- 1Department of Prosthodontics, College of Dentistry, Yonsei University, Seoul, Republic of Korea
- 2Graduate School of Artificial Intelligence, Pohang University of Science and Technology, Pohang, Republic of Korea
- 3Tufts University School of Medicine, Boston, MA, United States
- 4Department of Life Sciences, Pohang University of Science and Technology, Pohang, Republic of Korea
Abstract
Determining genotype–phenotype correlations in patients with hypodontia is important for understanding disease pathogenesis, although only a few studies have elucidated it. We aimed to identify genetic variants linked to non-syndromic bilateral mandibular second premolar hypodontia in a Korean population for the first time by specifying the phenotype of hypodontia. Twenty unrelated individuals with non-syndromic bilateral mandibular second premolar hypodontia were enrolled for whole-exome sequencing. Using a tooth agenesis gene set panel consisting of 112 genes based on literature, potential candidate variants were screened through variant filtering and prioritization. We identified 13 candidate variants in 12 genes, including a stop-gain variant (c.4750C>T) in LAMA3. Through the functional enrichment analysis of the prioritized genes, several terms related to tooth development were enriched in a protein–protein interaction network of candidate genes for mandibular premolar hypodontia. The hypodontia group also had approximately 2-fold as many mutated variants in all four genes related to these key terms, which are CDH1, ITGB4, LAMA3, LAMB3, as those in the 100 healthy control group individuals. The relationship between enriched terms and pathways and mandibular premolar hypodontia was also investigated. In addition, we identified some known oligodontia variants in patients with hypodontia, strengthening the possibility of synergistic effects in other genes. This genetic investigation may be a worthwhile preliminary attempt to reveal the pathogenesis of tooth agenesis and sets a background for future studies.
1 Introduction
Congenital tooth agenesis, which is a failure in tooth development, is the most common dental anomaly in humans (De Coster et al., 2009). Tooth agenesis can alter masticatory efficiency, aesthetic appearance, and speech. Studies have found that significant facial changes can occur with congenital tooth agenesis (Taju et al., 2018). Since teeth tend to migrate towards edentulous regions, missing teeth may result in poor positioning of the remaining teeth, making it difficult to manage proper oral rehabilitation. Early diagnosis of tooth agenesis is crucial so that long-term treatment plans, such as the conservation of deciduous teeth or the maintenance of appropriate space for lost teeth for dental implants, can be established for the patients (Nunn et al., 2003).
Different terms describe tooth agenesis based on the number of missing teeth, excluding third molars. When less than six teeth fail to develop, tooth agenesis is referred to as “hypodontia.” When six or more teeth are missing, tooth agenesis is called “oligodontia,” and “anodontia” refers to the developmental failure of all teeth. Hypodontia is associated with approximately 150 genetic syndromes, including cleft lip and/or cleft palate (Vieira, 2003); however, non-syndromic tooth agenesis (NSTA), in which only dentition is affected as familial or sporadic forms, can also occur without systemic genetic syndromes. Hypodontia can be caused by environmental factors, such as infection, trauma, or early radiation therapy, but is mainly associated with genetic mutations (Ye and Attaie, 2016).
The overall prevalence of hypodontia is 6.4%, but this varies within different continents and racial groups (Khalaf et al., 2014). Mandibular second premolars are the most commonly affected teeth, and there are no specific studies on the prevalence of bilateral mandibular second premolar hypodontia. However, we can assume the occurrence to be approximately 1% from a meta-analysis report, considering that approximately 21% of tooth agenesis patients have missing bilateral mandibular premolars, and approximately 67% of patients with more than one missing tooth have two missing teeth (Polder et al., 2004).
Teeth develop via a series of complex signaling interactions between the oral epithelium and neural crest-derived mesenchyme through epithelial thickening (initiation) and bud, cap, and bell stages (Tucker and Sharpe, 2004). These molecular signaling pathways are under strict genetic control during development (Yin and Bian, 2015). Numerous factors, such as nuclear factor (NF)-κB, wingless-related integration site (Wnt), bone morphogenic protein (Bmp), fibroblast growth factor (FGF), and sonic hedgehog (Shh) families, are considerably involved in the regulation of tooth development. Any alteration in these factors can potentially contribute to hypodontia (Galluccio et al., 2012). Since mutations in the MSX1 and PAX9 were found to cause NSTA (Vastardis et al., 1996; Stockton et al., 2000), more than 18 genes have been identified as additional causes (AlFawaz et al., 2015; Bonczek et al., 2017; Williams and Letra, 2018). A recent systematic review aimed to investigate the NSTA phenotypes associated with gene mutations and revealed patterns of tooth agenesis based on the identity of mutated genes. For example, mutations in MSX1 and PITX2 were reported to be responsible for premolar and third molar agenesis, whereas PAX9 mutations resulted in agenesis in all molars and mandibular central incisors (Fournier et al., 2018). Two WNT10A variants cause maxillary lateral incisor agenesis (Mostowska et al., 2015). In addition, rs15705 and rs3178250 variants of BMP2 increase the risk of mandibular incisor agenesis (Lu et al., 2016). Despite accumulating evidence for different hypodontia phenotype pathogeneses, research in this field is very limited. Hypodontia shows variable expressivity due to the numerous ways in which 28 unique, permanent teeth could be absent or positioned. Additionally, mutations in several genes contribute to diverse phenotypes with the possibility of oligogenic inheritance. A study that performed genotype–phenotype analysis in patients with WNT10A mutations reported that WNT10A mutations were associated with the number and type of tooth agenesis, such as biallelic WNT10A mutations associated with absence of maxillary and mandibular molars as well as mandibular central incisors. However, one limitation of this study was the candidate gene analysis (Arzoo et al., 2014).
Recent developments in next-generation sequencing have enabled rapid and efficient analysis of personal DNA genome sequences. Whole-exome sequencing (WES) has received particular attention for the identification of genetic variants of diseases. Exomes refer to all exons within the genome and represent approximately 1.5% of the human genome. WES has been proven effective because up to 90%–85% of known disease-related variants are contained in exons. Earlier, traditional research on non-syndromic hypodontia focused on finding variants in candidate genes, but current trends using WES have revealed new mutated genes for non-syndromic hypodontia (Zeng et al., 2017; Dinckan et al., 2018; Zhang et al., 2019), including OPN3, which was previously thought to be irrelevant to tooth development (Inagaki et al., 2021). Genotype–phenotype correlation in hypodontia is important not only for understanding the pathogenesis of the disease but also for the specific tooth regeneration required for replacement eventually (Chhabra et al., 2014; Williams and Letra, 2018). By specifying the phenotype of non-syndromic hypodontia as agenesis of the bilateral mandibular second premolar and applying WES and bioinformatics in the Korean population, in this study, we aimed to better understand the underlying mechanisms of hypodontia by identifying commonalities between hypodontia-causing variants.
