- 1Agricultural College, Yanbian University, Yanji, Jilin, China
- 2Jilin Academy of Vegetable and Flower Science, Changchun, Jilin, China
The dirigent (DIR) gene is a key player in environmental stress response and has been identified in many multidimensional tube plant species. However, there are few studies on the StDIR gene in potato. In this study, we used genome-wide identification to identify 31 StDIR genes in potato. Among the 12 potato chromosomes, the StDIR gene was distributed on 11 chromosomes, among which the third chromosome did not have a family member, while the tenth chromosome had the most members with 11 members. 22 of the 31 StDIRs had a classical DIR gene structure, with one exon and no intron. The conserved DIR domain accounts for most of the proteins in the 27 StDIRs. The structure of the StDIR gene was analyzed and ten different motifs were detected. The StDIR gene was divided into three groups according to its phylogenetic relationship, and 22 duplicate genes were identified. In addition, four kinds of cis-acting elements were detected in all 31 StDIR promoter regions, most of which were associated with biotic and abiotic stress. The findings demonstrated that the StDIR gene exhibited specific responses to cold stress, salt stress, ABA, and drought stress. This study provides new candidate genes for improving potato’s resistance to stress.
1 Introduction
Solanum tuberosum, commonly known as potato, represents a crucial non-cereal food crop cultivated across approximately 19.3 million hectares globally, yielding an annual production of nearly 400 million metric tons, ranking only behind rice, wheat, and maize (Halterman et al., 2016; Liu et al., 2023). In recent years, people’s demand for potato is increasing day by day, and the demand for planting areas is also gradually increasing, but the resulting planting problems are also increasing significantly. These consequent problems caused a large reduction in potato production, resulting in great economic losses. To cope with severe biotic and abiotic stress, plants also spontaneously evolved a series of defense measures to quickly respond to the complex environment, reduce the damage and strive for more survival resources (Atkinson and Urwin, 2012; Zulfiqar and Ashraf, 2022). Plants respond to complex environments by activating many different genes, including the DIR protein (Arasan et al., 2013; Lopez-Zaplana et al., 2022; Meher et al., 2022).
The inception of dirigent (DIR) genes dates back to 1985, when they were initially characterized in pea (Pisum sativum). But the function of DIR proteins were first identified in Forsythia intermedia in 1997, and then was later reported in Arabidopsis (Arabidopsis thaliana) (Ralph et al., 2006; Cheng et al., 2018; Zhang et al., 2022; Luo et al., 2022). DIR belongs to a multi-gene family. DIR protein is relatively conserved, among which the conserved domain accounts for a large part of DIR protein, and usually does not contain introns in the DIR gene (Corbin et al., 2018). DIR genes are found in almost all vascular plants, including ferns, gymnosperms, and angiosperms (Arasan et al., 2013; Song and Peng, 2019; Ma et al., 2021). Furthermore, DIR has been demonstrated to play a guiding role in lignin synthesis (Cheng et al., 2018; Dong et al., 2021; Liu et al., 2021). Research has illustrated the congruence between the localization sites of DIR proteins and the locations of lignin deposition and biosynthesis (Davin and Lewis, 2000; Burlat et al., 2001). Thus, DIR proteins play a crucial role in directing the correct formation of lignin. Lignin is mainly deposited in terminally differentiated cells, which is an important part of the cell wall. Lignin is essential for resistance to adverse external environments, serving as the first physical barrier to safeguard plant growth and development. The involvement of DIRs in the biosynthesis of lignin units and lignin has attracted the scientific community to discover the functions of these proteins in various stress tolerance mechanisms.
DIR exhibits responsiveness to a wide array of biotic and abiotic stresses. At the same time, there have been many studies on DIR, most of them related to disease resistance and tolerance. DIR genes have been found in a variety of plants, and all of them play important roles. The results showed that overexpression of GhDIR1 in cotton would lead to the increase of lignin content, which could improve the resistance of cotton to Verticillium dahliae (Shi et al., 2012). The loss of CaDIR7 function reduced root activity after salt stress and inhibited the induction of stress-related genes in CaDIR7 silenced plants. CaDIR7 gene silencing in pepper significantly reduced the tolerance of pepper to pathogens and salt stress (Khan et al., 2018). There is evidence of the overexpression of GmDIR22 in soybean can increase total lignan accumulation and enhance plant resistance to Phytophthora sojae (Li et al., 2017). The high expression of DIR in flax leads to pinoresinol accumulation after the pathogen invasion of plants (Corbin et al., 2018). In terms of resistance to insect and pathogen stresses induced by weevils or mechanical damage, six DIR genes from Sitka spruce (Picea sitchensis) bark can be expressed rapidly and strongly (up to 500-fold) (Ralph et al., 2007). Hormonal pathways hold significance within plant regulatory networks, and investigations have demonstrated that DIR likewise exhibits responsiveness to hormones. An earlier report showed that a dirigent gene (BhDIR1) from Boea hygrometrica was involved in response to plant dehydration status, applied phytohormones, signaling molecules and temperature stresses (Wu et al., 2009).
Currently, research on the potato DIR gene family is limited. Furthermore, investigations into DIR have insufficiently acknowledged the contribution of potato DIR genes in responding to both biotic and abiotic stresses. The study of the StDIR gene in potato can provide important information for the molecular breeding of potato. In this experiment, we analyzed the gene structure, chromosomal location, phylogenetic relationship family evolutionary tree and transcriptomic-based expression data of the potato StDIR gene family and constructed the expression map of this gene in different tissues under different biotic and abiotic stresses. This study conducted a detailed and comprehensive analysis of the DIR gene family, which could provide a reference for the research on the role of the DIR gene family in the process of exploring potato’s response to biotic and abiotic stresses.
2 Materials and methods
2.1 Plant materials and treatment
Germinated seed tubers (10 g) of potato variety Youjin were planted in a greenhouse of Yanbian University with day/night time is 16/8 h, and a day/night temperature of 23/17°C. The 50-days-old potato plants were treated with cold, salt, PEG, and ABA stress for 0, 24, 48, 72, and 96 h. The methods of treatment are followed: Cold: The culture temperature was 4°C, Salt: Place potato seedlings in a 200 mM NaCl solution, Drought: Place potato seedlings in a 10% polyethylene glycol (PEG, polyethylene glycol) solution and ABA (Abscisic Acid): The 0.1 mM ABA solution was applied evenly to the surface of the potato seedlings until the solution was about to drip from the edge of the leaves. The top leaves of each potato plant were taken and stored in a −80°C refrigerator for later use, and each treatment groups contain three biological replicates.
2.2 StDIR gene identification and chromosomal distribution analysis
First of all, the total proteins sequence file, the genome annotation file (GFF3 format) and the genome sequences file of potato (Solanum tuberosum) were downloaded from the Ensembl plants database (http://plants.ensembl.org/index.html). The amino acid sequence of 25 Arabidopsis DIR proteins was downloaded from TAIR (The Arabidopsis Information Resource, https://www.arabidopsis.org/) database and was used as blast queries against the total proteins file of potato. Then, the hidden Markov model (HMM) file of conserved DIR-like protein domain PF03018 was downloaded from the Pfam database (https://www.ebi.ac.uk/Tools/hmmer/search/phmmer). All the proteins sequence of the above two methods results were submitted to Pfam and CDD (Conserved Domains Database, https://www.ncbi.nlm.nih.gov/cdd) to confirm they contain the conserved domain of DIR protein. The gene ID, genomic location, and amino acid number were obtained from the genome annotation file. ProtParam software (https://web.expasy.org/protparam/) was used to analyze the molecular weight (MolWt) of proteins and the theoretical isoelectric point (pI). In the potato genome annotation file, the length information of potato chromosomes and the position information of all potato DIR genes on chromosomes were obtained. At the same time, TBtools was used to draw the chromosome localization map of potato DIR genes, which was used to represent the position information and distance relationship of all potato DIR gene family members on chromosomes (Chen et al., 2020).
