Artificial Intelligence-Aid Colonoscopy Vs. Conventional Colonoscopy for Polyp and Adenoma Detection: A Systematic Review of 7 Discordant Meta-Analyses
- 1Department of Endoscopy, Shanghai Jiangong Hospital, Shanghai, China
- 2Endoscopy Center, Zhongshan Hospital, Fudan University, Shanghai, China
- 3Department of Gastroenterology, Shanghai Jiangong Hospital, Shanghai, China
- 4Department of Surgery, Shanghai Jiangong Hospital, Shanghai, China
Objectives: Multiple meta-analyses which investigated the comparative efficacy and safety of artificial intelligence (AI)-aid colonoscopy (AIC) vs. conventional colonoscopy (CC) in the detection of polyp and adenoma have been published. However, a definitive conclusion has not yet been generated. This systematic review selected from discordant meta-analyses to draw a definitive conclusion about whether AIC is better than CC for the detection of polyp and adenoma.
Methods: We comprehensively searched potentially eligible literature in PubMed, Embase, Cochrane library, and China National Knowledgement Infrastructure (CNKI) databases from their inceptions until to April 2021. Assessment of Multiple Systematic Reviews (AMSTAR) instrument was used to assess the methodological quality. Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) checklist was used to assess the reporting quality. Two investigators independently used the Jadad decision algorithm to select high-quality meta-analyses which summarized the best available evidence.
Results: Seven meta-analyses met our selection criteria finally. AMSTAR score ranged from 8 to 10, and PRISMA score ranged from 23 to 26. According to the Jadad decision algorithm, two high-quality meta-analyses were selected. These two meta-analyses suggested that AIC was superior to CC for colonoscopy outcomes, especially for polyp detection rate (PDR) and adenoma detection rate (ADR).
Conclusion: Based on the best available evidence, we conclude that AIC should be preferentially selected for the route screening of colorectal lesions because it has potential value of increasing the polyp and adenoma detection. However, the continued improvement of AIC in differentiating the shape and pathology of colorectal lesions is needed.
Introduction
According to the latest statistics from the World Health Organization (WHO), colorectal cancer (CRC) remains one of the leading reasons of cancer-related death (1). Colonoscopy has been regarded as the preferred modality for early detection and removal of premalignant lesions (2, 3). Evidence revealed that colonoscopy was not only associated with a reduced incidence of CRC, but was also associated with reduced risk of mortality of CRC (4). However, colonoscopy has been found to have high adenoma miss rate (5). As one of the most important measures of colonoscopy quality, studies estimated that for every 1% increase in the rate of adenoma detection, there is a 3% decrease in CRC-caused mortality and a 5% decrease in incidence of interval CRC (6). It is therefore imperative that adenoma miss rate during colonoscopy continue to be decreased (5, 7).
Currently, published studies have explored possible factors which may have an impact on performance of colonoscopy and demonstrated multiple factors which were associated with increased adenoma miss rate, including quality of bowel preparation before colonoscopy, blind spots of colon, recognition of colorectal lesions, and features of polyp and adenoma (8–10). Enormous efforts have been exerted to increase the detection of polyp and adenoma through the improvement of endoscopic techniques and quality of bowel preparation (9), however high miss rate for polyp and adenoma is still not significantly decreased. Moreover, diminutive colorectal polyp and adenoma is difficult to be detected visually, and even under the assistance of magnified and high-resolution endoscopy (11). Evidence suggested that repeated colonoscopy performed by a second endoscopist increased polyp detection rate (PDR) and adenoma detection rate (ADR) (12, 13). Therefore, more and more attention has been paid to the improvement of endoscopist's cognitive aspect and recognition for colorectal lesions (14–16).
With the development of artificial intelligence (AI), several real-time automatic polyp and adenoma detection systems based on convolutional neural network (CNN) have been developed and validated (17, 18), which are considered to have the ability of assisting endoscopist to efficiently and accurately detect colorectal lesions during colonoscopy (13). Several randomized controlled trials (RCTs) have investigated the value of these systems for assisting the detection of colorectal lesions in real world (9, 19–24). Meanwhile, on the basis of published RCTs, multiple meta-analyses have also been performed (25–31). Unfortunately, a definitive conclusion has not yet been generated from published meta-analyses. The comparative detection performance of AIC vs. conventional colonoscopy (CC) for polyps and adenomas remains under debate. We therefore performed this systematic review: (a) to systematically structure meta-analyses which compared AIC with CC; (b) to objectively select high-quality meta-analyses out from discordant meta-analyses; and (c) to determine the preferred colonoscopy modality of CRC screening using the currently available evidence.
Materials and Methods
We developed the framework of the present systematic review according to the methodological framework recommended by the Cochrane handbook (32). We reported results according to the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) guidelines (33, 34).
Study Identification
We systematically searched potentially eligible studies in PubMed, Embase, Cochrane Library, and China National Knowledgement Infrastructure (CNKI) databases from their inceptions until to April 2021. We used the following search terms to construct search strategies: colonoscopy, artificial intelligence, convolutional neural network. Essential search strategy was modified to generate specific strategy according to the unique requirements of individual database. The search strategies of English databases are summarized in Supplementary Table 1. We also manually checked the references of all eligible studies in order to capture any eligible meta-analysis.
Selection Criteria
According to the selection criteria developed in previous systematic review of discordant meta-analyses, we defined the following inclusion criteria: any meta-analysis which compared AIC with CC for the detection of polyps and adenomas during colonoscopy were considered to be eligible for our criteria. Meanwhile, we defined the following exclusion criteria: (a) narrative review; (b) meta-analysis incorporating non-RCTs into analysis; (c) systematic review without meta-analysis; and (d) meta-analysis which did not report clinical outcomes.
