Upregulation of FGD6 Predicts Poor Prognosis in Gastric Cancer
- 1The Affiliated Hospital of Kunming University of Science and Technology, The First People's Hospital of Yunnan Province, Kunming, China
- 2The First Affiliated Hospital of Bengbu Medical College, Bengbu, China
- 3Medical College, Qingdao University, Qingdao, China
- 4Second Department of General Surgery, The First People's Hospital of Yunnan Province, The Affiliated Hospital of Kunming University of Science and Technology, Kunming, China
Background: The aim of this study was to investigate the prognostic significance of faciogenital dysplasia 6 (FGD6) in gastric cancer (GC).
Methods: The data of GC patients from The Cancer Genome Atlas (TCGA) database were used for the primary study. Then, our data were validated by the GEO database and RuiJin cohort. The relationship between the FGD6 level and various clinicopathological features was analyzed by logistic regression and univariate Cox regression. Multivariate Cox regression analysis was used to evaluate whether FGD6 was an independent prognostic factor for survival of patients with GC. The relationship between FGD6 and overall survival time was explored by the Kaplan–Meier method. In addition, gene set enrichment analysis (GSEA) was performed to investigate the possible biological processes of FGD6.
Results: The FGD6 level was significantly overexpressed in GC tissues, compared with adjacent normal tissues. The high expression of FGD6 was related to a high histological grade, stage, and T classification and poor prognosis of GC. Multivariate Cox regression analysis showed that FGD6 was an independent prognostic factor for survival of patients with GC. GSEA identified that the high expression of FGD6 was mainly enriched in regulation of actin cytoskeleton.
Conclusion: FGD6 may be a prognostic biomarker for predicting the outcome of patients with GC.
Introduction
Gastric cancer (GC) is the fifth incidence cancer, and it has become the fourth leading cause of cancer deaths in the world, which led to nearly 769,000 deaths in 2020 (1). The treatment strategies for GC include surgery, chemotherapy, radiotherapy, and molecular targeted therapy. Nevertheless, the prognosis remains poor, and the age-standardized 5-year net survival rate was only 20–40% (2), partially due to the majority of GC cases lacking typical symptoms and being diagnosed at an advanced stage with an experience of postsurgical disease relapse or metastasis (3). To this end, new methods of diagnosis or novel molecular biomarkers needed to be discovered for prognosis and reliable therapeutic targets of GC patients.
The modular architectures of faciogenital dysplasia (FGD) family proteins, encompassing FYVE (domain present in Fab1p, YOTB, Vac1p, and EEA1) domains, Dbl homology (DH), and two plectron homology (PH) domains, recognize some phospholipids and proteins such as phosphoinositide (4, 5), phosphatidylinositol 3-phosphate (6), and GTPases, to promote cellular development (7), whose act serves as Rho guanine nucleotide exchange factors (GEFs) (8). There are seven members of the FGD family, namely, FGD1, FGD2, FGD3, FGD4, FGD5, FGD6, and FRG (9). For example, several researches have revealed that FGD family proteins regulated culler functions by specifically activating Cdc42 (10–12).
FGD6, also known as ZFYVE24, is localized on the Homo sapiens chromosome 12q22 and consists of 1,400 amino acids. Previous studies have reported that variations in FGD6 increase individual susceptibility to polypoidal choroidal vasculopathy (13). Furthermore, it was also demonstrated that FGD6 coordinates cell polarity and endosomal membrane recycling in osteoclasts by regulating the assembly of different actin-based protein networks and activating Cdc42 at different locations (14). However, to date, little is investigated about the relationship between the expression level of FGD6 and the prognosis of GC.
In the present study, by using bioinformatics analysis in TCGA database, we identified that FGD6 was increased in GC tissues compared with adjacent non-cancerous tissues, which was also verified in paired GC tissues by using GSE63089. Herein, we found that GC patients with higher FGD6 in the tumor tissues had shorter overall survival and poorer clinical phenotypes. In addition, the gene set enrichment analysis (GSEA) was used to investigate the signaling pathways of FGD6 in patients with GC. All data presented that FGD6 may serve as a promising prognostic marker for GC.
