Gut Microbiota Dysbiosis Is a Crucial Player for the Poor Outcomes for COVID-19 in Elderly, Diabetic and Hypertensive Patients
- 1Laboratory of Inflammation, Oswaldo Cruz Institute, Oswaldo Cruz Foundation (Fiocruz), Rio de Janeiro, Brazil
- 2Laboratory on Thymus Research, Oswaldo Cruz Institute, Oswaldo Cruz Foundation (Fiocruz), Rio de Janeiro, Brazil
- 3National Institute of Science and Technology on Neuroimmunomodulation (INCT-NIM), Oswaldo Cruz Institute, Oswaldo Cruz Foundation (Fiocruz), Rio de Janeiro, Brazil
- 4Rio de Janeiro Research Network on Neuroinflammation (RENEURIN), Oswaldo Cruz Institute, Oswaldo Cruz Foundation (Fiocruz), Rio de Janeiro, Brazil
A new infectious disease, named COVID-19, caused by the coronavirus associated to severe acute respiratory syndrome (SARS-CoV-2) has become pandemic in 2020. The three most common pre-existing comorbidities associated with COVID-19-related death are elderly, diabetic, and hypertensive people. A common factor among these risk groups for the outcome of death in patients infected with SARS-CoV-2 is dysbiosis, with an increase in the proportion of bacteria with a pro-inflammatory profile. Due to this dysbiosis, elderly, diabetic, and hypertensive people present a higher propensity to mount an inflammatory environment in the gut with poor immune editing, culminating in a weakness of the intestinal permeability barrier and high bacterial product translocation to the bloodstream. This scenario culminates in a low-grade, persistent, and systemic inflammation. In this context, we propose here that high circulating levels of bacterial products, like lipopolysaccharide (LPS), can potentiate the SARS-CoV-2-induced cytokines, including IL-6, being crucial for development of the cytokine storm in the severe form of the disease. A better understanding on the possible correlation between gut dysbiosis and poor outcomes observed in elderly, diabetic, and hypertensive people can be useful for the development of new therapeutic strategies based on modulation of the gut microbiota.
Introduction
In early December 2019, a new infectious disease, caused by the coronavirus associated to severe acute respiratory syndrome (SARS-CoV-2), emerged in Wuhan, China (1). The disease caused by this infection, COVID-19, spread very rapidly in many other countries reaching pandemic proportions (2, 3). By 24 May 2021, there were 166,814,851 individuals diagnosed with COVID-19, including 3,458,905 fatal cases, as shown in the WHO data center (4). In severe COVID-19 patients, 93% of deaths result from respiratory failure caused by acute respiratory distress syndrome (ARDS). Besides, the storm of cytokines and symptoms of sepsis, with failure of some vital organs, including heart and kidney, derived by the primary viral infection and/or secondary infections were observed in 70% of fatal cases (5). No specific effective therapeutics are so far available for COVID-19 and the management of the disease includes physical distancing, mask wearing, supportive medical care, and vaccines (4). Herein, we propose a role of gut dysbiosis in the worse prognosis of COVID-19 in elderly people and in patients with Diabetes mellitus (DM) or hypertension.
COVID-19 and Gut Microbiota
The human microbiota is made up of microorganisms, including bacteria, fungi, archaea, viruses, and protozoa, that colonize particular locations of the human body such as skin, as well as respiratory and gastrointestinal tracts (6, 7). The gut microbiota refers specifically to a complex bacterial community situated in the gastrointestinal tract (8). Although approximately 40% of patients infected with SARS-CoV-2 showed a high concentration of viral genetic material in the anal swab, and various patients reported nausea, vomiting, and diarrhea (9, 10) little has been so far discussed on the role of the gut in the pathophysiology of COVID-19, especially envisioning microbiota as being responsible for the greatest risk factor to develop the severe form of the disease.
It is well known that the membrane angiotensin I converting enzyme 2 (ACE2) is the pathway of entry into the target cells (11). Human mature enterocytes located in the small intestine express membrane ACE2, and SARS-CoV-2 is able to infect those cells in a process facilitated by TMPRSS2 and TMPRSS4 proteases (12). The infection of enterocytes with SARS-CoV-2 may promote a significant reduction of enteric ACE2 integrity/functionality. The decrease of ACE2 expression leads to an upregulation of other renin-angiotensin system components, including angiotensin (Ang) II (13). Remarkably, increased Ang II levels can modify gut microbial composition and metabolomics in a sex-specific manner (14). In addition, the SARS-CoV-2 infection-induced reduction of ACE2 function may also culminate in gut dysbiosis through a decrease in the mTOR-mediated synthesis of AMPs independently of RAS (15).
The possibility that SARS-CoV-2 infection of enterocytes modify gut microbiota is supported by the fact that some patients with COVID-19 present intestinal dysbiosis (16, 17). There is evidence that hospitalized COVID-19 patients exhibit a significant reduction in gut microbiome diversity with depletion of beneficial bacterial symbionts and enrichment of opportunistic pathogens, including Actinomyces, Rothia, and Streptococcus (17, 18). Patients infected with SARS-CoV-2 also showed a decrease in the relative abundance of Faecalibacterium prausnitzii and Bifidobacterium bifidum, which are bacteria responsible for the production of butyrate (17, 19). Butyrate is a short-chain fatty acid (SCFA) that influences both the proliferation and differentiation of epithelial intestinal cells, by enhancing the renewal and integrity of the epithelial barrier function (20). Moreover, patients undergoing allogeneic hematopoietic cell transplantation showing greater abundance of butyrate-producing bacteria have five-fold protection against the development of viral lower respiratory tract infection (21).
Interestingly, there are several pathologies in which the gut microbiota is modified and in some of them a direct relationship has been found with the severity of COVID-19, including elderly, diabetes, hypertension, obesity, periodontitis, and kidney diseases, as summarized in Table 1. Among these conditions, aging, diabetes, and hypertension stand out, since they are the main cause of COVID-19-related death (95–99). Yet, before getting into this point, it seemed worthwhile to discuss basic aspects of the gut microbiota, as well as the dysbiosis seen in aging and disease, particularly diabetes and hypertension.
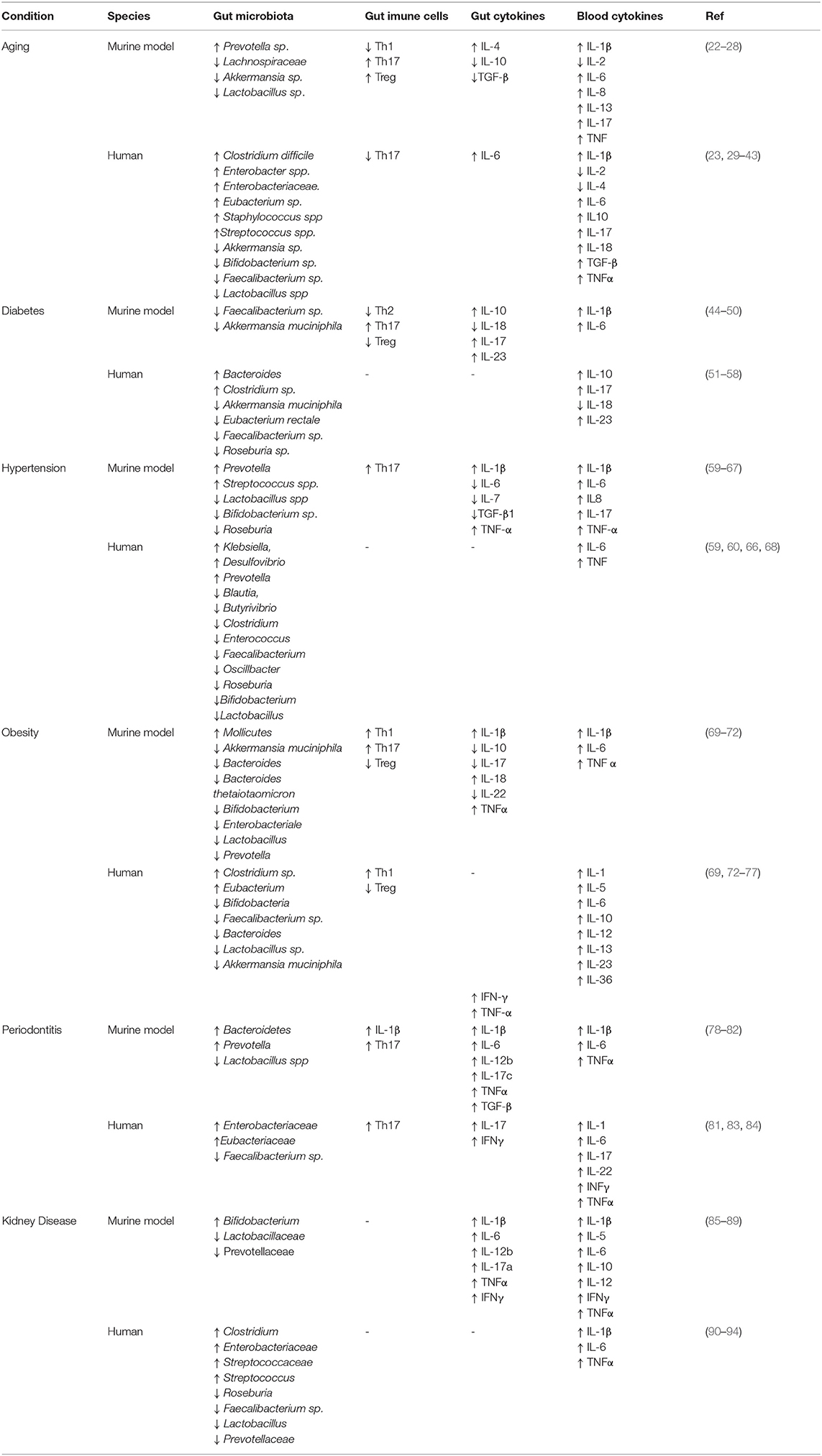
Table 1. Summary of the alterations in the gut microbiota, gut immune cells, blood and gut cytokine profiles in main groups at risk for COVID-19.