Selection of Studies
Two reviewers independently selected studies. Any meta-analysis with full-text was selected if it met our selection criteria. Two reviewers independently extracted the following information from each eligible meta-analysis, including the name of the first author, publication year, journal of publication with the latest impact factor, level of evidence, time duration of literature search, selection criteria, database searched, design of primary studies, accumulated number of each eligible meta-analysis, software applied, additional analysis including heterogeneity analysis, sensitivity analysis, subgroup analysis, trail sequential analysis or the Grading of Recommendations Assessment, Development and Evaluation (GRADE). All outcomes of each meta-analysis were extracted.
Assessment of Methodological Quality
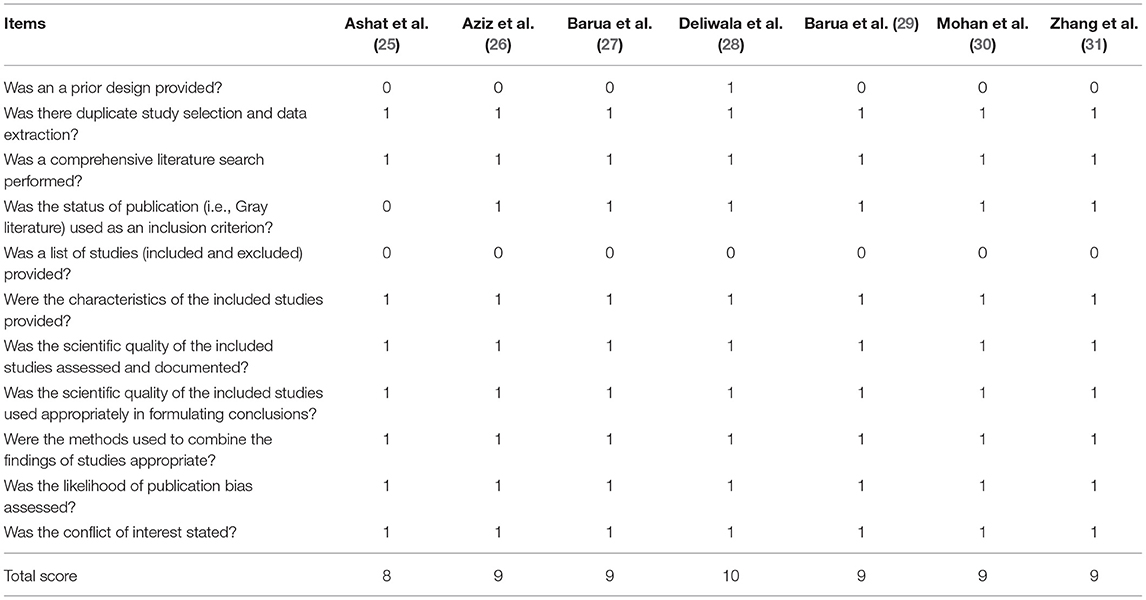
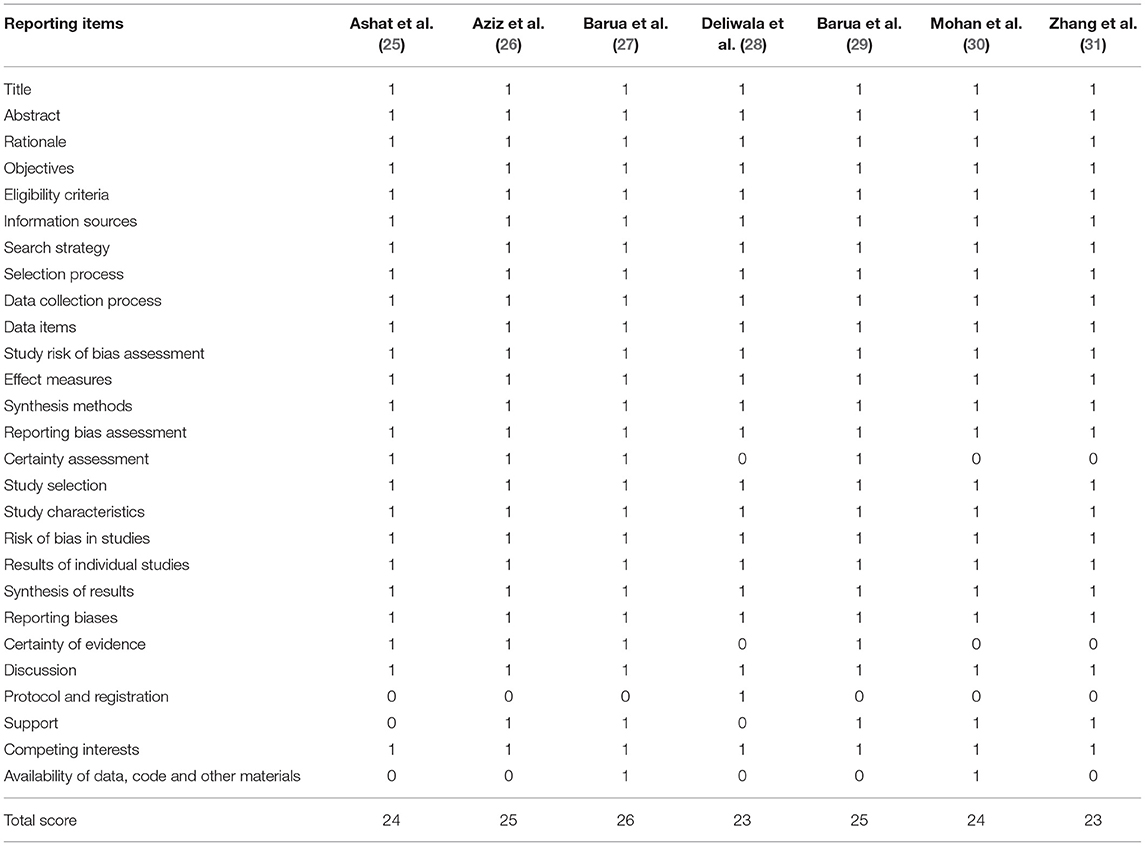
Two independent reviewers assessed the methodological quality of each meta-analysis by using the Assessment of Multiple Systematic Reviews (AMSTAR) instrument (35). In AMSTAR instrument, the overall level of each meta-analysis is identified according to the matching level between actual information and 11 items. We also assess the quality of each meta-analysis by using the Oxford Evidence-based Medicine Levels of Evidence (36). Meanwhile, we used the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) to measure the level of reporting quality.