2 Materials and methods
2.1 Participants
Twenty unrelated individuals were included in the study. The participants were selected based on the following criteria: congenital absence of bilateral mandibular second premolars, presence of all other permanent teeth (excluding the third molars), and absence of any other genetic syndromes. They were examined by one dentist for case eligibility using radiographic and clinical examinations and were excluded if they showed any other craniofacial anomaly or syndrome. Eligibility was confirmed after asking participants about their dental and medical histories, including pedigree data, to assess their family history. Panoramic radiographic images, clinical records, and written informed consent were obtained from all patients in this study. One hundred healthy exome sequencing data from 3,703 Ansan–Ansung cohort participants provided by the National Biobank of Korea, the Korea Disease Control and Prevention Agency, Republic of Korea (KBN-2021-063) were used as controls. This clinical study was approved by the Institutional Review Board of the Armed Forces Medical Command (AFMC-20085-IRB-20-085; approved on 3 November 2020). This clinical study was conducted in accordance with the Declaration of Helsinki.
2.2 DNA sample collection
A 2-mL saliva sample was obtained from each participant using an Oragene DNA self-collection kit (OG-500; DNA Genotek). Each saliva sample was mixed with a solution from the collection kit, which stabilized the sample at room temperature. Genomic DNA was extracted using a preplT-L2P DNA extraction kit (DNA Genotek) according to the manufacturer’s instructions. DNA Link inc. performed DNA collection, extraction, and whole-exome sequencing. The concentration and purity of the DNA samples were evaluated using agarose gel electrophoresis and a PicoGreen dsDNA Assay (Invitrogen).
2.3 Whole-exome sequencing (WES)
Briefly, exons were captured according to the manufacturer’s instructions using SureSelect Human All Exon V5 (Agilent Technologies), and 2 × 100 base pair reads were sequenced using an Illumina Novaseq 6,000 system (Illumina). FASTQC v0.11.5 was used to assess the quality of the read sequences. Each read was compared to the Genome Reference Consortium Human Build 37, and the Burrows–Wheeler Aligner package v0.7.12 was used to align the reads with the reference genome. The reads were realigned around indels, and base quality was recalibrated based on changes using GenomeAnalysisTK (GATK) v3.5. After these steps, alignment quality control was performed, and the variants were called using the GATK HaplotypeCaller gVCF mode. Raw variants were recalibrated using GATK v3.5, and finally, analysis-ready VCF files were produced using filtered variants.
2.4 Filtering of exomic variants and data analysis
We applied two different approaches. First, Fisher’s exact test was performed to compare the allele frequencies of each variant between the case-control groups. Odds ratios with 95% confidence intervals were used to evaluate the corresponding risks of variants and mandibular premolar hypodontia. PLINK v1.9 was used as the statistical software. Variants with an association p < 1 × 10−7 and odds ratios >1 were considered statistically significant. We then applied an additional filter with variants annotated as MODERATE or HIGH impact on SnpEff v 4.2 (Cingolani et al., 2012). Finally, we selected variants with a total allele frequency (AF) < 0.01 in the genome aggregation database (gnomAD) v2.1.1. GnomAD has the largest population variation collection in databases and is used to identify pathogenic variants (Karczewski et al., 2020). Data are available on the gnomAD website (https://gnomad.broadinstitute.org/) (Gudmundsson et al., 2022).
Second, we constructed a gene set panel for previously identified genes involved in syndromic tooth agenesis (STA) or NSTA, consisting of 112 genes from literature (Yin and Bian, 2015; Williams and Letra, 2018; Andersson et al., 2020). Whole gene names are listed in Supplementary Table S1. From the genetic variants in the gene set panel, we selected variants annotated as MODERATE or HIGH impact on SnpEff v 4.2 with total AFs under 0.01 in both the 1000G project and gnomAD v2.1.1.
2.5 Prioritization of mutated variants
To prioritize and elucidate the pathogenicity of mutated variants that may cause mandibular premolar hypodontia, we applied 18 in silico prediction tools: Mutation Taster, PROVEAN, SIFT, SIFT4G, Polyphen2, MetaSVM, REVEL, LIST-S2, LRT, M-CAP, Mutation Assessor, BayesDel noAF, MetaLR, DANN, EIGEN, EIGEN PC, FATHMM, and FATHMM-MKL. SnpEff was used to identify missense variants from SIFT and PolyPhen2. In addition, the scores of Combined Annotation-Dependent Depletion (CADD) were checked for candidate variants. CADD integrates multiple annotations, such as sequence conservation score and ENCODE project functional annotations, and expresses the variants’ deleteriousness as a number (Kircher et al., 2014). The CADD score can be considered a ranking, and the higher the score, the more deleterious the variant. We prioritized variants that were estimated as deleterious by at least half of the in silico prediction tools (more than medium in the case of Mutation Assessor) and had CADD scores above 20. Prioritized variants were compared with the Human Gene Mutation Database to check if they were previously reported in tooth agenesis. Prioritized variants were finally ruled out if they were classified as benign using the ClinVar database or the American College of Medical Genetics and Genomics (ACMG) guidelines. According to a recent report, Sanger sequencing validation was not performed, considering we only detected single nucleotide variants, and the coverage depth of WES obviated the need (Arteche-López et al., 2021).
2.6 In silico functional analysis of candidate genes related to the disease
We applied the computational tool STRING v11.5 (Szklarczyk et al., 2019) to examine the pathways and functional terms associated with the candidate genes of mandibular premolar hypodontia. We inputted 12 candidate genes obtained from WES with “homo sapiens” as the organism. We used the multiple proteins query and selected “full STRING network,” “medium confidence level,” and “medium false discovery rate (FDR) stringency.” A protein–protein interaction (PPI) network was visualized, and functional enrichment analysis was performed in the network. The following categories were analyzed for functional enrichment using Fisher’s exact test with Benjamini–Hochberg FDR correction: Gene Ontology (GO), STRING Clusters, KEGG Pathways, Reactome Pathways, Wiki Pathways, Disease-gene associations (DISEASES), Subcellular localization (COMPARTMENTS), Human Phenotype (Monarch), UniProt Annotated Keywords, and SMART protein domains. To compare the total number of mutations in the candidate genes between the case-control groups, the alternate/reference allele ratio of all variants’ sites was calculated. Both synonymous and non-synonymous variants were obtained and odds ratios with p-values were performed by Fisher’s exact test and Bonferroni’s correction.
3 Results
3.1 Patient characteristics
The 20 samples for WES analysis comprised 18 males and 2 females. The median age of the participants in the sample was 21 years (range: 19–65 years). After clinical and radiographic evaluations, all patients were diagnosed with non-syndromic bilateral mandibular second premolar hypodontia (Figure 1). No other signs of genetic syndromes were found in any of the patients.
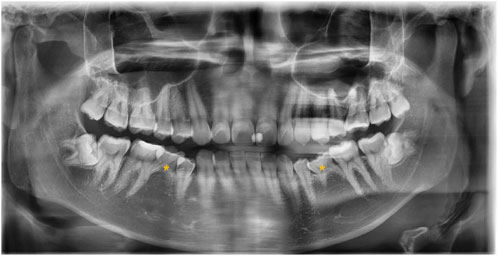
FIGURE 1. Panoramic radiograph of the subject with a congenital lack of mandibular second premolars (indicated with asterisks).
