The evolution and polymorphism of mono-amino acid repeats in androgen receptor and their regulatory role in health and disease
- 1VIB-VUB Center for Structural Biology, Vlaams Instituut voor Biotechnologie (VIB), Brussels, Belgium
- 2Structural Biology Brussels (SBB), Vrije Universiteit Brussel (VUB), Brussels, Belgium
- 3Research Centre for Natural Sciences (RCNS), ELKH, Budapest, Hungary
Androgen receptor (AR) is a key member of nuclear hormone receptors with the longest intrinsically disordered N-terminal domain (NTD) in its protein family. There are four mono-amino acid repeats (polyQ1, polyQ2, polyG, and polyP) located within its NTD, of which two are polymorphic (polyQ1 and polyG). The length of both polymorphic repeats shows clinically important correlations with disease, especially with cancer and neurodegenerative diseases, as shorter and longer alleles exhibit significant differences in expression, activity and solubility. Importantly, AR has also been shown to undergo condensation in the nucleus by liquid-liquid phase separation, a process highly sensitive to protein solubility and concentration. Nonetheless, in prostate cancer cells, AR variants also partition into transcriptional condensates, which have been shown to alter the expression of target gene products. In this review, we summarize current knowledge on the link between AR repeat polymorphisms and cancer types, including mechanistic explanations and models comprising the relationship between condensate formation, polyQ1 length and transcriptional activity. Moreover, we outline the evolutionary paths of these recently evolved amino acid repeats across mammalian species, and discuss new research directions with potential breakthroughs and controversies in the literature.
Introduction
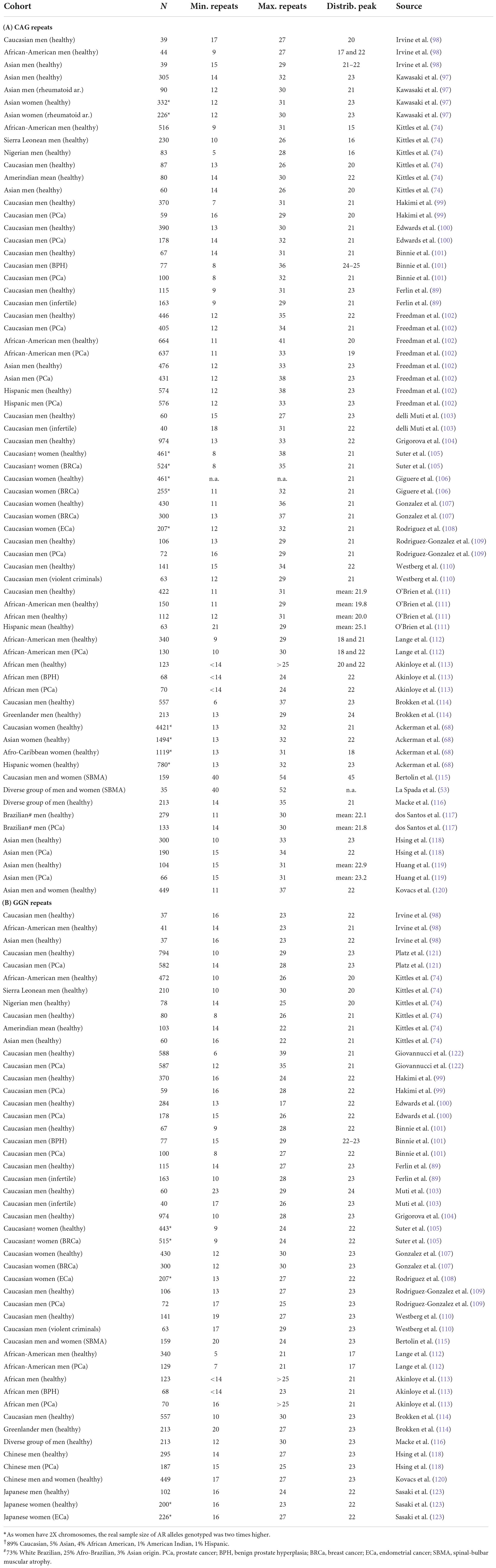
The protein family of nuclear hormone receptors (NHRs) includes several hormone-sensitive transcription factors (TFs), which were discovered and initially characterized as tissue-specific intracellular receptors whose functions are regulated by specific endocrine hormones (1, 2). Sequencing these NHRs [glucocorticoid receptor (GR), estrogen receptor (ER), thyroid hormone receptor, and retinoic acid receptor] revealed a common domain architecture and sequence homology that enabled the establishment of this class as a protein family, and also revealed a large set of orphan receptors with no identified activating hormones (1, 3) and among those mineralocorticoid receptor (MR), androgen receptor (AR), and progesterone receptor (PR) (Figure 1) as reveal later (4–6). The longest isoforms of NHRs generally have a hormone-sensitive ligand-binding domain (LBD), encoded by 5 exons whereas 2 exons encode two zinc-fingers that make up the DNA-binding domain (DBD). In addition, most commonly one exon encodes the N-terminal transactivation domain (NTD) with variable size characteristic to each NHR (Figure 1). In NTD of AR, there is a polymorphic glutamine repeat (polyQ1) and a polymorphic glycine repeat (polyG) with variable length in the human population (see Sections “The polymorphic polyglutamine regions of human androgen receptor” and “The polymorphic polyglycine region of human androgen receptor”), and also in other mammalian species (see Section “Evolution and phylogenetics of the polymorphic repeat regions”). This variability arises due to the slippage of the DNA polymerase during DNA replication caused by the presence of multiple copies of CAG and GGN codons on the template strand (7).
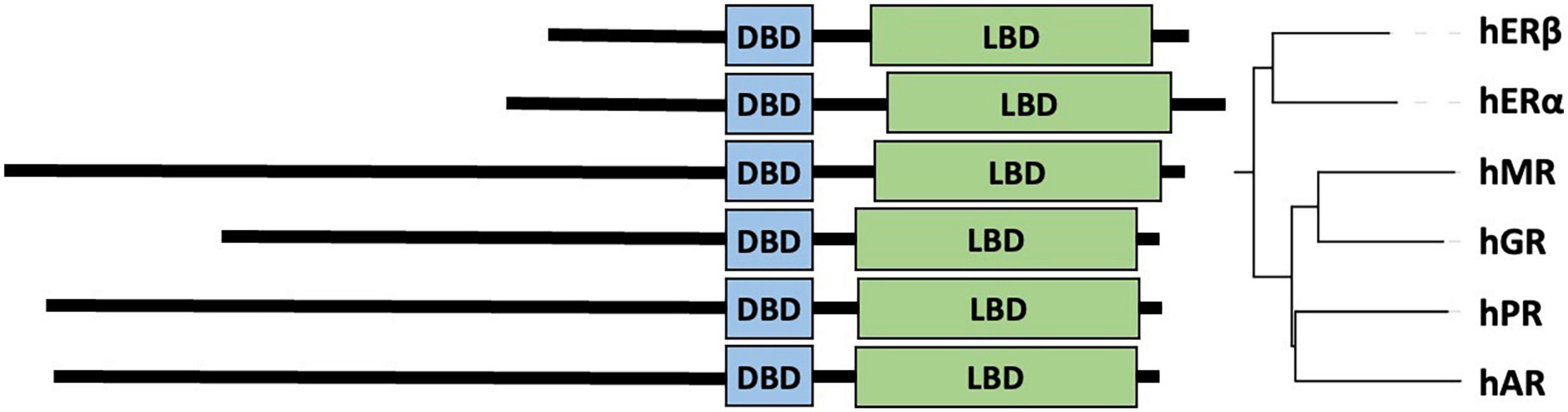
Figure 1. Similarity of type-1 nuclear hormone receptors. Domain architecture aligned by DBD (left) and neighbor-joining phylogenetic tree (right) of six steroid hormone receptor proteins globally aligned (32). UniProt accessions of the sequences (33) are human estrogen receptor beta (hERβ): Q92731, human estrogen receptor alpha (hERα): P03372, human mineralocorticoid receptor (hMR): P08235, human glucocorticoid receptor (hGR): P04150, human progesterone receptor (hPR): P06401, and human androgen receptor (hAR): P10275.
Most NHRs have multiple different isoforms that are shorter than the canonical isoforms. A shorter isoform of AR primarily expressed in castration-resistant prostate cancer (CRPC), ARv7, indicates poorer prognosis in prostate cancer (PCa), as it lacks the LBD and remains overly active even in absence of the hormone, the mechanism of which is still not fully understood (8). Our understanding of these regulatory mechanisms is also limited by the fact that the structure of full-length AR has not been fully determined at near-atomic resolution due to its high degree of flexibility (559 amino acid-long intrinsically disordered NTD) and large size (920 residues). The structures of human AR-LBD (PDB: 4oha) and rat AR-DBD (PDB: 1r4i) have been resolved by X-ray crystallography at 1.42 Å and 3.10 Å resolution, respectively (9, 10). AR-NTD has only been partially modeled (Tau5-R2/3) by nuclear magnetic resonance (NMR) chemical shift-reweighted ensembles (PED: PED00206) based on molecular dynamics simulations (11) and is made available in the Protein Ensemble Database (PED) (12). Recently, the first low-resolution cryo-electron microscopy structure (EMDB: EMD-22079/22080) was reported for transcriptionally active full-length AR, highlighting important conformations in the interdomain cross-talk in AR upon DNA binding and also in complex with Src3 and p300 (13).
In its inactive state, AR localizes in the cytoplasm sequestered by heat shock proteins (HSPs) (13). In a mechanistic structural study, it was shown that Hsp70 and Hsp40 inhibit the NTD-LBD interaction by binding a hydrophobic motif in the NTD (14). Upon binding androgen hormones by the LBD, AR dissociates from HSPs and undergoes homodimerization through LBD and DBD, and enters the nucleus where it binds specific DNA sequences, called androgen response elements (AREs) (13, 15, 16). However, ARv7, which lacks LBD, is constitutively active and resistant against regular cancer treatments that mainly target the LBD. Interestingly, ARv7 also shows constituent nuclear localization despite lacking the natural hinge region, which contains an important nuclear localization sequence (NLS) (17–19). The nuclear shuttling of ARv7 has been shown to occur with a different molecular mechanism compared to the full-length AR (20). In the NTD, there have been important regions proposed to regulate the efficient transcription activation function, including a transactivation domain AF1 (aa. 142-485). AF1 is responsible for recruiting different partners and co-factors to regulate transcription, such as SRC1, SRC3, p300, TFIIF via various motifs (21–23). Moreover, the highly conserved first 30 residue of NTD contains a hydrophobic binding motif (FQNLF), which interacts with the LBD or melanoma-associated antigen-11 (MAGE-A11), thereby regulating the NTD-LBD interaction (24). The same hydrophobic motif is responsible for binding Hsp40 and Hsp70 in the cytosol (14). Lastly, the length-polymorphic glutamine and glycine stretches (polyQ1/polyG) have also been shown to affect the transcriptional activity (25–27).
Intriguingly, besides physiological cytoplasmic sequestering, HSPs can also affect AR signaling in PCa by triggering the degradation of the receptor, thereby resulting in lower transactivation (28, 29). Even though ARv7 seemed to be resistant to first-generation HSP inhibitors (28, 30), a new, second-generation HSP90 inhibitor decreased ARv7 level through a different mechanism, affecting the mRNA splicing of this variant (31).
In this review, we initially focus on the structure-function relationship of AR’s polymorphic (polyQ1, polyG) and non-polymorphic (polyQ2, polyP) mono-amino acid repeat regions, the evolution and phylogenetic differences thereof, also highlighting current controversies in the literature. In the end, we list a few well-known and recently identified research gaps and propose future research directions with high potential for great breakthroughs.
Structure, function, and disease relevance of the androgen receptor’s mono-amino acid repeat regions
The polymorphic polyglutamine regions of human androgen receptor
Polyglutamine repeat tracts are frequent in the proteome from yeast to humans and they are over-represented in the activation domain of TFs (34). In yeast, even small modification in the number of glutamine residues in the polyQ region of the transcription factor Ssn6 (Cyc8) resulted in phenotypic differences between strains as well as different fitness under certain nutrient stress (34). The exact mechanism of action is still under debate. However, the most accepted explanation is that the length of the polyQ influences the solubility and conformation of the protein (35). Therefore, a difference in polyQ length can alter the interaction of the TF with its cofactors, hence affecting upregulation or downregulation of target genes.
The transactivation domains of TFs are usually intrinsically disordered and considered to be of low complexity (36–39). The NTD of AR has also been predicted to be disordered by all the prediction tools available, which has been also verified experimentally (40, 41). However, a study based on circular dichroism and fluorescence emission spectra suggested that the polyQ1 region has an alpha-helical propensity (42), which was later verified by in vitro NMR experiments. In this NMR study of the first 153 residues of the NTD, it was shown that the polyQ1 stretch has alpha-helical structure, while the rest of the sequence displayed no persistent secondary structure (43). Interestingly, when deleting the four leucines (55-LLLL-58) preceding polyQ1, helix formation was disrupted and the aggregation propensity of the construct highly increased (43). This study also revealed that the length of the polyQ1 correlates with the aggregation propensity of the fragment. Due to this, the authors concluded that the longest polyQ1 fragment that can be studied in vitro is 25Q. In a follow-up NMR study, the same group showed that the side chains of glutamines form H-bonds with the main chain (44). The strength of these H-bonds is determined by the H-bond acceptor, leucines being better acceptors than alanines, providing an explanation to the conservation of leucines preceding the polyQ1. Interestingly, in the case of polyQ2, there are two leucines, which also show a high level of conversation in mammals (see Section “Evolution and phylogenetics of the polymorphic repeat regions”). It is important to mention here that the polyQ region of huntingtin in Huntington’s disease also has an important polyP flanking region, which decreases aggregation propensity (45), highlighting the potential role of solubility-enhancing flanking regions as an evolutionary mechanism to mitigate cytotoxicity. In addition, a study in yeast also found that the flanking regions of polyQ repeats can profoundly alter their toxicity (45, 46).
In humans, nine proteins with polymorphic polyQ repeats–including AR–have pathological implications when their repeat lengths are out of the physiological range (47, 48). These proteins have been the subject of various studies to shed light on the molecular mechanism of the relationships between the length and biological effect (Table 1A). In the case of AR, there is a physiological range between 9 and 36 (26, 49). PolyQ1 stretches longer than 37 successive Qs have been reported to form neurotoxic aggregates (50), as according to the proposed mechanism, longer Q-repeats decrease solubility and hence allow for fibrillar aggregate formation (51, 52), leading to a disease called spinal-bulbar muscular atrophy (SBMA), also known as Kennedy’s disease (53). In case of the disease, the patients show androgen insensitivity worsening with age, typically affecting adult males at older age (54). Affected patients have an expanded polyQ1 tract between 38 and 62 glutamines (55) and similarly to other CAG repeat expansion related diseases, the length of expansion is inversely correlated with the age of onset, disease severity and progression (55, 56). There are multiple pathways along the transformation from physiological to pathological state. Due to the previously mentioned aggregation tendency of the expanded polyQ1 region, there is a gain-of-function toxicity that results in the loss of alteration of normal AR function (57). Moreover, the elimination of the misfolded AR is hindered by autophagy dysregulation (58). Lim and et al. (59) have recently shown that delivering a naturally occurring AR isoform–isoform 2 that lacks the polyQ1 harboring NTD–by adenovirus vector can rescue the neurotoxic phenotype in SBMA mice models. This provided proof-of-principle type of evidence of the role of AR with extended polyQ1 in disease, and a possible future therapeutic approach by gene therapy.
In addition to the intrinsic aggregation propensity of the polyQ1, a short, highly conserved sequence upstream from this repeat (235-KELCKAVSVSM-245) has recently been reported to undergo reversible amyloid fiber formation under mild oxidative conditions (60). In a follow-up study, the same group showed by atomic force microscopy that the oligomeric state of AR-NTD fragment was modulated by this amyloidogenic sequence, suggesting that this region can function as a nucleation center for subsequent aggregation of polyQ1 (61). However, they did not observe fibril formation for a polyQ of a length within the physiological range (22), only in case of aberrant length (45). Interestingly, this region partially overlaps with the binding site of the RNA polymerase-associated protein 74 subunit of the general transcription factor TFIIF, and mutation of conserved bulky hydrophobic residues in this sequence to smaller hydrophobic alanine significantly impaired transcriptional activity (62). Moreover, EPI-001, a compound that binds specifically to AR-NTD inhibiting transcriptional activity, showed weak chemical shift perturbation in this region by NMR titration (63). In this and a follow-up NMR study intermediate helical propensity was observed in this region (23, 63).
