- 1 National Institute of Cancer Research, National Health Research Institutes, Zhunan Town, Miaoli County, Taiwan
- 2 NRPGM Core Facilities for Proteomics Research, Institute of Biological Chemistry, Academia Sinica, Taipei, Taiwan
- 3 School of Dentistry, University of California at Los Angeles, Los Angeles, CA, USA
The replication and transcription activator (RTA) of Kaposi’s sarcoma-associated herpesvirus (KSHV), K-RTA, is a lytic switch protein that moderates the reactivation process of KSHV latency. By mass spectrometric analysis of affinity purified K-RTA, we showed that Thr-513 or Thr-514 was the primary in vivo phosphorylation site. Thr-513 and Thr-514 are proximal to the nuclear localization signal (527KKRK530) and were previously hypothesized to be target sites of Ser/Thr kinase hKFC. However, substitutions of Thr with Ala at 513 and 514 had no effect on K-RTA subcellular localization or transactivation activity. By contrast, replacement of Ser with Ala at Ser-634 and Ser-636 located in a Ser/Pro-rich region of K-RTA, designated as S634A/S636A, produced a polypeptide with ∼10 kDa shorter in molecular weight and reduced transactivation in a luciferase reporter assay relative to the wild type. In contrast to prediction, the decrease in molecular weight was not due to lack of phosphorylation because the overall Ser and Thr phosphorylation state in K-RTA and S634A/S636A were similar, excluding that Ser-634 or Ser-636 motif served as docking sites for consecutive phosphorylation. Interestingly, S634A/S636A lost ∼30% immuno-reactivity to MPM2, an antibody specific to pSer/pThr-Pro motif, indicating that 634SPSP637 motif was in vivo phosphorylated. By in vitro kinase assay, we showed that K-RTA is a substrate of CDK9, a Pro-directed Ser/Thr kinase central to transcriptional regulation. Importantly, the capability of K-RTA in associating with endogenous CDK9 was reduced in S634A/S636A, which suggested that Ser-634 and Ser-636 may be involved in CDK9 recruitment. In agreement, S634A/S636A mutant exhibited ∼25% reduction in KSHV lytic cycle reactivation relative to that by the wild type K-RTA. Taken together, our data propose that Ser-634 and Ser-636 of K-RTA are phosphorylated by host transcriptional kinase CDK9 and such a process contributes to a full transcriptional potency of K-RTA.
Introduction
In a cell latently infected with Kaposi’s sarcoma-associated herpesvirus (KSHV), the switch to viral lytic replication can be achieved via signals transmitted from interferon-γ, phorbol ester, HDAC inhibitors, or with the ectopic expression of KSHV replication and transcription activator, referred to as K-RTA (also known as ORF50 and Lyta, reviewed in Ganem, 2007). Consecutive expression of KSHV lytic genes induced by either chemicals or K-RTA has been previously demonstrated (Sarid et al., 1998; Lukac et al., 1999; Sun et al., 1999; Gradoville et al., 2000; Fakhari and Dittmer, 2002; Nakamura et al., 2003; Yoo et al., 2005). In addition, the combination of sodium butyrate plus ectopic K-RTA yielded synergistic effects in an HEK293 cell background (Vieira and O’Hearn, 2004). K-RTA interacts with various cellular molecules involved in the transcription process. These include transcription factors (POU2F1/Oct-1, Sakakibara et al., 2001; STAT3, Gwack et al., 2002; RBPJ/CSL, Liang and Ganem, 2003; C/EBPα, Wang et al., 2003; ZNF426/K-RBP, Yang and Wood, 2007) and chromatin modifiers/remodelers (CBP, HDAC1, MED12/TRAP230, SMARCA4; Gwack et al., 2001, 2003a). Depending on the cellular context and promoters, association of these host factors results in either activation or repression of K-RTA transactivation activity. It bears noting that K-RTA-mediated gene regulation is not limited to the KSHV genome; a number of cellular genes are also modulated by K-RTA, as evidenced by genome-wide transcriptome analysis (Chang et al., 2005; Brown et al., 2010). Either on viral or host genomes, K-RTA requires host RNA polymerase II (RNA Pol II) for mRNAs synthesis since no RNA polymerase homolog was found in the KSHV genome.
The process of RNA Pol II-directed transcription can be divided into four distinct phases: (1) pre-initiation complex formation, (2) promoter clearance (escape), (3) RNA Pol II stalling at a promoter-proximal pausing site, and (4) productive elongation (Peterlin and Price, 2006; Saunders et al., 2006). The advancement of the transcription machinery from one stage to the next is tightly regulated by protein phosphorylation. Specifically, promoter clearance depends on CDK7-mediated Ser-5 phosphorylation of the tandem heptapeptide repeats (YSPTSPS) located in the carboxyl terminal domain (CTD) of RNA Pol II. Next, Ser-5 phosphorylated CTD undergoes a conformational change that unmasks the Ser-2 in the heptapeptide repeats (YSPTSPS), which serves to recruit positive transcription elongation factor b (P-TEFb; Lolli, 2009). P-TEFb not only phosphorylates Ser-2 but also phosphorylates negative elongation factor complex (NELF and DSIF) by which RNA Pol II is released from the pausing site and enters the productive elongation stage. Accumulating evidence demonstrate that RNA Pol II pausing is enriched in the 5′ region of genes involved in central regulatory processes, including developmental switches that are required for embryogenesis and rapidly inducible molecules involved in cellular responses to stimuli (Zeitlinger et al., 2007; Gilchrist et al., 2008). Recently, RNA Pol II pausing was also documented in controlling the expressions of viral genes in the KSHV and Epstein–Barr virus (EBV) genomes (Kang and Lieberman, 2011; Palermo et al., 2011).
Because expression of the viral transcriptome is usually RNA Pol II dependent, hijacking P-TEFb activity by viral molecules could be advantageous for viral transcription. The first example was in human immunodeficiency virus type I (HIV-I). P-TEFb is composed of a catalytic subunit (CDK9) and a transcription regulatory cyclin (cyclin T or K). By modifying the substrate specificity of CDK9, HIV-1 Tat increased noticeably the efficiency of the elongation step during HIV-1 transcription (Garber et al., 2000; Zhou et al., 2000). By contrast, CDK9 dominant negative mutants severely impaired HIV-1 replication in cultured human cells (Foskett et al., 2001; Fujinaga et al., 2002). Furthermore, CDK9 was recently shown to complex with ICP22 of herpesvirus type 1 (Durand and Roizman, 2008). The CDK9-bound ICP22 was required for optimal expression of certain viral late genes transcribed by RNA Pol II (Durand and Roizman, 2008). In the case of human cytomegalovirus, recruitment of cyclin T1 and CDK9 by IE2 86 kDa protein was critical to establishing an active immediate-early transcriptome in the first 8 h after viral infection (Kapasi and Spector, 2008; Kapasi et al., 2009). Finally, viral latent promoters activated by nuclear protein of EBV EBNA2 are enriched with the CDK9-mediated, elongated form of RNA Pol II (Bark-Jones et al., 2006).
In addition to moderating components of the RNA Pol II apparatus, phosphorylation of viral factors also contributes to the expression of the viral lytic transcriptome. Phosphorylation of Ser-186 located in the DNA binding domain of EBV BZLF1/ZEBRA is required for the disruption of EBV latency (Francis et al., 1997). Such a phosphorylation event is essential to bind and then activate with high preference methylated EBV immediate-early promoter (Bhende et al., 2005). In addition, phosphorylation of Ser-167 and Ser-173 leads BZLF1/ZEBRA to function as a repressor in expression of one late gene (El-Guindy and Miller, 2004). In human cytomegalovirus, the phosphorylation status of a Ser-rich domain encompassing amino acids 258–275 of immediate-early protein IE2 orchestrates the temporal expression of distinctive viral genes (Barrasa et al., 2005). Furthermore, the immediate-early protein ICP0 of human herpesvirus type 1 contains eleven potential phosphorylation sites clustered into three regions adjacent to domains required for transactivation. Mutation of the first phosphorylation region in ICP0 altered its E3 ligase activity that in turn led to virus replication defect (Boutell et al., 2008).
As every regulator needs to be regulated, in the present study we focus on the role of phosphorylation in the latent-lytic switch activity of K-RTA. Previous studies by Lukac et al. (1999) established that K-RTA is a highly phosphorylated protein evidenced by the alleviation of ∼20 kDa molecular mass on SDS-PAGE by calf intestinal phosphatase treatment. To extend this finding further, we performed computational analysis of K-RTA amino acid sequence using NetPhosK (Blom et al., 2004) and GPS 2.0 (Xue et al., 2008) for potential phosphorylation sites, and, Blast search in various protein databases for homologous protein motifs. We found: (1) K-RTA is a Ser- and Thr-rich protein (122 of 691, 17.7%) with the most abundant region located at amino acids 500–550 (33%). (2) In addition to multiple PKC and casein kinase II consensus sites, K-RTA harbors seven potential CDK phosphorylation motifs (T449, T540, T628, S634, S636, S644, S650). (3) In the Human Proteinpedia database, the sequences between amino acids 633–652 share high homology with an in vivo phosphorylated tryptic peptide derived from negative elongation factor B (NELF-B; Beausoleil et al., 2004; Olsen et al., 2006). In addition, PredictNLS (Cokol et al., 2000) identified one nuclear localization signal (NLS) in K-RTA, which is equivalent to the NLS-2 designated by Lukac and colleagues (Lukac et al., 1998, 1999; Bu et al., 2008). Among these predictions, we showed that Ser-634 and Ser-636 located in the NELF-B homologous region are involved in CDK9 recruitment and phosphorylation. Substitutions of Ser with Ala at Ser-634 and Ser-636 impaired K-RTA transactivation activity and altered its electrophoretic mobility on SDS-PAGE. In addition, CDK9 inhibitors suppressed the expressions of various K-RTA target genes and KSHV viral production in a HEK293/rKSHV.219 cell model. Together, these results support an emerging notion that some regions of the latent KSHV genome are consistently associated with paused RNA Pol II whose activity can be acutely induced by positive elongation factors such as CDK9 in response to various stimuli.
