- Department of Healthcare Biotechnology, Neurobiology Research Laboratory, Atta-Ur-Rahman School of Applied Biosciences, National University of Sciences and Technology, Islamabad, Pakistan
Background: In recent years, microRNAs (miRNAs) have emerged as key players in the pathophysiology of multiple diseases including Alzheimer’s disease (AD). Messenger RNA (mRNA) targeting for regulation of gene expression by miRNAs has been implicated in the annotation of disease pathophysiology as well as in the explication of their starring role in contemporary therapeutic interventions. One such miRNA is miR-153 which mediates the survival of cortical neurons and inhibits plaque formation. However, the core mRNA targets of miR-153 have not been fully illustrated.
Objective: The present study aimed to elucidate the potential involvement of miR-153 in AD pathogenesis and to reveal its downstream targets.
Methods: miRanda was used to identify AD-associated targets of miR-153. TargetScan, PicTar, miRmap, and miRDB were further used to validate these targets. STRING 12 was employed to assess the protein-protein interaction network while Gene ontology (GO) analysis was carried out to identify the molecular functions exhibited by these gene targets.
Results: In silico analysis using miRanda predicted five important AD-related targets of miR-153, including APP, SORL1, PICALM, USF1, and PSEN1. All five target genes are negatively regulated by miR-153 and are substantially involved in AD pathogenesis. A protein interaction network using STRING 12 uncovered 30 potential interacting partners for SORL1, PICALM, and USF1. GO analysis revealed that miR-153 target genes play a critical role in neuronal survival, differentiation, exon guidance, amyloid precursor protein processing, and synapse formation.
Conclusion: These findings unravel the potential role of miR-153 in the pathogenesis of AD and provide the basis for forthcoming experimental studies.
1 Introduction
Alzheimer’s disease (AD) is a neurodegenerative disorder characterized by the formation of neurofibrillary tangles (NFTs) and Amyloid-beta (Aβ) plaques in sub-cortical brain regions that eventually lead to cognitive impairment (Amber et al., 2020). Various genetic, epigenetic, and environmental factors contribute to the development of AD therefore the identification of informative biomarkers remained a significant challenge. Since the last decade, epigenetic mechanisms gained widespread prominence as the regulators of various important biological processes, and central to these processes are microribonucleic acids (miRNAs) (Filipowicz et al., 2008). miRNAs belong to the class of small non-coding RNAs that modulate gene expression post-transcriptionally either by target mRNA degradation or translational inhibition (Pu et al., 2019). miRNA:mRNA duplex formation necessitates the complementarity between eight nucleotide seed regions within both sequences. The duplex is either directed toward polyribosomes to regulate the mRNA translational process or targeted to the P-bodies for storage/degradation (Filipowicz et al., 2008). miRNAs are known to control the expression of almost 60% of protein-coding genes, therefore, these are considered important biomarkers for early diagnosis of various disorders. Their potential as potent biomarkers can be derived from unique secretory properties as they regulate the expression of multiple genes in various cell types without cell-to-cell contact (Schwarzenbach et al., 2014). Apart from their presence in tissues, miRNAs are also secreted in extracellular fluids, blood plasma, and saliva and therefore can serve as potential non-invasive markers for disease diagnosis (François et al., 2019). The preliminary evidence about the involvement of miRNAs in human diseases originated from cancer studies. Various expression profiling studies revealed the abnormal expression of different miRNAs in cancer samples as compared to the control (Calin et al., 2002).
The miRNAs that were consistently found to be deregulated in AD include; miR-9, miR-29, miR-34, miR-107, miR-181, miR-186, miR-146a, miR-155 and miR-153 (Femminella et al., 2015). The miR-153 is implicated in various diseases such as hypertension, osteosarcoma, glioblastoma, and various other cancers. miR-153 contributes toward the hypertensive state via the downregulation of KCNQ4 (Carr et al., 2016). An increase in miR-153 expression elevated neurogenesis and improved cognition (Qiao et al., 2020). Moreover, a significant reduction in the expression levels of miR-153 is also observed in early, moderate, and severe AD cases as compared to age-matched control specimens. Additionally, an inverse correlation was observed between miR-153 and Aβ plaque burden making it a potential disease biomarker and novel drug target (Long et al., 2012). Ectopic expression of miR-153-3p induced inflammation by increasing the release of IL-1β, TNF-α, and IL-6 and decreased neural stem cell differentiation via regulating GPR55 expression (Dong et al., 2023). Increased expression of miR-153 disrupted synapsin 1 in the hippocampus and impaired glutamatergic vesicle transport thus causing chronic cerebral hypoperfusion in rats (Zhang et al., 2020).
Due to the substantial role of miR-153 in neuronal disorders including AD, it is vital to identify the molecular targets associated with this very same miRNA to elucidate the underlying mechanisms leading to the disease phenotype. The data regarding the regulatory and therapeutic role of miRNAs is scarce due to the limitations of current experimental procedures (Jaberi et al., 2024). Owing to the significance of miRNAs in disease-related processes the pace of miRNA target prediction needs to be improved. Various in silico algorithms are available to reveal the molecular targets of a large proportion of miRNAs with relative sensitivity and specificity (Hamzeiy et al., 2014). Therefore, this study aimed to investigate the important AD-related mRNA targets of miR-153 to improve the current understanding of disease at the molecular level. AD-associated mRNA targets of miR-153 are identified via the miRanda algorithm and results are cross-validated by four other publicly available algorithms, TargetScan, PicTar, miRmap, and miRDB.
