- MOH Key Laboratory of Systems Biology of Pathogens, Institute of Pathogen Biology, Chinese Academy of Medical Sciences and Peking Union Medical College, Beijing, China
The untranslated regions (UTRs) of mRNA contain important features that are relevant to the post-transcriptional and translational regulation of gene expression. Most studies of bacterial UTRs have focused on the 5′regions; however, 3′UTRs have recently emerged as a new class of post-transcriptional regulatory elements. 3′UTRs were found to regulate the decay and translation initiation in their own mRNAs. In addition, 3′UTRs constitute a rich reservoir of small regulatory RNAs, regulating target gene expression. In the current review, we describe several recently discovered examples of bacterial regulatory 3′UTRs, discuss their modes of action, and illustrate how they facilitate gene regulation in various environments.
Introduction
Environmental conditions experienced by free-living organisms, such as nutrient availability, temperature, oxygen, pH, and osmolarity, frequently change. To adapt to the ever-changing environment, bacteria have established an intricate network of regulators to accurately modulate gene expression. Regulation of gene expression in prokaryotes takes place primarily at the transcriptional level, i.e., by the activation or repression of transcription; however, post-transcriptional regulation of gene expression may play a critical role when bacteria must rapidly adjust to the changing environment.
mRNA functions primarily as a carrier of genetic information; however, its sequence, especially the 5′ and 3′untranslated regions (UTRs), contains many features that can modulate gene expression at the post-transcriptional level (Pesole et al., 2001). During the last few decades, significant progress has been made in the understanding of eukaryotic and prokaryotic 5′UTR-mediated gene regulation (Chen et al., 1991; Sonenberg, 1994; Agaisse and Lereclus, 1996; Oliva et al., 2015; Hinnebusch et al., 2016). RNA thermometers and riboswitches have been found in 5′UTRs and have been extensively studied in bacteria; these elements regulate gene in both transcriptional and translational levels, as well as mRNA stability (Henkin, 2008; Breaker, 2011; Kortmann and Narberhaus, 2012; Serganov and Patel, 2012; Krajewski and Narberhaus, 2014). In addition, RNA-binding proteins bind to 5′UTRs to regulate gene expression by modulating the accessibility of ribosome-binding sites (RBSs) on mRNAs (Babitzke et al., 2009; Van Assche et al., 2015).
In eukaryotes, 3′UTRs regulate gene transcription by modulating mRNA decay, translation, or localization, and these processes have been well studied (St Johnston, 1995; Pesole et al., 2001; Wilkie et al., 2003; Barreau et al., 2005). It was traditionally believed, however, that bacterial 3′UTRs mainly contain transcriptional terminators, which are either Rho-dependent or Rho-independent. Recently, 3′UTRs were found to be involved in post-transcriptional gene regulation in bacteria as well (Figure 1). 3′UTRs especially long 3′UTRs can be cleaved by ribonuclease to initiate mRNA decay (Figure 1A).
