Clinical and Biological Markers in Hypereosinophilic Syndromes
- Human Eosinophil Section, Laboratory of Parasitic Diseases, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Bethesda, MD, United States
Hypereosinophilic syndromes (HES) are rare, heterogeneous syndromes characterized by markedly elevated eosinophil counts in the blood and/or tissue and evidence of eosinophil-associated pathology. Although parasitic infections, drug hypersensitivity, and other disorders of defined etiology can present as HES (associated HES), treatment is directed at the underlying cause rather than the eosinophilia itself. A number of additional subtypes of HES have been described, based on clinical and laboratory features. These include (1) myeloid HES—a primary disorder of the myeloid lineage, (2) lymphocytic variant HES—eosinophilia driven by aberrant or clonal lymphocytes secreting eosinophil-promoting cytokines, (3) overlap HES—eosinophilia restricted to a single organ or organ system, (4) familial eosinophilia—a rare inherited form of HES, and (5) idiopathic HES. Since clinical manifestations, response to therapy, and prognosis all differ between HES subtypes, this review will focus on clinical and biological markers that serve as markers of disease activity in HES (excluding associated HES), including those that are likely to be useful only in specific clinical subtypes.
Introduction
Hypereosinophilic syndromes (HES) are defined by the presence of hypereosinophilia [absolute eosinophil count (AEC) > 1,500/μL or marked tissue eosinophilia] and eosinophil-associated clinical manifestations. Various clinical subtypes of HES have been described based on the etiology of the eosinophilia (primary, secondary, or unknown) and clinical features (systemic or organ-restricted) (1). Although HES can occur in the context of defined disorders, such as drug hypersensitivity, helminth infection, and neoplasia, for which specific treatment of the underlying secondary cause leads to resolution of the eosinophilia (associated HES), for the purposes of this review, HES refers to all clinical subtypes of HES with the exception of associated HES.
The development of standardized clinical assessments of disease activity, such as patient-reported outcomes (PROs) and clinician-reported outcomes (ClinROs), that can be used to guide treatment and serve as clinical trial endpoints has been complicated in HES due to the heterogeneity of disease across HES subtypes and organ systems and the rarity of the disorder itself. Although significant progress has been made in the development of these tools in organ-restricted eosinophilic disorders, such as eosinophilic esophagitis (EoE) (2–5), these subtype-specific PROs and ClinROs are not broadly applicable to the overall HES population. This has, in turn, hampered the development of surrogate markers of disease activity in HES.
Given the central role of eosinophils in HES, quantification of eosinophil numbers in the blood or tissue would seem the most logical method to monitor disease activity in HES and is, in fact, the most common biomarker used in clinical practice. Despite this, AEC has not been widely accepted as a surrogate of disease activity in clinical trials of HES, particularly those involving novel therapies that specifically target eosinophils but may or may not affect clinical outcomes. Clearly, additional biological markers are needed. This review is divided into two parts. The first section will focus on data pertaining to biomarkers related to eosinophilia and eosinophil activation as general indicators of disease activity in HES. This will be followed by a discussion of biomarkers relevant to selected subtypes of HES, but unlikely to be generalizable to HES as a whole.
General Biomarkers of Disease Activity in HES
Eosinophils are characterized by the presence of eosin-avid secondary granules containing cationic granule proteins [major basic protein (MBP), eosinophil cationic protein (ECP), eosinophil peroxidase (EPO), and eosinophil-derived neurotoxin (EDN)] and a wide array of cytokines and chemokines. When released into the tissues by activated eosinophils, these mediators, together with reactive oxygen species and lipid mediators, can lead to tissue damage and the end organ manifestations of HES. As mentioned earlier, a major controversy in clinical trial endpoint design in HES has been whether reduction in AEC is an appropriate surrogate marker of disease activity. The fact that some individuals with hypereosinophilia (AEC > 1,500/μL) are asymptomatic and do not develop end organ manifestations (6) has been cited as evidence that biomarkers of eosinophil activation or tissue infiltration might be more useful in this regard. Available data addressing this question are summarized below.
Absolute Eosinophil Count
The association between elevated peripheral eosinophil counts and clinical pathology was first noted at the turn of the century by Loeffler who described a characteristic form of endomyocardial fibrosis in association with blood eosinophilia (7). Subsequent case series, using persistent AEC > 1,500/μL as a defining criterion for HES, noted an association between extremely elevated AEC (white blood cell counts >100,000/μL) and poor prognosis (8, 9). Consistent with these findings, patients with PDGFRA-positive myeloid neoplasm, one of the most aggressive forms of HES, have higher AECs than patients with other clinical subtypes of HES and dramatic resolution of clinical manifestations following normalization of the AEC with imatinib therapy (10).
Despite these findings and the large body of circumstantial evidence from case reports, case series and clinical practice documenting an association between the resolution of clinical manifestations of HES and normalization of the AEC, assessment of the AEC as a surrogate marker of disease activity has not been studied directly in the context of clinical trials to date. That said, the efficacy of mepolizumab as a steroid-sparing agent was associated with reduction of AEC in two placebo-controlled, double-blind trials in HES [one in subjects with steroid-responsive HES (11) and the second in subjects with eosinophilic granulomatosis with polyangiitis (EGPA) (12)], suggesting that AEC is a useful marker of disease activity. The results of ongoing and recently completed trials (NCT02130882; NCT02101138) in HES using agents that selectively target eosinophils should provide additional support for the utility of the AEC as a biomarker of response.
