- 1Institute of Medicinal Plant Development, Peking Union Medical College and Chinese Academy of Medical Sciences, Beijing, China
- 2Key Laboratory of Bioactive Substances and Resources Utilization of Chinese Herbal Medicine, Ministry of Education, Institute of Medicinal Plant Development, Chinese Academy of Medical Sciences and Peking Union Medical College, Beijing, China
- 3Beijing Key Laboratory of Innovative Drug Discovery of Traditional Chinese Medicine (Natural Medicine) and Translational Medicine, Institute of Medicinal Plant Development, Peking Union Medical College and Chinese Academy of Medical Sciences, Beijing, China
- 4Key Laboratory for Research and Evaluation of Pharmacovigilance, Institute of Medicinal Plant Development, Peking Union Medical College and Chinese Academy of Medical Sciences, Beijing, China
Hepatocellular carcinoma (HCC) is a complex and heterogeneous malignancy with high incidence and poor prognosis. In addition, owing to the lack of diagnostic and prognostic markers, current multimodal treatment options fail to achieve satisfactory outcomes. Tumor immune microenvironment (TIME), angiogenesis, epithelial-mesenchymal transition (EMT), invasion, metastasis, metabolism, and drug resistance are important factors influencing tumor development and therapy. The intercellular communication of these important processes is mediated by a variety of bioactive molecules to regulate pathophysiological processes in recipient cells. Among these bioactive molecules, non-coding RNAs (ncRNAs), including microRNAs (miRNAs), long non-coding RNAs (lncRNAs), and circular RNAs (circRNAs), account for a large part of the human transcriptome, and their dysregulation affects the progression of HCC. The purpose of this review is to evaluate the potential regulatory mechanisms of ncRNAs in HCC, summarize novel biomarkers from somatic fluids (plasma/serum/urine), and explore the potential of some small-molecule modulators as drugs. Thus, through this review, we aim to contribute to a deeper understanding of the regulatory mechanisms, early diagnosis, prognosis, and precise treatment of HCC.
Introduction
Hepatocellular carcinoma (HCC) is the sixth most common malignancy and the third leading cause of cancer-related deaths globally, posing a serious threat to human health and life (1, 2). Until now, mainstay curative treatments have included surgical resection, liver transplantation, radiofrequency ablation (RFA), transarterial chemoembolization (TACE), transarterial embolization (TAE), and systemic treatment with molecular-targeted agents. Despite great breakthroughs, current treatments have failed to deliver satisfactory outcomes, with an overall 5-year survival rate of only 12% (3–5). This failure is attributed to high heterogeneity, frequent recurrence, and drug resistance of HCC (6, 7). In addition, owing to the lack of reliable biomarkers, most patients progress to the intermediate and advanced stages of HCC at the time of diagnosis, regrettably missing the optimal treatment window. Therefore, deepening the understanding of the molecular mechanism to develop new therapeutic strategies and identifying biomarkers that can be effectively monitored at an early-stage is crucial in the fight against HCC.
The development of HCC is a multifactorial process. In recent years, immunotherapy has attracted extensive attention and has emerged as the next-generation therapy since the approval of immune checkpoint inhibitors (ICIs). However, studies have shown that its efficacy is closely related to the state of the tumor immune microenvironment (TIME). Essentially, anti-tumor immune efficacy mainly depends on the status and function of the immune cells in the TIME. Thus, it is necessary to elucidate the immune microenvironment of HCC to select appropriate ICIs (7). Angiogenesis not only delivers oxygen and nutrients to the growing tumor but also transports tumor cells to the metastatic site (8). Epithelial-mesenchymal transition (EMT) is a complex phenotypic event that directly affects changes in the characteristic features of HCC, such as occurrence, migration, invasion, metastasis, and even drug resistance (9, 10). Following primary tumor growth, angiogenesis and EMT, tumors are more prone to invasion and metastasis, which plays a key role in limiting patient outcomes overall. Therefore, there is an urgent need to explore invasive-metastatic cascade response of HCC (11, 12). Dysregulation of tumor cell metabolic activity may impair anti-tumor response, whereas metabolic reprogramming secures energy and substrates for the tumor (13, 14). Even more disastrous is drug resistance to chemotherapeutic agents in HCC. According to reports, mortality due to drug resistance accounts for more than 90% of cancer-specific mortality (15). In short, the above-mentioned issues are key barriers to the successful treatment of HCC.
Non-coding RNAs (ncRNAs) are endogenous RNAs accounting for the majority (98%) of the transcribed genome. They were once regarded as “dark matter” because of their lack of ability to encode proteins. After years of exploration, they have been found to act as important signaling molecules in the regulation of key cellular pathways (16–18). They are abundant and stable and mainly include microRNAs (miRNAs), long non-coding RNAs (lncRNAs), and circular RNAs (circRNAs). Approximately 30% of genes in the human body are regulated by miRNAs, which are the most abundant and studied group of ncRNAs (19). miRNAs regulate gene expression by binding to DNA, RNA, or proteins, which further regulates various biological functions (20). LncRNAs are linear RNAs with a transcript length of > 200 nucleotides, they have more diverse modes of action than miRNAs as the roles of spatial and temporal lncRNAs in cell physiology and pathology have gradually become clear. They can act as signals, decoys, scaffolds, or guides. Even the same kind of lncRNAs can function via different mechanisms (21–23). Emerging evidence indicates that circRNAs are novel ncRNAs that are related to many pathological diseases. In contrast to the standard splicing of linear RNAs, circRNAs are closed-loop structures produced by back-splicing (24). CircRNAs exhibit high abundance, diversity, sequence conservation among species, stability, tissue specificity, and tumor stage-dependent characteristics (25, 26). They exert their functions by binding to RNA-binding proteins, sponging miRNAs, translating into peptides or proteins, regulating gene transcription, and competing with canonical splicing (26, 27). To date, studies have found that these functional molecules mediate intercellular communication, which plays a non-negligible role in the TIME, angiogenesis, EMT, invasion, metastasis, metabolism, and drug resistance (14, 28–30). ncRNAs can also be detected as circulating molecules in the serum/plasma/urine, indicating that they are of great significance in early diagnosis and prognosis. In addition, small-molecule modulators that target ncRNAs are of great use. Hence, ncRNAs hold great promise as potential biomarkers or therapeutic targets (31). Herein, we summarize the latest findings on ncRNAs (miRNAs, lncRNAs, and circRNAs) that affect various aspects of HCC, including TIME, angiogenesis, EMT, invasion, metastasis, metabolism, and drug resistance. The potential uses of ncRNAs in cancer diagnosis/prognosis and the therapeutic activity of small-molecule modulators that selectively target ncRNAs are also summarized.
Regulatory mechanisms of ncRNAs in HCC
Regulation of TIME
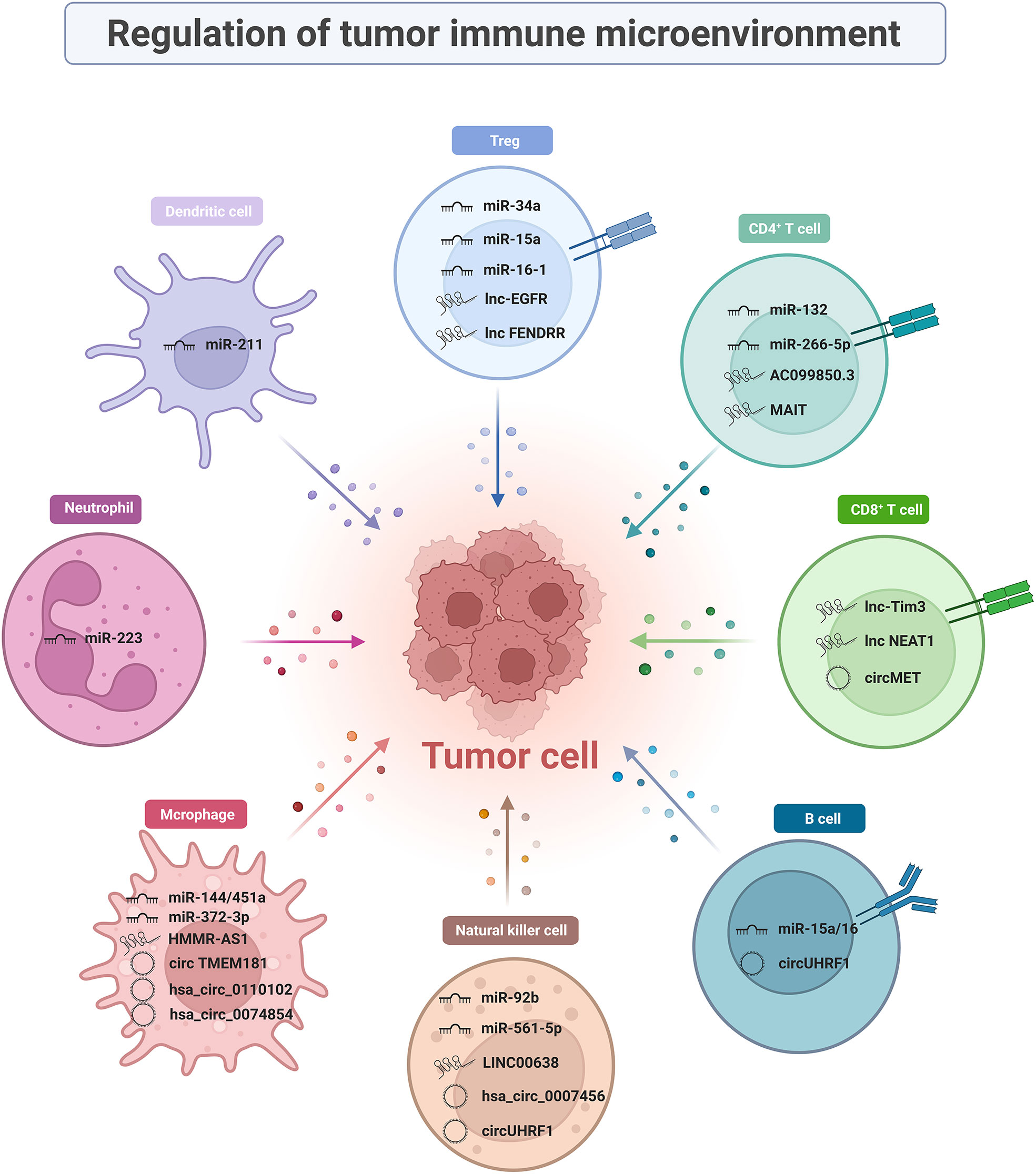
Immune cells are complex, heterogeneous cells with different developmental stages and functional subpopulations. ncRNAs present in the TIME can regulate immune cells and influence the development of tumor immune responses. The molecular mechanisms by which ncRNAs regulate immune cellular subpopulations in TIME and tumor immune response development are described in detail in this section (Figure 1).
T cells
CD8+ T cells
CD8+ T cells are key players that perform anti-tumor immune functions in the TIME and characteristic markers of good prognosis in HCC. They mediate target cell apoptosis by secreting perforin and granzyme or by expressing Fas ligand (FasL) (32, 33). Single-cell RNA sequencing indicated that the effector CD8+ T cells in advanced HCC patients were depleted and weakened in cytotoxicity compared to those in early-stage of HCC, which may lead to impaired anti-tumor function (34). Unfortunately, high expression of activation/depletion markers (notably PD1, TIM3, and LAG3) on the surface of CD8+ T cells shifts high-density cytotoxic T cells towards immune exclusion (35). This phenomenon raises a prerequisite condition for the application of immune checkpoints, that is, how to convert “cold tumors” into “hot tumors”, which is also a pressing issue to be addressed. Some studies have found that targeting ncRNAs can alter CD8+ T cell activity and restore anti-tumor immune function in the tumor microenvironment (TME). Since then, considerable research has been conducted that ncRNAs regulate the anti-tumor effects of CD8+ T cells.
Currently, multiple lncRNA biomarkers obtained by invasive procedures show a great capability in mediating the interaction between tumor cells and CD8+ T cells, which has generated great research interest. Tim-3 is a negatively regulated T-cell-dependent immune responses’ immune checkpoint that serves as a perfect target for next-generation immunotherapy owing to its precision and specificity. A previous study reported that Lnc-Tim3, which is highly expressed in tumor-infiltrating CD8+ T cells, specifically bond to Tim-3 and blocked the interaction with Bat3. This phenomenon inhibits downstream Lck/NFAT1/AP-1 signal transduction, thereby exacerbating CD8+ T lymphocyte exhaustion (36). Other clinical studies found that the expression of NEAT1 was upregulated in peripheral blood mononuclear cells (PBMCs) from patients with HCC and could interfere with Tim-3 expression by binding to miR-155. Downregulation of NEAT1 inhibits apoptosis in CD8+ T cell and enhances cytolytic activity, thereby inhibiting tumor growth (37).
CircMET is a widely studied circRNA that is aberrantly expressed in HCC tumors (38). Mice subcutaneously implanted with Hep1-6-circMET had a smaller tumor burden and a higher density of tumor-infiltrating CD8+ T cells than those implanted with Hep1-6-control cell lines. This indicates that circMET is detrimental to CD8+ T cell infiltration, and in-depth studies revealed that it achieved its goal through the miR-30-5p/Snail/dipeptidyl peptidase-4 (DPP4) axis. Based on the combined clinical approach of DPP4 inhibitor and anti-PD1 blocking immunotherapy, it further validated that the DPP4 inhibitor sitagliptin can enhance CD8+ T cell trafficking and increase infiltration levels, thereby improving the efficacy of PD1 blockade immunotherapy, supporting the clinical application of combining DPP4 inhibitors with anti-PD1 blocking immunotherapy.
CD4+ T cells
CD4+ T cells should not be underestimated because they perform multiple functions in the adaptive immune system. They are not only able to kill tumor cells directly (cytotoxic CD4+ T cells) but are also well-known for their indirect role in the TIME as T helper (Th) cells. They can coordinate the enhancement of other anti-tumor effector cell functions, such as CD8+ T cell function and macrophage phagocytosis. Differentiation into various subpopulations, such as Th1, Th2, Th17, and regulatory T (Treg) cells, is induced in different cytokine environments, with different on anti-tumor effects (39, 40). Since the study of Treg cells requires an in-depth review of available information, it was singled out for discussion. ncRNAs are an integral part of gene expression networks, which dynamically regulate CD4+ T cells’ differentiation and plasticity. Dysregulation of certain ncRNAs in cancer cells can increase the levels of immunosuppressive factors, which in turn contributes to immune privilege (41).
Interestingly, lncRNA AC099850.3 exerts oncogenic effects via the PRR11/PI3K/Akt signaling pathway. An immune infiltration analysis revealed that T follicular helper cells and CD4+ memory T cells were activated while CD8+ T cells and monocytes were suppressed when AC099850.3 was up-regulated, explaining the oncogenicity of AC099850.3 (42). LncRNA MAIT is mainly expressed in CD4+ T cells from HCC tumor tissues and paracancerous tissues. It is not only positively correlated with the level of CD4+ T cell infiltration, but also with immunosuppressive molecules, such as PD-1, PD-L1, and CTLA4 (43). miR-26b-5p targeting proviral integrations of moloney virus 2 (PIM2) can affect the secretion of tumor necrosis factor α (TNF-α), interferon-γ (IFN-γ), interleukin-6 (IL-6), and interleukin-2 (IL-2) in CD4+ T cells (44). Th17 cells mediate pro-inflammatory functions by secreting cytokines (such as IL-17, IL-21, and L-22), and they participate in many organ-specific autoimmune diseases. Furthermore, ncRNA functions are being actively explored in Th17 cells in the TIME. miR-132 expression is much higher in CD4 IL-17+ cells than in CD4 IL-17- cells. miR-132 mediates Th17 cell differentiation by promoting IL-22 expression, which in turn enhances hepatic stellate cell (HSC) activation and induces tumor migration (45).
Regulatory T cells (tregs)
Tregs, a regulatory subpopulation of infiltrating CD4+ T cells, are recognized as a major suppressive component of the immune system. They are extremely important for the formation of an immunosuppressive microenvironment in HCC (46, 47). Tregs are critical in maintaining self-tolerance and immune homeostasis and are co-opted by tumor cells to evade immune surveillance. They are up-regulated in tumor tissues and peripheral blood from patients with HCC or mice than in healthy individuals (48). High FOXP3 expression on the cell surface is a distinctive feature of these cells (47, 49). Several studies have reported that the biological behavior and function of Tregs are partially dependent on the regulation of ncRNAs. ncRNAs affect the expression of immune-related cytokines and growth factors (e.g., IL-2) by regulating the secretion of chemokines (e.g., CCL22), which further affects the function and differentiation of Tregs that accumulate in the TIME (50, 51). Several studies have provided evidence suggesting that miR-34a, miR-15a, miR-16-1, lnc-EGFR, and lncRNA FENDRR play a crucial role in affecting Tregs in the HCC microenvironment.
CCL22 is a chemokine required for Tregs to exceed CD8+ T cells. As early as 2012, miR-34a strongly supported the idea that ncRNAs affect the immunosuppressive function of TIME by regulating the secretion of CCL22. Yang et al. found that elevated activity of tumor growth factor-β (TGF-β) suppressed miR-34a expression and dose-dependently enhanced production of CCL22 in PVTT-1 cells. This blocks the strong binding of CCL2 to CCR4 on the surface of Tregs, resulting in attenuated Treg cell recruitment and immune escape suppression (52). Similarly, miR-15a and miR-16-1 directly target NF-κB to impair CCL22 transcription. Subsequently, activated NF-κB/CCL22 signaling attenuates the hepatic recruitment of Tregs. Such biological activity also upregulates CD80 expression in Kupfer cells (KCs) and CD28 in Tregs, facilitating communication between KCs and Tregs (53). A study confirmed the existence of a forward-feedback loop lnc-EGFR-EGFR-NF-AT1/AP1-lnc-EGFR in Tregs as a facilitating mechanism for HCC (54). In addition, loss of GADD45B can upregulate the number of Tregs. However, the tumor suppressor lncRNA FENDRR targets GADD45B as a miR-423-5p sponge to suppress the secretion of immune-related factors TGF-β, vascular endothelial growth factor (VEGF), IL-2, and IL-10, thereby suppressing Treg-mediated immune escape (55).
B cells
B lymphocytes are derived from pluripotent stem cells in the bone marrow. They play a dual role in tumor immunity by supporting or suppressing anti-tumor immunity (56). B cells may act as antigen-presenting cells to enhance humoral and cellular responses to tumors. However, the strong prognosis of tumor-infiltrating B cells (TIL-Bs) in cancer reject this idea. Therefore, it is difficult to determine the specific role of B cells (57). One claim is that miRNAs may influence the differentiation of the regulatory B cells (Bregs). Similar to Tregs, Bregs produce high levels of IL-10 and suppress the host immune response, thereby exerting a pro-tumor effect (58). An increased frequency of CD19+ Tim-1 cells and tumor growth have been observed in young miR-15a/16-/- mice transplanted with HCC cells. This phenomenon depends on the efficiency of microRNA cluster of miR-15a/16 that enhances STAT3 activity. Then STAT3 activation contributes to IL-10 production by CD19 Tim-1 cells, and finally promotes Bregs’ activity (59). Pseudogenes are a special type of lncRNAs. The expression of pseudogenes RP11-424C20.2 correlates with the level of tumor-infiltrating immunocytes, including B cells (60). It linked high levels of B cells with worse outcomes for thymoma (THYM) patients. Necessarily, further studies are needed to explore how other ncRNAs exert regulatory effects on B cells and their specific mechanisms.
