Diving on damage—the muscle transcriptome of parasitic infested harbor porpoises (Phocoena phocoena) hints at oxidative stress but not hypoxia
- 1Institute for Terrestrial and Aquatic Wildlife Research (ITAW), University of Hanover MedicineHanover Foundation, Büsum, Germany
- 2Institute of Animal Cell and Systems Biology (ICS), University of Hamburg, Hamburg, Germany
The only native cetacean in German waters, the harbor porpoise (Phocoena phocoena), is impacted by numerous pathological lesions in the respiratory tract mainly caused by parasites or bacteria. Although harbor porpoises have been observed to not use their complete lung volume, it has not been studied whether this insufficiency leads to lower oxygen uptake, impaired diving ability, and, ultimately, reduced foraging success. This project aims to analyze whether harbor porpoises developed novel molecular adaptations to compensate impairments in oxygen supply, thus remaining viable and competitive despite the high parasitic load. Here, initial comparative transcriptome RNA sequencing (NextSeq 2000, Illumina) was performed on muscles of harbor porpoises with a respiratory tract considered as healthy and of harbor porpoises that suffered from more severe lesions and parasitic infestations in the respiratory tract. Our findings suggest an elevated response to oxidative stress in the muscles of parasitic infested harbor porpoises compared with that of healthy animals. Higher antioxidant and antiapoptotic gene expression in the muscles of non-healthy harbor porpoises might function as a compensatory effect to enhanced reactive oxygen species production and accumulation in the muscles. Simultaneously enhanced selective proteasomal degradation and myogenesis suggest a tightly controlled, finely tuned switch of the intrinsic muscle response to stress. Lipid metabolism pathways and rate-limiting transcripts involved in glycolysis were upregulated and may uphold muscle energy supply for tissue function and energy-consuming regenerative and biosynthetic processes. These preliminary results hint at a defined response of the muscle to oxidative stress that may be caused by lung tissue with more severe pathological lesions and may indicate a possible adaptation in cetaceans.
Introduction
In the North and Baltic Seas, harbor porpoises (Phocoena phocoena) are increasingly impacted by ever-expanding human activities, leading to an endangered status (Carlén et al., 2021; Nachtsheim et al., 2021). Although disturbances by shipping and underwater noise during feeding and foraging are especially detrimental due to their high metabolic rate and restricted energy storage capacity (Read & Hohn, 1995; Wisniewska et al., 2016; Rojano-Doñate et al., 2018; Wisniewska et al., 2018), it has been observed that underwater noise even leads to behavioral changes (Kastelein et al., 2018). Moreover, not only is entanglement in fishing gear and bycatch affecting population sizes, but growing contaminant exposure is thought to reduce reproduction success in female harbor porpoises (Vinther & Larsen, 2004; Kesselring et al., 2017). Accumulating exposure to chemical waste and pollution as well as microplastic can compromise the immune system of the harbor porpoises (Sonne et al., 2020a; Sonne et al., 2020b; Philipp et al., 2021). Uptake through contaminated food sources allows for accumulation of long-lasting pollutants in sentinel species such as the harbor porpoise (Siebert et al., 2009; Sonne et al., 2020a). This can weaken the ability of the immune system to defend against illnesses and infections, thus leading to a higher susceptibility to more frequent and severe infections or infestations with parasites (Wünschmann et al., 2001; Siebert et al., 2009; Siebert et al., 2020). Compared with Scandinavian and Arctic populations, German harbor porpoises possess less blubber while also suffering from severe pathological lesions and inflammations (Wünschmann et al., 2001; Siebert et al., 2006; Siebert et al., 2009). Particularly in the respiratory tract and lungs, considerable injuries have been determined, which are mainly caused by parasites or bacteria (Siebert et al., 2001; Lehnert et al., 2014; Siebert et al., 2020). It has been proposed that large accumulations of parasites in the respiratory tract may negatively affect the diving and hunting ability of the harbor porpoises (Siebert et al., 2001; Ten Doeschate et al., 2017).
To effectively dive and hunt under water, marine mammals possess a plethora of adaptations that enable them to live in an aquatic environment (Hindle, 2020). They prefer to dive aerobically and possess a large amount of readily available oxygen stored in their blood, lungs, and muscles (Kooyman, 1973; Davis, 2014). The locomotor muscles of marine mammals consume vast amounts of oxygen during aerobic dives to produce propulsion and force (Pabst, 1993). A heterogenic distribution of oxygen-binding myoglobin and higher concentrations in muscles of deep-diving species has already been confirmed, strengthening the suggestion that high myoglobin concentrations in oxidative, high-energy tissues prolong the aerobic dive limit (Polasek & Davis, 2001). Whereas deep-diving species preferably use the muscle-stored oxygen, shallow-diving whales such as the harbor porpoise also rely on their lungs as an oxygen storage (Snyder, 1983; Fahlman et al., 2017). Although their dives are typically slow, short, and shallow (14–32 m; Westgate et al., 1995; Otani et al., 2001; Piscitelli et al., 2010; Nielsen et al., 2018), they can perform deeper foraging dives (226 m; Westgate et al., 1995; Nielsen et al., 2018). Moreover, porpoises of the Kattegat and Belt Sea have been observed trying to escape underwater noise with unplanned, deeper dives (Wisniewska et al., 2018). These prolonged dives of marine mammals are typically fueled by glycolysis following depletion of oxygen storages (Kooyman et al., 1980; Castellini et al., 1981; Arregui et al., 2021; Torres-Velarde et al., 2021). For small marine mammals like the harbor porpoise, it has been proposed that a high anaerobic buffering capacity of the locomotor muscles may extend their ability to dive aerobically (Noren, 2004). This could be true for fleeing or distressed harbor porpoises that, at birth, exhibit already 69% of the adult muscle anaerobic buffering capacity (Noren et al., 2014). In marine mammals, high baseline antioxidant capacity and levels have been confirmed in various tissues and species to protect from oxidative damage (García-Castañeda et al., 2017; Allen & Vázquez-Medina, 2019 Vázquez-Medina et al., 2006). Furthermore, growing evidence supports accelerated evolution or positive selection of antioxidative genes in marine mammals (Foote et al., 2015; Romano et al., 2002; Park et al., 2015; Li et al., 2021).
A recent study in belugas found that muscle oxygen storage capacity is correlated with overall body condition (Choy et al., 2019). However, the skeletal muscle is known to be a plastic tissue, swiftly adapting to changing conditions (Frontera & Ochala, 2015) with an elevated recovery capacity upon tissue injury or disease (Howard et al., 2020). Regeneration of muscles consists of a conserved three step response: degradation of affected cells, inflammation of the injured area, followed by tissue regeneration and remodeling (Grounds, 2014; Levine & Kroemer, 2019). Oxidative stress and hypoxia are known to induce muscle atrophy by increasing proteolysis and inhibiting translation of proteins (Lian et al., 2022). As a post-mitotic tissue, it is very prone to oxidative damage induced by reactive oxygen species (ROS) and can accumulate damage over time (Frontera & Ochala, 2015; Rom & Reznick, 2016). Multiple muscle transcriptome studies of stressed elephant seals have been published (Khudyakov et al., 2015a; Khudyakov et al., 2015b; Crocker et al., 2016; Hindle et al., 2019; Piotrowski et al., 2021; Torres-Velarde et al., 2021), and a specific response pattern with minimal catabolism and tissue function and suppressed energy-consuming processes such as proliferation and development has been observed (Khudyakov et al., 2015a). To date, mostly histological, pathologies describing or physiological studies of harbor porpoises muscles exist (Noren & Williams, 2000; Siebert et al., 2001; Noren, 2004; Sierra et al., 2013; Lehnert et al., 2014; Noren et al., 2014; Sierra et al., 2017; McDonald et al., 2018). This is the first transcriptomic study of the muscles of harbor porpoises.
Here, we have performed comparative muscle transcriptome analyses to investigate whether harbor porpoises developed molecular compensatory adaptations caused by damaged lungs and impaired lung function, thus remaining viable and competitive. We compared transcriptomes of the main locomotor muscle (Musculus longissimus dorsalis) from German harbor porpoises, with a respiratory tract regarded as either healthy or non-healthy. M. longissimus dorsalis is one of the most energy-reliant muscles in cetaceans and necessary for the upward stroke of the fluke (Pabst, 1993; Noren & Williams, 2000), therefore representing an appropriate target to analyze possible adaptations in the muscle to an impaired respiratory function. We conducted Gene Ontology (GO) term analyses to find indicators of possible adaptations in non-healthy harbor porpoises. Furthermore, we identified differentially expressed transcripts (DETs) in non-healthy harbor porpoises compared with that in healthy ones and verified selected upregulated candidate genes in a quantitative real-time (qRT)–PCR.
Material and methods
Animals and sampling
M. longissimus dorsalis samples of harbor porpoises (n = 14, Table 1; see Supplementary data) were opportunistically obtained. All harbor porpoises died of live stranding or have been by-caught between 2015 and 2022. Fresh tissue samples were collected by the necropsy team of the Institute of Terrestrial and Aquatic Wildlife Research (ITAW), University of Veterinary Medicine Hannover, Foundation, Büsum. Tissue samples were immediately preserved in RNA stabilization solution (NucleoProtect RNA, Macherey-Nagel, Düren, Germany) and stored at −80°C until subsequent usage. Full necropsies and further investigations were conducted on all individuals according to Siebert et al., 2001. On the basis of the summary of findings, animals were categorized into healthy and non-healthy individuals. Non-healthy animals displayed pathological lesions due to lungworm and bacterial infections and suffered or died from bronchopneumonia (Table 1; see Supplementary data).
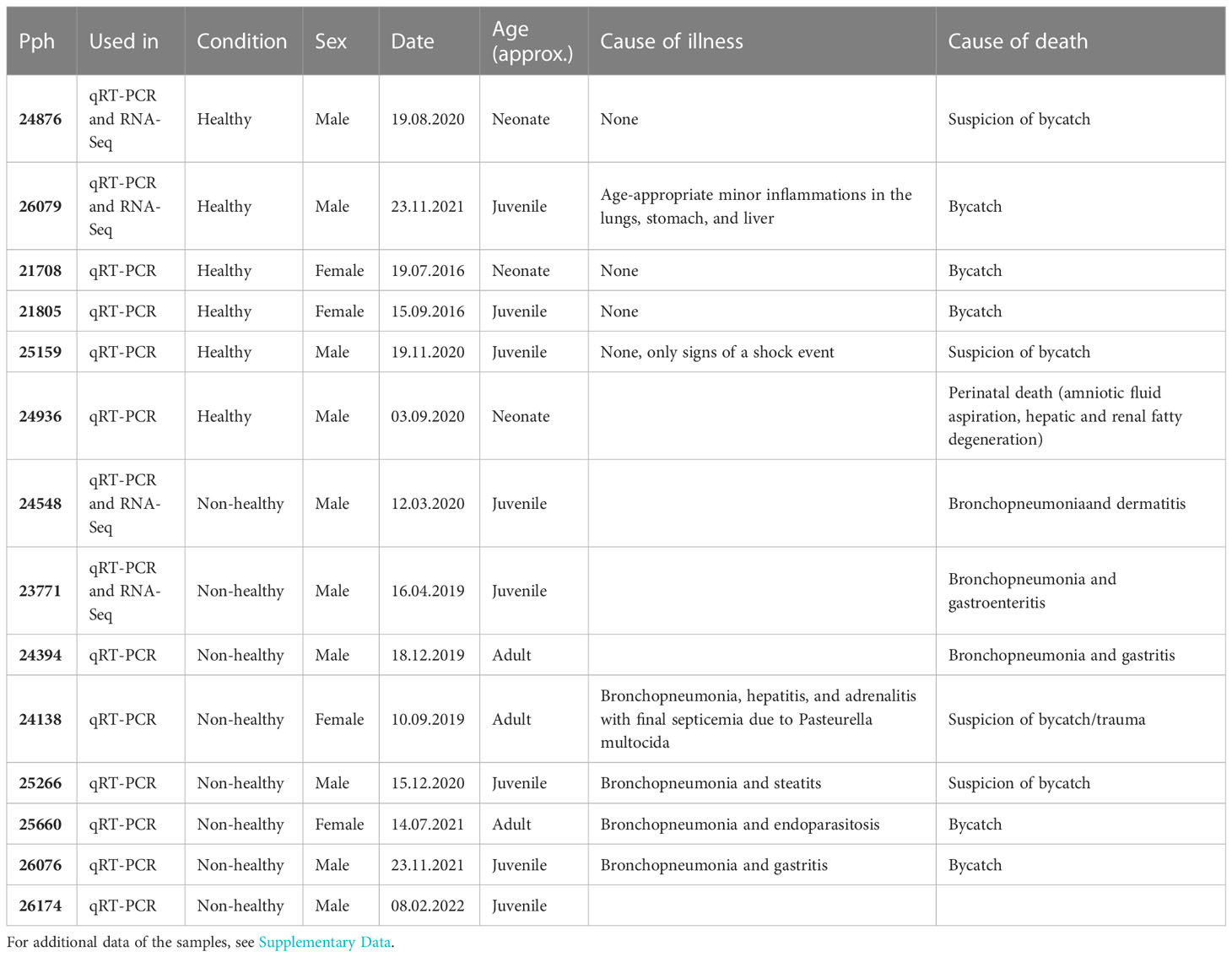
Table 1 Important data of the individual harbor porpoise samples and for which experiments samples were used.