Materials and Methods
Plasmids
pLenti4-FLAG-CPO is a gift from Dr. Dan Robinson at MCTP, University of Michigan (Ann Arbor, MI, USA). pLenti4-FLAG-CPO is a modified vector derived from pLenti4/TO/V5-DEST (Invitrogen). Briefly, the original attR1 site to V5 epitope region (nt 2405–4203) in pLenti4/TO/V5-DEST was replaced with an in-frame DNA fragment encoding Kozak sequence, ATG, FLAG tag, and a rare cutter CPO I site (5′CGGTCCG). Accordingly, pLenti4-FLAG-K-RTA was constructed by cloning the coding sequences of K-RTA (GeneID: 4961526) into the CPO I site of pLenti4-FLAG-CPO. The expression plasmid pLenti4-FLAG-NLSm was generated by replacing 527KKRK530 motif of K-RTA with AAAA in pLenti4-FLAG-K-RTA by QuikChange® site-directed mutagenesis kit (Stratagene). Similarly, expression plasmids for the other NLS and phosphorylation mutants, including NLSm1, NLSm2, T513A, T514A, T513A/T514A, S634A, S634T, S634D, S636A, S636T, S636D, S644A, S652A, and S634A/S636A were generated by using pLenti4-FLAG-K-RTA as template in the QuikChange® site-directed mutagenesis reactions. In luciferase reporter assays, the upstream sequences of PAN (nt 28159–28660 of U75698) and ORF57 (nt 81556–82005) were cloned into SacI–XhoI sites of pGL3-Basic (Promega), yielding pGL3-Basic-PANp, and pGL3-Basic-ORF57p, respectively. Expression plasmids for CDK9, dominant negative CDK9 (D167N), and cyclin T have been described previously (Chang and Li, 2008).
Establishment of 293TetKR and 293TetNLSm Conditional Expression Cell Lines
The Tetracycline-Regulated Expression HEK293 (TREx™, Invitrogen) cell line were infected with lentivirions harboring pLenti4-FLAG-K-RTA and pLenti4-FLAG-NLSm by using ViraPower™ system (Invitrogen). Forty-eight hour after infection, the cells were selected with 400 μg/ml zeocin (Invitrogen) for 2 weeks. Zeocin-resistance clones were subjected to doxycycline-inducibility test for the expression of desired genes by Western blot analysis using M2 anti-FLAG antibody (Sigma-Aldrich). For each conditional expression line, multiple positive clones with similar growth rate and expression level were pooled, collectively designated as 293TetKR and 293TetNLSm, respectively.
Cells and Cell Culture
Conditional expression cell lines 293TetKR and 293TetNLSm were maintained in DMEM supplemented with 10% Tet System Approved FBS (Clontech Laboratories), 5 μg/ml blasticidin-S-HCl (Invitrogen), and 200 μg/ml zeocin. HEK293 was maintained in DMEM containing 10% FBS. 293/rKSHV.219 was maintained in DMEM supplemented with 10% FBS and 660 ng/ml puromycin (Becton Dickinson). All cells were grown in a humidified 37°C incubator with 5% CO2.
Transfection and Luciferase Reporter Assay
Transfection was performed in 24-well plates. HEK293 cells (2.4 × 105) were seeded into each well to reach a 90% confluence the next day before transfection. Transfection was carried out using Lipofectamine™ 2000 (Invitrogen) conveying appropriate plasmids according to the manufacturer’s instruction. Twenty-four hour after transfection, cells were harvested for luciferase activity assay. Firefly and renilla luciferase activities were measured by using Dual-Glo Lucifearse Assay Kit (Promega). Transfection efficiency was normalized with a cotransfected renilla luciferase reporter (phRL-TK, Promega).
Protein Purification and Mass Spectrometric Analysis
Immuno-purification of FLAG-tagged proteins from 293Tet-inducible cell lines was carried out according to two previously described procedures (Barlev et al., 2003; Trester-Zedlitz et al., 2005) with minor modifications. Specifically, 108 cells with 80% confluence were treated with 50 ng/ml doxycycline for 10 h before cell harvest. Combined cell pellet (∼1 g) was solubilized in 10 ml of lysis buffer (50 mM Tris–HCl, pH 7.4, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, 1× protease inhibitors, 0.2 mM sodium orthovanadate) at 4°C for 40 min with gentle shaking. Protein extracts were clarified by centrifugation at 12,000 × g, 4°C for 15 min. Collected protein supernatant was further filtrated through a 0.45 micron filter. The filtrated protein supernatant was applied onto 400 μl anti-FLAG M2 affinity resin (A2220, Sigma-Aldrich) equilibrated with lysis buffer. The column eluate was re-loaded onto the column for two more cycles. The column was alternatively washed with one column volume of TBS (50 mM Tris–HCl, pH 7.4) followed by one column volume of high salt (500 mM NaCl)–TBS for three times. The column was washed with additional two column volumes of TBS buffer prior to elution. The FLAG-tagged proteins were eluted from the column by TBS buffer supplemented with 100 μg/ml FLAG peptide (F3290, Sigma-Aldrich) and 20% glycerol. For each purification, four eluted fractions were collected and samples were immediately frozen in −70°C for further studies. Approximately 500 ng affinity purified K-RTA were resolved in a SDS-PAGE gel, stained with SYPRO® Ruby (Invitrogen) followed by in-gel digestion. Phosphopeptides were enriched by C18-functionalized Fe3O4 nanoparticles as described in Hsiao et al. (2007) followed by LC/MS/MS analysis as described previously (Lin et al., 2008).
Western Blot Analysis and Co-Immunoprecipitation
Cells were lysed in RIPA buffer (50 mM Tris–HCl, pH 8, 150 mM NaCl, 0.1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% SDS, 1 mM phenylmethylsulfonyl fluoride, 200 μM sodium orthovanadate, 1× protease inhibitors, 1× phosphatase inhibitors). The protein concentration was measured spectrophotometrically at 562 nm using BCA protein assay reagent (Pierce, Rockford, IL, USA). For each co-immunoprecipitation assay, the cells were lysed in cold EBC lysis buffer (50 mM Tris–HCl, pH 7.4, 0.5% NP-40, 120 mM NaCl, 50 mM NaF, 1 mM phenylmethylsulfonyl fluoride, 200 μM sodium orthovanadate, 1× protease inhibitors, 1× phosphatase inhibitors). The protein extracts were incubated with M2 anti-FLAG affinity resin or anti-CDK9 bounded Dynabeads-protein G (Invitrogen) with continuous rocking at 4°C overnight. After centrifugation or magnetic separation, the resin, or beads were washed thoroughly with TBS (50 mM Tris–HCl, pH 7.4, 150 mM NaCl) three times followed by elution of immunocomplex in 1× sample buffer. Cell lysates or the eluted immunocomplex were subjected to SDS-PAGE separation and analyzed by Western blotting as described elsewhere (Chen et al., 2011). For IP-kinase assay (Figures 5A,B), the eluted immunocomplex were washed three times with TBS and once with kinase buffer (25 mM Tris–HCl, pH 7.5, 5 mM β-glycerophosphate, 2 mM DTT, 0.1 mM sodium orthovanadate, 10 mM MgCl2, 10 μM ATP). The immunocomplex were resuspended in 29 μl kinase buffer containing 1 μCi [γ-33P] ATP and incubated at 30°C for 30 min. After eluting proteins from resin, the proteins were resolved by SDS-polyacrylamide gel electrophoresis. The gel was dried, exposed to an X-ray film and developed by autoradiography.
Calf Intestinal Alkaline Phosphatase Treatment
Cells were lysed in EBC lysis buffer without phosphatase inhibitors or sodium orthovanadate. For each sample 50 μg of cell extract was incubated with 60 U calf alkaline phosphatase in buffer 3 (New England Biolabs) at 37°C for 45 min. Extracts were separated by SDS-PAGE followed by Western blot analysis.
Immunofluorescence Assay
In Figure 1B, 1 day before transfection, cells at a density of 105/0.4 ml/well were seeded on eight-well chamber slide (Nunc) coated with poly-D-Lysine (Sigma-Aldrich). The cells were transfected with indicated plasmids by using LipofectamineTM 2000 (Invitrogen) and incubated for 24 h. In Figure 2A, cells were pre-treated with doxycycline for 24 h before assay. Cells were fixed with acetone/methanol (v/v = 1:1) at −20°C for 20 min and permeablized with 0.4% Triton X-100 at RT for 5 min. The slides were blocked for 30 min in blocking buffer (PBS containing 1% FBS) and incubated with anti-FLAG antibody at RT for 2 h. The slides were washed with PBS three times for 5 min each. The slides were incubated with fluorescein isothiocyanate-conjugated anti-mouse IgG for 2 h and counterstained with DAPI (Sigma-Aldrich) for 5 min at RT. The slides were observed under a confocal fluorescence microscope.
