- 1Department of Ophthalmology, The First Affiliated Hospital of Zhengzhou University, Zhengzhou, China
- 2Department of Ophthalmology, the First Affiliated Hospital of Henan University of Chinese Medicine, Zhengzhou, China
Objective: To reveal the potential mechanisms of curcumin for the treatment of skin cutaneous melanoma (SKCM) and its identify novel prognostic biomarkers.
Methods: We searched the Cancer Genome Atlas and Traditional Chinese Medicine Systems Pharmacology database for the data on SKCM and curcumin. We conducted data analysis using R and online tools. The propagation and migration of SKCM cells were assessed with CCK-8 and scratch wound assays, respectively. We assessed apoptosis by TUNEL assay and western blot.
Results: The survival analysis revealed that the mRNA expressions of DPYD, DPYS, LYN, PRKCQ, and TLR1 were significantly related to a favorable overall survival in SKCM patients. Additionally, the mRNA expression level of DPYD was associated with GPI, LYN, PCSK9, PRKCQ, and TLR1 mRNAs. GSEA results showed that the prognostic hub genes were augmented with ultraviolet, apoptosis, and metastasis. Curcumin expressed proliferation and migration of SK-MEL-1 cells (p < 0.05), and induced apoptosis (p < 0.05) significantly.
Conclusion: Curcumin may have potential therapeutic effects in SKCM by inhibiting cell proliferation and migration and inducing apoptosis by regulating oxygen-related signaling pathways. The hub genes might be identified as novel biomarkers for SKCM.
1 Introduction
Skin cutaneous melanoma (SKCM) is one of the most vigorous and fatal skin cancer types. The worldwide incidence of SKCM increases faster annually than any other cancer (Ali et al., 2013). The latest research shows that about 95,830 new cases of SKCM in situ have been reported in the United States in 2019 (Siegel et al., 2019). Ultraviolet (UV) radiation may be the main environmental risk factor (Gilchrest et al., 1999). Although the pathogenesis and diagnostic methods of diseases have shown great progress, the morbidity and mortality of SKCM have increased over the past 50 years in developed countries (Lu et al., 2018). Moreover, the treatment strategies that are currently available for metastatic melanoma have shown a relatively poor rate of success, and most newly developed anti-melanoma treatments are associated with severe adverse reactions (Kalal et al., 2017; Kozar et al., 2019; Luther et al., 2019). For these reasons, people have started to gain interest in natural compounds. It has been found that phytochemicals have demonstrated anti-proliferation, apoptosis promoting, anti-invasion, and anti-angiogenesis properties in mouse models and melanoma cell lines, without obvious toxicity (Fontana et al., 2019).
Curcumin is a polyphenolic compound, and curcumin is derived from turmeric (Curcuma longa), as its primary bioactive component (Pisano et al., 2010). It is a dietary spice made from the rhizome of Curcuma longa and is commonly used in curry powder as well as for centuries in traditional Chinese medicine (Maheshwari et al., 2006; Yu et al., 2010). Research has identified that curcumin has various therapeutic properties via different biological functions and pharmacological effects. These therapeutic properties include anti-inflammatory, antioxidant, immunomodulatory, antimicrobial, anti-ischemic, anti-cancer, and antirheumatic activities (Sahebkar, 2010, 2013; Mirzaei et al., 2016; Momtazi et al., 2016; Pulido-Moran et al., 2016; Kunnumakkara et al., 2017). Curcumin can induce endoplasmic reticulum stress in SKCM by inhibiting classic signaling pathways, which included nuclear factor kappa B (NF-κB), signal transducer and activator of transcription 3 (STAT3), Akt/Mtor, and Wnt/β-catenin, the expression of reactive oxygen species (ROS)thus the enhancement of oxidative stress injury has been increased (Bakhshi et al., 2008; Liao et al., 2017; Siwak et al., 2005; Zhang et al., 2015; Zheng et al., 2004). However, no systematic study on the mechanism, target, and effect of curcumin in the course of SKCM has been published thus far.
Therefore, we used the resources in traditional Chinese medicine and tumor databases to analyze what role curcumin plays in the treatment of SKCM. Specifically, we aimed to identify the target genes of curcumin acting on SKCM and analyze whether these genes are related to pathogenesis, staging, and prognosis. Furthermore, the effects of varying concentrations of curcumin on proliferation, migration, and apoptosis of SKCM cells were assessed in vitro. The results of this study were the basis for future studies on the effects of curcumin on SKCM.
2 Materials and methods
2.1 Detection of potential target genes
We obtained the molecular formula of curcumin from the Traditional Chinese Medicine Systems Pharmacology Database and Analysis Platform (TCMSP, https://old.tcmsp-e.com/tcmsp.php, accessed on 25 June 2021) (Ru et al., 2014). The effectiveness of target genes was identified from the PharmMapper Server (http://www.lilab-ecust.cn/pharmmapper/, accessed on 25 June 2021) (Liu et al., 2010; Wang et al., 2016; Wang et al., 2017) through Druggable Pharmacophore Models. Besides, we obtained the SKCM-related target genes and all of the protein-coding genes from the GeneCards Human gene database (https://www.genecards.org/, accessed on 25 June 2021) (Harel et al., 2009; Stelzer et al., 2016). Thus, the common target genes of curcumin and SKCM were identified.
2.2 Gene ontology and kyoto encyclopedia of genes and genomes analyses
We conducted the GO and KEGG analyses based on these common target genes. Providing data about the gene expression of common target genes. The GO and KEGG analyses were conducted to elucidate which mechanisms were applied by curcumin in the treatment of SKCM using the KOBAS 3.0 web server (http://kobas.cbi.pku.edu.cn/index.php, accessed on 30 June 2021) (Wu et al., 2006; Xie et al., 2011) and STRING v11.0 (https://string-db.org/, accessed on 30 June 2021) (Szklarczyk et al., 2019). Additionally, the Biological Networks Gene Ontology (BiNGO) (Maere et al., 2005), a GO function analysis tool, was applied to predict the functionality of common target genes.
2.3 Establishing the Protein-Protein Interaction network
A PPI network provides systematic visual data on the relationships between drugs, target genes, and proteins. We obtained the data from the STRING protein query and utilized data to construct the PPI network. Medium confidence of 0.400 was selected as the threshold in the analysis. Some nodes that were disconnected from each other were not displayed. Cytoscape software 3.6.1 was used to visualize the PPI network.
2.4 The construction of Genetic Interaction network
A GI network shows the complex interaction between genes of interest, and it was generated in GeneMANIA (https://genemania.org/, accessed on 30 June 2021) (Warde-Farley et al., 2010). The common target genes were used as query terms and then the predicted ones were shown simultaneously.
2.5 Survival data preparation
We obtained the Cancer Genome Atlas (TCGA) data and survival rate/time data, including clinical information (ID, age at index, gender, race, vital status, tumor stage, treatment, and the mRNA expression of common target genes of SKCM patients) from OncoLnc (http://www.oncolnc.org/, accessed on 25 June 2021) (Anaya, 2016) and TCGA (https://cancergenome.nih.gov/, accessed on 1 July 2021). Briefly, all common target genes were registered in the database, then the patients with SKCM were categorized half based on the expression of every gene, and as a result, the survival data of these SKCM patients was obtained. Ultimately, the common target genes related to overall survival (OS) were identified as hub genes.