RNA isolation and quality control
The muscle samples (20–30 mg) were minced and homogenized by bead beating in 1 mL of Trifast reagent (PEQLAB, Erlangen, Germany). RNA was isolated from the homogenates by phase extraction using chloroform and ethanol (70%). Total RNA was extracted with the Crystal RNA Mini Kit (Biolab Products, Bebensee, Germany) in accordance with the manufacturer’s instructions. In addition, a 15-min on-column DNA I digest (Qiagen, Hilden, Germany) was conducted. RNA concentration and quality, indicated by the RNA integrity number (RIN), were assessed using the Agilent TapeStation System (Agilent Technology, Santa Clara, CA, USA). RIN scores of the muscle samples varied from 4.8 to 6.1.
Sequencing and quality trimming
The RNA sequencing (RNA-Seq) library preparation for paired-end sequencing of 2 × 150 nucleotides (nt) was generated from 5,000 ng of RNA of healthy (n = 2) and non-healthy (n = 2) harbor porpoises. Sequencing was performed on an Illumina NextSeq 2000 platform (StarSEQ, Mainz, Germany) with an output of 25 million reads per sample. Sequence quality control, mapping, and alignment were carried out on the Galaxy server (version 21.09) of the Department of Biology at the University of Hamburg. Quality control was assessed using the FastQC version 0.73 and MultiQC version 1.11 tool on Galaxy.
The first 20 5′-terminal nucleotides and Illumina adapter sequences (mismatch count = 2, internal match = 10) were cropped from the raw reads with Trimmomatic version 0.38.1. Reads below the length of 20 nt were discarded, and a minimum average quality value of 20 was required for consideration.
The raw sequence files are available at the National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA) from (SRA BioProject ID: PRJNA977857, see Supplementary Table 1 for SRA accession numbers).
Expression analysis via RNA-Seq
The trimmed sequences were mapped and aligned against the bottlenose dolphin genome (Tursiops truncatus, mTurTru1.mat.Y, released March 2020, RefSeq Accession: GCF_011762595.1) using HISAT version 2.2.1 and featurecounts version 1.6.4. Individual transcripts per million (TPM) for each transcript were counted with the Galaxy tool “Generate CPM, TPM, RPK” (version 0.4.0), and mean values for the healthy and the non-healthy animals were calculated.
Differential expression analysis
DETs were determined using DESeq2 version 2.11.40.7 on the Galaxy server. DETs were then filtered with cutoffs for up- and downregulated genes at an adjusted p-value (padj) ≤ 0.05, fold change (FClog2) ≥ 1 or ≤ −1, and mean TPMnon-healthy ≥ 5 (see Supplementary data).
Gene Ontology analysis
Analysis of GO Slim terms was performed with the PANTHER Overrepresentation Test (PANTHER version 17.0, released 2022_02) using the human as reference list. DETs with a padj ≤ 0.05, a FClog2 ≥ 1 or ≤ −1, and a mean TPMnon-healthy ≥ 5 were considered for the analysis. Overrepresentation was determined in the PANTHER “Pathways” and GO Slim categories “Biological Process”, “Molecular Function”, and “Cellular Component” categories. Fisher’s exact test and the False discovery rate (FDR) were used as correction for multiple testing.
Expression confirmation via qRT-PCR
Because of our small muscle sample size for the RNA-Seq, we verified the results in a larger subset of healthy (n = 6) and non-healthy (n = 8) harbor porpoises (Table 1). mRNA expressions of the selected transcripts chosen as candidate genes were analyzed in a qRT-PCR. Total RNA was isolated as previously stated. RNA quantity and quality were determined by spectrophotometry and gel electrophoresis. For complementary DNA (cDNA) synthesis, a maximum of 1,000 ng of total RNA and the RevertAid H–First Strand cDNA Synthesis Kit (Thermo Scientific, Germany) were used according to the manufacturer’s protocol. The synthetized cDNA was further diluted in 20 µl of Ribonuclease (RNAse)-free water, resulting in a total volume of 40 µl. qRT-PCR was conducted on the ABI 7500 real-time PCR system with the Power SYBR Green master mix (Applied Biosystems, Darmstadt, Germany). The qRT-PCR was performed with a protocol consisting of 40 cycles (95°C for 15 s, 58°C–60°C for 60 s, 72°C for 30 s). Species-specific primers are presented in Supplementary Table 2. Samples were applied as three technical replications. The qRT-PCR was used as a relative verification of the transcriptome analysis, so no recombinant plasmid was necessary as standard. RPLP0, RPLP1, and EF2 were selected as reference genes. Relative FCs were calculated according to the ΔΔCT method (Livak & Schmittgen, 2001).
Statistical analysis
All statistical analyses of the results qRT-PCR were conducted using GraphPad Prism version 9.5.1. Mean CT values of the technical replicates were calculated and tested for normal distribution. The Mann–Whitney U-test was performed for post-hoc analysis and testing for statistical significance.
Results
Transcriptomes of the muscles
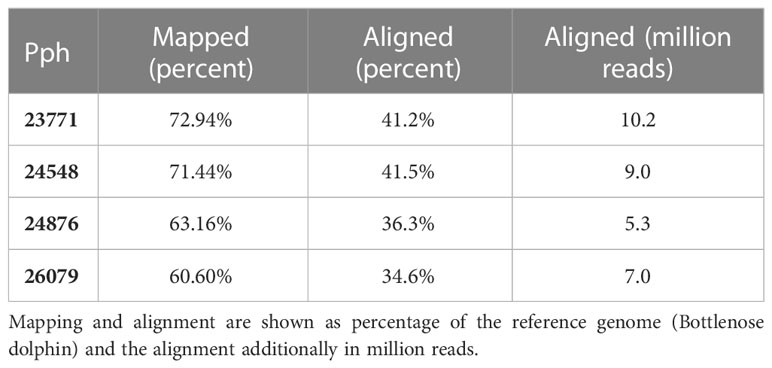
Per sample of non-healthy harbor porpoises, a total of ~43 million paired-end Illumina reads could be generated, and a total of ~25 million and ~36 million paired-end Illumina reads were generated per muscle sample of healthy harbor porpoises. The quality of the raw reads was sufficient (Phred score = 33). The mapping of the transcriptomes resulted in ~15 million ± 3 million mapped reads in healthy porpoises and ~21 million reads in non-healthy porpoises (67 ± 6%; Table 2). Alignment of the mapped reads against the bottlenose dolphin genome resulted in ~38.05 ± 3.45% assigned reads (Table 2). The analysis of DETs resulted in 1,594 upregulated and 1,286 downregulated DETs in muscles of non-healthy porpoises. For further analyses, only DETs with a p-valueadj ≤ 0.05, FClog2 ≥ 1 or −1, and a TPMnon-healthy ≥ 5 were considered, comprising 739 upregulated transcripts and 194 downregulated transcripts (see Supplementary data).
High expressed transcripts in GO Slim terms for muscle function, regeneration, and glucose metabolism
We performed an overrepresentation analysis using the PANTHER categories GO Slim “Biological Process,” “Molecular Function,” “Cellular Component,” and “Pathways” to identify overrepresented GO Slim terms. For this, we only considered transcripts of the filtered set with cutoffs FClog2 ≥ 1, p-valueFDR ≤ 0.05, and TPMnon-healthy ≥ 5 (see Supplementary data) with the human genome as reference (Fisher’s exact test with FDR-corrected p-value < 0.05). The top 10 overrepresented biological processes in muscles of non-healthy harbor porpoises compared with that of healthy porpoises are shown here (Figure 1). Many of the GO Slim terms were associated with skeletal muscle function and development (regulation of epithelial cell differentiation, Fold Enrichment (FE) = 18.38; myofibril assembly, FE = 8.36; muscle tissue development, FE = 7.29) as well as metabolic processes (glucose homeostasis, FE = 12.77; alpha–amino acid biosynthetic process, FE = 5.80; triglyceride metabolic process, FE = 6.13; fatty acid catabolic process, FE = 5.25). In addition, ribosome-related processes were found enriched (maturation of small subunit ribosomal RNA (SSU-rRNA) from tricistronic rRNA transcript, FE = 6.57; ribosomal large subunit biogenesis, FE = 7.07), and processes associated with cell death and clearance were enriched in the set of transcripts (autophagy of mitochondrion, FE = 6.57). The only identified, enriched GO Slim terms in the “Molecular Function” category were involved in “catalytic activity” and binding (snoRNA binding, transcription factor binding; Figure 1).
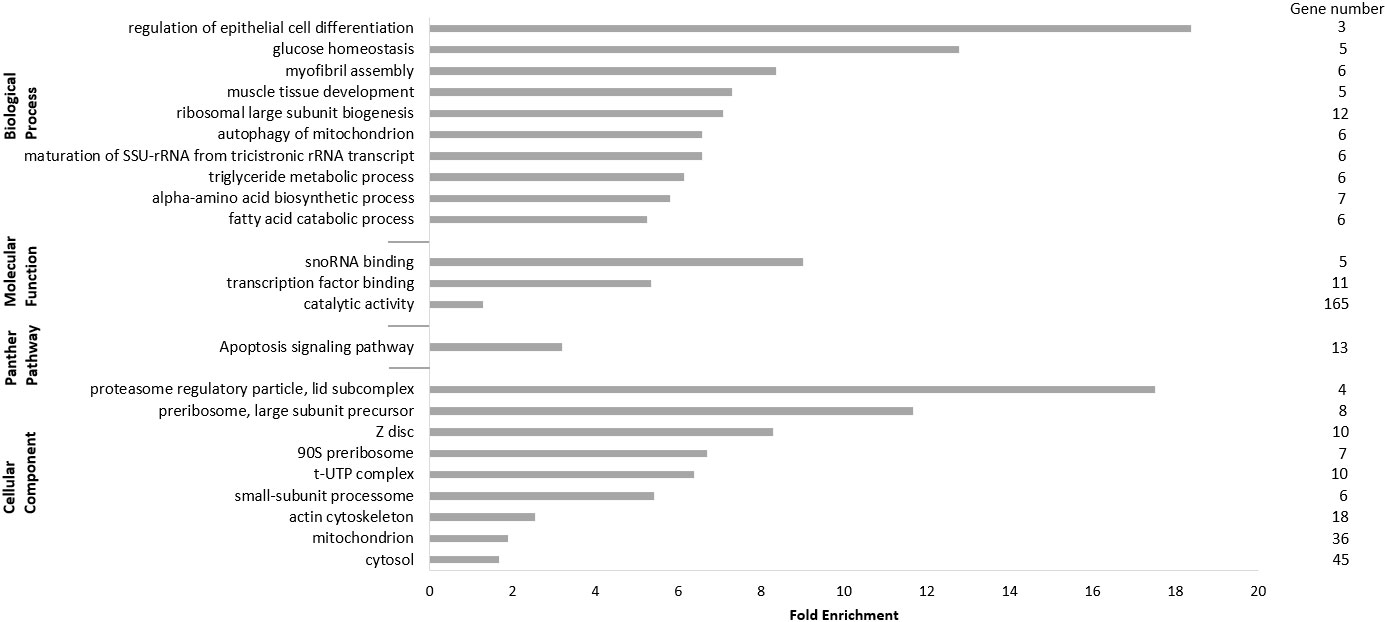
Figure 1 Panther analysis of the upregulated differentially expressed transcripts (DETs). Only DETs with the applied cutoffs fold change (FClog2) ≥ 1, FDR-corrected p-value (p-valueFDR) ≤ 0.05, and transcripts per million (TPMnon-healthy) ≥ 5 were considered for the analysis, resulting in 739 transcripts. Presented are the top 10 significantly upregulated terms of the GO Slim categories “Biological Processes”, “Molecular Function”, “Cellular Component”, and “Pathways” with fold enrichment and number of genes.
