- 1IHRC, Inc., Atlanta, GA, United States
- 2United States Department of Health and Human Services, Health Resources and Services Administration, Health Systems Bureau, National Hansen’s Disease Program, Baton Rouge, LA, United States
- 3Armauer Hansen Research Institute, Addis Ababa, Ethiopia
- 4Mycobacterial Research Laboratories, Anandaban Hospital, The Leprosy Mission Nepal, Kathmandu, Nepal
- 5Cebu Skin Clinic, Leonard Wood Memorial Center for Leprosy Research, Cebu, Philippines
Mycobacterium leprae is a slow-growing species of mycobacteria that cannot be cultured in axenic media. This presents a number of challenges for monitoring treatment efficacy and advancing new drugs and regimens for treating leprosy. We previously developed a molecular viability assay (MVA) which measures expression of hsp18 and esxA transcripts to determine viability of M. leprae directly from infected tissue. The objective of the current study was to determine the utility of the MVA for practical use on clinical specimens. Leprosy cases from the Philippines (N = 199), Ethiopia (N = 40), and Nepal (N = 200) were diagnosed by clinical examination, slit-skin smears (SSS) from index sites, and/or histopathology. Biopsy specimens for MVA were collected from an active lesion and stored in 70% ethanol. DNA and RNA were extracted from the tissue, and M. leprae were enumerated on the DNA fraction via RLEP qPCR. Based on this count, DNased RNA was normalized to the equivalent of 3x103 M. leprae per reverse transcription reaction, and hsp18 and esxA transcripts were amplified by PCR on the resulting cDNA. There was a strong correlation between RLEP enumeration on the specific biopsy specimen for MVA and the average SSS bacterial index (BI) in all three cohorts (p < 0.001). The MVA could be performed on most biopsies with an average SSS BI ≥ 2 and showed a decrease in M. leprae viability with increasing duration of leprosy multidrug therapy (R2 = 0.81, p < 0.001). The MVA also detected viable M. leprae in relapse patients where it showed significant correlation with the mouse footpad assay (p = 0.018). The MVA is a M. leprae-specific, sensitive, and relatively quick test. Clinically, the MVA would likely be most useful to monitor treatment, confirm suspected relapse cases, and determine efficacy of new leprosy drugs in clinical trials.
Introduction
Mycobacterium leprae, and the closely related Mycobacterium lepromatosis, are the causative agents of leprosy (1, 2). Both species have tropism for skin, mucous membranes, and Schwann cells in the peripheral nerves (3). When left untreated, leprosy can cause irreversible damage to the peripheral nerves. However, with timely implementation of leprosy multidrug therapy (MDT), physical disfigurement and other disabilities can be prevented and/or minimized.
Monitoring drug efficacy and developing improved treatment regimens is difficult as proof of M. leprae viability is an elusive detection factor. Although numerous attempts have been made, this organism has never been cultured on axenic medium and does not grow outside of host tissues. Moreover, M. leprae has an extremely long division time, requiring 12-14 days for a single replication cycle, so cultivation experiments can take months to complete (4). To compound matters further, M. leprae bacilli possess a dense lipid cell wall (5), and it can take months to years for the immune system to clear dead bacilli from tissues following effective drug therapy. Because of these unique characteristics, specialized techniques are required to ascertain the viability of this organism.
Over the past several decades, a variety of assays have been developed as indicators of M. leprae viability (reviewed in (6)). One of the initial tests used on clinical specimens was the highly subjective morphological index, or ratio of solid staining to beaded bacteria in a sample. Subsequently, various viability stains and metabolic assays were developed that were quite accurate. These assays, however, require high numbers of freshly prepared and purified bacilli, and the numbers of M. leprae bacilli in clinical samples are generally too low for their use. The gold standard for determining M. leprae viability is the mouse footpad (MFP) assay, in which the M. leprae bacilli in question are inoculated into the footpads of mice, and bacterial multiplication is measured over several months. While fairly sensitive, this procedure takes up to one year of culture before data acquisition and can handicap timely clinical applications. Furthermore, few clinics have access to laboratories that can conduct MFP culture and most cannot easily transfer fresh biopsies to those centers within the requisite 48-72 hours necessary for optimum viability retention. Therefore, a sensitive, field-feasible assay that can rapidly and accurately detect viable M. leprae in tissues would have immediate clinical applications in strategically placed referral laboratories involved in drug resistance monitoring, relapse detection and validation, or new drug evaluation.
Our laboratory previously developed a molecular viability assay (MVA) for M. leprae that is based on the expression of the mycobacterial esxA and hsp18 transcripts (7, 8). Since expression of these transcripts is normalized to the number of M. leprae present in the sample and compared to a standard curve, an absolute viability measurement is obtained that eliminates the need for a paired control. Additionally, the MVA can determine M. leprae viability in fixed tissue samples in less than one week and with smaller sampling requirements. It has proven useful for experimental studies of leprosy immunology and new drug evaluation as these transcripts are highly expressed by viable M. leprae, but expression rapidly decreases upon immunological (7–9) or drug-mediated (7, 10) killing of the bacilli. Detection of these transcripts has also shown promise for leprosy transmission studies (11).
The purpose of the current study was to evaluate the performance of the MVA on clinical biopsy specimens. These samples were from three distinct populations, and included patients who were newly diagnosed, relapsed, and currently undergoing treatment with MDT.
Methods
Ethics statements
Leprosy lesion biopsies for the MVA were collected after signed informed consent under specific country approvals (CLTRFI/LWM-IEC 2015-004; AAERC PO04/16; NHRC 108/2015) that conform to the standards set forth by the Declaration of Helsinki. A parent or legal guardian signed the consent form for any participant under the age of 18 years. Protocols for the MFP assay were reviewed and carried out according to Nepal guidelines. Mouse footpad tissues used as controls for the MVA were obtained in accordance with the United States Public Health Service Policy on the Humane Care and Use of Laboratory Animals. The NHDP Institutional Animal Care and Use Committee (Assurance #D16-00019 [A3032-01]) reviewed and approved the protocol. Animal experiments were conducted in accordance with The Guide for the Care and Use of Laboratory Animals, Eighth Edition.
Clinical assessment and diagnosis
This study included a total of 439 leprosy patients from three different sites: Cebu Skin Clinic (CSC), Leonard Wood Memorial Center for Leprosy Research, Cebu, Philippines (N = 199); Armauer Hansen Research Institute (AHRI), Addis Ababa, Ethiopia (N = 40); and Anandaban Hospital (AH), Kathmandu, Nepal (N = 200). Experienced leprologists diagnosed all cases utilizing accepted standards for evaluation of clinical signs and symptoms. Diagnosis was supported and confirmed by bacterial enumeration of slit-skin smear (SSS) samples and/or histopathology of a lesion biopsy. All cases examined from AHRI-Ethiopia were new untreated cases, as were the majority of cases from the CSC-Philippines and AH-Nepal. All samples from new cases were collected before commencement of leprosy therapy. The CSC-Philippines and AH-Nepal cohorts also contained relapse and suspected relapse cases. Relapse cases were diagnosed by the emergence of new skin lesions and an increase in a single site BI ≥ 2+ among any of the SSS sites (12). Regarding suspected relapse cases, the second diagnosis was made at the contributing facility, but the first diagnosis, which may have been many years before, was made at another clinic so definitive records were not always available. For the purposes of this study, suspected relapse and confirmed relapse cases were grouped together as relapse cases. The AH-Nepal cohort also contained cases currently undergoing treatment with WHO MDT.
Slit-skin smears (SSS)
SSS were collected from 3-6 sites. CSC-Philippines SSS were collected from both earlobes plus four active sites. If there were no lesions on the elbows and knees, SSS were collected where active lesions were located. AHRI-Ethiopia SSS were collected from each of the earlobes and one from either of the eyebrows. AH-Nepal SSS were collected from one earlobe, elbow, knee, and any suspect lesion. The SSS smears were stained with Ziehl-Neelsen carbol fuchsin, decolorized with 2% acid-alcohol, and counterstained with methylene blue. Results were expressed as the average SSS bacillary index (BI) calculated by the method of Ridley (13).
Histopathology
Leprosy lesion specimens for histopathology were collected using a 4mm punch biopsy, fixed in buffered formalin, and embedded in paraffin. Sections were stained with hematoxylin and eosin and Fite-Faraco stain for Ridley-Jopling classification (14).
Molecular viability assay (MVA)
Samples for the MVA were 4mm punch biopsies collected from an active lesion. All MVA specimens were stored in 70% ethanol at -20°C until shipment to the National Hansen’s Disease Programs (NHDP) in Baton Rouge, Louisiana, USA. Upon arrival at NHDP, all samples were stored at -20°C until processed. RNA and DNA were extracted from the fixed biopsy specimens using a chloroform/TRIzol extraction and a vertical homogenizer as previously described (8). Quantitative PCR (qPCR) by means of Taqman technology, and primers and probe specific for the M. leprae repetitive element (RLEP) were used to enumerate M. leprae on the DNA fraction of each specimen. Based on this count, the RNA from each specimen was normalized by adding the equivalent of 3 x 103 M. leprae to the reverse transcription (RT) reaction. The molecular viability of M. leprae was ascertained by PCR for the esxA and hsp18 target sequences on the resulting cDNA. All PCR results were compared to a standard curve of dilutions of a known concentration of M. leprae. RT-PCR of M. leprae 16S was run on each normalized sample as an RNA extraction quality control. A “mock” RT reaction containing no reverse transcriptase was run to ensure no contaminating DNA was present in the RNA fraction. Mouse footpad tissue infected with viable M. leprae was run as a positive RT-PCR control.
Mouse footpad (MFP) assay
The MFP assay was performed at AH-Nepal as previously described by Sapkota, et al. (15). Briefly, a homogenate was prepared from a fresh leprosy lesion and adjusted to contain 1 x 104 M. leprae per 0.03ml. Swiss albino mice were inoculated in both hind footpads with a volume of 0.03 ml per footpad. After 12 months, the footpads were harvested and the M. leprae were counted by the method of Shepard and McRae (16). M. leprae counts of ≥ 1 x 105 bacteria per footpad was considered growth and deemed positive in the MFP assay.
Statistical methods
The correlation between SSS and RLEP enumeration was calculated by Spearman’s rank order correlation test. A chi-square test of independence was used to determine an association between an average SSS BI ≥ 2 and the recovery of sufficient M. leprae in the lesion biopsies. The association between MVA data and MFP data was calculated using Fisher’s Exact test. Linear regression analysis was used to determine the effect of treatment duration on SSS, RLEP enumeration, and MVA. All calculations were performed with SigmaPlot 14.0 software. p < 0.05 was considered statistically significant.