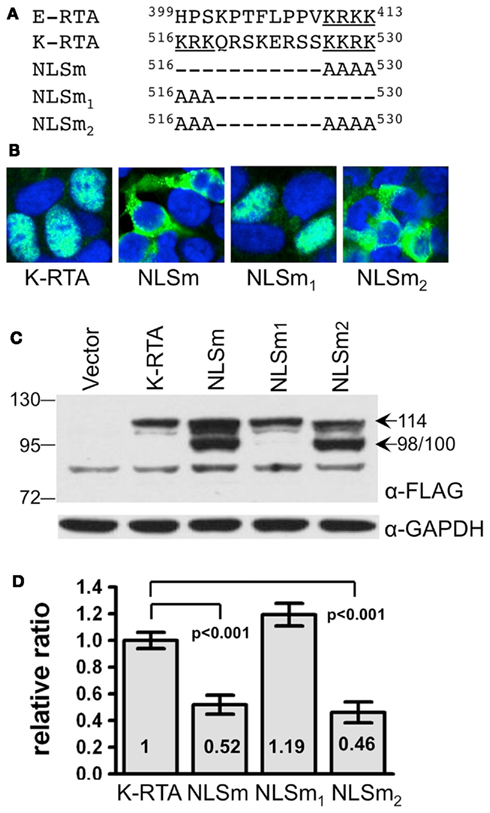
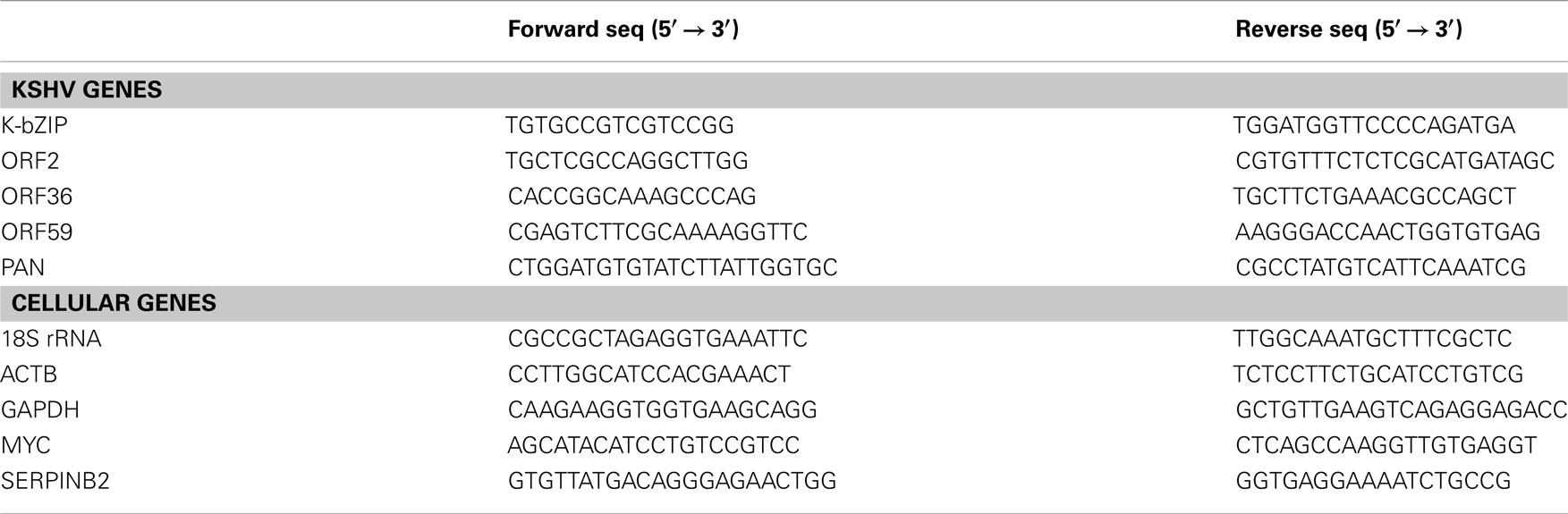
Figure 1. Characterization of a prototypic bipartite nuclear localization signal (NLS) of K-RTA locating at amino acids 516–530. (A) Alignment of NLS from EBV Rta (E-RTA), K-RTA, NLSm, NLSm1, and NLSm2. The KRKK motif required for E-RTA nuclear localization (Hsu et al., 2005) and the two clusters of basic amino acids located in the bipartite NLS of K-RTA are underlined. (B) Subcellular localization of K-RTA and the three NLS mutants in HEK293 cells revealed by an immunofluorescence assay using M2 FLAG antibody followed by confocal microscopy. Only mutations locating at the second basic motif affect the nuclear targeting (NLSm and NLSm2). (C) Western blot analysis of HEK293 cells transiently transfected with K-RTA or each of three NLS mutants. An additional 98/100 doublet was detected only in the extract of variants harboring the second basic motif mutation (NLSm and NLSm2). GAPDH served as a loading control. (D) Luciferase reporter assay of KSHV ORF57 promoter responding to K-RTA and NLS mutants transiently expressed in HEK293 cells. Each firefly luciferase value was normalized to an internal control derived from cotransfected plasmid phRL-TK. The value in K-RTA was set to 1. Data are presented as means ± SD from triplicate transfections in an experiment. Statistical evaluations were performed by using Student’s t-test. Three independent experiments were performed; a representative result is shown.
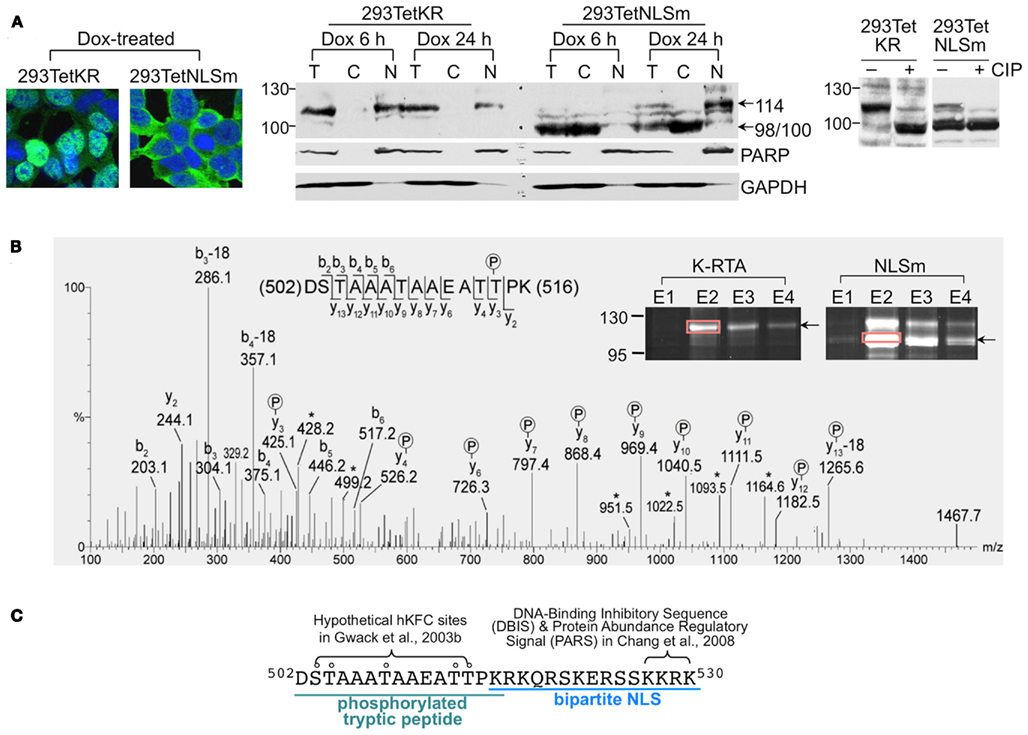
Figure 2. Thr-513 and Thr-514 of K-RTA is the primary in vivo phosphorylation motif identified by mass spectrometry. (A) (Left) distinct subcellular localization of K-RTA and NLSm was revealed in doxycycline (Dox, 50 ng/ml)-treated 293TetKR and 293TetNLSm cells by an immunofluorescence assay using M2 FLAG antibody. (Middle) subcellular fractionation of protein lysates prepared from 6 to 24 h Dox-treated 293TetKR and 293TetNLSm cells. T, total lysate; N, nuclear fraction; C, cytosolic fraction. PARP and GAPDH served as the nuclear and cytosolic indicators, respectively. (Right) protein lysate of the 293TetKR and 293TetNLSm was preincubated with (+) or without (−) calf intestinal alkaline phosphatase (CIP) at 37°C for 45 min followed by Western blot analysis. (B) MS/MS spectrum on [M + 2H]2+ (m/z 743.31) ion for the one phosphorylation-modified peptide DSTAAATAAEATTPK from K-RTA (rectangle). The product ion y3 that carries a phosphate indicated that Thr-514 was phosphorylated but its low intensity among other background/unassigned peaks does not preclude the alternative Thr-513 site. A single phosphorylation on either site was unambiguously supported by additional phosphorylated y ions. Product ions marked with starts resulted from elimination of H2O. Of note, the same spectrum was observed for 100 kDa NLSm (not shown). (C) Summary of various features identified in amino acids 502–530.
Subcellular Fractionation
Subcellular fractionation was conducted according to a previously described procedure (Wang et al., 2005). Briefly, cells were incubated with hypotonic buffer (10 mM HEPES, pH 7.4, 10 mM KCl, 1.5 mM MgCl2, 0.34 M Sucrose, 10% glycerol, 0.1% Triton X-100, 1 mM DTT, 1× protease inhibitors) on ice for 10 min. Cells were centrifuged at 1300 × g for 4 min at 4°C. The resulting pellet was the nuclear fraction. The supernatant was further centrifuged at 20,000 × g for 15 min at 4°C to remove debris. The resulting supernatant was the cytosolic fraction.
Flow Cytometric Analysis
After transfection, the cells were harvested by trypsinization and resuspended in 500 μl PBS. A total of 5,000 cells were acquired by using a flow cytometer (FACSCalibur, Becton Dickinson) and analyzed using the WinMDI v2.8 software. GFP and RFP signals were detected at 488 and 540 nm, respectively.
Titration of KSHV Viral Particles
Filtrated (0.45 μm) viral supernatant (160 μl) was incubated with 2 U DNase I (Invitrogen) at 37°C for 30 min followed by extraction of encapsidated KSHV DNA using QIAamp MinElute virus spin kit (QIAGEN). Each comparative quantitative PCR reaction was composed of 2 μl diluted viral DNA, 5 μl Power SYBR Green Master Mix (Applied Biosystems), and 3 μl primer mix (0.66 μM). The primers used for detecting KSHV genome are ORF9-forward (5′-CCAACATCATCCAATGCCTC-3′) and ORF9-reverse (5′-GGGAAAAGTCACGGGAATG-3′). Known copy numbers of serially diluted cosmid GB11 DNA encompassing KSHV genome nt 1–35,022 (U75698) were used as standards in titrating KSHV viral particles. The reaction was conducted and detected by StepOnePlusTM Real-Time PCR system (Applied Biosystems).
Quantitative Reverse Transcription-Polymerase Chain Reaction
Total RNA was extracted from cells by using RNeasy kit (Qiagen). Reverse transcription of 2.5 μg RNA was performed in a 10 μl SuperScript™ III reaction mixtures (Invitrogen) according to the manufacturer’s instructions. One percent of the resulting cDNAs were used for each real-time PCR reaction composed of 2 μl diluted cDNA, 5 μl Power SYBR Green Master Mix (Applied Biosystems), and 3 μl primer mix (0.66 μM). The primers used in the present study are listed in Table A1 in Appendix. The reaction was conducted and detected by StepOnePlus™ Real-Time PCR system (Applied Biosystems).
Antibodies
Monoclonal and polyclonal anti-K-RTA antibodies were kindly provided by Drs. Keiji Ueda (Osaka University Graduate School of Medicine, Japan) and Yoshihiro Izumiya (University of California at Davis, Sacramento, CA, USA), respectively. Other antibodies used in this study are M2 FLAG (F1804, Sigma), β-actin (A5441, Sigma), mouse monoclonal CDK9 (sc-13130, Santa Cruz), PARP (sc-8007, Santa Cruz), rabbit monoclonal CDK9 (#2316, Cell Signaling), p44/42 MAPK (#9107, Cell Signaling), phospho-p44/42 MAPK (#4376, Cell Signaling), MPM2 (#05-368, Upstate), α-tubulin (05-829, Millipore), phospho-serine (AB1603, Millipore), phospho-threonine (AB1607, Millipore), GAPDH (#54593, AnaSpec, Inc.).