Aging and Gut Microbiota
Aging is usually accompanied by a progressive decline of physiological functions determined by (epi) genetic, stochastic, and environmental processes (100). The elderly population has an increasing tendency to multimorbidity, fragility and disability. One of the biological systems most compromised by senility is the gastrointestinal tract (101). Along with aging, there is a degeneration of the enteric nervous system (ENS), alteration of intestinal motility, and changes in the intestinal mucous barrier, decreasing the defense function and favoring the development of gastrointestinal disorders (101, 102).
A mutual characteristic of aging in tissues and aging-related diseases is the inflammaging, which is the low-grade, persistent and systemic inflammation, even in the absence of infection, culminating in tissue degeneration and chronic diseases (101, 103). In addition, other hallmarks of immunosenescence are represented by a decrease in the capacity to respond to new antigens and the accumulation of memory T cells (103, 104). In aging, the gut dysbiosis leads, at least partly, to immune dysfunction, culminating in a more inflammatory environment with poor immune editing (29, 105). It is important to know that although the gut microbiota does not age its profile changes during aging. Furthermore, the maintenance of a “youthful” or “healthy” gut microbiota architecture throughout aging may postpone or limit immunosenescence (22).
During aging, the gut microbiota is characterized by an increase in the expression of proteolytic genes and a decrease in saccharolytic ones leading to the growth of pathogens, which in turn intensify inflammation (29). The most striking change in the microbiota of elderly individuals is the change in the relative proportions of Firmicutes and Bacteroidetes; the elderly having a higher proportion of Bacteroidetes, while in young adults the Firmicutes prevail (30). Moreover, the production of anti-inflammatory factors by the microbiota of elderly individuals is reduced, including butyrate (29). All these alterations observed in the gut microbiota during aging enhance a more pro-inflammatory environment, contributing to inflammaging.
Aging-associated gut dysbiosis induces a weakening of the intestinal barrier (102). Therefore, it is possible to observe high levels of bacterial products in the bloodstream such as LPS (31, 103), which could lead to an increase in the production of pro-inflammatory mediators. Indeed, elderly people have a rise in the amount of circulating cytokines as well as a decrease in the lymphocyte response, natural killer cells, and phagocytic activity (32, 103). Furthermore, aging animals have increased inflammatory cytokines in the plasma and an augmentation in the intestinal permeability compared to young animals (33). This pro-inflammatory status seems to be related causally to the microbiota profile, since aged GF animals do not present inflammaging status. In addition, when both aged and young GF animals received the microbiota from aging wild type (WT) animals, they exhibited an increase in the circulating contents of inflammatory cytokines and intestinal permeability. Aging animals also showed an increase in the LPS-induced inflammatory cell infiltration and IL-6 levels compared to young animals, indicating the development of ARDS that is one of the most prevalent morbidities associated with aging. Nevertheless, old GF mice presented less LPS-evoked inflammatory infiltrates in the lungs compared to WT animals (33). Therefore, the microbiota of aging animals is important to the development of inflammaging.
Diabetes and Gut Microbiota
Diabetes Mellitus is a group of metabolic diseases characterized by hyperglycemia. Usually, DM is classified as type 1 and type 2 and related to low production and failure of insulin action, respectively (106). Nevertheless, this simple subdivision is not accurate, because it does not take into account the intermediate forms of DM with overlapping features. The “double diabetes” or type 1.5 diabetes is a disease with metabolic characteristics of type 2 DM with autoantibodies for β-cells typical of type 1 DM (107). Another intermediate form of DM is the Latent Autoimmune Diabetes in Adults (LADA), which shares autoimmune destruction of β-cells and insulin resistance, although to a lesser extent than type 1 DM (108). The hyperglycemia noted in diabetic patients is accompanied by the presence of cytokines such as IL-1β, IL-6, and TNF-α, characterizing a low-grade inflammation status (109).
A common change in all types of DM patients is the dysbiosis (110, 111). Although there is a controversy about which bacterial phyla is altered in the gut microbiota of diabetic patients, it is a consensus that the relationship between Firmicutes and Bacteroidetes is unbalanced in these patients (51, 112, 113). Besides, diabetic animals treated with probiotics containing the Lactobacillus rhamnosus NCDC17 improved the parameters regarding oral glucose tolerance test and led to an increase in plasma insulin, together with decreased the inflammatory cytokines IL-6 and TNF in the epididymal fat (114). Therefore, the absence or excessive proliferation of some bacteria could be one of the mechanisms of intestinal barrier dysfunction observed in diabetic models, leading to increased permeability of bacterial content to the bloodstream, as LPS (110). Replacement with Faecalibacterium sp. in diabetic animals improved the intestinal barrier integrity and circulating LPS levels (115).
Interestingly, the gut microbiota of non-obese diabetic mice changed before the onset of diabetes (52). Alterations observed included reduction of bacteria abundance and diversity, and one of the most affected groups was the butyrate-producing bacteria (53). Butyrate regulates the permeability of the intestinal barrier by inducing mucin production and decreasing the transit of bacteria, oxidative stress, as well as local and systemic inflammation (54). Accordingly, the increased permeability of the intestinal barrier observed in diabetic patients can be attributed, at least partly, to the reduction of butyrate-producing bacteria (55). Thus, it is plausible to think that butyrate replacement in diabetic patients, through direct administration or ingestion of prebiotics, may reduce intestinal permeability and low-grade inflammation triggered by gut microbiota products translocated into the bloodstream.
Hypertension and Gut Microbiota
Hypertension is a progressive cardiovascular syndrome whose early markers are usually present even before the sustained increase of blood pressure (BP). The progression of hypertension may be represented as stages 1, 2, and 3. In stage 1, patients present occasional or intermittent BP elevations, early cardiovascular disease, and no target organ disease. In stage 2, patients exhibit sustained BP elevations or progressive cardiovascular disease and early signs of target organ disease. In stage 3, the patients show marked and sustained BP elevations or advanced cardiovascular disease and overtly present target organ disease (116). Unfortunately, despite advances in awareness about lifestyle improvements, new therapies, and intensive medical interventions, around a third of hypertensive patients do not obtain control of BP when prescribed three or more antihypertensive drugs, presenting the so-called “treatment-resistant” hypertension (59).
Although the etiology of hypertension seems to depend on both genetic and environmental factors, the exact cause remains unknown. Several pieces of evidence suggest that hypertension can result from intestinal dysbiosis. For instance, treatment with antibiotics produces an increase in BP, indicating the participation of gut microbiota in the control of BP (60). Furthermore, GF mice showed lower BP as compared to conventional ones and present attenuation of BP increase in response to infused angiotensin II (61). Also, metabolites of gut microbiota are involved in the control of BP, including trimethylamine N-oxide, hydrogen sulfate, and SCFAs (117).