3.2 Candidate variants found in WES analysis and prioritization
A total of 112,487 variants were found in the 20 samples after performing WES. Of these, 853 variants were considered statistically significant after the case-control group comparison, but none met the prioritization criteria. From the gene set panel, 13 variants in 12 genes were found in 11 patients. Of these, 12 variants were missense, and 1 variant was stop-gain, and all 13 variants were heterozygous. No variant was identified in the tooth agenesis gene set panel from the other nine patients. We divided these variants into three categories: known variants, candidate variants in NSTA genes, and candidate variants in STA genes.
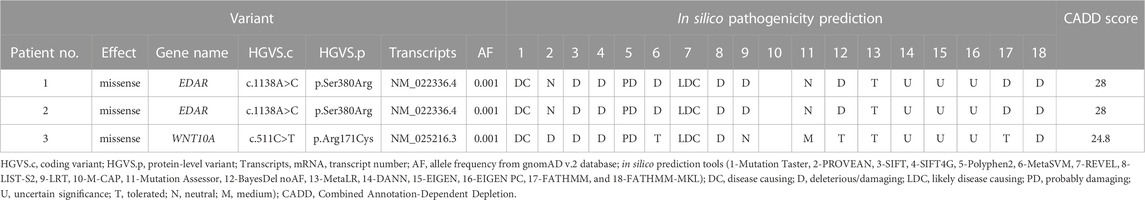
3.3 Known variants
Two variants from three patients were identified from samples previously reported in oligodontia patients (Table 1). A known missense variant (c.1138A>C) in EDAR was found in two patients (Zeng et al., 2017). This variant has been reported pathogenic for non-syndromic oligodontia but benign for ectodermal dysplasia in the ClinVar database. Another known missense variant (c.511C>T) in WNT10A was also found in these samples (Song et al., 2014; Zeng et al., 2017; Park et al., 2019; Zhao et al., 2019; Zhou et al., 2021; Xie et al., 2022).
3.4 Candidate variants in NSTA genes
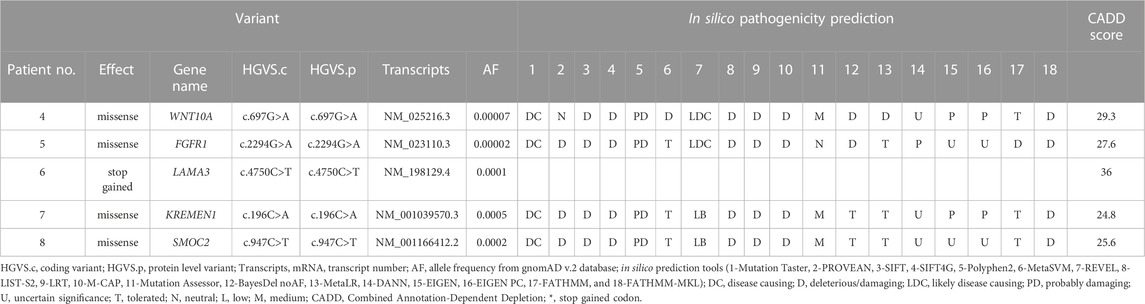
Five candidate variants were identified in the five NSTA genes (Table 2). A missense variant (c.697G>A) in WNT10A has not been previously reported, but a different nucleotide change in the same codon (c.697G>T) was identified in a previous study (Dinckan et al., 2018). No variant was found in MSX1, PAX9, AXIN2, PITX2, EDA, or EDARRD.
3.5 Candidate variants in STA genes
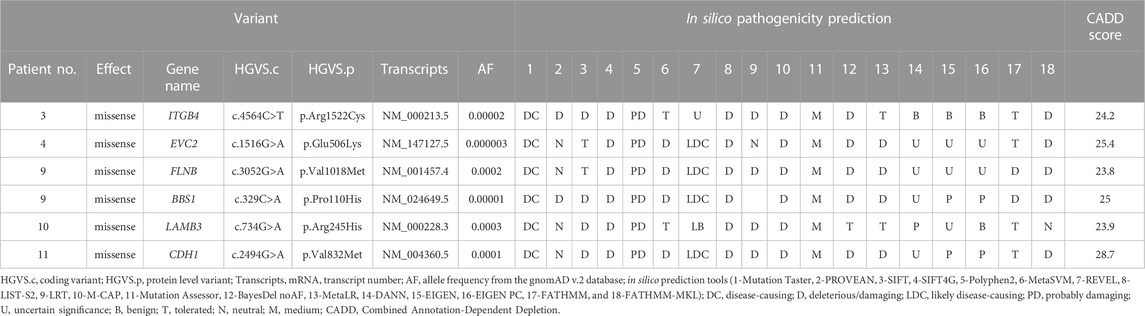
Six variants in six STA genes were identified from the samples (Table 3). Patient 9 had a missense variant (c.3052G>A) in FLNB with a missense variant (c.329C>A) in BBS1. The FLNB variant has conflicting interpretations as three submissions as uncertain significance and one as benign, and the BBS1 variant was of uncertain significance in the ClinVar database. FLNB mutation caused abnormal cranium morphology in a mouse model (Lu et al., 2007). Patient 11 had a missense variant (c.2494G>A) in CDH1. A member of the cadherin superfamily, CDH1 is a transmembrane adhesion protein (Taneyhill, 2008). By regulating cell–cell adhesion or interacting with Wnt intracellular signaling, cadherins serve critical roles in craniofacial morphogenesis and dental development (Schambony et al., 2004; Bienz, 2005; Brembeck et al., 2006; Di Benedetto et al., 2015).
3.6 Predicted effect of a stop-gain variant in LAMA3
We focused on a stop-gain variant (c.4750C>T) in LAMA3 found in patient 6. The transcript domain was located in the laminin IV type A domain, causing termination at the protein position 1,584 instead of 3,333 (Figure 2). This variant can lead to functional consequences due to production of a truncated protein or degradation of the transcript by Nonssense-Mediated Decay (NMD). Mutated LAMA3 protein loses coiled-coil and globular laminin domains, which have adhesion, signaling, differentiation, and gene expression functions. The CADD score was very high at 36, and the variant’s clinical significance was conflicting interpretations of pathogenicity with two uncertain significances for junctional epidermolysis bullosa and one likely benign in the ClinVar database (accession number VCV000551372.4).
