- 1Department of Population and Quantitative Health Sciences, Cleveland Institute for Computational Biology, Case Western Reserve University, Cleveland, OH, United States
- 2Department of Nutrition, Case Western Reserve University School of Medicine, Cleveland, OH, United States
- 3Department of Genomic Medicine, Lerner Research Institute, Cleveland Clinic Foundation, Cleveland, OH, United States
- 4Department of Molecular Medicine, Case Western Reserve University School of Medicine, Cleveland, OH, United States
- 5Department of Medicine, Vanderbilt University School of Medicine, Nashville, TN, United States
- 6Department of Psychiatry, HIV Neurobehavioral Research Center, University of California, San Diego, San Diego, CA, United States
- 7Departments of Medicine and Psychiatry, University of California, San Diego, San Diego, CA, United States
- 8Departments of Neurosciences and Psychiatry, University of California, San Diego, San Diego, CA, United States
A common two-exon deletion distinguishes the gene encoding the free hemoglobin capturing protein—haptoglobin (HP)–into two alleles: HP1 and HP2. To evaluate the impact of this copy number variant (CNV) on neurocognitive impairment (NCI) in people living with HIV, we imputed this variant in 432 European-descent (EUR) and 491 African-descent (AFR) participants from the CNS HIV Antiretroviral Therapy Effects Research Study using an optimized imputation pipeline and evaluated its associations with NCI. At baseline, in AFR, the HP2 allele decreased the odds of NCI (defined by a global deficit score, GDS,
1 Introduction
Neurocognitive disorders have long been complications of HIV infection. Combination antiretroviral therapy (cART) has reduced morbidity and mortality in people living with HIV (PLWH) and remarkably prolonged their life expectancy (Grant et al., 2014; Watkins and Treisman, 2015). However, as PLWH age, neurologic complications like HIV-associated neurocognitive disorders (HAND) have become increasingly prevalent in both African-descent (AFR) and European-descent (EUR) populations (Antinori et al., 2007; Heaton et al., 2010; Grant et al., 2014). Ranked according to the severity of neurocognitive impairment (NCI), as established by Frascati criteria, the most severe yet uncommon form of HAND is HIV-associated dementia (HAD), followed by milder forms, including mild neurocognitive disorder (MND) and asymptomatic neurocognitive impairment (ANI) (Grant et al., 2014). Altogether, the overall prevalence of HAND is about 30–50% among randomly selected PLWH (Heaton et al., 2010; Jia et al., 2017). Typical phenotypes include mental slowing, memory loss, difficulties with complex tasks requiring executive function, and motor disorders (Simioni et al., 2010). Individuals also have behavioral abnormalities including apathy and a decrease in spontaneity or emotional responses (Simioni et al., 2010). NCI is the defining feature of HAND. It affects one or more of seven cognitive function domains, including verbal fluency, speed of information processing (SIP), learning, memory, motor function, attention and working memory, and executive function (Woods et al., 2004). The Global Deficit Score (GDS) represents an overall measure of neurocognitive performance based on a comprehensive neuropsychological test battery, and is widely used to rate HIV-associated NCI (Blackstone et al., 2012). Ranging from zero (unimpaired) to five (maximum severity) the GDS has been shown to be able to detect milder, HIV-associated cognitive impairment across multiple domains (Blackstone et al., 2012). It was previously shown that defining NCI as GDS
HIV infection and its induced chronic neuroinflammation are key factors found to contribute towards the development of HIV-associated NCI. HIV virus infects the CNS within days of acute infection, causes neuropathological changes in the basal ganglia and the white matter, and leads to high rates of delirium, depression, opportunistic CNS infections, and dementia; long-term HIV replication occurs in astrocytes and microglia and compromises neuronal function (Simioni et al., 2010; Lutgen et al., 2020; Valdebenito et al., 2021). Elevated markers of immune activation and inflammation are commonly detected in cerebrospinal fluid (CSF) from PLWH who have HAND (Gannon et al., 2011). Infected monocytes crossing the blood-brain barrier (BBB) can infect perivascular and other microglia in the brain, a process which is further enhanced by inflammatory mediators released by these cells (Strazza et al., 2011; Saylor et al., 2016). Increases in activated monocytes in the peripheral blood also have been shown to be associated with HAND (Strazza et al., 2011). Moreover, the BBB is disrupted by HIV infection both at the beginning of the infection and after virus entry into the CNS through infected monocyte-macrophages and this may lead to generation of reactive oxygen species (ROS) in CNS and damage brain tissues (Strazza et al., 2011). Other established risk factors for NCI in PLWH include age, nadir CD4+ T cell count, anemia, possibly female sex and comorbidities (e.g., cardiometabolic disorders, substance abuse, hepatitis C) (Ellis et al., 2011; Nightingale et al., 2014; Watkins and Treisman, 2015; Kallianpur et al., 2016; Rubin and Maki, 2019). Furthermore, genetic studies have shown that host genetic variations also play an important role in NCI and its progression (Kallianpur and Levine, 2014; Jia et al., 2017; Olivier et al., 2018).
The plasma glycoprotein Haptoglobin (HP, with Ensembl ID: ENSG00000257017 and Entrez Gene ID: 3240) has potential functional relationships with NCI. HP is mostly generated in the liver and secreted into the blood where its major function is to bind free hemoglobin (Hb), a highly reactive oxygen carrier molecule, and facilitate its clearance (Schaer and Alayash, 2010; Ratanasopa et al., 2013; MacKellar and Vigerust, 2016). In the brain, oligodendrocytes can also synthesize HP, although HP is not synthesized in the brain under normal conditions (Zhao et al., 2009; Bulters et al., 2018). Higher CSF HP levels were found to be associated with NCI and HAND in PLWH who had minimal comorbidity from a study including both AFR and EUR populations (Kallianpur et al., 2019). HP has also been associated with the pathogenesis of other neurocognitive diseases, such as Alzheimer’s disease (AD) (Yerbury et al., 2009; Spagnuolo et al., 2014; Song et al., 2015; MacKellar and Vigerust, 2016). In addition, HP reduces the oxidation of apolipoprotein E (APOE), rendering APOE more soluble and better able to clear plasma lipids, thereby promoting its function in cholesterol homeostasis (Salvatore et al., 2009; Spagnuolo et al., 2014).
A common copy number variation (CNV) that spans 2 tandem exons of the HP gene distinguishes alleles HP1 (one copy of exons 3 and 4) and HP2 (two copies of exons 3 and 4) in humans (Boettger et al., 2016). Boettger et al. hypothesized in their work that the HP2 allele is ancestral (based on comparisons with Neanderthal and Denisova genomes), and that the HP1 allele arose due to multiple recurrent deletions across different human populations. The evolution of these alleles is also thought to follow different tracks in both AFR and EUR populations due to migration and potential natural selection (Boettger et al., 2016). This variant is not detected by typical genotyping methods but can be inferred from a group of single nucleotide polymorphisms (SNPs) within the HP gene region with high accuracy using genotype imputation (r2 = 0.94 for EUR and r2 = 0.92 for AFR) (Boettger et al., 2016). The HP CNV affects HP protein structure and function. Western blot experiments and electronic microscopy images have shown that the HP1-1 (both alleles are HP1, i.e., only HP1 protein is available) only forms a functional dimer, while the HP1-2 and HP2-2 can form multimers with linear and circular conformations (Bulters et al., 2018). Although all forms have similar Hb binding affinity, compared to HP1-1, the HP2-2 proteins have larger sizes, which lower binding capacity and result in lower efficiency in clearing Hb, thereby reducing protection against free-Hb-mediated oxidative damage (Melamed-Frank et al., 2001; MacKellar and Vigerust, 2016). The HP CNV was also reported to be associated with HIV outcomes: Caucasian PLWH with HP2-2 had a higher mortality rate, with a reduction in median survival of approximately 4 years, compared to PLWH with HP1-1 and HP1-2 (Delanghe et al., 1998). EUR PLWH who have HP2-2 also have higher HIV viral load (Delanghe et al., 1998; MacKellar and Vigerust, 2016).
