- 1IRMB, Univ Montpellier, INSERM, CHU Montpellier, Montpellier, France
- 2Immunology Department, CHU Montpellier, Montpellier, France
- 3Firalis SA, Molecular Diagnostics, Huningue, France
MicroRNAs (miRNAs) are small non-coding single-stranded RNAs that represent important posttranscriptional regulators of protein-encoding genes. In particular, miRNAs play key roles in regulating cellular processes such as proliferation, migration, and cell differentiation. Recently, miRNAs emerged as critical regulators of osteoclasts (OCs) biology and have been involved in OCs pathogenic role in several disorders. OCs are multinucleated cells generated from myeloid precursors in the bone marrow, specialized in bone resorption. While there is a growing number of information on the cytokines and signaling pathways that are critical to control the differentiation of osteoclast precursors (OCPs) into mature OCs, the connection between OC differentiation steps and miRNAs is less well-understood. The present review will first summarize our current understanding of the miRNA-regulated pathways in the sequential steps required for OC formation, from the motility and migration of OCPs to the cell-cell fusion and the final formation of the actin ring and ruffled border in the functionally resorbing multinucleated OCs. Then, considering the difficulty of working on primary OCs and on the generation of robust data we will give an update on the most recent advances in the detection technologies for miRNAs quantification and how these are of particular interest for the understanding of OC biology and their use as potential biomarkers.
Introduction
MicroRNAs (miRNAs) are key regulatory molecules that control cellular processes such as proliferation, migration and cell differentiation small. As shown in Figure 1, in the canonical pathway, miRNAs are transcribed by RNA polymerase II as large RNA precursors called primary (pri-) miRNAs that will be cleaved in the nucleus by the microprocessors complex into short hairpin precursors (pre-miRNAs) of about 70-nucleotides in length (1, 2). Pre-miRNAs are subsequently exported to the cytoplasm to be processed by DICER and yield mature miRNA duplexes (~22 nucleotides long) prior their loading onto the Argonaute-containing RNA-induced silencing complex (RISC). They bind through imperfect complementarity, mostly to the 3′-UTR regions of their target mRNAs, and lead to translation inhibition or degradation (3). These single-stranded RNAs thus modulate gene expression mostly at a posttranscriptional level. Current database describes more than 1,917 miRNA genes, which can contain 3 and 5p miRNAs. MiRNAs are recognized as crucial regulators of the expression of more than 60% of mammalian genes. The number of encoded miRNAs is limited compared to mRNAs and proteins expressed; however, one miRNA may regulate hundreds of mRNAs/lncRNAs and, as a result, may have substantial effects on gene expression networks. Although a lot of miRNAs have conserved sequences between species, the targeted mRNA sequences may be poorly conserved and biological effects are difficult to predict. In silico analyses using updated databases are thus useful to screen for putative targets according to the species and to find the miRNA sequence homology. In vitro functional studies are however needed to validate the miRNA targets, which may provide clues on the biological effects of the miRNA. In addition, available and freely accessible algorithms have been designed to identify potential miRNA-promoter interactions conserved between species that could represent additional clues to further push toward experimental validation (4). In vivo studies add indeed robustness to the biological role of miRNA-mediated regulation in pathophysiological conditions.
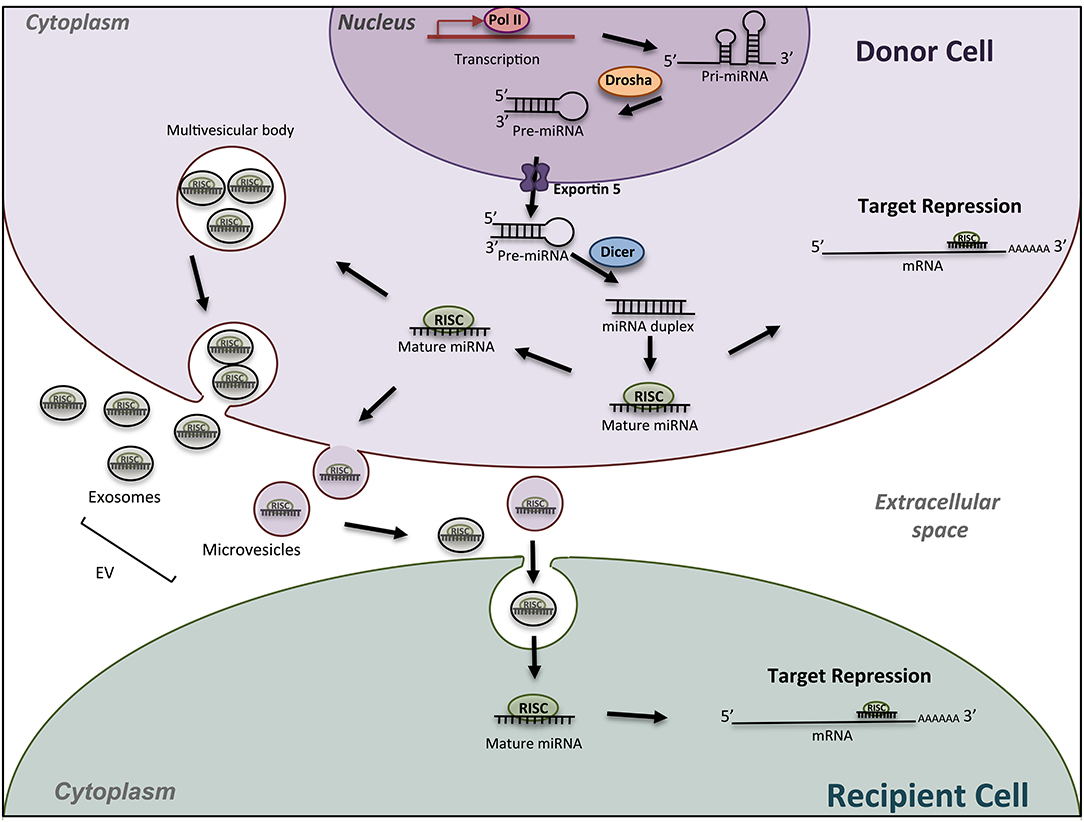
Figure 1. Schematic miRNA biogenesis and mode of action. miRNA biogenesis begins in the nucleus with transcription of miRNA gene into a pri-miRNA, followed by the action of the enzyme Drosha to produce pre-miRNA hairpins. After exportation into the cytosol, pre-miRNA, are processed into an intermediary miRNA duplex by Dicer. One miRNA strand is loaded onto the RNA-induced silencing complex (RISC) to form mature miRNA, which can regulate the expression of target mRNAs. The miRNA/RISC complex can also be incorporated into extracellular vesicles such as exosomes or microvesicle bodies, to be released into extracellular space. Then, miRNAs can be found in body fluids and travel across the entire body till elimination, or can be incorporated into a recipient cell and specifically regulate the expression of target genes into this new cell. Drosha, RNase III-type endonuclease family protein; Dicer, endoribonuclease; RISC; RNA- used silencing complex; EV, extracellular vesicles.
As to many other biological processes, miRNAs act as fine modulators to maintain bone homeostasis. Key evidence that miRNAs are essential to osteoclastogenesis is provided by genetic studies deleting one enzyme essential for their biogenesis, DICER1. DICER deficient mouse and osteoclast-specific DICER gene deficiency both lead to impaired osteoclast (OC) formation and activity (5, 6). Since then, the field of bone biology has regularly reviewed the role of miRNAs in OC biology or bone remodeling, mostly in the context of osteoporosis (7–13). A growing interest in miRNA-based therapeutic strategies has also emerged in bone-related disorders [for review see (14)]. Although the role of miRNAs in the OC lineage and ontogeny is a current hot topic, it remains poorly studied.
OCs are multinucleated cells specialized in bone resorption and derived from myeloid precursors that differentiate in situ in the bone marrow (15). The commitment of myeloid precursors in osteoclastic differentiation is controlled by micro-environmental factors to maintain bone homeostasis. Among them, osteoblasts, osteocytes and bone marrow stromal cells stimulate OC differentiation through the production of receptor activator of nuclear factor kappa-B ligand (RANKL), which binds to its receptor RANK. The growth factor macrophage colony-stimulating factor (MCSF) is also required to initiate the differentiation of osteoclastic precursors by binding its receptor CSF1R (colony stimulating factor 1 receptor). The Wnt-5a ligand is another pro-osteoclastic factor secreted by osteoblasts and OCs themselves (16, 17). In addition to these pro-osteoclastogenic factors, there are several inhibitory regulators including the osteoprotegerin (OPG), a soluble protein secreted by stromal cells that binds soluble and membrane forms of RANKL, thus preventing the activation of the RANK/RANKL signaling pathway. Indirectly, estrogens repress bone resorption by stimulating the production of OPG. Other environmental factors such as cytokines and lipid mediators impact on the osteoclastogenesis (18, 19). In addition to these well-known regulatory mechanisms, other elements could influence the commitment of myeloid precursors to the osteoclastic lineage. Instead of being differentiated into OCs, the myeloid precursors can also be differentiated into macrophages, especially in the presence of MCSF. Indeed, the osteoclastic and macrophagic lineages are thought to originate from an immediate bipotent precursor (20), demonstrating their close proximity. OCs retain the phagocytic potential and the ability to present the antigen of macrophages (21) but are the only cells capable of bone resorption. Among critical regulators of the polarization toward the OC vs. macrophage lineage, the mitochondrial metabolism has been involved (22). Although the role of miRNAs in the commitment of hematopoietic (23) and osteoblastic (24) lineages has been reviewed, only few studies have addressed the involvement of miRNAs in the commitment of the osteoclastic lineage.
The initiation of osteoclastogenesis requires the major signaling pathways RANK/RANKL and CSF1R/MCSF. The transcription factor NFATC1 (nuclear factor of activated T cells 1) is the cornerstone of the early phase of osteoclastogenesis. An amplification loop of NFATC1 induces the expression of many genes of the late phase, such as ACP5 (acid phosphatase 5, tartrate resistant), CTSK (Cathepsin K), and DCSTAMP (dendrocyte expressed seven transmembrane protein) [reviewed in (25)].
