- 1State Key Laboratory for Diagnosis and Treatment of Infectious Diseases, Collaborative Innovation Center for Diagnosis and Treatment of Infectious Diseases, First Affiliated Hospital of Medical School, College of Medicine, Zhejiang University, Hangzhou, China
- 2Department of Infectious Diseases, Zhejiang Provincial People's Hospital, People's Hospital of Hangzhou Medical College, Hangzhou, China
- 3Division of Hepatobiliary and Pancreatic Surgery, Department of Surgery, First Affiliated Hospital, School of Medicine, Zhejiang University, Hangzhou, China
Transmissible colistin resistance mediated by the mcr gene has been reported worldwide, but clinical isolates of mcr-negative colistin-resistant Escherichia coli are rarely reported. The aim of this study was to evaluate the mechanism of colistin resistance among mcr-positive and mcr-negative E. coli clinical isolates by performing a molecular epidemiological surveillance. For the first time ever, we show nearly the same isolation ratio for mcr-negative and mcr-positive colistin-resistant clinical isolates (47.5 and 52.5%, respectively), with no demonstrable nosocomial transmission. We provide evidence for the prevalence of the mcr-positive IncX4 plasmid and its high potential for horizontal transfer, with no obvious sequence type (ST) preference. In addition, the minimal inhibitory concentrations (MICs) of colistin of the mcr-negative E. coli isolates were obviously higher than those of mcr-positive isolates. Apart from the usually detected genes, i.e., pmrAB, phoPQ, and mgrB, other genes may be associated with the colistin resistance in mcr-negative E. coli. To the best of our knowledge, this is the first paper to report the molecular epidemiological surveillance and the proper mechanism of colistin resistance in mcr-negative E. coli clinical isolates. Together, the results show that colistin resistance was prevalent not only in the mcr-positive clinical E. coli isolates but also in the mcr-negative isolates.
Introduction
Polymyxin, recently reintroduced in human medicine practice, constitutes one of the last resorts for the treatment of multidrug-resistant Gram-negative bacteria (Nation et al., 2014). Colistin (polymyxin E), with pronounced antimicrobial activity against Gram-negative bacteria, is a cyclic polycationic peptide that interacts with anionic lipopolysaccharide (LPS) molecules (Olaitan et al., 2014). Even though LPS is the initial target, the exact mechanism underpinning the effect of polymyxins on Gram-negative bacteria remains unclear (Poirel et al., 2017).
A plasmid-mediated colistin resistance gene, mcr-1, harbored by Escherichia coli and Klebsiella pneumoniae isolated from animals and hospital inpatients, was first reported in China (Liu et al., 2016). Then it became popular all over the world (Rapoport et al., 2016; Rolain et al., 2016; Sonnevend et al., 2016), demonstrating a horizontal transfer mechanism for colistin resistance. Additional novel plasmid-encoded colistin resistance genes were identified as well: mcr-2, identified in E. coli isolates and sharing 76.7% of nucleotide identity with mcr-1 (Liassine et al., 2016; Xavier et al., 2016); and mcr-3, identified in porcine E. coli isolates and sharing 45.0% of nucleotide sequence identity with mcr-1 (Yin et al., 2017). These genes encode a phosphoethanolamine transferase family protein that modifies the lipid A component of LPS (Liassine et al., 2016; Yin et al., 2017) and confers a low level of colistin resistance (MIC = 4–8 mg/L) (Liu et al., 2016).
Of note, the mcr-1 gene was found to co-occur with other antibiotic resistance genes, such as ESBL and NDM (Yang R. S. et al., 2016; Yang Y. Q. et al., 2016; Zheng et al., 2016), raising the likelihood of a serious bacterial infection that would be difficult to cure and contain. More importantly, a transposon element identified in mcr-1–harboring plasmids may expedite its mobilization to different plasmid backbones (Bernasconi et al., 2016; Doumith et al., 2016), and between bacterial strains and species, further propelling drug resistance in Gram-negative bacteria. Thus, far, the mcr-1 gene has been found in different plasmid incompatibility (Inc) groups, including IncI2, X4, F, HI2, and P (Fernandes et al., 2016; Sun et al., 2016; Yang Q. E. et al., 2016). In pHNSHP45, the first reported mcr-1–harboring plasmid, ISApl1 is inserted directly upstream of the mcr-1 gene (Petrillo et al., 2016). However, ISApl1 is absent in several contigs on IncI2, IncX4, and IncP plasmids, while different plasmid backbones contain a horizontally-transferred 2.6-kb sequence nearly identical to mcr-1-pap2 (Li et al., 2016).
Before the discovery of mcr, clinical polymyxin resistance mechanisms were reported to be associated with chromosomal mutations or IS sequence insertions. A qualitative modification of LPS involves a large panel of genes and operons, including genes and operons that are directly involved in LPS modifications, i.e., the pmrC and pmrE genes; regulatory genes involved in two-component systems, i.e., the pmrAB and phoPQ genes; and regulators of these two-component systems, i.e., the mgrB gene (also called yobG), whose product negatively regulates the PhoPQ system (Poirel et al., 2017). It has been suggested that PmrAB or PhoPQ two-component systems that are associated with LPS modification may play an important regulatory role in colistin or polymyxin B resistance in several Gram-negative pathogens (Adams et al., 2009; Schurek et al., 2009). Moreover, amino acid polymorphisms of PmrA and/or PmrB have been observed in clinical isolates of Acinetobacter baumannii and K. pneumoniae (Poirel et al., 2017). In addition, MgrB alteration is a common mechanism of colistin resistance among KPC-K. pneumoniae in clinical settings. Quesada et al. showed that mutations in PmrAB were associated with colistin or polymyxin B resistance in E. coli isolated from poultry and swine; they found two unique amino acid substitutions in colistin-resistant E. coli strains. Nevertheless, mcr-negative colistin-resistant clinical isolates of E. coli are rarely reported. Chromosomal mutations of the pmrAB, phoPQ, and mgrB genes that result in lipid A modification might be harbored by clinical isolates of E. coli, and could be responsible for their resistance.