It remains unclear whether the HP CNV is associated with NCI in PLWH, including individuals receiving suppressive cART. We address this question by imputing HP genotypes for the AFR and EUR participants in the CNS HIV Antiretroviral Therapy Effects Research (CHARTER) Study, a large, observational HIV cohort with comprehensive neurocognitive assessments and previously measured CSF HP protein levels, and evaluating the associations between HP genotype and NCI at baseline and over time.
2 Methods
2.1 CHARTER Study Population and Neurocognitive Assessments
The CHARTER Study is a prospective, observational study of neurocognitive outcomes in PLWH. Ambulatory, PLWH were enrolled at six medical centers in the U.S., as described previously (Heaton et al., 2010). Detailed, structured interviews and comprehensive neurocognitive examinations, as well as laboratory assessments were conducted to collect information on HIV disease and treatment-related factors from participants at baseline and 6-months follow-up visits according to a protocol that was standardized across sites. Details of CHARTER study eligibility and assessment protocols have been published before (Heaton et al., 2010). For participants who consented, CSF samples were also obtained by lumbar puncture. To assess the GDS, participants underwent a comprehensive test battery that involved seven neurocognitive domains and were assigned test scores which were then converted to demographically corrected standard scores (T-scores) (Heaton et al., 2010). A single T-score was calculated for each of the cognitive domains by averaging the T-scores for each of the tests in that domain. The domain impairment is determined when an individual’s T-score is below one standard deviation from the mean (Antinori et al., 2007). A composite GDS was then derived, as a continuous measurement, with deficit scores converted from standard domain T-scores, using a published objective algorithm (Carey et al., 2004; Heaton et al., 2010). Participants’ NCI status was also determined by applying a GDS cutoff of 0.5, with ‘Normal/Not impaired’ defined by a GDS
2.2 Genotyping, Measurements of CSF HP Levels, and Known Factors Influencing NCI
Plasma HIV RNA (viral load) was determined by reverse transcriptase PCR (Heaton et al., 2010). The CD4+ nadir was obtained by self-report and confirmed by documented prior measurements in a subset of CHARTER Study participants (Ellis et al., 2011). CSF HP protein levels were quantified in 405 participants using multiplex bead-based suspension array immunoassays (Kallianpur et al., 2019). Detailed methods for quantification of CSF HP and other markers were published previously (Heaton et al., 2010; Ellis et al., 2011; Kallianpur et al., 2019). Genomic DNA was extracted from peripheral blood mononuclear cells collected at the baseline CHARTER visit using PUREGENE (GentraSystems, Inc., Minneapolis, MN). Genotyping was conducted using the Affymetrix Genome-Wide Human SNP Array 6.0TM by the Vanderbilt Technologies for Advanced Genomics (VANTAGE) at Vanderbilt University in two batches: n = 576 samples were genotyped before 2009 and n = 506 (six repeated for QC) were genotyped in 2012, due to funding reasons only (Jia et al., 2017). Nevertheless, only minor changes were observed from explicit testing of batch effects (Jia et al., 2017). Due to limited accuracy for imputing APOE, the APOE genotypes were determined for a subset (n = 401) of CHARTER participants by genotyping of rs7412 and rs429358, using TaqMan predesigned SNP genotyping assays (C_904973_10 and C_30846793_20; Applied Biosystems, Foster City, CA) as described in a previous publication (Morgan et al., 2013).
2.3 Quality Control
The QC and basic data cleaning pipeline used for CHARTER genomic data was published previously (Jia et al., 2017); here we describe the additional QC work that was conducted for HP imputation. Since the HP reference panels are separated for AFR and EUR, we performed all the additional QC steps respectively for AFR and EUR participants. The overall genotyping call rate was checked for each study participant and we found the overall call rate was
2.4 HP Genotype Imputation
We adopted published HP CNV imputation references for AFR and EUR (Boettger et al., 2016). An HP marker that was collapsed from all four HP subtype markers was added to the imputation reference with “0” represents HP1 and “1” represents HP2. We performed in silico validation and found the HP genotype we obtained from the HP marker 100% identical to the genotype we obtained from subtype markers. The full lengths of chromosome 16 of the individuals in the HP imputation reference were extracted from 1,000 Genomes (1KG) data and pre-phased using SHAPEITv2 (Delaneau et al., 2013) software. The HP imputation markers were then extracted from the pre-phased chromosome 16 to obtain the phased HP imputation reference panels. Further validation of this imputation strategy and optimization of the IMPUTEv2 (Howie et al., 2009) software settings were conducted using: 1. HP-genotype-removed imputation reference as data input with an accuracy metric and 2. the GTEx sequencing and expression data by comparing the imputed HP genotype and the read count of the exon 4 & 5 junction (unique to HP2) from RNA sequencing. The HP region of all CHARTER Study participants was imputed for HP genotypes, using both of the phased references and samples that were extracted based on the reference population; i.e., EUR participants were extracted from the imputation using European reference and AFR participants were extracted from the imputation using African reference. The HP imputation was conducted using the IMPUTEv2 software.
2.5 Association Analyses
The imputed dosages were hardcalled using a 0.9 threshold; in other words, dosages
3 Results
3.1 Haptoglobin HP2 Allele Associates With Lower CSF HP Levels
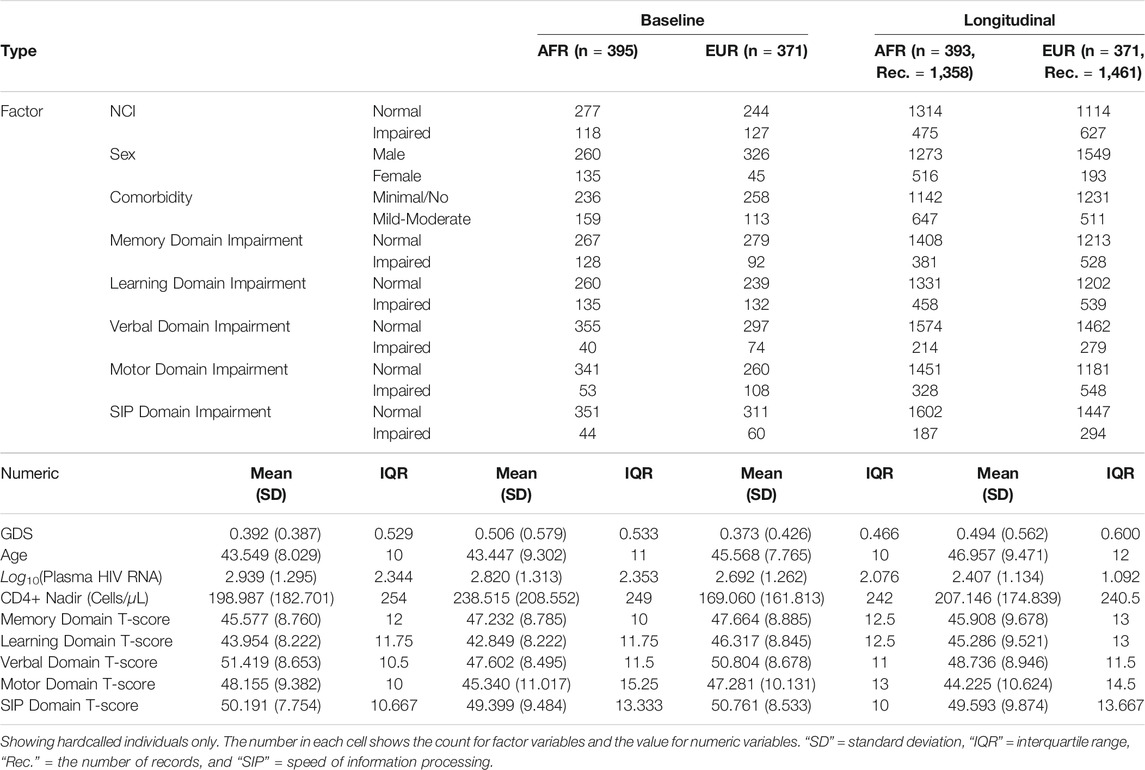
Statistics of all the study variables are in Table 1. Due to the complex evolutionary history of HP alleles in different populations, all analyses were conducted in AFR and EUR ancestral groups separately as well as jointly. The HP imputation process was validated by comparing the imputed HP genotypes and exon 4 & 5 junction counts from the Genotype-Tissue Expression (GTEx) RNA sequencing data (Supplementary Figure S1). HP genotypes were then imputed in the CHARTER AFR and EUR participants with high certainty (IMPUTEv2 info metric = 0.831 for AFR, and 0.830 for EUR). A low imputation dosage indicates a low certainty of the imputed genotype, thus, we filtered out the low confidence genotypes (hardcalls) with a stringent threshold, 0.9, to ensure that we had high quality genotypes for further analyses (Supplementary Figure S2). We obtained 371 hardcalls (86% of 432 samples) and 395 hardcalls (80% of 491 samples) in EUR and AFR, respectively. After hardcalling, Hardy-Weinberg Equilibrium was tested in both populations and no significant deviations were found.
