- 1Human Leukocyte Antigen Typing Laboratory, Blood Center of Zhejiang Province, Hangzhou, China
- 2Human Leukocyte Antigen Typing Laboratory, Key Laboratory of Blood Safety Research, Hangzhou, China
In order to treat the alloimmunization platelet transfusion refractoriness (PTR), human leukocyte antigen (HLA)-type and/or human platelet antigen (HPA)-type matched platelets between donors and patients are usually used. Therefore, genotyping of HLA-A and HLA-B loci, as well as HPA systems, for donors and patients, is of great significance. However, there is a rare report of genotyping for HLA-A and HLA-B loci as well as HPA systems at the same time. In this study, a high-throughput method for simultaneous genotyping of HLA-A and HLA-B loci, as well as HPA genotyping, was developed. A RNA capture probe panel was designed covering all exon sequences of the GP1BA, GP1BB, ITGA2, CD109, ITGB3, and ITGA2B genes and HLA-A and HLA-B loci. The HLA-A, HLA-B, and 34 HPA systems were genotyped using a targeted next-generation sequencing (NGS) method. The genotypes of the HLA-A and HLA-B loci, as well as the HPA, were assigned based on the nucleotides in the polymorphism sites. Using the NGS method, 204 unrelated blood specimens were successfully genotyped for all 34 HPA systems as well as HLA-A and HLA-B loci. The accuracy of the NGS method was 100%. Only HPA-2, HPA-3, HPA-5, HPA-6w, HPA-15, and HPA-21w showed polymorphism with frequencies of 0.9412, 0.6863, 0.9853, 0.9779, 0.4314, and 0.9951 for a allele, respectively. Thirty-two single nucleotide variants (SNVs) were detected. Of them, 12 SNVs can lead to amino acid change. HLA-A*11:01 and HLA-B*46:01 are the most common alleles for HLA-A and HLA-B loci. A targeted next-generation sequencing method for simultaneously genotyping HPA systems and HLA-A and HLA-B loci was first established, which could be used to create a database of HLA-typed and/or HPA-typed unrelated donors.
Introduction
There are a variety of alloantigen expression on the human platelet membrane surface, including some erythrocyte blood group antigens, human leukocyte antigen (HLA) class I proteins, human platelet antigens (HPA), and CD36 antigen (1–4). Nonetheless, the blood group system antigens, HLA class I antigen, and CD36 antigen in the human platelet membrane are shared with other cells (5–7). Classical HLA class I genes consist of HLA-A, HLA-B, and HLA-C loci. It has been reported that the HLA-A, HLA-B, and HLA-C loci are highly polymorphic, and their distribution in various populations is different (8). To date, 35 HPA have been identified and officially nominated by the International Platelet Immunology Nomenclature Committee of the International Society of Blood Transfusion (ISBT) (9). All HPA are located on the GPIIb/IIIa, GPIb/V/IX, GPIa/IIa, and CD109 platelet membrane glycoprotein complexes (1, 10). The antigens of the HPA systems showed a certain degree of polymorphism, but HPA a and b antigens were only found in HPA-1 to HPA-6w, HPA-15, and HPA-21w in the Chinese Han population (11).
Due to the difference in platelets’ alloantigen expression in the individuals, it can trigger a corresponding immune response to form antibodies through transfusion, pregnancy pathways, and allotransplantation. Antibodies against HLA-A and HLA-B, HPA, and CD36 antigens are responsible for some clinical syndromes and transfusion-related conditions, such as platelet transfusion refractoriness (PTR), posttransfusion purpura (PTP), and neonatal alloimmune thrombocytopenia (NAIT) (1, 12, 13). Currently, the transfusion of platelets for both prophylaxis and treatment of bleeding is relevant to all areas of clinical medicine. PTR is a common complication of patients receiving multiple transfusions, which is defined as an unsatisfactory posttransfusion platelet count increment. PTR can be separated into immune and nonimmune causes (14). In the alloimmunization PTR, most cases can be found with anti-HLA, anti-HPA, and/or anti-CD36 (15–17). In order to treat and prevent the alloimmunization of PTR, a common method was used using HLA-typed and/or HPA-typed matched platelets (18, 19). However, this needs to establish a database for HLA-typed and/or HPA-typed unrelated donors. It shows that 5,000, 18,000, and 25,000 donor candidates would be necessary to find at least five completely compatible donors in the Japanese, European Caucasoid, and North American Caucasoid populations, respectively (19). Although platelets’ membrane surfaces express the HLA-A, HLA-B, and HLA-C molecules, platelet donors are not routinely matched for HLA-C antigen during the HLA match platelet procedure (20).