Results
The KKRK Motif of the Bipartite Nuclear Localization Signal Contributes to Nuclear Targeting and Transactivation Activity of K-RTA
Computer-assisted identification of the NLS sequence in K-RTA was achieved using PredictNLS, a program serving to identify in silico potential NLS present in a given sequence (Cokol et al., 2000). Accordingly, a prototypic bipartite NLS located at amino acids 516–530 (KRKQRSKERSSKKRK) was identified with 97.92% probability (Figure 1A). This subclass of bipartite NLS can be found in 193 nuclear proteins collected in the PredictNLS database as of September 2011. In agreement, functional domains encompassing amino acids 516–530 of K-RTA were recently shown to be a bona fide NLS (Bu et al., 2008) or to possess inhibitory effects on DNA binding (Chang et al., 2008). Noteworthy, Chen et al. (2000) showed that amino acids 6–12 of K-RTA, designated as NLS-1 in Lukac’s et al. (1998) publication, was demonstrated to be a functional NLS when fused to β-Gal protein, and, is required for efficient nucleus-targeting since product of genomic ORF50 devoid of NLS-1 tends to localize to both the nucleus and the cytoplasm. This finding suggests that NLS-1 can serve as a cryptic NLS and may participate in correct folding of K-RTA that is required for the exposure of the C-terminal bipartite NLS.
Alignment of amino acids 516–530 with the NLS derived from EBV Rta (Hsu et al., 2005), a functionally homologous protein to K-RTA, revealed little similarity except for the last four basic residues (Figure 1A). To characterize further which of the two basic motifs is responsible for nuclear localization, the Lys and Arg located at amino acids 516–518 and 527–530, respectively, were mutated to Ala (Figure 1A). Subcellular localization of K-RTA and NLS mutants was inspected by an immunofluorescence assay. As shown in Figure 1B, NLSm and NLSm2 were clearly retained in the cytoplasm, indicating that the mutation of the second basic motif composed of KKRK is the determinant of nuclear targeting. By contrast, modification of the first basic motif from KRK to AAA did not seem to interfere with the nuclear targeting of K-RTA, as represented by NLSm1. In addition, compared with the wild type that was estimated to be 114 kDa in size, mutation of the second basic motif consistently yielded more abundant but shorter 98/100 kDa doublet bands in Western blot analysis (Figure 1C). To assess whether mutations affect the transactivation capability of K-RTA, a luciferase reporter assay using ORF57 promoter sequences was performed. Again, mutation in the second (NLSm and NLSm2), but not in the first (NLSm1) basic motif showed a dramatic decrease (50–60%) in transactivation activity (Figure 1D). A similar result was obtained in a PAN promoter-driven luciferase reporter assay (data not shown). Collectively, these results revealed that the second basic motif KKRK of the bipartite NLS is responsible for the targeting of K-RTA to the nucleus and the full potency of transactivation, and is involved in regulating protein abundance. Because NLSm requires the least changes in altering subcellular localization, it was used as the cytoplasmic version of K-RTA in the following studies.
K-RTA is Phosphorylated in both Cytoplasm and Nucleus
The most probable explanation for the 14 kDa difference between K-RTA and NLSm was that it is caused by different extents of phosphorylation taking place in different compartments of the cell. To corroborate this hypothesis, cell lines conditionally expressing K-RTA and NLSm were established by using the Virapower system (Invitrogen). Briefly, TREx-293TM cells were stably transduced with a lentiviral-based plasmid expressing FLAG-tagged K-RTA or NLSm under the control of a doxycycline (Dox)-inducible promoter, CMVTetO2™. The resulting cell lines were designated as 293TetKR and 293TetNLSm, respectively. As expected, upon Dox treatment, K-RTA was found predominantly in the nucleus of 293TetKR cells using an immunofluorescence assay, while NLSm was largely confined to the cytoplasm of 293TetNLSm cells (Figure 2A, left). The protein extracts of Dox-treated 293TetKR and 293TetNLSm cells were subjected to subcellular fractionation followed by Western blot analysis. Clearly, the 114 kDa polypeptide and the 98/100 kDa protein bands existed in distinct fractions: the 114 kDa was exclusively in the nucleus and the 98/100 kDa doublet in the cytoplasm (Figure 2A, middle). Further, when protein extract was pre-treated with calf intestinal alkaline phosphatase (CIP), which should remove most of the phosphate groups attached to K-RTA and NLSm, both molecules were converted to a single 98 kDa species (Figure 2A, right). These results indicated that the non-phosphorylated forms of K-RTA and NLSm were similar, yet compartmental phosphorylation processes contribute to their individual molecular masses. Thereby, we hypothesized that K-RTA is mildly phosphorylated in the cytoplasm as indicated by the smaller, 98/100 kDa doublet polypeptides, and is further phosphorylated once it enters the nucleus as indicated by the mature 114 kDa protein.
Identification by Mass Spectrometry of Thr-513 and Thr-514 as the Primary in vivo Phosphorylation Motif in K-RTA
In order to conclusively identify which residues are phosphorylated in vivo, affinity purified 114 kDa K-RTA and 100 kDa NLSm derived from Dox-treated 293TetKR and 293TetNLSm cells, respectively, were subjected to LC/MS/MS analysis (Figure 2B). It was expected that there would be more phosphorylated residues identified in the 114 kDa nuclear K-RTA than in the 100 kDa cytoplasmic NLSm. Disappointedly, the same results were obtained from both analyses: Thr-513 or Thr-514 located in the tryptic peptides encompassing amino acids 502–516 (DSTAAATAAEATTPK) of K-RTA and NLSm were the only amino acids with phosphate potentially conjugated to them. Of these two residues, Thr-513 is more likely to serve as an alternative position for phosphate modification (Figure 2B, detailed in the legend). From these results, we can conclude that Thr-513 or Thr-514 is preferentially phosphorylated in the cytoplasm (NLSm), and this modified status is retained in the nucleus (K-RTA).
Next, the role of phosphorylated Thr-513 or Thr-514 in K-RTA activity was assessed by biological assays using three Ala substitution mutants, which mimic the non-phosphorylated state of 513 and 514. The three variants were primarily localized in the nucleus and the protein expression levels and molecular weights of the three variants were comparable to those of the wild type K-RTA by Western blot analysis (data not shown). A luciferase reporter assay using upstream sequences of KSHV PAN RNA was performed to quantitate the transactivation activity of each Ala variant. No significant difference was observed between any Ala mutant and the wild type K-RTA (not shown). These results suggested that that phosphorylation of Thr-513 and Thr-514 might play only an ancillary role in K-RTA activity. Interestingly, Gwack et al. (2003b) noted previously that Ser-503, Thr-504, Thr-508, Thr-513, and Thr-514 are five potential phosphorylation sites for a Ste20-like kinase hKFC, and that phosphorylation of K-RTA by hKFC resulted in a negative effect in K-RTA-mediated transactivation. Thus, our results excluded the possibility that Thr-513 or Thr-514 is an hKFC target site. Taken together, although located proximal to NLS and a DNA binding inhibitory/protein abundance regulatory signal (Figure 2C), defective in phosphorylation of Thr-513 or Thr-514 has little influence in K-RTA nuclear localization, transcriptional activation, or protein abundance.
Identification of an NELF-B Homologous Motif in the C-Terminus of K-RTA
In order to explore the existence of phosphorylated Ser or Thr residues that had been missed by mass spectrometry (MS; discussed below), computer-assisted Blast search was employed to look for known phosphorylated tryptic peptides that share sequence similarity with K-RTA. Briefly, amino acid sequences of several large tryptic peptides derived from K-RTA were used as queries to find hits using a pre-calculated position-specific score matrix in the NCBI Conserved Domain database as described by Marchler-Bauer et al. (2005). Amino acids 633–652 were found to share significant homology with an in vivo phosphorylated tryptic peptide encompassing amino acids 554–573 of NELF-B (Figure 3A). NELF-B is one of the five negative elongation factors that control the processivity of RNA Pol II on an active promoter. The in vivo phosphorylation sites of NELF-B have been identified previously to be located at Ser-557, Thr-564, and Ser-573 (Beausoleil et al., 2004; Olsen et al., 2006), which were aligned to K-RTA Ser-636, Ser-644, and Ser-652, respectively (Figure 3A). Although the role of phosphorylated Ser-557, Thr-564, and Ser-573 in NELF-B is still unclear, it was tempting to presume that the conserved residues in K-RTA are also phosphorylated. To test our hypothesis, Ser to Ala point mutation was individually introduced into K-RTA Ser-636, Ser-644, and Ser-652 (Figure 3A). In addition, we initially included S634A as a control variant since no conserved Ser was matched in the corresponding site of NELF-B. We first confirmed that all four Ser to Ala mutants correctly resided in the nucleus of HEK293 cells (data not shown). Next, the protein expression level of each mutant was compared by Western blot analysis in parallel with that of wild type K-RTA. Noteworthy, the major bands in S634A and S636A, but not in S644A and S652A were estimated to be 104 kDa, approximately 10 kDa shorter than that of the wild type (Figure 3B, compare lanes 1, 2, 5, 8, 9). To investigate further, Ser to Thr, Ser to Asp, and double Ser to Ala substitutions were introduced into amino acids 634 and 636, yielding S634T, S634D, S636T, S636D, and S634A/S636A. Similarly, each mutant was shown by an immunofluorescence assay to localize in the nucleus (data not shown). Interestingly, the discrepancy in molecular mass in S634A and S636A could be partially or completely overcome by substitution of a functionally conserved residue (e.g., S634T and S636T) or with a phosphorylation mimetic residue (e.g., S634D and S636D), suggesting that phosphorylation of Ser-634 or Ser-636 is involved in migration mobility of K-RTA (Figure 3B, lanes 3, 4, 6, 7). To corroborate the molecular mass discrepancy with transactivation potential, a PAN promoter-driven luciferase reporter assay was performed to compare transactivation activity of K-RTA and the nine Ser-634 and Ser-636 variants. As summarized in Figure 3B (lower panel), the transactivation activities were 8–27% lower in S634A, S636A, and S634A/S636A relative to that of the wild type and other variants. Although the reductions were small, these results mirrored those observed for migration mobility, namely substitution of Ser with Ala at 634 or 636 produced a 104 kDa species of K-RTA that is less potent. Because S634A/S636A yielded the most apparent changes in migration pattern and transactivation activity, this mutant was chosen for further investigation.