Furthermore, we sorted the transcripts by their FC (FClog2) and annotated the 10 most highly expressed DETs in non-healthy harbor porpoises compared with that in healthy porpoises (Table 3). The most highly expressed transcript was TRIM63 (FClog2 = 5.65), a marker gene for muscle atrophy that, under amino acid starvation, induces proteasomal degradation of muscle protein (Rom & Reznick, 2016). Three transcripts were identified with their main function in metabolism (SLC2A1, FClog2 = 4.83; AMPD3, FClog2 = 4.40; CA7, FClog2 = 4.68). CA7 also has a role in oxidative stress and gluconeogenesis (Monti et al., 2017; Di Fiore et al., 2018). One transcript in endocytosis and monocyte adhesion (SORL1, FClog2 = 4.58), one transcript in ribosomal biogenesis (RRS1, FClog2 = 4.50), and two transcripts were involved in regeneration processes (CSRP3, FClog2 = 4.43; LYPD3, FClog2 = 4.47). Multiple transcripts (TNFRSF12A, FClog2 = 4.37; GADD45G, FClog2 = 4.40) were associated with response to various cell stressors.
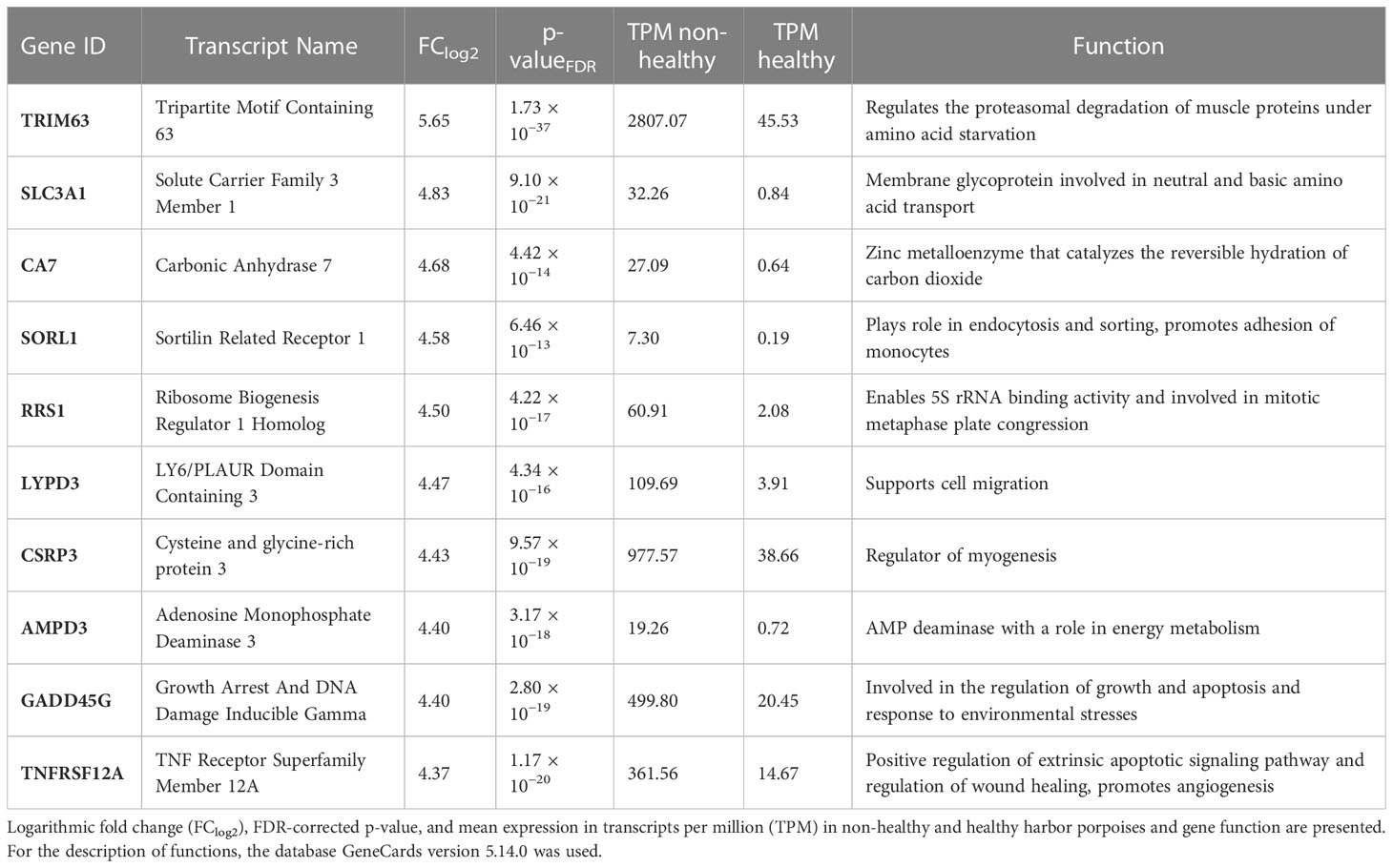
Table 3 Top 10 of the differentially expressed transcripts with the highest fold change in the muscles of non-healthy harbor porpoises compared with that in the muscles of healthy harbor porpoises.
Downregulated transcripts in non-healthy harbor porpoise muscles are involved in cell adhesion and cell organization
For the analysis of enriched terms within the lowly expressed transcript set in muscles of non-healthy harbor porpoises compared with that of healthy porpoises, only DETs with FClog2 ≤ −1, p-valueFDR ≤ 0.05, and TPMnon-healthy ≥ 5 were considered (Table 4; see Supplementary data). Here, we could identify development-related biological processes (anatomical structure development, FE = −2.48) and molecular functions associated with adhesion and binding (collagen binding, FE = −40.85; actin binding, FE = −4.93) significantly less enriched in muscles of non-healthy harbor porpoises compared with those of healthy porpoises (Figure 2).
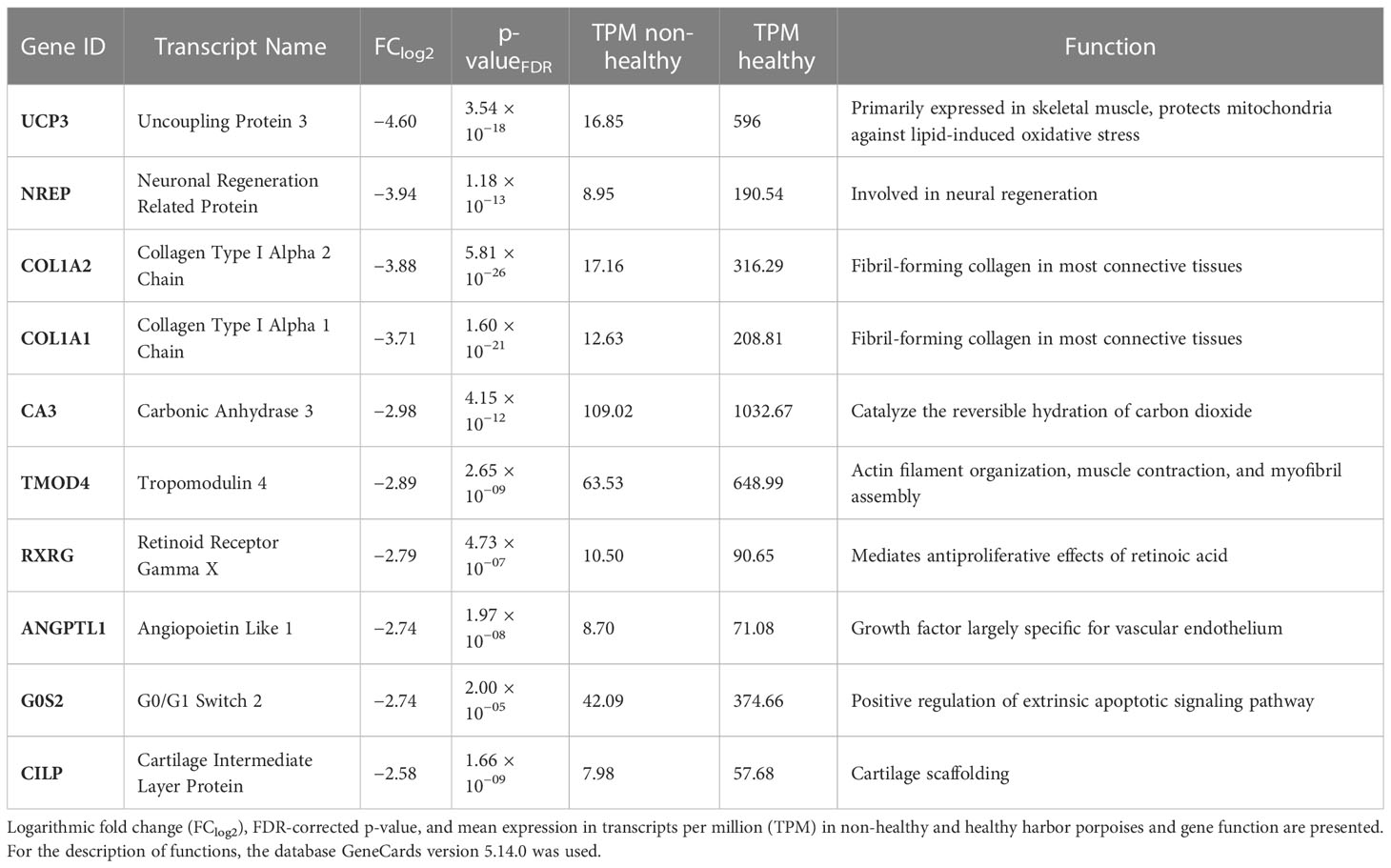
Table 4 Top 10 of the differentially expressed transcripts with the lowest fold change in the muscles of non-healthy harbor porpoises compared with that in the muscles of healthy harbor porpoises.
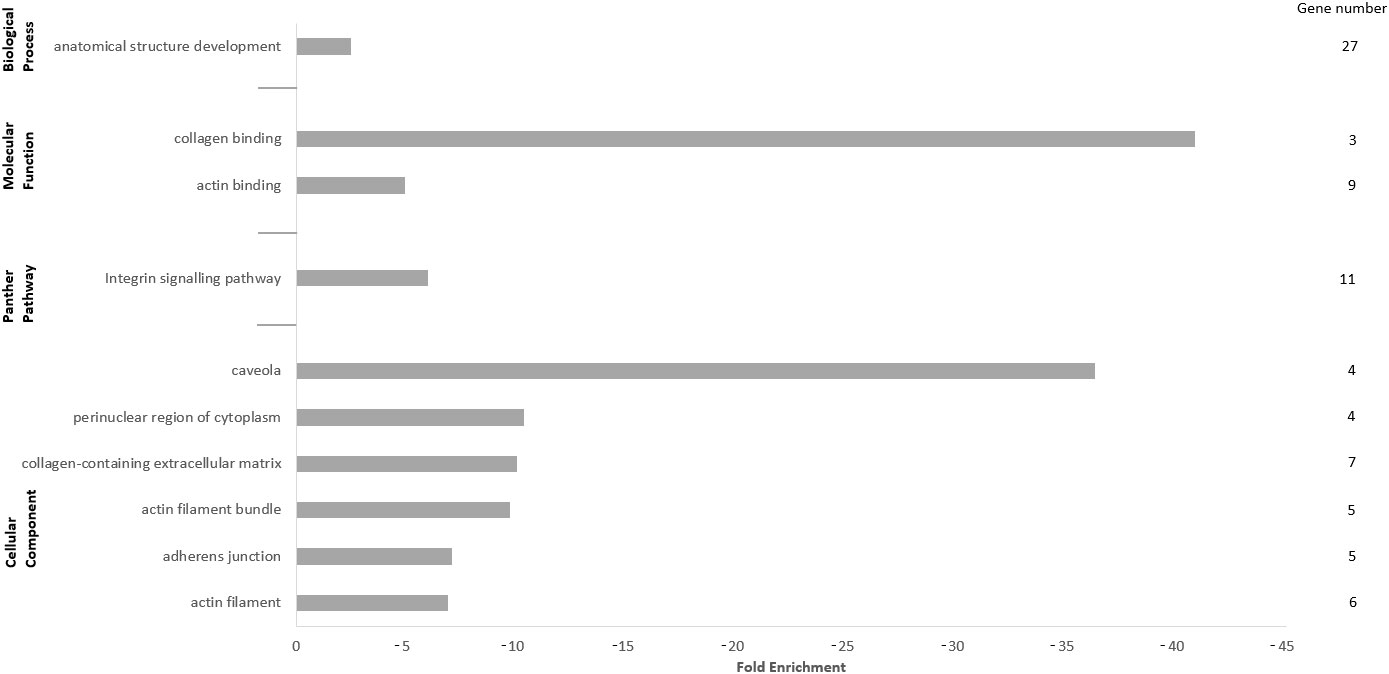
Figure 2 Panther analysis of the downregulated differentially expressed transcripts (DETs). Only DETs with the applied cutoffs fold change (FClog2) ≤ −1, FDR-corrected p-value (p-valueFDR) ≤ 0.05, and transcripts per million (TPMnon-healthy) ≥ 5 were considered for the analysis, comprising 194 genes. Presented are the top 10 significantly upregulated terms of the GO Slim categories “Biological Processes”, “Molecular Function”, “Cellular Component”, and “Pathways” with fold enrichment and number of genes.