2 Methods
2.1 Targets prediction of miR-153
Web-based bioinformatic algorithm miRanda (Oliveira et al., 2017) was assessed to predict the mRNA targets of miR-153 and the mirSVR scores were assigned to each predicted target site. The sequence of miR-153 is available in the NCBI database (>LM608503.1 TPA: Homo sapiens microRNA hsa-mir-153precursor CTCACAGCTGCCAGTGTCATTTTTGTGATCTGCAGCTAGTATTCTCACTCCAGTTGCATAGTCACAAAAG TGATCATTGGCAGGTGTGGC).
The miRanda algorithm is developed for the prediction of mRNA targets and expression profiles of miRNAs available at MicroRNA.org (http://www.microrna.org); while mirSVR score is a regression model that reveals contextual features and sequence of the predicted miRNA:mRNA duplex and is directly correlated to the downregulation of miRNA and target sites of interest. Homo sapiens was selected as a species of choice and all the search was performed using default parameters (MFE threshold: −20 kcal/mol, scaling parameter: 4∙00, score threshold: 140.00, gap open and extend penalty: −4∙000 and −9.000 respectively).
2.2 Validation of results by different algorithms
The mRNA targets obtained from miRanda were further validated by four other publicly available algorithms, i.e., TargetScan, PicTar, miRDB, and MiRmap. In the TargetScan database, (Release 8, http://www.targetscan.org/), humans were selected as the species of choice. Furthermore, there were two options to find the target, i.e., by entering the gene name or miRNA name. The miRNA-153 was entered as a query and it gave two options such as miR-153–3p and miR-153–5p. Both options were explored for the target genes (Huang et al., 2020).
In the PicTar database, “PicTar target prediction in vertebrates” was selected. Following that, vertebrates was chosen as a species and then, miR-153 was selected from the dropdown menu. (http://pictar.mdc-berlin.de/) (Xue et al., 2020). In miRmap, human was selected as a species and then miR-153 was selected from the dropdown menu (https://mirmap.ezlab.org/) (Vejnar and Zdobnov, 2012).
In miRDB, humans were selected as the species of choice. Furthermore, there were two options to find the target, i.e., by entering the gene name or miRNA name. The miRNA-153 was entered as a query and it gave two selections such as miR-153–3p and miR-153–5p. Both options were explored for the target genes (http://mirdb.org/miRDB/) (Wong and Wang, 2015).
2.3 Protein association, functional enrichment, and post-translational modification analysis
Targets predicted by miRanda were submitted to STRING v.12 (Szklarczyk et al., 2017) (http://string-db.org/) database to explore the functional association networks of target proteins using UniProt accession numbers. Homo sapiens was selected from the given list of species. Biological processes, cellular localization, molecular functions, and miRNA targets of the specific miR-153 affected proteins were investigated by GO analysis and microRNA target analysis using the WEB-based Gene SeTAnaLysis Toolkit (WEBGESTALT) (Wang et al., 2017). Swiss-Prot accession numbers of miR-153 target proteins were employed for enrichment analysis.
The phosphorylation modification sites were predicted for the identified target proteins, using NetPhos 3.1 server (www.cbs.dtu.dk/services/NetPhos-3.1) (Arshad et al., 2018) while S-nitrosylation, and N and O glycosylation sites were predicted using GPS-SNO http://sno.biocuckoo.org (Mazina et al., 2023), NetNGlyc 1.0 www.cbs.dtu.dk/services/NetNGlyc/(Azevedo et al., 2018), and NetOGlyc 4.0 www.cbs.dtu.dk/services/NetOGlyc/(Kwan et al., 2021), respectively. Default settings were used for the analysis of posttranslational modification (PTM) sites and the predictions having output scores above 0.5 were only selected to avoid the possibility of false positive results. The FASTA sequence of the targeted proteins was acquired from the NCBI protein database (https://www.ncbi.nlm.nih.gov/pubmed/).
3 Results
3.1 Targets prediction of miR-153
miRanda algorithm returned 5,810 targets for miR-153 which were further screened to identify the targets involved in AD pathophysiology using a literature search. Five of the 5,810 targets found to be most relevant with AD include; sortilin-related receptor 1 (SORL1), amyloid precursor protein (APP), phosphatidylinositol binding clathrin assembly protein (PICALM), upstream stimulatory factor 1 (USF1) and presenilin-1 (PSEN1). miSVR scores indicated that miR-153 downregulates all the target genes. The results were cross-validated by four different freely accessible software TargetScan, PicTar, miRmap, and miRDB. It is observed that all five targets were not predicted by all the software (Table 1). APP and PSEN1 are already reported to be affected by miR-153 so we used SORL1, PICALM, and USF1 for further analysis.
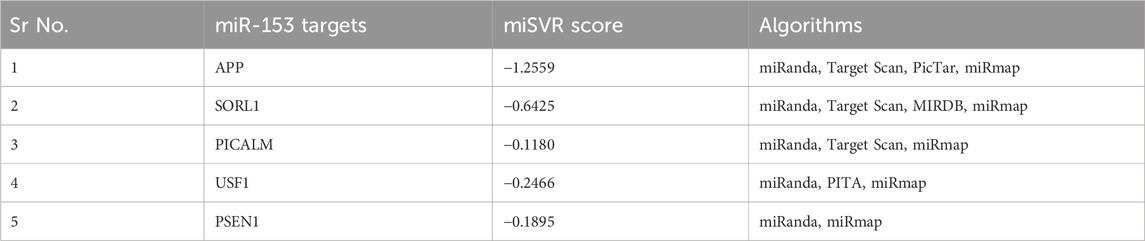
TABLE 1. miR-153 targets and their miSVR scores predicted by miRanda and validated by different software.