Natural killer cells
NK cells can exert their effects as an essential component of innate immunity even without prior stimulation, which constitutes the first line of host immunological defense against cancer cell invasion. NK cells lead to target cell apoptosis by secreting perforin and granzyme, expressing FasL, and mediating antibody-dependent cellular cytotoxicity (ADCC) (61, 62). Changes in the phenotypes and functions of NK cells have been detected both in patients with aggressive human liver cancer and transgenic mouse models (63, 64). Single-cell RNA sequencing and flow cytometry of innate lymphoid cells (ILCs) revealed that NK cells lose their cytotoxic profile as they transition into NK-like-ILC3 cells (65). Several therapies of NK cell-mediated ADCC have been evaluated in clinical trials. Undoubtedly, NK cells are promising candidates for the development of advanced cancer immunotherapy. Evidence collected so far suggested that multiple ncRNAs mediate interactions between NK and HCC cells.
CD69 is an NK cell activation marker that mediates NK cell cytotoxicity. When transferred to NK cells, miR-92b causes CD69 downregulation and cytotoxic damage (66). Chen et al. found that miR-137, miR-149-5p, and miR-561-5p are associated with the innate immune response, especially miR-561-5p. Additional evidence has shown some differences in chemotaxis and function among different NK cell subsets. miR-561-5p attenuated the anti-tumor response by downregulating CX3CL1 messenger RNA (mRNA) to reduce the function of CX3CR1 NK cells (67).
Recent studies have found that the LINC00638 is mainly enriched in eight signaling pathways, in which NK cell-mediated cytotoxic pathway is highly correlated with immune infiltration. In HCC tumor tissues, overexpression of ULBP1 can recruit NK cells to the tumor and leads to immune escape when accompanied by PD-L1 expression. Mechanistically, LINC00638 can achieve this goal by acting as a sponge for miR-4732-3p and eliminating the inhibition of ULBP1 expression (68).
Notably, attention has gradually been focused on circRNAs in HCC immunity regulation. One of the most well-known examples is circUHRF1 (hsa_circ_0048677). Highly expressed circUHRF1 sponges miR-449c-5p to upregulate the expression of the downstream target gene, T-cell immunoglobulin mucin 3 (TIM-3), thereby reducing the secretion of TNF-α and IFN-γ, which ultimately promote the immune escape of HCC cells (69). Moreover, the downregulation of hsa_circ_0007456 reduces NK cell susceptibility and attenuates their binding by the downstream miR-6852-3p/ICAM-1 axis, thus promoting the immune escape of HCC cells (70).
Tumor-associated macrophages
Macrophages are key mediators of tissue homeostasis. They can directly kill tumor cells by phagocytizing massive pathogens (71). Additionally, they are vital antigen-presenting cells that activate endogenous anti-tumor T cell responses. They are highly plastic and can be classified into two subtypes: classical pro-inflammatory activation (M1-like macrophages) and alternative anti-inflammatory activation (M2-like macrophages) (72). The tumor recruits them into the TIME and induces the formation of TAMs, which are crucial for promoting the immunosuppressive microenvironment, tumor cell invasion, angiogenic switch, and immune escape of malignant cells.
Previous studies have shown roles of miRNAs in M1/M2 macrophage polarization, which mediates the onset and growth of HCC. Zhao et al. reported that the CpG island deletion (ΔCpG) of the miR-144/miR-451a promoter induces chromatin conformational remodeling. It increases the expression of miR-144/miR-451a while decreasing the expression of hepatocyte growth factor (HGF) and macrophage migration inhibitory factor (MIF), conferring the paracrine activation of macrophage M1-like repolarization (73).
Many studies have revealed the mechanism of action of lncRNAs in TAM polarization regulation. Overexpression of PART1 promotes macrophage M2-like polarization by affecting the miR-372-3p/TLR4 axis (74). Similarly, promoting competitive adsorption of miR-147a by lncRNA HMMR-AS1 in a hypoxic environment affected ARID3A-mediated macrophage polarization (75). In addition, it has been found that LINC00662, lncRNA TUC339, lncRNA MALAT1, PCED1B-AS1, lnc-Ma301, and lncRNA cox-2 can promote the differentiation of TAMs into the M2 phenotype and inhibit the anti-tumor response (76–81).
Other important studies have confirmed the crucial role of circRNAs in orchestrating macrophage polarization in HCC progression. The use of macrophage-specific CD39-knockout mice showed that circTMEM181 upregulates CD39 expression in macrophages and CD73 expression in HCC cells. The cooperation between CD39 and CD73 triggers eATP-adenosine activation, thereby promoting immunosuppression (82). Hsa_circ_0110102 may act as a sponge for miR-580-5p to regulate target gene function. It regulates the secretion of CCL2 into the TME by decreasing PPARα expression. The release of pro-inflammatory cytokines from macrophages was inhibited by modulating the COX-2/PGE2 pathway. Thus, hsa_circ_0074854 may be a potential prognostic predictor or therapeutic target for HCC (83). Besides, the knockdown of hsa_circ_0074854 can inhibit M2 macrophage polarization both in vitro and in vivo. Mechanistically, the downregulation of hsa_circ_0074854 inhibits macrophage M2 polarization by interacting with human antigen R (HuR), thereby inhibiting the migration and invasion of HCC cells (84).
Other immune cells
In addition to the above-mentioned immune cells, ncRNAs exert regulatory effects on other immune cells, such as tumor-associated neutrophils (TANs) and dendritic cells (DCs). TAN is the most abundant circulating leukocyte in humans that mediates tumor growth and progression. It has two distinct phenotypes-N1 (anti-tumor) and N2 (pro-tumor), exhibiting functional heterogeneity in response to different stimuli (85). Therefore, it is a potent modulator of TIME. The deregulation of miR-223 may play a role in a range of liver diseases by affecting neutrophil infiltration. In addition, miR-223 expression positively correlates with the differentiation of granulocyte-monocyte progenitor cells into granulocytes (86). As antigen-presenting cells, DCs can induce primary immune responses (87). Wu pointed out that lncRNA ASB16-AS1 negatively correlated with dendritic cells and neutrophils as validated in five HCC cell lines (88). The mechanism by which ncRNAs regulate immune cells in the TIME is still in its infancy and needs to be supported by further studies.
Regulation of tumor angiogenesis
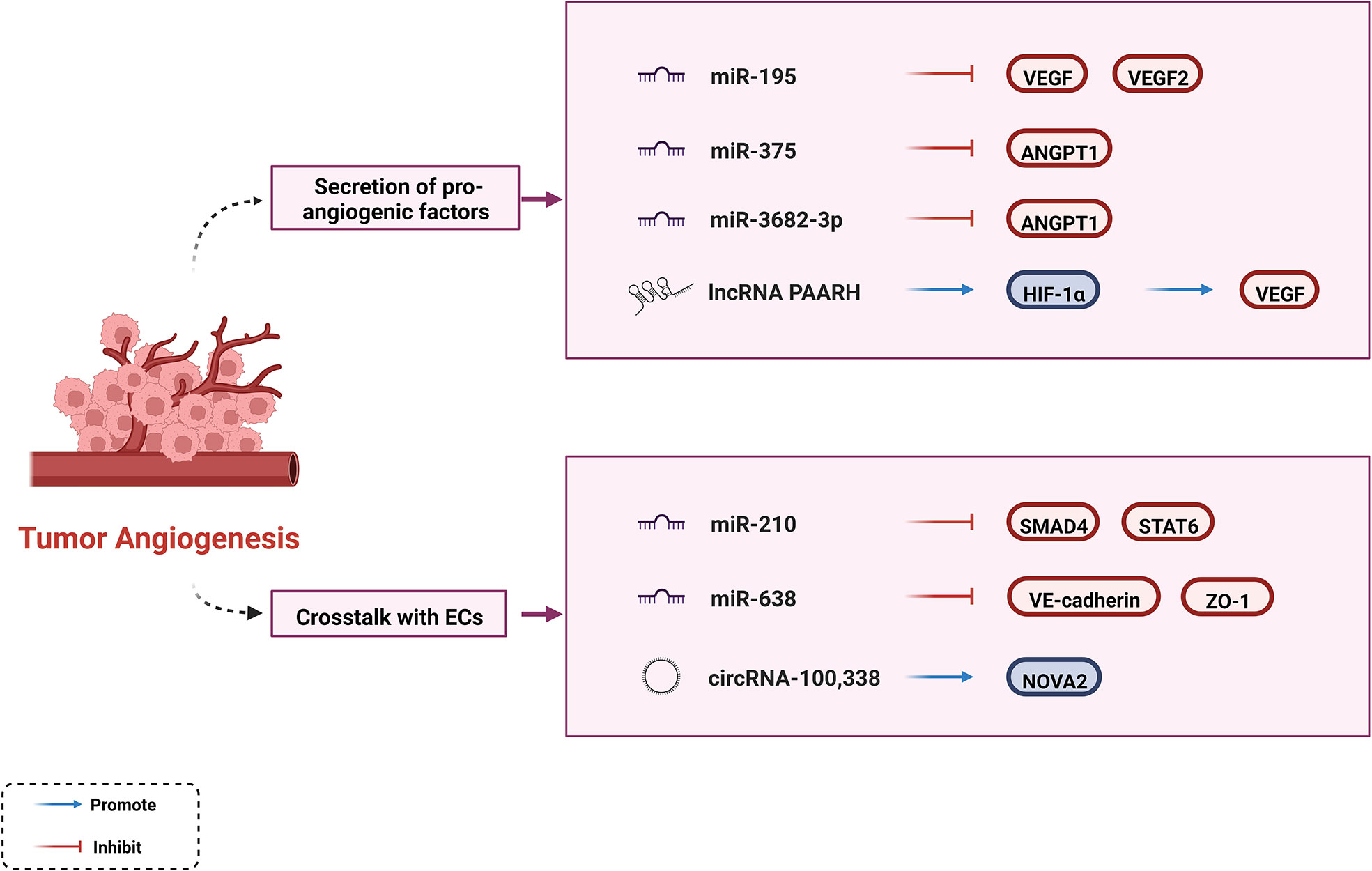
In the tumor growth environment, there is a dynamic imbalance between pro- and anti-angiogenic factors. As a hypervascular tumor, HCC tends to induce the secretion of proangiogenic factors such as VEGF, angiopoietin-1 (ANGPT1), platelet-derived growth factor (PDGF), and basic fibroblast growth factor (bFGF). This phenomenon contributes to angiogenesis, enabling tumors to receive adequate nutritional support and expel metabolic waste and carbon dioxide, leading to continuous tumor growth and progression (8, 89). Overexpression of VEGF contributes to vascular network development, endothelial cell proliferation, and tube formation (90, 91). Current anti-angiogenesis drugs, such as bevacizumab and ramucirumab, mostly target the VEGF signaling pathways. Accumulating evidence suggests that ncRNAs regulate tumor progression by interacting with VEGF in HCC. A previous study showed that miR-195 is negatively correlated with angiogenesis which directly inhibits VEGF levels and VEGF receptor 2 signaling in endothelial cells (92). Another study showed that the lncRNA PAARH positively correlated with vascular invasion in HCC tissues, upregulated VEGF expression and microvascular density. PAARH facilitated the recruitment of HIF-1α to the VEGF promoter, which caused high VEGF expression (93). In addition to VEGF, ncRNAs interact with other growth factors. By targeting ANGPT1, miR-375 suppresses proangiogenic activity and miR-3682-3p weakens angiogenesis both affecting tumor progression (94, 95).
ncRNAs promote angiogenesis through crosstalk with cancer-associated endothelial cells (ECs), affecting tube formation (30). It has been shown that miR-210 targets SMAD4 and STAT6 to stimulate EC tubulogenesis in vitro and angiogenesis in vivo. STAT6 alleviates the inhibitory effects of IL-13 on human coronary artery EC migration and tube formation; however, the mechanism of action of SMAD4 on ECs remains unknown (96). In contrast, ncRNAs inhibit the permeability of ECs to mediate cancer cell proliferation, which are potential targets for anti-angiogenic therapies. For example, miR-638 can disrupt endothelial tight junctions and enhance the permeability of FITC-dextran by downregulating VE-cadherin and ZO-1 expression in non-cancerous regions of the liver (97). Another study has explained the specific mechanism by which circRNA-100,338 regulates endothelial permeability. CircRNA-100,338 affects cell permeability and vasculogenic mimicry (VM) capability by interacting with NOVA2 and inhibits HCC growth by binding to IFN-α (98).
Collectively, ncRNAs can regulate angiogenic activity through different pathways (Figure 2). ncRNAs may be emerging targets for anti-angiogenic therapies. The clinical benefits of multi-kinase inhibitors (such as Sorafenib and Lenvatinib) that target VEGF and its receptors have not been as good as initially expected, perhaps partly because of the off-target effect of anti-tumor agents (99). Synergistic biological effects exist between antiangiogenic agents and ICIs, and their toxicity profiles do not overlap (99). Based on these characteristics, some experts have focused on promoting the ncRNA-targeting combinations of ICI inhibitors with Sorafenib or other anti-angiogenic drugs, which could contribute through the aforementioned mechanism.
Regulation of EMT
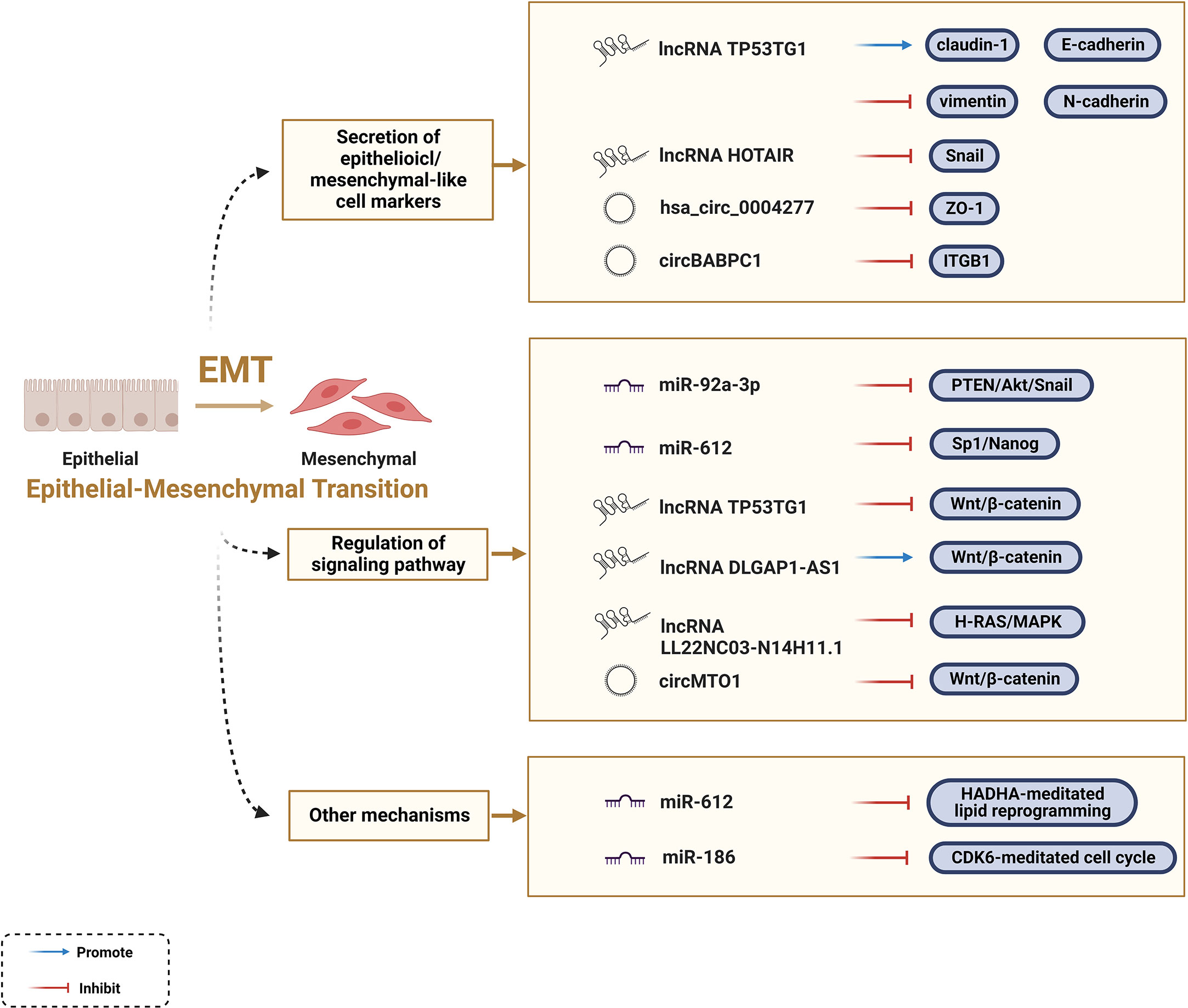
EMT is indispensable at all life stages from embryonic development to death, and it is essential for the acquisition of the invasive and metastatic characteristics of malignant tumors. Featured with reversibility, plasticity, and heterogeneity, EMT can alter multiple phenotypic changes including the loss of cell polarity, dissolution of intracellular junctions, and basement membrane detachment (9, 10). Epithelioid cell markers, such as β-catenin, E-cadherin, ZO-1, and claudin-1, were negatively correlated with the EMT process, while mesenchymal-like cell markers, such as Snail, twist, vimentin, N-cadherin, ZEB, and α-SMA, were positively correlated with the EMT process. Besides, certain signaling pathways facilitate the EMT process, such as the Wnt/β-catenin signaling pathway. An increasing number of studies have shown that ncRNAs act as mediators affecting the EMT process in tumor cells (28).
ZO-1 regulates cell material transport and maintains epithelial polarity. Circ-0004277 competitively binds to HuR and blocks the binding of ZO-1 and HuR, thereby stimulating EMT progression and promoting HCC (100). LncRNA TP53TG1 is a tumor suppressor gene that negatively correlates with N-calmodulin and vimentin, and positively correlates with E-cadherin and claudin-1 at the protein and RNA levels (101). Snail acts as a major player in EMT, whereas HOTAIR negatively regulates the EMT process (102). The results confirmed that HOTAIR-sbid, a HOTAIR deletion mutant, can impair the interaction between Snail and EZH2, thus affecting the capacity of Snail to convey EZH2 to specific epithelial target sites (i.e., HNF4α, E-calmodulin, and HNF1α). The design of such dominant-negative mutants opens new perspectives for highly specific future RNA therapeutics to counteract tumor progression (103). A recent investigation showed that the tumor-suppressive function of circPABPC1 is manifested by promoting the degradation of β1 integrin (ITGB1), a pivotal member of the integrin family, thereby reducing cell adhesion between cells and the extracellular matrix (104). In contrast, lncRNA TP53TG1 interacts with peroxiredoxin-4 (PRDX4) to promote its ubiquitin-mediated degradation, subsequently downregulating the levels of proteins involved in the Wnt/β-catenin pathway, thereby slowing down the EMT process (101). Notably, a growing body of information is available on different ncRNAs, such as miR-1246, lncRNA DLGAP1-AS1, and circMTO1, which regulate the Wnt/β-catenin pathway to affect the EMT process (105, 106). In addition, miR-92a-3p inhibits the PTEN/Akt/Snail pathway, miR-612 inhibits the Sp1/Nanog signaling pathway, and lncRNA LL22NC03-N14H11.1 activates the H-RAS/MAPK pathway to induce mitochondrial fission, all of which can explain the key role of ncRNAs in EMT progression (107–109).
Furthermore, scientists have elucidated how ncRNAs regulate the EMT process in HCC from some new perspectives. In addition to the Sp1/Nanog signaling pathway mentioned above, Liu et al. found that miR-612 regulates the EMT process through direct downstream target HADHA-mediated lipid reprogramming. Mechanistically, miR-612 affects invadopodia formation through HADHA-mediated alterations in the β-oxidation of fatty acids, cholesterol biosynthesis, and cell membrane fluidity (110). Evidence also indicates that cell cycle-related genes are involved in the regulation of ncRNAs in EMT; for example, miR-186 induces apoptosis by directly targeting cyclin-dependent kinase 6 (CDK6) (111). In conclusion, ncRNAs are key factors regulating the EMT process in HCC (Figure 3); however, their regulatory functions and underlying mechanisms remain to be elucidated.