Results
Patient demographics
Biopsies were collected from 439 leprosy cases at the CSC-Philippines, AHRI-Ethiopia, and AH-Nepal. Based on the WHO classification for multibacillary (MB) leprosy (17) as any case with the presence of acid-fast bacilli in a SSS regardless of the number of lesions, nerve involvement, or ≥ 6 lesions, the cohorts were heavily skewed towards MB cases: 99.0% (197/199) from CSC-Philippines, 95.0% (38/40) from AHRI-Ethiopia, and 96.0% (192/200) from AH-Nepal (Table 1). For all three cohorts, the majority of cases occurred in males and were in the 21 - 40 years age group. Most patients had had symptoms for 1 – 5 years prior to seeking care, and the majority were not in reaction.
Enumeration of M. leprae in case specimens
The bacterial load in each case was estimated using the average SSS BI method. The individual average SSS BI ranged from 0 to 5+ in the CSC-Philippines cohort and 0 to 6+ in the AHRI-Ethiopia and AH-Nepal cohorts. The number of M. leprae in the biopsy specimen used for the MVA was determined by RLEP enumeration. Overall, RLEP PCR was positive in 98.5% (196/199) of CSC-Philippines cases, 95% (38/40) of AHRI-Ethiopia cases, and 96% (192/200) of AH-Nepal cases. Moreover, RLEP PCR was positive in 75.0% (9/12 CSC-Philippines), 87.5% (14/16 AHRI-Ethiopia), and 81.1% (30/37 AH-Nepal) of lesions collected from patients with an average SSS BI < 1. Spearman’s rank order correlation was computed to assess the relationship between the average SSS BI count of an individual patient and the number of M. leprae recovered from the leprosy lesion biopsy sent for the MVA. CSC-Philippines specimens showed a significant positive correlation between these two variables (r = 0.70, p < 0.001). Likewise, AHRI-Ethiopia (r = 0.81, p < 0.001) and AH-Nepal (r = 0.73, p < 0.001) specimens showed a significant positive correlation as well.
The MVA is designed such that 2 x 104 M. leprae must be recovered from a tissue specimen to properly normalize the reverse transcription reaction (8). Therefore, we determined whether the average SSS BI could predict the likelihood of sufficient bacterial recovery from a lesion biopsy to perform the MVA. The distribution of the average SSS BI versus the percentage of MVA biopsies from which ≥ 2 x 104 M. leprae (red bars) or < 2 x 104 M. leprae (green bars) were recovered is depicted for the CSC-Philippines (Figure 1A), AHRI-Ethiopia (Figure 1C), and AH-Nepal (Figure 1E) cohorts. Chi-square analyses determined that there was a significant association (p < 0.001) between an average SSS BI ≥ 2 and the recovery of sufficient M. leprae in the lesion biopsies from the CSC-Philippines (Figure 1B), AHRI-Ethiopia (Figure 1D), and AH-Nepal (Figure 1F). Furthermore, this association held when all samples were analyzed together (Figures 1G, H). These data support that an average SSS BI ≥ 2 is a good indicator of attaining enough M. leprae from a lesion to run the MVA.
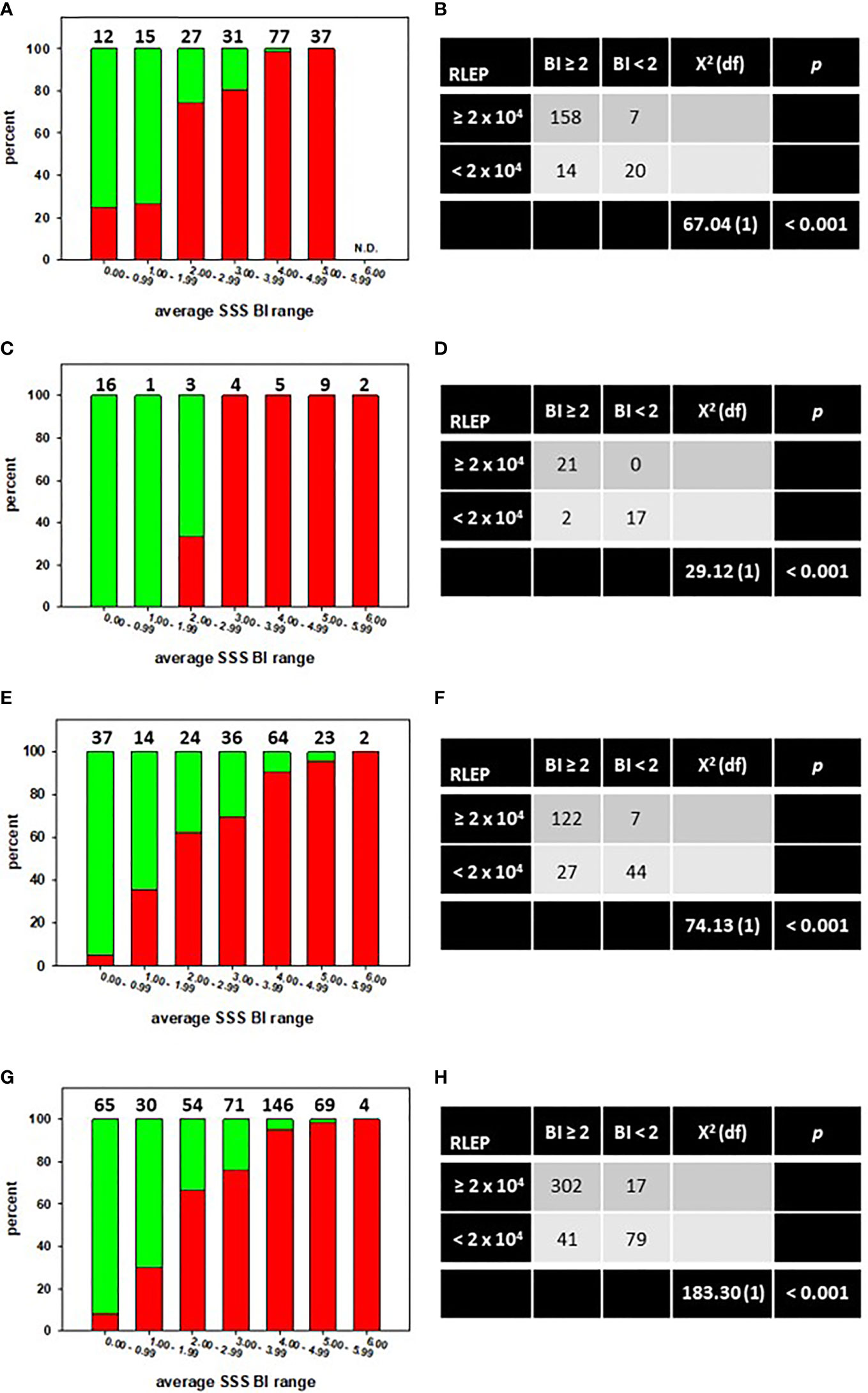
Figure 1 An average SSS BI ≥ 2 is associated with recovery of sufficient M. leprae from a skin biopsy to run the MVA. The distribution of the average SSS BI versus the percentage of MVA biopsies from which ≥ 2 x 104 M. leprae (red bars) or < 2 x 104 M. leprae (green bars) were recovered in specimens from CSC-Philippines (A), AHRI-Ethiopia (C), AH-Nepal (E), or a Composite of all specimens combined (G). N.D. = none determined. Numbers above the bars indicate the number of cases per average SSS BI group. Chi-square tests of independence showed a significant association between an average SSS BI ≥ 2 and recovery of sufficient M. leprae for MVA in CSC-Philippines (B), AHRI-Ethiopia (D), AH-Nepal (F), or a Composite of all specimens combined (H).
Determination of M. leprae viability
Overall, 317 lesion biopsies contained enough M. leprae for normalization and viability determination. Satisfactory RNA preparations, as determined by 16S rRNA expression, were obtained from 269 (84.9%) of these specimens.
CSC-Philippines new cases
Of the 199 specimens from the CSC-Philippines, 148 samples satisfied the M. leprae number (≥ 2 x 104) and RNA extraction criteria. Of these, 142 were new cases (Figure 2A). These new cases were evaluated for M. leprae viability via the MVA using both hsp18 and esxA transcripts as targets. 66.9% (95/142) of the new cases expressed hsp18 (Figure 2B) while 59.9% (85/142) expressed esxA (Figure 2C). Overall, viable M. leprae (MVA[+], expression of one or both transcripts) were detected in 75.4% (107/142) of the new cases (Figure 2D). Expression of both transcripts was detected in 69.2% (74/107), while only hsp18 was detected in 19.6% (21/107) cases and only esxA was detected in 11.2% (12/107) cases.