3.2 Protein association network and functional analysis
STRING 12 analysis exhibited a strong association (score >0.7) of miR-153 target proteins with various other proteins, i.e., SORL1 exhibited strong interaction with GGA1, GGA2, APOE, ABCA7, CLU, APP, VPS35, VPS26A, LRPAP1, and NTS; PICALM is strongly associated with CLINT1, AP2A1, EPS15, RPS27A, CLTC, EPN2, EPN3, UBA52, UBB and UBC. Similarly, USF1 also showed significant interaction with ten different proteins such as SP1, ESR1, SMARCD3, EP300, FOSL1, USF2, MED1, RFX5, TAF7, and GTF2I (Figure 1). The functions and complete names of all the interacting partners are listed in Table 2.
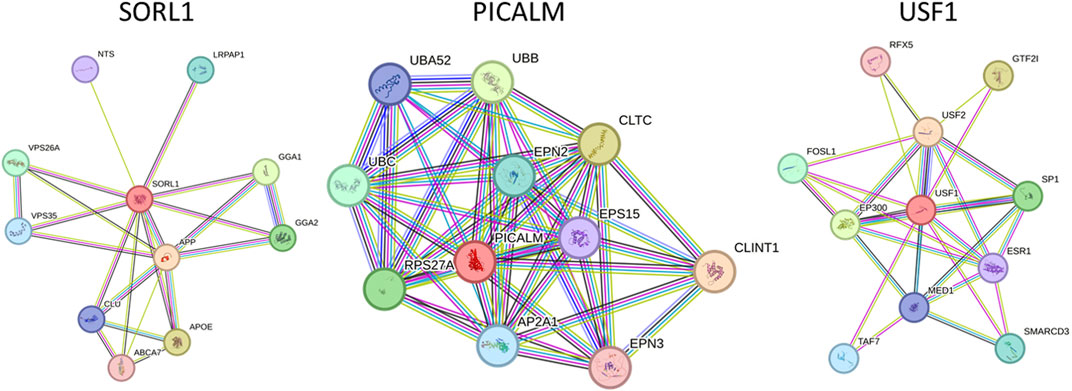
FIGURE 1. Functional association network of miR-153 target proteins. High-confidence protein-protein interaction network of identified proteins derived from the STRING database. Each protein is represented as a node with edged interactions.
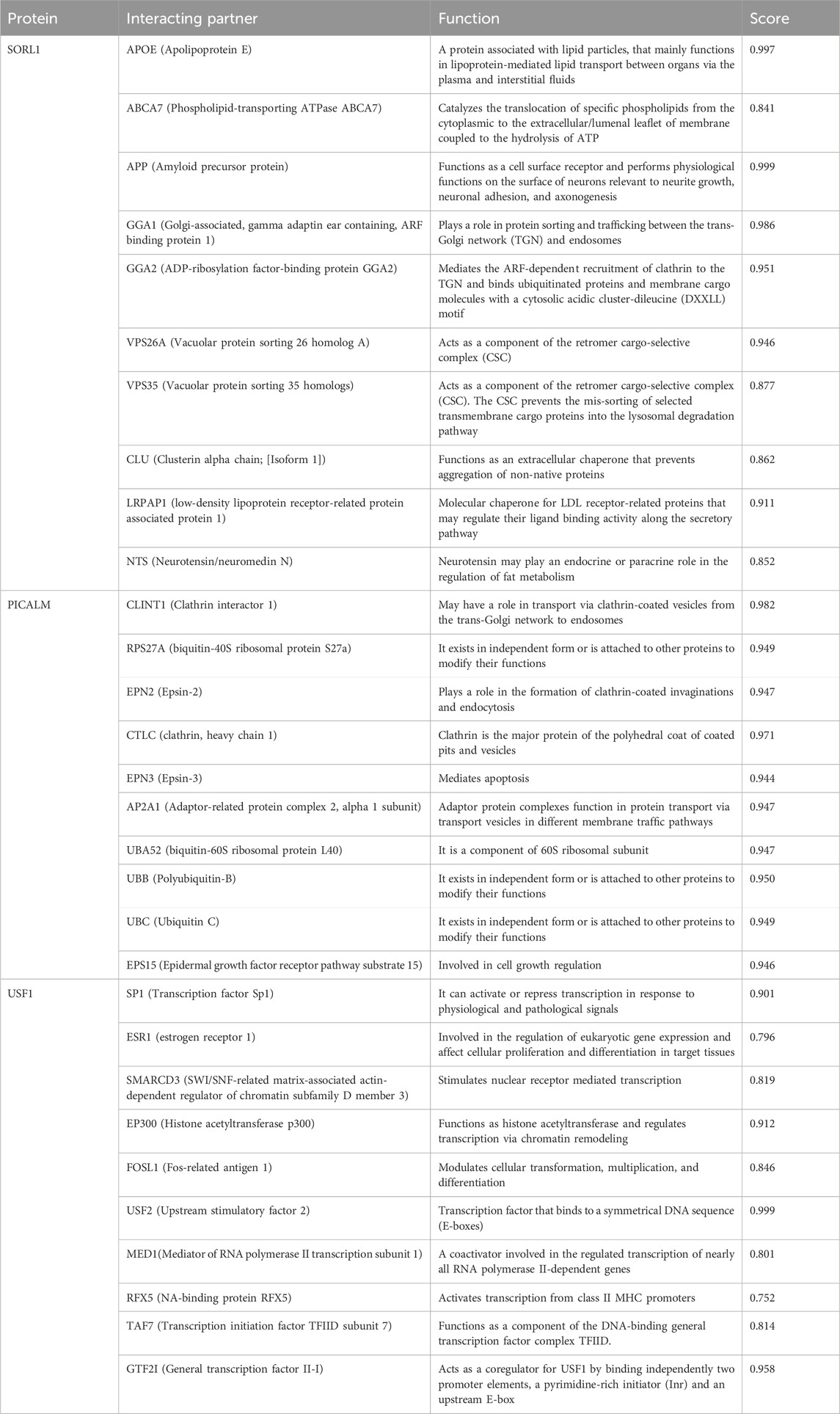
TABLE 2. Functional association of SORL1, PICALM, and USF1 along with interacting partner derived from the STRING database.
