Environmental DNA metabarcoding reveals spatial and seasonal patterns in the fish community in the Venice Lagoon
- 1Department of Biology, Università di Padova, Padova, Italy
- 2Department of Biology, Università di Pisa, Pisa, Italy
- 3Department of Biology, Università di Firenze, Firenze, Italy
- 4Consorzio Nazionale Interuniversitario per le Scienze del Mare, Roma, Italy
- 5Institute of Marine Science, National Research Council (CNR ISMAR), Venezia, Italy
- 6National Biodiversity Future Center (NBFC), Palermo, Italy
- 7Department of Research Infrastructures for Marine Biological Resources, Stazione Zoologica Anton Dohrn, Naples, Italy
- 8Department of Life Sciences, Università di Trieste, Trieste, Italy
Environmental DNA (eDNA) is an emerging tool for assessing biodiversity and understanding spatial and temporal community patterns and processes, directly from DNA sequencing of environmental samples such as air, water, and sediments. We applied eDNA methods to monitor bony fish communities, detecting as well locally allochthonous species, and to reveal seasonal patterns at two sites in the Venice Lagoon. We analyzed 17 water samples collected over 12 months at two ecologically distinct sites by using available primers for teleosts and High Throughput Illumina sequencing. We identified 1,289 amplicon sequence variants (ASVs) assigned to 62 fish taxa. Most of the species known to inhabit or to enter the Venice Lagoon were detected, with eDNA data reflecting differences in fish communities between the internal (freshwater associated) and the external (sea associated) part of the lagoon. Moreover, seasonal trends of migration have been portrayed, highlighting the most involved species and disclosing possible clashes between migration events and the temporary interruption of sea-lagoon connectivity due to MOSE (MOdulo Sperimentale Elettromeccanico). Of interest, the first-time detection of Oceanic puffer (Lagocephalus lagocephalus) DNA in the Venice Lagoon provides evidence of the further northward expansion of this species in the high Adriatic Sea. eDNA successfully profiled fish communities by season and habitat in the Venice Lagoon. Our results support routine application of eDNA to monitor potential ecological consequences of MOSE closures in this World Heritage site.
Introduction
Environmental DNA (eDNA) is an emerging and non-invasive technique which is considered highly promising for species monitoring studies (Bohmann et al., 2014). eDNA is a mixture of intracellular and/or extracellular nucleic acids, commonly released by organisms in the environment through feces, shred tissue, mucus, and/or blood, which can be analyzed by purification from different substrates (e.g., water, sediments, air, ice) (Taberlet et al., 2018). By combining co-amplification of small portions of barcode genes from purified eDNA with high throughput sequencing technologies (HTS), the eDNA analysis is highly effective in simultaneously identifying and differentiating multiple species (Deiner et al., 2017), thus providing “snap-shots” of different ecosystems (DiBattista et al., 2020). The analysis of environmental samples eases the detection of rare and endangered species in large systems (Bergman et al., 2016; Eva et al., 2016; Anderson et al., 2018). Besides being non-invasive, eDNA metabarcoding is also cost- and time-effective and has higher sensitivity compared with traditional approaches, allowing species detection at any life stage (Shu et al., 2020).
In the last decade, eDNA has been successfully applied, giving higher priority to fish macro-fauna (Tsuji et al., 2019), as support to conventional monitoring approaches, in species surveys of aquatic environments to obtain more detailed inventories of species distribution in time and space (Stoeckle et al., 2017; Priè et al., 2020; Aglieri et al., 2021). However, the studies that focused on aquatic ecosystems rarely considered transitional environments like lagoons (Suarez-Menendez et al., 2020; Oka et al., 2021; DiBattista et al., 2022). These are shallow coastal waters, intermittently subjected to variation in salinity and other parameters. As seen by their high productivity, they usually host a remarkable number of species by providing many habitat types, nurseries, and feeding grounds (Malavasi et al., 2005; Basset et al., 2006; Elliott and Whitfield, 2011; Sigovini, 2011; Tagliapietra et al., 2012; Whitfield et al., 2012; Basset et al., 2013). Due to their direct economic role in intensive aquaculture and fisheries, transitional environments also suffer from overexploitation and many other human threats, including water pollution, climate change, and introduction of alien species, raising the need for their protection (Giupponi et al., 1999; Solidoro et al., 2010; Occhipinti-Ambrogi et al., 2011).
We applied eDNA metabarcoding and set up sampling, filtration, and amplification protocols to analyze water samples in transitional environments. Short fragments of the 12S mitochondrial gene have been selected as the gold standard for eDNA detection of fishes. We analyzed samples that were collected in the Venice Lagoon, a wide coastal environment located in the northern Adriatic Sea. For the incredible cultural patrimony, Venice and its lagoon have been included in the UNESCO World Heritage Site List. The Venice Lagoon is separated from the sea by islands that can be temporarily connected by a huge complex of mobile dams (MOSE - MOdulo Sperimentale Elettromeccanico), whose function is to prevent the lagoon water level from flooding Venice during extreme high tides. After a long stalemate, the MOSE infrastructure has been nearly completed and tested over the few past years and it is currently working in cases of tides over 130 cm (https://www.mosevenezia.eu/). From a long-term perspective, an increase of the number of closures per year and an even larger and more rapid increase of the duration of closures is likely in the future (Mel et al., 2021) due to the high tide phenomenon that has strongly impacted the lagoon area for many years and has recently been increasing in frequency, which is most likely because of global climate change. If this occurs, the connectivity between the sea and the lagoon will alter, leading to ecological consequences.
The objectives of the present study were: 1) to obtain a fish biodiversity inventory in the Venice Lagoon, observing the resolution capacity of the Tele02 metabarcoding primer pair by comparing the detected amplicon sequence variants (ASVs) to what is known from traditional species surveys of the lagoon; 2) to examine fish communities, thus providing baseline information on which species are present in the lagoon (autochthonous and allochthonous) before the complete activation of MOSE, with a particular focus on the marine migratory component that is expected to be the most vulnerable to sea-lagoon connectivity perturbations, which can help anticipate ecological consequences; 3) to assess if, through eDNA metabarcoding, it is possible to portray differences in fish communities between two different areas of the same lagoon, a sea-associated one and a freshwater-associated one; and 4) to test if eDNA analysis can be useful in identifying patterns of seasonality in migratory fishes.
The present study tests a tool to enhance the lagoon species inventory and aims at providing the basis for a future systematic survey that might be crucial in understanding how healthy the functioning of the Venice Lagoon ecosystem will be after the connectivity perturbations of the MOSE infrastructure.
Materials and methods
Study area, eDNA sampling procedure, and extraction
The sampling was carried out in the Venice Lagoon (Italy), whose surface extends 500 km2 into the north of the Adriatic Sea (Umgiesser et al., 2004). The lagoon environment is linked with the sea by three inlets (from north to south): Lido, Malamocco, and Chioggia (Figure 1). A total of 17 sampling campaigns were performed from November 2018 to December 2019 at two sites 31.39 km apart, one in the northern basin of the lagoon, close to Torcello (45°29.952’N 12°25.043’E), and the other in the southern part, close to the inlet of Chioggia (45°13.938’N 12°17.184’E) (Figure 1). The northern site is approximately 7.5 km from the closest connection to the sea (Lido inlet) and is part of the Long Term Ecological Research network (LTER_EU_IT_016, station 5). The southern site (station 15) lies within proximity to the southern inlet (Chioggia inlet). During each sampling campaign, at least two replicates of superficial water were collected in plastic tanks of about 10 L.
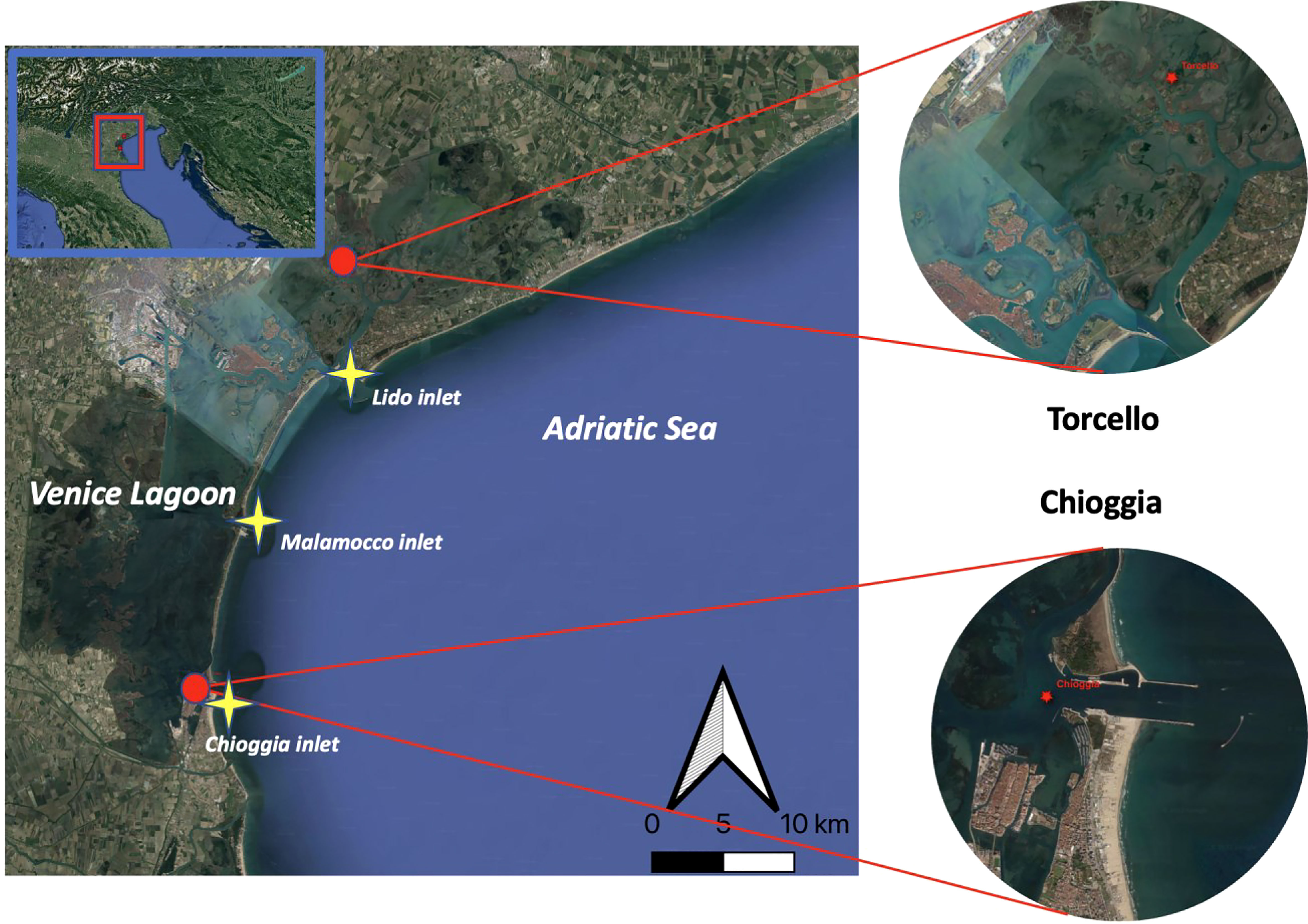
Figure 1 Sampling sites for eDNA collection. Represented is the Venice Lagoon, with red dots indicating sampling stations of Torcello (station 5) and Chioggia (station 15). The three lagoon inlets are indicated by yellow stars. Map created using the Free and Open Source QGIS from Google Earth satellite images.