Under physiological conditions, the selective expression of miRNAs promoting and repressing the generation of OCs relies on the regulation of their own promoter (a shared promoter in case of miRNA clusters) and is thus closely linked to the sequential signaling pathways involved in the different steps of OC differentiation. MiRNAs are thus essential in a lot of biological feedback loops, including in the regulation of the OC biology.
Our present review will focus on the miRNA-mediated regulation of the sequential steps of OC formation. We will also report on the most recent advances in technologies used for the quantification of miRNAs. Finally, we will discuss the contribution of these technologies to the field of OC biology and the questions that remain to be asked.
miRNAs and the Commitment of Progenitors Toward OCs
There are very few studies available on the miRNA-mediated regulation of OC commitment. Osteoclastogenesis is repressed by miR-155-mediated control of the transcription factor MITF (melanocyte inducing transcription factor) that is involved in the differentiation of monocytes toward macrophages (26). Conversely, miR-29 family (miR-29a, b, c) guides the commitment of bone marrow precursors toward the OC lineage by inhibiting GPR85 and CD93, two molecules involved in macrophage engagement (27). Authors showed that the transfection of the mouse monocytic cell line RAW264.7 with an inhibitor of miR-29 promotes macrophage differentiation, as evidenced by an up-regulation of the F4/80 surface marker and of the phagocytosis, even in presence of the major pro-osteoclastogenic cytokine RANKL.
miRNAs in the Early Phase of OCs Generation
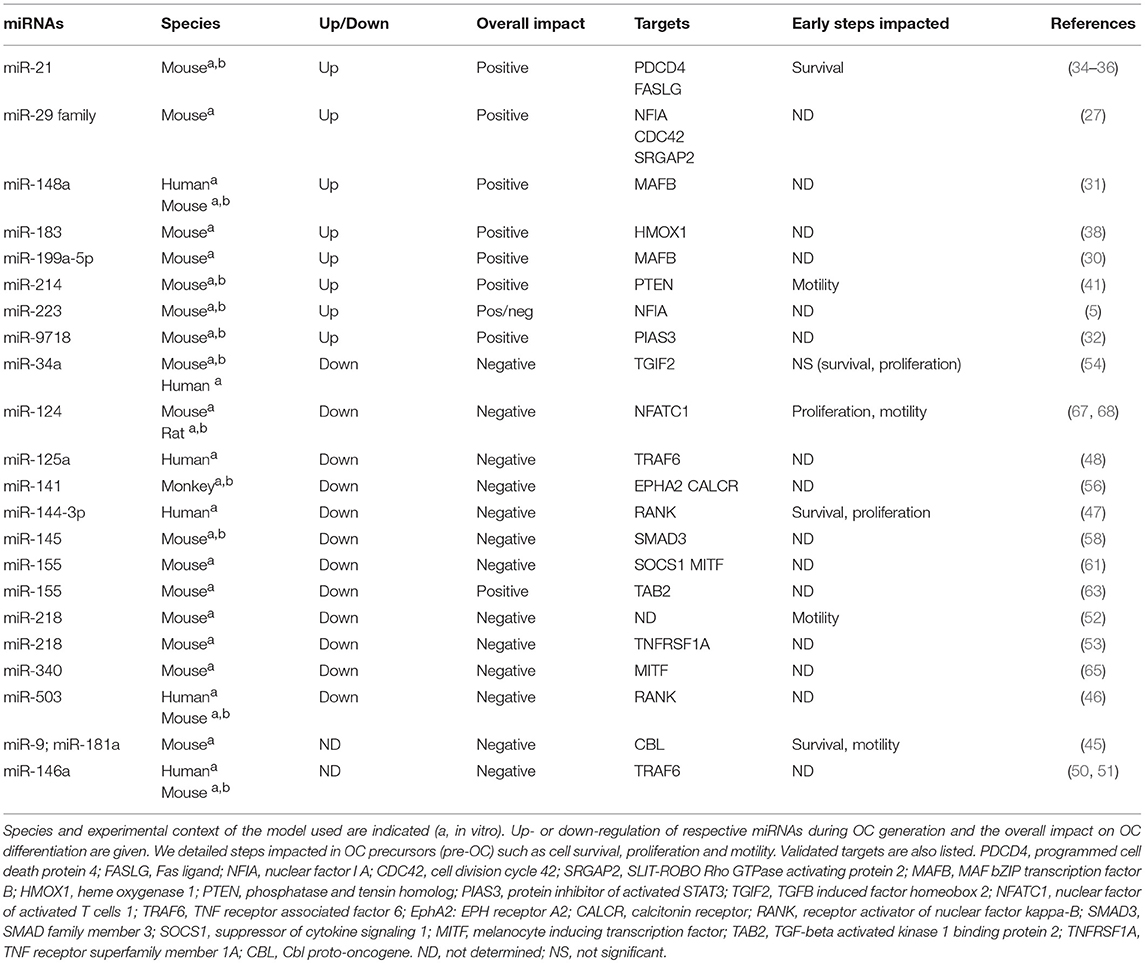
Majority of the studies have described the global impact of miRNAs on the terminal OC formation, as evidenced by OC number and bone resorption activity. Few studies addressed beyond this final step which cellular processes are impacted. In Figure 2, we have summarized the miRNAs involved in the functional steps paving the generation of OCs, including cell survival, proliferation and motility of OC precursors. We also provide an updated list of their validated target genes (Table 1).
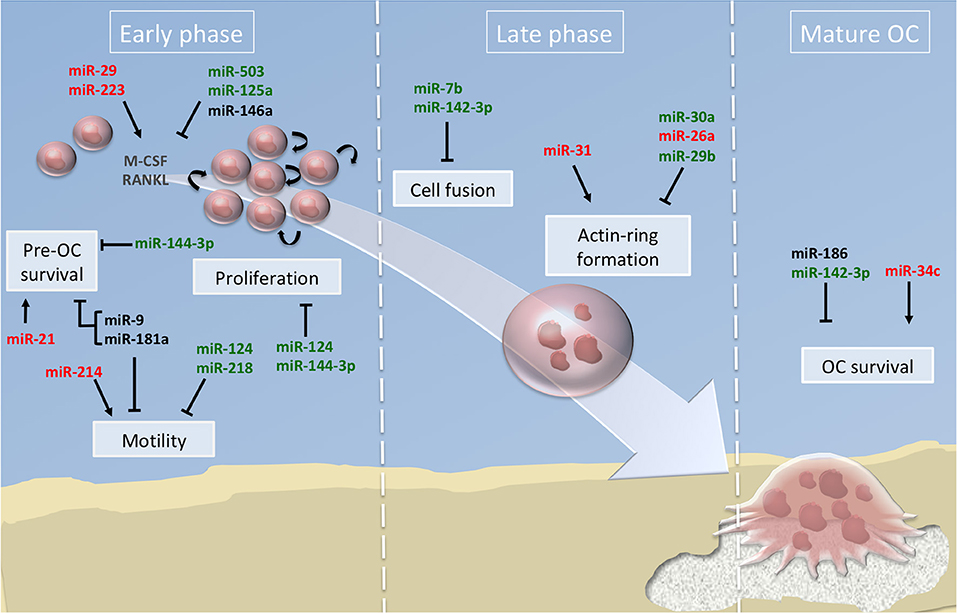
Figure 2. miRNA regulation of osteoclast differentiation. Illustration of the 3 phases of osteoclastogenesis. The early phase is associated with pre-osteoclast (OC) survival, motility, and proliferation; the late phase focused on pre-OC cell fusion and OC acting-ring formation; and the mature OC phase consists in the degradation of the mineralized matrix by mature OC. Green, red and black colors indicate down, up and normal miRNA expression, respectively. Arrows and bars indicate positive or negative effects on osteoclastogenesis respectively. M-CSF, macrophage colony-stimulating factor; RANKL, Receptor activator of NF-κB ligand.
Pro-Osteoclastogenic miRNAs
In addition to its role in the OC commitment of precursors, miR-29 family may promote migration of precursors since miR-29 neutralization inhibits the migration RAW264.7 cells (27). Furthermore, miR-29 family is involved in early phase of osteoclastogenesis by targeting NFIA (nuclear factor I A), a negative regulator of CSF1R (27). CSF1R is also indirectly induced by miR-223 through NFIA targeting as a positive feedback loop enhanced by the transcription factor PU.1 induced downstream of the MCSF/CSF1R (5). Same authors however previously reported contradictory findings using the same RAW264.7 cell line, as overexpression of miRNA-223 suppressed TRAP-positive OC formation (28). These later data were in agreement with a work performed on human peripheral blood mononuclear cells (PBMCs) (29). Bone loss is enhanced in arthritic condition due to activation of OC differentiation, and miR-223 is intensely expressed in rheumatoid arthritis (RA) synovium, particularly in monocyte/macrophage and CD4+ T-cell subsets (29). All these findings suggest an important role of miR-223 in the early phase of osteoclastogenesis, but an in-depth evaluation of the expression level of miR-223 under physiological osteoclastogenesis requires further studies.
The early phase of OC generation triggers increased NFATC1 expression, together with reduced expression of its three negative regulators MAFB (MAF bZIP transcription factor B), IRF8 (interferon regulatory factor 8), and BCL6 (BCL6, transcription repressor) (25). MAFB is a relevant target of miRNAs in OC precursors. Up-regulation of miR-199a-5p and miR-148a promotes the amplification loop of NFATC1 and the formation of resorbing OCs by targeting MAFB in RAW264.7 cells (30) and in CD14+ PBMCs (31), respectively. MiR-9718 is a newly described miRNA specifically expressed in the OC lineage, which promotes OC differentiation by targeting PIAS3 (protein inhibitor of activated STAT3) (32), another negative modulator of NFATC1 (33). Finally, the injection of molecules neutralizing miR-148a or miR-9718 in ovariectomy-induced osteoporotic mice increases total bone mass and decreases the OC number and activity (31, 32).