3.3 Functional enrichment and plausible port translational modifications analysis
The identified miR-153 target proteins were functionally annotated using WEBGESTALT and Uniprot (www.uniprot.org/). Target proteins were classified based on molecular function, biological process, and cellular localization (Table 3). All three proteins are actively involved in different biological processes. SORL1 is a neuronal apolipoprotein E receptor and its gene is predominantly expressed in the central nervous system (CNS) and is involved in beta-amyloid binding, vesicle-mediated transport, cholesterol metabolic process, negative regulation of neurogenesis, and various other important cellular pathways. PICALM plays an important role in clathrin-mediated endocytosis, vesicle-mediated transport, axonogenesis, neuron projection development, neuronal differentiation, dendrite development, and many other different processes. USF1 acts as a transcription factor and belongs to the basic helix-loop-helix leucine zipper family which is known to regulate the macromolecule metabolic process, cellular metabolic process, coagulation, hypoxia, glucose homeostasis, fibrinolysis, and nutrient levels.
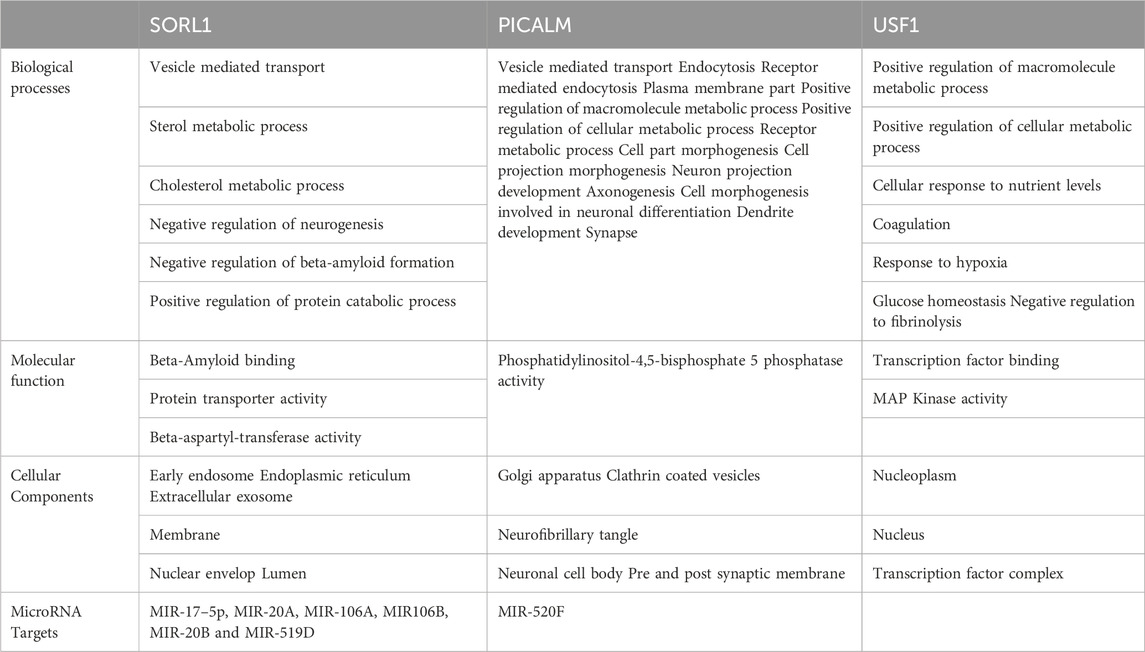
TABLE 3. Functional distribution of SORL1, PICALM and USF1 on the basis of biological process, molecular function and cellular compartment.
A total of 49 serine, tyrosine, and threonine sites were predicted as plausible phosphorylation sites for USF1, 29 for PICALM, and 368 sites for SORL1. The S-nitrosylation prediction analysis revealed 2 cysteine residue sites at positions 229 and 248 for USF1, while 3 and 4 sites were predicted for PICALM and SORL1, respectively. The cysteine modifications for PICALM were predicted at positions 27, 48, and 230 whereas positions 942, 1,042, 1,502, and 1,593 of SORL1 are susceptible to cysteine modifications. The N and O glycosylation analysis for the target proteins also showed significant susceptibility for these PTMs. A total of 6 sites were predicted as plausible sites for N-glycosylation in the PICALM sequence at positions 69, 105, 384, 445, 505, and 513. The O-glycosylation was also predicted for 58 sites of PICALM For USF1, 43 sites were predicted for O-glycosylation while no plausible sites were identified for N-glycosylation. The SORL1 has 28 predicted sites for N-glycosylation while 314 sites were found to be susceptible to O-glycosylation (Supplementary Data).