Regulation of tumor invasion and metastasis
Tumor invasion and metastasis are complex processes of multi-stage evolution and hallmarks of malignancy, and can lead to a low survival rate in patients with distal metastasis. Even more problematic is that the current clinical situation is not conducive for detecting dormant cancer cells and micro-metastasis. This further exacerbates the difficulty of treatment, which requires a deeper understanding of tumor invasive and metastatic mechanisms. The processes remodeling the extracellular microenvironment that ncRNA-induced, such as angiogenesis and EMT, are niches for tumor metastasis. Those have been described in detail in the previous two sections. Further, malignant cells’ ability to distant movement is a favorable niche for tumor metastasis (112). Research on ncRNAs in the complex invasive-metastatic cascade response has provided new insights into the molecular mechanisms involved in hepatocellular carcinoma.
Upregulation of miR-1251-5p in tissues of HCC patients is significantly associated with clinical stage, high tumor lymph node metastasis (TNM), and poor prognosis. miR-1251-5p overexpression promoted cell invasion in vitro, whereas miR-1251-5p silencing inhibited HCC cell invasion in vivo. A-kinase anchor protein 12 (AKAP12) exerts anti-tumor effects by acting as a scaffolding protein in signal transduction. The luciferase report showed that miR-1251-5p could directly target AKAP12 for oncogenic effects, and this mechanism was validated by AKAP12 knockdown rescuing the miR-1251-5p knockdown-attenuated cell invasion (113). miRNAs have been studied for a long time and there is now a wealth of data on their broad and critical role in tumor metastasis through target genes and signaling pathways, such as miR-195, miR-122, miR-103a-3p (92, 114, 115). LncRNAs regulate the expression of target genes by mediating miRNA activity. Metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) is a widely studied lncRNA in cancer and is important for cancer-related pathway regulation. The luciferase reporter assay confirmed that MALAT1 negatively regulates miR-200a expression and is involved in the proliferation and invasion of Hep3B cells under hypoxia (116). It is well established that lncRNA is a key regulator of tumorigenesis. Liu’s research cascaded the exact correlation of this lncRNA to hepatocyte invasion and metastasis and revealing downstream regulatory pathways. The potential miRNA binding site of lncRNA BACE1-AS, miR-377-3p, was identified by a raw letter prediction and experimentally confirmed method. Further, the analysis showed that miR-377-3p negatively regulates CELF1, an RNA-binding protein that is a relative marker of malignancy (117). MMP9, a member of the zinc-dependent endopeptidase family, is a vital role in metastasis, especially in degrading ECM. Mouse xenograft models and mouse lung metastasis models confirmed the pro-tumor growth and lung metastasis role of circUBAP2. It negatively regulates HCC by acting as a competing endogenous RNAs(ceRNAs) for miR-194-3p and inhibiting MMP9 (118). In conclusion, complex biochemical and biological alterations in the tumor cells themselves and the associated stroma contribute to this aggressive phenotype. The involvement of ncRNAs in this process also expands the hope for finding new regulatory key nodes and therapeutic strategies for metastatic tumors.
Regulation of tumor metabolism
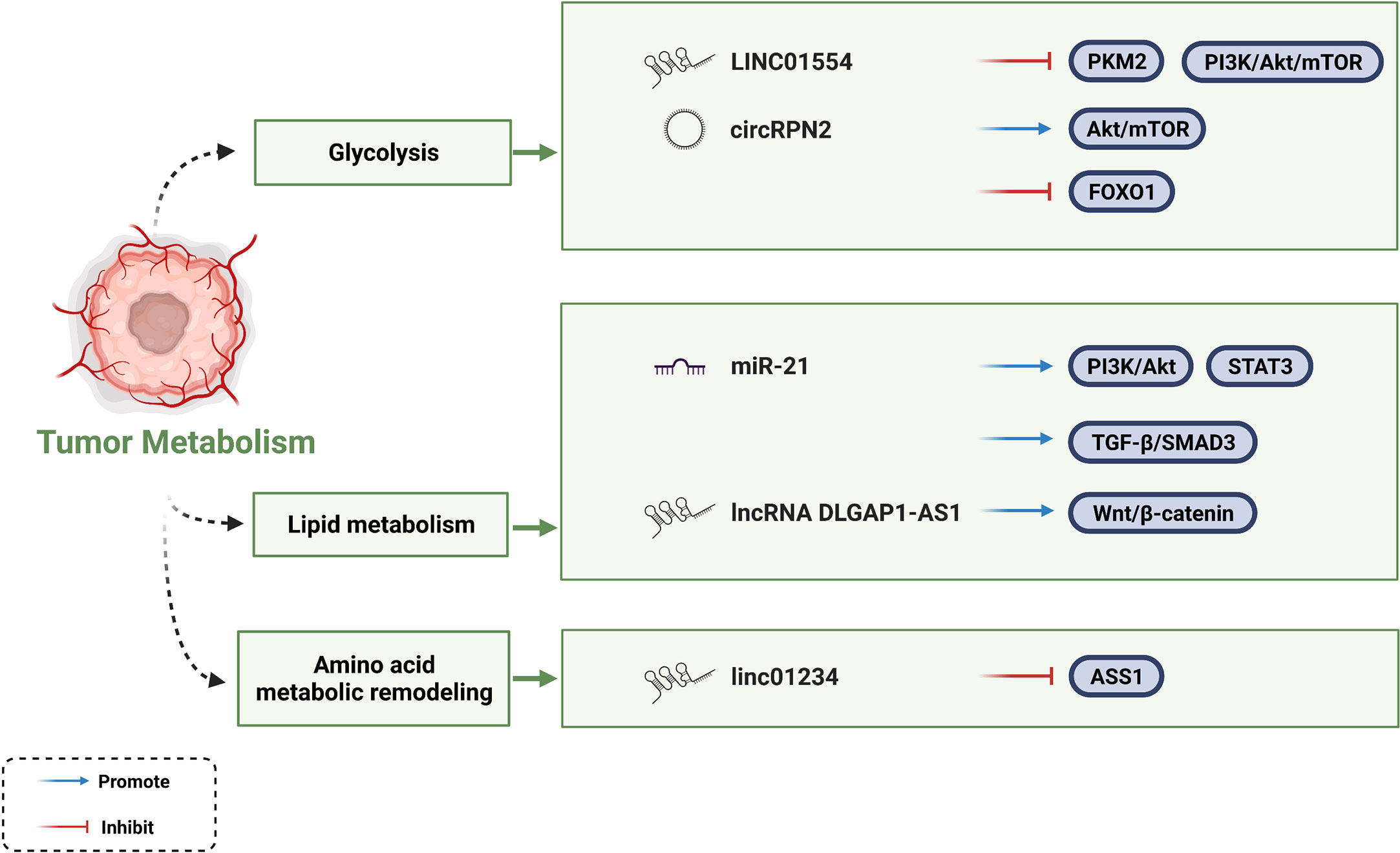
Vasculature-restricted glucose and oxygen delivery is insufficient to supply uncontrolled proliferation of cancer cells. Therefore, cancer cells undergo metabolic reprogramming as a survival strategy. Cancer cells generate energy through aerobic glycolysis and lactic acid fermentation in a process rather than the TCA cycle to meet the energy demand of rapid cancer cell proliferation. This process is known as the Warburg effect. In this way, additional substrates and energy requirements can be provided for the biosynthesis of macromolecules (14, 119, 120). Several ncRNAs have been shown to rewire glycolytic networks (14). Zheng et al. found that LINC01554 negatively regulates the key rate-limiting enzyme pyruvate kinase isozyme M2 (PKM2) in the aerobic glycolysis pathway of HCC. However, whether this mechanism is due to ubiquitin-mediated degradation or as a scaffold remains to be further investigated. In contrast, LINC01554 inhibits the PI3K/Akt/mTOR signaling pathway, which is a central signaling pathway coordinating glucose uptake, glycogen synthesis, and tumorigenesis (121). Consistent with LINC01554, circRPN2 also facilitates glycolytic reprogramming through the Akt/mTOR pathway (122). In addition, it inhibits aerobic glycolysis by regulating the miR-183-5p/FOXO1 axis. Moreover, studies have verified that lncRNA TUG1, lncRNA RAET1K, and circFBLIM1 play a role in metabolic reprogramming by regulating glycolysis (123–125).
In addition to their roles in glycolytic metabolism, miR-21 and miR-122 play central roles in hepatic lipid metabolism and cholesterol synthesis relying on a complex lipogenic transcriptional network (126, 127). miR-21 regulates lipid metabolism through at least three pathophysiological pathways in a zebrafish model. One of the approaches is that miR-21 promotes hepatic lipid accumulation via the PTS (PI3K/Akt, TGF-β/SMAD3, STAT3) signaling networks. As for miR-122, free fatty acid (FFA) increases miR-122 secretion and transportation by activating retinoic acid-related orphan receptor alpha (ROR-α). In turn, miR-122 reduces the mRNAs levels of genes (Agpat1 and Dgat1) involved in triglyceride synthesis.
Amino acid metabolic remodeling is another powerful proponent of cancer cell malignant activities. Mechanism can be attributed to the interaction between lncRNA and rate-limiting enzyme of amino acid synthesis pathway. Chen et al. found that LINC01234 bound to the promoter of ASS1 to inhibit its transcriptional activation, thereafter leading to increased aspartate levels and activation of rapamycin pathway targets (128). As the understanding of tumor cell metabolism in TME continues to advance, people have discovered multiple strategies to target tumor metabolism. Currently, fatty acid synthesis inhibitors (FAS), especially targeting fatty acid synthase (FASN), have been focused on as potential strategies for cancer treatment. The molecular mechanisms by which the ncRNAs mentioned in this section affecting HCC metabolism are summarized in Figure 4.
Regulation of drug resistance
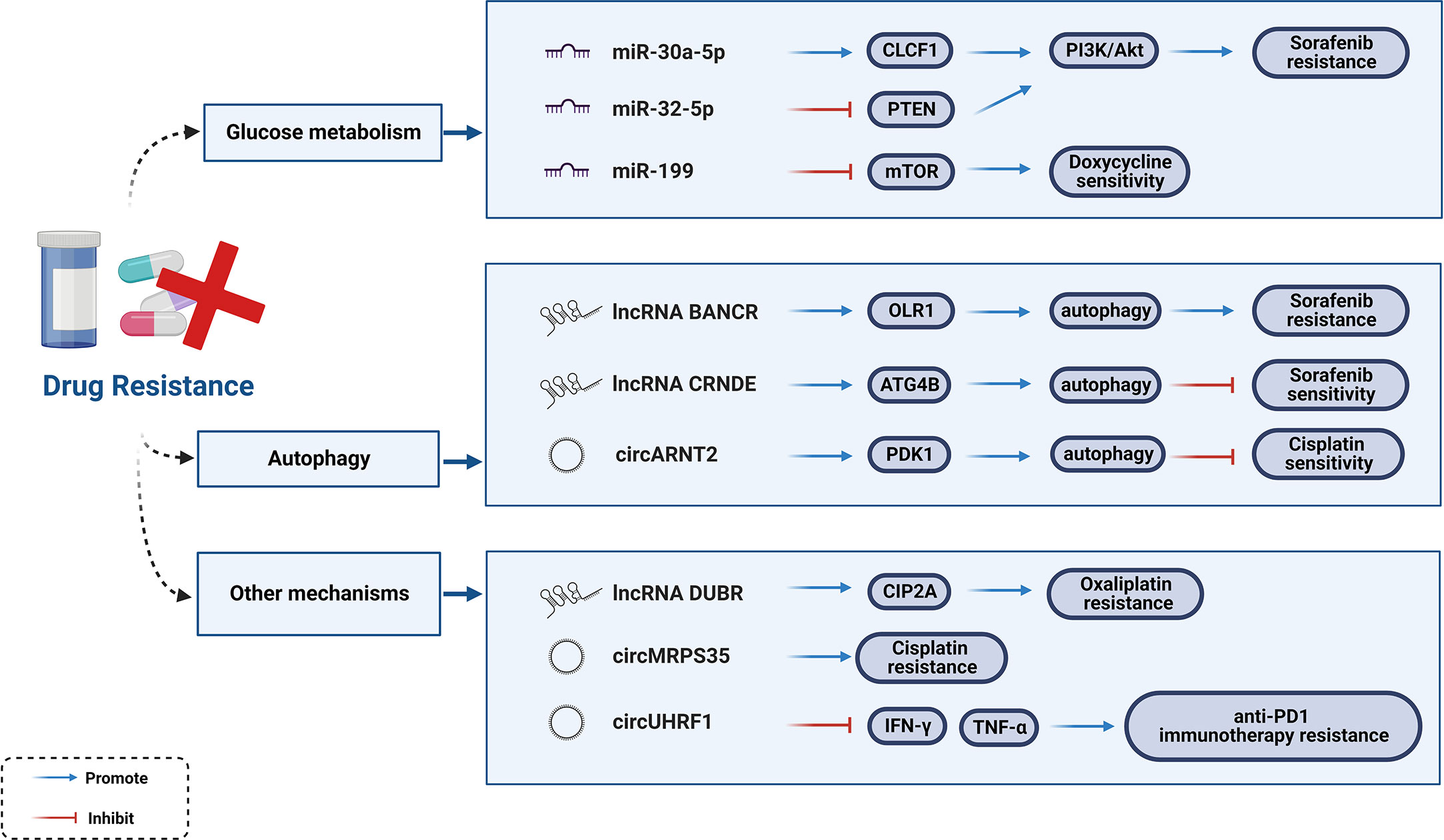
For unresectable HCC, the main treatment approaches include chemotherapy (e.g., Cisplatin and 5-Fluorouracil) and molecular targeted therapies (e.g., multi-kinase inhibitors, monoclonal antibodies, and ICIs). Multidrug resistance (MDR) is inevitable in tumor cells with their rapid development. Statistics indicate that only 30% of HCC patients show an increase in overall survival (OS) by 3 months after treatment with Sorafenib (129). Moreover, Sorafenib resistance was observed within 6 months of treatment, with adverse events of varying degrees, such as gastrointestinal reactions, hand-foot syndrome, and hypertension (130, 131). To further understand the emerging function and mechanism axis of ncRNAs in HCC, chemoresistance has become a hot research topic.
Aberrantly expressed miRNAs are universal features of HCC. From the perspective of clinical medication (Sorafenib, 5-Fluorouracil, Cisplatin), researchers have systematically elaborated on miRNAs modulating HCC drug resistance as well as the underlying mechanisms (132). Glucose metabolism has been implicated in the maintenance of Sorafenib resistance in HCC cells (133). Activation of the PI3K/Akt signaling pathway enhances glycolysis, and the expression of downstream glycolysis genes cause reactions in Sorafenib-resistant HCC cells. Studies verified that both miR-30a-5p and miR-32-5p were abnormally expressed in drug-resistant tissues, and may be targets for reversing Sorafenib resistance in HCC. miR-30a-5p induces MDR to activate the PI3K/Akt signaling pathway by upregulating CLCF1, while miR-32-5p activates the PI3K/Akt signaling pathway by downregulating PTEN (133, 134). Intravenous injection of miR-199-modified vehicle suppress mTOR signaling (135, 136). Overactivation of the mTOR signaling pathway promotes tumorigenesis and tumor progression, which significantly sensitizes HCC cells to Doxycycline (137).
Autophagy plays a dual role in drug resistance. Few lncRNAs influence Sorafenib-induced chemoresistance and sensitivity through autophagy mechanism. A previous study showed that Rutin treatment reduced the number of autophagosomes in Sorafenib-resistant cells, while reducing the expression of the lncRNA BANCR (BANCR knockdown increases the sensitivity of Sorafenib-resistant cells to Sorafenib). The lncRNA BANCR inhibits autophagy through the BANCR/miRNA-590-5P/OLR1 axis (138). Another study reported that the lncRNA CRNDE has the opposite effect as it enhances the stability of ATG4B primarily by isolating miR-543, thereby triggering autophagy (139). The autophagy mechanism of drug resistance is regulated not only by lncRNAs but also by circRNAs. Li et al. identified 968 dysregulated circRNAs in Cisplatin-resistant HCC tissues and reported that circARNT2 competes with miR-155-5p to upregulate PDK1-induced autophagy, ultimately enhancing the sensitivity of HCC cells to Cisplatin (140).
In addition to autophagy, circRNAs function through many other mechanisms. For instance, circMED27, circMEMO1, circFOXM1, circFN1, and circRNA-SORE act as ceRNAs targeting corresponding miRNAs to affect the drug resistance or sensitivity of HCC cells to chemotherapy drugs (140–144). Existing studies have confirmed the molecular mechanisms associated with resistance to the first-line chemotherapy drug, Oxaliplatin (OXA), in which cancer stem cells (CSCs) play an important role (145, 146). The lncRNA DUBR is highly expressed in liver CSCs and functions as one of the factors that promote OXA resistance. Liu et al. found that the specificity protein 1 (SP1)-induced lncRNA DUBR upregulates CIP2A expression via the E2F1 protein, promoting stemness and OXA resistance. They also identified another mechanism by which lncRNA DUBR acts as a ceRNA to upregulate CIP2A, which in turn stabilizes the E2F1 protein, thereby activating the Notch1 signaling pathway (147). Additionally, circMRPS35-encoding peptide is significantly induced by chemotherapeutic agents and promotes Cisplatin resistance, suggesting that circMRPS35 may be a possible mediator of Cisplatin resistance (148). Interestingly, emerging evidence suggests that ncRNAs are involved in the resistance to immune checkpoint blockers. A newly reported example is that where circUHRF1 can impair the function of NK cells and induce an exhausted phenotype that cannot secrete IFN-γ and TNF-α. Hence, circUHRF1 could be regarded as a therapeutic target for anti-PD1 immunotherapy and drug resistance (69). In conclusion, clarifying the mechanisms of drug resistance (Figure 5) and targeting these dysregulated ncRNAs would be a fatal step forward in HCC treatment.