Application of the Jadad Decision Algorithm
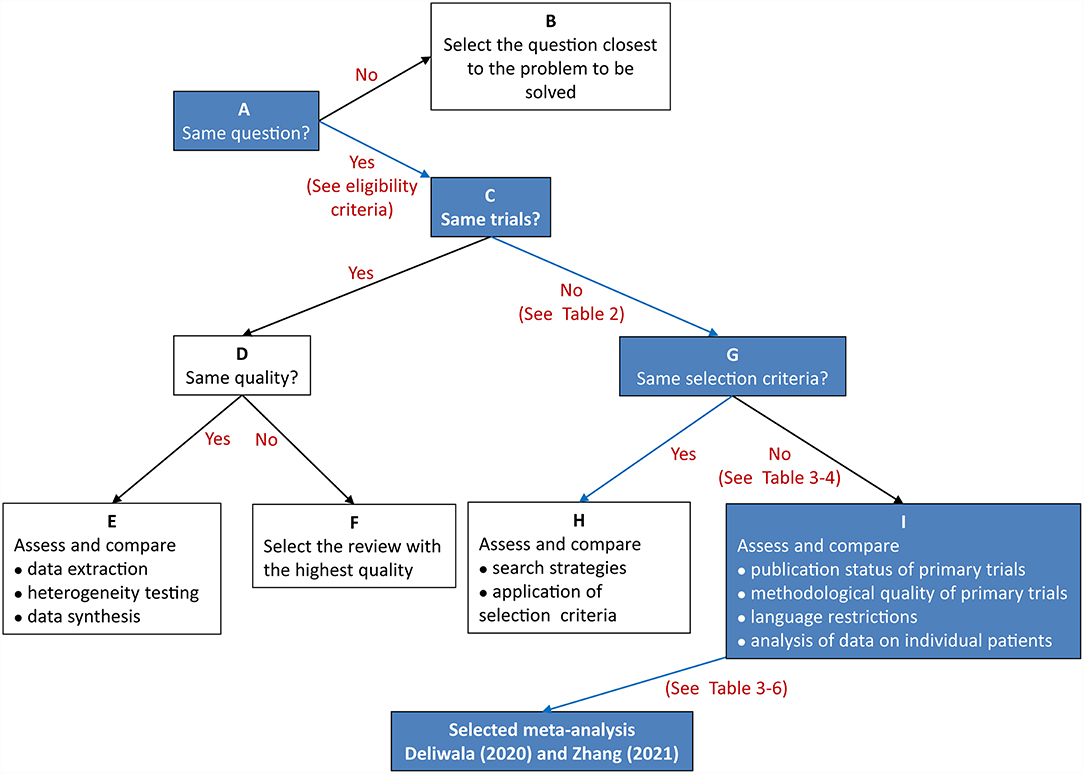
We applied the Jadad decision algorithm to assist selecting high-quality meta-analyses from multiple discordant meta-analyses (37). Jadad decision algorithm is a useful tool for differentiating discordant meta-analyses. The Jadad decision algorithm was designed based on following four questions: (a) Do the meta-analyses ask the same question? (b) Do the meta-analyses include the same primary studies? (c) Do the meta-analyses incorporating the same primary trials achieve the same methodological quality? (c) Do the discordant meta-analyses incorporating different primary trials apply the same selection criteria? Two reviewers independently applied this decision algorithm, and then their results were cross-checked to ensure selecting meta-analysis with the highest quality of evidence to develop recommendations.
Results
Search Results
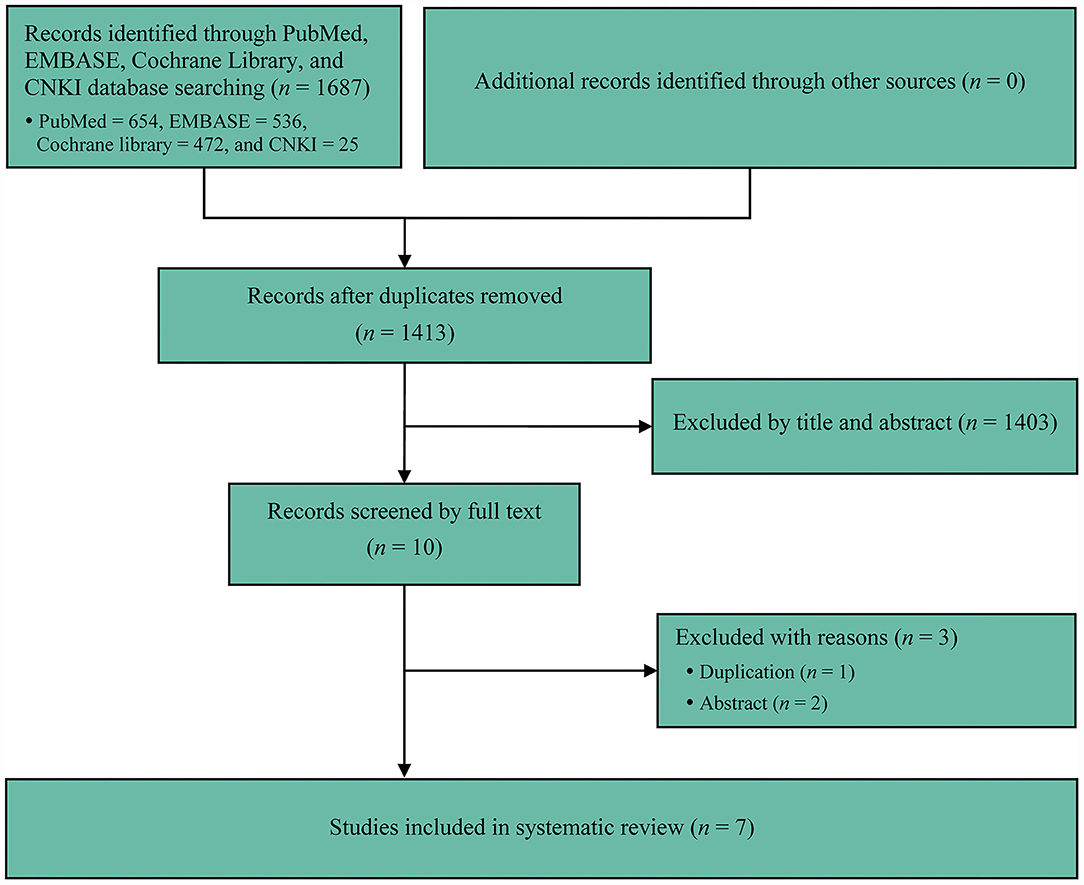
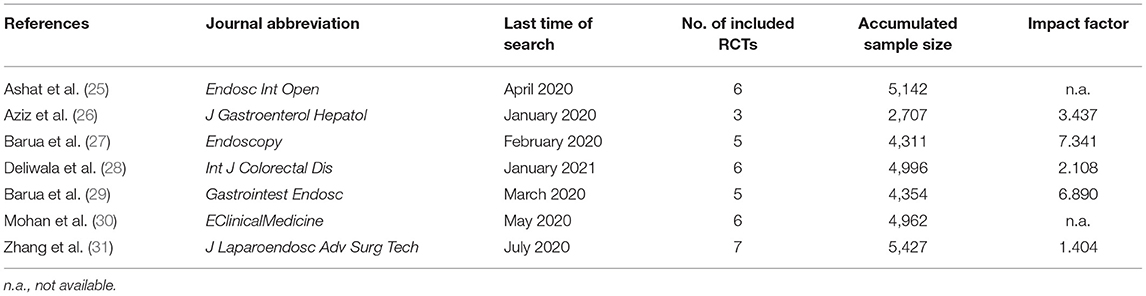