2.3 StDIR genes structure, conserved domains and conserved motifs analysis
All StDIR protein sequences of the potato were obtained from the total protein sequence file of the potato and used as input files. Conserved motif analysis was performed using MEME v4.12 software. The parameters were set as follows: the number of conserved motifs was 10, the amino acid length of the smallest motif was 6, the amino acid length of the largest motif was 100, and the number of searches was 10,000. Gene structure analysis was performed using TBtools to identify the coding sequence (CDS) and untranslated region (UTR) of the StDIR gene. The conserved domain information of StDIR proteins was obtained from the CDD database Finally, TBtools was used to visualize the conserved motifs, conserved domains and gene structure of the StDIR protein.
2.4 Phylogenetic analysis
To characterize the phylogenetic relationships of StDIR proteins, the amino acid sequences of DIR proteins from many plant species were downloaded from the TAIR database and Ensembl Plants database, including Arabidopsis and potato (S. tuberosum). In addition, a phylogenetic tree was also constructed in MEGA 7.0 with the neighbor-joining method. The Poisson model was selected and the boot value was set to 1,000.
2.5 Analysis of cis-acting elements in promoter regions of StDIR genes
The PlantCARE database (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) was used to predict the cis-acting elements in the promoter regions of StDIR genes. The promoter regions were analyzed using sequences 1.5 kb upstream of the initiation codon ATG. The cis-acting elements were classified into different groups based on their potential functions. Use the Python program to count the predicted results of cis-acting elements and use R v4.2 for visualization.
2.6 Duplication and Ka/Ks analysis of StDIR gene family
There are two values synonymous substitution rate (Ks) and nonsynonymous substitution rate (Ka) to describe the evolutionary relationship between genes (Hurst, 2002). The MCScanX-transposed software was used to identify the relationship between the StDIR gene family by using the CDS (coding sequence) sequences of 31 StDIR genes (Koch et al., 2000). The simple Ka/Ks calculator module of TBtools was used to calculate the value of Ka and Ks. At the same time, filter the results by the length of aligned genes was greater than 70% of the longer gene, and the similarity between the two genes was greater than 70%; the distance between the two genes was less than 100 kb (Vatansever et al., 2016). The diversity time was calculated by T = Ks/2r, and the r of dicotyledonous plants was taken to be 1.5 × 10−8 synonymous substitutions per site per year (Koch et al., 2000). The advanced circos module of TBtools was used to visualize collinear StDIR gene family members in the potato genome.
2.7 Synteny analysis of DIR genes in potato, arabidopsis, rice and tomato
To explore the synteny relationship of DIR genes between potato and Arabidopsis, rice, tomato, the genome sequence and genomic annotation file of Arabidopsis, rice and tomato were downloaded from the Ensembl plants database, and then the One Step MCScanX module of TBtools was used to comparative genomic analysis. The synteny relationship between the potato and the other three species was visualized by TBtools.
2.8 Gene expression analysis
In order to explore the response of the StDIR gene to high-temperature stress, the transcriptome data was used to analyze the expression profile of StDIR genes. The transcriptome data of accession: PRJNA577000 was downloaded from the NCBI (National Center for Biotechnology Information) database (https://www.ncbi.nlm.nih.gov/sra/). HISAT software was used to mapping the raw data to the potato genome (http://daehwankimlab.github.io/hisat2/), and the Rsubread package in R (version “4.3”) was used for the quantification of the gene (Mo et al., 2021). The expression of StDIR genes under high-temperature stress was analyzed by TBtools.
2.9 qRT-PCR analysis
Ten genes were selected to verify RNAseq. At the same time, after different stress treatments, we also carried out q-PCR of these ten genes to analyze the response of StDIR family members to stress. Primer 5 software was used for primer design (Table 1). Primers were ordered from Sangon Bioscience Co., Ltd. and the dye used for qRT-PCR was purchased from Biorun (BioRun Biosciences Co., Ltd., Wuhan, China). The system of qRT-PCR is referred to Mo’s study (Mo et al., 2021).
3 Results
3.1 StDIR gene identification and chromosomal distribution analysis
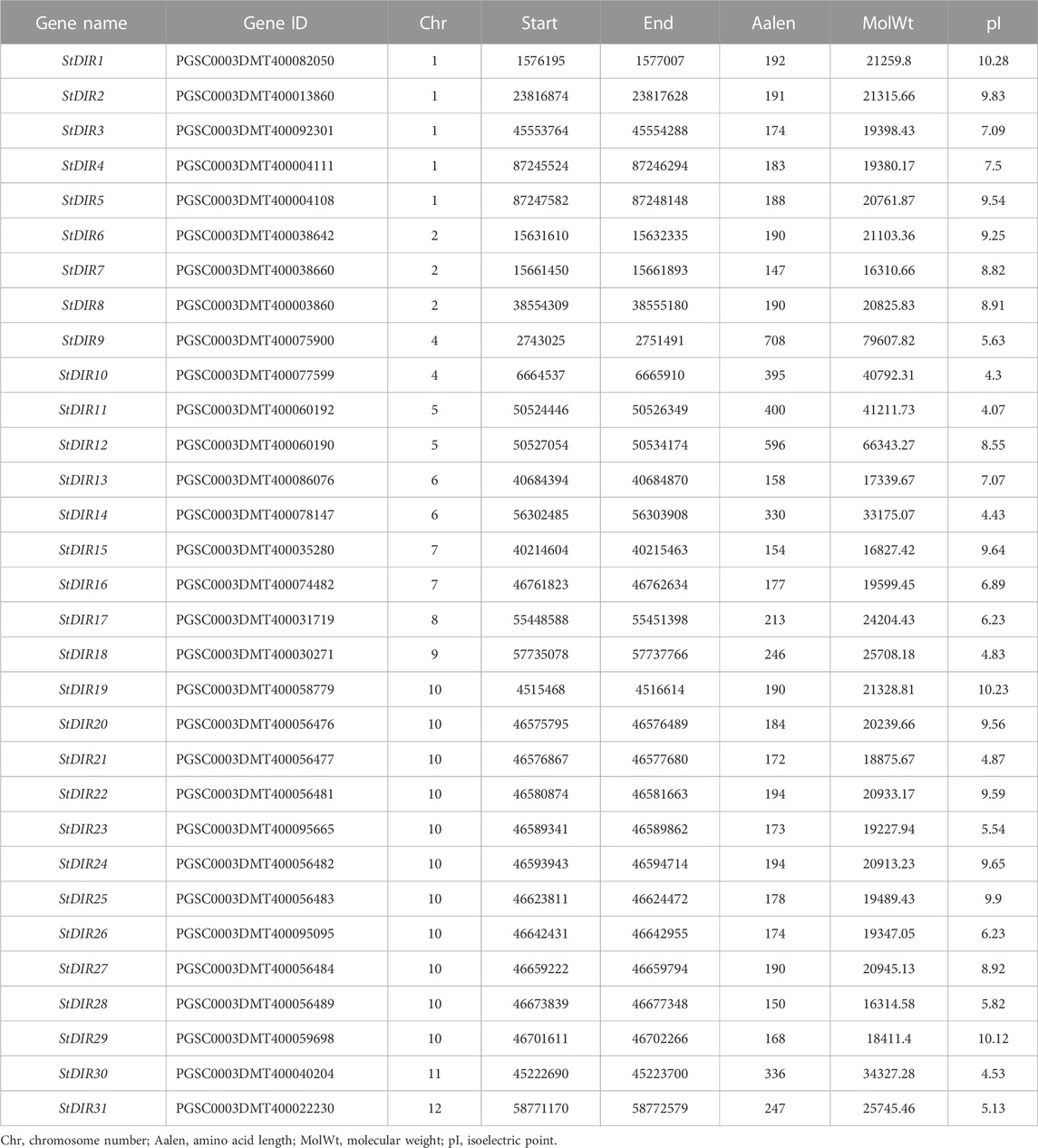
To search for potato DIR proteins, the amino acid sequences of 25 Arabidopsis DIR proteins were used as blast queries against the total proteins sequence of potato. HMMER was used to analyze whether the protein contain the conserved DIR domain with the HMM file of PF03018. A total of 31 StDIR genes were identified in the potato genome (Table 2). The genome length of StDIR ranges from 443 (StDIR7) to 8,466 bp (StDIR9), and the genome length of 26 StDIR genes is less than 2,000 bp. The number of amino acids varies from 147 (StDIR7) to 708 (StDIR9), with 22 of the 31 StDIR proteins having less than 200 amino acids, suggesting that most StDIRs are small proteins. The molecular weight of the StDIR protein ranged from 16.31 kDa (StDIR7) to 79.60 KDa (StDIR9), and the theoretical pI ranged from 4.07 (StDIR11) to 10.28 (StDIR1). In addition, 18 StDIR proteins were predicted to be alkaline proteins (pI > 7.0) and 13 were predicted to be acidic proteins (pI < 7.0).