Tissue Eosinophilia
Although tissue eosinophilia would seem to be a more specific indicator of disease activity in HES, the utility of eosinophil quantification in tissue biopsies to monitor disease activity is hampered by the difficulty in obtaining samples, the patchy nature of eosinophilic tissue infiltration, and the fact that intact eosinophils may be absent despite clear evidence of their involvement by immunohistochemical staining for eosinophil granule proteins (EGPs) (13–16). To date, the best data associating tissue eosinophil numbers with clinical symptomatology come from EoE where the numbers of eosinophils in normal tissue have been defined (17), and suppression of tissue eosinophil counts has been associated with improved long-term prognosis (18). Unfortunately, despite encouraging data from a small open-label trial (19), randomized placebo-controlled trials using anti-IL-5 antibody therapy (mepolizumab and reslizumab) have not demonstrated an association between reduction in tissue eosinophilia and improved symptoms (20–22). Potential explanations for the lack of symptomatic improvement include incomplete depletion of tissue eosinophilia, involvement of other cell types and/or structural changes due to fibrosis and remodeling that may require a longer time frame for resolution.
Eosinophil Granule Proteins
Released during eosinophil activation and deposited in tissue in sites of eosinophilic inflammation, EGPs are attractive candidate biomarkers for the monitoring of disease activity in HES (23). They can be detected and quantified in the blood (24–26), body fluids (27), and tissue (13–15, 28–31) using various immunoassays, and blood and/or body fluid levels have been shown to correlate with tissue deposition of EGP in a wide range of HES, including EoE in the absence of peripheral eosinophilia (24).
There are a several biologically relevant differences between EDN, EPO, MBP, and ECP. MBP is the predominant protein in the core of the eosinophil secondary granule. It exists as two highly basic homologs, MBP-1 and MBP-2 (26), both of which circulate as neutral pH pro-proteins. Of note, most immunoassays do not distinguish between pro-MBP and MBP. Whereas EPO is quite specific for the eosinophil lineage, MBP-1, EDN, and ECP are also present in neutrophils and/or basophils albeit at lower levels (32, 33). This does not appear to affect their ability to be used as a proxy for eosinophil-associated tissue pathology in most settings but deserves mention.
Immunohistochemical staining of tissue for EGP has been extremely useful in clarifying the role of eosinophils in the pathogenesis of HES when intact eosinophils are not detectable. Moreover, EGP staining has been shown to correlate with disease activity in some settings. For example, in one study, serial skin biopsies from patients with episodic angioedema with eosinophilia demonstrated EGP staining only when symptoms were present (15, 34). Unfortunately, the utility of EGP tissue staining as a biomarker of disease activity is limited by the need for serial tissue sampling. To address this issue in EoE, a number of novel and less invasive techniques have been developed. These include the esophageal string test (35) and the cytosponge (36). Both techniques involve swallowing a string (in the case of the cytosponge, this is attached to a gelatin capsule containing a mesh) from which EGP can be eluted and quantified. Good correlation between eluted EGP levels and immunohistochemical staining of matched biopsies has been confirmed for both techniques (37). Finally, ultrasound visualization of granule protein density using MBP-1 labeled-insulin particles has been demonstrated in ex vivo monkey esophagi and may ultimately provide a third non-invasive tool for the measurement of tissue EGP in EoE (38). The applicability of these or similar techniques to other tissues remains to be seen.
The utility of measuring circulating levels of EGPs to monitor disease activity in HES has been somewhat controversial in large part due to the lack of standardization of sample collection (eosinophil lysis could lead to falsely elevated levels) and differing assay parameters between studies. Nevertheless, there are some data to suggest that circulating EGP levels have value in the monitoring of disease activity in HES. An interesting observation in this regard has been the association of elevated EDN and EPO levels with clinical manifestations rather than peak eosinophil count in patients with episodic angioedema and eosinophilia (24). A similar association between clinical disease and elevated EDN levels was seen in a study comparing subjects with asymptomatic familial eosinophilia to subjects with active HES (39).
Data from clinical treatment trials have been more difficult to interpret. Whereas decreases in serum EDN levels were reported in subjects who received active drug in a placebo-controlled trial of mepolizumab in HES (11) and ECP levels decreased in response to mepolizumab therapy in three patients with HES and eosinophilic dermatitis (40), AEC also decreased in both studies making it difficult to assess the added benefit of measuring EGP levels. Moreover, in a recent study of non-invasive biomarkers of EoE, AEC, but not serum levels of ECP, was predictive of residual disease following topical steroid therapy (41).
Measurement of EGP in body fluids, such as urine, that do not normally contain eosinophils, has the theoretical advantage of eliminating false positive results due to eosinophil lysis. Although there are no studies examining urine levels of EGP in HES to date, several small studies in atopic dermatitis (42, 43) demonstrated a correlation between urine levels of ECP and clinical disease severity. By contrast, no such relationship was noted in children with asthma (44).
Eosinophil Surface Receptors
A wide variety of eosinophil surface markers are reported to be up- or downregulated on activated eosinophils (23, 45), Many of these, including IL-5Rα, CD69, and CD44, have been shown to have altered expression on eosinophils from patients with HES, but also in patients with HEUS (39). Despite this, there are little longitudinal data assessing changes in expression of these activation markers in response to therapy in patients with HES. In a single study in EoE, expression of activation markers on blood eosinophils was unchanged by topical steroid therapy, although the effect of therapy on esophageal eosinophilia was not reported (46).