Materials and Methods
The Extraction Process of the GC Patients' Data
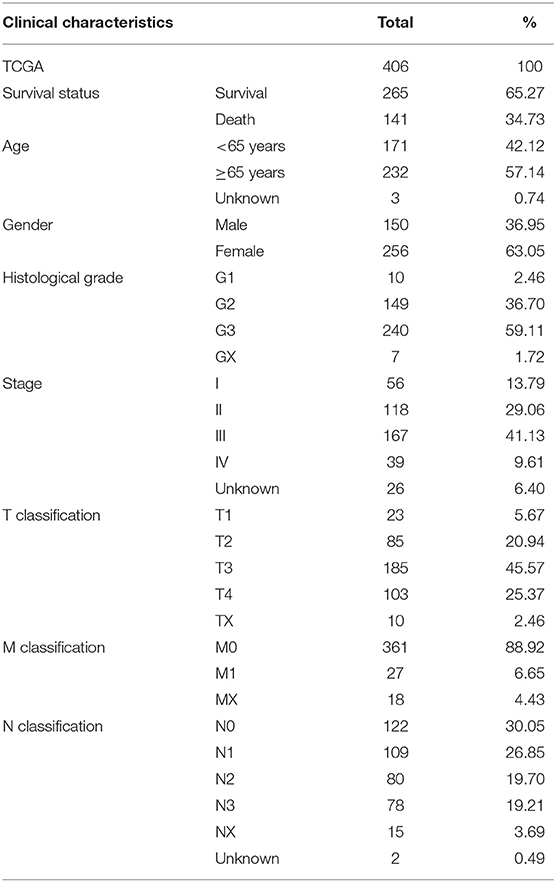
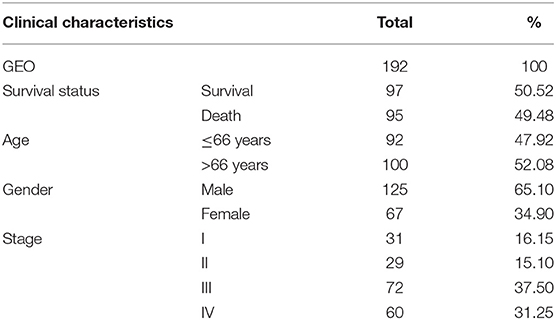
The RNA sequencing data with 373 cases, which include 343 GC patients and 30 adjacent normal tissues, and the clinical information with 406 GC patients were downloaded from the TCGA Genomic Data Commons data portal (https://portal.gdc.cancer.gov/repository). The different expressions of FGD6 were analyzed by the R package limma function, and its prognosis-related values were analyzed by survival analysis. The expression data of GSE63089 with no clinical data in the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo/) were analyzed to validate the FGD6 expression level by the R package limma function. The details included survival status, age, gender, histological grade, stage, T classification, N classification, and M classification (Table 1). The GSE15459 dataset was used to analyze the clinicopathological factors and the relationship between FGD6 and overall survival time. The clinicopathological data included sex, age, stage, and survival status (Table 2). Patients who were followed up for <30 days and unknown or incomplete clinical information were excluded from TCGA database.
Clinical Specimens
Twenty cases of GC tissues and normal tissues were collected from RuiJin Hospital, Shanghai Jiaotong University, and have been approved by the Ethics Committee of RuiJin Hospital, Shanghai Jiaotong University. Tissues were preserved in a −80°C refrigerator after surgical excision. All patients had given written informed consent before the study. None of the patients had received preoperative radiotherapy or chemotherapy before surgical resection.
Quantitative Real-Time Polymerase Chain Reaction Analysis
The total RNA was extracted from GC tissues and normal tissues by a TRIzol reagent (Invitrogen, USA), following the manufacturer's instructions. Reverse transcription was performed to obtain cDNA by 5*fastking, and qRT-PCR was performed with the SYBR Green Master Mix (Thermo Fisher Scientific, USA). Sequences were as follows: for FGD6, forward 5′-CAGCCTGGTCGGGTTTTTCT-3′ and reverse 5′-CAGCCAGTGAGAGCATGTTG-3′; for GAPDH, forward 5′-GACTCATGACCACAGTCCATGC-3′ and reverse 5′-AGAGGCAGGGATGATGTTCTG-3′. The relative mRNA expression level was calculated by the 2−ΔCT method.
GSEA
GSEA 4.1.0 was used to detect the potential mechanism of FGD6 and showed statistically significant differential expression between high and low expression groups based on the median expression of FGD6. GESA was conducted according to the default weighted enrichment statistics for 1,000 times. Gene sets with normalized (NOM) p-value < 0.05 and false discovery rate (FDR) < 0.25 were considered significantly enriched.
Statistical Analysis
All statistical analyses in our study were carried out using R software (V.3.5.3) and IBM SPSS statistical software (version 22.0). The differences between groups were analyzed using the Wilcoxon test and logistic regression. Clinicopathological characteristics related to overall survival in GC patients were verified using Cox regression and the Kaplan–Meier method. Multivariate Cox analysis was used to evaluate the relationship between the clinicopathological features and FGD6 expression. The cutoff value for FGD6 expression was determined based on the median values. The scatter plot of qRT-PCR results in this study was performed using GraphPad Prism 8 software. A p-value < 0.05 was considered statistically significant.
Results
Overexpression of FGD6 in GC Tissues
Results of the data from TCGA database, as shown in Figure 1A, revealed that the mRNA expression of FGD6 was significantly upregulated in GC tissues compared with adjacent normal stomach tissues (p = 7.249e−8). In addition, the mRNA expression of FGD6 was compared in 30 pairs of tumor and adjacent normal tissues, revealing its expression enhanced in tumor tissues compared with the adjacent normal tissues (p = 0.008) (Figure 1B). Similarly, the result of FGD6 mRNA expression also showed the same phenomenon by using the GSE63089 dataset (p = 9.436e−10) (Figure 1C). Consistently, qRT-PCR analysis showed that FGD6 expression at the mRNA level was higher in GC tissues relative to that in corresponding non-cancerous tissues (P = 0.01) (Figure 1D).
