- 1Radio-Oncology Department, School of Medicine, Shiraz University of Medical Sciences, Shiraz, Iran
- 2Department of Immunology, School of Medicine, Shiraz University of Medical Sciences, Shiraz, Iran
- 3Department of Bacteriology and Immunology, University of Helsinki and Diagnostic Center of the Helsinki University Hospital, Helsinki, Finland
- 4Autoimmune Diseases Research Center, Shiraz University of Medical Sciences, Shiraz, Iran
The current coronavirus pandemic (COVID-19), caused by SARS-CoV-2, has had devastating effects on the global health and economic system. The cellular and molecular mediators of both the innate and adaptive immune systems are critical in controlling SARS-CoV-2 infections. However, dysregulated inflammatory responses and imbalanced adaptive immunity may contribute to tissue destruction and pathogenesis of the disease. Important mechanisms in severe forms of COVID-19 include overproduction of inflammatory cytokines, impairment of type I IFN response, overactivation of neutrophils and macrophages, decreased frequencies of DC cells, NK cells and ILCs, complement activation, lymphopenia, Th1 and Treg hypoactivation, Th2 and Th17 hyperactivation, as well as decreased clonal diversity and dysregulated B lymphocyte function. Given the relationship between disease severity and an imbalanced immune system, scientists have been led to manipulate the immune system as a therapeutic approach. For example, anti-cytokine, cell, and IVIG therapies have received attention in the treatment of severe COVID-19. In this review, the role of immunity in the development and progression of COVID-19 is discussed, focusing on molecular and cellular aspects of the immune system in mild vs. severe forms of the disease. Moreover, some immune- based therapeutic approaches to COVID-19 are being investigated. Understanding key processes involved in the disease progression is critical in developing therapeutic agents and optimizing related strategies.
1 Background
The coronavirus disease 2019 (COVID-19), caused by the severe acute respiratory syndrome- coronavirus- 2 (SARS-CoV-2), is a viral infectious disease spread worldwide (1). The outbreak of COVID-19 has led to extensive global morbidity and mortality (2). The Coronaviridae family contains α and β coronaviruses capable of causing both mild and severe respiratory diseases in wild animals and humans. SARS-CoV, MERS-CoV and SARS-CoV-2 are positive- sense single-stranded RNA β-coronaviruses causing severe respiratory tract infections in humans (3, 4).
SARS-CoV-2 infection can have a variable clinical picture ranging from asymptomatic to severe. In about 5% of patients, there is a risk of death, while the overall mortality is about 1% (5). With increasing vaccine coverage the numbers are getting smaller. SARS-CoV-2 can enter and infect epithelial, and possibly endothelial cells expressing the receptor angiotensin- converting enzyme 2 receptor (ACE2) and, in certain cells, neuropilin-1 (6, 7).
During the initial exposure, the virus infects epithelial cells in the upper respiratory tract through the ACE2 receptor. At this stage, the innate and adaptive immunity try to inhibit the virus at the mucosal sites and prevent the progression of the disease by various mechanisms. These include the production of mucus and antiviral metabolites, interferons and other cytokines, as well as IgA, when there is preexisting immunity to the virus.
SARS-CoV-2 is able to inhibit innate and adaptive immune responses in various ways. It can replicate unchecked in the respiratory tract and eventually spread to the lungs (8, 9). COVID-19 patients may develop a systemic disease with extrapulmonary manifestations. Systemic manifestations during a later stage of COVID-19 are linked to lymphopenia, increased amounts of neutrophils, cytokine release syndrome (CRS) or cytokine storm, activation of complement and coagulation cascades and elevated levels of inflammatory chemokines and alarmins (10, 11). Several studies have shown that male gender, higher age, obesity, diabetes, hematological cancer, autoimmune diseases (e.g., psoriasis), hypertension (especially pulmonary arterial hypertension), and cardiovascular, cerebrovascular and chronic kidney diseases are risk factors closely related to the severity and outcomes of COVID-19 infection (12–16).
Although innate and adaptive immunities are linked, they both also have different cell types with varying functions (17). The innate immune system includes monocytes, macrophages, dendritic cells (DC), myeloid-derived suppressor cells (MDSC) and innate lymphoid cells (ILCs). ILCs can be divided into different groups: natural killer (NK) cells, 3 subsets of helper like ILCs (ILC1, 2 and 3), fetal lymphoid tissue- inducer (LTi) cells and a population of innate intraepithelial lymphocytes (IELs) in the intestine that express T cell markers (18). In addition, the innate response includes a number of molecules, e.g. the entire complement system with more than 40 different soluble or cell-bound components. Innate immune cells expressing pattern-recognition receptors (PRRs) can detect pathogen-associated molecular patterns (PAMPs) of invading viruses to develop and maintain innate immune responses by secreting interferons (IFNs) of type I (IFN-α and IFN-β) and type III (IFN-λ), as well as pro-inflammatory cytokines and chemokines (19, 20). The adaptive immune system comprises B cells, CD4+ T helper and CD8+ cytotoxic T cells. CD4+ T cells orchestrate the immune response by producing cytokines and providing help to other immune cells. CD8+ T cells kill virally infected cells, and B cells develop and produce antigen-specific IgM, IgG and IgA antibodies (21). Larger amounts of specific antibodies are produced by plasma cells that locate themselves into niches in the bone marrow.
Given that appropriate innate and adaptive immune responses are required to control and contain COVID-19 infection and prevent the disease in the long term, it is critical to recognize both types of responses to SARS-CoV-2. This review aims to shed light on the molecular and cellular players of the immune system in COVID-19 pathogenesis. Accordingly, the molecular and cellular compartments involved in the innate and adaptive immune responses to SARS-CoV-2 are thoroughly reviewed (A brief comparison of immune responses in mildly versus critically ill COVID-19 patients has been provided in Table 2). Special emphasis is on delineating differences that distinguish severe disease forms from those of milder forms. Why do certain individuals suffer from serious lung complications and systemic effects, and what is the contribution of the immune system itself in a vicious circle of inflammation in response to a viral infection?.
2 The relationship between the innate immune system and the progression of COVID-19
2.1 Neutrophils
SARS-CoV-2 infection is marked by neutrophil migration into pulmonary capillaries and alveolar spaces, leading to acute capillaritis, fibrin deposition and neutrophilic mucositis (22). Neutrophils display antiviral activity through different mechanisms, including direct, antibody or complement-mediated recognition of the virus and phagocytosis and secretion of cytokines, myeloperoxidase (MPO), elastase and defensins. They can also form neutrophil extracellular traps (NET) and modulate adaptive antiviral immunity (23–25). Recent studies have found increased neutrophil counts in the peripheral blood of non-surviving and severely ill COVID-19 patients compared to mildly ill cases. In addition, increased numbers of low-density neutrophils have been found in the blood of acute COVID-19 patients (26, 27). Different populations of granulocytes in the blood could reflect emergency myelopoiesis. Some of the granulocytes (N2-type) could be immunosuppressive and cause impaired lymphocyte responses during acute disease (28). Previous studies have also observed a high neutrophil- to- lymphocyte ratio (NLR) in critically ill COVID-19 patients compared to cases with mild disease. Therefore, NLR has been introduced as an independent risk factor for severe disease (27, 29, 30). High levels of MPO and citrullinated histone H3 (Cit-H3), serum markers of NETs and indicators of neutrophil activation have also been described in severe COVID-19 (31–33). It has thus been concluded that neutrophilia and excessive formation of NETs, as well as the release of reactive oxygen species (ROS), in severe COVID-19 trigger cytokine release and respiratory failure, contributing to disease severity (22, 34, 35). The changes in innate immune cells in COVID-19 are summarized in Figure 1.

Figure 1 The frequency and function of innate immune cells in severe COVID-19. SARS-CoV-2 infects cells that present the surface receptor ACE2. It is recognized by tissue-resident immune cells in the lungs, which provoke a local immune response. More innate immune cells are recruited from the blood by chemokines and complement activation products. The amounts of neutrophils, CD16+ monocytes, M1 macrophages and MDSCs are bigger in severe COVID-19 patients, while the percentages of basophils, eosinophils, M2 macrophages, classical monocytes, DCs, ILCs, NK cells and unconventional T cells are reduced. Activated neutrophils can amplify inflammation by secretion of pro-inflammatory cytokines, MPO, elastase and defensins. They can also promote NET formation and Th17 cell differentiation. Activated basophils and eosinophils appear to predispose the development of ARDS by releasing inflammatory and vasoactive substances. Peripheral CD16+ monocytes and M1 macrophages are involved in a hyper-inflammatory response and cytokine storm during the inflammatory phase in severe COVID-19. Significant decrease in viral sensing and IFN signaling as well as suppressed antigen presentation by both pDC and cDC have been reported in severe COVID-19. Aberrant or ineffective responses of ILCs (ILC1, ILC2, ILC3 and NK cells), MDSCs and unconventional T cells (MAIT cells, γδ T cells) also have role in promoting inflammation in severe COVID-19. ACE2, Angiotensin-converting enzyme 2; MDSCs, Myeloid-derived suppressor cells; DC, Dendritic cell; ILCs, Innate-like lymphocyte; NK cell, Natural killer cell; MPO, Myeloperoxidase; NET, Neutrophil extracellular trap; Th, T helper; ARDS, Acute respiratory distress syndrome; MAS, Macrophage activation syndrome; PD-L1, Programmed cell death ligand 1; IFN: Interferon; cDC, Classical DC; pDC, Plasmacytoid DCiNOS, Inducible nitric oxide synthase; PMN-MDSCs, Polymorphonuclear-MDSCs; M-MDSCs, Monocytic-MDSCs; ADCC, antibody-dependent cellular cytotoxicity; NKG2A, NK cell lectin-like receptor subfamily C member 1. Figure was created by BioRender (Toronto, ON, Canada).
2.2 Monocytes
Monocytes are a pivotal component of the innate immune response. They participate in inflammatory responses, phagocytosis, antigen presentation and other immune functions (36, 37). Human monocytes are classified into classical (CD14high CD16−), intermediate (CD14highCD16+) and non-classical (CD14low CD16+) subsets (38). Intermediate and non-classical monocytes are known as the “inflammatory” subsets due to their potent pro-inflammatory activity. They contribute to various conditions such as asthma, coronary artery disease, Crohn’s disease, sepsis and hepatitis B (39). Recent studies have demonstrated that inflammatory monocytes are involved in the hyper-inflammatory response and cytokine storm observed during the infectious and inflammatory phases of severe COVID-19 (40). The results indicate that the absolute numbers of all blood and CD16- classical monocytes decrease in severely ill COVID-19 patients, while the levels of the inflammatory subsets increase in patients in comparison to the healthy controls (41). One study showed an increased percentage of inflammatory monocytes expressing S100A8+ S100A9 (calprotectin), CD14 and CD62L, possibly correlating with a profibrotic differentiation pattern in severe COVID-19 as compared to mild disease (42). Furthermore, a significant increase in the frequency of CD14high CD16+ monocytes producing IL-6 was reported in the peripheral blood of COVID-19 patients in the ICU compared to those without ICU hospitalization (43, 44). An increased number of inflammatory monocytes in patients with mild disease has also been reported in several studies. This could be considered as an indicator of viral control in such patients (45, 46).
2.3 Macrophages
The function of alveolar macrophages in the lungs is to preserve homeostasis by removing dead cells and invading pathogens (47). Under steady-state, two distinct populations of macrophages, the interstitial macrophages (IMs) and alveolar macrophages (AMs), are present in the lungs (48, 49). Interstitial macrophages are characterized as CD11b+CD11c- cells localized in the parenchyma, while AMs are identified as CD11b-CD11c+ macrophages residing in the air space lumen (50). In inflammation and infectious diseases, alveolar macrophages are polarized into the M1 or M2 subgroup (47). M1 macrophages cooperate with T helper (Th) 1 cell to produce pro-inflammatory cytokines and clear pathogens. They also induce the migration of other immune cells to the lung parenchyma and alveoli. M2 macrophages are associated with Th2 cells to increase the secretion of anti-inflammatory cytokines and clearance of apoptotic or senescent cells. Thus, the M2 cells restrict inflammation and promote tissue remodeling (47, 51).
Based on recent data, macrophages are considered promoters of the cytokine storm during infection by SARS-CoV-2. They are associated with severe COVID-19 with a high risk of death (52, 53). Moreover, the cytokine storm, mainly macrophage activation syndrome (MAS), in addition to complement and neutrophil activation, contributes to COVID-19-associated adult respiratory distress syndrome (ARDS) and its exacerbation (54, 55). Another study reported a significant correlation between SARS-CoV-2 infection of macrophages in the spleen and lymph nodes and severe lymphocyte apoptosis in COVID-19 patients (27). Recent studies by single-cell RNA sequencing of samples recovered from bronchoalveolar lavage fluid of mildly and severely ill COVID-19 patients have revealed that SARS-CoV-2 infection leads to the recruitment of an inflammatory macrophage subset with down-regulation of type I IFN genes, which is related to disease severity (56–58). In conclusion, macrophages sense SARS-CoV-2 and respond to its threat by secreting pro-inflammatory cytokines and chemokines. However, an uncontrolled macrophage response could trigger MAS, ARDS and an exacerbation of COVID-19 (42).
2.4 Dendritic cells
Among other places, DCs are distributed in the respiratory tract, where they can recognize invading microbes. SARS-CoV-2 can gain entry into DCs, particularly into interstitial lung DCs, via ACE2, CD147 and dendritic cell-specific intracellular adhesion molecule-grabbing non-integrin (DC-SIGN) receptors (59). Classical myeloid DCs (cDCs) and plasmacytoid DCs (pDCs) are two distinct DC subsets in humans. The former serve as potent antigen- presenting cells (APCs), while the latter is a specialized subset that can produce type 1 IFN (60, 61). It has been found that pDC along with viral sensing, interferon signaling and antigen presentation are more defective in severe versus mild COVID-19 (62–68). Further impairment in DC maturation and function in severe vs. mild disease may be associated with a worse disease (69). In addition, another study reported that the frequency of DCs in the blood of COVID-19 patients decreased compared to healthy controls. It was found that DCs from individuals with acute COVID-19 were functionally impaired in maturation and in the ability to activate T cells (43, 70). Two recent studies found both types of DCs (cDCs and pDCs) to be much less abundant in the blood samples of people with severe COVID-19 than in those with mild COVID-19 (71, 72).
2.5 Innate-like lymphocytes
ILCs are effector cells that detect environmental stimuli and contribute to early immune responses by producing large amounts of cytokines (73). They are categorized into four main groups: NK cells, ILC1, ILC2, and ILC3 (74, 75). Transcriptionally and functionally, the ILC1, ILC2, and ILC3 resemble Th1, Th2, and Th17 cells, respectively, while NK cells resemble CD8+ T cells (73). However, little is known about the distribution and function of different ILC subsets in COVID-19. Recent studies have reported a reduced percentage and lower absolute counts of ILCs in the blood of COVID-19 patients (73). Accordingly, results have shown a significant decrease in the frequency of ILC2 in severe compared to moderate COVID-19 (73). Furthermore, previous studies have observed that the frequency of ILC2s in COVID-19 patients negatively correlates with the level of the coagulation factor fibrinogen-derived D-dimer and the development of severe disease (76, 77). ILC2s could thus have a protective role in the disease.
Previously, it has been shown that ILC2 plays an important role in lung tissue repair during influenza A infection in mice (78). Therefore, it can be hypothesized that low ILC2 levels in COVID-19 patients correlate with a more severe disease outcome. One further investigation reported a decreased frequency of all ILCs, and particularly of the ILC2 subpopulation, in severe COVID-19. Also, compared to mildly diseased patients, it was observed that the expression of NKG2D+, the activating C-type lectin-like molecule, on ILC2s was significantly reduced in patients requiring mechanical ventilation (79). Therefore, an increase in NKG2D+ ILC2s, along with elevated levels of anti-inflammatory mediators, might be related to a better prognosis in severe COVID-19 (Figure 1).
2.6 Natural killer cells
NK cells are the most important innate immune cells in fighting viral infection. This is because they can induce direct lysis of target cells by producing perforin and granzyme B, secrete inflammatory cytokines, contribute to antibody-dependent cell cytotoxicity (ADCC) and interact with other immune cells such as monocytes (80). A significant decrease in the absolute number of NK cells in the peripheral blood of patients with severe COVID-19, as well as in those admitted to ICU, has been reported recently (81–83). According to Leem et al., unconventional CD56dimCD16neg NK cells, associated with decreased cytotoxic activity, expanded in both severe and mild forms of the disease. These changes were reversed more rapidly in mild vs. severe COVID-19 (84). Another study showed that the percentage of perforin+ NK cells in COVID-19 patients, who were in the ICU, was reduced as compared with non-ICU patients and healthy controls (85). Previous studies have shown an increased expression of the inhibitory receptor NKG2A on NK cells in severely ill COVID-19 patients, as compared to mildly ill patients and healthy subjects (86). NKG2A expression along with decreased IFN-γ, granzyme B, and TNF-α production is associated with NK cell exhaustion and disease progression in COVID-19 patients. Further, the percentage of NK cells expressing NKG2A is reduced in COVID-19 patients, who recovered after receiving proper therapy (27, 87). Hajeer et al. found an association between two NK cell immunoglobulin-like receptors (KIR), named KIR2DS4 and KIR3DL1, with an inhibitory and activating function, respectively, and an increased risk of severe disease (88). Another study reported a significant decrease in the inhibitory KIR receptors in COVID-19 patients compared to healthy controls and a considerable increase in activating KIR receptors in patients with a severe disease (89) (Figure 1).
2.7 Basophils
Basophils are a type of granulocyte representing less than 1% of peripheral blood leukocytes (90). As they are involved in tissue repair and production of anti-coagulant factors, their depletion results in increased pneumonitis in COVID-19 (91). In recent studies, a significant decrease has been observed in the frequency and the absolute numbers of basophils in patients with severe COVID-19 (82, 92, 93). Moreover, a stronger expression of programmed cell death ligand 1 (PD-L1), an inhibitor of T cell activation, has been found on basophils in severe vs. mild COVID-19, correlating with poor disease outcomes (89).