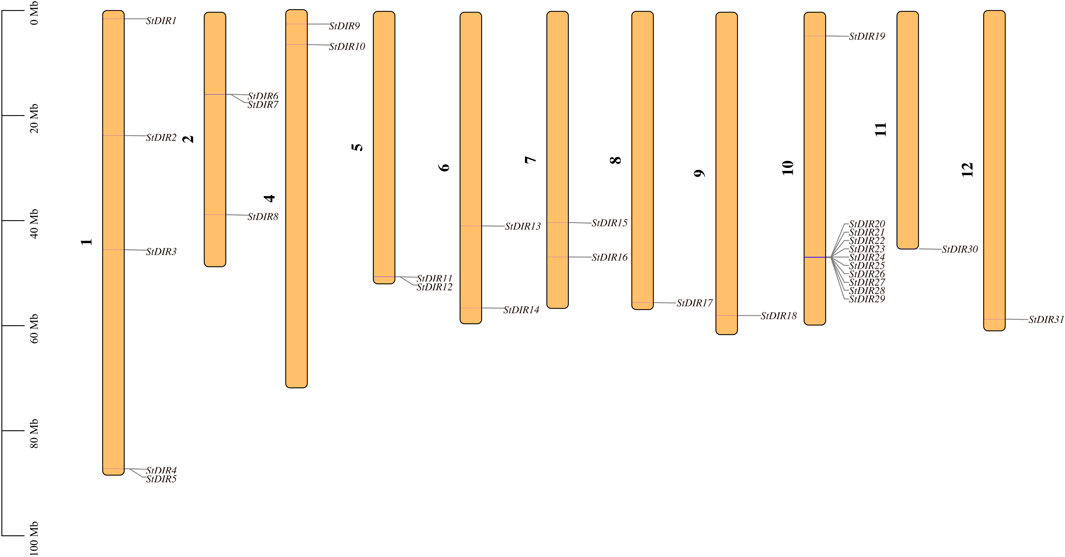
A total of 31 StDIR genes were identified on 12 potato chromosomes and their distribution on chromosomes was uneven. Except for chromosome 3, all 11 chromosomes contained the StDIR genes (Figure 1). Among these chromosomes, chromosome 10 contains the most StDIR genes, with 11 members, and the positions of these genes on chromosome 10 are very close, except for StDIR19. Chromosome 1 is followed by four members. The distribution of these 31 StDIR genes on chromosomes is not obvious, some are distributed in the middle of the chromosome (near the centromere), and some are distributed at both ends of the chromosome.
3.2 Phylogenetic relationships among StDIR genes
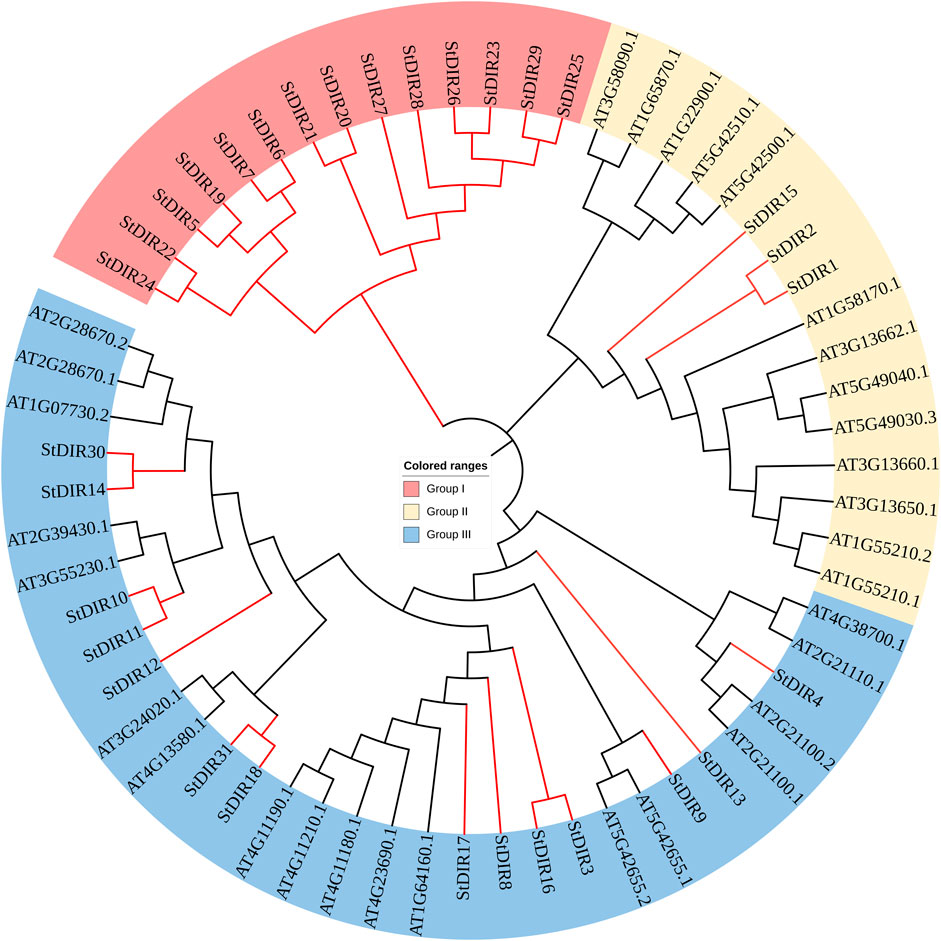
To group StDIR genes into different subfamilies and predict the functions of StDIRs from well-studied DIR homologous genes in other species, the amino acid sequences of DIR proteins from Arabidopsis were used to construct a phylogenetic tree using MEGA 7.0. Our analyses showed that DIR gene families were divided into three major subclades, of which group III had the most DIR proteins, while Group I all belonged to potato, with a total of 14 proteins. 3 potato proteins and 13 Arabidopsis proteins belong to Group II, and 14 potato proteins and 18 Arabidopsis proteins belong to Group III (Figure 2). It is worth noting that the phylogenetic tree found that Group III contains a large number of StDIR and AtDIR proteins, indicating that they are highly conserved during evolution and may have similar functions.
3.3 Conserved motifs, conserved domain and gene structure analyses of StDIR genes
To study the exon-intron organization of the StDIR genes, we used the gene Structure Display Server program to analyze the StDIR genome and coding sequence. Of the 31 StDIR genes, 22 had classical DIR gene structure, which has one exon and no intron. The exception is StDIR31, StDIR18, StDIR14, StDIR12, StDIR17, StDIR9, StDIR19, StDIR21, and StDIR28, there are one or more introns (Figure 3). In addition, most StDIR genes contain UTRs, 17 in total, and StDIR13, StDIR30, StDIR12, StDIR17, StDIR9, StDIR3, StDIR5, StDIR19, StDIR7, StDIR21, StDIR23, StDIR26, StDIR28 and StDIR27 has no UTRs (Figure 3). The conserved DIR domain accounts for the majority of proteins in most StDIR except StDIR9, StDIR10, StDIR11 and StDIR12. Then, we used the MEME tool to analyze the conserved motif of the StDIR protein. A total of 10 distinct motifs were detected in all 31 StDIR proteins. All 31 StDIR proteins contain the conserved DIR domains: Dirigent domain. The difference in motifs reflects the diversity of StDIR proteins. For example, motif 9 only exists in StDIR10 and StDIR11, motif 10 only exists in StDIR12, and motif 8 only exists in StDIR12, StDIR14, StDIR18, StDIR30 and StDIR31, indicating that the StDIR protein has various functions.