It is important to note that polyQ1 displays not only an increased aggregation propensity upon pathological expansion (64), it also has a negative correlation with transcriptional activity of AR (65–67). It has been suggested that the length of polyQ1 influences NTD–LBD interaction, which can affect the activity of AR (26). There is also a difference in the polyQ1 length among ethnicities (Table 1A): African people have the shortest, Asian people the longest and Caucasian people in between (68). Furthermore, shorter repeats are associated with higher PCa propensity (25, 69, 70). The commonly accepted hypothesis is that the length of the polyQ1 and the transcriptional activity of AR are inversely correlated, and long-term exposure of prostate cells to elevated AR activity can increase proliferation and trigger oncogenic transformation. This further supports the argument that males with African ancestry have shorter CAG repeats on average (Table 1A) in comparison to non-hispanic Caucasian and Asian people (71–74) and have higher mortality caused by PCa than Caucasian and Asian people (75–77). However, detailed comparison of these studies led us to pinpoint various controversies, which we are going to dissect in more detail in Section “Controversies.” In addition, a recent review summarizes additional biological risk factors based on new genome-wide association studies as well as environmental and social risk factors with regards to African or European ancestry (78).
It has been well-established in the literature that Wnt signaling pathway is often misregulated in disease, especially in cancers like PCa, where it drives oncogenic proliferation (79, 80). Elevated β-catenin expression in the nucleus enhances tumorigenesis in the prostate (81–83) promoting a very aggressive form of PCa with poor survival (84). Conversely, in patients with early onset PCa with very severe tumor growth (84, 85) and in many African American PCa patients (86), both Wnt and androgen signaling are significantly upregulated. In a recent study, He et al. (87) highlighted the involvement of polyQ1 in this misregulation. Using compound mice with humanized AR sequence bearing different polyQ1 lengths (12, 21, and 48 glutamines), they found that short polyQ1–compared to the longer counterpart–displayed an earlier onset of oncogenic transformation along with accelerated and more aggressive tumor development in the prostate. These results provide further explanation to the already existing hypothesis (i.e., short polyQ1 results in higher activity) and highlight the complexity of PCa tumorigenesis.
Androgen receptor activity is essential for spermatogenesis (88), however, most men with aberrant spermatogenesis have normal serum androgen levels (66). Therefore, researchers explored the possible involvement of the polymorphic regions of AR, as they can modulate AR level and activity independently of the androgen serum level (as mentioned before). In an early study, Tut et al. (66) analyzed samples from patients (N = 153) with normal androgen serum level, and found a significant correlation between the length of polyQ1 and defective sperm production. They found that longer polyQ1 repeats (≥28) increased the risk of impaired spermatogenesis four fold. However, later many more studies came to conflicting results, some showing correlation while others don’t (89). Ferlin et al. (89) also failed to confirm the link and only observed some association when the two polymorphic regions polyQ1 and polyG were analyzed separately, and only in the case of a few individuals (Table 1). It is important to mention that most of the studies were performed in different parts of the world and usually on a subset of the local population.
Because COVID-19-associated intensive care admission as well as mortality is higher in men than in women, researchers started to explore the possible explanations (90, 91). AR regulates the transcription of transmembrane protease serine 2 (TMPRSS2) (92), which primes the spike protein of the virus, therefore the spike can bind to the receptor of the host cell and enter (93). Mohamed et al. (94) in a recent review proposed that shorter CAG repeats confer higher AR activity, therefore higher TMPRSS2 transcription which causes higher risk of severe disease outcome. However, later, two independent studies with patient samples came to similar conclusions arguing for opposing trends. One of these studies observed that European males from Italy and Spain with longer CAG repeat (≥23) had worse clinical outcomes due to severe COVID-19 than patients with shorter CAG repeat (≤22) (95). The other study also concluded that longer CAG repeat (≥22) has conferred worse COVID-19 outcome in males (96).
Finally, another disease-linked correlation regarding the length of the polyQ1 was reported by Kawasaki et al. (97) who found that short polyQ increases the risk of early onset rheumatoid arthritis in males younger than 55.
The polymorphic polyglycine region of human androgen receptor
The polymorphic polyglycine (polyG) region is also located within the intrinsically disordered NTD of AR, and is encoded by three GGT triplets followed by one GGG and two more GGT triplets, and a variable number of GGC triplets. It has been shown that the length of this region has an effect on the translation of the protein itself (124, 125) and potentially also on the transcriptional activity of AR (27, 125, 126). For example, recombinantly expressed AR constructs with only 10 GGN repeats decreased relative AR activity to 40–68% of the wild-type (27, 126), while longer GGN repeats with a glycine stretch of 27 also exhibited reduced activity of 37–78% (126). In case of genetically engineered AR constructs with shorter and longer GGC repeats, protein abundance was found to be inversely correlated with polyG length, and it is hypothesized that the longer GGC repeats form a more stable hairpin structure in the mRNA that interferes more with translation (124, 125).
Moreover, it has been established that across certain races and ethnic groups the range of polyG/GGN repeat variation exhibits differences (Table 1B), and hence, this factor has to be considered during study design. In the following, we summarize the most significant findings on the relationship between diseases and polyG length.
In light of the molecular details mentioned above, it is not surprising that the polymorphic length of GGN repeats, and consequently the polyG tract, is a risk factor in certain cancer types (99, 123, 127) correlating with progression and/or severity of the disease, or the outlook for relapse-free periods (100, 108, 109). However, it is important to note that there are still significant controversies in the literature on the importance of the polyG length in particular cancer types (see the “Controversies” Section for details and Table 1B).
In an early study, Hakimi et al. (99) found that both polyG ≤ 14 and polyQ ≤ 17 are more common in the general Caucasian male population with clinical PCa diagnosis (N = 59), and patients with any of the two allele types have higher odds of developing malignancy, although the frequencies of the polymorphisms seem to be independent of each other. A large meta-analysis on the relationship between PCa and AR polymorphisms in the Caucasian population showed that short polyG of max. 16 repeats imposed the same amount of risk for PCa than a short polyQ with less than 22 repeats, while the combination of both short polyQ and polyG doubles the odds ratio of PCa risk (95% CI: 1.29–3.29) (127). Edwards et al. (100) found that long GGC repeats of more than 16 significantly increased the risk of relapse and risk of death in British Caucasian men (N = 178) from around 33 months after PCa diagnosis; furthermore, long GGC repeats were associated with a worse prognosis and survival at all disease levels of stage and grade. In men from the Canary Islands (N = 72), an immunohistochemistry study showed that the polyG length was negatively correlated to prostate specific antigen (PSA) staining intensity, especially in samples with simultaneously shorter polyQ or from the more severe type of PCa with Gleason score of at least 7 (109).
On the other hand, longer polyG repeats also come with a risk for women (Table 1B): Based on a study on a Japanese cohort (N = 226), longer GGC repeats (≥17 GGC) are more frequently associated with endometrial cancer (ECa) as compared to the control population (123). In women from the Canary Islands (N = 207), shorter polyG was found to be more frequently associated with benign type of ECa with slower cancer progression and better outcomes (108). In a cohort from the USA with 89% Caucasian study participants, longer GGC repeats were associated with reduced risks of breast cancer (BRCa) (105, 108). Gonzalez et al. found that the combination of long polyQ (>22) and long polyG (≥24) is more common in female BRCa patients from the Canary Islands (N = 257) than average polyQ (>22) (105, 107, 108).
As AR has a key role in androgen insensitivity syndrome (AIS), a disease often leading to defects in virilization and infertility, investigating the role of mutations, including the polymorphic GGN/polyG alleles, are of high importance. Grigorova et al. (104) found that those with decreased sperm counts more commonly had longer GGN repeats. Although polyG length alone was not found to be prognostic to infertility, it may further tune the effects of other mutations or polymorphisms. In accord, lowest sperm counts were found in individuals with both longer GGN and longer CAG repeats. Another study carried out a detailed analysis of polymorphic CAG/GGN alleles and also found that min. 21 CAG and min. 24 GGN repeats simultaneously significantly increase the relative risk of sterility (severe hypospermatogenesis) by a factor of 1.6 (89). A smaller survey also showed worse sperm motility in case of longer CAG and GGN repeats (103). On the other hand, Brokken et al. (114) examined fertile Caucasian men (N = 557), and found that those with shorter than 23 GGN repeats (N = 44) had higher concentration of inhibin B, higher levels of progressive sperm and of correct morphology, and a higher fraction of Fas-positive sperm. Men with min. 24 GGN (N = 153) or min. 25 CAG (N = 118) both had higher estradiol levels, while those with 23 or fewer CAG had higher sperm DNA fragmentation (114).
GGN/polyG polymorphisms of AR may also correlate with certain measures of cognitive performance and risks of neurological conditions. For example, in a Chinese cohort of healthy individuals, a significant association was detected between polyG length and verbal memory of women (120). While in a Swedish cohort, a significant relationship was found between AR repeat polymorphisms and neuroticism or somatic anxiety, with an overrepresentation of people having short polyQ and long polyG repeat regions simultaneously (110).
The non-polymorphic polyproline region of human androgen receptor
Androgen receptor-NTD also contains a polyproline stretch (polyP at aa. 374-381) of eight amino acids, consecutively, which is conserved from human to rodents (Supplementary File 1). Substitution of prolines in polyP is relatively rare, however leucines, alanines and histidines do occur in more than one species, while serine is only observed in spotted hyena and threonine in the last position of polyP in rodents.
Structurally, polyP sequences are known to fold into polyproline helices, and AlphaFold (128) does model this specific region of AR as a polyproline helix as seen in the AlphaFold Protein Structure Database (129). Functionally, proline-rich regions are known to be recognized by SH3 domains. Migliaccio et al. (130) showed that the SH3 domain of Src can actually bind polyP of AR, while interaction between AR and Src lacking the SH3 was barely detectable, and binding between AR lacking the polyP region and Src was undetectable. Deleting the C-terminus of polyP and its flanking region only exhibited very weak activation of Src (131). Furthermore, titration with this synthesized peptide (Ac-PPPHPHARIK-NH2) could also inhibit the AR-Src interaction (131). Moreover, another SH3-containing protein SH3YL was also proposed as a binding partner of AR’s polyP. Blessing and coworkers used a phage display to confirm the interaction with SH3YL1 and also concluded that the disruption of AR-NTD’s polyP reduced the hormone-dependent proliferation and migration of PCa cells (132).
It is also noteworthy that huntingtin, the protein involved in Huntington’s disease, also has mono-amino acid repeats of both polyQ and polyP. For this protein, it was demonstrated that the polyP region chaperones the polyQ region, and without polyP the polyQ repeat is more prone to aggregation (45, 133–136). It definitely would be interesting to study if the short polyP region of AR also chaperones its polyQ region, affecting its aggregation propensity.
The important role of the polyP region of AR is also highlighted by its sensitivity to mutations. The P380R substitution is cataloged in AIS (137), causing partial androgen insensitivity with ambiguous genitalia and sexual underdevelopment (138). Using luciferase reporter assays, it was demonstrated that the P380R substitution significantly reduces the hormone-induced transactivation of AR to ∼20% of the wild-type, thereby highlighting the mechanistic details of how this mutation causes AIS (139).
Evolution and phylogenetics of the polymorphic repeat regions
Type 1 NHRs are a major and well-studied group of NHRs that bind bipartite hormone elements in homodimeric form (140). They evolved in a way that AR, PR, GR, and MR diverged from ER alpha and beta (141–143). Of the four type 1 steroid receptors, AR seems to be the most distant from ER-alpha (Id = 15.6%) (141, 143). The DBDs and LBDs of nucleic hormone receptors are well-conserved, most of their divergence arises from the intrinsically disordered NTDs that differ both in length and sequence (Figure 1, Supplementary File 1), which is not surprising for regions with structural disorder (144–149).
Across mammalian species, AR’s DBD and LBD are fully conserved with only a single amino acid (glutamate) insertion in the sheep DBD, two mismatches in the DBD and one mismatch in the LBD of spotted hyena (Supplementary File 1). These positions either correspond to the C-terminal end of the DBD fold and probably enable flexible motion of the domain with respect to the hinge region and LBD or represent part of the DBD fold but are not in the close proximity of DNA (hyena’s S614P using human AR numbering/S596P using rat AR numbering). While in case of the hyena LBD, the mismatch E838D (human AR numbering) is also distant from the steroid hormone binding pocket. However, E838 is located in a druggable cleft of AR-LBD, for example flufenamic acid and tiratricol interacts with it (150).
Going further in evolution toward vertebrates, there is a high degree of conservation of DBD, which can be explained by its function to bind to conserved DNA recognition elements (151). Mutation on the ARE site and/or the DBD could result in a disrupted signaling cascade (152). Therefore, the DBD remained practically unchanged for at least 500 million years (152, 153). The mutations in humans compared to fishes in the DBD resulted in AR being able to bind to other hormone response elements as well as increasing transcriptional activity (154). LBD is less conserved than DBD in a longer evolutionary context, still it is highly conserved and diverged significantly less during evolution than the LBD of other SRs (142).
Androgen receptor-NTD conservation across mammalian species is also relatively high with the exception of the polymorphic polyQ1 and polyG regions, however, non-mammalian vertebrates have significantly lower conservation of the NTD (142, 155), and are completely devoid of polyQ and polyG regions (Figure 2). Within mammals, the most conserved parts of the NTD is the extreme N-terminal 35 residues preceding the polyQ1 stretch and region 231–255 (human AR numbering) harboring a putative CRM1 nuclear export signal (144, 156, 157). This region is also responsible for the interaction with Hsp70 (158) and has been reported to be amyloidogenic (60, 61).
