Transcriptomic Analysis and Time to Hatch Visual Prediction of Embryo Development in the Ornate Spiny Lobster (Panulirus ornatus)
- 1Centre for Bioinnovation, University of the Sunshine Coast, Maroochydore, QLD, Australia
- 2School of Science and Engineering, University of the Sunshine Coast, Maroochydore, QLD, Australia
- 3Institute for Marine and Antarctic Studies, University of Tasmania, Hobart, TAS, Australia
The ability to conduct closed life-cycle culture of tropical spiny lobsters, Panulirus ornatus, at the Institute for Marine and Antarctic Studies (IMAS) provides a unique opportunity to investigate specific developmental stages during embryogenesis. The production of closed life-cycle lobsters provides access to embryos at defined time points, yet physical staging is challenging due to their small size and environmental factors impacting their development. A transcriptome comprising 11 distinct stages across the 30-day P. ornatus embryonal development period allowed the establishment of the most detailed transcriptomic library of embryogenesis across decapods. A series of key genes across the 11 stages of embryonal development were characterized. The expression of neuropeptides was reported across P. ornatus embryos, suggesting they are primarily active during the later stages of embryogenesis as the nervous system develops and the animal prepares to hatch. Gastrula-specific genes, nanos and brachyury, presented an expression profile indicating gastrulation occurs early in embryogenesis. In addition to the molecular tools used to characterize embryo development, certain developmental characteristics, such as eye spot development, provide a measurable indicator that can be visualised. Hatch prediction models based on visual characteristics were shown to be an accurate method to predict the timing of the hatch for P. ornatus embryos kept at 26°C. The combination of morphological and visual measures with transcriptomics can be used to further define and establish the groundwork for future characterisation and staging of P. ornatus embryogenesis.
Introduction
Tropical spiny lobsters support coastal communities’ livelihoods across the globe through commercial and recreational fisheries. The ornate spiny lobster (Panulirus ornatus) is a prime candidate for aquaculture production. P. ornatus possesses several enviable traits, including consistently high market value and strong commercial demand unmet by fully exploited fisheries (Jeffs, 2010; Sachlikidis et al., 2010). Whilst there has been considerable interest in the development of sustainable closed life cycle spiny lobster aquaculture, until recently their complex and extended larval cycle, specialised feed requirements and need for high quality water posed significant challenges. Enhancing our understanding of lobster embryology may provide additional biological insights into the early life-history of lobsters.
Female P. ornatus can produce up to one million eggs in a single spawning event (Musbir et al., 2018). Once spawned and fertilized, the eggs are carried on the female’s abdomen attached to the pleopods, where they undergo embryogenesis lasting 3-5 weeks, with duration defendant on ambient water temperatures (Sachlikidis et al., 2010). At colder temperatures, embryogenesis is significantly longer than at warmer temperatures and expectedly, embryonal development lasts longer in temperate and sub-temperate spiny lobster species. At 20°C, the incubation period for the sub-temperate eastern spiny lobster, Sagmariasus verreauxi, is approximately 55 days (Moss et al., 2004). In P. ornatus, embryogenesis lasted 36 days when kept at 24°C and reduced to 22 days at 30°C (Sachlikidis et al., 2010), a timeline confirmed by Liang and He (2012). Previous work in S. verreauxi showed that when reared at higher temperatures, eggs produce smaller larvae upon hatching, which did not survive as long when food was withheld, in comparison to eggs reared at lower temperatures with a longer incubation period (Moss et al., 2004). The control of environmental systems at the Institute for Marine and Antarctic Studies (IMAS), particularly temperature, provide a unique opportunity to sample P. ornatus embryos according to a defined timeline.
There is little understanding of egg morphological development and categorization of stages in spiny lobsters. With decapod embryogenesis research both diverse and scant in topic and availability (Helluy and Beltz, 1991), inferring from one genus to another is challenging. In the model arthropod Drosophila melanogaster, a syncytial blastoderm is formed as nuclei migrate to the egg perimeter early in embryogenesis. Approximately 4 hours following fertilization, nuclei accumulate within the cytoplasm and move towards the surface where they form cells surrounding the yolk sac resulting in a cellular blastoderm (Campos-Ortega and Hartenstein, 1997). Gastrulation then occurs, creating the germ layers: endoderm, mesoderm and ectoderm. Cells are rearranged as the germ band elongates and distinct cells appear, embryo segments then become visible, forming an arrangement along the anterior-posterior axis. This is followed by segmentation and the formation of the nervous system and internal organs (Campos-Ortega and Hartenstein, 1997). However, these stages are not as clearly partitioned in spiny lobsters, which makes defining stages difficult (Scholtz and Wolff, 2013).
In the American lobster, Homarus americanus, superficial cleavage results in blastoderm formation where the central mass of yolk remains intact (Helluy and Beltz, 1991). Following cleavage and during blastoderm, yolk pyramids are formed when the yolk is divided into wedge-shaped sections (Brena and Akam, 2012). After gastrulation, where the embryo transforms from a single layer of epithelial cells (blastula) into a multilayered structure (Pijuan-Sala et al., 2019), yolk pyramids are formed from the midgut cells, forming compartments dividing the yolk (Scholtz and Wolff, 2013). The formation of the germ disc, which will give rise to the three germ layers, leads to the disappearance of the yolk pyramids. As the germ disc grows longitudinally, it becomes the germ band extending down one side of the egg, and is comprised of blastoderm cells (Scholtz and Wolff, 2013). Based on several examples, it is concluded that the germ band, formed from the blastoderm thickening, is related to yolk quantity. The germ band stretches across the ventral side of the embryo, which is where body segments begin to form (Scholtz and Wolff, 2013). Typically, in arthropods, the body segments are formed one at a time from the anterior to posterior. After several days, naupliar organization is apparent in H. americanus. This is characterized by the presence of the median eye as well as three pairs of appendages (the antennulae, antennae and mandibles) (Helluy and Beltz, 1991). The differentiation and growth of the post-mandibular appendages results in the metanauplius. H. americanus hatch as a mature metanauplius that rapidly molts into first stage larvae (Helluy and Beltz, 1991), whereas spiny lobsters hatch as naupliosoma, undertaking a molt to first stage phyllosoma within minutes (Moss et al., 2004).
While it has been difficult to obtain visual confirmation of the various embryogenesis phases in some arthropods, including spiny lobsters, there are key genes involved in embryogenesis which are well described and express at a consistently timed manner across the phylum. The analysis of transcriptomic changes could therefore provide an alternate solution to defining embryogenesis in spiny lobsters. Moreover, in the case of P. ornatus, detailed transcriptomic libraries are available for larval stages (Hyde et al., 2019) and adult tissues (Ventura et al., 2020) that can assist to further resolve tissue and stage specific expressions. Decapod transcriptomics have primarily focused on larval and adult stages over the past decade, addressing key challenges faced by the aquaculture industry (Nguyen, Jung, et al., 2018). A limited number of transcriptomic investigations addressed embryonal development in decapods, all addressing only one to three embryonal stages at most (Table 1).
Genes associated with different stages of embryonal development were uncovered in several decapod species. In the marine prawn Penaeus japonicus, embryos were collected immediately after spawning and developed to late gastrulation prior to RNA isolation (Sellars et al., 2015). The gene brachyury was expressed specifically during gastrulation, consistent with invagination that occurs in this stage (Sellars et al., 2015). Samples of oosperms (zygote) and embryos at the 2-4 cell stage were taken from the Chinese mitten crab Eriocheir sinensis, and 432 differentially expressed genes (DEGs) were identified between the two stages with results suggesting that embryonal development events begin prior to cleavage in crabs (Hui et al., 2017). Genes upregulated in the 2-4 cell stage embryos of E. sinensis include Nanos, which is associated with sex differentiation of the germline, as well as genes involved in segmentation such as segmentation protein paired, hunchback and bric-a-brac. Embryos at 20 and 27 days following fertilization were collected for transcriptomic analysis from the red claw crayfish Cherax quadricarinatus, demonstrating large morphological changes, with 20 day embryos corresponding with the formation of eye pigments and short abdominal appendages without segmentation and day 27 embryos corresponding to the pre-hatching stage as the body and appendages were segmented (Wang et al., 2020). Fundamental development genes were highly expressed in the day 20 embryos, including HtrA2, Caspase 8, hedgehog, and MAPKm (Wang et al., 2020). Establishing a detailed P. ornatus embryo transcriptome would allow for genes associated with embryonal development to be mapped across embryogenesis and enable more accurate staging of the P. ornatus embryonal stages.
Neuropeptides play key roles across the developmental process in P. ornatus. The expression of neuropeptides has been examined across later P. ornatus life stages, with distinct expression profiles observed, particularly between phyllosoma and puerulus. Differential expression analysis revealed that towards the end of the phyllosoma phase, there is wide downregulation of the neuropeptides in the 24 hours prior to metamorphosis (gut retraction) and in the post-molt puerulus (Hyde et al., 2020b). In adults, neuropeptides have been examined in a variety of decapod species, including the Norway lobster, Nephrops norvegicus (Nguyen, Rotllant, et al., 2018), P. ornatus (Ventura et al., 2020), and C. quadricarinatus (Nguyen et al., 2016) where characterization was focused on biological processes such as sexual development and largely based on expression profiles across the tissues.
In this study we developed a reference P. ornatus embryo transcriptome across 11 embryonal developmental stages, presenting a detailed description regarding the expression of key genes across embryogenesis in decapods in a species for which we currently have a limited understanding of embryonal development. Differential expression analysis uncovered key genes with differential expression profiles across the 11 stages of embryonal development, including maternally inherited genes, stage-specific genes involved with gastrulation, and crustacean neuropeptides. Two hatch prediction models, based on visual characteristics of P. ornatus embryos, were examined to accurately predict hatching. This provides a basis for further characterization and staging of embryo development in P. ornatus.