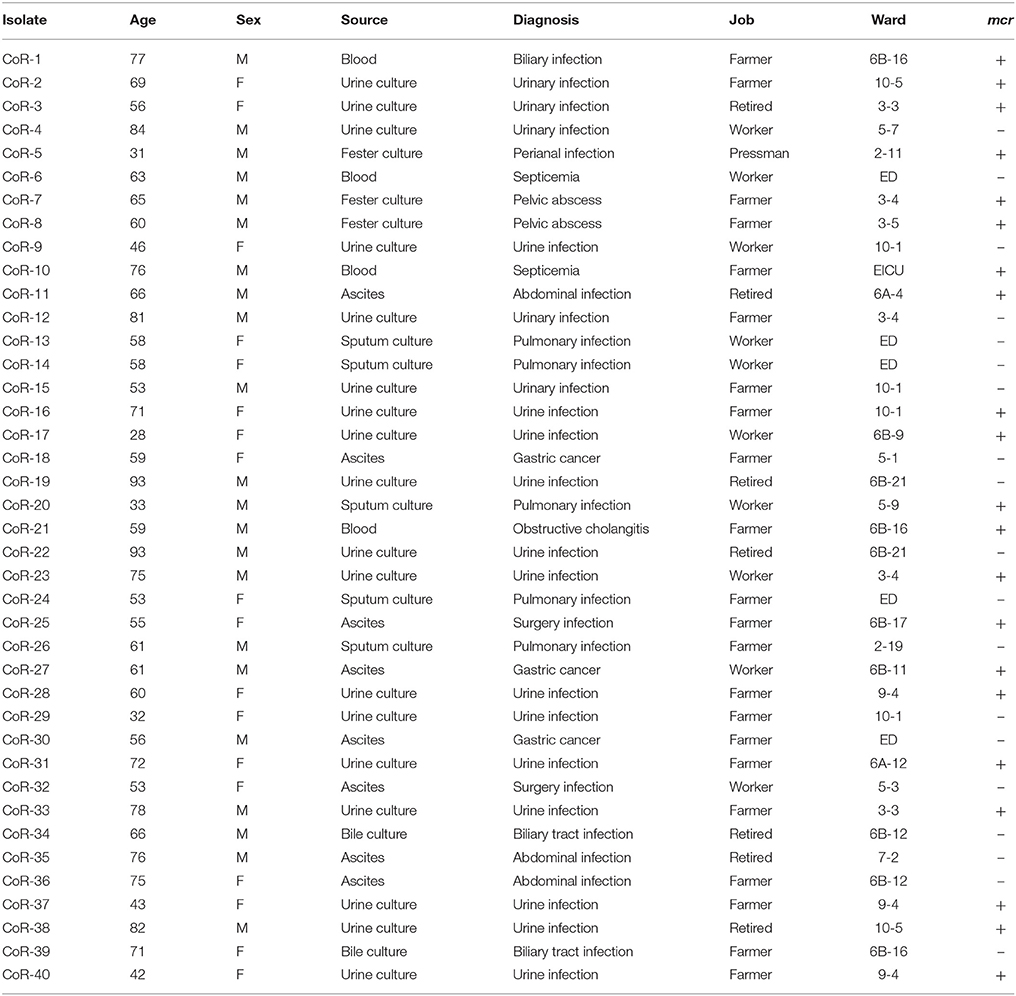
In the current study, we performed molecular epidemiological surveillance of colistin resistance in clinical isolates of E. coli, not only in mcr-positive isolates but also in mcr-negative isolates, and determined the mechanisms of resistance. We report a high incidence of medical records detailing colistin-resistant E. coli in the hospital. We identified 40 (3.15%) colistin-resistant isolates, including 21 mcr-positive genetically diverse isolates that all harbored mcr-1, but not mcr-2 or mcr-3; the mcr-1 gene was carried on plasmids of various sizes (ca. 30–270 kb), which were all readily transferrable to a recipient strain in vitro. Earlier studies presented evidence for the transmission of mcr genes from animals to human (Brauer et al., 2016; Liu et al., 2016). Here, we discovered that blaCTX−M−55 was the most common ESBL gene among the ESBL-producing mcr-positive isolates, which is another indication of a possible transmission of colistin resistance from animal to human. Among the ESBL-positive isolates unambiguously shown to harbor mcr-positive plasmid replicon groups, we demonstrated the prevalence and high potential for horizontal transfer of IncX4 plasmids of similar size and with the same genetic structure of mcr-positive elements. More significantly, 47.5% (19/40) of colistin-resistant E. coli isolates did not possess mcr, while their minimal inhibitory concentrations (MICs) for colistin were higher than those of an E. coli trans-conjugant harboring an mcr-positive plasmid. In our study, although five unique amino acid substitutions were identified, four in PmrB and one in PhoQ, two isolates did not have amino acids in PmrAB, PhoPQ, or MgrB that differed from those in sensitive isolates, indicating that other novel mechanisms were responsible for the colistin resistance of these isolates. Furthermore, the core genome phylogeny tree revealed that the mcr-positive and mcr-negative isolates were genotypically unrelated.
To the best of our knowledge, this study is the first-ever to report that a similar incidence of mcr-negative and mcr-positive colistin-resistant isolates (47.5 and 52.5%, respectively), with no demonstrated nosocomial transmission. In addition, we also report for the first time the molecular epidemiological surveillance for colistin resistance of mcr-negative E. coli clinical isolates as well as the mechanisms responsible for this resistance.
Materials and Methods
Patients and Bacterial Isolates
In total, 1,270 E. coli isolates were obtained from the First Affiliated Hospital (Zhejiang University, Hangzhou, China) between January, 2014, and March, 2015. The patients were from different counties. The isolates were identified using the MALDI-TOF technique. The work was compliant with the ethical guidelines of the Declaration of Helsinki, and written informed consent was obtained from all patients prior to enrollment in this study. All procedures involving human subjects were approved by the medical ethics committee of the First Affiliated Hospital (College of Medicine, Zhejiang University).
Clinical Data and MIC Testing
Patients who harbored colistin-resistant E. coli all dwell in Zhejiang Province; none of the patients were treated with colistin as an antimicrobial drug and none of the patients has ever traveled abroad (Table 1). Susceptibility testing to a wide range of antibiotics was routinely undertaken or performed using Vitek II (2017 CLSI). Colistin susceptibility was screened via the agar dilution method to determine MICs (Dafopoulou et al., 2015). The ESBL phenotype was confirmed by a standard double disc synergy test, as recommended by the CLSI. E. coli ATCC 25922 and K. pneumoniae ATCC 700603 were used as quality controls.
PCR Amplification, Amino Acid Variants and MLST
The mcr-harboring isolates were screened by PCR (using primers for mcr-1, mcr-2, and mcr-3; Table S1), and were validated by sequencing. The entire pmrAB, mgrB, and phoPQ genes were amplified and sequenced using specific oligonucleotide primers listed in the appendix (Table S1). Amino acid sequences of mcr-negative colistin-resistant isolates were compared with those of colistin-sensitive clinical isolates, and the reference strain E. coli K-12 MG1655. Sorting Intolerant From Tolerant (SIFT) scores (http://sift.jcvi.org) were calculated to predict whether amino acid substitutions in MgrB, PmrAB, and PhoPQ affect protein function. In addition, SMART analysis (http://smart.embl.de/) was performed to determine the domain architectures of PmrA, PmrB, PhoP, PhoQ, and MgrB. E. coli MLST was performed using the Warwick database (http://mlst.warwick.ac.uk/mlst/dbs/Ecoli). PCR amplification was employed to detect β-lactamase genes. The primers for PCR detection and sequencing were as described in a previous study (Zhang et al., 2016). The basic local alignment search tool (BLAST) was used to analyze the sequencing results.
Plasmid Analysis, Southern Hybridization, and PFGE
Conjugation assays were performed largely according to a method described previously (Wang et al., 2003). mcr-1–harboring E. coli was used as the donor, while E. coli J53 (sodium azide-resistant) served as the recipient strain. Trans-conjugants were selected on Mueller-Hinton agar supplemented with sodium azide (100 mg/L) and colistin (2 mg/L). PCR and DNA sequencing were used to detect the presence of mcr-1 gene in trans-conjugants; the colistin MICs of the conjugants were then determined. For the transformation experiments, plasmid DNA from mcr-negative isolates was prepared by alkaline lysis; the plasmid DNA was then electroporated into E. coli strain DH5α, and the bacteria plated on media containing 2 μg/mL of colistin.
To estimate the sizes of mcr-1 plasmids, S1-PFGE and Southern hybridization were performed. Briefly, bacterial whole cell DNA was prepared in agarose plugs and digested with S1 nuclease (TaKaRa, Dalian, China). The DNA was separated using the CHEF-MAPPER PFGE system (Bio-Rad) under the following conditions: 14°C, 6 V/cm, and a 120° pulse angle for 16 h, with the initial and final pulses conducted for 2.16 and 63.8 s, respectively. The separated DNA fragments were transferred to nylon membranes, hybridized with digoxigenin-labeled mcr-1–specific probes (primer mcr-1-SB-F/R; Table S1) and detected using the NBT/BCIP color detection kit (Roche, cat. no. 11745832910).
PFGE of mcr-positive isolates and their trans-conjugants were performed to exclude the occurrence of spontaneous sodium azide resistant mutants. Briefly, whole cell DNA of clinical strains and markers (Salmonella enterica serovar Braenderup H9812) in agar gels were digested by XbaI and then separated by pulsed field electrophoresis, under the same conditions as S1-PFGE.