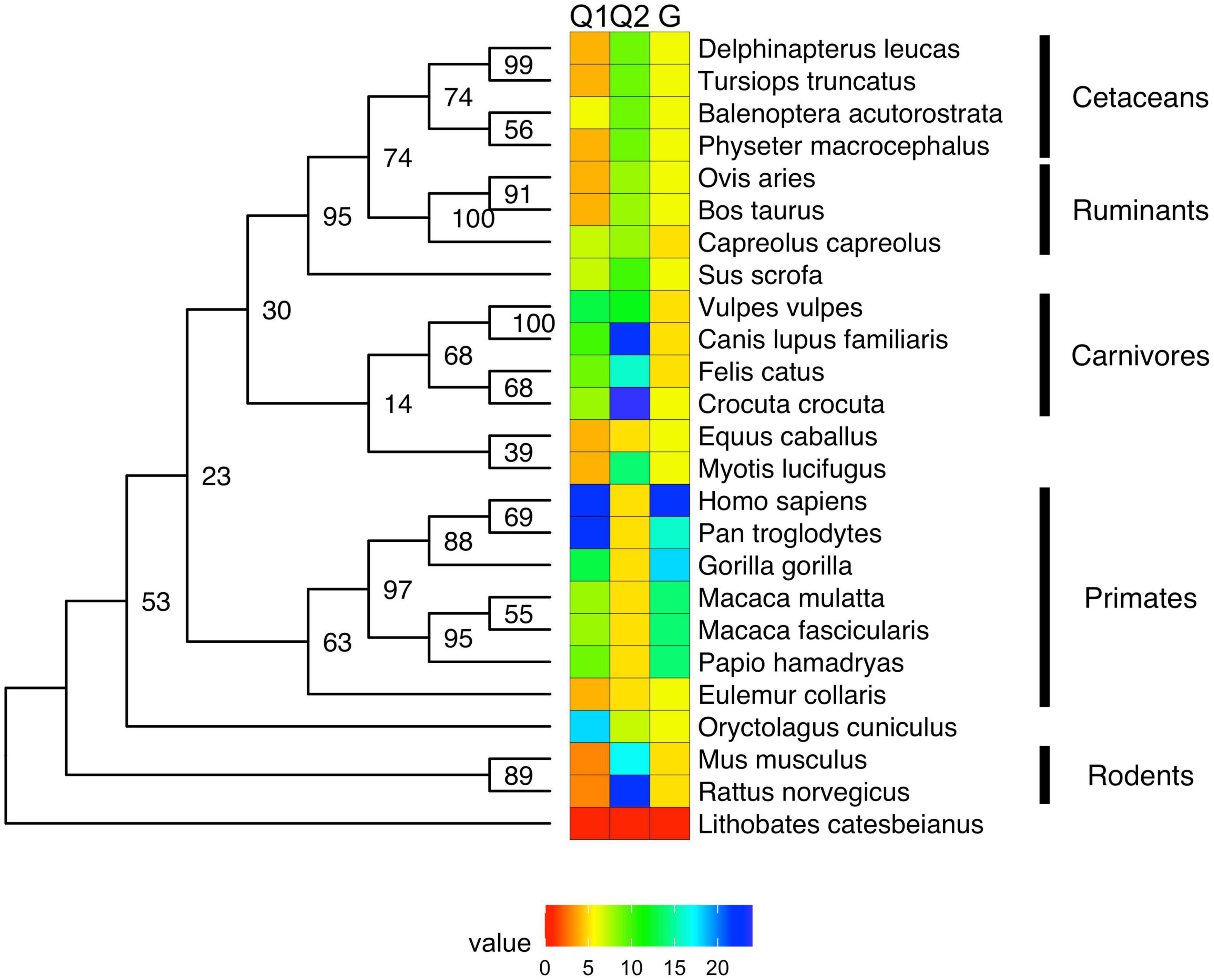
Figure 2. Maximum likelihood (ML) phylogenetic tree of androgen receptor with heat map showing the length of the polyQ and polyG repeats. The ML phylogenetic tree (here shown as a cladogram) is displayed for the full-length UniProt sequences of AR (33). Sequences were aligned by MAFFT (159) and refined by RAxML (160, 161), and numeric values on the tree of the aligned AR sequences (159) indicate bootstrap percentages from 2000 iterations (160, 161) that can be regarded as a confidence score of local tree topology.
In this article, we present a multiple sequence alignment (Supplementary File 1) and corresponding phylogenetic tree showing the evolution of polymorphic regions in mammalian species (Figures 2, 3). The overall topology of the maximum likelihood (ML) protein tree (Figure 2) reveals some divergence from nuclear phylogenies of mammals (163, 164). Rodents (Mus musculus, Rattus norvegicus) and rabbits (Oryctolagus cuniculus) occupy a basal position, decisively distant from the primates’ clade, to which it is closely related in the nuclear phylogenies. Another notable exception is the bat’s AR, which appears to be closest to horses, and is positioned in a very nested position within the phylogeny. It is also worth mentioning that bat (Myotis lucifugus) appears to have the longest branch length among all the species considered, suggesting that its AR amino acid sequence has diverged the most compared to all the other orthologs considered, based on its position on Figure 3.
Figure 3. Evolution of mono-amino acid repeat lengths in androgen receptor. The same ML tree as on Figure 2 (here shown as a phylogram) was colored according to the reconstructed length of polyQ1 (panel A), polyQ2 (panel B), and polyG (panel C) repeats using the phytools R package (162) and its fastAnc and conMap commands.
With this considered, one can appreciate that a significant part of these small changes among mammalian ARs occur around polymorphic regions. The polymorphic polyglutamine repeat (polyQ1) features in higher primates, being shorter or interrupted with other residues or even absent from mammals to frog and fish, which is also the case for other polyQ-containing and neurodegenerative disease related proteins (42). Two early studies tried to shed light on the timeline of divergence of polymorphic regions in mammals (165, 166). Despite the importance of these pioneering works, it cannot be ignored that the sample size of primates was very small, and apes were compared to rats as mammalian controls, which leads to biased conclusions as the rodents have shown to be outliers by the phylogenetic tree.
The conservation of the 22/23-residue-long polyQ1 region is quite poor, i.e., polyQ1 stretches of at least 14 glutamines can only be found in human and chimpanzee AR-NTD (considering the most common allele) (Figure 2). Interestingly, only the sequence of the human and of a few apes (chimpanzee, gorilla) have 3–4 leucines as N-terminal flanking “gatekeeper residues” of the polyQ1 region, the other mammals with shorter CAG repeats have a single leucine as flank, suggesting that the length of the leucine-stretch correlates with the polyQ1 length in evolution. After apes, carnivores have the next longest polyQ1 regions (8–10), followed by Old World monkeys (8–9) and then New World monkeys (4) (Figure 2). Pigs have longer polyQ1 region (7) than New World monkeys, but odd-toed ungulates, ruminants, cetaceans and bats generally have 4-residue-long polyQ1. The shortest polyQ1 region is in rodents with a length of 2 (Figure 2). The CDK phosphorylation site XX([ST])P[RK] immediately adjacent to the polyQ1 (ETSPR) is also well-conserved in ARs with only little variation across mammalian species (Supplementary File 1). Following the phosphorylation site, most mammals have 4–8 more glutamines, with the exceptions of cats that have 11, and rodents that only have 2 glutamines and the rest of the glutamines are mutated to arginine or histidine. The outgroup species considered for the calibration of our trees (the frog species Lithobates catesbeianus) completely lacks the polyQ1 repeats (Figure 2), highlighting a trend of acquiring the polyQ1 region and increasing its length in mammals, particularly in the apes’ clade. This is clearly elucidated through ancestral reconstruction displayed on the tree in Figure 3A. The ancestral state is predicted to have polyQ1 length of 2, which disappeared in amphibians after divergence. Although in rodents the ancestral length is maintained, an overall increase of polyQ1 is observable in the rest of the species, with an average of ∼12 repeats. While carnivores retain this state, a few reversals (i.e., subsequent decrease in polyQ length) are observed: in the case of bat (M. lucifugus), horse (Equus caballus) and the Artiodactylian clade (Cetaceans and Ruminants), which show 4–10 polyQ1 repeats. In the primate clade, a range of repeat lengths is observed, from the 2 of the basal lemur (Eulemur fulvus collaris) to the 23 repeats in Homo sapiens, the latter representing the highest value observed in this set of sequences (Figure 3A). An interesting case is the one of rabbit (O. cuniculus), which shows a long polyQ1 repeat (∼15) despite its early divergence in the phylogeny (Figure 2).
Polyglutamine repeat 2 is located C-terminally ∼115 amino acids away from polyQ1. An opposite trend is observed in the evolution of these repeats (Figures 3A,B). The most ancient mammalian ancestor reconstructed here is predicted to possess a polyQ2 repeat of ∼12 residues (Figure 3B). Bullfrog (L. catesbeianus) lacks the polyQ2 repeat, while the ancestral state is maintained throughout most of the mammalian evolution (Figure 3B). In contrast to polyQ1, basal clades like rodents show the longest polyQ2 repeats (22–24) and primates bear the shortest ones (∼5 glutamines), and the two polyglutamine stretches are slightly (inversely) correlated (Spearman’s R = −0.16). Interestingly, carnivores and rats have the longest (min. 20 residues) polyQ2, but often interrupted by arginines or histidines (Supplementary File 1).
The polyG region of AR is ∼86 amino acids N-terminally from AR-DBD, and it is longest in humans (∼23 residues), followed by apes (17–22 residues), Old World monkeys (15 residues), New World monkeys (8–14 residues) and other non-primate species (<10 residues) in order (Figure 2). Its evolution shows a similar trend to polyQ1, with an ancestral condition of ∼4 repeats, no polyG in bullfrog (L. catesbeianus), a basal state of 5–10 repeats for most of the mammalian species (Figure 3C). The exception is the primate order, which shows a rapid increase in the number of glycine repeats (22 in human). It is of note again, that glycines of polyG are also sometimes mutated to other small amino acids (serines, threonines, and alanines). In most mammals, the polyG region is flanked N-terminally by a cysteine and C-terminally by a glutamate (Supplementary File 1). In mouse and rat, the cysteine is missing or substituted by glycine (5 uninterrupted glycines in a row); moreover, the glutamate is replaced by aspartate, while the second half of the polyG is mutated to 451-SSSPS-455 (rat AR numbering). This highlights how far most mammalian AR evolved from those of rodents. Interestingly, we also confirmed the significant correlation between polyG and polyQ1 length (Spearman’s R = 0.47), furthermore significant inverse correlation between polyG and polyQ2 length (Spearman’s R = −0.54) across mammalian species.
In summary, mono-amino acid repeats do not occur in non-mammalian species, not even the non-polymorphic polyP despite its high conservation of sequence (max. 2 substitutions) and length (eight amino acids) across mammals (Figure 2).
Repeat polymorphisms across individuals within a species was not found to be unique to the human AR (Table 1), polyQ1 and polyG length also varies in other primates. In chimpanzees (N = 89) the polyG ranges between 14 and 22 repeats with 17–19 being the most common, while in bonobos (N = 54) only alleles with 18 and 19 repeats were found with 87 and 13% frequency (167, 168). Two independent studies have concluded that polyG and polyQ1 lengths are inversely correlated in chimpanzees (167, 169). In common squirrel monkeys (N = 10) polyG ranges between 21 and 24, with 21 being the most frequent allele, while polyQ length stays invariantly 4 + 5 (polyQ1 + polyQ2) (169). In tufted capuchin monkeys (N = 47) polyG varies between 11 and 14, and similarly the fewest repeats (11) being the most frequent allele; by contrast, the length of polyQ regions does not vary much and stays 5 + 5 or less frequently 5 + 4 (polyQ1 + polyQ2) (169). Surprisingly, both squirrel monkeys and tamarins were found to have a significantly higher number of GGA glycine codons (29 and 42%) in polyG in comparison to Old World monkeys and apes that are devoid of this codon in their polyG region (169). The polymorphic polyG and polyQ1 length did not (inversely) correlate in New World monkeys the same way as it did in chimpanzees (169).
Polyglutamine repeat polymorphism in AR outside primates has also been discovered in a few carnivores. In the polyQ1 of red foxes (N = 181), most frequent CAG allele had 10 repeats, both in males and females (65.85 and 57.39%, respectively), followed by 10T (24.39 and 31.25%)–meaning one CAG was mutated to CAT–, then 13 repeats (7.32 and 9.09%) and finally 12 repeats (2.44 and 2.27%, respectively) in order (170). Interestingly, uninterrupted CAG10 was more common in aggressive female foxes than in curious females, while CAT/His interrupted CAG10 was less common in aggressive female foxes than in curious females (170). In the polyQ1 and polyQ2 of healthy dogs (N = 172), three polyQ1 alleles (with 10, 11, and 12 CAG) and three polyQ2 alleles (with 11, 12, and 13 CAG) were discovered with 11 being the most common in both of them (48.8 and 75.6%, respectively) (171). Interestingly, in the Doberman dog breed (N = 31) polyQ1 with 10 CAG was way more common (67.7%) than 11 CAG (32.3%), and 12 CAG was not represented at all despite the 18.6% expected occurrence (171). Doberman was the only guard dog breed in this present study, which raises the question whether the shorter polyQ1 region contributed to making this dog breed fearless, zealous, and fierce. Similarly to men, genotyped dogs with canine PCa (N = 31) had a tendency for shorter polyQ1 length with 10 or 11 CAG repeats (54.8 and 45.2%, respectively), while none of the dogs with PCa had 12 CAG in polyQ1 (171). Ochiai et al. (172) tested recombinant canine AR with polyQ1 of 9–12 glutamines in PC3 cells and found that constructs with shorter polyQ1 had significantly higher activities than those with longer polyQ1 (luciferase assays). As most male dogs are castrated when young, PCa cases not responding to hormonal androgen ablation and AR antagonists are very common, hence prognosis is as poor as in human CRPC (172), and radio- or chemotherapy and radical prostatectomy remain as last resorts.
Controversies
Although the relationship between fewer CAG repeats in polyQ1 and increased risk of PCa is seemingly established, it is still debated (173). For example, in a large multi-ethnic cohort of more than 4,000 men, Freedman et al. (102) did not find a significant relationship between CAG repeat length and the risk of PCa, similarly to smaller studies on the Caucasian population (100, 101, 121) (Table 1A). Controversies also exist with regards to the GGC repeats of the polyG region: for instance, the GGC repeats alone did not exhibit correlation with PCa risk in Scottish men (101), in the African American population (112), in Nigerian men (113), and in the Turkish population (174) (Table 1B).
Another point of debate is the negative correlation between the polymorphic CAG and GGC repeats, as various studies confirmed it (98, 112, 115), while others could not provide conclusive evidence (99, 109). The correlation was also demonstrated in chimpanzees (167, 169), however no correlation was observed in New World monkeys (169).
Yet another disputed area is the transcription activity of AR, which is generally thought to be inversely correlated with the length of both CAG repeats and GGN repeats. Nonetheless, some early studies were unable to detect these differences with varying CAG repeats (175, 176). Although in each of these studies, the template of the AR gene for further cellular experiments was isolated from two patients diagnosed with SBMA, the cell lines used in the experiments were neither prostate nor even human cells. Increased PSA levels are claimed to be reflective of overstimulated transcription activity of AR with shorter polyQ1/CAG repeats (109, 113, 177), however this correlation was not detected by others (117, 178). Interestingly, Bennett et al. (73) found almost three times higher PSA concentration in African-American than in Caucasian PCa patients, and in the same time median length of 20 vs. 22 CAG repeats, respectively.
Furthermore, more research is clearly needed to clarify whether CAG repeats encoding for polyQ1 can influence cognitive function. An interesting study assessed cognitive impairment (problems with thinking, communication, understanding and memory) by Mini-Mental State Examination (MMSE) in predominantly Caucasian elderly men, and found an association between longer CAG repeats and poorer performance (179). However, Kovacs et al. (120) found no relationship between CAG repeats and memory function in the Chinese population. On the other hand, the latter study surprisingly reported that GGN repeat length may affect verbal memory, which was not tested by the MMSE study on elderly female subjects. Notwithstanding, the lack of consistency in results may stem from the difference in cohort subjects with regards to age, ethnicity, and gender.
Current research gaps
In this review, we summarized efforts in determining the risk of length variations in the polymorphic regions of AR to certain types of cancer. These studies have mostly focused on a specific population in a single country, aside from a few exceptions, including the studies by Ackerman et al. (68) and Kittles et al. (74). As elucidated in the previous sections, these may explain controversies and ongoing debates about associations detected by some studies but not by others. In the coming years, it would be important to clarify the source of these differences by larger and more diverse collaborative or consortium-led surveys to resolve which exact polymorphism (and combination of alleles) have what effect. The advantage would be a great deal of control over the methodology (no arbitrary grouping, no limitation due to small sample size, same comparisons, tests, and metrics), the clinical parameters and biomarkers measured could be harmonized, and the diverse cohort could reveal new differences in ethnicity, age and gender. Furthermore, it is important to emphasize that anonymized raw data was very rarely shared along with the publication (link to data, Supplementary material), which would be largely beneficial for smooth accessibility, reproducibility, and reusability for meta-data analyses.
Studies exploring the polymorphic nature of polyQ1 and polyG repeats in certain subpopulations, and the clinical associations thereof are dominant in the literature, nonetheless efforts should also focus on the exact mechanisms how these regions function. It is still not totally clear, how longer GGC repeats result in lower protein abundance, and whether shorter than average GGC repeat length could result in higher intracellular concentration. PolyG length certainly varies across species from short to long (see Section “Evolution and phylogenetics of the polymorphic repeat regions”), which tempts us to wonder if AR abundance is again higher in those animals in comparison to humans. Moreover, what is the interplay between polyG/GGC repeats and polyQ1/CAG repeats that makes them correlate throughout evolution? Does the polyG/GGC stretch affect the structure and function of AR on the protein level, or it only regulates the translation efficiency on transcript level? For example, the polyG region is adjacent to the binding segments of the ralaniten-like drug candidates (11, 63, 180). It would be interesting to know if polyG length has an effect on the binding of the compound. Also, polyQ length was shown to readily modulate the NTD-LBD interaction (26), but more research should be dedicated to explore its effect on binding other macromolecular partners, as well (21, 24, 181).