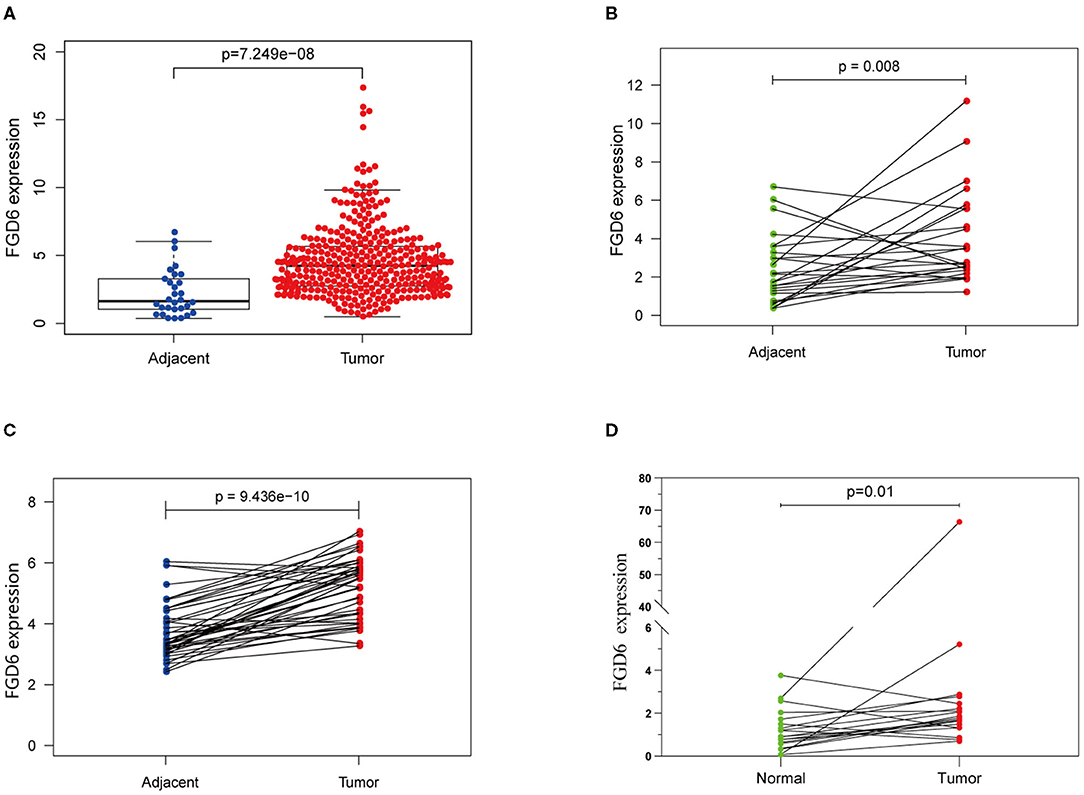
Figure 1. The expression of FGD6 in GC tissues and adjacent normal tissues. (A) FGD6 expression is upregulated in TCGA cohort; (B) FGD6 is overexpressed in GC tissue compared to para-cancerous tissues in TCGA cohort; (C) FGD6 expression is elevated in GC tissues in the GSE63089 dataset; (D) FGD6 expression was significantly upregulated in GC compared to normal tissues by qRT-PCR.
Correlation Between Expression of FGD6 and Clinical Features of GC Patients
As shown in Figure 2, the clinical parameters of GC patients from TCGA were analyzed, showing that the mRNA expression of FGD6 was related to histological grade (p = 0.013), stage (p = 0.009), and T classification (p = 0.002). Logistic regression analysis revealed that overexpression of FGD6 in GC was significantly correlated with high histological grade (OR = 8.385 for G3 vs. G1, p = 0.049), stage (OR = 2.730 for II vs. I, p = 0.006; OR = 2.667 for III vs. I, p = 0.006; OR = 2.625 for IV vs. I, p = 0.037), and high T classification (OR = 4.293 for T2 vs. T1, p = 0.030; OR = 5.684 for T3 vs. T1, p = 0.007; OR = 7.619 for T4 vs. T1, p = 0.002) (Table 3). These findings indicated that GC patients with high FGD6 expression have more chances to develop an advanced GC than GC patients with low FGD6 expression.
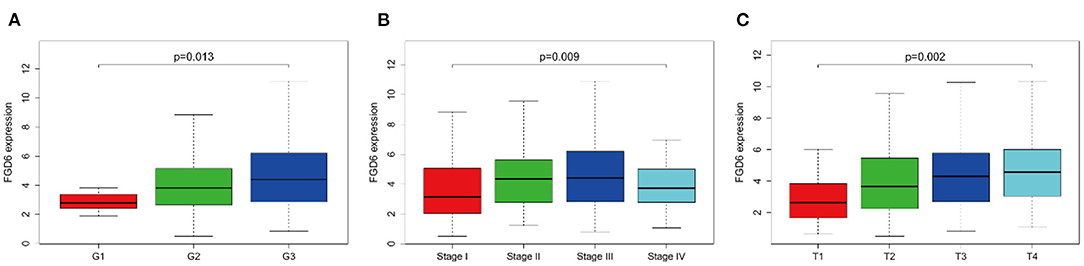
Figure 2. The link between FGD6 mRNA expression and clinicopathologic features in GC patients. (A) Histological grade, (B) Clinical stage, and (C) T classification.