3.4 Duplication analysis of potato DIR genes
As shown in Table 3, there are a total of 22 StDIR gene family members with four duplicate relationships: proximal duplication (3 gene pairs, 5 StDIR genes), segmental duplication (7 gene pairs, 10 StDIR genes), tandem duplication (4 gene pairs, 8 StDIR genes) and transposed duplication (2 gene pairs, 3 StDIR genes) have been identified in StDIR gene family. The value of Ka/Ks were between 0.0987 (StDIR10 and StDIR11) and 0.5101 (StDIR28 and StDIR29), all the StDIR duplicate genes with a Ka/Ks value less than 1, means all the genes in Table 3 were purifying selection. And the divergence time of StDIR duplicate genes range from 19.6013 Mya to 1320.6681 Mya. The genomic position of all the StDIR genes with duplication relationship were shown in Figure 4.
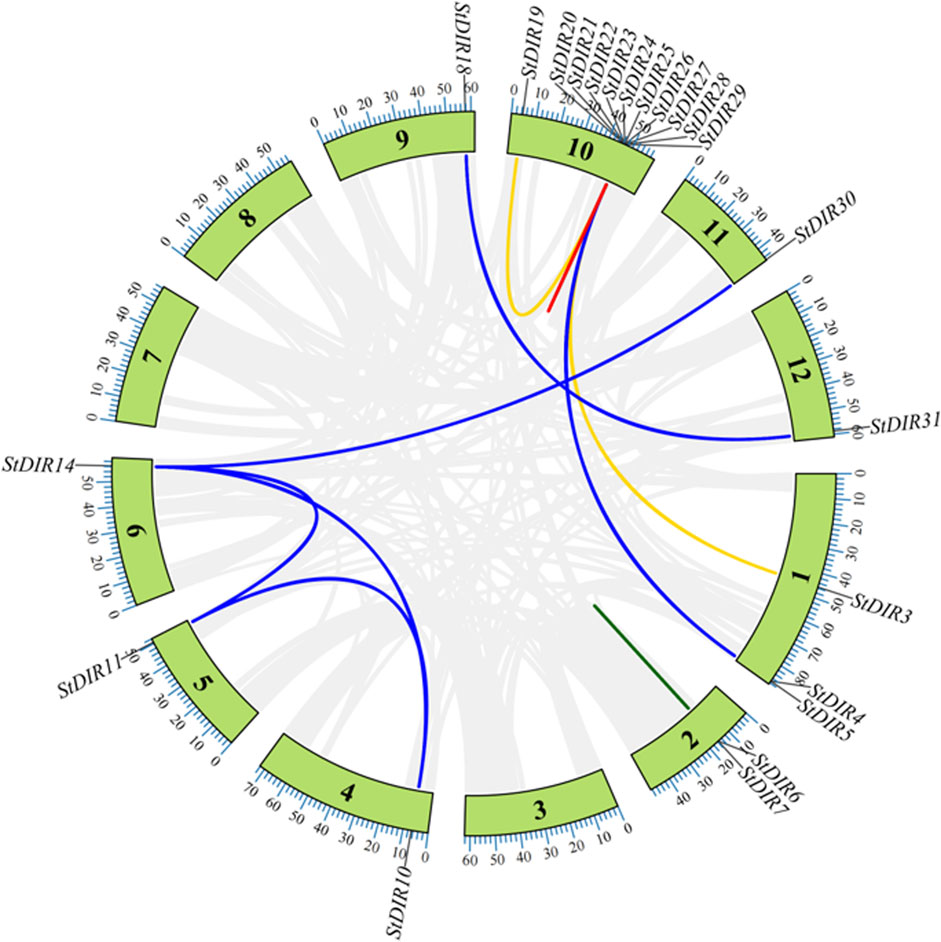
FIGURE 4. The duplication genes in the StDIR gene family. Proximal duplication gene pairs are inked by the red line, proximal duplication gene pairs are linked by the blue line, tandem duplication gene pairs are linked by the green line and transposed duplication gene pairs are linked by the yellow link. And there are too many pairs in Chr 10, so there only one line can be shown in the figure.
3.5 Promoter cis-element analysis of StDIR genes
To reveal the diversity of cis-elements in the promoter region of StDIR genes, we selected the 1500 bp upstream StDIR gene sequence and submitted it to the PlantCARE database for promoter cis-element analysis. R package pheatmap was used to create heatmaps to visualize the four classes of cis-components (Figure 5). Among them, 5 components were related to plant development, 4 components were related to light response, and 3 components were related to biotic stress. The largest number of elements is related to abiotic stresses, with a total of 11 elements. This indicates that the StDIR gene family is mainly involved in the regulation of abiotic stress in plants. For example, LTR is a group of low-temperature response elements, and MBS is a drought response element. Among the abiotic stress-related elements, the number of members of the StDIR family containing MYB is the largest. Among the 31 members, only StDIR2 does not have a MYB element. The MYB element has a wide range of functions and responds to abiotic stresses such as high temperature, drought, low temperature and high salt. In addition, the number of abiotic stress elements identified in the promoter region of StDIR was second only to biotic stress, including CGTCA-motif, W box and WRE2. A total of 26 family members contained 68 biotic stress elements, indicating that StDIR family members also play an important role in plant biotic stress. There are also a large number of light response elements and plant development elements in the StDIR family, but the number is far less than biotic stress and abiotic stress elements. This elucidates the pivotal role of StDIR in orchestrating crucial responses to both biotic and abiotic stresses in plants.
3.6 Synteny analysis of DIR genes between potato and arabidopsis, rice, tomato
To explore the synteny relationship between DIR genes in different species, three species were selected, containing the model and dicotyledons plant: Arabidopsis, monocotyledon and crop plant: rice and the Solanaceae plant: tomato. As shown in Figure 6, there are 18 DIR gene pairs (involving 11 StDIR genes) between potato and Arabidopsis, 5 DIR gene pairs (involving 5 StDIR genes) between potato and rice and 28 DIR gene pairs (involving 20 StDIR genes) between potato and tomato.
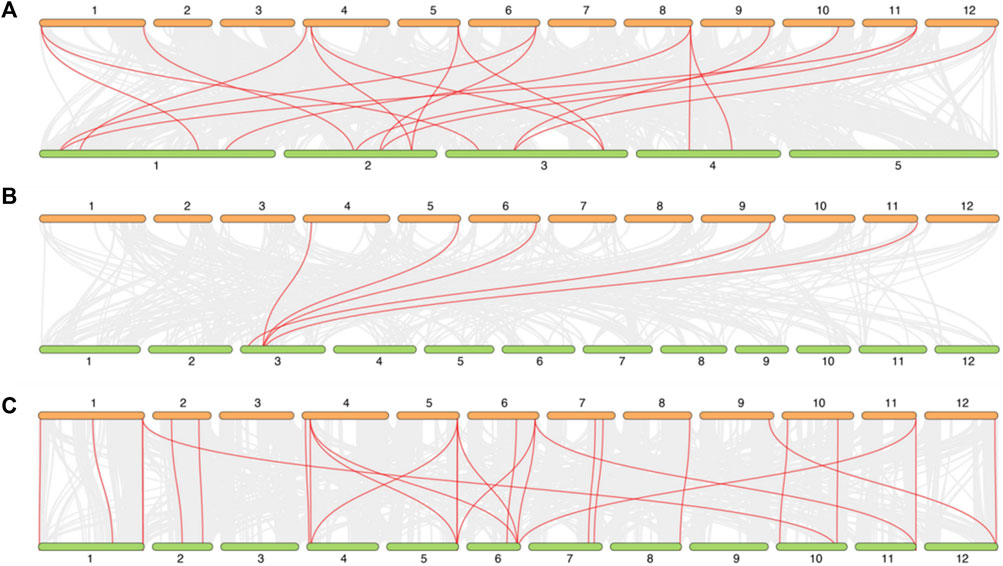
FIGURE 6. The synteny relationship between potato and Arabidopsis, rice, tomato. (A) potato and Arabidopsis, (B) potato and rice, (C) potato and tomato. All the synteny relationship DIR genes between different species were linked by the red line, the collinear blocks within the potato and other plant genomes were linked by the gray lines in the background.