Prior to sampling, the equipment was carefully sterilized by bleaching (1:10 solution) and thoroughly rinsing with MilliRo water. Afterwards, the samples were transported to the laboratory (Department of Biology at UNIPD) and stored at 8°C until filtration. Filtration took place within 24 hours of sampling, using a vacuum pump Edwards 5 two-stage. Given that most studies reported particles of a size up to 10 µm as the most common fish DNA molecules found in water samples (Shu et al., 2020), filtration was conducted for each water sample using both glass fiber filters GF/C (Whatman®) with a pore size of 1.2 µm and cellulose acetate filters (Sartorius®) with a 0.45 µm pore size, until the filters became clogged. Prior to filtration of each sample, all equipment, components of the filtration device, and surfaces were cleaned with 10% bleach. After each cleaning, a minimum of 1.5 L of pure water (MilliRo) was filtered, and these samples were used as filtration “blanks” to estimate the level of contamination, as suggested in Taberlet et al. (2018).
Two different filter types were used to identify the most appropriate for the specific protocol. However, since the results obtained didn’t show significant differences between the kind of filters (Martino, 2022), they are considered replicates for the purpose of this study. Full details on samples and filters are reported in Table 1.
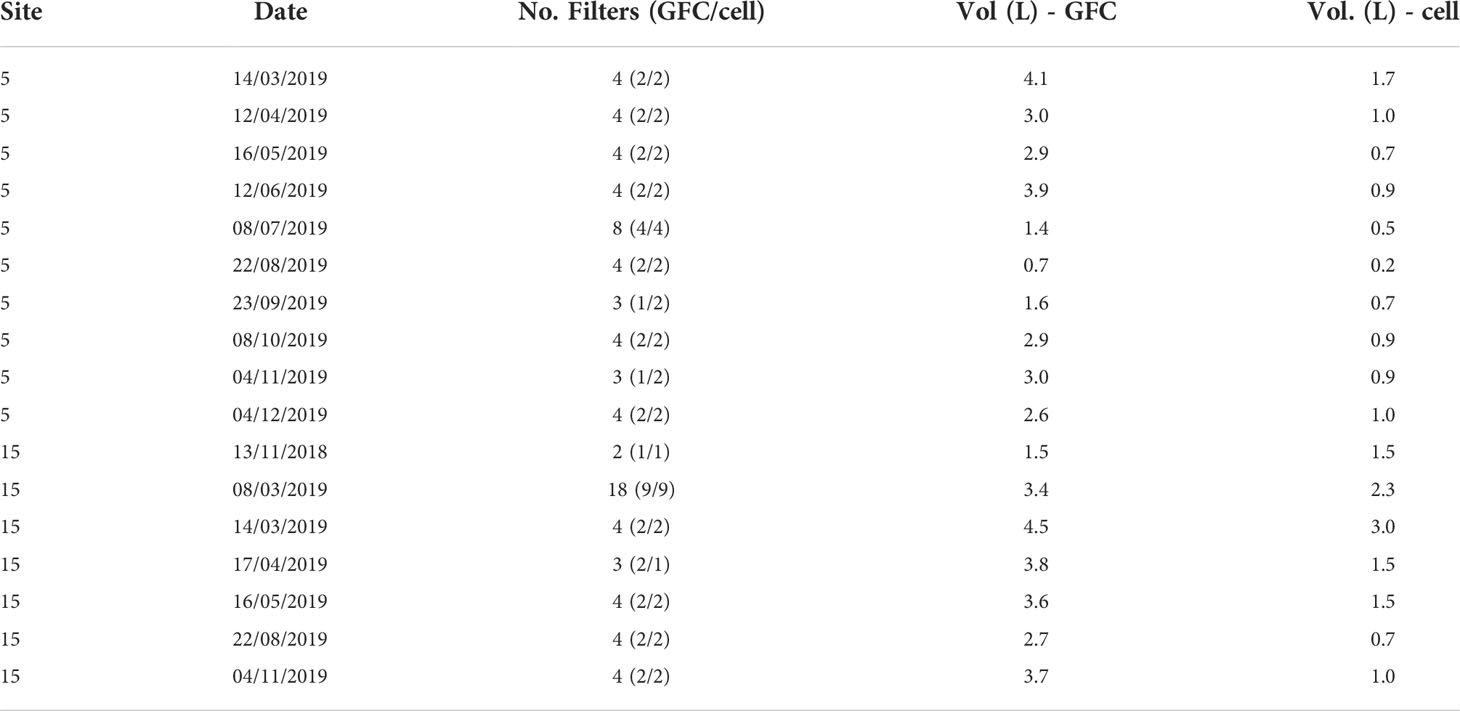
Table 1 Sample collection table reporting the site, the date of sampling, the total number of filters analyzed (between parentheses, the number of GF/C and cellulose acetate filters used), and the average volume (in liters) of filtered water for each filter type.
A total of 81 filters, 42 from station 5 and 39 from station 15, were obtained from filtering lagoon water samples and stored at -80°C until the DNA extraction.
DNA extraction was carried out using the DNeasy Blood & Tissue kit (Qiagen®) in a separated pre-PCR environment equipped with positive air pressure. The amount of ATL buffer (Qiagen®) was increased until it completely covered the filters during the first incubation, and the volume of Proteinase K was modified accordingly. After incubation, 200 µl of the solution containing DNA was processed following the manufacturer’s protocol. At the end, the elution buffer was halved to increase the DNA concentration and the final DNA product was eluted in 100 μl of AE buffer (Qiagen®). For each day of extraction, two extraction blanks were processed under the same conditions, by simulating an extraction without any filter (De Barba et al., 2014; Taberlet et al., 2018; Aglieri et al., 2021). Extracted eDNA samples were stored at -20°C.
Library amplification
For eDNA fish amplification, we used the primer pair Tele02 described in Taberlet et al. (2018), targeting, based on in-silico validation, a short fragment of about 130-209 bp of the 12S rRNA mtDNA gene of bony fishes.
Since 18 filters, from September 2019 to December 2019, out of the 81 starting filters, were processed twice, and one filter from March 2019 was amplified three times, the total number of eDNA libraries processed in this study was 101.
The eDNA samples were amplified using a single-step PCR protocol, as this minimized the possibility of contaminations by reducing the protocol steps, and it is reported to be the most effective in amplifying low-concentration templates (Taberlet et al., 2018; however, see Ushio et al., 2022, for potential disadvantages of this approach, such as the increase in the cost for primers and the variation among tags). To be multiplexed, Tele02 forward and reverse primers were tailed with an 8 bp-barcode and up to four random nucleotides at the 5’ end (e.g., 5’NNNN-8bpBarcode-Primer-3’). This step allowed for tagging each amplicon by a unique and identifiable double-tag combination. The complete list of barcodes used in this study is given in Table S1; the 36 barcodes used were retrieved from Taberlet et al. (2018).
In addition to the eDNA samples, positive controls represented by DNA of the black goby, Gobius niger, were included in the PCR reaction to assess the success of amplification, sequencing, and taxonomic identification. This species was excluded from later analysis because of its use as a positive control during the PCR step. Moreover, PCR blanks were added, using water as a template, as well as sequencing blanks (one for each plate column and row, according to Taberlet et al., 2018) consisting of plate tubes with just the enzyme and primers tagged with unique tags. The latter blanks allow us to estimate the amount of “tag-jumping” (Schnell et al., 2015). PCR reactions were performed using a thermocycler (SimpliAmp™, Applied Biosystems®) in triplicate to reduce amplification stochasticity. Final PCR volume was 10-20 μl, containing AmpliTaq Gold™ 360 MasterMix 1X (Life Technologies), each primer at the final concentration of 0.5 μM, and 2 μl of template (extracted eDNA or blank). The amplification thermal profile started with 10 min at 95°C, followed by 35 cycles of 30 s at 95°C, 1 min at 50°C, 30 s/1 min at 72°C, and finally 7 min at 72°C. Extractions and PCR reactions were set up in a pre-PCR environment, exclusively dedicated to eDNA, using filtered pipette tips.
The presence of PCR amplicons was checked on 1.8% agarose gel with a transilluminator (Gel DocTM XR+, Bio-Rad) for all reactions. PCR products were mixed to produce two different pools, the first including 96 and the second 48 samples (comprising eDNA samples, replicates, negative and positive controls). The amplicon pools were purified with the MinElute PCR Purification Kit (Qiagen®), with a final elution volume of 16 μl, and the purification success was checked on 1.8% agarose gel. To reduce stochasticity, six purification replicates of each pool were carried out. Finally, after combining the six purification replicates, the two purified pools were quantified using Nanodrop 2000c (ThermoFisher), and Qubit™ 4 Fluorometer (ThermoFisher). All the steps were performed in a post-PCR environment with the use of pipettes with filtered tips.
Afterwards, 20 μl of each purified pool was sent to a sequencing service (Norwegian Sequencing Center, Oslo, or BMR Genomics S.R.L., Padova) for adding Illumina-adapters, and for sequencing with Illumina Technologies 150 bp paired-end. The first pool was sequenced on an HiSeq4000 platform, starting from a sequencing library produced by the NEBNext® Ultra™ DNA Library Prep Kit, and splitting the sequencing library on two half-lanes. The second pool was sequenced on a single MiSeq run, starting from a library obtained with the NEBNext® Ultra™ II DNA Library Prep Kit. Two different sequencing platforms were used due to the unavailability of the HiSeq sequencing facility, and because the preliminary analysis of HiSeq data showed that the MiSeq throughput was adequate for our level of complexity, as indeed confirmed by the growing literature that used this latter platform (e.g., Miya et al., 2020).