2.8 Eosinophils
Eosinophils are multifunctional granulocytes involved in various inflammatory contexts, including helminth, bacterial and viral infections, tissue injury, tumor immunity and allergic diseases. They secrete pro-inflammatory mediators, express adhesion molecules and support smooth muscle cells (94, 95). Significantly decreased eosinophil counts have been found in COVID-19 patients compared to healthy controls or patients with mild disease (82, 96). A decrease in eosinophil counts is related to fever, fatigue, shortness of breath and poor outcome in COVID-19 patients (96, 97).
2.9 Myeloid-derived suppressor cells
In humans, MDSCs are classified into polymorphonuclear-MDSCs (PMN-MDSCs) and monocytic-MDSCs (M-MDSCs), which control inflammation and T cell responses through a high-level expression of inducible nitric oxide synthase (iNOS), arginase-1 (Arg-1), nicotinamide adenine dinucleotide phosphate oxidase (NOX2) and transforming growth factor-beta (TGF-β) (98). In viral infections, MDSC-mediated immunosuppression causes viral persistence and disease progression (99). A recent study showed a significantly higher frequency of PMN-MDSCs in the peripheral blood of patients with severe COVID-19 and in those who required ICU compared to healthy blood donors (98). This correlated with decreased antigen-specific T cell responses and poor disease outcomes (98). Another study demonstrated an increased frequency of MDSCs in both mild and severe COVID-19. According to this study, MDSC expansion during the early phase of acute infection was protective by reducing T cell hyperactivation and inflammation but could also decrease the protective immune response (100). The percentage of M-MDSCs correlated positively with neutrophil count, C-reactive protein (CRP) and D-dimer levels, hospital stay and viral RNA load and negatively with lymphocyte count and serum albumin in COVID-19 patients admitted to ICU compared to healthy controls and non-ICU patients (101). An increase in MDSC recruitment and expansion along with impaired T cell function was observed in severe COVID-19. This was related to IL-10, arginase-1 and indoleamine 2,3-dioxygenase (IDO) production by MDSCs (102, 103).
2.10 Unconventional T cells
Unconventional T or innate-like T cells, such as natural killer T (NKT), mucosa-associated invariant T (MAIT) and γδ T cells, exhibit characteristics of both innate and adaptive cells. They express T cell receptors (TCRs) with limited diversity and recognize alternative microbial antigens in major histocompatibility complex (MHC)-unrestricted compartments (74). Most responses of NKT and γδ T cells seem to be stimulated against pathogenic agents. In particular, the potent cytotoxic responses of these cells in patients infected with influenza A viruses, herpes viruses or lentiviruses may be essential for suppressing viral replication and regulating immunosuppressive MDSCs (104–110). Recent studies have shown a reduced frequency of NKT cells in patients with severe COVID-19 and a small PaO2/FiO2 (P/F) ratio (82, 111). An increased expansion of CD160+ NKT cells has been reported in mild COVID-19, possibly supporting recovery from infection by direct cytotoxicity (112). Furthermore, decreases in the levels of circulating NKT cells and IFN-γ production are observed in severe COVID-19. However, the expression levels of CD69 and PD-1 are increased in NKT cells, and a strong PD-1 expression in NKT cells continues in patients in ICU on day 15 (113). Another study showed that while the total percentage of γδ T cells decreased in COVID-19 patients, the percentage of CD4+γδ T cells significantly increased (114). It was suggested that the latter subset might be associated with antigen presentation and activation of adaptive immune cells (114). Based on another investigation, increased frequencies and absolute numbers of naïve-like (CD45RA+CD62L+) γδ T cells and a decreased frequency of effector-like (CD45RA-CD62L−) γδT cells were observed both in mild and severe COVID-19. This study suggested that effector-like γδ T cells might be localized in the lungs of COVID-19 patients to contribute to the immune response against the infecting virus (115) (Figure 1).
2.11 Complement system
The complement system belongs mostly to innate immunity. It can respond to invading microbes and other foreign materials (22, 116). It may recognize SARS-CoV-2 directly or with the help of antibodies. Additionally, tissue damage caused by the virus can activate complement. Complement activation can lead to acute or chronic inflammation, thrombus formation, endothelial cell dysfunction and intravascular coagulation, thus ultimately contributing to multiple organ failure (MOF) and death (117). A recent study showed prominent complement activation in the lungs, skin and blood of severely ill COVID-19 patients compared to cases with mild disease. Some individuals treated with a complement inhibitor recovered with no adverse reactions (118). Complement activation generates the pro-inflammatory polypeptides, C3a and C5a, which can recruit and activate neutrophils and monocytes. Mannose-binding lectin (MBL) interacts with SARS-CoV-2 and could consequently activate complement C3 via the lectin pathway (119). Through neutrophil extracellular trap (NETs) formation, neutrophils activate the alternative complement pathway and may engage an inflammatory feedback loop. Recently, a significant correlation has been found between the activation of complement component C3 and COVID-19 severity (120, 121). Similarly, an investigation showed higher levels of C3a and C5a as well as C3-fragment deposition in the lung biopsy samples of severe COVID-19 patients (122). Furthermore, in a further study, high levels of soluble C5a and an increased expression of the C5a receptor in blood and pulmonary myeloid cells were found in severe COVID-19 with acute respiratory distress syndrome compared to mildly diseased patients and controls (123). Thus, multiple features of severe COVID-19 point to the importance of complement activation in its pathogenesis, mainly during inflated inflammatory responses and effects on vascular endothelial cells, including promotion of blood coagulation and increased capillary permeability.
3 The relationship between the adaptive immune system and the progression of COVID-19
3.1 Cell-mediated immunity
CD4+ and CD8+ T cells play important roles in resolving acute viral infections and providing subsequent protection against reinfection. DCs induce polarization and maturation of naive T cells via the presentation of peptides in the context of MHC along with the secretion of chemokines and cytokines. Subsequently, activated T cells migrate to the site of infection, producing antiviral cytokines, chemokines and cytotoxic molecules (124). Acute and severe SARS-CoV-2 infections are associated with lymphopenia and remarkable loss of CD4+ and CD8+ T cells, which is reversed by disease recovery (125–127). Transient lymphopenia may be associated with a direct effect of SARS-CoV-2 infection on the production and/or differentiation of lymphocytes in primary lymphoid organs and their release into the circulation (115, 128). High levels of IL-6, IL-10 or TNF-alpha directly promote thymic dysfunction and indirectly promote T cell apoptosis (127, 129–132). In early studies on COVID-19, impairment in function and increased expression of markers, which are hallmarks for activation and/or exhaustion of CD4+ and CD8+ T cells, were observed in patients. The expression levels of Ki-67, PD-1, perforin and granzyme B in CD4+ or CD8+ T cells were found to be high in patients with severe disease (133, 134). Some studies have considered the PD-1/PDL-1 axis as a severity-associated biomarker that can inform about lymphocyte depletion/exhaustion (135, 136). However, it seems that the stronger expression of T cell inhibitory molecules (PD-1 and Tim-3) in severe vs. mild COVID-19 might not only indicate T cell exhaustion but could also suggest the existence of antigen-specific T cells (77, 137, 138). Recent studies, including more than 1000 COVID-19 cases, have indicated that a reduced frequency of CD4+ T cells is related to disease severity (139, 140). The lower proportion of SARS-CoV-2-specific CD4+ CD154+ T cells, which are unable to secrete IL-2 and IFN-γ following stimulation with S- protein, and weaker humoral immune responses have been reported in asymptomatic individuals with SARS-CoV-2 infection. In the symptomatic group, the higher frequency of SARS-CoV-2-specific CD4+ CD154+ T cells was significantly correlated with higher serum levels of SARS-CoV-2-specific IgA, IgG and IgM antibodies (141). Different subpopulations of CD4+ T cells, such as Th1, Th2, Th17 and regulatory T cells (Treg), have been found to accumulate in COVID-19 patients. In the following, the subpopulations of CD4+T cells, as well as CD8+ T cells and memory T cell subsets in COVID-19 inflammatory conditions, will be discussed.
3.2 Th1 cells
Previous studies have indicated that Th1 responses could play protective roles in the early phase of SARS-CoV-2 infection (142). Indeed, highly functional and terminally differentiated effector Th1 cells that eliminate infected target cells have been identified in the early phase of COVID-19 (143, 144). A decreased expression of IFN-γ on CD4+ T cells has also been shown in severe COVID-19 cases (145). Moreover, Th1 hypo-activation is associated with a poor prognosis of the disease, and a lower percentage of Th1 cells is observed in severe COVID-19 patients compared to mild patients (146, 147). Earlier investigations suggested that in comparison to mild COVID-19, increased expansion of peripheral neutrophils in severe COVID-19 potentially suppresses Th1 cell differentiation and triggers Th17 cell polarization (148) (Figure 2).
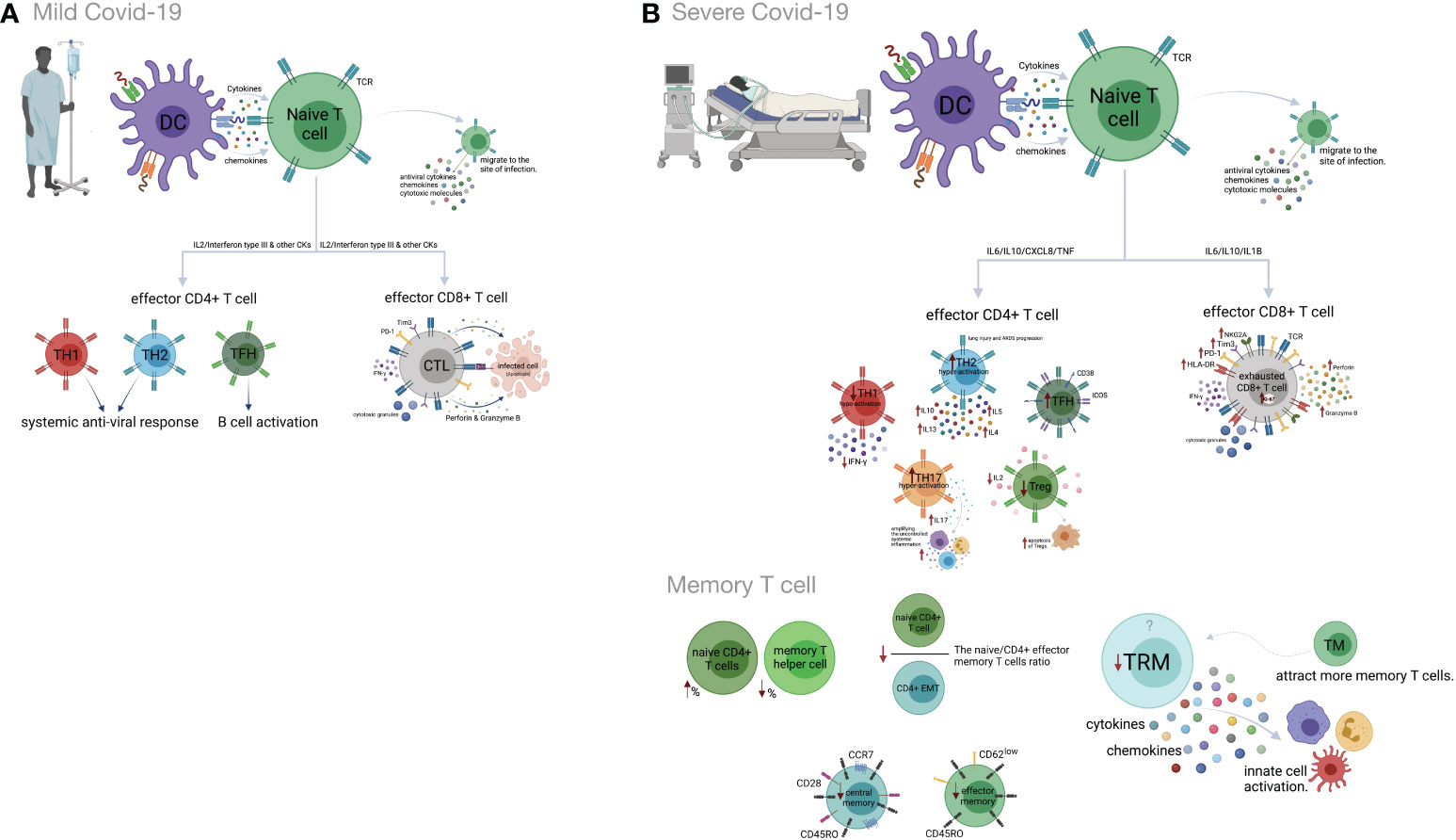
Figure 2 Cell-mediated immune responses during COVID-19 progression in mild versus severe disease. (A) Dendritic cells (DCs) promote activation of T cell responses by presentation of SARS-CoV-2 antigenic peptides in major histocompatibility complex (MHC) molecules along with costimulatory interactions and the secretion of chemokines and cytokines. T cell responses can mainly be polarized into T helper (Th) cells and effector cytotoxic cells (CTLs). In mild COVID-19, a higher proportion of Th1 cells plays an important role in defense against the virus by producing interleukin-2 (IL-2) and interferon-gamma (IFN-γ). Th1 cells and the cytokines they secrete activate macrophages, CTLs and natural killer (NK) cells. The Th2 cells stimulate the humoral response and activate eosinophils, basophils and mast cells through IL-4 and IL-5 secretion. The T follicular helper (Tfh) cells are essential for the formation of germinal centers (GCs), B cell maturation and immunoglobulin (Ig) production. CD8+ T cells, which coordinate the protective antiviral activities in COVID-19, secrete cytotoxic granules containing perforin and granzyme B as well as IFN-γ in patients who developed mild disease. (B) Lymphopenia is a hallmark feature of patients with severe COVID-19. This may be the result of the migration of enormous numbers of T cells to the lungs and other sites of inflammation. Both CD4+ and CD8+ T cells express strongly CD69, CD38, CD45RO, CD44, HLA-DR, programmed cell death protein-1 (PD-1), T cell immunoglobulin domain and mucin domain-3 (TIM-3) and killer cell lectin-like receptor subfamily C member 1 (NKG2A), which can represent hyperactivated, exhausted or a hypoactivated state of T cells in severe patients. Furthermore, the numbers and functions of regulatory T cells (Treg) are significantly reduced in these patients. Smaller proportions of CD4+ and/or CD8+ effector cells (CD45RO+ CD45RA− CCR7−CD28− CD62Llow), central memory cells (CD45RO+ CD45RA− CCR7+ CD28+) and tissue-resident memory (CD69+ CD103+ CD49a+) T cells were demonstrated in severe COVID-19 patients. Figure was created by BioRender (Toronto, ON, Canada).
3.3 Th2 cells
Th2 cells usually mount an appropriate immune response against extracellular pathogens. A recent study showed that Th2 hyper-activation and higher plasma levels of Th2-produced cytokines, including IL-4, -5, -10, and -13, significantly correlated with COVID-19 severity and its related mortality (146, 147). Some studies have highlighted the possible role of Th2 responses in immune-driven lung injury and ARDS progression (142, 143). The association between an increased percentage of Th2 cells and critical care, as well as the poor prognosis, is reported in COVID-19 patients (149) (Figure 2).
Th17 cells secrete IL-17 and induce effector cells and inflammatory cytokine production that amplifies uncontrolled systemic inflammation and triggers tissue injury as well as multi-organ failure and death. Several studies have demonstrated that increased frequency and hyper-activation of Th17 cells and subsequent production of pro-inflammatory cytokines are related to the poor outcomes of COVID-19 (35, 142, 145, 148, 150–156). Recently, a higher percentage of GM-CSF+ IL-6+ CCR6+ Th17 cells has been detected in the blood circulation of COVID-19 patients (43). According to new research, Th17 hyperactivation and signaling are significantly correlated with a critical form of COVID-19 (148, 153).
3.4 Treg cells
Current evidence indicates smaller amounts of Treg cells in correlation with disease severity in the peripheral blood of COVID-19 patients (115, 157, 158). The frequency of this subset increases during the recovery phase of the disease (159). The decrease in the frequency of Tregs in the peripheral blood of COVID-19 cases might be related to Treg migration into the lungs to resolve lung injury. Besides, there is some evidence of enhanced apoptosis of Tregs due to reduced IL-2 levels in severe compared to mild cases (139, 154, 160–162). A higher frequency of Treg cells in the early phase and a lower frequency in the late phase of SARS-CoV-2 infection could be associated with poor outcomes of COVID-19 (139, 154, 160–162). Accordingly, a decrease in Treg cells along with an increase in Th17 cells is associated with the deregulated pro-inflammatory cytokine secretion in COVID-19 patients (163) (Figure 2).
3.5 CD8+ T cells
CD8+ T cells exert their antiviral activity by killing or inducing apoptosis of infected cells. This occurs by release of cytotoxic granules (154) and secretion of cytokines (164). Several studies reported lower CD8+ T cell counts in patients, even with mild or moderate, but especially with severe or critical COVID-19 (160, 165–168). A decreased frequency of CD8+ T cells in non-survivors was reported until death (169). The percentage of HLA-DR+ PD-1+ Tim-3+ NKG2A+ activated CD8+ T cells has been reported to be higher in patients with severe than mild COVID-19 (154, 170, 171). These findings suggest that hyper-active CD8+ T cells may play a protective role in the early phase of COVID-19, but rather a pathogenic role in the late phases of the disease due to reduced cytotoxic function and increased cytokine production (160).