Associations between the HP CNV and CSF HP protein levels were determined using an ANOVA F-test among the participants who had CSF HP levels measured (n = 283, after hardcalling) (Kallianpur et al., 2019). In both populations, the presence of additional HP2 alleles decreased the CSF HP protein levels, as shown in Figure 1A, (p = 2.17 × 10–5 in n = 131 AFR participants, p = 1.70 × 10–9 in n = 146 EUR participants, and p = 7.85 × 10–12 when combined).
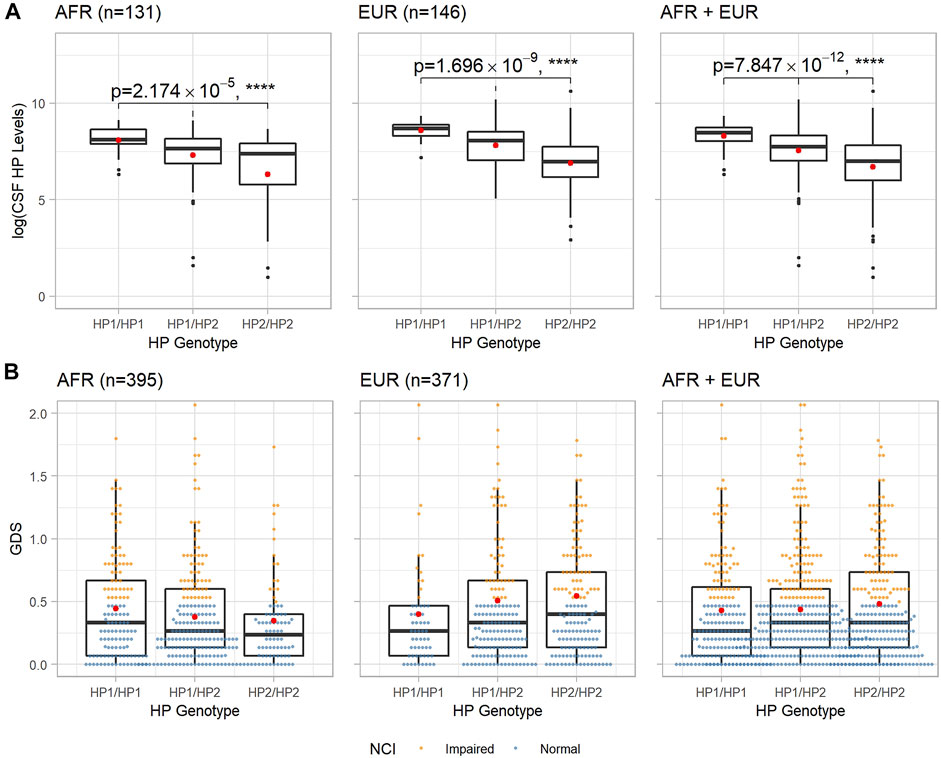
FIGURE 1. Boxplots of CSF HP protein levels and baseline GDS of each HP genotype in AFR and EUR ancestry groups. (A): CSF HP protein levels in logarithmic scale vs. HP genotypes. p_values and asterisks indicating level of significance are from the ANOVA F-test. (B): GDS at baseline vs. HP genotypes. In both panels, within each box, the black horizontal bar indicates the median value, and the red dot shows the mean value.
3.2 Haptoglobin CNV Associates With Baseline NCI Differently Across Ancestry Groups
The distributions of baseline GDS for each HP genotype in AFR and EUR populations are shown in Figure 1B; we observed an almost linear relationship between the mean GDS of different HP genotypes and the number of HP2 alleles in both AFR and EUR. However, neither ANOVA nor linear regression showed a statistically significant association between the HP CNV and the GDS. We then looked at the NCI as a dichotomous trait, defined by GDS
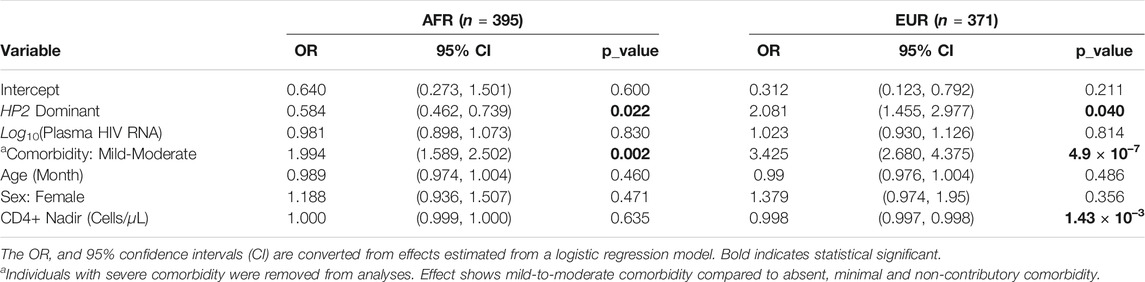
We adjusted for known risk factors for the dichotomous NCI in PLWH, including age, sex, comorbidity conditions, plasma HIV RNA (viral load), and CD4+ nadir in a logistic regression model as covariates (Heaton et al., 2010). In contrast to the consistent effects on CSF HP levels seen across both ancestry groups, we observed opposite directions of effect of the HP2 allele on NCI in AFR and EUR individuals. As shown in Table 2, in 395 AFR participants, the HP2 allele was associated with decreased odds of NCI (Odds Ratio, OR = 0.584, p = 0.022), while in 371 EUR participants, the HP2 allele was associated with increased odds of NCI (OR = 2.081, p = 0.040).
To account for uncertainty in genotype imputation estimations, we also tested the association between NCI and HP2 using directly imputed dosages rather than hardcalls. In EUR, we were able to detect a significant additive effect of HP2 dosage with OR = 1.605, (p = 0.002), which is similar to what we obtained using hardcalls. No significant associations were found in AFR using directly imputed allele dosages.
We also found that the effect of HP2 on NCI is independent from its association with CSF HP protein levels in both EUR and AFR PLWH. Although the sample size was limited, we performed sensitivity tests with the CSF HP protein levels to see if the effect of the HP CNV is driven by changes in the HP protein levels. Adjusting for the CSF HP levels did not change the significance of the HP2 alleles in either EUR or AFR PLWH. From Supplementary Table S1, we could see that the CSF HP levels is not significantly associated with NCI risk in either AFR or EUR individuals. In fact, this adjustment strengthened the HP association in EUR (Supplementary Table S1).