Figure 3. Ser to Ala substitutions at Ser-634 and Ser-636 impaired K-RTA transactivation potency and reduced KSHV reactivation. (A) Alignment of a Ser-rich region (amino acids 633–652, 30% Ser) located in the C-terminus of K-RTA with an in vivo phosphorylated peptide derived from negative elongation factor B (NELF-B). In vivo phosphorylated residues in NELF-B identified previously are denoted by circled P (Beausoleil et al., 2004; Olsen et al., 2006). Accordingly, a series of K-RTA variants with mutations in Ser-634, -636, -644, and -652 were generated. The four Ser to Ala mutants and Ser-634 and Ser-636 double mutants are denoted. (B) (Upper) the expression levels and patterns of K-RTA and the nine phosphorylation site mutants were compared in parallel by Western blot analysis using M2 FLAG antibody. α-tubulin served as a loading control. (Lower) luciferase reporter assay of KSHV PAN promoter responding to the wild type and the phosphorylation mutants transiently expressed in HEK293 cells. Data are presented as the means ± SD for quadruplicate assays. Three independent experiments were performed; a representative result is shown. (C) (Upper) comparison by flow cytometry of potency for viral latency disruption of K-RTA and S634A/S636A. 293/rKSHV.219 cells transfected with K-RTA or S634A/S636A for 72 h were analyzed on a fluorescence-activated cell sorter followed by scoring the percentage of double positive (GFP and RFP) cells in each population, as indicated in the upper right of each panel. Untreated cells (mock) served as a control. (Lower left) results derived from four independent experiments are summarized in the schematic chart. Data are presented as the means ± SD with K-RTA group set to 1. Statistical evaluation was performed with Student’s t-test. (Lower right) induction of expression of lytic protein K-bZIP by K-RTA and S634A/S636A in 293/rKSHV.219 cells was compared in parallel by Western blot analysis. β-actin served as a loading control. (D) (Left) total RNAs of 293/rKSHV.219 cells transfected with pLenti-FLAG-CPO (vector control), pLenti-FLAG-K-RTA, and pLenti-FLAG-S634A/S636A for 48 h were analyzed by quantitative RT-PCR for gene expressions of K-bZIP, ORF59, ORF2, and ORF36. Results from each reaction were normalized to cellular GAPDH. Statistical evaluations were performed by using Student’s t-test. (Right) KSHV viral particles released from K-RTA or S634A/S636A-expressing cells at 72 h were determined by comparative quantitative PCR of KSHV DNA polymerase genes (ORF9). Data are presented as means ± SD for three PCR assays in an experiment. Similar results were obtained from two independent experiments; one set of data is shown.
To extend these studies to cells infected with KSHV, K-RTA, and S634A/S636A, respectively, were ectopically expressed in 293/rKSHV.219 cells (Vieira and O’Hearn, 2004), followed by scoring of the percentage of GFP and RFP double positive cells, an indicator of KSHV lytic reactivation (Vieira and O’Hearn, 2004). The results showed that lytic reactivation induced by K-RTA was impaired when Ser-634 and Ser-636 were replaced with Ala, either by scoring of PAN promoter activity or by detecting K-bZIP expression (Figure 3C). This defect in lytic cycle reactivation of S634A/S636A was further validated by quantitative RT-PCR analysis of additional lytic genes (ORF2, ORF36, and ORF59) and determination of viral particles released in the culture media (Figure 3D). Thereby, we provide evidence indicating that phosphorylation of Ser-634 and Ser-636 contributes to the full potency of K-RTA in lytic cycle reactivation and virus production.
Taken together, we found that amino acids 633–652 of K-RTA shares 70% homology with a phosphorylated tryptic peptide derived from NELF-B. Among the three conserved Ser residues between K-RTA and NELF-B, Ser-636 seemed to play a role in K-RTA migration mobility and transactivation activity. In conjunction with Ser-634, Ser to Ala substitutions resulted in ∼25% decrement in K-RTA activity and was ∼10 kDa smaller in molecular weight.
K-RTA 634SPSP637 is a Potential CDK9 Target Site
At first, we hypothesized that the 10 kDa decrease in molecular mass of S634A/S636A was caused by a deficiency in phosphorylation, namely Ser-634 or Ser-636 may serve as a priming site to “dock” kinase(s), after which a successive, extensive phosphorylation is initiated. Thus, given a phosphate group is 80 Da, the 10 kDa difference could be attributed to as many as 125 phosphates conjugating onto K-RTA, but not to the S634A/S636A mutant; that is, the overall phosphorylation state in K-RTA should be higher than that in S634A/S636A. To validate this hypothesis, protein extracts of HEK293 cells transiently expressing FLAG-tagged K-RTA or S634A/S636A were treated with CIP and analyzed against their untreated counterparts by SDS-PAGE and Western blot analysis. As expected, CIP converted the majority of K-RTA and S634A/S636A to a 98 kDa species (Figure 4A). To determine the overall phosphorylation state in K-RTA vs. S634A/S636A, protein extracts were immunoprecipitated with M2 FLAG resin followed by Western blot analysis using anti-phospho Ser or anti-phospho Thr antibodies (Figure 4B). After normalized with the total protein (α-FLAG), the overall phosphorylation status of K-RTA and S634A/S636A was very similar. Taken together, these results refuted our hypothesis that the 10 kDa difference was caused by a deficiency in the degree of phosphorylation in S634A/S636A.
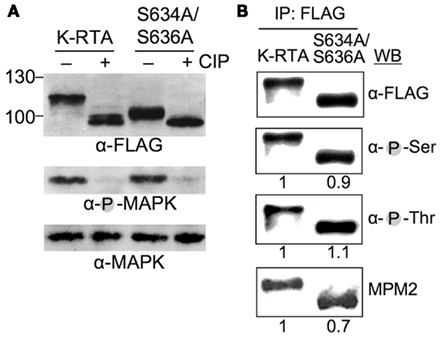
Figure 4. The 10-kDa difference between K-RTA and S634A/S636A is not due to distinct global phosphorylation state. (A) Protein extract from HEK293 cells transfected with plasmids expressing K-RTA or S634A/S636A for 24 h was incubated with (+) or without (−) calf intestinal phosphatase (CIP) at 37°C for 45 min. Each reaction mixture was analyzed in parallel by Western blot analysis using specified antibodies. Phosphorylated MAPK served as a control for completeness of CIP treatment. (B) The overall Ser phosphorylation (α-pSer), Thr phosphorylation (α-pThr), and MPM2 reactivity of K-RTA and S634A/S636A were revealed by immunoprecipitation (IP)-Western blot analysis. Because the expression abundance of each species were varied, the intensity of each band was first normalized with the intensity of its corresponding FLAG polypeptide (α-FLAG), followed by comparing to the normalized K-RTA value that was set to 1.
On the other hand, we noticed that 634SPSP637 of K-RTA represents a canonical motif of MPM2, an antibody that recognizes the phosphorylated Ser-Pro or phosphorylated Thr-Pro motifs present on a polypeptide, we tested whether mutation of this motif (634APAP637 in S634A/S636A) would influence its immuno-reactivity to MPM2 antibody. Indeed, as shown in Figure 4B, S634A/S636A retained ∼70% immuno-reactivity of MPM2 antibody relative to K-RTA by IP-Western blot analysis. This result suggested that 634SPSP637 could be an in vivo phosphorylation site. Because 634SPSP637 also represents a typical motif for Pro-directed kinases including various CDKs, an in vitro translation (IVT) coupled IP-kinase assay was performed to identify potential CDKs that phosphorylate Ser-634 or Ser-636. As a result, phosphorylation of K-RTA was detectable when CDK1, 2, or 9 were used as kinase source (not shown). Among these, we were particularly interested in CDK9 because it belongs to transcriptional CDK families that tightly control the conformation and processivity of RNA Pol II and negative elongation factor complex, NELF, and DSIF (Lolli, 2009).
First, by in vitro IP-kinase assay, we confirmed that CDK9, but not its dominant negative variant (CDK9-D167N) specifically phosphorylated K-RTA in vitro (Figure 5A). To inspect whether Ser-634 or Ser-636 were the target sites of CDK9, IP-kinase assays of K-RTA, and S634A/S636A were performed in parallel. As depicted in Figure 5B, although CDK9 phosphorylated both species, CDK9 phosphorylated higher fraction of K-RTA than that of S634A/S636A (1:0.4), strongly indicating that the 634SPSP637 motif could be one of the multiple CDK9 sites present in K-RTA. If this were true, we envisioned that in vivo, interaction between CDK9 and K-RTA would be stronger than that between CDK9 and S634A/S636A. As anticipated, results of IP-Western blot analysis revealed that higher fraction of K-RTA associated with CDK9 relative to that of S634A/S636A (1: 0.4, Figure 5C). The involvement of CDK9 in Ser-634 and Ser-636 phosphorylation was further supported by using DRB (5,6-dichloro-1-beta-D-ribofuranosylbenzimidazole) and roscovitine in a PAN promote-driven luciferase reporter assay. DRB and roscovitine are well-characterized CDK9 inhibitors (Taylor et al., 2004; Kapasi et al., 2009). When used as a dosage that did not significantly influence general transcription (evidenced by negligible changes in rellina activities of transfection controls, expressions of FLAG protein and β-actin between the control and the drugged groups, not shown), S634A/S636A was less sensitive to CDK9 inhibitors compared to K-RTA (Figure 5D), reinforcing that 634SPSP637 is an in vivo target site of CDK9. To extend these studies to cells infected with KSHV, K-RTA expressing 293/rKSHV.219 cells were treated with vehicle control (DMSO), DRB, or roscovitine for 42 h before harvest. RNAs of each group were subjected to quantitative RT-PCR analysis for various K-RTA target genes. As shown in Figure 5E, while the expressions of β-actin or RNA Pol III-directed 18S ribosomal RNA were insensitive to CDK9 inhibitors, the expression of MYC was significantly suppressed by DRB or roscovitine treatment, which is consistent with previous finding that MYC is a prototypic RNA Pol II pausing gene whose expression is regulated by CDK9-related pathway (Romano and Giordano, 2008). Notably, DRB and roscovitine also down-regulated the expressions of known K-RTA target genes (cellular SERPINB1; Brown et al., 2010, PAN, K-bZIP, and ORF59) and decreased the amount of viral particles released in the culture media, reinforcing the positive regulatory role of CDK9 in K-RTA-mediated transactivation and virus production. Taken together, we showed that although both K-RTA and S634A/S636A were phosphorylated in vitro by CDK9, S634A/S636A only retained 40% intensity of phosphorylation compared to that in K-RTA; both K-RTA and S634A/S636A associated with CDK9 in vivo, yet S634A/S636A only preserved 40% recruitment capacity relative to that of K-RTA; CDK9 inhibitors, DRB, and roscovitine, displayed higher inhibition potency on K-RTA than that of S634A/S636A. We conclude that there are multiple CDK9 target sites on K-RTA and 634SPSP637 motif is highly likely to be one of them.