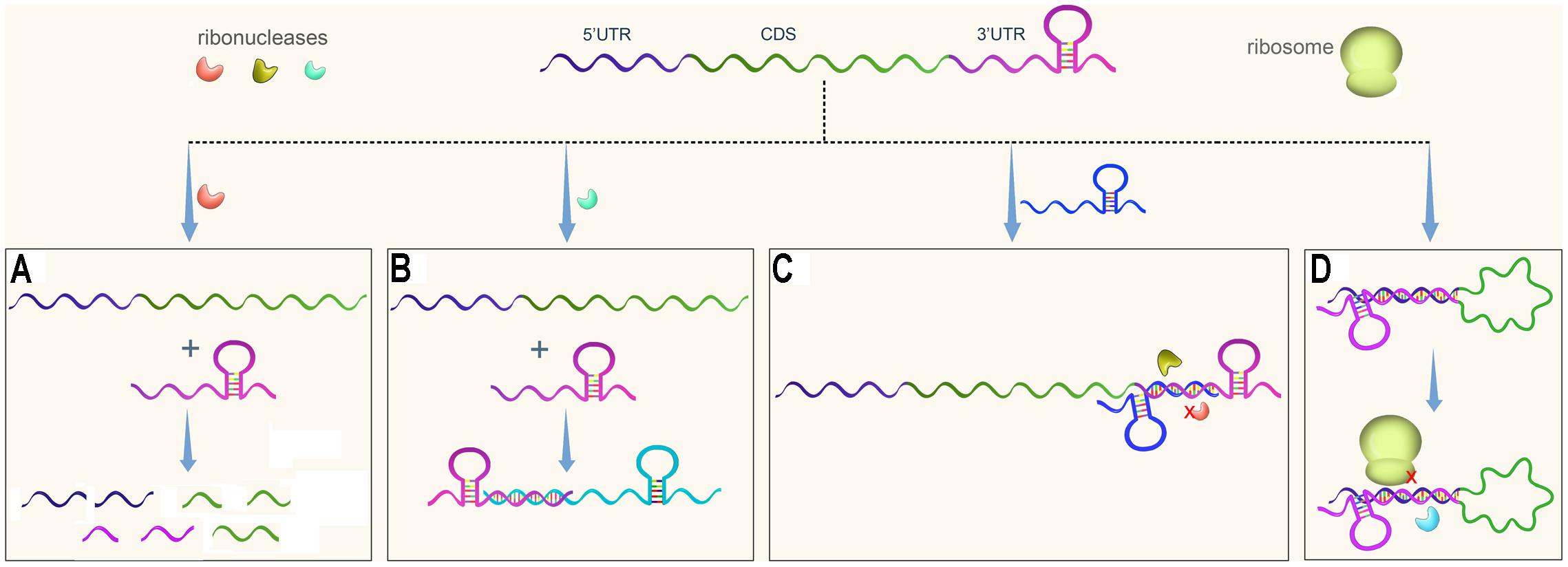
FIGURE 1. Regulatory pathways of 3′UTRs in bacteria. An mRNA usually contains three regions, the 5′UTR, the coding sequence (CDS), and the 3′UTR. (A) A 3′UTR is recognized and cleaved by ribonuclease to initiate mRNA decay. (B) A 3′UTR-derived sRNA regulates expression of its target gene. (C) A 3′UTR functions as the target of regulatory sRNA, resulting in protection of the 3′UTR from degradation or promoting its degradation. (D) A 3′ UTR interacts with the 5′ region of its own gene, potentially affecting mRNA stability and translational initiation.
3′UTRs are a rich reservoir of small regulatory RNAs and regulate target gene expression (Figure 1B). In addition, 3′UTRs are a target of regulatory sRNAs (Figure 1C). Finally, 3′UTRs interact with 5′UTRs to regulate translation initiation (Figure 1D). This paper summarizes several papers published in the last several years that suggest that 3′UTRs function as new post-transcriptional regulators. The regulatory mechanisms of 3′UTRs and how these genetic elements facilitate gene regulation in response to the changing environment are discussed.
3′UTRs Undergo Ribonuclease Cleavage
Regulation of mRNA decay is an important mechanism of post-transcriptional control of gene expression. In bacteria, mRNA decay depends on the rate-determining initial step (Lalaouna et al., 2013; Laalami et al., 2014). Following an initial endonucleolytic cleavage, mRNA fragments are subsequently degraded by exoribonucleases. Ribosomes shield mRNA from degradation by endonucleolytic enzymes (Deneke et al., 2013). Hence, available (untranslated) mRNA regions are more likely to be attacked by ribonucleases than translated mRNA regions. Selinger et al. (2003) analyzed the positional patterns of transcript degradation in Escherichia coli using subgenic-resolution oligonucleotide microarrays; they found that, although 5′UTRs are generally less stable than 3′UTRs, some 3′UTRs were also unstable. This indicated that mRNA decay might be initiated from the 3′UTRs as well (Selinger et al., 2003). Consistent with these microarray data, 3′UTR-mediated mRNA decay was reported recently (Maeda and Wachi, 2012; Lopez-Garrido et al., 2014; Liu et al., 2016; Zhu et al., 2016).
3′UTRs may be attacked by an endonuclease, initiating mRNA decay. One early example of this was the C. glutamicum aceA 3′UTR (Maeda and Wachi, 2012). The aceA gene encodes isocitratelyase, which catalyzes the cleavage of isocitrate to succinate and glyoxylate (Gerstmeir et al., 2003). The aceA 3′UTR is involved in negative regulation of its own gene expression and the expression of lacZ fusion (Maeda and Wachi, 2012). Further analysis showed that the aceA 3′UTR contains a single-stranded AU-rich region, which might constitute the cleavage target of RNase E/G (Maeda and Wachi, 2012). RNase E/G- and 3′UTR-mediated degradation of aceA mRNA may enable instantaneous adjustment of cellular metabolism (Maeda and Wachi, 2012).