3.3 The HP CNV Influences NCI Over Time
Given the longitudinal study design of CHARTER, we were also able to examine the effect of the HP CNV on NCI as the study participants age. For the AFR group, the median follow-up time is 6 months and the mean is 1.406 years. While in EUR, the median follow-up time is 6 months and the mean is 1.679 years.
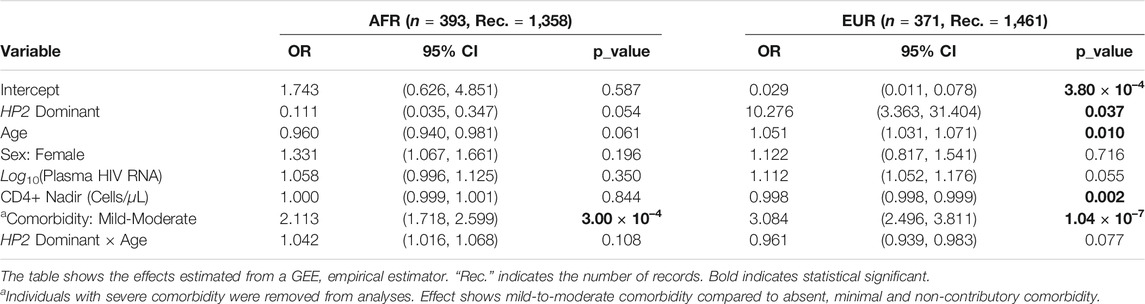
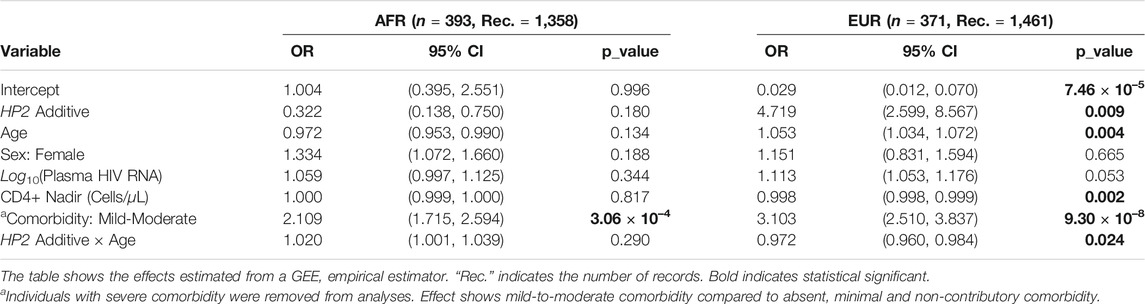
As shown in Table 3, EUR PLWH having HP2 were at 10.276 higher OR (p = 0.037) of developing NCI than PLWH that do not have HP2 (Figure 2A). This OR decreased by 0.961 per year (p = 0.077, Figure 2B). This decrease reflects a cumulative change in risk; for example, a decrease of 0.961 means the individuals probability of NCI equals the probability of the previous year × 0.961. We also noticed that though the risk of NCI for EUR HP1/HP1 individuals was lower than HP2 individuals at the beginning, it increased faster and became greater than HP2 individuals after approximately 58 years of age. In contrast, though only marginally significant, AFR PLWH with HP2 had lower risk (OR = 0.111, p = 0.054) of developing NCI compared to people without HP2 (Figure 2A). The directions of the HP2 effect on NCI were consistent with the baseline models within each population. These effects are visualized in Figure 2. We see a dramatic change in the trend of predicted NCI probability over age in both AFR and EUR between HP2 dominant and HP1/HP1 individuals. These effects were not observed from the analyses that combines the AFR and EUR participants as the opposing effects of the HP alleles in each population cancel each other out (Figure 2). We also detected an additive effect of HP2 on NCI in EUR as shown in Table 4. In EUR, each copy of HP2 allele was associated with a 4.719 increase in the OR (p = 0.009) of developing NCI (Figure 2C). This OR decreased by 0.972 per HP2 allele per year (p = 0.024, Figure 2D). These effects are not statistically significant in the AFR or combined analyses (Table 4).
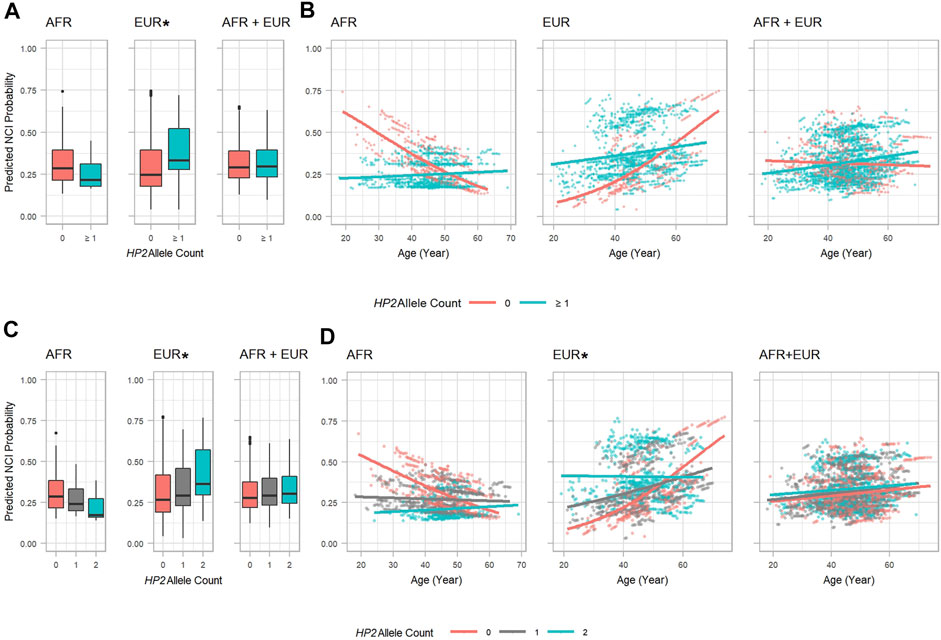
FIGURE 2. HP2 Dominant Effect on Predicted NCI Probability in the AFR and EUR Participants. (A, C): Boxplots showing the main effect of HP2 on predicted NCI probabilities. (B, D): Scatter plots with trendlines showing the effect of age on predicted NCI probabilities. Probabilities in (A, B) are from models shown in Table 3. Probabilities in (C, D) are from models shown in Table 4. Plots are colored by HP2 status. Additional adjustment of the first 3 PCs was used in the AFR + EUR analysis. “∗” indicates statistical significance.
HIV RNA levels were significantly different by HP genotype (Supplementary Figures S3–S6), suggesting that the risk for NCI may be influenced by altering viral load. To examine the potential mediation effect of the HIV RNA, we re-ran the analyses in the virus suppressed subgroup [⩽ lower limit of quantitation (LLQ)]. Despite changes in significance levels in some tests due to dramatic decreases in sample sizes, the directions of the effects remained consistent in the longitudinal analyses (Supplementary Tables S2, S3). Further analyses adjusting for the ⩽ LLQ status showed that the ⩽ LLQ status did not significantly contribute to NCI risk (Supplementary Tables S4, S5).