3.6 Memory T cells
Memory T cells display rapid responses against subsequent infection with similar or related pathogens (172). SARS-CoV-2-specific memory T cell responses in the early recovery phase have been reported recently (138). Compared to mild disease, an increased percentage of naïve CD4+T cells and a decreased percentage of memory T helper cells are observed in severe COVID-19 (173). Also, reduced frequencies of CD4+ and/or CD8+ effector and central memory T cells have been observed in severe compared to mild COVID-19. The naive/effector CD4+ memory T cell ratio, an indicator of the differentiation from naive to memory T cells, is reduced in severe cases due to severe impairment of adaptive immunity (14, 174). Tissue-resident memory T cells secrete cytokines and chemokines, resulting in the activation of innate cells and an increase in the recruitment of memory T cells from the periphery (175, 176). Although airway-resident memory T cells mediate protective immune responses against emerging respiratory coronaviruses, the role of this memory subset in the immunopathogenesis of SARS-Cov2 infection should be analyzed further (177). Accordingly, studies on COVID-19 patients, who required ICU treatment, have revealed a lower frequency of tissue-resident memory T cells than effector-memory T cells (178). Likewise, tissue-resident memory-like Th17 cells (TRM17 cells), characterized by an aberrant cytokine profile, are found in the lungs of recovered cases (179). Despite these findings, some studies state that the enrichment of virus-specific CD8+ TRM cells in the patients´ respiratory system is positively correlated with the degree of lung damage and even contributes to damage after the recovery (180) (Figure 2).
3.7 Humoral immunity
In addition to antibody production, B cells display antiviral immune responses through the secretion of inflammatory mediators and T cell activation through antigen presentation (181–183). These cells are a pivotal source of neutralizing antibodies (Abs) that interfere with viral infections, including SARS, MERS, HIV and Ebola. They bind to the virions and prevent their entry into the host cells. Antibodies may activate the complement system and kill infected cells through antibody-dependent cellular cytotoxicity (ADCC) (126, 184, 185). On the other hand, antibodies could also facilitate viral infections by increasing virion uptake via Fc or complement receptors. The former is referred to as antibody-dependent enhancement (ADE). ADE has been observed in dengue virus, HIV, influenza, RSV and Ebola, as well as SARS-CoV-2 infections (181, 186–188).
Impaired cellular and humoral immune responses have been reported in COVID-19 patients (5, 189). B cell immune responses to SARS-CoV-2 infection have been found to be stronger in severe than in mild COVID-19. Accordingly, increased clonal expansion of B cells, and frequencies of plasmablasts, complement activation and phagocytosis have been reported in severe cases (190, 191). Similarly to chronic hepatitis B infection, the overall amounts of B cells are large in severe COVID-19 cases. However, the B cell responses are deficient due to decreased clonal diversity. In addition to decreased T cell activity, this is another cause for improper immune responses against infectious agents in elderly individuals (192, 193). Conversely, several studies have reported decreased absolute numbers and frequencies of B cells, as well as lower expression of genes associated with BCR activation signaling, in severe vs. mild COVID-19. This might be due to the consumption of B cells by SARS-CoV-2 in order to inhibit immune activity (2, 83, 154, 194–198).
Also memory B cells are produced after infection with SARS-CoV-2. They include classical CD24+ class-switched memory B cells, activated CD24- and natural unswitched CD27+ IgD+ IgM+ subsets. Recently, reduced frequencies of switched and unswitched memory B cells have been observed in severe COVID-19 patients (83, 190, 197, 199–201). In contrast, some studies have reported higher percentages of SARS-CoV2-S-specific memory B cells in severely ill compared to mildly ill patients. The memory B cell levels remain stable several months after disease resolution and provide protection against future exposure (202–206). In one study, a higher proportion of double-negative B cells (IgD- CD27-) was reported in severe COVID-19 (199, 207, 208). These cells exacerbate disease progression, possibly by cytokine secretion (209–212). Increased frequencies of CD21+and CD21low/- transitional B cells have also been observed in mild to moderate and severe forms of COVID-19 (190, 199).
Many studies have shown high titers of antibodies in severe COVID-19 that do not correlate to disease resolution (83, 154, 178, 191, 213–216). Indeed, it has been shown that anti-spike IgG could promote pro-inflammatory responses in M1 macrophages, disruption of endothelial barriers, and microvascular thrombosis (217). This type of thrombotic microangiopathy could be mediated by complement activation. Moreover, anti-SARS-CoV-2 antibodies could enhance virus entry into immune cells through the ADE mechanism (166, 218, 219). In severely ill patients, increased production of autoantibodies, such as antinuclear (ANAs), anti-phospholipid and anti-cardiolipin antibodies, have been associated with poor outcomes (219, 220). They could be due to polyclonal B cell activation or be induced by tissue damage. Also, several studies have reported higher titers of anti-S1- and anti-N- specific IgG and IgM antibodies, as well as neutralizing antibodies (nAbs) in severe vs. mild COVID-19 (199, 221–223). High titers of anti-N Abs have been found to be related to increased virus replication and more severe disease (221) (Figure 3).
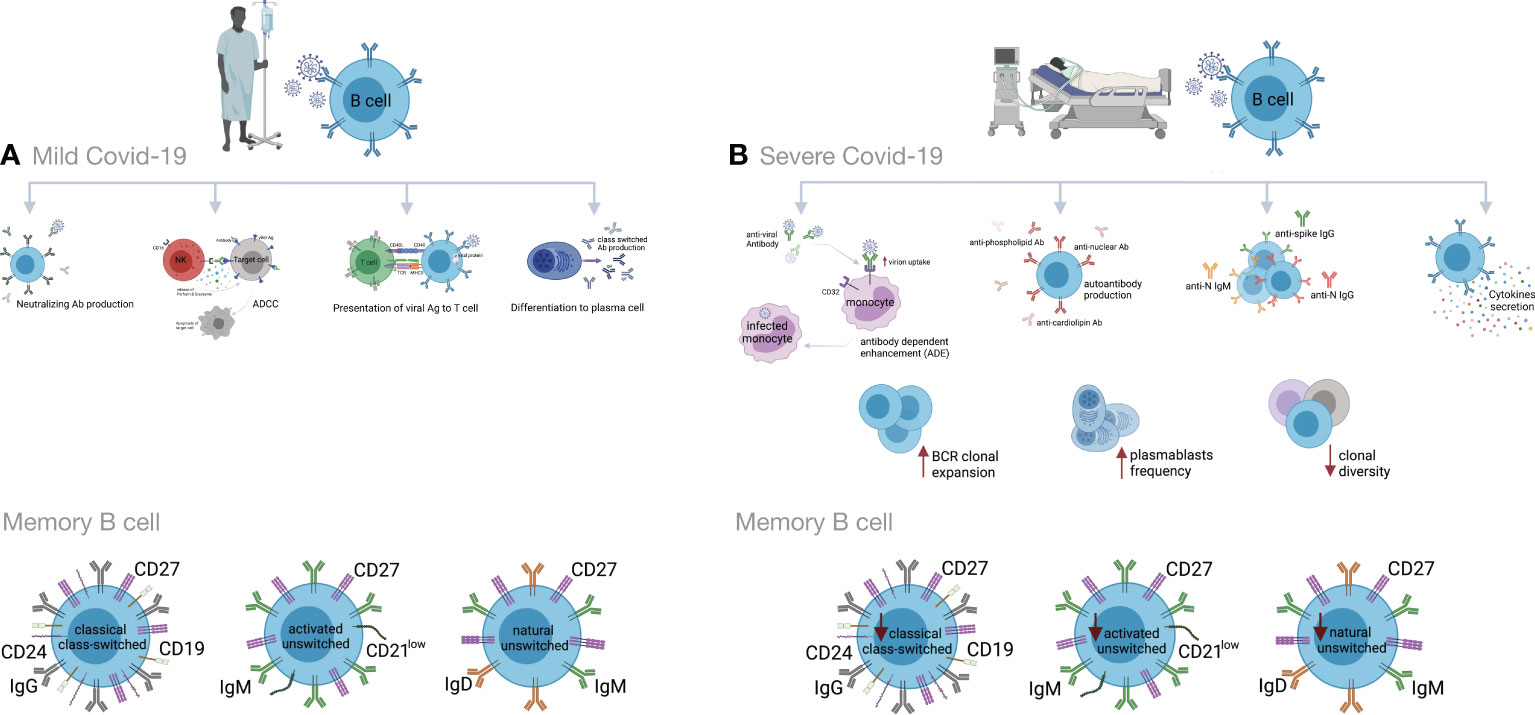
Figure 3 Humoral immune responses during COVID-19 progression in mild versus severe disease. (A) During mild COVID-19, B cells contribute to antiviral immune responses in many ways. The first is through production of neutralizing antibodies (Abs), which will bind to the virion and block its entry to the host cells, thus impeding further infection. The second is through a process called antibody-dependent cellular cytotoxicity (ADCC), in which Fc receptor-bearing effector cells can recognize and kill antibody-coated target cells expressing pathogen-derived antigens. The third is through antigen presentation to T cells and activation of cell-mediated immunity. B cells also produce some cytokines to regulate inflammation and activation of adaptive responses. Lastly, B cells can differentiate into antibody-secreting plasma cells, which produce high affinity and class-switched Abs. (B) B cell immune responses in severe COVID-19 may exacerbate disease progression by excessive secretion of cytokines and production of autoantibodies. As an adverse effect anti-viral antibodies can facilitate the entry of virus into monocytes/macrophages and granulocytic cells through interaction with Fc and/or complement receptors in a process that is called antibody dependent enhancement (ADE). In addition, increased clonal expansion of B cells, higher frequency of plasmablasts as well as reduced frequency of CD24+ switched and CD24-CD27+ IgD+ IgM+ unswitched memory B cells have been reported in severe COVID-19 cases. Figure was created by BioRender (Toronto, ON, Canada).
4 Cytokines, interferons and the progression of COVID-19
Cytokines play a major role in many physiological and pathological processes. In the inflammatory context, different immune and non-immune cells, such as monocytes, macrophages, T cells and platelets, as well as endothelial cells, epithelial cells and fibroblasts, produce various cytokines. A cytokine storm, CRS, is defined as an overproduction of cytokines due to an uncontrolled host inflammatory response to different triggers. The triggers could include infections, tissue damage, malignancy, rheumatic disorders and certain drugs. Several studies on severely ill COVID-19 patients have identified significant variations in the serum or plasma levels of cytokines associated with COVID-19 pathogenicity (224, 225). It has been demonstrated that cytokine storm plays an important role in the severity of SARS-CoV-2 infection and lung damage, possibly resulting in ARDS and patient referral to ICU (226, 227). Overproduction of cytokines, including IL-17, IL-7, IL-1β, IL-9, IL-2, IL-10, TNF-α, GM-CSF, G-CSF, IFN-γ, MCP1, MIP1A, MIP1B, CXCL10 and CXCL8, by monocytes and macrophages has been reported in severe COVID-19 cases (182, 228, 229). Moreover, in a study conducted in a group of 52 patients with severe conditions, high plasma concentrations of GM-CSF were reported in COVID-19 patients who required ICU, suggesting a role for GM-CSF in disease severity (182). A study conducted in Taiwan in 2021 compared 40 cytokines in COVID-19 patients with mild, moderate, severe or critical diseases. It was found that the levels of 22 cytokines gradually increased in critical and severe cases compared to moderate ones (230). Evidence indicates that macrophages are the main promoters of cytokine storm during SARS-CoV-2 infection, leading to deleterious clinical manifestations and acute mortality in critically ill patients with COVID-19 (37, 231).
As a broad class of cytokines, interferons (IFNs) are a group of related proteins produced by a variety of cells against viral infection (232). IFNs are classified into three types: type І, type П, and type Ш. Plasmacytoid dendritic cells (pDC) are the main producers of type І IFNs. By binding to their receptors, the interferons induce the expression of IFN-stimulated genes (ISGs), thereby generating an antiviral state by inhibiting virus replication in cells under infection threat (233–235). Several studies have reported that both IFN-β1b and IFN-β1a exert an inhibitory effect on SARS-CoV-2 (236–239). In this regard, an impaired type I IFN response is reported in patients with severely and critically ill COVID-19 compared to those with mild infections. Indeed, patients with lower plasma levels of type I IFN have higher levels of TNF- α and IL-6, increased viral load, and extensive NFκB-driven inflammatory responses (240). A recent study showed that SARS-CoV-2 mRNA-encoded proteins, ORF3b and ORF6, inhibit IFN production and signaling (241). Another study found that high IFN-λ levels during the early stages of SARS-CoV-2 infection were associated with a lower viral load in bronchial aspirates and a better outcome in severely ill COVID-19 patients. In late stages in critically ill COVID-19 patients, however, the serum levels of IFN-α and IFN-λ were found to be decreased compared to those with mild disease (242).
5 Therapeutic approaches to COVID-19
Considering that SARS-CoV-2 infection and imbalanced immune responses are the two main drivers of COVID-19 pathogenesis, glucocorticoids and antiviral drugs have been the most commonly used treatments to treat COVID-19 and its complications (243, 244). Safe strategies to limit morbidity and mortality and to improve the efficacy of treatments by finding new therapeutic targets are still urgently required. Current studies have focused on specific means to interfere with SARS-CoV-2 infection (245, 246). Nowadays, several approaches are followed to explore candidates to decrease or subvert inflammatory responses against primary SARS-CoV-2 and mutant strains. Below, the most promising potential targets are reviewed, and an update on the progress of treatments for COVID-19 is provided (Table 1).
5.1 Anti-cytokine therapy
One approach to treat cytokine storm in COVID-19 is to use traditional anti-inflammatory drugs such as corticosteroids, chloroquine and colchicine (258, 259). However, except for corticosteroids, their use has remained limited. Recently, recombinant cytokines and antibodies or inhibitor molecules against different cytokines and their signaling pathways have been tried or are in the pipeline for production. Below, some of the approaches that are currently or could potentially be applied to manage more serious forms of COVID-19 are described. For the less serious, flu-like illnesses, rest and symptomatic treatment are usually sufficient.
Recombinant human IL-7 (rhIL-7). According to a recent study, recombinant human IL-7 (rhIL-7) administration significantly rescued the immune function in a 74-year-old ICU patient with severe COVID-19 (260). In another study, higher lymphocyte counts without widespread inflammation or pulmonary injury were observed in COVID-19 cases who received IL-7 (247). It has been found that administration of dexamethasone with IL-7 leads to IL7 receptor (IL-7Rα) upregulation and increased IL-7 activity in the more severe stage of COVID-19 (261–263). Another clinical trial (NCT04379076) is currently investigating the effect of CYT107, a commercial derivative of rhIL-7, on the clinical picture and immune reconstitution in severe COVID-19 patients. Accordingly, these results suggest that appropriate administration of IL-7 with or without other agents could be applied to critically ill COVID-19 patients with severe lymphopenia. Because of its lymphopoiesis- stimulating effect, IL-7 has also been suggested as a potential vaccine adjuvant.
5.2 Targeting IL-6 signaling
A high concentration of the acute phase response stimulating cytokine IL-6 is related to an increased risk for severe COVID-19. Thus, targeting IL-6 signaling may provide a therapeutic approach for the prevention of aggravated inflammation in SARS-CoV-2 infection (264–266). A retrospective clinical trial showed that treatment with tocilizumab, a monoclonal antibody against IL-6R, reduced oxygen requirements, serum level of CRP, and hospital stays, as well as improving lymphocyte recovery and clinical outcomes in severe or critical COVID-19 (217). Another cohort study demonstrated a decreased mortality in ICU COVID-19 patients who received tocilizumab immediately after ICU admission as compared to those who did not receive early tocilizumab intervention (249). A recent study showed that the treatment of severe COVID-19 patients with sarilumab, a monoclonal antibody against IL-6R, promoted the recovery of cases with mild lung disease (267). In addition, according to a retrospective clinical trial, sarilumab treatment improved clinical symptoms and reduced serum CRP concentrations in most COVID-19 patients (268). A controlled cohort study (NCT04322188) evaluated the impact of siltuximab, a monoclonal antibody against IL-6, on the mortality rate of 30 COVID-19 patients requiring ventilator support. The results showed that siltuximab treatment, in combination with optimal supportive care, reduced the mortality rate of COVID-19 cases as compared to control patients who received only optimal supportive care (269).
5.3 Targeting IL-1 signaling
Given the important role of IL-1β in the cytokine storm, several agents targeting IL-1β signaling, including canakinumab and anakinra, have been introduced to clinical COVID-19 treatment trials (250, 270, 271). A retrospective clinical trial among 22 severe/critical COVID-19 patients indicated that >8 days of treatment with anakinra (IL-1 receptor antagonist) led to a reduced requirement for mechanical ventilation, decreased serum CRP levels, and improved clinical conditions in patients compared to the control group (251). Another cohort study found that anakinra treatment reduced several clinical parameters, including temperature, white blood cell count, and plasma levels of ferritin, creatinine, procalcitonin, and bilirubin in COVID-19 patients (272). According to a retrospective study, subcutaneous administration of canakinumab (anti-IL-1β) decreased hyperinflammation and improved PaO2/FiO2 ratio in COVID-19 patients (250). Another cohort study on non-ICU patients with mild or severe COVID-19 who received subcutaneous canakinumab showed that canakinumab treatment significantly increased the PaO2/FiO2 ratio and reduced inflammation (273). Furthermore, other studies have reported the positive effects of canakinumab during SARS-CoV-2 infection (274–276), and six other clinical trials (NCT04348448, NCT04476706, NCT04362813, NCT04365153, NCT04510493, NCT04278404) have been registered to evaluate their potential therapeutic effects in SARS-CoV-2 infection (252).
5.4 Targeting TNF-α
Since TNF-α is an initial driver of NF-κB activation and involved in the expression of several pro-inflammatory and anti-apoptotic genes in SARS-CoV-2 infection, its inhibition can attenuate excessive cytokine release and hyperinflammation in COVID-19 (28, 197, 277, 278). Recently, it has been observed that the temporary use of etanercept, a soluble TNF-α receptor fusion protein, reduces hyperinflammation in severe COVID-19 (28). A case report study demonstrated that subcutaneous etanercept administration in a 60-year-old man induced a rapid recovery from COVID-19 and did not cause any signs of respiratory failure (279). A total of four clinical trials of infliximab (anti-TNF-α) (NCT04425538, NCT04734678, NCT04593940, NCT04344249) and two clinical trials with adalimumab, another anti-TNF-α antibody (ChiCTR2000030089, NCT04705844) are ongoing to evaluate its effects in COVID-19 (252).
