- College of Animal Sciences and Technology, Henan Agricultural University, Zhengzhou, China
Introgression of genetic features from European pigs into Chinese pigs was reported possibly contributing to improvements in productivity traits, such as feed conversion efficiency and body size. However, the genomic differences from European pigs and the potential role of introgression in Henan indigenous pigs remains unclear. In this study, we found significant introgression from European pigs into the genome of Chinese indigenous pigs, especially in Henan indigenous pigs. The introgression in Henan indigenous pigs, particularly in the Nanyang black pig, was mainly derived from Duroc pigs. Most importantly, we found that the NR6A1, GPD2, and CSRNP3 genes were introgressed and reshaped by artificial selection, and these may have contributed to increases in pig body size and feed conversion efficiency. Our results suggest that human-mediated introgression and selection have reshaped the genome of Henan pigs and improved several of their desired traits. These findings contribute to our understanding of the history of Henan indigenous pigs and provide insights into the genetic mechanisms affecting economically important traits in pig populations.
Introduction
Livestock animals were domesticated from wild ancestors for human use, subsequently undergoing contiguous breeding and improvement through the generation of breeds with distinct phenotypes. Crossbreeding between breeds or subspecies is often conducted during the breeding process to improve livestock's productivity or desired appearance (Bosse et al., 2014). Examples of this selective breeding includes yellow skin, penciled feathers, and spotted comb in domesticated chickens resulting from introgressions from Gallus sonneratii, Gallus varius, and Gallus lafayettii (Morejohn, 1968; Eriksson et al., 2008; Fallahshahroudi et al., 2019). Human-mediated introgression of alleles plays an important role in altering the genome of domesticated animals, including pigs (Bosse et al., 2014).
Divergence between Asian wild and European wild pigs can been traced to approximately half a million years ago, prior to independent domestication (~10,000 years ago) (Giuffra et al., 2000; Larson et al., 2005; Groenen et al., 2012). Subsequently, many domesticated pig populations were formed, each with distinct phenotypes. Since then, hybridization and introgression have reshaped the modern pig genome (Giuffra et al., 2000; Goedbloed et al., 2013). According to historical records, the Chinese pig was imported into Europe in the mid-to-late 18th century (Chen et al., 2020). This introgression was likely to have played an important role in improving the reproductive traits of the Large White (LW) pig (Chen et al., 2020). Moreover, Chinese pigs were imported into America to improve the production performance of American indigenous pigs through crossbreeding, and this accounts for about 33% of the genome on average (Fang and Andersson, 2006). Several studies based on genetic data have revealed evidence for introgression between sub-populations (Yang et al., 2009, 2014; Jiang et al., 2012; Wang et al., 2020). However, the genomic differences and functions introduced by introgression are still not well understood. Henan is a central province of China with a rich diversity of indigenous pig breeds. Three indigenous pig breeds, the Nanyang Black pig (NY), the Huainan pig (HN), and the Queshan black pig (QS), have been named based on the classification scheme from the National Commission of Animal Genetics resource book (2011). These three pig breeds exhibited strong disease-resistance, high meat yield and early maturation (Qiao et al., 2019). Low density SNP analysis revealed that there has been admixture between Henan indigenous pigs and European commercial pigs (Qiao et al., 2019). However, the genomic differences and potential role of introgression in Henan indigenous pigs is still unclear.
Here, whole genome data of Henan indigenous pigs were generated and analyzed together with data for European pigs and other Chinese indigenous pigs. Furthermore, admixture and population structures were investigated. Subsequently, introgression from European pigs into Henan indigenous pigs was determined by multiple statistical analyses, and the proportion of introgression was estimated. Most importantly, common introgressed regions and selection regions were identified, which were possibly associated with high feed conversion efficiency and larger body size.
Results and Discussion
Whole-Genome Resequencing of Henan Indigenous Pigs
A set of whole-genome sequencing data comprising 102 individuals was collected, from nine Chinese indigenous pig breeds (CD), four European commercial pig breeds (EUD), one Chinese wild pig population (CW), one European wild pig population (EUW), and one Java warty pig (Sus verrucosus, an outgroup) (Supplementary Table 1). Of these, we sequenced 30 pigs from three Henan indigenous pigs (CDh), generating whole-genome data with an average ~15x sequencing depth. Approximately 66.3 million autosomal biallelic SNPs were identified. After filtering minor allelic frequencies <0.01 and call rates >0.9, about 29.9 million autosomal biallelic SNPs were ultimately retained and used in subsequent analyses.
Inference of Population Structure
Divergence between Asian wild and European wild pigs was traced to approximately half a million years prior to subsequent independent domestication (~10,000 years ago) (Giuffra et al., 2000; Larson et al., 2005). Evidence from principal component analysis (PCA) revealed obvious differentiation between Chinese pigs and European pigs, representing the genetic differences between these sub-species (Figure 1A). The first principal component (PC1) captured 17.6% of the total eigenvalue, dividing individuals into two clades (European pigs and Chinese pigs). The second principal component (PC2) captured 7.57% of the total eigenvalue, dividing Chinese pigs into Southwestern indigenous pigs (the Tibetan, TT, and Wuzhishan pigs, WZS), Chinese wild pigs, and other indigenous pigs. Interestingly, we found that a cluster of three Henan indigenous pig (CDh) breeds showed a closer relatedness to European pigs (Figure 1A). When rooting using the Java warty pig (Sus verrucosus), a phylogenetic tree based on a GTR model demonstrated that different subgroups formed obvious clades (Figure 1B). Interestingly, we also observed that Henan indigenous pigs (CDh) clustered with European pigs. These results imply admixture between Henan indigenous pigs (CDh) and European pigs. According to the methods found in Evanno et al. 2005, the best estimate for the number of clusters (K-value) was equal to 2 (Figure 1C). When K = 2 in the admixture graph, two ancestral lineages separated Chinese pigs and European pigs from the population in which admixture occurred (Figure 1C). Obviously, CDh and two CD pigs (HTDE and MIN) showed significant admixture with a European lineage, especially in Nanyang pigs (NY). Collectively, these findings revealed the clear signature of admixture of Chinese indigenous pigs with European pigs, especially in Henan indigenous pigs (CDh). This result was also consistent with previous evidence of European pig introgression with MIN pigs (MIN), Laiwu black pigs (LWH), Hetao pigs (HTDE), and Tibetan pigs (TT) (Chen et al., 2020; Wang et al., 2020).