Serum Cytokines, Chemokines, and Soluble Receptors
Despite its clear role in the production, activation and regulation of eosinophils, IL-5 has been disappointing as a biomarker of disease activity in HES. Although IL-5 levels correlate with AEC overall, serum IL-5 is undetectable in some patients with untreated HES [Figure 1, unpublished data from Ref. (47)]. In this regard, serum IL-5 levels do not contribute additional information when the AEC is known. In addition, increased serum IL-5 levels have been reported in the setting of clinical and hematologic remission following administration of several different biologics designed to target eosinophils, including mepolizumab and benralizumab (48, 49). The reasons for this are likely multifactorial and include measurement of IL-5/anti-IL-5 immune complexes (mepolizumab) and antibody blocking of IL-5 binding to its receptor (benralizumab). Soluble IL-5R is measurable in the serum of most, if not all, patients with HES, and levels are correlated with serum IL-5 levels (47). Whether this would provide a better biomarker of disease activity, owing to its reliable detection in serum in contrast to IL-5, remains to be seen. Finally, a number of studies have looked at other serum cytokines and chemokines as biomarkers of disease activity in HES. Of these, mediators of potential interest have been identified mostly in EGPA and include IL-25 (50), serum CCL17/thymus and activation-regulated chemokine (TARC) levels (51, 52), and CCL26/eotaxin-3 (53, 54). Interestingly, despite tissue data implicating CCL26/eotaxin-3 in the pathogenesis of EoE, serum levels of these mediators were not increased in EoE patients and were not altered by therapy (55). Finally, although some authors have reported elevated IL-3 in the plasma of patients with eosinophilia in conjunction with intracellular staining in CD8+ T cells (56), IL-3 is not universally detected in serum (57) of patients with HES, and the role of IL-3 as a biomarker in HES remains to be explored.
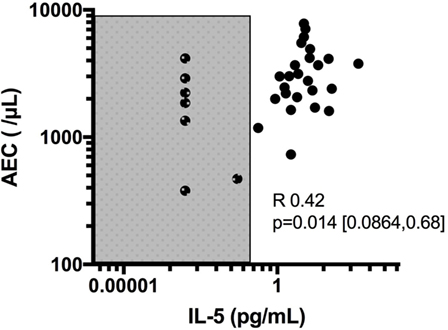
Figure 1. Correlation of absolute eosinophil count (AEC) with IL-5 levels and demonstration of subjects with undetectable IL-5 despite elevated eosinophil counts. Shaded area denotes values below the limit of detection of IL-5 (0.1 pg/mL) in the assay used.
Omics
An exciting advance in biomarker development has been the use of molecular profiling techniques to identify patterns of expression that can be used to follow disease activity. This approach has been used successfully in EoE, where patterns of gene expression in esophageal biopsies have led to the development of an EoE molecular diagnostic panel (EDP) (58, 59) which is further discussed in the accompanying review, and a microRNA signature (60) that correlate with disease activity and response to therapy.
Clinical Subtype-Specific Biomarkers in HES
Over the past decade, it has become increasingly apparent that there are distinct clinical subtypes of HES that differ in their etiologies, clinical manifestations, and responses to therapy. These are more extensively discussed in the companion article by Lefevre (61). Whereas the biomarkers discussed herein are relevant to eosinophilic disorders and HES in general, additional markers have been described that have utility restricted to a particular HES clinical subtype.
Lymphocytic Variant HES (LHES)
LHES is defined by the presence of a clonal or aberrant phenotypic T cell that secretes type 2 cytokines driving the eosinophilia and elevated serum IgE levels seen in this clinical variant (62, 63). Whereas the most common aberrant immunophenotype is CD3−CD4+, various aberrant immunophenotypes have been described (64), and some patients have cytokine-secreting clonal T-cell populations despite an apparently normal immunophenotype. Dermatologic manifestations, including angioedema, nodules, eczematous dermatitis, and erythroderma, are common in patients with LHES (65), and aberrant T cells can often be detected in skin biopsies from affected areas (66). Patients with LHES are often glucocorticoid responsive but typically require moderately high doses (67). Consequently, glucocorticoid-sparing agents with effects on T cells, such as interferon-alpha and cyclosporine, are frequently used. Although LHES is considered a benign lymphoproliferative disorder, a small proportion of LHES patients eventually develop a lymphoid malignancy, often heralded by expansion of the aberrant clonal T-cell population (62, 68). Conversely, regression of the aberrant T-cell population can be seen in response to effective therapy (Figure 2). Elevations in serum CCL17/TARC are more frequent in patients with LHES (69, 70), but information is lacking on the use of CCL17/TARC levels to monitor disease activity.
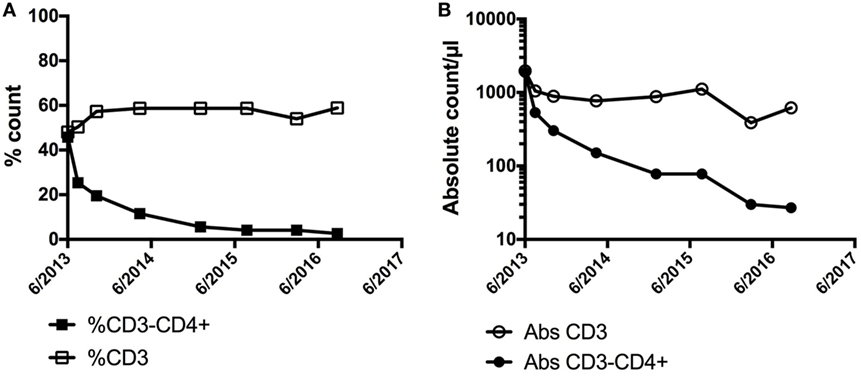
Figure 2. Demonstration of a preferential decline in the (A) % of CD3−CD4+ and (B) absolute count of CD3−CD4+ cells in a patient with LHES after initiation of interferon-alpha with associated clinical improvement in skin involvement. No significant change was noted in the % or absolute counts of CD3+ cells overall.