Among the other miRNAs up-regulated in OCs, the pro-osteoclastogenic role of miR-21 was demonstrated in vivo using the miR-21 KO mouse that display a slight increase in the trabecular bone mass and a reduced OC number and bone resorption (34). The expression of miR-21 is induced by c-Fos, which activation upon RANKL treatment of mouse bone marrow-derived macrophages (BMMs) operates a positive feedback loop by targeting the programmed cell death protein 4 (PDCD4) (35). The binding of c-Fos to miR-21 promoter is strikingly diminished by estrogen E2 treatment in RANKL-induced osteoclastogenesis. Estrogen attenuate miR-21 biogenesis, leading to increased FasL protein level and caspase-3 activity in mouse BMMs precursors (36). These results suggest that miR-21 expression is important in the development of OCs, particularly by controlling pre-osteoclast survival.
The activation of mitogen activated protein kinases (MAPKs) downstream of the early RANKL pro-osteoclastogenic signaling cascade is supported by the reactive oxygen species (ROS) produced by RANK-NADPH oxidase 1-dependant pathway (25). ROS production by mouse BMMs is regulated by heme oxygenase 1 (HMOX1), which attenuates osteoclastogenesis, specifically during the early phase of OC formation (37). MiR-183 is up-regulated by RANKL and targets HMOX1, thus promoting the early phase of osteoclastogenesis (38).
The PI3K/AKT pathway is induced by RANKL signaling and promotes cell survival (39). By reversing the action of PI3K (phosphoinositide 3-kinase), PTEN (phosphatase and tensin homolog) negatively impacts on OC precursor motility in the early phase of osteoclastogenesis (40). It was shown that miR-214 enhances the OC precursor differentiation via the PTEN/PI3K/AKT pathway, downstream of RANK signaling in RAW264.7 and primary mouse BMMs. In vivo, OC-specific miR-214 transgenic mice exhibit reduced expression of PTEN, increased OC resorption activity, and reduced bone mineral density (41). Since a miR-214/PTEN axis has been involved in cell proliferation and invasion of various cancer cells (42–44), it would be of particular interest to investigate the specific role of miR-214 on cell proliferation, survival and motility in the context of OC lineage.
The over-expression of miR-9 and miR-181a diminishes the migration of RAW264.7 cells and primary mouse OC survival by repressing the expression of the proto-oncogene Cbl, which enhances the amount of the pro-apoptotic protein Bim (45). This was the first study reporting a functional role for miR-9 and miR-181a by targeting proteins belonging to the apoptosis pathway. Further experiments are needed to confirm the potential role of these miRNAs on the precursor survival during osteoclastogenesis.
miRNAs With an Inhibitory Role on OC Precursors
The initiation of the OC precursor differentiation is largely mediated by RANK/RANKL signaling. The expression density of RANK at the cell surface conditions the efficacy of RANK trimerization and downstream signal transduction. In human CD14+ precursors, miR-503 targets RANK mRNA in the coding sequence (CDS) region, leading to reduced RANK protein level, OC numbers and cell density in vitro (46). In vivo, treatment of OVX mice with anti-miR-503 reduced bone resorption (46). The 3′ untranslated region (UTR) of RANK is also targeted by miR-144-3p in CD14+ precursors, controlling OC formation, proliferation and survival of OC precursors (47).
The binding of RANKL to RANK induces the recruitment of the adaptor protein TRAF6 (TNF Receptor Associated Factor 6). This important signaling adaptor for RANK is targeted by miR-125a and miR-146a in human PBMCs (48, 49). The expression of miR-125a is controlled by NFATC1, which directly binds to the promoter of miR-125a during osteoclastogenesis and reduces its expression (48). MiR-146a has been extensively studied in monocytes in pathological conditions, including pathologies associated with bone erosion such as RA. The expression of miR-146a is induced by LPS, TNFα, or IL1β signaling cascades, through the activation of NF-κB (nuclear factor kappa B), which directly binds to the miR-146 promoter (49). It was shown that miR-146a inhibits OC formation from human PBMCs in a dose-dependent manner and that the treatment of collagen-induced arthritic mice with miR-146 mimics attenuates bone resorption (50). We recently demonstrated that the reduced expression of miR-146a in the Ly6Chigh monocyte subset of arthritic mice is involved in the pathogenic bone erosion, and that it can be rescued by specific delivery of miR-146 mimics to Ly6Chigh monocytes (51). Taken together, these findings evidence a negative regulation of osteoclastogenesis by NFκB-induced miR-146a to partly counterbalance the deregulated differentiation of OC precursors in inflammatory disorders.
The RANK/TRAF6 signaling cascade activates the NFκB and MAPK pathways, which may represent additional miRNA targets. Indeed, miR-218 over-expression inhibits osteoclastogenesis by controling the p38MAPK pathway in mouse BMMs (52). Interestingly, miR-218 negatively impacts on the migration of RANKL-treated BMMs. Although authors also reported a decrease of actin-ring formation, it was most probably a consequence than a cause of the decreased OC number. The putative targets of miR-218 were not explored in this study. A recent study confirmed the negative regulation of osteoclastogenesis by miR-218, and show that it was mediated by targeting TNFRSF1A (TNF receptor superfamily member 1A), which leads to the inhibition of the NFκB pathway activation in RAW264.7 cells (53). Overall, miR-218 may act on both NFκB and MAPK pathways in OC precursors to control OC precursor differentiation, and further studies are required to unravel the molecular mechanisms involved.
The early RANKL signaling is mediated by two transcription factors, NFκB and AP1, which are essential to the initiation of the NFATC1 amplification loop. AP1 components such as c-Jun and c-Fos could be critical targets to modulate NFATC1 activation in OC precursors. The transcriptional regulator TGIF2 (TGFB induced factor homeobox 2) is induced by NFATC1, c-Fos and c-Jun and potentiates the activity of NFATC1 and c-Jun in turn, promoting the osteoclastogenesis in a positive feedback loop (54). Interestingly, TGIF2 is a direct target of miR-34a, and OC-specific miR-34a transgenic mice exhibit lower bone resorption and higher bone mass, with no alteration of OC precursor survival and proliferation (54). Finally, miR-34a seems to negatively regulate the NFATC1 pathway during OC differentiation, mostly in the early phase upon RANKL signaling.
RANKL signaling enhances another co-stimulatory signal mediated by the ephrinA2-EphA2 interaction at the cell surface of OC precursors. EphrinA2 expression is rapidly induced in a c-Fos-dependent manner and cleaved by metalloproteinases to release an active soluble form able to interact with its receptor EphA2, enhancing osteoclastogenesis (55). In rhesus monkey BMMs EphA2 is a potential target of miR-141 (56). OC differentiation and bone resorption are suppressed in vitro by miR-141, and in vivo using repeated injections of an OC-targeted delivery system into aged monkeys (56). The calcitonin receptor (CALCR) is also a target of miR-141 in rhesus OCs. A down-regulation of CALCR is however expected to suppress the negative effect of calcitonin on OC differentiation and to enhance bone resorption, whereas miR-141 globally inhibits OC differentiation and activity, both in vitro and in vivo. One can speculate that the targeting of CALCR by miR-141 may represent a minor part of the effects of miR-141 functions in rhesus OCs.
Another critical interaction in RANKL-induced osteoclast ogenesis is the cooperation between Smad complex and c-Fos, which leads to NFATC1 transcription (57). Recently, miR-145 was shown to target Smad3, thus reducing the formation of p-Smad2/3 complex, repressing c-Fos and NFATC1 transcription in mouse OC precursors, and decreasing OC number (58). Smad proteins are induced by members of the transforming growth factor beta (TGFβ) super family, and Smad pathway has become of particular interest in inflammatory disorders [reviewed in (59)]. TGFβ1/Smad4 signaling directly induces miR-155 expression, a negative regulator of osteoclastogenesis (60). In addition, miR-155 is induced by interferon (IFN)-β and mediates its suppressive effect on OC differentiation by targeting the pro-osteoclastogenic gene SOCS1 (suppressor of cytokine signaling 1) in OC precursors (61). These data were suggestive of a suppressive role of miR-155 in osteoclastogenesis. In physiological condition, miR-155 is downregulated during osteoclastogenesis. Nevertheless, miR-155 is up-regulated in activated immune cells, such as lymphocyte B-cells, T-cells and dendritic cells, promoting the inflammatory response and thus aggravating the inflammatory-induced arthritis and bone erosion in vivo through the indirect immune-mediated activation of OCs [reviewed in (62)]. In lipopolysaccharide (LPS)-induced inflammatory condition, miR-155 directly induces autophagy in OCs as well as OC differentiation and activity by targeting TGFβ-activated kinase 1-binding protein 2 (TAB2) (63). A fine modulation of the TAB2 expression level may promote the destabilization of the inactive complex TAB2/Beclin1, leading to (i) the release of Beclin1 and induction of autophagy and (ii) the interaction of the adaptor proteins TAB2 and TAK1 that activates the RANK/TRAF6/NFκB pathway. These findings illustrate the variable role of miR-155 in osteoclastogenesis depending on the microenvironment, i.e., according to the presence of LPS, IFNβ, or TGFβ-mediated inflammatory signals. To make things even more complex, miR-155 also targets the transcription factor MITF that is up-regulated upon RANKL signaling (60, 61). MITF plays a critical role in the OC differentiation by collaborating with NFATC1 in the early phase of osteoclastogenesis (64). In summary, miR-155 seems to globally inhibit the OC generation in physiological condition by acting both in the commitment of myeloid precursors (26) and in osteoclastogenesis (61) through the regulation of MITF expression. MITF is also targeted by miR-340, which inhibits the OC differentiation of mouse BMMs (65). This reinforces the idea that MITF is a key target for miRNAs in osteoclastogenesis.