Whole Genome Sequencing and Bioinformatics Analysis
Crude DNA extracts were prepared using QIAamp DNA mini kit (QIAGEN, cat. no. 51304) from overnight cultures of E. coli. The DNA was sequenced using the Illumina Hi-seq platform after library preparation. The quality-trimmed raw sequence data were assembled using Velvet 1.2.7. Prokka v 1.10 was used for the annotation of plasmid genes; PlasmidFinder (https://cge.cbs.dtu.dk/services/PlasmidFinder/) was used to detect plasmid Inc groups; Parsnp (https://github.com/marbl/parsnp) was used to align the core genome. The resistance genes were identified by performing BLASTn against the ResFinder 2.1 database (https://cge.cbs.dtu.dk/services/ResFinder/). Bioinformatics tools used in this work are available at the following web platforms: NCBI [National Center for Biotechnological Information, i.e., BRIG (BLAST Ring Image Generator)], SMS (Sequence Manipulation Suite), and EBI (European Bioinformatics Institute).
Nucleotide Sequence Accession Numbers
The whole genome sequences described in this paper have been deposited in DDBJ/ENA/GenBank under the accession numbers MKFK00000000-MKFS00000000, MKIH00000000-MKIL00000000, and IRU00000000- NISB00000000, and were in a processing queue (BioProject: PRJNA344524). The versions described in this paper are MKFK01000000-MKFS01000000, MKIH01000000-MKIL01000000, and NIRU00000000-NISB00000000.
Results
Detection of Colistin-Resistant Clinical Isolates and Their Clinical Characteristics
From 1,270 clinical E. coli isolates, we identified 40 isolates that exceeded the colistin resistance breakpoint (>2 mg/mL) using the agar dilution method (Table 1). Of these, 21 mcr-positive isolates were identified using the mcr-1–specific primers and were clearly colistin resistant. Full gene sequencing confirmed that all these 21 strains encoded MCR-1. The other 19 colistin-resistant isolates were found to have no mcr-1, mcr-2, or mcr-3 genes using specific primers (Table S1). These mcr-negative isolates had higher colistin MICs than those of the mcr-positive isolates (Table 2).
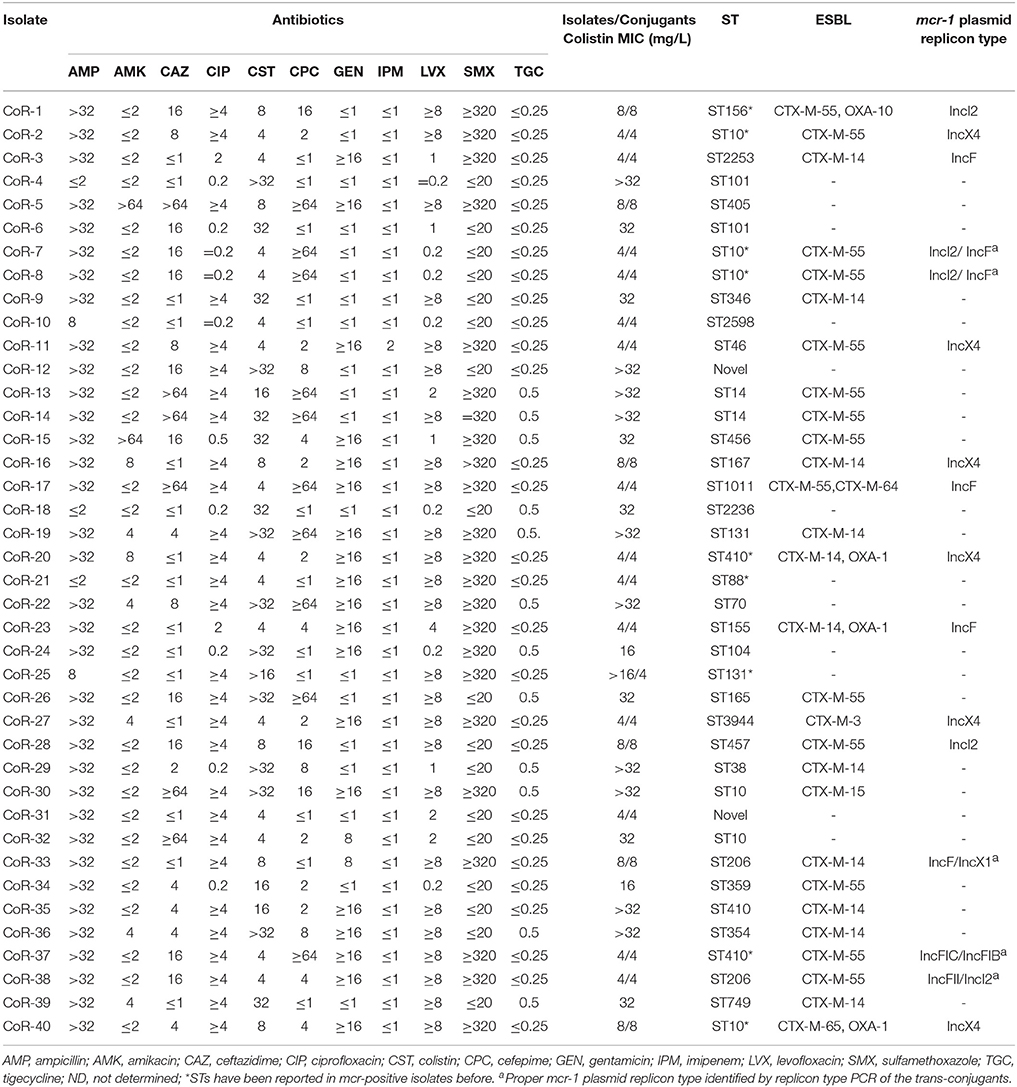
Table 2. Antibiotic susceptibilities (mg/L), sequence types (ST), ESBL genes, and mcr-1-harboring plasmid replicon types of E. coli isolates.
According to the epidemiological data of the 40 corresponding patients (Table 1), the patients ranged in age from 28 to 93 years. These samples obtained from culture included urine (n = 18), ascites (n = 8), blood (n = 4), sputum (n = 5), and other secretions (n = 5; Table 1). Sample sources were not distinguishably different between mcr-positive and -negative isolates. The isolates were obtained from in 16 wards and the patients were from multiple cities in the Zhejiang Province, China. Medical records were reviewed, and patient histories confirmed that they had not been prescribed polymyxin or traveled abroad previously. With the exception of two patients who died of abdominal infection and respiratory failure, all the other patients recovered. The patients received empirical antibiotic treatment during their hospitalization, including levofloxacin, amikacin, piperacillin/tazobactam, cefixime, imipenem, and meropenem.
Characteristics of the Antibiotic Susceptibility, ESBL Phenotypes, and Sequence Types (STs)
The MICs of the mcr-positive colistin-resistant isolates were 4–8 mg/L, except for CoR-25 (>16 mg/L) (Table 2). However, for the mcr-negative colistin-resistant isolates, the MICs were ≥16 mg/L, and for more than half of them (10/19), they were >32 mg/L (Table 2). Further, 28 of the 40 isolates (70%) were ESBL-positive, while none of them was carbapenem resistant. Only one strain, CoR-11, showed intermediate sensitivity to imipenem, while the rest of the isolates were susceptible. Gentamicin resistance was seen in 19/40 (47.5%) of the isolates, with two isolates displaying a high level of amikacin resistance (Table 2). High resistance ratios to ciprofloxacin (70%) and levofloxacin (62.5%) were seen in these isolates. The colistin resistant isolates retained susceptibility to tigecycline (Table 2). The observed antibiotic susceptibilities of mcr-positive and mcr-negative colistin-resistant isolates were not markedly different. Further, sequence analysis revealed that the ESBL genes were present in a wide variety of E. coli STs, and most ESBL-producing E. coli produced more than one type of β-lactamase. The most common ESBL gene was blaCTX−M−55 in the CTX-M-1 group (14/28, 50.0%), followed by blaCTX−M−14 (11/28, 39.3%) in the CTX-M-9 group.