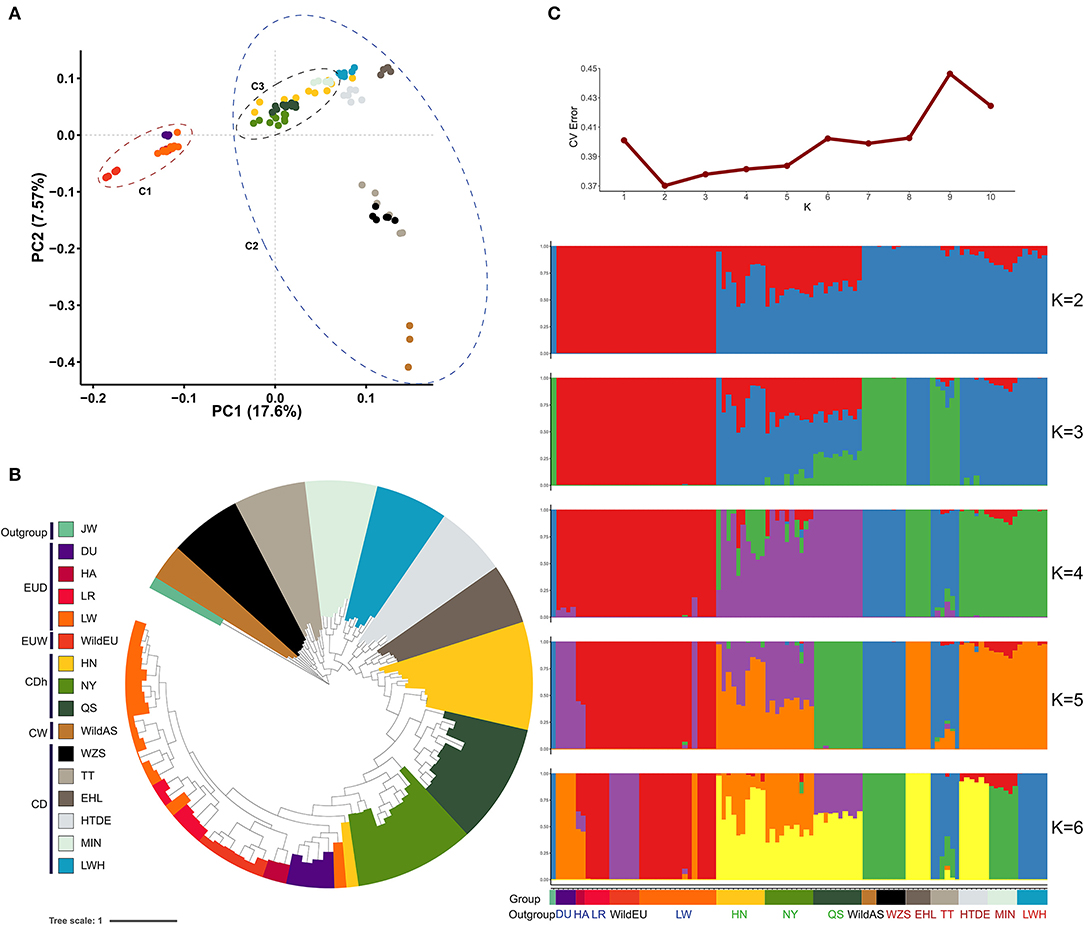
Figure 1. Population structure and relationships. (A) Principal component analysis of Chinese and European pigs. C1, European pigs; C2, Chinese pigs; C3, Henan indigenous pigs. (B) Phylogenetic tree of Chinese and European pigs based on the GTR model. Group colors used in (A) are the same as (B). (C) Population structure of Chinese and European pigs revealed by admixture analysis. JW, Java warty pig; DU, Duroc pig; HA, Hampshire pig; LR, Landrace pig; LW, Large White pig; WildEU, European Wild pig; HN, Huainan pig; NY, Nanyang pig; QS, Queshan pig; WildAS, Chinese wild pig; WZS, Wuzhishan pig; TT, Tibetan pig; EHL, Erhualian pig; HTDE, Hetao daer pig; MIN, Min pig; and LWH, Laiwuhei pig. All pigs were also classified as Chinese indigenous pig breeds (CD), European commercial pig breeds (EUD), Chinese wild pig populations (CW), European wild pigs (EUW), or Java warty pigs (Sus verrucosus, the outgroup).
Gene Flow of Duroc Pigs to Henan Indigenous Pigs
To obtain direct introgression evidence, f4 statistics and D-statistics were calculated for each combination of European and Chinese indigenous pigs. As expected, we found evident gene flow of European pigs in seven Chinese indigenous pig breeds according to f4 statistics, with the exceptions of WZS and EHL pigs (Figure 2A, Supplementary Figure 1). The order of introgression intensity from high to low was NY > QS > HN > MIN > HTDE > LWH > TT. Evidence from D-statistics showed the same pattern as f4 statistics did. We also calculated f3 statistics (CD; European pig, CW) to validate the admixture of Chinese indigenous pigs from European pigs and Chinese wild pigs. We only observed significant admixture for NY pigs, whereas HN and QS pigs were close to the threshold value (Figure 2, Supplementary Figure 1). This implied that f3 statistics (CD, CW, European pigs) were insensitive to introgression at low proportions. To identify which breed of European pig was responsible for the introgression detected, the outgroup-f3 statistics were explored. The Duroc (DU) pig was more genetically similar to the NY, QS, and HN pigs than to European pigs, while LW pigs were closer to other Chinese indigenous pigs. This result might be explained by historical evidence indicating that Chinese pigs were introduced into European breeds to improve the fertility and immunity of LW pigs (Chen et al., 2020). Alternatively, introgression of LW into some Chinese indigenous pigs could not be ruled out.