3.4 HP CNV Associations to NCI Are Driven by Changes in Specific Domains
We further decomposed the global model of NCI risk by investigating which cognitive domains are significantly affected by the HP CNV by conducting longitudinal analyses using domain impairments as outcomes. The higher risk of NCI in EUR with HP2 is likely driven by an increased risk in the learning domain impairment. This effect is slightly (but significantly) offset by a significant decrease in risk for verbal domain impairment based on an additive model of the HP alleles. These two effects, combined with more modest (non-significant) effects in other domains together create an overall increase in risk for NCI (Figure 3, Supplementary Tables S6, S7). In EUR, PLWH with HP2 had a higher OR of impairment in the learning domain (OR = 14.526, p = 0.019, Supplementary Table S6 and Figure 3A) with the risk reducing over time (by OR = 0.951, p = 0.030 per year of age, Supplementary Table S6). These over-time effects are reflected in Figure 3B where we observed a dramatic difference in slope between HP2 dominant and HP1 individuals. No statistically significant effects were noted in the AFR and combined (AFR + EUR) analyses (Figure 3B). We also found that with each HP2 allele, EUR PLWH have a significant decrease in risk of verbal domain impairment (OR = 0.259, p = 0.019, Supplementary Table S7 and Figure 3E). This effect is smaller than the contrasting effect of HP2 on the learning domain, and over time this risk modestly increases by OR = 1.020 per year (p = 0.023, Supplementary Table S7 and Figure 3F). In AFR, the HP2 allele was associated with a lower risk of impairment in the speed of information processing domain (Figure 3 and Supplementary Table S8) with this risk increasing by OR = 1.072, per year (p = 0.038, Supplementary Table S8 and Figure 3D).
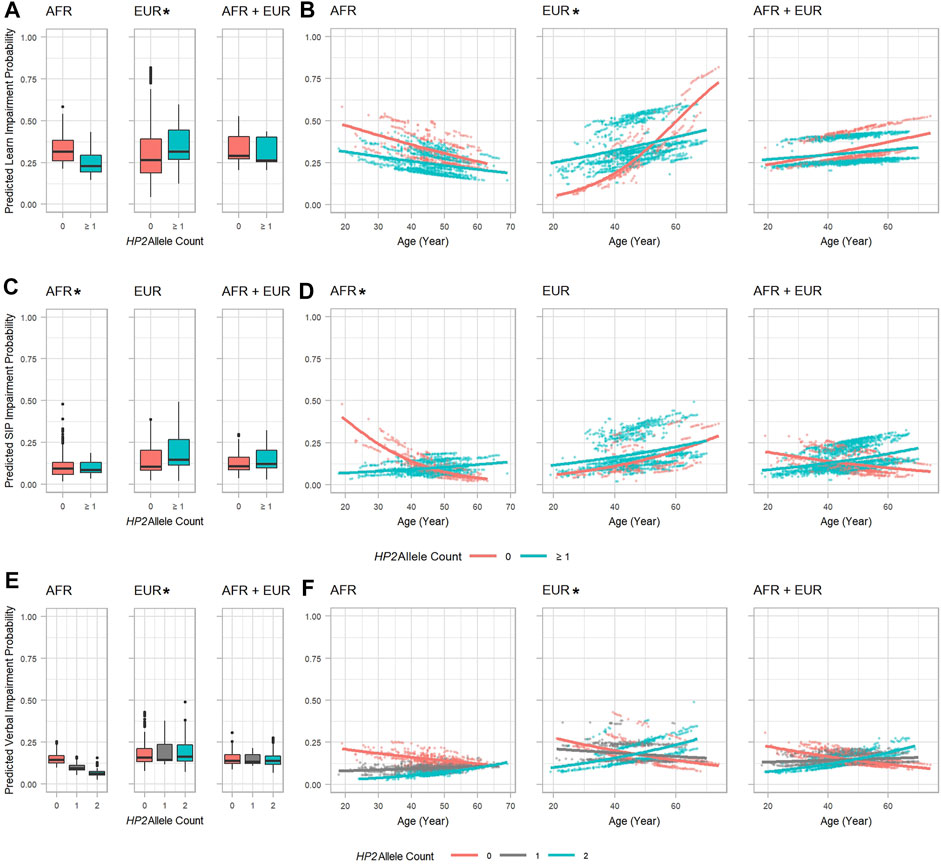
FIGURE 3. HP2 effect on predicted specific cognitive domain impairment probability in the AFR and EUR participants. (A, C) and (E): Boxplots showing the effect of HP2 on predicted learning, SIP, and verbal domain impairment probabilities, respectively. (B, D), and (F): Scatter plots with trendlines showing the effect of age on predicted learning, SIP, and verbal domain impairment probabilities, respectively. (A, B) are from the model in Supplementary Table S6. (C, D) are from the model in Supplementary Table S8. (E, F) are from the model in Supplementary Table S7. Plots are colored by HP2 status. Additional adjustment of the first 3 PCs was used in AFR + EUR analyses. “∗” indicates statistical significance.
From Figure 3B, similar to the global NCI risk, we also observed an intersection point of the trend lines between 50 and 60 years of age, after which the risk of learning impairment for EUR HP1/HP1 individuals became higher than HP2 individuals. This alteration was also found in the verbal and speed of information processing domains. In both EUR and AFR, HP is associated with some additional changes to cognitive domain T-scores that did not impact impairment (see Supplementary Tables S9, S10).
4 Discussion
We report that a functional CNV in the HP gene is associated with NCI in PLWH. We found that both AFR and EUR individuals with more HP2 alleles in our study sample had significantly lower HP levels. Despite the consistent direction of the relationship between CSF HP and HP2 alleles in these two populations, however, the HP CNV showed differing directions of association with NCI at baseline in AFR and EUR populations. At baseline, having one or more copies of the HP2 allele was protective in AFR, but detrimental in EUR participants. In longitudinal analyses, this effect became non-significant in AFR while it became more significant in EUR plus a significant over-time effect. Though not significant globally, in AFR, HP variation still impacted the speed of information processing cognitive domain. In EUR, the HP2 allele affected NCI through the learning and verbal cognitive domains. The consistent association between CSF HP protein levels and the HP2 variation in both the EUR and AFR groups provides additional support for our HP imputation process, and for some similarity in the basic biological impact of the alleles, even though their ultimate effects on cognitive impairment may differ.
Our finding of reduced cognitive decline among AFR individuals with HIV and HP2 alleles is consistent with findings from a cohort study of 466 HIV-negative African-American adults with type 2 diabetes, which found that HP1-1 individuals had poorer cognitive function at baseline and smaller cognitive decline over time compared to other HP genotypes adjusting for key demographic and cardiovascular risk factors (Beeri et al., 2018). Our finding of elevated risk of NCI in EUR also, to some extent, agrees with the previous findings in 653 Caucasian PLWH that individuals with HP2-2 had a higher mortality rate and reduced median survival compared to HP1-1 and HP1-2 (Delanghe et al., 1998). The disparate effects between AFR and EUR individuals noted in this study is not uncommon. The evolution of the HP alleles have progressed differently in these populations, which have been exposed to very different selection pressures and disease risks, as is described by Boettger et al. and others (Gichohi-Wainaina et al., 2016; Rametta et al., 2020). With regard to the HP alleles, divergent HP1-1 associations with cognitive function have been reported to differ between Ashkenazi Jews and non-Ashkenazi Jews with type 2 diabetes (Gichohi-Wainaina et al., 2016). Thus, the opposing effects in the AFR and EUR populations are likely due to the interactions of these alleles with other genetic, environmental, and socio-cultural factors that influence the complex clinical trajectory of NCI in PLWH.
Haptoglobin plays an intricate role in iron homeostasis and the inflammatory response within the CNS (Bulters et al., 2018) which likely influences a variety of body functions across the lifespan. HP in the CSF can protect neurons from being damaged by the products of red blood cell lysis including Hb and iron-mediated ROS. HP may also assist in the stable formation of β-amyloid (Aβ) and APOE complex, thus, helping the clearance of Aβ (Spagnuolo et al., 2014). Prior work has shown HP2 has less efficient antioxidative activity relative to HP1. Since HP also serves as an antioxidant for APOE, HP2 may lead to enhanced accumulation of Aβ and further the deterioration of the BBB (Montagne et al., 2020). Multiple biomarkers (such as Aβ42, tau, etc.) have been explored or used for clinical diagnosis of AD, and of these, S100β is a promising biomarker for BBB damage whose increase in serum indicates potential leakage of BBB (Marchi et al., 2003; Blennow and Zetterberg, 2018; Kadry et al., 2020). Future studies of these biomarkers in PLWH may reveal additional insights into the mechanism of HP CNV on NCI. On the other hand, HP2 may be more active in promoting tissue repair in chronic inflammatory conditions (Cid et al., 1993), which may have specific ramifications in the context of neuroinflammation in PLWH. Thus, over time, individuals with HP2 could display a symptomatic palliation of NCI.