The aceA 3′UTR contains only 63 nt (Maeda and Wachi, 2012); however, the subsequently identified 3′UTRs that are involved in mRNA decay are usually longer. The 3′UTR of hilD mRNA (encoding a transcriptional regulator of S. enterica pathogenicity island 1) contains 310 nt (Lopez-Garrido et al., 2014). The presence of the hilD 3′UTR reduces the expression of its own gene and the gfp reporter gene, indicating that the hilD 3′UTR functions as an independent module regulating gene expression (Lopez-Garrido et al., 2014). Further analysis showed that the hilD 3′UTR is a target during hilD mRNA degradation by a degradosome containing RNase E and PNPase (Lopez-Garrido et al., 2014).
Another example of mediation of gene expression by a long 3′UTR is the hmsT 3′UTR, which contains 283 nt (Zhu et al., 2016). The hmsT gene encodes a diguanylate cyclase that stimulates biofilm formation in Y. pestis by synthesizing the secondary messenger c-di-GMP (Kirillina et al., 2004). The hmsT 3′UTR negatively modulates hmsT mRNA decay, in which PNPase is involved. This 3′UTR strongly represses gene expression at 37°C, but only weakly affects gene expression at 21°C, suggesting that the temperature is a signal that can be sensed by the hmsT 3′UTR to regulate its gene expression under changing environmental conditions (Zhu et al., 2016).
Both hilD and aceA possess a Rho-independent terminator (Maeda and Wachi, 2012; Lopez-Garrido et al., 2014), while hmsT has a Rho-dependent terminator (Zhu et al., 2016). Recently, it was reported that Rho-dependent termination is required for PNPase-mediated turnover of slrA mRNA in Bacillus subtilis (Liu et al., 2016). Replacement of the Rho-dependent terminator by a Rho-independent terminator in the slrA gene eliminated the PNPase-mediated turnover of mRNA, indicating that the slrA 3′UTR might also be involved in the regulation of mRNA decay. The 3′UTRs of hilD and aceA, containing Rho-independent terminators, possess a specific AU-rich regulatory region. The AU-rich region might be recognized and cleaved by endonuclease to initiate mRNA decay (Maeda and Wachi, 2012; Lopez-Garrido et al., 2014). However, the 3′UTRs of hmsT and slrA, containing Rho-dependent terminators, lack a specific regulatory region, and, in this situation, PNPase is primarily responsible for efficient mRNA turnover from the 3′ ends (Liu et al., 2016; Zhu et al., 2016).
There is a unique advantage to initiating transcript decay from the 3′UTRs rather than the 5′UTRs. Although initiating transcript decay from the 5′UTRs can rapidly inactivate a functional mRNA by removing the RBS, it cannot prevent translation that has already started from producing a full-length protein. Initiating the decay of a transcript from the 3′UTRs can rapidly halt translation by removing nearby encoding sequences. Hence, although 3′UTR-mediated mRNA decay is not an economical method of regulating gene expression, it might allow for a quicker response to the changing environment. Thus, mediation of mRNA turnover via the 3′UTRs might be useful for rapid control of gene expression in bacteria.
3′UTRs Interact with 5′UTRs
Translational control in eukaryotes is largely conferred by specific cis-acting sequences located in the 3′UTRs that serve as binding sites for the associated trans-acting factors (Sonenberg, 1994; Mazumder et al., 2003; Wilkie et al., 2003). By contrast, translational control in bacteria is mainly modulated through the 5′UTRs, which contains the Shine–Dalgarno (SD) sequence (Babitzke et al., 2009; Geissmann et al., 2009; Nakamoto, 2009; Waters and Storz, 2009). Recently, it was reported that the 3′UTR of icaR mRNA can interact with the 5′UTR to affect mRNA stability and translation in Staphylococcus aureus (Ruiz de los Mozos et al., 2013). A UCCCCUG motif is located at the 3′UTR of the icaR transcript, and is complementary to the SD region in the 5′UTR. On one hand, this interaction provides a double-stranded RNA substrate for RNase III cleavage to promote mRNA decay. On the other hand, it inhibits ribosome binding and hinders the formation of a translational complex, thus repressing the translation. This study illustrates that the bacterial 3′UTRs can interact with the SD region in the 5′UTRs of the same transcript leading to post-transcriptional regulation of gene expression (Ruiz de los Mozos et al., 2013). There are two additional examples for the modulation of bacterial translation by the interaction between 3′UTRs and 5′UTRs. It has been reported that the 3′ end of a full-length hok mRNA folds back to pair with its translational region to form a closed structure (Thisted et al., 1995). The formation of the closed structure renders the translation of the hok gene (Thisted et al., 1995). Another example is RNAIII (514 nt), the most studied regulatory RNA of S. aureus, which actually encodes a small peptide and contains a long 3′UTRs (354 nt) (Balaban and Novick, 1995; Felden et al., 2011). The RNAIII forms several regulatory structures including structures that facilitate interactions between the 5′ end and the 3′ end that regulate the expression of different target genes (Novick et al., 1993; Felden et al., 2011). Less is known about whether the formation of these structures regulates the translation of its own gene product. It has been reported that deletion of the 3′UTRs abolishes the temporal delay between transcription and translation of RNAIII (Balaban and Novick, 1995). The mechanism was not clarified, but it was proposed that the 3′UTRs might interact with the 5′UTRs to inhibit translation (Balaban and Novick, 1995).
