- 1ICAR-National Bureau of Plant Genetic Resources, New Delhi, India
- 2DBT-National Institute of Plant Genome Research, New Delhi, India
- 3Shivaji College, University of Delhi, New Delhi, India
- 4Alliance of Bioversity International and CIAT, India Office, National Agricultural Science Complex, New Delhi, India
Legumes play a significant role in food and nutritional security and contribute to environmental sustainability. Although legumes are highly beneficial crops, it has not yet been possible to enhance their yield and production to a satisfactory level. Amid a rising population and low yield levels, per capita average legume consumption in India has fallen by 71% over the last 50 years, and this has led to protein-related malnutrition in a large segment of the Indian population, especially women and children. Several factors have hindered attempts to achieve yield enhancement in grain legumes, including biotic and abiotic pressures, a lack of good ideotypes, less amenability to mechanization, poorer responsiveness to fertilizer input, and a poor genetic base. Therefore, there is a need to mine the approximately 0.4 million ex situ collections of legumes that are being conserved in gene banks globally for identification of ideal donors for various traits. The Indian National Gene Bank conserves over 63,000 accessions of legumes belonging to 61 species. Recent initiatives have been undertaken in consortia mode with the aim of unlocking the genetic potential of ex situ collections and conducting large-scale germplasm characterization and evaluation analyses. We assume that large-scale phenotyping integrated with omics-based science will aid the identification of target traits and their use to enhance genetic gains. Additionally, in cases where the genetic base of major legumes is narrow, wild relatives have been evaluated, and these are being exploited through pre-breeding. Thus far, >200 accessions of various legumes have been registered as unique donors for various traits of interest.
1 Introduction
Legumes of the family Fabaceae are among the most important plant groups on planet Earth. While legumes are an important source of food and nutrition, they also play an important role in improving soil health and ecosystem sustainability. Legume grains are often considered to be “the poor man’s meat,” as the vegetarian human population is highly dependent on legume grains for its protein needs (Roy et al., 2017). The “green revolution” has helped several countries to attain self-sufficiency in food, which can primarily be attributed to a manyfold increase in the production of cereals, particularly rice, wheat, and maize. However, similar advances in grain legume production have not been achieved (Figure 1), probably because legumes are less amenable to the adoption of green revolution technologies. Over 200 species of legumes are cultivated worldwide. Of these, we list the major grain legume crops, with their production and yield status and taxonomic information, in Table 1.
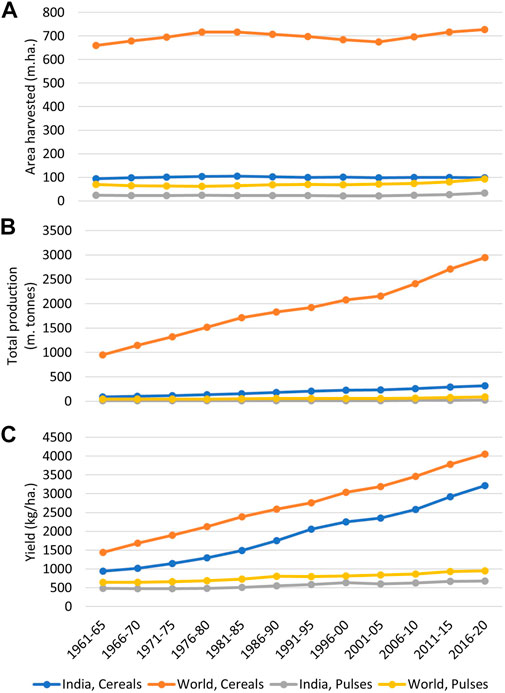
FIGURE 1. A graphical comparison of cereals and pulses in terms of total area harvested (A), total production (B), and yield (C) in India and the world. The graph indicates how the onset of the green revolution has tremendously enhanced the production of cereals in India and worldwide, which can be primarily attributed to yield improvement in these crops. By comparison, yield and production improvements in pulses have remained insignificant during this period (Data source: FAOSTAT, 2022).
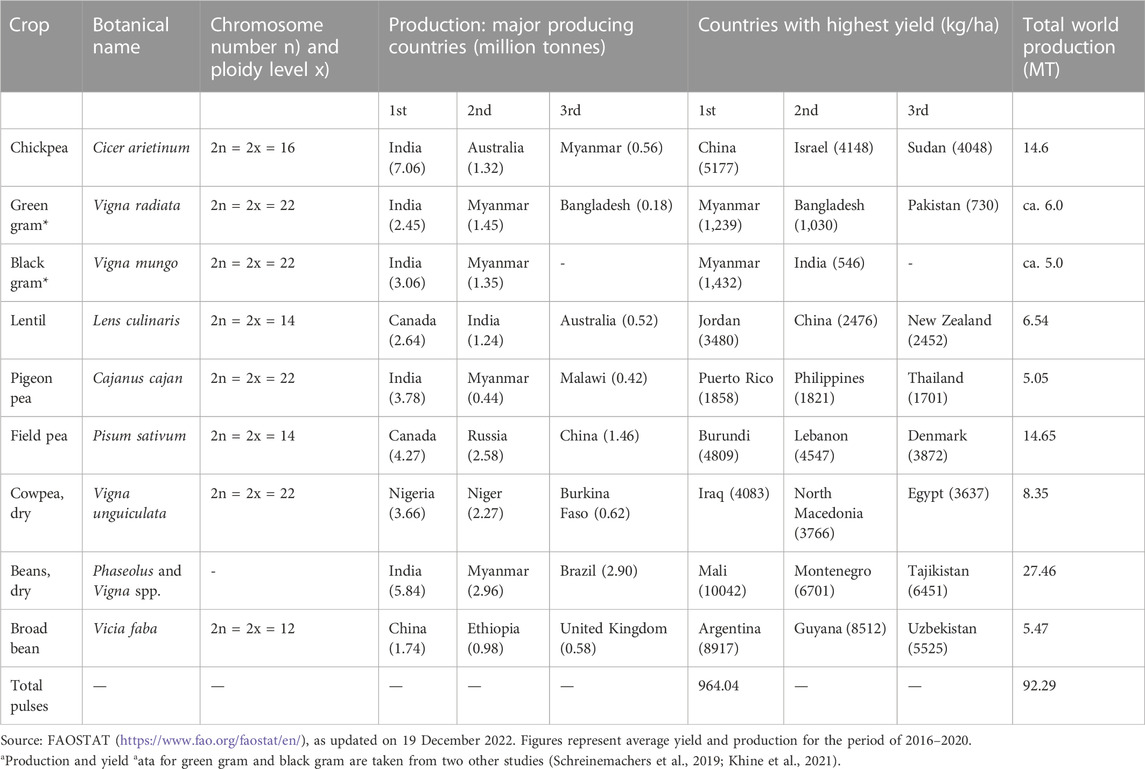
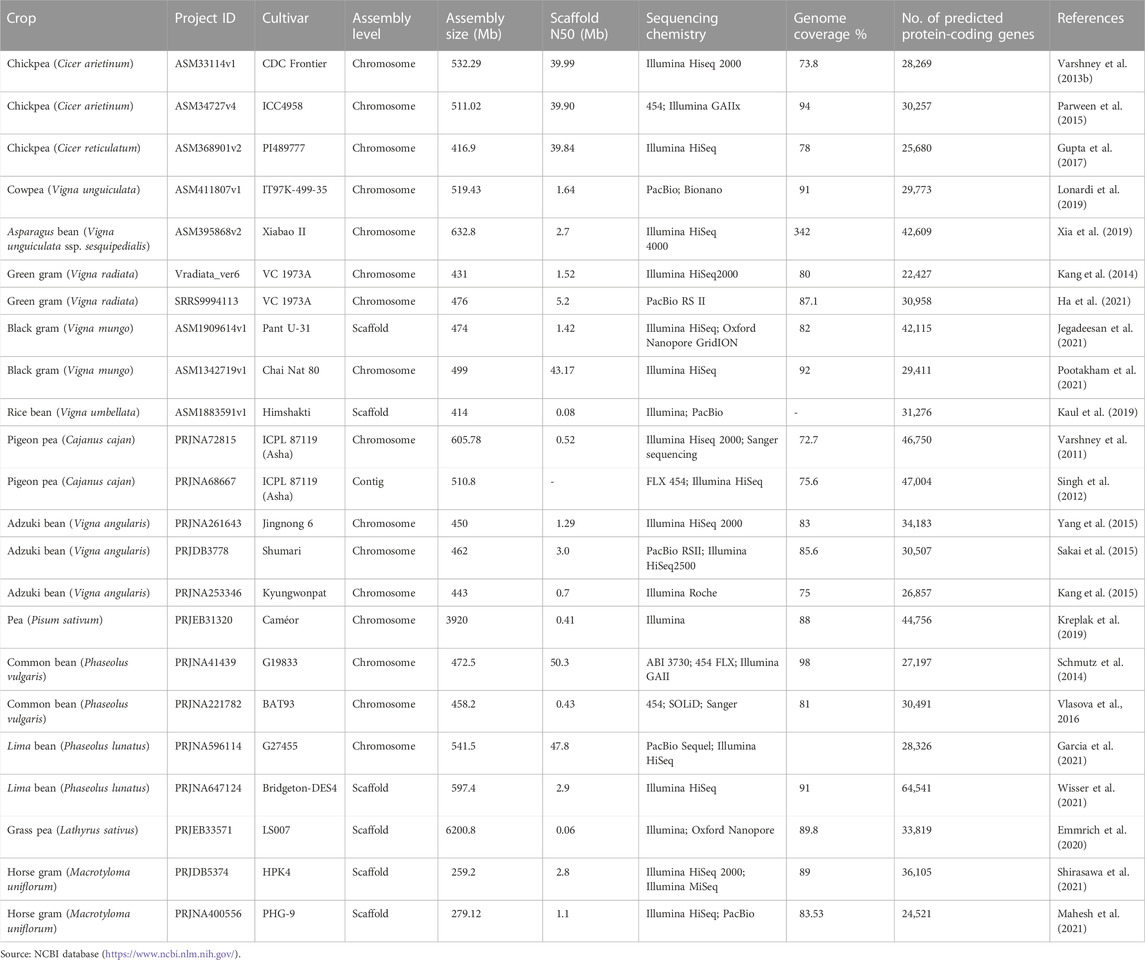
TABLE 1. Information on the production and yield status of the major grain legume crops cultivated worldwide, along with their botanical names and chromosome numbers.
India is the largest producer and consumer of grain legumes globally. India’s contribution constitutes around 28.12% of global grain legume production (ca. 23.37 million tonnes), and this is the output of ca. 29 million ha of cultivated land (Department of Economics and Statistics, Department of Agriculture and Farmers Welfare, Ministry of Agriculture and Farmers Welfare, GoI). Of this total, around 34% of the cultivated land (9.89 mha) is covered by cultivation of chickpea alone; this is followed by black gram (4.81 mha), pigeon pea (4.72 mha), green gram (4.61 mha), lentil (1.43 mha), and field pea (0.75 mha). Other minor legumes cultivated in India are green gram, cowpea, moth bean, grass pea, and horse gram. In 2020, global grain legume production was approximately 83.1 million tonnes; this figure is the sum of the production values of grain legumes grown globally, specifically beans (dry), chickpeas, peas (dry), lentils, cowpeas, pigeon peas, Bambara beans, and other minor pulses (www.fao.org/faostat/en). The primary contributors to this total (at 42.12 million tonnes) were the five major grain legume producing countries of India (28.12%), Canada (9.24%), Myanmar (4.84%), Nigeria (4.47%), and the Russian Federation (4.01%). The major producing countries for each crop are given in Table 1.
Globally, over 3,800 improved cultivars of grain legumes have been released, with improvements to traits such as yield, crop duration, and nutritional qualities (Pratap et al., 2022). However, between 1961 and 2020, only a 1.5-fold increase in grain legume productivity was achieved, from 637 kg/ha to 964 kg/ha (Pratap et al., 2022; Figure 1). This can be primarily attributed to various factors, such as a narrow genetic base in cultivated gene pools, poor plant ideotype, high susceptibility to insect pests and diseases, a lack of robust seed systems, and frequent stresses from drought, heat, and flooding.
Over 850 high-yielding varieties of food legumes have been developed in India, and these are now playing a vital role in food legume production (Chauhan et al., 2016). However, the foundation of any crop breeding program is based on only a small number of parental lines, which has led to a narrow genetic base in these cultivated varieties. In a pedigree analysis, it was found that 41% of chickpea varieties had PB 7 as one of its ancestors; in pigeon pea, T 1 and T 190 appeared in 34% of varieties; and T 9 and T 1 appeared in 64% and 35% of varieties of black gram and green gram, respectively (Kumar et al., 2004). Furthermore, in the process of rigorous selection in the development of a variety, alleles conferring defense mechanisms are also lost. This is one of the reasons that the actual yield of most food legume crops is half their potential yield. Recently, drastic climatic change, to which abrupt temperature rises, erratic and heavy rainfall, frequent droughts, episodes of flooding, and rapid pest and pathogen evolution can be attributed, has exceeded the adaptation capability of modern varieties (Guo, 2022). As a result, the breakdown of resistance to biotic stress has become rather common in modern cultivars (Sharma et al., 1999; Burdon et al., 2014; Rex Consortium, 2016; Mbinda and Masaki, 2021; Hu et al., 2022; Van de Wouw et al., 2022). Therefore, ex situ collections are now being utilized to increase genetic variability in modern cultivars in order to improve their climate resilience, including via genetic gains in breeding programs. Advances in genomics, phenomics, and breeding methods are playing an important role and exerting a significant impact on legume improvement by accelerating genetic gains via enhancements to selection efficiency and the advancement of desired genotypes with high precision.
It is well understood that, in terms of enhancing the variability of a crop gene pool, landraces are the primary resource; the desired traits need to be sought out among these, as they are easy to cross and their use significantly reduces the chances of linkage drag as compared to the use of wild species. Additionally, landraces are well adapted to microclimatic niches and have several superior traits in terms of nutritional value. In this study, we have focused on the identification of desired genes and traits and their utilization in the improvement of legume crops. We also propose a comprehensive strategy for the enhancement of genetic gains (Figure 2).
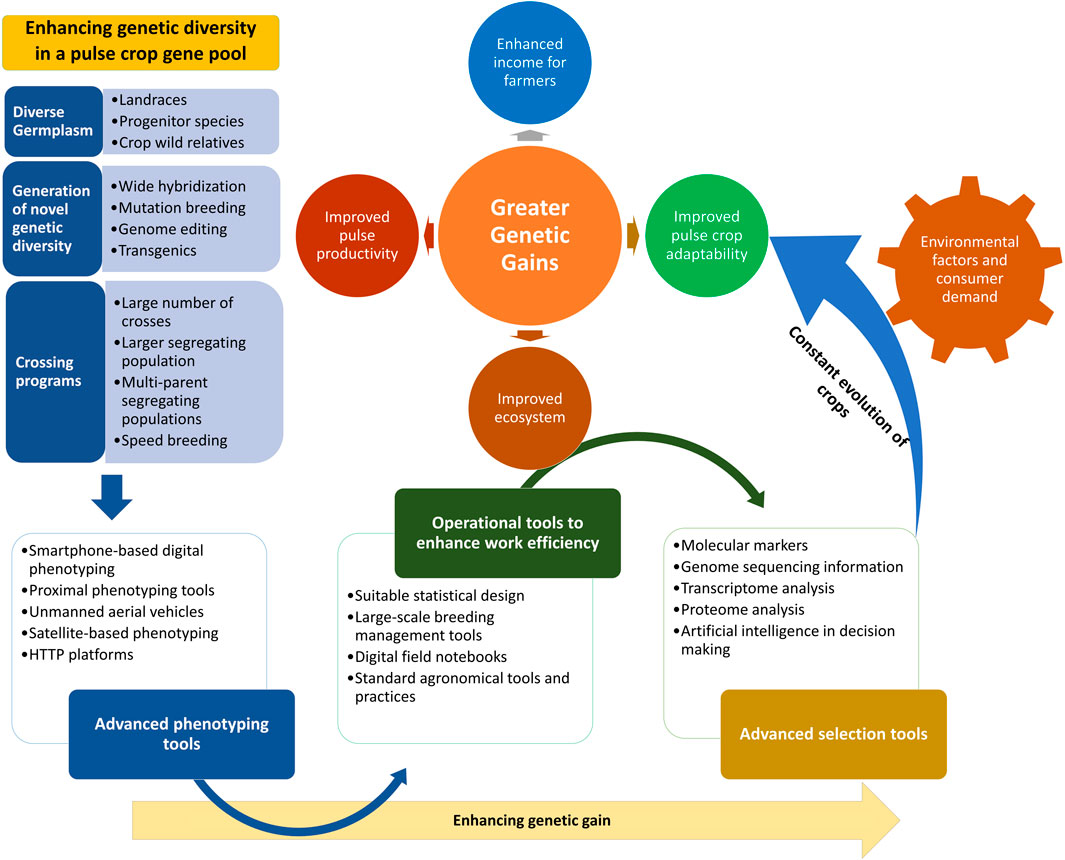
FIGURE 2. Strategy to enhance genetic gains through utilization of advanced phenotyping tools, efficient operational tools, and advanced selection methods and technologies. A strategy to achieve higher genetic gains by broadening the genetic base through the infusion of increasing levels of variability from diverse sources into the target breeding populations is illustrated. The integration of improved crossing program strategies and advanced tools for phenotyping, operations, and desired genotype selection will further enhance the genetic gains made. This strategy will help with the attainment of greater genetic gains along with enhanced crop adaptability to changing climatic conditions.
2 Legume germplasm collections in the Indian national gene bank
The collection, conservation, and selection of germplasm are the primary components of the crop domestication process. Wild species were initially brought under cultivation and improved through selection for their agronomic traits, and the practice is still being followed by farmers and breeders. Diverse environments of crop cultivation, including rainfed, dryland, and coastal areas, flood-prone areas, and areas at high altitude, as well as disease hotspots and human preferences in terms of nutritional qualities, aesthetics, and cultural values, have played important roles in the development and deployment of diverse germplasm. Although diversity has been continually developing and has been sustained through traditional practices over the last several thousand years, crop diversity has recently come under threat due to increasing pressure arising from demographic, sociocultural, and technological changes.
India is rich center of diversity for several cultivated crops, including important legumes such as chickpea (Cicer arietinum), moth bean (Vigna aconitifolia), rice bean (Vigna umbellata), cowpea (Vigna unguiculata), yard-long bean (Vigna unguiculata subsp. sesquipedalis), green gram (V. radiata), black gram (V. mungo), horse gram (Macrotyloma uniflorum), and dolichos bean (Lablab purpureus) (Zeven and Zhukovsky, 1975; Hawkes, 1983). The development of extensive and organized germplasm collections and conservation activity in India began only after the establishment of the National Bureau of Plant Genetic Resources (NBPGR) in 1976 (Rana et al., 2016). Since then, around 63,000 accessions of legumes have been collected and conserved in ex situ conditions (Table 2). The organization is continuously enriching its collections based on gap analysis with respect to earlier collections established within India and also introducing accessions from abroad. Globally, over 0.7 million legume germplasms, including their crop wild relatives (CWRs), are conserved in 276 gene banks distributed worldwide (WIEWS, 2022).
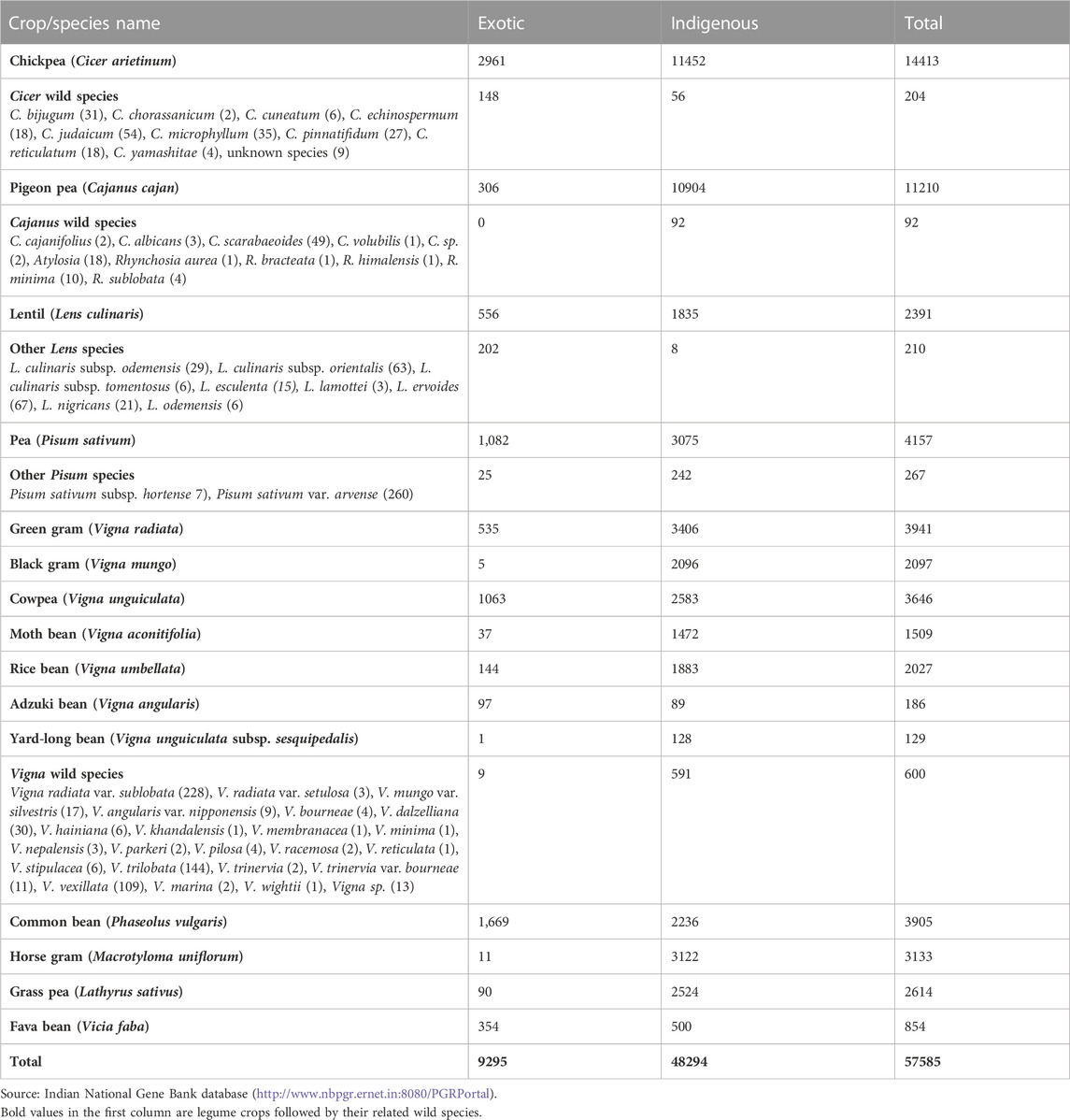
TABLE 2. Status of collections of grain legume crops and their wild relatives available in the Indian National Gene Bank.