Causative evidence for the role of gut dysbiosis in the genesis of hypertension came since transfection of dysbiotic fecal samples from hypertensive patients to GF mice raised BP in the recipients (22). A study carried out in pre-hypertensive and hypertensive patients detected a lower richness and diversity of the intestinal microbiota as compared to healthy individuals. Hypertensive patients presented an increase of gram-negative groups and an elevation of the ratio between Firmicutes and Bacteroidetes (22, 34, 35).
Gut microbiota and their metabolites reduce the epithelium barrier integrity during hypertension, and this is linked to the downregulation of tight junction protein expression (118, 119). Hypertensive rats also presented a higher intestinal permeability to trimethylamine (TMA), a microbiota metabolite precursor of trimethylamine N-oxide, which is a marker of cardiovascular mortality. Furthermore, spontaneously hypertensive rats (SHR) showed suppression of components of T cell receptor signaling cascade in the colonic epithelium compared to Wistar Kyoto (WKY) normotensive rats, including glycoprotein CD3 gamma chain and lymphocyte cytosolic protein 2 (Lcp2). SHR animals also presented a decrease in the expression of IL-6, IL-7, and TGF-β1 in the colonic epithelium, related to marked lower production of alkaline phosphatase in the intestinal epithelial cells (120). Together, these alterations in the colonic epithelium of SHRs characterize changes in the gut immune response and epithelial layer in hypertension.
It is well known that one of the major triggers of hypertension is the imbalanced diet with high salt content (121, 122). Such high salt environment induces Th17 cells (62, 123), which are pro-inflammatory; being also involved with the development of hypertension (63, 68). Mice and humans exposed to a high salt challenge showed depletion of Lactobacillus spp. in the gut microbiome along with the rise of Th17 cells and BP (35), indicating an association of Th17 cells produced by gut microbiota and the generation of hypertension. Of note, an increase in pro-inflammatory cytokines was also reported in hypertensive rats (64). In particular, IL-6 is a central cytokine in the regulation of BP, since it is responsive to angiotensin II to raise BP regardless of baseline values (65). Furthermore, a study carried out in hypertensive patients found an increase in pro-inflammatory cytokines in peripheral blood samples associated with changes in the profile of intestinal microbiota (124).
Can Gut Microbiota Dysbiosis Be Important To SARS-CoV-2-Induced Immune Hyperresponsiveness and SARS Development in Elderly, Diabetic, and Hypertensive Individuals?
The main groups at risk for the COVID-19-related death are aging, DM, and hypertension. These conditions have a key point in common, which is dysbiosis that results in high intestinal permeability, translocation of bacterial contents to the bloodstream, and the development of basal inflammation. Therefore, a central question arises from this observation: can dysbiosis and the consequent pro-inflammatory status be critical for development of COVID-19 severity in aging, DM, and hypertensive individuals, similar to SARS and hyper-immune response also referred as a cytokine storm? Likely yes is the answer.
Some TLR4-activated danger-associated molecular pattern (DAMP) signals, including oxidized 1-palmitoyl-2-arachidonoyl-phosphatidylcholine (OxPAPC) and high-mobility group box 1 (HMGB1), are increased in the acute lung injury (ALI) caused by respiratory viruses such as the influenza virus (125, 126). It is important to note that influenza-triggered ALI seems to occur secondary to the cytokine storm induced by the activation of TLR4 by host-derived DAMPs such as OxPAPC and HMGB1 (125, 127). Notably, TLR4−/− mice have been protected against influenza A virus-provoked lethality, and the therapeutic treatment with TLR4 antagonists, Eritoran and FP7, inhibited influenza virus-induced cytokine production, ALI, and mortality in wild-type mice (127–129).
Interestingly, low doses of LPS exacerbate the TLR3 activation-induced inflammatory response in human monocytes in vitro (130). Furthermore, macrophages infected with Influenza A and stimulated with low concentrations of LPS showed increased levels of cytokines compared to macrophages that were infected only with the virus. The authors proposed that LPS enhances the release of bioactive cytokines by infected macrophages, which can lead to a decompensated increase in inflammatory metabolites (131, 132). These data reinforce the idea that weakness of intestinal permeability and consequent translocation of LPS in the elderly, diabetic and hypertensive individuals can be relevant to the severity of COVID-19 in these populations.
In a clinical setting involving 48 subjects, the expression of TLR4 and its downstream signaling molecules as well as S100A9 (TLR4 ligand) were significantly upregulated in PBMCs from severe COVID-19 patients as compared to those from healthy controls. Furthermore, S100A9 amplified the recombinant S2 protein of SARS-CoV-2-induced IL-1β mRNA expression in PBMCs in vitro (133), suggesting that activation of TLR4 by LPS from the gut microbiota of elderly, diabetic, and hypertensive individuals may be related to the severity of COVID-19. In keeping with these results, respiratory syncytial virus infection induced an increase of TLR4 expression in the airway epithelial cells in vitro, and activation of these cells with LPS potentiated the release of IL-6 and IL-8 induced by the virus (134).
Since severe COVID-19 patients show high expression of TLR4 in PBMCs (133), we can speculate that the activation of this receptor by LPS derived from the gut microbiota of elderly, diabetic, and hypertensive individuals would also potentiate the production of IL-6 induced by SARS-CoV-2. In this respect, it should be pointed out that, among all increased cytokines, the rise of IL-6 circulating levels predicted mechanical ventilation, intensive care unit admission, shock, and death in severe patients with COVID-19 (18, 135, 136). Furthermore, a follow-up with 21 individuals with several or critical COVID-19 revealed that a single dose of tocilizumab, an anti-IL-6 receptor drug, recovered 90% of patients (137).
Conclusion
In conclusion, we postulate that the gut dysbiosis may be responsible for COVID-19-related death in elderly individuals as well as diabetic and hypertensive patients, since these subjects show a change in the profile of gut microbiota followed by low-grade inflammation, especially with high circulating levels of IL-6. The possibility does exist that augmentation of pro-inflammatory bacteria in the gut may alter the intestinal immune repertoire with consequent weakness of epithelium-intestinal permeability and increased LPS translocation into the bloodstream. We believe that the hyperactivation of TLR4 induced by gut microbiota products, translocated into the circulation, strongly contributes to the cytokine storm, worsening the prognosis of COVID-19 in the elderly, diabetic and hypertensive individuals (Figure 1). In this respect, new therapeutic strategies based on prebiotics or bacterial metabolites, as butyrate, appear as potentially practical approaches for adjuvant treatment of these patients.
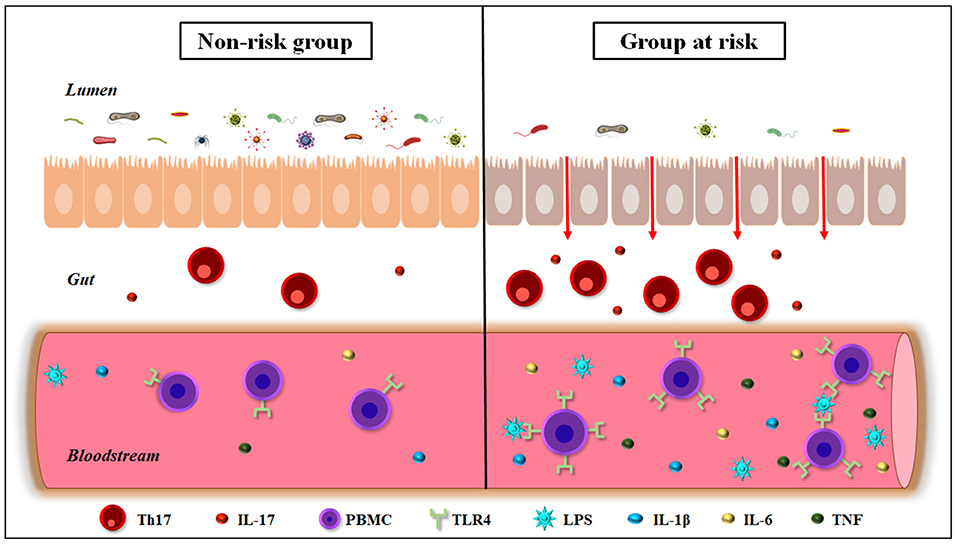
Figure 1. Gut-immune interactions in elderly, diabetic, and hypertensive individuals. These conditions are the three most COVID-19-related death risk factors, and show a decrease in the diversity of the gut microbiota, leading to dysbiosis and weakness of the intestinal barrier permeability. In addition, people belonging to risk groups for COVID-19-related death show hyperimmune activation in the intestine, increasing Th17+ T cells and IL-17 production. These individuals also exhibit a rise in the circulating levels of bacterial endotoxins such as LPS, as well as pro-inflammatory cytokines, as IL-1β, IL-6, and TNF. Furthermore, the elderly, diabetic, and hypertensive individuals show an increase in the expression of TLR4 in peripheral blood mononuclear cells (PBMCs). IL-17, interleukin-17; IL-1 β, interleukin-1β; IL-6, interleukin-6; LPS, lipopolysaccharide; Th17, T helper 17; TLR4, Toll-like receptor 4; TNF, Tumor necrosis factor.