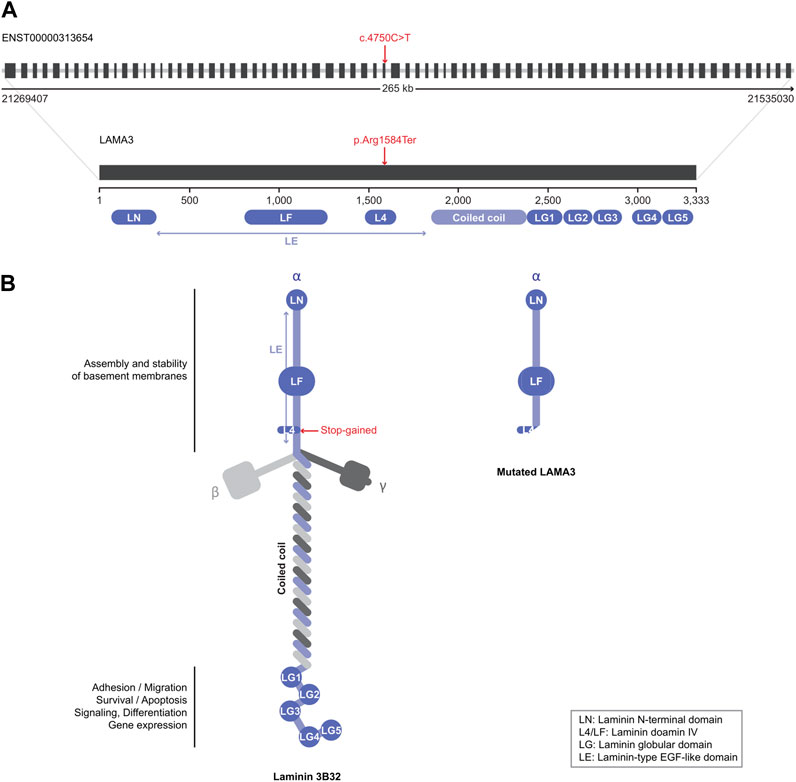
FIGURE 2. Schematic view of the LAMA3 structure. (A) Introns are presented as gray lines and coding exons as black boxes (upper structure). Variants identified in this study are indicated by red characters. (B) Native (left) and mutated (right) LAMA3 protein structures. Each domain’s name and role are explained in the figure.
3.7 In silico functional study of the candidate genes
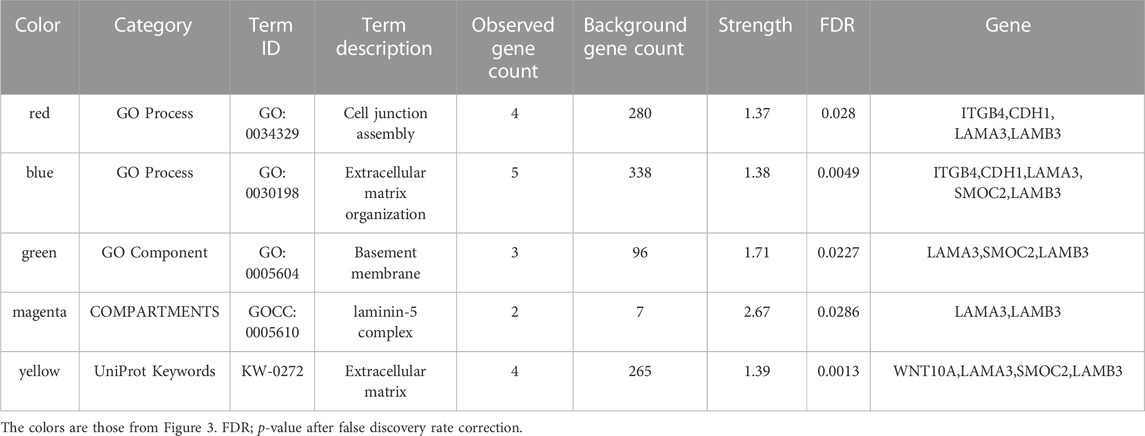
The PPI network for our candidate genes for mandibular premolar hypodontia was visualized using STRING (Figure 3). In total, 343 terms were functionally enriched. Of these, 13 were enriched in GO, 8 in KEGG Pathways, 9 in Reactome Pathways, 7 in Wiki Pathways, 12 in DISEASES, 1 in COMPARTMENTS, 280 in Monarch, 12 in UniProt Annotated Keywords, and 1 in SMART. Whole functional enrichment analysis results are shown in Supplementary Table S2. The terms were mainly related to “cell junction,” “extracellular matrix,” “laminin,” and “basement membrane.” We color-coded the key terms in the PPI network, and these are elaborated on in Table 4. Eight out of the 12 candidate genes were found to be significant in case-control comparisons of alternate/reference allele ratio. In particular, hypodontia groups had mutated variants approximately 2-fold as much in all four genes related to “cell junction assembly” and “extracellular matrix organization” (Table 5).

FIGURE 3. The protein–protein interaction network of variants found in mandibular premolar hypodontia predicted by STRING. Network nodes and edges represent proteins and protein–protein associations, respectively.
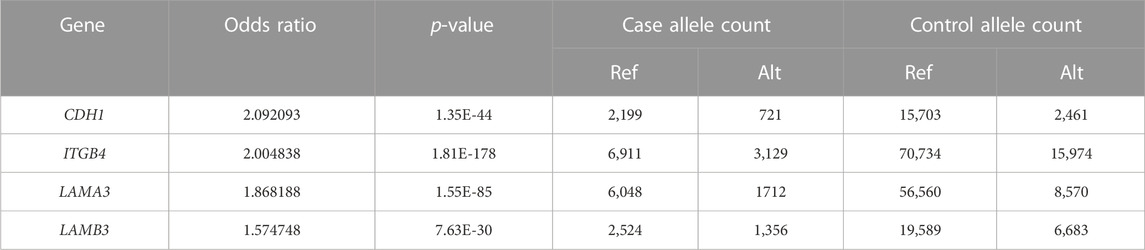
TABLE 5. Fisher’s exact test results of the alternate/reference allele ratio of candidate genes related to key functional terms between the case-control groups.
4 Discussion
To the best of our knowledge, this is the first study to reveal the underlying genetic variants in a specific phenotype of hypodontia using WES and bioinformatics analyses. We selected bilateral mandibular second premolar hypodontia as a phenotype for the following reasons. First, because rodents do not have premolars, it is difficult to unveil the pathogenesis of premolar agenesis using experiments. Second, maxillary and mandibular teeth are reported to be developed by different genetic programs (Ferguson et al., 2000; Cobourne and Sharpe, 2003), and extrinsic factors could be involved in unilateral hypodontia (De Coster et al., 2009).
While a meta-analysis of the prevalence of dental agenesis was conducted (Polder et al., 2004), there may still exist a considerable number of individuals with dental agenesis who are unaware of their condition. Moreover, since there is no specific research for the prevalence of bilateral mandibular second premolar hypodontia, we had to speculate the prevalence from a meta-analysis result. These facts may have affected the outcomes and be a limitation for the study.