3 Utilization of grain legume germplasm for crop improvement
Crop evolution in early times was based entirely on appearance and performance in terms of agro-morphological traits, and these are still the primary focus of plant breeders and researchers. During the domestication process and subsequent structured breeding programs, genotypes with greater biotic and abiotic stress tolerance are often unintentionally selected, but agronomic traits have always been the prime target for selection. Landraces, which are locally adapted cultivars with a high level of genetic variability developed by farmers over the years, are the primary source of such traits in modern breeding programs. However, in terms of the utilization of germplasm from gene banks, it has become difficult to identify a manageable number of accessions with the desired levels of variability and traits. Recognizing this challenge, Frankel (1984) proposed the concept of a core collection, a minimum number of representative accessions representing maximum variability across the entire collection. Since then, several crop-specific diverse core sets have been developed (Table 3), and this has accelerated the utilization of gene bank collections. A number of significant studies conducted to date in the area of trait identification and utilization are discussed below, presented in crop-wise fashion, and promising trait-specific accessions are summarized in Table 4 (biotic stress resistance) and Table 5 (abiotic stress tolerance). We also find that in the process of breeding modern varieties, the focus on yield per se has eventually led to a gradual decrease in the nutritional qualities of new varieties. Comparative studies on the nutritional composition of landraces and traditional cultivars in various crops, such as vegetables and fruits (Davis et al., 2004), wheat (Fan et al., 2008), the potato (White et al., 2009), the common bean (Celmeli et al., 2018), and green gram (Ebert et al., 2017), have indicated that the improved varieties are poorer than the older varieties in terms of nutritional value. Therefore, recognizing the significance of nutritional value and of the availability of nutritional variability in germplasm, we also discuss the important nutritional characteristics of each legume crop in the following sections.
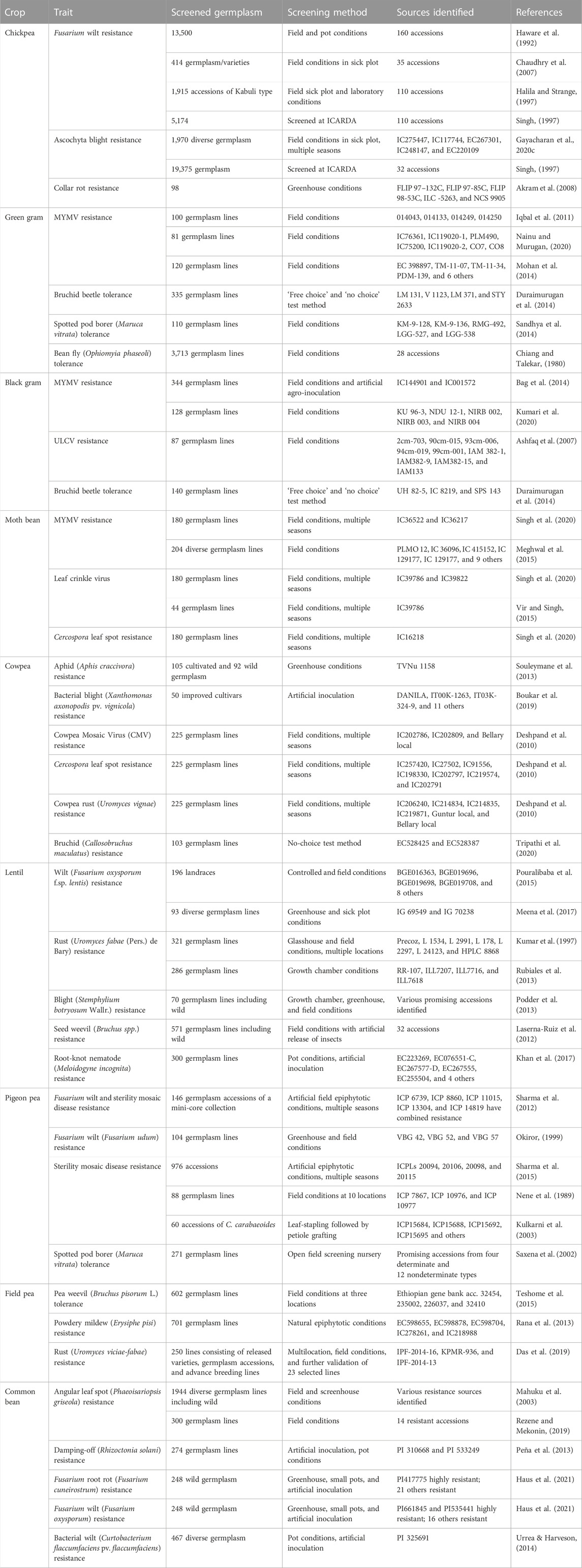
TABLE 4. List of important resistance sources identified for various important biotic stresses in grain legume crops.
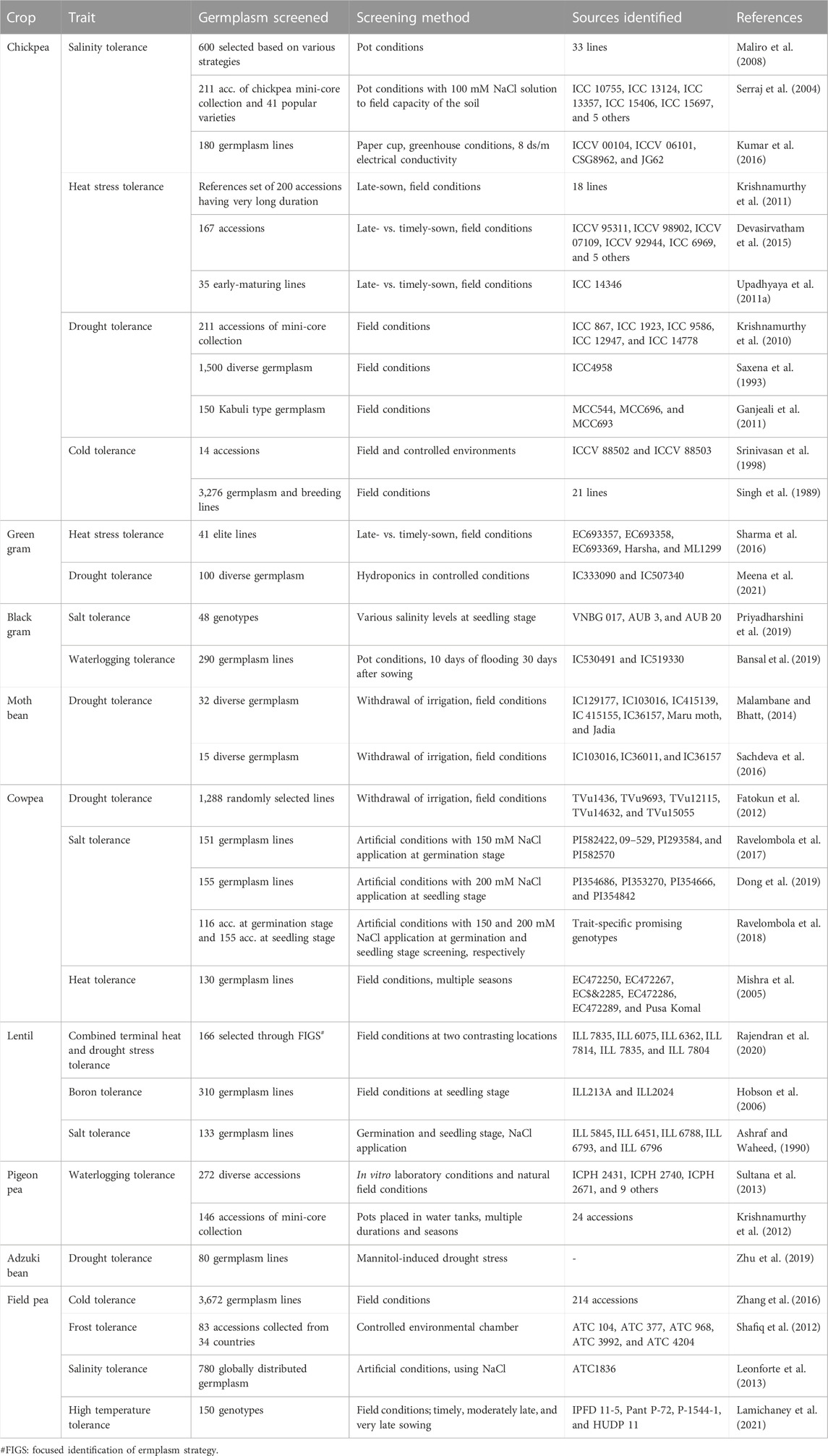
TABLE 5. List of important resistance sources identified for various important abiotic stresses in grain legume crops.
3.1 Chickpea
3.1.1 Agronomic traits
Development of the first core collection in the domain of legumes was reported by Hannan et al. (1994) with the objective of making use of chickpea collections. In this study, a diverse set of 505 chickpea accessions was designated as a core set; this was derived from 7,613 accessions conserved in the Western Regional Plant Introduction Station (WRPIS), USDA. Later, the International Crops Research Institute for the Semi-Arid Tropics (ICRISAT) developed a core set of 1,956 accessions based on information on the geographic origins and on 13 morphological traits for 16,991 accessions (Upadhyay and Ortiz, 2001). Following this exercise, Upadhyay et al. (2007a) identified 28 early-maturing chickpea germplasm lines having wide geographical distribution. Based on multi-location trials of the core set, ICC 16641, ICC 16644, ICC 11040, ICC 11180, and ICC 12424 were further identified as extra-early maturing lines, while ICC 14648, ICC 16641, and ICC 16644 were identified as having higher seed weight. Additionally, in an evaluation of 1,956 accessions of the chickpea core set on 14 agronomic traits, several superior accessions were identified in terms of early flowering, pods/plant, seed yield, and seed weight (Upadhyaya et al., 2007b). Furthermore, in order to reduce the size of the core collections, a mini-core set of 211 accessions was developed based on more extensive phenotypic data and a suitable statistical approach (Upadhyaya and Ortiz, 2001); this has been extensively utilized for the evaluation and identification of important traits (Upadhyaya et al., 2010). Promising accessions for traits such as water use efficiency (ICC 16374, ICC 1422, ICC 4958, ICC 10945, ICC 16374, ICC 16903) and biotic-abiotic stresses were identified (Upadhyaya et al., 2010). Erect type chickpea lines suitable for mechanical harvesting were also identified (Upadhyaya et al., 2017). A similar approach was followed at the Indian National Gene Bank to accelerate the utilization of chickpea germplasm; there, the gene bank’s entire chickpea collection (14,651 accessions) was characterized and evaluated for agronomic traits in 2012, and several promising accessions in terms of agronomic traits were identified (Archak et al., 2016). The characterization of a large number of accessions also provides the opportunity to identify rare and unique morphotypes, which sometimes turns out to be very useful. For example, in the study carried out by Archak et al. (2016), accession IC486088 was found to have upright podding behavior, which makes it a potential donor that could be used in altering chickpea plant type (Singh et al., 2013) (Figure 3). To enhance the utilization of such unique germplasm of economic or scientific value in crop improvement programs, these are registered with a national germplasm registration facility, i.e., the Germplasm Registration and Information System (GRIS; http://www.nbpgr.ernet.in:8080/registration/AboutUs.aspx). As of December 2022, a total of 28 unique accessions of chickpea have been registered by this facility. These unique traits help in the development of plant types and/or high-yielding cultivars. For example, a unique determinate phenotype was identified in BGD 9971; this is considered an important trait in the alteration of chickpea plant type (Hegde, 2011; Ambika et al., 2021). In the F2 line of an inter-specific cross, ICC 5783 (C. arietinum) × ICCW 9 (C. reticulatum), 3 to 9 flowers per flowering node were observed; this is an important trait for improving chickpea plant type and yield (Gaur and Gour, 2002). Finally, to reduce harvesting and threshing time and cost, chickpea genotypes with the erect plant type were identified and are being used to develop chickpea cultivars suitable for mechanical harvesting (Vishnu et al., 2020). GBM 2 and NBeG 47 (Dheera) are the first two such chickpea varieties that have been released (http://dpd.gov.in/Varieties/Chickpea%20varieties.pdf) (Figure 3).

FIGURE 3. Highlights of various important agro-morphological variations. Genotype (ICC16358) with a large number of branches per plant (A); genotype (IC486088) having upright peduncle and pods (B); genotype (ICC15559) with two to three flowers/peduncle (C); genotype (EC398937) with greater pod length (>15 cm) and a higher number of seeds/pod (D); leaflet size variation (E); leaf size variation (F); genotype with short internode length and compact phenotype (G); genotype (IC24417) with erect and tall growth habit (H); an erect genotype (NBeG 47) in chickpea (I); early-maturing (IC347181) (J) and erect type (VLG 39) (K) genotypes of horse gram; sona mung with bright yellow seeds having superior visual appeal (L) in green gram; a common bean germplasm (IC341862) having pea-shaped, bright white-colored seed with superior visual appeal (M); and a pigeon pea genotype with determinate growth habit (N).
3.1.2 Biotic stress
In chickpea, the major diseases are fusarium wilt (Fusarium oxysporum f. sp. ciceris), ascochyta blight (Ascochyta rabiei (Pass.) Lab.), collar rot (Sclerotium rolfsii Sacc.), dry root rot (Rhizoctonia bataticola (Taub.) Butler), and botrytis gray mold (Botrytis cinerea Pers. Ex. Fr.), and the major pest is pod borer (Helicoverpa armigera Hubner). Chickpea germplasm screening programs have identified rather plentiful instances of donor germplasm resistant to fusarium wilt. However, robust resistant donor sources for dry root rot, botrytis gray mold, collar rot, and pod borer are lacking; thus, germplasm use could result in the identification of moderately resistant donors for these diseases (Pandey et al., 2004; Sharma M et al., 2015; Reddy et al., 2016). The ICAR–National Bureau of Plant Genetic Resources (ICAR-NBPGR) has evaluated over 2,500 accessions for resistance to botrytis gray mold, collar rot, and dry root rot under artificial inoculation and field conditions, but only a few moderately resistant accessions have been identified, such as IC244185, IC251727, ICC6881, and IC350842 for BGM; IC270930, IC95064, IC350829, IC95100, IC209375, IC83805, IC487359, and IC83991 for collar rot; and IC413984, IC397375, IC487359, IC506915, and ICC4295 for dry root rot (unpublished data). This is a sign of the narrow genetic base of the cultivated germplasm. Similarly, no resistance sources have yet been identified for pod borer. Gayacharan U. et al. (2020) have identified several robust resistance sources (viz., IC275447, IC117744, EC267301, IC248147, and EC220109) for ascochyta blight disease using the sick plot method following artificial inoculation in multiple environments and seasons; these are now being utilized in national chickpea breeding programs. Pande et al. (2006) have also identified several other promising chickpea accessions (viz., ICC 17211, IG 69986, IG 70030, IG 70037, and IG 70038), which have shown combined tolerance against ascochyta blight and botrytis gray mold diseases. Finally, Singh (1997) has listed several of the important sources identified at the International Center for Agricultural Research in the Dry Areas (ICARDA) and at the ICRISAT. Various important sources of biotic stress resistance are listed by Singh et al. (2022) and are also presented in Table 4 and Figure 4.

FIGURE 4. Several promising newly identified resistance donors: IC486215 (A) for resistance against dry root rot in chickpea; IC275447 (B) for resistance against ascochyta blight in chickpea; IC118998 (C) for yellow mosaic disease (YMD) resistance in green gram; and IC278261 (D) for powdery mildew resistance in field pea.
3.1.3 Abiotic stress
In chickpea, the major abiotic stresses are terminal drought, terminal heat stress, and low temperatures during the late vegetative stage. The Northern Plains of India, which was once the most favorable zone for chickpea production, has faced a drastic decline in production of this crop due to a sharp rise in the minimum night temperature (Basu et al., 2009). Terminal heat stress is also a major challenge in the expansion of chickpea cultivation to rice-fallow land, of which there are around 11.7 million ha in the country (Singh N et al., 201a). Therefore, in order to tackle this problem, new sources of tolerance are being sought to enable the development of short-duration and heat-tolerant varieties. Basu and coworkers (2009) screened chickpea germplasm and identified several highly heat-tolerant chickpea lines, viz., ICCV 92944 (JG14), ICCV 37, ICC67, JKG 1, GCP 101, and PG 12. A reference collection for heat stress tolerance has also been developed and screened at the reproductive stage (Krishnamurthy et al., 2011). The researchers observed broad genetic variation in heat-responsive traits, and later identified 10 heat stress tolerance lines under field conditions (Devasirvatham et al., 2015). A mini-core collection has also been screened for resistance to drought stress; five highly tolerant accessions (namely, ICC867, ICC 1923, ICC9586, ICC12947, and ICC14778) were identified (Krishnamurthy, et al., 2010). A germplasm line (ICC4958) developed by Saxena et al. (1993) has been extensively used for breeding drought-tolerant varieties. In-depth molecular analysis of the ICC4958 line has identified a QTL-hotspot region that harbors several traits related to drought tolerance (Bharadwaj et al., 2021). Certain other drought stress tolerant lines have been identified under field conditions (Ganjeali et al., 2011). A mini-core set was also screened for salinity stress resistance under pot conditions and artificial application of saline water (100 mM), which led to the identification of 10 highly tolerant accessions (Serraj et al., 2004). Genotypes ICCV 00104, ICCV 06101, CSG8962, and JG62 have also been identified as promising in terms of salinity tolerance (Kumar et al., 2016). Additionally, a total of 3,276 germplasm lines of chickpea were evaluated against cold stress at the ICARDA, Tel Hadya, Syria, between 1981 and 1987; 21 lines were found to be tolerant of cold stress (Singh et al., 1989). Choudhary and coworkers (2018) list several popular donors that represent the major sources for improvement of chickpea tolerance to abiotic stress. An extensive list of chickpea germplasm lines that have been identified as promising in relation to various abiotic stresses is also given in Table 5.
3.1.4 Nutritional Quality
The chickpea is well known for its nutritionally rich grains, which are widely used as an alternative source of supplementary nutrients. Chickpea grains contain 63% total carbohydrate, 21% protein, and 2.70%–6.48% total fat (Wang et al., 2021). The prominent minerals are K (1.2 g/100 g in desi type, 1.1 g/100 g in Kabuli type), P (0.38 g/100 g in desi, 0.5 g/100 g in Kabuli), Mg (169 mg/100 g in desi, 178 mg/100 g in Kabuli), and Ca (162 mg/100 g in desi, 107 mg/100 g in Kabuli; Wang et al., 2021). Chickpea grains are also a good source of vitamins C, B2, B3, B5, γ–tocopherol, E (α–tocopherol), and folic acid.
Large-scale nutritional profiling has not yet been carried out for legumes, primarily due to a lack of high-throughput nutritional profiling platforms. However, nutrient-specific donors with high mineral content have been identified, such as for Zn (MG–13, MG–17), Ca (PI518255, PI358934), and P (PI339154), and these can be used for biofortification of modern chickpea cultivars (Constantini et al., 2021). In an analysis of 79 accessions, one (LEGCA728) was identified as having high lutein content (28.32 μg g−1), and distinct morphotypes were identified as superior in terms of high concentration of specific nutrients (Serrano et al., 2017). In this study, it was observed that nutritional variation is associated with seed morphology. Black and brown seeded varieties were found to have higher dietary fiber content, ranging from 18.0 to 22.1 g 100 g−1, and higher polyunsaturated fatty acid (PUFA) content (67.0 g 100 g−1 of total fatty acids; Summo et al., 2019). Accessions with brown coloring also have high water absorption capacity (1.9 g water g−1 of flour), which makes these varieties suitable for mixing with cereal flours to produce nutritionally rich cereal-based food products. The vitamins, minerals, and fibers present in chickpea grains promote their utilization for many health benefits. Finally, carotenoid concentration (with the exception of lycopene) has been found to be higher in wild germplasm as compared to cultivated types (Jukanti et al., 2012).
3.2 Lentil
3.2.1 Agronomic traits
Lentil is one of the eight founder crops of agriculture (Ambika et al., 2022) and the most nutritious cool season legume cultivated in many farming systems worldwide. Lentil is divided into two categories based on seed size, i.e., microsperma (seed diameter 2–6 mm) and macrosperma (seed diameter 6–9 mm), with 100 seed weight ranging from 1.5 to 8.0 g. In order to identify new germplasm for various agro-morphological traits, extensive germplasm exploration, characterization, and evaluation programs have been undertaken globally. As a result, several trait-specific donors have been identified and used to develop improved varieties. High biomass, good plant standing, higher seed weight, and number of pods/peduncle are considered to be crucial traits for yield enhancement in lentil. With the availability of such genotypes having tall (>30 cm), erect growth habits and good standing ability with good ground clearance (>15 cm), high-yielding varieties that are suitable for mechanical harvesting have been developed, such as ILL590, ILL1005, ILL6037, ILL6212, ILL6994, ILL7155, and ILL7947 (Sarker and Erskine, 2006; Kumar et al., 2013). Germplasm lines for early flowering (IC560333, IC559639, IC560111, and IC560148), high biomass (IC559744, IC559608, IC559767, and IC560040), a large number of primary branches (IC559870, IC318881, IC398688, and IC560182), and high yield (IC398094, IC560212, IC560332, and IC560206) have also been identified through gene bank germplasm characterization (Gautam et al., 2013). Mishra et al. (2022b) identified PMF-1, PMF-2, PMF-3, and PMF-4 as producing multiple flowers per peduncle, which is an important trait in lentil breeding. The GRIS portal indicates the registration of accessions for a range of important unique traits, such as extended funiculus, which helps with rapid water uptake (IC317520); multiple flowers and pods per peduncle (IC241473); early flowering and maturity (IC241532); and extra bold seeds (EC499760). A core set of 287 accessions was developed for lentil using diversity documentation on 3,068 accessions conserved at the WRPIS, USDA, by Simon and Hannan (1995). Promising germplasm lines have been identified for various agronomic traits, such as seedling vigor, earliness, number of pods/peduncle, number of pods/plant, and seed weight (Singh., 1995). In another large-scale characterization conducted at the ICAR–NBPGR, accessions were characterized on 26 agro-morphological traits, and a core set of 170 accessions was developed (Tripathi et al., 2021a). Kumar et al. (2013) have also highlighted important lines for traits such as winter hardiness, short duration type, mechanical harvesting, and higher seed weight.
3.2.2 Biotic stress
The major diseases in the lentil crop are fusarium wilt (Fusarium oxysporum f. sp. lentis (Fol)), root rot complex, rust (Uromyces viciae-fabae), stemphylium blight (Stemphylium botryosum), powdery mildew (Erysiphe spp.), and ascochyta blight (Ascochyta lentis); the major pests are pod borer (Etiella zinkenella), aphids (Aphis craccivora), and seed weevil (Bruchus spp.). Several studies have been conducted to identify resistant donors, some of which are listed in Table 4. For example, 12 accessions were identified as resistant to fusarium wilt out of 196 landraces screened under both field and controlled conditions (Pouralibaba et a., 2015). In another study, 93 accessions were screened under three different screening conditions (specifically, a hotspot location, field sick plot, and artificial greenhouse conditions) for resistance to wilt, and two highly resistant germplasm lines (viz., IG 69549 and IG 70238) were identified (Meena et al., 2017). Sources for fusarium disease resistance, such as ILL5883, ILL5588, ILL4400, and ILL590, and for resistance to other important diseases, such as rust (ILL358, ILL4605, ILL5604, ILL6002, and ILL6209), ascochyta blight (Indianhead, ILL358, ILL857, ILL5562, ILL5588, ILL5684, ILL5883, and ILL6024), and Stemphylium blight (ILL 4605), have been highlighted by Kumar et al. (2013). Additionally, 4 lines (RR–107, ILL7207, ILL7716, and ILL7618) have been identified as resistant to rust (Uromyces fabae (Pers.) de Bary) out of 286 accessions screened under controlled conditions (Rubiales et al., 2013). Blight (Stemphylium botryosum Wallr.) resistance has also been identified in wild lentil germplasm (Podder et al., 2013). Seed weevil (Bruchus spp.) is another major threat to lentil grains; therefore, 571 accessions of lentil originating from 27 different countries were evaluated under natural field conditions in central Spain, with wide variation (0%–70%) being observed in infestation rate in the lentil germplasm (Laserna–Ruiz et al., 2012). In this study, a total of 32 accessions with lower levels of infestation were identified. In a separate screening of 300 lentil accessions against root-knot nematode (Meloidogyne incognita), 9 accessions were identified as tolerant (Khan et al., 2017; Table 4). Furthermore, these have been registered with the GRIS portal to enhance the utilization of such important sources in lentil breeding programs. Examples of such accessions include IC296883 for multiple resistance against Meloidoyne incognita, M. javanica, Botrytis gray mold, and pod borer; IC567650 for rust resistance; and IC559673 and IC559890 for nematode resistance.