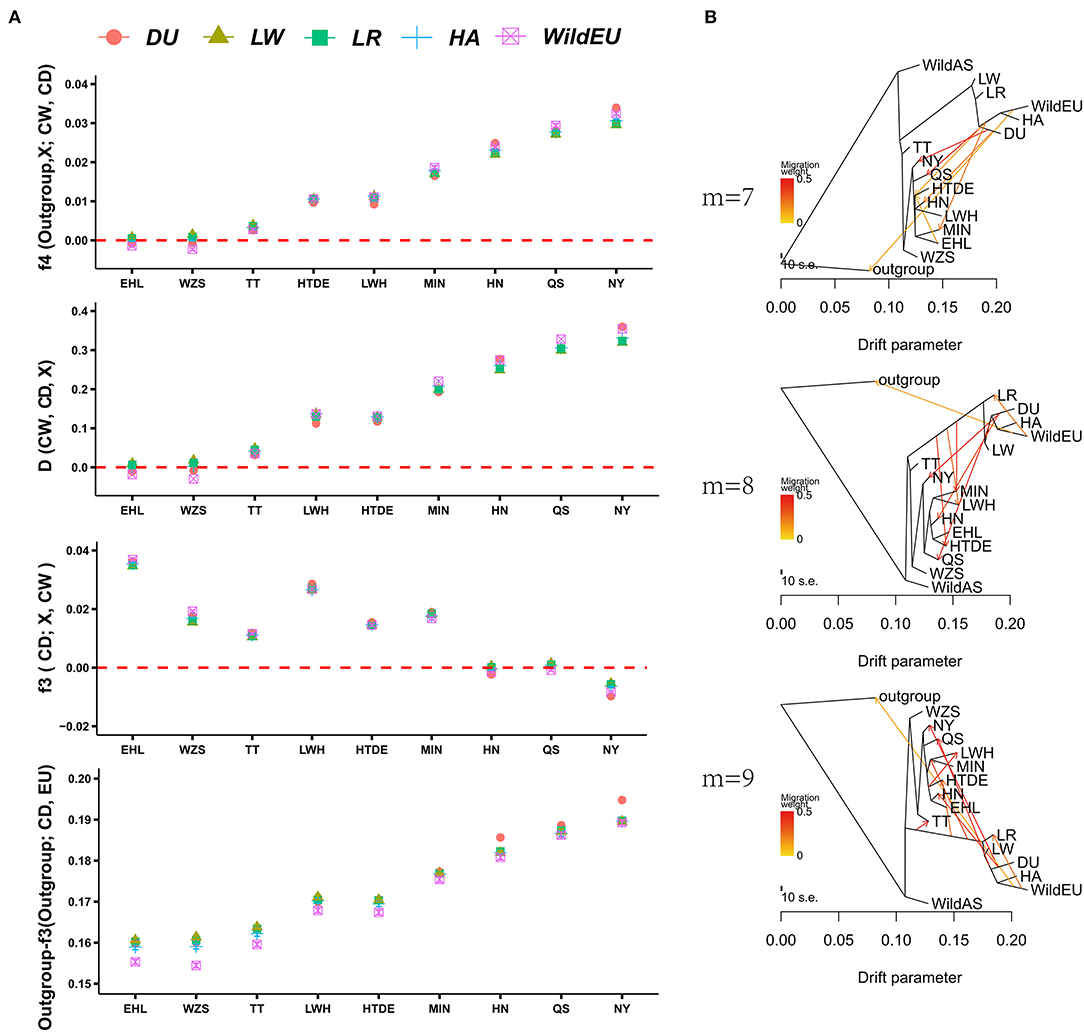
Figure 2. Admixture of Chinese indigenous pigs and European pigs. (A) Evidence from f4 statistics, D-statistics, f3 statistics, and outgroup-f3 statistics showing admixture of Chinese and European pigs. (B) TreeMix analysis revealing gene flow from European to Chinese pigs.
To obtain more detailed introgression evidence, TreeMix analysis was conducted. The TreeMix analysis presented the same results as above, i.e., that admixture and division between European and Chinese pigs had occurred (Figure 2B). Increasing the number of migration events marginally enhanced the fitness of the model testing. When m = 7, the model explained 0.9936 of the variance for the autosomal SNP data, while when m = 8 or 9, the model explained 0.9957 or 0.9963 of the variance, respectively. Evidence from TreeMix also indicated significant gene flow from DU into CDh (HN, QS, and NY), consistent with the results of our outgroup-f3 analysis. We also observed weak introgression from DU to MIN, HTDE, TT, and LWH, which was consistent with the previous introgression results based on SNP chip data (Wang et al., 2020).
Identification of Introgression and Selective Regions in Henan Indigenous Pigs
To locate the detailed genomic regions for the gene flow from DU to CDh, we calculated the f-statistic (f_d) value for each combination of DU and CDh (HN, QS, and NY) by the sliding-window method. After Z-transformation, the top 5% of the windows were considered to be outlier introgression regions (Figure 3, Supplementary Table 2). Totals of 506, 487, and 522 regions passed this threshold (Supplementary Table 2). Combining the overlapping regions, these identified introgression regions accounted for 12.53% of NY, 10.61% of HN, and 9.43% of the QS genome. The overlapping regions of CDh were located on three chromosomes, comprised of chr1:262,974,058-267,766,091, chr9:123,030,904-123,611,236, and chr15:61,503,802-74,093,823. Genes located in these overlapped regions were retrieved from the Ensemble database and are shown in Supplementary Table 3. Of these, the NR6A1, CSRNP3, and GPD2 genes were considered to be putative important introgression genes (Figure 3).