We initially captured 1,687 records from target databases, and seven meta-analyses (2–31) were considered to meet our selection criteria eventually. We designed Figure 1 to delineate the process of retrieval and selection of eligible literature. Meanwhile, the reasons of excluding ineligible studies were also described in Figure 1. All included meta-analyses were published between 2020 and 2021 (Table 1). Five meta-analyses (26–31) were published in high-impact journals, which were defined to have an impact factor released by Web of Science (Table 1). The accumulated number of included primary RCTs in individual meta-analysis ranged from 3 to 7 (Tables 1, 2). The accumulated sample size of individual meta-analysis ranged from 2707 to 5427 (Table 1).
Search Methodology
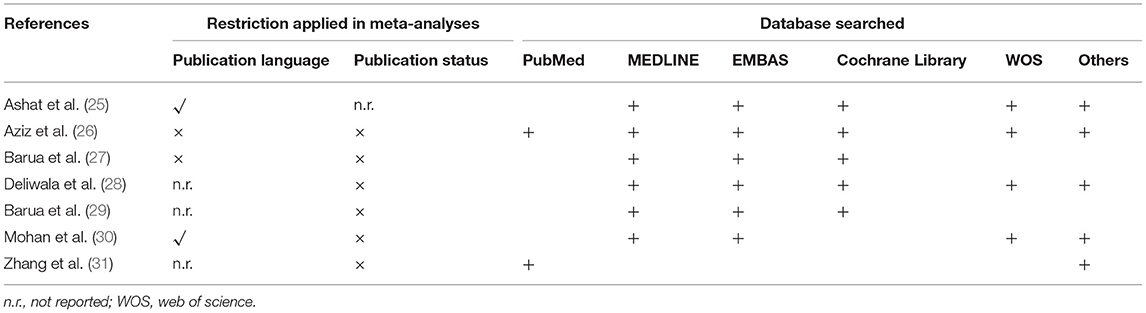
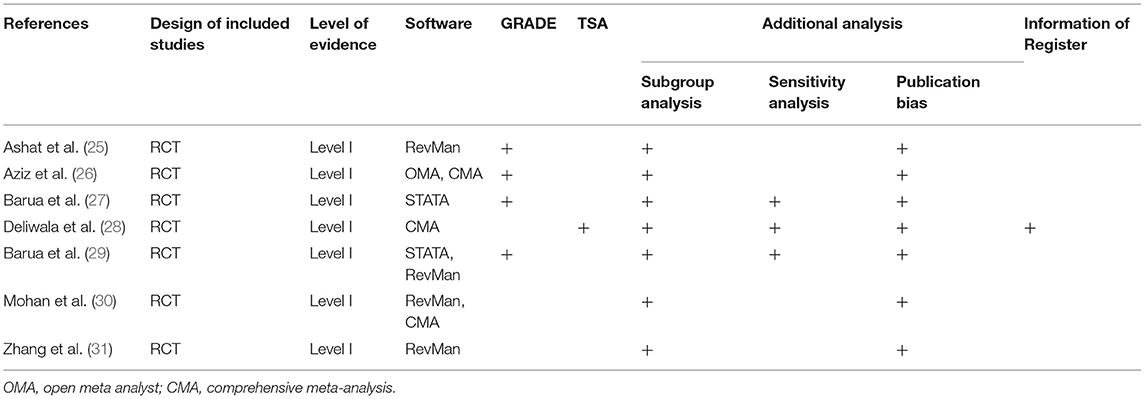
All studies comprehensively searched databases. The number of databases which were searched in each meta-analysis ranged from 2 to 6. It is noted that Medline, EMBASE and the Cochrane Library were searched for capturing eligible primary studies among most meta-analyses (25–29). Unfortunately, other databases such as Scopus and Ovid platform were differentially selected by each meta-analysis. We summarized details of the search methodology in Table 3.