A key missing area to further explore, is the modulatory role of polyQ1 and polyG and the effect of their length, respectively, as well as in certain combinations on biomolecular condensation, i.e., the formation of nuclear foci, by AR. It has been known for a long time that many TFs have a non-homogeneous distribution in the nucleus and form foci (or also termed nuclear puncta) at the DNA target site (182–186). Liquid-liquid phase separation (LLPS), a recently emerged phenomenon, provides a mechanistic explanation to the formation of these biological condensates, which has been detailed in recent reviews (187–189). LLPS is a thermodynamically driven reversible phenomenon present from bacteria to humans, and also in plants, reported to be involved in many biological processes and diseases (190–193). Upon LLPS, two separate phases of substantially different concentration and viscosity form, giving rise to a low concentration dilute phase and a high concentration condensed phase (194). Many TFs–including nuclear receptors GR, ER and AR–have been indicated to undergo LLPS (195–197). Moreover, other important transcriptional machinery proteins (e.g., MED1) were demonstrated to drive condensate formation, while others such as RNA polymerase II were shown to be recruited to the condensates–in both cases via their low-complexity intrinsically disordered regions (IDRs)–suggesting that LLPS have an important role in transcriptional regulations (196, 198). Due to their multivalency, IDRs are often considered to be potential drivers of condensate formation (199). In case of AR an early access preprint manuscript reported that only full-length AR can undergo LLPS upon ligand binding, and ARv7, which contains the unstructured low-complexity NTD but lacks the globular LBD, did not show condensate formation (200). They also showed that upon disruption of condensate formation the transcription activity was inhibited as well, suggesting that it has a crucial role in the regulation of AR activity. Another study verified that ARv7 and AF1 (aa. 144-488) were unable to undergo LLPS alone or in the presence of RNA mimic polyU (201). However, the AR-DBD was identified as a minimal region capable of driving LLPS in the presence of polyU (201). Another recent study showed that the length of the polyQ affects nuclear localization and hence the transcriptional activity of AR (202). This suggests that despite AR-NTD being insufficient for driving LLPS alone, it still has a regulatory role, probably by determining solubility via the length of the polyQ1 and recruiting co-factors that can alter the LLPS propensity. However, this research direction is still poorly understood, although there is increasing attention in the cell biology field to explore this new modality for regulation of certain molecular functions. For example, AR can constitute part of enhanceosomes (200, 202–204), hence overactivation of transcription in cancer should also be studied in the contexts of liquid-like phase separated condensate-state. The importance of LLPS raises the mechanistic question of the role of different domains (driver, regulatory or passive region), and polymorphic and splice variants, of AR in biomolecular condensate formation (transcriptional condensates, enhanceosomes) in late stages of PCa.
Another interesting, yet undiscovered area is related to long non-coding RNAs (lncRNAs). These RNAs comprise a huge part of the human transcriptome (205) and are subject of intense research due to their indication in many important cellular regulatory processes and cancer implications (206). AR has been reported to interact with several PCa-related lncRNAs, such as HOTAIR, PCAT1, HOXA11-AS-203, SOCS2-AS1, LBCS, GAS5 with a poorly understood mechanism (207, 208). In advanced PCa cell lines many of these lncRNAs are either upregulated or downregulated, further strengthening the relevance of the need to understand the molecular mechanism of these interactions. PCa-related lncRNAs have been summarized in a recent review in detail by Yang et al. (207). It is of high relevance, that the interaction between SLCNR1, a melanoma-related lncRNAs and AR was reported recently (209, 210). The authors identified a pyridine-rich motif that they proposed as a canonical AR-NTD binding motif, as it exists in other AR interacting lncRNAs, such as HOTAIR and HOXA11-AS-203 (211). In a follow up study, the same group successfully targeted the binding motif by oligonucleotides sterically blocking the interaction and thereby attenuating SLNCR1-mediated melanoma invasion (211). The NTD used by the authors in the studies contained only the most frequent polyQ1 and polyG length. It would be interesting to compare the binding of NTD with different polymorphic variants to lncRNAs to shed light on the possible direct or allosteric effect of mono-amino acid repeat length.
RNAs often facilitate LLPS (212), which has already been reported regarding the DBD of AR (201). Furthermore, many lncRNAs form ribonucleoprotein condensates, which are important in transcription (213, 214). Therefore, it would be important to study the effect of lncRNAs on AR’s LLPS behavior with different lengths of the polymorphic regions in pathophysiology, as it could shed light on future therapeutic windows to target these interactions.
Hopefully, addressing these research gaps will enable potential breakthroughs in understanding these polymorphisms and their cross-talk, with implications in diagnostics of patients with AR alleles representing moderate to high risk to certain diseases and in developing therapeutics that are not affected by these polymorphisms or therapeutics that counterbalance the effect of overly long or short alleles.
Future directions and potential breakthroughs in the field
Given the pace with which molecular and cell biology, genetics, diagnostics, and drug discovery develop, one can foresee a number of potential breakthroughs in the field focusing on better understanding and modulating of AR. It would be crucial to understand the molecular mechanism of the LLPS behavior of AR with regards to its activity. Further systematic in vitro and in vivo studies are required to elucidate the contribution of the different domains as well as the two polymorphic regions to the condensate formation. Including co-factors and other crucial partners to these future studies could enable better understanding of the transition from physiological to pathological states, and explaining some of the controversies around these regions. Within NTD, elucidating the mechanism by which polyQ1 and polyG affects the functional repertoire of AR, would also enable the therapeutic targeting of these protein segments. Effect of polyQ1 length of AR on PCa and neurodegenerative disease like SBMA has been confirmed but there is a need for validation on the effect of polyG and polyP and their interplay before targeting. There are different possibilities to target repeat associated diseases for AR at DNA, RNA and protein level.
On RNA level one way of targeting these polymorphic regions is by antisense oligonucleotides (ASOs) and stabilized miRNA analogs, which inhibit the translation of mRNA, this represents a fast-developing modality of drug design for repeat-associated diseases like Huntington’s disease (215–217), myotonic dystrophy type-1 (218–220) and amyotrophic lateral sclerosis (221–223). ASO stability and delivery have been ongoing problems, but now there is a growing number of new technologies for delivery, like liposomes, to mitigate these difficulties, which make them very attractive for targeting repeat-associated diseases (224).
At the DNA level, another approach could be a CRISPR/Cas9-based therapy for genetic engineering to restore the wild-type repeat number of the polymorphic regions, a method that is already developed for other repeat-associated diseases like Huntington’s disease (225–227), Duchenne muscular dystrophy (228), myotonic dystrophy type-1 (229, 230), spinocerebellar ataxia type-3 (231), Friedreich’s ataxia (231, 232) and amyotrophic lateral sclerosis (233). Off-target effects of this technology has been initially a challenge, but there are intensive efforts on reducing it, e.g., by dual CRISPR/Cas9 technology (234).
Inhibiting intramolecular or intermolecular interactions of AR-NTD is yet another way of interfering with its pathogenic malfunctioning. However, this is particularly challenging due to the intrinsically disordered nature of the NTD. IDRs have been considered to be undruggable for a long while, although new success stories of upcoming molecules targeting IDRs have mostly dissolved this dogma (235–237), including the development of ralaniten and its further optimized versions (11, 63, 180). As a subcategory of small molecule targeting, induced degradation of AR, especially its pathological isoforms and alleles, by proteolysis targeting chimeric compounds (PROTACs), molecular glues and autophagosome-tethering compounds is also expected to lead to potential breakthroughs (238–242). This strategy enables to lower the intracellular concentration of AR, therefore downregulating downstream transcriptional signaling.
Recent advances in understanding phase separation of AR provide opportunities to modulate condensates, thus targeting enhanceosomes and transcriptional condensates of AR variants may hold the future for drug discovery (200, 203, 204, 243–245). Currently, condensate modulators are being conceived to inhibit LLPS, re-solubilize the condensates or dissolve the aggregates formed from condensate foci, or on the contrary to harden condensate for inactivation (246–250).
Conclusion
Mono-amino acid repeats are present in many organisms including animals, plants, and fungi. For example, polyQ regions with increasing length affect solubility, stability, and abundance of proteins. Changes in hydrophobicity and secondary structure could result in oligomer formation, which potentially leads to condensation and aggregation.
Polyglutamine repeat regions are also located in AR-NTD with flanking regions exerting inhibitory effects on aggregation for both polyQ1 and polyQ2. PolyQ1 flanking region contains four leucines, while polyQ2 contains 2 leucines that act as aggregation gatekeepers. Mutation in leucine and/or polymorphism in polyQ1 can induce structural changes, which can result in different diseases. For example, shorter polyQ1 is associated with increased activity of AR, which can cause PCa and rheumatoid arthritis (87, 251). A longer length of polyQ1 results in aggregation and is associated with the neurological disease SBMA (252). PolyQ1 polymorphism and its effect on protein aggregation has been studied at the molecular level by different groups. It was shown that the N-terminus of AR can even form amyloids in vitro, and in cellulo aggregates in SBMA (61, 251).
Recently, it was reported that AR forms condensates in the nucleus, and elevated nuclear localization was observed despite decreased transcriptional activity with increasing length of polyQ1 (202). This work is very preliminary, and further studies will be needed to elaborate whether increase in polyQ1 length results in phase separation and/or aggregation. It would also be important to explore the effect of different lengths of polyQ1 on the dynamic liquid-like properties of these condensates, and how that affects the recruitment of different binding partners and resulting downstream signaling. Moreover, it would be also worth studying whether the length of the polyG has any effect on condensate formation.
It has been shown that length of polyG is associated with a decrease in its own translation and consequently transcriptional activity. Polymorphism in polyG length has been proposed to be associated with diseases including prostate and ECa, AIS, and neurological diseases. PolyG polymorphism has also been studied in relation to co-occurrence with polyQ1 repeats in pathology, and polyG ≤ 14 and polyQ1 ≤ 17 were found to be associated with PCa in the Caucasian population. Risk of being sterile increases for men with min. 21 CAG and min. 24 GGN repeats.
Even though the length polymorphism of polyG in AR has been studied at population level very intensively–although with controversial results –, there has not been a lot of work performed at the molecular level. Shorter polyG negatively correlated with PSA staining, especially in the more severe type of PCa with higher Gleason scores, but it would be important to investigate how polyG interplays with polyQ1, and how changes in lengths affect the phase separation and protein aggregation properties and consequently the resulting phenotype. Answering these questions will help understand the mechanism by which these polymorphic repeats function from shorter to longer alleles of the population.
It is little discussed that AR-NTD also contains an 8-amino acid-long stretch of polyP. It has been shown that disruption of the polyP–SH3YL1 interaction results in reduced hormone-dependent proliferation. It is of note that in cases of other proteins, e.g., huntingtin, it was shown that the polyP segment can chaperone the adjacent polyQ region. However, further research is needed to test if such chaperoning also applies to AR.
Overall, there are several knowledge gaps that hinder the understanding of these repeats in AR, and also of their crosstalk, i.e., how this interplay at molecular level brings changes at population level. These need to be explored further and studying this will provide opportunities to find ways of targeting diseases and potentially also to transfer this knowledge to other repeat-rich proteins with similar build-up like AIB1/NCOA3, SK3 and huntingtin.
Targeting of AR-NTD is especially challenging due to the intrinsically disordered nature of the region and the limited coverage of NTD with detailed structural characterization. So far, only ralaniten and its further developed successors has been shown to bind AR-NTD (covalently or with sufficient affinity) with properties compatible with drug development. One can expect to see more studies directly addressing the influence of the adjacent polyG region with length polymorphism, or the effect of the partially interacting polyQ1 region on the drug binding properties of transactivation unit AR-Tau5 in the coming years. Alternative to protein-protein/DNA interaction inhibitors, targeted protein degradation inducing compounds offer a complementary approach to interfere with the overactive signaling or aggregating oligomers of a protein. To the best of our knowledge PROTACs against AR are all based on drugs interacting with LBD, however one could also envision degraders developed from ralaniten-like compounds. Moreover, therapeutic targeting of AR can also concentrate on the DNA and mRNA level by CRISPR/Cas9-based technologies and ASOs, which methods have a relatively lower entry barrier for drug development but well-known challenges in delivery and stability. However, as AR phase separates in the nucleus to form transcriptional condensates (e.g., overactive enhanceosomes in PCa) it might be an important property for drugs to be able to partition into these condensates to exert their effects. Alternatively, condensate modulators can also be applied to hinder LLPS formation or dissolve condensates and thereby inhibit the constitutive transcriptional activation. As the number of investigational condensate modulators are rapidly growing, it is not far-fetched to expect these compounds to come of age in the near future.
Author contributions
TL, AM, JA, and GR: conceptualization, investigation, and writing – original draft. GR and TL: visualization. TL: project administration. TL and PT: supervision. TL, AM, JA, GR, and PT: writing – review and editing. PT: funding acquisition. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by Vrije Universiteit Brussel, Spearhead grant–grant number: SRP51; National Research, Development and Innovation Office (NKFIH, Hungary)–grant numbers: K131702 and K124670; EC H2020-MSCA-RISE Action “IDPfun”–grant number: 778247; EC H2020-WIDESPREAD-2020-5 Twinning grant–grant number: 952334. JA and GR are Ph.D. fellows (SB) of FWO–grant numbers: FWOSB77 and FWOSB72.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmed.2022.1019803/full#supplementary-material
Supplementary File 1 | Multiple sequence alignment of mammalian androgen receptor protein sequences and androgen receptor of American bullfrog (Lithobates catesbeianus) as an outgroup. All sequences are publicly available from UniProt with the accession code preceding the sequences.
Abbreviations
AIS, androgen insensitivity syndrome; AR, androgen receptor; ARE, androgen response element; ARv7, androgen receptor splice variant 7; ASO, antisense oligonucleotide; BPH, benign prostate hyperplasia; BRCa, breast cancer; CRPC, castration-resistant prostate cancer; DBD, DNA-binding domain; ECa, endometrial cancer; ER, estrogen receptor; GR, glucocorticoid receptor; HSP, heat shock protein; IDP, intrinsically disordered protein; IDR, intrinsically disordered region; LBD, ligand-binding domain; LLPS, liquid-liquid phase separation; lncRNA, long non-coding RNA; ML, maximum likelihood; MR, mineralocorticoid receptor; NHR, nuclear hormone receptor; NMR, nuclear magnetic resonance; NTD, N-terminal domain; PCa, prostate cancer; polyG, polyglycine repeat; polyP, polyproline repeat; polyQ, polyglutamine repeat; PR, progesterone receptor; PROTAC, proteolysis targeting chimeric compounds; PSA, prostate specific antigen; SBMA, spinal-bulbar muscular atrophy; SH3, Src Homology 3 domain; TF, transcription factor.
References
1. Evans RM, Mangelsdorf DJ. Nuclear receptors, RXR, and the big bang. Cell. (2014) 157:255–66. doi: 10.1016/j.cell.2014.03.012
3. Germain P, Staels B, Dacquet C, Spedding M, Laudet V. Overview of nomenclature of nuclear receptors. Pharmacol Rev. (2006) 58:685–704.
4. Arriza JL, Weinberger C, Cerelli G, Glaser TM, Handelin BL, Housman DE, et al. Cloning of human mineralocorticoid receptor complementary DNA: structural and functional kinship with the glucocorticoid receptor. Science. (1987) 237:268–75. doi: 10.1126/science.3037703
5. Trapman J, Klaassen P, Kuiper GG, van der Korput JA, Faber PW, van Rooij HC, et al. Cloning, structure and expression of a cDNA encoding the human androgen receptor. Biochem Biophys Res Commun. (1988) 153:241–8.
6. Loosfelt H, Atger M, Misrahi M, Guiochon-Mantel A, Meriel C, Logeat F, et al. Cloning and sequence analysis of rabbit progesterone-receptor complementary DNA. Proc Natl Acad Sci USA. (1986) 83:9045–9.