Myeloid HES (MHES)
Myeloid HES refers to the subgroup of patients with HES in the setting of a primary myeloid disorder. Although most of these patients have detectable molecular abnormalities (most commonly the fusion gene FIP1L1-PDGFRA), others have a similar clinical phenotype of unknown cause. Clinical and laboratory features associated with MHES include dysplastic eosinophils, anemia and/or thrombocytopenia, elevated serum tryptase and B12 levels, and bone marrow features suggestive of a myeloid neoplasm (10). Before the availability of imatinib (a tyrosine kinase inhibitor with activity against PDGFRA and PDGFRB), mortality rates in patients with MHES were extremely high, due primarily to endomyocardial fibrosis and thromboembolic events. Currently, remission rates on imatinib therapy approach 100% in patients with PDGFR-associated disease and up to 50% in patients with other forms of MHES.
Although the AEC normalizes with effective therapy in MHES and can be used to monitor disease activity, data from chronic myelogenous leukemia and drug interruption trials in PDGFRA-associated HES suggest that molecular monitoring is preferable when possible since molecular relapse may precede hematologic (and clinical) relapse by several months (71). This is particularly important in view of recent data demonstrating sustained remission after imatinib discontinuation in some patients (72–74).
Overlap HES
Overlap HES includes single organ and/or defined disorders that are characterized by eosinophilia and eosinophil-associated pathogenesis, including eosinophilic gastrointestinal disorders and EGPA. These disorders have distinct clinical presentations and complications and, for this reason, are often approached differently than other forms of HES. Although potential biomarkers for these conditions include the previously discussed general markers of eosinophilia and eosinophil activation, additional disease-specific issues are discussed below.
Eosinophilic Esophagitis
Despite significant advances in the development of biomarkers for EoE, the lack of correlation between the number of eosinophils in tissue and clinical symptoms remains a problem, particularly with regard to clinical trial design. Consequently, there has been increasing interest in the development of additional objective measures to assess improvement of long-term sequelae. One such tool is EndoFLIP® (endolumenal functional lumen imaging probe), an inflatable balloon that measures the cross-sectional area and intraluminal pressure of the esophagus while under distension (as if a solid bolus was present). Using this technique, reduced distensibility was demonstrated in patients with dysphagia or a history of impaction as compared with healthy controls (75). Interestingly, decreased distensibility did not correlate with mucosal eosinophilia (75) but did correlate with ring severity and impactions (76).
Eosinophilic Granulomatosis with Polyangiitis
Several studies have examined the use of standard laboratory markers of inflammation, including erythrocyte sedimentation rate and C-reactive protein, in EGPA. Although these markers have been shown to be elevated in active disease at the population level, a longitudinal study using a validated ClinRO as the gold standard found that they were affected by disease severity and treatment status, limiting their success in predicting disease activity and relapse at the individual patient level (77).
Conclusion
With the advent of targeted therapies that reduce blood eosinophilia but may have varied effects on tissue eosinophilia and eosinophil-related end organ manifestations, there is an increasing need for reliable, non-invasive markers of disease activity in HES. Although some progress has been made in select subtypes of HES, including EoE and PDGFRA-positive myeloid neoplasm, generally applicable, validated biomarkers in HES are lacking. This is likely due, at least in part, to the heterogeneity of clinical manifestations, lack of understanding of the factors driving the varied HES subtypes and paucity of longitudinal studies addressing this issue. Although not validated as a surrogate marker for disease activity in HES, the AEC remains a key laboratory test that is used by experts to assess disease activity and response to therapy in all HES subtypes. Soluble mediators that correlate with active disease, including serum levels of EGP, have been identified, although increased predictive value compared to the AEC has not been demonstrated in most cases and none have been validated in prospective double-blind clinical trials to date. Finally, tissue-based markers, including tissue eosinophilia, granule protein deposition, and transcriptome analysis, have demonstrated utility in monitoring disease activity in some settings but are limited by the availability of appropriate tissue samples. While the development of novel non-invasive sampling methods and global approaches to biomarker discovery (“omics”) are exciting, carefully designed clinical trials are clearly needed to validate existing and novel biomarkers for accurate monitoring and assessment of therapeutic interventions.
Ethics Statement
The data presented in this manuscript were collected under research protocol NCT00001406. This study was carried out in accordance with the recommendations of the Belmont Report with written informed consent from all subjects. All subjects gave written informed consent in accordance with the Declaration of Helsinki. The protocol was approved by the NIAID Institutional Review Board.
Author Contributions
AK, PK, and MM each contributed to the writing of the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer PA declared a past collaboration with the authors to the handling Editor.
Funding
This study was funded by the Division of Intramural Research, NIAID, NIH.