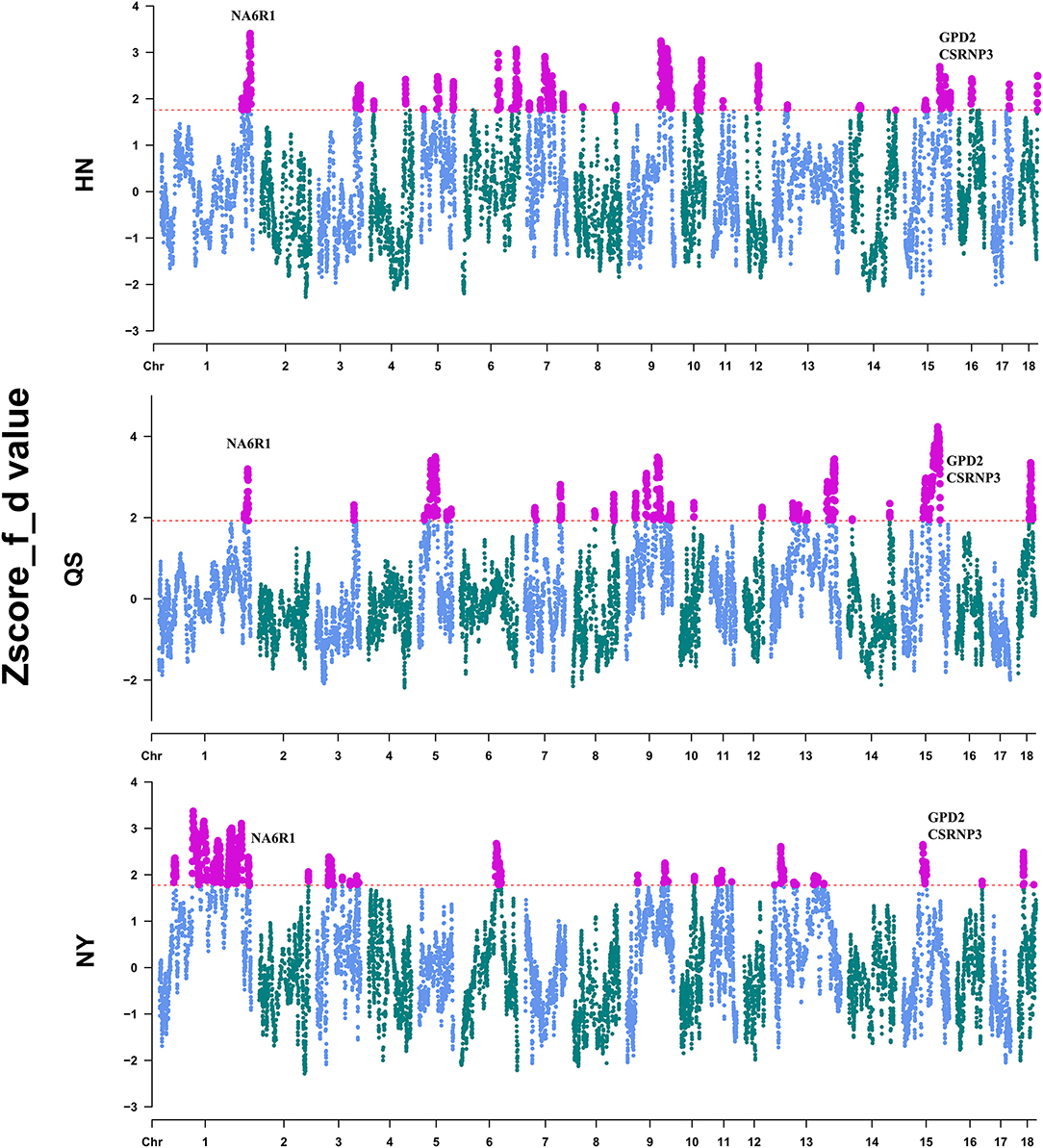
Figure 3. Identification of the introgression region of Duroc pigs to Henan indigenous pigs. The red dashed line shows the 5% cutoff used to define the introgression region.
To assess the importance of domestication and post-domestication selection on Henan indigenous pigs in the face of gene flow, we conducted a whole-genome scan for selective signatures via computing the XP-CLR of each CDh relative to Chinese wild boars (Supplementary Figure 2). A total of 342 selective regions were commonly identified in these three CDh (Supplementary Table 4). Genes located in the selective regions are shown in Supplementary Table 5. Two of three introgression regions were identified as being under selection and were divided into seven small genome regions: (chr1: 264,350,001–264,500,000, chr1: 265,300,001–265,600,000, chr1: 266,150,001–266,250,000, chr15: 63,100,001–63,250,000, chr15: 63,700,001–63,800,000, chr15: 64,600,001–64,700,000, and chr15: 72,100,001–72,200,000). A total of 10 coding genes and 2 non-coding genes were included, in these regions, consisting of NR6A1, NR5A1, MAPKAP1, OLFML2A, DENND1A, CSRNP3, GPD2, ENSSSCG00000042870, ENSSSCG00000047320, ENSSSCG00000043788, ssc-miR-181a-2, and ssc-miR-181b-2.
Introgression and Selection Reshaped the NR6A1 Locus of Henan Indigenous Pigs
The NR6A1 gene, nuclear receptor subfamily six group A member 1, encodes an orphan nuclear receptor. It has been widely reported to have been under selection during domestication and was shown to be associated with the number of vertebrae (Ribani et al., 2019; Zhang et al., 2019, 2020). A variant allele of p.Pro192Leu of NR6A1 was correlated with an increase in the number of pig thoracic and dorsal vertebrae, from an average of 19 to 21–23 (Mikawa et al., 2007). An increased number of vertebrae plays a positive role in increasing body length and overall meat production (Burgos et al., 2015). Variation in the NR6A1 gene is under selection and nearly fixed in European commercial pigs relative to European wild boars (Rubin et al., 2012). This implies the positive effect of the NR6A1 gene was associated with this human desired production trait. In this study, we found that a region encompassing the NR6A1 gene was significantly introgressed in Henan indigenous pigs from Duroc pigs based on f_d values (Figure 3). Moreover, we observed a remarkably high XP-CLR value in CDh pigs relative to CD pigs (Figure 4). Results from genotyping of European pigs and Chinese pigs for SNPs in the NR6A1 region also showed that CDh pigs showed significant differences from Chinese pigs and greater similarity to EUD pigs. This region was reported to be fixed in European commercial pigs relative to European wild pigs and Chinese pigs represented by Meishan pigs (Rubin et al., 2012). Additionally, the NR6A1 locus in European pigs was also reported to have introgressed into Chinese Licha pigs and has been associated with increasing vertebral number (Yang et al., 2009). Therefore, it can be inferred that this region was first introgressed into Henan indigenous pigs from European commercial pigs and was then subjected to conscious or unconscious artificial selection, since the NR6A1 gene functions in increasing body length and meat production that are human-desired traits.
