Identification of the Gene sxtA (Domains sxtA1 and sxtA4) in Mexican Strains of Gymnodinium catenatum (Dinophyceae) and Their Evolution
- 1Departamento de Plancton y Ecología Marina, Centro Interdisciplinario de Ciencias Marinas, Instituto Politécnico Nacional, La Paz, Mexico
- 2CONACyT, Departamento de Plancton y Ecología Marina, Centro Interdisciplinario de Ciencias Marinas, Instituto Politécnico Nacional, La Paz, Mexico
- 3Departamento de Biotecnología Marina, Centro de Investigación Científica y de Educación Superior de Ensenada, Ensenada, Mexico
- 4Centro de Investigaciones Biológicas del Noroeste, La Paz, Mexico
Saxitoxin (STX) and its analogs are a broad group of natural neurotoxic alkaloids, commonly known as paralytic shellfish toxins. SxtA is the initial gene in the biosynthesis of saxitoxin. It has been proposed that the genes for STX biosynthesis had a bacterial origin and were acquired in the dinoflagellates by a horizontal gene transfer (HGT). In Gymnodinium catenatum, the origin of the STX genes is not well established. In this paper, we sequenced sxtA gene (domains sxtA1 and sxtA4) and determined the gene copy number in the genome in four Mexican strains of G. catenatum. We compare them with sequences of G. catenatum, Pyrodinium bahamense, and Alexandrium spp. from other geographic regions, and non-toxic producing dinoflagellates. Amplifications were performed for domains sxtA1 and sxtA4 from strains of G. catenatum and the phylogenetic analyses was done by maximum likelihood and Bayesian inference. The copy number determination was carried out using qPCR. The phylogenetic tree of domain sxtA4 showed the formation of two clades where G. catenatum sequences separated from the Alexandrium/Pyrodinium clade. The domain sxtA1 formed a higher number of clades than sxtA4. Sequences of G. catenatum were grouped together with sequences of Alexandrium. Dinoflagellates sequences that do not produce saxitoxin formed a separate clade. The gene copy number was 64 ± 30 and 110 ± 50 copies of sxtA1 and sxtA4 respectively. The identification of the gene sxtA of G. catenatum shows that the sequences are similar to those of Alexandrium species with low variations between species. These results may indicate that the acquisition of the gene sxtA was an early HGT event in the evolution of dinoflagellates. The possible loss of the ability to produce STX in some species suggests that the HGT from Alexandrium species toward G. catenatum is not possible.
Introduction
Saxitoxin (STX) and its analogs are a broad group of natural neurotoxic alkaloids, commonly known as paralytic shellfish toxins (PSTs). PSTs are the causative agents of paralytic shellfish poisoning (PSP) and are mostly associated with marine dinoflagellates (eukaryotes) and freshwater cyanobacteria (prokaryotes), which form extensive blooms around the world (Wiese et al., 2010). The biosynthetic pathway and genes responsible for STX synthesis have been characterized in the cyanobacteria Cylindrospermopsis raciborskii (Woloszynska) Seenayya & Subba Raju T3. This biosynthesis pathway is encoded by a fragment larger than 35 kb, comparative sequences analysis assigns 30 catalytic functions that correspond to 26 proteins (Kellmann and Neilan, 2007; Kellmann et al., 2008). Subsequently, the STX gene cluster was characterized in the cyanobacteria Anabaena circinalis Rabenhorst ex Bornet & Flahault, Aphanizomenon sp. Morren ex Bornet & Flahault, Rhaphidiopsis brookii Hill and Lyngbya wollei Farlow ex Gomont (Mihali et al., 2009, 2011; Moustafa et al., 2009; Murray et al., 2011).
The origin of the STX genes in cyanobacteria involved multiple events of horizontal gene transfer (HGT) from different sources, followed presumably by the coordination of the expression of foreign and native genes in the common ancestor. The possible ancestral source for some STX genes was a proteobacteria; suggesting that the key PSP-toxin biosynthesis gene may have evolved in a prokaryotic organism (Kellmann et al., 2008; Moustafa et al., 2009).
SxtA is the unique starting gene of STX-synthesis in cyanobacteria. This gene has a polyketide synthase (PKS)-like structure characterized by four catalytic domains with activities of a S-adenosyl-methionine- (SAM) dependent methyltransferase (sxtA1), GCN5-related N-acetyltransferase (sxtA2), acyl carrier protein (sxtA3), and a class II aminotransferase (sxtA4) (Kellmann et al., 2008).
Some dinoflagellate species of the genus Alexandrium Halim, as well as Pyrodinium bahamense Plate and Gymnodinium catenatum Graham, are known for producing PST (Scholin et al., 1995; Blackburn et al., 2001). Homologs of the genes putatively associated for the STX-synthesis have been identified in A. tamarense (Lebour) Balech Group IV, A. fundyense Balech, and A. minutum Halim (Stüken et al., 2011; Hackett et al., 2013). The transcripts of A. tamarense and A. minutum show that the dinoflagellate transcripts of sxtA have the same domain structure as the cyanobacterial sxtA genes. In contrast to the bacterial homologs, the dinoflagellate transcripts are monocistronic, have a higher guanine–cytosine (GC) content, occur in multiple copies, contain typical dinoflagellate spliced-leader sequences, and eukaryotic polyA-tails (Stüken et al., 2011).
The genes responsible for the STX-synthesis were not exchanged directly between cyanobacteria and dinoflagellates. The strong monophyly of sxtA-related proteins from STX+ dinoflagellates, and separately from cyanobacteria, supports the hypothesis that precursors of these sxtA genes were acquired independently by each linage (Hackett et al., 2013). Based on the sxtA data, the origin of the STX gene cluster within the dinoflagellates may have occurred between an ancestral STX+ bacterium and the common ancestor of Alexandrium and Pyrodinium (Orr et al., 2013).
Gymnodinium catenatum, a chain-forming dinoflagellate is the only species that can produce PST within the genus (Attaran-Fariman et al., 2007; Negri et al., 2007; Hallegraef et al., 2012; Gu et al., 2013). The origin of SXT genes in this species is not well established, but the distribution of the sxtA and sxtG genes suggest that G. catenatum acquired STX genes independently from a secondary dinoflagellate-dinoflagellate transfer (Orr et al., 2013). Another hypothesis, proposing that a single horizontal transfer event occurred early in dinoflagellate evolution, is the most parsimonious explanation for the origin of STX in dinoflagellates (Murray et al., 2015).