MLST differentiated the 21 representative mcr-positive E. coli strains into 15 STs and one unknown ST (untypable). The most common ST was ST10 (n = 4), followed by ST410 (n = 2), and ST206 (n = 2), and then by single ST type isolates. MLST also differentiated the mcr-negative colistin-resistant isolates into many different STs, with no obvious dominant ST (Table 2). PFGE analysis of the colistin-resistant isolates was also performed to evaluate any clonal relationships between the mcr-positive and mcr-negative resistant strains. PFGE indicated that the isolates were not genetically related, and not nosocomially transmitted (Figure S1). Hence, colistin-resistant clinical isolates comprised a variety of STs and were therefore genetically different, with the nosocomial transmission excluded.
Plasmid Analysis and Southern Blotting
All plasmids harboring the mcr-1 gene were transferable to E. coli J53. The trans-conjugants were resistant to colistin, with the MIC of 4 or 8 mg/L, which was consistent with their mother strains, except for CoR-25 (4 mg/L, Table 2). The MIC of the CoR-25 conjugant was lower than that of CoR-25, a finding that might indicate that chromosomal PhoQ mutation contributed to the colistin resistance. In order to exclude the occurrence of spontaneous sodium azide resistant mutants in the trans-conjugants, PFGE analysis of mcr-positive isolates vs. the recipient cells were performed (Figure S1). The results indicated that the trans-conjugants were all from the recipient cells.
On the basis of the total DNA S1-PFGE and corresponding Southern hybridization, we determined that the mcr-1 gene was exclusively located on diverse plasmids, with plasmid sizes ranging from ca. 30 to 270 kb (Figure 1). Southern hybridization of CoR-11 and CoR-21 was unsuccessful for unknown reasons; however, the presence of mcr-1 was confirmed by PCR and the strains' mcr-positive plasmids were transferable and conferred colistin resistance. Further, 16 of the 21 E. coli isolates that carried mcr-1 were ESBL-positive (Table 2), and 14 of them were selected for whole genome sequencing. Whole genome analysis revealed three replicon types (IncX4, IncF, and IncI2) in the ESBL-producing isolates, which unambiguously confirmed mcr-1–harboring plasmid replicon groups (Table 2). IncX4 accounted for more than half of these plasmids (6/11), indicating the prevalence of mcr-1–harboring IncX4 in Zhejiang, China.
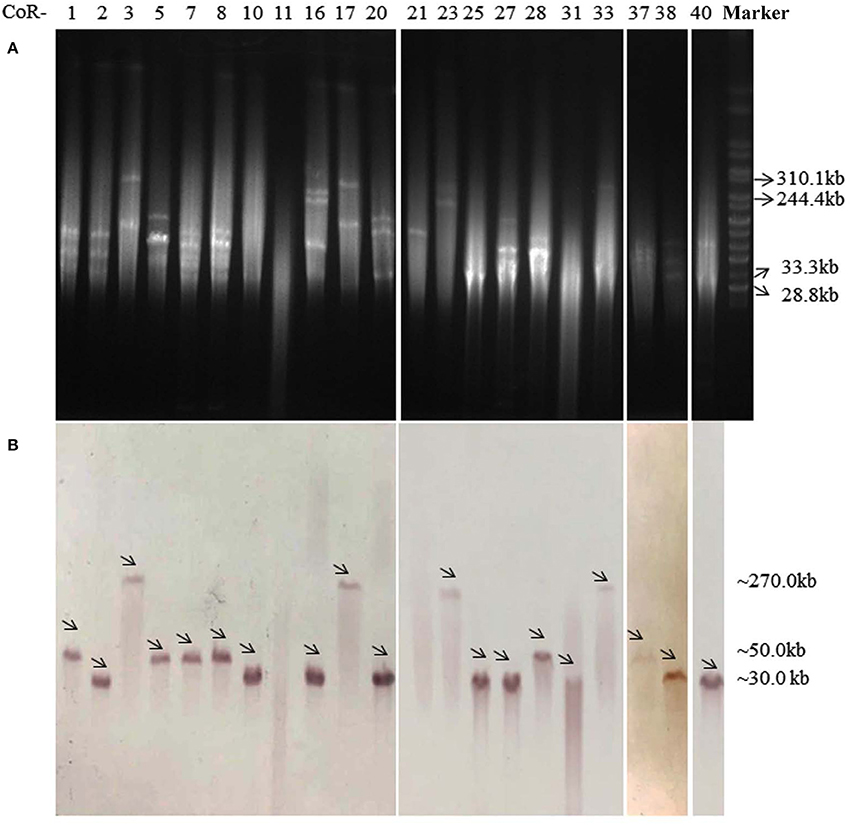
Figure 1. S1-PFGE profiles (A) and Southern blot (B) analysis with an mcr-1–specific probe of 21 mcr-1–harboring E. coli isolates, with Salmonella enterica serovar Branderup as a molecular mass marker (the last lane). The names of the isolates (“CoR-”) were shown in the first line of (A). The arrows in (B) indicated the locations of mcr-1-harboring plasmids according to Southern blot experiment. The mcr-1 Southern blot hybridization primers (mcr-1-SB-F/R) are shown in Table S1.
The Genetic Structure of mcr-1 in the mcr-Positive Isolates
The open reading frames (ORFs) in mcr-1–harboring plasmid contigs (ca. 9.3 kb) from CoR-2, CoR-11, CoR-16, CoR-20, CoR-27, and CoR-40 were all identical and belonging to the IncX4 incompatibility group. Sequence alignment revealed that these contigs were similar to those of other IncX4 plasmids, including pICBEC72Hmcr (Fernandes et al., 2016), from clinical isolates in Brazil, and pESTMCR (Brauer et al., 2016), isolated from a pig slurry in Estonia (Figure 2). The Southern hybridization data indicated that these IncX4 plasmids were all ca. 30 kb in length, suggesting that they were possibly from the same plasmid. The IncX4 plasmid was present in six phylogenetically different STs E. coli isolates, suggesting horizontal transfer, and no obvious ST preference. Interestingly, the insertion element ISApl1, initially found to be associated with mcr-1 in pHNSHP45, was not present in IncX4 plasmids in the current study.
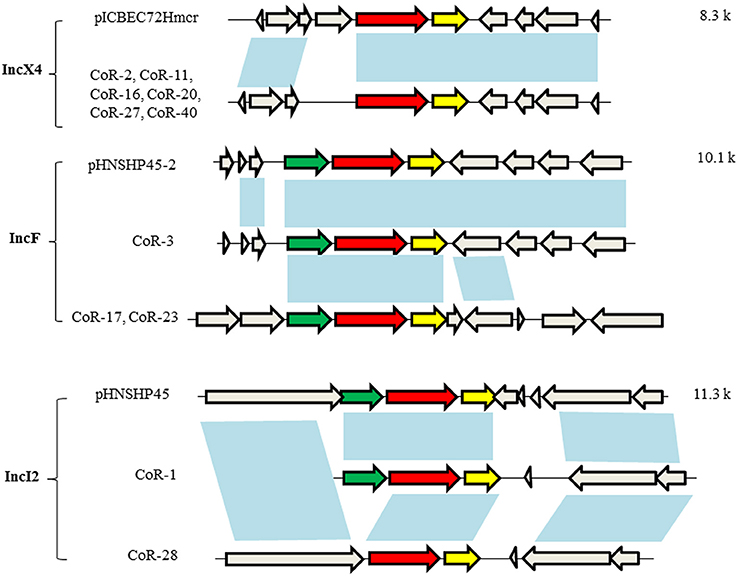
Figure 2. Genetic structures of the mcr-1–harboring elements of selected plasmids. Plasmid incompatibility groups are noted on the left, along with the plasmids and/or strains harboring the depicted genetic structure. Sequence lengths of the depicted areas are noted on the right. The arrows denote open reading frames (ORFs), with the red, yellow, green, and gray arrows denoting mcr-1, pap2, ISApl1, and the neighboring genes, respectively. The blue shading indicates the same ORF.