Methodology
Sample Preparation and RNA Extraction
Broodstock P. ornatus (6 mature females and 1 mature male) were reared at the IMAS aquaculture facility, Tasmania where they were maintained in 5600 L fiberglass tanks kept at a consistent temperature of 26°C and photoperiod (14: 10 L: D) control. The environmental water conditions were maintained by a recirculating aquaculture system, with water quality parameters monitored daily to keep conditions consistent: salinity 34-35 ppt, dissolved oxygen 100%, pH 8.1-8.2, NH4-N <1.0 mg L-1, NH3 <0.2 mg L-1 and NO2 – H <5.0 mg L-1. Embryo samples (approximately 250-500 individual embryos per extractions) were taken from three mature spawning females (spawning is considered day 0 of embryo development) and then every three days until hatch on day thirty (Figure 1). Samples were snap frozen in liquid nitrogen and kept frozen at -80°C prior to RNA extraction. Total RNA was extracted from a total of 33 pooled embryo samples (n = 3 for each of the 11 stages), weighing 50 - 100 mg each. The frozen samples were submerged in 500 μl RNAzol (Molecular Research Center) containing 10 μl β-mercapto-ethanol and homogenized using a pellet pestle motor for 60 seconds until no visible tissue remained. Following homogenization, 500 μl RNAzol was added and centrifuged at 12,000 g for 5 minutes at 4°C to remove excess lipids. The liquid beneath the lipid layer was transferred to 400 μl RNase free H20, and the homogenate mixed vigorously using a vortex for 15 seconds. The samples were incubated at room temperature for 15 minutes and then centrifuged at 12,000 g for 15 minutes at 4°C. One mL of supernatant was collected and transferred to a new tube. Four hundred μl 75% pure grade ethanol were added to the tubes which were inverted, then incubated for 10 minutes at 4°C. Following incubation, the samples were centrifuged at 12,000 g for 8 minutes at 4°C. The supernatant was discarded, and the pellet was washed with 500 μl 75% ethanol, vortexed briefly and centrifuged at 8,000 g for 3 minutes at 4°C. The supernatant was discarded, and the wash repeated twice. Following the final wash, the supernatant was discarded, and the tube spun down briefly to collect residual ethanol at the bottom of the tube, which was removed by pipetting. The tubes were left open in the fumehood for 10 minutes to allow evaporation of the excess ethanol from the pellet. Fifty μl molecular grade RNase free water was added, the tubes were then vortexed briefly and left for 10 minutes to dissolve the pellet at room temperature. The extracted RNA samples were then quantified using NanoDrop 2000 (ThermoFisher, Australia) and kept in -80°C.
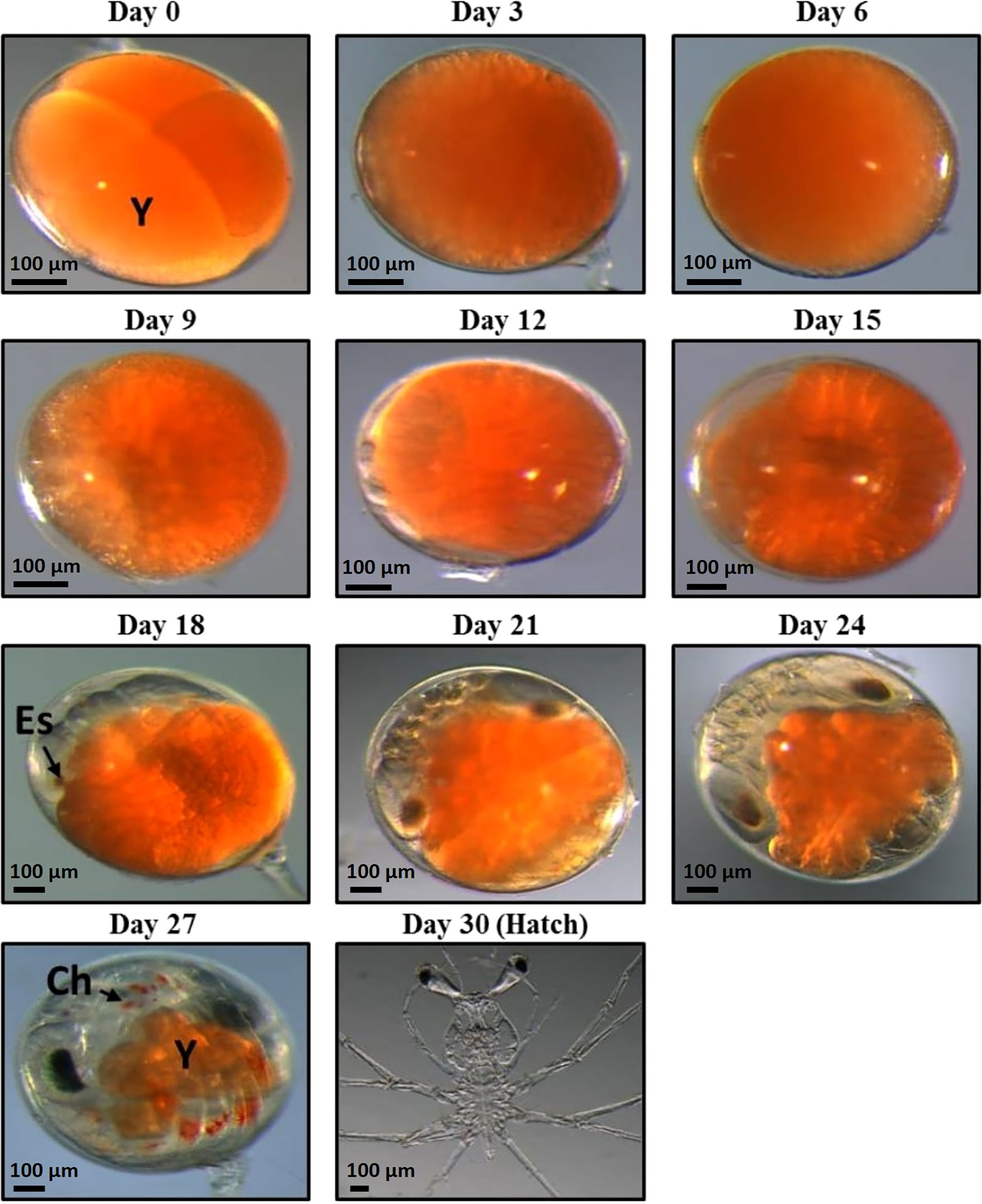
Figure 1 11 stages of Panulirus ornatus embryo development samples were taken for RNA extraction. Embryos were sampled every 3 days from day zero until hatch at day 30 (photos taken by Larnie Linton). Day 30 (hatch) photo is stage I phyllosoma, rather than hatched naupliosoma, which were unable to be photographed. There are 11 stages of embryonic development of P. ornatus; fertilized egg, cleavage, blastula, gastrula, egg-nauplius, embryo with 7 appendage pairs, embryo with 9 appendage pairs, embryo with 11 appendage pairs, compound eye pigments, pre-hatching and hatching (Liang and He, 2012) Abbreviations are as follows: Y, yolk; Es, eye spot; Ch, chromatophore.
Sequencing and Transcriptome Assembly
A minimum of 3 μg clean RNA (integrity of RNA assessed by bioanalyzer; Supplementary Material 1) from each of the 33 pooled samples were rehydrated with RNAstable® LD (Sigma-Aldrich) following the manufacturer’s protocol and sent to Novogene (Hong-Kong) for quality control using Bioanalyzer, library preparation (TrueSeq) and Illumina RNA Sequencing (HiSeq2500, 150 paired end reads). A minimum of 20 million clean reads were generated/sample. Clean reads were de novo assembled using Trinity. Hierarchical clustering was performed by Corset. The assembly was based on clean reads from a total of 41 fastq files, including 11 stages, 12 samples from across metamorphosis (Hyde et al., 2019) and 18 adult tissues (Ventura et al., 2020), generating a comprehensive reference transcriptome forming a library of transcripts expressed across the P. ornatus life cycle. One replicate per embryo stage (n=11) was chosen for the assembly to reduce the computational load and minimize the generation of redundancy in predicted isoforms. The 12 metamorphosis samples were based on RNA extracted from whole animals (7 phyllosoma, 3 puerulus and 2 juveniles) homogenized in RNAzol RT (MRC) with 1% β–mercaptoethanol, and then following manufacturer guidelines (Hyde et al., 2019). Eighteen adult tissues were dissected from male and female adult P. ornatus and RNA was extracted from up to 100 mg of tissue using RNAzol (MRC) (Ventura et al., 2020). The expression of the embryo-derived transcripts was digitally quantified using DeSeq2 in each of the samples relative to the library size, calculated as reads per kilobase per million reads (RPKM). Functional annotation was inferred based on seven databases: NR, NT, KO, Swissprot, PFAM, GO, and KOG.
Gene Expression Analysis
The RPKM of the transcripts were used to compare the 11 stages of P. ornatus embryonal development. A principal component analysis (PCA) was performed on the top 1000 expressed transcripts using ClustVis (Metsalu and Vilo, 2015). RPKM values were used to identify differentially expressed genes (DEGs) across the 11 stages. A list of genes associated with different aspects of embryonal development was curated based on previous studies of decapod embryonal development. A BLAST search using the protein sequences derived from the embryonal development-associated genes was conducted against the embryo transcriptome database to identify P. ornatus transcripts encoding these genes. The RPKM of the P. ornatus derived transcripts were retrieved and plotted against the 11 stages of embryo development.