Interferon therapy: Some patients with life-threatening COVID-19 have been shown to have IgG autoantibodies against type I interferon. Defects, both genetic and acquired, within the IFN pathways have also been shown to predispose to severe COVID-19. Therefore, IFNs have been introduced into clinical trials in an attempt to decrease COVID-19 morbidity and mortality (280). The results of a clinical trial demonstrated that the early triple combination of IFN-1β + lopinavir/ritonavir + ribavirin could reduce clinical symptoms and hospital stay in patients with mild-to-moderate COVID-19 (238). A previous study in China found that lopinavir/ritonavir in combination with IFN-1α reduced the duration of SARS-CoV-2 shedding (281). Other clinical trials and case reports have shown that IFN-α1b and -α2b positively affect cytokine levels in the blood, virus clearance, and clinical symptoms (254, 255). On the other hand, IFNs can also induce adverse effects such as flu-like symptoms, headaches, gastrointestinal reactions, and rashes. In addition, the sustained presence of IFNs could be involved in maintaining local and systemic inflammation and causing tissue damage. The results described above suggest that interferon therapy with antiviral drugs early in the course of infection could help people with mild COVID-19; however, its practicality still remains to be seen.
5.5 Cell therapy
While current treatment options for COVID-19 are mainly nonspecific, e.g., the use of dexamethasone or anti-inflammatory agents with significant side effects, the accessibility of other approaches such as remdesivir and tocilizumab (anti-IL-6 receptor) is limited (282). These therapeutic agents have no effects on regenerating damaged tissue structures or functions. Thus, the relevance of cell therapies in the treatment of COVID-19 has received considerable attention (283). Although, particular cell types used in cell therapy for COVID-19 are mesenchymal stem cells (MSCs), NK cells, and T cells, early apoptotic cells, and other cell types are being investigated (284). MSCs exhibit strong immunomodulatory and pluripotential properties that can suppress CD3, CD8, and CD4 T cells and attenuate cytokine secretion (285, 286). MSCs have been used in several clinical trials, including graft vs. host disease (GvHD) (287), type 2 diabetes (288), autoimmune diseases (289), and spinal cord injury (290). Considering that MSCs do not express ACE2 receptors, MSC therapy can accomplish immunomodulatory effects in SARS-CoV-2 infection (291). Previous clinical trials have shown that the intravenous injection of human umbilical cord-derived MSCs is related to an attenuated cytokine storm and improves outcomes in severely ill COVID-19 patients (256). Treatment of severely ill patients who were refractory to steroids with human umbilical cord MSCs results in the healing of pulmonary lesions (292). It has also been reported that a single- dose injection of MSCs in mild, severe, and critical COVID-19 leads to improved clinical outcomes (256). A recent study indicated that COVID-19 patients were extubated approximately 7 days after injection of adipose-derived MSCs (293). Furthermore, a phase 1/2a randomized trial (NCT04355728) found that administration of umbilical cord derived-MSCs for COVID-19 patients with ARDS improved survival following therapy (294).
5.6 Intravenous immunoglobulin therapy
IVIG is a natural immunoglobulin pool prepared from the sera of healthy donors. The major component of IVIG is the serum IgG fraction, mainly IgG1 and IgG2 subclasses (295). It is well known that anti-cytokine autoantibodies, including IL-1, IL-6, and IFN-γ autoantibodies, are present in the IVIG of healthy individuals. These antibodies may be related to the anti-inflammatory effects of IVIG in inflammatory and autoimmune disorders (296–298). Previously, the promising effect of IVIG therapy has been reported in SARS and the 2009 H1N1 influenza pandemic (299, 300). Recently, the possible positive effects of IVIG administration during the early phase of COVID-19 have been reported (301). A meta-analysis in three groups of patients with non-severe, severe, or critical illness, including 825 hospitalized COVID-19 patients, indicated that IVIG might be associated with a reduced mortality rate in critically ill patients (302). Further, a significant correlation between IVIG treatment and increased survival rate and decreased COVID-19 progression has been shown (283). Moreover, the potential benefit of IVIG administration along with antiviral drugs and mechanical ventilation in severe COVID-19 patients has been reported (257). Conversely, IVIG administration, along with hydroxychloroquine and lopinavir/ritonavir, in the treatment of severe COVID-19 cases, has not been supported by recent findings (301).
6 Conclusion
At this point in the pandemic, further understanding of the SARS-CoV-2 biology and systemic host immune responses can provide information on the processes and mechanisms involved in immune-mediated viral clearance and define specific targets for the treatment of COVID-19. Recent research on the immune mechanisms in COVID-19 refer to the initiation of infection by SARS-CoV-2 accompanied by cellular immune responses, including specifically poly-functional CD4+ and CD8+ T cell responses, which may become chronic along with immune inflammation. Recovery from tissue injury, prolonged inflammation and deviations in adaptive immune activity may play a role in the postinfectious complications, termed post-COVID. The consequences of deviated immune processes also depend on genetic make-up and environmental risk factors. Due to the SARS-CoV-2 contagiousness, the increased risk of death and the need for an ICU in severe cases of the disease, there is an urgent need for long-term follow-up of the molecular and cellular mechanisms in virus-host communication at all stages of the disease. This information is needed for delineating the optimal management of infected patients in order to prevent the progression to severe forms of the disease. Currently, ongoing large clinical trials based on antiviral and immune-based treatments may soon provide efficient therapeutic agents for COVID-19 patients. In any case, vaccination still remains the best means for preventing severe illness.
Author contributions
HN, SM and KK conceived the study and wrote the manuscript. HN contributed to the final revision of the manuscript. AT, ZS, MV, PH, NK and MN participated in preparing the first draft and were involved in the final revision of the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by Academy of Finland (Grant No 336431), the Helsinki University Hospital Funds (TYH2022315,TYH2023322) and Sigrid Jusélius Foundation.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Jesenak M, Brndiarova M, Urbancikova I, Rennerova Z, Vojtkova J, Bobcakova A, et al. Immune parameters and covid-19 infection – associations with clinical severity and disease prognosis. Front Cell Infect Microbiol (2020) 10:364(364). doi: 10.3389/fcimb.2020.00364
2. Yang L, Liu S, Liu J, Zhang Z, Wan X, Huang B, et al. Covid-19: Immunopathogenesis and immunotherapeutics. Signal Transduct Target Ther (2020) 5(1):1–8. doi: 10.1038/s41392-020-00243-2
3. Wartecki A, Rzymski P. On the coronaviruses and their associations with the aquatic environment and wastewater. Water (2020) 12(6):1598. doi: 10.3390/w12061598
4. Roussel Y, Giraud-Gatineau A, Jimeno M-T, Rolain J-M, Zandotti C, Colson P, et al. Sars-Cov-2: Fear versus data. Int J Antimicrob Agents (2020) 55(5):105947. doi: 10.1016/j.ijantimicag.2020.105947
5. Guan W-j, Ni Z-y, Hu Y, Liang W-h, Ou C-q, He J-x, et al. Clinical characteristics of coronavirus disease 2019 in China. N Engl J Med (2020) 382(18):1708–20. doi: 10.1056/NEJMoa2002032
6. Daly JL, Simonetti B, Klein K, Chen K-E, Williamson MK, Antón-Plágaro C, et al. Neuropilin-1 is a host factor for sars-Cov-2 infection. Science (2020) 370(6518):861–5. doi: 10.1126/science.abd3072
7. Cantuti-Castelvetri L, Ojha R, Pedro LD, Djannatian M, Franz J, Kuivanen S, et al. Neuropilin-1 facilitates sars-Cov-2 cell entry and infectivity. Science (2020) 370(6518):856–60. doi: 10.1126/science.abd2985
8. Smith N, Goncalves P, Charbit B, Grzelak L, Beretta M, Planchais C, et al. Distinct systemic and mucosal immune responses during acute sars-Cov-2 infection. Nat Immunol (2021) 22(11):1428–39. doi: 10.1038/s41590-021-01028-7
9. Ziegler CGK, Miao VN, Owings AH, Navia AW, Tang Y, Bromley JD, et al. Impaired local intrinsic immunity to sars-Cov-2 infection in severe covid-19. Cell (2021) 184(18):4713–33.e22. doi: 10.1016/j.cell.2021.07.023
10. Zhou Z, Zhao Z, Shi S, Wu J, Li D, Li J, et al. Model-based cellular kinetic analysis of sars-Cov-2 infection: Different immune response modes and treatment strategies. arXiv preprint arXiv:210104477 (2021), Available at: https://arxiv.org/abs/2101.04477v1
11. Mahler M, Meroni PL, Infantino M, Buhler KA, Fritzler MJ. Circulating calprotectin as a biomarker of covid-19 severity. Expert Rev Clin Immunol (2021) 17(5):431–43. doi: 10.1080/1744666x.2021.1905526
12. Chen T, Wu D, Chen H, Yan W, Yang D, Chen G, et al. Clinical characteristics of 113 deceased patients with coronavirus disease 2019: Retrospective study. BMJ (2020) 368:m1091. doi: 10.1136/bmj.m1091
13. Wu C, Chen X, Cai Y, Xia J, Zhou X, Xu S, et al. Risk factors associated with acute respiratory distress syndrome and death in patients with coronavirus disease 2019 pneumonia in wuhan, China. JAMA Internal Med (2020) 180(7):934–43. doi: 10.1001/jamainternmed.2020.0994
14. Kalpakci Y, Hacibekiroglu T, Trak G, Karacaer C, Demirci T, Kocayigit H, et al. Comparative evaluation of memory T cells in covid-19 patients and the predictive role of Cd4+ Cd8+ double positive T lymphocytes as a new marker. Rev Assoc Med Bras (2020) 66:1666–72. doi: 10.1590/1806-9282.66.12.1666
15. Zhang H, Zhou P, Wei Y, Yue H, Wang Y, Hu M, et al. Histopathologic changes and sars-Cov-2 immunostaining in the lung of a patient with covid-19. Ann Internal Med (2020) 172(9):629–32. doi: 10.7326/m20-0533
16. Zhang JJ, Dong X, Liu GH, Gao YD. Risk and protective factors for covid-19 morbidity, severity, and mortality. Clin Rev Allergy Immunol (2022), 1–18. doi: 10.1007/s12016-022-08921-5
17. Sette A, Crotty S. Adaptive immunity to sars-Cov-2 and covid-19. Cell (2021) 184(4):861–80. doi: 10.1016/j.cell.2021.01.007
18. Guia S, Narni-Mancinelli E. Helper-like innate lymphoid cells in humans and mice. Trends Immunol (2020) 41(5):436–52. doi: 10.1016/j.it.2020.03.002
19. Kawai T, Akira S. The role of pattern-recognition receptors in innate immunity: Update on toll-like receptors. Nat Immunol (2010) 11(5):373–84. doi: 10.1038/ni.1863
20. Amigues I, Pearlman AH, Patel A, Reid P, Robinson PC. Coronavirus disease 2019: Investigational therapies in the prevention and treatment of hyperinflammation. Expert Rev Clin Immunol (2020) 16(12):1185–204. doi: 10.1080/1744666x.2021.1847084
21. Newton AH, Cardani A, Braciale TJ. The host immune response in respiratory virus infection: Balancing virus clearance and immunopathology. Semin immunopathol (2016) 38(4):471–82. doi: 10.1007/s00281-016-0558-0
22. Zuo Y, Estes SK, Ali RA, Gandhi AA, Yalavarthi S, Shi H, et al. Prothrombotic antiphospholipid antibodies in covid-19. medRxiv preprint server Health Sci (2020) 2020.06.15.20131607. doi: 10.1101/2020.06.15.20131607
23. Naumenko V, Turk M, Jenne CN, Kim S-J. Neutrophils in viral infection. Cell Tissue Res (2018) 371(3):505–16. doi: 10.1007/s00441-017-2763-0
24. Mohamadzadeh M, Coberley SS, Olinger GG, Kalina WV, Ruthel G, Fuller CL, et al. Activation of triggering receptor expressed on myeloid cells-1 on human neutrophils by marburg and Ebola viruses. J Virol (2006) 80(14):7235–44. doi: 10.1128/JVI.00543-06
25. Hashimoto Y, Moki T, Takizawa T, Shiratsuchi A, Nakanishi Y. Evidence for phagocytosis of influenza virus-infected, apoptotic cells by neutrophils and macrophages in mice. J Immunol (2007) 178(4):2448–57. doi: 10.4049/jimmunol.178.4.2448
26. Wang D, Hu B, Hu C, Zhu F, Liu X, Zhang J, et al. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in wuhan, China. Jama (2020) 323(11):1061–9. doi: 10.1001/jama.2020.1585
27. Du RH, Liang LR, Yang CQ, Wang W, Cao TZ, Li M, et al. Predictors of mortality for patients with covid-19 pneumonia caused by sars-Cov-2: A prospective cohort study. Eur Respir J (2020) 55(5); 2000524. doi: 10.1183/13993003.00524-2020
28. Schulte-Schrepping J, Reusch N, Paclik D, Baßler K, Schlickeiser S, Zhang B, et al. Severe covid-19 is marked by a dysregulated myeloid cell compartment. Cell (2020) 182(6):1419–40.e23. doi: 10.1016/j.cell.2020.08.001
29. Liu J, Li S, Liu J, Liang B, Wang X, Wang H, et al. Longitudinal characteristics of lymphocyte responses and cytokine profiles in the peripheral blood of sars-Cov-2 infected patients. EBioMedicine (2020) 55:102763. doi: 10.1016/j.ebiom.2020.102763
30. Fu J, Kong J, Wang W, Wu M, Yao L, Wang Z, et al. The clinical implication of dynamic neutrophil to lymphocyte ratio and d-dimer in covid-19: A retrospective study in suzhou China. Thromb Res (2020) 192:3–8. doi: 10.1016/j.thromres.2020.05.006
31. Middleton EA, He XY, Denorme F, Campbell RA, Ng D, Salvatore SP, et al. Neutrophil extracellular traps contribute to immunothrombosis in covid-19 acute respiratory distress syndrome. Blood (2020) 136(10):1169–79. doi: 10.1182/blood.2020007008
32. Zuo Y, Yalavarthi S, Shi H, Gockman K, Zuo M, Madison JA, et al. Neutrophil extracellular traps in covid-19. JCI Insight (2020) 5(11): e138999. doi: 10.1172/jci.insight.138999
33. Saheb Sharif-Askari N, Saheb Sharif-Askari F, Mdkhana B, Hussain Alsayed HA, Alsafar H, Alrais ZF, et al. Upregulation of oxidative stress gene markers during sars-Cov-2 viral infection. Free Radical Biol Med (2021) 172:688–98. doi: 10.1016/j.freeradbiomed.2021.06.018
34. Laforge M, Elbim C, Frère C. Tissue damage from neutrophil-induced oxidative stress in covid-19. Nat Rev Immunol (2020) 20(9):515–6. doi: 10.1038/s41577-020-0407-1
35. Shibabaw T. Inflammatory cytokine: Il-17a signaling pathway in patients present with covid-19 and current treatment strategy. J Inflammation Res (2020) 13:673–80. doi: 10.2147/jir.s278335
36. Sampath P, Moideen K, Ranganathan UD, Bethunaickan R. Monocyte subsets: Phenotypes and function in tuberculosis infection. Front Immunol (2018) 9:1726. doi: 10.3389/fimmu.2018.01726
37. Pence BD. Severe covid-19 and aging: Are monocytes the key? GeroScience (2020) 42(4):1051–61. doi: 10.1007/s11357-020-00213-0
38. Boyette LB, Macedo C, Hadi K, Elinoff BD, Walters JT, Ramaswami B, et al. Phenotype, function, and differentiation potential of human monocyte subsets. PloS One (2017) 12(4):e0176460. doi: 10.1371/journal.pone.0176460
39. Ong S-M, Hadadi E, Dang T-M, Yeap W-H, Tan CT-Y, Ng T-P, et al. The pro-inflammatory phenotype of the human non-classical monocyte subset is attributed to senescence. Cell Death Dis (2018) 9(3):1–12. doi: 10.1038/s41419-018-0327-1