Sequence analyses
Sequences were demultiplexed with Cutadapt (Martin, 2011), independently processed through QIIME2 pipeline (Bolyen et al., 2019), and denoised with DADA2 algorithm (Callahan et al., 2016). Chimeras were removed through the DADA2 algorithm (–p-chimera-method ‘consensus’).
Taxonomic assignments were performed by using the Mitohelper genetic database (Lim and Thompson, 2021), a QIIME2 compatible fish specific database. The whole pipeline is deposited in Github at the following link https://github.com/Slide95/combinatorial-dual-indexes-metabarcoding. Taxonomic assignation of ASVs was curated manually through BLAST (Altschul et al., 1990) when ASVs were assigned to controversial species (species whose presence in the study area is not documented) or in case of clear mis-assignments (ASVs assigned to species whose presence in the study area is not possible). We removed ASVs that have been assigned to taxa with a taxonomic resolution lower than family (i.e., order, class etc.). As a general rule, if BLAST top matches had a percent identity lower than 97%, the ASV was discarded; if the top matches consisted of congeneric species with a very close percent identity, the ASV was assigned to the genus; if they were non-congeneric species but belonged to the same family, the ASV was assigned to the family. In the case of two similar congeneric species, if only one was reported in the Atlantic Ocean or the Mediterranean Sea, the ASV was assigned to this species. The ASVs that were assigned to the same taxon were at this point clustered into Operational Taxonomic Units (OTUs) for the subsequent analyses. OTUs assigned to the same taxon were merged, and their reads were summed.
To account for the different throughput of the two sequencing platforms used, read counts were normalized. To this end, we first calculated the ratio between the number of HiSeq and MiSeq retained reads, and then multiplied, for each sample and OTU, the MiSeq reads count by this correction factor. Then, OTU tables from the two sequencing outputs (HiSeq and MiSeq) were merged. Consequently, for each sample, read counts lower than 10 were removed to increase the reliability of the results such as in Adamo et al. (2020) and Lopes et al. (2021). Finally, data were cleaned of contaminations through R (R Development Core Team, 2021). To this purpose, we first calculated, for each OTU, the distribution of the number of reads observed in the negative controls (filtration, extraction, PCR, and sequencing blanks), and then we subtracted the value of the third quartile of the distribution from the number of reads assigned to the corresponding OTU of each sample. To evaluate if our normalization and filtering procedure was successful in removing the bias associated with different sequencing depths, we performed a correlation test between the two vectors of normalized OTUs counts obtained from one technical replicate, consisting of a sample sequenced on both HiSeq and MiSeq platforms. The test was performed using the cor.test function with Pearson’s method in R package version 4.1.2. In addition, as an alternative to normalization, we performed a coverage-based rarefaction (Chao and Jost, 2012) of the HiSeq and MiSeq retained reads, using the function phyloseq_coverage_raref of the metagMisc package (available at https://rdrr.io/github/vmikk/metagMisc/man/phyloseq_coverage_raref.html), and we cross-checked the results of the ecological analyses obtained, starting from normalized and rarefied data.
Statistical and ecological analyses
For statistical and ecological analyses, the R Vegan Package (Oksanen et al., 2007) and QIIME2 were used. To account for eDNA performance, a species table was built with species names, sites in which they were detected by eDNA, IUCN status, and ecological guild. The table was then compared to fish distribution data retrieved from the literature (Cavraro et al., 2017). To investigate community dissimilarity between north and south, and seasonal clusterization, pairwise PERMANOVA analyses were performed through QIIME2 (“qiime diversity” function) starting from a Bray-Curtis dissimilarity matrix built from the refined OTU table of normalized abundances. This approach was selected because it was used in similar studies (Turon et al., 2020; DiBattista et al., 2022). However, since the Bray-Curtis distance is based on counts (number of reads in metabarcoding studies), and a quantitative interpretation of eDNA is still being debated, we checked to see if the relevant results were affected by our choice by performing PERMANOVA, using a Jaccard distance, after conversion of our matrix of normalized OTUs abundances to presence/absence data. In addition, considering that our dataset was comprised of different numbers of replicates for each site and sampling date (Table 1), we checked to see if these differences affected the PERMANOVA results by repeating this analysis on a different matrix. For this purpose, we subsampled our dataset to select only strictly comparable samples, i.e., those with the same number of replicates that were collected in the same month and year at the two sites. This resulted in keeping 38 of the original 101 samples and, indeed, also controlled for the inclusion of different sampling months at the two sites in the original dataset.
Differences between sites and seasons were plotted by non-metric Multidimensional Scaling (nMDS), using the metaMDS function in Vegan. To identify singular patterns of species habitat selection between the two study areas, the number of reads were normalized for the two sites by dividing the read counts of each OTU of each sample by the average number of reads assigned to the corresponding site (north and south), and the ratios of normalized number (NAR) of reads corresponding to each species for both sites were compared. To identify the species which mostly contributes to the observed seasonality pattern, the “similarity percentage breakdown” procedure (SIMPER; Clarke et al., 2014) was performed using the Bray-Curtis dissimilarity matrix. Since the results can be skewed by overrepresented species (in metabarcoding, the species with the highest number of reads), the complete OTU table was normalized by dividing the read counts by the mean number of reads assigned to the OTUs in the relative sample, and species overrepresented in terms of reads were excluded before the SIMPER analysis. From the results, the first two species with a contribution higher than 5% and a p-value smaller than 0.05 were selected for ecological inferences.
Results
HiSeq and MiSeq sequencing (Illumina) produced, respectively, approximately 240 x 106 and 12 x 106 raw reads, with demultiplexing resulting in 77,487,725 (32.3%) and 8,780,314 (73%) reads. Denoising of reads, using the DADA2 pipeline, was performed independently for the two sequencing outputs. After filtration, denoising, paired-end merging, and chimera-removal, the number of reads retained for HiSeq and MiSeq outputs were, respectively, 53,409,947 (69% of demultiplexed reads) and 3,937,504 (45% of demultiplexed reads; complete denoising stats for every sample available in supplementary material, Table S2). Since two different sequencing platforms (HiSeq and MiSeq) were used at different times before proceeding to the ecological analyses, we estimated the stability of the data across different sequencing depths comparing, for a sample sequenced twice, its vector of OTU abundances obtained with HiSeq with the vector of its normalized MiSeq OTU abundances, as described in the Materials and Methods section. The high and significant correlation (rho = 0.989; cor.test p-value < 2.2e-16) supports the soundness of our approach and also, indirectly, the robustness of our results as, indeed, does the concordance of the ecological results obtained from the normalized data and from the coverage-based rarefied data (see below and Figures S1, S2).
eDNA detection of fish biodiversity
The analysis of 17 water samples from the Venice Lagoon allowed us to detect 42,888 ASVs, of which 1,289 were assigned to 62 taxa (2 families, 6 genera, 54 species) that are potentially present in this environment (Cavraro et al., 2017); the distribution of each species from south to north is reported in Table S3. Each taxon was assigned, based on its behavioral habits, to a given ecological guild, following Potter et al. (2015) and Cavraro et al. (2017). A graphic representation of the overall proportions of ecological guilds detected in our samples is reported in Figure 2.
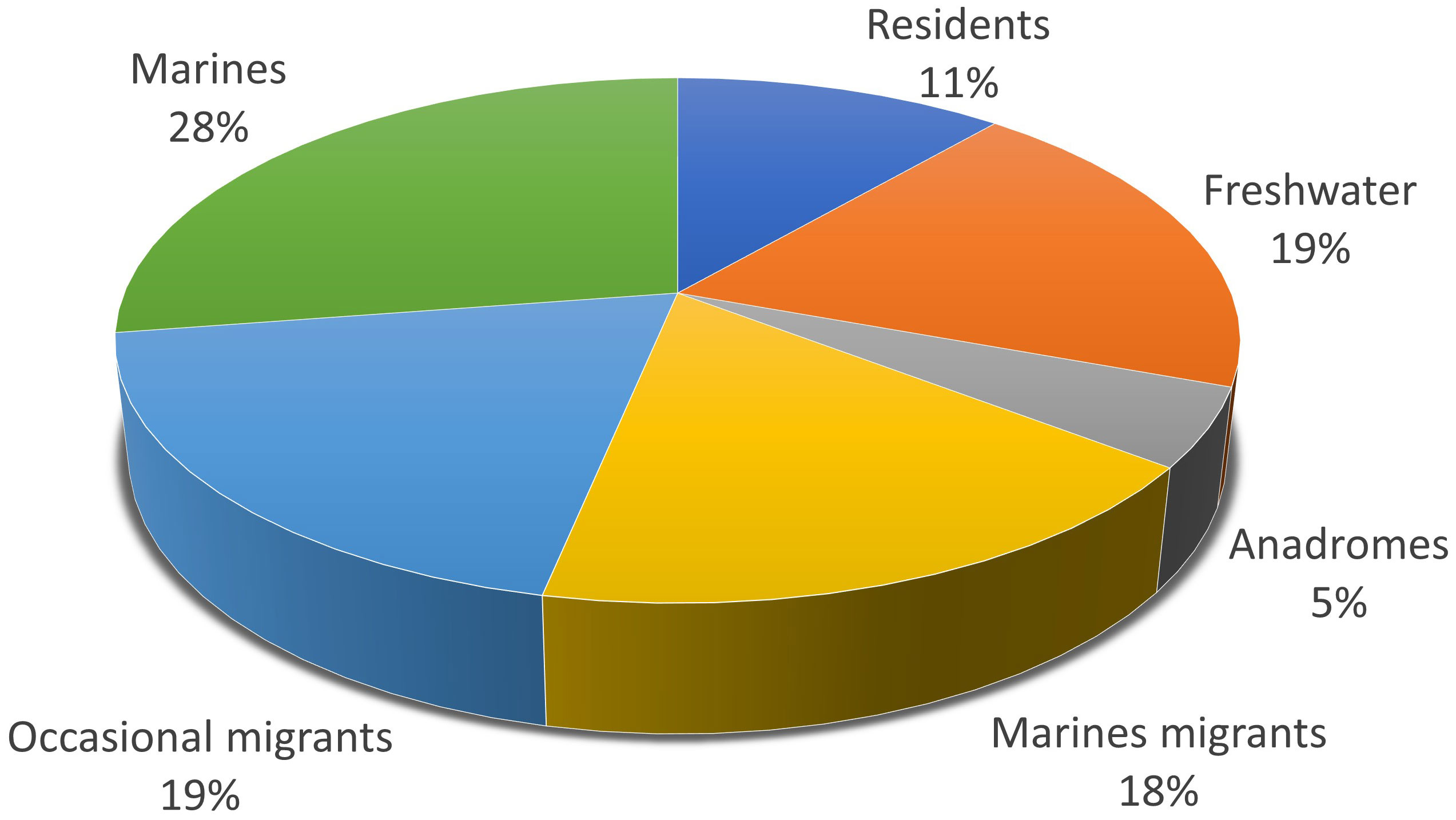
Figure 2 Pie-chart showing the percentage proportions of ecological guilds (Potter et al., 2015) of the species detected in the Venice Lagoon through eDNA metabarcoding.