No significant variants were found in the case-control group comparisons. We were unable to identify the specific variant that may affect the agenesis of the mandibular second premolars. This might be due to the small sample, but considering the genetic heterogeneity of tooth agenesis, it is probable that a single critical variant for the specific phenotype of the disease may not exist. Instead, considering oligodontia is a rare disease (almost 0.1% prevalence) and a severe form of tooth agenesis, it is interesting that the same variants reported to cause oligodontia were also found in these hypodontia patients. Missense variants (c.1138A>C) and (c.511C>T) in EDAR and WNT10A, respectively, have been previously reported in oligodontia patients. The results of recent WES investigations in tooth agenesis proposed the idea of mutational load with oligogenic inheritance and multilocus variation models (Posey et al., 2017; Dinckan et al., 2018; Andersson et al., 2020). This variable expressivity reinforces the theory that variations in several genes, working either alone or in concert with other genes, may determine the severity of NSTA (Arte et al., 2013; Dreesen et al., 2014; Williams and Letra, 2018).
Notably, LAMA3, LAMB3, and ITGB4 are involved in junctional epidermolysis bullosa (Fine et al., 2008). These proteins are involved in basement membrane-mediated cell adhesion and are critical for proper tooth development (Aberdam et al., 1994; Asaka et al., 2009). Key terms from functional enrichment analysis included “cell junction,” “extracellular matrix,” “basement membrane,” and “laminin.” Moreover, the hypodontia group had approximately 2-fold as many mutated variants in all four genes related to these key terms as the control group. Basement membranes are specialized extracellular matrices composed primarily of different types of laminin, type IV collagen, perlecan, and nidogen (Timpl, 1996). During early development, they act as a physical barrier and regulatory structure for reciprocal signaling between the oral epithelium and mesenchyme layers. These basement membrane matrices regulate proliferation, polarity, and adhesion, as well as the size and morphology of tooth germs (Yoshizaki and Yamada, 2013). Early in the development of the mouse embryo, type IV collagen and laminin molecules are evenly distributed in the dental basement membranes, and they vanish as the membranes are broken down by enzymes (Lesot et al., 1981; Kjoelby et al., 1994). The basement membrane also becomes discontinuous and vanishes at the beginning of the dentin mineralization process (Kawashima and Okiji, 2016). Considering the critical roles of these key terms and the importance of fine balancing between complex networks of signaling pathways during the early odontogenic process, enriched terms may be involved in the pathogenesis of tooth agenesis.
Pathway-related enriched terms included “PI3K-Akt signaling pathway” and “α6β4 signaling pathway”. Several pathways act early in development to determine the differential location, identity, shape, and size of teeth. Even if no specific gene is responsible for each tooth shape, these enriched terms are more likely to be related to molar regions rather than incisor regions. BMP4 and FGF8 are the first molecular signals that initiate differential tooth morphogenesis (Laugel-Haushalter et al., 2013). Fgf8 is expressed in the proximal (presumptive molar region) oral epithelium and promotes Barx1 expression, whereas Bmp4 is initially expressed in the distal (presumptive incisor) epithelium (Tucker et al., 1998). The PI3K-Akt signaling pathway has been associated with cell growth, cell cycle regulation, and cell survival. Recently, it has been proposed to function as an intracellular pathway for transducing FGF8 signals into nuclei in dental mesenchyme, preventing cell apoptosis (Lin et al., 2022). Integrin α6β4 is thought to mediate laminin-10/11-induced cell spreading and filopodia formation of the dental epithelium, implying that these interactions play an important role in determining the size and shape of tooth germs. The interaction between laminin-10/11 and integrin α6β4 is also thought to be required for PI3K/Akt pathway activation in the dental epithelium (Fukumoto et al., 2006). Furthermore, integrin α6β4 stimulates Rac1 and RhoA activation, which modulate cuspal shape decisions by coordinating adhesion junctions, actin distribution, and fibronectin localization to trigger inner dental epithelium invagination (Stewart and O’Connor, 2015; Li et al., 2016).
PITX1, which encodes a novel bicoid-related family of homeoproteins that is differentially expressed between the upper and lower molars, is one of the prime candidate genes for controlling maxillary/mandibular tooth identity (Laugel-Haushalter et al., 2013). Pitx1 is expressed in the proximal mesenchyme of the developing mandible, hindlimb, oral epithelium, developing teeth, and pituitary gland. Inactivation of the Pitx1 gene in mice impacts mandibular tooth morphogenesis (Mitsiadis and Drouin, 2008). Top-ranked terms from functional analyses of Pitx1-dependent genes have included the extracellular matrix during hindlimb development, and differentially expressed gene analysis between control and PITX1-overexpressing osteosarcoma genes also categorized the extracellular matrix and PI3K/Akt in the top 20 KEGG pathways (Nemec et al., 2017; Wang et al., 2018; Zhang et al., 2023).
Laminin α3 (the LAMA3 protein) is an element of laminin-5 (α3β3γ2) that regulates epithelial cell anchoring and motility via the integrins α6β4 and α3β1, respectively (Fukumoto and Yamada, 2005). In an animal study, Lama3-targeted knockout mice showed abnormalities in ameloblast differentiation (Ryan et al., 1999). LAMA3 mutations have also been previously reported in non-syndromic hypodontia patients (Dinckan et al., 2018). We also identified a stop-gain variant (c.4750C>T) in LAMA3 in our patients. To our knowledge, this is the third report of the LAMA3 mutation in non-syndromic tooth agenesis. The exact mechanism by which LAMA3 mutations lead to hypodontia is not fully understood, but it is thought to be related to the role of laminin-5 in tooth bud development and the attachment of teeth to the surrounding tissues. Further, other candidate genes sharing similar functions and enriched terms to LAMA3 may also have a similar role in mandibular premolar hypodontia.
In a genome-wide association study (GWAS) of NSTA, including hypodontia and oligodontia, the rs917412-T variant was reported to be associated with the agenesis of mandibular second premolars (Jonsson et al., 2018). However, none of our samples had this variant possibly because of differences in allele frequency between different ethnicities. The allele frequency of rs917412-T is 0.0007704 in East Asians, which is significantly lower than the overall allele frequency of 0.2054 in the gnomAD database.
Regeneration of teeth is the most ideal treatment in dentistry and there are many advances such as tooth regenerative medicine strategies using targeted molecular therapy (Takahashi et al., 2020). But still many challenges are ahead such as the precise differentiation of dental stem cells into a specific tooth. Increased effort in expanding our comprehensive understanding through WES and bioinformatics is required for the future regenerative therapy.
There are some limitations to this pilot study. First, the sample size was small. Therefore, we attempted to overcome this shortcoming by specifying the phenotype of the disease. Second, the development of teeth is very complex, and interpretation of the impact variants have on the disease is challenging. Simple case-control comparisons may not be able to explain the pathogenicity of tooth agenesis. Pathways and signaling interactions that intersect and interplay during tooth development may be the key to the genotype–phenotype correlation. Third, we investigated genetic variants of unrelated individuals that share the same disease phenotype. Therefore, there is a need for segregation analysis to further verify candidate gene variant impact. Lastly, hypodontia may arise due to the mutations in non-coding regions such as gene regulatory domains. The application of whole-genome sequencing analysis may help further elucidate the pathogenesis of hypodontia.