40. Martinez FO, Combes TW, Orsenigo F, Gordon S. Monocyte activation in systemic covid-19 infection: Assay and rationale. EBioMedicine (2020) 59:102964. doi: 10.1016/j.ebiom.2020.102964
41. Qin S, Jiang Y, Wei X, Liu X, Guan J, Chen Y, et al. Dynamic changes in monocytes subsets in covid-19 patients. Hum Immunol (2021) 82(3):170–6. doi: 10.1016/j.humimm.2020.12.010
42. Liao M, Liu Y, Yuan J, Wen Y, Xu G, Zhao J, et al. Single-cell landscape of bronchoalveolar immune cells in patients with covid-19. Nat Med (2020) 26(6):842–4. doi: 10.1038/s41591-020-0901-9
43. Zhou Y, Fu B, Zheng X, Wang D, Zhao C, Qi Y, et al. Pathogenic T-cells and inflammatory monocytes incite inflammatory storms in severe covid-19 patients. Natl Sci Rev (2020) 7(6):998–1002. doi: 10.1093/nsr/nwaa041
44. Zhang D, Guo R, Lei L, Liu H, Wang Y, Wang Y, et al. Covid-19 infection induces readily detectable morphologic and inflammation-related phenotypic changes in peripheral blood monocytes. J Leukoc Biol (2020) 109 (1):13–22. doi: 10.1002/JLB.4HI0720-470R
45. Silvin A, Chapuis N, Dunsmore G, Goubet AG, Dubuisson A, Derosa L, et al. Elevated calprotectin and abnormal myeloid cell subsets discriminate severe from mild covid-19. Cell (2020) 182(6):1401–18.e18. doi: 10.1016/j.cell.2020.08.002
46. Sanchez-Cerrillo I, Landete P, Aldave B, Sanchez-Alonso S, Sanchez-Azofra A, Marcos-Jimenez A, et al. Differential redistribution of activated monocyte and dendritic cell subsets to the lung associates with severity of covid-19. medRxiv preprint server Health Sci (2020) 2020.05.13.20100925. doi: 10.1101/2020.05.13.20100925
47. Meidaninikjeh S, Sabouni N, Marzouni HZ, Bengar S, Khalili A, Jafari R. Monocytes and macrophages in covid-19: Friends and foes. Life Sci (2021), 269:119010. doi: 10.1016/j.lfs.2020.119010
48. Hussell T, Bell TJ. Alveolar macrophages: Plasticity in a tissue-specific context. Nat Rev Immunol (2014) 14(2):81–93. doi: 10.1038/nri3600
49. Franke-Ullmann G, Pförtner C, Walter P, Steinmüller C, Lohmann-Matthes ML, Kobzik L. Characterization of murine lung interstitial macrophages in comparison with alveolar macrophages in vitro. J Immunol (1996) 157(7):3097–104. doi: 10.4049/jimmunol.157.7.3097
50. Helms J, Kremer S, Merdji H. Neurologic features in severe sars-Cov-2 infection. N Engl J Med (2020) 382(23):2268–70. doi: 10.1056/NEJMc2008597
51. Nemati M, Malla N, Yadav M, Khorramdelazad H, Jafarzadeh A. Humoral and T cell–mediated immune response against trichomoniasis. Parasite Immunol (2018) 40(3):e12510. doi: 10.1111/pim.12510
52. Wu A, Peng Y, Huang B, Ding X, Wang X, Niu P, et al. Genome composition and divergence of the novel coronavirus (2019-ncov) originating in China. Cell Host Microbe (2020) 27(3):325–8. doi: 10.1016/j.chom.2020.02.001
53. Kosyreva A, Dzhalilova D, Lokhonina A, Vishnyakova P, Fatkhudinov T. The role of macrophages in the pathogenesis of sars-Cov-2-Associated acute respiratory distress syndrome. Front Immunol (2021) 12:682871. doi: 10.3389/fimmu.2021.682871
54. Jafarzadeh A, Chauhan P, Saha B, Jafarzadeh S, Nemati M. Contribution of monocytes and macrophages to the local tissue inflammation and cytokine storm in covid-19: Lessons from sars and mers, and potential therapeutic interventions. Life Sci (2020), 257:118102. doi: 10.1016/j.lfs.2020.118102
55. Tay MZ, Poh CM, Rénia L, MacAry PA, Ng LF. The trinity of covid-19: Immunity, inflammation and intervention. Nat Rev Immunol (2020) 20(6):363–74. doi: 10.1038/s41577-020-0311-8
56. Wang Y, Ding Q, Lu YC, Cao SY, Liu QX, Zhang L. Interferon-stimulated gene 15 enters posttranslational modifications of P53. J Cell Physiol (2019) 234(5):5507–18. doi: 10.1002/jcp.27347
57. Nile SH, Nile A, Qiu J, Li L, Jia X, Kai G. Covid-19: Pathogenesis, cytokine storm and therapeutic potential of interferons. Cytokine Growth factor Rev (2020) 53:66–70. doi: 10.1016/j.cytogfr.2020.05.002
58. Bost P, Giladi A, Liu Y, Bendjelal Y, Xu G, David E, et al. Host-viral infection maps reveal signatures of severe covid-19 patients. Cell (2020) 181(7):1475–88.e12. doi: 10.1016/j.cell.2020.05.006
59. Borges RC, Hohmann MS, Borghi SM. Dendritic cells in covid-19 immunopathogenesis: Insights for a possible role in determining disease outcome. Int Rev Immunol (2021) 40(1-2):108–25. doi: 10.1080/08830185.2020.1844195
60. Banchereau J, Steinman RM. Dendritic cells and the control of immunity. Nature (1998) 392(6673):245–52. doi: 10.1038/32588
61. Pollara G, Kwan A, Newton PJ, Handley ME, Chain BM, Katz DR. Dendritic cells in viral pathogenesis: Protective or defective? Int J Exp Pathol (2005) 86(4):187–204. doi: 10.1111/j.0959-9673.2005.00440.x
62. Wack A. Monocyte and dendritic cell defects in covid-19. Nat Cell Biol (2021) 23(5):445–7. doi: 10.1038/s41556-021-00685-y
63. Bencze D, Fekete T, Pázmándi K. Correlation between type I interferon associated factors and covid-19 severity. Int J Mol Sci (2022) 23(18): 10968. doi: 10.3390/ijms231810968
64. Wang X, Bai H, Ma J, Qin H, Zeng Q, Hu F, et al. Identification of distinct immune cell subsets associated with asymptomatic infection, disease severity, and viral persistence in covid-19 patients. Front Immunol (2022) 13:812514. doi: 10.3389/fimmu.2022.812514
65. Rajamanickam A, Kumar NP, Pandiaraj AN, Selvaraj N, Munisankar S, Renji RM, et al. Restoration of dendritic cell homeostasis and type I/Type iii interferon levels in convalescent covid-19 individuals. BMC Immunol (2022) 23(1):51. doi: 10.1186/s12865-022-00526-z
66. Falck-Jones S, Österberg B, Smed-Sörensen A. Respiratory and systemic monocytes, dendritic cells, and myeloid-derived suppressor cells in covid-19: Implications for disease severity. J Internal Med (2022) 293 (2):130–143. doi: 10.1111/joim.13559
67. Winheim E, Rinke L, Lutz K, Reischer A, Leutbecher A, Wolfram L, et al. Impaired function and delayed regeneration of dendritic cells in covid-19. PloS Pathog (2021) 17(10):e1009742. doi: 10.1371/journal.ppat.1009742
68. Marongiu L, Protti G, Facchini FA, Valache M, Mingozzi F, Ranzani V, et al. Maturation signatures of conventional dendritic cell subtypes in covid-19 reflect direct viral sensing. bioRxiv (2021) 52 (1):109–122. doi: 10.1101/2021.03.03.433597
69. Marongiu L, Protti G, Facchini FA, Valache M, Mingozzi F, Ranzani V, et al. Maturation signatures of conventional dendritic cell subtypes in covid-19 suggest direct viral sensing. Eur J Immunol (2022) 52(1):109–22. doi: 10.1002/eji.202149298
70. Borcherding L, Teksen AS, Grosser B, Schaller T, Hirschbühl K, Claus R, et al. Impaired dendritic cell homing in covid-19. Front Med (2021) 8:761372. doi: 10.3389/fmed.2021.761372
71. Kulkarni-Munje A, Palkar S, Shrivastava S, Lalwani S, Mishra AC, Arankalle VA. Disease-duration based comparison of subsets of immune cells in sars cov-2 infected patients presenting with mild or severe symptoms identifies prognostic markers for severity. Immun Inflammation Dis (2021); 9 (2):419–434. doi: 10.1002/iid3.402
72. Matic S, Popovic S, Djurdjevic P, Todorovic D, Djordjevic N, Mijailovic Z, et al. Sars-Cov-2 infection induces mixed M1/M2 phenotype in circulating monocytes and alterations in both dendritic cell and monocyte subsets. PloS One (2020) 15(12):e0241097. doi: 10.1371/journal.pone.0241097
73. García M, Kokkinou E, Carrasco García A, Parrot T, Palma Medina LM, Maleki KT, et al. Innate lymphoid cell composition associates with covid-19 disease severity. Clin Trans Immunol (2020) 9(12):e1224. doi: 10.1002/cti2.1224
74. Hildreth AD, O’Sullivan TE. Tissue-resident innate and innate-like lymphocyte responses to viral infection. Viruses (2019) 11(3):272. doi: 10.3390/v11030272
75. Vabret N, Britton GJ, Gruber C, Hegde S, Kim J, Kuksin M, et al. Immunology of covid-19: Current state of the science. Immunity (2020) 52 (6):910–941. doi: 10.1016/j.immuni.2020.05.002
76. Velavan TP, Meyer CG. Mild versus severe covid-19: Laboratory markers. Int J Infect Dis (2020) 95:304–7. doi: 10.1016/j.ijid.2020.04.061
77. Zhou F, Yu T, Du R, Fan G, Liu Y, Liu Z, et al. Clinical course and risk factors for mortality of adult inpatients with covid-19 in wuhan, China: A retrospective cohort study. Lancet (2020) 395(10229):1054–62. doi: 10.1016/S0140-6736(20)30566-3
78. Monticelli LA, Sonnenberg GF, Abt MC, Alenghat T, Ziegler CG, Doering TA, et al. Innate lymphoid cells promote lung-tissue homeostasis after infection with influenza virus. Nat Immunol (2011) 12(11):1045–54. doi: 10.1031/ni.2131
79. Gomez-Cadena A, Spehner L, Kroemer M, Khelil MB, Bouiller K, Verdeil G, et al. Severe covid-19 patients exhibit an Ilc2 Nkg2d+ population in their impaired ilc compartment. Cell Mol Immunol (2021) 18(2):484–6. doi: 10.1038/s41423-020-00596-2
80. van Eeden C, Khan L, Osman MS, Cohen Tervaert JW. Natural killer cell dysfunction and its role in covid-19. Int J Mol Sci (2020) 21(17): 6351. doi: 10.3390/ijms21176351
81. Song J-W, Zhang C, Fan X, Meng F-P, Xu Z, Xia P, et al. Immunological and inflammatory profiles in mild and severe cases of covid-19. Nat Commun (2020) 11(1):1–10. doi: 10.1038/s41467-020-17240-2
82. Mazzoni A, Salvati L, Maggi L, Capone M, Vanni A, Spinicci M, et al. Impaired immune cell cytotoxicity in severe covid-19 is il-6 dependent. J Clin Invest (2020) 130(9):4694–703. doi: 10.1172/jci138554
83. Carsetti R, Zaffina S, Piano Mortari E, Terreri S, Corrente F, Capponi C, et al. Different innate and adaptive immune responses to sars-Cov-2 infection of asymptomatic, mild, and severe cases. Front Immunol (2020) 11:610300. doi: 10.3389/fimmu.2020.610300
84. Leem G, Cheon S, Lee H, Choi SJ, Jeong S, Kim ES, et al. Abnormality in the nk-cell population is prolonged in severe covid-19 patients. J Allergy Clin Immunol (2021) 148(4):996–1006.e18. doi: 10.1016/j.jaci.2021.07.022
85. Bordoni V, Sacchi A, Cimini E, Notari S, Grassi G, Tartaglia E, et al. An inflammatory profile correlates with decreased frequency of cytotoxic cells in coronavirus disease 2019. Clin Infect Dis (2020) 71(16):2272–5. doi: 10.1093/cid/ciaa577
86. Malengier-Devlies B, Filtjens J, Ahmadzadeh K, Boeckx B, Vandenhaute J, De Visscher A, et al. Severe covid-19 patients display hyper-activated nk cells and nk cell-platelet aggregates. Front Immunol (2022), 13:5004. doi: 10.3389/fimmu.2022.861251
87. Di Vito C, Calcaterra F, Coianiz N, Terzoli S, Voza A, Mikulak J, et al. Natural killer cells in sars-Cov-2 infection: Pathophysiology and therapeutic implications. Front Immunol (2022) 13:888248. doi: 10.3389/fimmu.2022.888248
88. Hajeer A, Jawdat D, Massadeh S, Aljawini N, Abedalthagafi MS, Arabi YM, et al. Association of kir gene polymorphisms with covid-19 disease. Clin Immunol (2022) 234:108911. doi: 10.1016/j.clim.2021.108911
89. Maruthamuthu S, Rajalingam K, Kaur N, Morvan MG, Soto J, Lee N, et al. Individualized constellation of killer cell immunoglobulin-like receptors and cognate hla class I ligands that controls natural killer cell antiviral immunity predisposes covid-19. Front Genet (2022) 13. doi: 10.3389/fgene.2022.845474
90. Siracusa MC, Kim BS, Spergel JM, Artis D. Basophils and allergic inflammation. J Allergy Clin Immunol (2013) 132(4):789–801. doi: 10.1016/j.jaci.2013.07.046
91. Negro F. Facts and fictions of hcv and comorbidities: Steatosis, diabetes mellitus, and cardiovascular diseases. J Hepatol (2014) 61(1 Suppl):S69–78. doi: 10.1016/j.jhep.2014.08.003
92. Kazancioglu S, Bastug A, Ozbay BO, Kemirtlek N, Bodur H. The role of haematological parameters in patients with covid-19 and influenza virus infection. Epidemiol Infect (2020) 148: e272. doi: 10.1017/S095026882000271X
93. Vitte J, Diallo AB, Boumaza A, Lopez A, Michel M, Allardet-Servent J, et al. A granulocytic signature identifies covid-19 and its severity. J Infect Dis (2020) 222(12):1985–96. doi: 10.1093/infdis/jiaa591
94. Hogan SP, Rosenberg HF, Moqbel R, Phipps S, Foster PS, Lacy P, et al. Eosinophils: Biological properties and role in health and disease. Clin Exp Allergy (2008) 38(5):709–50. doi: 10.1111/j.1365-2222.2008.02958.x
95. Gao Y-d, Agache I, Akdis M, Nadeau K, Klimek L, Jutel M, et al. The effect of allergy and asthma as a comorbidity on the susceptibility and outcomes of covid-19. Int Immunol (2022) 34(4):177–88. doi: 10.1093/intimm/dxab107
96. Xie G, Ding F, Han L, Yin D, Lu H, Zhang M. The role of peripheral blood eosinophil counts in covid-19 patients. Allergy (2021) 76(2):471–82. doi: 10.1111/all.14465
97. Eggert LE, He Z, Collins W, Lee AS, Dhondalay G, Jiang SY, et al. Asthma phenotypes, associated comorbidities, and long-term symptoms in covid-19. Allergy (2022) 77(1):173–85. doi: 10.1111/all.14972
98. Sacchi A, Grassi G, Bordoni V, Lorenzini P, Cimini E, Casetti R, et al. Early expansion of myeloid-derived suppressor cells inhibits sars-Cov-2 specific T-cell response and may predict fatal covid-19 outcome. Cell Death Dis (2020) 11(10):1–9. doi: 10.1038/s41419-020-03125-1
99. Wang C, Zhang N, Qi L, Yuan J, Wang K, Wang K, et al. Myeloid-derived suppressor cells inhibit T follicular helper cell immune response in Japanese encephalitis virus infection. J Immunol (2017) 199(9):3094–105. doi: 10.4049/jimmunol.1700671
100. Agrati C, Sacchi A, Bordoni V, Cimini E, Notari S, Grassi G, et al. Expansion of myeloid-derived suppressor cells in patients with severe coronavirus disease (Covid-19). Cell Death Differ (2020) 27(11):3196–207. doi: 10.1038/s41418-020-0572-6
101. Xue G, Jiang M, Zhao R, Le A, Li J. Elevated frequencies of Cd14(+)Hla-Dr(Lo/Neg) mdscs in covid-19 patients. Aging (2021) 13(5):6236–46. doi: 10.18632/aging.202571
102. Reizine F, Lesouhaitier M, Gregoire M, Pinceaux K, Gacouin A, Maamar A, et al. Sars-Cov-2-Induced Ards associates with mdsc expansion, lymphocyte dysfunction, and arginine shortage. J Clin Immunol, 41(3):1–11. doi: 10.1007/s10875-020-00920-5
103. Dean MJ, Ochoa JB, Sanchez-Pino MD, Zabaleta J, Garai J, Del Valle L, et al. Severe covid-19 is characterized by an impaired type I interferon response and elevated levels of arginase producing granulocytic myeloid derived suppressor cells. Front Immunol (2021) 12:695972. doi: 10.3389/fimmu.2021.695972
104. De Santo C, Salio M, Masri SH, Lee LY-H, Dong T, Speak AO, et al. Invariant nkt cells reduce the immunosuppressive activity of influenza a virus–induced myeloid-derived suppressor cells in mice and humans. J Clin Invest (2008) 118(12):4036–48. doi: 10.1172/JCI36264
105. Johnson TR, Hong S, Van Kaer L, Koezuka Y, Graham BS. Nk T cells contribute to expansion of Cd8+ T cells and amplification of antiviral immune responses to respiratory syncytial virus. J Virol (2002) 76(9):4294–303. doi: 10.1128/jvi.76.9.4294-4303.2002
106. Chen N, McCarthy C, Drakesmith H, Li D, Cerundolo V, McMichael AJ, et al. Hiv-1 down-regulates the expression of Cd1d Via nef. Eur J Immunol (2006) 36(2):278–86. doi: 10.1002/eji.200535487
107. Miura S, Kawana K, Schust DJ, Fujii T, Yokoyama T, Iwasawa Y, et al. Cd1d, a sentinel molecule bridging innate and adaptive immunity, is downregulated by the human papillomavirus (Hpv) E5 protein: A possible mechanism for immune evasion by hpv. J Virol (2010) 84(22):11614–23. doi: 10.1128/JVI.01053-10