Clinical significance of ncRNAs in HCC
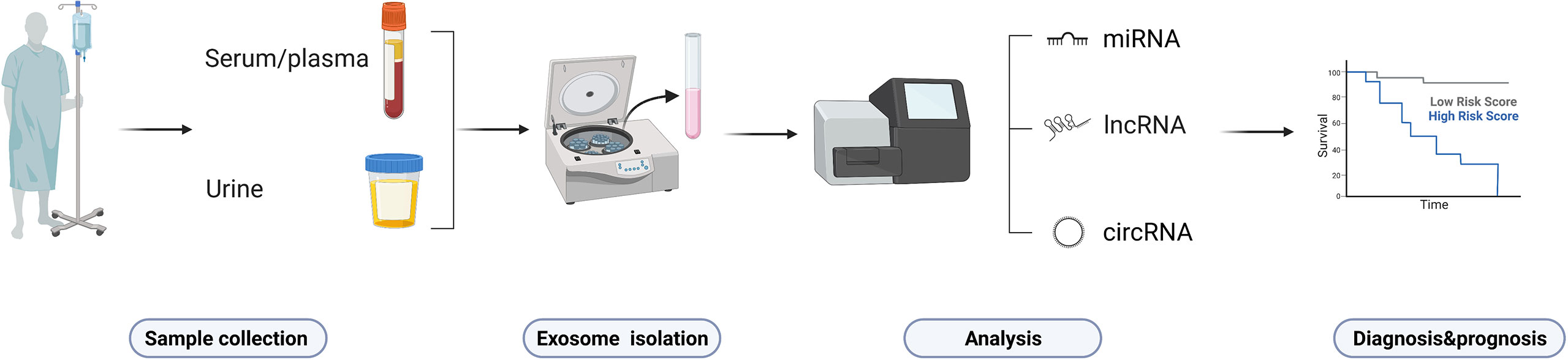
Most HCC patients are often diagnosed too late or have a high recurrence rate, that is why exploring predictive/prognostic biomarkers in early-stage of HCC is relevant for physicians to develop precise treatment strategies (149). Evaluation of tissues from liver biopsies and surgical specimens are both invasive approaches that too painful for the patient. α-fetoprotein (AFP) is the “gold standard” but with suboptimal sensitivity and specificity of only 39-64% and 76-91% (150, 151). Moreover, AFP appears as an unanticipated false positive that is elevated in some patients with chronic liver diseases (e.g., cirrhosis, viral hepatitis, etc.). More reliable biomarkers are urgently needed to monitor and diagnose HCC and improve patient prognosis. The secretion of circulating ncRNAs has been identified in biological fluids (e.g., serum, plasma, and urine) of patients (the process is shown in Figure 6), where changes in levels or expression indicate cancer status. Owing to their abundance, accessibility, non-invasiveness, easy reproducibility, and disease specificity, ncRNAs are considered ideal non-invasive diagnostic biomarkers for HCC (152). As a non-invasive liquid biopsy, ncRNAs have entered the spotlight for the development of diagnostic markers in oncology. Several ncRNAs in body fluids have been the most promising biomarkers to date. This section summarizes the main diagnostic/prognostic markers currently under investigation.
ncRNAs act as diagnostic biomarkers
miRNAs
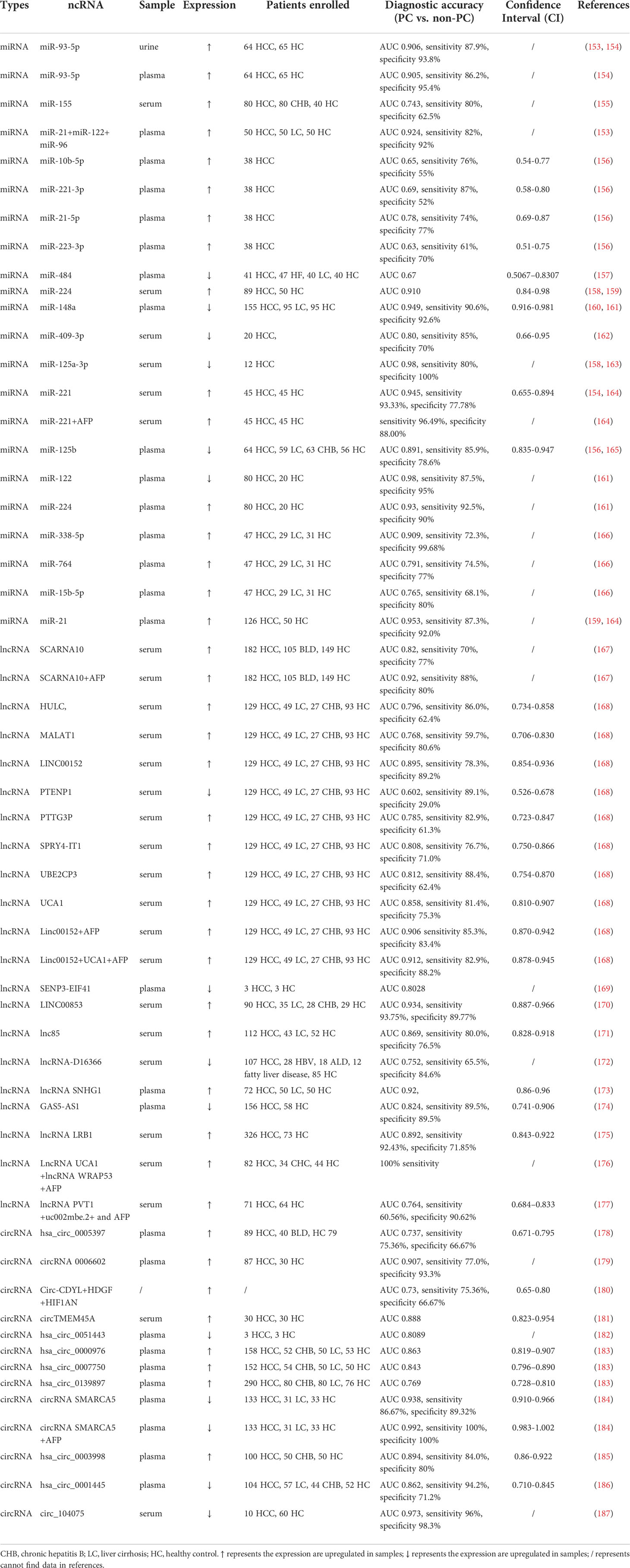
In recent years, several research groups have evaluated the potential of miRNAs as diagnostic, prognostic, and therapeutic responsiveness biomarkers for liver disease using clinical trial databases or clinical trials. In addition, the clinical utility of circulating miRNAs in patients with HCC has been explored, such as miR-21, miR-122, miR-96, miR-194/192, and miR-484; more information is summarized in Table 1. The plasma oncogenic factor miR-21 has been explored as a biochemical marker for HCC considering its important role in HCC progression. The potential of miR-21 as a biomarker for HCV patients complicated by HCC was explored by collecting trials registered in the clinicaltrial.gov database, enrolling a total of 100 participants aged 31-67 years in Egypt (NCT05449847). miR-21 can differentiate between uncomplicated and complicated HCV patients. It has been suggested that multiple miRNAs or a combination of miRNAs with the already widely used AFP may be more desirable diagnostic modalities. For instance, the combination of miR-21, miR-122, and miR-96 in serum showed a much higher diagnostic accuracy in the cirrhotic group than AFP or miR-21 alone (153). A combination of miRNAs has shown a higher area under the curve (AUC) value of 0.924. Thus, miR-21 was not only superior to AFP alone, but also showed better differentiation in combination with AFP. Studies have confirmed that miR-221 is also upregulated in patients with HCC. The AUC of the combination with AFP for diagnosis is 0.945, sensitivity is 93.33%, specificity is 77.78%, accuracy is 90.9%, and thus the combination has a better performance than individual detection (164). Clinical trials of miR-221 have also been conducted, and details can be found by querying NCT02928627. Shohda identified liver-specific miR-484 as an early diagnostic marker for HCC. miR-484 has shown great sensitivity in distinguishing HCC from non-HCC, with an AUC of 0.67. Moreover, miR-484 signatured across various stages of HCV-mediated hepatic disease progression, revealing promising performance in staging, prognosis, and early diagnosis (157).
LncRNAs
LncRNA UCA1 and lncRNA WRAP53 act as natural p53 single transcripts and are effective in regulating the expression of corresponding sense genes. The role of lncRNA UCA1 in bladder cancer and breast cancer has been previously identified, while the role for HCC remains elusive. In a study, both lncRNAs were found to be highly expressed in the sera of HCV patients with HCC, with AUC-ROC of 0.76 and 0.87, respectively. Interestingly, the combination of lncRNA UCA1, lncRNA WRAP53, and AFP resulted in a high diagnostic sensitivity of 100%, strongly confirming the diagnostic efficacy of the combination of miRNA and AFP (176). To validate these two lncRNAs as potential biomarkers for HCC diagnosis, a clinical trial has been published, enrolling a total of 80 participants. This work is nearing completion and we will wait to see the results of the validation (NCT05088811). In another study, Huang discussed the diagnostic efficacy of eight lncRNAs alone and in combination with AFP, in which the combination of LINC00152 and AFP in serum had the highest accuracy in predicting HCC. The AUC and sensitivity of LINC00152 alone are limited and correspond to an increase when used in combination with AFP. Next, researchers have investigated the diagnostic ability of the combinations of various lncRNAs and AFP, and the combination of LINC00152, UCA1, and AFP has shown the most reliable predictive ability, with an AUC of 0.912, sensitivity of 82.9%, and specificity of 88.2% (168). The expression of SENP3-EIF4A1 in patients with HCC was significantly lower than that in healthy controls, with an AUC of 0.8028 obtained by ROC analysis. Simultaneously, its expression is associated with tumor size, tumor stage, and lymph node metastasis (169). The ROC curve showed that LINC00853 exhibited excellent discriminatory ability in the diagnosis of all-stage HCC (AUC = 0.934, 95% CI = 0.887-0.966). On comparing the diagnostic performance with that of AFP with a 14-fold cut off value, LINC00853 showed sensitivity, specificity, and positive predictive values of 93.75%, 89.77%, and 76.92% respectively. These parameters all far exceed the sensitivity of 9.38%, specificity of 72.73%, and positive predictive value of 11.11% exhibited by AFP for diagnosis of early-stage HCC (170). The combination of lncRNA PVT1, uc002mbe.2, and AFP also far exceeded either the lncRNA or AFP assessed alone. Surprisingly, this combination distinguished patients with HCC from healthy controls, regardless of whether the patients were infected with HBV (177).
CircRNAs
Compared to miRNA and lncRNA, the number of studies on circRNA as a diagnostic biomarker is relatively few. Plasma circSMARCA5 can differentiate liver disease progression with high accuracy (AUC = 0.938, sensitivity of 0.853, specificity of 0.711). More excitingly, it has significant implications in diagnosing HCC patients with low AFP levels, especially for those with serum levels below 200 ng/ml, and is a better serum predictor for patients with poorly diagnosed AFP (184). Circ_104075 is highly expressed in patients with HCC and levels decreased after curative surgery. It is positively correlated with stage of HCC and was able to predict the development of disease. It has an AUC of 0.973, sensitivity of 96.0%, and specificity of 98.3%, surpassing the classical protein biomarkers AFP, α-fetoprotein-L3 (AFP-L3), and des-carboxy-prothrombin (DCP) (187). Likewise, circRNA applies to this combination. Circ-CDYL, the most significantly upregulated circRNA in a ncRNA network from a validated tumor tissue, interacts with the target genes encoding hepatoma-derived growth factor (HDGF) and hypoxia-inducible factor asparagine hydroxylase (HIF1AN). These proteins are highly and specifically expressed in early-stage of HCC. Compared with early-stage HCC patients, the combination of Circ-CDYL, HDGF, and HIF1AN increased the AUC, sensitivity, and specificity to 0.73, 75.36%, and 66.67% respectively. These results confirmed that the diagnostic efficiency of circ-CDYL or circ-CDYL in combination with HDGF and HIF1AN was higher than that of AFP alone, however this does not apply to advanced HCC (180). Along with the proliferation of research on circRNAs, circRNAs that can be used as diagnostic biomarkers also containing circTMEM45A, hsa_circ_0051443, hsa_circ_0000976, hsa_circ_0007750, and hsa_circ_0139897 (181–183).
ncRNAs act as prognostic biomarkers
HCC is known for its high recurrence rate and poor prognosis. In addition to their diagnostic values, circulating ncRNAs have been identified as valid prognostic markers. When changes occur in the human body, the altered levels of these ncRNAs can be used as a feature and a “beacon” for cancer prognosis. Studies on the application of ncRNAs as prognostic biomarkers in hepatocellular carcinoma are summarized in Table 2.
miRNAs
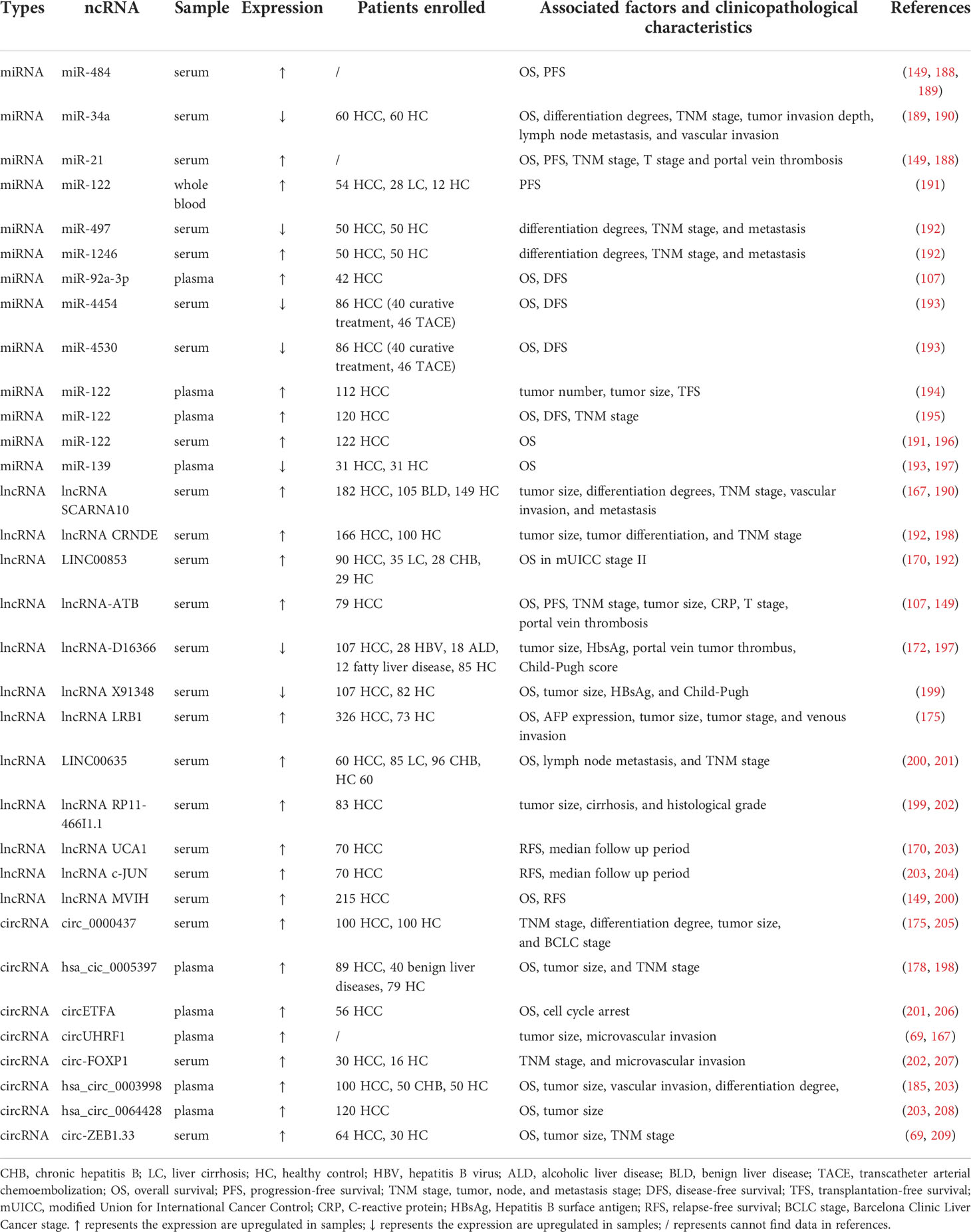
Differentially expressed serum/plasma miRNAs are helpful in the prognosis of HCC. Circulating miR-21 has long been shown to be an oncogenic factor, with its high serum levels predicting poor prognosis. Extensive studies have associated it with poor survival, high levels of recurrence, and an increased risk of disease progression. According to Lee’s study, miR-21 not only correlates with OS and progression-free survival (PFS), but also relates to multiple prognostic factors such as tumor, nodes, and metastases (TNM) stage, T stage, and portal vein thrombosis (149) (188). When using the TNM stage system, miR-21 expression was higher in stages III and IV than in stages I and II; when using the BCLC staging system, miR-21 expression was higher in late BCLC stages C-D than in early BCLC stages 0-B (149). Another study showed that elevated miR-484 in serum could predict HCV-induced liver lesion progression and is strongly associated with shorter OS and PFS (189). In addition, increased levels of miR-122 in serum, predict a longer survival rate (196). By testing blood samples from 50 each of HCC patients, cirrhotic patients and healthy volunteers, miR-122 was found to be less expressed in HCC patients, while miR-21 and miR-96 were opposite. The expression of all three affected the survival time of patients. Interestingly, the combination of the three was better than each miRNA alone in predicting the survival time of patients (153). Unlike the majority of reports published, miR-21 were measured in whole blood samples rather than in serum or plasma in Pelizzaro’s study (191). In a study, a KM survival analysis showed that high expression of miR-4454 and miR-4530 was significantly associated with improved OS (193). The above evidence confirms that miRNAs are potential prognostic markers of HCC.
LncRNAs
LncRNA MVIH, a microvascular invasion-associated lncRNA, is highly expressed in HCC. Sheng et al. first identified the relationship between up-regulated lncRNA MVIH expression and specific clinicopathological characteristics. Kaplan-Meier’s analyses of correlations between lncRNA MVIH expression and RFS and OS of 215 HCC patients after hepatectomy indicated that lncRNA MVIH is an independent risk factor for RFS and OS (200). The expression of lncRNA X91348 in patients with HCC was significantly lower than that in healthy individuals. A total of 107 HCC patients and 82 age- and sex-matched healthy volunteers were included in this study. Clinicopathological characteristics such as tumor size, HBsAg, and Child-Pugh could be observed influence the expression of X91348. The relationship between X91348 expression and survival was assessed after 5 years of follow-up. A median follow-up rate of 31.02 ± 15.11 months for patients with HCC was obtained, and the OS of patients with high X91348 expression was longer than that for patients with low expression. In conclusion, X91348 has a satisfactory prognostic ability (199). LINC00853 serves not only as a diagnostic biomarker but also as a prognostic marker in this cohort. High LINC00853 expression predicted lower OS in modified Union for International Cancer Control (mUICC) stage II and was independent of other stages of HCC (170). High expression of HOTTIP in the blood is an independent prognostic factor for tumor recurrence after liver transplantation, suggesting a short OS (204). Higher serum levels of circulating lncRNA-ATB could be associated with OS, PFS, TNM stage, tumor size, C-reactive protein (CRP), T stage, and portal vein thrombosis (149, 195). Moreover, a variety of lncRNAs have been investigated in Table 2.
CircRNAs
Stably expressed in the plasma, hsa_cic_0005397 has an intact closed-loop structure. According to a survival analysis based on follow-up data, hsa_cic_0005397 may serve as an independent prognostic marker for OS, as evidenced by the positive correlation between tumor size and TNM stage (178). Regarding circRNAs, the high expression of circUHRF1 in the plasma is associated with large tumor size and high microvascular invasion, which indicates the potential of circUHRF1 in predicting poor prognosis (69). Clinically, high expression of circ-FOXP1 and Circ-ZEB1.33 in serum is strongly associated with large tumors, advanced TNM, and poor prognosis (207, 209). In vitro experiments have also confirmed that circ-ZEB1.33 and circETFA affect tumor cell proliferation by regulating the cell cycle, likely serving as a novel prognostic marker (206). Circ_0000437 expression was positively correlated with TNM classification, differentiation degree, tumor size, and Barcelona Clinic Liver Cancer (BCLC) stage; hsa_circ_0003998 expression positively correlated with serum AFP level, tumor diameter, and microvascular invasion, whereas hsa_circ_0064428 expression was negatively correlated with patient’s survival, tumor size and metastasis. To date, fewer circRNAs derived from plasma or serum have been used as prognostic markers compared with those obtained from tissues. In summary, the prognostic information for circRNAs derived from body fluids remains to be explored.
In summary, ncRNAs can be valuable biomarkers for the diagnosis and prognosis of HCC. However, the current problem is that studies about ncRNAs are scarce as diagnostic and prognostic biomarkers (especially those derived from blood and urine) (149). The heterogeneity of tumors in different populations is a significant challenge for their application. Therefore, future multicenter and large-scale diagnostic/prognostic nested case–control studies are required to validate their utility. Fortunately, institutions have begun to validate the potential of certain ncRNAs as markers clinically, such as miR-21, miR-221, lncRNA UCA1, and lncRNA WRAP53 in the clinical trials NCT05449847, NCT02928627, NCT05088811 mentioned above. Although some trials are not yet completed, these are “milestones” for ncRNAs as clinical biomarkers for HCC. It is also believed that more hospitals, research institutions and companies will be involved in the exploration and development of clinical biomarkers.