3.2.3 Abiotic stress
Drought, heat, cold, frost, salinity, and waterlogging are the major abiotic stresses affecting lentil cultivation around the world. Several studies have been conducted to identify germplasm tolerant to these stresses (Table 5). In one such study, 166 lentil accessions were screened under field conditions, and six lines (ILL 7835, ILL 6075, ILL 6362, ILL 7814, ILL 7835, and ILL 7804) were identified for combined heat and drought stress tolerance (Rajendran et al., 2020). The Focused Identification of Germplasm Strategy (FIGS) was used to select 162 accessions for screening against heat and combined heat–drought stresses under field conditions at two locations (El Haddad et al., 2020); one germplasm line (IC621470) has been registered for drought tolerance on the GRIS portal. Based on a salt stress tolerance index, several promising accessions (ILL 5845, ILL 6451, ILL6788, ILL 6793, and ILL 6796) were identified in a screening of 133 accessions under artificial conditions (Ashraf and Waheed, 1990). Accessions ILL52, ILL465, ILL 1878, ILL 1918, ILL7115, ILL7155, ILL468, ILL590, ILL662, ILL669, ILL780, ILL857, ILL975, WA8649041, and WA8649090 have been selected for winter hardiness (Erskine et al., 1981; Hamdi et al., 1996). Additionally, Stoddard et al. (2006) have identified ILL5865 and ILL1878 as lines with good levels of tolerance to freezing. In Australia, in order to expand lentil cultivation, 310 accessions were screened in soils with a high boron concentration; accessions originating from Afghanistan and Ethiopia were found to perform comparatively well under these conditions. Boron-tolerant accessions ILL213A and ILL2024 were also recorded as having higher biomass than boron-intolerant accessions (Hobson et al., 2006).
3.2.4 Nutritional quality
Lentil has a high protein content (20%–27%; Zaccardelli et al., 2012) and contains 2%–3% fat, 50%–65% starch, and 8%–9% soluble sugars (Jood et al., 1998). Lentil protein is considered to be among the most beneficial, as it has good Leu/Ile and Leu/Lys ratios (1.24–1.98 and 1.08–2.03, respectively), high digestibility (∼83%), and strong potential for use in food products (Jarpa-Parra, 2018). Among pulses, lentil is also one of the richest sources of Zn and Fe. A screening of over 2000 cultivated and wild germplasm has revealed a wide range of variation in Fe (42–168 ppm) and Zn (22–101 ppm; Mehra et al., 2018), with accessions EC78933 and EC 78414 found to have particularly high Fe and Zn content, respectively. In one of the experiments conducted by Kumar et al. (2014), 41 genotypes were examined for stability of Zn and Fe content over three locations; L 4704 (136.91 mg/kg grain) and VL 141 (81.542 mg/kg grain) were found to be promising in relation to Fe and Zn, respectively. A germplasm line (IC317520) with an extended funicle has also been identified; this is expected to be associated with shorter cooking time (Tripathi et al., 2021b). Several genotypes have been registered with GRIS: IC208329 and IC208326 for high protein content (27.4%–28.5%), and IC0616579 for high iron 136.91 (mg/kg grain) and zinc (71.69 mg/kg grain) content.
3.3 Common bean
3.3.1 Agronomic traits
Common bean is an economically important legume and is cultivated worldwide. In order to assess phenotypic variability in the ex-situ collections of the Indian National Gene Bank, 4,274 accessions were characterized on 22 traits, and a good range of variation was observed in leaf length, leaf width, pod length, number of pods per plant, number of seeds per pod, and seed weight (Rana et al., 2015). Promising accessions were identified for early flowering (IC370764), pod length (IC328871, EC271552), pods/plant (EC500299), early maturity (EC0944456), a large number of seeds/pod (IC383008), etc. In another study, 203 accessions of a core collection were examined for seed quality traits and to identify promising germplasm lines (Saba et al., 2016). The Andean Diversity Panel (ADP), a regional core collection comprising 396 accessions, with the majority originating from the Andean region, was established in order to enhance germplasm utilization in the region’s common bean improvement program (Cichy et al., 2015a). The ADP consists primarily of popular cultivars, breeding lines, and landraces. The CIAT gene bank conserves over 40,000 common bean accessions, making it the largest collection in the world. In an evaluation of a core set of 1,414 accessions, Amirul et al. (2006) observed wide variability in their morphological, biochemical, and nutritional traits. Through 12 multi-environment trials, a recent study has also identified four specific germplasm from 481 breeding lines with notable agronomic traits; the authors also developed a model to predict genotypic performance under different environmental conditions (Keller et al., 2020).
3.3.2 Biotic stress
The common bean is affected by many bacterial, fungal, and viral diseases, as well as insect pests. Several studies have been conducted to identify resistant sources in common bean germplasm (Table 4). A recent study has identified 14 accessions resistant to angular leaf spot (Phaeoisariopsis griseola) under field conditions (Rezene and Mekonin, 2019). Peña et al. (2013) screened 274 germplasm lines under artificial and pot conditions and identified two lines (PI 310668 and PI 533249) showing resistance against damping-off disease (Rhizoctonia solani). A set of 248 accessions of wild bean (Phaseolus spp.) were screened under greenhouse, pot, and artificial conditions against fusarium root rot (Fusarium cuneirostrum) and fusarium wilt (Fusarium oxysporum), resulting in the identification of 21 and 16 lines resistant to fusarium root rot and fusarium wilt, respectively (Haus et al., 2021). Urrea and Harveson (2014) carried out screening of 467 germplasm lines against bacterial wilt (Curtobacterium flaccumfaciens pv. flaccumfaciens) under pot and artificial conditions, and identified PI 325691 as a resistant line to the disease. The GRIS portal also indicates that several accessions have been registered as resistant to important diseases, such as anthracnose (IC0341862, IC635031, and IC635032), white mold disease (EC271515 and IC278744), and bean common mosaic virus (IC340947 and IC0360831).
3.3.3 Abiotic stress
The common bean is severely affected by abiotic stresses, such as cold, drought, heat, and salinity, and not a great deal of work has been carried out to identify trait-specific donors, with a few exceptions. Urrea and Porch (2009) screened 277 accessions of P. vulgaris and P. acutifolius under terminal drought stress conditions at Mitchell. The G35346 line has been identified for aluminum (Al) tolerance and used to transfer Al tolerance to common bean varieties (Butare et al., 2012). Tepary bean (P. acutifolius Gray), a relative of common bean, is known to have comparatively better tolerance for drought and sub-zero temperatures; on this basis and through preliminary screening of tepary bean accessions, one accession (W6 15578) has been identified as a potential donor for tolerance of both these stresses (Souter et al., 2017). Additionally, Dasgan and Koc (2009) screened 64 lines at 125 mM NaCl to identify salt-tolerant donor lines; a good level of variation was observed, and five highly tolerant genotypes were identified: Yalova 5, TR68587, Kibris Amerikan, Magnum, and Yerhammadisi.
3.3.4 Nutritional quality
Common bean is an excellent source of protein, dietary fiber, vitamins, and minerals. Its grains are a rich source of water-soluble vitamins, particularly thiamin, riboflavin, niacin, and folic acid. Analysis of a Chilean bean core collection of 246 accessions revealed protein content ranging from 183.5 to 259.7 g kg−1, Fe content from 68.9 to 152.4 mg kg−1, and Zn content from 27.9 to 40.7 mg kg−1 (Paredes et al., 2009). Kaur et al. (2009) also studied the physicochemical, hydration, textural, and cooking properties of common bean, observing a wide range of variation in terms of seed density (0.51–2.15 g/ml), hydration capacity (0.03–0.62 g/seed), hydration index (0.16–0.97), swelling capacity (1.24–1.93 ml/seed), cooking time (50–120 min), and amylose content (0.09%–5.02%). Another study revealed the ranges of variation in common bean for antioxidant activity (5.5%–44.9%), starch content (17.4%–40.7%), size of starch granules (1.64–176 μm), rapidly digestible starch (11.1%–19.5%), slowly digestible starch (8.5%–17.3%), and resistant starch (63.9%–76.1%; Sharma et al., 2015). Common bean is well known for rich diversity in seed coat color, and this color plays a major role in the selection, taste, and palatability of particular genotypes. Therefore, to investigate the relationship between color and protein and mineral content, a study was conducted in 100 genotypes having carioca, black, and other grain color patterns (Silva et al., 2012). The results indicated that black-colored beans are richer in protein, iron, and zinc; carioca grains are richer in manganese and magnesium; and grains of other colors are rich in calcium. Ciat-A-257, Bolinha, Iapar 81, Linea 29, and Roxo PV were found to be rich in protein (28.95%–30.40%). Additionally, 206 accessions from the Andean Diversity Panel were evaluated on cooking time, and five accessions (ADP0367, ADP0521, ADP0469, ADP0518, and ADP0452) were identified as promising in terms of shorter cooking time (Cichy et al., 2015b). Germplasm was also compared on nutritional composition and cooking characteristics with its closely related cultivated species, the tepary bean (Phaseolus acutifolius), in order to identify superior donors, as the latter species is highly tolerant to abiotic stresses (Porch et al., 2017). The results of this study indicated that there were no species-level differences on most nutritional parameters, with the exception of shorter cooking times for tepary bean accessions (Porch et al., 2017).
3.4 Pigeon pea
3.4.1 Agronomic traits
Pigeon pea is a legume of Indian origin (Ambika et al., 2022), and India remains its largest producer and consumer (Bohra et al., 2012). Pigeon pea is a resource-rich crop in terms of genetic and genomic resources, whole genome sequencing information, availability of trait-specific germplasm, genetic stocks, etc. The largest collection of pigeon pea germplasm is currently conserved at the ICRISAT gene bank (13,632 acc.), followed by the Indian National Gene Bank, ICAR–NBPGR (11,210 acc.); these collections are the major resources for trait identification and crop improvement. To enhance germplasm utilization, a set of 1,290 pigeon pea accessions (Reddy et al., 2005) has been developed, followed by a mini-core set of 146 accessions (Upadhyaya et al., 2006) and a composite core set of 1,000 accessions, plus a reference set of the most diverse 300 accessions (Upadhyaya et al., 2011a). An exceptionally good level of phenotypic variation has been observed for traits such as pods/plant, number of racemes, plant height, seed yield/plant, and days to maturity (Reddy et al., 2005). Promising accessions included in the pigeon pea composite core set have been listed for important economic traits, such as early flowering, a large number of pods/plant, seed weight, and yield/plant (Upadhyaya et al., 2011b). A vast amount of variability in flowering period has been observed, and a number of genotypes have been reported to show exceptionally short and long flowering durations. ICPL 90011 is reported to be an extra-short duration genotype with the lowest photoperiod sensitivity (Silim et al., 2007). Diverse trait-specific germplasms have been identified for use as potential sources for improvement programs (Upadhyaya et al., 2007c; Mir et al., 2014; Yohane et al., 2020). As of December 2022, 55 pigeon pea germplasms have been registered with the GRIS portal for a range of unique traits, including genetic male sterility (IC296750), cytoplasmic genetic male sterility (IC471860, IC471861, IC296590, IC296592, IC555904, etc.), cytoplasmic male sterility (IC296625, IC296623, etc.), fertility restoration (IC296805, IC296806, IC296807, etc.), early maturity (IC0587711, IC0587712), open flower (IC0573418, IC0573419, IC0573420), determinate growth habit (IC296589), and several other important traits.
3.4.2 Biotic stress
Pigeon pea production is adversely affected by many insects and diseases, such as wilt (Fusarium udum Butler), sterility mosaic virus (PPSMV) disease, phytophthora blight (Phytophthora drechsleri f. sp. cajani), Gram pod borer (Helicoverpa armigera), pod fly [Melanagromyza obtusa (Malloch)], and spotted pod borer [Maruca vitrata (Geyer)]. In one experiment, Saxena et al. (2002) evaluated 271 accessions under natural field conditions, and found that disease severity scores in pigeon pea germplasm ranged from 3 to 9. Screening against fusarium wilt and sterility mosaic disease (SMD) was carried out under artificial conditions for multiple seasons, resulting in the identification of several resistant accessions, viz., ICP 6739, ICP 8860, ICP 11015, ICP 13304, and ICP 14819 (Sharma et al., 2012). Accession ICPW 94 of the wild species C. scarabaeoides has been identified as resistant to all isolates of SMD, and is used in crossing programs (Hema et al., 2014). Earlier similar studies were also conducted using petiole grafting and artificial conditions in search of donors for SMD resistance in pigeon pea (Kulkarni et al., 2003; Sharma et al., 2015). Several sources of resistance for various biotic stresses in pigeon pea are listed in a review by Sultana et al. (2021) and in Table 4.
3.4.3 Abiotic stress
Pigeon pea is considered to be a drought-tolerant crop due to its deep root system and wide range in maturity period, which allows it to fit into a wide range of environments and cropping systems (Choudhary et al., 2011). Major abiotic stresses limiting pigeon pea productivity are waterlogging, drought, low temperatures (<10°C), and photoperiod sensitivity. Through several germplasm evaluation programs in pigeon pea, a number of popular donors have been identified; these are major sources for abiotic stress tolerance (Choudhary et al., 2018). In one study, 96 pigeon pea accessions were identified for early flowering; these are considered potential sources for the breeding of early-maturing pigeon pea varieties in order to avoid terminal drought and heat stress (Upadhyaya et al., 2011b). Sultana et al. (2013) screened 272 pigeon pea lines for waterlogging stress tolerance under laboratory and field conditions, and identified 12 lines tolerant to waterlogging. Similarly, in another study conducted under pot conditions for multiple seasons, 24 pigeon pea accessions were identified as waterlogging-tolerant (Krishnamurthy et al., 2012). Various other sources for resistance to important abiotic stresses are also listed in a review by Sultana et al. (2021) and in Table 5.
3.4.4 Nutritional quality
Pigeon pea contains approximately 86.6%–88.0% dry matter, 19.0%–21.7% crude protein, 1.2%–1.3% crude fat, 9.8%–13.0% crude fiber, and 3.9%–4.3% ash content (Amarteifio et al., 2002). Pigeon pea mineral content (mg/100 g dry matter) ranges are as follows: 1845–1941 K, 163–293 P, 120–167 Ca, 113–127 Mg, 11.3–12.0 Na, 7.2–8.2 Zn, 2.5–4.7 Fe, and 1.6–1.8 Cu. However, these values vary with genotype and across different studies (Talari and Shakappa, 2018). Biochemical evaluation of a total of 55 genotypes comprising advanced lines, improved cultivars, and landraces resulted in the identification of variation in four parameters: crude protein content (16.7%–28.4%), total phenol (21.9–84.4 mg/100 g), total flavonoid (16.4–33.4 mg/100 g), and total antioxidant activity (19.2–82.5 mg/100 g) (Cheboi et al., 2019).
3.5 Field pea
3.5.1 Agronomic traits
Field pea is cultivated in over 100 countries for fresh and dry grains and for fodder. Over 31,000 germplasm accessions of Pisum are conserved ex situ in various gene banks, including the Australian Grains Genebank, Australia; the Western Regional Plant Introduction Station, USDA, United States of America; the Leibniz Institute of Plant Genetics and Crop Plant Research, Germany; and the ICAR–NBPGR, New Delhi, India. Although a limited number of large-scale studies have been conducted on the agro-morphological characterization of field pea and for trait identification, several studies nevertheless indicate a substantial amount of phenotypic variability on qualitative as well as quantitative traits, such as days to 50% flowering, seed weight, plant height, and number of pods/plant (Azmat et al., 2011; Bhuvaneswari et al., 2017). The accessions IPF–5–19, EC 8495, HUDP–15, and DDR–30 have been found to show promise in terms of seed yield (Bhuvaneswari et al., 2017). Singh et al. (2010) evaluated 71 accessions on agronomic performance and seed and flower characteristics, identifying promising accessions in terms of early flowering (IC279013), early maturity (IC394017), a large number of pods/cluster (IC279195), longer pods (IC279013), pods/plant (IC219027), seed yield (IC279082), and seeds/pod (IC394028). Genotypes with five flowers per peduncle (VRPM–901–5) and three flowers per peduncle at multiple flowering nodes have been reported in garden pea, which could be highly useful in field pea improvement (Devi et al., 2018). Several unique trait-specific pea accessions have been registered in the GRIS portal, such as IC296677 (leafletless, dual purpose, and high-yielding), IC296678 (dwarf, leafletless), IC296737 (male sterile line governed by a single gene), IC279125 (bold seed with 50.14 g 100 seed weight), IC0610501 and IC630592 (≥ three pods/peduncle), IC636671 and IC640781 (extra-early flowering), and EC414478 (extended funicle).
3.5.2 Biotic stress
The major biotic stresses affecting field pea are powdery mildew (Erysiphe pisi), rust (Uromyces viciae-fabae), ascochyta blight (complex of Ascochyta spp.), white rot (Sclerotinia sclerotiorum (Lib) de Bary), wilt (Fusarium oxysporum f. sp. pisi), root rot (many pathogenic fungi), and collar rot (Sclerotium rolfsii). Screening against pea weevil (Bruchus pisorum L.) in 602 field pea lines, primarily from the Ethiopian Institute of Biodiversity (EIB), Addis Ababa, Ethiopia, resulted in the identification of four resistant lines: 32454, 235002, 226037, and 32410 (Teshome et al., 2015). Large-scale germplasm screening against powdery mildew disease under natural epiphytotic conditions has also been carried out, with the germplasm lines EC598655, EC598878, EC598704, IC278261, and IC218988 being identified as promising (Rana et al., 2013). Nisar et al. (2006) also reported three germplasm lines (Fallon, PS99102238, and PS0010128) to be highly resistant against powdery mildew.
3.5.3 Abiotic stress
Cold, frost, salinity, and heat stresses are the major sources of abiotic stress in field pea crop production. Many studies have taken up the aim of developing lines tolerant to abiotic stresses. In one such study, five field pea germplasm (ATC 104, ATC 377, ATC 968, ATC 3992, and ATC 4204) were identified as frost-tolerant at the reproductive stage through screening of 84 accessions under controlled environmental conditions (Shafiq et al., 2012). Screening of 3,672 pea germplasm lines under field conditions led to the identification of 214 cold-tolerant lines (Zhang et al., 2016). Additionally, 780 accessions were screened for salinity stress tolerance under artificial conditions (Leonforte et al., 2013). Finally, in a recent study, IPFD 11–5, Pant P–72, P–1544–1, and HUDP 11 were identified as heat-tolerant lines based on evaluation under timely- and late-sown field conditions (Lamichaney et al., 2021).
3.5.4 Nutritional quality
Field peas in general have lower protein content (∼25%), very low fat content (∼0.1%), and very high carbohydrate content (∼70%). Major yield-attributing traits in field pea are pods/plant, number of grains/pod, and seed weight. In one study, 94 pea genotypes were examined for pea carotenoid content; higher carotenoid content (10–27 μg/g) was observed in accessions with green cotyledons, and comparatively low carotenoid content (5–17 μg/g) in accessions with yellow cotyledons (Ashokkumar et al., 2015). Pea grains have comparatively higher antioxidant activity than chickpeas. Promising field pea accessions have also been identified in terms of mineral content, such as Zn (IG52442, IG134828), Cu (IG116297, IG52442) and Ca (IG51520, IG52442), by Costantini et al. (2021). Additionally; Singh et al. (2010) have identified lines with shorter cooking time (IC260344) and observed that the genotypes that absorb more water and swell more during soaking require less cooking time. The authors have also identified IC320964 as superior in terms of ash content (3.73%), and several other accessions as promising in terms of their physicochemical properties. In a nutritional analysis of 96 accessions from diverse collections at the USDA National Germplasm Center, Pullman, WA, a wide range of variation was observed in mineral micronutrient content (Hacisalihoglu et al., 2021). An atypical morphotype having extended funicle (EC0414478) was identified in pea germplasm, and this accession was found to be associated with faster water uptake in comparison to the checks included (Tripathi et al., 2021b); this is likely to help with the development of pea cultivars with shorter cooking times.
3.6 Cowpea
3.6.1 Agronomic traits
Cowpea is a multi-purpose grain legume (yielding grains, green pods, and leaves) and is widely cultivated in Asia, Africa, and America. It is considered to be one of the best suited crops for hotter, semi-arid agro-climatic conditions, as it requires less water and also grows well in sandy soils. The germplasm conserved in various gene banks has exhibited a good amount of genetic variability, which enables it to grow in various agro-climatic regions and in various soil types. To enhance the utilization of cowpea germplasm, over 12,000 accessions of cowpea were characterized on 28 agro–botanical descriptors at the International Institute of Tropical Agriculture (IITA), Ibadan, and a core set of 2,062 accessions was developed (Mahalakshmi et al., 2007). In another study, 4,000 accessions were characterized in multi-location trials by the ICAR-National Bureau of Plant Genetic Resources (unpublished records). A great deal of variability was observed in plant and seed morphology. Gerrano et al. (2015) identified germplasm lines having desirable grain yield characteristics, such as Fahari, IT93K129-4, Glenda, and vegetable cowpea dakama cream; Nkhoma et al. (2020) identified lines Bubebe, CP411, CP421, CP645, Chimponogo, and MS1–8–1-4 as high-yielding and genetically divergent among 90 genotypes studied, making them ideal parental lines. Cowpea genotypes IT96D-604, 93K-619-1, IT97K-569-9, and IT99K-1060 have also been identified as high-yielding (Goa et al., 2022).
3.6.2 Biotic stress
The major diseases affecting cowpea are cowpea mosaic virus (CpMV) disease, Cercospora leaf spot (CLS), brown blotch (Colletotrichum capsici), and bacterial blight (Xanthomonas axonopodis pv. vignicola), while the major pests are pod borer, aphids, thrips and bruchids. The severity of these biotic factors varies with agro-climatic zone and growing conditions. Although cowpea is one of the more prominent legume crops and the largest of the Vigna group, it has not received commensurate research attention. As a result, cowpea improvement has suffered from a lack of reliable donors for resistance to many of these biotic factors. Nonetheless, efforts have recently been undertaken in this direction, and several important and promising donors for resistance to a small number of these biotic stresses have been identified; these are listed in Table 4. In a study that aimed to identify resistant donors for aphid (Aphis craccivora), cultivated germplasm (105 accessions) and wild germplasm (92 accessions) were screened under greenhouse conditions; only a single accession (TVNu 1,158) was identified as a resistant line (Souleymane et al., 2013). The findings of this study also indicated that both the cultivated and the wild relatives of this crop have poor genetic bases. Boukar et al. (2019) identified 14 lines having resistance to bacterial blight (Xanthomonas axonopolis pv. vignicola) under artificial inoculation. In another study, 225 germplasm lines were screened against CpMV, CLS, and cowpea rust (Uromyces vignae), resulting in the identification of promising accessions for resistance to these pathogens (Deshpand et al., 2010; Table 4). Finally, Tripathi et al. (2020) identified EC528425 and EC528387 as tolerant to bruchid (Callosobruchus maculatus) through the screening of 103 cowpea lines using a ‘no-choice’ test method.