2.6 Survival analysis
We used the 50% limitation as standard for each hub gene’s mRNA to divide the patients into groups with high- or low expression. Based on OS, the log-rank test and Kaplan-Meier estimator were applied in the survival analysis to calculate the log-rank p-value and identify the OS of hub genes. Subsequently, a Cox regression analysis was conducted to identify any associations between clinical information and the risk score, for which a nomogram was produced. R v3.6 was used to create the survival curves and nomogram.
2.7 mRNA expression levels and correlation analyses
The Gene Expression Profiling Interactive Analysis (GEPIA: http://gepia.cancer-pku.cn/, accessed on 1 July 2021) dataset (Tang et al., 2017) was applied to create a boxplot in which the hub gene mRNA’s expression levels were demonstrated. We calculated the mRNA expression levels through the retrieved TCGA data. Furthermore, we defined the expression of hub genes as high and low based on the median value. The high expression group referred to patients with expression values that were higher than the median values of the specific hub genes. Besides, the low expression group referred to patients with expression values that were lower than the median values of the specific hub genes. R v3.6 was used to perform Pearson correlation coefficient analysis by which the co-expression relationship among hub genes was assessed.
2.8 Gene Set Enrichment Analysis
GSEA (http://software.broadinstitute.org/gsea/index.jsp; accessed on 1 July 2021) (Subramanian et al., 2005) was conducted to identify which potential mechanisms were responsible for the effect that the risk score has on SKCM prognosis. The Molecular Signatures Databases (MSigDB) c2 (c2. cp.kegg.v6.1. symbols.GMT) and c5 (c5. all.v6.1. symbols.GMT) were used to investigate the crucial functions and pathways that could affect SKCM on the basis of prognosis-related hub gene mRNAs. We defined the significance as a nominal p-value < 0.01 and false discovery rate (FDR) < 0.25 for the sets of enrichment genes in the GSEA. The nine most significant gene sets were selected for this study and six of the eight prognosis-related genes (DPYD, GPI, LYN, MMP2, PRKCQ, and TLR1) were included in the GSEA due to the limitation of the dataset.
2.9 Cell line and drugs
The human SKCM cell line (SK-MEL-1) was retrieved from Procell Life Science & Technology (Wuhan, China). The curcumin was obtained from Solarbio Life Sciences (Beijing, China). The cells were cultured in Dulbecco’s Modified Eagle Medium (DMEM) added with 20% FBS and then incubated at 37°C.
2.10 Cell proliferation assay
We used a Cell Counting Kit-8 (CCK-8) assay (Beyotime Biotechnology, Shanghai, China) to measure the spectrophotometric absorbance, with which the proliferation of cells at 24 h was estimated. The SK-MEL-1 cells were cultured on plastic 96-well culture plates at a concentration of 5 × 103 cells/well. All experiments were performed as instructed by the manufacturer.
2.11 Cell migration assay
Cell migration was detected through a scratch wound assay. The SK-MEL-1 cells confluence in culture plates was scratched with a sterile pipette tip to produce a space free of cells. Serum-free DMEM was used to first rinse all the cells and then they were photographed to document the width of the wound at 0 h. Then, we used the serum-free medium to culture the negative control group of cells for up to 24 h, and the other two groups were treated with 20 and 30 μM curcumin, respectively. Photographs of the marked wound location were taken again at 6, 16, and 24 h to measure the migration of cells.
2.12 TUNEL assay
The rate of apoptosis in cultured SK-MEL-1 cells was measured with the One Step TUNEL Apoptosis Assay Kit (Dalian Meilun Biotechnology, Dalian, China) as described by the manufacturer’s instructions. SK-MEL-1 cells cultured in plastic 6-well culture plates were treated with proteinase K (20 μg/ml) and stained as recommended. The apoptotic index of cells was calculated by observing the TUNEL-positive cells in six fields that did not overlap under ×200 magnification.
2.13 Western blot
Cell apoptosis was also evaluated on protein level by western blot. The primary antibodies (GAPDH, Bcl-2, Cleaved Caspase 3, and Bax) were obtained from Cell Signaling Technology Inc. (MA, United States). The extracts of protein were stored at −80°C until use. Identical quantities of denatured protein were exposed to 10% SDS-PAGE and the separated proteins were placed on polyvinylidene difluoride (PVDF) membranes (Solarbio Life Sciences, Beijing, China). An LI-COR automatic chemiluminescence image analysis system was used to visualize the protein bands. The Odyssey Fc Imaging System was used to quantify the Western blot signals.
2.14 Statistical analyses
We used R v3.6 to obtain the correlation plot, survival curves, nomogram, and visualization of data. A p-value < 0.05 was deemed statistically significant. A workflow diagram is shown in Figure 1. GraphPad Prism (GraphPad Software, San Diego, United States) was used to perform one-way and two-way ANOVA.
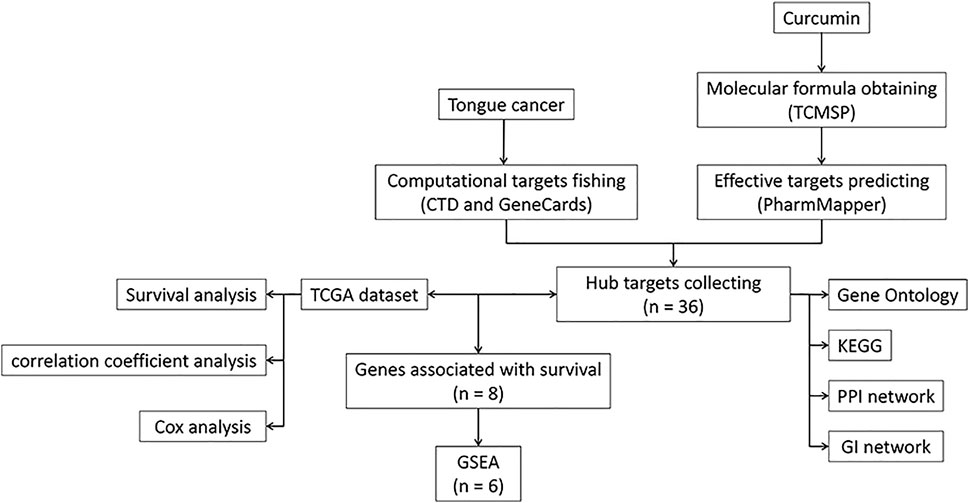
FIGURE 1. A workflow diagram. TCMSP, Traditional Chinese Medicine Systems Pharmacology Database, and Analysis Platform; KEGG, Kyoto encyclopedia of genes and genomes; PPI, Protein-Protein Interaction; GI, Genetic Interaction; TCGA, The Cancer Genome Atlas; GSEA, Gene Set Enrichment Analysis.
3 Results
3.1 The identification and functional analyses of target genes
The molecular formula of curcumin was obtained from TCMSP and 158 human target genes were collected (Figure 2A), of which 36 were identified as common targets (Figure 2B). The significant (the cut-off criterion for statistic difference was corrected to p < 0.0001) GO categories are shown in Supplementary Table S1. The common target genes were mainly enriched in the cytoplasm (GO:0005737), catabolic process (GO:0009056), response to endogenous stimulus (GO:0009719), and other terms. The KEGG analysis showed that the hub genes were mainly enriched in carbon metabolism (hsa01200), metabolic pathways (hsa01100), and NF-κB signaling pathway (hsa04064), and other associated pathways (Supplementary Figure S1). These results were consistent with the BiNGO outcomes (Supplementary Figure S2).