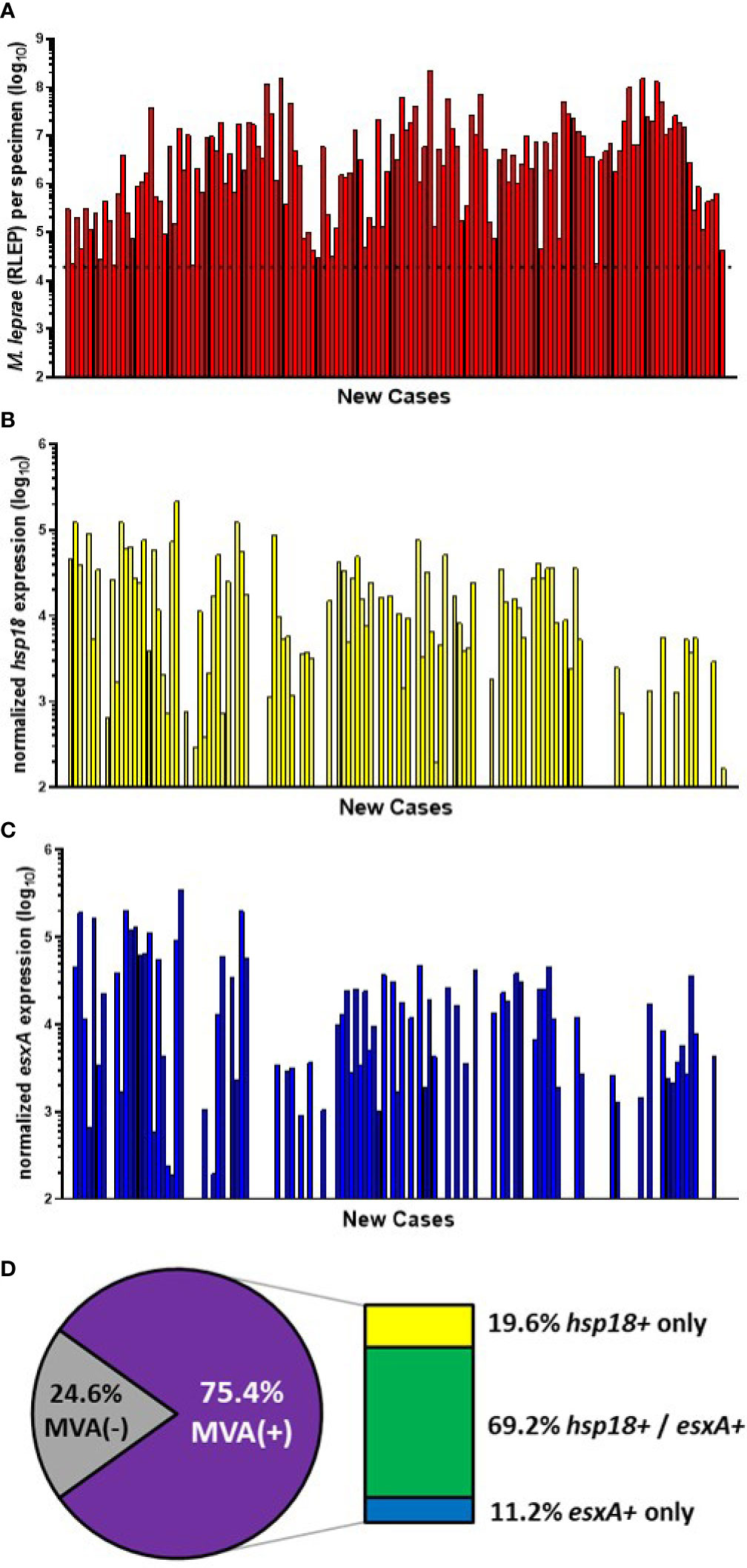
Figure 2 M. leprae enumeration and viability in new cases from CSC-Philippines. DNA and RNA were extracted from biopsy specimens. M. leprae were enumerated (A) by RLEP PCR on the DNA fraction. The dotted line represents the minimum number of M. leprae that must be present in a sample to progress to the viability step. Based on the RLEP count, each DNased RNA was normalized to 3 x 103 M. leprae equivalents and reverse transcribed. Viability was determined on the resulting cDNA by hsp18 (B) and esxA (C) PCR. (D) The percentage of MVA(+) samples (expression of one or both transcripts) and MVA(-) samples (no expression of either transcript), and the percentage of MVA(+) samples expressing each transcript.
CSC-Philippines relapse cases
The remaining 6 CSC-Philippines cases were relapse cases (Figure 3A). 100% (6/6) of these cases showed expression of hsp18 (Figure 3B) and 83.3% (5/6) showed expression of esxA (Figure 3C). Overall (Figure 3D), there was a 100% (6/6) detection of viable M. leprae in the relapse cases with 83.3% (5/6) expressing both transcripts and 16.7% (1/6) expressing only hsp18. None of the cases expressed only esxA.
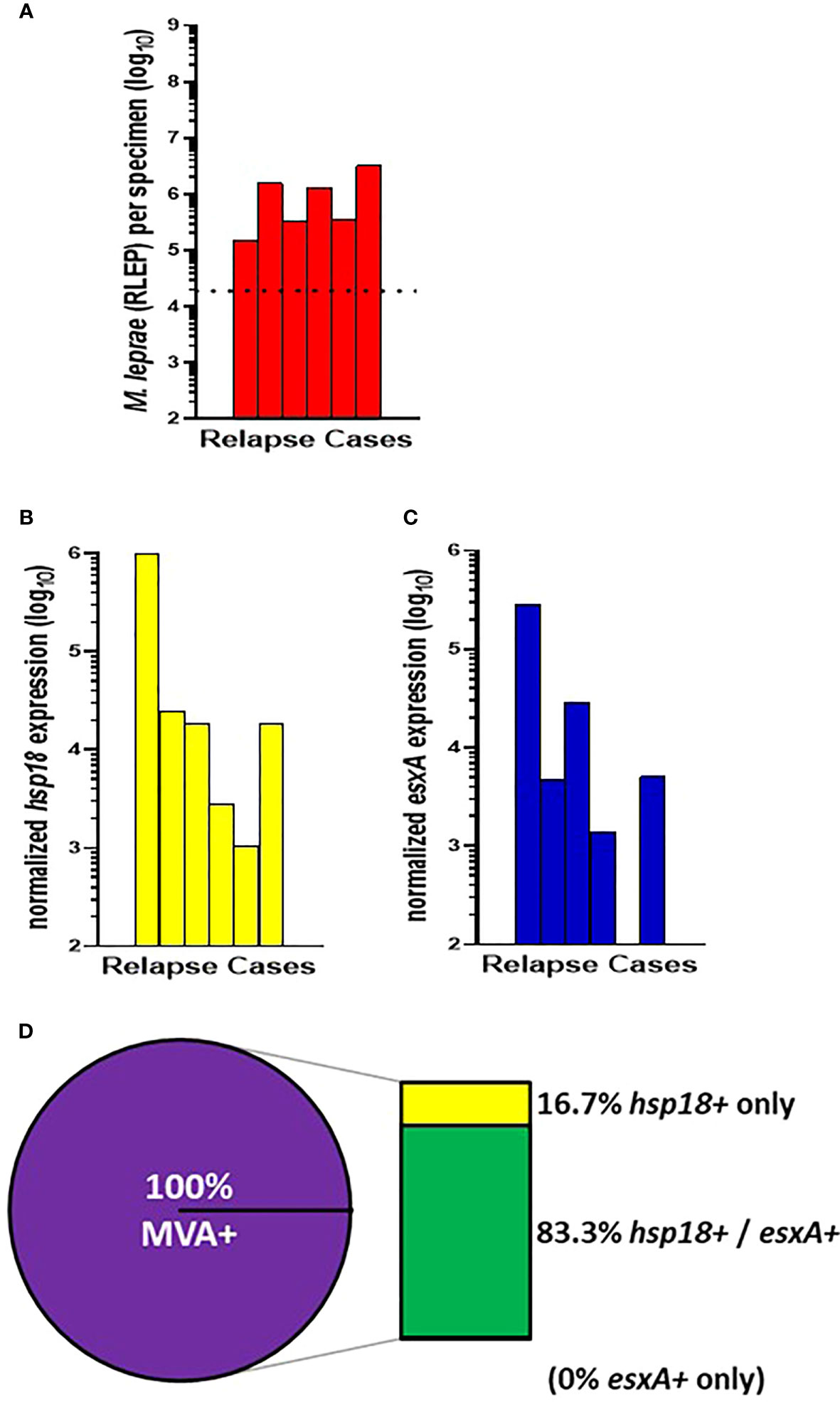
Figure 3 M. leprae enumeration and viability in relapse cases from CSC-Philippines. DNA and RNA were extracted from biopsy specimens. M. leprae were enumerated (A) by RLEP PCR on the DNA fraction. The dotted line represents the minimum number of M. leprae that must be present in a sample to progress to the viability step. Based on the RLEP count, each DNased RNA was normalized to 3 x 103 M. leprae equivalents and reverse transcribed. Viability was determined on the resulting cDNA by hsp18 (B) and esxA (C) PCR. (D) The percentage of MVA(+) samples (expression of one or both transcripts) and MVA(-) samples (no expression of either transcript), and the percentage of MVA(+) samples expressing each transcript.
AHRI-Ethiopia new cases
All 40 of the AHRI-Ethiopia biopsy specimens were from new cases. This cohort contained a high percentage of very low BI cases (Figures 1C, D); therefore, 18 cases progressed to the MVA (Figure 4A). 77.8% (14/18) of the cases showed expression of hsp18 (Figure 4B) while 38.9% (7/18) showed expression of esxA (Figure 4C). Expression of both transcripts was detected in 50% (7/14) of the MVA(+) samples, but 50% (7/14) expressed only hsp18, yielding an overall viability detection rate of 77.8% (14/18) in the new cases (Figure 4D). There were no samples in the AHRI-Ethiopian cohort that expressed only esxA.
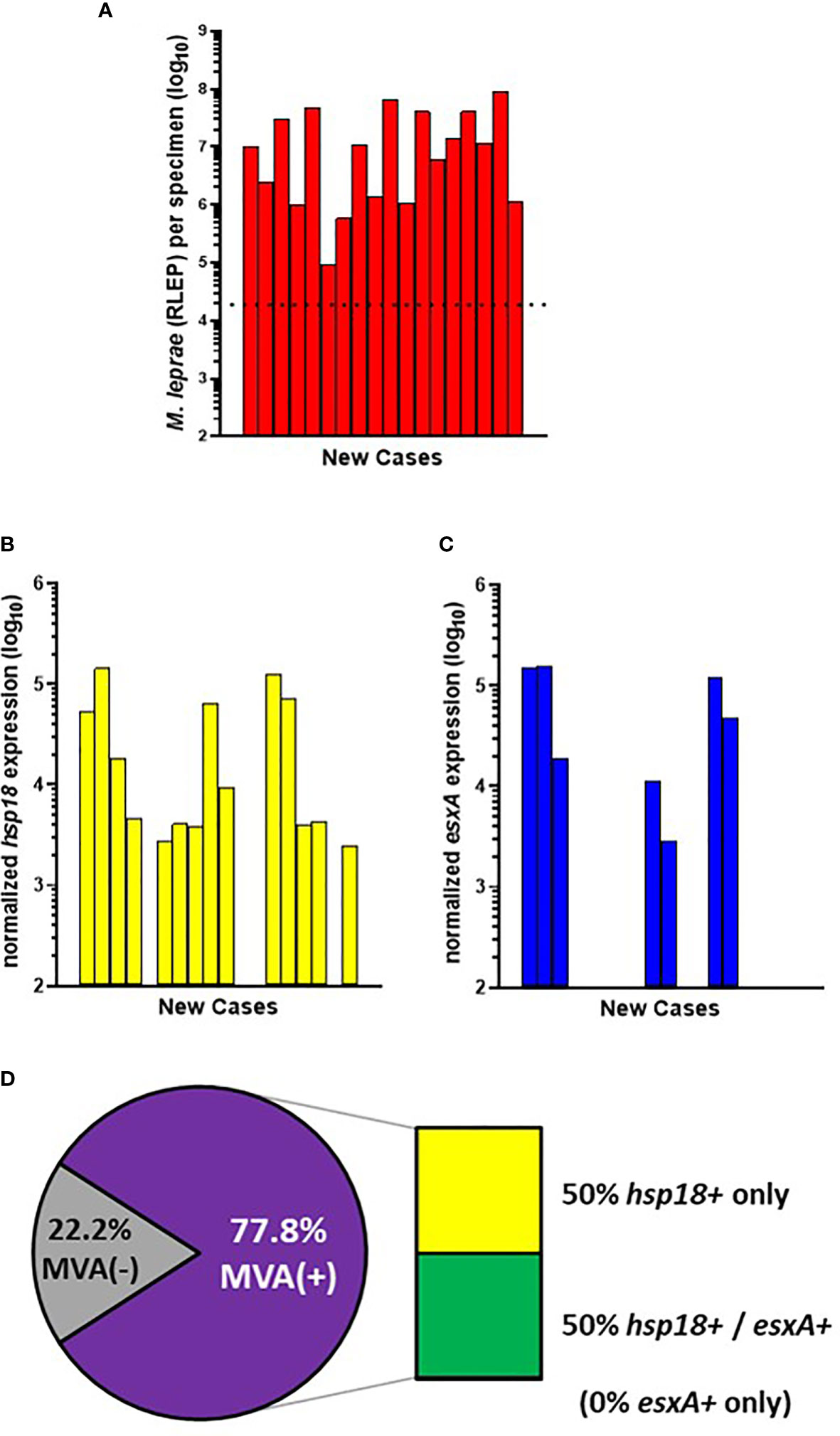
Figure 4 M. leprae enumeration and viability in new cases from AHRI-Ethiopia. DNA and RNA were extracted from biopsy specimens. M. leprae were enumerated (A) by RLEP PCR on the DNA fraction. The dotted line represents the minimum number of M. leprae that must be present in a sample to progress to the viability step. Based on the RLEP count, each DNased RNA was normalized to 3 x 103 M. leprae equivalents and reverse transcribed. Viability was determined on the resulting cDNA by hsp18 (B) and esxA (C) PCR. (D) The percentage of MVA(+) samples (expression of one or both transcripts) and MVA(-) samples (no expression of either transcript), and the percentage of MVA(+) samples expressing each transcript.