108. Rao P, Pham HT, Kulkarni A, Yang Y, Liu X, Knipe DM, et al. Herpes simplex virus 1 glycoprotein b and Us3 collaborate to inhibit Cd1d antigen presentation and nkt cell function. J Virol (2011) 85(16):8093–104. doi: 10.1128/JVI.02689-10
109. Sanchez DJ, Gumperz JE, Ganem D. Regulation of Cd1d expression and function by a herpesvirus infection. J Clin Invest (2005) 115(5):1369–78. doi: 10.1172/JCI24041
110. Webb TJR, Litavecz RA, Khan MA, Du W, Gervay-Hague J, Renukaradhya GJ, et al. Inhibition of Cd1d1-mediated antigen presentation by the vaccinia virus B1r and H5r molecules. Eur J Immunol (2006) 36(10):2595–600. doi: 10.1002/eji.200636024
111. Zingaropoli MA, Perri V, Pasculli P, Dezza FC, Nijhawan P, Savelloni G, et al. Major reduction of nkt cells in patients with severe covid-19 pneumonia. Clin Immunol (2021) 222:108630. doi: 10.1016/j.clim.2020.108630
112. Zhang JY, Wang XM, Xing X, Xu Z, Zhang C. Single-cell landscape of immunological responses in patients with covid-19. Nat Immunol (2020) 21(9):1107–18. doi: 10.1038/s41590-020-0762-x
113. Jouan Y, Guillon A, Gonzalez L, Perez Y, Boisseau C, Ehrmann S, et al. Phenotypical and functional alteration of unconventional T cells in severe covid-19 patients. J Exp Med (2020) 217(12):e20200872. doi: 10.1084/jem.20200872
114. Lei L, Qian H, Yang X, Zhang X, Zhang D, Dai T, et al. The phenotypic changes of Γδ T cells in covid-19 patients. J Cell Mol Med (2020) 24(19):11603–6. doi: 10.1111/jcmm.15620
115. Odak I, Barros-Martins J, Bošnjak B, Stahl K, David S, Wiesner O, et al. Reappearance of effector T cells is associated with recovery from covid-19. EBioMedicine (2020) 57:102885. doi: 10.1016/j.ebiom.2020.102885
116. Mastellos DC, Ricklin D, Lambris JD. Clinical promise of next-generation complement therapeutics. Nat Rev Drug Discovery (2019) 18(9):707–29. doi: 10.1038/s41573-019-0031-6
117. Noris M, Benigni A, Remuzzi G. The case of complement activation in covid-19 multiorgan impact. Kidney Int (2020) 98(2):314–322. doi: 10.1016/j.kint.2020.05.013
118. Lo MW, Kemper C, Woodruff TM. Covid-19: Complement, coagulation, and collateral damage. J Immunol (2020) 205(6):1488–95. doi: 10.4049/jimmunol.2000644
119. Ip WK, Chan KH, Law HK, Tso GH, Kong EK, Wong WH, et al. Mannose-binding lectin in severe acute respiratory syndrome coronavirus infection. J Infect Dis (2005) 191(10):1697–704. doi: 10.1086/429631
120. Java A, Apicelli AJ, Liszewski MK, Coler-Reilly A, Atkinson JP, Kim AH, et al. The complement system in covid-19: Friend and foe? JCI Insight (2020) 5(15):e140711. doi: 10.1172/jci.insight.140711
121. Zinellu A, Mangoni AA. Serum complement C3 and C4 and covid-19 severity and mortality: A systematic review and meta-analysis with meta-regression. Front Immunol (2021) 12:696085. doi: 10.3389/fimmu.2021.696085
122. Risitano AM, Mastellos DC, Huber-Lang M, Yancopoulou D, Garlanda C, Ciceri F, et al. Complement as a target in covid-19? Nat Rev Immunol (2020) 20(6):343–4. doi: 10.1038/s41577-020-0320-7
123. Carvelli J, Demaria O, Vély F, Batista L, Benmansour NC, Fares J, et al. Association of covid-19 inflammation with activation of the C5a–C5ar1 axis. Nature (2020), 588(7836): 1–5. doi: 10.1038/s41586-020-2600-6
124. Channappanavar R, Zhao J, Perlman S. T Cell-mediated immune response to respiratory coronaviruses. Immunol Res (2014) 59(1):118–28. doi: 10.1007/s12026-014-8534-z
125. Xie L-X. Interpretation of the 7th edition of the “Diagnosis and treatment guidelines of coronavirus disease 2019 in china”: Progress and challenges. Chronic Dis Transl Med (2020) 6(2):75. doi: 10.1016/j.cdtm.2020.04.001
126. Yan L, Zhang H-T, Goncalves J, Xiao Y, Wang M, Guo Y, et al. An interpretable mortality prediction model for covid-19 patients. Nat Mach Intell (2020) 2(5):283–8. doi: 10.1038/s42256-020-0180-7
127. Dai W, Zhong A, Qiao Q, Wu J, Li W, Wu Q, et al. Characteristics of lymphocyte subset alterations in covid-19 patients with different levels of disease severity. Virol J (2022) 19(1):1–14. doi: 10.1186/s12985-022-01926-8
128. Rahimmanesh I, Kouhpayeh S, Azizi Y, Khanahmad H. Conceptual framework for Sars-Cov-2–related lymphopenia. Adv Biomed Res (2022), 11:16. doi: 10.4103/abr.abr_303_20
129. Carbajosa S, Gea S, Chillón-arinas C, Poveda C, del Carmen Maza M, Fresno M, et al. Altered bone marrow lymphopoiesis and interleukin-6-Dependent inhibition of thymocyte differentiation contribute to thymic atrophy during trypanosoma cruzi infection. Oncotarget (2017) 8(11):17551. doi: 10.18632/oncotarget.14886
130. Diao B, Wang C, Tan Y, Chen X, Liu Y, Ning L, et al. Reduction and functional exhaustion of T cells in patients with coronavirus disease 2019 (Covid-19). Front Immunol (2020) 11:827. doi: 10.3389/fimmu.2020.00827
131. Urra J, Cabrera C, Porras L, Ródenas I. Selective Cd8 cell reduction by sars-Cov-2 is associated with a worse prognosis and systemic inflammation in covid-19 patients. Clin Immunol (2020) 217:108486. doi: 10.1016/j.clim.2020.108486
132. Niedźwiedzka-Rystwej P, Majchrzak A, Kurkowska S, Małkowska P, Sierawska O, Hrynkiewicz R, et al. Immune signature of covid-19: In-depth reasons and consequences of the cytokine storm. Int J Mol Sci (2022) 23(9):4545. doi: 10.3390/ijms23094545
133. Kang CK, Han G-C, Kim M, Kim G, Shin HM, Song K-H, et al. Aberrant hyperactivation of cytotoxic T-cell as a potential determinant of covid-19 severity. Int J Infect Dis (2020) 97:313–21. doi: 10.1016/j.ijid.2020.05.106
134. Georg P, Astaburuaga-García R, Bonaguro L, Brumhard S, Michalick L, Lippert LJ, et al. Complement activation induces excessive T cell cytotoxicity in severe covid-19. Cell (2022) 185(3):493–512.e25. doi: 10.1016/j.cell.2021.12.040
135. Beserra DR, Alberca RW, Branco ACCC, Oliveira LM, de Souza Andrade MM, Gozzi-Silva SC, et al. Upregulation of pd-1 expression and high spd-L1 levels associated with covid-19 severity. J Immunol Res (2022) 2022: 9764002. doi: 10.1155/2022/9764002
136. BenMohamed L, Srivastava R, Dhanushkodi N, Prakash S, Coulon P-G, Vahed H, et al. High frequencies of phenotypically and functionally senescent and exhausted Cd56+ Cd57+ pd-1+ natural killer cells, sars-Cov-2-Specific memory Cd4+ and Cd8+ T cells associated with severe disease in unvaccinated covid-19 patients. bioRxiv (2022) 2022.07.26.501655. doi: 10.1101/2022.07.26.501655
137. Ronit A, Berg RM, Bay JT, Haugaard AK, Ahlström MG, Burgdorf KS, et al. Compartmental immunophenotyping in covid-19 Ards: A case series. J Allergy Clin Immunol (2021) 147(1):81–91. doi: 10.1016/j.jaci.2020.09.009
138. Sekine T, Perez-Potti A, Rivera-Ballesteros O, Strålin K, Gorin J-B, Olsson A, et al. Robust T cell immunity in convalescent individuals with asymptomatic or mild covid-19. Cell (2020) 183(1):158–68. e14. doi: 10.1016/j.cell.2020.08.017
139. Qin C, Zhou L, Hu Z, Zhang S, Yang S, Tao Y, et al. Dysregulation of immune response in patients with coronavirus 2019 (Covid-19) in wuhan, China. Clin Infect Dis (2020) 71(15):762–8. doi: 10.1093/cid/ciaa248
140. Liu Z, Long W, Tu M, Chen S, Huang Y, Wang S, et al. Lymphocyte subset (Cd4+, Cd8+) counts reflect the severity of infection and predict the clinical outcomes in patients with covid-19. J infect (2020) 81(2):318–56. doi: 10.1016/j.jinf.2020.03.054
141. Mazzoni A, Maggi L, Capone M, Spinicci M, Salvati L, Colao MG, et al. Cell-mediated and humoral adaptive immune responses to sars-Cov-2 are lower in asymptomatic than symptomatic covid-19 patients. Eur J Immunol (2020) 50(12):2013–24. doi: 10.1002/eji.202048915
142. Martonik D, Parfieniuk-Kowerda A, Rogalska M, Flisiak R. The role of Th17 response in covid-19. Cells (2021) 10(6):1550. doi: 10.3390/cells10061550
143. Neidleman J, Luo X, Frouard J, Xie G, Gill G, Stein ES, et al. Sars-Cov-2-Specific T cells exhibit phenotypic features of helper function, lack of terminal differentiation, and high proliferation potential. Cell Rep Med (2020) 1(6):100081. doi: 10.1016/j.xcrm.2020.100081
144. Verhagen J, van der Meijden ED, Lang V, Kremer AE, Völkl S, Mackensen A, et al. Human Cd4+ T cells specific for dominant epitopes of sars-Cov-2 spike and nucleocapsid proteins with therapeutic potential. Clin Exp Immunol (2021) 205 (3):363–378. doi: 10.1111/cei.13627
145. Sadeghi A, Tahmasebi S, Mahmood A, Kuznetsova M, Valizadeh H, Taghizadieh A, et al. Th17 and treg cells function in sars-Cov2 patients compared with healthy controls. J Cell Physiol (2021) 236(4):2829–39. doi: 10.1002/jcp.30047
146. Gil-Etayo FJ, Suàrez-Fernández P, Cabrera-Marante O, Arroyo D, Garcinuño S, Naranjo L, et al. T-Helper cell subset response is a determining factor in covid-19 progression. Front Cell Infect Microbiol (2021) 11:624483. doi: 10.3389/fcimb.2021.624483
147. Aleebrahim-Dehkordi E, Molavi B, Mokhtari M, Deravi N, Fathi M, Fazel T, et al. T Helper type (Th1/Th2) responses to sars-Cov-2 and influenza a (H1n1) virus: From cytokines produced to immune responses. Transplant Immunol (2022) 70:101495. doi: 10.1016/j.trim.2021.101495
148. Parackova Z, Bloomfield M, Klocperk A, Sediva A. Neutrophils mediate Th17 promotion in covid-19 patients. J Leukoc Biol (2021) 109(1):73–6. doi: 10.1002/JLB.4COVCRA0820-481RRR
149. Wei Ll, Wang Wj, Chen Dx, Xu B. Dysregulation of the immune response affects the outcome of critical covid-19 patients. J Med Virol (2020) 92(11):2768–76. doi: 10.1002/jmv.26181
150. Muyayalo KP, Huang DH, Zhao SJ, Xie T, Mor G, Liao AH. Covid-19 and Treg/Th17 imbalance: Potential relationship to pregnancy outcomes. Am J Reprod Immunol (2020) 84(5):e13304. doi: 10.1111/aji.13304
151. Wu D, Yang XO. Th17 responses in cytokine storm of covid-19: An emerging target of Jak2 inhibitor fedratinib. J Microbiol Immunol Infect (2020) 53(3):368–70. doi: 10.1016/j.jmii.2020.03.005
152. Tahmasebi S, El-Esawi MA, Mahmoud ZH, Timoshin A, Valizadeh H, Roshangar L, et al. Immunomodulatory effects of nanocurcumin on Th17 cell responses in mild and severe covid-19 patients. J Cell Physiol (2021) 236(7):5325–38. doi: 10.1002/jcp.30233
153. Orlov M, Wander PL, Morrell ED, Mikacenic C, Wurfel MM. A case for targeting Th17 cells and il-17a in sars-Cov-2 infections. J Immunol (2020) 205(4):892–8. doi: 10.4049/jimmunol.2000554
154. Zheng M, Gao Y, Wang G, Song G, Liu S, Sun D, et al. Functional exhaustion of antiviral lymphocytes in covid-19 patients. Cell Mol Immunol (2020) 17(5):533–5. doi: 10.1038/s41423-020-0402-2
155. Wang J, Li Q, Qiu Y, Lu H. Covid-19: Imbalanced cell-mediated immune response drives to immunopathology. Emerg Microbes Infect (2022) 11(1):2393–404. doi: 10.1080/22221751.2022.2122579
156. Choto TA, Makupe I, Cakana AZ, Sibanda EN, Mduluza T. Excessive neutrophil recruitment promotes typical T-helper 17 responses in coronavirus disease 2019 patients. PloS One (2022) 17(8):e0273186. doi: 10.1371/journal.pone.0273186
157. Tahmasebi S, Saeed BQ, Temirgalieva E, Yumashev AV, El-Esawi MA, Navashenaq JG, et al. Nanocurcumin improves treg cell responses in patients with mild and severe sars-Cov2. Life Sci (2021) 276:119437. doi: 10.1016/j.lfs.2021.119437
158. Alahyari S, Rajaeinejad M, Jalaeikhoo H, Amani D. Regulatory T cells in immunopathogenesis and severity of covid-19: A systematic review. Arch Iranian Med (2022) 25(2):127–32. doi: 10.34172/aim.2022.22
159. Carissimo G, Xu W, Kwok I, Abdad MY, Chan Y-H, Fong S-W, et al. Whole blood immunophenotyping uncovers immature neutrophil-to-Vd2 T-cell ratio as an early marker for severe covid-19. Nat Commun (2020) 11(1):1–12. doi: 10.1038/s41467-020-19080-6
160. Chen G, Wu D, Guo W, Cao Y, Huang D, Wang H, et al. Clinical and immunological features of severe and moderate coronavirus disease 2019. J Clin Invest (2020) 130(5):2620–9. doi: 10.1172/JCI137244
161. Kalfaoglu B, Almeida-Santos J, Tye CA, Satou Y, Ono M. T-Cell hyperactivation and paralysis in severe covid-19 infection revealed by single-cell analysis. Front Immunol (2020) 11:589380. doi: 10.3389/fimmu.2020.589380
162. Chu H, Zhou J, Wong BH-Y, Li C, Chan JF-W, Cheng Z-S, et al. Middle East respiratory syndrome coronavirus efficiently infects human primary T lymphocytes and activates the extrinsic and intrinsic apoptosis pathways. J Infect Dis (2016) 213(6):904–14. doi: 10.1093/infdis/jiv380
163. Neurath MF. Covid-19 and immunomodulation in ibd. Gut (2020) 69(7):1335–42. doi: 10.1136/gutjnl-2020-321269
164. Taylor GA, Feng CG, Sher A. P47 gtpases: Regulators of immunity to intracellular pathogens. Nat Rev Immunol (2004) 4(2):100–9. doi: 10.1038/nri1270
165. Peng Y, Mentzer AJ, Liu G, Yao X, Yin Z, Dong D, et al. Broad and strong memory Cd4+ and Cd8+ T cells induced by sars-Cov-2 in uk convalescent individuals following covid-19. Nat Immunol (2020) 21(11):1336–45. doi: 10.1038/s41590-020-0782-6
166. Cao X. Covid-19: Immunopathology and its implications for therapy. Nat Rev Immunol (2020) 20(5):269–70. doi: 10.1038/s41577-020-0308-3
167. Zhang W, Zhao Y, Zhang F, Wang Q, Li T, Liu Z, et al. The use of anti-inflammatory drugs in the treatment of people with severe coronavirus disease 2019 (Covid-19): The perspectives of clinical immunologists from China. Clin Immunol (2020) 214:108393. doi: 10.1016/j.clim.2020.108393
168. Ma Y, Qiu F, Deng C, Li J, Huang Y, Wu Z, et al. Integrating single-cell sequencing data with gwas summary statistics reveals Cd16+ monocytes and memory Cd8+ T cells involved in severe covid-19. Genome Med (2022) 14(1):1–21. doi: 10.1186/s13073-022-01021-1
169. Heeb LE, Egholm C, Boyman O. Evolution and function of interleukin-4 receptor signaling in adaptive immunity and neutrophils. Genes Immun (2020) 21(3):143–9. doi: 10.1038/s41435-020-0095-7
170. Moon C. Fighting covid-19 exhausts T cells. Nat Rev Immunol (2020) 20(5):277–8. doi: 10.1038/s41577-020-0304-7
171. Kaneko N, Boucau J, Kuo H-H, Perugino C, Mahajan VS, Farmer JR, et al. Temporal changes in T cell subsets and expansion of cytotoxic Cd4+ T cells in the lungs in severe covid-19. Clin Immunol (2022) 237:108991. doi: 10.1016/j.clim.2022.108991
172. Wherry EJ, Ahmed R. Memory Cd8 T-cell differentiation during viral infection. J Virol (2004) 78(11):5535–45. doi: 10.1128/JVI.78.11.5535-5545.2004
173. Qin C, Zhou L, Hu Z, Zhang S, Yang S, Tao Y, et al. Dysregulation of immune response in patients with covid-19 in wuhan, China. Clin Infect Dis (2020) 71 (15):762–768. doi: 10.1093/cid/ciaa248
174. Zuin J, Fogar P, Musso G, Padoan A, Piva E, Pelloso M, et al. T Cell senescence by extensive phenotyping: An emerging feature of covid-19 severity. Lab Med (2022) 53(6):609–13. doi: 10.1093/labmed/lmac048
175. Strutt TM, McKinstry KK, Dibble JP, Winchell C, Kuang Y, Curtis JD, et al. Memory Cd4+ T cells induce innate responses independently of pathogen. Nat Med (2010) 16(5):558–64. doi: 10.1038/nm.2142
176. Schenkel JM, Fraser KA, Vezys V, Masopust D. Sensing and alarm function of resident memory Cd8+ T cells. Nat Immunol (2013) 14(5):509–13. doi: 10.1038/ni.2568