In this paper, we analyse the identification, the genomic copies number and the evolution of the sxtA gene in G. catenatum using Mexican strains and comparing them with sequences of G. catenatum, P. bahamense, and Alexandrium species from other geographic regions, as well other non-toxic producing dinoflagellates.
Materials and Methods
Strains and Culture Maintenance
The strains of G. catenatum in our study were isolated from several localities along the Mexican Pacific and the Gulf of California (Figure 1, Table 1). The strains were grown in modified GSe media (Bustillos-Guzmán et al., 2015) at 24 ± 1°C under a 12:12 h light:dark photoperiod and a photon irradiance of 150 μE m−2s−1 from white cool fluorescent lights.
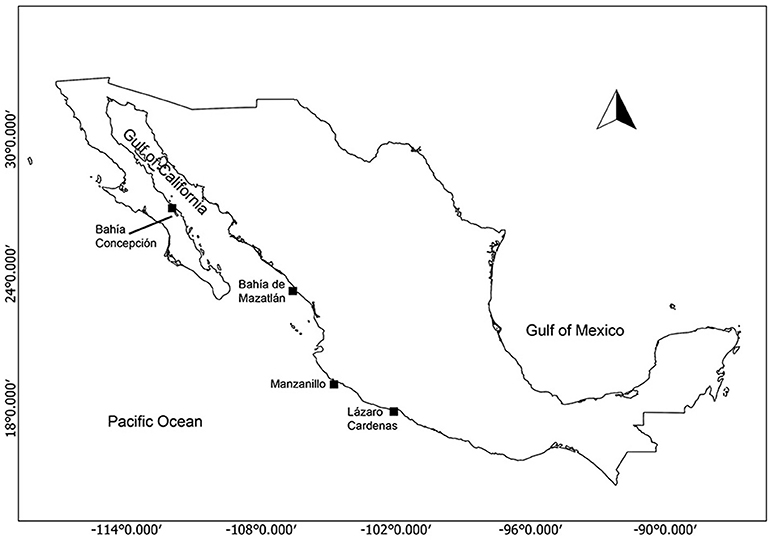
Figure 1. Isolation localities of Gymnodinium catenatum strains in the Gulf of California and the Mexican Pacific.
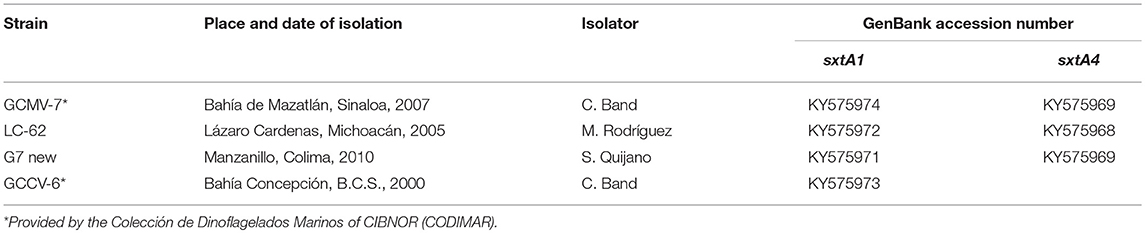
Table 1. Strains, place and year of isolation, and GenBank accession number of the Mexican Gymnodinium catenatum.
DNA Extraction and PCR
For the DNA extraction, 30 mL of each strain were harvested during the exponential growth phase by centrifugation at 1,000 × g for 10 min; the supernatant was removed by decantation. The pellet was frozen until extraction of DNA, using the CTAB method (Band-Schmidt et al., 2012). Quantity and quality of DNA were determined using nanodrop instrument (NanoDrop, Thermo Fisher Scientific, Waltham, MA) and by agarose gel electrophoresis.
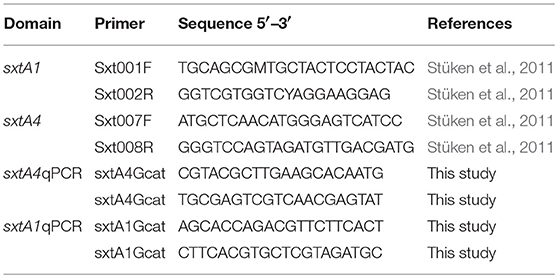
The genomic DNA was amplified by PCR in a PCR thermal-cycler system (MJ Mini, Bio-Rad Laboratories, Hercules, CA). The PCR reactions to amplify sxtA1 and sxtA4 were carried out in a final volume of 25 μl, the reaction mix consisted of 1 x PCR buffer, 0.5% DMSO, 1 mM MgCl2, 0.5 μM of each primer (Table 2), 200 μM dNTP's, and 1 U Taq DNA polymerase (Invitrogen, Carlsbad, CA). The sxtA1 was amplified as follows: 1 cycle of 5 min at 94°C; 35 cycles of 30 s at 94°C, 30 s at 61°C, and 1 min at 72°C, and 1 cycle of 7 min at 72°C. The sxtA4 was amplified as follows: 1 cycle of 5 min at 94°C; 35 cycles of 30 s at 94°C, 30 s at 58°C, and 1 min at 72°C, and 1 cycle of 7 min at 72°C. The fragments were visualized on an agarose gel by electrophoresis.
Sequencing and Phylogenetic Analyses
PCR products were purified by precipitating with ethanol and sent for sequencing in Macrogen (Seoul, South Korea) using an automatic DNA sequencer (ABI 3730 × 1).
For the phylogenetic analysis of the domain sxtA1 the EST from different species of dinoflagellates deposited in the Marine Eukaryote Transcriptome Sequencing Project were used. To obtain the domain from these sequences BLAST searches were done using the software BLAST+ 2.7.1 (National Center for Biotechnology Information, USA).
Local databases were created using the peptides sequences and comparing them with Alexandrium protein sequence as a query, for the searches blastp was used, hits with e-value < 1e−3 were included in the phylogenetic analyses. Sequences of toxic and non-toxic Alexandrium species deposited in GenBank were also included in the analyses.