The top 10 downregulated DETs were also further examined (Table 4). Within the lowest expressed transcripts in muscles of non-healthy harbor porpoises compared with those of healthy animals, the least expressed transcript was UCP3 (FClog2 = −4.60). It is primarily expressed in skeletal muscle and protects from lipid-induced oxidative stress in mitochondria. One transcript involved in neural regeneration (NREP, FClog2 = −3.94) was found less expressed. Several transcripts that are associated with extracellular matrix organization and remodeling and membrane structure (COL1A2, FClog2 = −3.88; COL1A1, FClog2 = −3.71; TMOD4, FClog2 = −2.89) were among the top 10 downregulated DETs. CA3 (FClog2 = −2.98), an isoform of CA7, was identified and also catalyzes the hydration of carbon dioxide, although with a lower efficiency than CA7 (Monti et al., 2017). ANGPTL1 (FClog2 = −2.74) showed a low expression and is the only known vascular endothelium growth factor. One pro-apoptotic (G0S2, FClog2 = −2.74), one anti-proliferative transcript interacting with retinoid acid (RXRG, FClog2 = −2.80), and one involved in cartilage scaffolding and insulin signaling (CILP, FClog2 = −2.58) were identified among the lowest expressed transcripts.
Moreover, important transcripts involved in gluconeogenesis and glycolytic metabolism were found differentially expressed in the filtered set of highly and less expressed transcripts (Figure 3; see Supplementary data). Upregulated transcripts played key roles in catalyzation of acetyl-coA (ACSS1, FClog2 = 1.10), production of glucose from lactate (PCK2, FClog2 = 2.39), and initiation of the first step of the glucose metabolism (HK2, FClog2 = 1.90). Downregulated transcripts were involved in regulation of gluconeogenesis (FBP2, FClog2 = −1.78), glycolytic production of ATP (PGK1, FClog2 = −1.40), breakdown and synthesis of glucose (PGM1, FClog2 = −1.64), or key steps of glycolysis (ALDOA, FClog2 = −1.52).
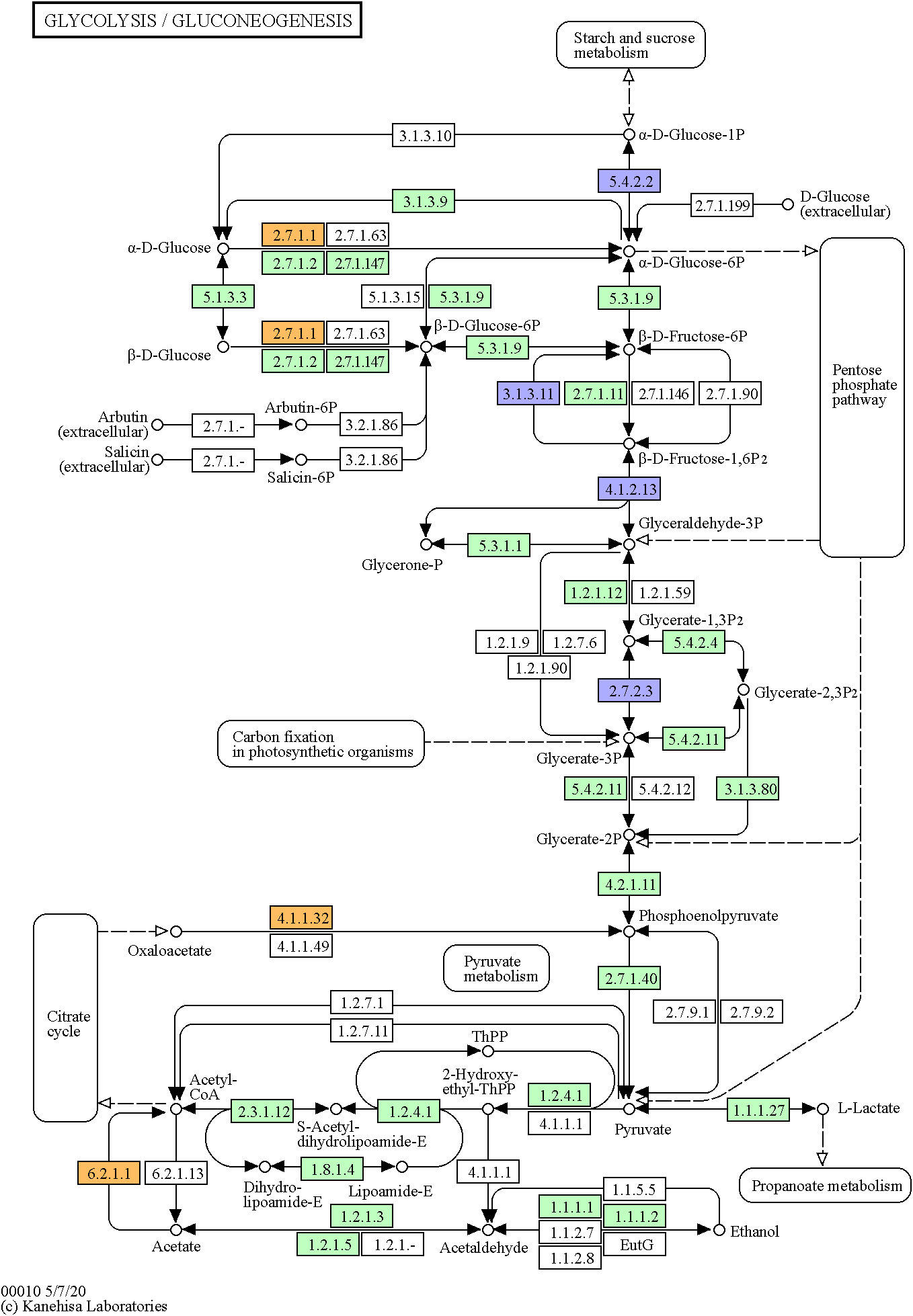
Figure 3 Glycolysis/gluconeogenesis Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway of the human (hsa00010). Marked in orange are identified upregulated DETs in muscles of non-healthy harbor porpoises (2.7.1.1, HK2; 4.1.1.32, PCK2; 6.2.11, ACSS1), and marked in violet are here identified downregulated DETs (5.4.2.2, PGM1; 3.1.3.11, FBP2; 4.1.2.13, ALDOA; 2.7.2.3, PGK1). The green colored genes stand for identified genes of the pathway in the human.
Verification of transcriptome analysis in qRT-PCR of selected candidate genes
Because of the limited sample set of 2 per group for the transcriptome analysis, we verified the differential gene expression results by qRT-PCR in additional muscle samples of non-healthy and healthy harbor porpoises. A total of 18 candidate genes were chosen on the basis of differences in expression between the two conditions (upregulation): abundance of transcripts and involvement in GO Slim “Biological Process” terms (Supplementary Table 3). Only upregulated and higher expressed transcripts were considered as candidate genes as they are likely involved in the active response to the possible lack of oxyen and resulting negative consequences. For the normalization of the qRT-PCR data, we tested three reference genes, namely, RPLP1, RPLP0, and EF2, of which RPLP1 was the most stable reference gene and selected for normalization. In addition, few of the candidate genes have been mentioned in other whale or seal studies, hinting at a possible aquatic adaptation in marine mammals but have not been examined in more detail (FOSL1, GADD45G, GLUL, HIPK2, IGFR1, NPY, PCK2, SIK1, TOLLIP, and TNFRSF12A). Of the chosen candidate genes, one could not be verified in qRT-PCR (TNFRSF12A). Because of a high content of Guanine/Cytosine (GC) and a high number of repetitive bases, it was not possible to generate an adequate primer pair. All candidate genes that were found upregulated in the RNA-Seq also had a higher relative FC (rel. FC) in non-healthy harbor porpoises in the qRT-PCRs (Figure 4; see Supplementary data).
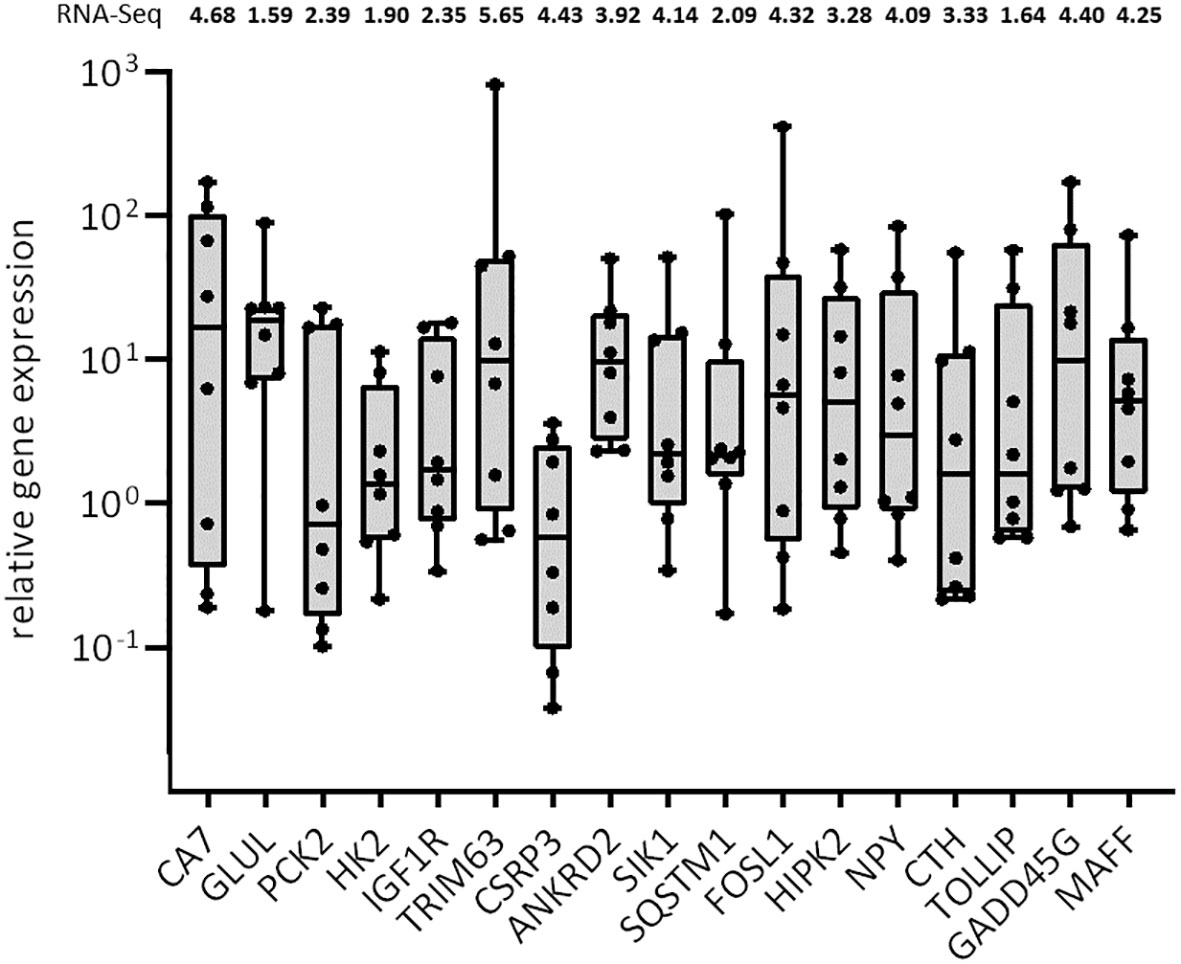
Figure 4 Boxplot with median of the relative gene expression of selected candidate genes in muscles of non-healthy harbor porpoises compared with that of healthy animals. The gene expression of the individual non-healthy harbor porpoises is represented by the dots (n = 8). The vertical y-axis is presented logarithmically due to the high inter-individual variation of the harbor porpoises. The fold changes (FClog2) of the RNA-Seq for each transcript are indicated above.