In order to genotype for HLA-typed and/or HPA-typed unrelated donors, it needs to provide the corresponding methods for HLA-A and HLA-B loci, and HPA systems. Currently, many methods have been developed to detect HLA-A and HLA-B loci and HPA systems, including PCR sequence-specific primers (PCR-SSP), PCR sequence-based typing (PCR-SBT), and real-time PCR (11, 21–23). However, these methods always detect single gene separately and need too many wells and multiple amplification for the test. Now, next-generation sequencing (NGS) has been widely used for genotyping HLA loci. When compared with PCR-SBT, it has some advantages in terms of throughput and cost (24–26). Besides short-read sequencing, long-read sequencing technologies have been reported, such as single-molecule real-time sequencing (SMRT; Pacific Biosciences (PacBio)) and nanopore sequencing (Oxford Nanopore Technologies (ONT)) (27–29). Compared with short-read sequencing, these two long-read technologies provide lower per-read accuracy and require high data processing equipment. Furthermore, some studies have reported that the NGS method was used for HPA genotyping (30–34). Vorholt et al. reported an amplicon-based approach to genotyping for HPA-1 to HPA-5, HPA-9w, HPA-10w, HPA-16w, HPA-19w, HPA-27w, and HPA-34w, which required being amplified into 12 different fragments in each specimen (33). Davey et al. developed a targeted enrichment, high-sensitivity HaloPlex assay for 29 HPA systems, and 47 samples were sequenced (34). However, there is a rare report of simultaneously genotyping HLA-A and HLA-B loci as well as HPA systems. Here, in order to establish a database for HLA-typed and/or HPA-typed unrelated donors, a simultaneous genotyping method for HLA-A and HLA-B loci and HPA-1–HPA-30 and HPA-32–HPA-35 systems was established based on the target enrichment technology. The frequencies of genotypes and alleles of 34 HPA systems and HLA-A and HLA-B loci were also analyzed in the Chinese platelet blood donors.
Materials and methods
Study specimens
In total, 204 unrelated blood specimens were collected from healthy platelet blood donors who have at least donated the platelet three times at the Blood Center of Zhejiang Province, China. The ethnic background of all individuals is Zhejiang Han. Informed consent was obtained from all participants. This study was approved by the Ethical Scientific Committee of the Blood Center of Zhejiang Province, China (2020-005). From each platelet donor, 5 ml peripheral blood with EDTA anticoagulant was collected. To validate the accuracy of the method, nine HPA reference specimens from the 14th, 16th, and 20th Platelet Immunology Workshop of ISBT have been chosen for detection, which contained HPA-1ab, HPA-1bb, and HPA-4ab genotypes, respectively.
Genomic DNA extraction
The genomic DNA was extracted using a commercial DNA extraction reagent kit (RBC Bioscience, Taiwan) and an automatic Magcore nucleic acid extraction instrument (RBC Bioscience, Taiwan) according to the manufacturer’s instructions. The OD260/280 ratio of the DNA was 1.6 to 1.8. The final DNA concentration was adjusted to 30 ng/μl.
The probe panel design
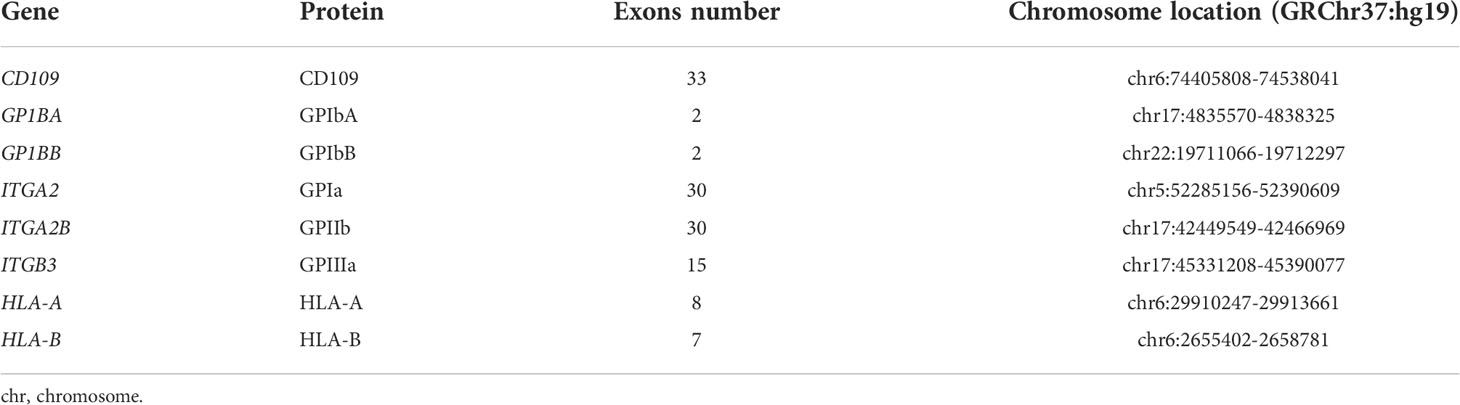
A RNA capture probe panel covering all exon sequences of the GP1BA, GP1BB, ITGA2, CD109, ITGB3, and ITGA2B genes and HLA-A and HLA-B loci was designed based on the GRCh37 reference sequence and synthesized by a commercial company (Lianchuan Biotechnology Co. Ltd., Hangzhou, China). This panel included 498 nonoverlapping probes, covering 97.88% of target regions. The length of each probe was 120 bp, and all probes were designed by end-to-end tiling to match the reference sequence. The 5′ end of all capture probes is coupled with biotin and then all probes were mixed. The information on the covered region in the gene for HPA and HLA-A and HLA-B genotyping are listed in Table 1. The sequences of all probes are listed in Supplementary Table S1.
Genomic DNA fragmentation and amplification
DNA fragmentation and amplification were prepared from genomic DNA using the VariantBaits™ Target Enrichment System (Lianchuan Biotechnology Co., Ltd., Hangzhou, China) according to the manufacturer’s instructions. Briefly, a total of 200 ng genomic DNA for each specimen was sheared to 200–250 bp by a focused ultrasonicator (M220-Covaris, Auburn, MA, USA) at 4°C, followed by end-repairing, A-tailing, index ligation, and purification. The purification DNA was then amplified in a 50 µl reaction system, including 20 µl DNA fragments, 5 µl primers, and 25 µl DNA polymerase master mixture, by initial denaturation at 98°C for 45 s, followed by 7 cycles at 98°C for 15 s, 60°C for 30 s, 72°C for 30 s, and final extension at 72°C for 1 min. The PCR amplicons with 200–500 bp length were obtained by size selection using purification magnetic beads (ThermoFisher, San Jose, CA, USA). The PCR amplicon concentration and quality were determined by the Qubit instrument (ThermoFisher, San Jose, CA, USA) and the Agilent 4200 Bioanalyzer (Agilent Technologies Inc., Santa Clara, CA, USA).