For the phylogenetic analyses of the sxtA4 domain, only sequences of Alexandrium species and Pyrodinium bahamense deposited in GenBank were included (Table 3). In both cases (sxtA1 and sxtA4) the sequences were compared with cyanobacterial sequences of the gen sxtA.
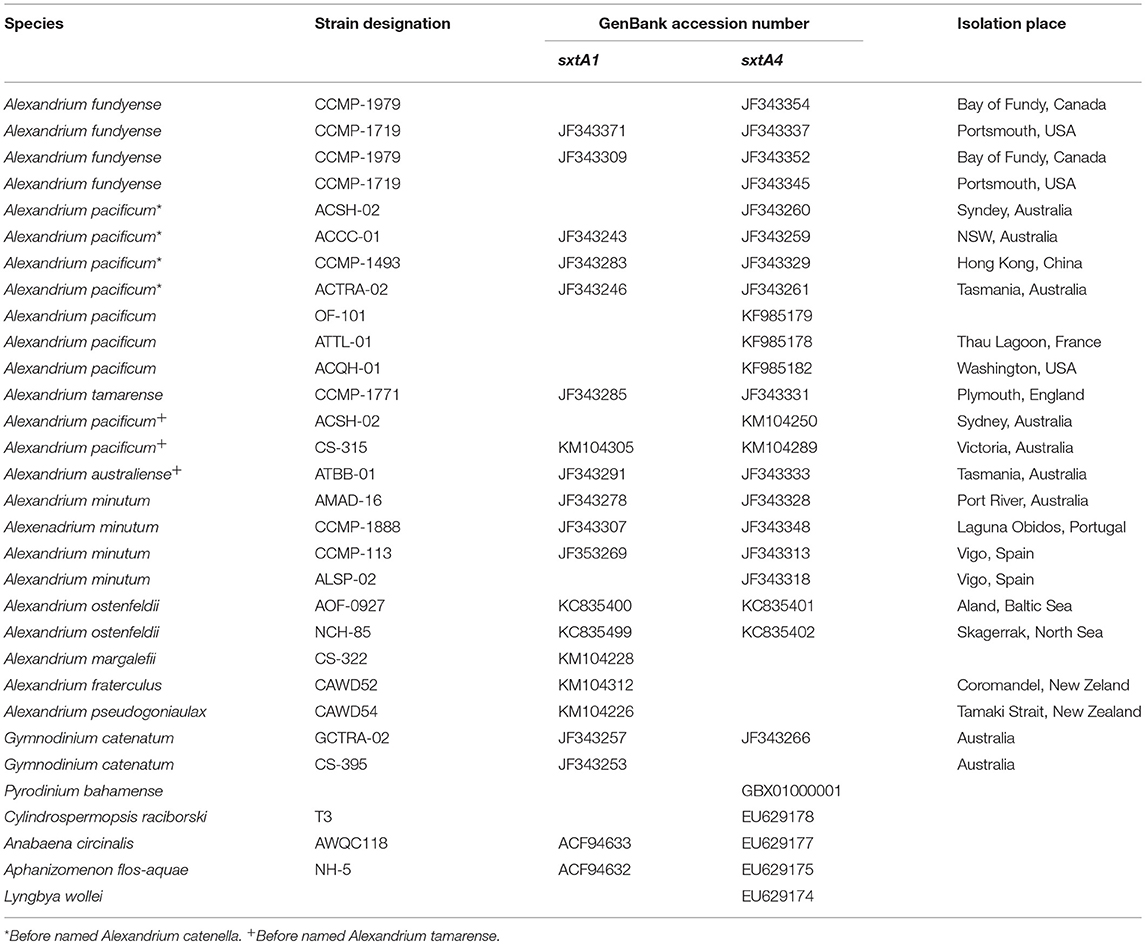
Table 3. Strains and GenBank accession numbers of species used in phylogenetic analyses for the sxtA1 and sxtA4 domains.
The sequences were examined and edited with Sequencher 4.1.4 software (Gene Codes, Ann Arbor, MI, USA). The sequences of domain sxtA1 of G. catenatum, P. bahamense and Alexandrium was translated to amino acid sequences using the program CLC Sequence Viewer 7 (CLC Bio, Qiagen, Denmark). The dinoflagellate amino acid sequences were aligned with MAFFTv7.313 (Yamada et al., 2016) with the L-INS-I model using the default parameters. Poorly aligned positions of amino acid alignments were removed with Gblocks set to the least stringent trimming options.
The best fit model for the amino acid sequences was selected using MEGA6 (Tamura et al., 2013); that WAG model was the optimal evolutionary model. Maximum Likelihood analyses were performed with RAxMLGUI (Silvestro and Michalak, 2012), PROTWAG model was used, with 100 bootstrap replicates. Bayesian inference was performed with the software MrBayes 3.2.6 (Huelsenbeck and Ronquist, 2001) using the same model; one million generations were run until the standard deviation of split sequences was less than 0.01.
The nucleotide sequences of domain sxtA4 were aligned with MAFFTv7.313 with the G-INS-I model using the default parameters. Poorly aligned positions of nucleotide alignments were removed with Gblocks. Maximum Likelihood analyses were performed with RAxMLGUI (Silvestro and Michalak, 2012). The GTR model with gamma distribution was used for the analysis with 500 bootstrap replicates. The Bayesian Inference was performed using GTR model; one million generations were run until the deviation standard was lower than 0.01. Trees were sampled every 200 generations with a burning of 1000 trees.
Copy Number Determination of the Domains sxtA1 and sxtA4
For the determination of the copy number G. catenatum (strain 62L) we cultivated the strain under the culture conditions previously described, using batch cultures in triplicate in 250 mL Erlenmeyer flasks with 150 mL of media. For DNA extraction 100 mL were harvested by centrifugation in the lag phase, 50 mL in the early exponential phase and 30 mL in the late exponential and stationary phase; in each phase a 2 mL sample was taken to determine the cell abundance using a 1 mL Sedgewick–Rafter chamber, the counts were done in an inverted microscope Carl Zeiss Axio Vert.A1. For cell counts cells were fixed with lugol.