AH-Nepal new cases
Of the 200 specimens from AH-Nepal, 103 progressed to the MVA. Of these, 60 were new cases (Figure 5A). 73.3% (44/60) of the new cases showed expression of hsp18 (Figure 5B) while 35.0% (21/60) showed expression of esxA (Figure 5C). Of the total MVA(+) cases, expression of both transcripts was detected in 44.5% (20/45), while only hsp18 was detected in 53.3% (24/45) and only esxA was detected in 2.2% (1/45) cases. This yielded an overall viability detection rate of 75.0% (45/60) in the AH-Nepal new cases (Figure 5D).
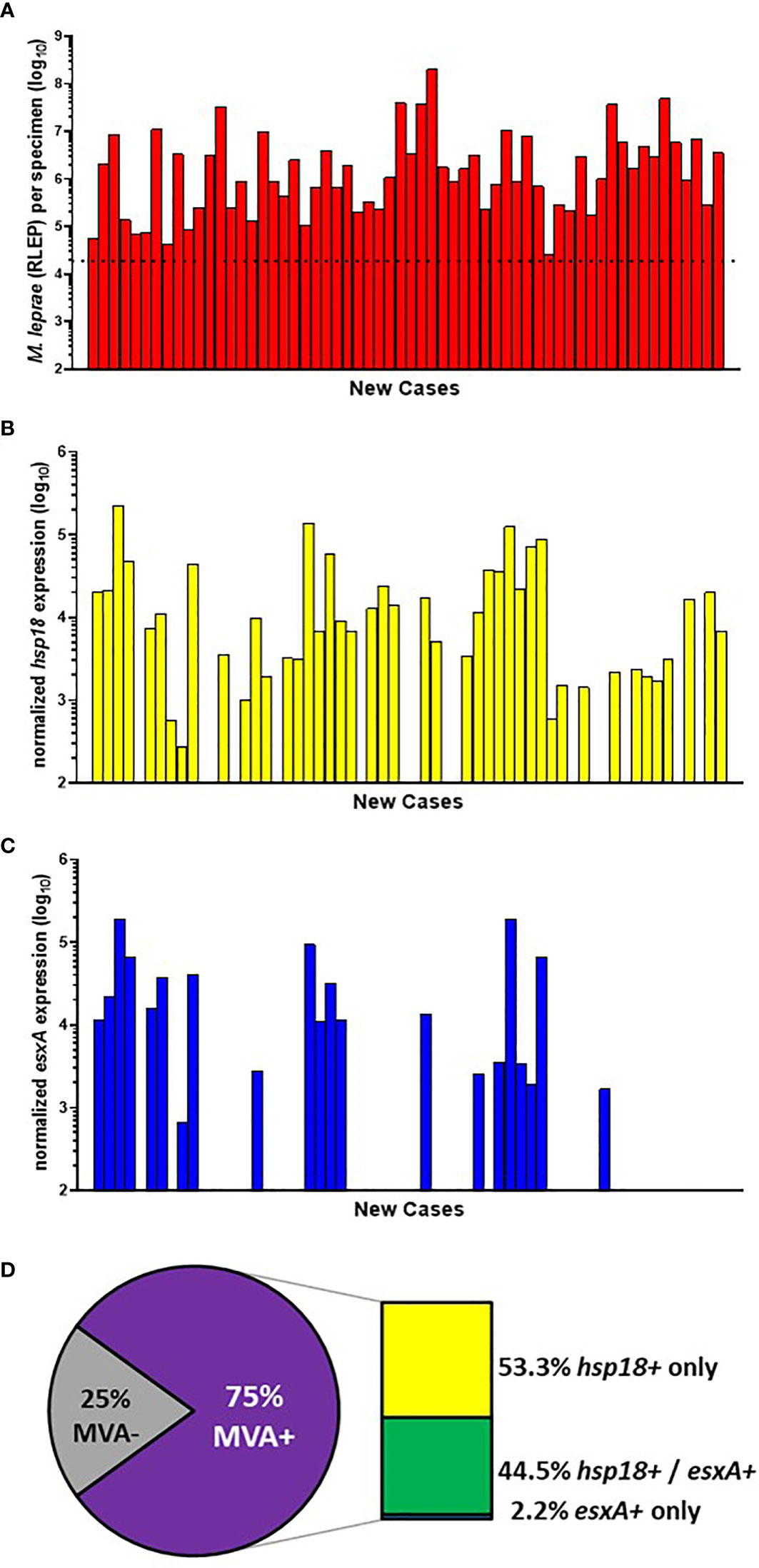
Figure 5 M. leprae enumeration and viability in new cases from AH-Nepal. DNA and RNA were extracted from biopsy specimens. M. leprae were enumerated (A) by RLEP PCR on the DNA fraction. The dotted line represents the minimum number of M. leprae that must be present in a sample to progress to the viability step. Based on the RLEP count, each DNased RNA was normalized to 3 x 103 M. leprae equivalents and reverse transcribed. Viability was determined on the resulting cDNA by hsp18 (B) and esxA (C) PCR. (D) The percentage of MVA(+) samples (expression of one or both transcripts) and MVA(-) samples (no expression of either transcript), and the percentage of MVA(+) samples expressing each transcript.
AH-Nepal relapse cases
29 of the AH-Nepal cases tested were relapse cases (Figure 6A). 55.2% (16/29) of these cases showed expression of hsp18 (Figure 6B) and 20.7% (6/29) showed expression of esxA (Figure 6C). Overall (Figure 6D), there was a 58.6% (17/29) detection of viable M. leprae in the relapse cases. Of these MVA(+) cases, 64.7% (11/17) expressed only hsp18, 29.4% (5/17) expressed both transcripts, and 5.9% (1/17) expressed only esxA.
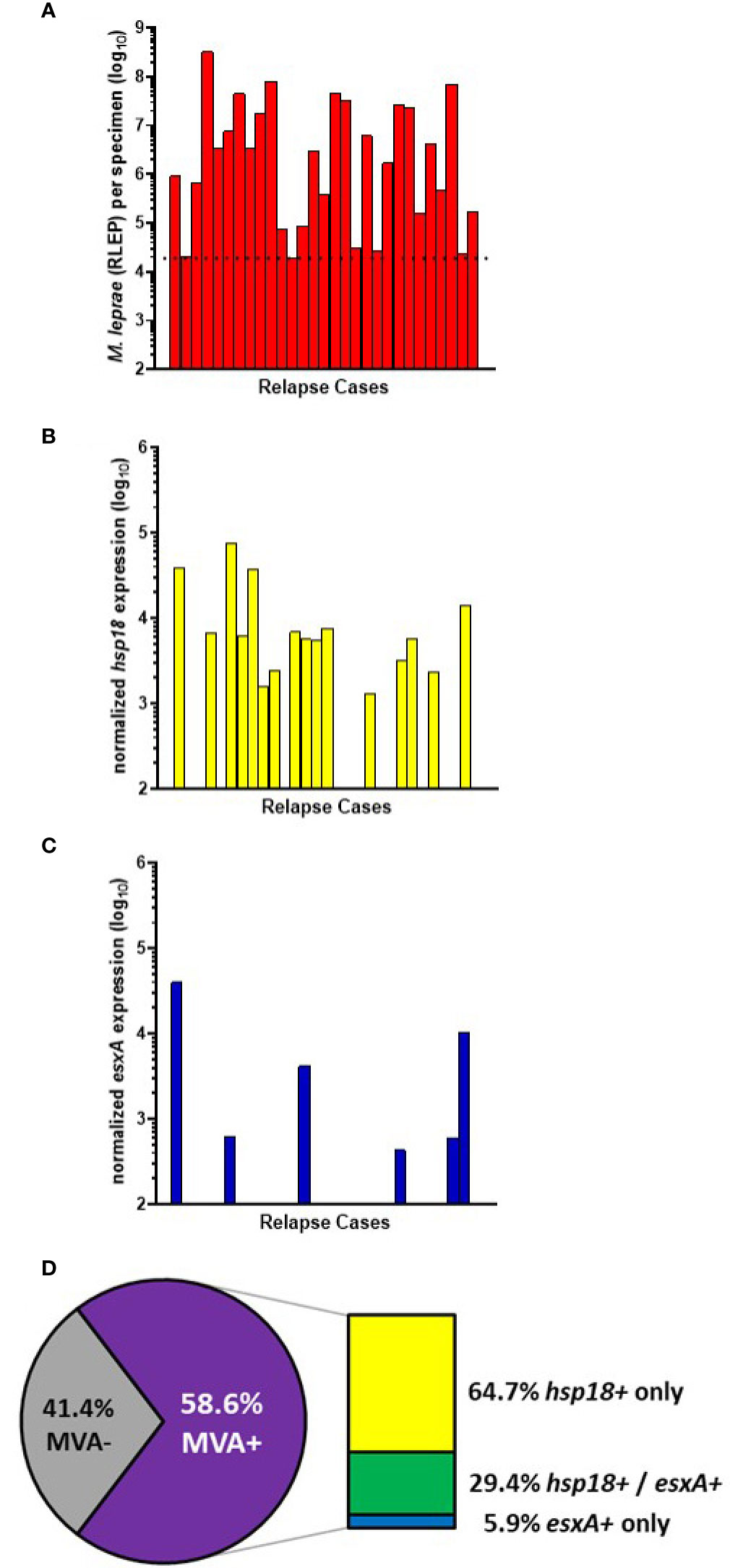
Figure 6 M. leprae enumeration and viability in relapse cases from AH-Nepal. DNA and RNA were extracted from biopsy specimens. M. leprae were enumerated (A) by RLEP PCR on the DNA fraction. The dotted line represents the minimum number of M. leprae that must be present in a sample to progress to the viability step. Based on the RLEP count, each DNased RNA was normalized to 3 x 103 M. leprae equivalents and reverse transcribed. Viability was determined on the resulting cDNA by hsp18 (B) and esxA (C) PCR. (D) The percentage of MVA(+) samples (expression of one or both transcripts) and MVA(-) samples (no expression of either transcript), and the percentage of MVA(+) samples expressing each transcript.