4 Discussion
By regulating the expression of target genes, miRNAs mediate various biological processes. Different miRNAs are reported to associate with AD, however, miR-153 plays a crucial role in regulating the expression of amyloid precursor protein (APP). Its expression is significantly downregulated in early and late-stage AD as observed in the APPswe/PSΔE9 murine model (Liang et al., 2012). SNHG1-mediated suppression of miR-153 increases neurotoxicity in SH-SY5Y cells (Zhao et al., 2020). Inversely, increased expression of miR-153 protects the neurons from cellular death via the upregulation of PRX5 (Xu et al., 2019). Similarly, miR-153–3p reduces LPS-induced neuroinflammation and subsequently cell death by inhibiting the NF-κB signaling pathway (Choi et al., 2022).
miR-153 obstructs APP production in neurons therefore its deregulation may drive over-expression of APP and subsequently leads to AD progression. Apart from APP miR-153 also reduced the expression of APLP2, an (APP homolog), in human fetal brain cultures therefore, it was hypothesized that it may target some of the other critical genes linked to neurodegeneration and AD development (Long et al., 2012). In this study, five main culprits of AD pathogenesis were found to be negatively regulated by miR-153 that include; APP, SORL1, PICALM, USF1, and PSEN1. The relationship between miR-153 and APP expression is well established while PSEN1 is predicted by just one algorithm hence we primarily focused on SORL1, PICALM, and USF1.
Apart from the direct role of these genes in AD, the complex interaction with various important disease-promoting/alleviating entities is revealed by the STRING database. The interaction network exhibits that complex multi-dimensional regulation takes place between key AD players, such as APP, SORL1, PICALM, USF1, PSEN1, and other disease-causing agents. The predicted genes/proteins are significant to neuroprotection, synapse formation, memory and learning, intellectual abilities, and neurodegeneration (Chandrasekaran and Bonchev, 2016). Neuronal sortilin receptor-related gene (SORL1) mediates the intracellular trafficking of APP and dysregulation of the particular process leads to Aβ accumulation and subsequently neuronal apoptosis. The exact underlying mechanisms determining the influence of SORL1 on APP trafficking and export are not explicitly studied therefore opening new avenues to investigate AD from a different perspective (Lee et al., 2008). SORL1 exhibited strong interaction with various proteins, such as GGA1, GGA2, APOE, ABCA7, CLU, APP, VPS35, VPS26A, LRPAP1, and NTS. Apolipoprotein E (APOE) modulates lipid metabolism and is implicated in AD pathogenesis. Lower levels of APOE are linked with a decline in cognitive abilities. Genetic variations in the APOE region alter the plasma expression levels of this gene and increase the risk for AD (Aslam et al., 2023). APOE ε4 allele leads to poor cognitive abilities and increased amyloid beta burden in the brain. Moreover, it alters the microglial immune response by downregulating innate immunity (lysosomal and complement pathways) and inducing stress-like responses (Liu et al., 2023). Apolipoproteins mediate cholesterol metabolism mainly via ABCA1 (ATP-binding cassette transporter A1) (Chen et al., 2013). ABCA1 is widely present in neurons and astrocytes and maintains cholesterol homeostasis in the brain. A recent study reported that amyloid beta-mediated dysfunctional ABCA1 in astrocytes altered the transport of cholesterol from astrocytes to the neurons. It subsequently led to impairment of cholesterol metabolism, a prominent feature of AD pathogenesis (Azizidoost et al., 2022). Clusterin (CLU) plays a protective role in the brain however, mutations in CLU increase the risk of developing AD. The rs11136000C mutation in CLU causes dysregulation in GABAergic signaling thus promoting AD pathogenesis (Chen et al., 2023).
Phosphatidylinositol-binding clathrin assembly protein (PICALM) is associated with clathrin-mediated endocytosis (Kyriazis et al., 2008). It is predominantly situated in neurons, oligodendrocytes, astrocytes, and endothelial cells where it recruits the adaptor protein 2 (AP-2) and clathrin to the plasma membrane to encapsulate the target proteins (Yao et al., 2003). The clathrin-coated vesicles are further processed in endosomes or lysosomes to be removed from the cell. PICALM is also associated with the removal of Aβ from the cells, therefore, minimizing the plaque burden and preventing AD pathology. Altered PICALM expression levels are reported in AD brain tissues however, it is yet to be determined whether it affects the Aβ transport or is influenced by Aβ levels (Baig et al., 2010). PICALM is strongly associated with various other proteins and alterations in its expression may influence the biological activities of target proteins correspondingly. The interacting partners of PICALM include; CLINT1, AP2A1, EPS15, RPS27A, CLTC, EPN2, EPN3, UBA52, UBB and UBC. RPS27A is a fusion protein consisting of ubiquitin and S27a (ribosomal protein) (Sayers et al., 2018). An in silico analysis revealed the potential role of RPS27A in neurodegenerative disorders by modulating the expression of Il-18 and Cx3cl1 (Khayer et al., 2020). The role of other target proteins is still unclear in AD and needs further research.
Upstream transcription factor 1 (USF1), a ubiquitously expressed gene encodes a transcription factor that stimulates the transcription of various lipid and glucose-metabolizing genes (Lee et al., 2006) including APOE (Salero et al., 2003). USF1 plays a significant role in abnormal lipid aggregation (Guo et al., 2018), neuronal differentiation, and synaptic plasticity, moreover activates the APP promoter thereby affecting Aβ production and processing (Isotalo et al., 2012). USF1 strongly interacts with various other proteins such as SP1, ESR1, SMARCD3, EP300, FOSL1, USF2, MED1, RFX5, TAF7, and GTF2I. ESR1 (Estrogen receptor 1) is implicated in AD progression and it is described that ESR1 mutant (rs9340803) may lead to AD by perturbing cholesterol metabolism and accumulating amyloid beta in the brain. Nevertheless, further studies on larger cohorts are required to confirm the role of the ESR1 variant in AD (Li et al., 2018).
The post-translational modification data for the target proteins revealed a significant number of predicted sites with susceptibility towards phosphorylation, S-nitrosylation, and N and O-glycosylation. There is ample evidence that PTMs play a crucial role in AD pathology (Marcelli et al., 2018). Phosphorylation of tau and amyloid beta is detected in AD mouse models and these modifications affect the functions of microtubules and synapses, respectively (Wang et al., 2023).