8. Roggero CM, Jin L, Cao S, Sonavane R, Kopplin NG, Ta HQ, et al. A detailed characterization of stepwise activation of the androgen receptor variant 7 in prostate cancer cells. Oncogene. (2021) 40:1106–17. doi: 10.1038/s41388-020-01585-5
9. Hsu C-L, Liu J-S, Wu P-L, Guan H-H, Chen Y-L, Lin A-C, et al. Identification of a new androgen receptor (AR) co-regulator BUD31 and related peptides to suppress wild-type and mutated AR-mediated prostate cancer growth via peptide screening and X-ray structure analysis. Mol Oncol. (2014) 8:1575–87. doi: 10.1016/j.molonc.2014.06.009
10. Shaffer PL, Jivan A, Dollins DE, Claessens F, Gewirth DT. Structural basis of androgen receptor binding to selective androgen response elements. Proc Natl Acad Sci USA. (2004) 101:4758–63.
11. Zhu J, Salvatella X, Robustelli P. Small molecules targeting the disordered transactivation domain of the androgen receptor induce the formation of collapsed helical states. bioRxiv. [Preprint]. (2021). doi: 10.1101/2021.12.23.474012
12. Lazar T, Martínez-Pérez E, Quaglia F, Hatos A, Chemes LB, Iserte JA, et al. PED in 2021: a major update of the protein ensemble database for intrinsically disordered proteins. Nucleic Acids Res. (2021) 49:D404–11. doi: 10.1093/nar/gkaa1021
13. Yu X, Yi P, Hamilton RA, Shen H, Chen M, Foulds CE, et al. Structural insights of transcriptionally active, full-length androgen receptor coactivator complexes. Mol Cell. (2020) 79:812–23.e4. doi: 10.1016/j.molcel.2020.06.031
14. Eftekharzadeh B, Banduseela VC, Chiesa G, Martínez-Cristóbal P, Rauch JN, Nath SR, et al. Hsp70 and Hsp40 inhibit an inter-domain interaction necessary for transcriptional activity in the androgen receptor. Nat Commun. (2019) 10:3562.
15. Wärnmark A, Treuter E, Wright APH, Gustafsson J-A. Activation functions 1 and 2 of nuclear receptors: molecular strategies for transcriptional activation. Mol Endocrinol. (2003) 17:1901–9.
16. Nadal M, Prekovic S, Gallastegui N, Helsen C, Abella M, Zielinska K, et al. Structure of the homodimeric androgen receptor ligand-binding domain. Nat Commun. (2017) 8:14388.
17. Cao B, Qi Y, Zhang G, Xu D, Zhan Y, Alvarez X, et al. Androgen receptor splice variants activating the full-length receptor in mediating resistance to androgen-directed therapy. Oncotarget. (2014) 5:1646–56.
18. Sun S, Sprenger CCT, Vessella RL, Haugk K, Soriano K, Mostaghel EA, et al. Castration resistance in human prostate cancer is conferred by a frequently occurring androgen receptor splice variant. J Clin Invest. (2010) 120:2715–30. doi: 10.1172/JCI41824
19. Hu R, Dunn TA, Wei S, Isharwal S, Veltri RW, Humphreys E, et al. Ligand-independent androgen receptor variants derived from splicing of cryptic exons signify hormone-refractory prostate cancer. Cancer Res. (2009) 69:16–22. doi: 10.1158/0008-5472.CAN-08-2764
20. Chan SC, Li Y, Dehm SM. Androgen receptor splice variants activate androgen receptor target genes and support aberrant prostate cancer cell growth independent of canonical androgen receptor nuclear localization signal. J Biol Chem. (2012) 287:19736–49. doi: 10.1074/jbc.M112.352930
21. He B, Kemppainen JA, Voegel JJ, Gronemeyer H, Wilson EM. Activation Function 2 in the Human Androgen Receptor Ligand Binding Domain Mediates Interdomain Communication with the NH2-terminal Domain. J Biol Chem. (1999) 274:37219–25. doi: 10.1074/jbc.274.52.37219
22. Heery DM, Kalkhoven E, Hoare S, Parker MG. A signature motif in transcriptional co-activators mediates binding to nuclear receptors. Nature. (1997) 387:733–6.
23. De Mol E, Szulc E, Di Sanza C, Martínez-Cristóbal P, Bertoncini CW, Fenwick RB, et al. Regulation of androgen receptor activity by transient interactions of its transactivation domain with general transcription regulators. Structure. (2018) 26:145–52.e3.
24. Bai S, He B, Wilson EM. Melanoma antigen gene protein MAGE-11 regulates androgen receptor function by modulating the interdomain interaction. Mol Cell Biol. (2005) 25:1238–57. doi: 10.1128/mcb.25.4.1238-1257.2005
25. Stanford JL, Just JJ, Gibbs M, Wicklund KG, Neal CL, Blumenstein BA, et al. Polymorphic repeats in the androgen receptor gene: molecular markers of prostate cancer risk. Cancer Res. (1997) 57:1194–8.
26. Buchanan G, Yang M, Cheong A, Harris JM, Irvine RA, Lambert PF, et al. Structural and functional consequences of glutamine tract variation in the androgen receptor. Hum Mol Genet. (2004) 13:1677–92. doi: 10.1093/hmg/ddh181
27. Werner R, Holterhus P-M, Binder G, Schwarz H-P, Morlot M, Struve D, et al. The A645D mutation in the hinge region of the human androgen receptor (AR) gene modulates AR activity, depending on the context of the polymorphic glutamine and glycine repeats. J Clin Endocrinol Metab. (2006) 91:3515–20. doi: 10.1210/jc.2006-0372
28. He S, Zhang C, Shafi AA, Sequeira M, Acquaviva J, Friedland JC, et al. Potent activity of the Hsp90 inhibitor ganetespib in prostate cancer cells irrespective of androgen receptor status or variant receptor expression. Int J Oncol. (2013) 42:35–43.
29. Solit DB, Rosen N. Hsp90: a novel target for cancer therapy. Curr Top Med Chem. (2006) 6:1205–14.
30. Shafi AA, Cox MB, Weigel NL. Androgen receptor splice variants are resistant to inhibitors of Hsp90 and FKBP52, which alter androgen receptor activity and expression. Steroids. (2013) 78:548–54. doi: 10.1016/j.steroids.2012.12.013
31. Ferraldeschi R, Welti J, Powers MV, Yuan W, Smyth T, Seed G, et al. Second-Generation HSP90 Inhibitor Onalespib Blocks mRNA Splicing of Androgen Receptor Variant 7 in Prostate Cancer Cells. Cancer Res. (2016) 76:2731–42. doi: 10.1158/0008-5472.CAN-15-2186
32. Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. (2004) 32:1792–7.
33. UniProt Consortium. UniProt: the universal protein knowledgebase in 2021. Nucleic Acids Res. (2021) 49:D480–9. doi: 10.1093/nar/gkaa1100
34. Gemayel R, Chavali S, Pougach K, Legendre M, Zhu B, Boeynaems S, et al. Variable Glutamine-Rich Repeats Modulate Transcription Factor Activity. Mol Cell. (2015) 59:615–27. doi: 10.1016/j.molcel.2015.07.003
35. Schaffar G, Breuer P, Boteva R, Behrends C, Tzvetkov N, Strippel N, et al. Cellular toxicity of polyglutamine expansion proteins: mechanism of transcription factor deactivation. Mol Cell. (2004) 15:95–105. doi: 10.1016/j.molcel.2004.06.029
36. Ferreira ME, Hermann S, Prochasson P, Workman JL, Berndt KD, Wright APH. Mechanism of transcription factor recruitment by acidic activators. J Biol Chem. (2005) 280:21779–84.
37. Vise PD, Baral B, Latos AJ, Daughdrill GW. NMR chemical shift and relaxation measurements provide evidence for the coupled folding and binding of the p53 transactivation domain. Nucleic Acids Res. (2005) 33:2061–77. doi: 10.1093/nar/gki336
38. Uesugi M, Nyanguile O, Lu H, Levine AJ, Verdine GL. Induced alpha helix in the VP16 activation domain upon binding to a human TAF. Science. (1997) 277:1310–3. doi: 10.1126/science.277.5330.1310
39. Sanborn AL, Yeh BT, Feigerle JT, Hao CV, Townshend RJL, Aiden EL, et al. Simple biochemical features underlie transcriptional activation domain diversity and dynamic, fuzzy binding to mediator. Elife. (2021) 10:e68068. doi: 10.7554/elife.68068
40. Reid J, Kelly SM, Watt K, Price NC, McEwan IJ. Conformational analysis of the androgen receptor amino-terminal domain involved in transactivation. Influence of structure-stabilizing solutes and protein-protein interactions. J Biol Chem. (2002) 277:20079–86. doi: 10.1074/jbc.M201003200
41. Lavery DN, McEwan IJ. Functional characterization of the native NH2-terminal transactivation domain of the human androgen receptor: binding kinetics for interactions with TFIIF and SRC-1a. Biochemistry. (2008) 47:3352–9. doi: 10.1021/bi702220p
42. Davies P, Watt K, Kelly SM, Clark C, Price NC, McEwan IJ. Consequences of poly-glutamine repeat length for the conformation and folding of the androgen receptor amino-terminal domain. J Mol Endocrinol. (2008) 41:301–14. doi: 10.1677/JME-08-0042
43. Eftekharzadeh B, Piai A, Chiesa G, Mungianu D, García J, Pierattelli R, et al. Sequence context influences the structure and aggregation behavior of a PolyQ tract. Biophys J. (2016) 110:2361–6. doi: 10.1016/j.bpj.2016.04.022
44. Escobedo A, Topal B, Kunze MBA, Aranda J, Chiesa G, Mungianu D, et al. Side chain to main chain hydrogen bonds stabilize a polyglutamine helix in a transcription factor. Nat Commun. (2019) 10:2034. doi: 10.1038/s41467-019-09923-2
45. Bhattacharyya A, Thakur AK, Chellgren VM, Thiagarajan G, Williams AD, Chellgren BW, et al. Oligoproline effects on polyglutamine conformation and aggregation. J Mol Biol. (2006) 355:524–35. doi: 10.1016/j.jmb.2005.10.053
46. Duennwald ML, Jagadish S, Muchowski PJ, Lindquist S. Flanking sequences profoundly alter polyglutamine toxicity in yeast. Proc Natl Acad Sci USA. (2006) 103:11045–50. doi: 10.1073/pnas.0604547103
47. Orr HT, Zoghbi HY. Trinucleotide repeat disorders. Annu Rev Neurosci. (2007) 30:575–621. doi: 10.1146/annurev.neuro.29.051605.113042
48. Costa MD, Maciel P. Modifier pathways in polyglutamine (PolyQ) diseases: from genetic screens to drug targets. Cell Mol Life Sci. (2022) 79:274. doi: 10.1007/s00018-022-04280-8
49. Callewaert L, Christiaens V, Haelens A, Verrijdt G, Verhoeven G, Claessens F. Implications of a polyglutamine tract in the function of the human androgen receptor. Biochem Biophys Res Commun. (2003) 306:46–52.
50. Li M, Miwa S, Kobayashi Y, Merry DE, Yamamoto M, Tanaka F, et al. Nuclear inclusions of the androgen receptor protein in spinal and bulbar muscular atrophy. Ann Neurol. (1998) 44:249–54.
51. Chen S, Ferrone FA, Wetzel R. Huntington’s disease age-of-onset linked to polyglutamine aggregation nucleation. Proc Natl Acad Sci USA. (2002) 99:11884–9.
52. Yang W, Dunlap JR, Andrews RB, Wetzel R. Aggregated polyglutamine peptides delivered to nuclei are toxic to mammalian cells. Hum Mol Genet. (2002) 11:2905–17. doi: 10.1093/hmg/11.23.2905
53. Spada ARL, La Spada AR, Wilson EM, Lubahn DB, Harding AE, Fischbeck KH. Androgen receptor gene mutations in X-linked spinal and bulbar muscular atrophy. Nature. (1991) 352:77–9. doi: 10.1038/352077a0
54. Dejager S, Bry-Gauillard H, Bruckert E, Eymard B, Salachas F, LeGuern E, et al. A comprehensive endocrine description of Kennedy’s disease revealing androgen insensitivity linked to CAG repeat length. J Clin Endocrinol Metab. (2002) 87:3893–901. doi: 10.1210/jcem.87.8.8780
55. Orafidiya FA, McEwan IJ. Trinucleotide repeats and protein folding and disease: the perspective from studies with the androgen receptor. Future Sci OA. (2015) 1:FSO47. doi: 10.4155/fso.15.47
56. Igarashi S, Tanno Y, Onodera O, Yamazaki M, Sato S, Ishikawa A, et al. Strong correlation between the number of CAG repeats in androgen receptor genes and the clinical onset of features of spinal and bulbar muscular atrophy. Neurology. (1992) 42:2300–2. doi: 10.1212/wnl.42.12.2300
57. Thomas PS, Fraley GS, Damien V, Woodke LB, Zapata F, Sopher BL, et al. Loss of endogenous androgen receptor protein accelerates motor neuron degeneration and accentuates androgen insensitivity in a mouse model of X-linked spinal and bulbar muscular atrophy. Hum Mol Genet. (2006) 15:2225–38. doi: 10.1093/hmg/ddl148
58. Cortes CJ, Miranda HC, Frankowski H, Batlevi Y, Young JE, Le A, et al. Polyglutamine-expanded androgen receptor interferes with TFEB to elicit autophagy defects in SBMA. Nat Neurosci. (2014) 17:1180–9. doi: 10.1038/nn.3787
59. Lim WF, Forouhan M, Roberts TC, Dabney J, Ellerington R, Speciale AA, et al. Gene therapy with AR isoform 2 rescues spinal and bulbar muscular atrophy phenotype by modulating AR transcriptional activity. Sci Adv. (2021) 7:eabi6896. doi: 10.1126/sciadv.abi6896
60. Asencio-Hernández J, Ruhlmann C, McEwen A, Eberling P, Nominé Y, Céraline J, et al. Reversible amyloid fiber formation in the N terminus of androgen receptor. Chembiochem. (2014) 15:2370–3. doi: 10.1002/cbic.201402420
61. Oppong E, Stier G, Gaal M, Seeger R, Stoeck M, Delsuc M-A, et al. An amyloidogenic sequence at the N-terminus of the androgen receptor impacts polyglutamine aggregation. Biomolecules. (2017) 7:44. doi: 10.3390/biom7020044
62. Betney R, McEwan IJ. Role of conserved hydrophobic amino acids in androgen receptor AF-1 function. J Mol Endocrinol. (2003) 31:427–39. doi: 10.1677/jme.0.0310427
63. De Mol E, Fenwick RB, Phang CTW, Buzón V, Szulc E, de la Fuente A, et al. EPI-001, a compound active against castration-resistant prostate cancer, targets transactivation unit 5 of the androgen receptor. ACS Chem Biol. (2016) 11:2499–505. doi: 10.1021/acschembio.6b00182
64. Jochum T, Ritz ME, Schuster C, Funderburk SF, Jehle K, Schmitz K, et al. Toxic and non-toxic aggregates from the SBMA and normal forms of androgen receptor have distinct oligomeric structures. Biochim Biophys Acta. (2012) 1822:1070–8. doi: 10.1016/j.bbadis.2012.02.006
65. Chamberlain NL, Driver ED, Miesfeld RL. The length and location of CAG trinucleotide repeats in the androgen receptor N-terminal domain affect transactivation function. Nucleic Acids Res. (1994) 22:3181–6. doi: 10.1093/nar/22.15.3181
66. Tut TG, Ghadessy FJ, Trifiro MA, Pinsky L, Yong EL. Long polyglutamine tracts in the androgen receptor are associated with reduced trans-activation, impaired sperm production, and male infertility. J Clin Endocrinol Metab. (1997) 82:3777–82. doi: 10.1210/jcem.82.11.4385
67. Irvine RA, Ma H, Yu MC, Ross RK, Stallcup MR, Coetzee GA. Inhibition of p160-mediated coactivation with increasing androgen receptor polyglutamine length. Hum Mol Genet. (2000) 9:267–74. doi: 10.1093/hmg/9.2.267
68. Ackerman CM, Lowe LP, Lee H, Hayes MG, Dyer AR, Metzger BE, et al. Hapo Study Cooperative Research Group. Ethnic variation in allele distribution of the androgen receptor (AR) (CAG)n repeat. J Androl. (2012) 33:210–5. doi: 10.2164/jandrol.111.013391
69. Buchanan G, Irvine RA, Coetzee GA, Tilley WD. Contribution of the androgen receptor to prostate cancer predisposition and progression. Cancer Metastasis Rev. (2001) 20:207–23.