Finally, NFATC1 has hardly been described as a direct target of miRNAs so far. The activity of NFATC1 is regulated by phosphorylation, which retains NFATC1 in the cytosol compartment and inhibits its translocation into the nucleus. Epigenetic controls of the NFATC1 gene have been reported [reviewed in (66)]. To date, only miR-124 is predicted to bind NFATC1 in its 3′ UTR in OC precursors (67). The targeting of the 3′UTR of both rat and human NFATC1 mRNAs by miR-124 was confirmed using luciferase reporter assays (68). Functionally, miR-124 represses NFATC1 expression in mouse BMMs and diminishes the migration and proliferation of OC precursors (67), without impact on their survival (69).
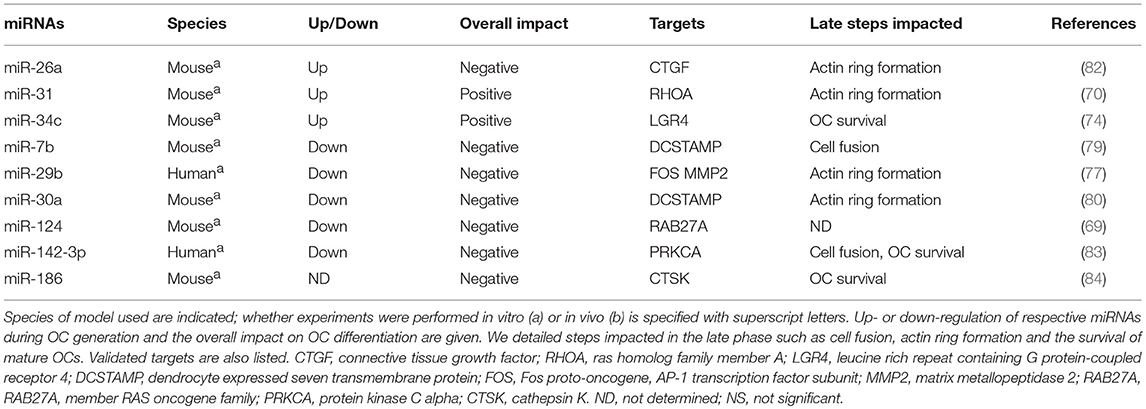
miRNAs in the Late Phase of OC Generation
To achieve OC maturation, the key transcription factors MITF, PU.1, and NFATC1 lead to the expression of several osteoclastogenic genes involved in the cytoskeleton organization, the cell fusion and actin-ring formation. MiRNAs also regulate the late stage of OC formation by targeting Rho GTPases, DCSTAMP, CSTK, and the RANKL-receptor inhibitor LGR4 (Table 2).
Pro-Osteoclastogenic miRNAs
One of the most up-regulated miRNAs during osteoclastogenesis in mouse BMMs is miR-31. It specifically acts on the actin-ring formation (70). Neutralization of miR-31 impairs the matrix resorption and the ring-shaped OC formation, while cell-fusion is conserved. Increased RhoA activity and protein expression level are also observed in miR-31-deficient OC precursors, suggesting that RhoA is targeted by miR-31 in the OC lineage (70). Small GTPases of the Rho family (Rac1, Rac2, CDC42, RhoA, and RhoU) play important roles in the cell-fusion of OC precursors, podosome organization, migration, and polarization of mature OCs [reviewed in (71)]. RhoA controls the polymerization of actin, the turnover of podosomes, and the migration of OCs through the bone matrix (72). While a moderate level of RhoA activity is required to allow both stability of the sealing zone and bone resorption, RhoA over-activation or inhibition cause disassembly of the podosomes and thus impair the OC activity [reviewed in (73)]. Finally, miR-31 seems essential to OC maturation by finely modulating RhoA activity.
Another relevant miRNA in mature OC biology is miR-34c, which promotes the OC survival at the end of maturation by targeting the R-spondins receptor LGR4 (leucine rich repeat containing G protein-coupled receptor 4), also known as GPR48 (74). During the OC maturation, NFATC1 induces the expression of LGR4 at the cell surface to negatively regulate RANK/RANKL-signaling by a direct competition with RANK. The binding of RANKL to LGR4 activates the NFκB-inhibitor GSK3β, which results in OC apoptosis (75). These data suggest that miR-34c sustains the OC formation.
miRNAs Displaying an Inhibitory Role in OC Maturation
Based on the study of Franceschetti et al. (27), we reviewed above the role of miR-29 family in the OC commitment and in the early phase of OC generation. The authors however also showed a late up-regulation of miR-29 (a, b, c) family members at the third day of RANKL-induced OC differentiation of mouse BMMs, suggesting another predominant role of miR-29 in the late phase of osteoclastogenesis. They found two targets involved in the cytoskeleton organization, CDC42 and SRGAP2 (SLIT-ROBO Rho GTPase Activating Protein 2), which present opposite effects. Indeed, SRGAP2 belongs to GAP family, which inactivates Rho GTPases such as CDC42 by increasing the intrinsic GTPase activity of Rho proteins (76). Rho GTPases are essential in the polarization and the podosome belt/sealing zone formation of functional OCs [reviewed in (17, 71)].The neutralization of miR-29 family has no effect on the formation of the actin-ring in mature RAW-derived OCs. A role for miR-29 family in the OC maturation remains thus questionable. Another study showed a down-regulation of miR-29b during OC generation, and demonstrated an inhibitory role of miR-29b in resorption and actin-ring formation in human CD14+-derived OCs (77). Considering only the multinucleated cells for their analysis, authors observed disarranged nodular actin spots in OCs over-expressing miR-29b, leading to a failure to form actin-rings. Although the authors did not perform a validation of the known targets of miR-29b in the OC lineage, they observed a significant down-regulation of c-Fos and MMP2 expression in miR-29b-transfected OCs at the end of the differentiation. The apparent discrepancies on the role of miR-29 family members and miR-29b could be partially explained by the chosen approach in the different studies, which rely either on gain-of function of miR-29b (77) or loss-of function of miR-29 family (27), as well as on the nature of OC precursors used (cell line vs. primary cells).
In addition to its negative effect on cell proliferation and motility in the early phase of osteoclatogenesis (67), miR-124 seems to act on the late phase of the differentiation by targeting Rab27a (69), a protein belonging to the small Rab GTPase family and involved in vesicle trafficking and resorbing activity of OCs (78). Notably, miR-124 is under-expressed in the OVX mouse model, which displays a deregulated resorbing activity (69).
DCSTAMP is a key protein involved in cell fusion. DCSTAMP is targeted by miR-7b and miR-30a in mouse BM precursors (79, 80). Mimics of miR-7b and miR-30a repress the expression level of DCSTAMP, decrease the OC number and inhibit matrix resorption. Conversely, anti-miR-7b promotes the OC formation and increase nuclei number in mature OC, suggesting an increase of cell fusion events (79). By measuring the membrane merge rate, the authors confirmed that overexpression of miR-7b in mouse BMMs significantly abrogates OC fusion (81). Anti-miR-30a enhances the actin-ring formation (80). Similarly, miR-26a attenuates the actin-ring formation and resorption in mouse BMMs. MiR-26a targets the connective tissue growth factor (CTGF), which induces and interacts with DCSTAMP (82). Contrary to miR-7b and miR-30a, miR-26a is upregulated in the late phase of osteoclastogenesis, suggesting a physiological regulation of multinucleation in the OC lineage. Thus, it will be of particular interest to evaluate the role of miR-30a and miR-26a on cell fusion.
During OC formation from human CD14+ progenitors, the enforced expression of miR-142-3p inhibits cell-to-cell contact, clustering and fusion events associated with the induction of OC apoptosis upon RANKL stimulation (83). The negative effect of miR-142-3p on OC fusion could be partially explained by the silencing of PKCα. Indeed, PKCα is involved in the microtubule and actin networks and is predicted as a putative target of miR-142-3p by prediction software. A decreased expression of anti-apoptotic factors downstream of PKCα might also explain the induction of cell death (83), however it has not been functionally explored yet.
Finally, miR-186 was newly described as a negative regulator of mature OC survival. Mimics of miR-186 induce caspase-3/7 activity and OC apoptosis in transfected RAW264.7-derived mature OCs (84). Moreover, miR-186 targets the CTSK gene (84) and probably represses the resorbing function of OCs, although it remains to be explored in functional assays.
Advances in Technologies for the Quantification of miRNAs in Biological Tissues
MiRNAs are classically studied from the total mRNA (including small RNAs) extracted from the tissue of interest. As miRNAs are expressed in various cell types and tissues, it is necessary to well define the targeted sample and to control the purity of the elicited source of miRNAs. The study of miRNAs in primary OCs is challenging because of the localization of OCs into bone cavities, and thus a purified extract of primary OCs is very hard. Finally, primary OC precursors from blood sample or bone marrow are used to derive OCs in in vivo experiments. Nevertheless, the OC differentiation in culture is only partial without reaching a pure OC population. The obtained population includes all stages from the precursor to the mature OC. The study of miRNAs in such heterogeneous cell culture is not satisfactory and gives a lot of variations between samples that could impact on the reliability of the results, particularly in “end-point” studies without a kinetic expression of miRNAs during the OC differentiation. Some studies are based on purified OCs using chemical methods to eliminate non- and poor-adherent cells, as mature OCs are extremely adherent. Very recently, a novel method of purification based on the OC sorting has been described, allowing a standardized approach to better characterize the miRNA profiling in mature OCs (85).
Other biological tissues are become novel sources of study in the OC biology and the bone remodeling. Indeed, miRNAs can be exported from a donor cell to a recipient via exosomes and microvesicles (Figure 1), and thus participate to the cell communication between osteocytes, osteoblasts and osteoclasts in the bone micro-environment (86–89). Circulating miRNAs can also be used as biomarkers in pathophysiological conditions, reaching liquid biopsies as potential interesting samples in clinical application.