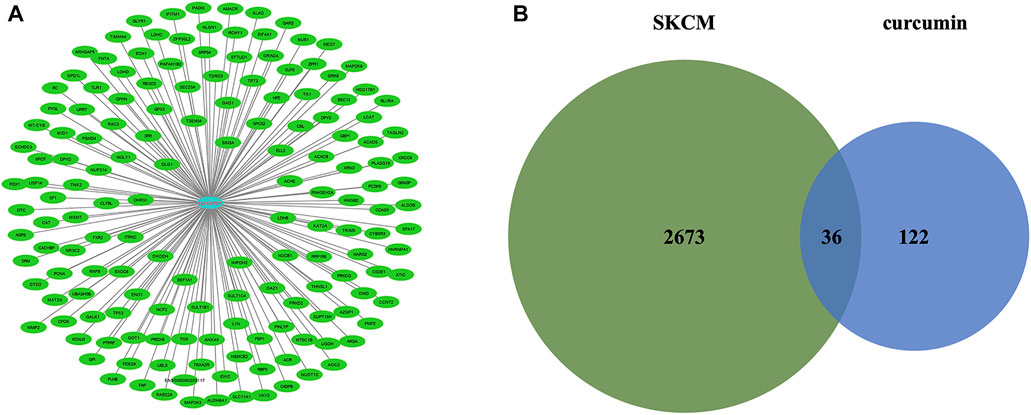
FIGURE 2. (A) Targets for curcumin. (B) Venn diagram summarizing differentially expressed targets for SKCM and curcumin. SKCM, skin cutaneous melanoma. The data are derived from PharmMapper.
3.2 The construction of PPI and GI network
The PPI network was constructed using the STRING online tool, and the Cytoscape 3.6.1 software was used for visualization. Identification of the most significant genes using a network constructed from common target genes (Supplementary Figure S3A). As shown in Supplementary Figure S3A, the tumor protein 53 (TP53), catalase (CAT), and enolase 1 (ENO1) were evidently at the PPI network’s center. The GeneMANIA online tool was used to construct the GI network, which shows the interaction among the 36 common target genes and predicted genes (Supplementary Figure S3B).
3.3 Survival analysis
The results of the log-rank test and Kaplan-Meier estimator indicated that the following eight out of 36 common target genes were significantly associated to the OS of SKCM patients: matrix metallopeptidase 2 (MMP2, p = 0.001), toll like receptor 1 (TLR1, p = 0.00052), dihydropyrimidine dehydrogenase (DPYD, p < 0.0001), proprotein convertase subtilisin/kexin type 9 (PCSK9, p = 0.011), protein kinase C theta (PRKCQ, p = 0.0013), glucose-6-phosphate isomerase (GPI, p = 0.0032), dihydropyrimidinase (DPYS, p = 0.00047), and the LYN proto-oncogene, Src family tyrosine kinase (LYN, p = 0.0061) (Figure 3). Based on the clinical information of SKCM patients, race (p = 0.0072), age at index (p < 0.0001), and tumor stage (p < 0.0001) were all correlated to OS. Figure 4 shows the generated nomogram and the c-index of this model was 0.669.
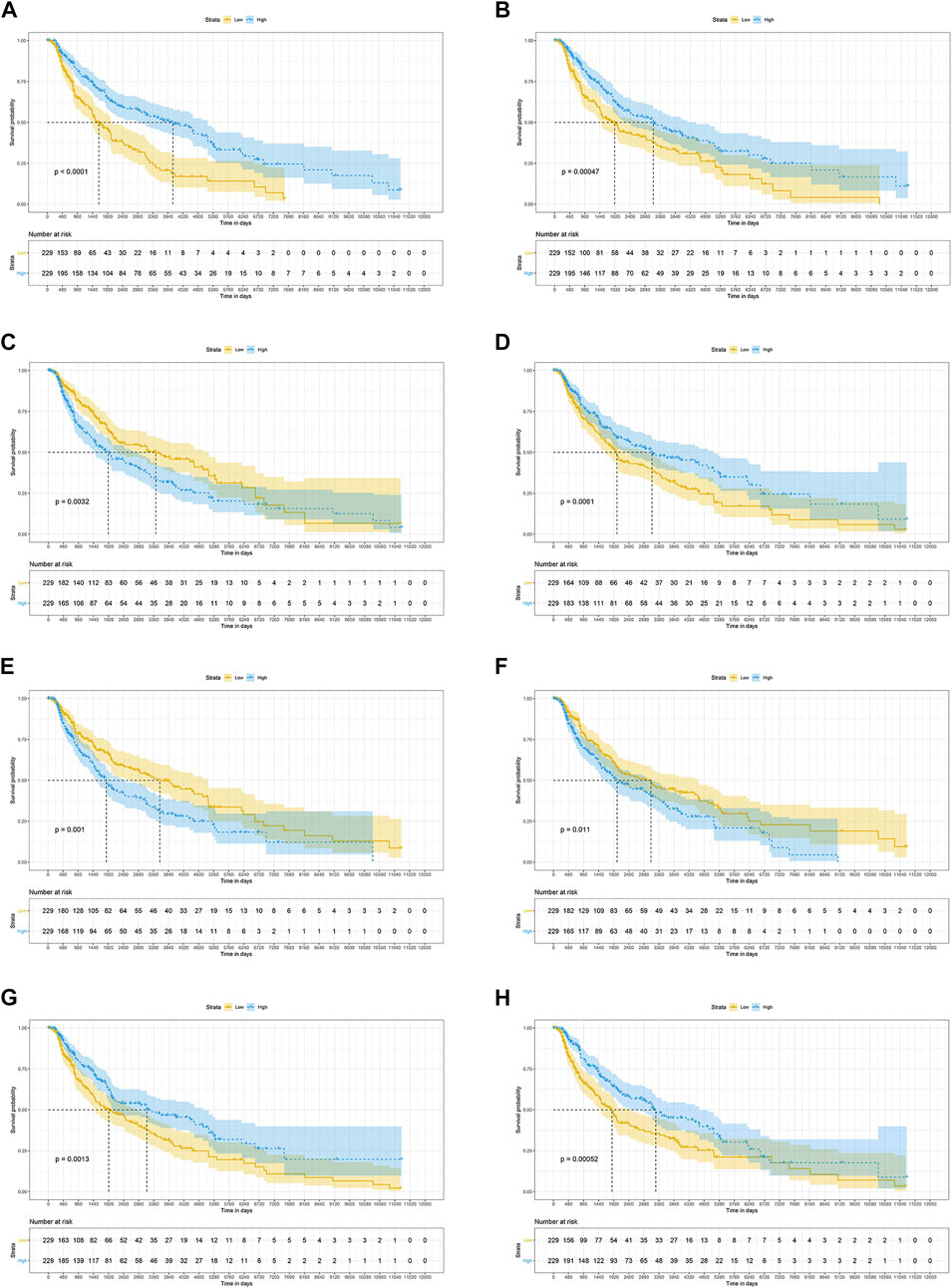
FIGURE 3. The prognostic significance of common targets for the OS of SKCM patients (A–H) Kaplan-Meier survival curves for all SKCM patients based on DPYD (A), DPYS (B), GPI (C), LYN (D), MMP2 (E), PCSK9 (F) PRKCQ (G), and TLR1 (H) expression (n = 458). OS, overall survival; SKCM, skin cutaneous melanoma.
