiolE–PsrfJ) was used to spray 3-week-old tomato plants grown in sterile conditions. Plants were germinated in ½ MS for 1 week and grown in ¼ MS media during 2 weeks. Bacteria were grown 1 day before the infection in LM plates. Three tomato plants were spray-inoculated with bacteria suspended in 10 mM MgCl2 at OD600 0.1. Tomato plants were imaged 2 days p.i. with an X-ray film exposed for 48 h.
Contribution of SrfJ to Survival of Salmonella in Plants and Activation of Plant Defense Responses
The expression of srfJ in plants suggested that the product of this gene could be relevant during Salmonella colonization of these alternative hosts. To test this hypothesis, we compared the survival of wild-type S. Typhimurium with the survival of the srfJ mutants in leaves of lettuce and tomato. Leaves were syringe infiltrated with bacterial suspensions and the CFU were counted at different time points. Interestingly, the srfJ null mutant showed a significantly improved survival in leaves of both plants 14 days post-inoculation (Figures 8A,B). Since expression of srfJ in plants depends specifically on the PiolE promoter, we also tested survival in plant leaves of a S. Typhimurium mutant with an intact coding sequence of srfJ but with a deletion of PiolE. As shown in Figures 8A,B, this mutant confirmed the results obtained with the srfJ mutant. These results suggest that SrfJ could be involved in the modulation of plant defense responses that could limit bacterial growth. To test this hypothesis, the expression of five genes known to be involved in tomato defense responses was studied after inoculation of tomato plants with Salmonella wild-type or srfJ mutant using qRT-PCR. Monitored genes encode an acidic extracellular chitinase (CHI3), a basic intracellular chitinase (CHI9), an acidic extracellular β-1,3-glucanase (GLUA), a basic intracellular β-1,3-glucanase (GLUB), and a PR-1 protein isoform PR-P6 (PR-1a) (Joosten and De Wit, 1989; Enkerli et al., 1993; Uehara et al., 2010). Interestingly, the expression of these genes was significantly lower 6 h and/or 12 h after inoculation with the srfJ mutant compared to the wild-type (Figure 8C).
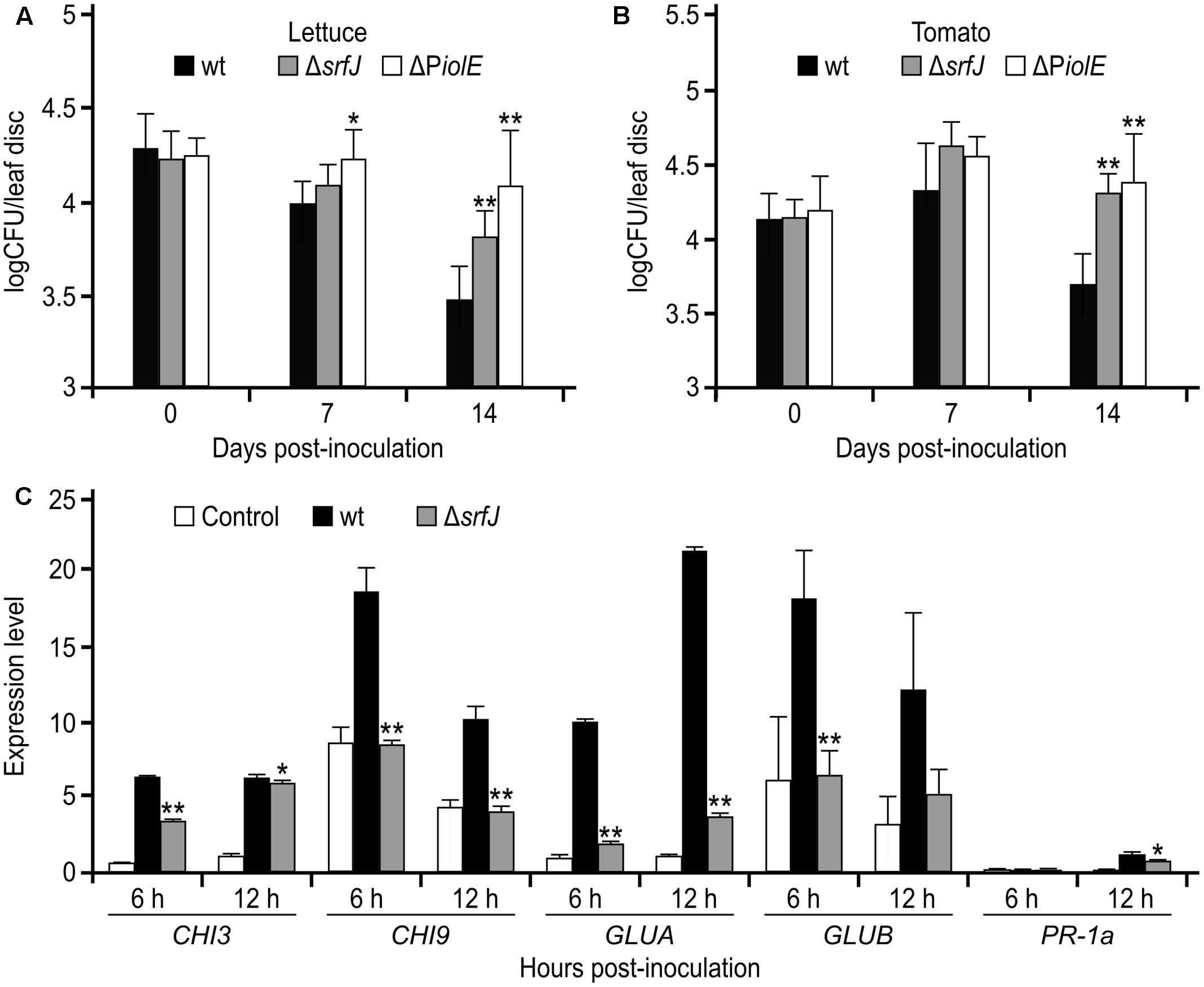
FIGURE 8. Survival of Salmonella srfJ mutants and activation of defense response genes in plants. Salmonella wt strain 14028 (wt), srfJ mutant, and PiolE mutant were syringe infiltrated onto leaves of lettuce (A) or tomato (B). The infiltrated leaves were sampled 0, 7, and 14 days after infiltration to determine the number of CFU of Salmonella. The results shown are the means from six experiments. Error bars represent standard deviations. Asterisks indicate significant differences of mutants compared to wt the same day post-inoculation by Student’s t-test (∗P < 0.05, ∗∗P < 0.01). (C) Salmonella wt and srfJ mutant were used to spray 3-week-old tomato plants grown in sterile conditions. Plants were sampled 6 and 12 h post-inoculation. Relative expression levels of CHI3, CHI9, GLUA, GLUB, and PR-1a were assessed using quantitative RT-PCR and normalized to the expression of the house keeping gene for actin. Data are presented as mean values + standard deviations of three replicates. Asterisks indicate significant differences of mutants compared to wt by Student’s t-test (∗P < 0.05, ∗∗P < 0.01).