MVA versus MFP
MFP were inoculated with M. leprae derived from biopsy material from 54 of the 60 AH-Nepal new cases and 27 of the 29 AH-Nepal relapse cases. Biopsies for the MFP assay were collected before commencement of treatment, but not necessarily at the same time as the MVA biopsy collection or from the same lesion. Regarding the 54 new cases (Figures 7A, B), 79.6% (43/54) were MVA(+) and 63.0% (34/54) exhibited growth in the mouse footpad (MFP[+]). Thirty cases were both MVA(+) and MFP(+). Seven cases exhibited no viability in the molecular assay (MVA[-]) and no growth in the footpads (MFP[-]). This yielded an overall agreement between the MVA and the MFP of 68.5% (p = 0.077). There were 4 cases that were MVA (–) but MFP(+), whereas there were 13 cases that were MVA(+) but MFP(-). Regarding the 27 relapse cases, (Figures 7C, D), 63.0% (17/27) were MVA(+) and 51.9% (14/27) were MFP(+). Twelve of these cases were both MVA(+) and MFP(+), while eight cases were MVA(-) and MFP(-) negative for an overall agreement of 74.1% (p = 0.018). There were 2 cases that were MVA(-) but MFP(+) and 5 cases that were MVA(+) but MFP(-).
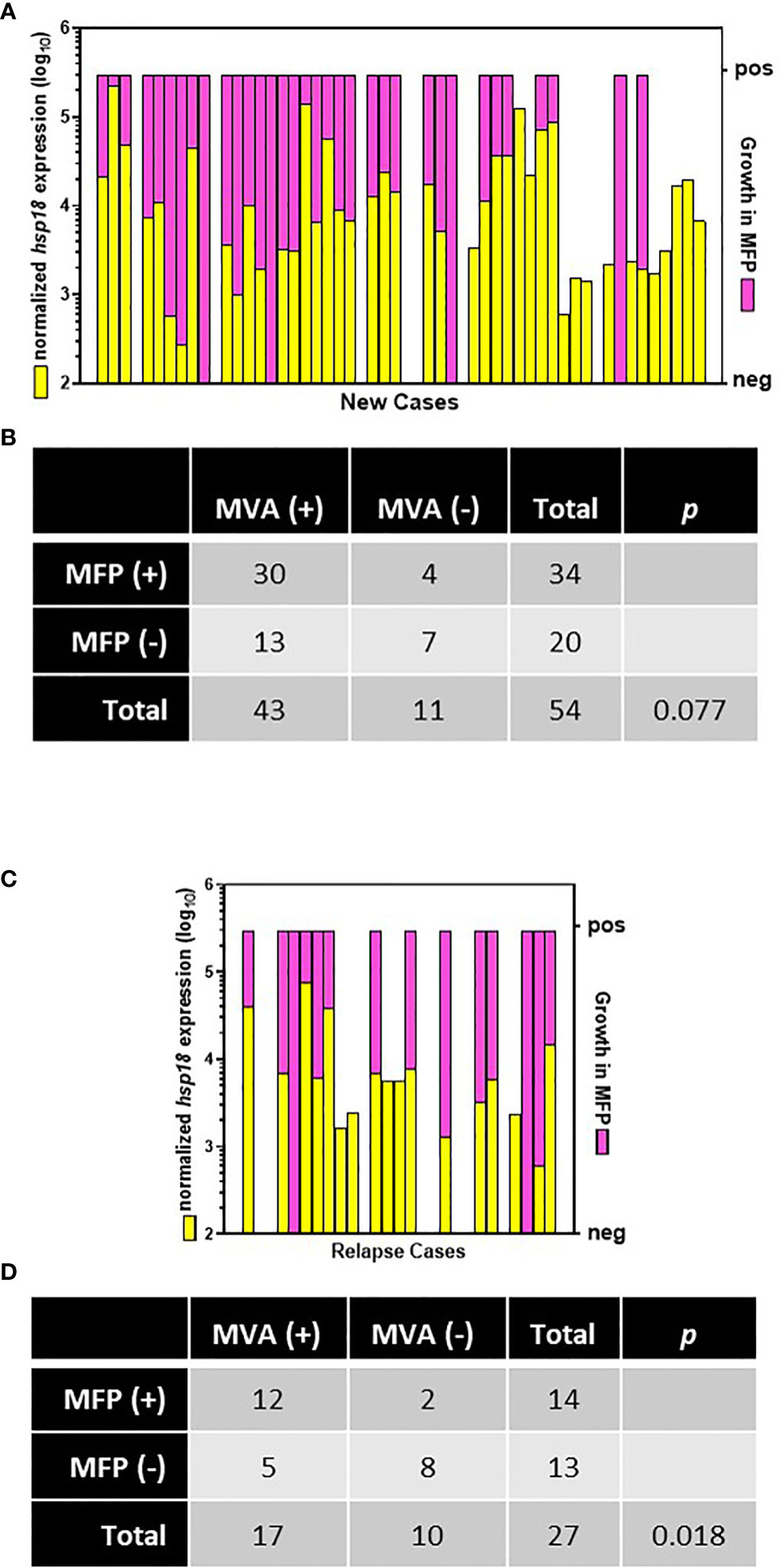
Figure 7 Comparison of MVA and MFP assay. Biopsies from new (A) and relapse (C) cases from AH-Nepal were tested for normalized expression of hsp18 (yellow bars) in the MVA and growth in MFP (pink bars). Multiplication of M. leprae in footpads to ≥ 1 x 105 bacilli was scored as growth (pos). Recovery of < 1 x 105 bacilli was scored as no growth (neg). The association between the MVA and the MFP assay in new (B) and relapse (D) cases was analyzed using Fisher’s Exact test.
AH-Nepal cases currently on MDT
Ten of the AH-Nepal cases were currently undergoing treatment with leprosy MDT. Four of these cases had received one month of MDT, two cases had received three months of MDT, and four cases had received ≥ 9 months of MDT treatment (Figure 8A). Again, there was a positive correlation between the average SSS BI and the number of M. leprae per biopsy sample (r[8] = 0.708, p = 0.019). Duration of treatment had no effect on either M. leprae counts (R2 = 0.001, F (1,8) = 0.008, p = 0.93) or average SSS BI (R2 = 0.04, F (1,8) = 0.3, p = 0.601). However, as shown in Figure 8B, MDT treatment had a significant effect on M. leprae viability as measured by the MVA (R2 = 0.81, F (1,8) = 35.02, p < 0.001). Viable M. leprae were detected in all 4 cases that had received only one month of MDT, in one of two cases that had received 3 months of MDT, but in none of the cases treated with ≥ 9 months of MDT.
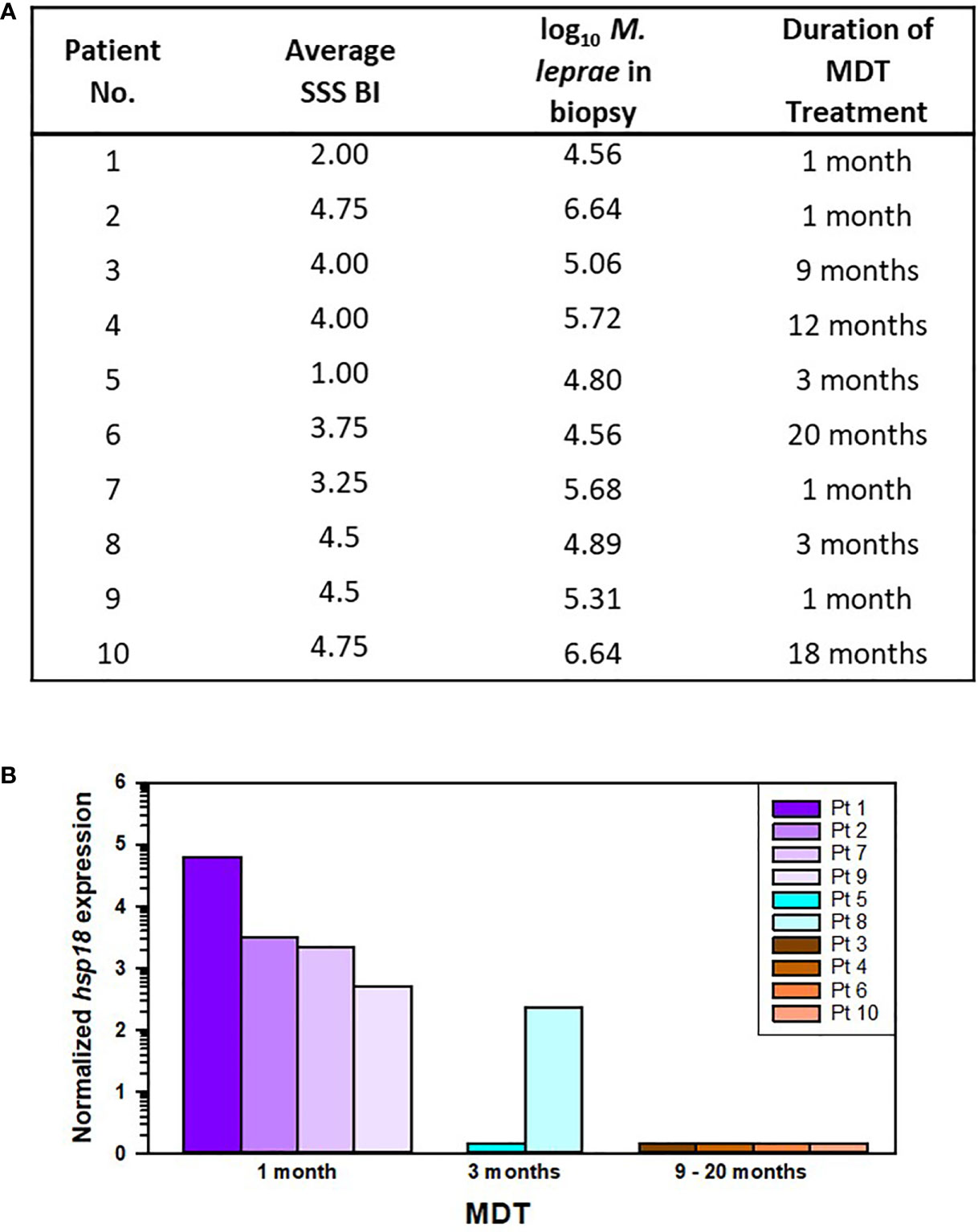
Figure 8 M. leprae enumeration and viability in patients from AH-Nepal currently on leprosy MDT. (A) The table depicts the average SSS BI, number of M. leprae (log10) in the biopsy specimen used for MVA, and the duration of leprosy MDT. (B) The graph shows the normalized hsp18 expression in patients treated with MDT for 1 month (purple bars), 3 months (blue bars), and > 9 months (brown bars).