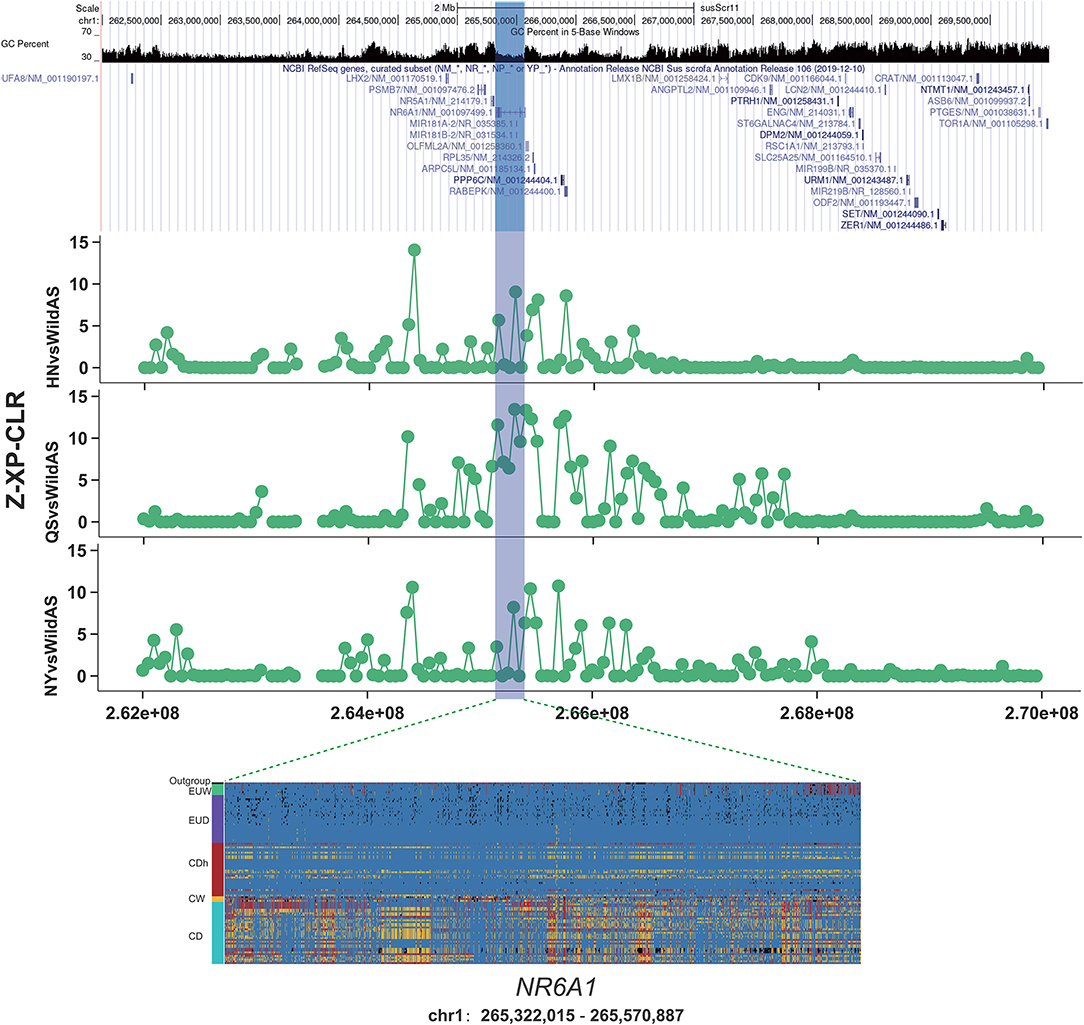
Figure 4. Introgression at the NR6A1 locus. XP-CLR showed a significant selection signature located in the NR6A1 gene in Henan indigenous pigs compared to Chinese wild pigs. Results from genotyping a diverse panel of European and Chinese pigs for SNPs in the putative selective sweeps containing NR6A1.
Introgression and Selection Reshaped the GPD2 and CSRNP3 Loci of Henan Indigenous Pigs
European commercial pigs demonstrated significantly better production performance than Chinese indigenous pigs in traits such as higher feed conversion efficiency and larger body size. In this study, we found that the loci GPD2 and CSRNP3 were introgressed from Duroc pigs into three Henan indigenous pig breeds (Figure 3). Moreover, these regions presented significant selection signals in Henan indigenous pig breeds relative to Chinese wild pigs (Figure 5). GPD2, encoding glycerol-3-phosphate dehydrogenase, catalyzes the unidirectional conversion of glycerol-3-phosphate to dihydroxyacetone phosphate and eventually to glycolysis (Gerbitz et al., 1996). The GPD2 gene was shown to be significantly associated with residual feed intake traits, and this locus was significantly down-regulated in individuals with lower residual feed intake relative to those with higher residual feed intake (Lkhagvadorj et al., 2010). CSRNP3 encodes a transcription factor, cysteine and serine rich nuclear protein 3, and the expression level of CSRNP3 was associated with pig residual feed intake and feed conversion ratio traits (Vincent et al., 2015; Messad et al., 2019). In cattle, SNPs located in CSRNP3 were significantly associated with chest width, hip width, or rump width (Doyle et al., 2020). We also compared these genotypes between European and Chinese pigs, and these showed obvious similarity to European commercial pigs (EUD) compared to Chinese wild pigs and other Chinese indigenous pigs (Figure 5). As is well-known, European commercial pigs have a higher feed conversion efficiency and a larger body size than Chinese native pigs. Functions of the CSRNP3 and GPD2 genes in feed conversion efficiency and body size may have driven the alleles from European commercial pigs to near fixation after introgression into Henan indigenous pigs under human subjective selection.
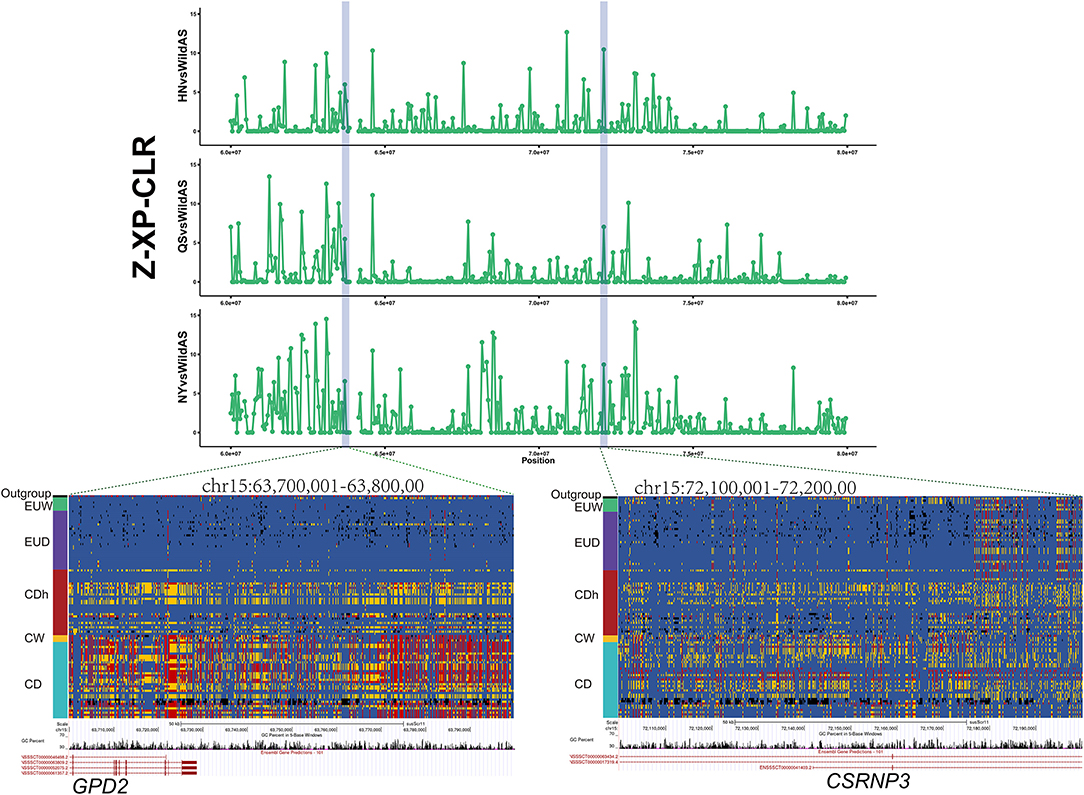
Figure 5. Introgression at the GPD2 and CSRNP3 loci. XP-CLR showed significant selection signatures located in the GPD2 and CSRNP3 genes in Henan indigenous pigs compared to Chinese wild pigs. Results from genotyping a diverse panel of European and Chinese pigs for SNPs in the putative selective sweeps containing GPD2 or CSRNP3.