References
1. Klion AD. How I treat hypereosinophilic syndromes. Blood (2015) 126:1069–77. doi:10.1182/blood-2014-11-551614
2. Martin LJ, Franciosi JP, Collins MH, Abonia JP, Lee JJ, Hommel KA, et al. Pediatric Eosinophilic Esophagitis Symptom Scores (PEESS v2.0) identify histologic and molecular correlates of the key clinical features of disease. J Allergy Clin Immunol (2015) 135:1519–28.e8. doi:10.1016/j.jaci.2015.03.004
3. Franciosi JP, Hommel KA, Bendo CB, King EC, Collins MH, Eby MD, et al. PedsQL eosinophilic esophagitis module: feasibility, reliability, and validity. J Pediatr Gastroenterol Nutr (2013) 57:57–66. doi:10.1097/MPG.0b013e31828f1fd2
4. Safroneeva E, Coslovsky M, Kuehni CE, Zwahlen M, Haas NA, Panczak R, et al. Eosinophilic oesophagitis: relationship of quality of life with clinical, endoscopic and histological activity. Aliment Pharmacol Ther (2015) 42:1000–10. doi:10.1111/apt.13370
5. Safroneeva E, Straumann A, Coslovsky M, Zwahlen M, Kuehni CE, Panczak R, et al. Symptoms have modest accuracy in detecting endoscopic and histologic remission in adults with eosinophilic esophagitis. Gastroenterology (2016) 150:581–90.e4. doi:10.1053/j.gastro.2015.11.004
6. Chen Y-YK, Khoury P, Ware JM, Holland-Thomas NC, Stoddard JL, Gurprasad S, et al. Marked and persistent eosinophilia in the absence of clinical manifestations. J Allergy Clin Immunol (2014) 133:1195–202. doi:10.1016/j.jaci.2013.06.037
8. Fauci AS, Harley JB, Roberts WC, Ferrans VJ, Gralnick HR, Bjornson BH. NIH conference. The idiopathic hypereosinophilic syndrome. Clinical, pathophysiologic, and therapeutic considerations. Ann Intern Med (1982) 97:78–92. doi:10.7326/0003-4819-97-1-78
9. Chusid MJ, Dale DC, West BC, Wolff SM. The hypereosinophilic syndrome: analysis of fourteen cases with review of the literature. Medicine (1975) 54:1–27. doi:10.1097/00005792-197501000-00001
10. Klion AD, Noel P, Akin C, Law MA, Gilliland DG, Cools J, et al. Elevated serum tryptase levels identify a subset of patients with a myeloproliferative variant of idiopathic hypereosinophilic syndrome associated with tissue fibrosis, poor prognosis, and imatinib responsiveness. Blood (2003) 101:4660–6. doi:10.1182/blood-2003-01-0006
11. Rothenberg ME, Klion AD, Roufosse FE, Kahn JE, Weller PF, Simon H-U, et al. Treatment of patients with the hypereosinophilic syndrome with mepolizumab. N Engl J Med (2008) 358:1215–28. doi:10.1056/NEJMoa070812
12. Wechsler ME, Akuthota P, Jayne D, Khoury P, Klion A, Langford CA, et al. Mepolizumab or placebo for eosinophilic granulomatosis with polyangiitis. N Engl J Med (2017) 376:1921–32. doi:10.1056/NEJMoa1702079
13. Tai PC, Ackerman SJ, Spry CJ, Dunnette S, Olsen EG, Gleich GJ. Deposits of eosinophil granule proteins in cardiac tissues of patients with eosinophilic endomyocardial disease. Lancet (1987) 1:643–7. doi:10.1016/S0140-6736(87)90412-0
14. Wichman A, Buchthal F, Pezeshkpour GH, Fauci AS. Peripheral neuropathy in hypereosinophilic syndrome. Neurology (1985) 35:1140–5. doi:10.1212/WNL.35.8.1140
15. Gleich GJ, Schroeter AL, Marcoux JP, Sachs MI, O’Connell EJ, Kohler PF. Episodic angioedema associated with eosinophilia. N Engl J Med (1984) 310:1621–6. doi:10.1056/NEJM198406213102501
16. Peterson KA, Cobell WJ, Clayton FC, Krishnamurthy C, Ying J, Pease LF, et al. Extracellular eosinophil granule protein deposition in ringed esophagus with sparse eosinophils. Dig Dis Sci (2015) 60:2646–53. doi:10.1007/s10620-015-3665-1
17. Furuta GT, Liacouras CA, Collins MH, Gupta SK, Justinich C, Putnam PE, et al. Eosinophilic esophagitis in children and adults: a systematic review and consensus recommendations for diagnosis and treatment. Gastroenterology (2007) 133:1342–63. doi:10.1053/j.gastro.2007.08.017
18. Greuter T, Bussmann C, Safroneeva E, Schoepfer AM, Biedermann L, Vavricka SR, et al. Long-term treatment of eosinophilic esophagitis with swallowed topical corticosteroids: development and evaluation of a therapeutic concept. Am J Gastroenterol (2017) 112:1527–35. doi:10.1038/ajg.2017.202
19. Stein ML, Collins MH, Villanueva JM, Kushner JP, Putnam PE, Buckmeier BK, et al. Anti-IL-5 (mepolizumab) therapy for eosinophilic esophagitis. J Allergy Clin Immunol (2006) 118:1312–9. doi:10.1016/j.jaci.2006.09.007
20. Spergel JM, Rothenberg ME, Collins MH, Furuta GT, Markowitz JE, Fuchs G, et al. Reslizumab in children and adolescents with eosinophilic esophagitis: results of a double-blind, randomized, placebo-controlled trial. J Allergy Clin Immunol (2012) 129:456–63, 463.e1–3. doi:10.1016/j.jaci.2011.11.044
21. Assa’ad AH, Gupta SK, Collins MH, Thomson M, Heath AT, Smith DA, et al. An antibody against IL-5 reduces numbers of esophageal intraepithelial eosinophils in children with eosinophilic esophagitis. Gastroenterology (2011) 141:1593–604. doi:10.1053/j.gastro.2011.07.044