Discussion
Leprosy is primarily a clinical diagnosis that is based on signs and symptoms such as loss of sensation in a pale or reddish lesion, peripheral nerve enlargement with loss of sensation or muscle weakness, and/or the presence of acid-fast bacilli in a SSS (17–19). Histopathological examination of lesion biopsies and bacteriological analysis of SSS are used for support and confirmation as they correlate well with clinical diagnosis (20–25). Histopathology enables the confirmation of acid-fast bacilli in a nerve and the classification of disease in the Ridley-Jopling spectrum (3). The bacterial load in a patient is generally estimated by counting the number of acid-fast bacilli present in SSS obtained from 3 - 6 sites and calculating the average BI. The BI is a semi-logarithmic scale that ranges from 0 (no bacilli in 100 oil immersion fields) to 6 (1000 bacilli in 1 oil immersion field) (13). Although it is acknowledged to have limited sensitivity, the SSS is often times the primary metric used to guide treatment and monitor patient responses to drug therapy. Currently, the presence of bacilli in a SSS classifies a patient as MB, regardless of the number of lesions (17).
PCR tests have also been investigated by numerous researchers for use as diagnostic tools (26). The repetitive sequence, RLEP (27), is a highly sensitive and specific marker for M. leprae, and is superior to many other markers (28–30). Moreover, most PCR tests correlate well with clinical diagnosis, SSS, and histopathology (31–34). As PCR tests become more standardized (35, 36) and affordable, this tool will likely become routine for the diagnosis of leprosy, especially for paucibacillary cases. It is important to note, however, that RLEP PCR only detects M. leprae DNA and does not differentiate between live and dead bacilli.
In the study presented here, RLEP PCR detected M. leprae DNA in 97% (426/439) of the biopsies overall, including 80.3% (53/66) from cases with an average SSS BI < 1. This underscores the exquisite sensitivity of RLEP PCR for use as a diagnostic tool. However, the primary purpose of running RLEP PCR in this study was to enumerate M. leprae in the biopsy specimen sent for viability testing in the MVA. A tissue sample must meet a specific threshold of bacilli present based on RLEP molecular enumeration on the DNA fraction (2 x 104 M. leprae per specimen) in order to normalize the RNA fraction for reverse transcription (8). Since PCR is not routinely used in most laboratories to diagnose leprosy or to estimate bacterial load in a patient, we first determined if there was a correlation between the average SSS BI calculated from samples collected from 3 - 6 index sites with the number of M. leprae recovered from the leprosy lesion biopsy sent for MVA. Our data showed a significant positive correlation between these two variables in all three cohorts. More importantly, we found that an average SSS BI of ≥ 2 is predictive of recovering sufficient M. leprae to run the MVA. Ultimately, it is important that a viability assay be sensitive so that it can be applicable to as many patients as possible, including those with low BI. On the other hand, for MVA testing one should avoid taking a biopsy from a patient that would not likely yield enough M. leprae for the assay. Therefore, this value will serve as a guide to predict the likelihood of a successful viability determination and reduce the number of unnecessary biopsies performed.
An ideal molecular viability indicator should be highly expressed by viable M. leprae but rapidly degraded once the bacteria are no longer viable. Several gene transcripts have been evaluated for expression in M. leprae-infected tissues as determinants of bacterial viability, including hsp18, hsp71, 16S, SodA, and esxA (7, 29, 37–44). Transcriptomics analyses of M. leprae are also contributing to the search for better transcripts (45, 46). We currently use the expression of hsp18 and esxA transcripts for viability assessment. hsp18 belongs to the small heat-shock family of proteins and encodes the M. leprae-specific 18kd protein. This protein is expressed by M. leprae in macrophages and functions in intracellular survival and growth (47). It is a molecular chaperone and has been shown to bind ATP, prevent thermal inactivation of enzymes, and offer protection from aggregation and killing (48). esxA codes for ESAT-6, the early secreted antigenic target-6. It is a member of the type VII ESX secretion and virulence system in mycobacteria (49). It may play a role in cell adherence, invasion, phagosomal rupture (50) and translocation of the mycobacteria from the phagosome to the cytosol (51, 52). Both hsp18 and esxA transcripts are highly expressed by viable M. leprae in the mouse model (7, 8, 45, 46). Interestingly, in the current study we saw some variance among the different cohorts in the expression of these transcripts. Overall, hsp18 was expressed by 72.7% ± 5.5% of the new cases across the three cohorts. In contrast, esxA was expressed by nearly 60% of the CSC-Philippines new cases but < 40% of the AHRI-Ethiopia and AH-Nepal cases. The differential expression of esxA among these diverse population groups is intriguing and worthy of further study.
When new leprosy cases were examined for M. leprae viability in the lesion biopsies, there was a MVA-positivity rate of 76.1 ± 1.5% (Figures 2D, 4D, and 5D). The precision of this data across the three diverse population groups is striking and supports the robustness of the MVA. The current gold standard for determining M. leprae viability in biopsy material is the MFP assay. Prior studies across decades of investigation have shown a recovery of viable M. leprae in the MFP assay of 48% - 87% when inoculated with MB biopsy-derived M. leprae (53–59). Our current MVA data falls toward the upper end of this range. This lends support to the value of the MVA for human biopsies. It must be acknowledged that 23.9 ± 1.5% of biopsies from untreated patients, even some with a high number of bacilli, were MVA(-). These results could be due to the age of the lesions. In nude MFP, M. leprae show a sigmoidal growth curve and eventually reach a stationary phase. Although bacterial numbers increase over several months, a higher percentage of metabolically active M. leprae are obtained early in infection compared to those derived from older, larger footpads (60, 61), emphasizing that a high number of M. leprae does not necessarily indicate viability. As with the MFP assay, it is important to take steps to optimize detection of viable M. leprae in the MVA, such as selecting an active lesion, taking the biopsy sample from the leading edge of the lesion, and immediately fixing the samples for preservation of nucleic acids.
Regarding the MFP data from AH-Nepal patients in the current study, it was somewhat serendipitous that these data were available. Comparing the MVA data to this MFP data, especially when the biopsy samples were collected at different times and likely from different lesions, is an extremely high standard to set for the MVA. Nevertheless, there was a 68.5% agreement between the MVA and MFP assay in new patients and a 74.1% agreement in relapse patients. While the correlation between the MVA and the MFP assay did not reach significance in the new patients (p = 0.077), it was highly significant among relapse patients (p = 0.018). A possible explanation for this difference may be that a relapse case is often diagnosed earlier than a new case of leprosy as a former patient will recognize the symptoms and will have been encouraged to seek diagnosis and treatment without delay if symptoms return (59). A new lesion in a relapse case likely contains metabolically active viable M. leprae that can be detected by both assays. Interestingly, for both new cases and relapse cases the main discrepancy between the two assays was that the MVA detected viable M. leprae in more specimens than the MFP assay, i.e MVA(+) and MFP(-). This would suggest that the MVA is more sensitive than the MFP assay. Alternatively, one could argue that the difference may result from “false positives” in the MVA. We have evidence (unpublished data) that hsp18 and esxA transcripts can be detected in M. leprae-infected mouse footpads 1 day after drug treatment, but detection is negligible within 4 weeks. This indicates that once bacilli are killed the transcripts are degraded in < 1 month, but there may be a short window of low-level, non-degraded transcript detection. Likewise, samples that were MVA(-) and MFP(+) contained M. leprae populations with an extremely low percent viability. Theoretically, one viable M. leprae can grow up in the MFP (62) and this may have been missed by the MVA, which can detect ~30 viable bacilli in a tissue specimen (8). Lastly, some biopsies from untreated patients were negative in both assays, despite the extensive experience of the respective laboratories in biopsy collection, MFP assay, and MVA. Running both the MVA and the MFP assay on the same biopsy specimen, while difficult, could resolve these issues.
Finally, our data show that the MVA can be used to monitor killing of M. leprae following drug treatment in patients. The long-term tissue persistence of dead M. leprae in tissues after completion of MDT can be troubling for physicians, raising suspicions of non-compliance or drug resistance. In this study, the duration of MDT treatment had no effect on either the average SSS BI or the number of bacilli in the biopsy. This data corroborates the extremely slow clearance of bacilli from the lesions and emphasizes that decreasing bacterial counts is a tediously prolonged and inadequate measure of drug efficacy. However, there was a significant decrease in M. leprae viability, as measured by the MVA, with duration of treatment. Although the number of cases presented here was small, this data supports the need for further studies on the use of a viability assay to monitor drug efficacy and perhaps even enabling the shortening of therapy or development of more personalized therapy, especially in cases involving adverse drug effects.