As indicated by a wealth of neuroimaging and neurobehavioral studies, age is a strong risk factor for the development of neurocognitive decline and NCI among PLWH (Watkins and Treisman, 2015). In PLWH, accelerated aging was found and associated with HAND, a disorder with severe NCI (Levine et al., 2016). AD and Parkinson’s disease (PD) related pathological changes are also observed in ART-treated PLWH including elevated hyperphosphorylated tau protein in the hippocampus and Aβ deposition in the frontal cortex and hippocampus (Gannon et al., 2011). Thus, some researchers think HAND is associated with accelerated aging. Other researchers argue that HAND symptoms are induced by HIV infection and the use of antiretroviral therapy. Studies found that the HIV viral load is significantly higher in participants that develop NCI later yet age remains a risk modifier (Becker et al., 2004). Other mechanisms are also possible. However, given the current longevity of PLWH, we may not be able to fully understand the combined role of cART and HIV (Watkins and Treisman, 2015). Overall, NCI is a complex phenotype, and its relationship with HP is also complicated, and our associations show effects that are not easily delineated mechanistically.
To sum up on our findings, in addition to the existing effects of HIV viral load, HIV duration, and aging from previous publications (Ellis et al., 1997; Robertson et al., 1998; Becker et al., 2004; Simioni et al., 2010; Gannon et al., 2011; Watkins and Treisman, 2015; Olivier et al., 2018), the HP alleles are associated with NCI in PLWH, especially with an increased risk of NCI in EUR as well as a significantly alteration on the change of NCI risk from aging. Furthermore, it is unlikely that the effect of HP2 on NCI is exclusively mediated by the HIV RNA because: 1. the analyses restricted to virally suppressed individuals illustrated that HP has an independent effect on NCI, 2. the HP association on NCI is robust after adjustment of HIV RNA. Given the known interaction of HP and APOE, an interaction effect of HP and APOE alleles may also exist. However, the nature of the CHARTER study data limited our analyses. Several key variables were collected only within different subgroups and the number of samples that have two or more of those variables available is even smaller. Thus, due to limited testing of APOE genotype status in CHARTER Study participants we were unable to draw a clear conclusion involving APOE as the sample size drops substantially for APOE genotype stratified analyses. We also tried to impute the CHARTER APOE status. However, the two SNPs required to infer APOE genotypes were imputed with low r2 values: r2 = 0.664 using the Haplotype Reference Consortium (HRC) reference and r2 = 0.439 14 using the 1KG reference for rs429358, and r2 = 0.658 using HRC and r2 = 0.460 using 1KG for rs7412, and could not be used. Like the APOE, serum HP protein levels were also tested only within a limited subgroup that have HP hardcalls (n = 25 in AFR, and n = 48 in EUR), and we have limited power to assess the association between the HP genotype and serum HP protein levels. The overall genotyping call rate was
Code Availability
Code for the analyses of this study could be found at: https://github.com/bushlab-genomics/Haptoglobin-CHARTER.
Data Availability Statement
The data analyzed in this study is subject to the following licenses/restrictions: With regards to access to the data, the authors cannot make the data publicly available as they have obtained it from a third party, the CHARTER group. Requests to access these datasets should be directed to https://nntc.org/content/requests.
Ethics Statement
The studies involving human participants were reviewed and approved by the Institutional Review Boards of all participating institutions of the CHARTER study. The patients/participants provided their written informed consent to participate in this study. Instead, only de-identified data was used in the present analysis.
Author Contributions
HB designed study, conducted statistical analyses, interpreted data, etc. AK and HK assisted with interpretation of results and writing of manuscript. TH, RE, DF, and SL revised manuscript and provided expert opinions and interpretation for the CHARTER cohort.
Funding
This work was funded by the National Institute of Mental Health R01 MH107345 (Letendre, Heaton), R01 MH095621 (Hulgan, Kallianpur), R01 MH124530-01A1 (Kallianpur), and the National Institute on Aging R01 AG061351 (Below, Naj, Bush).
Conflict of Interest
SL has received support for research projects from Abbott, Merck, Tibotec, Schering-Plough, and GlaxoSmithKline and has received honoraria for speaking from Abbott, GlaxoSmithKline, and Tibotec. RE gave sponsored talks and received honoraria for serving on the scientific advisory board of GlaxoSmithKline.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We gratefully acknowledge the contributions of all participants in CHARTER Study. We also acknowledge the Genotype-Tissue Expression Project. The Genotype-Tissue Expression Project was supported by the Common Fund of the Office of the Director of the National Institutes of Health, and by NCI, NHGRI, NHLBI, NIDA, NIMH, and NINDS. The data used for the analyses described in this manuscript were obtained from the GTEx Portal on 03/08/2 018 and dbGaP accession number phs000424.vN.pN on 03/08/2 018.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2021.756685/full#supplementary-material
Abbreviations
Aβ, β-amyloid; AFR, African-descent; ANI, Asymptomatic neurocognitive impairment; APOE, apolipoprotein E; BBB, blood-brain barrier; AR1, first-order autoregressive; cART, Combination antiretroviral therapy; CNV, copy number variant; CNS, central nervous system; CSF, cerebrospinal fluid; CHARTER, CNS HIV antiretroviral therapy effects research; EUR, European-descent; GDS, global deficit score; GTEx, genotype-tissue expression; GEE, generalized estimating equations; HIV, human immunodeficiency virus; HP, haptoglobin; HRC, Haplotype reference consortium; HAND, HIV-associated neurocognitive disorders; HAD, HIV-associated dementia; LLQ, lower limit of quantitation; MND, mild neurocognitive disorder; NCI, neurocognitive impairment; OR, odds ratio; PLWH, people living with HIV; PC, principal components; QC, quality control; ROS, Reactive oxygen species; SIP, speed of information processing; 1KG, 1,000 Genomes.