Author Contributions
NM contributed to the conception and design of the study, wrote the manuscript, discussed the content and contributed to the manuscript revision. PS, MM, and WS discussed the content and contributed to the manuscript revision. VC contributed to the conception and design of the study, wrote the manuscript, discussed the content and contributed to the manuscript revision. All authors reviewed and/or edited the manuscript prior submission.
Funding
This work was supported by Oswaldo Cruz Institute, Oswaldo Cruz Foundation (Fiocruz), Ministry of Health, Brazil.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We would like to thank the PrInt Fiocruz-CAPES Program; Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq); Programa Fiocruz de Fomento a Inovação (INOVA-FIOCRUZ) and Fundação Carlos Chagas de Amparo à Pesquisa do Estado do Rio de Janeiro (FAPERJ); Mercosur Fund for Structural Convergence (FOCEM/Mercosur) for the financial support. This work was developed in the context of the Brazilian National Institute of Science and Technology on Neuroimmunomodulation and the Rio de Janeiro Research Network on Neuroinflammation. This article is dedicated to Juliana de Meis, young researcher in the Laboratory on Thymus Research (Fiocruz, Rio de Janeiro), who passed way on July 16th 2021, due to COVID-19.
Abbreviations
ACE2, angiotensin I converting enzyme 2; ALI, acute lung injury; AMP, anti-microbial peptides; ARDS, acute respiratory distress syndrome; BP, blood pressure; CD3, cluster of differentiation 3; COV, coronavirus; DAMP, danger-associated molecular pattern; DM, diabetes mellitus; ENS, enteric nervous system; GF, germ-free; HMGB1, high-mobility group box 1; IFN-γ, interferon gamma γ; IgA, immunoglobulin A; IL-10, interleukin 10; IL-17, interleukin 17; IL-1β, interleukin 1β; IL-6, interleukin 6; IL-7, interleukin 7; IL-8, interleukin 8; LADA, Latent Autoimmune Diabetes in Adults; Lcp2, lymphocyte cytosolic protein 2; LPS, lipopolysaccharide; mRNA, messenger ribonucleic acid; OxPAPC, oxidized 1-palmitoyl-2-arachidonoyl-phosphatidylcholine; PBMC, peripheral blood mononuclear cells; SARS, severe acute respiratory syndrome; SCFA, short-chain fatty acids; SHR, hypertensive rats; TGF-β1, Transforming growth factor beta 1; Th17, T helper 17; TLR4, toll-like receptor 4; TMA, Trimethylamine; TNF, Tumor necrosis factor; WKY, Wistar Kyoto; WT, wild type.
References
1. Guo YR, Cao QD, Hong ZS, Tan YY, Chen SD, Jin HJ, et al. The origin, transmission and clinical therapies on coronavirus disease 2019 (COVID-19) outbreak - an update on the status. Mil Med Res. (2020) 7:11. doi: 10.1186/s40779-020-00240-0
2. Gonzalez-Reiche AS, Hernandez MM, Sullivan MJ, Ciferri B, Alshammary H, Obla A, et al. Introductions and early spread of SARS-CoV-2 in the New York City area. Science. (2020) 369:297–301. doi: 10.1126/science.abc1917
3. Khosrawipour V, Lau H, Khosrawipour T, Kocbach P, Ichii H, Bania J, et al. Failure in initial stage containment of global COVID-19 epicenters. J Med Virol. (2020) 92:863–7. doi: 10.1002/jmv.25883
4. World Health Organization. Coronavirus Disease (COVID-19) Dashboard. (2021). Available online at: https://covid19.who.int/. (accessed May 24, 2021).
5. Zhou F, Yu T, Du R, Fan G, Liu Y, Liu Z, et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. (2020) 395:1054–62. doi: 10.1016/S0140-6736(20)30566-3
6. Belkaid Y, Harrison OJ. Homeostatic immunity and the microbiota. Immunity. (2017) 46:562–76. doi: 10.1016/j.immuni.2017.04.008
7. Cong J, Zhang X. How human microbiome talks to health and disease. Eur J Clin Microbiol Infect Dis. (2018) 37:1595–601. doi: 10.1007/s10096-018-3263-1
8. Landman C, Quevrain E. Gut microbiota: description, role and pathophysiologic implications. Rev Med Interne. (2016) 37:418–23. doi: 10.1016/j.revmed.2015.12.012
9. Chen W, Lan Y, Yuan X, Deng X, Li Y, Cai X, et al. Detectable 2019-nCoV viral RNA in blood is a strong indicator for the further clinical severity. Emerg Microbes Infect. (2020) 9:469–73. doi: 10.1080/22221751.2020.1732837
10. Kotfis K, Skonieczna-Zydecka K. COVID-19: gastrointestinal symptoms and potential sources of SARS-CoV-2 transmission. Anaesthesiol Intensive Ther. (2020) 52:171–2. doi: 10.5114/ait.2020.93867
11. Gheblawi M, Wang K, Viveiros A, Nguyen Q, Zhong JC, Turner AJ, et al. Angiotensin-converting enzyme 2: SARS-CoV-2 receptor and regulator of the renin-angiotensin system: celebrating the 20th anniversary of the discovery of ACE2. Circ Res. (2020) 126:1456–74. doi: 10.1161/CIRCRESAHA.120.317015
12. Zang R, Gomez Castro MF, McCune BT, Zeng Q, Rothlauf PW, Sonnek NM, et al. TMPRSS2 and TMPRSS4 promote SARS-CoV-2 infection of human small intestinal enterocytes. Sci Immunol. (2020) 5. doi: 10.1101/2020.04.21.054015
13. Beyerstedt S, Casaro EB, Rangel EB. COVID-19: angiotensin-converting enzyme 2 (ACE2) expression and tissue susceptibility to SARS-CoV-2 infection. Eur J Clin Microbiol Infect Dis. (2021) 40:905–19. doi: 10.1007/s10096-020-04138-6
14. Cheema MU, Pluznick JL. Gut microbiota plays a central role to modulate the plasma and fecal metabolomes in response to angiotensin II. Hypertension. (2019) 74:184–93. doi: 10.1161/HYPERTENSIONAHA.119.13155
15. Hashimoto T, Perlot T, Rehman A, Trichereau J, Ishiguro H, Paolino M, et al. ACE2 links amino acid malnutrition to microbial ecology and intestinal inflammation. Nature. (2012) 487:477–81. doi: 10.1038/nature11228
16. Gu S, Chen Y, Wu Z, Chen Y, Gao H, Lv L, et al. Alterations of the Gut Microbiota in Patients With Coronavirus Disease 2019 or H1N1 Influenza. Clin Infect Dis. (2020) 71:2669–78. doi: 10.1093/cid/ciaa709
17. Zuo T, Zhang F, Lui GCY, Yeoh YK, Li AYL, Zhan H, et al. Alterations in Gut Microbiota of Patients With COVID-19 During Time of Hospitalization. Gastroenterology. (2020) 159:944–55 e8. doi: 10.1053/j.gastro.2020.05.048
18. Gubernatorova EO, Gorshkova EA, Polinova AI, Drutskaya MS. IL-6: Relevance for immunopathology of SARS-CoV-2. Cytokine Growth Factor Rev. (2020) 53:13–24. doi: 10.1016/j.cytogfr.2020.05.009
19. Yeoh YK, Zuo T, Lui GC, Zhang F, Liu Q, Li AY, et al. Gut microbiota composition reflects disease severity and dysfunctional immune responses in patients with COVID-19. Gut. (2021) 70:698–706. doi: 10.1136/gutjnl-2020-323020
20. Valdes AM, Walter J, Segal E, Spector TD. Role of the gut microbiota in nutrition and health. BMJ. (2018) 361:k2179. doi: 10.1136/bmj.k2179
21. Haak BW, Littmann ER, Chaubard JL, Pickard AJ, Fontana E, Adhi F, et al. Impact of gut colonization with butyrate-producing microbiota on respiratory viral infection following allo-HCT. Blood. (2018) 131:2978–86. doi: 10.1182/blood-2018-01-828996