Discussion
We previously showed that SrfJ can be secreted through the T3SS2 of S. Typhimurium and that its expression is positively regulated by PhoP and SsrB, and negatively regulated by RcsB and IolR (Cordero-Alba et al., 2012). In this work, we show that these regulators act through two different promoters: a proximal promoter, PsrfJ, that responds to PhoP, SsrB, and RcsB, and a distal promoter, PiolE, that responds to IolR (Figure 2).
The proximal promoter (PsrfJ) initiate the transcription in an adenine located 33 bases upstream the srfJ start codon (Figure 3C). The three regulators that control expression from PsrfJ are well-known relevant regulators of Salmonella virulence. (i) SsrB is the response regulator of a Salmonella-specific two-component regulatory system where the kinase SsrA detects low pH in the host vacuole through a histidine-rich periplasmic sensor domain (Mulder et al., 2015) and phosphorylates SsrB. Phosphorylated SsrB activates transcription of target genes (Deiwick et al., 1999). Positive regulation by SsrB is a common feature of SPI2 genes and other genes encoding effectors specifically secreted through T3SS2 (Xu and Hensel, 2010). (ii) The ancestral PhoQ/PhoP regulatory system is a master two-component system that regulates more than 100 genes (Zwir et al., 2005; Tran et al., 2016) in response to environmental signals including low Mg2+, acidic pH, and cationic antimicrobial peptides (Chamnongpol et al., 2003; Bader et al., 2005; Prost et al., 2007). Since PhoP regulates expression of ssrA and ssrB (Bijlsma and Groisman, 2005), it also regulates expression of genes in the SsrB regulon. (iii) Finally, the Rcs phosphorelay system has been shown to play an important role in virulence in mice, in particular during systemic infections (Detweiler et al., 2003; Domínguez-Bernal et al., 2004; García-Calderón et al., 2005; Erickson and Detweiler, 2006). Repression of PsrfJ by RcsB correlates with previous reports suggesting that high level of activation of the system negatively regulates expression of SPI1 and SPI2 genes (Wang et al., 2007). Thus, this pattern of regulation indicates that PsrfJ functions as a typical promoter of a T3SS2-related gene that responds specifically to intravacuolar signals. Consistent with this, transcription from this promoter was induced in LPM at pH 5.8, a medium that mimics intracellular conditions (Figure 1). More importantly, it was also induced in bacteria phagocytized by macrophages (Figure 4), which are conditions known to induce the expression of the T3SS2.
Regulation by IolR and MI was more puzzling. Nonetheless, we were able to show that these act through a different, distal promoter, PiolE. The transcription start site corresponding to this promoter is an adenine located 99 bases upstream of iolE (Figure 3C) and 2130 bases upstream of srfJ. Two lines of evidence support the existence of an operon encompassing iolE, iolG1, and srfJ: (i) RT-PCR carried out with appropriate oligonucleotide pairs on an RNA sample obtained from an iolR-mutant strain yielded products of the expected size (Figure 3B) if the three genes are transcriptionally linked. (ii) PiolE can drive expression of the reporter lux operon in response to an iolR mutation (Figure 2) or MI supplementation (not shown) from a proximal (pSB377-PiolE) and a distal position (pSB377-PiolE–PsrfJ).
In contrast to PsrfJ, PiolE does not respond to intravacuolar signals but to MI. This carbohydrate, produced by most of the plants, is important for plant growth and development (Loewus and Loewus, 1983): oxidation of MI is an important pathway in cell wall polysaccharide biogenesis (Loewus and Murthy, 2000; Loewus, 2006); inositol and derived molecules are involved in stress-related responses (Loewus and Murthy, 2000); and MI is used as a precursor of inositol-containing signaling molecules including phosphatidylinositol and phosphoinositides (Gillaspy, 2011). The presence of this carbohydrate in plant extracts explains expression of srfJ from PiolE in LM and TM (Figure 6). The observation that this expression is detected 24 h but not 8 h after the inoculation of the media is in agreement with a previously reported extended lag phase during the growth of Salmonella in the presence of MI as the sole carbon source (Kröger and Fuchs, 2009). The authors of this report exclude catabolite repression as an explanation but suggest that the iol genes in Salmonella could be under a tight repression or under the action of an additional unknown regulatory factor. Interestingly, our results suggest that expression of srfJ from the MI responsive promoter PiolE could be important during the plant colonization by Salmonella (Figure 7). This could also explain the chromosomal location of srfJ inside the MI island from an evolutionary point of view.