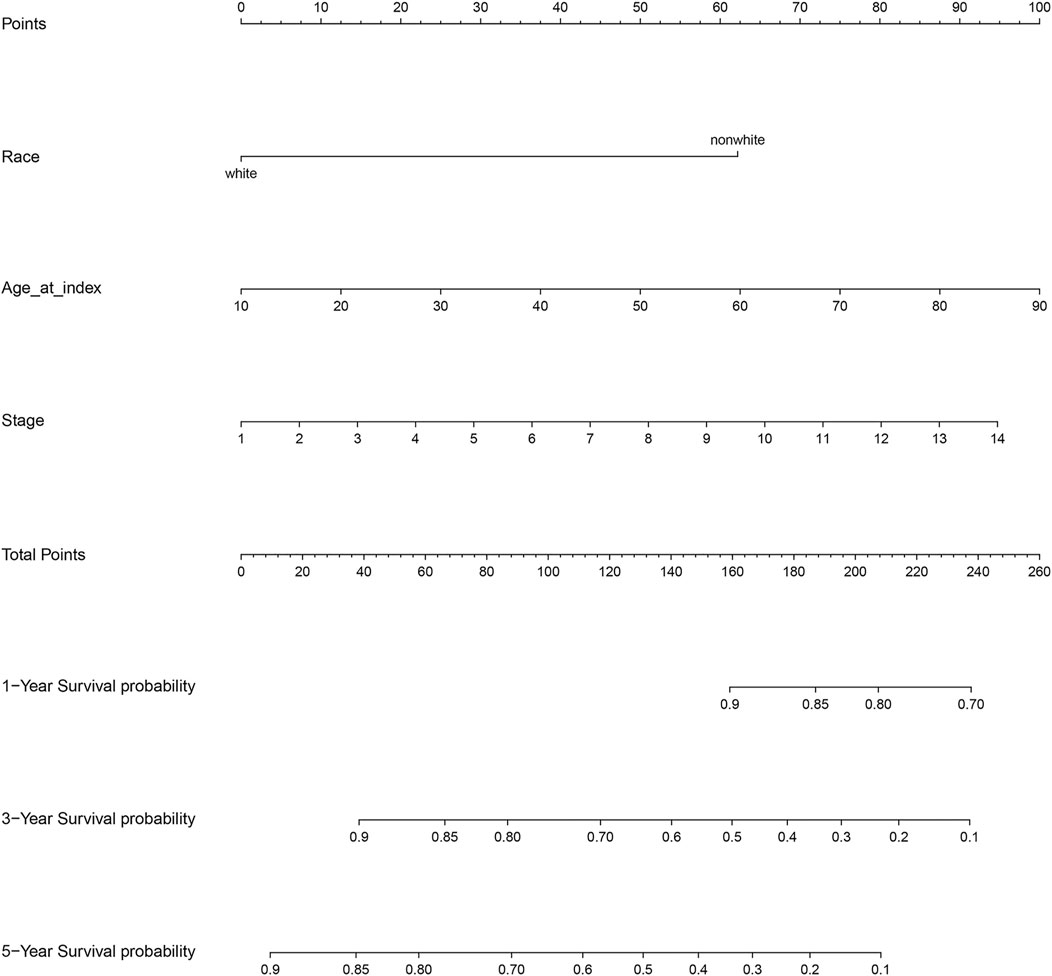
FIGURE 4. Nomogram for the relationship between clinical data and risk score. Stage 1 for I/II nos, 2 for 0, 3 for I, 4 for I a, 5 for I b, 6 for II, 7 for II a, 8 for II b, 9 for II c, 10 for III, 11 for III a, 12 for III b, 13 for III c, and 14 for IV.
3.4 mRNA expression levels and correlation analyses
As shown in boxplots, significant differences were discovered between the mRNA expression levels of hub genes found in normal tissues and SKCM tissues. Furthermore, the mRNA expression of DPYS, GPI, TLR1, PRKCQ, and LYN (Supplementary Figures S4B–D,G,H) in SKCM tissues were greater compared to those of normal tissues, of which the difference was statistically significant in GPI, PRKCQ, and LYN (all p-value < 0.01, Supplementary Figures S4C,G,H). In contrast, the mRNA expression of DPYD, MMP2, and PCSK9 in SKCM tissues was lower in comparison to that of normal tissues (Supplementary Figures S4A,E,F), of which the difference was only statistically significant in MMP2 (p-value < 0.01, Supplementary Figure S4F).
The correlation between the mRNA expression levels of hub genes was determined by Pearson correlation coefficient analysis. The results have shown that the mRNA expression level of DPYD was correlated with most of the hub gene mRNAs (GPI, LYN, PCSK9, PRKCQ, and TLR1) (all p-value < 0.01, Supplementary Figure S5 ; Supplementary Table S2).
3.5 GSEA analysis
GSEA was performed to calculate an enrichment score (ES) by going through the list of genes. The green line, representing a running-sum statistic, was enhanced when a gene was part of the gene set and decreased when it was not. A positive ES indicated enrichment of the gene set at the top of the ranking list and a negative ES indicated enrichment at the bottom. The horizontal bar’s red part and blue part represented positive and negative ES, respectively.
The results of the GSEA indicated that DPYD was primarily enriched in apoptosis, the JAK/STAT and MAPK signaling pathways, and the response to UV and cell adhesion functions (Supplementary Figure S6). For GPI, the AKT1 signaling pathway and the response to ultraviolet, glucose metabolism, oxidative phosphorylation, and electron transport chain were mainly enriched (Supplementary Figure S7). LYN was enriched in MAPK, JAK/STAT, and apoptotic-related signaling pathways. Additionally, LYN was also associated with DNA damage by UV and cell adhesion (Supplementary Figure S8). According to the results, MMP2 was highly related to cell migration and adhesion, apoptosis, and skin cancer progression (Supplementary Figure S9). As demonstrated, PRKCQ was mainly enriched in the JAK/STAT and MAPK signaling pathways, as well as apoptosis, DNA damage by UV, and cell adhesion (Supplementary Figure S10). TLR1 was mainly associated with the Toll-like receptor, JAK/STAT and AKT1 signaling pathways, and UV-induced apoptosis and DNA damage (Supplementary Figure S11).
Overall, the hub prognosis-related genes were mainly associated with cell adhesion and UV-related functions and participated in JAK/STAT and MAPK signaling pathways. Notably, MMP2 was associated with skin cancer progression.
3.6Curcumin inhibits SK-MEL-1 proliferation
The anti-proliferative effect of curcumin on SK-MEL-1 cells was evaluated in vitro by a CCK-8 assay. SK-MEL-1 cells were treated with different concentrations of curcumin, and the results showed that the inhibition of cell proliferation was more pronounced at 30 than at 20 μM (p < 0.001, Figure 5). Curcumin showed potential antiproliferative effects in SK-MEL-1 cells.