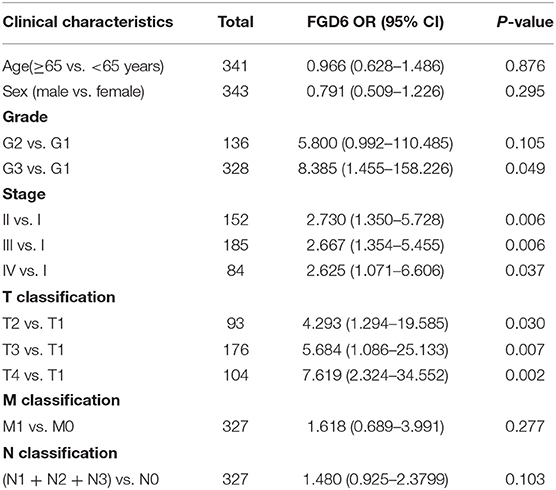
Table 3. FGD6 expression associated with clinical pathological characteristics (logistic regression).
High Expression of FGD6 Predicts Poor Overall Survival in GC
To evaluate the diagnostic value of FGD6, univariate and multivariate Cox regressions were used to analyze the impact of FGD6 expression and other clinicopathological factors on survival (Table 4). In univariate analysis, age (HR = 1.026, p = 0.009), stage (HR = 1.483, p = 0.001), N classification (HR = 1.244, p = 0.018), and FGD6 (HR = 1.440, p = 0.022) were related to overall survival in patients with GC. In multivariate analysis, FGD6 expression (HR = 1.406, p = 0.036) was significantly associated with poor overall survival, similar to age (HR = 1.034, p = 0.002) and stage (HR = 1.634, p = 0.013) (Figure 3). Furthermore, univariate Cox regression analysis showed that stage (HR = 2.789, p = 3.150e−14) and FGD6 expression (HR = 1.567, p = 0.001) were associated with poorer survival in the GSE15459 dataset. Multivariate Cox regression analysis of the above factors found that FGD6 expression (HR = 1.374, p = 0.027) and stage (HR = 2.764, p = 1.500e−13) were independent prognostic factors of GC in the GSE15459 dataset (Table 5). Kaplan–Meier survival curves showed that patients with high expression of FGD6 exhibited poorer survival time compared with those with low expression of FGD6 (Figure 4A, p = 0.044). In the same way, this result was further validated in the GSE15459 dataset (Figure 4B, p = 0.011).
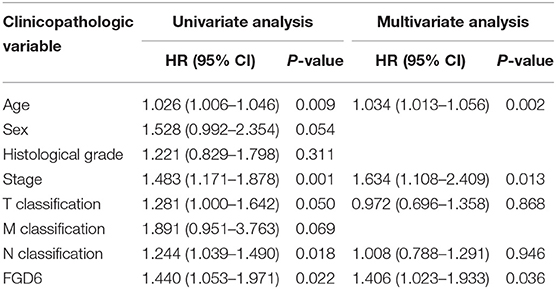
Table 4. Univariate and multivariate Cox regression analyses of the correlation of prognostic factors with overall survival among GC patients by TCGA database.
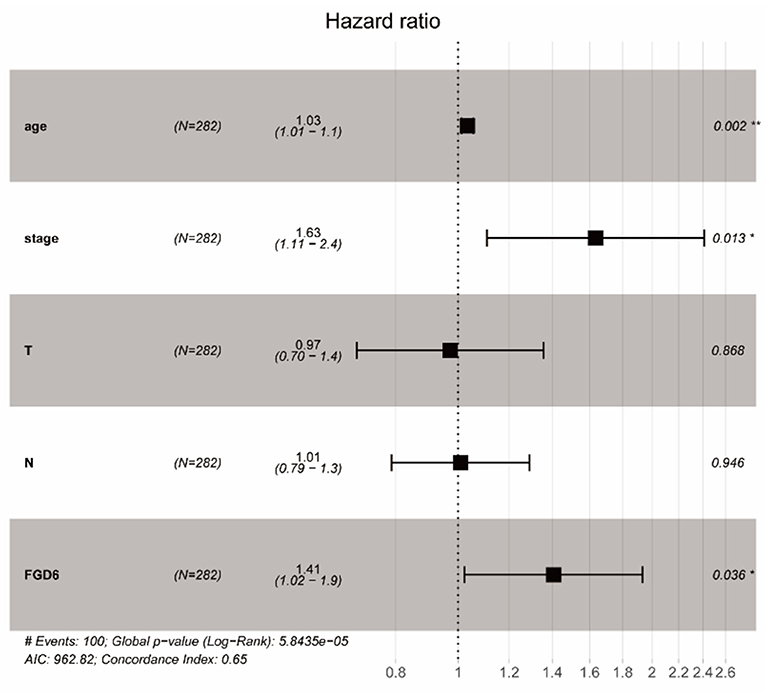
Figure 3. Forest plot of the correlation between FGD6 expression and GC patient overall survival. *P < 0.05, **P < 0.01.