Molecular technologies are used to detect and quantify miRNAs, and some panels were developed to determine the miRNA profiling. Here we provide a global and detailed view of these technologies and their application in the quantification of miRNAs in biological tissues.
Screening Methods for the miRNA Profiling
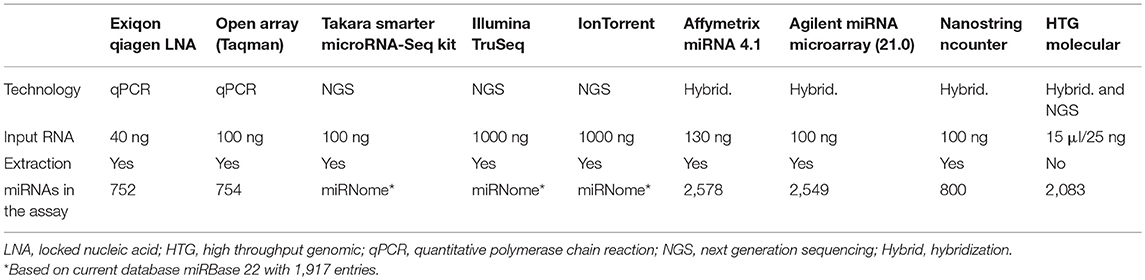
In recent years, technological advances in research tools including qPCR, microarrays, and next generation sequencing (NGS), have enabled sensitive detection of miRNAs. Typically, miRNA biomarkers are measured with quantitative polymerase chain reaction (qPCR) after RNA extraction and conversion into complementary DNA (cDNA). Many other methods are available for large screening of the miRNome to understand pathologies or discovery of biomarkers. Mestdagh et al., have extensively analyzed analytical parameters of many solutions available for miRNA measurement based on qPCR, hybridization platforms and sequencing technologies (90). Recently, new technologies for miRNAs measurement or optimization of existing methods have emerged. Currently, the miRbase 22 includes 1917 miRNA genes. With the constant evolution of the miRbase, the different miRNA detection platforms need to adapt their product. This flexibility however depends on the technic used. New technologies that are more sensitive, or extraction free chemistry, are definitively modifying the miRNA measurement landscape (Table 3).
Among these technologies, we will focus on the most innovative technologies for miRNA detection. These technologies provide real advance and alternative with the direct use of matrices avoiding preprocessing step or optimized library preparation for RNA-Seq method for the profiling of miRNAs. For the measurement of single miRNAs, new methods using microfluidic detection by laminar flow (91) or absolute quantification of signal by RCA-FRET technology (92) are in development. These last technologies may simplify the adoption of miRNA testing in clinical laboratories.
Screening methods such as the microarrays of Affymetrix (version 4.1) and Agilent (v21) now include 2,578 and 2,549 miRNAs, respectively, thus offering a more comprehensive dataset. In addition to these updated microarray versions, the company Takara Bio have recently developed the SMARTer miRNA-Seq kit, which uses Mono-Adapter ligation and Intramolecular Circularization (MAGIC) technology to efficiently capture miRNA species with reduced bias inherent to other approaches. Measurement of equimolar mixture of 963 miRNAs shows that >70% of the miRNAs are accurately represented, whereas other competitors have 49–79% of the miRNAs underrepresented Nevertheless, besides having less bias than the competitors (including TruSeq and NEXTFlex), this kit also produces large amounts of side products and as a result did not perform better for the detection of biological miRNAs (93). Since several years now, a new technology of HTG Molecular (High Throughput Genomic) called EdgeSeq provides miRNAs measurement without RNA extraction. The technology is a combination of hybridization with nuclease protection assay and next generation sequencing. Based on an extraction free chemistry, the technology1 allows the direct measurement of miRNAs from as little as 15 μl of body fluid. This last critical point avoids biases associated to the extraction protocols (94) and increases the sensitivity of the measure. Correlation between replicates is very similar to those shown previously (90) for hybridization and sequencing technologies, whereas accuracy of the gradient of miRNA measure is superior to other hybridization and sequencing platform2. Nevertheless, cross reactivity is higher compared to qPCR assays2. It has been shown that HTG EdgeSeq results were closest to the RNASeq results with >95% concordance on tissue samples (95). The technology also shows very good correlation with the PCR on plasma samples, with Pearson's coefficient of 0.93 and 0.94 for qPCR and digital PCR (dPCR) data, respectively (96).
Circulating miRNAs as Biomarkers
In the past decade, the search for circulating miRNA for functional studies and biomarkers research has yielded numerous associations between miRNAs and different types of disease. However, many of these relations could not be replicated in subsequent studies under similar experimental conditions. Although this lack of reproducibility may be explained by variations in experimental design and analytical methods, guidelines of the most appropriate design and methods of analysis are scarce. MiRNAs have significant promise as biomarkers for diseases, due to their regulatory role in many cellular processes, and their stability in samples such as plasma and serum. Circulating miRNAs are moreover easily accessible. Biomarker experiments generally consist of a discovery phase and a validation phase. In the discovery phase, typically hundreds of miRNAs are simultaneously measured to identify candidates. Because of the costs of such high-throughput experiments, numbers of subjects are often too small, which can lead to false positives and negatives. In validation phase, a small number of identified candidates are measured in a large cohort, generally using quantitative PCR (qPCR). Although qPCR is a sensitive method to measure miRNAs in the circulation, experimental design, and qPCR data analysis remain challenging with many sources of biases. The MIQE guidelines are useful to stress on the most important biases in qPCR experiments and to give some elements to improve experimental practice (97). There is still a need for standardization or development of new methods to reach the clinic. Thus, choosing the right tools is critical for a successful miRNA-based experiment.
Despite new advances and evolution of technologies, many challenges remain unmet. For example, the impact of RNA isolation, which is known to induce biases (94), but also the lack of standardization of miRNA measurement or normalization of miRNAs data from plasma/serum samples, are key factors of variation, making more challenging the translation of biomarker discovery to diagnostic tool. Indeed, while many reports are describing miRNAs as potential biomarkers since many years, miRNA-based diagnostics have many difficulties to enter to the clinic and to get IVD approval. Moreover, these kinds of tests are likely best suited to a companion role. In contrast to RNA or DNA-based tests, especially that indicate the presence of SNP or a specific expression profile (98), miRNA tests produce results that are more difficult to interpret. While many miRNAs were reported as biomarkers in many reports (99), most miRNAs are expressed widely in a non-cell-specific manner, and their levels of expression are not differing drastically between patients' group and controls. For liquid biopsies such as blood, urine, or other body fluids, miRNA levels are very sensitive to pre-processing and post-processing factors. As a result, despite being very stable, miRNA-based tests are often based on a combination of miRNAs associated with an algorithm, and strict standardization of the entire process, from obtaining and processing the sample to results reporting, is mandatory and is key for reproducible results, no matter what technology is used.
Point of care diagnostics requires short detection time, small sample volume and portability of the device. These requests are often not compatible with miRNA measurement technologies since they are requesting many steps and devices. New methods based on microfluidic chip and laminar flow assisted dentritic amplification is currently under development, with a promise of time to result of only 20 min (91). Sensitivity and accuracy have still to be increased, but this attractive solution could potentially allow the compatibility of miRNAs to short diagnostic delay. Without such solution able to reduce time to results, use of miRNAs in diagnostic is only possible for non-urgent test. Most used platforms remain qPCR, with various technologies available such as LNA/Taqman/SybrGreen PCR, which request high level of standardization to provide accurate and stable results. Another novel technology that could facilitate miRNA transfer to clinic is called RCA-FRET, which is a combination of rolling circle amplification (RCA) and Förster resonance energy transfer (FRET) (92). Key advantage of the technology is a simple workflow, device needed and material for absolute quantification results. This last point is of key importance since normalization of miRNA data is a true hurdle.
Indeed, accurate quantification of miRNAs using qPCR is largely dependent on proper normalization techniques, the absence of which can lead to misinterpretation of data and incorrect conclusions (100). The goal of most miRNA experiments using qPCR is to identify differences in expression between two groups of samples. In this case, cell-free miRNAs from biofluids are emerging as important noninvasive biomarkers because of their stability. Many works are describing miRNAs as potential blood biomarkers. One challenge in studying miRNAs from serum and plasma is their relatively low abundance and lack of reliable endogenous controls. It has been shown that hsa-miR-24, hsamiR-126, hsa-miR-484 (101), hsa-miR-16-5p, hsa-miR-93-5p (102), hsamiR-484, and hsa-miR-191-5p (103) are stable normalizers in serum. Nevertheless, some reports indicate also these miRNAs not as normalizers but as biomarkers, as for miR-16-5p described in the progression of gastric cancer for example (104). In the absence of reliable endogenous controls in serum/plasma, exogenous or spike-in controls can be used to normalize miRNA expression data. Exogenous controls can also be used to monitor extraction efficiency or sample input amount for difficult samples (e.g., serum, plasma, or other biofluids).
In addition to most common used matrices, extracellular vesicles (EVs) are emerging and even more challenging to get reliable results. EVs are nanometer-scale particles, which include exosomes, microvesicles, and apoptotic bodies. EVs are intercellular communicators released by most cell types with key functions in physiological and pathological processes (86, 105). EVs deliver specific proteins, microRNAs and other cellular components. One of the most important aspects of EV research is analyzing their nucleic acid cargo, particularly miRNAs. These are commonly quantified by RT-qPCR or, increasingly, by comprehensive transcriptomic profiling using NGS or hybridization technologies. One critical issue is the methods for RNA extraction that can influence downstream analyses by yielding non-identical, kit-specific results. This is of particular challenge since typical concentration of this type of sample is low, RNA integrity is decreased and associated to high individual variability. Several kits are on the market and tested (106) but optimal isolation methodology is mainly dependent on the respective research setting and downstream analyses.