Primers for qPCR were designed from conserved regions of G. catenatum to amplify partial sequences sxtA1 (150 bp) and sxtA4 (180 pb) using the software Primer 3 (Untergasser et al., 2012). The qPCR reactions were performed in triplicate in 10 μL reactions mixtures containing, 0.5 μM of each primer, 2.5 mM MgCl2, 1x PCR buffer, 200 μM dNTPs, 1U Taq Platinum (Invitrogen), and 1 × EvaGreen dye. Amplifications were carried out in a CFX96 Touch Real-Time PCR Detection System (Bio-Rad), with an initial denaturing step at 95°C for 4 minutes and 45 cycles of 95°C for 15 s, 56 or 55°C (to sxtA1 and sxtA4 primers, respectively) for 30 s. A melting-curve analysis was performed at the end of each cycle to confirm amplification specificity.
For both domains standard curves were constructed, from a 10-fold dilution series using fresh PCR product of each domain, ranging from 5 to 5 × 10−5 ng. The molecules of PCR product were determined: (A × 6.022 × 1023) × (660 × B)−1, where A is the concentration of the PCR product, 6.022 × 1023 Avogadro's number, 660: average molecular weight per base pair and B is the length of the PCR product.
Results
Phylogenetic Analysis Domain stxA1
The sequences length of domain sxtA1 of G. catenatum were among 525 and 541 bp, with an agreement between predicted and observed PCR amplifications. The GC content of the sequences was 64%. The search performed in BLAST, shows that the amplified sequences of G. catenatum correspond to sequences of the same species and domain (98% identities and 0.0 e-value). The second best match was related to Alexandrium fundyense and gene sxtA (91% identities and 0.0 e-value).
The phylogenetic inference of domain sxtA1 showed the separation of the cyanobacteria group, named cyanobacteria clade. Dinoflagellates were included in a separate group, named dinoflagellate clade. With this clade two well-defined branches were observed, one branch grouping dinoflagellates from different genera and some non-PSP producer species of Alexandrium (A. ostenfeldii, A. monilatum, and A. fratercolum) and the second branch including toxic and non-PSP producer species of Alexandrium, as well strains of G. catenatum (Figure 2).
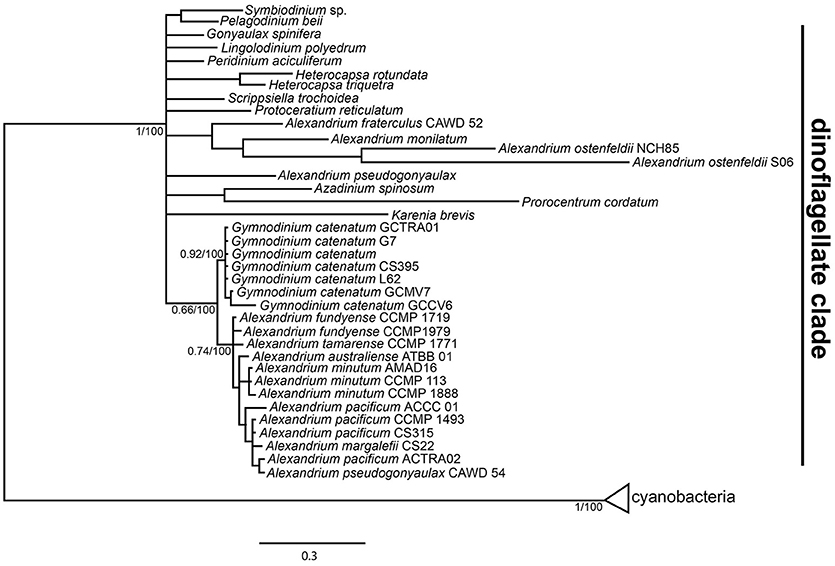
Figure 2. Phylogenetic tree of the domain sxtA1 from amino acids sequences of dinoflagellates and cyanobacteria. The tree is reconstructed with Bayesian Inference, the numbers in the internal nodes represent the posterior probabilities/boostrap values.
Within the dinoflagellate clade two subclades were formed. One contained sequences of G. catenatum strains from Australia and Mexico, without any differentiation as a result of the low differences between G. catenatum strains (0.2%), and another subclade formed by Alexandrium spp. sequences. The sequences used in the phylogenetic analysis of Alexandrium generally were distributed without species- or strain-related patterns. The pairwise distances between Alexandrium subclade and Gymnodinium subclade were between 7.1 and 10.7%.
Phylogenetic Analysis Domain sxtA4
PCR amplification of the domain sxtA4 of G. catenatum strains resulted in one product of approximately 658 bp; there was agreement between predicted and observed PCR amplifications with a GC content of the sequences of 60%. For this domain, sequences from the three dinoflagellate genera/species that produce STX (Alexandrium, G. catenatum, and P. bahamense) were used. BLAST searches indicated that the amplified sequences of Mexican strains of G. catenatum aligned in the first place with the sequence of the sxtA4 domain of a strain of G. catenatum (99% identity); the second alignment in BLAST corresponded to the species A. fundyense (90% identity).
The cyanobacterial sequences formed a well-supported clade separate from the dinoflagellate clade (Figure 3A). The phylogenetic tree constructed by maximum-likelihood and Bayesian inference showed that G. catenatum sequences form a fully supported cluster (100 bootstrap value and 1.0 posterior probabilities), separated from Alexandrium and Pyrodinium species. In this case, within the Gymnodinium clade one Mexican strain was grouped with the Australian strain and the other two Mexican strains were grouped in a second branch (Figure 3B). The differences within Gymnodinium strains $was 3%.