The mcr-1–harboring plasmid contigs from CoR-3, CoR-17, and CoR-23 belonged to the IncF group, i.e., the same plasmid incompatibility group as pHNSHP45-2. Sequence alignment indicated that the ca. 160-kb contig of the CoR-3 IncF plasmid shared 99% identity with pHNSHP45-2 (Figure 2). A comparison of plasmids from CoR-3, CoR-17, and CoR-23, and the reported plasmid pHNSHP45-2 is shown in Figure S2. Moreover, the ca. 177-kb contig from the CoR-17 IncF plasmid and the ca. 97-kb contig from the CoR-23 IncF plasmid were identical. In addition, ISApl1 was present in IncF plasmids (Figure 2). The mcr-1–harboring plasmid contigs from CoR-1 and CoR-28, belonging to the IncI2 group, were the same in the plasmid incompatibility group as pHNSHP45. A comparison of the mcr-1 genetic structure of CoR-1 and CoR-28 with that of pHNSHP45 revealed ORF differences (Figure 2, Figure S3). The mcr-1–associated ISApl1 element was not detected in the IncI2 plasmid from CoR-28 but it was present in the IncI2 plasmid from CoR-1 (Figure 2). Most importantly, an approximately 2.6-kb mcr-1–pap2 element was found to be shared by all mcr-1–harboring plasmids.
Amino Acid Variations of PmrAB, PhoPQ, and MgrB in mcr-Negative Isolates
To determine the exact colistin resistant mechanism in mcr-negative resistant isolates, we first had to eliminate the transferable resistant genes mediating colistin resistance. Therefore, conjugation and transformation experiments were done using the mcr-negative resistant isolates. We found that colistin-resistant phenotypes of all these strains were not transferable to E. coli J53. In the absence of a plasmid bearing the mcr gene, the mcr-negative colistin-resistant isolates may have acquired colistin resistance via chromosomal mutations in the mgrB, phoPQ, and, pmrAB genes that might confer lipid A modifications.
We next determined whether PmrAB, PhoPQ, and MgrB mutations conferred colistin resistance in the 10 mcr-negative isolates with high-MIC values (MICs > 32 mg/L). We randomly selected nine colistin-sensitive E. coli clinical isolates (S1–S9, Table 3) as negative controls, and used K-12 MG1655 as a reference. Many nucleotide variations were found, but most of them were synonymous; amino acid alterations were found at four sites in PmrA; 9 sites in PmrB; one site in PhoP; six sites of PhoQ; and one site of MgrB. While 11 amino acid substitutions were found in both colistin-resistant and sensitive isolates, only five amino acid substitutions were identified in the 10 mcr-negative resistant isolates with high MICs (Table 3). Most of the amino acid substitutions unique to the high-MIC isolates were found in PmrB; these were A118T, E123D, Y315F, and Y358N, located in the histidine kinases phosphatases domain (HAMP) and histidine kinase-like ATPases domain (HATPase_c), according to SMART analysis. N346K in the HATPase_c domain of PhoQ was also identified in one colistin-resistant isolate.
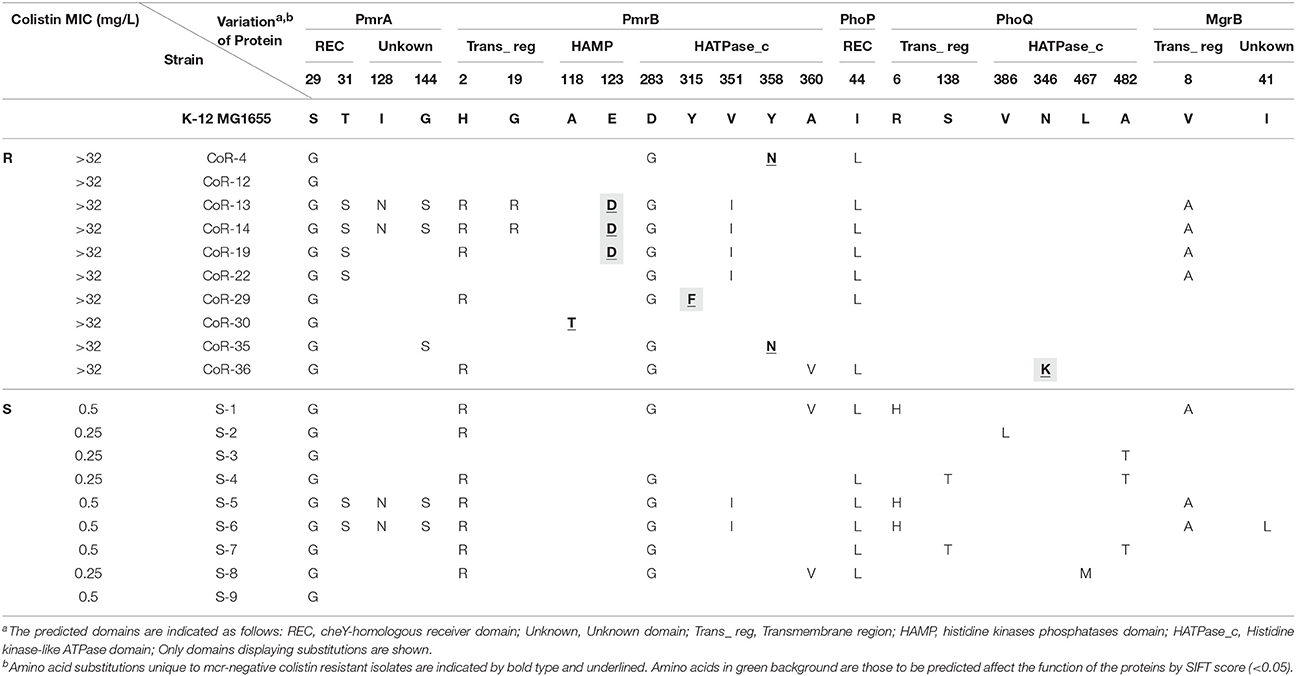
Table 3. Amino acid variations in functional domains of PmrAB, PhoPQ, and MgrB in clinical E. coli isolates.
Mutations in PmrB and PhoQ May Contribute to the Colistin-Resistant Phenotype of the mcr-Negative Isolates
Mutations in both PmrB and PhoQ might have affected phosphate transfer between the two system components since all of them were located in the kinase and phosphate-related domains. The E123D and Y315F substitutions in PmrA, and N346K in PhoQ were predicted to affect the function of proteins encoded by these genes, according to the SIFT score. However, no unique amino acid substitutions were identified in PmrA, PhoP, and MgrB in the 10 high-MIC value (MICs > 32 mg/L) colistin-resistant isolates, and neither frameshift mutations nor deletions were identified. Two of the 10 high-MIC value mcr-negative resistant isolates (CoR-12 and CoR-22) did not harbor amino acid variations in these proteins that could be used to differentiate them from the sensitive isolates. This indicated that a change in gene expression or other gene mutations contribute to colistin resistance in E. coli.
Interestingly, the phoQ allele of CoR-25 contained a frameshift mutation (deletion of A at nucleotide position 9) corresponding to a change of Lys to Asn, resulting in a premature stop at codon 24, with truncation of the PhoQ protein (Figure 3). This PhoQ frameshift mutation may have contributed to the high-level colistin resistance of the mcr-1–harboring isolate CoR-25.