The top 250 expressed genes in the ovary of adult P. ornatus were analysed. Those that were specific to the ovary or ovary-enriched (> 5-fold increase compared to other tissues) were then identified in the 11 embryonal development samples. The expression of the ovary-enriched transcripts was mapped across the adult tissues and embryogenesis in the form of a heat map (Babicki et al., 2016). Amino acid sequences from previously known embryo stage-specific genes, including vasa, nanos and brachyury, were subjected to a BLAST search across the embryo development transcriptome. Phylogenetic analysis confirmed the identities of the P. ornatus transcripts. The reads for the embryo samples were mapped onto the brachyury gene to confirm the reliability of the quantification. The transcripts were plotted against the 11 stages of embryogenesis to confirm stage-specific expression in P. ornatus. The crustacean neuropeptides were identified based on sequence similarity with related decapods including P. ornatus (Hyde et al., 2020c) and digital expression profiles mapped the P. ornatus expression patterns across embryogenesis.
Hatch Prediction Models for Developing Panulirus ornatus Embryos
P. ornatus embryos, attached to abdomen of adult females kept at 26°C were sampled every day from spawning (day 0) to hatch (day 30). Measurements of the eye index and yolk percentage were taken using LASv4 measuring program. The eye index was calculated as the length of the eye + the width of the eye (the widest section) divided by the number of measurements taken. The yolk percentage was measured as the percentage that the yolk contributed to the embryo. Measurements were plotted against the number of days until hatch to define hatch prediction models.
Results
Transcriptome Sequencing of Panulirus ornatus Embryo Development Stages
Assembly of 41 RNA-Seq FASTQ files (11 embryo stages, 12 metamorphic stages and 18 tissues, as described in the methodology) generated a total of 253,700 transcripts, with 245,416 transcripts in the embryo samples. More than fifty percent of the transcripts were less than 1 kbp. The assembled embryo-derived transcripts were annotated based on seven databases: NR (74,986 genes annotated), NT (22,559 genes annotated), KO (26,290 genes annotated), Swissprot (52,025 genes annotated), Pfam (28,095 genes annotated), GO (26,102 genes annotated), and KOG (26,372 genes annotated), with 31.52% of all transcripts annotated in at least one database. The 33 embryo FASTQ files were uploaded to the NCBI SRA database (PRJNA761502). Principal component analysis displayed varying levels of clustering of the biological replicates (Figure 2; see Supplementary Material 2). This was based on the top 1000 expressed transcripts which were calculated to equate 52.68% of the total expression in the embryos, representing the majority of expression profiles in these samples. The top 1000 expressed transcripts across the 11 stages of embryogenesis were plotted against the main two principal components, showing that the day 0 embryos cluster tightly together and far away from the remainder of the samples which cluster within biological replicates in sequential order from day 3 to day 30 embryos. This pattern remained when the day 0 embryos were removed from analysis, with day 3 embryos clustering tightly and away from the remaining embryo stages. When the day 0 and day 3 embryos were removed from the analysis, the clustering of replicates was less distinct.
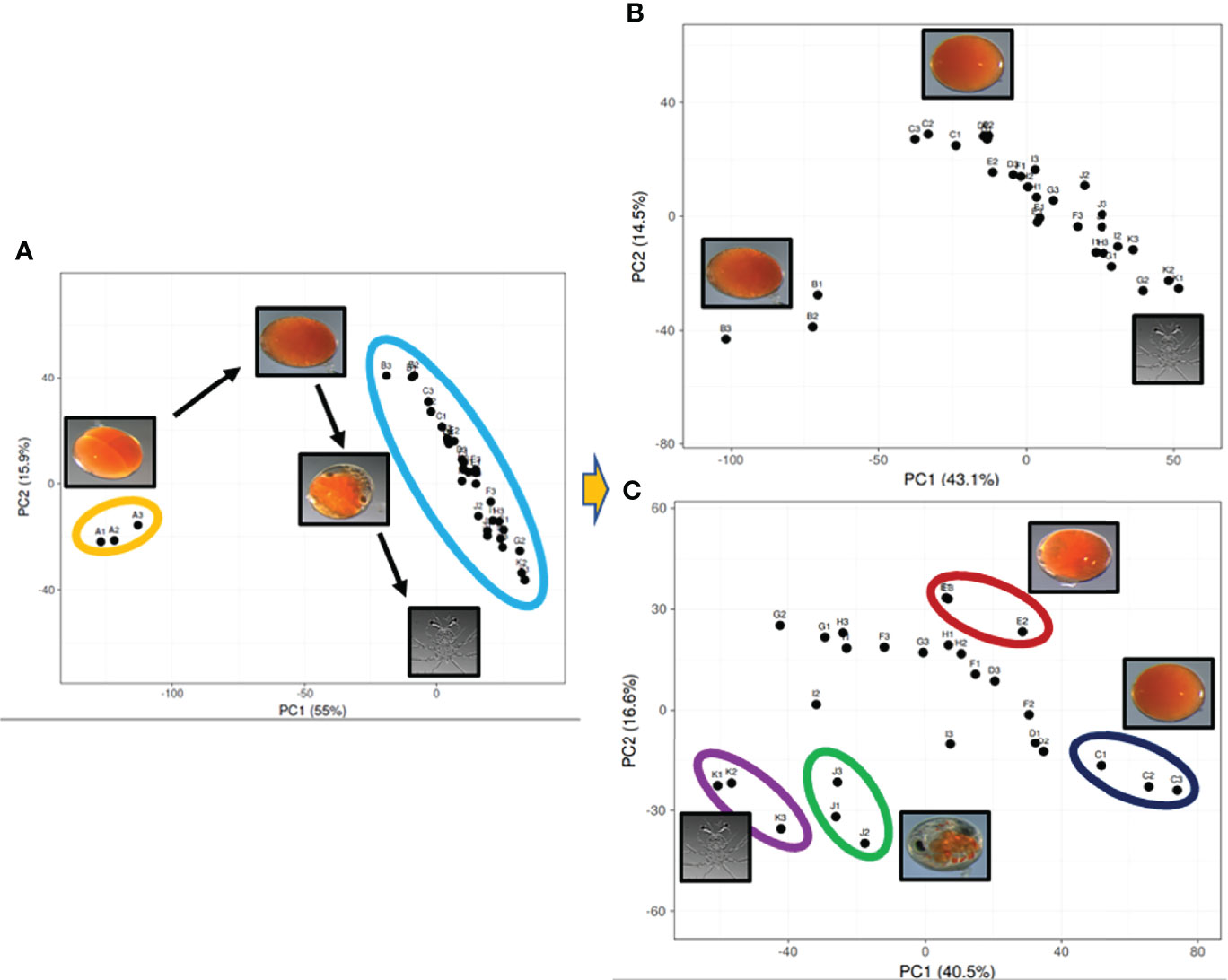
Figure 2 Principal component analysis visualizing the top 1000 expressed genes across the 11 stages of embryo development derived from the Panulirus ornatus transcriptome. The top 1000 genes are the same across all three plots, the number of samples and stages is receding with 33 (11) (A), then 30 (10) (B), 27 (9) (C), where the two main principal components explain diminishing, yet significant variation at 55%, 43.1% and 40.5% for PC1 and 15.9%, 14.5% and 16.6% for PC2.
Differential Expression of Genes
The RPKM values of the top 250 expressed transcripts in the P. ornatus ovary were compared to the RPKM values of the other adult tissues. Those transcripts that were specific, or enriched in the ovary (at least a 5-fold increase in the ovary) were further analysed. Seventy-two transcripts demonstrated ovary-specificity or ovary-enrichment. These 72 transcripts were then identified in the P. ornatus embryo. The expression of the 72 transcripts was then mapped across the adult tissues and embryogenesis in the form of a heat map (Figure 3). This revealed that the maternally-inherited genes were expressed predominantly in the day 0 embryos, where expression was the highest, and for the majority of the transcripts there is little to no expression in the other embryonal development stages. Thirty-eight transcripts were provided with annotation including, gametocyte-specific factor, lipid storage droplet protein, carcinin-like protein, papilin, AN1-type zinc finger protein, allantoicase, and coactosin-like protein. Of the 72 ovary-enriched transcripts, 34 were not provided with annotation from the seven databases, including those with the highest expression. A BLAST search across the NCBI database determined that these transcripts were mitochondrial in origin, but the majority did not have annotation (see Supplementary Material 3). Whilst the expression was high across the six ovary samples, the expression of all 72 transcripts was consistently higher in the developed ovaries of the adult females in comparison with the immature ovaries in the juveniles.