We could confirm upregulation of transcripts involved in oxidative stress response and antioxidant defense in muscles of non-healthy harbor porpoises (HIPK2, mean rel. FCnon-healthy = 14.60; CTH, mean rel. FCnon-healthy = 10.05; GADD45G, mean rel. FCnon-healthy = 36.93; MAFF, mean rel. FCnon-healthy = 13.86) of which two also possess antimicrobial properties (NPY, mean rel. FCnon-healthy = 17.23; TOLLIP, mean rel. FCnon-healthy = 12.34). Muscles of non-healthy harbor porpoises were confirmed to express higher degradation-associated genes (TRIM63, mean rel. FCnon-healthy = 116.88; SQSTM1, mean rel. FCnon-healthy = 15.67). Multiple candidate genes with a role in muscle regeneration and development were also found highly expressed in non-healthy harbor porpoises compared with that in healthy individuals (CSRP3, mean rel. FCnon-healthy = 1.22; ANKRD2, mean rel. FCnon-healthy = 14.69; SIK1, mean rel. FCnon-healthy = 10.93; FOSL1, mean rel. FCnon-healthy = 60.64). We lastly tested candidate genes with a role in anaerobic and lipid metabolism in a larger subset of samples. Upregulation of transcripts involved in gluconeogenesis and glycolysis (CA7, mean rel. FCnon-healthy = 48.58; PCK2, mean rel. FCnon-healthy = 7.36; HK2, mean rel. FCnon-healthy = 3.23), amino acid metabolism (GLUL, mean rel. FCnon-healthy = 23.37), and fatty acid metabolic pathways (IGF1R, mean rel. FCnon-healthy = 5.95; CA7) could be confirmed in qRT-PCR. The gene expression varied strongly between the single individuals for most analyzed candidate genes (Figure 4). The highest interindividual variation was found for TRIM63 (rel. FC max = 815.48, rel. FC min = 0.56), FOSL1 (rel. FC max = 410.41, rel. FC min = 0.18), CA7 (rel. FC max = 171.86, rel. FC min = 0.18), and GADD45G (rel. FC max = 171.29, rel. FC min = 0.69), whereas the gene expression of CSRP3 was the most homogeneous among the non-healthy harbor porpoises (rel. FC max = 3.60, rel. FC min = 0.04).
Discussion
Harbor porpoises from the North and Baltic Seas show a higher level of parasitic infections and associated lesions in the respiratory tract compared with porpoises from Arctic waters (Wünschmann et al., 2001; Siebert et al., 2006; Siebert et al., 2020). This may lead to an impaired oxygen uptake and available oxygen for high energy- and oxygen-consuming organs such as the brain, heart, and muscles. The skeletal musculature is especially essential for the fully-aquatic living marine mammals. They not only enable locomotion and diving ability but also play an important role in oxygen and nutrient storage in cetaceans. To analyze whether and how muscles are affected, we generated transcriptomes of the muscles from two non-healthy harbor porpoises and compared them with the muscle transcriptomes of healthy porpoises. We performed GO analyses and identified significantly differentially regulated transcripts in non-healthy animals. Last, we selected a set of 18 transcripts as candidate genes for verification of the results of the RNA-Seq in a larger subset of samples and for further analyses.
Enhanced transcripts involved in response to oxidative stress but not to hypoxia in muscles of non-healthy porpoises
The skeletal musculature is necessary for locomotion and consumes large amounts of energy and oxygen, which is taken up and distributed from the lungs. We hypothesized that the observed lung damages of the harbor porpoises reduce oxygen uptake and supply to the muscles and thus cause hypoxic conditions in the tissue. Contrarily to this, we could not find a defined response to hypoxia. We found transcripts (NPY) upregulated in non-healthy harbor porpoises that are assumed to enhance tissue perfusion (Mirman et al., 2020) and thus may help secure adequate oxygen distribution in the tissue and alleviate possible hypoxic states. In non-healthy harbor porpoises, we confirmed an elevated oxidative stress response (HIPK2, GADD45G, MAFF, CTH, and SLC3A1; Figures 1, 4; Table 3; Supplementary Table 3). Oxidative stress not only can be induced by free radicals such as ROS and xenobiotics but also form naturally during metabolic processes (Sies & Jones, 2020; Sies et al., 2022). Excessive concentrations of ROS can lead to apoptosis (Sies & Jones, 2020). HIPK2 is thought to act as a transcriptional switch, deciding between apoptosis or survival of the cell (de la Vega et al., 2012) and increases survival signaling when overexpressed (Torrente et al., 2017; Li et al., 2018). GADD45G, which can act as a stress sensor and is associated with oxidative stress, was also upregulated in muscles of non-healthy harbor porpoises, but has been found decreased in blubber of Northern elephant seals after prolonged stress conditions (Khudyakov et al., 2017; Turner et al., 2019). The opposite regulation observed here may be due to different functions and, hence, regulation in the respective tissues. Three highly expressed transcripts in non-healthy harbor porpoises were associated with synthesis or regulation of the antioxidant glutathione (MAFF, CTH, and SLC3A1; Figure 1; Table 3; Supplementary Table 3). Whereas MAFF regulates glutathione concentrations (Wang et al., 2020), CTH and SLC3A1 are involved with the synthesis of glutathione by maintaining levels of its precursors cystine and cysteine (McBean, 2017; Cha et al., 2018; Wu et al., 2020). Glutathione is an important factor of the antioxidant defense by effectively clearing excessive reactive oxygen or nitrogen species and xenobiotics (Wilhelm Filho et al., 2002; Cantú-Medellín et al., 2011; García-Castañeda et al., 2017). Therefore, it may be more likely elevated due to high oxidative stress in the tissue and, additionally, exposure to pollutants of the porpoises (Sies & Jones, 2020). Glutathione can exist in two states, namely, reduced (GSH) or oxidized (GSSG). The ratio between GSH to GSSG can be used as indicator of oxidative stress, with increased GSSG implying higher oxidative stress (Owen & Butterfield, 2010), which should be considered for future studies.
Transcriptome analysis hints at enhanced clearance and regeneration of muscle cells in non-healthy harbor porpoises
Oxidative stress and hypoxia are known to induce muscle atrophy by increasing proteolysis and inhibiting translation of proteins (Lian et al., 2022). However, multiple studies indicate that autophagy is indispensable for muscle recovery after injury, for timely clearance of cell debris and after intense exercise (Call & Nichenko, 2020; Xia et al., 2021). In muscles of non-healthy harbor porpoises, we found processes and transcripts upregulated that are associated with infections, degradation, and atrophic flux (Figure 1; Table 3). Several transcripts (NPY and TOLLIP; Figure 4) with antimicrobial properties were found (Mancia et al., 2012; Anderson et al., 2022). Although it is not possible to determine the specific cause, the upregulation may protect from an easily contractible, secondary bacterial infection due to an impaired immune system or act as a reaction to an already existing bacterial infection (Siebert et al., 2001; Mancia et al., 2012). Still, the muscle function of TOLLIP has not been fully described and remains partly hidden, highlighting it as a possible novel adaptation for further research (Boursereau et al., 2017). The qRT-PCR results (Figure 4) could confirm high expression in non-healthy harbor porpoises of muscle atrophy-associated transcript TRIM63 (Rom & Reznick, 2016; Thoma & Lightfoot, 2018) and of SQSTM1, which may be indicative of autophagic cargo flux (Puissant et al., 2012). Together, these result hint at a high degradation rate of debris, defective cells or misfolded proteins in non-healthy harbor porpoises. Interestingly, we found elevated ribosome biogenesis (RRS1, Table 3; “ribosomal large subunit biogenesis” and “maturation of SSU-rRNA from tricistronic rRNA transcript”, “snoRNA binding”, Figure 1), despite it being one of the first inhibited processes upon injury, as it is highly energy-demanding (Shah et al., 2013). Furthermore, we found enhanced regenerative processes and confirmed elevated expression of associated transcripts (CSRP3, ANKRD2, SORL1, FOSL1, and SIK1; Table 3; Figure 4; see Supplementary data). Two highly expressed transcripts were found to promote myogenesis (SIK1) and function as master regulator of muscle function and development (CSRP3; Stewart et al., 2013; Mutryn et al., 2015; Williams et al., 2021). CSRP3 can also interact with Ankyrin Repeat Domains (ANKRD2, Supplementary Table 3), which are transcriptional stress responders and upregulated under oxidative stress (Belgrano et al., 2011; Tsompanidis et al., 2016; Cenni et al., 2019). FOSL1, an early response gene that mediates muscle injury, has also been found upregulated in the brain of hooded seals after reoxygenation (Hoff et al., 2017), so it may support perfusion of the muscle tissue and prevention of ROS. Although we could confirm elevated degradation processes in muscles of non-healthy harbor porpoises, we could also confirm unexpectedly enhanced regeneration and development, which may point to an adaptive mechanism in harbor porpoises. A precautionary high baseline expression of transcripts involved in muscle regeneration and development may ensure fast replacement of degraded cells, support tissue function, and prevent tissue loss observed in other mammals (Nixon et al., 2016; Yadava et al., 2016; Xia et al., 2021). However, protein turnover is a very costly process, and, therefore, its elevation may strongly affect the maintenance of the muscles and overall energy balance of the porpoises.
Dysregulation of lipid and glucose metabolism for high energy processes in muscles of non-healthy harbor porpoises
Stress such as hypoxia and ROS can affect metabolism pathways that have been continuously suggested for and investigated in marine mammals (Fair & Becker, 2000; Horscroft & Murray, 2014; Khudyakov et al., 2015a). Whales rely heavily on carbohydrate and lipid energy production, changing to aerobically metabolized lipids as preferred energy source under stress and reduced food uptake (Kanatous et al., 2008; Velten et al., 2013; Chicco et al., 2014; Sierra et al., 2015). In muscles of non-healthy harbor porpoises, we identified differently regulated lipid and anaerobic metabolic pathways like “glucose homeostasis”, “triglyceride metabolic process”, and “fatty acid catabolic process” (Figure 1; Table 3). Transcripts indicating a dysregulation of glucose and lipid metabolism were identified and confirmed in non-healthy porpoises (AMPD3 and SQSTM1, Table 3; Supplementary Table 3; Hong et al., 2017; Calvo-Garrido et al., 2019; Caspi et al., 2020). In concert with this, we found Insulin like growth factor (IGF) receptor IGF1R upregulated, which enhances survival and affects lipid homeostasis (Houser et al., 2013; Li et al., 2022; O’Neill et al., 2016). In addition, transcripts protecting from lipid-induced oxidative injury were found downregulated in non-healthy harbor porpoises (UPC3, Table 4), potentially hinting at a higher capacity of aerobic lipid metabolism (Polasek et al., 2006; Burns et al., 2010). Moreover, gluconeogenic transcripts were found downregulated, whereas mostly glycolytic transcripts were found upregulated (Figure 2). Interestingly, ALDOA, which can indicate glycolytic metabolism rates (Hoff et al., 2017), was found downregulated (Figure 3; Supplementary data). However, other metabolic rate-limiting transcripts were confirmed to be highly expressed (Figures 3, 4) and are also involved in production of glucose from lactate (PCK2 and HK2, Figure 3; Champagne et al., 2012; Nedvedova et al., 2018), in glutamate/glutamine synthesis, which can be used for high energy processes (GLUL, see Supplementary data; de Theije et al., 2018; Rogeri et al., 2020) and with a function in lipogenesis and gluconeogenesis (CA7, Table 3; Supplementary data; Monti et al., 2017; Di Fiore et al., 2018). Although, CA7 is the most efficient catalytic carbonic anhydrases, it is still one of the least researched isoforms and has not been studied extensively in marine mammals (Tower and Young, 1973; Chegwidden and Carter, 2000; Yang et al., 2000; Cincinelli et al., 2011). A high reliance on glycolysis has also been observed in marine mammals under long-term stress (Champagne et al., 2012; Park et al., 2015; Fabrizius et al., 2016; Tian et al., 2017; Torres-Velarde et al., 2021), which may also be true in response to ROS stress and maintenance of high energy functions such as translation and regeneration of energy-consuming muscles observed in this study.