In the whole dataset (Table S4), the most abundant OTUs with a taxonomic resolution at the species level, based on the number of reads assigned, were the big-scale sand smelt, Atherina boyeri, (10% of total reads assigned to OTUs), and the grass goby, Zosterisessor ophiocephalus (48%). This result was expected because the two species belong to the guild of the resident fishes in the Venice Lagoon. Overall, we identified seven species of lagoon residents (R) and ten species of marine migratory fishes (MM), the latter corresponded to fish species that tend to migrate from the sea to the lagoon during a specific time of year seeking food or that aggregate for the breeding season. The catadromous guild, here represented exclusively by Anguilla anguilla, was assimilated to marine migrants for these kinds of analyses. In addition, twelve species belonging to the freshwater guild (FW), twelve occasional migratory species (MO), as well as seventeen exclusively marine species (EM) have been detected. Based on the Italian Committee IUCN (International Union for Conservation of Nature) Red List (Battistoni et al., 2013), critically endangered species were found, such as Anguilla anguilla and Squalus acanthias, together with endangered or vulnerable species such as Mustelus mustelus and Alosa fallax (Table S3). Most of the other detected species were not currently in danger.
Detection of locally allochthonous species
Besides the alien species that are already known to be present in the Venice Lagoon, such as Silurus glanis (Corro, 2020), Oncorhynchus mykiss (Sicuro et al., 2016), and Gambusia holbrooki (Monti et al., 2021), eDNA analysis found traces of the presence of the oceanic pufferfish, Lagocephalus lagocephalus, a species that has never been detected in the lagoon or in the northern portion of the Adriatic Sea. The strong signal represented by the number of reads (more than 7000, see Table S4), and the absence of this species in the list of possible contaminants obtained by negative controls indicate that the detection of the eDNA of this species may be an early signal of its presence. Indeed, the presence of eDNA of the pufferfish was detected at the South site in March 2019 and at the North site in September and October of the same year.
Differences in northern and southern lagoon biodiversity
Retained reads, totals after the normalization and merging of the HiSeq and MiSeq data, were then assigned to the sites. Respectively, 9,134,931 reads were attributed to Torcello and 10,080,623 to Chioggia. Through eDNA metabarcoding, 54 taxa were assigned to the southern and 48 to the northern lagoon.
Prior to the ecological analyses, we excluded from the dataset species that had never been documented before in the Mediterranean Sea Clupea harengus, Pleuronectes platessa, Salmo salar, and Sebastes mentella because their detection was probably the result of contamination. A pairwise PERMANOVA analysis between the two sampling sites was performed on a Bray-Curtis matrix built from a refined OTU table, obtained from the ASV table with realistic taxonomic assignments (see sequence analyses for details), and it returned highly significant results (Pseudo-F = 7.97; p = 0.001). Consequently, a nMDS ordination was performed on the distance matrix, and its results (k = 3, stress = 0.171) were plotted in a two-dimensional scatterplot, showing a clear differentiation among the northern and southern sites (Figure 3, see also Figure S1 for coverage-based rarefied data). The existence of differences between these sites was confirmed by using presence/absence data with a Jaccard distance matrix (PERMANOVA Pseudo-F = 9.44; p = 0.001) and by the analysis of a reduced dataset comprising of only samples with the same number of replicates collected in the same month and year at the two different sites (PERMANOVA Pseudo-F = 2.43; p = 0.005).
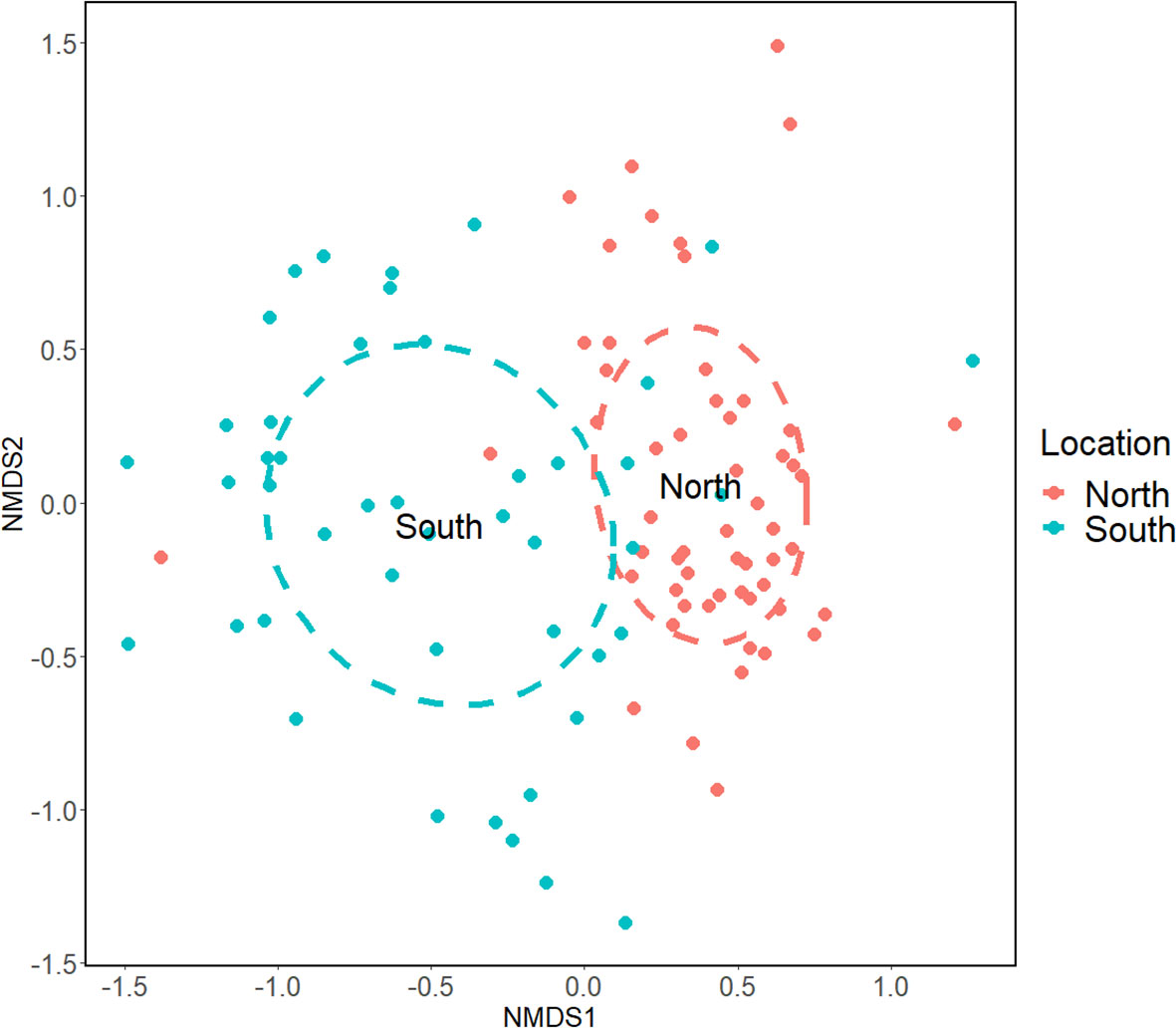
Figure 3 Non-metric Multidimensional Scaling of Venice Lagoon fish communities by sites, according to eDNA metabarcoding. Graph produced starting with a Bray-Curtis distance matrix calculated from the normalized OTU table. Southern lagoon samples (Chioggia) are indicated in blue and northern lagoon samples (Torcello) in red (k = 3; Stress = 0.171).
Eight species were detected exclusively in the northern lagoon, while fourteen were unique to the southern site. Of the species uniquely assigned to Torcello, five (62.5%) were freshwater fishes (“FW” ecological guild), while eight (57.1%) species, which were uniquely detected in Chioggia, were exclusively marine fishes (“EM” ecological guild). Moreover, to account not only for exclusivity but also for a clear disproportionality of the read numbers between the two sites in some OTUs, a ratio between normalized abundances (NAR) was calculated and used to produce a graphic representation of site selection for species. Even though most of the species were detected in both sites of the study, some of those were clearly more abundant in one site compared to the other one in terms of actual number of sequences detected (Figure 4). Species of the freshwater guild were mostly present in the internal site, the one closer to freshwater inputs. For example, Cyprinus carpio and Rhodeus sericeus, two freshwater species, were clearly more present in the northern site of the lagoon (Figure 4). On the other hand, species such as Conger conger and Sardinella aurita, exclusively marine, were more abundant in the site of Chioggia, which is the nearest site to the sea.