3′UTRs that Function as sRNA Targets
It is reported that 3′UTRs might overlap with adjacent transcripts encoded on the opposite DNA strand (Rasmussen et al., 2009; Lasa et al., 2011). These overlapping 3′UTRs may be targeted by neighboring genes (usually encoding sRNAs) via a cis-acting antisense RNA mechanism. One example is a pair of toxin-antitoxin modules, TxpA and RatA (Silvaggi et al., 2005). RatA, an RNA antitoxin, is a sRNA (222 nt long) that inhibits the accumulation of mRNA of the toxic gene txpA. The orientations of txpA and ratA genes are convergent, and the genes overlap by ca. 75 nt; thus the 3′end of txpA mRNA is complementary to the sRNA ratA, thereby triggering txpA degradation (Silvaggi et al., 2005). Another example is the regulation of gadX by sRNA GadY. GadX is involved in the regulation of acid resistance in E. coli (Opdyke et al., 2004). The sRNA GadY overlaps with the 3′UTR of the gadX gene, and this overlap region is necessary for the regulation of gadX by gadY. Unlike other 3′UTRs, the interaction of the gadX 3′UTR with GadY increases the mRNA stability (Opdyke et al., 2004). The above examples illustrate mRNA crosstalk, whereby 3′UTRs act as sRNA targets to influence their own gene expression by positively or negatively modulating mRNA stability.
3′UTRs that Function as sRNA Reservoirs
sRNAs are usually associated with the RNA-binding protein Hfq in Gram negative bacteria, which facilities their regulatory function and protects sRNAs from degradation (De Lay et al., 2013; Wagner, 2013). Recently, 3′ regions of many mRNAs have been co-immunoprecipitated with Hfq, suggesting that the cellular functions of these 3′UTRs are independent of the role of the protein encoded by their parental mRNA, and might indeed function as sRNA reservoirs (Chao et al., 2012; Gossringer and Hartmann, 2012; Tree et al., 2014). CpxQ, an sRNA derived from the 3′ end of cpxP mRNA, is an excellent example of a 3′UTR acting as a sRNA reservoir (Chao and Vogel, 2016). CpxP, a periplasmic protein, combats envelope stress by tagging misfolded membrane proteins for degradation (Danese and Silhavy, 1998). The 3′UTR-derived sRNA CpxQ (60-nt-long) is generated during mRNA decay by RNase E and functions as an Hfq-dependent repressor of multiple mRNAs encoding extracytoplasmic proteins in E. coli (Chao and Vogel, 2016). Thus, CpxQ may analogously reduce synthesis of problematic proteins to combat envelope stress (Chao and Vogel, 2016). Another example is the sodF 3′UTR-derived sRNA in Streptomyces coelicolor (Kim et al., 2014). This sRNA (90-nt-long) is derived from the sodF 3′UTR and represses the expression of sodN, thereby shutting off the synthesis of Ni-SOD during nickel starvation (Kim et al., 2014). In addition, a 75 nt sRNA SorX, generated by RNase E cleavage of the 3′UTR of RSs2461 mRNA in Rhodobacter sphaeroides, represses the expression of potA (Peng et al., 2016). PotA is involved in the uptake of spermidine, which affects the sensitivity of R. sphaeroides to organic hydroperoxides. Hence, the 3′UTR-derived sRNA SorX can repress the import of spermidine to counteract oxidative stress (Peng et al., 2016). Altogether, these examples suggest that 3′UTRs can function as a sRNA reservoir that can post-transcriptionally regulate the expression of physically unlinked genes in response to changing environments (Miyakoshi et al., 2015). A remaining issue that needs to be addressed is whether the cleavage of mRNA during maturation of 3′UTR derived sRNAs affects the mRNA decay of its own mRNA.