22. Straumann A, Conus S, Grzonka P, Kita H, Kephart G, Bussmann C, et al. Anti-interleukin-5 antibody treatment (mepolizumab) in active eosinophilic oesophagitis: a randomised, placebo-controlled, double-blind trial. Gut (2010) 59:21–30. doi:10.1136/gut.2009.178558
23. Metcalfe DD, Pawankar R, Ackerman SJ, Akin C, Clayton F, Falcone FH, et al. Biomarkers of the involvement of mast cells, basophils and eosinophils in asthma and allergic diseases. World Allergy Organ J (2016) 9:7. doi:10.1186/s40413-016-0094-3
24. Makiya MA, Herrick JA, Khoury P, Prussin CP, Nutman TB, Klion AD. Development of a suspension array assay in multiplex for the simultaneous measurement of serum levels of four eosinophil granule proteins. J Immunol Methods (2014) 411:11–22. doi:10.1016/j.jim.2014.05.020
25. Kim H-R, Jun C-D, Lee Y-J, Yang S-H, Jeong E-T, Park S-D, et al. Levels of circulating IL-33 and eosinophil cationic protein in patients with hypereosinophilia or pulmonary eosinophilia. J Allergy Clin Immunol (2010) 126:880–2.e6. doi:10.1016/j.jaci.2010.06.038
26. Plager DA, Loegering DA, Checkel JL, Tang J, Kephart GM, Caffes PL, et al. Major basic protein homolog (MBP2): a specific human eosinophil marker. J Immunol (2006) 177:7340–5. doi:10.4049/jimmunol.177.10.7340
27. Ochkur SI, Kim JD, Protheroe CA, Colbert D, Condjella RM, Bersoux S, et al. A sensitive high throughput ELISA for human eosinophil peroxidase: a specific assay to quantify eosinophil degranulation from patient-derived sources. J Immunol Methods (2012) 384:10–20. doi:10.1016/j.jim.2012.06.011
28. Saffari H, Hoffman LH, Peterson KA, Fang JC, Leiferman KM, Pease LF, et al. Electron microscopy elucidates eosinophil degranulation patterns in patients with eosinophilic esophagitis. J Allergy Clin Immunol (2014) 133:1728–34.e1. doi:10.1016/j.jaci.2013.11.024
29. Armengot M, Garín L, Carda C. Eosinophil degranulation patterns in nasal polyposis: an ultrastructural study. Am J Rhinol Allergy (2009) 23:466–70. doi:10.2500/ajra.2009.23.3357
30. Wright BL, Leiferman KM, Gleich GJ. Eosinophil granule protein localization in eosinophilic endomyocardial disease. N Engl J Med (2011) 365:187–8. doi:10.1056/NEJMc1103005
31. Dellon ES, Speck O, Woodward K, Covey S, Rusin S, Gebhart JH, et al. Markers of eosinophilic inflammation for diagnosis of eosinophilic esophagitis and proton pump inhibitor-responsive esophageal eosinophilia: a prospective study. Clin Gastroenterol Hepatol (2014) 12:2015–22. doi:10.1016/j.cgh.2014.06.019
32. Sur S, Glitz DG, Kita H, Kujawa SM, Peterson EA, Weiler DA, et al. Localization of eosinophil-derived neurotoxin and eosinophil cationic protein in neutrophilic leukocytes. J Leukoc Biol (1998) 63:715–22.
33. Abu-Ghazaleh RI, Dunnette SL, Loegering DA, Checkel JL, Kita H, Thomas LL, et al. Eosinophil granule proteins in peripheral blood granulocytes. J Leukoc Biol (1992) 52:611–8.
34. Khoury P, Herold J, Alpaugh A, Dinerman E, Holland-Thomas N, Stoddard J, et al. Episodic angioedema with eosinophilia (Gleich syndrome) is a multilineage cell cycling disorder. Haematologica (2015) 100:300–7. doi:10.3324/haematol.2013.091264
35. Furuta GT, Kagalwalla AF, Lee JJ, Alumkal P, Maybruck BT, Fillon S, et al. The oesophageal string test: a novel, minimally invasive method measures mucosal inflammation in eosinophilic oesophagitis. Gut (2013) 62:1395–405. doi:10.1136/gutjnl-2012-303171
36. Katzka DA, Geno DM, Ravi A, Smyrk TC, Lao-Sirieix P, Miremadi A, et al. Accuracy, safety, and tolerability of tissue collection by cytosponge vs endoscopy for evaluation of eosinophilic esophagitis. Clin Gastroenterol Hepatol (2015) 13:77–83.e2. doi:10.1016/j.cgh.2014.06.026
37. Katzka DA, Smyrk TC, Alexander JA, Geno DM, Beitia RA, Chang AO, et al. Accuracy and safety of the cytosponge for assessing histologic activity in eosinophilic esophagitis: a two-center study. Am J Gastroenterol (2017) 112:1538–44. doi:10.1038/ajg.2017.244
38. Saffari H, Kennedy A, Peterson KA, Gleich GJ, Pease LF. Non-invasive ultrasound to identify eosinophil granule proteins in eosinophilic esophagitis. Ultrasound Med Biol (2015) 41:884–9. doi:10.1016/j.ultrasmedbio.2014.09.017
39. Klion AD, Law MA, Riemenschneider W, McMaster ML, Brown MR, Horne M, et al. Familial eosinophilia: a benign disorder? Blood (2004) 103:4050–5. doi:10.1182/blood-2003-11-3850
40. Plötz S-G, Simon H-U, Darsow U, Simon D, Vassina E, Yousefi S, et al. Use of an anti-interleukin-5 antibody in the hypereosinophilic syndrome with eosinophilic dermatitis. N Engl J Med (2003) 349:2334–9. doi:10.1056/NEJMoa031261