70. Kumar R, Atamna H, Zakharov MN, Bhasin S, Khan SH, Jasuja R. Role of the androgen receptor CAG repeat polymorphism in prostate cancer, and spinal and bulbar muscular atrophy. Life Sci. (2011) 88:565–71.
71. Sartor O, Zheng Q, Eastham JA. Androgen receptor gene CAG repeat length varies in a race-specific fashion in men without prostate cancer. Urology. (1999) 53:378–80. doi: 10.1016/s0090-4295(98)00481-6
72. Coetzee GA, Ross RK. Re: prostate cancer and the androgen receptor. J Natl Cancer Inst. (1994) 86:872–3.
73. Bennett CL, Price DK, Kim S, Liu D, Jovanovic BD, Nathan D, et al. Racial variation in CAG repeat lengths within the androgen receptor gene among prostate cancer patients of lower socioeconomic status. J Clin Oncol. (2002) 20:3599–604. doi: 10.1200/JCO.2002.11.085
74. Kittles RA, Young D, Weinrich S, Hudson J, Argyropoulos G, Ukoli F, et al. Extent of linkage disequilibrium between the androgen receptor gene CAG and GGC repeats in human populations: implications for prostate cancer risk. Hum Genet. (2001) 109:253–61. doi: 10.1007/s004390100576
75. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. (2018) 68:394–424.
76. Resnick MJ, Canter DJ, Guzzo TJ, Brucker BM, Bergey M, Sonnad SS, et al. Does race affect postoperative outcomes in patients with low-risk prostate cancer who undergo radical prostatectomy? Urology. (2009) 73:620–3. doi: 10.1016/j.urology.2008.09.035
77. DeSantis CE, Siegel RL, Sauer AG, Miller KD, Fedewa SA, Alcaraz KI, et al. Cancer statistics for African Americans, 2016: progress and opportunities in reducing racial disparities. CA Cancer J Clin. (2016) 66:290–308. doi: 10.3322/caac.21340
78. Lewis DD, Cropp CD. The impact of African ancestry on prostate cancer disparities in the era of precision medicine. Genes. (2020) 11:1471. doi: 10.3390/genes11121471
79. Verras M, Sun Z. Roles and regulation of Wnt signaling and beta-catenin in prostate cancer. Cancer Lett. (2006) 237:22–32.
80. Robinson D, Van Allen EM, Wu Y-M, Schultz N, Lonigro RJ, Mosquera J-M, et al. Integrative clinical genomics of advanced prostate cancer. Cell. (2015) 162:454.
81. Chesire DR, Isaacs WB. Ligand-dependent inhibition of beta-catenin/TCF signaling by androgen receptor. Oncogene. (2002) 21:8453–69.
82. Bierie B, Nozawa M, Renou J-P, Shillingford JM, Morgan F, Oka T, et al. Activation of beta-catenin in prostate epithelium induces hyperplasias and squamous transdifferentiation. Oncogene. (2003) 22:3875–87. doi: 10.1038/sj.onc.1206426
83. Gounari F, Signoretti S, Bronson R, Klein L, Sellers WR, Kum J, et al. Stabilization of beta-catenin induces lesions reminiscent of prostatic intraepithelial neoplasia, but terminal squamous transdifferentiation of other secretory epithelia. Oncogene. (2002) 21:4099–107. doi: 10.1038/sj.onc.1205562
84. Lee SH, Luong R, Johnson DT, Cunha GR, Rivina L, Gonzalgo ML, et al. Androgen signaling is a confounding factor for β-catenin-mediated prostate tumorigenesis. Oncogene. (2016) 35:702–14. doi: 10.1038/onc.2015.117
85. Weischenfeldt J, Simon R, Feuerbach L, Schlangen K, Weichenhan D, Minner S, et al. Integrative genomic analyses reveal an androgen-driven somatic alteration landscape in early-onset prostate cancer. Cancer Cell. (2013) 23:159–70.
86. Wang B-D, Yang Q, Ceniccola K, Bianco F, Andrawis R, Jarrett T, et al. Androgen receptor-target genes in african american prostate cancer disparities. Prostate Cancer. (2013) 2013:763569. doi: 10.1155/2013/763569
87. He Y, Mi J, Olson A, Aldahl J, Hooker E, Yu E-J, et al. Androgen receptor with short polyglutamine tract preferably enhances Wnt/β-catenin-mediated prostatic tumorigenesis. Oncogene. (2020) 39:3276–91. doi: 10.1038/s41388-020-1214-7
88. Zirkin BR, Santulli R, Awoniyi CA, Ewing LL. Maintenance of advanced spermatogenic cells in the adult rat testis: quantitative relationship to testosterone concentration within the testis. Endocrinology. (1989) 124:3043–9. doi: 10.1210/endo-124-6-3043
89. Ferlin A, Bartoloni L, Rizzo G, Roverato A, Garolla A, Foresta C. Androgen receptor gene CAG and GGC repeat lengths in idiopathic male infertility. Mol Hum Reprod. (2004) 10:417–21.
90. Wadman M. Sex hormones signal why virus hits men harder. Science. (2020) 368:1038–9. doi: 10.1126/science.368.6495.1038
91. Pivonello R, Auriemma RS, Pivonello C, Isidori AM, Corona G, Colao A, et al. Sex disparities in COVID-19 severity and outcome: are men weaker or women stronger? Neuroendocrinology. (2021) 111:1066–85.
92. Lin B, Ferguson C, White JT, Wang S, Vessella R, True LD, et al. Prostate-localized and androgen-regulated expression of the membrane-bound serine protease TMPRSS2. Cancer Res. (1999) 59:4180–4.
93. Hoffmann M, Kleine-Weber H, Schroeder S, Krüger N, Herrler T, Erichsen S, et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. (2020) 181:271–80.e8.
94. Mohamed MS, Moulin TC, Schiöth HB. Sex differences in COVID-19: the role of androgens in disease severity and progression. Endocrine. (2021) 71:3–8.
95. Baldassarri M, Picchiotti N, Fava F, Fallerini C, Benetti E, Daga S, et al. Shorter androgen receptor polyQ alleles protect against life-threatening COVID-19 disease in European males. EBioMedicine. (2021) 65:103246.
96. McCoy J, Wambier CG, Herrera S, Vaño-Galván S, Gioia F, Comeche B, et al. Androgen receptor genetic variant predicts COVID-19 disease severity: a prospective longitudinal study of hospitalized COVID-19 male patients. J Eur Acad Dermatol Venereol. (2021) 35:e15–7. doi: 10.1111/jdv.16956
97. Kawasaki T, Ushiyama T, Ueyama H, Inoue K, Mori K, Ohkubo I, et al. Polymorphic CAG repeats of the androgen receptor gene and rheumatoid arthritis. Ann Rheum Dis. (1999) 58:500–2.
98. Irvine RA, Yu MC, Ross RK, Coetzee GA. The CAG and GGC microsatellites of the androgen receptor gene are in linkage disequilibrium in men with prostate cancer. Cancer Res. (1995) 55:1937–40.
99. Hakimi JM, Schoenberg MP, Rondinelli RH, Piantadosi S, Barrack ER. Androgen receptor variants with short glutamine or glycine repeats may identify unique subpopulations of men with prostate cancer. Clin Cancer Res. (1997) 3:1599–608.
100. Edwards SM, Badzioch MD, Minter R, Hamoudi R, Collins N, Ardern-Jones A, et al. Androgen receptor polymorphisms: association with prostate cancer risk, relapse and overall survival. Int J Cancer. (1999) 84:458–65. doi: 10.1002/(sici)1097-0215(19991022)84:53.0.co;2-y
101. Binnie MC, Alexander FE, Heald C, Habib FK. Polymorphic forms of prostate specific antigen and their interaction with androgen receptor trinucleotide repeats in prostate cancer. Prostate. (2005) 63:309–15. doi: 10.1002/pros.20178
102. Freedman ML, Pearce CL, Penney KL, Hirschhorn JN, Kolonel LN, Henderson BE, et al. Systematic evaluation of genetic variation at the androgen receptor locus and risk of prostate cancer in a multiethnic cohort study. Am J Hum Genet. (2005) 76:82–90. doi: 10.1086/427224
103. Delli Muti N, Agarwal A, Buldreghini E, Gioia A, Lenzi A, Boscaro M, et al. Have androgen receptor gene CAG and GGC repeat polymorphisms an effect on sperm motility in infertile men? Andrologia. (2014) 46:564–9. doi: 10.1111/and.12119
104. Grigorova M, Punab M, Kahre T, Ivandi M, Tõnisson N, Poolamets O, et al. The number of CAG and GGN triplet repeats in the androgen receptor gene exert combinatorial effect on hormonal and sperm parameters in young men. Andrology. (2017) 5:495–504. doi: 10.1111/andr.12344
105. Suter NM, Malone KE, Daling JR, Doody DR, Ostrander EA. Androgen receptor (CAG)n and (GGC)n polymorphisms and breast cancer risk in a population-based case-control study of young women. Cancer Epidemiol Biomarkers Prev. (2003) 12:127–35.
106. Giguère Y, Dewailly E, Brisson J, Ayotte P, Laflamme N, Demers A, et al. Short polyglutamine tracts in the androgen receptor are protective against breast cancer in the general population. Cancer Res. (2001) 61:5869–74.
107. González A, Javier Dorta F, Rodriguez G, Brito B, Rodríguez MADC, Cabrera A, et al. Increased risk of breast cancer in women bearing a combination of large CAG and GGN repeats in the exon 1 of the androgen receptor gene. Eur J Cancer. (2007) 43:2373–80. doi: 10.1016/j.ejca.2007.07.001
108. Rodríguez G, Bilbao C, Ramírez R, Falcón O, León L, Chirino R, et al. Alleles with short CAG and GGN repeats in the androgen receptor gene are associated with benign endometrial cancer. Int J Cancer. (2006) 118:1420–5. doi: 10.1002/ijc.21516
109. Rodríguez-González G, Cabrera S, Ramírez-Moreno R, Bilbao C, Díaz-Chico JC, Serra L, et al. Short alleles of both GGN and CAG repeats at the exon-1 of the androgen receptor gene are associated to increased PSA staining and a higher Gleason score in human prostatic cancer. J Steroid Biochem Mol Biol. (2009) 113:85–91. doi: 10.1016/j.jsbmb.2008.11.010
110. Westberg L, Henningsson S, Landén M, Annerbrink K, Melke J, Nilsson S, et al. Influence of androgen receptor repeat polymorphisms on personality traits in men. J Psychiatry Neurosci. (2009) 34:205–13.
111. O’Brien TG, Guo Y, Visvanathan K, Sciulli J, McLaine M, Helzlsouer KJ, et al. Differences in ornithine decarboxylase and androgen receptor allele frequencies among ethnic groups. Mol Carcinog. (2004) 41:120–3. doi: 10.1002/mc.20047
112. Lange EM, Sarma AV, Ray A, Wang Y, Ho LA, Anderson SA, et al. The androgen receptor CAG and GGN repeat polymorphisms and prostate cancer susceptibility in African-American men: results from the Flint Men’s Health Study. J Hum Genet. (2008) 53:220–6. doi: 10.1007/s10038-007-0240-4
113. Akinloye O, Gromoll J, Simoni M. Variation in CAG and GGN repeat lengths and CAG/GGN haplotype in androgen receptor gene polymorphism and prostate carcinoma in Nigerians. Br J Biomed Sci. (2011) 68:138–42. doi: 10.1080/09674845.2011.11730341
114. Brokken LJS, Rylander L, Jönsson BA, Spanò M, Pedersen HS, Ludwicki JK, et al. Non-linear association between androgen receptor CAG and GGN repeat lengths and reproductive parameters in fertile European and Inuit men. Mol Cell Endocrinol. (2013) 370:163–71. doi: 10.1016/j.mce.2013.03.005
115. Bertolin C, Querin G, Da Re E, Sagnelli A, Bello L, Cao M, et al. No effect of AR polyG polymorphism on spinal and bulbar muscular atrophy phenotype. Eur J Neurol. (2016) 23:1134–6. doi: 10.1111/ene.13001
116. Macke JP, Hu N, Hu S, Bailey M, King VL, Brown T, et al. Sequence variation in the androgen receptor gene is not a common determinant of male sexual orientation. Am J Hum Genet. (1993) 53:844–52.
117. Santos MLD, Sarkis AS, Nishimoto IN, Nagai MA. Androgen receptor CAG repeat polymorphism in prostate cancer from a Brazilian population. Cancer Detect Prev. (2003) 27:321–6.
118. Hsing AW, Gao YT, Wu G, Wang X, Deng J, Chen YL, et al. Polymorphic CAG and GGN repeat lengths in the androgen receptor gene and prostate cancer risk: a population-based case-control study in China. Cancer Res. (2000) 60:5111–6.
119. Huang S-P, Chou Y-H, Chang W-SW, Wu M-T, Yu C-C, Wu T, et al. Androgen receptor gene polymorphism and prostate cancer in Taiwan. J Formos Med Assoc. (2003) 102:680–6.
120. Kovacs D, Vassos E, Liu X, Sun X, Hu J, Breen G, et al. The androgen receptor gene polyglycine repeat polymorphism is associated with memory performance in healthy Chinese individuals. Psychoneuroendocrinology. (2009) 34:947–52. doi: 10.1016/j.psyneuen.2009.01.007
121. Platz EA, Giovannucci E, Dahl DM, Krithivas K, Hennekens CH, Brown M, et al. The androgen receptor gene GGN microsatellite and prostate cancer risk. Cancer Epidemiol Biomarkers Prev. (1998) 7:379–84.
122. Giovannucci E, Stampfer MJ, Krithivas K, Brown M, Dahl D, Brufsky A, et al. The CAG repeat within the androgen receptor gene and its relationship to prostate cancer. Proc Natl Acad Sci USA. (1997) 94:3320–3.
123. Sasaki M, Karube A, Karube Y, Watari M, Sakuragi N, Fujimoto S, et al. GGC and StuI polymorphism on the androgen receptor gene in endometrial cancer patients. Biochem Biophys Res Commun. (2005) 329:100–4. doi: 10.1016/j.bbrc.2005.01.104
124. Ding D, Xu L, Menon M, Reddy GPV, Barrack ER. Effect of GGC (glycine) repeat length polymorphism in the human androgen receptor on androgen action. Prostate. (2005) 62:133–9. doi: 10.1002/pros.20128
125. Brockschmidt FF, Nöthen MM, Hillmer AM. The two most common alleles of the coding GGN repeat in the androgen receptor gene cause differences in protein function. J Mol Endocrinol. (2007) 39:1–8. doi: 10.1677/JME-06-0072
126. Lundin KB, Giwercman A, Dizeyi N, Giwercman YL. Functional in vitro characterisation of the androgen receptor GGN polymorphism. Mol Cell Endocrinol. (2007) 264:184–7. doi: 10.1016/j.mce.2006.11.008
127. Weng H, Li S, Huang J-Y, He Z-Q, Meng X-Y, Cao Y, et al. Androgen receptor gene polymorphisms and risk of prostate cancer: a meta-analysis. Sci Rep. (2017) 7:40554.
128. Jumper J, Evans R, Pritzel A, Green T, Figurnov M, Ronneberger O, et al. Highly accurate protein structure prediction with AlphaFold. Nature. (2021) 596:583–9.