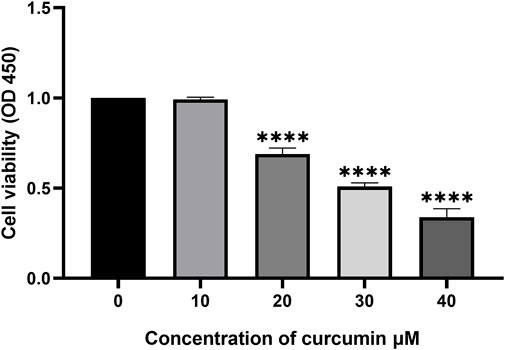
FIGURE 5. Study of cytotoxicity by using CCK-8 assay. The SK-MEL-1 cells were treated with curcumin in different concentrations. The cell proliferation was significantly decreased under the stimulation of 20 and 30 μM curcumin compared to that of 10 μM ***p < 0.001 and ****p < 0.0001, one-way ANOVA.
3.7 Curcumin inhibits SK-MEL-1 cell migration
The anti-migration effect of curcumin on SK-MEL-1 cells was measured in vitro by performing the scratch wound assay. The results (Figure 6) showed that there was no significant difference at 6 h (p > 0.05) in terms of migration distance observed among the 3 groups. A concentration of 100 μM curcumin significantly suppresses the migration of SK-MEL-1 cells at 16 h (p < 0.01) and 24 h (p < 0.001) in comparison to the control group. In addition, compared with the control group, curcumin at a concentration of 50 μM significantly inhibited the migration of SK-MEL-1 cells at 24 h (p < 0.01). The results indicated that curcumin had a potential anti-migratory effect in SK-MEL-1 cells.
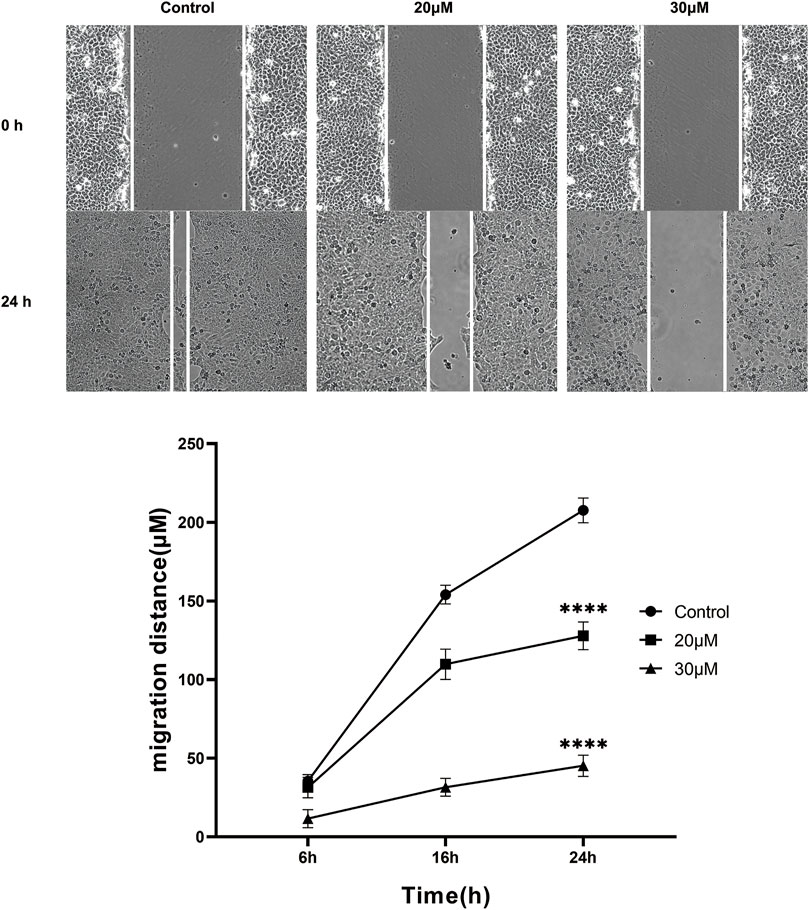
FIGURE 6. Cell migration evaluation by scratch wound assay. The SK-MEL-1 cells were treated with curcumin in concentrations of 0, 20, and 30 μM. Cell migration was significantly suppressed in curcumin-treated groups in comparison to that of the control group. **p < 0.01 and ***p < 0.001, two-way ANOVA.
3.8 Curcumin promotes apoptosis in SK-MEL-1 cells
We conducted a TUNEL assay to evaluate the pro-apoptosis effect of curcumin on SK-MEL-1 cells. The results of TUNEL (Figure 7) showed that the apoptosis rate and the apoptosis rate of SK-MEL-1 cells in the curcumin-treated group were significantly higher than those in the control group (p < 0.0001). The results of the western blot were consistent with that of the TUNEL assay (Figure 8).
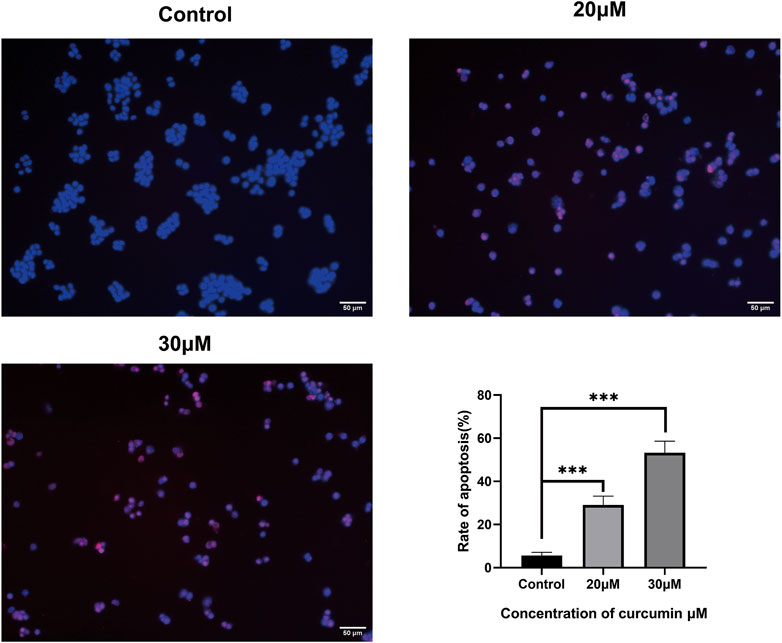
FIGURE 7. Evaluation of cell apoptosis by TUNEL assay. The SK-MEL-1 cells were treated with curcumin in concentrations of 0, 20, and 30 μM. Cell apoptosis was significantly promoted in curcumin-treated groups in comparison to that of the control group. TUNEL-positive cells were stained in red color, and the nucleus stained by DAPI was in blue. *p < 0.05 and ****p < 0.0001, one-way ANOVA.
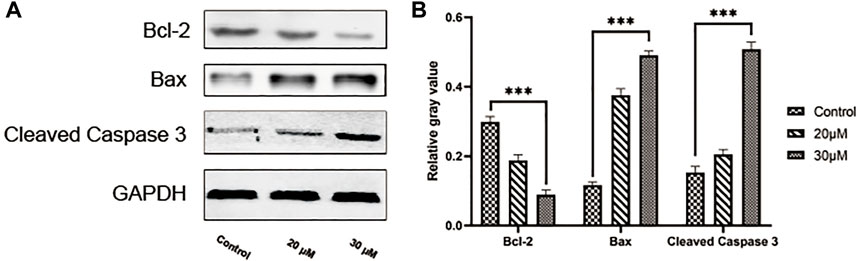
FIGURE 8. Evaluation of cell apoptosis by western blot. Blots showing proteins in the SK-MEL-1 cells treated with curcumin in different concentrations (A) and quantification (B). **p < 0.01, ***p < 0.001 and ****p < 0.0001, one-way ANOVA.