41. Min SB, Nylund CM, Baker TP, Ally M, Reinhardt B, Chen Y-J, et al. Longitudinal evaluation of noninvasive biomarkers for eosinophilic esophagitis. J Clin Gastroenterol (2017) 51:127–35. doi:10.1097/MCG.0000000000000621
42. Breuer K, Kapp A, Werfel T. Urine eosinophil protein X (EPX) is an in vitro parameter of inflammation in atopic dermatitis of the adult age. Allergy (2001) 56:780–4. doi:10.1034/j.1398-9995.2001.056008780.x
43. Pucci N, Lombardi E, Novembre E, Farina S, Bernardini R, Rossi E, et al. Urinary eosinophil protein X and serum eosinophil cationic protein in infants and young children with atopic dermatitis: correlation with disease activity. J Allergy Clin Immunol (2000) 105:353–7. doi:10.1016/S0091-6749(00)90087-3
44. Lönnkvist K, Hellman C, Lundahl J, Halldén G, Hedlin G. Eosinophil markers in blood, serum, and urine for monitoring the clinical course in childhood asthma: impact of budesonide treatment and withdrawal. J Allergy Clin Immunol (2001) 107:812–7. doi:10.1067/mai.2001.114246
45. Bochner BS. Systemic activation of basophils and eosinophils: markers and consequences. J Allergy Clin Immunol (2000) 106:S292–302. doi:10.1067/mai.2000.110164
46. Lingblom C, Bergquist H, Johnsson M, Sundström P, Quiding-Järbrink M, Bove M, et al. Topical corticosteroids do not revert the activated phenotype of eosinophils in eosinophilic esophagitis but decrease surface levels of CD18 resulting in diminished adherence to ICAM-1, ICAM-2, and endothelial cells. Inflammation (2014) 37:1932–44. doi:10.1007/s10753-014-9926-x
47. Wilson TM, Maric I, Shukla J, Brown M, Santos C, Simakova O, et al. IL-5 receptor α levels in patients with marked eosinophilia or mastocytosis. J Allergy Clin Immunol (2011) 128:1086–92.e1–3. doi:10.1016/j.jaci.2011.05.032
48. Stein ML, Villanueva JM, Buckmeier BK, Yamada Y, Filipovich AH, Assa’ad AH, et al. Anti-IL-5 (mepolizumab) therapy reduces eosinophil activation ex vivo and increases IL-5 and IL-5 receptor levels. J Allergy Clin Immunol (2008) 121:1473–83, 1483.e1–4. doi:10.1016/j.jaci.2008.02.033
49. Pham T-H, Damera G, Newbold P, Ranade K. Reductions in eosinophil biomarkers by benralizumab in patients with asthma. Respir Med (2016) 111:21–9. doi:10.1016/j.rmed.2016.01.003
50. Terrier B, Bièche I, Maisonobe T, Laurendeau I, Rosenzwajg M, Kahn J-E, et al. Interleukin-25: a cytokine linking eosinophils and adaptive immunity in Churg-Strauss syndrome. Blood (2010) 116:4523–31. doi:10.1182/blood-2010-02-267542
51. Dallos T, Heiland GR, Strehl J, Karonitsch T, Gross WL, Moosig F, et al. CCL17/thymus and activation-related chemokine in Churg-Strauss syndrome. Arthritis Rheum (2010) 62:3496–503. doi:10.1002/art.27678
52. Khoury P, Zagallo P, Talar-Williams C, Santos CS, Dinerman E, Holland NC, et al. Serum biomarkers are similar in Churg-Strauss syndrome and hypereosinophilic syndrome. Allergy (2012) 67:1149–56. doi:10.1111/j.1398-9995.2012.02873.x
53. Zwerina J, Bach C, Martorana D, Jatzwauk M, Hegasy G, Moosig F, et al. Eotaxin-3 in Churg-Strauss syndrome: a clinical and immunogenetic study. Rheumatology (Oxford) (2011) 50:1823–7. doi:10.1093/rheumatology/keq445
54. Polzer K, Karonitsch T, Neumann T, Eger G, Haberler C, Soleiman A, et al. Eotaxin-3 is involved in Churg-Strauss syndrome – a serum marker closely correlating with disease activity. Rheumatology (Oxford) (2008) 47:804–8. doi:10.1093/rheumatology/ken033
55. Dellon ES, Rusin S, Gebhart JH, Covey S, Higgins LL, Beitia R, et al. Utility of a noninvasive serum biomarker panel for diagnosis and monitoring of eosinophilic esophagitis: a prospective study. Am J Gastroenterol (2015) 110:821–7. doi:10.1038/ajg.2015.57
56. Stoeckle C, Simon HU. CD8(+) T cells producing IL-3 and IL-5 in non-IgE-mediated eosinophilic diseases. Allergy (2013) 68:1622–5. doi:10.1111/all.12311
57. Koike T, Enokihara H, Arimura H, Ninomiya H, Yamashiro K, Tsuruoka N, et al. Serum concentrations of IL-5, GM-CSF, and IL-3 and the production by lymphocytes in various eosinophilia. Am J Hematol (1995) 50:98–102. doi:10.1002/ajh.2830500205
58. Wen T, Stucke EM, Grotjan TM, Kemme KA, Abonia JP, Putnam PE, et al. Molecular diagnosis of eosinophilic esophagitis by gene expression profiling. Gastroenterology (2013) 145:1289–99. doi:10.1053/j.gastro.2013.08.046
59. Wen T, Rothenberg ME. Clinical applications of the eosinophilic esophagitis diagnostic panel. Front Med (2017) 4:108. doi:10.3389/fmed.2017.00108
60. Lu TX, Sherrill JD, Wen T, Plassard AJ, Besse JA, Abonia JP, et al. MicroRNA signature in patients with eosinophilic esophagitis, reversibility with glucocorticoids, and assessment as disease biomarkers. J Allergy Clin Immunol (2012) 129:1064–75.e9. doi:10.1016/j.jaci.2012.01.060
61. Kahn JE, Groh M, Lefèvre G. (A Critical Appraisal of) Classification of Hypereosinophilic Disorders. Front. Med (2017) 4:216