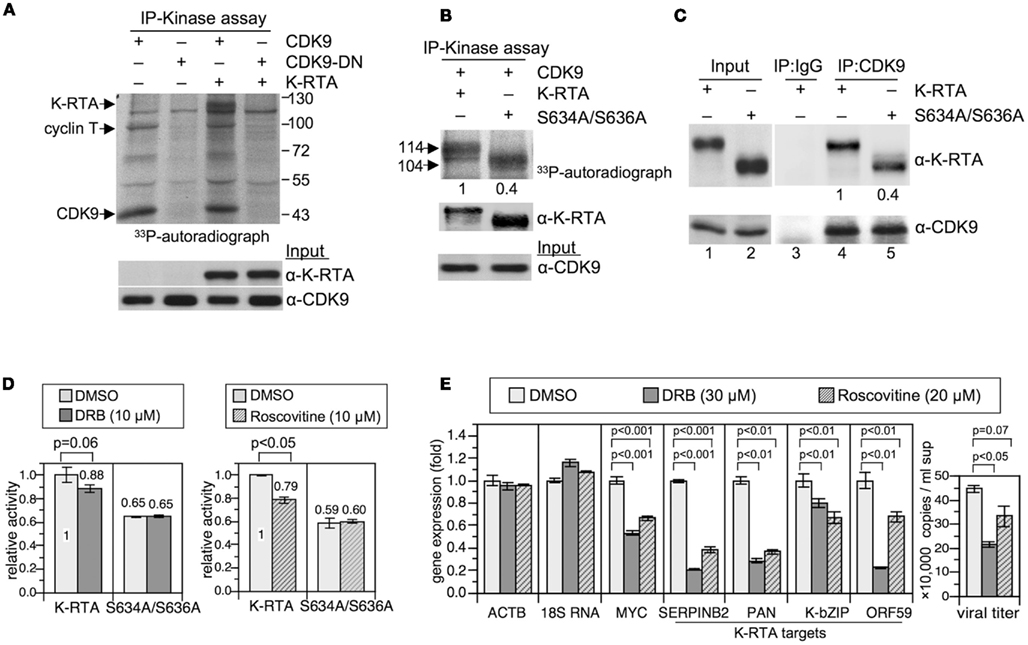
Figure 5. CDK9 phosphorylated K-RTA in vitro and CDK9 associated with K-RTA in vivo. (A) Protein extracts of 293T cells expressing indicated plasmids were harvested and subjected to immunoprecipitation (IP)-kinase assay using M2 FLAG affinity resin. The resulting mixtures were resolved by 8% SDS-PAGE and revealed by autoradiography. A distinct 114 kDa band is detected only when both K-RTA and CDK9 are present, indicating that K-RTA is a CDK9 substrate. CDK9-DN, CDK9 dominant negative. (B) Quantitation of CDK9 activities toward K-RTA vs. S634A/S636A. The band intensity in the autoradiograph film was normalized with the band intensity in α-K-RTA followed by setting K-RTA’s value to be 1. Although more S634A/S636A was captured in the immunocomplex, only 40% γ-33P-ATP was labeled onto S634A/S636A relative to that in K-RTA, suggesting that wild type K-RTA is a better substrate for CDK9. (C) Phosphorylation of Ser-634 and Ser-636 may play a role in CDK9 recruitment. Association of K-RTA and S634A/S636A with cellular CDK9 was determined by IP-Western blot analysis using α-CDK9 bounded Dynabeads-protein G. After normalized with the input proteins (lanes 1 and 2), S634A/S636A only retained 40% in CDK9 association ability, suggesting that Ser-634 ad Ser-636 are involved in CDK9 recruitment. (D) The effects of DRB (5,6-dichloro-1-beta-D-ribofuranosylbenzimidazole) and roscovitine on K-RTA-mediated transactivation of PAN promoter were determined by a luciferase reporter assay. HEK293 cells were cotransfected with plasmids expressing K-RTA or S634A/S636A together with PANp-driven luciferase reporter plasmid for 6 h, followed by inhibitor treatments for 18 h. Each firefly luciferase value was normalized to an internal control plasmid phRL-TK. The normalized value of the untreated group in K-RTA was set to 1. Data are presented as the means ± SD for quadruplicate transfections. Similar pattern was observed in two independent experiments. One set of data is shown. Statistical evaluations were performed with Student’s t-test. (E) (Left) quantitative RT-PCR analysis for gene expressions of β-actin (ACTB), 18S ribosomal RNA (18S RNA), known RNA Pol II pausing gene (MYC), and K-RTA target genes (cellular SERPINB2, KSHV PAN, K-bZIP, ORF59) in K-RTA expressing 293/rKSHV.219 cells treated with indicated inhibitors for 42 h. GADPH-normalized RNA value was quantified as fold change over the DMSO group. Statistical evaluations were performed by using Student’s t-test. (Right) KSHV viral particles released in the culture media from each treated groups were determined by comparative quantitative PCR of KSHV ORF9, as described in Figure 3D. Data are presented as means ± SD for three PCR assays in an experiment. Similar results were obtained from two independent experiments; one set of data is shown.
Discussion
In the present study, we demonstrated that phosphorylation of K-RTA can be subsidiary to (at Thr-513 and Thr-514) or influential in (at Ser-634 and Ser-636) the biological activity of K-RTA. We showed that mutation of 634SPSP637 to 634APAP637 in K-RTA reduced its capacity in CDK9 recruitment and potency in viral lytic cycle reactivation. Because CDK9 is central to regulating the RNA Pol II processivity on active promoters, we propose that similar to HIV Tat, by recruiting CDK9 to the viral genome K-RTA has evolved to potentiate the host transcription machinery for a robust viral transcriptome expression during lytic cycle replication.
Among the multiple mechanisms that control RNA Pol II activity in a transcription cycle, promoter-proximal pausing/stalling of RNA Pol II is considered as a rate-limiting step in the activation of hundreds of genes responding to dynamic environmental and developmental cues (Muse et al., 2007; Zeitlinger et al., 2007). In addition, genome-wide epigenetic analysis of human cells revealed that although only ∼30–45% of known genes have detectable transcripts, the majority of protein-coding genes are with active chromatin marks (K3K4me3) and bound with low level of RNA Pol II in the regions surrounding their promoters (Guenther et al., 2007). Interestingly, recent epigenetic studies revealed similar phenomena in the KSHV genome that it is also dynamically marked with both active and suppressive chromatin marks during latent infection, indicating that the KSHV genome is transcriptionally poised during latency, but can be rapidly activated in response to lytic cycle inducers (Gunther and Grundhoff, 2010; Toth et al., 2010).
The negative elongation factor (NELF) is a transcription regulatory complex comprises four subunits, NELF-A, B, C/D, and E. NELF exerts to interact stably with RNA Pol II and other elongation regulatory factors within the promoter-proximal region, which prevents further transcription elongation. Recently, a novel role of NELF was revealed: by residing at the pausing site, NELF prevents nucleosome formation and maintains a permissive chromatin architecture that allows a rapid transcription process upon induction. Thus, NELF-mediated RNA Pol II pausing can provide both negative and positive effects on gene expression (Gilchrist et al., 2008). Regardless of which effect, phosphorylation of NELF by CDK9 enables the disengagement of NELF from the promoter-proximal region that is followed by a productive elongation process.
Here, we found that amino acids 633–652 of K-RTA share sequence homology with amino acids 554–573 of NELF-B, which is located in an in vivo phosphorylated tryptic peptide (554–580). Although the kinase that phosphorylates this tryptic peptide in NELF-B has not been characterized, we found that Ser-634 and Ser-636 in K-RTA are involved in CDK9 recruitment/phosphorylation and the expressions of multiple K-RTA target genes were impaired when CDK9 inhibitors were present. In addition, de-phosphorylated Ser-634 and Ser-636 mimetic, namely S634A/S636A, is 10 kDa shorter than the wild type, however, this decrement in molecular mass was not due to deficiency in global Ser- or Thr-phosphorylation, which raises a possibility that phosphorylation-mediated conformational change might be involved. Along the same vein, S634A/S636A was less immunoreactive to MPM2 antibody, suggesting that phosphorylation of 634SPSP637 could be targeted by PIN1, a peptidyl-prolyl cis/trans isomerase that regulates the conformation and function of numerous MPM2-recognized proteins (Stukenberg and Kirschner, 2001; Lu and Zhou, 2007; Shaw, 2007). More experiments are required to prove this hypothesis. Taken together, it is envisioned that K-RTA possesses functional domain similar to those in NELF, which serves to recruit CDK9 and assures a productive gene expression program on the viral genome.
Although K-RTA has been known as a nuclear protein, the full maturation process of K-RTA may involve multiple cellular compartments. To distinguish various compartment-specific phosphorylation processes, we constructed a nuclear localization mutant of K-RTA, referred to as NLSm. While primarily located in the cytoplasm by immunofluorescence assays, NLSm retained 50% wild type transactivation activity (Figure 1D). There are two possibilities to account for this result. First, a proportion of NLSm may “sneak into” the nucleus in the overwhelming liposomal transfection used in the luciferase reporter assay. This possibility is supported by the presence of the 114 kDa nuclear polypeptide in protein extracts of long term (24 h) Dox-treated NLSm cells (Figure 2A, middle). Alternatively, NLSm could exert some yet-to-be-identified actions in the cytosol. For example, EBV Rta was documented to be functional in the cytoplasm (Hsu et al., 2005).
Originally, we anticipated to see more phosphorylated residues in the 114 kDa nuclear form of K-RTA because of its apparently larger molecular mass. To our surprise, the same Thr-513 and Thr-514 were shown to be the only phosphorylated amino acids identified in both K-RTA and NLSm by LC/MS/MS. Two possibilities could contribute to this result. One is that our MS analysis may exclude some larger tryptic peptides because of their size (e.g., amino acids 531–633, approximately 10.6 kDa, contain 25 Ser/Thr). The other possibility is that we did not obtain adequate quantity of phosphorylated K-RTA in the first place. We believe that further improvement of the purification procedure, or the use of alternative proteases such as chymotrypsin to supplement the trypsin digestion, will help to identify more phosphorylated sites in the mature 114 kDa K-RTA by MS. Nonetheless, in the present study we show that Thr-513 or Thr-514 is the preferred phosphorylation site in K-RTA that is modified in the cytoplasmic compartment and retained in the nucleus.