22. Vaiserman AM, Koliada AK, Marotta F. Gut microbiota: a player in aging and a target for anti-aging intervention. Ageing Res Rev. (2017) 35:36–45. doi: 10.1016/j.arr.2017.01.001
23. Forsey RJ, Thompson JM, Ernerudh J, Hurst TL, Strindhall J, Johansson B, et al. Plasma cytokine profiles in elderly humans. Mech Ageing Dev. (2003) 124:487–93. doi: 10.1016/S0047-6374(03)00025-3
24. Palmeri M, Misiano G, Malaguarnera M, Forte GI, Vaccarino L, Milano S, et al. Cytokine serum profile in a group of Sicilian nonagenarians. J Immunoassay Immunochem. (2012) 33:82–90. doi: 10.1080/15321819.2011.601781
25. Luo D, Chen K, Li J, Fang Z, Pang H, Yin Y, et al. Gut microbiota combined with metabolomics reveals the metabolic profile of the normal aging process and the anti-aging effect of FuFang Zhenshu TiaoZhi(FTZ) in mice. Biomed Pharmacother. (2020) 121:109550. doi: 10.1016/j.biopha.2019.109550
26. Man AL, Bertelli E, Rentini S, Regoli M, Briars G, Marini M, et al. Age-associated modifications of intestinal permeability and innate immunity in human small intestine. Clin Sci (Lond). (2015) 129:515–27. doi: 10.1042/CS20150046
27. Dillon SM, Liu J, Purba CM, Christians AJ, Kibbie JJ, Castleman MJ, et al. Age-related alterations in human gut CD4 T cell phenotype, T helper cell frequencies, and functional responses to enteric bacteria. J Leukoc Biol. (2020) 107:119–32. doi: 10.1002/JLB.5A0919-177RR
28. Hor YY, Lew LC, Jaafar MH, Lau AS, Ong JS, Kato T, et al. Lactobacillus sp. improved microbiota and metabolite profiles of aging rats. Pharmacol Res. (2019) 146:104312. doi: 10.1016/j.phrs.2019.104312
29. Ctoi AF, Corina A, Katsiki N, Vodnar DC, Andreicut AD, Stoian AP, et al. Gut microbiota and aging-A focus on centenarians. Biochim Biophys Acta Mol Basis Dis. (2020) 1866:165765. doi: 10.1016/j.bbadis.2020.165765
30. Maynard C, Weinkove D. The gut microbiota and ageing. Subcell Biochem. (2018) 90:351–71. doi: 10.1007/978-981-13-2835-0_12
31. Ghosh S, Lertwattanarak R, Garduno Jde J, Galeana JJ Li J, Zamarripa F, et al. Elevated muscle TLR4 expression and metabolic endotoxemia in human aging. J Gerontol A Biol Sci Med Sci. (2015) 70:232–46. doi: 10.1093/gerona/glu067
32. Biagi E, Nylund L, Candela M, Ostan R, Bucci L, Pini E, et al. Through ageing, and beyond: gut microbiota and inflammatory status in seniors and centenarians. PLoS ONE. (2010) 5:e10667. doi: 10.1371/journal.pone.0010667
33. Thevaranjan N, Puchta A, Schulz C, Naidoo A, Szamosi JC, Verschoor CP, et al. Age-associated microbial dysbiosis promotes intestinal permeability, systemic inflammation, and macrophage dysfunction. Cell Host Microbe. (2017) 21:455–66 e4. doi: 10.1016/j.chom.2017.03.002
34. Yang T, Santisteban MM, Rodriguez V, Li E, Ahmari N, Carvajal JM, et al. Gut dysbiosis is linked to hypertension. Hypertension. (2015) 65:1331–40. doi: 10.1161/HYPERTENSIONAHA.115.05315
35. Wilck N, Matus MG, Kearney SM, Olesen SW, Forslund K, Bartolomaeus H, et al. Salt-responsive gut commensal modulates TH17 axis and disease. Nature. (2017) 551:585–9. doi: 10.1038/nature24628
36. Rea MC, O'Sullivan O, Shanahan F, O'Toole PW, Stanton C, Ross RP, et al. Clostridium difficile carriage in elderly subjects and associated changes in the intestinal microbiota. J Clin Microbiol. (2012) 50:867–75. doi: 10.1128/JCM.05176-11
37. Askarova S, Umbayev B, Masoud AR, Kaiyrlykyzy A, Safarova Y, Tsoy A, et al. The links between the gut microbiome, aging, modern lifestyle and alzheimer's disease. Front Cell Infect Microbiol. (2020) 10:104. doi: 10.3389/fcimb.2020.00104
38. Odamaki T, Kato K, Sugahara H, Hashikura N, Takahashi S, Xiao JZ, et al. Age-related changes in gut microbiota composition from newborn to centenarian: a cross-sectional study. BMC Microbiol. (2016) 16:90. doi: 10.1186/s12866-016-0708-5
39. Collado MC, Derrien M, Isolauri E, de Vos WM, Salminen S. Intestinal integrity and Akkermansia muciniphila, a mucin-degrading member of the intestinal microbiota present in infants, adults, and the elderly. Appl Environ Microbiol. (2007) 73:7767–70. doi: 10.1128/AEM.01477-07
40. Salazar N, Gonzalez S, Nogacka AM, Rios-Covian D, Arboleya S, Gueimonde M, et al. Microbiome: effects of ageing and diet. Curr Issues Mol Biol. (2020) 36:33–62. doi: 10.21775/cimb.036.033
41. Ferrucci L, Corsi A, Lauretani F, Bandinelli S, Bartali B, Taub DD, et al. The origins of age-related proinflammatory state. Blood. (2005) 105:2294–9. doi: 10.1182/blood-2004-07-2599
42. Bruunsgaard H, Andersen-Ranberg K, Hjelmborg J, Pedersen BK, Jeune B. Elevated levels of tumor necrosis factor alpha and mortality in centenarians. Am J Med. (2003) 115:278–83. doi: 10.1016/S0002-9343(03)00329-2
43. Rea IM, Gibson DS, McGilligan V, McNerlan SE, Alexander HD, Ross OA. Age and age-related diseases: role of inflammation triggers and cytokines. Front Immunol. (2018) 9:586. doi: 10.3389/fimmu.2018.00586
44. Rodrigues KF, Pietrani NT, Bosco AA, Campos FMF, Sandrim VC, Gomes KB. IL-6, TNF-alpha, and IL-10 levels/polymorphisms and their association with type 2 diabetes mellitus and obesity in Brazilian individuals. Arch Endocrinol Metab. (2017) 61:438–46. doi: 10.1590/2359-3997000000254
45. Pickup JC, Chusney GD, Thomas SM, Burt D. Plasma interleukin-6, tumour necrosis factor alpha and blood cytokine production in type 2 diabetes. Life Sci. (2000) 67:291–300. doi: 10.1016/S0024-3205(00)00622-6
46. Naito Y, Uchiyama K, Takagi T A. next-generation beneficial microbe: akkermansia muciniphila. J Clin Biochem Nutr. (2018) 63:33–5. doi: 10.3164/jcbn.18-57
47. Furet JP, Kong LC, Tap J, Poitou C, Basdevant A, Bouillot JL, et al. Differential adaptation of human gut microbiota to bariatric surgery-induced weight loss: links with metabolic and low-grade inflammation markers. Diabetes. (2010) 59:3049–57. doi: 10.2337/db10-0253
48. He C, Shan Y, Song W. Targeting gut microbiota as a possible therapy for diabetes. Nutr Res. (2015) 35:361–7. doi: 10.1016/j.nutres.2015.03.002
49. Miranda MCG, Oliveira RP, Torres L, Aguiar SLF, Pinheiro-Rosa N, Lemos L, et al. Frontline Science: Abnormalities in the gut mucosa of non-obese diabetic mice precede the onset of type 1 diabetes. J Leukoc Biol. (2019) 106:513–29. doi: 10.1002/JLB.3HI0119-024RR
50. Alam C, Valkonen S, Palagani V, Jalava J, Eerola E, Hanninen A. Inflammatory tendencies and overproduction of IL-17 in the colon of young NOD mice are counteracted with diet change. Diabetes. (2010) 59:2237–46. doi: 10.2337/db10-0147