3.7 Expression pattern analysis of potato StDIR gene family members under stress
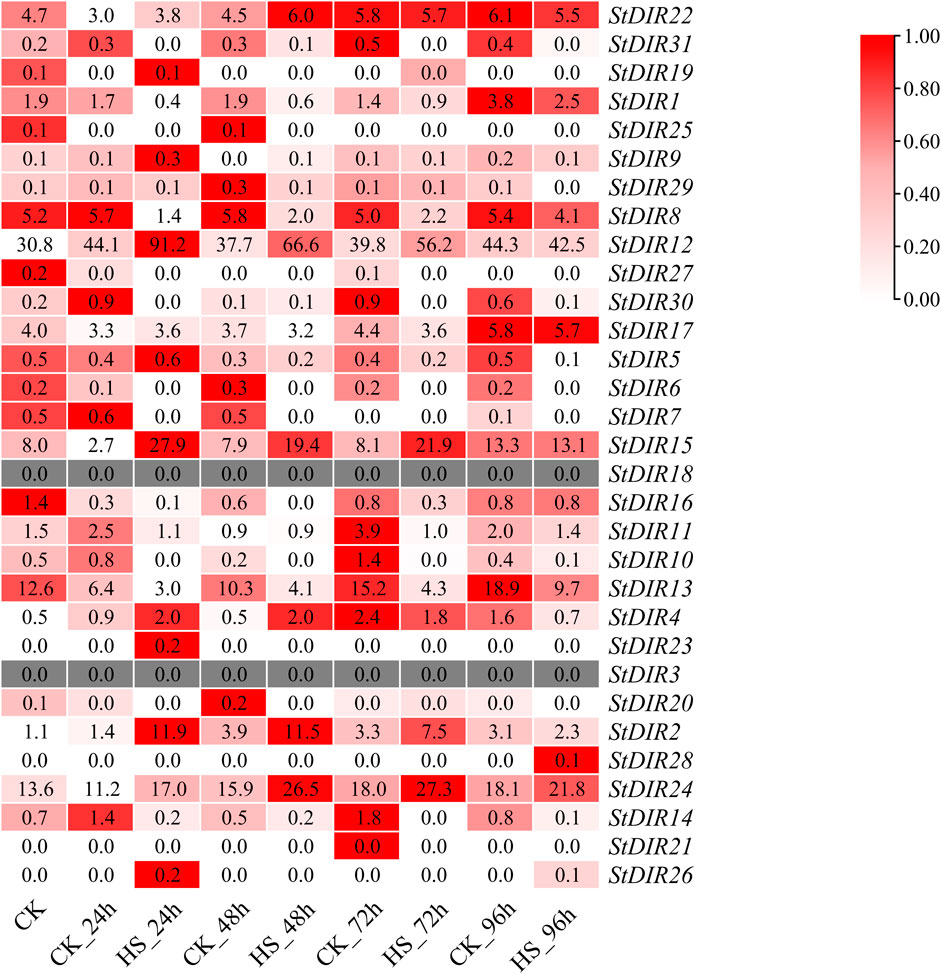
In the transcriptome data, StDIR12 had the highest expression level after 24 h of heat stress. Subsequent to heat stress, the StDIR family members primarily exhibited two distinct trends: an initial decrease followed by an increase, and a gradual increase (Figure 7). Among them, StDIR22, StDIR31, StDIR1, StDIR30, StDIR17, StDIR11, StDIR13 and StDIR14 showed a trend of decreasing first and then increasing, while StDIR12, StDIR15, StDIR4, StDIR2 and StDIR24 were significantly increased after heat stress. Concurrently, StDIR3 and StDIR18 were not discernible within the transcriptomic dataset. These results indicated that most members of the StDIR family responded to heat stress to some extent, indicating that the StDIR gene family may play an important role in plant response to heat stress.
To verify the accuracy of RNA-Seq data and study the response of the StDIR gene family to abiotic stress in potato, ten StDIR family members, StDIR1, StDIR2, StDIR4, StDIR8, StDIR11, StDIR13, StDIR15, StDIR17, StDIR22 and StDIR24 were selected in this study. As shown in Figure 8, the chosen genes exhibited an initial decrement followed by an increment under heat stress, aligning seamlessly with the transcriptome data findings.

FIGURE 8. The expression of the ten genes selected in the transcriptomic data. HS means high temperature stress.
Then, we treated potato plants with cold stress, salt (NaCl) stress, drought (PEG) stress, and ABA treatment and selected nine genes from different subclades of the phylogenetic tree to analyze their responses (Figure 9). Under cold stress, StDIR22, StDIR25 and StDIR1 were significantly downregulated, while StDIR2, StDIR11 and StDIR15 was significantly upregulated. Under salt stress, StDIR2, StDIR11, StDIR15, StDIR22 and StDIR25 were significantly upregulated but had no obvious trend, while StDIR1 was significantly downregulated. After ABA and PEG treatment, the overall trend of the selected genes increased first and then decreased. The foregoing outcomes signify the responsiveness of StDIR family constituents to a majority of stresses or treatments, further substantiating the crucial role the gene family may play in plants' reaction to diverse abiotic stresses.
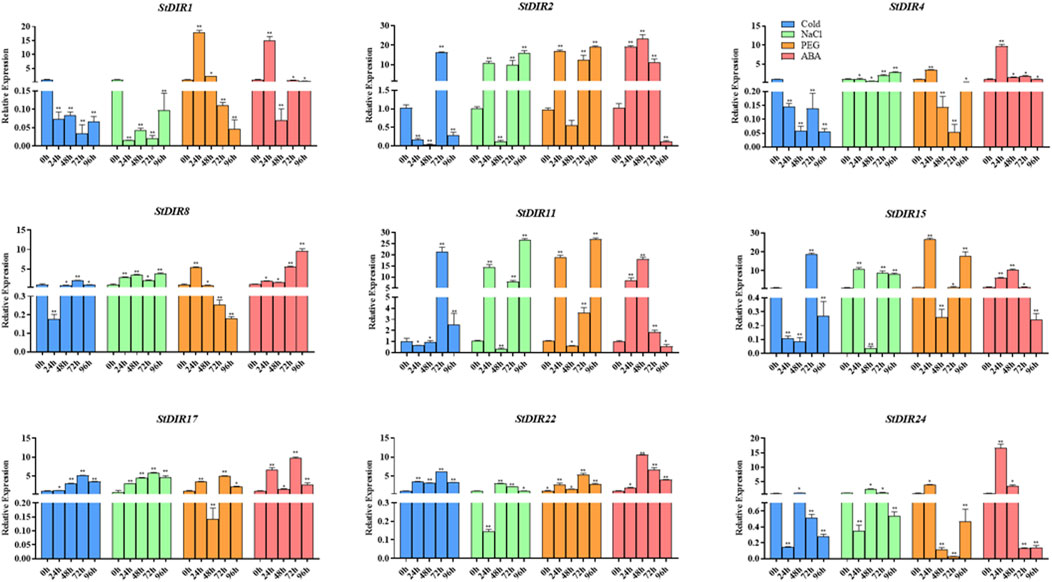
FIGURE 9. Expression levels of nine selected StDIR genes in response to cold stress, salt stress, drought stress and ABA treatment.
4 Discussion
Abiotic stress is an environmental adversity that affects the normal growth and development of plants, including drought, high salt and extreme temperature, and is the main factor affecting crop yield. Abiotic stress inhibits plant photosynthesis, affects chloroplast stability and induces chloroplast degradation, thus causing premature senescence of plants and ultimately affecting crop yield (Khatun et al., 2017; Ogden et al., 2020; Liu et al., 2022). The damage caused by inhospitable environmental conditions has severely limited the yield of potato, so the mining of excellent resistance genes in plants has become one of the main methods of researchers today.