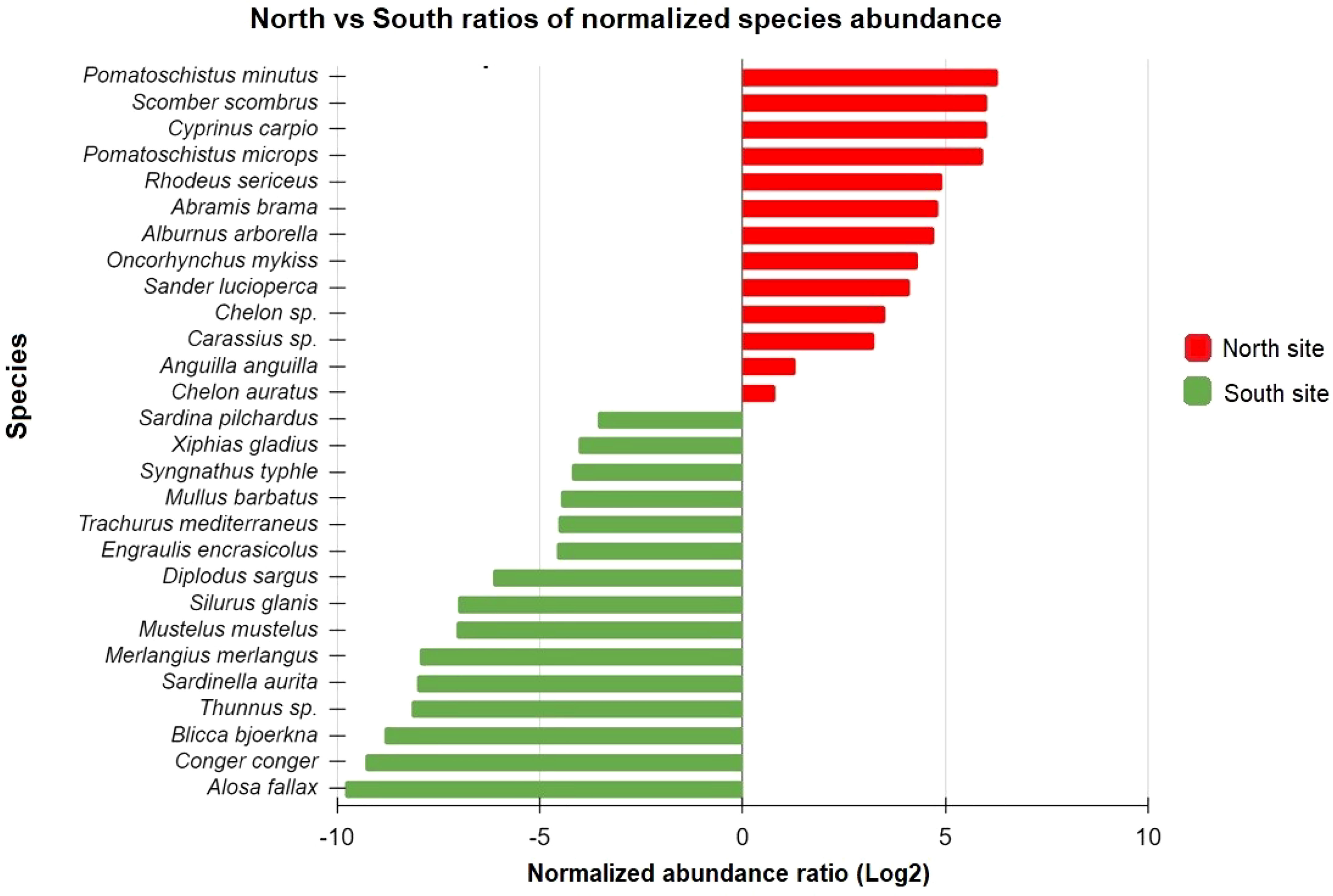
Figure 4 Overrepresented species at the North and South sites. On the x-axis, the ratio between North and South normalized abundances is reported in Log2 scale; positive values indicate species overrepresented in the northern site, negative values indicate species overrepresented in the southern site. Only species both present in north and south sites have been considered in this graph and reported on the y-axis.
Seasonality
PERMANOVA pairwise analysis results are illustrated in Table 2. As before, Atlantic species known to be absent in the Venice Lagoon were excluded from this analysis. Significant differences were detected between all seasons, with a particularly clear distinction between Fall and Winter samples with respect to the other seasons, as also shown by nMDS (Figure 5, see also Figure S2 for coverage-based rarefied data). Similar to what was reported for the differences between sites, PERMANOVA also confirmed the differences between seasons when using presence/absence data (Table S5) and when analyzing only strictly comparable samples (Table S6), though in this latter case some of the pairwise differences were smaller due to the use of a reduced dataset.
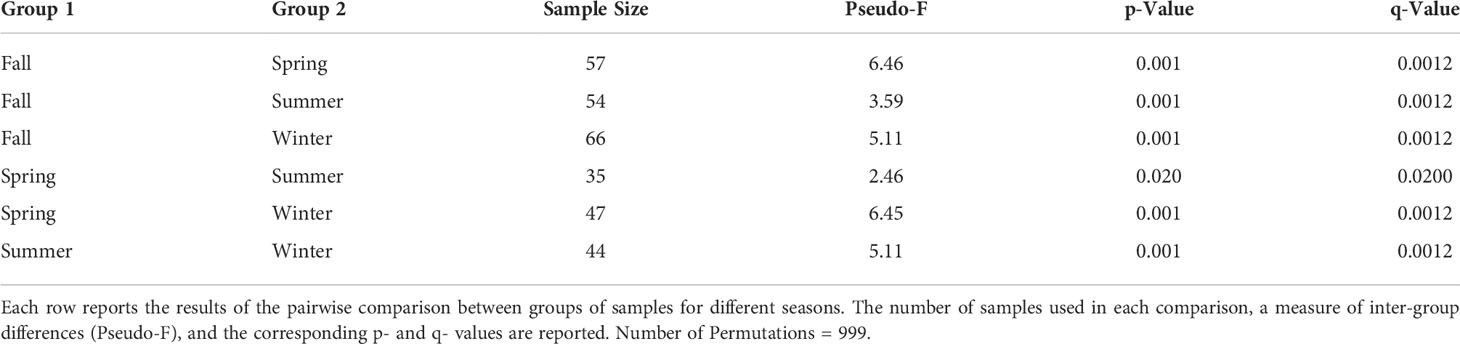
Table 2 Summary of pairwise PERMANOVA results for seasonality, based on Bray-Curtis dissimilarity and 101 samples.
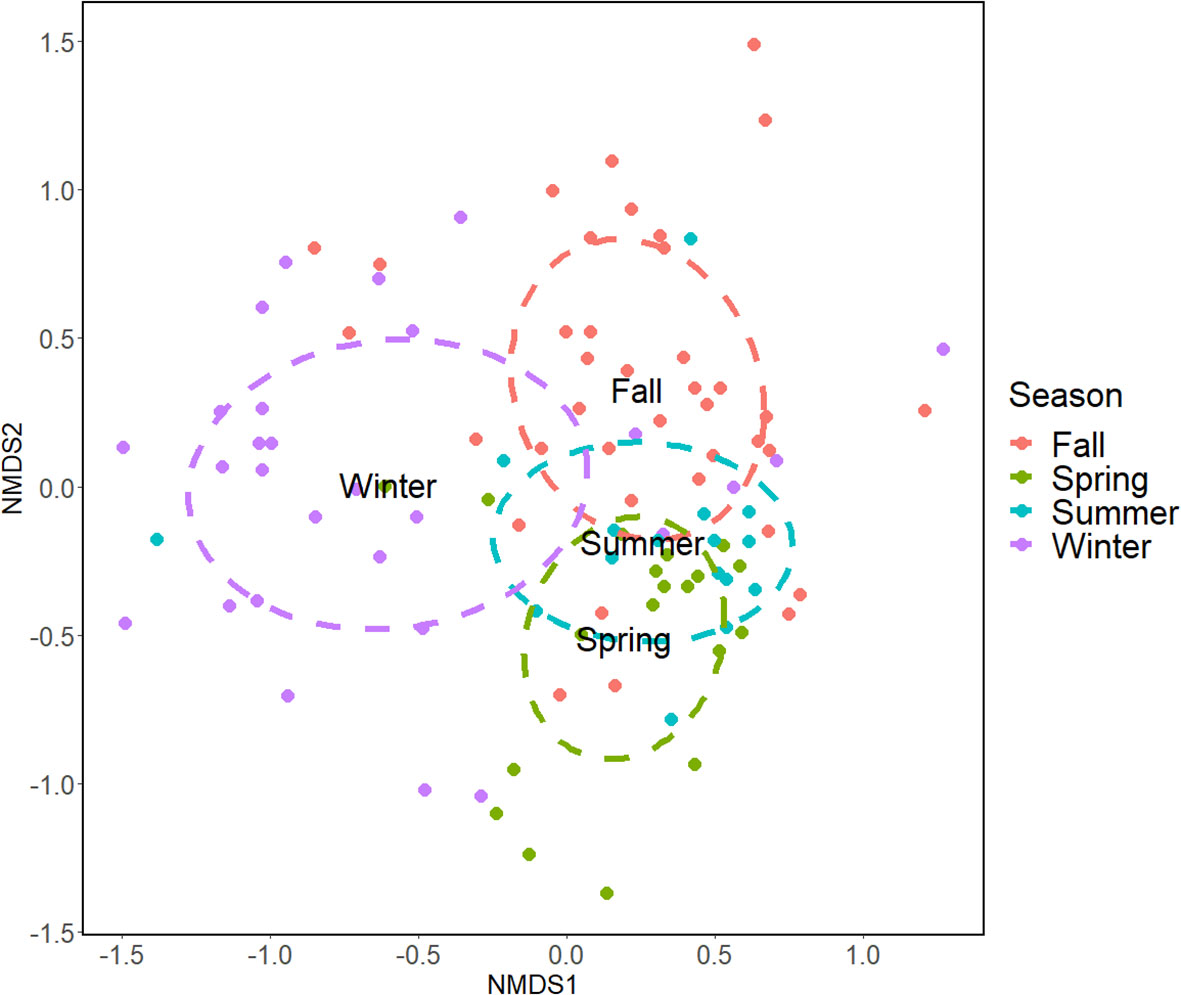
Figure 5 Non-metric Multidimensional Scaling of Venice Lagoon fish communities by seasons, according to eDNA metabarcoding. Graph produced starting with a Bray-Curtis distance matrix and calculated from the normalized OTU table. Samples collected in different seasons are shown in different colors (k = 3; Stress = 0.171).