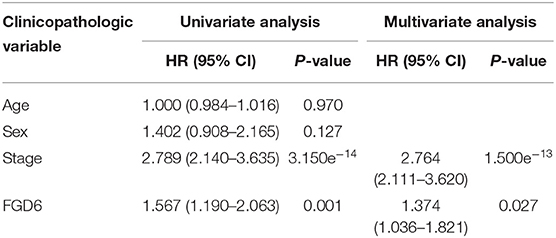
Table 5. Univariate and multivariate Cox regression analyses of the correlation of prognostic factors with overall survival among GC patients by the GSE15459 dataset.
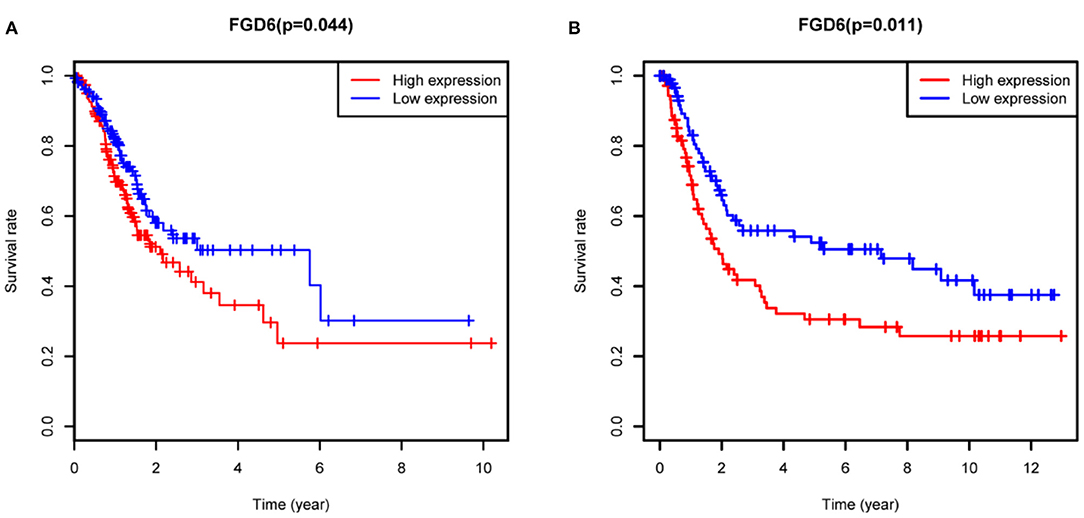
Figure 4. Kaplan-Meier analysis for overall survival of GC patients with FGD6 expression. (A) TCGA cohort. (B) GSE15459 dataset.
Signaling Pathways Associated With FGD6 Expression
In order to investigate the potential signaling pathways and promising biological function of FGD6 in GC, we explored high and low FGD6 expression datasets though GSEA based on the median expression of FGD6. As shown in Figure 5, GSEA mainly regulated the JAK/STAT signaling pathway, regulation of actin cytoskeleton, Toll-like receptor signaling pathway, focal adhesion, apoptosis, NOD-like receptor signaling pathway, natural killer cell-mediated cytotoxicity, T-cell receptor signaling pathway, and chemokine signaling pathway, which were obviously enriched in the FGD6 high-expression phenotype.
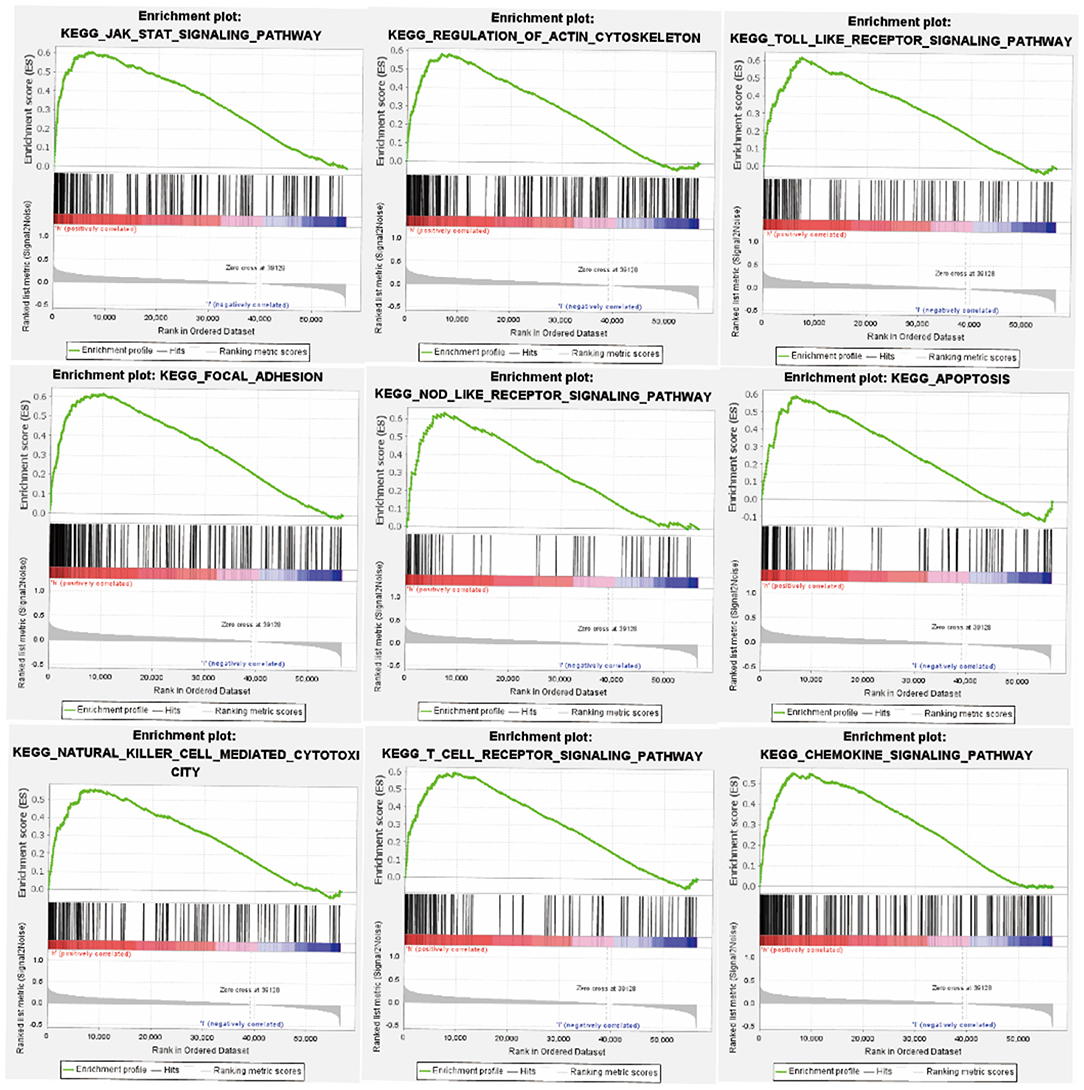
Figure 5. GSEA enrichment plots. The GSEA results suggested that the significantly enriched signaling pathways were the JAK/STAT signaling pathway, regulation of actin cytoskeleton, Toll-like receptor signaling pathway, focal adhesion, apoptosis, NOD-like receptor signaling pathway, natural killer cell-mediated cytotoxicity, T-cell receptor signaling pathway, and chemokine signaling pathway.
Discussion
Prognostic biomarkers play an important role in the prognosis of GC, and there has been a lot of literature on the biomarkers of GC (15). However, the current conventional prediction models for GC rely on clinical prognostic factors such as age, TNM stage, and pathological grade. Prognostic assessment of patients with GC remains a challenging issue. A non-invasive or less-invasive test with tumor-specific biomarkers to facilitate diagnosis, recurrence assessment, and personalized chemotherapy for GC patients is urgently required.
GEFs are associated with numerous cellular responses such as proliferation, differentiation, and motility though activating Rho-family GTPases (16). Rho-family GTPases are activated by GEFs, which leads to aberrant signaling from growth factor receptors such as transmembrane receptor tyrosine kinases and G protein-coupled receptors in cancer (17). The FGD family contains the DH and PH1 domains that can reconcile activation of Rho GTPases by acting as GEFs (7). Rho GTPases function as binary molecular switches converted between an active, GTP-bound state and an inactive, GDP-bound state, where the transformation of GDP to GTP is mediated by GEFs (16), and it also was involved in cancer cell invasion and metastasis (18). CDC42 belongs to a