Overall, it is not easy to give a simple answer to a complex problem. Many methods are available for miRNAs measurement and generate complex data that need to be validated. Real efforts have to be done on standardization and analytical validation until one can consider to successfully translate biomarker discovery to the clinic.
Conclusion
In the present review we have shown the complexity of understanding OC biology and pointed out key miRNAs that have been involved in OC differentiation as key regulators, with very specific roles in the different phases progressing from early progenitors toward fully mature OC. We also stressed the challenge to get reliable data from miRNA measurement either for supporting the knowledge of OC regulation or the translation into biomarker tools for clinical application.
Data Availability
All datasets generated for this study are included in the manuscript and/or the supplementary files.
Author Contributions
CL, ID-R, HF, ES, and FA wrote the review. CL, ID-R, and ES designed the figures and tables.
Funding
INSERM, ANR-16-CE14-0030-02, ANR-15-RAR3-0013-05, University of Montpellier, and H2020 project RABIOPRED (#666798).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
1. ^From www.takarabio.com/learning-centers/next-generation-sequencing/technical-notes/accurate-mirna-representation-in-microrna-seq,2018
2. ^From the poster presented at ESHG, 2017, available on https://www.htgmolecular.com/assets/htg/publications/PO-17-Aissaoui-EHSG-miRNA-Metrics.pdf
References
1. Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. (1998) 391:806–11. doi: 10.1038/35888
2. Winter J, Jung S, Keller S, Gregory RI, Diederichs S. Many roads to maturity: microRNA biogenesis pathways and their regulation. Nat Cell Biol. (2009) 11:228–34. doi: 10.1038/ncb0309-228
3. Ha M, Kim VN. Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol. (2014) 15:509–24. doi: 10.1038/nrm3838
4. Piriyapongsa J, Bootchai C, Ngamphiw C, Tongsima S. microPIR2: a comprehensive database for human-mouse comparative study of microRNA-promoter interactions. Database. (2014) 2014:bau115. doi: 10.1093/database/bau115
5. Sugatani T, Hruska KA. Impaired Micro-RNA pathways diminish osteoclast differentiation and function. J Biol Chem. (2009) 284:4667–78. doi: 10.1074/jbc.M805777200
6. Mizoguchi F, Izu Y, Hayata T, Hemmi H, Nakashima K, Nakamura T, et al. Osteoclast-specific Dicer gene deficiency suppresses osteoclastic bone resorption. J Cell Biochem. (2010) 109:866–75. doi: 10.1002/jcb.22228
7. Xia Z, Chen C, Chen P, Xie H, Luo X. MicroRNAs and their roles in osteoclast differentiation. Front Med. (2011) 5:414–9. doi: 10.1007/s11684-011-0168-0
8. Tang P, Xiong Q, Ge W, Zhang L. The role of microRNAs in osteoclasts and osteoporosis. RNA Biol. (2014) 11:1355–63. doi: 10.1080/15476286.2014.996462
9. Chen J, Qiu M, Dou C, Cao Z, Dong S. MicroRNAs in bone balance and osteoporosis. Drug Dev Res. (2015) 76:235–45. doi: 10.1002/ddr.21260
10. Sun M, Zhou X, Chen L, Huang S, Leung V, Wu N, et al. The regulatory roles of MicroRNAs in bone remodeling and perspectives as biomarkers in osteoporosis. Biomed Res Int. (2016) 2016:1652417. doi: 10.1155/2016/1652417
11. Ji X, Chen X, Yu X. MicroRNAs in osteoclastogenesis and function: potential therapeutic targets for osteoporosis. Int J Mol Sci. (2016) 17:349. doi: 10.3390/ijms17030349
12. Gennari L, Bianciardi S, Merlotti D. MicroRNAs in bone diseases. Osteoporos Int. (2017) 28:1191–13. doi: 10.1007/s00198-016-3847-5
13. Feng Q, Zheng S, Zheng J. The emerging role of microRNAs in bone remodeling and its therapeutic implications for osteoporosis. Biosci Rep. (2018) 38:BSR20180453. doi: 10.1042/BSR20180453
14. Yang Y, Fang S. Small non-coding RNAs-based bone regulation and targeting therapeutic strategies. Mol Cell Endocrinol. (2017) 456:16–35. doi: 10.1016/j.mce.2016.11.018
15. Boyle WJ, Simonet WS, Lacey DL. Osteoclast differentiation and activation. Nature. (2003) 423:337–42. doi: 10.1038/nature01658
16. Maeda K, Kobayashi Y, Udagawa N, Uehara S, Ishihara A, Mizoguchi T, et al. Wnt5a-Ror2 signaling between osteoblast-lineage cells and osteoclast precursors enhances osteoclastogenesis. Nat Med. (2012) 18:405–12. doi: 10.1038/nm.2653
17. Uehara S, Udagawa N, Kobayashi Y. Non-canonical Wnt signals regulate cytoskeletal remodeling in osteoclasts. Cell Mol Life Sci. (2018) 75:3683–92. doi: 10.1007/s00018-018-2881-1
18. Oikawa T, Kuroda Y, Matsuo K. Regulation of osteoclasts by membrane-derived lipid mediators. Cell Mol Life Sci. (2013) 70:3341–53. doi: 10.1007/s00018-012-1238-4
19. Amarasekara DS, Yun H, Kim S, Lee N, Kim H, Rho J. Regulation of osteoclast differentiation by cytokine networks. Immune Netw. (2018) 18:e8. doi: 10.4110/in.2018.18.e8
20. Xiao Y, Palomero J, Grabowska J, Wang L, de Rink I, van Helvert L, et al. Macrophages and osteoclasts stem from a bipotent progenitor downstream of a macrophage/osteoclast/dendritic cell progenitor. Blood Adv. (2017) 1:1993–2006. doi: 10.1182/bloodadvances.2017008540
21. Li H, Hong S, Qian J, Zheng Y, Yang J, Yi Q. Cross talk between the bone and immune systems: osteoclasts function as antigen-presenting cells and activate CD4+ and CD8+ T cells. Blood. (2010) 116:210–7. doi: 10.1182/blood-2009-11-255026
22. Jin Z, Wei W, Yang M, Du Y, Wan Y. Mitochondrial complex I activity suppresses inflammation and enhances bone resorption by shifting macrophage-osteoclast polarization. Cell Metab. (2014) 20:483–98. doi: 10.1016/j.cmet.2014.07.011
23. Undi RB, Kandi R, Gutti RK. MicroRNAs as haematopoiesis regulators. Adv Hematol. (2013) 2013:695754. doi: 10.1155/2013/695754
24. Valenti MT, Dalle Carbonare L, Mottes M. Role of microRNAs in progenitor cell commitment and osteogenic differentiation in health and disease (Review). Int J Mol Med. (2018) 41:2441–9. doi: 10.3892/ijmm.2018.3452
25. Park JH, Lee NK, Lee SY. Current understanding of RANK signaling in osteoclast differentiation and maturation. Mol Cells. (2017) 40:706–13. doi: 10.14348/molcells.2017.0225
26. Mann M, Barad O, Agami R, Geiger B, Hornstein E. miRNA-based mechanism for the commitment of multipotent progenitors to a single cellular fate. Proc Natl Acad Sci USA. (2010) 107:15804–9. doi: 10.1073/pnas.0915022107
27. Franceschetti T, Kessler CB, Lee S-K, Delany AM. miR-29 promotes murine osteoclastogenesis by regulating osteoclast commitment and migration. J Biol Chem. (2013) 288:33347–60. doi: 10.1074/jbc.M113.484568
28. Sugatani T, Hruska KA. MicroRNA-223 is a key factor in osteoclast differentiation. J Cell Biochem. (2007) 101:996–9. doi: 10.1002/jcb.21335
29. Shibuya H, Nakasa T, Adachi N, Nagata Y, Ishikawa M, Deie M, et al. Overexpression of microRNA-223 in rheumatoid arthritis synovium controls osteoclast differentiation. Mod Rheumatol. (2013) 23:674–85. doi: 10.3109/s10165-012-0710-1
30. Guo K, Zhang D, Wu H, Zhu Q, Yang C, Zhu J. MiRNA-199a-5p positively regulated RANKL-induced osteoclast differentiation by target Mafb protein. J Cell Biochem. (2018). doi: 10.1002/jcb.27968. [Epub ahead of print].