Exploitation of 3′UTR-Mediated Control of Gene Expression
Regulation of mRNA stability is a common mechanism used by bacteria to regulate gene expression. mRNA stability is strongly affected by the initial endonucleolytic cleavage of mRNA (Laalami et al., 2014). Thus, the addition of a regulatory sequence at the 3′UTRs could be used to artificially control gene expression. A ribozyme is usually composed of a 50–150 nt RNA motif with intrinsic RNA cleavage activity (Doherty and Doudna, 2000). Felletti et al. (2016) reported that the incorporation of a ligand-dependent ribozyme in the 3′UTR region induces the cleavage of the 3′UTRs in a ligand-dependent manner, which could be employed to control gene expression. In addition, 3′UTRs engineering has been used to improve the soluble expression of heterologous enzymes, and thus can be used to fine-tune enzyme activity in microbial cells (Song et al., 2016). The above studies suggest that 3′UTRs could serve as targets for post-transcriptional regulation of gene expression, a feature that could be exploited by biotechnology, synthetic biology, and metabolic engineering.
Conclusion and Perspectives
It was traditionally believed that bacterial 3′UTRs mainly contain a transcriptional terminator. Recently, long 3′UTRs (>100 nt) were identified by transcriptomic analysis in many bacterial transcripts (Rasmussen et al., 2009; Toledo-Arana et al., 2009; ten Broeke-Smits et al., 2010; Ruiz de los Mozos et al., 2013). Since ca.40–50 nt is a sufficient length for a transcriptional terminator (Ruiz de los Mozos et al., 2013), additional regulatory elements are predicted to exist in these long 3′UTRs. Consistent with this hypothesis, 3′UTRs were found to regulate mRNA decay and translation and to act as sRNA targets or reservoirs for adaptation to various environmental changes, such as changes in temperature, pH, and nutrition availability (Opdyke et al., 2004; Kim et al., 2014; Peng et al., 2016; Chao and Vogel, 2016; Zhu et al., 2016). Although these 3′UTRs employ different mechanisms to regulate their own gene expression or targeted gene expression, one common feature is the regulation of gene transcription in a post-transcriptional manner.
A number of functional 3′UTRs have been identified. However, it is very likely that many more 3′UTRs function as post-transcriptional regulators, given that long 3′UTRs are widely present in bacteria. Furthermore, additional mechanisms of 3′UTR-regulated gene expression are expected to be identified; for example, RNA-binding proteins might bind to 3′UTRs to regulate gene expression. A group of mRNA termed ‘cutoRNAs’, which have long 3′UTRs that overlap with a downstream, convergently transcribed gene, has recently been reported (Moody et al., 2013). The regulatory function of these cutoRNAs remains unclear. Identification of new functional 3′UTRs and their regulatory mechanisms will lead to a better understanding of how bacteria use 3′UTRs as post-transcriptional regulators to respond to changes in the environment, and how such 3′UTRs regulation interacts with other transcriptional or post-transcriptional regulators. This knowledge will provide insights for new or improved biotechnological applications. Further, since many 3′UTRs play an important role in the regulation of virulence gene expression, interfering with the roles of these 3′UTRs might comprise an interesting alternative strategy for controlling bacterial pathogens.
Author Contributions
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
Funding
This work was supported by the National Natural Science Foundation of China (31670139) and (81501723), the National Basic Research Program of China (973 Program) (2015CB554200), the National Major Research & Development Program of China (2016YFC1202600) and CAMS Initiative for Innovative Medicine (2016-I2M-1-013).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Agaisse, H., and Lereclus, D. (1996). STAB-SD: a shine-dalgarno sequence in the 5′ untranslated region is a determinant of mRNA stability. Mol. Microbiol. 20, 633–643. doi: 10.1046/j.1365-2958.1996.5401046.x
Babitzke, P., Baker, C. S., and Romeo, T. (2009). Regulation of translation initiation by RNA binding proteins. Annu. Rev. Microbiol. 63, 27–44. doi: 10.1146/annurev.micro.091208.073514
Balaban, N., and Novick, R. P. (1995). Translation of RNAIII, the Staphylococcus aureus agr regulatory RNA molecule, can be activated by a 3′-end deletion. FEMS Microbiol. Lett. 133, 155–161. doi: 10.1016/0378-1097(95)00356-a
Barreau, C., Paillard, L., and Osborne, H. B. (2005). AU-rich elements and associated factors: are there unifying principles? Nucleic Acids Res. 33,7138–7150. doi: 10.1093/nar/gki1012
Breaker, R. R. (2011). Prospects for riboswitch discovery and analysis. Mol. Cell. 43, 867–879. doi: 10.1016/j.molcel.2011.08.024
Chao, Y., Papenfort, K., Reinhardt, R., Sharma, C. M., and Vogel, J. (2012). An atlas of Hfq-bound transcripts reveals 3′ UTRs as a genomic reservoir of regulatory small RNAs. EMBO J. 31, 4005–4019. doi: 10.1038/emboj.2012.229
Chao, Y., and Vogel, J. (2016). A 3′ UTR-derived small RNA provides the regulatory noncoding arm of the inner membrane stress response. Mol. Cell 61, 352–363. doi: 10.1016/j.molcel.2015.12.023
Chen, L. H., Emory, S. A., Bricker, A. L., Bouvet, P., and Belasco, J. G. (1991). Structure and function of a bacterial mRNA stabilizer: analysis of the 5′ untranslated region of ompA mRNA. J. Bacteriol. 173, 4578–4586. doi: 10.1128/jb.173.15.4578-4586.1991
Danese, P. N., and Silhavy, T. J. (1998). CpxP, a stress-combative member of the Cpx regulon. J. Bacteriol. 180, 831–839.