3.6.3 Abiotic stress
The major abiotic stresses are drought, heat stresses, and poor soil fertility, especially in sub-Saharan Africa (SSA), where cowpea is grown as a major crop, as well as soil salinity in almost all irrigated areas worldwide (Horn and Shimelis, 2020). Several studies have been conducted to identify resistant donors (see Table 5). Five lines with superior drought stress tolerance (viz., TVu1436, TVu9693, TVu12115, TVu14632, and TVu15055) have been identified using the water withdrawal method under field conditions (Fatokun et al., 2012), while Dagupan Pangasinan, UCR 369, and Negro have been identified as tolerant to waterlogging at the seedling stage (Olorunwa et al., 2022). Accessions EC472250, EC472267, EC472285, EC472286, EC472289, and Pusa Komal have been identified as tolerant to heat stress through screening in multiple seasons under field conditions (Mishra et al., 2005). Accessions PI582422, 09–529, PI293584, and PI582570 have been identified as tolerant to salinity stress under artificial screening conditions through imposition of salinity stress (150 mM NaCl) at the seed germination stage (Ravelombola et al., 2017). Other similar studies have also identified lines tolerant to salt stress using different NaCl concentrations (150 mM and 120 mM) at the germination and seedling stages (Ravelombola et al., 2018; Dong et al., 2019). Based on screening of 155 cowpea lines in 200 mM NaCl, several promising lines were identified as salt tolerant, i.e., PI354686, PI353270, PI354666, PI354842 PI548785, PI582466, PI339599, and 09-697 (Dong et al., 2019).
3.6.4 Nutritional quality
Cowpea is a major source of nutrition in sub-Saharan Africa, Asia, and Latin America. Based on nutritional profiling of 100 breeding lines on a dry weight basis, a significant range of variation has been observed in terms of protein content (22.9%–32.5%), ash content (2.9%–3.9%), fat content (1.4%–2.7%), and carbohydrate content (59.7%–71.6%; Nielsen et al., 1993). Genotypes also vary in 50% cooking time, which ranges from 21.1 to 61.9 min, and promising donors have been identified, such as IT83S–872 for protein content, IT84S–2085 and IT86D–466 for ash content, and IT85F–2805 for shortest cooking time (Nielsen et al., 1993). In a study aiming to investigate nutritional variability in immature cowpea pods, 22 genotypes were analyzed on various nutritional composition parameters; genotypes such as ITOOK-1060, TVU-14196, and 98K–5301 were found to be superior on such parameters as Mg, Na, Mn, Boron, Al, Zn, Cu, K, P, and protein (Gerrano et al., 2017). The fresh young leaves of cowpea are also consumed in several countries; therefore, analyses have been conducted of the nutritional composition of 15 varieties and the sensory attributes of 10 varieties (Ahenkora et al., 1998). In this study, nutrient concentration in cowpea leaves on a dry weight basis was found to range from 303.8 to 468.9 mg/100 g for phosphorus, from 33.5 to 148.0 mg/100 g for ascorbic acid, and from 27.1% to 34.7% for protein.
3.7 Black gram
3.7.1 Agronomic traits
Black gram is a grain legume of Indian origin, primarily cultivated in South Asian regions. Although black gram is an important legume, its productivity level is very poor compared to that of other legumes, which can mainly be attributed to a lack of good plant ideotypes and resistance sources for major diseases in its cultivated gene pool (Kumar and Singh, 2014; Shanthi et al., 2019; Subramaniyan et al., 2022). Therefore, to identify donors for desired agro-morphological traits, 484 accessions have been characterized on qualitative and quantitative traits; a good deal of variation was observed in flowering and maturity period and in yield (Ghafoor et al., 2001). Recently, 840 accessions of black gram were also characterized, resulting in the identification of promising germplasm lines in terms of early flowering (IC343936, IC436615), synchronous flowering (IC73523, IC396032, IC485444), pod length (IC438379), number of seeds/pod (IC472051_2, IC565238), and seed weight (IC485605_2, IC485588) (Gayacharan et al., 2022). For novel trait generation in black gram, gamma-irradiated mutants were generated using black gram cultivars ADT 3, Co 6, and TU 17-9, which have exhibited high plant yield (Dhasarathan et al., 2021). Additionally, RBU1012 and Pant U-19 have been found to be the most stable genotypes in terms of yield when evaluated under field conditions (Singh N. P et al., 2016). The GRIS portal indicates that unique germplasms of black gram such as IC296878 (dwarf with ground pod bearing habit), IC553269 (brown pods with yellow seeds), IC594172 (male sterile flowers with protruded stigma and crumpled petals), IC594173 (sympodial pod-bearing habit), IC426765 (photosensitive), and IC636672 (extra-early maturing) have been registered for important traits.
3.7.2 Biotic stress
Urdbean leaf crinkle disease (ULCD) and mungbean yellow mosaic disease (MYMD) are the two major diseases of the black gram crop. Yield losses may reach or exceed 60%, depending on the susceptibility of the host plant, if the crop is affected in its early vegetative stage. Nevertheless, unlike green gram, black gram has a high level of resistance against MYMD in its cultivated gene pool, as revealed in a large-scale evaluation under field and artificial conditions conducted during 2019 and 2020 (unpublished records). Several black gram sources of MYMD resistance, identified on the basis of field screening, are highlighted in a review published by Mishra et al. (2020a). Urdbean leaf crinkle virus (ULCV) disease has spread across all the cropping systems in India, and yield losses can reach 100% if the disease outbreak occurs at the early growth stage under favorable weather and host genotype conditions (Biswas et al., 2009). Resistance sources for ULCV have been reported by several researchers (Ashfaq et al., 2007; Gautam et al., 2016); several such sources for this and other diseases are listed in Table 4. In the GRIS database, accessions IC0570267, IC0570268, IC0570269, IC11613, IC636672, IC0144901, and IC485638 are registered as MYMD resistant, and IC0585931 as bruchid resistant. Powdery mildew (Erysiphe polygoni) and Cercospora leaf spot (Cercospora canescens) are the other major diseases of black gram. The major pests affecting this crop include spotted pod borer (Maruca testulalis r), whitefly (Bemicia tabaci), bruchids (Callosobruchus chinensis. and C. maculatus), and nematodes (Meloodogyne incognita, M. javanica, and Heterodera cajani), for which reliable sources of resistance are lacking. Bruchids begin infesting the crop during the pod maturity stage, and they are the cause of up to 90% of produce losses (Soundararajan et al., 2012).
3.7.3 Abiotic stress
The crop is grown in a rainfed environment under tropical and sub-tropical climatic conditions. Therefore, terminal drought and heat, as well as waterlogging, are the major constraints on black gram production. Salinity is another problem, particularly in arid and semi-arid regions. Only a small number of studies have examined the potential for improvement of black gram in terms of resistance to abiotic stresses. Saline-tolerant lines, such as BARI Mash-1 (Hasan et al., 2017), VNBG 017, AUB 3, and AUB 20 (Priyadharshini et al., 2019), have been identified as promising under artificial screening conditions. Under natural waterlogging conditions during a germplasm characterization program, a small number of germplasm lines have been identified as tolerant; these were further evaluated under artificial waterlogging conditions, and accessions IC530491 and IC519330 were found to be tolerant to waterlogging (Bansal et al., 2019). In another study, 26 genotypes were analyzed under waterlogging stress; a large amount of variation was observed in various quantitative traits, and BU Acc 25, BU Acc 17, and BU Acc 24 were identified as the strongest performers in terms of yield (Rana et al., 2019). In terms of drought stress tolerance, cultivars VBN4 and K1 have been identified as promising based on protein and biochemical analyses (Sai and Chidambaranathan, 2019).
3.7.4 Nutritional quality
Black gram grains are a rich source of protein (22%–26%) and moderately high in calories (ca. 350 cal/100 g), carbohydrates (ca. 56.6%), and fat (1.1%–1.2%) (Panhwar, 2005; Suneja et al., 2011). They also contain vitamins, viz., Vit. B1 (0.42 mg/100 g), Vit. B2 (0.37 mg/100 g), Niacin (2 mg/100 g), and minerals, viz., Ca (185 mg/100 g), Fe (8.7 mg/100 g), and P (345 mg/100 g) (Panhwar, 2005). However, a limited amount of germplasm has been nutritionally profiled for the identification of nutrient-rich lines. Black gram is reported to exhibit a substantial amount of variation in nutrient content between the whole grain and its milled fraction (Girish et al., 2012). A small number of genotypes among 26 investigated, such as Shekhar 2, have been found to have high Fe and Zn content, and genotypes Yakubpur, PU 31, IPU 99–200, and PDU 1 have been found to have high polyphenol content (Singh J et al., 2017). There is a need for large-scale nutritional profiling to develop an understanding of nutritional variability in the germplasm and to identify superior genotypes with minimal levels of anti-nutritional factors to enhance the palatability of the crop.
3.8 Green gram
3.8.1 Agronomic traits
Green gram is a highly nutritious and palatable grain legume cultivated in Asia, primarily for its grains. Green gram cultivation faces constraints such as a narrow genetic base in the cultivated gene pool, a lack of ideal plant type, and many biotic and abiotic stresses. In order to identify new donors, 1,532 ex situ collections of green gram conserved in the Indian National Gene Bank were characterized, potential donors for certain agro-morphological traits were identified, and a core set of 152 accessions was also developed (Bisht et al., 1998). A good level of variation was observed in branch length, nodulation, number of pods bearing a peduncle, number of pods per plant, and yield per plant. The World Vegetable Center, Taiwan, holds over 6,700 accessions of green gram, which have been utilized for the development of a core set of 1,481 accessions based on geographic stratification and clustering of genotypes on eight phenotypic traits (Schafleitner et al., 2015). This core set was genotyped using 20 microsatellite markers, and a mini-core set of 289 accessions was developed; this is now extensively utilized for trait identification. In another large-scale characterization and preliminary evaluation of green gram germplasm, promising germplasm lines were identified in terms of early flowering (EC398944, EC398883), synchronous flowering (EC396115, IC76414, and IC488968), greater pod length (EC398937), seed weight (EC398903, EC398884, and EC396413), etc. (Gayacharan K. et al., 2020). Recently, the entire green gram ex situ collection of the Indian National Gene Bank has been characterized in multi-location trials, and a diverse core set of 400 accessions has been developed (unpublished records). Photoperiod-insensitive genotypes (EC 318985–319057) have also been identified in green gram (Pratap et al., 2014; Pratap et al., 2019). The GRIS portal lists a number of accessions with unique traits, such as a photosensitive nature (IC546478), high seed weight (IC418452), early maturity (IC0589309, IC589310, IC39289, and IC639796), and penta-foliate leaves (IC296679).
3.8.2 Biotic stress
Green gram production is affected by biotic stresses such as yellow mosaic disease (YMD), pod borers, and storage pests. YMD is a comparatively new disease in green gram and is spreading rapidly into new areas, which is a cause for concern. In YMD-susceptible genotypes of green gram, yield losses up to 85% are reported (Karthikeyan et al., 2014), but it has been observed that losses may reach 100% if the crop is infected at seedling stage. Resistance sources are lacking in the entire cultivated gene pool of the crop, as revealed in a field screening of 4,100 accessions at New Delhi (a YMD hotspot location). However, variability in the severity of the disease is observed according to multiple factors, such as genotypic constitution, vector population load, weather conditions, presence of multiple virus strains, etc. (unpublished record). Similar reports have also made by other researchers based on germplasm screening (Shad et al., 2006). Several resistant sources for YMD are listed in a review (Mishra et al., 2020a). There are also several reports of YMD resistance in green gram under field conditions (Iqbal et al., 2011; Mohan et al., 2014; Nainu and Murugan, 2020). Aside from YMD, Duraimurugan et al. (2014) identified four lines (viz. LM 131, V 1123, LM 371, and STY 2633) as resistant against bruchid beetle based on a ‘free choice’ and ‘no choice’ test method. Spotted pod borer (Maruca vitrata) also causes severe damage to the crop, if not controlled at the appropriate stage of crop growth, and there are no resistant sources available for this pest. Sandhya et al. (2014) have reported that KM–9–128, KM–9–136, RMG–492, LGG–5, and LGG–538 are tolerant to Maruca vitrata following field screening of 110 genotypes.
3.8.3 Abiotic stress
Green gram is primarily grown under rainfed conditions; thus, abiotic stresses such as drought, waterlogging, heat, and salinity affect crop production (Singh and Singh, 2011). In general, reliable tolerant donors for these abiotic stresses are lacking in this crop. Forty-one elite lines were screened for heat stress tolerance under late-sown conditions; of these, five lines (viz., EC693357, EC693358, EC693369, Harsha, and ML1299) showed heat stress tolerance (Sharma et al., 2016). Additionally, IC333090 and IC507340 were found to be drought tolerant, out of 100 lines screened under hydroponics conditions (Meena et al., 2021). Mung bean lines OBGG-2013-9 and OBGG-2013-14 have also been reported to exhibit cold tolerance (Kabi et al., 2017).
3.8.4 Nutritional quality
Green gram is nutrient-rich and possesses various health benefits, such as antioxidant, anti-cancer, anti-inflammatory and hypolipidemic activity (Sudhakaran and Bukkan, 2021). Because of its high nutritional value, green gram is regarded as “green pearl” (Nair et al., 2013). It contains approximately 19.7%–29.1% protein (Bartwal et al., 2022), 61%–63% carbohydrates, 1.1%–2.3% fat, 3.2%–4.2% ash, 0.03–0.06 g Fe kg−1, and 0.02–0.04 g Zn kg−1 (Nair et al., 2013; Sudhakaran and Bukkan, 2021). The nutritional composition of green gram and black gram is very similar, but green gram is reported to have higher moisture, fat, and protein content (Shaheen et al., 2012). A small number of accessions with particularly high nutritional value are listed in the GRIS portal; these could potentially function as donors for nutritional improvement of the crop. Specifically, accessions IC296771 (27.8%) and IC573456 (25.8%) are registered for high protein content, and IC573449, IC573450, IC573451, IC573453, and IC573454 for high Fe and Zn content.
3.9 Horse gram
3.9.1 Agronomic traits
Horse gram is one of the least utilized and least studied legumes. The crop is known for its nutritional and therapeutic value, and is primarily cultivated in hill states and dry areas of southern India. This crop has failed to attract the attention of breeders and researchers due to a lack of ideal ideotypes and morphological variation. A small number of characterization and evaluation studies have been conducted, indicating comparatively wide variation in terms of plant height, pod length, seed test weight, and pods per plant (Singh et al., 2019). Additionally, genotypes CRHG-6 and CRHG-8 are of the non-shattering type, which has been developed through mutation breeding (Salini et al., 2014). In a characterization study examining seven qualitative and quantitative traits in 66 horse gram genotypes, a good amount of variability was observed for pod length and pods per plant (Gomashe et al., 2018). Priyanka et al. (2021) studied 12 quantitative traits across 252 horse gram genotypes, reporting that the highest yield was 65.61 g per plant. Another characterization and evaluation study of 51 accessions led to the identification of several promising accessions (viz., S44/L23, S56/L29, S8/L4, S96/L49, and S29/L14) in terms of early flowering and maturity (Kaundal and Kumar, 2021). In the GRIS portal, only one accession (IC587788) is registered for high fodder yield.
3.9.2 Biotic stress
Horse gram is susceptible to various biotic stresses and still lacks resistant donors for use in crop improvement programs. Only a small number of studies have been carried out to identify resistant donors for selected diseases and pests, such as YMD, wilt, anthracnose, and storage pests. Parimala et al. (2011) identified horse gram accessions AK-38, HG-14, HG-52, HG-59, HG-63, HG-75 as having resistance against horse gram YMD, and AK-38 and HG-46 as resistant to powdery mildew disease. In another study, accessions HG 63, HG 58, HG 50, and Palem 2 were identified as resistant to wilt disease (Durg, 2012). Accession IC470275 has also been identified as resistant to anthracnose disease (Colletotrichum dematium) (Sankar et al., 2015). Finally, horse gram lines Palem-1, Palem-2, AK-21, and NSB-27 have been identified as resistant against Callosobruchus chinensis, a storage pest (Divya et al., 2012; Divya et al., 2013). Accession IC587786 is registered on the GRIS portal as resistant to anthracnose disease.
3.9.3 Abiotic stress
Horse gram germplasm have been screened against abiotic stresses, such as drought, salinity, moisture, and heavy metal stress. Several germplasm lines, such as M-249 and HPK-4, have shown resistance against drought stress (Bhardwaj and Yadav, 2021). Yasin et al. (2014) identified a line tolerant to moisture stress, namely D13. The variety Paiyur-2 was found to have high proline and glycine betaine content and lower lipid peroxidation under salinity stress (Kanagaraj and Sathish, 2017). This genotype was further tested for antioxidant activity, and was found to exhibit enhanced antioxidant activity under salinity stress (Desingh and Kanagaraj, 2019). Separately, the same Paiyur-2 variety was found to be promising for salinity tolerance, and heavy metal tolerance was observed in Madhu (for chromium) and in HGR-4 (for nickel; Dhali et al., 2021; Edulamudi et al., 2021). Based on a screening of 88 germplasm lines for biochemical parameters, accessions TCR491, IC110286, IC56145, and IC53641 were identified as suitable for environments imposing drought stress (Sharma and Chahota, 2022).
3.9.4 Nutritional quality
Horse gram is used as a food and fodder crop and is known for its medicinal and therapeutic uses. It provides protein (17.9%–25.3%), carbohydrates (51.9%–60.9%), lipids (0.58%–2.06%), and vitamins, such as riboflavin, niacin, and vitamin C (Jha et al., 2022). The protein content of horse gram is relatively high compared to that of green gram, black gram, dry peas, the kidney bean, chickpea, or pigeon pea (Longvah et al., 2017). As consumers have become more health conscious, consumption of sprouted horse gram seeds has increased. In the seeds, albumin–globulin is a major contributor (∼75.27–78.76%) to the total protein content. The seeds are low in fatty acid content but rich in dietary fiber, required for proper functioning of the lower intestine (Kawale et al., 2005). Horse gram seeds also exhibit antioxidant activity and radical scavenging activity. In a recent study, metabolic profiling was conducted for 96 accessions of horse gram, which were derived from 700 accessions spread across the entirety of India (Gautam and Chahota, 2022). Tremendous variability in protein content was observed, with the lowest protein content (13%) being found in IC120837 and TCR-1439, while a related wild species (Macrotyloma sar-gharwalensis) had the highest protein content (40%). Accessions IC280031 and IC139356 were found to be most nutritive, as the largest number of metabolites (44) was observed for these among the 96 lines selected in an analysis using 1H NMR spectroscopy (Gautam and Chahota, 2022). An earlier study had also identified the species Macrotyloma sar-gharwalensis (IC212722) as containing 34.88% protein (Yadav et al., 2004).
3.10 Moth bean
3.10.1 Agronomic traits
Moth bean is considered to be a hardy crop suitable for hot, arid regions. In addition to this, it also helps to reduce soil erosion, particularly in sandy deserts, due to its extensive root system and profuse foliage cover. Most local cultivars continue to have wild traits, such as pod-shattering, a trailing and spreading growth habit, asynchronous maturity, and a photo-sensitive nature. Few studies have been conducted to explore the existing variability in the gene pool. However, a good amount of phenotypic variability in moth bean was reported by Chaudhary et al. (2021) in an evaluation of 40 genotypes on 10 morphological traits. In another study, accessions IC 36607, IC 39675, IC 251908, IC 36563, and IC 36245 exhibited higher seed yield as compared to checks (Vir and Singh, 2015). Similarly, 50 genotypes of moth bean were phenotyped on 12 quantitative traits and exhibited high levels of variability (Sahoo et al., 2019). Additionally, Singh S et al. (2017) have developed a variety (RMO 257) with superior agronomic traits, such as plant height, dry matter accumulation, and seed yield. A small number of moth bean genotypes are registered in the GRIS portal for early maturity (IC432859 and IC120963) and for single stem formation (IC432859).
3.10.2 Biotic stress
The major biotic stresses are YMD, leaf crinkle disease (LCD), bacterial leaf spot (Xanthomonas phaseoli), Cercospora leaf spot (Cercospora dolichi), charcoal rot (Macrophomina phaseolina Tassi), pod borer, and bruchids. Accessions IC36522 and IC36217 have been identified as YMD resistant as a result of screening under field conditions in multiple seasons (Singh et al., 2020). Meghwal et al. (2015) screened 204 germplasm lines and identified 14 accessions resistant to YMD. Resistance to leaf crinkle virus and Cercospora leaf spot has also been reported in the crop (Singh et al., 2020). Vir and Singh (2015) identified LCD resistance in moth bean under field conditions in multiple seasons.
3.10.3 Abiotic stress
Moth bean is one of the best-suited crops for arid and semi-arid environments, and is highly tolerant to drought and heat stress. Although only a small number of moth bean accessions have been investigated, a good amount of variation in the germplasm has been revealed in terms of resistance to drought and heat stress; this could be exploited for crop improvement in order to sustain its productivity amid climate change (Vir and Singh, 2015; Pal et al., 2020). Accessions such as IC129177, IC103016, IC415139, IC 415155, IC36157, Maru moth, and Jadia, which have been identified as tolerant to drought stress, can serve as donors for crop improvement (Malambane and Bhatt, 2014). Additionally, in a separate study, lines IC103016, IC36011, and IC36157 have been identified as promising for drought tolerance (Sachdeva et al., 2016).
4 Role of genomics in enhancing grain legume germplasm utilization and attaining higher genetic gains
The first step toward enhancing the utilization of grain legume crop germplasm accessions for trait discovery and subsequent genetic improvement requires thorough and extensive genotypic and phenotypic characterization of such accessions using large-scale data (Rasheed et al., 2017). The numerous germplasm resources (including landraces, wild accessions, cultivated varieties, and breeding lines) available for diverse grain legume crop species, representing diverse agro-climatic regions of the world, have been stored efficiently at various national and international gene banks and repository centers. Considering the difficulties involved in genotypic and phenotypic characterization of this vast set of available germplasm resources, efforts are currently being made to develop core and mini-core collections in the case of several legumes by identifying the greatest amount of genetic diversity that can be represented with a minimal number of accessions (Table 3). This is where the crucial role of genomics comes into play, especially as a means of producing realistic estimates of the level of molecular diversity existing among germplasm accessions, which enables efficient screening of unduplicated authentic accessions in the process of constructing core collections of grain legumes.
Tremendous technological advances made over the last decade in sequencing and other high-throughput sequence- and array-based genotyping assays have supplied much-needed momentum to germplasm characterization. Draft and reference whole genomes, resequencing information, and global transcriptome information have now been decoded for many important grain legume crop plants using first-generation Sanger sequencing and next-generation sequencing (NGS)-based second-generation short read and third-generation long read sequencing assays; the results of these are now publicly accessible (Table 6; Michael and Jackson, 2013; http://www.embl.de; http://www.ebi.ac.uk; http://www.ddbj.nig.ac.jp; http://www.phytozome.org). Grain legumes were once considered to be resource-poor crops, but recent efforts by national and international organizations has altered this trend. As a result, a vast amount of genome sequence information, including whole genome sequences, is available in public databases (Table 6). This sequence information has since been used to understand the genomic features and evolutionary characteristics of the crops in question, and also to develop a vast range of genomic resources, including molecular markers (Garg et al., 2022). Among several sequence-based molecular markers that have been made available, simple sequence repeats (SSRs) and single-nucleotide polymorphisms (SNPs) have occupied a central position, finding extensive use in allelic diversity screening and other genomics-assisted crop improvement programs due to their genome-wide distribution, multi/bi-allelic nature, and amenability to high-throughput detection and genotyping assays (Lateef, 2015). The availability of several high-throughput genotyping platforms and the rapid evolution of the chemical techniques that they employ has enhanced the precision and pace of large-scale mining and genotyping of SSR and SNP markers (Kujur et al., 2015). High-throughput genotyping of SSR and SNP markers in a larger set of germplasm accessions and core or mini-core collections of grain legumes has been expedited via the use of various array-based and NGS assays, especially automated fragment analyzer (ABI3730xl automated DNA sequencer), Illumina GoldenGate, and Infinium assays; the Fluidigm dynamic array; KASP (KBioScience Allele-Specific Polymorphism) profiling; MALDI–TOF; the Affymetrix GeneTitan SNP Chip array; and Genotyping-By-Sequencing (GBS) assay (Varshney et al., 2013a; Jain et al., 2013). Among these, the MALDI–TOF, Illumina GoldenGate, and Infinium assays, SNP Chip Array, and KASP profiling have come to be considered highly advantageous and are utilized widely for high-throughput genotyping of previously mined SNP markers in many crop plants (Gaur et al., 2012; Hiremath et al., 2012). In particular, GBS assay has been extensively utilized for simultaneous genome-wide discovery and genotyping of SNPs in diverse plant species (Sonah et al., 2013; Spindel et al., 2013). Its development has thus expedited the mining of novel functional allelic variants and their large-scale validation and genotyping at the whole genome level for efficient trait association mapping of diverse small- and large-genome grain legume crop plants.