4 Discussion
From the point of view of molecular biological networks, traditional Chinese medicine network pharmacology provides a systematic research method in which the application of available traditional Chinese medicine compounds in various diseases can be evaluated. Previous research has shown that curcumin has great potential in preventing and treating various cancers (Ghalaut et al., 2012; Golombick et al., 2012; Ryan et al., 2013). In this study, we demonstrated that the molecular targets of curcumin on SKCM cells can be used as biomarkers of diagnosis and prognosis. In addition, the possible targets and concentrations of curcumin on SKCM were also analyzed. Analysis of clinical survival data has indicated that the survival rate was higher in SKCM patients with a high expression of DYPD, DYPS, LYN, PRKCQ, and TLR1, while the survival rate was lower in patients with a high expression of GPI, MMP2, and PCSK9. In the detection of tumor tissue and normal tissue, it was shown that the expression of GPI, LYN, and TLR1 was enhanced, whereas the expression of MMP2 was reduced. The targets of curcumin in the treatment of SKCM discovered in this study can be used as prognostic features and provide a theoretical basis for curcumin in the treatment of SKCM.
Co-expression analysis has indicated that these genes were not highly co-expressed with one another at the gene as well as protein levels. However, the regulation of these genes can influence the expression of TP53 at both gene and protein levels. TP53 has an important role as a tumor suppressor gene in humans, associated with the induction or inhibition of the cell cycle regulation, apoptosis regulation, DNA repair, and cell senescence-related gene expression after activation (Giaccia and Kastan, 1998). In subsequent in vitro experiments, we found that curcumin could upregulate the expression of Caspase3 and Bax in SK-MEL-1 cells and significantly reduce Bcl-2’s expression. Thus, TP53 may serve as the therapeutic target of curcumin in SKCM treatment. In addition, the GO analysis showed that these target genes were significantly enriched in the regulation of the apoptotic process, positive regulation of cell death, and regulation of the macromolecule metabolic process. The results of the KEGG analysis indicated that curcumin may play a role in SKCM by regulating metabolic pathways in tumor-related signaling pathways and the biosynthesis of amino acids and proteoglycans.
Based on GSEA analysis and literature review, we believe that these important genes/proteins are closely related to the occurrence of SKCM. DYPD can affect SKCM by regulating the cell response to UV, MAPK signaling pathway, JAK-STAT signaling pathway, apoptosis signaling pathway, and affecting cell-to-cell adhesion. DPYD is a vital enzyme in the metabolic pathway and is related to the drug response to 5-fluorouracil chemotherapy (Kristensen et al., 2010; Kimura et al., 2011). In addition, GPI function by regulating UV, tumor metastasis, and NF-κB signaling pathways. GPI is a glycolysis enzyme that plays a biological role through cell secretion. Its overexpression has been related to increased invasive phenotypes and mortality in many types of cancer (Lyu et al., 2016; Ma et al., 2018). Therefore, the survival rate of patients with elevated GPI is decreased, which was also observed in our study. Moreover, LYN, MMP2, PRKCQ, and TLR1 can affect SKCM by influencing tumor cell metastasis, UV-induced cell injury, the NF-κB signaling pathway, and apoptosis. LYN can regulate proliferation, differentiation, migration, and apoptosis, and over-expression of LYN plays a vital role in solid tumors (Tabariès et al., 2015; Q. Zhang et al., 2019). Previous research has indicated that MMP2 can produce the key pre-invasion factor induced by carcinogenic inflammation, and promotes tumor growth and invasion (Winerdal et al., 2018; W. Zhang et al., 2006). PRKCQ is widely expressed in the entire hematopoietic system and can induce the production and migration of breast tumors (Vyas et al., 2001; Byerly et al., 2016). TLR1 mediates local inflammation by affecting NF-κB. In pancreatic ductal carcinoma, the strong expression of TLR1 indicates a good prognosis, while the negative expression of TLR1 is a sign of poor prognosis (Lanki et al., 2019). We further investigated the effects of curcumin on the proliferation and apoptosis of human SKCM cells in vitro.
The cell wound assay demonstrated inhibited SK-MEL-1 cell growth by curcumin in a dose-dependent manner, and TUNEL staining showed that apoptosis increased significantly. In addition, after treatment with curcumin, the western blot demonstrated that curcumin could stimulate the activation of Caspase3 and downregulate the ratio of BCL2 and Bax. These findings are consistent with previously reported results of curcumin-induced apoptosis in osteosarcoma cells (Khodaei et al., 2022). Living cells possess higher levels of Bcl-2, which leads to the inhibition of apoptosis. Bcl-2 regulates the cellular activities of proteins related to cell proliferation or apoptosis, such as Caspase3 and Bax36. The first stage of the intrinsic apoptotic pathway relies on the activity of Bax, which modulates cellular fidelity to the pathway by altering mitochondrial physiology. Therefore, the effect of apoptosis induction by curcumin in SK-MEL-1 cells can be elucidated by detecting the expression of Caspase3 and the ratio of Bcl-2 to Bax. Thus, these target genes may act together on SKCM and may be potential therapeutic targets in SKCM.
5 Conclusion
We applied a network pharmacology method to identify the potential mechanisms of curcumin for the SKCM treatment methods. The common target genes might participate in the regulation of the inflammatory microenvironment of the tumor, thereby affecting the occurrence and metastasis of the tumor and improving the prognosis of SKCM patients. The in vitro experiments have indicated that curcumin has anti-proliferative and pro-apoptotic effects in SK-MEL-1 cells. Based on the functions that these hub genes occupy in cell proliferation and apoptosis; further in vitro research is necessary to clarify the specific anti-SKCM effect of curcumin.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author contributions
CM designed the outline of the study. All authors (LL, SL, and CM.) conducted experiments and data analysis. LL was involved in the preparation and writing of the manuscript. CM supervised the study and contributed to data interpretation and manuscript revision. All authors have read and agreed to the published version of the manuscript.