In summary, we specified a phenotype for non-syndromic hypodontia as agenesis of the bilateral mandibular second premolars in a Korean population and attempted to reveal the pathogenesis of the disease using WES and bioinformatics. We found some known oligodontia gene variants in hypodontia patients, strengthening the possibility of additive effects in other genes and oligogenic inheritance in tooth agenesis. Furthermore, we identified some functionally enriched terms important to mandibular molar development and explored the possible common features of the etiology of mandibular premolar hypodontia. This study is a viable preliminary attempt to reveal the pathogenesis of tooth agenesis and may help design future tooth agenesis studies. Since there are various types of teeth, further studies with other phenotypes and larger populations are required to compare and understand the disease in the future.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, PRJNA983431.
Ethics statement
The studies involving humans were approved by Institutional Review Board of the Armed Forces Medical Command (AFMC-20085-IRB-20-085; approved on 3 November 2020). The studies were conducted in accordance with the local legislation and institutional requirements. The participants provided their written informed consent to participate in this study.
Author contributions
SL: Conceptualization, Methodology, Clinical observation, Data collection, Writing–original draft. HA: Conceptualization, Methodology, Formal analysis, Visualization, Writing–original draft. HK: Writing–Review and Editing, Investigation. KL: Validation, Formal analysis. SK: Supervision, Writing–Review and Editing. JL: Supervision, Writing–Review and Editing, Project administration. All authors contributed to the article and approved the submitted version.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported in part by the National Research Foundation of Korea (grant numbers 2020R1A6A1A03047902 and 2021R1A2B5B01001903).
Acknowledgments
We appreciate the participation of the patients in this study. We would like to thank Editage (www.editage.co.kr) for English language editing.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2023.1248326/full#supplementary-material
References
Aberdam, D., Aguzzi, A., Baudoin, C., Galliano, M. F., Ortonne, J. P., and Meneguzzi, G. (1994). Developmental expression of nicein adhesion protein (laminin-5) subunits suggests multiple morphogenic roles. Cell Adhes. Commun. 2, 115–129. doi:10.3109/15419069409004431
AlFawaz, S., Plagnol, V., Wong, F. S., and Kelsell, D. P. (2015). A novel frameshift MSX1 mutation in a Saudi family with autosomal dominant premolar and third molar agenesis. Arch. Oral Biol. 60, 982–988. doi:10.1016/j.archoralbio.2015.02.023
Andersson, K., Malmgren, B., Åström, E., Nordgren, A., Taylan, F., and Dahllöf, G. (2020). Mutations in COL1A1/A2 and CREB3L1 are associated with oligodontia in osteogenesis imperfecta. Orphanet J. Rare Dis. 15, 80. doi:10.1186/s13023-020-01361-4
Arte, S., Parmanen, S., Pirinen, S., Alaluusua, S., and Nieminen, P. (2013). Candidate gene analysis of tooth agenesis identifies novel mutations in six genes and suggests significant role for WNT and EDA signaling and allele combinations. PLOS ONE 8, e73705. doi:10.1371/journal.pone.0073705
Arteche-López, A., Ávila-Fernández, A., Romero, R., Riveiro-Álvarez, R., López-Martínez, M. A., Giménez-Pardo, A., et al. (2021). Sanger sequencing is no longer always necessary based on a single-center validation of 1109 NGS variants in 825 clinical exomes. Sci. Rep. 11, 5697. doi:10.1038/s41598-021-85182-w
Arzoo, P. S., Klar, J., Bergendal, B., Norderyd, J., and Dahl, N. (2014). WNT10A mutations account for ¼ of population-based isolated oligodontia and show phenotypic correlations. Am. J. Med. Genet. A 164A, 353–359. doi:10.1002/ajmg.a.36243
Asaka, T., Akiyama, M., Domon, T., Nishie, W., Natsuga, K., Fujita, Y., et al. (2009). Type XVII collagen is a key player in tooth enamel formation. Am. J. Pathol. 174, 91–100. doi:10.2353/ajpath.2009.080573
Bienz, M. (2005). Beta-catenin: A pivot between cell adhesion and Wnt signalling. Curr. Biol. 15, R64–R67. doi:10.1016/j.cub.2004.12.058
Bonczek, O., Balcar, V. J., and Šerý, O. (2017). PAX9 gene mutations and tooth agenesis: A review. Clin. Genet. 92, 467–476. doi:10.1111/cge.12986
Brembeck, F. H., Rosário, M., and Birchmeier, W. (2006). Balancing cell adhesion and Wnt signaling, the key role of beta-catenin. Curr. Opin. Genet. Dev. 16, 51–59. doi:10.1016/j.gde.2005.12.007
Chhabra, N., Goswami, M., and Chhabra, A. (2014). Genetic basis of dental agenesis—Molecular genetics patterning clinical dentistry. Med. Oral Patol. Oral Cir. Bucal. 19, e112–e119. doi:10.4317/medoral.19158
Cingolani, P., Platts, A., le Wang, L., Coon, M., Nguyen, T., Wang, L., et al. (2012). A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly. (Austin) 6, 80–92. doi:10.4161/fly.19695
Cobourne, M. T., and Sharpe, P. T. (2003). Tooth and jaw: molecular mechanisms of patterning in the first branchial arch. Arch. Oral Biol. 48, 1–14. doi:10.1016/s0003-9969(02)00208-x
De Coster, P. J., Marks, L. A., Martens, L. C., and Huysseune, A. (2009). Dental agenesis: genetic and clinical perspectives. J. Oral Pathol. Med. 38, 1–17. doi:10.1111/j.1600-0714.2008.00699.x
Di Benedetto, A., Brunetti, G., Posa, F., Ballini, A., Grassi, F. R., Colaianni, G., et al. (2015). Osteogenic differentiation of mesenchymal stem cells from dental bud: role of integrins and cadherins. Stem Cell Res. 15, 618–628. doi:10.1016/j.scr.2015.09.011
Dinckan, N., Du, R., Petty, L. E., Coban-Akdemir, Z., Jhangiani, S. N., Paine, I., et al. (2018). Whole-exome sequencing identifies novel variants for tooth agenesis. J. Dent. Res. 97, 49–59. doi:10.1177/0022034517724149