Several reports suggest the important role of Salmonella T3SS during plant colonization. Salmonella lacking T3SS1 and T3SS2, prgH, and ssaV mutants, respectively, showed reduced proliferation in syringe-infiltrated leaves of Arabidopsis thaliana (Schikora et al., 2011). Symptoms caused by these mutants were more pronounced in comparison with the wild-type strain, indicating that T3SSs are involved in suppressing the hypersensitive response (HR), an induced, localized cell death, which limits the spread of pathogens. Furthermore, transcriptome analyses showed that a prgH mutant induced stronger defense gene expression than wild-type bacteria in Arabidopsis seedlings (Schikora et al., 2011; Garcia et al., 2014). Similarly, experiments in Nicotiana tabacum have shown that the T3SS1 is essential for the active suppression of defense mechanisms by Salmonella (Shirron and Yaron, 2011). Interestingly, the response was different in other plants or even in different organs of the same plants: mutants lacking T3SS1 (sipB or spaS) colonized Medicago sativa roots, A. thaliana roots, and Triticum aestivum roots and seedlings in significantly greater numbers than the wild-type strain 14028 (Iniguez et al., 2005). Because the sipB mutation did not enhance colonization in a npr1 Arabidopsis mutant, which is defective in both salicylic acid (SA)-dependent and SA-independent defense responses (Ton et al., 2002), the authors concluded that T3SS1 is involved in the induction of both kinds of plant responses. In contrast, another study concluded that T3SS1 and T3SS2 were not involved in suppression of plant defenses in Nicotiana benthamiana leaves (Meng et al., 2013). These discrepant results indicate that the exploration of a variety of experimental conditions and host models will be necessary to ascertain the role of Salmonella T3SSs and particular effectors in plants. Interestingly, although Salmonella-mediated delivery of effector proteins into plant cells have not been shown yet (García and Hirt, 2014), effectors SseF and SspH2 were able to trigger cell death through resistance-gene-mediated signaling in N. benthamiana when heterologously delivered using Agrobacterium tumefaciens or Xanthomonas campestris (Üstün et al., 2012; Bhavsar et al., 2013).
An important aspect of this work was the analysis of the contribution of SrfJ to the survival of Salmonella inside animal and plant hosts. Our results suggest that the expression of this effector at the appropriate time is a relevant factor in the interaction of Salmonella with mice macrophages and with lettuce and tomato leaves: (i) The results obtained with the iolR mutant and the iolR srfJ double mutant indicate that the ectopic production of SrfJ caused by the absence of the IolR repressor decreases survival/proliferation of Salmonella inside RAW264.7 macrophages (Figure 5). (ii) The lack of production of SrfJ in plants caused by a deletion of the coding sequence of srfJ or a deletion of the distal promoter PiolE leads to an improved survival of Salmonella in plants (Figure 8). This result is in line with the reduced defense gene activation displayed by the srfJ mutant compared to the wild-type (Figure 8C) and suggests that SrfJ could act in this system as an avirulence protein (Mansfield, 2009). These proteins are effectors of plant pathogens that, in the course of plant-pathogen co-evolution, have been recognized by plant receptors to activate defense responses. As such, SrfJ could have a virulent role in other sensitive plant strains or species.
Additional experiments will be needed to ascertain if SrfJ expressed in Salmonella during plant colonization can be secreted through T3SS2. In this context, it should be noted that in our experimental model PsrfJ was not active during plant colonization with Salmonella. This result suggests that, under these conditions, the SsrB regulon, including, SPI2, would not be expressed and therefore SrfJ, although expressed from PiolE, would not be secreted through T3SS2. Nevertheless, the phenotype of the ssaV mutant noted above (Schikora et al., 2011) argues for the expression of this system at some point of the colonization of plants. Our previous results showed that translocation into macrophages was specifically T3SS2-dependent (Cordero-Alba et al., 2012), and here we have shown that PsrfJ is the only promoter that drives expression of srfJ inside these cells (Figure 4). However, secretion of SrfJ through T3SS1 is another interesting possibility that could be explored in different cell types or hosts. Several effectors can be secreted through both systems. For example, for PipB2, considered as a T3SS2 effector, we have previously shown the possibility of translocation through T3SS1 into a variety of cell types (Baisón-Olmo et al., 2012). Specificity of secretion is achieved, at least for some effectors, simply by coexpression between the particular T3SS and its effectors. An example is SseK1: when expressed from a constitutive promoter it can be secreted through T3SS1 at earlier time points p.i. than when expressed from its own promoter (Baisón-Olmo et al., 2015). Coexpression of T3SS1 and srfJ from PiolE may take place in plants due to the presence of MI, making it possible the delivery of the effector through this way. A recent report suggests that S. Typhimurium is unable to translocate T3SS effectors into cells of beet roots or pepper leaves (Chalupowicz et al., 2017). However, there is the possibility of translocation in other plant systems. Alternatively, some effectors could be secreted and exert their action in the apoplast rather than inside plant cells.