Hybridization in solution and target-capture
Every four indexed PCR amplicons (500 ng) were pooled into one tube for hybridization and subsequently added 7.5 µl of different blocking solutions. The tube was evaporated in a vacuum freeze drier (Heto-Holten, ThermoFisher Scientific, San Jose, CA, USA) at 40°C. In total, 7.5 μl 2× hybridization buffer and 3 μl blocking solution B were added to the dry tube and incubated at 95°C for 5 min and 65°C for 5 min. After, 4.5 µl VariantBaits™ biotinylated probes (Lianchuan Biotechnology Co. Ltd., Hangzhou, China) were added, and the hybridization was performed at 65°C for 16 to 24 h. In this step, PCR amplicons that contained the target sequences of the HPA systems and HLA-A and HLA-B loci would be specifically hybridized into biotinylated capture probes. In the next reaction, 40 µl of Dynabeads™ MyOne™ Streptavidin T1 magnetic beads (ThermoFisher, San Jose, CA, USA) was used to separate and purify the captured product from the above hybridization system, and the product was resuspended using 20 µl nuclease-free water. The 20 µl separated DNA was amplified with a 50 µl reaction mix containing 5 µl primer mixture and 25 µl DNA polymerase master mixture, performed as follows: 98°C for 45 s; 16 cycles of 98°C for 15 s, 60°C for 30 s, and 72°C for 30 s; and a final extension at 72°C for 1 min. The PCR products were purified with magnetic beads, and the quality was detected by the Agilent 4200 Bioanalyzer.
Sequencing
The aforementioned PCR products from different tubes were pooled into one new tube equally, which formed the DNA libraries. The quality and size of the pooled library were detected by the Agilent Bioanalyzer 4200. The concentration of the library was determined by the Qubit instrument, prepared to a final concentration of 12 pmol/L, and then sequenced using a MiSeq instrument (Illumina, San Diego, CA, USA) with a standard v2 Reagent Kit (2*318 cycle; Illumina, San Diego, CA, USA).
Data analysis
The FASTQ files generated by the MiSeq instrument were analyzed for all exon sequences, including the nucleotides in the polymorphism sites of the HPA systems using CLC benchwork 23.11 (Qiagen Company, Stockach, Germany) according to the manufacturer’s instructions. The sequences of the GP1BA (NG_008767), GP1BB (NG_007974), ITGA2 (NG_008330), CD109 (NG_0033971), ITGB3 (NG_008332), and ITGA2B (NG_008331) were set as the reference sequences, respectively. The FASTQ files were aligned with the different reference sequences using CLC benchwork 23.11. Sequence data with quality over Q30 and depth of coverage over 30× were accepted. The genotypes for HPA-1 to HPA-35 (except for HPA-31) were assigned manually according to the nucleotides in the polymorphism sites from the versiti-HPA database (www.versiti.org). The genotypes of HLA-A and HLA-B loci were assigned using the TypeStream Visual Software version 2.0 (One Lambda Inc., Canoga Park, CA, USA).
Genotyping HPA-1–HPA-35 systems by PCR-SBT
In order to validate the results of the established NGS method, the specimens were also genotyped for HPA-1–HPA-28 systems using a PCR-SBT method according to our previous report (11). Three new primer pairs were added for genotyping HPA-29 to HPA-35 systems, and the procedure is the same as in our previous report except for the primers. In brief, 21 of the specific primers were divided into seven groups; therefore, one group has three primer pairs and mixed into one well. The nucleotide sequence of each HPA system was amplified using the primer mixture, and the amplicons were then Sanger sequenced using a Big Dye Terminator v3.1 cycle sequencing kit (ThermoFisher Scientific, Shanghai, China). The genotypes for HPA systems were assigned according to the nucleotides in the polymorphism sites of the HPA systems.
Genotyping HLA-A and HLA-B loci using a commercial NGS method
The HLA-A and HLA-B loci were also genotyped using an AllType NGS 9-Loci Amplification Kit (One Lambda Inc.) according to the manufacturer’s instructions. The HLA-A and HLA-B genotypes were assigned using the TypeStream Visual Software version 2.0 with the IMGT/HLA Database version 3.46.0.0 (One Lambda Inc.) as previously reported (35).