Identification and validation of these predicted PTM sites and their pathological correlation with miR-153 targets will also provide substantial data that will be helpful in further elucidation of molecular mechanisms involved in AD pathology.
In this study, bioinformatics analysis predicted some of the important AD-related targets of miR-153. The gene ontology (GO) analysis of putative miR-153 targets revealed their important functions relevant to AD such as regulation of Aβ formation, negative regulation of neurogenesis, neuronal projection development, synapse formation, and NFTs formation. miRNAs perform their regulatory functions by affecting the target genes therefore it is crucial to study the potential targets and their underlying effects. This approach will facilitate the identification of novel regulatory networks of various miRNAs in different disease-related processes.
5 Conclusion
Our findings may aid the understanding of different molecular mechanisms and identification of effective therapeutic targets for AD. Further experimental studies may provide additional insights into the regulatory role of miR-153 and its targets in the development of AD and other neurodegenerative disorders.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author contributions
SA: Conceptualization, Data curation, Formal Analysis, Investigation, Methodology, Writing–original draft. SZ: Project administration, Resources, Supervision, Validation, Writing–review and editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. The authors are highly obliged to the research facilities of Atta-ur-Rahman School of Applied Biosciences (ASAB), National University of Sciences and Technology (NUST).
Acknowledgments
Authors are grateful to Sadia Nazir, PhD scholar, ASAB-NUST for her help in database search.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmolb.2023.1325588/full#supplementary-material
References
Amber, S., Mirza, F. J., Asif, M., Hassan, D., Ahmed, T., Zahid, S., et al. (2020). Amyloid-beta induced neurotoxicity impairs cognition and adult hippocampal neurogenesis in a mouse model for Alzheimer’s disease. Curr. Alzheimer Res. 17 (11), 1033–1042. doi:10.2174/1567205017666201224162730
Arshad, M., Bhatti, A., and John, P. (2018). Identification and in silico analysis of functional SNPs of human TAGAP protein: a comprehensive study. PloS one 13 (1), e0188143. doi:10.1371/journal.pone.0188143
Aslam, M. M., Fan, K. H., Lawrence, E., Bedison, M. A., Snitz, B. E., DeKosky, S. T., et al. (2023). Genome-wide analysis identifies novel loci influencing plasma apolipoprotein E concentration and Alzheimer’s disease risk. Mol. Psychiatry 5, 1–2. doi:10.1038/s41380-023-02170-4
Azevedo, R., Silva, A. M. N., Reis, C. A., Santos, L. L., and Ferreira, J. A. (2018). In silico approaches for unveiling novel glycobiomarkers in cancer. J. Proteomics 171, 95–106. doi:10.1016/j.jprot.2017.08.004
Azizidoost, S., Babaahmadi-Rezaei, H., Nazeri, Z., Cheraghzadeh, M., and Kheirollah, A. (2022). Amyloid beta increases ABCA1 and HMGCR protein expression, and cholesterol synthesis and accumulation in mice neurons and astrocytes. Biochimica Biophysica Acta (BBA)-Molecular Cell Biol. Lipids 1867 (1), 159069. doi:10.1016/j.bbalip.2021.159069
Baig, S., Joseph, S., Tayler, H., Abraham, R., Owen, M., Williams, J., et al. (2010). Distribution and expression of picalm in Alzheimer disease. J. Neuropathol. Exp. Neurol. 69, 1071–1077. doi:10.1097/NEN.0b013e3181f52e01
Calin, G. A., Dumitru, C. D., Shimizu, M., Bichi, R., Zupo, S., Noch, E., et al. (2002). Frequent deletions and down-regulation of micro-RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc. Natl. Acad. Sci. 99, 15524–15529. doi:10.1073/pnas.242606799
Carr, G., Barrese, V., Stott, J. B., Povstyan, O. V., Jepps, T. A., Figueiredo, H. B., et al. (2016). MicroRNA-153 targeting of KCNQ4 contributes to vascular dysfunction in hypertension. Cardiovasc Res. 112, 581–589. doi:10.1093/cvr/cvw177
Chandrasekaran, S., and Bonchev, D. (2016). Network topology analysis of post-mortem brain microarrays identifies more alzheimer’s related genes and micrornas and points to novel routes for fighting with the disease. PloS One 11, e0144052. doi:10.1371/journal.pone.0144052
Chen, C., Tang, X., Lan, Z., Chen, W., Su, H., Li, W., et al. (2023). GABAergic signaling abnormalities in a novel CLU mutation Alzheimer's disease mouse model. Transl. Res. 260, 32–45. doi:10.1016/j.trsl.2023.05.003
Chen, J., Zhang, X., Kusumo, H., Costa, L. G., and Guizzetti, M. (2013). Cholesterol efflux is differentially regulated in neurons and astrocytes: implications for brain cholesterol homeostasis. Biochimica Biophysica Acta (BBA)-Molecular Cell Biol. Lipids 1831 (2), 263–275. doi:10.1016/j.bbalip.2012.09.007
Choi, H. R., Ha, J. S., Kim, E. A., Cho, S. W., and Yang, S. J. (2022). MiR-30a-5p and miR-153-3p regulate LPS-induced neuroinflammatory response and neuronal apoptosis by targeting NeuroD1. BMB Rep. 55 (9), 447–452. doi:10.5483/BMBRep.2022.55.9.061
Dong, X., Wang, H., Zhan, L., Li, Q., Li, Y., Wu, G., et al. (2023). miR-153-3p suppresses the differentiation and proliferation of neural stem cells via targeting GPR55. Aging (Albany NY) 15 (16), 8518–8527. doi:10.18632/aging.204002
Femminella, G. D., Ferrara, N., and Rengo, G. (2015). The emerging role of micrornas in alzheimer's disease. Front. Physiol. 6, 40. doi:10.3389/fphys.2015.00040
Filipowicz, W., Bhattacharyya, S. N., and Sonenberg, N. (2008). Mechanisms of post-transcriptional regulation by micrornas: are the answers in sight? Nat. Rev. Genet. 9, 102–114. doi:10.1038/nrg2290
François, M., Bull, C. F., Fenech, M. F., and Leifert, W. R. (2019). Current state of saliva biomarkers for aging and alzheimer's disease. Curr. Alzheimer Res. 16, 56–66. doi:10.2174/1567205015666181022094924
Guo, J., Fang, W., Chen, X., Lin, Y., Hu, G., Wei, J., et al. (2018). Upstream stimulating factor 1 suppresses autophagy and hepatic lipid droplet catabolism by activating mTOR. FEBS Lett. 592 (16), 2725–2738. doi:10.1002/1873-3468.13203
Hamzeiy, H., Allmer, J., and Yousef, M. (2014). “Computational methods for MicroRNA target prediction,” in miRNomics: MicroRNA biology and computational analysis. Methods in molecular biology (methods and protocols). Editors M. Yousef, and J. Allmer (Totowa, NJ: Humana Press), 207–221.