The function of SrfJ inside the host is presently unknown. Its amino acid sequence and its structure (Kim et al., 2009) suggest that it may have glucosylceramidase activity. This enzymatic activity catalyzes hydrolysis of glucosylceramide into glucose and ceramide, the simplest member of the family of sphingolipids. These lipids not only represent essential structural elements of membranes, but several members of this family, including ceramide, are also secondary messenger molecules that regulate intra- and intercellular processes (Ilan, 2016). Ceramides can affect cellular proliferation, differentiation, cell death, insulin resistance, oxidative stress, and inflammation (Pandey et al., 2007; Saddoughi and Ogretmen, 2013; Kogot-Levin and Saada, 2014; Kuzmenko and Klimentyeva, 2016). Glycosphingolipids are membrane components that can affect numerous cellular events including homeostasis, adhesion growth, motility, apoptosis, proliferation, stress, and inflammatory responses (Ilan, 2016). Interestingly, glucosylceramide is the only glycosphingolipid that plants, fungi, and animals have in common (Warnecke and Heinz, 2003). Glucosylceramide is important in animals for the activation of antigen-presenting cells, induction of Th1 and Th7 responses, and neutrophil recruitment (Pandey et al., 2012). There is less information about the functions of glucosylceramide in plants, but it has been suggested to be necessary for normal Golgi-mediated protein trafficking (Melser et al., 2010, 2011). A more recent report has shown that null mutants for glucosylceramide synthase failed to develop beyond the seedling stage and that glucosylceramide is critical for cell-type differentiation and organogenesis (Msanne et al., 2015). Future research should focus on the relevance and consequences of the putative effect of SrfJ on this important lipid target both in animal and plant cells.
Author Contributions
All authors listed above have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
Funding
This work was supported by grants SAF2013-46229-R and SAF2016-75365-R from Spanish Ministerio de Economía, Industria y Competitividad, and the European Regional Development Fund. The work of AZ was supported by the DAAD. JA-H was supported by a short-term EMBO fellowship.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors thank Anja Wiechmann and Paul Williams for providing pSB377, and Modesto Carballo, Laura Navarro, and Cristina Reyes of the Servicio de Biología (CITIUS, Universidad de Sevilla) for help in experiments performed at the facility.
References
Bader, M. W., Sanowar, S., Daley, M. E., Schneider, A. R., Cho, U., Xu, W., et al. (2005). Recognition of antimicrobial peptides by a bacterial sensor kinase. Cell 122, 461–472. doi: 10.1016/j.cell.2005.05.030
Baisón-Olmo, F., Cardenal-Muñoz, E., and Ramos-Morales, F. (2012). PipB2 is a substrate of the Salmonella pathogenicity island 1-encoded type III secretion system. Biochem. Biophys. Res. Commun. 423, 240–246. doi: 10.1016/j.bbrc.2012.05.095
Baisón-Olmo, F., Galindo-Moreno, M., and Ramos-Morales, F. (2015). Host cell type-dependent translocation and PhoP-mediated positive regulation of the effector SseK1 of Salmonella enterica. Front. Microbiol. 6:396. doi: 10.3389/fmicb.2015.00396
Bakowski, M. A., Braun, V., and Brumell, J. H. (2008). Salmonella-containing vacuoles: directing traffic and nesting to grow. Traffic 9, 2022–2031. doi: 10.1111/j.1600-0854.2008.00827.x
Bensing, B. A., Meyer, B. J., and Dunny, G. M. (1996). Sensitive detection of bacterial transcription initiation sites and differentiation from RNA processing sites in the pheromone-induced plasmid transfer system of Enterococcus faecalis. Proc. Natl. Acad. Sci. U.S.A. 93, 7794–7799. doi: 10.1073/pnas.93.15.7794
Bhavsar, A. P., Brown, N. F., Stoepel, J., Wiermer, M., Martin, D. D. O., Hsu, K. J., et al. (2013). The Salmonella type III effector SspH2 specifically exploits the NLR co-chaperone activity of SGT1 to subvert immunity. PLOS Pathog. 9:e1003518. doi: 10.1371/journal.ppat.1003518
Bijlsma, J. J. E., and Groisman, E. A. (2005). The PhoP/PhoQ system controls the intramacrophage type three secretion system of Salmonella enterica. Mol. Microbiol. 57, 85–96. doi: 10.1111/j.1365-2958.2005.04668.x
Boyle, E. C., Brown, N. F., and Finlay, B. B. (2006). Salmonella enterica serovar Typhimurium effectors SopB, SopE, SopE2 and SipA disrupt tight junction structure and function. Cell. Microbiol. 8, 1946–1957. doi: 10.1111/j.1462-5822.2006.00762.x
Chalupowicz, L., Nissan, G., Brandl, M. T., McClelland, M., Sessa, G., Popov, G., et al. (2017). Assessing the ability of Salmonella enterica to translocate type III effectors into plant cells. Mol. Plant. Microbe Interact. doi: 10.1094/MPMI-07-17-0166-R [Epub ahead of print].
Chamnongpol, S., Cromie, M., and Groisman, E. A. (2003). Mg2+ sensing by the Mg2+ sensor PhoQ of Salmonella enterica. J. Mol. Biol. 325, 795–807.
Chan, R. K., Botstein, D., Watanabe, T., and Ogata, Y. (1972). Specialized transduction of tetracycline resistance by phage P22 in Salmonella typhimurium. II. Properties of a high-frequency-transducing lysate. Virology 50, 883–898.