31. Cheng P, Chen C, He H-B, Hu R, Zhou H-D, Xie H, et al. miR-148a regulates osteoclastogenesis by targeting V-maf musculoaponeurotic fibrosarcoma oncogene homolog B. J Bone Miner Res. (2013) 28:1180–90. doi: 10.1002/jbmr.1845
32. Liu T, Qin A-P, Liao B, Shao H-G, Guo L-J, Xie G-Q, et al. A novel microRNA regulates osteoclast differentiation via targeting protein inhibitor of activated STAT3 (PIAS3). Bone. (2014) 67:156–65. doi: 10.1016/j.bone.2014.07.004
33. Kim K, Lee J, Kim JH, Jin HM, Zhou B, Lee SY, et al. Protein inhibitor of activated STAT 3 modulates osteoclastogenesis by down-regulation of NFATc1 and osteoclast-associated receptor. J Immunol. (2007) 178:5588–94. doi: 10.4049/jimmunol.178.9.5588
34. Hu C-H, Sui B-D, Du F-Y, Shuai Y, Zheng C-X, Zhao P, et al. miR-21 deficiency inhibits osteoclast function and prevents bone loss in mice. Sci Rep. (2017) 7:43191. doi: 10.1038/srep43191
35. Sugatani T, Vacher J, Hruska KA. A microRNA expression signature of osteoclastogenesis. Blood. (2011) 117:3648–57. doi: 10.1182/blood-2010-10-311415
36. Sugatani T, Hruska KA. Down-regulation of miR-21 biogenesis by estrogen action contributes to osteoclastic apoptosis. J Cell Biochem. (2013) 114:1217–22. doi: 10.1002/jcb.24471
37. Florczyk-Soluch U, Józefczuk E, Stȩpniewski J, Bukowska-Strakova K, Mendel M, Viscardi M, et al. Various roles of heme oxygenase-1 in response of bone marrow macrophages to RANKL and in the early stage of osteoclastogenesis. Sci Rep. (2018) 8:10797. doi: 10.1038/s41598-018-29122-1
38. Ke K, Sul O-J, Rajasekaran M, Choi H-S. MicroRNA-183 increases osteoclastogenesis by repressing heme oxygenase-1. Bone. (2015) 81:237–46. doi: 10.1016/j.bone.2015.07.006
39. Teitelbaum SL, Ross FP. Genetic regulation of osteoclast development and function. Nat Rev Genet. (2003) 4:638–49. doi: 10.1038/nrg1122
40. Sugatani T, Alvarez U, Hruska KA. PTEN regulates RANKL- and osteopontin-stimulated signal transduction during osteoclast differentiation and cell motility. J Biol Chem. (2003) 278:5001–8. doi: 10.1074/jbc.M209299200
41. Zhao C, Sun W, Zhang P, Ling S, Li Y, Zhao D, et al. miR-214 promotes osteoclastogenesis by targeting Pten/PI3k/Akt pathway. RNA Biol. (2015) 12:343–53. doi: 10.1080/15476286.2015.1017205
42. Penna E, Orso F, Taverna D. miR-214 as a key hub that controls cancer networks: small player, multiple functions. J Invest Dermatol. (2015) 135:960–9. doi: 10.1038/jid.2014.479
43. Xin R, Bai F, Feng Y, Jiu M, Liu X, Bai F, et al. MicroRNA-214 promotes peritoneal metastasis through regulating PTEN negatively in gastric cancer. Clin Res Hepatol Gastroenterol. (2016) 40:748–54. doi: 10.1016/j.clinre.2016.05.006
44. Liu J, Chen W, Zhang H, Liu T, Zhao L. miR-214 targets the PTEN-mediated PI3K/Akt signaling pathway and regulates cell proliferation and apoptosis in ovarian cancer. Oncol Lett. (2017) 14:5711–8. doi: 10.3892/ol.2017.6953
45. Wang S, Tang C, Zhang Q, Chen W. Reduced miR-9 and miR-181a expression down-regulates Bim concentration and promote osteoclasts survival. Int J Clin Exp Pathol. (2014) 7:2209–18.
46. Chen C, Cheng P, Xie H, Zhou H-D, Wu X-P, Liao E-Y, et al. MiR-503 regulates osteoclastogenesis via targeting RANK. J Bone Miner Res. (2014) 29:338–47. doi: 10.1002/jbmr.2032
47. Wang C, He H, Wang L, Jiang Y, Xu Y. Reduced miR-144-3p expression in serum and bone mediates osteoporosis pathogenesis by targeting RANK. Biochem Cell Biol. (2018) 96:627–35. doi: 10.1139/bcb-2017-0243
48. Guo L-J, Liao L, Yang L, Li Y, Jiang T-J. MiR-125a TNF receptor-associated factor 6 to inhibit osteoclastogenesis. Exp Cell Res. (2014) 321:142–52. doi: 10.1016/j.yexcr.2013.12.001
49. Taganov KD, Boldin MP, Chang K-J, Baltimore D. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc Natl Acad Sci USA. (2006) 103:12481–6. doi: 10.1073/pnas.0605298103
50. Nakasa T, Shibuya H, Nagata Y, Niimoto T, Ochi M. The inhibitory effect of microRNA-146a expression on bone destruction in collagen-induced arthritis. Arthritis Rheum. (2011) 63:1582–90. doi: 10.1002/art.30321
51. Ammari M, Presumey J, Ponsolles C, Roussignol G, Roubert C, Escriou V, et al. Delivery of miR-146a to Ly6C high monocytes inhibits pathogenic bone erosion in inflammatory arthritis. Theranostics. (2018) 8:5972–85. doi: 10.7150/thno.29313
52. Qu B, Xia X, Yan M, Gong K, Deng S, Huang G, et al. miR-218 is involved in the negative regulation of osteoclastogenesis and bone resorption by partial suppression of p38MAPK-c-Fos-NFATc1 signaling: potential role for osteopenic diseases. Exp Cell Res. (2015) 338:89–96. doi: 10.1016/j.yexcr.2015.07.023
53. Wang W, Yang L, Zhang D, Gao C, Wu J, Zhu Y, et al. MicroRNA-218 negatively regulates osteoclastogenic differentiation by repressing the nuclear factor-κB signaling pathway and targeting tumor necrosis factor receptor 1. Cell Physiol Biochem. (2018) 48:339–47. doi: 10.1159/000491740
54. Krzeszinski JY, Wei W, Huynh H, Jin Z, Wang X, Chang T-C, et al. miR-34a blocks osteoporosis and bone metastasis by inhibiting osteoclastogenesis and Tgif2. Nature. (2014) 512:431–5. doi: 10.1038/nature13375
55. Irie N, Takada Y, Watanabe Y, Matsuzaki Y, Naruse C, Asano M, et al. Bidirectional signaling through ephrinA2-EphA2 enhances osteoclastogenesis and suppresses osteoblastogenesis. J Biol Chem. (2009) 284:14637–44. doi: 10.1074/jbc.M807598200
56. Yang S, Zhang W, Cai M, Zhang Y, Jin F, Yan S, et al. Suppression of bone resorption by miR-141 in aged rhesus monkeys. J Bone Miner Res. (2018) 33:1799–812. doi: 10.1002/jbmr.3479
57. Omata Y, Yasui T, Hirose J, Izawa N, Imai Y, Matsumoto T, et al. Genomewide comprehensive analysis reveals critical cooperation between Smad and c-Fos in RANKL-induced osteoclastogenesis. J Bone Miner Res. (2015) 30:869–77. doi: 10.1002/jbmr.2418
58. Yu F, Xie C, Sun J, Peng W, Huang X. Overexpressed miR-145 inhibits osteoclastogenesis in RANKL-induced bone marrow-derived macrophages and ovariectomized mice by regulation of Smad3. Life Sci. (2018) 202:11–20. doi: 10.1016/j.lfs.2018.03.042
59. Fennen M, Pap T, Dankbar B. Smad-dependent mechanisms of inflammatory bone destruction. Arthritis Res Ther. (2016) 18:279. doi: 10.1186/s13075-016-1187-7
60. Zhao H, Zhang J, Shao H, Liu J, Jin M, Chen J, et al. Transforming growth factor β1/Smad4 signaling affects osteoclast differentiation via regulation of miR-155 expression. Mol Cells. (2017) 40:211–21. doi: 10.14348/molcells.2017.230
61. Zhang J, Zhao H, Chen J, Xia B, Jin Y, Wei W, et al. Interferon-β-induced miR-155 inhibits osteoclast differentiation by targeting SOCS1 and MITF. FEBS Lett. (2012) 586:3255–62. doi: 10.1016/j.febslet.2012.06.047
62. Su L-C, Huang A-F, Jia H, Liu Y, Xu W-D. Role of microRNA-155 in rheumatoid arthritis. Int J Rheum Dis. (2017) 20:1631–7. doi: 10.1111/1756-185X.13202
63. Sul O-J, Sung Y-B, Rajasekaran M, Ke K, Yu R, Back S-H, et al. MicroRNA-155 induces autophagy in osteoclasts by targeting transforming growth factor β-activated kinase 1-binding protein 2 upon lipopolysaccharide stimulation. Bone. (2018) 116:279–89. doi: 10.1016/j.bone.2018.08.014
64. Lu S-Y, Li M, Lin Y-L. Mitf regulates osteoclastogenesis by modulating NFATc1 activity. Exp Cell Res. (2014) 328:32–43. doi: 10.1016/j.yexcr.2014.08.018
65. Zhao H, Zhang J, Shao H, Liu J, Jin M, Chen J, et al. miRNA-340 inhibits osteoclast differentiation via repression of MITF. Biosci Rep. (2017) 37:BSR20170302. doi: 10.1042/BSR20170302
66. Kim JH, Kim N. Regulation of NFATc1 in osteoclast differentiation. J Bone Metab. (2014) 21:233–41. doi: 10.11005/jbm.2014.21.4.233
67. Lee Y, Kim HJ, Park CK, Kim Y-G, Lee H-J, Kim J-Y, et al. MicroRNA-124 regulates osteoclast differentiation. Bone. (2013) 56:383–9. doi: 10.1016/j.bone.2013.07.007
68. Nakamachi Y, Ohnuma K, Uto K, Noguchi Y, Saegusa J, Kawano S. MicroRNA-124 inhibits the progression of adjuvant-induced arthritis in rats. Ann Rheum Dis. (2016) 75:601–8. doi: 10.1136/annrheumdis-2014-206417
69. Tang L, Yin Y, Liu J, Li Z, Lu X. MiR-124 attenuates osteoclastogenic differentiation of bone marrow monocytes via targeting Rab27a. Cell Physiol Biochem. (2017) 43:1663–72. doi: 10.1159/000484027
70. Mizoguchi F, Murakami Y, Saito T, Miyasaka N, Kohsaka H. miR-31 controls osteoclast formation and bone resorption by targeting RhoA. Arthritis Res Ther. (2013) 15:R102. doi: 10.1186/ar4282
71. Strzelecka-Kiliszek A, Mebarek S, Roszkowska M, Buchet R, Magne D, Pikula S. Functions of Rho family of small GTPases and Rho-associated coiled-coil kinases in bone cells during differentiation and mineralization. Biochim Biophys Acta Gen Subj. (2017) 1861:1009–23. doi: 10.1016/j.bbagen.2017.02.005
72. Chellaiah MA, Soga N, Swanson S, McAllister S, Alvarez U, Wang D, et al. Rho-A is critical for osteoclast podosome organization, motility, and bone resorption. J Biol Chem. (2000) 275:11993–2002. doi: 10.1074/jbc.275.16.11993
73. Georgess D, Machuca-Gayet I, Blangy A, Jurdic P. Podosome organization drives osteoclast-mediated bone resorption. Cell Adh Migr. (2014) 8:191–204. doi: 10.4161/cam.27840
74. Cong F, Wu N, Tian X, Fan J, Liu J, Song T, et al. MicroRNA-34c promotes osteoclast differentiation through targeting LGR4. Gene. (2017) 610:1–8. doi: 10.1016/j.gene.2017.01.028
75. Luo J, Yang Z, Ma Y, Yue Z, Lin H, Qu G, et al. LGR4 is a receptor for RANKL and negatively regulates osteoclast differentiation and bone resorption. Nat Med. (2016) 22:539–46. doi: 10.1038/nm.4076
76. Heasman SJ, Ridley AJ. Mammalian Rho GTPases: new insights into their functions from in vivo studies. Nat Rev Mol Cell Biol. (2008) 9:690–701. doi: 10.1038/nrm2476
77. Rossi M, Pitari MR, Amodio N, Di Martino MT, Conforti F, Leone E, et al. miR-29b negatively regulates human osteoclastic cell differentiation and function: implications for the treatment of multiple myeloma-related bone disease. J Cell Physiol. (2013) 228:1506–15. doi: 10.1002/jcp.24306
78. Shimada-Sugawara M, Sakai E, Okamoto K, Fukuda M, Izumi T, Yoshida N, Tsukuba T. Rab27A regulates transport of cell surface receptors modulating multinucleation and lysosome-related organelles in osteoclasts. Sci Rep. (2015) 5:9620. doi: 10.1038/srep09620
79. Dou C, Zhang C, Kang F, Yang X, Jiang H, Bai Y, et al. MiR-7b directly targets DC-STAMP causing suppression of NFATc1 and c-Fos signaling during osteoclast fusion and differentiation. Biochim Biophys Acta. (2014) 1839:1084–96. doi: 10.1016/j.bbagrm.2014.08.002
80. Yin Y, Tang L, Chen J, Lu X. MiR-30a attenuates osteoclastogenesis via targeting DC-STAMP-c-Fos-NFATc1 signaling. Am J Transl Res. (2017) 9:5743–53.