Dreesen, K., Swinnen, S., Devriendt, K., and Carels, C. (2014). Tooth agenesis patterns and phenotype variation in a cohort of Belgian patients with hypodontia and oligodontia clustered in 79 families with their pedigrees. Eur. J. Orthod. 36, 99–106. doi:10.1093/ejo/cjt021
Ferguson, C. A., Tucker, A. S., and Sharpe, P. T. (2000). Temporospatial cell interactions regulating mandibular and maxillary arch patterning. Development 127, 403–412. doi:10.1242/dev.127.2.403
Fine, J. D., Eady, R. A., Bauer, E. A., Bauer, J. W., Bruckner-Tuderman, L., Heagerty, A., et al. (2008). The classification of inherited epidermolysis bullosa (EB): report of the third international consensus Meeting on diagnosis and classification of EB. J. Am. Acad. Dermatol. 58, 931–950. doi:10.1016/j.jaad.2008.02.004
Fournier, B. P., Bruneau, M. H., Toupenay, S., Kerner, S., Berdal, A., Cormier-Daire, V., et al. (2018). Patterns of dental agenesis highlight the nature of the causative mutated genes. J. Dent. Res. 97, 1306–1316. doi:10.1177/0022034518777460
Fukumoto, S., Miner, J. H., Ida, H., Fukumoto, E., Yuasa, K., Miyazaki, H., et al. (2006). Laminin alpha5 is required for dental epithelium growth and polarity and the development of tooth bud and shape. J. Biol. Chem. 281, 5008–5016. doi:10.1074/jbc.M509295200
Fukumoto, S., and Yamada, Y. (2005). Review: extracellular matrix regulates tooth morphogenesis. Connect. Tissue Res. 46, 220–226. doi:10.1080/03008200500344017
Galluccio, G., Castellano, M., and La Monaca, C. (2012). Genetic basis of non-syndromic anomalies of human tooth number. Arch. Oral Biol. 57, 918–930. doi:10.1016/j.archoralbio.2012.01.005
Gudmundsson, S., Singer-Berk, M., Watts, N. A., Phu, W., Goodrich, J. K., Solomonson, M., et al. (2022). Variant interpretation using population databases: lessons from gnomAD. Hum. Mutat. 43, 1012–1030. doi:10.1002/humu.24309
Inagaki, Y., Ogawa, T., Tabata, M. J., Nagata, Y., Watanabe, R., Kawamoto, T., et al. (2021). Identification of OPN3 as associated with non-syndromic oligodontia in a Japanese population. J. Hum. Genet. 66, 769–775. doi:10.1038/s10038-021-00903-3
Jonsson, L., Magnusson, T. E., Thordarson, A., Jonsson, T., Geller, F., Feenstra, B., et al. (2018). Rare and common variants conferring risk of tooth agenesis. J. Dent. Res. 97, 515–522. doi:10.1177/0022034517750109
Karczewski, K. J., Francioli, L. C., Tiao, G., Cummings, B. B., Alföldi, J., Wang, Q., et al. (2020). The mutational constraint spectrum quantified from variation in 141,456 humans. Nature 581, 434–443. doi:10.1038/s41586-020-2308-7
Kawashima, N., and Okiji, T. (2016). Odontoblasts: specialized hard-tissue-forming cells in the dentin-pulp complex. Congenit. Anom. (Kyoto). 56, 144–153. doi:10.1111/cga.12169
Khalaf, K., Miskelly, J., Voge, E., and Macfarlane, T. V. (2014). Prevalence of hypodontia and associated factors: A systematic review and meta-analysis. J. Orthod. 41, 299–316. doi:10.1179/1465313314Y.0000000116
Kircher, M., Witten, D. M., Jain, P., O’Roak, B. J., Cooper, G. M., and Shendure, J. (2014). A general framework for estimating the relative pathogenicity of human genetic variants. Nat. Genet. 46, 310–315. doi:10.1038/ng.2892
Kjoelby, M., Thesleff, I., Sahlberg, C., Fejerskov, O., and Josephsen, K. (1994). Degradation of the dental basement membrane during mouse tooth development in vitro. Int. J. Dev. Biol. 38, 455–462.
Laugel-Haushalter, V., Paschaki, M., Thibault-Carpentier, C., Dembelé, D., Dollé, P., and Bloch-Zupan, A. (2013). Molars and incisors: show your microarray IDs. BMC Res. Notes. 6, 113. doi:10.1186/1756-0500-6-113
Lesot, H., Osman, M., and Ruch, J. V. (1981). Immunofluorescent localization of collagens, fibronectin, and laminin during terminal differentiation of odontoblasts. Dev. Biol. 82, 371–381. doi:10.1016/0012-1606(81)90460-7
Li, L., Tang, Q., Nakamura, T., Suh, J. G., Ohshima, H., and Jung, H. S. (2016). Fine tuning of Rac1 and RhoA alters cuspal shapes by remolding the cellular geometry. Sci. Rep. 6, 37828. doi:10.1038/srep37828
Lin, C., Ruan, N., Li, L., Chen, Y., Hu, X., Chen, Y., et al. (2022). FGF8-mediated signaling regulates tooth developmental pace during odontogenesis. J. Genet. Genomics. 49, 40–53. doi:10.1016/j.jgg.2021.08.009
Lu, J., Lian, G., Lenkinski, R., De Grand, A., Vaid, R. R., Bryce, T., et al. (2007). Filamin B mutations cause chondrocyte defects in skeletal development. Hum. Mol. Genet. 16, 1661–1675. doi:10.1093/hmg/ddm114
Lu, Y., Qian, Y., Zhang, J., Gong, M., Wang, Y., Gu, N., et al. (2016). Genetic variants of BMP2 and their association with the risk of non-syndromic tooth agenesis. PLOS ONE 11, e0158273. doi:10.1371/journal.pone.0158273
Mitsiadis, T. A., and Drouin, J. (2008). Deletion of the Pitx1 genomic locus affects mandibular tooth morphogenesis and expression of the Barx1 and Tbx1 genes. Dev. Biol. 313, 887–896. doi:10.1016/j.ydbio.2007.10.055
Mostowska, A., Biedziak, B., Zadurska, M., Matuszewska-Trojan, S., and Jagodziński, P. P. (2015). WNT10A coding variants and maxillary lateral incisor agenesis with associated dental anomalies. Eur. J. Oral Sci. 123, 1–8. doi:10.1111/eos.12165
Nemec, S., Luxey, M., Jain, D., Huang Sung, A., Pastinen, T., and Drouin, J. (2017). Pitx1 directly modulates the core limb development program to implement hindlimb identity. Development 144, 3325–3335. doi:10.1242/dev.154864
Nunn, J. H., Carter, N. E., Gillgrass, T. J., Hobson, R. S., Jepson, N. J., Meechan, J. G., et al. (2003). The interdisciplinary management of hypodontia: background and role of paediatric dentistry. Br. Dent. J. 194, 245–251. doi:10.1038/sj.bdj.4809925
Park, H., Song, J. S., Shin, T. J., Hyun, H. K., Kim, Y. J., and Kim, J. W. (2019). WNT10A mutations causing oligodontia. Arch. Oral Biol. 103, 8–11. doi:10.1016/j.archoralbio.2019.05.007
Polder, B. J., Van’t Hof, M. A., Van der Linden, F. P., and Kuijpers-Jagtman, A. M. (2004). A meta-analysis of the prevalence of dental agenesis of permanent teeth. Community Dent. Oral Epidemiol. 32, 217–226. doi:10.1111/j.1600-0528.2004.00158.x