62. Roufosse F, Cogan E, Goldman M. Lymphocytic variant hypereosinophilic syndromes. Immunol Allergy Clin North Am (2007) 27:389–413. doi:10.1016/j.iac.2007.07.002
63. Roufosse F, Schandené L, Sibille C, Willard-Gallo K, Kennes B, Efira A, et al. Clonal Th2 lymphocytes in patients with the idiopathic hypereosinophilic syndrome. Br J Haematol (2000) 109:540–8. doi:10.1046/j.1365-2141.2000.02097.x
64. Simon HU, Plötz SG, Dummer R, Blaser K. Abnormal clones of T cells producing interleukin-5 in idiopathic eosinophilia. N Engl J Med (1999) 341:1112–20. doi:10.1056/NEJM199910073411503
65. Simon HU, Plötz SG, Simon D, Dummer R, Blaser K. Clinical and immunological features of patients with interleukin-5-producing T cell clones and eosinophilia. Int Arch Allergy Immunol (2001) 124:242–5. doi:10.1159/000053723
66. Carruthers MN, Park S, Slack GW, Dalal BI, Skinnider BF, Schaeffer DF, et al. IgG4-related disease and lymphocyte-variant hypereosinophilic syndrome: a comparative case series. Eur J Haematol (2017) 98:378–87. doi:10.1111/ejh.12842
67. Khoury P, Abiodun AO, Holland-Thomas N, Fay MP, Klion AD. Hypereosinophilic syndrome subtype predicts responsiveness to glucocorticoids. J Allergy Clin Immunol Pract (2017). doi:10.1016/j.jaip.2017.06.006
68. Roufosse F, de Leval L, van Krieken H, van Deuren M. Lymphocytic variant hypereosinophilic syndrome progressing to angioimmunoblastic T-cell lymphoma. Leuk Lymphoma (2015) 56:1891–4. doi:10.3109/10428194.2014.976823
69. de Lavareille A, Roufosse F, Schandené L, Stordeur P, Cogan E, Goldman M. Clonal Th2 cells associated with chronic hypereosinophilia: TARC-induced CCR4 down-regulation in vivo. Eur J Immunol (2001) 31:1037–46. doi:10.1002/1521-4141(200104)31:4<1037::AID-IMMU1037>3.0.CO;2-#
70. de Lavareille A, Roufosse F, Schmid-Grendelmeier P, Roumier A-S, Schandené L, Cogan E, et al. High serum thymus and activation-regulated chemokine levels in the lymphocytic variant of the hypereosinophilic syndrome. J Allergy Clin Immunol (2002) 110:476–9. doi:10.1067/mai.2002.127003
71. Klion AD, Robyn J, Maric I, Fu W, Schmid L, Lemery S, et al. Relapse following discontinuation of imatinib mesylate therapy for FIP1L1/PDGFRA-positive chronic eosinophilic leukemia: implications for optimal dosing. Blood (2007) 110:3552–6. doi:10.1182/blood-2007-07-100164
72. Khoury P, Desmond R, Pabon A, Holland-Thomas N, Ware JM, Arthur DC, et al. Clinical features predict responsiveness to imatinib in platelet-derived growth factor receptor-alpha-negative hypereosinophilic syndrome. Allergy (2016) 71:803–10. doi:10.1111/all.12843
73. Helbig G, Soja A, Świderska A, Hus M, Kyrcz-Krzemień S. Imatinib discontinuation for hypereosinophilic syndrome harboring theFIP1L1-PDGFRA transcript. Leuk Lymphoma (2016) 57:708–10. doi:10.3109/10428194.2015.1065983
74. Legrand F, Renneville A, Macintyre E, Mastrilli S, Ackermann F, Cayuela JM, et al. The spectrum of FIP1L1-PDGFRA-associated chronic eosinophilic leukemia: new insights based on a survey of 44 cases. Medicine (2013) 92:e1–9. doi:10.1097/MD.0b013e3182a71eba
75. Kwiatek MA, Hirano I, Kahrilas PJ, Rothe J, Luger D, Pandolfino JE. Mechanical properties of the esophagus in eosinophilic esophagitis. Gastroenterology (2011) 140:82–90. doi:10.1053/j.gastro.2010.09.037
76. Nicodème F, Hirano I, Chen J, Robinson K, Lin Z, Xiao Y, et al. Esophageal distensibility as a measure of disease severity in patients with eosinophilic esophagitis. Clin Gastroenterol Hepatol (2013) 11:1101–7.e1. doi:10.1016/j.cgh.2013.03.020
77. Grayson PC, Monach PA, Pagnoux C, Cuthbertson D, Carette S, Hoffman GS, et al. Value of commonly measured laboratory tests as biomarkers of disease activity and predictors of relapse in eosinophilic granulomatosis with polyangiitis. Rheumatology (Oxford) (2015) 54:1351–9. doi:10.1093/rheumatology/keu427
Keywords: eosinophil, biomarkers, hypereosinophilic syndrome, eosinophilic esophagitis, eosinophilic granulomatosis with polyangiitis, eosinophilia, eosinophilic disorders
Citation: Khoury P, Makiya M and Klion AD (2017) Clinical and Biological Markers in Hypereosinophilic Syndromes. Front. Med. 4:240. doi: 10.3389/fmed.2017.00240
Received: 14 September 2017; Accepted: 13 December 2017;
Published: 22 December 2017
Edited by:
Florence Emmanuelle Roufosse, Université libre de Bruxelles, BelgiumReviewed by:
Hans-Uwe Simon, University of Bern, SwitzerlandAnastasios G. Kriebardis, Technological Educational Institute of Athens, Greece
Praveen Akuthota, University of California, San Diego, United States
Copyright: © 2017 Khoury, Makiya and Klion. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Amy D. Klion, aklion@niaid.nih.gov
 Paneez Khoury
Paneez Khoury Michelle Makiya
Michelle Makiya Amy D. Klion
Amy D. Klion