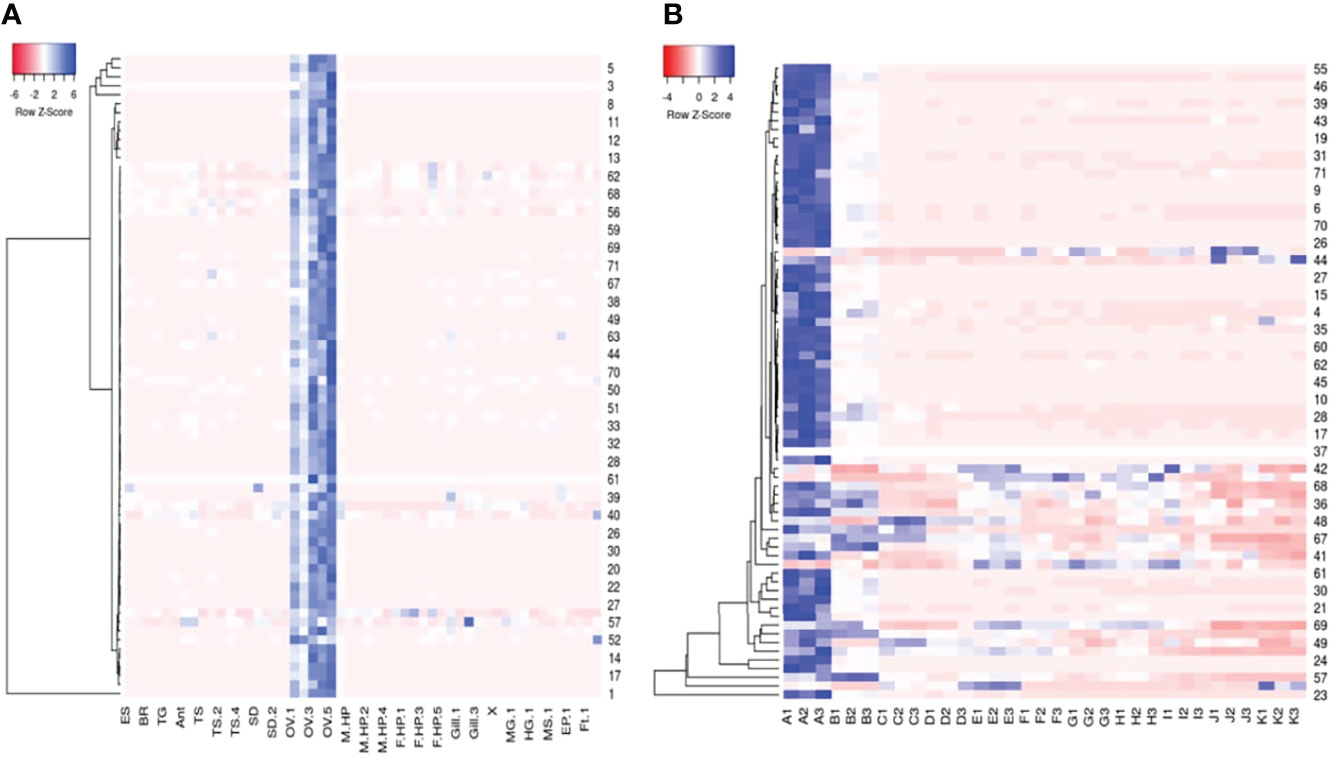
Figure 3 Heatmaps of seventy-two Panulirus ornatus maternally-inherited transcripts identified in adult tissues (A) and 11 stages of embryonal development (B) made using Heatmapper (Babicki et al., 2016). The intensity of the transcript expression, represented as RPKM are displayed as colours ranging from red (lowest expression) to blue (highest expression). ES, eyestalk; BR, brain; TG, thoracic ganglia; Ant, antennal gland; TS, testes; SD, sperm duct; OV, ovary; M HP, male hepatopancreas, F HP, female hepatopancreas; MG, midgut (stomach); HG, hindgut (intestine); MS, muscle; EP, epidermis; Ft, fat (A). The 11 P. ornatus embryonal stages from fertilization to hatch are represented in triplicates as follows; A – day 0, B – day 3, C – day 6, D – day 9, E – day 12, F – day 15, G – day 18, H – day 21, I – day 24, J – day 27, K – day 30.
A search for the gastrula associated genes, vasa, nanos, and brachyury, retrieved six P. ornatus transcripts; 3 vasa, 2 nanos and 1 brachyury. Vasa expression remained insignificant across the 11 embryogenesis stages, whilst nanos and brachyury demonstrate specificity to day 3 embryos (Figure 4). The reads from day 3 embryo samples (triplicates) were mapped onto the brachyury sequence derived from the P. ornatus embryo transcriptome, to demonstrate the reliability of the quantification used for the expression of genes across P. ornatus embryogenesis (Supplementary Material 4). The P. ornatus brachyury sequence presents several SNPs and demonstrates that this is a partial sequence.
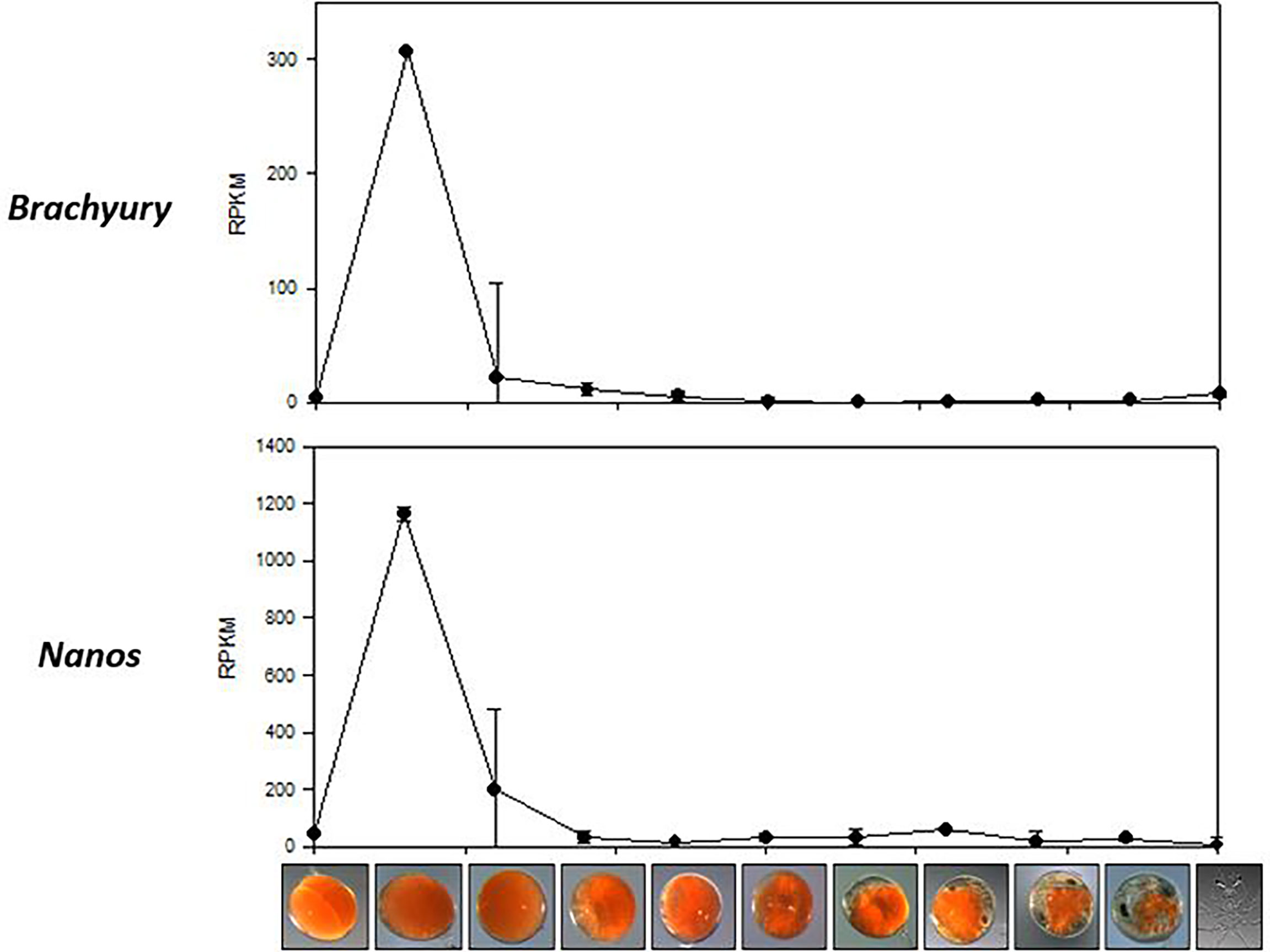
Figure 4 Expression of two gastrula-specific genes, Brachyury and Nanos across the 11 embryonal development stages of Panulirus ornatus, represented as RPKM.
A search for 67 neuropeptides previously identified in P. ornatus (Hyde et al., 2020b) found 58 putative neuropeptides across the 11 stages of embryonal development. The majority of the putative neuropeptides have relatively low expression with a gradual increase in expression as the embryos develop (Figure 5; see Supplementary Material 5). The digital expression patterns show two clear expression profiles. Forty-nine of the neuropeptides demonstrate minimal expression in the earlier stages of embryonal development, with a significant increase in the later stages, including PDH2, calcitonin-like, NPF1a, orcokinin, RPCJ, sNPF, and sulfakinin. The majority of the neuropeptides following this expression profile peak on day 30. Nine of the neuropeptides have expression predominantly in the earlier embryonal stages with a decrease in expression during the later stages of development. Five of these demonstrate significantly elevated expression in day 0 embryos – Diuretic hormone (DH44), GPB5, NFT, NP-4, and SIFamide.