Heterogeneity Assessment
Among all included meta-analyses, statistical heterogeneity was measured using the Cochrane Q and inconsistent index (I2 statistic). Total 4 softwares including RevMan, STATA, open meta-analyst (OMA), and comprehensive meta-analysis (CMA) were used for statistical analysis (Table 4). All meta-analyses (25–31) performed publication bias test and subgroup analysis, and three meta-analyses (27–29) performed sensitivity analysis (Table 4). Four meta-analyses (25–27, 29) used GRADE method to grade the level of evidence. Only one meta-analysis (28) registered protocol at public platform and used TSA method to test the robustness of pooled results (Table 4).
Study Quality and Validity
All eligible meta-analyses were performed based on RCTs. The AMSTAR score ranged from 8 to 10 points, which is documented in Table 5. According to the Oxford Levels of Evidence, all meta-analyses (25–31) were assessed as Level I evidence (Table 1). According to the PRISMA criteria, the score of individual meta-analysis was between 23 and 26, indicating a high-level reporting quality (Table 6).
Results of the Jadad Decision Algorithm
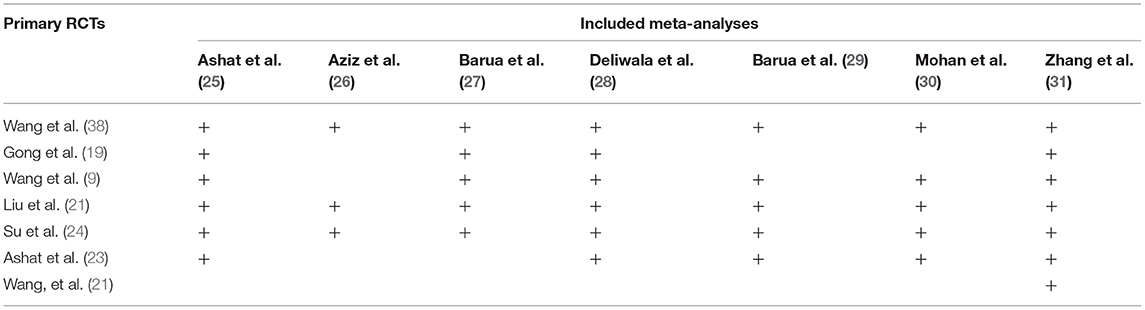
Two reviewers independently performed the same flow path according to the Jadad decision algorithm. Because (a) all included meta-analyses investigated the same question (comparing AIC with CC for polyp and adenoma detection), (b) the included meta-analyses did not include the same RCTs, and (c) they did not design the same selection criteria, we selected the meta-analysis with the highest quality based on the following factors: publication status of the primary studies, methodology of the primary studies, language restrictions and the analysis of data on individual patients (37). Two eligible meta-analyses imposed language restriction, however all eligible meta-analyses did not restrict the publication status of primary studies. Therefore, we firstly excluded two meta-analyses performed by Ashat (25) and Mohan (30), respectively. Another three meta-analyses (26, 27, 29) were also excluded because of they did not completely include primary studies. Finally, two reviewers selected the remaining meta-analyses by Deliwala (28, 31), respectively, as the meta-analyses providing the best available evidence (Figure 2).
Among the selected meta-analyses (Figure 3), PDR and ADR were all significantly increased in the AIC group compared to CC group, which was all confirmed by the results from TSA which was reported in the study performed by Deliwala (28). Meanwhile, these two meta-analyses consistently suggested that AIC was associated with increased number of polyps and adenomas detected per colonoscopy. Zhang and colleagues found that AIC benefits to detect benign lesions compared to CC, however CC may be better than AIC in detecting advanced adenoma (31). Meta-analysis by Deliwala indicated that AIC benefits to achieve target withdrawal time (6 min) compared to CC (28). For the detection of diminutive polyp and adenoma and small to large polyps, two meta-analyses reported conflicting results. Two meta-analyses suggested that AIC increased the detection of polyp in the transverse colon, however detected more polyps in rectum and pedicle adenomas. For detection of adenoma in transverse colon and pedicle and flat polyps, two meta-analyses reported conflicting results.
Discussion
Currently, several RCTs (9, 19–24, 38) which investigated the comparative efficacy and safety of AIC vs. CC for the detection of polyps and adenomas during colonoscopy have been reported. Unfortunately, conflicting results were reported in these primary studies. Meanwhile, multiple meta-analyses (25–31) based on published RCTs have also been performed to systematically evaluate the role of AI-aid automatic detection system in assisting the detection of polyp and adenoma. Published meta-analyses comprehensively searched potentially eligible studies in main databases such as PubMed, EMBASE and Cochrane library, however they enrolled different RCTs and obtained conflicting conclusions, which has confused decision makers to appropriately make recommendations.