In summary, we have utilized a MVA that ascertains M. leprae viability by measuring the normalized mRNA expression levels of the bacterial genes, esxA and hsp18, and applied this assay to 3 cohorts of clinical cases. Despite the diversity of these cohorts, there was excellent agreement in the detection of viable M. leprae from biopsy specimens. Nevertheless, one may question if there is a need for a M. leprae viability assay in clinical medicine as new cases of leprosy will continue to be diagnosed clinically and treated with the recommended leprosy therapy. The use of a rapid molecular viability assay could be warranted, however, under certain circumstances. First, treatment efficacy could be verified following drug therapy, especially as new shorter drug regimens may be approved. In addition, detection of viable M. leprae following completion of therapy would justify drug susceptibility testing and could contribute to transmission studies. Second, such an assay would likely be useful to confirm relapse cases. Moreover, as molecular epidemiological (63) and immunological (64, 65) assays become more discerning, determination of M. leprae viability together with these assays may help differentiate relapse from reinfection and relapse from reaction. Finally, a rapid molecular viability test would greatly aid clinical trials for new leprosy drugs by replacing the time-intensive requirement for MFP passage of biopsy material from treated patients. Our MVA is specific for M. leprae, sensitive to an average SSS BI of ≥ 2, can be performed in < 4 days, is field-friendly with regard to fixation, storage, and transport of tissue specimens, and is the only assay validated with known viable M. leprae bacteria.
Author contributions
All authors listed have made a substantial, direct, and intellectual contribution to the work and approved it for publication.
Funding
This work was supported by funding from the Leprosy Research Initiative (LRI) and the Turing Foundation under LRI Grant No. 703.15.43.
Acknowledgments
The authors wish to thank Dr Mahesh Shah, Mr Kapil Neupane, Ms Jeni Maharjan, and Mr. Nischal Pokhrel.
Conflict of interest
Author SL was employed by company IHRC, Inc.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Author disclaimer
The views expressed in this article are solely the opinions of the authors and do not necessarily reflect the official policies of the U. S. Department of Health and Human Services or the Health Resources and Services Administration, nor does mention of the department or agency names imply endorsement by the U.S. Government.
References
1. White C, Franco-Paredes C. Leprosy in the 21st century. Clin Microbiol Rev (2015) 28(1):80–94. doi: 10.1128/CMR.00079-13
2. Deps P, Collin SM. Mycobacterium lepromatosis as a second agent of Hansen's Disease. Front Microbiol (2021) 12:698588. doi: 10.3389/fmicb.2021.698588
3. Scollard DM. Pathogenesis and pathology of leprosy, chapter 2.4. In: Scollard DM, Gillis TP, editors. International textbook of leprosy. Greenville: American Leprosy Missions (2016). Available at: www.internationaltextbookofleprosy.org.
4. McRae DH, Shepard CC. Relationship between the staining quality of Mycobacterium leprae and infectivity for mice. Infect Immun (1971) 3(1):116–20. doi: 10.1128/iai.3.1.116-120.1971
5. Brennan PJ, Spencer JS. The physiology of Mycobacterium leprae, chapter 5.1. In: Scollard DM, Gillis TP, editors. International textbook of leprosy. Greenville: American Leprosy Missions (2019). Available at: www.internationaltextbookofleprosy.org.
6. Lahiri R, Adams LB. Cultivation and viability determination of Mycobacterium leprae, chapter 5.3. In: Scollard DM, Gillis TP, editors. International textbook of leprosy. Greenville: American Leprosy Missions (2016). Available at: www.internationaltextbookofleprosy.org/.
7. Davis GL, Ray NA, Lahiri R, Gillis TP, Krahenbuhl JL, Williams DL, et al. Molecular assays for determining Mycobacterium leprae viability in tissues of experimentally infected mice. PloS Negl Trop Dis (2013) 7(8):e2404. doi: 10.1371/journal.pntd.0002404
8. Collins JH, Lenz SM, Ray NA, Balagon MF, Hagge DA, Lahiri R, et al. A sensitive and quantitative assay to enumerate and measure Mycobacterium leprae viability in clinical and experimental specimens. Curr Protoc (2022) 2(2):e359. doi: 10.1002/cpz1.359
9. Hagge DA, Scollard DM, Ray NA, Marks VT, Deming AT, Spencer JS, et al. IL-10 and NOS2 modulate antigen-specific reactivity and nerve infiltration by T cells in experimental leprosy. PloS Negl Trop Dis (2014) 8(9):e3149. doi: 10.1371/journal.pntd.0003149
10. Lahiri R, Adams LB, Thomas SS, Pethe K. Sensitivity of Mycobacterium leprae to telacebec. Emerg Infect Dis (2022) 28(3):749–51. doi: 10.3201/eid2803.210394
11. Tongluan N, Shelton LT, Collins JH, Ingraffia P, McCormick G, Pena M, et al. Mycobacterium leprae infection in ticks and tick-derived cells. Front Microbiol (2021) 12:761420. doi: 10.3389/fmicb.2021.761420
12. Balagon MF, Cellona RV, Cruz E, Burgos JA, Abalos RM, Walsh GP, et al. Long-term relapse risk of multibacillary leprosy after completion of 2 years of multiple drug therapy (WHO-MDT) in cebu, Philippines. Am J Trop Med Hyg (2009) 81(5):895–9. doi: 10.4269/ajtmh.2009.09-0189
13. Ridley DS. Therapeutic trials in leprosy using serial biopsies. Lepr Rev (1958) 29(1):45–52. doi: 10.5935/0305-7518.19580004
14. Ridley DS, Jopling WH. Classification of leprosy according to immunity. a five-group system. Int J Lepr Other Mycobact Dis (1966) 34(3):255–73.
15. Sapkota BR, Ranjit C, Neupane KD, Macdonald M. Development and evaluation of a novel multiple-primer PCR amplification refractory mutation system for the rapid detection of mutations conferring rifampicin resistance in codon 425 of the rpoB gene of Mycobacterium leprae. J Med Microbiol (2008) 57(Pt 2):179–84. doi: 10.1099/jmm.0.47534-0
16. Shepard CC, McRae DH. A method for counting acid-fast bacteria. Int J Lepr Other Mycobact Dis (1968) 36(1):78–82.
17. WHO Bulletin. Guidelines for the diagnosis, treatment and prevention of leprosy. New Delhi: World Health Organization, Regional Office for South-East Asia (2017).
18. Kumar D, Uprety S, Dogra S. Clinical diagnosis of leprosy, chapter 2.1. In: Scollard DM, Gillis TP, editors. International textbook of leprosy. Greenville: American Leprosy Missions (2017). Available at: www.internationaltextbookofleprosy.org.
19. Palit A, Ragunatha S, Inamadar A, Girdhar B. History taking and methods of clinical and nerve examination. In: Kumar B, Narang T, Dongre V, Samanta S, editors. IAL handbook of leprosy, 1st ed, Mumbai: Bhalani Publishing House (2018). p. 60–82.
20. Ponnighaus JM, Lienhardt C, Lucas S, Fine PE, Sterne JA. Comparison of bacillary indexes in slit-skin smears, skin and nerve biopsies; a study from Malawi. Int J Lepr Other Mycobact Dis (1997) 65(2):211–6.
21. Giridhar M, Arora G, Lajpal K, Singh Chahal K. Clinicohistopathological concordance in leprosy - a clinical, histopathological and bacteriological study of 100 cases. Indian J Lepr (2012) 84(3):217–25.
22. Kumaran SM, Bhat IP, Madhukara J, Rout P, Elizabeth J. Comparison of bacillary index on slit skin smear with bacillary index of granuloma in leprosy and its relevance to present therapeutic regimens. Indian J Dermatol (2015) 60(1):51–4. doi: 10.4103/0019-5154.147791
23. Kakkad K, Padhi T, Pradhan K, Agrawal KC. A study of clinical, bacteriological & histopathological correlation in leprosy cases attending a government medical college in Western Odisha: Some observations. Indian J Lepr (2016) 88(2):97–103.
24. Premalatha P, Renuka IV, Meghana A, Devi SI, Charyulu P, Sampoorna G. Utility of bacillary index in slit skin smears in correlation with clinical and histopathological alterations in Hansen's Disease: An attempt to revive a simple useful procedure. Ann Med Health Sci Res (2016) 6(3):181–4. doi: 10.4103/2141-9248.183936
25. Naik SM, More SA, Joshi SR. Correlation of histomorphological findings with bacteriological index in leprosy patients. Iran J Pathol (2022) 17(1):48–55. doi: 10.30699/ijp.2021.534122.2682
26. Martinez AN, Talhari C, Moraes MO, Talhari S. PCR-based techniques for leprosy diagnosis: from the laboratory to the clinic. PloS Negl Trop Dis (2014) 8(4):e2655. doi: 10.1371/journal.pntd.0002655
27. Woods SA, Cole ST. A family of dispersed repeats in Mycobacterium leprae. Mol Microbiol (1990) 4(10):1745–51. doi: 10.1111/j.1365-2958.1990.tb00552.x
28. Truman RW, Andrews PK, Robbins NY, Adams LB, Krahenbuhl JL, Gillis TP. Enumeration of Mycobacterium leprae using real-time PCR. PloS Negl Trop Dis (2008) 2(11):e328. doi: 10.1371/journal.pntd.0000328
29. Turankar RP, Pandey S, Lavania M, Singh I, Nigam A, Darlong J, et al. Comparative evaluation of PCR amplification of RLEP, 16S rRNA, rpoT and sod a gene targets for detection of M. leprae DNA from clinical and environmental samples. Int J Mycobacteriol (2015) 4(1):54–9. doi: 10.1016/j.ijmyco.2014.11.062
30. Braet S, Vandelannoote K, Meehan CJ, Brum Fontes AN, Hasker E, Rosa PS, et al. The repetitive element RLEP is a highly specific target for detection of Mycobacterium leprae. J Clin Microbiol (2018) 56(3):e01924–17. doi: 10.1128/JCM.01924-17
31. Yoon KH, Cho SN, Lee MK, Abalos RM, Cellona RV, Fajardo TT Jr., et al. Evaluation of polymerase chain reaction amplification of Mycobacterium leprae-specific repetitive sequence in biopsy specimens from leprosy patients. J Clin Microbiol (1993) 31(4):895–9. doi: 10.1128/jcm.31.4.895-899.1993
32. Banerjee S, Biswas N, Kanti Das N, Sil A, Ghosh P, Hasanoor Raja AH, et al. Diagnosing leprosy: revisiting the role of the slit-skin smear with critical analysis of the applicability of polymerase chain reaction in diagnosis. Int J Dermatol (2011) 50(12):1522–7. doi: 10.1111/j.1365-4632.2011.04994.x
33. Siwakoti S, Rai K, Bhattarai NR, Agarwal S, Khanal B. Evaluation of polymerase chain reaction (PCR) with slit skin smear examination (SSS) to confirm clinical diagnosis of leprosy in Eastern Nepal. PloS Negl Trop Dis (2016) 10(12):e0005220. doi: 10.1371/journal.pntd.0005220
34. Azevedo MC, Ramuno NM, Fachin LR, Tassa M, Rosa PS, Belone AF, et al. qPCR detection of Mycobacterium leprae in biopsies and slit skin smear of different leprosy clinical forms. Braz J Infect Dis (2017) 21(1):71–8. doi: 10.1016/j.bjid.2016.09.017
35. Sharma R, Singh P, McCoy RC, Lenz SM, Donovan K, Ochoa MT, et al. Isolation of Mycobacterium lepromatosis and development of molecular diagnostic assays to distinguish Mycobacterium leprae and M. lepromatosis. Clin Infect Dis (2020) 71(8):e262–e9. doi: 10.1093/cid/ciz1121
36. Manta FSN, Jacomasso T, Rampazzo RCP, Moreira SJM, Zahra NM, Cole ST, et al. Development and validation of a multiplex real-time qPCR assay using GMP-grade reagents for leprosy diagnosis. PloS Negl Trop Dis (2022) 16(2):e0009850. doi: 10.1371/journal.pntd.0009850
37. Patel BK, Banerjee DK, Butcher PD. Determination of Mycobacterium leprae viability by polymerase chain reaction amplification of 71-kDa heat-shock protein mRNA. J Infect Dis (1993) 168(3):799–800. doi: 10.1093/infdis/168.3.799
38. Kurabachew M, Wondimu A, Ryon JJ. Reverse transcription-PCR detection of Mycobacterium leprae in clinical specimens. J Clin Microbiol (1998) 36(5):1352–6. doi: 10.1128/JCM.36.5.1352-1356.1998
39. Chae GT, Kim MJ, Kang TJ, Lee SB, Shin HK, Kim JP, et al. DNA-PCR and RT-PCR for the 18-kDa gene of Mycobacterium leprae to assess the efficacy of multi-drug therapy for leprosy. J Med Microbiol (2002) 51(5):417–22. doi: 10.1099/0022-1317-51-5-417
40. Jadhav RS, Kamble RR, Shinde VS, Edward S, Edward VK. Use of reverse transcription polymerase chain reaction for the detection of Mycobacterium leprae in the slit-skin smears of leprosy patients. Indian J Lepr (2005) 77(2):116–27.