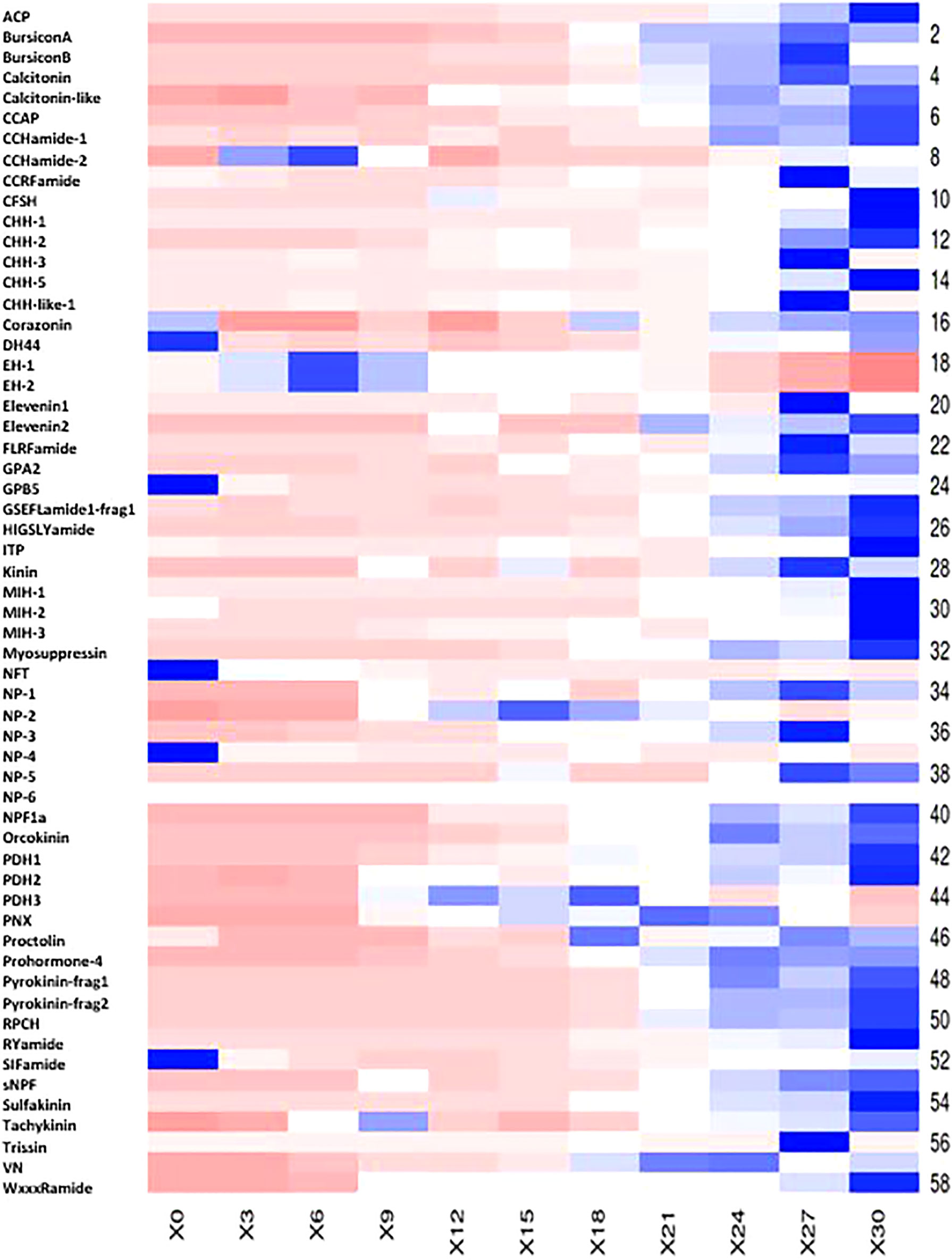
Figure 5 Heatmap of 58 putative Panulirus ornatus neuropeptides across 11 stages of embryogenesis, generated using Heatmapper (Babicki et al., 2016). The intensity of the transcript expression, represented as RPKM are displayed as colours ranging from red (lowest expression) to blue (highest expression). X axis presents the stage of embryonal development, represented as days post-fertilization.
Hatch Prediction of Developing P. ornatus Embryos
Two measurements were used for P. ornatus embryos hatch prediction. The eye index and the yolk percentage of developing embryos were measured every day from fertilization until hatch (Figure 6; see Supplementary Material 6). Based on the eye index hatch prediction model, there is a clear relationship between the eye index and hatching of the embryos. The size of the eye increases as the embryo develops and gets closer to hatching. The earliest the eye index could be measured was 14 days prior to hatch in the 30-day embryonal development period at 26°C, with the eye measuring 15 μm. As the embryos develop, the eye index increases linearly. At 8 days until hatch, the eye index of two embryos was calculated at 71 μm and 78 μm. At 2 days until hatch, three measurements were taken, with an eye index range of 117 μm, to 146 μm. The measurements taken earlier in embryogenesis demonstrate less variance in the size of the eye, indicating that this model can be used to reliability predict hatching.
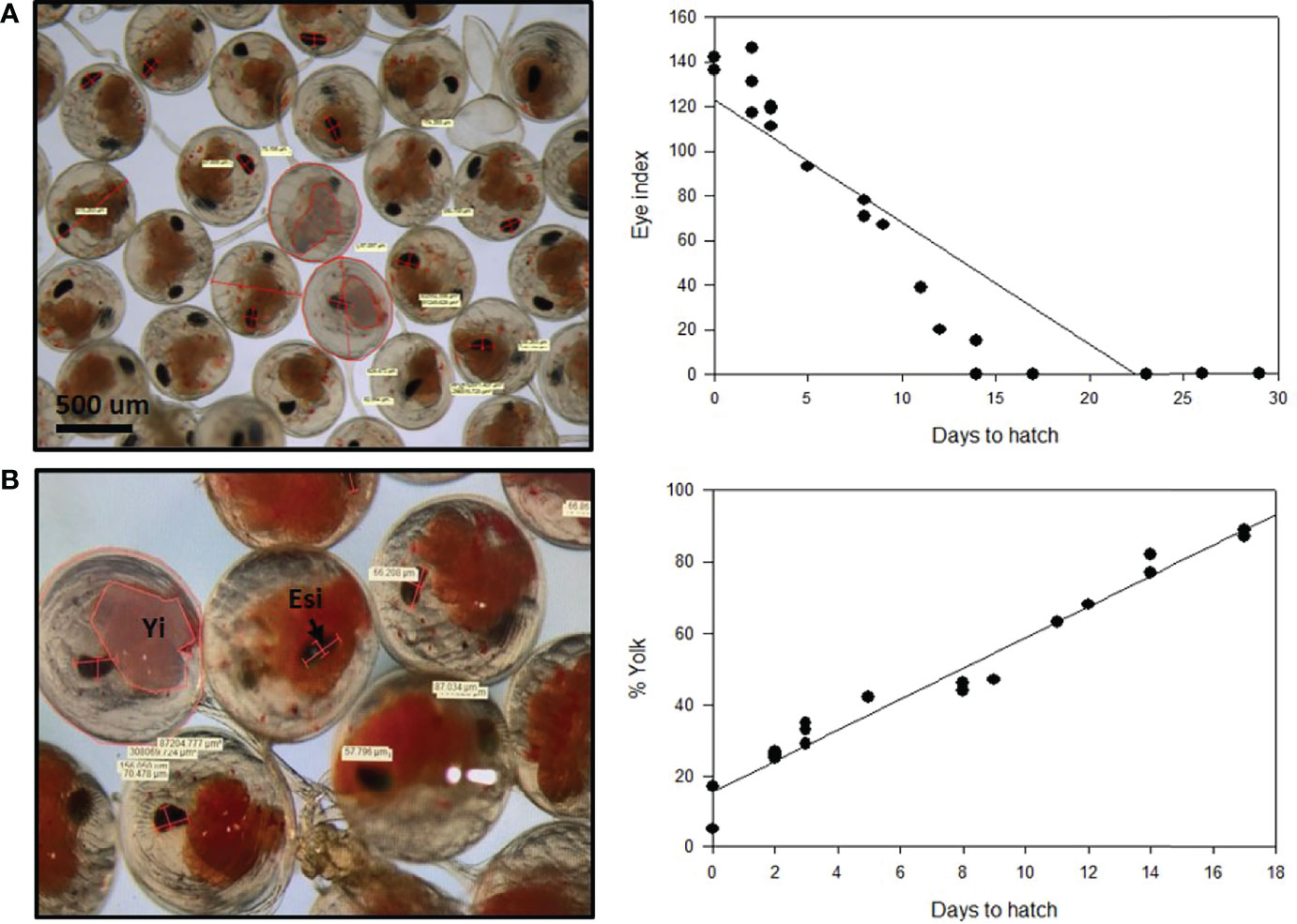
Figure 6 Hatch prediction models for developing Panulirus ornatus embryos based on eye index (A) and yolk percentage (B). Embryos were kept at 26°C at the Institute for Marine and Antarctic Studies (IMAS). Eye index = length of eye + width of eye (widest section)/number of measurements taken. Photos and measurements taken by Larnie Linton.
There is an inverse relationship between the yolk percentage and hatching, with the yolk percentage steadily decreasing as the embryos go through development and prepare to hatch. The percentage of the egg that the yolk encompassed was measured across the 30 days of embryogenesis and plotted against the length of time until the embryos hatched. This showed that the yolk percentage was also directly related to the length of time until hatching occurred. The percentage of yolk consistently decreased as the embryos developed and got closer towards hatch. The yolk percentage was first measured on approximately day 13 of embryo development, with 17 days remaining until the embryos hatched. Measurements were taken from two embryos, with 87% yolk and 89%. The measurements taken remained consistent throughout development, with 44% and 46% at 8 days until hatch, 29%, 33% and 35% at 3 days until hatch and 25%, 26% and 27% 2 days prior to hatching.
Discussion
Embryogenesis is a vital aspect of rearing spiny lobsters in culture, yet there are many unknowns about the embryonal development process. The consistent environmental culture conditions, particularly temperature, reduce compounding factors and provides a reliable supply of embryos known to hatch approximately 30 days from fertilization. This study created the first transcriptomic library detailing 11 stages across embryonal development in P. ornatus. Previous transcriptomic work in decapods have encompassed three or less stages of embryos (summarized in Table 1). This is the first study to look at such a large number of distinct stages across decapod embryonal development. The triplicates of each of the 11 embryo stages, from three different egg development batches, allows more robust quantification of transcripts expression across the P. ornatus embryonal development. The transcripts can be easily identified and their quantity across life stages and tissues of P. ornatus visualized using CrustyBase.org.