Over the past few decades, several studies have characterized the DIR gene, and this work has greatly increased our understanding of plant responses to stress in many species. Many studies of DIR gene expression have associated its involvement with biological and abiotic stresses. However, until now, the DIR gene family has not been reported in potato, which has hindered the research on the function of the potato DIR gene to some extent. Based on this, a systematic bioinformatics analysis of the potato DIR gene family was carried out in this study. In this study, 31 DIR genes were identified from potato, more than those from pepper (Capsicum annuum) (Khan et al., 2018) and Brassica (Brassica rapa) (Arasan et al., 2013), indicating that potato contains more family members, and their expression patterns under heat, cold, salt and drought stress were analyzed. The 31 StDIR genes in the potato genome have an average molecular weight of 25.8 KDa and an average pI of 7.51 (Table 2). Meanwhile, we also carried out chromosome localization of the potato StDIR gene family, constructed the evolutionary tree of potato and Arabidopsis, and explored the phylogenetic relationship of the potato DIR gene family with Arabidopsis as a reference. In addition, four classes of cis-acting elements were identified in the analysis of the StDIR gene family, which were related to plant development, light response, abiotic stress and biological stress. Among these elements, most elements are associated with abiotic stress. Wu et al. found that in Ulva prolifera (Ulvophyceae, Chlorophyta), both HSE and LTR are indispensable under cold stress, as the deletion results in a complete loss of promoter activity. In algae domestication and artificial breeding, HSE and LTR elements may be potential molecular targets for temperature domestication (Wu et al., 2019; Puchtel, 2022). It was reported that 65% of MYB genes expressed in seedlings were differentially regulated under drought stress in rice (Katiyar et al., 2012; Peng et al., 2022; Qu et al., 2022). An even higher percentage is observed in Arabidopsis: transcriptomic data collected in the GENEVESTIGATOR database (Zimmermann et al., 2004a; Zimmermann et al., 2004b) show that 51% of AtMYB genes are upregulated by drought and 41% are downregulated (Katiyar et al., 2012). In the analysis of cis-acting elements, elements related to biological stress such as WRE3, CGTCA-motif and W Box also accounted for a large part. The existence of such a large number of biological and abiotic stress response elements further proves that the StDIR gene family plays a key role in potato coping with biological and abiotic stress.
Gene duplication plays an important role in gene diversity, StDIR25 can be found in proximal duplication, segmental duplication and transposed duplication with a total of 5 times, StDIR25 may be a key gene in the gene duplicate events (Figure 4).
The synteny relationship genes analysis between different species can show that the diverge of genes followed by the diverge of species. In Figure 6, monocotyledon plant rice had a long divergence time with potato, so there are only 5 pair homologous, 18 pair homologous in dicotyledons Arabidopsis, and there 28 pair homologous between Solanaceae plant potato and tomato, The results indicated that the DIR genes are more homologous and conserved in dicotyledons than monocotyledons, and Solanaceae than dicotyledons. Besides, there are not only the number of homologous is the most in potato and tomato, but also the homologous have the parallelism between DIR genes on chromosomes, such as chromosome 2.
Subsequently, we analyzed the expression levels of the potato StDIR gene family after heat stress by transcriptomic data and selected four genes for q-PCR. The results showed that the q-PCR trend of the selected genes was the same as that of the transcriptome. This also verified the authenticity and reliability of transcriptome data. In the whole transcriptome, most members of the StDIR gene family showed a response to heat stress. Most members of the StDIR gene family showed an increasing trend after plants were treated with heat stress. We then treated potatoes with cold, salt, ABA and PEG, and selected some genes for qRT-PCR detection to further explore the response of the StDIR gene family to different stresses. Through the above experiments, we once again verified that the StDIR gene family has a certain degree of response to many abiotic stresses, indicating that the members of the StDIR gene family play a key role in the process of potato abiotic stress. It has been shown that the participation of AtDP1/AtDIR12 and AtLAC5 in the biosynthesis of new lignans through SC/FC has a strong protective effect on Arabidopsis seeds (Yonekura-Sakakibara et al., 2021). Furthermore, both the stress tolerance from ScDIR-expressed E. coli and its increased expression in sugarcane suggest that the ScDIR gene is involved in the response to abiotic stresses of drought, salt and oxidant (Guo et al., 2012; Li et al., 2022). Ralph et al. revealed a role for spruce DIR genes in constitutive and induced phenolic defense mechanisms against stem-boring insects (Ralph et al., 2006).
DIR not only plays an important role in a variety of biological and abiotic stresses, but also as a key gene in many other aspects due to its special gene function. The stone cell is a special cell of pear, which is a kind of dead cell formed by the lignification of the thin-walled cell wall in the pulp (Lu et al., 2015; Zeng et al., 2022). It can seriously affect the pear quality and economic benefits (Thompson et al., 2003; Sugino et al., 2018; Zhang et al., 2022; Wang et al., 2022). Therefore, regulating the content of stone cells is the key to improving the pear quality. Cheng et al. found that PbDIR may also be involved in stone cell development and lignin metabolism of pear (Jiang et al., 2020). Keiko et, al. found that AtDP1/AtDIR12 and AtLAC5 are involved in neolignan biosynthesis via SC/FC. A tetrazolium penetration assay showed that seed coat permeability increased in atdp1 mutants, suggesting a protective role of neolignans in Arabidopsis seeds (Yonekura-Sakakibara et al., 2021). Studies have also shown that the DIR1 gene promoter could be activated by MeJA, NaCl, and D-mannitol, and inhibited by ABA (Roh et al., 2010; Li et al., 2021). By analyzing the response of DIR-promoter to different hormones in different species, Li et, al. revealed the regulatory mechanism of the DIR gene to a certain extent, which played a certain reference role in the study of the specific mechanism of DIR gene family members. Lignin is closely related to DIR. Modern agricultural and industrial processes generate a significant amount of lignin-based waste, most of this involving use of fungal-mediated bio-catalysis (Lo et al., 2021; Maldhure et al., 2022; Pongchaiphol et al., 2022). Siarhei studied the DIR family of bacteria and suggested that bacteria may be better at degrading lignin than traditional fungi due to differences in DIR family members (Dabravolski, 2020). The DIR gene family has prominent functions in many aspects. Bioinformatics analysis of the DIR gene family and analysis of its gene structure can provide a strong reference for an in-depth exploration of its functions.
5 Conclusion
In summary, this study identified StDIR gene family members in potatoes for the first time. Through bioinformatics analysis and qRT-PCR analysis under abiotic stress, it revealed that StDIR gene family members in potatoes may play an important role in potato growth and development, stress and hormone response. This study provides ideas for studying the response of the StDIR gene in potato to stress and also provides a basis for further studies on the role of the StDIR gene family in potato breeding and resistance improvement.
Data availability statement
Publicly available datasets were analyzed in this study. This data can be found here: NCBI (National Center for Biotechnology Information) database (https://www.ncbi.nlm.nih.gov/sra/), PRJNA577000.