Statistical analysis
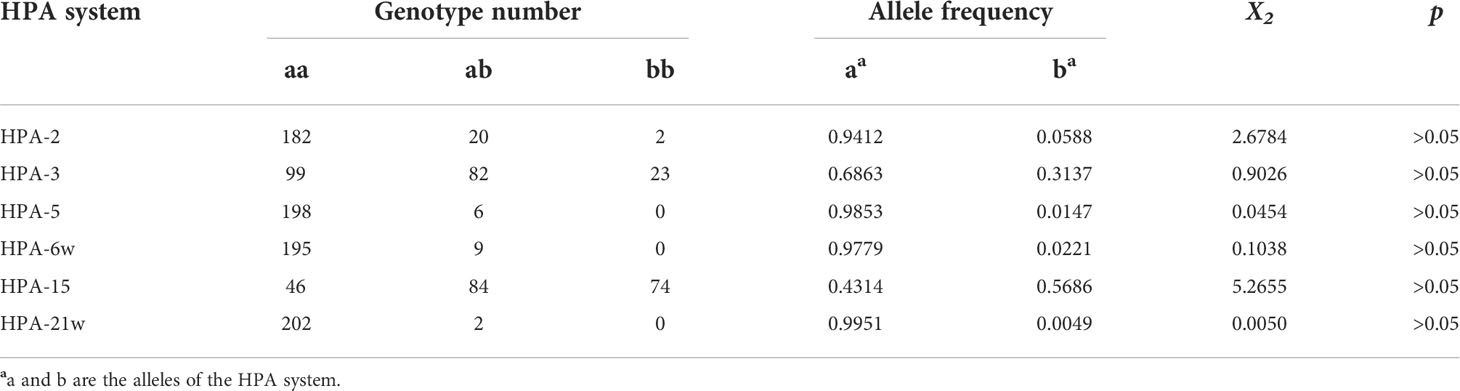
A Hardy–Weinberg equilibrium (HWE) was determined for each HPA system using the Chi-square test. p-values of less than 0.05 were considered statistically significant.
Results
Validate the results of the established NGS method
A total of 204 specimens from the blood donors were detected by the NGS method. The results of the HLA-A and HLA-B loci and HPAs in all specimens consisted of those of the commercial NGS or PCR-SBT in-house. The accuracy of the NGS method for HLA and HPA genotyping was 100%. The HPA genotypes in the nine reference specimens by the NGS method were in concordance with the reference results and demonstrated in Supplementary Table S2.
Some quality parameters of the NGS procedure were as below. All the observed sequence lengths were 150 bps, and the distribution of GC-content fits normal distribution, and the relative GC-content of a sequence of R1 and R2 in 20%–80% were 99.81% and 99.54%, respectively. The relative N-content of a sequence less than 1% of R1 and R2 were 99.95% and 99.99%. In addition, the distribution of average sequence quality scores over 30 of R1 and R2 was 98.78% and 98.08%, respectively. The mean value of the reading depth in the exon regions for each gene is shown in Supplementary Table S3. Meanwhile, the mean reading depth frequency value of minor alleles of single nucleotide variants (SNVs) in the heterozygous positions is shown in Supplementary Table S4.
The distribution of 34 HPA systems by the NGS method
The genotype distributions of the 34 HPA systems (HPA-31w was not analyzed by the NGS method) were fitted with Hardy–Weinberg equilibrium (p > 0.05). Among these HPA systems, only HPA-2, HPA-3, HPA-5, HPA-6bw, HPA-15bw, and HPA-21bw systems observed polymorphism in this study. However, a/a homozygote individuals were found in the other 28 HPA systems. The genotypes and allele frequencies of the systems with polymorphism are listed in Table 2. Among these 34 HPA systems, HPA-3 and HPA-15 systems showed high polymorphisms, with frequencies of 0.6863 and 0.3137 for HPA-3a and HPA-3b alleles and 0.4314 and 0.5686 for HPA-15a and HPA-15b alleles, respectively.
Variants detected in the ITGB3, ITGA2B, ITGA2, CD109, and GP1BA genes
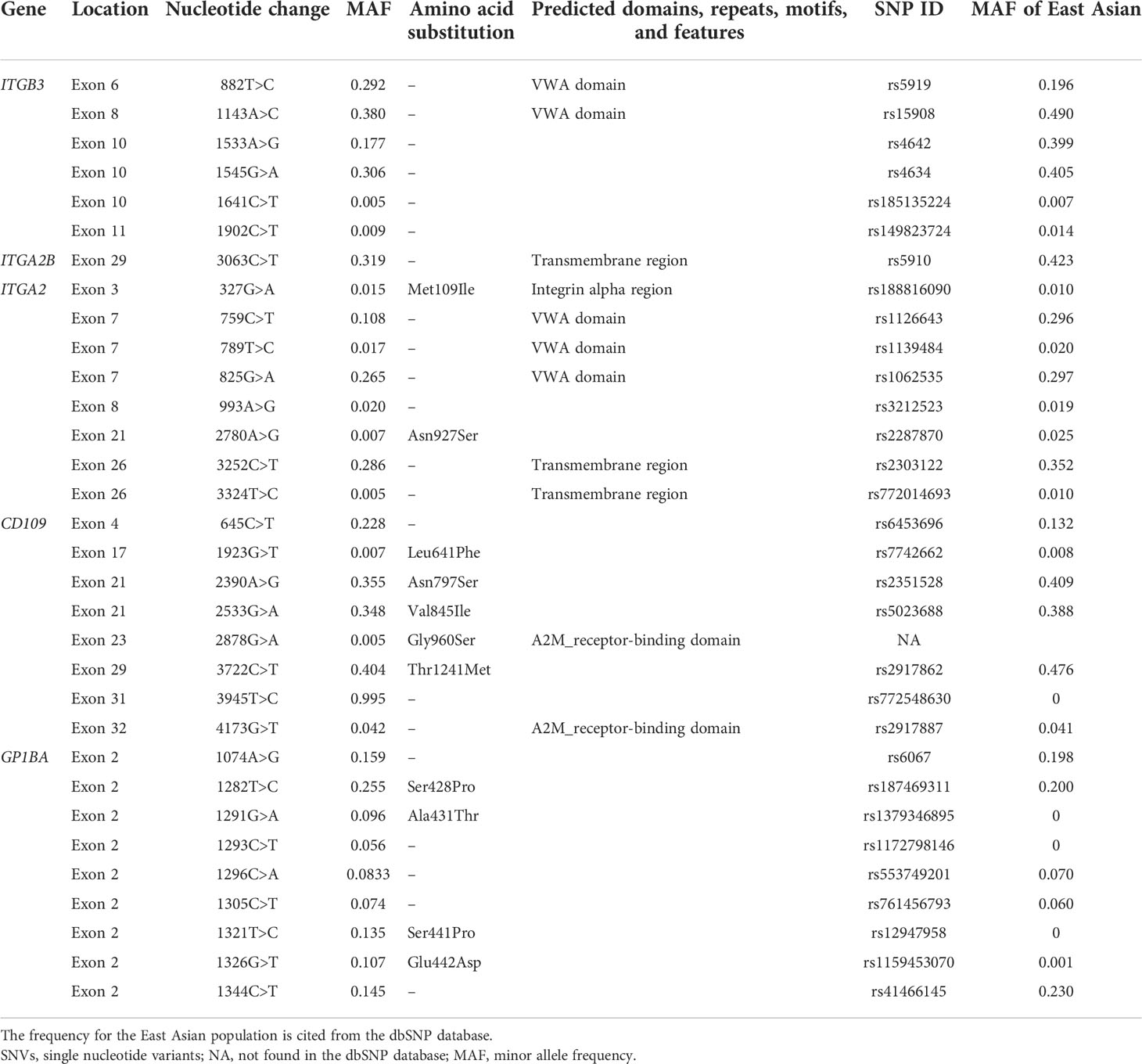
Thirty-two SNVs were detected in the exon regions of the ITGB3, ITGA2B, ITGA2, CD109, and GP1BA genes. Only one SNV has not existed in the dbSNP database. No SNV was found in the GP1BB gene. The position of these SNVs and allele frequencies are shown in Table 3. Of these 32 SNVs identified, 12 SNVs can result in an amino acid change, while the other SNVs are synonymous changes. A new SNV site, c.2878G>A located on the exon 23 of CD109, can result in an amino acid change, p.Gly960Ser, the novel SNV has been submitted to Genbank, the Accession number is OP434394. The amino acid was located in the A-macroglobulin receptor-binding domain according to the Simple Modular Architecture Research Tool (SMART) analysis based on the CD109 antigen (Q6YHK3-4). The probably damaging effect was predicted for this SNV using the PolyPhen-2 software (v2.2.3r406) in silico, and the score was 1.000 (score 0.000–0.452 = benign, 0.453–0.956 = possibly damaging, 0.957–1.000 = probably damaging).