member of the Rho family of small GTPases that have been involved in the regulation of multiple signaling pathways (19), and it plays a pivotal role in epithelial-to-mesenchymal transition, cell-cycle progression, migration, invasion, tumor growth, angiogenesis, and oncogenic transformation (20). Researches have demonstrated that FGD6 can activate CDC42 through its PH domains, binding phosphatidylinositol in the vicinity of ligand-bound integrins (14). It was reported that knockdown of CDC42 inhibited the proliferation, migration, and invasion of GC cells (21). Moreover, studies have found that FARP1 binds to integrin αvβ5 and promotes GC cell migration and invasion through the activation of CDC42 (22). Recently, a study has shown that aberrant expression of CDC42 in colorectal cancer can cause the migration, invasion, and metastasis through activation of the VEGF/NRP1 axis and is closely related to the prognosis of the patients (23).
In this study, bioinformatic analysis was performed in RNA-sequencing data of GC, and established the prognostic relevance of FGD6 mRNA expression level to GC patient clinical outcomes based on TCGA database. We identified and verified that FGD6 was highly expressed in GC tissues relative to para-cancerous tissues, as well as in the GSE63089 dataset of GEO database; the FGD6 expression level was significantly overexpressed in GC tissues as compared to the normal gastric tissues, as verified by qRT-PCR, and its expression level was correlated with clinicopathological parameters and GC patient survival. In addition, GSEA suggested that the main functions of high FGD6 expression were enriched in the JAK/STAT signaling pathway, Regulation of actin cytoskeleton, Toll-like receptor signaling pathway, focal adhesion, apoptosis, NOD-like receptor signaling pathway, natural killer cell-mediated cytotoxicity, T-cell receptor signaling pathway, and chemokine signaling pathway.
However, the regulatory mechanism of FGD6 needs to be further illustrated. The actin cytoskeleton assembles with microtubules and intermediate filaments to form branching networks or bundles that polarize cells, support intercellular junctions, and promote cell adhesion and migration (24). For example, it was reported that neuropilin and tolloid-like 1 modulate the expression of cytoskeletal enhanced migration and invasive potential of epithelial ovarian cancer (25). In parallel, actin cytoskeleton reorganization was also associated with cell migration and motility in human breast cancer (18). Earlier reports had identified that Rho GTPases promote cell adhesion and migration via reassembling actin cytoskeletons (26–29). Besides, we speculated that FGD6 may provide us with a new direction in invasion and migration of GC.