De Lay, N., Schu, D. J., and Gottesman, S. (2013). Bacterial small RNA-based negative regulation: Hfq and its accomplices. J. Biol. Chem. 288, 7996–8003. doi: 10.1074/jbc.R112.441386
Deneke, C., Lipowsky, R., and Valleriani, A. (2013). Effect of ribosome shielding on mRNA stability. Phys. Biol. 10:046008. doi: 10.1088/1478-3975/10/4/046008
Doherty, E. A., and Doudna, J. A. (2000). Ribozyme structures and mechanisms. Annu. Rev. Biochem. 69, 597–615. doi: 10.1146/annurev.biochem.69.1.597
Felden, B., Vandenesch, F., Bouloc, P., and Romby, P. (2011). The Staphylococcus aureus RNome and its commitment to virulence. PLoS Pathog. 7:e1002006. doi: 10.1371/journal.ppat.1002006
Felletti, M., Bieber, A., and Hartig, J. S. (2016). The 3′-untranslated region of mRNAs as a site for ribozyme cleavage-dependent processing and control in bacteria. RNA Biol. doi: 10.1080/15476286.2016.1240141 [Epub ahead of print].
Geissmann, T., Marzi, S., and Romby, P. (2009). The role of mRNA structure in translational control in bacteria. RNA Biol. 6, 153–160. doi: 10.4161/rna.6.2.8047
Gerstmeir, R., Wendisch, V. F., Schnicke, S., Ruan, H., Farwick, M., Reinscheid, D., et al. (2003). Acetate metabolism and its regulation in Corynebacterium glutamicum. J. Biotechnol. 104, 99–122. doi: 10.1016/S0168-1656(03)00167-6
Gossringer, M., and Hartmann, R. K. (2012). 3′-UTRs as a source of regulatory RNAs in bacteria. EMBO J. 31, 3958–3960. doi: 10.1038/emboj.2012.269
Henkin, T. M. (2008). Riboswitch RNAs: using RNA to sense cellular metabolism. Genes Dev. 22, 3383–3390. doi: 10.1101/gad.1747308
Hinnebusch, A. G., Ivanov, I. P., and Sonenberg, N. (2016). Translational control by 5′-untranslated regions of eukaryotic mRNAs. Science 352, 1413–1416. doi: 10.1126/science.aad9868
Kim, H. M., Shin, J. H., Cho, Y. B., and Roe, J. H. (2014). Inverse regulation of Fe- and Ni-containing SOD genes by a fur family regulator Nur through small RNA processed from 3′UTR of the sodF mRNA. Nucleic Acids Res. 42, 2003–2014. doi: 10.1093/nar/gkt1071
Kirillina, O., Fetherston, J. D., Bobrov, A. G., Abney, J., and Perry, R. D. (2004). HmsP, a putative phosphodiesterase, and HmsT, a putative diguanylate cyclase, control Hms-dependent biofilm formation in Yersinia pestis. Mol. Microbiol. 54, 75–88. doi: 10.1111/j.1365-2958.2004.04253.x
Kortmann, J., and Narberhaus, F. (2012). Bacterial RNA thermometers: molecular zippers and switches. Nat. Rev. Microbiol. 10, 255–265. doi: 10.1038/nrmicro2730
Krajewski, S. S., and Narberhaus, F. (2014). Temperature-driven differential gene expression by RNA thermosensors. Biochim. Biophys. Acta 1839, 978–988. doi: 10.1016/j.bbagrm.2014.03.006
Laalami, S., Zig, L., and Putzer, H. (2014). Initiation of mRNA decay in bacteria. Cell Mol. Life. Sci. 71, 1799–1828. doi: 10.1007/s00018-013-1472-4
Lalaouna, D., Simoneau-Roy, M., Lafontaine, D., and Masse, E. (2013). Regulatory RNAs and target mRNA decay in prokaryotes. Biochim. Biophys. Acta 1829, 742–747. doi: 10.1016/j.bbagrm.2013.02.013
Lasa, I., Toledo-Arana, A., Dobin, A., Villanueva, M., de los Mozos, I. R., Vergara-Irigaray, M., et al. (2011). Genome-wide antisense transcription drives mRNA processing in bacteria. Proc. Natl. Acad. Sci. U.S.A. 108, 20172–20177. doi: 10.1073/pnas.1113521108