The distribution for HLA-A and HLA-B loci, respectively
The numbers of HLA-A and HLA-B alleles were 23 and 49 in the 204 specimens, respectively. The HLA-A*11:01 allele and HLA-B*46:01 allele are the most common alleles for each locus, with a frequency of 22.55% and 14.22%, respectively. The allele distributions of the HLA-A and HLA-B loci are listed in Supplementary Table S5.
Discussion
Thrombocytopenic patients with immune-based PTR would have significantly increased the risk of a major spontaneous or life-threatening bleed. In order to treat these patients, one of the common methods is to provide cross-match-compatible or HLA-matched/compatible platelet units (15, 18). In order to provide matching platelets, it needs a large number of donors with known HLA and/or HPA genotypes (18, 19). At present, many DNA-based methods have been used for HLA and/or HPA genotyping in platelet donors (20–28, 30, 31). It has been reported that PCR-SBT and NGS methods have been routinely used for HLA genotyping in the laboratory (24–26, 35, 36). The NGS methods for HLA genotyping are divided into targeted and amplicon-based methods (34, 35). Classical HLA-A, HLA-B, and HLA-C molecules were all expressed in the platelet, but the HLA-C molecule was expressed at a low level. Therefore, only HLA-A and HLA-B loci were routinely analyzed to search for HLA-matched/compatible platelet units (20). The HPA genotyping methods included PCR-SSP, TaqMan assay, PCR-SBT, and NGS (22, 23, 33, 34). However, there is a rare study for HPA and HLA genotyping simultaneously. Now genotyping for all HPA systems and HLA-A and HLA-B loci needs multiple PCR amplifications and/or sequencing reactions using the PCR-SSP and PCR-SBT methods, which do not simultaneously detected the all HPA systems and HLA-A and HLA-B loci. Here, we report an NGS method for simultaneously genotyping HPA-1~35 systems (except HPA-31w) and HLA-A and HLA-B loci in platelet donors.
In this study, target-enrichment panel probes for the HLA-A, HLA-B, ITGB3, ITGA2B, ITGA2, CD109, GP1BA, and GP1BB genes were designed based on GRCh37 as a reference. Due to the HPA-31w polymorphism, the site is located in the GP9 gene and it was not nominated when we initiated this study (10). Therefore, the target probes were not for the GP9 gene, and the protocol cannot assign the HPA-31w system. The design probes overlapped one by one to cover all exon regions. Therefore, all known human platelet antigens except for HPA-31w and novel variations in the encoding regions can be detected. In the panel, the probes are RNA-based, which has better hybridization efficiency and stability to DNA complexes compared to DNA probes (37, 38). However, the RNA probes are expensive and require a lower temperature to store. In the hybridization reaction, four tubes were pooled into one to assure the volume was less than 31 μl after pooling; otherwise, it will affect hybridization efficiency. In order to validate the results of the established NGS method, the specimens were genotyped for all HPAs by a PCR-SBT method in-house and for HLA-A and HLA-B loci using a commercial NGS method. It showed that the accuracy of the established NGS method for HLA and HPA was 100%.