Fortunately, those possible sources of causing discordant results among meta-analyses have been deeply interpreted and clearly reported by Jadad and colleagues (37). Based on those possible sources, Jadad and colleagues developed a decision algorithm to assist researchers and practitioners selecting high-quality evidence from multiple discordant meta-analyses. To date, Jadad decision algorithm has been extensively applied to select the currently best available evidence from discordant meta-analyses. In the present systematic review, we eventually identified 7 meta-analyses (25–31) to meet our selection criteria. Unfortunately, among these 7 meta-analyses, three studies (26, 27, 29) were excluded because of they missed some eligible primary studies, which greatly impair the reliability of pooled results (32) although they used GRADE method to grade the level of evidence. As a gold method of pooling the currently best evidence, meta-analysis has been deeply developed and extensively used in route practice. However, it is critically important to capture all potentially eligible studies without restriction of language and publication status to generate robust and reliable pooled estimates (32). Unfortunately, another two meta-analyses (25, 30) were subsequently excluded because of Ashat and Mohan imposed language restriction in their meta-analyses. Although meta-analysis by Deliwala (28) missed one eligible primary study, TSA method was applied to examine the robustness of pooled results. We therefore selected it as candidate of providing currently best evidence. Meanwhile, meta-analysis by Zhang (31) included the most number of eligible primary studies and did not have significant drawbacks in terms of methodology, and thus it was also selected to provide currently best evidence for decision-making.
As one of the most common quality metrics of colonoscopy (6), PDR and ADR were reported to have significant improvement in AIC group compared to CC group in our selected two high-quality meta-analyses (28, 31). Meanwhile, Deliwala and colleagues applied TSA method to further confirm the reliability and robustness of pooled results of PDR and ADR (28). It must be noted that missed colorectal lesions especially sessile polyp and adenoma have been found to be associated with increased risk of interval CRC (7, 24), and thus it is critically important to greatly increase the detection of possible colorectal lesions during colonoscopy. Actually, several methods have been used to increase ADR in clinical practice, and the relative efficacy of these methods have also been comprehensively investigated in two published network meta-analyses currently (39, 40). However, in the present systematic review of discordant meta-analyses, we aimed to further determine the performance of AIC in the improvement of ADR compared with CC. Among selected meta-analyses, average numbers of polyps and adenomas detected per colonoscopy were all significantly increased in the AIC group related to the CC group. Blind spot of the colon is also important factor of reducing detection of colorectal lesions (8, 9). Automatic real-time polyp and adenoma detection system based on deep CNN has been found to have ability of offsetting powerless of the human eye for blind spot (15, 17). Selected meta-analyses also suggested that AIC increase the detection of polyps and adenoma in the transverse colon. Moreover, the size, shape and pathology of polyp and adenoma were all proved to be closely related to the canceration (29, 30). However, selected meta-analyses did not report consistent results for these outcomes (28, 31). So, these two meta-analyses concluded that AIC could significantly increase the PDR and ADR, especially for diminutive polyps and adenomas, however recognition for shape and pathology through AIC should be further improved.
Our current study has some strength: First, it is the first systematic review to differentiate multiple discordant meta-analyses investigating the comparative efficacy and safety of AIC vs. CC for the detection of polyp and adenoma. Second, our systematic review selects the currently best high-quality evidence from multiple discordant meta-analyses for determining a preferred colonoscopy modality during CRC screening. Our study is a systematic review of discordant meta-analyses, which is utilized to appraise the methodological quality and quality of reporting of meta-analyses. It is distinct from traditional systematic reviews that analyze primary studies. In this systematic review of discordant meta-analyses, we selected the best one from the currently published meta-analyses and provided high-level evidence to decision makers. Therefore, the results of our present study are more strength than that from previous meta-analyses or RCTs.
Certainly, we must acknowledge that our systematic review has also several limitations. First and foremost, prospective registration of protocol is the prerequisite of objectively and transparently conducting a systematic review and subsequently reporting results. However, we did not register our protocol on a public platform, which may negatively affect the confidence of our findings. Second, although we selected a meta-analysis incorporating TSA method to provide currently best evidence, all meta-analyses incorporating GRADE method was excluded, and thus we could not determine the confidence for each outcome. Third, we selected two meta-analyses to provide the best available evidence for decision makers finally, however one with the maximum number of primary studies did not register corresponding protocol on public platform (PROSPERO, an international prospective register of systematic reviews). As mentioned previously, prospective registration could guarantee the objectivity and transparent transparency of a meta-analysis, and thus results from that one without registration should be cautiously interpreted.
Conclusions
In conclusion, this systematic review of discordant meta-analyses concludes that AI-based polyp detection systems during colonoscopy increase the detection of polyps and adenomas, especially for diminutive and small, non-advanced polyps and adenomas. However, the potential value of AIC in recognizing the shape and pathology of colorectal lesions should be further improved.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
Author Contributions
HP and MC conceived the study and revised the initial manuscript. QL, YJ, and YL captured and selected citations. XZ and YY designed the data extraction table and extracted data. HP, MC, QL, YJ, and YL performed statistical analyses and prepared for the manuscript draft. All authors approved the final version of the manuscript.