Funding
Joint construction project of Henan Medical Science and technology (LHGJ20220370).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2022.983943/full#supplementary-material
References
Ali, Z., Yousaf, N., and Larkin, J. (2013). Melanoma epidemiology, biology and prognosis. EJC Suppl. 11 (2), 81–91. doi:10.1016/j.ejcsup.2013.07.012
Anaya, J. (2016). OncoLnc: Linking TCGA survival data to mRNAs, miRNAs, and lncRNAs. PeerJ Comput. Sci. 2, e67. doi:10.7717/peerj-cs.67
Bakhshi, J., Weinstein, L., Poksay, K. S., Nishinaga, B., Bredesen, D. E., and Rao, R. V. (2008). Coupling endoplasmic reticulum stress to the cell death program in mouse melanoma cells: Effect of curcumin. Apoptosis 13 (7), 904–914. doi:10.1007/s10495-008-0221-x
Byerly, J., Halstead-Nussloch, G., Ito, K., Katsyv, I., and Irie, H. Y. (2016). PRKCQ promotes oncogenic growth and anoikis resistance of a subset of triple-negative breast cancer cells. Breast Cancer Res. 18 (1), 95. doi:10.1186/s13058-016-0749-6
Fontana, F., Raimondi, M., Di Domizio, A., Moretti, R. M., Montagnani Marelli, M., and Limonta, P. (2019). Unraveling the molecular mechanisms and the potential chemopreventive/therapeutic properties of natural compounds in melanoma. Semin. Cancer Biol. 59, 266–282. doi:10.1016/j.semcancer.2019.06.011
Ghalaut, V. S., Sangwan, L., Dahiya, K., Ghalaut, P. S., Dhankhar, R., and Saharan, R. (2012). Effect of imatinib therapy with and without turmeric powder on nitric oxide levels in chronic myeloid leukemia. J. Oncol. Pharm. Pract. 18 (2), 186–190. doi:10.1177/1078155211416530
Giaccia, A. J., and Kastan, M. B. (1998). The complexity of p53 modulation: Emerging patterns from divergent signals. Genes Dev. 12 (9), 2973–2983. doi:10.1101/gad.12.19.2973
Gilchrest, B. A., Eller, M. S., Geller, A. C., and Yaar, M. (1999). The pathogenesis of melanoma induced by ultraviolet radiation. N. Engl. J. Med. 320, 1341–1348. doi:10.1056/NEJM199904293401707
Golombick, T., Diamond, T. H., Manoharan, A., and Ramakrishna, R. (2012). Monoclonal gammopathy of undetermined significance, smoldering multiple myeloma, and curcumin: A randomized, double-blind placebo-controlled cross-over 4g study and an open-label 8g extension study. Am. J. Hematol. 87 (5), 455–460. doi:10.1002/ajh.23159
Harel, A., Inger, A., Stelzer, G., Strichman-Almashanu, L., Dalah, I., Safran, M., et al. (2009). GIFtS: Annotation landscape analysis with GeneCards. BMC Bioinforma. 10, 348. doi:10.1186/1471-2105-10-348
Kalal, B. S., Upadhya, D., and Pai, V. R. (2017). Chemotherapy resistance mechanisms in advanced skin cancer. Oncol. Rev. 11 (1), 326. doi:10.4081/oncol.2017.326
Khodaei, A., Jahanmard, F., Madaah Hosseini, H. R., Bagheri, R., Dabbagh, A., Weinans, H., et al. (2022). Controlled temperature-mediated curcumin release from magneto-thermal nanocarriers to kill bone tumors. Bioact. Mat. 11, 107–117. doi:10.1016/j.bioactmat.2021.09.028
Kimura, M., Kuwabara, Y., Mitsui, A., Ishiguro, H., Sugito, N., Tanaka, T., et al. (2011). Thymidylate synthetase and dihydropyrimidine dehydrogenase mRNA levels in esophageal cancer. Oncol. Lett. 2, 297–301. doi:10.3892/ol.2010.227
Kozar, I., Margue, C., Rothengatter, S., Haan, C., and Kreis, S. (2019). Many ways to resistance: How melanoma cells evade targeted therapies. Biochim. Biophys. Acta. Rev. Cancer 1871 (2), 313–322. doi:10.1016/j.bbcan.2019.02.002
Kristensen, M. H., Pedersen, P. L., Melsen, G. V., Ellehauge, J., and Mejer, J. (2010). Variants in the dihydropyrimidine dehydrogenase, methylenetetrahydrofolate reductase and thymidylate synthase genes predict early toxicity of 5-fluorouracil in colorectal cancer patients. J. Int. Med. Res. 38 (3), 870–883. doi:10.1177/147323001003800313
Kunnumakkara, A. B., Bordoloi, D., Padmavathi, G., Monisha, J., Roy, N. K., Prasad, S., et al. (2017). Curcumin, the golden nutraceutical: Multitargeting for multiple chronic diseases. Br. J. Pharmacol. 174 (11), 1325–1348. doi:10.1111/bph.13621
Lanki, M., Seppanen, H., Mustonen, H., Hagstrom, J., and Haglund, C. (2019). Toll-like receptor 1 predicts favorable prognosis in pancreatic cancer. PloS One 14 (7), e0219245. doi:10.1371/journal.pone.0219245
Liao, W., Xiang, W., Wang, F.-F., Wang, R., and Ding, Y. (2017). Curcumin inhibited growth of human melanoma A375 cells via inciting oxidative stress. Biomed. Pharmacother. 95, 1177–1186. doi:10.1016/j.biopha.2017.09.026
Liu, X., Ouyang, S., Yu, B., Liu, Y., Huang, K., Gong, J., et al. (2010). PharmMapper server: A web server for potential drug target identification using pharmacophore mapping approach. Nucleic Acids Res. 38, W609–W614. doi:10.1093/nar/gkq300
Lu, X., Zhang, Q., Wang, Y., Zhang, L., Zhao, H., Chen, C., et al. (2018). Molecular classification and subtype-specific characterization of skin cutaneous melanoma by aggregating multiple genomic platform data. J. Cancer Res. Clin. Oncol. 144 (9), 1635–1647. doi:10.1007/s00432-018-2684-7
Luther, C., Swami, U., Zhang, J., Milhem, M., and Zakharia, Y. (2019). Advanced stage melanoma therapies: Detailing the present and exploring the future. Crit. Rev. Oncol. Hematol. 133, 99–111. doi:10.1016/j.critrevonc.2018.11.002
Lyu, Z., Chen, Y., Guo, X., Zhou, F., Yan, Z., Xing, J., et al. (2016). Genetic variants in glucose-6-phosphate isomerase gene as prognosis predictors in hepatocellular carcinoma. Clin. Res. Hepatol. Gastroenterol. 40 (6), 698–704. doi:10.1016/j.clinre.2016.05.001
Ma, Y. T., Xing, X. F., Dong, B., Cheng, X. J., Guo, T., Du, H., et al. (2018). Higher autocrine motility factor/glucose-6-phosphate isomerase expression is associated with tumorigenesis and poorer prognosis in gastric cancer. Cancer Manag. Res. 10, 4969–4980. doi:10.2147/CMAR.S177441
Maere, S., Heymans, K., and Kuiper, M. (2005). BiNGO: A Cytoscape plugin to assess overrepresentation of gene Ontology categories in biological networks. Bioinformatics 21 (16), 3448–3449. doi:10.1093/bioinformatics/bti551
Maheshwari, R. K., Singh, A. K., Gaddipati, J., and Srimal, R. C. (2006). Multiple biological activities of curcumin: A short review. Life Sci. 78 (18), 2081–2087. doi:10.1016/j.lfs.2005.12.007
Mirzaei, H., Naseri, G., Rezaee, R., Mohammadi, M., Banikazemi, Z., Mirzaei, H. R., et al. (2016). Curcumin: A new candidate for melanoma therapy? Int. J. Cancer 139 (8), 1683–1695. doi:10.1002/ijc.30224