Chen, H.-M., Wang, Y., Su, L.-H., and Chiu, C.-H. (2013). Nontyphoid Salmonella infection: microbiology, clinical features, and antimicrobial therapy. Pediatr. Neonatol. 54, 147–152. doi: 10.1016/j.pedneo.2013.01.010
Chen, M. H., Takeda, S., Yamada, H., Ishii, Y., Yamashino, T., and Mizuno, T. (2001). Characterization of the RcsC– > YojN– > RcsB phosphorelay signaling pathway involved in capsular synthesis in Escherichia coli. Biosci. Biotechnol. Biochem. 65, 2364–2367.
Cirillo, D. M., Valdivia, R. H., Monack, D. M., and Falkow, S. (1998). Macrophage-dependent induction of the Salmonella pathogenicity island 2 type III secretion system and its role in intracellular survival. Mol. Microbiol. 30, 175–188.
Cordero-Alba, M., Bernal-Bayard, J., and Ramos-Morales, F. (2012). SrfJ, a Salmonella type III secretion system effector regulated by PhoP, RcsB, and IolR. J. Bacteriol. 194, 4226–4236. doi: 10.1128/JB.00173-12
Costa, L. F., Paixão, T. A., Tsolis, R. M., Bäumler, A. J., and Santos, R. L. (2012). Salmonellosis in cattle: advantages of being an experimental model. Res. Vet. Sci. 93, 1–6. doi: 10.1016/j.rvsc.2012.03.002
Datsenko, K. A., and Wanner, B. L. (2000). One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U.S.A. 97, 6640–6645. doi: 10.1073/pnas.120163297
Deiwick, J., Nikolaus, T., Erdogan, S., and Hensel, M. (1999). Environmental regulation of Salmonella pathogenicity island 2 gene expression. Mol. Microbiol. 31, 1759–1773. doi: 10.1046/j.1365-2958.1999.01312.x
Deng, W., Marshall, N. C., Rowland, J. L., McCoy, J. M., Worrall, L. J., Santos, A. S., et al. (2017). Assembly, structure, function and regulation of type III secretion systems. Nat. Rev. Microbiol. 15, 323–337. doi: 10.1038/nrmicro.2017.20
Detweiler, C. S., Monack, D. M., Brodsky, I. E., Mathew, H., and Falkow, S. (2003). virK, somA and rcsC are important for systemic Salmonella enterica serovar Typhimurium infection and cationic peptide resistance. Mol. Microbiol. 48, 385–400. doi: 10.1046/j.1365-2958.2003.03455.x
Domínguez-Bernal, G., Pucciarelli, M. G., Ramos-Morales, F., García-Quintanilla, M., Cano, D. A., Casadesús, J., et al. (2004). Repression of the RcsC-YojN-RcsB phosphorelay by the IgaA protein is a requisite for Salmonella virulence. Mol. Microbiol. 53, 1437–1449. doi: 10.1111/j.1365-2958.2004.04213.x
Drecktrah, D., Knodler, L. A., Ireland, R., and Steele-Mortimer, O. (2006). The mechanism of Salmonella entry determines the vacuolar environment and intracellular gene expression. Traffic 7, 39–51. doi: 10.1111/j.1600-0854.2005.00360.x
Enkerli, J., Gisi, U., and Mösinger, E. (1993). Systemic acquired resistance to Phytophthora infestans in tomato and the role of pathogenesis related proteins. Physiol. Mol. Plant Pathol. 43, 161–171. doi: 10.1006/pmpp.1993.1048
Erickson, K. D., and Detweiler, C. S. (2006). The Rcs phosphorelay system is specific to enteric pathogens/commensals and activates ydeI, a gene important for persistent Salmonella infection of mice. Mol. Microbiol. 62, 883–894. doi: 10.1111/j.1365-2958.2006.05420.x
Fass, E., and Groisman, E. A. (2009). Control of Salmonella pathogenicity island-2 gene expression. Curr. Opin. Microbiol. 12, 199–204. doi: 10.1016/j.mib.2009.01.004
Fink, S. L., and Cookson, B. T. (2007). Pyroptosis and host cell death responses during Salmonella infection. Cell. Microbiol. 9, 2562–2570. doi: 10.1111/j.1462-5822.2007.01036.x
Foley, S. L., Lynne, A. M., and Nayak, R. (2007). Salmonella challenges: prevalence in swine and poultry and potential pathogenicity of such isolates. J. Anim. Sci. 86, E149–E162. doi: 10.2527/jas.2007-0464
Fornefeld, E., Schierstaedt, J., Jechalke, S., Grosch, R., Schikora, A., and Smalla, K. (2017). Persistence of Salmonella Typhimurium LT2 in soil enhanced after growth in lettuce medium. Front. Microbiol. 8:757. doi: 10.3389/fmicb.2017.00757
Galán, J. E., and Curtiss, R. (1989). Cloning and molecular characterization of genes whose products allow Salmonella typhimurium to penetrate tissue culture cells. Proc. Natl. Acad. Sci. U.S.A. 86, 6383–6387.