The accuracy of the NGS method depends on some influence factors. Davey et al. reported the results of the NGS method for HPA were in concordance 100% with historical HPA genotypes (34). However, Vorholt et al. reported that the concordance with TaqMan assay and NGS results for HPAs was 99.8%, and the discrepancies between them were due to allele dropout in real-time PCR (33). It is generally considered that a depth of coverage of over 30× is necessary for some SNV detection (39). Due to the depth of coverage being different among the genes in most NGS methods, it needs to assure a depth of coverage of over 30× for each gene. The accuracy can be improved with increased coverage (40). However, the coverage can also be influenced by the GC content, which may affect the efficiency of PCR. There was also evidence that the same volume had more coverage than the same mole (33), but the specimens that were mixed in the same mole were better than the same volume in our experiment. In addition, we found a repetitive sequence in exon 2 of the GP1BA gene, which can affect the analysis accuracy when using the CLC benchwork 23.11. Therefore, the SNVs in the GP1BA gene by the NGS method were also confirmed by PCR-SBT in-house. Besides the depth of coverage, the breadth of coverage also helps to ensure that sensitivity and specificity are sufficient for supporting variant detection. However, the coverage profiles may not be uniform and a lack of coverage in key regions, such as exons, may affect the accuracy of the typing. Although software analysis programs will usually have built-in filters to define the minimum coverage required for accurate typing, certain circumstances may permit going below this threshold, such as when the polymorphisms of two alleles of a locus are phased or when the region with low coverage does not affect the genotyping (i.e., introns, untranslated regions). Furthermore, besides the depth of coverage, for HLA typing data analysis, the assessment of adequate allele balance is important for detecting issues due to allele dropout.
The distribution of HLA-A, HLA-B, and HPA systems was similar to those of our previous reports (11, 35). Furthermore, some SNVs were detected in the coding region, which is located in the structural domain of the platelet membrane glycoproteins (Table 3). c.882T>C and c.1143A>C in the ITGB3 were both located in the VWA domain (41), which are synonymous changes. c.3063C>T in the ITGA2B is located in the transmembrane region (42), a hydrophobic region of the protein. c.327G>A in the ITAG2 is located in the integrin alpha region of the GPIa (43); c.759C>T, c.789T>C, c.825G>A, and c.993A>G in the ITAG2 are located in the VWA domain of the GPIa; and c.3252C>T and c.3324T>C are located in the transmembrane region of the GPIa. c.4173G>T in the CD109 is located in the A2M_receptor-binding domain, which is the receptor-binding domain (RBD) of alpha-2-macroglobulin proteins (44, 45). Therefore, some SNVs may affect the domain function, but this needs further study.
In summary, a targeted NGS method can be successfully established to genotype simultaneously 34 HPA systems as well as HLA-A and HLA-B loci. Its advantages include high throughput and simultaneous testing for HLA-A, HLA-B, and HPA genotypes. The method can be used to establish a database bank of HLA-typed and/or HPA-typed unrelated donors, which can help provide matching platelets for PTR, PTP, and NAIT patients.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics statement
Written informed consent was obtained from the individual(s) for the publication of any potentially identifiable images or data included in this article.
Author contributions
JW, XY and YH performed the experiments, collected the data, and wrote the rough draft. XH and JH analyzed the data. ST and FZ interpreted the results and drafted the paper. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Science Research Foundation of Zhejiang Healthy Bureau (2021KY137 and WKJ-ZJ-2021).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2022.945994/full#supplementary-material
Abbreviations
HPA, human platelet antigen; PCR-SBT, polymerase chain reaction sequence-based typing; PTR, platelet transfusion refractoriness; PTP, posttransfusion purpura; NAIT, neonatal alloimmune thrombocytopenia; NGS, next-generation sequencing.
References
1. Curtis BR, McFarland JG. Human platelet antigens - 2013. Vox Sang (2014) 106(2):93–102. doi: 10.1111/vox.12085
2. Curtis BR, Edwards JT, Hessner MJ, Klein JP, Aster RH. Blood group a and b antigens are strongly expressed on platelets of some individuals. Blood (2000) 96(4):1574–81. doi: 10.1182/blood.V96.4.1574
3. Santoso S. Human platelet alloantigens. Transfus Apher Sci (2003) 28(3):227–36. doi: 10.1016/S1473-0502(03)00040-5
4. Armesilla AL, Vega MA. Structural organization of the gene for human CD36 glycoprotein. J Biol Chem (1994) 269(29):18985–91. doi: 10.1016/S0021-9258(17)32263-9
5. Ravn V, Dabelsteen E. Tissue distribution of histo-blood group antigens. APMIS (2000) 108(1):1–28. doi: 10.1034/j.1600-0463.2000.d01-1.x
6. Garrido F. HLA class-I expression and cancer immunotherapy. Adv Exp Med Biol (2019) 1151:79–90. doi: 10.1007/978-3-030-17864-2_3
7. Greenwalt DE, Lipsky RH, Ockenhouse CF, Ikeda H, Tandon NN, Jamieson GA. Membrane glycoprotein CD36: a review of its roles in adherence, signal transduction, and transfusion medicine. Blood (1992) 80(5):1105–15. doi: 10.1182/blood.V80.5.1105.1105
8. Hurley CK, Kempenich J, Wadsworth K, Sauter J, Hofmann JA, Schefzyk D, et al. Common, intermediate and well-documented HLA alleles in world populations: CIWD version 3.0.0. HLA (2020) 95(6):516–31. doi: 10.1111/tan.13811