Huang, J., Weng, Q., Shi, Y., Mao, W., Zhao, Z., Wu, R., et al. (2020). MicroRNA-155-5p suppresses PD-L1 expression in lung adenocarcinoma. FEBS Open Bio 10 (6), 1065–1071. doi:10.1002/2211-5463.12853
Isotalo, K., Kok, E. H., Luoto, T. M., Haikonen, S., Haapasalo, H., Lehtimäki, T., et al. (2012). Upstream transcription factor 1 (USF1) polymorphisms associate with Alzheimer's disease-related neuropathological lesions: tampere Autopsy Study. Brain Pathol. 22, 765–775. doi:10.1111/j.1750-3639.2012.00586.x
Jaberi, K. R., Alamdari-Palangi, V., Jaberi, A. R., Esmaeli, Z., Shakeri, A., Hayat, S. M., et al. (2024). The regulation, functions, and signaling of miR-153 in neurological disorders, and its potential as a biomarker and therapeutic target. Curr. Mol. Med. 23 (9), 863–875. doi:10.2174/1566524023666220817145638
Khayer, N., Mirzaie, M., Marashi, S. A., and Jalessi, M. (2020). Rps27a might act as a controller of microglia activation in triggering neurodegenerative diseases. Plos one 15 (9), e0239219. doi:10.1371/journal.pone.0239219
Kwan, S. H., Wan-Ibrahim, W. I., Juvarajah, T., Fung, S. Y., and Abdul-Rahman, P. S. (2021). Isolation and identification of O-and N-linked glycoproteins in milk from different mammalian species and their roles in biological pathways which support infant growth. Electrophoresis 42 (3), 233–244. doi:10.1002/elps.202000142
Kyriazis, G. A., Wei, Z., Vandermey, M., Jo, D. G., Xin, O., Mattson, M. P., et al. (2008). Numb endocytic adapter proteins regulate the transport and processing of the amyloid precursor protein in an isoform-dependent manner implications for alzheimer disease pathogenesis. J. Biol. Chem. 283, 25492–25502. doi:10.1074/jbc.M802072200
Lee, J. C., Lusis, A. J., and Pajukanta, P. (2006). Familial combined hyperlipidemia: upstream transcription factor 1 and beyond. Curr. Opin. Lipidol. 17, 101–109. doi:10.1097/01.mol.0000217890.54875.13
Lee, J. H., Barral, S., and Reitz, C. (2008). The neuronal sortilin-related receptor gene sorl1 and late-onset alzheimer’s disease. Curr. Neurol. Neurosci. Rep. 8, 384–391. doi:10.1007/s11910-008-0060-8
Li, X., Zhu, X., Zhang, W., Yang, F., Hui, J., Tan, J., et al. (2018). The etiological effect of a new low-frequency ESR1 variant on Mild Cognitive Impairment and Alzheimer’s Disease: a population-based study. Aging (Albany NY) 10 (9), 2316–2337. doi:10.18632/aging.101548
Liang, C., Zhu, H., Xu, Y., Huang, L., Ma, C., Deng, W., et al. (2012). MicroRNA-153 negatively regulates the expression of amyloid precursor protein and amyloid precursor-like protein 2. Brain Res. 1455, 103–113. doi:10.1016/j.brainres.2011.10.051
Liu, C. C., Wang, N., Chen, Y., Inoue, Y., Shue, F., Ren, Y., et al. (2023). Cell-autonomous effects of APOE4 in restricting microglial response in brain homeostasis and Alzheimer’s disease. Nat. Immunol. 19, 1854–1866. doi:10.1038/s41590-023-01640-9
Long, J. M., Ray, B., and Lahiri, D. K. (2012). MicroRNA-153 physiologically inhibits expression of amyloid-β precursor protein in cultured human fetal brain cells and is dysregulated in a subset of Alzheimer disease patients. J. Biol. Chem. 287, 31298–31310. doi:10.1074/jbc.M112.366336
Marcelli, S., Corbo, M., Iannuzzi, F., Negri, L., Blandini, F., Nistico, R., et al. (2018). The involvement of post-translational modifications in Alzheimer's disease. Curr. Alzheimer Res. 15, 313–335. doi:10.2174/1567205014666170505095109
Mazina, A., Shumilina, J., Gazizova, N., Repkin, E., Frolov, A., and Minibayeva, F. (2023). S-nitrosylated proteins involved in autophagy in Triticum aestivum roots: a bottom-up proteomics approach and in silico predictive algorithms. Life 13 (10), 2024. doi:10.3390/life13102024
Oliveira, A. C., Bovolenta, L. A., Nachtigall, P. G., Herkenhoff, M. E., Lemke, N., and Pinhal, D. (2017). Combining results from distinct microRNA target prediction tools enhances the performance of analyses. Front. Genet. 16 (8), 59. doi:10.3389/fgene.2017.00059