Liu, B., Kearns, D. B., and Bechhofer, D. H. (2016). Expression of multiple Bacillus subtilis genes is controlled by decay of slrA mRNA from Rho-dependent 3′ ends. Nucleic Acids Res. 44, 3364–3372. doi: 10.1093/nar/gkw069
Lopez-Garrido, J., Puerta-Fernandez, E., and Casadesus, J. (2014). A eukaryotic-like 3′ untranslated region in Salmonella enterica hilD mRNA. Nucleic Acids Res. 42, 5894–5906. doi: 10.1093/nar/gku222
Maeda, T., and Wachi, M. (2012). 3′ Untranslated region-dependent degradation of the aceA mRNA, encoding the glyoxylate cycle enzyme isocitrate lyase, by RNase E/G in Corynebacterium glutamicum. Appl. Environ. Microbiol. 78, 8753–8761. doi: 10.1128/AEM.02304-12
Mazumder, B., Seshadri, V., and Fox, P. L. (2003). Translational control by the 3′-UTR: the ends specify the means. Trends Biochem. Sci. 28, 91–98. doi: 10.1016/S0968-0004(03)00002-1
Miyakoshi, M., Chao, Y., and Vogel, J. (2015). Regulatory small RNAs from the 3′ regions of bacterial mRNAs. Curr. Opin. Microbiol. 24, 132–139. doi: 10.1016/j.mib.2015.01.013
Moody, M. J., Young, R. A., Jones, S. E., and Elliot, M. A. (2013). Comparative analysis of non-coding RNAs in the antibiotic-producing Streptomyces bacteria. BMC Genomics 14:558. doi: 10.1186/1471-2164-14-558
Nakamoto, T. (2009). Evolution and the universality of the mechanism of initiation of protein synthesis. Gene 432, 1–6. doi: 10.1016/j.gene.2008.11.001
Novick, R. P., Ross, H. F., Projan, S. J., Kornblum, J., Kreiswirth, B., and Moghazeh, S. (1993). Synthesis of staphylococcal virulence factors is controlled by a regulatory RNA molecule. EMBO J. 12, 3967–3975.
Oliva, G., Sahr, T., and Buchrieser, C. (2015). Small RNAs, 5′ UTR elements and RNA-binding proteins in intracellular bacteria: impact on metabolism and virulence. FEMS Microbiol. Rev. 39, 331–349. doi: 10.1093/femsre/fuv022
Opdyke, J. A., Kang, J. G., and Storz, G. (2004). GadY, a small-RNA regulator of acid response genes in Escherichia coli. J. Bacteriol. 186, 6698–6705. doi: 10.1128/JB.186.20.6698-6705.2004
Peng, T., Berghoff, B. A., Oh, J. I., Weber, L., Schirmer, J., Schwarz, J., et al. (2016). Regulation of a polyamine transporter by the conserved 3′ UTR-derived sRNA SorX confers resistance to singlet oxygen and organic hydroperoxides in Rhodobacter sphaeroides. RNA Biol. 13, 988–999. doi: 10.1080/15476286.2016.1212152
Pesole, G., Mignone, F., Gissi, C., Grillo, G., Licciulli, F., and Liuni, S. (2001). Structural and functional features of eukaryotic mRNA untranslated regions. Gene 276, 73–81. doi: 10.1016/S0378-1119(01)00674-6
Rasmussen, S., Nielsen, H. B., and Jarmer, H. (2009). The transcriptionally active regions in the genome of Bacillus subtilis. Mol. Microbiol. 73, 1043–1057. doi: 10.1111/j.1365-2958.2009.06830.x
Ruiz de los Mozos, I., Vergara-Irigaray, M., Segura, V., Villanueva, M., Bitarte, N., Saramago, M., et al. (2013). Base pairing interaction between 5′- and 3′-UTRs controls icaR mRNA translation in Staphylococcus aureus. PLoS Genet. 9:e1004001. doi: 10.1371/journal.pgen.1004001
Selinger, D. W., Saxena, R. M., Cheung, K. J., Church, G. M., and Rosenow, C. (2003). Global RNA half-life analysis in Escherichia coli reveals positional patterns of transcript degradation. Genome Res. 13, 216–223. doi: 10.1101/gr.912603