Posey, J. E., Harel, T., Liu, P., Rosenfeld, J. A., James, R. A., Coban Akdemir, Z. H., et al. (2017). Resolution of disease phenotypes resulting from multilocus genomic variation. N. Engl. J. Med. 376, 21–31. doi:10.1056/NEJMoa1516767
Ryan, M. C., Lee, K., Miyashita, Y., and Carter, W. G. (1999). Targeted disruption of the LAMA3 gene in mice reveals abnormalities in survival and late stage differentiation of epithelial cells. J. Cell Biol. 145, 1309–1323. doi:10.1083/jcb.145.6.1309
Schambony, A., Kunz, M., and Gradl, D. (2004). Cross-regulation of Wnt signaling and cell adhesion. Differentiation 72, 307–318. doi:10.1111/j.1432-0436.2004.07207002.x
Song, S., Zhao, R., He, H., Zhang, J., Feng, H., and Lin, L. (2014). WNT10A variants are associated with non-syndromic tooth agenesis in the general population. Hum. Genet. 133, 117–124. doi:10.1007/s00439-013-1360-x
Stewart, R. L., and O’Connor, K. L. (2015). Clinical significance of the integrin α6β4 in human malignancies. Lab. Invest. 95, 976–986. doi:10.1038/labinvest.2015.82
Stockton, D. W., Das, P., Goldenberg, M., D’Souza, R. N., and Patel, P. I. (2000). Mutation of PAX9 is associated with oligodontia. Nat. Genet. 24, 18–19. doi:10.1038/71634
Szklarczyk, D., Gable, A. L., Lyon, D., Junge, A., Wyder, S., Huerta-Cepas, J., et al. (2019). STRING v11: protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 47, D607–D613. doi:10.1093/nar/gky1131
Taju, W., Sherriff, M., Bister, D., and Shah, S. (2018). Association between severity of hypodontia and cephalometric skeletal patterns: A retrospective study. Eur. J. Orthod. 40, 200–205. doi:10.1093/ejo/cjx049
Takahashi, K., Kiso, H., Murashima-Suginami, A., Tokita, Y., Sugai, M., Tabata, Y., et al. (2020). Development of tooth regenerative medicine strategies by controlling the number of teeth using targeted molecular therapy. Inflamm. Regen. 40, 21. doi:10.1186/s41232-020-00130-x
Taneyhill, L. A. (2008). To adhere or not to adhere: the role of cadherins in neural crest development. Cell adh. Migr. 2, 223–230. doi:10.4161/cam.2.4.6835
Timpl, R. (1996). Macromolecular organization of basement membranes. Curr. Opin. Cell Biol. 8, 618–624. doi:10.1016/s0955-0674(96)80102-5
Tucker, A. S., Al Khamis, A., and Sharpe, P. T. (1998). Interactions between Bmp-4 and Msx-1 act to restrict gene expression to odontogenic mesenchyme. Dev. Dyn. 212, 533–539. doi:10.1002/(SICI)1097-0177(199808)212:4<533:AID-AJA6>3.0.CO;2-I
Tucker, A., and Sharpe, P. (2004). The cutting-edge of mammalian development; how the embryo makes teeth. Nat. Rev. Genet. 5, 499–508. doi:10.1038/nrg1380
Vastardis, H., Karimbux, N., Guthua, S. W., Seidman, J. G., and Seidman, C. E. (1996). A human MSX1 homeodomain missense mutation causes selective tooth agenesis. Nat. Genet. 13, 417–421. doi:10.1038/ng0896-417
Vieira, A. R. (2003). Oral clefts and syndromic forms of tooth agenesis as models for genetics of isolated tooth agenesis. J. Dent. Res. 82, 162–165. doi:10.1177/154405910308200303
Wang, J. S., Infante, C. R., Park, S., and Menke, D. B. (2018). PITX1 promotes chondrogenesis and myogenesis in mouse hindlimbs through conserved regulatory targets. Dev. Biol. 434, 186–195. doi:10.1016/j.ydbio.2017.12.013
Williams, M. A., and Letra, A. (2018). The changing landscape in the genetic etiology of human tooth agenesis. Genes (Basel). 9, 255. doi:10.3390/genes9050255
Xie, W., Zeng, B., Li, P., Xu, D., Yu, D., and Zhao, W. (2022). Two novel mutations in ectodysplasin-A identified in syndromic tooth agenesis. J. Coll. Physicians Surg. Pak. 32, 570–574. doi:10.29271/jcpsp.2022.05.570
Ye, X., and Attaie, A. B. (2016). Genetic basis of nonsyndromic and syndromic tooth agenesis. J. Pediatr. Genet. 5, 198–208. doi:10.1055/s-0036-1592421
Yin, W., and Bian, Z. (2015). The gene network underlying hypodontia. J. Dent. Res. 94, 878–885. doi:10.1177/0022034515583999
Yoshizaki, K., and Yamada, Y. (2013). Gene evolution and functions of extracellular matrix proteins in teeth. Orthod. Waves. 72, 1–10. doi:10.1016/j.odw.2013.01.040
Zhang, T., Zhao, X., Hou, F., Sun, Y., Wu, J., Ma, T., et al. (2019). A novel PAX9 mutation found in a Chinese patient with hypodontia via whole exome sequencing. Oral Dis. 25, 234–241. doi:10.1111/odi.12982
Zeng, B., Zhao, Q., Li, S., Lu, H., Lu, J., Ma, L., et al. (2017). Novel EDA or EDAR mutations identified in patients with X-linked hypohidrotic ectodermal dysplasia or non-syndromic tooth agenesis. GenesGenes (Basel). 8, 259. doi:10.3390/genes8100259
Zhang, Y., Chen, Y., Chen, C., Guo, H., Zhou, C., Wang, H., et al. (2023). PITX1 suppresses osteosarcoma metastasis through exosomal LINC00662-mediated M2 macrophage polarization. Clin. Exp. Metastasis. 40, 79–93. doi:10.1007/s10585-022-10192-5
Zhao, K., Lian, M., Zou, D., Huang, W., Zhou, W., Shen, Y., et al. (2019). Novel mutations identified in patients with tooth agenesis by whole-exome sequencing. Oral Dis. 25, 523–534. doi:10.1111/odi.13002
Keywords: hypodontia, genotype-phenotype correlation, genetic association studies, bioinformatics, extracellular matrix
Citation: Lee S, Ahn H, Kim H, Lee K, Kim S and Lee JH (2023) Identification of potential key variants in mandibular premolar hypodontia through whole-exome sequencing. Front. Genet. 14:1248326. doi: 10.3389/fgene.2023.1248326
Received: 11 July 2023; Accepted: 29 August 2023;
Published: 08 September 2023.
Edited by:
Vaidutis Kucinskas, Vilnius University, LithuaniaReviewed by:
Cybel Mehawej, Lebanese American University, LebanonKhalda Amr, National Research Centre, Egypt
Hiroshi Kurosaka, Osaka University, Japan
Copyright © 2023 Lee, Ahn, Kim, Lee, Kim and Lee. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jae Hoon Lee, jaehoon115@yuhs.ac; Sanguk Kim, sukim@postech.ac.kr
†These authors have contributed equally to this work and share first authorship
 Shinyeop Lee1†
Shinyeop Lee1† Hyunsoo Ahn
Hyunsoo Ahn Jae Hoon Lee
Jae Hoon Lee