Pu, M., Chen, J., Tao, Z., Miao, L., Qi, X., Wang, Y., et al. (2019). Regulatory network of miRNA on its target: coordination between transcriptional and post-transcriptional regulation of gene expression. Cell. Mol. Life Sci. 76, 441–451. doi:10.1007/s00018-018-2940-7
Qiao, J., Zhao, J., Chang, S., Sun, Q., Liu, N., Dong, J., et al. (2020). MicroRNA-153 improves the neurogenesis of neural stem cells and enhances the cognitive ability of aged mice through the notch signaling pathway. Cell Death Differ. 27 (2), 808–825. doi:10.1038/s41418-019-0388-4
Salero, E., Giménez, C., and Zafra, F. (2003). Identification of a non-canonical E-box motif as a regulatory element in the proximal promoter region of the apolipoprotein E gene. Biochem. J. 370, 979–986. doi:10.1042/BJ20021142
Sayers, E. W., Agarwala, R., Bolton, E. E., Brister, J. R., Canese, K., Connor, R., et al. (2018). Database resources of the national center for biotechnology information. Nucleic acids Res. 46, D8–D13. doi:10.1093/nar/gkx1095
Schwarzenbach, H., Nishida, N., Calin, G. A., and Pantel, K. (2014). Clinical relevance of circulating cell-free microRNAs in cancer. Nat. Rev. Clin. Oncol. 11, 145–156. doi:10.1038/nrclinonc.2014.5
Szklarczyk, D., Morris, J. H., Cook, H., Kuhn, M., Wyder, S., Simonovic, M., et al. (2017). The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 45, D362–D368. doi:10.1093/nar/gkw937
Vejnar, C. E., and Zdobnov, E. M. (2012). MiRmap: comprehensive prediction of microRNA target repression strength. Nucleic acids Res. 40 (22), 11673–11683. doi:10.1093/nar/gks901
Wang, J., Vasaikar, S., Shi, Z., Greer, M., and Zhang, B. (2017). WebGestalt 2017: a more comprehensive, powerful, flexible and interactive gene set enrichment analysis toolkit. Nucleic Acids Res. 45, W130–W137. doi:10.1093/nar/gkx356
Wang, Q., Xia, C., Zhu, A., Bao, Y., Lu, J., Chen, Y., et al. (2023). Discrepancy of synaptic and microtubular protein phosphorylation in the hippocampus of APP/PS1 and MAPT× P301S transgenic mice at the early stage of Alzheimer’s disease. Metab. Brain Dis. 9, 1983–1997. doi:10.1007/s11011-023-01209-3
Wong, N., and Wang, X. (2015). miRDB: an online resource for microRNA target prediction and functional annotations. Nucleic Acids Res. 43, D146–D152. doi:10.1093/nar/gku1104
Xu, C., Wang, C., Meng, Q., Gu, Y., Wang, Q., Xu, W., et al. (2019). miR-153 promotes neural differentiation in the mouse hippocampal HT-22 cell line and increases the expression of neuron-specific enolase. Mol. Med. Rep. 20 (2), 1725–1735. doi:10.3892/mmr.2019.10421
Xue, W. X., Zhang, M. Y., Li, R., Liu, X., Yin, Y. H., and Qu, Y. Q. (2020). Serum miR-1228-3p and miR-181a-5p as noninvasive biomarkers for non-small cell lung cancer diagnosis and prognosis. BioMed Res. Int. 2020, 1–13. doi:10.1155/2020/9601876
Yao, P., Zhang, P., Mattson, M., and Furukawa, K. (2003). Heterogeneity of endocytic proteins: distribution of clathrin adaptor proteins in neurons and glia. Neuroscience 121, 25–37. doi:10.1016/s0306-4522(03)00431-7
Zhang, S., Yan, M. L., Yang, L., An, X. B., Zhao, H. M., Xia, S. N., et al. (2020). MicroRNA-153 impairs hippocampal synaptic vesicle trafficking via downregulation of synapsin I in rats following chronic cerebral hypoperfusion. Exp. Neurol. 332, 113389. doi:10.1016/j.expneurol.2020.113389
Keywords: amyloid precursor protein, neurodegeneration, MicroRNAs, Alzheimer’s disease, miR-153
Citation: Amber S and Zahid S (2024) An in silico approach to identify potential downstream targets of miR-153 involved in Alzheimer’s disease. Front. Genet. 15:1271404. doi: 10.3389/fgene.2024.1271404
Received: 02 August 2023; Accepted: 08 January 2024;
Published: 16 January 2024.
Edited by:
Yadong Zheng, Zhejiang Agriculture and Forestry University, ChinaReviewed by:
Erik Josef Behringer, Loma Linda University, United StatesIsabel Duarte, University of Algarve, Portugal
Natasha Andressa Jorge, Leipzig University, Germany
Copyright © 2024 Amber and Zahid. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Saadia Zahid, saadiazahid@hotmail.com, saadia.zahid@asab.nust.edu.pk
 Sanila Amber
Sanila Amber Saadia Zahid
Saadia Zahid