Conclusion
This study is the first describing the muscle transcriptomes of the harbor porpoise. Here, we investigated the muscle transcriptomes of two German harbor porpoises suffering from lesions in the respiratory tract and bronchopneumonia and compared them with muscle transcriptomes of healthy porpoises to analyze adaptations to reduced oxygen levels. We could find hints at adaptations like a possibly finely tuned switch between swift degradation of cell debris or misfolded proteins and tissue regeneration. Glutathione-associated transcripts have been found upregulated in muscles of the non-healthy harbor porpoises and may point to a high reliance on this antioxidative gene. Hints at a higher capacity for glycolysis and higher reliance on lipid metabolism have been found in muscles of non-healthy harbor porpoises. This may be utilized by the high-energy consuming muscle to ensure additional highly consuming processes like regeneration and translation. Our findings suggest that harbor porpoises do not suffer from hypoxic conditions in the muscles exacerbated by pathological lesions in the lung but may experience elevated oxidative stress caused by excessive ROS. Although the function of the identified transcripts has to be further analyzed in future studies, this study presents an important step to better understand the adaptations to stresses of cetaceans on a molecular level.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics statement
Ethical review and approval were not required for the study since the investigated animals were collected once they were dead by stranding or by-caught within the German stranding network. The German stranding network conducts work such as collecting and holding carcasses and samples from European protected species following appropriate licenses from the relevant authorities.
Author contributions
ED, US, and AF conceived the research idea. US provided the samples and additional data. ED and AF derived the experimental procedure. ED performed the experiments and data analysis and wrote the manuscript with input from all authors. All the authors contributed to the article and approved the submitted version.
Funding
This work was partly supported by the EU Horizon SATURN project, which has received funding from the European Union’s Horizon 2020 research and innovation programme under grant agreement No 101006443. This Open Access publication was funded by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) - 491094227 “Open Access Publication Funding” and the University of Veterinary Medicine Hanover, Foundation.
Acknowledgments
The authors thank the necropsy team at the ITAW who sampled the harbor porpoises and collected and provided additional data of the animals.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2023.1232305/full#supplementary-material
References
Allen K. N., Vázquez-Medina J. P. (2019). Natural tolerance to ischemia and hypoxemia in diving mammals: a review. Front. Physiol. 10, 1199. doi: 10.3389/fphys.2019.01199
Anderson Z. T., Dawson A. D., Slominski A. T., Harris M. L. (2022). Current insights into the role of neuropeptide Y in skin physiology and pathology. Front. Endocrinol. 13. doi: 10.3389/fendo.2022.838434
Arregui M., Singleton E. M., Saavedra P., Pabst D. A., Moore M. J., Sierra E., et al. (2021). Myoglobin concentration and oxygen stores in different functional muscle groups from three small cetacean species. Animals 11 (2), 451. doi: 10.3390/ani11020451
Belgrano A., Rakicevic L., Mittempergher L., Campanaro S., Martinelli V. C., Mouly V., et al. (2011). Multi-tasking role of the mechanosensing protein Ankrd2 in the signaling network of striated muscle. PloS One 6 (10), e25519. doi: 10.1371/journal.pone.0025519
Boursereau R., Abou-Samra M., Lecompte S., Noel L., Brichard S. M. (2017). New targets to alleviate skeletal muscle inflammation: role of microRNAs regulated by adiponectin. Sci. Rep. 7 (1), 43437. doi: 10.1038/srep43437
Burns J. M., Skomp N., Bishop N., Lestyk K., Hammill M. (2010). Development of aerobic and anaerobic metabolism in cardiac and skeletal muscles from harp and hooded seals. J. Exp. Biol. 213 (5), 740–748. doi: 10.1242/jeb.037929
Call J. A., Nichenko A. S. (2020). Autophagy: an essential but limited cellular process for timely skeletal muscle recovery from injury. Autophagy 16 (7), 1344–1347. doi: 10.1080/15548627.2020.1753000
Calvo-Garrido J., Maffezzini C., Schober F. A., Clemente P., Uhlin E., Kele M., et al. (2019). SQSTM1/p62-directed metabolic reprogramming is essential for normal neurodifferentiation. Stem Cell Rep. 12 (4), 696–711. doi: 10.1016/j.stemcr.2019.01.023
Cantú-Medellín N., Byrd B., Hohn A., Vázquez-Medina J. P., Zenteno-Savín T. (2011). Differential antioxidant protection in tissues from marine mammals with distinct diving capacities. Shallow/short vs. deep/long divers. Comp. Biochem. Physiol. Part A: Mol. Integr. Physiol. 158 (4), 438–443. doi: 10.1016/j.cbpa.2010.11.029
Carlén I., Nunny L., Simmonds M. P. (2021). Out of sight, out of mind: how conservation is failing European porpoises. Front. Mar. Sci. 8 (February). doi: 10.3389/fmars.2021.617478
Caspi T., Straw S., Cheng C., Garnham J. O., Scragg J. L., Smith J., et al. (2020). Unique transcriptome signature distinguishes patients with heart failure with myopathy. J. Am. Heart Assoc. 9 (18), e017091. doi: 10.1161/JAHA.120.017091
Castellini M. A., Somero G. N., Kooyman G. L. (1981). Glycolytic enzyme activities in tissues of marine and terrestrial mammals. Physiol. Zool. 54 (2), 242–252. doi: 10.1086/physzool.54.2.30155826
Cenni V., Kojic S., Capanni C., Faulkner G., Lattanzi G. (2019). Ankrd2 in mechanotransduction and oxidative stress response in skeletal muscle: New cues for the pathogenesis of muscular laminopathies. Oxid. Med. Cell. Longevity 2019. doi: 10.1155/2019/7318796
Cha Y. J., Kim E.-S., Koo J. S. (2018). Amino acid transporters and glutamine metabolism in breast cancer. Int. J. Mol. Sci. 19 (3), 907. doi: 10.3390/ijms19030907
Champagne C. D., Houser D. S., Fowler M. A., Costa D. P., Crocker D. E. (2012). Gluconeogenesis is associated with high rates of tricarboxylic acid and pyruvate cycling in fasting northern elephant seals. Am. J. Physiol. - Regul. Integr. Comp. Physiol. 303 (3), 340–352. doi: 10.1152/ajpregu.00042.2012
Chegwidden W. R., Carter N. D. (2000). Introduction to the carbonic anhydrases. In: Chegwidden W.R., Carter N.D., Edwards Y.H. (eds). Carbonic Anhydrases: New Horizons. Birkhäuser, Basel. 90, 13–28. doi: 10.1007/978-3-0348-8446-4_2
Chicco A. J., Le C. H., Schlater A., Nguyen A., Kaye S., Beals J. W., et al. (2014). High fatty acid oxidation capacity and phosphorylation control despite elevated leak and reduced respiratory capacity in northern elephant seal muscle mitochondria. J. Exp. Biol. 217 (16), 2947–2955. doi: 10.1242/jeb.105916
Choy E. S., Campbell K. L., Berenbrink M., Roth J. D., Loseto L. L. (2019). Body condition impacts blood and muscle oxygen storage capacity of free-living beluga whales (Delphinapterus leucas). J. Exp. Biol. 222 (11). doi: 10.1242/jeb.191916
Cincinelli A., Martellini T., Innocenti A., Scozzafava A., Supuran C. T. (2011). Purification and inhibition studies with anions and sulfonamides of an α-carbonic anhydrase from the Antarctic seal Leptonychotes weddellii. Bioorganic Medicinal Chem. 19 (6), 1847–1851. doi: 10.1016/j.bmc.2011.02.015
Crocker D. E., Khudyakov J. I., Champagne C. D. (2016). Oxidative stress in northern elephant seals: Integration of omics approaches with ecological and experimental studies. Comp. Biochem. Physiol. Part A: Mol. Integr. Physiol. 200, 94–103. doi: 10.1016/j.cbpa.2016.02.011
Davis R. W. (2014). A review of the multi-level adaptations for maximizing aerobic dive duration in marine mammals: from biochemistry to behavior. J. Comp. Physiol. B 184, 23–53. doi: 10.1007/s00360-013-0782-z
de la Vega L., Grishina I., Moreno R., Krüger M., Braun T., Schmitz M. L. (2012). A redox-regulated SUMO/acetylation switch of HIPK2 controls the survival threshold to oxidative stress. Mol. Cell 46 (4), 472–483. doi: 10.1016/j.molcel.2012.03.003
de Theije C. C., Schols A. M. W. J., Lamers W. H., Ceelen J. J. M., van Gorp R. H., Hermans J. J. R., et al. (2018). Glucocorticoid receptor signaling impairs protein turnover regulation in hypoxia-induced muscle atrophy in male mice. Endocrinology 159 (1), 519–534. doi: 10.1210/en.2017-00603
Di Fiore A., Monti D. M., Scaloni A., De Simone G., Monti S. M. (2018). Protective role of carbonic anhydrases III and VII in cellular defense mechanisms upon redox unbalance. Oxid. Med. Cell. Longevity 2018. doi: 10.1155/2018/2018306
Fabrizius A., Hoff M. L. M., Engler G., Folkow L. P., Burmester T. (2016). When the brain goes diving: transcriptome analysis reveals a reduced aerobic energy metabolism and increased stress proteins in the seal brain. BMC Genomics 17 (1), 1–11. doi: 10.1186/s12864-016-2892-y
Fahlman A., Moore M. J., Garcia-Parraga D. (2017). Respiratory function and mechanics in pinnipeds and cetaceans. J. Exp. Biol. 220 (10), 1761–1773. doi: 10.1242/jeb.126870
Fair P. A., Becker P. R. (2000). Review of stress in marine mammals. J. Aquat. Ecosystem Stress Recovery 7 (4), 335–354. doi: 10.1023/A:1009968113079
Foote A. D., Liu Y., Thomas G. W. C., Vinař T., Alföldi J., Deng J., et al. (2015). Convergent evolution of the genomes of marine mammals. Nat. Genet. 47 (3), 272–275. doi: 10.1038/ng.3198
Frontera W. R., Ochala J. (2015). Skeletal muscle: a brief review of structure and function. Calcified Tissue Int. 96, 183–195. doi: 10.1007/s00223-014-9915-y
García-Castañeda O., Gaxiola-Robles R., Kanatous S., Zenteno-Savín T. (2017). Circulating glutathione concentrations in marine, semiaquatic, and terrestrial mammals. Mar. Mammal Sci. 33 (3), 738–747. doi: 10.1111/mms.12391
Grounds M. D. (2014). The need to more precisely define aspects of skeletal muscle regeneration. Int. J. Biochem. Cell Biol. 56, 56–65. doi: 10.1016/j.biocel.2014.09.010
Hindle A. G. (2020). Diving deep: understanding the genetic components of hypoxia tolerance in marine mammals. J. Appl. Physiol. 128 (5), 1439–1446. doi: 10.1152/japplphysiol.00846.2019