51. Santos-Marcos JA, Perez-Jimenez F, Camargo A. The role of diet and intestinal microbiota in the development of metabolic syndrome. J Nutr Biochem. (2019) 70:1–27. doi: 10.1016/j.jnutbio.2019.03.017
52. Krych L, Nielsen DS, Hansen AK, Hansen CH. Gut microbial markers are associated with diabetes onset, regulatory imbalance, and IFN-gamma level in NOD mice. Gut Microbes. (2015) 6:101–9. doi: 10.1080/19490976.2015.1011876
53. Ganesan K, Chung SK, Vanamala J, Xu B. Causal relationship between diet-induced gut microbiota changes and diabetes: a novel strategy to transplant faecalibacterium prausnitzii in preventing diabetes. Int J Mol Sci. (2018) 19:3720. doi: 10.3390/ijms19123720
54. Guilloteau P, Martin L, Eeckhaut V, Ducatelle R, Zabielski R, Van Immerseel F. From the gut to the peripheral tissues: the multiple effects of butyrate. Nutr Res Rev. (2010) 23:366–84. doi: 10.1017/S0954422410000247
55. de Groot PF, Belzer C, Aydin O, Levin E, Levels JH, Aalvink S, et al. Distinct fecal and oral microbiota composition in human type 1 diabetes, an observational study. PLoS ONE. (2017) 12:e0188475. doi: 10.1371/journal.pone.0188475
56. Santiago AF, Alves AC, Oliveira RP, Fernandes RM, Paula-Silva J, Assis FA, et al. Aging correlates with reduction in regulatory-type cytokines and T cells in the gut mucosa. Immunobiology. (2011) 216:1085–93. doi: 10.1016/j.imbio.2011.05.007
57. Abdel-Moneim A, Bakery HH, Allam G. The potential pathogenic role of IL-17/Th17 cells in both type 1 and type 2 diabetes mellitus. Biomed Pharmacother. (2018) 101:287–92. doi: 10.1016/j.biopha.2018.02.103
58. Fatima N, Faisal SM, Zubair S, Ajmal M, Siddiqui SS, Moin S, et al. Role of pro-inflammatory cytokines and biochemical markers in the pathogenesis of type 1 diabetes: correlation with age and glycemic condition in diabetic human subjects. PLoS ONE. (2016) 11:e0161548. doi: 10.1371/journal.pone.0161548
59. Writing Group M, Mozaffarian D, Benjamin EJ, Go AS, Arnett DK, Blaha MJ, et al. Executive summary: heart disease and stroke statistics−2016 update: a report from the american heart association. Circulation. (2016) 133:447–54. doi: 10.1161/CIR.0000000000000366
60. Honour J. The possible involvement of intestinal bacteria in steroidal hypertension. Endocrinology. (1982) 110:285–7. doi: 10.1210/endo-110-1-285
61. Karbach SH, Schonfelder T, Brandao I, Wilms E, Hormann N, Jackel S, et al. Gut microbiota promote angiotensin ii-induced arterial hypertension and vascular dysfunction. J Am Heart Assoc. (2016) 5:e003698. doi: 10.1161/JAHA.116.003698
62. Kleinewietfeld M, Manzel A, Titze J, Kvakan H, Yosef N, Linker RA, et al. Sodium chloride drives autoimmune disease by the induction of pathogenic TH17 cells. Nature. (2013) 496:518–22. doi: 10.1038/nature11868
63. Norlander AE, Saleh MA, Kamat NV, Ko B, Gnecco J, Zhu L, et al. Interleukin-17A regulates renal sodium transporters and renal injury in angiotensin II-induced hypertension. Hypertension. (2016) 68:167–74. doi: 10.1161/HYPERTENSIONAHA.116.07493
64. Tanase DM, Gosav EM, Radu S, Ouatu A, Rezus C, Ciocoiu M, et al. Arterial hypertension and interleukins: potential therapeutic target or future diagnostic marker? Int J Hypertens. (2019) 2019:3159283. doi: 10.1155/2019/3159283
65. De Miguel C, Rudemiller NP, Abais JM, Mattson DL. Inflammation and hypertension: new understandings and potential therapeutic targets. Curr Hypertens Rep. (2015) 17:507. doi: 10.1007/s11906-014-0507-z
66. Muller PA, Koscso B, Rajani GM, Stevanovic K, Berres ML, Hashimoto D, et al. Crosstalk between muscularis macrophages and enteric neurons regulates gastrointestinal motility. Cell. (2014) 158:300–13. doi: 10.1016/j.cell.2014.04.050
67. Li J, Zhao F, Wang Y, Chen J, Tao J, Tian G, et al. Gut microbiota dysbiosis contributes to the development of hypertension. Microbiome. (2017) 5:14. doi: 10.1186/s40168-016-0222-x
68. Madhur MS, Lob HE, McCann LA, Iwakura Y, Blinder Y, Guzik TJ, et al. Interleukin 17 promotes angiotensin II-induced hypertension and vascular dysfunction. Hypertension. (2010) 55:500–7. doi: 10.1161/HYPERTENSIONAHA.109.145094
69. Stephens RW, Arhire L, Covasa M. Gut microbiota: from microorganisms to metabolic organ influencing obesity. Obesity (Silver Spring). (2018) 26:801–9. doi: 10.1002/oby.22179
70. Araujo JR, Tomas J, Brenner C, Sansonetti PJ. Impact of high-fat diet on the intestinal microbiota and small intestinal physiology before and after the onset of obesity. Biochimie. (2017) 141:97–106. doi: 10.1016/j.biochi.2017.05.019
71. Garidou L, Pomie C, Klopp P, Waget A, Charpentier J, Aloulou M, et al. The gut microbiota regulates intestinal CD4 T cells expressing RORgammat and controls metabolic disease. Cell Metab. (2015) 22:100–12. doi: 10.1016/j.cmet.2015.06.001
72. Winer DA, Luck H, Tsai S, Winer S. The intestinal immune system in obesity and insulin resistance. Cell Metab. (2016) 23:413–26. doi: 10.1016/j.cmet.2016.01.003
73. Crovesy L, Masterson D, Rosado EL. Profile of the gut microbiota of adults with obesity: a systematic review. Eur J Clin Nutr. (2020) 74:1251–62. doi: 10.1038/s41430-020-0607-6
74. Gomes AC, Hoffmann C, Mota JF. The human gut microbiota: metabolism and perspective in obesity. Gut Microbes. (2018) 9:308–25. doi: 10.1080/19490976.2018.1465157
75. Zak-Golab A, Kocelak P, Aptekorz M, Zientara M, Juszczyk L, Martirosian G, et al. Gut microbiota, microinflammation, metabolic profile, and zonulin concentration in obese and normal weight subjects. Int J Endocrinol. (2013) 2013:674106. doi: 10.1155/2013/674106
76. Scheithauer TPM, Rampanelli E, Nieuwdorp M, Vallance BA, Verchere CB, van Raalte DH, et al. Gut microbiota as a trigger for metabolic inflammation in obesity and type 2 diabetes. Front Immunol. (2020) 11:571731. doi: 10.3389/fimmu.2020.571731
77. Schmidt FM, Weschenfelder J, Sander C, Minkwitz J, Thormann J, Chittka T, et al. Inflammatory cytokines in general and central obesity and modulating effects of physical activity. PLoS ONE. (2015) 10:e0121971. doi: 10.1371/journal.pone.0121971
78. Xue L, Zou X, Yang XQ, Peng F, Yu DK, Du JR. Chronic periodontitis induces microbiota-gut-brain axis disorders and cognitive impairment in mice. Exp Neurol. (2020) 326:113176. doi: 10.1016/j.expneurol.2020.113176
79. Arimatsu K, Yamada H, Miyazawa H, Minagawa T, Nakajima M, Ryder MI, et al. Oral pathobiont induces systemic inflammation and metabolic changes associated with alteration of gut microbiota. Sci Rep. (2014) 4:4828. doi: 10.1038/srep04828