SIMPER analysis was performed after also excluding, in addition to Atlantic species, OTUs assigned to Atherina boyeri and to Zosterisessor ophiocephalus because these species are overrepresented (Table S4) and possible sources of bias. Species that contributed the most to the differences between Summer and Fall were Dicentrarchus labrax (10.5%, p = 0.010) and Chelon auratus (9.7%, p = 0.033). Fish that primarily contributed to the Winter-Summer dissimilarity were Chelon auratus (10.7%, p = 0.009) and Diplodus sargus (5.0%, p = 0.001), while the ones that contributed to the Spring-Summer were Chelon auratus (18.3%, p = 0.001) and Pomatoschistus minutus (6.5%, p = 0.001). Finally, when including in the analysis only the samples of the northern site, also Anguilla anguilla resulted as a significant driver of the Summer-Winter diversity (6.6%, p = 0.031). In general, species found to influence the most seasonality patterns belonged mostly to the migratory ecological guilds, such as migratory fishes Dicentrarchus labrax, Diplodus sargus and Chelon auratus.
Discussion
Like in other eDNA studies metabarcoding fish diversity in the Mediterranean Sea by using Tele02 primers (Aglieri et al., 2021; Maiello et al., 2022), a high number of taxa was detected, and an elevated taxonomic resolution was achieved. In addition to bony fishes, three species of cartilaginous fishes were identified, indicating the applicability of this marker to simultaneously detect Actinopterygii and Chondrichthyes. The species richness retrieved in this study using eDNA metabarcoding was comparable to that reported in species checklists of the Venice Lagoon (Franzoi et al., 2010; Cavraro et al., 2017) and confirmed the high power of metabarcoding reported by Schroeder et al. (2020), who analyzed zooplankton samples collected at five sites in the Venice Lagoon, including LTER_EU_IT_016, station 5, one of the two chosen for our study.
Using eDNA, we detected species belonging to different ecological guilds (defined following Potter et al., 2015): occasional migrants (MO), marine migrants (MM), lagoon resident fishes (RL), freshwater species (FW), exclusive marine species (EM), and anadromes (AN). This variety of ecological guilds shows that surface water samples, taken from the Venice Lagoon, can produce a realistic picture of the functional ecological diversity along with the quantitative diversity represented by the number of species detected. The freshwater species detected (Abramis brama, Alburnus alborella, Blicca bjoerkna, Cyprinus carpio, Gambusia holbrooki, Padogobius martensii, Pseudorasbora parva, Rhodeus sericeus, Sander lucioperca, Silurus glanis, and Squalius cephalus) have different tolerances to salinity. Consequently, the detection of species less tolerant to low salinity could be due to transport of eDNA with river discharge, while the detection of tolerant species can be, more confidently, attributed to the presence of a local community. Interestingly, the RL guild, composed of species adapted to live in transitional environments, was represented by the highest number of reads. The MM and MO guilds were also remarkable, and they deserve particular attention due to the predictable impact that the MOSE system can have on these kinds of species, limiting or interrupting their movement between the lagoon and the sea. In this perspective, the fact that migrators were detected in this study in the same periods when MOSE was first activated (https://www.mosevenezia.eu/il-mose-in-funzione/#mvbtab_61894462f0019-1) highlights the potential impact of this infrastructure on these species.
Regarding unexpected species, this study is the first, to the best of our knowledge, to detect the oceanic puffer Lagocephalus lagocephalus along the upper Adriatic coasts (http://www.iucn.it/scheda.php?id=-2081245894), though caution is needed since only eDNA was detected. The literature on the oceanic puffer mostly focuses on its strong poison (Saoudi et al., 2008; Pinto et al., 2019), whereas studies on the congeneric species Lagocephalus sceleratus also focus on ecological factors and the ongoing invasion (Coro et al., 2018). The oceanic puffer is diffused throughout the oceanic tropical and subtropical waters, and it was first reported in the Mediterranean Sea, around Sicily, by Doderlain (1878). The species is considered present in the Mediterranean Sea by Tortonese (1986), except for the Adriatic and Black seas. Several recent “first detections” have been recently reported for areas in the Mediterranean Sea, where L. lagocephalus was never recorded before (see for instance: Erguden et al., 2017; Alshawy et al., 2019), suggesting that the species is currently expanding its range. In addition, according to Zava et al. (2005), starting from 1999, the number of detections of the oceanic puffer near the Sicilian coasts has increased. In the Adriatic Sea, the oceanic puffer was first spotted in 2004, with one specimen captured near the southern Adriatic Croatian coast (Dulcic and Pallaoro, 2006), and then, in 2015, two new records were reported further north along the Croatian coasts, with one specimen from the Kornati archipelago and one near to the island of Rab (Tsiamis et al., 2015). The detection of the oceanic pufferfish DNA in the Venice Lagoon adds a further support for an ongoing range expansion of this species in the Mediterranean Sea and raises concerns for the Adriatic Italian coast.
The northern and internal site (Torcello) and the southern sea-associated one (Chioggia), surveyed in this study, are both inside the Venice Lagoon but 31.39 km apart. They represent two different habitats inside a transitional ecosystem: Torcello is about 7.5 km from the closest lagoon inlet, while the Chioggia site is directly next to the Chioggia inlet. Based on this, our strategy has been to retrieve, through eDNA metabarcoding in both sites, the portrait of the ichthyological communities in both high salinity and low salinity environments (see https://issos.ve.ismar.cnr.it/for salinity maps). Our results indicate the presence of statistically significant differences and a clear clustering between samples belonging to the two sites. Moreover, the list of species (Table S3) and the proportion of the normalized reads abundance (Figure 4) show that exclusive marine species are predominant in Chioggia and freshwater fishes in Torcello. This, as other studies such as DiBattista et al. (2022), shows that eDNA metabarcoding can portray changes in fish communities caused by salinity in a lagoon environment. As a note of caution, differences between these two sites could be due to the proximity to bias factors. In fact, eDNA samples from Chioggia contained species known to be absent in the lagoon, such as the swordfish Xiphias gladius or the tuna Thunnus sp., whose DNA could have been detected due to the fact that they are traded, as a consequence of fish market discards. Such a hypothesis is supported by the presence of one of the largest fish-markets in Italy in the city of Chioggia, about 1.5 km from the Chioggia sampling site. Moreover, it cannot be excluded that the detection of eDNA belonging to these species at the Chioggia site is due to transport from off-shore because of hydrodynamics.
In addition to investigating differences between sites, our study aimed at picturing seasonal trends of marine migratory fishes through eDNA, in line with other works (Sigsgaard et al., 2017; Djurhuus et al., 2020; DiBattista et al., 2022). In our case, these trends are important because the MOSE infrastructure, a huge complex of dams that will alter the sea-lagoon connectivity, will inevitably condition the marine migratory ichthyological guild. Statistical analyses detected significant differences between seasons, particularly when comparing winter and fall with the other seasons. Migrators such as Chelon auratus, Diplodus sargus, and Dicentrarchus labrax contributed most to these differences, as evidenced by the SIMPER analysis. These species are characterized by marine reproduction and spend the early stages, as well as other periods of their lives, in shallow coastal waters or lagoons (Rossi, 1986; Franzoi et al., 1989; Franzoi and Trisolini, 1991; Franco et al., 2006). Abundance peaks of these species in the Venice Lagoon have been documented before in late winter/early spring and at the end of the summer/start of fall (Rossi, 1986), coherent to the SIMPER results obtained in this study.
Importantly, also the catadromous species Anguilla anguilla was detected in the Venice Lagoon, almost exclusively in eDNA for the fall and winter seasons (Table S4). Although it was not one of the species that contributed most to the seasonality patterns when the SIMPER analysis was performed on the whole dataset, it resulted as a significant driver of the summer-winter dissimilarity when considering only the more freshwater associated northern site. Individuals of this species are known to leave rivers to migrate to the Sargasso Sea for reproduction (Cresci, 2020), and this species is temporarily abundant in the Venice Lagoon in fall, according to catches of fyke net surveys (Scapin et al., 2022). The MOSE infrastructure, according to official data, have been activated 33 times in the period of October 2020 - October 2022, exclusively in fall and winter (https://www.mosevenezia.eu/il-mose-in-funzione/#mvbtab_61894462f0019-1). A correct management of this alteration in the future will be important as Anguilla anguilla is classified by the IUCN as a critically endangered species, and its severe demographic decrease is attributed to many causes, among which is the migratory barrier establishment (Starkie, 2003).
Conclusions
In this work, we obtained an ichthyological list of the Venice Lagoon species by using eDNA metabarcoding. The list of species is comparable to available checklists obtained by traditional monitoring. Beyond the qualitative result, functional and ecological diversity was observed, through the detection of six different ecological guilds. Two distinct communities were observed, Torcello with a strong component of freshwater fishes’ eDNA, and Chioggia with a dominant component of exclusive marine fishes. In addition, eDNA metabarcoding unveiled seasonality trends, and species most involved in this pattern were catadromous or migratory fishes. Migration periods of several species do coincide with the functioning of MOSE, the huge complex of dams that is currently working to preserve Venice from high tides. This structure, despite its importance for the protection of the historical and cultural patrimony that Venice represents, could function as a barrier for migrators, thus impacting the life strategy of these species, many of which are of strong commercial importance at the local scale. We highlight the urgence to account for this phenomenon and the need to perform extensive biodiversity monitoring, particularly for migratory species. Considering that the expected frequency of extreme events in the future, together with the projected sea level rise, will increase the number and duration of MOSE closures, we believe that eDNA should be routinely used to document ongoing changes in lagoon biodiversity and to provide input to stakeholders useful to move toward a “regulated lagoon”.
Data availability statement
The datasets presented in this study can be found in online repositories. The name of the repository and accession numbers can be found below: NCBI; PRJNA878955, BioSamples SAMN30788935 and SAMN30788936.
Author contributions
GC, IG, FM, EB, IAMM, and LZ conceived and designed the experiments. GC, IG, and FM performed the experiments. GC, IG, FM, and TL analyzed the data. EC, MP, AS, and LC contributed sampling/reagents/materials/analysis tools. GC, IG, FM, TL, EB, EC, LC, IAMM, MP, AS, and LZ critically discussed results; GC, IG, FM, TL, and LZ wrote the paper, which was approved by all the coauthors.
Funding
This scientific activity was performed within the Research Program Venezia 2021, Research Line 1.1, PI Luca Zaggia, coordinated by CORILA, and funded by the “Ministero delle Infrastrutture e della Mobilità Sostenibili - Provveditorato Interregionale per le Opere Pubbliche del Veneto - Trentino Alto Adige - Friuli Venezia Giulia” through the concessionary Consorzio Venezia Nuova. A part of the bioinformatic analyses have been performed on the CAPRI high performance infrastructure, thanks to the University of Padova Strategic Research Infrastructure Grant 2017: “CAPRI: Calcolo ad Alte Prestazioni per la Ricerca e l’Innovazione”.