Funding
This study was support by Shanghai Youth Medical Talents-Specialist Program (2019[72]), Zhongshan Hospital Youth Medical Talents (2019ZSYQ11) and National Key Research and Development Plan of China (No.2017YFC1308802).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We would wish to express our sincere appreciation to authors who performed all eligible studies included in the present study.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmed.2021.775604/full#supplementary-material
References
1. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. (2021) 71:209–49. doi: 10.3322/caac.21660
2. Bibbins-Domingo K, Grossman DC, Curry SJ, Davidson KW, Epling Jr JW, García FAR, et al. Screening for colorectal cancer: US preventive services task force recommendation statement. JAMA. (2016) 315:2564–75. doi: 10.1001/jama.2016.5989
3. Rex DK, Boland CR, Dominitz JA, Giardiello FM, Johnson DA, Kaltenbach T, et al. Colorectal cancer screening: recommendations for physicians and patients from the U.S. Multi-society task force on colorectal cancer. Am J Gastroenterol. (2017) 112:1016–30. doi: 10.1038/ajg.2017.174
4. Doubeni CA, Corley DA, Quinn VP, Jensen CD, Zauber AG, Goodman M, et al. Effectiveness of screening colonoscopy in reducing the risk of death from right and left colon cancer: a large community-based study. Gut. (2018) 67:291–8. doi: 10.1136/gutjnl-2016-312712
5. Zhao S, Wang S, Pan P, Xia T, Chang X, Yang X, et al. Risk factors, and factors associated with adenoma miss rate of tandem colonoscopy: a systematic review and meta-analysis. Gastroenterology. (2019) 156:1661–74.e11. doi: 10.1053/j.gastro.2019.01.260
6. Corley DA, Jensen CD, Marks AR, Zhao WK, Lee JK, Doubeni CA, et al. Adenoma detection rate and risk of colorectal cancer and death. N Engl J Med. (2014) 370:1298–306. doi: 10.1056/NEJMoa1309086
7. Cheung D, Evison F, Patel P, Trudgill N. Factors associated with colorectal cancer occurrence after colonoscopy that did not diagnose colorectal cancer. Gastroint Endoscopy. (2016) 84:287–95.e1. doi: 10.1016/j.gie.2016.01.047
8. Mahmood F, Durr NJ. Deep learning and conditional random fields-based depth estimation and topographical reconstruction from conventional endoscopy. Med Image Anal. (2018) 48:230–43. doi: 10.1016/j.media.2018.06.005
9. Wang P, Liu X, Berzin TM, Glissen Brown JR, Liu P, Zhou C, et al. Effect of a deep-learning computer-aided detection system on adenoma detection during colonoscopy (CADe-DB trial): a double-blind randomised study. Lancet Gastroenterol Hepatol. (2020) 5:343–51. doi: 10.1016/S2468-1253(19)30411-X
10. Guo X, Zhang N, Guo J, Zhang H, Hao Y, Hang J. Automated polyp segmentation for colonoscopy images: a method based on convolutional neural networks and ensemble learning. Med Phys. (2019) 46:5666–76. doi: 10.1002/mp.13865
11. Kataoka S, Kudo SE, Misawa M, Nakamura H, Takeda K, Toyoshima N, et al. Endocytoscopy with NBI has the potential to correctly diagnose diminutive colorectal polyps that are difficult to diagnose using conventional NBI. Endoscopy Int Open. (2020) 8:E360–e7. doi: 10.1055/a-1068-9228
12. Aslanian HR, Shieh FK, Chan FW, Ciarleglio MM, Deng Y, Rogart JN, et al. Nurse observation during colonoscopy increases polyp detection: a randomized prospective study. Am J Gastroenterol. (2013) 108:166–72. doi: 10.1038/ajg.2012.237
13. Byrne MF, Shahidi N, Rex DK. Will computer-aided detection and diagnosis revolutionize colonoscopy? Gastroenterology. (2017) 153:1460–64.e1. doi: 10.1053/j.gastro.2017.10.026
14. Kader R, Brandao P, Ahmad OF, Hussein M, Islam S, De Carvalho T, et al. Computer aided characterisation of colorectal polyps using artificial intelligence. Unit Eur Gastroenterol J. (2020) 8:772. doi: 10.1055/s-0041-1724968
15. Misawa M, Kudo SE, Mori Y, Hotta K, Ohtsuka K, Matsuda T, et al. Development of a computer-aided detection system for colonoscopy and a publicly accessible large colonoscopy video database (with video). Gastroint Endoscopy. (2021) 93:960–967.e3. doi: 10.1016/j.gie.2020.07.060
16. Xue P, Tang C, Li Q, Li Y, Shen Y, Zhao Y, et al. Development and validation of an artificial intelligence system for grading colposcopic impressions and guiding biopsies. BMC Med. (2020) 18:406. doi: 10.1186/s12916-020-01860-y
17. Tajbakhsh N, Gurudu SR, Liang J. Automated polyp detection in colonoscopy videos using shape and context information. IEEE Trans Med Imag. (2016) 35:630–44. doi: 10.1109/TMI.2015.2487997
18. Misawa M, Kudo SE, Mori Y, Cho T, Kataoka S, Yamauchi A, et al. Artificial intelligence-assisted polyp detection for colonoscopy: initial experience. Gastroenterology. (2018) 154:2027–29.e3. doi: 10.1053/j.gastro.2018.04.003
19. Gong D, Wu L, Zhang J, Mu G, Shen L, Liu J, et al. Detection of colorectal adenomas with a real-time computer-aided system (ENDOANGEL): a randomised controlled study. Lancet Gastroenterol Hepatol. (2020) 5:352–61. doi: 10.1016/S2468-1253(19)30413-3
20. Luo Y, Zhang Y, Liu M, Lai Y, Liu P, Wang Z, et al. Artificial intelligence-assisted colonoscopy for detection of colon polyps: a prospective, randomized cohort study. J Gastroint Surgery. (2020) 8:2011–18. doi: 10.1007/s11605-020-04802-4
21. Wang P, Liu P, Glissen Brown JR, Berzin TM, Zhou G, Lei S, et al. Lower adenoma miss rate of computer-aided detection-assisted colonoscopy vs routine white-light colonoscopy in a prospective tandem study. Gastroenterology. (2020) 159:1252–61.e5. doi: 10.1053/j.gastro.2020.06.023