Methods for detecting ncRNAs in clinical samples
Detecting tumors in their early stage is a long-standing goal of researchers. In the past few years, the study of ncRNAs in body fluids as potential biomarkers has grown exponentially. However, no ncRNAs have yet been able to be used clinically to detect hepatocellular carcinogenesis and predict prognosis. The limitations of circulating ncRNA measurement techniques are the main reason for this dilemma. There are several main assays available, corresponding to different advantages and limitations, to detect miRNA as an example: (1) qRT-RCR has the advantage of being widely available and highly sensitive, but has the disadvantage of having several biological and technical limitations. First, it is limited to quantifying only a defined set of miRNAs and cannot be used for high-throughput analysis. Second, because of the low abundance in biofluids, genomic DNA contamination needs to be removed prior to the reverse transcription step (210, 211). (2) Assessing the relative number of miRNAs by comparing the fluorescence intensity emitted by microarrays with hundreds of thousands of probes with labeled miRNAs has the advantage of high throughput, but the resulting disadvantage of a fixed range and the inability to detect new unannotated miRNAs. Therefore, this is only suitable for preliminary screening (212, 213). (3) Next Generation Sequencing (NGS) based on deep sequencing to detect circulating miRNAs not only overcomes the shortcomings of microarrays that can only detect known miRNAs, but also greatly increases in the detection order of magnitude. The cost of sequencing is reduced, while more information can be harvested. However, bioinformatics expertise is required for interpretation, and the technology is expensive with long turnaround time (214, 215). For ncRNAs, firstly their short sequences, showing higher levels of homology pose a significant challenge for accurate identification. Secondly, the low abundance predicts that the detection needs to span four orders of magnitude dynamic range with high sensitivity and accuracy requirements. Then, different detection methods correspond to different strengths and weaknesses, requiring researchers to break through and validate with newer studies. Notably, further efforts are required in this field.
Therapeutic potential of ncRNAs in HCC
Protein-like targets with stable structures and conformations have occupied an absolute leadership position in human diseases as mainstream targets for drug development (216). However, less than 2% of RNAs are translated into proteins, and most proteins are “non-druggable” (217, 218). With the elucidation of the mechanisms by which ncRNAs affect diseases, the feasibility of targeting RNA continues to be demonstrated. ncRNA can regulate signaling pathways and affect different enzymes and genes. Having such a large and fine regulatory mechanism, ncRNAs are therefore at the core of a multi-target regulatory network. Data show that representative small-molecule drugs, such as Bisphenol-A, Mitoxantrone, and Enoxacin, act against different cancers by targeting ncRNAs, thus providing new insights for drug development (216).
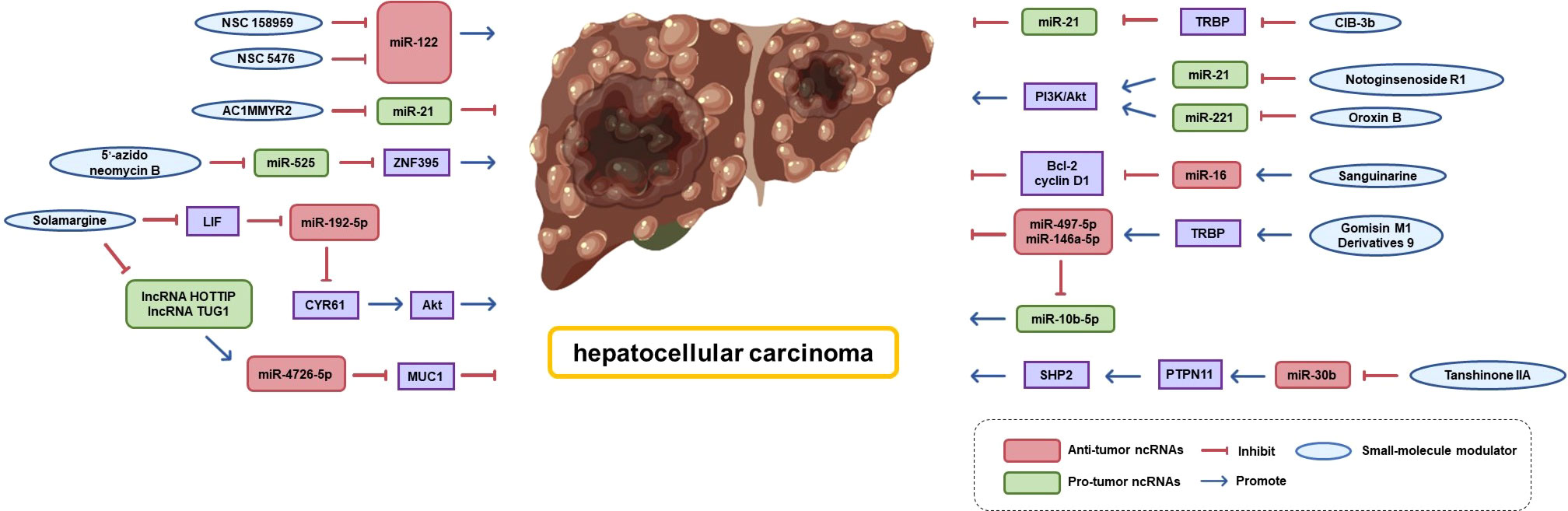
As shadow pioneers of ncRNAs, many small-molecule modulators that target miRNAs have been found to exhibit therapeutic activity against HCC (shown in Figure 7). Several small-molecule modulators that block miRNA biogenesis by directly binding to Drosha/Dicer-binding sites in pri/pre-miRNAs have been identified. For example, Douglas reported the first small-molecule modulator of miR-122 (the most abundant miRNA in the liver), demonstrating that small-molecule modulators of miRNA function have therapeutic potential. They found that two small-molecule inhibitors (NSC 158959 and NSC 5476) accelerated the processing and maturation of pri-miR-122 to miR-122 and were able to reduce hepatitis C virus replication (219). miR-525 confers invasive properties to HCC cells, while 5′-azido-neomycin B binds the Drosha processing site in pre-miR-525 to inhibit the production of mature miR-525 and salvage the expression of ZNF395 (220). Shi et al. screened AC1MMYR2, a small-molecule inhibitor of miR-21, using a high-throughput approach. They found that it blocks miR-21 maturation. AC1MMYR2 can reverse EMT and inhibit tumor growth, invasion, and metastasis without causing significant tissue cytotoxicity (221). Similarly, phenyl oxazole derivatives CIB-3b, a regulator of miR-21 biogenesis, interferes with TRBP2 to induce dissociation from Ago2 and Dicer (222).
Many natural products and their derivatives act as novel pathways for cancer therapy by specifically targeting ncRNAs (223). Gomisin M1 is a natural dibenzocyclooctadiene lignan isolated from Schisandra chinensis. Its derivatives are thought to be novel TRBP2 modulators that promote the binding of TRBP to Dicer and regulate miR-497-5p, miR-146a-5p, and miR-10b-5p maturation, thereby inhibiting HCC cell proliferation, migration, and invasion (224). Solamargine, a natural alkaloid extracted from Solanaceae plants, downregulates the expression of lncRNA HOTTIP and lncRNA TUG1 and subsequently upregulates the expression of miR-4726-5p that directly targets MUC1. The combination of Solamargine and Sorafenib significantly showed a significantly synergetic effect on MUC1 protein expression, providing a potential strategy for HCC treatment (225). Moreover, Solamargine can induce apoptosis and autophagy in HCC cells through the LIF/miR-192-5p/CYR61/Akt signaling pathway or by stimulating the TIME (226). Sanguinarine is a potent activator of miR-16 expressing wild-type or mutated p53. In Zhang’s research, Sanguinarine activated miR-16-2 expression by increasing p53 occupancy on the miR-16-2 promoter and decreased the expression of miR-16 target genes Bcl-2 and cyclin D1. This effect was validated by anti-miR-16 inhibitor treatment silencing (223).
In addition, we aimed to elucidate the potential mechanisms underlying the anti-cancer properties of various natural active compounds derived from traditional Chinese herbal medicines from the perspective of epigenetic modifications. Notoginsenoside R1 reduces miR-21 expression and subsequently inhibits the PI3K/Akt pathway, thereby exerting anti-HCC activity (227). Oroxin B, a flavonoid monomer compound in the traditional Chinese medicine Oroxylum indicum (L.) Vent has a same role (228). Tanshinone IIA may induce hepatoma cell death by downregulating miR-30b transcription and subsequently upregulating PTPN11 levels, which in turn stimulates the SHP2 pathway (229).
In conclusion, RNA-targeted small-molecule modulators have been emerging for almost 20 years, but the FDA has not yet approved miRNA therapies for HCC (230). It is true that miRNAs, which are the pioneers of ncRNAs, still less for lncRNAs and circRNAs. For small-molecule modulators targeting ncRNAs, the feasibility of their therapeutic application in HCC has been proven, but there is still a long way to go before they can become drugs. Their development is facing several hurdles, such as, first, identify specific RNA targets and binding sites and elucidating their mechanisms of action. These challenges all require the development of new design strategies (high-throughput screening, small-molecule microarray screening, structure-based designing, phenotype-based screening, etc.) to screen ncRNAs (216). Second, multi-targeting of endogenous RNAs may be risky for a therapeutic process, and the resulting off-target effects need to be addressed (231). Third, appropriate in vivo and in vitro models need to be established to confirm the structure-function relationships, potency, and specificity. In conclusion, small-molecule modulators targeting ncRNAs can greatly broaden the range of druggable targets, thus representing a new frontier for drug development. Continued research may lead to breakthrough discoveries while addressing the above-mentioned issues.
Conclusion
HCC is a multifactorial, multistage malignancy for which it is difficult to obtain satisfactory survival with current treatment options. ncRNAs play an important role in the initiation and progression of HCC and are associated with clinical diagnostic and prognostic properties. This review article focuses on the oncogenic or tumor-suppressive properties of ncRNAs, detailing how ncRNAs regulate key processes (TIME, angiogenesis, EMT, invasion, metastasis, metabolism, and drug resistance) in HCC and their specific mechanisms. Further, we discuss the promising approaches and potential roles of ncRNAs in the field of cancer diagnosis and therapy. Although ncRNAs have shown unique advantages as biomarkers and potential therapeutic targets, they still face challenges. For instance, there remains a vast uncharted territory in ncRNA research, and exploring the emerging role requires advances in next-generation sequencing technologies. Standardized and efficient techniques for rapid and large-scale extraction of ncRNAs have not yet been established. Meanwhile, improved targeting methods and delivery systems are needed to detect and reduce off-target effects. In terms of experimental techniques, off-target effects in knockdown, FISH, and pull-down technologies are difficult to avoid or eliminate completely. Moreover, while the current research on ncRNAs is still carried out at the cellular or animal level, research in clinical settings needs to be advanced to validate key ncRNA functions. Finally, ncRNA-based therapies require interdisciplinary cooperation among various fields, including immunology, molecular biology, pharmacology, and nanotechnology. These findings and challenges reveal the unexpected complexity of ncRNA regulatory mechanisms, which provides many answers but raises more questions. It is believed that, in the near future, ncRNAs can be developed as promising tools for targeted therapies alone or in combination with other therapies.
Author contributions
QH collected the related paper and drafted the manuscript. MW, XD provided valuable comments. FW, YL and XS supported the funding, designed the manuscript, and revised the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This study was financially supported by CAMS Innovation Fund for Medical Sciences (CIFMS) (No. 2021-I2M-1-031) and National Key Research and Development Plan Program (2019YFC1710504).
Acknowledgments
Figures were created with BioRender software (https://biorender.com/ (accessed on 17 September 2022).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin (2021) 71(3):209–49. doi: 10.3322/caac.21660
2. Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2021. CA Cancer J Clin (2021) 71(1):7–33. doi: 10.3322/caac.21654
3. Huang A, Yang XR, Chung WY, Dennison AR, Zhou J. Targeted therapy for hepatocellular carcinoma. Signal Transduct Target Ther (2020) 5(1):146. doi: 10.1038/s41392-020-00264-x
4. Hilmi M, Neuzillet C, Calderaro J, Lafdil F, Pawlotsky JM, Rousseau B. Angiogenesis and immune checkpoint inhibitors as therapies for hepatocellular carcinoma: current knowledge and future research directions. J Immunother Cancer (2019) 7(1):333. doi: 10.1186/s40425-019-0824-5
5. Yang JD, Hainaut P, Gores GJ, Amadou A, Plymoth A, Roberts LR. A global view of hepatocellular carcinoma: trends, risk, prevention and management. Nat Rev Gastroenterol Hepatol (2019) 16(10):589–604. doi: 10.1038/s41575-019-0186-y
6. Chen Z, Xie H, Hu M, Huang T, Hu Y, Sang N, et al. Recent progress in treatment of hepatocellular carcinoma. Am J Cancer Res (2020) 10(9):2993–3036.
7. Oura K, Morishita A, Tani J, Masaki T. Tumor immune microenvironment and immunosuppressive therapy in hepatocellular carcinoma: A review. Int J Mol Sci (2021) 22(11):5801. doi: 10.3390/ijms22115801
8. De Bock K, Mazzone M, Carmeliet P. Antiangiogenic therapy, hypoxia, and metastasis: risky liaisons, or not? Nat Rev Clin Oncol (2011) 8(7):393–404. doi: 10.1038/nrclinonc.2011.83
9. Ge X, Yao Y, Li J, Li Z, Han X. Role of LncRNAs in the epithelial-mesenchymal transition in hepatocellular carcinoma. Front Oncol (2021) 11:690800. doi: 10.3389/fonc.2021.690800
10. Li X, Li C, Zhang L, Wu M, Cao K, Jiang F, et al. The significance of exosomes in the development and treatment of hepatocellular carcinoma. Mol Cancer (2020) 19(1):1. doi: 10.1186/s12943-019-1085-0
11. Ge Y, Mu W, Ba Q, Li J, Jiang Y, Xia Q, et al. Hepatocellular carcinoma-derived exosomes in organotropic metastasis, recurrence and early diagnosis application. Cancer Lett (2020) 477:41–8. doi: 10.1016/j.canlet.2020.02.003
12. Lin YL, Li Y. Study on the hepatocellular carcinoma model with metastasis. Genes Dis (2020) 7(3):336–50. doi: 10.1016/j.gendis.2019.12.008
13. Mossenta M, Busato D, Dal Bo M, Toffoli G. Glucose metabolism and oxidative stress in hepatocellular carcinoma: Role and possible implications in novel therapeutic strategies. Cancers (Basel) (2020) 12(6):1688. doi: 10.3390/cancers12061668
14. Lin X, Wu Z, Hu H, Luo ML, Song E. Non-coding RNAs rewire cancer metabolism networks. Semin Cancer Biol (2021) 75:116–26. doi: 10.1016/j.semcancer.2020.12.019
15. Liang Y, Liang Q, Qiao L, Xiao F. MicroRNAs modulate drug resistance-related mechanisms in hepatocellular carcinoma. Front Oncol (2020) 10:920. doi: 10.3389/fonc.2020.00920
16. Di Martino MT, Riillo C, Scionti F, Grillone K, Polerà N, Caracciolo D, et al. miRNAs and lncRNAs as novel therapeutic targets to improve cancer immunotherapy. Cancers (Basel) (2021) 13(7):1587. doi: 10.3390/cancers13071587
17. Deepak KGK, Vempati R, Nagaraju GP, Dasari VR, Nagini S, Rao DN, et al. Tumor microenvironment: Challenges and opportunities in targeting metastasis of triple negative breast cancer. Pharmacol Res (2020) 153:104683. doi: 10.1016/j.phrs.2020.104683
18. Liu J, Ren L, Li S, Li W, Zheng X, Yang Y, et al. The biology, function, and applications of exosomes in cancer. Acta Pharm Sin B (2021) 11(9):2783–97. doi: 10.1016/j.apsb.2021.01.001
19. Paul P, Chakraborty A, Sarkar D, Langthasa M, Rahman M, Bari M, et al. Interplay between miRNAs and human diseases. J Cell Physiol (2018) 233(3):2007–18. doi: 10.1002/jcp.25854
20. Tan S, Xia L, Yi P, Han Y, Tang L, Pan Q, et al. Exosomal miRNAs in tumor microenvironment. J Exp Clin Cancer Res (2020) 39(1):67. doi: 10.1186/s13046-020-01570-6
21. Atianand MK, Fitzgerald KA. Long non-coding RNAs and control of gene expression in the immune system. Trends Mol Med (2014) 20(11):623–31. doi: 10.1016/j.molmed.2014.09.002
22. Xie C, Li SY, Fang JH, Zhu Y, Yang JE. Functional long non-coding RNAs in hepatocellular carcinoma. Cancer Lett (2021) 500:281–91. doi: 10.1016/j.canlet.2020.10.042
23. Chen Y, Li Z, Chen X, Zhang S. Long non-coding RNAs: From disease code to drug role. Acta Pharm Sin B (2021) 11(2):340–54. doi: 10.1016/j.apsb.2020.10.001
24. Aishanjiang K, Wei XD, Fu Y, Lin X, Ma Y, Le J, et al. Circular RNAs and hepatocellular carcinoma: New epigenetic players with diagnostic and prognostic roles. Front Oncol (2021) 11:653717. doi: 10.3389/fonc.2021.653717
25. Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. Rna (2013) 19(2):141–57. doi: 10.1261/rna.035667.112
26. Tang X, Ren H, Guo M, Qian J, Yang Y, Gu C. Review on circular RNAs and new insights into their roles in cancer. Comput Struct Biotechnol J (2021) 19:910–28. doi: 10.1016/j.csbj.2021.01.018
27. Zhang Y, Wang Y. Circular RNAs in hepatocellular carcinoma: Emerging functions to clinical significances. Front Oncol (2021) 11:667428. doi: 10.3389/fonc.2021.667428
28. Dongre A, Weinberg RA. New insights into the mechanisms of epithelial-mesenchymal transition and implications for cancer. Nat Rev Mol Cell Biol (2019) 20(2):69–84. doi: 10.1038/s41580-018-0080-4
29. Moeng S, Son SW, Lee JS, Lee HY, Kim TH, Choi SY, et al. Extracellular vesicles (EVs) and pancreatic cancer: From the role of EVs to the interference with EV-mediated reciprocal communication. Biomedicines (2020) 8(8):267. doi: 10.3390/biomedicines8080267
30. Wu Q, Zhou L, Lv D, Zhu X, Tang H. Exosome-mediated communication in the tumor microenvironment contributes to hepatocellular carcinoma development and progression. J Hematol Oncol (2019) 12(1):53. doi: 10.1186/s13045-019-0739-0
31. Winkle M, El-Daly SM, Fabbri M, Calin GA. Noncoding RNA therapeutics - challenges and potential solutions. Nat Rev Drug Discovery (2021) 20(8):629–51. doi: 10.1038/s41573-021-00219-z
32. Zhao Q, Wongpoomchai R, Chariyakornkul A, Xiao Z, Pilapong C. Identification of gene-set signature in early-stage hepatocellular carcinoma and relevant immune characteristics. Front Oncol (2021) 11:740484. doi: 10.3389/fonc.2021.740484
33. Zhou SL, Zhou ZJ, Hu ZQ, Huang XW, Wang Z, Chen EB, et al. Tumor-associated neutrophils recruit macrophages and T-regulatory cells to promote progression of hepatocellular carcinoma and resistance to sorafenib. Gastroenterology (2016) 150(7):1646–58.e17. doi: 10.1053/j.gastro.2016.02.040
34. Song G, Shi Y, Zhang M, Goswami S, Afridi S, Meng L, et al. Global immune characterization of HBV/HCV-related hepatocellular carcinoma identifies macrophage and T-cell subsets associated with disease progression. Cell Discovery (2020) 6(1):90. doi: 10.1038/s41421-020-00214-5
35. Crespo J, Sun H, Welling TH, Tian Z, Zou W. T Cell anergy, exhaustion, senescence, and stemness in the tumor microenvironment. Curr Opin Immunol (2013) 25(2):214–21. doi: 10.1016/j.coi.2012.12.003
36. Ji J, Yin Y, Ju H, Xu X, Liu W, Fu Q, et al. Long non-coding RNA lnc-Tim3 exacerbates CD8 T cell exhaustion via binding to Tim-3 and inducing nuclear translocation of Bat3 in HCC. Cell Death Dis (2018) 9(5):478. doi: 10.1038/s41419-018-0528-7
37. Yan K, Fu Y, Zhu N, Wang Z, Hong JL, Li Y, et al. Repression of lncRNA NEAT1 enhances the antitumor activity of CD8(+)T cells against hepatocellular carcinoma via regulating miR-155/Tim-3. Int J Biochem Cell Biol (2019) 110:1–8. doi: 10.1016/j.biocel.2019.01.019
38. Huang XY, Zhang PF, Wei CY, Peng R, Lu JC, Gao C, et al. Circular RNA circMET drives immunosuppression and anti-PD1 therapy resistance in hepatocellular carcinoma via the miR-30-5p/snail/DPP4 axis. Mol Cancer (2020) 19(1):92. doi: 10.1186/s12943-020-01213-6
39. Miggelbrink AM, Jackson JD, Lorrey SJ, Srinivasan ES, Waibl-Polania J, Wilkinson DS, et al. CD4 T-cell exhaustion: Does it exist and what are its roles in cancer? Clin Cancer Res (2021) 27(21):5742–52. doi: 10.1158/1078-0432.CCR-21-0206
40. Accogli T, Bruchard M, Végran F. Modulation of CD4 T cell response according to tumor cytokine microenvironment. Cancers (Basel) (2021) 13(3):373. doi: 10.3390/cancers13030373
41. Pagani M, Rossetti G, Panzeri I, de Candia P, Bonnal RJ, Rossi RL, et al. Role of microRNAs and long-non-coding RNAs in CD4(+) T-cell differentiation. Immunol Rev (2013) 253(1):82–96. doi: 10.1111/imr.12055
42. Zhong F, Liu S, Hu D, Chen L. LncRNA AC099850.3 promotes hepatocellular carcinoma proliferation and invasion through PRR11/PI3K/AKT axis and is associated with patients prognosis. J Cancer (2022) 13(3):1048–60. doi: 10.7150/jca.66092.