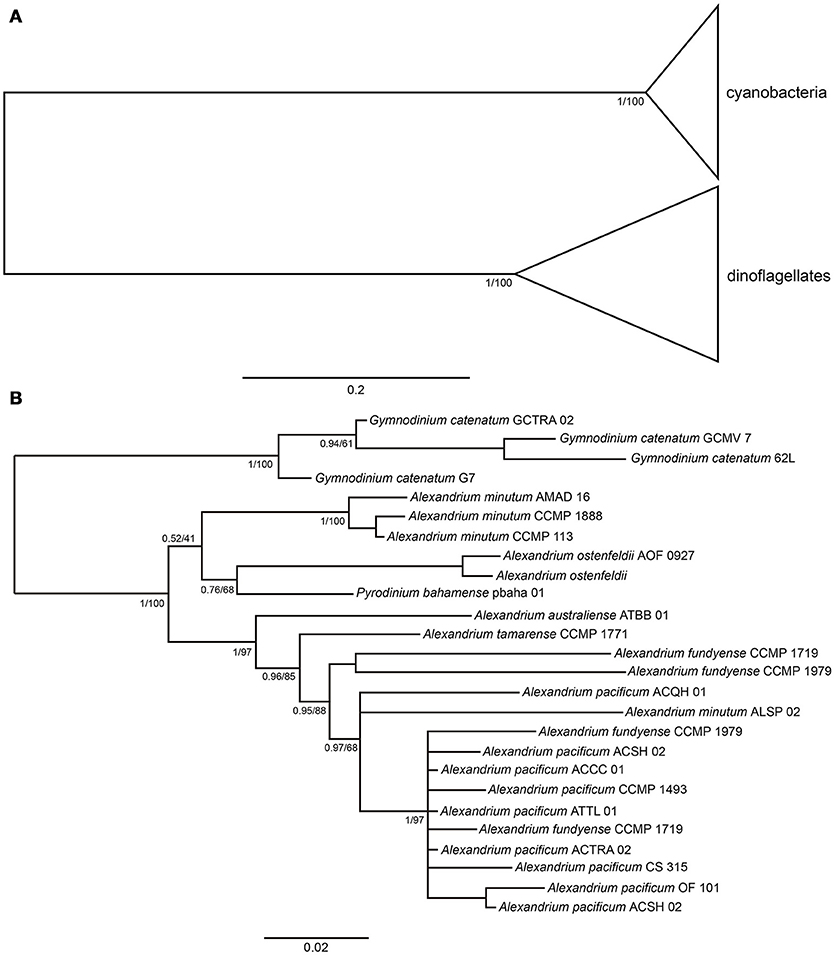
Figure 3. (A) Phylogenetic tree of sxtA4 domain of cyanobacteria and dinoflagellates, the node labels indicate the posterior probabilities and bootstrap values. (B) Phylogenetic tree of sxtA4 domain of dinoflagellates, the tree was reconstructed by Bayesian Inference, the node labels indicate the posterior probabilities and bootstrap values.
The only available sequence of P. bahamense was placed together with Alexandrium species, forming their own cluster (100-bootstrap value and 1.0 posterior probabilities), which was designated as the Alexandrium/Pyrodinium cluster. Similar to the sxtA1 domain, the sequences of Alexandrium do not show a species- or strains-related pattern. The differences between the sequences of Alexandrium and G. catenatum were of 13%.
Copy Number Determination
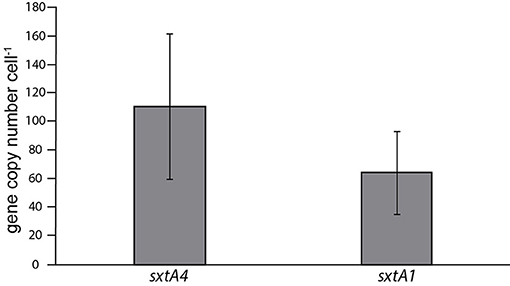
Between 75 and 32 genomic copies of sxtA1, and 143–74 genomic copies of sxtA4 in G. catenatum were found in triplicate batch cultures of strain 62L collected in different phases of the growth curve, based on the qPCR assay (Figure 4). There was no difference in the number of genomic copies among the different growth phases of the culture.
Discussion
Horizontal Gene Transfer (HGT) is a common process in eukaryotes and plays an important role in the genome evolution in these organisms (Andersson, 2005; Keeling and Palmer, 2008). In dinoflagellates, HGT has been reported in various species, especially in genes involved in photosynthesis and the Calvin Cycle. The source of these genes differs, being transferred mainly between eukaryotes (Takishita et al., 2003, 2008; Nosenko et al., 2006; Patron et al., 2006; Minge et al., 2010). In these cases the HGT correspond to tertiary endosymbiosis where the endosymbionts transferred the genes to the genome of the host.
Murray et al. (2015) propose that the most parsimonious explanation for the origin of STX in dinoflagellates is a single horizontal transfer event that occurred early in dinoflagellate evolution. The presence of paralogs of the sxtA1 domain in species that do not produce STX may support this hypothesis. If HGT occurred in an early stage in the evolution of dinoflagellates, the horizontal transfer from Alexandrium to G. catenatum proposed by Orr et al. (2013) may not have been possible.
Ingestion of prey or phagocytosis has been described as the mechanism of foreign gene acquisition in eukaryotes (Doolittle, 1998). If Alexandrium was a prey of G. catenatum and transferred its genes, it would be expected that G. catenatum was included within the same clade with Alexandrium and Pyrodinium species. Since G. catenatum in both domains form a well-supported clade, that separates from Alexandrium and Pyrodinium species, this could indicate that the HGT from Alexandrium to Gymnodinium probably did not take place. It is possible that HGT occurred during early evolution of dinoflagellates, with loss of genes in other species of the genus. Moreover, G. catenatum can only feed on small phytoplankton species <12μm (Jeong et al., 2005). Feeding of G. catenatum on Alexandrium spp. has hitherto not been reported.
The genus Alexandrium originated around the late Cretaceous 77 MYA (John et al., 2003), while G. catenatum, may have evolved around of 150–140 MYA from Gymnodinium microreticulate complex (Bolch, 1999). The presence of paralogs of the sxtA1 domain in two species of the order Gymnodiniales (Karenia brevis and Pelagodinium beii), suggest that HGT occurred before the divergence of Gymnodiniales and Gonyaulacales, with the subsequent loss in the capacity of PST biosynthesis in other dinoflagellates in the order Gymnodiniales; especially the loss of the domain sxtA4, which is present only in species that can produce PST.
In cyanobacteria, STX production genes originated from multiple HGT. The sxtA gene has a chimeric origin with the fusion of two different sources in one gene (Kellmann et al., 2008; Moustafa et al., 2009). The presence of a single domain of the sxtA gene in the dinoflagellates that do not produce STX can be explained by the loss of the domain that is found in the STX-producing species or that there were multiple transfers and in this case the transfer of the sxtA4 domain in non-toxic dinoflagellates has not occurred.