Garai, P., Gnanadhas, D. P., and Chakravortty, D. (2012). Salmonella enterica serovars Typhimurium and Typhi as model organisms: revealing paradigm of host-pathogen interactions. Virulence 3, 377–388. doi: 10.4161/viru.21087
Garcia, A. V., Charrier, A., Schikora, A., Bigeard, J., Pateyron, S., de Tauzia-Moreau, M.-L., et al. (2014). Salmonella enterica flagellin is recognized via FLS2 and activates PAMP-triggered immunity in Arabidopsis thaliana. Mol. Plant 7, 657–674. doi: 10.1093/mp/sst145
García, A. V., and Hirt, H. (2014). Salmonella enterica induces and subverts the plant immune system. Front. Microbiol. 5:141. doi: 10.3389/fmicb.2014.00141
García-Calderón, C. B., Casadesús, J., and Ramos-Morales, F. (2007). Rcs and PhoPQ regulatory overlap in the control of Salmonella enterica virulence. J. Bacteriol. 189, 6635–6644. doi: 10.1128/JB.00640-07
García-Calderón, C. B., García-Quintanilla, M., Casadesús, J., Ramos-morales, F., Garcı, M., Garcı, C. B., et al. (2005). Virulence attenuation in Salmonella enterica rcsC mutants with constitutive activation of the Rcs system. Microbiology 151, 579–588. doi: 10.1099/mic.0.27520-0
Gillaspy, G. E. (2011). The cellular language of myo-inositol signaling. New Phytol. 192, 823–839. doi: 10.1111/j.1469-8137.2011.03939.x
Hanahan, D. (1983). Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166, 557–580. doi: 10.1016/S0022-2836(83)80284-8
Holden, N. J., Jackson, R. W., and Schikora, A. (2015). Editorial on plants as alternative hosts for human and animal pathogens. Front. Microbiol. 6:397. doi: 10.3389/fmicb.2015.00397
Ilan, Y. (2016). Compounds of the sphingomyelin-ceramide-glycosphingolipid pathways as secondary messenger molecules: new targets for novel therapies for fatty liver disease and insulin resistance. Am. J. Physiol. Gastrointest. Liver Physiol. 310, G1102–G1117. doi: 10.1152/ajpgi.00095.2016
Iniguez, A. L., Dong, Y., Carter, H. D., Ahmer, B. M. M., Stone, J. M., and Triplett, E. W. (2005). Regulation of enteric endophytic bacterial colonization by plant defenses. Mol. Plant. Microbe Interact. 18, 169–178. doi: 10.1094/MPMI-18-0169
Joosten, M. H., and De Wit, P. J. (1989). Identification of several pathogenesis-related proteins in tomato leaves inoculated with Cladosporium fulvum (syn. Fulvia fulva) as 1,3-beta-glucanases and chitinases. Plant Physiol. 89, 945–951.
Kim, Y.-G., Kim, J.-H., and Kim, K. (2009). Crystal structure of the Salmonella enterica serovar Typhimurium virulence factor SrfJ, a glycoside hydrolase family enzyme. J. Bacteriol. 191, 6550–6554. doi: 10.1128/JB.00641-09
Knodler, L. A., and Steele-Mortimer, O. (2003). Taking possession: biogenesis of the Salmonella-containing vacuole. Traffic 4, 587–599.
Kogot-Levin, A., and Saada, A. (2014). Ceramide and the mitochondrial respiratory chain. Biochimie 100, 88–94. doi: 10.1016/j.biochi.2013.07.027
Kröger, C., and Fuchs, T. M. (2009). Characterization of the myo-inositol utilization island of Salmonella enterica serovar Typhimurium. J. Bacteriol. 191, 545–554. doi: 10.1128/JB.01253-08
Kuzmenko, D. I., and Klimentyeva, T. K. (2016). Role of ceramide in apoptosis and development of insulin resistance. Biochemistry 81, 913–927. doi: 10.1134/S0006297916090017
Liao, A. P., Petrof, E. O., Kuppireddi, S., Zhao, Y., Xia, Y., Claud, E. C., et al. (2008). Salmonella type III effector AvrA stabilizes cell tight junctions to inhibit inflammation in intestinal epithelial cells. PLOS ONE 3:e2369. doi: 10.1371/journal.pone.0002369
Lin, Z., Zhang, Y.-G., Xia, Y., Xu, X., Jiao, X., and Sun, J. (2016). Salmonella enteritidis effector AvrA stabilizes intestinal tight junctions via the JNK pathway. J. Biol. Chem. 291, 26837–26849. doi: 10.1074/jbc.M116.757393
Liss, V., and Hensel, M. (2015). Take the tube: remodelling of the endosomal system by intracellular Salmonella enterica. Cell. Microbiol. 17, 639–647. doi: 10.1111/cmi.12441
Loewus, F. A. (2006). “Inositol and plant cell wall polysaccharide biogenesis,” in Biology of Inositols and Phosphoinositides, eds A. Lahiri Majumder and B. B. Biswas (New York, NY: Springer), 21–45.
Loewus, F. A., and Loewus, M. W. (1983). myo-Inositol: its biosynthesis and metabolism. Annu. Rev. Plant Physiol. 34, 137–161. doi: 10.1146/annurev.pp.34.060183.001033
Loewus, F. A., and Murthy, P. P. N. (2000). myo-Inositol metabolism in plants. Plant Sci. 150, 1–19. doi: 10.1016/S0168-9452(99)00150-8
Maloy, S. R. (1990). Experimental Techniques in Bacterial Genetics. Burlington, MA: Jones & Bartlett Learning.