Hindle A. G., Allen K. N., Batten A. J., Hückstädt L. A., Turner-Maier J., Schulberg S. A., et al. (2019). Low guanylyl cyclase activity in Weddell seals: implications for peripheral vasoconstriction and perfusion of the brain during diving. Am. J. Physiology-Regulatory Integr. Comp. Physiol. 316 (6), R704–R715. doi: 10.1152/ajpregu.00283.2018
Hoff M. L. M., Fabrizius A., Czech-Damal N. U., Folkow L. P., Burmester T. (2017). Transcriptome analysis identifies key metabolic changes in the hooded seal (Cystophora cristata) brain in response to hypoxia and reoxygenation. PloS One 12 (1), e0169366. doi: 10.1371/journal.pone.0169366
Hong S., Zhou W., Fang B., Lu W., Loro E., Damle M., et al. (2017). Dissociation of muscle insulin sensitivity from exercise endurance in mice by HDAC3 depletion. Nat. Med. 23 (2), 223–234. doi: 10.1038/nm.4245
Horscroft J. A., Murray A. J. (2014). Skeletal muscle energy metabolism in environmental hypoxia: climbing towards consensus. Extreme Physiol. Med. 3 (1), 1–17. doi: 10.1186/2046-7648-3-19
Houser D. S., Champagne C. D., Crocker D. E. (2013). ). A non-traditional model of the metabolic syndrome: The adaptive significance of insulin resistance in fasting-adapted seals. Front. Endocrinol. 4 (NOV). doi: 10.3389/fendo.2013.00164
Howard E. E., Pasiakos S. M., Blesso C. N., Fussell M. A., Rodriguez N. R. (2020). Divergent roles of inflammation in skeletal muscle recovery from injury. Frontiers in Physiology. 11 87. doi: 10.3389/fphys.2020.00087
Kanatous S. B., Hawke T. J., Trumble S. J., Pearson L. E., Watson R. R., Garry D. J., et al. (2008). The ontogeny of aerobic and diving capacity in the skeletal muscles of Weddell seals. J. Exp. Biol. 211 (16), 2559–2565. doi: 10.1242/jeb.018119
Kastelein R. A., Van de Voorde S., Jennings N. (2018). Swimming speed of a harbor porpoise (Phocoena phocoena) during playbacks of offshore pile driving sounds. Aquat. Mammals 44 (1), 92–99. doi: 10.1578/AM.44.1.2018.92
Kesselring T., Viquerat S., Brehm R., Siebert U. (2017). Coming of age: - Do female Harbour porpoises (Phocoena phocoena) from the North Sea and Baltic Sea have sufficient time to reproduce in a human influenced environment? PloS One 12 (10). doi: 10.1371/journal.pone.0186951
Khudyakov J. I., Champagne C. D., Meneghetti L. M., Crocker D. E. (2017). Blubber transcriptome response to acute stress axis activation involves transient changes in adipogenesis and lipolysis in a fasting-adapted marine mammal. Sci. Rep. 7 (1), 1–12. doi: 10.1038/srep42110
Khudyakov J. I., Champagne C. D., Preeyanon L., Ortiz R. M., Crocker D. E. (2015a). Muscle transcriptome response to ACTH administration in a free-ranging marine mammal. Physiol. Genomics 47 (8), 318–330. doi: 10.1152/physiolgenomics.00030.2015
Khudyakov J. I., Preeyanon L., Champagne C. D., Ortiz R. M., Crocker D. E. (2015b). Transcriptome analysis of northern elephant seal (Mirounga angustirostris) muscle tissue provides a novel molecular resource and physiological insights. BMC Genomics 16 (1), 1–11. doi: 10.1186/s12864-015-1253-6
Kooyman G. L. (1973). Respiratory adaptations in marine mammals. Am. Zoologist 13 (2), 457–468. doi: 10.1093/icb/13.2.457
Kooyman G. L., Wahrenbrock E. A., Castellini M. A., Davis R. W., Sinnett E. E. (1980). Evidence of preferred pathways from blood chemistry and behavior. J. Comp. Physiol. 138, 335–346. doi: 10.1007/BF00691568
Lehnert K., Seibel H., Hasselmeier I., Wohlsein P., Iversen M., Nielsen N. H., et al. (2014). Increase in parasite burden and associated pathology in Harbour porpoises (Phocoena phocoena) in West Greenland. Polar Biol. 37, 321–331. doi: 10.1007/s00300-013-1433-2
Levine B., Kroemer G. (2019). Biological functions of autophagy genes: a disease perspective. Cell 176 (1–2), 11–42. doi: 10.1016/j.cell.2018.09.048
Li B., Feng L., Wu X., Cai M., Yu J. J., Tian Z. (2022). Effects of different modes of exercise on skeletal muscle mass and function and IGF-1 signaling during early aging in mice. J. Exp. Biol. 225 (21), jeb244650.
Li F., Qiao Z., Duan Q., Nevo E. (2021). Adaptation of mammals to hypoxia. Anim. Models Exp. Med. 4 (4), 311–318. doi: 10.1002/ame2.12189
Li R., Shang J., Zhou W., Jiang L., Xie D., Tu G. (2018). Overexpression of HIPK2 attenuates spinal cord injury in rats by modulating apoptosis, oxidative stress, and inflammation. Biomedicine Pharmacother. 103, 127–134. doi: 10.1016/j.biopha.2018.03.117
Lian D., Chen M.-M., Wu H., Deng S., Hu X. (2022). The role of oxidative stress in skeletal muscle myogenesis and muscle disease. Antioxidants 11 (4), 755. doi: 10.3390/antiox11040755
Livak K. J., Schmittgen T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2– ΔΔCT method. Methods 25 (4), 402–408. doi: 10.1006/meth.2001.1262
Mancia A., Ryan J. C., Chapman R. W., Wu Q., Warr G. W., Gulland F. M. D., et al. (2012). Health status, infection and disease in California sea lions (Zalophus californianus) studied using a canine microarray platform and machine-learning approaches. Dev. Comp. Immunol. 36 (4), 629–637. doi: 10.1016/j.dci.2011.10.011
McBean G. J. (2017). Cysteine, glutathione, and thiol redox balance in astrocytes. Antioxidants 6 (3), 62. doi: 10.3390/antiox6030062
McDonald B. I., Johnson M., Madsen P. T. (2018). Dive heart rate in Harbour porpoises is influenced by exercise and expectations. J. Exp. Biol. 221 (1), jeb168740. doi: 10.1242/jeb.168740
Mirman B., Ikeda I., Zhang Z., Liu Y., Yu L., Ehsan A., et al. (2020). Effects of neuropeptide Y on the microvasculature of human skeletal muscle. Surgery 168 (1), 155–159. doi: 10.1016/j.surg.2020.04.020
Monti D. M., De Simone G., Langella E., Supuran C. T., Di Fiore A., Monti S. M. (2017). Insights into the role of reactive sulfhydryl groups of carbonic anhydrase III and VII during oxidative damage. J. Enzyme Inhibition Medicinal Chem. 32 (1), 5–12. doi: 10.1080/14756366.2016.1225046
Mutryn M. F., Brannick E. M., Fu W., Lee W. R., Abasht B. (2015). Characterization of a novel chicken muscle disorder through differential gene expression and pathway analysis using RNA-sequencing. BMC Genomics 16 (1), 1–19. doi: 10.1186/s12864-015-1623-0
Nachtsheim D. A., Viquerat S., Ramírez-Martínez N. C., Unger B., Siebert U., Gilles A. (2021). Small cetacean in a human high-use area: trends in harbor porpoise abundance in the North Sea over two decades. Front. Mar. Sci. 7 (January). doi: 10.3389/fmars.2020.606609
Nedvedova I., Kolar D., Elsnicova B., Hornikova D., Novotny J., Kalous M., et al. (2018). Mitochondrial genome modulates myocardial Akt/Glut/HK salvage pathway in spontaneously hypertensive rats adapted to chronic hypoxia. Physiol. Genomics 50 (7), 532–541. doi: 10.1152/physiolgenomics.00040.2017
Nielsen N. H., Teilmann J., Sveegaard S., Hansen R. G., Sinding M.-H. S., Dietz R., et al. (2018). Oceanic movements, site fidelity and deep diving in Harbour porpoises from Greenland show limited similarities to animals from the North Sea. Mar. Ecol. Prog. Ser. 597, 259–272. doi: 10.3354/meps12588
Nixon M., Stewart-Fitzgibbon R., Fu J., Akhmedov D., Rajendran K., Mendoza-Rodriguez M. G., et al. (2016). Skeletal muscle salt inducible kinase 1 promotes insulin resistance in obesity. Mol. Metab. 5 (1), 34–46. doi: 10.1016/j.molmet.2015.10.004
Noren S. R. (2004). Buffering capacity of the locomotor muscle in cetaceans: Correlates with postpartum development, dive duration, and swim performance. Mar. Mammal Sci. 20 (4), 808–822. doi: 10.1111/j.1748-7692.2004.tb01194.x
Noren S. R., Noren D. P., Gaydos J. K. (2014). Living in the fast lane: rapid development of the locomotor muscle in immature harbor porpoises (Phocoena phocoena). J. Comp. Physiol. B 184 (8), 1065–1076. doi: 10.1007/s00360-014-0854-8
Noren S. R., Williams T. M. (2000). Body size and skeletal muscle myoglobin of cetaceans: adaptations for maximizing dive duration. Comp. Biochem. Physiol. Part A: Mol. Integr. Physiol. 126 (2), 181–191. doi: 10.1016/S1095-6433(00)00182-3
O’Neill B. T., Lee K. Y., Klaus K., Softic S., Krumpoch M. T., Fentz J., et al. (2016). Insulin and IGF-1 receptors regulate FoxO-mediated signaling in muscle proteostasis. J. Clin. Invest. 126 (9), 3433–3446. doi: 10.1172/JCI86522
Otani S., Naito Y., Kato A., Kawamura A. (2001). Oxygen consumption and swim speed of the harbor porpoise Phocoena phocoena. Fisheries Sci. 67 (5), 894–898. doi: 10.1046/j.1444-2906.2001.00338.x
Owen J. B., Butterfield D. A. (2010). Measurement of oxidized/reduced glutathione ratio. In: Bross P., Gregersen N. (eds) Protein Misfolding and Cellular Stress in Disease and Aging. Methods Mol. Biol. Totowa, NJ:Humana Press. 648, 269–277. doi: 10.1007/978-1-60761-756-3_18
Pabst D. A. (1993). Intramuscular morphology and tendon geometry of the epaxial swimming muscles of dolphins. J. Zool. 230 (1), 159–176. doi: 10.1111/j.1469-7998.1993.tb02679.x
Park J. Y., An Y.-R., Kanda N., An C.-M., An H. S., Kang J.-H., et al. (2015). Cetaceans evolution: insights from the genome sequences of common minke whales. BMC Genomics 16 (1), 1–8. doi: 10.1186/s12864-015-1213-1
Philipp C., Unger B., Ehlers S. M., Koop J. H. E., Siebert U. (2021). First evidence of retrospective findings of microplastics in Harbour porpoises (Phocoena phocoena) from German waters. Front. Mar. Sci. 8. doi: 10.3389/fmars.2021.682532
Piotrowski E. R., Tift M. S., Crocker D. E., Pearson A. B., Vázquez-Medina J. P., Keith A. D., et al. (2021). Ontogeny of carbon monoxide-related gene expression in a deep-diving marine mammal. Front. Physiol. 1841. doi: 10.3389/fphys.2021.762102
Piscitelli M. A., McLellan W. A., Rommel S. A., Blum J. E., Barco S. G., Ann Pabst D. (2010). Lung size and thoracic morphology in shallow- and deep-diving cetaceans. J. Morphol. 271 (6), 654–673. doi: 10.1002/jmor.10823
Polasek L. K., Davis R. W. (2001). Heterogeneity of myoglobin distribution in the locomotory muscles of five cetacean species. J. Exp. Biol. 204 (2), 209–215. doi: 10.1242/jeb.204.2.209
Polasek L. K., Dickson K. A., Davis R. W. (2006). ). Metabolic indicators in the skeletal muscles of harbor seals (Phoca vitulina). Am. J. Physiology-Regulatory Integr. Comp. Physiol. 290 (6), R1720–R1727. doi: 10.1152/ajpregu.00080.2005
Puissant A., Fenouille N., Auberger P. (2012). When autophagy meets cancer through p62/SQSTM1. Am. J. Cancer Res. 2 (4), 397.