References
Antinori, A., Arendt, G., Becker, J. T., Brew, B. J., Byrd, D. A., Cherner, M., et al. (2007). Updated Research Nosology for HIV-Associated Neurocognitive Disorders. Neurology. 69, 1789–1799. doi:10.1212/01.WNL.0000287431.88658.8b
Becker, J. T., Lopez, O. L., Dew, M. A., and Aizenstein, H. J. (2004). Prevalence of Cognitive Disorders Differs as a Function of Age in HIV Virus Infection. Aids. 18 (Suppl. 1), 11–18. doi:10.1097/00002030-200418001-00003
Beeri, M. S., Lin, H.-M., Sano, M., Ravona-Springer, R., Liu, X., Bendlin, B. B., et al. (2018). Association of the Haptoglobin Gene Polymorphism With Cognitive Function and Decline in Elderly African American Adults With Type 2 Diabetes. JAMA Netw. Open. 1, e184458–11. doi:10.1001/jamanetworkopen.2018.4458
Blackstone, K., Moore, D. J., Franklin, D. R., Clifford, D. B., Collier, A. C., Marra, C. M., et al. (2012). Defining Neurocognitive Impairment in HIV: Deficit Scores Versus Clinical Ratings. The Clin. neuropsychologist. 26, 894–908. doi:10.1080/13854046.2012.694479
Blennow, K., and Zetterberg, H. (2018). Biomarkers for Alzheimer's Disease: Current Status and Prospects for the Future. J. Intern. Med. 284, 643–663. doi:10.1111/joim.12816
Boettger, L. M., Salem, R. M., Handsaker, R. E., Peloso, G. M., Kathiresan, S., Hirschhorn, J. N., et al. (2016). Recurring Exon Deletions in the HP (Haptoglobin) Gene Contribute to Lower Blood Cholesterol Levels. Nat. Genet. 48, 359–366. doi:10.1038/ng.3510
Bulters, D., Gaastra, B., Zolnourian, A., Alexander, S., Ren, D., Blackburn, S. L., et al. (2018). Haemoglobin Scavenging in Intracranial Bleeding: Biology and Clinical Implications. Nat. Rev. Neurol. 14, 416–432. doi:10.1038/s41582-018-0020-0
Carey, C. L., Woods, S. P., Gonzalez, R., Conover, E., Marcotte, T. D., Grant, I., et al. (2004). Predictive Validity of Global Deficit Scores in Detecting Neuropsychological Impairment in HIV Infection. J. Clin. Exp. Neuropsychol. 26, 307–319. doi:10.1080/13803390490510031
Chang, C. C., Chow, C. C., Tellier, L. C. A. M., Vattikuti, S., Purcell, S. M., and Lee, J. J. (2015). Second-Generation PLINK: Rising to the Challenge of Larger and Richer Datasets. GigaScience 4 (1). doi:10.1186/s13742-015-0047-8
Cid, M. C., Grant, D. S., Hoffman, G. S., Auerbach, R., Fauci, A. S., and Kleinman, H. K. (1993). Identification of Haptoglobin as an Angiogenic Factor in Sera from Patients with Systemic Vasculitis. J. Clin. Invest. 91, 977–985. doi:10.1172/JCI116319
Delaneau, O., Zagury, J.-F., and Marchini, J. (2013). Improved Whole-Chromosome Phasing for Disease and Population Genetic Studies. Nat. Methods. 10, 5–6. doi:10.1038/nmeth.2307
Delanghe, J. R., Langlois, M. R., Boelaert, J. R., Van Acker, J., Van Wanzeele, F., van der Groen, G., et al. (1998). Haptoglobin Polymorphism, Iron Metabolism and Mortality in HIV Infection. Aids. 12, 1027–1032. doi:10.1097/00002030-199809000-00010
Ellis, R. J., Badiee, J., Vaida, F., Letendre, S., Heaton, R. K., Clifford, D., et al. (2011). CD4 Nadir Is a Predictor of HIV Neurocognitive Impairment in the Era of Combination Antiretroviral Therapy. Aids. 25, 1747–1751. doi:10.1097/QAD.0b013e32834a40cd
Ellis, R. J., Hsia, K., Spector, S. A., Nelson, J. A., Heaton, R. K., Wallace, M. R., et al. (1997). Cerebrospinal Fluid Human Immunodeficiency Virus Type 1 RNA Levels Are Elevated in Neurocognitively Impaired Individuals With Acquired Immunodeficiency Syndrome. Ann. Neurol. 42, 679–688. doi:10.1002/ana.410420503
Gannon, P., Khan, M. Z., and Kolson, D. L. (2011). Current Understanding of HIV-Associated Neurocognitive Disorders Pathogenesis. Curr. Opin. Neurol. 24, 275–283. doi:10.1097/WCO.0b013e32834695fb
Gichohi-Wainaina, W. N., Tanaka, T., Towers, G. W., Verhoef, H., Veenemans, J., Talsma, E. F., et al. (2016). Associations Between Common Variants in Iron-Related Genes With Haematological Traits in Populations of African Ancestry. PLoS ONE. 11, e0157996–17. doi:10.1371/journal.pone.0157996
Grant, I., Franklin, D. R., Deutsch, R., Woods, S. P., Vaida, F., Ellis, R. J., et al. (2014). Asymptomatic HIV-Associated Neurocognitive Impairment Increases Risk for Symptomatic Decline. Neurology. 82, 2055–2062. doi:10.1212/WNL.0000000000000492
Halekoh, U., Højsgaard, S., and Yan, J. (2006). TheRPackagegeepackfor Generalized Estimating Equations. J. Stat. Soft. 15, 1–11. doi:10.18637/jss.v015.i02
Heaton, R. K., Clifford, D. B., Franklin, D. R., Woods, S. P., Ake, C., Vaida, F., et al. (2010). HIV-associated Neurocognitive Disorders Persist in the Era of Potent Antiretroviral Therapy: CHARTER Study. Neurology. 75, 2087–2096. doi:10.1212/WNL.0b013e318200d727
Howie, B. N., Donnelly, P., and Marchini, J. (2009). A Flexible and Accurate Genotype Imputation Method for the Next Generation of Genome-Wide Association Studies. Plos Genet. 5, e1000529. doi:10.1371/journal.pgen.1000529
Jia, P., Zhao, Z., Hulgan, T., Bush, W. S., Samuels, D. C., Bloss, C. S., et al. (2017). Genome-Wide Association Study of HIV-Associated Neurocognitive Disorder (HAND): A CHARTER Group Study. Am. J. Med. Genet. 174, 413–426. doi:10.1002/ajmg.b.32530
Kadry, H., Noorani, B., and Cucullo, L. (2020). A Blood-Brain Barrier Overview on Structure, Function, Impairment, and Biomarkers of Integrity. Fluids Barriers CNS. 17, 1–24. doi:10.1186/s12987-020-00230-3
Kallianpur, A. R., Gittleman, H., Gittleman, H., Letendre, S., Ellis, R., Barnholtz-Sloan, J. S., et al. (2019). Cerebrospinal Fluid Ceruloplasmin, Haptoglobin, and Vascular Endothelial Growth Factor Are Associated With Neurocognitive Impairment in Adults With HIV Infection. Mol. Neurobiol. 56, 3808–3818. doi:10.1007/s12035-018-1329-9
Kallianpur, A. R., and Levine, A. J. (2014). Host Genetic Factors Predisposing to HIV-Associated Neurocognitive Disorder. Curr. Hiv/aids Rep. 11, 336–352. doi:10.1007/s11904-014-0222-z
Kallianpur, A. R., Wang, Q., Jia, P., Hulgan, T., Zhao, Z., Letendre, S. L., et al. (2016). Anemia and Red Blood Cell Indices Predict HIV-Associated Neurocognitive Impairment in the Highly Active Antiretroviral Therapy Era. J. Infect. Dis. 213, 1065–1073. doi:10.1093/infdis/jiv754
Levine, A. J., Quach, A., Moore, D. J., Achim, C. L., Soontornniyomkij, V., Masliah, E., et al. (2016). Accelerated Epigenetic Aging in Brain Is Associated With Pre-mortem HIV-Associated Neurocognitive Disorders. J. Neurovirol. 22, 366–375. doi:10.1007/s13365-015-0406-3
Lutgen, V., Narasipura, S. D., Barbian, H. J., Richards, M., Wallace, J., Razmpour, R., et al. (2020). HIV Infects Astrocytes In Vivo and Egresses From the Brain to the Periphery. Plos Pathog. 16, e1008381–32. doi:10.1371/journal.ppat.1008381
MacKellar, M., and Vigerust, D. J. (2016). Role of Haptoglobin in Health and Disease: A Focus on Diabetes. Clin. Diabetes. 34, 148–157. doi:10.2337/diaclin.34.3.148
Marchi, N., Fazio, V., Cucullo, L., Kight, K., Masaryk, T., Barnett, G., et al. (2003). Serum Transthyretin Monomer as a Possible Marker of Blood-To-CSF Barrier Disruption. J. Neurosci. 23, 1949–1955. doi:10.1523/jneurosci.23-05-01949.2003
Marchini, J., Howie, B., Myers, S., McVean, G., and Donnelly, P. (2007). A New Multipoint Method for Genome-Wide Association Studies by Imputation of Genotypes. Nat. Genet. 39, 906–913. doi:10.1038/ng2088
Melamed-Frank, M., Lache, O., Enav, B. I., Szafranek, T., Levy, N. S., Ricklis, R. M., et al. (2001). Structure-Function Analysis of the Antioxidant Properties of Haptoglobin. Blood. 98, 3693–3698. doi:10.1182/blood.v98.13.3693
Montagne, A., Nation, D. A., Sagare, A. P., Barisano, G., Sweeney, M. D., Chakhoyan, A., et al. (2020). APOE4 Leads to Blood-Brain Barrier Dysfunction Predicting Cognitive Decline. Nature. 581, 71–76. doi:10.1038/s41586-020-2247-3
Morgan, E. E., Woods, S. P., Woods, S. P., Letendre, S. L., Franklin, D. R., Bloss, C., et al. (2013). Apolipoprotein E4 Genotype Does Not Increase Risk of HIV-Associated Neurocognitive Disorders. J. Neurovirol. 19, 150–156. doi:10.1007/s13365-013-0152-3
Nightingale, S., Winston, A., Letendre, S., Michael, B. D., McArthur, J. C., Khoo, S., et al. (2014). Controversies in HIV-Associated Neurocognitive Disorders. The Lancet NeurologyNeurology. 13, 1139–1151. doi:10.1016/S1474-4422(14)70137-1
Olivier, I., Cacabelos, R., and Naidoo, V. (2018). Risk Factors and Pathogenesis of HIV-Associated Neurocognitive Disorder: The Role of Host Genetics. Int. J. Mol. Sci. 19, 3594. doi:10.3390/ijms19113594
Purcell, S., and Chang, C. (2019). PLINK 1.9. Available at: www.cog-genomics.org/plink/1.9/.