9. Versiti. HPA database (2020). Available at: https://www.versiti.org/hpa.
10. Jallu V, Beranger T, Bianchi F, Casale C, Chenet C, Ferre N, et al. Cab4b, the first human platelet antigen carried by glycoprotein IX discovered in a context of severe neonatal thrombocytopenia. J Thromb Haemost (2017) 15(8):1646–54. doi: 10.1111/jth.13744
11. Hong X, Chen S, Ying Y, Liu Y, Xu X, He J, et al. Simultaneous genotyping of human platelet alloantigen-1 to 28bw systems by multiplex polymerase chain reaction sequence-based typing. Vox Sang (2017) 112(4):360–6. doi: 10.1111/vox.12507
12. de Vos TW, Winkelhorst D, de Haas M, Lopriore E, Oepkes D. Epidemiology and management of fetal and neonatal alloimmune thrombocytopenia. Transfus Apher Sci (2020) 59(1):102704. doi: 10.1016/j.transci.2019.102704
13. Solves Alcaina P. Platelet transfusion: And update on challenges and outcomes. J Blood Med (2020) 11:19–26. doi: 10.2147/JBM.S234374
14. Belizaire R, Makar RS. Non-alloimmune mechanisms of thrombocytopenia and refractoriness to platelet transfusion. Transfus Med Rev (2020) 34(4):242–9. doi: 10.1016/j.tmrv.2020.09.002
15. Schmidt AE, Refaai MA, Coppage M. HLA-mediated platelet refractoriness. Am J Clin Pathol (2019) 151(4):353–63. doi: 10.1093/ajcp/aqy121
16. Inoue H, Sakamoto R, Nishimiya H, Sakamoto H, Terasu S, Aminaka R, et al. Minor impact of patient alloantibodies against human platelet antigen (HPA)-15 in the effectiveness of platelet transfusion: A pilot study. Transfusion (2021) 61(3):738–43. doi: 10.1111/trf.16181
17. Schmidt AE, Sahai T, Refaai MA, Sullivan M, Curtis BR. Severe platelet transfusion refractoriness in association with antibodies against CD36. Lab Med (2020) 51(5):540–4. doi: 10.1093/labmed/lmz091
18. Saris A, Pavenski K. Human leukocyte antigen alloimmunization and alloimmune platelet refractoriness. Transfus Med Rev (2020) 34(4):250–7. doi: 10.1016/j.tmrv.2020.09.010
19. Bub CB, Torres MA, Moraes ME, Hamerschlak N, Kutner JM. Determination of an unrelated donor pool size for human leukocyte antigen-matched platelets in Brazil. Rev Bras Hematol Hemoter (2016) 38(1):1–6. doi: 10.1016/j.bjhh.2015.10.005
20. Saito S, Ota S, Seshimo H, Yamazaki Y, Nomura S, Ito T, et al. Platelet transfusion refractoriness caused by a mismatch in HLA-c antigens. Transfusion (2002) 42(3):302–8. doi: 10.1046/j.1537-2995.2002.00051.x
21. Madden K, Chabot-Richards D. HLA testing in the molecular diagnostic laboratory. Virchows Arch (2019) 474(2):139–47. doi: 10.1007/s00428-018-2501-3
22. Jovanovic S Srzentic, Lilic M, Vavic N, Radovic I, Djilas I. Genotyping of eight human platelet antigen systems in Serbian blood donors: Foundation for platelet apheresis registry. Transfus Med Hemother (2021) 48(4):228–33. doi: 10.1159/000514487
23. Azizi SG, Samiee S, Shaiegan M, Zadsar M. Genotyping of human platelet antigen-1 to -5 and -15 by polymerase chain reaction with sequence-specific primers (PCR-SSP) and real-time PCR in Azeri blood donors. Iran J Allergy Asthma Immunol (2021) 20(3):350–63. doi: 10.18502/ijaai.v20i3.6333
24. Segawa H, Kukita Y, Kato K. HLA genotyping by next-generation sequencing of complementary DNA. BMC Genomics (2017) 18(1):914. doi: 10.1186/s12864-017-4300-7
25. Ingram KJ, O’Shields EF, Kiger DF, Gautreaux MD. NGS and HLA: The long road ahead. Hum Immunol (2020) 81(6):280–4. doi: 10.1016/j.humimm.2020.03.001
26. Alizadeh M, Walencik A, Frassati C, Moskovtchenko P, Lafarge X, Verite F, et al. Evidence for a higher resolution of HLA genotyping by a new NGS-based approach. Transfus Clin Biol (2017) 24(3):120–3. doi: 10.1016/j.tracli.2017.05.011
27. Flusberg BA, Webster DR, Lee JH, Travers KJ, Olivares EC, Clark TA, et al. Direct detection of DNA methylation during single-molecule, real-time sequencing. Nat Methods (2010) 7(6):461–5. doi: 10.1038/nmeth.1459
28. Lu H, Giordano F, Ning Z. Oxford Nanopore MinION sequencing and genome assembly. Genomics Proteomics Bioinf (2016) 14(5):265–79. doi: 10.1016/j.gpb.2016.05.004
29. Johansson T, Koskela S, Yohannes DA, Partanen J, Saavalainen P. Targeted RNA-based Oxford nanopore sequencing for typing 12 classical HLA genes. Front Genet (2021) 12:635601. doi: 10.3389/fgene.2021.635601
30. Orzińska A, Guz K, Uhrynowska M, Dębska M, Mikula M, Ostrowski J, et al. Noninvasive prenatal HPA-1 typing in HPA-1a negative pregnancies selected in the polish PREVFNAIT screening program. Transfusion (2018) 58(11):2705–11. doi: 10.1111/trf.14963