Mansfield, J. W. (2009). From bacterial avirulence genes to effector functions via the hrp delivery system: an overview of 25 years of progress in our understanding of plant innate immunity. Mol. Plant Pathol. 10, 721–734. doi: 10.1111/j.1364-3703.2009.00576.x
Melser, S., Batailler, B., Peypelut, M., Poujol, C., Bellec, Y., Wattelet-Boyer, V., et al. (2010). Glucosylceramide biosynthesis is involved in Golgi morphology and protein secretion in plant cells. Traffic 11, 479–490. doi: 10.1111/j.1600-0854.2009.01030.x
Melser, S., Molino, D., Batailler, B., Peypelut, M., Laloi, M., Wattelet-Boyer, V., et al. (2011). Links between lipid homeostasis, organelle morphodynamics and protein trafficking in eukaryotic and plant secretory pathways. Plant Cell Rep. 30, 177–193. doi: 10.1007/s00299-010-0954-1
Meng, F., Altier, C., and Martin, G. B. (2013). Salmonella colonization activates the plant immune system and benefits from association with plant pathogenic bacteria. Environ. Microbiol. 15, 2418–2430. doi: 10.1111/1462-2920.12113
Msanne, J., Chen, M., Luttgeharm, K. D., Bradley, A. M., Mays, E. S., Paper, J. M., et al. (2015). Glucosylceramides are critical for cell-type differentiation and organogenesis, but not for cell viability in Arabidopsis. Plant J. 84, 188–201. doi: 10.1111/tpj.13000
Mulder, D. T., McPhee, J. B., Reid-Yu, S. A., Stogios, P. J., Savchenko, A., and Coombes, B. K. (2015). Multiple histidines in the periplasmic domain of the Salmonella enterica sensor kinase SsrA enhance signaling in response to extracellular acidification. Mol. Microbiol. 95, 678–691. doi: 10.1111/mmi.12895
Neumann, C., Fraiture, M., Hernàndez-Reyes, C., Akum, F. N., Virlogeux-Payant, I., Chen, Y., et al. (2014). The Salmonella effector protein SpvC, a phosphothreonine lyase is functional in plant cells. Front. Microbiol. 5:548. doi: 10.3389/fmicb.2014.00548
Pandey, M. K., Rani, R., Zhang, W., Setchell, K., and Grabowski, G. A. (2012). Immunological cell type characterization and Th1–Th17 cytokine production in a mouse model of Gaucher disease. Mol. Genet. Metab. 106, 310–322. doi: 10.1016/j.ymgme.2012.04.020
Pandey, S., Murphy, R. F., and Agrawal, D. K. (2007). Recent advances in the immunobiology of ceramide. Exp. Mol. Pathol. 82, 298–309. doi: 10.1016/j.yexmp.2006.07.009
Perrett, C. A., and Zhou, D. (2013). Salmonella type III effector SopB modulates host cell exocytosis. Emerg. Microbes Infect. 2:e32. doi: 10.1038/emi.2013.31
Prost, L. R., Daley, M. E., Le Sage, V., Bader, M. W., Le Moual, H., Klevit, R. E., et al. (2007). Activation of the bacterial sensor kinase PhoQ by acidic pH. Mol. Cell 26, 165–174. doi: 10.1016/j.molcel.2007.03.008
Ramos-Morales, F. (2012). Impact of Salmonella enterica Type III secretion system effectors on the eukaryotic host cell. ISRN Cell Biol. 2012:787934. doi: 10.5402/2012/787934
Ruiz-Albert, J., Yu, X.-J., Beuzón, C. R., Blakey, A. N., Galyov, E. E., and Holden, D. W. (2002). Complementary activities of SseJ and SifA regulate dynamics of the Salmonella typhimurium vacuolar membrane. Mol. Microbiol. 44, 645–661.
Saddoughi, S. A., and Ogretmen, B. (2013). Diverse functions of ceramide in cancer cell death and proliferation. Adv. Cancer Res. 117, 37–58. doi: 10.1016/B978-0-12-394274-6.00002-9
Saito, S., Kakeshita, H., and Nakamura, K. (2009). Novel small RNA-encoding genes in the intergenic regions of Bacillus subtilis. Gene 428, 2–8. doi: 10.1016/j.gene.2008.09.024
Schikora, A., Garcia, A. V., and Hirt, H. (2012). Plants as alternative hosts for Salmonella. Trends Plant Sci. 17, 245–249. doi: 10.1016/j.tplants.2012.03.007
Schikora, A., Virlogeux-payant, I., Bueso, E., Garcia, A. V., Nilau, T., Pelletier, S., et al. (2011). Conservation of Salmonella infection mechanisms in plants and animals. PLOS ONE 6:e24112. doi: 10.1371/journal.pone.0024112
Schmieger, H. (1972). Phage P22-mutants with increased or decreased transduction abilities. Mol. Gen. Genet. 119, 75–88.
Segura, I., Casadesús, J., and Ramos-Morales, F. (2004). Use of mixed infections to study cell invasion and intracellular proliferation of Salmonella enterica in eukaryotic cell cultures. J. Microbiol. Methods 56, 83–91. doi: 10.1016/j.mimet.2004.03.013
Shirron, N., and Yaron, S. (2011). Active suppression of early immune response in tobacco by the human pathogen Salmonella Typhimurium. PLOS ONE 6:e18855. doi: 10.1371/journal.pone.0018855
Steele-Mortimer, O. (2008). The Salmonella-containing vacuole: moving with the times. Curr. Opin. Microbiol. 11, 38–45. doi: 10.1016/j.mib.2008.01.002
Stout, V., and Gottesman, S. (1990). RcsB and RcsC: a two-component regulator of capsule synthesis in Escherichia coli. J. Bacteriol. 172, 659–669.
Ton, J., De Vos, M., Robben, C., Buchala, A., Métraux, J.-P., Van Loon, L. C., et al. (2002). Characterization of Arabidopsis enhanced disease susceptibility mutants that are affected in systemically induced resistance. Plant J. 29, 11–21.