Acknowledgments
The CORILA staff and particularly Cristina Dabalà and the Director Pierpaolo Campostrini provided constant and precious support. We are thankful to Andrea Sambo and Cristina Breggion (University of Padova) for their logistic support, and to Maria Gabriella Marin and Valerio Matozzo (University of Padova) for their generous offer of lab space and filtration equipment. We are deeply indebted to Stefania Bortoluzzi and Alessia Buratin for statistical advice (University of Padova). Carlotta Mazzoldi, Alessandro Vezzi, Annalisa Scapolatiello, Enrico Negrisolo (University of Padova), Alberto Pallavicini (University of Trieste), and Bella Galil (The Steinhardt Museum of Natural History, Tel Aviv University) provided useful discussions that helped to interpret the results. We would also like to thank the three reviewers for their very helpful and constructive comments.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer GA declared a shared affiliation with the author MP to the handling editor at the time of review.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2022.1009490/full#supplementary-material
References
Adamo M., Voyron S., Chialva M., Marmeisse R., Girlanda M. (2020). Metabarcoding on both environmental DNA and RNA highlights differences between fungal communities sampled in different habitats. PloS One 15 (12), e0244682. doi: 10.1371/journal.pone.0244682
Aglieri G., Baillie C., Mariani S., Cattano C., Calò A., Turco G., et al. (2021). Environmental DNA effectively captures functional diversity of coastal fish communities. Mol. Ecol. 30, 3127–3139. doi: 10.1111/mec.15661
Alshawy F., Ibrahim A., Hussein C., Lahlah M. (2019). First record of the oceanic puffer Lagocephalus lagocephalus (Linnaeus 1758) from the Syrian marine waters (eastern Mediterranean). Mar. Biodivers. Rec. 12, 1–4. doi: 10.1186/s41200-019-0170-9
Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. doi: 10.1016/S0022-2836(05)80360-2
Anderson J. T., Schumer G., Anders P. J., Horvath K., Merz J. E. (2018). Confirmed observation: A north American green sturgeon Acipenser medirostris recorded in the Stanislaus river, California. J. Fish. Wildl. Manage. 9, 624–630. doi: 10.3996/012018-JFWM-006
Basset A., Elliott M., West R. J., Wilson J. G. (2013). Estuarine and lagoon biodiversity and their natural goods and services. Estuar. Coast. Shelf Sci. 132, 1–4. doi: 10.1016/j.ecss.2013.05.018
Basset A., Sabetta L., Fonnesu A., Mouillot D., Chi T. D., Viaroli P., et al. (2006). Typology in Mediterranean transitional waters: New challenges and perspectives. Aquat. Conserv.: Mar. Freshw. Ecosyst. 16, 441–455. doi: 10.1002/aqc.767
Battistoni A., Peronace V., Rondinini C., Teofili C. (2013). Lista rossa dei vertebrati italiani. (Roma, IT: Comitato Italiano IUCN e Ministero dell’Ambiente e della Tutela del Territorio e del Mare)
Bergman P. S., Schumer G., Blankenship S., Campbell E. (2016). Detection of adult green sturgeon using environmental DNA analysis. PloS One 11 (4), e0153500. doi: 10.1371/journal.pone.0153500
Bohmann K., Evans A., Gilbert M. T. P., Carvalho G. R., Creer S., Knapp M., et al. (2014). Environmental DNA for wildlife biology and biodiversity monitoring. Trends Ecol. Evol. 29, 358–367. doi: 10.1016/j.tree.2014.04.003
Bolyen E., Rideout J. R., Dillon M. R., Bokulich N. A., Abnet C. C., Al-Ghalith G. A., et al. (2019). Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 37, 852–857. doi: 10.1038/s41587-019-0190-3
Callahan B. J., McMurdie P. J., Rosen M. J., Han A. W., Johnson A. (2016). JA, Holmes SP. 2016. DADA2: high-resolution sample inference from illumina amplicon data. Nat. Methods 13, 581–583. doi: 10.1038/nmeth.3869
Cavraro F., Redolfi Bristol S., Georgalas V., Torricelli P., Zucchetta M., Franzoi P. (2017). “Ingresso e distribuzione di uova, larve e giovanili di teleostei marini in laguna di venezia: connettività mare-laguna e funzione di nursery,” in Il controllo ambientale della costruzione del MOSE. Eds. Campostrini P., Dabalà C., Del Negro P., Tosi L., (Venice, IT: CORILA) 375–409.
Chao A., Jost L. (2012). Coverage-based rarefaction and extrapolation: standardizing samples by completeness rather than size. Ecology 93, 12, 2533–2547. doi: 10.1890/11-1952.1
Clarke K. R., Tweedley J. R., Valesini F. J. (2014). Simple shade plots aid better long-term choices of data pre-treatment in multivariate assemblage studies. J. Mar. Biol. Assoc. UK 94, 1–16. doi: 10.1017/S0025315413001227
Coro G., Vilas L. G., Magliozzi C., Ellenbroek A., Scarponi P., Pagano P. (2018). Forecasting the ongoing invasion of Lagocephalus sceleratus in the Mediterranean Sea. Ecol. Modell. 371, 37–49. doi: 10.1016/j.ecolmodel.2018.01.007
Corro G. (2020). Fish fauna in the oligohaline wetland habitats of the Venice lagoon: The case of Valle Averto (Padova: Università degli Studi di Padova).
Cresci A. (2020). A comprehensive hypothesis on the migration of European glass eels (Anguilla anguilla). Biol. Rev. 95, 1273–1286. doi: 10.1111/brv.12609
De Barba M., Miquel C., Boyer F., Mercier C., Rioux D., Coissac E., et al. (2014). DNA Metabarcoding multiplexing and validation of data accuracy for diet assessment: Application to omnivorous diet. Mol. Ecol. Resour. 14, 306–323. doi: 10.1111/1755-0998.12188
Deiner K., Bik H. M., Mächler E., Seymour M., Lacoursière-Roussel A., Altermatt F., et al. (2017). Environmental DNA metabarcoding: Transforming how we survey animal and plant communities. Mol. Ecol. 26, 5872–5895. doi: 10.1111/mec.14350
DiBattista J. D., Fowler A. M., Riley I. J., Reader S., Hay A., Parkinson K., et al. (2022). The use of environmental DNA to monitor impacted coastal estuaries. Mar. Pollut. Bull. 181, 113860. doi: 10.1016/j.marpolbul.2022.113860
DiBattista J. D., Reimer J. D., Stat M., Masucci G. D., Biondi P., De Brauwer M., et al. (2020). Environmental DNA can act as a biodiversity barometer of anthropogenic pressures in coastal ecosystems. Sci. Rep. 10, 1–15. doi: 10.1038/s41598-020-64858-9
Djurhuus A., Closek C. J., Kelly R. P., Pitz K. J., Michisaki R. P., Starks H. A., et al. (2020). Environmental DNA reveals seasonal shifts and potential interactions in a marine community. Nat. Commun. 11, 1–9. doi: 10.1038/s41467-019-14105-1
Doderlein P. (1878). Prospetto metodico delle varie specie di pesci riscontrate sin’ora nelle acque marine e fluviali della sicilia (Tipografia del Giornale di Sicilia: Palermo).
Dulcic J., Pallaoro A. (2006). First record of the oceanic puffer (Lagocephalus lagocephalus Linnaeus 1758), for the Adriatic Sea. J. Appl. Ichthyol. 22, 94–95. doi: 10.1111/j.1439-0426.2006.00699.x
Elliott M., Whitfield A. K. (2011). Challenging paradigms in estuarine ecology and management. Estuar. Coast. Shelf Sci. 94, 306–314. doi: 10.1016/j.ecss.2011.06.016
Erguden D., Gurlek M., Turan C. (2017). First occurrence of the oceanic puffer, Lagocephalus lagocephalus (Linnaeus 1758) in iskenderun bay, north-eastern Mediterranean, Turkey. J. Appl. Ichthyol. 33, 801–803. doi: 10.1111/jai.13363
Eva B., Harmony P., Thomas G., Francois G., Alice V., Claude M., et al. (2016). Trails of river monsters: detecting critically endangered Mekong giant catfish Pangasianodon gigas using environmental DNA. Global Ecol. Conserv. 7, 148–156. doi: 10.1016/j.gecco.2016.06.007
Franco A., Franzoi P., Malavasi S., Riccato F., Torricelli P., Mainardi D. (2006). Use of shallow water habitats by fish assemblages in a Mediterranean coastal lagoon. Estuarine Coast. Shelf Sci. 66 (1-2), 67–83. doi: 10.1016/j.ecss.2005.07.020
Franzoi P., Franco A., Torricelli P. (2010). Fish assemblage diversity and dynamics in the Venice lagoon. Rendiconti Lincei. 21, 269–281. doi: 10.1007/s12210-010-0079-z
Franzoi P., Trisolini R., Carrieri A., Rossi R. (1989). Caratteristiche ecologiche del popolamento ittico ripario della Sacca di Scardovari (Delta del Po). Nova Thalassia 10 (Suppl 1), 399–405.
Giupponi C., Eiselt B., Ghetti P. F. (1999). A multicriteria approach for mapping risks of agricultural pollution for water resources: the Venice lagoon watershed case study. J. Environ. Manage. 56, 259–269. doi: 10.1006/jema.1999.0283
Lim S. J., Thompson L. R. (2021). Mitohelper: A mitochondrial reference sequence analysis tool for fish eDNA studies. Environ. DNA 3, 706–715. doi: 10.14806/ej.17.1.200
Lopes C. M., Baêta D., Valentini A., Lyra M. L., Sabbag A. F., Gasparini J. L., et al. (2021). Lost and found: Frogs in a biodiversity hotspot rediscovered with environmental DNA. Mol. Ecol. 30, 3289–3298. doi: 10.1111/mec.15594
Maiello G., Talarico L., Carpentieri P., De Angelis F., Franceschini S., Harper L. R., et al. (2022). Little samplers, big fleet: eDNA metabarcoding from commercial trawlers enhances ocean monitoring. Fish. Res. 249, 106259. doi: 10.1016/j.fishres.2022.106259
Malavasi S., Franco A., Fiorin R., Franzoi P., Torricelli P., Mainardi D. (2005). The shallow water gobiid assemblage of the Venice lagoon: abundance, seasonal variation and habitat partitioning. J. Fish. Biol. 67, 146–165. doi: 10.1111/j.0022-1112.2005.00919.x
Martin M. (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal 17, 10–12. doi: 10.14806/ej.17.1.200
Martino F. (2022). Analisi del DNA ambientale di echinodermi ed altri invertebrati presenti in laguna di venezia (Padova: Università degli Studi di Padova).