Whether other genetic mutations cause uncontrolled proliferation of GC was considered, and FGD6 is coincidentally increased. TCGA mutation data were analyzed by the Perl language. Then, the top 30 mutated genes were selected in the GC samples, and the correlation between FGD6 and these genes was analyzed by using the correlation analysis function of the GEPIA2 (http://gepia2.cancer-pku.cn/). The results are shown in Supplementary Data Sheet 1. In future analyses, the specific molecular mechanism of FGD6 regulation of GC is still unclear, which requires some experiments to be conducted in vitro and in vivo. In summary, our study revealed that the overexpression of FGD6 was a prognostic biomarker and a potentially viable therapeutic agent of GC patients.
Data Availability Statement
Publicly available datasets were analyzed in this study. This data can be found here: The Cancer Genome Atlas (https://portal.gdc.cancer.gov/). NCBI Gene Expression Omnibus (GSE63089 and GSE15459).
Ethics Statement
The studies involving human participants were reviewed and approved by RuiJin hospital, Shanghai Jiaotong University. The patients/participants provided their written informed consent to participate in this study.
Author Contributions
JZ wrote the manuscript and provided the design idea of this study. ML and HS analyzed the data and supplemented the ideas. JG provided some comments for this paper. All authors read and approved the final manuscript.
Funding
This work was supported by Priority Union Foundation of Yunnan Provincial Science and Technology Department and Kunming Medical University [2019FE001(-175)] and Ten Thousand Person Plan for Famous Doctors of Yunnan Province (YNWR-MY-2018-015).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We sincerely appreciate the RuiJin Hospital for providing samples.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmed.2021.672595/full#supplementary-material
References
1. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. (2021). 71:209–49. doi: 10.3322/caac.21660
2. Allemani C, Matsuda T, Di Carlo V, Harewood R, Matz M, Nikšić M, et al. Global surveillance of trends in cancer survival 2000–14 (CONCORD-3): analysis of individual records for 37 513 025 patients diagnosed with one of 18 cancers from 322 population-based registries in 71 countries. Lancet. (2018) 391:1023–75. doi: 10.1016/S0140-6736(17)33326-3
3. Smyth EC, Nilsson M, Grabsch HI, van Grieken NCT, Lordick F. Gastric cancer. Lancet. (2020) 396:635–48. doi: 10.1016/S0140-6736(20)31288-5
4. Overduin M, Cheever ML, Kutateladze TG. Signaling with phosphoinositides: better than binary. Mol Interv. (2001) 1:150–9.
5. Sankaran VG, Klein DE, Sachdeva MM, Lemmon MA. High-affinity binding of a FYVE domain to phosphatidylinositol 3-phosphate requires intact phospholipid but not FYVE domain oligomerization. Biochemistry. (2001) 40:8581–7. doi: 10.1021/bi010425d
6. Kutateladze T, Overduin M. Structural mechanism of endosome docking by the FYVE domain. Science. (2001) 291:1793–6. doi: 10.1126/science.291.5509.1793
7. Eitzen G, Smithers CC, Murray AG, Overduin M. Structure and function of the Fgd family of divergent FYVE domain proteins (1). Biochem Cell Biol. (2019) 97:257–64. doi: 10.1139/bcb-2018-0185
8. Snyder JT, Worthylake DK, Rossman KL, Betts L, Pruitt WM, Siderovski DP, et al. Structural basis for the selective activation of Rho GTPases by Dbl exchange factors. Nat Struct Biol. (2002) 9:468–75. doi: 10.1038/nsb796
9. Kurogane Y, Miyata M, Kubo Y, Nagamatsu Y, Kundu RK, Uemura A, et al. FGD5 mediates proangiogenic action of vascular endothelial growth factor in human vascular endothelial cells. Arterioscler Thromb Vasc Biol. (2012) 32:988–96. doi: 10.1161/ATVBAHA.111.244004
10. Egorov MV, Capestrano M, Vorontsova OA, Di Pentima A, Egorova AV, Mariggio S, et al. Faciogenital dysplasia protein (FGD1) regulates export of cargo proteins from the golgi complex via Cdc42 activation. Mol Biol Cell. (2009) 20:2413–27. doi: 10.1091/mbc.e08-11-1136
11. Huber C, Martensson A, Bokoch GM, Nemazee D, Gavin AL. FGD2, a CDC42-specific exchange factor expressed by antigen-presenting cells, localizes to early endosomes and active membrane ruffles. J Biol Chem. (2008) 283:34002-12. doi: 10.1074/jbc.M803957200