In summary, hijacking of host transcription machinery, including CDK9, by the virus to express its own transcriptome is increasingly documented (Garber et al., 2000; Zhou et al., 2000; Bark-Jones et al., 2006; Durand and Roizman, 2008; Kapasi and Spector, 2008; Kapasi et al., 2009). Our finding that K-RTA-mediated transcriptional activation is impaired by two CDK9 inhibitors provides a new example of this paradigm. Interestingly, the CDK inhibitor R-roscovitine (also known as seliciclib and CYC202) was used recently to reduce tumor size and plasma EBV DNA in patients with nasopharyngeal carcinoma (Hsieh et al., 2009). Thus, the application of CDK9 inhibitors to disturb virus replication may provide a promising direction for treatment of viral malignancies including Kaposi’s sarcoma.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank the Khoo Lab (Institute of Biological Chemistry, Academia Sinica, Taiwan) for inspiring discussion and the initiation of this project. We are grateful to Dr. Dan Robinson (MCTP, University of Michigan, Ann Arbor) for providing pLenti4-FLAG-CPO related plasmids, Dr. Jeffrey Vieira (University of Washington, WI) for rKSHV.219, Dr. Keiji Ueda (Osaka University Graduate School of Medicine, Japan), and Dr. Yoshihiro Izumiya (University of California at Davis, USA) for K-RTA antibodies. The work was supported by Taiwan NHRI CA-101-PP17, DOH101-TD-C-111-004, and NSC100-2325-B-002-067.
References
Bark-Jones, S. J., Webb, H. M., and West, M. J. (2006). EBV EBNA 2 stimulates CDK9-dependent transcription and RNA polymerase II phosphorylation on serine 5. Oncogene 25, 1775–1785.
Barlev, N. A., Emelyanov, A. V., Castagnino, P., Zegerman, P., Bannister, A. J., Sepulveda, M. A., Robert, F., Tora, L., Kouzarides, T., Birshtein, B. K., and Berger, S. L. (2003). A novel human Ada2 homologue functions with Gcn5 or Brg1 to coactivate transcription. Mol. Cell. Biol. 23, 6944–6957.
Barrasa, M. I., Harel, N. Y., and Alwine, J. C. (2005). The phosphorylation status of the serine-rich region of the human cytomegalovirus 86-kilodalton major immediate-early protein IE2/IEP86 affects temporal viral gene expression. J. Virol. 79, 1428–1437.
Beausoleil, S. A., Jedrychowski, M., Schwartz, D., Elias, J. E., Villen, J., Li, J., Cohn, M. A., Cantley, L. C., and Gygi, S. P. (2004). Large-scale characterization of HeLa cell nuclear phosphoproteins. Proc. Natl. Acad. Sci. U.S.A. 101, 12130–12135.
Bhende, P. M., Seaman, W. T., Delecluse, H. J., and Kenney, S. C. (2005). BZLF1 activation of the methylated form of the BRLF1 immediate-early promoter is regulated by BZLF1 residue 186. J. Virol. 79, 7338–7348.
Blom, N., Sicheritz-Ponten, T., Gupta, R., Gammeltoft, S., and Brunak, S. (2004). Prediction of post-translational glycosylation and phosphorylation of proteins from the amino acid sequence. Proteomics 4, 1633–1649.
Boutell, C., Everett, R., Hilliard, J., Schaffer, P., Orr, A., and Davido, D. (2008). Herpes simplex virus type 1 ICP0 phosphorylation mutants impair the E3 ubiquitin ligase activity of ICP0 in a cell type-dependent manner. J. Virol. 82, 10647–10656.
Brown, H. J., Peng, L., Harada, J. N., Walker, J. R., Cole, S., Lin, S. F., Zack, J. A., Chanda, S. K., and Sun, R. (2010). Gene expression and transcription factor profiling reveal inhibition of transcription factor cAMP-response element-binding protein by gamma-herpesvirus replication and transcription activator. J. Biol. Chem. 285, 25139–25153.
Bu, W., Palmeri, D., Krishnan, R., Marin, R., Aris, V. M., Soteropoulos, P., and Lukac, D. M. (2008). Identification of direct transcriptional targets of the Kaposi’s sarcoma-associated herpesvirus Rta lytic switch protein by conditional nuclear localization. J. Virol. 82, 10709–10723.
Chang, H., Gwack, Y., Kingston, D., Souvlis, J., Liang, X., Means, R. E., Cesarman, E., Hutt-Fletcher, L., and Jung, J. U. (2005). Activation of CD21 and CD23 gene expression by Kaposi’s sarcoma-associated herpesvirus RTA. J. Virol. 79, 4651–4663.
Chang, P. C., and Li, M. (2008). Kaposi’s sarcoma-associated herpesvirus K-cyclin interacts with Cdk9 and stimulates Cdk9-mediated phosphorylation of p53 tumor suppressor. J. Virol. 82, 278–290.
Chang, P. J., Shedd, D., and Miller, G. (2008). A mobile functional region of Kaposi’s sarcoma-associated herpesvirus ORF50 protein independently regulates DNA binding and protein abundance. J. Virol. 82, 9700–9716.
Chen, J., Ueda, K., Sakakibara, S., Okuno, T., and Yamanishi, K. (2000). Transcriptional regulation of the Kaposi’s sarcoma-associated herpesvirus viral interferon regulatory factor gene. J. Virol. 74, 8623–8634.
Chen, Y. J., Tsai, W. H., Chen, Y. L., Ko, Y. C., Chou, S. P., Chen, J. Y., and Lin, S. F. (2011). Epstein-Barr virus (EBV) Rta-mediated EBV and Kaposi’s Sarcoma-associated herpesvirus lytic reactivations in 293 cells. PLoS ONE 6, e17809. doi:10.1371/journal.pone.0017809
Cokol, M., Nair, R., and Rost, B. (2000). Finding nuclear localization signals. EMBO Rep. 1, 411–415.
Durand, L. O., and Roizman, B. (2008). Role of cdk9 in the optimization of expression of the genes regulated by ICP22 of herpes simplex virus 1. J. Virol. 82, 10591–10599.
El-Guindy, A. S., and Miller, G. (2004). Phosphorylation of Epstein-Barr virus ZEBRA protein at its casein kinase 2 sites mediates its ability to repress activation of a viral lytic cycle late gene by Rta. J. Virol. 78, 7634–7644.
Fakhari, F. D., and Dittmer, D. P. (2002). Charting latency transcripts in Kaposi’s sarcoma-associated herpesvirus by whole-genome real-time quantitative PCR. J. Virol. 76, 6213–6223.
Foskett, S. M., Ghose, R., Tang, D. N., Lewis, D. E., and Rice, A. P. (2001). Antiapoptotic function of Cdk9 (TAK/P-TEFb) in U937 promonocytic cells. J. Virol. 75, 1220–1228.
Francis, A. L., Gradoville, L., and Miller, G. (1997). Alteration of a single serine in the basic domain of the Epstein-Barr virus ZEBRA protein separates its functions of transcriptional activation and disruption of latency. J. Virol. 71, 3054–3061.
Fujinaga, K., Irwin, D., Geyer, M., and Peterlin, B. M. (2002). Optimized chimeras between kinase-inactive mutant Cdk9 and truncated cyclin T1 proteins efficiently inhibit Tat transactivation and human immunodeficiency virus gene expression. J. Virol. 76, 10873–10881.
Ganem, D. (2007). “Kaposi’s sarcoma-associated herpesvirus,” in Fields Virology, eds D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (Philadelphia, PA: Lippincott Williams and Wilkins), 2847–2888.
Garber, M. E., Mayall, T. P., Suess, E. M., Meisenhelder, J., Thompson, N. E., and Jones, K. A. (2000). CDK9 autophosphorylation regulates high-affinity binding of the human immunodeficiency virus type 1 tat-P-TEFb complex to TAR RNA. Mol. Cell. Biol. 20, 6958–6969.
Gilchrist, D. A., Nechaev, S., Lee, C., Ghosh, S. K., Collins, J. B., Li, L., Gilmour, D. S., and Adelman, K. (2008). NELF-mediated stalling of Pol II can enhance gene expression by blocking promoter-proximal nucleosome assembly. Genes Dev. 22, 1921–1933.
Gradoville, L., Gerlach, J., Grogan, E., Shedd, D., Nikiforow, S., Metroka, C., and Miller, G. (2000). Kaposi’s sarcoma-associated herpesvirus open reading frame 50/Rta protein activates the entire viral lytic cycle in the HH-B2 primary effusion lymphoma cell line. J. Virol. 74, 6207–6212.
Guenther, M. G., Levine, S. S., Boyer, L. A., Jaenisch, R., and Young, R. A. (2007). A chromatin landmark and transcription initiation at most promoters in human cells. Cell 130, 77–88.
Gunther, T., and Grundhoff, A. (2010). The epigenetic landscape of latent Kaposi sarcoma-associated herpesvirus genomes. PLoS Pathog. 6, e1000935. doi:10.1371/journal.ppat.1000935
Gwack, Y., Baek, H. J., Nakamura, H., Lee, S. H., Meisterernst, M., Roeder, R. G., and Jung, J. U. (2003a). Principal role of TRAP/mediator and SWI/SNF complexes in Kaposi’s sarcoma-associated herpesvirus RTA-mediated lytic reactivation. Mol. Cell. Biol. 23, 2055–2067.
Gwack, Y., Nakamura, H., Lee, S. H., Souvlis, J., Yustein, J. T., Gygi, S., Kung, H. J., and Jung, J. U. (2003b). Poly(ADP-ribose) polymerase 1 and Ste20-like kinase hKFC act as transcriptional repressors for gamma-2 herpesvirus lytic replication. Mol. Cell. Biol. 23, 8282–8294.
Gwack, Y., Byun, H., Hwang, S., Lim, C., and Choe, J. (2001). CREB-binding protein and histone deacetylase regulate the transcriptional activity of Kaposi’s sarcoma-associated herpesvirus open reading frame 50. J. Virol. 75, 1909–1917.
Gwack, Y., Hwang, S., Lim, C., Won, Y. S., Lee, C. H., and Choe, J. (2002). Kaposi’s Sarcoma-associated herpesvirus open reading frame 50 stimulates the transcriptional activity of STAT3. J. Biol. Chem. 277, 6438–6442.
Hsiao, H. H., Hsieh, H. Y., Chou, C. C., Lin, S. Y., Wang, A. H., and Khoo, K. H. (2007). Concerted experimental approach for sequential mapping of peptides and phosphopeptides using C18-functionalized magnetic nanoparticles. J. Proteome Res. 6, 1313–1324.
Hsieh, W. S., Soo, R., Peh, B. K., Loh, T., Dong, D., Soh, D., Wong, L. S., Green, S., Chiao, J., Cui, C. Y., Lai, Y. F., Lee, S. C., Mow, B., Soong, R., Salto-Tellez, M., and Goh, B. C. (2009). Pharmacodynamic effects of seliciclib, an orally administered cell cycle modulator, in undifferentiated nasopharyngeal cancer. Clin. Cancer Res. 15, 1435–1442.
Hsu, T. Y., Chang, Y., Wang, P. W., Liu, M. Y., Chen, M. R., Chen, J. Y., and Tsai, C. H. (2005). Reactivation of Epstein-Barr virus can be triggered by an Rta protein mutated at the nuclear localization signal. J. Gen. Virol. 86, 317–322.