Using the aforementioned high-throughput marker-based genotyping strategies, along with large-scale multi-environment phenotyping information, sets of 211, 146, 184, and 289 germplasm accessions belonging to core or mini-core collections of chickpea, pigeon pea, groundnut, and green gram, respectively, have been developed. These have been collated based on the 16,991, 13,632, 15,490, and 6700 accessions available for these respective crop species as a result of the efforts of international institutes such as the IRRI, ICRISAT, USDA, and the World Vegetable Center (Upadhyaya et al., 2001; 2002; Zhang et al., 2011; Schafleitner et al., 2015). These core or mini-core germplasm resources, readily available for many grain legume crop plants, are the primary sources of trait discovery once these collections have been thoroughly characterized genotypically and phenotypically for diverse traits of agronomic importance, including yield, (a)biotic stress tolerance, and nutritional quality traits. Under this perspective, candidate gene-based association mapping and genome-wide association studies (GWAS) relying on the large-scale genotyping of informative SNP markers and robust field phenotyping information on these core or mini-core germplasm lines (i.e., association panels) are now considered to be an effective approach for the identification of major and minor genes/QTLs and alleles regulating both simple qualitative and complex quantitative traits in grain legume crop plants (Varshney et al., 2011). Candidate gene-based association mapping, which is carried out using genotyping information from SNPs in various coding and regulatory sequence components of genes in a trait-specific association panel, plays a significant role in the identification of genes/QTLs controlling yield, nutritional quality, and stress tolerance traits in grain legume crops. With the availability of high-throughput genome-wide SSR and SNP marker-based genotyping information on germplasm lines belonging to an association panel, the GWAS has now become a routine approach for high-resolution scanning of the whole genome to identify target genomic regions, including genes/QTLs (both major and minor) associated with traits of agricultural importance in many grain legume crops (Varshney et al., 2017; Varshney et al., 2021). The trait-influencing molecular signatures once identified using trait association mapping are significant for their potential utilization for genomics- (marker-)assisted crop improvement programs.
The delineated molecular signatures regulating traits of agronomic importance have been utilized for introgression, combining, and pyramiding into selected grain legume crop genotypes of interest through traditional and advanced genomics-assisted breeding approaches in order to develop superior crop varieties in terms of yield and stress tolerance (Varshney, 2016). Recently, a chickpea cultivar ‘Pusa JG16’ has been released in India as a drought-hardy cultivar; this was developed through genomics-assisted breeding utilizing a QTL-hotspot region from ICC4958 (Bhardwaj et al., 2021). Introgressions of functional natural genetic variations and of favorable genes, QTLs, alleles, and chromosomal segments identified from a larger set of grain legume germplasm accessions (including landraces and wild species), particularly for yield and stress component traits, have been transferred into the cultivated genetic background for improvement of the relevant crop through the use of such approaches as introgression lines (ILs), advanced-backcross QTL (AB–QTL) analysis, association genetics, and multi-parent advanced generation intercross (MAGIC) populations (Roorkiwal et al., 2020; Bohra et al., 2021). For example, the ‘Geletu’ chickpea variety was developed through marker-assisted back-crossing (MABC) and released in Ethiopia; it provided a yield advantage of 15% over the check variety ‘Teketay’ and 78% over a local check (https://www.icrisat.org/). The MABC technique has been used in the development of introgression of QTLs into elite cultivars in order to develop introgressed lines (Bharadwaj and Yadav, 2012; Varshney et al., 2013b; Barmukh et al., 2022). The ‘Pusa Chickpea 20211’ variety is another example in which resistance genes for multiple races (foc 1,2,3,4, and 5) of fusarium wilt have been stacked through MABC in the mega desi chickpea variety ‘Pusa 391’ (Bharadwaj et al., 2022). Molecular tags associated with major effects on qualitative and quantitative trait regulation have now been transferred into diverse grain legume crop genotypes for their genetic enhancement through marker-assisted selection (MAS), including MABC and marker-assisted foreground and background selection. The identification of a QTL-hotspot region in linkage group 4 (CaLG04) in chickpea that harbors major QTLs for multiple drought adaptive traits, followed by its introgression into elite chickpea cultivars, is an excellent example of genomics-assisted breeding (Barmukh et al., 2022). This region accounts for 58.2% of explained phenotypic variation and a 16% yield enhancement under drought conditions in introgressed lines, which is primarily attributed to improvements in root traits, such root length, density, surface area, and volume (Varshney et al., 2013b; Roorkiwal et al., 2020; Bharadwaj et al., 2021).
Complications in the domain of genetic background effects/epistasis and linkage drag of QTLs, as well as minor effects of both minor and major QTLs/genes on complex trait regulation, have impeded the use of the traditional MAS (QTL–MAS) approach in the genetic enhancement of grain legume crops on complex quantitative traits. To overcome these complexities, many novel advanced genomics-assisted breeding approaches are currently emerging, such as marker-assisted recurrent selection (MARS), MAGIC, and genomic/genome-wide (haplotype) selection. These involve the transferal and pyramiding of the favorable alleles of minor-effect genes/QTLs controlling complex quantitative traits for the genetic enhancement of grain legume crop plants in terms of yield, nutritional quality, and stress tolerance. Varshney et al. (2021) have identified superior haplotypes through whole genome sequencing (WGS) of 3,336 accessions, both cultivated (3,171) and wild (195), for important traits relating to yield enhancement; these can then be introgressed into elite chickpea cultivars. The study also identified target genomic regions for the purging of deleterious alleles, which can be achieved through genomics-assisted breeding and/or gene editing (Varshney et al., 2021). Similarly, superior haplotypes have been identified in pigeon pea based on the 292 pigeon pea genotypes of a reference set that included breeding lines, landraces, and wild species (Sinha et al., 2020). In this study, haplotype–phenotype association analysis for drought-responsive traits resulted in the identification of promising haplotypes (C. cajan_23080-H2, C. cajan_30211-H6, C. cajan_26230-H11, and C. cajan_26230-H5) for three genes regulating five drought component traits (Sinha et al., 2020). Genomic selection (GS) and integrated genomic–enviromic prediction (iGEP) are other promising strategies that can be used to improve genetic gain in legume crops (Heffner et al., 2009; Xu et al., 2022). GS uses a smaller training population that is well genotyped and phenotyped, while iGEP uses additional data on genotype–environment interactions to build a prediction model. These models are then used to predict the true breeding values of selecting particular candidates based on multi-omics data, big data technology, and artificial intelligence (Xu et al., 2022). Thus, genomics plays an integral role in improving genetic gain in modern agricultural practices and has immense potential to expedite future grain legume crop improvement programs. The traditional and novel genomics-assisted breeding approaches that are now available provide clues to the quantitative dissection of complex trait regulation, and thus have potential to expedite studies of the genetic enhancement of complex traits in diverse grain legume crops.
5 Future outlook
Grain legumes are a major source of food and nutrition globally. However, it has not been possible to enhance gain legume production to the required level, primarily due to the narrow genetic base of most of the legume crops, coupled with changing climatic conditions. Most grain legumes are lacking in desired plant ideotypes and resistance sources for various biotic and abiotic stresses. The genetic diversity conserved in gene banks globally is a major resource for crop breeding programs, but it is utilized only marginally. Therefore, in order to broaden the genetic base of grain legume crops and enhance the genetic gains made in improvement programs, conventional approaches and modern scientific tools should be integrated in a phased and carefully judged manner. The first phase should focus on the search for desired traits and the infusion of diversity into the cultivated gene pool through use of landraces and CWRs. The second phase should focus on the utilization of advanced selection tools, such as genomics, high-throughput precision phenotyping, and artificial intelligence, to exploit the hidden potential of the available genetic diversity; and the third phase should involve technologies such as mutational breeding, genome editing, and transgenic technologies to improve, modify, or introgress any novel or alien traits that are not available in the entire crop gene pool. We presume that enrichment of the genetic diversity of cultivated grain legume gene pools, along with simultaneous improvements in their yield and plant type with the aid of advanced scientific tools, will enhance grain legume crop yields to the required level.
Author contributions
JR conceptualized the topic addressed in the manuscript. Gayacharan, SP, HV, and RY collated the literature and wrote the sections on their respective areas of expertise. NM extended her assistance in grammatical correction, reference editing, and the creation of figures. Gayacharan compiled the contributions received from all authors, and finally the manuscript was edited by JR and Gayacharan. All authors reviewed the final draft of the manuscript and gave their approval.
Funding
The authors thank the GEF project “Mainstreaming agricultural biodiversity conservation and utilization in agricultural sector to ensure ecosystem services and reduce vulnerability,” implemented in India by the UN Environment (Project Code: A 1265), for providing funding support.
Acknowledgments
The authors duly acknowledge the ICAR-National Bureau of Plant Genetic Resources, New Delhi; the DBT–National Institute of Plant Genome Research, New Delhi; Shivaji College, University of Delhi; and the Alliance of Biodiversity International and CIAT, India Office, NASC Complex, New Delhi for providing the necessary support.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Ahenkora, K., AduDapaah, H. K., and Agyemang, A. (1998). Selected nutritional components and sensory attributes of cowpea (Vigna unguiculata [L.] Walp) leaves. Plant Foods Hum. Nutr. 52, 221–229. doi:10.1023/a:1008019113245
Akram, A., Iqbal, S. M., Rauf, C. A., and Aleem, R. (2008). Detection of resistant sources for collar rot disease in chickpea germplasm. Pak. J. Bot. 40, 2211–2215.
Amarteifio, J., Munthali, D., Karikari, S., and Morake, T. K. (2002). The composition of pigeon peas (Cajanus cajan (L. Millsp.) grown in Botswana. Plant Foods Hum. Nutr. 57, 173–177. doi:10.1023/A:1015248326920
Ambika, , Gayacharan, , Hamwieh, A., Talukdar, A., Kumar, S., Sharma, B. B., et al. (2022). Unraveling origin, history, genetics, and strategies for accelerated domestication and diversification of food legumes. Front. Genet. 13, 932430. doi:10.3389/fgene.2022.932430
Ambika, , Hegde, V., Nimmy, M. S., Bharadwaj, C., Tripathi, S., Singh, R. K., et al. (2021). Unraveling genetics of semi-determinacy and identification of markers for indeterminate stem growth habit in chickpea (Cicer arietinum L.). Sci. Rep. 11 (1), 21837–21838. doi:10.1038/s41598-021-01464-3
Amirul, I. F. M., Basford, K. E., Redden, R. J., and Beebe, S. (2006). Preliminary evaluation of the common bean core collection at CIAT. Plant Genet. Resour. newslett. (IPGRI/FAO) 145, 29.
Archak, S., Tyagi, R. K., Harer, P. N., Mahase, L. B., Singh, N., Dahiya, O. P., et al. (2016). Characterization of chickpea germplasm conserved in the Indian National Genebank and development of a core set using qualitative and quantitative trait data. Crop J 4, 417–424. doi:10.1016/j.cj.2016.06.013
Ashfaq, M., Khan, M. A., Mughal, S. M., Javed, N., Mukhtar, T., and Bashir, M. (2007). Evaluation of urdbean germplasm for resistance against urdbean leaf crinkle virus. Pak. J. Bot. 39, 2103.
Ashokkumar, K., Diapari, M., Jha, A. B., Tar’an, B., Arganosa, G., and Warkentin, T. D. (2015). Genetic diversity of nutritionally important carotenoids in 94 pea and 121 chickpea accessions. J. Food Compos. Anal. 43, 49–60. doi:10.1016/j.jfca.2015.04.014
Ashraf, M., and Waheed, A. (1990). Screening of local/exotic accessions of lentil (Lens culinaris Medic.) for salt tolerance at two growth stages. Plant Soil 128, 167–176. doi:10.1007/bf00011106
Azmat, M. A., Nawab, N. N., Khan, A. A., Ashraf, M., Niaz, S., and Mahmood, K. (2011). Characterization of pea germplasm. Int. J. Veg. Sci. 17 (3), 246–258. doi:10.1080/19315260.2010.544380
Bag, M. K., Gautam, N. K., Prasad, T. V., Pandey, S., Dutta, M., and Roy, A. (2014). Evaluation of an Indian collection of blackgram germplasm and identification of resistance sources to Mungbean yellow mosaic virus. Crop Prot. 61, 92–101. doi:10.1016/j.cropro.2014.03.021
Bansal, R., Sharma, S., Tripathi, K., Gayacharan, , and Kumar, A. (2019). Waterlogging tolerance in blackgram [Vigna mungo (L.) Hepper] is associated with chlorophyll content and membrane integrity. Indian J. biochem. Biophys. 5, 81.
Barmukh, R., Roorkiwal, M., Dixit, G. P., Bajaj, P., Kholova, J., Smith, M. R., et al. (2022). Characterization of ‘QTL-hotspot’ introgression lines reveals physiological mechanisms and candidate genes associated with drought adaptation in chickpea. J. Exp. Bot. 73 (22), 7255–7272. doi:10.1093/jxb/erac348
Bartwal, A., John, R., Padhi, S. R., Suneja, P., Bhardwaj, B., Gayacharan, , et al. (2022). NIR spectra processing for developing efficient protein prediction model in mungbean. J. Food Compos. Anal. 105087, 105087. doi:10.1016/j.jfca.2022.105087
Basu, P. S., Ali, M., and Chaturvedi, S. K. (2009). “Terminal heat stress adversely affects chickpea productivity in Northern India–strategies to improve thermotolerance in the crop under climate change,” in W3 workshop proceedings: Impact of climate change on agriculture, 189–193.
Bharadwaj, C., Jorben, J., Rao, A., Roorkiwal, M., Patil, B. S., Ahammed, S. K., et al. (2022). Development of high yielding Fusarium wilt resistant cultivar by pyramiding of “genes” through marker-assisted backcrossing in chickpea (Cicer arietinum L.). Front. Genet. 1747, 924287. doi:10.3389/fgene.2022.924287
Bharadwaj, C., Tripathi, S., Soren, K. R., Thudi, M., Singh, R. K., Sheoran, S., et al. (2021). Introgression of “QTL-hotspot” region enhances drought tolerance and grain yield in three elite chickpea cultivars. Plant Genome 14 (1), e20076. doi:10.1002/tpg2.20076
Bhardwaj, J., and Yadav, S. K. (2021). Comparative study on biochemical parameters and antioxidant enzymes in a drought tolerant and a sensitive variety of Horsegram (Macrotylomauniflorum) under drought stress. Am. J. Plant Physiol. 7 (1), 17–29. doi:10.3923/ajpp.2012.17.29
Bhuvaneswari, S., Sharma, S. K., Punitha, P., Shashidhar, K. S., Naveenkumar, K. L., and Prakash, N. (2017). Evaluation of morphological diversity of field pea [Pisum sativum subsp. arvense (L.)] germplasm under sub–tropical climate of Manipur. Legume Res. 40 (2), 215. doi:10.18805/lr.v0iof.10756
Bisht, I. S., Mahajan, R. K., and Patel, D. P. (1998). The use of characterisation data to establish the Indian mungbean core collection and assessment of genetic diversity. Genet. Resour. Crop Evol. 45, 127–133. doi:10.1023/a:1008670332570
Biswas, K. K., Tarafdar, A., Kumar, A., Dikshit, H. K., and Malathi, V. G. (2009). Multiple infection in urdbean (Vigna mungo) in natural condition by begomovirus, tospovirus and urdbean leaf crinkle virus complex. Indian Phytopathol. 62 (1), 75–82.
Bohra, A., Kilian, B., Sivasankar, S., Caccamo, M., Mba, C., McCouch, S. R., et al. (2021). Reap the crop wild relatives for breeding future crops. Trends Biotechnol. 40, 412–431. doi:10.1016/j.tibtech.2021.08.009
Bohra, A., Saxena, R. K., Gnanesh, B. N., Saxena, K., Byregowda, M., Rathore, A., et al. (2012). An intra-specific consensus genetic map of pigeonpea [Cajanus cajan (L.) Millspaugh] derived from six mapping populations. Theor. Appl. Genet. 125 (6), 1325–1338. doi:10.1007/s00122-012-1916-5
Boukar, O., Belko, N., Chamarthi, S., Togola, A., Batieno, J., Owusu, E., et al. (2019). Cowpea (Vigna unguiculata): Genetics, genomics and breeding. Plant Breed. 138, 415–424. doi:10.1111/pbr.12589
Burdon, J. J., Barrett, L. G., Rebetzke, G., and Thrall, P. H. (2014). Guiding deployment of resistance in cereals using evolutionary principles. Evol. Appl. 7 (6), 609–624. doi:10.1111/eva.12175
Butare, L., Rao, I., Lepoivre, P., Cajiao, C., Polania, J., Cuasquer, J., et al. (2012). Phenotypic evaluation of interspecific recombinant inbred lines (RILs) of Phaseolus species for aluminium resistance and shoot and root growth response to aluminium–toxic acid soil. Euphytica 186 (3), 715–730. doi:10.1007/s10681-011-0564-1
Celmeli, T., Sari, H., Canci, H., Sari, D., Adak, A., Eker, T., et al. (2018). The nutritional content of common bean (Phaseolus vulgaris L.) landraces in comparison to modern varieties. Agronomy 8, 166. doi:10.3390/agronomy8090166
Chaudhary, N. B., Patel, A. M., Chaudhari, R. H., Viradiyam, Y. A., and Patel, H. S. (2021). Genetic analysis for yield and quality related traits in moth bean [Vigna aconitifolia (Jacq.) Marechal]. Int. J. Chem. Stud. 9 (1), 2658–2661. doi:10.22271/chemi.2021.v9.i1ak.11629
Chaudhry, M. A., Muhammad, F., and Afzal, M. (2007). Screening of chickpea germplasm against Fusarium wilt. J. Agric. Res. 44 (4), 307.
Chauhan, J. S., Singh, B. B., and Gupta, S. (2016). Enhancing pulses production in India through improving seed and variety replacement rates. Indian J. Genet. Plant Breed. 76 (4), 410–10. doi:10.5958/0975-6906.2016.00060.2
Cheboi, J. J., Kinyua, M. G., Kimurto, P. K., Kiplagat, O. K., Nganga, F., and Ghimire, S. R. (2019). Biochemical composition of pigeonpea genotypes in Kenya. Aust. J. Crop Sci. 13 (11), 1848–1855. doi:10.21475/ajcs.19.13.11.p1886
Chiang, H. S., and Talekar, N. S. (1980). Identification of sources of resistance to the beanfly and two other agromyzid flies in soybean and mungbean. J. Econ. Entomol. 73 (2), 197–199. doi:10.1093/jee/73.2.197
Choudhary, A. K., Sultana, R., Pratap, A., Nadarajan, N., and Jha, U. C. (2011). Breeding for abiotic stresses in pigeonpea. J. Food Legum. 24 (3), 165.
Choudhary, A. K., Sultana, R., Vales, M. I., Saxena, K. B., Kumar, R. R., and Ratnakumar, P. (2018). Integrated physiological and molecular approaches to improvement of abiotic stress tolerance in two pulse crops of the semi–arid tropics. Crop J 6 (2), 99–114. doi:10.1016/j.cj.2017.11.002
Cichy, K. A., Porch, T. G., Beaver, J. S., Cregan, P., Fourie, D., Glahn, R. P., et al. (2015a). A Phaseolus vulgaris diversity panel for Andean bean improvement. Crop Sci. 55 (5), 2149–2160. doi:10.2135/cropsci2014.09.0653
Cichy, K. A., Wiesinger, J. A., and Mendoza, F. A. (2015b). Genetic diversity and genome–wide association analysis of cooking time in dry bean (Phaseolus vulgaris L.). Theor. Appl. Genet. 128 (8), 1555–1567. doi:10.1007/s00122-015-2531-z
Costantini, M., Summo, C., Centrone, M., Rybicka, I., D’Agostino, M., Annicchiarico, P., et al. (2021). Macro– and micro–nutrient composition and antioxidant activity of chickpea and pea accessions. Pol. J. Food Nutr. Sci. 71 (2), 177–185. doi:10.31883/pjfns/135813
Das, A., Parihar, A. K., Saxena, D., Singh, D., Singha, K. D., Kushwaha, K. P. S., et al. (2019). Deciphering genotype-by-environment interaction for targeting test environments and rust resistant genotypes in field pea (Pisum sativum L.). Front. Plant Sci. 10, 825. doi:10.3389/fpls.2019.00825
Dasgan, H. Y., and Koc, S. (2009). Evaluation of salt tolerance in common bean genotypes by ion regulation and searching for screening parameters. J. Food Agric. Environ. 7 (2), 363.
Davis, D. R., Epp, M. D., and Riordan, H. D. (2004). Changes in USDA food composition data for 43 garden crops, 1950 to 1999. J. Am. Coll. Nutr.Vigna Unguiculata. 231 (64), 669. doi:10.1080/07315724.2004.10719409
Deshpand, K. S., Patil, B. R., Salimath, P. M., Nidagundi, J. M., and Karthigeyan, S. (2010). Evaluation of native and collected germplasm for earliness seed traits and resistance to rust, CMV and leaf spot in cowpea [Vigna unguiculata (L.) Walp]. Electron. J. Plant Breed. 1, 384
Desingh, R., and Kanagaraj, G. (2019). Differential responses to salt stress on antioxidant enzymatic activity of two horse gram [Macrotyloma uniflorum (Lam.) Verdc] varieties. Int. J. Res. Anal. Rev. 6, 425.
Devasirvatham, V., Gaur, P. M., Raju, T. N., Trethowan, R. M., and Tan, D. K. Y. (2015). Field response of chickpea (Cicer arietinum L.) to high temperature. Field Crops Res. 172, 59–71. doi:10.1016/j.fcr.2014.11.017
Devi, J., Mishra, G. P., Sanwal, S. K., Dubey, R. K., Singh, P. M., and Singh, B. (2018). Development and characterization of penta-flowering and triple-flowering genotypes in garden pea (Pisum sativum L. var. hortense). PloS one 13 (7), e0201235. doi:10.1371/journal.pone.0201235
Dhali, S., Pradhan, M., Sahoo, R. K., Mohanty, S., and Pradhan, C. (2021). Alleviating Cr(VI) stress in horse gram (Macrotyloma uniflorum Var. Madhu) by native Cr-tolerant nodule endophytes isolated from contaminated site of Sukinda. Environ. Sci. Pollut. Res. 28, 31717–31730. doi:10.1007/s11356-021-13009-2
Dhasarathan, M., Geetha, S., Karthikeyan, A., Sassikumar, D., and Meenakshiganesan, N. (2021). Development of novel blackgram (Vigna mungo (L.) Hepper) mutants and deciphering Genotype × Environment interaction for yield–related traits of mutants. Agronomy 11, 1287. doi:10.3390/agronomy11071287
Divya, P., Durga, K. K., and Udayababu, P. (2013). Relative susceptibility of horse gram (Macrotyloma uniflorum) accessions to the attack of Callosobruchus chinensis (Fab.) (Coleoptera: Buchidae). Madras Agric. J. 100, 597.