12. Park S, Guo Y, Negre J, Preto J, Smithers CC, Azad AK, et al. Fgd5 is a Rac1-specific Rho GEF that is selectively inhibited by aurintricarboxylic acid. Small GTPases. (2019) 12:1–14. doi: 10.1080/21541248.2019.1674765
13. Huang L, Zhang H, Cheng CY, Wen F, Tam PO, Zhao P, et al. A missense variant in FGD6 confers increased risk of polypoidal choroidal vasculopathy. Nat Genet. (2016) 48:640–7. doi: 10.1038/ng.3546
14. Steenblock C, Heckel T, Czupalla C, Espirito Santo AI, Niehage C, Sztacho M, et al. The Cdc42 guanine nucleotide exchange factor FGD6 coordinates cell polarity and endosomal membrane recycling in osteoclasts. J Biol Chem. (2014) 289:18347–59. doi: 10.1074/jbc.M113.504894
15. Matsuoka T, Yashiro M. Biomarkers of gastric cancer: current topics and future perspective. World J Gastroenterol. (2018) 24:2818–32. doi: 10.3748/wjg.v24.i26.2818
16. Rossman KL, Der CJ, Sondek J. GEF means go: turning on RHO GTPases with guanine nucleotide-exchange factors. Nat Rev Mol Cell Biol. (2005) 6:167-80. doi: 10.1038/nrm1587
17. Vigil D, Cherfils J, Rossman KL, Der CJ. Ras superfamily GEFs and GAPs: validated and tractable targets for cancer therapy? Nat Rev Cancer. (2010) 10:842–57. doi: 10.1038/nrc2960
18. Zeng Y, Cao Y, Liu L, Zhao J, Zhang T, Xiao L, et al. SEPT9_i1 regulates human breast cancer cell motility through cytoskeletal and RhoA/FAK signaling pathway regulation. Cell Death Dis. (2019) 10:720. doi: 10.1038/s41419-019-1947-9
19. Melendez J, Grogg M, Zheng Y. Signaling role of Cdc42 in regulating mammalian physiology. J Biol Chem. (2011) 286:2375–81. doi: 10.1074/jbc.R110.200329
20. Maldonado MDM, Dharmawardhane S. Targeting Rac and Cdc42 GTPases in Cancer. Cancer Res. (2018) 78:3101-11. doi: 10.1158/0008-5472.CAN-18-0619
21. Du DS, Yang XZ, Wang Q, Dai WJ, Kuai WX, Liu YL, et al. Effects of CDC42 on the proliferation and invasion of gastric cancer cells. Mol Med Rep. (2016) 13:550–4. doi: 10.3892/mmr.2015.4523
22. Hirano T, Shinsato Y, Tanabe K, Higa N, Kamil M, Kawahara K, et al. FARP1 boosts CDC42 activity from integrin alphavbeta5 signaling and correlates with poor prognosis of advanced gastric cancer. Oncogenesis. (2020) 9:13. doi: 10.1038/s41389-020-0190-7
23. Ma LL, Guo LL, Luo Y, Liu GL, Lei Y, Jing FY, et al. Cdc42 subcellular relocation in response to VEGF/NRP1 engagement is associated with the poor prognosis of colorectal cancer. Cell Death Dis. (2020) 11:171. doi: 10.1038/s41419-020-2370-y
24. Smith MA, Hoffman LM, Beckerle MC. LIM proteins in actin cytoskeleton mechanoresponse. Trends Cell Biol. (2014) 24:575–83. doi: 10.1016/j.tcb.2014.04.009
25. Xu Y, Wang W, Chen J, Mao H, Liu Y, Gu S, et al. High neuropilin and tolloid-like 1 expression associated with metastasis and poor survival in epithelial ovarian cancer via regulation of actin cytoskeleton. J Cell Mol Med. (2020) 24:9114–24. doi: 10.1111/jcmm.15547
26. Genda T, Sakamoto M, Ichida T, Asakura H, Kojiro M, Narumiya S, et al. Cell motility mediated by rho and Rho-associated protein kinase plays a critical role in intrahepatic metastasis of human hepatocellular carcinoma. Hepatology. (1999) 30:1027–36. doi: 10.1002/hep.510300420
27. Itoh K, Yoshioka K, Akedo H, Uehata M, Ishizaki T, Narumiya S. An essential part for Rho-associated kinase in the transcellular invasion of tumor cells. Nat Med. (1999) 5:221–5. doi: 10.1038/5587
28. Wei L, Surma M, Shi S, Lambert-Cheatham N, Shi J. Novel insights into the roles of rho kinase in cancer. Arch Immunol Ther Exp. (2016) 64:259–78. doi: 10.1007/s00005-015-0382-6
Keywords: FGD6, gastric cancer, TCGA, prognosis, GSEA
Citation: Zeng J, Li M, Shi H and Guo J (2021) Upregulation of FGD6 Predicts Poor Prognosis in Gastric Cancer. Front. Med. 8:672595. doi: 10.3389/fmed.2021.672595
Received: 26 February 2021; Accepted: 01 June 2021;
Published: 05 July 2021.
Edited by:
Mohammad H. Derakhshan, University of Glasgow, United KingdomReviewed by:
Nazri Mustaffa, Universiti Sains Malaysia (USM), MalaysiaMaya Gulubova, Trakia University, Bulgaria
Copyright © 2021 Zeng, Li, Shi and Guo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jianhui Guo, guojianhuikm@163.com
 Jianmin Zeng
Jianmin Zeng Man Li2
Man Li2  Huasheng Shi
Huasheng Shi