Read A. J., Hohn A. A. (1995). Life in the fast lane: the life history of harbor porpoises from the Gulf of Maine. Mar. Mammal Sci. 11 (4), 423–440. doi: 10.1111/j.1748-7692.1995.tb00667.x
Rogeri P. S., Gasparini S. O., Martins G. L., Costa L. K. F., Araujo C. C., Lugaresi R., et al. (2020). Crosstalk between skeletal muscle and immune system: which roles do IL-6 and glutamine play? Front. Physiol. 11, 582258. doi: 10.3389/fphys.2020.582258
Rojano-Doñate L., McDonald B. I., Wisniewska D. M., Johnson M., Teilmann J., Wahlberg M., et al. (2018). High field metabolic rates of wild Harbour porpoises. J. Exp. Biol. 221 (23), jeb185827. doi: 10.1242/jeb.185827
Rom O., Reznick A. Z. (2016). The role of E3 ubiquitin-ligases MuRF-1 and MAFbx in loss of skeletal muscle mass. Free Radical Biol. Med. 98, 218–230. doi: 10.1016/j.freeradbiomed.2015.12.031
Romano T. A., Olschowka J. A., Felten S. Y., Quaranta V., Ridgway S. H., Felten D. L. (2002). Immune response, stress, and environment: Implications for cetaceans. Cell Mol. Biol. Mar. Mammals; CJ Pfeiffer Ed. Krieger Publishing Co. Inc. 253–279.
Shah A. N., Alam M. M., Cao T., Zhang L. (2013). The role of ribosomes in mediating hypoxia response and tolerance in Eukaryotes. Mol. Mech. UNDERLYING HYPOXIA-TOLERANCE AND RESPONSE IN YEAST, 81.
Siebert U., Pawliczka I., Benke H., von Vietinghoff V., Wolf P., Pilāts V., et al. (2020). Health assessment of Harbour porpoises (Phocoena phocoena) from Baltic area of Denmark, Germany, Poland and Latvia. Environ. Int. 143. doi: 10.1016/j.envint.2020.105904
Siebert U., Prenger-Berninghoff E., Weiss R. (2009). Regional differences in bacterial flora in Harbour porpoises from the North Atlantic: environmental effects? J. Appl. Microbiol. 106 (1), 329–337. doi: 10.1111/j.1365-2672.2008.04006.x
Siebert U., Tolley K., Víkingsson G. A., Ólafsdottir D., Lehnert K., Weiss R., et al. (2006). Pathological findings in Harbour porpoises (Phocoena phocoena) from Norwegian and Icelandic waters. J. Comp. Pathol. 134 (2–3), 134–142. doi: 10.1016/j.jcpa.2005.09.002
Siebert U., Wünschmann A., Weiss R., Frank H., Benke H., Frese K. (2001). Post-mortem findings in Harbour porpoises (Phocoena phocoena) from the German North and Baltic Seas. J. Comp. Pathol. 124 (2–3), 102–114. doi: 10.1053/jcpa.2000.0436
Sierra E., Espinosa de los Monteros A., Fernández A., Díaz-Delgado J., Suárez-Santana C., Arbelo M., et al. (2017). Muscle pathology in free-ranging stranded cetaceans. Veterinary Pathol. 54 (2), 298–311. doi: 10.1177/0300985816660747
Sierra E., Fernández A., De Los Monteros A. E., Arbelo M., De Quirós Y. B., Herráez P. (2013). Muscular senescence in cetaceans: Adaptation towards a slow muscle fibre phenotype. Sci. Rep. 3, 1–8. doi: 10.1038/srep01795
Sierra E., Fernández A., Espinosa De Los Monteros A., Díaz-Delgado J., Bernaldo De Quirós Y., García-Álvarez N., et al. (2015). Comparative histology of muscle in free ranging cetaceans: Shallow versus deep diving species. Sci. Rep. 5. doi: 10.1038/srep15909
Sies H., Belousov V. V., Chandel N. S., Davies M. J., Jones D. P., Mann G. E., et al. (2022). Defining roles of specific reactive oxygen species (ROS) in cell biology and physiology. Nat. Rev. Mol. Cell Biol. 23 (7), 499–515. doi: 10.1038/s41580-022-00456-z
Sies H., Jones D. P. (2020). Reactive oxygen species (ROS) as pleiotropic physiological signalling agents. Nat. Rev. Mol. Cell Biol. 21 (7), 363–383. doi: 10.1038/s41580-020-0230-3
Snyder G. K. (1983). Respiratory adaptations in diving mammals. Respiration Physiol. 54 (3), 269–294. doi: 10.1016/0034-5687(83)90072-5
Sonne C., Lakemeyer J., Desforges J.-P., Eulaers I., Persson S., Stokholm I., et al. (2020a). A review of pathogens in selected Baltic Sea indicator species. Environ. Int. 137, 105565. doi: 10.1016/j.envint.2020.105565
Sonne C., Siebert U., Gonnsen K., Desforges J. P., Eulaers I., Persson S., et al. (2020b). Health effects from contaminant exposure in Baltic Sea birds and marine mammals: A review. Environ. Int. 139. doi: 10.1016/j.envint.2020.105725
Stewart R., Akhmedov D., Robb C., Leiter C., Berdeaux R. (2013). Regulation of SIK1 abundance and stability is critical for myogenesis. Proc. Natl. Acad. Sci. 110 (1), 117–122. doi: 10.1073/pnas.1212676110
Ten Doeschate M. T. I., Ijsseldijk L. L., Hiemstra S., De Jong E. A., Strijkstra A., Gröne A., et al. (2017). Quantifying parasite presence in relation to biological parameters of Harbour porpoises phocoena phocoena stranded on the Dutch coast. Dis. Aquat. Organisms 127 (1), 49–56. doi: 10.3354/dao03182
Thoma A., Lightfoot A. P. (2018). NF-kB and inflammatory cytokine signalling: role in skeletal muscle atrophy. In: Xiao J. (eds) Muscle Atrophy. Adv. Exp. Med. Biol. Singapore: Springer. 1088, 267–279. doi: 10.1007/978-981-13-1435-3_12
Tian R., Yin D., Liu Y., Seim I., Xu S., Yang G. (2017). Adaptive evolution of energy metabolism-related genes in hypoxia-tolerant mammals. Front. Genet. 8, 205. doi: 10.3389/fgene.2017.00205
Torrente L., Sanchez C., Moreno R., Chowdhry S., Cabello P., Isono K., et al. (2017). Crosstalk between NRF2 and HIPK2 shapes cytoprotective responses. Oncogene 36 (44), 6204–6212. doi: 10.1038/onc.2017.221
Torres-Velarde J. M., Kolora S. R. R., Khudyakov J. I., Crocker D. E., Sudmant P. H., Vázquez-Medina J. P. (2021). Elephant seal muscle cells adapt to sustained glucocorticoid exposure by shifting their metabolic phenotype. Am. J. Physiology-Regulatory Integr. Comp. Physiol. 321 (3), R413–R428. doi: 10.1152/ajpregu.00052.2021
Tower D. B., Young O. M. (1973). The activities of butyrylcholinesterase and carbonic anhydrase, the rate of anaerobic glycolysts, and the question of a constant density of glial cells in cerebral cortices of various mamMalian species from mouse to whale. J. Neurochem. 20 (2), 269–278. doi: 10.1111/j.1471-4159.1973.tb12126.x
Tsompanidis A., Vafiadaki E., Blüher S., Kalozoumi G., Sanoudou D., Mantzoros C. S. (2016). Ciliary neurotrophic factor upregulates follistatin and Pak1, causes overexpression of muscle differentiation related genes and downregulation of established atrophy mediators in skeletal muscle. Metabolism 65 (6), 915–925. doi: 10.1016/j.metabol.2016.03.005
Turner D. C., Seaborne R. A., Sharples A. P. (2019). Comparative transcriptome and methylome analysis in human skeletal muscle anabolism, hypertrophy and epigenetic memory. Sci. Rep. 9 (1), 4251. doi: 10.1038/s41598-019-40787-0
Vázquez-Medina J. P., Zenteno-Savín T., Elsner R. (2006). Antioxidant enzymes in ringed seal tissues: potential protection against dive-associated ischemia/reperfusion. Comp. Biochem. Physiol. Part C: Toxicol. Pharmacol. 142 (3–4), 198–204. doi: 10.1016/j.cbpc.2005.09.004
Velten B. P., Dillaman R. M., Kinsey S. T., McLellan W. A., Pabst D. A. (2013). Novel locomotor muscle design in extreme deep-diving whales. J. Exp. Biol. 216 (10), 1862–1871. doi: 10.1242/jeb.081323
Vinther M., Larsen F. (2004). Updated estimates of Harbour porpoise (Phocoena phocoena) bycatch in the Danish North Sea bottom-set gillnet fishery. J. Cetacean Res. Manage. 6 (1), 19–24. doi: 10.47536/jcrm.v6i1.785
Wang X., Zhang Y., Wan X., Guo C., Cui J., Sun J., et al. (2020). Responsive expression of MafF to β-amyloid-induced oxidative stress. Dis. Markers 2020. doi: 10.1155/2020/8861358
Westgate A. J., Read A. J., Koopaaan H. N., Caskin D. E. (1995). Diving behaviour of Harbour porpoises, Phocoena phocoena. Can. J. Fisheries Aquat. Sci. 52 (5), 1064–1073. doi: 10.1139/f95-104
Wilhelm Filho D., Sell F., Ribeiro L., Ghislandi M., Carrasquedo F., Fraga C. G., et al. (2002). Comparison between the antioxidant status of terrestrial and diving mammals. Comp. Biochem. Physiol. Part A: Mol. Integr. Physiol. 133 (3), 885–892. doi: 10.1016/S1095-6433(02)00253-2
Williams Z. J., Velez-Irizarry D., Gardner K., Valberg S. J. (2021). Integrated proteomic and transcriptomic profiling identifies aberrant gene and protein expression in the sarcomere, mitochondrial complex I, and the extracellular matrix in Warmblood horses with myofibrillar myopathy. BMC Genomics 22 (1), 1–20. doi: 10.1186/s12864-021-07758-0
Wisniewska D. M. M., Johnson M., Teilmann J., Rojano-Doñate L., Shearer J., Sveegaard S., et al. (2016). Ultra-high foraging rates of harbor porpoises make them vulnerable to anthropogenic disturbance. Curr. Biol. 26 (11), 1441–1446. doi: 10.1016/j.cub.2016.03.069
Wisniewska D. M., Johnson M., Teilmann J., Siebert U., Galatius A., Dietz R., et al. (2018). High rates of vessel noise disrupt foraging in wild Harbour porpoises (Phocoena phocoena). Proc. R. Soc. B: Biol. Sci. 285 (1872), 20172314. doi: 10.1098/rspb.2017.2314
Wu K. C., Reisman S. A., Klaassen C. D. (2020). Tissue distribution, hormonal regulation, ontogeny, diurnal expression, and induction of mouse cystine transporters Slc3a1 and Slc7a9. Free Radical Res. 54 (7), 525–534. doi: 10.1080/10715762.2020.1812597
Wünschmann A., Frese K., Müiller G., Baumgärtner W., Siebert U., Weiss R., et al. (2001). Evidence of infectious diseases in Harbour porpoises (Phocoena phocoena) hunted in the waters of Greenland and by-caught in the German North Sea and Baltic Sea. Veterinary Rec. 148 (23), 715–720. doi: 10.1136/vr.148.23.715
Xia Q., Huang X., Huang J., Zheng Y., March M. E., Li J., et al. (2021). The role of autophagy in skeletal muscle diseases. Front. Physiol. 12, 638983. doi: 10.3389/fphys.2021.638983
Yadava R. S., Foff E. P., Yu Q., Gladman J. T., Zheng T. S., Mahadevan M. S. (2016). TWEAK regulates muscle functions in a mouse model of RNA toxicity. PloS One 11 (2), e0150192. doi: 10.1371/journal.pone.0150192
Keywords: cetacea, hypoxia, oxidative stress, muscle, transcriptome, harbor porpoise, marine mammals
Citation: Dönmez EM, Siebert U and Fabrizius A (2023) Diving on damage—the muscle transcriptome of parasitic infested harbor porpoises (Phocoena phocoena) hints at oxidative stress but not hypoxia. Front. Mar. Sci. 10:1232305. doi: 10.3389/fmars.2023.1232305
Received: 31 May 2023; Accepted: 20 July 2023;
Published: 10 August 2023.
Edited by:
Jose Carlos Báez, Spanish Institute of Oceanography (IEO), SpainReviewed by:
Lene H. Petersen, Texas A&M University at Galveston, United StatesStephen T. Kinsey, University of North Carolina Wilmington, United States
Copyright © 2023 Dönmez, Siebert and Fabrizius. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Eda Merve Dönmez, eda.doenmez@uni-hamburg.de
 Eda Merve Dönmez
Eda Merve Dönmez Ursula Siebert
Ursula Siebert Andrej Fabrizius
Andrej Fabrizius