Tran, T.-K., Han, Q.-Q., Shi, Y., and Guo, L. (2016). A comparative proteomic analysis of Salmonella typhimurium under the regulation of the RstA/RstB and PhoP/PhoQ systems. Biochim. Biophys. Acta 1864, 1686–1695. doi: 10.1016/j.bbapap.2016.09.003
Uehara, T., Sugiyama, S., Matsuura, H., Arie, T., and Masuta, C. (2010). Resistant and susceptible responses in tomato to cyst nematode are differentially regulated by salicylic acid. Plant Cell Physiol. 51, 1524–1536. doi: 10.1093/pcp/pcq109
Üstün, Ş., Müller, P., Palmisano, R., Hensel, M., Börnke, F., Ustün, S., et al. (2012). SseF, a type III effector protein from the mammalian pathogen Salmonella enterica, requires resistance-gene-mediated signalling to activate cell death in the model plant Nicotiana benthamiana. New Phytol. 194, 1046–1060. doi: 10.1111/j.1469-8137.2012.04124.x
Wall, D. M., Nadeau, W. J., Pazos, M. A., Shi, H. N., Galyov, E. E., and McCormick, B. A. (2007). Identification of the Salmonella enterica serotype Typhimurium SipA domain responsible for inducing neutrophil recruitment across the intestinal epithelium. Cell. Microbiol. 9, 2299–2313. doi: 10.1111/j.1462-5822.2007.00960.x
Wang, Q., and Harshey, R. M. (2009). Rcs signalling-activated transcription of rcsA induces strong anti-sense transcription of upstream fliPQR flagellar genes from a weak intergenic promoter: regulatory roles for the anti-sense transcript in virulence and motility. Mol. Microbiol. 74, 71–84. doi: 10.1111/j.1365-2958.2009.06851.x
Wang, Q., Zhao, Y., McClelland, M., and Harshey, R. M. (2007). The RcsCDB signaling system and swarming motility in Salmonella enterica serovar typhimurium: dual regulation of flagellar and SPI-2 virulence genes. J. Bacteriol. 189, 8447–8457. doi: 10.1128/JB.01198-07
Warnecke, D., and Heinz, E. (2003). Recently discovered functions of glucosylceramides in plants and fungi. Cell. Mol. Life Sci. 60, 919–941. doi: 10.1007/s00018-003-2243-4
Wiedemann, A., Virlogeux-Payant, I., Chaussé, A.-M., Schikora, A., and Velge, P. (2014). Interactions of Salmonella with animals and plants. Front. Microbiol. 5:791. doi: 10.3389/fmicb.2014.00791
Winfield, M. D., and Groisman, E. A. (2003). Role of nonhost environments in the lifestyles of Salmonella and Escherichia coli. Appl. Environ. Microbiol. 69, 3687–3694.
Winson, M. K., Swift, S., Hill, P. J., Sims, C. M., Griesmayr, G., Bycroft, B. W., et al. (1998). Engineering the luxCDABE genes from Photorhabdus luminescens to provide a bioluminescent reporter for constitutive and promoter probe plasmids and mini-Tn5 constructs. FEMS Microbiol. Lett. 163, 193–202. doi: 10.1016/S0378-1097(98)00173-6
Worley, M. J., Ching, K. H. L., and Heffron, F. (2000). Salmonella SsrB activates a global regulon of horizontally acquired genes. Mol. Microbiol. 36, 749–761. doi: 10.1046/j.1365-2958.2000.01902.x
Xu, X., and Hensel, M. (2010). Systematic analysis of the SsrAB virulon of Salmonella enterica. Infect. Immun. 78, 49–58. doi: 10.1128/IAI.00931-09
Zhang, Y., Higashide, W. M., McCormick, B. A., Chen, J., and Zhou, D. (2006). The inflammation-associated Salmonella SopA is a HECT-like E3 ubiquitin ligase. Mol. Microbiol. 62, 786–793. doi: 10.1111/j.1365-2958.2006.05407.x
Zhang, Y., Wu, S., Ma, J., Xia, Y., Ai, X., and Sun, J. (2015). Bacterial protein AvrA stabilizes intestinal epithelial tight junctions via blockage of the C-Jun N-terminal kinase pathway. Tissue Barriers 3:e972849. doi: 10.4161/21688362.2014.972849
Keywords: Salmonella, SrfJ, type III secretion, myo-inositol, macrophages, plants, bioluminescence
Citation: Aguilera-Herce J, Zarkani AA, Schikora A and Ramos-Morales F (2017) Dual Expression of the Salmonella Effector SrfJ in Mammalian Cells and Plants. Front. Microbiol. 8:2410. doi: 10.3389/fmicb.2017.02410
Received: 21 July 2017; Accepted: 21 November 2017;
Published: 06 December 2017.
Edited by:
Xihui Shen, Northwest A&F University, ChinaReviewed by:
Dipshikha Chakravortty, Indian Institute of Science, IndiaMiguel A. De la Cruz, Instituto Mexicano de Seguridad Social (IMSS), Mexico
Eleonora Garcia Vescovi, Institute of Molecular and Cellular Biology of Rosario (IBR), Argentina
Copyright © 2017 Aguilera-Herce, Zarkani, Schikora and Ramos-Morales. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Francisco Ramos-Morales, framos@us.es
 Julia Aguilera-Herce
Julia Aguilera-Herce Azhar A. Zarkani2
Azhar A. Zarkani2 Adam Schikora
Adam Schikora Francisco Ramos-Morales
Francisco Ramos-Morales