Figure 3. Partial sequence alignment of the PhoQ protein in CoR-25 and the control strains. The controls share the same amino acid sequence; CoR-25 PhoQ harbors a K to N substitution at position 3, which results in a premature stop codon at codon 24 and truncation of the PhoQ protein.
The mcr-Positive and mcr-Negative Isolates Are Genotypically Unrelated
Parsnp was employed to compare the core genomes of the two groups or isolates (the mcr-positive and mcr-negative colistin-resistant isolates). The final Parsnp multiple alignment contained all SNPs, Indel scores, and structural variations within the core genome; 13 mcr-positive colistin-resistant isolates and 8 mcr-negative colistin-resistant isolates were selected for core genome alignments and phylogeny reconstruction (one of the 14 sequenced mcr-positive isolate genomes was so remote from the genomes of these isolates that we could not add it to the phylogenetic tree). The core genome phylogeny tree revealed that the mcr-positive and mcr-negative isolates were genotypically unrelated (Figure 4). The mcr-negative isolates were remotely related while 8 of the 13 mcr-positive isolates were clustered together, indicating that E. coli backgrounds enabling the acquisition of mcr-harboring plasmids might be to some extent core genome-specific, which is an observation worthy of further study.
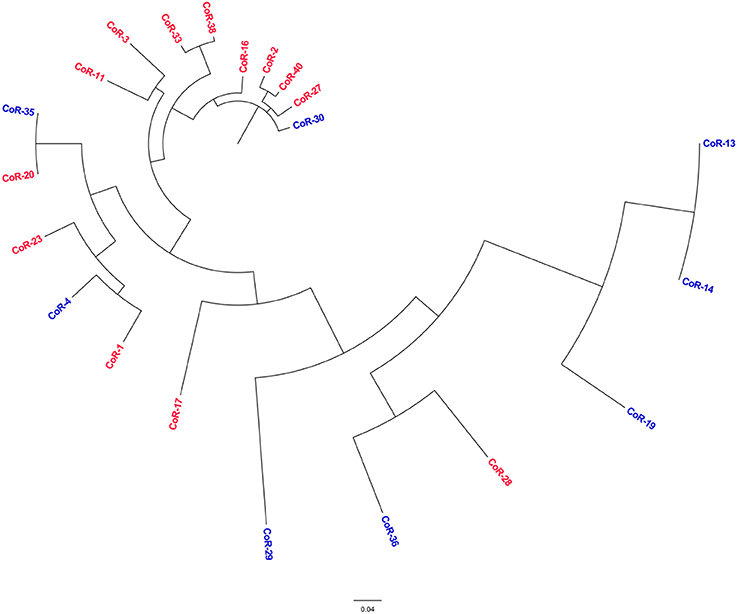
Figure 4. Phylogenetic tree analysis. The phylogeny of colistin-resistant clinical E. coli isolates based on the genotype at core genome SNPs. The characters in red correspond to the mcr-positive isolates, and in blue to the mcr-negative isolates.
Discussion
In China, where colistin is not used in the clinic for the treatment of infectious disease, the extensive use of antibiotics in food animal production, including swine and poultry farming, has unfortunately increased the risk of transfer of the resistant bacteria to human. In the current study, no patients had a history of colistin treatment. The spread of mcr-1 might be associated with the occupations of the patients, as more than half of the mcr-1–harboring patients were farmers. In China, some farmers use colistin to increase the quality of food animals, which they raise in their backyards. More importantly, the five reported mcr-1–harboring STs identified in the current study (156, 10, 410, 88, and 131) had been previously isolated from poultry and swine (Bai et al., 2016; Ewers et al., 2016; Falgenhauer et al., 2016; Yang R. S. et al., 2016). This included ST10, the most prevalent ST in the current study, which has also been isolated from travelers returning from India (Bernasconi et al., 2016) and from European animals (El Garch et al., 2016). The ST diversity of the mcr-1–harboring E. coli isolates in the present study indicated a scattered and non-clonal prevalence of mcr-1–harboring bacteria. Furthermore, blaCTX−M−55 is the dominant CTX-M-1 group gene in E. coli isolated from the environment and swine in China (Hu et al., 2013), while the CTX-M-15 and CTX-M-14 genes are harbored by the majority of clinical E. coli isolates in China (Liu et al., 2015). The high incidence of CTX-M-55 in the mcr-1–harboring clinical isolates observed in the current study provides further evidence for the possible transmission between the human and animals, via a mcr-1 gene–harboring pathogen or the gene itself. Therefore, the use of colistin could result in selective pressure in the farm environment and transmission of mcr genes to human.
Here, the genetic structures of the IncX4 contigs from six phylogenetically different strains were exactly the same, and the IncX4 plasmids had the same size, as determined by Southern hybridization. These findings may indicate that these plasmids have the same origin. However, the six isolates belonged to five different STs, implying high potential for horizontal transfer of the IncX4 plasmid. The discovery of mcr-1 gene associated with ISApl1 on a conjugative plasmid may signify its transfer to different plasmid backbones, as well as between different bacterial species and genera. Of note, in the current study, a nearly 2.6-kb mcr-1–pap2 element was found to be shared by all the sequenced mcr-1–carrying plasmids. Nevertheless, further investigation is needed to identify the mcr-1 transfer mechanism. Interestingly, 8 of the 13 mcr-positive colistin-resistant isolates clustered together, indicating that E. coli backgrounds that favor the acquisition of mcr-harboring plasmids may have some core genome in common. This should be further investigated with a large sample size.
In this study, none of the patients had a history of colistin treatment. This might indicate that the increasing use of colistin or polymyxin B in agriculture, livestock, and aquaculture has increased polymyxin resistance acquired via diverse mechanisms, i.e., not only by a horizontal transfer of mcr-positive plasmids but also by chromosomal mutations that confer high-MIC resistance in the absence of mcr. Overall, the colistin resistance of the clinical isolates was not only associated with mcr-1 but also with chromosomal mutations, although the patient histories confirmed that they had not previously received polymyxin treatment. It is of note that the mcr-negative colistin-resistant isolates were characterized by higher MICs than the mcr-positive isolates, posing more challenging in clinical treatment. The mcr-negative colistin-resistant isolates had higher MICs than the mcr-positive isolates, which will undoubtedly render clinical treatment more challenging. Although, chromosomal mutations cannot be horizontally transferred, the high incidence of mcr-negative colistin-resistant isolates cannot be ignored.
Mutations leading to the addition of cationic groups to lipid A result in a less anionic lipid A and changes leading to inactivation of lipid A biosynthesis result in a complete loss of LPS; consequently, this leads to reduced colistin binding or loss of the colistin target (Poirel et al., 2017). Several chromosomally encoded genes and operons are associated with colistin resistance in Gram-negative bacteria, such as PmrA/PmrB and PhoP/PhoQ two-component systems and the mgrB gene, which encodes a negative regulator of PhoPQ. The mechanisms of colistin resistance can be determined by sequencing these genes to detect mutations. Specific mutations in the PmrAB and PhoPQ two-component systems are associated with colistin resistance in some bacteria, e.g., P. aeruginosa. Inactivation of the MgrB regulator was reported to be associated with colistin resistance of K. pneumoniae. In the current study, we identified 19 highly colistin-resistant mcr-1 negative isolates (MIC ≥16 mg/L). The high level of resistance detected among clinical isolates in this study suggests that the resistance was a consequence of chromosomal mutations. Quesada et al. showed that mutations in PmrAB are associated with colistin or polymyxin B resistance in E. coli isolated from poultry and swine. The authors found two unique amino acid mutations in colistin-resistant E. coli strains, R81S in PmrA and V161G in PmrB. In this study, we identified five amino acid substitutions unique to mcr-negative colistin-resistant isolates, four in PmrB and one in PhoQ. These mutations have not been identified in previous studies. Mutations in both PmrB and PhoQ might have affected phosphate transfer between the two system components since all of them were located in the kinase and phosphate-related domains.