Divya, P., Durga, K. K., Sunil, N., Rajasri, M., and Udayababu, P. (2012). Screening of horse gram accessions against pulse beetle, Callosobruchus chinensis. Indian J. Plant Prot. 40, 280.
Dong, L., Ravelombola, W., Weng, Y., Qin, J., Bhattarai, G., Zia, B., et al. (2019). Seedling salt tolerance for above ground–related traits in cowpea (Vigna unguiculata (L.) Walp). Euphytica 215 (3), 53–22. doi:10.1007/s10681-019-2379-4
Duraimurugan, P., Pratap, A., Singh, S. K., and Gupta, S. (2014). Evaluation of screening methods for bruchid beetle (Callosobruchus chinensis) resistance in greengram (Vigna radiata) and blackgram (Vigna mungo) genotypes and influence of seed physical characteristics on its infestation. Vegetos 27 (1), 60–67. doi:10.5958/j.2229-4473.27.1.011
Durga, K. K. (2012). Variability and divergence in horse gram (Dolichos uniflorus). J. Arid. Land 4 (1), 71–76. doi:10.3724/sp.j.1227.2012.00071
Ebert, A. W., Chang, C. H., Yan, M. R., and Yang, R. Y. (2017). Nutritional composition of mungbean and soybean sprouts compared to their adult growth stage. Food Chem. 237, 15–22. doi:10.1016/j.foodchem.2017.05.073
Edulamudi, P., Masilamani, A. J. A., Vanga, U. R., Divi, V. R. S. G., and Konada, V. M. (2021). Nickel tolerance and biosorption potential of rhizobia associated with horse gram [Macrotyloma uniflorum (Lam.) Verdc.]. Int. J. Phytoremediation 23 (11), 1184–1190. doi:10.1080/15226514.2021.1884182
El–Haddad, N., Rajendran, K., Smouni, A., Es–Safi, N. E., Benbrahim, N., Mentag, R., et al. (2020). Screening the FIGS set of lentil (Lens culinaris Medikus) germplasm for tolerance to terminal heat and combined drought–heat stress. Agronomy 10 (7), 1036. doi:10.3390/agronomy10071036
Emmrich, P. M., Sarkar, A., Njaci, I., Kaithakottil, G. G., Ellis, N., Moore, C., et al. (2020). A draft genome of grass pea (Lathyrus sativus), a resilient diploid legume. Biorxiv [Preprint]. Available at:Accessed May 15, 2022) https://www.biorxiv.org/content/10.1101/2020.04.24.058164v1.
Erskine, W., Meyveci, K., and Izgin, N. (1981). Screening a world lentil collection for cold tolerance. LENS Newsl. 8, 5.
Fan, M. S., Zhao, F. J., Fairweather–Tait, S. J., Poulton, P. R., Dunham, S. J., and McGrath, S. P. (2008). Evidence of decreasing mineral density in wheat grain over the last 160 years. J. Trace Elem. Med. Biol. 22 (4), 315–324. doi:10.1016/j.jtemb.2008.07.002
Fatokun, C. A., Boukar, O., and Muranaka, S. (2012). Evaluation of cowpea (Vigna unguiculata (L.) Walp.) germplasm lines for tolerance to drought. Plant Genet. Resour. 10 (3), 171–176. doi:10.1017/s1479262112000214
Frankel, O. H. (1984). “Genetic perspective of germplasm conservation,” in Genetic manipulations: Impact on man and society. Editor W. Arber (England: Cambridge Univ. Press), 161–170.
Ganjeali, A., Porsa, H., and Bagheri, A. (2011). Assessment of Iranian chickpea (Cicer arietinum L.) germplasms for drought tolerance. Agric. Water Manag. 98 (9), 1477–1484. doi:10.1016/j.agwat.2011.04.017
Garcia, T., Duitama, J., Zullo, S. S., Gil, J., Ariani, A., Dohle, S., et al. (2021). Comprehensive genomic resources related to domestication and crop improvement traits in Lima bean. Nat. Commun. 12 (1), 702–717. doi:10.1038/s41467-021-20921-1
Garg, V., Dudchenko, O., Wang, J., Khan, A. W., Gupta, S., Kaur, P., et al. (2022). Chromosome-length genome assemblies of six legume species provide insights into genome organization, evolution, and agronomic traits for crop improvement. J. Adv. Res. 42, 315–329. doi:10.1016/j.jare.2021.10.009
Gaur, P. M., and Gour, V. K. (2002). A gene producing one to nine flowers per flowering node in chickpea. Euphytica 128 (2), 231–235. doi:10.1023/a:1020845815319
Gaur, R., Azam, S., Jeena, G., Khan, A. W., Choudhary, S., Jain, M., et al. (2012). High–throughput SNP discovery and genotyping for constructing a saturated linkage map of chickpea (Cicer arietinum L.). DNA Res. 19, 357–373. doi:10.1093/dnares/dss018
Gautam, M., and Chahota, R. K. (2022). Metabolite profiling and protein quantification to a large library of 96 horsegram (Macrotyloma uniflorum) germplasm. Sci. Rep. 12 (1), 7865–7869. doi:10.1038/s41598-022-11962-7
Gautam, N. K., Kumar, K., and Prasad, M. (2016). Leaf crinkle disease in urdbean (Vigna mungo L. Hepper): An overview on causal agent, vector and host. Protoplasma 253 (3), 729–746. doi:10.1007/s00709-015-0933-z
Gautam, N. K., Singh, M., Khan, Z., Roy, A., Akhtar, J., and Ram, B. (2013). Assessment of lentil germplasm with respect to agromonic performance and major biotic stress. Legume Res. 36 (3), 214–219.
Gayacharan, , Gupta, K., Gupta, V., Tyagi, V., and Singh, K. (2020a). “Mungbean genetic resources and utilization,” in The mungbean genome, compendium of plant genomes. Editors R. Nair, R. Schafleitner, and S. H. Lee (Cham: Springer), 9–25. doi:10.1007/978–3–030–20008–4_2
Gayacharan, , Rani, U., Singh, S., Basandrai, A. K., Rathee, V. K., Tripathi, K., et al. (2020b). Identification of novel resistant sources for ascochyta blight (Ascochyta rabiei) in chickpea. Plos One 15 (10), e0240589. doi:10.1371/journal.pone.0240589
Gayacharan, , Tripathi, K., Aski, M. S., Singh, N., Kumar, A., and Lal, H. (2022). Understanding genetic diversity in blackgram [Vigna mungo (L.) Hepper] collections of Indian National Genebank. Genet. Resour. Crop Evol. 69 (3), 1229–1245. doi:10.1007/s10722-021-01301-6
Gerrano, A. S., Adebola, P. O., Rensburg, W. S. J., and Laurie, S. M. (2015). Genetic variability in cowpea (Vigna unguiculata (L.) Walp.) genotypes. S. Afr. J. Plant Soil 32 (3), 165–174. doi:10.1080/02571862.2015.1014435
Gerrano, A. S., Rensburg, W. S. J., Adebola, P. O., et al. (2017). Nutritional composition of immature pods in selected cowpea (Vigna unguiculata (L.) Walp.) genotypes in South Africa. Aust. J. Crop Sci. 11 (2), 134–141. doi:10.21475/ajcs.17.11.02.p72
Ghafoor, A., Sharif, A., Ahmad, Z., Zahid, M. A., and Rabbani, M. A. (2001). Genetic diversity in blackgram (Vigna mungo L. Hepper). Field Crops Res. 69 (2), 183–190. doi:10.1016/s0378-4290(00)00141-6
Girish, T. K., Pratape, V. M., and Rao, U. P. (2012). Nutrient distribution, phenolic acid composition, antioxidant and alpha–glucosidase inhibitory potentials of blackgram (Vigna mungo L.) and its milled by–products. Food Res. Int. 46 (1), 370–377. doi:10.1016/j.foodres.2011.12.026
Goa, Y., Mohammed, H., Worku, W., and Urage, E. (2022). Genotype by environment interaction and yield stability of cowpea (Vigna unguiculata (L.) Walp.) genotypes in moisture limited areas of Southern Ethiopia. Heliyon 8 (3), e09013. doi:10.1016/j.heliyon.2022.e09013
Gomashe, S. S., Dikshit, N., Chand, D., and Shingane, S. N. (2018). Assessment of genetic diversity using morpho-agronomical traits in horse gram. Int. J. Curr. Microbiol. Appl. Sci. 7 (5), 2095–2103. doi:10.20546/ijcmas.2018.705.244
Guo, Y. (2022). Disentangling the breakdown of resistance. Nat. Food 3 (8), 566. doi:10.1038/s43016-022-00582-z
Gupta, S., Nawaz, K., Parween, S., Roy, R., Sahu, K., Kumar, A., et al. (2017). Draft genome sequence of Cicer reticulatum L., the wild progenitor of chickpea provides a resource for agronomic trait improvement. DNA Res. 24 (1), 1–10. doi:10.1093/dnares/dsw042
Ha, J., Satyawan, D., Jeong, H., Lee, E., Cho, K. H., Kim, M. Y., et al. (2021). A near-complete genome sequence of mungbean (Vigna radiata L.) provides key insights into the modern breeding program. Plant Genome 14 (3), e20121. doi:10.1002/tpg2.20121
Hacisalihoglu, G., Beisel, N. S., and Settles, A. M. (2021). Characterization of pea seed nutritional value within a diverse population of Pisum sativum. PloS One 16 (11), e0259565. doi:10.1371/journal.pone.0259565
Halila, M. H., and Strange, R. N. (1997). Screening of Kabuli chickpea germplasm for resistance to Fusarium wilt. Euphytica 96 (2), 273–279. doi:10.1023/a:1003051100683
Hamdi, A., Kusmenoglu, I., and Erskine, W. (1996). Sources of winter-hardiness in wild lentil. Genet. Resour. Crop Evol. 43, 63–67. doi:10.1007/bf00126942
Hannan, R. M., Kaiser, W. J., and Muehlbauer, F. J. (1994). Development and utilization of the USDA chickpea germplasm core collection. Madison, WI: Agronomy AbstractsASA, 217.
Hasan, M. K., Sabagh, A., Sikdar, M. S. I., Alam, M. J., Ratnasekera, D., Barutcular, C., et al. (2017). Comparative adaptable agronomic traits of blackgram and mungbean for saline lands. Plant Arch. 17 (1), 589–593.
Haus, M. J., Pierz, L. D., Jacobs, J. L., Wiersma, A. T., Awale, H. E., Chilvers, M. I., et al. (2021). Preliminary evaluation of wild bean (Phaseolus spp.) germplasm for resistance to Fusarium cuneirostrum and Fusarium oxysporum. Crop Sci. 61 (5), 3264–3274. doi:10.1002/csc2.20495
Haware, M. P., Nene, Y. L., Pundir, R. P. S., and Rao, J. N. (1992). Screening of world chickpea germplasm for resistance to fusarium wilt. Field crop. Res. 30, 147–154. doi:10.1016/0378-4290(92)90063-f
Hawkes, J. G. (1983). The diversity of crop plants. Cambridge, Massachusetts: Harvard University Press.
Heffner, E. L., Sorrells, M. E., and Jannink, J. L. (2009). Genomic selection for crop improvement. Crop Sci. 49 (1), 1–12. doi:10.2135/cropsci2008.08.0512
Hegde, V. S. (2011). Morphology and genetics of a new found determinate genotype in chickpea. Euphytica 182 (1), 35–42. doi:10.1007/s10681-011-0447-5
Hema, M., Sreenivasulu, P., Patil, B. L., Kumar, P. L., and Reddy, D. V. (2014)., 90. Academic Press, 431–505.Tropical food legumes: Virus diseases of economic importance and their controlAdv. virus Res.
Hiremath, P. J., Kumar, A., Penmetsa, R. V., Farmer, A., Schlueter, J. A., Chamarthi, S. K., et al. (2012). Large-scale development of cost-effective SNP marker assays for diversity assessment and genetic mapping in chickpea and comparative mapping in legumes. Plant Biotechnol. J. 10, 716–732. doi:10.1111/j.1467-7652.2012.00710.x
Hobson, K., Armstrong, R., Nicolas, M., Connor, D., and Materne, M. (2006). Response of lentil (Lens culinaris) germplasm to high concentrations of soil boron. Euphytica 151 (3), 371–382. doi:10.1007/s10681-006-9159-7
Horn, Lydia N., and Shimelis, H. (2020). Production constraints and breeding approaches for cowpea improvement for drought prone agro-ecologies in Sub-Saharan Africa. Ann. Agric. Sci. 651, 83–91. doi:10.1016/j.aoas.2020.03.002
Hu, Z. J., Huang, Y. Y., Lin, X. Y., Feng, H., Zhou, S. X., Xie, Y., et al. (2022). Loss and natural variations of blast fungal avirulence genes breakdown rice resistance genes in the sichuan basin of China. Front. Plant Sci. 13, 788876. doi:10.3389/fpls.2022.788876
Iqbal, U., Iqbal, S. M., Afzal, R., Jamal, A., Farooq, M. A., and Zahid, A. (2011). Screening of mungbean germplasm against mungbean yellow mosaic virus (MYMV) under field conditions. Pak. J. Phytopathol. 23 (1), 48–51.
Jain, M., Misra, G., Patel, R. K., Priya, P., Jhanwar, S., Khan, A. W., et al. (2013). A draft genome sequence of the pulse crop chickpea (Cicer arietinum L.). Plant. J. 74, 715–729. doi:10.1111/tpj.12173
Jarpa-Parra, M. (2018). Lentil protein: A review of functional properties and food application. An overview of lentil protein functionality. Int. J. Food Sci. Technol. 53 (4), 892–903. doi:10.1111/ijfs.13685
Jegadeesan, S., Raizada, A., Dhanasekar, P., and Suprasanna, P. (2021). Draft genome sequence of the pulse crop blackgram [Vigna mungo (L.) Hepper] reveals potential R-genes. Sci. Rep. 11, 11247. doi:10.1038/s41598-021-90683-9
Jha, U. C., Nayyar, H., Parida, S. K., and Siddique, K. H. (2022). Horse gram, an underutilized climate–resilient legume: Breeding and genomic approach for improving future genetic gain” in developing climate resilient grain and forage legumes. Singapore: Springer, 167.
Jood, S., Bishnoi, S., and Sharma, A. (1998). Chemical analysis and physico-chemical properties of chickpea and lentil cultivars. Food/Nahrung 42 (02), 71–74. doi:10.1002/(sici)1521-3803(199804)42:02<71::aid-food71>3.3.co;2-u
Jukanti, A. K., Gaur, P. M., Gowda, C. L. L., and Chibbar, R. N. (2012). Nutritional quality and health benefits of chickpea (Cicer arietinusm L.): A review. Br. J. Nutr. 108 (S1), S11–S26. doi:10.1017/S0007114512000797
Kabi, M., Das, T. R., and &Baisakh, B. (2017). Screening of superior genotypes for cold tolerance and MYMV resistance in greengram (Vigna radiata). Int. J. Currmicrobiol. Sci. 12, 2270. doi:10.20546/ijcmas
Kanagaraj, G., and Sathish, C. (2017). Salt Stress induced changes in growth, pigments and protein contents in two horse gram varieties. J. Sci. Agric. 08.
Kang, Y. J., Kim, S. K., Kim, M. Y., Lestari, P., Kim, K. H., Ha, B. K., et al. (2014). Genome sequence of mungbean and insights into evolution within Vigna species. Nat. Commun. 5 (1), 5443–5449. doi:10.1038/ncomms6443
Kang, Y. J., Satyawan, D., Shim, S., Lee, T., Lee, J., Hwang, W. J., et al. (2015). Draft genome sequence of adzuki bean, Vigna angularis. Sci. Rep. 5, 8069. doi:10.1038/srep08069
Karthikeyan, A., Shobhana, V. G., Sudha, M., Raveendran, M., Senthil, N., Pandiyan, M., et al. (2014). Mungbean yellow mosaic virus (MYMV): A threat to greengram (Vigna radiata) production in Asia. Int. J. Pest Manag. 60 (4), 314–324. doi:10.1080/09670874.2014.982230
Kaul, T., Eswaran, M., Thangaraj, A., Meyyazhagan, A., Nehra, M., Raman, N. M., et al. (2019). Rice bean (Vigna umbellata) draft genome sequence: Unravelling the late flowering and unpalatability related genomic resources for efficient domestication of this underutilized crop. BioRxiv (Preprint)Available at: https://www.biorxiv.org/content/10.1101/816595v1.abstract (Accessed May 15, 2022).
Kaundal, S. P., and Kumar, R. (2021). Preliminary evaluation for most promising accessions from procured germplasm of forgotten medicinal legume Horsegram {Macrotyloma uniflorum (Lam.) Verdc} in Jalandhar Region of Punjab. J. Res. Agric. Anim. Sci. 8 (12), 09.
Kaur, S., Singh, N., Sodhi, N. S., Rana, J. C., Kawale, S. B., Kadam, S. S., et al. (2009). Diversity in properties of seed and flour of kidney bean germplasmEffect of processing on insoluble dietary fiber and resistant starch in kidney bean and horse gram. Food Chem. Food Sci. Technol. 11742 (2), 282361.
Keller, B., Ariza–Suarez, D., de la Hoz, J., Aparicio, J. S., Portilla–Benavides, A. E., Buendia, H. F., et al. (2020). Genomic prediction of agronomic traits in common bean (Phaseolus vulgaris L.) under environmental stress. Front. Plant Sci. 11, 1001. doi:10.3389/fpls.2020.01001
Khan, Z., Gautam, N. K., Gawade, B. H., and Dubey, S. C. (2017). Screening of lentil (Lens culinarisMedik.) germplasm for resistance to root–knot nematode, Meloidogyneincognita. Indian J. Genet. 77 (3), 408–413. doi:10.5958/0975-6906.2017.00055.4
Khine, N. A., Kundu, K. K., Malik, D. P., and Devi, M. (2021). Production and trade performance of blackgram (Vigna mungo) and greengram (Vigna radiata) in India and Myanmar. Asian J. Agric. Ext. Econ. Sociol. 39 (10), 231–243. doi:10.9734/ajaees/2021/v39i1030687
Kreplak, J., Madoui, M. A., Cápal, P., Novák, P., Labadie, K., Aubert, G., et al. (2019). A reference genome for pea provides insight into legume genome evolution. Nat. Genet. 51 (9), 1411–1422. doi:10.1038/s41588-019-0480-1
Krishnamurthy, L., Gaur, P. M., Basu, P. S., Chaturvedi, S. K., Tripathi, S., Vadez, V., et al. (2011). Large genetic variation for heat tolerance in the reference collection of chickpea (Cicer arietinum L.) germplasm. Plant Genet. Resour. 9 (1), 59–69. doi:10.1017/s1479262110000407
Krishnamurthy, L., Kashiwagi, J., Gaur, P. M., Upadhyaya, H. D., and Vadez, V. (2010). Sources of tolerance to terminal drought in the chickpea (Cicer arietinum L.) minicore germplasm. Field crop. Res. 119, 322–330. doi:10.1016/j.fcr.2010.08.002
Krishnamurthy, L., Upadhyaya, H. D., Saxena, K. B., and Vadez, V. (2012). Variation for temporary waterlogging response within the mini core pigeonpea germplasm. J. Agric. Sci. 150 (3), 357–364. doi:10.1017/s0021859611000682
Kujur, A., Bajaj, D., Upadhyaya, H. D., Das, S., Ranjan, R., Shree, T., et al. (2015). Employing genome-wide SNP discovery and genotyping strategy to extrapolate the natural allelic diversity and domestication patterns in chickpea. Front. Plant Sci. 6, 162. doi:10.3389/fpls.2015.00162
Kulkarni, N. K., Reddy, A. S., Kumar, P. L., Vijaynarasimha, J., Rangaswamy, K. T., Muniyappa, V., et al. (2003). Broad–based resistance to pigeonpea sterility mosaic disease in accessions of Cajanus scarabaeoides (L.) Benth. Indian J. Plant Prot. 31 (1), 6–11.
Kumar, H., Dikshit, H. K., Singh, A., Jain, N., Kumari, J., Singh, A. M., and Prabhu, K. V. (2014). Characterization of grain iron and zinc in lentil ('Lens culinaris' Medikus' culinaris') and analysis of their genetic diversity using SSR markers. Aust. J. Crop Sci. 8 (7), 1005.
Kumar, M., and Singh, P. S. (2014). Screening of blackgram genotypes against major insect pests. Indian J. Entomology 76 (1), 84.
Kumar, N., Bharadwaj, C., Pal, M., Satyavathi, C. T., Kumar, T., Singhal, T., et al. (2016). Screening of chickpea (cicer arietinum L.) genotypes for seedling salinity tolerance. J. Trop. Agric. 34 (3), 667.
Kumar, S., Barpete, S., Kumar, J., Gupta, P., and Sarker, A. (2013). Global lentil production: Constraints and strategies. SATSA Mukhapatra–Annual Tech. 17, 1.
Kumar, S., Gupta, S., Chandra, S., and Singh, B. B. (2004). “How wide is the genetic base of pulse crops?” in Pulses in new perspective. Editors M. Ali, B. B. Singh, S. Kumar, and V. Dhar (Kanpur, India: Indian Institute of Pulse Research), 211.
Kumar, V., Singh, B. M., Singh, S., and Sugha, S. K. (1997). Evaluation of lentil germplasm for resistance to rust. Lentil Exp. News Serv. 24 (1–2), 21–22.
Kumari, S., Sahni, S., and Prasad, B. D. (2020). Screening of urdbean germplasm for high seed yield coupled with resistance to Mungbean yellow mosaic virus (MYMV). Curr. Appl. Sci. Technol. 39 (1), 100–106. doi:10.9734/cjast/2020/v39i130485
Lamichaney, A., Parihar, A. K., Hazra, K. K., Dixit, G. P., Katiyar, P. K., Singh, D., et al. (2021). Untangling the influence of heat stress on crop phenology, seed set, seed weight, and germination in field pea (Pisum sativum L.). Front. Plant Sci. 12, 635868. doi:10.3389/fpls.2021.635868
Laserna–Ruiz, I., De–Los, M. P., Santana–Méridas, O., Sánchez–Vioque, R., and Rodríguez–Conde, M. F. (2012). Screening and selection of lentil (Lens Miller) germplasm resistant to seed bruchids (Bruchus spp.). Euphytica 188 (2), 153–162. doi:10.1007/s10681-012-0752-7
Lateef, D. D. (2015). DNA marker technologies in plants and applications for crop improvements. J. Biosci. Med. 3 (05), 7–18. doi:10.4236/jbm.2015.35002
Leonforte, A., Forster, J. W., Redden, R. J., Nicolas, M. E., and Salisbury, P. A. (2013). Sources of high tolerance to salinity in pea (Pisum sativum L.). Euphytica 189 (2), 203–216. doi:10.1007/s10681-012-0771-4
Logozzo, G., Donnoli, R., Macaluso, L., Papa, R., Knuppfer, H., and Zeuli, P. S. (2007). Analysis of the contribution of Mesoamerican and Andean gene pools to European common bean (Phaseolus vulgaris L.) germplasm and strategies to establish a core collection. Genet. Resour. Crop Evol. 54 (8), 1763–1779. doi:10.1007/s10722-006-9185-2
Lonardi, S., Muñoz-Amatriaín, M., Liang, Q., Shu, S., Wanamaker, S., Lo, S., et al. (2019). The genome of cowpea (Vigna unguiculata [L.] Walp.). Plant J 98 (5), 767–782. doi:10.1111/tpj.14349
Longvah, T., Ananthan, R., Bhaskarachary, K., and Venkaiah, K. (2017). Indian food composition tables. Hyderabad: National Institute of Nutrition Indian Council of Medical Research Department of Health Research Ministry of Health and Family Welfare, Government of India, 501.