81. Dou C, Ding N, Luo F, Hou T, Cao Z, Bai Y, et al. Graphene-based MicroRNA transfection blocks preosteoclast fusion to increase bone formation and vascularization. Adv Sci. (2018) 5:1700578. doi: 10.1002/advs.201700578
82. Kim K, Kim JH, Kim I, Lee J, Seong S, Park Y-W, et al. MicroRNA-26a regulates RANKL-induced osteoclast formation. Mol Cells. (2015) 38:75–80. doi: 10.14348/molcells.2015.2241
83. Fordham JB, Guilfoyle K, Naqvi AR, Nares S. MiR-142-3p is a RANKL-dependent inducer of cell death in osteoclasts. Sci Rep. (2016) 6:24980. doi: 10.1038/srep24980
84. Ma Y, Yang H, Huang J. Icariin ameliorates dexamethasone-induced bone deterioration in an experimental mouse model via activation of microRNA-186 inhibition of cathepsin K. Mol Med Rep. (2018) 17:1633–41. doi: 10.3892/mmr.2017.8065
85. Madel M-B, Ibáñez L, Rouleau M, Wakkach A, Blin-Wakkach C. A novel reliable and efficient procedure for purification of mature osteoclasts allowing functional assays in mouse cells. Front Immunol. (2018) 9:2567. doi: 10.3389/fimmu.2018.02567
86. Liu M, Sun Y, Zhang Q. Emerging role of extracellular vesicles in bone remodeling. J Dent Res. (2018) 97:859–68. doi: 10.1177/0022034518764411
87. Xie Y, Chen Y, Zhang L, Ge W, Tang P. The roles of bone-derived exosomes and exosomal microRNAs in regulating bone remodelling. J Cell Mol Med. (2017) 21:1033–41. doi: 10.1111/jcmm.13039
88. Yuan F-L, Wu Q-Y, Miao Z-N, Xu M-H, Xu R-S, Jiang D-L, et al. Osteoclast-derived extracellular vesicles: novel regulators of osteoclastogenesis and osteoclast-osteoblasts communication in bone remodeling. Front Physiol. (2018) 9:628. doi: 10.3389/fphys.2018.00628
89. Tao S-C, Guo S-C. Extracellular vesicles in bone: “dogrobbers” in the “eternal battle field.” Cell Commun Signal. (2019) 17:6. doi: 10.1186/s12964-019-0319-5
90. Mestdagh P, Hartmann N, Baeriswyl L, Andreasen D, Bernard N, Chen C, et al. Evaluation of quantitative miRNA expression platforms in the microRNA quality control (miRQC) study. Nat Methods. (2014) 11:809–15. doi: 10.1038/nmeth.3014
91. Ishihara R, Hasegawa K, Hosokawa K, Maeda M. Multiplex MicroRNA detection on a power-free microfluidic chip with laminar flow-assisted dendritic amplification. Anal Sci. (2015) 31:573–6. doi: 10.2116/analsci.31.573
92. Wu X, Zhu S, Huang P, Chen Y. Highly specific quantification of microRNA by coupling probe-rolling circle amplification and Förster resonance energy transfer. Anal Biochem. (2016) 502:16–23. doi: 10.1016/j.ab.2016.03.001
93. Dard-Dascot C, Naquin D, d'Aubenton-Carafa Y, Alix K, Thermes C, van Dijk E. Systematic comparison of small RNA library preparation protocols for next-generation sequencing. BMC Genomics. (2018) 19:118. doi: 10.1186/s12864-018-4491-6
94. Rice J, Roberts H, Burton J, Pan J, States V, Rai SN, et al. Assay reproducibility in clinical studies of plasma miRNA. PLoS ONE. (2015) 10:0121948. doi: 10.1371/journal.pone.0121948
95. Yeri A, Courtright A, Danielson K, Hutchins E, Alsop E, Carlson E, et al. Evaluation of commercially available small RNASeq library preparation kits using low input RNA. BMC Genomics. (2018) 19:331. doi: 10.1186/s12864-018-4726-6
96. Songia P, Chiesa M, Valerio V, Moschetta D, Myasoedova VA, D'Alessandra Y, et al. Direct screening of plasma circulating microRNAs. RNA Biol. (2018) 15:1268–72. doi: 10.1080/15476286.2018.1526538
97. Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, et al. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem. (2009) 55:611–22. doi: 10.1373/clinchem.2008.112797
98. Deng MC. The AlloMap™ genomic biomarker story: 10 years after. Clin Transplant. (2017) 31:e12900. doi: 10.1111/ctr.12900
99. Haider BA, Baras AS, McCall MN, Hertel JA, Cornish TC, Halushka MK. A critical evaluation of microRNA biomarkers in non-neoplastic disease. PLoS ONE. (2014) 9:e89565. doi: 10.1371/journal.pone.0089565
100. Peltier HJ, Latham GJ. Normalization of microRNA expression levels in quantitative RT-PCR assays: identification of suitable reference RNA targets in normal and cancerous human solid tissues. RNA. (2008) 14:844–52. doi: 10.1261/rna.939908
101. Marabita F, de Candia P, Torri A, Tegnér J, Abrignani S, Rossi RL. Normalization of circulating microRNA expression data obtained by quantitative real-time RT-PCR. Brief Bioinform. (2016) 17:204–12. doi: 10.1093/bib/bbv056
102. Song J, Bai Z, Han W, Zhang J, Meng H, Bi J, et al. Identification of suitable reference genes for qPCR analysis of serum microRNA in gastric cancer patients. Dig Dis Sci. (2012) 57:897–904. doi: 10.1007/s10620-011-1981-7
103. Hu Z, Dong J, Wang L-E, Ma H, Liu J, Zhao Y, et al. Serum microRNA profiling and breast cancer risk: the use of miR-484/191 as endogenous controls. Carcinogenesis. (2012) 33:828–34. doi: 10.1093/carcin/bgs030
104. Zhang J, Song Y, Zhang C, Zhi X, Fu H, Ma Y, et al. Circulating MiR-16-5p and MiR-19b-3p as two novel potential biomarkers to indicate progression of gastric cancer. Theranostics. (2015) 5:733–45. doi: 10.7150/thno.10305
105. Luo H, Li X, Li T, Zhao L, He J, Zha L, et al. Exosomes/microvesicles microRNA-423-3p derived from cardiac fibroblasts mediates the cardioprotective effects of ischemic postconditioning. Cardiovasc Res. (2018) doi: 10.1093/cvr/cvy231. [Epub ahead of print].
Keywords: microRNA, osteoclast, differentiation, regulation, detection, biomarker
Citation: Lozano C, Duroux-Richard I, Firat H, Schordan E and Apparailly F (2019) MicroRNAs: Key Regulators to Understand Osteoclast Differentiation? Front. Immunol. 10:375. doi: 10.3389/fimmu.2019.00375
Received: 08 December 2018; Accepted: 14 February 2019;
Published: 07 March 2019.
Edited by:
Teun J. De Vries, VU University Amsterdam, NetherlandsReviewed by:
Toshio Kukita, Kyushu University, JapanFrédéric Velard, Université de Reims Champagne-Ardenne, France
Jeroen Van De Peppel, Erasmus University Rotterdam, Netherlands
Copyright © 2019 Lozano, Duroux-Richard, Firat, Schordan and Apparailly. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Florence Apparailly, florence.apparailly@inserm.fr
 Claire Lozano
Claire Lozano Isabelle Duroux-Richard
Isabelle Duroux-Richard Hüseyin Firat3
Hüseyin Firat3 Eric Schordan
Eric Schordan Florence Apparailly
Florence Apparailly