22. Liu WN, Zhang YY, Bian XQ, Wang LJ, Yang Q, Zhang XD, et al. Study on detection rate of polyps and adenomas in artificial-intelligence-aided colonoscopy. Saudi J Gastroenterol. (2020) 26:13–9. doi: 10.4103/sjg.SJG_377_19
23. Repici A, Badalamenti M, Maselli R, Correale L, Radaelli F, Rondonotti E, et al. Efficacy of real-time computer-aided detection of colorectal neoplasia in a randomized trial. Gastroenterology. (2020) 159:512–20.e7. doi: 10.1053/j.gastro.2020.04.062
24. Su JR, Li Z, Shao XJ, Ji CR, Ji R, Zhou RC, et al. Impact of a real-time automatic quality control system on colorectal polyp and adenoma detection: a prospective randomized controlled study (with videos). Gastroint Endoscopy. (2020) 91:415–24.e4. doi: 10.1016/j.gie.2019.08.026
25. Ashat M, Klair JS, Singh D, Murali AR, Krishnamoorthi R. Impact of real-time use of artificial intelligence in improving adenoma detection during colonoscopy: a systematic review and meta-analysis. Endoscopy Int Open. (2021) 9:E513–21. doi: 10.1055/a-1341-0457
26. Aziz M, Fatima R, Dong C, Lee-Smith W, Nawras A. The impact of deep convolutional neural network-based artificial intelligence on colonoscopy outcomes: a systematic review with meta-analysis. J Gastroenterol Hepatol. (2020) 35:1676–83. doi: 10.1111/jgh.15070
27. Barua I, Vinsard DG, Jodal HC, Løberg M, Kalager M, Holme Ø, et al. Artificial intelligence for polyp detection during colonoscopy: a systematic review and meta-analysis. Endoscopy. (2021) 53:277–84. doi: 10.1055/a-1201-7165
28. Deliwala SS, Hamid K, Barbarawi M, Lakshman H, Zayed Y, Kandel P, et al. Artificial intelligence (AI) real-time detection vs. routine colonoscopy for colorectal neoplasia: a meta-analysis and trial sequential analysis. Int J Color Dis. (2021) 11:2291–2303. doi: 10.1007/s00384-021-03929-3
29. Hassan C, Spadaccini M, Iannone A, Maselli R, Jovani M, Chandrasekar VT, et al. Performance of artificial intelligence in colonoscopy for adenoma and polyp detection: a systematic review and meta-analysis. Gastroint Endoscopy. (2021) 93:77–85.e6. doi: 10.1016/j.gie.2020.06.059
30. Mohan BP, Facciorusso A, Khan SR, Chandan S, Kassab LL, Gkolfakis P, et al. Real-time computer aided colonoscopy versus standard colonoscopy for improving adenoma detection rate: a meta-analysis of randomized-controlled trials. EClin Med. (2020) 30:100622. doi: 10.1016/j.eclinm.2020.100622
31. Zhang Y, Zhang X, Wu Q, Gu C, Wang Z. Artificial intelligence-aided colonoscopy for polyp detection: a systematic review and meta-analysis of randomized clinical trials. J Laparoend Adv Surg Techn Part A. (2021) 10:1143–49. doi: 10.1089/lap.2020.0777
32. Higgins JPT, Thomas J, Chandler J, Cumpston M, Li T, Page MJ. Cochrane Handbook for Systematic Reviews of Interventions version 6.2. (2021). Available online at: www.training.cochrane.org/handbook (accessed February 2021).
33. Page MJ, McKenzie JE, Bossuyt PM, Boutron I, Hoffmann TC, Mulrow CD, et al. The PRISMA 2020 statement: an updated guideline for reporting systematic reviews. BMJ. (2021) 372:n71. doi: 10.1136/bmj.n71
34. Page MJ, Moher D, Bossuyt PM, Boutron I, Hoffmann TC, Mulrow CD, et al. PRISMA 2020 explanation and elaboration: updated guidance and exemplars for reporting systematic reviews. BMJ. (2021) 372:n160. doi: 10.1136/bmj.n160
35. Shea BJ, Grimshaw JM, Wells GA, Boers M, Andersson N, Hamel C, et al. Development of AMSTAR: a measurement tool to assess the methodological quality of systematic reviews. BMC Med Res Methodol. (2007) 7:10. doi: 10.1186/1471-2288-7-10
36. Wright JG, Swiontkowski MF, Heckman JD. Introducing levels of evidence to the journal. The Journal of bone and joint surgery. Am Vol. (2003) 85:1–3. doi: 10.2106/00004623-200301000-00001
37. Jadad AR, Cook DJ, Browman GP. A guide to interpreting discordant systematic reviews. CMAJ. (1997) 156:1411–6.
38. Wang P, Berzin TM, Glissen Brown JR, Bharadwaj S, Becq A, Xiao X, et al. Real-time automatic detection system increases colonoscopic polyp and adenoma detection rates: a prospective randomised controlled study. Gut. (2019) 68:1813–9. doi: 10.1136/gutjnl-2018-317500
39. Facciorusso A, Del Prete V, Buccino RV, Della Valle N, Nacchiero MC, Monica F, et al. Comparative efficacy of colonoscope distal attachment devices in increasing rates of adenoma detection: a network meta-analysis. Clin Gastroenterol Hepatol. (2018) 16:1209–19.e9. doi: 10.1016/j.cgh.2017.11.007
Keywords: colonoscopy, artificial intelligence, polyp detection, adenoma detection, discordant meta-analysis, Jadad algorithm
Citation: Pan H, Cai M, Liao Q, Jiang Y, Liu Y, Zhuang X and Yu Y (2022) Artificial Intelligence-Aid Colonoscopy Vs. Conventional Colonoscopy for Polyp and Adenoma Detection: A Systematic Review of 7 Discordant Meta-Analyses. Front. Med. 8:775604. doi: 10.3389/fmed.2021.775604
Received: 14 September 2021; Accepted: 20 December 2021;
Published: 13 January 2022.
Edited by:
William Karnes, University of California, Irvine, United StatesReviewed by:
Antonio Facciorusso, University of Foggia, ItalyIoannis Papanikolaou, National and Kapodistrian University of Athens, Greece
Copyright © 2022 Pan, Cai, Liao, Jiang, Liu, Zhuang and Yu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hui Pan, Jennyp730@163.com; Mingyan Cai, Cai.mingyan@qq.com
†These authors have contributed equally to this work and share first authorship
 Hui Pan1*†
Hui Pan1*†  Mingyan Cai
Mingyan Cai