41. Hirawati, Katoch K, Chauhan DS, Singh HB, Sharma VD, Singh M, et al. Detection of M. leprae by reverse transcription- PCR in biopsy specimens from leprosy cases: a preliminary study. J Commun Dis (2006) 38(3):280–7.
42. Lini N, Shankernarayan NP, Dharmalingam K. Quantitative real-time PCR analysis of Mycobacterium leprae DNA and mRNA in human biopsy material from leprosy and reactional cases. J Med Microbiol (2009) 58(Pt 6):753–9. doi: 10.1099/jmm.0.007252-0
43. Sharma R, Lavania M, Katoch K, Chauhan DS, Gupta AK, Gupta UD, et al. Development and evaluation of real-time RT-PCR assay for quantitative estimation of viable Mycobacterium leprae in clinical samples. Indian J Lepr (2008) 80(4):315–21.
44. Martinez AN, Lahiri R, Pittman TL, Scollard D, Truman R, Moraes MO, et al. Molecular determination of Mycobacterium leprae viability by use of real-time PCR. J Clin Microbiol (2009) 47(7):2124–30. doi: 10.1128/JCM.00512-09
45. Williams DL, Torrero M, Wheeler PR, Truman RW, Yoder M, Morrison N, et al. Biological implications of Mycobacterium leprae gene expression during infection. J Mol Microbiol Biotechnol (2004) 8(1):58–72. doi: 10.1159/000082081
46. Ojo O, Williams DL, Adams LB, Lahiri R. Mycobacterium leprae transcriptome during in vivo growth and ex vivo stationary phases. Front Cell Infect Microbiol (2021) 11:817221. doi: 10.3389/fcimb.2021.817221
47. Dellagostin OA, Esposito G, Eales LJ, Dale JW, McFadden J. Activity of mycobacterial promoters during intracellular and extracellular growth. Microbiol (1995) 141(Pt 8):1785–92. doi: 10.1099/13500872-141-8-1785
48. Nandi SK, Panda AK, Chakraborty A, Rathee S, Roy I, Barik S, et al. Role of ATP-small heat shock protein interaction in human diseases. Front Mol Biosci (2022) 9:844826. doi: 10.3389/fmolb.2022.844826
49. Roltgen K, Pluschke G, Spencer JS, Brennan PJ, Avanzi C. The immunology of other mycobacteria: M. ulcerans, M. leprae. Semin Immunopathol (2020) 42(3):333–53. doi: 10.1007/s00281-020-00790-4
50. Bao Y, Wang L, Sun J. A small protein but with diverse roles: A review of esxA in Mycobacterium-host interaction. Cells (2021) 10(7):1645. doi: 10.3390/cells10071645
51. van der Wel N, Hava D, Houben D, Fluitsma D, van Zon M, Pierson J, et al. M. tuberculosis and M. leprae translocate from the phagolysosome to the cytosol in myeloid cells. Cell (2007) 129(7):1287–98. doi: 10.1016/j.cell.2007.05.059
52. van der Niet S, van Zon M, de Punder K, Grootemaat A, Rutten S, Moorlag S, et al. IL-1R1-dependent signals improve control of cytosolic virulent mycobacteria in vivo. mSphere (2021) 6(3):e00153–21. doi: 10.1128/mSphere.00153-21
53. Shepard CC. Multiplication of Mycobacterium leprae in the foot-pad of the mouse. Int J Lepr (1962) 30:291–306.
54. Levy L, Murray LP, Shepard CC. A comparative study of mouse foot pad inoculation of skin biopsy specimens from patients with lepromatous leprosy in San Francisco and Atlanta. Int J Lepr Other Mycobact Dis (1970) 38(1):54–9.
55. Gupta UD, Katoch K, Sharma RK, Singh HB, Natragan M, Singh D, et al. Analysis of quantitative relationship between viability determination in leprosy by MFP, ATP bioluminescence and gene amplification assay. Int J Lepr Other Mycobact Dis (2001) 69(4):328–34.
56. Wakade AV, Shetty VP. Isolation of Mycobacterium leprae from untreated borderline tuberculoid, mid-borderline and indeterminate cases using the mouse foot pad technique–a study of 209 cases. Lepr Rev (2006) 77(4):366–70. doi: 10.47276/lr.77.4.366
57. Agrawal VP, Shetty VP. Comparison of radiorespirometric budemeyer assay with ATP assay and mouse foot pad test in detecting viable Mycobacterium leprae from clinical samples. Indian J Med Microbiol (2007) 25(4):358–63. doi: 10.1016/S0255-0857(21)02051-X
58. Agrawal VP, Shetty VP. Comparison of the mouse foot pad test with a buddemeyer type radiorespirometric assay in detecting viable Mycobacterium leprae in human lesional biopsies. Indian J Dermatol Venereol Leprol (2007) 73(6):384–8. doi: 10.4103/0378-6323.37054
59. Rajan L, Das M, Ponnaiya J, Ebenezer M. Clinical profile of relapse in leprosy and laboratory correlation, including mouse footpad inoculation and molecular tests, in a tertiary leprosy institution. Lepr Rev (2021) 92:260–8. doi: 10.47276/lr.92.3.260
60. Truman RW, Krahenbuhl JL. Viable M. leprae as a research reagent. Int J Lepr Other Mycobact Dis (2001) 69(1):1–12.
61. Lahiri R, Randhawa B, Krahenbuhl JL. Effects of purification and fluorescent staining on viability of Mycobacterium leprae. Int J Lepr Other Mycobact Dis (2005) 73(3):194–202.
62. Welch TM, Gelber RH, Murray LP, Ng H, O'Neill SM, Levy L. Viability of Mycobacterium leprae after multiplication in mice. Infect Immun (1980) 30(2):325–8. doi: 10.1128/iai.30.2.325-328.1980
63. Avanzi C, Singh P, Truman RW, Suffys PN. Molecular epidemiology of leprosy: An update. Infect Genet Evol (2020) 86:104581. doi: 10.1016/j.meegid.2020.104581
64. Duthie MS, Roferos FO, Abellana JF, Taborada T, Sanchez R, Maghanoy A, et al. Utility and limitations of serodiagnostic tests in monitoring the response to treatment of leprosy patients. Diagn Microbiol Infect Dis (2020) 96(4):114984. doi: 10.1016/j.diagmicrobio.2019.114984
Keywords: Leprosy, Mycobacterium leprae (M. leprae), Mycobacterium leprae hsp18, leprosy (clinical), drug trial research, Mycobacterium leprae esxA, Molecular viability assay (MVA), Mouse footpad (MFP) assay
Citation: Lenz SM, Ray NA, Lema T, Collins JH, Thapa R, Girma S, Balagon M, Bobosha K, Hagge DA, Williams DL, Scollard DM, Lahiri R and Adams LB (2022) Utility of a Mycobacterium leprae molecular viability assay for clinical leprosy: An analysis of cases from the Philippines, Ethiopia, and Nepal. Front. Trop. Dis. 3:967351. doi: 10.3389/fitd.2022.967351
Received: 12 June 2022; Accepted: 21 July 2022;
Published: 24 August 2022.
Edited by:
Lucio Vera-Cabrera, Universidad Autonoma de Nuevo León, MexicoReviewed by:
Masanori Matsuoka, Retired, Tokyo, JapanMary Fafutis-Morris, University of Guadalajara, Mexico
Copyright © 2022 Lenz, Ray, Lema, Collins, Thapa, Girma, Balagon, Bobosha, Hagge, Williams, Scollard, Lahiri and Adams. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Linda B. Adams, ladams@hrsa.gov
†Present address: Shannon M. Lenz, MedPace, Inc., Cincinnati, OH, United States
 Shannon M. Lenz1†
Shannon M. Lenz1† J. Hunter Collins
J. Hunter Collins Ruby Thapa
Ruby Thapa Selfu Girma
Selfu Girma Deanna A. Hagge
Deanna A. Hagge David M. Scollard
David M. Scollard Ramanuj Lahiri
Ramanuj Lahiri Linda B. Adams
Linda B. Adams