Within the G. catenatum strains, the Mexican and Australian sequences group together, but the evolutionary distance between the strains of both regions was greater in the sxtA4 domain than the sxtA1 domain. One possible evolutionary scenario is that both domains have different mutation rates. Between Alexandrium and G. catenatum a similar pattern of evolutionary distance was observed.
The difference in the sequences within the G. catenatum strain in each domain may be explained since these domains are present in multiple copies inside the genome, and this modification corresponds to the variance between the copies without a relationship within their geographic origin. One characteristic of the dinoflagellates is that they have a large genome and most of the genes families are present in multiple copies (Bachvaroff and Place, 2008; Lee et al., 2014) and they normally occur in 30–5000 copies per genome, indicating that a high gene copy number is widespread in dinoflagellates (Hou and Lin, 2009). In Alexandrium species, there are differences in the sequences in clones of the same strain, which may result from the presence of multiple copies of the same gene (Stüken et al., 2011; Wiese et al., 2014; Murray et al., 2015). This is not only observed in STX genes, but also by the presence of many copies of the actin gene inside the genome of Amphidinium and the changes between copies (Bachvaroff and Place, 2008).
The genomic copy number of the domain sxtA4 of G. catenatum is lower than the reported for A. catenella from 100 to 280 cell−1 (Murray et al., 2011; Stüken et al., 2011); and higher than the values reported for A. minutum and A. ostenfeldii with 1.5–10.8 and 1–15 copies cell−1 respectively (Stüken et al., 2015; Savela et al., 2016). There is no information about the genomic copy number of the domain sxtA1 in species of the genus Alexandrium, but in this study the gene copy number of G. catenatum of domain sxtA1 is lower than number for the sxtA4 domain. Stüken et al. (2015) proposed that gene copy number are not related to the genome size in A. minutum when comparing many strains of the species. Within the Alexandrium species, A. catenella and A. ostenfeldii have a similar genome size; 100 pg cell−1 and 115 pg cell−1 respectively (Figueroa et al., 2010), where the difference in the sxA4 copy number is great. Within dinoflagellates the genome size exhibits a positive correlation with cell size (Lin, 2011). The size of G. catenatum varies from 31 to 41 μm long and from 27 to 33 μm wide (Blackburn et al., 1989), while A. catenella size varies from 21 to 23.5 μm long and from 23 to 25 μm wide (Kim et al., 2002). Although the genome size of G. catenatum is unknow; it can be assumed that G. catenatum has a higher genome than A. catenella. The copy number in sxtA4 domain is higher than the copy number in G. catenatum, therefore, it could be suggested that the genome size is not related to the sxtA4 copy number in dinoflagellates.
Here it is shown for the first time the gene copy number of the domain sxtA4 of G. catenatum, as well as for the domain sxtA1, and a variation in the domain sxtA4 with respect to Alexandrium species. Murray et al. (2015) mention that following the acquisition of the genes trough HGT, a duplication process of the genes occurred. However, this duplication process was not the same within dinoflagellates that produce STX when the genes were acquired, and this could be the reason of the difference in gene copy numbers within dinoflagellates, and the selection pressure can act in various ways over the STX genes.
This study establishes that HGT of the gene sxtA did not occur from Alexandrium to G. catenatum, and that the HGT in G. catenatum was an early evolutionary event that probably occurred before this species diverged. The posterior loss of the capacity to produce STX in other members of the microreticulate complex, like in the Alexandrium genus, resulted in a the unique species of the genus Gymnodinium that has the capacity to produce STX.
If the HGT occurred in an ancestor of G. catenatum and Alexandrium, it is important to observe the presence of the paralogs of the sxtA1 domain in more species of the order Gymnodiniales; especially in the Gymnodinium microreticulate complex, in order to gain a better understanding of how the transfer and losses of STX genes has been within the complex, and how the evolution of these genes occurred in dinoflagellates.
Author Contributions
AM-F carried out all experimental work, acquired, analyzed, and interpreted data and drafted the manuscript. IL-V, CB-S participated in drafting the manuscript and analyzed and interpretation of phylogenetic data and supervised the overall progress of this project. CG-S participated in the experimental design of the qPCR analysis and drafting the manuscript. JB-G contributed with strains and commented and approved the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This research was funded by Instituto Politécnico Nacional (IPN grants SIP 2018-0662), and Consejo Nacional de Ciencia y Tecnología (CONACyT grant 248468, FORDECyT grant 260040). AM-F. is a recipient of fellowship from CONACyT (grant 268184) and BEIFI - IPN, CB-S. is a COFFA – IPN and EDI fellow. I. Fogel and D. W. Johnson for the English language editing.
References
Andersson, J. O. (2005). Lateral gene transfer in eukaryotes. Cell. Mol. Life Sci. 62, 1182–1197. doi: 10.1007/s00018-005-4539-z
Attaran-Fariman, G., de Salas, M. F., Negri, A. P., and Bloch, C. J. S. (2007). Morphology and phylogeny of Gymnodinium trapeziforme sp. nov. (Dinophyceae): a new dinoflagellate from the southeast coast of Iran that forms microreticulate resting cysts. Phycologia 46, 644–656. doi: 10.2216/07-05.1
Bachvaroff, T. R., and Place, R. (2008). From stop to start: tandem gene arrangement, copy number and trans-splicing sites in the dinoflagellate Amphidinium caterae. PLoS ONE 3:e2929. doi: 10.1371/journal.pone.0002929
Band-Schmidt, C. J., Martínez-López, A., Bustillos-Guzmán, J. J., Carreón-Palau, L., Morquecho, L., Olguín-Monroy, N., et al. (2012). Morphology, biochemistry, and growth of raphidophyte strains from the Gulf of California. Hydrobiologia 693, 81–97. doi: 10.1007/s10750-012-1088-y
Blackburn, S. I., Bolch, C. J. S., Haskard, K. A., and Hallegraeff, G. M. (2001). Reproductive compatibility among four global populations of the toxic dinoflagellate Gymnodinium catenatum (Diniohyceae). Phycologia 40, 78–87. doi: 10.2216/i0031-8884-40-1-78.1
Blackburn, S. I., Hallegraeff, G. M., and Bolch, C. J. (1989). Vegetative reproduction and sexual life cycle of the toxic dinoflagellate Gymnodinium catenatum from Tasmania, Australia. J. Phycol. 25, 577–590.