PCA of the top 1000 expressed genes across embryonal development indicated differences in the genes expressed across the 11 stages. It was expected that the beginning and end stages of embryogenesis cluster tightly and away from other stages as the developing embryos are the most morphologically different at these stages and likely have very different gene expression profiles. It was confirmed that the day 0 and day 3, and day 27 and day 30 embryos had very different gene expression from the embryos sampled across the mid-stages of embryonal development. The day 0 and day 3 embryos are initiating the embryonal development pathways, such as gastrulation, which requires a large quantity of genes to be expressed at the appropriate times to ensure correct growth and development. The day 27 and day 30 embryos are approaching the end of embryogenesis, preparing for hatch and the subsequent metamorphosis into the larvae which requires significant gene expression changes to regulate the morphological transition. In Macrobrachium acanthurus, a study investigating the morphology and chronology of the 16-day embryonic development period reported that there were significant morphological changes from day 1 to 3 in relation to cell division and the initialization of embryo shape (MÜLler et al., 2007). Significant embryonic events were visualised in the latter half of the development period with the organization of complex structures (MÜLler et al., 2007). The highest expressed genes in the day 0 embryos share more similarity with the genes expressed in the ovary of adult female P. ornatus. It is possible that these transcripts encode maternal RNA (Winata and Korzh, 2018), which are expected to have elevated expression in the mature ovary and early embryos. This supports the idea that significant gene expression differences would be present in the earlier stages of P. ornatus embryogenesis relating to initializing embryonic development.
Seventy-two ovary-enriched transcripts suggested to be maternally-inherited genes, were mapped across the 11 stages of embryogenesis, demonstrating specificity to the very early stages of development. Of the 72 maternally-inherited genes, 38 were provided with annotation including gametocyte-specific factor, lipid storage droplet protein, carcinin-like protein, papilin, AN1-type zinc finger protein, allantoicase, and coactosin-like protein. Gametocyte-specific factor is responsible for suppressing retrotransposon transcription in male germ cells and is considered a marker for gamete developmental competence in mammalian embryos (Huntriss et al., 2017). Carcinin is an antimicrobial protein, similar to defensins in invertebrates (Brockton et al., 2007). The glycoprotein, papilin, is associated with gastrulation, where it is essential for the arrangement of embryonic structures (Kramerova et al., 2000). Without sufficient expression of papilin at the appropriate time points, embryogenesis is impaired leading to inappropriate muscle cell differentiation, and malformations of body structures (Kramerova et al., 2000). Images taken of the P. ornatus embryo stages provide minimal detail of the precise morphological changes occurring within the egg, due to the presence of yolk. Day 0 is established as the 8-cell stage due to presence of visible cells inside the egg. There is little characterization until the eye pigmentation becomes visible on day 18 and eye spots develop on day 21. Prior to the development of eye pigmentation, characterisation is largely based on time (Helluy and Beltz, 1991). Characterisation of embryo development can assist in detailing the changes occurring within an embryo that cannot be visualised.
Eight previous studies with an emphasis on transcriptomic profiling of decapod embryo development curated a short list of genes associated with specific aspects of embryonal development. Three genes, vasa, nanos and brachyury were specific to the gastrula phase. Vasa belongs to the DEAD-box family that is exclusively expressed in the germline cells (Aflalo et al., 2007), nanos is essential for abdominal segmentation in D. melanogaster embryogenesis (Wang et al., 1994), and brachyury is a T-box transcription factor with a key role in regulating gastrulation including specifying cell identity in other invertebrates, but hasn’t yet been confirmed in crustaceans (Hertzler et al., 2018). Three predicted vasa transcripts, 2 nanos transcripts and 1 brachyury transcript were derived from the P. ornatus embryo transcriptome. While there is minimal vasa expression across P. ornatus embryogenesis, both nanos and brachyury expression is elevated exclusively in day 3 embryos. Based on the 30-day embryo development period of P. ornatus kept at 26°C, day 3 embryos can be considered at approximately 10% development. Based on a percent-staging method that has been used to characterize other lobster species (Helluy and Beltz, 1991), the gastrula phase occurs at approximately 10% embryonal development, which is consistent with the gene expression of the gastrula-specific nanos and brachyury.
Neuropeptides are a diverse group of key signalling messengers comprised of short chains of amino acids with a range of roles across physiological processes (Mykles et al., 2010; Nguyen et al., 2016). Of the 67 predicted neuropeptides in P. ornatus (Hyde et al., 2020b), 58 were identified in the embryonal development transcriptome. The majority of the transcripts demonstrate very little expression in the early stages, with increasing expression as the embryos become more developed. It was expected that the expression of the predicted neuropeptides would increase as the embryos develop their central nervous system and neural tissues, towards the end of embryonal development. Unexpectedly, nine of the neuropeptides showed the inverse expression profile, with higher expression levels occurring earlier during embryonal development, with a decrease in expression during later embryonal development. This group of neuropeptides includes CCHamide-2, corazonin, DH44, EH-1, EH-2, GPB5, NFT, NP-4 and SIFamide. Corazonin has the lowest expression, with minimal expression on day 0, day 18, day 24, day 27 and day 30. In male decapods, corazonin has a key role in the regulation of the masculinizing insulin-like androgenic gland hormone as well as other physiological processes including stress response, heartbeat, pigment production and reducing silk production in insects (Farhadi et al., 2021). In D. melanogaster, corazonin inhibits vitellogenin and egg-laying (Gospocic et al., 2017). It is possible that the adult female is expressing corazonin to inhibit further egg production, and residual corazonin is being detected in the day 0 embryos as a result. DH44 has previously been characterised as a phyllosoma-specific neuropeptide that regulates a pathway specific to this stage (Hyde et al., 2020a). In P. ornatus embryos, DH44 expression increases in the last few days before hatch, possibly establishing some of these pathways in preparation for the larval phase. However, across embryogenesis, DH44 expression is highest on day 0, which suggests a role outside of processes specific to P. ornatus larvae. In D. melanogaster, DH44 expression was detected at low levels during embryonal development, increasing by the third larval instar and remaining elevated in the pupal brain and adult ventral nerve chord (Cannell et al., 2016). In the bug Rhodnius prolixus, DH44 is released into the hemolymph following feeding (Nässel and Wu, 2021). Glycoprotein B5 (GPB5) has been shown to regulate the reabsorption of sodium in the hindgut of the mosquito, Aedes aegypti (Paluzzi et al., 2014) . In P. ornatus embryos, GPB5 expression is exclusive to day 0 embryos, suggesting that it is only present during the initialization period of embryo development. GPB5 is expressed in the mature ovary of several decapod species, as well as the neural tissues, and is suggested to have a link to the maturation of the gonads (Ventura et al., 2020). NFT demonstrates elevated expression in day 0 embryos, with very little expression throughout the remainder of embryonal development in P. ornatus. NP-4 is specific to the day 0 embryos and is the only neuroparsin with expression early in the embryo. SIFamide has the highest expression of all the predicted P. ornatus neuropeptides, peaking on day 0. SIFamide is conserved in decapods, with multiple functions described, including roles in the flow of food between the foregut and midgut, sexual behavior control and olfactory system modulation (Dickinson et al., 2008). It is secreted from neurons in Macrobrachium rosenbergii and is implicated in aggression modulation, whilst in insects it regulates feeding and reproductive behaviors (Martelli et al., 2017). In P. ornatus phyllosoma, SIFamide expression decreased in late-stage phyllosoma and increased consistently throughout puerulus and juvenile development (Hyde et al., 2020a).
Two neuropeptides show elevated expression in the mid-stages of embryogenesis. The expression of PDH3, a pigment dispersing hormone (Christie et al., 2010) , remains low until day 9 with elevated expression from day 9 to day 18 and then remaining low for the remainder of embryonal development. NP-2, a neuroparsin, follows a similar expression profile, with low expression until day 12, elevated expression from day 12 to day 21 and then reduced expression until hatch. Neuroparsins are a family of neuropeptides well-conserved within arthropods, known to be involved in reproduction in insects, however there is little known about their role in crustacea (Yang et al., 2014). In P. ornatus, the 6 neuroparsins identified differ in their expression across the 11 stages of embryogenesis.
Many of the neuropeptides followed the expected expression profile, with minimal expression early in development and peaking on day 27 or day 30. PDH2 had the highest expression of all P. ornatus neuropeptides in this expression profile. Crustacean hyperglycemic hormone (CHH), molt inhibiting hormone (MIH), and red pigment concentrating hormone (RPCH) are associated with sexual development in P. ornatus, with elevated expression in the eyestalk and significantly reduced expression in other adult tissues (Ventura et al., 2020). In the P. ornatus embryo, CHH, MIH and RPCH do not have meaningful expression until the later stages of embryonal development. CHH expression increases on day 27, MIH expression begins to increase on day 24, and RPCH expression begins to increase on day 21, all peaking on day 30 when the embryos hatch. BursiconA, bursiconB, and crustacean cardioactive neuropeptide (CCAP) are associated with molting events. In P. ornatus embryos, bursiconA and bursiconB show no expression until day 21 and peaking at day 27 and CCAP does not express until day 24 and peaks on day 30. This indicates that bursiconA, bursiconB and CCAP are associated with preparing the embryo for hatching to the naupliosoma and the subsequent first true molting event, to the phyllosoma body form within minutes of hatching. Currently, there is limited understanding of the roles of neuropeptides across embryos in decapods, however this study provides a basis for further characterization of neuropeptides in P. ornatus and other decapods.