Kang, H., and Lieberman, P. M. (2011). Mechanism of glycyrrhizic acid inhibition of Kaposi’s sarcoma-associated herpesvirus: disruption of CTCF-cohesin-mediated RNA polymerase II pausing and sister chromatid cohesion. J. Virol. 85, 11159–11169.
Kapasi, A. J., Clark, C. L., Tran, K., and Spector, D. H. (2009). Recruitment of cdk9 to the immediate-early viral transcriptosomes during human cytomegalovirus infection requires efficient binding to cyclin T1, a threshold level of IE2 86, and active transcription. J. Virol. 83, 5904–5917.
Kapasi, A. J., and Spector, D. H. (2008). Inhibition of the cyclin-dependent kinases at the beginning of human cytomegalovirus infection specifically alters the levels and localization of the RNA polymerase II carboxyl-terminal domain kinases cdk9 and cdk7 at the viral transcriptosome. J. Virol. 82, 394–407.
Liang, Y., and Ganem, D. (2003). Lytic but not latent infection by Kaposi’s sarcoma-associated herpesvirus requires host CSL protein, the mediator of Notch signaling. Proc. Natl. Acad. Sci. U.S.A. 100, 8490–8495.
Lin, S. Y., Chen, Y. Y., Fan, Y. Y., Lin, C. W., Chen, S. T., Wang, A. H., and Khoo, K. H. (2008). Precise mapping of increased sialylation pattern and the expression of acute phase proteins accompanying murine tumor progression in BALB/c mouse by integrated sera proteomics and glycomics. J. Proteome Res. 7, 3293–3303.
Lolli, G. (2009). Binding to DNA of the RNA-polymerase II C-terminal domain allows discrimination between Cdk7 and Cdk9 phosphorylation. Nucleic Acids Res. 37, 1260–1268.
Lu, K. P., and Zhou, X. Z. (2007). The prolyl isomerase PIN1: a pivotal new twist in phosphorylation signalling and disease. Nat. Rev. Mol. Cell Biol. 8, 904–916.
Lukac, D. M., Kirshner, J. R., and Ganem, D. (1999). Transcriptional activation by the product of open reading frame 50 of Kaposi’s sarcoma-associated herpesvirus is required for lytic viral reactivation in B cells. J. Virol. 73, 9348–9361.
Lukac, D. M., Renne, R., Kirshner, J. R., and Ganem, D. (1998). Reactivation of Kaposi’s sarcoma-associated herpesvirus infection from latency by expression of the ORF 50 transactivator, a homolog of the EBV R protein. Virology 252, 304–312.
Marchler-Bauer, A., Anderson, J. B., Cherukuri, P. F., DeWeese-Scott, C., Geer, L. Y., Gwadz, M., He, S., Hurwitz, D. I., Jackson, J. D., Ke, Z., Lanczycki, C. J., Liebert, C. A., Liu, C., Lu, F., Marchler, G. H., Mullokandov, M., Shoemaker, B. A., Simonyan, V., Song, J. S., Thiessen, P. A., Yamashita, R. A., Yin, J. J., Zhang, D., and Bryant, S. H. (2005). CDD: a conserved domain database for protein classification. Nucleic Acids Res. 33, D192–D196.
Muse, G. W., Gilchrist, D. A., Nechaev, S., Shah, R., Parker, J. S., Grissom, S. F., Zeitlinger, J., and Adelman, K. (2007). RNA polymerase is poised for activation across the genome. Nat. Genet. 39, 1507–1511.
Nakamura, H., Lu, M., Gwack, Y., Souvlis, J., Zeichner, S. L., and Jung, J. U. (2003). Global changes in Kaposi’s sarcoma-associated virus gene expression patterns following expression of a tetracycline-inducible Rta transactivator. J. Virol. 77, 4205–4220.
Olsen, J. V., Blagoev, B., Gnad, F., Macek, B., Kumar, C., Mortensen, P., and Mann, M. (2006). Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell 127, 635–648.
Palermo, R. D., Webb, H. M., and West, M. J. (2011). RNA polymerase II stalling promotes nucleosome occlusion and pTEFb recruitment to drive immortalization by Epstein-Barr virus. PLoS Pathog. 7, e1002334. doi:10.1371/journal.ppat.1002334
Peterlin, B. M., and Price, D. H. (2006). Controlling the elongation phase of transcription with P-TEFb. Mol. Cell 23, 297–305.
Romano, G., and Giordano, A. (2008). Role of the cyclin-dependent kinase 9-related pathway in mammalian gene expression and human diseases. Cell Cycle 7, 3664–3668.
Sakakibara, S., Ueda, K., Chen, J., Okuno, T., and Yamanishi, K. (2001). Octamer-binding sequence is a key element for the autoregulation of Kaposi’s sarcoma-associated herpesvirus ORF50/Lyta gene expression. J. Virol. 75, 6894–6900.
Sarid, R., Flore, O., Bohenzky, R. A., Chang, Y., and Moore, P. S. (1998). Transcription mapping of the Kaposi’s sarcoma-associated herpesvirus (human herpesvirus 8) genome in a body cavity-based lymphoma cell line (BC-1). J. Virol. 72, 1005–1012.
Saunders, A., Core, L. J., and Lis, J. T. (2006). Breaking barriers to transcription elongation. Nat. Rev. Mol. Cell Biol. 7, 557–567.
Shaw, P. E. (2007). Peptidyl-prolyl cis/trans isomerases and transcription: is there a twist in the tail? EMBO Rep. 8, 40–45.
Stukenberg, P. T., and Kirschner, M. W. (2001). Pin1 acts catalytically to promote a conformational change in Cdc25. Mol. Cell 7, 1071–1083.
Sun, R., Lin, S. F., Staskus, K., Gradoville, L., Grogan, E., Haase, A., and Miller, G. (1999). Kinetics of Kaposi’s sarcoma-associated herpesvirus gene expression. J. Virol. 73, 2232–2242.
Taylor, S. L., Kinchington, P. R., Brooks, A., and Moffat, J. F. (2004). Roscovitine, a cyclin-dependent kinase inhibitor, prevents replication of varicella-zoster virus. J. Virol. 78, 2853–2862.
Toth, Z., Maglinte, D. T., Lee, S. H., Lee, H. R., Wong, L. Y., Brulois, K. F., Lee, S., Buckley, J. D., Laird, P. W., Marquez, V. E., and Jung, J. U. (2010). Epigenetic analysis of KSHV latent and lytic genomes. PLoS Pathog. 6, e1001013. doi:10.1371/journal.ppat.1001013
Trester-Zedlitz, M., Burlingame, A., Kobilka, B., and von Zastrow, M. (2005). Mass spectrometric analysis of agonist effects on posttranslational modifications of the beta-2 adrenoceptor in mammalian cells. Biochemistry 44, 6133–6143.
Vieira, J., and O’Hearn, P. M. (2004). Use of the red fluorescent protein as a marker of Kaposi’s sarcoma-associated herpesvirus lytic gene expression. Virology 325, 225–240.
Wang, J. T., Yang, P. W., Lee, C. P., Han, C. H., Tsai, C. H., and Chen, M. R. (2005). Detection of Epstein-Barr virus BGLF4 protein kinase in virus replication compartments and virus particles. J. Gen. Virol. 86, 3215–3225.
Wang, S. E., Wu, F. Y., Yu, Y., and Hayward, G. S. (2003). CCAAT/enhancer-binding protein-alpha is induced during the early stages of Kaposi’s sarcoma-associated herpesvirus (KSHV) lytic cycle reactivation and together with the KSHV replication and transcription activator (RTA) cooperatively stimulates the viral RTA, MTA, and PAN promoters. J. Virol. 77, 9590–9612.
Xue, Y., Ren, J., Gao, X., Jin, C., Wen, L., and Yao, X. (2008). GPS 2.0, a tool to predict kinase-specific phosphorylation sites in hierarchy. Mol. Cell. Proteomics 7, 1598–1608.
Yang, Z., and Wood, C. (2007). The transcriptional repressor K-RBP modulates RTA-mediated transactivation and lytic replication of Kaposi’s sarcoma-associated herpesvirus. J. Virol. 81, 6294–6306.
Yoo, S. M., Zhou, F. C., Ye, F. C., Pan, H. Y., and Gao, S. J. (2005). Early and sustained expression of latent and host modulating genes in coordinated transcriptional program of KSHV productive primary infection of human primary endothelial cells. Virology 343, 47–64.
Zeitlinger, J., Stark, A., Kellis, M., Hong, J. W., Nechaev, S., Adelman, K., Levine, M., and Young, R. A. (2007). RNA polymerase stalling at developmental control genes in the Drosophila melanogaster embryo. Nat. Genet. 39, 1512–1516.
Zhou, M., Halanski, M. A., Radonovich, M. F., Kashanchi, F., Peng, J., Price, D. H., and Brady, J. N. (2000). Tat modifies the activity of CDK9 to phosphorylate serine 5 of the RNA polymerase II carboxyl-terminal domain during human immunodeficiency virus type 1 transcription. Mol. Cell. Biol. 20, 5077–5086.
Appendix
Keywords: Kaposi’s sarcoma-associated herpesvirus, replication and transcription activator, phosphorylation, negative elongation factor B, CDK9
Citation: Tsai W-H, Wang P-W, Lin S-Y, Wu I-L, Ko Y-C, Chen Y-L, Li M and Lin S-F (2012) Ser-634 and Ser-636 of Kaposi’s sarcoma-associated herpesvirus RTA are involved in transactivation and are potential Cdk9 phosphorylation sites. Front. Microbio. 3:60. doi: 10.3389/fmicb.2012.00060
Received: 01 December 2011;
Paper pending published: 19 December 2011;
Accepted: 05 February 2012;
Published online: 22 February 2012.
Edited by:
Keiji Ueda, Osaka University Graduate School of Medicine, JapanReviewed by:
Keiji Ueda, Osaka University Graduate School of Medicine, JapanYoshihiro Izumiya, University of California Davis, USA
Sathish Narayanan, Indian Institute of Science Education and Research Bhopal, India
Copyright: © 2012 Tsai, Wang, Lin, Wu, Ko, Chen, Li and Lin. This is an open-access article distributed under the terms of the Creative Commons Attribution Non Commercial License, which permits non-commercial use, distribution, and reproduction in other forums, provided the original authors and source are credited.
*Correspondence: Su-Fang Lin, National Institute of Cancer Research, National Health Research Institutes, No. 35, Keyan Road, Zhunan Town, Miaoli County, Taiwan. e-mail: sflin1@gmail.com
 Wan-Hua Tsai1
Wan-Hua Tsai1