177. Zhao J, Zhao J, Mangalam AK, Channappanavar R, Fett C, Meyerholz DK, et al. Airway memory Cd4+ T cells mediate protective immunity against emerging respiratory coronaviruses. Immunity (2016) 44(6):1379–91. doi: 10.1016/j.immuni.2016.05.006
178. Oja AE, Saris A, Ghandour CA, Kragten NA, Hogema BM, Nossent EJ, et al. Divergent sars-Cov-2-Specific T-and b-cell responses in severe but not mild covid-19 patients. Eur J Immunol (2020) 50(12):1998–2012. doi: 10.1002/eji.202048908
179. Zhao Y, Kilian C, Turner J-E, Bosurgi L, Roedl K, Bartsch P, et al. Clonal expansion and activation of tissue-resident memory-like Th17 cells expressing gm-csf in the lungs of severe covid-19 patients. Sci Immunol (2021) 6(56): eabf6692. doi: 10.1126/sciimmunol.abf6692
180. Chen L, Wei B, Di D. A narrative review of tissue-resident memory T cells and their role in immune surveillance and covid-19. Eur Rev Med Pharmacol Sci (2022) 26(12):4486–96. doi: 10.26355/eurrev_202206_29088
181. Lam JH, Smith FL, Baumgarth N. B cell activation and response regulation during viral infections. Viral Immunol (2020) 33(4):294–306. doi: 10.1089/vim.2019.0207
182. Jin Y, Yang H, Ji W, Wu W, Chen S, Zhang W, et al. Virology, epidemiology, pathogenesis, and control of covid-19. Viruses (2020) 12(4):372. doi: 10.3390/v12040372
183. Wildner NH, Ahmadi P, Schulte S, Brauneck F, Kohsar M, Lütgehetmann M, et al. B cell analysis in sars-Cov-2 versus malaria: Increased frequencies of plasmablasts and atypical memory b cells in covid-19. J Leukoc Biol (2021) 109(1):77–90. doi: 10.1002/JLB.5COVA0620-370RR
184. Davis CW, Jackson KJ, McElroy AK, Halfmann P, Huang J, Chennareddy C, et al. Longitudinal analysis of the human b cell response to Ebola virus infection. Cell (2019) 177(6):1566–82. e17. doi: 10.1016/j.cell.2019.04.036
185. Butler AL, Fischinger S, Alter G. The antibodiome–mapping the humoral immune response to hiv. Curr HIV/AIDS Rep (2019) 16(2):169–79. doi: 10.1007/s11904-019-00432-x
186. Siracusano G, Pastori C, Lopalco L. Humoral immune responses in covid-19 patients: A window on the state of the art. Front Immunol (2020) 11:1049. doi: 10.3389/fimmu.2020.01049
187. Kulkarni R. Antibody-dependent enhancement of viral infections. Dyn Immune Activation Viral Dis Ed Bramhachari P (2020), (Springer, Singapore). doi: 10.1007/978-981-15-1045-8_2
188. Yang Y, Xu F. Evolving understanding of antibody-dependent enhancement (ADE) of SARS-CoV-2. Front Immunol (2022), 13: 1008285. doi: 10.3389/fimmu.2022.1008285
189. Yao C, Bora SA, Parimon T, Zaman T, Friedman OA, Palatinus JA, et al. Cell-Type-Specific immune dysregulation in severely ill covid-19 patients. Cell Rep (2021) 34(1):108590. doi: 10.1016/j.celrep.2020.108590
190. Sosa-Hernández VA, Torres-Ruíz J, Cervantes-Díaz R, Romero-Ramírez S, Páez-Franco JC, Meza-Sánchez DE, et al. B cell subsets as severity-associated signatures in covid-19 patients. Front Immunol (2020) 11:611004. doi: 10.3389/fimmu.2020.611004
191. Bastard P, Rosen LB, Zhang Q, Michailidis E, Hoffmann H-H, Zhang Y, et al. Autoantibodies against type I ifns in patients with life-threatening covid-19. Science (2020) 370(6515): eabd4585. doi: 10.1126/science.abd4585
192. Tabibian-Keissar H, Hazanov L, Schiby G, Rosenthal N, Rakovsky A, Michaeli M, et al. Aging affects b-cell antigen receptor repertoire diversity in primary and secondary lymphoid tissues. Eur J Immunol (2016) 46(2):480–92. doi: 10.1002/eji.201545586
193. Hong B, Wang L, Huang C, Hong X, Liu A, Li Q, et al. Decrease of clone diversity in igm repertoires of hbv chronically infected individuals with high level of viral replication. Front Immunol (2021) 11:615669. doi: 10.3389/fmicb.2020.615669
194. Kaneko N, Kuo H-H, Boucau J, Farmer JR, Allard-Chamard H, Mahajan VS, et al. Loss of bcl-6-Expressing T follicular helper cells and germinal centers in covid-19. Cell (2020) 183(1):143–57.e13. doi: 10.1016/j.cell.2020.08.025
195. Wang F, Nie J, Wang H, Zhao Q, Xiong Y, Deng L, et al. Characteristics of peripheral lymphocyte subset alteration in covid-19 pneumonia. J Infect Dis (2020) 221(11):1762–9. doi: 10.1093/infdis/jiaa150
196. Shuwa HA, Shaw TN, Knight S, McClure FA, Wemyss K, Prise I, et al. Long-lasting alterations in T and b cell function in convalescent covid-19 patients. (2020). doi: 10.2139/ssrn.3720301
197. Feldmann M, Maini RN, Woody JN, Holgate ST, Winter G, Rowland M, et al. Trials of anti-tumour necrosis factor therapy for covid-19 are urgently needed. Lancet (London England) (2020) 395(10234):1407–9. doi: 10.1016/s0140-6736(20)30858-8
198. Stephenson E, Reynolds G, Botting RA, Calero-Nieto FJ, Morgan MD, Tuong ZK, et al. Single-cell multi-omics analysis of the immune response in covid-19. Nat Med (2021) 27(5):904–16. doi: 10.1038/s41591-021-01329-2
199. Woodruff MC, Ramonell RP, Nguyen DC, Cashman KS, Saini AS, Haddad NS, et al. Extrafollicular b cell responses correlate with neutralizing antibodies and morbidity in covid-19. Nat Immunol (2020) 21(12):1506–16. doi: 10.1038/s41590-020-00814-z
200. Woodruff M, Ramonell R, Cashman K, Nguyen D, Saini A, Haddad N, et al. Dominant extrafollicular b cell responses in severe covid-19 disease correlate with robust viral-specific antibody production but poor clinical outcomes. medRxiv preprint server Health Sci (2020) 21 (12):1506–1516. doi: 10.1101/2020.04.29.20083717
201. Hoehn KB, Ramanathan P, Unterman A, Sumida TS, Asashima H, Hafler DA, et al. Cutting edge: Distinct b cell repertoires characterize patients with mild and severe covid-19. J Immunol (2021) 206(12):2785–90. doi: 10.4049/jimmunol.2100135
202. Gao L, Zhou J, Yang S, Wang L, Chen X, Yang Y, et al. The dichotomous and incomplete adaptive immunity in covid-19 patients with different disease severity. Signal Transduct Target Ther (2021) 6(1):1–10. doi: 10.1038/s41392-021-00525-3
203. Sakharkar M, Rappazzo CG, Wieland-Alter WF, Hsieh C-L, Wrapp D, Esterman ES, et al. Prolonged evolution of the human b cell response to sars-Cov-2 infection. Sci Immunol (2021) 6(56): eabg6916. doi: 10.1126/sciimmunol.abg6916
204. Rodda LB, Netland J, Shehata L, Pruner KB, Morawski PA, Thouvenel CD, et al. Functional sars-Cov-2-Specific immune memory persists after mild covid-19. Cell (2021) 184(1):169–83.e17. doi: 10.1016/j.cell.2020.11.029
205. Golovkin A, Kalinina O, Bezrukikh V, Aquino A, Zaikova E, Karonova T, et al. Imbalanced immune response of T-cell and b-cell subsets in patients with moderate and severe covid-19. Viruses (2021) 13(10):1966. doi: 10.3390/v13101966
206. Laidlaw BJ, Ellebedy AH. The germinal centre b cell response to sars-Cov-2. Nat Rev Immunol (2022) 22(1):7–18. doi: 10.1038/s41577-021-00657-1
207. Hasan A, Al-Ozairi E, Al-Baqsumi Z, Ahmad R, Al-Mulla F. Cellular and humoral immune responses in covid-19 and immunotherapeutic approaches. ImmunoTargets Ther (2021) 10:63. doi: 10.2147/ITT.S280706
208. Röltgen K, Boyd SD. Antibody and b cell responses to sars-Cov-2 infection and vaccination. Cell Host Microbe (2021) 29(7):1063–75. doi: 10.1016/j.chom.2021.06.009
209. Bulati M, Buffa S, Martorana A, Gervasi F, Camarda C, Azzarello DM, et al. Double negative (Igg+ igd– Cd27–) b cells are increased in a cohort of moderate-severe alzheimer's disease patients and show a pro-inflammatory trafficking receptor phenotype. J Alzheimers Dis (2015) 44(4):1241–51. doi: 10.3233/JAD-142412
210. You X, Zhang R, Shao M, He J, Chen J, Liu J, et al. Double negative b cell is associated with renal impairment in systemic lupus erythematosus and acts as a marker for nephritis remission. Front Med (Lausanne) (2020) 7:85. doi: 10.3389/fmed.2020.00085
211. Taefehshokr N, Taefehshokr S, Heit B. Mechanisms of dysregulated humoral and cellular immunity by sars-Cov-2. Pathogens (2020) 9(12):1027. doi: 10.3390/pathogens9121027
212. Ruschil C, Gabernet G, Lepennetier G, Heumos S, Kaminski M, Hracsko Z, et al. Specific induction of double negative b cells during protective and pathogenic immune responses. Front Immunol (2020) 11:606338. doi: 10.3389/fimmu.2020.606338
213. Kuri-Cervantes L, Pampena MB, Meng W, Rosenfeld AM, Ittner CA, Weisman AR, et al. Comprehensive mapping of immune perturbations associated with severe covid-19. Sci Immunol (2020) 5(49): eabd7114. doi: 10.1126/sciimmunol.abd7114
214. Zhao J, Yuan Q, Wang H, Liu W, Liao X, Su Y, et al. Antibody responses to sars-Cov-2 in patients with novel coronavirus disease 2019. Clin Infect Dis (2020) 71(16):2027–34. doi: 10.1093/cid/ciaa344
215. Farris AD, Guthridge JM. Overlapping b cell pathways in severe covid-19 and lupus. Nat Immunol (2020) 21(12):1478–80. doi: 10.1038/s41590-020-00822-z
216. Guthmiller JJ, Stovicek O, Wang J, Changrob S, Li L, Halfmann P, et al. Sars-Cov-2 infection severity is linked to superior humoral immunity against the spike. Mbio (2021) 12(1): e02940-20. doi: 10.1128/mBio.02940-20
217. Hoepel W, Chen H-J, Allahverdiyeva S, Manz X, Aman J, Bonta P, et al. Anti-Sars-Cov-2 igg from severely ill covid-19 patients promotes macrophage hyper-inflammatory responses. BioRxiv (2020). doi: 10.1101/2020.07.13.190140
218. Karthik K, Senthilkumar TMA, Udhayavel S, Raj GD. Role of antibody-dependent enhancement (Ade) in the virulence of sars-Cov-2 and its mitigation strategies for the development of vaccines and immunotherapies to counter covid-19. Hum Vaccin Immunother (2020) 16(12):3055–60. doi: 10.1080/21645515.2020.1796425
219. Lu L, Zhang H, Zhan M, Jiang J, Yin H, Dauphars DJ, et al. Antibody response and therapy in covid-19 patients: What can be learned for vaccine development? Sci China Life Sci (2020), 63 (12):1833–1849. doi: 10.1007/s11427-020-1859-y
220. Pascolini S, Vannini A, Deleonardi G, Ciordinik M, Sensoli A, Carletti I, et al. Covid-19 and immunological dysregulation: Can autoantibodies be useful? Clin Transl Sci (2020); 14 (2):502–508. doi: 10.1111/cts.12908
221. Hashem AM, Algaissi A, Almahboub SA, Alfaleh MA, Abujamel TS, Alamri SS, et al. Early humoral response correlates with disease severity and outcomes in covid-19 patients. Viruses (2020) 12(12):1390. doi: 10.3390/v12121390
222. Chakraborty S, Gonzalez J, Edwards K, Mallajosyula V, Buzzanco AS, Sherwood R, et al. Proinflammatory igg fc structures in patients with severe covid-19. Nat Immunol (2021) 22(1):67–73. doi: 10.1038/s41590-020-00828-7
223. Yang Y, Yang M, Peng Y, Liang Y, Wei J, Xing L, et al. Longitudinal analysis of antibody dynamics in covid-19 convalescents reveals neutralizing responses up to 16 months after infection. Nat Microbiol (2022) 7(3):423–33. doi: 10.1038/s41564-021-01051-2
224. Ye Q, Wang B, Mao J. The pathogenesis and treatment of the `Cytokine Storm' in COVID-19. J Infect (2020); 80(6):607–613. doi: 10.1016/j.jinf.2020.03.037
225. Mistry P, Barmania F, Mellet J, Peta K, Strydom A, Viljoen IM, et al. Sars-Cov-2 variants, vaccines, and host immunity. Front Immunol (2021) 12:809244. doi: 10.3389/fimmu.2021.809244
226. Wiersinga WJ, Rhodes A, Cheng AC, Peacock SJ, Prescott HC. Pathophysiology, transmission, diagnosis, and treatment of coronavirus disease 2019 (Covid-19): A review. Jama (2020) 324(8):782–93. doi: 10.1001/jama.2020.12839
227. İnandıklıoğlu N, Akkoc T. Immune Responses to SARS-CoV, MERS-CoV and SARS-CoV-2. Cell Biol Trans Med Vol 9: Adv Exp Med Biol (2020), 1288:5–12. doi: 10.1007/5584_2020_549
228. Yang X, Yu Y, Xu J, Shu H, Liu H, Wu Y, et al. Clinical course and outcomes of critically ill patients with sars-Cov-2 pneumonia in wuhan, China: A single-centered, retrospective, observational study. Lancet Respir Med (2020) 8(5):475–81. doi: 10.1016/S2213-2600(20)30079-5
229. Eskilsson A, Mirrasekhian E, Dufour S, Schwaninger M, Engblom D, Blomqvist A. Immune-induced fever is mediated by il-6 receptors on brain endothelial cells coupled to Stat3-dependent induction of brain endothelial prostaglandin synthesis. J Neurosci (2014) 34(48):15957–61. doi: 10.1523/JNEUROSCI.3520-14.2014
230. Hsu RJ, Yu WC, Peng GR, Ye CH, Hu S, Chong PCT, et al. The role of cytokines and chemokines in severe acute respiratory syndrome coronavirus 2 infections. Front Immunol (2022) 13:832394. doi: 10.3389/fimmu.2022.832394
231. Bhaskar S, Sinha A, Banach M, Mittoo S, Weissert R, Kass JS, et al. Cytokine storm in Covid-19–immunopathological mechanisms, clinical considerations, and therapeutic approaches: The reprogram consortium position paper. Front Immunol (2020) 11:1648. doi: 10.3389/fimmu.2020.01648
232. Samuel CE. Antiviral actions of interferons. Clin Microbiol Rev (2001) 14(4):778–809. doi: 10.1128/CMR.14.4.778-809.2001
233. Takeuchi O, Akira S. Innate immunity to virus infection. Immunol Rev (2009) 227(1):75–86. doi: 10.1111/j.1600-065X.2008.00737.x
234. Katze MG, He Y, Gale M. Viruses and interferon: A fight for supremacy. Nat Rev Immunol (2002) 2(9):675–87. doi: 10.1038/nri888
235. Tang F, Du Q, Liu Y-J. Plasmacytoid dendritic cells in antiviral immunity and autoimmunity. Sci China Life Sci (2010) 53(2):172–82. doi: 10.1007/s11427-010-0045-0
236. Felgenhauer U, Schoen A, Gad HH, Hartmann R, Schaubmar AR, Failing K, et al. Inhibition of sars-Cov-2 by type I and type iii interferons. J Biol Chem (2020) 295(41):13958–64. doi: 10.1074/jbc.AC120.013788
237. Huang CT, Chao TL, Kao HC, Pang YH, Lee WH, Hsieh CH, et al. Enhancement of the ifn-B-Induced host signature informs repurposed drugs for covid-19. Heliyon (2020) 6(12):e05646. doi: 10.1016/j.heliyon.2020.e05646
238. Hung IF, Lung KC, Tso EY, Liu R, Chung TW, Chu MY, et al. Triple combination of interferon beta-1b, lopinavir-ritonavir, and ribavirin in the treatment of patients admitted to hospital with covid-19: An open-label, randomised, phase 2 trial. Lancet (London England) (2020) 395(10238):1695–704. doi: 10.1016/s0140-6736(20)31042-4
239. Khamis F, Al Naabi H, Al Lawati A, Ambusaidi Z, Al Sharji M, Al Barwani U, et al. Randomized controlled open label trial on the use of favipiravir combined with inhaled interferon beta-1b in hospitalized patients with moderate to severe covid-19 pneumonia. Int J Infect Dis (2021) 102:538–43. doi: 10.1016/j.ijid.2020.11.008
240. Hadjadj J, Yatim N, Barnabei L, Corneau A, Boussier J, Smith N, et al. Impaired type I interferon activity and inflammatory responses in severe covid-19 patients. Science (2020) 369(6504):718–24. doi: 10.1126/science.abc6027
241. Kopecky-Bromberg SA, Martínez-Sobrido L, Frieman M, Baric RA, Palese P. Severe acute respiratory syndrome coronavirus open reading frame (Orf) 3b, orf 6, and nucleocapsid proteins function as interferon antagonists. J Virol (2007) 81(2):548–57. doi: 10.1128/JVI.01782-06
242. Galani I-E, Rovina N, Lampropoulou V, Triantafyllia V, Manioudaki M, Pavlos E, et al. Untuned antiviral immunity in covid-19 revealed by temporal type I/Iii interferon patterns and flu comparison. Nat Immunol (2021) 22(1):32–40. doi: 10.1038/s41590-020-00840-x
243. Yousefifard M, Zali A, Mohamed Ali K, Madani Neishaboori A, Zarghi A, Hosseini M, et al. Antiviral therapy in management of covid-19: A systematic review on current evidence. Arch Acad Emergency Med (2020) 8(1):e45.