80. Kobayashi R, Ogawa Y, Hashizume-Takizawa T, Kurita-Ochiai T. Oral bacteria affect the gut microbiome and intestinal immunity. Pathog Dis. (2020) 78:ftaa024. doi: 10.1093/femspd/ftaa024
81. Hajishengallis G, Chavakis T. Local and systemic mechanisms linking periodontal disease and inflammatory comorbidities. Nat Rev Immunol. (2021) 21:426–40. doi: 10.1038/s41577-020-00488-6
82. Suarez LJ, Garzon H, Arboleda S, Rodriguez A. Oral Dysbiosis and Autoimmunity: From Local Periodontal Responses to an Imbalanced Systemic Immunity. A Review Front Immunol. (2020) 11:591255. doi: 10.3389/fimmu.2020.591255
83. Li J, Lu H, Wu H, Huang S, Chen L, Gui Q, et al. Periodontitis in elderly patients with type 2 diabetes mellitus: impact on gut microbiota and systemic inflammation. Aging. (2020) 12:25956–80. doi: 10.18632/aging.202174
84. Olsen I, Yamazaki K. Can oral bacteria affect the microbiome of the gut? J Oral Microbiol. (2019) 11:1586422. doi: 10.1080/20002297.2019.1586422
85. He LX, Abdolmaleky HM, Yin S, Wang Y, Zhou JR. Dietary fermented soy extract and oligo-lactic acid alleviate chronic kidney disease in mice via inhibition of inflammation and modulation of gut microbiota. Nutrients. (2020) 12:2376. doi: 10.3390/nu12082376
86. Yang J, Lim SY, Ko YS, Lee HY, Oh SW, Kim MG, et al. Intestinal barrier disruption and dysregulated mucosal immunity contribute to kidney fibrosis in chronic kidney disease. Nephrol Dial Transplant. (2019) 34:419–28. doi: 10.1093/ndt/gfy172
87. Sumida K, Kovesdy CP. The gut-kidney-heart axis in chronic kidney disease. Physiol Int. (2019) 106:195–206. doi: 10.1556/2060.106.2019.19
88. Andres-Hernando A, Dursun B, Altmann C, Ahuja N, He Z, Bhargava R, et al. Cytokine production increases and cytokine clearance decreases in mice with bilateral nephrectomy. Nephrol Dial Transplant. (2012) 27:4339–47. doi: 10.1093/ndt/gfs256
89. Hung TV, Suzuki T. Dietary Fermentable Fibers Attenuate Chronic Kidney Disease in Mice by Protecting the Intestinal Barrier. J Nutr. (2018) 148:552–61. doi: 10.1093/jn/nxy008
90. Stanford J, Charlton K, Stefoska-Needham A, Ibrahim R, Lambert K. The gut microbiota profile of adults with kidney disease and kidney stones: a systematic review of the literature. BMC Nephrol. (2020) 21:215. doi: 10.1186/s12882-020-01805-w
91. Amdur RL, Feldman HI, Gupta J, Yang W, Kanetsky P, Shlipak M, et al. Inflammation and progression of CKD: the CRIC study. Clin J Am Soc Nephrol. (2016) 11:1546–56. doi: 10.2215/CJN.13121215
92. Hobby GP, Karaduta O, Dusio GF, Singh M, Zybailov BL, Arthur JM. Chronic kidney disease and the gut microbiome. Am J Physiol Renal Physiol. (2019) 316:F1211–F7. doi: 10.1152/ajprenal.00298.2018
93. Castillo-Rodriguez E, Fernandez-Prado R, Esteras R, Perez-Gomez MV, Gracia-Iguacel C, Fernandez-Fernandez B, et al. Impact of altered intestinal microbiota on chronic kidney disease progression. Toxins. (2018) 10:300. doi: 10.3390/toxins10070300
94. Mihai S, Codrici E, Popescu ID, Enciu AM, Albulescu L, Necula LG, et al. Inflammation-related mechanisms in chronic kidney disease prediction, progression, and outcome. J Immunol Res. (2018) 2018:2180373. doi: 10.1155/2018/2180373
95. Williamson EJ, Walker AJ, Bhaskaran K, Bacon S, Bates C, Morton CE, et al. Factors associated with COVID-19-related death using OpenSAFELY. Nature. (2020) 584:430–6. doi: 10.1038/s41586-020-2521-4
96. Bajgain KT, Badal S, Bajgain BB, Santana MJ. Prevalence of comorbidities among individuals with COVID-19: A rapid review of current literature. Am J Infect Control. (2021) 49:238–46. doi: 10.1016/j.ajic.2020.06.213
97. Li J, Huang DQ, Zou B, Yang H, Hui WZ, Rui F, et al. Epidemiology of COVID-19: A systematic review and meta-analysis of clinical characteristics, risk factors, and outcomes. J Med Virol. (2021) 93:1449–58. doi: 10.1002/jmv.26424
98. Izcovich A, Ragusa MA, Tortosa F, Lavena Marzio MA, Agnoletti C, Bengolea A, et al. Prognostic factors for severity and mortality in patients infected with COVID-19: A systematic review. PLoS ONE. (2020) 15:e0241955. doi: 10.1371/journal.pone.0241955
99. Wolff D, Nee S, Hickey NS, Marschollek M. Risk factors for Covid-19 severity and fatality: a structured literature review. Infection. (2021) 49:15–28. doi: 10.1007/s15010-020-01509-1
100. Urtamo A, Jyvakorpi SK, Strandberg TE. Definitions of successful ageing: a brief review of a multidimensional concept. Acta Biomed. (2019) 90:359–63. doi: 10.23750/abm.v90i2.8376
101. Mangiola F, Nicoletti A, Gasbarrini A, Ponziani FR. Gut microbiota and aging. Eur Rev Med Pharmacol Sci. (2018) 22:7404–13. doi: 10.26355/eurrev-201811-16280
102. Nagpal R, Mainali R, Ahmadi S, Wang S, Singh R, Kavanagh K, et al. Gut microbiome and aging: physiological and mechanistic insights. Nutr Healthy Aging. (2018) 4:267–85. doi: 10.3233/NHA-170030
103. Amsterdam D, Ostrov BE. The impact of the microbiome on immunosenescence. Immunol Invest. (2018) 47:801–11. doi: 10.1080/08820139.2018.1537570
104. Boren E, Gershwin ME. Inflamm-aging: autoimmunity, and the immune-risk phenotype. Autoimmun Rev. (2004) 3:401–6. doi: 10.1016/j.autrev.2004.03.004
105. Prata L, Ovsyannikova IG, Tchkonia T, Kirkland JL. Senescent cell clearance by the immune system: Emerging therapeutic opportunities. Semin Immunol. (2018) 40:101275. doi: 10.1016/j.smim.2019.04.003
106. Thomas CC, Philipson LH. Update on diabetes classification. Med Clin North Am. (2015) 99:1–16. doi: 10.1016/j.mcna.2014.08.015
107. Khawandanah J. Double or hybrid diabetes: a systematic review on disease prevalence, characteristics and risk factors. Nutr Diabetes. (2019) 9:33. doi: 10.1038/s41387-019-0101-1
108. Carlsson S. Etiology and pathogenesis of latent autoimmune diabetes in adults (LADA) compared to type 2 diabetes. Front Physiol. (2019) 10:320. doi: 10.3389/fphys.2019.00320
109. Popko K, Gorska E, Stelmaszczyk-Emmel A, Plywaczewski R, Stoklosa A, Gorecka D, et al. Proinflammatory cytokines Il-6 and TNF-alpha and the development of inflammation in obese subjects. Eur J Med Res. (2010) 15:120–2. doi: 10.1186/2047-783X-15-S2-120
110. Durazzo M, Ferro A, Gruden G. Gastrointestinal microbiota and type 1 diabetes mellitus: the state of art. J Clin Med. (2019) 8:1843. doi: 10.3390/jcm8111843
111. Munoz-Garach A, Diaz-Perdigones C, Tinahones FJ. Gut microbiota and type 2 diabetes mellitus. Endocrinol Nutr. (2016) 63:560–8. doi: 10.1016/j.endoen.2016.07.004
112. Ahmad A, Yang W, Chen G, Shafiq M, Javed S, Ali Zaidi SS, et al. Analysis of gut microbiota of obese individuals with type 2 diabetes and healthy individuals. PLoS ONE. (2019) 14:e0226372. doi: 10.1371/journal.pone.0226372