Bolch, C. J. S. (1999). Evolution, Species Resolution and Molecular Population Genetics of the Gymnodinium Catenatum Toxic Dinoflagellate Species Complex: Tracing Global Dispersal and Population Dynamics. Doctoral thesis, University of Tasmania.
Bustillos-Guzmán, J. J., Band-Schmidt, C. J., Durán-Riveroll, L. M., Hernández-Sandoval, F. E., López-Cortés, D. J., Núñez-Vázquez, E. J., et al. (2015). Paralytic toxin profile of the marine dinoflagellate Gymnodinium catenatum Graham from the Mexican Pacific as revealed by LC-MS/MS. Food. Addit. Contam. A 32, 381–394. doi: 10.1080/19440049.2014.1000978
Doolittle, W. F. (1998). You are what you eat: a gene transfer ratchet could account for bacterial genes in eukaryotic nuclear genomes. Trends. Genet. 14, 307–311.
Figueroa, R. I., Garcés, E., and Bravo, I. (2010). The use of flow cytometry for species identification and life-cycle studies in dinoflagellates. Deep Sea Res. II Top. stud. Oceanogr. 57, 301–307. doi: 10.1016/j.dsr2.2009.09.008
Gu, H., Liu, T., Vale, P., and Luo, Z. (2013). Morphology, phylogeny and toxin profiles of Gymnodinium inusitatum sp. nov., Gymnodinum catenatum and Gymnodinium microreticulatum (Dinophyceae) from the Yellow Sea, China. Harm. Algae 28, 97–107. doi: 10.1016/j.hal.2013.06.001
Hackett, J. D., Wisecaver, J. H., Brosnahan, M. L., Kulis, D. M., Anderson, D. M., Bhattacharya, D., et al. (2013). Evolution of saxitoxin synthesis in cyanobacteria and dinoflagellates. Mol. Biol. Evol. 30, 70–78. doi: 10.1093/molbev/mss142
Hallegraef, G. M., Blackburn, S. I., Doblin, M. A., and Bolch, C. J. S. (2012). Global toxicology, ecophysiology and population relationship of the chainforming PST dinoflagellate Gymnodinium catenatum. Harm. Algae 14, 130–143. doi: 10.1016/j.hal.2011.10.018
Hou, Y., and Lin, S. (2009). Distinct gene number-genome size relationships for eukaryotes and non-eukaryotes: gene content estimation for dinoflagellate genomes. PLoS ONE 4:e6978 doi: 10.1371/journal.pone.0006978
Huelsenbeck, J. P., and Ronquist, F. (2001). MrBayes: Bayesian inference of phylogenetic trees. Bioinformatics 17, 754–755. doi: 10.1093/bioinformatics/17.8.754
Jeong, H. J., Yoo, Y. Y., Park, J. Y., Song, J. Y., Kim, S. T., Lee, S. H., et al. (2005). Feeding by phototrophic red-tide dinoflagellates: five species newly revealed and six species previously known to be mixotrophic. Aquat. Microb. Ecol. 40, 133–150. doi: 10.3354/ame040133
John, U., Fensome, R. A., and Medlin, L. K. (2003). The application of a molecular clock based on molecular sequences and the fossil record to explain biogeographic distributions within the Alexandrium tamarense “species complex” (Dinophyceae). Mol. Biol. Evol. 20, 1015–1027. doi: 10.1093/molbev/msg105
Keeling, P. J., and Palmer, J. D. (2008). Horizontal gene transfer in eukaryotic evolution. Nat. Rev. Genet. 9, 605–618. doi: 10.1038/nrg2386
Kellmann, R., Mihali, T. K., Jeon, Y. J., Pickford, R., Pomati, F., and Neilan, B. A. (2008). Biosynthetic intermediate analysis and functional homology reveal a saxitoxin gene cluster in cyanobacteria. Appl. Environ. Microb. 74, 4044–4053. doi: 10.1128/AEM.00353-08
Kellmann, R., and Neilan, B. A. (2007). Biochemical characterization of paralytic shellfish toxin biosynthesis in vitro. J. Phycol. 43, 497–508. doi: 10.1111/j.1529-8817.2007.00351.x
Kim, K. Y., Yoshida, M., Fukuyo, Y., and Kim, C. H. (2002). Morphological observation of Alexandrium tamarense (Lebour) Balech, A. catenella (Whedon et Kofoid) Balech and related morphotype (Dinophyceae) in Korea. Algae 17, 11–19. doi: 10.4490/ALGAE.2002.17.1.011
Lee, R., Lai, H., Banoo-Malik, S. B., Saldarriaga, J. F., Keeling, P. J., and Slamovits, C. H. (2014). Analysis of EST data of the marine protist Oxyrrhis marina, an emerging model for alveolate biology and evolution. BMC Genomics 15:122. doi: 10.1186/1471-2164-15-122
Lin, S. (2011). Genomic understanding of dinoflagellates. Res. Microbiol. 162, 551–569. doi: 10.1016/j.resmic.2011.04.006
Mihali, T. K., Carmichael, W. W., and Neilan, B. A. (2011). A putative gene cluster from a Lynbya wollei bloom that encodes paralytic shellfish toxin biosynthesis. PLoS ONE 6:e14657. doi: 10.1371/journal.pone.0014657
Mihali, T. K., Kellmann, R., and Neilan, B. A. (2009). Characterization of the paralytic shellfish toxin biosynthesis gene clusters in Anabaena circinalis AWQC131C and Aphanizomenon sp. NH-5. BMC Biochem. 10:8. doi: 10.1186/1471-2091-10-8
Minge, M. A., Shalchian-Tabrizi, K., Torresen, O. K., Takishita, K., Probert, I., Inagaki, Y., et al. (2010). A phylogenetic mosaic plastid proteome and unusual plastid-targeting signals in the green-colored dinoflagellate Lepidodinium chlorophorum. BMC Evol. Biol. 10:191. doi: 10.1186/1471-2148-10-191
Moustafa, A., Loram, J. E., Hackett, J. D., Anderson, D. M., Plumley, F. G., and Bhattacharya, D. (2009). Origin of saxitoxin biosynthetic genes in cyanobacteria. PLoS ONE 4:e5758. doi: 10.1371/journal.pone.0005758