43. Peng L, Chen Y, Ou Q, Wang X, Tang N. LncRNA MIAT correlates with immune infiltrates and drug reactions in hepatocellular carcinoma. Int Immunopharmacol (2020) 89(Pt A):107071. doi: 10.1016/j.intimp.2020.107071
44. Han W, Li N, Liu J, Sun Y, Yang X, Wang Y. MicroRNA-26b-5p enhances T cell responses by targeting PIM-2 in hepatocellular carcinoma. Cell Signal (2019) 59:182–90. doi: 10.1016/j.cellsig.2018.11.011
45. Feng R, Cui Z, Liu Z, Zhang Y. Upregulated microRNA-132 in T helper 17 cells activates hepatic stellate cells to promote hepatocellular carcinoma cell migration in vitro. Scand J Immunol (2021) 93(5):e13007. doi: 10.1111/sji.13007
46. Gao Y, You M, Fu J, Tian M, Zhong X, Du C, et al. Intratumoral stem-like CCR4+ regulatory T cells orchestrate the immunosuppressive microenvironment in HCC associated with hepatitis b. J Hepatol (2022) 76(1):148–59. doi: 10.1016/j.jhep.2021.08.029
47. Li C, Jiang P, Wei S, Xu X, Wang J. Regulatory T cells in tumor microenvironment: new mechanisms, potential therapeutic strategies and future prospects. Mol Cancer (2020) 19(1):116. doi: 10.1158/1557-3125.HIPPO19-B11
48. Mailloux AW, Young MR. Regulatory T-cell trafficking: from thymic development to tumor-induced immune suppression. Crit Rev Immunol (2010) 30(5):435–47. doi: 10.1615/CritRevImmunol.v30.i5.30
49. Wing JB, Tanaka A, Sakaguchi S. Human FOXP3(+) regulatory T cell heterogeneity and function in autoimmunity and cancer. Immunity (2019) 50(2):302–16. doi: 10.1016/j.immuni.2019.01.020
50. Cho HJ, Cheong JY. Role of immune cells in patients with hepatitis b virus-related hepatocellular carcinoma. Int J Mol Sci (2021) 22(15):8011. doi: 10.3390/ijms22158011
51. Sullivan JA, Tomita Y, Jankowska-Gan E, Lema DA, Arvedson MP, Nair A, et al. Treg-Cell-Derived IL-35-Coated extracellular vesicles promote infectious tolerance. Cell Rep (2020) 30(4):1039–51.e5. doi: 10.1016/j.celrep.2019.12.081
52. Yang P, Li QJ, Feng Y, Zhang Y, Markowitz GJ, Ning S, et al. TGF-β-miR-34a-CCL22 signaling-induced treg cell recruitment promotes venous metastases of HBV-positive hepatocellular carcinoma. Cancer Cell (2012) 22(3):291–303. doi: 10.1016/j.ccr.2012.07.023
53. Liu N, Chang CW, Steer CJ, Wang XW, Song G. MicroRNA-15a/16-1 prevents hepatocellular carcinoma by disrupting the communication between kupffer cells and regulatory T cells. Gastroenterology (2022) 162(2):575–89. doi: 10.1053/j.gastro.2021.10.015
54. Jiang R, Tang J, Chen Y, Deng L, Ji J, Xie Y, et al. The long noncoding RNA lnc-EGFR stimulates T-regulatory cells differentiation thus promoting hepatocellular carcinoma immune evasion. Nat Commun (2017) 8:15129. doi: 10.1038/ncomms15129
55. Yu Z, Zhao H, Feng X, Li H, Qiu C, Yi X, et al. Long non-coding RNA FENDRR acts as a miR-423-5p sponge to suppress the treg-mediated immune escape of hepatocellular carcinoma cells. Mol Ther Nucleic Acids (2019) 17:516–29. doi: 10.1016/j.omtn.2019.05.027
56. Michaud D, Steward CR, Mirlekar B, Pylayeva-Gupta Y. Regulatory b cells in cancer. Immunol Rev (2021) 299(1):74–92. doi: 10.1111/imr.12939
57. Engelhard V, Conejo-Garcia JR, Ahmed R, Nelson BH, Willard-Gallo K, Bruno TC, et al. B cells and cancer. Cancer Cell (2021) 39(10):1293–6. doi: 10.1016/j.ccell.2021.09.007
58. Han Q, Zhao H, Jiang Y, Yin C, Zhang J. HCC-derived exosomes: Critical player and target for cancer immune escape. Cells (2019) 8(6):558. doi: 10.3390/cells8060558
59. Jia X, Liu H, Xu C, Han S, Shen Y, Miao X, et al. MiR-15a/16-1 deficiency induces IL-10-producing CD19(+) TIM-1(+) cells in tumor microenvironment. J Cell Mol Med (2019) 23(2):1343–53. doi: 10.1111/jcmm.14037
60. Yang J, Zhang Y, Song H. A disparate role of RP11-424C20.2/UHRF1 axis through control of tumor immune escape in liver hepatocellular carcinoma and thymoma. Aging (Albany NY) (2019) 11(16):6422–39. doi: 10.18632/aging.102197
61. Sivori S, Vacca P, Del Zotto G, Munari E, Mingari MC, Moretta L. Human NK cells: surface receptors, inhibitory checkpoints, and translational applications. Cell Mol Immunol (2019) 16(5):430–41. doi: 10.1038/s41423-019-0206-4
62. Robinson MW, Harmon C, O’Farrelly C. Liver immunology and its role in inflammation and homeostasis. Cell Mol Immunol (2016) 13(3):267–76. doi: 10.1038/cmi.2016.3
63. Mantovani S, Oliviero B, Varchetta S, Mele D, Mondelli MU. Natural killer cell responses in hepatocellular carcinoma: Implications for novel immunotherapeutic approaches. Cancers (Basel) (2020) 12(4):926. doi: 10.3390/cancers12040926
64. Coulouarn C, Factor VM, Conner EA, Thorgeirsson SS. Genomic modeling of tumor onset and progression in a mouse model of aggressive human liver cancer. Carcinogenesis (2011) 32(10):1434–40. doi: 10.1093/carcin/bgr133
65. Heinrich B, Gertz EM, Schäffer AA, Craig A, Ruf B, Subramanyam V, et al. The tumour microenvironment shapes innate lymphoid cells in patients with hepatocellular carcinoma. Gut (2022) 71(6):1161–75. doi: 10.1136/gutjnl-2021-325288
66. Nakano T, Chen IH, Wang CC, Chen PJ, Tseng HP, Huang KT, et al. Circulating exosomal miR-92b: Its role for cancer immunoediting and clinical value for prediction of posttransplant hepatocellular carcinoma recurrence. Am J Transplant (2019) 19(12):3250–62. doi: 10.1111/ajt.15490
67. Chen EB, Zhou ZJ, Xiao K, Zhu GQ, Yang Y, Wang B, et al. The miR-561-5p/CX(3)CL1 signaling axis regulates pulmonary metastasis in hepatocellular carcinoma involving CX(3)CR1(+) natural killer cells infiltration. Theranostics (2019) 9(16):4779–94. doi: 10.7150/thno.32543
68. Qi F, Du X, Zhao Z, Zhang D, Huang M, Bai Y, et al. Tumor mutation burden-associated LINC00638/miR-4732-3p/ULBP1 axis promotes immune escape via PD-L1 in hepatocellular carcinoma. Front Oncol (2021) 11:729340. doi: 10.3389/fonc.2021.729340
69. Zhang PF, Gao C, Huang XY, Lu JC, Guo XJ, Shi GM, et al. Cancer cell-derived exosomal circUHRF1 induces natural killer cell exhaustion and may cause resistance to anti-PD1 therapy in hepatocellular carcinoma. Mol Cancer (2020) 19(1):110. doi: 10.1186/s12943-020-01222-5
70. Shi M, Li ZY, Zhang LM, Wu XY, Xiang SH, Wang YG, et al. Hsa_circ_0007456 regulates the natural killer cell-mediated cytotoxicity toward hepatocellular carcinoma via the miR-6852-3p/ICAM-1 axis. Cell Death Dis (2021) 12(1):94. doi: 10.1038/s41419-020-03334-8
71. Lecoultre M, Dutoit V, Walker PR. Phagocytic function of tumor-associated macrophages as a key determinant of tumor progression control: a review. J Immunother Cancer (2020) 8(2):e001408. doi: 10.1136/jitc-2020-001408
72. Murray PJ, Allen JE, Biswas SK, Fisher EA, Gilroy DW, Goerdt S, et al. Macrophage activation and polarization: nomenclature and experimental guidelines. Immunity (2014) 41(1):14–20. doi: 10.1016/j.immuni.2014.06.008
73. Zhao J, Li H, Zhao S, Wang E, Zhu J, Feng D, et al. Epigenetic silencing of miR-144/451a cluster contributes to HCC progression via paracrine HGF/MIF-mediated TAM remodeling. Mol Cancer (2021) 20(1):46. doi: 10.1186/s12943-021-01343-5
74. Zhou J, Che J, Xu L, Yang W, Zhou W, Zhou C. Tumor-derived extracellular vesicles containing long noncoding RNA PART1 exert oncogenic effect in hepatocellular carcinoma by polarizing macrophages into M2. Dig Liver Dis (2022) 54(4):543–53. doi: 10.1016/j.dld.2021.07.005
75. Wang X, Zhou Y, Dong K, Zhang H, Gong J, Wang S. Exosomal lncRNA HMMR-AS1 mediates macrophage polarization through miR-147a/ARID3A axis under hypoxia and affects the progression of hepatocellular carcinoma. Environ Toxicol (2022) 37(6):1357–72. doi: 10.1002/tox.23489
76. Tian X, Wu Y, Yang Y, Wang J, Niu M, Gao S, et al. Long noncoding RNA LINC00662 promotes M2 macrophage polarization and hepatocellular carcinoma progression via activating wnt/β-catenin signaling. Mol Oncol (2020) 14(2):462–83. doi: 10.1002/1878-0261.12606
77. Li X, Lei Y, Wu M, Li N. Regulation of macrophage activation and polarization by HCC-derived exosomal lncRNA TUC339. Int J Mol Sci (2018) 19(10):2958. doi: 10.3390/ijms19102958
78. Hou ZH, Xu XW, Fu XY, Zhou LD, Liu SP, Tan DM. Long non-coding RNA MALAT1 promotes angiogenesis and immunosuppressive properties of HCC cells by sponging miR-140. Am J Physiol Cell Physiol (2020) 318(3):C649–c63. doi: 10.1152/ajpcell.00510.2018
79. Fan F, Chen K, Lu X, Li A, Liu C, Wu B. Dual targeting of PD-L1 and PD-L2 by PCED1B-AS1 via sponging hsa-miR-194-5p induces immunosuppression in hepatocellular carcinoma. Hepatol Int (2021) 15(2):444–58. doi: 10.1007/s12072-020-10101-6
80. Luo HL, Luo T, Liu JJ, Wu FX, Bai T, Ou C, et al. Macrophage polarization-associated lnc-Ma301 interacts with caprin-1 to inhibit hepatocellular carcinoma metastasis through the Akt/Erk1 pathway. Cancer Cell Int (2021) 21(1):422. doi: 10.1186/s12935-021-02133-1
81. Ye Y, Xu Y, Lai Y, He W, Li Y, Wang R, et al. Long non-coding RNA cox-2 prevents immune evasion and metastasis of hepatocellular carcinoma by altering M1/M2 macrophage polarization. J Cell Biochem (2018) 119(3):2951–63. doi: 10.1002/jcb.26509
82. Lu JC, Zhang PF, Huang XY, Guo XJ, Gao C, Zeng HY, et al. Amplification of spatially isolated adenosine pathway by tumor-macrophage interaction induces anti-PD1 resistance in hepatocellular carcinoma. J Hematol Oncol (2021) 14(1):200. doi: 10.1186/s13045-021-01207-x
83. Wang X, Sheng W, Xu T, Xu J, Gao R, Zhang Z. CircRNA hsa_circ_0110102 inhibited macrophage activation and hepatocellular carcinoma progression via miR-580-5p/PPARα/CCL2 pathway. Aging (Albany NY) (2021) 13(8):11969–87. doi: 10.18632/aging.202900
84. Wang Y, Gao R, Li J, Tang S, Li S, Tong Q, et al. Downregulation of hsa_circ_0074854 suppresses the migration and invasion in hepatocellular carcinoma via interacting with HuR and via suppressing exosomes-mediated macrophage M2 polarization. Int J Nanomed (2021) 16:2803–18. doi: 10.2147/IJN.S284560
85. Zhao Y, Rahmy S, Liu Z, Zhang C, Lu X. Rational targeting of immunosuppressive neutrophils in cancer. Pharmacol Ther (2020) 212:107556. doi: 10.1016/j.pharmthera.2020.107556
86. Ye D, Zhang T, Lou G, Liu Y. Role of miR-223 in the pathophysiology of liver diseases. Exp Mol Med (2018) 50(9):1–12. doi: 10.1038/s12276-018-0153-7
87. Hinshaw DC, Shevde LA. The tumor microenvironment innately modulates cancer progression. Cancer Res (2019) 79(18):4557–66. doi: 10.1158/0008-5472.CAN-18-3962
88. Wu L, Liao W, Wang X, Zhao Y, Pang J, Chen Y, et al. Expression, prognosis value, and immune infiltration of lncRNA ASB16-AS1 identified by pan-cancer analysis. Bioengineered (2021) 12(2):10302–18. doi: 10.1080/21655979.2021.1996054
89. Zhu AX, Duda DG, Sahani DV, Jain RK. HCC and angiogenesis: possible targets and future directions. Nat Rev Clin Oncol (2011) 8(5):292–301. doi: 10.1038/nrclinonc.2011.30
90. Jain RK. Antiangiogenesis strategies revisited: from starving tumors to alleviating hypoxia. Cancer Cell (2014) 26(5):605–22. doi: 10.1016/j.ccell.2014.10.006
91. Raskopf E, Vogt A, Sauerbruch T, Schmitz V. siRNA targeting VEGF inhibits hepatocellular carcinoma growth and tumor angiogenesis in vivo. J Hepatol (2008) 49(6):977–84. doi: 10.1016/j.jhep.2008.07.022
92. Wang R, Zhao N, Li S, Fang JH, Chen MX, Yang J, et al. MicroRNA-195 suppresses angiogenesis and metastasis of hepatocellular carcinoma by inhibiting the expression of VEGF, VAV2, and CDC42. Hepatology (2013) 58(2):642–53. doi: 10.1002/hep.26373
93. Wei H, Xu Z, Chen L, Wei Q, Huang Z, Liu G, et al. Long non-coding RNA PAARH promotes hepatocellular carcinoma progression and angiogenesis via upregulating HOTTIP and activating HIF-1α/VEGF signaling. Cell Death Dis (2022) 13(2):102. doi: 10.1038/s41419-022-04505-5
94. Li D, Wang T, Sun FF, Feng JQ, Peng JJ, Li H, et al. MicroRNA-375 represses tumor angiogenesis and reverses resistance to sorafenib in hepatocarcinoma. Cancer Gene Ther (2021) 28(1-2):126–40. doi: 10.1038/s41417-020-0191-x
95. Dong SS, Dong DD, Yang ZF, Zhu GQ, Gao DM, Chen J, et al. Exosomal miR-3682-3p suppresses angiogenesis by targeting ANGPT1 via the RAS-MEK1/2-ERK1/2 pathway in hepatocellular carcinoma. Front Cell Dev Biol (2021) 9:633358. doi: 10.3389/fcell.2021.633358
96. Lin XJ, Fang JH, Yang XJ, Zhang C, Yuan Y, Zheng L, et al. Hepatocellular carcinoma cell-secreted exosomal MicroRNA-210 promotes angiogenesis in vitro and in vivo. Mol Ther Nucleic Acids (2018) 11:243–52. doi: 10.1016/j.omtn.2018.02.014
97. Yokota Y, Noda T, Okumura Y, Kobayashi S, Iwagami Y, Yamada D, et al. Serum exosomal miR-638 is a prognostic marker of HCC via downregulation of VE-cadherin and ZO-1 of endothelial cells. Cancer Sci (2021) 112(3):1275–88. doi: 10.1111/cas.14807
98. Huang XY, Huang ZL, Huang J, Xu B, Huang XY, Xu YH, et al. Exosomal circRNA-100338 promotes hepatocellular carcinoma metastasis via enhancing invasiveness and angiogenesis. J Exp Clin Cancer Res (2020) 39(1):20. doi: 10.1186/s13046-020-1529-9
99. Yi M, Jiao D, Qin S, Chu Q, Wu K, Li A. Synergistic effect of immune checkpoint blockade and anti-angiogenesis in cancer treatment. Mol Cancer (2019) 18(1):60. doi: 10.1186/s12943-019-0974-6
100. Zhu C, Su Y, Liu L, Wang S, Liu Y, Wu J. Circular RNA hsa_circ_0004277 stimulates malignant phenotype of hepatocellular carcinoma and epithelial-mesenchymal transition of peripheral cells. Front Cell Dev Biol (2021) 8:585565. doi: 10.3389/fcell.2020.585565
101. Chen B, Lan J, Xiao Y, Liu P, Guo D, Gu Y, et al. Long noncoding RNA TP53TG1 suppresses the growth and metastasis of hepatocellular carcinoma by regulating the PRDX4/β-catenin pathway. Cancer Lett (2021) 513:75–89. doi: 10.1016/j.canlet.2021.04.022
102. Battistelli C, Sabarese G, Santangelo L, Montaldo C, Gonzalez FJ, Tripodi M, et al. The lncRNA HOTAIR transcription is controlled by HNF4α-induced chromatin topology modulation. Cell Death Differ (2019) 26(5):890–901. doi: 10.1038/s41418-018-0170-z
103. Battistelli C, Garbo S, Riccioni V, Montaldo C, Santangelo L, Vandelli A, et al. Design and functional validation of a mutant variant of the LncRNA HOTAIR to counteract snail function in epithelial-to-Mesenchymal transition. Cancer Res (2021) 81(1):103–13. doi: 10.1158/0008-5472.CAN-20-1764
104. Shi L, Liu B, Shen DD, Yan P, Zhang Y, Tian Y, et al. A tumor-suppressive circular RNA mediates uncanonical integrin degradation by the proteasome in liver cancer. Sci Adv (2021) 7(13):eabe5043. doi: 10.1126/sciadv.abe5043
105. Lin Y, Jian Z, Jin H, Wei X, Zou X, Guan R, et al. Long non-coding RNA DLGAP1-AS1 facilitates tumorigenesis and epithelial-mesenchymal transition in hepatocellular carcinoma via the feedback loop of miR-26a/b-5p/IL-6/JAK2/STAT3 and wnt/β-catenin pathway. Cell Death Dis (2020) 11(1):34. doi: 10.1038/s41419-019-2188-7
106. Li D, Zhang J, Yang J, Wang J, Zhang R, Li J, et al. CircMTO1 suppresses hepatocellular carcinoma progression via the miR-541-5p/ZIC1 axis by regulating wnt/β-catenin signaling pathway and epithelial-to-mesenchymal transition. Cell Death Dis (2021) 13(1):12. doi: 10.1038/s41419-021-04464-3
107. Yang B, Feng X, Liu H, Tong R, Wu J, Li C, et al. High-metastatic cancer cells derived exosomal miR92a-3p promotes epithelial-mesenchymal transition and metastasis of low-metastatic cancer cells by regulating PTEN/Akt pathway in hepatocellular carcinoma. Oncogene (2020) 39(42):6529–43. doi: 10.1038/s41388-020-01450-5
108. Liu Y, Liu DL, Dong LL, Wen D, Shi DM, Zhou J, et al. miR-612 suppresses stem cell-like property of hepatocellular carcinoma cells by modulating Sp1/Nanog signaling. Cell Death Dis (2016) 7(9):e2377. doi: 10.1038/cddis.2016.282
109. Yi T, Luo H, Qin F, Jiang Q, He S, Wang T, et al. LncRNA LL22NC03-N14H11.1 promoted hepatocellular carcinoma progression through activating MAPK pathway to induce mitochondrial fission. Cell Death Dis (2020) 11(10):832. doi: 10.1038/s41419-020-2584-z
110. Liu Y, Lu LL, Wen D, Liu DL, Dong LL, Gao DM, et al. MiR-612 regulates invadopodia of hepatocellular carcinoma by HADHA-mediated lipid reprogramming. J Hematol Oncol (2020) 13(1):12. doi: 10.1186/s13045-019-0841-3
111. Lu J, Zhao Z, Ma Y. miR-186 represses proliferation, migration, invasion, and EMT of hepatocellular carcinoma via directly targeting CDK6. Oncol Res (2020) 28(5):509–18. doi: 10.3727/096504020X15954139263808