Author contributions
KL designed the research strategy and conceived and supervised the project; WJ wrote the manuscript; YX analyzed the data; ML, SZ, and ZH performed the experiments and drew the figure. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Jilin Province Science and Technology Development Plan (No. 20210202114NC, No. 20230203167SF), The National Natural Science Foundation of China (No. 32160702), and The National System of Modern Agriculture Industrial Technology of China (No. CARS-09-ES07).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Arasan, S. K. T., Park, J. I., Ahmed, N. U., Jung, H. J., Hur, Y., Kang, K. K., et al. (2013). Characterization and expression analysis of dirigent family genes related to stresses in Brassica. Plant Physiol. Bioch 67, 144–153. doi:10.1016/j.plaphy.2013.02.030
Atkinson, N. J., and Urwin, P. E. (2012). The interaction of plant biotic and abiotic stresses: from genes to the field. J. Exp. Bot. 63 (10), 3523–3543. doi:10.1093/jxb/ers100
Burlat, V., Kwon, M., Davin, L. B., and Lewis, N. G. (2001). Dirigent proteins and dirigent sites in lignifying tissues. Phytochemistry 57 (6), 883–897. doi:10.1016/s0031-9422(01)00117-0
Chen, C. J., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y. H., et al. (2020). TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13 (8), 1194–1202. doi:10.1016/j.molp.2020.06.009
Cheng, X., Su, X., Muhammad, A., Li, M., Zhang, J., Sun, Y., et al. (2018). Molecular characterization, evolution, and expression profiling of the dirigent (DIR) family genes in Chinese white pear (pyrus bretschneideri). Front. Genet. 9, 136. doi:10.3389/fgene.2018.00136
Corbin, C., Drouet, S., Markulin, L., Auguin, D., Laine, E., Davin, L. B., et al. (2018). A genome-wide analysis of the flax (Linum usitatissimum L) dirigent protein family: from gene identification and evolution to differential regulation. Plant Mol. Biol. 97 (1-2), 73–101. doi:10.1007/s11103-018-0725-x
Dabravolski, S. A. (2020). The resurgence of dirigent story: time for a bacterial chapter. Curr. Microbiol. 77 (4), 517–521. doi:10.1007/s00284-019-01809-2
Davin, L. B., and Lewis, N. G. (2000). Dirigent proteins and dirigent sites explain the mystery of specificity of radical precursor coupling in lignan and lignin biosynthesis. Plant Physiol. 123 (2), 453–462. doi:10.1104/pp.123.2.453
Dong, Y. Q., Qiang, T. Y., Liu, J. S., Li, B., Wei, X. P., Qi, Y. D., et al. (2021). Identification and characterization of DIR gene family in Schisandra chinensis. Zhongguo Zhong Yao Za Zhi 46 (20), 5270–5277. doi:10.19540/j.cnki.cjcmm.20210723.101
Guo, J. L., Xu, L. P., Fang, J. P., Su, Y. C., Fu, H. Y., Que, Y. X., et al. (2012). A novel dirigent protein gene with highly stem-specific expression from sugarcane, response to drought, salt and oxidative stresses. Plant Cell Rep. 31 (10), 1801–1812. doi:10.1007/s00299-012-1293-1
Halterman, D., Guenthner, J., Collinge, S., Butler, N., and Douches, D. (2016). Biotech potatoes in the 21st century: 20 Years since the first biotech potato. Am. J. Potato Res. 93 (1), 1–20. doi:10.1007/s12230-015-9485-1
Hurst, L. D. (2002). The ka/ks ratio: diagnosing the form of sequence evolution. Trends Genet. 18 (9), 486. doi:10.1016/s0168-9525(02)02722-1
Jiang, W. Q., Geng, Y. P., Liu, Y. K., Chen, S. H., Cao, S. L., Li, W., et al. (2020). Genome-wide identification and characterization of SRO gene family in wheat: molecular evolution and expression profiles during different stresses. Plant Physiol. Bioch 154, 590–611. doi:10.1016/j.plaphy.2020.07.006
Katiyar, A., Smita, S., Lenka, S. K., Rajwanshi, R., Chinnusamy, V., and Bansal, K. C. (2012). Genome-wide classification and expression analysis of MYB transcription factor families in rice and Arabidopsis. Bmc Genomics 13, 544. doi:10.1186/1471-2164-13-544
Khan, A., Li, R. J., Sun, J. T., Ma, F., Zhang, H. X., Jin, J. H., et al. (2018). Genome-wide analysis of dirigent gene family in pepper (Capsicum annuum L) and characterization of CaDIR7 in biotic and abiotic stresses. Sci. Rep. 8 (1), 5500. doi:10.1038/s41598-018-23761-0
Khatun, K., Robin, A. H. K., Park, J. I., Nath, U. K., Kim, C. K., Lim, K. B., et al. (2017). Molecular characterization and expression profiling of tomato GRF transcription factor family genes in response to abiotic stresses and phytohormones. Int. J. Mol. Sci. 18 (5), 1056. doi:10.3390/ijms18051056
Koch, M. A., Haubold, B., and Mitchell-Olds, T. (2000). Comparative evolutionary analysis of chalcone synthase and alcohol dehydrogenase loci in Arabidopsis, Arabis, and related genera (Brassicaceae). Mol. Biol. Evol. 17 (10), 1483–1498. doi:10.1093/oxfordjournals.molbev.a026248
Li, N., Zhao, M., Liu, T., Dong, L., Cheng, Q., Wu, J., et al. (2017). A novel soybean dirigent gene GmDIR22 contributes to promotion of lignan biosynthesis and enhances resistance to Phytophthora sojae. Front. Plant Sci. 8, 1185. doi:10.3389/fpls.2017.01185
Li, X. F., Liu, Z. L., Zhao, H. Y., Deng, X. L., Su, Y. Z., Li, R., et al. (2022). Overexpression of sugarcane ScDIR genes enhances drought tolerance in nicotiana benthamiana. Int. J. Mol. Sci. 23 (10), 5340. doi:10.3390/ijms23105340
Li, Z. Y., Li, B., Zhao, Y. C., and Zhao, D. G. (2021). Cloning and characterization of the DIR1 promoter from Eucommia ulmoides Oliv and its response to hormonal and abiotic stress. Plant Cell Tiss. Org. 146 (2), 313–322. doi:10.1007/s11240-021-02070-x
Liu, D., Shen, Z. P., Zhuang, K. Q., Qiu, Z. W., Deng, H. M., Ke, Q. L., et al. (2022). Systematic annotation reveals CEP function in tomato root development and abiotic stress response. Cells-Basel 11 (19), 2935. doi:10.3390/cells11192935
Liu, H. J., Zhu, P. F., Li, Z. G., Li, J. P., Tchuenbou-Magaia, F., and Ni, J. H. (2023). Thermo-biomechanical coupling analysis for preventing tomato fruit cracking during ripening. J. Food Eng. 341, 111336. doi:10.1016/j.jfoodeng.2022.111336
Liu, Z., Wang, X., Sun, Z., Zhang, Y., Meng, C., Chen, B., et al. (2021). Evolution, expression and functional analysis of cultivated allotetraploid cotton DIR genes. Bmc Plant Biol. 21 (1), 89. doi:10.1186/s12870-021-02859-0
Lo, C. C., Chang, Y. W., Chen, Y. L., Liu, Y. L., Wu, H. S., and Sun, Y. M. (2021). Lignin recovery from rice straw biorefinery solid waste by soda process with ethylene glycol as co-solvent. J. Taiwan Inst. Chem. E 126, 50–57. doi:10.1016/j.jtice.2021.07.030
Lopez-Zaplana, A., Martinez-Garcia, N., Carvajal, M., and Barzana, G. (2022). Relationships between aquaporins gene expression and nutrient concentrations in melon plants (Cucumis melo L) during typical abiotic stresses. Environ. Exp. Bot. 195, 104759. doi:10.1016/j.envexpbot.2021.104759
Lu, G. L., Li, Z. J., Zhang, X. F., Wang, R., and Yang, S. L. (2015). Expression analysis of lignin-associated genes in hard end pear (pyrus pyrifolia whangkeumbae) and its response to calcium chloride treatment conditions. J. Plant Growth Regul. 34 (2), 251–262. doi:10.1007/s00344-014-9461-x
Luo, R. H., Pan, W. Q., Liu, W. Q., Tian, Y., Zeng, Y., Li, Y. H., et al. (2022). The barley DIR gene family: an expanded gene family that is involved in stress responses. Front. Genet. 13, 1042772. doi:10.3389/fgene.2022.1042772