129. Varadi M, Anyango S, Deshpande M, Nair S, Natassia C, Yordanova G, et al. AlphaFold protein structure database: massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. (2022) 50:D439–44. doi: 10.1093/nar/gkab1061
130. Migliaccio A, Castoria G, Di Domenico M, de Falco A, Bilancio A, Lombardi M, et al. Steroid-induced androgen receptor-oestradiol receptor beta-Src complex triggers prostate cancer cell proliferation. EMBO J. (2000) 19:5406–17. doi: 10.1093/emboj/19.20.5406
131. Migliaccio A, Varricchio L, De Falco A, Castoria G, Arra C, Yamaguchi H, et al. Inhibition of the SH3 domain-mediated binding of Src to the androgen receptor and its effect on tumor growth. Oncogene. (2007) 26:6619–29. doi: 10.1038/sj.onc.1210487
132. Blessing AM, Ganesan S, Rajapakshe K, Ying Sung Y, Reddy Bollu L, Shi Y, et al. Identification of a Novel Coregulator, SH3YL1, That Interacts With the Androgen Receptor N-Terminus. Mol Endocrinol. (2015) 29:1426–39. doi: 10.1210/me.2015-1079
133. Darnell G, Orgel JPRO, Pahl R, Meredith SC. Flanking polyproline sequences inhibit beta-sheet structure in polyglutamine segments by inducing PPII-like helix structure. J Mol Biol. (2007) 374:688–704. doi: 10.1016/j.jmb.2007.09.023
134. Darnell GD, Derryberry J, Kurutz JW, Meredith SC. Mechanism of cis-inhibition of polyQ fibrillation by polyP: PPII oligomers and the hydrophobic effect. Biophys J. (2009) 97:2295–305. doi: 10.1016/j.bpj.2009.07.062
135. Crick SL, Ruff KM, Garai K, Frieden C, Pappu RV. Unmasking the roles of N- and C-terminal flanking sequences from exon 1 of huntingtin as modulators of polyglutamine aggregation. Proc Natl Acad Sci USA. (2013) 110:20075–80. doi: 10.1073/pnas.1320626110
136. Zhang L, Kang H, Perez-Aguilar JM, Zhou R. Possible co-evolution of polyglutamine and polyproline in huntingtin protein: proline-rich domain as transient folding chaperone. J Phys Chem Lett. (2022) 13:6331–41. doi: 10.1021/acs.jpclett.2c01184
137. Audi L, Fernández-Cancio M, Carrascosa A, Andaluz P, Torán N, Piró C, et al. Novel (60%) and recurrent (40%) androgen receptor gene mutations in a series of 59 patients with a 46,XY disorder of sex development. J Clin Endocrinol Metab. (2010) 95:1876–88. doi: 10.1210/jc.2009-2146
138. Bermejo-Costa F, Lloreda-García JM, Donate-Legaz JM. Partial androgen insensitivity syndrome with persistent müllerian remnants. A case report. Endocrinol Nutr. (2015) 62:469–71. doi: 10.1016/j.endoen.2015.11.001
139. Tadokoro-Cuccaro R, Davies J, Mongan NP, Bunch T, Brown RS, Audi L, et al. Promoter-dependent activity on androgen receptor N-terminal domain mutations in androgen insensitivity syndrome. Sex Dev. (2014) 8:339–49. doi: 10.1159/000369266
140. Mangelsdorf DJ, Thummel C, Beato M, Herrlich P, Schütz G, Umesono K, et al. The nuclear receptor superfamily: the second decade. Cell. (1995) 83:835–9.
141. Detera-Wadleigh SD, Fanning TG. Phylogeny of the steroid receptor superfamily. Mol Phylogenet Evol. (1994) 3:192–205.
142. Thornton JW, Kelley DB. Evolution of the androgen receptor: structure-function implications. Bioessays. (1998) 20:860–9. doi: 10.1002/(SICI)1521-1878(199810)20:10<860::AID-BIES12>3.0.CO;2-S
143. Kelley ST, Thackray VG. Phylogenetic analyses reveal ancient duplication of estrogen receptor isoforms. J Mol Evol. (1999) 49:609–14. doi: 10.1007/pl00006582
144. McEwan IJ, Lavery D, Fischer K, Watt K. Natural disordered sequences in the amino terminal domain of nuclear receptors: lessons from the androgen and glucocorticoid receptors. Nucl Recept Signal. (2007) 5:e001. doi: 10.1621/nrs.05001
145. Tompa P. Intrinsically unstructured proteins evolve by repeat expansion. Bioessays. (2003) 25:847–55.
146. Brown CJ, Johnson AK, Daughdrill GW. Comparing models of evolution for ordered and disordered proteins. Mol Biol Evol. (2010) 27:609–21.
147. Davey NE, Cyert MS, Moses AM. Short linear motifs - ex nihilo evolution of protein regulation. Cell Commun Signal. (2015) 13:43. doi: 10.1186/s12964-015-0120-z
148. Varadi M, Guharoy M, Zsolyomi F, Tompa P. DisCons: a novel tool to quantify and classify evolutionary conservation of intrinsic protein disorder. BMC Bioinformatics. (2015) 16:153. doi: 10.1186/s12859-015-0592-2
149. Jemth P, Karlsson E, Vögeli B, Guzovsky B, Andersson E, Hultqvist G, et al. Structure and dynamics conspire in the evolution of affinity between intrinsically disordered proteins. Sci Adv. (2018) 4:eaau4130. doi: 10.1126/sciadv.aau4130
150. Estébanez-Perpiñá E, Arnold LA, Nguyen P, Rodrigues ED, Mar E, Bateman R, et al. A surface on the androgen receptor that allosterically regulates coactivator binding. Proc Natl Acad Sci USA. (2007) 104:16074–9.
151. Wilson S, Qi J, Filipp FV. Refinement of the androgen response element based on ChIP-Seq in androgen-insensitive and androgen-responsive prostate cancer cell lines. Sci Rep. (2016) 6:32611. doi: 10.1038/srep32611
152. Helsen C, Kerkhofs S, Clinckemalie L, Spans L, Laurent M, Boonen S, et al. Structural basis for nuclear hormone receptor DNA binding. Mol Cell Endocrinol. (2012) 348:411–7.
153. Schuppe ER, Miles MC, Fuxjager MJ. Evolution of the androgen receptor: perspectives from human health to dancing birds. Mol Cell Endocrinol. (2020) 499:110577. doi: 10.1016/j.mce.2019.110577
154. Monge A, Jagla M, Lapouge G, Sasorith S, Cruchant M, Wurtz J-M, et al. Unfaithfulness and promiscuity of a mutant androgen receptor in a hormone-refractory prostate cancer. Cell Mol Life Sci. (2006) 63:487–97. doi: 10.1007/s00018-005-5471-y
155. Ogino Y, Katoh H, Kuraku S, Yamada G. Evolutionary history and functional characterization of androgen receptor genes in jawed vertebrates. Endocrinology. (2009) 150:5415–27. doi: 10.1210/en.2009-0523
156. Dar JA, Masoodi KZ, Eisermann K, Isharwal S, Ai J, Pascal LE, et al. The N-terminal domain of the androgen receptor drives its nuclear localization in castration-resistant prostate cancer cells. J Steroid Biochem Mol Biol. (2014) 143:473–80. doi: 10.1016/j.jsbmb.2014.03.004
157. Kumar M, Michael S, Alvarado-Valverde J, Mészáros B, Sámano-Sánchez H, Zeke A, et al. The eukaryotic linear motif resource: 2022 release. Nucleic Acids Res. (2022) 50:D497–508. doi: 10.1093/nar/gkab975
158. He B, Bai S, Hnat AT, Kalman RI, Minges JT, Patterson C, et al. An androgen receptor NH2-terminal conserved motif interacts with the COOH terminus of the Hsp70-interacting protein (CHIP). J Biol Chem. (2004) 279:30643–53. doi: 10.1074/jbc.m403117200
159. Katoh K, Misawa K, Kuma K-I, Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. (2002) 30:3059–66. doi: 10.1093/nar/gkf436
160. Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. (2014) 30:1312–3. doi: 10.1093/bioinformatics/btu033
161. Eaton DAR, Overcast I. ipyrad: interactive assembly and analysis of RADseq datasets. Bioinformatics. (2020) 36:2592–4. doi: 10.1093/bioinformatics/btz966
162. Revell LJ. Two new graphical methods for mapping trait evolution on phylogenies. Methods Ecol Evol. (2013) 4:754–9. doi: 10.1111/2041-210x.12066
163. Song S, Liu L, Edwards SV, Wu S. Resolving conflict in eutherian mammal phylogeny using phylogenomics and the multispecies coalescent model. Proc Natl Acad Sci USA. (2012) 109:14942–7.
164. Tsagkogeorga G, Parker J, Stupka E, Cotton JA, Rossiter SJ. Phylogenomic analyses elucidate the evolutionary relationships of bats. Curr Biol. (2013) 23:2262–7.
165. Djian P, Hancock JM, Chana HS. Codon repeats in genes associated with human diseases: fewer repeats in the genes of nonhuman primates and nucleotide substitutions concentrated at the sites of reiteration. Proc Natl Acad Sci USA. (1996) 93:417–21. doi: 10.1073/pnas.93.1.417
166. Choong CS, Kemppainen JA, Wilson EM. Evolution of the primate androgen receptor: a structural basis for disease. J Mol Evol. (1998) 47:334–42.
167. Hong K-W, Hibino E, Takenaka O, Hayasaka I, Murayama Y, Ito S, et al. Comparison of androgen receptor CAG and GGN repeat length polymorphism in humans and apes. Primates. (2006) 47:248–54.
168. Garai C, Furuichi T, Kawamoto Y, Ryu H, Inoue-Murayama M. Androgen receptor and monoamine oxidase polymorphism in wild bonobos. Meta Gene. (2014) 2:831–43. doi: 10.1016/j.mgene.2014.10.005
169. Hiramatsu C, Paukner A, Kuroshima H, Fujita K, Suomi SJ, Inoue-Murayama M. Short poly-glutamine repeat in the androgen receptor in New World monkeys. Meta Gene. (2017) 14:105–13. doi: 10.1016/j.mgene.2017.08.006
170. Gronek P, Przysiecki P, Nowicki S, Kalak R, Juzwa W, Szalata M, et al. Is G→T substitution in the sequence of CAG repeats within the androgen receptor gene associated with aggressive behaviour in the red foxVulpes vulpes? Acta Theriol. (2008) 53:17–25. doi: 10.1007/bf03194275
171. Lai C-L, L’Eplattenier H, van den Ham R, Verseijden F, Jagtenberg A, Mol JA, et al. Androgen receptor CAG repeat polymorphisms in canine prostate cancer. J Vet Intern Med. (2008) 22:1380–4. doi: 10.1111/j.1939-1676.2008.0181.x
172. Ochiai K, Sutijarit S, Uemura M, Morimatsu M, Michishita M, Onozawa E, et al. The number of glutamines in the N-terminal of the canine androgen receptor affects signalling intensities. Vet Comp Oncol. (2021) 19:399–403. doi: 10.1111/vco.12663
173. Coetzee G, Irvine R. Size of the androgen receptor CAG repeat and prostate cancer: does it matter? J Clin Oncol. (2002) 20:3572–3. doi: 10.1200/jco.2002.20.17.3572
174. Alptekin D, Izmirli M, Bayazit Y, Luleyap HU, Yilmaz MB, Soyupak B, et al. Evaluation of the effects of androgen receptor gene trinucleotide repeats and prostate-specific antigen gene polymorphisms on prostate cancer. Genet Mol Res. (2012) 11:1424–32. doi: 10.4238/2012.May.18.1
175. Choong CS, Kemppainen JA, Zhou ZX, Wilson EM. Reduced androgen receptor gene expression with first exon CAG repeat expansion. Mol Endocrinol. (1996) 10:1527–35.
176. Neuschmid-Kaspar F, Gast A, Peterziel H, Schneikert J, Muigg A, Ransmayr G, et al. CAG-repeat expansion in androgen receptor in Kennedy’s disease is not a loss of function mutation. Mol Cell Endocrinol. (1996) 117:149–56.
177. Xue WM, Coetzee GA, Ross RK, Irvine R, Kolonel L, Henderson BE, et al. Genetic determinants of serum prostate-specific antigen levels in healthy men from a multiethnic cohort. Cancer Epidemiol Biomarkers Prev. (2001) 10:575–9.
178. Hardy DO, Scher HI, Bogenreider T, Sabbatini P, Zhang ZF, Nanus DM, et al. Androgen receptor CAG repeat lengths in prostate cancer: correlation with age of onset. J Clin Endocrinol Metab. (1996) 81:4400–5.
179. Yaffe K, Edwards ER, Lui L-Y, Zmuda JM, Ferrell RE, Cauley JA. Androgen receptor CAG repeat polymorphism is associated with cognitive function in older men. Biol Psychiatry. (2003) 54:943–6.
180. Sadar MD. Discovery of drugs that directly target the intrinsically disordered region of the androgen receptor. Expert Opin Drug Discov. (2020) 15:551–60. doi: 10.1080/17460441.2020.1732920
181. Wang H, Jiang M, Cui H, Chen M, Buttyan R, Hayward SW, et al. The stress response mediator ATF3 represses androgen signaling by binding the androgen receptor. Mol Cell Biol. (2012) 32:3190–202. doi: 10.1128/MCB.00159-12
182. Alvarez M, Rhodes SJ, Bidwell JP. Context-dependent transcription: all politics is local. Gene. (2003) 313:43–57. doi: 10.1016/s0378-1119(03)00627-9
183. Jolly C, Morimoto R, Robert-Nicoud M, Vourc’h C. HSF1 transcription factor concentrates in nuclear foci during heat shock: relationship with transcription sites. J Cell Sci. (1997) 110:2935–41. doi: 10.1242/jcs.110.23.2935
184. Pombo A, Cuello P, Schul W, Yoon JB, Roeder RG, Cook PR, et al. Regional and temporal specialization in the nucleus: a transcriptionally-active nuclear domain rich in PTF, Oct1 and PIKA antigens associates with specific chromosomes early in the cell cycle. EMBO J. (1998) 17:1768–78. doi: 10.1093/emboj/17.6.1768
185. Elbi C, Misteli T, Hager GL. Recruitment of dioxin receptor to active transcription sites. Mol Biol Cell. (2002) 13:2001–15.
186. Arnett-Mansfield RL, Graham JD, Hanson AR, Mote PA, Gompel A, Scurr LL, et al. Focal subnuclear distribution of progesterone receptor is ligand dependent and associated with transcriptional activity. Mol Endocrinol. (2007) 21:14–29. doi: 10.1210/me.2006-0041
187. Sabari BR, Dall’Agnese A, Young RA. Biomolecular condensates in the nucleus. Trends Biochem Sci. (2020) 45:961–77.
188. Sołtys K, Ożyhar A. Transcription regulators and membraneless organelles challenges to investigate them. Int J Mol Sci. (2021) 22:12758. doi: 10.3390/ijms222312758
189. Guo C, Luo Z, Lin C. Phase separation properties in transcriptional organization. Biochemistry. (2022). [Epub ahead of print]. doi: 10.1021/acs.biochem.2c00220
190. Wang B, Zhang L, Dai T, Qin Z, Lu H, Zhang L, et al. Liquid–liquid phase separation in human health and diseases. Signal Transduct Target Ther. (2021) 6:290. doi: 10.1038/s41392-021-00678-1
191. Tong X, Tang R, Xu J, Wang W, Zhao Y, Yu X, et al. Liquid-liquid phase separation in tumor biology. Signal Transduct Target Ther. (2022) 7:221.
192. Darling AL, Shorter J. Combating deleterious phase transitions in neurodegenerative disease. Biochim Biophys Acta Mol Cell Res. (2021) 1868:118984.
193. Webber CJ, Lei SE, Wolozin B. The pathophysiology of neurodegenerative disease: disturbing the balance between phase separation and irreversible aggregation. Prog Mol Biol Transl Sci. (2020) 174:187–223. doi: 10.1016/bs.pmbts.2020.04.021
194. Banani SF, Lee HO, Hyman AA, Rosen MK. Biomolecular condensates: organizers of cellular biochemistry. Nat Rev Mol Cell Biol. (2017) 18:285–98.