Conclusions
Collectively, we found significant introgression from European pigs into Chinese indigenous pigs, especially in Henan indigenous pigs. The introgression derived from Duroc pigs was captured for Henan indigenous pigs, especially in Nanyang black pigs. Most importantly, we found that the NR6A1, GPD2, and CSRNP3 genes were introgressed and selected factors that may contribute to pig body size and feed conversion ratio traits. This suggests that human-mediated introgression and selection could have reshaped the pig genome and improved several desired traits. These findings further deepen our understanding of the history of Henan indigenous pigs and provide insights into the genetic mechanisms affecting economically important pig traits.
Materials and Methods
Ethics Statement
Ear tissues of three indigenous pigs from Henan province were collected in strict accordance with protocols approved by the Institutional Animal Care and Use Committee (IACUC) at Henan Agricultural University.
Sample Collection
Genome resequencing data for 102 pigs was collected in this study, of which 30 individuals were derived from three Henan indigenous breeds and 10 Yorkshire pigs were generated in our previous study (Li et al., 2020). We downloaded 68 accessions chosen from recent studies from the National Center for Biotechnology Information (NCBI) Sequence Read Archive database, which included from 13 breeds (Supplementary Table 1). Multiple statistics were used to improve the accuracy of the results in this study, but we could not rule out sampling error caused by differences in sample size.
Whole-Genome Resequencing and SNP Calling
The procedures for DNA extraction and library construction were described in our previous studies (Li et al., 2020; Shang et al., 2020). Paired end reads were generated using the BGISEQ-500 platform (BGI Genomics Co., Ltd.). Raw reads were filtered using the Trimmomatic software (seed mismatches: palindrome: simple clip −2:30:10) (Bolger et al., 2014). Filtered reads were aligned to a reference genome (Sscrofa v11.1) using the Burrows–Wheeler alignment (BWA) tool with default parameters (Li and Durbin, 2009). SAM files were merged and sorted using SAMtools (Li et al., 2009). Subsequently, duplicate reads were marked using the MarkDuplicates function of Picard Tools (http://broadinstitute.github.io/picard/). SNP calling for each individual was implemented using the HaplotypeCaller program of the GATK4 software and further joint calling using the GenomicsDBImport and GenotypeGVCFs programs of GATK4 (v4.1.2.0) (McKenna et al., 2010). Finally, we filtered our SNP panel using the Variant Filtration program of GATK4 with the parameters “QD<2.0 || FS > 200.0 || SOR > 10.0 || MQRankSum < –12.5 || ReadPosRankSum < –8.0.”
Population Structure Analysis
Biallelic SNPs located on autosomes were identified and retained using the VCFtools software (0.1.16) (http://vcftools.github.io/) (Danecek et al., 2011). Subsequently, variants with a call rate >90% and MAF >0.01 were identified and retained (http://www.cog-genomics.org/plink2/) using PLINK (v1.9) (Chang et al., 2015). Principal Component Analysis (PCA) was also performed using PLINK. Scatter distributions for each individual were plotted using the first principal component. Population assignment analysis was conducted using the Admixture software (Kalyaanamoorthy et al., 2017), and the best number of clusters (K) was determined by the method from Evanno et al. (2005). A phylogenetic tree was constructed using the IQ-TREE software (v 1.6.12) with 1,000 bootstrap replicates based on the GTR model (Kalyaanamoorthy et al., 2017) and visualized using the iTOL online web server with a root at the outgroup (Letunic and Bork, 2019). The Java warty pig (Sus verrucosus) was defined as the outgroup for these analyses.
Whole-Genome Analysis of Genomic Introgression
To investigate the admixture and splitting of domesticated pigs, the separation and admixture were inferred using the TreeMix software (v. 1.13) (Pickrell and Pritchard, 2012). The threepop and fourpop programs of TreeMix were used to identify the introgression from European pigs to Chinese pigs. The f4 (outgroup, EUD; CW, and CD) statistics were calculated to measure the shared drift between European and CD (Chinese domestic pigs) based on allelic frequency (Patterson et al., 2012). For the f4 statistics, positive values indicated gene flow between European pigs and CD pigs under the hypothesis that there was no admixture of the outgroup into any group. The Java warty pig (Sus verrucosus) was defined as the outgroup in this study. The outgroup-f3 (outgroup; CW, EUD) statistics measured the branch lengths from the outgroup to the common ancestor of Chinese domestic pigs and European pigs, and higher values indicated greater genetic similarity. Another f3 statistic, f3 (CD; CW, ED), was computed to identify the admixture of CD with CW and EUD pigs. The pattern's D-statistic was also computed using the Dsuite software (Patterson et al., 2012). The f_d statistics for each window were also computed to investigate the introgression region with sliding windows containing 10,000 SNP and 1,000 SNP steps. A Z-transformation of f_d statistics was treated using the “scale” function in R, of which the top 5% were defined as significant introgression regions. Genes located within the introgression regions were retrieved using the Biomart program of the ENSEMBLE web server.
Signatures of Selection
Selection signatures across the whole genome were scanned in a comparison between Henan indigenous pigs and Chinese wild pigs. The XP-CLR (cross-population composite likelihood ratio) was calculated using a 100-kb sliding window with a step size of 50 kb (https://github.com/hardingnj/xpclr). Usually, a genetic distance of one cM is empirically equal to one Mb of physical distance. The regions with top 5% normalized xp-clr values were classified as significantly selected between each comparison. Genes located within the selective regions were retrieved using the Biomart program of the ENSEMBLE web server.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://bigd.big.ac.cn, CRA004121.