Ma, X. F., Xu, W. Y., Liu, T., Chen, R. Y., Zhu, H., Zhang, H. Y., et al. (2021). Functional characterization of soybean (Glycine max) DIRIGENT genes reveals an important role of GmDIR27 in the regulation of pod dehiscence. Genomics 113 (1), 979–990. doi:10.1016/j.ygeno.2020.10.033
Maldhure, A., Waghela, A., Nagarnaik, P., Khadse, G., and Labhasetwar, P. (2022). Microwave treated activated carbon from industrial waste lignin for denitrification of surface water. Int. J. Environ. Sci. Te 19 (6), 4987–4996. doi:10.1007/s13762-021-03361-8
Meher, P. K., Begam, S., Sahu, T. K., Gupta, A., Kumar, A., Kumar, U., et al. (2022). ASRmiRNA: abiotic stress-responsive miRNA prediction in plants by using machine learning algorithms with pseudo K-tuple nucleotide compositional features. Int. J. Mol. Sci. 23 (3), 1612. doi:10.3390/ijms23031612
Mo, F. L., Zhang, N. A., Qiu, Y. W., Meng, L. J., Cheng, M. Z., Liu, J. Y., et al. (2021). Molecular characterization, gene evolution and expression analysis of the F-box gene family in tomato (Solanum lycopersicum). Genes-Basel 12 (3), 417. doi:10.3390/genes12030417
Ogden, A. J., Bhatt, J. J., Brewer, H. M., Kintigh, J., Kariuki, S. M., Rudrabhatla, S., et al. (2020). Phloem exudate protein profiles during drought and recovery reveal abiotic stress responses in tomato vasculature. Int. J. Mol. Sci. 21 (12), 4461. doi:10.3390/ijms21124461
Peng, Y., Tang, N., Zou, J., Ran, J., and Chen, X. B. (2022). Rice MYB transcription factor OsMYB1R1 negatively regulates drought resistance. Plant Growth Regul. 99, 515–525. doi:10.1007/s10725-022-00922-w
Pongchaiphol, S., Suriyachai, N., Hararak, B., Raita, M., Laosiripojana, N., and Champreda, V. (2022). Physicochemical characteristics of organosolv lignins from different lignocellulosic agricultural wastes. Int. J. Biol. Macromol. 216, 710–727. doi:10.1016/j.ijbiomac.2022.07.007
Puchtel, I. S. (2022). Re-Os isotope and HSE abundance systematics of the 2.9 Ga komatiites and basalts from the sumozero-kenozero greenstone belt, SE fennoscandian shield: implications for the mixing rates of the mantle. Petrology+ 30 (6), 548–566. doi:10.1134/s0869591122060054
Qu, X. X., Zou, J. J., Wang, J. X., Yang, K. Z., Wang, X. Q., and Le, J. (2022). A rice r2r3-type MYB transcription factor OsFLP positively regulates drought stress response via OsNAC. Int. J. Mol. Sci. 23 (11), 5873. doi:10.3390/ijms23115873
Ralph, S., Park, J. Y., Bohlmann, J., and Mansfield, S. D. (2006). Dirigent proteins in conifer defense: gene discovery, phylogeny, and differential wound- and insect-induced expression of a family of DIR and DIR-like genes in spruce (picea spp). Plant Mol. Biol. 60 (1), 21–40. doi:10.1007/s11103-005-2226-y
Ralph, S. G., Jancsik, S., and Bohlmann, J. (2007). Dirigent proteins in conifer defense II: extended gene discovery, phylogeny, and constitutive and stress-induced gene expression in spruce (picea spp). Phytochemistry 68 (14), 1975–1991. doi:10.1016/j.phytochem.2007.04.042
Roh, H. S., Jung, Y., Koo, K. Y., Jung, U. H., Seo, Y. S., and Yoon, W. L. (2010). Steam reforming of methane over highly active and KOH-resistant Ni/γ-Al2O3 catalysts for direct internal reforming (DIR) in a molten carbonate fuel cell (MCFC). Appl. Catal. a-Gen 383 (1-2), 156–160. doi:10.1016/j.apcata.2010.05.037
Shi, H., Liu, Z., Zhu, L., Zhang, C., Chen, Y., Zhou, Y., et al. (2012). Overexpression of cotton (Gossypium hirsutum) dirigent1 gene enhances lignification that blocks the spread of Verticillium dahliae. Acta Biochim. Biophys. Sin. (Shanghai) 44 (7), 555–564. doi:10.1093/abbs/gms035
Song, M., and Peng, X. Y. (2019). Genome-wide identification and characterization of DIR genes in medicago truncatula. Biochem. Genet. 57 (4), 487–506. doi:10.1007/s10528-019-09903-7
Sugino, T., Taguchi, K., Unno, R., Hamamoto, S., Ando, R., Okada, A., et al. (2018). Transdifferentiation from white adipocytes to beige cells prevents kidney stone formation. J. Urol. 199 (4), E294–E295.
Thompson, M., Wilkins, S. J., Compton, R. G., and Viles, H. A. (2003). Channel flow cell studies on the evaluation of surface pretreatments using phosphoric acid or polymaleic acid for calcite stone protection. J. Colloid Interf. Sci. 259 (2), 338–345. doi:10.1016/S0021-9797(02)00224-2
Vatansever, R., Koc, I., Ozyigit, , , Sen, U., Uras, M. E., Anjum, N. A., et al. (2016). Genome-wide identification and expression analysis of sulfate transporter (SULTR) genes in potato (Solanum tuberosum L). Planta 244 (6), 1167–1183. doi:10.1007/s00425-016-2575-6
Wang, T. Z., Zhang, Y. X., Liu, Y. Y., Zhang, Z. J., and Yan, T. B. (2022). Intelligent evaluation of stone cell content of korla fragrant pears by vis/NIR reflection spectroscopy. Foods 11 (16), 2391. doi:10.3390/foods11162391
Wu, C. H., Zheng, C. Y., Ji, G. S., and Jiang, P. (2019). Synergistic effects of HSE and LTR elements from hsp70 gene promoter of Ulva prolifera (Ulvophyceae, Chlorophyta) upon temperature induction(1). J. Phycol. 55 (3), 738–743. doi:10.1111/jpy.12854
Wu, R. H., Wang, L., Wang, Z., Shang, H. H., Liu, X., Zhu, Y., et al. (2009). Cloning and expression analysis of a dirigent protein gene from the resurrection plant Boea hygrometrica. Prog. Nat. Sci-Mater 19 (3), 347–352. doi:10.1016/j.pnsc.2008.07.010
Yonekura-Sakakibara, K., Yamamura, M., Matsuda, F., Ono, E., Nakabayashi, R., Sugawara, S., et al. (2021). Seed-coat protective neolignans are produced by the dirigent protein AtDP1 and the laccase AtLAC5 in Arabidopsis. Plant Cell 33 (1), 129–152. doi:10.1093/plcell/koaa014
Zeng, M., Zhang, Y., Zhang, X. L., Zhang, W. L., Yu, Q., Zeng, W. Y., et al. (2022). Two birds with one stone: YQSSF regulates both proliferation and apoptosis of bone marrow cells to relieve chemotherapy-induced myelosuppression. J. Ethnopharmacol. 289, 115028. doi:10.1016/j.jep.2022.115028
Zhang, H., Gao, S. Y., Wang, T. Y., Xu, M. Y., Li, X. Y., and Du, G. D. (2022b). Ca2+ mediates transcription factor PuDof2.5 and suppresses stone cell production in pear fruits. Front. Plant Sci. 13, 976977. doi:10.3389/fpls.2022.976977
Zhang, K. J., Xing, W. J., Sheng, S. A., Yang, D. K., Zhen, F. X., Jiang, H. K., et al. (2022a). Genome-wide identification and expression analysis of eggplant DIR gene family in response to biotic and abiotic stresses. Horticulturae 8 (8), 732. doi:10.3390/horticulturae8080732
Zimmermann, P., Hirsch-Hoffmann, M., Hennig, L., and Genevestigator, G. W. (2004b). Arabidopsis microarray database and analysis toolbox (vol 136, pg 2621, 2004). Plant Physiol. 136 (4), 4335.
Zimmermann, P., Hirsch-Hoffmann, M., Hennig, L., and Gruissem, W. (2004a). GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiol. 136 (1), 2621–2632. doi:10.1104/pp.104.046367
Keywords: abiotic stress, bioinformatics, DIR, gene family, potato
Citation: Jia W, Xiong Y, Li M, Zhang S, Han Z and Li K (2023) Genome-wide identification, characterization, evolution and expression analysis of the DIR gene family in potato (Solanum tuberosum). Front. Genet. 14:1224015. doi: 10.3389/fgene.2023.1224015
Received: 17 May 2023; Accepted: 14 August 2023;
Published: 23 August 2023.
Edited by:
Ertugrul Filiz, Duzce University, TürkiyeReviewed by:
Bin Chen, Hebei Agricultural University, ChinaFirat Kurt, Mus Alparslan University, Türkiye
Copyright © 2023 Jia, Xiong, Li, Zhang, Han and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kuihua Li, khli@ybu.edu.cn
 Wenqi Jia1
Wenqi Jia1 Kuihua Li
Kuihua Li