Serganov, A., and Patel, D. J. (2012). Metabolite recognition principles and molecular mechanisms underlying riboswitch function. Annu. Rev. Biophys. 41, 343–370. doi: 10.1146/annurev-biophys-101211-113224
Silvaggi, J. M., Perkins, J. B., and Losick, R. (2005). Small untranslated RNA antitoxin in Bacillus subtilis. J. Bacteriol. 187, 6641–6650. doi: 10.1128/JB.187.19.6641-6650.2005
Sonenberg, N. (1994). mRNA translation: influence of the 5′ and 3′ untranslated regions. Curr. Opin. Genet. Dev 4, 310–315. doi: 10.1016/S0959-437X(05)80059-0
Song, J. W., Woo, J. M., Jung, G. Y., Bornscheuer, U. T., and Park, J. B. (2016). 3′-UTR engineering to improve soluble expression and fine-tuning of activity of cascade enzymes in Escherichia coli. Sci. Rep. 6:29406. doi: 10.1038/srep29406
St Johnston, D. (1995). The intracellular localization of messenger RNAs. Cell 81, 161–170. doi: 10.1016/0092-8674(95)90324-0
ten Broeke-Smits, N. J., Pronk, T. E., Jongerius, I., Bruning, O., Wittink, F. R., Breit, T. M., et al. (2010). Operon structure of Staphylococcus aureus. Nucleic Acids Res. 38, 3263–3274. doi: 10.1093/nar/gkq058
Thisted, T., Sorensen, N. S., and Gerdes, K. (1995). Mechanism of post-segregational killing: secondary structure analysis of the entire Hok mRNA from plasmid R1 suggests a fold-back structure that prevents translation and antisense RNA binding. J. Mol. Biol. 247, 859–873. doi: 10.1006/jmbi.1995.0186
Toledo-Arana, A., Dussurget, O., Nikitas, G., Sesto, N., Guet-Revillet, H., Balestrino, D., et al. (2009). The Listeria transcriptional landscape from saprophytism to virulence. Nature 459, 950–956. doi: 10.1038/nature08080
Tree, J. J., Granneman, S., McAteer, S. P., Tollervey, D., and Gally, D. L. (2014). Identification of bacteriophage-encoded anti-sRNAs in pathogenic Escherichia coli. Mol. Cell 55, 199–213. doi: 10.1016/j.molcel.2014.05.006
Van Assche, E., Van Puyvelde, S., Vanderleyden, J., and Steenackers, H. P. (2015). RNA-binding proteins involved in post-transcriptional regulation in bacteria. Front. Microbiol. 6:141. doi: 10.3389/fmicb.2015.00141
Waters, L. S., and Storz, G. (2009). Regulatory RNAs in bacteria. Cell 136, 615–628. doi: 10.1016/j.cell.2009.01.043
Wilkie, G. S., Dickson, K. S., and Gray, N. K. (2003). Regulation of mRNA translation by 5′- and 3′-UTR-binding factors. Trends Biochem. Sci. 28, 182–188. doi: 10.1016/S0968-0004(03)00051-3
Keywords: 3′untranslated region, mRNA decay, mRNA translation initiation, bacteria, post-transcriptional regulation
Citation: Ren G-X, Guo X-P and Sun Y-C (2017) Regulatory 3′ Untranslated Regions of Bacterial mRNAs. Front. Microbiol. 8:1276. doi: 10.3389/fmicb.2017.01276
Received: 10 April 2017; Accepted: 26 June 2017;
Published: 10 July 2017.
Edited by:
Shihua Wang, Fujian Agriculture and Forestry University, ChinaReviewed by:
Jin He, Huazhong Agricultural University, ChinaAntonio Juárez, University of Barcelona, Spain
Copyright © 2017 Ren, Guo and Sun. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yi-Cheng Sun, sunyc@ipbcams.ac.cn
 Gai-Xian Ren
Gai-Xian Ren Xiao-Peng Guo
Xiao-Peng Guo Yi-Cheng Sun
Yi-Cheng Sun