Author Contributions
KW, XinL, and XH participated in the design of the study and obtained project funding. KW, XinL, XH, LZ, DD, RQ, and XiuL collected the samples. KW carried out the analysis, interpretation of data, and initiated and drafted and revised the manuscript. XinL and XH revised the manuscript. All of the authors have read and approved the final manuscript.
Funding
This work was supported by the Pig Industry Technology System Innovation Team Project of Henan Province (S2012-06-G03), the National Natural Science Foundation of China (32002142), the Grand Science and Technology Special Project in Tibet (2021), and Henan Key Research & Development Program (Grant Nos. 192102110067, 212102110003, and 202102110242).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank LetPub (www.letpub.com) for its linguistic assistance during the preparation of this manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2021.705803/full#supplementary-material
Supplementary Figure 1. Z-transformation of f4 (outgroup, X; CW, CD) and f3 (outgroup; CW, CD). X indicates one of the European pig breeds.
Supplementary Figure 2. Z-XP-CLR values calculated for each combination between Henan indigenous pigs (CDh) and Chinese wild pigs (CW). The red dashed lines represent the 5% cutoff values used to define selective signatures.
Supplementary Table 1. Sample information used in this study.
Supplementary Table 2. Introgression genomic regions in Henan indigenous pigs from Duroc pigs.
Supplementary Table 3. Genes located in the common regions of introgression from Duroc to Henan indigenous pigs.
Supplementary Table 4. Common significantly selective regions of Henan indigenous pigs.
Supplementary Table 5. Genes located in the common selective regions of Henan indigenous pigs.
References
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Bosse, M., Megens, H. J., Frantz, L. A., Madsen, O., Larson, G., Paudel, Y., et al. (2014). Genomic analysis reveals selection for Asian genes in European pigs following human-mediated introgression. Nat. Commun. 5:4392. doi: 10.1038/ncomms5392
Burgos, C., Latorre, P., Altarriba, J., Carrodeguas, J. A., Varona, L., and Lopez-Buesa, P. (2015). Allelic frequencies of NR6A1 and VRTN, two genes that affect vertebrae number in diverse pig breeds: a study of the effects of the VRTN insertion on phenotypic traits of a Duroc × Landrace–Large White cross. Meat. Sci. 100, 150–155. doi: 10.1016/j.meatsci.2014.09.143
Chang, C. C., Chow, C. C., Tellier, L. C., Vattikuti, S., Purcell, S. M., and Lee, J. J. (2015). Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience 4:7. doi: 10.1186/s13742-015-0047-8
Chen, H., Huang, M., Yang, B., Wu, Z., Deng, Z., Hou, Y., et al. (2020). Introgression of Eastern Chinese and Southern Chinese haplotypes contributes to the improvement of fertility and immunity in European modern pigs. Gigascience 9:giaa014. doi: 10.1093/gigascience/giaa014
Danecek, P., Auton, A., Abecasis, G., Albers, C. A., Banks, E., DePristo, M. A., et al. (2011). The variant call format and VCFtools. Bioinformatics 27, 2156–2158. doi: 10.1093/bioinformatics/btr330
Doyle, J. L., Berry, D. P., Veerkamp, R. F., Carthy, T. R., Walsh, S. W., Evans, R. D., et al. (2020). Genomic regions associated with skeletal type traits in beef and dairy cattle are common to regions associated with carcass traits, feed intake and calving difficulty. Front. Genet. 11:20. doi: 10.3389/fgene.2020.00020
Eriksson, J., Larson, G., Gunnarsson, U., Bed'hom, B., Tixier-Boichard, M., Stromstedt, L., et al. (2008). Identification of the yellow skin gene reveals a hybrid origin of the domestic chicken. PLoS Genet. 4:e1000010. doi: 10.1371/journal.pgen.1000010
Evanno, G., Regnaut, S., and Goudet, J. (2005). Detecting the number of clusters of individuals using the software structure: a simulation study. Mol. Ecol. 14, 2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x
Fallahshahroudi, A., Sorato, E., Altimiras, J., and Jensen, P. (2019). The domestic BCO2 allele buffers low-carotenoid diets in chickens: possible fitness increase through species hybridization. Genetics 212, 1445–1452. doi: 10.1534/genetics.119.302258
Fang, M., and Andersson, L. (2006). Mitochondrial diversity in European and Chinese pigs is consistent with population expansions that occurred prior to domestication. Proc. Biol. Sci. 273, 1803–1810. doi: 10.1098/rspb.2006.3514
Gerbitz, K. D., Gempel, K., and Brdiczka, D. (1996). Mitochondria and diabetes. genetic, biochemical, and clinical implications of the cellular energy circuit. Diabetes 45, 113–126. doi: 10.2337/diabetes.45.2.113
Giuffra, E., Kijas, J. M., Amarger, V., Carlborg, O., Jeon, J. T., and Andersson, L. (2000). The origin of the domestic pig: independent domestication and subsequent introgression. Genetics 154, 1785–1791. doi: 10.1093/genetics/154.4.1785
Goedbloed, D. J., Megens, H. J., Van Hooft, P., Herrero-Medrano, J. M., Lutz, W., Alexandri, P., et al. (2013). Genome-wide single nucleotide polymorphism analysis reveals recent genetic introgression from domestic pigs into Northwest European wild boar populations. Mol. Ecol. 22, 856–866. doi: 10.1111/j.1365-294X.2012.05670.x
Groenen, M. A., Archibald, A. L., Uenishi, H., Tuggle, C. K., Takeuchi, Y., Rothschild, M. F., et al. (2012). Analyses of pig genomes provide insight into porcine demography and evolution. Nature 491, 393–398. doi: 10.1038/nature11622
Jiang, Y. Z., Zhu, L., Tang, G. Q., Li, M. Z., Jiang, A. A., Cen, W. M., et al. (2012). Carcass and meat quality traits of four commercial pig crossbreeds in China. Genet. Mol. Res. 11, 4447–4455. doi: 10.4238/2012.September.19.6