Remarkably, since a loss-of-function mutation of phoQ can contribute to high-level polymyxin resistance in clinical strains (Miller et al., 2011), the PhoQ frameshift mutation might have contributed to the high colistin resistance of CoR-25, one of the mcr-1–harboring isolates that exhibited high colistin resistance. To the best of our knowledge, this is the first-ever description of a mcr-positive E. coli where a chromosome mutation most likely confers a high level of colistin resistance.
Although PmrAB and PhoPQ may be responsible for most polymyxin resistance in Gram-negative pathogens, the mechanisms of polymyxin resistance appear to be highly diverse. Among the 10 high-MIC mcr-negative resistant isolates, two isolates (CoR-12 and CoR-22) did not harbor amino acid variations in PmrAB, PhoPQ, or MgrB that could be used to differentiate them from the sensitive isolates. Thus, amino acid changes in PmrAB, PhoPQ, and MgrB might not be essential for colistin resistance in E. coli. However, many chromosomally-encoded mechanisms of resistance remain to be identified in E. coli. In A. baumannii, colistin resistance can be induced in vitro by serial passages in media containing increasing colistin concentrations and may be mediated by diverse mechanisms. However, it is difficult to be sure that the amino acid substitutions found in genes involved in LPS biosynthesis in clinical isolates with complicated genomic backgrounds lead to resistance. On the other hand, resistance could depend on the expression levels of the genes involved in colistin resistance in E. coli. Therefore, we explored the relative expression levels of pmrAB, phoPQ, and mgrB genes in 10 high-MIC mcr-negative resistant isolates and compared them with those of 9 mcr-negative colistin sensitive clinical isolates (S1–S9). Unfortunately, the expression levels varied considerably, and there were no differences that could be used to distinguish the colistin resistant isolates from the colistin sensitive isolates (data not shown). Thus, the gene expression changes that might contribute to colistin resistance in E. coli are difficult to determine in clinical isolates but not in the parental strain that underwent serial passages in media with increasing concentrations of colistin. Other mechanisms of colistin resistance are conceivable and will be investigated in a future study.
Author Contributions
Conceived and designed the experiments: QL, YX, and LL. Performed the experiments: QL, WY, CH, HX, PS, HL, and SX. Analyzed the data: QL, KZ, and LG. Wrote the paper: QL.
Funding
This work was supported by the 973 National Key Basic Research Programs of China (2015CB554201), National Natural Science Foundation of China 81361138021, Mega-projects of Science Research of China (2013ZX10004217) and the Open Project Program of Jiangsu Key Laboratory of Zoonosis (No. 1606). Medical health science and technology plan projects in Zhejiang province (2017KY343).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Yunbo Chen, Jinru Ji and Yunhui Fang, for the support of technical support and quality control.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2017.02262/full#supplementary-material
References
Adams, M. D., Nickel, G. C., Bajaksouzian, S., Lavender, H., Murthy, A. R., Jacobs, M. R., et al. (2009). Resistance to colistin in Acinetobacter baumannii associated with mutations in the PmrAB two-component system. Antimicrob. Agents Chemother. 53, 3628–3634. doi: 10.1128/AAC.00284-09
Bai, L., Hurley, D., Li, J., Meng, Q., Wang, J., Fanning, S., et al. (2016). Characterisation of multidrug-resistant Shiga toxin-producing Escherichia coli cultured from pigs in China: co-occurrence of extended-spectrum β-lactamase- and mcr-1-encoding genes on plasmids. Int. J. Antimicrob. Agents 48, 445–448. doi: 10.1016/j.ijantimicag.2016.06.021
Bernasconi, O. J., Kuenzli, E., Pires, J., Tinguely, R., Carattoli, A., Hatz, C., et al. (2016). Travelers can import colistin-resistant enterobacteriaceae, including those possessing the plasmid-mediated mcr-1 gene. Antimicrob. Agents Chemother. 60, 5080–5084. doi: 10.1128/AAC.00731-16
Brauer, A., Telling, K., Laht, M., Kalmus, P., Lutsar, I., Remm, M., et al. (2016). Plasmid with colistin resistance gene mcr-1 in ESBL-producing Escherichia coli strains isolated from pig slurry in Estonia. Antimicrob. Agents Chemother. 60, 6933–6936. doi: 10.1128/AAC.00443-16
Dafopoulou, K., Zarkotou, O., Dimitroulia, E., Hadjichristodoulou, C., Gennimata, V., Pournaras, S., et al. (2015). Comparative evaluation of colistin susceptibility testing methods among carbapenem-nonsusceptible Kiebsiella pneumoniae and Acinetobacter baumannii clinical isolates. Antimicrob. Agents Chemother. 59, 4625–4630. doi: 10.1128/AAC.00868-15
Doumith, M., Godbole, G., Ashton, P., Larkin, L., Dallman, T., Day, M., et al. (2016). Detection of the plasmid-mediated mcr-1 gene conferring colistin resistance in human and food isolates of Salmonella enterica and Escherichia coli in England and Wales. J. Antimicrob. Chemother. 71, 2300–2305. doi: 10.1093/jac/dkw093
El Garch, F., Sauget, M., Hocquet, D., Lechaudee, D., Woehrle, F., and Bertrand, X. (2016). mcr-1 is borne by highly diverse Escherichia coli isolates since 2004 in food-producing animals in Europe. Clin. Microbiol. Infect. 23, 51.e1–51.e4. doi: 10.1016/j.cmi.2016.08.033
Ewers, C., Göttig, S., Bülte, M., Fiedler, S., Tietgen, M., Leidner, U., et al. (2016). Genome sequence of avian Escherichia coli strain IHIT25637, an extraintestinal pathogenic E. coli strain of ST131 encoding colistin resistance determinant MCR-1. Genome Announc. 4, e00863–16 doi: 10.1128/genomeA.00863-16
Falgenhauer, L., Waezsada, S. E., Gwozdzinski, K., Ghosh, H., Doijad, S., Bunk, B., et al. (2016). Chromosomal locations of mcr-1 and bla CTX-M-15 in fluoroquinolone-resistant Escherichia coli ST410. Emerg. Infect. Dis. 22, 1689–1691. doi: 10.3201/eid2209.160692
Fernandes, M. R., McCulloch, J. A., Vianello, M. A., Moura, Q., Perez-Chaparro, P. J., Esposito, F., et al. (2016). First report of the globally disseminated IncX4 plasmid carrying the mcr-1 gene in a colistin-resistant Escherichia coli ST101 isolated from a human infection in Brazil. Antimicrob. Agents Chemother. 60, 6415–6417. doi: 10.1128/AAC.01325-16
Hu, Y. Y., Cai, J. C., Zhou, H. W., Chi, D., Zhang, X. F., Chen, W. L., et al. (2013). Molecular typing of CTX-M-producing Escherichia coli isolates from environmental water, swine feces, specimens from healthy humans, and human patients. Appl. Environ. Microbiol. 79, 5988–5996. doi: 10.1128/AEM.01740-13