244. Mattos-Silva P, Felix NS, Silva PL, Robba C, Battaglini D, Pelosi P, et al. Pros and cons of corticosteroid therapy for covid-19 patients. Respir Physiol Neurobiol (2020) 280:103492. doi: 10.1016/j.resp.2020.103492
245. Krumm ZA, Lloyd GM, Francis CP, Nasif LH, Mitchell DA, Golde TE, et al. Precision therapeutic targets for covid-19. Virol J (2021) 18(1):66. doi: 10.1186/s12985-021-01526-y
246. Zhou YW, Xie Y. Therapeutic targets and interventional strategies in covid-19: Mechanisms and clinical studies. Signal Transduct Target Ther (2021) 6(1):317. doi: 10.1038/s41392-021-00733-x
247. Laterre PF, François B, Collienne C, Hantson P, Jeannet R, Remy KE, et al. Association of interleukin 7 immunotherapy with lymphocyte counts among patients with severe coronavirus disease 2019 (Covid-19). JAMA netw Open (2020) 3(7):e2016485. doi: 10.1001/jamanetworkopen.2020.16485
248. Monneret G, de Marignan D, Coudereau R, Bernet C, Ader F, Frobert E, et al. Immune monitoring of interleukin-7 compassionate use in a critically ill covid-19 patient. Cell Mol Immunol (2020) 17(9):1001–3. doi: 10.1038/s41423-020-0516-6
249. Gupta S, Wang W, Hayek SS, Chan L, Mathews KS, Melamed ML, et al. Association between early treatment with tocilizumab and mortality among critically ill patients with covid-19. JAMA Internal Med (2021) 181(1):41–51. doi: 10.1001/jamainternmed.2020.6252
250. Ucciferri C, Auricchio A, Di Nicola M, Potere N, Abbate A, Cipollone F, et al. Canakinumab in a subgroup of patients with covid-19. Lancet Rheumatol (2020) 2(8):e457–ee8. doi: 10.1016/s2665-9913(20)30167-3
251. Cauchois R, Koubi M. Early il-1 receptor blockade in severe inflammatory respiratory failure complicating covid-19. Proc Natl Acad Sci U.S.A. (2020) 117(32):18951–3. doi: 10.1073/pnas.2009017117
252. Yang L, Xie X, Tu Z, Fu J, Xu D, Zhou Y. The signal pathways and treatment of cytokine storm in covid-19. Signal Transduct Target Ther (2021) 6(1):255. doi: 10.1038/s41392-021-00679-0
253. Duret PM, Spielmann L, Messer L. Response to: 'Correspondence on recovery from covid-19 in a patient with spondyloarthritis treated with tnf-alpha inhibitor etanercept. a report on a covid-19 patient with psoriatic arthritis receiving ustekinumab' by Messina et al. Ann Rheum Dis (2021) 80(5):e80. doi: 10.1136/annrheumdis-2020-218147
254. Zhou Q, Chen V, Shannon CP, Wei XS, Xiang X, Wang X, et al. Interferon-A2b treatment for covid-19. Front Immunol (2020) 11:1061. doi: 10.3389/fimmu.2020.01061
255. Xie X, Jiang Y, Zeng Y, Liu H. Combination antiviral therapy with Lopinavir/Ritonavir, arbidol and interferon-A1b for covid-19. Antiviral Ther (2020) 25(4):233–9. doi: 10.3851/imp3362
256. Leng Z, Zhu R, Hou W, Feng Y, Yang Y, Han Q, et al. Transplantation of Ace2(-) mesenchymal stem cells improves the outcome of patients with covid-19 pneumonia. Aging Dis (2020) 11(2):216–28. doi: 10.14336/ad.2020.0228
257. Suzuki Y, Tanino Y, Nikaido T, Minemura H, Umeda T, Rikimaru M, et al. Severe coronavirus disease 2019 that recovered from respiratory failure by treatment that included high-dose intravenous immunoglobulin. Internal Med (Tokyo Japan) (2021) 60(3):457–61. doi: 10.2169/internalmedicine.6326-20
258. Liu Q, Zhou YH, Yang ZQ. The cytokine storm of severe influenza and development of immunomodulatory therapy. Cell Mol Immunol (2016) 13(1):3–10. doi: 10.1038/cmi.2015.74
259. Lee DW, Gardner R, Porter DL, Louis CU, Ahmed N, Jensen M, et al. Current concepts in the diagnosis and management of cytokine release syndrome. Blood (2014) 124(2):188–95. doi: 10.1182/blood-2014-05-552729
260. Kaplon H, Reichert JM. Antibodies to watch in 2021. MAbs (2021) 13(1):1860476. doi: 10.1080/19420862.2020.1860476
261. Clark IA. Background to new treatments for COVID-19, including its chronicity, through altering elements of the cytokine storm. Rev Med Virol (2020); 31 (5):1–13. doi: 10.1002/rmv.2210
262. Sterne JAC, Murthy S, Diaz JV, Slutsky AS, Villar J, Angus DC, et al. Association between administration of systemic corticosteroids and mortality among critically ill patients with covid-19: A meta-analysis. Jama (2020) 324(13):1330–41. doi: 10.1001/jama.2020.17023
263. Johnson RM, Vinetz JM. Dexamethasone in the management of covid -19. Bmj (2020) 370:m2648. doi: 10.1136/bmj.m2648
264. Lin P. Targeting interleukin-6 for noninfectious uveitis. Clin Ophthalmol (Auckland NZ) (2015) 9:1697–702. doi: 10.2147/opth.s68595
265. Karkhur S, Hasanreisoglu M, Vigil E, Halim MS, Hassan M, Plaza C, et al. Interleukin-6 inhibition in the management of non-infectious uveitis and beyond. J ophthal Inflammation infect (2019) 9(1):17. doi: 10.1186/s12348-019-0182-y
266. Tang L, Yin Z, Hu Y, Mei H. Controlling cytokine storm is vital in covid-19. Front Immunol (2020) 11:570993. doi: 10.3389/fimmu.2020.570993
267. Della-Torre E, Campochiaro C. Interleukin-6 blockade with sarilumab in severe covid-19 pneumonia with systemic hyperinflammation: An open-label cohort study. Ann Rheum Dis (2020) 79(10):1277–85. doi: 10.1136/annrheumdis-2020-218122
268. Benucci M, Giannasi G, Cecchini P, Gobbi FL, Damiani A, Grossi V, et al. Covid-19 pneumonia treated with sarilumab: A clinical series of eight patients. J Med Virol (2020) 92(11):2368–70. doi: 10.1002/jmv.26062
269. Gritti G, Raimondi F, Ripamonti D, Riva I, Landi F, Alborghetti L, et al. Il-6 signalling pathway inactivation with siltuximab in patients with covid-19 respiratory failure: An observational cohort study. medRxiv preprint server Health Sci (2020). doi: 10.1101/2020.04.01.20048561
270. Cavalli G, De Luca G, Campochiaro C, Della-Torre E, Ripa M, Canetti D, et al. Interleukin-1 blockade with high-dose anakinra in patients with covid-19, acute respiratory distress syndrome, and hyperinflammation: A retrospective cohort study. Lancet Rheumatol (2020) 2(6):e325–e31. doi: 10.1016/s2665-9913(20)30127-2
271. Dimopoulos G, de Mast Q, Markou N, Theodorakopoulou M, Komnos A, Mouktaroudi M, et al. Favorable anakinra responses in severe covid-19 patients with secondary hemophagocytic lymphohistiocytosis. Cell Host Microbe (2020) 28(1):117–23.e1. doi: 10.1016/j.chom.2020.05.007
272. Kooistra EJ, Waalders NJB, Grondman I, Janssen NAF, de Nooijer AH, Netea MG, et al. Anakinra treatment in critically ill covid-19 patients: A prospective cohort study. Crit Care (2020) 24(1):688. doi: 10.1186/s13054-020-03364-w
273. Katia F, Myriam DP, Ucciferri C. Efficacy of canakinumab in mild or severe covid-19 pneumonia. Immun Inflammation Dis (2021) 9(2):399–405. doi: 10.1002/iid3.400
274. Generali D, Bosio G, Malberti F, Cuzzoli A, Testa S, Romanini L, et al. Canakinumab as treatment for covid-19-Related pneumonia: A prospective case-control study. Int J Infect Dis (2021) 104:433–40. doi: 10.1016/j.ijid.2020.12.073
275. Sheng CC, Sahoo D, Dugar S, Prada RA, Wang TKM, Abou Hassan OK, et al. Canakinumab to reduce deterioration of cardiac and respiratory function in sars-Cov-2 associated myocardial injury with heightened inflammation (Canakinumab in covid-19 cardiac injury: The three c study). Clin Cardiol (2020) 43(10):1055–63. doi: 10.1002/clc.23451
276. Landi L, Ravaglia C, Russo E, Cataleta P, Fusari M, Boschi A, et al. Blockage of interleukin-1β with canakinumab in patients with covid-19. Sci Rep (2020) 10(1):21775. doi: 10.1038/s41598-020-78492-y
277. Kunisaki R, Tsukiji J, Kudo M. Potential inhibition of covid-19-Driven pneumonia by immunosuppressive therapy and anti-tnfα antibodies: A case report. J Crohn's colitis (2020) 14 (12):1786–1787. doi: 10.1093/ecco-jcc/jjaa105
278. Guo Y, Hu K, Li Y, Lu C, Ling K, Cai C, et al. Targeting tnf-A for covid-19: Recent advanced and controversies. Front Public Health (2022) 10:833967. doi: 10.3389/fpubh.2022.833967
279. Duret PM, Sebbag E, Mallick A, Gravier S, Spielmann L. Recovery from covid-19 in a patient with spondyloarthritis treated with tnf-alpha inhibitor etanercept. Ann Rheum Dis (2020) 79(9):1251–2. doi: 10.1136/annrheumdis-2020-217362
280. Calabrese LH, Lenfant T, Calabrese C. Interferon therapy for covid-19 and emerging infections: Prospects and concerns. Cleveland Clinic J Med (2020). doi: 10.3949/ccjm.87a.ccc066
281. Zuo Y, Liu Y, Zhong Q, Zhang K, Xu Y, Wang Z. Lopinavir/Ritonavir and interferon combination therapy may help shorten the duration of viral shedding in patients with covid-19: A retrospective study in two designated hospitals in anhui, China. J Med Virol (2020) 92(11):2666–74. doi: 10.1002/jmv.26127
282. Mehta N, Mazer-Amirshahi M, Alkindi N, Pourmand A. Pharmacotherapy in covid-19; a narrative review for emergency providers. Am J Emergency Med (2020) 38(7):1488–93. doi: 10.1016/j.ajem.2020.04.035
283. Saleh M, Vaezi AA, Aliannejad R, Sohrabpour AA, Kiaei SZF, Shadnoush M, et al. Cell therapy in patients with covid-19 using wharton's jelly mesenchymal stem cells: A phase 1 clinical trial. Stem Cell Res Ther (2021) 12(1):410. doi: 10.1186/s13287-021-02483-7
284. Zaki MM, Lesha E. Cell therapy strategies for covid-19: Current approaches and potential applications. Sci Adv (2021) 7(33): eabg5995. doi: 10.1126/sciadv.abg5995
285. Hashmi S, Ahmed M, Murad MH, Litzow MR, Adams RH, Ball LM, et al. Survival after mesenchymal stromal cell therapy in steroid-refractory acute graft-Versus-Host disease: Systematic review and meta-analysis. Lancet Haematol (2016) 3(1):e45–52. doi: 10.1016/s2352-3026(15)00224-0
286. Aggarwal S, Pittenger MF. Human mesenchymal stem cells modulate allogeneic immune cell responses. Blood (2005) 105(4):1815–22. doi: 10.1182/blood-2004-04-1559
287. Fisher SA, Cutler A, Doree C, Brunskill SJ, Stanworth SJ, Navarrete C, et al. Mesenchymal stromal cells as treatment or prophylaxis for acute or chronic graft-Versus-Host disease in haematopoietic stem cell transplant (Hsct) recipients with a haematological condition. Cochrane Database syst Rev (2019) 1(1):Cd009768. doi: 10.1002/14651858.CD009768.pub2
288. Päth G, Perakakis N, Mantzoros CS, Seufert J. Stem cells in the treatment of diabetes mellitus - focus on mesenchymal stem cells. Metab: Clin Exp (2019) 90:1–15. doi: 10.1016/j.metabol.2018.10.005
289. Weiss ARR, Dahlke MH. Immunomodulation by mesenchymal stem cells (Mscs): Mechanisms of action of living, apoptotic, and dead mscs. Front Immunol (2019) 10:1191. doi: 10.3389/fimmu.2019.01191
290. Shende P, Subedi M. Pathophysiology, mechanisms and applications of mesenchymal stem cells for the treatment of spinal cord injury. Biomed pharmacother = Biomed pharmacother (2017) 91:693–706. doi: 10.1016/j.biopha.2017.04.126
291. Wang SH, Shetty AK, Jin K, Chunhua Zhao R. Combating covid-19 with mesenchymal Stem/Stromal cell therapy: Promise and challenges. Front Cell Dev Biol (2020) 8:627414. doi: 10.3389/fcell.2020.627414
292. Liang B, Chen J, Li T, Wu H, Yang W, Li Y, et al. Clinical remission of a critically ill covid-19 patient treated by human umbilical cord mesenchymal stem cells: A case report. Med (Baltimore) (2020) 99(31):e21429. doi: 10.1097/md.0000000000021429
293. Sánchez-Guijo F, García-Arranz M, López-Parra M, Monedero P, Mata-Martínez C, Santos A, et al. Adipose-derived mesenchymal stromal cells for the treatment of patients with severe sars-Cov-2 pneumonia requiring mechanical ventilation. A Proof Concept Study EClinicalMed (2020) 25:100454. doi: 10.1016/j.eclinm.2020.100454
294. Lanzoni G, Linetsky E, Correa D, Messinger Cayetano S, Alvarez RA, Kouroupis D. Umbilical cord mesenchymal stem cells for covid-19 acute respiratory distress syndrome: A double-blind, phase 1/2a, randomized controlled trial. Stem Cells Transl Med (2021) 10(5):660–73. doi: 10.1002/sctm.20-0472
295. Schwab I, Nimmerjahn F. Intravenous immunoglobulin therapy: How does igg modulate the immune system? Nat Rev Immunol (2013) 13(3):176–89. doi: 10.1038/nri3401
296. Ross C, Svenson M, Hansen MB, Vejlsgaard GL, Bendtzen K. High avidity ifn-neutralizing antibodies in pharmaceutically prepared human igg. J Clin Invest (1995) 95(5):1974–8. doi: 10.1172/jci117881
297. Mohan PV, Tarnow-Mordi W, Stenson B, Brocklehurst P, Haque K, Cavendish V, et al. Can polyclonal intravenous immunoglobulin limit cytokine mediated cerebral damage and chronic lung disease in preterm infants? Arch Dis childhood Fetal neonatal edition (2004) 89(1):F5–8. doi: 10.1136/fn.89.1.f5
298. Watanabe M, Uchida K, Nakagaki K, Trapnell BC, Nakata K. High avidity cytokine autoantibodies in health and disease: Pathogenesis and mechanisms. Cytokine Growth factor Rev (2010) 21(4):263–73. doi: 10.1016/j.cytogfr.2010.03.003
299. Hung IFN, To KKW, Lee CK, Lee KL, Yan WW, Chan K, et al. Hyperimmune iv immunoglobulin treatment: A multicenter double-blind randomized controlled trial for patients with severe 2009 influenza a(H1n1) infection. Chest (2013) 144(2):464–73. doi: 10.1378/chest.12-2907
300. Chong PY, Chui P, Ling AE, Franks TJ, Tai DY, Leo YS, et al. Analysis of deaths during the severe acute respiratory syndrome (Sars) epidemic in Singapore: Challenges in determining a sars diagnosis. Arch Pathol Lab Med (2004) 128(2):195–204. doi: 10.5858/2004-128-195-aoddts
301. Tabarsi P, Barati S, Jamaati H, Haseli S, Marjani M, Moniri A, et al. Evaluating the effects of intravenous immunoglobulin (Ivig) on the management of severe covid-19 cases: A randomized controlled trial. Int Immunopharmacol (2021) 90:107205. doi: 10.1016/j.intimp.2020.107205
Keywords: SARS-CoV-2, COVID-19, immune response, cytokine storm, T cells, antibodies, inflammation, therapeutic targets
Citation: Nasrollahi H, Talepoor AG, Saleh Z, Eshkevar Vakili M, Heydarinezhad P, Karami N, Noroozi M, Meri S and Kalantar K (2023) Immune responses in mildly versus critically ill COVID-19 patients. Front. Immunol. 14:1077236. doi: 10.3389/fimmu.2023.1077236
Received: 22 October 2022; Accepted: 12 January 2023;
Published: 30 January 2023.
Edited by:
Edwin Bölke, Heinrich Heine University of Düsseldorf, GermanyReviewed by:
Chang Li, Chinese Academy of Agricultural Sciences (CAAS), ChinaZhanbo Zhu, Heilongjiang Bayi Agricultural University, China
Copyright © 2023 Nasrollahi, Talepoor, Saleh, Eshkevar Vakili, Heydarinezhad, Karami, Noroozi, Meri and Kalantar. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kurosh Kalantar, Kalantark@sums.ac.ir; Seppo Meri, seppo.meri@helsinki.fi
 Hamid Nasrollahi
Hamid Nasrollahi Atefe Ghamar Talepoor
Atefe Ghamar Talepoor Zahra Saleh2
Zahra Saleh2 Mahsa Eshkevar Vakili
Mahsa Eshkevar Vakili Paria Heydarinezhad
Paria Heydarinezhad Narges Karami
Narges Karami Seppo Meri
Seppo Meri Kurosh Kalantar
Kurosh Kalantar