Kalyaanamoorthy, S., Minh, B. Q., Wong, T. K. F., von Haeseler, A., and Jermiin, L. S. (2017). Model finder: fast model selection for accurate phylogenetic estimates. Nat. Methods 14, 587–589. doi: 10.1038/nmeth.4285
Larson, G., Dobney, K., Albarella, U., Fang, M., Matisoo-Smith, E., Robins, J., et al. (2005). Worldwide phylogeography of wild boar reveals multiple centers of pig domestication. Sci. 307, 1618–1621. doi: 10.1126/science.1106927
Letunic, I., and Bork, P. (2019). Interactive tree of life (iTOL) v4: recent updates and new developments. Nucleic Acids Res. 47, W256–W259. doi: 10.1093/nar/gkz239
Li, H., and Durbin, R. (2009). Fast and accurate short read alignment with burrows-wheeler transform. Bioinformatics 25, 1754–1760. doi: 10.1093/bioinformatics/btp324
Li, H., Handsaker, B., Wysoker, A., Fennell, T., Ruan, J., Homer, N., et al. (2009). The sequence alignment/map format and SAM tools. Bioinformatics 25, 2078–2079. doi: 10.1093/bioinformatics/btp352
Li, X., Ye, J., Han, X., Qiao, R., Li, X., Lv, G., et al. (2020). Whole-genome sequencing identifies potential candidate genes for reproductive traits in pigs. Genomics 112, 199–206. doi: 10.1016/j.ygeno.2019.01.014
Lkhagvadorj, S., Qu, L., Cai, W., Couture, O. P., Barb, C. R., Hausman, G. J., et al. (2010). Gene expression profiling of the short-term adaptive response to acute caloric restriction in liver and adipose tissues of pigs differing in feed efficiency. Am. J. Physiol. Regul. Integr. Comp. Physiol. 298, R494–R507. doi: 10.1152/ajpregu.00632.2009
McKenna, A., Hanna, M., Banks, E., Sivachenko, A., Cibulskis, K., Kernytsky, A., et al. (2010). The genome analysis toolkit: a map reduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303. doi: 10.1101/gr.107524.110
Messad, F., Louveau, I., Koffi, B., Gilbert, H., and Gondret, F. (2019). Investigation of muscle transcriptomes using gradient boosting machine learning identifies molecular predictors of feed efficiency in growing pigs. BMC Genomics 20:659. doi: 10.1186/s12864-019-6010-9
Mikawa, S., Morozumi, T., Shimanuki, S., Hayashi, T., Uenishi, H., Domukai, M., et al. (2007). Fine mapping of a swine quantitative trait locus for number of vertebrae and analysis of an orphan nuclear receptor, germ cell nuclear factor (NR6A1). Genome Res. 17, 586–593. doi: 10.1101/gr.6085507
Morejohn, G. V. (1968). Study of plumage of the four species of the genus gallus. The Condor 70, 56–65. doi: 10.2307/1366508
Patterson, N., Moorjani, P., Luo, Y., Mallick, S., Rohland, N., Zhan, Y., et al. (2012). Ancient admixture in human history. Genetics 192, 1065–1093. doi: 10.1534/genetics.112.145037
Pickrell, J. K., and Pritchard, J. K. (2012). Inference of population splits and mixtures from genome-wide allele frequency data. PLoS Genet. 8:e1002967. doi: 10.1371/journal.pgen.1002967
Qiao, R., Li, X., Han, X., Wang, K., Lv, G., Ren, G., et al. (2019). Population structure and genetic diversity of four Henan pig populations. Anim. Genet. 50, 262–265. doi: 10.1111/age.12775
Ribani, A., Utzeri, V. J., Geraci, C., Tinarelli, S., Djan, M., Velickovic, N., et al. (2019). Signatures of de-domestication in autochthonous pig breeds and of domestication in wild boar populations from MC1R and NR6A1 allele distribution. Anim. Genet. 50, 166–171. doi: 10.1111/age.12771
Rubin, C. J., Megens, H. J., Martinez Barrio, A., Maqbool, K., Sayyab, S., Schwochow, D., et al. (2012). Strong signatures of selection in the domestic pig genome. Proc. Natl. Acad. Sci. U.S.A. 109, 19529–19536. doi: 10.1073/pnas.1217149109
Shang, P., Li, W., Tan, Z., Zhang, J., Dong, S., Wang, K., et al. (2020). Population genetic analysis of ten geographically isolated tibetan pig populations. Animals 10:1297. doi: 10.3390/ani10081297
Vincent, A., Louveau, I., Gondret, F., Trefeu, C., Gilbert, H., and Lefaucheur, L. (2015). Divergent selection for residual feed intake affects the transcriptomic and proteomic profiles of pig skeletal muscle. J. Anim. Sci. 93, 2745–2758. doi: 10.2527/jas.2015-8928
Wang, J., Liu, C., Chen, J., Bai, Y., Wang, K., Wang, Y., et al. (2020). Genome-wide analysis reveals human-mediated introgression from western pigs to indigenous chinese breeds. Genes 11:275. doi: 10.3390/genes11030275
Yang, G., Ren, J., Zhang, Z., and Huang, L. (2009). Genetic evidence for the introgression of Western NR6A1 haplotype into Chinese Licha breed associated with increased vertebral number. Anim. Genet. 40, 247–250. doi: 10.1111/j.1365-2052.2008.01820.x
Yang, S., Li, X., Li, K., Fan, B., and Tang, Z. (2014). A genome-wide scan for signatures of selection in Chinese indigenous and commercial pig breeds. BMC Genet. 15:7. doi: 10.1186/1471-2156-15-7
Zhang, S., Zhang, K., Peng, X., Zhan, H., Lu, J., Xie, S., et al. (2020). Selective sweep analysis reveals extensive parallel selection traits between large white and Duroc pigs. Evol. Appl. 13, 2807–2820. doi: 10.1111/eva.13085
Keywords: pig, whole-genome resequencing, introgression, selection, genetic
Citation: Wang K, Zhang L, Duan D, Qiao R, Li X, Li X and Han X (2021) Genomic Analysis Reveals Human-Mediated Introgression From European Commercial Pigs to Henan Indigenous Pigs. Front. Genet. 12:705803. doi: 10.3389/fgene.2021.705803
Received: 06 May 2021; Accepted: 24 May 2021;
Published: 18 June 2021.
Edited by:
Hai Xiang, Foshan University, ChinaReviewed by:
Lifan Zhang, Nanjing Agricultural University, ChinaYunlong Ma, Huazhong Agricultural University, China
Copyright © 2021 Wang, Zhang, Duan, Qiao, Li, Li and Han. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xinjian Li, lxjlongfei@163.com; Xuelei Han, hxl014@126.com
 Kejun Wang
Kejun Wang Lige Zhang
Lige Zhang