Hatch prediction models are beneficial in aquaculture, as they can allow the control of hatch timing and provide adequate preparation prior to larval rearing. Uncertainty surrounding hatch timing and the variability of hatched larvae is not ideal and can be disruptive to production schedules (Sachlikidis et al., 2010). This has led to several studies investigating hatching prediction techniques based on visual appearance and percentage of development. Previous hatching models have had considerable success in temperate species with longer embryonal development periods (Moss et al., 2004, Tong et al., 2000). IMAS rears P. ornatus, a tropical species, all year round, providing the ideal opportunity to investigate models of hatch prediction in a tropical lobster. In this study, two models were measured, based on eye index and yolk percentage measured against the length of time until the embryos hatched. A direct correlation between the eye index and time remaining until hatch was identified. This was anticipated since the eyes increase in size as the embryo develops and prepares to hatch into larvae. At two days prior to hatch, eye index was calculated for three individual embryos, measuring at 117 μm, 131 μm and 146 μm. This difference may be due to a difference in the development of some of the embryos with hatching often occurring over one to three days. Measurement bias is another possibility for this substantial difference in eye index. The measurements are calculated using a two-dimensional microscope; however the developing embryo eye is a three-dimensional structure. The larger the eye grows, the more prone it is to measurement bias. The yolk percentage steadily decreased as the embryo developed, beginning at 90% of the egg in day 12 embryos and decreasing to less than 20% immediately prior to hatching. The decrease in yolk percentage is due to the depletion of the yolk as it is used as an energy source for the developing embryo (García-Guerrero, 2010). As these energy reserves are utilized to form the animal, the yolk decreases in size and color, providing a visual indicator of embryo development progression. This method of hatch prediction can be used as an indicator of development expectations and serve as a warning if embryonic milestones are not reached at the expected time points. This is the first presentation of eye index and yolk percentage to predict the time to hatch in the tropical spiny lobster species P. ornatus.
Conclusion
Embryogenesis of decapods is an area of research that has not received significant attention. Improving the understanding of the morphological and gene expression changes occurring throughout embryonal development in P. ornatus will greatly improve the understanding of the physiological processes occurring within the embryo and inform the best culture practices. A detailed transcriptome of 11 stages of embryonal development across a 30-day embryonal development period in P. ornatus was constructed. Key genes associated with embryonal development were characterized across the 11 distinct stages of embryonal development. Genes enriched in the ovary of adult females were found to be almost exclusively present in the initial phase of embryogenesis immediately following fertilization and characterized as maternally-inherited genes. Gastrulation was defined for the first time in P. ornatus embryogenesis, based on the characterization of two genes, nanos and brachyury, shown to be specific to the gastrula phase in decapod embryos. Crustacean neuropeptides were mapped across the P. ornatus embryos and associated with key physiological processes such as reproduction and molt pathways. Hatch prediction models were shown to be a successful method to accurately predict hatching times for P. ornatus. The study contributes resources towards the characterization and staging of embryogenesis and the morphological and molecular processes occurring within P. ornatus and other tropical lobster species.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: NCBI, accession: PRJNA761502.
Author Contributions
CLL conducted the research and wrote the manuscript. CLL and TV revised the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by the Australian Government with funding from the Australian Research Council (http://www.arc.gov.au/) Industrial Transformation Research Hub (project number IH190100014). The views expressed herein are those of the authors and are not necessarily those of the Australian Government or Australian Research Council.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
This research was conducted at USC in collaboration with IMAS, UTas. Photos of P. ornatus embryos and hatch prediction models were kindly contributed by Ms Larnie Linton.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2022.889317/full#supplementary-material
References
Aflalo E. D., Bakhrat A., Raviv S., Harari D., Sagi A., Abdu U. (2007). “Characterization of a Vasa-Like Gene From the Pacific White Shrimp Litopenaeus Vannamei and its Expression During Oogenesis.” Mol. Reprod. Dev. 74 (2), 172–177. doi: 10.1002/mrd.20622
Babicki S., Arndt D., Marcu A., Liang Y., Grant J. R., Maciejewski A., et al. (2016). Heatmapper: Web-Enabled Heat Mapping for All. Nucleic Acids Res. 44 (W1), W147–W153. doi: 10.1093/nar/gkw419
Brena C., Akam M. (2012). The Embryonic Development of the Centipede Strigamia Maritima. Dev. Biol. 363 (1), 290–307. doi: 10.1016/j.ydbio.2011.11.006
Brockton V., Hammond J. A., Smith V. J. (2007). “Gene Characterisation, Isoforms and Recombinant Expression of Carcinin, an Antibacterial Protein From the Shore Crab, Carcinus Maenas.” Mol. Immunol. 44 (5), 943–949. doi: 10.1016/j.molimm.2006.03.017
Campos-Ortega José A., Hartenstein V. (1997). “Stages of Drosophila Embryogenesis.” in The Embryonic Development of Drosophila Melanogaster. Eds. Campos-Ortega José A., Hartenstein V. (Berlin, Heidelberg: Springer Berlin Heidelberg), 9–102.
Cannell E., Dornan A. J., Halberg K. A., Terhzaz S., Dow J. A.T., Davies S.- A. (2016). The Corticotropin-Releasing Factor-Like Diuretic Hormone 44 (DH44) and Kinin Neuropeptides Modulate Desiccation and Starvation Tolerance in Drosophila Melanogaster. Peptides 80, 96–107. doi: 10.1016/j.peptides.2016.02.004
Christie A. E., Stemmler E. A., Dickinson P. S. (2010). Crustacean Neuropeptides. Cell. Mol. Life Sci. 67 (24), 4135–4169. doi: 10.1007/s00018-010-0482-8
Dickinson P. S., Stemmler E. A., Cashman C. R., Brennan H. R., Dennison B., Huber K. E., et al. (2008). SIFamide Peptides in Clawed Lobsters and Freshwater Crayfish (Crustacea, Decapoda, Astacidea): A Combined Molecular, Mass Spectrometric and Electrophysiological Investigation. Gen. Comp. Endocrinol. 156 (2), 347–360. doi: 10.1016/j.ygcen.2008.01.011
Farhadi A., Cui W., Zheng H., Li S., Zhang Y., Ikhwanuddin M., et al. (2021). The Regulatory Mechanism of Sexual Development in Decapod Crustaceans. Front. Mar. Sci. 8 (477). doi: 10.3389/fmars.2021.679687
García-Guerrero M. U. (2010). Effect of Temperature on Consumption Rate of Main Yolk Components During Embryo Development of the Prawn Macrobrachium Americanum (Crustacea: Decapoda: Palaemonidae). J. World Aquac. Soc 41 (s1), 84–92. doi: 10.1111/j.1749-7345.2009.00336.x
Gospocic J., Shields E. J., Glastad K. M., Lin Y. P., Penick C. A., Yan H., et al. (2017). The Neuropeptide Corazonin Controls Social Behavior and Caste Identity in Ants. Cell 170 (4), 748. doi: 10.1016/j.cell.2017.07.014
Helluy S. M., Beltz B. S. (1991). Embryonic Development of the American Lobster (Homarus Americanus): Quantitative Staging and Characterization of an Embryonic Molt Cycle. Biol Bull. 180 (3), 355–371. doi: 10.2307/1542337
Hertzler P. L., Wei J., Droste A. P., Yuan J., Xiang J. (2018). Penaeid Shrimp Brachyury: Sequence Analysis and Expression During Gastrulation. Dev. Genes Evol. 228 (5), 219–225. doi: 10.1007/s00427-018-0618-7
Hua L., JianGuo H. (2012). Study on the Artificial Propagation and Embryonic Development of Panulirus Ornatus. Acta Microbiol. Sin. 36 (2), 236–245.
Huerlimann R., Wade N. M., Gordon L., Montenegro J. D., Goodall J., McWilliam S., et al. (2018). De Novo Assembly, Characterization, Functional Annotation and Expression Patterns of the Black Tiger Shrimp (Penaeus Monodon) Transcriptome. Sci. Rep. 8 (1), 13553. doi: 10.1038/s41598-018-31148-4
Hui M., Cui Z. X., Liu Y., Song C. W. (2017). Transcriptome Profiles of Embryos Before and After Cleavage in Eriocheir Sinensis: Identification of Developmental Genes at the Earliest Stages. Chin. J. Oceanol. Limnol. 35 (4), 770–781. doi: 10.1007/s00343-017-5364-6
Huntriss J., Lu J., Hemmings K., Bayne R., Anderson R., Rutherford A., et al. (2017). Isolation and Expression of the Human Gametocyte-Specific Factor 1 Gene (GTSF1) in Fetal Ovary, Oocytes, and Preimplantation Embryos. J. Assist. Reprod. Genet. 34, 23–31. doi: 10.1007/s10815-016-0795-0
Hyde C. J., Fitzgibbon Q. P., Elizur A., Smith G. G., Ventura T. (2019). Transcriptional Profiling of Spiny Lobster Metamorphosis Reveals Three New Additions to the Nuclear Receptor Superfamily. BMC Genomics 20, 1–14. doi: 10.1186/s12864-019-5925-5
Hyde C. J., Nguyen T., Fitzgibbon Q. P., Elizur A., Smith G. G., Ventura T. (2020a). Neural Remodelling in Spiny Lobster Larvae Is Characterized by Broad Neuropeptide Suppression. Gen. Comp. Endocrinol. 294, 1–10. doi: 10.1016/j.ygcen.2020.113496
Hyde C. J., Nguyen T., Fitzgibbon Q. P., Elizur A., Smith G. G., Ventura T. (2020b). Neural Remodelling in Spiny Lobster Larvae Is Characterized by Broad Neuropeptide Suppression. Gen. Comp. Endocrinol. 294, 113496. doi: 10.1016/j.ygcen.2020.113496
Hyde C. J., Nguyen T., Fitzgibbon Q. P., Elizur A., Smith G. G., Ventura T. (2020c). Neural Remodelling in Spiny Lobster Larvae is Characterized by Broad Neuropeptide Suppression. Gen. Comp. Endocrinol. 294, 113496. doi: 10.1016/j.ygcen.2020.113496
Jaramillo M. L., Guzman F., Paese C. L., Margis R., Nazari E. M., Ammar D., et al. (2016). Exploring Developmental Gene Toolkit and Associated Pathways in a Potential New Model Crustacean Using Transcriptomic Analysis. Dev. Genes Evol. 226 (5), 325–337. doi: 10.1007/s00427-016-0551-6
Jeffs A. (2010). Status and Challenges of Advancing Lobster Aquaculture Globally. J. Mar. Biol. Assoc. India 52, 320–326.