113. Pascale A, Marchesi N, Govoni S, Coppola A, Gazzaruso C. The role of gut microbiota in obesity, diabetes mellitus, and effect of metformin: new insights into old diseases. Curr Opin Pharmacol. (2019) 49:1–5. doi: 10.1016/j.coph.2019.03.011
114. Singh S, Sharma RK, Malhotra S, Pothuraju R, Shandilya UK. Lactobacillus rhamnosus NCDC17 ameliorates type-2 diabetes by improving gut function, oxidative stress and inflammation in high-fat-diet fed and streptozotocintreated rats. Benef Microbes. (2017) 8:243–55. doi: 10.3920/BM2016.0090
115. Xu J, Liang R, Zhang W, Tian K, Li J, Chen X, et al. Faecalibacterium prausnitzii-derived microbial anti-inflammatory molecule regulates intestinal integrity in diabetes mellitus mice via modulating tight junction protein expression. J Diabetes. (2020) 12:224–36. doi: 10.1111/1753-0407.12986
116. Giles TD, Materson BJ, Cohn JN, Kostis JB. Definition and classification of hypertension: an update. J Clin Hypertens (Greenwich). (2009) 11:611–4. doi: 10.1111/j.1751-7176.2009.00179.x
117. Richards EM, Pepine CJ, Raizada MK, Kim S. The gut, its microbiome, and hypertension. Curr Hypertens Rep. (2017) 19:36. doi: 10.1007/s11906-017-0734-1
118. Santisteban MM Qi Y, Zubcevic J, Kim S, Yang T, Shenoy V, et al. Hypertension-linked pathophysiological alterations in the gut. Circ Res. (2017) 120:312–23. doi: 10.1161/CIRCRESAHA.116.309006
119. Zhang S, Zhang Y, Ahsan MZ, Yuan Y, Liu G, Han X, et al. Atorvastatin attenuates cold-induced hypertension by preventing gut barrier injury. J Cardiovasc Pharmacol. (2019) 74:143–51. doi: 10.1097/FJC.0000000000000690
120. Yang T, Li H, Oliveira AC, Goel R, Richards EM, Pepine CJ, et al. Transcriptomic signature of gut microbiome-contacting cells in colon of spontaneously hypertensive rats. Physiol Genomics. (2020) 52:121–32. doi: 10.1152/physiolgenomics.00087.2019
121. O'Donnell M, Mente A, Rangarajan S, McQueen MJ, Wang X, Liu L, et al. Urinary sodium and potassium excretion, mortality, and cardiovascular events. N Engl J Med. (2014) 371:612–23. doi: 10.1056/NEJMx140049
122. Mozaffarian D, Fahimi S, Singh GM, Micha R, Khatibzadeh S, Engell RE, et al. Global sodium consumption and death from cardiovascular causes. N Engl J Med. (2014) 371:624–34. doi: 10.1056/NEJMoa1304127
123. Wu C, Yosef N, Thalhamer T, Zhu C, Xiao S, Kishi Y, et al. Induction of pathogenic TH17 cells by inducible salt-sensing kinase SGK1. Nature. (2013) 496:513–7. doi: 10.1038/nature11984
124. Silveira-Nunes G, Durso DF, Jr L, Cunha EHM, Maioli TU, Vieira AT, et al. Hypertension is associated with intestinal microbiota dysbiosis and inflammation in a brazilian population. Front Pharmacol. (2020) 11:258. doi: 10.3389/fphar.2020.00258
125. Imai Y, Kuba K, Neely GG, Yaghubian-Malhami R, Perkmann T, van Loo G, et al. Identification of oxidative stress and Toll-like receptor 4 signaling as a key pathway of acute lung injury. Cell. (2008) 133:235–49. doi: 10.1016/j.cell.2008.02.043
126. Patel MC, Shirey KA, Boukhvalova MS, Vogel SN, Blanco JCG. Serum high-mobility-group box 1 as a biomarker and a therapeutic target during respiratory virus infections. mBio. (2018) 9:e00246–18. doi: 10.1128/mBio.00246-18
127. Shirey KA, Lai W, Scott AJ, Lipsky M, Mistry P, Pletneva LM, et al. The TLR4 antagonist Eritoran protects mice from lethal influenza infection. Nature. (2013) 497:498–502. doi: 10.1038/nature12118
128. Nhu QM, Shirey K, Teijaro JR, Farber DL, Netzel-Arnett S, Antalis TM, et al. Novel signaling interactions between proteinase-activated receptor 2 and Toll-like receptors in vitro and in vivo. Mucosal Immunol. (2010) 3:29–39. doi: 10.1038/mi.2009.120
129. Perrin-Cocon L, Aublin-Gex A, Sestito SE, Shirey KA, Patel MC, Andre P, et al. TLR4 antagonist FP7 inhibits LPS-induced cytokine production and glycolytic reprogramming in dendritic cells, and protects mice from lethal influenza infection. Sci Rep. (2017) 7:40791. doi: 10.1038/srep40791
130. Monguio-Tortajada M, Franquesa M, Sarrias MR, Borras FE. Low doses of LPS exacerbate the inflammatory response and trigger death on TLR3-primed human monocytes. Cell Death Dis. (2018) 9:499. doi: 10.1038/s41419-018-0520-2
131. Bender A, Sprenger H, Gong JH, Henke A, Bolte G, Spengler HP, et al. The potentiating effect of LPS on tumor necrosis factor-alpha production by influenza A virus-infected macrophages. Immunobiology. (1993) 187:357–71. doi: 10.1016/S0171-2985(11)80350-5
132. Peschke T, Bender A, Nain M, Gemsa D. Role of macrophage cytokines in influenza A virus infections. Immunobiology. (1993) 189:340–55. doi: 10.1016/S0171-2985(11)80365-7
133. Sohn KM, Lee SG, Kim HJ, Cheon S, Jeong H, Lee J, et al. COVID-19 patients upregulate toll-like receptor 4-mediated inflammatory signaling that mimics bacterial sepsis. J Korean Med Sci. (2020) 35:e343. doi: 10.3346/jkms.2020.35.e343
134. Xie XH, Law HK, Wang LJ Li X, Yang XQ, Liu EM. Lipopolysaccharide induces IL-6 production in respiratory syncytial virus-infected airway epithelial cells through the toll-like receptor 4 signaling pathway. Pediatr Res. (2009) 65:156–62. doi: 10.1203/PDR.0b013e318191f5c6
135. Herold T, Jurinovic V, Arnreich C, Lipworth BJ, Hellmuth JC, von Bergwelt-Baildon M, et al. Elevated levels of IL-6 and CRP predict the need for mechanical ventilation in COVID-19. J Allergy Clin Immunol. (2020) 146:128–36 e4. doi: 10.1016/j.jaci.2020.05.008
136. Luo M, Liu J, Jiang W, Yue S, Liu H, Wei S. IL-6 and CD8+ T cell counts combined are an early predictor of in-hospital mortality of patients with COVID-19. JCI Insight. (2020) 5:1–11. doi: 10.1172/jci.insight.139024
Keywords: COVID19, aging, diabetes, gut microbiota, hypertension, SARS-CoV-2
Citation: Magalhães NS, Savino W, Silva PMR, Martins MA and Carvalho VF (2021) Gut Microbiota Dysbiosis Is a Crucial Player for the Poor Outcomes for COVID-19 in Elderly, Diabetic and Hypertensive Patients. Front. Med. 8:644751. doi: 10.3389/fmed.2021.644751
Received: 21 December 2020; Accepted: 20 July 2021;
Published: 11 August 2021.
Edited by:
Hu Zhang, Sichuan University, ChinaReviewed by:
Yuji Naito, Kyoto Prefectural University of Medicine, JapanJose A. Uranga, Rey Juan Carlos University, Spain
Copyright © 2021 Magalhães, Savino, Silva, Martins and Carvalho. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Vinicius Frias Carvalho, viniciusfrias@hotmail.com
 Nathalia Santos Magalhães
Nathalia Santos Magalhães Wilson Savino
Wilson Savino Patrícia Machado Rodrigues Silva
Patrícia Machado Rodrigues Silva Marco Aurélio Martins
Marco Aurélio Martins Vinicius Frias Carvalho
Vinicius Frias Carvalho