31. Orzińska A, Guz K, Mikula M, Kluska A, Balabas A, Ostrowski J, et al. Prediction of fetal blood group and platelet antigens from maternal plasma using next-generation sequencing. Transfusion (2019) 59(3):1102–7. doi: 10.1111/trf.15116
32. Orzińska A, Guz K, Mikula M, Kulecka M, Kluska A, Balabas A, et al. A preliminary evaluation of next-generation sequencing as a screening tool for targeted genotyping of erythrocyte and platelet antigens in blood donors. Blood Transfus (2018) 16(3):285–92. doi: 10.2450/2017.0253-16
33. Vorholt SM, Hamker N, Sparka H, Enczmann J, Zeiler T, Reimer T, et al. High-throughput screening of blood donors for twelve human platelet antigen systems using next-generation sequencing reveals detection of rare polymorphisms and two novel protein-changing variants. Transfus Med Hemother (2020) 47(1):33–44. doi: 10.1159/000504894
34. Davey S, Navarrete C, Brown C. Simultaneous human platelet antigen genotyping and detection of novel single nucleotide polymorphisms by targeted next-generation sequencing. Transfusion (2017) 57(6):1497–504. doi: 10.1111/trf.14092
35. Wang F, Dong L, Wang W, Chen N, Zhang W, He J, et al. The polymorphism of HLA-a, -c, -b, -DRB3/4/5, -DRB1, -DQB1 loci in zhejiang han population, China using NGS technology. Int J Immunogenet (2021) 48(6):485–9. doi: 10.1111/iji.12554
36. Schöfl G, Lang K, Quenzel P, Böhme I, Sauter J, Hofmann JA, et al. Lange V. 2.7 million samples genotyped for HLA by next generation sequencing: lessons learned. BMC Genomics (2017) 18(1):161. doi: 10.1186/s12864-017-3575-z
37. Lesnik EA, Freier SM. Relative thermodynamic stability of DNA, RNA, and DNA:RNA hybrid duplexes: relationship with base composition and structure. Biochemistry (1995) 34(34):10807–15. doi: 10.1021/bi00034a013
38. Kozarewa I, Armisen J, Gardner AF, Slatko BE, Hendrickson CL. Overview of target enrichment strategies. Curr Protoc Mol Biol (2015) 112:1–23. doi: 10.1002/0471142727.mb0721s112
39. Hou R, Yang Z, Li M, Xiao H. Impact of the next-generation sequencing data depth on various biological result inferences. Sci China Life Sci (2013) 56(2):104–9. doi: 10.1007/s11427-013-4441-0
40. Meynert AM, Ansari M, FitzPatrick DR, Taylor MS. Variant detection sensitivity and biases in whole genome and exome sequencing. BMC Bioinf (2014) 15(1):247. doi: 10.1186/1471-2105-15-247
41. Zimrin AB, Eisman R, Vilaire G, Schwartz E, Bennett JS, Poncz M. Structure of platelet glycoprotein IIIa. a common subunit for two different membrane receptors. J Clin Invest (1988) 81(5):1470–5. doi: 10.1172/JCI113478
42. Poncz M, Eisman R, Heidenreich R, Silver SM, Vilaire G, Surrey S, et al. Structure of the platelet membrane glycoprotein IIb. homology to the alpha subunits of the vitronectin and fibronectin membrane receptors. J Biol Chem (1987) 262(18):8476–82. doi: 10.1111/j.1749-6632.1987.tb30994.x
43. Takada Y, Hemler ME. The primary structure of the VLA-2/collagen receptor alpha 2 subunit (platelet GPIa): homology to other integrins and the presence of a possible collagen-binding domain. J Cell Biol (1989) 109(1):397–407. doi: 10.1083/jcb.109.1.397
44. Xiao T, DeCamp DL, Sprang SR. Structure of a rat α1-macroglobulin receptor-binding domain dimer. Protein Sci (2000) 9(10):1889–97. doi: 10.1110/ps.9.10.1889
Keywords: human leukocyte antigen, human platelet antigens, platelet transfusion refractoriness, next-generation sequencing, platelet blood donors
Citation: Wang J, You X, He Y, Hong X, He J, Tao S and Zhu F (2022) Simultaneous genotyping for human platelet antigen systems and HLA-A and HLA-B loci by targeted next-generation sequencing. Front. Immunol. 13:945994. doi: 10.3389/fimmu.2022.945994
Received: 17 May 2022; Accepted: 30 August 2022;
Published: 29 September 2022.
Edited by:
Christophe Picard, Établissement Français du Sang (EFS), FranceReviewed by:
Antonio Balas, Transfusion Center of the Community of Madrid, SpainBin Li, Shanghai Jiao Tong University, China
Copyright © 2022 Wang, You, He, Hong, He, Tao and Zhu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Faming Zhu, zfm00@hotmail.com; Sudan Tao, taosudan@126.com
 Jielin Wang
Jielin Wang Xuan You1,2
Xuan You1,2 Xiaozhen Hong
Xiaozhen Hong Ji He
Ji He Faming Zhu
Faming Zhu