Rametta, R., Meroni, M., and Dongiovanni, P. (2020). From Environment to Genome and Back: A Lesson From HFE Mutations. Int. J. Mol. Sci. 21, 3505. doi:10.3390/ijms21103505
Ratanasopa, K., Chakane, S., Ilyas, M., Nantasenamat, C., and Bulow, L. (2013). Trapping of Human Hemoglobin by Haptoglobin: Molecular Mechanisms and Clinical Applications. Antioxid. Redox Signaling. 18, 2364–2374. doi:10.1089/ars.2012.4878
Robertson, K., Fiscus, S., Kapoor, C., Robertson, W., Schneider, G., Shepard, R., et al. (1998). CSF, Plasma Viral Load and HIV Associated Dementia. J. Neurovirol. 4, 90–94. doi:10.3109/13550289809113485
Rubin, L. H., and Maki, P. M. (2019). Neurocognitive Complications of HIV Infection in Women: Insights From the WIHS Cohort. Curr. Top. Behav. Neurosci., 175–191. (Epub ahead of print), August 9. doi:10.1007/7854_2019_101
Salvatore, A., Cigliano, L., Carlucci, A., Bucci, E. M., and Abrescia, P. (2009). Haptoglobin Binds Apolipoprotein e and Influences Cholesterol Esterification in the Cerebrospinal Fluid. J. Neurochem. 110, 255–263. doi:10.1111/j.1471-4159.2009.06121.x
Samuels, D. C., Kallianpur, A. R., Ellis, R. J., Bush, W. S., Letendre, S., Franklin, D., et al. (2016). European Mitochondrial DNA Haplogroups Are Associated With Cerebrospinal Fluid Biomarkers of Inflammation in HIV Infection. Pai. 1, 330–351. doi:10.20411/pai.v1i2.156
Saylor, D., Dickens, A. M., Sacktor, N., Haughey, N., Slusher, B., Pletnikov, M., et al. (2016). HIV-Associated Neurocognitive Disorder - Pathogenesis and Prospects for Treatment. Nat. Rev. Neurol. 12, 234–248. doi:10.1038/nrneurol.2016.27
Schaer, D. J., and Alayash, A. I. (2010). Clearance and Control Mechanisms of Hemoglobin From Cradle to Grave. Antioxid. Redox Signaling. 12, 181–184. doi:10.1089/ars.2009.2923
Simioni, S., Cavassini, M., Annoni, J.-M., Rimbault Abraham, A., Bourquin, I., Schiffer, V., et al. (2010). Cognitive Dysfunction in HIV Patients Despite Long-Standing Suppression of Viremia. AIDS (London, England). 24, 1243–1250. doi:10.1097/QAD.0b013e3283354a7b
Song, I.-U., Kim, Y.-D., Chung, S.-W., and Cho, H.-J. (2015). Association Between Serum Haptoglobin and the Pathogenesis of Alzheimer's Disease. Intern. Med. 54, 453–457. doi:10.2169/internalmedicine.54.2876
Spagnuolo, M. S., Maresca, B., La Marca, V., Carrizzo, A., Veronesi, C., Cupidi, C., et al. (2014). Haptoglobin Interacts With Apolipoprotein E and Beta-Amyloid and Influences Their Crosstalk. ACS Chem. Neurosci. 5, 837–847. doi:10.1021/cn500099f
Strazza, M., Pirrone, V., Wigdahl, B., and Nonnemacher, M. R. (2011). Breaking Down the Barrier: The Effects of HIV-1 on the Blood-Brain Barrier. Brain Res. 1399, 96–115. doi:10.1016/j.brainres.2011.05.015
Valdebenito, S., Castellano, P., Ajasin, D., and Eugenin, E. A. (2021). Astrocytes Are HIV Reservoirs in the Brain: A Cell Type With Poor HIV Infectivity and Replication but Efficient Cell‐to‐Cell Viral Transfer. J. Neurochem. 158, 429–443. doi:10.1111/jnc.15336
Watkins, C., and Treisman, G. (2015). Cognitive Impairment in Patients With Aids – Prevalence and Severity. Hiv. 7, 35–47. doi:10.2147/HIV.S39665
Woods, S. P., Rippeth, J. D., Frol, A. B., Levy, J. K., Ryan, E., Soukup, V. M., et al. (2004). Interrater Reliability of Clinical Ratings and Neurocognitive Diagnoses in HIV. J. Clin. Exp. Neuropsychol. 26, 759–778. doi:10.1080/13803390490509565
Yan, J., and Fine, J. (2004). Estimating Equations for Association Structures. Statist. Med. 23, 859–874. doi:10.1002/sim.1650
Yan, J. (2002). Geepack: Yet Another Package for Generalized Estimating Equations. R-News. 2/3, 12–14.
Yerbury, J. J., Kumita, J. R., Meehan, S., Dobson, C. M., and Wilson, M. R. (2009). α2-Macroglobulin and Haptoglobin Suppress Amyloid Formation by Interacting with Prefibrillar Protein Species. J. Biol. Chem. 284, 4246–4254. doi:10.1074/jbc.M807242200
Keywords: haptoglobin, neurocognitive impairment, HIV, CHARTER, longitudinal, age, HIV-associated neurocognitive disorder, genetic imputation
Citation: Bai H, Kaur H, Kallianpur AR, Hulgan T, Franklin DR, Letendre SL, Ellis RJ and Bush WS (2021) A Haptoglobin Exon Copy Number Variant Associates With HIV-Associated Neurocognitive Impairment in European and African-Descent Populations. Front. Genet. 12:756685. doi: 10.3389/fgene.2021.756685
Received: 10 August 2021; Accepted: 24 November 2021;
Published: 22 December 2021.
Edited by:
Brian Wigdahl, Drexel University, United StatesReviewed by:
Paul B. Higgins, Letterkenny Institute of Technology, IrelandEliseo A. Eugenin, University of Texas Medical Branch at Galveston, United States
Copyright © 2021 Bai, Kaur, Kallianpur, Hulgan, Franklin, Letendre, Ellis and Bush. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: William S. Bush, wsb36@case.edu
 Haimeng Bai
Haimeng Bai Harpreet Kaur
Harpreet Kaur Asha R. Kallianpur
Asha R. Kallianpur Todd Hulgan5
Todd Hulgan5 Scott L. Letendre
Scott L. Letendre William S. Bush
William S. Bush