Li, A., Yang, Y., Miao, M., Chavda, K. D., Mediavilla, J. R., Xie, X., et al. (2016). complete sequences of mcr-1-harboring plasmids from extended-spectrum-beta-lactamase- and carbapenemase-producing Enterobacteriaceae. Antimicrob. Agents Chemother. 60, 4351–4354. doi: 10.1128/AAC.00550-16
Liassine, N., Assouvie, L., Descombes, M. C., Tendon, V. D., Kieffer, N., Poirel, L., et al. (2016). Very low prevalence of MCR-1/MCR-2 plasmid-mediated colistin resistance in urinary tract Enterobacteriaceae in Switzerland. Int. J. Infect. Dis. 51, 4–5. doi: 10.1016/j.ijid.2016.08.008
Liu, H., Wang, Y., Wang, G., Xing, Q., Shao, L., Dong, X., et al. (2015). The prevalence of Escherichia coli strains with extended spectrum beta-lactamases isolated in China. Front. Microbiol. 6:335. doi: 10.3389/fmicb.2015.00335
Liu, Y. Y., Wang, Y., Walsh, T. R., Yi, L. X., Zhang, R., Spencer, J., et al. (2016). Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect. Dis. 16, 161–168. doi: 10.1016/S1473-3099(15)00424-7
Miller, A. K., Brannon, M. K., Stevens, L., Johansen, H. K., Selgrade, S. E., Miller, S. I., et al. (2011). PhoQ mutations promote lipid A modification and polymyxin resistance of Pseudomonas aeruginosa found in colistin-treated cystic fibrosis patients. Antimicrob. Agents Chemother. 55, 5761–5769. doi: 10.1128/AAC.05391-11
Nation, R. L., Velkov, T., and Li, J. (2014). Colistin and polymyxin B: peas in a pod, or chalk and cheese? Clin. Infect. Dis. 59, 88–94. doi: 10.1093/cid/ciu213
Olaitan, A. O., Morand, S., and Rolain, J. M. (2014). Mechanisms of polymyxin resistance: acquired and intrinsic resistance in bacteria. Front. Microbiol. 5:643. doi: 10.3389/fmicb.2014.00643
Petrillo, M., Angers-Loustau, A., and Kreysa, J. (2016). Possible genetic events producing colistin resistance gene mcr-1. Lancet Infect. Dis. 16, 280. doi: 10.1016/S1473-3099(16)00005-0
Poirel, L., Jayol, A., and Nordmann, P. (2017). Polymyxins: antibacterial activity, susceptibility testing, and resistance mechanisms encoded by plasmids or chromosomes. Clin. Microbiol. Rev. 30, 557–596. doi: 10.1128/CMR.00064-16
Rapoport, M., Faccone, D., Pasteran, F., Ceriana, P., Albornoz, E., Petroni, A., et al. (2016). First description of mcr-1-mediated colistin resistance in human infections caused by Escherichia coli in Latin America. Antimicrob. Agents Chemother. 60, 4412–4413. doi: 10.1128/AAC.00573-16
Rolain, J. M., Kempf, M., Leangapichart, T., Chabou, S., Olaitan, A. O., Le Page, S., et al. (2016). Plasmid-mediated mcr-1 gene in colistin-resistant clinical isolates of Klebsiella pneumoniae in France and Laos. Antimicrob. Agents Chemother. 60, 6994–6995. doi: 10.1128/AAC.00960-16
Schurek, K. N., Sampaio, J. L. M., Kiffer, C. R. V., Sinto, S., Mendes, C. M. F., and Hancock, R. E. W. (2009). Involvement of pmrAB and phoPQ in polymyxin B adaptation and inducible resistance in non-cystic fibrosis clinical isolates of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 53, 4345–4351. doi: 10.1128/AAC.01267-08
Sonnevend, Á., Ghazawi, A., Alqahtani, M., Shibl, A., Jamal, W., Hashmey, R., et al. (2016). Plasmid-mediated colistin resistance in Escherichia coli from the Arabian Peninsula. Int. J. Infect. Dis. 50, 85–90. doi: 10.1016/j.ijid.2016.07.007
Sun, J., Li, X. P., Yang, R. S., Fang, L. X., Huo, W., Li, S. M., et al. (2016). Complete nucleotide sequence of an IncI2 plasmid coharboring blaCTX-M-55 and mcr-1. Antimicrob. Agents Chemother. 60, 5014–5017. doi: 10.1128/AAC.00774-16
Wang, M., Tran, J. H., Jacoby, G. A., Zhang, Y., Wang, F., and Hooper, D. C. (2003). Plasmid-mediated quinolone resistance in clinical isolates of Escherichia coli from Shanghai, China. Antimicrob. Agents Chemother. 47, 2242–2248. doi: 10.1128/AAC.47.7.2242-2248.2003
Xavier, B. B., Lammens, C., Ruhal, R., Kumar-Singh, S., Butaye, P., Goossens, H., et al. (2016). Identification of a novel plasmid-mediated colistin-resistance gene, mcr-2, in Escherichia coli, Belgium, June 2016. Euro Surveill. 21:30280. doi: 10.2807/1560-7917.ES.2016.21.27.30280
Yang, Q. E., Walsh, T. R., Liu, B. T., Zou, M. T., Deng, H., Fang, L. X., et al. (2016). Complete sequence of the FII plasmid p42-2, carrying blaCTX-M-55, oqxAB, fosA3, and floR from Escherichia coli. Antimicrob. Agents Chemother. 60, 4336–4338. doi: 10.1128/AAC.00475-16
Yang, R. S., Feng, Y., Lv, X. Y., Duan, J. H., Chen, J., Fang, L. X., et al. (2016). Emergence of NDM-5 and MCR-1-producing Escherichia coli clone ST648 and ST156 from a single muscovy duck (Cairina moschata). Antimicrob. Agents Chemother. 60, 6899–6902. doi: 10.1128/AAC.01365-16
Yang, Y. Q., Zhang, A. Y., Ma, S. Z., Kong, L. H., Li, Y. X., Liu, J. X., et al. (2016). Co-occurrence of mcr-1 and ESBL on a single plasmid in Salmonella enterica. J. Antimicrob. Chemother. 71, 2336–2338. doi: 10.1093/jac/dkw243
Yin, W., Li, H., Shen, Y., Liu, Z., Wang, S., Shen, Z., et al. (2017). Novel plasmid-mediated colistin resistance gene mcr-3 in Escherichia coli. MBio 8, e00543–17 doi: 10.1128/mBio.00543-17
Zhang, J., Zhou, K., Zheng, B., Zhao, L., Shen, P., Ji, J., et al. (2016). High prevalence of ESBL-producing Klebsiella pneumoniae causing community-onset infections in China. Front. Microbiol. 7:1830. doi: 10.3389/fmicb.2016.01830
Keywords: colistin, mcr-positive, mcr-negative, resistant mechanism, molecular epidemiology, clinical isolated E. coli
Citation: Luo Q, Yu W, Zhou K, Guo L, Shen P, Lu H, Huang C, Xu H, Xu S, Xiao Y and Li L (2017) Molecular Epidemiology and Colistin Resistant Mechanism of mcr-Positive and mcr-Negative Clinical Isolated Escherichia coli. Front. Microbiol. 8:2262. doi: 10.3389/fmicb.2017.02262
Received: 30 June 2017; Accepted: 02 November 2017;
Published: 17 November 2017.
Edited by:
Manuela Caniça, Istituto Nacional de Saúde, PortugalReviewed by:
Isabelle Kempf, Agence Nationale de Sécurité Sanitaire de l'Alimentation, de l'Environnement et du Travail (ANSES), FranceRaffaele Zarrilli, Department of Public Health, University of Naples Federico II, Italy
Copyright © 2017 Luo, Yu, Zhou, Guo, Shen, Lu, Huang, Xu, Xu, Xiao and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yonghong Xiao, xiao-yonghong@163.com
 Qixia Luo
Qixia Luo Wei Yu1,2
Wei Yu1,2 Kai Zhou
Kai Zhou Lihua Guo
Lihua Guo Hao Xu
Hao Xu Yonghong Xiao
Yonghong Xiao Lanjuan Li
Lanjuan Li