Mahalakshmi, V., Ng, Q., Lawson, M., and Ortiz, R. (2007). Cowpea [Vigna unguiculata (L.) Walp.] core collection defined by geographical, agronomical and botanical descriptors. Plant Genet. Resour. 5 (3), 113–119. doi:10.1017/s1479262107837166
Mahesh, H. B., Prasannakumar, M. K., Manasa, K. G., Perumal, S., Khedikar, Y., Kagale, S., et al. (2021). Genome, transcriptome, and germplasm sequencing uncovers functional variation in the warm–season grain legume Horsegram Macrotyloma uniflorum (Lam.) Verdc. Front. Plant Sci. 12, 758119. doi:10.3389/fpls.2021.758119
Mahuku, G. S., Jara, C., Cajiao, C., and Beebe, S. (2003). Sources of resistance to angular leaf spot (Phaeoisariopsisgriseola) in common bean core collection, wild Phaseolus vulgaris and secondary gene pool. Euphytica 130 (3), 303–313. doi:10.1023/a:1023095531683
Malambane, G., and Bhatt, K. V. (2014). Agro–morphological performance of moth bean (Vigna aconitifolia) lines in severe drought stressed and rain–fed condition. Direct Res. J. Agric. Food Sci. 2, 107.
Maliro, M. F., McNeil, D., Redden, B., Kollmorgen, J. F., and Pittock, C. (2008). Sampling strategies and screening of chickpea (Cicer arietinum L.) germplasm for salt tolerance. Genet. Resour. Crop Evol. 55 (1), 53–63. doi:10.1007/s10722-007-9214-9
Mbinda, W., and Masaki, H. (2021). Breeding strategies and challenges in the improvement of blast disease resistance in finger millet. A current review. Front. Plant Sci. 11, 602882. doi:10.3389/fpls.2020.602882
McClean, P. E., Terpstra, J., McConnell, M., White, C., Lee, R., and Mamidi, S. (2012). Population structure and genetic differentiation among the USDA common bean (Phaseolus vulgaris L.) core collection. Genet. Resour. Crop Evol. 59 (4), 499–515. doi:10.1007/s10722-011-9699-0
Meena, J. K., Singh, A., Dikshit, H. K., Mishra, G. P., Aski, M., Srinivasa, N., et al. (2017). Screening of lentil (Lens culinarisMedikus sub sp. culinaris) germplasm against Fusarium wilt (Fusarium oxysporum f. sp. lentis). Int. J. Curr. Microbiol. App. Sci. 6 (11), 2533–2541. doi:10.20546/ijcmas.2017.611.298
Meena, S. K., Pandey, R., Sharma, S., Kumar, T., Singh, M. P., Dikshit, H. K., et al. (2021). Physiological basis of combined stress tolerance to low phosphorus and drought in a diverse set of mungbean germplasm. Agronomy 11 (1), 99. doi:10.3390/agronomy11010099
Meghwal, R., Joshi, U., Kumar, S., and Sharma, R. (2015). Screening of moth bean (Vigna aconitifolia) core collection against yellow mosaic virus. Indian J. agri. Sci. 85, 571–575.
Mehra, R., Sarker, A., Dixit, H. K., Aski, M., Khandia, R., and Munjal, A. (2018). Genetic diversity iron and zinc content in lentil (Lens culinaris Medikus subsp. culinaris) as assessed by SSR marker. Life Sci. Inf. Publ. 4, 440.
Michael, T. P., and Jackson, S. (2013). The first 50 plant genomes. Plant Genome 6. doi:10.3835/plantgenome2013.03.0001in
Mir, R. R., Kudapa, H., Srikanth, S., Saxena, R. K., Sharma, A., Azam, S., et al. (2014). Candidate gene analysis for determinacy in pigeonpea (Cajanus spp.). Theor. Appl. Genet. 127 (12), 2663–2678. doi:10.1007/s00122-014-2406-8
Mishra, G. P., Dikshit, H. K., Kumari, J., Tripathi, K., Devi, J., Aski, M., et al. (2020a). Identification and characterization of novel penta-podded genotypes in the cultivated lentil. Crop Sci. 60 (4), 1974–1985. doi:10.1002/csc2.20156
Mishra, G. P., Dikshit, H. K., Sv, R., Tripathi, K., Kumar, R. R., Aski, M., et al. (2020b). Yellow mosaic disease (YMD) of mungbean (Vigna radiata (L.) Wilczek): Current status and management opportunities. Front. Plant Sci. 11, 1064. doi:10.3389/fpls.2020.01064
Mishra, S. K., Singh, B. B., Daya, C., and Meena, K. N. (2005). Sources for high temperature tolerant germplasm in cowpea (Vigna unguiculata L. Walp.). J. Arid. Legum. 2 (1), 78.
Mohan, S., Sheeba, A., Murugan, E., and Ibrahim, S. M. (2014). Screening of mungbean germplasm for resistance to mungbean yellow mosaic virus under natural condition. Indian. J. Sci. Technol. 7 (7), 891–896. doi:10.17485/ijst/2014/v7i7.1
Nainu, A., and Murugan, S. (2020). Screening of mungbean [Vigna radiata (L.) Wilczek] genotypes under field condition for resistance against mungbean yellow mosaic virus (MYMV). Plant Arch. 20 (2), 6867.
Nair, R. M., Yang, R. Y., Easdown, W. J., Thavarajah, D., Thavarajah, P., Hughes, J. D. A., et al. (2013). Biofortification of mungbean (Vigna radiata) as a whole food to enhance human health. J. Sci. Food Agric. 93 (8), 1805–1813. doi:10.1002/jsfa.6110
Nene, Y. L., Reddy, M. V., Beniwal, S. P. S., Mahmood, M., Zote, K. K., Singh, R. N., et al. (1989). Multilocational testing of pigeonpea for broad–based resistance to sterility mosaic in India. Indian Phytopathol. 42 (3), 444.
Nielsen, S. S., Brandt, W. E., and Singh, B. B. (1993). Genetic variability for nutritional composition and cooking time of improved cowpea lines. Crop Sci. 33 (3), 469–472. doi:10.2135/cropsci1993.0011183x003300030010x
Nisar, M., Ghafoor, A., Khan, M. R., and Qureshi, A. S. (2006). Screening of Pisum sativum L. Germplasm against Erysiphe pisi syd. Acta Biol. Crac. Ser. Bot. 48 (2), 33.
Nkhoma, N., Shimelis, H., Laing, M. D., Shayanowako, A., and Mathew, I. (2020). Assessing the genetic diversity of cowpea [Vigna unguiculata (L.) Walp.] germplasm collections using phenotypic traits and SNP markers. BMC Genet. 21 (1), 110–116. doi:10.1186/s12863-020-00914-7
Okiror, M. A. (1999). Evaluation of pigeonpea (Cajanus cajan) germplasm for resistance to Fusarium wilt. Indian J. Agric. Sci. 69 (8), 600.
Olorunwa, O. J., Adhikari, B., Shi, A., and &Barickman, T. C. (2022). Screening of cowpea (Vigna unguiculata (L.) Walp.) genotypes for waterlogging tolerance using morpho-physiological traits at early growth stage. Plant Sci. 315, 111136. doi:10.1016/j.plantsci.2021.111136
Pal, S., Sharma, N. K., Sharma, A. K., Kumar, A., and Meena, S. (2020). Study of genetic variability parameters for seed yield and component traits in moth bean [Vigna aconitifolia (Jacq) Marechal]. Int. J. Chem. Stud. 8 (4), 2638–2641. doi:10.22271/chemi.2020.v8.i4ae.10038
Pande, S., Kishore, G. K., and Rao, J. N. (2004). Evaluation of chickpea lines for resistance to dry root rot caused by Rhizoctonia. Int. Chickpea PigeonpeaNewslett. 11 (2), 37.
Pande, S., Ramgopal, D., Kishore, G. K., Mallikarjuna, N., Sharma, M., Pathak, M., et al. (2006). Evaluation of wild Cicer species for resistance to Ascochyta blight and Botrytis graymold in controlled environment at ICRISAT, Patancheru, India. J. Sat. Agric. Res. 2 (1), 1.
Panhwar, F. (2005). Chemistry and biochemistry of legume, with cultural activities in Sindh”. Editor L. Chem (Germany: Digitalverlag GmbH).
Paredes, M., Becerra, V., and Tay, J. (2009). Inorganic nutritional composition of common bean (Phaseolus vulgaris L.) genotypes race Chile. Chil. J. Agric. Res. 69 (4), 486. doi:10.4067/s0718-58392009000400002
Parimala, K., Meenakumari, K. V. S., Sudhakar, R., and Durga, K. K. (2011). Screening of horsegram genotypes against yellow mosaic virus and powdery mildew diseases. Indian J. Plant. Prot. 39, 160.
Parween, S., Nawaz, K., Roy, R., Pole, A. K., Venkata, S., B., Misra, G., et al. (2015). An advanced draft genome assembly of a desi type chickpea (Cicer arietinum L.). Sci. Rep. 5 (1), 12806–12814. doi:10.1038/srep12806
Peña, P. A., Steadman, J. R., Eskridge, K. M., and Urrea, C. A. (2013). Identification of sources of resistance to damping–off and early root/hypocotyl damage from Rhizoctonia solani in common bean (Phaseolus vulgaris L.). Crop Prot. 54, 92–99. doi:10.1016/j.cropro.2013.04.014
Pengelly, B. C., and Maass, B. L. (2001). Lablab purpureus (L.) Sweet–diversity, potential use and determination of a core collection of this multi–purpose tropical legume. Genet. Resour. Crop Evol. 48 (3), 261. doi:10.1023/A:1011286111384
Podder, R., Banniza, S., and Vandenberg, A. (2013). Screening of wild and cultivated lentil germplasm for resistance to stemphylium blight. Plant Genet. Resour. 11 (1), 26–35. doi:10.1017/s1479262112000329
Pootakham, W., Nawae, W., Naktang, C., Sonthirod, C., Yoocha, T., Kongkachana, W., et al. (2021). A chromosome-scale assembly of the blackgram (Vigna mungo) genome. Mol. Ecol. Resour. 21 (1), 238–250. doi:10.1111/1755-0998.13243
Porch, T. G., Cichy, K., Wang, W., Brick, M., Beaver, J. S., Santana–Morant, D., et al. (2017). Nutritional composition and cooking characteristics of tepary bean (Phaseolus acutifolius Gray) in comparison with common bean (Phaseolus vulgaris L.). Genet. Resour. Crop Evol. 64 (5), 935–953. doi:10.1007/s10722-016-0413-0
Pouralibaba, H. R., Rubiales, D., and Fondevilla, S. (2015). Identification of resistance to Fusarium oxysporumf. sp. lentis in Spanish lentil germplasm. Eur. J. Plant Pathol. 143 (2), 399–405. doi:10.1007/s10658-015-0692-x
Pratap, A., Basu, P. S., Gupta, S., Malviya, N., Rajan, N., Tomar, R., et al. (2014). Identification and characterization of sources for photo-and thermo-insensitivity in Vigna species. Plant Breed. 133 (6), 756–764. doi:10.1111/pbr.12215
Pratap, A., Gupta, S., Basu, P. S., Tomar, R., Dubey, S., Rathore, M., et al. (2019). “Towards development of climate smart mungbean: Challenges and opportunities,” in Genomic designing of climate–smart pulse crops. Editor C. Kole (Cham): Springer), 235–264.
Pratap, A., Kumar, S., Polowick, P. L., Blair, M. W., and Baum, M. (2022). Editorial: Accelerating genetic gains in pulses. Front. Plant Sci. 13, 879377. doi:10.3389/fpls.2022.879377
Priyadharshini, B., Vignesh, M., Prakash, M., and Anandan, R. (2019). Evaluation of blackgram genotypes for saline tolerance at seedling stage. Indian J. Agric. Res. 53 (1), 83. doi:10.18805/ijare.a-5118
Priyanka, S., Sudhagar, R., Vanniarajan, C., and Ganesamurthy, K. (2021). Assessment of genetic divergence in horse gram (Macrotylum uniflorum (Lam) Verdc.) using quantitative traits. Legume Res. 44, 261–267. doi:10.18805/LR–4103
Rajendran, K., Smouni, A., Es–Safi, N. E., Benbrahim, N., Mentag, R., Nayyar, H., et al. (2020). Screening the FIGS set of lentil (Lens culinaris Medikus) germplasm for tolerance to terminal heat and combined drought–heat stress. Agronomy 10 (7), 1036. doi:10.3390/agronomy10071036
Rana, J. C., Banyal, D. K., Sharma, K. D., Sharma, M. K., Gupta, S. K., and Yadav, S. K. (2013). Screening of pea germplasm for resistance to powdery mildew. Euphytica 189 (2), 271–282. doi:10.1007/s10681-012-0798-6
Rana, J. C., Gautam, N. K., GayacharanSingh, M., Yadav, R., Tripathi, K., Yadav, S. K., et al. (2016). Genetic resources of pulse crops in India: An overview. Indian J. Genet. Pl. Breed. 76 (4), 420–436. doi:10.5958/0975-6906.2016.00061.4
Rana, J. C., Sharma, T. R., Tyagi, R. K., Chahota, R. K., Gautam, N. K., Singh, M., et al. (2015). Characterisation of 4274 accessions of common bean (Phaseolus vulgaris L.) germplasm conserved in the Indian gene bank for phenological, morphological and agricultural traits. Euphytica 205 (2), 441–457. doi:10.1007/s10681-015-1406-3
Rana, M., Hossain, M., Urmi, T., Ahmed, S., Haque, M., and Islam, M. (2019). Evaluation of Blackgram (Vigna mungo L.) genotypes for their tolerance to flooding. Agric. 17 (1–2), 89–101. doi:10.3329/agric.v17i1–2.44699
Rasheed, A., Hao, Y., Xia, X., Khan, A., Xu, Y., Varshney, R. K., et al. (2017). Crop breeding chips and genotyping platforms: Progress, challenges, and perspectives. Mol. plant 10 (8), 1047–1064. doi:10.1016/j.molp.2017.06.008
Ravelombola, W., Shi, A., Weng, Y., Mou, B., Motes, D., Clark, J., et al. (2018). Association analysis of salt tolerance in cowpea (Vigna unguiculata (L.) Walp) at germination and seedling stages. Theor. Appl. Genet. 131 (1), 79–91. doi:10.1007/s00122-017-2987-0
Ravelombola, W. S., Shi, A., Weng, Y., Clark, J., Motes, D., Chen, P., et al. (2017). Evaluation of salt tolerance at germination stage in cowpea [Vigna unguiculata (L.) Walp]. HortSci 52 (9), 1168–1176. doi:10.21273/hortsci12195-17
Reddy, A. T., Gowda, J. E., Ravikumar, R., Rao, A. M., Ramesh, S., and Saifulla, M. (2016). Resistance source identification for dry root rot disease in chickpea. Adv. Life Sci. 5 (21), 9767.
Reddy, L. J., Upadhyaya, H. D., Gowda, C. L. L., and Singh, S. (2005). Development of core collection in pigeonpea [Cajanus cajan (L.) Millspaugh] using geographic and qualitative morphological descriptors. Genet. Resour.CropEvol. 52 (8), 1049–1056. doi:10.1007/s10722-004-6152-7
Rex Consortium (2016). Combining selective pressures to enhance the durability of disease resistance genes. Front. Plant Sci. 7, 1916. doi:10.3389/fpls.2016.01916
Rezene, Y., and Mekonin, S. (2019). Screening common bean (Phaseolus vulgaris L.) germplasm for resistance against angular leaf spot (Pseudocercosporagriseola) disease under field condition. J. Plant Stud. 8 (1), 30. doi:10.5539/jps.v8n1p30
Rodiño, A. P., Santalla, M., De Ron, A. M., and Singh, S. P. (2003). A core collection of common bean from the Iberian peninsula. Euphytica 131 (2), 165–175.
Roorkiwal, M., Bharadwaj, C., Barmukh, R., Dixit, G. P., Thudi, M., Gaur, P. M., et al. (2020). Integrating genomics for chickpea improvement: Achievements and opportunities. Theor. Appl. Genet. 133 (5), 1703–1720. doi:10.1007/s00122-020-03584-2
Roy, D., Joshi, P. K., and Chandra, R. (2017). Pulses for nutrition in India: Changing patterns from farm to fork. Intl. Food Policy Res. Inst.
Rubiales, D., Rojas-Molina, M. M., and Sillero, J. C. (2013). Identification of pre-and posthaustorial resistance to rust (Uromycesviciae fabae) in lentil (Lens culinaris) germplasm. Plant Breed. 132 (6), 676–680. doi:10.1111/pbr.12096
Saba, I., Sofi, P. A., Zeerak, N. A., Bhat, M. A., and Mir, R. R. (2016). Characterisation of a core set of common bean (Phaseolus vulgaris L.) germplasm for seed quality traits. SABRAO J. Breed. Genet. 48 (3), 359.
Sachdeva, S., Sharma, V., Bhat, K. V., and Sen, S. (2016). Morphological and physiological response studies of moth bean (Vigna Aconitifolia L.) genotypes under drought stress. Int. J. Appl. Sci. Eng. 5 (7).
Sahoo, S., Sanadya, S. K., Kumari, N., and Baranda, B. (2019). Estimation of the various genetic variability parameters for seed yield and its component traits in Mothbean germplasm [Vigna aconitifolia (Jacq.) Marechal]. J. Pharmcog. Phytochem SP3, 49.
Sai, C. B., and Chidambaranathan, P. (2019). Reproductive stage drought tolerance in blackgram is associated with role of antioxidants on membrane stability. Plant Physiol. Rep. 24 (3), 399–409. doi:10.1007/s40502-019-00471-x
Sakai, H., Naito, K., Ogiso–Tanaka, E., Takahashi, Y., Iseki, K., Muto, C., et al. (2015). The power of single molecule real–time sequencing technology in the de novo assembly of a eukaryotic genome. Sci. Rep. 5 (1), 16780–16813. doi:10.1038/srep16780
Salini, K., Maruthi, V., Maheswari, M., and Sarkar, B. (2014). “Genetic improvement of horse gram through mutation breeding,” in 3rd international conference on agriculture and horticulture (India: Hyderabad International Convention Centre). October 27–29.
Sandhya, R. C., Rao, G. R., Chalam, M. S. V., Kumar, P. A., and Rao, V. S. (2014). Field screening of greengram genotypes against Marucavitrata in Summer. J. Agric. Sci. 1, 53.
Sankar, A. U., Anitha, K., Sivaraj, N., Kumari, K. V. S., Sunil, N., and Chakrabarty, S. K. (2015). Screening of horsegram germplasm collected from Andhra Pradesh against anthracnose. Legume Res.–An Int. J. 38 (6), 753–757. doi:10.18805/lr.v38i6.6719
Sarker, A., and Erskine, W. (2006). Recent progress in the ancient lentil. J. Agric. Sci. 144 (1), 19–29. doi:10.1017/s0021859605005800
Saxena, K. B., Chandrasena, G. D. S. N., Hettiarachchi, K., Iqbal, Y. B., Fonseka, H. H. D., and Jayasekera, S. J. B. A. (2002). Evaluation of pigeonpea accessions and selected lines for reaction to Maruca. Crop Sci. 42 (2), 615–618. doi:10.2135/cropsci2002.6150
Saxena, N. P., Krishnamurthy, L., and Johansen, C. (1993). Registration of a drought–resistant chickpea germplasm. Crop Sci. 33 (6), 1424. doi:10.2135/cropsci1993.0011183x003300060088x
Schafleitner, R., Nair, R. M., Rathore, A., Wang, Y. W., Lin, C. Y., Chu, S. H., et al. (2015). The AVRDC–The World Vegetable Center mungbean (Vigna radiata) core and mini core collections. BMC Genomics 16, 344. doi:10.1186/s12864-015-1556-7
Schmutz, J., McClean, P. E., Mamidi, S., Wu, G. A., Cannon, S. B., Grimwood, J., et al. (2014). A reference genome for common bean and genome–wide analysis of dual domestications. Nat. Genet. 46 (7), 707–713. doi:10.1038/ng.3008
Schreinemachers, P., Sequeros, T., Rani, S., Rashid, M. A., Gowdru, N. V., Rahman, M. S., et al. (2019). Counting the beans: Quantifying the adoption of improved mungbean varieties in South Asia and Myanmar. Food secur. 11 (3), 623–634. doi:10.1007/s12571-019-00926-x
Serraj, R., Krishnamurthy, L., and Upadhyaya, H. D. (2004). Screening chickpea mini–core germplasm for tolerence to soil salinity. Int. Chickpea PigeonpeaNewslett. 11, 29–32.
Serrano, C., Carbas, B., Castanho, A., Soares, A., Patto, M. C. V., and Brites, C. (2017). Characterisation of nutritional quality traits of a chickpea (Cicer arietinum) germplasm collection exploited in chickpea breeding in Europe. Crop Pasture Sci. 68 (11), 1031. doi:10.1071/CP17129
Shad, N., Mughal, S. M., Farooq, K., and Bashir, M. (2006). Evaluation of mungbean germplasm for resistance against mungbean yellow mosaic begomovirus. Pak. J. Bot. 38 (2), 449.
Shafiq, S., Mather, D. E., Ahmad, M., and Paull, J. G. (2012). Variation in tolerance to radiant frost at reproductive stages in field pea germplasm. Euphytica 186 (3), 831–845. doi:10.1007/s10681-012-0625-0
Shaheen, S., Harun, N., Khan, F., Hussain, R. A., Ramzan, S., Rani, S., et al. (2012). Comparative nutritional analysis between Vigna radiata and Vigna mungo of Pakistan. Afr. J. Biotechnol. 11 (25), 6694. doi:10.5897/AJB11.3496
Shanthi, P., Ganesan, K. N., Manivannan, N., and Natarajan, C. (2019). Correlation and path analysis in Blackgram (Vigna mungo L.). Electron. J. Plant Breed. 10 (3), 1218–1222. doi:10.5958/0975-928x.2019.00154.6
Sharma, A., and Chahota, R. K. (2022). Comparative analysis of biochemical parameters in response to stress in horsegram (Macrotyloma uniflorum L.) germplasm. Himachal J. Agric. Res. 47 (2), 143–148.