Momtazi, A. A., Derosa, G., Maffioli, P., Banach, M., and Sahebkar, A. (2016). Role of microRNAs in the therapeutic effects of curcumin in non-cancer diseases. Mol. Diagn. Ther. 20 (4), 335–345. doi:10.1007/s40291-016-0202-7
Pisano, M., Pagnan, G., Dettori, M. A., Cossu, S., Caffa, I., Sassu, I., et al. (2010). Enhanced anti-tumor activity of a new curcumin-related compound against melanoma and neuroblastoma cells. Mol. Cancer 9, 137. doi:10.1186/1476-4598-9-137
Pulido-Moran, M., Moreno-Fernandez, J., Ramirez-Tortosa, C., and Ramirez-Tortosa, M. (2016). Curcumin and health. Molecules 21 (3), 264. doi:10.3390/molecules21030264
Ru, J., Li, P., Wang, J., Zhou, W., Li, B., Huang, C., et al. (2014). Tcmsp: A database of systems pharmacology for drug discovery from herbal medicines. J. Cheminform. 6, 13. doi:10.1186/1758-2946-6-13
Ryan, J. L., Heckler, C. E., Ling, M., Katz, A., Williams, J. P., Pentland, A. P., et al. (2013). Curcumin for radiation dermatitis: A randomized, double-blind, placebo-controlled clinical trial of thirty breast cancer patients. Radiat. Res. 180 (1), 34–43. doi:10.1667/RR3255.1
Sahebkar, A. (2010). Molecular mechanisms for curcumin benefits against ischemic injury. Fertil. Steril. 94 (5), e75–76. doi:10.1016/j.fertnstert.2010.07.1071
Sahebkar, A. (2013). Why it is necessary to translate curcumin into clinical practice for the prevention and treatment of metabolic syndrome? Biofactors 39 (2), 197–208. doi:10.1002/biof.1062
Siegel, R. L., Miller, K. D., and Jemal, A. (2019). Cancer statistics, 2019. Ca. Cancer J. Clin. 69 (1), 7–34. doi:10.3322/caac.21551
Siwak, D. R., Shishodia, S., Aggarwal, B. B., and Kurzrock, R. (2005). Curcumin-induced antiproliferative and proapoptotic effects in melanoma cells are associated with suppression of IkappaB kinase and nuclear factor kappaB activity and are independent of the B-Raf/mitogen-activated/extracellular signal-regulated protein kinase pathway and the Akt pathway. Cancer 104 (4), 879–890. doi:10.1002/cncr.21216
Stelzer, G., Rosen, N., Plaschkes, I., Zimmerman, S., Twik, M., Fishilevich, S., et al. (2016). The GeneCards suite: From gene data mining to disease genome sequence analyses. Curr. Protoc. Bioinforma. 54, 131–3031. doi:10.1002/cpbi.5
Subramanian, A., Tamayo, P., Mootha, V. K., Mukherjee, S., Ebert, B. L., Gillette, M. A., et al. (2005). Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U. S. A. 102 (43), 15545–15550. doi:10.1073/pnas.0506580102
Szklarczyk, D., Gable, A. L., Lyon, D., Junge, A., Wyder, S., Huerta-Cepas, J., et al. (2019). STRING v11: Protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 47 (D1), D607–D613. doi:10.1093/nar/gky1131
Tabariès, S., Annis, M. G., Hsu, B. E., Tam, C. E., Savage, P., Park, M., et al. (2015). Lyn modulates Claudin-2 expression and is a therapeutic target for breast cancer liver metastasis. Oncotarget 6 (11), 9476–9487. doi:10.18632/oncotarget.3269
Tang, Z., Li, C., Kang, B., Gao, G., Li, C., and Zhang, Z. (2017). Gepia: A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 45 (W1), W98–W102. doi:10.1093/nar/gkx247
Vyas, Y. M., Mehta, K. M., Morgan, M., Maniar, H., Butros, L., Jung, S., et al. (2001). Spatial organization of signal transduction molecules in the NK cell immune synapses during MHC class I-regulated noncytolytic and cytolytic interactions. J. Immunol. 167 (8), 4358–4367. doi:10.4049/jimmunol.167.8.4358
Wang, X., Pan, C., Gong, J., Liu, X., and Li, H. (2016). Enhancing the enrichment of pharmacophore-based target prediction for the polypharmacological profiles of drugs. J. Chem. Inf. Model. 56 (6), 1175–1183. doi:10.1021/acs.jcim.5b00690
Wang, X., Shen, Y., Wang, S., Li, S., Zhang, W., Liu, X., et al. (2017). PharmMapper 2017 update: A web server for potential drug target identification with a comprehensive target pharmacophore database. Nucleic Acids Res. 45 (W1), W356–w360. doi:10.1093/nar/gkx374
Warde-Farley, D., Donaldson, S. L., Comes, O., Zuberi, K., Badrawi, R., Chao, P., et al. (2010). The GeneMANIA prediction server: Biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 38, W214–W220. doi:10.1093/nar/gkq537
Winerdal, M. E., Krantz, D., Hartana, C. A., Zirakzadeh, A. A., Linton, L., Bergman, E. A., et al. (2018). Urinary bladder cancer tregs suppress MMP2 and potentially regulate invasiveness. Cancer Immunol. Res. 6 (5), 528–538. doi:10.1158/2326-6066.CIR-17-0466
Wu, J., Mao, X., Cai, T., Luo, J., and Wei, L. (2006). KOBAS server: A web-based platform for automated annotation and pathway identification. Nucleic Acids Res. 34, W720–W724. doi:10.1093/nar/gkl167
Xie, C., Mao, X., Huang, J., Ding, Y., Wu, J., Dong, S., et al. (2011). Kobas 2.0: A web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 39, W316–W322. doi:10.1093/nar/gkr483
Yu, T., Li, J., and Sun, H. (2010). C6 ceramide potentiates curcumin-induced cell death and apoptosis in melanoma cell lines in vitro. Cancer Chemother. Pharmacol. 66 (5), 999–1003. doi:10.1007/s00280-010-1374-1
Zhang, Q., Meng, X., Qin, G., Xue, X., and Dang, N. (2019). Lyn kinase promotes the proliferation of malignant melanoma cells through inhibition of apoptosis and autophagy via the PI3K/Akt signaling pathway. J. Cancer 10 (5), 1197–1208. doi:10.7150/jca.28908
Zhang, W., Matrisian, L. M., Holmbeck, K., Vick, C. C., and Rosenthal, E. L. (2006). Fibroblast-derived MT1-MMP promotes tumor progression in vitro and in vivo. BMC Cancer 6 (1), 52–59. doi:10.1186/1471-2407-6-52
Zhang, Y. P., Li, Y. Q., Lv, Y. T., and Wang, J. M. (2015). Effect of curcumin on the proliferation, apoptosis, migration, and invasion of human melanoma A375 cells. Genet. Mol. Res. 14 (1), 1056–1067. doi:10.4238/2015.February.6.9
Keywords: curcumin, skin cutaneous melanoma, biomarker, proliferation, apoptosis, migration
Citation: Li L, Lu S and Ma C (2022) Anti-proliferative and pro-apoptotic effects of curcumin on skin cutaneous melanoma: Bioinformatics analysis and in vitro experimental studies. Front. Genet. 13:983943. doi: 10.3389/fgene.2022.983943
Received: 01 July 2022; Accepted: 23 August 2022;
Published: 12 September 2022.
Edited by:
Fu Wang, Xi’an Jiaotong University, ChinaReviewed by:
Chengyong Xie, Guizhou University, ChinaEswar Shankar, The Ohio State University, United States
Copyright © 2022 Li, Lu and Ma. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chao Ma, gmchao219@163.com
 Long Li1
Long Li1 Chao Ma
Chao Ma