195. Bouchard JJ, Otero JH, Scott DC, Szulc E, Martin EW, Sabri N, et al. Cancer mutations of the tumor suppressor SPOP disrupt the formation of active, phase-separated compartments. Mol Cell. (2018) 72:19–36.e8. doi: 10.1016/j.molcel.2018.08.027
196. Boija A, Klein IA, Sabari BR, Dall’Agnese A, Coffey EL, Zamudio AV, et al. Transcription factors activate genes through the phase-separation capacity of their activation domains. Cell. (2018) 175:1842–55.e16. doi: 10.1016/j.cell.2018.10.042
197. Stortz M, Pecci A, Presman DM, Levi V. Unraveling the molecular interactions involved in phase separation of glucocorticoid receptor. BMC Biol. (2020) 18:59. doi: 10.1186/s12915-020-00788-2
198. Lu H, Yu D, Hansen AS, Ganguly S, Liu R, Heckert A, et al. Phase-separation mechanism for C-terminal hyperphosphorylation of RNA polymerase II. Nature. (2018) 558:318–23. doi: 10.1038/s41586-018-0174-3
199. Kato M, Han TW, Xie S, Shi K, Du X, Wu LC, et al. Cell-free formation of RNA granules: low complexity sequence domains form dynamic fibers within hydrogels. Cell. (2012) 149:753–67. doi: 10.1016/j.cell.2012.04.017
200. Zhang F, Wong S, Lee J, Lingadahalli S, Wells C, Saxena N, et al. Dynamic phase separation of the androgen receptor and its coactivators to regulate gene expression. bioRxiv. [Preprint]. (2021). doi: 10.1101/2021.03.27.437301
201. Ahmed J, Meszaros A, Lazar T, Tompa PDNA. -binding domain as the minimal region driving RNA-dependent liquid–liquid phase separation of androgen receptor. Protein Sci. (2021) 30:1380–92. doi: 10.1002/pro.4100
202. Roggero CM, Esser V, Duan L, Rice AM, Ma S, Raj GV, et al. Poly-glutamine-dependent self-association as a potential mechanism for regulation of androgen receptor activity. PLoS One. (2022) 17:e0258876. doi: 10.1371/journal.pone.0258876
203. Wen S, He Y, Wang L, Zhang J, Quan C, Niu Y, et al. Aberrant activation of super enhancer and choline metabolism drive antiandrogen therapy resistance in prostate cancer. Oncogene. (2020) 39:6556–71. doi: 10.1038/s41388-020-01456-z
204. Stortz M, Presman DM, Pecci A, Levi V. Phasing the intranuclear organization of steroid hormone receptors. Biochem J. (2021) 478:443–61. doi: 10.1042/BCJ20200883
205. Iyer MK, Niknafs YS, Malik R, Singhal U, Sahu A, Hosono Y, et al. The landscape of long noncoding RNAs in the human transcriptome. Nat Genet. (2015) 47:199–208.
206. Li J, Xuan Z, Liu C. Long non-coding RNAs and complex human diseases. Int J Mol Sci. (2013) 14:18790–808.
207. Yang Y, Liu KY, Liu Q, Cao Q. Androgen Receptor-Related Non-coding RNAs in Prostate Cancer. Front Cell Dev Biol. (2021) 9:660853. doi: 10.3389/fcell.2021.660853
208. Hudson WH, Pickard MR, de Vera IMS, Kuiper EG, Mourtada-Maarabouni M, Conn GL, et al. Conserved sequence-specific lincRNA-steroid receptor interactions drive transcriptional repression and direct cell fate. Nat Commun. (2014) 5:5395. doi: 10.1038/ncomms6395
209. Schmidt K, Carroll JS, Yee E, Thomas DD, Wert-Lamas L, Neier SC, et al. The lncRNA SLNCR recruits the androgen receptor to EGR1-bound genes in melanoma and inhibits expression of tumor suppressor p21. Cell Rep. (2019) 27:2493–507.e4. doi: 10.1016/j.celrep.2019.04.101
210. Schmidt K, Joyce CE, Buquicchio F, Brown A, Ritz J, Distel RJ, et al. The lncRNA SLNCR1 mediates melanoma invasion through a conserved SRA1-like region. Cell Rep. (2016) 15:2025–37. doi: 10.1016/j.celrep.2016.04.018
211. Schmidt K, Weidmann CA, Hilimire TA, Yee E, Hatfield BM, Schneekloth JS Jr., et al. Targeting the oncogenic long non-coding RNA SLNCR1 by blocking its sequence-specific binding to the androgen receptor. Cell Rep. (2020) 30:541–54.e5. doi: 10.1016/j.celrep.2019.12.011
212. Roden C, Gladfelter AS. RNA contributions to the form and function of biomolecular condensates. Nat Rev Mol Cell Biol. (2021) 22:183–95. doi: 10.1038/s41580-020-0264-6
213. Luo J, Qu L, Gao F, Lin J, Liu J, Lin A. LncRNAs: architectural scaffolds or more potential roles in phase separation. Front Genet. (2021) 12:626234. doi: 10.3389/fgene.2021.626234
214. Ribeiro DM, Zanzoni A, Cipriano A, Delli Ponti R, Spinelli L, Ballarino M, et al. Protein complex scaffolding predicted as a prevalent function of long non-coding RNAs. Nucleic Acids Res. (2018) 46:917–28. doi: 10.1093/nar/gkx1169
215. Skotte NH, Southwell AL, Østergaard ME, Carroll JB, Warby SC, Doty CN, et al. Allele-specific suppression of mutant huntingtin using antisense oligonucleotides: providing a therapeutic option for all Huntington disease patients. PLoS One. (2014) 9:e107434. doi: 10.1371/journal.pone.0107434
216. Ferguson MW, Kennedy CJ, Palpagama TH, Waldvogel HJ, Faull RLM, Kwakowsky A. Current and possible future therapeutic options for Huntington’s Disease. J Cent Nerv Syst Dis. (2022) 14:11795735221092517.
217. Doxakis E. Therapeutic antisense oligonucleotides for movement disorders. Med Res Rev. (2021) 41:2656–88.
218. López Castel A, Overby SJ, Artero R. MicroRNA-based therapeutic perspectives in myotonic dystrophy. Int J Mol Sci. (2019) 20:5600. doi: 10.3390/ijms20225600
219. Hu N, Kim E, Antoury L, Li J, González-Pérez P, Rutkove SB, et al. Antisense oligonucleotide and adjuvant exercise therapy reverse fatigue in old mice with myotonic dystrophy. Mol Ther Nucleic Acids. (2021) 23:393–405. doi: 10.1016/j.omtn.2020.11.014
220. Ait Benichou S, Jauvin D, De Serres-Bérard T, Pierre M, Ling KK, Bennett CF, et al. Antisense oligonucleotides as a potential treatment for brain deficits observed in myotonic dystrophy type 1. Gene Ther. (2022). [Epub ahead of print]. doi: 10.1038/s41434-022-00316-7
221. Ly CV, Miller TM. Emerging antisense oligonucleotide and viral therapies for amyotrophic lateral sclerosis. Curr Opin Neurol. (2018) 31:648–54.
222. Sathyaprakash C, Manzano R, Varela MA, Hashimoto Y, Wood MJA, Talbot K, et al. Development of LNA gapmer oligonucleotide-based therapy for ALS/FTD caused by the C9orf72 repeat expansion. Methods Mol Biol. (2020) 2176:185–208. doi: 10.1007/978-1-0716-0771-8_14
223. Suzuki N, Nishiyama A, Warita H, Aoki M. Genetics of amyotrophic lateral sclerosis: seeking therapeutic targets in the era of gene therapy. J Hum Genet. (2022). [Epub ahead of print]. doi: 10.1038/s10038-022-01055-8
224. Roberts TC, Langer R, Wood MJA. Advances in oligonucleotide drug delivery. Nat Rev Drug Discov. (2020) 19:673–94.
225. Monteys AM, Ebanks SA, Keiser MS, Davidson BL. CRISPR/Cas9 editing of the mutant huntingtin allele in vitro and in vivo. Mol Ther. (2017) 25:12–23. doi: 10.1016/j.ymthe.2016.11.010
226. Shin JW, Kim K-H, Chao MJ, Atwal RS, Gillis T, MacDonald ME, et al. Permanent inactivation of Huntington’s disease mutation by personalized allele-specific CRISPR/Cas9. Hum Mol Genet. (2016) 25:4566–76. doi: 10.1093/hmg/ddw286
227. Ekman FK, Ojala DS, Adil MM, Lopez PA, Schaffer DV, Gaj T. CRISPR- Cas9-mediated genome editing increases lifespan and improves motor deficits in a Huntington’s disease mouse model. Mol Ther Nucleic Acids. (2019) 17:829–39. doi: 10.1016/j.omtn.2019.07.009
228. Chey YCJ, Arudkumar J, Aartsma-Rus A, Adikusuma F, Thomas PQ. CRISPR applications for Duchenne muscular dystrophy: from animal models to potential therapies. WIREs Mech Dis. (2022). [Epub ahead of print]. doi: 10.1002/wsbm.1580
229. Raaijmakers RHL, Ripken L, Ausems CRM, Wansink DG. CRISPR/Cas applications in myotonic dystrophy: expanding opportunities. Int J Mol Sci. (2019) 20:3689. doi: 10.3390/ijms20153689
230. Cardinali B, Provenzano C, Izzo M, Voellenkle C, Battistini J, Strimpakos G, et al. Time-controlled and muscle-specific CRISPR/Cas9-mediated deletion of CTG-repeat expansion in the gene. Mol Ther Nucleic Acids. (2022) 27:184–99. doi: 10.1016/j.omtn.2021.11.024
231. He Q, Jantac Mam-Lam-Fook C, Chaignaud J, Danset-Alexandre C, Iftimovici A, Gradels Hauguel J, et al. Influence of polygenic risk scores for schizophrenia and resilience on the cognition of individuals at-risk for psychosis. Transl Psychiatry. (2021) 11:518. doi: 10.1038/s41398-021-01624-z
232. Rocca CJ, Rainaldi JN, Sharma J, Shi Y, Haquang JH, Luebeck J, et al. CRISPR-Cas9 gene editing of hematopoietic stem cells from patients with Friedreich’s ataxia. Mol Ther Methods Clin Dev. (2020) 17:1026–36. doi: 10.1016/j.omtm.2020.04.018
233. Ababneh NA, Scaber J, Flynn R, Douglas A, Barbagallo P, Candalija A, et al. Correction of amyotrophic lateral sclerosis related phenotypes in induced pluripotent stem cell-derived motor neurons carrying a hexanucleotide expansion mutation in C9orf72 by CRISPR/Cas9 genome editing using homology-directed repair. Hum Mol Genet. (2020) 29:2200–17. doi: 10.1093/hmg/ddaa106
234. Park SB, Uchida T, Tilson S, Hu Z, Ma CD, Leek M, et al. A dual conditional CRISPR-Cas9 system to activate gene editing and reduce off-target effects in human stem cells. Mol Ther Nucleic Acids. (2022) 28:656–69. doi: 10.1016/j.omtn.2022.04.013
235. Ruan H, Sun Q, Zhang W, Liu Y, Lai L. Targeting intrinsically disordered proteins at the edge of chaos. Drug Discov Today. (2019) 24:217–27. doi: 10.1016/j.drudis.2018.09.017
236. Biesaga M, Frigolé-Vivas M, Salvatella X. Intrinsically disordered proteins and biomolecular condensates as drug targets. Curr Opin Chem Biol. (2021) 62:90–100.
237. Whitfield JR, Soucek L. The long journey to bring a Myc inhibitor to the clinic. J Cell Biol. (2021) 220:e202103090. doi: 10.1083/jcb.202103090
238. Liu L, Shi L, Wang Z, Zeng J, Wang Y, Xiao H, et al. Targeting Oncoproteins for Degradation by Small Molecule-Based Proteolysis-Targeting Chimeras (PROTACs) in Sex Hormone-Dependent Cancers. Front Endocrinol. (2022) 13:839857. doi: 10.3389/fendo.2022.839857
239. Li Z, Zhu C, Ding Y, Fei Y, Lu B. ATTEC: a potential new approach to target proteinopathies. Autophagy. (2020) 16:185–7. doi: 10.1080/15548627.2019.1688556
240. Békés M, Langley DR, Crews CM. PROTAC targeted protein degraders: the past is prologue. Nat Rev Drug Discov. (2022) 21:181–200. doi: 10.1038/s41573-021-00371-6
241. Spradlin JN, Zhang E, Nomura DK. Reimagining druggability using chemoproteomic platforms. Acc Chem Res. (2021) 54:1801–13. doi: 10.1021/acs.accounts.1c00065
242. Huang J, Lin B, Li B. Anti-androgen receptor therapies in prostate cancer: a brief update and perspective. Front Oncol. (2022) 12:865350. doi: 10.3389/fonc.2022.865350
243. Baumgart SJ, Nevedomskaya E, Lesche R, Newman R, Mumberg D, Haendler B. Darolutamide antagonizes androgen signaling by blocking enhancer and super-enhancer activation. Mol Oncol. (2020) 14:2022–39. doi: 10.1002/1878-0261.12693
244. Guo H, Wu Y, Nouri M, Spisak S, Russo JW, Sowalsky AG, et al. Androgen receptor and MYC equilibration centralizes on developmental super-enhancer. Nat Commun. (2021) 12:7308. doi: 10.1038/s41467-021-27077-y
245. Qiao X, van der Zanden SY, Wander DPA, Borràs DM, Song J-Y, Li X, et al. Uncoupling DNA damage from chromatin damage to detoxify doxorubicin. Proc Natl Acad Sci USA. (2020) 117:15182–92. doi: 10.1073/pnas.1922072117
246. Basu S, Mackowiak SD, Niskanen H, Knezevic D, Asimi V, Grosswendt S, et al. Unblending of transcriptional condensates in human repeat expansion disease. Cell. (2020) 181:1062–79.e30. doi: 10.1016/j.cell.2020.04.018
247. Pakravan D, Orlando G, Bercier V, Van Den Bosch L. Role and therapeutic potential of liquid-liquid phase separation in amyotrophic lateral sclerosis. J Mol Cell Biol. (2021) 13:15–28.
248. Wheeler RJ, Lee HO, Poser I, Pal A, Doeleman T, Kishigami S, et al. Small molecules for modulating protein driven liquid-liquid phase separation in treating neurodegenerative disease. bioRxiv. [Preprint]. (2019). doi: 10.1101/721001
249. Wheeler RJ. Therapeutics—how to treat phase separation-associated diseases. Emerg Top Life Sci. (2020) 4:331–42. doi: 10.1042/etls20190176
250. Risso-Ballester J, Galloux M, Cao J, Le Goffic R, Hontonnou F, Jobart-Malfait A, et al. A condensate-hardening drug blocks RSV replication in vivo. Nature. (2021) 595:596–9. doi: 10.1038/s41586-021-03703-z
251. Nihei Y, Ito D, Okada Y, Akamatsu W, Yagi T, Yoshizaki T, et al. Enhanced aggregation of androgen receptor in induced pluripotent stem cell-derived neurons from spinal and bulbar muscular atrophy. J Biol Chem. (2013) 288:8043–52. doi: 10.1074/jbc.m112.408211
Keywords: androgen receptor, polyQ, amino acid repeats, polymorphism, phylogenetics, cancer, aggregation, phase separation
Citation: Meszaros A, Ahmed J, Russo G, Tompa P and Lazar T (2022) The evolution and polymorphism of mono-amino acid repeats in androgen receptor and their regulatory role in health and disease. Front. Med. 9:1019803. doi: 10.3389/fmed.2022.1019803
Received: 15 August 2022; Accepted: 30 September 2022;
Published: 20 October 2022.
Edited by:
Anders Strom, University of Houston, United StatesReviewed by:
Iain J. McEwan, University of Aberdeen, United KingdomYongfeng He, Cornell University, United States
Copyright © 2022 Meszaros, Ahmed, Russo, Tompa and Lazar. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Peter Tompa, peter.tompa@vub.be; Tamas Lazar, tamas.lazar@vub.be
 Attila Meszaros
Attila Meszaros Junaid Ahmed
Junaid Ahmed Giorgio Russo1,2
Giorgio Russo1,2  Peter Tompa
Peter Tompa Tamas Lazar
Tamas Lazar