Sharma, H. C., Mukuru, S. Z., Manyasa, E., and Were, J. W. (1999). Breakdown of resistance to sorghum midge, Stenodiplosissorghicola. Euphytica 109 (2), 131–140. doi:10.1023/a:1003724217514
Sharma, L., Priya, M., Bindumadhava, H., Nair, R. M., and Nayyar, H. (2016). Influence of high temperature stress on growth, phenology and yield performance of mungbean [Vigna radiata (L.) Wilczek] under managed growth conditions. Sci. Hortic. 213, 379–391. doi:10.1016/j.scienta.2016.10.033
Sharma, M., Ghosh, R., and Pande, S. (2015). Dry root rot (Rhizoctonia bataticola (taub.) butler): An emerging disease of chickpea–where do we stand? Arch. Phytopathol. Plant Prot. 48, 797–812. doi:10.1080/03235408.2016.1140564
Sharma, M., Rathore, A., Mangala, U. N., Ghosh, R., Sharma, S., Upadhyay, H. D., et al. (2012). New sources of resistance to Fusarium wilt and sterility mosaic disease in a mini–core collection of pigeonpea germplasm. Eur. J. Plant Pathol. 133 (3), 707–714. doi:10.1007/s10658-012-9949-9
Sharma, M, M., Telangre, R., Ghosh, R., and Pande, S. (2015). Multi–environment field testing to identify broad, stable resistance to sterility mosaic disease of pigeonpea. J. Gen. Plant Pathol. 81 (3), 249–259. doi:10.1007/s10327-015-0585-z
Sharma, S, S., Singh, N., Virdi, A. S., and Rana, J. C. (2015). Himalayan kidney bean germplasm: Grain-flour characteristics, structural-functional properties and in-vitro digestibility of starches. Food Res. Int. 77, 498–505. doi:10.1016/j.foodres.2015.08.030
Shirasawa, K., Chahota, R., Hirakawa, H., Nagano, S., Nagasaki, H., Sharma, T., et al. (2021). A Chromosome–scale draft genome sequence of horsegram (Macrotyloma uniflorum). Gigabyte 2021, 1–23. doi:10.46471/gigabyte.30
Silim, S. N., Gwataa, E. T., Coeb, R., and Omanga, P. A. (2007). Response of pigeonpea genotypes of differrent maturity duration to temperature and photoperiod in Kenya. Afr. Crop Sci. J. 15 (2). doi:10.4314/acsj.v15i2.54420
Silva, C. A., Abreu, Â. D. F. B., Ramalho, M. A. P., and Maia, L. G. S. (2012). Chemical composition as related to seed color of common bean. Crop Breed. Appl. Biotechnol. 12, 132–137. doi:10.1590/s1984-70332012000200006
Simon, C. J., and Hannan, R. M. (1995). Development and use of core subsets of cool–season food legume germplasm collections. Hort. Sci. 30 (4), 907C–907C. doi:10.21273/hortsci.30.4.907c
Singh, D. P. (1995). Lentil germplasm: Evaluation and utilization, a review. Indian J. Genet. Plant Breed. 55 (02), 173.
Singh, D. P., and Singh, B. B. (2011). Breeding for tolerance to abiotic stresses in mungbean. J. Food Leg. 24 (2), 83.
Singh, K. B. (1997). Chickpea (Cicer arietinum L.). Field Crops Res. 53, 161–170. doi:10.1016/s0378-4290(97)00029-4
Singh, K. B., Malhotra, R. S., and Saxena, M. C. (1989). Chickpea evaluation for cold tolerance under field conditions. Crop Sci. 29 (2), 282–285. doi:10.2135/cropsci1989.0011183x002900020009x
Singh, K., Shekhawat, N., Singh, O. V., Choudhary, M., and Ram, D. (2020). Identification for sources of resistance to biotic stresses in mothbean [Vigna aconitifolia (Jaeq.) Marcchal] Germplasm. Int. J. Curr. Microbiol. Appl. Sci. 9 (5), 1243–1249. doi:10.20546/ijcmas.2020.905.138
Singh, M., Dahiya, O. P., Singh, N., Nizar, A., Dutta, M., Tyagi, R. K., et al. (2013). Identification of an upright peduncle and podding genotype in chickpea germplasm conserved in the National Genebank. Indian J. Plant Genet. Resour. 26 (2), 166–168.
Singh, N., Kaur, N., Rana, J. C., and Sharma, S. K. (2010). Diversity in seed and flour properties in field pea (Pisum sativum) germplasm. Food Chem. 122 (3), 518–525. doi:10.1016/j.foodchem.2010.02.064
Singh, N. K., Gupta, D. K., Jayaswal, P. K., Mahato, A. K., Dutta, S., Singh, S., et al. (2012). The first draft of the pigeonpea genome sequence. J. Plant Biochem. Biotechnol. 21 (1), 98–112. doi:10.1007/s13562-011-0088-8
Singh, R. K., Singh, C., Chandana, B. S., Mahto, R. K., Patial, R., Gupta, A., et al. (2022). Exploring chickpea germplasm diversity for broadening the genetic base utilizing genomic resourses. Front. Genet. 13, 905771. doi:10.3389/fgene.2022.905771
Singh, R., Salam, J. L., Mandavi, N. C., Saxena, R. R., and Rao, A. (2019). Genetic diversity estimation in horsegram [Macrotyloma uniflorum (L) Verdcout] genotypes collected from Bastar plateau. Int. J. Curr. Microbiol. App. Sci. 8 (12), 613–620. doi:10.20546/ijcmas.2019.812.080
Singh, J, J., Kanaujia, R., Sivastava, A. K., Dixit, G. P., and Singh, N. P. (2017). Genetic variability for iron and zinc as well as antinutrients affecting bioavailability in blackgram (Vigna mungo (L.) Hepper). J. Food Sci. Technol. 54 (4), 1035–1042. doi:10.1007/s13197-017-2548-1
Singh, N, N., Nongthombam, R., SeyieKonjengbamKhoyumthem, K. N. P., and Naorem, B. (2016). Stability analysis of high yielding varieties of blackgram (Vigna Mungo L. Hepper). Int. J. Agric. Environ. Biotechnol. 9, 17–24. doi:10.5958/2230–732X.2016.00003.6
Singh, N. P, N. P., Praharaj, C. S., and Sandhu, J. S. (2016). Utilizing untapped potential of rice fallow of East and North–east India through pulse production. Indian J. Genet. Plant Breed. 76 (4), 388–398. doi:10.5958/0975-6906.2016.00058.4
Singh, S, S., Gupta, V., Singh, S. P., and Yadava, N. S. (2017). Growth and productivity of Moth bean [Vigna aconitifolia (Jacq.) Marechal] in response to different varieties and phosphorus levels. J. Pharm. Phytochem. 6 (3), 811. doi:10.1007/s13197-017-2548-1
Sinha, P., Singh, V. K., Saxena, R. K., Khan, A. W., Abbai, R., Chitikineni, A., et al. (2020). Superior haplotypes for haplotype-based breeding for drought tolerance in pigeonpea (Cajanus cajan L.). Plant Biotechnol. J. 18 (12), 2482–2490. doi:10.1111/pbi.13422
Sonah, H., Bastien, M., Iquira, E., Tardivel, A., Legare, G., Boyle, B., et al. (2013). An improved genotyping by sequencing (GBS) approach offering increased versatility and efficiency of SNP discovery and genotyping. PLoS One 8, e54603. doi:10.1371/journal.pone.0054603
Souleymane, A., Aken’Ova, M. E., Fatokun, C. A., and Alabi, O. Y. (2013). Screening for resistance to cowpea aphid (Aphis craccivora Koch) in wild and cultivated cowpea (Vigna unguiculata L. Walp.) accessions. Int. J. Sci. Environ. Technol. 2, 611.
Soundararajan, R. P., Chitra, N., Geetha, S., and Poorani, J. (2012). Biological control of bruchid Callosobruchus maculatus (F.) in blackgram. J. Biopestic. 5, 192.
Souter, J. R., Gurusamy, V., Porch, T. G., and Bett, K. E. (2017). Successful introgression of abiotic stress tolerance from wild tepary bean to common bean. Crop Sci. 57 (3), 1160–1171. doi:10.2135/cropsci2016.10.0851
Spindel, J., Wright, M., Chen, C., Cobb, J., Gage, J., Harrington, S., et al. (2013). Erratum to: Bridging the genotyping gap: Using genotyping by sequencing (GBS) to add high-density SNP markers and new value to traditional bi-parental mapping and breeding populations. Theor. Appl. Genet. 126, 201–202. doi:10.1007/s00122-015-2618-6
Srinivasan, A., Johansen, C., and Saxena, N. P. (1998). Cold tolerance during early reproductive growth of chickpea (Cicer arietinum L.): Characterization of stress and genetic variation in pod set. Field Crops Res. 57 (2), 181–193. doi:10.1016/s0378-4290(97)00118-4
Stoddard, F. L., Balko, C., Erskine, W., Khan, H. R., Link, W., and Sarker, A. (2006). Screening techniques and sources of resistance to abiotic stresses in cool-season food legumes. Euphytica 147 (1), 167–186. doi:10.1007/s10681-006-4723-8
Subramaniyan, R., Narayana, M., Krishnamoorthy, I., Natarajan, G., and Gandhi, K. (2022). Novel and stable QTL regions conferring resistance to MYMV disease and its inheritance in blackgram (Vigna mungo (L.) Hepper). J. Genet. 101 (1), 18–27. doi:10.1007/s12041-022-01359-w
Sudhakaran, S. M. N., and Bukkan, D. S. (2021). A review on nutritional composition, antinutritional components and health benefits of greengram (Vigna radiata (L.) Wilczek). J. Food Biochem. 45 (6), e13743. doi:10.1111/jfbc.13743
Sultana, R., Saxena, K. B., Kumar, R. R., Kumar, D., and Kirti, M. (2021). Pigeonpea” in the Beans and the peas. Woodhead Publishing, 217–240.
Sultana, R., Vales, M. I., Saxena, K. B., Rathore, A., Rao, S., Rao, S. K., et al. (2013). Waterlogging tolerance in pigeonpea (Cajanus cajan (L.) millsp.): Genotypic variability and identification of tolerant genotypes. J. Agric. Sci. 151 (5), 659–671. doi:10.1017/s0021859612000755
Summo, C., De Angelis, D., Ricciardi, L., Caponio, F., Lotti, C., Pavan, S., et al. (2019). Nutritional, physico–chemical and functional characterization of a global chickpea collection. J.Food Compos. Anal. 84, 103306. doi:10.1016/j.jfca.2019.103306
Suneja, Y., Kaur, S., Gupta, A. K., and Kaur, N. (2011). Levels of nutritional constituents and antinutritional factors in blackgram (Vigna mungo L. Hepper). Food Res. Int. 44 (2), 621–628. doi:10.1016/j.foodres.2010.12.020
Talari, A., and Shakappa, D. (2018). Role of pigeon pea (Cajanus cajan L.) in human nutrition and health: A review. Asian J. Dairy Food Res. doi:10.18805/AJDFR.DR-1379
Teshome, A., Mendesil, E., Geleta, M., Andargie, D., Anderson, P., Rämert, B., et al. (2015). Screening the primary gene pool of field pea (Pisum sativum L. subsp. sativum) in Ethiopia for resistance against pea weevil (Bruchuspisorum L.). Genet. Resour. Crop Evol. 62 (4), 525–538. doi:10.1007/s10722-014-0178-2
Tripathi, K., Gore, P. G., Bansal, R., Gayacharan, C., Shubha, K., Kumar, V., et al. (2021b). Identification and revealing the potential traits of the unique germplasm with extended funiculus in pea (Pisum sativum L.). Genet. Res. Crop Evol. 68 (8), 3125–3132. doi:10.1007/s10722-021-01275-5
Tripathi, K., Kumari, J., Gore, P. G., Mishra, D. C., Singh, A. K., Mishra, G. P., et al. (2021a). Agro–morphological characterization of lentil germplasm of Indian National genebank and development of a core set for efficient utilization in lentil improvement programs. Front. Plant Sci. 12, 751429. doi:10.3389/fpls.2021.751429
Tripathi, K., Prasad, T. V., Bhardwaj, R., Jha, S. K., Semwal, D. P., Gore, P. G., et al. (2020). Evaluation of diverse germplasm of cowpea [Vigna unguiculata (L.) Walp.] against bruchid [Callosobruchus maculatus (Fab.)] and correlation with physical and biochemical parameters of seed. Plant Genet. Resour. 18 (3), 120–129. doi:10.1017/s1479262120000180
Upadhyaya, H. D., Bajaj, D., Srivastava, R., Daware, A., Basu, U., Tripathi, S., et al. (2017). Genetic dissection of plant growth habit in chickpea. Funct. Integr. Genomics 17 (6), 711–723. doi:10.1007/s10142-017-0566-8
Upadhyaya, H. D., Bramel, P. J., and Singh, S. (2001). Development of a chickpea core subset using geographic distribution and quantitative traits. Crop Sci. 41 (1), 206–210. doi:10.2135/cropsci2001.411206x
Upadhyaya, H. D., Dronavalli, N., Gowda, C. L. L., and Singh, S. (2011a). Identification and evaluation of chickpea germplasm for tolerance to heat stress. Crop Sci. 51 (5), 2079–2094. doi:10.2135/cropsci2011.01.0018
Upadhyaya, H. D., Dwivedi, S. L., Gowda, C. L. L., and Singh, S. (2007b). Identification of diverse germplasm lines for agronomic traits in a chickpea (Cicer arietinum L.) core collection for use in crop improvement. Field Crops Res. 100 (2–3), 320–326. doi:10.1016/j.fcr.2006.08.008
Upadhyaya, H. D., and Ortiz, R. (2001). A mini core subset for capturing diversity and promoting utilization of chickpea genetic resources in crop improvement. Theor. Appl. Genet. 102 (8), 1292–1298. doi:10.1007/s00122-001-0556-y
Upadhyaya, H. D., Ortiz, R., Bramel, P. J., Singh, S., et al. (2002). Phenotypic diversity for morphological and agronomic characteristics in chickpea core collection. Euphytica 123, 333–342. doi:10.1023/a:1015088417487
Upadhyaya, H. D., Reddy, K. N., Sastry, D. V. S. S. R., and Gowda, C. L. L. (2007c). Identification of photoperiod insensitive sources in the world collection of pigeonpea at ICRISAT. J. Sat. Agric. Res. 3 (1), 1.
Upadhyaya, H. D., Reddy, K. N., Sharma, S., Varshney, R. K., Bhattacharjee, R., Singh, S., et al. (2011b). Pigeonpea composite collection and identification of germplasm for use in crop improvement programmes. Plant Genet. Resour. 9 (1), 97–108. doi:10.1017/s1479262110000419
Upadhyaya, H. D., Reddy, L. J., Gowda, C. L. L., Reddy, K. N., and Singh, S. (2006). Development of a mini core subset for enhanced and diversified utilization of pigeonpea germplasm resources. Crop Sci. 46 (5), 2127–2132. doi:10.2135/cropsci2006.01.0032
Upadhyaya, H. D., Salimath, P. M., Gowda, C. L. L., and Singh, S. (2007a). New early–maturing germplasm lines for utilization in chickpea improvement. Euphytica 157 (1), 195–208. doi:10.1007/s10681-007-9411-9
Upadhyaya, H. D., Yadav, D., Dronavalli, N., Gowda, C. L. L., and Singh, S. (2010). Mini core germplasm collections for infusing genetic diversity in plant breeding programs. Electron. J. Plant Breed. 1 (4), 1294.
Urrea, C. A., and Harveson, R. M. (2014). Identification of sources of bacterial wilt resistance in common bean (Phaseolus vulgaris). Plant Dis. 98 (7), 973–976. doi:10.1094/PDIS-04-13-0391-RE
Urrea, C. A., and Porch, T. (2009). Phenotypic evaluation of a subset of the Phaseolusvulgaris core collections and the P. acutifolius germplasm collection, and advanced common bean lines for drought tolerance in Nebraska. Cooperative 104
Van de Wouw, A. P., Sheedy, E. M., Ware, A. H., Marcroft, S. J., and &Idnurm, A. (2022). Independent breakdown events of the Brassica napus Rlm7 resistance gene including via the off-target impact of a dual-specificity avirulence interaction. Mol. Plant Pathol. 23, 997–1010. doi:10.1111/mpp.13204
Varshney, R. K., Bohra, A., Yu, J., Graner, A., Zhang, Q., and Sorrells, M. K. (2021). Designing future crops: Genomics-assisted breeding comes of age. Trends Plant Sci. 26 (6), 621–630. doi:10.1016/j.tplants.2005.10.004
Varshney, R. K., Chen, W., Li, Y., Bharti, A. K., Saxena, R. K., Schlueter, J. A., et al. (2011). Draft genome sequence of pigeonpea (Cajanus cajan), an orphan legume crop of resource–poor farmers. Nat. Biotechnol. 30 (1), 83–89. doi:10.1038/nbt.2022
Varshney, R. K. (2016). Exciting journey of 10 years from genomes to fields and markets: Some success stories of genomics-assisted breeding in chickpea, pigeonpea and groundnut. Plant Sci. 242, 98–107. doi:10.1016/j.plantsci.2015.09.009
Varshney, R. K., Gaur, P. M., Chamarthi, S. K., Krishnamurthy, L., Tripathi, S., Kashiwagi, J., et al. (2013b). Fast-track introgression of “QTL-hotspot” for root traits and other drought tolerance traits in JG 11, an elite and leading variety of chickpea. Plant Genome 6 (3), plantgenome2013–07. doi:10.3835/plantgenome2013.07.0022
Varshney, R. K., Roorkiwal, M., Sun, S., Bajaj, P., Chitikineni, A., Thudi, M., et al. (2021). A chickpea genetic variation map based on the sequencing of 3, 366 genomes. Nature 599 (7886), 622–627. doi:10.1038/s41586-021-04066-1
Varshney, R. K., Saxena, R. K., Upadhyaya, H. D., Khan, A. W., Yu, Y., Kim, C., et al. (2017). Whole-genome resequencing of 292 pigeonpea accessions identifies genomic regions associated with domestication and agronomic traits. Nat. Genet. 49, 1082–1088. doi:10.1038/ng.3872
Varshney, R. K., Song, C., Saxena, R. K., Azam, S., Yu, S., Sharpe, A. G., et al. (2013a). Draft genome sequence of chickpea (Cicer arietinum) provides a resource for trait improvement. Nat. Biotechnol. 1 (3), 240–246. doi:10.1038/nbt.2491
Vir, O., and Singh, A. K. (2015). Moth bean [Vigna aconitifolia (Jacq.) Marechal] germplasm: Evaluation for genetic variability and inter characters relationship in hot arid climate of Western Rajasthan, India. Legume Res. 38 (6), 748–752.
Vishnu, B., Jayalakshmi, V., Rani, M. S., Vlasova, A., Capella–Gutiérrez, S., Rendón–Anaya, M., et al. (2020). Genetic diversity studies among chickpea (Cicer arietinum L.) genotypes under rainfed and irrigated conditions for yield attributing and traits related to mechanical harvestingGenome and transcriptome analysis of the Mesoamerican common bean and the role of gene duplications in establishing tissue and temporal specialization of genes. Legume Research-An Int. JournalGenome Biol. 4317 (21), 321–19418. doi:10.1186/s13059-016-0883-6
Wang, J., Li, Y., Li, A., Liu, R. H., Gao, X., Li, D., et al. (2021). Nutritional constituent and health benefits of chickpea (Cicer arietinum L.): A review. Food Res. Int. 150, 110790. doi:10.1016/j.foodres.2021.110790
White, P. J., Bradshaw, J. E., Finlay, M., Dale, B., Ramsay, G., Hammond, J. P., et al. (2009). Depolarization-activated calcium channels shape the calcium signatures induced by low-temperature stress. HortSci 44 (1), 6–8. doi:10.1111/j.1469-8137.2009.02857.x
WIEWS (2022). WIEWS - World Information and Early Warning System on Plant Genetic Resources for Food and Agriculture Available at: https://www.fao.org/wiews/data/ex–situ–sdg–251/search/en/?no_cache=1 (Accessed on February 2, 2022).
Wisser, R. J., Oppenheim, S. J., Ernest, E. G., Mhora, T. T., Dumas, M. D., Gregory, N. F., et al. (2021). Genome assembly of a mesoamerican derived variety of lima bean: A foundational cultivar in the mid–atlantic USA. G3 11 (11), jkab207. doi:10.1093/g3journal/jkab207
Xia, Q., Pan, L., Zhang, R., Ni, X., Wang, Y., Dong, X., et al. (2019). The genome assembly of asparagus bean, Vigna unguiculata ssp. Sci. Data 6 (1), 124. doi:10.1038/s41597-019-0130-6
Xu, H., Jing, T., Tomooka, N., Kaga, A., Isemura, T., and Vaughan, D. A. (2008). Genetic diversity of the azuki bean (Vigna angularis (Willd.) Ohwiand Ohashi) gene pool as assessed by SSR markers. Genome 51 (9), 728–738. doi:10.1139/G08-058
Xu, Y., Zhang, X., Li, H., Zheng, H., Zhang, J., Olsen, M. S., et al. (2022). Smart breeding driven by big data, artificial intelligence and integrated genomic-enviromic prediction. Mol. Plant 15, 1664–1695. doi:10.1016/j.molp.2022.09.001
Xu–Xiao, Z., Jian–Ping, G., Wang, S., Liu, Q., Redden, R. R., and Ford, R. (2008). Genetic diversity and core collection of alien Pisum sativum L. germplasm. Acta Agron. Sin. 34 (9), 1518–1528. doi:10.1016/s1875-2780(09)60003-1
Yadav, S., Negi, K. S., and Mandal, S. (2004). Protein and oil rich wild horsegram. Genet. Resour. Crop Evol. 51 (6), 629–633. doi:10.1023/b:gres.0000024650.44318.2d
Yang, K., Tian, Z., Chen, C., Luo, L., Zhao, B., Wang, Z., et al. (2015). Genome sequencing of adzuki bean (Vigna angularis) provides insight into high starch and lowfat accumulation and domestication. Proc. Natl. Acad. Sci. 112 (43), 13213–13218. doi:10.1073/pnas.1420949112
Yasin, J. K., Nizar, M. A., Rajkumar, S., Verma, M., Verma, N., Pandey, S., et al. (2014). Existence of alternate defense mechanisms for combating moisture stress in horse gram [Macrotyloma uniflorum (Lam.) Verdc.]. Legume Res. Int. J. 37 (2), 145–154. doi:10.5958/j.0976-0571.37.2.022
Yohane, E. N., Shimelis, H., Laing, M., Mathew, I., and Shayanowako, A. (2020). Phenotypic divergence analysis in pigeonpea [Cajanus cajan (L.) Millspaugh] germplasm accessions. Agronomy 10 (11), 1682. doi:10.3390/agronomy10111682
Zaccardelli, M., Lupo, F., Piergiovanni, A. R., Laghetti, G., Sonnante, G., Daminati, M. G., et al. (2012). Characterization of Italian lentil (Lens culinarisMedik.) germplasm by agronomic traits, biochemical and molecular markers. Genet. Resour. Crop Evol. 59 (5), 727–738. doi:10.1007/s10722-011-9714-5
Zeven, A. C., and Zhukovsky, P. M. (1975). Dictionary of cultivated plants and their centres of diversity. Wageningen: PUDOC.
Zhang, H., Zhang, D., Wang, M., Sun, J., Qi, Y., Li, J., et al. (2011). A core collection and mini core collection of Oryza sativa L. in China. Theor. Appl. Genet. 122, 49–61. doi:10.1007/s00122-010-1421-7
Zhang, X., Wan, S., Hao, J., Hu, J., Yang, T., and Zong, X. (2016). Large–scale evaluation of pea (Pisum sativum L.) germplasm for cold tolerance in the field during winter in Qingdao. Crop J 4 (5), 377–383. doi:10.1016/j.cj.2016.06.016
Keywords: pulse production, crop domestication, biotic and abiotic stresses, legume genomics, legume collections
Citation: Gayacharan , Parida SK, Mondal N, Yadav R, Vishwakarma H and Rana JC (2023) Mining legume germplasm for genetic gains: An Indian perspective. Front. Genet. 14:996828. doi: 10.3389/fgene.2023.996828
Received: 18 July 2022; Accepted: 05 January 2023;
Published: 23 January 2023.
Edited by:
Aamir Raina, Aligarh Muslim University, IndiaReviewed by:
Parvaze Sofi, Sher-e-Kashmir University of Agricultural Sciences and Technology of Kashmir, IndiaMomina Hussain, National Institute for Biotechnology and Genetic Engineering, Pakistan
Copyright © 2023 Gayacharan, Parida, Mondal, Yadav, Vishwakarma and Rana. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jai C. Rana, j.rana@cgiar.org
 Gayacharan
Gayacharan Swarup K. Parida
Swarup K. Parida Nupur Mondal
Nupur Mondal Rashmi Yadav
Rashmi Yadav Harinder Vishwakarma
Harinder Vishwakarma Jai C. Rana
Jai C. Rana