112. Xie F, Zhou X, Fang M, Li H, Su P, Tu Y, et al. Extracellular vesicles in cancer immune microenvironment and cancer immunotherapy. Adv Sci (Weinh) (2019) 6(24):1901779. doi: 10.1002/advs.201901779
113. Han S, Wang L, Sun L, Wang Y, Yao B, Chen T, et al. MicroRNA-1251-5p promotes tumor growth and metastasis of hepatocellular carcinoma by targeting AKAP12. BioMed Pharmacother (2020) 122:109754. doi: 10.1016/j.biopha.2019.109754
114. Chun KH. Molecular targets and signaling pathways of microRNA-122 in hepatocellular carcinoma. Pharmaceutics (2022) 14(7):1380. doi: 10.3390/pharmaceutics14071380
115. Fang JH, Zhang ZJ, Shang LR, Luo YW, Lin YF, Yuan Y, et al. Hepatoma cell-secreted exosomal microRNA-103 increases vascular permeability and promotes metastasis by targeting junction proteins. Hepatology (2018) 68(4):1459–75. doi: 10.1002/hep.29920
116. Zhao ZB, Chen F, Bai XF. Long noncoding RNA MALAT1 regulates hepatocellular carcinoma growth under hypoxia via sponging MicroRNA-200a. Yonsei Med J (2019) 60(8):727–34. doi: 10.3349/ymj.2019.60.8.727
117. Liu C, Wang H, Tang L, Huang H, Xu M, Lin Y, et al. LncRNA BACE1-AS enhances the invasive and metastatic capacity of hepatocellular carcinoma cells through mediating miR-377-3p/CELF1 axis. Life Sci (2021) 275:119288. doi: 10.1016/j.lfs.2021.119288
118. Liu B, Tian Y, Chen M, Shen H, Xia J, Nan J, et al. CircUBAP2 promotes MMP9-mediated oncogenic effect via sponging miR-194-3p in hepatocellular carcinoma. Front Cell Dev Biol (2021) 9:675043. doi: 10.3389/fcell.2021.675043
119. Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell (2011) 144(5):646–74. doi: 10.1016/j.cell.2011.02.013
120. DeBerardinis RJ, Lum JJ, Hatzivassiliou G, Thompson CB. The biology of cancer: metabolic reprogramming fuels cell growth and proliferation. Cell Metab (2008) 7(1):11–20. doi: 10.1016/j.cmet.2007.10.002
121. Zheng YL, Li L, Jia YX, Zhang BZ, Li JC, Zhu YH, et al. LINC01554-mediated glucose metabolism reprogramming suppresses tumorigenicity in hepatocellular carcinoma via downregulating PKM2 expression and inhibiting Akt/mTOR signaling pathway. Theranostics (2019) 9(3):796–810. doi: 10.7150/thno.28992
122. Li J, Hu ZQ, Yu SY, Mao L, Zhou ZJ, Wang PC, et al. CircRPN2 inhibits aerobic glycolysis and metastasis in hepatocellular carcinoma. Cancer Res (2022) 82(6):1055–69. doi: 10.1158/0008-5472.CAN-21-1259
123. Lu L, Huang J, Mo J, Da X, Li Q, Fan M, et al. Exosomal lncRNA TUG1 from cancer-associated fibroblasts promotes liver cancer cell migration, invasion, and glycolysis by regulating the miR-524-5p/SIX1 axis. Cell Mol Biol Lett (2022) 27(1):17. doi: 10.1186/s11658-022-00309-9
124. Lai Z, Wei T, Li Q, Wang X, Zhang Y, Zhang S. Exosomal circFBLIM1 promotes hepatocellular carcinoma progression and glycolysis by regulating the miR-338/LRP6 axis. Cancer Biother Radiopharm (2020). doi: 10.1089/cbr.2020.3564
125. Zhou Y, Huang Y, Hu K, Zhang Z, Yang J, Wang Z. HIF1A activates the transcription of lncRNA RAET1K to modulate hypoxia-induced glycolysis in hepatocellular carcinoma cells via miR-100-5p. Cell Death Dis (2020) 11(3):176. doi: 10.1038/s41419-020-2366-7
126. Lai CY, Yeh KY, Lin CY, Hsieh YW, Lai HH, Chen JR, et al. MicroRNA-21 plays multiple oncometabolic roles in the process of NAFLD-related hepatocellular carcinoma via PI3K/AKT, TGF-β, and STAT3 signaling. Cancers (Basel) (2021) 13(5):940. doi: 10.3390/cancers13050940
127. Chai C, Rivkin M, Berkovits L, Simerzin A, Zorde-Khvalevsky E, Rosenberg N, et al. Metabolic circuit involving free fatty acids, microRNA 122, and triglyceride synthesis in liver and muscle tissues. Gastroenterology (2017) 153(5):1404–15. doi: 10.1053/j.gastro.2017.08.013
128. Chen M, Zhang C, Liu W, Du X, Liu X, Xing B. Long noncoding RNA LINC01234 promotes hepatocellular carcinoma progression through orchestrating aspartate metabolic reprogramming. Mol Ther (2022) 30(6):2354–69. doi: 10.1016/j.ymthe.2022.02.020
129. Cheng AL, Kang YK, Chen Z, Tsao CJ, Qin S, Kim JS, et al. Efficacy and safety of sorafenib in patients in the Asia-pacific region with advanced hepatocellular carcinoma: a phase III randomised, double-blind, placebo-controlled trial. Lancet Oncol (2009) 10(1):25–34. doi: 10.1016/S1470-2045(08)70285-7
130. Tang W, Chen Z, Zhang W, Cheng Y, Zhang B, Wu F, et al. The mechanisms of sorafenib resistance in hepatocellular carcinoma: theoretical basis and therapeutic aspects. Signal Transduct Target Ther (2020) 5(1):87. doi: 10.1038/s41392-020-0187-x
131. Chen J, Jin R, Zhao J, Liu J, Ying H, Yan H, et al. Potential molecular, cellular and microenvironmental mechanism of sorafenib resistance in hepatocellular carcinoma. Cancer Lett (2015) 367(1):1–11. doi: 10.1016/j.canlet.2015.06.019
132. Wei L, Wang X, Lv L, Liu J, Xing H, Song Y, et al. The emerging role of microRNAs and long noncoding RNAs in drug resistance of hepatocellular carcinoma. Mol Cancer (2019) 18(1):147. doi: 10.1186/s12943-019-1086-z
133. Fu X, Liu M, Qu S, Ma J, Zhang Y, Shi T, et al. Exosomal microRNA-32-5p induces multidrug resistance in hepatocellular carcinoma via the PI3K/Akt pathway. J Exp Clin Cancer Res (2018) 37(1):52. doi: 10.1186/s13046-018-0677-7
134. Zhang Z, Tan X, Luo J, Yao H, Si Z, Tong JS. The miR-30a-5p/CLCF1 axis regulates sorafenib resistance and aerobic glycolysis in hepatocellular carcinoma. Cell Death Dis (2020) 11(10):902. doi: 10.1038/s41419-020-03123-3
135. Zhang K, Shao CX, Zhu JD, Lv XL, Tu CY, Jiang C, et al. Exosomes function as nanoparticles to transfer miR-199a-3p to reverse chemoresistance to cisplatin in hepatocellular carcinoma. Biosci Rep (2020) 40(7):BSR20194026. doi: 10.1042/BSR20194026
136. Lou G, Chen L, Xia C, Wang W, Qi J, Li A, et al. MiR-199a-modified exosomes from adipose tissue-derived mesenchymal stem cells improve hepatocellular carcinoma chemosensitivity through mTOR pathway. J Exp Clin Cancer Res (2020) 39(1):4. doi: 10.1186/s13046-019-1512-5
137. Yu XN, Chen H, Liu TT, Wu J, Zhu JM, Shen XZ. Targeting the mTOR regulatory network in hepatocellular carcinoma: Are we making headway? Biochim Biophys Acta Rev Cancer (2019) 1871(2):379–91. doi: 10.1016/j.bbcan.2019.03.001
138. Zhou M, Zhang G, Hu J, Zhu Y, Lan H, Shen X, et al. Rutin attenuates sorafenib-induced chemoresistance and autophagy in hepatocellular carcinoma by regulating BANCR/miRNA-590-5P/OLR1 axis. Int J Biol Sci (2021) 17(13):3595–607. doi: 10.7150/ijbs.62471
139. Chen L, Sun L, Dai X, Li T, Yan X, Zhang Y, et al. LncRNA CRNDE promotes ATG4B-mediated autophagy and alleviates the sensitivity of sorafenib in hepatocellular carcinoma cells. Front Cell Dev Biol (2021) 9:687524. doi: 10.3389/fcell.2021.687524
140. Li Y, Zhang Y, Zhang S, Huang D, Li B, Liang G, et al. circRNA circARNT2 suppressed the sensitivity of hepatocellular carcinoma cells to cisplatin by targeting the miR-155-5p/PDK1 axis. Mol Ther Nucleic Acids (2021) 23:244–54. doi: 10.1016/j.omtn.2020.08.037
141. Dong ZR, Ke AW, Li T, Cai JB, Yang YF, Zhou W, et al. CircMEMO1 modulates the promoter methylation and expression of TCF21 to regulate hepatocellular carcinoma progression and sorafenib treatment sensitivity. Mol Cancer (2021) 20(1):75. doi: 10.1186/s12943-021-01361-3
142. Weng H, Zeng L, Cao L, Chen T, Li Y, Xu Y, et al. circFOXM1 contributes to sorafenib resistance of hepatocellular carcinoma cells by regulating MECP2 via miR-1324. Mol Ther Nucleic Acids (2021) 23:811–20. doi: 10.1016/j.omtn.2020.12.019
143. Yang C, Dong Z, Hong H, Dai B, Song F, Geng L, et al. circFN1 mediates sorafenib resistance of hepatocellular carcinoma cells by sponging miR-1205 and regulating E2F1 expression. Mol Ther Nucleic Acids (2020) 22:421–33. doi: 10.1016/j.omtn.2020.08.039
144. Xu J, Wan Z, Tang M, Lin Z, Jiang S, Ji L, et al. N(6)-methyladenosine-modified CircRNA-SORE sustains sorafenib resistance in hepatocellular carcinoma by regulating β-catenin signaling. Mol Cancer (2020) 19(1):163. doi: 10.1186/s12943-020-01281-8
145. Zhou J, Kang Y, Chen L, Wang H, Liu J, Zeng S, et al. The drug-resistance mechanisms of five platinum-based antitumor agents. Front Pharmacol (2020) 11:343. doi: 10.3389/fphar.2020.00343
146. Wang MY, Qiu YH, Cai ML, Zhang CH, Wang XW, Liu H, et al. Role and molecular mechanism of stem cells in colorectal cancer initiation. J Drug Targeting (2020) 28(1):1–10. doi: 10.1080/1061186X.2019.1632317
147. Liu S, Bu X, Kan A, Luo L, Xu Y, Chen H, et al. SP1-induced lncRNA DUBR promotes stemness and oxaliplatin resistance of hepatocellular carcinoma via E2F1-CIP2A feedback. Cancer Lett (2022) 528:16–30. doi: 10.1016/j.canlet.2021.12.026
148. Li P, Song R, Yin F, Liu M, Liu H, Ma S, et al. circMRPS35 promotes malignant progression and cisplatin resistance in hepatocellular carcinoma. Mol Ther (2022) 30(1):431–47. doi: 10.1016/j.ymthe.2021.08.027
149. Lee YR, Kim G, Tak WY, Jang SY, Kweon YO, Park JG, et al. Circulating exosomal noncoding RNAs as prognostic biomarkers in human hepatocellular carcinoma. Int J Cancer (2019) 144(6):1444–52. doi: 10.1002/ijc.31931
150. Huang Z, Zhou JK, Peng Y, He W, Huang C. The role of long noncoding RNAs in hepatocellular carcinoma. Mol Cancer (2020) 19(1):77. doi: 10.1186/s12943-020-01188-4
151. Oura K, Morishita A, Masaki T. Molecular and functional roles of MicroRNAs in the progression of hepatocellular carcinoma-a review. Int J Mol Sci (2020) 21(21):8362. doi: 10.3390/ijms21218362
152. Jiang L, Gu Y, Du Y, Liu J. Exosomes: Diagnostic biomarkers and therapeutic delivery vehicles for cancer. Mol Pharm (2019) 16(8):3333–49. doi: 10.1021/acs.molpharmaceut.9b00409
153. Wang S, Yang Y, Sun L, Qiao G, Song Y, Liu B. Exosomal MicroRNAs as liquid biopsy biomarkers in hepatocellular carcinoma. Onco Targets Ther (2020) 13:2021–30. doi: 10.2147/OTT.S232453
154. Zhou G, Zeng Y, Luo Y, Guo S, Bao L, Zhang Q. Urine miR-93-5p is a promising biomarker for early detection of HBV-related hepatocellular carcinoma. Eur J Surg Oncol (2022) 48(1):95–102. doi: 10.1016/j.ejso.2021.06.015
155. Mohamed AA, Omar AAA, El-Awady RR, Hassan SMA, Eitah WMS, Ahmed R, et al. MiR-155 and MiR-665 role as potential non-invasive biomarkers for hepatocellular carcinoma in Egyptian patients with chronic hepatitis c virus infection. J Transl Int Med (2020) 8(1):32–40. doi: 10.2478/jtim-2020-0006
156. Ghosh S, Bhowmik S, Majumdar S, Goswami A, Chakraborty J, Gupta S, et al. The exosome encapsulated microRNAs as circulating diagnostic marker for hepatocellular carcinoma with low alpha-fetoprotein. Int J Cancer (2020) 147(10):2934–47. doi: 10.1002/ijc.33111
157. El-Maraghy SA, Adel O, Zayed N, Yosry A, El-Nahaas SM, Gibriel AA. Circulatory miRNA-484, 524, 615 and 628 expression profiling in HCV mediated HCC among Egyptian patients; implications for diagnosis and staging of hepatic cirrhosis and fibrosis. J Adv Res (2020) 22:57–66. doi: 10.1016/j.jare.2019.12.002
158. Cui Y, Xu HF, Liu MY, Xu YJ, He JC, Zhou Y, et al. Mechanism of exosomal microRNA-224 in development of hepatocellular carcinoma and its diagnostic and prognostic value. World J Gastroenterol (2019) 25(15):1890–8. doi: 10.3748/wjg.v25.i15.1890
159. Tomimaru Y, Eguchi H, Nagano H, Wada H, Kobayashi S, Marubashi S, et al. Circulating microRNA-21 as a novel biomarker for hepatocellular carcinoma. J Hepatol (2012) 56(1):167–75. doi: 10.1016/j.jhep.2011.04.026
160. Han J, Li J, Qian Y, Liu W, Liang J, Huang Z, et al. Identification of plasma miR-148a as a noninvasive biomarker for hepatocellular carcinoma. Clin Res Hepatol Gastroenterol (2019) 43(5):585–93. doi: 10.1016/j.clinre.2018.12.008
161. Amr KS, Elmawgoud Atia HA, Elazeem Elbnhawy RA, Ezzat WM. Early diagnostic evaluation of miR-122 and miR-224 as biomarkers for hepatocellular carcinoma. Genes Dis (2017) 4(4):215–21. doi: 10.1016/j.gendis.2017.10.003
162. Weis A, Marquart L, Calvopina DA, Genz B, Ramm GA, Skoien R. Serum MicroRNAs as biomarkers in hepatitis c: Preliminary evidence of a MicroRNA panel for the diagnosis of hepatocellular carcinoma. Int J Mol Sci (2019) 20(4):864. doi: 10.3390/ijms20040864
163. Oura K, Fujita K, Morishita A, Iwama H, Nakahara M, Tadokoro T, et al. Serum microRNA-125a-5p as a potential biomarker of HCV-associated hepatocellular carcinoma. Oncol Lett (2019) 18(1):882–90. doi: 10.3892/ol.2019.10385
164. Li F, Wang F, Zhu C, Wei Q, Zhang T, Zhou YL. miR-221 suppression through nanoparticle-based miRNA delivery system for hepatocellular carcinoma therapy and its diagnosis as a potential biomarker. Int J Nanomed (2018) 13:2295–307. doi: 10.2147/IJN.S157805
165. Chen S, Chen H, Gao S, Qiu S, Zhou H, Yu M, et al. Differential expression of plasma microRNA-125b in hepatitis b virus-related liver diseases and diagnostic potential for hepatitis b virus-induced hepatocellular carcinoma. Hepatol Res (2017) 47(4):312–20. doi: 10.1111/hepr.12739
166. Chen Y, Chen J, Liu Y, Li S, Huang P. Plasma miR-15b-5p, miR-338-5p, and miR-764 as biomarkers for hepatocellular carcinoma. Med Sci Monit (2015) 21:1864–71. doi: 10.12659/MSM.893082
167. Han Y, Jiang W, Wang Y, Zhao M, Li Y, Ren L. Serum long non-coding RNA SCARNA10 serves as a potential diagnostic biomarker for hepatocellular carcinoma. BMC Cancer (2022) 22(1):431. doi: 10.1186/s12885-022-09530-3
168. Huang J, Zheng Y, Xiao X, Liu C, Lin J, Zheng S, et al. A circulating long noncoding RNA panel serves as a diagnostic marker for hepatocellular carcinoma. Dis Markers (2020) 2020:5417598. doi: 10.1155/2020/5417598
169. Wang J, Pu J, Zhang Y, Yao T, Luo Z, Li W, et al. Exosome-transmitted long non-coding RNA SENP3-EIF4A1 suppresses the progression of hepatocellular carcinoma. Aging (Albany NY) (2020) 12(12):11550–67. doi: 10.18632/aging.103302
170. Kim SS, Baek GO, Ahn HR, Sung S, Seo CW, Cho HJ, et al. Serum small extracellular vesicle-derived LINC00853 as a novel diagnostic marker for early hepatocellular carcinoma. Mol Oncol (2020) 14(10):2646–59. doi: 10.1002/1878-0261.12745
171. Huang X, Sun L, Wen S, Deng D, Wan F, He X, et al. RNA Sequencing of plasma exosomes revealed novel functional long noncoding RNAs in hepatocellular carcinoma. Cancer Sci (2020) 111(9):3338–49. doi: 10.1111/cas.14516
172. Chao Y, Zhou D. lncRNA-D16366 is a potential biomarker for diagnosis and prognosis of hepatocellular carcinoma. Med Sci Monit (2019) 25:6581–6. doi: 10.12659/MSM.915100
173. Gao S, Xu X, Wang Y, Zhang W, Wang X. Diagnostic utility of plasma lncRNA small nucleolar RNA host gene 1 in patients with hepatocellular carcinoma. Mol Med Rep (2018) 18(3):3305–13. doi: 10.3892/mmr.2018.9336
174. Wang Y, Jing W, Ma W, Liang C, Chai H, Tu J. Down-regulation of long non-coding RNA GAS5-AS1 and its prognostic and diagnostic significance in hepatocellular carcinoma. Cancer biomark (2018) 22(2):227–36. doi: 10.3233/CBM-170781
175. Wang ZF, Hu R, Pang JM, Zhang GZ, Yan W, Li ZN. Serum long noncoding RNA LRB1 as a potential biomarker for predicting the diagnosis and prognosis of human hepatocellular carcinoma. Oncol Lett (2018) 16(2):1593–601. doi: 10.3892/ol.2018.8825
176. Kamel MM, Matboli M, Sallam M, Montasser IF, Saad AS, El-Tawdi AHF. Investigation of long noncoding RNAs expression profile as potential serum biomarkers in patients with hepatocellular carcinoma. Transl Res (2016) 168:134–45. doi: 10.1016/j.trsl.2015.10.002
177. Yu J, Han J, Zhang J, Li G, Liu H, Cui X, et al. The long noncoding RNAs PVT1 and uc002mbe.2 in sera provide a new supplementary method for hepatocellular carcinoma diagnosis. Med (Baltimore) (2016) 95(31):e4436. doi: 10.1097/MD.0000000000004436
178. Liu R, Li Y, Wu A, Kong M, Ding W, Hu Z, et al. Identification of plasma hsa_circ_0005397 and combined with serum AFP, AFP-L3 as potential biomarkers for hepatocellular carcinoma. Front Pharmacol (2021) 12:639963. doi: 10.3389/fphar.2021.639963
179. Guo S, Hu C, Zhai X, Sun D. Circular RNA 0006602 in plasma exosomes: a new potential diagnostic biomarker for hepatocellular carcinoma. Am J Transl Res (2021) 13(6):6001–15.