Murray, S. A., Diwan, R., Orr, R. J., Kohli, G. S., and John, U. (2015). Gene duplication, loss and the selection in the evolution of saxitoxin biosynthesis in alveolates. Mol. Phylogenet. Evol. 92, 165–180. doi: 10.1016/j.ympev.2015.06.017
Murray, S. A., Mihali, T. K., and Neilan, B. A. (2011). Extraordinary conservation, gene loss, and positive selection in the evolution of an ancient neurotoxin. Mol. Biol. Evol. 28, 1173–1182. doi: 10.1093/molbev/msq295
Negri, A. P., Bolch, C. S., Geier, S., Green, D. H., Park, T. G., and Blackburn, S. J. (2007). Widespread presence of hydrophobic paralytic shellfish toxins in Gymnodinium catenatum. Harm. Algae 6, 774–780. doi: 10.1016/j.hal.2007.04.001
Nosenko, T., Lidie, K. L., Van Dolah, F. M., Lindquist, E., Cheng, J. F., US Department of Energy-Joint Genome Institute, et al. (2006). Chimeric plastid proteome in the Florida “red tide” dinoflagellate Karenia brevis. Mol. Biol. Evol. 23, 2026–2038. doi: 10.1093/molbev/msl074
Orr, R. J., Stüken, A., Murray, S. A., and Jakobsen, K. S. (2013). Evolution and distribution of saxitoxin biosynthesis in dinoflagellates. Mar. Drugs 11, 2814–2828. doi: 10.3390/md11082814
Patron, N. J., Waller, R. F., and Keeling, P. J. (2006). A tertiary plastid uses genes from two endosymbionts. J. Mol. Biol. 357, 1373–1382. doi: 10.1016/j.jmb.2006.01.084
Savela, H., Harju, K., Spoof, L., Lindehoff, E., Meriluoto, J., Vehniäinen, M., et al. (2016). Quantity of the dinoflagellate toxin production in Alexandrium ostenfeldii blooms. Harm. Algae 52, 1–10. doi: 10.1016/j.hal.2015.10.018
Scholin, C. A., Hallegraeff, G. M., and Anderson, D. M. (1995). Molecular evolution of the Alexandrium tamarense “species complex” (Dinophyceae): dispersal in the North American and West Pacific regions. Phycologia 34, 472–485.
Silvestro, D., and Michalak, I. (2012). raxmlGUI: a graphical front-end for RAxML. Org. Divers. Evol. 12, 335–337. doi: 10.1007/s13127-011-0056-0
Stüken, A., Orr, R. J., Kellman, R., Murray, S. A., Neilan, B. A., and Jakobsen, K. S. (2011). Discovery of nuclear-encoded genes for the neurotoxin saxitoxin in dinoflagellates. PLoS ONE 6:e20096. doi: 10.1371/journal.pone.0020096
Stüken, A., Riobó, P., Franco, J., Jakobsen, K. S., Guillou, L., and Figueroa, R. I. (2015). Paralityc shellfish toxin content is related to genomic sxtA4 copy number in Alexandrium minutum strains. Front. Microbiol. 6:404. doi: 10.3389/fmicb.2015.00404
Takishita, K., Ishida, K., and Maruyama, T. (2003). An enigmatic GAPDH gene in the symbiotic dinoflagellate genus Symbiodinium and its related species (the order Suessiales): possible lateral gene transfer between two eukaryotic algae, dinoflagellate and euglenophyte. Protist 154, 443–454. doi: 10.1078/143446103322454176
Takishita, K., Kawachi, M., Noël, M. H., Matsumoto, T., Kakizoe, N., Watanabe, M. M., et al. (2008). Origins of plastid and glyceraldehyde-3-phosphate dehydrogenase genes in the green-colored dinoflagellate Lepidodinium chlorophorum. Gene. 410, 26–36. doi: 10.1016/j.gene.2007.11.008
Tamura, K., Stecher, G., Peterson, D., Filipski, A., and Kumar, S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. doi: 10.1093/molbev/mst197
Untergasser, A., Cutcutache, I., Koressaar, T., Ye, J., Faircloth, B. C., Remm, M., et al. (2012). Primer3 – new capabilities and interfaces. Nucleic Acids Res. 40:e115. doi: 10.1093/nar/gks596
Wiese, M., D'angostino, P. M., Mihali, T. K., Moffitt, M. C., and Neilan, B. A. (2010). Neurotoxic alkaloids: saxitoxin and its analogs. Mar. Drugs 8, 2185–2211. doi: 10.3390/md8072185
Wiese, M., Murray, S. A., Alvin, A., and Neilan, B. A. (2014). Gene expression and molecular evolution of sxtA4 in a saxitoxin producing dinoflagellate Alexandrium catenella. Toxicon 92, 102–112. doi: 10.1016/j.toxicon.2014.09.015
Keywords: dinoflagellate, Gymnodinium catenatum, horizontal gene transfer, paralytic shellfish toxins, gene copy number
Citation: Mendoza-Flores A, Leyva-Valencia I, Band-Schmidt CJ, Galindo-Sánchez CE and Bustillos-Guzmán JJ (2018) Identification of the Gene sxtA (Domains sxtA1 and sxtA4) in Mexican Strains of Gymnodinium catenatum (Dinophyceae) and Their Evolution. Front. Mar. Sci. 5:289. doi: 10.3389/fmars.2018.00289
Received: 29 April 2018; Accepted: 27 July 2018;
Published: 29 August 2018.
Edited by:
Marius Nils Müller, Universidade Federal de Pernambuco, BrazilReviewed by:
Shauna Murray, University of Technology Sydney, AustraliaPaulo Sergio Salomon, Universidade Federal do Rio de Janeiro, Brazil
Copyright © 2018 Mendoza-Flores, Leyva-Valencia, Band-Schmidt, Galindo-Sánchez and Bustillos-Guzmán. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Armando Mendoza-Flores, armando.mf6@gmail.com; amendozaf1200@alumno.ipn.mx
 Armando Mendoza-Flores
Armando Mendoza-Flores Ignacio Leyva-Valencia2
Ignacio Leyva-Valencia2  Clara E. Galindo-Sánchez
Clara E. Galindo-Sánchez José J. Bustillos-Guzmán
José J. Bustillos-Guzmán