Kramerova I. A., Kawaguchi N., Fessler L. I., Nelson R. E., Chen Y., Kramerov A. A., et al. (2000). Papilin in Development; a Pericellular Protein With a Homology to the ADAMTS Metalloproteinases. Development 127 (24), 5475–5485. doi: 10.1242/dev.127.24.5475
Liang H., He J. (2012). Study on the Artificial Propagation and Embryonic Development of Panulirus Ornatus. Acta Microbiol. Sin. 36 (2), 236–245.
Li P., Wang Y., Wang Y., Guo H. (2017). Whole Transcriptome Sequencing and Analysis of Gastrula Embryos of Marsupenaeus Japonicus (Bate 1888). Madridge. J. Aquacult. Res. Dev. 1, 1–7. doi: 10.18689/mjard-1000101
Martelli C., Pech U., Kobbenbring S., Pauls D., Bahl B., Sommer M. V., et al. (2017). SIFamide Translates Hunger Signals Into Appetitive and Feeding Behavior in Drosophila. Cell Rep. 20 (2), 464–478. doi: 10.1016/j.celrep.2017.06.043
Metsalu T., Vilo J. (2015). ClustVis: A Web Tool for Visualizing Clustering of Multivariate Data Using Principal Component Analysis and Heatmap. Nucleic Acids Res. 43 (W1), W566–W570. doi: 10.1093/nar/gkv468
Moss G. A., James P. J., Allen S. E., Bruce M. P. (2004). Temperature Effects on the Embryo Development and Hatching of the Spiny Lobster Sagmariasus Verreauxi. N. Z. J. Mar. Freshw. Res. 38 (5), 795–801. doi: 10.1080/00288330.2004.9517278
MÜLler Y., Pacheco C., SimÕEs-Costa M., Ammar D., Nazari E. (2007). Morphology and Chronology of Embryonic Development in Macrobrachium Acanthurus (Crustacea, Decapoda). Invertebr. Reprod. Dev. 50, 67–74. doi: 10.1080/07924259.2007.9652229
Musbir M., Sudirman S., Mallawa A., Bohari R. (2018). Egg Quantity of Wild Breeders of Spiny Lobster (Panulirus Ornatus) Caught From Southern Coastal Waters of Bulukumba, South Sulawesi, Indonesia. AACL. Bioflux. 11, 295–300.
Mykles D. L., Adams M. E., Gäde G., Lange A. B., Marco H. G., Orchard I. (2010). Neuropeptide Action in Insects and Crustaceans. Physiol. Biochem. Zool. 83 (5), 836–846. doi: 10.1086/648470
Nässel D. R., Wu S.-F. (2021). Leucokinins: Multifunctional Neuropeptides and Hormones in Insects and Other Invertebrates. Int. J. Mol. Sci. 22 (4), 1531. doi: 10.3390/ijms22041531
Nguyen T. V., Cummins S. F., Elizur A., Ventura T. (2016). Transcriptomic Characterization and Curation of Candidate Neuropeptides Regulating Reproduction in the Eyestalk Ganglia of the Australian Crayfish, Cherax Quadricarinatus. Sci. Rep. 6, 38658. doi: 10.1038/srep38658
Nguyen T. V., Jung H., Rotllant G., Hurwood D., Mather P., Ventura T. (2018). Guidelines for RNA-Seq Projects: Applications and Opportunities in Non-Model Decapod Crustacean Species. Hydrobiologia 825 (1), 5–27. doi: 10.1007/s10750-018-3682-0
Nguyen T. V., Rotllant G. E., Cummins S. F., Elizur A., Ventura T. (2018). Insights Into Sexual Maturation and Reproduction in the Norway Lobster (Nephrops Norvegicus) via in Silico Prediction and Characterization of Neuropeptides and G Protein-Coupled Receptors. Front. Endocrinol. 9. doi: 10.3389/fendo.2018.00430
Paluzzi J. P., Vanderveken M., O’Donnell M. J. (2014). The Heterodimeric Glycoprotein Hormone, GPA2/GPB5, Regulates Ion Transport Across the Hindgut of the Adult Mosquito, Aedes Aegypti. PLoS One 9 (1), e86386. doi: 10.1371/journal.pone.0086386
Pijuan-Sala B., Griffiths J. A., Guibentif C., Hiscock T. W., Jawaid W., Calero-Nieto F. J., et al. (2019). A Single-Cell Molecular Map of Mouse Gastrulation and Early Organogenesis. Nature 566 (7745), 490–495. doi: 10.1038/s41586-019-0933-9
Sachlikidis N. G., Jones C. M., Seymour J. E. (2010). The Effect of Temperature on the Incubation of Eggs of the Tropical Rock Lobster Panulirus Ornatus. Aquaculture 305 (1-4), 79–83. doi: 10.1016/j.aquaculture.2010.04.015
Scholtz G., Wolff C. (2013). “Arthropod Embryology: Cleavage and Germ Band Development.” in Arthropod Biology and Evolution: Molecules, Development, Morphology. Eds. Minelli A., Boxshall G., Fusco G. (Berlin, Heidelberg: Springer Berlin Heidelberg), 63–89.
Sellars M. J., Trewin C., Sean M., Glaves M. R.S.E., Hertzler P. L. (2015). Transcriptome Profiles of Penaeus (Marsupenaeus) Japonicus Animal and Vegetal Half-Embryos: Identification of Sex Determination, Germ Line, Mesoderm, and Other Developmental Genes. Mar. Biotechnol. 17 (3), 252–265. doi: 10.1007/s10126-015-9613-4
Tong L. J., Moss G. A., Pickering T. D., Paewai M. P. (2000). Temperature Effects on Embryo and Early Larval Development of the Spiny Lobster Jasus Edwardsii, and Description of a Method to Predict Larval Hatch Times. Mar. Freshw. Res. 51 (3), 243–248. doi: 10.1071/Mf99049
Ventura T., Chandler J. C., Nguyen T. V., Hyde C. M. J., Elizur A., Fitzgibbon Q. P., et al. (2020). Multi-Tissue Transcriptome Analysis Identifies Key Sexual Development-Related Genes of the Ornate Spiny Lobster (Panulirus Ornatus). Genes 11 (10), 1–17. doi: 10.3390/genes11101150
Wang C., Dickinson L. K., Lehmann R. (1994). Genetics of Nanos Localization in Drosophila. Dev. Dyn. 199 (2), 103–115. doi: 10.1002/aja.1001990204
Wang Y., Wang B. J., Shao X. Q., Liu M., Jiang K. Y., Wang M. Q., et al. (2020). A Comparative Transcriptomic Analysis in Late Embryogenesis of the Red Claw Crayfish Cherax Quadricarinatus. Mol. Genet. Genom. 295 (2), 299–311. doi: 10.1007/s00438-019-01621-4
Wei J., Glaves R. S. E., Sellars M. J., Xiang J., Hertzler P. L. (2016). Expression of the Prospective Mesoderm Genes Twist, Snail, and Mef2 in Penaeid Shrimp. Dev. Genes Evol. 226 (4), 317–324. doi: 10.1007/s00427-016-0544-5
Wei J., Zhang X., Yang Y., Huang H., Li F., Xiang J. (2014). Comparative Transcriptomic Characterization of the Early Development in Pacific White Shrimp Litopenaeus Vannamei. PLoS One 9 (9), e106201–e106201. doi: 10.1371/journal.pone.0106201
Winata C. L., Korzh V. (2018). The Translational Regulation of Maternal mRNAs in Time and Space. FEBS Lett. 592 (17), 3007–3023. doi: 10.1002/1873-3468.13183
Keywords: Panulirus ornatus, transcriptomics, embryogenesis, gastrula, neuropeptides, hatch prediction
Citation: Lewis CL, Fitzgibbon QP, Smith GG, Elizur A and Ventura T (2022) Transcriptomic Analysis and Time to Hatch Visual Prediction of Embryo Development in the Ornate Spiny Lobster (Panulirus ornatus). Front. Mar. Sci. 9:889317. doi: 10.3389/fmars.2022.889317
Received: 04 March 2022; Accepted: 23 May 2022;
Published: 12 July 2022.
Edited by:
Xiaotong Wang, Ludong University, ChinaReviewed by:
Hongyu Ma, Shantou University, ChinaKhor Waiho, University of Malaysia Terengganu, Malaysia
Copyright © 2022 Lewis, Fitzgibbon, Smith, Elizur and Ventura. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tomer Ventura, tventura@usc.edu.au
 Courtney L. Lewis
Courtney L. Lewis Quinn P. Fitzgibbon
Quinn P. Fitzgibbon Gregory G. Smith
Gregory G. Smith Abigail Elizur
Abigail Elizur Tomer Ventura
Tomer Ventura