180. Wei Y, Chen X, Liang C, Ling Y, Yang X, Ye X, et al. A noncoding regulatory RNAs network driven by circ-CDYL acts specifically in the early stages hepatocellular carcinoma. Hepatology (2020) 71(1):130–47. doi: 10.1002/hep.30795
181. Zhang T, Jing B, Bai Y, Zhang Y, Yu H. Circular RNA circTMEM45A acts as the sponge of MicroRNA-665 to promote hepatocellular carcinoma progression. Mol Ther Nucleic Acids (2020) 22:285–97. doi: 10.1016/j.omtn.2020.08.011
182. Chen W, Quan Y, Fan S, Wang H, Liang J, Huang L, et al. Exosome-transmitted circular RNA hsa_circ_0051443 suppresses hepatocellular carcinoma progression. Cancer Lett (2020) 475:119–28. doi: 10.1016/j.canlet.2020.01.022
183. Yu J, Ding WB, Wang MC, Guo XG, Xu J, Xu QG, et al. Plasma circular RNA panel to diagnose hepatitis b virus-related hepatocellular carcinoma: A large-scale, multicenter study. Int J Cancer (2020) 146(6):1754–63. doi: 10.1002/ijc.32647
184. Li Z, Zhou Y, Yang G, He S, Qiu X, Zhang L, et al. Using circular RNA SMARCA5 as a potential novel biomarker for hepatocellular carcinoma. Clin Chim Acta (2019) 492:37–44. doi: 10.1016/j.cca.2019.02.001
185. Qiao GL, Chen L, Jiang WH, Yang C, Yang CM, Song LN, et al. Hsa_circ_0003998 may be used as a new biomarker for the diagnosis and prognosis of hepatocellular carcinoma. Onco Targets Ther (2019) 12:5849–60. doi: 10.2147/OTT.S210363
186. Zhang X, Zhou H, Jing W, Luo P, Qiu S, Liu X, et al. The circular RNA hsa_circ_0001445 regulates the proliferation and migration of hepatocellular carcinoma and may serve as a diagnostic biomarker. Dis Markers (2018) 2018:3073467. doi: 10.1155/2018/3073467
187. Zhang X, Xu Y, Qian Z, Zheng W, Wu Q, Chen Y, et al. circRNA_104075 stimulates YAP-dependent tumorigenesis through the regulation of HNF4a and may serve as a diagnostic marker in hepatocellular carcinoma. Cell Death Dis (2018) 9(11):1091. doi: 10.1038/s41419-018-1132-6
188. Pelizzaro F, Cardin R, Penzo B, Pinto E, Vitale A, Cillo U, et al. Liquid biopsy in hepatocellular carcinoma: Where are we now? Cancers (Basel) (2021) 13(9):2274. doi: 10.3390/cancers13092274
189. Jia YZ, Liu J, Wang GQ, Song ZF. miR-484: A potential biomarker in health and disease. Front Oncol (2022) 12:830420. doi: 10.3389/fonc.2022.830420
190. Chen S, Mao Y, Chen W, Liu C, Wu H, Zhang J, et al. Serum exosomal miR-34a as a potential biomarker for the diagnosis and prognostic of hepatocellular carcinoma. J Cancer (2022) 13(5):1410–7. doi: 10.7150/jca.57205
191. Pelizzaro F, Cardin R, Sartori A, Imondi A, Penzo B, Aliberti C, et al. Circulating MicroRNA-21 and MicroRNA-122 as prognostic biomarkers in hepatocellular carcinoma patients treated with transarterial chemoembolization. Biomedicines (2021) 9(8):890. doi: 10.3390/biomedicines9080890
192. Chen S, Fu Z, Wen S, Yang X, Yu C, Zhou W, et al. Expression and diagnostic value of miR-497 and miR-1246 in hepatocellular carcinoma. Front Genet (2021) 12:666306. doi: 10.3389/fgene.2021.666306
193. Pratama MY, Visintin A, Crocè LS, Tiribelli C, Pascut D. Circulatory miRNA as a biomarker for therapy response and disease-free survival in hepatocellular carcinoma. Cancers (Basel) (2020) 12(10):2810. doi: 10.3390/cancers12102810
194. Kim SS, Nam JS, Cho HJ, Won JH, Kim JW, Ji JH, et al. Plasma micoRNA-122 as a predictive marker for treatment response following transarterial chemoembolization in patients with hepatocellular carcinoma. J Gastroenterol Hepatol (2017) 32(1):199–207. doi: 10.1111/jgh.13448
195. Cho HJ, Kim JK, Nam JS, Wang HJ, Lee JH, Kim BW, et al. High circulating microRNA-122 expression is a poor prognostic marker in patients with hepatitis b virus-related hepatocellular carcinoma who undergo radiofrequency ablation. Clin Biochem (2015) 48(16-17):1073–8. doi: 10.1016/j.clinbiochem.2015.06.019
196. Xu Y, Bu X, Dai C, Shang C. High serum microRNA-122 level is independently associated with higher overall survival rate in hepatocellular carcinoma patients. Tumour Biol (2015) 36(6):4773–6. doi: 10.1007/s13277-015-3128-5
197. Li T, Yin J, Yuan L, Wang S, Yang L, Du X, et al. Downregulation of microRNA-139 is associated with hepatocellular carcinoma risk and short-term survival. Oncol Rep (2014) 31(4):1699–706. doi: 10.3892/or.2014.3032
198. Wang T, Zhu H, Xiao M, Zhou S. Serum exosomal long noncoding RNA CRNDE as a prognostic biomarker for hepatocellular carcinoma. J Clin Lab Anal (2021) 35(11):e23959. doi: 10.1002/jcla.23959
199. Zeng Z, Dong J, Li Y, Dong Z, Liu Z, Huang J, et al. The expression level and clinical significance of lncRNA X91348 in hepatocellular carcinoma. Artif Cells Nanomed Biotechnol (2019) 47(1):3067–71. doi: 10.1080/21691401.2019.1640228
200. Yuan SX, Yang F, Yang Y, Tao QF, Zhang J, Huang G, et al. Long noncoding RNA associated with microvascular invasion in hepatocellular carcinoma promotes angiogenesis and serves as a predictor for hepatocellular carcinoma patients’ poor recurrence-free survival after hepatectomy. Hepatology (2012) 56(6):2231–41. doi: 10.1002/hep.25895
201. Xu H, Chen Y, Dong X, Wang X. Serum exosomal long noncoding RNAs ENSG00000258332.1 and LINC00635 for the diagnosis and prognosis of hepatocellular carcinoma. Cancer Epidemiol Biomarkers Prev (2018) 27(6):710–6. doi: 10.1158/1055-9965.EPI-17-0770
202. Zhang J, Zhang D, Zhao Q, Qi J, Li X, Qin C. A distinctively expressed long noncoding RNA, RP11-466I1.1, may serve as a prognostic biomarker in hepatocellular carcinoma. Cancer Med (2018) 7(7):2960–8.
203. El-Tawdi AH, Matboli M, El-Nakeep S, Azazy AE, Abdel-Rahman O. Association of long noncoding RNA and c-JUN expression in hepatocellular carcinoma. Expert Rev Gastroenterol Hepatol (2016) 10(7):869–77. doi: 10.1080/17474124.2016.1193003
204. Wu L, Yang Z, Zhang J, Xie H, Zhou L, Zheng S. Long noncoding RNA HOTTIP expression predicts tumor recurrence in hepatocellular carcinoma patients following liver transplantation. Hepatobil Surg Nutr (2018) 7(6):429–39. doi: 10.21037/hbsn.2018.10.07
205. Chen G, Xie D, Zhang P, Zhou H. Circular RNA hsa_circ_0000437 may be used as a new indicator for the diagnosis and prognosis of hepatocellular carcinoma. Bioengineered (2022) 13(6):14118–24. doi: 10.1080/21655979.2022.2081458
206. Lu C, Rong D, Hui B, He X, Jiang W, Xu Y, et al. CircETFA upregulates CCL5 by sponging miR-612 and recruiting EIF4A3 to promote hepatocellular carcinoma. Cell Death Discovery (2021) 7(1):321. doi: 10.1038/s41420-021-00710-x
207. Wang W, Li Y, Li X, Liu B, Han S, Li X, et al. Circular RNA circ-FOXP1 induced by SOX9 promotes hepatocellular carcinoma progression via sponging miR-875-3p and miR-421. BioMed Pharmacother (2020) 121:109517. doi: 10.1016/j.biopha.2019.109517
208. Weng Q, Chen M, Li M, Zheng YF, Shao G, Fan W, et al. Global microarray profiling identified hsa_circ_0064428 as a potential immune-associated prognosis biomarker for hepatocellular carcinoma. J Med Genet (2019) 56(1):32–8. doi: 10.1136/jmedgenet-2018-105440
209. Gong Y, Mao J, Wu D, Wang X, Li L, Zhu L, et al. Circ-ZEB1.33 promotes the proliferation of human HCC by sponging miR-200a-3p and upregulating CDK6. Cancer Cell Int (2018) 18:116. doi: 10.1186/s12935-018-0602-3
210. Kroh EM, Parkin RK, Mitchell PS, Tewari M. Analysis of circulating microRNA biomarkers in plasma and serum using quantitative reverse transcription-PCR (qRT-PCR). Methods (2010) 50(4):298–301. doi: 10.1016/j.ymeth.2010.01.032
211. Precazzini F, Detassis S, Imperatori AS, Denti MA, Campomenosi P. Measurements methods for the development of MicroRNA-based tests for cancer diagnosis. Int J Mol Sci (2021) 22(3):1176. doi: 10.3390/ijms22031176
212. Li W, Ruan K. MicroRNA detection by microarray. Anal Bioanal Chem (2009) 394(4):1117–24. doi: 10.1007/s00216-008-2570-2
213. Wang X, He Y, Mackowiak B, Gao B. MicroRNAs as regulators, biomarkers and therapeutic targets in liver diseases. Gut (2021) 70(4):784–95. doi: 10.1136/gutjnl-2020-322526
214. Li S, Mao M. Next generation sequencing reveals genetic landscape of hepatocellular carcinomas. Cancer Lett (2013) 340(2):247–53. doi: 10.1016/j.canlet.2012.09.027
215. Chen M, Zhao H. Next-generation sequencing in liquid biopsy: cancer screening and early detection. Hum Genomics (2019) 13(1):34. doi: 10.1186/s40246-019-0220-8
216. Zhao R, Fu J, Zhu L, Chen Y, Liu B. Designing strategies of small-molecule compounds for modulating non-coding RNAs in cancer therapy. J Hematol Oncol (2022) 15(1):14. doi: 10.1186/s13045-022-01230-6
217. Dang CV, Reddy EP, Shokat KM, Soucek L. Drugging the ‘undruggable’ cancer targets. Nat Rev Cancer (2017) 17(8):502–8. doi: 10.1038/nrc.2017.36
218. Costales MG, Childs-Disney JL, Haniff HS, Disney MD. How we think about targeting RNA with small molecules. J Med Chem (2020) 63(17):8880–900. doi: 10.1021/acs.jmedchem.9b01927
219. Young DD, Connelly CM, Grohmann C, Deiters A. Small molecule modifiers of microRNA miR-122 function for the treatment of hepatitis c virus infection and hepatocellular carcinoma. J Am Chem Soc (2010) 132(23):7976–81. doi: 10.1021/ja910275u
220. Childs-Disney JL, Disney MD. Small molecule targeting of a MicroRNA associated with hepatocellular carcinoma. ACS Chem Biol (2016) 11(2):375–80. doi: 10.1021/acschembio.5b00615
221. Shi Z, Zhang J, Qian X, Han L, Zhang K, Chen L, et al. AC1MMYR2, an inhibitor of dicer-mediated biogenesis of oncomir miR-21, reverses epithelial-mesenchymal transition and suppresses tumor growth and progression. Cancer Res (2013) 73(17):5519–31. doi: 10.1158/0008-5472.CAN-13-0280
222. Peng T, He Y, Wang T, Yu J, Ma X, Zhou Z, et al. Discovery of a novel small-molecule inhibitor disrupting TRBP-dicer interaction against hepatocellular carcinoma via the modulation of microRNA biogenesis. J Med Chem (2022) 65(16):11010–33. doi: 10.1021/acs.jmedchem.2c00189
223. Zhang B, Wang X, Deng J, Zheng H, Liu W, Chen S, et al. p53-dependent upregulation of miR-16-2 by sanguinarine induces cell cycle arrest and apoptosis in hepatocellular carcinoma. Cancer Lett (2019) 459:50–8. doi: 10.1016/j.canlet.2019.05.042
224. Zhou Z, Li Y, Ma X, Cao B, Peng T, Sheng Y, et al. Identification of a novel TAR RNA-binding protein 2 modulator with potential therapeutic activity against hepatocellular carcinoma. J Med Chem (2021) 64(11):7404–21. doi: 10.1021/acs.jmedchem.1c00018
225. Tang Q, Li X, Chen Y, Long S, Yu Y, Sheng H, et al. Solamargine inhibits the growth of hepatocellular carcinoma and enhances the anticancer effect of sorafenib by regulating HOTTIP-TUG1/miR-4726-5p/MUC1 pathway. Mol Carcinog (2022) 61(4):417–32. doi: 10.1002/mc.23389
226. Yin S, Jin W, Qiu Y, Fu L, Wang T, Yu H. Solamargine induces hepatocellular carcinoma cell apoptosis and autophagy via inhibiting LIF/miR-192-5p/CYR61/Akt signaling pathways and eliciting immunostimulatory tumor microenvironment. J Hematol Oncol (2022) 15(1):32. doi: 10.1186/s13045-022-01248-w
227. Li Y, Li Z, Jia Y, Ding B, Yu J. In vitro anti-hepatoma activities of notoginsenoside R1 through downregulation of tumor promoter miR-21. Dig Dis Sci (2020) 65(5):1364–75. doi: 10.1007/s10620-019-05856-4
228. Li N, Men W, Zheng Y, Wang H, Meng X. Oroxin b induces apoptosis by down-regulating MicroRNA-221 resulting in the inactivation of the PTEN/PI3K/AKT pathway in liver cancer. Molecules (2019) 24(23):4384. doi: 10.3390/molecules24234384
229. Ren X, Wang C, Xie B, Hu L, Chai H, Ding L, et al. Tanshinone IIA induced cell death via miR30b-p53-PTPN11/SHP2 signaling pathway in human hepatocellular carcinoma cells. Eur J Pharmacol (2017) 796:233–41. doi: 10.1016/j.ejphar.2016.11.046
230. Zhang S, Zhou Y, Wang Y, Wang Z, Xiao Q, Zhang Y, et al. The mechanistic, diagnostic and therapeutic novel nucleic acids for hepatocellular carcinoma emerging in past score years. Brief Bioinform (2021) 22(2):1860–83. doi: 10.1093/bib/bbaa023
Keywords: hepatocellular carcinoma, ncRNA, mechanism, biomarker, small-molecule modulator
Citation: Han Q, Wang M, Dong X, Wei F, Luo Y and Sun X (2022) Non-coding RNAs in hepatocellular carcinoma: Insights into regulatory mechanisms, clinical significance, and therapeutic potential. Front. Immunol. 13:985815. doi: 10.3389/fimmu.2022.985815
Received: 04 July 2022; Accepted: 23 September 2022;
Published: 10 October 2022.
Edited by:
Jian Song, University Hospital Münster, GermanyReviewed by:
Yuan Liu, Shanghai Jiao Tong University, ChinaYefei Zhu, Nanjing Medical University, China
Lipeng Qiu, Jiangsu University, China
Copyright © 2022 Han, Wang, Dong, Wei, Luo and Sun. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yun Luo, ly20040423@126.com; Xiaobo Sun, sun_xiaobo163@163.com
 Qin Han
Qin Han Mengchen Wang
Mengchen Wang Xi Dong1,2,3,4
Xi Dong1,2,3,4 Xiaobo Sun
Xiaobo Sun