Mel R., Carniello L., D’Alpaos L. (2021). How long the Mo.S.E. barriers will be effective in protecting all urban settlements within the Venice lagoon? the wind setup constraint. Coast. Eng. 168, 103923. doi: 10.1016/j.coastaleng.2021.103923
Miya M., Gotoh R. O., Sado T. (2020). MiFish metabarcoding: a high-throughput approach for simultaneous detection of multiple fish species from environmental DNA and other samples. Fish. Sci. 86 (6), 939–970. doi: 10.1007/s12562-020-01461-x
Monti F., Marcelli M., Fastelli P., Fattorini N. (2021). Pushed to the edge: Environmental factors drive ecological responses of Aphanius fasciatus when in sympatry with invasive Gambusia holbrooki. Aquat. Conserv.: Mar. Freshw. Ecosyst. 31, 2547–2559. doi: 10.1002/aqc.3600
Occhipinti-Ambrogi A., Marchini A., Cantone G., Castelli A., Chimenz C., Cormaci M., et al. (2011). Alien species along the Italian coasts: an overview. Biol. Invasions 13, 215–237. doi: 10.1007/s10530-010-9803-y
Oka S. I., Doi H., Miyamoto K., Hanahara N., Sado T., Miya M. (2021). Environmental DNA metabarcoding for biodiversity monitoring of a highly diverse tropical fish community in a coral reef lagoon: Estimation of species richness and detection of habitat segregation. Environ. DNA 3, 55–69. doi: 10.1002/edn3.132
Oksanen J., Kindt R., Legendre P., O’Hara B., Stevens M. H. H., Oksanen M. J., et al. (2007). The vegan package. Community Ecol. Package 10, 631–637.
Pinto E. P., Rodrigues S. M., Gouveia N., Timóteo V., Costa P. R. (2019). Tetrodotoxin and saxitoxin in two native species of puffer fish, Sphoeroides marmoratus and Lagocephalus lagocephalus, from NE Atlantic Ocean (Madeira island, Portugal). Mar. Environ. Res. 151, 104780. doi: 10.1016/j.marenvres.2019.104780
Potter I. C., Tweedley J. R., Elliott M., Whitfield A. K. (2015). The ways in which fish use estuaries: a refinement and expansion of the guild approach. Fish. 16, 230–239. doi: 10.1111/faf.12050
Prié V., Lopes-Lima M., Taberlet P., Valentini A., Poulet N., Jean P., et al. (2020). Large-Scale monitoring of freshwater bivalves: an eDNA point of view on species distribution and conservation. Authorea. doi: 10.22541/au.158888143.31089837
R Development Core Team (2021). “R: A language and environment for statistical computing, reference index version 4.2.1.,” in (Vienna: R Foundation for Statistical Computing). Available at: http://www.R-project.org/.
Rossi R. (1986). Occurrence, abundance and growth of fish fry in Scardovari bay, a nursery ground of the Po river delta (Italy). Arch. Oceanogr. Limnol. 20, 259–280.
Saoudi M., Abdelmouleh A., Kammoun W., Ellouze F., Jamoussi K., El Feki A. (2008). Toxicity assessment of the puffer fish Lagocephalus lagocephalu from the Tunisian coast. C. R. Biol. 331, 611–616. doi: 10.1016/j.crvi.2008.05.005
Scapin L., Zucchetta M., Pranovi F., Franzoi P. (2022). Influence of seascape on coastal lagoon fisheries: The role of habitat mosaic in the Venice lagoon. Estuar. Coasts 45, 793–811. doi: 10.1007/s12237-021-00986-3
Schnell I. B., Bohmann K., Gilbert M. T. P. (2015). Tag jumps illuminated–reducing sequence-to-sample misidentifications in metabarcoding studies. Mol. Ecol. Resour. 15, 1289–1303. doi: 10.1111/1755-0998.12402
Schroeder A., Stanković D., Pallavicini A., Gionechetti F., Pansera M., Camatti E. (2020). DNA Metabarcoding and morphological analysis - assessment of zooplankton biodiversity in transitional waters. Mar. Environ. Res. 160, 104946. doi: 10.1016/j.marenvres.2020.104946
Shu L., Ludwig A., Peng Z. (2020). Standards for methods utilizing environmental DNA for detection of fish species. Genes 11, 296. doi: 10.3390/genes11030296
Sicuro B., Tarantola M., Valle E. (2016). Italian Aquaculture and the diffusion of alien species: costs and benefits. Aquac. Res. 47, 3718–3728. doi: 10.1111/are.12997
Sigovini M. (2011). Multiscale dynamics of zoobenthic communities and relationships with environmental factors in the lagoon of Venice (Venezia: Università Ca’ Foscari Venezia).
Sigsgaard E. E., Nielsen I. B., Carl H., Krag M. A., Knudsen S. W., Xing Y., et al. (2017). Seawater environmental DNA reflects seasonality of a coastal fish community. Mar. Biol. 164, 1–15. doi: 10.1007/s00227-017-3147-4
Solidoro C., Bandelj V., Camatti E., Ciavatta S., Cossarini G., Facca C., et al. (2010). “Response of Venice lagoon ecosystem to natural and anthropogenic pressures over the last 50 years,” in Coastal lagoons: Critical habitats of environmental change. Eds. Pearl H., Kennish M. (Boca Raton, FL, USA: CRC Press, Taylor & Francis Group), 483–511.
Starkie A. (2003). Management issues relating to the European eel, Anguilla anguilla. Fish. Manage. Ecol. 10, 361–364. doi: 10.1111/j.1365-2400.2003.00351.x
Stoeckle M. Y., Soboleva L., Charlop-Powers Z. (2017). Aquatic environmental DNA detects seasonal fish abundance and habitat preference in an urban estuary. PloS One 12, e0175186. doi: 10.3389/fevo.2020.00009
Suarez-Menendez M., Planes S., Garcia-Vazquez E., Ardura A. (2020). Early alert of biological risk in a coastal lagoon through eDNA metabarcoding. Frontiers in Ecology and Evolution, 8, 9. doi: 10.3389/fevo.2020.00009
Taberlet P., Bonin A., Zinger L., Coissac E. (2018). Environmental DNA: For biodiversity research and monitoring (Oxford, UK: Oxford University Press).
Tagliapietra D., Keppel E., Sigovini M., Lambert G. (2012). First record of the colonial ascidian Didemnum vexillum KOTT 2002 in the Mediterranean: lagoon of Venice (Italy). BioInvasions Rec. 1, 247–254. doi: 10.3391/bir.2012.1.4.02
Tortonese E. (1986). “Tetraodontidae,” in Fishes of the north-eastern Atlantic and the Mediterranean, vol. 3 . Eds. Whitehead P. J. P., Bauchot M. L., Hureau J. C., Nielsen J., Tortonese E. (Paris, FR: UNESCO), 1341–1347.
Tsiamis K., Aydogan O., Bailly N., Balistreri P., Bariche M., Carden-noad S., et al. (2015). New Mediterranean biodiversity records (July 2015). Mediterr. Mar. Sci. 16, 472–488. doi: 10.12681/mms.1440
Tsuji S., Takahara T., Doi H., Shibata N., Yamanaka H. (2019). The detection of aquatic macroorganisms using environmental DNA analysis — a review of methods for collection, extraction, and detection. Environ. DNA 1, 99–108. doi: 10.1002/edn3.21
Turon M., Angulo-Preckler C., Antich A., Præbel K., Wangensteen O. S. (2020). More than expected from old sponge samples: A natural sampler DNA metabarcoding assessment of marine fish diversity in Nha Trang Bay (Vietnam). Front. Mar. Sci. 7, 605148. doi: 10.3389/fmars.2020.605148
Umgiesser G., Canu D. M., Cucco A., Solidoro C. (2004). A finite element model for the Venice lagoon. development, set up, calibration and validation. J. Mar. Syst. 51, 123–145. doi: 10.1016/j.jmarsys.2004.05.009
Ushio M., Furukawa S., Murakami H., Masuda R., Nagano A. J. (2022). An efficient early-pooling protocol for environmental DNA metabarcoding. Environ. DNA 00, 1–17. doi: 10.1002/edn3.337
Whitfield A. K., Elliott M., Basset A., Blaber S. J. M., West R. J. (2012). Paradigms in estuarine ecology–a review of the remane diagram with a suggested revised model for estuaries. Estuar. Coast. Shelf Sci. 97, 78–90. doi: 10.1016/j.ecss.2011.11.026
Keywords: environmental DNA, metabarcoding, fish community, Tele02, alien species, oceanic pufferfish, Venice, lagoons
Citation: Cananzi G, Gregori I, Martino F, Li T, Boscari E, Camatti E, Congiu L, Marino IAM, Pansera M, Schroeder A and Zane L (2022) Environmental DNA metabarcoding reveals spatial and seasonal patterns in the fish community in the Venice Lagoon. Front. Mar. Sci. 9:1009490. doi: 10.3389/fmars.2022.1009490
Received: 02 August 2022; Accepted: 28 October 2022;
Published: 17 November 2022.
Edited by:
Stefano Piraino, University of Salento, ItalyReviewed by:
Giorgio Aglieri, Stazione Zoologica Anton Dohrn Napoli, ItalyMark Stoeckle, The Rockefeller University, United States
Masayuki Ushio, Hong Kong University of Science and Technology, Hong Kong SAR, China
Copyright © 2022 Cananzi, Gregori, Martino, Li, Boscari, Camatti, Congiu, Marino, Pansera, Schroeder and Zane. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lorenzo Zane, lorenzo.zane@unipd.it
†These authors have contributed equally to this work
 Gabriele Cananzi
Gabriele Cananzi Irene Gregori
Irene Gregori Francesco Martino
Francesco Martino Tianshi Li
Tianshi Li Elisa Boscari
Elisa Boscari Elisa Camatti
Elisa Camatti Leonardo Congiu
Leonardo Congiu Ilaria Anna Maria Marino
Ilaria Anna Maria Marino Marco Pansera
Marco Pansera Anna Schroeder
Anna Schroeder Lorenzo Zane
Lorenzo Zane